- 1Department of Plant, Soil and Microbial Sciences, Center for Microbial Pathogenesis, Michigan State University, East Lansing, MI, United States
- 2Department of Plant Pathology and Ecology, The Connecticut Agricultural Experiment Station, New Haven, CT, United States
Type III secretion (TTS) chaperones are critical for the delivery of many effector proteins from Gram-negative bacterial pathogens into host cells, functioning in the stabilization and hierarchical delivery of the effectors to the type III secretion system (TTSS). The plant pathogen Erwinia amylovora secretes at least four TTS effector proteins: DspE, Eop1, Eop3, and Eop4. DspE specifically interacts with the TTS chaperone protein DspF, which stabilizes the effector protein in the cytoplasm and promotes its efficient translocation through the TTSS. However, the role of E. amylovora chaperones in regulating the delivery of other secreted effectors is unknown. In this study, we identified functional interactions between the effector proteins DspE, Eop1, and Eop3 with the TTS chaperones DspF, Esc1 and Esc3 in yeast. Using site-directed mutagenesis, secretion, and translocation assays, we demonstrated that the three TTS chaperones have additive roles for the secretion and translocation of DspE into plant cells whereas DspF negatively affects the translocation of Eop1 and Eop3. Collectively, these results indicate that TTS chaperone proteins exhibit a cooperative behavior to orchestrate the effector secretion and translocation dynamics in E. amylovora.
Introduction
The delivery of effector proteins via the type III secretion system (TTSS) is a critical step for pathogenesis of many Gram-negative bacteria (Büttner and He, 2009; Raymond et al., 2013; Galán et al., 2014). The secretion signal is typically localized in the N-terminal domain of the effector or within the 5′ region of the effector mRNA (Miao and Miller, 2000; Mudgett et al., 2000; Blaylock et al., 2008). Loosely conserved sequence or structural patterns in this region can be used in computational approaches for in silico prediction of type III secreted substrates (Guttman et al., 2002; Petnicki-Ocwieja et al., 2002). The translocation efficiency of many effector proteins also depends on a physical association with cytoplasmic type III secretion (TTS) chaperone proteins (Luo et al., 2001; Parsot et al., 2003; Matsumoto and Young, 2009; Triplett et al., 2009). TTS chaperones are typically low molecular weight acidic proteins that remain within the bacterial cytoplasm and form dimeric or hexameric structures that bind to their target effectors (Thomas et al., 2012; Tsai et al., 2015). TTS chaperones are categorized into three groups: class I TTS chaperones bind to effector proteins, class II TTS chaperones bind to type III pore-forming (translocon) proteins, and class III TTS chaperones bind to needle proteins (Cornelis and Gijsegem, 2000; Parsot et al., 2003). Two subclasses are found within class I TTS chaperones: class IA chaperones bind to single effector proteins, and class IB chaperones bind to multiple effectors (reviewed in Thomas et al., 2012). Despite exhibiting low overall amino acid residue similarity, TTS chaperones share a conserved three-dimensional structure and similar modes of interaction with their effector partners (Ghosh, 2004; Cornelis, 2006). These similarities have been used to model the three-dimensional structure of chaperone proteins from plant pathogens, as well as to identify critical residues in the interaction with their cognate effectors (Triplett et al., 2010).
Type III secretion chaperone binding may directly protect effectors from degradation by the Lon or other proteases in permissive conditions for TTS (Losada and Hutcheson, 2005). In addition, TTS chaperones in diverse bacteria interact with ATPases associated with the TTSS. This interaction induces the docking, unfolding and release of the effector protein to the secretion system (Akeda and Galán, 2005; Lorenz and Buttner, 2009; Cooper et al., 2010). The TTS chaperone HpaB from Xanthomonas campestris pv. vesicatoria establishes a secretion hierarchy that allows the secretion of TTSS components prior to that of effector proteins (Lorenz et al., 2008). TTS chaperones may also interact with non-secreted proteins, such as transcription factors, in order to upregulate the expression of effector genes and facilitate the global regulation of the TTS (Darwin and Miller, 2001).
Erwinia amylovora, the causal agent of fire blight disease of rosaceous plants including apple and pear (Malnoy et al., 2012) secretes at least four effector proteins: DspA/E (DspE henceforth), Eop1, AvrRpt2Ea/Eop4 (Eop4 henceforth) and Eop3 (Bogdanove et al., 1998; Zhao et al., 2006; Nissinen et al., 2007). Among these, only DspE is required for pathogenicity, multiplication in planta, and for disease promotion by the alteration of host defenses, inducing cell death in both host and non-host plants (Gaudriault et al., 1997; Boureau et al., 2006). DspE interacts with the TTS chaperone protein DspF, which stabilizes the effector and prevents its degradation in the cytoplasm, and promotes its efficient translocation through the TTSS (Gaudriault et al., 2002). However, a dspF mutant does not lack pathogenic ability, but exhibits reduced aggressiveness and is still able to translocate the N terminal region of DspE (Triplett et al., 2009; Oh et al., 2010), suggesting that other proteins may be involved in the secretion of this effector protein in the absence of or in addition to DspF. The effector protein Eop1, a member of the YopJ family of proteins, is also translocated via the TTSS. Like dspE, the eop1 gene is located adjacent to a TTS chaperone gene, named orfA (Oh and Beer, 2005). The orfA product interacts not only with Eop1 but also with DspE in yeast (Asselin et al., 2006), suggesting that TTS chaperones in E. amylovora may be involved in the translocation of several effectors. The roles of chaperones other than DspF in the regulation of E. amylovora effector translocation are unknown.
Understanding the dynamic roles of TTS chaperones during plant pathogenesis is challenging due to the large number of TTS effectors in many model bacterial pathogens. Conversely, the small number of effectors in E. amylovora makes it well-suited for understanding the global secretory roles of TTS chaperones in plant pathogens. In this report, we investigated the effect of TTS chaperones on all known effector proteins of E. amylovora. We identified novel functional interactions between the effector proteins DspE, Eop1, and Eop3 with their cognate and non-cognate predicted TTS chaperones. We then analyzed the individual and collective effects of these chaperones on secretion, host translocation, and pathogenicity, and demonstrated that TTS chaperones act cooperatively in the regulation of E. amylovora effector translocation dynamics.
Materials and Methods
Bacterial Strains, Plasmids, Growth Conditions, and Genetic Techniques
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown at 28°C in Luria-Bertani (LB) broth and agar unless otherwise noticed. Media were amended with ampicillin (Amp; 50 mg L-1), chloramphenicol (Cm; 10 mg L-1), gentamicin (Gm; 10 mg L-1) or kanamycin (Km; 25 mg L-1) as necessary. PCR, restriction digestions, gene cloning and gel electrophoresis were performed according to standard methods (Sambrook et al., 2001).
Mutant Construction
Chromosomal mutants were constructed using an adaptation of the λ red recombinase system as previously described (Datsenko and Wanner, 2000; Zhao et al., 2005; Triplett et al., 2009). Briefly, Cm and Km resistance cassettes were amplified from template plasmids pKD3 and pKD4 using primers with 50 bp overhangs, homologous with the gene of interest. PCR products were purified and electroporated into E. amylovora wild-type (WT) strain Ea1189 expressing the genes of red, β, λ and exo recombinases from the pKD46 plasmid. Resultant colonies were screened for antibiotic resistance, and gene disruption was verified by PCR and sequencing. In order to create triple and quadruple mutants, pKD46 was cured from Ea1189ΔdspF/esc3 and Ea1189ΔdspF/esc1/esc3 strains by repetitive growth cycles without antibiotic selection and including heat shock at 37°C. Cured strains were transformed with pCP20 in order to resolve antibiotic resistances by the thermo-inducible resolvase encoded in this plasmid. Transformants were tested for Amp, Cm and Km sensitivity prior to initiating the next round of mutagenesis.
Pathogenicity Assays
Strain pathogenicity was evaluated using immature pear fruit assays as previously described (Zhao et al., 2005; Koczan et al., 2011). Briefly, bacterial suspensions were grown overnight and adjusted to ≈ 1 × 104 CFU mL-1 in 0.5x sterile phosphate-buffered saline (PBS). Three microliters of the bacterial suspension were inoculated on previously stab-wounded surface-sterilized immature pears and incubated at 28°C. ImageJ software (National Institutes of Health; Bethesda, MD, United States) was used to quantify lesion area at 4 days post-inoculation (dpi). Pear assays were done in triplicate, and each experiment was repeated at least three times. For evaluation of hypersensitive-like cell death, overnight bacterial suspensions were adjusted to ≈ 1 × 107 CFU mL-1 in 0.5x PBS and infiltrated into 8-week old Nicotiana tabacum cv. Samsun leaves, using a needleless syringe. Cell collapse was evaluated 24 h post-infiltration (hpi). This assay was done in triplicate and each experiment was repeated at least three times. Statistical analyses were done using a one-way analysis of variance, and mean separation was accomplished using the Tukey–Kramer HDS test using JMP 12 (Cary, NC, United States).
Yeast Two-Hybrid Assays
dspE, eop3, eop4, and eop1 (full-gene and fragments) were cloned in fusion with the LexA binding domain into the bait vector pGilda (Clontech; Mountain View, CA, United States) using BamHI and XhoI restriction sites. esc1, esc3, and hrpN were digested with BamHI and EcoRI and cloned into the prey vector pB42AD. Prey and bait constructs were co-transformed into Saccharomyces cerevisiae EGY48 (pLacZ) using the Frozen-EZ Yeast Transformation II Kit (Zymo Research Corporation; Irvine, CA, United States). Transformants were selected on minimal synthetic dropout (SD)-galactose/raffinose medium amended with -Ura/-His/-Trp dropout. Positive interaction was screened on SD-galactose medium amended with -Ura/-His/-Trp/-Leu dropout and 5-bromo-4-chloro-3-indolyl-β-galactopyranoside (X-gal; 80 mg L-1). A yeast strain co-transformed with pLexA-53, containing p53, and pB42AD-T, containing SV40 large T-antigen, was used as a positive control for protein interaction.
CyaA Translocation Assay
eop1, eop3, and eop4 genes were amplified by PCR and ligated into the XbaI and SmaI restriction sites of pMJH20 in fusion with the catalytic domain of CyaA from Bordetella pertussis (Miao and Miller, 2000). Bacterial strains were transformed with the CyaA fusion constructs, and stable expression was confirmed by immunoblot using anti-CyaA antibody (Santa Cruz Biotechnology; Santa Cruz, CA, United States). Overnight suspensions were adjusted to ≈5 × 108 CFU mL-1 in PBS and were syringe-infiltrated into fully expanded leaves of 8-week-old N. tabacum cv. Samsun plants. Nine hours after inoculation, leaf disks of 1 cm in diameter were collected from the infiltrated areas, and were immediately frozen in liquid nitrogen. Samples were processed for cAMP quantification as previously described (Schechter et al., 2004). Briefly, frozen leaf disks were ground and resuspended in 250 μL of 0.1M HCl. Suspensions were centrifuged 5 min at 3000 g and cAMP levels in supernatants were measured using the cAMP EIA Kit (Cayman Chemical; Ann Arbor, MI, United States) according to the manufacturer’s instructions. Protein concentration in samples was determined using the BCA Protein Assay Kit (Thermo Fisher Scientific; Rockfort, IL, United States). Statistical analyses were done using a one-way analysis of variance, and mean separation was accomplished using the Tukey–Kramer HDS test using JMP 12 (Cary, NC, United States).
Protein Secretion Assay
Liquid cultures of the WT strain Ea1189 and mutant strains were grown overnight in 50 mL LB medium at 28°C. Cell pellets were resuspended in 40 mL of Hrp-inducing minimal medium (HrpMM), pH 5.7 (Huynh et al., 1989) and induced for 48 h at 24°C. Induced cultures were pelleted and both pellet and supernatants treated with 0.5 mM phenyl methylsulfonyl fluoride (PMSF) and concentrated (300x) using the Amicon Ultra-15 Centrifugal Filter Unit (30 kDa molecular cut-off; Millipore; Billerica, MA, United States). Protein concentrations were measured using the bicinchoninic acid (BCA) Protein Assay Kit. Ten micrograms of proteins in pellet and supernatant were analyzed by SDS-PAGE using a MiniPROTEAN3 system (Bio-Rad, Hercules, CA, United States), and gels were stained with the PierceTM Silver Stain Kit (Thermo Fisher Scientific; Rockfort, IL, United States).
Results
E. amylovora TTS Chaperones Interact with Multiple Effector Proteins in Yeast
TTS chaperone genes have often been found as short open reading frames (ORFs) located adjacent to the genes encoding their cognate effector proteins (Frithz-Lindsten et al., 1995; Cornelis, 2006). Kim and Beer (1998) reported the presence of two putative TTS chaperone genes named orfA and orfC located adjacent to eop1 and the harpin gene hrpW, respectively (Figure 1A). Similarly, Nissinen et al. (2007) reported the presence of a gene downstream of a hrpL-regulated promoter and located within an operon with the effector gene eop3, which shares homology with the TTS chaperone gene shcF from P. syringae pv. tomato (Shan et al., 2004) (Figure 1A). Further analyses of this putative TTS chaperone gene including prediction of the secondary structure for this 137-amino acid chaperone protein using the software 3D-Jury (Ginalski et al., 2003) and the Phyre server (Kelley and Sternberg, 2009) indicated the presence of three α-helical motifs and an acidic pI, both characteristic of TTS chaperones supporting a role as secretion chaperone for the protein encoded by this gene. Conversely, the secondary structure predicted for the protein encoded by orfC did not match the structural characteristics of TTS chaperones. In addition, sequence annotation and secondary structure analyses of genes surrounding the secreted effector Eop4 and the putative effector gene hopPtoCEa (Zhao et al., 2005) did not reveal the presence of any ORF with the characteristics of a TTS chaperone gene. These results indicate that in addition to DspE, two other effector proteins in E. amylovora are encoded adjacent to confirmed or putative chaperone genes. Because these effector proteins are named Eop1 and Eop3, we propose the putative genes encoding chaperone proteins be named esc1 and esc3 for Erwinia secretion chaperones 1 and 3, respectively.
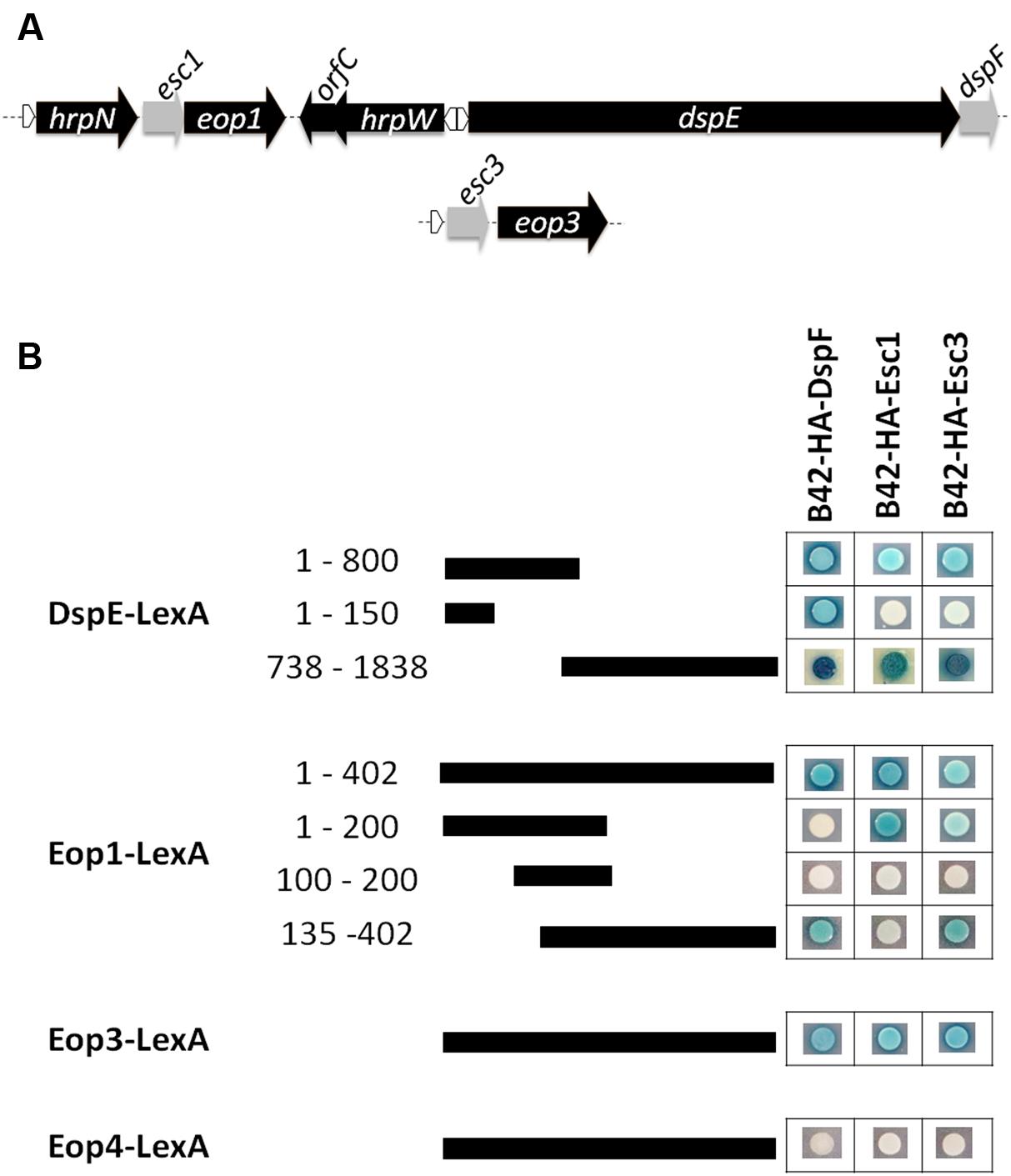
FIGURE 1. Interactions of TTS chaperones and effector proteins in E. amylovora. (A) Schematic diagram of gene organization of effector genes dspE, eop1, and eop3 with some adjacent genes and their putative chaperone partners. Depiction of these regions based on analysis of the E. amylovora strain ATCC49946 genome (accession number NC_013971.1). Gray-labeled ORFs are confirmed (dspF) or predicted to encode putative TTS chaperone proteins (esc1 and esc3). White triangles indicate the presence of a hrpL-regulated promoter. (B) Yeast two-hybrid interactions between prey fusions of DspF, the putative chaperones Esc1 and Esc3 in the pB42AD vector, and bait fusions of the effector proteins Eop1, Eop3, Eop4, N- terminal portions of DspE in the pGilda vector. Pairs of prey and bait fusions were transformed in the EGY48 yeast strain and selected on SD-galactose/raffinose medium amended with -Ura/-His/-Trp/-Leu dropout and Xgal (80 mg L-1). Positive interactions were identified when a yeast colony harboring a particular prey/bait fusion pair turned blue.
Similar to other TTS chaperone proteins, DspF has been shown to interact with more than one effector protein in yeast two-hybrid experiments (Asselin et al., 2006). In order to assess whether DspF, Esc1, and Esc3 interact with multiple TTS effector proteins in E. amylovora, we performed a series of yeast two hybrid analyses. All of the evaluated chaperone proteins fused with a B42-hemagglutinin (HA) tag interacted with fusions of the N-terminal portion of DspE with the LexA binding domain [DspE(1-800)-LexA], the C-terminal portion of DspE (DspE(738-1838)-LexA), Eop1-LexA, and Eop3-LexA, but did not interact with Eop4-LexA (Figure 1B). In contrast with DspF, which interacts with residues 51- 100 of DspE as previously reported (Triplett et al., 2009; Oh et al., 2010), B42-HA-Esc1 and B42-HA-Esc3 did not interact with the DspF-binding domain in the N terminal region of DspE-LexA (Figure 1B), indicating that the interaction domain for these chaperones is not shared with DspF and is located elsewhere in the effector. Indeed, a strong interaction of DspE(738-1838)-LexA with B42-HA-Esc1was detected, in agreement with similar results observed by Oh and collaborators with a DspE780-1838-LexA fusion (Oh et al., 2010), and with B42-HA-Esc3 as well. Interestingly, an interaction of DspF with the C-terminal portion of DspE (residues 738–1838) was detected, suggesting that this chaperone protein has multiple binding regions along the effector protein. The chaperone binding domains (CBD) of the Eop1 effector were mapped with further yeast studies. While the N-terminal 200 residues of Eop1 interacted strongly with its partner chaperone B42-HA-Esc1, no interaction with B42-HA-DspF and B42-HA-Esc3 was observed. Conversely, interaction of residues 135 – 402 in the C terminus of Eop1 with B42-HA-DspF and B42-HA-Esc3 was evidenced, while no interaction with B42-HA-Esc1 was observed (Figure 1B).
Simultaneous Expression of dspF, esc1, and esc3 Genes Is Required for Full Secretion of DspE and Translocation of a DspE(1-737)-CyaA Reporter
We previously used an adenylate cyclase reporter (CyaA) to demonstrate that the N-terminal CBD of DspE, is stably expressed and translocated into tobacco cells by the WT strain E. amylovora Ea1189 (Triplett et al., 2009). To assess the importance of DspF, Esc1, and Esc3 for the successful delivery of DspE into host cells, we compared the secretome of the WT Ea1189 and the different mutant strains when grown under hrpL-inducing conditions, and the translocation levels of the DspE(1-737)-CyaA fusion from the same strains to tobacco plants, as described in the methods section.
A reduction in DspE intracellular accumulation in the absence of DspF has been previously reported (Gaudriault et al., 2002). Moreover, secretion profiling revealed that, although DspE was secreted by all the strains tested in this study, seen by the presence of a previously characterized unique 198 kDa band (Gaudriault et al., 2002; Nissinen et al., 2007), secretion of this effector was apparently reduced in the double mutants Ea1189ΔdspF/esc1 and Ea1189ΔdspF/esc3, and in the triple chaperone gene mutant Ea1189ΔdspF/esc1/esc3, when compared with the single Ea1189ΔdspF mutant (Figure 2A). Secretion of DspE was not impaired in single mutants Ea1189Δesc1 and Ea1189Δesc3 when compared with the WT strain. Furthermore, while cAMP accumulation due to translocation of DspE(1-737)-CyaA from the Δesc1 and Δesc3 single mutants was not significantly different from the Ea1189 WT, significantly reduced levels of cAMP were observed for Ea1189ΔdspF and for both double mutants deleted in dspF and esc1 or esc3 (Figure 2B). The lowest level of translocation observed was for the Ea1189ΔdspF/esc1/esc3 triple mutant which was not significantly different from the DspE(1-15)-CyaA fusion, previously reported to be a non-translocated fragment of DspE (Triplett et al., 2009). Overall, these results indicate that TTS chaperones in E. amylovora act additively in order to effectively secrete DspE to the extracellular milieu and to translocate this effector protein into the host cytoplasm through the TTSS.
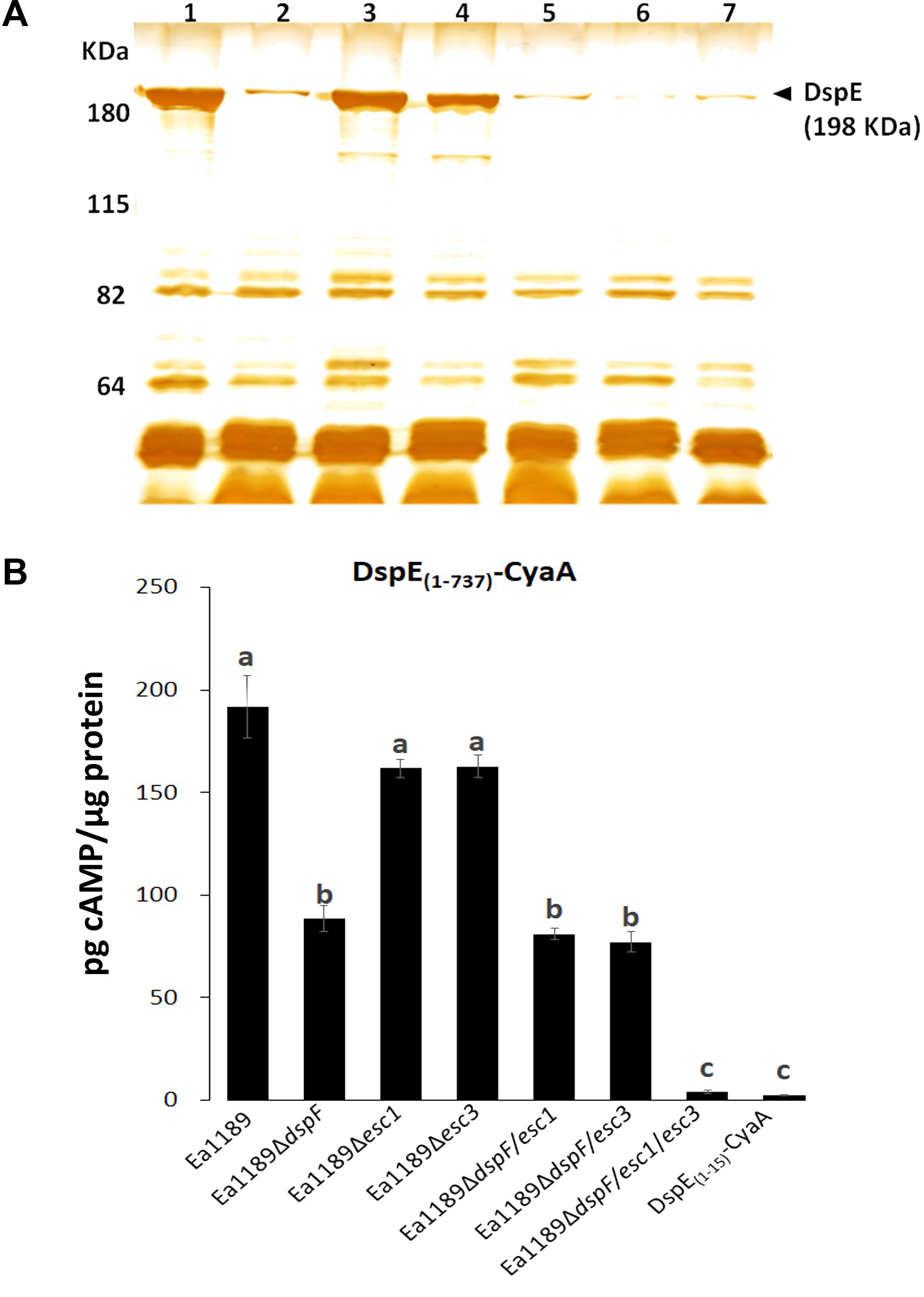
FIGURE 2. Effect of TTS chaperones in secretion of DspE and translocation of DspE(1-737)-CyaA. (A) Supernatant protein profiles of Ea1189 (1), ΔdspF (2), Δesc1 (3), Δesc3 (4), ΔdspF/esc1 (5), ΔdspF/esc3 (6), and ΔdspF/esc1/esc3 (7) after 48 h of growth in HrpMM, to induce the expression of the TTSS. Supernatant fractions were separated from pellets by centrifugation and equal amounts of protein (10 micrograms) were analyzed by SDS-PAGE. The assay was repeated twice with similar results. (B) cAMP accumulation in tobacco leaves inoculated with Ea1189, and mutant strains expressing DspE(1-737)-CyaA at 9 hpi. Ea1189 expressing DspE(1-15)-CyaA was used as negative control. Leaf samples were collected using a 1 cm diameter core borer and immediately frozen in liquid nitrogen for posterior processing as described in Section “Materials and Methods.” Results are the means and error bars represent the SED. Different letters above bars denote statistically significant differences (Tukey–Kramer HDS test, P < 0.05). The assay was done twice with similar results.
Esc1 and Esc3 Do Not Affect E. amylovora Pathogenicity
The pathogenicity of E. amylovora has been reported to be dramatically reduced in a dspF deletion mutant when compared with the WT strain (Gaudriault et al., 2002; Triplett et al., 2009). However, a small amount of the N-terminal portion of DspE is translocated in the absence of this chaperone protein, and a deletion mutant of dspF, although less aggressive than the WT, is still pathogenic (Triplett et al., 2009). To determine whether the additional TTS chaperone proteins Esc1 and Esc3 have an additive effect in the efficient translocation of DspE and hence, an additive impact on the pathogenicity phenotype of E. amylovora, a series of mutant strains was constructed and evaluated in an immature pear disease model and for induction of hypersensitive-like cell death in tobacco leaves. While inoculation with the Ea1189 WT, Ea1189Δesc1, and Ea1189Δesc3 resulted in severe tobacco leaf collapse at 24 hpi, inoculation with Ea1189ΔdspF and all Ea1189ΔdspF-derived double and triple chaperone mutants triggered no signs of necrosis (Figure 3A). Similarly, single deletions of esc1 and esc3 did not have a significant effect on pathogenicity on immature pears, whereas double deletion mutants Ea1189ΔdspF/esc1 and Ea1189ΔdspF/esc3 showed a reduction of aggressiveness that was statistically equivalent with the reduction in aggressiveness in the dspF mutant background (Figures 3B,C). Interestingly, a mutant strain lacking the three TTS chaperone genes still caused disease at the same level as double deletion mutants.
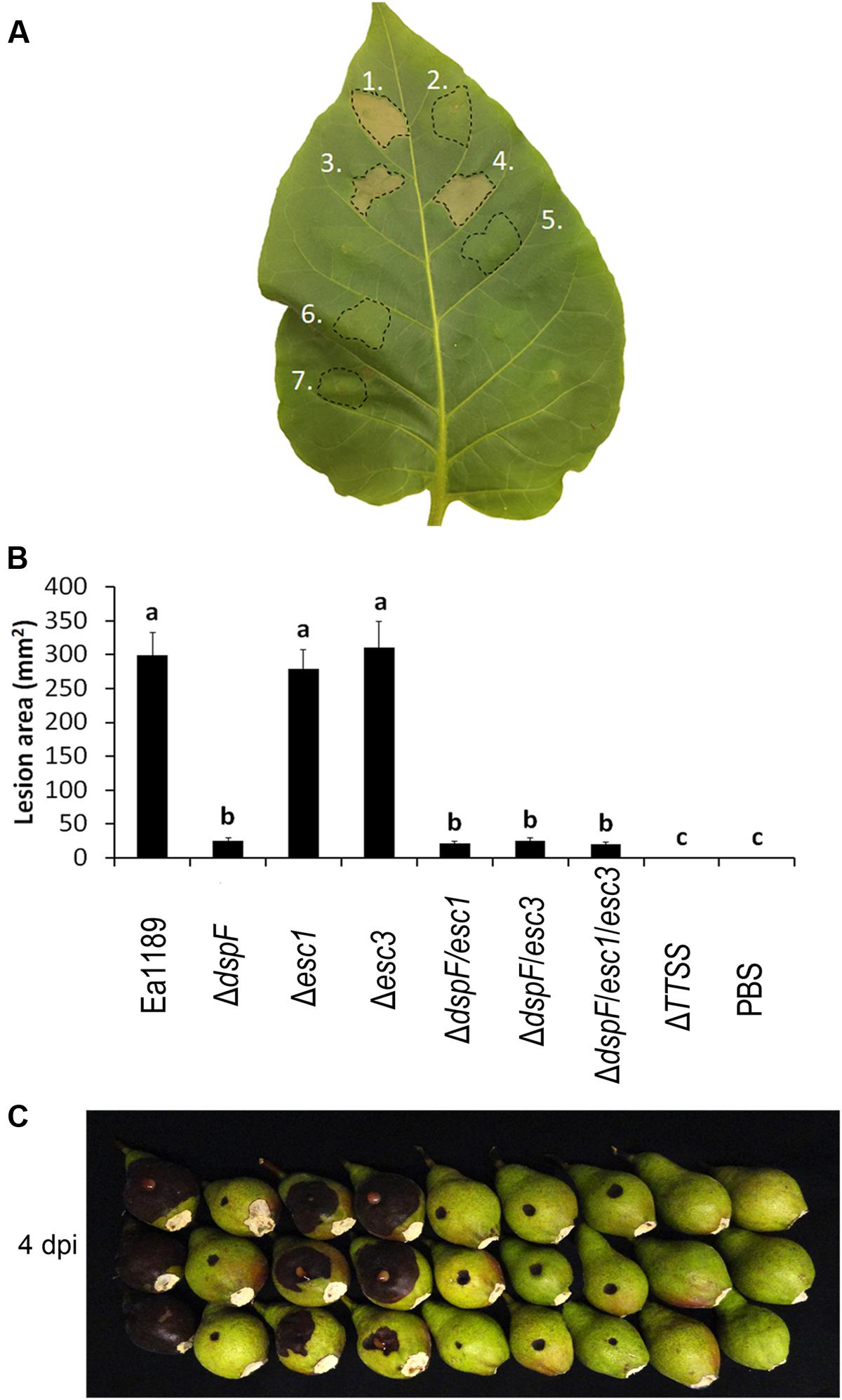
FIGURE 3. Effect of the TTS chaperones DspF, Esc1 and Esc3 on the virulence in E. amylovora. (A) Tobacco leaf cell death 24 h after infiltration with Ea1189 (1), Ea1189ΔdspF (2), Ea1189Δesc1 (3), Ea1189Δesc3 (4), Ea1189ΔdspF/esc1 (5), Ea1189ΔdspF/esc3 (6), and Ea1189ΔdspF/esc1/esc3 (7). (B) Lesion size on immature pears inoculated with 3 μL of the same strains at ≈1 × 104 CFU mL-1. Ea1189ΔTTSS strain was used as negative control. Lesions were photographed and the area was quantified at 4 dpi using ImageJ software. The experiment was repeated three times with similar results. Results are the means and error bars represent the SED. Different letters above bars denote statistically significant differences (Tukey–Kramer HDS test, P < 0.05). (C) Symptom development on stab-wounded immature pears at 4 dpi with Ea1189 and mutant strains.
DspF Negatively Affects the Translocation of Eop1-CyaA and Eop3-CyaA, But Not Eop4-CyaA
To determine whether DspF affects the translocation of the effector proteins Eop1, Eop3 and Eop4, accumulation of cAMP in tobacco leaf fusions was evaluated after infiltration of Ea1189ΔdspF containing either Eop1-CyaA, Eop3-CyaA, or Eop4-CyaA. Surprisingly, translocation of Eop1-CyaA and Eop3-CyaA was significantly increased from Ea1189ΔdspF compared to the WT Ea1189 or to the Ea1189ΔdspF/dspF complemented strain (Figures 4A,B). On the other hand, cAMP accumulation due to Eop4-CyaA translocation was not affected by the deletion of dspF (Figure 4C). These observations suggest that DspF might play a regulatory role in the translocation of Eop1 and Eop3.
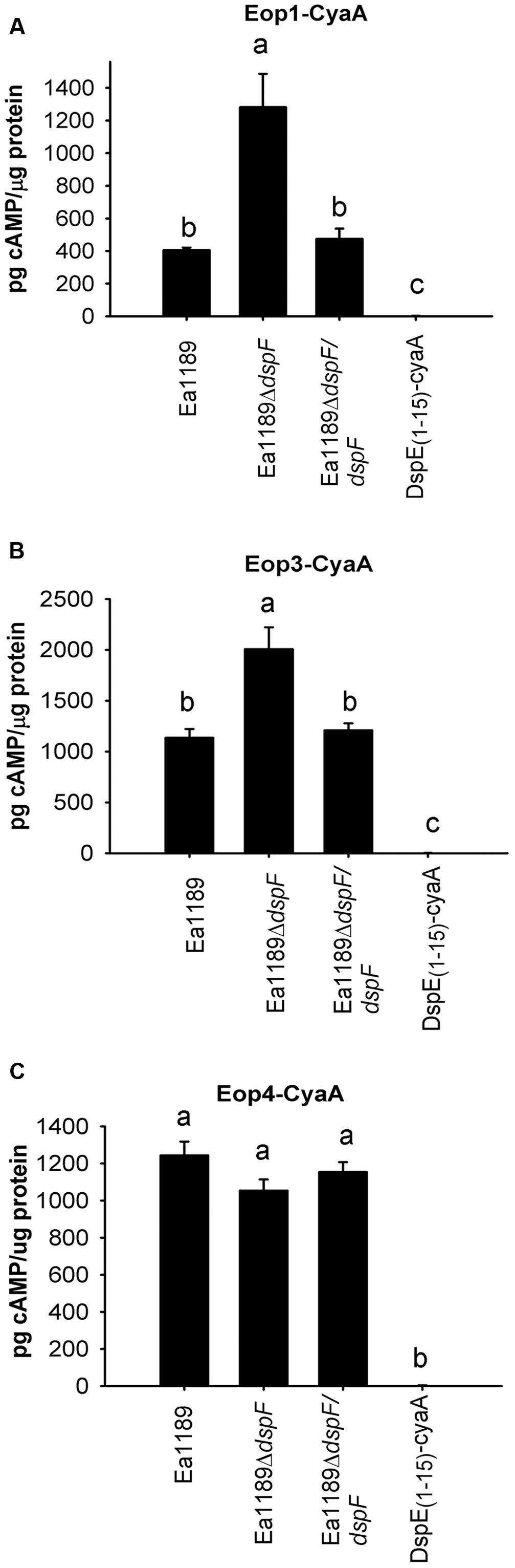
FIGURE 4. Effect of the TTS chaperones DspF on the traslocation of Eop1-CyaA, Eop3 -CyaA and Eop4-CyaA fusion proteins. cAMP accumulation was measured at 9 hpi on tobacco leaves infiltrated with Ea1189, Ea1189ΔdspF and Ea1189ΔdspF/dspF, expressing Eop1-CyaA (A), Eop3-CyaA (B) and Eop4-CyaA (C). Ea1189 expressing DspE(1-15)-CyaA was used as negative control. Leaf samples were collected using a 1 cm diameter core borer and immediately frozen in liquid nitrogen for posterior processing Results represent the means and error bars represent the SED. Different letters above bars denote statistically significant differences (Tukey–Kramer HDS test, P < 0.05). The experiment was done twice with similar results.
Discussion
The TTSS exhibits various mechanisms of regulation at every stage in the assembly and translocation processes (Cornelis, 2006). In fact, a hierarchical organization in effector protein delivery through the TTSS has been demonstrated for several animal pathogens (Mills et al., 2008; Lara-Tejero et al., 2011; Portaliou et al., 2017). However, mechanisms regulating TTSS assembly and the translocation of pre-formed proteins in plant pathogenic bacteria are still poorly understood. In this study, we examined the roles of DspF and two other TTS chaperones, Esc1 and Esc3, for interactions with effector proteins, effects on secretion and translocation of effectors, and effects on bacterial pathogenicity.
Although many TTS chaperones interact with a single effector protein (class IA), class IB TTS chaperones that bind to multiple target effectors (multiple cargo) have been described in several animal pathogenic bacteria. Examples of chaperones that can bind multiple effectors include SrcA and InvB from Salmonella enterica serovar Typhimurium and CesT from enteropathogenic Escherichia coli (Bronstein et al., 2000; Creasey et al., 2003; Ehrbar et al., 2004; Thomas et al., 2005; Cooper et al., 2010). Plant pathogen examples include HpaB from X. campestris pv. vesicatoria, and ShcS1 and ShcO1 from P. syringae pv. tomato (Büttner et al., 2004; Kabisch et al., 2005; Büttner et al., 2006). Our yeast two-hybrid studies suggest that DspF, Esc1, and Esc3 belong to the class IB TTS chaperone category, as they bind not only to their cognate effector partner, but also seem to be functioning as multi-cargo chaperones. In the case of DspE, these TTS chaperones function cooperatively in DspE cellular trafficking and translocation into the plant cell. This finding is consistent with previous studies in Chlamydia pneumoniae showing that the TTS chaperones Ssc1 and Ssc4 bind forming a complex that interacts with the N-terminal region of the effector protein CopN, promoting CopN secretion through the TTSS (Silva-Herzog et al., 2011). Similarly, the TTS chaperones EscH and EscS from Edwardsiella piscicida have be demonstrated to interact with the effector protein EseK, enhancing secretion and translocation into host cells (Cao et al., 2017).
In a previous report, we mapped a CBD for DspF to residues 51- 100 in the N terminus of DspE (Triplett et al., 2009). Interestingly, yeast two-hybrid results suggest that, in addition to the N terminal-localized CBS, DspF interacts with at least one additional domain of DspE. Since one the main roles of TTS chaperones is the stabilization of the cognate effector within the bacterial cytoplasm, it is not surprising that DspF might bind to multiple regions along the length of DspE, especially given the large size of this effector protein (1838 residues). Moreover, our results suggest that the CBDs for Esc1 and Esc3 are not located in the N-terminal portion of DspE, but are located elsewhere in the effector protein, ruling out the possibility of heterodimerization with DspF for binding in this specific location of the effector. The presence of CBDs in non-N-terminal effector regions has been reported previously including in P. syringae pv. tomato for the TTS chaperones ShcO1, ShcS1, and ShcS2, which bind to the middle third portion of HopO1-1 (Guo et al., 2005), and for CT548, a TTS chaperone from Chlamydia trachomatis, that binds to the central region of CT082, a type III substrate (Pais et al., 2013).
Echoing the specificity of DspE N-terminal CBD for the cognate chaperone DspF, the CBD in residues 1- 100 of the effector Eop1 were only bound by the cognate chaperone Esc1, while DspF and Esc3 binding sites are likely located within the last 200 residues of this effector. Although it has been previously reported that DspF is indispensable for stable expression of DspE in E. amylovora cells and for secretion to the extracellular milieu, as this effector protein was not detected by immunoblot analyses in whole cell lysates or culture supernatants of a dspF mutant strain (Gaudriault et al., 2002), our studies indicated that the full-length DspE can be expressed and secreted in the absence of DspF, at lower levels than the WT strain (Figure 3A). This discrepancy can be explained by the differences between the approaches used to detect the protein and their detection thresholds. Moreover, the fact that a dpsF mutant strain retains some pathogenicity while a dspE mutant does not (Gaudriault et al., 2002; Triplett et al., 2009), supports our observation that DspE can be expressed, secreted, and translocated in a DspF-independent fashion.
The capacity of the N-terminal region of DspE for DspF-independent translocation previously observed (Triplett et al., 2009), and the interaction of LexA-DspE(1-800) and LexA-DspE(738-1838) with B42-HA-Esc1 and B42-HA-Esc3 observed in this study, led us to hypothesize that TTS chaperone proteins other than DspF might also be involved in the efficient translocation of DspE into the host cell. While deletions of esc1 or esc3 do not have a significant effect on pathogenicity, our secretion and translocation assays indicated that the activity of the TTS chaperones on DspE secretion and translocation is additive, as secretion of DspE was visibly diminished from the double mutants Ea1189ΔdspF/esc1 and Ea1189ΔdspF/esc3 and the ΔdspF/esc1/esc3 triple mutant, and the ΔdspF/esc1/esc3 triple mutant strain permits less translocation of DspE(1-737)-CyaA translocation than single or double chaperone mutants. It should be noted that for all of our translocation studies we used an N-terminal portion of DspE rather than the full-length protein, and that the translocation efficiency of the N-terminal reporter could differ from that of the intact protein. Our results present primary evidence of TTS chaperone cooperative behavior for the translocation of DspE, and further studies with the full-length effector would complement these findings.
In contrast to DspE(1-737)-CyaA and Eop4-CyaA, our experiments indicated that translocation of Eop1-CyaA and Eop3-CyaA is negatively affected by DspF. These results suggest that DspF might play an antagonistic role, delaying the translocation of effectors other than DspE, and establishing a hierarchy for effector export. In a recent study, Portaliou et al. (2017) demonstrated that the TTS chaperone association of SepD with the effector protein SepL in enteropathogenic E. coli is critical for the temporal regulation of TTS substrate passage through the translocase channel. Furthermore, the multi-cargo chaperone HpaB in X. campestris pv. vesicatoria has been determined to function as a regulator of the recognition of translocation signals independently of its TTS chaperone role (Scheibner et al., 2017). The mechanism of DspF-dependent regulation of translocation remains unknown, and further studies would be helpful in determining if this regulation involves differences in chaperone-effector affinities or regulation at the transcriptional, translational or post-translational levels. In addition, several studies have postulated Eop1 and Eop3 as effector proteins exhibiting avirulence functions (Asselin et al., 2011; Bocsanczy et al., 2012) which might explain the antagonistic role of DspF on these effector proteins.
In this study we took advantage of the small number of TTS effector and chaperone proteins produced by E. amylovora in order to investigate the interactions that mediate effector cellular trafficking and extracellular export and their implications in bacterial pathogenicity. We determined that the TTS chaperones DspF, Esc1 and Esc3 exhibit features of multi-cargo and that cooperation exists between them in order to efficiently deliver the TTS effector DspE into plant cells by E. amylovora. Moreover, our findings suggest that in addition to enhancing DspE delivery to the host cell through the TTSS, DspF exerts additional regulatory roles on other effectors proteins, delaying their translocation and thus modulating the timing of effector export. Further studies are needed to determine how E. amylovora orchestrates hierarchical secretion and translocation of effectors to colonize its host and cause disease.
Author Contributions
LC, LT, and GS conceived and designed the experiments. LC and LT performed the experiments and analyzed the data. LC, LT, and GS wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was supported by the Agriculture and Food Research Initiative Competitive Grants Program Grant no. 2015-67013-23068 from the USDA National Institute of Food and Agriculture and Michigan State University AgBioResearch.
References
Akeda, Y., and Galán, J. E. (2005). Chaperone release and unfolding of substrates in type III secretion. Nature 437, 911–915.
Asselin, J., Oh, C., Nissinen, R., and Beer, S. (2006). The secretion of EopB from Erwinia amylovora. Acta Hortic. 704, 409–416.
Asselin, J. E., Bonasera, J. M., Kim, J. F., Oh, C. S., and Beer, S. V. (2011). Eop1 from a Rubus strain of Erwinia amylovora functions as a host-range limiting factor. Phytopathology 101, 935–944. doi: 10.1094/PHYTO-12-10-0339
Blaylock, B., Sorg, J. A., and Schneewind, O. (2008). Yersinia enterocolitica type III secretion of YopR requires a structure in its mRNA. Mol. Microbiol. 70, 1210–1222. doi: 10.1111/j.1365-2958.2008.06474.x
Bocsanczy, A. M., Schneider, D. J., DeClerck, G. A., Cartinhour, S., and Beer, S. V. (2012). HopX1 in Erwinia amylovora functions as an avirulence protein in apple and is regulated by HrpL. J. Bacteriol. 194, 553–560. doi: 10.1128/JB.05065-11
Bogdanove, A. J., Bauer, D. W., and Beer, S. V. (1998). Erwinia amylovora secretes DspE, a pathogenicity factor and functional AvrE homolog, through the Hrp (type III secretion) pathway. J. Bacteriol. 180, 2244–2247.
Boureau, T., ElMaarouf-Bouteau, H., Garnier, A., Brisset, M.-N., Perino, C., Pucheu, I., et al. (2006). DspA/E, a type III effector essential for Erwinia amylovora pathogenicity and growth in planta, induces cell death in host apple and nonhost tobacco plants. Mol. Plant Microbe Interact. 19, 16–24. doi: 10.1094/mpmi-19-0016
Bronstein, P. A., Miao, E. A., and Miller, S. I. (2000). InvB is a type III secretion chaperone specific for SspA. J. Bacteriol. 182, 6638–6644. doi: 10.1128/jb.182.23.6638-6644.2000
Burse, A., Weingart, H., and Ullrich, M. S. (2004). The phytoalexin-inducible multidrug efflux pump AcrAB contributes to virulence in the fire blight pathogen, Erwinia amylovora. Mol. Plant Microbe Interact. 17, 43–54. doi: 10.1094/mpmi.2004.17.1.43
Büttner, D., Gürlebeck, D., Noël, L. D., and Bonas, U. (2004). HpaB from Xanthomonas campestris pv. vesicatoria acts as an exit control protein in type III-dependent protein secretion. Mol. Microbiol. 54, 755–768. doi: 10.1111/j.1365-2958.2004.04302.x
Büttner, D., and He, S. Y. (2009). Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 150, 1656–1664.
Büttner, D., Lorenz, C., Weber, E., and Bonas, U. (2006). Targeting of two effector protein classes to the type III secretion system by a HpaC- and HpaB-dependent protein complex from Xanthomonas campestris pv. vesicatoria. Mol. Microbiol. 59, 513–527. doi: 10.1111/j.1365-2958.2005.04924.x
Cao, H., Yang, C., Quan, S., Hu, T., Zhang, L., Zhang, Y., et al. (2017). Novel T3SS effector EseK in Edwardsiella piscicida is chaperoned by EscH and EscS to express virulence. Cell. Microbiol. 20:e12790. doi: 10.1111/cmi.12790
Cherepanov, P. P., and Wackernagel, W. (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14. doi: 10.1016/0378-1119(95)00193-a
Cooper, C. A., Zhang, K., Andres, S. N., Fang, Y., Kaniuk, N. A., Hannemann, M., et al. (2010). Structural and biochemical characterization of SrcA, a multi-cargo type III secretion chaperone in Salmonella required for pathogenic association with a host. PLOS Pathog. 6:e1000751. doi: 10.1371/journal.ppat.1000751
Cornelis, G. R., and Gijsegem, F. V. (2000). Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54, 735–774. doi: 10.1146/annurev.micro.54.1.735
Creasey, E. A., Delahay, R. M., Bishop, A. A., Shaw, R. K., Kenny, B., Knutton, S., et al. (2003). CesT is a bivalent enteropathogenic Escherichia coli chaperone required for translocation of both Tir and Map. Mol. Microbiol. 47, 209–221. doi: 10.1046/j.1365-2958.2003.03290.x
Darwin, K. H., and Miller, V. L. (2001). Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20, 1850–1862.
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Ehrbar, K., Hapfelmeier, S., Stecher, B., and Hardt, W.-D. (2004). InvB Is required for type III-dependent secretion of SopA in Salmonella enterica serovar Typhimurium. J. Bacteriol. 186, 1215–1219. doi: 10.1128/jb.186.4.1215-1219.2004
Frithz-Lindsten, E., Rosqvist, R., Johansson, L., and Forsberg, Å (1995). The chaperone-like protein YerA of Yersinia pseudotuberculosis stabilizes YopE in the cytoplasm but is dispensible for targeting to the secretion loci. Mol. Microbiol. 16, 635–647. doi: 10.1111/j.1365-2958.1995.tb02426.x
Galán, J. E., Lara-Tejero, M., Marlovits, T. C., and Wagner, S. (2014). Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 68, 415–438. doi: 10.1146/annurev-micro-092412-155725
Gaudriault, S., Malandrin, L., Paulin, J. P., and Barny, M. A. (1997). DspA, an essential pathogenicity factor of Erwinia amylovora showing homology with AvrE of Pseudomonas syringae, is secreted via the Hrp secretion pathway in a DspB-dependent way. Mol. Microbiol. 26, 1057–1069. doi: 10.1046/j.1365-2958.1997.6442015.x
Gaudriault, S., Paulin, J.-P., and Barny, M.-A. (2002). The DspB/F protein of Erwinia amylovora is a type III secretion chaperone ensuring efficient intrabacterial production of the Hrp-secreted DspA/E pathogenicity factor. Mol. Plant Pathol. 3, 313–320. doi: 10.1046/j.1364-3703.2002.00124.x
Ghosh, P. (2004). Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68, 771–795. doi: 10.1128/mmbr.68.4.771-795.2004
Ginalski, K., Elofsson, A., Fischer, D., and Rychlewski, L. (2003). 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics 19, 1015–1018. doi: 10.1093/bioinformatics/btg124
Guo, M., Chancey, S. T., Tian, F., Ge, Z., Jamir, Y., and Alfano, J. R. (2005). Pseudomonas syringae type III chaperones ShcO1, ShcS1, and ShcS2 facilitate translocation of their cognate effectors and can substitute for each other in the secretion of HopO1-1. J. Bacteriol. 187, 4257–4269. doi: 10.1128/jb.187.12.4257-4269.2005
Guttman, D. S., Vinatzer, B. A., Sarkar, S. F., Ranall, M. V., Kettler, G., and Greenberg, J. T. (2002). A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295, 1722–1726. doi: 10.1126/science.295.5560.1722
Huynh, T., Dahlbeck, D., and Staskawicz, B. (1989). Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245, 1374–1377. doi: 10.1126/science.2781284
Kabisch, U., Landgraf, A., Krause, J., Bonas, U., and Boch, J. (2005). Type III secretion chaperones ShcS1 and ShcO1 from Pseudomonas syringae pv. tomato DC3000 bind more than one effector. Microbiology 151, 269–280. doi: 10.1099/mic.0.27491-0
Kelley, L. A., and Sternberg, M. J. E. (2009). Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371. doi: 10.1038/nprot.2009.2
Kim, J. F., and Beer, S. V. (1998). HrpW of Erwinia amylovora, a new harpin that contains a domain homologous to pectate lyases of a distinct class. J. Bacteriol. 180, 5203–5210.
Koczan, J. M., Lenneman, B. R., McGrath, M. J., and Sundin, G. W. (2011). Cell surface attachment structures contribute to biofilm formation and xylem colonization by Erwinia amylovora. Appl. Environ. Microbiol. 77, 7031–7039. doi: 10.1128/AEM.05138-11
Lara-Tejero, M., Kato, J., Wagner, S., Liu, X., and Galán, J. E. (2011). A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331, 1188–1191. doi: 10.1126/science.1201476
Lorenz, C., and Buttner, D. (2009). Functional characterization of the type III secretion ATPase HrcN from the plant pathogen Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 191, 1414–1428. doi: 10.1128/JB.01446-08
Lorenz, C., Kirchner, O., Egler, M., Stuttmann, J., Bonas, U., and Büttner, D. (2008). HpaA from Xanthomonas is a regulator of type III secretion. Mol. Microbiol. 69, 344–360. doi: 10.1111/j.1365-2958.2008.06280.x
Losada, L. C., and Hutcheson, S. W. (2005). Type III secretion chaperones of Pseudomonas syringae protect effectors from Lon-associated degradation. Mol. Microbiol. 55, 941–953. doi: 10.1111/j.1365-2958.2004.04438.x
Luo, Y., Bertero, M. G., Frey, E. A., Pfuetzner, R. A., Wenk, M. R., Creagh, L., et al. (2001). Structural and biochemical characterization of the type III secretion chaperones CesT and SigE. Nat. Struct. Mol. Biol. 8, 1031–1036.
Malnoy, M., Martens, S., Norelli, J. L., Barny, M.-A., Sundin, G. W., Smits, T. H. M., et al. (2012). Fire blight: applied genomic insights of the pathogen and host. Annu. Rev. Phytopathol. 50, 475–494. doi: 10.1146/annurev-phyto-081211-172931
Matsumoto, H., and Young, G. M. (2009). Essential role of the SycP chaperone in type III secretion of the YspP effector. J. Bacteriol. 191, 1703–1715. doi: 10.1128/JB.01021-08
Miao, E. A., and Miller, S. I. (2000). A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. U.S.A. 97, 7539–7544. doi: 10.1073/pnas.97.13.7539
Mills, E., Baruch, K., Charpentier, X., Kobi, S., and Rosenshine, I. (2008). Real-time analysis of effector translocation by the type III secretion system of enteropathogenic Escherichia coli. Cell Host Microbe 3, 104–113. doi: 10.1016/j.chom.2007.11.007
Mudgett, M. B., Chesnokova, O., Dahlbeck, D., Clark, E. T., Rossier, O., Bonas, U., et al. (2000). Molecular signals required for type III secretion and translocation of the Xanthomonas campestris AvrBs2 protein to pepper plants. Proc. Natl. Acad. Sci. U.S.A. 97, 13324–13329. doi: 10.1073/pnas.230450797
Nissinen, R. M., Ytterberg, A. J., Bogdanove, A. J., Van Wijk, K. J., and Beer, S. V. (2007). Analyses of the secretomes of Erwinia amylovora and selected hrp mutants reveal novel type III secreted proteins and an effect of HrpJ on extracellular harpin levels. Mol. Plant Pathol. 8, 55–67. doi: 10.1111/j.1364-3703.2006.00370.x
Oh, C., and Beer, S. V. (2005). Molecular genetics of Erwinia amylovora involved in the development of fire blight. FEMS Microbiol. Lett. 253, 185–192. doi: 10.1016/j.femsle.2005.09.051
Oh, C., Carpenter, S. C. D., Hayes, M. L., and Beer, S. V. (2010). Secretion and translocation signals and DspB/F-binding domains in the type III effector DspA/E of Erwinia amylovora. Microbiology 156, 1211–1220. doi: 10.1099/mic.0.027144-0
Pais, S. V., Milho, C., Almeida, F., and Mota, L. J. (2013). Identification of novel type III secretion chaperone-substrate complexes of Chlamydia trachomatis. PLOS ONE 8:e56292. doi: 10.1371/journal.pone.0056292
Parsot, C., Hamiaux, C., and Page, A.-L. (2003). The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 6, 7–14. doi: 10.1016/S1369-5274(02)00002-4
Petnicki-Ocwieja, T., Schneider, D. J., Tam, V. C., Chancey, S. T., Shan, L., Jamir, Y., et al. (2002). Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U.S.A. 99, 7652–7657. doi: 10.1073/pnas.112183899
Portaliou, A. G., Tsolis, K. C., Loos, M. S., Balabanidou, V., Rayo, J., Tsirigotaki, A., et al. (2017). Hierarchical protein targeting and secretion is controlled by an affinity switch in the type III secretion system of enteropathogenic Escherichia coli. EMBO J. 36, 3517–3531. doi: 10.15252/embj.201797515
Raymond, B., Young, J. C., Pallett, M., Endres, R. G., Clements, A., and Frankel, G. (2013). Subversion of trafficking, apoptosis, and innate immunity by type III secretion system effectors. Trends Microbiol. 21, 430–441. doi: 10.1016/j.tim.2013.06.008
Sambrook, J., Fritsch, E. F., and Maniatis, T. (2001). Molecular Cloning. New York, NY: Cold Spring Harbor Laboratory Press.
Schechter, L. M., Roberts, K. A., Jamir, Y., Alfano, J. R., and Collmer, A. (2004). Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186, 543–555. doi: 10.1128/jb.186.2.543-555.2004
Scheibner, F., Hartmann, N., Hausner, J., Lorenz, C., Hoffmeister, A.-K., and Büttner, D. (2017). The type III secretion chaperone HpaB controls the translocation of effector and noneffector proteins from Xanthomonas campestris pv. vesicatoria. Mol. Plant Microbe Interact. 31, 61–74. doi: 10.1094/MPMI-06-17-0138-R
Shan, L., Oh, H.-S., Chen, J., Guo, M., Zhou, J., Alfano, J. R., et al. (2004). The HopPtoF locus of Pseudomonas syringae pv. tomato DC3000 encodes a type III chaperone and a cognate effector. Mol. Plant Microbe Interact. 17, 447–455. doi: 10.1094/MPMI.2004.17.5.447
Silva-Herzog, E., Joseph, S. S., Avery, A. K., Coba, J. A., Wolf, K., Fields, K. A., et al. (2011). Scc1 (CP0432) and Scc4 (CP0033) function as a type III secretion chaperone for CopN of Chlamydia pneumoniae. J. Bacteriol. 193, 3490–3496. doi: 10.1128/JB.00203-11
Thomas, N. A., Deng, W., Puente, J. L., Frey, E. A., Yip, C. K., Strynadka, N. C. J., et al. (2005). CesT is a multi-effector chaperone and recruitment factor required for the efficient type III secretion of both LEE- and non-LEE-encoded effectors of enteropathogenic Escherichia coli. Mol. Microbiol. 57, 1762–1779. doi: 10.1111/j.1365-2958.2005.04802.x
Thomas, N. A., Ma, I., Prasad, M. E., and Rafuse, C. (2012). Expanded roles for multicargo and class 1B effector chaperones in type III secretion. J. Bacteriol. 194, 3767–3773. doi: 10.1128/jb.00406-12
Triplett, L. R., Melotto, M., and Sundin, G. W. (2009). Functional analysis of the N terminus of the Erwinia amylovora secreted effector DspA/E reveals features required for secretion, translocation, and binding to the chaperone DspB/F. Mol. Plant Microbe Interact. 22, 1282–1292. doi: 10.1094/MPMI-22-10-1282
Triplett, L. R., Wedemeyer, W. J., and Sundin, G. W. (2010). Homology-based modeling of the Erwinia amylovora type III secretion chaperone DspF used to identify amino acids required for virulence and interaction with the effector DspE. Res. Microbiol. 161, 613–618. doi: 10.1016/j.resmic.2010.05.015
Tsai, C.-L., Burkinshaw, B. J., Strynadka, N. C. J., and Tainer, J. A. (2015). The Salmonella type III secretion system virulence effector forms a new hexameric chaperone assembly for export of effector/chaperone complexes. J. Bacteriol. 197, 672–675. doi: 10.1128/jb.02524-14
Zhao, Y., Blumer, S. E., and Sundin, G. W. (2005). Identification of Erwinia amylovora genes induced during infection of immature pear tissue. J. Bacteriol. 187, 8088–8103. doi: 10.1128/jb.187.23.8088-8103.2005
Keywords: type III secretion, type III chaperones, class IB type III chaperone proteins, effector translocation, Erwinia amylovora
Citation: Castiblanco LF, Triplett LR and Sundin GW (2018) Regulation of Effector Delivery by Type III Secretion Chaperone Proteins in Erwinia amylovora. Front. Microbiol. 9:146. doi: 10.3389/fmicb.2018.00146
Received: 20 November 2017; Accepted: 23 January 2018;
Published: 08 February 2018.
Edited by:
Mahmut Tör, University of Worcester, United KingdomReviewed by:
Kubilay Kurtulus Bastas, Selçuk University, TurkeyJohn Mansfield, Imperial College London, United Kingdom
Copyright © 2018 Castiblanco, Triplett and Sundin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: George W. Sundin, c3VuZGluQG1zdS5lZHU=
†These authors have contributed equally to this work.
 Luisa F. Castiblanco
Luisa F. Castiblanco Lindsay R. Triplett
Lindsay R. Triplett George W. Sundin
George W. Sundin