
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 05 June 2018
Sec. Infectious Agents and Disease
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01205
Many cystic fibrosis (CF) airway infections are considered to be polymicrobial and microbe–microbe interactions may play an important role in disease pathology. Pseudomonas aeruginosa and Aspergillus fumigatus are the most prevalent bacterial and fungal pathogens isolated from the CF airway, respectively. We have previously shown that patients co-colonized with these pathogens had comparable outcomes to those chronically colonized with P. aeruginosa. Our objective was to examine the interactions between A. fumigatus and P. aeruginosa, specifically the effects of co-colonization on biofilm formation, virulence and host pro-inflammatory responses. Our findings suggest that co-infections of A. fumigatus and P. aeruginosa in the Galleria mellonella acute infection model showed that pre-exposure of larvae to sub-lethal inocula of A. fumigatus increased the mortality caused by subsequent P. aeruginosa infection. Co-infection of human bronchial epithelial cells (CFBE41o-) with both pathogens did not enhance IL-6 and IL-8 production beyond the levels observed following single infections. In addition, both pathogens stimulated cytokine secretion via the same two mitogen-activated protein kinases (MAPKs) signaling pathways, ERK and p38. Mixed species biofilms showed overall reduced biofilm development with crystal violet staining. Quantification by species-specific qPCR revealed that both pathogens had mutually antagonistic effects on each other. A. fumigatus supernatants showed strong anti-Pseudomonal activity and gliotoxin was the main active agent. Gliotoxin resulted in varying levels of anti-biofilm activity toward other bacteria commonly found in the CF airways. Gliotoxin produced by A. fumigatus colonizing the CF airways may have a significant impact on the CF airway microbiome composition with potential clinical implications.
Cystic fibrosis (CF) is the most common inherited life-shortening condition, characterized by mutations in the CF transmembrane conductance regulator (CFTR) gene. The resulting defective chloride secretion, altered airway surface liquid and impaired mucociliary secretion results in recurrent chronic respiratory infections (Haq et al., 2016). CF airway disease is characterized by a continuous cycle of persistent infection and inflammation contributing to morbidity and mortality in CF. Early detection and eradication of these infections remains a major challenge in CF lung disease management. P. aeruginosa is the leading cause of infection in patients with CF with approximately 28.4% being colonized by 71 months (Douglas et al., 2009). Prevalence in adults varies from 31 to 49.6% in recent reports (Salsgiver et al., 2016; Reece et al., 2017). Early intervention is paramount as P. aeruginosa is very difficult to eradicate once it has colonized the airways. P. aeruginosa is independently linked to worsened prognosis for CF patients; they have a decreased life expectancy of 30 years, compared with 40 years in non-colonized patients, experiencing a more rapid decline in pulmonary function with more frequent hospitalizations (Kosorok et al., 2001; Li et al., 2005). P. aeruginosa is a very accomplished bacterium and can adapt to life in the CF airway (Friman et al., 2013; Cullen and McClean, 2015).
Cystic fibrosis airway microbiology has recently been revolutionized by the discovery of complex communities of bacteria and fungi co-existing in the lungs of adults and children with CF (Sibley et al., 2011; Delhaes et al., 2012). These studies have highlighted a number of new and emerging microorganisms in CF airway disease (Bittar et al., 2008; Spicuzza et al., 2009) but have also verified the importance of common CF pathogens, such as P. aeruginosa (Cox et al., 2010; Klepac-Ceraj et al., 2010; Coburn et al., 2015) and Aspergillus fumigatus (Delhaes et al., 2012). Perhaps most importantly these metagenomic studies have confirmed that important CF pathogens do not colonize the airways in isolation. Exacerbation and disease progression in patients may be influenced by the interactions between these microorganisms in the CF airway. P. aeruginosa was identified in 54.1% of CF patients with persistent A. fumigatus infection and these co-colonized patients showed decreased lung function compared with patients clear of both pathogens (Amin et al., 2010). Direct contact between these organisms or indirect signaling may influence microbial pathogenicity (Duan et al., 2003).
Several studies have shown that P. aeruginosa interacts with other bacteria and fungi, inhibiting Burkholderia cenocepacia (Costello et al., 2014), Candida albicans (Brand et al., 2008) and Scedosporium aurantiacum (Kaur et al., 2015). P. aeruginosa increases its virulence when in co-culture with Gram-positive bacteria in Drosophila (Korgaonkar et al., 2013). P. aeruginosa pathogenicity was enhanced in a rat lung infection model by the presence of oropharyngeal flora and the increased virulence may be due to interspecies communication via autoinducer-2 (AI-2) mediated signaling (Duan et al., 2003). Both filamentation and biofilm formation of A. fumigatus was inhibited by P. aeruginosa through direct cell contact and by secreted molecules. Recently it was demonstrated that P. aeruginosa and A. fumigatus interact with each other via volatile communication mediators and that stimulation of A. fumigatus growth by P. aeruginosa did not require direct contact (Briard et al., 2016). P. aeruginosa culture filtrates inhibited and damaged A. fumigatus biofilms via metacaspase activation (Shirazi et al., 2016). P. aeruginosa elastase production is enhanced in the presence of A. fumigatus and may play a role in the damaging pathology associated with the CF lung and may explain why, at least in part, co-colonized patients have a poorer prognosis (Smith et al., 2015).
We have recently shown that 3.1% of Irish CF patients registered with the CF registry of Ireland were intermittently co-colonized with A. fumigatus and P. aeruginosa (Reece et al., 2017). In addition, co-colonization with both pathogens (even intermittently) resulted in comparable levels of hospitalizations, respiratory exacerbations and lung function reduction as in patients that were chronically colonized with P. aeruginosa alone which emphasizes the clinical significance of co-colonization with these microorganisms. The potential consequences of co-colonization with these two pathogens on infection and inflammation are poorly understood. We now report our investigations on the interactions between A. fumigatus and P. aeruginosa in terms of their virulence and the effect co-colonization has on the CF host immune system.
A number of clinical isolates and reference strains were used in this study and are listed in Table 1. All experiments were performed in a SafeFAST Class II Biosafety cabinet (Mason Technology, Dublin, Ireland), in compliance with BSL-II containment. Two P. aeruginosa isolates (PAco1 and PAco2) were isolated from the sputum of a CF patient co-colonized with A. fumigatus (AFco1 and AFco2). Sputum was homogenized by adding Sputasol 1:1 (Thermo Fisher Scientific) and incubated at 35°C for 15–20 min shaking. A volume of 50 μl was plated on CHROMAgar Pseudomonas (CHROMAgarTM) (LIP Diagnostics, Galway, Ireland) and incubated at 35°C for 48 h. Any blue/green colonies were picked and identity confirmed using Mass Spectrometry (Biomerieux Vitek). The AFref1, ΔgliG and complement ΔgliGc strains were previously described (Davis et al., 2011). A. fumigatus strains were routinely cultured on Malt Extract agar (MEA) plates (LIP Diagnostics, Galway, Ireland) for 48 h at 37°C and conidia were harvested using phosphate buffer saline (PBS) (Sigma, Wicklow, Ireland) containing 0.1% Tween 80 (PBST) (Sigma, Wicklow, Ireland). Conidia were enumerated on a Fast Read 102 counting slide (Biosigma, Cona, Italy). Liquid cultures were grown in Malt extract broth and sterile filtered through a 0.22 μM Whatman filter (Sigma, Wicklow, Ireland) to harvest culture supernatants (CSNs). A. fumigatus CSNs were plated on MEA and cultured overnight to confirm the absence of conidia. All P. aeruginosa isolates were grown on Blood agar plates (LIP Diagnostics) and cultured in Luria-Bertani (LB) (Sigma, Wicklow, Ireland) broth at 37°C. Overnight cultures in LB broth were diluted to obtain an OD600nm of 0.6 and sterile filtered through a 0.22 μM Whatman filter (Sigma, Wicklow, Ireland) to harvest CSNs. P. aeruginosa CSNs were plated on blood agar and cultured overnight to confirm absence of bacterial cells.
In order to establish bioburden to be injected, 10-fold serial dilutions of A. fumigatus strains were prepared in PBS and conidia counted on MEA plates. P. aeruginosa strains were cultured in LB broth for 24 h at 37°C, diluted to OD600nm 0.6, pelleted and resuspended and 10-fold serial dilutions prepared of the neat sample (2 × 107 CFU/ml) in 10 mM MgSO4 (Sigma, Wicklow, Ireland) and 50 μl aliquots spread on blood agar to confirm the bioburden. Prior to co-infection studies, kill curves (CFU/ml versus larval survival) for each P. aeruginosa or A. fumigatus strain were established by injecting serial dilutions of each P. aeruginosa or A. fumigatus strain into individual groups of larvae. The LD50 (CFU that resulted in 50% larval death after 24 h) of each strain was determined. For co-infection studies, groups of six healthy G. mellonella larvae (Livefoods Direct, Sheffield, United Kingdom) in the final instar larval stage weighing 0.25–0.35 g were selected and 10 μl of serial dilutions were injected into the hind left pro-leg using a 29-gauge needle (BD Biosciences, Oxford, United Kingdom) as described by Cullen et al. (2015). To establish the effect of P. aeruginosa infection in an A. fumigatus colonized model, G. mellonella were inoculated with non-lethal doses of A. fumigatus strains at 102 conidia/10 μl (Supplementary Figure 2) 24 h prior to subsequent inoculation with P. aeruginosa strains at a concentration of 2 × 105 CFU/10 μl. For experiments that required a second injection the hind right pro-leg was used. In all experiments two control groups were used, a group with no manipulation and a vehicle control group. Larvae were incubated at 37°C for the duration of each experiment and assessed every 24 h up to 7 days for viability as determined by their response to touch. The LD50 dose was determined from the percentage survival in three independent experiments.
CFBE41o- bronchial epithelial cells (CFBEs) (Gruenert et al., 1988) were cultured in T-75 flasks coated with 1% (v/v) fibronectin, 1% (v/v) collagen, and 10% (v/v) bovine serum albumin with minimum essential media (MEM) supplemented with 10% fetal bovine serum (FBS), 1% (v/v) L-glutamine, 1% (v/v) non-essential amino acids and 1% (v/v) penicillin-streptomycin at 37°C in 5% CO2. Culture media was replenished every 2 days and the cells were sub-cultured by trypsin digestion when growth had reached 80–90% confluence. All culture reagents were supplied by Sigma (Wicklow, Ireland).
CFBE41o- bronchial epithelial cells were seeded (4 × 105 cells/ml) onto coated 24-well plates and incubated for 24 h at 37°C in 5% CO2 overnight. The media was changed to serum free media and the cells incubated for a further 24 h. Overnight cultures of P. aeruginosa were inoculated into 100 ml of LB broth and incubated at 37°C until OD600nm 0.6 was reached. The CFU/ml was determined by plating on blood agar. The cultures were centrifuged at 4,500 × g for 10 min, resuspended in 1 ml MEM and applied to CFBE cells at 2 × 107 CFU/ml [multiplicity of infection (MOI) of 50:1] in triplicate and incubated at 37°C with 5% CO2 (Costello et al., 2014). A. fumigatus cultures centrifuged at 4,500 × g for 10 min were resuspended in 1 ml MEM and conidia applied to CFBE cells at a concentration of 8 × 105 conidia/ml (MOI of 2:1) in triplicate. The effect of P. aeruginosa and A. fumigatus infection on the viability of CFBE cells was assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide MTT Cell Proliferation Assay Kit (Cayman Chemical, Ann Arbor, MI, United States) according to the manufacturer’s instructions. The CFBE cells infected with P. aeruginosa PAref1 (MOI of 30:1, 50:1, 80:1, and 100:1) or A. fumigatus AFref1 (MOI of 1:1, 2:1, 5:1, 10:1) in triplicate for 24 h at 37°C in 5% CO2. Control cells were incubated with MEM only.
CFBE41o- bronchial epithelial cells were co-infected with P. aeruginosa and A. fumigatus in the presence MAPK inhibitors as follows: 10 μM of SP600125 (JNK inhibitor), SB203580 (p38 inhibitor) or PD 98,059 (ERK inhibitor) (Sigma, Wicklow, Ireland) or combinations of these for 1 h at 37°C. The cells were subsequently infected with P. aeruginosa strains (MOI of 50:1) or A. fumigatus stains (MOI of 2:1) in triplicate. After 24 h infection the plates were centrifuged and the supernatant removed and stored at -80°C for the pro-inflammatory response assays. Experiments were carried out in triplicate on three independent occasions.
IL-6 and IL-8 were measured by sandwich ELISA (BD Biosciences, Oxford, United Kingdom) according to manufacturer’s instructions. Briefly, the wells were coated with 100 μl/well of anti-human IL-6 or IL-8 monoclonal antibody and blocked for 1 h at room temperature with 200 μl/well assay diluent (PBS with 10% FBS). Standards (100 μl/well recombinant human IL-6 or IL-8) were prepared at a concentration range of 0–300 pg/ml for IL-6 and 0–200 pg/ml for IL-8 and plates were incubated for 2 h at room temperature. The detection antibody and Streptavidin-HRP reagent were added and incubated for a further 1 h at room temperature. The plates were washed with PBS with 0.05% Tween-20 (wash buffer) seven times, before addition of TMB substrate and incubation for 30 min in the dark. The stop solution (1M H3PO4) at 50 μl/well was added and the absorbance was read at 450 nm using Gen 5 plate reader (Bio-Tek). Experiments were carried out in triplicate on three independent occasions.
Well diffusion assays were performed on MEA plates with 4 × 105 CFU/100 μl lawns of P. aeruginosa and 4 × 105 CFU/100 μl A. fumigatus inoculated in the wells and vice versa. The zones of inhibition were examined after 48 h incubation at 37°C.
In vitro biofilm formation was measured in round bottom plastic 96-well plates using crystal violet to measure the biomass using a modification of a previously described method (O’Toole and Kolter, 1998). Briefly, P. aeruginosa and A. fumigatus cells/conidia were seeded into a 96-well plate at 1 × 106 CFU/ml per well. Plates were incubated at 37°C for 48 h. The effect of A. fumigatus conidia on P. aeruginosa biofilm formation was determined by adding A. fumigatus conidia to P. aeruginosa biofilms at time zero. The biomass of the mixed biofilm was then compared to the biomass of the sum of the P. aeruginosa and A. fumigatus untreated biofilm controls. P. aeruginosa cells were also seeded with 72 h CSNs from A. fumigatus. Sterile LB media was used in negative controls. Fresh media was added to the wells every 24 h to replenish nutrients. Unattached bacterial/fungal cells were removed by rinsing with 250 μl of distilled water five times. The wells were air dried for 45 min. The adherent cells were stained with 1% (w/v) Crystal Violet (Sigma, Wicklow, Ireland) at room temperature for 30 min and wells washed again. The dye was solubilized with 95% ethanol containing 0.05% (l/v) Triton X-100 (Sigma, Wicklow, Ireland). Biofilm formation was determined by reading the absorbance at 590 nm (Synergy H1 Multi-Mode Reader, Bio-Tek). Each experiment was carried out on three independent occasions, with six strain replicates per 96 well plate.
Pseudomonas aeruginosa strains and A. fumigatus strains were each seeded at 1 × 106 CFU/ml in a 6-well round bottom plate for 0, 24, 48, and 72 h. Co-cultured biofilms were created by seeding wells with 1 × 106 CFU/ml of an A. fumigatus strain together with 1 × 106 CFU/ml of a P. aeruginosa strain. P. aeruginosa biofilms were also seeded with 24, 48, and 72 h A. fumigatus CSNs. Wells containing sterile LB media were used as negative controls. The plates were incubated at 37°C and spent LB broth replenished every 24 h. At 24, 48, and 72 h the broth was aspirated and the biofilms removed by scraping and transferred to a fresh tube. All biofilm experiments were performed in triplicate on three independent occasions. The biofilms were vortexed and 50 μM propidium monoazide (PMA) (Biotium, Fremont, CA, United States) was added prior to incubation in the dark for 30 min. The samples were irradiated with light at 464–476 nm using the phAST Blue activation system (GenUIL, Barcelona, Spain) for 15 min to activate the PMA and exclude DNA from non-viable cells (Nocker et al., 2006). The samples were then centrifuged at 5,000 × g for 10 min and pellets resuspended in 500 μl PBS before centrifuging again at 5,000 × g for 10 min.
Pelleted cells were resuspended in 200 μl molecular grade water (Sigma, Wicklow, Ireland) before transferring to sterile screw-capped tube containing 0.3 g sterile acid washed glass beads (Sigma, Wicklow, Ireland) and vortexing in a horizontal multi-vortexer for 180 s at 2500 rpm. The tubes were centrifuged at 420 × g for 1 min, supernatants transferred to fresh centrifuge tube and the DNA eluted using the High Pure PCR Template Preparation kit (Roche, Clare, Ireland) according to the manufacturer’s instructions. A negative extraction control was performed for each batch of DNA extractions where the entire DNA extraction protocol was performed on 200 μl molecular grade water.
Pseudomonas aeruginosa was specifically quantified by multiplex qPCR with primers for the ecfX and gyrB genes and hydrolysis probes ecfX-TM and gyrB-TM (Qin et al., 2003; Anuj et al., 2009). A. fumigatus was quantified using primers and probed for the internal transcribed spacer 1 (ITS1) region (Walsh et al., 2011) (Supplementary Table 1) All primers and probes were supplied by Eurofins Genomics, Ebersberg, Germany. The master mix was composed of 10 μl of TaqMan Gene Expression (Biosciences, Dublin, Ireland), 0.4 μM of each primer, 0.16 μM of each hydrolysis probe, and 4 μl of DNA in a final reaction volume of 20 μl. Cycling was performed on QuantStudioTM 5 Real Time PCR System (Applied Biosystems, Dublin, Ireland), with an initial hold at 95°C for 15 min, followed by 45 cycles at 95°C for 15 s, and 60°C for 1 min, with a CT of 35 being the threshold. A negative control consisting of master mix without DNA was included in each qPCR run alongside the negative extraction control. DNA extracted from 10-fold serial dilutions (101–106 CFU/ml) of either AFref1 or PAref1 was used to generate a standard curve and to act as a positive control. The standard curves of different reference concentrations were plotted against the corresponding CT values and used to determine the concentrations of unknown samples. A ctrA plasmid was amplified in duplicate on each qPCR run (Supplementary Table 1) as a reaction control. Each sample was run in duplicate and samples were considered as positive if both duplicates showed CT values below 35. The amplification efficiency of the qPCR was calculated from the slope of the standard curve using the following equation; E = 10 (-1/slope).
Pseudomonas aeruginosa, Staphylococcus aureus, Stenotrophomonas maltophilia, B. cenocepacia, Acinetobacter baumannii, H. influenzae, and Escherichia coli were seeded at 1 × 106 CFU/ml per well in a 96 well plate and a range of concentrations of gliotoxin (Gt) (0.2–1 μM) were added to each well and incubated at 37°C for 48 h. HPLC analysis showed the concentration of gliotoxin produced by A. fumigatus at 72 h equates to approximately 0.8 μM commercial Gt (Coughlan et al., 2012), which was therefore used in this study. Fresh media was added to the wells every 24 h to replenish nutrients. Crystal violet assays were carried out to determine if Gt reduced the biomass of the biofilms of each of these pathogens.
Two-way ANOVAs with Bonferroni multiple comparisons tests were used to analyze the cytokine and MAPK assays. One-way ANOVAs with Dunnett’s multiple comparisons tests were used to analyze the crystal violet, the MTT and qPCR assays. LD50 values were compared via Student t-tests to determine statistical significance. P-values < 0.05 were considered significant. All results were analyzed using GraphPad Prism 6 and 7. The qPCR data was analyzed using the Quantstudio Design and Analysis software (Applied Biosystems).
As an initial visual evaluation of the potential for the two pathogens to interact with each other, we evaluated inhibition of growth in well diffusion assays. PA27 and PA27N inhibited A. fumigatus strains with zone of inhibition (ZOI) apparent around the wells (Supplementary Figures 1A–D). The two clinical P. aeruginosa strains (PAco1 and PAco2) which were isolated from patients co-colonized with A. fumigatus also caused ZOIs in the growth of their respective co-colonizing strains AFco1 and AFco2 (Supplementary Figures 1E,F).
In order to examine whether these two pathogens alter their virulence when co-infecting with the other, their virulence in the G. mellonella acute infection model was evaluated. A non-lethal dose for A. fumigatus was first determined. All A. fumigatus strains caused larval mortality in a dose dependent fashion (Supplementary Figure 2) and day 4 was identified as the optimal time-point for distinguishing subtle differences between strains. There was 100% survival of the G. mellonella after 3 days when injected with 102 conidia/ml for all A. fumigatus strains. Therefore, 102 conidia/ml of A. fumigatus were considered a non-lethal dose after 3 days in future experiments.
P. aeruginosa reference strain PAref, showed an LD50 of 1 CFU/ml. Pre-exposure of larvae to a non-lethal dose of A. fumigatus strain AF1 significantly reduced the LD50 of P. aeruginosa PAref (p = 0.012) compared to PAref alone indicating an apparent increase in virulence of PAref (Figure 1A). Consistent with this, the LD50s of the PAco1 strain was also reduced in the presence of AFref (p = 0.007) and AF1 (p = 0.003) (Figure 1B). The mucoid P. aeruginosa strain, PA27, showed a much higher LD50 than its non-mucoid conversion isolate and pre-exposure of larvae to all but one A. fumigatus strain significantly reduced LD50s of P. aeruginosa (p = 0.0002 for all three comparisons) compared to PA27 alone (Figure 1C). The LD50s of the non-mucoid P. aeruginosa strain, PA27N, reduced in the presence of AFref (p = 0.0018), AFco1 (p = 0.0082), AF1 (p = 0.001) and AF2 (p = 0.008) (Figure 1D). Interestingly pre-exposure to the co-colonizing strain, AFco1 did not reduce the LD50 of PA27, PAref or PAco1 (Figures 1A–C). Overall pre-exposure of larvae to a non-lethal dose of the majority of A. fumigatus isolates resulted in an apparent increase in virulence of the individual P. aeruginosa isolates when co-infecting in G. mellonella and this increase seems to be strain specific.
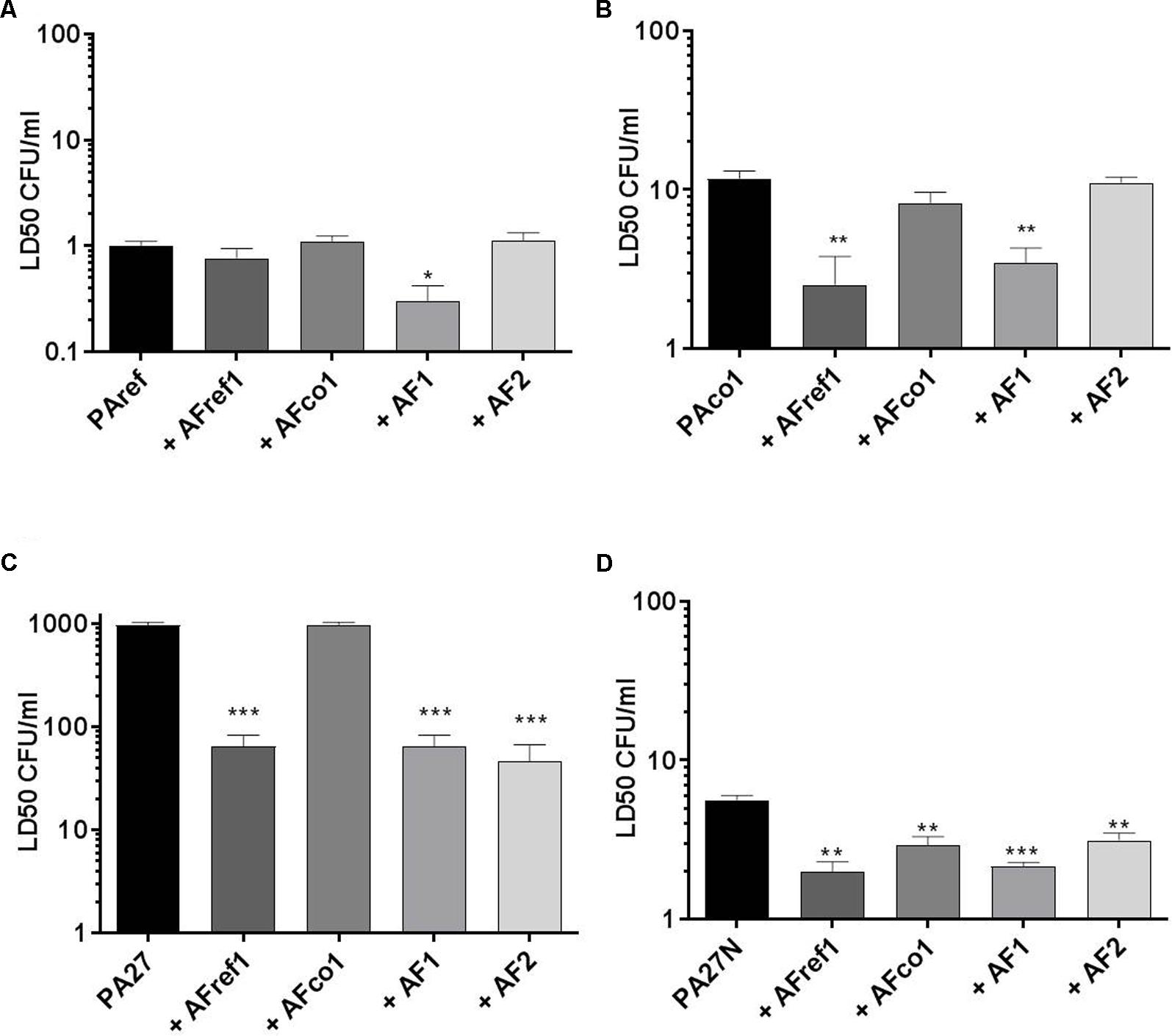
FIGURE 1. Virulence of P. aeruginosa and A. fumigatus co-infections in the G. mellonella infection model. LD50 of P. aeruginosa (A) PAref, (B) PAco1, (C) PA27, and (D) PA27N following pre-exposure to non-lethal doses of A. fumigatus strains in the G. mellonella infection model. Experiments were carried out on three independent occasions and error bars represent standard error. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001.
Given that both P. aeruginosa and A. fumigatus elicit a profound pro-inflammatory response at the lung epithelium, we wanted to examine the effects of co-colonization on the inflammatory response of CFBE cells relative to A. fumigatus or P. aeruginosa isolates alone. Only two combinations resulted in significantly increased IL-6 and IL-8 secretion (Figure 2). There was a significant increase in IL-6 production from CFBE cells when infected with co-cultures of PA27 and AFco1 compared to PA27 or AFco1 alone (p ≤ 0.0001 and p ≤ 0.0001, respectively) (Figure 2B). An increase in IL-8 release was also observed when CFBE cells were co-infected with PA27N and AFco1 compared to PA27N (p = 0.013) or AFco1 alone (p < 0.0001) (Figure 2F). In contrast, IL-6 and IL-8 production from CFBE cells when infected with all other co-cultures showed the cytokine released did not increase compared to either P. aeruginosa or A. fumigatus alone (Figures 2A,C–E). The lack of enhancement of pro-inflammatory responses for the majority of tested co-infections suggests that co-infections of A. fumigatus and P. aeruginosa may not generally further exacerbate the inflammatory response. Uninfected controls produced on average of 74 pg/ml IL-6 and 53 pg/ml IL-8. The CFU of P. aeruginosa and A. fumigatus used to infect CFBE cells (MOI 50:1 and 2:1, respectively) were not cytotoxic to the CFBE cells (Supplementary Figure 3) ensuring that the lack of enhancement of cytokine release was not due to a loss in viability of the CFBE cells.
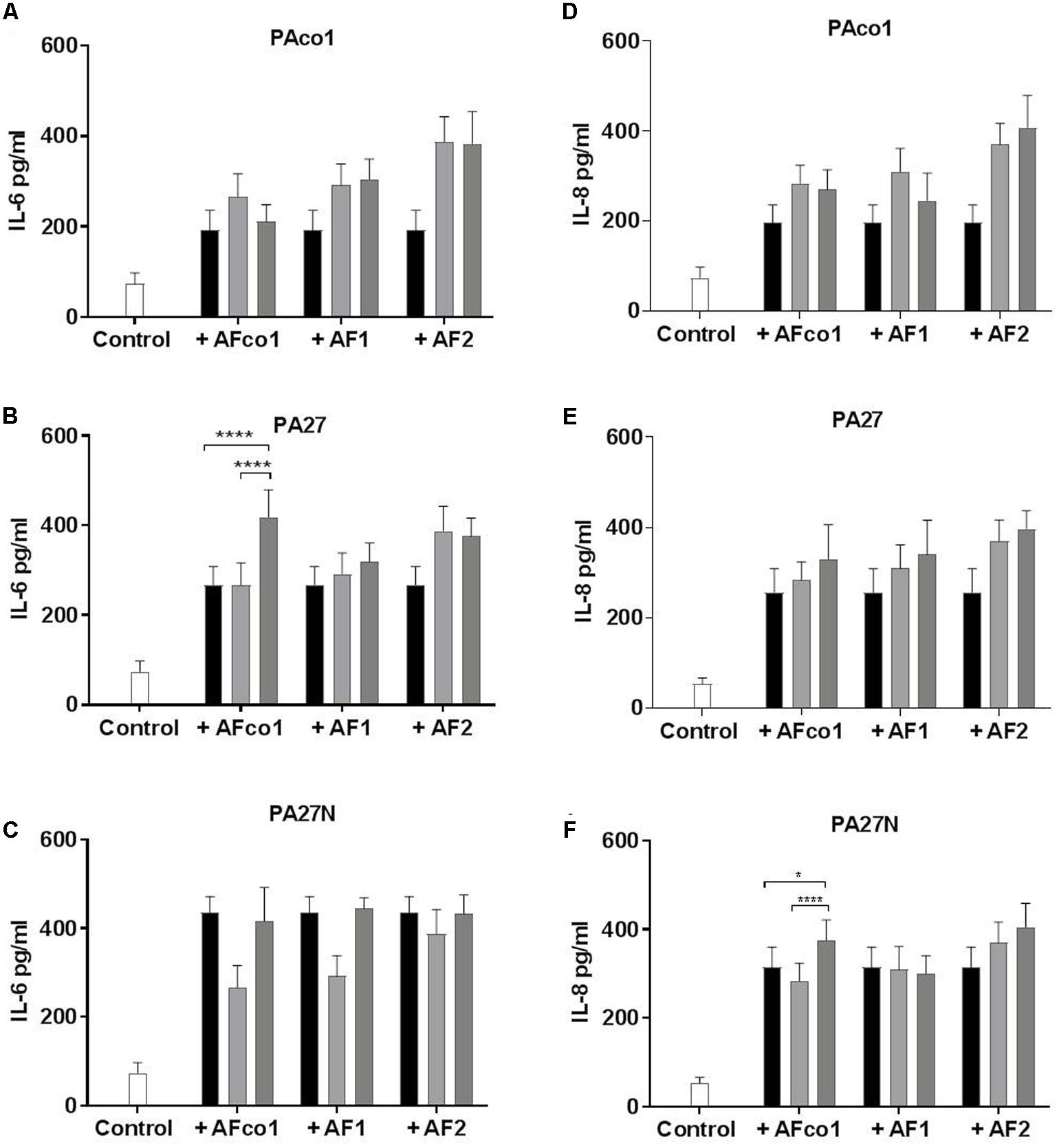
FIGURE 2. IL-6 and IL-8 secretion from CFBE cells in response to co-infection of P. aeruginosa cells and A. fumigatus conidia. Secretion of IL-6 pg/ml (A–C) and IL-8 pg/ml (D–F) from CFBE cells in response to co-infections of P. aeruginosa cells PAco1 (A,D), PA27 (B,E), PA27N (C,F) and A. fumigatus conidia. Black bars represent P. aeruginosa alone, light gray bars A. fumigatus alone and dark gray bars co-exposure of the CFBEs. Experiments were carried out on three independent occasions in triplicate and error bars represent standard deviation. ∗p ≤ 0.05, ∗∗∗∗p ≤ 0.0001.
In order to further investigate why the majority of co-infections did not have an additive effect on the secretion of IL-6 and IL-8, we examined the signaling mechanisms of both pathogens. All four co-cultures elicited reduced IL-6 secretion in the presence of either ERK or p38 inhibitors and these levels were further reduced to that of the uninhibited controls when ERK and p38 inhibitors were used in combination (p ≤ 0.0001 in all four co-cultures). These reductions in IL-6 secretion varied from 16 to 26% relative to the corresponding uninhibited samples, suggesting the co-culture of both P. aeruginosa and A. fumigatus induce an IL-6 response via the ERK and p38 MAPK pathways (Supplementary Figures 4A–D).
In all co-culture combinations there was also a significant decrease in IL-8 secretion in the presence of the combination of ERK and p38 inhibitors. The IL-8 secretion was reduced to 11–19% (p ≤ 0.0001) of the uninhibited samples which suggests that the IL-8 secretion elicited by the co-cultures of P. aeruginosa and A. fumigatus also signals via the ERK and p38 pathways (Supplementary Figures 4E–H). When the CFBEs were infected with the co-cultures PA27 and AF1 in the presence of JNK inhibitors there was only a slight decrease in IL-8 production to 89% (p = 0.04) (Supplementary Figure 4E). Overall the impact of the JNK pathway was minimal.
Initial well diffusion assays suggested that there was a degree of antagonism between the two pathogens, but this was not quantifiable. In order to quantitate the overall level of antagonism mixed biofilms of A. fumigatus conidia and P. aeruginosa cells were prepared. It was apparent the resulting biofilm biomass (as measured by crystal violet) was significantly reduced to 60–79% relative to the sum of single species biofilm controls for all isolates tested (range p = 0.0001–0.011) (Table 2). These results suggest that when P. aeruginosa and A. fumigatus were cultured together in a co-cultured biofilm, biofilm formation was significantly impaired relative to that formed by individual species. Overall mixed biofilms between any of the P. aeruginosa strains and any of the A. fumigatus strains showed reduced total biofilm formation, compared with the individual biofilm on its own.
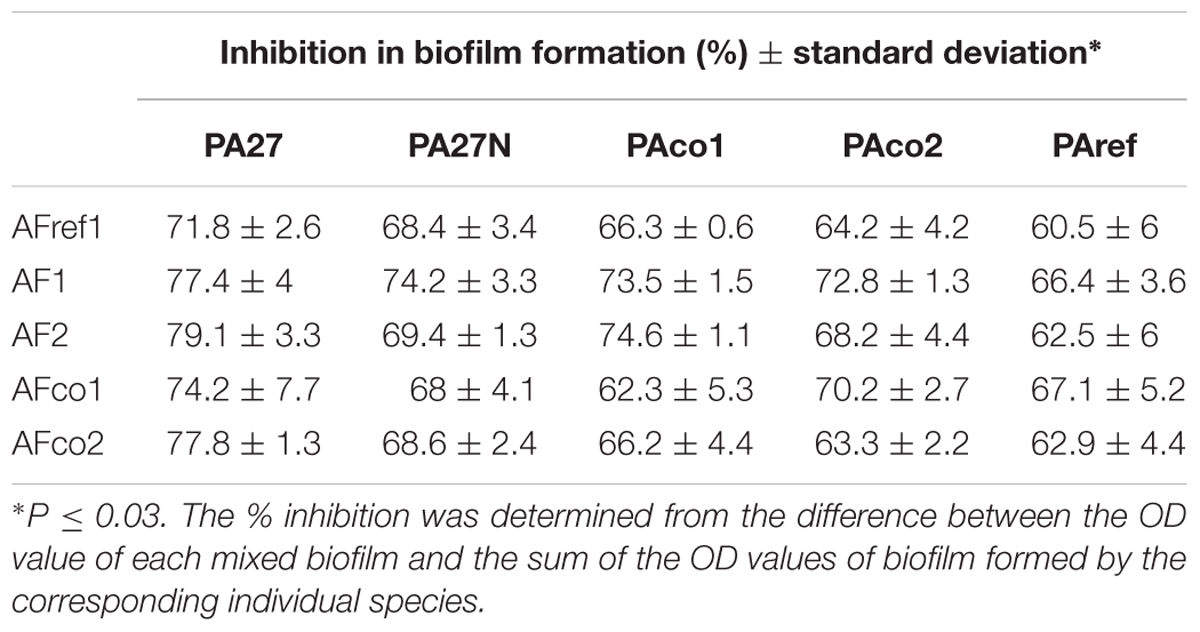
TABLE 2. Reduction in mixed species biofilm formation in comparison to single species biofilms as measured by the crystal violet assay.
In order to quantify the inhibitory effects at the species level, species-specific qPCR was carried out to determine the reduction of A. fumigatus biofilms by P. aeruginosa and vice versa in the mixed biofilms. Optimization of qPCR resulted in amplification efficiencies of P. aeruginosa and A. fumigatus of 105.3% (+4.97% and ranged between 90.6 and 112.2% efficiency). The R2 values of the standard curves varied from 0.9807 to 0.9985 and all duplicates were within 0.5 Ct values of each other, indicating high consistency across replicates.
Aspergillus fumigatus-specific qPCR confirmed that A. fumigatus AFref1 biofilm increased over time from seeding at 0 to 72 h culminating in an estimated 3.8 × 108 equivalent conidia/ml. In the mixed biofilm PAref did not cause any reduction in AFref1 biomass up to 48 h, however, at 72 h A. fumigatus was significantly reduced in the co-cultured biofilm from 3.8 × 108 equivalent conidia/ml to 3.9 × 104 CFU/ml (p ≤ 0.0001) (Figure 3A), demonstrating that the presence of P. aeruginosa in a co-cultured biofilm with A. fumigatus inhibited the viable fungal biofilm.
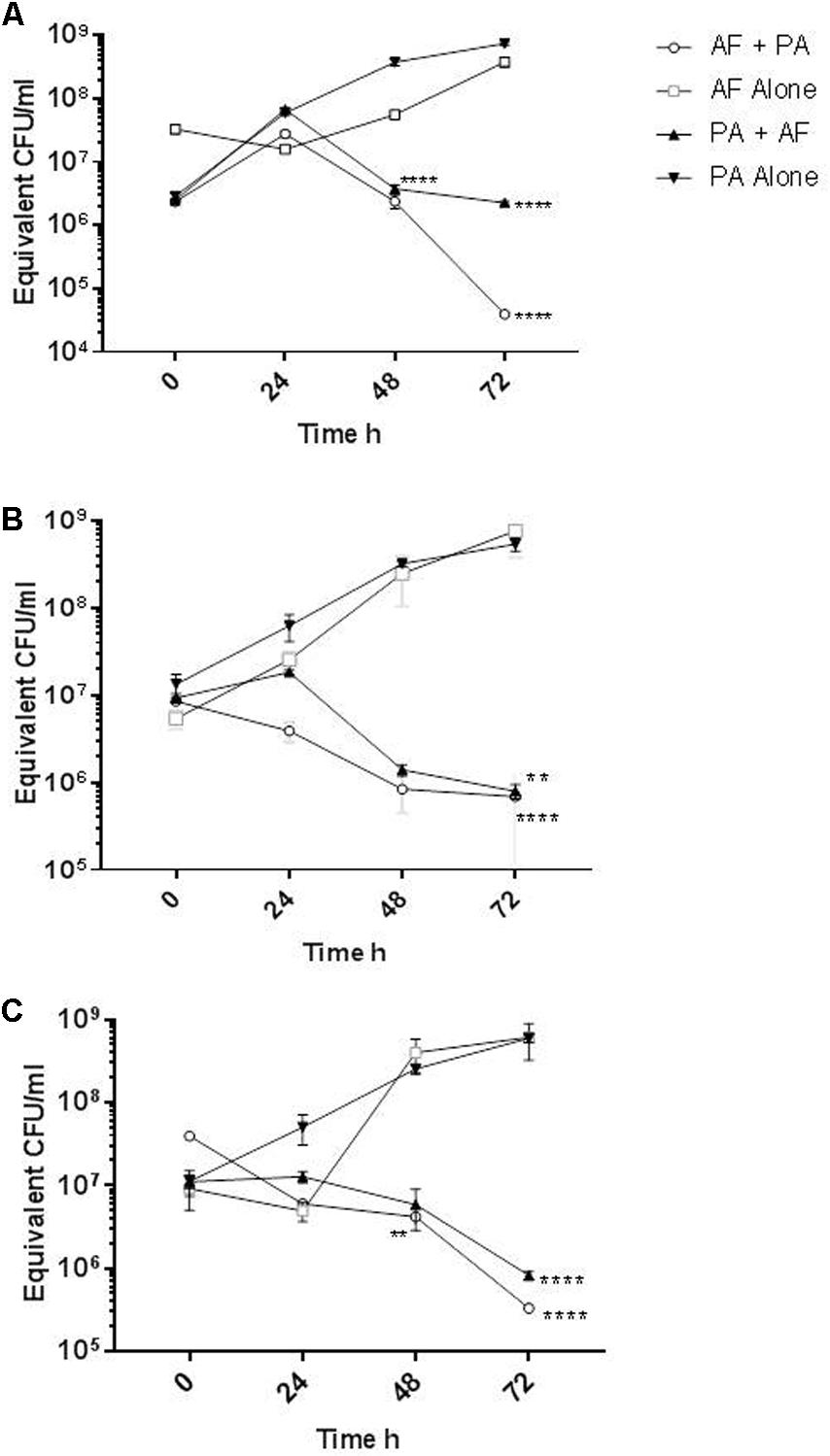
FIGURE 3. Reduction of A. fumigatus and P. aeruginosa biofilm when in mixed species biofilms. Quantification of biofilm biomass as measured by qPCR of A. fumigatus biofilm alone (open squares) and when in a co-cultured biofilm with P. aeruginosa (open circles) or P. aeruginosa biofilm alone (inverted triangles) and when in a co-cultured biofilm with A. fumigatus (upright triangles). (A) AFref1 and PAref individually and in co-cultured biofilm; (B) AFco1 and PAco1 individually and in co-cultured biofilm; (C) AFco2 and PAco2 individually and in co-cultured biofilm. Experiments were carried out on three independent occasions and error bars represent standard error. ∗∗p ≤ 0.01, ∗∗∗∗p ≤ 0.0001.
The biofilm formed by P. aeruginosa reference strain PAref in isolation increased over 72 h culminating in 7.5 × 108 equivalent CFU/ml. P. aeruginosa biofilm formation was significantly reduced when in a mixed biofilm with AFref1 at 48 h by over 2 log (p ≤ 0.0001) and at 72 h (p ≤ 0.0001) (Figure 3A), demonstrating that the presence of A. fumigatus in a co-cultured biofilm with P. aeruginosa inhibited the viable bacterial biofilm.
Similar results were observed with mixed biofilms of the co-colonizing clinical strains. A. fumigatus AFco1 biofilm formation increased over 72 h culminating in an estimated 7.6 × 108 equivalent conidia/ml. At 72 h A. fumigatus was significantly lower in the co-cultured biofilm at 6.8 × 105 equivalent conidia/ml (p ≤ 0.0001) (Figure 3B). The biofilm of PAco1 increased over time culminating in 5.4 × 108 equivalent CFU/ml at 72 h. P. aeruginosa biofilm formation was significantly less when in a mixed biofilm with AFref1 at 72 h at 8 × 105 equivalent CFU/ml (p = 0.0024) (Figure 3B). A. fumigatus AFco2 biofilm increased over 72 h culminating in an estimated 6.1 × 108 equivalent conidia/ml. In the mixed biofilm AFco2 was significantly reduced when in a mixed biofilm with PAco2 at 48 h and at 72 h (p = 0.0085, Figure 3C). The biofilm of PAco1 increased over time culminating in 5.9 × 108 equivalent CFU/ml at 72 h. P. aeruginosa biofilm formation was significantly reduced when in a mixed biofilm with AFref1 at 72 h to 8.2 × 105 equivalent CFU/ml (p ≤ 0.0001) (Figure 3C). It is apparent that there is a strong mutual antagonistic relationship present between A. fumigatus and P. aeruginosa in a mixed biofilm.
Pseudomonas aeruginosa biofilms were treated with 72 h CSNs from A. fumigatus and the biomass measured by crystal violet. The bacterial biomass was reduced to 72–88% of the untreated biofilm controls (Table 3), indicating that A. fumigatus CSN can significantly decrease bacterial biofilm growth.
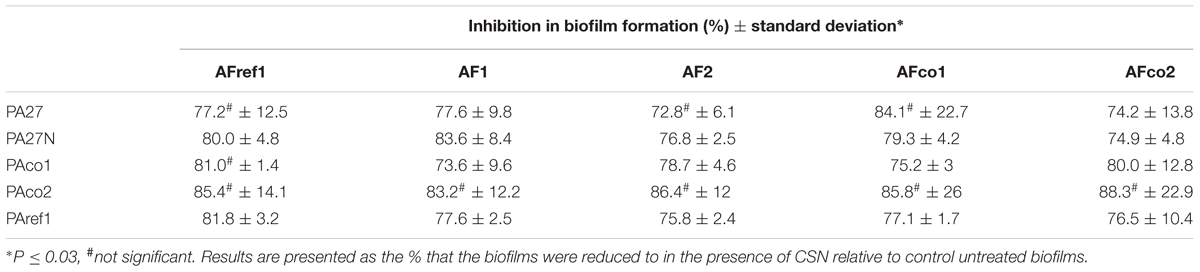
TABLE 3. Reduction of P. aeruginosa biofilm formation when treated with 72 h CSNs from A. fumigatus.
In order to quantify the reduction of P. aeruginosa biofilm formation by A. fumigatus CSN, qPCR was utilized and the bacterial cells were treated with 24, 48, and 72-h-old A. fumigatus CSN and grown for 48 h. When PAco1 biofilms (1.01 × 109 CFU/ml) were treated with 24-h-old AFco1 CSN there was no biofilm decrease, however, when the bacterial biofilm was exposed to 48 h and 72 h AFco1 CSN the viable P. aeruginosa biofilm decreased to 8.6 × 105 CFU/ml (p = 0.0183), and 1.9 × 106 CFU/ml (p = 0.0184), respectively. Figure 4A, suggesting 48-h and 72-h-old AFco1 CSNs inhibited the growth of P. aeruginosa viable biofilms.
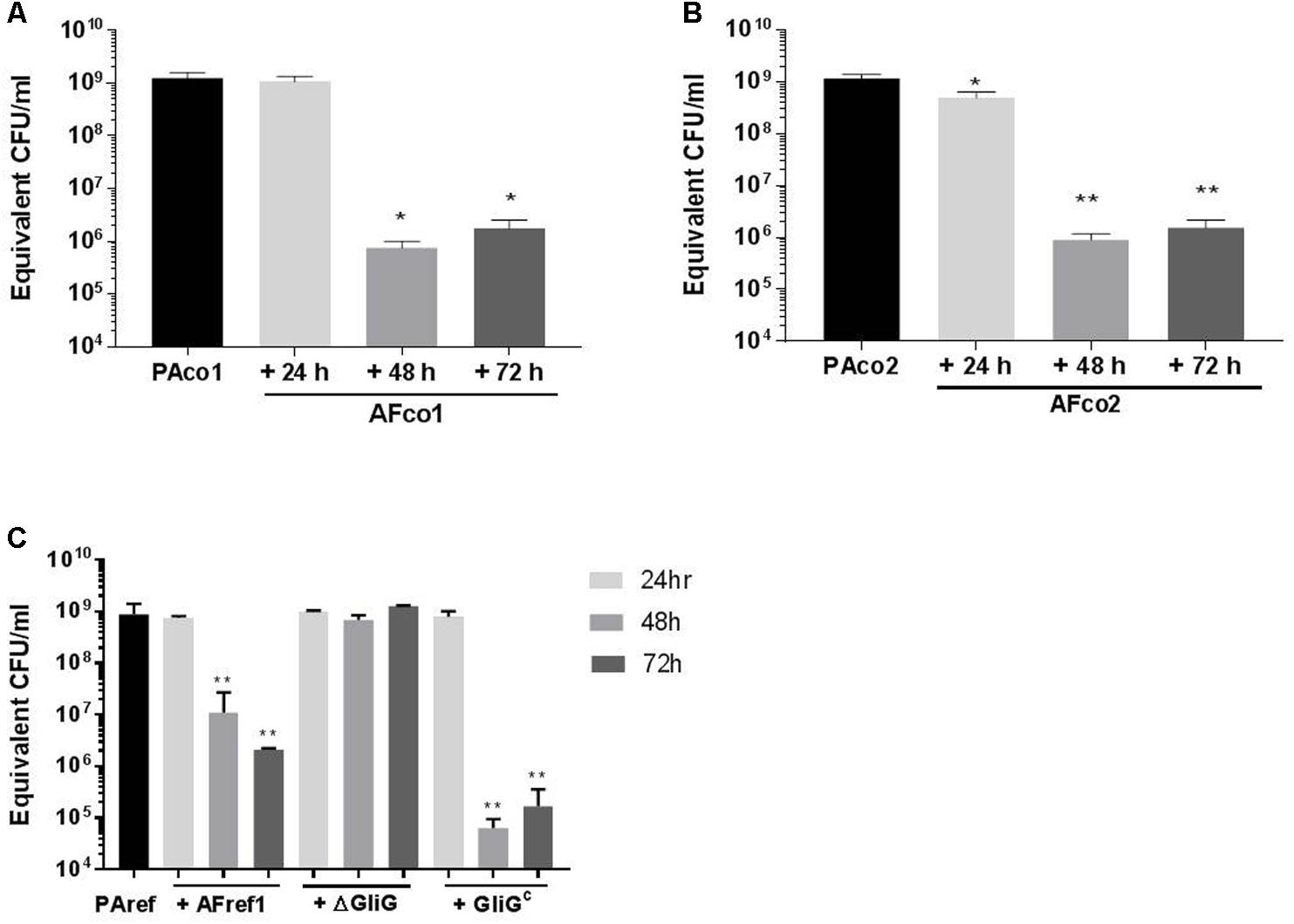
FIGURE 4. Reduction of P. aeruginosa biofilm when exposed to A. fumigatus CSN. Quantification of 48 h P. aeruginosa (A) PAco1, (B) PAco2, (C) PAref1 biofilms when exposed to (A) AFco1 (B) AFco2 (C) AFref1, AFref1 ΔGliG and AFref1 ΔGliGc 24, 48, and 72-h-old CSNs as determined by qPCR. Experiments were carried out on three independent occasions and error bars represent standard deviation. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001.
When PAco2 biofilms were treated with 24-h-old AFco2 CSN the biofilm decreased from 1.1 × 109 CFU/ml to 8.5 × 108 CFU/ml (p = 0.028) and when the bacterial biofilm was exposed to 48 h and 72 h AFco2 CSN the viable biofilm decreased to 6.7 × 105 CFU/ml (p = 0.001) and 2.2 × 106 CFU/ml (p = 0.001), respectively (Figure 4B), suggesting CSN from A. fumigatus inhibited growth of P. aeruginosa viable biofilms. PAref biofilms decreased from 7.55 × 108 CFU/ml to 1.1 × 107 CFU/ml and 2.1 × 106 CFU/ml (p = 0.0012 and p = 0.0011, respectively) when treated with 48 h and 72 h AFref1 CSNs (Figure 4C) suggesting 48-h and 72-h-old AFref1 CSNs inhibited growth P. aeruginosa viable biofilms.
Gt is known to be produced by A. fumigatus in high quantities in 48 h and 72 h CSNs but in relatively low concentration at 24 h (Reeves et al., 2004) leading us to investigate whether Gt was causing this anti-Pseudomonal and anti-biofilm effect. P. aeruginosa cells were treated with CSNs from the ΔgliG mutant AFref1 strain and viable P. aeruginosa biofilms were quantified by qPCR after 48 h. The CSNs from the ΔgliG mutant strain did not inhibit P. aeruginosa biofilm formation suggesting Gt is the main inhibitory agent secreted at 48 h and 72 h by A. fumigatus and has anti-pseudomonal and anti-biofilm effects. Exposure to CSN from the complemented strain AFref1ΔgliGc strain restored inhibition (Figure 4C).
In order to examine whether the gliotoxin inhibitory action was specific to P. aeruginosa or it had general anti-biofilm effect, a number of bacterial biofilms that are commonly found in the CF airways were treated with Gt at concentrations of 0.2–1 μM for 48 h. P. aeruginosa biofilms decreased significantly when treated with Gt at concentrations of 1–0.4 μM (p = 0.0001–0.0002) (Figure 5A). The biomass of S. maltophilia decreased significantly when treated with Gt at concentrations of 1–0.4 μM (p = 0.0012–0.0046) (Figure 5B). Gt also significantly decreased the biofilms of S. aureus (p = 0.024), H. influenzae (p = 0.0015–0.0146), A. baumannii (p = 0.0218–0.0426) and B. multivorans (p = 0.0003–0.0403) (Figures 5C–F). These results indicate gliotoxin has broad-spectrum antibacterial activity.
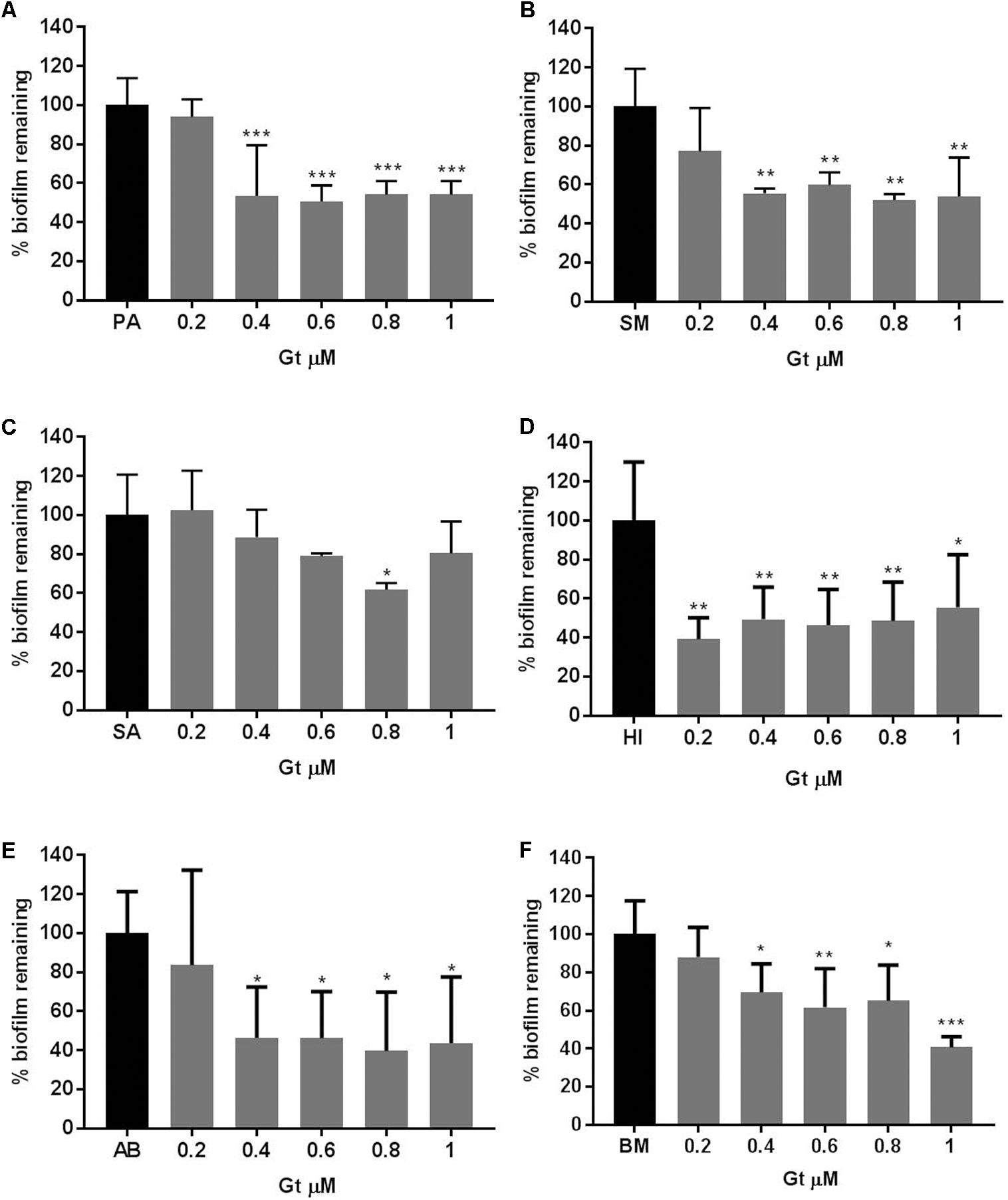
FIGURE 5. Reduction of bacterial biofilm formation when treated with gliotoxin. Percentage reduction of (A) PA: P. aeruginosa, (B) SM: S. maltophilia, (C) SA: S. aureus, (D) HI: H. influenzae, (E) AB: A. baumannii, (F) BM: B. multivorans biofilms when treated with gliotoxin (Gt) at concentrations of 0.2–1 μM measured by crystal violet assay. Experiments were carried out on three independent occasions and error bars represent standard deviation. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001.
The airways of patients with CF lung diseases host a diverse and complex community of microorganisms (Beck et al., 2012; Renwick et al., 2014). Inter-, intra-species and cross-kingdom competition and/or cooperation within the airway microbiome is likely to influence disease progression (Sibley et al., 2009; Drescher et al., 2014). P. aeruginosa and A. fumigatus are isolated together from the sputum of CF patients and it has been well established that P. aeruginosa inhibits A. fumigatus in vitro (Kerr et al., 1999; Mowat et al., 2010; Ferreira et al., 2015; Shirazi et al., 2016; Anand et al., 2017) In contrast, much less is known about the impact of A. fumigatus on P. aeruginosa. We now report that P. aeruginosa and A. fumigatus have a mutually antagonistic relationship both in planktonic and biofilm growth forms and provide evidence that this may contribute to increased mortality in the G. mellonella infection model and reduced inflammation in CFBE epithelial cells. In particular, we report for the first time the anti-pseudomonal and anti-biofilm effects of A. fumigatus on P. aeruginosa.
We studied a combination of eleven clinical isolates and reference strains, including co-colonizing clinical strains AFco1, AFco2 with their respective co-colonizers, PAco1 and PAco2 as it is widely accepted that different strains of the same species can be phenotypically quite different. For instance, there is a broad-strain dependent variation of the immune response elicited to A. fumigatus infection indicating that different isolates have diverse virulence (Rizzetto et al., 2013). P. aeruginosa mucoid conversion isolates also behave very differently (Ryall et al., 2014). The mucoid phenotype showed enhanced microcolony formation and tobramycin resistance compared to the non-mucoid form of P. aeruginosa (Hentzer et al., 2001). We now show that all of the strains caused varying degrees of antagonism, virulence, and inflammation and demonstrate the strain-specific interactions between organisms in microbial communities. It is vital for studies of this nature to include a variety of clinical strains with reference strains in order to get a thorough picture of the nature of species interaction.
The well-diffusion assay, although relatively basic, provided a good starting point for testing relationships between microorganisms and showed that all the P. aeruginosa strains tested inhibited the A. fumigatus strains, with zones of clearance apparent around all wells. Antagonism is defined as the action of any organism that suppresses or interferes with the normal growth and activity of another organism. Antagonism was evident in biofilm growth patterns where different strain combinations caused varying degrees of disruption to overall biofilm biomass generation. The crystal violet biofilm stain is widely published as a measure of total biomass (O’Toole and Kolter, 1998); however, when studying polymicrobial biofilms, tracking growth of each species individually is essential. We performed species-specific qPCR to distinguish between A. fumigatus and P. aeruginosa in co-cultured biofilms and found that both A. fumigatus and P. aeruginosa were negatively impacted by the interaction revealing a mutually antagonistic relationship. We have confirmed this across a range of reference strains and clinical isolates. It is worth highlighting that this PMA qPCR method is a very useful method for studying polymicrobial biofilms. The inclusion of a PMA-treatment step is important for accurate qPCR based enumeration, removing non-viable A. fumigatus and P. aeruginosa from biofilms and resulting in a more accurate count of viable pathogens. The qPCR-PMA+ assays offer a valuable diagnostic tool and could prove useful in monitoring the pathogen bioburden during/post antibiotic treatment. Overall, these reductionist approaches would be enhanced via microscopic analysis of co-cultured biofilms of these two organisms in a flow cell system (Murphy and Caraher, 2015).
We observed that A. fumigatus CSNs inhibited P. aeruginosa biofilm formation and this has not been shown previously. A. fumigatus hyphae produce a number of toxins including helvolic acid, fumagillin and gliotoxin (Amitani et al., 1995; Hogan et al., 1996). Increased concentrations of these toxins in 48 h and 72 h CSN may account for the increased inhibitory effect on P. aeruginosa biofilms. Manavathu and Vazquez (2014) examined the antibacterial activity of A. fumigatus CSN on P. aeruginosa planktonic growth and showed that at 48 h A. fumigatus CSN contained a heat stable compound which inhibited P. aeruginosa growth in a bacteriostatic fashion (Manavathu and Vazquez, 2014). We demonstrate that A. fumigatus produces an anti-pseudomonal and anti-biofilm agent at 48 h and 72 h of growth. Gt is a redox-active metabolite that is produced by A. fumigatus at 48 h and 72 h (Reeves et al., 2004), and its biosynthesis is directed by the gli gene cluster. Employing a gli gene mutant strain of A. fumigatus, we showed that CSNs from the mutant strain did not possess any inhibitory qualities against P. aeruginosa and exposure to CSNs from the complement strain restored inhibition suggesting gliotoxin is the main inhibitory agent secreted by A. fumigatus.
Gt belongs to the epipolythiodioxopiperazine class of secondary metabolites produced by A. fumigatus and it is characterized by a disulfide bridge known to be essential for activity (Chai and Waring, 2000). Gt displays immunosuppressive properties (Sutton et al., 1994; Schlam et al., 2016) and was previously considered for treatment of bone marrow transplant patients (Müllbacher et al., 1987; Mullbacher et al., 1988). Gt can also inhibit macrophage function (Bertout et al., 2002), induce apoptotic cell death in macrophages (Waring, 1990), block T and B cell activation and generate cytotoxic cells (Müllbacher and Eichner, 1984; Eichner et al., 1986; Sutton et al., 1996). Gt and its analogs possess antimicrobial activity against a number of microorganisms including methicillin-resistant S. aureus, extended-spectrum beta-lactamase-producing E. coli, vancomycin-resistant Enterococcus faecalis and C. albicans (Li et al., 2006; Svahn et al., 2012). While Gt would be too toxic for use as an anti-pseudomonal, Gt derivatives that maintain anti-biofilm activity without host cell toxicity should be evaluated. Although Gt is unlikely as a potential therapeutic, its antimicrobial effects against multiple species demonstrated here suggests that A. fumigatus colonization and Gt production in the airways may cause disruption of the airway microbiome and this is yet to be fully explored.
Co-infections with these mutual antagonists caused greater G. mellonella mortality than single P. aeruginosa infections. G. mellonella possess an innate immune system homologous to that of mammals (Bergin et al., 2005). Pre-exposure of larvae to non-lethal doses of A. fumigatus conidia increased the mortality of G. mellonella when co-infected with P. aeruginosa above that caused by P. aeruginosa alone. A. fumigatus produces many immunoregulatory toxins that could suppress the immune system of G. mellonella, making the larvae more susceptible to P. aeruginosa infection. Gt inhibits G. mellonella hemocyte phagocytosis and ROS formation (Renwick et al., 2007). Alternatively, the interaction between A. fumigatus and P. aeruginosa could cause a more virulent infection. Interestingly the isolates, which had originally co-colonized in the same patient showed no increase in larvae mortality when co-infecting, suggesting that these isolates may have adapted to co-exist in the same environment. Overall, these findings reveal bacterial–fungal interactions may contribute to increased pathogenicity in a strain-specific manner.
Pseudomonas aeruginosa is widely accepted to stimulate inflammation via MAP kinase activation (Beaudoin et al., 2013). A. fumigatus has been reported to have both pro-inflammatory and immunosuppressive effects (Margalit and Kavanagh, 2015; King et al., 2016; Zhang et al., 2017). The A. fumigatus strains examined in this study stimulated the secretion of pro-inflammatory cytokines from CFBE cells. While it might be expected that the combination of both pro-inflammatory pathogens would have an additive effect on inflammation, we have shown no additive inflammation when cells were exposed to both A. fumigatus and P. aeruginosa for the majority of combinations tested. We acknowledge that these polymicrobial effects on host immune dysregulation are likely to be quite complex, thus we attempted to explore possible pathways for the lack of additional inflammatory cytokine secretion. Mutual antagonism between the pathogens may reduce the bioburden that CF lung cells were exposed to potentially providing a benefit to the co-colonizers and/or it may be an adaptation of the co-colonizers allowing them to co-colonize and evade the immune system of the host and survive in the airways. Another likely scenario is that both pathogens saturate the ERK and p38 MAP kinase pathway for activating cytokine production. MAPKs facilitate signaling associated with a number of cellular activities such as cell survival, cell death, proliferation, differentiation, and inflammation (Cowan and Storey, 2003). In this study, we found that P. aeruginosa and A. fumigatus both signal cytokine production in CF epithelial cells via activation of the ERK and p38 MAP kinase pathways. Therefore the lack of synergistic inflammatory response observed due to co-infection may be due to saturation of signaling pathways. The complexity of these multifactorial interactions, will require much more examination to elucidate these pathways and identify other potential mechanisms by which these pathogens are able to evade the host immune system, establish favorable conditions and chronically persist in the CF airways.
Overall, we show for the first time that P. aeruginosa and A. fumigatus have mutually antagonistic effects at the formation stage of biofilm development in the mixed species biofilm. Other studies have noted the anti-fumigatus effects of P. aeruginosa in planktonic (Briard et al., 2016) and biofilm (Mowat et al., 2010; Briard et al., 2015; Ferreira et al., 2015; Shirazi et al., 2016) forms of growth, however, it has not been previously shown that P. aeruginosa also suffers from this cross-kingdom interaction. Gt secreted by A. fumigatus is anti-pseudomonal and anti-biofilm and could be investigated with a view to finding safer analogs as therapeutics against antibiotic resistant biofilms in CF airway infections and many other infections. Bacterial–fungal interactions may also contribute to increased pathogenicity, which could have a significant impact on the treatment of these pathogens in patients with CF. These co-infecting microbes contribute to an altered inflammatory response that could aid evasion of the immune system and chronic colonization resulting in detrimental effects on a CF patient’s health status.
ER was involved in study design, data analysis, data interpretation, and writing and editing the manuscript. SM was involved in study design, data interpretation, and writing and editing the manuscript. JR was involved in study design, data interpretation, and editing the manuscript. SD was involved in study design, provision of gliotoxin mutant and editing the manuscript. PG was involved in study design, data interpretation, and manuscript editing.
This work was funded by the National Children’s Hospital, Tallaght Hospital, Dublin, Ireland (Grant numbers: 1318, 1509, 1608, and 1714). The funding body had no role in designing the study or in collecting, analysing, and interpreting the data or in writing the manuscript.
PG has served on advisory boards and received honoraria from Vertex, Novartis, Pharmaxis, and PTC pharmaceuticals in the past however none of these organizations have contributed financially or in any other way to this work.
The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AP and handling Editor declared their shared affiliation.
We would like to thank the National Children’s Hospital for funding this research (http://thenationalchildrenshospital.ie/). We also thank the Centre for Applied Science for Health at Institute of Technology Tallaght and the Meath Foundation Research Laboratory in Tallaght Hospital for use of facilities. We are very grateful to the CF patients who contributed samples.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01205/full#supplementary-material
Amin, R., Dupuis, A., Aaron, S. D., and Ratjen, F. (2010). The effect of chronic infection with Aspergillus fumigatus on lung function and hospitalization in patients with cystic fibrosis. Chest 137, 171–176. doi: 10.1378/chest.09-1103
Amitani, R., Murayama, T., Nawada, R., Lee, W. J., Niimi, A., Suzuki, K., et al. (1995). Aspergillus culture filtrates and sputum sols from patients with pulmonary aspergillosis cause damage to human respiratory ciliated epithelium in vitro. Eur. Respir. J. 8, 1681–1687. doi: 10.1183/09031936.95.08101681
Anand, R., Clemons, K. V., and Stevens, D. A. (2017). Effect of Anaerobiasis or Hypoxia on Pseudomonas aeruginosa Inhibition of Aspergillus fumigatus Biofilm. Arch. Microbiol. 199, 881–890. doi: 10.1007/s00203-017-1362-5
Anuj, S. N., Whiley, D. M., Kidd, T. J., Bell, S. C., Wainwright, C. E., Nissen, M. D., et al. (2009). Identification of Pseudomonas aeruginosa by a duplex real-time polymerase chain reaction assay targeting the ecfX and the gyrB genes. Diagn. Microbiol. Infect. Dis. 63, 127–131. doi: 10.1016/j.diagmicrobio.2008.09.018
Beaudoin, T., Lafayette, S., Roussel, L., Bérubé, J., Desrosiers, M., Nguyen, D., et al. (2013). The level of p38α mitogen-activated protein kinase activation in airway epithelial cells determines the onset of innate immune responses to planktonic and biofilm Pseudomonas aeruginosa. J. Infect. Dis. 207, 1544–1555. doi: 10.1093/infdis/jit059
Beck, J. M., Young, V. B., and Huffnagle, G. B. (2012). The microbiome of the lung. Transl. Res. 160, 258–266. doi: 10.1038/nrmicro2539.Modulation
Bergin, D., Reeves, E. P., Renwick, J., Wientjes, F. B., and Kavanagh, K. (2005). Superoxide production in Galleria mellonella hemocytes: identification of proteins homologous to the NADPH oxidase complex of human neutrophils. Infect. Immun. 73, 4161–4170. doi: 10.1128/IAI.73.7.4161-4170.2005
Bertout, S., Badoc, C., Mallié, M., Giaimis, J., and Bastide, J. M. (2002). Spore diffusate isolated from some strains of Aspergillus fumigatus inhibits phagocytosis by murine alveolar macrophages. FEMS Immunol. Med. Microbiol. 33, 101–106. doi: 10.1016/S0928-8244(02)00281-X
Bittar, F., Richet, H., Dubus, J. C., Reynaud-Gaubert, M., Stremler, N., Sarles, J., et al. (2008). Molecular detection of multiple emerging pathogens in sputa from cystic fibrosis patients. PLoS One 3:e2908. doi: 10.1371/journal.pone.0002908
Brand, A., Barnes, J. D., Mackenzie, K. S., Odds, F. C., and Gow, N. A. (2008). Cell wall glycans and soluble factors determine the interactions between the hyphae of Candida albicans and Pseudomonas aeruginosa. FEMS Microbiol. Lett. 287, 48–55. doi: 10.1111/j.1574-6968.2008.01301.x
Briard, B., Bomme, P., Lechner, B. E., Mislin, G. L., Lair, V., Prévost, M.-C., et al. (2015). Pseudomonas aeruginosa manipulates redox and iron homeostasis of its microbiota partner Aspergillus fumigatus via phenazines. Sci. Rep. 5:8220. doi: 10.1038/srep08220
Briard, B., Heddergott, C., and Latgé, J. (2016). Volatile compounds emitted by Pseudomonas aeruginosa stimulate growth of the fungal pathogen Aspergillus fumigatus. mBio 7:e00219-16. doi: 10.1128/mBio.00219-16
Chai, C. L., and Waring, P. (2000). Redox sensitive epidithiodioxopiperazines in biological mechanisms of toxicity. Redox Rep. 5, 257–264. doi: 10.1179/135100000101535799
Coburn, B., Wang, P. W., Diaz Caballero, J., Clark, S. T., Brahma, V., Donaldson, S., et al. (2015). Lung microbiota across age and disease stage in cystic fibrosis. Sci. Rep. 5:10241. doi: 10.1038/srep10241
Costello, A., Reen, F. J., O’Gara, F., Callaghan, M., and McClean, S. (2014). Inhibition of co-colonizing cystic fibrosis-associated pathogens by Pseudomonas aeruginosa and Burkholderia multivorans. Microbiology 160, 1474–1487. doi: 10.1099/mic.0.074203-0
Coughlan, C. A., Chotirmall, S. H., Renwick, J., Hassan, T., Low, T. B., Bergsson, G., et al. (2012). The effect of Aspergillus fumigatus infection on vitamin D receptor expression in cystic fibrosis. Am. J. Respir. Crit. Care Med. 186, 999–1007. doi: 10.1164/rccm.201203-0478OC
Cowan, K. J., and Storey, K. B. (2003). Mitogen-activated protein kinases: new signaling pathways functioning in cellular responses to environmental stress. J. Exp. Biol. 206, 1107–1115. doi: 10.1242/jeb.00220
Cox, M. J., Allgaier, M., Taylor, B., Baek, M. S., Huang, Y. J., Daly, R. A., et al. (2010). Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One 5, doi: 10.1371/journal.pone.0011044
Cullen, L., and McClean, S. (2015). Bacterial adaptation during chronic respiratory infections. Pathogens 4, 66–89. doi: 10.3390/pathogens4010066
Cullen, L., Weiser, R., Olszak, T., Maldonado, R. F., Moreira, A. S., Slachmuylders, L., et al. (2015). Phenotypic characterization of an international Pseudomonas aeruginosa reference panel: strains of cystic fibrosis (cf) origin show less in vivo virulence than non-cf strains. Microbiology 161, 1961–1977. doi: 10.1099/mic.0.000155
Davis, C., Carberry, S., Schrettl, M., Singh, I., Stephens, J. C., Barry, S. M., et al. (2011). The role of glutathione S-transferase GliG in gliotoxin biosynthesis in Aspergillus fumigatus. Chem. Biol. 18, 542–552. doi: 10.1016/j.chembiol.2010.12.022
De Soyza, A., Hall, A. J., Mahenthiralingam, E., Drevinek, P., Kaca, W., Drulis-Kawa, Z., et al. (2013). Developing an international Pseudomonas aeruginosa reference panel. Microbiologyopen 2, 1010–1023. doi: 10.1002/mbo3.141
Delhaes, L., Monchy, S., Fréalle, E., Hubans, C., Salleron, J., Leroy, S., et al. (2012). The airway microbiota in cystic fibrosis: a complex fungal and bacterial community—implications for therapeutic management. PLoS One 7:e36313. doi: 10.1371/journal.pone.0036313
Douglas, T. A., Brennan, S., Gard, S., Berry, L., Gangell, C., Stick, S. M., et al. (2009). Acquisition and eradication of P. aeruginosa in young children with cystic fibrosis. Eur. Respir. J. 33, 305–311. doi: 10.1183/09031936.00043108
Drescher, K., Nadell, C. D., Stone, H. A., Wingreen, N. S., and Bassler, B. L. (2014). Solutions to the public goods dilemma in bacterial biofilms. Curr. Biol. 24, 50–55. doi: 10.1016/j.cub.2013.10.030
Duan, K., Dammel, C., Stein, J., Rabin, H., and Surette, M. G. (2003). Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 50, 1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x
Eichner, R. D., Al Salami, M., Wood, P. R., and Müllbacher, A. (1986). The effect of gliotoxin upon macrophage function. Int. J. Immunopharmacol. 8, 789–797. doi: 10.1016/0192-0561(86)90016-0
Ferreira, J. A., Penner, J. C., Moss, R. B., Haagensen, J. A., Clemons, K. V., Spormann, A. M., et al. (2015). Inhibition of Aspergillus fumigatus and its biofilm by Pseudomonas aeruginosa is dependent on the source, phenotype and growth conditions of the bacterium. PLoS One 10:e0134692. doi: 10.1371/journal.pone.0134692
Friman, V. P., Ghoul, M., Molin, S., Johansen, H. K., and Buckling, A. (2013). Pseudomonas aeruginosa adaptation to lungs of cystic fibrosis patients leads to lowered resistance to phage and protist enemies. PLoS One 8:e75380. doi: 10.1371/journal.pone.0075380
Gruenert, D. C., Basbaum, C. B., Welsh, M. J., Li, M., Finkbeiner, W. E., and Nadel, J. A. (1988). Characterization of human tracheal epithelial cells transformed by an origin-defective simian virus 40. Proc. Natl. Acad. Sci. U.S.A. 85, 5951–5955. doi: 10.1073/pnas.85.16.5951
Haq, I. J., Gray, M. A., Garnett, J. P., Ward, C., and Brodlie, M. (2016). Airway surface liquid homeostasis in cystic fibrosis: Pathophysiology and therapeutic targets. Thorax 71, 284–287. doi: 10.1136/thoraxjnl-2015-207588
Hentzer, M., Teitzel, G. M., Balzer, G. J., Molin, S., Givskov, M., Matthew, R., et al. (2001). Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183, 5395–5401. doi: 10.1128/JB.183.18.5395
Hogan, L. H., Klein, B. S., and Levitz, S. M. (1996). Virulence factors of medically important fungi. Clin. Microbiol. Rev. 9, 469–488.
Kaur, J., Pethani, B. P., Kumar, S., Kim, M., Sunna, A., Kautto, L., et al. (2015). Pseudomonas aeruginosa inhibits the growth of Scedosporium aurantiacum, an opportunistic fungal pathogen isolated from the lungs of cystic fibrosis patients. Front. Microbiol. 6:866. doi: 10.3389/fmicb.2015.00866
Kerr, J. R., Taylor, G. W., Rutman, A., Høiby, N., Cole, P. J., and Wilson, R. (1999). Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J. Clin. Pathol. 52, 385–387. doi: 10.1136/jcp.52.5.385
King, J., Brunel, S. F., and Warris, A. (2016). Aspergillus infections in cystic fibrosis. J. Infect. 72, S50–S55. doi: 10.1016/j.jinf.2016.04.022
Klepac-Ceraj, V., Lemon, K. P., Martin, T. R., Allgaier, M., Kembel, S. W., Knapp, A. A., et al. (2010). Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ. Microbiol. 12, 1293–1303. doi: 10.1111/j.1462-2920.2010.02173.x
Korgaonkar, A., Trivedi, U., Rumbaugh, K. P., and Whiteley, M. (2013). Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl. Acad. Sci. U.S.A. 110, 1059–1064. doi: 10.1073/pnas.1214550110
Kosorok, M. R., Zeng, L., West, S. E., Rock, M. J., Splaingard, M. L., Laxova, A., et al. (2001). Acceleration of lung disease in children with cystic fibrosis after Pseudomonas aeruginosa acquisition. Pediatr. Pulmonol. 32, 277–287. doi: 10.1002/ppul.1120
Li, X., Kim, S.-K., Nam, K. W., Kang, J. S., Choi, H. D., and Son, B. W. (2006). A new antibacterial dioxopiperazine alkaloid related to gliotoxin from a marine isolate of the fungus Pseudallescheria. J. Antibiot. 59, 248–250. doi: 10.1038/ja.2006.35
Li, Z., Kosorok, M. R., Farrell, P. M., Laxova, A., West, S. E., Green, C. G., et al. (2005). Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. JAMA 293, 581–588. doi: 10.1001/jama.293.5.581
Manavathu, E. K., and Vazquez, J. A. (2014). Antibacterial activities of stationary phase culture filtrates of Aspergillus fumigatus. Microbiol. Exp. 1, 2–4. doi: 10.15406/jmen.2014.01.00020
Margalit, A., and Kavanagh, K. (2015). The innate immune response to Aspergillus fumigatus at the alveolar surface. FEMS Microbiol. Rev. 39, 670–687. doi: 10.1093/femsre/fuv018
Mowat, E., Rajendran, R., Williams, C., McCulloch, E., Jones, B., Lang, S., et al. (2010). Pseudomonas aeruginosa and their small diffusible extracellular molecules inhibit Aspergillus fumigatus biofilm formation. FEMS Microbiol. Lett. 313, 96–102. doi: 10.1111/j.1574-6968.2010.02130.x
Müllbacher, A., and Eichner, R. D. (1984). Immunosuppression in vitro by a metabolite of a human pathogenic fungus. Proc. Natl. Acad. Sci. U.S.A. 81, 3835–3837. doi: 10.1073/pnas.81.12.3835
Müllbacher, A., Hume, D., Braithwaite, A. W., Waring, P., and Eichner, R. D. (1987). Selective resistance of bone marrow-derived hemopoietic progenitor cells to gliotoxin. Proc. Natl. Acad. Sci. U.S.A. 84, 3822–3825. doi: 10.1073/pnas.84.11.3822
Mullbacher, A., Moreland, A. F., Waring, P., Sjaarda, A., and Eichner, R. D. (1988). Prevention of graft-versus-host disease by treatment of bone marrow with gliotoxin in fully allogeneic chimeras and their cytotoxic T cell repertoire. Transplantation 46, 120–125. doi: 10.1097/00007890-198807000-00022
Murphy, M. P., and Caraher, E. (2015). Residence in biofilms allows Burkholderia cepacia complex (Bcc) bacteria to evade the antimicrobial activities of neutrophil-like dHL60 cells. Pathog. Dis. 73:ftv069. doi: 10.1093/femspd/ftv069
Nocker, A., Cheung, C.-Y., and Camper, A. K. (2006). Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 67, 310–320. doi: 10.1016/j.mimet.2006.04.015
O’Toole, G. A., and Kolter, R. (1998). Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28, 449–461. doi: 10.1046/j.1365-2958.1998.00797.x
Qin, X., Emerson, J., Stapp, J., Stapp, L., Abe, P., and Burns, J. L. (2003). Use of real-time PCR with multiple targets to identify Pseudomonas aeruginosa and other nonfermenting gram-negative bacilli from patients with cystic fibrosis. J. Clin. Microbiol. 41, 4312–4317. doi: 10.1128/JCM.41.9.4312
Reece, E., Segurado, R., Jackson, A., McClean, S., Renwick, J., and Greally, P. (2017). Co-colonisation with Aspergillus fumigatus and Pseudomonas aeruginosa is associated with poorer health in cystic fibrosis patients: an Irish registry analysis. BMC Pulm. Med. 17:70. doi: 10.1186/s12890-017-0416-4
Reeves, E. P., Messina, C. G., Doyle, S., and Kavanagh, K. (2004). Correlation between gliotoxin production and virulence of Aspergillus fumigatus in Galleria mellonella. Mycopathologia 158, 73–79. doi: 10.1023/B:MYCO.0000038434.55764.16
Renwick, J., McNally, P., John, B., DeSantis, T., Linnane, B., and Murphy, P. (2014). The microbial community of the cystic fibrosis airway is disrupted in early life. PLoS One 9:e109798. doi: 10.1371/journal.pone.0109798
Renwick, J., Reeves, E. P., Wientjes, F. B., and Kavanagh, K. (2007). Translocation of proteins homologous to human neutrophil p47phox and p67phox to the cell membrane in activated hemocytes of Galleria mellonella. Dev. Comp. Immunol. 31, 347–359. doi: 10.1016/j.dci.2006.06.007
Rizzetto, L., Giovannini, G., Bromley, M., Bowyer, P., Romani, L., and Cavalieri, D. (2013). Strain dependent variation of immune responses to A. fumigatus: definition of pathogenic species. PLoS One 8:e56651. doi: 10.1371/journal.pone.0056651
Ryall, B., Carrara, M., Zlosnik, J. E. A., Behrends, V., Lee, X., Wong, Z., et al. (2014). The mucoid switch in Pseudomonas aeruginosa represses quorum sensing systems and leads to complex changes to stationary phase virulence factor regulation. PLoS One 9:e96166. doi: 10.1371/journal.pone.0096166
Salsgiver, E. L., Fink, A. K., Knapp, E. A., LiPuma, J. J., Olivier, K. N., Marshall, B. C., et al. (2016). Changing epidemiology of the respiratory bacteriology of patients with cystic fibrosis. Chest 149, 390–400. doi: 10.1378/chest.15-0676
Schlam, D., Canton, J., Carreño, M., Kopinski, H., Freeman, S. A., Grinstein, S., et al. (2016). Gliotoxin suppresses macrophage immune function by subverting phosphatidylinositol 3,4,5-trisphosphate homeostasis. mBio 7:e02242-15. doi: 10.1128/mBio.02242-15
Shirazi, F., Ferreira, J. A., Stevens, D. A., Clemons, K. V., and Kontoyiannis, D. P. (2016). Biofilm filtrates of pseudomonas aeruginosa strains isolated from cystic fibrosis patients inhibit preformed Aspergillus fumigatus biofilms via apoptosis. PLoS One 11:e0150155. doi: 10.1371/journal.pone.0150155
Sibley, C. D., Grinwis, M. E., Field, T. R., Eshaghurshan, C. S., Faria, M. M., Dowd, S. E., et al. (2011). Culture enriched molecular profiling of the cystic fibrosis airway microbiome. PLoS One 6:e22702. doi: 10.1371/journal.pone.0022702
Sibley, C. D., Parkins, M. D., Rabin, H. R., and Surette, M. G. (2009). The relevance of the polymicrobial nature of airway infection in the acute and chronic management of patients with cystic fibrosis. Curr. Opin. Investig. Drugs 10, 787–794.
Smith, K., Rajendran, R., Kerr, S., Lappin, D. F., Mackay, W. G., Williams, C., et al. (2015). Aspergillus fumigatus enhances elastase production in Pseudomonas aeruginosa co-cultures. Med. Mycol. 53, 645–655. doi: 10.1093/mmy/myv048
Spicuzza, L., Sciuto, C., Vitaliti, G., Di Dio, G., Leonardi, S., and La Rosa, M. (2009). Emerging pathogens in cystic fibrosis: ten years of follow-up in a cohort of patients. Eur. J. Clin. Microbiol. Infect. Dis. 28, 191–195. doi: 10.1007/s10096-008-0605-4
Sutton, P., Newcombe, N. R., Waring, P., and Mullbacher, A. (1994). In vivo immunosuppressive activity of gliotoxin, a metabolite produced by human pathogenic fungi. Infect. Immun. 62, 1192–1198.
Sutton, P., Waring, P., and Mullbacher, A. (1996). Exacerbation of invasive aspergillosis by the immunosuppressive fungal metabolite, gliotoxin. Immunol. Cell Biol. 74, 318–322. doi: 10.1038/icb.1996.57
Svahn, K. S., Göransson, U., El-Seedi, H., Bohlin, L., Larsson, D. G. J., Olsen, B., et al. (2012). Antimicrobial activity of filamentous fungi isolated from highly antibiotic-contaminated river sediment. Infect. Ecol. Epidemiol. 2, 6–11. doi: 10.3402/iee.v2i0.11591
Walsh, T. J., Wissel, M. C., Grantham, K. J., Petraitiene, R., Petraitis, V., Kasai, M., et al. (2011). Molecular detection and species-specific identification of medically important Aspergillus species by real-time PCR in experimental invasive pulmonary aspergillosis. J. Clin. Microbiol. 49, 4150–4157. doi: 10.1128/JCM.00570-11
Waring, P. (1990). DNA fragmentation induced in macrophages by gliotoxin does not require protein synthesis and is preceded by raised inositol triphosphate levels. J. Biol. Chem. 265, 14476–14480.
Keywords: co-colonization, cystic fibrosis, Aspergillus fumigatus, Pseudomonas aeruginosa, biofilm, inflammation, gliotoxin
Citation: Reece E, Doyle S, Greally P, Renwick J and McClean S (2018) Aspergillus fumigatus Inhibits Pseudomonas aeruginosa in Co-culture: Implications of a Mutually Antagonistic Relationship on Virulence and Inflammation in the CF Airway. Front. Microbiol. 9:1205. doi: 10.3389/fmicb.2018.01205
Received: 18 December 2017; Accepted: 17 May 2018;
Published: 05 June 2018.
Edited by:
Giovanni Di Bonaventura, Università degli Studi “G. d’Annunzio” Chieti-Pescara, ItalyReviewed by:
Arianna Pompilio, Università degli Studi “G. d’Annunzio” Chieti-Pescara, ItalyCopyright © 2018 Reece, Doyle, Greally, Renwick and McClean. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siobhán McClean, c2lvYmhhbi5tY2NsZWFuQHVjZC5pZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.