- 1Institute of Microbiology, Christian-Albrechts University of Kiel, Kiel, Germany
- 2Institute of Zoology, Christian-Albrechts University of Kiel, Kiel, Germany
- 3GEOMAR Helmholtz Centre for Ocean Research Kiel, Kiel, Germany
Most eukaryotic species are colonized by a microbial community – the microbiota – that is acquired during early life stages and is critical to host development and health. Much research has focused on the microbiota biodiversity during the host life, however, empirical data on the basic ecological principles that govern microbiota assembly is lacking. Here we quantify the contribution of colonizer order, arrival time and colonization history to microbiota assembly on a host. We established the freshwater polyp Hydra vulgaris and its dominant colonizer Curvibacter as a model system that enables the visualization and quantification of colonizer population size at the single cell resolution, in vivo, in real time. We estimate the carrying capacity of a single Hydra polyp as 2 × 105 Curvibacter cells, which is robust among individuals and time. Colonization experiments reveal a clear priority effect of first colonizers that depends on arrival time and colonization history. First arriving colonizers achieve a numerical advantage over secondary colonizers within a short time lag of 24 h. Furthermore, colonizers primed for the Hydra habitat achieve a numerical advantage in the absence of a time lag. These results follow the theoretical expectations for any bacterial habitat with a finite carrying capacity. Thus, Hydra colonization and succession processes are largely determined by the habitat occupancy over time and Curvibacter colonization history. Our experiments provide empirical data on the basic steps of host-associated microbiota establishment – the colonization stage. The presented approach supplies a framework for studying habitat characteristics and colonization dynamics within the host–microbe setting.
Introduction
Most eukaryotic organisms are colonized by a microbial community that is known to have diverse functions in health and development of their host (McFall-Ngai et al., 2013; Sommer and Bäckhed, 2013; Sommer et al., 2017). Yet, the rules governing the assembly of host-associated microbial communities are only beginning to be understood. A recent hypothesis posits that the microbiota constitutes an ecological community utilizing the host as an ecological habitat (Costello et al., 2012; Christian et al., 2015; Coyte et al., 2015). Under this view, a host can be described as a finite habitat for bacterial colonization. As such, it is characterized by multiple abiotic and biotic parameters with its carrying capacity as a prominent property. Habitat carrying capacity is defined as the maximum number of individuals that an environment, here the host, can sustain. The ecological carrying capacity has an evolutionary significance since population size is a well-known determinant of the population evolutionary dynamics (reviewed in Lanfear et al., 2014).
To study host colonization and carrying capacity in detail, we established a system to follow spatial and temporal dynamics of colonizer population size of the freshwater cnidarian Hydra vulgaris strain AEP (hereafter Hydra) and its dominant bacterial colonizer. Under constant laboratory conditions Hydra reproduces clonally; hence, individual polyps are assumed isogenic. Hydra stably maintains a highly specific microbiota, including three main families, whose composition under laboratory conditions is similar to that in nature (Fraune and Bosch, 2007; Fraune et al., 2015). The bacteria inhabit the outer layer of the ectodermal epithelial cells – termed glycocalyx (Fraune and Bosch, 2007; Fraune et al., 2015). The glycocalyx layer is considered as a physicochemical interface for Hydra–microbiota interactions (Schröder and Bosch, 2016). The microbial community residing in the glycocalyx has been observed to have an anti-fungal activity (Fraune et al., 2015), however, the role of specific community members remains unknown. The ability to generate germ-free polyps and to culture the microbiota members (Franzenburg et al., 2013; Fraune et al., 2015), makes it an ideal model system for the study of microbiota colonization dynamics over time. One bacterium, Curvibacter sp., was identified as the main colonizer of Hydra. The Gram-negative β-proteobacterium represents about 76% of the bacterial community stably associated with Hydra (Fraune et al., 2015).
The quantification of host carrying capacity provides an estimate for the total host-associated microbiota population size that in turn provides information about ecological and evolutionary trajectories of the inhabiting bacteria. Much less is known about the colonizer population size dynamics during the initial colonization phase from single colonizer cells to maximum population size (i.e., carrying capacity). If the host, here the Hydra polyp, is indeed viewed as a habitat, we expect the colonization dynamics to follow the same rules of establishment as in any ecological habitat [e.g., soil (Nemergut et al., 2006) or particles (Datta et al., 2016)]. Here we present a one host – one microbe system to study host colonization dynamics. Using genetically labeled Curvibacter strains, we characterize the colonizers’ population dynamics during initial colonization and quantify the Hydra habitat carrying capacity. The labeled Curvibacter strains are thus considered as a proxy for the total microbiota community (in which Curvibacter constitutes the majority). Furthermore, we test for priority effects in host colonization, and whether those are directly dependent on the time lag between competing colonizers.
Materials and Methods
Culturing of Hydra and Generation of Germ-Free Polyps
Hydra vulgaris (strain AEP) was routinely cultured according to standard procedures at 18°C (Lenhoff and Brown, 1970). Germ-free polyps were prepared by incubation in an antibiotic solution containing 50 μg ml-1 each of ampicillin, rifampicillin, streptomycin, and neomycin with daily exchange of the medium as previously described (Hemmrich et al., 2007). After 1 week of treatment, the polyps were transferred into antibiotic-free medium for recovery (1 week). The absence of bacteria was verified by plating on Reasoner’s 2A (R2A) medium agar plates. During antibiotic treatment and during re-colonization experiments, polyps were starved.
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are provided in Table S1, Supplementary File 1. Curvibacter sp. AEP1.3 was routinely grown in pure culture at 30°C with aeration in R2A medium or on R2A agar plates. When required, antibiotics were added to the medium at the following concentrations: Kanamycin (Km), 3 or 5 μg ml-1 and gentamicin (Gm), 2 μg ml-1. The Curvibacter sp. strains described in Table S1, Supplementary File 1 were constructed using standard molecular biology techniques including plasmid delivery via bi- and triparental mating using E. coli MFDpir (Ferrières et al., 2010) as plasmid donor. A mobilizable derivate of the gene targeting plasmid vector pGT41 (Kickstein et al., 2007) was used for the construction of the substitution/deletion mutation of the putative flgC gene involved in flagellar motility. A chromosomal mini-Tn7 insertion (attTn7::miniTn7(aacC1)) was constructed using a transposon vector developed by Crepin et al. (2012). A detailed description of the plasmid and strain constructions is provided in the electronic Supplementary Material (Supplementary File 1).
Colonization Assays
Germ-free Hydra polyps were divided in multiple pools with a defined number of animals in each group and kept in 50 ml Hydra medium (Lenhoff and Brown, 1970) throughout all experiments. The polyps were re-incubated with approximately 100 or 104 cells per polyp for 24 h with cells from overnight cultures of either Curvibacter wild type or Curvibacter carrying either pRL153-GFP or pTW1-mCherry. After 24 h the medium was replaced in order to remove free cells. Every 24 h, three animals from one pool per time point were sampled, pooled and homogenized in an Eppendorf tube using a sterile pestle. Serial dilutions of the homogenate were plated on R2A agar plates or R2A agar plates supplemented with 5 μg ml-1 kanamycin to determine the CFUs per polyp and to distinguish between wild type and labeled (i.e., antibiotics resistant) populations. Experiments were carried out either for 168, 288 h or until a maximum of 312 h.
For experiments with Hydra polyps colonized with their natural resident microbiota, polyps were incubated with approximately 100 cells per polyp of Curvibacter carrying pRL153-GFP and treated as described above.
After acquiring the Curvibacter on-host population size (i.e., CFUs per polyp), non-linear least-squares minimization was used to fit a logistic growth model to the results and estimate the carrying capacity using the R package growthcurver (Sprouffske and Wagner, 2016).
Fluorescence Microscopy of Colonized Hydra Polyps
Hydra polyps colonized by GFP- and mCherry-expressing Curvibacter cells were visualized with an epifluorescence microscope (Zeiss Axio Imager 2, Plan-Apochromat 63×/1.40 Oil DIC M27 objective). Single polyps were transferred on concave microscopy glass slides and subsequently cooled down on ice for 10 min prior to imaging.
Priming Experiments
Two replicate populations of Curvibacter carrying plasmid pRL153-GFP were independently incubated on Hydra polyp pools for 168 h. After homogenization of the polyps, Curvibacter cells were carefully pelleted by centrifugation, recovered and serially diluted in Hydra medium. In parallel, two populations of Curvibacter wild type were incubated in R2A medium with daily transfers (1:100) for 168 h. Host-primed Curvibacter cells (obtained from the homogenized Hydra pools) and Curvibacter grown in R2A medium were used to initiate on-host-competition experiments. The experiments were conducted with approximately 100 cells per polyp of both Curvibacter strains. The number of both competitors per polyp was determined as described for the colonization assay.
Results
Establishing a Host–Microbe Model System: A Genetic Toolset for Curvibacter
In order to visualize and follow bacterial growth and population dynamics on Hydra we equipped Curvibacter with the mobilizable RSF1010 derivative (IncQ) pRL153-GFP (carrying nptII with its original Tn5 promoter and GFP expressed from the promoter Ptrc) (Tolonen et al., 2006). In biparental mating experiments with E. coli MFDpir, pRL153-GFP was effectively transferred to Curvibacter (frequency per recipients after 2 h: 5 × 10-5, n = 2, SEM = 2 × 10-5). For expression of the second fluorophore, we replaced GFP with mCherry (pTW1-mCherry). Infecting germ-free Hydra with marked Curvibacter lines enabled us to visualize Curvibacter populations on Hydra in real time at a single cell resolution (Figures 1A,B). We observed no impact of plasmid carriage on the fitness of Curvibacter compared to the plasmid-free wild type in pure culture (Table S3, Supplementary File 2). Consequently, the observed pattern of Curvibacter proliferation on Hydra cannot be explained by fluorophore expression or plasmid carriage.
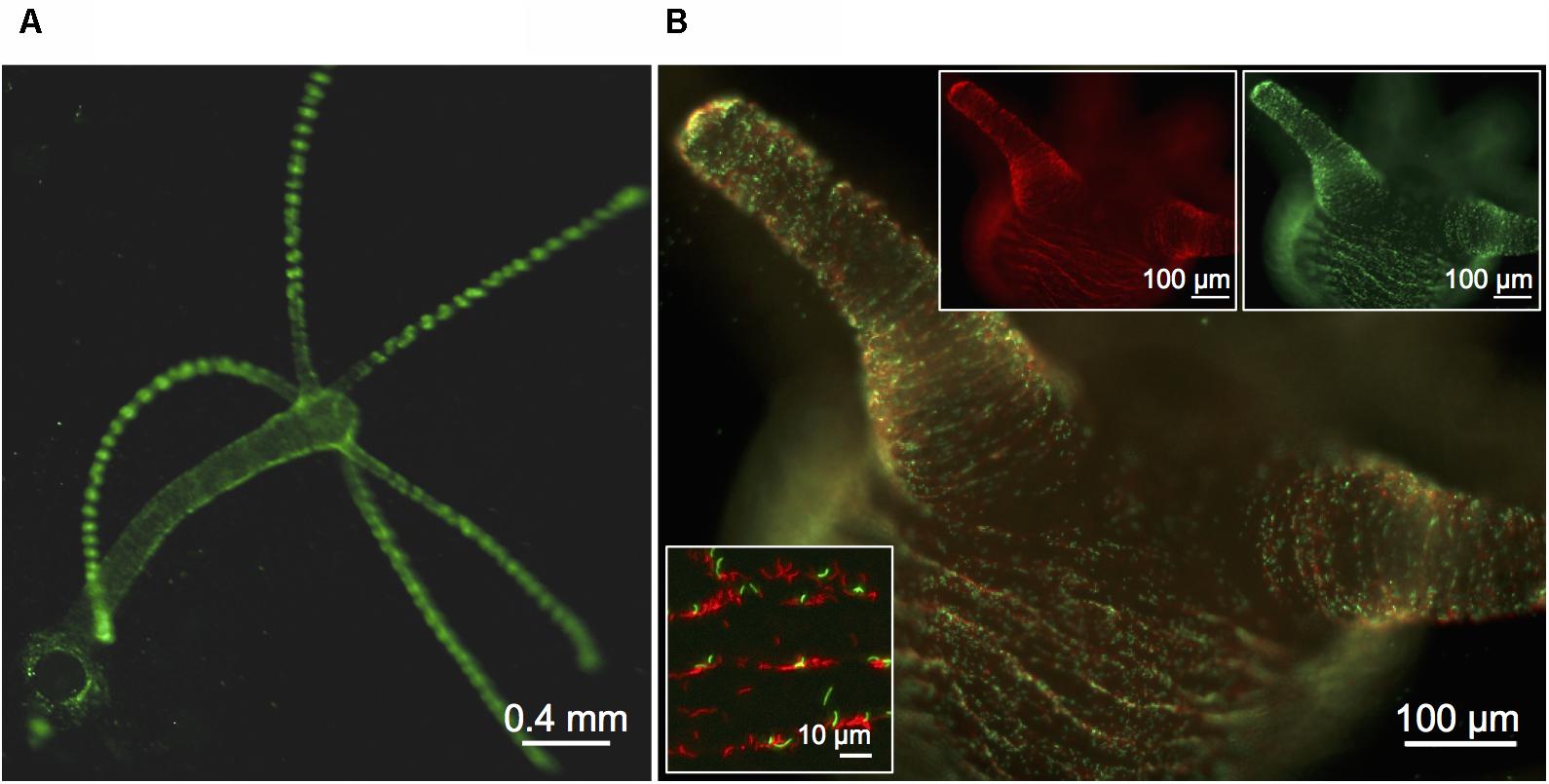
FIGURE 1. Visualization of fluorescently labeled Curvibacter. (A,B) Micrographs of a previously sterile polyp visualized 72 h post-infection with 6 × 103 cells per polyp of Curvibacter carrying plasmid pRL153-GFP (green) or plasmid pTW1-mCherry (red). (A) Overview of a Hydra polyp densely colonized by GFP-expressing Curvibacter. (B) Side-view on the Hydra head region with mixed populations of GFP- and mCherry-expressing Curvibacter. White boxes: Fluorescence signals of the GFP- (top right) and mCherry-labeled population (top left). Close-up of Curvibacter cells densely settling in the proximity of the polyp’s folding molds (bottom left).
Based on our observation that pRL153-GFP could be efficiently activated for conjugation, we developed a mobilizable derivate of the previously described ColE-derived gene targeting plasmid vector pGT41 (Kickstein et al., 2007; Overballe-Petersen et al., 2013; Hülter et al., 2017) that can be used for the generation of chromosomal deletion mutations in Curvibacter (see Supplementary File 1 for details).
Visualization of Curvibacter Host Colonization
A pool of germ-free Hydra polyps was infected with an isogenic inoculum consisting of approximately 100 fluorophore-expressing Curvibacter cells per polyp. After 24 h polyps were washed and single Curvibacter cells were observed inhabiting Hydra. After 48 h patches of Curvibacter cells were observed on the Hydra epithelium and at 72 h the cells densely covered the polyp (Figure 2A). The Curvibacter cells pattern on the polyp revealed a gradual increase of the Curvibacter population size on Hydra over time. Furthermore, our results reveal that Curvibacter settles in neat lines that follow the structure of the epithelium, which is determined by the shape of individual epithelium muscle cells (Anton-Erxleben et al., 2009; Figure 2B). This distinct spatial pattern suggests that the continuous constriction movement of Hydra has an impact on the Curvibacter spatial distribution. After 72 h the highest density of Curvibacter was observed in the Hydra tentacles region, hence Curvibacter distribution on Hydra is not homogeneous rather it is biased toward specific body parts (Figure 2A), as recently shown (Augustin et al., 2017).
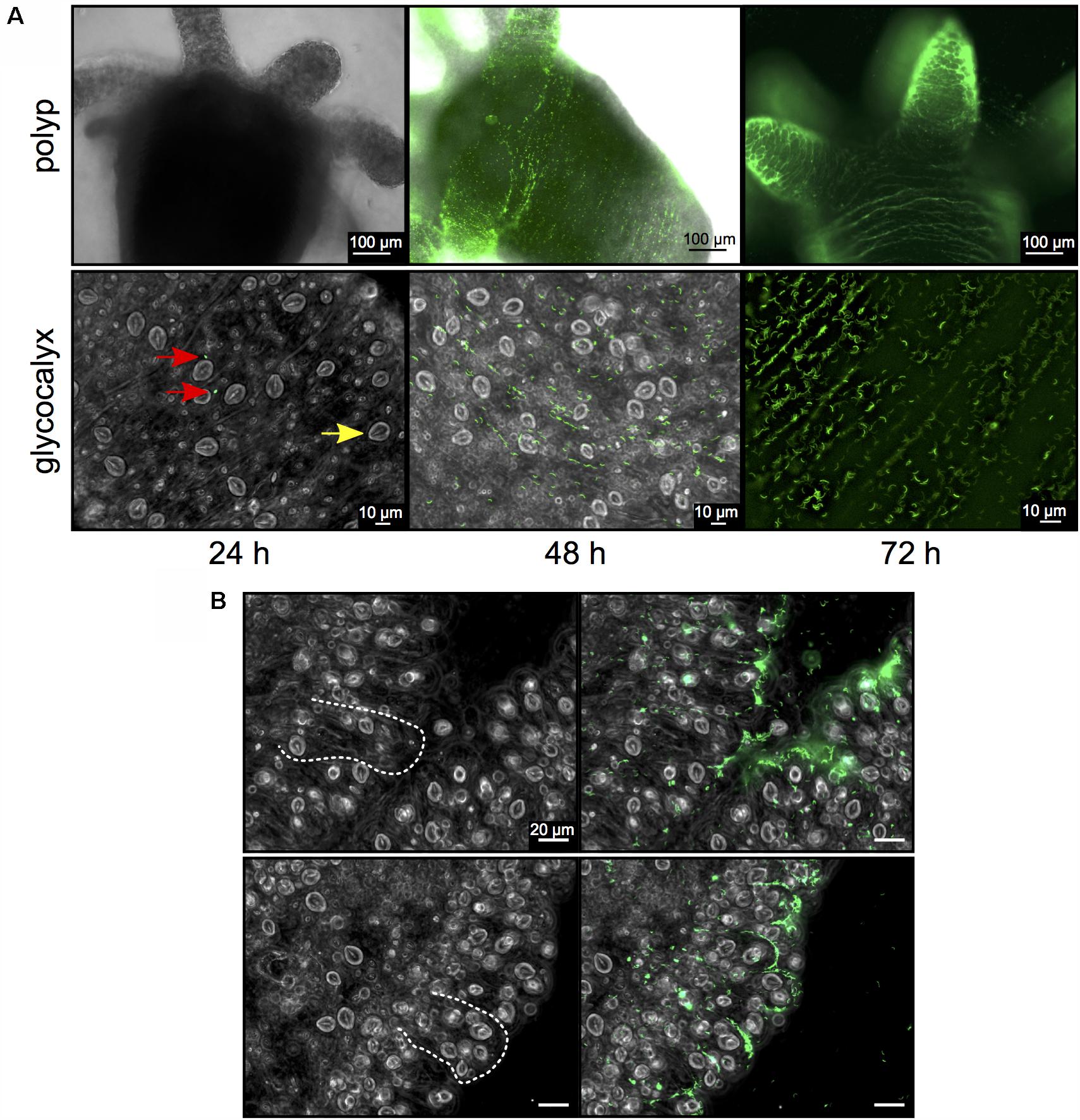
FIGURE 2. Curvibacter population size dynamics on Hydra. (A) Twenty-four hours post-infection, single Curvibacter cells (red arrows) are attached to the polyp (yellow arrow indicates a host nematocyst). Proliferation of Curvibacter leads to a dense layer of cells after 48 and 72 h. (B) Bright field micrographs of Hydra epithelium. Dashed lines indicate the shape of the epithelial battery cells filled with nematocyst in the tentacles of Hydra (left). Curvibacter carrying pRL153-GFP colonizing the Hydra epithelium along the shape of the epithelial muscle cells (right).
Quantification of Host Carrying Capacity and Colonization Dynamics
To further study Curvibacter colonization dynamics, we established a pipeline to quantify Curvibacter population size on Hydra over time (Figure 3A). We began by infecting two independent large germ-free Hydra polyp pools with Curvibacter (either wild type, or labeled). Batches of three polyps from each animal pool were sampled at consecutive time points and homogenized. The homogenate was plated on non-selective R2A plates. The number of colony forming units (CFUs) served as a direct estimate of the total bacterial population size on Hydra (Figure 3A). The quantification of CFUs per polyp for Curvibacter carrying pRL153-GFP or pTW1-mCherry was conducted by plating on non-selective R2A plates and R2A plates supplemented with kanamycin in parallel. No reduction in the plating efficiency of plasmid-carrying Curvibacter was observed. Hence, the CFU proxy was not affected by the marker presence. Furthermore, the plasmids pRL153-GFP and pTW1-mCherry were stable in Curvibacter under non-selective conditions. Only fluorescent colonies were found on non-selective R2A plates, hence the wild type CFU counts were not biased by the emergence of plasmid-free segregants.
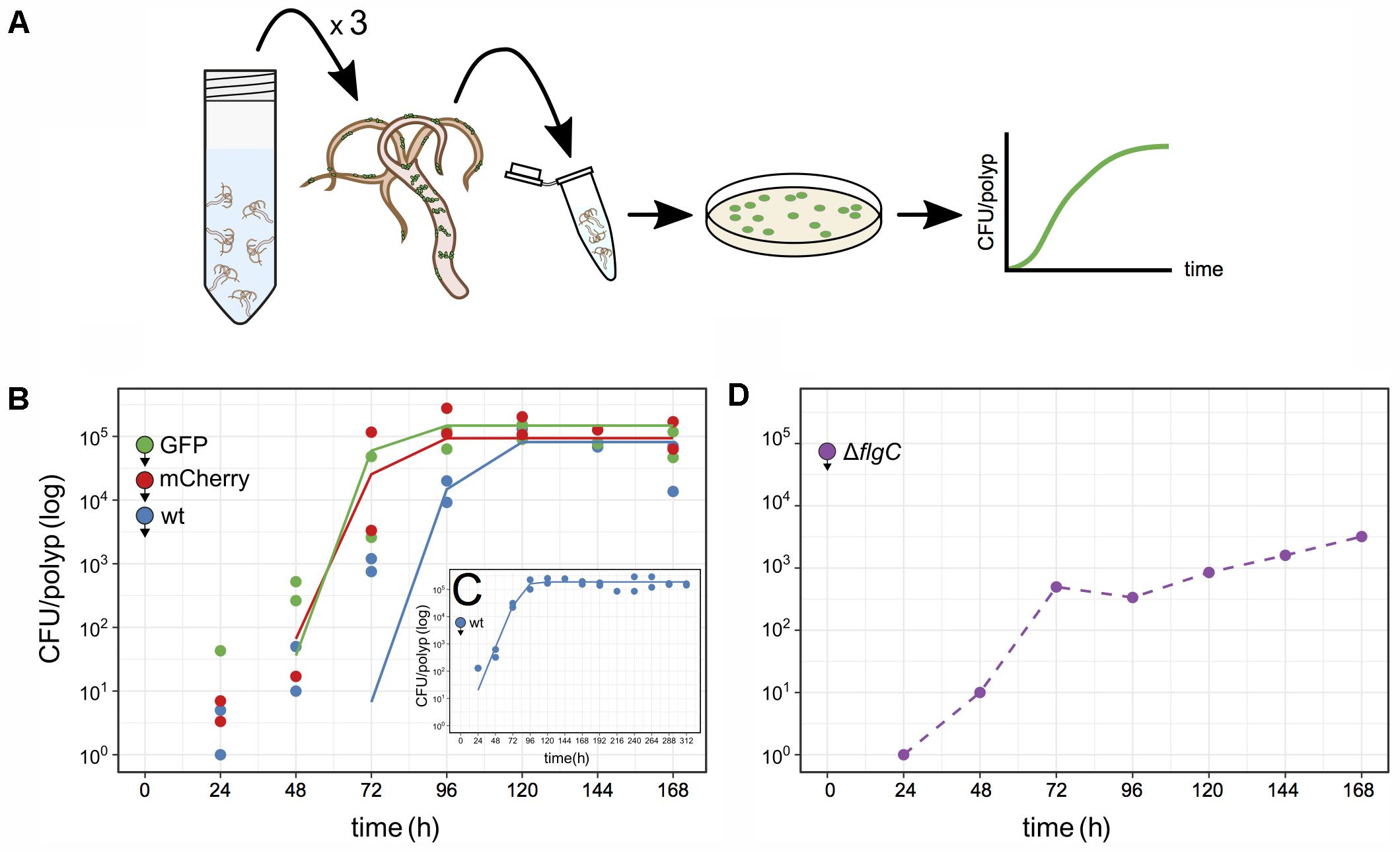
FIGURE 3. Quantification of Curvibacter population size on Hydra. (A) Schematic of the assay pipeline to follow colonization dynamics over time. Every 24 h, three polyps were sampled, homogenized, and the homogenate plated. (B–D) Population size of Curvibacter on Hydra over time. Hydra pools were incubated with 100 (B,C) or 104 (D) cells per polyp (see section “Materials and Methods” for details). Filled circles with arrowheads indicate the bacterial strain and its arrival time on Hydra. (B) Population size of three differently labeled but otherwise isogenic populations [n = 2, wild type (wt), pRL153-GFP, and pTW1-mCherry] estimated from independent Hydra pools over time. Solid lines represent the best fit of a logistic growth model (wt P-value = 1.7 × 10-4, GFP P-value = 9.0 × 10-6, mCherry P-value = 5.4 × 10-5). (C) Long-time population size dynamic of Curvibacter on Hydra over 312 h shows logistic growth (n = 2, P-value = 4 × 10-11). (D) Population size of a motility impaired Curvibacter flgC deletion mutant over time (dashed line).
Using the described pipeline, we followed the colonization dynamics of three lines, including the wild type, GFP, and mCherry lines in independent Hydra pools. At 24 h post-infection the Curvibacter population attached to Hydra was smaller than the inoculum size and reached a stable maximum after 96–120 h (Figure 3B). The gradual increase in CFU counts conforms to the increasing density of bacteria visually observed on Hydra over 72 h (Figure 2A). The wild type and the marked lines reached a similar stable maximum around 2 × 105 cells per polyp 168 h post-infection. We note that the CFU per polyp dynamics were similar among the marked and non-marked lines, hence there is no difference in their relative fitness. Repeating the experiment for a longer time of 312 h showed that a maximum of about 2 × 105 CFU per polyp was reached after 96 h and remained stable 312 h post-infection (Figure 3C). Conducting the colonization experiment with an inoculum size of 104 cells per polyp showed that a similar stable maximum was rapidly reached 48–72 h post-infection (Supplementary Figure S1). In order to confirm that the source of the observed Curvibacter cells was indeed only from Hydra-associated bacteria we performed two control experiments. In the first we plated supernatant sampled from the polyp pool 48 h post-infection. No bacterial colonies could be observed in this experiment (less than two cells per milliliter), indicating that the Curvibacter colonies originate only from Hydra polyps. In addition, we incubated Curvibacter in Hydra medium supplemented with glucose and plated aliquots of the medium after 24 h. No colonies were observed on non-selective plates in this experiment, indicating that Curvibacter was not persisting in the Hydra medium. Consequently, we conclude that the colony counts in our pipeline truly reflect the abundance of Hydra-associated bacteria.
We further tested our approach to document colonization dynamics by investigating whether swimming motility plays a role in the early colonization process. Curvibacter is highly motile during early and mid-logarithmic growth phase and the presence of a full flagellum gene repertoire indicates that Curvibacter motility is flagellum-driven. For the purpose of this experiment we constructed a deletion mutant where a putative flgC gene was replaced by a marker cassette (strain Fm11; Table S1, Supplementary File 1). The flgC gene encodes a flagellar body rod protein and its absence is known to hamper bacterial motility (Shippy et al., 2014; Qin et al., 2016). Using our pipeline, we examined the host colonization dynamics of the ΔflgC strain (Figure 3D). Starting with an inoculum of about 100 cells per polyp revealed no colonies 24–168 h post-infection. Increasing the initial inoculum to 104 cells per polyp improved the colonization success. About 500 cells per polyp were observed 72 h post-infection but only a maximum abundance of about 5 × 103 cells was reached 168 h post-infection (Figure 3D). A comparison of these results to the wild type colonization experiment, which was done in parallel (e.g., Figure 3B), indicates that Curvibacter motility is an important factor for the colonization initiation and maintenance.
Overall, we find robust growth characteristics of Curvibacter in Hydra host colonization, namely, consistent growth kinetics of Curvibacter among hosts that are characterized by a simple logistic growth model (Figures 3B,C). We show that the Hydra carrying capacity amounts to about 2 × 105 Curvibacter cells. This capacity is reached rapidly within 24–72 h, it is stable over long time, and it has robust characteristics among Hydra individuals.
Colonizing Population Disturbance and Re-colonization
To test the robustness of Hydra colonization and its carrying capacity we studied the colonization dynamics after an artificial disturbance of the colonizing Curvibacter population. Curvibacter is sensitive to gentamicin [adding 2 μg ml-1 gentamicin to a pure culture (108 cells) resulted in no detectable CFU after 2 h of incubation]. Thus, we disturbed the host-inhabiting Curvibacter population by dosing the colonized Hydra polyps with gentamicin. In this experiment, we disturbed the Curvibacter population by treating the Hydra pool with a dose of 2 μg ml-1 gentamicin. The first treatment was applied at 96 h post-infection and was repeated every 24 h (without changing the medium) until no colonies could be observed. Overall, four gentamicin treatments were required to eliminate the total Curvibacter population (Figure 4). To test the Hydra carrying capacity robustness, we re-infected the Hydra pool with a gentamicin resistant Curvibacter (strain AEP1.3-Gm; Table S1, Supplementary File 1). The first re-infection was performed 24 h after the last gentamicin treatment (at 192 h) with the same inoculum size as before. After 24 h, no CFUs were detected; consequently we repeated the re-infection twice in time lags of 24 h until CFUs could be detected (at 264 h; Figure 4). Within 72 h the Curvibacter population size of the re-colonization phase reached the typical Hydra carrying capacity (∼2 × 105). This experiment demonstrates that the Hydra carrying capacity is a robust characteristic of the host habitat that is independent of disturbance events. Additionally, we show that a decline in the existing colonizer population opens up ecological opportunities for new incoming colonizer populations.
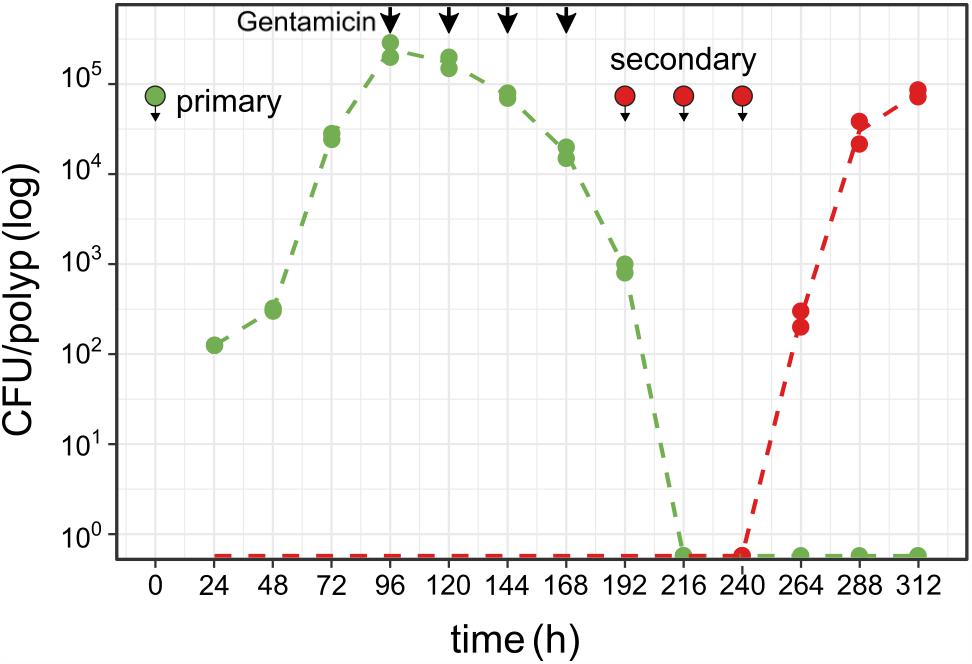
FIGURE 4. Colonizer population disturbance and re-colonization on Hydra. Curvibacter population size of first (green, pRL153-GFP) and second [red, attTn7::miniTn7(aacC1)] colonizer populations on Hydra (n = 2; dashed line represents the mean population size) during colonization and after colonization disruption by antibiotic treatment (black arrows, see section “Results” for details). Symbols indicate the time points of infection with Curvibacter populations.
The Consequences of Host Carrying Capacity for Consecutive Colonization
Using the Curvibacter–Hydra model we tested the consequences of preemptive competition in the Hydra habitat during colonization. To quantify the impact of colonization order and time, we manipulated the time lag between two consecutive colonizer populations. Using our pipeline (Figure 3A), Curvibacter wild type served as the primary colonizer while Curvibacter carrying plasmid pRL153-GFP was used as the secondary colonizer. The secondary colonizer (100 cells per polyp) was added at different time lags of 24, 48, or 72 h post-primary infection. We evaluated the colonizer population size by plating simultaneously on non-selective and selective media. The growth characteristics and Hydra carrying capacity of both colonizers were estimated using a logistic growth model.
In the first time-lag experiment the secondary colonizer population was added 24 h post-primary infection. The secondary colonizer population size was slightly fluctuating over time but resulted in a similar density as the primary colonizer 288 h post-infection. The total Hydra carrying capacity in this experiment was similar to the observed for a single colonizer population (Table 1 and Figure 5) and the carrying capacity of both colonizers remained stable over time (Figure 5A). Nevertheless, the secondary colonizer reached a slightly higher carrying capacity in comparison to the primary colonizer and constituted about 60% of the final colonizer population (Table 1 and Figure 5). This suggests that the secondary colonizer had a slight advantage over the primary colonizer on Hydra under these conditions despite the later arrival.
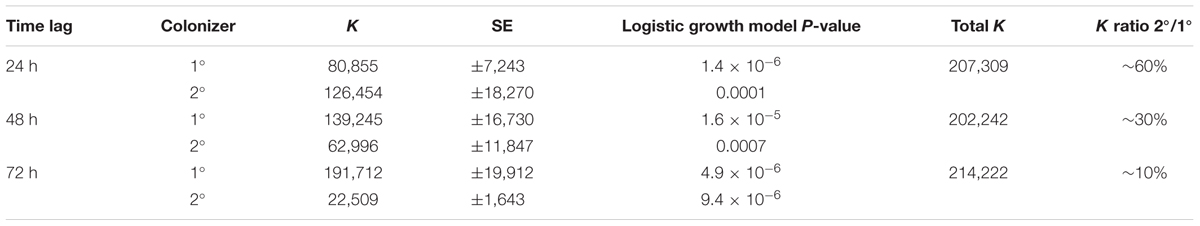
TABLE 1. Carrying capacity (K) for consecutive Curvibacter colonization in the host habitat Hydra fitted to a logistic growth model.
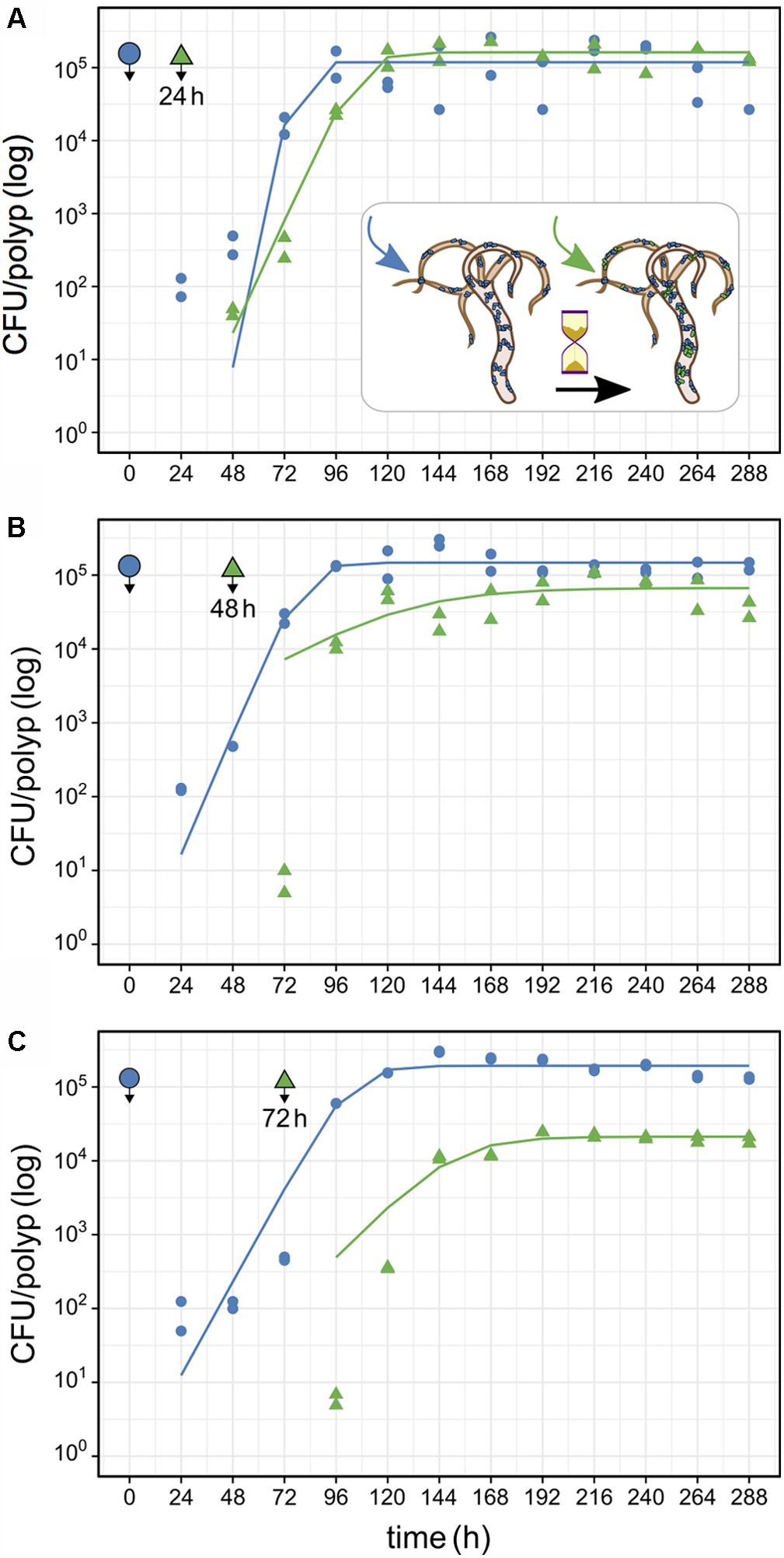
FIGURE 5. Population size of primary and secondary Curvibacter colonizers on Hydra. After infection with the primary colonizer (blue, wild type), the second colonizer (green, pRL153-GFP) was added after a time lag of (A) 24 h, (B) 48 h or (C) 72 h. All experiments were conducted with two biological replicates. Solid lines represent the best fit of a logistic growth model.
In the next experiment we applied a time lag of 48 h prior to infection of Hydra with the secondary colonizer. Shortly after the re-infection, secondary colonizers were observed inhabiting the host while their population size slightly fluctuated around a maximum of 2.5 × 104 cells per polyp. Together, the two colonizers reached a stable population size after 96 h with a total population size (∼2 × 105) similar to the estimates obtained from previous experiments. However, the secondary colonizer contributed to around 30% of the total colonizer population size only (Table 1 and Figure 5B). At this time lag, the primary colonizer had a significant numerical advantage over the secondary colonizer that remained stable over time. Thus, in contrast to a time lag of 24 h, a time lag of 48 h between primary and secondary colonization results in different carrying capacity for the two colonizers (Table 1 and Figure 5B).
The third experiment was performed with an extended time lag of 72 h between primary and secondary infection. Here, the secondary colonizer contributed about 10% to the total bacterial population size on Hydra (Table 1). This means that the carrying capacity for the consecutive colonizer was strongly reduced due to the high numerical advantage of the primary colonizer (Table 1 and Figure 5C). Thus, with 48 and 72 h time lag the Hydra carrying capacity of the secondary colonizer is significantly lower in comparison to the primary colonizer, whereas the total carrying capacity is similar to the previous experiments and is stable over time (Table 1 and Figure 5C).
To further test the consequences of host occupancy on the success of an incoming colonizer we performed a colonization experiment with Hydra individuals that were fully colonized by their natural microbiota. Our results revealed no CFU of the incoming Curvibacter colonizer over 168 h, hence the incoming Curvibacter population completely failed to colonize a previously populated Hydra.
Our results demonstrate that the time lag between consecutive colonization is a decisive determinant of their population size on Hydra. Since the Hydra carrying capacity is finite, the carrying capacity of an incoming colonizer largely depends on the Hydra habitat occupancy. Host occupancy is thus an important determinant of the establishment success of incoming colonizers.
Colonization Dynamics of Primed and Naïve Colonizers
To assess the source of advantage of the first colonizer we tested the effect of colonization history on the colonization success by performing a direct competition between primed and naïve colonizers. Primed colonizers (pRL153-GFP) were sampled from Curvibacter populations that were cultivated on Hydra prior to the experiment, while naïve colonizers (wild type) were sampled from Curvibacter populations maintained in liquid R2A medium (as in previous colonization experiments). A pool of germ-free Hydra polyps was inoculated with primed and naïve colonizers simultaneously at an equal density of approximately 100 cells per polyp. Using our pipeline, we documented changes in the primed and naïve colonizer population size by plating on non-selective and selective media. Our results reveal that 24 h post-infection both colonizers reach similar cell densities on the Hydra polyps. After 48 h the primed colonizer population size was slightly larger than that of the naïve colonizer, whereas at 72 h post-infection the primed colonizer largely outcompeted the naïve colonizer. Throughout the remaining experiment time, the naïve colonizer remained at low cell density of about 100 cells per polyp with a ratio of approximately 1:100 naïve to primed colonizer cells (Figure 6). To test whether this result was due to a general fitness benefit of the primed Curvibacter strain, we compared the fitness of the primed relative to the naïve strain in the absence of the host. A competition experiment between these two strains in liquid culture revealed no advantage of the primed strain after 24 or 48 h (Table S3, Supplementary File 2). Hence, the fitness advantage of the primed Curvibacter manifests itself only in the presence of the host. These experiments reveal that Curvibacter colonization history plays a major role in the host colonization success.
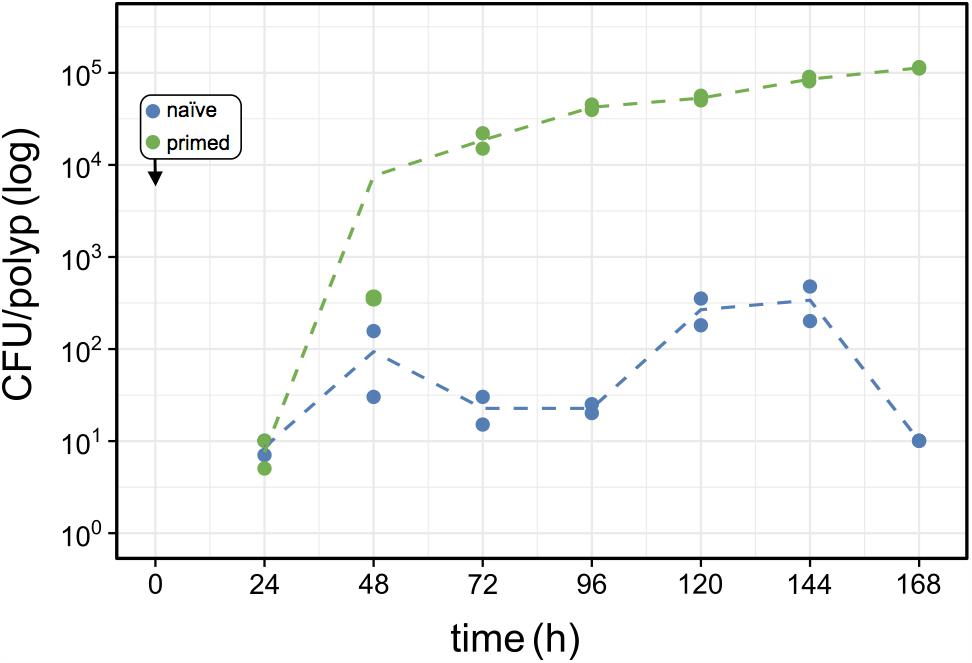
FIGURE 6. Effects of host-priming on Curvibacter fitness. Population size of primed Curvibacter carrying pRL153-GFP (green) and a naïve Curvibacter wild type (blue) on Hydra over time. Both Curvibacter strains were added simultaneously to the Hydra pool (box) (n = 2; dashed line represents the mean population size).
Discussion
In our experiments we aimed to follow the total Hydra host colonization process over time and to test whether basic ecological concepts of habitat occupation (i.e., finite carrying capacity, robustness, and priority effects) hold true in a reductionist one host – one colonizer system. The ability to genetically modify Curvibacter enables the quantification of host colonization dynamics from single colonizer cells to growth-limited populations in real time. Our finding that IncQ plasmids are stably maintained in Curvibacter enables us to visualize Curvibacter colonization on Hydra and follow the emergence of colonization patterns over time. Our results confirm the hypothesis that the carrying capacity of Hydra as a habitat is finite, stable over time and has little variation among individual polyps (coefficient of variance between pairs of biological replicates did not exceed 23%). The carrying capacity we observed for Curvibacter is comparable to the total microbiota load on Hydra (Pietschke et al., 2017). Hence, our reductionist model appears realistic with regards to bacterial population size in the Hydra habitat. We demonstrate that the carrying capacity remains robust even after disturbance of the colonizer population. Furthermore, we show that the presence of colonizing bacteria in the host habitat decreases the carrying capacity of late colonizing bacteria. Thus, habitat occupancy is a determinant of colonizer carrying capacity, similarly to other ecological habitats. We show that the first arriving colonizer has a numerical advantage over late arrivals that is corresponding to the time lag to the second colonizer, a phenomenon that has been termed priority effect (reviewed in De Meester et al., 2016). The priority effect of first arriving Curvibacter is not only due to a numerical advantage as colonizers primed for Hydra have an advantage over naïve colonizers. This indicates that the priority effect in Curvibacter colonization of Hydra is enhanced by colonization history. Similarly to any other habitat, priority effects may be widespread in host–microbe interactions and might play an important role in host colonization. For example, a study comparing the colonization success of first and late arriving pathogenic Borrelia burgdorferi strains showed that the first arriving colonizer had a strong priority effect on the late arriving colonizer in a double host habitat comprising mice and ticks (Devevey et al., 2015). Nonetheless, the importance of colonization history in microbial community assembly was so far shown only for abiotic habitats (e.g., Gomez et al., 2016).
The colonization visualization with fluorescence microscopy revealed that Curvibacter cells are localized perpendicular to Hydra’s muscle cells, which suggests that the host constriction movements are the cause for Curvibacter spatial distribution on Hydra. The clear enrichment of Curvibacter in the tentacle area suggests that different Hydra body parts constitute distinct niches. It is thus likely that the host strongly influences bacterial cell localization and distribution in the host habitat. This observation is in agreement with a recent finding of a Hydra neuropeptide that is involved in patchy bacteria distribution on Hydra (Augustin et al., 2017). Additionally, the polyp movement appears to be an important determinant of bacterial population’s dynamics on the host and may strongly influence the colonization outcome. Similarly to Hydra, gut movements of the larval zebrafish were shown to significantly alter the colonization success of two colonizing populations (Wiles et al., 2016).
The robust characteristics of host habitat carrying capacity can be studied by a comparison of habitat characteristics among isogenic host individuals. In contrast to our results on Hydra, a study of bacteria residing in the gut of zebrafish larvae showed that the microbiota carrying capacity was variable among different individuals (Jemielita et al., 2014). This could be attributed to the strong gut motility leading to stochastic differences among individuals (Jemielita et al., 2014; Wiles et al., 2016). Furthermore, the host carrying capacity may be heterogeneous among different niches that correspond to sites or organs of the hosting organism. A study of plant microbiota of green snap beans (Phaseolus vulgaris) showed that leaf colonization by epiphytic bacteria occurs at multiple colonization sites having different carrying capacities (Remus-Emsermann et al., 2012). The patchiness in local carrying capacity could be attributed to a heterogeneous distribution of nutrients in the leaf habitat. Here, the total habitat carrying capacity of the green snap bean leaf is the sum of several local carrying capacities (Remus-Emsermann et al., 2012). The visualization of the unequal colonization of Curvibacter on Hydra may reveal a similar effect highlighting that a host can be comprised of different habitat niches.
Our results provide insights into the basic principles that govern host-associated bacterial colonization. As such, our data supports the idea that the microbiota is an ecological community utilizing the finite resources of a host habitat (Costello et al., 2012; Douglas and Werren, 2016). As it has been pointed out before, this does not rule out past or present co-evolution, as is the case in any other ecological community (Thompson and Cunningham, 2002; Thrall et al., 2007). The observed dominance and competitiveness of early colonizers constitute an extreme instance of ecological drift that is coupled with a strong population genetic bottleneck that is a consequence of the finite carrying capacity (Hu et al., 2006). An alternative mode of symbiont population bottleneck has been observed in the Squid–Vibrio symbiosis where the number of bacterial cells entering the light organ is limited and the community diversification occurs after the initial colonization (Wollenberg and Ruby, 2009; Sun et al., 2016).
A priority effect that is further enhanced by adaptation can lead to intraspecific genetic diversification of colonizers in distinct habitats (e.g., Gomez et al., 2016), where local adaptation and exclusion further promote the population divergence (Knope et al., 2012; Orsini et al., 2013). Using the Hydra–Curvibacter model we demonstrate the importance of colonization history, which may depend on specific host-mediated modifications of Curvibacter quorum-sensing signals (Pietschke et al., 2017), in host colonization and suggest ongoing adaptation that may lead to diversification and speciation. Thus, the phylogenetic relations of microbial populations reflect their ecological and evolutionary history. The application of ecological theory to explain host-associated microbiota establishment and colonization patterns is only just beginning. Our study provides first empirical data on the basic steps of host-associated microbiota establishment – the colonization stage in real time.
Author Contributions
TW, TD, and NH designed the study. TW and NH performed the experiments. TW, TD, TR, and NH analyzed the data. SF and TB contributed Hydra polyps and wild type Curvibacter sp. AEP1.3 strain. TW, TD, SF, TB, TR, and NH wrote the manuscript.
Funding
TW, NH, and TD acknowledge support from the European Research Council (Grant No. 281357 to TD). TD, SF, TB, and TR acknowledge support from the CRC1182 Origin and Function of Metaorganisms.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer TM and handling Editor declared their shared affiliation.
Acknowledgments
We thank Elie Jami, Judith Ilhan, Giddy Landan, Maxime Godfroid, and Julia Weissenbach for critical comments on the manuscript and Doris Willoweit-Ohl for the preparation of germ-free polyps.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00443/full#supplementary-material
References
Anton-Erxleben, F., Thomas, A., Wittlieb, J., Fraune, S., and Bosch, T. C. G. (2009). Plasticity of epithelial cell shape in response to upstream signals: a whole-organism study using transgenic Hydra. Zoology 112, 185–194. doi: 10.1016/j.zool.2008.09.002
Augustin, R., Schröder, K., Rincón, A. P. M., Fraune, S., Anton-Erxleben, F., Herbst, E.-M., et al. (2017). A secreted antibacterial neuropeptide shapes the microbiome of Hydra. Nat. Commun. 8:698. doi: 10.1038/s41467-017-00625-1
Christian, N., Whitaker, B. K., and Clay, K. (2015). Microbiomes: unifying animal and plant systems through the lens of community ecology theory. Front. Microbiol. 6:869. doi: 10.3389/fmicb.2015.00869
Costello, E. K., Stagaman, K., Dethlefsen, L., Bohannan, B. J. M., and Relman, D. A. (2012). The application of ecological theory toward an understanding of the human microbiome. Science 336, 1255–1262. doi: 10.1126/science.1224203
Coyte, K. Z., Schluter, J., and Foster, K. R. (2015). The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666. doi: 10.1126/science.aad2602
Crepin, S., Harel, J., and Dozois, C. M. (2012). Chromosomal complementation using Tn7 transposon vectors in Enterobacteriaceae. Appl. Environ. Microbiol. 78, 6001–6008. doi: 10.1128/AEM.00986-12
Datta, M. S., Sliwerska, E., Gore, J., Polz, M. F., and Cordero, O. X. (2016). Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat. Commun. 7:11965. doi: 10.1038/ncomms11965
De Meester, L., Vanoverbeke, J., Kilsdonk, L. J., and Urban, M. C. (2016). Evolving perspectives on monopolization and priority effects. Trends Ecol. Evol. 31, 136–146. doi: 10.1016/j.tree.2015.12.009
Devevey, G., Dang, T., Graves, C. J., Murray, S., and Brisson, D. (2015). First arrived takes all: inhibitory priority effects dominate competition between co-infecting Borrelia burgdorferi strains. BMC Microbiol. 15:61. doi: 10.1186/s12866-015-0381-0
Douglas, A. E., and Werren, J. H. (2016). Holes in the hologenome: Why host-microbe symbioses are not holobionts. mBio 7:e2099-15. doi: 10.1128/mBio.02099-15
Ferrières, L., Hémery, G., Nham, T., Guérout, A.-M., Mazel, D., Beloin, C., et al. (2010). Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J. Bacteriol. 192, 6418–6427. doi: 10.1128/JB.00621-10
Franzenburg, S., Fraune, S., Altrock, P. M., Künzel, S., Baines, J. F., Traulsen, A., et al. (2013). Bacterial colonization of Hydra hatchlings follows a robust temporal pattern. ISME J. 7, 781–790. doi: 10.1038/ismej.2012.156
Fraune, S., Anton-Erxleben, F., Augustin, R., Franzenburg, S., Knop, M., Schröder, K., et al. (2015). Bacteria–bacteria interactions within the microbiota of the ancestral metazoan Hydra contribute to fungal resistance. ISME J. 9, 1543–1556. doi: 10.1038/ismej.2014.239
Fraune, S., and Bosch, T. C. G. (2007). Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl. Acad. Sci. U.S.A. 104, 13146–13151. doi: 10.1073/pnas.0703375104
Gomez, P., Paterson, S., De Meester, L., Liu, X., Lenzi, L., Sharma, M. D., et al. (2016). Local adaptation of a bacterium is as important as its presence in structuring a natural microbial community. Nat. Commun. 7, 1–8. doi: 10.1038/ncomms12453
Hemmrich, G., Anokhin, B., Zacharias, H., and Bosch, T. C. G. (2007). Molecular phylogenetics in Hydra, a classical model in evolutionary developmental biology. Mol. Phyl. Evol. 44, 281–290. doi: 10.1016/j.ympev.2006.10.031
Hu, X.-S., He, F., and Hubbell, S. P. (2006). Neutral theroy in macroecology and population genetics. OIKOS 113, 548–556.
Hülter, N., Sørum, V., Borch-Pedersen, K., Liljegren, M. M., Utnes, A. L. G., Primicerio, R., et al. (2017). Costs and benefits of natural transformation in Acinetobacter baylyi. BMC Microbiol. 17:34. doi: 10.1186/s12866-017-0953-2
Jemielita, M., Taormina, M. J., Burns, A. R., Hampton, J. S., Rolig, A. S., Guillemin, K., et al. (2014). Spatial and temporal features of the growth of a bacterial species colonizing the zebrafish Gut. mBio 5:e1751-14. doi: 10.1128/mBio.01751-14
Kickstein, E., Harms, K., and Wackernagel, W. (2007). Deletions of recBCD or recD influence genetic transformation differently and are lethal together with a recJ deletion in Acinetobacter baylyi. Microbiology 153, 2259–2270. doi: 10.1099/mic.0.2007/005256-0
Knope, M. L., Forde, S. E., and Fukami, T. (2012). Evolutionary history, immigration history, and the extent of diversification in community assembly. Front. Microbiol. 2:273. doi: 10.3389/fmicb.2011.00273
Lanfear, R., Kokko, H., and Eyre-Walker, A. (2014). Population size and the rate of evolution. Trends Ecol. Evol. 29, 33–41. doi: 10.1016/j.tree.2013.09.009
Lenhoff, H. M., and Brown, R. D. (1970). Mass culture of Hydra: an improved method and its application to other aquatic invertebrates. Lab. Anim. 4, 139–154. doi: 10.1258/002367770781036463
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C. G., Carey, H. V., Domazet-Losot, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
Nemergut, D. R., Anderson, S. P., Cleveland, C. C., Martin, A. P., Miller, A. E., Seimon, A., et al. (2006). Microbial community succession in an unvegetated, recently deglaciated soil. Microb. Ecol. 53, 110–122. doi: 10.1007/s00248-006-9144-7
Orsini, L., Vanoverbeke, J., Swillen, I., Mergeay, J., and De Meester, L. (2013). Drivers of population genetic differentiation in the wild: isolation by dispersal limitation, isolation by adaptation and isolation by colonization. Mol. Ecol. 22, 5983–5999. doi: 10.1111/mec.12561
Overballe-Petersen, S., Harms, K., Orlando, L. A. A., Mayar, J. V. M., Rasmussen, S., Dahl, T. W., et al. (2013). Bacterial natural transformation by highly fragmented and damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 110, 19860–19865. doi: 10.1073/pnas.1315278110
Pietschke, C., Treitz, C., Forêt, S., Schultze, A., Künzel, S., Tholey, A., et al. (2017). Host modification of a bacterial quorum-sensing signal induces a phenotypic switch in bacterial symbionts. Proc. Natl. Acad. Sci. U.S.A. 114, E8488–E8497. doi: 10.1073/pnas.1706879114
Qin, Y., Lin, G., Chen, W., Xu, X., and Yan, Q. (2016). Flagellar motility is necessary for Aeromonas hydrophila adhesion. Microb. Pathog. 98, 160–166. doi: 10.1016/j.micpath.2016.07.006
Remus-Emsermann, M. N. P., Tecon, R., Kowalchuk, G. A., and Leveau, J. H. J. (2012). Variation in local carrying capacity and the individual fate of bacterial colonizers in the phyllosphere. ISME J. 6, 756–765. doi: 10.1038/ismej.2011.209
Schröder, K., and Bosch, T. C. G. (2016). The origin of mucosal immunity: Lessons from the holobiont Hydra. mBio 7:e1184-16. doi: 10.1128/mBio.01184-16
Shippy, D. C., Eakley, N. M., Mikheil, D. M., and Fadl, A. A. (2014). Role of the flagellar basal-body protein, FlgC, in the binding of Salmonella enterica Serovar Enteritidis to host cells. Curr. Microbiol. 68, 621–628. doi: 10.1007/s00284-014-0521-z
Sommer, F., Anderson, J. M., Bharti, R., Raes, J., and Rosenstiel, P. (2017). The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 15, 630–638. doi: 10.1038/nrmicro.2017.58
Sommer, F., and Bäckhed, F. (2013). The gut microbiota — masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238. doi: 10.1038/nrmicro2974
Sprouffske, K., and Wagner, A. (2016). Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics 17:172. doi: 10.1186/s12859-016-1016-7
Sun, Y., LaSota, E. D., Cecere, A. G., LaPenna, K. B., Larios-Valencia, J., Wollenberg, M. S., et al. (2016). Intraspecific competition impacts Vibrio fischeri strain diversity during initial colonization of the squid light organ. Appl. Environ. Microbiol. 82, 3082–3091. doi: 10.1128/AEM.04143-15
Thompson, J. N., and Cunningham, B. M. (2002). Geographic structure and dynamics of coevolutionary selection. Nature 417, 735–738. doi: 10.1038/nature00810
Thrall, P. H., Hochberg, M. E., Burdon, J. J., and Bever, J. D. (2007). Coevolution of symbiotic mutualists and parasites in a community context. Trends Ecol. Evol. 22, 120–126. doi: 10.1016/j.tree.2006.11.007
Tolonen, A. C., Liszt, G. B., and Hess, W. R. (2006). Genetic manipulation of Prochlorococcus strain MIT9313: green fluorescent protein expression from an RSF1010 plasmid and Tn5 transposition. Appl. Environ. Microbiol. 72, 7607–7613. doi: 10.1128/AEM.02034-06
Wiles, T. J., Jemielita, M., Baker, R. P., Schlomann, B. H., Logan, S. L., Ganz, J., et al. (2016). Host gut motility promotes competitive exclusion within a model intestinal microbiota. PLoS Biol. 14:e1002517. doi: 10.1371/journal.pbio.1002517
Keywords: Curvibacter, microbial ecology, host–microbe interactions, metaorganisms, Hydra
Citation: Wein T, Dagan T, Fraune S, Bosch TCG, Reusch TBH and Hülter NF (2018) Carrying Capacity and Colonization Dynamics of Curvibacter in the Hydra Host Habitat. Front. Microbiol. 9:443. doi: 10.3389/fmicb.2018.00443
Received: 09 November 2017; Accepted: 26 February 2018;
Published: 14 March 2018.
Edited by:
Iliana B. Baums, Pennsylvania State University, United StatesReviewed by:
Tim Miyashiro, Pennsylvania State University, United StatesCatherine Maree Burke, University of Technology Sydney, Australia
Kimberly B. Ritchie, University of South Carolina Beaufort, United States
Copyright © 2018 Wein, Dagan, Fraune, Bosch, Reusch and Hülter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nils F. Hülter, bmh1ZWx0ZXJAaWZhbS51bmkta2llbC5kZQ==
 Tanita Wein
Tanita Wein Tal Dagan
Tal Dagan Sebastian Fraune
Sebastian Fraune Thomas C. G. Bosch
Thomas C. G. Bosch Thorsten B. H. Reusch
Thorsten B. H. Reusch Nils F. Hülter
Nils F. Hülter