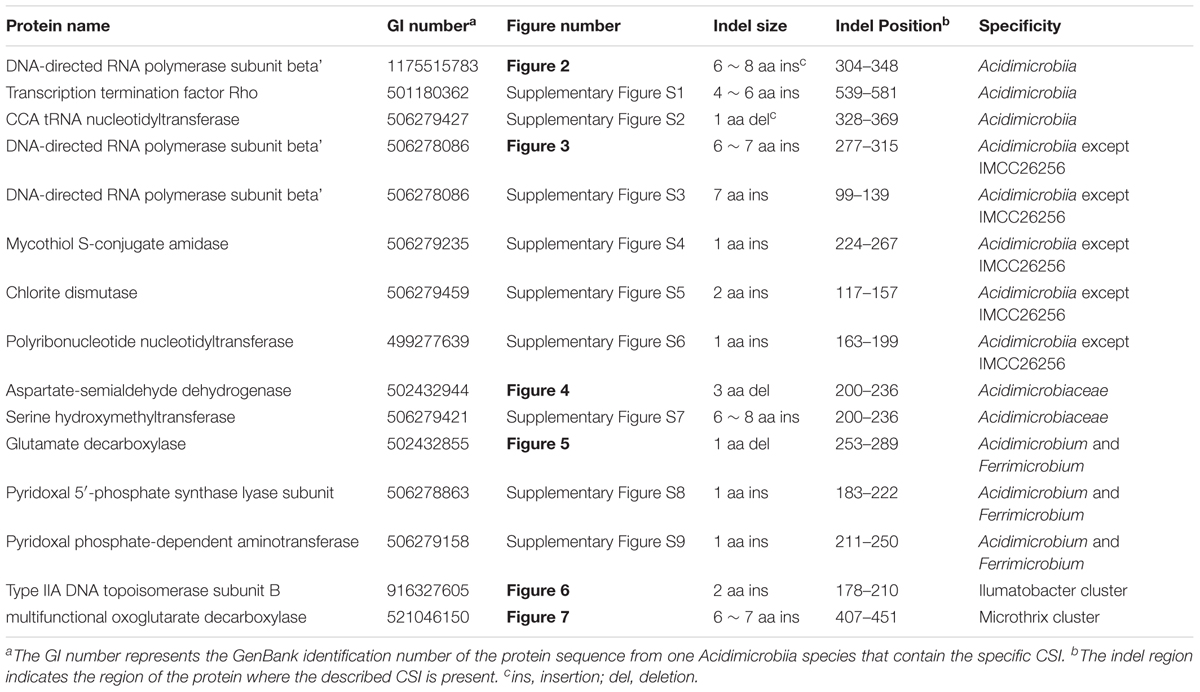
- 1CAS Key Laboratory of Tropical Marine Bio-resources and Ecology, Guangdong Key Laboratory of Marine Materia Medica, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
- 2University of Chinese Academy of Sciences, Beijing, China
Recent metagenomic surveys of microbial community suggested that species associated with the class Acidimicrobiia are abundant in diverse aquatic environments such as acidic mine water, waste water sludge, freshwater, or marine habitats, but very few species have been cultivated and characterized. The current taxonomic framework of Acidimicrobiia is solely based on 16S rRNA sequence analysis of few cultivable representatives, and no molecular, biochemical, or physiological characteristics are known that can distinguish species of this class from the other bacteria. This study reports the phylogenomic analysis for 20 sequenced members of this class and reveals another three major lineages in addition to the two recognized families. Comparative analysis of the sequenced Acidimicrobiia species identified 15 conserved signature indels (CSIs) in widely distributed proteins and 26 conserved signature proteins (CSPs) that are either specific to this class as a whole or to its major lineages. This study represents the most comprehensive phylogenetic analysis of the class Acidimicrobiia and the identified CSIs and CSPs provide useful molecular markers for the identification and delineation of species belonging to this class or its subgroups.
Introduction
The class Acidimicrobiia is a deep-rooting lineage within the phylum Actinobacteria. This class is comprised of few cultivable representatives that were mostly isolated from extremely acidic environments (Zhi et al., 2009; Gao and Gupta, 2012b; Ludwig et al., 2012). Four type species of this class namely, Acidimicrobium ferrooxidans, Acidithrix ferrooxidans, Ferrimicrobium acidiphilum, and Ferrithrix thermotolerans are extremely acidophilic, with optimal growth pH at around 2.0, and are able to oxidize ferrous iron at relatively fast rates (Cleaver et al., 2007; Ludwig et al., 2012; Norris, 2012). These species were mainly isolated from acidic mine waters or geothermal sites, and were responsible for the regeneration of ferric iron within the acidic ecosystem (Clark and Norris, 1996; Johnson et al., 2009; Jones and Johnson, 2015). In contrast, other members of this class were not acidophiles and inhabited more diverse aquatic environments. For example, neutrophilic Iamia majanohamensis was isolated from the abdominal epidermis of a sea cucumber, filamentous “Candidatus Microthrix parvicella” (henceforth called M. parvicella) from wastewater sludge, while members of the genus Ilumatobacter from estuary sediment or seashore sand (Kurahashi et al., 2009; Matsumoto et al., 2009; Mcllroy et al., 2013).
In spite of few cultivable Acidimicrobiia species, metagenomic analyses have revealed that there were many uncultured actinobacterial species belonging to the class Acidimicrobiia in freshwater and marine samples (Rheims et al., 1996; Jensen and Lauro, 2008; Ghai et al., 2014). Warnecke et al. (2004) analyzed actinobacterial 16S rRNA genes from freshwater habitats and suggested four most prominent lineages, one of which “acIV lineage” is associated with the order Acidimicrobiales. An extensive microbial community composition survey of northwestern Sargasso Sea identified that a marine clade closely related to M. parvicella were abundant in the deep chlorophyll maximum (DCM), with occasional blooms during summer stratification period (Treusch et al., 2009). More recently, Chen et al. (2016) investigated the actinobacterial diversity in the deep sea along the Southwest Indian Ridge and discovered that Acidimicrobiales is one of the two most widely distributed and abundant actinobacterial orders in all nine samples from deep sea environments. In addition to the species diversity analyses based on 16S rRNA sequences, further genomic data mining of uncultured Acidimicrobiia species suggested that the ecological and metabolic diversity of this class is far underestimated by the culture-dependent species characterization. A metagenomic analysis of Mediterranean DCM assembled four nearly complete genomes for marine Acidimicrobiales, and pathway analysis indicated that these species have the capability to assimilate C2 compounds and also derive energy from dimethylsulfoniopropionate, sulfonate, and carbon monoxide (Mizuno et al., 2015). In addition, one of the genomes encodes acidirhodopsin, a novel rhodopsin clade related to freshwater actinorhodopsins (Mizuno et al., 2015).
Although the metagenomic data greatly expanded our knowledge of the species diversity of Acidimicrobiia, the current taxonomic frame of this class contains only one order Acidimicrobiales, and two families Acidimicrobiaceae and Iamiaceae with few genera (Ludwig et al., 2012). The taxonomic ranks were determined solely based on 16S rRNA gene sequence analyses and taxon-specific 16S rRNA signature nucleotides using limited representative isolates. To date, despite the availability of seven complete genomes from cultivated Acidimicrobiia species and many incomplete genomes from metagenomic data, there is no comprehensive phylogenetic analysis performed to examine the evolutionary relationship within this class. As such, no detailed evolutionary relationship of the uncultured species can be assigned with relation to the known type species of the class Acidimicrobiia. (Warnecke et al., 2004; Ghai et al., 2014; Mizuno et al., 2015). In addition, except the branching pattern of these species in phylogenetic trees, no molecular, biochemical or physiological characteristics are known that can clearly distinguish Acidimicrobiia species from other Actinobacteria (Norris, 2012).
Comparative genomic analyses can lead to discovery of molecular markers that are specific to different higher taxon (e.g., genus level and above), which cannot be easily derived from culture-dependent phenotypic characterization (Gupta and Gao, 2010; Gao and Gupta, 2012a). One important category of these molecular markers is conserved signature indels (CSIs) that are uniquely found in the genes/proteins homologs from a specific group of organisms. Another type of molecular markers are conserved signature proteins (CSPs) that are uniquely shared by a monophyletic group of prokaryotes. The two molecular marker types represent highly reliable characteristics of specific groups of organisms, and they provide novel methods for the identification or delineation of prokaryotic taxonomic units in clear molecular terms (Gao and Gupta, 2012b; Ho et al., 2016; Zhang et al., 2016; Alnajar and Gupta, 2017).
In the present work, a robust phylogenetic tree was constructed for 20 sequenced members of class Acidimicrobiia based on 30 universal conserved proteins. The tree clearly showed another three major clusters in addition to the two recognized families within this class, and these clusters may comprise separate families. Besides, comparative analysis of the sequenced Acidimicrobiia species identified 15 CSIs in universal proteins and 26 CSPs, which are either specific for this class as a whole or to its major lineages. This study represents the most comprehensive phylogenetic analysis of the class Acidimicrobiia and the identified CSIs and CSPs provide useful molecular markers for the identification and demarcation of the members belonging to this class or its subgroups.
Materials and Methods
Phylogenetic Analysis
A phylogenetic tree for 20 genome-sequenced members of class Acidimicrobiia (Supplementary Table S1) was constructed based on the concatenation of 30 protein sequences, selected from a set of 92 single copy orthologous proteins (Na et al., 2018) and can be retrieved for the most assembled genomes of this class (Supplementary Table S2). Sequences from Rubrobacter radiotolerans was used as outgroup to root the tree. Multiple sequence alignments for each protein were performed using the Clustal X 2.1 program (Larkin et al., 2007) and concatenated to produce a single alignment file. The poorly aligned regions of the sequence alignment were removed by the Gblocks 0.91b program (Talavera and Castresana, 2007). The resulting alignment containing 7600 aligned amino acids was used for phylogenetic analysis. A maximum-likelihood (ML) tree was constructed by MEGA 6.0 (Tamura et al., 2013) with the Whelan and Goldman substitution model based on 1000 bootstrap replicates. Another ML tree including more assembled genomes from freshwater Acidimicrobiia was constructed based on concatenation of 10 ribosomal protein sequences (Supplementary Table S3). The method applied here was the same as done earlier and the final combined protein alignment used for phylogenetic analysis include 1814 amino acids.
A neighbor-joining (NJ) tree based on sequence alignment of 16S rRNA gene sequences was constructed for the representative strains of cultured Acidimicrobiia and some assembled genomes. Full length 16S rRNA sequences were retrieved from Ribosomal Database Project (Cole et al., 2014) or NCBI GenBank, and accession number of each 16S rRNA sequences were summarized in Supplementary Table S4. To root the tree, sequences from three Rubrobacter species were used as outgroup. The tree was constructed by MEGA 6.0 using the Kimura 2_parameter model with 1000 bootstrap replicates.
Identification of CSIs
Conserved signature indels were identified as previously described (Gupta, 2014; Zhang et al., 2016). Briefly, Blastp searches were carried out on all proteins from the genome of A. ferrooxidans DSM 10331 (Accession number NC_013124.1) (Clum et al., 2009) against all sequences in the GenBank non-redundant database. Multiple sequence alignments were created for homologs of all available Acidimicrobiia species and few other bacteria. These sequence alignments were inspected for any conserved insertions or deletions that were restricted to Acidimicrobiia species only and also flanked by at least 5–6 identical or conserved residues in the neighboring 30 ~ 40 amino acids on each side. The indels, whose flanking regions were not conserved, were not further considered and removed. To assess the specificity of the identified indels, detailed BLASTp searches were carried out with a short sequence segment containing the indel and the flanking conserved regions (60–100 amino acids long) against the GenBank database. To further confirm that the identified signatures are restricted to Acidimicrobiia homologs, the top 500 BLAST hits with the highest similarity to the query sequence were examined for the presence or absence of these CSIs. Signature files were then created by two programs Sig_Create and Sig_Style (available from Gleans.net) (Gupta, 2014). Due to space limitation, indels containing sequence alignment in all figures and supplementary figures only include those that are found in all Acidimicrobiia sequences and few sequences from representative strains of other bacterial groups. It should also be noted that a number of CSIs and CSPs described here are also observed in the assembled genome of endosymbiont cyanobacterium TDX16 deposited by Hebei University of Technology (Accession number NDGV01000834.1, no publication available). We suspect that this unclassified “Cyanobacteria” genome assemble is not from DNA of pure culture but rather contamination from multiple bacterial strains since BLASTp searches of individual protein from this assembled genome returns top 5 hits from diverse bacteria, none of which belongs to Cyanobacteria and the most frequent best hits are from Planctomycetes species. Therefore, sequences from cyanobacterium TDX16 genome is not considered in our analysis.
Identification of CSPs
BLASTp searches were performed on individual protein from the genome of A. ferrooxidans DSM 10331 to identify proteins that are restricted to Acidimicrobiia species. These searches were executed against all sequences in the NCBI non-redundant database and the results were then examined manually for proteins with significant hits present only in Acidimicrobiia genomes following the same criteria as described in earlier work (Gao et al., 2006; Gao and Gupta, 2012b).
Results and Discussion
Phylogenetic Analysis of the Class Acidimicrobiia Based on Combined Protein Dataset and 16S rRNA Trees
Previous phylogenomic analyses of Acidimicrobidae considered two or three fully sequenced species (A. ferrooxidans, M. parvicella, and Ilumatobacter coccineum) and not more than six assembled genomes from metagenomic sequences (Hugerth et al., 2015; Mizuno et al., 2015). As a result, except confirming the association of these assembled genomes with Acidimicrobiia, no detailed evolutionary relationship among these uncultured species and known species can be concluded. In order to get a comprehensive overview of the phylogeny of class Acidimicrobiia, a phylogenetic tree was constructed for 7 completely sequenced species of this class and additional 13 assembled genomes from metagenomic data, whose genome information is nearly complete (Supplementary Table S1). The tree was constructed by ML analysis based on concatenation of 30 universally distributed orthologous protein sequences that are mainly involved in translation and transcription (Supplementary Table S2). To date, this tree represents the most comprehensive phylogenetic analysis of the class Acidimicrobiia (Figure 1A).
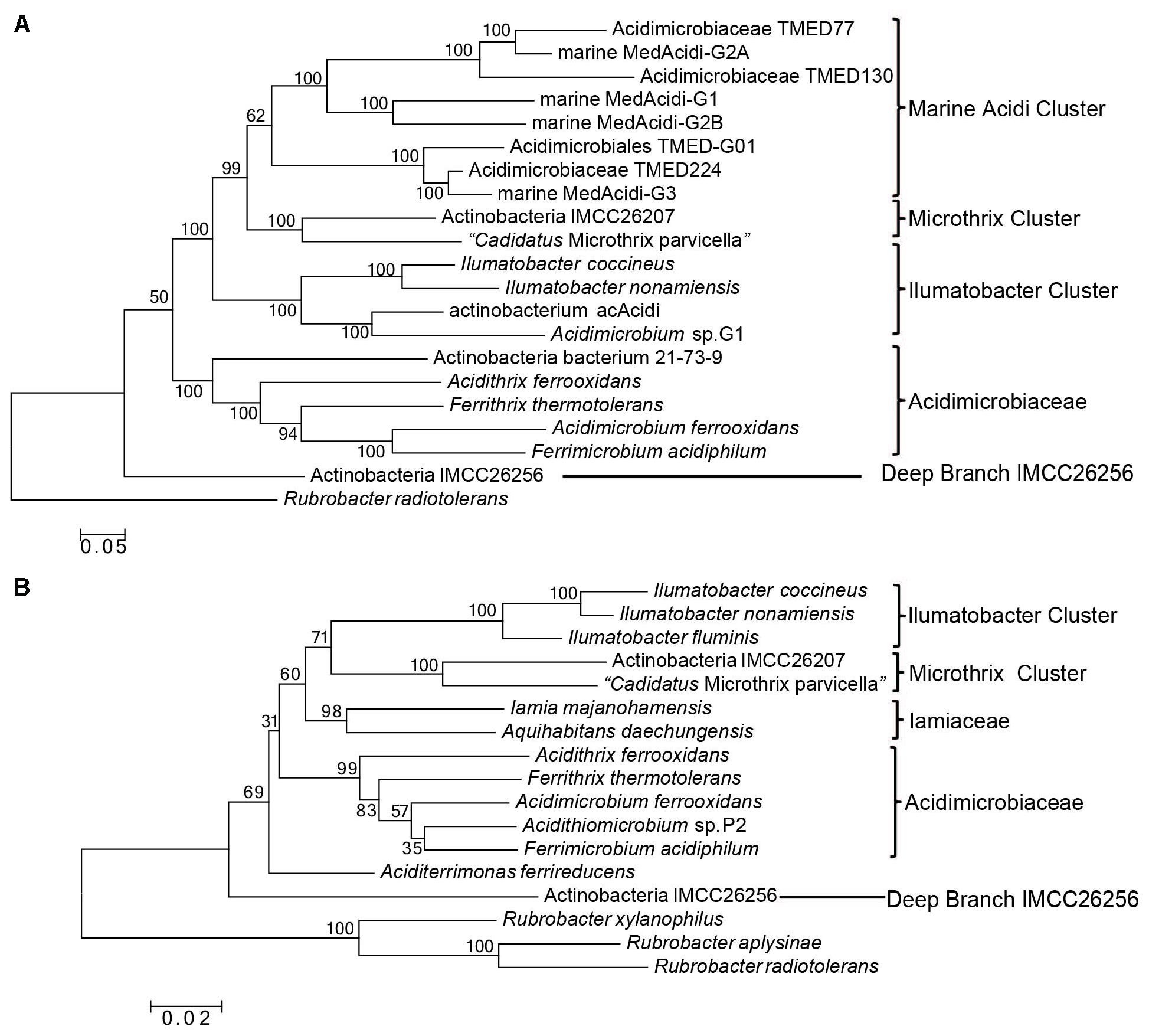
FIGURE 1. Phylogenetic tree analysis of class Acidimicrobiia. (A) ML tree for 20 Acidimicrobiia species based upon concatenated sequences of 30 conserved proteins. (B) Neighbor-joining tree based on full length 16S rRNA gene sequences of all type species within the class Acidimicrobiia. Bootstrap values (%) are shown at each node and different clusters that are consistently observed in both phylogenetic trees are marked.
In this combined protein tree, an assembled genome “Actinobacteria bacterium IMCC26256” from freshwater sample forms the deepest branch, clearly separated from other Acidimicrobiia species. Four type species of the family Acidimicrobiaceae together with an assembled genome “Actinobacteria bacterium 21-73-9” from mine drainage metagenome form a well-defined cluster. Based on their branching pattern in the tree and their similar isolation environment, Actinobacteria bacterium 21-73-9 should be affiliated with the family Acidimicrobiaceae. The rest in the tree formed three distinctive clusters, and were named after the cultured type species if available found in each cluster namely the “Ilumatobacter Cluster” and “Microthrix Cluster.” A third cluster comprised of assembled genomes from different marine metagenomes is named as “Marine Acidimicrobiia Cluster.”
Since Iamia, one of the only two families within this class, do not have any genome sequenced and cannot be used as reference in the combined protein tree analysis, we constructed another phylogenetic tree based on 16S rRNA gene sequences (Figure 1B). In this analysis, we try to include all the named species of this class and two assembled genomes from the combined protein tree analysis since the full length 16S rRNA sequences cannot be retrieved from GenBank for the rest of assembled genomes. Consistent with the combined protein tree, Actinobacteria bacterium IMCC26256 formed the deepest branch in the 16S rRNA gene tree of the class Acidimicrobiia. In addition, four clusters can be distinguished from each other with high bootstrap scores at each branch node, namely Acidimicrobiaceae, Iamiaceae, Microthrix Cluster, and Ilumatobacter Cluster. In both trees shown in Figure 1, M. parvicella branched with Actinobacteria bacterium IMCC26207 from freshwater metagenome, distinctive from Acidimicrobiaceae and Iamiaceae. A recent 16S rRNA analysis of identified Acidimicrobiia species and many uncultured environmental clones also indicated that strain IMCC26207 and M. parvicella form a clade clearly separated from Acidimicrobiaceae and Iamiaceae (Kim et al., 2017). In addition, although the current taxonomic outline placed Ilumatobacter within the Acidimicrobiaceae, our phylogenetic tree analysis based on both combined protein dataset and 16S rRNA sequences suggest that they are not monophyletic with Acidimicrobiaceae species. Hence, in view of the distinctive clustering pattern of Microthrix Cluster and Ilumatobacter Cluster from the two identified families, these two clusters may warrant assignment of novel families within this class. Certainly, this assignment requires additional molecular markers to support the monophyletic relationship of individual cluster.
Molecular Markers Specific for the Class Acidimicrobiia
The availability of complete and nearly complete assembled genomes from class Acidimicrobiia provide great resources to explore genomic characteristics that are unique to this class or subgroups within it. CSIs in genes/proteins sequences are important rare genetic changes for understanding bacterial phylogeny (Gao and Gupta, 2007; Gupta and Gao, 2009). The CSIs that serve as useful molecular markers are generally of defined size and their flanking residues are very conserved to ensure their reliability (Gupta, 2014, 2016). Because of the rarity and highly specific nature of such genetic changes, it is less likely that they could arise independently by either convergent or parallel evolution. Most likely, the genetic change responsible for a specific CSI occurred once in a common ancestor of the specific group of species and then passed on vertically to the various descendants. Therefore, CSIs that are restricted to particular clade(s) have generally provided very good phylogenetic markers for evolutionary studies.
Comparative analyses of protein sequence alignment from species of class Acidimicrobiia and other bacterial groups led to the identification of three CSIs in different conserved proteins that are uniquely shared by all Acidimicrobiia species sequenced till date. As shown in Figure 2, a 6 ~ 7 amino acids (aa) insertion in a highly conserved region of DNA-directed RNA polymerase subunit beta’ was found to be specific to seven completely sequenced Acidimicrobiia species and assembled Acidimicrobiia genomes but not present in any other bacteria outside this class. Additionally, a 4 ~ 6 aa insertion in transcription termination factor Rho and a 1 aa deletion in CCA tRNA nucleotidyltransferase were exclusively present in members of class Acidimicrobiia (Supplementary Figures S1, S2). For all these CSI containing proteins, homolog sequences of assembled genomes from metagenomic data that belong to class Acidimicrobiia were included in the alignment, and all including the deepest branch Actinobacteria bacterium IMCC26256, were found to contain the same CSIs. Thus, these three CSIs constituted distinctive characteristics of the class Acidimicrobiia and can be used as molecular markers to define and distinguish species belonging to this class.
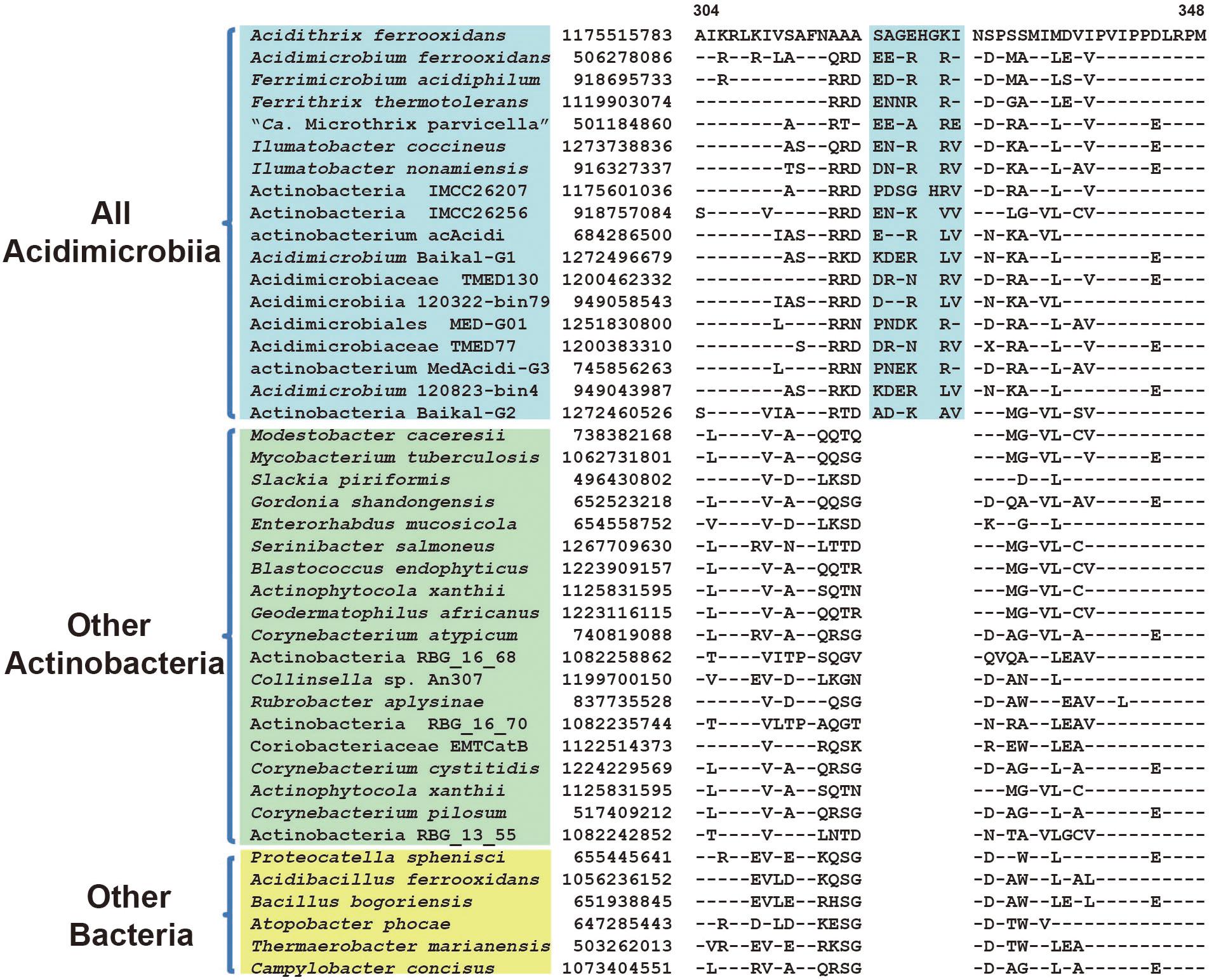
FIGURE 2. Conserved signature indel (CSI) specific to all Acidimicrobiia species. Partial sequence alignment of the protein DNA-directed RNA polymerase subunit beta’ showing a 6 ~ 8aa insertion in a conserved region that is specific for members of the class Acidimicrobiia. The dashes in this alignment as well as all other alignments indicate identity with the amino acid on the top line. The GenBank identification numbers of the protein sequences are shown, and the topmost numbers indicate the position of this sequence in the species shown on the top line. Information for other CSIs that are also specific to the class Acidimicrobiia are presented in Table 1 and Supplementary Figures S1,S2.
In addition to the above CSIs, BLASTp searches of each protein from the genome of type species A. ferrooxidans DSM 10331 were carried out to search for CSPs that are unique to class Acidimicrobiia. Five proteins were found in almost all sequenced Acidimicrobiia genomes including the deepest branch IMCC26256 but not present in any other bacteria outside this class (Table 2). Similar to CSIs, these CSPs provide additional molecular markers for class Acidimicrobiia. These five proteins identified as hypothetical proteins with unknown function, and due to their specificity, functional studies of these proteins may reveal characteristics that are likely to be unique to members of this class.

TABLE 2. Conserved signature proteins (CSPs) that are uniquely found in the Acidimicrobiia and its subgroups.
Molecular Signatures for Some of the Subclades of Acidimicrobiia
As mentioned earlier, uncultivated Actinobacteria bacterium IMCC26256 formed the deepest branch in both phylogenetic trees based on combined protein dataset and 16S rRNA, which suggest that this species might be the earliest branch within known Acidimicrobiia species to date. In our analysis, we have identified five CSIs in four different widely distributed conserved proteins that are uniquely shared by all members of class Acidimicrobiia except strain IMCC26256. Although missing in IMCC26256 genome, these CSIs are not found in any other non-Acidimicrobiia species. One example of these CSIs is shown in Figure 3. In a highly conserved region of DNA-directed RNA polymerase subunit beta’, a 7 aa insert is unique to all Acidimicrobiia species but missing in IMCC26256 genome. Additional four CSIs showing similar specificity are presented in Supplementary Figures S3–S6. The absence of the identified CSIs in homologs of IMCC26256 genome are not due to lateral gene transfer since the best BLASTp hit of these CSIs containing proteins in IMCC26256 genome are homologous sequences of Acidimicrobiia species rather than other bacterial groups. There are two possible explanations for the presence of these five CSIs. First, these CSIs evolved in a common ancestor of all Acidimicrobiia but subsequently lost in IMCC26256 genome; second, these CSIs were introduced in the common ancestor of other Acidimicrobiia lineages after the branch of Actinobacteria bacterium IMCC26256. Although we cannot discriminate which of the two evolutionary scenarios account for the absence of these five CSIs in IMCC26256 genome, the unique presence of these CSIs in the rest of Acidimicrobiia species indicated that they constitute distinctive characteristics of this class.
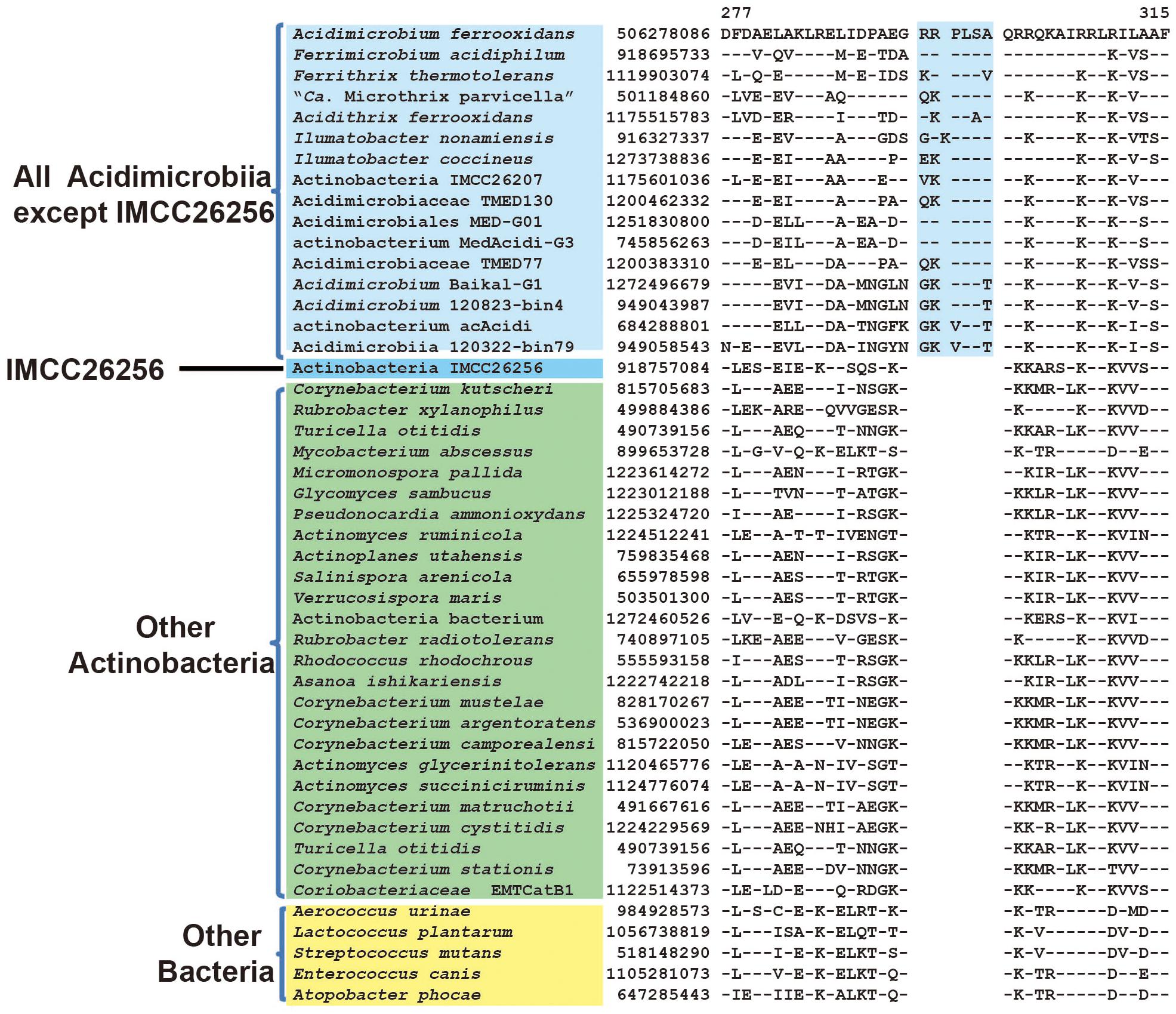
FIGURE 3. Conserved signature indel specific to all Acidimicrobiia species except strain IMCC26256. Partial sequence alignment of DNA-directed RNA polymerase subunit beta’ showing a 6 ~ 7 aa insertion that is specific for all Acidimicrobiia except Actinobacteria bacterium IMCC26256. Information for other CSIs specific for this clade are presented in Table 1 and Supplementary Figures S3–S6.
Our analysis also identified two CSIs that are specifically shared by members of Acidimicrobiaceae, namely A. ferrooxidans, F. acidiphilum, F. thermotolerans, and Acidithrix ferrooxidans. These CSIs include a 3 aa deletion in aspartate-semialdehyde dehydrogenase (Figure 4) and a 6 ~ 8 aa insertion in serine hydroxymethyltransferase (Supplementary Figure S7). They are exclusively present in the above four species belonging to the family Acidimicrobiaceae but not found in any other species. In addition, we also identified eight CSPs that are unique to these four species (Table 2). In contrast, no CSIs or CSPs were found that are uniquely shared by these species and Ilumatobacter species, which are currently assigned under the family Acidimicrobiaceae. These results suggest that most likely Ilumatobacter and the above four species are not monophyletic, consistent with the results from phylogenetic tree analysis. Therefore, Ilumatobacter should not be placed under the family Acidimicrobiaceae. Moreover, these two CSIs and eight CSPs provide distinctive molecular markers for the family Acidimicrobiaceae that can be used to define and delineate species belonging to this family.

FIGURE 4. Conserved signature indel specific to the family Acidimicrobiaceae. Partial alignment of the protein aspartate-semialdehyde dehydrogenase showing a 3 aa deletion that is specific for the family Acidimicrobiaceae. Information for other CSIs that are specific for the family Acidimicrobiaceae are presented in Table 1 and Supplementary Figure S7.
Among members of the family Acidimicrobiaceae, A. ferrooxidans, and F. acidiphilum formed a clade in phylogenetic trees, thereby indicating a more closer relationship among the two than from the other species of this family (Figure 1). Supporting this relationship, we have identified three CSIs in different proteins that were present only in A. ferrooxidans and F. acidiphilum. These CSIs include: 1 aa deletion in glutamate decarboxylase (Figure 5), a 1 aa insertion in pyridoxal 5′-phosphate synthase lyase subunit PdxS, and a 1 aa insertion in pyridoxal phosphate-dependent aminotransferase (Supplementary Figures S8, S9). Besides, 13 CSPs were identified as unique proteins shared by both A. ferrooxidans and F. acidiphilum (Table 2).
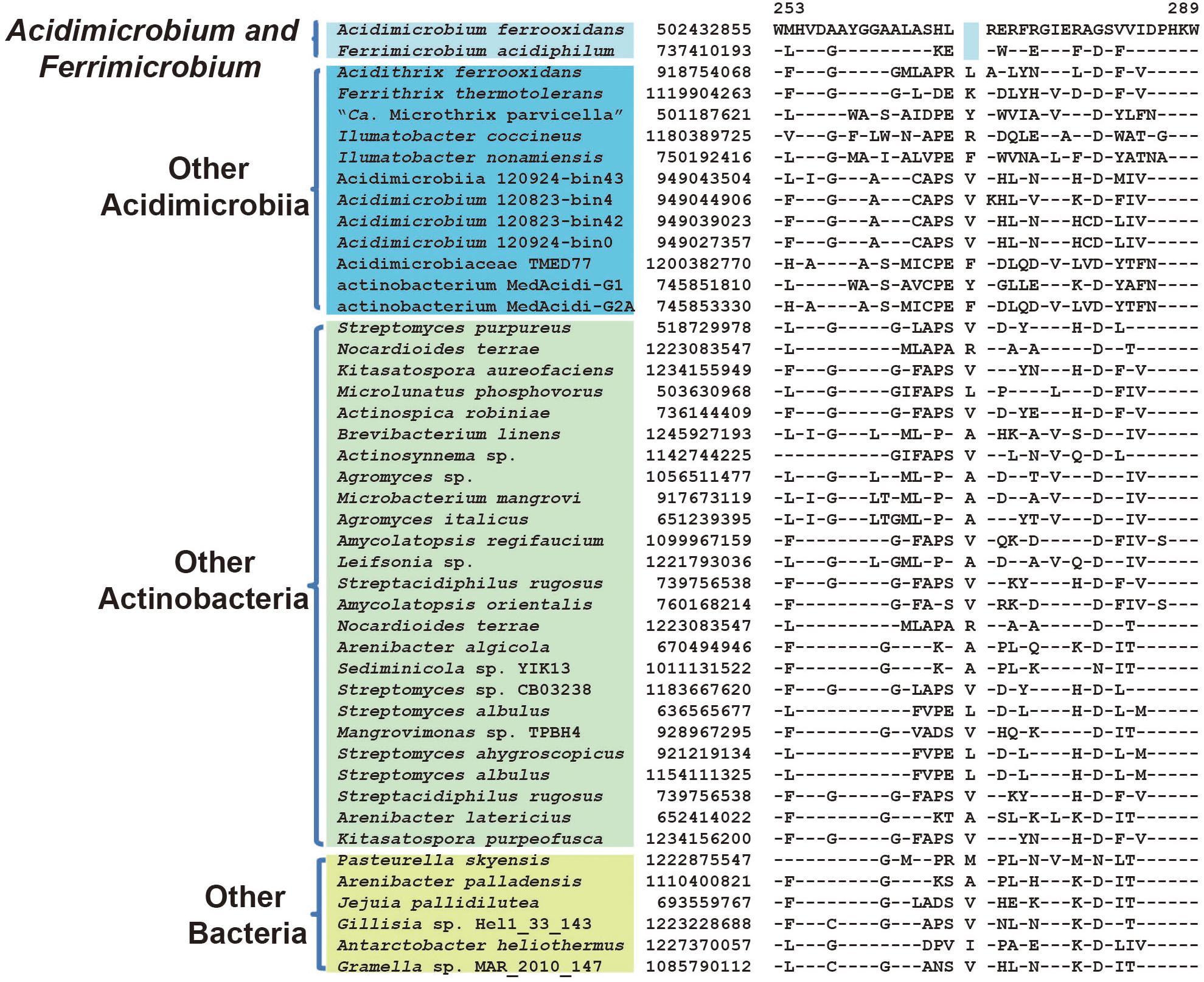
FIGURE 5. Conserved signature indel specific to genera Acidimicrobium and Ferrimicrobium. A 1 aa deletion in the protein glutamate decarboxylase that is uniquely shared by A. ferrooxidans and F. acidiphilum. Information of other CSIs specific for this cluster is present in Table 1, and Supplementary Figures S8, S9.
As revealed by phylogenetic tree analysis, Ilumatobacter species branched together with assembled genome “actinobacterium acAcidi” from freshwater and “Acidimicrobium sp. Baikal-G1” from lake water. Two other phylogenetic trees based on 16S rRNA and combined protein dataset with limited Acidimicrobiia species also indicate that Ilumatobacter formed a cluster with acIV freshwater lineage (Hugerth et al., 2015; Mizuno et al., 2015). In our CSIs searches, we identified one CSI, a 2 aa insertion in type IIA DNA topoisomerase subunit B, that are uniquely shared by Ilumatobacter species and multiple assembled Acidimicrobiia genomes from freshwater samples (Figure 6). To further confirm the relationship of numbers of the genus Ilumatobacter and additional assembled freshwater Acidimicrobiia genomes, we constructed another phylogenetic tree based on ten ribosomal proteins for which sequences can be retrieved from the incomplete genomes of freshwater species (Supplementary Figure S10). Indeed, Ilumatobacter species formed a well-defined cluster with freshwater Acidimicrobiia species supported by high bootstrap score at the branch node. Taken together, the identified CSI and phylogenetic tree analysis suggested that members of the genus Ilumatobacter genus showed more close relationship with freshwater Acidimicrobiia lineage, and they should be assigned as an independent family different from that of the parent family Acidimicrobiaceae.

FIGURE 6. Conserved signature indel specific to Ilumatobacter cluster. A 2 aa CSI in the protein type IIA DNA topoisomerase subunit B that is specific for the Ilumatobacter cluster.
Microthrix parvicella was frequently retrieved from activated sludge wastewater treatment plants and had characteristic long unbranched filamentous morphology (Rossetti et al., 2005). Previous phylogenetic analyses indicated that M. parvicella belonged to Acidimicrobiia but formed a separate branch from the other type species of this class (Mizuno et al., 2015). In our phylogenetic trees based on both 16S rRNA and combined protein dataset (Figure 1), a recently published assembled genome of freshwater isolate, strain IMCC26207 with proposed species name “Candidatus Limnosphaera aquatica” (Kim et al., 2017), formed a distinctive clade with M. parvicella supported by high bootstrap score. This is the most closely related genome for M. parvicella reported to date (Kim et al., 2017). A 6 ~ 7 aa insertion in a highly conserved region of multifunctional oxoglutarate decarboxylase was identified to be specific to M. parvicella and IMCC26207 (Figure 7), which provide a potential molecular marker for Microthrix cluster but awaits confirmation with more homologous sequences from closely related species.
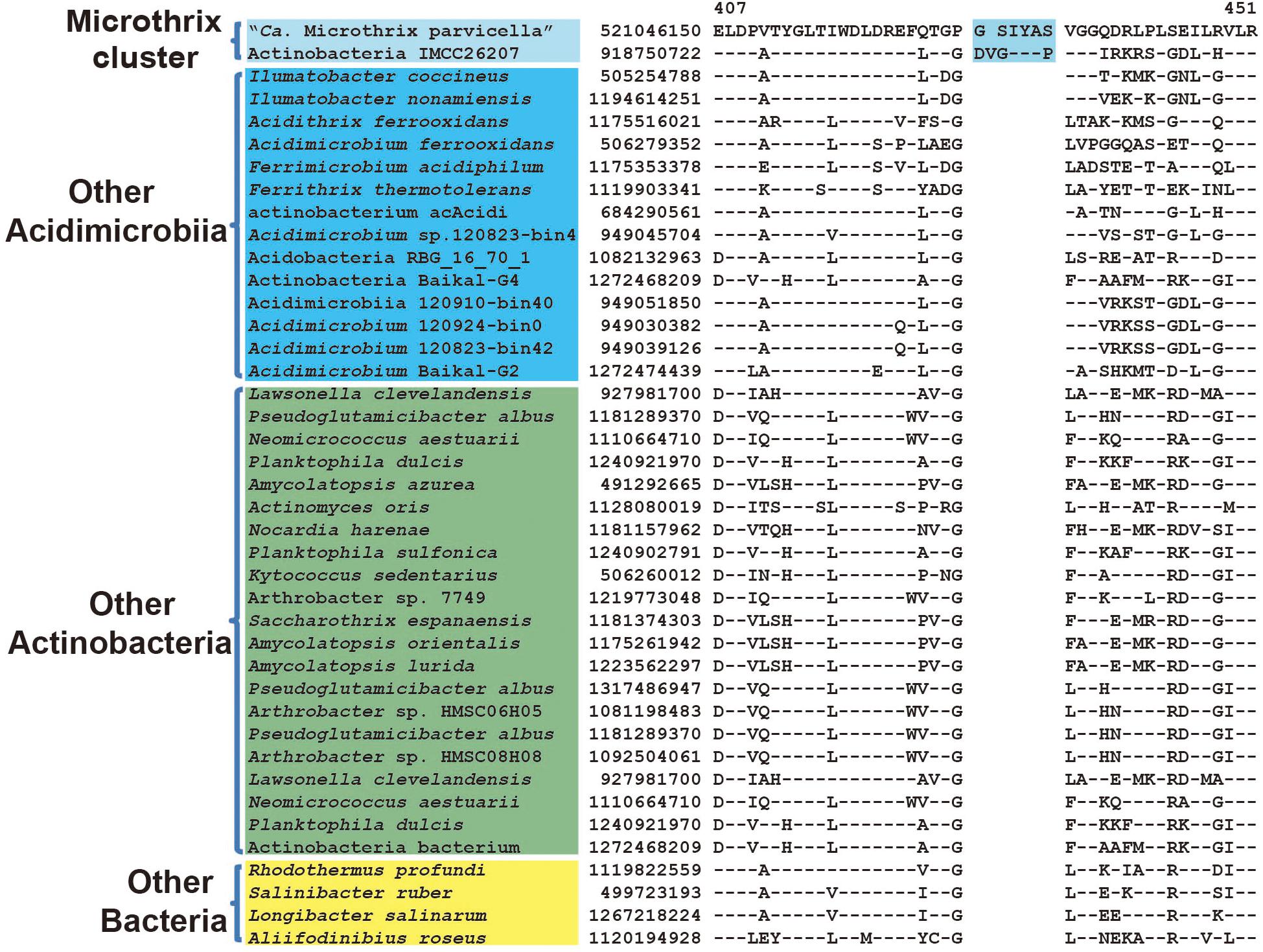
FIGURE 7. Conserved signature indel specific to Microthrix cluster. Partial sequence alignment of multifunctional oxoglutarate decarboxylase showing a 6 ~ 7aa insertion that is specific for the Microthrix cluster comprising of M. parvicella and strain IMCC26207.
Conclusion
In spite of the abundance of Acidimicrobiia in diverse aquatic habitats and their important role in biogeochemical cycling, presently there is limited study on the phylogeny of this deep branch class of the phylum Actinobacteria. The current taxonomic framework based on few cultivated species need to be updated to serve as guide map for increasing metagenomic investigation of species diversity of this class. In the present work, we have performed detailed phylogenomic analysis of sequenced Acidimicrobiia species and assembled genomes, which revealed three distinctive clusters namely Ilumatobacter cluster, Microthrix Cluster and Marine Acidic Cluster, in addition to the only two recognized families. More importantly, we have identified multiple CSIs in different proteins and CSPs that are specific to either class Acidimicrobiia or certain lineages within it. These genomic signature sequences can be used as molecular markers to define or delineate class Acidimicrobiia or its subgroups at higher taxonomic ranks, in addition to the current standard based on 16S rRNA alone. The class Acidimicrobiia currently only consists of two families-Acidimicrobiaceae and Iamiaceae, the latter of which has no genomes sequenced, and thus no CSIs/CSPs can be identified. In total, we have discovered two CSIs and eight CSPs specific for four species of the family Acidimicrobiaceae, but not present in species of the genera Microthrix and Ilumatobacter genera. Based on the clustering pattern of phylogenetic trees presented in Figure 1 and the identified CSIs for Ilumatobacter Cluster and Microthrix Cluster, these two clades are not monophyletic with type species of Acidimicrobiaceae and should be defined as independent families. Furthermore, according to our phylogenomic analysis, Acidimicrobiia species from marine environments formed a cluster distinct from the other cultured type species, suggesting that these marine Acidimicrobiia might share unique genotypic and phenotypic characteristics. Hence, it is of much interest to identify molecular markers that are uniquely shared by marine Acidimicrobiia in the future.
Finally, both CSI-containing proteins and CSPs perform different functions in the bacterial cells, although the function of most of these molecular markers are unknown at present. Due to their specificity, the function of these CSIs and CSPs might be some characteristics unique to the specific taxon that contain them. For example, one Actinobacteria-specific CSP, ParJ (SCO1662), was functionally characterized as regulating the polymerization of ParA protein and affecting chromosome segregation and cell division during Streptomyces sporulation (Ditkowski et al., 2010). The Acidimicrobiia-specific CSIs and CSPs presented here provide novel targets for functional studies, which may reveal yet undiscovered features that are unique to species of this diverse class.
Author Contributions
DH carried out comparative analyses of the Acidimicrobiia genomes to identify signatures reported here, DH and GC constructed the phylogenetic trees. BG and DH were responsible for the writing and editing of the manuscript. All of the work was carried out under the direction of BG.
Funding
This work was supported by Strategic Priority Research Program of the Chinese Academy of Sciences (XDA13020300), National Natural Science Foundation of China (31570011), and Natural Science Foundation of Guangdong Province (2015A030306039). BG is also a scholar of the “100 Talents Project” of the Chinese Academy of Sciences.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer SN and handling Editor declared their shared affiliation.
Acknowledgments
The authors want to thank Dr. Radhey S. Gupta from McMaster University for generously providing the “SIG_CREATE” and “SIG_STYLE” programs.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00987/full#supplementary-material
References
Alnajar, S., and Gupta, R. S. (2017). Phylogenomics and comparative genomic studies delineate six main clades within the family Enterobacteriaceae and support the reclassification of several polyphyletic members of the family. Infect. Genet. Evol. 54, 108–127. doi: 10.1016/j.meegid.2017.06.024
Chen, P., Zhang, L., Guo, X., Dai, X., Liu, L., Xi, L., et al. (2016). Diversity, biogeography, and biodegradation potential of actinobacteria in the deep-sea sediments along the southwest Indian ridge. Front. Microbiol. 7:1340. doi: 10.3389/fmicb.2016.01340
Clark, D. A., and Norris, P. R. (1996). Acidimicrobium ferrooxidans gen nov, sp nov: mixed-culture ferrous iron oxidation with Sulfobacillus species. Microbiology 142, 785–790. doi: 10.1099/00221287-142-4-785
Cleaver, A. A., Burton, N. P., and Norris, P. R. (2007). A novel Acidimicrobium species in continuous cultures of moderately thermophilic, mineral-sulfide-oxidizing acidophiles. Appl. Environ. Microbiol. 73, 4294–4299. doi: 10.1128/AEM.02658-06
Clum, A., Nolan, M., Lang, E., Glavina Del Rio, T., Tice, H., Copeland, A., et al. (2009). Complete genome sequence of Acidimicrobium ferrooxidans type strain (ICP). Stand. Genomic Sci. 1, 38–45. doi: 10.4056/sigs.1463
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., Mcgarrell, D. M., Sun, Y., et al. (2014). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642. doi: 10.1093/nar/gkt1244
Ditkowski, B., Troc, P., Ginda, K., Donczew, M., Chater, K. F., Zakrzewska-Czerwinska, J., et al. (2010). The actinobacterial signature protein ParJ (SCO1662) regulates ParA polymerization and affects chromosome segregation and cell division during Streptomyces sporulation. Mol. Microbiol. 78, 1403–1415. doi: 10.1111/j.1365-2958.2010.07409.x
Gao, B., and Gupta, R. S. (2007). Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis. BMC Genomics 8:86. doi: 10.1186/1471-2164-8-86
Gao, B., and Gupta, R. S. (2012a). Microbial systematics in the post-genomics era. Antonie Van Leeuwenhoek 101, 45–54. doi: 10.1007/s10482-011-9663-1
Gao, B., and Gupta, R. S. (2012b). Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol. Mol. Biol. Rev. 76, 66–112. doi: 10.1128/MMBR.05011-11
Gao, B., Paramanathan, R., and Gupta, R. S. (2006). Signature proteins that are distinctive characteristics of Actinobacteria and their subgroups. Antonie Van Leeuwenhoek G 90, 69–91. doi: 10.1007/s10482-006-9061-2
Ghai, R., Mizuno, C. M., Picazo, A., Camacho, A., and Rodriguez-Valera, F. (2014). Key roles for freshwater Actinobacteria revealed by deep metagenomic sequencing. Mol. Ecol. 23, 6073–6090. doi: 10.1111/mec.12985
Gupta, R. S. (2014). “Identification of conserved indels that are useful for classification and evolutionary studies,” in Methods in Microbiology, eds M. Goodfellow, I. Sutcliffe, and J. Chun (Oxford: Academic Press), 153–182.
Gupta, R. S. (2016). Impact of genomics on the understanding of microbial evolution and classification: the importance of Darwin’s views on classification. FEMS Microbiol. Rev. 40, 520–553. doi: 10.1093/femsre/fuw011
Gupta, R. S., and Gao, B. (2009). Phylogenomic analyses of clostridia and identification of novel protein signatures that are specific to the genus Clostridium sensu stricto (cluster I). Int. J. Syst. Evol. Microbilo. 59, 285–294. doi: 10.1099/ijs.0.001792-0
Gupta, R. S., and Gao, B. (2010). “Recent advances in understanding microbial systematics,” in Microbial Population Genetics, ed. J. P. Xu (Norfolk: Caister Academic Press), 1–14.
Ho, J., Adeolu, M., Khadka, B., and Gupta, R. S. (2016). Identification of distinctive molecular traits that are characteristic of the phylum “Deinococcus-Thermus” and distinguish its main constituent groups. Syst. Appl. Microbiol. 39, 453–463. doi: 10.1016/j.syapm.2016.07.003
Hugerth, L. W., Larsson, J., Alneberg, J., Lindh, M. V., Legrand, C., Pinhassi, J., et al. (2015). Metagenome-assembled genomes uncover a global brackish microbiome. Genome Biol. 16:279. doi: 10.1186/s13059-015-0834-7
Jensen, P. R., and Lauro, F. M. (2008). An assessment of actinobacterial diversity in the marine environment. Antonie Van Leeuwenhoek 94, 51–62. doi: 10.1007/s10482-008-9239-x
Johnson, D. B., Bacelar-Nicolau, P., Okibe, N., Thomas, A., and Hallberg, K. B. (2009). Ferrimicrobium acidiphilum gen. nov., sp. nov. and Ferrithrix thermotolerans gen. nov., sp. nov.: heterotrophic, iron-oxidizing, extremely acidophilic actinobacteria. Int. J. Syst. Evol. Microbiol. 59, 1082–1089. doi: 10.1099/ijs.0.65409-0
Jones, R. M., and Johnson, D. B. (2015). Acidithrix ferrooxidans gen. nov., sp. nov.; a filamentous and obligately heterotrophic, acidophilic member of the Actinobacteria that catalyzes dissimilatory oxido-reduction of iron. Res. Microbiol. 166, 111–120. doi: 10.1016/j.resmic.2015.01.003
Kim, S., Kang, I., and Cho, J. C. (2017). Genomic analysis of a freshwater Actinobacterium, “Candidatus limnosphaera aquatica” strain IMCC26207, isolated from lake soyang. J. Microbiol. Biotechnol. 27, 825–833. doi: 10.4014/jmb.1701.01047
Kurahashi, M., Fukunaga, Y., Sakiyama, Y., Harayama, S., and Yokota, A. (2009). Iamia majanohamensis gen. nov., sp. nov., an actinobacterium isolated from sea cucumber Holothuria edulis, and proposal of Iamiaceae fam. nov. Int. J. Syst. Evol. Microbiol. 59, 869–873. doi: 10.1099/ijs.0.005611-0
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Ludwig, W., Euzeby, J., Schumann, P., Busse, H. J., Trujillo, M. E., Kampfer, P., et al. (2012). “Road map of the phylum Actinobacteria,” in Bergey’s Manual of Systematic Bacteriology, The Actinobacteria, 2 Edn, Vol. 5, eds W. Whitman, P. Kampfer, H. J. Busse, M. E. Trujillo, and W. Ludwig (New York, NY: Springer), 1–28.
Matsumoto, A., Kasai, H., Matsuo, Y., Omura, S., Shizuri, Y., and Takahashi, Y. (2009). Ilumatobacter fluminis gen. nov., sp. nov., a novel actinobacterium isolated from the sediment of an estuary. J. Gen. Appl. Microbiol. 55, 201–205. doi: 10.2323/jgam.55.201
Mcllroy, S. J., Kristiansen, R., Albertsen, M., Karst, S. M., Rossetti, S., Nielsen, J. L., et al. (2013). Metabolic model for the filamentous ‘Candidatus Microthrix parvicella’ based on genomic and metagenomic analyses. ISME J. 7, 1161–1172. doi: 10.1038/ismej.2013.6
Mizuno, C. M., Rodriguez-Valera, F., and Ghai, R. (2015). Genomes of planktonic Acidimicrobiales: widening horizons for marine Actinobacteria by metagenomics. mBio 6:e2083-14. doi: 10.1128/mBio.02083-14
Na, S. I., Kim, Y. O., Yoon, S. H., Ha, S. M., Baek, I., and Chun, J. (2018). UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 281–285. doi: 10.1007/s12275-018-8014-6
Norris, P. R. (2012). “Class II. Acidimicrobiia class. nov,” in Bergey’s Manual of Systematic Bacteriology, The Actinobacteria, 2 Edn, Vol. 5, eds W. Whitman, P. Kampfer, H. J. Busse, M. E. Trujillo, and W. Ludwig (New York, NY: Springer), 1968–1975.
Rheims, H., Sproer, C., Rainey, F. A., and Stackebrandt, E. (1996). Molecular biological evidence for the occurrence of uncultured members of the actinomycete line of descent in different environments and geographical locations. Microbiology 142, 2863–2870. doi: 10.1099/13500872-142-10-2863
Rossetti, S., Tomei, M. C., Nielsen, P. H., and Tandoi, V. (2005). “Microthrix parvicella”, a filamentous bacterium causing bulking and foaming in activated sludge systems: a review of current knowledge. FEMS Microbiol. Rev. 29, 49–64. doi: 10.1016/j.femsre.2004.09.005
Talavera, G., and Castresana, J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. doi: 10.1080/10635150701472164
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Treusch, A. H., Vergin, K. L., Finlay, L. A., Donatz, M. G., Burton, R. M., Carlson, C. A., et al. (2009). Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J. 3, 1148–1163. doi: 10.1038/ismej.2009.60
Warnecke, F., Amann, R., and Pernthaler, J. (2004). Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ. Microbiol. 6, 242–253. doi: 10.1111/j.1462-2920.2004.00561.x
Zhang, G., Gao, B., Adeolu, M., Khadka, B., and Gupta, R. S. (2016). Phylogenomic analyses and comparative studies on genomes of the Bifidobacteriales: identification of molecular signatures specific for the order Bifidobacteriales and its different subclades. Front. Microbiol. 7:978. doi: 10.3389/fmicb.2016.00978
Zhi, X. Y., Li, W. J., and Stackebrandt, E. (2009). An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int. J. Syst. Evol. Microbiol. 59, 589–608. doi: 10.1099/ijs.0.65780-0
Keywords: Acidimicrobiia, marine Acidimicrobiia, phylogenomics, molecular signatures, conserved signature indels, conserved signature proteins
Citation: Hu D, Cha G and Gao B (2018) A Phylogenomic and Molecular Markers Based Analysis of the Class Acidimicrobiia. Front. Microbiol. 9:987. doi: 10.3389/fmicb.2018.00987
Received: 12 February 2018; Accepted: 27 April 2018;
Published: 15 May 2018.
Edited by:
Wen-Jun Li, Sun Yat-sen University, ChinaReviewed by:
Xiao-Yang Zhi, Yunnan University, ChinaSalam Nimaichand, Sun Yat-sen University, China
Jing Li, Ocean University of China, China
Copyright © 2018 Hu, Cha and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beile Gao, gaob@scsio.ac.cn
 Danyu Hu
Danyu Hu Guihong Cha1,2
Guihong Cha1,2 Beile Gao
Beile Gao