
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 17 May 2018
Sec. Evolutionary and Genomic Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01017
This article is part of the Research Topic Non-clinical Environments as Evolutionary Cradles of Bacterial Virulence View all 6 articles
Most opportunistic pathogens transit in the environment between hosts and the environment plays a significant role in the evolution of protective traits. The coincidental evolution hypothesis suggests that virulence factors arose as a response to other selective pressures rather for virulence per se. This idea is strongly supported by the elucidation of bacterial-protozoal interactions. In response to protozoan predation, bacteria have evolved various defensive mechanisms which may also function as virulence factors. In this review, we summarize the dual role of factors involved in both grazing resistance and human pathogenesis, and compare the traits using model intracellular and extracellular pathogens. Intracellular pathogens rely on active invasion, blocking of the phagosome and lysosome fusion and resistance to phagocytic digestion to successfully invade host cells. In contrast, extracellular pathogens utilize toxin secretion and biofilm formation to avoid internalization by phagocytes. The complexity and diversity of bacterial virulence factors whose evolution is driven by protozoan predation, highlights the importance of protozoa in evolution of opportunistic pathogens.
Many bacterial pathogens are able to survive in non-clinical environments, where environmental conditions play an important role in the persistence and infectivity of those bacteria. In fact, most opportunistic pathogens are not transmitted host to host but rather transit in the environment between hosts for significant amounts of time. Thus, the environment impacts not only their survival but also potential infectivity. For example, global warming has been suggested to be responsible for increased outbreaks of disease caused by Vibrio spp., as rising sea surface temperatures promote their growth and virulence in aquatic environments (Vezzulli et al., 2013, 2015). The environmental persistence of bacterial pathogens also depends on their ability to adapt to various stresses. Predation by bacterivorous protists is one of the key mortality factors faced by bacteria in the environment. In response, bacteria have evolved many anti-protozoal mechanisms, such as formation of filaments, increases in swimming speed, surface masking, toxin production and biofilm formation, etc. (Matz and Kjelleberg, 2005). Some mechanisms providing resistance to protozoan grazing may also provide advantages during infection of human and animal hosts. As proposed in the review by Erken et al. (2013), protozoan grazing is “a factor driving the evolution of human pathogens in the environment.” This review summarizes the mechanisms of bacterial pathogens involved in both resistance to predation and human pathogenesis, with an emphasis on advances made in the past 5 years. We compare these mechanisms using examples of intracellular and extracellular pathogens.
Protists, or protozoa, are single-celled eukaryotes that range in size from 2 to 2000 μm. Protozoa are grouped based on morphology and mechanisms of feeding and locomotion into three groups, flagellates, ciliates, and amoebae. Heterotrophic flagellates and ciliates feed by sweeping food particles into a mouth-like cytostome, while amoebae engulf food particles using pseudopodia. After ingestion, food vacuoles (phagosomes) are trafficked through the phagocytic pathway and subsequently fuse with lysosomes (phagolysosome) enabling digestion of food particles (Fenchel, 1987).
Predation by bacterivorous protozoa is a well-known mechanism for top down control of bacterial communities due to their high feeding rates (Sherr and Sherr, 2002) and their ability to predate on surface-associated bacterial biofilms (Parry, 2004). However, there are also studies demonstrating that many bacteria, including pathogenic species, benefit from interactions with protozoa. Protozoa have been called the “Trojan horses of the microbial world” for their ability to promote the survival of pathogenic bacteria in the environment (Barker and Brown, 1994). For example, pathogenic bacteria that have been shown to have increased survival in the environment after interactions with protozoa include Campylobacter jejuni (Trigui et al., 2016; Reyes-Batlle et al., 2017), Francisella tularensis (Buse et al., 2017), Listeria monocytogenes (Fieseler et al., 2014), Legionella pneumophila (Cervero-Arago et al., 2014, 2015), Mycobacterium leprae (Wheat et al., 2014), Stenotrophomonas maltophilia (Cateau et al., 2014), and Yersinia enterocolitica (Lambrecht et al., 2013). Although the mechanisms of increased bacterial survival are not fully understood, it has been suggested that bacteria can obtain nutrients from protozoa. Furthermore, protozoa are not only ubiquitous in the environment, but are also members of gut microbiota in many organisms. The interactions between protozoa and other gut microorganisms can be beneficial or harmful to host health (Burgess et al., 2017; Chabe et al., 2017).
In addition, some protozoal species are able to form dormant cysts that are resistant to a variety of stresses, and these cysts provide encased bacteria a protective niche against adverse conditions. For example, Escherichia coli, L. monocytogenes, Salmonella enterica, and Y. enterocolitica demonstrated increased resistance to antibiotics and low pH when in Acanthamoeba castellanii cysts (Lambrecht et al., 2015). L. pneumophila within cysts of Acanthamoeba polyphaga was also reported to be resistant to chlorine treatment (Kilvington and Price, 1990). Therefore, protozoa and cysts generated by protozoa can be vectors for pathogenic bacteria and facilitate disease transmission (Denoncourt et al., 2014; Balczun and Scheid, 2017).
In some cases, interactions of bacteria with protozoa induce the expression of virulence traits, and thus, environmental amoebae have been referred to as “training grounds” for bacteria virulence (Molmeret et al., 2005). This is due to the fact that the bactericidal mechanisms used by amoeba and human immune cells, such as macrophage, are conserved. This includes the cell biology of phagocytosis and phagosome maturation. The mechanisms used by macrophage and amoeba to kill engulfed bacterial cells are similar, including H+–ATPase related acidification of the phagosome, oxidative burst from reactive oxygen and nitrogen species, use of metal transporters for iron and manganese efflux and copper and zinc influx, nutrient deprivation and the battery of antimicrobial proteins and lysosomal hydrolases expressed (Siddiqui and Khan, 2012; German et al., 2013). Many bacteria have evolved anti-digestion mechanisms that allow them to survive inside of protozoa, and some of these mechanisms also contribute to their virulence during infection of human and animal hosts (Gong et al., 2016; Paquet and Charette, 2016). Consequently, intracellular pathogens that inhabit phagosomal compartments interfere with their maturation and/or are resistant to host killing mechanisms. Thus, bacteria that have evolved to escape the bactericidal mechanisms of amoeba will be better protected (or more virulent) when encountering immune cells. Protozoan grazing has been shown to shape phenotypic and genotypic composition of bacteria community (Jurgens and Matz, 2002). Although there is limited experimental data showing that long-term protozoan grazing results in genotypic changes, it has been reported that protozoa can induce the expression of bacterial virulence in the short term, probably due mainly to changes in gene expression. For example, Mycobacterium ulcerans when co-cultured with A. polyphaga infected the footpads of mice much faster than the M. ulcerans only controls (Azumah et al., 2017). S. enterica serotype Typhimurium was shown to enter a hyperinvasive state after passage through A. castellanii (Carlson et al., 2007), and intracellular growth in A. castellanii also induced the invasion and virulence of L. pneumophila in mouse infection models (Cirillo et al., 1999).
The micronutrients, Fe, Mn, Zn, and Cu play important roles in the innate immune system and in antimicrobial activity of macrophages in defense against invading microbial pathogens (Hood and Skaar, 2012; Samanovic et al., 2012). Both protozoa and macrophage use the toxicity of Cu and Zn to kill bacteria in mature phagosomes. Some bacteria are able to avoid damage induced by these metals by the use of efflux pumps. Thus, copper resistance increases not only the environmental survival of bacterial pathogens, but also contributes to virulence in vivo (German et al., 2013). E. coli strains with mutations in Cu(I)-translocating P1B-type ATPase (copA), iron uptake transporters (feoAB and entC) and manganese uptake transporters (mntH), as well as Pseudomonas aeruginosa mutants in Cu(I)-translocating P1B-type ATPase (cueA) showed reduced grazing resistance against Dictyostelium discoideum (Hao et al., 2016). Efflux systems are also involved in antibiotic resistance. For example, the C. jejuni RND-type efflux pump, CmeABC which is associated with multidrug resistance, may also be involved in virulence as well as survival in A. polyphaga (Vieira et al., 2017). These data highlight the role of these “virulence factors” in persistence and survival in the environment and likely evolved to protect bacteria from predation rather than for protection against antibiotics, given the long history of co-evolution of protozoa and bacteria.
Horizontal gene transfer (HGT) contributes to bacterial adaptation and evolution. For bacterial pathogens, HGT is involved in resistance to antibiotics and virulence during infection (Juhas, 2015) and protozoan grazing has been shown to play an important role in HGT. Due to the indiscriminate feeding of many protozoa, it is likely that there will be a mix of different bacteria encased in a single food vacuole. This would facilitate the interactions of these bacteria in an enclosed system. For example, E. coli strains engulfed by the ciliate, Tetrahymena pyriformis, exhibited increased rates of conjugation (Schlimme et al., 1997). After a full digestion cycle, the frequency of conjugation increased 2000- to 4000-fold. Tetrahymena thermophila has also been reported to cause an increase in HGT by accumulating phage and susceptible bacteria in the phagocytic vacuoles (Aijaz and Koudelka, 2017). Prophages have been shown to be induced after ingestion due to exposure of the bacterium to oxidative stress in the phagosome, resulting in a switch to the lytic cycle and release of free phage particles. The frequency of lysogen formation was shown to increase sixfold as a consequence of being encased within the phagosome. In both cases, the accumulation of bacteria, phage and DNA in protozoan food vacuoles is proposed to be responsible for the increase in HGT. Protozoan grazing also stimulates biofilm formation by grazing resistant bacteria (Matz et al., 2004, 2005) and it has been proposed that the high cell density in bacterial biofilms is also responsible for increased HGT (Madsen et al., 2012).
L. pneumophila is the causative agent of Legionnaires’ disease and is one of the best-studied intracellular pathogens. In the environment, L. pneumophila interacts with diverse protozoan hosts, which is critical for its persistence as this organism is not transmitted from person to person (Boamah et al., 2017). This further highlights that environmental protozoa are the true hosts for this pathogen. The L. pneumophila life cycle within amoebae and macrophage has been described in detail in numerous reviews (Escoll et al., 2013; Richards et al., 2013; Hoffmann et al., 2014). Briefly, L. pneumophila (1) invades the host cell after phagocytosis, forming Legionella-containing vacuoles (LCVs), (2) uses the Dot/Icm type IV secretion system (T4SS) to inject approximately 300 effector proteins into the host cell, (3) differentiates into the replicative form and proliferates when nutrients are present, (4) when nutrients become limiting, L. pneumophila differentiates into the mature infectious form and enters the cytosol, (5) cells are finally released as free swimming cells after host cell lysis (Figure 1). Recently, it has been proposed that the L. pneumophila life cycle contains even more developmental stages and cell forms (Robertson et al., 2014).
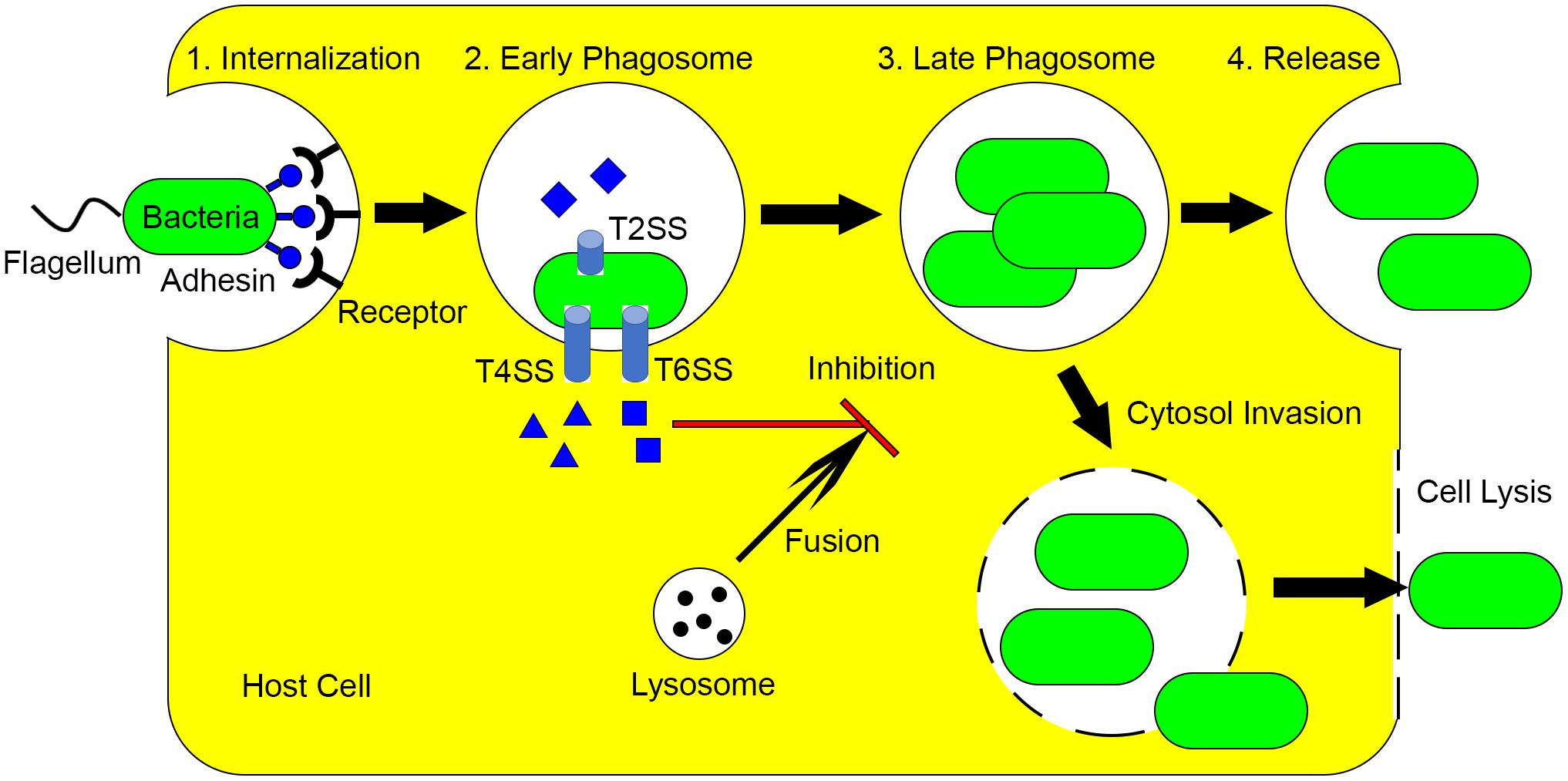
FIGURE 1. Mechanisms utilized by intracellular pathogens to avoid killing by macrophage and protozoa. (1) Intracellular pathogens utilize flagella and adhesins to invade host cells. (2) After internalization, various effectors are produced through multiple secretion systems, which inhibits the lysosome phagosome fusion. (3) Depending on nutrient conditions, bacteria replicate in phagosome and/or enter cytosol. (4) The intracellular pathogens are released by exocytosis or lysis of host cells.
Host cell invasion by L. pneumophila is mainly mediated by surface factors like the flagellum and pilus. L. pneumophila has a single monopolar flagellum composed of one major subunit, FlaA. A mutation of FlaA reduces the efficiency of invasion of A. castellanii and macrophage due to reduced motility (Dietrich et al., 2001; Schell et al., 2016a). In addition, PilY1 is responsible for the biogenesis of the type IV pili, which is important for twitching motility, surface attachment and host cell invasion (Hoppe et al., 2017). PilY1 is also involved in inhibition of fusion of the lysosome with the phagosome in the social amoebae D. discoideum and a mutant of PilY1 demonstrated reduced intracellular growth in macrophage (Shevchuk et al., 2014). The mechanisms facilitating host cell invasion are common in other intracellular pathogens. For example, a pathogenicity island in Mycobacterium avium encodes proteins inducing actin polymerization of host cells and mutants of this pathogenicity island are deficient in invasion of both macrophage and A. castellanii (Danelishvili et al., 2007).
After internalization, the intracellular survival of L. pneumophila largely depends on effectors secreted by the T4SS that have similar functions in amoebae and macrophage, probably due to the high similarity of mechanisms of phagocytic trafficking expressed by these two host cells. Most of the effectors work in a redundant manner to block phagosome-lysosome fusion and promote L. pneumophila intracellular survival. This has been summarized in earlier reviews (Finsel and Hilbi, 2015; So et al., 2015). Recently, one of the effectors, a cell membrane-localized iron transporter, IroT, was reported to be involved in ferrous iron acquisition and the mutant is defective in intracellular growth in A. castellanii and macrophage (Portier et al., 2015). Iron-dependent virulence is widely distributed among bacterial pathogens (Reinhart and Oglesby-Sherrouse, 2016; Butt and Thomas, 2017; Ramakrishnan, 2017). Although there are limited evidences showing that iron-regulated virulence factors contribute to grazing resistance, it has been reported that E. coli strains encoding genes for iron uptake survived better with D. discoideum than avirulent strains lacking these genes (Adiba et al., 2010). A recent report demonstrates that E. coli strains with mutations in iron uptake genes are attenuated for intracellular survival in D. discoideum (Hao et al., 2016). Another L. pneumophila T4SS effector, RidL, binds with the retromer (protein complex responsible for recycling transmembrane receptors from endosomes to the trans-Golgi network) of host cells and inhibits retrograde trafficking, resulting in better intracellular growth in both D. discoideum and macrophage (Finsel et al., 2013). In contrast, the Shiga toxin produced by Shigella dysenteriae, the cholera toxin produced by Vibrio cholerae and the exotoxin produced by P. aeruginosa rely on functional retrograde trafficking to enter the eukaryotic cell cytosol (Johannes and Popoff, 2008), indicating that the intracellular and extracellular pathogens have different strategies for interactions with host cells. Another effector, LegG1, activates the host cell GTPase, Ran on the LCV membrane, causing polymerization of microtubules, allowing the LCV to move along the microtubules in host cells. A mutant of LegG1 exhibited compromised intracellular growth (Rothmeier et al., 2013; Hilbi et al., 2014). Interestingly, although the L. pneumophila infection generally inhibits chemotaxis of amoebae and macrophage, this inhibition is relieved by LegG1 expression. This may help host cells acquire nutrients from the environment, which then benefits L. pneumophila allowing for intracellular multiplication (Simon et al., 2014). Furthermore, many T4SS effectors encoded by the L. pneumophila genome are eukaryotic-like proteins which are believed to have been acquired during residence in protozoa, and these eukaryotic-like proteins are used as virulence factors to modulate protist host cell functions (Lurie-Weinberger et al., 2010; Gomez-Valero et al., 2011, 2014). Most of these proteins encode motifs that are involved in protein–protein interactions and mimic host proteins, thereby allowing control of host machinery and intracellular replication. These motifs include F-box, U-box, ankyrin and serine threonine protein kinase motifs.
In addition to the T4SS, the type II secretion system (T2SS) is also important for intracellular survival of L. pneumophila in protozoa and macrophage (Rossier et al., 2004) and the more than 25 exoproteins secreted by the T2SS are effective in a host specific manner (Cianciotto, 2009). For example, a T2SS secreted chitinase is responsible for persistence during lung infection in a mouse model of infection, but is not involved in survival in the amoeba, Hartmannella vermiformis or human macrophage-like cells (DebRoy et al., 2006). A mutation in a novel T2SS-dependant exoprotein, NttA, reduces intracellular survival in A. castellanii, but not other host cells, while the RNase, SrnA, acyltransferase, PlaC and metalloprotease, ProA promote multiplication of L. pneumophila in H. vermiformis and Naegleria lovaniensis only (Tyson et al., 2013).
L. pneumophila has a quorum sensing (QS) system consisting of an autoinducer synthase, LqsA, two sensor kinases, LqsS and LqsT, and a response regulator, LqsR, that controls virulence, motility, filament production, natural competence and the switch between the replicative and infectious forms (Kessler et al., 2013; Schell et al., 2014, 2016b; Personnic et al., 2017). A mutant of LqsR is defective for intracellular growth in A. castellanii, D. discoideum and macrophage (Tiaden et al., 2007). Interestingly, the autoinducer produced by LqsA, as well as its homologous cholera autoinducer-1 (CAI-1), inhibits the cell migration of D. discoideum and macrophage in a T4SS-independent manner, suggesting that these autoinducers play a role in interkingdom signaling (Simon et al., 2015).
Mycobacterium spp., including the causal agent of tuberculosis, Mycobacterium tuberculosis, uses similar strategies to infect and replicate in bacteria-containing vacuoles, such as inhibition of phagosome maturation (Awuh and Flo, 2017). It has been reported that Mycobacterium marinum can inhibit phagosome-lysosome fusion by inducing host WASH complex dependent actin polymerization in D. discoideum and macrophage, although the bacterial effector is still unknown (Kolonko et al., 2014). In S. enterica serovar Typhimurium, virulence factors [i.e., waaL responsible for ligation of O-antigen to LPS, invA and ssaD essential structural components of type 3 secretion systems (T3SS), clpV chaperone essential for protein secretion by type VI secretion systems (T6SS), and PhoP/PhoQ two-component system] were shown to be involved in intracellular survival in D. discoideum (Riquelme et al., 2016). Interestingly, F. tularensis, the causal agent of tularemia, exhibits different survival mechanisms in macrophage and amoebae (Ozanic et al., 2015). In macrophages, F. tularensis escapes from acidified phagosomes to the cytosol and then proliferates, while in H. vermiformis, it replicates in non-acidified phagosomes (Santic et al., 2011). The Francisella pathogenicity island (FPI) encoding the T6SS is critical for survival in both hosts. One of the FPI proteins, IglC, a homolog of the T6SS Hcp protein (de Bruin et al., 2011), is responsible for escape from phagosome to cytosol (Santic et al., 2005) and invasion of non-phagocytic epithelial cells (Law et al., 2014). Mutants of IglC and the regulator MglA are defective for intracellular survival in both macrophage and A. castellanii (Lauriano et al., 2004).
Taken together, the intracellular pathogens possess many dual role factors that mediate their interactions with both protozoa and human phagocytes. Most of the similarities are found between amoebae and macrophage due to the conservative phagocytic process. Intracellular pathogens use surface factors to facilitate invasion and then rely on various effectors delivered by secretion systems to inhibit phagosome maturation, where many effectors are under precise regulation. These dual role mechanisms more likely evolved during the long-term interactions with protozoa in the environment, rather than during infections of human and animals. The temporal and spatial distribution of opportunistic pathogens in the environment between hosts, provides greater chances for them to face grazing pressure by bacterivorous protozoa. As a result, humans and animals become the accidental victims of coincidental evolution and further support the coincidental evolution hypothesis.
V. cholerae is the causal agent of cholera and is generally recognized as a model for non-invasive diarrheal disease (Sack et al., 2004). V. cholerae uses surface masking, toxin secretion and biofilm formation to defend against protozoan grazing and these mechanisms are also important for colonization and infection of humans (Figure 2) (Lutz et al., 2013). More than 200 serogroups classified by the lipopolysaccharide (LPS) O-antigen of V. cholerae exist, but only O1 and O139 have caused pandemics to date (Chatterjee and Chaudhuri, 2003). The V. cholerae LPS, or endotoxin, is an important virulence factor that mediates intestinal attachment and modulates host immunological responses (Chatterjee and Chaudhuri, 2006). The lipid-A of LPS has been proposed to inhibit phagocytosis by T. pyriformis (Kovacs et al., 1986). Similarly, production of LPS, capsular polysaccharide and outer membrane proteins protects Klebsiella pneumoniae from phagocytosis by D. discoideum and macrophage. Mutant strains of K. pneumoniae defective in the LPS core, lipid A, palmitoylation, OmpA and OmpK36 are susceptible to phagocytosis (March et al., 2013).
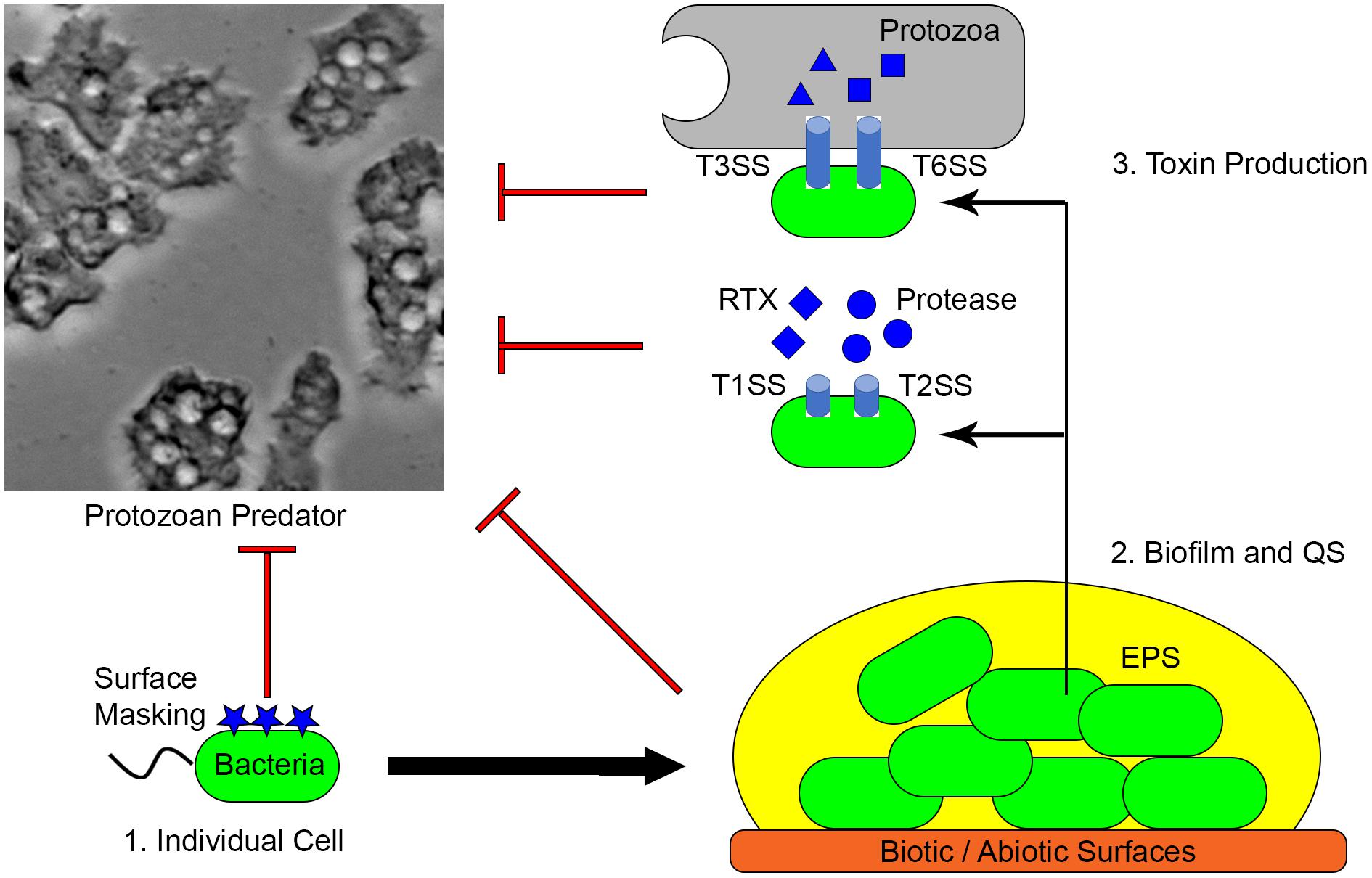
FIGURE 2. Mechanisms utilized by extracellular pathogens. (1) Surface masking facilitates bacterial cells avoiding phagocytosis. (2) Extracellular pathogens form biofilms on various surfaces. EPS production and QS-regulated activities protect bacteria from predation. (3) Toxins produced by the biofilm community are important for both grazing resistance and animal infection.
V. cholerae possesses multiple secretion systems that are important for pathogenesis and ecological fitness. The type I secretion system (T1SS) is responsible for delivery of RtxA, a multifunctional autoprocessing repeats-in-toxin (MARTX) toxin, which is involved in the colonization of the small intestine (Olivier et al., 2007) and destruction of host cell actin cytoskeleton (Prochazkova et al., 2009). MARTX is also secreted by the T1SS of Vibrio vulnificus and has cytopathogenic activity toward host cells, leading to rounding, cell death and interference of internalization by host phagocytes (Lee et al., 2007; Kim et al., 2008). V. vulnificus RtxA1 can cause damage in a variety of ways, including generation of reactive oxygen species (ROS) (Chung et al., 2010), cytoskeletal rearrangement (Kim et al., 2008), apoptotic cell death (Lee et al., 2008), and interference with the cytosolic Ca2+ flux to inhibit phagocytosis (Kuo et al., 2015). V. vulnificus produces at least four different types of MARTX (types I–IV), where the plasmid-encoded MARTX type III (RtxA13) is structurally and evolutionarily different to MARTX types I and II (Kwak et al., 2011; Roig et al., 2011). MARTX type III of V. vulnificus biotype 2 is involved in the lysis of a wide range of eukaryotic cells, including amoebae, erythrocytes, epithelial cells, and phagocytes. For example, the amoebae Neoparamoeba pemaquidensis isolated and purified from turbot (Scophthalmus maximus) gills, grew significantly less in the presence of the wild-type strain compared to the rtxA13 mutant (Lee et al., 2013).
The T2SS of V. cholerae is involved in the secretion of cholera toxin (Sandkvist et al., 1997). A recent study has reported that V. cholerae protease, PrtV, is transported across the outer membrane by T2SS and then packed into outer membrane vesicles (Rompikuntal et al., 2015). V. cholerae PrtV, which causes instant cytotoxic effects during infection in human intestinal cell lines, is also toxic to predators such as the ciliate T. pyriformis and the flagellate Cafeteria roenbergensis (Vaitkevicius et al., 2006).
Although there are limited studies of T3SS-dependent virulence factors in V. cholerae, including VopE (Suzuki et al., 2014), VopF (Tam et al., 2007), and VopX (Alam et al., 2011), and the role of V. cholerae T3SS in grazing resistance has not been reported, T3SS has been shown to contribute to anti-protozoal activities in E. coli (Siddiqui et al., 2011), P. aeruginosa (Matz et al., 2008) and Vibrio parahaemolyticus (Matz et al., 2011). Interestingly, pathogenic E. coli strains (EHEC or STEC) encode a second T3SS, which may not be a functional secretion system, but is still important for virulence during infection and may be associated with Shiga toxin (Zhou et al., 2014). Shiga toxin is the major virulence factor in EHEC strains, but its role in grazing resistance is under debate. It has been reported that Shiga toxins of E. coli O157:H7 provides a survival advantage in T. pyriformis food vacuoles (Steinberg and Levin, 2007) and is cytotoxic to A. castellanii (Arnold and Koudelka, 2014) and T. thermophila (Stolfa and Koudelka, 2012). In contrast, there are also studies showing that Shiga toxin is not important in association of EHEC strains with A. castellanii (Chekabab et al., 2012) and may even decrease survival in the presence of A. castellanii (Chekabab et al., 2013). Virulence of EHEC strains depends on the induction of Shiga toxin-converting prophage. Bacteriophage-mediated lysis of Shiga toxin-encoding bacteria is necessary for toxicity in protozoa whereas digestion of bacteria by T. thermophila is harmless to the cell. Shiga toxin-converting prophage can be induced via treatment with H2O2. EHEC strains with mutations in bacterial SOS response genes exhibit increased susceptibility to grazing by T. thermophila (Lainhart et al., 2009; Stolfa and Koudelka, 2012). Oxidative stress as a consequence of H2O2 excretion by either neutrophils during infection or protist predators could potentially induce the production of Shiga toxin-converting prophage (Licznerska et al., 2016). A phage encoded toxin is also important for the anti-protozoal activity of Corynebacterium diphtheriae, as the diphtheria toxin kills A. castellanii during co-culturing (Arnold and Koudelka, 2014).
The T6SS of V. cholerae injects effector proteins into target cells by the action of a structure analogous to an intracellular membrane-attached contractile phage tail sheath (Basler et al., 2012; Ho et al., 2014). In V. cholerae non-O1/non-O139 strains, the T6SS has been shown to have toxic effects toward D. discoideum as well as mammalian macrophages (Pukatzki et al., 2006, 2007; Miyata et al., 2011). In addition to the direct role of T6SS in eukaryotic host pathogenesis by the injection of toxins responsible for actin crosslinking, the T6SS also has an indirect role in competition with neighboring bacteria (Dong et al., 2013; Ho et al., 2014).
Biofilm formation is important for both pathogenesis and environmental survival. Colonization of intestinal epithelial cells allows V. cholerae to deliver virulence factors effectively and provides resistance against various stresses, such as bile acids and antimicrobial peptides (Silva and Benitez, 2016). Biofilm formation also protects V. cholerae against predation while planktonic cells are eliminated. In addition, predation by protozoa promotes V. cholerae biofilm formation and induces a smooth to rugose morphotypic shift due to increased extracellular polysaccharide (EPS) production (Matz et al., 2005). EPS as the major component of the biofilm matrix, shields V. cholerae from predation by the surface feeding nanoflagellate, Rhynchomonas nasuta and A. castellanii (Sun et al., 2013). High cell densities within biofilms also allow accumulation of bioactive compounds. For example, production of pyomelanin plays a role in virulence factor expression and colonization by V. cholerae (Valeru et al., 2009). Pyomelanin production in V. cholerae biofilm also leads to increased ROS levels, which promotes resistance to predation by A. castellanii (Noorian et al., 2017).
The high cell density of V. cholerae biofilms enables QS, which regulates a number of virulence factors including motility, protease secretion, toxin production via the master regulator ToxR and biofilm formation (Zhu et al., 2002). Both PrtV and the T6SS are QS-regulated. A mutant in the V. cholerae QS master response regulator, hapR, resulted in reduced grazing resistance in comparison to the wild-type when exposed to R. nasuta (Matz et al., 2005). When biofilms of mixed V. cholerae wild-type and QS-deficient strains were exposed to predation, the QS mutant was selectively removed, indicating that QS also regulates V. cholerae surface components recognized by protozoa (Sun et al., 2013).
Similar to V. cholerae, biofilm formation increases P. aeruginosa resistance to host defenses and chemotherapy (Mulcahy et al., 2014). In addition, EPS and QS-regulated inhibitors are also responsible for grazing resistance of P. aeruginosa biofilms (Matz et al., 2004; Weitere et al., 2005). A newly identified P. aeruginosa modulator, GTPase TypA, is responsible for surface attachment and biofilm formation, resistance to antibiotics, and T3SS-dependent virulence. A mutant of TypA showed reduced resistance to D. discoideum grazing and increased uptake by macrophage (Neidig et al., 2013).
In contrast to intracellular pathogens, extracellular pathogens use surface factors to avoid internalization and produce various toxins to inhibit protozoan predators. Biofilm formation provides physical protection and accumulates anti-protozoal compounds. These dual role factors also enable extracellular pathogens to colonize epithelial surfaces and deliver toxins to host cells in an effective manner. Notably, the host cell receptors and targets of many dual role toxins are not always conserved between protozoa and human phagocytes, but nonetheless, are effective in both instances. Thus, toxin production can lead to enhanced virulence during infections.
The coincidental evolution hypothesis suggests that virulence factors arose as a response to other selective pressures, such as predation, rather than for virulence per se (Levin, 1996; Adiba et al., 2010; Brown et al., 2012; Erken et al., 2013). There are many examples of virulence factors or their homologs that exist in environmental microorganisms that potentially play another functional role in the environment, including but not limited to grazing resistance. In the case of V. cholerae, it has been demonstrated that the T6SS of V. cholerae is functionally activated under high osmolarity and low temperature conditions, suggesting that the system may be important for the survival of the bacterium in the environment (Ishikawa et al., 2012). Furthermore, the cold shock gene, cspV, which controls biofilm formation through modulation of the second messenger cyclic-di-GMP, also regulates T6SS-mediated interspecies killing in a temperature-dependent manner (Townsley et al., 2016). A mutant in cspV showed significant defects for attachment and T6SS-mediated killing on the surface of the aquatic crustacean Daphnia magna (Townsley et al., 2016). In addition, chitin colonization is important for the long-term environmental persistence of V. cholerae (Pruzzo et al., 2008), where V. cholerae colonization factor GbpA (Kirn et al., 2005; Wong et al., 2012) and toxin co-regulated pilus (Reguera and Kolter, 2005) are involved in attachment to both chitin and epithelial surfaces.
The evolution of virulence driven by protozoan grazing has also been reported for fungi and viruses. Cryptococcus neoformans is a yeast that can cause lung infections in immunocompromised people and invades the brain by using host monocytes as “Trojan horses” (Charlier et al., 2009). Its ability to survive intracellularly has been proposed to have evolved for protection against amoeba predation (Steenbergen et al., 2001; Casadevall, 2012). Although the mechanisms of association are still unknown, A. castellanii has been reported to be the carrier or vectors for at least 2 human viruses, adenovirus (Scheid and Schwarzenberger, 2012) and coxsackie B3 virus (Mattana et al., 2006). For coxsackie B3 virus, an enrichment on A. castellanii surfaces was observed and the virus was eventually located inside the trophozoite (Mattana et al., 2006).
In some cases, grazing resistance mechanisms benefit the bacterium in association with a broad range of hosts. The methionine sulfoxide reductase can repair ROS-damaged proteins and are important for many bacterial pathogens (Sasindran et al., 2007). Mutation of methionine sulfoxide reductases in Aeromonas hydrophila reduces resistance to predation by T. thermophila and infection of zebrafish (Pang et al., 2016). Hemagglutinin protease which destroys host cell receptors for several different V. cholerae adhesins, is also involved in the degradation of chironomid egg masses (Halpern et al., 2003). Furthermore, QS-regulated production of hemagglutinin protease is important for defense against phages within high cell density bacterial populations (Hoque et al., 2016).
The bacterial–protozoal interaction has been suggested to be one of the oldest interactions between prokaryotic and eukaryotic organisms (Cavalier-Smith, 2002). During their long history of interaction, many virulence factors of bacterial pathogens evolved as adaptations to grazing pressure, rather than for pathogenesis to human and animal hosts. For example, a genome analysis of a Chlamydia-related amoebal symbiont indicates that many traits for intracellular survival, including the T3SS, existed in the bacterial genome 700 million years ago, 500 million years earlier than the appearance of Mammalia (Horn et al., 2004).
Our understanding of the pathogenesis of intracellular and extracellular pathogens is distorted due to our tendency to focus on the pathogen in the human host rather than in their natural environmental niche. The functions of virulence genes or their homologs in the environment have for the most part, not been fully explored. Here, we summarized the bacterial mechanisms providing grazing resistance and virulence in model intracellular and extracellular pathogens. For intracellular pathogens, active invasion, blocking of the phagosome lysosome fusion and resistance to phagocytic digestion are critical traits for infecting host cells. Extracellular pathogens, in contrast, rely on toxin secretion and biofilm formation as important mechanisms of protection against predation and pathogenesis. Some bacteria possess traits common in both intracellular and extracellular pathogens. For example, L. pneumophila can develop biofilms on environmental surfaces (Abdel-Nour et al., 2013) and V. cholerae has been found to proliferate inside of amoebae (Abd et al., 2005; Van der Henst et al., 2015). Since bacterial surface components are involved in the initial contact with host cells, possession of factors mediating active invasion may be used as a key feature to distinguish intracellular and extracellular pathogens. In conclusion, evolution of mechanisms that allow for survival within protozoa may have selected for traits that also allow bacteria to escape that harmful effects of phagocytes.
SS, PN, and DM contributed to the writing of the first draft of the manuscript. SS, PN, and DM contributed to manuscript revision, read and approved the submitted version.
This work was supported by The Australian Research Council Discovery Project grants DP170100453 and DP1096481, the Singapore Centre for Environmental Life Sciences Engineering, Nanyang Technological University, and the ithree Institute, University of Technology Sydney.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abd, H., Weintraub, A., and Sandstrom, G. (2005). Intracellular survival and replication of Vibrio cholerae O139 in aquatic free-living amoebae. Environ. Microbiol. 7, 1003–1008. doi: 10.1111/j.1462-2920.2005.00771.x
Abdel-Nour, M., Duncan, C., Low, D. E., and Guyard, C. (2013). Biofilms: the stronghold of Legionella pneumophila. Int. J. Mol. Sci. 14, 21660–21675. doi: 10.3390/ijms141121660
Adiba, S., Nizak, C., van Baalen, M., Denamur, E., and Depaulis, F. (2010). From grazing resistance to pathogenesis: the coincidental evolution of virulence factors. PLoS One 5:e11882. doi: 10.1371/journal.pone.0011882
Aijaz, I., and Koudelka, G. B. (2017). Tetrahymena phagocytic vesicles as ecological micro-niches of phage transfer. FEMS Microbiol. Ecol. 93:fix030. doi: 10.1093/femsec/fix030
Alam, A., Miller, K. A., Chaand, M., Butler, J. S., and Dziejman, M. (2011). Identification of Vibrio cholerae type III secretion system effector proteins. Infect. Immun. 79, 1728–1740. doi: 10.1128/IAI.01194-10
Arnold, J. W., and Koudelka, G. B. (2014). The Trojan Horse of the microbiological arms race: phage-encoded toxins as a defence against eukaryotic predators. Environ. Microbiol. 16, 454–466. doi: 10.1111/1462-2920.12232
Awuh, J. A., and Flo, T. H. (2017). Molecular basis of mycobacterial survival in macrophages. Cell. Mol. Life Sci. 74, 1625–1648. doi: 10.1007/s00018-016-2422-8
Azumah, B. K., Addo, P. G., Dodoo, A., Awandare, G., Mosi, L., Boakye, D. A., et al. (2017). Experimental demonstration of the possible role of Acanthamoeba polyphaga in the infection and disease progression in Buruli Ulcer (BU) using ICR mice. PLoS One 12:e0172843. doi: 10.1371/journal.pone.0172843
Balczun, C., and Scheid, P. L. (2017). Free-living amoebae as hosts for and vectors of intracellular microorganisms with public health significance. Viruses 9:E65. doi: 10.3390/v9040065
Barker, J., and Brown, M. R. (1994). Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology 140(Pt 6), 1253–1259. doi: 10.1099/00221287-140-6-1253
Basler, M., Pilhofer, M., Henderson, G. P., Jensen, G. J., and Mekalanos, J. J. (2012). Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186. doi: 10.1038/nature10846
Boamah, D. K., Zhou, G., Ensminger, A. W., and O’Connor, T. J. (2017). From many hosts, one accidental pathogen: the diverse protozoan hosts of Legionella. Front. Cell. Infect. Microbiol. 7:477. doi: 10.3389/fcimb.2017.00477
Brown, S. P., Cornforth, D. M., and Mideo, N. (2012). Evolution of virulence in opportunistic pathogens: generalism, plasticity, and control. Trends Microbiol. 20, 336–342. doi: 10.1016/j.tim.2012.04.005
Burgess, S. L., Gilchrist, C. A., Lynn, T. C., and Petri, W. A. (2017). Parasitic protozoa and interactions with the host intestinal microbiota. Infect. Immun. 85:e00101-17. doi: 10.1128/IAI.00101-17
Buse, H. Y., Schaefer, F. W. III, and Rice, E. W. (2017). Enhanced survival but not amplification of Francisella spp. in the presence of free-living amoebae. Acta Microbiol. Immunol. Hung. 64, 17–36. doi: 10.1556/030.63.2016.015
Butt, A. T., and Thomas, M. S. (2017). Iron acquisition mechanisms and their role in the virulence of Burkholderia species. Front. Cell. Infect. Microbiol. 7:460. doi: 10.3389/fcimb.2017.00460
Carlson, S. A., Sharma, V. K., McCuddin, Z. P., Rasmussen, M. A., and Franklin, S. K. (2007). Involvement of a Salmonella genomic island 1 gene in the rumen protozoan-mediated enhancement of invasion for multiple-antibiotic-resistant Salmonella enterica serovar Typhimurium. Infect. Immun. 75, 792–800. doi: 10.1128/IAI.00679-06
Casadevall, A. (2012). Amoeba provide insight into the origin of virulence in pathogenic fungi. Adv. Exp. Med. Biol. 710, 1–10. doi: 10.1007/978-1-4419-5638-5_1
Cateau, E., Maisonneuve, E., Peguilhan, S., Quellard, N., Hechard, Y., and Rodier, M. H. (2014). Stenotrophomonas maltophilia and Vermamoeba vermiformis relationships: bacterial multiplication and protection in amoebal-derived structures. Res. Microbiol. 165, 847–851. doi: 10.1016/j.resmic.2014.10.004
Cavalier-Smith, T. (2002). The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 52, 297–354. doi: 10.1099/00207713-52-2-297
Cervero-Arago, S., Rodriguez-Martinez, S., Puertas-Bennasar, A., and Araujo, R. M. (2015). Effect of common drinking water disinfectants, chlorine and heat, on free Legionella and amoebae-associated Legionella. PLoS One 10:e0134726. doi: 10.1371/journal.pone.0134726
Cervero-Arago, S., Sommer, R., and Araujo, R. M. (2014). Effect of UV irradiation (253.7 nm) on free Legionella and Legionella associated with its amoebae hosts. Water Res. 67, 299–309. doi: 10.1016/j.watres.2014.09.023
Chabe, M., Lokmer, A., and Segurel, L. (2017). Gut protozoa: friends or foes of the human gut microbiota? Trends Parasitol. 33, 925–934. doi: 10.1016/j.pt.2017.08.005
Charlier, C., Nielsen, K., Daou, S., Brigitte, M., Chretien, F., and Dromer, F. (2009). Evidence of a role for monocytes in dissemination and brain invasion by Cryptococcus neoformans. Infect. Immun. 77, 120–127. doi: 10.1128/IAI.01065-08
Chatterjee, S. N., and Chaudhuri, K. (2003). Lipopolysaccharides of Vibrio cholerae. I. Physical and chemical characterization. Biochim. Biophys. Acta 1639, 65–79. doi: 10.1016/j.bbadis.2003.08.004
Chatterjee, S. N., and Chaudhuri, K. (2006). Lipopolysaccharides of Vibrio cholerae: III. Biological functions. Biochim. Biophys. Acta 1762, 1–16. doi: 10.1016/j.bbadis.2005.08.005
Chekabab, S. M., Daigle, F., Charette, S. J., Dozois, C. M., and Harel, J. (2012). Survival of enterohemorrhagic Escherichia coli in the presence of Acanthamoeba castellanii and its dependence on Pho regulon. Microbiologyopen 1, 427–437. doi: 10.1002/mbo3.40
Chekabab, S. M., Daigle, F., Charette, S. J., Dozois, C. M., and Harel, J. (2013). Shiga toxins decrease enterohaemorrhagic Escherichia coli survival within Acanthamoeba castellanii. FEMS Microbiol. Lett. 344, 86–93. doi: 10.1111/1574-6968.12158
Chung, K.-J., Cho, E.-J., Kim, M. K., Kim, Y. R., Kim, S.-H., Yang, H.-Y., et al. (2010). RtxA1-induced expression of the small GTPase Rac2 plays a key role in the pathogenicity of Vibrio vulnificus. J. Infect. Dis. 201, 97–105. doi: 10.1086/648612
Cianciotto, N. P. (2009). Many substrates and functions of type II secretion: lessons learned from Legionella pneumophila. Future Microbiol. 4, 797–805. doi: 10.2217/fmb.09.53
Cirillo, J. D., Cirillo, S. L., Yan, L., Bermudez, L. E., Falkow, S., and Tompkins, L. S. (1999). Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67, 4427–4434.
Danelishvili, L., Wu, M., Stang, B., Harriff, M., Cirillo, S. L., Cirillo, J. D., et al. (2007). Identification of Mycobacterium avium pathogenicity island important for macrophage and amoeba infection. Proc. Natl. Acad. Sci. U.S.A. 104, 11038–11043. doi: 10.1073/pnas.0610746104
de Bruin, O. M., Duplantis, B. N., Ludu, J. S., Hare, R. F., Nix, E. B., Schmerk, C. L., et al. (2011). The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB and DotU suggest roles in type VI secretion. Microbiology 157, 3483–3491. doi: 10.1099/mic.0.052308-0
DebRoy, S., Dao, J., Soderberg, M., Rossier, O., and Cianciotto, N. P. (2006). Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc. Natl. Acad. Sci. U.S.A. 103, 19146–19151. doi: 10.1073/pnas.0608279103
Denoncourt, A. M., Paquet, V. E., and Charette, S. J. (2014). Potential role of bacteria packaging by protozoa in the persistence and transmission of pathogenic bacteria. Front. Microbiol. 5:240. doi: 10.3389/fmicb.2014.00240
Dietrich, C., Heuner, K., Brand, B. C., Hacker, J., and Steinert, M. (2001). Flagellum of Legionella pneumophila positively affects the early phase of infection of eukaryotic host cells. Infect. Immun. 69, 2116–2122. doi: 10.1128/IAI.69.4.2116-2122.2001
Dong, T. G., Ho, B. T., Yoder-Himes, D. R., and Mekalanos, J. J. (2013). Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 110, 2623–2628. doi: 10.1073/pnas.1222783110
Erken, M., Lutz, C., and McDougald, D. (2013). The rise of pathogens: Predation as a factor driving the evolution of human pathogens in the environment. Microb. Ecol. 65, 860–868. doi: 10.1007/s00248-013-0189-0
Escoll, P., Rolando, M., Gomez-Valero, L., and Buchrieser, C. (2013). From amoeba to macrophages: exploring the molecular mechanisms of Legionella pneumophila infection in both hosts. Curr. Top. Microbiol. Immunol. 376, 1–34. doi: 10.1007/82_2013_351
Fenchel, T. (1987). Ecology of Protozoa: The Biology of Free-Living Phagotropic Protists. Berlin: Springer.
Fieseler, L., Doyscher, D., Loessner, M. J., and Schuppler, M. (2014). Acanthamoeba release compounds which promote growth of Listeria monocytogenes and other bacteria. Appl. Microbiol. Biotechnol. 98, 3091–3097. doi: 10.1007/s00253-014-5534-9
Finsel, I., and Hilbi, H. (2015). Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell. Microbiol. 17, 935–950. doi: 10.1111/cmi.12450
Finsel, I., Ragaz, C., Hoffmann, C., Harrison, C. F., Weber, S., van Rahden, V. A., et al. (2013). The Legionella effector RidL inhibits retrograde trafficking to promote intracellular replication. Cell Host Microbe 14, 38–50. doi: 10.1016/j.chom.2013.06.001
German, N., Doyscher, D., and Rensing, C. (2013). Bacterial killing in macrophages and amoeba: do they all use a brass dagger? Future Microbiol. 8, 1257–1264. doi: 10.2217/fmb.13.100
Gomez-Valero, L., Rusniok, C., Cazalet, C., and Buchrieser, C. (2011). Comparative and functional genomics of Legionella identified eukaryotic like proteins as key players in host-pathogen interactions. Front. Microbiol. 2:208. doi: 10.3389/fmicb.2011.00208
Gomez-Valero, L., Rusniok, C., Rolando, M., Neou, M., Dervins-Ravault, D., Demirtas, J., et al. (2014). Comparative analyses of Legionella species identifies genetic features of strains causing Legionnaires’ disease. Genome Biol. 15:505.
Gong, J., Qing, Y., Zou, S., Fu, R., Su, L., Zhang, X., et al. (2016). Protist-bacteria associations: Gammaproteobacteria and Alphaproteobacteria are prevalent as digestion-resistant bacteria in ciliated protozoa. Front. Microbiol. 7:498. doi: 10.3389/fmicb.2016.00498
Halpern, M., Gancz, H., Broza, M., and Kashi, Y. (2003). Vibrio cholerae hemagglutinin/protease degrades chironomid egg masses. Appl. Environ. Microbiol. 69, 4200–4204. doi: 10.1128/AEM.69.7.4200-4204.2003
Hao, X., Luthje, F., Ronn, R., German, N. A., Li, X., Huang, F., et al. (2016). A role for copper in protozoan grazing - two billion years selecting for bacterial copper resistance. Mol. Microbiol. 102, 628–641. doi: 10.1111/mmi.13483
Hilbi, H., Rothmeier, E., Hoffmann, C., and Harrison, C. F. (2014). Beyond Rab GTPases Legionella activates the small GTPase Ran to promote microtubule polymerization, pathogen vacuole motility, and infection. Small GTPases 5, 1–6. doi: 10.4161/21541248.2014.972859
Ho, B. T., Dong, T. G., and Mekalanos, J. J. (2014). A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15, 9–21. doi: 10.1016/j.chom.2013.11.008
Hoffmann, C., Harrison, C. F., and Hilbi, H. (2014). The natural alternative: protozoa as cellular models for Legionella infection. Cell. Microbiol. 16, 15–26. doi: 10.1111/cmi.12235
Hood, M. I., and Skaar, E. P. (2012). Nutritional immunity: transition metals at the pathogen–host interface. Nat. Rev. Microbiol. 10, 525–537. doi: 10.1038/nrmicro2836
Hoppe, J., Unal, C. M., Thiem, S., Grimpe, L., Goldmann, T., Gassler, N., et al. (2017). Pily1 promotes Legionella pneumophila infection of human lung tissue explants and contributes to bacterial adhesion, host cell invasion, and twitching motility. Front. Cell. Infect. Microbiol. 7:63. doi: 10.3389/fcimb.2017.00063
Hoque, M. M., Naser, I. B., Bari, S. M. N., Zhu, J., Mekalanos, J. J., and Faruque, S. M. (2016). Quorum regulated resistance of Vibrio cholerae against environmental bacteriophages. Sci. Rep. 6:37956. doi: 10.1038/srep37956
Horn, M., Collingro, A., Schmitz-Esser, S., Beier, C. L., Purkhold, U., Fartmann, B., et al. (2004). Illuminating the evolutionary history of chlamydiae. Science 304, 728–730. doi: 10.1126/science.1096330
Ishikawa, T., Sabharwal, D., Bröms, J., Milton, D. L., Sjöstedt, A., Uhlin, B. E., et al. (2012). Pathoadaptive conditional regulation of the Type VI secretion system in Vibrio cholerae O1 strains. Infect. Immun. 80, 575–584. doi: 10.1128/IAI.05510-11
Johannes, L., and Popoff, V. (2008). Tracing the retrograde route in protein trafficking. Cell 135, 1175–1187. doi: 10.1016/j.cell.2008.12.009
Juhas, M. (2015). Horizontal gene transfer in human pathogens. Crit. Rev. Microbiol. 41, 101–108. doi: 10.3109/1040841X.2013.804031
Jurgens, K., and Matz, C. (2002). Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Van Leeuwenhoek 81, 413–434. doi: 10.1023/A:1020505204959
Kessler, A., Schell, U., Sahr, T., Tiaden, A., Harrison, C., Buchrieser, C., et al. (2013). The Legionella pneumophila orphan sensor kinase LqsT regulates competence and pathogen-host interactions as a component of the LAI-1 circuit. Environ. Microbiol. 15, 646–662. doi: 10.1111/j.1462-2920.2012.02889.x
Kilvington, S., and Price, J. (1990). Survival of Legionella pneumophila within cysts of Acanthamoeba polyphaga following chlorine exposure. J Appl. Bacteriol. 68, 519–525. doi: 10.1111/j.1365-2672.1990.tb02904.x
Kim, Y. R., Lee, S. E., Kook, H., Yeom, J. A., Na, H. S., Kim, S. Y., et al. (2008). Vibrio vulnificus RTX toxin kills host cells only after contact of the bacteria with host cells. Cell. Microbiol. 10, 848–862. doi: 10.1111/j.1462-5822.2007.01088.x
Kirn, T. J., Jude, B. A., and Taylor, R. K. (2005). A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438, 863–866. doi: 10.1038/nature04249
Kolonko, M., Geffken, A. C., Blumer, T., Hagens, K., Schaible, U. E., and Hagedorn, M. (2014). WASH-driven actin polymerization is required for efficient mycobacterial phagosome maturation arrest. Cell. Microbiol. 16, 232–246. doi: 10.1111/cmi.12217
Kovacs, G., Nagy, S. U., and Csaba, G. (1986). The effect of bacterial endotoxin of phagocytosis of Tetrahymena and serotonin induced imprinting. Acta Microbiol. Hung. 33, 301–304.
Kuo, S. Y., Chou, M. C., Lee, S. L., Wang, Y., Chen, C. L., Lin, P. T., et al. (2015). Vibrio vulnificus RtxA1 modulated calcium flux contributes reduced internalization in phagocytes. Life Sci. 132, 55–60. doi: 10.1016/j.lfs.2015.03.027
Kwak, J. S., Jeong, H.-G., and Satchell, K. J. F. (2011). Vibrio vulnificus rtxA1 gene recombination generates toxin variants with altered potency during intestinal infection. Proc. Natl. Acad. Sci. U.S.A. 108, 1645–1650. doi: 10.1073/pnas.1014339108
Lainhart, W., Stolfa, G., and Koudelka, G. B. (2009). Shiga toxin as a bacterial defense against a eukaryotic predator, Tetrahymena thermophila. J. Bacteriol. 191, 5116–5122. doi: 10.1128/JB.00508-09
Lambrecht, E., Bare, J., Chavatte, N., Bert, W., Sabbe, K., and Houf, K. (2015). Protozoan cysts act as a survival niche and protective shelter for foodborne pathogenic bacteria. Appl. Environ. Microbiol. 81, 5604–5612. doi: 10.1128/AEM.01031-15
Lambrecht, E., Bare, J., Van Damme, I., Bert, W., Sabbe, K., and Houf, K. (2013). Behavior of Yersinia enterocolitica in the presence of the bacterivorous Acanthamoeba castellanii. Appl. Environ. Microbiol. 79, 6407–6413. doi: 10.1128/AEM.01915-13
Lauriano, C. M., Barker, J. R., Yoon, S. S., Nano, F. E., Arulanandam, B. P., Hassett, D. J., et al. (2004). MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. U.S.A. 101, 4246–4249. doi: 10.1073/pnas.0307690101
Law, H. T., Sriram, A., Fevang, C., Nix, E. B., Nano, F. E., and Guttman, J. A. (2014). IglC and PdpA are important for promoting Francisella invasion and intracellular growth in epithelial cells. PLoS One 9:e104881. doi: 10.1371/journal.pone.0104881
Lee, B. C., Choi, S. H., and Kim, T. S. (2008). Vibrio vulnificus RTX toxin plays an important role in the apoptotic death of human intestinal epithelial cells exposed to Vibrio vulnificus. Microbes Infect. 10, 1504–1513. doi: 10.1016/j.micinf.2008.09.006
Lee, C. T., Pajuelo, D., Llorens, A., Chen, Y. H., Leiro, J. M., Padrós, F., et al. (2013). MARTX of Vibrio vulnificus biotype 2 is a virulence and survival factor. Environ. Microbiol. 15, 419–432. doi: 10.1111/j.1462-2920.2012.02854.x
Lee, J. H., Kim, M. W., Kim, B. S., Kim, S. M., Lee, B. C., Kim, T. S., et al. (2007). Identification and characterization of the Vibrio vulnificus rtxA essential for cytotoxicity in vitro and virulence in mice. J. Microbiol. 45, 146–152.
Levin, B. R. (1996). The evolution and maintenance of virulence in microparasites. Emerg. Infect. Dis. 2, 93–102. doi: 10.3201/eid0202.960203
Licznerska, K., Nejman-Faleńczyk, B., Bloch, S., Dydecka, A., Topka, G., Gąsior, T., et al. (2016). Oxidative stress in shiga toxin production by enterohemorrhagic Escherichia coli. Oxid. Med. Cell. Longev. 2016:8. doi: 10.1155/2016/3578368
Lurie-Weinberger, M. N., Gomez-Valero, L., Merault, N., Glockner, G., Buchrieser, C., and Gophna, U. (2010). The origins of eukaryotic-like proteins in Legionella pneumophila. Int. J. Med. Microbiol. 300, 470–481. doi: 10.1016/j.ijmm.2010.04.016
Lutz, C., Erken, M., Noorian, P., Sun, S., and McDougald, D. (2013). Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front. Microbiol. 4:375. doi: 10.3389/fmicb.2013.00375
Madsen, J. S., Burmolle, M., Hansen, L. H., and Sorensen, S. J. (2012). The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 65, 183–195. doi: 10.1111/j.1574-695X.2012.00960.x
March, C., Cano, V., Moranta, D., Llobet, E., Perez-Gutierrez, C., Tomas, J. M., et al. (2013). Role of bacterial surface structures on the interaction of Klebsiella pneumoniae with phagocytes. PLoS One 8:e56847. doi: 10.1371/journal.pone.0056847
Mattana, A., Serra, C., Mariotti, E., Delogu, G., Fiori, P. L., and Cappuccinelli, P. (2006). Acanthamoeba castellanii promotion of in vitro survival and transmission of coxsackie B3 viruses. Eukaryot Cell 5, 665–671. doi: 10.1128/EC.5.4.665-671.2006
Matz, C., Bergfeld, T., Rice, S. A., and Kjelleberg, S. (2004). Microcolonies, quorum sensing and cytotoxicity determine the survival of Pseudomonas aeruginosa biofilms exposed to protozoan grazing. Environ. Microbiol. 6, 218–226. doi: 10.1111/j.1462-2920.2004.00556.x
Matz, C., and Kjelleberg, S. (2005). Off the hook–how bacteria survive protozoan grazing. Trends Microbiol. 13, 302–307. doi: 10.1016/j.tim.2005.05.009
Matz, C., McDougald, D., Moreno, A. M., Yung, P. Y., Yildiz, F. H., and Kjelleberg, S. (2005). Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 102, 16819–16824. doi: 10.1073/pnas.0505350102
Matz, C., Moreno, A. M., Alhede, M., Manefield, M., Hauser, A. R., Givskov, M., et al. (2008). Pseudomonas aeruginosa uses type III secretion system to kill biofilm-associated amoebae. ISME J. 2, 843–852. doi: 10.1038/ismej.2008.47
Matz, C., Nouri, B., McCarter, L., and Martinez-Urtaza, J. (2011). Acquired type III secretion system determines environmental fitness of epidemic Vibrio parahaemolyticus in the interaction with bacterivorous protists. PLoS One 6:e20275. doi: 10.1371/journal.pone.0020275
Miyata, S. T., Kitaoka, M., Brooks, T. M., McAuley, S. B., and Pukatzki, S. (2011). Vibrio cholerae requires the Type VI Secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect. Immun. 79, 2941–2949. doi: 10.1128/IAI.01266-10
Molmeret, M., Horn, M., Wagner, M., Santic, M., and Abu Kwaik, Y. (2005). Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71, 20–28. doi: 10.1128/AEM.71.1.20-28.2005
Mulcahy, L. R., Isabella, V. M., and Lewis, K. (2014). Pseudomonas aeruginosa biofilms in disease. Microb. Ecol. 68, 1–12. doi: 10.1007/s00248-013-0297-x
Neidig, A., Yeung, A. T., Rosay, T., Tettmann, B., Strempel, N., Rueger, M., et al. (2013). TypA is involved in virulence, antimicrobial resistance and biofilm formation in Pseudomonas aeruginosa. BMC Microbiol. 13:77. doi: 10.1186/1471-2180-13-77
Noorian, P., Hu, J., Chen, Z., Kjelleberg, S., Wilkins, M. R., Sun, S., et al. (2017). Pyomelanin produced by Vibrio cholerae confers resistance to predation by Acanthamoeba castellanii. FEMS Microbiol. Ecol. 93:fix147. doi: 10.1093/femsec/fix147
Olivier, V., Salzman, N. H., and Satchell, K. J. (2007). Prolonged colonization of mice by Vibrio cholerae El Tor O1 depends on accessory toxins. Infect. Immun. 75, 5043–5051. doi: 10.1128/IAI.00508-07
Ozanic, M., Marecic, V., Abu Kwaik, Y., and Santic, M. (2015). The divergent intracellular lifestyle of Francisella tularensis in evolutionarily distinct host cells. PLoS Pathog. 11:e1005208. doi: 10.1371/journal.ppat.1005208
Pang, M., Lin, X., Liu, J., Guo, C., Gao, S., Du, H., et al. (2016). Identification of Aeromonas hydrophila genes preferentially expressed after phagocytosis by Tetrahymena and involvement of methionine sulfoxide reductases. Front. Cell. Infect. Microbiol. 6:199. doi: 10.3389/fcimb.2016.00199
Paquet, V. E., and Charette, S. J. (2016). Amoeba-resisting bacteria found in multilamellar bodies secreted by Dictyostelium discoideum: social amoebae can also package bacteria. FEMS Microbiol. Ecol. 92:fiw025. doi: 10.1093/femsec/fiw025
Parry, J. D. (2004). Protozoan grazing of freshwater biofilms. Adv. Appl. Microbiol. 54, 167–196. doi: 10.1016/S0065-2164(04)54007-8
Personnic, N., Striednig, B., and Hilbi, H. (2017). Legionella quorum sensing and its role in pathogen-host interactions. Curr. Opin. Microbiol. 41, 29–35. doi: 10.1016/j.mib.2017.11.010
Portier, E., Zheng, H., Sahr, T., Burnside, D. M., Mallama, C., Buchrieser, C., et al. (2015). IroT/mavN, a new iron-regulated gene involved in Legionella pneumophila virulence against amoebae and macrophages. Environ. Microbiol. 17, 1338–1350. doi: 10.1111/1462-2920.12604
Prochazkova, K., Shuvalova, L. A., Minasov, G., Voburka, Z., Anderson, W. F., and Satchell, K. J. (2009). Structural and molecular mechanism for autoprocessing of MARTX toxin of Vibrio cholerae at multiple sites. J. Biol. Chem. 284, 26557–26568. doi: 10.1074/jbc.M109.025510
Pruzzo, C., Vezzulli, L., and Colwell, R. R. (2008). Global impact of Vibrio cholerae interactions with chitin. Environ. Microbiol. 10, 1400–1410. doi: 10.1111/j.1462-2920.2007.01559.x
Pukatzki, S., Ma, A. T., Revel, A. T., Sturtevant, D., and Mekalanos, J. J. (2007). Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513. doi: 10.1073/pnas.0706532104
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533. doi: 10.1073/pnas.0510322103
Ramakrishnan, G. (2017). Iron and Virulence in Francisella tularensis. Front. Cell. Infect. Microbiol. 7:107. doi: 10.3389/fcimb.2017.00107
Reguera, G., and Kolter, R. (2005). Virulence and the environment: a novel role for Vibrio cholerae toxin-coregulated pili in biofilm formation on chitin. J. Bacteriol. 187, 3551–3555. doi: 10.1128/JB.187.10.3551-3555.2005
Reinhart, A. A., and Oglesby-Sherrouse, A. G. (2016). Regulation of Pseudomonas aeruginosa virulence by distinct iron sources. Genes 7:E126. doi: 10.3390/genes7120126
Reyes-Batlle, M., Girbau, C., Lopez-Arencibia, A., Sifaoui, I., Liendo, A. R., Bethencourt Estrella, C. J., et al. (2017). Variation in Campylobacter jejuni culturability in presence of Acanthamoeba castellanii Neff. Exp. Parasitol. 183, 178–181. doi: 10.1016/j.exppara.2017.09.005
Richards, A. M., Von Dwingelo, J. E., Price, C. T., and Abu Kwaik, Y. (2013). Cellular microbiology and molecular ecology of Legionella-amoeba interaction. Virulence 4, 307–314. doi: 10.4161/viru.24290
Riquelme, S., Varas, M., Valenzuela, C., Velozo, P., Chahin, N., Aguilera, P., et al. (2016). Relevant genes linked to virulence are required for Salmonella Typhimurium to survive intracellularly in the social amoeba Dictyostelium discoideum. Front. Microbiol. 7:1305. doi: 10.3389/fmicb.2016.01305
Robertson, P., Abdelhady, H., and Garduno, R. A. (2014). The many forms of a pleomorphic bacterial pathogen-the developmental network of Legionella pneumophila. Front. Microbiol. 5:670. doi: 10.3389/fmicb.2014.00670
Roig, F. J., González-Candelas, F., and Amaro, C. (2011). Domain organization and evolution of multifunctional autoprocessing repeats-in-toxin (MARTX) toxin in Vibrio vulnificus. Appl. Environ. Microbiol. 77, 657–668. doi: 10.1128/AEM.01806-10
Rompikuntal, P. K., Vdovikova, S., Duperthuy, M., Johnson, T. L., Ahlund, M., Lundmark, R., et al. (2015). Outer membrane vesicle-mediated export of processed PrtV protease from Vibrio cholerae. PLoS One 10:e0134098. doi: 10.1371/journal.pone.0134098
Rossier, O., Starkenburg, S. R., and Cianciotto, N. P. (2004). Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires’ disease pneumonia. Infect. Immun. 72, 310–321. doi: 10.1128/IAI.72.1.310-321.2004
Rothmeier, E., Pfaffinger, G., Hoffmann, C., Harrison, C. F., Grabmayr, H., Repnik, U., et al. (2013). Activation of Ran GTPase by a Legionella effector promotes microtubule polymerization, pathogen vacuole motility and infection. PLoS Pathog. 9:e1003598. doi: 10.1371/journal.ppat.1003598
Sack, D. A., Sack, R. B., Nair, G. B., and Siddique, A. K. (2004). Cholera. Lancet 363, 223–233. doi: 10.1016/S0140-6736(03)15328-7
Samanovic, M. I., Ding, C., Thiele, D. J., and Darwin, K. H. (2012). Copper in microbial pathogenesis: meddling with the metal. Cell Host Microbe 11, 106–115. doi: 10.1016/j.chom.2012.01.009
Sandkvist, M., Michel, L. O., Hough, L. P., Morales, V. M., Bagdasarian, M., Koomey, M., et al. (1997). General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol. 179, 6994–7003. doi: 10.1128/jb.179.22.6994-7003.1997
Santic, M., Molmeret, M., Klose, K. E., Jones, S., and Kwaik, Y. A. (2005). The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7, 969–979. doi: 10.1111/j.1462-5822.2005.00526.x
Santic, M., Ozanic, M., Semic, V., Pavokovic, G., Mrvcic, V., and Kwaik, Y. A. (2011). Intra-vacuolar proliferation of F. novicida within H. vermiformis. Front. Microbiol. 2:78. doi: 10.3389/fmicb.2011.00078
Sasindran, S. J., Saikolappan, S., and Dhandayuthapani, S. (2007). Methionine sulfoxide reductases and virulence of bacterial pathogens. Future Microbiol. 2, 619–630. doi: 10.2217/17460913.2.6.619
Scheid, P., and Schwarzenberger, R. (2012). Acanthamoeba spp. as vehicle and reservoir of adenoviruses. Parasitol. Res. 111, 479–485. doi: 10.1007/s00436-012-2828-7
Schell, U., Kessler, A., and Hilbi, H. (2014). Phosphorylation signalling through the Legionella quorum sensing histidine kinases LqsS and LqsT converges on the response regulator LqsR. Mol. Microbiol. 92, 1039–1055. doi: 10.1111/mmi.12612
Schell, U., Simon, S., and Hilbi, H. (2016a). Inflammasome recognition and regulation of the Legionella flagellum. Curr. Top. Microbiol. Immunol. 397, 161–181. doi: 10.1007/978-3-319-41171-2_8
Schell, U., Simon, S., Sahr, T., Hager, D., Albers, M. F., Kessler, A., et al. (2016b). The alpha-hydroxyketone LAI-1 regulates motility, Lqs-dependent phosphorylation signalling and gene expression of Legionella pneumophila. Mol. Microbiol. 99, 778–793. doi: 10.1111/mmi.13265
Schlimme, W., Marchiani, M., Hanselmann, K., and Jenni, B. (1997). Gene transfer between bacteria within digestive vacuoles of protozoa. FEMS Microbiol. Ecol. 23, 239–247. doi: 10.1111/j.1574-6941.1997.tb00406.x
Sherr, E. B., and Sherr, B. F. (2002). Significance of predation by protists in aquatic microbial food webs. Antonie Van Leeuwenhoek 81, 293–308. doi: 10.1023/A:1020591307260
Shevchuk, O., Pagelow, D., Rasch, J., Dohrmann, S., Gunther, G., Hoppe, J., et al. (2014). Polyketide synthase (PKS) reduces fusion of Legionella pneumophila-containing vacuoles with lysosomes and contributes to bacterial competitiveness during infection. Int. J. Med. Microbiol. 304, 1169–1181. doi: 10.1016/j.ijmm.2014.08.010
Siddiqui, R., and Khan, N. A. (2012). Acanthamoeba is an evolutionary ancestor of macrophages: a myth or reality? Exp. Parasitol. 130, 95–97. doi: 10.1016/j.exppara.2011.11.005
Siddiqui, R., Malik, H., Sagheer, M., Jung, S. Y., and Khan, N. A. (2011). The type III secretion system is involved in Escherichia coli K1 interactions with Acanthamoeba. Exp. Parasitol. 128, 409–413. doi: 10.1016/j.exppara.2011.05.008
Silva, A. J., and Benitez, J. A. (2016). Vibrio cholerae biofilms and cholera pathogenesis. PLoS Negl. Trop. Dis. 10:e0004330. doi: 10.1371/journal.pntd.0004330
Simon, S., Schell, U., Heuer, N., Hager, D., Albers, M. F., Matthias, J., et al. (2015). Inter-kingdom signaling by the Legionella quorum sensing molecule LAI-1 modulates cell migration through an IQGAP1-Cdc42-ARHGEF9-dependent pathway. PLoS Pathog. 11:e1005307. doi: 10.1371/journal.ppat.1005307
Simon, S., Wagner, M. A., Rothmeier, E., Muller-Taubenberger, A., and Hilbi, H. (2014). Icm/Dot-dependent inhibition of phagocyte migration by Legionella is antagonized by a translocated Ran GTPase activator. Cell. Microbiol. 16, 977–992. doi: 10.1111/cmi.12258
So, E. C., Mattheis, C., Tate, E. W., Frankel, G., and Schroeder, G. N. (2015). Creating a customized intracellular niche: subversion of host cell signaling by Legionella type IV secretion system effectors. Can. J. Microbiol. 61, 617–635. doi: 10.1139/cjm-2015-0166
Steenbergen, J. N., Shuman, H. A., and Casadevall, A. (2001). Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc. Natl. Acad. Sci. U.S.A. 98, 15245–15250. doi: 10.1073/pnas.261418798
Steinberg, K. M., and Levin, B. R. (2007). Grazing protozoa and the evolution of the Escherichia coli O157:H7 Shiga toxin-encoding prophage. Proc. Biol. Sci. 274, 1921–1929. doi: 10.1098/rspb.2007.0245
Stolfa, G., and Koudelka, G. B. (2012). Entry and killing of Tetrahymena thermophila by bacterially produced Shiga toxin. mBio 4:e00416-12. doi: 10.1128/mBio.00416-12
Sun, S., Kjelleberg, S., and McDougald, D. (2013). Relative contributions of Vibrio polysaccharide and quorum sensing to the resistance of Vibrio cholerae to predation by heterotrophic protists. PLoS One 8:e56338. doi: 10.1371/journal.pone.0056338
Suzuki, M., Danilchanka, O., and Mekalanos, J. J. (2014). Vibrio cholerae T3SS effector VopE modulates mitochondrial dynamics and innate immune signaling by targeting Miro GTPases. Cell Host Microbe 16, 581–591. doi: 10.1016/j.chom.2014.09.015
Tam, V. C., Serruto, D., Dziejman, M., Brieher, W., and Mekalanos, J. J. (2007). A type III secretion system in Vibrio cholerae translocates a formin/spire hybrid-like actin nucleator to promote intestinal colonization. Cell Host Microbe 1, 95–107. doi: 10.1016/j.chom.2007.03.005
Tiaden, A., Spirig, T., Weber, S. S., Bruggemann, H., Bosshard, R., Buchrieser, C., et al. (2007). The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell. Microbiol. 9, 2903–2920. doi: 10.1111/j.1462-5822.2007.01005.x
Townsley, L., Sison Mangus, M. P., Mehic, S., and Yildiz, F. H. (2016). Response of Vibrio cholerae to low-temperature shifts: CspV regulation of type VI secretion, biofilm formation, and association with zooplankton. Appl. Environ. Microbiol. 82, 4441–4452. doi: 10.1128/AEM.00807-16
Trigui, H., Paquet, V. E., Charette, S. J., and Faucher, S. P. (2016). Packaging of Campylobacter jejuni into multilamellar bodies by the ciliate Tetrahymena pyriformis. Appl. Environ. Microbiol. 82, 2783–2790. doi: 10.1128/AEM.03921-15
Tyson, J. Y., Pearce, M. M., Vargas, P., Bagchi, S., Mulhern, B. J., and Cianciotto, N. P. (2013). Multiple Legionella pneumophila Type II secretion substrates, including a novel protein, contribute to differential infection of the amoebae Acanthamoeba castellanii, Hartmannella vermiformis, and Naegleria lovaniensis. Infect. Immun. 81, 1399–1410. doi: 10.1128/IAI.00045-13
Vaitkevicius, K., Lindmark, B., Ou, G., Song, T., Toma, C., Iwanaga, M., et al. (2006). A Vibrio cholerae protease needed for killing of Caenorhabditis elegans has a role in protection from natural predator grazing. Proc. Natl. Acad. Sci. U.S.A. 103, 9280–9285. doi: 10.1073/pnas.0601754103
Valeru, S. P., Rompikuntal, P. K., Ishikawa, T., Vaitkevicius, K., Sjöling,Å, Dolganov, N., et al. (2009). Role of melanin pigment in expression of Vibrio cholerae virulence factors. Infect. Immun. 77, 935–942. doi: 10.1128/IAI.00929-08
Van der Henst, C., Scrignari, T., Maclachlan, C., and Blokesch, M. (2015). An intracellular replication niche for Vibrio cholerae in the amoeba Acanthamoeba castellanii. ISME J. 10, 897–910. doi: 10.1038/ismej.2015.165
Vezzulli, L., Colwell, R. R., and Pruzzo, C. (2013). Ocean warming and spread of pathogenic vibrios in the aquatic environment. Microb. Ecol. 65, 817–825. doi: 10.1007/s00248-012-0163-2
Vezzulli, L., Pezzati, E., Brettar, I., Hofle, M., and Pruzzo, C. (2015). Effects of global warming on Vibrio ecology. Microbiol. Spectr. 3:VE-0004-2014. doi: 10.1128/microbiolspec.VE-0004-2014
Vieira, A., Ramesh, A., Seddon, A. M., and Karlyshev, A. V. (2017). CmeABC multidrug efflux pump contributes to antibiotic resistance and promotes Campylobacter jejuni survival and multiplication in Acanthamoeba polyphaga. Appl. Environ. Microbiol. 83:e01600-17. doi: 10.1128/AEM.01600-17
Weitere, M., Bergfeld, T., Rice, S. A., Matz, C., and Kjelleberg, S. (2005). Grazing resistance of Pseudomonas aeruginosa biofilms depends on type of protective mechanism, developmental stage and protozoan feeding mode. Environ. Microbiol. 7, 1593–1601. doi: 10.1111/j.1462-2920.2005.00851.x
Wheat, W. H., Casali, A. L., Thomas, V., Spencer, J. S., Lahiri, R., Williams, D. L., et al. (2014). Long-term survival and virulence of Mycobacterium leprae in amoebal cysts. PLoS Negl. Trop. Dis. 8:e3405. doi: 10.1371/journal.pntd.0003405
Wong, E., Vaaje-Kolstad, G., Ghosh, A., Hurtado-Guerrero, R., Konarev, P. V., Ibrahim, A. F. M., et al. (2012). The Vibrio cholerae colonization factor Gbpa possesses a modular structure that governs binding to different host surfaces. PLoS Pathog. 8:e1002373. doi: 10.1371/journal.ppat.1002373
Zhou, M., Guo, Z., Duan, Q., Hardwidge, P. R., and Zhu, G. (2014). Escherichia coli type III secretion system 2: a new kind of T3SS? Vet. Res. 45:32. doi: 10.1186/1297-9716-45-32
Keywords: pathogenicity, predation, Vibrio, evolution, virulence factors, coincidental evolution, protozoa
Citation: Sun S, Noorian P and McDougald D (2018) Dual Role of Mechanisms Involved in Resistance to Predation by Protozoa and Virulence to Humans. Front. Microbiol. 9:1017. doi: 10.3389/fmicb.2018.01017
Received: 14 March 2018; Accepted: 30 April 2018;
Published: 17 May 2018.
Edited by:
Niels Kroer, University of Copenhagen, DenmarkReviewed by:
Christopher Price, University of Louisville, United StatesCopyright © 2018 Sun, Noorian and McDougald. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Diane McDougald, ZGlhbmUubWNkb3VnYWxkQHV0cy5lZHUuYXU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.