Health-Associated Niche Inhabitants as Oral Probiotics: The Case of Streptococcus dentisani
- Division of Oral Microbiology and Immunology, Department of Operative and Preventive Dentistry and Periodontology, RWTH Aachen University Hospital, Aachen, Germany
A commentary on
Health-Associated Niche Inhabitants as Oral Probiotics: The Case of Streptococcus dentisani
by López-López, A., Camelo-Castillo, A., Ferrer, M. D., Simon-Soro, Á., and Mira, A. (2017). Front. Microbiol. 8:379. doi: 10.3389/fmicb.2017.00379
We comment on López-López et al. and their contributions on Streptococcus dentisani, especially the species-specific primers, buffering capacity, and prevalence in the oral cavity of healthy individuals (Camelo-Castillo et al., 2014; López-López et al., 2017), and its infective potential. S. dentisani was effectively reclassified as Streptococcus oralis subspecies dentisani by Jensen et al. (2016) and there exist two other closely related subspecies, S. oralis ssp. oralis and S. oralis ssp. tigurinus (Zbinden et al., 2012; Jensen et al., 2016; Conrads et al., 2017). López-López et al. studied the capacity of strains 7746 and 7747T for buffering low pH and thus preventing overgrowth of acidogenic and aciduric cariogenic species such as Streptococcus mutans. In addition, they developed two PCRs to quantify S. oralis ssp. dentisani in clinical specimens, one directed against the carbamate kinase gene arcC and one against ORF540, the latter coding for a bacteriocin-related protein. Primer specificity was evaluated with DNA from S. mutans, S. sobrinus, S. sanguinis, S. salivarius, S. mitis, S. pneumoniae, S. infantis, and S. oralis ATCC 35037T. They conclude that arcC-primers did not amplify any of the related streptococcal species and ORF540-primers cross-reacted with S. pneumoniae only.
With the intention of screening our own strain collection for the presence of S. oralis ssp. dentisani, we tested these PCR primers with the type strains of S. oralis ssp. dentisani 7747T, ssp. oralis ATCC 35037T, ssp. tigurinus Az3aT, six S. mitis strains (OMI 181-184, 187-188) and four S. pneumoniae strains (OMI 157-159, 186). The arcC-directed PCR (Figure 1A, CkSd-F/R) was indeed very specific but the product was very short (76 bp) and we designed the alternative CkSdAlt-F/R primer pair amplifying a longer fragment of 175 bp (Figure 1A). The ORF540-directed PCR was found to be more bacteriocin-type than species-directed. Furthermore, we designed a 16S rRNA-gene-directed PCR expanding the options for application (Figure 1A, SDent-16S-F/R). Applying the two Ck- and the 16S-directed PCR, we screened our strain collection of clade S. oralis strains. We found four S. oralis ssp. dentisani strains (SN39325, SN54787, SN54788, SN58364) among isolates of proven and epidemiologically unrelated cases of infective endocarditis (National Reference Center for Streptococci, RWTH Aachen University Hospital, Germany; described in Conrads et al., 2017), and two strains (OMI 214 and OMI 215) isolated from the tooth surface of healthy caries-free probands. Next, we characterized these strains genetically by sequencing the 16S-rRNA gene (Figure 1B) and phenotypically in-vitro for their buffering capacity (Figure 1C). The buffering phenotype is dependent on the arginine deiminase system, which is determined by arcA (arginine deiminase), arcB (ornithine carbamoyltransferase), and arcC (carbamate kinase). The system is activated by a pH drop and leads to ammonia formation and neutralization of the environment and is—besides bacteriocin production—the reason for the probiotic and anti-cariogenic nature. López-López et al. tested the buffer capacity of strain 7746 only under aerobic conditions (A. Mira, personal communication). However, the atmospheric conditions in the oral cavity and especially on tooth surfaces are rarely truly aerobic, but instead something between micro-aerobic, CO2-rich, and anaerobic. Thus, we reproduced the growth-experiments in the same arginine-enriched BHI-medium and applying the same protocol but this time under aerobic, CO2-rich (7%), and anaerobic conditions. Together with our six S. oralis ssp. dentisani strains, we used S. oralis ssp. dentisani (7746, 7747T) and S. gordonii strains (OMI 231, GH 355) as positive controls of a functional arginine deiminase system. S. oralis ssp. oralis and ssp. tigurinus, as well as un-inoculated BHI (plus/minus arginine), served as negative controls. Clearly, all our six isolates demonstrated buffering activity, even more strongly pronounced than with the strains 7746 and 7747T (Figure 1C). An increase of buffering activity was detected for S. oralis ssp. dentisani strains from CO2 (Δ pH 0.59–1.65, mean 1.36, median 1.43) to aerobic (Δ pH 0.56–2.22, mean 1.64, median 1.73) to anaerobic (Δ pH 1.41–1.85, mean 1.74, median 1.79) conditions. This increased activity under anaerobic conditions was significant for 5 of the 8 S. oralis ssp. dentisani strains tested. In time-curve experiments we confirmed that the alkalization process in arginine-supplemented BHI medium started between 6 and 8 h after inoculation but in the atmosphere of 7% CO2 it needed a pH as low as ≤ 5.5 to be induced. This finding has clinical impact since under microaerophilic, CO2-rich conditions, re-alkalization might start later than de-mineralization.
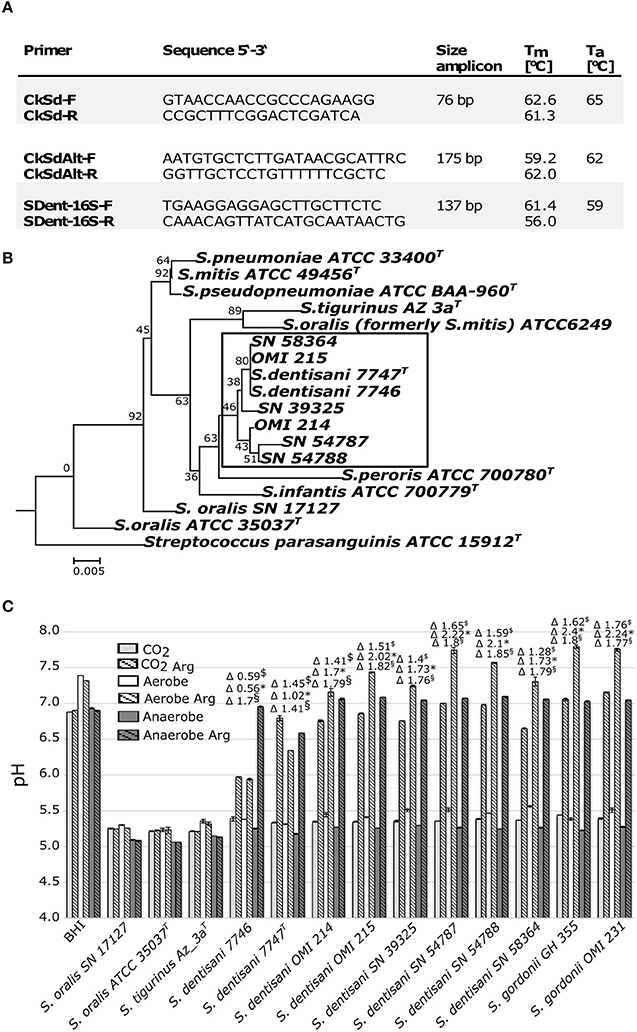
Figure 1. (A) Primer and conditions to specifically amplify S. oralis subspecies dentisani. (B) Neighbor-Joining phylogenetic tree based on the 16S rRNA-gene of 17 Mitis group streptococci including strains subjected in this study using MEGA6 with default settings and 200 bootstraps. SN-strains are endocarditis isolates of our collection. S. parasanguinis ATCC 15912T was used as outgroup. (C) Endpoint pH values and Δ pH values of S. oralis subspecies dentisani and related (sub-)species after growth over 16 h in BHI with and without the addition of arginine monohydrate (Arg, 5 gl−1) and under 7% CO2, aerobic and anaerobic conditions. $Δ pH 7% CO2, *Δ pH aerobic, §Δ pH anaerobic, BHI, Brain Heart Infusion medium control. The three S. oralis subspecies are abbreviated as S. dentisani, S. oralis, and S. tigurinus.
Finally, applying the arcC- and 16S-subspecies specific primers for real-time PCR-quantification, we subjected 10 saliva samples (1.5 ml freshly stimulated saliva processed to 100 μl DNA extract) of healthy, caries-free probands (aged 23–32, randomly selected from an ethically approved past study (Conrads et al., 2017). In principal, the arcC- and 16S-directed primers led to the same cell numbers which were between 1.2 × 101 and 1.2 × 105 cells per μl DNA extract or 8.0 × 102 and 8.1 × 106 per ml saliva. López-López et al. described much higher numbers (1.04 × 107 and 6.94 × 107) but they were derived from the pooled dental plaque of two individuals. Combined with the results of a universal bacterial 16S-directed PCR (modified from Nadkarni et al., 2002), we calculated the relative numbers of S. oralis ssp. dentisani within our saliva samples as between 0.01 and 10.46% (mean 1.73%, median 0.13%).
In conclusion, we support the findings of López-López et al. but with the following limitations: (i) the ORF450-directed primer showed low accuracy. We have designed alternative primers amplifying a longer arcC-fragment and a V1-V2-fragment of the 16S, expanding the applications; (ii) the induction and potential of alkalization of S. oralis ssp. dentisani is dependent on the atmosphere, with best results obtained under anaerobic conditions; (iii) as we found four endocarditis-associated strains in our collection and a Danish group recently even found six (Rasmussen et al., 2017), it must be indicated that some strains of S. oralis ssp. dentisani might bear the potential for causing infective endocarditis in immunocompromised patients. The challenge thus is to find—or produce—a strain with the most probiotic and the least infective potential.
Author Contributions
GC supervised the study, wrote the commentary, and is the corresponding author. JB performed experiments on the phenotypic characterization of S. dentisani isolates. CK performed experiments on the genotypic characterization of S. dentisani isolates and designed the PCR primers. MA checked genomic data and all results for plausibility and commented on and corrected the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank Mark van der Linden (National Reference Center for Streptococci, RWTH Aachen University Hospital, Aachen, Germany) for providing the IE-related strains and Mrs. Beate Melzer for her excellent technical assistance.
References
Camelo-Castillo, A., Benitez-Paez, A., Belda-Ferre, P., Cabrera-Rubio, R., and Mira, A. (2014). Streptococcus dentisani sp. nov., a novel member of the mitis group. Int. J. Syst. Evol. Microbiol. 64, 60–65. doi: 10.1099/ijs.0.054098-0
Conrads, G., Barth, S., Möckel, M., Lenz, L., van der Linden, M., and Henne, K. (2017). Streptococcus tigurinus is frequent among gtfR-negative Streptococcus oralis isolates and in the human oral cavity, but highly virulent strains are uncommon. J. Oral Microbiol. 9:1307079. doi: 10.1080/20002297.2017.1307079
Jensen, A., Scholz, C. F., and Kilian, M. (2016). Re-evaluation of the taxonomy of the Mitis group of the genus Streptococcus based on whole genome phylogenetic analyses, and proposed reclassification of Streptococcus dentisani as Streptococcus oralis subsp. dentisani comb. nov., Streptococcus tigurinus as Streptococcus oralis subsp. tigurinus comb. nov., and Streptococcus oligofermentans as a later synonym of Streptococcus cristatus. Int. J. Syst. Evol. Microbiol. 66, 4803–4820. doi: 10.1099/ijsem.0.001433
López-López, A., Camelo-Castillo, A., Ferrer, M. D., Simon-Soro, Á., and Mira, A. (2017). Health-associated niche inhabitants as oral probiotics: the case of Streptococcus dentisani. Front. Microbiol. 8:379. doi: 10.3389/fmicb.2017.00379
Nadkarni, M. A., Martin, F. E., Jacques, N. A., and Hunter, N. (2002). Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148, 257–266. doi: 10.1099/00221287-148-1-257
Rasmussen, L. H., Højholt, K., Dargis, R., Christensen, J. J., Skovgaard, O., Justesen, U. S., et al. (2017). In silico assessment of virulence factors in strains of Streptococcus oralis and Streptococcus mitis isolated from patients with infective endocarditis. J. Med. Microbiol. 66, 1316–1323. doi: 10.1099/jmm.0.000573
Keywords: Streptococcus oralis subspecies dentisani, probiotics, dental caries, pH buffering, arginolytic pathway, PCR detection
Citation: Conrads G, Bockwoldt JA, Kniebs C and Abdelbary MMH (2018) Commentary: Health-Associated Niche Inhabitants as Oral Probiotics: The Case of Streptococcus dentisani. Front. Microbiol. 9:340. doi: 10.3389/fmicb.2018.00340
Received: 16 November 2017; Accepted: 12 February 2018;
Published: 27 February 2018.
Edited by:
Giovanna Batoni, University of Pisa, ItalyReviewed by:
Marcelle Nascimento, University of Florida Health, United StatesCopyright © 2018 Conrads, Bockwoldt, Kniebs and Abdelbary. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Georg Conrads, Z2NvbnJhZHNAdWthYWNoZW4uZGU=
 Georg Conrads
Georg Conrads Julia A. Bockwoldt
Julia A. Bockwoldt Caroline Kniebs
Caroline Kniebs Mohamed M. H. Abdelbary
Mohamed M. H. Abdelbary