
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 23 March 2018
Sec. Food Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.00534
This work is a short review of the interactions between oenological yeasts and lactic acid bacteria (LAB), especially Oenococcus oeni, the main species carrying out the malolactic fermentation (MLF). The emphasis has been placed on non-Saccharomyces effects due to their recent increased interest in winemaking. Those interactions are variable, ranging from inhibitory, to neutral and stimulatory and are mediated by some known compounds, which will be discussed. One phenomena responsible of inhibitory interactions is the media exhaustion by yeasts, and particularly a decrease in L-malic acid by some non-Saccharomyces. Clearly ethanol is the main inhibitory compound of LAB produced by S. cerevisiae, but non-Saccharomyces can be used to decrease it. Sulfur dioxide and medium chain fatty acids (MCFAs) produced by yeasts can exhibit inhibitory effect upon LAB or even result lethal. Interestingly mixed fermentations with non-Saccharomyces present less MCFA concentration. Among organic acids derived as result of yeast metabolism, succinic acid seems to be the most related with MLF inhibition. Several protein factors produced by S. cerevisiae inhibiting O. oeni have been described, but they have not been studied in non-Saccharomyces. According to the stimulatory effects, the use of non-Saccharomyces can increase the concentration of favorable mediators such as citric acid, pyruvic acid, or other compounds derived of yeast autolysis such as peptides, glucans, or mannoproteins. The emergence of non-Saccharomyces in winemaking present a new scenario in which MLF has to take place. For this reason, new tools and approaches should be explored to better understand this new winemaking context.
Wine is the result of the alcoholic fermentation (AF) driven out by oenological yeasts in a complex microbial environment (Constantí et al., 1997; Beltran et al., 2002). Apart from Saccharomyces cerevisiae, recognized as the main agent of this process, other yeast species, known as non-Saccharomyces yeasts, such as Hanseniaspora/Kloeckera, Pichia, Candida, or Metschnikowia are implicated in early stages of the AF (Fleet et al., 1984). After the AF, the resultant wine can undergo the malolactic fermentation (MLF), which consists on a fairly simple reaction: a unique enzymatic decarboxylation of the L-malic acid to L-lactic acid (Liu, 2002). It is usually performed in red wines or high acidity white wines. This fermentation is carried out by lactic acid bacteria (LAB). Four LAB genera are usually found in wine: Lactobacillus, Pediococcus, Leuconostoc, and Oenococcus; and particularly, the main dominant species in wine is Oenococcus oeni (Wibowo et al., 1985; Lonvaud-Funel, 1999; Liu, 2002). MLF is related to a quality improvement in wine since this biotransformation leads to a pH increase, enhanced organoleptic properties and a microbial stabilization (Lonvaud-Funel, 1999). During MLF, LAB consume L-malic acid and other nutrients, impoverishing wine and avoiding the development of contaminant microorganisms.
In the last few years the interest on the use of non-Saccharomyces yeasts in winemaking has increased (Padilla et al., 2016; Petruzzi et al., 2017), due to the particular enzymatic activities that catalyze the liberation of aromas from their non-volatile precursors (Belda et al., 2017). Generally, these yeasts are inoculated to start the AF of must and later S. cerevisiae is inoculated to finish the process. This type of sequential inoculation with non-Saccharomyces undergoes chemical changes in wine which modulate the organoleptic profile of wines (Fleet, 2008; Padilla et al., 2016). What is more, this chemical modulation presents new scenery in which MLF may take place.
The purpose of this mini review is to summarize the current knowledge about the compounds responsible for the interactions that may take place between oenological yeasts and LAB during winemaking, highlighting the new scenery of non-Saccharomyces fermentations.
The performance of MLF by LAB is highly affected by the physicochemical intrinsic properties of wine, such as pH, ethanol, and SO2 (Carreté et al., 2002; Arnink and Henick-Kling, 2005). Moreover, since MLF takes place usually after the AF, it is also influenced by yeast metabolism. Those interactions range from inhibitory, to neutral and stimulatory. There is not much literature about this topic, but it is agreed that the type and impact of the interactions is dependent on several factors like (I) the initial must composition, (II) the yeast/bacteria strain combination, (III) the uptake and release of nutrients by yeasts, and (IV) the ability of yeasts to produce metabolites that affect somehow LAB (King and Beelman, 1986; Lonvaud-Funel et al., 1988; Alexandre et al., 2004; Du Plessis et al., 2017). There are some compounds which mediate these interactions (Figure 1) but, still the available information is not sufficient.
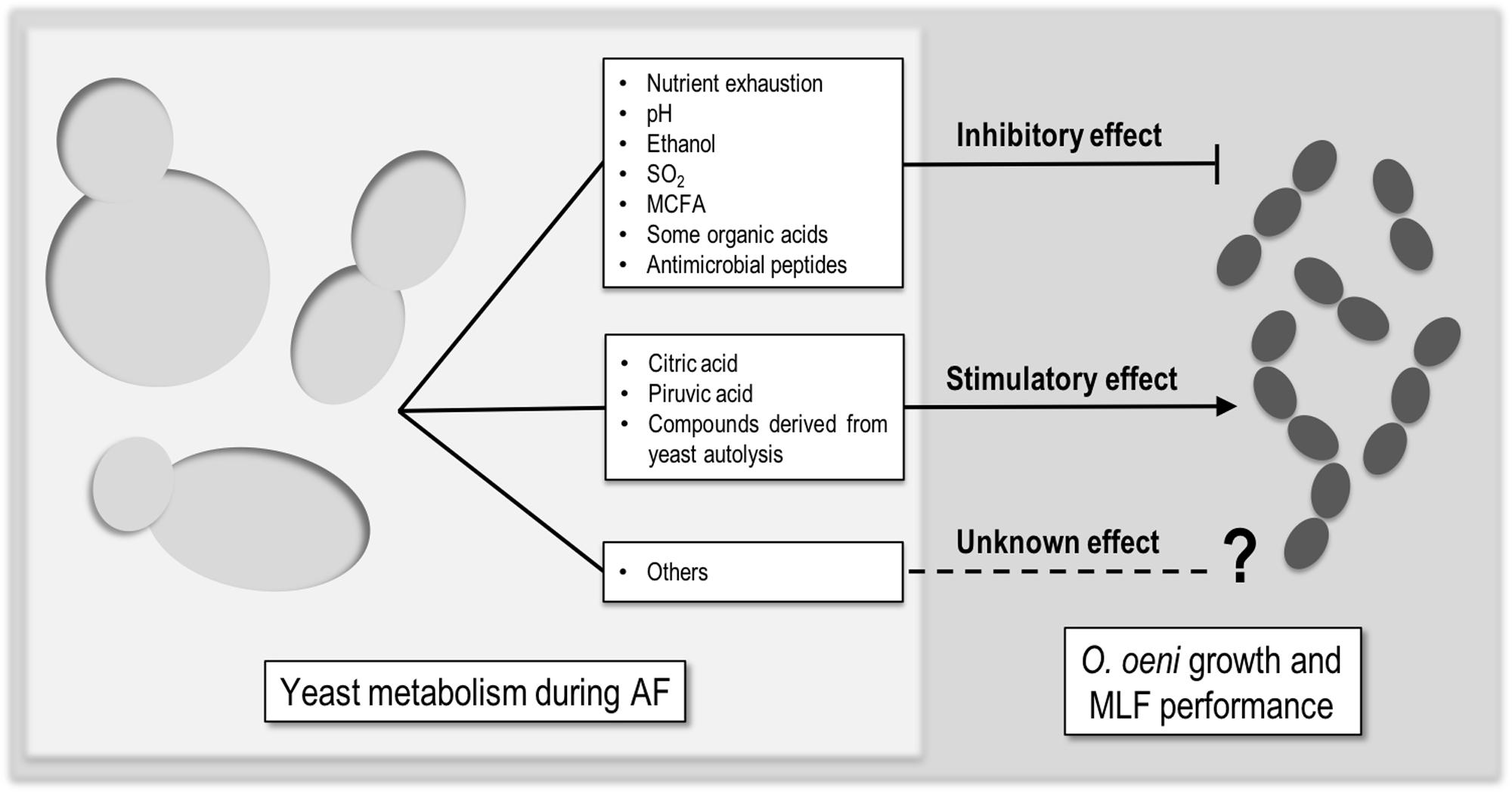
FIGURE 1. Compounds produced by yeast that can mediate inhibitory, stimulatory, or unknown effect in Oenococcus oeni growth or MLF performance.
Up to date, some strategies have been developed to mitigate the possible yeast- O. oeni inhibitory interactions (Sumby et al., 2014). Specifically, coinoculation of yeast and O. oeni has been proposed as a promising strategy to reduce the length of MLF (Izquierdo Cañas et al., 2014). In this way, the simultaneous AF and MLF co-immobilized in alginate beads is a technique currently in study (Bleve et al., 2016). Another classical approach to deal with the MLF difficulties is to select specific strains from the nature (Campbell-Sills et al., 2017; Petruzzi et al., 2017). The purpose of this selection is to identify the most relevant microorganisms related with the fermentation process in a particular area and use them as culture starters (Portillo et al., 2016; Franquès et al., 2017; Petruzzi et al., 2017).
Above the direct yeasts effect upon LAB and MLF performance, the must, and the winemaking practices, have a strong impact in how these interactions take place (Arnink and Henick-Kling, 2005; Tristezza et al., 2016).
Beyond the particular production of certain compounds (Table 1), yeast metabolism exhausts the nutrients of the medium. LAB have complex nutrient requirements (Garvie, 1967; Fourcassie et al., 1992; Terrade and Mira de Orduña, 2009), so their growth is highly dependent on the nutrients consumption during AF by yeasts (Ivey et al., 2013). The effect of these inhibitory interactions could be explained as the result of nutrient competition, such as yeast assimilable nitrogen (YAN) or amino acids (Costello et al., 2003). Therefore, yeast strains with complex nutrient requirements would exhibit an increased antagonistic relationship with LAB (Costello et al., 2003). In this way, it has been recently described that coinoculation of S. cerevisiae with other non-Saccharomyces yeasts result in a metabolic stimulation of glucose and nitrogen uptake by yeasts, which could lead to a more impoverished medium for LAB (Curiel et al., 2017).
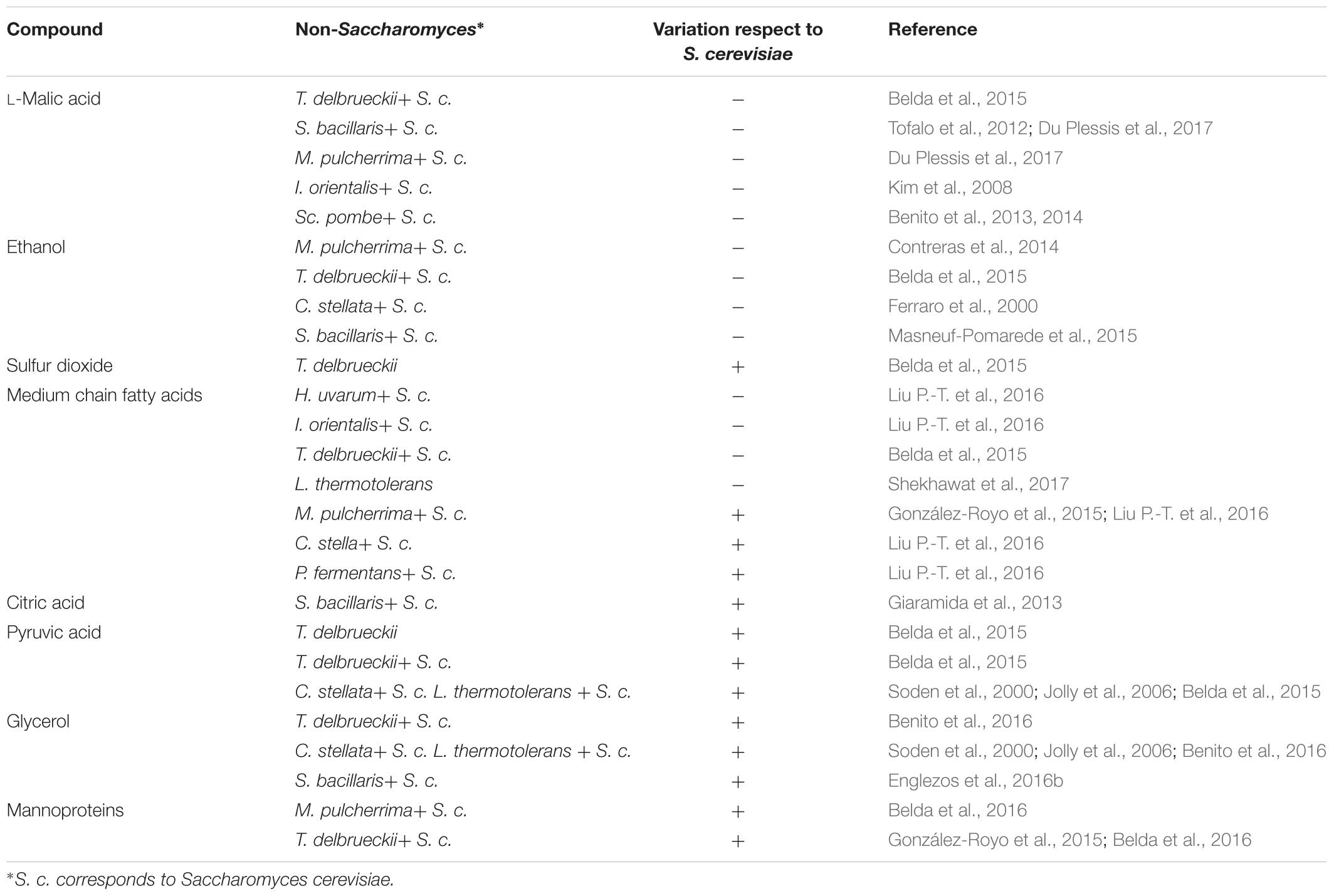
TABLE 1. Main compounds affected (variation in content, negative or positive) by the use of non-Saccharomyces in alcoholic fermentation regarding to S. cerevisiae as sole starter.
Moreover, it has been reported that the use of some yeast strains (Su et al., 2014) can cause a decrease in L-malic acid, the prior substrate of LAB in wine, which can negatively affect the MLF performance. Particularly, the use of non-Saccharomyces leads a higher consumption of L-malic acid, as it has been described with Torulaspora delbrueckii (Belda et al., 2015), Starmerella bacillaris (syn. Candida zemplinina) (Tofalo et al., 2012; Du Plessis et al., 2017), M. pulcherrima (Du Plessis et al., 2017), and Issatchenkia orientalis (Kim et al., 2008). There is also another non-Saccharomyces yeast that really consumes L-malic acid to dryness (Du Plessis et al., 2017). Schizosaccharomyces spp. can develop the maloalcoholic fermentation by consuming both sugars and L-malic acid (Benito et al., 2013, 2014).
Alcoholic fermentation of grape must undergoes deep chemical changes enhanced by ethanol and sulfur dioxide. Long ago, it is agreed that concentrations over 4% (v/v) of ethanol inhibit the growth of most LAB (Capucho and San Romao, 1994). Also, a more recent study reported the triad of ethanol, SO2 and medium chain fatty acids (MCFAs) as the main inhibitor compounds in the antagonism between yeast and O. oeni (Nehme et al., 2008). The main functional categories of genes affected by ethanol are metabolite transport and cell wall and membrane biogenesis (Olguín et al., 2015). Nowadays, some non-Saccharomyces yeasts are currently used in mixed fermentations to decrease the alcoholic content of wines (Giaramida et al., 2013; Loira et al., 2014; Ciani et al., 2016), such as M. pulcherrima (Contreras et al., 2014), T. delbrueckii (Belda et al., 2015), C. stellata (Ferraro et al., 2000) and S. bacillaris (Englezos et al., 2016a), possibly mitigating the negative effect of ethanol upon LAB growth.
The role of SO2 as an antimicrobial compound is known since ancient Romans that used to add this chemical to prevent food and beverage from spoilage. Its active mechanism affects O. oeni membrane and causes an ATPase activity decrease (Carreté et al., 2002), causing a delay or the failure of MLF (Lonvaud-Funel et al., 1988). It is customary to use this compound to control microbial communities since vineyard to wine in the winemaking. Moreover, yeasts are able to produce this compound as result of their metabolism (Wells and Osborne, 2011). The common amount of SO2 produced by S. cerevisiae strains is less than 30 mg/L, but some strains can produce more than 100 mg/L of this compound (Suzzi et al., 1985; Rankine, 1968). When it comes to non-Saccharomyces yeasts, there is no much information about their SO2 production since they are more affected by this compound (Jolly et al., 2014). However, it has to be pointed that the use of T. delbrueckii as sole starter increased the SO2 concentration of the final wine (Belda et al., 2015). Apart from the cited strain effect, the medium has great influence in the production of SO2 by yeasts. Higher concentration of YAN in must ends on higher amount of SO2 (Osborne and Edwards, 2006), as result of the metabolism of the sulfured amino acids.
During AF, yeasts produce different compounds as result of their growth metabolism that can inhibit O. oeni growth and MLF. MCFA (C8–C14) constitute a group of organic molecules that can limit O. oeni growth and even decrease their L-malic consumption (Edwards and Beelman, 1987; Lonvaud-Funel et al., 1988). It has to be mentioned the strong effect of winemaking practices in fatty acids metabolism by yeasts (Guilloux-Benatier et al., 1998). These authors related a fine MLF performance with a large pre fermentative maceration, possibly due to the high macromolecules concentration and long chain fatty acid extraction (Guilloux-Benatier et al., 1995, 1998). The effect of using non-Saccharomyces yeasts in the production of MCFA is variable. Strains belonging to M. pulcherrima, C. stella, and Pichia fermentans increase the final concentration of MCFA (Liu P.-T. et al., 2016). In contrast, mixed fermentations with H. uvarum, I. orientalis present the opposite behavior (Liu P.-T. et al., 2016). Also, a significant decrease in MCFA concentration has been reported by Lanchacea thermotolerans as sole starter (Shekhawat et al., 2017). Hu et al. (2018) reported a strong influence in MCFA concentration related with the inoculation timing of H. uvarum in mixed fermentation with S. cerevisiae. In this experiment inoculation timing seem to determine the increase or decrease in MCFA concentration regarding to S. cerevisiae traditional fermentation. Generally, C12 and C14, as free fatty acids, are the most toxic MCFA for O. oeni (Guilloux-Benatier et al., 1998). Moreover, the esterified forms are even more toxic than free fatty acids, being the most toxic esterified MCFA C10, C12, and C14 (Guilloux-Benatier et al., 1998). So, depending on the particular MCFA and its concentration, the inhibitory effect can become lethal to LAB (Edwards and Beelman, 1987).
Malolactic fermentation is the consequence of a unique enzymatic activity performed by the malolactic enzyme. Accordingly, structurally similar organic acids will act as competitive inhibitors for the active site of the malolactic enzyme (Lonvaud-Funel and Strasser de Saad, 1982) and probably they will delay the MLF duration. Early studies in this subject related this effect with succinic acid, fumaric acid, citric acid, and tartaric acid (Lonvaud-Funel and Strasser de Saad, 1982; Davis et al., 1985). Among these acids, succinic acid is the most studied since oenological yeasts can largely produce this compound. First studies related the inhibition of MLF by criotolerant S. cerevisiae strains which are characterized by high production of succinic acid and β-phenylethanol (Caridi and Corte, 1997). More recent studies agreed with the inhibition effect of succinic acid (Son et al., 2009), and not with its role as MLF extender.
Even though citric acid is considered as inhibitor of the malolactic enzyme (Lonvaud-Funel and Strasser de Saad, 1982), citric acid can be catabolized by LAB (Liu, 2002). This metabolic activity is found in some O. oeni strains as response to acidity or ethanol stress (Olguín et al., 2009). Due to the consumption of citric acid, diacetyl is produced (Swiegers et al., 2005). It is usually desirable to have strains which can consume citric acid due to the organoleptic complexity that is achieved (Lonvaud-Funel, 1999). In this way, a high concentration of diacetyl is reported as undesirable (Davis et al., 1985; Bartowsky and Henschke, 2004). Moreover, due to the citric acid metabolism, O. oeni increases the volatile acidity (Lonvaud-Funel, 1999; Liu, 2002). Even thought, citric acid increases the transmembrane gradient which generate energy in terms of proton-motive force for O. oeni (Liu Y. et al., 2016).
Anyway, since citric acid concentration is usually not very high, acetic acid does not increase very much. Citric acid production by yeast is highly species and strain dependent (Fleet, 2008). On the top of that, mixed fermentations with different non-Saccharomyces species exhibit particular citric acid production (Jussier et al., 2006; Giaramida et al., 2013; Izquierdo Cañas et al., 2014). For the moment the only mixed fermentation that clearly increased citric acid concentration is with S. bacillaris (Giaramida et al., 2013).
Pyruvic acid is an intermediary produced by yeast during the AF. This compound can improve MLF performance by O. oeni. It acts as external electron acceptor, facilitating the regeneration of NAD+ (Maicas et al., 2002). It can also promote diacetyl production (Mink et al., 2015). Related to increasing the concentration of this compound, Belda et al. (2015) reported higher production of pyruvic acid when T. delbrueckii was used as sole or mixed culture starter with S. cerevisiae. Benito et al. (2016) reported similar results using L. thermotolerans.
The production of glycerol is directly related with the activity of yeasts by the glyceropyruvic fermentation pathway (Ciani and Maccarelli, 1998). Glycerol can be assimilated and degraded by some spoiling Lactobacillus in wine (Liu, 2002). On the contrary, there is no literature that reports this behavior when it comes to O. oeni. It is unclear how can affect glycerol to O. oeni, since it does not assimilate it, neither degrade it. Usually, non-Saccharomyces yeasts exhibit higher metabolic activity of this pathway (Ciani and Maccarelli, 1998; Jolly et al., 2006, 2014). Specifically, T. delbrueckii (Belda et al., 2015) and C. stellata (Soden et al., 2000; Jolly et al., 2006) have been reported as big glycerol and pyruvic acid producers as result of their high glyceropyruvic fermentation activity. Also, mixed fermentations with S. bacillaris and L. thermotolerans exhibit higher production of glycerol in regards to a conventional S. cerevisiae fermentation (Benito et al., 2016; Englezos et al., 2016b).
One of the most known positive effects upon MLF performance is its development in presence of yeast lees (Guilloux-Benatier et al., 1995). It has been reported that the inhibitory interactions between yeasts and LAB is counteracted by the presence of yeast lees, and even more, the positive interactions are enlarged (Patynowski et al., 2002). During aging, yeasts undergo an autolytic process that result in the release of different compounds. Nitrogenated compounds, such as amino acids, peptides and proteins, are mainly released as result of yeast autolysis (Guilloux-Benatier et al., 1995; Martínez-Rodriguez et al., 2001). The release of such compounds can help to enrich the previously exhausted medium by yeasts (Costello et al., 2003), stimulating the growth of LAB and MLF performance (Guilloux-Benatier et al., 1995; Diez et al., 2010).
Other molecules like glucans and mannoproteins are also released due to this mentioned process and can stimulate LAB growth (Diez et al., 2010). These authors realized that the presence of mannoproteins only exhibited its positive effect on LAB growth when ethanol was present. O. oeni can catabolize these mannoproteins and release mannose, which can be substrate of the phosphotransferase system that helps the adaptation of O. oeni to the medium (Jamal et al., 2013). Besides this, the impact of the mannoproteins upon LAB was yeast-LAB strain dependent. Recently, it has been reported that some non-Saccharomyces strains belonging to M. pulcherrima and T. delbrueckii release more mannoproteins than S. cerevisiae (Belda et al., 2016). Moreover, these molecules could help hijack MCFA present in wine, stimulating LAB growth (Guilloux-Benatier et al., 1995). It has been also been reported that during AF those cited macromolecules are released, depending in the initial colloidal concentration (Guilloux-Benatier et al., 1995). Still, the same study states that the amount of macromolecules released during yeast growth is insignificant in regards to yeast autolysis.
Apart from the mentioned compounds, there are more released compounds during yeast autolysis, such as vitamins, nucleotides and long chain fatty acids, which could be also stimulatory to LAB (Alexandre et al., 2004). Unfortunately, there is no literature currently available about the possible effect of these compounds.
In regards to the possible incompatibility between oenological yeasts and LAB, apart from metabolite compounds, the production of antimicrobial proteinaceous compounds by some S. cerevisiae strains has been reported. Dick et al. (1992) firstly studied these compounds. They discovered two cationic proteins which were effective against LAB. More recently, another inhibitory protein fraction produced by S. cerevisiae CCMI 885 and active against LAB was identified (Branco et al., 2014). In this work, an exhaustive characterization was performed, which resulted in the identification of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) protein fragments. This newly identified antimicrobial peptides with 2–10 kDa size agreed with previously reported antimicrobial peptides (Comitini et al., 2005; Osborne and Edwards, 2007).
There are no studies about these compounds produced by non-Saccharomyces yeasts, but some species could present such antimicrobial compounds, like M. pulcherrima that produce pulcherrimic acid (Oro et al., 2014), active against other yeasts.
The increasing number of non-Saccharomyces species described as beneficial in winemaking demands further investigation of their metabolism. Many factors can influence the effect of non-Saccharomyces on wine composition. Besides the yeast species and strain characteristics, the time and the ratio of inoculation, with respect to S. cerevisiae, may alter notably the global effect on wine of the use of non-Saccharomyces. All these variables may also affect the development of O. oeni and MLF. Future research should contribute to a better knowledge of metabolic traits of a wider number of non-Saccharomyces strains and their influence on O. oeni performance. Among other possible approaches, metabolomics may be a powerful tool to elucidate how the new winemaking scenario of combined yeasts may change MLF evolution.
All authors conceived, drafted the manuscript, and approved the final version of the paper.
This work was supported by grant AGL2015-70378-R awarded by the Spanish Ministry of Economy and Competitiveness. The research leading to these results has received funding from “la Caixa” Foundation and Triptolemos Foundation. AiB was grateful to the predoctoral fellowship from the Universitat Rovira i Virgili.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
JB-G wished to express thanks to the Spanish Government for his postdoctoral research contract (Juan de la Cierva-incorporación).
Alexandre, H., Costello, P. J., Remize, F., Guzzo, J., and Guilloux-Benatier, M. (2004). Saccharomyces cerevisiae-Oenococcus oeni interactions in wine: current knowledge and perspectives. Int. J. Food Microbiol. 93, 141–154. doi: 10.1016/j.ijfoodmicro.2003.10.013
Arnink, K., and Henick-Kling, T. (2005). Influence of Saccharomyces cerevisiae and Oenococcus oeni strains on successful malolactic conversion in wine. Am. J. Enol. Vitic. 56, 228–237.
Bartowsky, E. J., and Henschke, P. A. (2004). The “buttery” attribute of wine - Diacetyl - Desirability, spoilage and beyond. Int. J. Food Microbiol. 96, 235–252. doi: 10.1016/j.ijfoodmicro.2004.05.013
Belda, I., Navascués, E., Marquina, D., Santos, A., Calderon, F., and Benito, S. (2015). Dynamic analysis of physiological properties of Torulaspora delbrueckii in wine fermentations and its incidence on wine quality. Appl. Microbiol. Biotechnol. 99, 1911–1922. doi: 10.1007/s00253-014-6197-2
Belda, I., Navascués, E., Marquina, D., Santos, A., Calderón, F., and Benito, S. (2016). Outlining the influence of non-conventional yeasts in wine ageing over lees. Yeast 33, 329–338. doi: 10.1002/yea.3165
Belda, I., Ruiz, J., Esteban-Fernández, A., Navascués, E., Marquina, D., Santos, A., et al. (2017). Microbial contribution to Wine aroma and its intended use for Wine quality improvement. Molecules 22, 1–29. doi: 10.3390/molecules22020189
Beltran, G., Torija, M. J., Novo, M., Ferrer, N., Poblet, M., Guillamón, J. M., et al. (2002). Analysis of yeast populations during alcoholic fermentation: a six year follow-up study. Syst. Appl. Microbiol. 25, 287–293. doi: 10.1078/0723-2020-00097
Benito, A., Calderon, F., Palomero, F., and Benito, S. (2016). Quality and composition of Airén wines fermented by sequential inoculation of Lachancea thermotolerans and Saccharomyces cerevisiae. Food Technol. Biotechnol. 54, 135–144. doi: 10.17113/ftb.54.02.16.4220
Benito, S., Palomero, F., Gálvez, L., Morata, A., Calderón, F., Palmero, D., et al. (2014). Quality and composition of red wine fermented with Schizosaccharomyces pombe as sole fermentative yeast, and in mixed and sequential fermentations with Saccharomyces cerevisiae. Food Technol. Biotechnol. 52, 376–382.
Benito, S., Palomero, F., Morata, A., Calderón, F., Palmero, D., and Suárez-Lepe, J. A. (2013). Physiological features of Schizosaccharomyces pombe of interest in making of white wines. Eur. Food Res. Technol. 236, 29–36. doi: 10.1007/s00217-012-1836-2
Bleve, G., Tufariello, M., Vetrano, C., and Mita Gand Grieco, F. (2016). Simultaneous alcoholic and malolactic fermentations by Saccharomyces cerevisiae and Oenococcus oeni cells co-immobilized in alginate beads. Front. Microbiol. 7:943. doi: 10.3389/fmicb.2016.00943
Branco, P., Francisco, D., Chambon, C., Hébraud, M., Arneborg, N., Almeida, M. G., et al. (2014). Identification of novel GAPDH-derived antimicrobial peptides secreted by Saccharomyces cerevisiae and involved in wine microbial interactions. Appl. Microbiol. Biotechnol. 98, 843–853. doi: 10.1007/s00253-013-5411-y
Campbell-Sills, H., El Khoury, M., Gammacurta, M., Miot-Sertier, C., Dutilh, L., Vestner, J., et al. (2017). Two different Oenococcus oeni lineages are associated to either red or white wines in Burgundy: genomics and metabolomics insights. OENO One 51, 309–322. doi: 10.20870/oeno-one.2017.51.4.1861
Capucho, I., and San Romao, M. V. (1994). Effect of ethanol and fatty acids on malolactic activity of Leuconostoc oenos. Appl. Microbiol. Biotechnol. 42, 391–395. doi: 10.1007/BF00902747
Caridi, A., and Corte, V. (1997). Inhibition of malolactic fermentation by cryotolerant yeasts. Biotechnol. Lett. 19, 723–726. doi: 10.1023/A:1018319705617
Carreté, R., Vidal, M. T., Bordons, A., and Constantí, M. (2002). Inhibitory effect of sulfur dioxide and other stress compounds in wine on the ATPase activity of Oenococcus oeni. FEMS Microbiol. Lett. 211, 155–159. doi: 10.1016/S0378-1097(02)00687-0
Ciani, M., and Maccarelli, F. (1998). Oenological properties of non-Saccharomyces yeasts associated with wine-making. World J. Microbiol. Biotechnol. 14, 199–203. doi: 10.1023/A:1008825928354
Ciani, M., Morales, P., Comitini, F., Tronchoni, J., Canonico, L., Curiel, J. A., et al. (2016). Non-conventional Yeast Species for Lowering Ethanol Content of Wines. Front. Microbiol. 7:642. doi: 10.3389/fmicb.2016.00642
Comitini, F., Ferretti, R., Clementi, F., Mannazzu, I., and Ciani, M. (2005). Interactions between Saccharomyces cerevisiae and malolactic bacteria: preliminary characterization of a yeast proteinaceous compound(s) active against Oenococcus oeni. J. Appl. Microbiol. 99, 105–111. doi: 10.1111/j.1365-2672.2005.02579.x
Constantí, M., Poblet, M., Arola, L., Mas, A., and Guillamón, J. M. (1997). Analysis of yeast populations during alcoholic fermentation in a newly established winery. Am. J. Enol. Vitic. 48, 339–344.
Contreras, A., Hidalgo, C., Henschke, P. A., Chambers, P. J., Curtin, C., and Varela, C. (2014). Evaluation of non-Saccharomyces yeasts for the reduction of alcohol content in wine. Appl. Environ. Microbiol. 80, 1670–1678. doi: 10.1128/AEM.03780-13
Costello, P. J., Henschke, P., and Markides, J. (2003). Standardised methodology for testing malolactic bacteria and wine yeast compatibility. Aust. J. Grape Wine Res. 9, 127–137. doi: 10.1111/j.1755-0238.2003.tb00263.x
Curiel, J. A., Morales, P., Gonzalez, R., and Tronchoni, J. (2017). Different non-Saccharomyces yeast species stimulate nutrient consumption in s. cerevisiae mixed cultures. Front. Microbiol. 8:2121. doi: 10.3389/fmicb.2017.02121
Davis, C. R., Wibowo, D., Eschenbruch, R., Lee, T. H., and Fleet, G. H. (1985). Practical implications of malolactic fermentation: a review. Am. J. Enol. Vitic. 36, 290–301.
Dick, K. J., Molan, P. C., and Eschembruch, R. (1992). The isolation from Saccharomyces cerevisiae of two antibacterial cationic proteins that inhibit malolactic bacteria. Vitis 31, 105–116.
Diez, L., Guadalupe, Z., Ayestarán, B., and Ruiz-Larrea, F. (2010). Effect of yeast mannoproteins and grape polysaccharides on the growth of wine lactic acid and acetic acid bacteria. J. Agric. Food Chem. 58, 7731–7739. doi: 10.1021/jf100199n
Du Plessis, H. W., du Toit, M., Hoff, J. W., Hart, R. S., Ndimba, B. K., and Jolly, N. P. (2017). Characterisation of non- Saccharomyces yeasts using different methodologies and evaluation of their compatibility with malolactic fermentation. S. Afr. J. Enol. Vitic. 38, 46–63. doi: 10.21548/38-1-819
Edwards, C. G., and Beelman, R. (1987). Inhibition of the malolactic bacterium Leuconostoc oenos (PSU-1) by decanoic acid and subsequent removal of the inhibition by yeast ghosts. Am. J. Enol. Vitic. 38, 239–242.
Englezos, V., Rantsiou, K., Cravero, F., Torchio, F., Ortiz-Julien, A., Gerbi, V., et al. (2016a). Starmerella bacillaris and Saccharomyces cerevisiae mixed fermentations to reduce ethanol content in wine. Appl. Microbiol. Biotechnol. 100, 5515–5526. doi: 10.1007/s00253-016-7413-z
Englezos, V., Torchio, F., Cravero, F., Marengo, F., Giacosa, S., Gerbi, V., et al. (2016b). Aroma profile and composition of Barbera wines obtained by mixed fermentations of Starmerella bacillaris (synonym Candida zemplinina) and Saccharomyces cerevisiae. LWT Food Sci. Technol. 73, 567–575. doi: 10.1016/j.lwt.2016.06.063
Ferraro, L., Fatichenti, F., and Ciani, M. (2000). Pilot scale vinification process using immobilized Candida stellata cells and Saccharomyces cerevisiae. Process. Biochem. 35, 1125–1129. doi: 10.1016/S0032-9592(00)00148-5
Fleet, G. H. (2008). Wine yeasts for the future. FEMS Yeast Res. 8, 979–995. doi: 10.1111/j.1567-1364.2008.00427.x
Fleet, G. H., Lafon-Lafourcade, S., and Ribereau-Gayon, P. (1984). Evolution of yeasts and lactic acid bacteria during fermentation and storage of Bordeaux wines. Appl. Environ. Microbiol. 48, 1034–1038.
Fourcassie, P., Belarbi, A., and Maujean, A. (1992). Growth, D-glucose utilization and malolactic fermentation by Leuconostoc śnos strains in 18 media deficient in one amino acid. J. Appl. Bacteriol. 73, 489–496. doi: 10.1111/j.1365-2672.1992.tb05010.x
Franquès, J., Araque, I., Palahí, E., Portillo, M. C., Reguant, C., and Bordons, A. (2017). Presence of Oenococcus oeni and other lactic acid bacteria in grapes and wines from Priorat (Catalonia, Spain). LWT Food Sci. Technol. 81, 326–334. doi: 10.1016/j.lwt.2017.03.054
Garvie, E. I. (1967). The growth factor and amino acid requirements of species of the genus Leuconostoc, including Leuconostoc paramesenteroides (sp. nov.) and Leuconostoc oenos. J. Gen. Microbiol. 48, 439–447. doi: 10.1099/00221287-48-3-439
Giaramida, P., Ponticello, G., Di Maio, S., Squadrito, M., Genna, G., Barone, E., et al. (2013). Candida zemplinina for production of wines with less alcohol and more glycerol. S. Afr. J. Enol. Vitic. 34, 204–211.
Guilloux-Benatier, M., Guerreau, J., and Feuillat, M. (1995). Influence of initial colloid content on yeast macromolecule production and on the metabolism of wine microorganisms. Am. J. Enol. Vitic. 46, 486–492.
González-Royo, E., Pascual, O., Kontoudakis, N., Esteruelas, M., Esteve-Zarzoso, B., Mas, A., et al. (2015). Oenological consequences of sequential inoculation with non-Saccharomyces yeasts (Torulaspora delbrueckii or Metschnikowia pulcherrima) and Saccharomyces cerevisiae in base wine for sparkling wine production. Eur. Food Res. Technol. 240, 999–1012. doi: 10.1007/s00217-014-2404-8
Guilloux-Benatier, M., Le Fur, Y., and Feuillat, M. (1998). Influence of fatty acids on the growth of wine microorganisms Saccharomyces cerevisiae and Oenococcus oeni. J. Ind. Microbiol. Biotechnol. 20, 144–149. doi: 10.1038/sj.jim.2900502
Hu, K., Jin, G.-J., Mei, W.-C., Li, T., and Tao, Y.-S. (2018). Increase of medium-chain fatty acid ethyl ester content in mixed H. uvarum/S. cerevisiae fermentation leads to wine fruity aroma enhancement. Food Chem. 239, 495–501. doi: 10.1016/j.foodchem.2017.06.151
Ivey, M., Massel, M., and Phister, T. G. (2013). Microbial interactions in food fermentations. Annu. Rev. Food Sci. Technol. 4, 141–162. doi: 10.1146/annurev-food-022811-101219
Izquierdo Cañas, P. M. I., Romero, E. G., Pérez-Martín, F., Seseña, S., and Palop, M. L. (2014). Sequential inoculation versus co-inoculation in Cabernet Franc wine fermentation. Food Sci. Technol. Int. 21, 203–212. doi: 10.1177/1082013214524585
Jamal, Z., Miot-Sertier, C., Thibau, F., Dutilh, L., Lonvaud-Funel, A., Ballestra, P., et al. (2013). Distribution and functions of phosphotransferase system genes in the genome of the lactic acid bacterium Oenococcus oeni. Appl. Environ. Microbiol. 79, 3371–3379. doi: 10.1128/AEM.00380-13
Jolly, N., Augustyn, O., and Pretorius, I. (2006). The role and use of non-Saccharomyces yeasts in wine production. S. Afr. J. Enol. Vitic. 27, 15–38.
Jolly, N. P., Varela, C., and Pretorius, I. S. (2014). Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 14, 215–237. doi: 10.1111/1567-1364.12111
Jussier, D., Morneau, A. D., and Mira de Orduña, R. (2006). Effect of simultaneous inoculation with yeast and bacteria on fermentation kinetics and key wine parameters of cool-climate chardonnay. Appl. Environ. Microbiol. 72, 221–227. doi: 10.1128/AEM.72.1.221
Kim, D. H., Hong, Y. A., and Park, H. D. (2008). Co-fermentation of grape must by Issatchenkia orientalis and Saccharomyces cerevisiae reduces the malic acid content in wine. Biotechnol. Lett. 30, 1633–1638. doi: 10.1007/s10529-008-9726-1
King, S., and Beelman, R. (1986). Metabolic interactions between Saccharomyces cerevisiae and Leuconostoc oenos in a model grape juice/wine system. Am. J. Enol. Vitic. 37, 53–60.
Liu, P.-T., Lu, L., Duan, C. Q., and Yan, G. L. (2016). The contribution of indigenous non-Saccharomyces wine yeast to improved aromatic quality of Cabernet Sauvignon wines by spontaneous fermentation. LWT Food Sci. Technol. 71, 356–363. doi: 10.1016/j.lwt.2016.04.031
Liu, S. Q. (2002). Malolactic fermentation in wine - Beyond deacidification. J. Appl. Microbiol. 92, 589–601. doi: 10.1046/j.1365-2672.2002.01589.x
Liu, Y., Forcisi, S., Harir, M., Deleris-Bou, M., Krieger-Weber, S., Lucio, M., et al. (2016). New molecular evidence of wine yeast-bacteria interaction unraveled by non-targeted exometabolomic profiling. Metabolomics 12, 1–16. doi: 10.1007/s11306-016-1001-1
Loira, I., Vejarano, R., Bañuelos, M. A., Morata, A., Tesfaye, W., Uthurry, C., et al. (2014). Influence of sequential fermentation with Torulaspora delbrueckii and Saccharomyces cerevisiae on wine quality. LWT Food Sci. Technol. 59, 915–922. doi: 10.1016/j.lwt.2014.06.019
Lonvaud-Funel, A. (1999). Lactic acid bacteria in the quality improvement and depreciation of wine. Antonie van Leeuwenhoek 76, 317–331. doi: 10.1023/A:1002088931106
Lonvaud-Funel, A., Joyeux, A., and Desens, C. (1988). Inhibition of malolactic fermentation of wines by products of yeast metabolism. J. Sci. Food Agric. 44, 183–191. doi: 10.1002/jsfa.2740440209
Lonvaud-Funel, A., and Strasser de Saad, A. M. (1982). Purification and properties of a malolactic enzyme from a strain of Leuconostoc mesenteroides isolated from grapes. Appl. Environ. Microbiol. 43, 357–361.
Maicas, S., Ferrer, S., and Pardo, I. (2002). NAD(P)H regeneration is the key for heterolactic fermentation of hexoses in Oenococcus oeni. Microbiology 148, 325–332. doi: 10.1099/00221287-148-1-325
Martínez-Rodriguez, A. J., Carrascosa, A. V., and Polo, M. C. (2001). Release of nitrogen compounds to the extracellular medium by three strains of Saccharomyces cerevisiae during induced autolysis in a model wine system. Int. J. Food Microbiol. 68, 155–160. doi: 10.1016/S0168-1605(01)00486-X
Masneuf-Pomarede, I., Juquin, E., Miot-Sertier, C., Renault, P., Laizen, Y., Salin, F., et al. (2015). The yeast Starmerella bacillaris (synonym Candida zemplinina) shows high genetic diversity in winemaking environments. FEMS Yeast Res. 15:fov045. doi: 10.1093/femsyr/fov045
Mink, R., Kölling, R., Sommer, S., Schmarr, H. G., and Scharfenberger-Schmeer, M. (2015). Diacetyl formation by Oenococcus oeni during winemaking induced by exogenous pyruvate. Am. J. Enol. Vitic. 66, 85–90. doi: 10.5344/ajev.2014.14056
Nehme, N., Mathieu, F., and Taillandier, P. (2008). Quantitative study of interactions between Saccharomyces cerevisiae and Oenococcus oeni strains. J. Ind. Microbiol. Biotechnol 35, 685–693. doi: 10.1007/s10295-008-0328-7
Olguín, N., Bordons, A., and Reguant, C. (2009). Influence of ethanol and pH on the gene expression of the citrate pathway in Oenococcus oeni. Food Microbiol. 26, 197–203. doi: 10.1016/j.fm.2008.09.004
Olguín, N., Champomier-Vergès, M., Anglade, P., Baraige, F., Cordero-Otero, R., Bordons, A., et al. (2015). Transcriptomic and proteomic analysis of Oenococcus oeni PSU-1 response to ethanol shock. Food Microbiol. 51, 87–95. doi: 10.1016/j.fm.2015.05.005
Oro, L., Ciani, M., and Comitini, F. (2014). Antimicrobial activity of Metschnikowia pulcherrima on wine yeasts. J. Appl. Microbiol. 116, 1209–1217. doi: 10.1111/jam.12446
Osborne, J. P., and Edwards, C. G. (2006). Inhibition of malolactic fermentation by Saccharomyces during alcoholic fermentation under low- and high-nitrogen conditions: a study in synthetic media. Aust. J. Grape Wine Res. 12, 69–78. doi: 10.1111/j.1755-0238.2006.tb00045.x
Osborne, J. P., and Edwards, C. G. (2007). Inhibition of malolactic fermentation by a peptide produced by Saccharomyces cerevisiae during alcoholic fermentation. Int. J. Food Microbiol. 118, 27–34. doi: 10.1016/j.ijfoodmicro.2007.05.007
Padilla, B., Gil, J. V., and Manzanares, P. (2016). Past and future of non-Saccharomyces yeasts: from spoilage microorganisms to biotechnological tools for improving wine aroma complexity. Front. Microbiol. 7:411. doi: 10.3389/fmicb.2016.00411
Patynowski, R. J., Jiranek, V., and Markides, A. J. (2002). Yeast viability during fermentation and sur lie ageing of a defined medium and subsequent growth of Oenococcus oeni. Aust. J. Grape Wine Res. 8, 62–69. doi: 10.1111/j.1755-0238.2002.tb00212.x
Petruzzi, L., Capozzi, V., Berbegal, C., Corbo, M. R., Bevilacqua, A., Spano, G., et al. (2017). Microbial resources and enological significance: opportunities and benefits. Front. Microbiol. 8:995. doi: 10.3389/fmicb.2017.00995
Portillo, M. C., Franquès, J., Araque, I., Reguant, C., and Bordons, A. (2016). Bacterial diversity of Grenache and Carignan grape surface from different vineyards at Priorat wine region (Catalonia, Spain). Int. J. Food Microbiol. 219, 56–63. doi: 10.1016/j.ijfoodmicro.2015.12.002
Rankine, B. C. (1968). The importance of yeasts in determining the composition and quality of wines. Vitis 7, 22–49.
Shekhawat, K., Bauer, F. F., and Setati, M. E. (2017). Impact of oxygenation on the performance of three non-Saccharomyces yeasts in co-fermentation with Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 101, 2479–2491. doi: 10.1007/s00253-016-8001-y
Soden, A., Fracis, I. L., Oakey, H., and Henschke, P. A. (2000). Effects of co-fermentation with Candida stellata and Saccharomyces cerevisiae on the aroma and composition of Chardonnay wine. Aust. J. Grape Wine Res. 6, 21–30. doi: 10.1111/j.1755-0238.2000.tb00158.x
Son, H. S., Hwang, G. S., Park, W. M., Hong, Y. S., and Lee, C. H. (2009). Metabolomic characterization of malolactic fermentation and fermentative behaviors of wine yeasts in grape wine. J. Agric. Food Chem. 57, 4801–4809. doi: 10.1021/jf9005017
Su, J., Wang, T., Wang, Y., Li, Y. Y., and Li, H. (2014). The use of lactic acid-producing, malic acid-producing, ormalic acid-degrading yeast strains for acidity adjustment in the wine industry. Appl. Microbiol. Biotechnol. 98, 2395–2413. doi: 10.1007/s00253-014-5508-y
Sumby, K. M., Grbin, P. R., and Jiranek, V. (2014). Implications of new research and technologies for malolactic fermentation in wine. Appl. Microbiol. Biotechnol. 98, 8111–8132. doi: 10.1007/s00253-014-5976-0
Suzzi, G., Romano, P., and Zambonelli, C. (1985). Saccharomyces strain selection in minimizing SO2 requirements during fermentation. Am. J. Enol. Vitic. 36, 199–202.
Swiegers, J. H., Bartowsky, E. J., Henschke, P. A., and Pretorius, I. S. (2005). Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 11, 139–173. doi: 10.1111/j.1755-0238.2005.tb00285.x
Terrade, N., and Mira de Orduña, R. (2009). Determination of the essential nutrient requirements of wine-related bacteria from the genera Oenococcus and Lactobacillus. Int. J. Food Microbiol. 133, 8–13. doi: 10.1016/j.ijfoodmicro.2009.03.020
Tofalo, R., Schirone, M., Torriani, S., Rantsiou, K., Cocolin, L., Perpetuini, G., et al. (2012). Diversity of Candida zemplinina strains from grapes and Italian wines. Food Microbiol. 29, 18–26. doi: 10.1016/j.fm.2011.08.014
Tristezza, M., di Feo, L., Tufariello, M., Grieco, F., Capozzi, V., Spano, G., et al. (2016). Simultaneous inoculation of yeasts and lactic acid bacteria: effects on fermentation dynamics and chemical composition of Negroamaro wine. LWT Food Sci. Technol. 66, 406–412. doi: 10.1016/j.lwt.2015.10.064
Wells, A., and Osborne, J. P. (2011). Production of SO2 and SO2 binding compounds by Saccharomyces cerevisiae during the alcoholic fermentation and the impact on wine lactic acid bacteria. S. Afr. J. Enol. Vitic. 32, 267–279.
Keywords: non-Saccharomyces, malolactic fermentation, Oenococcus, wine, microbial interactions
Citation: Balmaseda A, Bordons A, Reguant C and Bautista-Gallego J (2018) Non-Saccharomyces in Wine: Effect Upon Oenococcus oeni and Malolactic Fermentation. Front. Microbiol. 9:534. doi: 10.3389/fmicb.2018.00534
Received: 16 January 2018; Accepted: 08 March 2018;
Published: 23 March 2018.
Edited by:
Vittorio Capozzi, University of Foggia, ItalyReviewed by:
Lucía González-Arenzana, Instituto de Ciencias de la Vid y del Vino (ICVV), SpainCopyright © 2018 Balmaseda, Bordons, Reguant and Bautista-Gallego. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joaquín Bautista-Gallego, am9hcXVpbmJnQGlnLmNzaWMuZXM=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.