
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 01 May 2018
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.00825
Tachyplesin is a type of cationic β-hairpin antimicrobial peptide discovered in horseshoe crab approximately 30 years ago that is well known for both its potential antimicrobial activities against multidrug-resistant bacteria and its cytotoxicity to mammalian cells. Though its physical interactions with artificial membranes have been well studied, details of its physiological mechanism of action the physiological consequences of its action remain limited. By using the DNA-binding fluorescent dye propidium iodide to monitor membrane integrity, confocal microscopy to assess the intracellular location of FITC-tagged tachyplesin, and RNA sequencing of the differentially expressed genes in four Gram-negative bacteria (Escherichia coli, Acinetobacter baumannii, Klebsiella pneumoniae, and Pseudomonas aeruginosa) treated with lethal or sublethal concentrations of tachyplesin, we found that compared with levofloxacin-treated bacteria, tachyplesin-treated bacteria showed significant effects on the pathways underlying unsaturated fatty acid biosynthesis. Notably, RNA levels of the conserved and essential 3-ketoacyl carrier protein reductase in this pathway (gene FabG) were elevated in all of the four bacteria after tachyplesin treatment. In vitro tests including surface plasmon resonance and enzyme activity assays showed that tachyplesin could bind and inhibit 3-ketoacyl carrier protein reductase, which was consistent with molecular docking prediction results. As unsaturated fatty acids are important for membrane fluidity, our results provided one possible mechanism to explain how tachyplesin kills bacteria and causes cytotoxicity by targeting membranes, which may be helpful for designing more specific and safer antibiotics based on the function of tachyplesin.
Tachyplesin is a type of cationic β-hairpin antimicrobial peptide (AMP) discovered from horseshoe crab hemocytes approximately 30 years ago (Nakamura et al., 1988; Miyata et al., 1989; Kawano et al., 1990; Muta et al., 1990). Though it shows broad-spectrum and potent antimicrobial activity on pathogens, especially multidrug-resistant (MDR) pathogens, clinical application of tachyplesin is precluded by its highly hemolytic effects (Ramamoorthy et al., 2006; Cirioni et al., 2007; Liu et al., 2015; Edwards et al., 2017). More ominously, experimental induction of resistance to tachyplesin was reported due to increased proteolytic activity in Pseudomonas aeruginosa (Hong et al., 2016). A deep understanding of the unique mechanism of action by which tachyplesin kills bacteria may be helpful to overcome its shortcomings, including hemolysis and susceptibility to proteolysis, and may be helpful for the development of novel antibiotics.
Two models trying to explain how tachyplesin works emerged shortly after it was discovered. One proposed that tachyplesin interacted with lipid membranes and induced enhanced permeability of bacterial membranes, which may contribute to the death of the bacteria (Matsuzaki et al., 1991, 1993; Ohta et al., 1992; Katsu et al., 1993). The other proposed that the antiparallel beta-sheet structure formed by disulfides helped with tachyplesin binding to DNA and might play roles in bacteria killing (Yonezawa et al., 1992). Though the former model seems to be more prevalent, reports concerning this issue are mainly depictions of the physical interactions between this peptide and artificial lipid bilayers, and more biological details are still needed (Doherty et al., 2006, 2008). In any case, two conclusions can be drawn from these observations. The first is that compared with membrane rupture activities, the membrane translocation activity of tachyplesin is more relevant to its antimicrobial potential, which is demonstrated by the fact that linear analogs of tachyplesin without disulfide formation could cause more serious membrane disruptions than a cyclic peptide, which also formed pores during the translocation process but showed weaker membrane translocation ability and weaker antimicrobial activity (Matsuzaki et al., 1993, 1997; Doherty et al., 2006, 2008). The other conclusion is that membrane translocation activity is necessary but not sufficient for tachyplesin to kill bacteria, which is based on the reports that compared to tachyplesin alone, poly(ethylene glycol) (PEG)-grafted (PEGylated) tachyplesin showed similar membrane translocation activity but significantly weakened antimicrobial activity (Imura et al., 2007; Han and Lee, 2013). These phenomena imply that tachyplesin may play an intracellular role.
These speculations have been gradually strengthened by several recent reports. Research using confocal microscopy and flow cytometry to investigate the anti-tumor activities of tachyplesin showed that it induced controlled cell death by promoting apoptosis of the human erythroleukemia cell line K562 at lower concentrations and resulted in cell membrane disruption at higher concentrations (Paredes-Gamero et al., 2012). Proteomic profiling of the effects of tachyplesin on glioblastoma multiforme cell line U251 also implied that tachyplesin may cause cell death by affecting intracellular enzymes and by promoting apoptosis (Li et al., 2017). Regarding its antimicrobial effects, confocal microscopy and flow cytometry showed that bacteria died gradually in response to a sublethal concentration of tachyplesin and died instantaneously upon exposure to high concentrations of tachyplesin. Though this process was accompanied by the inactivation of intracellular esterases, no further details were reported (Hong et al., 2015).
A systemic view of how bacteria react to antibiotics by omics assay may reflect pathways with which these antibiotics interfere and is helpful to elucidate undefined mechanisms of novel antibiotics, including AMPs (Kohanski et al., 2010; Tan et al., 2014; Ho et al., 2016; Pulido et al., 2016; Shah et al., 2016). In the present study, we treated MDR clinically isolated pathogens with lethal and sublethal doses of tachyplesin, tested the membrane translocation activity of fluorescein isothiocyanate (FITC)-tagged tachyplesin with confocal microscopy, assessed the membrane rupture activity using DNA-binding fluorescent dye propidium iodide (PI), and further analyzed the differentially expressed RNA-sequencing data with standard bioinformatics analyses, such as Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. In vitro tests, including surface plasmon resonance and enzyme activity assays, molecular docking predictions, and RNA interference, were also performed to confirm the potential targets of tachyplesin identified by RNA sequencing data.
The animal experimental procedures were approved by the Ethics Committee of Animal Care and Welfare of the Institute of Medical Biology, Chinese Academy of Medical Sciences (CAMS) and Peking Union Medial College (PUMC) (Permit Number: SYXK (dian) 2010-0007). All efforts were made to minimize animal suffering.
These studies were carried out as we previously reported with slight modifications and briefly introduced as follows (Liu et al., 2017). Serum stability was assessed with increases over the minimal inhibitory concentration (MIC) of tachyplesin III (KWCFRVCYRGICYRKCR; cysteines with the same type font formed on disulfide; purity ≥ 95%, synthesized by GL Biochem Co., Ltd, Shanghai, China) on Escherichia coli strain DH5α after incubation in serum. Cytotoxicity in different cell lines was assessed using the Cell Proliferation Kit II (XTT) (Roche), and cytotoxicity in hemocytes was evaluated by the increase in the absorbance at 540 nm after peptide incubation. Membrane rupture was indicated by the entrance of the DNA-binding fluorescent dye PI into MDR clinical isolates of P. aeruginosa strain 1409. Membrane translocation of N-terminal FITC-conjugated tachyplesin III (purity ≥ 95%, synthesized by GL Biochem Co., Ltd, Shanghai, China) into MDR clinical isolates of Acinetobacter baumannii strain 1408 were detected by confocal laser-scanning microscopy.
The Vitek 32 system (bioMerieux, France) and 16S rDNA sequencing were used to verify the clinical isolates A. baumannii 1408, P. aeruginosa 1409, Klebsiella pneumoniae 5 and E. coli 513, which were grown overnight in Luria-Bertani (LB) medium at 37°C with constant shaking at 220 rpm to reach the middle of their logarithmic growth phase. A final concentration of 16 μg/ml tachyplesin III (1×, 2×, 1/2×, and 4× MICs for A. baumannii 1408, P. aeruginosa 1409, K. pneumoniae 5 and E. coli 513, respectively) and 32 μg/ml levofloxacin (1×, 2×, 2×, and 1× MICs for A. baumannii 1408, P. aeruginosa 1409, K. pneumoniae 5, and E. coli 513, respectively) were added, and the cultures were incubated at 37°C for 30 min. After the addition of 2 volumes of RNAprotect Bacteria Reagent (QIAGEN) to stabilized RNA, samples were vortexed for 5 s, incubated for 5 min at room temperature (15–25°C) and collected by centrifugation at 4°C for 5 min at 4000 g.
RNA isolation and sequencing were performed by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China) as previously described (Liu B. et al., 2014). Clean reads were mapped to the corresponding bacterial genomes using Bowtie 2 software (Langmead and Salzberg, 2012). Gene expression was calculated using the FPKM (expected number of fragments per kilobase of transcript sequence per millions of base pairs sequenced) method with HTSeq software (union model) (Anders et al., 2015). Differentially expressed genes were selected based on the DESeq R package with Padj values < 0.05 (Anders and Huber, 2010). KEGG pathway analysis was performed using KOBAS (2.0), and the corrected p value cut-off was set at 0.05 (Li et al., 2015).
Quantitative real-time PCR (RT-PCR) was performed following the 2-ΔΔCT method with RNAs from the aforementioned transcriptome analysis (Livak and Schmittgen, 2001; Liu C. et al., 2014). Untreated pathogen strains were used as the calibrator strains, and corresponding ribosomal rpsL genes were used as housekeeping reference genes with the primers shown in Supplementary Table S1 (Dumas et al., 2006).
Pseudomonas aeruginosa 1409 incubated overnight in LB medium at 37°C were re-incubated under the same conditions to reach an OD600 of 0.5–1.0, put on ice for 15 min to stop growth, washed three times with sterile deionized water and collected by centrifugation at 4°C for 15 min at 4000 × g. After washing with sterile 10% glycerol in water (v/v), cells were collected via the aforementioned centrifugation conditions and re-suspended in 10% glycerol in water (v/v) to obtain electrotransformation-competent P. aeruginosa cells.
With the RNA oligo pairs shown in Supplementary Table S2 (GenePharma Co., Ltd, Shanghai, China) targeting FabG from P. aeruginosa, RNAi was carried out with a GenePulser Xcell system using a 2 mm cuvette with the default settings for P. aeruginosa (2.5 kV/cm, 25 μF and 200 Ω). The transformants were either spread onto LB agar plates to calculate the number of bacterial clones or incubated in LB medium at 37°C for 40 min to determine FabG transcript levels by RT-PCR using the primers for P. aeruginosa shown in Supplementary Table S1.
N-terminal His-tagged FabG from E. coli and C-terminal His-tagged FabG from Staphylococcus aureus were cloned into the NcoI and XhoI cloning sites of the vector pET28a, expressed in BL21 DE3 E. coli and further purified with Ni resin (Kristan et al., 2009; Srinivas and Cronan, 2017). The binding interaction between tachyplesin III and FabG was analyzed by surface plasmon resonance (SPR) with a Biocore 3000 (Biocore, Piscataway, NJ, United States). Briefly, tachyplesin III was adsorbed onto a CM5 sensor chip using an amine-coupling kit (GE Healthcare, Little Chalfont, United Kingdom) to obtain approximately 1000 resonance units. For the analysis, different concentrations of FabG proteins in running buffer (HBS-N buffer, GE Healthcare) were injected at a flow rate of 20 μl/min for 6 min. The binding affinity of FabG for tachyplesin III was determined by BIAevaluation 3.0 software (Biacore) with a 1:1 Langmuir binding model for the kinetic calculation (Wei et al., 2013).
A 96-well plate with a final total volume of 200 μl per reaction was used for the FabG inhibition assay. Briefly, tachyplesin III (15, 7.5, and 3.75 μM) was incubated with 1.5 μM FabG from E. coli or 6 μM FabG from S. aureus at room temperature for 5 min, before the addition of 300 μM nicotinamide adenine dinucleotide phosphate (NADPH, Sigma) and then 150 μM acetoacetyl-CoA (Sigma) to initiate the reaction. The decrease in absorbance at 340 nm was recorded over 5 min to reflect the consumption of NADPH.
Tachyplesin III was processed with Chemdraw 11.0 according to the structure of tachyplesin I (PDB code 2RTV) and docked to the active site of FabG from E. coli (PDB code 1Q7B) or Staphylococcus aureus (PDB code 3SJ7) with AutoDock4 using the default settings (Price et al., 2004; Morris et al., 2009; Dutta et al., 2012; Kushibiki et al., 2014).
Tachyplesin III was stable after incubation in sterile deionized water at 37°C for nearly 48 h, reflected by the fact that the MIC against DH5α increased slightly from 4 mg/L to 8 mg/L after 24 h of incubation. A similar phenomenon was observed when tachyplesin III was incubated with mouse serum, which was tested during the first 6 h (Figure 1).
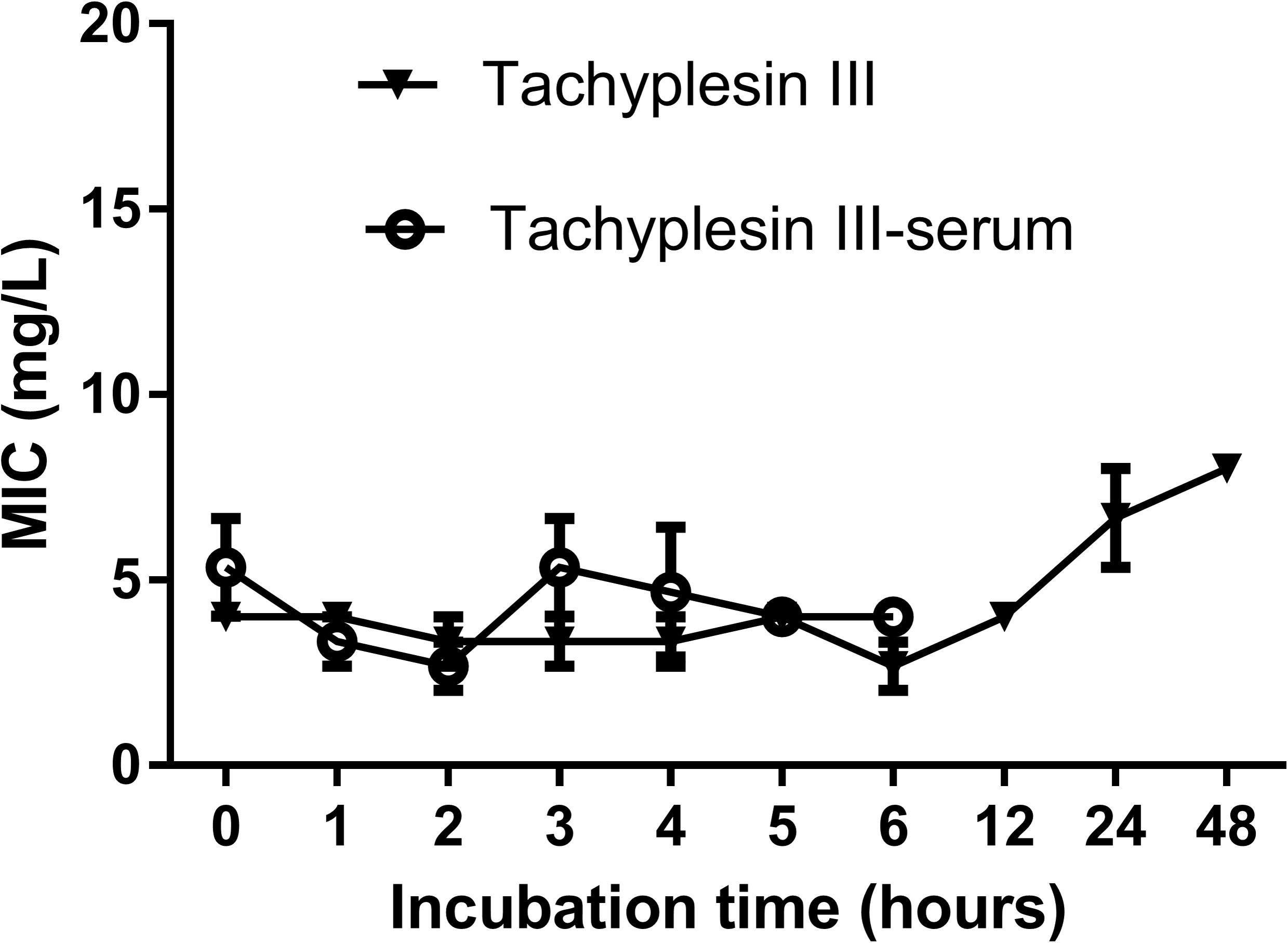
FIGURE 1. Serum stability of tachyplesin III. Tachyplesin III (2 mg/ml) was incubated in either sterile deionized water or mouse serum at 37°C for various lengths of time, aliquots were taken, and MICs against DH5α cells were tested. Stability of the MIC indicates the stability of tachyplesin III.
Approximately 1% and 2% hemolysis was observed when tachyplesin III was applied at concentrations of 50 and 100 mg/L, respectively, and hemolysis accelerated as the concentration of tachyplesin increased further (Figure 2A). Cell viability of L919 cells, the murine fibroblast cell line used for toxicity testing, declined sharply when the concentration of tachyplesin increased from 50 mg/L to 100 mg/L (Figure 2B). Dramatic decreases in viability were observed in 293FT human embryonic kidney cells when tachyplesin was increased from 100 mg/L to 200 mg/L (Figure 2C) and in A549 adenocarcinomic human alveolar basal epithelial cells when tachyplesin was increased 200 mg/L to 400 mg/L (Figure 2D).
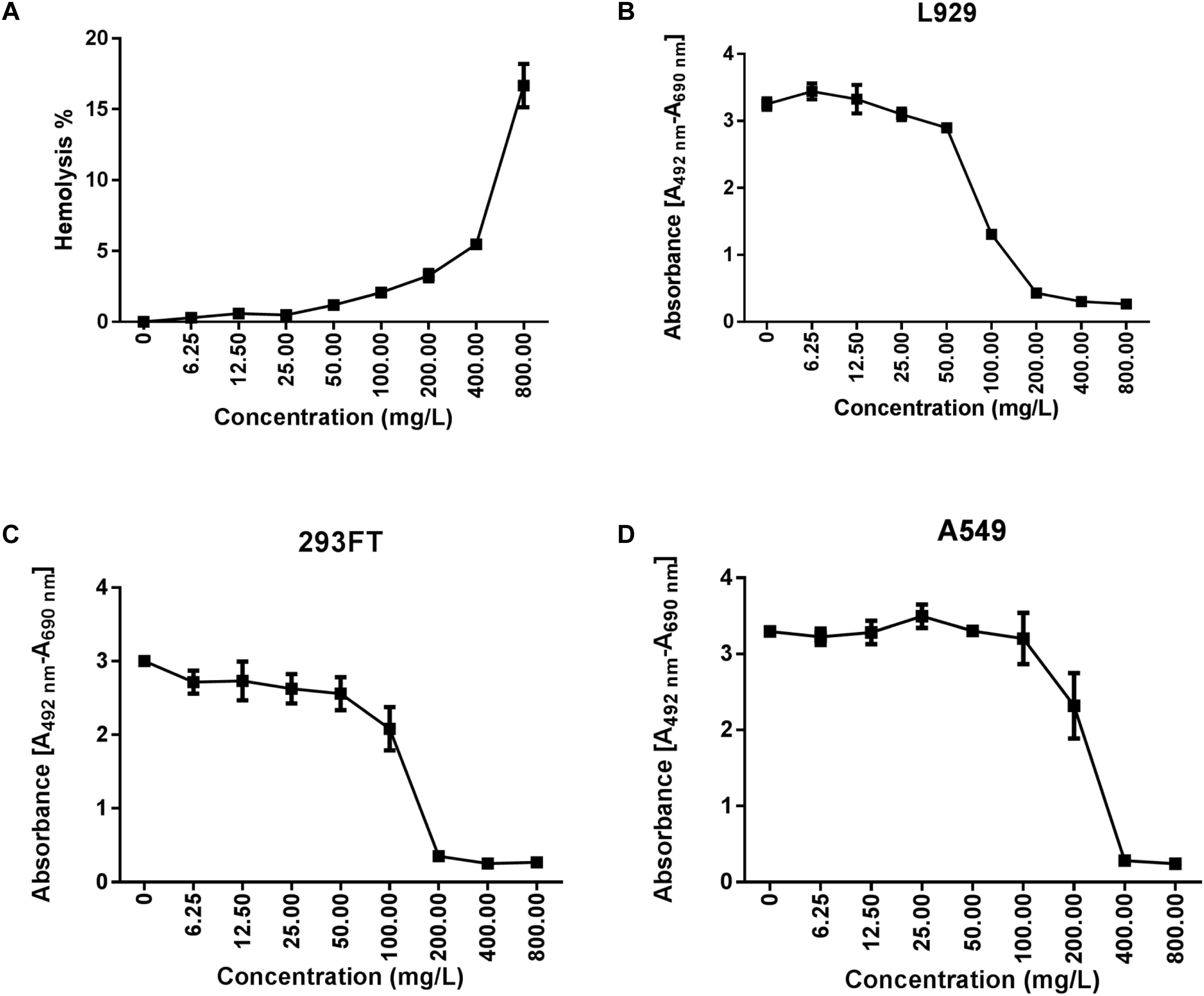
FIGURE 2. Cytotoxicity of tachyplesin III. Cytotoxicity of tachyplesin III was tested by hemolysis and cell viability assays. (A) Tachyplesin III dissolved in 0.9% saline was added to mouse erythrocytes that were diluted with the same buffer. After 30 min of incubation at 37°C and then centrifugation at 1000 g for 15 min, supernatants were diluted 1:4 with 0.9% saline, and absorbance at 540 nm was tested. With 1% Triton X-100 (v/v) to determine 100% hemolysis and 0.9% saline as the negative control, the hemolysis rate was calculated as [(Absorbancesample-Absorbancecontrol)/(Absorbance100%-Absorbancecontrol)] × 100. Cytotoxicity was assessed using the Cell Proliferation Kit II (XTT) (Roche) for (B) L929 (mouse fibroblast cell line), (C) 293FT (human embryonic kidney cell line) and (D) A549 (adenocarcinomic human alveolar basal epithelial cell line) cells. Absorbance [A492-A690 nm] was used to quantify viable cells.
PI is a DNA-binding fluorescent dye that can penetrate broken membranes but not intact membranes. As shown in Figure 3, while levofloxacin at low concentrations (1/4× MIC) did not affect the integrity of bacterial membranes, some of the bacterial membranes were broken slightly after treatment with levofloxacin at 4× MIC for 1 h at 25°C, as indicated by the emergence of weak red PI fluorescence inside these cells. Interestingly, tachyplesin at low concentrations (1/4× MIC) could also induce slight membrane breakage in a small proportion of the treated bacteria, while tachyplesin at high concentrations induced complete membrane breakage of nearly all of the treated bacteria.

FIGURE 3. Propidium iodide staining after treatment with levofloxacin or tachyplesin III at different concentrations. First, 1 × 107 MDR clinical isolates of P. aeruginosa strain 1409 were incubated with 1/4× MIC (4 μg/ml), 4× MIC (64 μg/ml) levofloxacin (Tokyo Chemical Industry Co. Ltd) or 1/4× MIC (2 μg/ml), 4× MIC (32 μg/ml) tachyplesin III in Luria-Bertani (LB) medium at 25°C for 1 h. After centrifugation at 3000 g for 10 min, cells were resuspended in the same volume of PBS (phosphate-buffered saline, 8 g/L NaCl, 0.2 g/L KCl, 1.44 g/L Na2HPO4, 0.24 g/L KH2PO4, pH 7.4). Then, PI was added to a final concentration of 10 μg/ml. After 30 min of incubation at 25°C, the cells were washed three times with PBS and immediately imaged with a fluorescence microscope.
Confocal laser-scanning microscopy showed more detailed results (Figure 4). After treatment with 1/4× MIC at 25°C for 1 h, most of the FITC-tagged tachyplesin (green in Figure 4F, left) co-localized with DNA (stained blue by Hoechst in Figure 4H, left) rather than with bacterial membranes (stained red by SynaptoRed C2 in Figure 4S, left). Treatment with tachyplesin at 4× MIC showed completely different results: tachyplesin was dispersed all over the bacteria (Figure 4F, right), including nuclear regions (Figure 4H, right) and bacterial membranes (Figure 4S, right). Notably, bacteria treated with tachyplesin at 4× MIC showed marked shape changes, including elongated nuclear regions (Figure 4H, right vs. Figure 4H, left) and enlarged cell bodies (Figure 4S right vs. Figure 4S left).
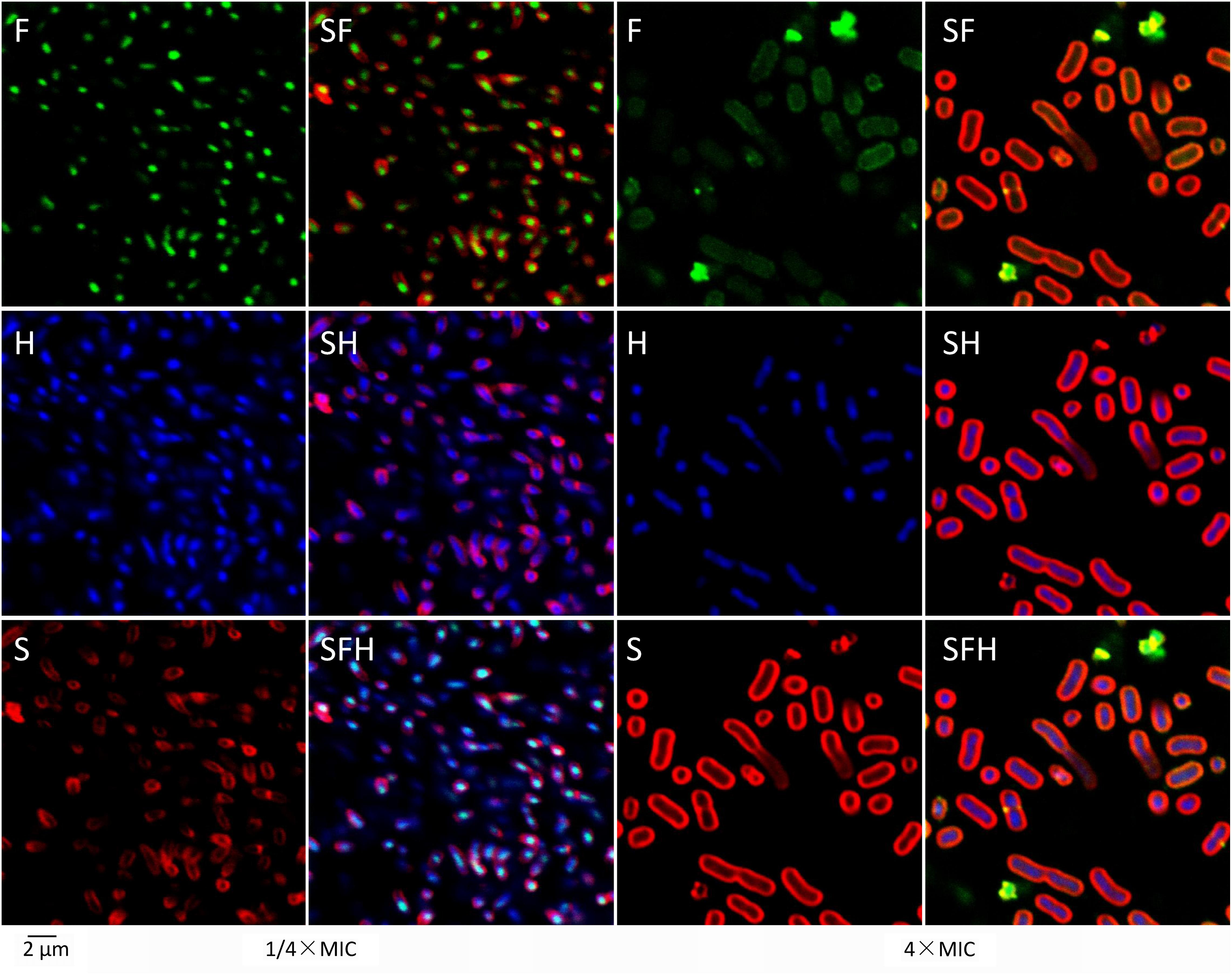
FIGURE 4. Localization of tachyplesin III with confocal laser-scanning microscopy. First, 1 × 107 MDR clinical isolates of A. baumannii strain 1408 were incubated with 1/4× MIC (2 μg/ml) or 4× MIC (32 μg/ml) N-terminally tagged tachyplesin III in LB medium at 25°C for 1 h. After centrifugation at 3000 g for 10 min, cells were resuspended with the same volume PBS and incubated with 20 μg/ml Hoechst (Sigma) at room temperature for 20 min. After another centrifugation at 3000 g for 10 min, cells were resuspended with the same volume of Hank’s solution (8 g/L NaCl, 0.4 g/L KCl, 1 g/L glucose, 60 mg/L KH2PO4, 47.5 mg/L Na2HPO4, pH 7.2) and incubated with 20 μg/ml SynaptoRed C2 (Tocris Bioscience) on ice for 1 min. Confocal laser-scanning microscopy was performed with excitation and emission wavelengths, respectively, of 488 nm and 530 nm for FITC, 352 nm and 461 nm for Hoechst, and 515 and 640 nm for SynaptoRed C2.
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive at the Beijing Institute of Genomics, Chinese Academy of Sciences, under accession number CRA000801 and are publicly accessible at http://bigd.big.ac.cn/gsa (Wang et al., 2017; Big Data Center Members, 2018). Unique read numbers for each expressed gene were calculated using FPKM and are shown in Supplementary Data Sheet S1. Fold changes of expressed genes and corresponding p-values are shown in Supplementary Data Sheet S2.
Among all of the KEGG pathways shared in all of the pathogens subjected to transcriptional analysis after tachyplesin treatment, the “biosynthesis of unsaturated fatty acids” pathway (KEGG ID 01040) was not enriched in any of the pathogens treated with levofloxacin (Table 1). Although the geraniol degradation pathway (KEGG ID 00281) was detected in the transcriptional analysis of tachyplesin-treated pathogens and undetected in levofloxacin-treated pathogens, such transcriptional changes were not shared by all of the pathogens tested, and this pathway was excluded from further analysis. The nicotinate and nicotinamide metabolism pathway, which was shared among all of the pathogens subjected to transcriptional analysis (and verified on the proteomic level based on unpublished data) after tachyplesin treatment, was excluded for similar reasons. Though transcriptional changes in this pathway were not detected after levofloxacin treatment, this pathway was affected at the proteomic level after levofloxacin treatment in both of the pathogens tested (A. baumannii 1408 and P. aeruginosa 1409, in unpublished data).
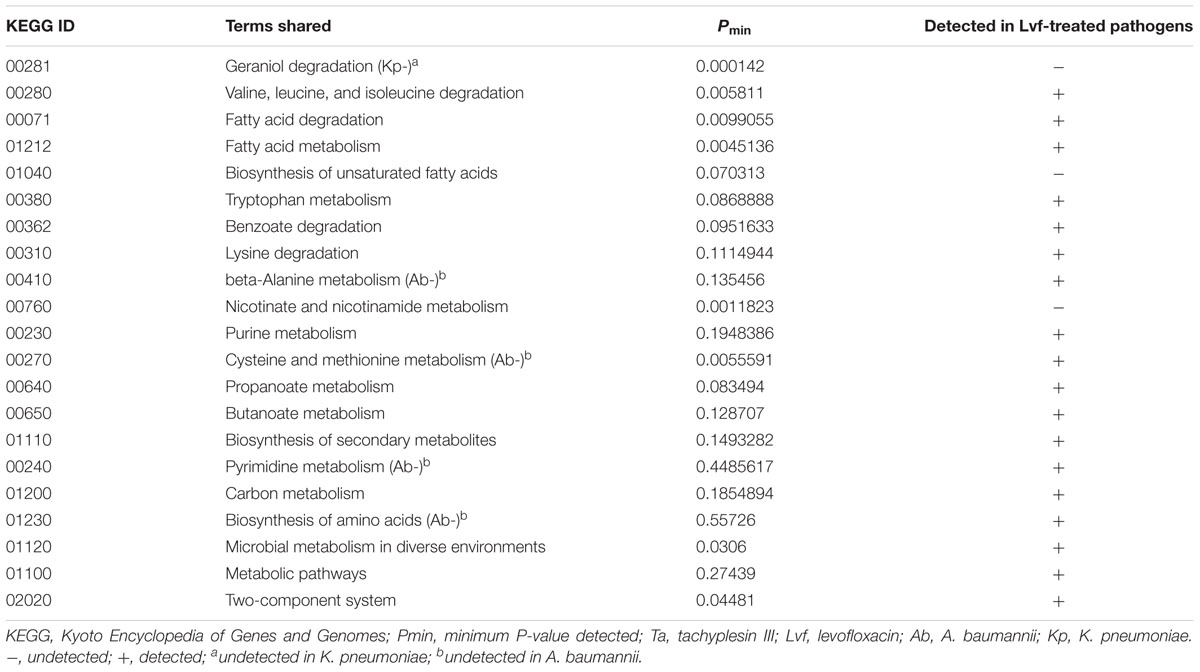
TABLE 1. Affected KEGG pathways shared among Ta-treated pathogens after transcriptome analysis and their existence in Lvf-treated pathogens.
Supplementary Figure S1 shows details of the biosynthetic pathways for unsaturated fatty acids of all the bacteria tested. Notably, the FabG gene, which encodes 3-ketoacyl-acyl carrier protein reductase, was the only gene with elevated expression detected after tachyplesin III treatment in all four bacteria (Table 2). These elevations after tachyplesin III treatment were also confirmed by quantitative real-time PCR (Figure 5).
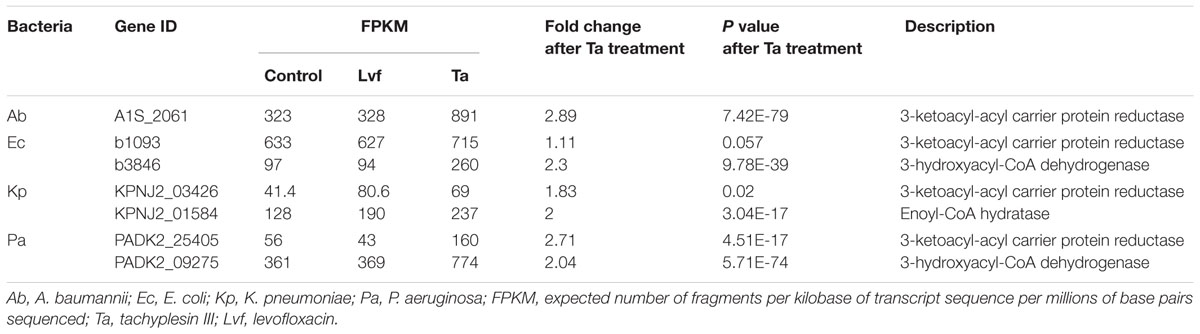
TABLE 2. Differentially expressed genes in the biosynthesis of unsaturated fatty acids pathways after treatment.
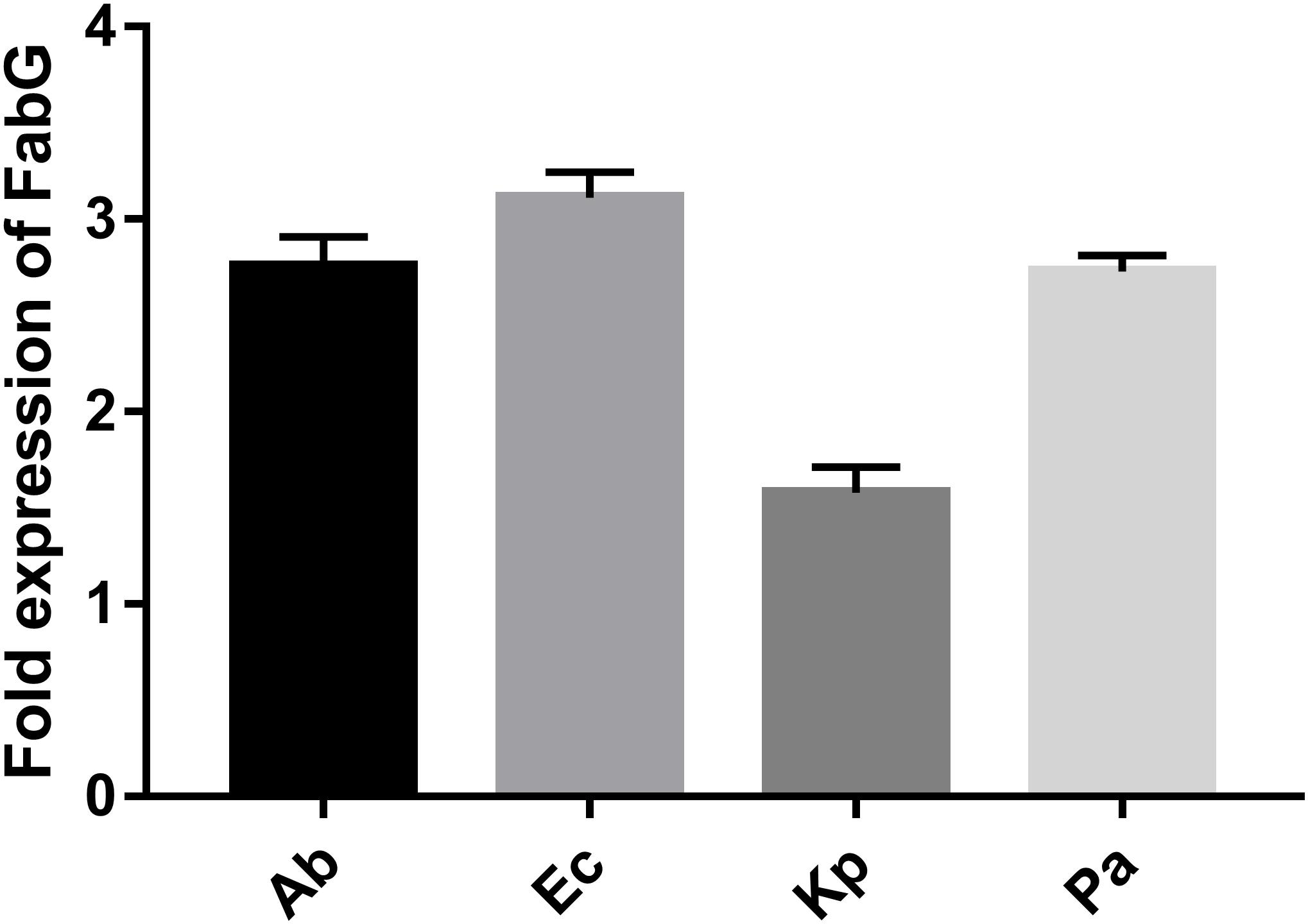
FIGURE 5. Expression of FabG increased after tachyplesin treatment. Quantitative real-time PCR was performed following the 2-ΔΔCT method. Untreated pathogen strains were used as the calibrator strains, and the corresponding ribosomal rpsL genes were used as housekeeping reference gene with the primers shown in Supplementary Table S1.
As shown in Figure 6A, three of the RNA oligos (1259, 1768, and 1430) that we selected worked well and significantly down-regulated the expression of FabG in P. aeruginosa (p < 0.01), which was accompanied by reduced bacterial colony survival (Figure 6B). However, for RNA oligo 1534, which was inefficient at down-regulating FabG in P. aeruginosa, transformation of this RNA oligo had no effect on the survival rate of bacterial colonies.
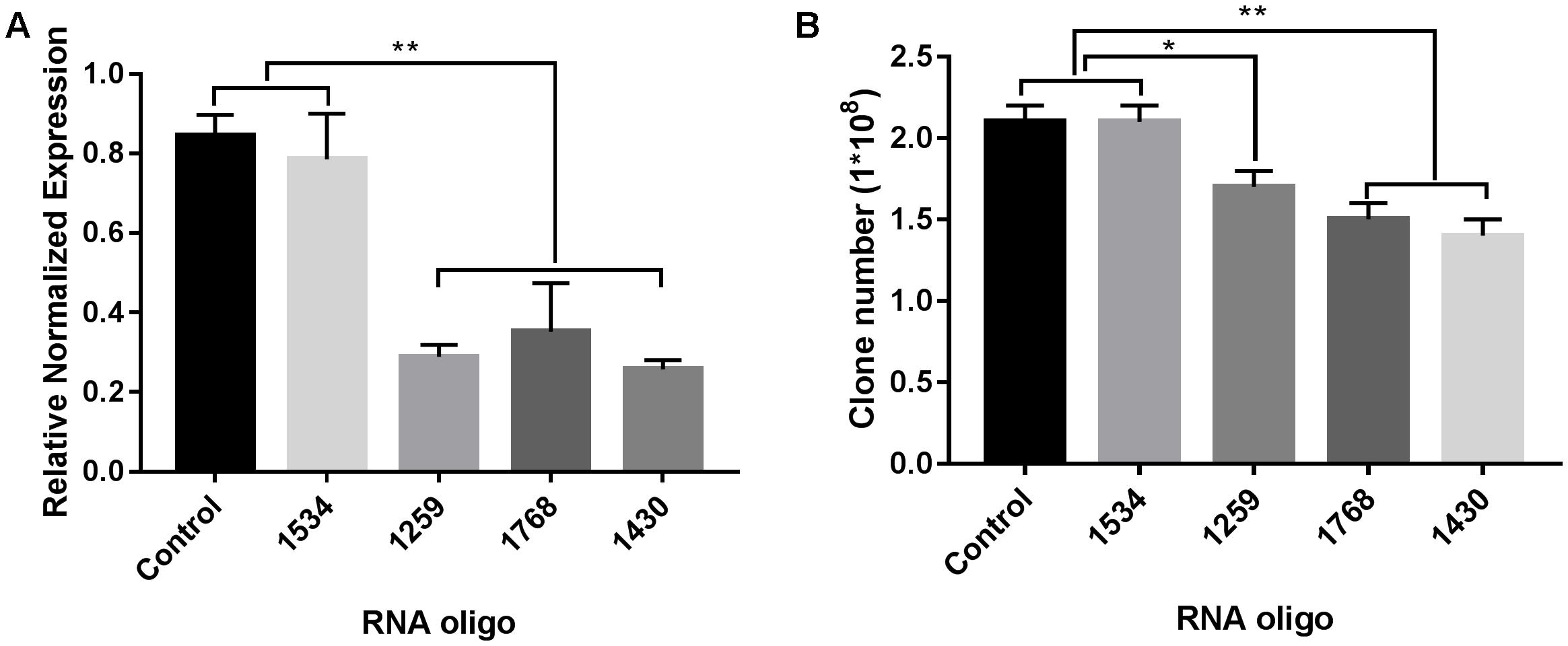
FIGURE 6. RNA silencing of FabG reduced bacterial colony counts. Using the RNA oligo pairs shown in Supplementary Table S2 (GenePharma Co., Ltd, Shanghai, China) to target FabG from P. aeruginosa, RNAi was carried out with a GenePulser Xcell system using a 2 mm cuvette with the default settings for electrotransformation-competent P. aeruginosa cells (2.5 kV/cm, 25 μF and 200 Ω). The transformants were either spread onto LB agar plates to calculate bacterial clones (B) or incubated in LB medium at 37°C for 40 min to determine FabG transcripts by RT-PCR (A) with the primers for P. aeruginosa shown in Supplementary Table S1. Statistical significance was analyzed by one-way ANOVA followed by the Newman–Keuls multiple-comparisons test, ∗∗p < 0.01, ∗p < 0.05.
Surface plasmon resonance analysis showed that tachyplesin III could bind with FabG from E. coli (Figure 7A) or S. aureus (Figure 7E) in a dose-dependent manner. This binding interaction between tachyplesin III and FabG is accompanied by inhibition of the enzyme activities of FabG (Figures 7B,F). While FabG from E. coli consumed 54% of NADPH after incubation for 5 min at room temperature, incubation with 15, 7.5 and 3.75 μM tachyplesin III reduced the consumption rates to 22, 29, and 34%, respectively (Figure 7B). The tests with FabG from S. aureus showed even more marked results. While FabG from S. aureus consumed 42% of NADPH after 5 min of incubation at room temperature, only 4% of NADPH was consumed after incubation with 3.75 μM tachyplesin III, and FabG activity was almost completely blocked after incubation with higher concentrations of tachyplesin III (Figure 7F).
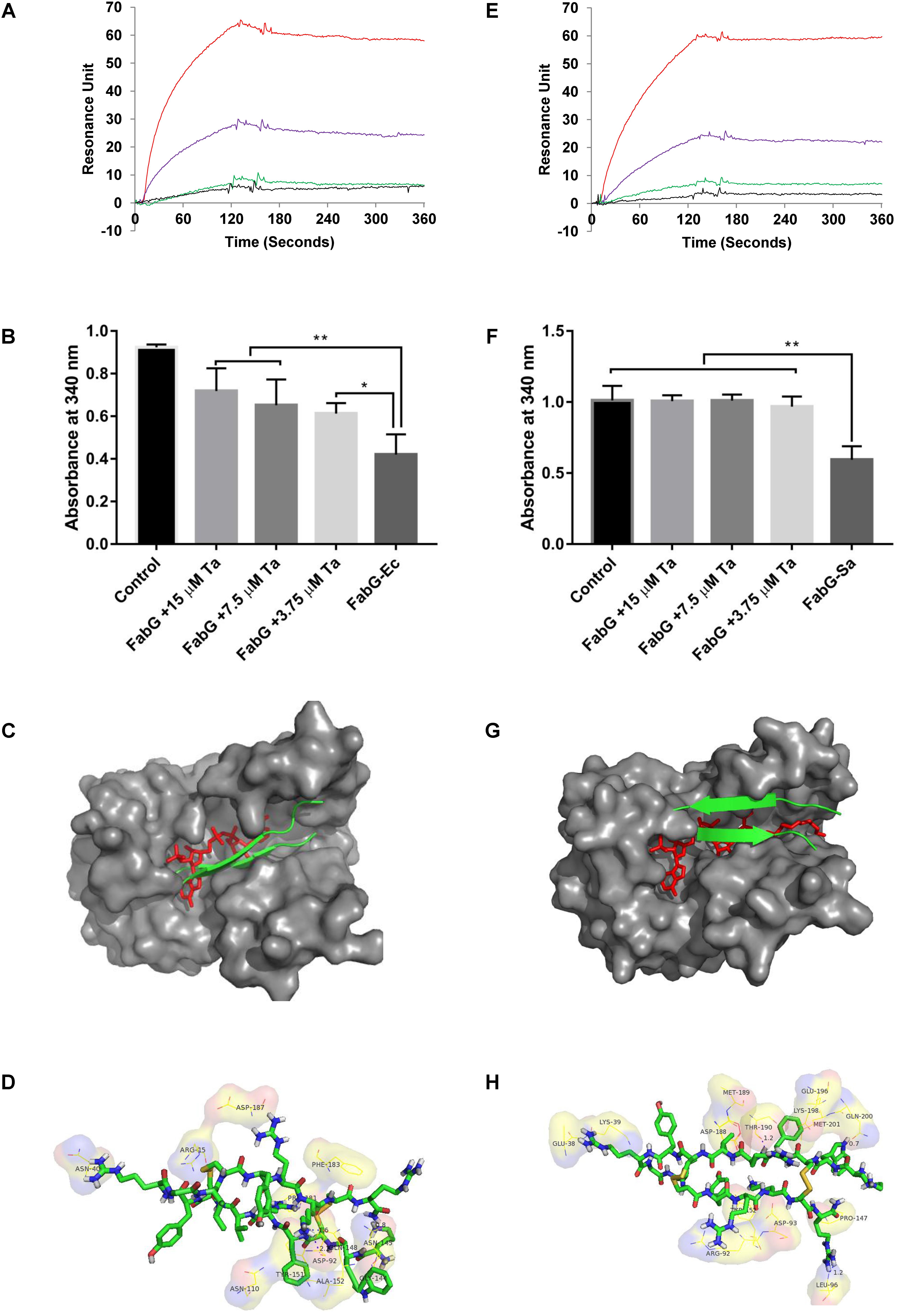
FIGURE 7. Tachyplesin III binding with FabG inhibits its enzyme activities in vitro, probably by blocking the active site. Binding interactions between tachyplesin III and FabG from E. coli (A) or from S. aureus (E) were analyzed by surface plasmon resonance (SPR) with a Biocore 3000 (Biocore, Piscataway, NJ, United States). Black line: 1.25 μg/mL FabG; green line: 2.5 μg/mL FabG; purple line: 5 μg/mL FabG; and red line: 10 μg/mL FabG. The inhibitory activity of tachyplesin III on FabG from E. coli (B) or from S. aureus (F) was evaluated based on the remaining NADPH (which shows absorbance at 340 nm at 𝜀340 = 6.3 × 103 mol-1) after FabG consumption for 5 min at room temperature. Tachyplesin was processed with Chemdraw 11.0 according to the structure of tachyplesin I (PDB code 2RTV) and then docked to the active site of FabG from E. coli (PDB code 1Q7B) or S. aureus (PDB code 3SJ7) with AutoDock4 using the default settings. Docking of tachyplesin III into E. coli is shown in (C), docking details are shown in (D). Docking of tachyplesin III into S. aureus is shown in (G), docking details are shown in (H). In (C,G), tachyplesin III is shown in green, and NADPH is shown in red. Statistical significance was analyzed by one-way ANOVA followed by the Newman–Keuls multiple-comparisons test, ∗∗p < 0.01, ∗p < 0.05.
As tachyplesin III could bind with FabG and inhibit its enzyme activities, we next predicted whether tachyplesin III could block the active site of FabG. Molecular docking analysis results showed that tachyplesin III could fit into the substrate-binding region of FabG proteins from both E. coli (Figure 7C) and S. aureus (Figure 7G), which is consistent with its efficacy against both Gram-negative and Gram-positive bacteria. Detailed analysis showed that tachyplesin could interact with the active site of FabG via hydrophobic interactions and van der Waals forces. Tachyplesin interacted with R15, N145, Y151, A152, N40, D92, and F183 and formed hydrogen bonds with N40, D92, N145, and Q148 in the active site of E. coli FabG (Figure 7D); in S. aureus, it interacted with K39, R92, D93, L96, P147, Y152, K198, and Q200 and formed hydrogen bonds with Leu96, Thr190, and Gln200 in the active site (Figure 7H).
Since the discovery of tachyplesin approximately 30 years ago, its potential and broad-spectrum activities against MDR bacteria have been repeatedly confirmed (Nakamura et al., 1988). Though experimental induction of resistance to tachyplesin was reported recently in Gram-negative bacteria, with the resistance mechanism of P. aeruginosa “not entirely dependent on extracellular proteolytic degradation of tachyplesin,” this resistance ability quickly disappeared after 6–9 passages without tachyplesin pressure because maintaining resistance may be a huge cost, similar to resistance to colistin (Fernandez-Reyes et al., 2009; Hong et al., 2015). Moreover, our results showed that tachyplesin III was stable in mouse serum for at least 6 h (Figure 1). As mouse serum is enriched in endogenous mammalian proteases, these results demonstrated that tachyplesin was resistant to proteolysis and may be stable in vivo.
Compared with serum stability issues, cytotoxicity seems to be the real inherent defect hindering the clinical application of tachyplesin (Edwards et al., 2016). At a final concentration of 100 mg/L, tachyplesin caused lysis of approximately 2% of hemocytes (Figure 2A), as well as a decline in the cell viability of 293FT human embryonic kidney cells (Figure 2C) and a sharp decline in the cell viability of L919 murine fibroblasts (Figure 2B), a cell line commonly used for toxicity testing. Interestingly, tumor cells such as A549 adenocarcinomic human alveolar basal epithelial cells seem to be more resistant to tachyplesin (Figure 2D), which made the reports of its anticancer effects somewhat controversial (Evans et al., 2017; Li et al., 2017).
A deep understanding of how tachyplesin works may help in developing novel effective antibiotics based on similar mechanisms while avoiding the problematic cytotoxicity. Compared with the DNA binding theory, the membrane rupture theory seems to be more likely as bacterial membrane rupture is an obvious phenomenon after tachyplesin treatment at both lethal and sublethal concentrations (Figure 3). It is natural to deduce that tachyplesin may act directly on membranes and form leakage pores to kill bacteria, and numerous studies have focused on the direct interactions between tachyplesin and membranes, mainly using artificial lipid bilayers (Matsuzaki et al., 1997; Doherty et al., 2006; Han and Lee, 2015). Interestingly, these studies have found that linear analogs of tachyplesin without disulfides also possess the membrane disruptive abilities of original tachyplesin but translocate through these bilayers inefficiently and accompanied by weaker antibacterial activities, implying that membrane translocation abilities instead of direct membrane rupture abilities are vital for its antibacterial activity (Matsuzaki et al., 1997). Furthermore, PEGylation of tachyplesin did not alter the toroidal pore formation and membrane translocation activities of tachyplesin but reduced its cytotoxicity and antimicrobial activities, implying that events inside the cell may determine both the cytotoxicity and the antimicrobial activities of tachyplesin (Imura et al., 2007).
Proteomic profiling showed that low concentrations of tachyplesin may promote apoptosis in the glioblastoma multiforme cell line U251 by affecting intracellular enzymes and cause cell death in E. coli by inactivating intracellular esterases (Hong et al., 2015; Li et al., 2017). However, treatment with a high concentration of tachyplesin caused the rapid death of K562 human erythroleukemia cells and E. coli, accompanied by complete cell membrane rupture in both cases (Paredes-Gamero et al., 2012; Hong et al., 2015). This phenomenon was also observed in our studies, in which sublethal concentrations of tachyplesin aggregated at specific regions within bacteria (Figure 4, 1/4× MIC) and caused slight membrane rupture in a small proportion of the bacteria (Figure 3, 1/4× MIC), whereas a lethal concentration of tachyplesin dispersed all over the bacteria (Figure 4, 4× MIC) and caused complete membrane rupture in nearly all of the treated bacteria (Figure 3, 4× MIC).
Two phenomena should be noted among these results. First, only some of the bacteria treated with lethal concentrations of levofloxacin showed membrane rupture, and the membrane rupture was minor in most cases, as demonstrated by the limited entrance of PI into treated bacteria and the weak fluorescent signals detected (Figure 3, 4× MIC for levofloxacin). As levofloxacin is known to disturb DNA separation and supercoiling by inhibiting bacterial topoisomerases, thereby killing the cells, the slight membrane rupture is presumably the result of cell death (Drlica and Zhao, 1997; Ferrandiz and de la Campa, 2014). By contrast, nearly all the bacteria treated with lethal concentrations of tachyplesin showed bright fluorescent signals (Figure 3, 4× MIC for tachyplesin), demonstrating that all of the bacteria killed by tachyplesin showed thorough membrane rupture and thus indicating that tachyplesin likely kills bacteria by targeting the membrane. To our surprise, existing theories concerning this topic are mainly based on the depiction of the physical interactions between this peptide and artificial lipid bilayers, and few biological details are offered. A dual effect theory emerged recently, suggesting that tachyplesin enter bacteria via the formation of non-lethal membrane pores, cause sub-lethal effects at low concentrations by inactivating intracellular enzymes, and cause cell membrane disruption at high concentrations. It may be difficult for a peptide to perform so many functions to kill bacteria, and some of those functions are unnecessary because they are not lethal. Moreover, no further biological details concerning membrane rupture have been reported (Paredes-Gamero et al., 2012; Hong et al., 2015). Second, bacteria treated with lethal concentrations of tachyplesin showed elongated nuclear regions (Figure 4H, right vs. Figure 4H, left) and enlarged cell bodies (Figure 4S, right vs. Figure 4S, left). This phenomenon was revealed by an approach called bacterial cytological profiling (BCP) that is quite helpful for characterizing the action mechanisms of antibiotics (Nonejuie et al., 2013). Though membrane lengths and DNA lengths of bacteria treated with lethal concentrations of tachyplesin were approximately twice those of bacteria treated with sublethal concentrations, which is consistent with the lipid biosynthesis inhibitors reported in this paper, we have no corresponding models or exact parameters for determining the mechanism of action with the BCP method.
As our laboratory could not conduct the BCP analysis, we turned to omics techniques to identify the effects of tachyplesin treatment on bacteria and to elucidate its action mechanisms (Kohanski et al., 2010; Pulido et al., 2016). Compared with levofloxacin-treated bacteria, all of the pathogens treated with tachyplesin III showed marked effects on the biosynthesis of unsaturated fatty acids, which is important for membrane fluidity, and this result was verified at the protein level (Table 1). In this pathway, the β-ketoacyl-acyl carrier protein (ACP) reductase FabG was up-regulated in all the bacteria tested (Figure 5 and Table 2). RNA silencing of FabG showed that efficient silencing of FabG by RNA oligos (Figure 6A) is accompanied by the reduced survival of bacterial colonies (Figure 6B), which is consistent with previous reports that FabG is a conserved and essential gene in various bacteria such that knockout of this enzyme results in bacterial death (Zhang et al., 2003; Lai and Cronan, 2004; Baba et al., 2006; Parish et al., 2007; Cukier et al., 2013).
The biosynthesis of unsaturated fatty acids occurs in the cytoplasm, and the membrane translocation activities of tachyplesin accompany its antimicrobial activities. FabG is essential for the growth of bacteria, and inhibition of FabG is associated with antimicrobial activity in both E. coli and S. aureus (Sohn et al., 2008). Thus, we propose that tachyplesin may kill bacteria by inhibiting FabG activities. Our surface plasmon resonance analysis results showed that tachyplesin III could bind with FabG from E. coli (Figure 7A) and S. aureus (Figure 7E) in a dose-dependent manner. This binding interaction between tachyplesin III and FabG is accompanied by inhibition of the enzyme activities of FabG (Figures 7B,F). Molecular docking predictions showed that tachyplesin III could not only enter the active site of FabG from E. coli by hydrophobic interactions and van der Waals forces but also block the NADPH binding site by interacting with the corresponding amino acid residues and forming stable hydrogen bonds (Figures 7C,D). A similar phenomenon was observed for the interaction of tachyplesin III and Gram-positive S. aureus (Figures 7G,H), which is consistent with the broad antimicrobial spectrum of tachyplesin.
Many antibiotics targeting bacterial pathways for fatty acid biosynthesis show some cytotoxicity to mammalian cells. For example, triclosan can kill bacteria by binding its enoyl-acyl carrier protein reductase (Nonejuie et al., 2013), but it also shows severe hemolytic activity (Singh et al., 2003). A similar phenomenon was observed for cerulenin, which can kill bacteria by binding bacterial fatty acid synthase (Nonejuie et al., 2013). Though no hemolytic activity was reported, cerulenin was reported to be cytotoxic to carcinoma cells (Pizer et al., 1996). Whether tachyplesin III may cause toxicity to mammalian cells via similar mechanism remains to be validated. Though the fatty acid synthetic pathway in bacteria (FAS II) consists of multiple individual enzymes, whereas the fatty acid synthetic pathway in mammals (FAS I) is a single protein with multifunctional polypeptides, sequence homologies may exist at similar active sites, and inhibitors may have similar effects on both the bacterial and mammalian fatty acid synthesis pathways. These phenomena remind us that more-specific inhibitors may be selected or designed to avoid cytotoxic effects when targeting these fatty acid synthases.
Efficient entrance into cells to act on FabGs that are located in the cytoplasm may be another strategy that we should adopt from tachyplesin. It was reported that inhibitors targeting an allosteric binding site of FabG showed effects only toward Gram-positive bacteria and not toward Gram-negative P. aeruginosa (Cukier et al., 2013). This may be attributable either to poor penetration activities of these inhibitors or to rapid efflux by P. aeruginosa. Though tachyplesin seems to target a different active site, designing inhibitors based on the action mechanism of tachyplesin and possessing penetration activity may be beneficial.
We find that tachyplesin may kill bacteria by targeting FabG, the conserved β-ketoacyl-acyl carrier protein reductase in unsaturated fatty acid biosynthesis. Targeting the homologous enzyme in mammalian cells may also result in instability or rupture of cell membranes, which manifested as cytotoxicity, including hemolysis. More-detailed research concerning the structural differences between bacterial and mammalian FabG proteins, as well as the selection of more specific inhibitory ligands, may be helpful for the development of safer antibiotics designed based on the mechanism of tachyplesin.
CL conceived and designed the experiments. CL, JQ, and BS performed the experiments. CL and JQ analyzed the data. BS contributed reagents, materials and analysis tools. CL and YM contributed to the writing of the manuscript.
This work was financially supported by the National Natural Science Foundation of China (grant numbers 81503117 and 81460322), the CAMS Initiative for Innovative Medicine (grant number 2017-I2M-3-022), the Foundation for Studying Abroad from State Administration of Foreign Experts Affairs China and China Scholarship Council (grant number 201808110121), the Fundamental Research Funds for the Central Universities of China (grant numbers 2016ZX350073 and 3332013084), and the Opening Foundation of the Key Laboratory of Bioactive Peptides of Yunnan Province (grant number AMHD-2017-1).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00825/full#supplementary-material
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 1–11. doi: 10.1038/msb4100050
Big Data Center Members (2018). Database resources of the BIG Data Center in 2018. Nucleic Acids Res. 46, D14–D20. doi: 10.1093/nar/gkx897
Cirioni, O., Ghiselli, R., Silvestri, C., Kamysz, W., Orlando, F., Mocchegiani, F., et al. (2007). Efficacy of tachyplesin III, colistin, and imipenem against a multiresistant Pseudomonas aeruginosa strain. Antimicrob. Agents Chemother. 51, 2005–2010. doi: 10.1128/AAC.01576-06
Cukier, C. D., Hope, A. G., Elamin, A. A., Moynie, L., Schnell, R., Schach, S., et al. (2013). Discovery of an allosteric inhibitor binding site in 3-Oxo-acyl-ACP reductase from Pseudomonas aeruginosa. ACS Chem. Biol. 8, 2518–2527. doi: 10.1021/cb4005063
Doherty, T., Waring, A. J., and Hong, M. (2006). Peptide-lipid interactions of the beta-hairpin antimicrobial peptide tachyplesin and its linear derivatives from solid-state NMR. Biochim. Biophys. Acta 1758, 1285–1291. doi: 10.1016/j.bbamem.2006.03.016
Doherty, T., Waring, A. J., and Hong, M. (2008). Dynamic structure of disulfide-removed linear analogs of tachyplesin-I in the lipid bilayer from solid-state NMR. Biochemistry 47, 1105–1116. doi: 10.1021/bi701390t
Drlica, K., and Zhao, X. (1997). DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61, 377–392.
Dumas, J. L., van Delden, C., Perron, K., and Kohler, T. (2006). Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. FEMS Microbiol. Lett. 254, 217–225. doi: 10.1111/j.1574-6968.2005.00008.x
Dutta, D., Bhattacharyya, S., and Das, A. K. (2012). Crystal structure and fluorescence studies reveal the role of helical dimeric interface of staphylococcal FabG1 in positive cooperativity for NADPH. Proteins 80, 1250–1257. doi: 10.1002/prot.24024
Edwards, I. A., Elliott, A. G., Kavanagh, A. M., Blaskovich, M. A. T., and Cooper, M. A. (2017). Structure-activity and -toxicity relationships of the antimicrobial peptide tachyplesin-1. ACS Infect. Dis. 3, 917–926. doi: 10.1021/acsinfecdis.7b00123
Edwards, I. A., Elliott, A. G., Kavanagh, A. M., Zuegg, J., Blaskovich, M. A., and Cooper, M. A. (2016). Contribution of amphipathicity and hydrophobicity to the antimicrobial activity and cytotoxicity of beta-hairpin peptides. ACS Infect. Dis. 2, 442–450. doi: 10.1021/acsinfecdis.6b00045
Evans, H. G., Guthrie, J. W., Jujjavarapu, M., Hendrickson, N., Eitel, A., Park, Y., et al. (2017). D-amino acid analogues of the antimicrobial peptide CDT exhibit anti- cancer properties in A549, a human lung adenocarcinoma cell line. Protein Pept. Lett. 24, 590–598. doi: 10.2174/0929866524666170621093647
Fernandez-Reyes, M., Rodriguez-Falcon, M., Chiva, C., Pachon, J., Andreu, D., and Rivas, L. (2009). The cost of resistance to colistin in Acinetobacter baumannii: a proteomic perspective. Proteomics 9, 1632–1645. doi: 10.1002/pmic.200800434
Ferrandiz, M. J., and de la Campa, A. G. (2014). The fluoroquinolone levofloxacin triggers the transcriptional activation of iron transport genes that contribute to cell death in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 58, 247–257. doi: 10.1128/AAC.01706-13
Han, E., and Lee, H. (2013). Effects of PEGylation on the binding interaction of magainin 2 and tachyplesin I with lipid bilayer surface. Langmuir 29, 14214–14221. doi: 10.1021/la4036985
Han, E., and Lee, H. (2015). Structural effects of tachyplesin I and its linear derivative on their aggregation and mobility in lipid bilayers. J. Mol. Graph. Model. 59, 123–128. doi: 10.1016/j.jmgm.2015.04.007
Ho, Y. H., Shah, P., Chen, Y. W., and Chen, C. S. (2016). Systematic analysis of intracellular-targeting antimicrobial peptides, bactenecin 7, hybrid of pleurocidin and dermaseptin, proline-arginine-rich peptide, and lactoferricin B, by Using Escherichia coli proteome microarrays. Mol. Cell. Proteomics 15, 1837–1847. doi: 10.1074/mcp.M115.054999
Hong, J., Guan, W., Jin, G., Zhao, H., Jiang, X., and Dai, J. (2015). Mechanism of tachyplesin I injury to bacterial membranes and intracellular enzymes, determined by laser confocal scanning microscopy and flow cytometry. Microbiol. Res. 170, 69–77. doi: 10.1016/j.micres.2014.08.012
Hong, J., Hu, J., and Ke, F. (2016). Experimental induction of bacterial resistance to the antimicrobial peptide tachyplesin I and investigation of the resistance mechanisms. Antimicrob. Agents Chemother. 60, 6067–6075. doi: 10.1128/AAC.00640-16
Imura, Y., Nishida, M., Ogawa, Y., Takakura, Y., and Matsuzaki, K. (2007). Action mechanism of tachyplesin I and effects of PEGylation. Biochim. Biophys. Acta 1768, 1160–1169. doi: 10.1016/j.bbamem.2007.01.005
Katsu, T., Nakao, S., and Iwanaga, S. (1993). Mode of action of an antimicrobial peptide, tachyplesin I, on biomembranes. Biol. Pharm. Bull. 16, 178–181. doi: 10.1248/bpb.16.178
Kawano, K., Yoneya, T., Miyata, T., Yoshikawa, K., Tokunaga, F., Terada, Y., et al. (1990). Antimicrobial peptide, tachyplesin I, isolated from hemocytes of the horseshoe crab (Tachypleus tridentatus). NMR determination of the beta-sheet structure. J. Biol. Chem. 265, 15365–15367.
Kohanski, M. A., Dwyer, D. J., and Collins, J. J. (2010). How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 8, 423–435. doi: 10.1038/nrmicro2333
Kristan, K., Bratkovic, T., Sova, M., Gobec, S., Prezelj, A., and Urleb, U. (2009). Novel inhibitors of beta-ketoacyl-ACP reductase from Escherichia coli. Chem. Biol. Interact. 178, 310–316. doi: 10.1016/j.cbi.2008.09.030
Kushibiki, T., Kamiya, M., Aizawa, T., Kumaki, Y., Kikukawa, T., Mizuguchi, M., et al. (2014). Interaction between tachyplesin I, an antimicrobial peptide derived from horseshoe crab, and lipopolysaccharide. Biochim. Biophys. Acta 1844, 527–534. doi: 10.1016/j.bbapap.2013.12.017
Lai, C. Y., and Cronan, J. E. (2004). Isolation and characterization of beta-ketoacyl-acyl carrier protein reductase (fabG) mutants of Escherichia coli and Salmonella enterica serovar Typhimurium. J. Bacteriol. 186, 1869–1878. doi: 10.1128/JB.186.6.1869-1878.2004
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, J., Giesy, J. P., Yu, L., Li, G., and Liu, C. (2015). Effects of Tris(1,3-dichloro-2-propyl) phosphate (TDCPP) in tetrahymena thermophila: targeting the ribosome. Sci. Rep. 5:10562. doi: 10.1038/srep10562
Li, X., Dai, J., Tang, Y., Li, L., and Jin, G. (2017). Quantitative proteomic profiling of tachyplesin I targets in U251 gliomaspheres. Mar. Drugs 15:20. doi: 10.3390/md15010020
Liu, B., Zhang, Y., and Zhang, W. (2014). RNA-Seq-based analysis of cold shock response in Thermoanaerobacter tengcongensis, a bacterium harboring a single cold shock protein encoding gene. PLoS One 9:e93289. doi: 10.1371/journal.pone.0093289
Liu, C., Shan, B., Qi, J., and Ma, Y. (2017). Systemic responses of multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii following exposure to the antimicrobial peptide cathelicidin-BF imply multiple intracellular targets. Front. Cell. Infect. Microbiol. 7:466. doi: 10.3389/fcimb.2017.00466
Liu, C., Yang, X., Yao, Y., Huang, W., Sun, W., and Ma, Y. (2014). Diverse expression levels of two codon-optimized genes that encode human papilloma virus type 16 major protein L1 in Hansenula polymorpha. Biotechnol. Lett. 36, 937–945. doi: 10.1007/s10529-014-1455-z
Liu, C. B., Shan, B., Bai, H. M., Tang, J., Yan, L. Z., and Ma, Y. B. (2015). Hydrophilic/hydrophobic characters of antimicrobial peptides derived from animals and their effects on multidrug resistant clinical isolates. Dongwuxue Yanjiu 36, 41–47. doi: 10.13918/j.issn.2095-8137.2015.1.41
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Matsuzaki, K., Fukui, M., Fujii, N., and Miyajima, K. (1991). Interactions of an antimicrobial peptide, tachyplesin I, with lipid membranes. Biochim. Biophys. Acta 1070, 259–264. doi: 10.1016/0005-2736(91)90173-6
Matsuzaki, K., Nakayama, M., Fukui, M., Otaka, A., Funakoshi, S., Fujii, N., et al. (1993). Role of disulfide linkages in tachyplesin-lipid interactions. Biochemistry 32, 11704–11710. doi: 10.1021/bi00094a029
Matsuzaki, K., Yoneyama, S., Fujii, N., Miyajima, K., Yamada, K., Kirino, Y., et al. (1997). Membrane permeabilization mechanisms of a cyclic antimicrobial peptide, tachyplesin I, and its linear analog. Biochemistry 36, 9799–9806. doi: 10.1021/bi970588v
Miyata, T., Tokunaga, F., Yoneya, T., Yoshikawa, K., Iwanaga, S., Niwa, M., et al. (1989). Antimicrobial peptides, isolated from horseshoe crab hemocytes, tachyplesin II, and polyphemusins I and II: chemical structures and biological activity. J. Biochem. 106, 663–668. doi: 10.1093/oxfordjournals.jbchem.a122913
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., et al. (2009). AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791. doi: 10.1002/jcc.21256
Muta, T., Fujimoto, T., Nakajima, H., and Iwanaga, S. (1990). Tachyplesins isolated from hemocytes of Southeast Asian horseshoe crabs (Carcinoscorpius rotundicauda and Tachypleus gigas): identification of a new tachyplesin, tachyplesin III, and a processing intermediate of its precursor. J. Biochem. 108, 261–266. doi: 10.1093/oxfordjournals.jbchem.a123191
Nakamura, T., Furunaka, H., Miyata, T., Tokunaga, F., Muta, T., Iwanaga, S., et al. (1988). Tachyplesin, a class of antimicrobial peptide from the hemocytes of the horseshoe crab (Tachypleus tridentatus). Isolation and chemical structure. J. Biol. Chem. 263, 16709–16713.
Nonejuie, P., Burkart, M., Pogliano, K., and Pogliano, J. (2013). Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc. Natl. Acad. Sci. U.S.A. 110, 16169–16174. doi: 10.1073/pnas.1311066110
Ohta, M., Ito, H., Masuda, K., Tanaka, S., Arakawa, Y., Wacharotayankun, R., et al. (1992). Mechanisms of antibacterial action of tachyplesins and polyphemusins, a group of antimicrobial peptides isolated from horseshoe crab hemocytes. Antimicrob. Agents Chemother. 36, 1460–1465. doi: 10.1128/AAC.36.7.1460
Paredes-Gamero, E. J., Martins, M. N., Cappabianco, F. A., Ide, J. S., and Miranda, A. (2012). Characterization of dual effects induced by antimicrobial peptides: regulated cell death or membrane disruption. Biochim. Biophys. Acta 1820, 1062–1072. doi: 10.1016/j.bbagen.2012.02.015
Parish, T., Roberts, G., Laval, F., Schaeffer, M., Daffe, M., and Duncan, K. (2007). Functional complementation of the essential gene fabG1 of Mycobacterium tuberculosis by Mycobacterium smegmatis fabG but not Escherichia coli fabG. J. Bacteriol. 189, 3721–3728. doi: 10.1128/JB.01740-06
Pizer, E. S., Wood, F. D., Pasternack, G. R., and Kuhajda, F. P. (1996). Fatty acid synthase (FAS): a target for cytotoxic antimetabolites in HL60 promyelocytic leukemia cells. Cancer Res. 56, 745–751.
Price, A. C., Zhang, Y. M., Rock, C. O., and White, S. W. (2004). Cofactor-induced conformational rearrangements establish a catalytically competent active site and a proton relay conduit in FabG. Structure 12, 417–428. doi: 10.1016/j.str.2004.02.008
Pulido, M. R., Garcia-Quintanilla, M., Gil-Marques, M. L., and McConnell, M. J. (2016). Identifying targets for antibiotic development using omics technologies. Drug Discov. Today 21, 465–472. doi: 10.1016/j.drudis.2015.11.014
Ramamoorthy, A., Thennarasu, S., Tan, A., Gottipati, K., Sreekumar, S., Heyl, D. L., et al. (2006). Deletion of all cysteines in tachyplesin I abolishes hemolytic activity and retains antimicrobial activity and lipopolysaccharide selective binding. Biochemistry 45, 6529–6540. doi: 10.1021/bi052629q
Shah, P., Hsiao, F. S., Ho, Y. H., and Chen, C. S. (2016). The proteome targets of intracellular targeting antimicrobial peptides. Proteomics 16, 1225–1237. doi: 10.1002/pmic.201500380
Singh, M. P., Kong, F., Janso, J. E., Arias, D. A., Suarez, P. A., Bernan, V. S., et al. (2003). Novel alpha-pyrones produced by a marine Pseudomonas sp. F92S91: taxonomy and biological activities. J. Antibiot. 56, 1033–1044. doi: 10.7164/antibiotics.56.1033
Sohn, M. J., Zheng, C. J., and Kim, W. G. (2008). Macrolactin S, a new antibacterial agent with FabG-inhibitory activity from Bacillus sp. AT28. J. Antibiot. 61, 687–691. doi: 10.1038/ja.2008.98
Srinivas, S., and Cronan, J. E. (2017). An eight-residue deletion in Escherichia coli FabG causes temperature-sensitive growth and lipid synthesis plus resistance to the calmodulin inhibitor trifluoperazine. J. Bacteriol. 199:e00074-17. doi: 10.1128/JB.00074-17
Tan, S. Y., Liu, Y., Chua, S. L., Vejborg, R. M., Jakobsen, T. H., Chew, S. C., et al. (2014). Comparative systems biology analysis to study the mode of action of the isothiocyanate compound Iberin on Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 58, 6648–6659. doi: 10.1128/AAC.02620-13
Wang, Y., Song, F., Zhu, J., Zhang, S., Yang, Y., Chen, T., et al. (2017). GSA: Genome Sequence Archive∗. Genomics Proteomics Bioinform. 15, 14–18. doi: 10.1016/j.gpb.2017.01.001
Wei, L., Wu, J., Liu, H., Yang, H., Rong, M., Li, D., et al. (2013). A mycobacteriophage-derived trehalose-6,6’-dimycolate-binding peptide containing both antimycobacterial and anti-inflammatory abilities. FASEB J. 27, 3067–3077. doi: 10.1096/fj.13-227454
Yonezawa, A., Kuwahara, J., Fujii, N., and Sugiura, Y. (1992). Binding of tachyplesin I to DNA revealed by footprinting analysis: significant contribution of secondary structure to DNA binding and implication for biological action. Biochemistry 31, 2998–3004. doi: 10.1021/bi00126a022
Keywords: tachyplesin, FabG, unsaturated fatty acids, membrane rupture, multidrug-resistant bacteria, cytotoxicity, hemolysis
Citation: Liu C, Qi J, Shan B and Ma Y (2018) Tachyplesin Causes Membrane Instability That Kills Multidrug-Resistant Bacteria by Inhibiting the 3-Ketoacyl Carrier Protein Reductase FabG. Front. Microbiol. 9:825. doi: 10.3389/fmicb.2018.00825
Received: 29 January 2018; Accepted: 11 April 2018;
Published: 01 May 2018.
Edited by:
Octavio Luiz Franco, Universidade Católica de Brasília, BrazilReviewed by:
Gergely Maroti, Hungarian Academy of Sciences, HungaryCopyright © 2018 Liu, Qi, Shan and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanbing Ma, eWFuYmluZ21hQDEyNi5jb20=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.