- 1Key Laboratory of Hebei Province for Plant Physiology and Molecular Pathology, Hebei Agricultural University, Baoding, China
- 2Mycotoxin and Molecular Plant Pathology Laboratory, Hebei Agricultural University, Baoding, China
A pathogenic mutant, BCG183, was obtained by screening the T-DNA insertion library of Botrytis cinerea. A novel pathogenicity-related gene BcKMO, which encodes kynurenine 3-monooxygenase (KMO), was isolated and identified via thermal asymmetric interlaced PCR, bioinformatics analyses, and KMO activity measurement. The mutant BCG183 grew slowly, did not produce conidia and sclerotia, had slender hyphae, and presented enhanced pathogenicity. The phenotype and pathogenicity of the BcKMO-complementing mutant (BCG183/BcKMO) were similar to those of the wild-type (WT) strain. The activities of polymethylgalacturonase, polygalacturonase, and toxins were significantly higher, whereas acid production was significantly decreased in the mutant BCG183, when compared with those in the WT and BCG183/BcKMO. Moreover, the sensitivity of mutant BCG183 to NaCl and KCl was remarkably increased, whereas that to fluconazole, Congo Red, menadione, H2O2, and SQ22536 and U0126 [cAMP-dependent protein kinase (cAMP) and mitogen-activated protein kinase (MAPK) signaling pathways inhibitors, respectively] were significantly decreased compared with the other strains. Furthermore, the key genes involved in the cAMP and MAPK signaling pathways, Pka1, Pka2, PkaR, Bcg2, Bcg3, bmp1, and bmp3, were significantly upregulated or downregulated in the mutant BCG183. BcKMO expression levels were also upregulated or downregulated in the RNAi mutants of the key genes involved in the cAMP and MAPK signaling pathways. These findings indicated that BcKMO positively regulates growth and development, but negatively regulates pathogenicity of B. cinerea. Furthermore, BcKMO was found to be involved in controlling cell wall degrading enzymes activity, toxins activity, acid production, and cell wall integrity, and participate in cAMP and MAPK signaling pathways of B. cinerea.
Introduction
Botrytis cinerea, a typical necrotrophic pathogenic fungus, infects more than 200 plant species worldwide, causing significant economic losses to agricultural production (Elad et al., 2007; Williamson et al., 2007). It is characterized by abundant conidia and sclerotia, and can overwinter through intact mycelia or sclerotia, which germinate in spring to produce conidia. The conidia are dispersed by wind and rainwater, and cause the next cycle of infections. Identification of the functional genes regulating the growth, development, and pathogenicity of B. cinerea is very important to understand the mechanism of this fungal dispersion. With the completion of genome sequencing of B. cinerea strains B05.10 and T4, B. cinerea has become a model organism in the field of developmental biology and molecular plant pathology.
Botrytis cinerea produces various virulence factors to penetrate the host surface, kill the host cells, and generate lesions (Choquer et al., 2007). Among them, multiple cell wall degrading enzymes (CWDEs) and non-specific pathotoxins secreted by B. cinerea are considered to be the major pathogenic factors. For instance, pectinase secreted by B. cinerea causes leaf rot, leading to the collapse of chloroplasts and mitochondrion of leaf cells. Several types of CWDEs are extracellularly secreted by B. cinerea at the stages of conidial germination and hyphal growth (Blanco-Ulate et al., 2014), among which polymethylgalacturonase (PMG) exhibits superior activity (Van Kan, 2006). Furthermore, it has been reported that the polygalacturonase (PG), BcPG1, contributes to penetration and is essential during colonization of B. cinerea (ten Have et al., 1998), whereas BcPG2 contributes only to penetration (Choquer et al., 2007; Patel et al., 2008). Moreover, inactivation of the pectin methylesterase (PME) gene, bcpme1, in B. cinerea has been found to result in a sharp reduction in the fungal virulence on several plant hosts, suggesting that PMEs might facilitate the action of PG (Valette-Collet et al., 2003). Cellulase (CX), hemicellulases and non-pectinolytic CWDEs also promote B. cinerea infection (Choquer et al., 2007). B. cinerea secretes nearly 20 types of non-specific pathotoxins, including polyketide botcinic acid and sesquiterpene botrydial, to support both penetration and colonization. Botrydial was first isolated and described in 1974, and has been reported to exhibit highest phytotoxic activity (Williamson et al., 2007). The genes involved in the botrydial biosynthetic pathway have been identified as a physical cluster, including BcBOT1, BcBOT2, BcBOT3, BcBOT4, and BcBOT5, among which BcBOT2 encodes sesquiterpene synthase that is responsible for the committed step in botrydial biosynthetic pathway (Pinedo et al., 2008). Sesquiterpene synthase converts farnesyl diphosphate to the precursor of botrydial, presilphiperfolan-8β-ol (PSP; the tricyclic sesquiterpene alcohol). PSP is converted to botrydial by the action of three cytochrome P450s (encoded by BcBOT1, BcBOT3, and BcBOT4) and an acetyl transferase (encoded by BcBOT5) (Wang et al., 2009). It has been reported that deletion of BcBOT2 eliminated the production of botrydial and resulted in strain-dependent loss of virulence (Pinedo et al., 2008).
In addition to CWDEs and pathotoxins, B. cinerea produces reactive oxygen species and oxalic acid during infection (Van Kan, 2006), and H2O2 accumulation has been noted in germinating spores, infection cushions, and in the early stages of B. cinerea infection. Deletion mutants of B. cinerea superoxide dismutase (BcSOD1; contributing to the accumulation of phytotoxic H2O2 levels) have been found to exhibit significantly retarded development of lesions, indicating that the H2O2-generating enzyme in B. cinerea is a virulence factor (Rolke et al., 2004). Furthermore, B. cinerea could produce non-host specific toxin, oxalic acid (OA), both in vivo and in vitro, which is often described as an important factor in the pathogenesis of Sclerotinia sclerotiorum (Walz et al., 2008). However, several reports have suggested that OA is a less important factor in B. cinerea infection, and that B. cinerea exploits the plant defense mechanisms for its growth and proliferation in the plant tissue.
Several genes of signal transduction pathways, such as the mitogen-activated protein kinase (MAPK), cAMP-dependent protein kinase (cAMP), and Ca2+/calcineurin pathways, have been characterized, and their effects on the growth, development, and pathogenicity of B. cinerea have been reported (Doehlemann et al., 2006; Choquer et al., 2007). The Slt2-type MAPK Bmp3 mutant has been reported to show significantly impaired conidiation, reduced vegetative growth, and loss of sclerotium formation (Rui and Hahn, 2007; Schamber et al., 2010), while the mutant of bos5 encoding a MAPK kinase has been found to exhibit severely impaired conidiation, reduced vegetative growth, and loss of pathogenicity (Yan et al., 2010). Furthermore, deletion mutants of the MAPK encoding gene, bcsak1, have been observed to present significantly impaired growth, conidia formation, sclerotia development, and pathogenicity (Segmuller et al., 2007; Heller et al., 2012), while in the BcPKA1 mutant, the catalytic subunit of the cAMP protein kinase A (PKA), demonstrated impaired growth, colony morphology, and virulence (Schumacher et al., 2008b). Moreover, deletion of BcCRZ1, a calcineurin-responsive zinc finger transcription factor, has been found to cause severely impaired growth, hyphal morphology, sclerotia and conidia formation, and pathogenicity (Schumacher et al., 2008a), and, so far, more than 100 genes involved in the growth, development, and pathogenicity of B. cinerea have been reported (Choquer et al., 2007).
Kynurenine 3-monooxygenase (KMO) is a NADPH-dependant flavoprotein hydroxylase and an important enzyme located at an important branch of the kynurenine pathway (Smith et al., 2016). Kynurenine pathway is the primary route of synthesis of the essential cellular cofactor NAD, and is responsible for >95% of tryptophan metabolism in mammals (Wilson et al., 2014). KMO belongs to the family of oxidoreductases, catalyzes kynurenine to generate 3-hydroxykynurenine in the kynurenine pathway. Currently, KMO is an attractive drug target for several immunological, neurodegenerative, and neuroinflammatory diseases, especially Alzheimer’s, Huntington’s, and Parkinson’s disease (Smith et al., 2016). The kynurenine pathway has long been considered to be specific for eukaryotic organisms only, but later, several genes of the kynurenine pathway were identified in several bacteria and shown to be functional (Kurnasov et al., 2003a,b). The kynurenine pathway was involved in the biosynthesis of a quinoline siderophore, actinomycin, and benzodiazepine in Pseudomonas siderophore, Streptomyces chrysomallus, and S. refuineus, respectively (Matthijs et al., 2004; Hu et al., 2007; Keller et al., 2010). So far, there is no report on the presence of kynurenine pathway and KMO in B. cinerea.
In the present study, a novel pathogenic mutant, BCG183, screened from the T-DNA insertion library of B. cinerea was described, and a newly identified locus, BcKMO (BC1G_07455), which encodes a hypothetical protein similar to kynurenine 3-monooxygenase (KMO), was examined. Furthermore, the CWDEs activity, toxins activity, and expression levels of pathogenicity-related genes were investigated in the B. cinerea wild-type (WT), BcKMO T-DNA insertion mutant (BCG183), and BcKMO-complementing mutant (BCG183/BcKMO). The findings of this study could contribute to better understanding of the regulatory mechanisms of BcKMO in the growth, development, and pathogenicity of B. cinerea.
Materials and Methods
Strains and Culture Conditions
The WT B. cinerea strain BC22 was isolated by the Mycotoxin and Molecular Plant Pathology Laboratory, Agriculture University of Hebei, China. The mutant BCG183 was obtained from the ATMT mutant library of the B. cinerea WT strain BC22. All the B. cinerea strains were cultured on potato dextrose agar (PDA) at 22°C. For DNA and RNA extraction, the mycelia were grown in potato dextrose (PD) medium for 5–7 days at 22°C. For protoplasts preparation, the mycelia from the mutant BCG183 were cultivated in malt/yeast medium for 24 h with shaking.
Identification of Mutant BCG183
Genomic DNA was isolated from the WT and the BCG183 mutant by cetyltrimethylammonium bromide method, and used as a PCR template along with the specific primers of hygromycin B phosphotransferase gene (hph). Southern blot was performed to confirm the presence of T-DNA in the genomic DNA of the mutant BCG183 by using specific fragment of the hph gene as the probe. The precise fragment of the hph gene was amplified with hph gene specific primers, purified, and labeled with digoxigenin (DIG) using a DIG Labeling Kit (Roche, Germany). The genomic DNA of the mutant BCG183 was isolated and digested by HindIII, electrophoretically separated on 0.8% (w/v) agarose gel, and transferred to a nylon membrane (Sangon Co., Ltd., China). Hybridization was performed for 16 h at 48°C in PerfectHyb plus hybridization buffer (Sigma-Aldrich, United States), and immunological detection was conducted overnight.
Identification of Mutant Gene in BCG183
The flanking sequence of the T-DNA insert was obtained by thermal asymmetric interlaced PCR (TAIL-PCR), as described previously (Mullins et al., 2001). The genomic DNA of the mutant BCG183 was used as a template along with AD4 primer and LB1/LB2/LB3 primer (Supplementary Table S1). The tertiary PCR products were electrophoresed on a 1.0% TAE-agarose gel and purified with QIAquick columns (TIANgel Midi Purification Kit, Cat. No. Dp209-03, Qiagen, Germany). The purified PCR products were cloned into the plasmid pMD-18 vector (Takara, Japan) and sequenced by Sangon Co., Ltd., China. The T-DNA insert locus was obtained using BLAST program with B. cinerea genome sequences in the B. cinerea genome database1.
Semi-quantitative RT-PCR was used to determine the expression levels of the T-DNA insert gene and confirm the presence of the mutant gene in the mutant BCG183. The total RNA was isolated from the WT and mutant BCG183 with a Total RNA Preparation Kit (Omega Bio-Tek, Inc., United States), according to the manufacturer’s instructions. Reverse transcription was performed with RevertAid M-MuLV Reverse Transcriptase (Sangon Co., Ltd., China). The synthesized cDNA served as the PCR template, whereas tubulin gene was used for equal loading.
Bioinformatics Analysis of the Mutant Gene BcKMO
The conserved domain of the BcKMO protein was obtained using the BLAST-based NCBI conserved domain search engine (Marchler-Bauer et al., 2017). Amino acid sequence homology alignment of the BcKMO protein was performed using BLAST programs of NCBI2. Phylogenetic analysis of BcKMO and related proteins was performed using MEGA7 software (Kumar et al., 2016).
KMO Activity Measurements
The purified BcKMO protein was obtained by prokaryotic expression analysis of BcKMO from B. cinerea, and the KMO activity was measured using Human KMO ELISA Kit (DLDEVELOP, Canada) based on sandwich enzyme immunoassay. The enzyme–substrate reaction was terminated by the addition of sulfuric acid solution, and the color change was measured spectrophotometrically at a wavelength of 450 ± 10 nm. The concentration of KMO in the samples was determined by comparing the optical density (OD) values of the samples to the standard curve.
Complementation of the BcKMO Gene
The BcKMO-coding region was amplified with BcKMO-specific primers and cloned into pBARKS1-eGFP using SacI and SmaI restriction sites, to obtain pBARKS1-BcKMO-eGFP (Supplementary Figure S3). The protoplasts of the mutant BCG183 were prepared and transformed with pBARKS1-BcKMO-eGFP as described previously (Levis et al., 1997). PCR of the transformant with GFP- and bar-specific primers showed that the constructs were integrated. Southern blot was used to confirm the integration of the construct with GFP-specific probe. Quantitative real-time PCR experiments established that the introduced construct was expressed in the transformant.
Phenotypic Analyses
For phenotypic analyses of the mutant BCG183 and complement mutant, the mycelia of the WT, mutant BCG183, and BcKMO-complementing mutant were, respectively, inoculated onto PDA plates and incubated for 10 days at 22°C. The color, morphology, and sclerotia formation of the colonies were observed. The morphology and septum spacing of the hyphae were determined under a microscope (Nikon Eclipse E-200). For conidiation assays, 10-day-old conidia of all the strains were collected with 7 mL of sterile water per PDA plate and counted under a microscope. For growth assays, 5 mm of mycelial blocks of all the strains were incubated at the center of PDA plates, and the colony diameters were measured every 24 h for 7 days. All experiments were performed in triplicate.
Pathogenicity Assay
Detached kidney beans and cucumber leaves were used for the pathogenicity assay. The leaves were washed 2–3 times with sterile water, sterilized with 75% alcohol for 3 min, and washed again with sterile water. Then, culture blocks of the WT and mutants were prepared and inoculated onto the intact leaves. The inoculated leaves were moisturized and incubated at 22°C, and observed daily. Lesion formation was examined at 4 days post-inoculation (dpi). The assay was performed in triplicate.
CWDEs Activity Assays
Cell wall degrading enzymes from B. cinerea were extracted and assayed based on a previously described method (Berto et al., 2001). The WT and mutants were, respectively, cultured for 10 days in liquid pectin and cellulose media at 22°C. Subsequently, the cultured media were filtered and centrifuged (8500 r/min) to remove the fungal mycelia, and the CWDEs were extracted and purified from the filtrates. PMG, PG, and CX activities were analyzed by the DNS method. The Hoffman method was used to determine the activities of polygalacturonic acid transeliminase (PGTE) and pectin methyl transelimination enzyme (PMTE). All the experiments were repeated thrice.
Toxins Activity Assay
The WT and mutants were, respectively, inoculated into Fries III medium, and cultured for 21 days at 22°C. Then, the culture filtrates of each strain were extracted thrice with ethyl acetate, and evaporated under reduced pressure to remove the solvent. The final extracts of each strain were dissolved in chromatographic methanol and inoculated into punctured tobacco leaves. The inoculated leaves were moisturized in a cylinder culture dish at 22°C and observed daily. The lesion areas were measured at 4 dpi.
Acid Production Assays
The WT and mutants were, respectively, inoculated onto PDA media supplemented with 0.05% (w/v) bromothymol blue, a pH indicator for weak acids and bases, and incubated in complete darkness at 22°C for 7 days. A change in the color of the media to yellow suggested that the pH of the media declined and the strains secreted acids. For pH detection, the WT and mutants were, respectively, inoculated in 100 mL of PD media and cultured for 7 days in darkness at 22°C. All the experiments were performed at least thrice.
Assessment of Mutant Cell Wall Integrity
To assess the mutant cell wall integrity, the WT and mutants were inoculated onto PDA media supplemented with NaCl, KCl, fluconazole, Congo Red, menadione, and H2O2 at optimal concentrations of 0.8 mol/L, 0.8 mol/L, 10 μg/mL, 2 mg/mL, 200 μmol/L, and 30 mmol/L, respectively, and were incubated in complete darkness at 22°C. The growth and inhibition rates of the WT and mutants were determined.
H2O2 Measurements
All samples from WT, BCG183 and BCG183/BcKMO mutant were homogenized in liquid nitrogen. Using the H2O2 Quantitative Assay Kit (Sangon Co., Ltd., China), we followed the manufacturer’s instructions to measure the quantity of H2O2 production in WT, BCG183 and BCG183/BcKMO.
Sensitivity of Mutants to Inhibitors of cAMP and MAPK Signaling Pathways
SQ22536 and U0126, the specific inhibitors of cAMP and MAPK signaling pathways, respectively, were used to detect the sensitivity of the WT and mutants to the inhibitors of cAMP and MAPK signaling pathways. The WT, BCG183, and BCG183/BcKMO were inoculated onto PDA media supplemented with 10 μmol/L SQ22536 or 10 μmol/L U0126, respectively, and the growth and inhibition rates of the strains were determined.
Analysis of cAMP Content in the Mutants
The cAMP in the WT, BCG183 and BCG183/BcKMO was extracted by bath method and detected by using Agilent 1100 series high-performance liquid chromatography (HPLC) system (Agilent, United States). HPLC was performed with Kromasil C18 anti-phase chromatographic column (250 mm × 4.6 mm, 5 μm) under the following conditions: flow rate, 0.8 mL/min; V (methanol):V (KH2PO4), 80:20; testing wave, 254 nm; column temperature, 30°C; and sample injection volume, 10 μL.
Construction of RNAi Mutants of Key Genes Involved in the cAMP and MAPK Signaling Pathways
The explicit fragments of the key genes involved in the cAMP and MAPK signaling pathways, including pka1, pka2, pkaR, bcg2, bcg3, bmp1, and bmp3, were amplified with specific primers using WT cDNA as the template, and linked into pCR8 vector to obtain the entry vectors. The appropriate entry vectors of the key genes involved in cAMP and MAPK signaling pathways were obtained by colony PCR screening and sequencing. The RNAi vectors of pka1, pka2, pkaR, bcg2, bcg3, bmp1, and bmp3 were obtained by LR reaction between the entry vectors and destination vector pK7GW1WG2. Then, by using the Agrobacterium-mediated method, the RNAi vectors of the key genes involved in the cAMP and MAPK signaling pathways were transferred into the WT, respectively. The RNAi mutants of pka1, pka2, pkaR, bcg2, bcg3, bmp1, and bmp3 were obtained through PCR, and real-time PCR identification with primers specific for the kan gene revealed that the RNAi vectors were integrated. Furthermore, real-time PCR with primers specific for pka1, pka2, pkaR, bcg2, bcg3, bmp1, and bmp3 confirmed that the expression levels of these genes were affected.
Quantitative Real-Time PCR Analysis
Quantitative real-time PCR was used to measure the transcript levels of BcKMO and key genes of the cAMP and MAPK signaling pathways, such as the PKA catalytic subunit genes, pka1 and pka2 (Schumacher et al., 2008b); the PKA regulatory subunit gene, pkaR (Schumacher et al., 2008b); subunits of heterotrimeric GTP binding protein encoding genes, bcg2 and bcg3 (Gronover et al., 2001); and MAPK genes including, Bmp1 and Bmp3 (Segmuller et al., 2007; Heller et al., 2012), using ABI SYBR Green PCR Master Mix (Applied Biosystems, United States). The total RNA of each strain was isolated by Total RNA Preparation Kit (Omega), and digested with DNase I (Promega) to remove genomic DNAs. The cDNAs were synthesized with a Revert Aid First-strand cDNA Synthesis Kit (Invitrogen), according to the manufacturer’s instructions; the tubulin gene was used as an internal standard for equal loading.
Results
BcKMO Encodes a KMO
A pathogenic mutant, BCG183, was isolated by screening the T-DNA insertion library of B. cinerea. The mutant gene of BCG183 was identified as BC1G_07455 by TAIL-PCR and semi-quantitative RT-PCR (Supplementary Figure S1). Sequence searches in the databases revealed that BC1G_07455 encodes a hypothetical protein similar to KMO with a monooxygenase flavin-adenine dinucleotide (FAD) binding domain and four aromatic-ring hydroxylase-like motifs (Figure 1A), indicating that BcKMO might have similar functions to its ortholog in animals. A phylogenetic tree was constructed to update the new functional clades using MEGA software (Figure 1B). BcKMO displayed high homology with the Rossmann-fold NAD(P)(+)-binding proteins of S. sclerotiorum and ubiquinone biosynthesis hydroxylase family proteins of S. borealis, which contain a conserved FAD-binding domain.

FIGURE 1. Bioinformatics analysis of BcKMO. (A) Conserved domains of BcKMO. The domain of BcKMO was predicted using NCBI’s conserved domain database. (B) Phylogenetic analysis of BcKMO and homologous proteins from other fungi. The full-length protein sequences were analyzed by MEGA 7.0 with unrooted neighbor-joining bootstrap (1000 replicates).
The purified BcKMO protein was obtained using a prokaryotic expression technique (Supplementary Figure S2), and its KMO activity was measured using the Human KMO ELISA Kit. Based on the concentration and OD of the standard, the standard curve of KMO activity was derived and the linear regression equation of the standard curve was calculated as y = 0.0171x + 0.0623 (Supplementary Figure S2E). The activity of the purified BcKMO protein was 2.56 U/L. These results suggested that BcKMO encodes a KMO in B. cinerea.
BcKMO Is Important for Growth and Development of B. cinerea
To determine the role of BcKMO in the growth and development of B. cinerea, colony morphology, mycelium morphology, growth rate, and conidial yield of the WT, BCG183, and BCG183/BcKMO strains were investigated. The WT and BCG183/BcKMO colonies were taupe-colored and produced large amounts of sclerotia. In contrast, the BCG183 colonies were gray and did not produce sclerotia (Figure 2A). Under optical microscope, the BCG183 mycelia were white and slender with shorter transverse septa, when compared with the WT and BCG183/BcKMO mycelia (Figure 2B). Furthermore, the growth rate of the mutant BCG183 was significantly lower than that of the WT and BCG183/BcKMO (Figure 2C). Conidiation assays showed that the BCG183 mutant did not produce conidia, whereas the WT and BCG183/BcKMO strains did (Figure 2D).
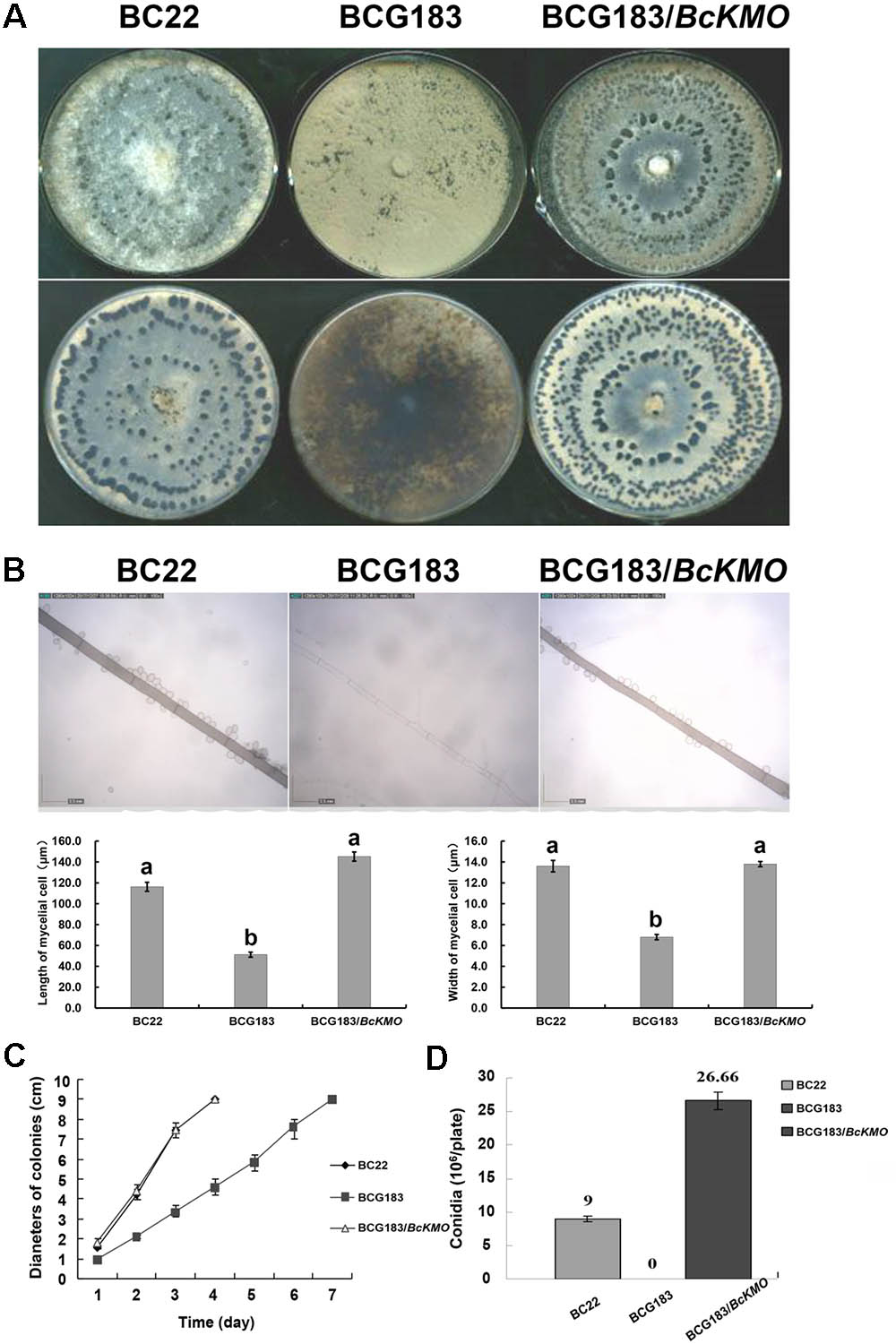
FIGURE 2. Phenotype analysis of BCG183 and BCG183/BcKMO mutants. (A) Colony morphology (10 dpi). (B) Mycelium morphology; CE: BcKMO-complementing BCG183/BcKMO mutant. Data in the table were averaged to evaluate statistical significances among the strains; a and b indicate significant differences at 0.05 probability level. (C) Detection of growth rates. (D) Determination of conidial yield.
BcKMO Is Vital for B. cinerea Pathogenicity
To determine the role of BcKMO in the pathogenicity of B. cinerea, culture blocks of the WT, BCG183, and BCG183/BcKMO strains were, respectively, inoculated onto detached bean and cucumber leaves, with blank PDA medium as the control. At 4 dpi, lesions appeared on the bean (Figure 3A) and cucumber (Figure 3B) leaves inoculated with the WT, BCG183, and BCG183/BcKMO strains, respectively, whereas the control did not exhibit any lesions. When compared with the WT and BCG183/BcKMO, the pathogenicity of BCG183 mutant on the beans (Figure 3A) and cucumber (Figure 3B) leaves dramatically increased. These results indicated that BcKMO is essential for the pathogenicity of B. cinerea.
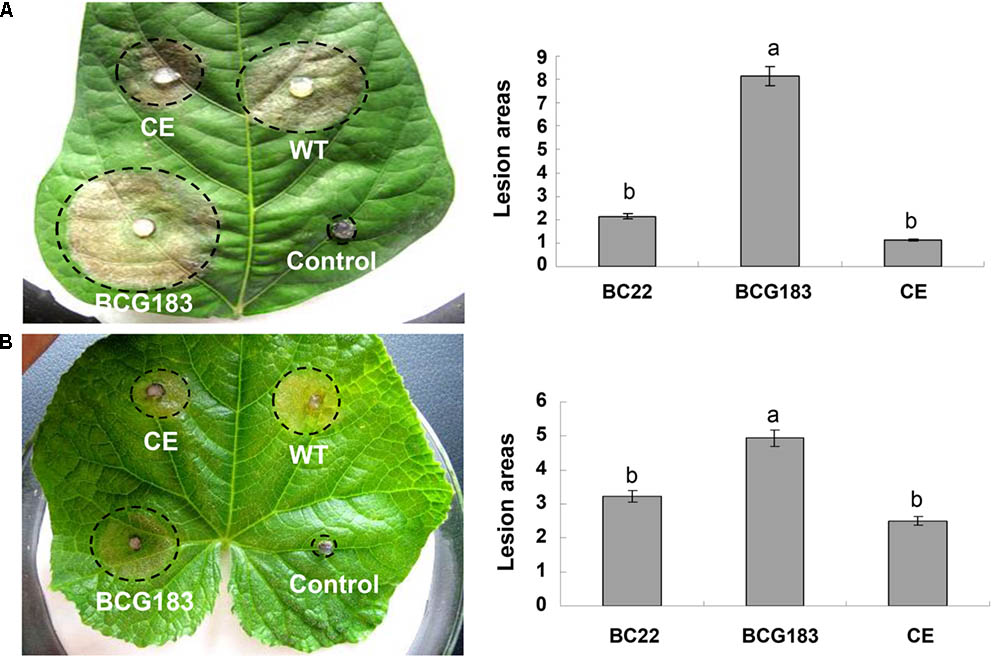
FIGURE 3. Pathogenicity analysis of BCG183 and BCG183/BcKMO mutants. (A) Pathogenicity analysis of mutants on kidney beans leaves. (B) Pathogenicity analysis of mutants on cucumber leaves. CE: BCG183/BcKMO mutant; a and b indicate significant differences at 0.05 probability level.
BcKMO Regulates the Activities of CWDEs, Toxins, and Acid Production
Cell wall degrading enzymes and toxins are important for the pathogenicity of B. cinerea. To investigate the mechanism underlying the increased pathogenicity of the BCG183 mutant, we examined the activities of several CWDEs and toxins in the WT, BCG183, and BCG183/BcKMO strains. When compared with the WT and BCG183/BcKMO, the mutant BCG183 exhibited significantly increased activities of PMG and PG, but showed no obvious difference in the activities of CX, PGTE, and PMTE (Figure 4A). Furthermore, at 4 dpi, lesions appeared on tobacco leaves inoculated with crude toxins from WT, BCG183, and BCG183/BcKMO, and the lesion areas of BCG183 mutant were significantly larger than those of the WT and BCG183/BcKMO (Figure 4B). These findings demonstrated that BcKMO is involved in the activities of CWDEs and toxins in B. cinerea.
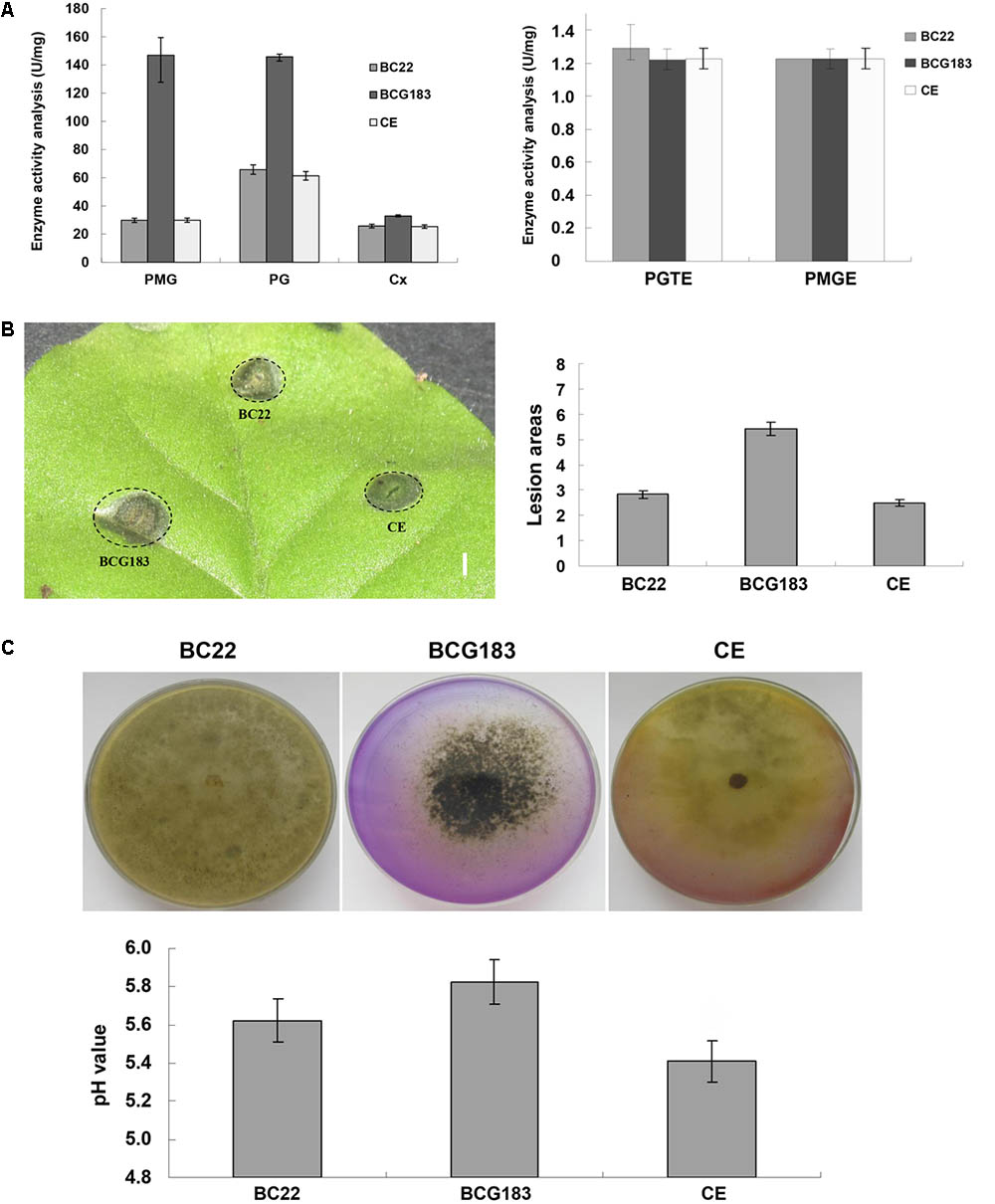
FIGURE 4. Activities of CWDEs and toxins of BCG183 and BCG183/BcKMO mutants. (A) CWDEs activity of BCG183 and BCG183/BcKMO mutants. (B) Toxins activity of BCG183 and BCG183/BcKMO mutants, reference bar stands for 1 cm. (C) Acid-producing ability of BCG183 and BCG183/BcKMO mutants. Acid production assays was performed on PDA medium with bromothymol blue. The pH of the PD medium inoculated with the WT, BCG183, and BCG183/BcKMO strains was measured.
Interactions between B. cinerea and plants produce acidic substances such as oxalic acid, salicylic acid, and jasmonic acid, which alter the signaling in host cells and induce host necrosis. In the present study, the PDA medium was supplemented with bromothymol blue to measure the acid production ability of the WT, BCG183, and BCG183/BcKMO strains. The color of the WT and BCG183/BcKMO media became yellow, but that of the BCG183 medium showed no obvious change (Figure 4C), suggesting that the pH of the WT and BCG183/BcKMO media declined. To confirm the changes in pH, the WT, BCG183, and BCG183/BcKMO strains were cultured in PD media supplemented with bromothymol blue. The medium pH of the WT, BCG183, and BCG183/BcKMO at 7 dpi was 5.62, 5.83, and 5.41, respectively (Figure 4C), indicating that the acid production of BCG183 was weaker than that of the WT and BCG183/BcKMO.
BcKMO Affects Hyphal Cell Walls
When the WT, BCG183, and BCG183/BcKMO strains were inoculated onto PDA medium with 0.8 mol/L NaCl or KCl (Figure 5A), the BCG183 mutant exhibited remarkably higher sensitivity to NaCl and KCl, when compared with the WT and BCG183/BcKMO strains, and grew slowly under identical cell wall stress conditions. These results indicated that BcKMO deficiency reduced the resistance of B. cinerea to osmotic stress, suggesting a positive regulatory role of this gene in osmotic stress resistance in B. cinerea. A similar trend was also noted with respect to the inhibition rate (Figure 5B).
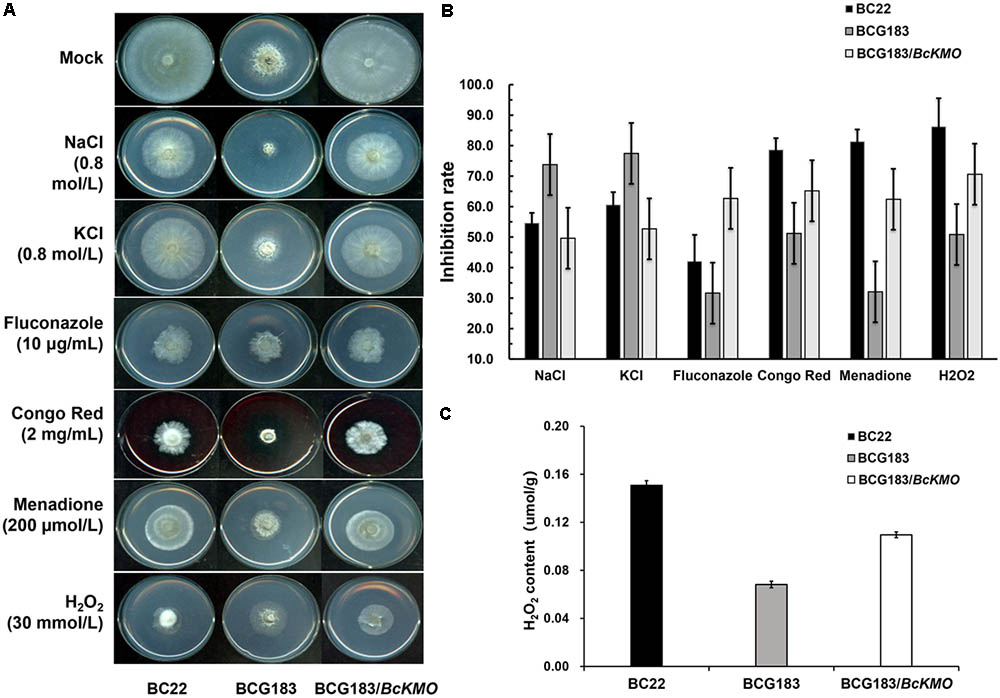
FIGURE 5. Sensitivity of BCG183 and BCG183/BcKMO mutants to cell wall stress. (A) The WT, BCG183, and BCG183/BcKMO strains were cultured on PDA plates with 0.8 mol/L NaCl, 0.8 mol/L KCl, 10 μg/mL fluconazole, 2 mg/mL Congo Red, 200 μmol/L menadione, and 30 mmol/L H2O2, respectively. (B) The inhibition rate of the three strains cultured on different media. Blank PDA was used as control. (C) H2O2 content of WT, BCG183, and BCG183/BcKMO strains.
When the strains were inoculated onto PDA medium with 10 μg/mL fluconazole, 2 mg/mL Congo Red, 200 μmol/L menadione, and 30 mmol/L H2O2, respectively (Figure 5A), the sensitivity of the mutant BCG183 to fluconazole, Congo Red, menadione, and H2O2 was significantly weaker, when compared with that of the WT and BCG183/BcKMO strains (Figure 5A). These findings revealed that the cellular integrity was enhanced in the mutant BCG183, suggesting that BcKMO regulates the cell wall integrity of B. cinerea. The inhibition rate also presented a similar trend (Figure 5B).
The quantitative assays of H2O2 were performed in WT, BCG183, and BCG183/BcKMO strains. We observed that H2O2 content was declined in the mutant BCG183, comparing with WT and BCG183/BcKMO strains (Figure 5C). This result suggested that BCG183 mutant may have stronger ability to clear hydrogen peroxide.
BcKMO Is Involved in cAMP and MAPK Signaling Pathways
To investigate the molecular mechanism of BcKMO in the regulation of the pathogenicity of B. cinerea, specific inhibitors of the cAMP and MAPK signaling pathways, SQ22536 and U0126, were used to detect the sensitivity of BCG183 and BCG183/BcKMO mutants to these inhibitors. The BCG183 mutant sensitivity to SQ22536 and U0126 was significantly weaker, when compared with that of the WT and BCG183/BcKMO strains (Figure 6A). Furthermore, the cAMP content in the mutant BCG183 was significantly lower, when compared with that in the WT and BCG183/BcKMO strains (Figure 6B). These results indicated that BcKMO is involved in cAMP and MAPK signaling pathways.
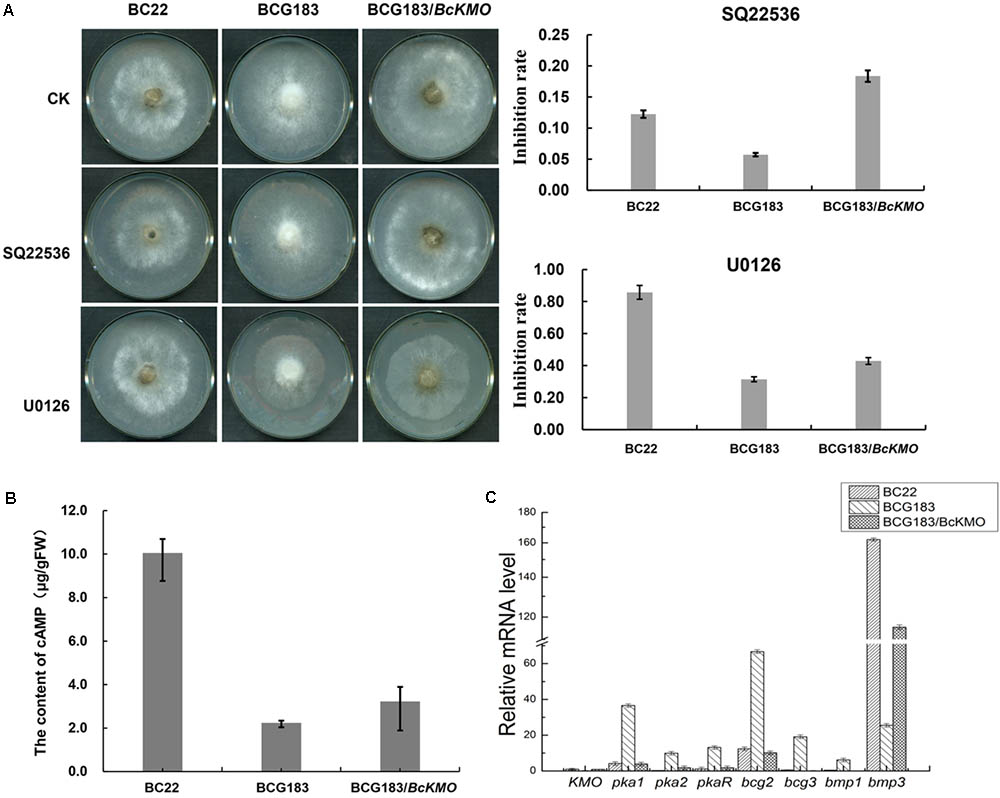
FIGURE 6. Role of BcKMO in the cAMP and MAPK signaling pathways. (A) Sensitivity of BCG183 and BCG183/BcKMO mutants to specific inhibitors of cAMP and MAPK signaling pathways. SQ22536, specific inhibitor of the cAMP signaling pathway; U0126, specific inhibitor of the MAPK signaling pathway. (B) Content of cAMP in BCG183 and BCG183/BcKMO mutants. (C) Expression levels of the key genes involved in the cAMP and MAPK signaling pathways in BCG183 and BCG183/BcKMO mutants.
Quantitative real-time PCR was used to examine the expression levels of the key genes involved in cAMP and MAPK signaling pathways in BCG183 and BCG183/BcKMO mutants. Pka1, Pka2, and PkaR in the cAMP-dependent pathway; subunits of heterotrimeric GTP-binding protein encoding genes, bcg2 and bcg3; and a key component gene in the MAPK signaling pathway, bmp1, were significantly upregulated in the mutant BCG183, whereas bmp3, another key gene in the MAPK signaling pathway was significantly downregulated in the BCG183 mutant (Figure 6C). These results demonstrated that BcKMO is involved in regulating the key genes involved in the cAMP and MAPK signaling pathways, namely, pka1, pka2, pkaR, bcg2, bcg3, bmp1, and bmp3.
Moreover, quantitative real-time PCR revealed that the expression levels of BcKMO in the RNAi mutants of pka1, bcg2, and bmp1 were significantly higher, whereas that in the RNAi mutants of pka2, pkaR, bcg3, and bmp3 were significantly lower, when compared with the BcKMO expression in the WT strain (Figure 7). These findings indicated that the key genes in the cAMP and MAPK signaling pathways, namely, pka1, pka2, pkaR, bcg2, bcg3, bmp1, and bmp3, regulate BcKMO gene expression.
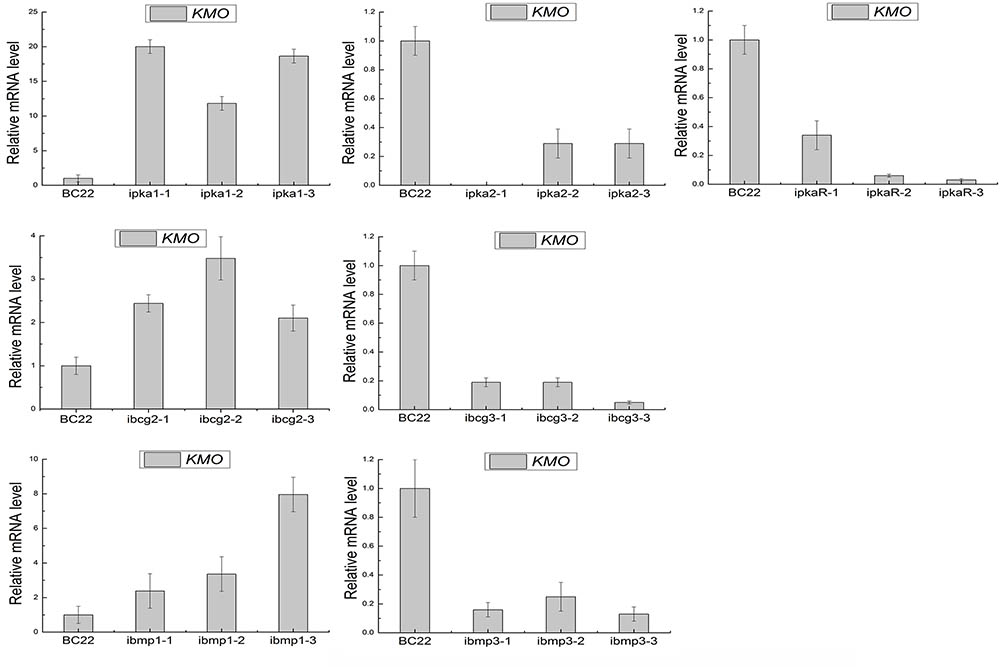
FIGURE 7. Expression levels of BcKMO in the RNAi mutants of the key genes involved in the cAMP and MAPK pathways.
Discussion
The identification and characterization of B. cinerea mutants have significantly contributed to the understanding of the regulatory mechanisms involved in fungal growth, development, and pathogenicity. In this study, a novel B. cinerea mutant BCG183, which grew slowly, did not produce conidia and sclerotia on PDA medium, and which presented enhanced pathogenicity, was described. Using TAIL-PCR, Southern blot, RT-PCR, and bioinformatics analysis, the mutant gene was determined to be BC1G_07455 (BcKMO), which encodes a hypothetical protein similar to KMO with a monooxygenase FAD-binding domain and four aromatic-ring hydroxylase-like motifs. The activity of the purified BcKMO protein was noted to be 2.56 U/L, suggesting that the BcKMO encoded a KMO and participated in kynurenine pathway in B. cinerea. Subsequently, complementation experiments were designed to investigate the role of BcKMO in the growth, development, and pathogenicity of B. cinerea. The BCG183 mutant did not produce conidia and sclerotia, and presented slow growth, slender hyphae, and strong pathogenicity. Moreover, the phenotype and pathogenicity of the WT stain and BcKMO-complementing mutant were similar. These results demonstrated that BcKMO positively regulates growth and development, and negatively regulates pathogenicity of B. cinerea. Meanwhile, these results suggested that the kynurenine pathway might be important to growth, development, and pathogenicity in B. cinerea.
Kynurenine 3-monooxygenase participates in tryptophan metabolism through the kynurenine pathway. KMO belongs to the family of oxidoreductases and catalyzes the hydroxylation of L-kynurenine through the insertion of molecular oxygen into the aromatic ring of kynurenine to produce 3-hydroxy-L-kynurenine (Amaral et al., 2013). Several genes of the kynurenine pathway were discovered in several bacterial including Bacillus cereus, Burkholderia fungorum, Ralstonia metallidurans, and Pseudomonas fluorescens (Kurnasov et al., 2003a,b). Kynurenine pathway of Pseudomonas aeruginosa, S. chrysomallus, and S. refuineus were reported to be involved in the biosynthesis of siderophore quinolobactin, actinomycin, and benzodiazepine, respectively (Matthijs et al., 2004; Hu et al., 2007; Keller et al., 2010). KMO in yeast was directly related to the pathophysiology of Huntington disease by a mechanism that may involve reactive oxygen species (ROS) (Giorgini et al., 2005). And ROS production also can mediate or activate MAPK pathways (McCubrey et al., 2006). In this study, we found that the mutant BCG183 sensitivity to menadione and H2O2 were remarkably weak compared to the WT and BCG183/BcKMO. In addition, H2O2 content was declined in the mutant BCG183, comparing with WT and BCG183/BcKMO strains. The results indicated that the BcKMO gene mutation enhanced the resistance to oxidative stress, suggesting that the BcKMO gene negatively regulates the resistance to oxidative stress, and then influences the MAPK signaling pathways in B. cinerea.
The conidia of B. cinerea play an important role in disease cycle and propagation. Therefore, it is generally believed that inhibition of formation and development of B. cinerea conidia could alleviate or effectively control the occurrence of gray mold. However, the relationship between the conidiation and pathogenicity of B. cinerea is not yet clear. In a previous study, a Δbcpka1 mutant was noted to sporulate in plants, and although its growth rate, conidiation, and conidial germination were not impaired, the mutant failed to produce soft rots of leaves (Schumacher et al., 2008b). Furthermore, deletion of genes encoding the MAPK, Bmp1 and BcSAK1, resulted in non-pathogenic mutants. It has been reported that the Δbmp1 mutant produced normal conidia and exhibited decreased growth rates (Zheng et al., 2000), whereas the Δbcsak1 mutant did not produce conidia and showed decreased growth rates (Gronover et al., 2001). Moreover, deletion of the MAP kinase gene, BMP3, has been found to result in reduced conidiation, growth rates, and virulence (Rui and Hahn, 2007). In the present study, the BCG183 mutant exhibited decreased growth rates and did not produce conidia, but presented obviously enhanced pathogenicity. These results suggest that conidiation and pathogenicity of B. cinerea are not necessarily correlated.
The major pathogenic mechanism of B. cinerea involves multiple CWDEs and pathogenic toxins (Juge, 2006; Choquer et al., 2007). Therefore, in the present study, the role of BcKMO in regulating CWDEs and pathogenic toxins activities was investigated. The activities of PMG, PG, and toxins in the BCG183 mutant were significantly higher, when compared with those in the WT and BCG183/BcKMO strains. In addition, inactivation of BcKMO upregulated the expression levels of Bcp1, god1, Bcpg1, and Sod1 (data not shown). These results suggest that BcKMO is involved in regulating CWDEs and toxins activities.
Oxalic acid is known to be an important factor in the pathogenesis of S. sclerotiorum (Walz et al., 2008), and favors plant cell wall degradation by decreasing the cellular pH close to the optimum for CWDE and removes Ca2+ ions bound to pectins. In addition, OA inhibits the plant oxidative burst and defense mechanisms and reduces plant programmed death (PCD), thereby increasing the pathogenicity of the pathogen. However, PCD has a clear role in promoting pathogen growth in some host–pathogen interactions (Greenberg and Yao, 2004). Studies on B. cinerea in tobacco and Arabidopsis suggested that the fungus may even need the hypersensitive reaction to achieve full pathogenicity (Govrin and Levine, 2000; Dickman et al., 2001). Indeed, Arabidopsis mutants with a delayed or reduced cell death response were generally more resistant to B. cinerea infection, whereas mutants in which cell death was accelerated were more susceptible (Van Baarlen et al., 2007). Depending on the timing, strength, and host, PCD has an important role in B. cinerea virulence. In the present study, the pathogenicity of the mutant on kidney beans and cucumber leaves was dramatically increased, whereas acid production of the BCG183 mutant significantly decreased, suggesting that the weak acid level in the mutant might have promoted oxidative burst and PCD resulting in increased pathogenicity. In contrast, the low pH in the WT and BCG183/BcKMO strains might have inhibited oxidative burst and PCD, thus limiting their pathogenicity.
Some signal transduction components of B. cinerea, especially those involved in the MAPK and cAMP-dependent pathways, are known to regulate the fungal development and pathogenicity. In addition, studies have demonstrated that ROS can induce or mediate the activation of the MAPK pathways (McCubrey et al., 2006). Therefore, in the present study, the mutant BCG183 sensitivity to menadione and H2O2 were remarkably weak compared to the WT and BCG183/BcKMO. The BcKMO-mediated pathway was further investigated by detecting the sensitivity of BCG183 and BCG183/BcKMO mutants to specific inhibitors of cAMP and MAPK signaling pathways, as well as by determining the cAMP content in these mutants. The sensitivity of the BCG183 mutant to signaling pathway specific inhibitors, SQ22536 and U0126, was remarkably weak (Figure 6A), and the content of cAMP was significantly reduced (Figure 6B). Thus, it can be concluded that BcKMO is involved in cAMP and MAPK signaling pathways. To further analyze the relationship between BcKMO and the key genes involved in the cAMP and MAPK signaling pathways, the expression levels of these genes in the BCG183 and BCG183/BcKMO mutants as well as the expression of BcKMO in the RNAi mutants of these genes were examined by quantitative real-time PCR. While pka1, pka2, pkaR, bcg2, bcg3, and bmp1 were significantly upregulated, bmp3 was significantly downregulated in the BCG183 mutant (Figure 6C), indicating that BcKMO negatively regulates pka1, pka2, pkaR, bcg2, bcg3, and bmp1 and positively regulates bmp3. The expression of BcKMO was obviously upregulated in the RNAi mutants of pka1, bcg2, and bmp1, but significantly downregulated in the RNAi mutants of pka2, pkaR, bcg3, and bmp3 (Figure 7), suggesting that BcKMO is negatively regulated by pka1, bcg2, and bmp1 and positively regulated by pka2, pkaR, bcg2, and bmp3. These findings provide a foundation for further examination of the molecular mechanisms of BcKMO in regulating the growth, development, and pathogenicity of B. cinerea.
Conclusion
The BcKMO gene, which encodes a KMO, was found to positively regulate growth and development, but to negatively control pathogenicity of B. cinerea. This gene was noted to be involved in the regulation of CWDEs activity, toxins activity, acid production, and cell wall integrity, and participate in cAMP and MAPK signaling pathways of B. cinerea.
Author Contributions
JX and DG conceived and designed the experiments. XY, JZ, and MW performed the experiments. KZ, XY, JZ, MW, FZ, PL, and HC analyzed the experimental data. KZ, XY, JH, and JX contributed reagents/materials/analysis tools. KZ, JX, and JD wrote the paper. All authors read and approved the final manuscript.
Funding
This study was supported by the Natural Science Foundation of Hebei Province (No. C2018204045), Higher Education Research Fund of Hebei (No. ZD2016001), and Scientific Research Foundation for Returned Chinese Scholars of Hebei (No. 0316012).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01039/full#supplementary-material
FIGURE S1 | Identification of T-DNA insertion site in the BCG183 mutant. (A) PCR identification of the BCG183mutant with the specific primers of hph gene. M: DNA marker; 1, 3: Mutant BCG183; 2: WT. A 800-bp fragment was amplified from the BCG183 mutant DNA, but not from the WT genomic DNA. (B) Southern hybridization analysis of T-DNA insertion in the BCG183 mutant. M: DNA marker; 1: Mutant BCG183. The specific fragment of hph gene was used as the DNA probe of Southern hybridization. A single band was detected by Southern blot of the BCG183 genomic DNA, suggesting a single-copy T-DNA insertion in the BCG183 genomic DNA. (C) Electrophoresis of TAIL-PCR products. M: DNA marker; 1, 2: BCG183 mutant. The flanking sequence of the T-DNA insertion site was amplified by TAIL-PCR from the BCG183 genomic DNA. (D) Illustration of the BC1G_07455 locus with the T-DNA insertion in the BCG183mutant. Alignment of the sequence of the TAIL-PCR product with Botrytis cinerea gene database revealed insertion in the 3′-end of the coding region of BC1G_07455. (E) Expression level of BC1G_07455 detected by semi-quantitative RT-PCR. The expression of BC1G_07455 in the BCG183 mutant was significantly lower than that in the WT strain.
FIGURE S2 | Identification of T-DNA insertion site in BCG183 mutant. (A) Construction of prokaryotic expression vector of BcKMO. M: DNA marker; 1: Identification of pGEX4T-1-BcKMO-GST by restriction enzyme digestion. (B) Relationship between different times of induction and expression levels of BcKMO-GST. M: Protein Marker; 1: Without IPTG; 2–6: Expression of BcKMO-GST with IPTG induction at 4, 8, 12, 16, and 20 h. (C) Purification of BcKMO-GST protein. M: Protein Marker; 1: Purified BcKMO-GST protein; 2: Supernatant before purification. (D) Western blot analysis of BcKMO-GST protein. M. Protein Marker; 1: Supernatant before purification; 2: Purified BcKMO-GST protein. (E) Standard curve of KMO activity measurement using the Human KMO ELISA Kit.
FIGURE S3 | Complementation of BcKMO. (A) Vector construction of pBARKS1-BcKMO-GFP. (B) Enzyme digestion identification of pBARKS1-BcKMO-eGFP; M: DNA marker. (C) PCR identification of the transformant (M: DNA marker; 1, 2: PCR amplification of the WT and transformant with bar-specific primers; 3, 4: PCR amplification of the WT and transformant with GFP-specific primers). (D) Southern blot identification of the transformant (M: DNA marker; 1: WT; 2, 3: transformant). (E) Real-time PCR identification of the transformant. BC22: WT; CE: BCG183/BcKMO mutant. Quantitative real-time PCR analysis of BcKMO expression was performed with BcKMO-specific primers. Transcript levels of BcKMO in the transformant were significantly higher than those in the WT strain.
TABLE S1 | Primers used in this study.
Footnotes
- ^http://www.broad.mit.edu/annotation/genome/botrytis_cinerea
- ^https://blast.ncbi.nlm.nih.gov/Blast.cgi
References
Amaral, M., Levy, C., Heyes, D. J., Lafite, P., Outeiro, T. F., Giorgini, F., et al. (2013). Structural basis of kynurenine 3-monooxygenase inhibition. Nature 496, 382–385. doi: 10.1038/nature12039
Berto, P., Jijakli, M. H., and Lepoivre, P. (2001). Possible role of colonization and cell wall-degrading enzymes in the differential ability of three Ulocladium atrum strains to control Botrytis cinerea on necrotic strawberry leaves. Phytopathology 91, 1030–1036. doi: 10.1094/PHYTO.2001.91.11.1030
Blanco-Ulate, B., Morales-Cruz, A., Amrine, K. C., Labavitch, J. M., Powell, A. L., and Cantu, D. (2014). Genome-wide transcriptional profiling of Botrytis cinerea genes targeting plant cell walls during infections of different hosts. Front. Plant Sci. 5:435. doi: 10.3389/fpls.2014.00435
Choquer, M., Becker, H. F., and Vidal-Cros, A. (2007). Identification of two group A chitinase genes in Botrytis cinerea which are differentially induced by exogenous chitin. Mycol. Res. 111(Pt 5), 615–625. doi: 10.1016/j.mycres.2007.03.011
Dickman, M. B., Park, Y. K., Oltersdorf, T., Li, W., Clemente, T., and French, R. (2001). Abrogation of disease development in plants expressing animal antiapoptotic genes. Proc. Natl. Acad. Sci. U.S.A. 98, 6957–6962. doi: 10.1073/pnas.091108998
Doehlemann, G., Berndt, P., and Hahn, M. (2006). Different signalling pathways involving a Galpha protein, cAMP and a MAP kinase control germination of Botrytis cinerea conidia. Mol. Microbiol. 59, 821–835. doi: 10.1111/j.1365-2958.2005.04991.x
Elad, Y., Williamson, B., Tudzynski, P., and Delen, N. (2007). “Botrytis spp. and diseases they cause in agricultural systems – An introduction,” in Botrytis: Biology, Pathology and Control, ed. Y. Elad (Dordrecht: Springer), 1–8. doi: 10.1007/978-1-4020-2626-3_1
Giorgini, F., Guidetti, P., Nguyen, Q. V., Bennett, S. C., and Muchowski, P. J. (2005). A genomic screen in yeast implicates kynurenine 3-monooxygenase as a therapeutic target for Huntington disease. Nat. Genet. 37, 526–531. doi: 10.1038/ng1542
Govrin, E. M., and Levine, A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10, 751–757. doi: 10.1016/S0960-9822(00)00560-1
Greenberg, J. T., and Yao, N. (2004). The role and regulation of programmed cell death in plant-pathogen interactions. Cell. Microbiol. 6, 201–211. doi: 10.1111/j.1462-5822.2004.00361.x
Gronover, C. S., Kasulke, D., Tudzynski, P., and Tudzynski, B. (2001). The role of G protein alpha subunits in the infection process of the gray mold fungus Botrytis cinerea. Mol. Plant Microbe Interact. 14, 1293–1302. doi: 10.1094/MPMI.2001.14.11.1293
Heller, J., Ruhnke, N., Espino, J. J., Massaroli, M., Collado, I. G., and Tudzynski, P. (2012). The mitogen-activated protein kinase BcSak1 of Botrytis cinerea is required for pathogenic development and has broad regulatory functions beyond stress response. Mol. Plant Microbe Interact. 25, 802–816. doi: 10.1094/MPMI-11-11-0299
Hu, Y., Phelan, V., Ntai, I., Farnet, C. M., Zazopoulos, E., and Bachmann, B. O. (2007). Benzodiazepine biosynthesis in Streptomyces refuineus. Chem. Biol. 14, 691–701. doi: 10.1016/j.chembiol.2007.05.009
Juge, N. (2006). Plant protein inhibitors of cell wall degrading enzymes. Trends Plant Sci. 11, 359–367. doi: 10.1016/j.tplants.2006.05.006
Keller, U., Lang, M., Crnovcic, I., Pfennig, F., and Schauwecker, F. (2010). The actinomycin biosynthetic gene cluster of Streptomyces chrysomallus: a genetic hall of mirrors for synthesis of a molecule with mirror symmetry. J. Bacteriol. 192, 2583–2595. doi: 10.1128/JB.01526-09
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kurnasov, O., Goral, V., Colabrov, K., Gerdes, S., Anantha, S., Osterman, A., et al. (2003a). NAD biosynthesis: identification of the tryptophan to quinolinate pathway in bacteria. Chem. Biol. 10, 1195–1204. doi: 10.1016/j.chembiol.2003.11.011
Kurnasov, O., Jablonski, L., Polanuyer, B., Dorrestein, P., Begley, T., and Osterman, A. (2003b). Aerobic tryptophan degradation pathway in bacteria: novel kynurenine formamidase. FEMS Microbiol. Lett. 227, 219–227. doi: 10.1016/S0378-1097(03)00684-0
Levis, C., Fortini, D., and Brygoo, Y. (1997). Transformation of Botrytis cinerea with the nitrate reductase gene (niaD) shows a high frequency of homologous recombination. Curr. Genet. 32, 157–162. doi: 10.1007/s002940050261
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Ress 45, D200–D203. doi: 10.1093/nar/gkw1129
Matthijs, S., Baysse, C., Koedam, N., Tehrani, K. A., Verheyden, L., Budzikiewicz, H., et al. (2004). The Pseudomonas siderophore quinolobactin is synthesized from xanthurenic acid, an intermediate of the kynurenine pathway. Mol. Microbiol. 52, 371–384. doi: 10.1111/j.1365-2958.2004.03999.x
McCubrey, J. A., Lahair, M. M., and Franklin, R. A. (2006). Reactive oxygen species-induced activation of the MAP kinase signaling pathways. Antioxid. Redox Signal. 8, 1775–1789. doi: 10.1089/ars.2006.8.1775
Mullins, E. D., Chen, X., Romaine, P., Raina, R., Geiser, D. M., and Kang, S. (2001). Agrobacterium-mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology 91, 173–180. doi: 10.1094/PHYTO.2001.91.2.173
Patel, R. M., van Kan, J. A., Bailey, A. M., and Foster, G. D. (2008). RNA-mediated gene silencing of superoxide dismutase (Bcsod1) in Botrytis cinerea. Phytopathology 98, 1334–1339. doi: 10.1094/PHYTO-98-12-1334
Pinedo, C., Wang, C.-M., Pradier, J.-M., Dalmais, B., Choquer, M., Le Pêcheur, P., et al. (2008). Sesquiterpene synthase from the botrydial biosynthetic gene cluster of the phytopathogen Botrytis cinerea. ACS Chem. Biol. 3, 791–801. doi: 10.1021/cb800225v
Rolke, Y., Liu, S., Quidde, T., Williamson, B., Schouten, A., Weltring, K.-M., et al. (2004). Functional analysis of H2O2-generating systems in Botrytis cinerea: the major Cu-Zn-superoxide dismutase (BCSOD1) contributes to virulence on French bean, whereas a glucose oxidase (BCGOD1) is dispensable. Mol. Plant Pathol. 5, 17–27. doi: 10.1111/j.1364-3703.2004.00201.x
Rui, O., and Hahn, M. (2007). The Slt2-type MAP kinase Bmp3 of Botrytis cinerea is required for normal saprotrophic growth, conidiation, plant surface sensing and host tissue colonization. Mol. Plant Pathol. 8, 173–184. doi: 10.1111/j.1364-3703.2007.00383.x
Schamber, A., Leroch, M., Diwo, J., Mendgen, K., and Hahn, M. (2010). The role of mitogen-activated protein (MAP) kinase signalling components and the Ste12 transcription factor in germination and pathogenicity of Botrytis cinerea. Mol. Plant Pathol. 11, 105–119. doi: 10.1111/j.1364-3703.2009.00579.x
Schumacher, J., de Larrinoa, I. F., and Tudzynski, B. (2008a). Calcineurin-responsive zinc finger transcription factor CRZ1 of Botrytis cinerea is required for growth, development, and full virulence on bean plants. Eukaryot. Cell 7, 584–601. doi: 10.1128/EC.00426-07
Schumacher, J., Kokkelink, L., Huesmann, C., Jimenez-Teja, D., Collado, I. G., Barakat, R., et al. (2008b). The cAMP-dependent signaling pathway and its role in conidial germination, growth, and virulence of the gray mold Botrytis cinerea. Mol. Plant Microbe Interact. 21, 1443–1459. doi: 10.1094/MPMI-21-11-1443
Segmuller, N., Ellendorf, U., Tudzynski, B., and Tudzynski, P. (2007). BcSAK1, a stress-activated mitogen-activated protein kinase, is involved in vegetative differentiation and pathogenicity in Botrytis cinerea. Eukaryot. Cell 6, 211–221. doi: 10.1128/EC.00153-06
Smith, J. R., Jamie, J. F., and Guillemin, G. J. (2016). Kynurenine-3-monooxygenase: a review of structure, mechanism, and inhibitors. Drug Discov. Today 21, 315–324. doi: 10.1016/j.drudis.2015.11.001
ten Have, A., Mulder, W., Visser, J., and van Kan, J. A. (1998). The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol. Plant Microbe Interact 11, 1009–1016. doi: 10.1094/MPMI.1998.11.10.1009
Valette-Collet, O., Cimerman, A., Reignault, P., Levis, C., and Boccara, M. (2003). Disruption of Botrytis cinerea pectin methylesterase gene Bcpme1 reduces virulence on several host plants. Mol. Plant Microbe Interact. 16, 360–367. doi: 10.1094/MPMI.2003.16.4.360
Van Baarlen, P., Woltering, E. J., Staats, M., and Van Kan, J. A. (2007). Histochemical and genetic analysis of host and non-host interactions of Arabidopsis with three Botrytis species: an important role for cell death control. Mol. Plant Pathol. 8, 41–54. doi: 10.1111/j.1364-3703.2006.00367.x
Van Kan, J. A. (2006). Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 11, 247–253. doi: 10.1016/j.tplants.2006.03.005
Walz, A., Zingen-Sell, I., Theisen, S., and Kortekamp, A. (2008). Reactive oxygen intermediates and oxalic acid in the pathogenesis of the necrotrophic fungus Sclerotinia sclerotiorum. Eur. J. Plant Pathol. 120, 317–330. doi: 10.1007/s10658-007-9218-5
Wang, C.-M., Hopson, R., Lin, X., and Cane, D. E. (2009). Biosynthesis of the sesquiterpene botrydial in Botrytis cinerea. Mechanism and stereochemistry of the enzymatic formation of presilphiperfolan-8β-ol. J. Am. Chem. Soc. 131, 8360–8361. doi: 10.1021/ja9021649
Williamson, B., Tudzynski, B., Tudzynski, P., and Van Kan, J. A. (2007). Botrytis cinerea: the cause of grey mould disease. Mol. Plant Pathol. 8, 561–580. doi: 10.1111/j.1364-3703.2007.00417.x
Wilson, K., Mole, D. J., Binnie, M., Homer, N. Z., Zheng, X., Yard, B. A., et al. (2014). Bacterial expression of human kynurenine 3-monooxygenase: solubility, activity, purification. Protein Expr. Purif. 95, 96–103. doi: 10.1016/j.pep.2013.11.015
Yan, L., Yang, Q., Sundin, G. W., Li, H., and Ma, Z. (2010). The mitogen-activated protein kinase kinase BOS5 is involved in regulating vegetative differentiation and virulence in Botrytis cinerea. Fungal Genet. Biol. 47, 753–760. doi: 10.1016/j.fgb.2010.06.002
Keywords: Botrytis cinerea, BcKMO, kynurenine 3-monooxygenase, growth, development, pathogenicity
Citation: Zhang K, Yuan X, Zang J, Wang M, Zhao F, Li P, Cao H, Han J, Xing J and Dong J (2018) The Kynurenine 3-Monooxygenase Encoding Gene, BcKMO, Is Involved in the Growth, Development, and Pathogenicity of Botrytis cinerea. Front. Microbiol. 9:1039. doi: 10.3389/fmicb.2018.01039
Received: 07 February 2018; Accepted: 02 May 2018;
Published: 18 May 2018.
Edited by:
Hector Mora Montes, Universidad de Guanajuato, MexicoReviewed by:
Chenggang Wang, University of Florida, United StatesHumberto Prieto, Instituto de Investigaciones Agropecuarias (INIA), Chile
Copyright © 2018 Zhang, Yuan, Zang, Wang, Zhao, Li, Cao, Han, Xing and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jihong Xing, eGluZ2ppaG9uZzIwMDBAMTI2LmNvbQ== Jingao Dong, ZG9uZ2ppbmdhb0AxMjYuY29t
†These authors have contributed equally to this work.
 Kang Zhang
Kang Zhang Xuemei Yuan1,2†
Xuemei Yuan1,2†