- 1Qingdao Key Laboratory of Mariculture Epidemiology and Biosecurity, Key Laboratory of Maricultural Organism Disease Control, Ministry of Agriculture, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao, China
- 2Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3School of Sciences and Medicine, Lake Superior State University, Sault Ste. Marie, MI, United States
- 4Department of Pathology, The Affiliated Hospital of Qingdao University, Qingdao, China
Viral covert mortality disease (VCMD), caused by covert mortality nodavirus (CMNV), is a newly emerging disease affecting most cultured shrimp and other crustaceans, but not fish. However, we discovered for the first time that Mugilogobius abei, a common marine fish collecting from shrimp farming ponds and surrounding coastal waters in China, was tested to be CMNV positive based on reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay. Further investigation based on the quantitative RT-LAMP assay indicated that 39% individuals of sampled M. abei were CMNV positive. Sequencing and alignment of sequences revealed that the partial RNA-dependent RNA polymerase gene of CMNV isolated from M. abei shared 98% homology with that from the original CMNV isolates. Histopathological analysis showed that CMNV infection in M. abei could induce extensive skeletal muscle necrosis, nervous tissue vacuolation in retina of eye and cerebellum of brain. Positive signals were verified in skeletal muscle, eye, brain and intestine by in situ hybridization (ISH) with CMNV probes. Under transmission electron microscope (TEM), CMNV particles were further visualized in the cytoplasm of neurogliocytes, granulocytes and myocytes in the CMNV positive samples diagnosed by ISH. All findings suggested that CMNV, a typical alphanodavirus originated from shrimp, could switch their hosts to fish by cross-species transmission. Meanwhile, the results reminded us to pay close attention to the high risk of CMNV to use fish as intermediate or new host as well as potentially spread or cause epidemic among cultured marine fish.
Introduction
Nodamura virus (NoV) was the first identified species in the Nodaviridae and it was originally isolated from mosquitoes (Culex tritaeniorhynchus) sampled from the village of Nodamura near Tokyo of Japan in 1956 (Scherer and Hurlbut, 1967; Scherer et al., 1968; Tesh, 1980). Till now, over 25 members have been identified, which belong to two Genus, Alphanodavirus and Betanodavirus (Andrew et al., 2011). All alphanodaviruses were isolated in nature from insects and NoV is the type species of the alphanodavirus genus (Johnson et al., 2003). The alphanodaviruses can infect insects, whereas NoV is a unique one that can also lethally infect mammals (Scherer et al., 1968; Johnson et al., 2004). All betanodaviruses were isolated from larvae, juvenile or adult marine fish, in which they cause “viral nervous necrosis” or “viral encephalopathy and retinopathy” associated with abnormal behavior and high mortalities (Andrew et al., 2011). Previous reports confirmed that betanodaviruses are pathogenic to fish and can result in significant problems for the marine fish aquaculture industry (Chi et al., 2001; Ucko et al., 2004; Walker and Winton, 2010).
Covert mortality nodavirus (CMNV), a new member of alphanodavirus, was identified to be the infectious agent of the viral covert mortality disease (VCMD) of farming shrimp (Zhang et al., 2014, 2017b). Studies had confirmed that CMNV can infect major cultured crustaceans including Penaeus vannamei, P. chinensis, Marsupenaeus japonicus, P. monodon, and Macrobrachium rosenbergii; and caused serious loss of farming crustaceans in recent years (Pooljun et al., 2016; Thitamadee et al., 2016; Zhang et al., 2017b).
Mugilogobius abei is a commonly distributed marine fish species in shrimp farming ponds and surrounding coastal waters in China (Jordan and Snyder, 1901; Jin et al., 2014; Liu et al., 2015). Recently, a sample of M. abei from a shrimp farming ponds attacked by VCMD were tested to be CMNV positive by reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay in our survey of CMNV natural host. CMNV positive of M. abei in level of nucleic acid detection clued that the M. abei might be infected naturally by CMNV. Up to now, alphanodaviruses have never been reported to be responsible to fish infection.
Following the clue of CMNV positive of the M. abei sample determined by RT-LAMP assay, we demonstrated the naturally infection of M. abei with CMNV based on a comprehensive investigation by using molecular histopathological, in situ hybridization (ISH) and transmission electron microscopic (TEM) assays in the present study. Our results provided the first evidence for supporting the host switching/extension of Alphanodavirus from shrimp to fish.
Materials and Methods
Sample Collection
Continuous monitoring of shrimp pathogens in a shrimp farming enterprise in Weifang City in China was executed by our laboratory from 2014 to 2018. The farming Penaeus vannamei, coexisting species of invertebrate and vertebrate in the ponds and the coastal water source of the enterprise, including M. abei, were free of CMNV before 2016. CMNV was introduced in several ponds of the enterprise through a starting offspring seeds infected by CMNV in May 2016. The farming P. vannamei in the ponds were attacked by VCMD from August 2016. So, M. abei samples (16∼21 mm in length), and live Penaeus vannamei samples (45∼98 mm in length), were collected from the VCMD attacked shrimp ponds and surrounding coastal water near the drainage channel of the enterprise. Nine M. abei individuals were sampled during 20th – 21st October 2016. Nine M. abei individuals and Five P. vannamei individuals were sampled in 15th August 2017 when the VCMD occurred again in the ponds. All the M. abei individuals looked normal in appearance, except some individuals showed abnormal swimming behaviors including vertically or horizontally spiraling movement.
Each individual sample was cut equally into two parts along the longitudinal axis and one part was preserved in 4% paraformaldehyde (4% PFA) solution (Sinopharm, Beijing, China). The other part was then divided into two parts and preserved respectively in RNAstore solution (Tiangen, Beijing, China) and 2.5% glutaraldehyde solution (Sinopharm, Beijing, China). These samples were applied for further analysis based on molecular, histopathological and electron microscopic approaches. The Ethics Committee of the Yellow Sea Fisheries Research Institute approved the use of animals and all procedure of operation in the study was complied with the national and institutional guidelines.
RNA Purification
Total RNA was extracted from approximately 30 mg muscle tissues of M. abei by TRIzol Reagent (Invitrogen, Carlsbad, CA, United States) using TaKaRa MiniBEST Universal RNA Extraction Kit (Takara, Dalian, China) according to the manufacture’s instruction. The concentration and purity of extracted RNA was measured by Nanodrop 2000 (Thermo Scientific, Waltham, MA, United States).
Detection of CMNV by Reverse Transcription Nested PCR and Sequencing
Total RNA samples of M. abei were submitted for diagnostic analysis of CMNV by reverse transcription nested PCR (RT-nPCR). Firstly, cDNA were synthesized from total RNA by using the SMART® MMLV Reverse Transcriptase (TAKARA) with the primer of CMNV-7R1 according to the recommended procedures. The first step PCR were conducted by using CloneAmp HiFi PCR Premix (Takara) with the primers sets of CMNV-7F1/R1 and annealing at 50°C. The second step PCR was carried out according to our previous report with a minor modification (Zhang et al., 2014). The expected CMNV target fragments would be 619 bp amplicon and 413 bp amplicon after the first and second step of the PCR amplifications, respectively. The PCR products were purified by a PCR purification kit (Tiangen, Beijing, China) and then subjected for sequencing by the commercial sequencing company of Shanghai SANGAN Chemical Trading Co. Ltd.
Phylogenetic Analysis
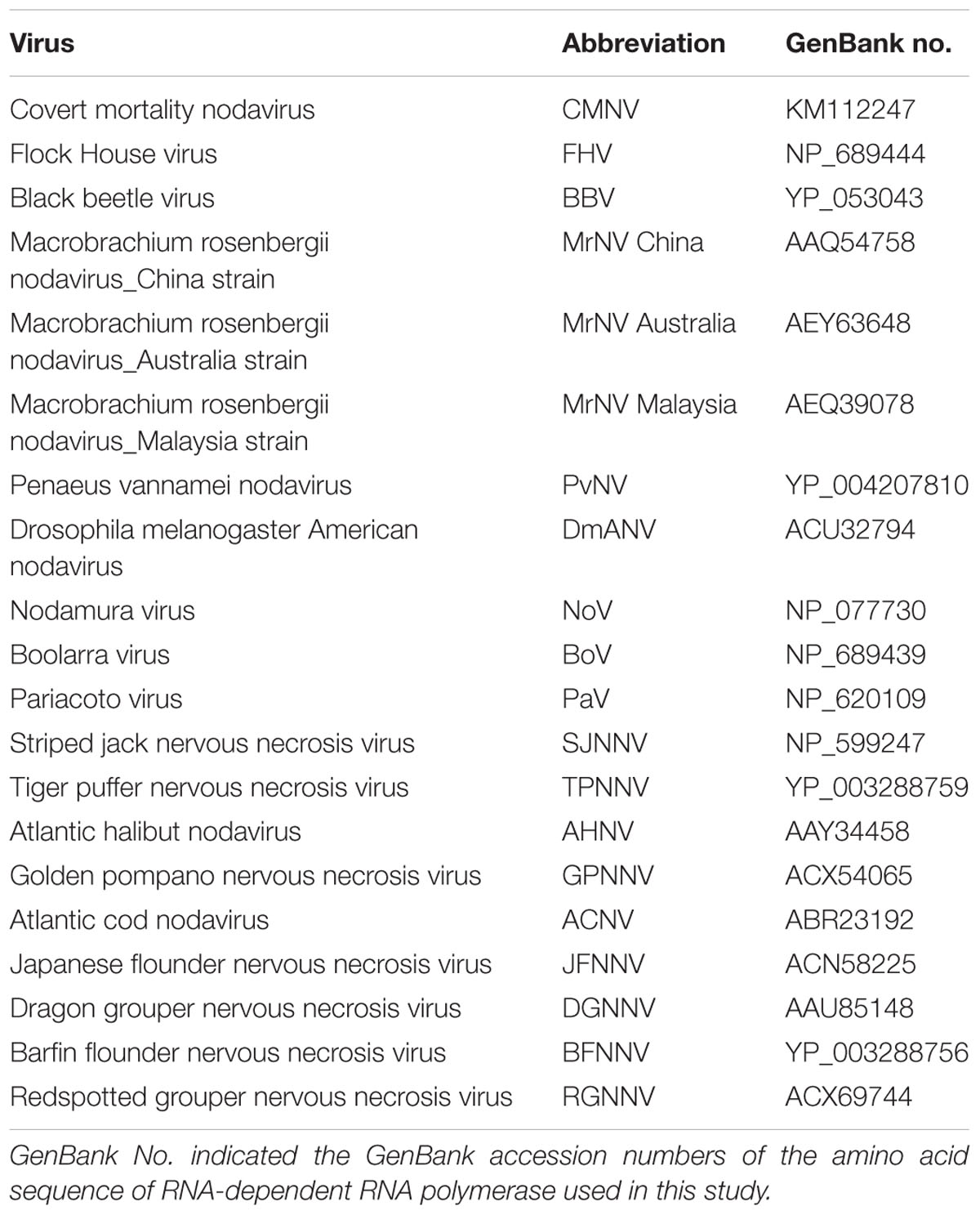
The target region (413 nt from nt no. 357 to 769) of CMNV RNA-dependent RNA polymerase (RdRp) gene of the second step PCR obtained in this study was aligned with the relevant sequences retrieved from the GeneBank database. Extra sequences were trimmed by comparing with the 413 nt CMNV RdRp gene sequence. All nucleotide sequences and deduced amino acid sequences were aligned using the ClustalW multiple alignment algorithm in the BioEdit 7.0. A phylogenetic tree was then constructed with bootstrap analysis (1000 replicates) using the software of MEGA 5.0 (Tamura et al., 2011).
Quantitative Analysis of CMNV Infection
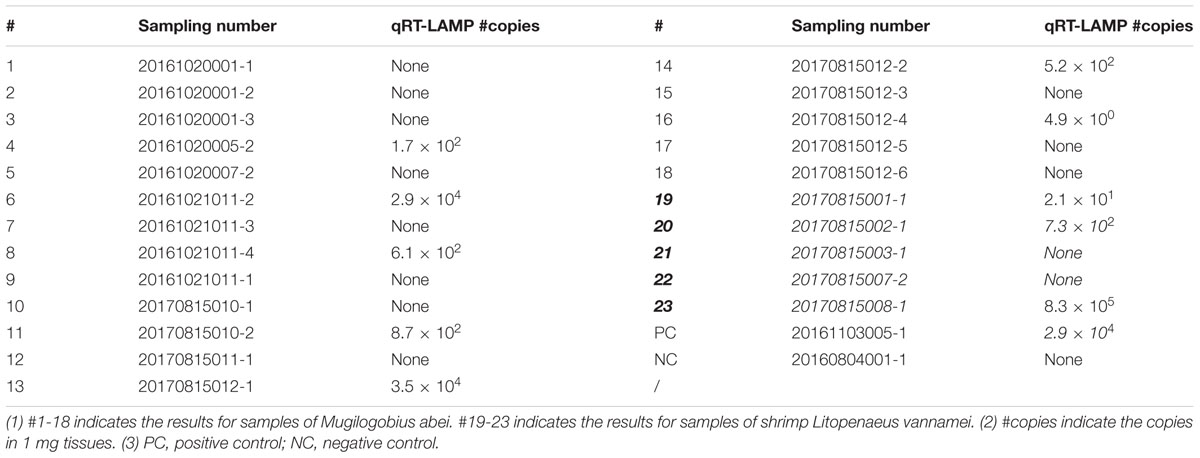
To quantify the CMNV copies in the M. abei samples, a quantitative reverse transcription loop-mediated isothermal amplification (qRT-LAMP) assay was applied to analyze the muscle tissues of M. abei according to the procedure previously described (Zhang et al., 2017a). A 10-fold serial dilution of the standard plasmid vector (pMD19-T) containing the target fragment from the CMNV RdRp gene was used as template to generate a standard curve for quantification. For the comparison, CMNV copies in the muscle tissues of L. vannamei were also quantified by using the same method.
Histopathological Section
The samples were firstly incubated in 4% PFA fixative for 24 h, then changed to 70% ethanol, and followed by embedding the samples in paraffin blocks as the histological method reported by Bell and Lightner (1988). Triplicate of paraffin sections (3 μm) were prepared for histological and ISH analysis. Sections were stained with routine hematoxylin and eosin-phloxine (H&E) according to previously described procedures (Lightner, 1996). After checking the H&E stained sections, the corresponding unstained sections were subjected to CMNV ISH assay with digoxigenin (DIG)-labeled RNA probe.
In Situ RNA Hybridization
A 244 bp DNA fragment of RdRp gene was amplified by using a set of primers with Hind III and Pst I recognition sites according to the previously reported protocol (Zhang et al., 2017b). The 244 bp DNA amplicons were digested to generate sticky ends of the dual-enzymes, and inserted into pBluescript II SK+ vector, followed by linearizing with Hind III and Pst I, respectively. The linearized vectors were used as template for transcription with T3 and T7 RNA polymerase for sense and antisense probes, respectively. In the procedure of transcription, DIG-NTP was added into the reaction to label the RNA probes. In situ RNA hybridization of each sample was conducted according to the protocols described previously (Piette et al., 2008; Chen et al., 2014). The sections post ISH were counterstained by using the Nuclear Fast Red solution as described elsewhere (Nuovo et al., 1999), and then visualized under the Nikon Eclipse E80i microscope (Nikon Co., Tokyo, Japan).
Transmission Electron Microscopy
The samples preserved in 2.5% glutaraldehyde solution was subjected to further fixation with 1% osmium tetroxide, and dehydrated in a graded ethanol series, then embedded in Spurr’s resin and prepared ultrathin sections of 50 nm in thickness. The sections were stained with uranyl acetate and lead citrate in accordance with the previously reported protocols (Graham and Orenstein, 2007; Panphut et al., 2011). Ultrathin sections were prepared on collodion coated grids, and examined by using JEOL JEM-1200 electron microscope.
Results
Detection of CMNV by RT-nPCR
A total of six M. abei were diagnosed for CMNV by RT-nPCR. Three fish were collected from the shrimp farming ponds suffering VCMD and another three from the coastal water of surrounding area. The expected 619 bp amplicon of the first step PCR and the 413 bp amplicon of the second step PCR were amplified by using the cDNA templates from total RNAs of six M. abei samples despite of their sources (Figure 1). These findings indicated that all 6 M. abei samples were CMNV positive despite of their resources (Figure 1).
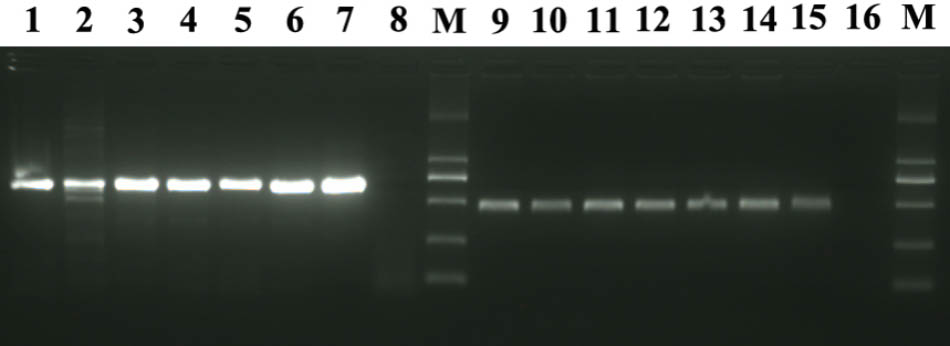
FIGURE 1. Electrophoretogram for detection of CMNV in Mugilogobius abei from shrimp farming ponds and coastal water by RT-nPCR. M: DL2000 molecular weight marker. Lane 1∼8 shows the result of the first step of RT-nPCR and Lane 9 ∼16 shows the result of the second step of RT-nPCR. Lane 1 and 9, lane 2 and 10, Lane 3 and 11 showed the amplification results of cDNAs from individuals collected from shrimp farming ponds. Lane 4 and 12, lane 5 and 13, Lane 6 and 14 showed the amplification results of cDNAs from individuals collected from coastal water near the ponds. Lane 7 and 15, lane 8 and 16 are the amplification results of cDNAs from the positive and negative control, respectively.
Sequencing and Phylogenetic Analyses
BLAST search results indicated that the target gene fragments of all CMNV positive samples shared as high as 98% sequence similarity with the known CMNV target gene (KM112247) (Figure 2). Phylogenetic analysis showed that the CMNV target gene fragments from four different samples were clustered tightly into the branch of the original CMNV isolate, which demonstrated higher similarity with flock house virus (FHV), black beetle virus (BBV), Drosophila melanogaster American nodavirus (DmANV) and Boolarra virus (BoV) rather than Macrobrachium rosenbergii nodavirus (MrNV) and Penaeus vannamei nodavirus (PvNV) in the branch of Alphanodavirus (Figure 3). Meanwhile, all members from Betanodavirus were clustered into the other independent branch in the phylogenetic tree.
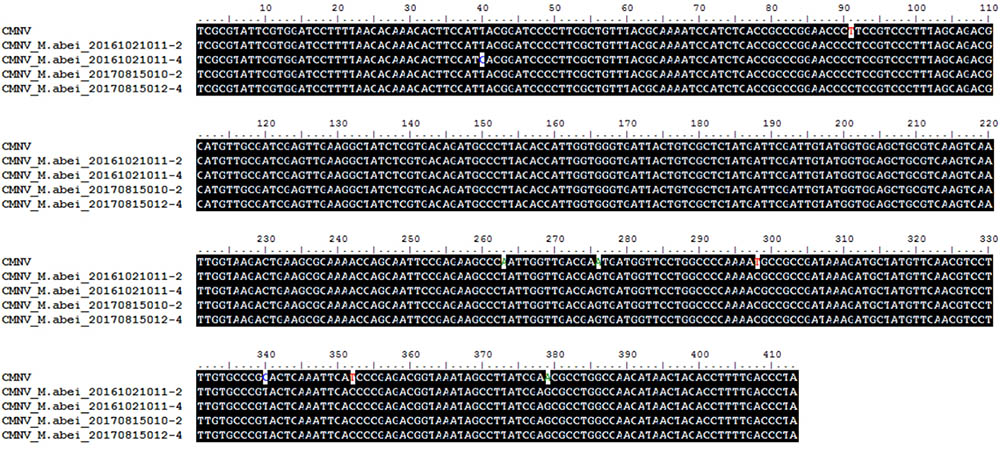
FIGURE 2. Alignments of the CMNV target gene from Mugilogobius abei with the corresponding gene (GenBank accession number KM112247) region of the original CMNV. The title of CMNV represented the RNA-dependent RNA polymerase (RdRp) gene from the original CMNV. The titles of CMNV_M. abei represented the RdRp gene from CMNV positive individuals of M. abei.
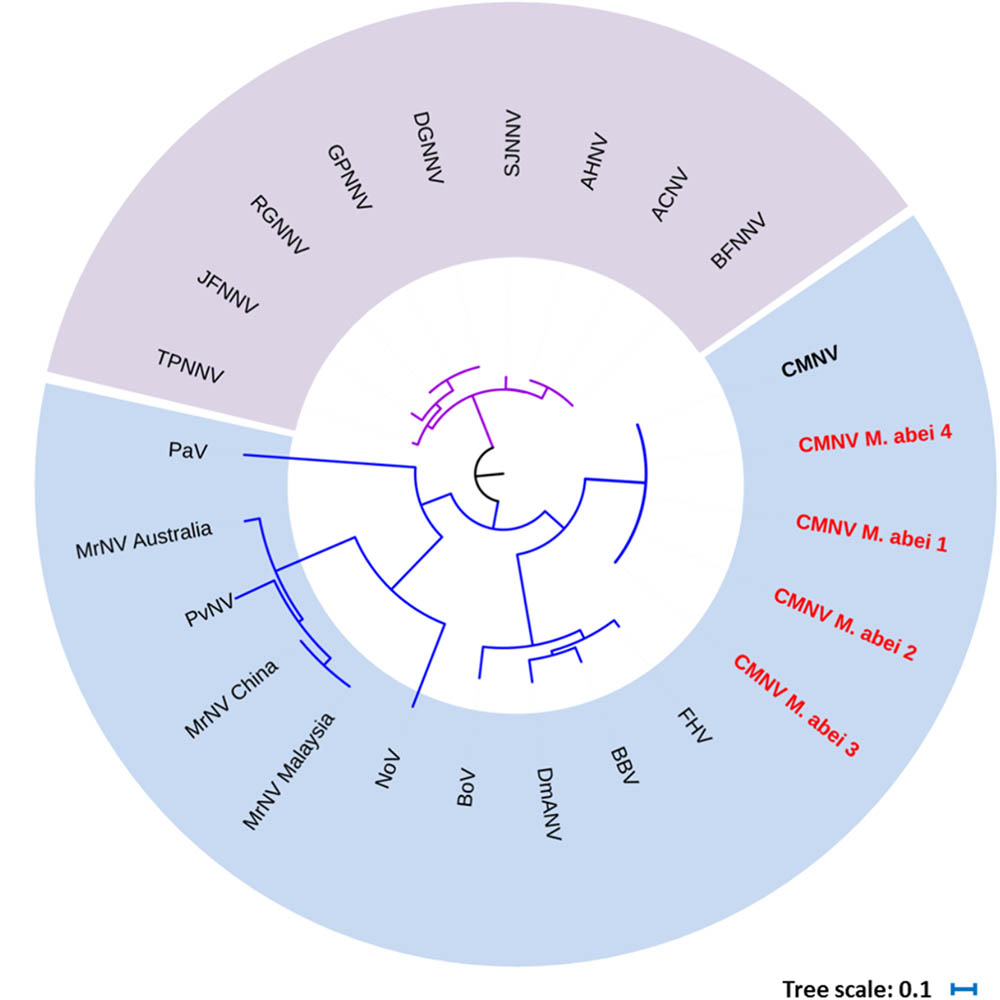
FIGURE 3. Phylogenetic tree based on the deduced amino acid sequences of RNA-dependent RNA polymerase (RdRp) gene from CMNV positive individuals of Mugilogobius abei and other nodaviruses (For virus abbreviations see Table 1). CMNV M. abei 1–2 were from individuals collected from shrimp farming ponds. CMNV M. abei 3–4 were from individuals collected from peripheral coastal water of ponds. The viral species of Alphanodavirus genus were indicated by baby blue background. The viral species of Betanodaviruses genus were indicated by lavender background. The tree was generated by the neighbor-joining method using the MEGA 5.0 program and the scale bar was 0.1.
Quantitation of CMNV Infection
Quantitation of viral copies in the muscle of eighteen M. abei samples and five L. vannamei samples were extrapolated based on the Ct value of the generated standard curve. The viral loads in the muscles of CMNV infected M. abei varied from 4.9 × 100 to 3.5 × 104 copies per mg tissues, which is lower than the viral loads in the muscles of L. vannamei (2.1 × 101 to 8.3 × 105). The quantitative RT-LAMP assay indicated that 39% individuals of sampled M. abei were CMNV positive (Table 2).
Histopathological Changes Due to CMNV Infection
Histological examination revealed obvious histopathological alteration and lesions in the skeletal muscle, retina and brain of the M. abei samples. Extensive muscular lysis, myonecrosis (Figures 4e,f), and haemocytic infiltration in fibromuscular stroma (Figures 5e,f) could be observed from the examinated samples. Meanwhile, moderate vacuolation was noticed in the retina of eye and cerebellum of M. abei (Figures 6e–f). Haemocytic infiltration could also be observed among enterocytes of intestine folds (Figures 7e–f).
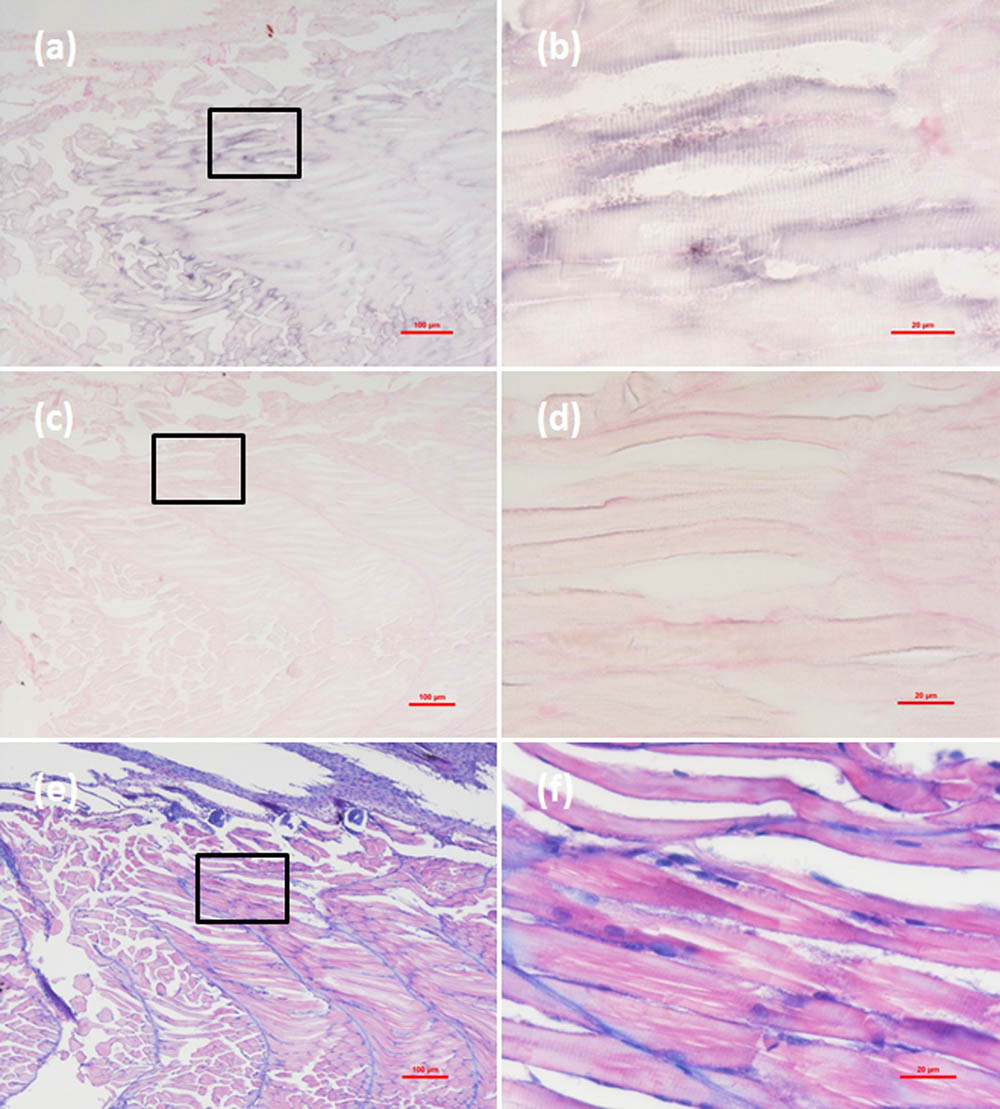
FIGURE 4. Micrographs of in situ hybridization (ISH) and H&E staining for necrotic skeletal muscle of the Mugilogobius abei naturally infected with CMNV. (a) Micrographs of ISH for skeletal muscle of CMNV-infected M. abei with the CMNV RNA probe. (b) Magnified micrograph of the area in the black frame in (a). Note the intense hybridization signal at the dissolved myoneme of myocyte which demonstrated apparent lysis. (c) Micrographs of ISH for skeletal muscle of CMNV-infected M. abei without the CMNV RNA probe. (d) Magnified micrograph of the area in the black frame in (c). (e) Micrographs of H&E staining for necrotic skeletal muscle of CMNV-infected M. abei. (f) Magnified micrograph of the area in the black frame in (e). Note the haemocytic infiltration in fibromuscular stroma. Scale bars = (a) 100 μm, (b) 20 μm, (c) 100 μm, (d) 20 μm, (e) 100 μm, and (f) 20 μm.
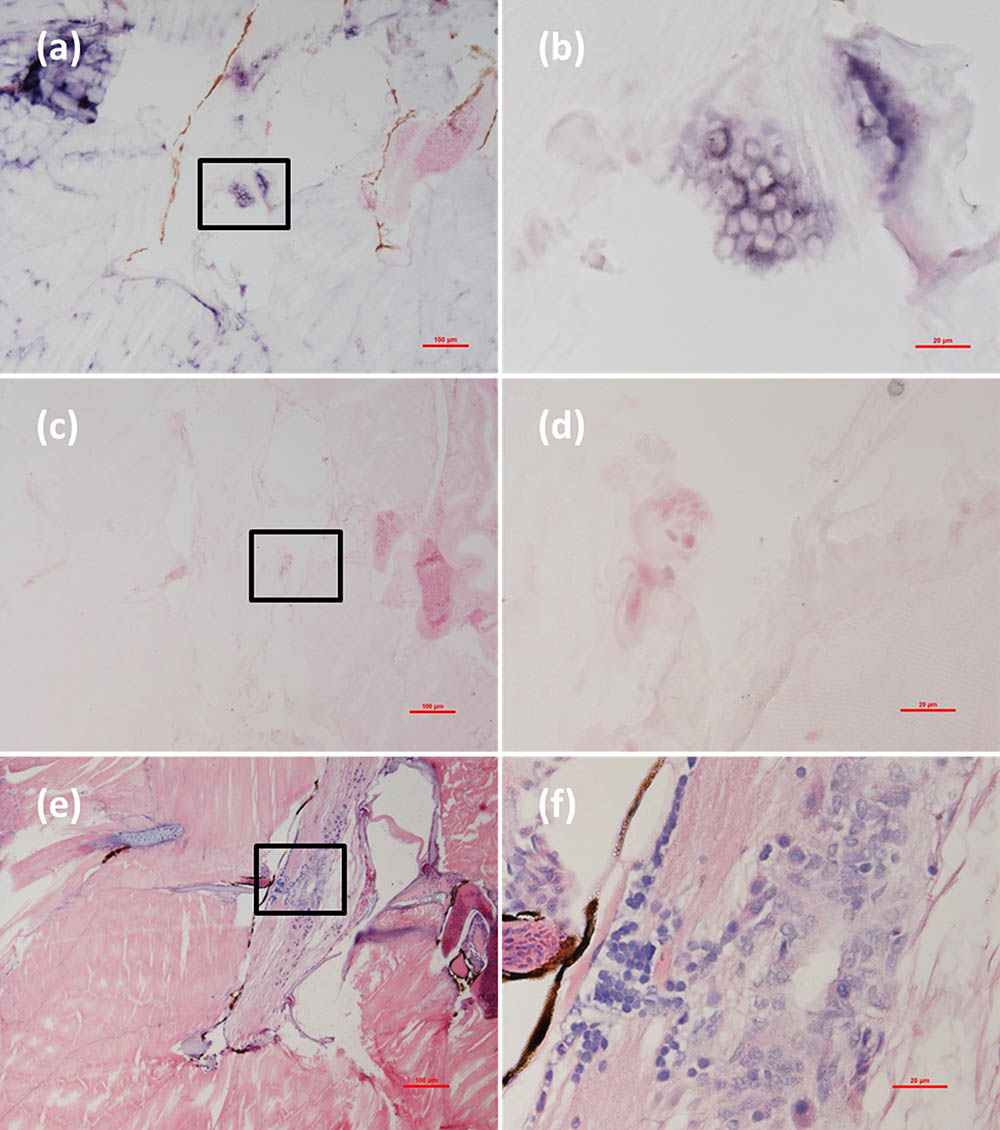
FIGURE 5. Micrographs of ISH and H&E staining for lesion of skeletal muscle with severe haemocytic infiltration of the Mugilogobius abei naturally infected with CMNV. (a) Micrographs of ISH for muscle lesion of CMNV-infected M. abei with the CMNV RNA probe. (b) Magnified micrograph of the area in the black frame in (a). Note the intense hybridization signal in infiltrating hemocytes in the lysed muscle. (c) Micrographs of ISH for muscle lesion of CMNV-infected M. abei without the CMNV RNA probe. (d) Magnified micrograph of the area in the black frame in (c). (e) Micrographs of H&E staining for muscle lesion of CMNV-infected M. abei. (f) Magnified micrograph of the area in the black frame in (e). Scale bars = (a) 100 μm, (b) 20 μm, (c) 100 μm, (d) 20 μm, (e) 100 μm, and (f) 20 μm.
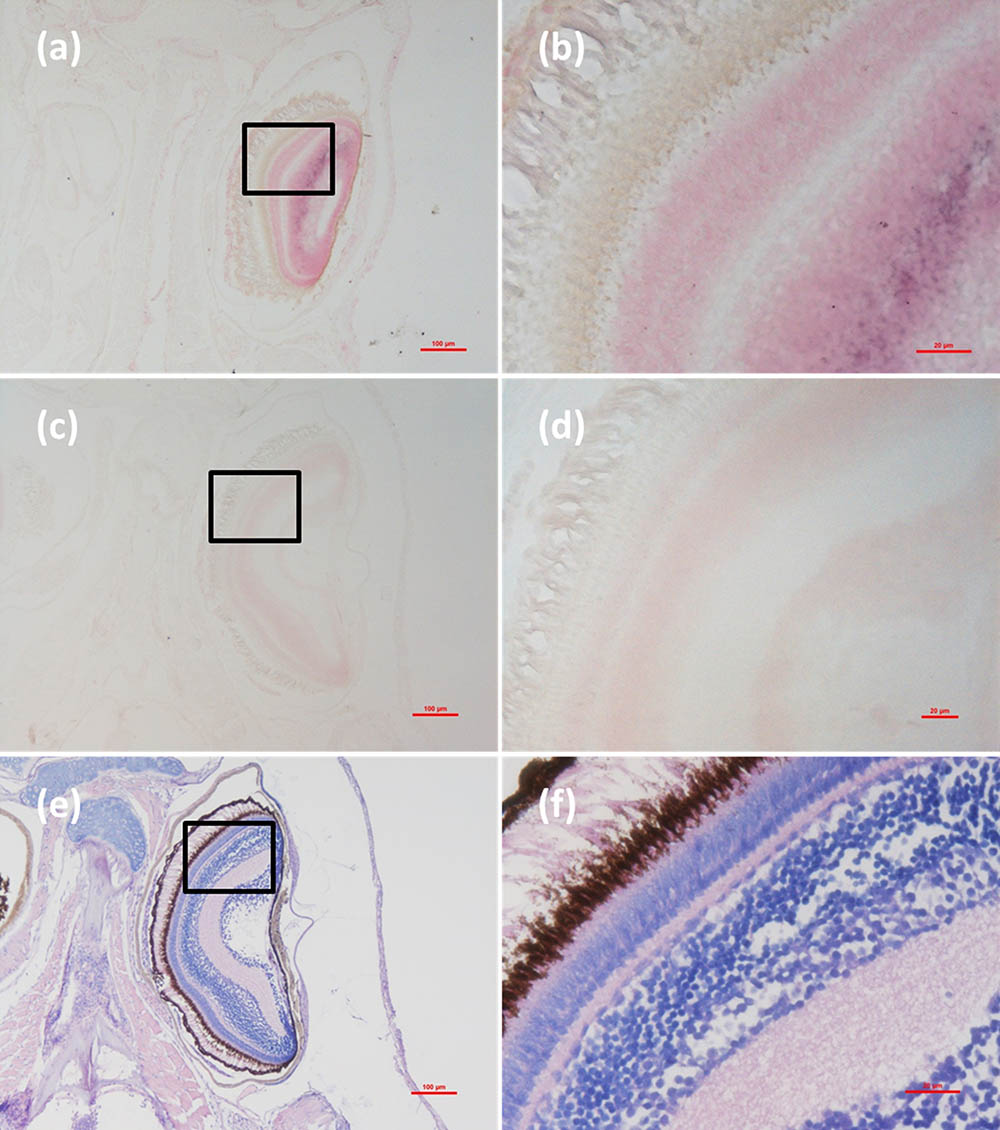
FIGURE 6. Micrographs of ISH and H&E staining for retina of the Mugilogobius abei naturally infected with CMNV. (a) Micrographs of ISH for retina of CMNV-infected M. abei with the CMNV RNA probe. (b) Magnified micrograph of the area in the black frame in (a). Note the purple hybridization signal in the outer plexiform layer of the retina. (c) Micrographs of ISH for retina of CMNV-infected M. abei without the CMNV RNA probe. (d) Magnified micrograph of the area in the black frame in (c). (e) Micrographs of H&E staining for the retina of eye from CMNV-infected M. abei. (f) Magnified micrograph of the area in the black frame in (e). Scale bars = (a) 100 μm, (b) 20 μm, (c) 100 μm, (d) 20 μm, (e) 100 μm, and (f) 20 μm.
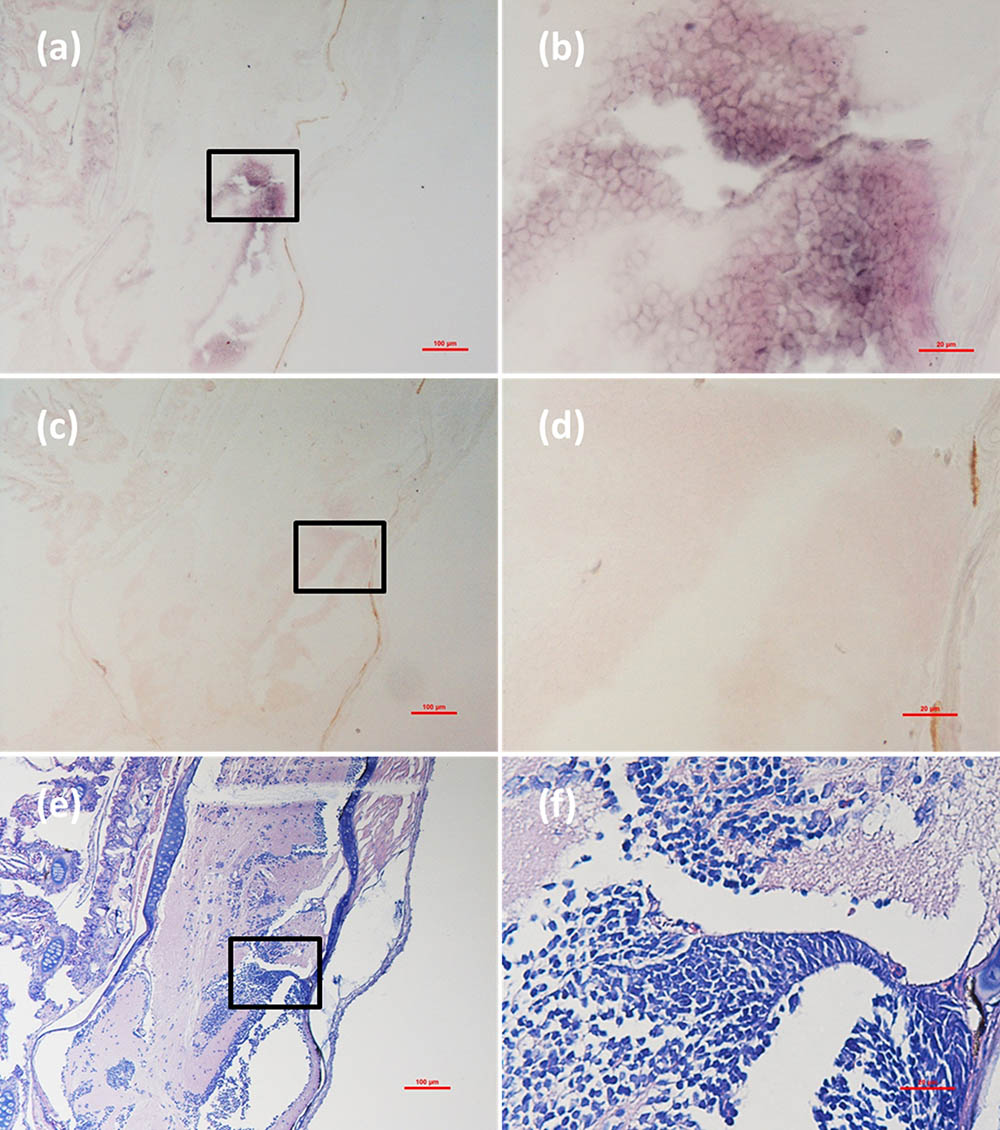
FIGURE 7. Micrographs of ISH and H&E staining for cerebellum of the Mugilogobius abei naturally infected with CMNV. (a) Micrographs of ISH for cerebellum of CMNV-infected M. abei with the CMNV RNA probe. (b) Magnified micrograph of the area in the black frame in (a). Note the purple hybridization signal in the granule cell layer of the cerebellum. (c) Micrographs of ISH for cerebellum of CMNV-infected M. abei without the CMNV RNA probe. (d) Magnified micrograph of the area in the black frame in (c). (e) Micrographs of H&E staining for the cerebellum from CMNV-infected M. abei. (f) Magnified micrograph of the area in the black frame in (e). Scale bars = (a) 100 μm, (b) 20 μm, (c) 100 μm, (d) 20 μm, (e) 100 μm, and (f) 20 μm.
Detection of CMNV in M. abei by ISH
Micrographs of ISH for M. abei samples showed that the intense positive hybridization signals presented in the necrotic skeletal muscle (Figure 4a). Purple hybridization signal was clearly colocalized at the dissolved myoneme of myocyte which appeared apparent lysis at high magnification (Figure 4b). Infiltrating hemocytes in the lysed muscle showed intense hybridization signal of CMNV RNA probe as well (Figures 5a,b). Purple hybridization signal could be observed in retina outer plexiform layer of the eye, cerebellum granule cell layer of the brain, and enterocytes of intestine (Figures 6, 8a,b), respectively. While, no positive hybridization signals appeared on the sections from both the same samples without CMNV RNA probe in the hybridization process (Figures 4–8c–f) and the CMNV negative sample determined by qRT-LAMP (see the Supplementary Data).
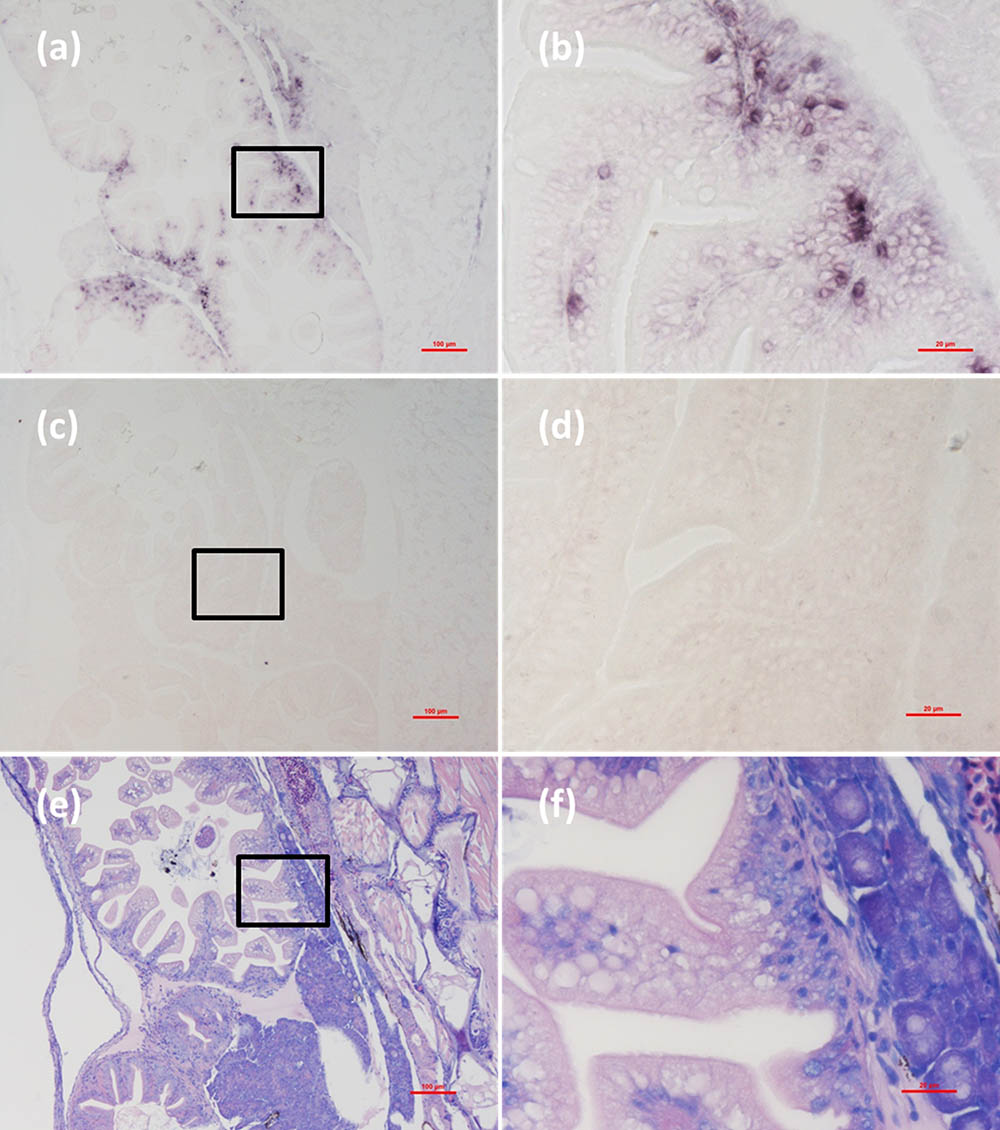
FIGURE 8. Micrographs of ISH and H&E staining for intestine of the Mugilogobius abei naturally infected with CMNV. (a) Micrographs of ISH for intestine of CMNV-infected M. abei with the CMNV RNA probe. (b) Magnified micrograph of the area in the black frame in (a). Note the intense hybridization signal in the enterocytes of intestine. (c) Micrographs of ISH for intestine of CMNV-infected M. abei without the CMNV RNA probe. (d) Magnified micrograph of the area in the black frame in (c). (e) Micrographs of H&E staining for the intestine from CMNV-infected M. abei. (f) Magnified micrograph of the area in the black frame in (e). Scale bars = (a) 100 μm, (b) 20 μm, (c) 100 μm, (d) 20 μm, (e) 100 μm, and (f) 20 μm.
Detection of CMNV in M. abei by TEM
Under the TEM, a group of unenveloped, virus-like particles were observed within the cytoplasm of myocyte, in which amounts of mitochondria surrounded the viral zone (Figures 9a,b). The ultrathin sections of skeletal muscle also revealed the presence of mass spherical CMNV-like particles with diameter about 24.7 ± 2.1 nm (n = 19) (Figures 9c,C,d) in the cytoplasm of granule cells. Conspicuous large vacuoles were noticed in the medulla oblongata of M. abei (Figures 10a,b) and numerous unenveloped CMNV-like particles could be found in the cytoplasm of adjacent neurogliocyte of the medulla oblongata (Figures 10b–d).
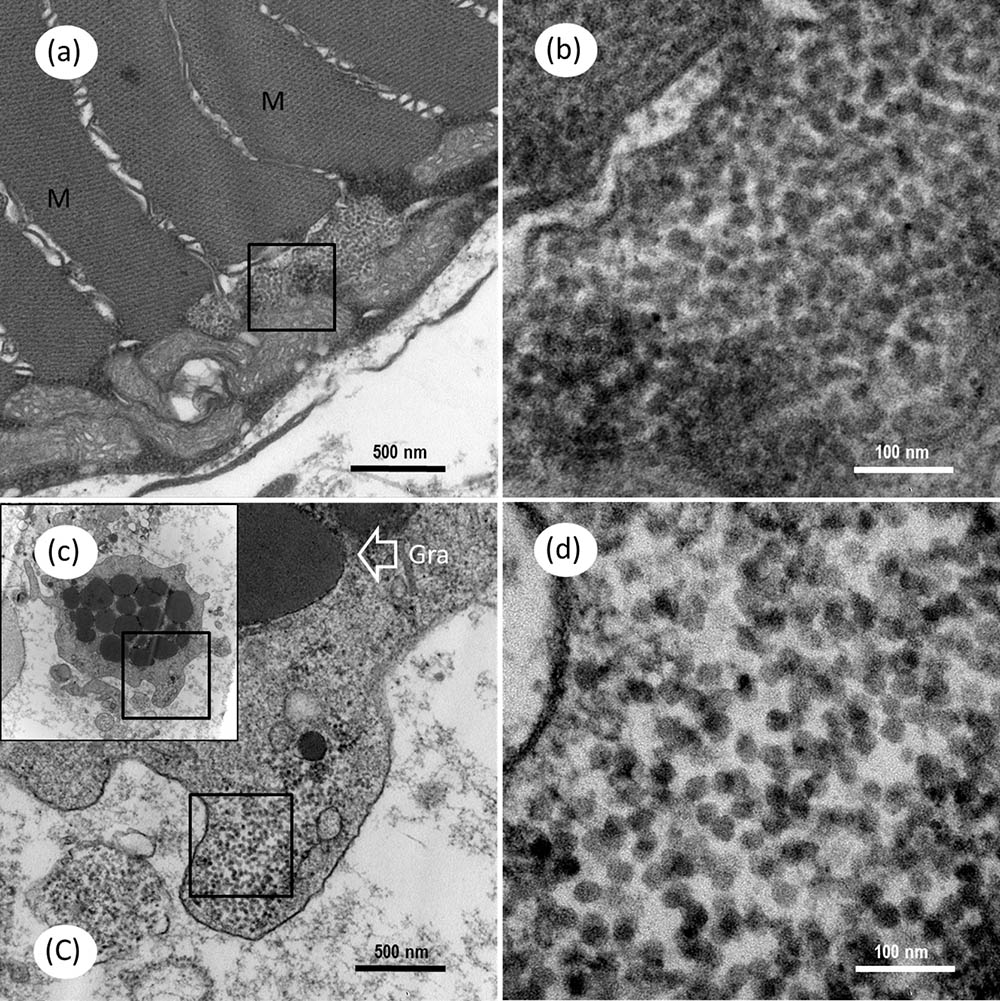
FIGURE 9. Transmission electron microscopic micrographs of ultrathin section for myocyte and granulocyte in skeletal muscle of Mugilogobius abei. (a) TEM of the myocyte of M. abei. (b) Magnified micrograph of the partial zone in the black frame in (a). Note that the scattering distribution of CMNV-like particles near the mitochondria can be observed. (c) TEM of one granulocyte of M. abei. (C) Magnified micrograph of the partial zone of the granulocyte in the black frame in (c). (d) Magnified micrograph of the partial zone of the granulocyte in the black frame in (C). Note the scattering distribution of CMNV-like particles. Gra, granulocyte; M, Muscle; Scale bars = (a) 500 nm, (b) 100 nm, (c) 500 nm, and (d) 100 nm.
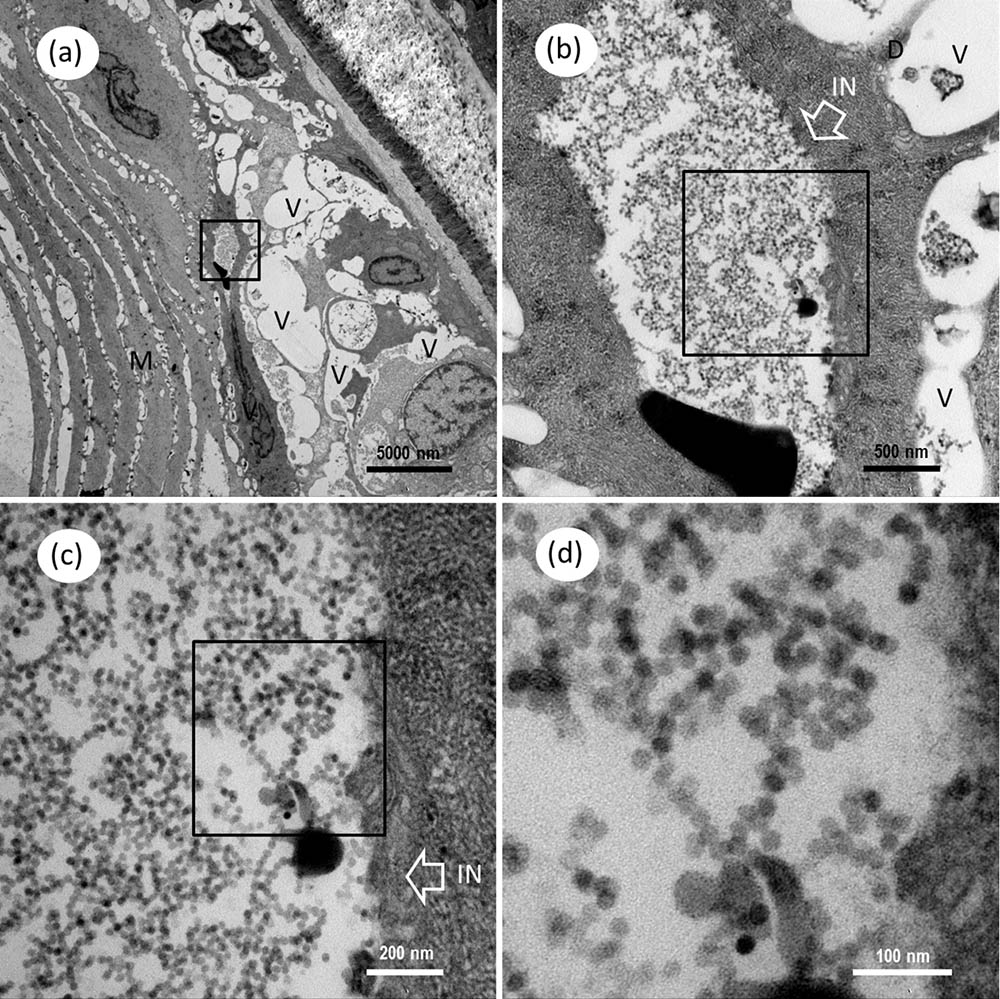
FIGURE 10. Transmission electron microscopic micrographs of ultrathin section for cerebellum of Mugilogobius abei. (a) TEM of the tissues of the cerebellum of M. abei. Conspicuous large vacuoles were noticed in the cerebellum of M. abei. (b) Magnified micrograph of the viral inclusion in the black frame in (a). Viral inclusion was clear in the ultrathin section of the cerebellum. The white box arrow white arrows indicated the inclusion. (c) Magnified micrograph of the partial zone of viral inclusion in the black frame in (b). (d) Magnified micrograph of the CMNV-like particles in the black frame in (c). Note that the scattering distribution of CMNV-like particles can be observed. IN, Inclusion; M, Muscle; V, vacuole; N, nucleus; Scale bars = (a) 5000 nm, (b) 500 nm, (c) 200 nm, and (d) 100 nm.
Discussion
Alphanodaviruses were originally isolated from insects and their host range appears to be restricted to insects with the exception of the type species of Nodamura virus (NoV) and the Flock house virus (FHV) (Selling et al., 1990; Stock et al., 2009). NoV was a unique species among the alphanodaviruses and able to kill both insects and mammals (Bailey and Scott, 1973; Bailey et al., 1975). NoV had been reported to be infectious to pigs and transferable to sucking mice via a mosquito vector (Ball and Johnson, 1999). FHV, another insect originated alphanodavirus, could replicate in many species of plants including barley Hordeum vulgare, cowpea Vigna sinensis, chenopodium Chenopodium hybridum, tobacco Nicotiana tabacum, and Nicotiana benthamiana (Selling et al., 1990). It seemed that NoV and FHV possess the distinct capability for host switching. However, the host ranges of betanodavirus were different from alphanodaviruses and betanodavirus mainly infect larvae, juvenile or adult marine fish (Iwamoto et al., 2004; Furusawa et al., 2007; Souto et al., 2015). Up to now, except Betanodavirus, no any virus in the family of Nordaviridae has been reported to be capable to infect fish whether naturally or artificially.
In the present study, we demonstrated for the first time that M. abei, a marine fish species, was naturally infected by CMNV, a member of alphanodavirus. Histopathological alteration and lesions, such as necrosis and heavy vacuolation, in the muscle, retina and cerebellum were very similar with that appeared in the target fishes infected by viral species of betanodavirus (Chi et al., 2001; Furusawa et al., 2007). Intense positive ISH signals in the necrotic myocyte indicated that CMNV could infect myocytes and caused muscular lysis in M. abei. The presence of CMNV particles in the cytoplasm of neurogliocytes, granule cells and skeletal muscle cells were further confirmed by TEM analysis. The result of natural infection of M. abei with CMNV provided the first clear evidence for that the viral member in alphanodaviruses can infect marine fish naturally (Figure 11).
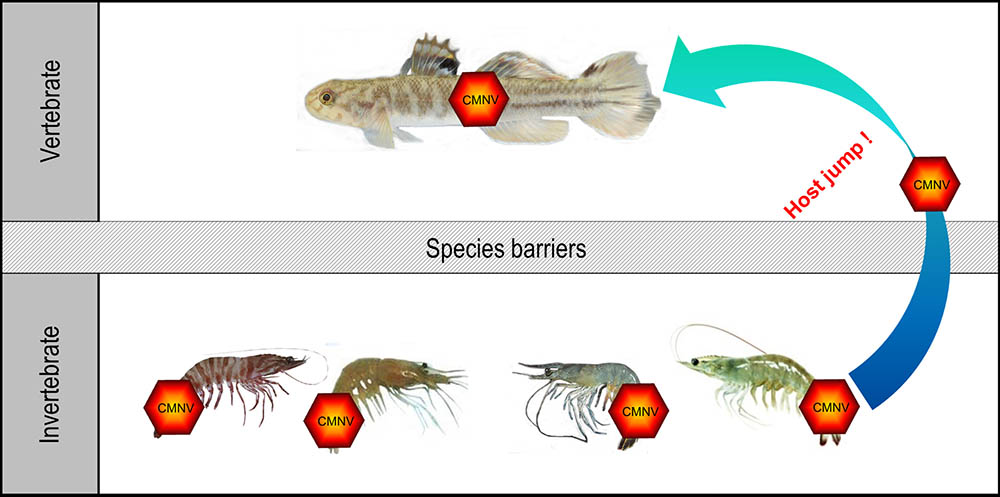
FIGURE 11. Host jump of CMNV from invertebrate to fish. The species of CMNV host in invertebrate included the Penaeus japonicus, Exopalaemon carinicauda, Penaeus vannamei, and Penaeus chinensis. This investigation revealed that CMNV, an emerging alphanodavirus originated from shrimp, possessed capability of crossing the species barriers and infecting Mugilogobius abei, a species of fish.
Emerging viruses usually had RNA genome and as such were capable of rapid mutation and selection of new variants in the face of environmental changes in host numbers and available target species (Howard and Fletcher, 2012). A few viruses could be transmitted to completely new host species that they had never infected previously. Emerging viral diseases were often the product of a host shift, where a pathogen jumped from its original host into a novel species (Longdon et al., 2014). From the point of view of taxonomy, CMNV belonged to alphanodaviruses and the host range of viruses in this genus appeared to be restricted to insects. Nevertheless, CMNV was proved to be the pathogenic agent of VCMD, an emerging disease of farming shrimp, recently. This study supplied the novel evidence for CMNV host jump from shrimp to the co-existing marine fish in farming ponds and coastal water (Figure 11). These host transfers might involve either increased exposure to virus of the new host organism or the acquisition of viral variations that allow the virus to overcome species barriers to infection of the new hosts. Similar virus cases of host jump were reported occasionally (Parrish et al., 2008; Kreuder et al., 2015) and several typical documentations included that a plant virus switching hosts to infect a vertebrate (Gibbs and Weiller, 1999), H7N9 avian influenza starting to infect humans (Shi et al., 2014) and permanent host shift of rabies virus from Chiroptera to Carnivora (Ding et al., 2017).
Mugilogobius abei was a species of fish in the family Gobiidae and mainly distributes in fresh, brackish and marine water of the Indo-Pacific region (Larson, 2001; Huang et al., 2015). It was the keystone species of bait fish in the Bohai Sea, the Yellow Sea, and the East China Sea and played important roles in the food web of the coastal water communities in the Sea (Jin et al., 2014; Liu et al., 2015, 2016). In this study, a total of 18 individuals of M. abei, collected either from shrimp farming ponds or surrounding coastal waters, were analyzed and seven individuals were CMNV positive based on RT-LAMP. The highest viral load in the CMNV positive samples was up to 3.5 × 104 viral copies per mg tissues. Natural infection of M. abei from coastal waters with CMNV, along with the fact of the high infection rate and infective dose of CMNV in M. abei samples, might alert CMNV as an emerging and significant hazards factor to the natural population of M. abei in coastal area. Recently, we identified CMNV from nearshore Chaeturichthys hexanema, another wild marine fish in the Yellow Sea, based on the RT-LAMP and RT-nPCR assay. All these results revealed the substantial risk of wide prevalence of CMNV in farmed and wild marine fish species.
Conclusion
Natural infection of M. abei with CMNV demonstrated by present study supplied the first evidence for alphanodavirus infecting fish. Our findings suggested that CMNV possessed the distinct capability of host jump and it could infect marine fish, M. abei, which was a comman species in shrimp farming ponds and a dominant species in coastal water in China. The result prompted that CMNV would be an important and emerging ecological risk for its infectious ability and pathogenicity to marine fish.
Author Contributions
QZ and SL designed, executed the experiments and analyzed the samples. SL conducted the in situ RNA hybridization assay. XL and TX did molecular and biological analysis. QZ conducted the TEM assay. XW and TX contributed to sampling. XB helped to identify the species of the fish samples. GF, SL, and JH prepared the histological sections. SS did the sequencing work. QZ and JL wrote the manuscript. All authors interpreted the data, critically revised the manuscript for important intellectual contents and approved the final version.
Funding
This work was supported by the National R&D Program of China (2017YFC1404503), Central Public-Interest Scientific Institution Basal Research Fund, YSFRI, CAFS (No. 20603022015003), National Natural Science Foundation of China (31672695), and Central Public-Interest Scientific Institution Basal Research Fund, CAFS (No. 2017HY-ZD0301).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank XW for his generous help in sampling.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01447/full#supplementary-material
References
Andrew, K., Elliot, L., Michael, A., Eric, C., and Carstens, E. (2011). “Nodaviridae,” Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses, (Amsterdam: Elsevier), 1019–1025.
Bailey, L., Newman, J. F., and Porterfield, J. S. (1975). The multiplication of Nodamura virus in insect and mammalian cell cultures. J. Gen. Virol. 26, 15–20.
Bailey, L., and Scott, H. A. (1973). The pathogenicity of Nodamura virus for insects. Nature 241:545.
Ball, L. A., and Johnson, K. L. (1999). Reverse genetics of nodaviruses. Adv. Virus Res. 53, 229–244.
Bell, T. A., and Lightner, D. V. (1988). A Handbook of Normal Penaeid Shrimp Histology. Baton Rouge, LA: World Aquaculture Society Press.
Chen, S., Zhang, G., Shao, C., Huang, Q., Liu, G., Zhang, P., et al. (2014). Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 46, 253–260. doi: 10.1038/ng.2890
Chi, S. C., Lo, C. F., and Lin, S. C. (2001). Characterization of grouper nervous necrosis virus. J. Fish Dis. 24, 313.
Ding, N. Z., Xu, D. S., Sun, Y. Y., He, H. B., and He, C. Q. (2017). A permanent host shift of rabies virus from Chiroptera to Carnivora associated with recombination. Sci. Rep. 7:289. doi: 10.1038/s41598-017-00395-2
Furusawa, R., Okinaka, Y., Uematsu, K., and Nakai, T. (2007). Screening of freshwater fish species for their susceptibility to a betanodavirus. Dis. Aquat. Organ. 77, 119–125.
Gibbs, M. J., and Weiller, G. F. (1999). Evidence that a plant virus switched hosts to infect a vertebrate and then recombined with a vertebrate-infecting virus. Proc. Natl. Acad. Sci. U.S.A. 96, 8022–8027.
Graham, L., and Orenstein, J. M. (2007). Processing tissue and cells for transmission electron microscopy in diagnostic pathology and research. Nat. Protoc. 2, 2439–2450.
Howard, C. R., and Fletcher, N. F. (2012). Emerging virus diseases: can we ever expect the unexpected? Emerg. Microbes Infect. 1:e46. doi: 10.1038/emi.2012.47
Huang, S. P., Shen, C. N., and Chen, I. S. (2015). The complete mitochondrial genome of the Abe’s mangrove goby Mugilogobius abei (Teleostei, Gobiidae). Mitochondrial DNA 26, 143–144. doi: 10.3109/19401736.2013.819494
Iwamoto, T., Okinaka, Y., Mise, K., Mori, K., Arimoto, M., Okuno, T., et al. (2004). Identification of host-specificity determinants in betanodaviruses by using reassortants between striped jack nervous necrosis virus and sevenband grouper nervous necrosis virus. J. Virol. 78, 1256–1262.
Jin, X. S., Qiu, S. Y., and Liu, X. Z. (2014). Foundation and Prospect of Fisheries Resources in Yellow Sea and Bohai Sea. Ottawa: Science Press.
Johnson, K. L., Price, B. D., and Ball, L. A. (2003). Recovery of infectivity from cDNA clones of nodamura virus and identification of small nonstructural proteins. Virology 305, 436–451.
Johnson, K. L., Price, B. D., Eckerle, L. D., and Andrew Ball, L. (2004). Nodamura virus nonstructural protein B2 can enhance viral RNA accumulation in both mammalian and insect cells. J. Virol. 78, 6698–6704.
Jordan, D. S., and Snyder, J. O. (1901). A review of the gobioid fishes of Japan, with descriptions of twenty-one new species. Proc. U.S. Nat. Mus. 24, 33–132. doi: 10.5479/si.00963801.24-1244.33
Kreuder, J. C., Hitchens, P. L., Smiley, E. T., Goldstein, T., Thomas, K., Clements, A., et al. (2015). Spillover and pandemic properties of zoonotic viruses with high host plasticity. Sci. Rep. 5:14830. doi: 10.1038/srep14830
Larson, H. K. (2001). A revision of the gobiid fish genus Mugilogobius (Teleostei: Gobioidei), and its systematic placement. Rec. West Aust. Mus. 62, 1–233.
Lightner, D. V. (1996). A Handbook of Shrimp Pathology and Diagnostic Procedures for Diseases of Cultured Penaeid Shrimp. Baton Rouge, LA: World Aquaculture Society Press.
Liu, S. H., Wang, J. H., Liu, C. C., Qin, Y. T., Liu, Z. G., Deng, B. P., et al. (2015). Inter-annual variation in pelagic fish egg, larval, and juvenile assemblages during summer in the Yangtze River Estuary, China. Acta Ecol. Sin. 35, 7191–7197.
Liu, Y. Y., Song, C., Zhao, F., Lv, Y., Yang, G., Zhang, L. Z., et al. (2016). Category composition and distributional patterns of fish larvae and juveniles in the Yangtze Estuary and adjacent waters. Mar. Fish. 38, 597–612.
Longdon, B., Brockhurst, M. A., Russell, C. A., Welch, J. J., and Jiggins, F. M. (2014). The evolution and genetics of virus host shifts. PLoS Pathog. 10:e1004395. doi: 10.1371/journal.ppat.1004395
Nuovo, G. J., Plaia, T. W., Belinsky, S. A., Baylin, S. B., and Herman, J. G. (1999). In situ detection of the hypermethylation-induced inactivation of the p16 gene as an early event in oncogenesis. Proc. Nat. Acad. Sci. U.S.A. 96, 12754–12759.
Panphut, W., Senapin, S., Sriurairatana, S., Withyachumnarnkul, B., and Flegel, T. W. (2011). A novel integrase-containing element may interact with Laem-Singh virus (LSNV) to cause slow growth in giant tiger shrimp. BMC Vet. Res. 7:18. doi: 10.1186/1746-6148-7-18
Parrish, C. R., Holmes, E. C., Morens, D. M., Park, E. C., Burke, D. S., Calisher, C. H., et al. (2008). Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 72, 457–470. doi: 10.1128/MMBR.00004-08
Piette, D., Hendrickx, M., Willems, E., Kemp, C. R., and Leyns, L. (2008). An optimized procedure for whole-mount in situ hybridization on mouse embryos and embryoid bodies. Nat. Protoc. 3, 1194–1201. doi: 10.1038/nprot.2008.103
Pooljun, C., Direkbusarakom, S., Chotipuntu, P., Hirono, I., and Wuthisuthimethaveea, S. (2016). Development of a TaqMan real-time RT-PCR assay for detection of covert mortality nodavirus (CMNV) in penaeid shrimp. Aquaculture 464, 445–450. doi: 10.1016/j.aquaculture.2016.06.044
Scherer, W. F., and Hurlbut, H. S. (1967). Nodamura virus from Japan: a new and unusual arbovirus resistant to diethyl ether and chloroform. Am. J. Epidemiol. 86, 271–285.
Scherer, W. F., Verna, J. E., and Richter, W. (1968). Nodamura virus, an ether- and chloroform-resistant arbovirus from Japan: physical and biological properties, with ecologic observations. Am. J. Trop. Med. Hyg. 17, 120–128.
Selling, B. H., Allison, R. F., and Kaesberg, P. (1990). Genomic RNA of an insect virus directs synthesis of infectious virions in plants. Proc. Nat. Acad. Sci. U.S.A. 87, 434–438.
Shi, Y., Wu, Y., Zhang, W., Qi, J., and Gao, G. F. (2014). Enabling the ’host jump’: structural determinants of receptor-binding specificity in influenza A viruses. Nat. Rev. Microbiol. 12, 822–831. doi: 10.1038/nrmicro3362
Souto, S., Lopez-Jimena, B., Alonso, M. C., Garcia-Rosado, E., and Bandin, I. (2015). Experimental susceptibility of European sea bass and Senegalese sole to different betanodavirus isolates. Vet. Microbiol. 177, 53–61. doi: 10.1016/j.vetmic.2015.02.030
Stock, S. P., Vandenburg, J., Glazer, I., and Boemare, N. (2009). Insect Pathogens: Molecular Approaches and Techniques. Wallingford: CABI Publishing press.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tesh, R. B. (1980). Infectivity and pathogenicity of Nodamura virus for mosquitoes. J. Gen. Virol. 48, 177–182.
Thitamadee, S., Prachumwat, A., Srisala, J., Jaroenlak, P., Salachan, P. V., Sritunyalucksana, K., et al. (2016). Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture 452, 69–87. doi: 10.1016/j.aquaculture.2015.10.028
Ucko, M., Colorni, A., and Diamant, A. (2004). Nodavirus infections in Israeli mariculture. J. Fish Dis. 27, 459–469.
Walker, P. J., and Winton, J. R. (2010). Emerging viral diseases of fish and shrimp. Vet. Res. 41:51. doi: 10.1051/vetres/2010022
Zhang, Q., Liu, Q., Liu, S., Yang, H., Liu, S., Zhu, L., et al. (2014). A new nodavirus is associated with covert mortality disease of shrimp. J. Gen. Virol. 95, 2700–2709. doi: 10.1099/vir.0.070078-0
Zhang, Q. L., Liu, S., Yang, H. L., Zhu, L. L., Wan, X. H., Li, X. P, et al. (2017a). Reverse transcription loop-mediated isothermal amplification for rapid and quantitative assay of covert mortality nodavirus in shrimp. J. Invertebr. Pathol. 150, 130–135. doi: 10.1016/j.jip.2015.09.001
Keywords: alphanodavirus, covert mortality nodavirus (CMNV), host jump, natural infection, Mugilogobius abei
Citation: Zhang QL, Liu S, Li J, Xu TT, Wang XH, Fu GM, Li XP, Sang SW, Bian XD and Hao JW (2018) Evidence for Cross-Species Transmission of Covert Mortality Nodavirus to New Host of Mugilogobius abei. Front. Microbiol. 9:1447. doi: 10.3389/fmicb.2018.01447
Received: 19 March 2018; Accepted: 11 June 2018;
Published: 09 July 2018.
Edited by:
Akio Adachi, Kansai Medical University, JapanReviewed by:
Fuhua Li, Institute of Oceanology (CAS), ChinaNajiah Musa, Universiti Malaysia Terengganu, Malaysia
Copyright © 2018 Zhang, Liu, Li, Xu, Wang, Fu, Li, Sang, Bian and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Li Zhang, emhhbmdxbEB5c2ZyaS5hYy5jbg==
 Qing Li Zhang
Qing Li Zhang Shuang Liu1,2
Shuang Liu1,2 Jun Li
Jun Li Song Wen Sang
Song Wen Sang