
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 19 June 2018
Sec. Microbial Physiology and Metabolism
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01300
Nicotinamide adenine dinucleotides (NAD(H)) play a vital role in various biological processes, including keeping the cellular redox balance. In this study, we investigate the regulatory responses of Streptococcus pneumoniae D39 to NADH and characterize the role of the Rex protein as a transcriptional repressor of the gapN, fba, pncB, adhB2, gap, and adhE genes. Transcriptomic analysis was used to observe the response of S. pneumoniae D39 to NADH. Our microarray studies revealed elevated expression of various genes/operons involved in transport and biosynthesis of niacin (gapN, fba, pncB, adhB2, gap, and adhE). Promoter lacZ-fusion assays and microarray studies with the rex mutant revealed the role of Rex as a transcriptional repressor of gapN, fba, pncB, adhB2, gap, and adhE involved in niacin uptake and biosynthesis, in the presence of NADH. We predict the operator site (5′-TTGTKAWAAWWTTCACAA-3′) of Rex in the regulatory regions of Rex-regulated genes that was subsequently validated by promoter mutational experiments.
Streptococcus pneumoniae is an important human nasopharyngeal pathogen and causes several infections leading to over a million deaths around the globe each year (Ispahani et al., 2004; O’Brien et al., 2009). Bacteria have to make use of the nutrients present in their surroundings, in addition to the virulence factors they possess, to successfully colonize and spread infections (Phillips et al., 1990; Titgemeyer and Hillen, 2002). Vitamins and co-factors are among the most important compounds needed for bacterial growth.
Nicotinamide Adenine Dinucleotide (NAD) participates in several cellular processes as an important cofactor for living organisms (Ishino et al., 1986; Chiarugi et al., 2012; Patel et al., 2014). NAD+ is an oxidized form of NAD, whereas NADH is its reduced form and niacin (nicotinic acid) acts as a precursor of NAD and NADP (Jurtshuk, 1996). The role of niacin in the regulatory mechanisms of NiaR and on global gene expression in S. pneumoniae has been revealed in our previous study (Afzal et al., 2017). Several genes, including nadC, niaX, fba, pnuC, rex, pncB, gapN, gap, adhB2, and adhE, were differentially expressed and role of NiaR as a transcriptional repressor of nadC, pnuC, and niaX was demonstrated in our study (Afzal et al., 2017).
There are several processes in bacteria to reduce NAD+ to NADH, for instance respiration, which involves either glycolysis and the tricarboxylic acid cycle or fermentation (Jurtshuk, 1996). The regeneration of NAD+ is an important process in bacteria for continued carbohydrate catabolism (Marquis, 1995; Higuchi et al., 1999). Aerobically, and when glucose is abundant, the intracellular concentration of fructose 1,6-bisphosphate increases, which induces the expression of lactate dehydrogenase (ldh) (Abbe et al., 1982). Elevated ldh expression levels keep the redox balance required for continuous glycolytic activity, as pyruvate gets converted into lactate and NADH is oxidized to NAD+ (Abbe et al., 1982). Bacteria also have the ability to use NAD+ in different forms of dehydrogenases when metabolizing aldehydes and alcohols (Temple et al., 1994; Nobelmann and Lengeler, 1996; Luong et al., 2015).
Rex was recognized in Streptomyces coelicolor as a redox-associated transcriptional regulator of nuoA-nuoN, hemACD, and cydABCD (Brekasis and Paget, 2003). Since then, the Rex protein has been identified in numerous other model microorganisms including Staphylococcus aureus, Bacillus subtilis, Enterococcus faecalis, and Streptococcus mutans. The Rex regulon has been experimentally characterized in the model hyperthermophilic bacterium Thermotoga maritima (Ravcheev et al., 2012). In Desulfovibrio vulgaris Hildenborough, a model Gram-negative bacterium for studying sulfate reduction, Rex has been described to be regulated by NADH and acts as a repressor of sulfate adenylyl transferase (Christensen et al., 2015). Moreover, Rex has been shown to be a global redox-sensing transcriptional regulator in Clostridium kluyveri and its role in the regulation of oxidative stress tolerance and alcohol production has been studied in Clostridium acetobutylicum (Zhang et al., 2014; Hu et al., 2016). Furthermore, Rex-associated transcriptional modulation has been attributed to species-specific genetic targets of various organisms and a putative Rex-binding motif and individual sites have been predicted previously (Ravcheev et al., 2012). Rex monitors the transcription of genes associated with fermentation, amino acid metabolism, and nitrate/nitrite respiration in S. aureus (Pagels et al., 2010). Rex regulates the transcription of genes related to oxidative stress response, biofilm formation, and fermentation in cariogenic S. mutans (Baker et al., 2014; Bitoun et al., 2011, 2012). Rex-deficiency in S. mutans leads to enhanced sensitivity to exogenous H2O2 and higher end-point pH values of stationary-phase culture medium (Baker et al., 2014; Bitoun et al., 2011, 2012). Moreover, absence of Rex results in diminished growth of aerated cultures, which can be restored by adding catalase to the medium. These observations suggest that a rex-deletion mutant has reduced capability to deal with oxidative stress (Mehmeti et al., 2011; Vesić and Kristich, 2013).
This study reveals the impact of NADH on the transcriptome of S. pneumoniae and characterizes the Rex regulon. We demonstrate that Rex represses the expression of several genes/operons involved in NADH uptake and consumption. Furthermore, we predict the putative operator site (5′- TTGTKAWAAWWTTCACAA -3′ such that W = A/T and K = G/T) of Rex in the promoter regions of its regulon genes that is also confirmed by promoter mutational experiments.
Bacterial strains and plasmids are tabulated in Supplementary Table S1 and primers in Supplementary Table S2. S. pneumoniae D39 chromosomal DNA was used to perform PCR amplification (Lanie et al., 2007). Growth of S. pneumoniae D39 and DNA manipulation techniques were performed as mentioned before (Kloosterman et al., 2006; Afzal et al., 2014). We used S. pneumoniae D39 derivatives grown in a chemically defined medium (CDM) supplemented either with 0.5 mg/ml NADH or without NADH for β-galactosidase assays. CDM was prepared without niacin. For pneumococcal selection, 2.5 μg/ml tetracycline was used and for Escherichia coli, 100 μg/ml ampicillin was used.
pORI280 was used to construct a marker-less rex mutant (MA1200) in S. pneumoniae D39 (Kloosterman et al., 2006). Left and right flanking regions of rex were generated using primer pairs Rex-KO-1/ Rex-KO-2 and Rex-KO-3/Rex-KO-4. The PCR fragment of the left flanking region contained EcoRI site at its 5′ end, whereas the PCR fragment of the right flanking region had BamHI at its 3′ end. These PCR fragments were joined together through overlap extension PCR and then cloned into the EcoRI/BamHI sites of pORI280 in E. coli EC1000, yielding pMA1200. The pOR1280 integrative vector carries an erythromycin resistance gene to select for chromosomal integrants and an expressed lacZ gene (colonies blue on X-Gal) to score the loss of the plasmid integrated into the chromosome. To achieve selection for erythromycin resistance, transformation of S. pneumoniae D39 was performed with pMA1200. pORI280 relies on RepA for replication. Therefore, the transformation of S. pneumoniae D39 results in single cross-over integration of the construct into the chromosome. Many LacZ-positive erythromycin-resistant integrants were plated on X-Gal medium (without antibiotic selection) as individual cultures for 30–50 generations (culturing two to four times until stationary phase). This led to the selection for clones that are now without the integration due to a second recombination event. Only few of the colonies were both white and erythromycin sensitive, which suggests that the plasmid had been excised from the chromosome. The required mutation was present in about eighty percent of these white erythromycin-sensitive colonies, which was then confirmed by colony PCR and DNA sequencing.
Transformation was performed by growing cells first at 37°C without shaking until an OD600 of ∼0.1. Afterwards, 100 ng/μl of CSP1, 0.2% BSA, and 1mM CaCl2 were added to 1ml of the grown culture in the 1.5 ml Eppendorf tube. After incubating cells at 37°C for 10–12 minutes, the DNA mixture was added to the incubated cells and the cells were grown for another 90–120 minutes at 37°C. The cell pellet was recovered by centrifugation for 1 minute at 7000 RPM. The cell pellet was dissolved in the 50–100 μl medium and plated out on blood agar plates.
We used primer pairs mentioned in Supplementary Table S2 to construct chromosomal transcriptional lacZ-fusions to PadhB2-lacZ, PgapN-lacZ, Pgap-lacZ, PpncB-lacZ, Pfba-lacZ, and PadhE-lacZ in pPP2 (Halfmann et al., 2007), resulting in strains listed in Supplementary Table S1. The above-mentioned promoter fusions were also transformed into the D39 Δrex strain, creating strains MA1201-MA1206, respectively. The following clones of PgapN-lacZ PpncB-lacZ, Pfba-lacZ, Pgap-lacZ, PadhE-R1-lacZ, and PadhE-R2-lacZ were constructed in pPP2 (Halfmann et al., 2007) with the primer pairs listed in Supplementary Table S2: Pfba-M, PpncB-M, Pgap-M, PadhE-M-R1, PadhE-M-R2, and PgapN-M, creating plasmids pMA1201-06, respectively. These lacZ-fusion constructs were transformed into the D39 wild-type, forming strains MA1207-12, respectively.
Beta-galactosidase assays were performed as mentioned previously using cells grown in CDM with or without added 0.5 mg/ml NADH and collected in the mid-log phase (Israelsen et al., 1995; Halfmann et al., 2007).
Microarray comparison of S. pneumoniae (in replicates) was performed by growing S. pneumoniae D39 wild-type in CDM having 0 mg/ml NADH and 0.5 mg/ml NADH. CDM was prepared without niacin, because niacin is a precursor of NAD and NAD+. Similar procedure was performed for Δrex where the global gene expression changes in Δrex were measured by comparing the S. pneumoniae D39 Δrex transcriptome against the S. pneumoniae D39 wild-type transcriptome both grown in replicates in complete CDM. Cells growing in the respective mid-log phase were collected for experiments. Data analysis and other procedures were adopted from previous studies (Shafeeq et al., 2011a,b; Afzal et al., 2015). A gene qualified to be differentially expressed if it satisfied a Bayesian p-value of <0.001 and a fold-change cut-off >1.5. Microarray data of this study submitted to Gene Expression Omnibus has been assigned the accession number GSE94573.
Niacin is a naturally existing vitamin B complex that forms important coenzymes NAD and NAD+, and also has a part in electron transfer in different metabolic activities (Wei et al., 2014). NAD is required in several enzymatic processes where it acts as an electron carrier either by being oxidized (NAD+) or reduced (NADH) (Belenky et al., 2007). Microarray comparison of S. pneumoniae was performed, where S. pneumoniae D39 wild-type was grown in CDM having 0 mg/ml NADH and 0.5 mg/ml NADH. Niacin was excluded from the CDM. Several genes/gene clusters exhibited differential expression under our experimental settings (Table 1). spd-0113-124 were considerably downregulated when there was no NADH in medium. Several genes of this putative gene cluster (spd-0113-124) were also differentially expressed in our previous study, where we had explored the global gene expression changes induced by niacin (Afzal et al., 2017). Further investigation of the role of these genes in the presence of NADH and niacin might be very interesting.
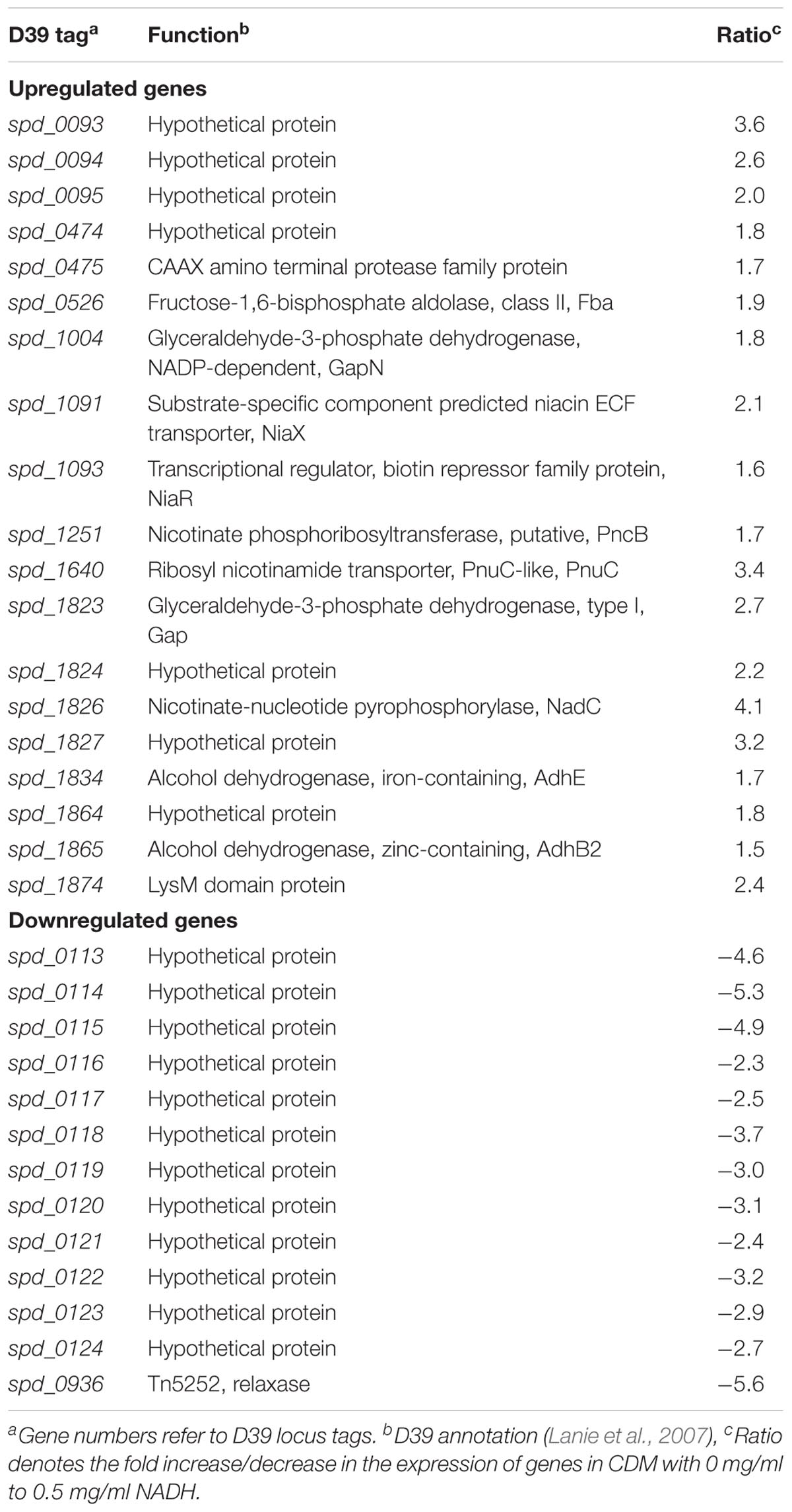
TABLE 1. List of genes differentially expressed in S. pneumoniae D39 wild-type grown in CDM with 0 mg/ml to 0.5 mg/ml NADH.
An increase in the expression of spd-0093-95 could be observed in the absence of NADH. This putative gene cluster codes for membrane proteins of unknown functions. Another gene coding for a putative fructose-1,6-bisphosphate aldolase was upregulated in our tested conditions. The genes, putatively involved in the transport and metabolism of niacin, were significantly upregulated in the absence of NADH. These genes are pnuC, niaX, and nadC, and code for a ribosyl nicotinamide transporter, a predicted substrate-specific niacin ECF transporter, and a nicotinate-nucleotide pyrophosphorylase, respectively. NadC has been anticipated to convert quinolinate made from tryptophan, aspartate, alanine, and glutamate metabolism into nicotinate D-ribonucleotide (Kanehisa et al., 2014). Many other genes showed altered expression in the absence of NADH. These genes include fba, pncB, gapN, gap, adhB2, and adhE. The expression of these genes was also altered in the presence of niacin (Afzal et al., 2017). These genes are proposed to be involved in the NADH pathway. Therefore, we further investigated the regulatory mode of these genes in the presence of NADH.
Beta-galactosidase activity for pncB, gapN, fba, niaX, pnuC, gap, adhB2, adhE, and nadC promoters lacZ-fusions made in S. pneumoniae D39 wild-type was measured using β-galactosidase assays. β-galactosidase assays results strengthened our microarray results and established that the expression of PpncB-lacZ, PgapN-lacZ, Pgap-lacZ, Pfba-lacZ, PniaX-lacZ, PpnuC-lacZ, PnadC-lacZ, PadhE-lacZ, and PadhB2-lacZ elevated notably without added NADH in the medium (Figure 1).
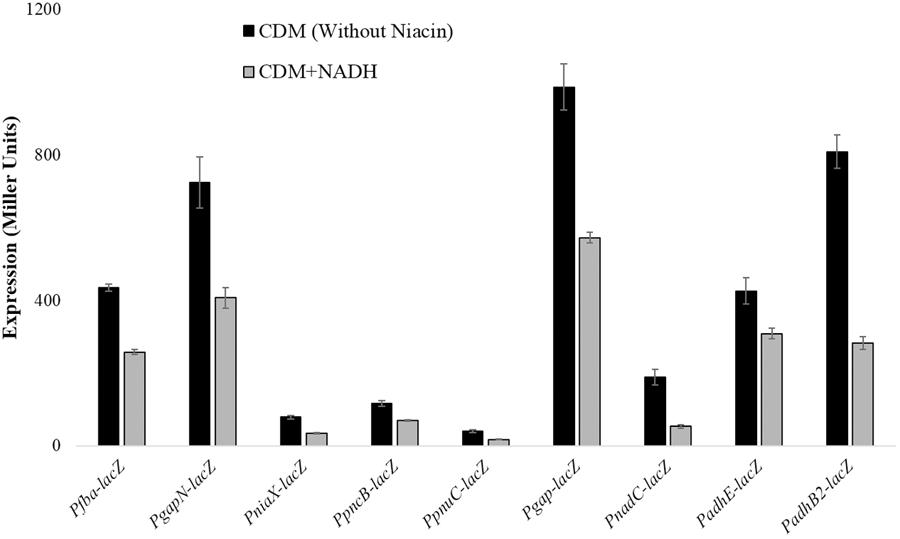
FIGURE 1. Expression levels (in Miller units) of PgapN-lacZ, Pfba-lacZ, PniaX-lacZ, PpnuC-lacZ, PpncB-lacZ, Pgap-lacZ, PadhB2-lacZ, PnadC-lacZ, and PadhE-lacZ in S. pneumoniae D39 wild-type grown in CDM with 0 mg/ml and 0.5 mg/ml NADH. CDM was prepared without niacin. Expression levels measurements were performed from three independent experiments and the values represented are means ± SD.
We further deleted the rex gene to examine its role in S. pneumoniae D39. Microarray study was performed, where S. pneumoniae D39 Δrex transcriptome was compared to that of D39 wild-type grown in complete CDM. Niacin is present at the concentration 8 μM in complete CDM, which is the normal concentration of niacin used in preparing CDM. Table 2 enlists the gene expression changes due to the absence of rex in S. pneumoniae D39. Expression of fba, pncB, gapN, gap, adhB2, and adhE, was significantly increased in D39 Δrex signifying the role of Rex as a transcriptional repressor of gapN, fba, pncB, adhE, adhB2, and gap in S. pneumoniae D39.
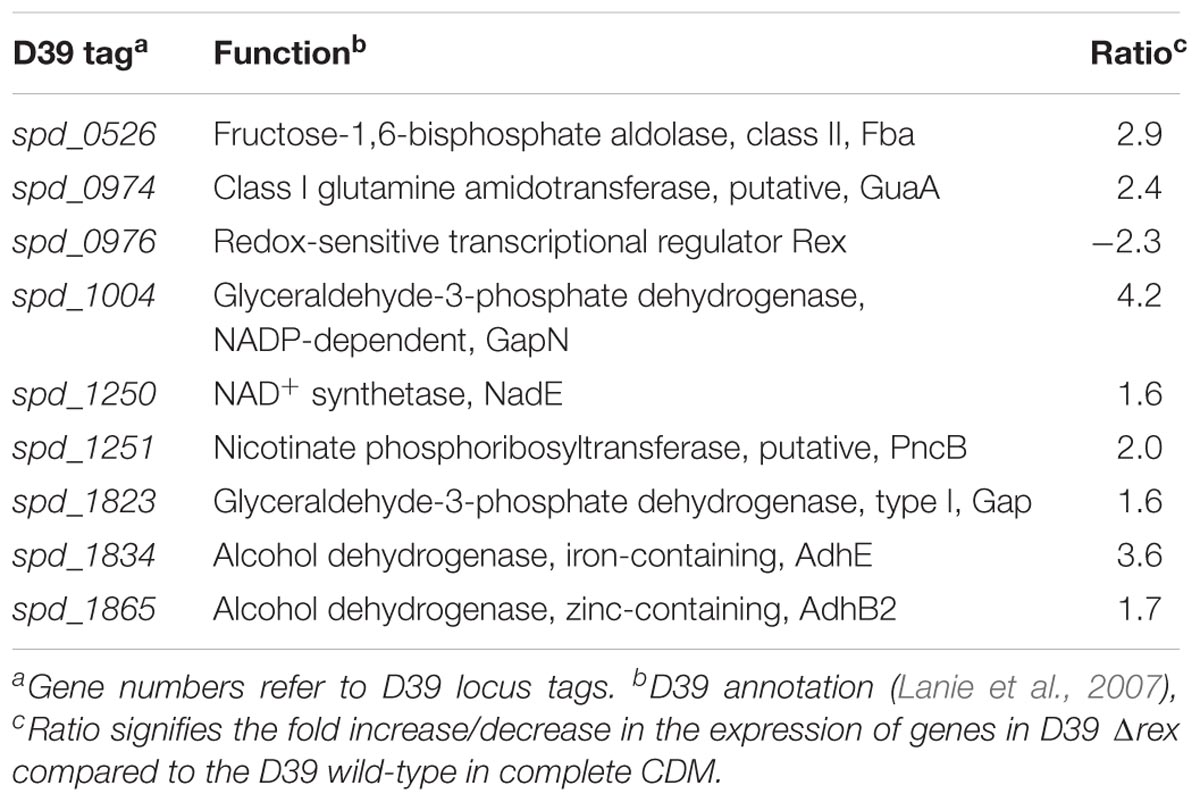
TABLE 2. Genes differentially expressed in S. pneumoniae D39 Δrex compared to the D39 wild-type grown in complete CDM.
To further investigate the role of Rex in the modulation of gapN, fba, pncB, adhE, adhB2, and gap, we constructed lacZ promoter fusions of gapN, fba, pncB, adhE, adhB2, and gap into S. pneumoniae D39 Δrex and measured β-galactosidase activity in complete CDM (Figure 2). Our enzyme assays results indicated an upregulation in the expression of all these promoters lacZ-fusions in D39 Δrex compared to the D39 wild-type, corroborating the role of Rex as a transcriptional repressor of gapN, fba, pncB, adhE, adhB2, and gap.
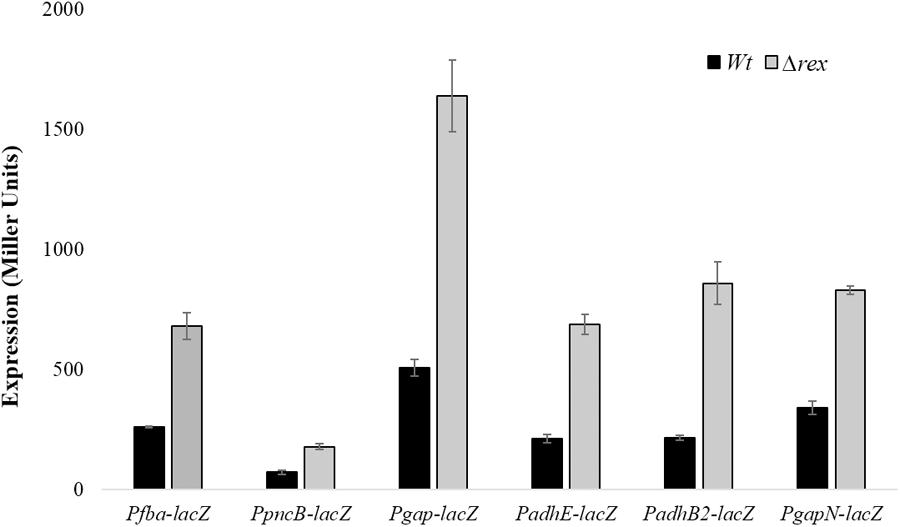
FIGURE 2. Expression levels (in Miller units) of Pfba-lacZ, PpncB-lacZ, Pgap-lacZ, PadhE-lacZ, PadhB2-lacZ, and PgapN-lacZ in S. pneumoniae D39 wild-type and D39 Δrex grown in complete CDM. Expression levels measurements were performed from three independent experiments and the values represented are means ± SD.
The Rex sites are predicted in bacteria using a comparative genomics approach (Ravcheev et al., 2012). The prediction identifies two other putative Rex targets (forT/spd_1075 and hemH/spd_0895) as well. A notable change in the expression of forT and hemH could not be established in our study neither in the presence of added NADH nor in the rex mutant. This might disprove the original prediction for these two genes. Moreover, Regprecise database has also speculated the Rex binding sites in S. pneumoniae (Novichkov et al., 2010). We analyzed the promoter regions of gapN, adhB2, fba, gap, pncB, and adhE using a software Genome-2D, and found an 18-bp palindromic-like sequence (5′- TTGTKAWAAWWTTCACAA -3′) similar to the Rex site projected by the Regprecise (Baerends et al., 2004; Novichkov et al., 2010). The Rex site found in the regulatory regions of gapN, adhB2 fba, gap, pncB, and adhE is shown in Figure 3.
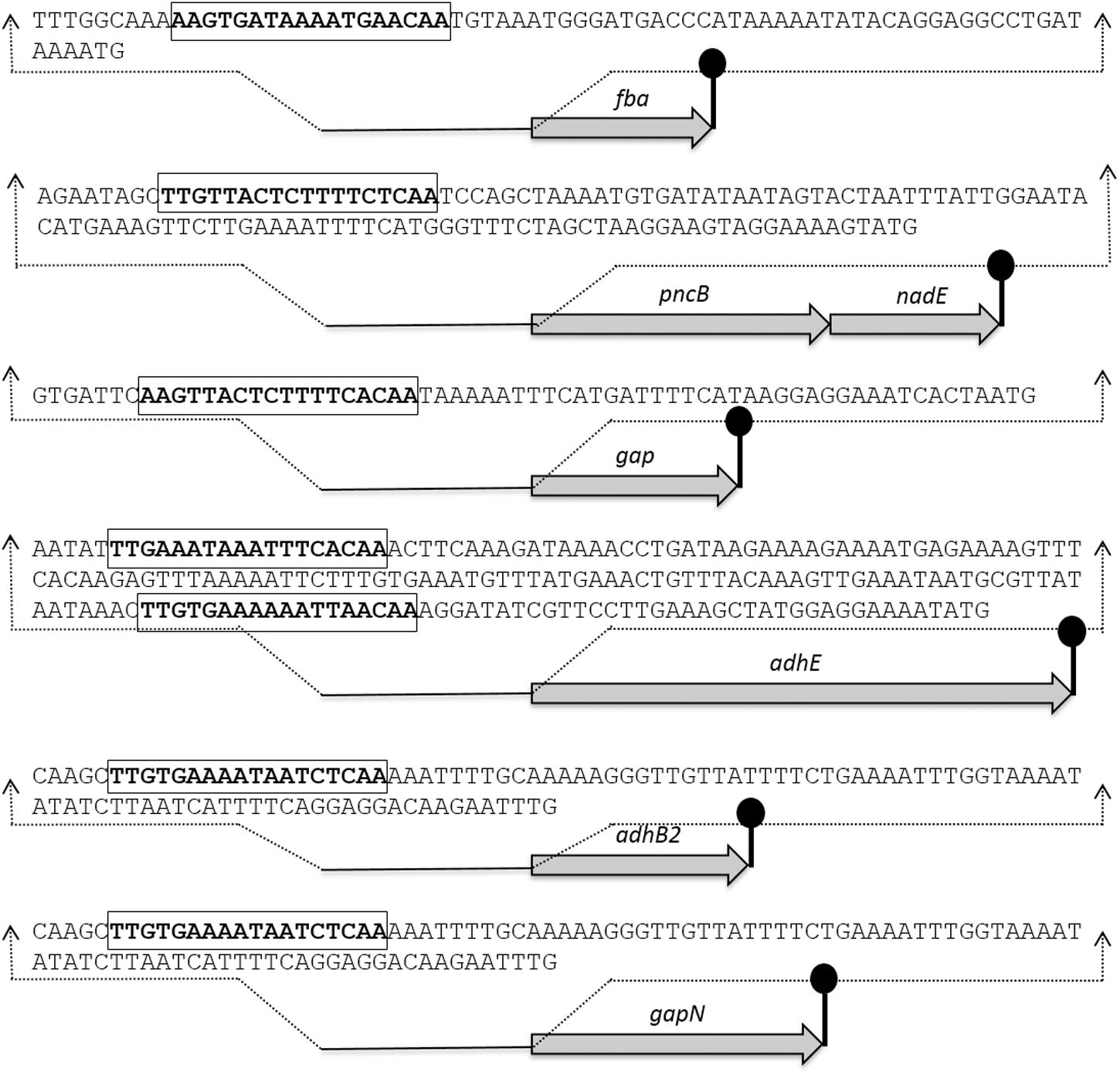
FIGURE 3. Detailed promoter sequences and organization of the Rex regulated genes in S. pneumoniae D39. Putative transcriptional terminators are denoted by the lollipop structures and the putative Rex operator sequences are in bold and rectangle.
To verify the Rex operator site situated in the regulatory sequences of gapN, adhB2, fba, gap, pncB, and adhE, we made transcriptional lacZ-fusions to Pfba, PpncB, Pgap, PadhE, and PgapN, in which consensus bases in the predicted Rex binding sites were mutated in Pfba, PpncB, Pgap, PadhE, and PgapN (Figure 4). We performed β-galactosidase assays in complete CDM. The expression of Pfba, PpncB, Pgap, and PgapN elevated considerably when Rex operator sites were mutated in the wild-type. This asserts that in S. pneumoniae D39, the predicted Rex sites in the regulatory areas of fba, pncB, gap, and gapN are intact and active (Figure 5). There are four possible operator sites for Rex in PadhE. Two of them were mutated individually in PadhE and β-galactosidase assays were performed. Derepression caused by Rex could only be seen when the Rex operator site R2 (5′- TTGTGAAAAAATTAACAA -3′) was mutated and we did not detect any significant change in the expression of PadhE due to mutation in Rex operator site R1 (5′- TTGAAATAAATTTCACAA -3′) (Figure 4). These data propose that operator site 2 (R2) is a functional operator site for Rex in PadhE (Figure 5). These data also suggest that the in silico predicted consensus sequences may not always be exactly correct and they must be confirmed by experiments.
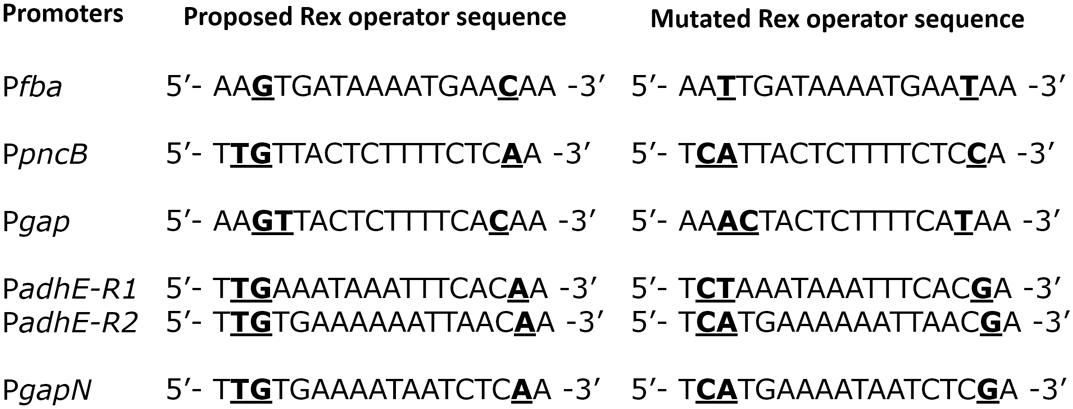
FIGURE 4. Aligned mutated and non-mutated Rex operator sequences in Pfba, PpncB, Pgap, PadhE, and PgapN.
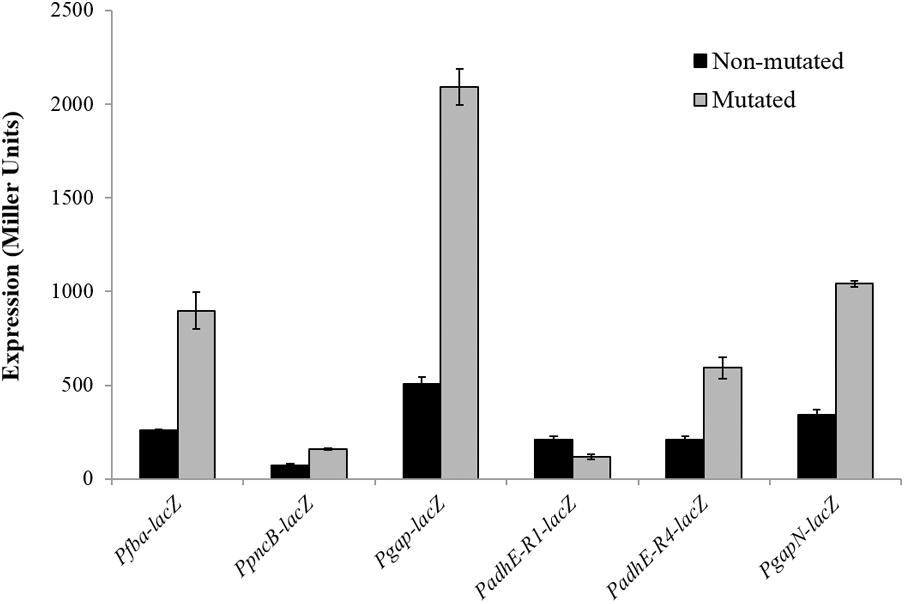
FIGURE 5. Expression levels (in Miller units) of Pfba-lacZ, PpncB-lacZ, Pgap-lacZ, PadhE-lacZ, and PgapN-lacZ with mutated and non-mutated Rex operator sites in S. pneumoniae D39 wild-type grown in complete CDM. Expression levels measurements were performed from three independent experiments and the values represented are means ± SD.
Based on the available knowledge, NAD metabolism can be best termed as a chain of events, none of which are absolutely preserved (Gazzaniga et al., 2009). Moreover, we note that the modulation mechanisms regulating the expression of specific gene units and link NAD absorption to vibrant cellular and ecological exercises, are vaguely known. Advances in computational techniques, together with molecular genetic methods developed recently can be employed to monitor how NAD metabolic gene products and enzymes are modulated to react to environmental cues and to manage microbiological tasks (Gazzaniga et al., 2009). This study sheds light on the regulatory role of Rex in the transcriptional regulation of adhB2, fba, pncB, gapN, adhE, and gap.
Transcriptomic analysis of D39 wild-type with 0 mg/ml to 0.5 mg/ml NADH exhibited an increase in the expression of niaX, pnuC, nadC, fba, pncB, gapN, gap, adhB2, and adhE. Promoter lacZ-fusions and microarray studies with a rex mutant showed that Rex acts as a transcriptional repressor of several genes/operons (gapN, fba, gap, pncB, adhB2, and adhE) involved in niacin uptake and biosynthesis. Fba (a fructose-bisphosphate aldolase encoded by fba) has been deemed essential for glycolysis and gluconeogenesis and is a potential drug target in Mycobacterium tuberculosis. Fba has been the center of structural, enzymatic, and drug developmental studies (Pegan et al., 2009; Puckett et al., 2014). gapN encodes a glyceraldehyde-3-phosphate dehydrogenase and NAD-dependent glyceraldehyde-3-phosphate dehydrogenase, and gap encodes a glyceraldehyde-3-phosphate dehydrogenase. Glycolysis and gluconeogenesis employ GAPs as the key players by enhancing the reversible oxidative phosphorylation of D-glyceraldehyde 3-phosphate to the energy-rich intermediate glyceraldehyde 1,3-bisphosphate. Moreover, GAP is widely known to have an extensive range of biological functions (Zhou et al., 2008; Sirover, 2012). Extracellular GAPs are described to have a role in pathogenesis of many bacteria (Seifert et al., 2003; Oliveira et al., 2012). AdhE (an iron-containing alcohol dehydrogenase coded by adhE has been shown to increase pneumolysin (Ply) during the ethanol stress conditions, therefore, enhancing pneumococcal virulence (Luong et al., 2015). AdhE has also been shown to regulate virulence in E. coli (Beckham et al., 2014). pncB encodes a nicotinate phosphoribosyl-transferase and adhB2 encodes a zinc-containing alcohol dehydrogenase. Involvement of the above-mentioned genes in virulence and their pivotal role for bacterial cell functions underscore the importance of studying their regulatory mode.
Our study demonstrates that in S. pneumoniae Rex is a transcriptional repressor of gapN, adhB2, fba, pncB, adhE, and gap. The Rex protein has been identified in numerous model microorganisms, including S. aureus, B. subtilis, E. faecalis, and S. mutans. In B. subtilis, Rex has been shown to repress expression of cydABCD (cytochrome bd oxidase), yjlC-ndh (NADH dehydrogenase), ywcJ (a formate-nitrate transporter), and lctP-ldh (NADH-linked fermentative lactate dehydrogenase) (Larsson et al., 2005; Gyan et al., 2006; Wang et al., 2008). Rex directly regulates at least 19 genes in S. aureus. It acts as a key transcriptional regulator of anaerobic metabolism leading to anaerobic NAD+ regeneration, which includes lactate, formate, and ethanol fermentation (adh1, adhE, lctP, ldh1, and pflBA) and nitrate respiration (narG, nirC, and nirR). Furthermore, in many organisms, Rex-mediated transcriptional regulation has been attributed to species-specific genetic targets (Ravcheev et al., 2012). Moreover, Rex regulates the transcription of genes involved in fermentation, oxidative stress response, and biofilm formation in cariogenic S. mutans (Bitoun et al., 2011, 2012; Baker et al., 2014). Based on RegPrecise database annotation1, it appears that the number of genes regulated by Rex can vary in different organisms. In addition to gap, pncB, and adhE, Rex may also be regulating pgk (phosphoglycerate kinase), ldh (lactate dehydrogenase), fba (Fructose-bisphosphate aldolase), eno (Enolase), and frdC (fumarate reductase) in S. mutans (Novichkov et al., 2010). Similarly, in addition to gap, pncB, and adhB1, Rex may be regulating tpi (triosephosphate isomerase), noxE (NADH oxidase), ldh (lactate dehydrogenase), ahpC (alkyl hydroperoxide reductase), forT (Formate/nitrite family of transporters), and eno (enolase) in Streptococcus pyogenes (Novichkov et al., 2010).
Interestingly, the binding sequence of Rex is highly conserved in Gram-positive bacteria. The reported consensus sequence in S. coelicolor (5′-TGTGAACNNNTTCACA-3′) (Brekasis and Paget, 2003), B. subtilis (5′-WWTGTGAANTNNTNNNCAAW-3′; W represents either A or T) (Wang et al., 2008), and S. aureus (5′-TTGTGAAWWWWTTCACAA-3′) (Pagels et al., 2010) are very similar. The binding site of Rex found in S. pneumoniae D39 (5′-TTGTKAWAAWWTTCACAA-3′) appears to be consistent with the above ones. Four putative Rex binding sites, 5′-TTGAAATAAATTTCACAA-3′, 5′-ATGAGAAAAGTTTCACAA-3′, 5′-TTATGAAACTGTTTACAA-3′ and 5′-TTGTGAAAAAATTAACAA-3′ were predicted to be preset in the adhE regulatory region. The first two sites and the last site are characterized by a high score (strong sites), whereas the third site is rather weak. The first and last sites were mutated in this study to check the functionality of the predicted sites and it was only the last site that appeared to be the functional operator site for Rex in the adhE regulatory region.
The expression of niaX, pnuC, nadC, gapN, fba, pncB, adhE, adhB2, and gap was also altered in our previous niacin microarray experiment, where we compared S. pneumoniae transcriptome with 0 mM to 10 mM niacin (Afzal et al., 2017). These genes are proposed to be involved in the NADH pathway, and niacin is the precursor of NADH. Proteins coded by two of the above-mentioned genes (NiaX and PnuC) are transporters (Johnson et al., 2015). We were unable to find a considerable difference in the expression of some other transporters, which might suggest that NiaX/PnuC might have a role in the transport of NADH. We are not sure whether NADH is potentially transported like niacin or whether it is first hydrolyzed to niacin extracellularly and then transported via NiaX/PnuC. One possible explanation might be that NADH is first converted into a form that is transportable by NiaX/PnuC. Although the data in literature show that NiaX exists as a preferred transporter for importing nicotinic acid and nicotinamide, whereas PnuC specifically imports nicotinamide ribosides (Johnson et al., 2015), NiaX and PnuC may be assisted by an extracellular protein having the ability to modify nicotinamide mononucleotide to an importable metabolite or pneumococcus might be possessing another import system (Johnson et al., 2015). The presence of another couple of putative niacin transporters (NiaY and NiaP) in the genomes of the Bacillus/Clostridium might support the theory of another possible niacin transporter in S. pneumoniae (Rodionov et al., 2008).
MA, SS, and OK substantially contributed to the conception or design of the work or the acquired, analyzed, or interpreted the data for the work; drafted the work or revised it critically for important intellectual content; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Government College University, Faisalabad, Pakistan, is supporting MA under the faculty development program of the Higher Education Commission Pakistan.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01300/full#supplementary-material
Abbe, K., Takahashi, S., and Yamada, T. (1982). Involvement of oxygen-sensitive pyruvate formate-lyase in mixed-acid fermentation by Streptococcus mutans under strictly anaerobic conditions. J. Bacteriol. 152, 175–182.
Afzal, M., Shafeeq, S., Henriques-Normark, B., and Kuipers, O. P. (2015). UlaR activates expression of the ula operon in Streptococcus pneumoniae in the presence of ascorbic acid. Microbiolology 161, 41–49. doi: 10.1099/mic.0.083899-0
Afzal, M., Shafeeq, S., and Kuipers, O. P. (2014). LacR is a repressor of lacABCD and LacT an activator of lacTFEG, constituting the lac-gene cluster in Streptococcus pneumoniae. Appl. Environ. Microbiol. 80, 5219–5230. doi: 10.1128/AEM.01370-14
Afzal, M., Shafeeq, S., and Kuipers, O. P. (2017). Niacin-mediated gene expression and role of NiaR as a transcriptional repressor of niaX, nadC and pnuC in Streptococcus pneumoniae. Front. Cell. Infect. Microbiol. 7:70. doi: 10.3389/fcimb.2017.00070
Baerends, R. J. S., Smits, W. K., de Jong, A., Hamoen, L. W., Kok, J., and Kuipers, O. P. (2004). Genome2D: a visualization tool for the rapid analysis of bacterial transcriptome data. Genome Biol. 5:R37. doi: 10.1186/gb-2004-5-5-r37
Baker, J. L., Derr, A. M., Karuppaiah, K., MacGilvray, M. E., Kajfasz, J. K., Faustoferri, R. C., et al. (2014). Streptococcus mutans NADH oxidase lies at the intersection of overlapping regulons controlled by oxygen and NAD+ levels. J. Bacteriol. 196, 2166–2177. doi: 10.1128/JB.01542-14
Beckham, K. S. H., Connolly, J. P. R., Ritchie, J. M., Wang, D., Gawthorne, J. A., Tahoun, A., et al. (2014). The metabolic enzyme AdhE controls the virulence of Escherichia coli O157:H7. Mol. Microbiol. 93, 199–211. doi: 10.1111/mmi.12651
Belenky, P., Bogan, K. L., and Brenner, C. (2007). NAD+ metabolism in health and disease. Trends Biochem. Sci. 32, 12–19. doi: 10.1016/j.tibs.2006.11.006
Bitoun, J. P., Liao, S., Yao, X., Xie, G. G., and Wen, Z. T. (2012). The redox-sensing regulator Rex modulates central carbon metabolism, stress tolerance response and biofilm formation by Streptococcus mutans. PLoS One 7:e44766. doi: 10.1371/journal.pone.0044766
Bitoun, J. P., Nguyen, A. H., Fan, Y., Burne, R. A., and Wen, Z. T. (2011). Transcriptional repressor Rex is involved in regulation of oxidative stress response and biofilm formation by Streptococcus mutans. FEMS Microbiol. Lett. 320, 110–117. doi: 10.1111/j.1574-6968.2011.02293.x
Brekasis, D., and Paget, M. S. B. (2003). A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2). EMBO J. 22, 4856–4865. doi: 10.1093/emboj/cdg453
Chiarugi, A., Dölle, C., Felici, R., and Ziegler, M. (2012). The NAD metabolome–a key determinant of cancer cell biology. Nat. Rev. Cancer 12, 741–752. doi: 10.1038/nrc3340
Christensen, G. A., Zane, G. M., Kazakov, A. E., Li, X., Rodionov, D. A., Novichkov, P. S., et al. (2015). Rex (encoded by DVU_0916) in Desulfovibrio vulgaris Hildenborough is a repressor of sulfate adenylyl transferase and is regulated by NADH. J. Bacteriol. 197, 29–39. doi: 10.1128/JB.02083-14
Gazzaniga, F., Stebbins, R., Chang, S. Z., McPeek, M. A., and Brenner, C. (2009). Microbial NAD metabolism: lessons from comparative genomics. Microbiol. Mol. Biol. Rev. 73, 529–541. doi: 10.1128/MMBR.00042-08
Gyan, S., Shiohira, Y., Sato, I., Takeuchi, M., and Sato, T. (2006). Regulatory loop between redox sensing of the NADH/NAD+ ratio by Rex (YdiH) and oxidation of NADH by NADH dehydrogenase Ndh in Bacillus subtilis. J. Bacteriol. 188, 7062–7071. doi: 10.1128/JB.00601-06
Halfmann, A., Hakenbeck, R., and Bruckner, R. (2007). A new integrative reporter plasmid for Streptococcus pneumoniae. FEMS Microbiol. Lett. 268, 217–224. doi: 10.1111/j.1574-6968.2006.00584.x
Higuchi, M., Yamamoto, Y., Poole, L. B., Shimada, M., Sato, Y., Takahashi, N., et al. (1999). Functions of two types of NADH oxidases in energy metabolism and oxidative stress of Streptococcus mutans. J. Bacteriol. 181, 5940–5947.
Hu, L., Huang, H., Yuan, H., Tao, F., Xie, H., and Wang, S. (2016). Rex in Clostridium kluyveri is a global redox-sensing transcriptional regulator. J. Biotechnol. 233, 17–25. doi: 10.1016/j.jbiotec.2016.06.024
Ishino, Y., Shinagawa, H., Makino, K., Tsunasawa, S., Sakiyama, F., and Nakata, A. (1986). Nucleotide sequence of the lig gene and primary structure of DNA ligase of Escherichia coli. Mol. Gen. Genet. 204, 1–7. doi: 10.1007/BF00330179
Ispahani, P., Slack, R. C. B., Donald, F. E., Weston, V. C., and Rutter, N. (2004). Twenty year surveillance of invasive pneumococcal disease in Nottingham: serogroups responsible and implications for immunisation. Arch. Dis. Child. 89, 757–762. doi: 10.1136/adc.2003.036921
Israelsen, H., Madsen, S. M., Vrang, A., Hansen, E. B., and Johansen, E. (1995). Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl. Environ. Microbiol. 61, 2540–2547.
Johnson, M. D. L., Echlin, H., Dao, T. H., and Rosch, J. W. (2015). Characterization of NAD salvage pathways and their role in virulence in Streptococcus pneumoniae. Microbiology 161, 2127–2136. doi: 10.1099/mic.0.000164
Jurtshuk, P. (1996). “Bacterial metabolism,” in Microbiology 161, 2127–2136. doi: 10.1099/mic.0.000164 Medical Microbiology, ed. S. Baron (Galveston, TX: University of Texas Medical Branch at Galveston).
Kanehisa, M., Goto, S., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2014). Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205. doi: 10.1093/nar/gkt1076
Kloosterman, T. G., Bijlsma, J. J. E., Kok, J., and Kuipers, O. P. (2006). To have neighbour’s fare: extending the molecular toolbox for Streptococcus pneumoniae. Microbiology 152, 351–359. doi: 10.1099/mic.0.28521-0
Lanie, J. A., Ng, W. L., Kazmierczak, K. M., Andrzejewski, T. M., Davidsen, T. M., Wayne, K. J., et al. (2007). Genome sequence of Avery’s virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189, 38–51. doi: 10.1128/JB.01148-06
Larsson, J. T., Rogstam, A., and von Wachenfeldt, C. (2005). Coordinated patterns of cytochrome bd and lactate dehydrogenase expression in Bacillus subtilis. Microbiology 151, 3323–3335. doi: 10.1099/mic.0.28124-0
Luong, T. T., Kim, E.-H., Bak, J. P., Nguyen, C. T., Choi, S., Briles, D. E., et al. (2015). Ethanol-induced alcohol dehydrogenase E (AdhE) potentiates pneumolysin in Streptococcus pneumoniae. Infect. Immun. 83, 108–119. doi: 10.1128/IAI.02434-14
Marquis, R. E. (1995). Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J. Ind. Microbiol. 15, 198–207. doi: 10.1007/BF01569826
Mehmeti, I., Jönsson, M., Fergestad, E. M., Mathiesen, G., Nes, I. F., and Holo, H. (2011). Transcriptome, proteome, and metabolite analyses of a lactate dehydrogenase-negative mutant of Enterococcus faecalis V583. Appl. Environ. Microbiol. 77, 2406–2413. doi: 10.1128/AEM.02485-10
Nobelmann, B., and Lengeler, J. W. (1996). Molecular analysis of the gat genes from Escherichia coli and of their roles in galactitol transport and metabolism. J. Bacteriol. 178, 6790–6795. doi: 10.1128/jb.178.23.6790-6795.1996
Novichkov, P. S., Laikova, O. N., Novichkova, E. S., Gelfand, M. S., Arkin, A. P., Dubchak, I., et al. (2010). RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38, D111–D118. doi: 10.1093/nar/gkp894
O’Brien, K. L., Wolfson, L. J., Watt, J. P., Henkle, E., Deloria-Knoll, M., McCall, N., et al. (2009). Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374, 893–902. doi: 10.1016/S0140-6736(09)61204-6
Oliveira, L., Madureira, P., Andrade, E. B., Bouaboud, A., Morello, E., Ferreira, P., et al. (2012). Group B streptococcus GAPDH is released upon cell lysis, associates with bacterial surface, and induces apoptosis in murine macrophages. PLoS One 7:e29963. doi: 10.1371/journal.pone.0029963
Pagels, M., Fuchs, S., Pané-Farré, J., Kohler, C., Menschner, L., Hecker, M., et al. (2010). Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol. Microbiol. 76, 1142–1161. doi: 10.1111/j.1365-2958.2010.07105.x
Patel, M. S., Nemeria, N. S., Furey, W., and Jordan, F. (2014). The pyruvate dehydrogenase complexes: structure-based function and regulation. J. Biol. Chem. 289, 16615–16623. doi: 10.1074/jbc.R114.563148
Pegan, S. D., Rukseree, K., Franzblau, S. G., and Mesecar, A. D. (2009). Structural basis for catalysis of a tetrameric class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis. J. Mol. Biol. 386, 1038–1053. doi: 10.1016/j.jmb.2009.01.003
Phillips, N. J., John, C. M., Reinders, L. G., Gibson, B. W., Apicella, M. A., and Griffiss, J. M. (1990). Structural models for the cell surface lipooligosaccharides of Neisseria gonorrhoeae and Haemophilus influenzae. Biomed. Environ. Mass Spectrom. 19, 731–745. doi: 10.1002/bms.1200191112
Puckett, S., Trujillo, C., Eoh, H., Marrero, J., Spencer, J., Jackson, M., et al. (2014). Inactivation of fructose-1,6-bisphosphate aldolase prevents optimal co-catabolism of glycolytic and gluconeogenic carbon substrates in Mycobacterium tuberculosis. PLoS Pathog. 10:e1004144. doi: 10.1371/journal.ppat.1004144
Ravcheev, D. A., Li, X., Latif, H., Zengler, K., Leyn, S. A., Korostelev, Y. D., et al. (2012). Transcriptional regulation of central carbon and energy metabolism in bacteria by redox-responsive repressor Rex. J. Bacteriol. 194, 1145–1157. doi: 10.1128/JB.06412-11
Rodionov, D. A., De Ingeniis, J., Mancini, C., Cimadamore, F., Zhang, H., Osterman, A. L., et al. (2008). Transcriptional regulation of NAD metabolism in bacteria: NrtR family of Nudix-related regulators. Nucleic Acids Res. 36, 2047–2059. doi: 10.1093/nar/gkn047
Seifert, K. N., McArthur, W. P., Bleiweis, A. S., and Brady, L. J. (2003). Characterization of group B streptococcal glyceraldehyde-3-phosphate dehydrogenase: surface localization, enzymatic activity, and protein-protein interactions. Can. J. Microbiol. 49, 350–356. doi: 10.1139/w03-042
Shafeeq, S., Kloosterman, T. G., and Kuipers, O. P. (2011a). Transcriptional response of Streptococcus pneumoniae to Zn2+ limitation and the repressor/activator function of AdcR. Metallomics 3, 609–618. doi: 10.1039/c1mt00030f
Shafeeq, S., Yesilkaya, H., Kloosterman, T. G., Narayanan, G., Wandel, M., Andrew, P. W., et al. (2011b). The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol. Microbiol. 81, 1255–1270. doi: 10.1111/j.1365-2958.2011.07758.x
Sirover, M. A. (2012). Subcellular dynamics of multifunctional protein regulation: mechanisms of GAPDH intracellular translocation. J. Cell. Biochem. 113, 2193–2200. doi: 10.1002/jcb.24113
Temple, L., Sage, A., Christie, G. E., and Phibbs, P. V. (1994). Two genes for carbohydrate catabolism are divergently transcribed from a region of DNA containing the hexC locus in Pseudomonas aeruginosa PAO1. J. Bacteriol. 176, 4700–4709. doi: 10.1128/jb.176.15.4700-4709.1994
Titgemeyer, F., and Hillen, W. (2002). Global control of sugar metabolism: a gram-positive solution. Antonie Van Leeuwenhoek 82, 59–71. doi: 10.1023/A:1020628909429
Vesić, D., and Kristich, C. J. (2013). A Rex family transcriptional repressor influences H2O2 accumulation by Enterococcus faecalis. J. Bacteriol. 195, 1815–1824. doi: 10.1128/JB.02135-12
Wang, E., Bauer, M. C., Rogstam, A., Linse, S., Logan, D. T., and von Wachenfeldt, C. (2008). Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex. Mol. Microbiol. 69, 466–478. doi: 10.1111/j.1365-2958.2008.06295.x
Wei, Z., Fu, Y., Zhou, E., Tian, Y., Yao, M., Li, Y., et al. (2014). Effects of niacin on Staphylococcus aureus internalization into bovine mammary epithelial cells by modulating NF-κB activation. Microb. Pathog. 7, 62–67. doi: 10.1016/j.micpath.2014.03.005
Zhang, L., Nie, X., Ravcheev, D. A., Rodionov, D. A., Sheng, J., Gu, Y., et al. (2014). Redox-responsive repressor Rex modulates alcohol production and oxidative stress tolerance in Clostridium acetobutylicum. J. Bacteriol. 196, 3949–3963. doi: 10.1128/JB.02037-14
Zhou, Y., Yi, X., Stoffer, J. B., Bonafe, N., Gilmore-Hebert, M., McAlpine, J., et al. (2008). The multifunctional protein glyceraldehyde-3-phosphate dehydrogenase is both regulated and controls colony-stimulating factor-1 messenger RNA stability in ovarian cancer. Mol. Cancer Res. 6, 1375–1384. doi: 10.1158/1541-7786.MCR-07-2170
Keywords: NADH, Rex, PncB, pneumococcus, GaPN
Citation: Afzal M, Shafeeq S and Kuipers OP (2018) NADH-Mediated Gene Expression in Streptococcus pneumoniae and Role of Rex as a Transcriptional Repressor of the Rex-Regulon. Front. Microbiol. 9:1300. doi: 10.3389/fmicb.2018.01300
Received: 03 January 2018; Accepted: 28 May 2018;
Published: 19 June 2018.
Edited by:
Biswarup Mukhopadhyay, Virginia Tech, United StatesReviewed by:
Dmitry A. Rodionov, Sanford-Burnham Institute for Medical Research, United StatesCopyright © 2018 Afzal, Shafeeq and Kuipers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oscar P. Kuipers, by5wLmt1aXBlcnNAcnVnLm5s
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.