
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 08 February 2018
Sec. Microbial Physiology and Metabolism
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.00159
This article is part of the Research Topic Using Genomics, Metagenomics and Other "Omics" to Assess Valuable Microbial Ecosystem Services and Novel Biotechnological Applications View all 53 articles
Osmotolerance is one of the critical factors for successful survival and colonization of microbes in saline environments. Nonetheless, information about these osmotolerance mechanisms is still inadequate. Exploration of the saline soil microbiome for its community structure and novel genetic elements is likely to provide information on the mechanisms involved in osmoadaptation. The present study explores the saline soil microbiome for its native structure and novel genetic elements involved in osmoadaptation. 16S rRNA gene sequence analysis has indicated the dominance of halophilic/halotolerant phylotypes affiliated to Proteobacteria, Actinobacteria, Gemmatimonadetes, Bacteroidetes, Firmicutes, and Acidobacteria. A functional metagenomics approach led to the identification of osmotolerant clones SSR1, SSR4, SSR6, SSR2 harboring BCAA_ABCtp, GSDH, STK_Pknb, and duf3445 genes. Furthermore, transposon mutagenesis, genetic, physiological and functional studies in close association has confirmed the role of these genes in osmotolerance. Enhancement in host osmotolerance possibly though the cytosolic accumulation of amino acids, reducing equivalents and osmolytes involving BCAA-ABCtp, GSDH, and STKc_PknB. Decoding of the genetic elements prevalent within these microbes can be exploited either as such for ameliorating soils or their genetically modified forms can assist crops to resist and survive in saline environment.
Soil is a rich and dynamic ecosystem, containing a vast number of microorganisms (van Veen et al., 1997). Geological activities like weathering of rocks, winds and poor agricultural practice are continuously increasing salt contents of the soils (Jiang et al., 2007; Canfora et al., 2014). Enhanced soil salinity modulates the microbial community structure and its physiological activity (Jiang et al., 2007; Canfora et al., 2014; Shrivastava and Kumar, 2015). The majority of microbes surviving in salt stress conditions demonstrate osmotolerance for varying duration, which may extend even to their entire lifespan (Roberts, 2005). The salt stress tolerance mechanisms are complex phenomena where pathways are coordinately linked (Culligan et al., 2012). These metabolic strengths to mitigate osmotic stress, seem to be genetically evolved through horizontal gene transfer (Koonin and Wolf, 2012; Yan et al., 2015; Gupta et al., 2017). Description of these osmotolerance mechanisms is crucial for comprehensive understanding of the biology of saline soil microbes, and exploiting them for their applications in improving soil quality and crop yields (Xiao and Roberts, 2010; Zhengbin et al., 2011; Culligan et al., 2012, 2013, 2014; Fernandes, 2014). A variety of culture dependent studies have been carried out to decode the gene(s) involved in osmotolerance within halophilic or halotolerant microbes (Zuleta et al., 2003; Klähn et al., 2009; Naughton et al., 2009; Meena et al., 2017). These studies have deciphered the role of proteins, Na+/H+ pumps, compatible solutes in salt stress tolerance (Sakamoto and Murata, 2002; Roberts, 2005). However, culture independent approach provides a vast opportunity for searching salt tolerant gene (s) (Singh J. et al., 2009; Mirete et al., 2015; Kumar J. et al., 2016; Chauhan et al., 2017; Gupta et al., 2017). Only a few studies have used metagenomic approach to decode the microbial salt stress tolerance mechanisms from various environments like pond water (Kapardar et al., 2010a,b), brines and moderate-salinity rhizosphere (Mirete et al., 2015), human gut microbiome (Culligan et al., 2012, 2013, 2014). Incidentally, the number of genes/pathways identified for salt stress tolerance are far below than the number of microbes which have been identified to reside within these environments (Humbert et al., 2009; Mirete et al., 2015). Hereby, the current study was proposed to identify the genetic machinery used by microbes as survival strategies in salt stress condition using functional metagenomic approach. The current study led to the identification of a number of osmotolerant genes that could be used to develop strategies to ensure survival of microbes under saline conditions.
Saline soil samples were collected in sterile containers after carefully removing the surface layer (up to 10 cm) from Village Malab, District Nuh situated at 28.0107°N, 77.0564°E. Metagenomic DNA was extracted from 5 g of the soil sample (Supplementary Methods).
Bacterial strains and plasmids used in the study are listed in Table 1. The oligonucleotides used in the study (GeNoRime, Shrimpex Biotech services Pvt. Ltd. India) are listed in Supplementary Table S1. Escherichia coli (DH10B) and E. coli (MKH13) strains were cultured in Luria-Bertani (LB) medium. Further, E. coli (DH10B) and E. coli (MKH13) strains containing pUC19 vector were cultured in LB medium supplemented with ampicillin (100 μg ml−1). All overnight cultures were grown in LB broth at 37°C with constant shaking at 200 rpm.
Saline soil metagenomic DNA was used to amplify the SSU rRNA gene (Supplementary Methods). The amplified product was used for next generation sequencing (NGS) with the aid of Roche 454 GS FLX+ platform (Morowitz et al., 2011; Gupta et al., 2017). Finally, Quantitative Insights Into Microbial Ecology (QIIME) 1.9.0 pipeline was implemented for SSU rRNA sequence data analysis (Caporaso et al., 2011). SSU rRNA gene sequence data was curated for quality, length and ambiguous bases as a quality filtering step. Each sample was pre-processed to remove sequences with length less than 200 nucleotides and more than 1,000 nucleotides and sequences with minimum average quality <25. Reads with ambiguities and barcode mismatch were discarded. Reads were assigned to operational taxonomic units (OTUs) using a closed reference OTU picking protocol using QIIME. The uclust was applied to search sequences against a subset of the Greengenes database, version 13_8 filtered at 97% sequence identity. The OTUs were classified taxonomically by using the Greengenes reference database at various taxonomic ranks (phylum, class, order, family, genus, and species).
Plasmid borne saline soil metagenomic library was prepared in E. coli DH10B using pUC19 vector (Supplementary Methods) (Chauhan et al., 2009, 2017) and manually screened for salt stress tolerant clones (Kapardar et al., 2010a,b). Salt stress resistant clones were screened by plating the soil metagenomic library (~165,000 clones with an average insert of 1.89 Kb) on LB agar medium supplemented with ampicillin (100 μg ml−1) and NaCl [5.8% (w/v)]. The 5.8% of NaCl (w/w) is a lethal concentration for E. coli DH10B cells and will allow the growth of only osmotolerant clones. RFLP analysis of salt stress tolerant clones was performed after digesting their recombinant plasmid DNA with EcoRI & HindIII at 37°C for 12 h. The minimum inhibitory concentration assay and growth inhibition studies were performed to analyze the salt stress tolerance property (Kapardar et al., 2010a,b). Growth inhibition assays of salt sensitive E. coli MKH13 clones were performed with 3% NaCl (w/v) & 3.7% KCl (w/v), while 5.8% NaCl (w/v) & 5.5 % of KCl (w/v) were used for E. coli DH10B clones. Graphs (created using Origin61) are presented as the average of triplicate experiments, with error bars being representative of the standard error of the mean.
The plasmid insert from salt resistant recombinant clones were sequenced using Sanger sequencing chemistry with primer walking approach at Eurofins Genomics India Pvt. Ltd (Bangalore, India). Sequence assembly was performed with Seq-Man sequence assembly software Lasergene package, version 5.07 (DNA Star, USA). Putative open reading frame (ORF) was predicted using an ORF finder tool at NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and checked for the database homology with Basic Local Alignment and Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/blast). Encoded protein sequences were analyzed for the presence of conserved domains (CDD) (Marchler-Bauer et al., 2014), topology prediction (HMMTOP) (Tusnády and Simon, 2001), phylogenetic analysis (MEGA7) (Kumar S. et al., 2016), and various physiological parameters (Tsirigos et al., 2015). Transposon mutagenesis of pSSR1, pSSR4, pSSR6, and pSSR21 was carried out with EZ-Tn5™ <Kan-2> Insertion kit (Epicenter Biotechnologies) following manufacturer's instructions. Transposon mutants of pSSR1, pSSR4, pSSR6, and pSSR21 were screened for the salt stress resistant and sensitive phenotypes to identify the active osmotolerant genomic regions within the cloned DNA fragment in pSSR1, pSSR4, pSSR6, and pSSR21. Salt tolerant active loci encoding putative BCAA-ABCtp, GSDH, STK_Pknb, and DUF3445 genes of pSSR1, pSSR4, pSSR6, and pSSR21 were subcloned in pUC19 vector (E. coli MKH13 host) using standard molecular cloning techniques. The growth studies of subclones were performed to analyze their salt stress maintenance property in the presence of salt stressors NaCl [3.0% (w/v)] and KCl [3.7% (w/v)]. All assays were performed in triplicates for calculation of standard deviation. A parametric t-test was used to calculate the p-value.
Elemental Quantification of intracellular Na+ in E. coli MKH13 carrying the empty vector (pUC19) and salt tolerant recombinant subclones (SSR1C1, SSR4C1, SSR6C1, SSR21C1) was measured with inductively coupled plasma spectroscopy-atomic emission spectroscopy (ICP-AES) analysis (Mirete et al., 2015) at SAIF, IIT Bombay, India. Results were expressed as mg of Na+ g−1 dry weight of cells. A parametric t-test was used to calculate the p-value.
Sequence reads generate in present study has been deposited in the NCBI SRA under accession number SRS2727172.
Physico-chemical properties of saline soil showed that the pH of soil was 9.0 ± 0.025, while its electrical conductivity (EC) was 6.5 ± 0.023. Elemental analysis of soil showed the excessive presence of salts, sodium (105 ppm), potassium (155 ppm), and lithium (188 ppm), confirming its moderate saline nature. A good quality (A260/280 >1.8), high molecular weight (>23 Kb) metagenomic DNA was extracted from the saline soil sample. Saline soil metagenomic DNA was used to analyze its SSU rRNA gene sequences to decode its native microbiome structure. Clustering of SSU rRNA gene identified a total of 487 OTUs distributed across seven microbial phyla (Figure 1). Out of 487 OTUs, we observed 153 unique OTUs (Supplementary Table S2). The inferred phylogeny of the soil microbiome based upon 153 unique OTUs (Figure 1) was comparable to the taxonomic classifications against with greengenes database, with most of the diversity of the microbiome being attributed to phyla Proteobacteria. The phylogeny was visualized by iTOL (Letunic and Bork, 2016). The dominant microbial phyla were Proteobacteria, Actinobacteria, Gemmatimonadetes, Bacteroidetes, Firmicutes, and Acidobacteria (Figure 1). Among these phyla, the majority of sequences were affiliated to Proteobacteria (43.7%) having a representation of Alphaproteobacteria (38.9%), Betaproteobacteria (7%), Deltaproteobacteria (10.7%), and Gammaproteobacteria (43.2%); followed by Actinobacteria (21.8%) showing presence of Acidimicrobiia (72.47%) and Nitriliruptoria (18.8%); Bacteroidetes (18.1%) having a proportionate representation of Rhodothermi (51.9%), Flavobacteriia (25.96%), Cytophagia (19.3%), and Gemmatimonadetes (11.3%) with a percentage representation of Gemm-2 (60.17%), Gemm-4 (12.3%), Gemm-1 (3.5%). Simultaneously, a minor fraction of sequences was affiliated to Acidobacteria (2.5%), Firmicutes (2.1%), and Nitrospirae (0.6%) microbial groups within saline soil microbiome. The taxonomic classification of saline soil microbiome confirms that a majority of microbial taxa belongs to phylum Proteobacteria (Figure 1) (Supplementary Table S2).
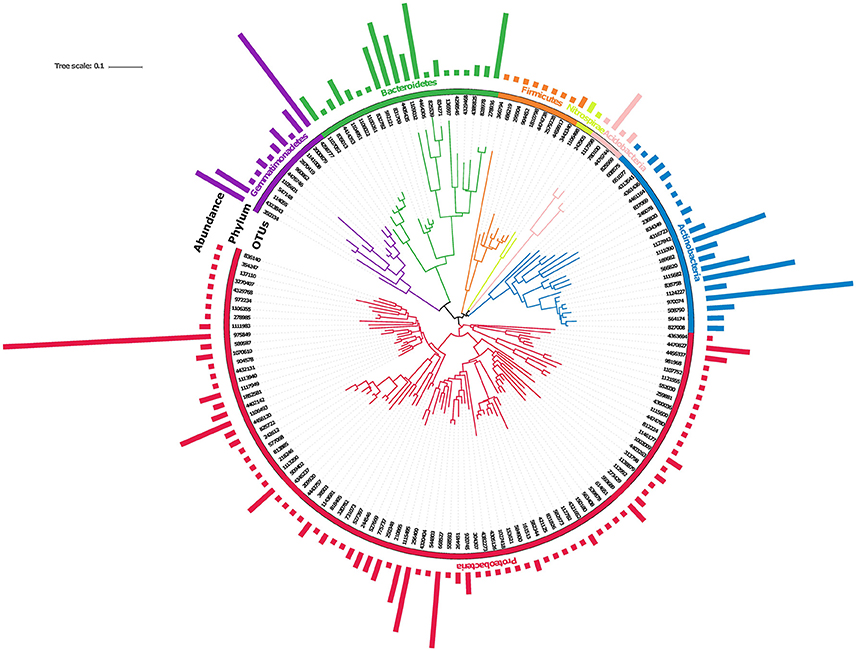
Figure 1. Cladogram of operational taxonomic units (OTUs) identified by SSU rRNA gene sequence analysis.
A saline soil metagenomic library was constructed with a total representation of 312 MB of cloned soil microbiome DNA. Primary screening of a saline soil metagenomic library at 5.8% NaCl (w/v) led to the identification of 24 salt stress tolerant clones. However, RFLP analysis indicated the presence of only four unique recombinant plasmids, labeled as pSSR1, pSSR4, pSSR6, and pSSR21. Minimum inhibitory concentration analysis showed almost two fold higher salt stress tolerance of SSR1, SSR4, SSR6, and SSR21 clones in comparison to the control E. coli (DH10B) (Supplementary Figure S1). The SSR1, SSR4, SSR6, and SSR21 also showed a statistically significant (P = 0.0009, P = 0.0003, P = 0.0014, P = 0.004) growth advantage in the presence of NaCl [4.0% (w/v)] (Figure 2A) and KCl [5.5% (w/v)] (P = 0.0045, P = 0.0008, P = 0.0486, P = 0.0022) as compared to E. coli (DH10B) strain carrying empty plasmid vector (pUC19) (Figure 2B), whereas no significant growth difference was observed between SSR1, SSR4, SSR6, SSR21, and native host E. coli (DH10B) carrying pUC19 in the presence of LB broth only (Figure 2C). Simultaneously pSSR1, pSSR4, pSSR6, and pSSR21 successfully complemented salt stress tolerance property within salt sensitive E. coli (MKH13) strain and showed a statistically significant growth advantage in the presence of NaCl [3.0% (w/v)] (P = 0.0007, P = 0.0002, P = 0.0003, P = 0.0001) (Figure 3A) and KCl [3.7% (w/v)] (P = 0.0003, P = 0.0008, P = 0.0003, P = 0.0023) (Figure 3B), as compared to E. coli (MKH13) strain carrying empty plasmid vector (pUC19). At the same time, no significant growth difference was observed between E. coli (MKH13) strain harboring pSSR1, pSSR4, pSSR6, pSSR21 as compared to E. coli (MKH13) strain carrying empty plasmid vector in the presence of LB broth only (Figure 3C).
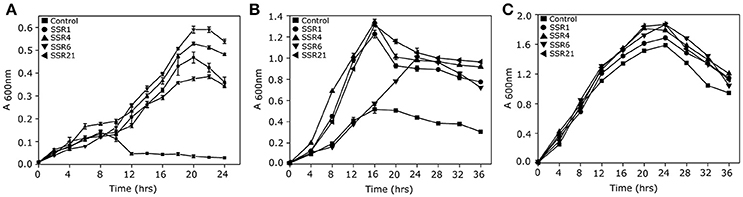
Figure 2. Characterization of salt stress tolerant clones for osmotolerance property. Growth of E. coli (DH10B) metagenomic clones SSR1 (•), SSR4 (▴), SSR6 (▾),SSR21(◂), and E. coli (DH10B) host strain carrying empty plasmid vector (■) in (A) LB broth supplemented with 4.0% NaCl (w/v), (B) LB broth supplemented with 5.5% KCl and (C) LB broth.
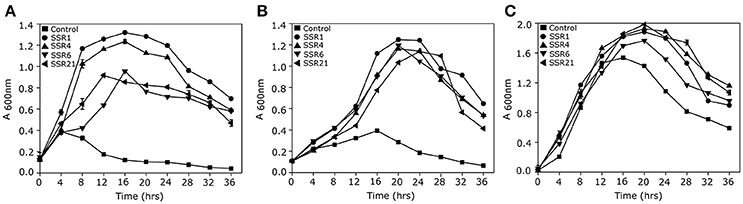
Figure 3. Complementation of osmotolerance property of salt stress tolerant clones. Growth of E. coli (MKH13) metagenomic clones harboring pSSR1 (•), pSSR4(▴), pSSR6(▾), pSSR21(◂), and E. coli (MKH13) host strain carrying empty plasmid vector (■) in (A) LB broth supplemented with 3.0% NaCl(w/v), (B) LB broth supplemented with 3.7% KCl,(C) LB broth.
Sequence assembly of pSSR1 resulted into a contig of 2,938 bp with a 66.87% G+C content. Cloned insert shared 76% homology with a halophilic proteobacterial lineage Haliangium ochraceum, indicating its plausible affiliation within proteobacterial clade. The gene prediction analysis indicated the presence of three complete and one truncated ORF, encoding proteins of 93, 114, 383, and 149aa respectively. Translated nucleotide sequences of these ORFs were subjected to BLASTP (maximum e-value cutoff of 1e-34) analysis to identify the homologous sequence in the database (Table 2). Transposon mutagenesis analysis confirmed functionally active locus for osmotolerance property encompassing ORF3 (positioned between 927 and 2,078 bp) (Figure 4). ORF3 encoded transmembrane protein shared homology with a transmembrane ABC transporter ATP-binding protein of Betaproteobacteria bacterium SG8_39 (74%) and branched-chain amino acid ABC transporter substrate-binding protein of Oceanibacterium hippocampi (71%). Pfam database search identified the presence of a periplasmic ligand-binding domain of the ABC (ATPase Binding Cassette) type active transport systems, known to be involved in the transport of three branched chain aliphatic amino acids (leucine, isoleucine and valine) (Davidson et al., 2008). STRING analysis also predicted ORF3 as part of an interactive periplasmic binding protein dependent transport system. Further, NsitePred web server identified strong nucleotide binding sites (ATP Binding site at Gly14 and ADP binding site at Gly12) within ORF3 encoded protein. This nucleotide binding site could be NBD, a common feature for ATP binding proteins, as predicted by its functional assignment. In consideration of physiological role and all structural features of ORF3 encoded protein, it is a type of ABC transporter ATP-binding protein involved in salt stress maintenance possibly through energy dependent interaction with ABC membrane transporters involved in the exchange of the solutes across membrane, thus labeled as putative branched chain amino acid (BCAA) ABC transporter gene (BCAA_ABCTP). The putative BCAA_ABCTP gene was subcloned (pSSR1C1) to confirm its osmotolerance property. Time dependent growth curve analysis of SSR1C1 harboring BCAA_ABCTP showed a significant growth advantage in the presence of NaCl [3.0% (w/v)] (P = 0.0006) (Figure 5A) and KCl [3.7 % (w/v)] (P = 0.0005) (Figure 5B) as compared to salt sensitive E. coli mutant MKH13 carrying only the empty vector (pUC19), while no significant difference was observed on LB only (Figure 5C). The intracellular elemental analysis in the presence of ionic stressor NaCl [3.0% (w/v)] showed that SSR1C1 has effectively reduced intracellular sodium ion concentration (P = 0.0134) in comparison to E. coli mutant MKH13 (Figure 5D). A reduced intracellular sodium concentration within SSR1C1 could be seen as a result of its transporter property, as predicated through genetic characterization. The growth pattern of SSR1C1 was found similar to the native SSR1 that confirms that salt tolerance of salt tolerant clone SSR1 was due to SSR1C1 cloned insert encoding a branched chain amino acid (BCAA) ABC transporter protein.
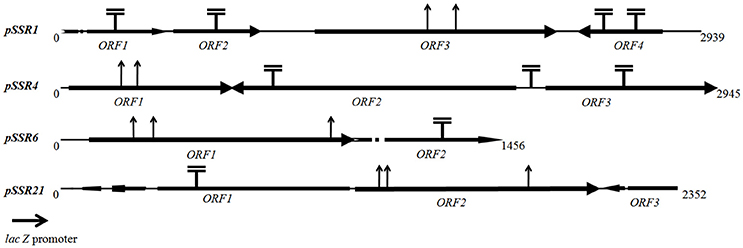
Figure 4. Transposon insertion map of pSSR1, pSSR4, pSSR6, and pSSR21. indicates a transposon insertion site identified within transposon positive mutants (no effect on plasmid derived osmotolerance property) while ↑ indicate transposon insertion site identified within transposon negative mutants (loss of plasmid derived osmotolerance property).
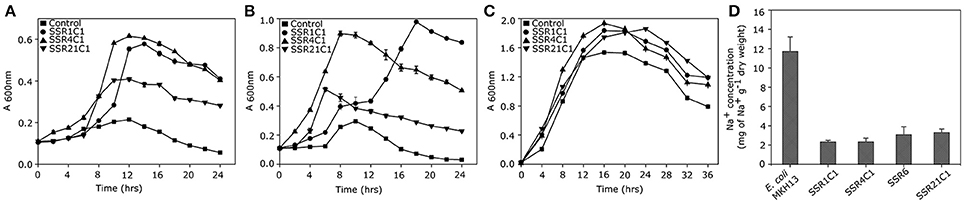
Figure 5. Physiological Characterization of BCAA_ABCTP, GSDH, STK_Pknb, and duf3445 for osmotolerance. Growth curve analysis of osmotolerant phenotype SSR1C1 (•), SSR4C1 (▴), SSR21C1 (▾), and E. coli (MKH13) host strain carrying empty plasmid vector (■) in (A) LB broth supplemented with 3% NaCl (w/v), (B) LB broth supplemented with 3.7 % KCl (w/v), and (C) LB only. Intracellular Na+ estimation in E. coli (MKH13) strain harboring pSSR1C1, pSSR4C1, pSSR6, and pSSR21C1 and E. coli (MKH13) host strain carrying empty plasmid vector (D).
The pSSR4 harbors a G+C rich (G+C% = 63.63) insert of 2,945 bp. The cloned sequence did not share any homology at the nucleotide level in existing database sequences. A total of three ORFs were predicted within the cloned insert, encoding proteins of 207, 507, and 210 amino acids respectively (Table 2). Transposon mutagenesis analysis identified the functionally active locus within the sequence region (235 bp), encompassing ORF1 (Figure 4). ORF1 encodes a cytosolic protein homologous to hypothetical protein of Anaerolineae bacterium SG8_19 and Glucose/sorbosone dehydrogenase-like protein of Pelobacter carbinolicus. A Pfam analysis of ORF1 encoded protein indicates it as a glucose/sorbosone dehydrogenase protein having conserved domains for the protein family GSDH. It indicates that the ORF1 possibly encodes a glucose/sorbosone dehydrogenase involved in salt stress tolerance, possibly through the cytosolic accumulation of reducing equivalents (NADPH and GSH). Predicted GSDH gene was amplified from pSSR4 and subcloned to validate its osmotolerance property. Time dependent growth curve assay of SSR4C1 showed a significant growth advantage NaCl [3.0% (w/v)] (P = 0.0004) (Figure 5A) and KCl [3.7 % (w/v)] (P = 0.0006) (Figure 5B) as compared to salt sensitive E. coli mutant MKH13 carrying the empty vector (pUC19), while no significant difference has been observed on LB only (Figure 5C). The growth pattern of SSR4C1 was similar to SSR4. It also confirmed that the salt tolerance property of salt tolerant clone pSSR4 was possibly due to GSDH gene encoding glucose/sorbosone dehydrogenase protein.
Sequence assembly of pSSR6 generated a G+C rich (65.66%) contig of 1,456 bp. The blastn analysis identified its low similarity to Betaproteobacteria GR16-43 genome sequences, indicating its affiliation from proteobacterial clade. The cloned sequence encodes only one complete ORF (ORF1), encoding a cytosolic protein of 345 amino acids with a G+C content of 65.79% (Table 2). Transposon mutagenesis analysis also confirmed the functionally active locus within ORF2 (Figure 4). Homologs of translated ORF2, corresponds to putative serine/threonine protein kinase of Woeseia oceani and Mycobacterium smegmatis str. MC2 155. A pfam database search of ORF2 encoded protein indicates it as a putative serine/threonine protein kinase having conserved domains for the protein family STKc_PknB, i.e., the catalytic domain of bacterial Serine/Threonine kinases, PknB family. Ser/Thr protein kinase homologs were found to be involved in osmosensory signaling in microbes (Hatzios et al., 2013). These identified proteins were important for the survival and in stress responses (Donat et al., 2009). The intracellular elemental analysis in presence of the ionic stressor NaCl [3.0% (w/v)] showed that SSR6 effectively reduced intracellular sodium ion concentration (P = 0.0200) in comparison to E. coli MKH13 (Figure 5D). A reduced intracellular sodium concentration within SSR6C1 could be due to enhanced ion transporter activity under the influence of signals generated by putative STKc_PknB of pSSR6 under the osmotic stress.
The pSSR21 was found to have a G+C rich (69.98%) insert of 2352 bp. The blastn analysis identified its homology with an Actinomycetes strain Allokutzneria albata. Gene prediction has indicated the presence of three ORFs in pSSR21, encoding proteins of 309, 337, and 84 amino acids respectively. Among three identified ORFs, only ORF2 was complete, while other two ORFs (ORF1 and 3) were truncated. Transposon mutagenesis analysis has identified the functionally active locus within ORF2 sequence region (1,220 and 1,330 bp). The database homologs of ORF2 corresponds to hypothetical protein A3F84_26310 of Candidatus Handelsmanbacteria bacterium (Table 2). The pfam analysis identified the presence of conserved domains in the protein family DUF3445, i.e., protein of unknown function (DUF3445). The G+C content of ORF2 was found to be 69.96% and the predicted functionally active region, ORF2 was subcloned. Time dependent growth curve assay of SSR21C1 showed a significant growth advantage NaCl [3.0% (w/v)] (P = 0.0005) (Figure 5A) and KCl [3.7% (w/v)] (P = 0.0045) (Figure 5B) as compared to salt sensitive E. coli mutant MKH13 carrying only the empty vector (pUC19), while no significant difference has been observed on LB only (Figure 5C). Elemental Quantification of intracellular Na+ in E. coli MKH13 carrying the empty vector and salt tolerant recombinant subclones SSR21C1 clearly showed that the cloned gene insert (duf3445) within SSR21C1 has significantly reduced the concentration of intracellular Na+ ion (P = 0.0174) (Figure 5D).
Metagenomics has the potential to advance our knowledge by studying the genetic components of uncultured microbes (Singh A. H. et al., 2009; Mirete et al., 2015; Chauhan et al., 2017; Yadav et al., 2017). Looking at the perspectives of metagenomics, it was used to explore soil microbiome for its composition and genetic/physiological mechanisms allowing successful adaptation of microbes in saline environments. Simultaneously, these genes could be utilized as potential candidate to develop ever demanding drought resistant transgenic crops (Zhengbin et al., 2011) or osmotolerant microbes for food processing (Fernandes, 2014) and waste water treatment applications (Xiao and Roberts, 2010). Metagenomic analysis based on SSU rRNA gene has identified dominance of Proteobacteria, Actinobacteria, Bacteroidetes, and Gemmatimonadetes in saline soil microbiome. These results are in parallel with the outcome of previous studies defining microbial community composition of saline soil environment (Zhang et al., 2003; Ma and Gong, 2013; Canfora et al., 2014; Kadam and Chuan, 2016). Canfora et al. has reported a correlation in abundance of a microbial group with respect to soil salinity. They had indicated a relative abundance of Proteobacteria with Bacteroidetes were positively and Acidobacteria was negatively correlated with salinity (Canfora et al., 2014). Similarly Actinobacteria was also been reported as another dominant microbial phylum in saline ecosystems (Kadam and Chuan, 2016). Gemmatimonadetes is another well-known hypersaline microbial phylum associated with biogeochemical transformations (Zhang et al., 2003). The existence of halophilic subcomponents within the identified microbial phylum, possibly making them capable to proliferate in a saline environment. These studies explain the abundance of Proteobacteria, Actinobacteria, Gemmatimonadetes, and Bacteroidetes in the studied ecosystem. Evolution of novel genetic features is a key to their successful survival in a changing environment (Gupta et al., 2017; Kumar Mondal et al., 2017). These saline microorganisms could have evolved salt stress tolerant genetic machinery to adapt and survive under salt induced osmotic stresses (Meena et al., 2017). A number of genetic elements were decoded for osmotolerance property from cultured and uncultured microbial representatives (Kapardar et al., 2010a,b; Culligan et al., 2012, 2013, 2014; Kim and Yu, 2012; Mirete et al., 2015). However, there is a disparity in a number of reported salt tolerance genes with a number of microorganisms from an ecosystem (Humbert et al., 2009; Mirete et al., 2015). This divergence could be allied with a number of factors like unculturability (Kim and Yu, 2012), lack of good quality genomic/metagenomic DNA (Kumar J. et al., 2016) and issues with foreign gene expression (Prakash and Taylor, 2012). An additional effort was made in the current study to decode osmotolerance genes prevalent in these saline soil microorganisms using functional metagenomics. It leads to the identification of four unique osmotolerant clones harboring DNA insert showing affiliation within halophile genomes. Genetic and physiological analysis has identified genes encoding putative proteins like membrane bound branched chain amino acid (BCAA) ABC transporter protein (BCAA_ABCTP), Glucose/sorbosone dehydrogenase, cytosolic STKc_PknB and DUF protein are responsible for osmotolerance property within salt stress tolerant clones. Among these, the role of the branched chain amino acid (BCAA) ABC transporter protein and glucose/sorbosone dehydrogenase in osmotolerance are well documented (Takami et al., 2002; Brosnan and Brosnan, 2006), while the scanty information is available about role for STKc_PknB and DUF3445 protein in stress maintenance (Hatzios et al., 2013).
ABC branched chain amino acid (BCAA) transporters are widely distributed in various marine microbes like Oceanibacillus iheyensis (Takami et al., 2002), Salinispora, Bacillus, and Roseobacter strains (Penn and Jensen, 2012). BCAA_ABCTP proteins are involved in the transport of branched chain aliphatic amino acids such as leucine, isoleucine and valine at high salt concentration. In the presence of 2-oxoglutarate and pyridoxal-5-phosphate, these branched chain amino acids are further converted to L-glutamate by branched chain amino acid transferase (Hutson, 2001). Accumulated glutamate acts as an osmoprotectant upon hyper-osmotic shock and activates sets of genes that allow the host to achieve long-term adaptation to high osmolarity (Gralla and Vargas, 2006). They also account for a significant proportion of the genes observed in the marine metagenome. Branched chain amino acid transporters are probably an important marine adaptation because accumulated glutamate may function as a counter ion for K+, which balances the electrical state of the cytoplasm (Penn and Jensen, 2012). Previous studies have reported a regulatory relationship between K+ and glutamate accumulation in response to osmotic stress in enteric bacteria and haloalkaliphilic archaea Natronococcus occultus (Kokoeva et al., 2002). Similarly, T. consotensis, a halotolerant bacterium accumulates glutamate to maintain electrical equilibrium within the cell in response to high salt concentrations (Rubiano-Labrador et al., 2015). This background information explains the possible physiological role of the BCAA_ABCTP gene of pSSR1 to increase host osmotolerance.
Glucose/Sorbosone dehydrogenase (GSDH) is responsible for the production of NADPH through oxidative cleavage of glucose (Oubrie et al., 1999). Under salt stress condition, NADPH acts as reducing potential for output of reduced glutathione (GSH) and involved in activity of membrane bound NADPH oxidase, which results in accumulation of hydrogen peroxide (H2O2) (Wang et al., 2008). H2O2 acts as a signal in regulating G6PDH activity and expression of this enzyme in the glutathione cycle through which the ability of GSH regeneration was increased under salt stress (Wang et al., 2008). Thus, G6PDH plays a critical role in maintaining cellular GSH levels under long-term salt stress conditions (Wang et al., 2008). It indicates that GSDH of pSSR4 is involved in salt stress tolerance possibly through the cytosolic accumulation of reducing equivalents (NADPH and GSH).
While in case of pSSR6 only one ORF was identified, sharing homology with Serine/Threonine kinases. This encoded protein also possesses two conserved domains for the protein family STKc_PknB, i.e. the catalytic domain of bacterial Serine/Threonine kinases, PknB and similar proteins; STKs and TOMM_kin_cyc, i.e., TOMM system kinase/cyclase fusion protein. STKs are well known for activating genetic locus concerned with osmosensing (Hatzios et al., 2013) and inducing topological changes such as DNA supercoiling (Gupta et al., 2014). These osmosensing signal and DNA supercoiling could initiate uptake of the osmolyte glycine betaine, proline (Csonka, 1989) or initiates the expression of genetic elements required to cope with osmotic stress (Higgins et al., 1988). This could be the possible mechanism by which putative STK_PknB of pSSR6 might be extending osmotolerance to the host E. coli.
The pSSR21 ORF2 encodes a protein sharing homology with a hypothetical protein of Candidatus Handelsmanbacteria possessing a conserved domain for DUF3445 superfamily, i.e., an uncharacterized protein family having conserved RLP sequence motif (Bateman et al., 2010). However, its physiological characterization and intracellular ion concentration analysis indicates that it extends host osmotolerance property by maintaining low intracellular ion concentration even in the presence of an ionic stressor. However, a detailed mechanism still needs to be elucidated.
In this study, the functional metagenomic approach was used to decipher salt stress tolerant genes in the saline soil microbiome. Identification of salt tolerant genes BCAA_ABCtp, GSDH STK_Pknb, and duf3445 has enriched our understanding about the survivability and adaptability of microbes in the highly saline soil ecosystem. These salt tolerant genes can be used for crop improvement and for producing bioactive molecules under high salt conditions, which reduces the chances of contamination by other microbes.
NC, MV: Designed the project; MV, VA: Performed experiments and NGS sequencing; SG, VM, and VA: Performed data analyses; MV, VM, and NC: Wrote the manuscript. All authors have read and approve the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Authors would like to thank Council of Scientific and Industrial Research (CSIR) and UGC Grant Commission for fellowships under the scheme 60(0099)/11/EMRII & F. 41-1256/2012 (SR). We are thankful to Dr. Tamara Hoffmann, Philip Universitat, Marburg for providing E. coli (MKH13) strain.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00159/full#supplementary-material
Bateman, A., Coggill, P., and Finn, R. D. (2010). DUFs: families in search of function. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 1148–1152. doi: 10.1107/S1744309110001685
Brosnan, J. T., and Brosnan, M. E. (2006). Branched-chain amino acids: enzyme and substrate regulation. J. Nutr. 136(Suppl. 1), 207S−211S. doi: 10.1093/jn/136.1.207S
Canfora, L., Bacci, G., Pinzari, F., Lo Papa, G., Dazzi, C., and Benedetti, A. (2014). Salinity and bacterial diversity: to what extent does the concentration of salt affect the bacterial community in a saline soil? PLoS ONE 9:e106662. doi: 10.1371/journal.pone.0106662
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chauhan, N. S., Nain, S., and Sharma, R. (2017). Identification of arsenic resistance genes from marine sediment metagenome. Indian J. Microbiol. 57, 299–306. doi: 10.1007/s12088-017-0658-0
Chauhan, N. S., Ranjan, R., Purohit, H. J., Kalia, V. C., and Sharma, R. (2009). Identification of genes conferring arsenic resistance to Escherichia coli from an effluent treatment plant sludge metagenomic library: Arsenic resistance genes from sludge metagenome. FEMS Microbiol. Ecol. 67, 130–139. doi: 10.1111/j.1574-6941.2008.00613.x
Csonka, L. N. (1989). Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 53, 121–147.
Culligan, E. P., Sleator, R. D., Marchesi, J. R., and Hill, C. (2012). Functional metagenomics reveals novel salt tolerance loci from the human gut microbiome. ISME J. 6, 1916–1925. doi: 10.1038/ismej.2012.38
Culligan, E. P., Sleator, R. D., Marchesi, J. R., and Hill, C. (2013). Functional environmental screening of a metagenomic library identifies stlA; a unique salt tolerance locus from the human gut microbiome. PLoS ONE 8:e82985. doi: 10.1371/journal.pone.0082985
Culligan, E. P., Sleator, R. D., Marchesi, J. R., and Hill, C. (2014). Metagenomic identification of a novel salt tolerance gene from the human gut microbiome which encodes a membrane protein with homology to a brp/blh-family β-carotene 15,15'-monooxygenase. PLoS ONE 9:e103318. doi: 10.1371/journal.pone.0103318
Davidson, A. L., Dassa, E., Orelle, C., and Chen, J. (2008). Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364. doi: 10.1128/MMBR.00031-07
Donat, S., Streker, K., Schirmeister, T., Rakette, S., Stehle, T., Liebeke, M., et al. (2009). Transcriptome and functional analysis of the eukaryotic-type serine/threonine kinase PknB in Staphylococcus aureus. J. Bacteriol. 191, 4056–4069. doi: 10.1128/JB.00117-09
Fernandes, P. (2014). Marine enzymes and food industry: insight on existing and potential interactions. Front. Mar. Sci. 1:46. doi: 10.3389/fmars.2014.00046
Gralla, J. D., and Vargas, D. R. (2006). Potassium glutamate as a transcriptional inhibitor during bacterial osmoregulation. EMBO J. 25, 1515–1521. doi: 10.1038/sj.emboj.7601041
Gupta, M., Sajid, A., Sharma, K., Ghosh, S., Arora, G., Singh, R., et al. (2014). HupB, a nucleoid-associated protein of Mycobacterium tuberculosis, is modified by serine/threonine protein kinases in vivo. J. Bacteriol. 196, 2646–2657. doi: 10.1128/JB.01625-14
Gupta, S., Kumar, M., Kumar, J., Ahmad, V., Pandey, R., and Chauhan, N. S. (2017). Systemic analysis of soil microbiome deciphers anthropogenic influence on soil ecology and ecosystem functioning. Int. J. Environ. Sci. Technol. 14, 2229–2238. doi: 10.1007/s13762-017-1301-7
Haardt, M., Kempf, B., Faatz, E., and Bremer, E. (1995). The osmoprotectant proline betaine is a major substrate for the binding-protein-dependent transport system ProU of Escherichia coli K-12. Mol. Gen. Genet. MGG 246, 783–786. doi: 10.1007/BF00290728
Hatzios, S. K., Baer, C. E., Rustad, T. R., Siegrist, M. S., Pang, J. M., Ortega, C., et al. (2013). Osmosensory signaling in Mycobacterium tuberculosis mediated by a eukaryotic-like Ser/Thr protein kinase. Proc. Natl. Acad. Sci. U.S.A. 110, E5069–E5077. doi: 10.1073/pnas.1321205110
Higgins, C. F., Dorman, C. J., Stirling, D. A., Waddell, L., Booth, I. R., May, G., et al. (1988). A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell 52, 569–584.
Humbert, J.-F., Dorigo, U., Cecchi, P., Le Berre, B., Debroas, D., and Bouvy, M. (2009). Comparison of the structure and composition of bacterial communities from temperate and tropical freshwater ecosystems. Environ. Microbiol. 11, 2339–2350. doi: 10.1111/j.1462-2920.2009.01960.x
Hutson, S. (2001). Structure and function of branched chain aminotransferases. Prog. Nucleic Acid Res. Mol. Biol. 70, 175–206. doi: 10.1016/S0079-6603(01)70017-7
Jiang, H., Dong, H., Yu, B., Liu, X., Li, Y., Ji, S., et al. (2007). Microbial response to salinity change in Lake Chaka, a hypersaline lake on Tibetan plateau. Environ. Microbiol. 9, 2603–2621. doi: 10.1111/j.1462-2920.2007.01377.x
Kadam, P. D., and Chuan, H. H. (2016). Erratum to: rectocutaneous fistula with transmigration of the suture: a rare delayed complication of vault fixation with the sacrospinous ligament. Int. Urogynecol. J. 27:505. doi: 10.1007/s00192-016-2952-5
Kapardar, R. K., Ranjan, R., Grover, A., Puri, M., and Sharma, R. (2010a). Identification and characterization of genes conferring salt tolerance to Escherichia coli from pond water metagenome. Bioresour. Technol. 101, 3917–3924. doi: 10.1016/j.biortech.2010.01.017
Kapardar, R. K., Ranjan, R., Puri, M., and Sharma, R. (2010b). Sequence analysis of a salt tolerant metagenomic clone. Indian J. Microbiol. 50, 212–215. doi: 10.1007/s12088-010-0041-x
Kim, M., and Yu, Z. (2012). Quantitative comparisons of select cultured and uncultured microbial populations in the rumen of cattle fed different diets. J. Anim. Sci. Biotechnol. 3:28. doi: 10.1186/2049-1891-3-28
Klähn, S., Marquardt, D. M., Rollwitz, I., and Hagemann, M. (2009). Expression of the ggpPS gene for glucosylglycerol biosynthesis from Azotobacter vinelandii improves the salt tolerance of Arabidopsis thaliana. J. Exp. Bot. 60, 1679–1689. doi: 10.1093/jxb/erp030
Kokoeva, M. V., Storch, K. F., Klein, C., and Oesterhelt, D. (2002). A novel mode of sensory transduction in archaea: binding protein-mediated chemotaxis towards osmoprotectants and amino acids. EMBO J. 21, 2312–2322. doi: 10.1093/emboj/21.10.2312
Koonin, E. V., and Wolf, Y. I. (2012). Evolution of microbes and viruses: a paradigm shift in evolutionary biology? Front. Cell. Infect. Microbiol. 2:119. doi: 10.3389/fcimb.2012.00119
Kumar, J., Kumar, M., Gupta, S., Ahmed, V., Bhambi, M., Pandey, R., et al. (2016). An improved methodology to overcome key issues in human fecal metagenomic DNA extraction. Genom. Proteom. Bioinform. 14, 371–378. doi: 10.1016/j.gpb.2016.06.002
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kumar Mondal, A., Kumar, J., Pandey, R., Gupta, S., Kumar, M., Bansal, G., et al. (2017). Comparative genomics of host–symbiont and free-living Oceanobacillus species. Genome Biol. Evol. 9, 1175–1182. doi: 10.1093/gbe/evx076
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Ma, B., and Gong, J. (2013). A meta-analysis of the publicly available bacterial and archaeal sequence diversity in saline soils. World J. Microbiol. Biotechnol. 29, 2325–2334. doi: 10.1007/s11274-013-1399-9
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2014). CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Meena, K. K., Sorty, A. M., Bitla, U. M., Choudhary, K., Gupta, P., Pareek, A., et al. (2017). Abiotic stress responses and microbe-mediated mitigation in plants: the omics strategies. Front. Plant Sci. 8:172. doi: 10.3389/fpls.2017.00172
Mirete, S., Mora-Ruiz, M. R., Lamprecht-Grandío, M., de Figueras, C. G., Rosselló-Móra, R., and González-Pastor, J. E. (2015). Salt resistance genes revealed by functional metagenomics from brines and moderate-salinity rhizosphere within a hypersaline environment. Front. Microbiol. 6:1121. doi: 10.3389/fmicb.2015.01121
Morowitz, M. J., Denef, V. J., Costello, E. K., Thomas, B. C., Poroyko, V., Relman, D. A., et al. (2011). Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc. Natl. Acad. Sci. U.S.A. 108, 1128–1133. doi: 10.1073/pnas.1010992108
Naughton, L. M., Blumerman, S. L., Carlberg, M., and Boyd, E. F. (2009). Osmoadaptation among Vibrio species and unique genomic features and physiological responses of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 75, 2802–2810. doi: 10.1128/AEM.01698-08
Oubrie, A., Rozeboom, H. J., Kalk, K. H., Olsthoorn, A. J., Duine, J. A., and Dijkstra, B. W. (1999). Structure and mechanism of soluble quinoprotein glucose dehydrogenase. EMBO J. 18, 5187–5194. doi: 10.1093/emboj/18.19.5187
Penn, K., and Jensen, P. R. (2012). Comparative genomics reveals evidence of marine adaptation in Salinispora species. BMC Genomics 13:86. doi: 10.1186/1471-2164-13-86
Prakash, T., and Taylor, T. D. (2012). Functional assignment of metagenomic data: challenges and applications. Brief. Bioinform. 13, 711–727. doi: 10.1093/bib/bbs033
Roberts, M. F. (2005). Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst. 1:5. doi: 10.1186/1746-1448-1-5
Rubiano-Labrador, C., Bland, C., Miotello, G., Armengaud, J., and Baena, S. (2015). Salt stress induced changes in the exoproteome of the halotolerant bacterium Tistlia consotensis deciphered by proteogenomics. PLoS ONE 10:e0135065. doi: 10.1371/journal.pone.0135065
Sakamoto, T., and Murata, N. (2002). Regulation of the desaturation of fatty acids and its role in tolerance to cold and salt stress. Curr. Opin. Microbiol. 5, 208–210. doi: 10.1016/S1369-5274(02)00306-5
Shrivastava, P., and Kumar, R. (2015). Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 22, 123–131. doi: 10.1016/j.sjbs.2014.12.001
Singh, A. H., Doerks, T., Letunic, I., Raes, J., and Bork, P. (2009). Discovering functional novelty in metagenomes: examples from light-mediated processes. J. Bacteriol. 191, 32–41. doi: 10.1128/JB.01084-08
Singh, J., Behal, A., Singla, N., Joshi, A., Birbian, N., Singh, S., et al. (2009). Metagenomics: concept, methodology, ecological inference and recent advances. Biotechnol. J. 4, 480–494. doi: 10.1002/biot.200800201
Takami, H., Takaki, Y., and Uchiyama, I. (2002). Genome sequence of Oceanobacillus iheyensis isolated from the Iheya Ridge and its unexpected adaptive capabilities to extreme environments. Nucleic Acids Res. 30, 3927–3935. doi: 10.1093/nar/gkf526
Tsirigos, K. D., Peters, C., Shu, N., Käll, L., and Elofsson, A. (2015). The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 43, W401–W407. doi: 10.1093/nar/gkv485
Tusnády, G. E., and Simon, I. (2001). The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850. doi: 10.1093/bioinformatics/17.9.849
van Veen, J. A., van Overbeek, L. S., and van Elsas, J. D. (1997). Fate and activity of microorganisms introduced into soil. Microbiol. Mol. Biol. Rev. 61, 121–135.
Wang, X., Ma, Y., Huang, C., Li, J., Wan, Q., and Bi, Y. (2008). Involvement of glucose-6-phosphate dehydrogenase in reduced glutathione maintenance and hydrogen peroxide signal under salt stress. Plant Signal. Behav. 3, 394–395. doi: 10.4161/psb.3.6.5404
Xiao, Y., and Roberts, D. J. (2010). A review of anaerobic treatment of saline wastewater. Environ. Technol. 31, 1025–1043. doi: 10.1080/09593331003734202
Yadav, M., Verma, M. K., and Chauhan, N. S. (2017). A review of metabolic potential of human gut microbiome in human nutrition. Arch Microbiol. doi: 10.1007/s00203-017-1459-x. [Epub ahead of print].
Yan, C., Zhang, D., Raygoza Garay, J. A., Mwangi, M. M., and Bai, L. (2015). Decoupling of divergent gene regulation by sequence-specific DNA binding factors. Nucleic Acids Res. 43, 7292–7305. doi: 10.1093/nar/gkv618
Zhang, H., Sekiguchi, Y., Hanada, S., Hugenholtz, P., Kim, H., Kamagata, Y., et al. (2003). Gemmatimonas aurantiaca gen. nov., sp. nov., a gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int. J. Syst. Evol. Microbiol. 53, 1155–1163. doi: 10.1099/ijs.0.02520-0
Zhengbin, Z., Ping, X., Hongbo, S., Mengjun, L., Zhenyan, F., and Liye, C. (2011). Advances and prospects: biotechnologically improving crop water use efficiency. Crit. Rev. Biotechnol. 31, 281–293. doi: 10.3109/07388551.2010.531004
Keywords: metagenome, halotolerance, SSU rRNA, soil microbiome, soil ecology
Citation: Ahmed V, Verma MK, Gupta S, Mandhan V and Chauhan NS (2018) Metagenomic Profiling of Soil Microbes to Mine Salt Stress Tolerance Genes. Front. Microbiol. 9:159. doi: 10.3389/fmicb.2018.00159
Received: 12 October 2017; Accepted: 23 January 2018;
Published: 08 February 2018.
Edited by:
Diana Elizabeth Marco, National Scientific Council (CONICET), ArgentinaReviewed by:
Richard Allen White III, RAW Molecular Systems (RMS) LLC, United StatesCopyright © 2018 Ahmed, Verma, Gupta, Mandhan and Chauhan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nar S. Chauhan, bnNjaGF1aGFuQG1kdXJvaHRhay5hYy5pbg==
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.