- Pathogen Molecular Genetics Section, Laboratory of Bacteriology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, United States
The Staphylococcus aureus enterotoxins are a superfamily of secreted virulence factors that share structural and functional similarities and possess potent superantigenic activity causing disruptions in adaptive immunity. The enterotoxins can be separated into two groups; the classical (SEA-SEE) and the newer (SEG-SElY and counting) enterotoxin groups. Many members from both these groups contribute to the pathogenesis of several serious human diseases, including toxic shock syndrome, pneumonia, and sepsis-related infections. Additionally, many members demonstrate emetic activity and are frequently responsible for food poisoning outbreaks. Due to their robust tolerance to denaturing, the enterotoxins retain activity in food contaminated previously with S. aureus. The genes encoding the enterotoxins are found mostly on a variety of different mobile genetic elements. Therefore, the presence of enterotoxins can vary widely among different S. aureus isolates. Additionally, the enterotoxins are regulated by multiple, and often overlapping, regulatory pathways, which are influenced by environmental factors. In this review, we also will focus on the newer enterotoxins (SEG-SElY), which matter for the role of S. aureus as an enteropathogen, and summarize our current knowledge on their prevalence in recent food poisoning outbreaks. Finally, we will review the current literature regarding the key elements that govern the complex regulation of enterotoxins, the molecular mechanisms underlying their enterotoxigenic, superantigenic, and immunomodulatory functions, and discuss how these activities may collectively contribute to the overall manifestation of staphylococcal food poisoning.
Introduction
Staphylococcus aureus is a dangerous human pathogen whose virulence potential predominantly relies on the production of an impressive catalog of protein toxins. These can work separately or in concert to cause a multitude of human diseases. Pneumonia, sepsis-related infections, toxic shock syndrome, and food poisoning are diseases that have traditionally been associated in particular with the production of enterotoxins (Lowy, 2003). However, recent studies suggest that the staphylococcal enterotoxins (SEs) have a broader role in the manifestation of a number of other human illnesses, including those associated with the respiratory tract (Pastacaldi et al., 2011; Huvenne et al., 2013) and the development of autoimmune diseases (Principato and Qian, 2014; Li et al., 2015). The SEs are powerful non-specific T-cell stimulators (superantigens) that cause unregulated activation of the immune response (for detailed reviews see Fraser and Proft, 2008; Spaulding et al., 2013). If this stimulation is sustained, a massive cytokine overload is produced preluding the clinical hallmarks of toxic shock syndrome, which is characterized by the fast onset of fever, organ failure and significant mortality (Lappin and Ferguson, 2009). Unlike the majority of other secreted toxins produced by S. aureus, the SEs require only minute quantities to be toxic in humans. Additionally, the SEs have a remarkable tolerance to extreme denaturing conditions, such as low pH (Schantz et al., 1965; Bergdoll, 1983), heating (Evenson et al., 1988; Asao et al., 2003) and proteolytic digestion (Humber et al., 1975; Regenthal et al., 2017). These combined qualities make the SEs, especially SEB, potential bioterrorism agents (Madsen, 2001). Notably, SEB is also classified as a Category B select agent by various United States federal agencies.
In addition to the toxic effects they have on the host, the SEs are potent emesis-inducing toxins. Reports of the involvement of enterotoxin-producing S. aureus in staphylococcal food poisoning (SFP) can be dated as far back as the 1900s. However, it was not until the 1930s that a link between the two were made (Dack, 1937). In healthy human individuals, SFP is an acute disease depicted by symptoms including nausea, vomiting, abdominal cramping, diarrhea, typically in the absence of fever, appearing within 3–9 h after the ingestion of food contaminated previously with enterotoxin-producing S. aureus. SFP is often self-limiting with recovery occurring 1–3 days after the onset of symptoms (Le Loir et al., 2003). However, symptoms may be more severe in the young, elderly and immunocompromised (Murray, 2005; Argudin et al., 2010). The SEs' ability to traverse the harsh acidic conditions within the gut to reach the intestine means that the advancement of SFP can also occur in the absence of live bacteria. Typically, only high nanogram to low microgram quantities of enterotoxins are needed to induce the symptoms of SFP (Larkin et al., 2009).
Next to E. coli, Shigella, Bacillus spp., and Clostridium spp., S. aureus is among the leading toxin-producing bacterial causative agents of food poisoning. S. aureus is also frequently mentioned in national foodborne illness estimates (Gkogka et al., 2011; Bennett et al., 2013; Thomas et al., 2013; Kirk et al., 2014; Mangen et al., 2015; Park et al., 2015; Van Cauteren et al., 2017), and is identified as a main player in major food poisoning outbreaks worldwide (Asao et al., 2003; Do Carmo et al., 2004; Chiang et al., 2008; Ostyn et al., 2010; Sato'o et al., 2014; Ercoli et al., 2017). In the US alone, it is estimated that S. aureus accounts for more than 240,000 foodborne illnesses per year (Scallan et al., 2011). However, considering that SFP can be resolved in individuals without hospitalization, it is not unusual for many cases to go unreported. While SFP rarely develops into a life-threatening disease, its frequency has a significant impact on the economy, resulting in a loss in productivity. It also represents a serious financial burden, especially for the food industry, catering businesses, and public healthcare systems. The implementation of traditional hygiene practices and proper food safety measures are key to preventing foodborne illness (Hussain and Dawson, 2013).
The Superfamily of Staphylococcal Enterotoxins; Proteins and Overview
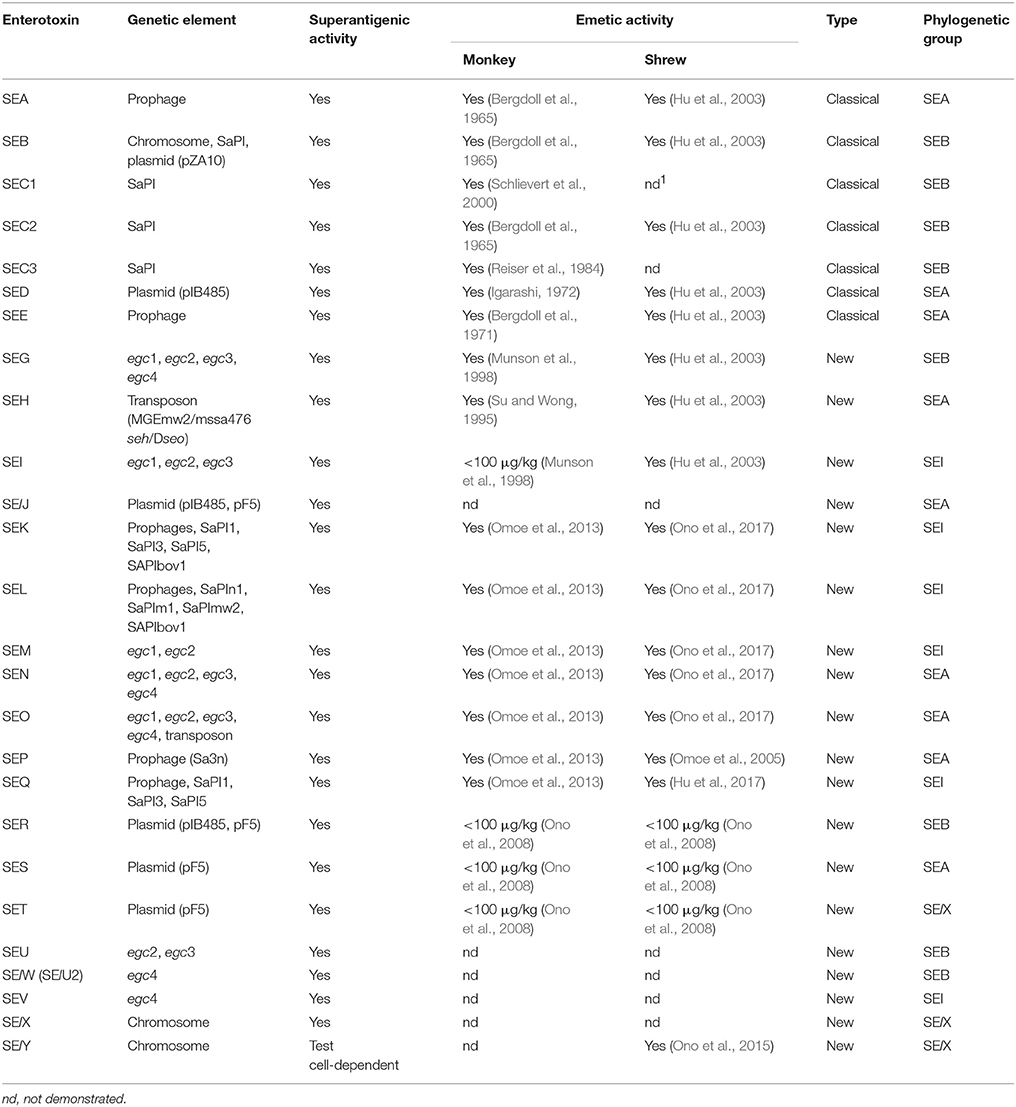
The superfamily of SEs and enterotoxin-like (SEls) proteins (Table 1) share many common features; they are non-glycosylated, antigenically distinct, low molecular weight (19–29 kDa) single-chain proteins that all fold into homologous globular structures (Thomas et al., 2007). Since the first characterization of the classical SEs (SEA to SEE) in S. aureus (Bergdoll et al., 1965, 1971, 1973; Casman et al., 1967; Marrack and Kappler, 1990), advancements in the area of molecular biology during the 1980s led to the identification of a new set of genes encoding closely-related proteins with superantigenic and emetic activities (Table 1). This sudden increase in the number of described SEs spurred a move to standardize their nomenclature (Lina et al., 2004). Only enterotoxins with demonstrated emetic potential in monkeys were designated “SE,” whereas enterotoxins that failed to do so or have not been evaluated in non-human primate models of emesis are designated enterotoxin like (SEl-) toxins (Table 1). The only exception to this rule is Toxic Shock Syndrome Toxin-1 (TSST-1), which was originally designated SEF (Bergdoll et al., 1981; Reiser et al., 1983). This toxin's apparent lack of emetic activity, possibly due it being less stable than other SEs (Edwin and Kass, 1989), prompted the name change to TSST-1, which has remained in place ever since. Joining TSST-1, SElJ is the only other tested SE that is non-emetic (Munson et al., 1998; Orwin et al., 2001, 2002). SElX, SElU, SElW, SElV, and SElY have yet to be tested for emetic activity in non-human primates.
The SE Genes Are Distributed Across a Variety of Different Genomic Locations
When considering the locations of the enterotoxin genes, selx (Wilson et al., 2011) and sely (Ono et al., 2015) are unique as they are found exclusively on the genome. The selx gene can be found in ~95% of S. aureus strains, whereas sely appears less frequently and has only been detected in a handful of strains thus far. In contrast, the other enterotoxin genes are sometimes found alone, but more commonly in groups, on a variety of large mobile segments of DNA called mobile genetic elements (MGEs) (Fraser and Proft, 2008; Argudin et al., 2010). These MGEs include prophages, plasmids, transposons, S. aureus pathogenicity islands (SaPIs), and the enterotoxin gene clusters (egc) (Table 1) (for a review on staphylococcal MGEs see, Malachowa and DeLeo, 2010). The egc locus is home to an operon of genes encoding SEG, SEI, SEM, SEN, SEO, and two pseudogenes, φent1 and φent2 (Jarraud et al., 2001; Monday and Bohach, 2001). Deletion, duplication and recombination events within this cluster make it a major hub for the generation of new types of SEs and variants (Letertre et al., 2003b; Thomas et al., 2006). The acquisition of MGEs generally has a significant impact on core genomes by causing striking differences in genome size and structure. In S. aureus, a comparison of the presence of SE genes from several major lineages shows that SE gene composition is strongly linked to specific genetic backgrounds, emphasizing the importance of vertical transmission, rather than horizontal transmission, of SE-encoding MGEs (Goerke et al., 2009). Around 80% of S. aureus isolates, including commensal, clinical, and food-poisoning isolates, carry an average of 5–6 SE genes (Jarraud et al., 2001; Baba et al., 2002; Becker et al., 2003; Holtfreter et al., 2004, 2007; Hait et al., 2014; Lv et al., 2014; Umeda et al., 2017).
The Enterotoxins Can be Further Separated Based on Nucleotide and Amino Acid Sequences
The 24 currently identified SEs and SEls, can be further separated into several evolutionary groups based on a comparison of their nucleotide and amino acid sequences; the SEA group (SEA, SED, SEE, SElJ, SEH, SEN, SEO, SEP, SES), the SEB group (SEB, SECs, SEG, SER, SElU, SElW, previously known as SElU2), the SEI group (SEI, SEK, SEL, SEQ, SEM, SElV), and the SElX group (TSST-1, SET, SElX, SElY and members of another group of staphylococcal exotoxins called superantigen-like (SSL) toxins) (for reviews, see Fraser and Proft, 2008; Ono et al., 2015) (Table 1). A fifth group, which is not produced by staphylococci, but only represented by a group of functionally and structurally similar superantigenic toxins produced by streptococci, will not be discussed further.
The presence or absence of two specific structural features predominantly defines the superantigenic and enterotoxigenic properties of the SEs and explains differences in activity between the evolutionary groups. First, enterotoxins belonging to the SElX and SEB groups only possess one low affinity α-chain major histocompatibility complex (MHC) II binding site, whereas enterotoxins from the SEA and SEI groups contain one low affinity α-chain MHC II and a second, high affinity β-chain MHC II binding site, which generally equates to superior superantigenic activity (Kozono et al., 1995). Additionally, differences in amino acid composition have given rise to variants of SEB (Kohler et al., 2012), SEC (Bohach and Schlievert, 1987; Couch and Betley, 1989; Marr et al., 1993), SED (Johler et al., 2016), SEG, SEI (Abe et al., 2000; Blaiotta et al., 2004), SEK (Aguilar et al., 2014), SEM, SEN, SEO, SElU, and SElV (Letertre et al., 2003b; Collery et al., 2009). Compared to the parent toxins, variants of SEB (Kohler et al., 2012) and SEC (Deringer et al., 1997) demonstrate altered species tropism or reduced superantigenic activities. The production of these mutations in SEs may be part of a broader strategy of S. aureus to adapt to different host species (Marr et al., 1993; Edwards et al., 1997; Johler et al., 2016).
Second, a separate and distinct loop comprising 9–19 varying amino acids flanked by 2 cysteine residues creating a disulfide bridge, was originally thought to be an essential feature of emesis-inducing SE members from the SEA and SEB evolutionary groups. However, mutational analyses of that loop demonstrated that only the disulfide bond between the two cysteine residues, rather than the loop itself, was required for emesis (Hovde et al., 1994). These data are consistent with experiments demonstrating that SEs that lack the loop can still induce emesis in primates (Omoe et al., 2013), leading to the conclusion that there are additional unidentified emesis-associated structural determinant(s) in the SEs.
S. aureus Has a Complex Network of Regulatory Pathways to Control Toxin Production
S. aureus responds to changes in the environment using a combination of quorum-sensing (QS) (Waters and Bassler, 2005) and other two-component systems (TCS), of which at least 16 have been discovered in S. aureus to date (Haag and Bagnoli, 2016), as well as many trans-acting regulatory proteins (Bronner et al., 2004). S. aureus relies on these systems to quickly make changes in the regulation of genes associated with important physiological features, including drug resistance, metabolism, immune evasion, and virulence. Each system can directly or indirectly control the transcription of specific sets of genes. However, the regulation of one gene may be influenced by multiple systems, leading to additional layers of regulation.
The accessory gene regulator (Agr) QS system, which is activated at high cell densities, is comprised of two transcriptional units transcribed in opposing directions; RNAII, which codes for four genes (agrA, agrB, agrC, and agrD) (Novick et al., 1995) and RNAIII, a regulatory RNA. These transcripts are controlled by the promoters P2 and P3, respectively. AgrD, which contains the sequence for the autoinducing peptide (AIP), is processed and exported out of the cell by the combined actions of the membrane-associated export protein, AgrB (Ji et al., 1995, 1997; Mayville et al., 1999) and a type I signal peptidase, SpsB (Kavanaugh et al., 2007). AIP acts as the ligand for the membrane bound histidine kinase, AgrC, leading to the phosphorylation of AgrA (Ji et al., 1995; Lina et al., 1998). Activated ArgA binds to the P2 and P3 promoters, resulting in the perpetuation of a positive feedback loop (Koenig et al., 2004).
Expression of agr is affected by various trans-activing regulators, such as the Sar family of regulatory proteins, (SarR, SarS, SarT, SarU, SarX, SarZ, SarV, MgrA, and Rot) (Cheung and Projan, 1994; Heinrichs et al., 1996; Cheung et al., 2008), σB (Lauderdale et al., 2009), and SrrAB (Staphylococcal respiratory response AB) (Yarwood et al., 2001; Pragman et al., 2004). Additionally, σB and Rot can affect another important two-component system called SaeRS (Li and Cheung, 2008; Kusch et al., 2011). Importantly, all these regulatory elements respond to various environmental stresses and stimuli; the SaeRS (S. aureus exoprotein expression) system responds to membrane attack by antimicrobial molecules produced by the innate host defense (Novick and Jiang, 2003; Kuroda et al., 2007; Geiger et al., 2008; Cho et al., 2015), SarA largely responds to changes in microenvironments (Cheung et al., 2004), σB responds to high temperature, catabolites, alkaline pH, high salinity (Betley et al., 1992; Wu et al., 1996; Kullik and Giachino, 1997; Kullik et al., 1998; Pané-Farré et al., 2006), whereas the SrrAB system has been shown to be particularly crucial for bacterial growth under anaerobic and hypoxic conditions (Yarwood et al., 2001; Pragman et al., 2007; Kinkel et al., 2013; Mashruwala and Boyd, 2017). Lastly, Rot, the global gene regulator (Saïd-Salim et al., 2003) is negatively regulated by RNAIII through an antisense mechanism (Geisinger et al., 2006; Boisset et al., 2007).
Regulation of the Classical Enterotoxins
It has been described early that there is unequal distribution of SE-associated MGEs among S. aureus isolates, and that thus, the host background has profound influences on enterotoxin production (Gaskill and Khan, 1988; Compagnone-Post et al., 1991). Surprisingly, our understanding of how the enterotoxins are regulated is still rather incomplete, but we do know that enterotoxin regulation is strongly dependent on the regulatory systems described above (Figure 1). Several Agr-controlled staphylococcal toxins, such as alpha-toxin (Morfeldt et al., 1995) and the family of phenol-soluble modulins (PSMs) (Queck et al., 2008) are produced between the early logarithmic and stationary phases. Early observations showing that the production of SEB (Czop and Bergdoll, 1974; Gaskill and Khan, 1988; Derzelle et al., 2009), SEC (Otero et al., 1990; Regassa et al., 1991), and SED (Bayles and Iandolo, 1989) also occurred between the exponential to stationary phases of bacterial growth (Gaskill and Khan, 1988; Regassa et al., 1991; Zhang and Stewart, 2000) suggested that they could be regulated by Agr. Indeed, isogenic S. aureus agr mutants showed significant decreases in SEC and SED production compared to the wild-type strain (Regassa et al., 1991). However, it was later shown that SEB, SEC, and SED is regulated indirectly by other factors. For instance, Agr-dependent regulation of SEB, SEC, and SED occurs via RNAIII-dependent inhibition of Rot (Regassa and Betley, 1993; Tseng et al., 2004; Tseng and Stewart, 2005). In addition to Rot, SEB is also negatively regulated by σB (Ziebandt et al., 2001, 2004; Pané-Farré et al., 2006; Rogasch et al., 2006).
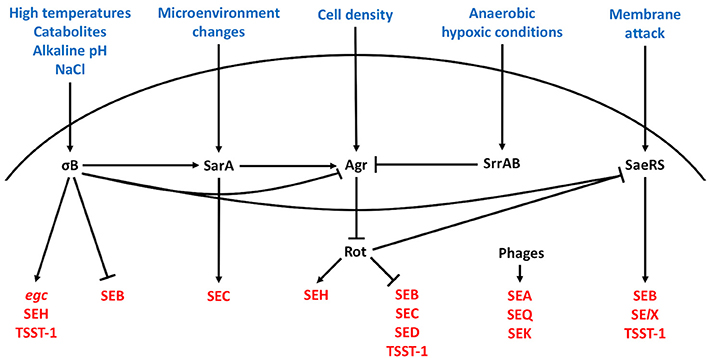
Figure 1. Regulation of staphylococcal enterotoxins. Harsh bacterial growth conditions, changes in the bacterial microenvironment, high cell density, hypoxia, and membrane changes direct enterotoxin expression through the alternative sigma factor, SarA protein family, Agr quorum sensing system, SrrAB protein, and SaeRS two-component system, respectively. The excitatory and inhibitory action of these systems on the other regulators and enterotoxins are summarized. Arrowheads represent upregulation and bars downregulation.
In contrast, the production of bacteriophage-associated SEA is generally constitutive (Thomas et al., 2007), although S. aureus strains with distinct high and low SEA expression patterns have been described (Borst and Betley, 1994; Wallin-Carlquist et al., 2010). Since the expression pattern of SEA was found to be different from that of SEB, SEC and SED, it was postulated and confirmed that SEA is regulated independently of Agr (Tremaine et al., 1993). The production of SEA was later discovered to be closely tied to the phage's life cycle (Cao et al., 2012) and to be inducible by bacterial stress (Zeaki et al., 2015).
Regulation of the Newer Enterotoxins
Information surrounding the regulation of the newer enterotoxins is only beginning to emerge. Unlike most of the classical enterotoxins, it appears that the regulation of several newer enterotoxins including SElJ (Zhang et al., 1998) and SEH (Lis et al., 2012), is Agr-independent. The expression of SFP-associated SEH, which is produced predominantly in the late exponential phase of bacterial growth (Sakai et al., 2008; Lis et al., 2012), was recently shown to be positively regulated by Rot, via direct binding to the seh promoter (Sato'o et al., 2015), σB (Kusch et al., 2011), several Sar homologs, and SaeR (Sato'o et al., 2015). Moreover, SaeRS appears to have a positive impact on SElX (Langley et al., 2017) and TSST-1 (Baroja et al., 2016) expression. In contrast, the production of enterotoxins encoded in the egc operon (SEG, SEI, SEM, SEN, SEO, and SElU) is highest in the earliest stages of exponential growth (Grumann et al., 2008) and dependent on σB (Kusch et al., 2011). Interestingly, one study showed that SEK production is dependent on the presence of SEB (Aguilar et al., 2014), whereas SEK and SEQ, which are also found on sea-associated phages, can be transcriptionally induced by mitomycin C (Sumby and Waldor, 2003). Taken together, the SEs are regulated by multiple regulatory elements that respond to a variety of different environmental signals. Likely, the delicate balance in enterotoxin expression facilitated by these regulatory elements has a profound impact on the commensal and pathogenic lifestyles of S. aureus.
Which Staphylococcal Enterotoxins Contribute to SFP?
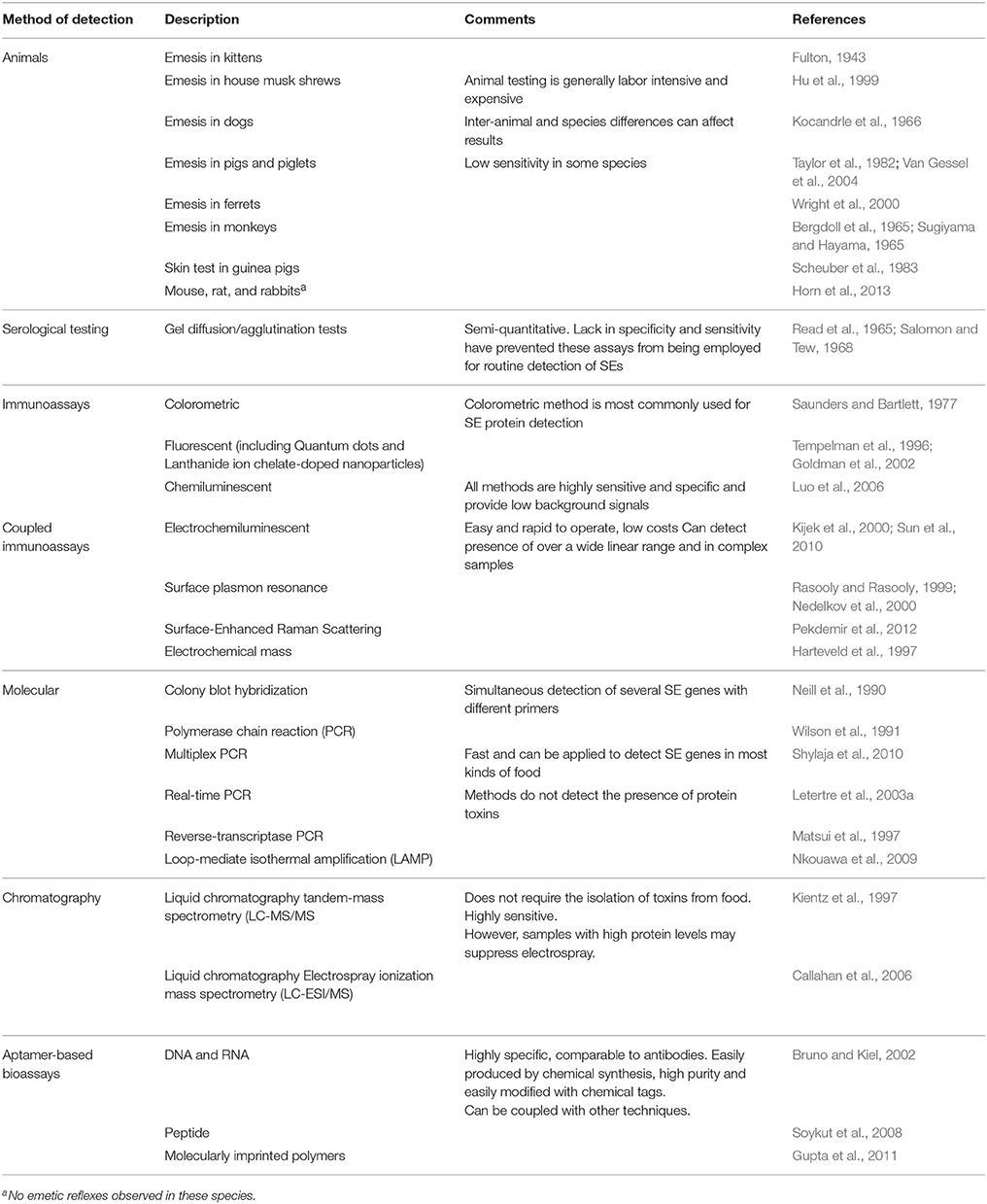
To control staphylococcal food poisoning and ensure food safety, the roles of both new and classical SEs must be considered. Although a wide variety of SE detection methods have been developed (Table 2), molecular detection of SE genes remains the most common method used for investigating the possible contribution of SEs toward SFP. Molecular studies spanning the last two decades have shown that egc-encoded genes (seg, seh, sei, or selj) are readily detected in S. aureus food poisoning isolates around the world (Blaiotta et al., 2004; Grumann et al., 2008; Yan et al., 2012; Viçosa et al., 2013; Chao et al., 2015; Johler et al., 2015; Cheng et al., 2016; Song et al., 2016; Shen et al., 2017; Umeda et al., 2017). Additionally, the detection of non egc-encoded enterotoxin genes, such as transposon associated-seh (McLauchlin et al., 2000; Ikeda et al., 2005; Jørgensen et al., 2005), plasmid-associated ser (Wattinger et al., 2012) and SaPI-associated seq (Chiang et al., 2008; Alibayov et al., 2014; Lv et al., 2014; Hu et al., 2017) suggest a role of these newer SEs in SFP.
While PCR is an invaluable tool, confirmation of the physical presence of toxin in food products suspected of contamination is needed to clearly verify their contribution to SFP. The immunological detection of the 5 classical SEs has helped to establish SEA as the top contributor (~80%) to SFP outbreaks (Pinchuk et al., 2010; Hennekinne et al., 2012), followed by SED, SEB, SEC, and SEE (Hu and Nakane, 2014). In contrast, due to the lack of sensitive detection methods, it has been impossible to draw such conclusions for the newer SEs. However, a steadily increasing number of immunological assays for the non-classical enterotoxins, such as SEG (Nagaraj et al., 2016), SEH (Su and Wong, 1996), SEI (Zhao et al., 2016b), SEK (Aguilar et al., 2014), SEM (Zhao et al., 2017), and SEQ (Hu et al., 2017) have been developed within the last decade. They indicated that one or more of the newer enterotoxins are potential causes of SFP outbreaks. Although few studies have examined the physical presence of multiple enterotoxins, it is most likely that multiple SEs contribute to SFP. The expansion of existing multiplex assays (Liang et al., 2015; Adhikari et al., 2016) would be the most efficient strategy to detect all SEs simultaneously. However, each platform has its advantages and disadvantages (Table 2; Wu et al., 2016 for review). An ideal platform to detect all SEs would have high sensitivity, low cross-reactivity, and universal adaptability. Although creating such a system is not impossible, it would be an extremely difficult task, requiring considerable resources, and vigorous testing.
Humans and Livestock Are Major Reservoirs for the Transmission of Enterotoxin-Producing S. aureus
S. aureus, a natural colonizer of humans, can be found on the skin (primarily on the hands, chest, and abdomen), gastrointestinal (GI) tract (Ridley, 1959; Armstrong-Esther, 1976; Wertheim et al., 2005), and nasopharyngeal cavities (Williams, 1963). All these sites represent possible reservoirs for the distribution of S. aureus causing human disease. Persistent colonization of the anterior nares with S. aureus, which currently is estimated to be around 20–30% of the population (Verhoeven et al., 2014), is believed to be the most important risk factor for infection, especially regarding health-care associated diseases (Von Eiff et al., 2001). While colonization of the GI tract by S. aureus has received significantly less attention, recent studies emphasize its underappreciated role in the association with and transmission of S. aureus disease (Nowrouzian et al., 2011, 2017; Senn et al., 2016; Gagnaire et al., 2017). With regards to SFP, studies investigating the presence of enterotoxin genes in S. aureus isolates sampled from the nose (Nashev et al., 2007; Collery et al., 2009; Wattinger et al., 2012; Ho et al., 2015) and gut (Lis et al., 2009; Shin et al., 2016) indicate that these two sites are important sources of enterotoxin-producing S. aureus.
S. aureus is particularly renowned for its ability to acquire and develop resistance to multiple antibiotics, which is a key factor contributing to the difficultly of treating infections caused by this pathogen. A majority of S. aureus infections are caused by methicillin-resistant strains (MRSA), which, historically, have been associated with disease in hospitalized patients in a variety of public healthcare settings [hospital-associated (HA)-MRSA]. However, in the early 1990s, a new breed of genetically distinct MRSA strains started to appear in the community [community-associated (CA)-MRSA] (Otto, 2010). Compared to the HA-MRSA strains, CA-MRSA strains are exceptionally pathogenic (Chambers, 2001; Cameron et al., 2011) because of the enhanced production and acquisition of a broad set of virulence factors that contribute to fitness, colonization and virulence (Otto, 2012). Additionally, MRSA infections in the community can be caused by strains initially associated with livestock [livestock-associated MRSA (LA-MRSA)] (Huijsdens et al., 2006; Lewis et al., 2008; Nemati et al., 2008). For instance, carriage, or infections caused by S. aureus in dairy cattle (e.g., mastitis) can lead to the contamination of dairy products and raw meat. In particular, unprocessed foods hold a substantial risk for the introduction of resistant microbes into the food chain, which can have a considerable economic impact, especially in countries with industrialized dairy sectors (Le Loir et al., 2003). Interestingly, epidemiological studies have indicated that LA-MRSA isolates belong to genetic lineages different from their HA- and CA-MRSA counterparts (for detailed reviews, see Fluit, 2012; Cuny et al., 2015; Smith, 2015) and harbor unique genes that are essential for host adaptation (Lowder et al., 2009; Guinane et al., 2010; Price et al., 2012).
Unsurprisingly, several recent studies reported high levels of multiple antibiotic resistance in LA-MRSA (Kérouanton et al., 2007; Ge et al., 2017; Sahibzada et al., 2017; Abdi et al., 2018; Suleiman et al., 2018), but unlike other enteric pathogens, such as Salmonella and E. coli, for which antimicrobial resistance can impose serious health risks in humans (Doyle, 2015), antibiotic resistance in HA-, CA-, or LA-MRSA isolates had little influence on the ability of S. aureus to cause SFP (Sergelidis and Angelidis, 2017). These observations are consistent with the notion that SFP is not a disease that is typically treated with antibiotics, since the enterotoxin-driven manifestation of SFP can progress in the absence of bacteria.
Mechanisms Underlying Enterotoxin-Induced Emetic and Diarrheal Activity
Progress in understanding the molecular mechanisms underlying the enterotoxigenic effects of the SEs has been hampered by a lack of relevant animal models. Small rodents, such as mice and rats, are non-emetic and generally less susceptible to the enterotoxigenic effects of the SEs (Bergdoll, 1988) whereas non-human primates, which are considered the gold standard for testing the emetic activity of enterotoxins, are costly and riddled with complex requirements in animal care and husbandry. However, the house musk shrew, Suncus murinus, was recently identified as a suitable animal model and an alternative to using monkeys (Hu et al., 2003). Studies in the shrew confirmed that a network of branched connections linking multiple organs of the body with the brain, called the vagus nerve, was an essential element for SE-induced emesis, recapitulating earlier observations from monkeys (Sugiyama and Hayama, 1965). Further studies in shrews revealed that the MHC II-independent release of 5-hydroxytryptamine (5-HT/Serotonin) from mast cell granules by SEs was crucial for SE-induced emesis (Ono et al., 2012). Other agonists involved in the emetic response have also been reported (Scheuber et al., 1987; Alber et al., 1989; Jett et al., 1990). In addition to mast cells, the SEs appear to have an affinity for epithelial cells (Hamad et al., 1997; Shupp et al., 2002; Danielsen et al., 2013; Zhao et al., 2016a) and goblet cells (Hirose et al., 2016). Unlike mast cells, SEs use epithelial cells (Danielsen et al., 2013) and mucus-producing goblet cells (Hirose et al., 2016) as gateways in order to traffic across the intestinal epithelia to reach other final targets. Importantly, the movement of enterotoxins through epithelial cells is thought to be a glycolipid-dependent transcytosis process that may be facilitated in the presence of other S. aureus virulence determinants (Edwards et al., 2012). Interestingly, a conserved stretch of 10-amino-acid peptides, located within the longest alpha-helical chain between the A and B domains of the enterotoxins, is an important structural determinant that promotes translocation (Shupp et al., 2002; Figure 2).
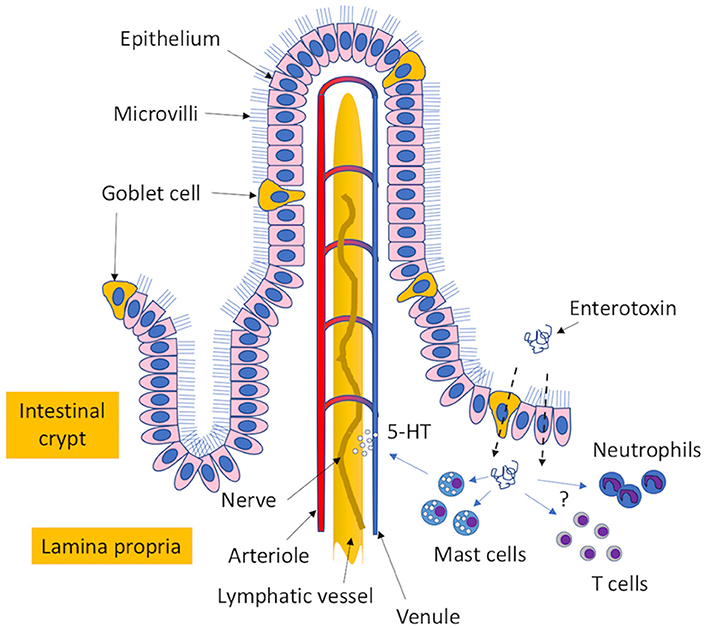
Figure 2. Proposed mechanism of enterotoxin-induced emesis. The enterotoxins transit through mucus-expelling goblet cells and epithelial cells in the intestinal epithelium to reach the lamina propria. Here, the enterotoxins can interact with mast cells to induce the release of 5-hydroxytryptamine (5-HT/serotonin precursor), which interacts with the vagus nerve to cause an emetic response. Additional cellular targets that may have possible roles in the induction of enterotoxigenic disease include different types of T cells and neutrophils.
In contrast to strong induction of emesis, the clinical symptoms of diarrhea are oftentimes less apparent in SFP, which may be in part due to the inability of some SEs, such as SEA and SEC, to cause fluid exudation and dilation of the intestinal segments (Maina et al., 2012). However, the symptoms of diarrhea sometimes observed with SEB intoxication may be due to the inhibition of water and electrolyte reabsorption in the small intestine (Sullivan, 1969; Sheahan et al., 1970). To this date, exactly how the SEs cause diarrhea is still far from understood. For a detailed review on other aspects of SE-induced emesis, see (Hu and Nakane, 2014).
The Superantigenic Activities of the Enterotoxins
The molecular details underlying the superantigenic activity of the SEs have been dissected by numerous X-ray crystallography, structural and mutational analyses. Unlike with conventional antigens, the non-specific activation of T cells by SEs occurs independently of antigen processing and presentation to the T cells by antigen-presenting cells (APCs). Instead, SEs act as a bridge between APCs and T cells. In the majority of cases, SEs first bind to the MHC class II molecules found on APCs and coordinate binding to one or more variable beta (Vβ) chain(s) of T-cell receptors (TCRs) (Kappler et al., 1989; White et al., 1989; Choi et al., 1990; Jarraud et al., 2001). However, these molecular interactions are not exclusive and other receptors have been described to be involved. For instance, the variable alpha (Vα) chain can be targeted by SEH (Saline et al., 2010). Moreover, maximal superantigenic activity of SEB is dependent on additional co-stimulatory receptors, CD28 and B7-2, on T cells and APCs, respectively (Arad et al., 2011; Levy et al., 2016). Interestingly, the same CD28 binding site can be found on other SEs, such as SEA and TSST-1 (Arad et al., 2011). Regardless of the mechanism of cross-linking, characteristic for SE activity is a polyclonal activation of a large pool of CD4+ and CD8+ T cells (~20% of the total T cells) (Marrack et al., 1990; Miethke et al., 1992; Leder et al., 1998) followed by a massive release of an assortment of T helper 1 (Th1) cytokines, such as tumor necrosis factor (TNF) α, interleukin 1 (IL-1), IL-2, and interferon (IFN) γ (Carlsson et al., 1988; Tiedemann and Fraser, 1996), all of which contribute to the SE superantigenic effect (for a detailed reviews, see Krakauer, 2013; Krakauer et al., 2016).
The Enterotoxins Are Immunomodulators of Multiple Immune Cell Types
The superantigenic and enterotoxigenic activities of the SEs are the best studied mechanisms underlying their pathogenicity. However, recent studies show that the SEs possess functions in addition to these conventional activities. For example, both TSST-1 and SElX (Wilson et al., 2011) show similarity to another family of staphylococcal exotoxins, called the staphylococcal superantigen-like (SSL) toxins (reviewed in Fraser and Proft, 2008). Although the SSL toxins lack the ability to induce Vβ-specific T-cell proliferation, they have diverse roles in immune evasion, including the ability to interfere with complement activation and neutrophil function (reviewed in Langley et al., 2010). Recently, it was discovered that SElX has a unique sialic acid-binding motif. This motif allows SElX to interact with adhesion molecules on neutrophils involved in immune recognition and cell activation (Langley et al., 2017; Tuffs et al., 2017). Importantly, the ability of SElX to bind neutrophils, which are considered the first line of defense against S. aureus (Spaan et al., 2013), was crucial for disease progression in a rabbit model of necrotizing pneumonia. Together, these studies describe an unusual member of the SE family that has both superantigenic and SSL functions.
Neutrophils are the latest among a growing list of immune cell types recognized to be targeted directly or indirectly by the SEs. Others include γδ T cells (Maeurer et al., 1995; Morita et al., 2001), invariant natural killer T (iNKT) cells (Rieder et al., 2011; Hayworth et al., 2012), B cells (Stohl et al., 1994), mast cells (Scheuber et al., 1987; Lotfi-Emran et al., 2017), and mucosa-associated invariant T (MAIT) cells (Shaler et al., 2017). Activation of these cell types by SEs can have a considerable impact on the immune system, which may lead to non-conventional overstimulation of the immune system, as exemplified by B cell proliferation and differentiation into plasma cells (Stohl et al., 1994). Additionally, excessive inflammation, as a result of the direct activation of iNKT cells and γδ T cells, can cause the production of SE-associated inflammatory disease in the lungs (Rieder et al., 2011) and systemic infection, as demonstrated in mouse infection models (Szabo et al., 2017).
In contrast to the overstimulation of the immune response by SEs, the activation of MAIT cells appears to have the opposite effect (Shaler et al., 2017). MAIT cells have significant roles in innate host defense against a variety of pathogens (Napier et al., 2015). Notably, the activation of MAIT cells by SEs was shown to be induced in a TCR-independent manner (Shaler et al., 2017). While direct activation of the MAIT cells by SEs could not be excluded, MAIT cell activation was mediated mostly by IL-12 and IL-18 released from the direct activation of conventional T cells by SEs (Shaler et al., 2017). Following a period of hyper-activation, these MAIT cells rapidly undergo exhaustion and are unable to respond further, leaving behind a suppressed and severely crippled arc of innate host defense.
Could Enterotoxicity be Dependent on T-Cell Immunomodulation?
Whether the superantigenic function is needed for the enterotoxigenic activity of the SEs is an interesting question. Shock and fever, hallmarks of superantigen-induced disease, is generally low or absent in patients with SFP (Dinges et al., 2000), arguing against the activation of a systemic immune response. However, it was shown that 5 times more of an SEA protein derivative, which lacked superantigenic but retained emetic activity, was required to induce emesis in a monkey model compared to unaltered SEA (Hoffman et al., 1996). This observation implies that both superantigenic and enterotoxicity activities are likely needed for a maximal emetic response.
Another aspect of immune interaction that may need to be further investigated is the potential role of T cells in SE enterotoxic activities. MAIT cells for example, which have been shown to have a protective role against GI bacterial disease (Powell and Macdonald, 2017; Salerno-Goncalves et al., 2017), represent ~10% of intestinal T cells (Treiner et al., 2003; Dusseaux et al., 2011) and ~50% of T cells in the intestines express γδ TCRs (Carding and Egan, 2002). Furthermore, γδ T cells that are present in the gut mucosa play an important role in mucosal immunity (Agace, 2008). Additionally, given that the SEs are highly potent at very low concentrations, enhanced expression of SEs may not be essential for the advancement of SE-mediated disease. In fact, when regulatory T cells (Tregs) are stimulated with lower concentrations of SEC, an immunosuppressed phenotype can be induced that may directly benefit S. aureus colonization and disease progression (Lee et al., 2017). In the healthy gut, Tregs play a crucial role in the maintenance of intestinal homeostasis by controlling inappropriate immune responses (Luu et al., 2017). Therefore, it is tempting to speculate that the combined targeting of MAIT cells, γδ T cells and Tregs in the gut by SEs may promote the pathogenesis of SFP. Whether MAIT, γδ T cells, and Tregs play any roles in SFP requires much more detailed investigation.
Conclusions
Although the classical enterotoxins have historically been considered the predominant contributors to SFP, a number of molecular studies suggest that many of the newer SEs also have a prominent role. However, in order to better determine which SEs are responsible for SFP, it is best for studies investigating SFP outbreaks to employ methods that can detect all SE genes as well as the physical presence of toxin in suspected contaminated foods. The ability to culture and accurately characterize SFP-causing S. aureus will significantly help understand true incidence and prevalence of SFP. It should also be noted that the inability to detect SEs in contaminated foods does not exclude that they contribute to SFP. Therefore, it is just as vital that we have a deeper understanding of what promotes SE production, especially in food environments. While it is accepted that multiple regulatory networks can have a significant impact on enterotoxin expression, it remains poorly understood how specific enterotoxins, especially the newer enterotoxins, are regulated.
In this review, we also provided an overview of the molecular mechanisms that contribute to SFP. Yet, compared to what we know about staphylococcal superantigen-associated disease, our comprehension of the structural elements and mechanisms by which SEs induce SFP has remained limited, especially considering that SFP is a common disease that continues to affect millions worldwide. A key gap in our knowledge is whether the superantigenicity of the SEs plays a pathogenic role in SFP. There is evidence that suggests that the manifestation of SFP does not solely rely on the enterotoxic function of SEs. Furthermore, we highlighted that different immune and non-immune cell types are susceptible to immunomodulation by the SEs. Any possible interaction between the SEs and these cell types, especially in the gut environment, is worth exploring. Overall, the molecular details involved in SE-mediated enterotoxigenic disease are slowly being uncovered; however, many basic questions remain. Future challenges therefore will consist of deciphering the series of events that lead to disease and whether there are other key cellular players, and identifying an appropriate animal model that is amenable to genetic manipulation.
Author Contributions
EF, GC, and MO: contributed to the drafting of the manuscript and approved the final version.
Funding
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, U.S. National Institutes of Health (grant ZIA AI000904).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abdi, R. D., Gillespie, B. E., Vaughn, J., Merrill, C., Headrick, S. I., Ensermu, D. B., et al. (2018). Antimicrobial resistance of Staphylococcus aureus isolates from dairy cows and genetic diversity of resistant isolates. Foodborne Pathog. Dis doi: 10.1089/fpd.2017.2362. [Epub ahead of print].
Abe, J., Ito, Y., Onimaru, M., Kohsaka, T., and Takeda, T. (2000). Characterization and distribution of a new enterotoxin-related superantigen produced by Staphylococcus aureus. Microbiol. Immunol. 44, 79–88. doi: 10.1111/j.1348-0421.2000.tb01250.x
Adhikari, R. P., Haudenschild, C., Sterba, P. M., Sahandi, S., Enterlein, S., Holtsberg, F. W., et al. (2016). Development of a novel multiplex electrochemiluminescent-based immunoassay for quantification of human serum IgG against 10 Staphylococcus aureus toxins. J. Immunol. Methods 430, 33–42. doi: 10.1016/j.jim.2016.01.013
Agace, W. W. (2008). T-cell recruitment to the intestinal mucosa. Trends Immunol. 29, 514–522. doi: 10.1016/j.it.2008.08.003
Aguilar, J. L., Varshney, A. K., Wang, X., Stanford, L., Scharff, M., and Fries, B. C. (2014). Detection and measurement of staphylococcal enterotoxin-like K (SEl-K) secretion by Staphylococcus aureus clinical isolates. J. Clin. Microbiol. 52, 2536–2543. doi: 10.1128/JCM.00387-14
Alber, G., Scheuber, P. H., Reck, B., Sailer-Kramer, B., Hartmann, A., and Hammer, D. K. (1989). Role of substance P in immediate-type skin reactions induced by staphylococcal enterotoxin B in unsensitized monkeys. J. Allergy Clin. Immunol. 84, 880–885. doi: 10.1016/0091-6749(89)90383-7
Alibayov, B., Zdenkova, K., Sykorova, H., and Demnerova, K. (2014). Molecular analysis of Staphylococcus aureus pathogenicity islands (SaPI) and their superantigens combination of food samples. J. Microbiol. Methods 107, 197–204. doi: 10.1016/j.mimet.2014.10.014
Arad, G., Levy, R., Nasie, I., Hillman, D., Rotfogel, Z., Barash, U., et al. (2011). Binding of superantigen toxins into the CD28 homodimer interface is essential for induction of cytokine genes that mediate lethal shock. PLoS Biol. 9:e1001149. doi: 10.1371/journal.pbio.1001149
Argudín, M. Á., Mendoza, M. C., and Rodicio, M. R. (2010). Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2, 1751–1773. doi: 10.3390/toxins2071751
Armstrong-Esther, C. A. (1976). Carriage patterns of Staphylococcus aureus in a healthy non-hospital population of adults and children. Ann. Hum. Biol. 3, 221–227. doi: 10.1080/03014467600001381
Asao, T., Kumeda, Y., Kawai, T., Shibata, T., Oda, H., Haruki, K., et al. (2003). An extensive outbreak of staphylococcal food poisoning due to low-fat milk in Japan: estimation of enterotoxin A in the incriminated milk and powdered skim milk. Epidemiol. Infect. 130, 33–40. doi: 10.1017/S0950268802007951
Baba, T., Takeuchi, F., Kuroda, M., Yuzawa, H., Aoki, K., Oguchi, A., et al. (2002). Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359, 1819–1827. doi: 10.1016/S0140-6736(02)08713-5
Baroja, M. L., Herfst, C. A., Kasper, K. J., Xu, S. X., Gillett, D. A., Li, J., et al. (2016). The SaeRS two-component system is a direct and dominant transcriptional activator of toxic shock syndrome toxin 1 in Staphylococcus aureus. J. Bacteriol. 198, 2732–2742. doi: 10.1128/JB.00425-16
Bayles, K. W., and Iandolo, J. J. (1989). Genetic and molecular analyses of the gene encoding staphylococcal enterotoxin D. J. Bacteriol. 171, 4799–4806. doi: 10.1128/jb.171.9.4799-4806.1989
Becker, K., Friedrich, A. W., Lubritz, G., Weilert, M., Peters, G., and Von Eiff, C. (2003). Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens. J. Clin. Microbiol. 41, 1434–1439. doi: 10.1128/JCM.41.4.1434-1439.2003
Bennett, S. D., Walsh, K. A., and Gould, L. H. (2013). Foodborne disease outbreaks caused by Bacillus cereus, Clostridium perfringens, and Staphylococcus aureus- United States, 1998-2008. Clin. Infect. Dis. 57, 425–433. doi: 10.1093/cid/cit244
Bergdoll, M. S. (1983). “Enterotoxins,” in Staphylococci and Staphylococcal Infections, eds C. S. F. Easton and C. Adlam (London: Academic Press), 559–598.
Bergdoll, M. S. (1988). Monkey feeding test for staphylococcal enterotoxin. Methods Enzymol. 165, 324–333. doi: 10.1016/S0076-6879(88)65048-8
Bergdoll, M. S., Borja, C. R., and Avena, R. M. (1965). Identification of a new enterotoxin as enterotoxin C. J. Bacteriol. 90, 1481–1485.
Bergdoll, M. S., Borja, C. R., Robbins, R. N., and Weiss, K. F. (1971). Identification of enterotoxin E. Infect. Immun. 4, 593–595.
Bergdoll, M. S., Crass, B. A., Reiser, R. F., Robbins, R. N., and Davis, J. P. (1981). A new staphylococcal enterotoxin, enterotoxin F, associated with toxic-shock-syndrome Staphylococcus aureus isolates. Lancet 1, 1017–1021. doi: 10.1016/S0140-6736(81)92186-3
Bergdoll, M. S., Robbins, R. N., Weiss, K., Borja, C. R., Huang, Y., and Chu, F. S. (1973). The staphylococcal enterotoxins: similarities. Contrib. Microbiol. Immunol. 1, 390–396.
Betley, M. J., Borst, D. W., and Regassa, L. B. (1992). Staphylococcal enterotoxins, toxic shock syndrome toxin and streptococcal pyrogenic exotoxins: a comparative study of their molecular biology. Chem. Immunol. 55, 1–35.
Blaiotta, G., Ercolini, D., Pennacchia, C., Fusco, V., Casaburi, A., Pepe, O., et al. (2004). PCR detection of staphylococcal enterotoxin genes in Staphylococcus spp. strains isolated from meat and dairy products. Evidence for new variants of seg and sei in S. aureus AB-8802. J. Appl. Microbiol. 97, 719–730. doi: 10.1111/j.1365-2672.2004.02349.x
Bohach, G. A., and Schlievert, P. M. (1987). Nucleotide sequence of the staphylococcal enterotoxin C1 gene and relatedness to other pyrogenic toxins. Mol. Gen. Genet. 209, 15–20. doi: 10.1007/BF00329830
Boisset, S., Geissmann, T., Huntzinger, E., Fechter, P., Bendridi, N., Possedko, M., et al. (2007). Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 21, 1353–1366. doi: 10.1101/gad.423507
Borst, D. W., and Betley, M. J. (1994). Promoter analysis of the staphylococcal enterotoxin A gene. J. Biol. Chem. 269, 1883–1888.
Bronner, S., Monteil, H., and Prévost, G. (2004). Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28, 183–200. doi: 10.1016/j.femsre.2003.09.003
Bruno, J. G., and Kiel, J. L. (2002). Use of magnetic beads in selection and detection of biotoxin aptamers by electrochemiluminescence and enzymatic methods. BioTechniques 32, 178–180, 182–183.
Callahan, J. H., Shefcheck, K. J., Williams, T. L., and Musser, S. M. (2006). Detection, confirmation, and quantification of staphylococcal enterotoxin B in food matrixes using liquid chromatography - mass spectrometry. Anal. Chem. 78, 1789–1800. doi: 10.1021/ac051292v
Cameron, D. R., Howden, B. P., and Peleg, A. Y. (2011). The interface between antibiotic resistance and virulence in Staphylococcus aureus and its impact upon clinical outcomes. Clin. Infect. Dis. 53, 576–582. doi: 10.1093/cid/cir473
Cao, R., Zeaki, N., Wallin-Carlquist, N., Skandamis, P. N., Schelin, J., and Rådström, P. (2012). Elevated enterotoxin A expression and formation in Staphylococcus aureus and its association with prophage induction. Appl. Environ. Microbiol. 78, 4942–4948. doi: 10.1128/AEM.00803-12
Carding, S. R., and Egan, P. J. (2002). Gammadelta T cells: functional plasticity and heterogeneity. Nat. Rev. Immunol. 2, 336–345. doi: 10.1038/nri797
Carlsson, R., Fischer, H., and Sjögren, H. O. (1988). Binding of staphylococcal enterotoxin A to accessory cells is a requirement for its ability to activate human T cells. J. Immunol. 140, 2484–2488.
Casman, E. P., Bennett, R. W., Dorsey, A. E., and Issa, J. A. (1967). Identification of a fourth staphylococcal enterotoxin, enterotoxin D. J. Bacteriol. 94, 1875–1882.
Chambers, H. F. (2001). The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 7, 178–182. doi: 10.3201/eid0702.010204
Chao, G., Bao, G., Cao, Y., Yan, W., Wang, Y., Zhang, X., et al. (2015). Prevalence and diversity of enterotoxin genes with genetic background of Staphylococcus aureus isolates from different origins in China. Int. J. Food Microbiol. 211, 142–147. doi: 10.1016/j.ijfoodmicro.2015.07.018
Cheng, J., Wang, Y., Cao, Y., Yan, W., Niu, X., Zhou, L., et al. (2016). The distribution of 18 enterotoxin and enterotoxin-like genes in Staphylococcus aureus strains from different sources in East China. Foodborne Pathog. Dis. 13, 171–176. doi: 10.1089/fpd.2015.1963
Cheung, A. L., and Projan, S. J. (1994). Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 176, 4168–4172. doi: 10.1128/jb.176.13.4168-4172.1994
Cheung, A. L., Bayer, A. S., Zhang, G., Gresham, H., and Xiong, Y. Q. (2004). Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40, 1–9. doi: 10.1016/S0928-8244(03)00309-2
Cheung, A. L., Nishina, K. A., Trotonda, M. P., and Tamber, S. (2008). The SarA protein family of Staphylococcus aureus. Int. J. Biochem. Cell Biol. 40, 355–361. doi: 10.1016/j.biocel.2007.10.032
Chiang, Y. C., Liao, W. W., Fan, C. M., Pai, W. Y., Chiou, C. S., and Tsen, H. Y. (2008). PCR detection of staphylococcal enterotoxins (SEs) N, O, P, Q, R, U, and survey of SE types in Staphylococcus aureus isolates from food-poisoning cases in Taiwan. Int. J. Food Microbiol. 121, 66–73. doi: 10.1016/j.ijfoodmicro.2007.10.005
Cho, H., Jeong, D. W., Liu, Q., Yeo, W. S., Vogl, T., Skaar, E. P., et al. (2015). Calprotectin increases the activity of the SaeRS two component system and murine mortality during Staphylococcus aureus infections. PLoS Pathog. 11:e1005026. doi: 10.1371/journal.ppat.1005026
Choi, Y. W., Herman, A., Digiusto, D., Wade, T., Marrack, P., and Kappler, J. (1990). Residues of the variable region of the T-cell-receptor beta-chain that interact with S. aureus toxin superantigens. Nature 346, 471–473. doi: 10.1038/346471a0
Collery, M. M., Smyth, D. S., Tumilty, J. J., Twohig, J. M., and Smyth, C. J. (2009). Associations between enterotoxin gene cluster types egc1, egc2 and egc3, agr types, enterotoxin and enterotoxin-like gene profiles, and molecular typing characteristics of human nasal carriage and animal isolates of Staphylococcus aureus. J. Med. Microbiol. 58, 13–25. doi: 10.1099/jmm.0.005215-0
Compagnone-Post, P., Malyankar, U., and Khan, S. A. (1991). Role of host factors in the regulation of the enterotoxin B gene. J. Bacteriol. 173, 1827–1830. doi: 10.1128/jb.173.5.1827-1830.1991
Couch, J. L., and Betley, M. J. (1989). Nucleotide sequence of the type C3 staphylococcal enterotoxin gene suggests that intergenic recombination causes antigenic variation. J. Bacteriol. 171, 4507–4510. doi: 10.1128/jb.171.8.4507-4510.1989
Cuny, C., Wieler, L. H., and Witte, W. (2015). Livestock-associated MRSA: the impact on humans. Antibiotics 4, 521–543. doi: 10.3390/antibiotics4040521
Czop, J. K., and Bergdoll, M. S. (1974). Staphylococcal enterotoxin synthesis during the exponential, transitional, and stationary growth phases. Infect. Immun. 9, 229–235.
Dack, G. M. (1937). Staphylococci in relation to food poisoning. Am. J. Public Health Nations. Health. 27, 440–443. doi: 10.2105/AJPH.27.5.440
Danielsen, E. M., Hansen, G. H., and Karlsdóttir, E. (2013). Staphylococcus aureus enterotoxins A- and B: binding to the enterocyte brush border and uptake by perturbation of the apical endocytic membrane traffic. Histochem. Cell Biol. 139, 513–524. doi: 10.1007/s00418-012-1055-8
Deringer, J. R., Ely, R. J., Monday, S. R., Stauffacher, C. V., and Bohach, G. A. (1997). Vbeta-dependent stimulation of bovine and human T cells by host-specific staphylococcal enterotoxins. Infect. Immun. 65, 4048–4054.
Derzelle, S., Dilasser, F., Duquenne, M., and Deperrois, V. (2009). Differential temporal expression of the staphylococcal enterotoxins genes during cell growth. Food Microbiol. 26, 896–904. doi: 10.1016/j.fm.2009.06.007
Dinges, M. M., Orwin, P. M., and Schlievert, P. M. (2000). Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13, 16–34. doi: 10.1128/CMR.13.1.16-34.2000
Do Carmo, L. S., Cummings, C., Linardi, V. R., Dias, R. S., De Souza, J. M., De Sena, M. J., et al. (2004). A case study of a massive staphylococcal food poisoning incident. Foodborne Pathog. Dis. 1, 241–246. doi: 10.1089/fpd.2004.1.241
Doyle, M. E. (2015). Multidrug-resistant pathogens in the food supply. Foodborne Pathog. Dis. 12, 261–279. doi: 10.1089/fpd.2014.1865
Dusseaux, M., Martin, E., Serriari, N., Péguillet, I., Premel, V., Louis, D., et al. (2011). Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood 117, 1250–1259. doi: 10.1182/blood-2010-08-303339
Edwards, L. A., O'neill, C., Furman, M. A., Hicks, S., Torrente, F., Pérez-Machado, M., et al. (2012). Enterotoxin-producing staphylococci cause intestinal inflammation by a combination of direct epithelial cytopathy and superantigen-mediated T-cell activation. Inflamm. Bowel Dis. 18, 624–640. doi: 10.1002/ibd.21852
Edwards, V. M., Deringer, J. R., Callantine, S. D., Deobald, C. F., Berger, P. H., Kapur, V., et al. (1997). Characterization of the canine type C enterotoxin produced by Staphylococcus intermedius pyoderma isolates. Infect. Immun. 65, 2346–2352.
Edwin, C., and Kass, E. H. (1989). Identification of functional antigenic segments of toxic shock syndrome toxin 1 by differential immunoreactivity and by differential mitogenic responses of human peripheral blood mononuclear cells, using active toxin fragments. Infect. Immun. 57, 2230–2236.
Ercoli, L., Gallina, S., Nia, Y., Auvray, F., Primavilla, S., Guidi, F., et al. (2017). Investigation of a staphylococcal food poisoning outbreak from a Chantilly cream dessert, in Umbria (Italy). Foodborne Pathog. Dis. 14, 407–413. doi: 10.1089/fpd.2016.2267
Evenson, M. L., Hinds, M. W., Bernstein, R. S., and Bergdoll, M. S. (1988). Estimation of human dose of staphylococcal enterotoxin A from a large outbreak of staphylococcal food poisoning involving chocolate milk. Int. J. Food Microbiol. 7, 311–316. doi: 10.1016/0168-1605(88)90057-8
Fluit, A. C. (2012). Livestock-associated Staphylococcus aureus. Clin. Microbiol. Infect. 18, 735–744. doi: 10.1111/j.1469-0691.2012.03846.x
Fraser, J. D., and Proft, T. (2008). The bacterial superantigen and superantigen-like proteins. Immunol. Rev. 225, 226–243. doi: 10.1111/j.1600-065X.2008.00681.x
Fulton, F. (1943). Staphylococcal enterotoxin - With special reference to the kitten test. Br. J. Exp. Pathol. 24, 65–72.
Gagnaire, J., Verhoeven, P. O., Grattard, F., Rigaill, J., Lucht, F., Pozzetto, B., et al. (2017). Epidemiology and clinical relevance of Staphylococcus aureus intestinal carriage: a systematic review and meta-analysis. Expert Rev. Anti Infect. Ther. 15, 767–785. doi: 10.1080/14787210.2017.1358611
Gaskill, M. E., and Khan, S. A. (1988). Regulation of the enterotoxin B gene in Staphylococcus aureus. J. Biol. Chem. 263, 6276–6280.
Ge, B., Mukherjee, S., Hsu, C. H., Davis, J. A., Tran, T. T. T., Yang, Q., et al. (2017). MRSA and multidrug-resistant Staphylococcus aureus in U.S. retail meats, 2010-2011. Food Microbiol. 62, 289–297. doi: 10.1016/j.fm.2016.10.029
Geiger, T., Goerke, C., Mainiero, M., Kraus, D., and Wolz, C. (2008). The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J. Bacteriol. 190, 3419–3428. doi: 10.1128/JB.01927-07
Geisinger, E., Adhikari, R. P., Jin, R., Ross, H. F., and Novick, R. P. (2006). Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 61, 1038–1048. doi: 10.1111/j.1365-2958.2006.05292.x
Gkogka, E., Reij, M. W., Havelaar, A. H., Zwietering, M. H., and Gorris, L. G. (2011). Risk-based estimate of effect of foodborne diseases on public health, Greece. Emerg. Infect. Dis. 17, 1581–1590. doi: 10.3201/eid1709.101766
Goerke, C., Pantucek, R., Holtfreter, S., Schulte, B., Zink, M., Grumann, D., et al. (2009). Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J. Bacteriol. 191, 3462–3468. doi: 10.1128/JB.01804-08
Goldman, E. R., Anderson, G. P., Tran, P. T., Mattoussi, H., Charles, P. T., and Mauro, J. M. (2002). Conjugation of luminescent quantum dots with antibodies using an engineered adaptor protein to provide new reagents for fluoroimmunoassays. Anal. Chem. 74, 841–847. doi: 10.1021/ac010662m
Grumann, D., Scharf, S. S., Holtfreter, S., Kohler, C., Steil, L., Engelmann, S., et al. (2008). Immune cell activation by enterotoxin gene cluster (egc)-encoded and non-egc superantigens from Staphylococcus aureus. J. Immunol. 181, 5054–5061. doi: 10.4049/jimmunol.181.7.5054
Guinane, C. M., Ben Zakour, N. L., Tormo-Mas, M. A., Weinert, L. A., Lowder, B. V., Cartwright, R. A., et al. (2010). Evolutionary genomics of Staphylococcus aureus reveals insights into the origin and molecular basis of ruminant host adaptation. Genome Biol. Evol. 2, 454–466. doi: 10.1093/gbe/evq031
Gupta, G., Bhaskar, A. S., Tripathi, B. K., Pandey, P., Boopathi, M., Rao, P. V., et al. (2011). Supersensitive detection of T-2 toxin by the in situ synthesized pi-conjugated molecularly imprinted nanopatterns. An in situ investigation by surface plasmon resonance combined with electrochemistry. Biosens. Bioelectron. 26, 2534–2540. doi: 10.1016/j.bios.2010.10.050
Haag, A. F., and Bagnoli, F. (2016). The role of two-component signal transduction systems in Staphylococcus aureus virulence regulation. Curr Top Microbiol. Immunol. 409, 145–198. doi: 10.1007/82_2015_5019
Hait, J., Tallent, S., Melka, D., Keys, C., and Bennett, R. (2014). Prevalence of enterotoxins and toxin gene profiles of Staphylococcus aureus isolates recovered from a bakery involved in a second staphylococcal food poisoning occurrence. J. Appl. Microbiol. 117, 866–875. doi: 10.1111/jam.12571
Hamad, A. R., Marrack, P., and Kappler, J. W. (1997). Transcytosis of staphylococcal superantigen toxins. J. Exp. Med. 185, 1447–1454. doi: 10.1084/jem.185.8.1447
Harteveld, J. L., Nieuwenhuizen, M. S., and Wils, E. R. (1997). Detection of staphylococcal enterotoxin B employing a piezoelectric crystal immunosensor. Biosens. Bioelectron. 12, 661–667. doi: 10.1016/S0956-5663(97)00024-9
Hayworth, J. L., Mazzuca, D. M., Maleki Vareki, S., Welch, I., Mccormick, J. K., and Haeryfar, S. M. (2012). CD1d-independent activation of mouse and human iNKT cells by bacterial superantigens. Immunol. Cell Biol. 90, 699–709. doi: 10.1038/icb.2011.90
Heinrichs, J. H., Bayer, M. G., and Cheung, A. L. (1996). Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J. Bacteriol. 178, 418–423. doi: 10.1128/jb.178.2.418-423.1996
Hennekinne, J. A., De Buyser, M. L., and Dragacci, S. (2012). Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol. Rev. 36, 815–836. doi: 10.1111/j.1574-6976.2011.00311.x
Hirose, S., Ono, H. K., Omoe, K., Hu, D. L., Asano, K., Yamamoto, Y., et al. (2016). Goblet cells are involved in translocation of staphylococcal enterotoxin A in the intestinal tissue of house musk shrew (Suncus murinus). J. Appl. Microbiol. 120, 781–789. doi: 10.1111/jam.13029
Ho, J., Boost, M., and O'donoghue, M. (2015). Prevalence of enterotoxin genes in Staphylococcus aureus colonising food handlers: does nasal carriage status matter? Eur. J. Clin. Microbiol. Infect. Dis. 34, 2177–2181. doi: 10.1007/s10096-015-2465-z
Hoffman, M., Tremaine, M., Mansfield, J., and Betley, M. (1996). Biochemical and mutational analysis of the histidine residues of staphylococcal enterotoxin A. Infect. Immun. 64, 885–890.
Holtfreter, S., Bauer, K., Thomas, D., Feig, C., Lorenz, V., Roschack, K., et al. (2004). egc-encoded superantigens from Staphylococcus aureus are neutralized by human sera much less efficiently than are classical staphylococcal enterotoxins or toxic shock syndrome toxin. Infect. Immun. 72, 4061–4071. doi: 10.1128/IAI.72.7.4061-4071.2004
Holtfreter, S., Grumann, D., Schmudde, M., Nguyen, H. T., Eichler, P., Strommenger, B., et al. (2007). Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. J. Clin. Microbiol. 45, 2669–2680. doi: 10.1128/JCM.00204-07
Horn, C. C., Kimball, B. A., Wang, H., Kaus, J., Dienel, S., Nagy, A., et al. (2013). Why can't rodents vomit? A comparative behavioral, anatomical, and physiological study. PLoS ONE 8:e60537. doi: 10.1371/journal.pone.0060537
Hovde, C. J., Marr, J. C., Hoffmann, M. L., Hackett, S. P., Chi, Y. I., Crum, K. K., et al. (1994). Investigation of the role of the disulphide bond in the activity and structure of staphylococcal enterotoxin C1. Mol. Microbiol. 13, 897–909. doi: 10.1111/j.1365-2958.1994.tb00481.x
Hu, D. L., and Nakane, A. (2014). Mechanisms of staphylococcal enterotoxin-induced emesis. Eur. J. Pharmacol. 722, 95–107. doi: 10.1016/j.ejphar.2013.08.050
Hu, D. L., Omoe, K., Shimoda, Y., Nakane, A., and Shinagawa, K. (2003). Induction of emetic response to staphylococcal enterotoxins in the house musk shrew (Suncus murinus). Infect. Immun. 71, 567–570. doi: 10.1128/IAI.71.1.567-570.2003
Hu, D. L., Omoe, K., Shimura, H., Ono, K., Sugii, S., and Shinagawa, K. (1999). Emesis in the shrew mouse (Suncus murinus) induced by peroral and intraperitoneal administration of staphylococcal enterotoxin A. J. Food Prot. 62, 1350–1353. doi: 10.4315/0362-028X-62.11.1350
Hu, D. L., Ono, H. K., Isayama, S., Okada, R., Okamura, M., Lei, L. C., et al. (2017). Biological characteristics of staphylococcal enterotoxin Q and its potential risk for food poisoning. J. Appl. Microbiol. 122, 1672–1679. doi: 10.1111/jam.13462
Huijsdens, X. W., Van Dijke, B. J., Spalburg, E., Van Santen-Verheuvel, M. G., Heck, M. E., Pluister, G. N., et al. (2006). Community-acquired MRSA and pig-farming. Ann. Clin. Microbiol. Antimicrob. 5:26. doi: 10.1186/1476-0711-5-26
Humber, J. Y., Denny, C. B., and Bohrer, C. W. (1975). Influence of pH on the heat inactivation of staphylococcal enterotoxin A as determined by monkey feeding and serological assay. Appl. Microbiol. 30, 755–758.
Hussain, M. A., and Dawson, C. O. (2013). Economic impact of food safety outbreaks on food businesses. Foods 2, 585–589. doi: 10.3390/foods2040585
Huvenne, W., Hellings, P. W., and Bachert, C. (2013). Role of staphylococcal superantigens in airway disease. Int. Arch. Allergy Immunol. 161, 304–314. doi: 10.1159/000350329
Igarashi, H. (1972). Staphylococcal enterotoxin D. Immunological identification with purified toxin. Jpn. J. Microbiol. 16, 483–491. doi: 10.1111/j.1348-0421.1972.tb00688.x
Ikeda, T., Tamate, N., Yamaguchi, K., and Makino, S. (2005). Mass outbreak of food poisoning disease caused by small amounts of staphylococcal enterotoxins A and H. Appl. Environ. Microbiol. 71, 2793–2795. doi: 10.1128/AEM.71.5.2793-2795.2005
Jarraud, S., Peyrat, M. A., Lim, A., Tristan, A., Bes, M., Mougel, C., et al. (2001). egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. 166, 669–677. doi: 10.4049/jimmunol.166.1.669
Jett, M., Brinkley, W., Neill, R., Gemski, P., and Hunt, R. (1990). Staphylococcus aureus enterotoxin B challenge of monkeys: correlation of plasma levels of arachidonic acid cascade products with occurrence of illness. Infect. Immun. 58, 3494–3499.
Ji, G., Beavis, R. C., and Novick, R. P. (1995). Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. U.S.A. 92, 12055–12059. doi: 10.1073/pnas.92.26.12055
Ji, G., Beavis, R., and Novick, R. P. (1997). Bacterial interference caused by autoinducing peptide variants. Science 276, 2027–2030. doi: 10.1126/science.276.5321.2027
Johler, S., Giannini, P., Jermini, M., Hummerjohann, J., Baumgartner, A., and Stephan, R. (2015). Further evidence for staphylococcal food poisoning outbreaks caused by egc-encoded enterotoxins. Toxins 7, 997–1004. doi: 10.3390/toxins7030997
Johler, S., Sihto, H. M., Macori, G., and Stephan, R. (2016). Sequence variability in staphylococcal enterotoxin genes seb, sec, and sed. Toxins. 8:169. doi: 10.3390/toxins8060169
Jørgensen, H. J., Mathisen, T., Løvseth, A., Omoe, K., Qvale, K. S., and Loncarevic, S. (2005). An outbreak of staphylococcal food poisoning caused by enterotoxin H in mashed potato made with raw milk. FEMS Microbiol. Lett. 252, 267–272. doi: 10.1016/j.femsle.2005.09.005
Kappler, J., Kotzin, B., Herron, L., Gelfand, E. W., Bigler, R. D., Boylston, A., et al. (1989). V beta-specific stimulation of human T cells by staphylococcal toxins. Science 244, 811–813. doi: 10.1126/science.2524876
Kavanaugh, J. S., Thoendel, M., and Horswill, A. R. (2007). A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol. Microbiol. 65, 780–798. doi: 10.1111/j.1365-2958.2007.05830.x
Kérouanton, A., Hennekinne, J. A., Letertre, C., Petit, L., Chesneau, O., Brisabois, A., et al. (2007). Characterization of Staphylococcus aureus strains associated with food poisoning outbreaks in France. Int. J. Food Microbiol. 115, 369–375. doi: 10.1016/j.ijfoodmicro.2006.10.050
Kientz, C. E., Hulst, A. G., and Wils, E. R. (1997). Determination of staphylococcal enterotoxin B by on-line (micro) liquid chromatography-electrospray mass spectrometry. J. Chromatogr. A. 757, 51–64. doi: 10.1016/S0021-9673(96)00661-9
Kijek, T. M., Rossi, C. A., Moss, D., Parker, R. W., and Henchal, E. A. (2000). Rapid and sensitive immunomagnetic-electrochemiluminescent detection of staphyloccocal enterotoxin B. J. Immunol. Methods. 236, 9–17. doi: 10.1016/S0022-1759(99)00234-3
Kinkel, T. L., Roux, C. M., Dunman, P. M., and Fang, F. C. (2013). The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. mBio. 4:e00696-13. doi: 10.1128/mBio.00696-13
Kirk, M., Ford, L., Glass, K., and Hall, G. (2014). Foodborne illness, Australia, circa 2000 and circa 2010. Emerging Infect. Dis. 20, 1857–1864. doi: 10.3201/eid2011.131315
Kocandrle, V., Houttuin, E., and Prohaska, J. V. (1966). Acute hemodynamic and gastrointestinal changes produced by staphylococcal exotoxin and enterotoxin in dogs. J. Surg. Res. 6, 50–57. doi: 10.1016/S0022-4804(66)80070-7
Koenig, R. L., Ray, J. L., Maleki, S. J., Smeltzer, M. S., and Hurlburt, B. K. (2004). Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J. Bacteriol. 186, 7549–7555. doi: 10.1128/JB.186.22.7549-7555.2004
Kohler, P. L., Greenwood, S. D., Nookala, S., Kotb, M., Kranz, D. M., and Schlievert, P. M. (2012). Staphylococcus aureus isolates encode variant staphylococcal enterotoxin B proteins that are diverse in superantigenicity and lethality. PLoS ONE 7:e41157. doi: 10.1371/journal.pone.0041157
Kozono, H., Parker, D., White, J., Marrack, P., and Kappler, J. (1995). Multiple binding sites for bacterial superantigens on soluble class II MHC molecules. Immunity 3, 187–196. doi: 10.1016/1074-7613(95)90088-8
Krakauer, T. (2013). Update on staphylococcal superantigen-induced signaling pathways and therapeutic interventions. Toxins 5, 1629–1654. doi: 10.3390/toxins5091629
Krakauer, T., Pradhan, K., and Stiles, B. G. (2016). Staphylococcal superantigens spark host-mediated danger signals. Front. Immunol. 7:23. doi: 10.3389/fimmu.2016.00023
Kullik, I., and Giachino, P. (1997). The alternative sigma factor sigmaB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch. Microbiol. 167, 151–159. doi: 10.1007/s002030050428
Kullik, I., Giachino, P., and Fuchs, T. (1998). Deletion of the alternative sigma factor sigmaB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180, 4814–4820.
Kuroda, H., Kuroda, M., Cui, L., and Hiramatsu, K. (2007). Subinhibitory concentrations of beta-lactam induce haemolytic activity in Staphylococcus aureus through the SaeRS two-component system. FEMS Microbiol. Lett. 268, 98–105. doi: 10.1111/j.1574-6968.2006.00568.x
Kusch, K., Hanke, K., Holtfreter, S., Schmudde, M., Kohler, C., Erck, C., et al. (2011). The influence of SaeRS and sigma(B) on the expression of superantigens in different Staphylococcus aureus isolates. Int. J. Med. Microbiol. 301, 488–499. doi: 10.1016/j.ijmm.2011.01.003
Langley, R. J., Ting, Y. T., Clow, F., Young, P. G., Radcliff, F. J., Choi, J. M., et al. (2017). Staphylococcal enterotoxin-like X (SElX) is a unique superantigen with functional features of two major families of staphylococcal virulence factors. PLoS Pathog. 13:e1006549. doi: 10.1371/journal.ppat.1006549
Langley, R., Patel, D., Jackson, N., Clow, F., and Fraser, J. D. (2010). Staphylococcal superantigen super-domains in immune evasion. Crit. Rev. Immunol. 30, 149–165. doi: 10.1615/CritRevImmunol.v30.i2.40
Lappin, E., and Ferguson, A. J. (2009). Gram-positive toxic shock syndromes. Lancet Infect. Dis. 9, 281–290. doi: 10.1016/S1473-3099(09)70066-0
Larkin, E. A., Carman, R. J., Krakauer, T., and Stiles, B. G. (2009). Staphylococcus aureus: the toxic presence of a pathogen extraordinaire. Curr. Med. Chem. 16, 4003–4019. doi: 10.2174/092986709789352321
Lauderdale, K. J., Boles, B. R., Cheung, A. L., and Horswill, A. R. (2009). Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 77, 1623–1635. doi: 10.1128/IAI.01036-08
Le Loir, Y., Baron, F., and Gautier, M. (2003). Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2, 63–76.
Leder, L., Llera, A., Lavoie, P. M., Lebedeva, M. I., Li, H., Sékaly, R. P., et al. (1998). A mutational analysis of the binding of staphylococcal enterotoxins B and C3 to the T cell receptor beta chain and major histocompatibility complex class II. J. Exp. Med. 187, 823–833. doi: 10.1084/jem.187.6.823
Lee, J., Park, N., Park, J. Y., Kaplan, B. L. F., Pruett, S. B., Park, J. W., et al. (2017). Induction of immunosuppressive CD8(+)CD25(+)FOXP3(+) regulatory T cells by suboptimal stimulation with staphylococcal enterotoxin C1. J. Immunol. 200, 669–680. doi: 10.4049/jimmunol.1602109
Letertre, C., Perelle, S., Dilasser, F., and Fach, P. (2003a). Detection and genotyping by real-time PCR of the staphylococcal enterotoxin genes sea to sej. Mol. Cell. Probes. 17, 139–147. doi: 10.1016/S0890-8508(03)00045-8
Letertre, C., Perelle, S., Dilasser, F., and Fach, P. (2003b). Identification of a new putative enterotoxin SEU encoded by the egc cluster of Staphylococcus aureus. J. Appl. Microbiol. 95, 38–43. doi: 10.1046/j.1365-2672.2003.01957.x
Levy, R., Rotfogel, Z., Hillman, D., Popugailo, A., Arad, G., Supper, E., et al. (2016). Superantigens hyperinduce inflammatory cytokines by enhancing the B7-2/CD28 costimulatory receptor interaction. Proc. Natl. Acad. Sci. U.S.A. 113, E6437–E6446. doi: 10.1073/pnas.1603321113
Lewis, H. C., Mølbak, K., Reese, C., Aarestrup, F. M., Selchau, M., Sørum, M., et al. (2008). Pigs as source of methicillin-resistant Staphylococcus aureus CC398 infections in humans, Denmark. Emerg. Infect. Dis. 14, 1383–1389. doi: 10.3201/eid1409.071576
Li, D., and Cheung, A. (2008). Repression of hla by rot is dependent on sae in Staphylococcus aureus. Infect. Immun. 76, 1068–1075. doi: 10.1128/IAI.01069-07
Li, J., Yang, J., Lu, Y. W., Wu, S., Wang, M. R., and Zhu, J. M. (2015). Possible role of staphylococcal enterotoxin B in the pathogenesis of autoimmune diseases. Viral Immunol. 28, 354–359. doi: 10.1089/vim.2015.0017
Liang, M., Zhang, T., Liu, X., Fan, Y., Xia, S., Xiang, Y., et al. (2015). Development of an indirect competitive enzyme-linked immunosorbent assay based on the multiepitope peptide for the synchronous detection of staphylococcal enterotoxin A and G proteins in milk. J. Food Prot. 78, 362–369. doi: 10.4315/0362-028X.JFP-14-323
Lina, G., Bohach, G. A., Nair, S. P., Hiramatsu, K., Jouvin-Marche, E., Mariuzza, R., et al. (2004). Standard nomenclature for the superantigens expressed by Staphylococcus. J. Infect. Dis. 189, 2334–2336. doi: 10.1086/420852
Lina, G., Jarraud, S., Ji, G., Greenland, T., Pedraza, A., Etienne, J., et al. (1998). Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol. Microbiol. 28, 655–662. doi: 10.1046/j.1365-2958.1998.00830.x
Lis, E., Korzekwa, K., Bystron, J., Zarczynska, A., Dabrowska, A., Molenda, J., et al. (2009). Enterotoxin gene content in Staphylococcus aureus from the human intestinal tract. FEMS Microbiol. Lett. 296, 72–77. doi: 10.1111/j.1574-6968.2009.01622.x
Lis, E., Podkowik, M., Bystron, J., Stefaniak, T., and Bania, J. (2012). Temporal expression of staphylococcal enterotoxin H in comparison with accessory gene regulator-dependent and -independent enterotoxins. J. Food Prot. 75, 238–244. doi: 10.4315/0362-028X.JFP-11-336
Lotfi-Emran, S., Ward, B. R., Le, Q. T., Pozez, A. L., Manjili, M. H., Woodfolk, J. A., et al. (2017). Human mast cells present antigen to autologous CD4(+) T cells. J. Allergy Clin. Immunol. 141, 311.e10–321.e10. doi: 10.1016/j.jaci.2017.02.048
Lowder, B. V., Guinane, C. M., Ben Zakour, N. L., Weinert, L. A., Conway-Morris, A., Cartwright, R. A., et al. (2009). Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 106, 19545–19550. doi: 10.1073/pnas.0909285106
Lowy, F. D. (2003). Antimicrobial resistance: the example of Staphylococcus aureus. J. Clin. Invest. 111, 1265–1273. doi: 10.1172/JCI18535
Luo, L. R., Zhang, Z. J., Chen, L. J., and Ma, L. F. (2006). Chemiluminescent imaging detection of staphylococcal enterotoxin C-1 in milk and water samples. Food Chem. 97, 355–360. doi: 10.1016/j.foodchem.2005.05.008
Luu, M., Steinhoff, U., and Visekruna, A. (2017). Functional heterogeneity of gut-resident regulatory T cells. Clin. Transl. Immunol. 6:e156. doi: 10.1038/cti.2017.39
Lv, G., Xu, B., Wei, P., Song, J., Zhang, H., Zhao, C., et al. (2014). Molecular characterization of foodborne-associated Staphylococcus aureus strains isolated in Shijiazhuang, China, from 2010 to 2012. Diagn. Microbiol. Infect. Dis. 78, 462–468. doi: 10.1016/j.diagmicrobio.2013.12.006
Madsen, J. M. (2001). Toxins as weapons of mass destruction. A comparison and contrast with biological-warfare and chemical-warfare agents. Clin. Lab. Med. 21, 593–605.
Maeurer, M., Zitvogel, L., Elder, E., Storkus, W. J., and Lotze, M. T. (1995). Human intestinal V delta 1+ T cells obtained from patients with colon cancer respond exclusively to SEB but not to SEA. Nat. Immun. 14, 188–197.
Maina, E. K., Hu, D. L., Tsuji, T., Omoe, K., and Nakane, A. (2012). Staphylococcal enterotoxin A has potent superantigenic and emetic activities but not diarrheagenic activity. Int. J. Med. Microbiol. 302, 88–95. doi: 10.1016/j.ijmm.2012.01.003
Malachowa, N., and DeLeo, F. R. (2010). Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 67, 3057–3071. doi: 10.1007/s00018-010-0389-4
Mangen, M. J., Bouwknegt, M., Friesema, I. H., Haagsma, J. A., Kortbeek, L. M., Tariq, L., et al. (2015). Cost-of-illness and disease burden of food-related pathogens in the Netherlands, 2011. Int. J. Food Microbiol. 196, 84–93. doi: 10.1016/j.ijfoodmicro.2014.11.022
Marr, J. C., Lyon, J. D., Roberson, J. R., Lupher, M., Davis, W. C., and Bohach, G. A. (1993). Characterization of novel type C staphylococcal enterotoxins: biological and evolutionary implications. Infect. Immun. 61, 4254–4262.
Marrack, P., and Kappler, J. (1990). The staphylococcal enterotoxins and their relatives. Science 248, 705–711. doi: 10.1126/science.2185544
Marrack, P., Blackman, M., Kushnir, E., and Kappler, J. (1990). The toxicity of staphylococcal enterotoxin B in mice is mediated by T cells. J. Exp. Med. 171, 455–464. doi: 10.1084/jem.171.2.455
Mashruwala, A. A., and Boyd, J. M. (2017). The Staphylococcus aureus SrrAB regulatory system modulates hydrogen peroxide resistance factors, which imparts protection to aconitase during aerobic growth. PLoS ONE 12:e0170283. doi: 10.1371/journal.pone.0170283
Matsui, S., Terabe, M., Mabuchi, A., Takahashi, M., Saizawa, M., Tanaka, S., et al. (1997). A unique response to staphylococcal enterotoxin B by intrahepatic lymphocytes and its relevance to the induction of tolerance in the liver. Scand. J. Immunol. 46, 230–234. doi: 10.1046/j.1365-3083.1997.d01-118.x
Mayville, P., Ji, G., Beavis, R., Yang, H., Goger, M., Novick, R. P., et al. (1999). Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. U.S.A. 96, 1218–1223. doi: 10.1073/pnas.96.4.1218
McLauchlin, J., Narayanan, G. L., Mithani, V., and O'neill, G. (2000). The detection of enterotoxins and toxic shock syndrome toxin genes in Staphylococcus aureus by polymerase chain reaction. J. Food Prot. 63, 479–488. doi: 10.4315/0362-028X-63.4.479
Miethke, T., Wahl, C., Heeg, K., Echtenacher, B., Krammer, P. H., and Wagner, H. (1992). T cell-mediated lethal shock triggered in mice by the superantigen staphylococcal enterotoxin B: critical role of tumor necrosis factor. J. Exp. Med. 175, 91–98. doi: 10.1084/jem.175.1.91
Monday, S. R., and Bohach, G. A. (2001). Genes encoding staphylococcal enterotoxins G and I are linked and separated by DNA related to other staphylococcal enterotoxins. J. Nat. Toxins. 10, 1–8.
Morfeldt, E., Taylor, D., Von Gabain, A., and Arvidson, S. (1995). Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 14, 4569–4577.
Morita, C. T., Li, H., Lamphear, J. G., Rich, R. R., Fraser, J. D., Mariuzza, R. A., et al. (2001). Superantigen recognition by gammadelta T cells: SEA recognition site for human Vgamma2 T cell receptors. Immunity 14, 331–344. doi: 10.1016/S1074-7613(01)00113-3
Munson, S. H., Tremaine, M. T., Betley, M. J., and Welch, R. A. (1998). Identification and characterization of staphylococcal enterotoxin types G and I from Staphylococcus aureus. Infect. Immun. 66, 3337–3348.
Murray, R. J. (2005). Recognition and management of Staphylococcus aureus toxin-mediated disease. Intern. Med. J. 35(Suppl. 2), S106–S119. doi: 10.1111/j.1444-0903.2005.00984.x
Nagaraj, S., Ramlal, S., Kingston, J., and Batra, H. V. (2016). Development of IgY based sandwich ELISA for the detection of staphylococcal enterotoxin G (SEG), an egc toxin. Int. J. Food Microbiol. 237, 136–141. doi: 10.1016/j.ijfoodmicro.2016.08.009
Napier, R. J., Adams, E. J., Gold, M. C., and Lewinsohn, D. M. (2015). The role of mucosal associated invariant T cells in antimicrobial immunity. Front. Immunol. 6:344. doi: 10.3389/fimmu.2015.00344
Nashev, D., Toshkova, K., Bizeva, L., Akineden, O., Lämmler, C., and Zschöck, M. (2007). Distribution of enterotoxin genes among carriage- and infection-associated isolates of Staphylococcus aureus. Lett. Appl. Microbiol. 45, 681–685. doi: 10.1111/j.1472-765X.2007.02254.x
Nedelkov, D., Rasooly, A., and Nelson, R. W. (2000). Multitoxin biosensor-mass spectrometry analysis: a new approach for rapid, real-time, sensitive analysis of staphylococcal toxins in food. Int. J. Food Microbiol. 60, 1–13. doi: 10.1016/S0168-1605(00)00328-7
Neill, R. J., Fanning, G. R., Delahoz, F., Wolff, R., and Gemski, P. (1990). Oligonucleotide probes for detection and differentiation of Staphylococcus aureus strains containing genes for enterotoxins A, B, and C and toxic shock syndrome toxin 1. J. Clin. Microbiol. 28, 1514–1518.
Nemati, M., Hermans, K., Lipinska, U., Denis, O., Deplano, A., Struelens, M., et al. (2008). Antimicrobial resistance of old and recent Staphylococcus aureus isolates from poultry: first detection of livestock-associated methicillin-resistant strain ST398. Antimicrob. Agents Chemother. 52, 3817–3819. doi: 10.1128/AAC.00613-08
Nkouawa, A., Sako, Y., Nakao, M., Nakaya, K., and Ito, A. (2009). Loop-mediated isothermal amplification method for differentiation and rapid detection of Taenia species. J. Clin. Microbiol. 47, 168–174. doi: 10.1128/JCM.01573-08
Novick, R. P., and Jiang, D. (2003). The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 149, 2709–2717. doi: 10.1099/mic.0.26575-0
Novick, R. P., Projan, S. J., Kornblum, J., Ross, H. F., Ji, G., Kreiswirth, B., et al. (1995). The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248, 446–458. doi: 10.1007/BF02191645
Nowrouzian, F. L., Dauwalder, O., Meugnier, H., Bes, M., Etienne, J., Vandenesch, F., et al. (2011). Adhesin and superantigen genes and the capacity of Staphylococcus aureus to colonize the infantile gut. J. Infect. Dis. 204, 714–721. doi: 10.1093/infdis/jir388
Nowrouzian, F. L., Lina, G., Hodille, E., Lindberg, E., Hesselmar, B., Saalman, R., et al. (2017). Superantigens and adhesins of infant gut commensal Staphylococcus aureus strains and association with subsequent development of atopic eczema. Br. J. Dermatol. 176, 439–445. doi: 10.1111/bjd.15138
Omoe, K., Hu, D. L., Ono, H. K., Shimizu, S., Takahashi-Omoe, H., Nakane, A., et al. (2013). Emetic potentials of newly identified staphylococcal enterotoxin-like toxins. Infect. Immun. 81, 3627–3631. doi: 10.1128/IAI.00550-13
Omoe, K., Imanishi, K., Hu, D. L., Kato, H., Fugane, Y., Abe, Y., et al. (2005). Characterization of novel staphylococcal enterotoxin-like toxin type P. Infect Immun. 73, 5540–5546. doi: 10.1128/IAI.73.9.5540-5546.2005
Ono, H. K., Hirose, S., Naito, I., Sato'o, Y., Asano, K., Hu, D. L., et al. (2017). The emetic activity of staphylococcal enterotoxins, SEK, SEL, SEM, SEN and SEO in a small emetic animal model, the house musk shrew. Microbiol. Immunol. 61, 12–16. doi: 10.1111/1348-0421.12460
Ono, H. K., Nishizawa, M., Yamamoto, Y., Hu, D. L., Nakane, A., Shinagawa, K., et al. (2012). Submucosal mast cells in the gastrointestinal tract are a target of staphylococcal enterotoxin type A. FEMS Immunol. Med. Microbiol. 64, 392–402. doi: 10.1111/j.1574-695X.2011.00924.x
Ono, H. K., Omoe, K., Imanishi, K., Iwakabe, Y., Hu, D. L., Kato, H., et al. (2008). Identification and characterization of two novel staphylococcal enterotoxins, types S and T. Infect. Immun. 76, 4999–5005. doi: 10.1128/IAI.00045-08
Ono, H. K., Sato'o, Y., Narita, K., Naito, I., Hirose, S., Hisatsune, J., et al. (2015). Identification and characterization of a novel staphylococcal emetic toxin. Appl. Environ. Microbiol. 81, 7034–7040. doi: 10.1128/AEM.01873-15
Orwin, P. M., Leung, D. Y., Donahue, H. L., Novick, R. P., and Schlievert, P. M. (2001). Biochemical and biological properties of staphylococcal enterotoxin K. Infect. Immun. 69, 360–366. doi: 10.1128/IAI.69.1.360-366.2001
Orwin, P. M., Leung, D. Y., Tripp, T. J., Bohach, G. A., Earhart, C. A., Ohlendorf, D. H., et al. (2002). Characterization of a novel staphylococcal enterotoxin-like superantigen, a member of the group V subfamily of pyrogenic toxins. Biochemistry 41, 14033–14040. doi: 10.1021/bi025977q
Ostyn, A., De Buyser, M. L., Guillier, F., Groult, J., Felix, B., Salah, S., et al. (2010). First evidence of a food poisoning outbreak due to staphylococcal enterotoxin type E, France, 2009. Euro Surveill. 15:19528
Otero, A., García, M. L., García, M. C., Moreno, B., and Bergdoll, M. S. (1990). Production of staphylococcal enterotoxins C1 and C2 and thermonuclease throughout the growth cycle. Appl. Environ. Microbiol. 56, 555–559.
Otto, M. (2010). Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu. Rev. Microbiol. 64, 143–162. doi: 10.1146/annurev.micro.112408.134309
Otto, M. (2012). MRSA virulence and spread. Cell Microbiol. 14, 1513–1521. doi: 10.1111/j.1462-5822.2012.01832.x
Pané-Farré, J., Jonas, B., Förstner, K., Engelmann, S., and Hecker, M. (2006). The sigmaB regulon in Staphylococcus aureus and its regulation. Int. J. Med. Microbiol. 296, 237–258. doi: 10.1016/j.ijmm.2005.11.011
Park, M. S., Kim, Y. S., Lee, S. H., Kim, S. H., Park, K. H., and Bahk, G. J. (2015). Estimating the burden of foodborne disease, South Korea, 2008-2012. Foodborne Pathog. Dis. 12, 207–213. doi: 10.1089/fpd.2014.1858
Pastacaldi, C., Lewis, P., and Howarth, P. (2011). Staphylococci and staphylococcal superantigens in asthma and rhinitis: a systematic review and meta-analysis. Allergy 66, 549–555. doi: 10.1111/j.1398-9995.2010.02502.x
Pekdemir, M. E., Ertürkan, D., Külah, H., Boyaci, I. H., Ozgen, C., and Tamer, U. (2012). Ultrasensitive and selective homogeneous sandwich immunoassay detection by Surface Enhanced Raman Scattering (SERS). Analyst 137, 4834–4840. doi: 10.1039/c2an35471c
Pinchuk, I. V., Beswick, E. J., and Reyes, V. E. (2010). Staphylococcal enterotoxins. Toxins 2, 2177–2197. doi: 10.3390/toxins2082177
Powell, N., and Macdonald, T. T. (2017). Recent advances in gut immunology. Parasite Immunol. 39:e12430. doi: 10.1111/pim.12430
Pragman, A. A., Ji, Y., and Schlievert, P. M. (2007). Repression of Staphylococcus aureus SrrAB using inducible antisense srrA alters growth and virulence factor transcript levels. Biochemistry 46, 314–321. doi: 10.1021/bi0603266
Pragman, A. A., Yarwood, J. M., Tripp, T. J., and Schlievert, P. M. (2004). Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 186, 2430–2438. doi: 10.1128/JB.186.8.2430-2438.2004
Price, L. B., Stegger, M., Hasman, H., Aziz, M., Larsen, J., Andersen, P. S., et al. (2012). Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio. 3:e00520-12. doi: 10.1128/mBio.00305-11
Principato, M., and Qian, B. F. (2014). Staphylococcal enterotoxins in the etiopathogenesis of mucosal autoimmunity within the gastrointestinal tract. Toxins 6, 1471–1489. doi: 10.3390/toxins6051471
Queck, S. Y., Jameson-Lee, M., Villaruz, A. E., Bach, T. H., Khan, B. A., Sturdevant, D. E., et al. (2008). RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell. 32, 150–158. doi: 10.1016/j.molcel.2008.08.005
Rasooly, L., and Rasooly, A. (1999). Real time biosensor analysis of staphylococcal enterotoxin A in food. Int. J. Food Microbiol. 49, 119–127. doi: 10.1016/S0168-1605(99)00053-7
Read, R. B. Jr., Bradshaw, J., Pritchard, W. L., and Black, L. A. (1965). Assay of staphylococcal enterotoxin from cheese. J. Dairy Sci. 48, 420–424. doi: 10.3168/jds.S0022-0302(65)88246-7
Regassa, L. B., and Betley, M. J. (1993). High sodium chloride concentrations inhibit staphylococcal enterotoxin C gene (sec) expression at the level of sec mRNA. Infect. Immun. 61, 1581–1585.
Regassa, L. B., Couch, J. L., and Betley, M. J. (1991). Steady-state staphylococcal enterotoxin type C mRNA is affected by a product of the accessory gene regulator (agr) and by glucose. Infect. Immun. 59, 955–962.
Regenthal, P., Hansen, J. S., Andre, I., and Lindkvist-Petersson, K. (2017). Thermal stability and structural changes in bacterial toxins responsible for food poisoning. PLoS ONE 12:e0172445. doi: 10.1371/journal.pone.0172445
Reiser, R. F., Robbins, R. N., Khoe, G. P., and Bergdoll, M. S. (1983). Purification and some physicochemical properties of toxic-shock toxin. Biochemistry 22, 3907–3912. doi: 10.1021/bi00285a028
Reiser, R. F., Robbins, R. N., Noleto, A. L., Khoe, G. P., and Bergdoll, M. S. (1984). Identification, purification, and some physicochemical properties of staphylococcal enterotoxin C3. Infect. Immun. 45, 625–630.
Ridley, M. (1959). Perineal carriage of Staph. aureus. Br. Med. J. 1, 270–273. doi: 10.1136/bmj.1.5117.270
Rieder, S. A., Nagarkatti, P., and Nagarkatti, M. (2011). CD1d-independent activation of invariant natural killer T cells by staphylococcal enterotoxin B through major histocompatibility complex class II/T cell receptor interaction results in acute lung injury. Infect. Immun. 79, 3141–3148. doi: 10.1128/IAI.00177-11
Rogasch, K., Rühmling, V., Pané-Farré, J., Höper, D., Weinberg, C., Fuchs, S., et al. (2006). Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J. Bacteriol. 188, 7742–7758. doi: 10.1128/JB.00555-06
Sahibzada, S., Abraham, S., Coombs, G. W., Pang, S., Hernández-Jover, M., Jordan, D., et al. (2017). Transmission of highly virulent community-associated MRSA ST93 and livestock-associated MRSA ST398 between humans and pigs in Australia. Sci. Rep. 7:5273. doi: 10.1038/s41598-017-04789-0
Saïd-Salim, B., Dunman, P. M., Mcaleese, F. M., Macapagal, D., Murphy, E., Mcnamara, P. J., et al. (2003). Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185, 610–619. doi: 10.1128/JB.185.2.610-619.2003
Sakai, F., Ihara, H., Aoyama, K., Igarashi, H., Yanahira, S., Ohkubo, T., et al. (2008). Characteristics of enterotoxin H-producing Staphylococcus aureus isolated from clinical cases and properties of the enterotoxin productivity. J. Food Prot. 71, 1855–1860. doi: 10.4315/0362-028X-71.9.1855
Salerno-Goncalves, R., Luo, D., Fresnay, S., Magder, L., Darton, T. C., Jones, C., et al. (2017). Challenge of humans with wild-type Salmonella enterica serovar Typhi elicits changes in the activation and homing characteristics of Mucosal-Associated Invariant T Cells. Front. Immunol. 8:398. doi: 10.3389/fimmu.2017.00398
Saline, M., Rodstrom, K. E., Fischer, G., Orekhov, V. Y., Karlsson, B. G., and Lindkvist-Petersson, K. (2010). The structure of superantigen complexed with TCR and MHC reveals novel insights into superantigenic T cell activation. Nat. Commun. 1:119. doi: 10.1038/ncomms1117
Salomon, L. L., and Tew, R. W. (1968). Assay of staphylococcal enterotoxin B by latex agglutination. Proc. Soc. Exp. Biol. Med. 129, 539–542. doi: 10.3181/00379727-129-33364
Sato'o, Y., Hisatsune, J., Nagasako, Y., Ono, H. K., Omoe, K., and Sugai, M. (2015). Positive regulation of staphylococcal enterotoxin H by Rot (Repressor of Toxin) protein and its importance in clonal complex 81 subtype 1 lineage-related food poisoning. Appl. Environ. Microbiol. 81, 7782–7790. doi: 10.1128/AEM.01936-15
Sato'o, Y., Omoe, K., Naito, I., Ono, H. K., Nakane, A., Sugai, M., et al. (2014). Molecular epidemiology and identification of a Staphylococcus aureus clone causing food poisoning outbreaks in Japan. J. Clin. Microbiol. 52, 2637–2640. doi: 10.1128/JCM.00661-14
Saunders, G. C., and Bartlett, M. L. (1977). Double-antibody solid-phase enzyme immunoassay for the detection of staphylococcal enterotoxin A. Appl. Environ. Microbiol. 34, 518–522.
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States - major pathogens. Emerging Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Schantz, E. J., Roessler, W. G., Wagman, J., Spero, L., Dunnery, D. A., and Bergdoll, M. S. (1965). Purification of staphylococcal enterotoxin B. Biochemistry 4, 1011–1016. doi: 10.1021/bi00882a005
Scheuber, P. H., Denzlinger, C., Wilker, D., Beck, G., Keppler, D., and Hammer, D. K. (1987). Staphylococcal enterotoxin B as a nonimmunological mast cell stimulus in primates: the role of endogenous cysteinyl leukotrienes. Int. Arch. Allergy Appl. Immunol. 82, 289–291. doi: 10.1159/000234209
Scheuber, P. H., Mossmann, H., Beck, G., and Hammer, D. K. (1983). Direct skin test in highly sensitized guinea pigs for rapid and sensitive determination of staphylococcal enterotoxin B. Appl. Environ. Microbiol. 46, 1351–1356.
Schlievert, P. M., Jablonski, L. M., Roggiani, M., Sadler, I., Callantine, S., Mitchell, D. T., et al. (2000). Pyrogenic toxin superantigen site specificity in toxic shock syndrome and food poisoning in animals. Infect. Immun. 68, 3630–3634. doi: 10.1128/IAI.68.6.3630-3634.2000
Senn, L., Clerc, O., Zanetti, G., Basset, P., Prod'hom, G., Gordon, N. C., et al. (2016). The stealthy superbug: the role of asymptomatic enteric carriage in maintaining a long-term hospital outbreak of ST228 methicillin-resistant Staphylococcus aureus. MBio. 7, e02039–e02015. doi: 10.1128/mBio.02039-15
Sergelidis, D., and Angelidis, A. S. (2017). Methicillin-resistant Staphylococcus aureus: a controversial food-borne pathogen. Lett. Appl. Microbiol. 64, 409–418. doi: 10.1111/lam.12735
Shaler, C. R., Choi, J., Rudak, P. T., Memarnejadian, A., Szabo, P. A., Tun-Abraham, M. E., et al. (2017). MAIT cells launch a rapid, robust and distinct hyperinflammatory response to bacterial superantigens and quickly acquire an anergic phenotype that impedes their cognate antimicrobial function: defining a novel mechanism of superantigen-induced immunopathology and immunosuppression. PLoS Biol. 15:e2001930. doi: 10.1371/journal.pbio.2001930
Sheahan, D. G., Jervis, H. R., Takeuchi, A., and Sprinz, H. (1970). The effect of staphylococcal enterotoxin on the epithelial mucosubstances of the small intestine of rhesus monkeys. Am. J. Pathol. 60, 1–18.
Shen, M., Li, Y., Zhang, L., Dai, S., Wang, J., Li, Y., et al. (2017). Staphylococcus enterotoxin profile of China isolates and the superantigenicity of some novel enterotoxins. Arch. Microbiol. 199, 723–736. doi: 10.1007/s00203-017-1345-6
Shin, E., Hong, H., Park, J., Oh, Y., Jung, J., and Lee, Y. (2016). Characterization of Staphylococcus aureus faecal isolates associated with food-borne disease in Korea. J. Appl. Microbiol. 121, 277–286. doi: 10.1111/jam.13133
Shupp, J. W., Jett, M., and Pontzer, C. H. (2002). Identification of a transcytosis epitope on staphylococcal enterotoxins. Infect. Immun. 70, 2178–2186. doi: 10.1128/IAI.70.4.2178-2186.2002
Shylaja, R., Murali, H. S., Batra, H. V., and Bawa, A. S. (2010). A novel multiplex PCR system for the detection of staphylococcal enterotoxin B, tsst, nuc and fem genes of Staphylococcus aureus in food system. J. Food Saf. 30, 443–454. doi: 10.1111/j.1745-4565.2010.00218.x
Smith, T. C. (2015). Livestock-associated Staphylococcus aureus: the United States experience. PLoS Pathog. 11:e1004564. doi: 10.1371/journal.ppat.1004564
Song, M., Shi, C., Xu, X., and Shi, X. (2016). Molecular typing and virulence gene profiles of enterotoxin gene cluster (egc)-positive Staphylococcus aureus isolates obtained from various food and clinical specimens. Foodborne Pathog. Dis. 13, 592–601. doi: 10.1089/fpd.2016.2162
Soykut, E. A., Dudak, F. C., and Boyaci, I. H. (2008). Selection of staphylococcal enterotoxin B (SEB)-binding peptide using phage display technology. Biochem. Biophys. Res. Commun. 370, 104–108. doi: 10.1016/j.bbrc.2008.03.065
Spaan, A. N., Surewaard, B. G., Nijland, R., and Van Strijp, J. A. (2013). Neutrophils versus Staphylococcus aureus: a biological tug of war. Annu. Rev. Microbiol. 67, 629–650. doi: 10.1146/annurev-micro-092412-155746
Spaulding, A. R., Salgado-Pabón, W., Kohler, P. L., Horswill, A. R., Leung, D. Y., and Schlievert, P. M. (2013). Staphylococcal and streptococcal superantigen exotoxins. Clin. Microbiol. Rev. 26, 422–447. doi: 10.1128/CMR.00104-12
Stohl, W., Elliott, J. E., and Linsley, P. S. (1994). Human T cell-dependent B cell differentiation induced by staphylococcal superantigens. J. Immunol. 153, 117–127.
Su, Y. C., and Wong, A. C. (1995). Identification and purification of a new staphylococcal enterotoxin, H. Appl. Environ. Microbiol. 61, 1438–1443.
Su, Y. C., and Wong, A. C. (1996). Detection of staphylococcal enterotoxin H by an enzyme-linked immunosorbent assay. J. Food Prot. 59, 327–330. doi: 10.4315/0362-028X-59.3.327
Sugiyama, H., and Hayama, T. (1965). Abdominal viscera as site of emetic action for staphylococcal enterotoxin in the monkey. J. Infect. Dis. 115, 330–336. doi: 10.1093/infdis/115.4.330
Suleiman, T. S., Karimuribo, E. D., and Mdegela, R. H. (2018). Prevalence of bovine subclinical mastitis and antibiotic susceptibility patterns of major mastitis pathogens isolated in Unguja island of Zanzibar, Tanzania. Trop. Anim. Health Prod. 50, 259–266. doi: 10.1007/s11250-017-1424-3
Sullivan, R. (1969). Effects of enterotoxin B on intestinal transport in vitro. Proc. Soc. Exp. Biol. Med. 131, 1159–1162. doi: 10.3181/00379727-131-34060
Sumby, P., and Waldor, M. K. (2003). Transcription of the toxin genes present within the staphylococcal phage phiSa3ms is intimately linked with the phage's life cycle. J. Bacteriol. 185, 6841–6851. doi: 10.1128/JB.185.23.6841-6851.2003
Sun, S., Yang, M., Kostov, Y., and Rasooly, A. (2010). ELISA-LOC: lab-on-a-chip for enzyme-linked immunodetection. Lab Chip. 10, 2093–2100. doi: 10.1039/c003994b
Szabo, P. A., Rudak, P. T., Choi, J., Xu, S. X., Schaub, R., Singh, B., et al. (2017). Invariant natural killer T Cells are pathogenic in the HLA-DR4-transgenic humanized mouse model of toxic shock syndrome and can be targeted to reduce morbidity. J. Infect. Dis. 215, 824–829. doi: 10.1093/infdis/jiw646
Taylor, S. L., Schlunz, L. R., Beery, J. T., Cliver, D. O., and Bergdoll, M. S. (1982). Emetic action of staphylococcal enterotoxin A on weanling pigs. Infect. Immun. 36, 1263–1266.
Tempelman, L. A., King, K. D., Anderson, G. P., and Ligler, F. S. (1996). Quantitating staphylococcal enterotoxin B in diverse media using a portable fiber-optic biosensor. Anal. Biochem. 233, 50–57. doi: 10.1006/abio.1996.0006
Thomas, D. Y., Jarraud, S., Lemercier, B., Cozon, G., Echasserieau, K., Etienne, J., et al. (2006). Staphylococcal enterotoxin-like toxins U2 and V, two new staphylococcal superantigens arising from recombination within the enterotoxin gene cluster. Infect. Immun. 74, 4724–4734. doi: 10.1128/IAI.00132-06
Thomas, D., Chou, S., Dauwalder, O., and Lina, G. (2007). Diversity in Staphylococcus aureus enterotoxins. Chem. Immunol. Allergy 93, 24–41. doi: 10.1159/000100856
Thomas, M. K., Murray, R., Flockhart, L., Pintar, K., Pollari, F., Fazil, A., et al. (2013). Estimates of the burden of foodborne illness in Canada for 30 specified pathogens and unspecified agents, circa 2006. Foodborne Pathog. Dis. 10, 639–648. doi: 10.1089/fpd.2012.1389
Tiedemann, R. E., and Fraser, J. D. (1996). Cross-linking of MHC class II molecules by staphylococcal enterotoxin A is essential for antigen-presenting cell and T cell activation. J. Immunol. 157, 3958–3966.
Treiner, E., Duban, L., Bahram, S., Radosavljevic, M., Wanner, V., Tilloy, F., et al. (2003). Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422, 164–169. doi: 10.1038/nature01433
Tremaine, M. T., Brockman, D. K., and Betley, M. J. (1993). Staphylococcal enterotoxin A gene (sea) expression is not affected by the accessory gene regulator (agr). Infect. Immun. 61, 356–359.
Tseng, C. W., and Stewart, G. C. (2005). Rot repression of enterotoxin B expression in Staphylococcus aureus. J. Bacteriol. 187, 5301–5309. doi: 10.1128/JB.187.15.5301-5309.2005
Tseng, C. W., Zhang, S., and Stewart, G. C. (2004). Accessory gene regulator control of staphyloccoccal enterotoxin D gene expression. J. Bacteriol. 186, 1793–1801. doi: 10.1128/JB.186.6.1793-1801.2004
Tuffs, S. W., James, D. B. A., Bestebroer, J., Richards, A. C., Goncheva, M. I., O'shea, M., et al. (2017). The Staphylococcus aureus superantigen SElX is a bifunctional toxin that inhibits neutrophil function. PLoS Pathog. 13:e1006461. doi: 10.1371/journal.ppat.1006461
Umeda, K., Nakamura, H., Yamamoto, K., Nishina, N., Yasufuku, K., Hirai, Y., et al. (2017). Molecular and epidemiological characterization of staphylococcal foodborne outbreak of Staphylococcus aureus harboring seg, sei, sem, sen, seo, and selu genes without production of classical enterotoxins. Int. J. Food Microbiol. 256, 30–35. doi: 10.1016/j.ijfoodmicro.2017.05.023
Van Cauteren, D., Le Strat, Y., Sommen, C., Bruyand, M., Tourdjman, M., Da Silva, N. J., et al. (2017). Estimated annual numbers of foodborne pathogen-associated illnesses, hospitalizations, and deaths, France, 2008-2013. Emerg. Infect. Dis. 23, 1486–1492. doi: 10.3201/eid2309.170081
Van Gessel, Y. A., Mani, S., Bi, S., Hammamieh, R., Shupp, J. W., Das, R., et al. (2004). Functional piglet model for the clinical syndrome and postmortem findings induced by staphylococcal enterotoxin B. Exp. Biol. Med. 229, 1061–1071. doi: 10.1177/153537020422901011
Verhoeven, P. O., Gagnaire, J., Botelho-Nevers, E., Grattard, F., Carricajo, A., Lucht, F., et al. (2014). Detection and clinical relevance of Staphylococcus aureus nasal carriage: an update. Expert Rev. Anti Infect. Ther. 12, 75–89. doi: 10.1586/14787210.2014.859985
Viçosa, G. N., Le Loir, A., Le Loir, Y., De Carvalho, A. F., and Nero, L. A. (2013). egc characterization of enterotoxigenic Staphylococcus aureus isolates obtained from raw milk and cheese. Int. J. Food Microbiol. 165, 227–230. doi: 10.1016/j.ijfoodmicro.2013.05.023
Von Eiff, C., Becker, K., Machka, K., Stammer, H., and Peters, G. (2001). Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N. Engl. J. Med. 344, 11–16. doi: 10.1056/NEJM200101043440102
Wallin-Carlquist, N., Cao, R., Márta, D., Da Silva, A. S., Schelin, J., and Rådström, P. (2010). Acetic acid increases the phage-encoded enterotoxin A expression in Staphylococcus aureus. BMC Microbiol. 10:147. doi: 10.1186/1471-2180-10-147
Waters, C. M., and Bassler, B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001
Wattinger, L., Stephan, R., Layer, F., and Johler, S. (2012). Comparison of Staphylococcus aureus isolates associated with food intoxication with isolates from human nasal carriers and human infections. Eur. J. Clin. Microbiol. Infect. Dis. 31, 455–464. doi: 10.1007/s10096-011-1330-y
Wertheim, H. F., Verveer, J., Boelens, H. A., Van Belkum, A., Verbrugh, H. A., and Vos, M. C. (2005). Effect of mupirocin treatment on nasal, pharyngeal, and perineal carriage of Staphylococcus aureus in healthy adults. Antimicrob. Agents Chemother. 49, 1465–1467. doi: 10.1128/AAC.49.4.1465-1467.2005
White, J., Herman, A., Pullen, A. M., Kubo, R., Kappler, J. W., and Marrack, P. (1989). The V beta-specific superantigen staphylococcal enterotoxin B: stimulation of mature T cells and clonal deletion in neonatal mice. Cell 56, 27–35. doi: 10.1016/0092-8674(89)90980-X
Williams, R. E. (1963). Healthy carriage of Staphylococcus aureus: its prevalence and importance. Bacteriol. Rev. 27, 56–71.
Wilson, G. J., Seo, K. S., Cartwright, R. A., Connelley, T., Chuang-Smith, O. N., Merriman, J. A., et al. (2011). A novel core genome-encoded superantigen contributes to lethality of community-associated MRSA necrotizing pneumonia. PLoS Pathog. 7:e1002271. doi: 10.1371/journal.ppat.1002271
Wilson, I. G., Cooper, J. E., and Gilmour, A. (1991). Detection of enterotoxigenic Staphylococcus aureus in dried skimmed milk: use of the polymerase chain reaction for amplification and detection of staphylococcal enterotoxin genes entB and entC1 and the thermonuclease gene nuc. Appl. Environ. Microbiol. 57, 1793–1798.
Wright, A., Andrews, P. L., and Titball, R. W. (2000). Induction of emetic, pyrexic, and behavioral effects of Staphylococcus aureus enterotoxin B in the ferret. Infect. Immun. 68, 2386–2389. doi: 10.1128/IAI.68.4.2386-2389.2000
Wu, S., de Lencastre, H., and Tomasz, A. (1996). Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 178, 6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996
Wu, S., Duan, N., Gu, H., Hao, L., Ye, H., Gong, W., et al. (2016). A review of the methods for detection of Staphylococcus aureus enterotoxins. Toxins 8:176. doi: 10.3390/toxins8070176
Yan, X., Wang, B., Tao, X., Hu, Q., Cui, Z., Zhang, J., et al. (2012). Characterization of Staphylococcus aureus strains associated with food poisoning in Shenzhen, China. Appl. Environ. Microbiol. 78, 6637–6642. doi: 10.1128/AEM.01165-12
Yarwood, J. M., Mccormick, J. K., and Schlievert, P. M. (2001). Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183, 1113–1123. doi: 10.1128/JB.183.4.1113-1123.2001
Zeaki, N., Susilo, Y. B., Pregiel, A., Rådström, P., and Schelin, J. (2015). Prophage-encoded staphylococcal enterotoxin A: regulation of production in Staphylococcus aureus strains representing different sea regions. Toxins 7, 5359–5376. doi: 10.3390/toxins7124889
Zhang, S., and Stewart, G. C. (2000). Characterization of the promoter elements for the staphylococcal enterotoxin D gene. J. Bacteriol. 182, 2321–2325. doi: 10.1128/JB.182.8.2321-2325.2000
Zhang, S., Iandolo, J. J., and Stewart, G. C. (1998). The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej). FEMS Microbiol. Lett. 168, 227–233. doi: 10.1111/j.1574-6968.1998.tb13278.x
Zhao, W., Li, Y., Liu, W., Ding, D., Xu, Y., Pan, L., et al. (2016a). Transcytosis, antitumor activity and toxicity of staphylococcal enterotoxin C2 as an oral administration protein drug. Toxins 8:185. doi: 10.3390/toxins8060185
Zhao, Y., Zhu, A., Tang, J., Tang, C., and Chen, J. (2017). Identification and measurement of staphylococcal enterotoxin M from Staphylococcus aureus isolate associated with staphylococcal food poisoning. Lett. Appl. Microbiol. 65, 27–34. doi: 10.1111/lam.12751
Zhao, Y., Zhu, A., Tang, J., Tang, C., Chen, J., and Liu, J. (2016b). Identification and measurement of staphylococcal enterotoxin-like protein I (SEll) secretion from Staphylococcus aureus clinical isolate. J. Appl. Microbiol. 121, 539–546. doi: 10.1111/jam.13181
Ziebandt, A. K., Becher, D., Ohlsen, K., Hacker, J., Hecker, M., and Engelmann, S. (2004). The influence of agr and sigmaB in growth phase dependent regulation of virulence factors in Staphylococcus aureus. Proteomics 4, 3034–3047. doi: 10.1002/pmic.200400937
Keywords: Staphylococcus aureus, superantigen, enterotoxins, food poisoning, regulation, virulence, emesis
Citation: Fisher EL, Otto M and Cheung GYC (2018) Basis of Virulence in Enterotoxin-Mediated Staphylococcal Food Poisoning. Front. Microbiol. 9:436. doi: 10.3389/fmicb.2018.00436
Received: 20 December 2017; Accepted: 26 February 2018;
Published: 13 March 2018.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Liguang Xu, Jiangnan University, ChinaAlison J. Carey, Drexel University, United States
Copyright © 2018 Fisher, Otto and Cheung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Otto, bW90dG9AbmlhaWQubmloLmdvdg==
 Emilie L. Fisher
Emilie L. Fisher Michael Otto
Michael Otto Gordon Y. C. Cheung
Gordon Y. C. Cheung