- 1Department of Medical Microbiology, Faculty of Health Sciences, University of Pretoria, Pretoria, South Africa
- 2Department of Medical Microbiology, Tshwane Academic Division, National Health Laboratory Service, Pretoria, South Africa
- 3Departments of Microbiology, Immunology, Infectious Diseases and Pathology and Laboratory Medicine, Cumming School of Medicine, University of Calgary, Calgary, AB, Canada
- 4Division of Microbiology, Calgary Laboratory Services, Calgary, AB, Canada
- 5Research Office, Faculty of Health Sciences, University of Pretoria, Pretoria, South Africa
Acinetobacter baumannii is an opportunistic pathogen that is increasingly responsible for hospital-acquired infections. The increasing prevalence of carbapenem resistant A. baumannii has left clinicians with limited treatment options. Last line antimicrobials (i.e., polymyxins and glycylcyclines) are often used as treatment options. The aim of this study was to determine the prevalence of selected β-lactamase genes from A. baumannii isolates obtained from patients with hospital-acquired infections and to determine the genetic relationship and epidemiological profiles among clinical A. baumannii isolates collected from two tertiary academic hospitals in the Tshwane region, South Africa (SA). Multiplex-PCR (M-PCR) assays were performed to detect selected resistance genes. The collected isolates’ genetic relatedness was determined by using pulsed field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). The acquired oxacillinase (OXA) genes, notably blaOXA-23-like were prevalent in the A. baumannii isolates. The M-PCR assays showed that the isolates collected from hospital A contained the OXA-23-like (96%; n = 69/72) genes and the isolates collected from hospital B contained the OXA-23-like (91%; n = 63/69) and OXA-58-like (4%; n = 3/69) genes. Colistin resistance was found in 1% of the isolates (n = 2/141) and tigecycline intermediate resistance was found in 6% of the isolates (n = 8/141). The A. baumannii isolates were genetically diverse. Molecular epidemiological data showed that specific sequence types (STs) (ST106, ST229, ST258 and ST208) were established in both hospitals, while ST848 was established in hospital A and ST502, ST339 and the novel ST1552 were established in hospital B. ST848 (established in hospital A) was predominately detected in ICU wards whereas ST208, ST339 and the novel ST1552 (established in hospital B) were detected in ICUs and the general wards. The origin of the A. baumannii isolates in the hospitals may be due to the dissemination and adaptation of a diverse group of successful clones. Poor infection control and prevention strategies and possibly the overuse of antimicrobials contributed to the establishment of these A. baumannii clones in the studied hospitals.
Introduction
The incidence of Acinetobacter baumannii infections increased over the last decade and unfortunately, so has this bacterium’s antimicrobial resistance (Jacobs et al., 2014). The main mechanisms of β-lactam resistance in A. baumannii are the production of carbapenemases, especially Ambler class D β-lactamases and to a lesser extent class B β-lactamases [metallo-β-lactamases (MBLs)] (Kuo et al., 2013; Salimizand et al., 2015; Ghaith et al., 2017). Most of the class D β-lactamases are not inhibited by β-lactamase inhibitors, but are inhibited in vitro by sodium chloride (NaCl) (Patel and Bonomo, 2013). The class D β-lactamases comprise of the intrinsic oxacillinase (OXA)-51-like as well as the acquired OXA-23- (OXA-27 and OXA-49); OXA-40- (OXA-25, OXA-26 and OXA-72); OXA-58- (OXA-96 and OXA-97); OXA-143- and OXA-235- (OXA-236 and OXA-237) like enzymes (Kuo et al., 2013; Potron et al., 2015; Ghaith et al., 2017). Insertion sequence (IS) elements play an important role in the mobilisation and expression of OXA-type β-lactamases and in the acquisition of resistance by A. baumannii (Evans and Amyes, 2014; Correa et al., 2017). Increased carbapenem resistance in isolates is often linked to ISAbaI, as the IS element provides an additional promoter that leads to the overexpression of determinants (Dijkshoorn et al., 2007; Peleg et al., 2008; Evans and Amyes, 2014).
Carbapenemase producing and other multidrug resistant (MDR) A. baumannii are extremely difficult to treat and have become a serious problem in healthcare settings due to the limited effective antimicrobial agents available and the association of these resistant strains with poor prognosis (Kuo et al., 2013; Lean et al., 2015). Colistin is often the last line of defence and is used either alone or in combination with other antimicrobials, such as tigecycline and meropenem (Cai et al., 2012; Sekyere, 2016). Colistin resistance among A. baumannii is still rare, but resistance and outbreaks have been reported in Italy, Korea and Spain (Agodi et al., 2014; Lean et al., 2014; Lee et al., 2014; Pournaras et al., 2014). Clinical laboratories have been advised to use the micro-broth dilution (MBD) assay to determine colistin susceptibility, since gradient diffusion and disc diffusion are unreliable for this drug class (EUCAST, 2016).
The establishment of clonal relationships among isolates through suitable molecular methods is fundamental for epidemiological surveillance and outbreak investigations as it permits the deployment of effective infection control measures (Cooper and Feil, 2004; Perez et al., 2007; Peleg et al., 2008; Hamouda et al., 2010; Field et al., 2014). The geographical distribution and evolution of the clonal strains can be determined (Grundmann et al., 1997; Field et al., 2014). Different methods can be used, which amongst others include pulsed field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). PFGE is highly discriminatory as minor mutations in outbreak strains can be detected (Fouad et al., 2013; Hu et al., 2013). As MLST is a sequence based method (sequence comparison of internal fragments of housekeeping genes), the global evolutionary relationships between clones can be determined (Cooper and Feil, 2004; Bartual et al., 2005; Fu et al., 2010; Hamouda et al., 2010; Yang et al., 2013; Hammoudi et al., 2014).
The aim of this study was to determine the prevalence of selected antimicrobial resistance genes from patients with hospital-acquired infections and to determine the genetic relationship and epidemiological profiles among clinical A. baumannii isolates collected from two tertiary academic hospitals in the Tshwane region, South Africa.
Materials and Methods
Isolate Collection, Study Setting and Ethics Statement
One hundred and fifty consecutive non-repeat clinical A. baumannii isolates were collected over an 8-month period (October 2013 to May 2014) at the diagnostic division of the Department of Medical Microbiology [Tshwane Academic Division, National Health Laboratory Service (NHLS)]. The laboratory processes specimens from tertiary academic hospitals as well as district hospitals and various clinics as part of standard care. Clinical A. baumannii isolates from two tertiary academic hospitals (75 isolates from each hospital) were selected for this study. Hospital A and B have 1 113 beds and 832 beds respectively. A total of nine isolates were excluded from this study, since the isolates tested negative for the OXA-51-like gene (six isolates) or were recovered from the same patient (three isolates). The results are reported on 72 isolates from hospital A and 69 isolates from hospital B (n = 141). All the collected isolates were from patients with hospital-acquired infections (conditions that developed 48 h after hospital admission).
This study received ethical approval from the Research Ethics Committee (REC), Faculty of Health Sciences, University of Pretoria (Protocol No. 71/2014). Informed consent was waivered by the REC, since the study was observational and patient care was not influenced.
Isolate Identification, Confirmation and Antimicrobial Susceptibility Profiles
The isolates were identified and tested for antimicrobial susceptibility using the VITEK® 2 automated system (bioMérieux, Marcy-l’Étoile, France) with the VITEK® 2 GN card and the VITEK® 2 AST-N255 card. Multidrug resistance was defined as bacteria non-susceptible to one or more antimicrobial agents in three or more antimicrobial categories (Magiorakos et al., 2012). All the isolates were screened for the intrinsic OXA-51-like gene (Feizabadi et al., 2008; Voets et al., 2011; Ghaith et al., 2017). The six excluded isolates that did not harbour the OXA-51-like gene were further analysed using matrix assisted laser desorption ionisation-time of flight mass spectrometry (MALDI-TOF). The VITEK® 2 (bioMérieux, Marcy-l’Étoile, France) results for colistin resistant isolates were confirmed by the clinical microbiology laboratory (TAD, NHLS) using a MBD assay (EUCAST, 2016; Clinical and Laboratory Standards Institute [CLSI], 2017).
Molecular Detection of Beta-Lactamase Resistance Genes
The genomic DNA of each of the 150 A. baumannii isolates was extracted as described elsewhere (Lowings et al., 2015). Multiplex PCR assays were performed for the detection of selected β-lactamase genes in A. baumannii. Primer concentrations used in the 10x primer mixture and the cycling conditions are shown in Supplementary Table S1.
Primers for ISAbaI as previously described by Turton et al. (2006) were used to detect the presence of ISAbaI, whereas the ISAbaI forward primer and the OXA-23-like, OXA-51-like and OXA-58-like reverse primers (Supplementary Table S1) were used to confirm the presence of ISAbaI upstream of the carbapenemase gene (Turton et al., 2006; Voets et al., 2011; Cicek et al., 2014).
Molecular Epidemiology of the Clinical A. baumannii Isolates
The PulseNet PFGE protocol (Ribot et al., 2006) was followed with some modifications as described elsewhere (Lowings et al., 2015). A distance matrix was constructed using the Dice coefficient and a dendrogram was constructed using the unweighted pair group method with arithmetic mean (UPGMA) (Fillaux et al., 2006). Pulsotype designation was based on isolates showing ≥80% relatedness, which corresponds to the Tenover criteria (possibly related 4 to 6 bands difference) (Tenover et al., 1995). Representatives from each major PFGE pulsotype (≥5 isolates) and selective minor pulsotypes (<5 isolates) with ≥80% similarity were chosen for MLST analyses.
Seven housekeeping genes were used for MLST as previously described1. All the amplicons were sequenced in both forward and reverse directions by Inqaba Biotechnical Industries, Pretoria, South Africa. The ABI files obtained were analysed using the CLC Main Workbench Version 6.0 (CLCbio, Waltham, MA, United States) software programme. The sequences were assigned to the corresponding allelic profiles and sequence types by using the pubMLST database.
Statistical Analysis
Analysis was performed with the STATA 14 package (StataCorp LP, College Station, TX, United States). The results were interpreted as follows: p-values of ≤ 0.05 were considered statistically significant and p-values of >0.1 were considered not significant.
Results
Isolate Information
The mean age of patients in hospital A and B were 40 and 41 years, respectively. There was no significant difference between male (M) and female (F) patients in both hospitals [p-value = 1.0; M (hospital A): 54% (n = 39/72) vs. M (hospital B): 54% (n = 37/69)]. More detail on the patient demographics and specimen collection sites can be found in Supplementary Tables S2.1, S2.2.
Susceptibility Profiles of A. baumannii Isolates Isolated From Two Tertiary Academic Hospitals
The susceptibility profiles of the A. baumannii isolates collected from both hospitals were determined with the VITEK® 2 automated system (bioMérieux, Marcy-l’Étoile, France) and are represented in Table 1. All the A. baumannii isolates were MDR and showed complete resistance toward nitrofurantoin. High levels of resistance toward ceftazidime (90%; n = 127/141), cefepime (81%; n = 114/141), imipenem (89%; n = 125/141), meropenem (87%; n = 123/141), gentamicin (74%; n = 105/141), ciprofloxacin (67%; n = 95/141) and trimethoprim/sulfamethoxazole (69%; n = 129/141) were also observed. The majority of the A. baumannii isolates collected from both hospitals were susceptible to tigecycline. However, seven isolates (10%; n = 7/72) from hospital A and one isolate (1%; n = 1/69) from hospital B showed intermediate resistance toward tigecycline. Two colistin resistant isolates (3%; n = 2/69) were isolated from two different patients in hospital B. The results were confirmed by MBD (patient 1: 16 μg mL-1 and patient 2: 64 μg mL-1).
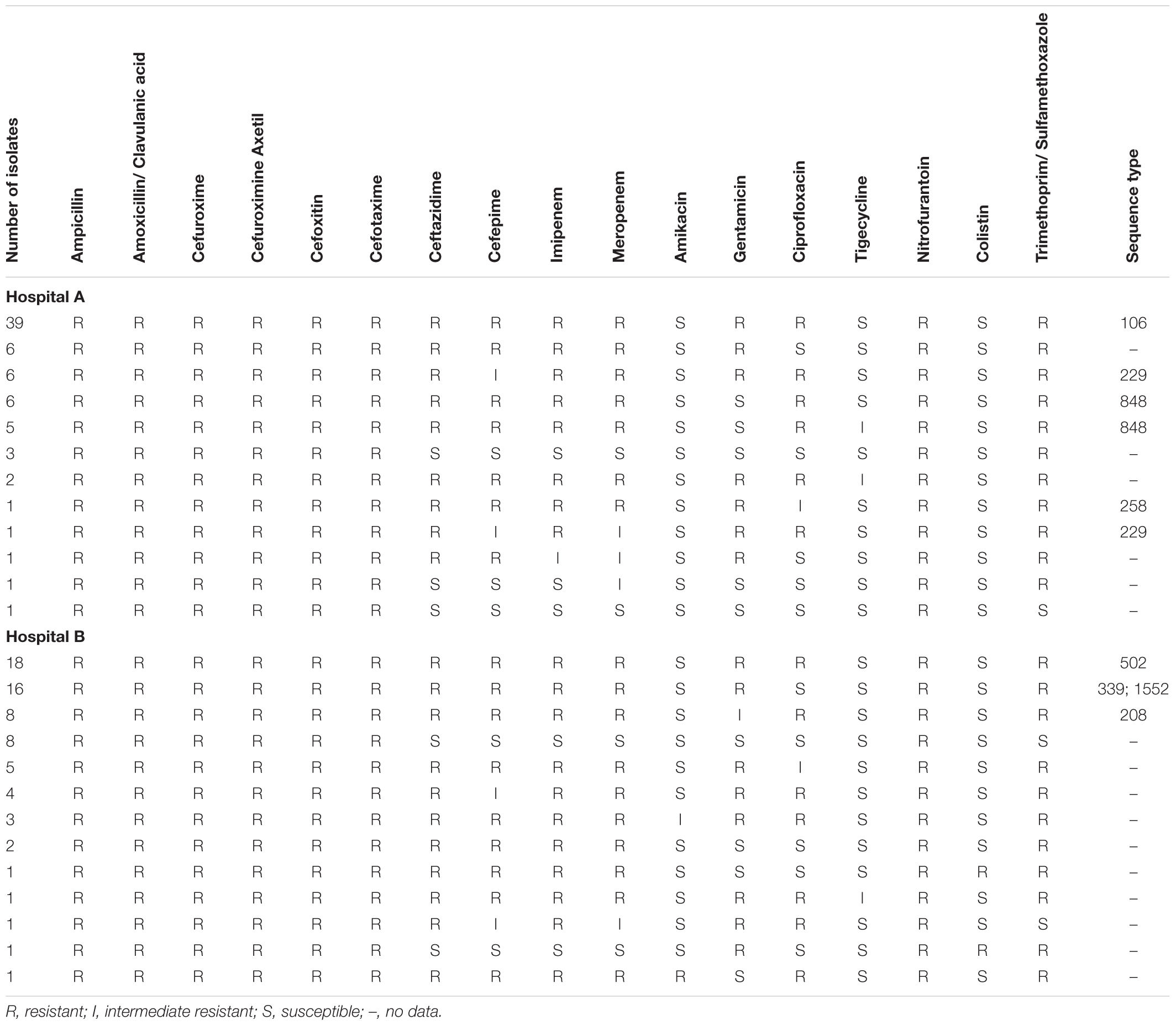
TABLE 1. Antimicrobial susceptibility profiles of A. baumannii isolates collected from hospital A and B.
Molecular Identification of Resistance Genes Found in A. baumannii Isolates Using M-PCR Assays
All the collected isolates were screened for the intrinsic OXA-51-like gene (Feizabadi et al., 2008; Voets et al., 2011; Ghaith et al., 2017). Six isolates were excluded that did not contain this intrinsic gene. The rest of the isolates all contained the OXA-51-like gene (hospital A: 100%; n = 72/72 and hospital B: 100%; n = 69/69). The isolates collected from hospital A contained the OXA-23-like (96%; n = 69/72) gene and the isolates collected from hospital B contained the OXA-23-like (91%; n = 63/69) and OXA-58-like (4%; n = 3/69) genes.
The 141 A. baumannii isolates were screened for ISAbaI and 100% (n = 72/72) of the isolates from hospital A and 88% (n = 61/69) of the isolates in hospital B contained this element. The detection of the ISAbaI was statistically significant [p-value = 0.003]. Further tests were performed to determine if the element was upstream of the OXA genes. Only 29% (n = 21/72) of the isolates in hospital A and 42% (n = 29/69) of the isolates in hospital B carried the ISAbaI upstream of the OXA-51-like gene. These findings were not statistically significant [p-value = 0.118] and no increased MICs for carbapenems were observed. The ISAbaI element was not detected upstream of either the OXA-23-like or the OXA-58-like genes in any of the isolates.
The Temoneira (TEM) gene was detected in 33% (n = 24/72) and 17% (n = 12/69) of the isolates collected from hospital A and B respectively. There was a significant association of the TEM gene between the hospitals [p-value = 0.035]. The A. baumannii isolates were negative for the following genes: Cefotaximase-Munich (CTX-M); Guiana extended-spectrum β-lactamase (GES); German imipenemase (GIM); Imipenem metallo-β-lactamase (IMP); Klebsiella pneumoniae carbapenemase (KPC); New Delhi metallo-β-lactamase (NDM), OXA-48; Pseudomonas extended resistance (PER); Sulfhydryl variant (SHV), Seoul imipenem metallo-β-lactamase (SIM-1); São Paulo metallo-β-lactamase (SPM), Vietnam extended-spectrum β-lactamase (VEB) and Verona integrin-encoded metallo-β-lactamase (VIM). The mechanisms of colistin and tigecycline resistance were not further investigated.
Determination of the Genetic Relationship and Global Epidemiology of Clinical A. baumannii Isolates
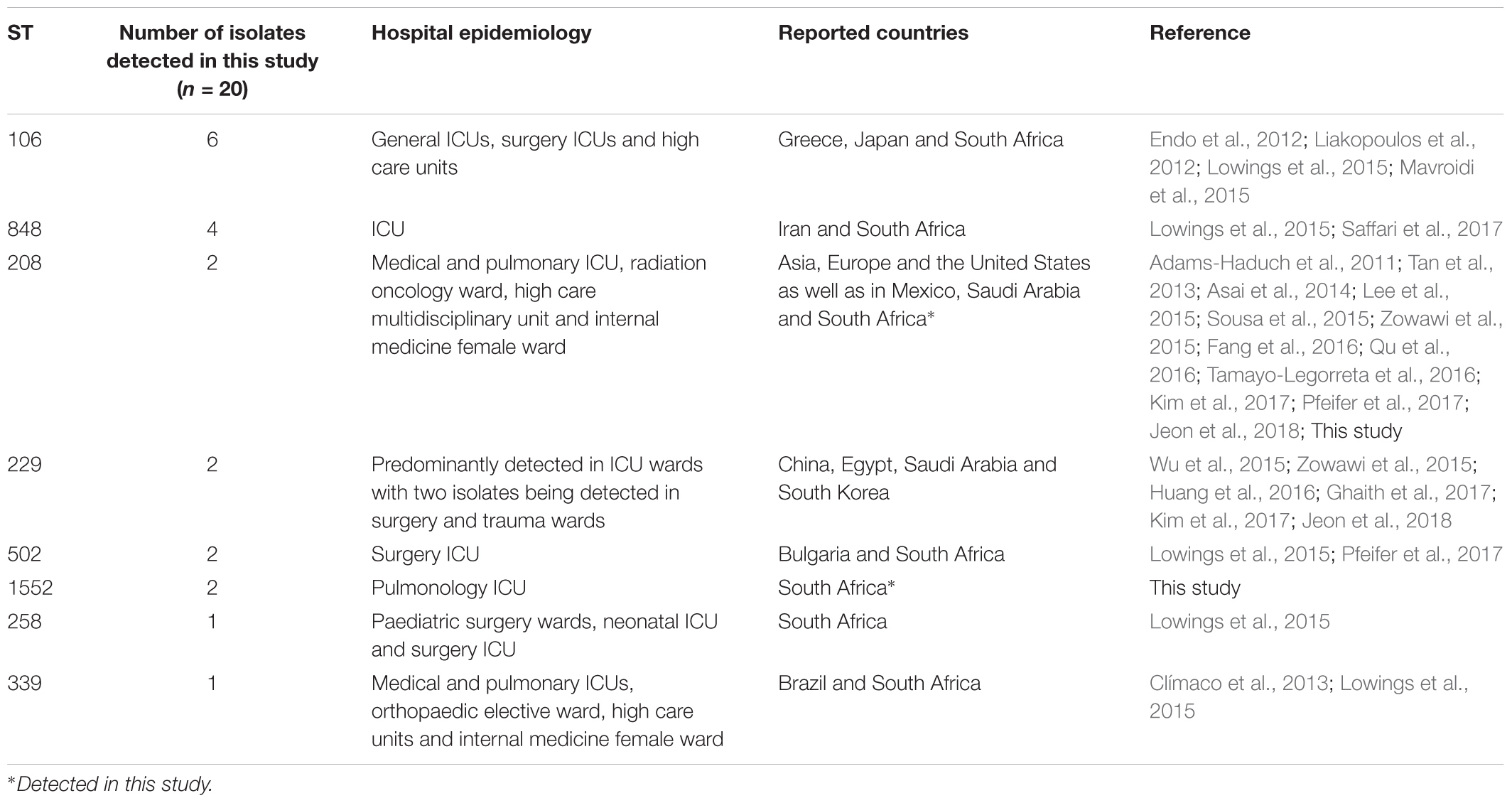
The PFGE analysis clustered the A. baumannii isolates into seven major pulsotypes (≥5 isolates) and several minor pulsotypes (<5 isolates) (Supplementary Figure S1). Twenty representative A. baumannii isolates were chosen from each major pulsotype (top and bottom isolates from each pulsotype) and selected minor pulsotypes. The following STs were identified according to the Oxford database: ST106 (1-1-1-1-1-98-6); ST208 (1-3-3-2-2-97-3); ST229 (1-15-2-28-1-107-32); ST258 (1-15-8-10-28-110-32); ST339 (44-73-4-11-44-121-4); ST502 (1-12-3-2-2-100-3); ST848 (1-15-3-2-2-142-3); and ST1552 (44-73-4-11-44-304-4). Molecular epidemiological data revealed that ST106, ST229, ST258 and ST208 are established in both hospitals. ST848 was only detected in hospital A, while ST502, ST339 and the novel ST1552 were only detected in hospital B.
Discussion
The MDR A. baumannii isolates pose great treatment challenges in the studied hospitals. When the data from this study is compared to that of previously published studies (Kock et al., 2013; Lowings et al., 2015) in the same hospitals, the data suggests that the problem is only set to increase. In Figure 1 the steady increase in antimicrobial resistance since 2008 in Tshwane, South Africa is depicted. The proportion of resistance increased alarmingly from 2008 to 2013; not much changed as similar resistance profiles were observed in 2014 (this study). The detection of colistin resistance and intermediate tigecycline resistance is worrisome as these are last resort antimicrobials. Resistance toward these antimicrobials are likely to increase in the years to come if it is not used as prescribed by local and international guidelines (Cai et al., 2012; Sekyere, 2016).
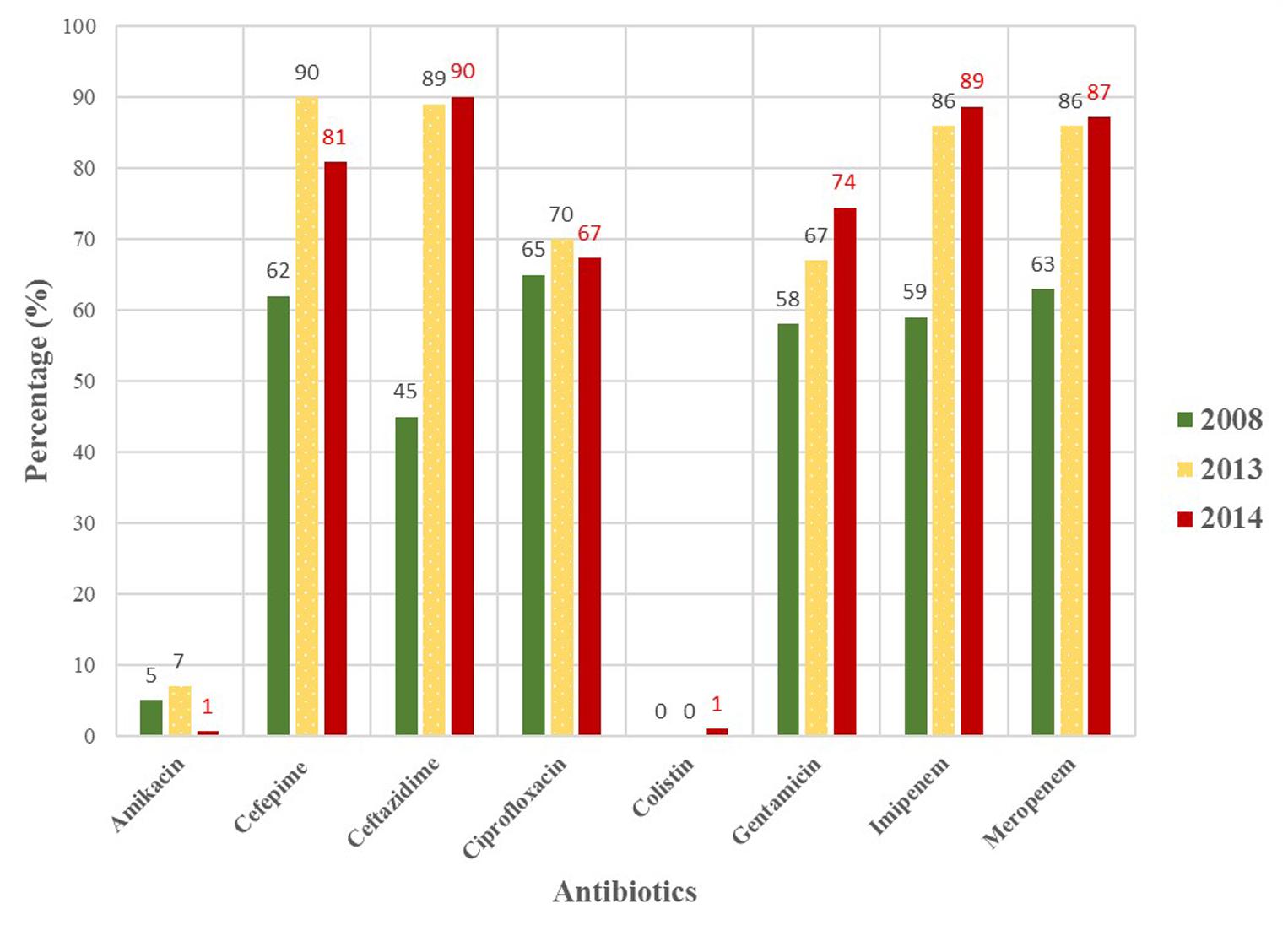
FIGURE 1. Antimicrobial susceptibility profiles of MDR Acinetobacter baumannii isolates collected from hospitals in the Tshwane region in 2008, 2013 and 2014 (Kock et al., 2013; Lowings et al., 2015).
Circulating antimicrobial resistance genes can be determined with molecular assays, such as PCR, which are relatively fast and cheap. The PCR results showed that the majority of the collected isolates were positive for the OXA-51-like gene except for six isolates that tested negative (these isolates were excluded from the study). These six isolates were identified by MALDI-TOF as: (i) Acinetobacter junii; (ii) A. pittii (x2); (iii) A. haemolyticus; (iv) A. nosocomialis and (v) Providencia rettgeri. The PCR and MALDI-TOF results have shown that the presence of the intrinsic/species-specific OXA-51-like gene can be used as a simple and reliable way to identify A. baumannii (Turton et al., 2006; Feizabadi et al., 2008; Voets et al., 2011; Ghaith et al., 2017). The prevalence of the OXA-23-like gene was very high in this study and have increased with 35% since 2008 to 2014 (Kock et al., 2013; Lowings et al., 2015). The drastic increase in the OXA-23 gene could be due to poor infection control practises in the studied hospitals. Isolates that harboured this gene showed higher resistance levels toward carbapenems, such as imipenem and meropenem. The high prevalence of the OXA-23 gene in this study is in agreement with other studies done in India (98%; n = 101/103), Colombia (97.5%; n = 118/121) and Egypt (96%; n = 48/50) (Vijayakumar et al., 2016; Correa et al., 2017; Ghaith et al., 2017). The prevalence of the OXA-58-like gene remains low in the studied hospitals and no drastic increase were observed over the years (Kock et al., 2013; Lowings et al., 2015). However, Mendes et al. (2009) and Liakopoulos et al. (2012) reported higher OXA-58 carriage rates [12% (Asia-Pacific nations) and 27.6% (Greece) respectively]. The TEM gene, classified as an extended spectrum β-lactamase (ESBL) from the Ambler class A group, was detected in some of the collected isolates. The TEM gene was the most frequently associated with ST848, followed by ST106 and ST208. The detection of the TEM gene in the studied hospitals shows how the A. baumannii clones are constantly evolving by gaining more genetic determinants from other Gram-negative bacteria.
Molecular typing methods, such as PFGE and MLST are useful in epidemiological studies to identify circulating clones (Villalón et al., 2011). A literature research was done on the eight clones detected in this study and the distributions of these clones are summarised in Table 2. Sequence type 106 was the most frequently detected clone followed by ST258 (both of these clones were only susceptible to amikacin, colistin and tigecycline). It appears that the ST258 clone is a South African clone (Lowings et al., 2015) as no other reports could be found. This is to our knowledge the first report of the presence of ST1552 – a single locus variant of ST339. This highlights the constant evolution and dissemination of A. baumannii clones in and between hospital settings. The global clone ST208 was also detected in this study (Adams-Haduch et al., 2011). These successful clones have increased virulence and survival factors and are adapted to survive in the harsh hospital environment. These circulating clones poses an increased threat to hospitalised patients and stricter adherence to infection control and prevention policies are needed to prevent further dissemination of these MDR clones. The general cleaning of the hospital environment should also be improved as it is possible for these clones to survive on abiotic surfaces, such as medical equipment, furniture, door handles and bed linen (Maragakis and Perl, 2008).
Limitations of this study include: (i) small sample size; (ii) only two tertiary hospitals in the Tshwane region were included and (iii) there is a lack of detailed clinical information, such as treatment received, clinical outcomes and other simultaneous co-morbidities and infections. The investigation of the different antimicrobial resistance mechanisms was not part of the scope of this study. Future studies, employing whole genome sequencing and other next generation technologies, should investigate the antimicrobial resistance mechanisms linked to the colistin and tigecycline resistance observed.
Conclusion
The high prevalence of MDR A. baumannii isolates is threatening the effectiveness of last resort antimicrobials. The detection of colistin resistance and intermediate tigecycline resistance is of concern, since no other treatment options are available if these drugs are rendered ineffective. A diverse group of highly resistant clones with the OXA-23-like gene is responsible for the spread and establishment of A. baumannii in the studied hospitals. The spread of A. baumannii clones between the studied hospitals could be due to patients being transferred from one hospital to the other (the hospitals are in close proximity) or it could be due to healthcare workers rotating between hospital A and B (both hospitals are part of the same academic institution). Future studies incorporating next generation sequencing methods focusing on a larger sample of A. baumannii isolates from various hospitals and lineages are necessary to better understand the rapid development of antimicrobial resistance in A. baumannii.
Author Contributions
ML, ME, and MK conceived and designed the study. MK was the principal investigator and budget owner. ML collected all the clinical isolates and performed the experimental work. ML, GP, and MK performed the data analysis. ML and FI collected all the clinical information. ML, ME, FI, GP, PB, JP, and MK wrote the manuscript with critical appraisal and contributions received from all of the authors. PB did the statistical analysis. All authors read and approved this version of the manuscript.
Funding
ML was supported by a National Research Foundation (NRF) grant. Opinions expressed and conclusions arrived at, are those of the authors and are not necessarily to be attributed to the NRF. The authors hereby acknowledge the NHLS Research Trust and RESCOM, UP for financial support.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer VG and handling Editor declared their shared affiliation.
Acknowledgments
The authors would like to thank the Department of Medical Microbiology and the NHLS for the use of their facilities and for the provided isolates. The authors also want to thank Dr. Anthony Smith, coordinator for PulseNet Africa for the donation of the Salmonella enterica subsp. enterica serovar Braenderup (ATCC BAA-664). MALDI-TOF analysis was performed by the molecular diagnostic reference centre at Ampath (Centurion, South Africa) and the MBD assay was performed by Dr. B. Mitton (Tshwane Academic Division, National Health Laboratory Service, Pretoria, South Africa).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01280/full#supplementary-material
Footnotes
References
Adams-Haduch, J. M., Onuoha, E. O., Bogdanovich, T., Tian, G. B., Marschall, J., Urban, C. M., et al. (2011). Molecular epidemiology of carbapenem-nonsusceptible Acinetobacter baumannii in the United States. J. Clin. Microbiol. 49, 3849–3854. doi: 10.1128/JCM.00619-11
Agodi, A., Voulgari, E., Barchitta, M., Quattrocchi, A., Bellocchi, P., Poulou, A., et al. (2014). Spread of a carbapenem- and colistin-resistant Acinetobacter baumannii ST2 clonal strain causing outbreaks in two Sicilian hospitals. J. Hosp. Infect. 86, 260–266. doi: 10.1016/j.jhin.2014.02.001
Asai, S., Umezawa, K., Iwashita, H., Ohshima, T., Ohashi, M., Sasaki, M., et al. (2014). An outbreak of blaOXA-51-like and blaOXA-66 positive Acinetobacter baumannii ST208 in the emergency intensive care unit. J. Med. Microbiol. 63, 1517–1523. doi: 10.1099/jmm.0.077503-0
Bartual, S. G., Seifert, H., Hippler, C., Luzon, M. A. D., Wisplinghoff, H., and Rodríguez-Valera, F. (2005). Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 49, 4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005
Cai, Y., Chai, D., Wang, R., Liang, B., and Bai, N. (2012). Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J. Antimicrob. Chemother. 67, 1607–1615. doi: 10.1093/jac/dks084
Cicek, A. C., Saral, A., Iraz, M., Ceylan, A., Duzgun, A. O., Peleg, A. Y., et al. (2014). OXA- and GES-type β-lactamases predominate in extensively drug-resistant Acinetobacter baumannii isolates form a Turkish university hospital. Clin. Microbiol. Infect. 20, 410–415. doi: 10.1111/1469-0691.12338
Clímaco, E. C., de Oliveira, M. L., Pitondo-Silva, A., Oliveira, M. G., Medeiros, M., Lincopan, N., et al. (2013). Clonal complexes 104, 109 and 113 playing a major role in the dissemination of OXA-carbapenemase-producing Acinetobacter baumannii in Southeast Brazil. Infect. Genet. Evol. 19, 127–133. doi: 10.1016/j.meegid.2013.06.024
Clinical and Laboratory Standards Institute [CLSI] (2017). Performance Standards for Antimicrobial Susceptibility Testing – Twenty Seven Edition. CLSI Supplement M100. Wayne, PA: CLSI.
Cooper, J. E., and Feil, E. J. (2004). Multilocus sequence typing – what is resolved? Trends Microbiol. 8, 373–377. doi: 10.1016/j.tim.2004.06.003
Correa, A., del Campo, R., Escandón-Vargas, K., Perenguez, M., Rodríguez-Baños, M., Hernández-Gómez, C., et al. (2017). Distinct genetic diversity of carbapenem-resistant Acinetobacter baumannii from Colombian hospitals. Microb. Drug Resist. 24, 48–54. doi: 10.1089/mdr.2016.0190
Dijkshoorn, L., Nemec, A., and Seifert, H. (2007). An increasing threat in hospitals: multidrug resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5, 939–951. doi: 10.1038/nrmicro1789
Endo, S., Yano, H., Hirakata, Y., Arai, K., Kanamori, H., Ogawa, M., et al. (2012). Molecular epidemiology of carbapenem-non-susceptible Acinetobacter baumannii in Japan. J. Antimicrob. Chemother. 67, 1623–1626. doi: 10.1093/jac/dks094
EUCAST (2016). Recommendations for Mic Determination of Colistin (Polymyxin E) as Recommended by the Joint CLSI-EUCAST Polymyxin Breakpoints Working Group. Available at: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf [accessed April 06, 2018].
Evans, B. A., and Amyes, S. G. B. (2014). OXA β-lactamases. Clin. Microbiol. Rev. 27, 241–263. doi: 10.1128/CMR.00117-13
Fang, Y., Quan, J., Hua, X., Feng, Y., Li, X., Wang, J., et al. (2016). Complete genome sequence of Acinetobacter baumannii XH386 (ST208), a multi-drug resistant bacteria isolated from pediatric hospital China. Genom. Data 7, 269–274. doi: 10.1016/j.gdata.2015.12.002
Feizabadi, M. M., Fathollahzadeh, B., Taherikalani, M., Rasoolinejad, M., Sadeghifard, N., Aligholi, M., et al. (2008). Antimicrobial susceptibility patterns and distribution of blaOXA genes among Acinetobacter spp. isolated from patients at Tehran hospitals. Jpn. J. Infect. Dis. 61, 274–278.
Field, N., Cohen, T., Struelens, M. J., Palm, D., Cookson, B., and Glynn, J. R. (2014). Strengthening the reporting of molecular epidemiology for infectious diseases (STROME-ID): an extension of the STROBE statement. Lancet Infect. Dis. 14, 341–352. doi: 10.1016/S1473-3099(13)70324-4
Fillaux, J., Dubouix, A., Concil, J. M., Laguerre, J., and Marty, N. (2006). Retrospective analysis of multidrug-resistant Acinetobacter baumannii strains isolated during a 4-year period in a university hospital. Infect. Control Hosp. Epidemiol. 27, 647–653. doi: 10.1086/507082
Fouad, M., Attia, A. S., Tawakkol, W. M., and Hashem, A. M. (2013). Emergence of carbapenem-resistant Acinetobacter baumannii harbouring the OXA-23 carbapenemase in intensive care units of Egyptian hospitals. Int. J. Infect. Dis. 17, e1252–e1254. doi: 10.1016/j.ijid.2013.07.012
Fu, Y., Zhou, J., Zhou, H., Yang, Q., Wei, Z., Yu, Y., et al. (2010). Wide dissemination of OXA-23-producing carbapenem-resistant Acinetobacter baumannii clonal complex 22 in multiple cities of China. J. Antimicrob. Chemother. 65, 644–650. doi: 10.1093/jac/dkq027
Ghaith, D. M., Zafer, M. M., Al-Agamy, M. H., Alyamani, E. J., Booq, R. Y., and Almoazzamy, O. (2017). The emergence of a novel sequence type of MDR Acinetobacter baumannii from the intensive care unit of an Egyptian tertiary care hospital. Ann. Clin. Microbiol. Antimicrob. 16:34. doi: 10.1186/s12941-017-0208-y
Grundmann, H. J., Towner, K. J., Dijkshoorn, L., Gerner-Smidt, P., Maher, M., Seifert, H., et al. (1997). Multicenter study is using standardized protocols and reagents for evaluation of reproducibility of PCR-based fingerprinting of Acinetobater spp. J. Clin. Microbiol. 35, 3071–3077.
Hammoudi, D., Moubareck, C. A., and Sarkis, D. K. (2014). How to detect carbapenemase producers? A literature review of phenotypic and molecular methods. J. Microb. Methods 107, 106–118. doi: 10.1016/j.mimet.2014.09.009
Hamouda, A., Evans, B. A., Towner, K. J., and Amyes, S. G. B. (2010). Characterization of epidemiologically unrelated Acinetobacter baumannii isolates from four continents by use of multilocus sequence typing, pulsed-field gel electrophoresis and sequence-based typing of blaOXA-51-like genes. J. Clin. Microbiol. 48, 2476–2483. doi: 10.1128/JCM.02431-09
Hu, Y., Li, B., Jin, D., Cui, Z., Tao, X., Zhang, B., et al. (2013). Comparison of multiple-locus variable-number tandem-repeat analysis with pulsed-field gel electrophoresis typing of Acinetobacter baumannii in China. J. Clin. Microbiol. 51, 1263–1268. doi: 10.1128/JCM.03108-12
Huang, G., Yin, S., Gong, Y., Zhao, X., Zou, L., Bei, J., et al. (2016). Multilocus sequence typing analysis of carbapenem-resistant Acinetobacter baumannii in a Chinese burns institute. Front. Microbiol. 7:1717. doi: 10.3389/fmicb.2016.01717
Jacobs, A. C., Thompson, M. G., Black, C. C., Kessler, J. L., Clark, L. P., McQueary, C. N., et al. (2014). AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio 5:e01076-14. doi: 10.1128/mBio.01076-14
Jeon, H., Kim, S., Kim, M. H., Kim, S. Y., Nam, D., Park, S. C., et al. (2018). Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates from Korean hospital that carry blaOXA-23. Infect. Genet. Evol. 58, 232–236. doi: 10.1016/j.meegid.2018.01.003
Kim, D. H., Jung, S., Kwon, K. T., and Ko, K. S. (2017). Occurrence of diverse AbGTI1-type genomic islands in Acinetobacter baumannii global clone 2 isolates from South Korea. Antimicrob. Agents Chemother. 61:e01972-16. doi: 10.1128/AAC.01972-16
Kock, M. M., Bellomo, A. N., Storm, N., and Ehlers, M. M. (2013). Prevalence of carbapenem resistance genes in Acinetobacter baumannii isolated from clinical specimens obtained from an academic hospital in South Africa. South Afr. J. Epidemiol. Infect. 28, 28–32. doi: 10.1080/10158782.2013.11441516
Kuo, S. C., Yang, S. P., Lee, Y. T., Chuang, H. C., Chen, C. P., Chang, C. L., et al. (2013). Dissemination of imipenem-resistant Acinetobacter baumannii with new plasmid-borne blaOXA-72 in Taiwan. BMC Infect. Dis. 13:319. doi: 10.1186/1471-2334-13-319
Lean, S., Suhaili, Z., Ismail, S., Rahman, N. I. A., Othman, N., Abdullah, F. H., et al. (2014). Prevalence and genetic characterization of carbapenem- and polymyxin-resistant Acinetobacter baumannii isolated from a tertiary hospital in Terengganu, Malaysia. ISRN Microbiol. 2014, 1–9. doi: 10.1155/2014/953417
Lean, S. S., Yeo, C. C., Suhaili, Z., and Thong, K. L. (2015). Whole-genome analysis of an extensively drug-resistant clinical isolate of Acinetobacter baumannii AC12: insights into the mechanisms of resistance of an ST195 cline from Malaysia. Int. J. Antimicrob. Agents 45, 178–182. doi: 10.1016/j.ijantimicag.2014.10.015
Lee, H., Huang, C., Chen, C., Wang, Y., Chang, C., and Chiu, C. (2015). Emergence in Taiwan of novel Imipenem-resistant Acinetobacter baumannii ST455 causing bloodstream infection in critical patients. J. Microbiol. Immunol. Infect. doi: 10.1016/j.jmii.2015.04.001 [Epub ahead of print].
Lee, S. Y., Shin, J. H., Park, K. H., Kim, J. H., Shin, M. G., Suh, S. P., et al. (2014). Identification, genotypic relation, and clinical features of colistin-resistant isolates of Acinetobacter genomic species 13BJ/14TU from bloodstreams of patients in a University hospital. J. Clin. Microbiol. 52, 931–939. doi: 10.1128/JCM.02868-13
Liakopoulos, A., Miriagou, V., Katsifas, E. A., Karagouni, A. D., Daikos, G. L., Tzouvelekis, L. S., et al. (2012). Identification of OXA-23-producing Acinetobacter baumannii in Greece, 2010 to 2011. Euro Surveill. 17:20117.
Lowings, M., Ehlers, M. M., Dreyer, A. W., and Kock, M. M. (2015). High prevalence of oxacillinases in clinical multidrug-resistant Acinetobacter baumannii isolates from the Tshwane region, South Africa – an update. BMC Infect. Dis. 15, 521–530. doi: 10.1186/s12879-015-1246-8
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an internal expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Maragakis, L. L., and Perl, T. M. (2008). Acinetobacter baumannii: epidemiology, antimicrobialresistance, and treatment options. Clin. Infect. Dis. 46, 1254–1263. doi: 10.1086/529198
Mavroidi, A., Likousi, S., Palla, E., Katsiari, M., Roussou, Z., Maguina, A., et al. (2015). Molecular identification of tigecycline- and colistin-resistant carbapenemase-producing Acinetobacter baumannii from a Greek hospital from 2011 to 2013. J. Med. Microbiol. 64, 993–997. doi: 10.1099/jmm.0.000127
Mendes, R. E., Bell, J. M., Turnidge, J. D., Castanheira, M., and Jones, R. N. (2009). Emergence and widespread dissemination of OXA-23, -24/40 and -58 carbapenemases among Acinetobacter spp. in Asia-Pacific nations: report from the SENTRY Surveillance Program. J. Antimicrob. Chemother. 63, 55–59. doi: 10.1093/jac/dkn434
Patel, G., and Bonomo, R. A. (2013). Stormy waters ahead”: global emergence of carbapenemases. Front. Microbiol. 4:48. doi: 10.3389/fmicb.2013.00048
Peleg, A. Y., Seifert, H., and Paterson, D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582. doi: 10.1128/CMR.00058-07
Perez, F., Hujer, A. M., Hujer, K. M., Decker, B. K., Rather, P. N., and Bonomo, R. A. (2007). Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51, 3471–3484. doi: 10.1128/AAC.01464-06
Pfeifer, Y., Trifonova, A., Pietsch, M., Brunner, M., Todorova, I., Gergova, I., et al. (2017). Clonal transmission of Gram-negative bacteria with carbapenemases NDM-1, VIM-1 and OXA-23/72 in a Bulgarian Hospital. Microb. Drug Resist. 23, 301–307. doi: 10.1089/mdr.2016.0059
Potron, A., Poirel, L., and Nordmann, P. (2015). Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int. J. Antimicrob. Agents 45, 568–585. doi: 10.1016/j.ijantimicag.2015.03.001
Pournaras, S., Poulou, A., Dafopoulou, K., Chabane, Y. N., Kristo, I., Makris, D., et al. (2014). Growth retardation, reduced invasiveness and impaired colistin-mediated cell death associated with colistin resistance development in Acinetobacter baumannii. Antimicrob. Agents Chemother. 58, 828–832. doi: 10.1128/AAC.01439-13
Qu, J., Du, Y., Yu, R., and Lü, X. (2016). The first outbreak caused by Acinetobacter baumannii ST208 and ST195 in China. Biomed Res. Int. 2016:9254907. doi: 10.1155/2016/9254907
Ribot, E. M., Fair, M. A., Gautom, R., Cameron, D. N., Hunter, S. B., Swaminathan, B., et al. (2006). Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella and Shigella for PulseNet. Foodborne Pathog. Dis. 3, 59–67. doi: 10.1089/fpd.2006.3.59
Saffari, F., Monsen, T., Karmostaji, A., Azimabad, F. B., and Winderström, M. (2017). Significant spread of extensively drug-resistant Acinetobacter baumannii genotypes of clonal complex 92 among intensive care unit patients in a university hospital in Southern Iran. J. Med. Microbiol. 66, 1656–1662. doi: 10.1099/jmm.0.000619
Salimizand, H., Noori, N., Meshkat, Z., Ghazvini, K., and Amel, S. J. (2015). Prevalence of Acinetobacter baumannii harbouring ISAba/blaOXA-23-like family in a burn center. Burns 41, 1100–1106. doi: 10.1016/j.burns.2014.12.008
Sekyere, J. O. (2016). Current state of resistance to antibiotics of last-resort in South Africa: a review from a public health perspective. Front. Public Health 4:209. doi: 10.3389/fpubh.2016.00209
Sousa, C., Botelho, J., Grosso, F., Silva, L., Lopes, J., and Peixe, L. (2015). Unsuitability of MALDO-TOF MS to discriminate Acinetobacter baumannii clones under routine experimental conditions. Front. Microbiol. 6:481. doi: 10.3389/fmicb.2015.00481
Tamayo-Legorreta, E., Turrubiartes-Martínez, E., Garza-Ramos, U., Niño-Moreno, P.,Barrios, H., Sánchez-Pérez, A., et al. (2016). Outbreak caused by blaOXA-72 producing Acinetobacter baumannii ST417 detected in clinical and environmental isolates. Microb. Drug Resist. 22, 129–133. doi: 10.1089/mdr.2015.0157
Tan, S. Y., Chua, S. L., Liu, Y., Høiby, N., Andersen, L. P., Givskov, M., et al. (2013). Comparative genomic analysis of rapid evolution of an extreme-drug-resistant Acinetobacter baumannii clone. Genome Biol. Evol. 5, 807–818. doi: 10.1093/gbe/evt047
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239.
Turton, J. F., Woodford, N., Glover, J., Yarde, S., Kaufmann, M. E., and Pitt, T. L. (2006). Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44, 2974–2976. doi: 10.1128/JCM.01021-06
Vijayakumar, S., Gopi, R., Gunasekaran, P., Bharathy, M., Walia, K., Anandan, S., et al. (2016). Molecular characterization of invasive carbapenem resistant Acinetobacter baumannii from a tertiary care hospital in South India. Infect. Dis. Ther. 5, 379–387. doi: 10.1007/s40121-016-0125-y
Villalón, P., Valdezate, S., Medina-Pascual, M. J., Rubio, V., Vindel, A., and Saez-Nieto, J. A. (2011). Clonal diversity of nosocomial epidemic Acinetobacter baumannii strains isolated in Spain. J. Clin. Microbiol. 49, 875–882. doi: 10.1128/JCM.01026-10
Voets, G. M., Fluit, A. C., Scharringa, J., Stuart, J. C., and Leverstein-Van Hall, M. A. (2011). A set of multiplex PCRs for genotypic detection of extended-spectrum β-lactamases, carbapenemases, plasmid-mediated AmpC β-lactamases and OXA β-lactamases. Int. J. Antimicrob. Agents 37, 356–359. doi: 10.1016/j.ijantimicag.2011.01.005
Wu, W., He, Y., Lu, J., Lu, Y., Wu, J., and Liu, Y. (2015). Transition of blaOXA-58-like to blaOXA-23-like in Acinetobacter baumannii clinical isolates in Southern China: an 8-year study. PLoS One 10:e0137174. doi: 10.1371/journal.pone.0137174
Yang, J. Y., Brooks, S., Meyer, J. A., Blakesley, R. R., Zelazny, A. M., Segre, J. A., et al. (2013). Pan-PCR, a computational method designing bacterium-typing assays based on whole-genome sequence data. J. Clin. Microbiol. 51, 752–758. doi: 10.1128/JCM.02671-12
Zowawi, H. M., Sartor, A. L., Sidjabat, H. E., Balkhy, H. H., Walsh, T. R., Al Johani, S. M., et al. (2015). Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates in the Gulf cooperation council states: dominance of OXA-23-type producers. J. Clin. Microbiol. 53, 896–903. doi: 10.1128/JCM.02784-14
Keywords: Acinetobacter baumannii, MDR, South Africa, PFGE, MLST, blaOXA-23-like
Citation: Lowe M, Ehlers MM, Ismail F, Peirano G, Becker PJ, Pitout JDD and Kock MM (2018) Acinetobacter baumannii: Epidemiological and Beta-Lactamase Data From Two Tertiary Academic Hospitals in Tshwane, South Africa. Front. Microbiol. 9:1280. doi: 10.3389/fmicb.2018.01280
Received: 15 February 2018; Accepted: 25 May 2018;
Published: 12 June 2018.
Edited by:
Jorge Blanco, Universidade de Santiago de Compostela, SpainReviewed by:
Vanesa García, Universidade de Santiago de Compostela, SpainJavier Fernández, Central University Hospital of Asturias, Spain
Copyright © 2018 Lowe, Ehlers, Ismail, Peirano, Becker, Pitout and Kock. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marleen M. Kock, marleen.kock@up.ac.za
 Michelle Lowe
Michelle Lowe Marthie M. Ehlers
Marthie M. Ehlers Farzana Ismail1,2
Farzana Ismail1,2 Gisele Peirano
Gisele Peirano Piet J. Becker
Piet J. Becker Johann D. D. Pitout
Johann D. D. Pitout Marleen M. Kock
Marleen M. Kock