- 1Department of Diagnostic Medicine and Pathobiology, Kansas State University, Manhattan, KS, United States
- 2Division of Molecular Biology, Center for Food Safety and Applied Nutrition, United States Food and Drug Administration, Laurel, MD, United States
- 3Veterinary Diagnostic Laboratory, Kansas State University, Manhattan, KS, United States
- 4Division of Foodborne, Waterborne, and Environmental Diseases, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, GA, United States
Escherichia coli O104:H4, a Shiga toxin-producing hybrid pathotype that was implicated in a major foodborne outbreak in Germany in 2011, has not been detected in cattle. However, serotypes of O104, other than O104:H4, have been isolated from cattle feces, with O104:H7 being the most predominant. In this study, we investigated, based on whole genome sequence analyses, the virulence potential of E. coli O104 strains isolated from cattle feces, since cattle are asymptomatic carriers of E. coli O104. The genomes of ten bovine E. coli O104 strains (six O104:H7, one O104:H8, one O104:H12, and two O104:H23) and five O104:H7 isolated from human clinical cases were sequenced. Of all the bovine O104 serotypes (H7, H8, H12, and H23) that were included in the study, only E. coli O104:H7 serotype possessed Shiga toxins. Four of the six bovine O104:H7 strains and one of the five human strains carried stx1c. Three human O104 strains carried stx2, two were of subtype 2a, and one was 2d. Genomes of stx carrying bovine O104:H7 strains were larger than the stx-negative strains of O104:H7 or other serotypes. The genome sizes were proportional to the number of genes carried on the mobile genetic elements (phages, prophages, transposable elements and plasmids). Both bovine and human strains were negative for intimin and other genes associated with the type III secretory system and non-LEE encoded effectors. Plasmid-encoded virulence genes (ehxA, epeA, espP, katP) were also present in bovine and human strains. All O104 strains were negative for antimicrobial resistance genes, except one human strain. Phylogenetic analysis indicated that bovine E. coli O104 strains carrying the same flagellar antigen clustered together and STEC strains clustered separately from non-STEC strains. One of the human O104:H7 strains was phylogenetically closely related to and belonged to the same sequence type (ST-1817) as the bovine O104:H7 STEC strains. This suggests that the bovine feces could be a source of human illness caused by E. coli O104:H7 serotype. Because bovine O104:H7 strains carried virulence genes similar to human clinical strains and one of the human clinical strains was phylogenetically related to bovine strains, the serotype has the potential to be a diarrheagenic pathogen in humans.
Introduction
Shiga toxin-producing Escherichia coli (STEC) O157 and six serogroups of non-O157 STEC, O26, O45, O103, O111, O121, and O145, are responsible for foodborne illnesses that range from mild to bloody diarrhea to serious complications of hemolytic uremic syndrome (HUS) in humans (Gould et al., 2013). Cattle are a major reservoir of O157 and non-O157 STEC serogroups, which harbor the organisms in their hindgut and shed them in the feces. The “top 7” STEC have been declared as adulterants in ground beef and non-intact raw beef products by the U.S. Department of Agriculture, Food Safety and Inspection Service (USDA-FSIS, 2011). In 2011, a novel serotype, E. coli O104:H4, was responsible for a large outbreak of hemorrhagic colitis and HUS in humans in Germany. The serotype was a hybrid pathotype carrying genes characteristic of enteroaggregative E. coli (EAEC) and STEC (Bielaszewska et al., 2011). Another serotype, E. coli O104:H21, was responsible for a small outbreak of hemorrhagic colitis in Helena, Montana in 1994 linked to consumption of contaminated milk (CDC, 1995). In addition, there are reports of sporadic cases of diarrhea associated with E. coli O104:H2, O104:H7, and O104:H12 (Delannoy et al., 2012a; Miko et al., 2013).
The German outbreak strain of E. coli O104:H4 has not been detected in cattle feces (Wieler et al., 2011; Auvray et al., 2012; Paddock et al., 2013; Shridhar et al., 2016). However, cattle do harbor and shed other serotypes of O104; including O104:H2, O104:H7, O104:H11, and O104:H21 (Paddock et al., 2013; Shridhar et al., 2016). Of the serotypes, O104:H7 appeared to be the dominant serotype and the only serotype that carried Shiga toxin gene (stx1) (Shridhar et al., 2016).
Shiga toxins (Stx) are major virulence factors of STEC and are responsible for life-threatening complications, such as HUS, associated with STEC infections (Karmali et al., 1985). Intimin, encoded by eae, a key adhesin expressed by enterohemorrhagic (EHEC), a subset of STEC, and enteropathogenic E. coli (EPEC), is responsible for the intimate attachment of bacteria to the host epithelial cells leading to attachment and effacement lesion (DeVinney et al., 1999). In addition, a number of other virulence factors, encoded by genes located on the chromosome or mobile genetic elements, are also involved in the pathogenesis of E. coli infections (Kaper et al., 1999). Therefore, determining the virulence gene profiles of E. coli O104 serotypes isolated from cattle is required to assess their potential to cause human illness. Whole genome sequence analysis provides a greater insight into the virulence potential and genetic diversity of strains within a serotype. The objectives of this study were to determine the virulence genes repertoire of E. coli O104 strains isolated from cattle by whole genome sequence analysis and to determine their phylogenetic relationship. Human clinical strains of E. coli O104:H7 were included for comparison.
Materials and Methods
Escherichia coli O104 Strains
Ten E. coli O104 strains isolated from rectal contents of feedlot cattle collected at a slaughter plant (Shridhar et al., 2016) and five E. coli O104 strains isolated from sporadic cases of diarrhea from human patients (Centers for Disease Control and Prevention, Atlanta, GA) were included in the study. Details on the study population, sampling and isolation procedures of cattle E. coli O104 strains have been described (Shridhar et al., 2016). Of the 10 cattle strains, six were identified to contain H7 flagellar antigen (Shridhar et al., 2016) and the other four were of unknown serotypes. All five human strains carried H7 flagellar type. The strains were cultured onto Tryptone soy agar (TSA; BD Difco, Sparks, MD) slants and shipped overnight to the Center for Food Safety and Applied Nutrition, Food and Drug Administration, Laurel, MD, for whole genome sequencing.
DNA Extraction and Whole Genome Sequencing
Escherichia coli O104 strains were streaked onto blood agar (Remel, Lenexa, KS). A single colony of each strain was inoculated into Luria Bertani (LB) broth and incubated on a shaker at 37°C. Genomic DNA was isolated from overnight culture using the Qiagen DNeasy blood and tissue kit (Qiagen, Inc., Valencia, CA). The purity of the DNA was assessed spectrophotometrically using the Nanodrop (Thermo Scientific, Waltham, MA). Genomic libraries of the strains were constructed using Nextera XT DNA Library Preparation kit and whole genome sequencing was performed on an Illumina MiSeq benchtop sequencer (Illumina, Inc., San Diego, CA) using the MiSeq version 2 reagent kit with 2 × 250 cycles. De novo assembly of quality-controlled trimmed sequenced reads was performed using the SPAdes genome assembler version 3.8.2 (http://cab.spbu.ru/software/spades/) (Bankevich et al., 2012).
Sequence Analysis
Draft genomes of E. coli O104 strains were initially annotated using RAST (Rapid Annotation using Subsystem Technology) server (Aziz et al., 2008). The RAST server also provides data on the distribution of genes in various categories. Serotypes, virulence genes and antimicrobial resistance genes of the strains were determined using SerotypeFinder 1.1 (Joensen et al., 2015), VirulenceFinder 1.5 (Joensen et al., 2014), and ResFinder 2.1 (Zankari et al., 2012), respectively, which are web-based tools developed by the Center for Genomic Epidemiology (CGE) at the Danish Technical University (DTU), Lyngby, Denmark (http://www.genomicepidemiology.org/). Different types of plasmid sequences were identified by PlasmidFinder 1.3 (https://cge.cbs.dtu.dk/services/PlasmidFinder/) tool (Carattoli, 2009). The total number of prophage sequences were determined using Phage Search Tool Enhanced Release (PHASTER; http://phaster.ca/). The tool identifies intact, questionable, and incomplete prophage sequences by scores of >90, 70–90, < 70, respectively (Zhou et al., 2011; Arndt et al., 2016). Clusters of regularly interspaced short palindromic repeats (CRISPR)-Cas system of bovine and human E. coli O104 strains were characterized based on annotation by CRISPRone, a web-based tool (http://omics.informatics.indiana.edu/CRISPRone). The tool provides class, type, and subtype of CRISPR-Cas system and number, length and nucleotide sequences of repeats and spacers (Zhang and Ye, 2017). The sequence types of the strains were determined in silico using MLST 1.8 (https://cge.cbs.dtu.dk/services/MLST/) (Wirth et al., 2006; Jaureguy et al., 2008). The Harvest Suite, a software package which includes tools such as Parsnp and Gingr, was used to determine the phylogenetic relationship among the E. coli O104 strains (Treangen et al., 2014). Parsnp v1.2 (http://harvest.readthedocs.io/en/latest/content/parsnp.html) was used to align the core genomes of human and bovine E. coli O104 strains, followed by the construction of maximum likelihood tree. Escherichia coli O104:H21, a human outbreak strain (Montana; GenBank accession no. CP009106.2) was also included in the phylogenetic analysis to determine the genetic relatedness to the bovine O104 strains. The phylogenetic tree was subsequently imported to FigTree 1.4.3 software (http://tree.bio.ed.ac.uk/software/figtree/) (Rambaut, 2016) for better visualization, and bootstrap values are reported for each branch.
Statistical Analysis
A single factor analysis of variance (ANOVA) test was performed to determine whether genome size and number of genes associated with different functional categories were significantly different between bovine STEC, non-STEC and human strains. If the means were significantly different (P < 0.01), then Tukey adjustment for multiple comparisons was performed using PROC GLIMMIX procedure of SAS version 9.4 (SAS Institute Inc., Cary, NC), to test each pairwise comparison.
Results
Serotypes of bovine (n = 6) and human O104:H7 (n = 5) strains were confirmed by SerotypeFinder 1.1 and the other four unknown serotypes from cattle were identified as O104:H8, O104:H12, and 104:H23 (n = 2).
Rapid Annotation Using Subsystem Technology
The genome size ranged from 5.2 to 5.3 Mb, and 4.7 to 5.0 Mb for STEC and non-STEC bovine strains, respectively. The genome size of human E. coli O104 strains ranged from 4.9 to 5.4 Mb. The average genome size of bovine STEC strains (5.3 Mb) were significantly (P < 0.01) larger than bovine non-STEC strains (4.9 Mb). The functional categorization of genes revealed that the number of genes associated with virulence, disease, and defense ranged from 109 to 116 in bovine strains and 110 to122 in human O104 strains. The number of genes carried on mobile elements such as phages, prophages, transposable elements and plasmids ranged from 60 to 201 in bovine O104 strains, and 80 to 201 in human O104 strains. The average number of genes associated with mobile genetic elements was significantly (P < 0.01) higher in bovine STEC strains (190) compared to non-STEC strains (71). The human strains had lower (P < 0.01) number of genes associated with mobile genetic elements than bovine O104:H7 STEC strains. There was no significant difference in the average number of genes associated with other subsystem categories (membrane transport, iron acquisition and metabolism, and stress response) between bovine and human strains (Table 1).
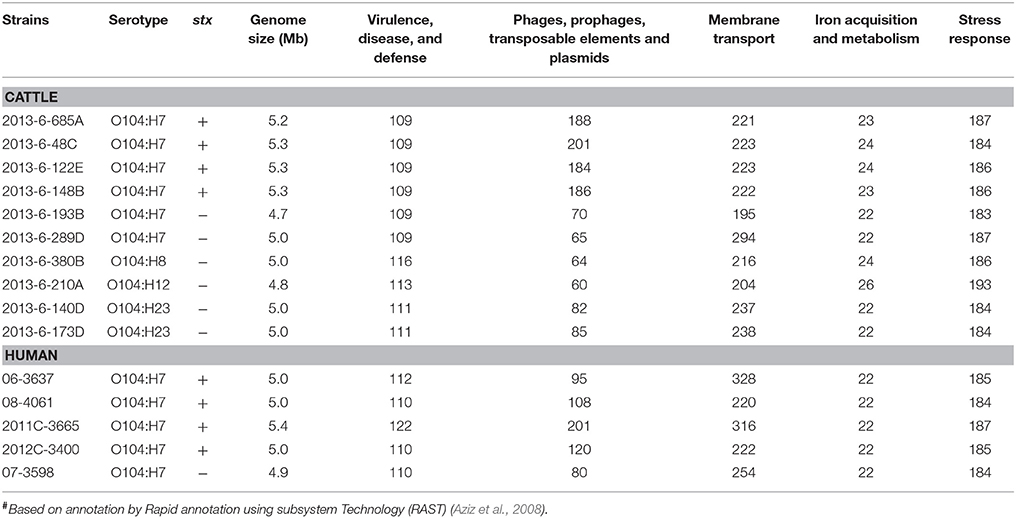
Table 1. Genome size and total number of major categories of genes in E. coli O104 strains isolated from cattle feces and human clinical cases#.
Virulence Genes
Four of the six bovine and one of the five human O104:H7 strains carried stx1 gene of subtype c. Three human STEC O104:H7 strains carried stx2, two were of subtype 2a and one was 2d (Table 2). The astA gene that codes for E. coli heat stable enterotoxin 1 was present in one bovine strain (2013-6-173D that belonged to the O104:H23 serotype). The gene subA (subtilase toxin subunit) was present in 4 of 5 human strains of O104:H7, but absent in all bovine strains. Both bovine and human O104 strains were negative for intimin (eae) and other genes associated with LEE Pathogenicity Island, and non-LEE encoded effectors. However, both bovine and human E. coli O104 strains carried other adhesins, such as irgA homolog adhesin (iha) and long polar fimbriae (lpfA).
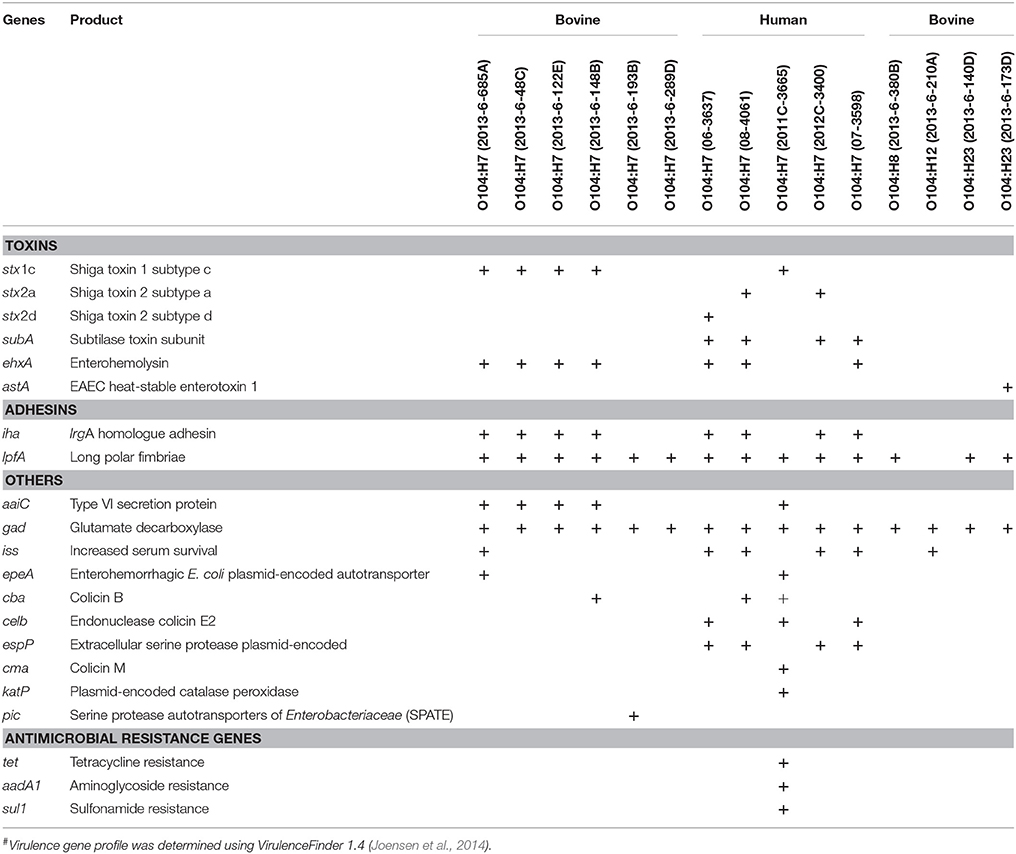
Table 2. Virulence genes and antimicrobial resistance genes present in E. coli O104 strains isolated from cattle feces (n = 10) and human clinical cases (n = 5)#.
Plasmid-encoded virulence genes (ehxA, epeA, espP, katP) were present in bovine and human strains. Four of the bovine strains, and three of the human strains carried ehxA (enterohemolysin). Enterohemorrhagic E. coli plasmid-encoded autotransporter (epeA) was present in one of the bovine strains (O104:H7; 2013-6-685A), and one human strain (O104:H7; 2011C-3665). Extracellular serine protease (espP; 4/5) and catalase peroxidase (katP; 1/5) were present only in human strains.
All bovine and human strains carried gad (Glutamate decarboxylase). Four bovine O104:H7 strains (2013-6-48C, 2013-6-122E, 2013-6-148B, and 2013-6-685A) and one human O104:H7 strain (2011C-3665) was positive for aaiC (Type VI secretory system protein). The gene encoding increased serum survival (iss) was present in two bovine (2013-6-685A and 2013-6-210A) and four human O104:H7 strains (06-3637, 07-3598, 08-3046, and 2012C-3400). One of the bovine strains (O104:H7; 2013-6-193B) carried pic (protein involved in intestinal colonization). Different types of genes that code for colicin, a bacteriocin, were also present in bovine and human E. coli O104 strains. The colicin B gene (cba) was present in one bovine strain (2013-6-148B) and two of the human strains (08-4061 and 2011C-3665), celb (endonuclease colicin E2) was present in three human strains (06-3637, 07-3598, and 2011C-3665), cma (Colicin M) was present only in one human strain (2011C-3665). Virulence gene profiles of bovine and human strains are provided in Table 2.
Antimicrobial Resistance Genes
Antimicrobial resistance genes, tet (tetracycline resistance), aadA1 (aminoglycoside resistance), and sul1 (sulfonamide resistance), were present only in one of the human strains (2011C-3665). All the bovine and other human strains were negative for antimicrobial resistance genes (Table 2).
Plasmid and Prophage Sequences
All the bovine STEC and one (2013-6-289D) of the non-STEC O104:H7 strains carried IncFIB and IncFII plasmid sequences (Table 3). IncH12 and IncH12A plasmid sequences were carried by all except one bovine non-STEC strain (O104:H7; 2013-6-193B). IncY was present in one (O104:H8; 2013-6-380B) of the bovine non-STEC strains and one human STEC strain (08-4061). IncB/O/K/Z and IncFIB plasmid sequences were present in all human strains except one (O104:H7; 2011C-3665). Col156 was present in two human STEC and one non-STEC strains. A complete list of plasmid sequences found in bovine and human E. coli O104 strains is provided in Table 3.
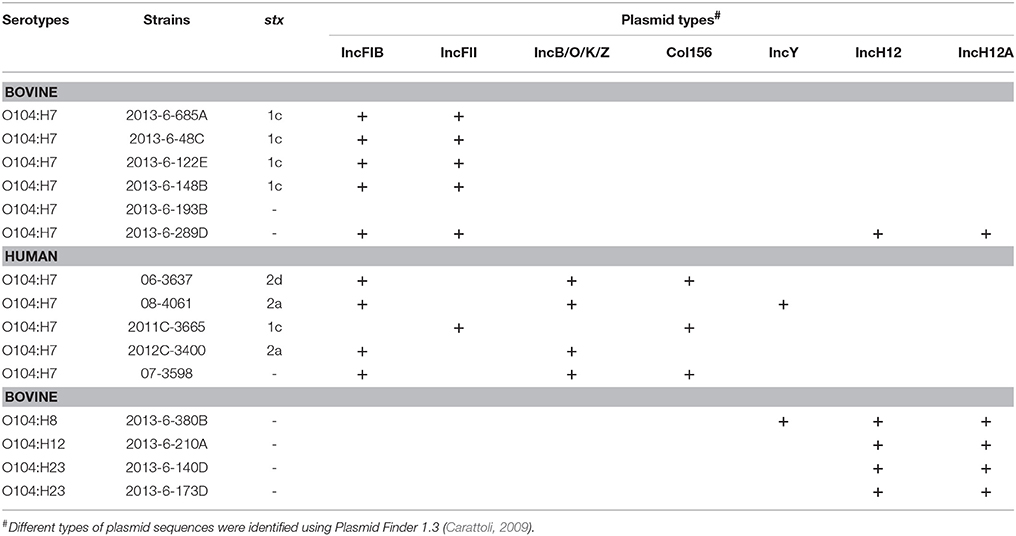
Table 3. Plasmid types in E. coli O104 strains isolated from cattle feces and human clinical cases using PlasmidFinder 1.3.
The four bovine STEC O104:H7 strains carried 8 intact prophage sequences, whereas the bovine non-STEC strains carried only 1 to 5 intact prophage sequences. However, the number of intact prophage sequences in human STEC O104:H7 strains ranged from 2 to 10, with 2011C-3665 strain carrying 10 intact prophage sequences. The number of incomplete prophage sequences ranged from 9 to 12, 1 to 5, and 4 to 7 for bovine STEC, bovine non-STEC, and human STEC strains, respectively. Total number of questionable prophage sequences ranged from 1 to 4, 0 to 1, and 1 to 4 for bovine STEC, bovine non-STEC, and human STEC strains, respectively. One of the human non-STEC strains (07-3598) carried four intact, six incomplete and zero questionable prophage sequences (Table 4).
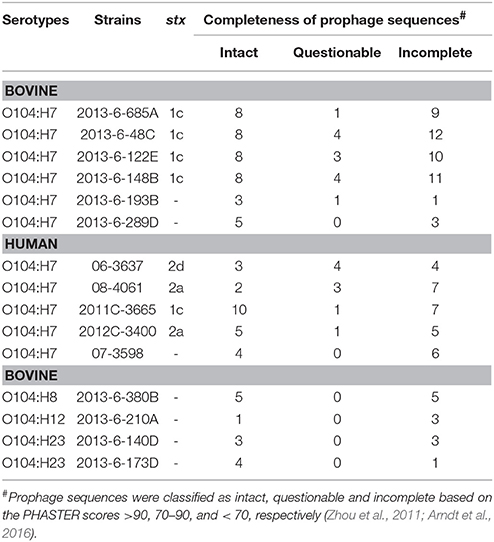
Table 4. Total number of prophage sequences in bovine and human E. coli O104 strains identified using PHASTER.
CRISPR-Cas System
The most common subtype of CRISPR-Cas system in most of the bovine and human E. coli O104 strains was I-A and I-E. Other subtypes identified were I-D (2011C-3665), I-F (2013-6-193B, 2013-6-289D), III-B (2013-6-148B), and VI-C (2013-6-148B). All the four bovine O104:H7 STEC strains and one the human O104:H7 STEC strain (2011C-3665) carried a single CRISPR locus, four human O104:H7 STEC strains and three stx-negative bovine strains carried two loci, and two stx-negative bovine strains carried four loci. One of the bovine O104:H12 strain (2013-6-210A) carried CRISPR region lacking cas genes. Total number, nucleotide sequence and average length of repeats and spacers were highly conserved across all stx-positive bovine O104:H7 STEC strains. Average length and nucleotide sequence of repeats and spacers of one of the human O104:H7 STEC strain (2011C-3665) was similar to bovine O104:H7 STEC strains, however, it carried fewer repeats and spacers compared to bovine O104:H7 STEC strains. Similarly, total number, nucleotide sequence and average length of repeats and spacers were highly conserved across all human O104:H7 STEC strains. Average length and nucleotide sequences of spacers and repeats were similar among bovine stx-negative O104:H7 strains, but they differed in number of repeats and spacer units. However, repeats and spacer units were not conserved among O104:H23 strains (Table 5).
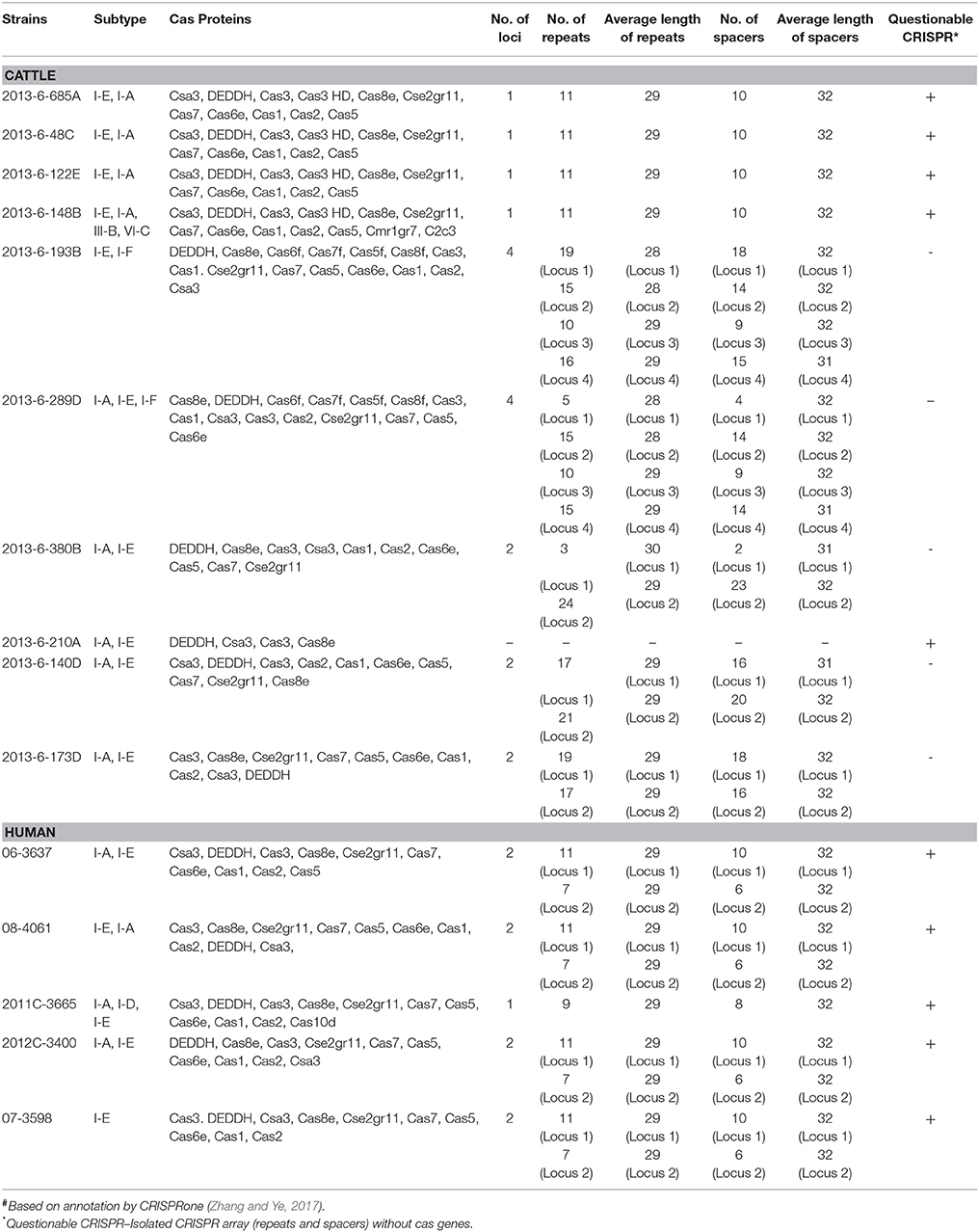
Table 5. Characteristic features of CRISPR-Cas system in E. coli O104 strains isolated from cattle feces and human clinical cases#.
Phylogenetic Relationship and Types (ST)
Bovine E. coli O104 strains carrying the same flagellar type clustered together (Figure 1). Among bovine O104:H7 strains, STEC strains clustered separately from non-STEC strains. Additionally, the O104:H8 strain (2013-6-380B) was more closely related to non-STEC O104:H7 strains (Figure 1). Escherichia coli O104:H12 strain (2013-6-210A) was distantly related to other O104 serotypes. All human O104:H7 strains, except one, clustered together. The human O104:H7 strain, 2011C-3665, which carried stx1c gene similar to bovine STEC O104:H7 strains clustered with the bovine O104:H7 STEC strains. The human Montana outbreak strain of O104:H21 (Montana outbreak) clustered more closely with one of the human (2011C-3665) and bovine O104:H7 STEC strains (Figure 1).

Figure 1. Phylogenetic tree and sequence types (ST) of stx-positive or –negative E. coli O104 strains isolated from cattle feces and human clinical strains constructed using Parsnp v1.2 and visualized using FigTree 1.4.3 (bootstrap values are provided for each branch).
The four bovine O104:H7 STEC strains and one of the four human O104:H7 strains (2011C-3665), which clustered with the bovine strains, belonged to ST-1817. The other 4 human O104:H7 strains that clustered separately belonged to ST-2773. One non-STEC bovine O104:H7 strain (2013-6-289D) belonged to ST-5014. The remaining bovine non-STEC O104 strains belonged to ST-678, ST-10, ST-6494, ST-129, and ST-939 (Figure 1).
Discussion
Shiga toxin-producing E. coli O104 serotypes, other than O104:H4 and O104:H21, have been reported to be associated with sporadic cases of diarrhea in humans (Hussein, 2007; Miko et al., 2013). Only one small outbreak of hemorrhagic colitis in Montana, USA, has been reported with the O104:H21 serotype (CDC, 1995). Several studies have characterized the strains belonging to various O104 serotypes by PCR-based detection of specific virulence targets (Miko et al., 2013; Shridhar et al., 2016). However, there has been one study on the genome-scale analysis of E. coli O104 serotypes, other than the German outbreak serotype of O104:H4 (Lambert et al., 2015; Yan et al., 2015). Genomic characterization of E. coli O104:H4 German outbreak strain has revealed that the hybrid pathotype evolved by gain and loss of chromosomal and plasmid encoded virulence genes (Mellmann et al., 2011; Rasko et al., 2011). Escherichia coli O104:H4 has been reported to be evolved from EAEC by acquisition of stx2 prophage and plasmid encoding CTX-M-15 extended spectrum beta lactamase (Rohde et al., 2011). Yan et al. (2015) sequenced the genomes of two human strains of E. coli O104:H21 (Montana outbreak) and an O104:H7 STEC strain isolated from cattle and compared them to E. coli O104:H4 (German and Central Africa outbreak strains) strains. They reported that the O104:H7 STEC strain was more similar to the Montana strain (O104:H21) than to the German strain (O104:H4). To our knowledge, this is the first study to analyze the whole genome sequences of human O104:H7 strains and to compare the virulence gene profiles of bovine O104:H7 strains.
Genomes of stx carrying bovine O104:H7 strains were larger than the stx-negative strains of O104:H7 or other serotypes. The genome sizes were proportional to the number of genes carried on the mobile genetic elements (phages, prophages, transposable elements and plasmids). Yan et al. (2015) have attributed the variations in the genomes of the three O104 strains (two H21 and one H7) analyzed to the gain or loss of mobile genetic elements (Yan et al., 2015). Although results of our study suggested that the variation in the size of genomes is due to variation in the mobile genetic elements, variations in sequencing coverage cannot be ruled out.
Of all the bovine O104 serotypes (H7, H8, H12, and H23) that were included in the study, only E. coli O104:H7 serotype possessed Shiga toxins. Two human O104:H7 strains carried stx2 subtype a, and one of the strains carried stx2 subtype d. Shiga toxin 2a and 2d have been reported to be most commonly associated with severe human illness and complications, such as HUS (Bielaszewska et al., 2006; Persson et al., 2007; Iyoda et al., 2014). The German outbreak strains of O104:H4 possessed stx2a (Scheutz et al., 2011), whereas the Montana strain O104:21 carried stx2d (Yan et al., 2015). Four of the bovine and one of the human O104:H7 strains carried stx1 subtype c. Friedrich et al. (2003) reported that stx1c carrying STEC strains were isolated from asymptomatic patients and patients with uncomplicated diarrhea (Friedrich et al., 2003). However, a stx1c carrying E. coli O78:H- strain was isolated from a 2-week old boy suffering from bacteremia and HUS and from family members who were asymptomatic (Lienemann et al., 2012).
All the bovine and human strains included in our study were negative for intimin and other LEE-encoded virulence genes, and non-LEE encoded effectors. Absence of eae has been reported to be a feature of O104 human outbreak strains (O104:H4 German outbreak and O104:H21 Montana outbreak strains) (Feng et al., 2001; Scheutz et al., 2011). A significant association between strains that carry stx1c and LEE-negative strains has been reported (Haugum et al., 2014). Shiga toxin-producing E. coli strains negative for LEE-encoded virulence genes have been isolated from humans with HUS (Bonnet et al., 1998; Paton et al., 1999). Both human and cattle E. coli O104 strains carried other adhesins such as iha and lpf A. Long polar fimbriae (lpf) and iha that encode for IrgA homologous adhesins were the most prevalent adhesins identified in LEE-negative E. coli strains isolated from cattle and human sources (Galli et al., 2010). These adhesins (iha and lpfA) were also carried by E. coli O104:H4 German outbreak strains and E. coli O104:H21 Montana outbreak strain (Bielaszewska et al., 2011; Yan et al., 2015). Additionally, none of the bovine and human strains investigated in our study was positive for efa1 (enterohemorrhagic E. coli factor for adherence), which is not surprising because previous studies have suggested a strong association between eae and efa1 genes (Galli et al., 2010). Human O104:H7 strains, and not bovine O104:H7, were positive for subtilase cytotoxin, which has been reported to be involved in damaging human microvascular endothelial cells (Amaral et al., 2013). The subA gene has also been detected in eae negative E. coli O103:H21 strains associated with sporadic cases of HUS (Paton et al., 1999). One of the stx negative bovine O104 strains was positive for astA gene, which encodes for EAEC heat-stable enterotoxin 1. Typical and atypical enteropathogenic E. coli strains carrying astA gene have been reported to be frequently associated with diarrhea (Silva et al., 2014).
Both bovine and human E. coli O104 strains included in the present study were positive for plasmid encoded virulence genes such as ehxA, espP, katP (encoded by pO157), and epeA (encoded by pO113). Enterohemolysin, encoded by ehxA, has been reported to be involved in increased production of IL-1β and cytotoxicity (Zhang et al., 2012). It has also been reported to be frequently associated with E. coli O111 STEC strains isolated from patients with HUS (Schmidt and Karch, 1996). Extracellular serine protease (espP) is responsible for mucosal hemorrhage observed in patients suffering from hemorrhagic colitis, due to cleavage of pepsin A and human coagulation factor V (Brunder et al., 1997). An autotransporter protease, epeA has been reported to be frequently found in association with LEE-negative STEC strains. The protease is involved in adherence and colonization of small intestine due to its mucinolytic activity (Leyton et al., 2003). One of the bovine non-STEC O104:H7 strain (2013-6-193B) carried pic (protein involved in intestinal colonization), which encodes for serine protease autotransporter. The protease, also secreted by Shigella flexneri and enteroaggregative E. coli, is involved in intestinal colonization (Henderson et al., 1999). The pic gene was also carried by E. coli O104:H4 (German outbreak strain) (Bielaszewska et al., 2011). Other genes characteristic of EAEC such as aggA (pilin subunit of aggregative adherence fimbriae I), agg3A (pilin subunit of aggregative adherence fimbriae III), aggR (transcriptional regulator), and serine protease autotransporter toxins (sepA; Shigella extracellular protein A, sigA; Shigella IgA protease-like homolog, aap; dispersin, aatPABCD; dispersin transporter) were present in E. coli O104:H4 German outbreak strains, but absent in E. coli O104:H21 Montana outbreak strain (Bielaszewska et al., 2011; Frank et al., 2011; Yan et al., 2015) and E. coli O104 strains investigated in our study.
IncF plasmids (1B and II) were present in all bovine O104 STEC strains, except one (stx negative 2013-6-193B). Antimicrobial resistance genes, such as blaTEM−1, blaOXA−1, and aac(6′)-Ib-cr, are carried on IncF plasmids (Carattoli, 2009; Cergole-Novella et al., 2010). However, none of the bovine strains analyzed in the study carried antimicrobial resistance genes according to the ResFinder data base. The human O104:H7 strain (2011C-3665) that carried the three antimicrobial resistance genes (tet, aadA1, and sul1) was negative for IncF1, but positive for IncFII plasmids. Tetracycline resistance was the most common resistance in E. coli O157 strains isolated from bovine and human sources (Wilkerson et al., 2004). Non-O157 STEC serogroups isolated from human, bovine and ovine sources were found to be most commonly resistant to streptomycin, sulfisoxazole, and tetracycline (Wang et al., 2016). IncH12 plasmid, which was present in bovine strains, has been shown to be involved in the dissemination of antimicrobial resistance genes (Fernández et al., 2007).
Mobile genetic elements have been reported to influence pathogenicity, survival and spread of O157 and non-O157 EHEC (Ogura et al., 2009). CRISPR-Cas system provides acquired immunity against viruses and plasmids by targeting foreign nucleic acids (Horvath and Barrangou, 2010). Plasmid and phage environment have been reported to influence spacer composition (Delannoy et al., 2015a). Hence, we also analyzed the CRISPR-Cas systems in addition to phage and plasmid profiles of the E. coli O104 strains. Analysis of CRISPR-Cas system in bovine and human E. coli O104 strains revealed that it was highly conserved among O104:H7 bovine and human STEC strains. It was also conserved among stx-negative bovine O104:H7 strains. These findings show that the CRISPR systems are specific for serotype, regardless of the presence or absence of stx. Similar findings have also been reported in previous studies (Delannoy et al., 2012a,b; Toro et al., 2014). However, additional strains should be analyzed to show the serotype specificity of CRISPR loci in other O104 serotypes investigated in this study.
Analysis of phylogenetic relationship of O104 strains revealed that O104:H21 strain was more closely related to stx1c-carrying O104:H7 strains of bovine and human origin. This is in agreement with the previous study (Yan et al., 2015). In our study, O104 strains with the same H-type clustered together. Escherichia coli serogroups carrying the same flagellar antigen have been hypothesized to share common ancestors (Ju et al., 2012). Miko et al. (2013) reported that the O104 STEC strains carrying the same H-types had similar virulence gene profiles and clustered together by PFGE analysis (Miko et al., 2013). The genetic diversity of different O104 serotypes could be attributed to gain and loss of mobile genetic elements (Yan et al., 2015). Additionally, bovine O104 STEC strains and one of the human STEC strain (2011C-3665) which clustered together belonged to same sequence type (ST-1817). This suggests that the bovine feces could be a source of human illness caused by E. coli O104:H7 serotype. One of the non-STEC O104 strains belonged to ST-678, which includes E. coli O104:H4 German outbreak strain (Mora et al., 2011).
Although, this study provides sufficient evidence for the virulence potential of bovine E. coli O104 strains, the conclusions are based on a limited number of strains. Hence, further analysis of additional O104 strains belonging to various serotypes and sources might provide strong evidence for the virulence potential and evolution of E. coli O104 strains. Also, the use of long read sequencing technologies such as PacBio will enable in-depth analysis of the virulence potential of the strains compared to short read technologies such as Illumina. A combination of short read and long read technologies have also shown to generate high quality sequences which allow in-depth analysis of strains (Delannoy et al., 2015b, 2017).
Conclusions
Whole genome analyses of E. coli O104 strains of different serotypes provide an insight into their genetic diversity and pathogenic potential. Strains of serotype O104:H7, could be of public health concern, because they carry Shiga toxin gene and several other genes encoding virulence factors responsible for human illness. Interestingly, the five O104:H7 strains of ST-1817 (1 human and 4 bovine) were all positive for the stx1c gene and clustered together phylogenetically. In contrast, four human O104:H7 strains of ST-2773, positive for either stx2a, stx2d or no Shiga toxin genes, clustered together, but were apart from the bovine ST-1817 strains. Similarly, CRISPR loci of four of the bovine O104:H7 STEC strains and one of the human O104:H7 STEC strain (2011C-3665) were conserved. These findings suggests that bovine feces could be a source of human O104:H7 STEC strains. The WGS-based analysis of virulence genes suggests that bovine O104:H7 STEC has the potential to be a diarrheagenic pathogen in humans.
Nucleotide Sequence Accession Numbers
The whole genome shotgun sequences have been deposited at DDBJ/ENA/GenBank under the accession numbers NEKE00000000 to NEKS00000000.
Author Contributions
TN, JB, CE, and NS conceived and designed the experiments. PS, IP, JG, LN, and XS performed the experiments. PS, IP, and JG analyzed the data. PS, TN, JB, and IP wrote the paper.
Funding
This material is based upon work that is supported by the National Institute of Food and Agriculture, U. S. Department of Agriculture, under award number 2012-68003-30155.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This is contribution no. 18-004-J of the Kansas Agricultural Experiment Station. This work is part of the doctoral dissertation (Shridhar, 2017).
References
Amaral, M. M., Sacerdoti, F., Jancic, C., Repetto, H. A., Paton, A. W., Paton, J. C., et al. (2013). Action of Shiga Toxin Type-2 and subtilase cytotoxin on human microvascular endothelial cells. PLoS ONE 8:e70431. doi: 10.1371/journal.pone.0070431
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Auvray, F., Dilasser, F., Bibbal, D., Kerouredan, M., Oswald, E., and Brugere, H. (2012). French cattle is not a reservoir of the highly virulent enteroaggregative Shiga toxin-producing Escherichia coli of serotype O104:H4. Vet. Microbiol. 158, 443–445. doi: 10.1016/j.vetmic.2012.02.029
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genom 9:75. doi: 10.1186/1471-2164-9-75
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., and Kulikov, A. S. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bielaszewska, M., Friedrich, A. W., Aldick, T., Schürk-Bulgrin, R., and Karch, H. (2006). Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin. Infect. Dis. 43, 1160–1167. doi: 10.1086/508195
Bielaszewska, M., Mellmann, A., Zhang, W., Köck, R., Fruth, A., Bauwens, A., et al. (2011). Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 11, 671–676. doi: 10.1016/S1473-3099(11)70165-7
Bonnet, R., Souweine, B., Gauthier, G., Rich, C., Livrelli, V., Sirot, J., et al. (1998). Non-O157:H7 Stx2-producing Escherichia coli strains associated with sporadic cases of hemolytic-uremic syndrome in adults. J. Clin. Microbiol. 36, 1777–1780.
Brunder, W., Schmidt, H., and Karch, H. (1997). EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 24, 767–778. doi: 10.1046/j.1365-2958.1997.3871751.x
Carattoli, A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238. doi: 10.1128/AAC.01707-08
CDC (1995). Outbreak of acute gastroenteritis attributable to Escherichia coli serotype O104:H21— helena, montana, 1994. JAMA 274, 529–530. doi: 10.1001/jama.1995.03530070027011
Cergole-Novella, M. C., Guth, B. E., Castanheira, M., Carmo, M. S., and Pignatari, A. C. (2010). First description of blaCTX−M−14- and blaCTX−M−15-producing Escherichia coli isolates in Brazil. Microb. Drug Resist. 16, 177–184. doi: 10.1089/mdr.2010.0008
Delannoy, S., Beutin, L., Burgos, Y., and Fach, P. (2012a). Specific detection of enteroaggregative hemorrhagic Escherichia coli O104:H4 strains by use of the CRISPR locus as a target for a diagnostic real-time PCR. J. Clin. Microbiol. 50, 3485–3492. doi: 10.1128/JCM.01656-12
Delannoy, S., Beutin, L., and Fach, P. (2012b). Use of clustered regularly interspaced short palindromic repeat sequence polymorphisms for specific detection of enterohemorrhagic Escherichia coli strains of serotypes O26:H11, O45:H2, O103:H2, O111:H8, O121:H19, O145:H28, and O157:H7 by Real-Time PCR. J. Clin. Microbiol. 50, 4035–4040. doi: 10.1128/JCM.02097-12
Delannoy, S., Mariani-Kurkdjian, P., Bonacorsi, S., Liguori, S., and Fach, P. (2015a). Characteristics of emerging human-pathogenic Escherichia coli O26:H11 Strains Isolated in France between 2010 and 2013 and Carrying the stx 2d gene only. J. Clin. Microbiol. 53, 486–492. doi: 10.1128/JCM.02290-14
Delannoy, S., Mariani-Kurkdjian, P., Bonacorsi, S., Liguori, S., Ison, S. A., and Fach, P. (2015b). Draft genome sequences of human-pathogenic Escherichia coli O26:H11 strains carrying the stx2 gene only and circulating in France. Genome Announc. 3, e00852–e00815. doi: 10.1128/genomeA.00852-15
Delannoy, S., Mariani-Kurkdjian, P., Webb, H. E., Bonacorsi, S., and Fach, P. (2017). The mobilome; a major contributor to Escherichia coli stx2-Positive O26:H11 strains intra-serotype diversity. Front. Microbiol. 8:1625. doi: 10.3389/fmicb.2017.01625
DeVinney, R., Stein, M., Reinscheid, D., Abe, A., Ruschkowski, S., and Finlay, B. B. (1999). Enterohemorrhagic Escherichia coli O157:H7 produces tir, which is translocated to the host cell membrane but is not tyrosine phosphorylated. Infect. Immun. 67, 2389–2398.
Feng, P., Weagant, S. D., and Monday, S. R. (2001). Genetic analysis for virulence factors in Escherichia coli O104:H21 that was implicated in an outbreak of hemorrhagic colitis. J. Clin. Microbiol. 39, 24–28. doi: 10.1128/JCM.39.1.24-28.2001
Fernández, A. G., Cloeckaert, A., Bertini, A., Praud, K., Doublet, B., Weill, F.-X., et al. (2007). Comparative Analysis of IncHI2 Plasmids Carrying blaCTX−M−2 or blaCTX−M−9 from Escherichia coli and Salmonella enterica strains isolated from poultry and humans. Antimicr. Agents Chemother. 51, 4177–4180. doi: 10.1128/AAC.00603-07
Frank, C., Werber, D., Cramer, J. P., Askar, M., Faber, M., an der Heiden, M., et al. (2011). Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N. Engl. J. Med. 365, 1771–1780. doi: 10.1056/NEJMoa1106483
Friedrich, A. W., Borell, J., Bielaszewska, M., Fruth, A., Tschape, H., and Karch, H. (2003). Shiga Toxin 1c-Producing Escherichia coli Strains: phenotypic and genetic characterization and association with human disease. J. Clin. Microbiol. 41, 2448–2453. doi: 10.1128/JCM.41.6.2448-2453.2003
Galli, L., Miliwebsky, E., Irino, K., Leotta, G., and Rivas, M. (2010). Virulence profile comparison between LEE-negative Shiga toxin-producing Escherichia coli (STEC) strains isolated from cattle and humans. Vet. Microbiol. 143, 307–313. doi: 10.1016/j.vetmic.2009.11.028
Gould, L. H., Mody, R. K., Ong, K. L., Clogher, P., Cronquist, A. B., Garman, K. N., et al. (2013). Increased recognition of non-O157 Shiga toxin-producing Escherichia coli infections in the United States during 2000-2010: epidemiologic features and comparison with E. coli O157 Infections. Foodborne Pathog. Dis. 10, 453–460. doi: 10.1089/fpd.2012.1401
Haugum, K., Johansen, J., Gabrielsen, C., Brandal, L. T., Bergh, K., Ussery, D. W., et al. (2014). Comparative genomics to delineate pathogenic potential in Non-O157 shiga toxin-producing Escherichia coli (STEC) from Patients with and without Haemolytic Uremic Syndrome (HUS) in Norway. PLoS ONE 9:e111788. doi: 10.1371/journal.pone.0111788
Henderson, I. R., Czeczulin, J., Eslava, C., Noriega, F., and Nataro, J. P. (1999). Characterization of Pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun. 67, 5587–5596.
Horvath, P., and Barrangou, R. (2010). CRISPR/Cas, the immune system of bacteria and archaea. Science 327, 167–170. doi: 10.1126/science.1179555
Hussein, H. S. (2007). Prevalence and pathogenicity of Shiga toxin-producing Escherichia coli in beef cattle and their products. J. Anim. Sci. 85, E63–E72. doi: 10.2527/jas.2006-421
Iyoda, S., Manning, S. D., Seto, K., Kimata, K., Isobe, J., Etoh, Y., et al. (2014). Phylogenetic Clades 6 and 8 of enterohemorrhagic Escherichia coli O157:H7 with particular stx subtypes are more frequently found in isolates from hemolytic uremic syndrome patients than from asymptomatic carriers. Open Forum Infect. Dis. 1:ofu061. doi: 10.1093/ofid/ofu061
Jaureguy, F., Landraud, L., Passet, V., Diancourt, L., Frapy, E., Guigon, G., et al. (2008). Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genomics 9:560. doi: 10.1186/1471-2164-9-560
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Joensen, K. G., Tetzschner, A. M., Iguchi, A., Aarestrup, F. M., and Scheutz, F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53, 2410–2426. doi: 10.1128/JCM.00008-15
Ju, W., Cao, G., Rump, L., Strain, E., Luo, Y., Timme, R., et al. (2012). Phylogenetic analysis of non-O157 shiga toxin-producing Escherichia coli strains by whole-genome sequencing. J. Clin. Microbiol. 50, 4123–4127. doi: 10.1128/JCM.02262-12
Kaper, J. B., Mellies, J. L., and Nataro, J. P. (1999). “Pathogenicity Islands and Other Mobile Genetic Elements of Diarrheagenic Escherichia coli,” in Pathogenicity Islands and Other Mobile Virulence Elements, eds J. B. Kaper and J. Hacker (Washington, DC: American Society of Microbiology; ASM Press), 33–58.
Karmali, M. A., Petric, M., Lim, C., Fleming, P. C., Arbus, G. S., and Lior, H. (1985). The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J. Infect. Dis. 151, 775–782. doi: 10.1093/infdis/151.5.775
Lambert, D., Carrillo, C. D., Koziol, A. G., Manninger, P., and Blais, B. W. (2015). GeneSippr: a rapid whole-genome approach for the identification and characterization of foodborne pathogens such as priority shiga toxigenic Escherichia coli. PLoS ONE 10:e0122928. doi: 10.1371/journal.pone.0122928
Leyton, D. L., Sloan, J., Hill, R. E., Doughty, S., and Hartland, E. L. (2003). Transfer region of pO113 from enterohemorrhagic Escherichia coli: similarity with R64 and identification of a novel plasmid-encoded autotransporter, EpeA. Infect. Immun. 71, 6307–6319. doi: 10.1128/IAI.71.11.6307-6319.2003
Lienemann, T., Salo, E., Rimhanen-Finne, R., Ronnholm, K., Taimisto, M., Hirvonen, J. J., et al. (2012). Shiga toxin-producing Escherichia coli serotype O78:H− in family, Finland, 2009. Emerg. Infect. Dis. 18, 577–581. doi: 10.3201/eid1804.111310
Mellmann, A., Harmsen, D., Cummings, C. A., Zentz, E. B., Leopold, S. R., Rico, A., et al. (2011). Prospective genomic characterization of the german enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS ONE 6:e22751. doi: 10.1371/journal.pone.0022751
Miko, A., Delannoy, S., Fach, P., Strockbine, N. A., Lindstedt, B. A., Mariani-Kurkdjian, P., et al. (2013). Genotypes and virulence characteristics of Shiga toxin-producing Escherichia coli O104 strains from different origins and sources. Int. J. Med. Microbiol. 303, 410–421. doi: 10.1016/j.ijmm.2013.05.006
Mora, A., Herrrera, A., Lopez, C., Dahbi, G., Mamani, R., Pita, J. M., et al. (2011). Characteristics of the Shiga-toxin-producing enteroaggregative Escherichia coli O104:H4 German outbreak strain and of STEC strains isolated in Spain. Int. Microbiol. 14, 121–141. doi: 10.2436/20.1501.01.142
Ogura, Y., Ooka, T., Iguchi, A., Toh, H., Asadulghani, M., Oshima, K., et al. (2009). Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 106, 17939–17944. doi: 10.1073/pnas.0903585106
Paddock, Z. D., Bai, J., Shi, X., Renter, D. G., and Nagaraja, T. G. (2013). Detection of Escherichia coli O104 in the feces of feedlot cattle by a multiplex PCR assay designed to target major genetic traits of the virulent hybrid strain responsible for the 2011 German outbreak. Appl. Environ. Microbiol. 79, 3522–3525. doi: 10.1128/AEM.00246-13
Paton, A. W., Woodrow, M. C., Doyle, R. M., Lanser, J. A., and Paton, J. C. (1999). Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J. Clin. Microbiol. 37, 3357–3361.
Persson, S., Olsen, K. E. P., Ethelberg, S., and Scheutz, F. (2007). Subtyping method for Escherichia coli Shiga Toxin (Verocytotoxin) 2 variants and correlations to clinical manifestations. J. Clin. Microbiol. 45, 2020–2024. doi: 10.1128/JCM.02591-06
Rasko, D. A., Webster, D. R., Sahl, J. W., Bashir, A., Boisen, N., Scheutz, F., et al. (2011). Origins of the E. coli strain causing an outbreak of hemolytic–uremic Syndrome in Germany. N. Engl. J. Med. 365, 709–717. doi: 10.1056/NEJMoa1106920
Rohde, H., Qin, J., Cui, Y., Li, D., Loman, N. J., Hentschke, M., et al. (2011). Open-source genomic analysis of shiga-toxin–producing E. coli O104:H4. N. Engl. J. Med. 365, 718–724. doi: 10.1056/NEJMoa1107643
Scheutz, F., Møller Nielsen, E., Frimodt-Møller, J., Boisen, N., Morabito, S., Tozzoli, R., et al. (2011). Characteristics of the enteroaggregative Shiga toxin/verotoxin-producing Escherichia coli O104:H4 strain causing the outbreak of haemolytic uraemic syndrome in Germany, May to June 2011. Eurosurveillance 16, 19889. doi: 10.2807/ese.16.24.19889-en
Schmidt, H., and Karch, H. (1996). Enterohemolytic phenotypes and genotypes of shiga toxin-producing Escherichia coli O111 strains from patients with diarrhea and hemolytic-uremic syndrome. J. Clin. Microbiol. 34, 2364–2367.
Shridhar, P. B. (2017). Detection, Quantification and Genetic Characterization of Six Major Non-O157 Shiga Toxinproducing Escherichia coli Serogroups and E. coli O104 in Feedlot Cattle Feces. Dissertation/Doctoral, Kansas State University.
Shridhar, P. B., Noll, L. W., Shi, X., Cernicchiaro, N., Renter, D. G., Bai, J., et al. (2016). Escherichia coli O104 in feedlot cattle feces: prevalence, isolation and characterization. PLoS ONE 11:e0152101. doi: 10.1371/journal.pone.0152101
Silva, L. E., Souza, T. B., Silva, N. P., and Scaletsky, I. C. (2014). Detection and genetic analysis of the enteroaggregative Escherichia coli heat-stable enterotoxin (EAST1) gene in clinical isolates of enteropathogenic Escherichia coli (EPEC) strains. BMC Microbiol. 14:135. doi: 10.1186/1471-2180-14-135
Toro, M., Cao, G., Ju, W., Allard, M., Barrangou, R., Zhao, S., et al. (2014). Association of clustered regularly interspaced short palindromic repeat (CRISPR) elements with specific serotypes and virulence potential of shiga toxin-producing Escherichia coli. Appl. Environ. Microbiol. 80, 1411–1420. doi: 10.1128/AEM.03018-13
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15, 524. doi: 10.1186/s13059-014-0524-x,
USDA-FSIS (2011). “USDA-FSIS. Shiga toxin producing E. coli in certain raw beef products,” in Federal Register (Washington, DC).
Wang, J., Stanford, K., McAllister, T. A., Johnson, R. P., Chen, J., Hou, H., et al. (2016). Biofilm formation, virulence gene profiles, and antimicrobial resistance of nine serogroups of non-O157 Shiga Toxin-Producing Escherichia coli. Foodborne Pathog. Dis. 13, 316–324. doi: 10.1089/fpd.2015.2099
Wieler, L. H., Semmler, T., Eichhorn, I., Antao, E. M., Kinnemann, B., Geue, L., et al. (2011). No evidence of the Shiga toxin-producing E. coli O104:H4 outbreak strain or enteroaggregative E. coli (EAEC) found in cattle faeces in northern Germany, the hotspot of the 2011 HUS outbreak area. Gut Pathog 3:17. doi: 10.1186/1757-4749-3-17
Wilkerson, C., Samadpour, M., van Kirk, N., and Roberts, M. C. (2004). Antibiotic resistance and distribution of tetracycline resistance genes in Escherichia coli O157:H7 isolates from humans and bovines. Antimicr. Agents Chemother. 48, 1066–1067. doi: 10.1128/AAC.48.3.1066-1067.2004
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x
Yan, X., Fratamico, P. M., Bono, J. L., Baranzoni, G. M., and Chen, C.-Y. (2015). Genome sequencing and comparative genomics provides insights on the evolutionary dynamics and pathogenic potential of different H-serotypes of Shiga toxin-producing Escherichia coli O104. BMC Microbiol. 15:83. doi: 10.1186/s12866-015-0413-9
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zhang, Q., and Ye, Y. (2017). Not all predicted CRISPR–Cas systems are equal: isolated cas genes and classes of CRISPR like elements. BMC Bioinformatics 18:92. doi: 10.1186/s12859-017-1512-4
Zhang, X., Cheng, Y., Xiong, Y., Ye, C., Zheng, H., Sun, H., et al. (2012). Enterohemorrhagic Escherichia coli specific enterohemolysin induced IL-1β in human macrophages and EHEC-Induced IL-1β required activation of NLRP3 inflammasome. PLoS ONE 7:e50288. doi: 10.1371/journal.pone.0050288
Keywords: E. coli O104, cattle feces, serotypes, whole genome sequencing, virulence genes
Citation: Shridhar PB, Patel IR, Gangiredla J, Noll LW, Shi X, Bai J, Elkins CA, Strockbine NA and Nagaraja TG (2018) Genetic Analysis of Virulence Potential of Escherichia coli O104 Serotypes Isolated From Cattle Feces Using Whole Genome Sequencing. Front. Microbiol. 9:341. doi: 10.3389/fmicb.2018.00341
Received: 15 December 2017; Accepted: 12 February 2018;
Published: 01 March 2018.
Edited by:
Jorge Blanco, Universidade de Santiago de Compostela, SpainReviewed by:
Patrick Fach, Agence Nationale de Sécurité Sanitaire de l'Alimentation, de l'Environnement et du Travail (ANSES), FranceJames Michael Fleckenstein, Washington University in St. Louis, United States
Copyright © 2018 Shridhar, Patel, Gangiredla, Noll, Shi, Bai, Elkins, Strockbine and Nagaraja. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: T. G. Nagaraja, dG5hZ2FyYWpAdmV0Lmstc3RhdGUuZWR1
 Pragathi B. Shridhar1
Pragathi B. Shridhar1 Isha R. Patel
Isha R. Patel Jayanthi Gangiredla
Jayanthi Gangiredla Jianfa Bai
Jianfa Bai Nancy A. Strockbine
Nancy A. Strockbine T. G. Nagaraja
T. G. Nagaraja