
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 14 March 2018
Sec. Infectious Agents and Disease
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.00437
This article is part of the Research TopicBacterial Colonization: Bystander Microbiota or the Initial Step of Chronic Disease?View all 15 articles
Streptococcus agalactiae or group B streptococcus (GBS) is a leading cause of serious neonatal infections. GBS is an opportunistic commensal constituting a part of the intestinal and vaginal physiologic flora and maternal colonization is the principal route of GBS transmission. GBS is a pathobiont that converts from the asymptomatic mucosal carriage state to a major bacterial pathogen causing severe invasive infections. At present, as many as 10 serotypes (Ia, Ib, and II–IX) are recognized. The aim of the current review is to shed new light on the latest epidemiological data and clonal distribution of GBS in addition to discussing the most important colonization determinants at a molecular level. The distribution and predominance of certain serotypes is susceptible to variations and can change over time. With the availability of multilocus sequence typing scheme (MLST) data, it became clear that GBS strains of certain clonal complexes possess a higher potential to cause invasive disease, while other harbor mainly colonizing strains. Colonization and persistence in different host niches is dependent on the adherence capacity of GBS to host cells and tissues. Bacterial biofilms represent well-known virulence factors with a vital role in persistence and chronic infections. In addition, GBS colonization, persistence, translocation, and invasion of host barriers are largely dependent on their adherence abilities to host cells and extracellular matrix proteins (ECM). Major adhesins mediating GBS interaction with host cells include the fibrinogen-binding proteins (Fbs), the laminin-binding protein (Lmb), the group B streptococcal C5a peptidase (ScpB), the streptococcal fibronectin binding protein A (SfbA), the GBS immunogenic bacterial adhesin (BibA), and the hypervirulent adhesin (HvgA). These adhesins facilitate persistent and intimate contacts between the bacterial cell and the host, while global virulence regulators play a major role in the transition to invasive infections. This review combines for first time epidemiological data with data on adherence and colonization for GBS. Investigating the epidemiology along with understanding the determinants of mucosal colonization and the development of invasive disease at a molecular level is therefore important for the development of strategies to prevent invasive GBS disease worldwide.
Streptococcus agalactiae or group B streptococcus (GBS) is a pathobiont that is often part of the normal microbiota found in the gastrointestinal and genitourinary tracts of healthy women (Verani et al., 2010). It can cause serious neonatal infections and adult infections. During the early 1930s, GBS was initially identified as a veterinary pathogen and a frequent source of bovine mastitis (Keefe, 1997). The first reported cases of fatal human GBS infections were investigated by Fry (1938). Severe perinatal GBS infections were originally described in the 1960s (Hood et al., 1961). Shortly afterward since the 1970s, GBS emerged as a leading cause of neonatal mortality and morbidity in the United States (Dermer et al., 2004).
The gastrointestinal tract is recognized as a reservoir for GBS and represents most probably the source of vaginal colonization (Meyn et al., 2009). Maternal colonization is the principal route of GBS transmission in early-onset infections as bacteria can spread either in utero by ascending infection or during birth through neonatal aspiration of contaminated amniotic or vaginal fluids (Maisey et al., 2008a; Rajagopal, 2009; Verani et al., 2010). About 30–70% of colonized mothers deliver GBS colonized newborns and 1–2% of these develop early-onset infections where heavy colonized mothers are more likely to transmit GBS to their offspring (Anthony et al., 1979; Barcaite et al., 2008; Melin, 2011; Melin and Efstratiou, 2013). However, the route for GBS acquisition in late-onset infections is less clear. It may develop through vertical transmission from mother to neonate, nosocomial transmission, contaminated breast milk or prematurity (Rajagopal, 2009; Le Doare and Kampmann, 2014; Zimmermann et al., 2017).
Group B streptococcus diseases in neonates which develop within the 1st week after birth are designated as early-onset disease (EOD). Late-onset infections (LOD) develop between the 7th day of birth and 2 or 3 months of age. Early-onset infections usually manifest as pneumonia and sepsis while meningitis is most common as a Late-onset event (Verani et al., 2010; Melin and Efstratiou, 2013). Newborns with EOD frequently suffer from respiratory failure which rapidly develops into bacteremia and septic shock. Infants surviving LOD meningitis will develop chronic neurologic sequelae including seizures, cognitive impairment, hearing loss and blindness in up to 50% (Schuchat, 1998; Maisey et al., 2008a; Libster et al., 2012; Melin and Efstratiou, 2013).
During the mid-1990s, the American College of Obstetricians and Gynecologists (ACOG), Centers for Disease Control and Prevention (CDC) and the American Academy of Pediatrics (AAP) recommended intrapartum antibiotic prophylaxis (IAP) to prevent perinatal GBS disease (Schuchat, 1998). This was followed by revised guidelines for the prevention of GBS disease issued in 2002 and the updated guidelines in 2010 (Verani et al., 2010; Schrag and Verani, 2013) which are currently in use. These recommendations included a universal culture-based screening for pregnant women at 35–37 weeks of pregnancy in order to limit IAP to a certain risk group. Widespread active implementation of an IAP program in the United States resulted in an outstanding decrease in the incidence of the disease. Invasive EOD GBS infections declined from 1.8 cases/1000 live births in the early 1990s to 0.26 cases/1000 live births in 2010 (Schrag and Verani, 2013). However, IAP had no impact on late-onset infections and thus LOD GBS diseases continue as the leading cause of neonatal morbidity and mortality (Verani et al., 2010; Schrag and Verani, 2013).
Asymptomatic GBS carriage is frequent and in general harmless in healthy women. GBS can, however, cause serious infections in pregnant women. Invasive maternal illness due to GBS in pregnant and postpartum women include bloodstream infections, meningitis, osteomyelitis, and endocarditis. Non-invasive maternal diseases manifest as bacteriuria, amnionitis, fasciitis, cellulitis, endometritis and wound infections associated with episiotomies or cesarean deliveries. In addition, GBS has been increasingly reported as being responsible for invasive disease in elderly and immunocompromised patients. In non-pregnant adults, GBS diseases include arthritis, endocarditis, pneumonia, bacteremia and urinary tract infections, as well as soft tissue, skin, and bone infections. Susceptibility to GBS is increased in the elderly and immunocompromised individuals with underlying conditions such as diabetes, cancer, and HIV (Schuchat, 1998; Farley, 2001; Phares et al., 2008; Sendi et al., 2008; Skoff et al., 2009). Overall neonatal as well as adult infections are, to a large percentage, endogenous infections with colonization of gastrointestinal and vaginal mucosal surfaces as a first crucial step. Nevertheless, in many cases, GBS persists as a commensal bacterium of the microbiota and does not proceed to cause invasive infections. During the last decades, hypervirulent clones have been identified and GBS regulators have been characterized that control the expression of major virulence factors, however, the switch from commensal to invasive pathogen is still incompletely deciphered. Understanding the determinants of GBS mucosal colonization as well as the different stages of the pathobiont lifestyle at a molecular level is therefore important for the development of prevention strategies to control invasive GBS disease. This review combines for first time epidemiological data with data on adherence and colonization for GBS. It covers the following aspects: GBS colonization rates and serotype distribution in different countries and the molecular epidemiology of GBS colonization, determined through MLST sequence types. Adherence properties of GBS will be summarized under the following headings, biofilm formation, and GBS colonization and adherence at a molecular level, which is be subdivided into (i) GBS adhesins to extracellular matrix proteins (ECM), (ii) GBS adhesins to cellular targets, and (iii) GBS pili. The last two sections describe the transition of GBS from commensal to pathogen and current efforts on immunoprophylaxis.
Since vaginal GBS colonization represents the most important risk factor for the development of invasive neonatal infections, colonization rates have been investigated in many different countries. Interestingly the GBS colonization rate among pregnant women in vagina and/or rectum is quite similar worldwide with some variations ranging from 10 to 30% in United States, 6.5% up to 36% in Europe, 7.1 to 16% in Asia, 9.1 to 25.3% in the Middle East, and 11.9 to 31.6% in Africa (Schuchat, 1998; Barcaite et al., 2008; Ippolito et al., 2010; Verani et al., 2010). The GBS colonization status, however, is intermittent and can be transient during pregnancy. Positive colonizers in early or mid-pregnancy may turn into negative colonizers at delivery (Hansen et al., 2004; Verani et al., 2010). Hence, timing of GBS screening and specimen collection is important to accurately predict colonization status at delivery. According to the CDC guidelines, the intrapartum colonization status is best determined in the late third trimester at no more than 5 weeks before delivery (Verani et al., 2010).
While GBS colonization rates may appear quite similar in different regions of the world, serotype prevalence and distribution is geographically distinct. Serotype classification is based on a sialic acid-rich capsular polysaccharide (CPS) which is one of the most important virulence factors in GBS that is involved in GBS persistence and survival within the host. CPS plays a critical role in immune evasion through its mimicry with the host carbohydrate epitopes. It also interferes with the complement-dependent defense pathways, dampens the phagocytic function of neutrophils and facilitates bacterial internalization and intracellular survival inside dendritic cells. Moreover, CPS has been reported to mediate biofilm formation of GBS in the presence of human plasma (Lemire et al., 2012; Pezzicoli et al., 2012; Chang et al., 2014a,b; Xia et al., 2015). At present, as many as 10 serotypes (Ia, Ib, and II–IX) are recognized (Slotved et al., 2007; Le Doare and Heath, 2013). The distribution and predominance of certain serotypes is susceptible to variations and can change over time. Serotypes Ia, Ib, II, III, and V are prevalent colonizers in the United States and Europe (Johri et al., 2006; Ippolito et al., 2010; Lamagni et al., 2013; Melin and Efstratiou, 2013; Florindo et al., 2014; Fabbrini et al., 2016). Serotypes VI and VIII are the most prevalent among pregnant women in Japan (Lachenauer et al., 1999; Matsubara et al., 2002) while serotypes IV and V predominate in the United Arab Emirates and Egypt, respectively (Amin et al., 2002; Shabayek et al., 2014). Serotype Ia is common in Mexico and is consistently higher in North America in comparison to other areas (Ippolito et al., 2010). The most recently characterized novel GBS serotype IX was reported from Denmark (Slotved et al., 2007). The geographical distribution of the most prevalent colonizing serotypes is shown in Table 1. On the other hand, serotype III is the most dominant invasive clone accounting for the majority of late-onset meningitis cases in neonates (Lamy et al., 2006; Tazi et al., 2010; Bellais et al., 2012; Florindo et al., 2014; Alhhazmi et al., 2016) while serotype Ia and V are dominant invasive isolates in non-pregnant cases (Phares et al., 2008; Alhhazmi et al., 2016). However, recent studies indicate the emergence of invasive serotype IV strains in neonates and adults (Ferrieri et al., 2013; Teatero et al., 2015; Alhhazmi et al., 2016) and among colonizing strains (Diedrick et al., 2010). A similar scenario could be expected for serotypes VI and VIII which are rarely reported outside Japan. In Malaysia, serotype VI was found among the dominant colonizing strains and the second most common isolate in adults with skin and soft tissue infections due to GBS (Karunakaran et al., 2009; Dhanoa et al., 2010). In Egypt, serotype VI was reported as a common colonizing serotype in women (Shabayek et al., 2014). And recently, Lin et al. (2016) reported a clonal dissemination of serotype VI among colonizing and invasive isolate in central Taiwan. Likewise, several studies identified sporadic strains of serotype VIII both as colonizing strains and as causative agents for invasive GBS disease (Paoletti et al., 1999; Hoshina et al., 2002; Ekelund et al., 2003; Karunakaran et al., 2009; Ippolito et al., 2010; Alhhazmi et al., 2016). Similar serotype emergence was earlier reported for serotype V in the United States and in Europe (Hickman et al., 1999; Flores et al., 2015). An increasing diversity of GBS serotypes, as well as serotype switching, represent powerful immune evasion strategies that may severely impair vaccine efforts, that currently rely on a conjugate vaccine incorporating a limited number of GBS serotypes. A close monitoring of the changing serotype distribution occurring in many different countries is therefore crucial in guiding GBS vaccination development.

TABLE 1. Geographical distribution of the most prevalent colonizing group B streptococcus (GBS) serotypes.
Capsular serotyping is the traditional tool used in GBS classification for epidemiological purposes. However, in the era of molecular epidemiology with the availability of numerous bacterial genomes and novel nucleotide sequencing-based tools our traditional picture of GBS epidemiology changed. In respect to corresponding genomes, gene content does not necessarily correlate with the assigned serotype. In 2003, a seven-gene multilocus sequence typing scheme (MLST) was introduced for GBS classification. First studies employing this MLST scheme revealed that capsular serotype is not restricted to a particular MLST sequence type (ST) and GBS strains with the same ST may have different serotypes (Jones et al., 2003). Several STs are grouped into clonal complexes (CC) when sharing six or seven matching alleles. The number of a particular CCs is designated after its ancestor ST or the predominant ST within this clone (Luan et al., 2005). By MLST the vast majority of the human GBS isolates can be grouped into six CCs which are CC1, CC10, CC17, CC19, CC23, and CC26 (Sørensen et al., 2010; Da Cunha et al., 2014). Interestingly, most of the bovine strains were exclusively designated as subtypes of the CC61 and CC67 which have never been reported for any of the human strains (Bisharat et al., 2004; Sukhnanand et al., 2005; Sørensen et al., 2010; Yang et al., 2013; Springman et al., 2014).
With the availability of MLST data, it became clear that GBS strains of certain CCs possess a higher potential to cause invasive disease, while other harbor mainly colonizing strains. MLST for a global GBS collection identified STs 1 and 19 to be significantly more associated with asymptomatic colonization and ST-23 was common for carriage and invasive GBS (Jones et al., 2003). This is in agreement with a study (Manning et al., 2008) that reported STs 1, 19, and 23 as the predominant colonizers in pregnant women and identified them as being well adapted to the vaginal mucosa with a poor invasion ability. Consistently, Teatero et al. (2017) found STs 1 and 23 the most frequent clones in colonized pregnant women. In general, the dominance of strains belonging to CCs 1, 23, and 19 among asymptomatic pregnant women was consistently reported (Jones et al., 2003; Luan et al., 2005; Springman et al., 2014), indicating that for these CCs being a mucosal commensal predominates.
A major clone responsible for a large proportion of invasive neonatal infections are the CC17 strains mostly belonging to serotype III. ST-17 strains first gained special interest due to their strong association with neonatal meningitis. Strains belonging to the CC17 are reported to be hypervirulent accounting for more than 80% of the GBS late-onset neonatal infections and are often but not exclusively associated with meningitis (Jones et al., 2003; Lamy et al., 2006; Manning et al., 2009; Tazi et al., 2010; Bellais et al., 2012; Florindo et al., 2014). Comparative phylogenetic analysis of human and bovine isolates revealed CC17 strains as a homogenous group with a recent origin and limited recombination in comparison to other CCs (Sørensen et al., 2010; Da Cunha et al., 2014). Moreover, evolutionary analysis between GBS isolates of human and bovine origin found ST-17 as the only human lineage that is clustered within the bovine population (Bisharat et al., 2004). The investigation suggested ST-17 to have bovine ancestors (Bisharat et al., 2004), which was supported by another study (Héry-Arnaud et al., 2007) providing evidence for a bovine origin of CC17 by investigating the prevalence of mobile genetic elements. However, a very close connection between the bovine CC67 and the human CC17 was also challenged by a more comprehensive genetic analysis of 238 bovine and human strains (Sørensen et al., 2010). The emergence of a highly virulent GBS clone, causing a large majority of neonatal infections, from a bovine ancestor provides intriguing phylogenetic aspects for placing GBS close to zoonotic diseases. Thus the question of how zoonotic GBS may be is currently controversially discussed and represents an exciting and ongoing scientific question to be solved.
For adult invasive infections, molecular clones other than CC17 are important. A significant percentage of GBS invasive disease in non-pregnant adults is resulting from strains belonging to serotype V (Phares et al., 2008; Lamagni et al., 2013; Teatero et al., 2014; Flores et al., 2015). Molecular epidemiology revealed a remarkable association of ST-1, a subtype of CC1, with serotype V GBS causing invasive disease. ST-1 invasive isolates from adults and neonates are mostly belonging to serotype V (Salloum et al., 2011). A recent analysis of a large cohort of GBS isolates of invasive serotype V strains from adults in United States and Canada (Flores et al., 2015). reported 92% of the serotype V strains as ST-1 whereas the most predominant sequence type among the non-ST-1 clones was ST-19. The authors regard the emergence of ST-1 strains as a leading cause of adult disease in the 1990s. Similar to human ST-17, the human ST-1 is suspected to originate from a bovine ancestor. Whole-genome sequencing recognized a 1992 ST-1 clone to be closely related to a 1970s Swedish strain causing cow mastitis. The ST-1 human GBS were found to possess mutations at loci involved in capsule production, pilus expression, and two-component regulatory systems (Flores et al., 2015) which all have been reported as key virulence factors in GBS (Maisey et al., 2008a; Rajagopal, 2009) and may thus represent the adaptation of this strain to human infections.
Particular interest should also be paid to the increasing emergence of type IV among colonizing and invasive GBS isolates. In several recent investigations, this serotype appears to be a hotspot for genetic recombination events, supported by the detection of serotype IV strains with different genetic backgrounds. Multiple studies demonstrated ST-196 as the most frequent sequence type of serotype IV isolates (Gherardi et al., 2007; Héry-Arnaud et al., 2007; Martins et al., 2007; van der Mee-Marquet et al., 2008; Lartigue et al., 2009; Diedrick et al., 2010). However, other investigations reported the STs 452 and 459, belonging to CCs 23 and 1, respectively, as the predominant colonizing type IV strains and found ST-196 less frequently (Diedrick et al., 2010; Ferrieri et al., 2013; Teatero et al., 2017). Comparing a collection of emerging type IV isolates in United States belonging to two distinct time periods from 1995 to 2000 and from 2004 to 2008, the older strains were more like a 1970s prototype reference strain with similar PFGE profiles whereas the more recent isolates showed PFGE profiles with considerable differences (Diedrick et al., 2010). Independent studies identified the invasive characteristics of type IV strains to be a result of capsular switching in CC17 strains accompanied with the acquisition of the HvgA adhesin, which has initially been isolated from hypervirulent CC17 serotype III strains (Teatero et al., 2014; Alhhazmi et al., 2016). Capsular switching events of CC17 strains have previously been observed (Da Cunha et al., 2014) leading to serotype IV ST-256 lineage.
For colonizing as well as invasive GBS strains dominant serotype-MLST genotypes associations have been observed such as serotype III with ST-17, serotype V with ST-1 and serotype IV with ST-196. Unusual associations are most likely to be due to capsular switching which resulted from recombination events around the capsular locus (Teatero et al., 2017). Investigation of the increase of type IV strains suggested the evolution of hybrid genomes including sequences from hypervirulent ST-17 strains (Teatero et al., 2016). Thus growing evidence points to an ongoing emergence of novel virulent GBS clones requiring continuous epidemiological surveillance.
Colonization and persistence in different host niches is dependent on the adherence capacity of GBS to host cells and tissues. This then facilitates bacterial cell aggregation and formation of sessile communities known as biofilms. Bacterial biofilms represent well-known virulence factors with a vital role in persistence and chronic infections. In the host environment, bacteria are often protected from the immune system by building sessile colonies embedded in an extracellular matrix of polysaccharides representing the biofilm. For GBS the bacterial capsule and type IIa pili have been demonstrated to play an important role in biofilm formation (Konto-Ghiorghi et al., 2009; Xia et al., 2015). Host environmental conditions are crucial determinants in developing bacterial biofilms (Costerton et al., 1999; Lewis, 2005; Nobbs et al., 2009; Rosini and Margarit, 2015). Contradictory data are available concerning the environmental cues favoring biofilm communities in GBS (Rosini and Margarit, 2015). As a normal inhabitant of the vagina, acidic pH seems to be optimal for GBS colonization. Early investigations reported enhanced GBS adherence to vaginal epithelial cells under low pH in comparison to neutral pH (Zawaneh et al., 1979; Tamura et al., 1994). In line with these observations, a significantly higher biofilm production of colonizing GBS isolates from pregnant women was demonstrated at pH 4.5 vs. pH 7 (Ho et al., 2013). Similarly, enhanced biofilm formation of GBS was shown under acidic pH conditions in comparison to neutral pH with the strongest biofilm producing GBS isolates belonged to the ST-17 sequence type. In respect to GBS origins, higher frequencies of strong biofilm producers were found among neonatal strains in comparison to colonizing strains (D’Urzo et al., 2014). However, a recent investigation reported invasive GBS belonging to CC17 and CC19 lineages as weak biofilm formers while GBS isolated from asymptomatic carriers were found to be strong biofilm producers (Parker et al., 2016). One possible explanation for this discrepancy is the experimental set up of the study since GBS biofilm formation was tested at neutral pH conditions and not under acidic pH. Furthermore, the presence of human plasma was shown to promote GBS biofilm formation (Xia et al., 2015).
In summary, biofilms allow long-term bacterial persistence and protect bacteria from recognition by the immune system. For GBS low pH and the presence of plasma appear as crucial environmental factors through controlling the expression of bacterial surface-associated structures, such as pili and the capsule, which are both involved in promoting bacterial biofilm formation.
As an opportunistic commensal constituting a part of the intestinal and vaginal physiologic flora, GBS colonization, persistence, translocation, and invasion of host barriers are largely dependent on their adherence abilities to host cells and ECM (Singh et al., 2012; Landwehr-Kenzel and Henneke, 2014). Functionally characterized adhesins mediating GBS adherence and/or invasion within the host are the fibrinogen-binding proteins (Fbs), the laminin-binding protein (Lmb), the group B streptococcal C5a peptidase (ScpB), the streptococcal fibronectin-binding protein A (SfbA), and the GBS immunogenic bacterial adhesin (BibA). In addition, surface-protruding structures comprised of multiples genes like pili are considered as essential adhesins in promoting GBS colonization, persistence, biofilm production, and central nervous system invasion. Major adhesins mediating GBS interaction with host cells are depicted in Figure 1.
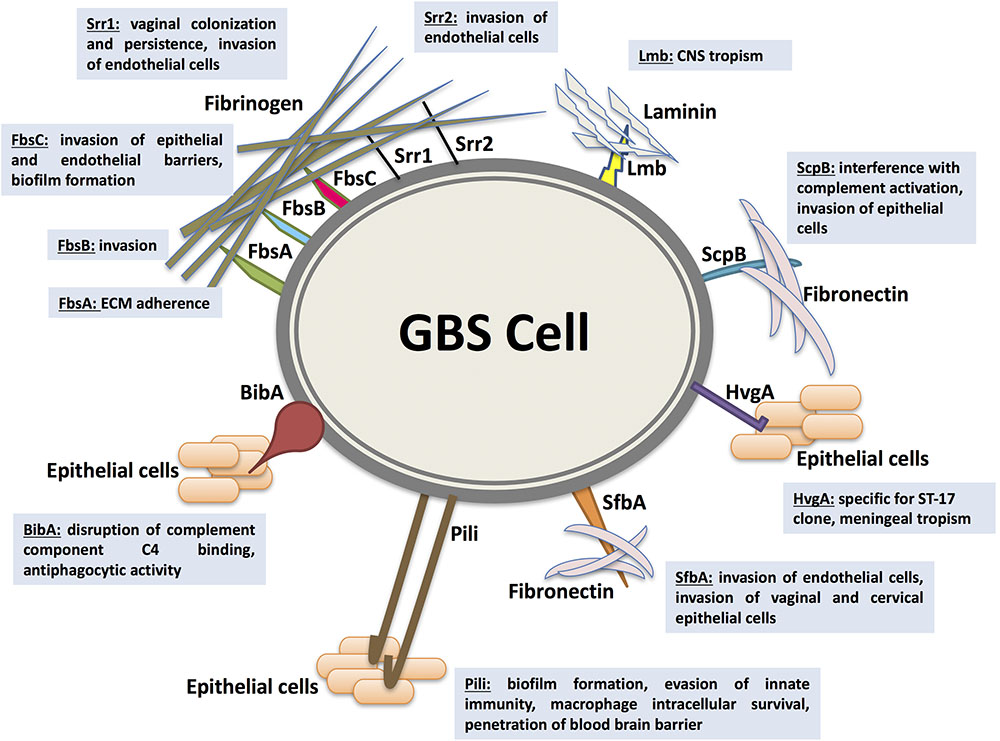
FIGURE 1. Major adhesins mediating Streptococcus agalactiae (GBS) interaction with host cells. GBS colonization, persistence, translocation, and invasion of host barriers are largely dependent on their ability to adhere to host cells and extracellular matrix proteins (ECM), an important step in breaching cellular barriers. The best characterized surface proteins mediating GBS adherence are the fibrinogen-binding proteins Fbs (including FbsA, FbsB, FbsC, or BsaB, the serine-rich repeat glycoproteins Srr1 and Srr2), the laminin-binding protein (Lmb), the Streptococcal C5a peptidase from group B (ScpB), the streptococcal fibronectin-binding protein A (SfbA), and the GBS immunogenic bacterial adhesin BibA. In addition, surface-protruding structures like pili are considered as essential adhesins in promoting GBS colonization, persistence, biofilm production, and central nervous system invasion. Associated virulence traits are illustrated for each adhesin as follows. FbsA was mainly shown to promote adherence whereas FbsB was shown to be required for invading human cells. Srr1 and Srr2 were reported to mediate invasion of microvascular endothelial cells. Additionally, Srr1 was demonstrated to promote vaginal colonization and persistence. FbsC was recently characterized to promote invasion of epithelial and endothelial barriers and biofilm formation. The Lmb adhesin appears to have a pronounced role in bacterial tropism of the central nervous system (CNS). ScpB interrupts complement activation through cleaving the neutrophil chemoattractant C5a. It is also involved in invasion of human epithelial cells. The SfbA adhesin is involved in human brain microvascular endothelial cells invasion. Furthermore, SfbA contributes to GBS invasion of vaginal and cervical epithelial cells and hence may take part in GBS colonization and niche establishment in the vagina. BibA was reported to aid GBS survival in human blood through interfering with the classic complement pathway by binding the C4-binding protein and by conferring anti-phagocytic activity against opsonophagocytic killing by human neutrophils. HvgA is specific for the hypervirulent clone ST-17. It was suggested to promote meningeal tropism in neonates. Pili in GBS have been shown to be primarily involved in epithelial cell colonization, biofilm formation, translocation, and invasion. PI-1 pili were also found to play an important role in evasion of innate immunity mechanism. The PI-2b protein, however, was demonstrated to increase the intracellular survival in macrophage. Pilus 2b was further identified as important for infection and penetration of the blood brain barrier.
Up to date, five Fbs have been characterized in GBS; FbsA (Schubert et al., 2004), FbsB (Gutekunst et al., 2004), the serine-rich repeat glycoproteins Srr1 and Srr2 (Seo et al., 2012, 2013), and recently FbsC or BsaB (Buscetta et al., 2014; Jiang and Wessels, 2014). In general, invasive GBS isolates display stronger fibrinogen-binding abilities in comparison to colonizing ones (Rosenau et al., 2007). FbsA was mainly shown to promote adherence (Schubert et al., 2004) whereas FbsB was shown to be required for invading human cells (Gutekunst et al., 2004). Srr1 and Srr2 were reported to mediate invasion of microvascular endothelial cells (Seo et al., 2012, 2013). Additionally, Srr1 was demonstrated to promote vaginal colonization and persistence, since a Srr1-deficient mutant displayed reduced persistence in a mouse GBS vaginal colonization model (Sheen et al., 2011). FbsC was recently characterized to promote invasion of epithelial and endothelial barriers. FbsC deletion mutant of GBS displayed a drastic reduction in abilities for adherence, invasion and biofilm formation. Besides, virulence abilities of FbsC deletion mutant were impaired in murine infection models (Buscetta et al., 2014). Interestingly, the fibrinogen-binding abilities of the hypervirulent CC17 clones are mainly attributable to FbsB more than FbsA. Deletion mutants of fbsB displayed 78–80% reduction in their binding abilities vs. 49–57% as encountered with fbsA deletion mutants of CC17 strains (Al Safadi et al., 2011). Accordingly, the relative transcription level of fbsB was up to 12.7-fold higher than fbsA gene in CC17 stains (Al Safadi et al., 2011). Moreover, Srr2 was highly expressed and exclusively detected in ST-17, however, Srr1 was absent (Seifert et al., 2006; Seo et al., 2013). Furthermore, CC17 strains are devoid of FbsC. The fbsC gene is not adequately expressed in CC17 strains because of a lineage-dependent frameshift mutation (Buscetta et al., 2014).
In addition to the Fbs family, the Lmb adhesin appears to have a pronounced role in bacterial tropism of the central nervous system. Spellerberg et al. (1999) reported Lmb to be essential for GBS colonization of damaged epithelium and subsequent translocation into the bloodstream. This role was later confirmed by Tenenbaum et al. (2007) as they demonstrated mutation of the lmb gene to result in a dramatic reduction in GBS invasion of the brain microvascular endothelial cells. In consistence, Al Safadi et al. (2010) displayed higher expression levels of Lmb in GBS strains associated with meningitis in comparison to other isolates whereas the expression levels of other ECM-binding proteins, such as ScpB mediating fibronectin binding ability, remained unchanged.
ScpB or the group B ScpB is a surface associated serine protease that both interrupts complement activation through splitting the neutrophil chemoattractant C5a and mediates bacterial binding to fibronectin (Chmouryguina et al., 1996; Bohnsack et al., 1997; Cheng et al., 2002; Lindahl et al., 2005). The fibronectin binding ability conferred by the scpB gene appears to be involved in cellular adherence and invasion. In frame deletion mutation of scpB gene significantly reduced invasion of human epithelial cells in vitro (Cheng et al., 2002). Strikingly, scpB and lmb genes were found to be encoded on a composite transposon where the scpB gene is positioned directly upstream of the lmb gene. The scpB-lmb intergenic region has been described as a hot spot for integration of the GBS mobile genetic elements GBSil and IS1548 which are located in the promoter region of the lmb gene (Franken et al., 2001; Granlund et al., 2001; Luan et al., 2003, 2005; Broker and Spellerberg, 2004). Al Safadi et al. (2010) reported a marked increase in the transcription levels of lmb gene for invasive GBS isolates carrying IS1548 in the scpB-lmb intergenic region associated with an increased laminin binding ability. However, no influence was observed on the scpB gene. Deletion mutation of IS1548 revealed IS1548 to act as an lmb gene up-regulator when compared to the wild-type parent strains. Interestingly the ability of GBS to colonize human mucosal surfaces seems to be closely linked to the presence of this composite transposon carrying scpB and lmb. In a large percentage of bovine strains, the encoded genes are absent while the presence in human colonizing strains, as well as invasive strains, is close to 100% (Franken et al., 2001; Sørensen et al., 2010; Rato et al., 2013).
More recently, a novel GBS fibronectin binding protein has been identified (Mu et al., 2014). It was designated as streptococcal fibronectin-binding protein A (SfbA) and reported to be highly conserved in GBS mediating cellular invasion but not adherence. SfbA was shown to be directly involved in fibronectin binding and human brain microvascular endothelial cells invasion. When expressed in recombinant non-pathogenic Lactococcus lactis, fibronectin binding ability was significantly greater in comparison to a SfbA negative control strain. The investigation also demonstrated SfbA to be primarily involved in brain microvascular endothelial cells invasion. Infection of mice with sfbA mutants resulted in a reduced ability to breach the blood brain barrier and subsequent meningitis. This is supported by a study showing SfbA to be crucial for invasion of astrocytes which are physically associated with the brain endothelial cells (Stoner et al., 2015). Furthermore, SfbA contributes to GBS invasion of vaginal and cervical epithelial cells and hence may take part in GBS colonization and niche establishment in the vagina (Mu et al., 2014). Another fibronectin binding protein was described in 2014 (Jiang and Wessels, 2014). BsaB or the bacterial surface adhesin of GBS is a fibronectin and laminin-binding protein which is involved in GBS binding to epithelial cells and in biofilm formation. Deletion of bsaB gene and a cotranscribed upstream region significantly abrogated GBS adherence to VK2 vaginal epithelial cells in vitro and immobilized fibronectin. However, genome and sequence analysis revealed BsaB and FbsC as identical proteins encoded by the same gene (Buscetta et al., 2014). The obtained results are in agreement with Jiang and Wessels (2014), except that FbsC or BsaB was found to mediate GBS attachment to fibrinogen instead of fibronectin. Hence, BsaB was renamed to FbsC.
The multitude of GBS adhesins allowing attachment to different ECM, stresses the importance of this step in GBS pathogenesis, which was confirmed in different in vivo models. In this regard, fibrinogen binding may play an especially important role as demonstrated by the presence of numerous fibrinogen binding proteins. These may represent a kind of “backup” system in cases where the primary fibrinogen adhesin was rendered non-functional.
Besides adherence to ECM, the adhesion to host cells plays an important role in the pathogenesis of GBS. An essential adhesin in this context is the GBS immunogenic bacterial adhesin (BibA). It is a cell wall-anchored protein which is well-conserved in GBS and is involved in bacterial binding to human epithelial cells (Santi et al., 2007, 2009b). A knockout mutant displayed impaired adherence capacity to the lung, intestinal, and cervical epithelial cells (Santi et al., 2007). Overexpression of BibA resulted in increased adherence to human epithelial cells in recombinant wild-type strains harboring a bibA plasmid (Santi et al., 2007). In addition, BibA was reported to aid GBS survival in human blood through interfering with the classic complement pathway by binding the C4-binding protein and by conferring anti-phagocytic activity against opsonophagocytic killing by human neutrophils (Santi et al., 2007, 2009b). A total of four variants of BibA (I, II, III, and IV) were described in GBS (Brochet et al., 2006; Santi et al., 2007, 2009b). Interestingly, variant IV, which was found to be highly similar to the bovine BibA counterparts, was exclusively associated with ST-17 strains (Lamy et al., 2006; Santi et al., 2009b). Thus, BibA seems to be a multifactorial virulence factor in regard to GBS as a pathobiont. It contributes to GBS mucosal colonization and adherence to host cells and then confers resistance to phagocytic killing at a stage when the switch to invasive GBS infection has occurred.
The GBS hypervirulent adhesin (HvgA) is a novel cell wall anchored protein that is specific for the hypervirulent clone ST-17. It was first described (Tazi et al., 2010) as being strongly associated with ST-17 causing neonatal meningitis in LOD. It was suggested to promote meningeal tropism in neonates through efficient intestinal colonization and subsequent translocation across the intestinal and the blood brain barriers. Bypassing intestinal colonization by intravenous infection resulted in a significant decrease in the amount of bacteria reaching the central nervous system. HvgA was required for intestinal colonization in orally infected mice for meningitis development. In addition, HvgA was found to mediate GBS adherence to intestinal epithelial cells, choroid epithelial cells, and microvascular endothelial cells (Tazi et al., 2010). Clones expressing HvgA exhibited greater adherence abilities than non-expressing ones. HvgA thus contributes to colonization as well as invasion of hypervirulent clones (Tazi et al., 2010).
Further structures that are crucial for GBS adhesion are pili. Different from their Gram-negative counterparts, pili in GBS have been shown to be primarily involved in epithelial cell colonization, biofilm formation, translocation, and invasion. Pili are cell-wall anchored appendages extending from the bacterial surface. They contain covalently linked multimeric motifs that are composed of three pilin proteins, the pilus shaft backbone protein (BP) or PilB subunits, and the two ancillary proteins AP1, AP2 located at the pilus tip (PilA subunit, the pilus-associated adhesin) and pilus base (PilC subunit, the pilus anchor), respectively, (Dramsi et al., 2006; Rosini et al., 2006; Maisey et al., 2007, 2008b; Cozzi et al., 2015). While PilB has been shown to be involved in bacterial invasion and paracellular translocation mediating resistance to phagocytic killing and virulence, PilA was found contributing to cellular adherence and colonization (Dramsi et al., 2006; Krishnan et al., 2007; Maisey et al., 2007, 2008b; Pezzicoli et al., 2008; Sheen et al., 2011). Three pilus variants named PI-1, PI-2a, PI-2b were reported in GBS representing two pilus islands (PI) where PI-2a and PI-2b are variants of the pilus island 2 (PI-2). All characterized GBS strains harbored at least one variant or a combination of two pilus islands (Rosini et al., 2006; Margarit et al., 2009; Springman et al., 2014). PI-1 pili were also found to play an important role in evasion of innate immunity mechanism. They diminished macrophage-mediated phagocytic killing of GBS by 50% with no influence on complement-promoted opsonophagocytic killing by neutrophils (Jiang et al., 2012). Strikingly, PI-1 pili do not appear to contribute to bacterial adhesion to lung, vaginal or cervical epithelial cells (Jiang et al., 2012). The PI-2a pili were found to have a specific involvement in adherence and biofilm formation and not PI or PI-2b (Konto-Ghiorghi et al., 2009; Rinaudo et al., 2010). The PI-2b protein, however, was demonstrated to increase the intracellular survival in macrophage (Chattopadhyay et al., 2011). In addition, a special role for pilus type 2b has been suggested in promoting strain invasiveness and bacterial host cell interactions. Mutants of pilus 2b possess less adherence and invasion capacities for epithelial and endothelial cells (Lazzarin et al., 2017). Pilus 2b was further identified as important for infection and penetration of the blood brain barrier. These results are supported by an investigation of the distribution of pilus islands among GBS strains belonging to ST lineages of human and bovine origin (Springman et al., 2014). In addition, the distribution of pili islands appears to determine the capacity for colonization or invasive infections. Invasive GBS were more likely to carry a combination of PI and one of the PI-2 variants in comparison to maternal colonizing isolates. Moreover, GBS causing invasive neonatal disease including all CC-17 strains were harboring PI-1 plus PI-2b. Earlier genomic studies showed pilus type 2b to be conserved in the ST-17 hypervirulent clone (Brochet et al., 2006). Interestingly PI-2b pilus variants are almost exclusively present in bovine GBS isolates. These bovine strains mostly lack PI-1, unlike the human isolates which commonly encode the pilus PI-1 in association with one of the PI-2 variants (Springman et al., 2014).
Group B streptococcus is a commensal bacterium of the vagina and the gastrointestinal tract of healthy adults. However, as a pathobiont, GBS can convert from the asymptomatic mucosal carriage state to a bacterial pathogen causing invasive infections. For neonatal infections that implies a switch from bacterial survival in the acidic host environment of the vagina to resistance of the immune system in the pH neutral environment of human blood. To address bacterial adaptation to these environmental changes, a genome-wide transcription analysis was performed in order to investigate GBS responses to acid stress in comparison to neutral pH conditions (Santi et al., 2009a). This study demonstrated the modulation of genes involved in GBS adaptation and vaginal persistence as well as virulence-related genes in response to fluctuating environmental pH which are mostly under the control of the CovRS two-component regulatory system (TCS). Among the adherence factors that were demonstrated to be under the control of CovRS are Fbs proteins, Lmb, C5a, BibA, HvgA, and pili (Lamy et al., 2004; Jiang et al., 2008, 2012; Santi et al., 2009a; Tazi et al., 2010; Park et al., 2012). These observations suggested that GBS translocation from acidic to neutral niches switches on virulence-related genes, favoring the transition from commensal to invasive bacterial pathogen. The CovRS system is a well-studied TCS in GBS and a major virulence regulator. The contribution of CovRS in coordinating GBS gene expression and virulence has been investigated in many studies (Lamy et al., 2004; Jiang et al., 2005, 2008; Di Palo et al., 2013; Faralla et al., 2014). Patras et al. (2013) conducted a global transcription profiling of human vaginal epithelial cells exposed to a CovR-deficient mutant in comparison to the wild-type strain and studied a mouse model of GBS vaginal colonization. Their major observation was that loss of CovR signaling promoted exaggerated host inflammatory reactions against GBS infection. Hence, the authors concluded that CovRS control of GBS virulence gene expression is crucial in maintaining GBS vaginal colonization by avoiding host immune responses (Patras et al., 2013). Apart from regulation through TCS, GBS genomic adaptations seem to take place during GBS transition from commensal to pathogen. Comparing the whole genomes of GBS pairs from infected newborns and their mothers genomic mutations were found with critical phenotypic impacts, including CovR mutations (Almeida et al., 2015). Genomic adaptations may also impact dynamics of maternal GBS colonization before and after delivery. Alterations in STs and serotypes can be observed in 18.3% of pregnant women who were positive colonizers before and after delivery (Manning et al., 2008). Definite STs appear to be related with GBS clearance and persistence. Not surprisingly capsule expression is linked to invasion capabilities of GBS. A significantly higher proportion of non-typeable strains are present among colonizing isolates in comparison to invasive isolates (Teatero et al., 2017). This may indicate that persistent colonizers undergo several lateral DNA exchanges leading to the abolishment of capsule expression since encapsulation is less important during carriage. Capsule loss has previously been suggested to favor GBS colonization and diminish virulence (Rubens et al., 1987) and capsule production is also regulated through CovRS. Thus the results of multiple independent investigations strengthen the role of CovRS as a central GBS regulator controlling the switch from mucosal colonization to invasive infection.
The ability of GBS to cause neonatal invasive infections is dependent on the maternal antibody titer. Early work done by Rebecca Lancefield in the 1930s (Lancefield, 1938) reported protection against GBS infection in mice by CPS-specific antiserum in rabbits. Baker and Kasper (1976) demonstrated an inverse association between the levels of maternal serotype-specific capsular antibodies and the increased susceptibility to invasive GBS disease in newborns. This association was confirmed in later investigations (Lin et al., 2001, 2004; Baker et al., 2014). Vaccination seems, therefore, to be a good alternative to peripartal antibiotic prophylaxis in order to prevent neonatal GBS infections. However, no licensed vaccines are available (Verani et al., 2010; Madhi and Dangor, 2017). The sialylated CPS has been the main target for GBS vaccine in most clinical trials. Phases I and II clinical trials demonstrated the safety and immunogenicity of monovalent CPS-conjugate vaccines among healthy women (Paoletti and Kasper, 2003). However, monovalent formulations with serotype-specific immune responses are not sufficient to provide protection against the different GBS serotypes found in invasive infections. Besides, possible capsular switching forced by the selective pressure exerted due to GBS monovalent vaccines can be expected. Multivalent formulations would provide broader vaccine coverage and overcome problems of serotype switching (Johri et al., 2006; Brueggemann et al., 2007; Nuccitelli et al., 2011; Heath et al., 2017). The Safety and immunogenicity of a trivalent GBS conjugate vaccine based on serotypes Ia, Ib, and III have has been evaluated in Phases I and II trials and there are considerations for a Phase III trial (Madhi et al., 2013; Donders et al., 2016; Heyderman et al., 2016; Kobayashi et al., 2016a,b; Leroux-Roels et al., 2016; Madhi et al., 2016). Pre-clinical studies of a pentavalent vaccine, based on serotypes Ia, Ib, II, III, V are currently under investigation (Kobayashi et al., 2016a). In addition to serotype-specificity, the development of a GBS vaccine with global relevance is a real challenge due to geographic serotype diversity and variations over time. Hence, new vaccine candidates based on conserved surface proteins have been investigated and these included ScpB, Lmb, surface immunogenic protein Sip, leucine rich repeat protein LrrG, Rib and tandem-repeat containing α and β components of the C protein (Wilkinson and Eagon, 1971; Russell-Jones et al., 1984; Michel et al., 1991; Madoff et al., 1992; Stålhammar-Carlemalm et al., 1993; Spellerberg et al., 1999; Cheng et al., 2001; Heath and Feldman, 2005; Seepersaud et al., 2005). A Phase I clinical trial revealed the immunogenicity of Rib and α-C surface proteins against invasive GBS disease (Madhi and Dangor, 2017). Based on reverse vaccinology utilizing genomics, proteomics, transcriptomics and in silico technologies, a panel of potential vaccine targets could be discovered (Johri et al., 2006). A similar approach also lead to the identification of GBS pili (Lauer et al., 2005). However, the development of a pilus-based vaccine has been hampered due to antigenic variations of pili in GBS (Margarit et al., 2009; Nuccitelli et al., 2011), but structural vaccinology may overcome this problem. Crystallographic structure of the BP-2a pilus subunit showed four IgG-like domains D1, D2, D3, and D4. Since the D3 domain elicited similar protection as the whole BP-2a component, a promising hybrid vaccine candidate consisting of 6xD3 of the D3 domains from six variant BP-2a pilus subunits was constructed. It is expected to confer protection against all six pilus variants found in GBS strains (Nuccitelli et al., 2011). Other promising vaccine targets that have been identified by reverse vaccinology include the Sip and BibA proteins (Maione et al., 2005; Santi et al., 2007, 2009b). It has been recently shown that a reduced likelihood of maternal GBS acquisition during pregnancy is directly associated with Sip and BibA induced antibodies (Dzanibe et al., 2017). The successful development and introduction of a GBS vaccine may, therefore, result in converting GBS into a predominantly harmless commensal of the mucosa.
Group B streptococcus adherence and colonization is a complex multifactorial process which determines the success of a pathobiont in the human ecosystems. GBS possess strict regulatory systems which are responsible for fine-tuning the expression of adhesins in order to optimize GBS fitness under the prevailing environmental conditions. Long-term colonization and persistence is largely dependent on the ability of GBS to avoid the host immune response. Over the last two decades, a detailed knowledge about molecular adhesion mechanisms was accumulated for GBS, which considerably enhances our understanding of disease development. These GBS adhesion determinants may represent novel targets for the development of innovative treatment and prophylaxis strategies aiming at control and prevention of invasive GBS disease in neonates as well as adult patients. To date, the sialic acid-containing CPS that underlies the serotyping system of GBS has been the main focus of most vaccine developments. GBS colonizers represent the silent reservoirs responsible for serotype diversity and evolutionary changes in virulence determinants. Investigation of GBS population structure is essential to monitor the circulating serotypes and identify changes in strains causing GBS disease. An aspect, which is especially important in view of a possible serotype-dependent vaccination in the near future.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Al Safadi, R., Amor, S., Hery-Arnaud, G., Spellerberg, B., Lanotte, P., Mereghetti, L., et al. (2010). Enhanced expression of lmb gene encoding laminin-binding protein in Streptococcus agalactiae strains harboring IS1548 in scpB-lmb intergenic region. PLoS One 5:e10794. doi: 10.1371/journal.pone.0010794
Al Safadi, R., Mereghetti, L., Salloum, M., Lartigue, M. F., Virlogeux-Payant, I., Quentin, R., et al. (2011). Two-component system RgfA/C activates the fbsB gene encoding major fibrinogen-binding protein in highly virulent CC17 clone group B Streptococcus. PLoS One 6:e14658. doi: 10.1371/journal.pone.0014658
Alhhazmi, A., Hurteau, D., and Tyrrell, G. J. (2016). Epidemiology of invasive group B streptococcal disease in Alberta. Canada, from 2003 to 2013. J. Clin. Microbiol. 54, 1774–1781. doi: 10.1128/jcm.00355-16
Almeida, A., Villain, A., Joubrel, C., Touak, G., Sauvage, E., Rosinski-Chupin, I., et al. (2015). Whole-genome comparison uncovers genomic mutations between group B streptococci sampled from infected newborns and their mothers. J. Bacteriol. 197, 3354–3366. doi: 10.1128/jb.00429-15
Amin, A., Abdulrazzaq, Y. M., and Uduman, S. (2002). Group B streptococcal serotype distribution of isolates from colonized pregnant women at the time of delivery in United Arab Emirates. J. Infect. 45, 42–46. doi: 10.1053/jinf.2001.0990
Anthony, B. F., Okada, D. M., and Hobel, C. J. (1979). Epidemiology of the group B streptococcus: maternal and nosocomial sources for infant acquisitions. J. Pediatr. 95, 431–436. doi: 10.1016/S0022-3476(79)80530-2
Baker, C. J., Carey, V. J., Rench, M. A., Edwards, M. S., Hillier, S. L., Kasper, D. L., et al. (2014). Maternal antibody at delivery protects neonates from early onset group B streptococcal disease. J. Infect. Dis. 209, 781–788. doi: 10.1093/infdis/jit549
Baker, C. J., and Kasper, D. L. (1976). Correlation of maternal antibody deficiency with susceptibility to neonatal group B streptococcal infection. N. Engl. J. Med. 294, 753–756. doi: 10.1056/nejm197604012941404
Barcaite, E., Bartusevicius, A., Tameliene, R., Kliucinskas, M., Maleckiene, L., and Nadisauskiene, R. (2008). Prevalence of maternal group B streptococcal colonisation in European countries. Acta Obstet. Gynecol. Scand. 87, 260–271. doi: 10.1080/00016340801908759
Bellais, S., Six, A., Fouet, A., Longo, M., Dmytruk, N., Glaser, P., et al. (2012). Capsular switching in group B Streptococcus CC17 hypervirulent clone: a future challenge for polysaccharide vaccine development. J. Infect. Dis. 206, 1745–1752. doi: 10.1093/infdis/jis605
Bisharat, N., Crook, D. W., Leigh, J., Harding, R. M., Ward, P. N., Coffey, T. J., et al. (2004). Hyperinvasive neonatal group B streptococcus has arisen from a bovine ancestor. J. Clin. Microbiol. 42, 2161–2167. doi: 10.1128/JCM.42.5.2161-2167.2004
Bohnsack, J. F., Widjaja, K., Ghazizadeh, S., Rubens, C. E., Hillyard, D. R., Parker, C. J., et al. (1997). A role for C5 and C5a-ase in the acute neutrophil response to group B streptococcal infections. J. Infect. Dis. 175, 847–855. doi: 10.1086/513981
Brochet, M., Couve, E., Zouine, M., Vallaeys, T., Rusniok, C., Lamy, M. C., et al. (2006). Genomic diversity and evolution within the species Streptococcus agalactiae. Microbes Infect. 8, 1227–1243. doi: 10.1016/j.micinf.2005.11.010
Broker, G., and Spellerberg, B. (2004). Surface proteins of Streptococcus agalactiae and horizontal gene transfer. Int. J. Med. Microbiol. 294, 169–175. doi: 10.1016/j.ijmm.2004.06.018
Brueggemann, A. B., Pai, R., Crook, D. W., and Beall, B. (2007). Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 3:e168. doi: 10.1371/journal.ppat.0030168
Buscetta, M., Papasergi, S., Firon, A., Pietrocola, G., Biondo, C., Mancuso, G., et al. (2014). FbsC, a novel fibrinogen-binding protein, promotes Streptococcus agalactiae-host cell interactions. J. Biol. Chem. 289, 21003–21015. doi: 10.1074/jbc.M114.553073
Chang, Y. C., Olson, J., Beasley, F. C., Tung, C., Zhang, J., Crocker, P. R., et al. (2014a). Group B Streptococcus engages an inhibitory Siglec through sialic acid mimicry to blunt innate immune and inflammatory responses in vivo. PLoS Pathog. 10:e1003846. doi: 10.1371/journal.ppat.1003846
Chang, Y. C., Olson, J., Louie, A., Crocker, P. R., Varki, A., and Nizet, V. (2014b). Role of macrophage sialoadhesin in host defense against the sialylated pathogen group B Streptococcus. J. Mol. Med. 92, 951–959. doi: 10.1007/s00109-014-1157-y
Chattopadhyay, D., Carey, A. J., Caliot, E., Webb, R. I., Layton, J. R., Wang, Y., et al. (2011). Phylogenetic lineage and pilus protein Spb1/SAN1518 affect opsonin-independent phagocytosis and intracellular survival of Group B Streptococcus. Microbes Infect. 13, 369–382. doi: 10.1016/j.micinf.2010.12.009
Cheng, Q., Carlson, B., Pillai, S., Eby, R., Edwards, L., Olmsted, S. B., et al. (2001). Antibody against surface-bound C5a peptidase is opsonic and initiates macrophage killing of group B streptococci. Infect. Immun. 69, 2302–2308. doi: 10.1128/iai.69.4.2302-2308.2001
Cheng, Q., Stafslien, D., Purushothaman, S. S., and Cleary, P. (2002). The group B streptococcal C5a peptidase is both a specific protease and an invasin. Infect. Immun. 70, 2408–2413. doi: 10.1128/IAI.70.5.2408-2413.2002
Chmouryguina, I., Suvorov, A., Ferrieri, P., and Cleary, P. P. (1996). Conservation of the C5a peptidase genes in group A and B streptococci. Infect. Immun. 64, 2387–2390.
Costerton, J. W., Stewart, P. S., and Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Cozzi, R., Malito, E., Lazzarin, M., Nuccitelli, A., Castagnetti, A., Bottomley, M. J., et al. (2015). Structure and assembly of group B streptococcus pilus 2b backbone protein. PLoS One 10:e0125875. doi: 10.1371/journal.pone.0125875
Da Cunha, V., Davies, M. R., Douarre, P. E., Rosinski-Chupin, I., Margarit, I., Spinali, S., et al. (2014). Streptococcus agalactiae clones infecting humans were selected and fixed through the extensive use of tetracycline. Nat. Commun. 5:4544. doi: 10.1038/ncomms5544
Dermer, P., Lee, C., Eggert, J., and Few, B. (2004). A history of neonatal group B streptococcus with its related morbidity and mortality rates in the United States. J. Pediatr. Nurs. 19, 357–363. doi: 10.1016/j.pedn.2004.05.012
Dhanoa, A., Karunakaran, R., and Puthucheary, S. D. (2010). Serotype distribution and antibiotic susceptibility of group B streptococci in pregnant women. Epidemiol. Infect. 138, 979–981. doi: 10.1017/s0950268809991105
Di Palo, B., Rippa, V., Santi, I., Brettoni, C., Muzzi, A., Metruccio, M. M., et al. (2013). Adaptive response of Group B streptococcus to high glucose conditions: new insights on the CovRS regulation network. PLoS One 8:e61294. doi: 10.1371/journal.pone.0061294
Diedrick, M. J., Flores, A. E., Hillier, S. L., Creti, R., and Ferrieri, P. (2010). Clonal analysis of colonizing group B Streptococcus, serotype IV, an emerging pathogen in the United States. J. Clin. Microbiol. 48, 3100–3104. doi: 10.1128/jcm.00277-10
Donders, G. G., Halperin, S. A., Devlieger, R., Baker, S., Forte, P., Wittke, F., et al. (2016). Maternal immunization with an investigational trivalent group B streptococcal vaccine: a randomized controlled trial. Obstet. Gynecol. 127, 213–221. doi: 10.1097/aog.0000000000001190
Dramsi, S., Caliot, E., Bonne, I., Guadagnini, S., Prevost, M. C., Kojadinovic, M., et al. (2006). Assembly and role of pili in group B streptococci. Mol. Microbiol. 60, 1401–1413. doi: 10.1111/j.1365-2958.2006.05190.x
D’Urzo, N., Martinelli, M., Pezzicoli, A., De Cesare, V., Pinto, V., Margarit, I., et al. (2014). Acidic pH strongly enhances in vitro biofilm formation by a subset of hypervirulent ST-17 Streptococcus agalactiae strains. Appl. Environ. Microbiol. 80, 2176–2185. doi: 10.1128/aem.03627-13
Dzanibe, S., Kwatra, G., Adrian, P. V., Kimaro-Mlacha, S. Z., Cutland, C. L., and Madhi, S. A. (2017). Association between antibodies against group B Streptococcus surface proteins and recto-vaginal colonisation during pregnancy. Sci. Rep. 7:16454. doi: 10.1038/s41598-017-16757-9
Ekelund, K., Slotved, H. C., Nielsen, H. U., Kaltoft, M. S., and Konradsen, H. B. (2003). Emergence of invasive serotype VIII group B streptococcal infections in Denmark. J. Clin. Microbiol. 41, 4442–4444. doi: 10.1128/JCM.41.9.4442-4444.2003
Fabbrini, M., Rigat, F., Rinaudo, C. D., Passalaqua, I., Khacheh, S., Creti, R., et al. (2016). The protective value of maternal group B Streptococcus antibodies: quantitative and functional analysis of naturally acquired responses to capsular polysaccharides and pilus proteins in European maternal sera. Clin. Infect. Dis. 63, 746–753. doi: 10.1093/cid/ciw377
Faralla, C., Metruccio, M. M., De Chiara, M., Mu, R., Patras, K. A., Muzzi, A., et al. (2014). Analysis of two-component systems in group B Streptococcus shows that RgfAC and the novel FspSR modulate virulence and bacterial fitness. mBio 5:e870-14. doi: 10.1128/mBio.00870-14
Farley, M. M. (2001). Group B streptococcal disease in nonpregnant adults. Clin. Infect. Dis. 33, 556–561. doi: 10.1086/322696
Ferrieri, P., Lynfield, R., Creti, R., and Flores, A. E. (2013). Serotype IV and invasive group B Streptococcus disease in neonates. Minnesota, USA, 2000-2010. Emerg. Infect. Dis. 19, 551–558. doi: 10.3201/eid1904.121572
Flores, A. R., Galloway-Pena, J., Sahasrabhojane, P., Saldana, M., Yao, H., Su, X., et al. (2015). Sequence type 1 group B Streptococcus, an emerging cause of invasive disease in adults, evolves by small genetic changes. Proc. Natl. Acad. Sci. U.S.A. 112, 6431–6436. doi: 10.1073/pnas.1504725112
Florindo, C., Damiao, V., Silvestre, I., Farinha, C., Rodrigues, F., Nogueira, F., et al. (2014). Epidemiological surveillance of colonising group B Streptococcus epidemiology in the Lisbon and Tagus Valley regions, Portugal (2005 to 2012): emergence of a new epidemic type IV/clonal complex 17 clone. Euro. Surveill. 19:20825. doi: 10.2807/1560-7917.ES2014.19.23.20825
Franken, C., Haase, G., Brandt, C., Weber-Heynemann, J., Martin, S., Lammler, C., et al. (2001). Horizontal gene transfer and host specificity of beta-haemolytic streptococci: the role of a putative composite transposon containing scpB and lmb. Mol. Microbiol. 41, 925–935. doi: 10.1046/j.1365-2958.2001.02563.x
Fry, R. M. (1938). Fatal infections by hemolytic streptococcus group B. Lancet 231, 199–201. doi: 10.1016/S0140-6736(00)93202-1
Gherardi, G., Imperi, M., Baldassarri, L., Pataracchia, M., Alfarone, G., Recchia, S., et al. (2007). Molecular epidemiology and distribution of serotypes, surface proteins, and antibiotic resistance among group B streptococci in Italy. J. Clin. Microbiol. 45, 2909–2916. doi: 10.1128/jcm.00999-07
Granlund, M., Michel, F., and Norgren, M. (2001). Mutually exclusive distribution of IS1548 and GBSi1, an active group II intron identified in human isolates of group B streptococci. J. Bacteriol. 183, 2560–2569. doi: 10.1128/jb.183.8.2560-2569.2001
Gutekunst, H., Eikmanns, B. J., and Reinscheid, D. J. (2004). The novel fibrinogen-binding protein FbsB promotes Streptococcus agalactiae invasion into epithelial cells. Infect. Immun. 72, 3495–3504. doi: 10.1128/iai.72.6.3495-3504.2004
Hansen, S. M., Uldbjerg, N., Kilian, M., and Sorensen, U. B. (2004). Dynamics of Streptococcus agalactiae colonization in women during and after pregnancy and in their infants. J. Clin. Microbiol. 42, 83–89. doi: 10.1128/JCM.42.1.83-89.2004
Heath, P. T., Culley, F. J., Jones, C. E., Kampmann, B., Le Doare, K., Nunes, M. C., et al. (2017). Group B streptococcus and respiratory syncytial virus immunisation during pregnancy: a landscape analysis. Lancet Infect. Dis. 17, e223–e234. doi: 10.1016/s1473-3099(17)30232-3
Heath, P. T., and Feldman, R. G. (2005). Vaccination against group B streptococcus. Expert Rev. Vaccines 4, 207–218. doi: 10.1586/14760584.4.2.207
Héry-Arnaud, G., Bruant, G., Lanotte, P., Brun, S., Picard, B., Rosenau, A., et al. (2007). Mobile genetic elements provide evidence for a bovine origin of clonal complex 17 of Streptococcus agalactiae. Appl. Environ. Microbiol. 73, 4668–4672. doi: 10.1128/aem.02604-06
Heyderman, R. S., Madhi, S. A., French, N., Cutland, C., Ngwira, B., Kayambo, D., et al. (2016). Group B streptococcus vaccination in pregnant women with or without HIV in Africa: a non-randomised phase 2, open-label, multicentre trial. Lancet Infect. Dis. 16, 546–555. doi: 10.1016/s1473-3099(15)00484-3
Hickman, M. E., Rench, M. A., Ferrieri, P., and Baker, C. J. (1999). Changing epidemiology of group B streptococcal colonization. Pediatrics 104, 203–209. doi: 10.1542/peds.104.2.203
Ho, Y. R., Li, C. M., Yu, C. H., Lin, Y. J., Wu, C. M., Harn, I. C., et al. (2013). The enhancement of biofilm formation in Group B streptococcal isolates at vaginal pH. Med. Microbiol. Immunol. 202, 105–115. doi: 10.1007/s00430-012-0255-0
Hood, M., Janney, A., and Dameron, G. (1961). Beta hemolytic streptococcus group B associated with problems of the perinatal period. Am. J. Obstet. Gynecol. 82, 809–818. doi: 10.1016/S0002-9378(16)36146-4
Hoshina, K., Suzuki, Y., Nishida, H., Kaneko, K., Matsuda, S., Kobayashi, M., et al. (2002). Trend of neonatal group B streptococcal infection during the last 15 years. Pediatr. Int. 44, 641–646. doi: 10.1046/j.1442-200X.2002.01638.x
Ippolito, D. L., James, W. A., Tinnemore, D., Huang, R. R., Dehart, M. J., Williams, J., et al. (2010). Group B streptococcus serotype prevalence in reproductive-age women at a tertiary care military medical center relative to global serotype distribution. BMC Infect. Dis. 10:336. doi: 10.1186/1471-2334-10-336
Jiang, S., Park, S. E., Yadav, P., Paoletti, L. C., and Wessels, M. R. (2012). Regulation and function of pilus island 1 in group B streptococcus. J. Bacteriol. 194, 2479–2490. doi: 10.1128/jb.00202-12
Jiang, S., and Wessels, M. R. (2014). BsaB, a novel adherence factor of group B Streptococcus. Infect. Immun. 82, 1007–1016. doi: 10.1128/iai.01014-13
Jiang, S. M., Cieslewicz, M. J., Kasper, D. L., and Wessels, M. R. (2005). Regulation of virulence by a two-component system in group B streptococcus. J. Bacteriol. 187, 1105–1113. doi: 10.1128/jb.187.3.1105-1113.2005
Jiang, S. M., Ishmael, N., Dunning Hotopp, J., Puliti, M., Tissi, L., Kumar, N., et al. (2008). Variation in the group B Streptococcus CsrRS regulon and effects on pathogenicity. J. Bacteriol. 190, 1956–1965. doi: 10.1128/jb.01677-07
Johri, A. K., Paoletti, L. C., Glaser, P., Dua, M., Sharma, P. K., Grandi, G., et al. (2006). Group B Streptococcus: global incidence and vaccine development. Nat. Rev. Microbiol. 4, 932–942. doi: 10.1038/nrmicro1552
Jones, N., Bohnsack, J. F., Takahashi, S., Oliver, K. A., Chan, M. S., Kunst, F., et al. (2003). Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41, 2530–2536. doi: 10.1128/JCM.41.6.2530-2536.2003
Karunakaran, R., Raja, N. S., Hafeez, A., and Puthucheary, S. D. (2009). Group B Streptococcus infection: epidemiology, serotypes, and antimicrobial susceptibility of selected isolates in the population beyond infancy (excluding females with genital tract- and pregnancy-related isolates) at the University Malaya Medical Centre, Kuala Lumpur. Jpn. J. Infect. Dis. 62, 192–194.
Kobayashi, M., Schrag, S. J., Alderson, M. R., Madhi, S. A., Baker, C. J., Sobanjo-Ter Meulen, A., et al. (2016a). WHO consultation on group B Streptococcus vaccine development: report from a meeting held on 27-28 April 2016. Vaccine. doi: 10.1016/j.vaccine.2016.12.029 [Epub ahead of print].
Kobayashi, M., Vekemans, J., Baker, C. J., Ratner, A. J., Le Doare, K., and Schrag, S. J. (2016b). Group B Streptococcus vaccine development: present status and future considerations, with emphasis on perspectives for low and middle income countries. F1000Res 5:2355. doi: 10.12688/f1000research.9363.1
Konto-Ghiorghi, Y., Mairey, E., Mallet, A., Dumenil, G., Caliot, E., Trieu-Cuot, P., et al. (2009). Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 5:e1000422. doi: 10.1371/journal.ppat.1000422
Krishnan, V., Gaspar, A. H., Ye, N., Mandlik, A., Ton-That, H., and Narayana, S. V. (2007). An IgG-like domain in the minor pilin GBS52 of Streptococcus agalactiae mediates lung epithelial cell adhesion. Structure 15, 893–903. doi: 10.1016/j.str.2007.06.015
Lachenauer, C. S., Kasper, D. L., Shimada, J., Ichiman, Y., Ohtsuka, H., Kaku, M., et al. (1999). Serotypes VI and VIII predominate among group B streptococci isolated from pregnant Japanese women. J. Infect. Dis. 179, 1030–1033. doi: 10.1086/314666
Lamagni, T. L., Keshishian, C., Efstratiou, A., Guy, R., Henderson, K. L., Broughton, K., et al. (2013). Emerging trends in the epidemiology of invasive group B streptococcal disease in England and Wales, 1991-2010. Clin. Infect. Dis. 57, 682–688. doi: 10.1093/cid/cit337
Lamy, M. C., Dramsi, S., Billoet, A., Reglier-Poupet, H., Tazi, A., Raymond, J., et al. (2006). Rapid detection of the “highly virulent” group B Streptococcus ST-17 clone. Microbes Infect. 8, 1714–1722. doi: 10.1016/j.micinf.2006.02.008
Lamy, M. C., Zouine, M., Fert, J., Vergassola, M., Couve, E., Pellegrini, E., et al. (2004). CovS/CovR of group B streptococcus: a two-component global regulatory system involved in virulence. Mol. Microbiol. 54, 1250–1268. doi: 10.1111/j.1365-2958.2004.04365.x
Lancefield, R. C. (1938). Two Serological Types Of Group B hemolytic streptococci with related. But not identical, type-specific substances. J. Exp. Med. 67, 25–40. doi: 10.1084/jem.67.1.25
Landwehr-Kenzel, S., and Henneke, P. (2014). Interaction of Streptococcus agalactiae and cellular innate immunity in colonization and disease. Front. Immunol. 5:519. doi: 10.3389/fimmu.2014.00519
Lartigue, M. F., Hery-Arnaud, G., Haguenoer, E., Domelier, A. S., Schmit, P. O., van der Mee-Marquet, N., et al. (2009). Identification of Streptococcus agalactiae isolates from various phylogenetic lineages by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 47, 2284–2287. doi: 10.1128/jcm.00175-09
Lauer, P., Rinaudo, C. D., Soriani, M., Margarit, I., Maione, D., Rosini, R., et al. (2005). Genome analysis reveals pili in Group B Streptococcus. Science 309:105. doi: 10.1126/science.1111563
Lazzarin, M., Mu, R., Fabbrini, M., Ghezzo, C., Rinaudo, C. D., Doran, K. S., et al. (2017). Contribution of pilus type 2b to invasive disease caused by a Streptococcus agalactiae ST-17 strain. BMC Microbiol. 17:148. doi: 10.1186/s12866-017-1057-8
Le Doare, K., and Heath, P. T. (2013). An overview of global GBS epidemiology. Vaccine 31(Suppl. 4), D7–D12. doi: 10.1016/j.vaccine.2013.01.009
Le Doare, K., and Kampmann, B. (2014). Breast milk and Group B streptococcal infection: vector of transmission or vehicle for protection? Vaccine 32, 3128–3132. doi: 10.1016/j.vaccine.2014.04.020
Lemire, P., Houde, M., Lecours, M. P., Fittipaldi, N., and Segura, M. (2012). Role of capsular polysaccharide in Group B Streptococccus interactions with dendritic cells. Microbes Infect. 14, 1064–1076. doi: 10.1016/j.micinf.2012.05.015
Leroux-Roels, G., Maes, C., Willekens, J., De Boever, F., de Rooij, R., Martell, L., et al. (2016). A randomized, observer-blind Phase Ib study to identify formulations and vaccine schedules of a trivalent Group B Streptococcus vaccine for use in non-pregnant and pregnant women. Vaccine 34, 1786–1791. doi: 10.1016/j.vaccine.2016.02.044
Lewis, K. (2005). Persister cells and the riddle of biofilm survival. Biochemistry 70, 267–274. doi: 10.1007/s10541-005-0111-6
Libster, R., Edwards, K. M., Levent, F., Edwards, M. S., Rench, M. A., Castagnini, L. A., et al. (2012). Long-term outcomes of group B streptococcal meningitis. Pediatrics 130, e8–e15. doi: 10.1542/peds.2011-3453
Lin, F. Y., Philips, J. B. III, Azimi, P. H., Weisman, L. E., Clark, P., Rhoads, G. G., et al. (2001). Level of maternal antibody required to protect neonates against early-onset disease caused by group B Streptococcus type Ia: a multicenter, seroepidemiology study. J. Infect. Dis. 184, 1022–1028. doi: 10.1086/323350
Lin, F. Y., Weisman, L. E., Azimi, P. H., Philips, J. B. III, Clark, P., Regan, J., et al. (2004). Level of maternal IgG anti-group B streptococcus type III antibody correlated with protection of neonates against early-onset disease caused by this pathogen. J. Infect. Dis. 190, 928–934. doi: 10.1086/422756
Lin, H. C., Chen, C. J., Chiang, K. H., Yen, T. Y., Ho, C. M., Hwang, K. P., et al. (2016). Clonal dissemination of invasive and colonizing clonal complex 1 of serotype VI group B Streptococcus in central Taiwan. J. Microbiol. Immunol. Infect. 49, 902–909. doi: 10.1016/j.jmii.2014.11.002
Lindahl, G., Stalhammar-Carlemalm, M., and Areschoug, T. (2005). Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin. Microbiol. Rev. 18, 102–127. doi: 10.1128/cmr.18.1.102-127.2005
Luan, S. L., Granlund, M., and Norgren, M. (2003). An inserted DNA fragment with plasmid features is uniquely associated with the presence of the GBSi1 group II intron in Streptococcus agalactiae. Gene 312, 305–312. doi: 10.1016/S0378-1119(03)00634-6
Luan, S. L., Granlund, M., Sellin, M., Lagergard, T., Spratt, B. G., and Norgren, M. (2005). Multilocus sequence typing of Swedish invasive group B streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 43, 3727–3733. doi: 10.1128/jcm.43.8.3727-3733.2005
Madhi, S. A., Cutland, C. L., Jose, L., Koen, A., Govender, N., Wittke, F., et al. (2016). Safety and immunogenicity of an investigational maternal trivalent group B streptococcus vaccine in healthy women and their infants: a randomised phase 1b/2 trial. Lancet Infect. Dis. 16, 923–934. doi: 10.1016/s1473-3099(16)00152-3
Madhi, S. A., and Dangor, Z. (2017). Prospects for preventing infant invasive GBS disease through maternal vaccination. Vaccine 35, 4457–4460. doi: 10.1016/j.vaccine.2017.02.025
Madhi, S. A., Dangor, Z., Heath, P. T., Schrag, S., Izu, A., Sobanjo-Ter Meulen, A., et al. (2013). Considerations for a phase-III trial to evaluate a group B Streptococcus polysaccharide-protein conjugate vaccine in pregnant women for the prevention of early- and late-onset invasive disease in young-infants. Vaccine 31(Suppl. 4), D52–D57. doi: 10.1016/j.vaccine.2013.02.029
Madoff, L. C., Michel, J. L., Gong, E. W., Rodewald, A. K., and Kasper, D. L. (1992). Protection of neonatal mice from group B streptococcal infection by maternal immunization with beta C protein. Infect. Immun. 60, 4989–4994.
Maione, D., Margarit, I., Rinaudo, C. D., Masignani, V., Mora, M., Scarselli, M., et al. (2005). Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science 309, 148–150. doi: 10.1126/science.1109869
Maisey, H. C., Doran, K. S., and Nizet, V. (2008a). Recent advances in understanding the molecular basis of group B streptococcus virulence. Expert Rev. Mol. Med. 10:e27. doi: 10.1017/s1462399408000811
Maisey, H. C., Hensler, M., Nizet, V., and Doran, K. S. (2007). Group B streptococcal pilus proteins contribute to adherence to and invasion of brain microvascular endothelial cells. J. Bacteriol. 189, 1464–1467. doi: 10.1128/jb.01153-06
Maisey, H. C., Quach, D., Hensler, M. E., Liu, G. Y., Gallo, R. L., Nizet, V., et al. (2008b). A group B streptococcal pilus protein promotes phagocyte resistance and systemic virulence. FASEB J. 22, 1715–1724. doi: 10.1096/fj.07-093963
Manning, S. D., Lewis, M. A., Springman, A. C., Lehotzky, E., Whittam, T. S., and Davies, H. D. (2008). Genotypic diversity and serotype distribution of group B streptococcus isolated from women before and after delivery. Clin. Infect. Dis. 46, 1829–1837. doi: 10.1086/588296
Manning, S. D., Springman, A. C., Lehotzky, E., Lewis, M. A., Whittam, T. S., and Davies, H. D. (2009). Multilocus sequence types associated with neonatal group B streptococcal sepsis and meningitis in Canada. J. Clin. Microbiol. 47, 1143–1148. doi: 10.1128/jcm.01424-08
Margarit, I., Rinaudo, C. D., Galeotti, C. L., Maione, D., Ghezzo, C., Buttazzoni, E., et al. (2009). Preventing bacterial infections with pilus-based vaccines: the group B streptococcus paradigm. J. Infect. Dis. 199, 108–115. doi: 10.1086/595564
Martins, E. R., Pessanha, M. A., Ramirez, M., and Melo-Cristino, J. (2007). Analysis of group B streptococcal isolates from infants and pregnant women in Portugal revealing two lineages with enhanced invasiveness. J. Clin. Microbiol. 45, 3224–3229. doi: 10.1128/jcm.01182-07
Matsubara, K., Katayama, K., Baba, K., Nigami, H., Harigaya, H., and Sugiyama, M. (2002). Seroepidemiologic studies of serotype VIII group B Streptococcus in Japan. J. Infect. Dis. 186, 855–858. doi: 10.1086/342411
Melin, P. (2011). Neonatal group B streptococcal disease: from pathogenesis to preventive strategies. Clin. Microbiol. Infect. 17, 1294–1303. doi: 10.1111/j.1469-0691.2011.03576.x
Melin, P., and Efstratiou, A. (2013). Group B streptococcal epidemiology and vaccine needs in developed countries. Vaccine 31(Suppl. 4), D31–D42. doi: 10.1016/j.vaccine.2013.05.012
Meyn, L. A., Krohn, M. A., and Hillier, S. L. (2009). Rectal colonization by group B Streptococcus as a predictor of vaginal colonization. Am. J. Obstet. Gynecol. 201, 76.e1–76.e7. doi: 10.1016/j.ajog.2009.02.011
Michel, J. L., Madoff, L. C., Kling, D. E., Kasper, D. L., and Ausubel, F. M. (1991). Cloned alpha and beta C-protein antigens of group B streptococci elicit protective immunity. Infect. Immun. 59, 2023–2028.
Mu, R., Kim, B. J., Paco, C., Del Rosario, Y., Courtney, H. S., and Doran, K. S. (2014). Identification of a group B streptococcal fibronectin binding protein, SfbA, that contributes to invasion of brain endothelium and development of meningitis. Infect. Immun. 82, 2276–2286. doi: 10.1128/iai.01559-13
Nobbs, A. H., Lamont, R. J., and Jenkinson, H. F. (2009). Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73, 407–450. doi: 10.1128/mmbr.00014-09
Nuccitelli, A., Cozzi, R., Gourlay, L. J., Donnarumma, D., Necchi, F., Norais, N., et al. (2011). Structure-based approach to rationally design a chimeric protein for an effective vaccine against Group B Streptococcus infections. Proc. Natl. Acad. Sci. U.S.A. 108, 10278–10283. doi: 10.1073/pnas.1106590108
Paoletti, L. C., and Kasper, D. L. (2003). Glycoconjugate vaccines to prevent group B streptococcal infections. Expert Opin. Biol. Ther. 3, 975–984. doi: 10.1517/14712598.3.6.975
Paoletti, L. J., Bradford, J., and Paoletti, L. C. (1999). A serotype VIII strain among colonizing group B streptococcal isolates in Boston, Massachusetts. J. Clin. Microbiol. 37, 3759–3760.
Park, S. E., Jiang, S., and Wessels, M. R. (2012). CsrRS and environmental pH regulate group B streptococcus adherence to human epithelial cells and extracellular matrix. Infect. Immun. 80, 3975–3984. doi: 10.1128/iai.00699-12
Parker, R. E., Laut, C., Gaddy, J. A., Zadoks, R. N., Davies, H. D., and Manning, S. D. (2016). Association between genotypic diversity and biofilm production in group B Streptococcus. BMC Microbiol. 16:86. doi: 10.1186/s12866-016-0704-9
Patras, K. A., Wang, N. Y., Fletcher, E. M., Cavaco, C. K., Jimenez, A., Garg, M., et al. (2013). Group B Streptococcus CovR regulation modulates host immune signalling pathways to promote vaginal colonization. Cell. Microbiol. 15, 1154–1167. doi: 10.1111/cmi.12105
Pezzicoli, A., Ruggiero, P., Amerighi, F., Telford, J. L., and Soriani, M. (2012). Exogenous sialic acid transport contributes to group B streptococcus infection of mucosal surfaces. J. Infect. Dis. 206, 924–931. doi: 10.1093/infdis/jis451
Pezzicoli, A., Santi, I., Lauer, P., Rosini, R., Rinaudo, D., Grandi, G., et al. (2008). Pilus backbone contributes to group B Streptococcus paracellular translocation through epithelial cells. J. Infect. Dis. 198, 890–898. doi: 10.1086/591182
Phares, C. R., Lynfield, R., Farley, M. M., Mohle-Boetani, J., Harrison, L. H., Petit, S., et al. (2008). Epidemiology of invasive group B streptococcal disease in the United States, 1999-2005. JAMA 299, 2056–2065. doi: 10.1001/jama.299.17.2056
Rajagopal, L. (2009). Understanding the regulation of group B streptococcal virulence factors. Future Microbiol. 4, 201–221. doi: 10.2217/17460913.4.2.201
Rato, M. G., Bexiga, R., Florindo, C., Cavaco, L. M., Vilela, C. L., and Santos-Sanches, I. (2013). Antimicrobial resistance and molecular epidemiology of streptococci from bovine mastitis. Vet. Microbiol. 161, 286–294. doi: 10.1016/j.vetmic.2012.07.043
Rinaudo, C. D., Rosini, R., Galeotti, C. L., Berti, F., Necchi, F., Reguzzi, V., et al. (2010). Specific involvement of pilus type 2a in biofilm formation in group B Streptococcus. PLoS One 5:e9216. doi: 10.1371/journal.pone.0009216
Rosenau, A., Martins, K., Amor, S., Gannier, F., Lanotte, P., van der Mee-Marquet, N., et al. (2007). Evaluation of the ability of Streptococcus agalactiae strains isolated from genital and neonatal specimens to bind to human fibrinogen and correlation with characteristics of the fbsA and fbsB genes. Infect. Immun. 75, 1310–1317. doi: 10.1128/iai.00996-06
Rosini, R., and Margarit, I. (2015). Biofilm formation by Streptococcus agalactiae: influence of environmental conditions and implicated virulence factors. Front. Cell. Infect. Microbiol. 5:6. doi: 10.3389/fcimb.2015.00006
Rosini, R., Rinaudo, C. D., Soriani, M., Lauer, P., Mora, M., Maione, D., et al. (2006). Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol. Microbiol. 61, 126–141. doi: 10.1111/j.1365-2958.2006.05225.x
Rubens, C. E., Wessels, M. R., Heggen, L. M., and Kasper, D. L. (1987). Transposon mutagenesis of type III group B Streptococcus: correlation of capsule expression with virulence. Proc. Natl. Acad. Sci. U.S.A. 84, 7208–7212. doi: 10.1073/pnas.84.20.7208
Russell-Jones, G. J., Gotschlich, E. C., and Blake, M. S. (1984). A surface receptor specific for human IgA on group B streptococci possessing the Ibc protein antigen. J. Exp. Med. 160, 1467–1475. doi: 10.1084/jem.160.5.1467
Salloum, M., van der Mee-Marquet, N., Valentin-Domelier, A. S., and Quentin, R. (2011). Diversity of prophage DNA regions of Streptococcus agalactiae clonal lineages from adults and neonates with invasive infectious disease. PLoS One 6:e20256. doi: 10.1371/journal.pone.0020256
Santi, I., Grifantini, R., Jiang, S. M., Brettoni, C., Grandi, G., Wessels, M. R., et al. (2009a). CsrRS regulates group B Streptococcus virulence gene expression in response to environmental pH: a new perspective on vaccine development. J. Bacteriol. 191, 5387–5397. doi: 10.1128/jb.00370-09
Santi, I., Maione, D., Galeotti, C. L., Grandi, G., Telford, J. L., and Soriani, M. (2009b). BibA induces opsonizing antibodies conferring in vivo protection against group B Streptococcus. J. Infect. Dis. 200, 564–570. doi: 10.1086/603540
Santi, I., Scarselli, M., Mariani, M., Pezzicoli, A., Masignani, V., Taddei, A., et al. (2007). BibA: a novel immunogenic bacterial adhesin contributing to group B Streptococcus survival in human blood. Mol. Microbiol. 63, 754–767. doi: 10.1111/j.1365-2958.2006.05555.x
Schrag, S. J., and Verani, J. R. (2013). Intrapartum antibiotic prophylaxis for the prevention of perinatal group B streptococcal disease: experience in the United States and implications for a potential group B streptococcal vaccine. Vaccine 31(Suppl. 4), D20–D26. doi: 10.1016/j.vaccine.2012.11.056
Schubert, A., Zakikhany, K., Pietrocola, G., Meinke, A., Speziale, P., Eikmanns, B. J., et al. (2004). The fibrinogen receptor FbsA promotes adherence of Streptococcus agalactiae to human epithelial cells. Infect. Immun. 72, 6197–6205. doi: 10.1128/iai.72.11.6197-6205.2004
Schuchat, A. (1998). Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11, 497–513.
Seepersaud, R., Hanniffy, S. B., Mayne, P., Sizer, P., Le Page, R., and Wells, J. M. (2005). Characterization of a novel leucine-rich repeat protein antigen from group B streptococci that elicits protective immunity. Infect. Immun. 73, 1671–1683. doi: 10.1128/iai.73.3.1671-1683.2005
Seifert, K. N., Adderson, E. E., Whiting, A. A., Bohnsack, J. F., Crowley, P. J., and Brady, L. J. (2006). A unique serine-rich repeat protein (Srr-2) and novel surface antigen (epsilon) associated with a virulent lineage of serotype III Streptococcus agalactiae. Microbiology 152, 1029–1040. doi: 10.1099/mic.0.28516-0
Sendi, P., Johansson, L., and Norrby-Teglund, A. (2008). Invasive group B Streptococcal disease in non-pregnant adults : a review with emphasis on skin and soft-tissue infections. Infection 36, 100–111. doi: 10.1007/s15010-007-7251-0
Seo, H. S., Minasov, G., Seepersaud, R., Doran, K. S., Dubrovska, I., Shuvalova, L., et al. (2013). Characterization of fibrinogen binding by glycoproteins Srr1 and Srr2 of Streptococcus agalactiae. J. Biol. Chem. 288, 35982–35996. doi: 10.1074/jbc.M113.513358
Seo, H. S., Mu, R., Kim, B. J., Doran, K. S., and Sullam, P. M. (2012). Binding of glycoprotein Srr1 of Streptococcus agalactiae to fibrinogen promotes attachment to brain endothelium and the development of meningitis. PLoS Pathog. 8:e1002947. doi: 10.1371/journal.ppat.1002947
Shabayek, S., Abdalla, S., and Abouzeid, A. M. (2014). Serotype and surface protein gene distribution of colonizing group B streptococcus in women in Egypt. Epidemiol. Infect. 142, 208–210. doi: 10.1017/s0950268813000848
Sheen, T. R., Jimenez, A., Wang, N. Y., Banerjee, A., van Sorge, N. M., and Doran, K. S. (2011). Serine-rich repeat proteins and pili promote Streptococcus agalactiae colonization of the vaginal tract. J. Bacteriol. 193, 6834–6842. doi: 10.1128/jb.00094-11
Singh, B., Fleury, C., Jalalvand, F., and Riesbeck, K. (2012). Human pathogens utilize host extracellular matrix proteins laminin and collagen for adhesion and invasion of the host. FEMS Microbiol. Rev. 36, 1122–1180. doi: 10.1111/j.1574-6976.2012.00340.x
Skoff, T. H., Farley, M. M., Petit, S., Craig, A. S., Schaffner, W., Gershman, K., et al. (2009). Increasing burden of invasive group B streptococcal disease in nonpregnant adults, 1990-2007. Clin. Infect. Dis. 49, 85–92. doi: 10.1086/599369
Slotved, H. C., Kong, F., Lambertsen, L., Sauer, S., and Gilbert, G. L. (2007). Serotype IX, a proposed new Streptococcus agalactiae serotype. J. Clin. Microbiol. 45, 2929–2936. doi: 10.1128/jcm.00117-07
Sørensen, U. B. S., Poulsen, K., Ghezzo, C., Margarit, I., and Kilian, M. (2010). Emergence and global dissemination of host-specific Streptococcus agalactiae clones. mBio 1:e00178-10. doi: 10.1128/mBio.00178-10
Spellerberg, B., Rozdzinski, E., Martin, S., Weber-Heynemann, J., Schnitzler, N., Lutticken, R., et al. (1999). Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin. Infect. Immun. 67, 871–878.
Springman, A. C., Lacher, D. W., Waymire, E. A., Wengert, S. L., Singh, P., Zadoks, R. N., et al. (2014). Pilus distribution among lineages of group b streptococcus: an evolutionary and clinical perspective. BMC Microbiol. 14:159. doi: 10.1186/1471-2180-14-159
Stålhammar-Carlemalm, M., Stenberg, L., and Lindahl, G. (1993). Protein rib: a novel group B streptococcal cell surface protein that confers protective immunity and is expressed by most strains causing invasive infections. J. Exp. Med. 177, 1593–1603. doi: 10.1084/jem.177.6.1593
Stoner, T. D., Weston, T. A., Trejo, J., and Doran, K. S. (2015). Group B streptococcal infection and activation of human astrocytes. PLoS One 10:e0128431. doi: 10.1371/journal.pone.0128431
Sukhnanand, S., Dogan, B., Ayodele, M. O., Zadoks, R. N., Craver, M. P., Dumas, N. B., et al. (2005). Molecular subtyping and characterization of bovine and human Streptococcus agalactiae isolates. J. Clin. Microbiol. 43, 1177–1186. doi: 10.1128/jcm.43.3.1177-1186.2005
Tamura, G. S., Kuypers, J. M., Smith, S., Raff, H., and Rubens, C. E. (1994). Adherence of group B streptococci to cultured epithelial cells: roles of environmental factors and bacterial surface components. Infect. Immun. 62, 2450–2458.
Tazi, A., Disson, O., Bellais, S., Bouaboud, A., Dmytruk, N., Dramsi, S., et al. (2010). The surface protein HvgA mediates group B streptococcus hypervirulence and meningeal tropism in neonates. J. Exp. Med. 207, 2313–2322. doi: 10.1084/jem.20092594
Teatero, S., Athey, T. B., Van Caeseele, P., Horsman, G., Alexander, D. C., Melano, R. G., et al. (2015). Emergence of serotype IV group B Streptococcus adult invasive disease in Manitoba and Saskatchewan. Canada, is driven by clonal sequence type 459 strains. J. Clin. Microbiol. 53, 2919–2926. doi: 10.1128/jcm.01128-15
Teatero, S., Ferrieri, P., and Fittipaldi, N. (2016). Serotype IV sequence type 468 group B Streptococcus neonatal invasive disease, Minnesota, USA. Emerg. Infect. Dis. 22, 1937–1940. doi: 10.3201/eid2211.152031
Teatero, S., Ferrieri, P., Martin, I., Demczuk, W., McGeer, A., and Fittipaldi, N. (2017). Serotype distribution, population structure, and antimicrobial resistance of group B Streptococcus strains recovered from colonized pregnant women. J. Clin. Microbiol. 55, 412–422. doi: 10.1128/jcm.01615-16
Teatero, S., McGeer, A., Low, D. E., Li, A., Demczuk, W., Martin, I., et al. (2014). Characterization of invasive group B streptococcus strains from the greater Toronto area, Canada. J. Clin. Microbiol. 52, 1441–1447. doi: 10.1128/jcm.03554-13
Tenenbaum, T., Spellerberg, B., Adam, R., Vogel, M., Kim, K. S., and Schroten, H. (2007). Streptococcus agalactiae invasion of human brain microvascular endothelial cells is promoted by the laminin-binding protein Lmb. Microbes Infect. 9, 714–720. doi: 10.1016/j.micinf.2007.02.015
van der Mee-Marquet, N., Fourny, L., Arnault, L., Domelier, A. S., Salloum, M., Lartigue, M. F., et al. (2008). Molecular characterization of human-colonizing Streptococcus agalactiae strains isolated from throat, skin, anal margin, and genital body sites. J. Clin. Microbiol. 46, 2906–2911. doi: 10.1128/jcm.00421-08
Verani, J. R., McGee, L., and Schrag, S. J. (2010). Prevention of perinatal group B streptococcal disease–revised guidelines from CDC, 2010. MMWR Recomm. Rep. 59, 1–36.
Wilkinson, H. W., and Eagon, R. G. (1971). Type-specific antigens of group B type Ic streptococci. Infect. Immun. 4, 596–604.
Xia, F. D., Mallet, A., Caliot, E., Gao, C., Trieu-Cuot, P., and Dramsi, S. (2015). Capsular polysaccharide of Group B Streptococcus mediates biofilm formation in the presence of human plasma. Microbes Infect. 17, 71–76. doi: 10.1016/j.micinf.2014.10.007
Yang, Y., Liu, Y., Ding, Y., Yi, L., Ma, Z., Fan, H., et al. (2013). Molecular characterization of Streptococcus agalactiae isolated from bovine mastitis in Eastern China. PLoS One 8:e67755. doi: 10.1371/journal.pone.0067755
Zawaneh, S. M., Ayoub, E. M., Baer, H., Cruz, A. C., and Spellacy, W. N. (1979). Factors influencing adherence of group B streptococci to human vaginal epithelial cells. Infect. Immun. 26, 441–447.
Keywords: Streptococcus agalactiae, colonization, molecular characteristics, epidemiology, serotype
Citation: Shabayek S and Spellerberg B (2018) Group B Streptococcal Colonization, Molecular Characteristics, and Epidemiology. Front. Microbiol. 9:437. doi: 10.3389/fmicb.2018.00437
Received: 29 November 2017; Accepted: 26 February 2018;
Published: 14 March 2018.
Edited by:
Bernd Kreikemeyer, University of Rostock, GermanyReviewed by:
Peter Valentin-Weigand, University of Veterinary Medicine Hannover, GermanyCopyright © 2018 Shabayek and Spellerberg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Barbara Spellerberg, YmFyYmFyYS5zcGVsbGVyYmVyZ0B1bmlrbGluaWstdWxtLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.