- 1Université Clermont Auvergne, INRA, UMR454 MEDiS, Clermont-Ferrand, France
- 2EA4043 Unité Bactéries Pathogènes et Santé, Châtenay-Malabry, France
- 3Micalis Institute, INRA, AgroParisTech, Université Paris-Saclay, Jouy-en-Josas, France
The cell envelope of parietal monoderm bacteria (archetypal Gram-positive bacteria) is formed of a cytoplasmic membrane (CM) and a cell wall (CW). While the CM is composed of phospholipids, the CW is composed at least of peptidoglycan (PG) covalently linked to other biopolymers, such as teichoic acids, polysaccharides, and/or polyglutamate. Considering the CW is a porous structure with low selective permeability contrary to the CM, the bacterial cell surface hugs the molecular figure of the CW components as a well of the external side of the CM. While the surfaceome corresponds to the totality of the molecules found at the bacterial cell surface, the proteinaceous complement of the surfaceome is the proteosurfaceome. Once translocated across the CM, secreted proteins can either be released in the extracellular milieu or exposed at the cell surface by associating to the CM or the CW. Following the gene ontology (GO) for cellular components, cell-surface proteins at the CM can either be integral (GO: 0031226), i.e., the integral membrane proteins, or anchored to the membrane (GO: 0046658), i.e., the lipoproteins. At the CW (GO: 0009275), cell-surface proteins can be covalently bound, i.e., the LPXTG-proteins, or bound through weak interactions to the PG or wall polysaccharides, i.e., the cell wall binding proteins. Besides monopolypeptides, some proteins can associate to each other to form supramolecular protein structures of high molecular weight, namely the S-layer, pili, flagella, and cellulosomes. After reviewing the cell envelope components and the different molecular mechanisms involved in protein attachment to the cell envelope, perspectives in investigating the proteosurfaceome in parietal monoderm bacteria are further discussed.
Introduction
As the interface of the cell with its surrounding, the bacterial cell surface plays a crucial role in all types of interactions. In the first instance, the diversity of the bacterial cell envelope is generally viewed as dichotomic, on the one hand, the Gram-positive bacteria, and on the other hand, the Gram-negative bacteria (Desvaux et al., 2004, 2009). This difference is based on the result of the Gram staining method originally developed by the Danish pharmacologist and physician Hans Christian Joachim Gram (Gram, 1884) and still routinely used worldwide to differentiate bacteria (Beveridge, 2001). With the development of microscopic techniques, it first appeared the difference in staining was the result of profound divergence in structural organisation of the bacterial cell envelope, where Gram-positive bacteria have a thick cell wall (CW) sitting atop of a cytoplasmic membrane (CM) (Silhavy et al., 2010). Later on, molecular analyses further revealed that Gram-positive bacteria corresponded to a phylogenetically coherent group within the domain Bacteria and belonged to only two phyla, namely the low G+C% Gram-positive bacteria of the phylum Firmicutes and the high G+C% Gram-positive bacteria of the phylum Actinobacteria (Woese, 1987; Woese et al., 1990). Over the years, though, it appears this terminology presents some ambiguity when considering the diversity of the domain Bacteria (Desvaux et al., 2009). Considering the term “Gram-positive bacteria,” it can refer to three distinct, and sometimes incompatible elements, i.e., a Gram staining result, a cell envelope architecture and/or a taxonomic group. For instance, bacteria of the class Mollicutes, comprising the genus Mycoplasma, cannot retain the Gram stain because they naturally lack a CW although the low G+C% content of their genomes and other molecular markers resemble those of Gram-positive bacteria of the phylum Firmicutes (Razin et al., 1998). Species of the genus Mycobacterium possess a peculiar cell envelope with a mycomembrane preventing Gram staining and thus require alternative staining methods called acid-fast (Somoskovi et al., 2001) but nonetheless belong to the high G+C% Gram-positive bacteria of the phylum Actinobacteria (Draper, 1998). In some deep branches of the phylum Firmicutes, some bacteria clearly exhibit Gram-negative cell envelope for which a new class was proposed, i.e., the Negativicutes (Marchandin et al., 2010).
Inspired by the research work of Gupta (1998a,b, 2000), the description of the bacterial cell envelope respective to the number of biological membranes appeared much more definite and was first reintroduced in the field of bacterial protein secretion (Desvaux et al., 2009). While monoderm bacteria refer to species exhibiting only one biological membrane, namely the CM, diderm bacteria correspond to species exhibiting two biological membranes, i.e., an inner membrane and an outer membrane. Monoderm bacteria can be further discriminated into (i) simple monoderm, lacking a CW (e.g., bacteria from the genus Mycoplasma), and (ii) parietal monoderm, exhibiting a CW (archetypal Gram-positive bacteria) (Sutcliffe, 2010; Gupta, 2011). As such, parietal monoderm bacteria include most Firmicutes, e.g., from the class Bacilli and Clostridia, but of course exclude the class Mollicutes and Negativicutes as well as the Actinobacteria exhibiting a mycolate outer membrane.
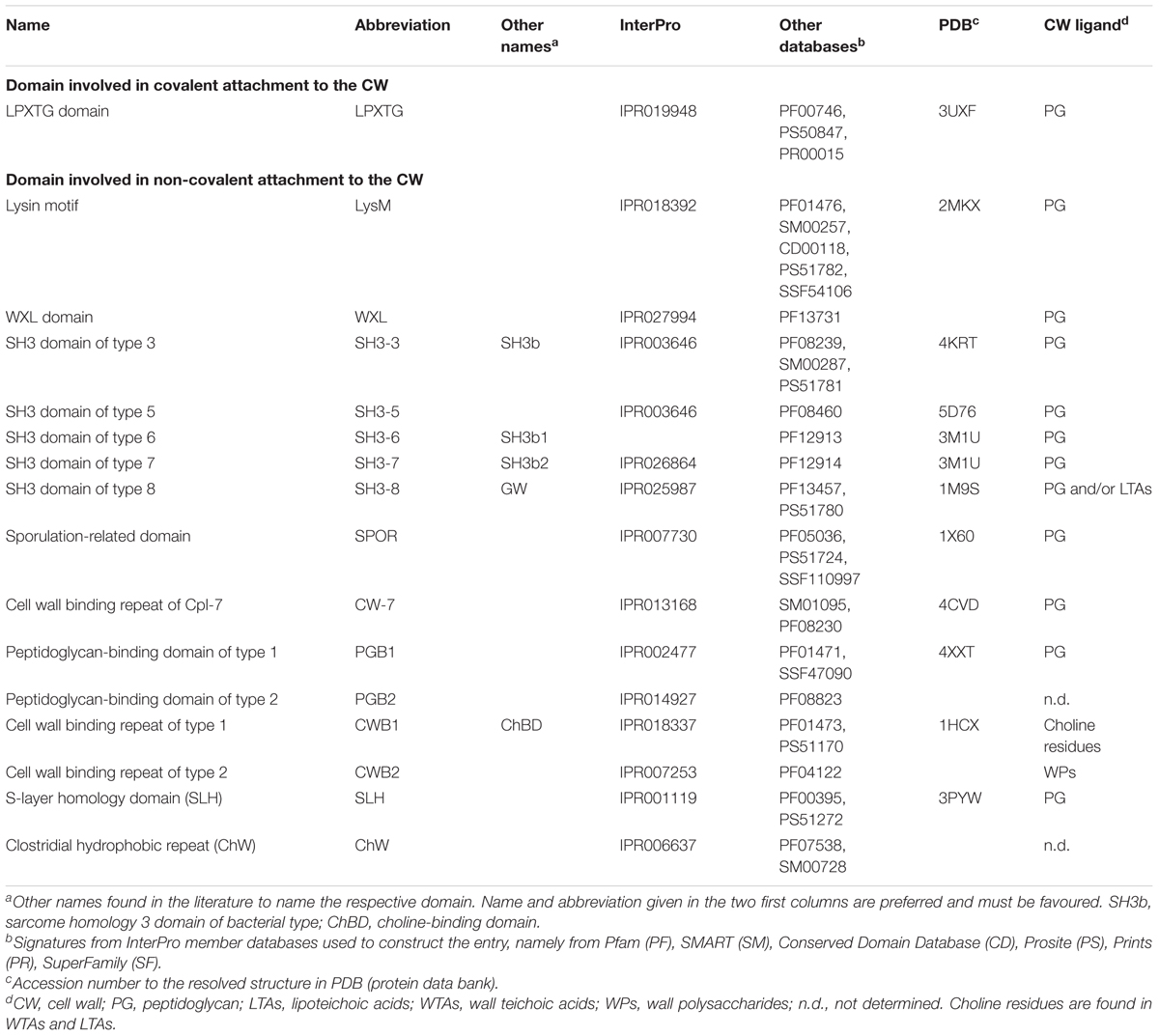
The CW of parietal monoderm bacteria is a complex structure composed at least of peptidoglycan (PG) covalently linked to other biopolymers, such as teichoic acids, polysaccharides, polyglutamate, or proteins (Shockman and Barrett, 1983; Figure 1). While constituting the outermost layer of the cell envelope of parietal monoderm bacteria, the CW is not impermeable but on the contrary a porous and penetrable structure. As such, cell envelope proteins are in contact with the external environment without ever having a domain protruding out the confines of the CW. Like for the fractal dimension of the protein surface (Richards, 1977; Banerji and Navare, 2013), the nature and definition of the bacterial cell surface strictly depends on the molecule considered, e.g., a water molecule or a globular protein, which can enter in contact, access, diffuse or penetrate differently the CW (Figure 2). To be exposed at the cell surface of parietal monoderm bacteria, proteins need to be first secreted across the CM. Several secretion systems allow protein translocation in parietal monoderm bacteria (Tjalsma et al., 2004; Desvaux et al., 2005; Desvaux and Hébraud, 2006; Sibbald et al., 2006; Chagnot et al., 2013), namely (i) the Sec (secretion), (ii) the Tat (twin-arginine translocation), (iii) ABC protein exporter, (iv) the FPE (fimbrilin-protein exporter), (v) the FEA (flagella export apparatus), and (vi) the ESX (ESAT-6 system), also called Wss (WXG100 secretion system). Of note, the status of the holins (hole forming) as protein secretion systems per se remain controversial (Desvaux, 2012). Proteins secreted via the Sec translocon generally possess a targeting signal called the signal peptide (SP) of type I (SP I), which is composed of three non-conserved domains, namely the n-domain (positively charged and at the N-terminus), the h-domain (a-helical hydrophobic core region), and the c-domain (cleavage site processed by a membrane-bound signal peptidase) (Fekkes and Driessen, 1999). While proteins secreted via Sec, Tat ABC exporter and FPE possess N-terminal SPs with some specificities, the signal targeting proteins to the FEA or ESX remain elusive. Besides transport across the CM, the transport and maturation of secreted proteins across the CW can be regulated by different mechanisms, such as the proteolytic maturation of secreted proenzymes, the requirement of divalent cations for activation or the post-translocational intervention of peptidyl-prolyl isomerase chaperones (Forster and Marquis, 2012).
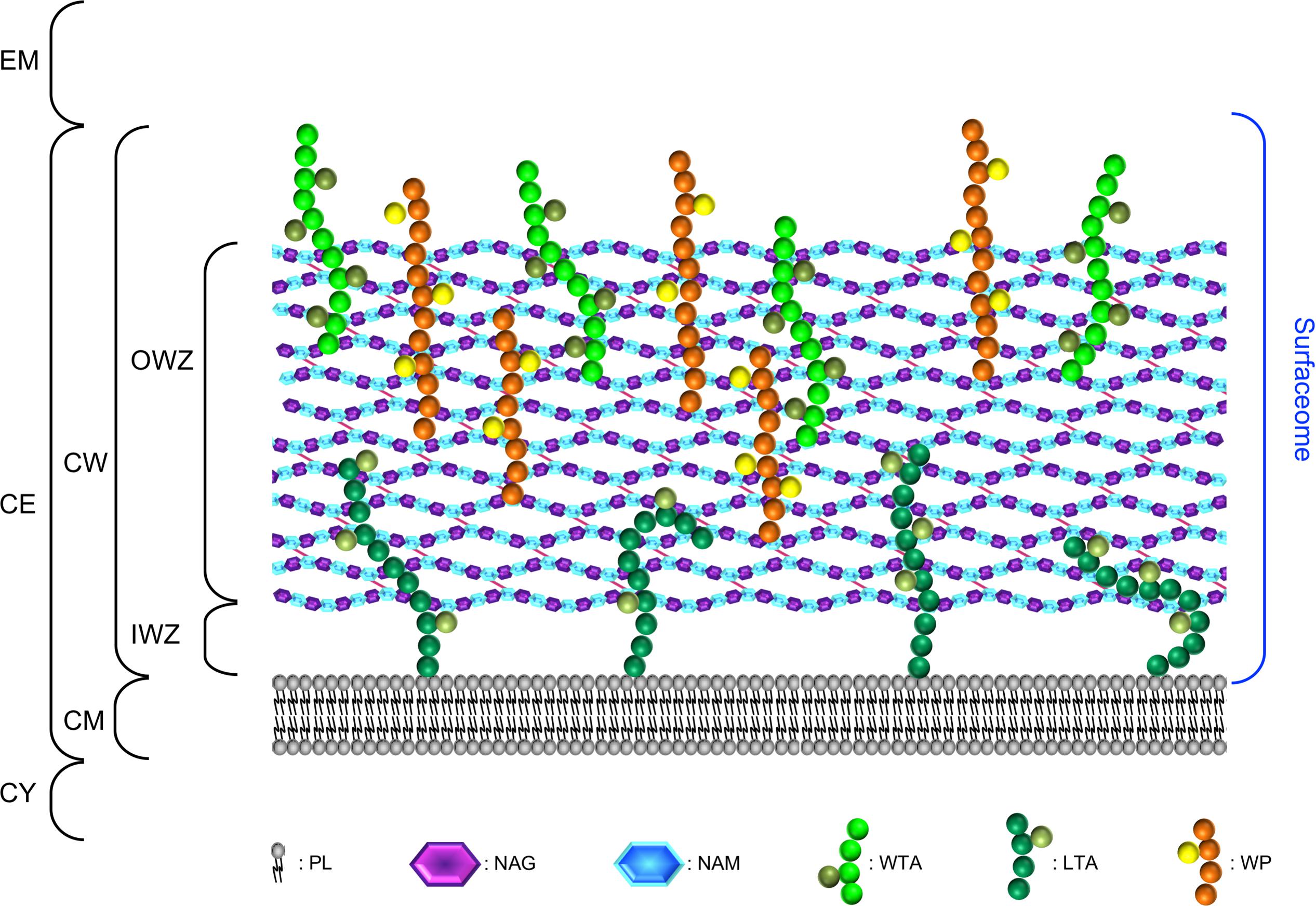
FIGURE 1. The surfaceome of parietal monoderm bacteria with respect of the organisation and composition of the cell envelope. The cell envelope (CE) of parietal monoderm bacteria is composed of a biological membrane acting as selective permeable barrier, i.e., the cytoplasmic membrane (CM) and a cell wall (CW) providing some resistance to mechanical stresses (e.g., internal turgor pressure) but also somehow acting as a philtre. While the CM is composed of phospholipids (PLs), the CW can be further subdivided into the inner wall zone (IWZ) and the outer wall zone (OWZ). The OWZ constitutes the main CW fabric. It is composed of N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM), both constituting the peptidoglycan (PG) with which wall teichoic acids (WTAs), and wall polysaccharides (WPs) are anchored. Lipoteichoic acids (LTAs) are anchored to the CM and protrude from the CM. As revealed by electron microscopy studies and contrary to the OWZ, the IWZ is a thinner zone of low density most certainly devoid of most cross-linked polymeric CW network, except LTAs and some proteins, e.g., lipoproteins (Matias and Beveridge, 2005, 2006); because this zone is not strictly bounded by two biological membranes like in diderm bacteria, the IWZ resembles but cannot be considered as a periplasm sensu stricto, i.e., it presents some analogies but no homology (Buist et al., 2008; Chagnot et al., 2013). In addition to the proteins present both at the CM and CW and that are not depicted here for clarity (see text and Figures 4–6), these different macromolecular molecules exposed on the external side of the CM constitute the surfaceome in parietal monoderm bacteria. CY, cytoplasm; EM, extracellular milieu.
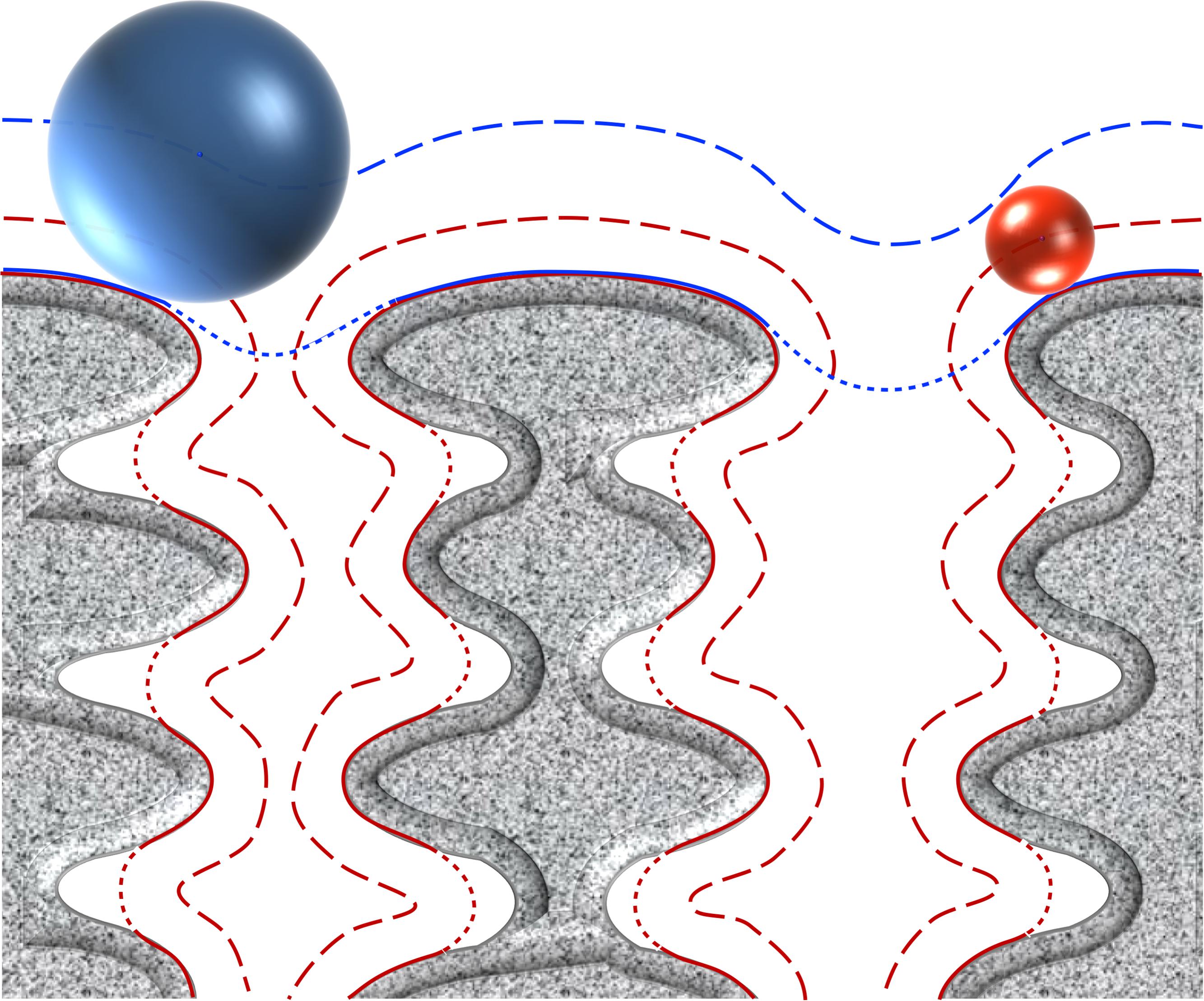
FIGURE 2. Concepts of molecular surface, contact surface, accessible surface, and reentrant surface to define the bacterial cell surface in parietal monoderm bacteria. Taking molecules of different sizes, their penetration in the cell envelope differs. The blue sphere represents a molecule of high molecular weight unable to penetrate the CW fabric (depicted in grey), whereas the red sphere represents a smaller molecule diffusing through. Depending on the molecules considered, the definition of the bacterial cell surface will also differ. The continuous lines represent the contact surface that is the molecular surface that actually comes in direct contact with the surface of the molecule considered. The dashed lines represent the accessible surface that is the continuous sheet referring to the centre of the molecule considered. The dotted lines correspond to the reentrant surface that is the interior-facing part of the molecule considered when it cannot come in direct contact with the molecular surface of the cell envelope. The definition of bacterial cell surface of parietal monoderm bacteria is thus very different when referring to the molecular surface of the cell envelope or the contact, accessible and reentrant surfaces with respect of the size of the molecule under consideration.
To explicitly describe the subcellular localization of proteins, the gene ontology (GO) respective to the cellular component is extremely useful (Ashburner et al., 2000; Chagnot et al., 2013). Indeed, secreted proteins can have different fate; they are either (i) associated to the CM (GO: 0005886), (ii) anchored to the CW (GO: 0009275), (iii) released in the extracellular milieu (GO: 0005576), the so-called exoproteins (extracellular proteins), or even (iv) injected into a prokaryotic or eukaryotic host cell. At the CM, proteins can be either integral (GO: 0031226), i.e., the IMPs (integral membrane proteins), or anchored to the membrane (GO: 0046658), i.e., the lipoproteins. At the CW, proteins can be covalently bound, i.e., the LPXTG-proteins, or bound through weak interactions, i.e., the CW binding proteins. It is worth stressing that all these extracytoplasmic proteins located at the cell envelope, wherever at the CM or the CW, can be considered as surface exposed. Besides monopolypeptides, some organelles can also be present and result from the assembly of protein subunits to form supramolecular structures, such as the well-known pili and flagella, but also the S-layer or cellulosome in some bacterial species.
Following the etymological meaning of the Greek suffix “-ome” (Lederberg and McCray, 2001), the totality of the molecules found at the bacterial cell surface corresponds to the surfaceome. Because of the spongy structure of the CW, it is misleading to restrict the surface of parietal monoderm bacteria to molecules strictly displayed at the outermost molecular layer of the CW. Instead, the cell surface of a parietal monoderm bacterium fits tightly to the molecular outline of the CW components and to the external side of the CM (Figure 2); as a biological membrane, the CM has a selective permeability contrary to the CW. The CW is not a rigid shell but constitutes a matrix, forming an elastic polyelectrolyte gel (Doyle and Marquis, 1994; Neuhaus and Baddiley, 2003), which would then acts like a sieve during the dynamic transit of solutes. The proteosurfaceome is the proteinaceous subset of the surfaceome found at the CW and totally or partially exposed on the external side of the CM.
The Surfaceome of Parietal Monoderm Bacteria
The cell envelope of parietal monoderm bacteria is composed of a CM and a CW, which can be divided into the inner wall zone (IWZ) and outer wall zone (OWZ) (Merchante et al., 1995; Matias and Beveridge, 2005; Zuber et al., 2006; Figure 1). The CW surrounding the CM is made of lipoteichoic acids (LTAs) and a thick layer of PG, decorated with wall teichoic acids (WTAs), wall polysaccharides (WPs), or/and polyglutamate. The CW also accommodates some proteins, including monopolypeptides and cell-surface supramolecular protein structures, namely pili, flagella, cellulosome, S-layer. Altogether these different macromolecular molecules and associated molecules constitute the surfaceome. This part focuses on the components of the cell envelope, excluding the proteinaceous compounds discussed in the subsequent part. Cell envelope proteins actually interact with some of these components for anchoring via different molecular mechanisms.
Composition and Organisation of the Cytoplasmic Membrane
The phospholipid bilayer of the membrane parietal monoderm bacteria is ∼90 Å thick and is composed of 10–40% lipids, 40–75% proteins, and 0.2–20% carbohydrates (Salton, 1967; Ghosh and Carroll, 1968; Bodman and Welker, 1969; Duda et al., 2006). Although membrane phospholipids vary from one species to another, the most commonly found in the CM are glycerophospholipids including phosphatidylglycerol, diphosphatidylglycerol (cardiolipin), and to some extend phosphatidylethanolamine and their amino acylated forms (Fischer et al., 1978; Roy, 2009; Malanovic and Lohner, 2016). Phospholipids vary also by their two fatty acid moieties, which impact on membrane fluidity (Mishra et al., 2012; Custer et al., 2014; Diomande et al., 2015; Malanovic and Lohner, 2016). Polyisoprenoid lipids are other important regulators of membrane fluidity. They constitute, together with cardiolipins and bacterial flotillins acting as scaffolding proteins, nanoscale functional membrane microdomains, which seem essential to the proper functioning of signal transduction cascades and protein transport in Bacillus subtilis and Staphylococcus aureus cells (Lopez and Kolter, 2010; Bramkamp and Lopez, 2015). By analogy with eukaryotic membranes, these microdomains are also referred to as lipid rafts. Consistently, membrane proteins or associated complexes constitute discrete focal sites in the CM and CW (Campo et al., 2004; Rosch et al., 2007; Lopez and Kolter, 2010; Kandaswamy et al., 2013). Biological significance of functional membrane microdomains could be to serve as platforms that control the assembly of membrane and CW proteins and multiprotein complexes involved in numerous cellular processes, such as cell division, protein trafficking, genetic transfer, or signal transduction (Lopez and Kolter, 2010; Schneider et al., 2015). Subcellular localization and spatiotemporal distribution of CM and CW proteins or supramolecular protein complexes are often intimately linked to their function and vary with the environmental conditions (Bierne and Dramsi, 2012; Mitra et al., 2016).
Composition and Organisation of the Cell Wall
The OWZ constitutes the main part of the CW. It is 15–30 nm thick and comprises the PG and WTA polymers (Navarre and Schneewind, 1999; Vollmer et al., 2008). The PG is made of N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) forming disaccharide glycan chains of various lengths that are cross-linked by peptides. PG composition depends on bacteria (Schleifer and Kandler, 1972). The glycan chain is uniform, whereas the peptide moiety and the cross-links are variable. The two major PGs in parietal monoderm bacteria have a meso-diaminopimelic acid (A2pm) or a lysin at the third position of the peptide. At this position, the cross-link occurs directly or through a penta-glycine bond, respectively. In B. subtilis, it is estimated that the glycan chain length is 1300 disaccharides in average, and that approximately 20% of the peptide chains are cross-linked (Ward, 1973; Atrih et al., 1998; Hayhurst et al., 2008). These glycan chains form helices of ∼50 nm width, and it was proposed that these cable-like structures coil around the narrow axis of the bacterium and are cross-linked by peptides (Hayhurst et al., 2008). The glycan chains of ovococcal bacteria, e.g., Streptococcus sp., are formed of more than 100 disaccharide units in average, whereas the glycan chains of cocci, e.g., Staphylococcus sp., are relatively short with 5–10 disaccharide units in average (Wheeler et al., 2011). The average effective mesh size in PG, i.e., the tessera, is estimated at 2.2 nm (Koch, 1990; Demchick and Koch, 1996; Figure 3). In other words, hydrophilic molecules of about 25 kDa (but also probably up to 50 kDa) can freely pass through a structured CW meshwork. Along with this, the CW network is actually not perfect, e.g., pseudo-tessera, and numerous PG defects cause increase in the porosity (Pink et al., 2000; Turner et al., 2013; Kim et al., 2015). Of note, though, the critical hole size in the CW beyond which lysis occurs, is estimated in the range of 15–24 nm (Mitchell et al., 2013). The OWZ of parietal monoderm bacteria is a very dynamic structure, as bacterial growth requires constant remodelling of the CW, which has a turnover rate of 50% per generation (Koch and Doyle, 1985). Remodelling is mediated by CW-anchored autolysins that are active on the outermost layer of the PG (Jolliffe et al., 1981).
FIGURE 3. Peptidoglycan organisation at the cell wall. The peptidoglycan is composed of N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) linked by β-1,4 bonds, where the NAM are further crosslinked via octapeptides either at the same plane or with the upper or lower layer (arrows represent peptides protruding up or down). The peptidoglycan is tiled with hexagonal tesserae, which constitute the structural unit of the CW fabric (one basic unit constituting a tessera is displayed inside the dotted frame). Two layers of tesserae are here schematically represented to highlight the network form by the peptide crosslinking. Of note, defects due to abnormal tesserae with more edges and larger area can also occur and resulting in the increase in porosity.
LTAs and WTAs are zwitterionic polymers anchored to the CM and CW, respectively. They are major polyanionic teichoic acids of the envelope of parietal monoderm bacteria. LTAs are localised in the IWZ at the interface of the CM and the CW (Neuhaus and Baddiley, 2003; Reichmann and Grundling, 2011; Schneewind and Missiakas, 2012; Percy and Grundling, 2014). The most common LTA structure found in Firmicutes and, referred as type I LTAs, consists in a polyglycerol phosphate polymer linked to a glycolipid anchor, often a diglucosyl-diacylglycerol (Glc2-DAG), anchored to the CM. Type II, III, IV, and V LTAs have more complex repeating units that contain glycosyl residues, e.g., in Streptococcus pneumoniae, type IV LTA is decorated with phosphocholine.
WTAs are covalently attached by the PG disaccharide unit via a phosphodiester linkage to NAM (Neuhaus and Baddiley, 2003; Brown et al., 2013). Although the structures of WTAs vary considerably between species, the most common ones are composed of glycerol-phosphate or ribitol-phosphate repeats. LTAs and WTAs are often modified with sugar moieties and D-alanine esters, which introduce positive charges to neutralise the negatively charged phosphates in the polymer backbone (Wooldridge and Williams, 1993; Xia et al., 2010; Schneewind and Missiakas, 2012; Percy and Grundling, 2014; Carvalho et al., 2015). In addition to their diversity between and within species, the degree of D-alanylation of teichoic acids is fine tuned in changing environments and thus likely influences the protein repertoire displayed at the CW. The zwitterionic WTA polymers potentially contribute to the sequestration of divalent cations within the OWZ, including Ca2+, Mg2+, and Fe2+ (Beveridge and Murray, 1980), and might thus influence the regulation of protein transport across the CW (Forster and Marquis, 2012).
WPs have various compositions, e.g., teichuronic acids in Bacillus (Ward, 1981) or highly diverse heteropolysaccharides in Lactococcus (Yasuda et al., 2011; Vinogradov et al., 2013; Ainsworth et al., 2014), which complexity and diversity can be even greater than expected as revealed by the ever increasing genome data regularly made available. The last and most external layer of the CW may be composed of a capsule, generally composed of WPs (Jones, 2005; Yother, 2011). Although the WP capsule structures are well documented, the anchoring was recently proposed to be at the β-D-N-acetylglucosamine of the PG via a direct glycosidic bond (Larson and Yother, 2017). In some cases, the capsule is composed of polyglutamate, e.g., in Bacillus anthracis (McLean et al., 1992; Candela and Fouet, 2006). Poly-γ-D-glutamate anchoring was reported to be covalent at the PG (Candela and Fouet, 2005; Candela et al., 2005). However, the exact anchoring mechanism is still controversial and may be either on the A2pm or on the PG glucosamine (Richter et al., 2009; Candela et al., 2014).
Overall, the CW of parietal monoderm bacteria is a complex structure that protects them from mechanical and osmotic lysis, and serves as a scaffold for anchoring proteins, glycopolymers, and cations that perform various functions (Navarre and Schneewind, 1999; Weidenmaier and Peschel, 2008). While WPs or WTAs can be essential for bacterial growth (Oh et al., 2017), WTAs have been shown to be dispensable in some other bacterial species (Chapot-Chartier and Kulakauskas, 2014; Mistou et al., 2016). However, wall rhamnose polysaccharides (RhaWPs) can be a functional counterpart of WTAs, as suggested in Streptococcus agalactiae and Streptococcus pyogenes (Caliot et al., 2012; van Sorge et al., 2014), where they appear to be covalently linked to PG NAM (Deng et al., 2000).
Cell-Surface Proteins Localised at the Cytoplasmic Membrane (Go: 0005737)
Cell-surface proteins specifically localised at the CM can either be integral to the CM (GO:0031226) or anchored to the CM (GO: 0046658). Besides, some proteins can interact by weak interactions with components of the CM surface and be extrinsic to the CM (GO:0019897).
Proteins Integral to the Cytoplasmic Membrane (GO: 0031226): The IMPs
As a common theme, all IMPs exhibit hydrophobic transmembrane α-helical domains (TMDs) enabling anchoring of the protein to the membrane (White and von Heijne, 2004). IMPs can be broadly discriminated between single-spanning IMPs (ss-IMPs) exhibiting a single TMD and multispanning-IMPs (ms-IMPs) with more than one TMD (Figure 4; Goder and Spiess, 2001; Higy et al., 2004). Whereas most IMPs are not synthesised with a cleavable N-terminal SP, some IMPs are (Facey and Kuhn, 2004). For the latters and after cleavage of the hydrophobic transmembrane α-helical SP by a signal peptidase (SPase), the ss-IMPs remain anchored to CM thanks to an additional hydrophobic TMD, i.e., the stop-transfer sequence also called signal domain of type I (SD1), which exhibits a Nout–Cin topology; as such, these ss-IMPs refer to the type I (ss-IMP1; Figure 4). Type II ss-IMPs (ss-IMP2) have a signal-anchor sequence also called signal domain of type II (SD2), with a Nin–Cout topology, which actually corresponds to an uncleavable SP. Type III ss-IMPs (ss-IMP3) have reverse signal-anchor sequence, i.e., a SD1 (TMD with a Nout–Cin topology); in the literature, they are sometimes described as ss-IMP1 without SP since the reverse signal-anchor sequence is a SD1. Of note, while the translocation mechanism of both type I and type II IMPs is in line with our current knowledge about the Sec/YidC translocation, i.e., involving an N-terminal SP (whenever cleavable or uncleavable) targeting the protein to CM, the mechanism for the translocation of type III IMPs in the absence of a SP remain unclear. In ms-IMPs, the type I (Nout–Cin TMD topology) and type II (Nin–Cout TMD topology) signals alternate along the protein sequence. Based on topology of the most N-terminal TMD enabling anchoring of the ms-IMP to the CM, the three types mentioned here above can be discriminated (Figure 4).
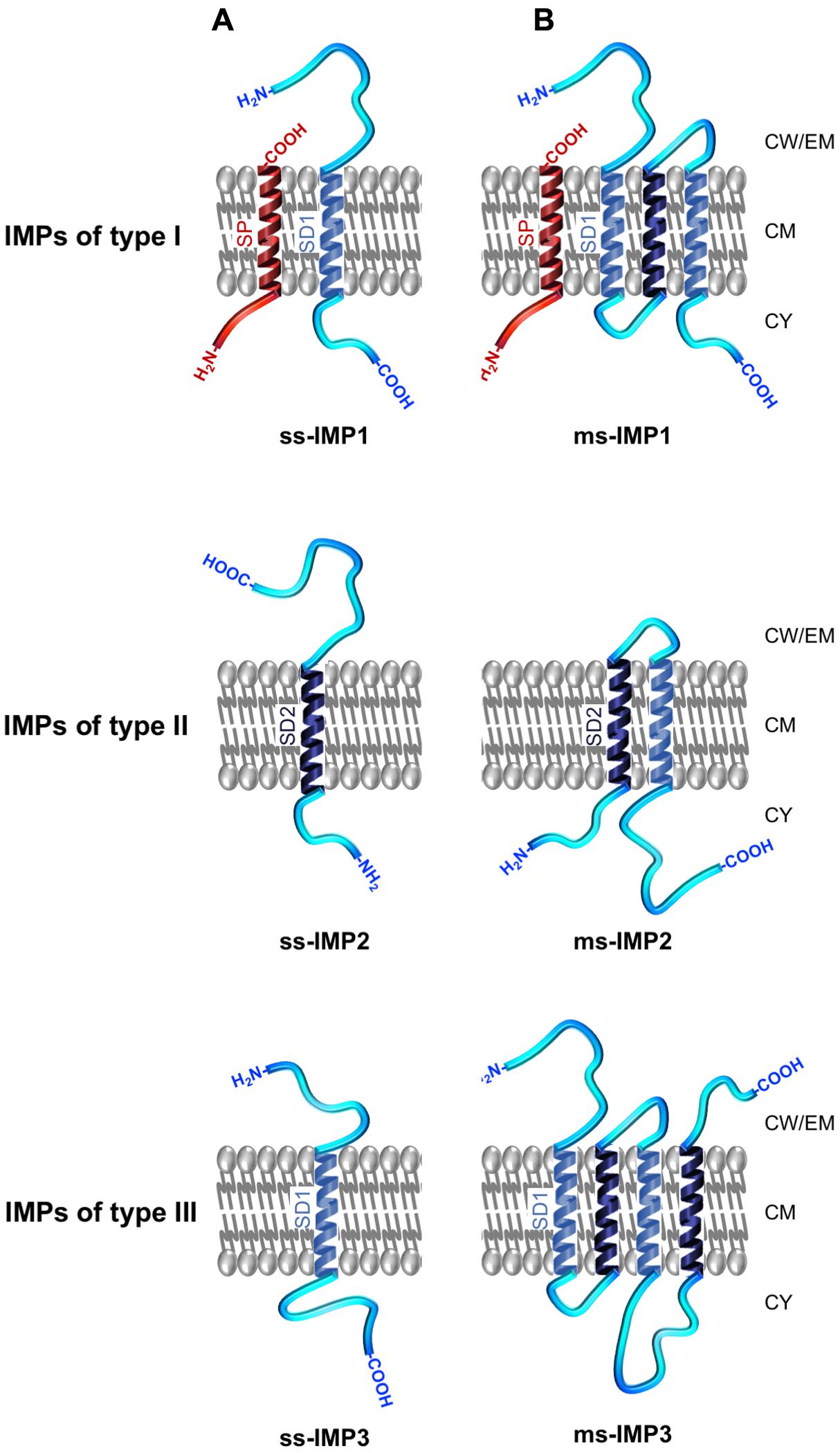
FIGURE 4. Topology and nomenclature of IMPs. IMPs are primarily categorised into (A) single-spanning IMPs (ss-IMPs) and (B) multi-spanning IMPs (ms-IMPs). Indeed, IMPs are anchored to the CM via hydrophobic transmembrane α-helical peptide domains (TMDs); when a TMD has a Nout–Cin topology, it is called a signal domain of type I (SD1; depicted in light blue), whereas a TMD with Nin–Cout topology is called a signal domain of type II (SD2; depicted dark blue) (White and von Heijne, 2004). In ss-IMPs, only one TMD is present, whereas at least two TMDs are present in ms-IMPs. Whenever ss-IMPs or ms-IMPs, they are further subcategorised into three types. A ss-IMP of type I (ss-IMP1) possesses a cleavable N-terminal signal peptide (SP; depicted in red) and are actually anchored to the CM by a SD1 (TMD with a Cin–Nout topology). A ss-IMP of type II (ss-IMP2) is anchored to the CM by a SD2 (TMD with a Nin–Cout topology). Like for a ss-IMP1, a ss-IMP of type III (ss-IMP3) is anchored to the CM by a SD1 but it did not originally exhibit a SP. For ms-IMPs, the classification is similar and based on the most N-terminal TMD anchoring the ms-IMP to the CM. As such, a ms-IMP of type I (ms-IMP1) has a cleavable SP followed by a SD1. A ms-IMP of type II (ms-IMP2) has a SD2 as the most N-terminal TMD. A ms-IMP of type III (ms-IMP3) has a SD1 as the most N-terminal TMD (and no cleavable SP). Of note, the TMD of a cleavable SP actually corresponds to a SD2; as such, a SD2 in IMPs of type II can be referred as an uncleavable SP. In ms-IMPs, a SD1 necessarily alternates with a SD2 along the polypeptide chain, and vice versa. Except for the TMDs, other regions of the IMPs can be in contact with the IWZ but also the OWZ or the extracellular milieu.
IMP biogenesis in lipopolysaccharidic-diderm bacteria (archetypal Gram-negative bacteria) involves an integrase known as YidC (Scotti et al., 2000). Up to two paralogues of the integrase YidC have been uncovered in parietal monoderm bacteria, namely SpoIIIJ and YqjG (Tjalsma et al., 2000; van Wely et al., 2001). While both SpoIIIJ and YqjG are involved IMP biogenesis and are essential for cell viability (Murakami et al., 2002; Tjalsma et al., 2003), SpoIIIJ is required for sporulation in B. subtilis but not YqjG (Errington et al., 1992; Murakami et al., 2002). Lately, these proteins have been renamed YidC1 and YidC2, respectively, in parietal monoderm bacteria (Funes et al., 2009; Wang and Dalbey, 2011; Palmer et al., 2012). In E. coli, YidC is associated to the Sec translocase enabling insertion of all IMPs to the CM in a SRP (signal-recognition particle) dependent mechanism (Scotti et al., 2000; Fröderberg et al., 2003; Ziehe et al., 2017). In this species, the YidC pathway is quite versatile since integration of IMPs to the CM can be SecA-, SecB-, and/or Sec-independent (Samuelson et al., 2000; Beck et al., 2001; Yen et al., 2002; Fröderberg et al., 2003; White and von Heijne, 2004). Moreover, flotillin-like proteins could contribute to the insertion of IMPs (Dempwolff et al., 2016). So far, these aspects have been poorly investigated in parietal monoderm bacteria.
Cell-Surface Proteins Anchored to the Cytoplasmic Membrane (GO: 0046658): The Lipoproteins
In parietal monoderm bacteria, lipoproteins are synthesised as pre-prolipoproteins that are exported by the Sec pathway and exposed on the outer face of the CM (Hutchings et al., 2009; Figure 5). The pre-prolipoproteins exhibit a SP of type II (SP II) that is harbouring a conserved lipobox motif at the cleavage site (Sutcliffe and Harrington, 2002). The consensus sequence for the lipobox is [LVI]-3-[ASTVI]-2-[GAS]-1-[C]+1 (Sutcliffe and Harrington, 2002; Babu et al., 2006). Once translocated across the CM, the lipoprotein maturation pathway in parietal monoderm bacteria is a two-step process. First, the lipobox motif is recognised by a prolipoprotein diacylglyceryl transferase (Lgt), which transfers of a diacylglyceryl moiety from a glycerophospholipid onto the thiol group of the conserved cysteine, giving rise to the prolipoprotein. Then, the SP II of the prolipoprotein is cleaved off by a lipoprotein signal peptidase (Lsp), generating a mature lipoprotein. The lipoprotein is consequently covalently bound to the acyl moiety of two fatty acids from the diacylglyceride by a cysteine residue at position 1 of the N-terminal end (Lai et al., 1980). Besides this classical form of lipid-modified cysteine for lipoprotein anchoring to the CM, intensive mass spectrometry analyses revealed three novel forms of mature lipoproteins in parietal monoderm bacteria (Nakayama et al., 2012; Figure 5). The N-acylated triacyl form of lipoproteins containing N-acyl-S-diacyl-glyceryl-cysteine was identified in S. aureus and S. epidermidis (Kurokawa et al., 2009; Asanuma et al., 2011). The N-acetyl form of lipoproteins identified in different Bacillaceae contains N-acetyl-S-diacyl-glyceryl-cysteine (Kurokawa et al., 2012b). The lyso-form of lipoproteins containing an N-acyl-S-monoacyl-glyceryl-cysteine was identified in Bacillus cereus, Enterococcus faecalis, Lactobacillus bulgaricus, and Streptococcus sanguinis (Asanuma et al., 2011). It further appeared that environmental conditions influenced the ratio between diacyl and triacyl forms of lipoproteins in S. aureus, with an accumulation of the diacyl lipoprotein form at high temperatures and high salt concentrations (Kurokawa et al., 2012a). Together, these recent findings are suggestive of uncharacterised non-canonical pathways for differential lipoprotein lipidation in parietal monoderm bacteria, analogous to the N-acylation of the lipidated cysteine by the apolipoprotein N-acyltransferase (Lnt) in lipopolysaccharidic-diderm bacteria. Actually, the lipoprotein intramolecular transacylase (Lit) involved in N-lyso-form biosynthesis was recently identified in E. faecalis and B. cereus (Armbruster and Meredith, 2017). If N-acylation is likely to involve acyltransferases adapted to specific phospholipids as acyl-donor substrates, novel enzymes and maybe pathways are to be discovered to explain how these alternative N-acetyl lipoprotein forms are biosynthesised in parietal monoderm bacteria.
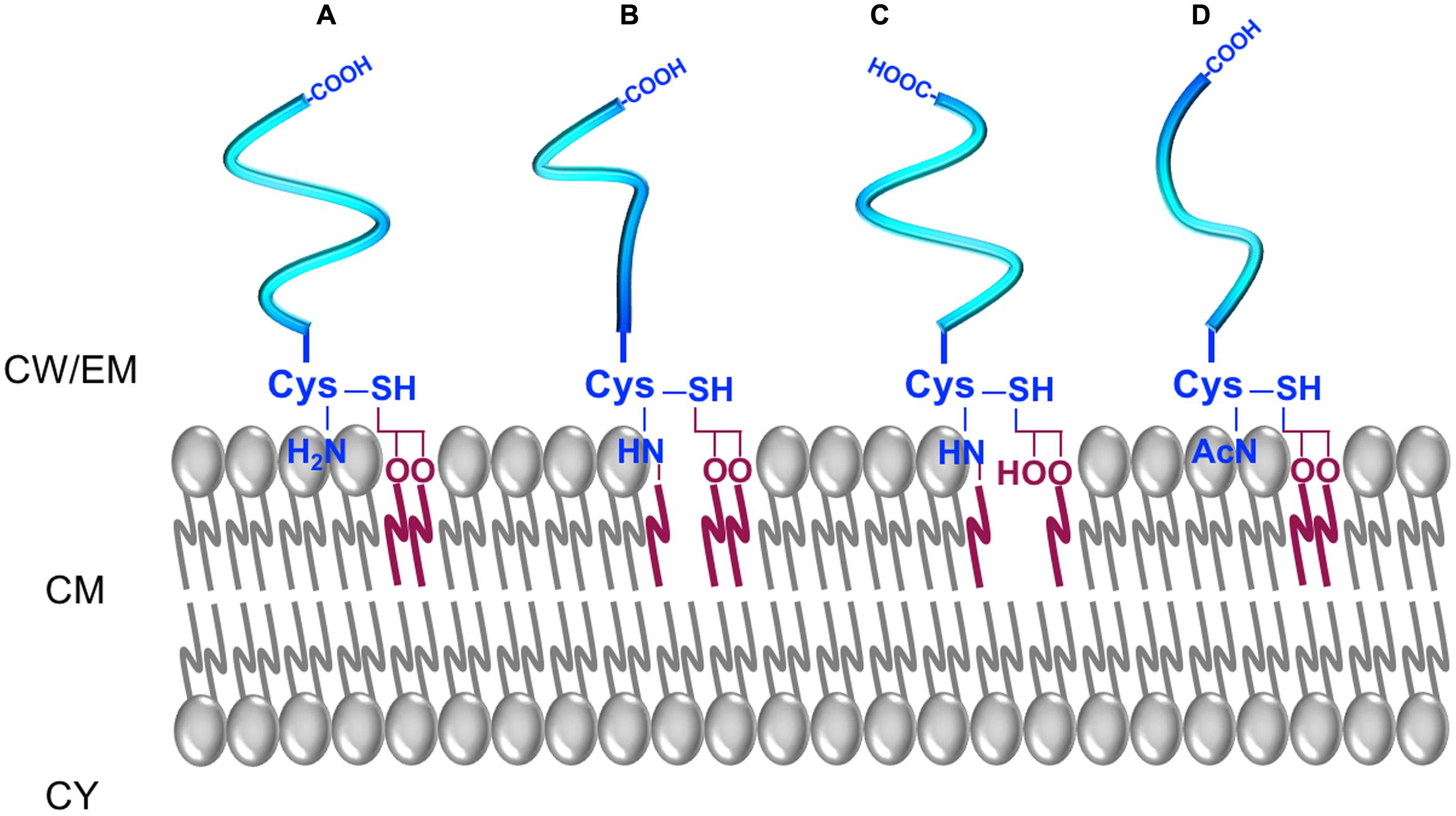
FIGURE 5. The different forms of lipoproteins in parietal monoderm bacteria. (A) A diacyl-lipoprotein contains an N-acyl-S-diacylated cysteine residue. (B) A N-acylated-triacyl-lipoprotein contains an N-acyl-S-triacylated cysteine residue. (C) A lyso-lipoprotein contains an N-acyl-S-monoacyl-glyceryl-cysteine. (D) A N-acetyl-form contains a N-acetyl-S-diacyl-glyceryl-cysteine.
Cell-Surface Proteins Localised at the Cell Wall (Go: 0009275)
The first surface associated proteins were described because of their activities on the bacterial CW. Most of them were autolysins or proteases. PG-binding domains were thereafter observed thanks to the sequencing data and bioinformatic analyses. Indeed, amino acid repetitions involved in the surface binding were highlighted. Most of the characterised and conserved domains are registered and classified by bioinformatic resources, especially InterPro (IPR; Zdobnov and Apweiler, 2001; Finn et al., 2017) regrouping several databases for motif signatures, such as Pfam (Soohammer et al., 1997; Finn et al., 2016), Prosite (Hulo et al., 2006; Sigrist et al., 2013), or SMART (Schultz et al., 1998; Letunic et al., 2015) (Table 1). Of note, the use of underscore (“_”), as given in the name of domains in databases, must be abstained by reminding the readers this sign is primarily designed for bioinformatics purpose when a space cannot be used due to command line constraints but are meant to be replaced by a space (“ ”) or a dash (“-”) in textbook. These binding domains allow protein subcellular location at the CW and are therefore often crucial for their activity on the surface structure and organisation (Figure 6). They can be divided into three main classes: domains that are (i) covalently attached to the PG, (ii) non-covalently bound to the PG, and (iii) non-covalently bound to WPs (Figure 6). Besides, the CW components targeted by some domains remain uncertain. These proteins are generally secreted by the Sec translocon and possess a SP I.
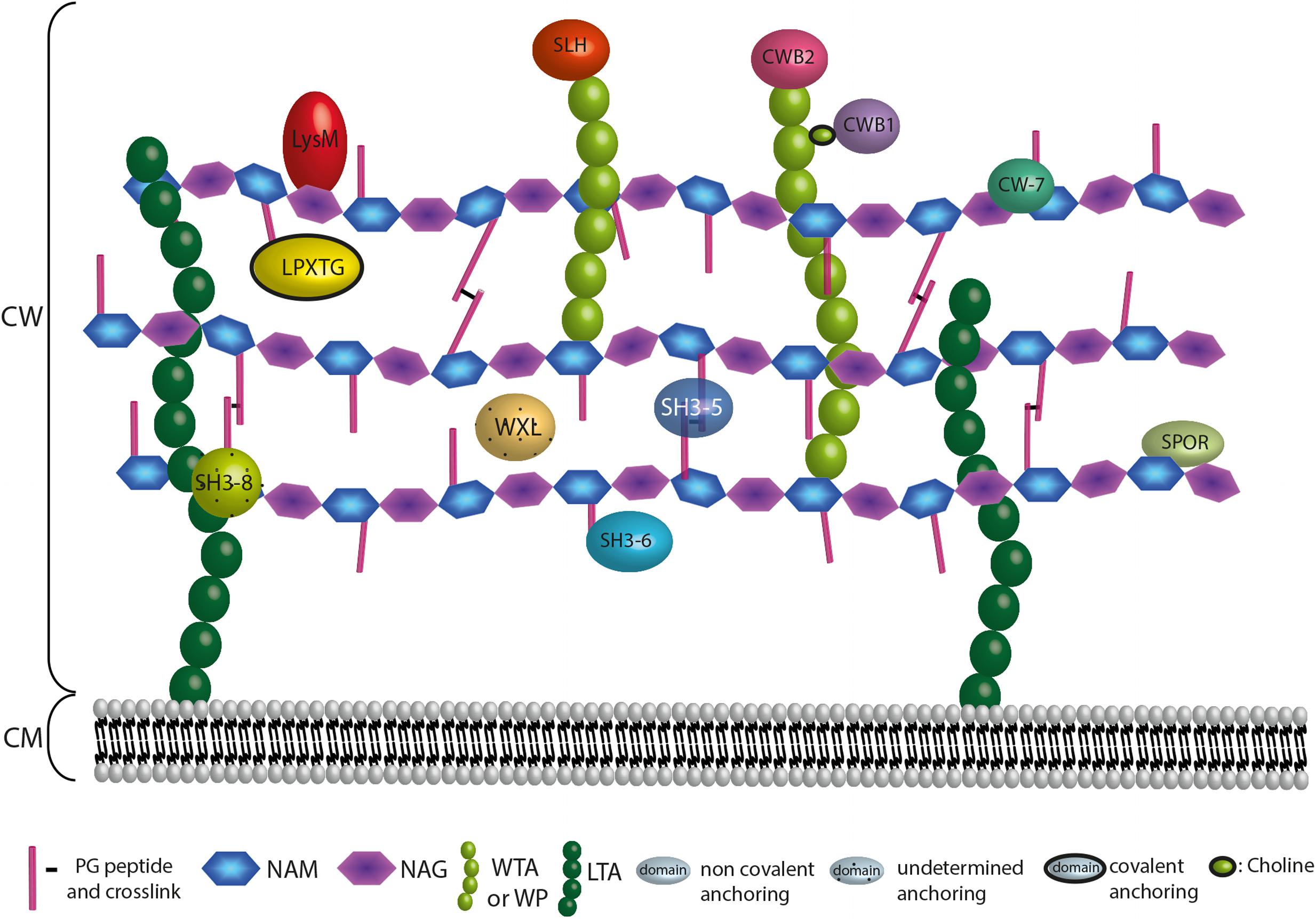
FIGURE 6. Anchoring localization of protein domains interacting with the CW. The localization of the CW proteins depends on their domains. Domains are interacting covalently or not at the bacterial CW through interaction with surface structures that are the PG, the WTAs, the WPs or the LTAs. LPXTG proteins are covalently attached to the A2pm or K residue of the PG. Proteins harbouring a LysM, SH3 of type 5 (SH3-5), SH3 of type 6 (SH3-6), SPOR, or CW-7 domain interact non-covalently with the PG. WXL interacts with PG but the precise anchoring region is undetermined. Proteins possessing a CWB2 or SLH domain are localised at the WTAs or WPs extremities, whereas proteins harbouring a CWB1 domain interact with the WTAs through a choline. For SH3 of type 8 (SH3-8), the CW target remains controversial.
Cell-Surface Proteins Covalently Bound to the Peptidoglycan: The LPXTG- Proteins
Covalent binding of LPXTG-proteins to the CW has been the subject of intensive studies and is certainly one of the best characterised molecular mechanisms for protein anchoring to the PG (Fischetti et al., 1990; Schneewind et al., 1992). Here, we review the major mechanism of anchoring. In parietal monoderm bacteria, a range of proteins called LPXTG (IPR019948) is covalently linked to the PG by enzymes named sortases. Among LPXTG-proteins are found colonising factors, toxins and proteases. In parietal monoderms, the LPXTG motif was identified in both classes of Actinobacteria and Firmicutes, especially in the orders of Coriobacteriales, Streptomycetales, Propionibacteriales, Bifidobacteriales, Micrococcales, and Corynebacteriales for the former, and the orders of Erysipelotrichales, Clostridiales, Lactobacillales, Bacillales, and Tissierellales for the latter. This is a C-terminal motif composed of the LPXTG sequence where X represents any amino acids, followed by a hydrophobic domain and a short positively charged tail. Several variations around this motif were reported, e.g., NP(Q/K)TN, but the overall motifs remain homologous and are included for profile search (Boekhorst et al., 2005). In any case, the motif is recognised by sortases that are classified into six classes from A to F (Dramsi and Bierne, 2017; Siegel et al., 2017). Sortase A anchors a wide range of LPXTG-proteins, whereas sortase B recognises the NP(Q/K)TN related motif. Sortase C allows the pilus assembly (see below), whereas sortases D, E, and F have been much less characterised. Sortases anchor the LPXTG-proteins on the nascent PG through their transpeptidase activity, by cleaving between T and G (or N) and transferring the protein on the PG. Depending on the PG nature, molecular binding can occur at the pentaglycine crossbridge (Marraffini and Schneewind, 2005) or at the A2pm (Budzik et al., 2008).
Cell-Surface Proteins Non-covalently Bound to the Peptidoglycan
Besides covalent binding to the PG, some proteins exhibit conserved motifs enabling specific binding to the CW components via weak interactions, such as van der Waals interactions, hydrogen or ions bonds.
Lysin Motif Domain
LysM (lysin motif) domain was first reported in a protein encoded by gene 15 of B. subtilis bacteriophage 364 ϕ29 (Garvey et al., 1986) and exhibiting lysozyme activity that is involved in PG degradation. This conserved domain is found across all kingdoms and is widely distributed among bacteria, although mainly found in Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes. A LysM domain (IPR018392) consists of 43–50 amino acids including the first 16 residues that are highly conserved. Multiple LysM domains are often separated by linkers that are rich in S, T, and N residues. From 1 up to 12 LysM domains can be found in a single protein. In bacteria, LysM domains are shown to bind directly the PG in a non-covalent manner (Mesnage et al., 2014). In E. faecalis, the LysM domains of AtlA interact with the N-acetyl group of the NAG with a minimum of two PG disaccharides NAG-x-NAG (where x corresponds to 1/4 of NAG or NAM). Interestingly, AtlA binds chitin with a higher affinity than PG (Mesnage et al., 2014). This may explain that the presence of a WTA covalently linked at the C6 position of NAM prevents interaction between PG and LysM domains (Steen et al., 2003; Frankel and Schneewind, 2012). Three LysM domains are sufficient for proper binding of AcmA, the major N-acetylglucosaminidase of Lactococcus lactis (Steen et al., 2005). However, multiple LysM domains are not forming a quaternary structure. In contrast, each LysM domain has a different affinity for the glucide interaction and is thought to bind glycan chains in a cooperative manner (Wong et al., 2014).
WXL Domain
The WXL domain (IPR027994) comprises two highly conserved sequence motifs Trp-X-Leu (WXL) including the distal motif YXXX(L/I/V)TWXLXXXP within the last ∼120 to 190 C-terminal of extracellular proteins (Siezen et al., 2006; Brinster et al., 2007). Initially observed in Lactobacillus plantarum (Kleerebezem et al., 2003), Lactobacillus coryniformis (Schachtsiek et al., 2004), and Lactobacillus sakei (Chaillou et al., 2005), extracellular proteins with a C-terminal WXL domain are predicted mainly in the orders of Lactobacillales and Bacillales, such as B. cereus, Listeria monocytogenes, Lactococcus garvieae, Lactobacillus rhamnosus, Lactobacillus casei (Siezen et al., 2006; Brinster et al., 2007; Dumas et al., 2008; Morita et al., 2009, 2011; Renier et al., 2012; Toh et al., 2013). The WXL domain was demonstrated to direct proteins to the bacterial cell surface by non-covalent binding to PG (Brinster et al., 2007). Consistently, WXL-proteins localise both at the cell surface and in the culture medium and bind to the surface of parietal monoderm bacteria in trans (Schachtsiek et al., 2004; Brinster et al., 2007). Genes encoding WXL-proteins are often organised in clusters (Siezen et al., 2006; Brinster et al., 2007; Galloway-Pena et al., 2015). The hypothesis that proteins of WXL clusters could form multicomponent complexes at the bacterial surface was recently substantiated by the interaction of two Enterococcus faecium WXL-proteins with their cognate transmembrane protein in vitro (Galloway-Pena et al., 2015). WXL-proteins remain poorly characterised at the experimental level (Siezen et al., 2006; Brinster et al., 2007; Cortes-Perez et al., 2015; Galloway-Pena et al., 2015). Functional, structural, and biochemical analyses of these proteins are urgently required to elucidate their architectural and biological properties.
SH3b Domains
SH3 [sarcoma (src) homology-3] domains were first described in eukaryotic proteins. They consist of 60 amino acids in average. In eukaryotes, SH3 domains are mainly involved in protein–protein interactions (Kaneko et al., 2008). In bacteria, SH3-like domains are named SH3b. However, in most articles authors named them indifferently SH3, SH3b, or with other names, e.g., GW. To avoid the promulgation of confusing statements in the scientific literature, they were here named according to the Pfam classification. Accordingly, five subgroups of SH3b domains are reported in parietal monoderm bacteria: SH3 of type 3 (PF08239), SH3 of type 5 (PF08460), SH3 of type 6 (or SH3b1; PF12913), SH3 of type 7 (or SH3b2; PF12914), and SH3 of type 8 (or GW; PF13457). These different SH3 domains allow recognition and binding to PG, but some would also be involved in protein–protein interactions, as suggested for the SH3 domain of type 3 (Rudolf et al., 2015). Of note, the SLAP domain (IPR024968) found in some bacterial cell surface proteins (Boot et al., 1995) may be distantly related to SH3 but further phylogenetic as well as experimental evidences of its implication in CW binding are most required.
The SH3 of type 5 (SH3-5) is a domain of 63 amino acids and is mainly found among Firmicutes, especially Streptococcus and Lactobacillus genera. This domain described in lysostaphin and Ale-1 proteins binds the pentaglycine peptide bridges of PG (Grundling and Schneewind, 2006; Lu et al., 2006). The SH3 of type 5 could be divided in two subgroups that bind PG with either low or high affinity (Becker et al., 2009).
The SH3 of type 6 and of type 7 were identified in a major class of CW endopeptidases, the NlpC/P60 hydrolases that cleave the linkage between D-Glu and A2pm (or K residue; Xu et al., 2015). The SH3 of type 6 (SH3-6) is suggested to bind the crossed-linked stem peptide of the PG. In contrast, SH3 of type 7 does not bind directly the cell surface but may be involved in the interaction between the SH3 of type 6 and the other protein domains (Xu et al., 2015).
The most well studied SH3-like domain is the SH3 of type 8 (SH3-8), also well-known as the GW (Glycine-Tryptophan rich) domain (Braun et al., 1997). In parietal monoderm bacteria, this domain of approximately 80 amino acids is mainly found among Firmicutes, especially Bacillus, Listeria, Lactobacillus, and Staphylococcus genera. In InlB from L. monocytogenes, the SH3 of type 8 was first described to be required for the non-covalent anchoring to the cell surface through an interaction with LTAs (Jonquieres et al., 1999). More recently, however, it was demonstrated to allow non-covalent anchoring directly to the PG (Percy et al., 2016). In the autolysin Atl from S. epidermidis, this domain was shown to be responsible for the direct binding to the PG (Biswas et al., 2006), but later, it was proposed to be responsible for the binding to LTAs (Zoll et al., 2010, 2012). Interestingly, in Lactobacilli, this domain is exclusively present in those proteins that harboured a S-layer (Johnson et al., 2015). These domains are also involved in the binding to the host cell receptors, or heparan sulphate proteoglycans (Jonquieres et al., 2001; Marino et al., 2002). They also have been reported to trigger MET phosphorylation and cellular phenotype and to bind Human Thrombospondin 1 and Vitronectin (Bleymuller et al., 2016). Overall, no consensus on the binding ligand for this domain is proposed, which would require further investigations. Nonetheless, a protein with less than two SH3-8 domains cannot bind to the CW (Braun et al., 1997; Jonquieres et al., 1999; Marino et al., 2002; Desvaux et al., 2010; Renier et al., 2012). In some proteins, the designated SH3b domain is not detected by InterPro/Pfam profiles; for instance, and in addition to a choline binding domain CWB1 (cell wall binding repeat of type 1, see below), LytB from S. pneumoniae exhibits a SH3b-like domain suggested to be involved in PG recognition (Bai et al., 2014). As this SH3b-like domain does not belong to any of the different types of SH3 domain described above, it suggests that novel types of SH3 domains remain to be uncovered.
Sporulation-Related Domain
The sporulation-related (SPOR) domain (IPR007730) was first described in the CwlC of B. subtilis (Mishima et al., 2005). CwlC is a CW amidase involved in PG hydrolysis of the mother cell allowing the release of the spore. This hydrolase property led to the name of SPOR domain. A SPOR domain consists of two repeats of 35 amino acid residues; from one to five SPOR domains can be found in a single protein. Among parietal monoderm bacteria, this domain was mainly identified in Firmicutes, especially in Clostridiales and Bacillales. This conserved domain binds the glycan part of PG and binding occurs in a cooperative manner (Mishima et al., 2005). Proteins harbouring a SPOR domain are essentially involved in sporulation or in cell-division processes (Yahashiri et al., 2015, 2017). For example, CwlC is a PG amidase secreted during sporulation and that hydrolyses the mother cell PG. It was proposed that SPOR-proteins, involved in the division process are preferentially localised at the septum, where amidases remove the stem peptides from the PG glycan chains. Thus, protein localization may be due to the binding of SPOR domains on naked PG, i.e., glycan strands lacking stem peptides, which are more abundant at the bacterial septum (Yahashiri et al., 2015).
Cell Wall Binding Repeat of Cpl-7
The cell wall binding repeat of Cpl-7 (CW-7; IPR013168) was originally found in the lysin encoded by the S. pneumoniae bacteriophage Cp-7 (Bustamante et al., 2010). This domain can be as single or up to three repeats in tandem, essentially in CW hydrolases. CW-7 was further shown to bind specifically to PG, with the CW muropeptide GlcNAc-MurNAc-Ala-isoGln as recognised CW target (Bustamante et al., 2012).
Other Domains Involved in PG Non-covalent Binding
Some other domains are described as potentially involved in recognition and non-covalent binding to PG. Among them, many phage lysins targeting the PG of L. casei harbour a novel type of PG-binding domain that is highly specific for amidated d-Asp Cross-bridge (Regulski et al., 2013). Other putative domains, mostly found in some Firmicutes, such as PG-binding domain of type 1 (PGB1; IPR002477; Layec et al., 2008) as well as PGB2 (IPR014927) were reported and would require further in-depth characterizations. In S. pneumoniae, LytB further exhibits a putative chitin binding domain (WW) domain, which was also proposed to be involved in PG binding (Bai et al., 2014).
Cell-Surface Proteins Bound to Cell Wall Polysaccharides
Cell Wall Binding Repeat of Type 1
The CWB1 (IPR018337) is also called choline-binding (ChBD) or CW binding repeat; for clarity and in echo to the cell wall binding repeat of type 2 (CWB2) reviewed here below, the CWB1 is preferred and favoured. This conserved domain is approximately 20 amino acids long. In parietal monoderm bacteria, CWB1 is mainly found among Firmicutes, especially in the families of the Lachnospiraceae, Ruminococcaceae, Clostridiaceae, Lactobacillaceae, and Streptococcaceae but also in some Actinobacteria, e.g., the Coriobacteriia and Bifidobacteriales orders. It was hypothesised that S. pneumoniae possessed an autolysin able to interact with phosphatidyl choline residues of the WTAs (Holtje and Tomasz, 1975). More than 10 years later, a glycosyltransferase, able to bind WPs through a repeated unit of amino acids was reported in Streptococcus sobrinus (Ferretti et al., 1987). This report was just followed by the demonstration that similar repeats in the lytic proteins of S. pneumoniae phage were involved in the recognition of choline-containing CWs (Garcia et al., 1998). Several surface proteins, including LytA from S. pneumoniae, were described to possess such a domain that was named glucan-binding domains (GBDs) and eventually CWB1 (Giffard and Jacques, 1994). LytA was the most characterised enzyme because it mediates indirectly virulence by lysis, allowing the release of toxins. The four LytA CWB1 domains were co-crystallised with choline (Fernandez-Tornero et al., 2001). Four choline interacting CWB1 sites are found in LytA, implying that at least three CWB1 are needed to form an interaction with one molecule of choline. It was then suggested that proteins harbouring less than three CWB1 are not expected to have affinity for CW choline residues.
Cell Wall Binding Repeat of Type 2
The CWB2 domain (IPR007253) was identified in CwlB of B. subtilis (Kuroda and Sekiguchi, 1991). In this species, CwlB is the major amidase. The CWB2 domain is approximately 90 amino acids long. In parietal monoderm bacteria, the CWB2 domain is found in the class of Actinobacteria, especially in the Micrococcales order, and the class of Firmicutes, especially in the orders of Clostridiales and Bacillales. Most of proteins carrying the CWB2 domains are reported with triple adjacent domains, more rarely with one or two (Fagan et al., 2011), e.g., the 29 Cwps (CW proteins) of Clostridium difficile all harboured three CWB2 domains. Among them, SlpA is the main S-layer protein of the C. difficile. Other Cwps were assigned with different potential functions, including amidase and protease (Fagan et al., 2011). This organisation may be due to the three-dimensional architecture; the formation of CWB2 trimer was indeed proposed to interact with the CW via a non-covalent linkage with the polysaccharide II (PSII; Willing et al., 2015). In C. difficile, the PSII is covalently anchored via a phosphodiester bond to the PG. In Cwp6 (CW protein 6) and Cwp8 from C. difficile, the trimer structure was recently solved by crystallography (Usenik et al., 2017). This structure revealed that 12 conserved residues were located between two domain interfaces. Moreover, using docking experiments, the structure formed by the CWB2 trimer was confirmed to be compatible with an interaction with the six monosaccharides that composed the PSII (Usenik et al., 2017). Two conserved surface R residues that may interact with the PSII are also found in the S-layer homology (SLH) trimers (see below; Kern et al., 2011). This result in combination with the SLH organisation in trimer and a similar function of polysaccharide anchoring suggests a common or convergent evolutionary origin (Kern et al., 2011).
S-Layer Homology Motif
The SLH domain (IPR001119) was first reported in three proteins of Clostridium thermocellum (Fujino et al., 1993). This domain was later named SLH after comparison of the S-layer protein sequences of Acetogenium kivui, C. thermocellum, and Bacillus brevis (Ebisu et al., 1990; Lupas et al., 1994). This domain consists of an approximately 55-amino acid-long sequence with a group of five highly conserved residues (ITRAE). In parietal monoderm bacteria, it is identified in some species of the class Actinobacteria, such as in the order Coriobacteriales, Bifidobacteriales, or Micrococcales, but mainly among Firmicutes, including Clostridia and Bacilli. Three SLH domains were shown to be sufficient for the anchoring at the CW surface of B. anthracis, but only two are necessary for the CW interaction (Mesnage et al., 1999; Huber et al., 2005). Moreover, SLH proteins from C. thermocellum are able to bind the CW of B. anthracis and vice versa (Chauvaux et al., 1999). SLH domains are shown to bind WP in a non-covalent manner. The WP fraction is pyruvylated by CsaB and this WP modification is essential for the SLH protein binding (Mesnage et al., 2000). The Sap structure, a B. anthracis S-layer protein, confirmed the potential interaction between WP and the three SLH motifs (Kern et al., 2011). Pyruvate was later found to be placed at the distal end of each WP (Forsberg et al., 2012). In Bacillus, it is proposed that the N-acetyl mannose of the WP is pyruvylated (Forsberg et al., 2012). In some cases, the SLH domains may be not sufficient for WP interaction. For instance, in the SbsB of Bacillus sphaericus, the C-terminal domain together with the SLH domains is needed for the WP interaction (Huber et al., 2005). Direct and exclusive binding of SLH to the PG or together with the WP is still subject to discussion and remains controversial (Zhao et al., 2005, 2006; Janesch et al., 2013).
Cell-Surface Proteins Bound to the CW by Unknown Mechanism
The ChW (clostridial hydrophobic repeat with a conserved W residue) domain (IPR006637) was first identified in Clostridium acetobutylicum and was predicted to be involved in cell surface anchoring or in protein–protein interaction (Nölling et al., 2001; Desvaux, 2005a). This domain is constituted of highly conserved GW dipeptide motifs and is about 50 amino acids long. A single protein can harbour between one and 12 ChW domains. It was suggested that the ChW domains are associated in triplet for the surface interaction but the biochemical nature of the CW ligand remains unknown (Sullivan et al., 2007). In parietal monoderm bacteria, the ChW domain is essentially found in some Firmicutes, especially of the class Clostridia, but also in some Erysipelotrichia and Bacilli, e.g., in the genera Lactococcus, Streptococcus, and Enterococcus, as well as in some bacteria of the phyla Actinobacteria, especially in the genus Streptomycetes. ChW-proteins are mostly endolysins suggesting the importance of this domain for CW interaction and enzymatic activity (Oliveira et al., 2013).
Cell-Surface Proteins with Uncharacterised Cell-Envelope Interacting Domain: The Moonlighting Proteins
Parietal monoderm bacteria have some surface-exposed proteins that lack a canonical signal sequence and a CW interacting domain. Although not sharing any domain or sequence homology, they share the ability to interact with fibronectin or extracellular matrix (ECM)-components. The most common are cytoplasmic enzymes or proteins, referred as moonlighting proteins. They include the ubiquitous glycolysis enzymes glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Pancholi and Fischetti, 1992), enolase, phosphoglycerate kinase, the glutamine synthetase (GlnS), and the translation elongation factor Ef-Tu (Amblee and Jeffery, 2015). While most act as adhesins by interacting with components of the host ECM (plasminogen, fibronectin, laminin, or mucin), some like Ef-Tu interact also with neuropeptides at the membrane level (Fulde et al., 2013; Mijouin et al., 2013; Amblee and Jeffery, 2015; N’Diaye et al., 2016a,b, 2017). Beside the anticipated lack of an SP and cell surface association domain, bioinformatic analysis of 98 experimentally reported intracellular proteins having a moonlighting cell surface function, failed to identify specific features shared by these proteins (Amblee and Jeffery, 2015). The domain interacting with plasminogen is frequently localised at the C-terminus of the protein, however, no conserved domain could be identified (Bergmann et al., 2003; Ehinger et al., 2004). Ionic bonds and low pH were shown to contribute to the association of cytoplasmic proteins with the cell surface of Lactobacillus crispatus (Antikainen et al., 2007). Conversely, GAPDH and enolase have been shown to bind LTA on the bacterial cell surface by ionic bonds (Antikainen et al., 2007; Kinoshita et al., 2008). Reversely, GAPDH of S. pneumoniae did not bind synthetic LTAs or TAs and direct binding to PG was observed (Terrasse et al., 2015). Moonlighting proteins occur in all bacteria and are thus involved in a large range of unrelated functions including colonisation, modulation of the host response and virulence (Kainulainen and Korhonen, 2014).
Other non-classical proteins exposed on the cell surface of parietal monoderm bacteria are known as fibronectin-binding proteins (FBPs) characterised by two adjacent conserved domains: the about first 400 amino acids (PF05833) of which 89 residues associate with fibronectin-binding activity (Courtney et al., 1994) followed by the conserved domain of unknown function DUF814 (IPR008532) of ∼100 amino acid residues including conserved motif (D/E)X(W/Y)XH. First identified in the fibronectin-binding protein FBP54 of S. pyogenes (Courtney et al., 1994), these domains have been reported in PavA of S. pneumoniae (Holmes et al., 2001), FbpA of Streptococcus gordonii (Christie et al., 2002), Fbp68 of C. difficile (Hennequin et al., 2003), FbpB of Clostridium perfringens (Katayama et al., 2009), EfbA in E. faecalis (Torelli et al., 2012), Fnm in E. faecium (Somarajan et al., 2015), YloA in B. subtilis (Rodriguez Ayala et al., 2017) and FbpA in Weissella cibaria (Wang et al., 2017). Consistently with impaired binding capacity to fibronectin upon nested deletions of the C-terminal part of S. pneumoniae PavA (Holmes et al., 2001), structural and functional analyses of FBPS of Streptococcus suis revealed that the C-terminal half of FBPs mediates binding to fibronectin whereas the N-terminal half interacts specifically with the surface of Streptococcus suis (Musyoki et al., 2016). The fact that N-terminal half of FBPs does not bind to S. pneumoniae nor S. agalactiae cells suggests an interaction with a specific CW component that remains to be identified. Despite their contribution to fibronectin binding and overall virulence in several pathogens, the exact role of these FBPs is still unclear (Kawabata et al., 2001; Dramsi et al., 2004; Pracht et al., 2005; Torelli et al., 2012; Somarajan et al., 2015).
How these proteins are released outside from the cell and attached to the cell surface remains poorly understood. Several lines of evidence indicate that the release of GAPDH of S. agalactiae, S. aureus, and S. pneumoniae bacterial involves autolysis (Pasztor et al., 2010; Oliveira et al., 2012; Terrasse et al., 2015). Consistently, moonlighting proteins localise preferentially at the septum. However, this issue is still debated as not all cytoplasmic proteins are detected at the CW (Ebner et al., 2016). Interestingly, based on indirect evidence using an inhibitor of a mechanosensitive channel it was recently proposed that EF-Tu and DnaK of S. epidermidis could be exported through the large mechanosensitive channel (N’Diaye et al., 2016b). Variations of channel diameter between bacterial species may explain differences between patterns of moonlighting proteins.
Cell-Surface Supramolecular Protein Structures
Besides monopolypeptides, some surface proteins form complex surface organelles. In parietal monoderm bacteria, such supramolecular protein structures include the S-layer, flagellum, various pili and cellulosome.
S-Layer
S-layer is a proteinaceous two-dimensional crystalline array constituting the outermost CW layer in the absence of a capsule (Fagan and Fairweather, 2014). Located above the PG, this surface supramolecular structure is not a common theme in parietal monoderm bacteria, e.g., it is present in numerous Bacillus or Clostridium species but absent from Listeria and Staphylococcus genera. Usually, a S-layer is formed by the auto-assembly of a unique protein that may be glycosylated. The S-layer proteins are usually rich in hydrophobic and acidic amino acids (Sára and Sleytr, 2000). The interactions between the S-layer subunits are stronger than surface interactions (Messner and Sleytr, 1992). Most often bacteria with an S-layer possess a single S-layer and in very rare cases two (Kuen et al., 1997). Most of the S-layer proteins are non-covalently anchored through SLH or CWP domains at the bacterial surface. Of note and as mentioned above, a protein harbouring a SLH or a CWP domain is not necessarily an S-layer protein.
The function of the S-layer remains unclear but it is generally suggested to act either as a scaffold, a sieve or a shield to some environmental stresses (Sára and Sleytr, 2000; Fagan and Fairweather, 2014; Gerbino et al., 2015). While cited as a virulent factor or adhesion factor, such a role has not been convincingly demonstrated in any parietal monoderm bacteria. In B. anthracis, Sap was suggested to be the receptor of the phage AP50c (Plaut et al., 2014). Investigating its contribution to colonisation processes, S-layer was negatively correlated with biofilm formation in B. cereus (Auger et al., 2009). Consistently, a C. difficile mutant strain lacking the Cwp84 protease, which plays a key role in the maturation of the S-layer protein SlpA, forms a biofilm 72-fold more important than the wild type strain (Pantaleon et al., 2015). Except for C. difficile (Merrigan et al., 2013), the bacterial S-layer is considered as non-essential. The S-layer proteins can account for up to 15% of total bacterial proteins, also the absence of common physiological functions among bacteria is intriguing. Undoubtedly, this call for in-depth investigations under conditions more relevant to the ecophysiology of the bacterial species considered.
Flagellum
The bacterial flagellum is secreted and assembled via the FEA. Several transmembrane components constitute the translocon (FlhAB-FliOPQR) and form the translocase together with the ATPase FliI (Macnab, 2003, 2004). The flagella per se is composed of a basal body, the hook, the junction and the filament proteins, which are secreted and assembled by the FEA (Evans et al., 2014). These proteins do not exhibit a SP and the signal necessary for targeting is still controversial (Aldridge and Hughes, 2001, 2002). While most knowledge about the assembly and regulation mechanisms results from investigations in different LPS-diderm bacteria, information related to parietal monoderm bacteria remains restricted to fewer bacterial species, e.g., B. subtilis (Mukherjee and Kearns, 2014; Rossez et al., 2015).
Of course the flagellum is a well-known motility factor that can be involved in swimming but also swarming (Henrichsen, 1972; Belas, 2014). Swarming is especially relevant for surface colonisation processes, including adhesion and biofilm formation (Beeby, 2015; Chaban et al., 2015). Mechanosensing by flagella and chemotaxis further allow the bacteria to switch developmental programmes and adapt in response to changes in their environment. Glycosylation of the flagella has now been demonstrated in several parietal monoderm bacteria (Schirm et al., 2004, 2005; Twine et al., 2008, 2009; Kajikawa et al., 2016) and they further appeared to play a role in pathogenesis and biofilm formation (Valguarnera et al., 2016; Valiente et al., 2016).
Pili
Pili are tubular cell-surface appendages, which size, diameter, and shape can be extremely variable depending on the type of appendage considered. In parietal monoderm bacteria, three main types of pili can be encountered, (i) the pili made of covalently linked subunits involved in colonisation and host interaction, (ii) the type 4 pili (T4P) involved in transformation, motility and adherence, and the most recently uncovered (iii) pili made of amyloids. In parietal monoderm bacteria, beside the evidence the formation of a DNA translocation channel, no conjugative pili has been formally demonstrated.
Covalently Assembled Pili
First reported and studied in the mycolic-diderm Actinobacteria and Corynebacteria, pili made of covalently linked pilins are assembled and anchored to the PG by sortases (Yanagawa et al., 1968; Ton-That and Schneewind, 2003). Since then they have been described in various parietal monoderm bacteria, e.g., including some bacilli, enterococci, streptococci, lactococci, lactobacilli, and bifidobacteria (Ton-That and Schneewind, 2003; Kankainen et al., 2009; Hendrickx et al., 2011; O’Connell Motherway et al., 2011; Oxaran et al., 2012; Murphy et al., 2014). Pili are all composed of a major pilin that forms the shaft and a minor tip pilin that is located at the tip of the pilus. Genes encoding pili are organised in operon of two or three prepilin genes and one or two pilin-specific sortase enzymes (Hendrickx et al., 2011). All prepilins contain an N-terminal SP for secretion and a C-terminal LPXTG domain for covalent binding to PG or formation of intermolecular bonds between pilins. In addition, they exhibit tandem Ig-like domains, also referred as CnaB domains (PF16569), contributing to pili integrity, stability, and biomechanical properties through self-generated intramolecular bonds between a lysine and an asparagine residue (Kang et al., 2007; Budzik et al., 2009; Kang and Baker, 2012; Echelman et al., 2016). The YPNK motif is typical of major and basal pilins and provides the K residue to form the intermolecular isopeptide bond with another molecule of pilin. Besides, major pilins have a conserved glutamate residue in an E-box motif (consensus YXLXETXAPXGY) that contributes to the autocatalytic formation of intramolecular isopeptide bonds (Budzik et al., 2009; Kang et al., 2009; Alegre-Cebollada et al., 2010). Basal pilins are usually smaller and have a proline-rich C-terminal tail involved in CW anchoring (Krishnan et al., 2007; Linke et al., 2010). Pilins are assembled by sequential transpeptidation reactions involving sortases. Successively, the threonine of the LPXTG sorting signal of the minor tip pilin is covalently linked to the conserved K residue of the YPKN pilin motif of the major pilin by a pilus-specific class C sortase (Budzik et al., 2008). Subunits of the major pilin are then successively polymerised head-to-tail by the pilus-specific sortase. High resolution transmission electron microscopy and pilin structural studies confirmed that these pili were heteropolymers of two to three head-to-tail covalently linked pilins (Kang et al., 2009). Once assembled and depending on the species, the pilus is generally anchored to the PG by the housekeeping sortase A either directly or through the incorporation of the minor basal pilin (Dramsi et al., 2006; Budzik et al., 2007; Mandlik et al., 2008; Necchi et al., 2011; Shaik et al., 2014). Tip pilins do not exhibit YPKN motif, but consistently with their adhesive function they harbour adhesion domains, e.g., vWFA (von Willebrand factor A) domain, in addition to classical IgG-like folds (Krishnan et al., 2007; Linke et al., 2010). However, several exceptions to this general picture have been reported, e.g., the pilin motif YPKN can be restrain to a single lysine (Kang et al., 2007; Cozzi et al., 2015), tip and basal pilins can spread along the pilus shaft (Dramsi et al., 2006; Kankainen et al., 2009; Reunanen et al., 2012; Yu et al., 2015), sortase A can be dispensable for pilus anchoring to the CW (LeMieux et al., 2008; Lazzarin et al., 2015).
Covalently assembled pili are essentially involved in colonisation processes, especially sessile development (Nallapareddy et al., 2006; Krishnan et al., 2007; Manetti et al., 2007; Konto-Ghiorghi et al., 2009; Pointon et al., 2010; Rinaudo et al., 2010; Sillanpaa et al., 2010, 2013; Danne et al., 2011; Oxaran et al., 2012). Zipper-like interactions favoured by multiple SpaC distributed along the pilus were suggested a major contributor to biofilm formation (Tripathi et al., 2013). These pili can also play key roles in bacterial adhesion to ECM proteins, e.g., fibronectin, collagens or mucins, via the tip pilin (Schwarz-Linek et al., 2003; Hilleringmann et al., 2008; von Ossowski et al., 2011). Covalent intra- and intermolecular bonds of covalently assembled pili confer remarkable spring-like biomechanical properties, which can withstand physiological shear forces. In addition to specific heterophilic interactions with mucin and collagen, tip pilin SpaC mediates homophilic interactions involved in bacterial aggregation (Tripathi et al., 2013). In S. pyogenes, the N-terminal thioester domain of the pilus adhesin Cpa was demonstrated to form covalent bonds with the polyamine spermidine, suggesting these pili could be involved in covalent attachment to host cells (Linke-Winnebeck et al., 2014).
Non-covalently Assembled Pili: The Type 4 Pili
Initially described and thoroughly studied in LPS-diderm bacteria, type 4 pili (T4P) are thin flexible filaments (5–8 nm) of several microns in length composed of thousands of copies of a major pilin (Craig et al., 2003). T4P pili are helical polymers of a major, which consists in a conserved α-helix at the N-terminus followed by a C-terminal β-sheet domain (Craig et al., 2004). The cohesion of the filament relies on hydrophobic interactions between the N-terminal helices amino acids N-terminal α-helices in the centre of the fibre. The presence of filaments resembling T4P in parietal monoderm bacteria was first observed in Ruminococcus albus (Rakotoarivonina et al., 2002). Since then, clusters of genes associated to T4P formation, have been detected in many genomes of Firmicutes (Imam et al., 2011; Berry and Pelicic, 2015). In parietal monoderm bacteria, components of the T4P are secreted and assembled by the FPE. According to B. subtilis nomenclature, the FPE system is composed of the ATPase ComGA, the IMP ComGB and the type 4 prepilin peptidase ComC, whereas ComGC is the major pilin, ComGD, ComGE, ComGF, and ComGG are minor pilins (Chen et al., 2006; Desvaux and Hébraud, 2006, 2009). In C. perfringens and S. sanguinis, the FPE comprises a retraction ATPase (IPR006321) and two conserved proteins involved in pili assembly (IPR005883 and IPR007813) in addition to the assembly ATPase ComGA (IPR001482), the IMP ComGB (IPR003004) and the type 4 prepilin peptidase ComC (IPR000045) (Melville and Craig, 2013; Berry and Pelicic, 2015). Besides, the T4P is composed of two major and three minor pilins (IPR012902). Based on models derived from LPS-diderm bacteria where the T4P is secreted and assembled by a type II secretion system (Peabody et al., 2003; Desvaux et al., 2009; Chagnot et al., 2013), the prepilins of <200 amino acid residues exhibit a SP with a conserved type 4 prepilin motif including a glutamate at position 5 of the mature protein (Tjalsma et al., 2000; Desvaux and Hébraud, 2006, 2009). Prepilins are processed by the prepilin peptidase cleaving the SP between the n- and h-domain. Polymerization of the mature pilins involves the assembly ATPase and integral membrane and accessory proteins. When present, the retraction ATPase mediates depolymerization of pilin subunits and subsequent pilus retraction (Melville and Craig, 2013; Berry and Pelicic, 2015). Structural characterization of PilA1, the major pilin of the T4P in C. difficile, confirms general structural conservation with an N-terminal α-helix, followed by a helical αβ-loop and a four-stranded anti-parallel β sheet, instead of the typical the C-terminal disulfide bond of type 4 pilins (Piepenbrink et al., 2015). Amino acid sequence variation in the C-terminal part of PilA1 between strains revealed alternative stabilising hydrogen bonds between the β loops, highlighting that T4P of parietal monoderms rely on specific mechanisms in spite structural and function conservation with those of LPS-diderm bacteria. Interestingly, S. sanguinis encodes an additional T4P, which proteins are orthologous to proteins involved in the assembly of the T4P in B. subtilis and S. pneumoniae (Xu et al., 2007; Gurung et al., 2016; Gurung et al., 2017).
T4P are generally involved in twitching motility, DNA uptake during conjugation and transformation, adherence to host cells and biofilm formation (Giltner et al., 2012). In parietal monoderm bacteria, the involvement of T4P in bacterial motility have been reported in C. perfringens and S. sanguinis (Varga et al., 2006; Gurung et al., 2016), whereas transformation by T4P has been experimentally demonstrated in S. pneumoniae (Laurenceau et al., 2013, 2015). It was evidenced that DNA fragments are too large to go through the T4P and exogenous double-stranded DNA would actually be captured by the pilus before being guided to the Com (competence development) uptake machinery (Dubnau, 1999; Dubnau and Provvedi, 2000; Chen and Dubnau, 2004; Johnston et al., 2014; Laurenceau et al., 2015). In R. albus, T4P is specifically involved in adherence to cellulose (Rakotoarivonina et al., 2002).
Amyloid Pili
Among macromolecular structures displayed at the surface of parietal monoderm bacteria, amyloid pili remain poorly characterised. Reminiscent of the curli in Enterobacteriaceae, these amyloid fibres are quaternary structure of peptide or protein aggregates forming parallel β-sheets perpendicular to the fibre axis (Rambaran and Serpell, 2008). Initially reported in B. subtilis, amyloid pili have so far mainly been involved in biofilm formation (Taglialegna et al., 2016a). Their biogenesis relies on different steps depending on the precursor protein, but always leads to stable β-sheet aggregates. Secreted by the Sec pathway, the B. subtilis amyloid protein TasA forms amyloid fibres of variable length and 10–15 nm in width (Romero et al., 2010). The co-encoded dedicated signal peptidase SipW and TapA are required to process and produce functional TasA fibres, respectively (Romero et al., 2011, 2014). Like for other amyloidogenic precursors, acidic pH promotes aggregation of TasA (Chai et al., 2013). However, the mechanism of TasA amyloïd fibre biogenesis is still unknown. In S. aureus, amyloids fibres are made of secreted peptides known as phenol-soluble modulins (Schwartz et al., 2012, 2014; Marinelli et al., 2016; Tayeb-Fligelman et al., 2017). The LPXTG-cell wall anchored adhesins P1 (AgII) and WapA in Streptococcus mutans and BapA in S. aureus have the ability to form amyloid fibres (Oli et al., 2012; Taglialegna et al., 2016b; Besingi et al., 2017). BapA is a member of biofilm-associated protein (Bap) family defined as high-molecular-weight CW anchored LPXTG proteins involved in biofilm formation (Shankar et al., 1999; Cucarella et al., 2001; Lembre et al., 2014). In BapA, the domain B self-assembles into amyloid fibres in acidic pH and low calcium concentration (Taglialegna et al., 2016b). Although domain B of BapA orthologue of Staphylococcus saprophyticus is amyloidogenic, other peptides may be involved in the biogenesis of Bap amyloid fibres since a short amyloidogenic peptide derived from the imperfect tandem repeats (C-repeats) in Bap proteins of other Staphylococcus species has been identified (Lembre et al., 2014). The amyloidogenic moiety is located in the C-terminal region of P1 and central part for WapA (Besingi et al., 2017). The amyloidogenic moiety of P1 also associates with covalently attached full-length P1 suggesting that P1 may serve as the platform for amyloidogenesis (Heim et al., 2014). Although much less characterised, the amyloidogenic moiety of WapA encompasses a collagen-binding domain (IPR008456) composed of two antiparallel β-sheets and two short α-helices. Occurring in specific conditions, amyloidogenesis can be viewed as a conformational adaptation of cell surface proteins with dual functions.
Conjugative Pili
Consistently with their ability to exchange DNA, several species of parietal monoderm bacteria have in their genomes mobile genetic elements that encode conjugative systems reminiscent of type IV secretion system (T4SS) in LPS-diderm bacteria (Guglielmini et al., 2013, 2014). Although incompletely understood, the best characterised conjugative elements in Firmicutes are the transposon Tn916 of E. faecalis, the plasmids pIP501 of S. agalactiae, pCF10 of E. faecalis and pCW3 of C. perfringens, the integrative and conjugative elements ICEBs1 of B. subtilis and ICESt1 of Streptococcus thermophilus (Alvarez-Martinez and Christie, 2009; Bhatty et al., 2013; Goessweiner-Mohr et al., 2013; Bellanger et al., 2014; Auchtung et al., 2016; Wisniewski and Rood, 2017). In contrast to conjugation in LPS-diderm bacteria, conjugation in parietal monoderm bacteria would not require pili formation (Andrup and Andersen, 1999). However, considering the identification of several proteins homologous to the T4SS and the analogous situation with the FPE in B. subtilis where only pseudo-pilus would be formed in parietal monoderm bacteria, much deeper investigations in that direction would be required to categorically exclude the formation of conjugative pili in any parietal monoderm bacteria. Not to forget that just a couple of decades ago, the presence of any pili in parietal monoderm bacteria was not even considered.
Cellulosome
A cellulosome is a supramolecular multienzymatic complex present at the bacterial surface of some parietal monoderm bacteria and dedicated to degradation of plant CW polysaccharides (Bayer et al., 2004; Fontes and Gilbert, 2010). It is organised around a scaffolding which assembles different catalytic subunits. Cellulosomes are only found in some parietal monoderm bacteria of the families Lachnospiraceae and Clostridiaceae (Guedon et al., 2000; Desvaux and Petitdemange, 2001; Desvaux, 2005a). In C. thermocellum, the scaffolding CipA (cellulosome-integrating protein A) is composed of a CBM (carbohydrate-binding module), a DocII (dockerin domain of type II), and 9 CohI (cohesin domains of type I) (Béguin and Lemaire, 1996; Smith and Bayer, 2013). Whereas different types of CBM allows binding to different carbohydrate-polymers with various affinities, CohI acts as a receptor domain for a dockerin domain of type I (DocI) harboured by cellulosomal enzymes. CipA can display up to nine different cellulosomal enzymes thanks to the 9 CohI and its anchor to the bacterial cell surface via DocII, which interacts with a CohII (type II cohesin domain) presents in a cell-surface protein, such as SdbA (scaffolding dockerin binding A) (Stern et al., 2016). Depending on the bacterial species, the scaffolding can harbour more or less cohesion domains (Schwarz, 2001). An even higher level of complexity can even be reached when different scaffoldings assemble one with another from complex cellulosomes like in Pseudobacteroides cellulosolvens or Acetivibrio cellulolyticus (Xu et al., 2003, 2004; Hamberg et al., 2014). The assembly of several cellulosomes to form a polycellulosome would occur via DocI/CohI interactions (Carvalho et al., 2003). Cellulosome components exhibit SP and would be secreted by the Sec pathway, even so the mechanism for the assembly of the different subunits at the cell surface appears quite complex (Desvaux, 2005a,b; Bras et al., 2016; Bule et al., 2017; Smith et al., 2017). The cellulosome is generally exposed at the bacterial cell surface following cohesin-dockerin interaction with cell-surface proteins, themselves anchored in the CW via SLH domains.
Conclusion
By reviewing the components of the cell envelope of parietal monoderm bacteria, this review stressed the difference between the surfaceome, i.e., the totality of molecules present at the bacterial cell surface, and the proteosurfaceome, the proteinaceous subset of the surfaceome. Besides, the concept of bacterial cell surface must be carefully balanced and considered with regards of the idea of scale and the notions of molecular, accessible, contact and reentrant surfaces. Considering both the CM and CW as well as monopolypeptides and supramolecular protein structures, this review provides an overview of the mechanisms of protein anchoring to the cell envelope of parietal monoderm bacteria. Nonetheless, it can hardly be considered as exhaustive. Indeed, some additional domains have been described in some bacterial species but have not been registered in InterPro as yet and/or would demand further characterization. For instance, the C-terminal WrY domain of Sbi (second binding protein for immunoglobulins) from S. aureus binds the LTAs (Smith et al., 2012). In Aap (accumulation associated proteins) from S. aureus, bioinformatic analyses strongly suggest the G5 domain could be involved in NAG binding but experimental evidences supporting this function are still awaited (Bateman et al., 2005). This is also the case for several S-layer proteins, which do not harbour SLH domains but are clearly attached to the CW (Chami et al., 1997; Schäffer et al., 1999; Steindl et al., 2002; Avall-Jaaskelainen and Palva, 2005). At the opposite, some domains reported in InterPro would still require further characterization since their first report to ascertain their involvement in protein attachment to the CW, e.g., the PGB2 (IPR014927) (Foster, 1991). Of note, no protein domain involved in the interaction with components of the capsule, such as the polyglutamate, has been uncovered so far. Bacteriophages often use cell surface polysaccharides as receptors. Exploration of the diversity of receptor-binding proteins of bacteriophages may help to identify novel WP binding domains although their multimeric state required for efficient binding will imply structural analyses (Regulski et al., 2013; McCabe et al., 2015; Koc et al., 2016).
It can also be stressed that some cell-surface proteins can exhibit several anchoring domains, e.g., some IMPs can also be lipoproteins or some LPXTG-proteins can also have additional CWBDs. Even for the well-characterised domains presented in this review, there is still some work to be done to refine their biochemical properties, especially to decipher in detail the interactions of a given domain with its CW ligand(s) and define their three-dimensional structure, which have been resolved only in a handful of them. For more complex structures, e.g., pili, tomography by cryo-EM is certainly one of the method of choice to reveal their molecular details (Li and Thanassi, 2009). While considered rare or even absent a decade ago, it appears bacterial proteins can be glycosylated, especially those cell-surface exposed (Lu et al., 2015; Schaffer and Messner, 2017). However, the extent to which glycosylation contributes to protein anchoring to the cell envelope of parietal monoderm bacteria remains an open question. All-in-all, it can be bet that novel cell-envelope binding domains will be uncovered in the years to comes and increase the repertoire of known surface proteins in parietal monoderm bacteria. Most of the protein domains interact non-covalently with the CW. Consistently with bacterial growth, the surfaceome is extremely dynamic and in constant renewing. Together with other processes, labile interactions participate to the flexibility and the spatio-temporal remodelling of the surfaceome in response to physiological or environmental changes (Bierne and Dramsi, 2012; Mitra et al., 2016). Undoubtedly, the composition of proteosurfaceome is of great importance for the colonisation of various environments, including bacterial adhesion and biofilm formation ability of parietal monoderm bacteria (Planchon et al., 2009; Renier et al., 2011; Chagnot et al., 2012, 2013).
By providing a comprehensive view of mechanisms of protein anchoring to the cell envelope of parietal monoderm bacteria, this review should be helpful for scientists and researchers involved in global approaches, especially genomics, transcriptomics and proteomics. Predicting the subcellular localization of genome encoded proteins is a key step in comprehending the physiology of a given micro-organism but also applied research dedicated to the mining of new degradative enzymes, adhesins or antigens. Respective to the proteosurfaceome, several individual tools allow predicting the presence of SP, e.g., SignalP (Nielsen, 2017), cell-envelope anchoring domain, e.g., InterProScan (Jones et al., 2014), or even the final subcellular location, e.g., PSORT (Peabody et al., 2016), but an integrated and combining approach based on the secretome concept, which considers the biology of protein secretion by including the protein secretion systems, post-translational and post-translocational modifications as well as retention signals, proved much more powerful than individual predictors (Renier et al., 2012). While genomics is useful for initial mapping of the secretome, which further allows defining the proteosurfaceome (i.e., the cell surface complement of the secretome), proteomics remains the ultimate method of choice to ascertain that proteins are effectively expressed and located as predicted (Planchon et al., 2007; Solis and Cordwell, 2011, 2016). Besides proteomics, which by definition focuses on the whole protein content, glycomics and lipidomics can also be used to investigate globally the polysaccharidic and lipid fractions of the bacterial surfaceome (Chessa et al., 2008; Kondakova et al., 2015). So far, however, such approaches have not been broadly applied in parietal monoderm bacteria but it is certainly a promise in the years to come to define more comprehensively the surfaceome of these bacteria, which are for some of them important pathogens. As a primary target, such research directions on the surfaceome are a prerequisite for the development of novel antibacterial agents or therapeutics.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by INRA (Institut National de la Recherche Agronomique).
References
Ainsworth, S., Sadovskaya, I., Vinogradov, E., Courtin, P., Guerardel, Y., Mahony, J., et al. (2014). Differences in lactococcal cell wall polysaccharide structure are major determining factors in bacteriophage sensitivity. mBio 5:e00880-14. doi: 10.1128/mBio.00880-14
Aldridge, P., and Hughes, K. T. (2001). How and when are substrates selected for Type III secretion? Trends Microbiol. 9, 209–214.
Aldridge, P., and Hughes, K. T. (2002). Regulation of flagellar assembly. Curr. Opin. Microbiol. 5, 160–165. doi: 10.1016/S1369-5274(02)00302-8
Alegre-Cebollada, J., Badilla, C. L., and Fernandez, J. M. (2010). Isopeptide bonds block the mechanical extension of pili in pathogenic Streptococcus pyogenes. J. Biol. Chem. 285, 11235–11242. doi: 10.1074/jbc.M110.102962
Alvarez-Martinez, C. E., and Christie, P. J. (2009). Biological diversity of prokaryotic Type IV secretion systems. Microbiol. Mol. Biol. Rev. 73, 775–808. doi: 10.1128/MMBR.00023-09
Amblee, V., and Jeffery, C. J. (2015). Physical features of intracellular proteins that moonlight on the cell surface. PLOS ONE 10:e0130575. doi: 10.1371/journal.pone.0130575
Andrup, L., and Andersen, K. (1999). A comparison of the kinetics of plasmid transfer in the conjugation systems encoded by the F plasmid from Escherichia coli and plasmid pCF10 from Enterococcus faecalis. Microbiology 145(Pt 8), 2001–2009. doi: 10.1099/13500872-145-8-2001
Antikainen, J., Kuparinen, V., Lahteenmaki, K., and Korhonen, T. K. (2007). pH-dependent association of enolase and glyceraldehyde-3-phosphate dehydrogenase of Lactobacillus crispatus with the cell wall and lipoteichoic acids. J. Bacteriol. 189, 4539–4543. doi: 10.1128/JB.00378-07
Armbruster, K. M., and Meredith, T. C. (2017). Identification of the lyso-form N-acyl intramolecular transferase in low-GC Firmicutes. J. Bacteriol. 199:e00099-17. doi: 10.1128/JB.00099-17
Asanuma, M., Kurokawa, K., Ichikawa, R., Ryu, K. H., Chae, J. H., Dohmae, N., et al. (2011). Structural evidence of alpha-aminoacylated lipoproteins of Staphylococcus aureus. FEBS J. 278, 716–728. doi: 10.1111/j.1742-4658.2010.07990.x
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Atrih, A., Zollner, P., Allmaier, G., Williamson, M. P., and Foster, S. J. (1998). Peptidoglycan structural dynamics during germination of Bacillus subtilis 168 endospores. J. Bacteriol. 180, 4603–4612.
Auchtung, J. M., Aleksanyan, N., Bulku, A., and Berkmen, M. B. (2016). Biology of ICEBs1, an integrative and conjugative element in Bacillus subtilis. Plasmid 86, 14–25. doi: 10.1016/j.plasmid.2016.07.001
Auger, S., Ramarao, N., Faille, C., Fouet, A., Aymerich, S., and Gohar, M. (2009). Biofilm formation and cell surface properties among pathogenic and nonpathogenic strains of the Bacillus cereus group. Appl. Environ. Microbiol. 75, 6616–6618. doi: 10.1128/AEM.00155-09
Avall-Jaaskelainen, S., and Palva, A. (2005). Lactobacillus surface layers and their applications. FEMS Microbiol. Rev. 29, 511–529. doi: 10.1016/j.fmrre.2005.04.003
Babu, M. M., Priya, M. L., Selvan, A. T., Madera, M., Gough, J., Aravind, L., et al. (2006). A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J. Bacteriol. 188, 2761–2773. doi: 10.1128/JB.188.8.2761-2773.2006
Bai, X. H., Chen, H. J., Jiang, Y. L., Wen, Z., Huang, Y., Cheng, W., et al. (2014). Structure of pneumococcal peptidoglycan hydrolase LytB reveals insights into the bacterial cell wall remodeling and pathogenesis. J. Biol. Chem. 289, 23403–23416. doi: 10.1074/jbc.M114.579714
Banerji, A., and Navare, C. (2013). Fractal nature of protein surface roughness: a note on quantification of change of surface roughness in active sites, before and after binding. J. Mol. Recognit. 26, 201–214. doi: 10.1002/jmr.2264
Bateman, A., Holden, M. T., and Yeats, C. (2005). The G5 domain: a potential N-acetylglucosamine recognition domain involved in biofilm formation. Bioinformatics 21, 1301–1303. doi: 10.1093/bioinformatics/bti206
Bayer, E. A., Belaich, J. P., Shoham, Y., and Lamed, R. (2004). The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58, 521–554. doi: 10.1146/annurev.micro.57.030502.091022
Beck, K., Eisner, G., Trescher, D., Dalbey, R. E., Brunner, J., and Muller, M. (2001). YidC, an assembly site for polytopic Escherichia coli membrane proteins located in immediate proximity to the SecYE translocon and lipids. EMBO Rep. 2, 709–714. doi: 10.1093/embo-reports/kve154
Becker, S. C., Foster-Frey, J., Stodola, A. J., Anacker, D., and Donovan, D. M. (2009). Differentially conserved staphylococcal SH3b_5 cell wall binding domains confer increased staphylolytic and streptolytic activity to a streptococcal prophage endolysin domain. Gene 443, 32–41. doi: 10.1016/j.gene.2009.04.023
Beeby, M. (2015). Motility in the epsilon-proteobacteria. Curr. Opin. Microbiol. 28, 115–121. doi: 10.1016/j.mib.2015.09.005
Béguin, P., and Lemaire, M. (1996). The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit. Rev. Biochem. Mol. Biol. 31, 201–236. doi: 10.3109/10409239609106584
Belas, R. (2014). Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol. 22, 517–527. doi: 10.1016/j.tim.2014.05.002
Bellanger, X., Payot, S., Leblond-Bourget, N., and Guedon, G. (2014). Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol. Rev. 38, 720–760. doi: 10.1111/1574-6976.12058
Bergmann, S., Wild, D., Diekmann, O., Frank, R., Bracht, D., Chhatwal, G. S., et al. (2003). Identification of a novel plasmin(ogen)-binding motif in surface displayed alpha-enolase of Streptococcus pneumoniae. Mol. Microbiol. 49, 411–423. doi: 10.1046/j.1365-2958.2003.03557.x
Berry, J. L., and Pelicic, V. (2015). Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives. FEMS Microbiol. Rev. 39, 134–154. doi: 10.1093/femsre/fuu001
Besingi, R. N., Wenderska, I. B., Senadheera, D. B., Cvitkovitch, D. G., Long, J. R., Wen, Z. T., et al. (2017). Functional amyloids in Streptococcus mutans, their use as targets of biofilm inhibition and initial characterization of SMU_63c. Microbiology 163, 488–501. doi: 10.1099/mic.0.000443
Beveridge, T. J. (2001). Use of the gram stain in microbiology. Biotech. Histochem. 76, 111–118. doi: 10.1080/bih.76.3.111.118
Beveridge, T. J., and Murray, R. G. (1980). Sites of metal deposition in the cell wall of Bacillus subtilis. J. Bacteriol. 141, 876–887.
Bhatty, M., Laverde Gomez, J. A., and Christie, P. J. (2013). The expanding bacterial type IV secretion lexicon. Res. Microbiol. 164, 620–639. doi: 10.1016/j.resmic.2013.03.012
Bierne, H., and Dramsi, S. (2012). Spatial positioning of cell wall-anchored virulence factors in Gram-positive bacteria. Curr. Opin. Microbiol. 15, 715–723. doi: 10.1016/j.mib.2012.10.010
Biswas, R., Voggu, L., Simon, U. K., Hentschel, P., Thumm, G., and Gotz, F. (2006). Activity of the major staphylococcal autolysin Atl. FEMS Microbiol. Lett. 259, 260–268. doi: 10.1111/j.1574-6968.2006.00281.x
Bleymuller, W. M., Lammermann, N., Ebbes, M., Maynard, D., Geerds, C., and Niemann, H. H. (2016). MET-activating residues in the B-repeat of the Listeria monocytogenes invasion protein InlB. J. Biol. Chem. 291, 25567–25577. doi: 10.1074/jbc.M116.746685
Bodman, H., and Welker, N. E. (1969). Isolation of spheroplast membranes and stability of spheroplasts of Bacillus stearothermophilus. J. Bacteriol. 97, 924–935.
Boekhorst, J., De Been, M. W., Kleerebezem, M., and Siezen, R. J. (2005). Genome-wide detection and analysis of cell wall-bound proteins with LPXTG-like sorting motifs. J. Bacteriol. 187, 4928–4934. doi: 10.1128/JB.187.14.4928-4934.2005
Boot, H. J., Kolen, C. P., and Pouwels, P. H. (1995). Identification, cloning, and nucleotide sequence of a silent S-layer protein gene of Lactobacillus acidophilus ATCC 4356 which has extensive similarity with the S-layer protein gene of this species. J. Bacteriol. 177, 7222–7230. doi: 10.1128/jb.177.24.7222-7230.1995
Bramkamp, M., and Lopez, D. (2015). Exploring the existence of lipid rafts in bacteria. Microbiol. Mol. Biol. Rev. 79, 81–100. doi: 10.1128/MMBR.00036-14
Bras, J. L., Pinheiro, B. A., Cameron, K., Cuskin, F., Viegas, A., Najmudin, S., et al. (2016). Diverse specificity of cellulosome attachment to the bacterial cell surface. Sci. Rep. 6:38292. doi: 10.1038/srep38292
Braun, L., Dramsi, S., Dehoux, P., Bierne, H., Lindahl, G., and Cossart, P. (1997). InlB: an invasion protein of Listeria monocytogenes with a novel type of surface association. Mol. Microbiol. 25, 285–294. doi: 10.1046/j.1365-2958.1997.4621825.x
Brinster, S., Furlan, S., and Serror, P. (2007). C-terminal WXL domain mediates cell wall binding in Enterococcus faecalis and other Gram-positive bacteria. J. Bacteriol. 189, 1244–1253. doi: 10.1128/JB.00773-06
Brown, S., Santa Maria, J. P. Jr., and Walker, S. (2013). Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 67, 313–336. doi: 10.1146/annurev-micro-092412-155620
Budzik, J. M., Marraffini, L. A., and Schneewind, O. (2007). Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol. Microbiol. 66, 495–510. doi: 10.1111/j.1365-2958.2007.05939.x
Budzik, J. M., Marraffini, L. A., Souda, P., Whitelegge, J. P., Faull, K. F., and Schneewind, O. (2008). Amide bonds assemble pili on the surface of bacilli. Proc. Natl. Acad. Sci. U.S.A. 105, 10215–10220. doi: 10.1073/pnas.0803565105
Budzik, J. M., Poor, C. B., Faull, K. F., Whitelegge, J. P., He, C., and Schneewind, O. (2009). Intramolecular amide bonds stabilize pili on the surface of bacilli. Proc. Natl. Acad. Sci. U.S.A. 106, 19992–19997. doi: 10.1073/pnas.0910887106
Buist, G., Steen, A., Kok, J., and Kuipers, O. P. (2008). LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 68, 838–847. doi: 10.1111/j.1365-2958.2008.06211.x
Bule, P., Alves, V. D., Israeli-Ruimy, V., Carvalho, A. L., Ferreira, L. M., Smith, S. P., et al. (2017). Assembly of Ruminococcus flavefaciens cellulosome revealed by structures of two cohesin-dockerin complexes. Sci. Rep. 7:759. doi: 10.1038/s41598-017-00919-w
Bustamante, N., Campillo, N. E., Garcia, E., Gallego, C., Pera, B., Diakun, G. P., et al. (2010). Cpl-7, a lysozyme encoded by a pneumococcal bacteriophage with a novel cell wall-binding motif. J. Biol. Chem. 285, 33184–33196. doi: 10.1074/jbc.M110.154559
Bustamante, N., Rico-Lastres, P., Garcia, E., Garcia, P., and Menendez, M. (2012). Thermal stability of Cpl-7 endolysin from the Streptococcus pneumoniae bacteriophage Cp-7; cell wall-targeting of its CW_7 motifs. PLOS ONE 7:e46654. doi: 10.1371/journal.pone.0046654
Caliot, E., Dramsi, S., Chapot-Chartier, M. P., Courtin, P., Kulakauskas, S., Pechoux, C., et al. (2012). Role of the group B antigen of Streptococcus agalactiae: a peptidoglycan-anchored polysaccharide involved in cell wall biogenesis. PLOS Pathog. 8:e1002756. doi: 10.1371/journal.ppat.1002756
Campo, N., Tjalsma, H., Buist, G., Stepniak, D., Meijer, M., Veenhuis, M., et al. (2004). Subcellular sites for bacterial protein export. Mol. Microbiol. 53, 1583–1599. doi: 10.1111/j.1365-2958.2004.04278.x
Candela, T., Balomenou, S., Aucher, W., Bouriotis, V., Simore, J. P., Fouet, A., et al. (2014). N-acetylglucosamine deacetylases modulate the anchoring of the gamma-glutamyl capsule to the cell wall of Bacillus anthracis. Microb. Drug Resist. 20, 222–230. doi: 10.1089/mdr.2014.0063
Candela, T., and Fouet, A. (2005). Bacillus anthracis CapD, belonging to the gamma-glutamyltranspeptidase family, is required for the covalent anchoring of capsule to peptidoglycan. Mol. Microbiol. 57, 717–726. doi: 10.1111/j.1365-2958.2005.04718.x
Candela, T., and Fouet, A. (2006). Poly-gamma-glutamate in bacteria. Mol. Microbiol. 60, 1091–1098. doi: 10.1111/j.1365-2958.2006.05179.x
Candela, T., Mock, M., and Fouet, A. (2005). CapE, a 47-amino-acid peptide, is necessary for Bacillus anthracis polyglutamate capsule synthesis. J. Bacteriol. 187, 7765–7772. doi: 10.1128/JB.187.22.7765-7772.2005
Carvalho, A. L., Dias, F. M. V., Prates, J. A. M., Nagy, T., Gilbert, H. J., Davies, G. J., et al. (2003). Cellulosome assembly revealed by the crystal structure of the cohesin-dockerin complex. Proc. Natl. Acad. Sci. U.S.A. 100, 13809–13814. doi: 10.1073/pnas.1936124100
Carvalho, F., Atilano, M. L., Pombinho, R., Covas, G., Gallo, R. L., Filipe, S. R., et al. (2015). L-Rhamnosylation of Listeria monocytogenes wall teichoic acids promotes resistance to antimicrobial peptides by delaying interaction with the membrane. PLOS Pathog. 11:e1004919. doi: 10.1371/journal.ppat.1004919
Chaban, B., Hughes, H. V., and Beeby, M. (2015). The flagellum in bacterial pathogens: for motility and a whole lot more. Semin. Cell Dev. Biol. 46, 91–103. doi: 10.1016/j.semcdb.2015.10.032
Chagnot, C., Listrat, A., Astruc, T., and Desvaux, M. (2012). Bacterial adhesion to animal tissues: protein determinants for recognition of extracellular matrix components. Cell. Microbiol. 14, 1687–1696. doi: 10.1111/cmi.12002
Chagnot, C., Zorgani, M. A., Astruc, T., and Desvaux, M. (2013). Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective. Front. Microbiol. 4:303. doi: 10.3389/fmicb.2013.00303
Chai, L., Romero, D., Kayatekin, C., Akabayov, B., Vlamakis, H., Losick, R., et al. (2013). Isolation, characterization, and aggregation of a structured bacterial matrix precursor. J. Biol. Chem. 288, 17559–17568. doi: 10.1074/jbc.M113.453605
Chaillou, S., Champomier-Verges, M. C., Cornet, M., Crutz-Le Coq, A. M., Dudez, A. M., Martin, V., et al. (2005). The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 23, 1527–1533. doi: 10.1038/nbt1160
Chami, M., Bayan, N., Peyret, J. L., Gulik-Krzywicki, T., Leblon, G., and Shechter, E. (1997). The S-layer protein of Corynebacterium glutamicum is anchored to the cell wall by its C-terminal hydrophobic domain. Mol. Microbiol. 23, 483–492. doi: 10.1046/j.1365-2958.1997.d01-1868.x
Chapot-Chartier, M. P., and Kulakauskas, S. (2014). Cell wall structure and function in lactic acid bacteria. Microb. Cell Fact. 13(Suppl. 1):S9. doi: 10.1186/1475-2859-13-S1-S9
Chauvaux, S., Matuschek, M., and Beguin, P. (1999). Distinct affinity of binding sites for S-layer homologous domains in Clostridium thermocellum and Bacillus anthracis cell envelopes. J. Bacteriol. 181, 2455–2458.
Chen, I., and Dubnau, D. (2004). DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2, 241–249. doi: 10.1038/nrmicro844
Chen, I., Provvedi, R., and Dubnau, D. (2006). A macromolecular complex formed by a pilin-like protein in competent Bacillus subtilis. J. Biol. Chem. 281, 21720–21727. doi: 10.1074/jbc.M604071200
Chessa, D., Dorsey, C. W., Winter, M., and Baumler, A. J. (2008). Binding specificity of Salmonella plasmid-encoded fimbriae assessed by glycomics. J. Biol. Chem. 283, 8118–8124. doi: 10.1074/jbc.M710095200
Christie, J., Mcnab, R., and Jenkinson, H. F. (2002). Expression of fibronectin-binding protein FbpA modulates adhesion in Streptococcus gordonii. Microbiology 148, 1615–1625. doi: 10.1099/00221287-148-6-1615
Cortes-Perez, N. G., Dumoulin, R., Gaubert, S., Lacoux, C., Bugli, F., Martin, R., et al. (2015). Overexpression of Enterococcus faecalis elr operon protects from phagocytosis. BMC Microbiol. 15:112. doi: 10.1186/s12866-015-0448-y
Courtney, H. S., Li, Y., Dale, J. B., and Hasty, D. L. (1994). Cloning, sequencing, and expression of a fibronectin/fibrinogen-binding protein from group A streptococci. Infect. Immun. 62, 3937–3946.
Cozzi, R., Malito, E., Lazzarin, M., Nuccitelli, A., Castagnetti, A., Bottomley, M. J., et al. (2015). Structure and assembly of group B streptococcus pilus 2b backbone protein. PLOS ONE 10:e0125875. doi: 10.1371/journal.pone.0125875
Craig, L., Pique, M. E., and Tainer, J. A. (2004). Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378. doi: 10.1038/nrmicro885
Craig, L., Taylor, R. K., Pique, M. E., Adair, B. D., Arvai, A. S., Singh, M., et al. (2003). Type IV pilin structure and assembly: x-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol. Cell 11, 1139–1150. doi: 10.1016/S1097-2765(03)00170-9
Cucarella, C., Solano, C., Valle, J., Amorena, B., Lasa, I., and Penades, J. R. (2001). Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183, 2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001
Custer, J. E., Goddard, B. D., Matter, S. F., and Kaneshiro, E. S. (2014). The relative proportions of different lipid classes and their fatty acid compositions change with culture age in the cariogenic dental pathogen Streptococcus mutans UA159. Lipids 49, 543–554. doi: 10.1007/s11745-014-3904-9
Danne, C., Entenza, J. M., Mallet, A., Briandet, R., Debarbouille, M., Nato, F., et al. (2011). Molecular characterization of a Streptococcus gallolyticus genomic island encoding a pilus involved in endocarditis. J. Infect. Dis. 204, 1960–1970. doi: 10.1093/infdis/jir666
Demchick, P., and Koch, A. L. (1996). The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J. Bacteriol. 178, 768–773. doi: 10.1128/jb.178.3.768-773.1996
Dempwolff, F., Schmidt, F. K., Hervas, A. B., Stroh, A., Rosch, T. C., Riese, C. N., et al. (2016). Super resolution fluorescence microscopy and tracking of bacterial flotillin (Reggie) paralogs provide evidence for defined-sized protein microdomains within the bacterial membrane but absence of clusters containing detergent-resistant proteins. PLOS Genet. 12:e1006116. doi: 10.1371/journal.pgen.1006116
Deng, L., Kasper, D. L., Krick, T. P., and Wessels, M. R. (2000). Characterization of the linkage between the type III capsular polysaccharide and the bacterial cell wall of group B Streptococcus. J. Biol. Chem. 275, 7497–7504. doi: 10.1074/jbc.275.11.7497
Desvaux, M. (2005a). The cellulosome of Clostridium cellulolyticum. Enzyme Microb. Technol. 37, 373–385. doi: 10.1016/j.enzmictec.2004.04.025
Desvaux, M. (2005b). Clostridium cellulolyticum: model organism of mesophilic cellulolytic clostridia. FEMS Microbiol. Rev. 29, 741–764.
Desvaux, M. (2012). Contribution of holins to protein trafficking: secretion, leakage or lysis? Trends Microbiol. 20, 259–261. doi: 10.1016/j.tim.2012.03.008
Desvaux, M., Dumas, E., Chafsey, I., Chambon, C., and Hébraud, M. (2010). Comprehensive appraisal of the extracellular proteins from a monoderm bacterium: theoretical and empirical exoproteomes of Listeria monocytogenes EGD-e by secretomics. J. Proteome Res. 9, 5076–5092. doi: 10.1021/pr1003642
Desvaux, M., and Hébraud, M. (2006). The protein secretion systems in Listeria: inside out bacterial virulence. FEMS Microbiol. Rev. 30, 774–805. doi: 10.1111/j.1574-6976.2006.00035.x
Desvaux, M., and Hébraud, M. (2009). “Chapter 14: Listeria monocytogenes,” in Bacterial Secreted Proteins: Secretory Mechanisms and Role in Pathogenesis, ed. K. Wooldridge (Norwich: Caister Academic Press), 313–345.
Desvaux, M., Hébraud, M., Talon, R., and Henderson, I. R. (2009). Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol. 17, 139–145. doi: 10.1016/j.tim.2009.01.004
Desvaux, M., Khan, A., Scott-Tucker, A., Chaudhuri, R. R., Pallen, M. J., and Henderson, I. R. (2005). Genomic analysis of the protein secretion systems in Clostridium acetobutylicum ATCC824. Biochim. Biophys. Acta 1745, 223–253. doi: 10.1016/j.bbamcr.2005.04.006
Desvaux, M., Parham, N. J., and Henderson, I. R. (2004). The autotransporter secretion system. Res. Microbiol. 155, 53–60. doi: 10.1016/j.resmic.2003.10.002
Desvaux, M., and Petitdemange, H. (2001). Flux analysis of the metabolism of Clostridium cellulolyticum grown in cellulose-fed continuous culture on a chemically defined medium under ammonium-limited conditions. Appl. Environ. Microbiol. 67, 3846–3851. doi: 10.1128/AEM.67.9.3846-3851.2001
Diomande, S. E., Nguyen-The, C., Guinebretiere, M. H., Broussolle, V., and Brillard, J. (2015). Role of fatty acids in Bacillus environmental adaptation. Front. Microbiol. 6:813. doi: 10.3389/fmicb.2015.00813
Doyle, R. J., and Marquis, R. E. (1994). Elastic, flexible peptidoglycan and bacterial cell wall properties. Trends Microbiol. 2, 57–60. doi: 10.1016/0966-842X(94)90127-9
Dramsi, S., and Bierne, H. (2017). Spatial organization of cell wall-anchored proteins at the surface of gram-positive bacteria. Curr. Top. Microbiol. Immunol. 404, 177–201. doi: 10.1007/82_2016_4
Dramsi, S., Bourdichon, F., Cabanes, D., Lecuit, M., Fsihi, H., and Cossart, P. (2004). FbpA, a novel multifunctional Listeria monocytogenes virulence factor. Mol. Microbiol. 53, 639–649. doi: 10.1111/j.1365-2958.2004.04138.x
Dramsi, S., Caliot, E., Bonne, I., Guadagnini, S., Prevost, M. C., Kojadinovic, M., et al. (2006). Assembly and role of pili in group B streptococci. Mol. Microbiol. 60, 1401–1413. doi: 10.1111/j.1365-2958.2006.05190.x
Draper, P. (1998). The outer parts of the mycobacterial envelope as permeability barriers. Front. Biosci. 3, D1253–D1261. doi: 10.2741/A360
Dubnau, D. (1999). DNA uptake in bacteria. Annu. Rev. Microbiol. 53, 217–244. doi: 10.1146/annurev.micro.53.1.217
Dubnau, D., and Provvedi, R. (2000). Internalizing DNA. Res. Microbiol. 151, 475–480. doi: 10.1016/S0923-2508(00)00166-2
Duda, V. I., Suzina, N. E., Severina, L. O., Bogdanova, T. I., Tsaplina, I. A., and Karavaiko, G. I. (2006). Ultrastructural organization of Alicyclobacillus tolerans strain K1T cells. Arch. Microbiol. 185, 63–68. doi: 10.1007/s00203-005-0070-8
Dumas, E., Meunier, B., Berdagué, J. L., Chambon, C., Desvaux, M., and Hébraud, M. (2008). Comparative analysis of extracellular and intracellular proteomes of Listeria monocytogenes strains reveals a correlation between protein expression and serovar. Appl. Environ. Microbiol. 74, 7399–7409. doi: 10.1128/AEM.00594-08
Ebisu, S., Tsuboi, A., Takagi, H., Naruse, Y., Yamagata, H., Tsukagoshi, N., et al. (1990). Conserved structures of cell wall protein genes among protein-producing Bacillus brevis strains. J. Bacteriol. 172, 1312–1320. doi: 10.1128/jb.172.3.1312-1320.1990
Ebner, P., Rinker, J., and Gotz, F. (2016). Excretion of cytoplasmic proteins in Staphylococcus is most likely not due to cell lysis. Curr. Genet. 62, 19–23. doi: 10.1007/s00294-015-0504-z
Echelman, D. J., Alegre-Cebollada, J., Badilla, C. L., Chang, C., Ton-That, H., and Fernandez, J. M. (2016). CnaA domains in bacterial pili are efficient dissipaters of large mechanical shocks. Proc. Natl. Acad. Sci. U.S.A. 113, 2490–2495. doi: 10.1073/pnas.1522946113
Ehinger, S., Schubert, W. D., Bergmann, S., Hammerschmidt, S., and Heinz, D. W. (2004). Plasmin(ogen)-binding alpha-enolase from Streptococcus pneumoniae: crystal structure and evaluation of plasmin(ogen)-binding sites. J. Mol. Biol. 343, 997–1005. doi: 10.1016/j.jmb.2004.08.088
Errington, J., Appleby, L., Daniel, R. A., Goodfellow, H., Partridge, S. R., and Yudkin, M. D. (1992). Structure and function of the spoIIIJ gene of Bacillus subtilis: a vegetatively expressed gene that is essential for sigma G activity at an intermediate stage of sporulation. J. Gen. Microbiol. 138, 2609–2618. doi: 10.1099/00221287-138-12-2609
Evans, L. D., Hughes, C., and Fraser, G. M. (2014). Building a flagellum outside the bacterial cell. Trends Microbiol. 22, 566–572. doi: 10.1016/j.tim.2014.05.009
Facey, S. J, and Kuhn, A. (2004). Membrane integration of E. coli model membrane proteins. Biochim. Biophys. Acta 1694, 55–66. doi: 10.1016/j.bbamcr.2004.03.012
Fagan, R. P., and Fairweather, N. F. (2014). Biogenesis and functions of bacterial S-layers. Nat. Rev. Microbiol. 12, 211–222. doi: 10.1038/nrmicro3213
Fagan, R. P., Janoir, C., Collignon, A., Mastrantonio, P., Poxton, I. R., and Fairweather, N. F. (2011). A proposed nomenclature for cell wall proteins of Clostridium difficile. J. Med. Microbiol. 60, 1225–1228. doi: 10.1099/jmm.0.028472-0
Fekkes, P., and Driessen, A. J. (1999). Protein targeting to the bacterial cytoplasmic membrane. Microbiol. Mol. Biol. Rev. 63, 161–173.
Fernandez-Tornero, C., Lopez, R., Garcia, E., Gimenez-Gallego, G., and Romero, A. (2001). A novel solenoid fold in the cell wall anchoring domain of the pneumococcal virulence factor LytA. Nat. Struct. Biol. 8, 1020–1024. doi: 10.1038/nsb724
Ferretti, J. J., Gilpin, M. L., and Russell, R. R. (1987). Nucleotide sequence of a glucosyltransferase gene from Streptococcus sobrinus MFe28. J. Bacteriol. 169, 4271–4278. doi: 10.1128/jb.169.9.4271-4278.1987
Finn, R. D., Attwood, T. K., Babbitt, P. C., Bateman, A., Bork, P., Bridge, A. J., et al. (2017). InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 45, D190–D199. doi: 10.1093/nar/gkw1107
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Fischer, W., Nakano, M., Laine, R. A., and Bohrer, W. (1978). On the relationship between glycerophosphoglycolipids and lipoteichoic acids in b Gram-positive bacteria. I. The occurrence of phosphoglycolipids. Biochim. Biophys. Acta 528, 288–297. doi: 10.1016/0005-2760(78)90018-8
Fischetti, V. A., Pancholi, V., and Schneewind, O. (1990). Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol. Microbiol. 4, 1603–1605. doi: 10.1111/j.1365-2958.1990.tb02072.x
Fontes, C. M., and Gilbert, H. J. (2010). Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu. Rev. Biochem. 79, 655–681. doi: 10.1146/annurev-biochem-091208-085603
Forsberg, L. S., Abshire, T. G., Friedlander, A., Quinn, C. P., Kannenberg, E. L., and Carlson, R. W. (2012). Localization and structural analysis of a conserved pyruvylated epitope in Bacillus anthracis secondary cell wall polysaccharides and characterization of the galactose-deficient wall polysaccharide from avirulent B. anthracis CDC 684. Glycobiology 22, 1103–1117. doi: 10.1093/glycob/cws080
Forster, B. M., and Marquis, H. (2012). Protein transport across the cell wall of monoderm Gram-positive bacteria. Mol. Microbiol. 84, 405–413. doi: 10.1111/j.1365-2958.2012.08040.x
Foster, S. J. (1991). Cloning, expression, sequence analysis and biochemical characterization of an autolytic amidase of Bacillus subtilis 168 trpC2. J. Gen. Microbiol. 137, 1987–1998. doi: 10.1099/00221287-137-8-1987
Frankel, M. B., and Schneewind, O. (2012). Determinants of murein hydrolase targeting to cross-wall of Staphylococcus aureus peptidoglycan. J. Biol. Chem. 287, 10460–10471. doi: 10.1074/jbc.M111.336404
Fröderberg, L., Houben, E., Samuelson, J. C., Chen, M., Park, S. K., Phillips, G. J., et al. (2003). Versatility of inner membrane protein biogenesis in Escherichia coli. Mol. Microbiol. 47, 1015–1027. doi: 10.1046/j.1365-2958.2003.03346.x
Fujino, T., Beguin, P., and Aubert, J. P. (1993). Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in attachment of the cellulosome to the cell surface. J. Bacteriol. 175, 1891–1899. doi: 10.1128/jb.175.7.1891-1899.1993
Fulde, M., Steinert, M., and Bergmann, S. (2013). Interaction of streptococcal plasminogen binding proteins with the host fibrinolytic system. Front. Cell. Infect. Microbiol. 3:85. doi: 10.3389/fcimb.2013.00085
Funes, S., Hasona, A., Bauerschmitt, H., Grubbauer, C., Kauff, F., Collins, R., et al. (2009). Independent gene duplications of the YidC/Oxa/Alb3 family enabled a specialized cotranslational function. Proc. Natl. Acad. Sci. U.S.A. 106, 6656–6661. doi: 10.1073/pnas.0809951106
Galloway-Pena, J. R., Liang, X., Singh, K. V., Yadav, P., Chang, C., La Rosa, S. L., et al. (2015). The identification and functional characterization of WxL proteins from Enterococcus faecium reveal surface proteins involved in extracellular matrix interactions. J. Bacteriol. 197, 882–892. doi: 10.1128/JB.02288-14
Garcia, J. L., Sanchez-Beato, A. R., Medrano, F. J., and Lopez, R. (1998). Versatility of choline-binding domain. Microb. Drug Resist. 4, 25–36. doi: 10.1089/mdr.1998.4.25
Garvey, K. J., Saedi, M. S., and Ito, J. (1986). Nucleotide sequence of Bacillus phage ϕ 29 genes 14 and 15: homology of gene 15 with other phage lysozymes. Nucleic Acids Res. 14, 10001–10008. doi: 10.1093/nar/14.24.10001
Gerbino, E., Carasi, P., Mobili, P., Serradell, M. A., and Gomez-Zavaglia, A. (2015). Role of S-layer proteins in bacteria. World J. Microbiol. Biotechnol. 31, 1877–1887. doi: 10.1007/s11274-015-1952-9
Ghosh, B. K., and Carroll, K. K. (1968). Isolation, composition, and structure of membrane of Listeria monocytogenes. J. Bacteriol. 95, 688–699.
Giffard, P. M., and Jacques, N. A. (1994). Definition of a fundamental repeating unit in streptococcal glucosyltransferase glucan-binding regions and related sequences. J. Dent. Res. 73, 1133–1141. doi: 10.1177/00220345940730060201
Giltner, C. L., Nguyen, Y., and Burrows, L. L. (2012). Type IV pilin proteins: versatile molecular modules. Microbiol. Mol. Biol. Rev. 76, 740–772. doi: 10.1128/MMBR.00035-12
Goder, V., and Spiess, M. (2001). Topogenesis of membrane proteins: determinants and dynamics. FEBS Lett. 504, 87–93. doi: 10.1016/S0014-5793(01)02712-0
Goessweiner-Mohr, N., Arends, K., Keller, W., and Grohmann, E. (2013). Conjugative Type IV secretion systems in Gram-positive bacteria. Plasmid 70, 289–302. doi: 10.1016/j.plasmid.2013.09.005
Gram, H. C. J. (1884). Über die isolirte färbung der Schizomyceten in schnitt- und trockenpräparaten. Fortschr. Med. 2, 185–189.
Grundling, A., and Schneewind, O. (2006). Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. J. Bacteriol. 188, 2463–2472. doi: 10.1128/JB.188.7.2463-2472.2006
Guedon, E., Payot, S., Desvaux, M., and Petitdemange, H. (2000). Relationships between cellobiose catabolism, enzyme levels, and metabolic intermediates in Clostridium cellulolyticum grown in a synthetic medium. Biotechnol. Bioeng. 67, 327–335. doi: 10.1002/(SICI)1097-0290(20000205)67:3<327::AID-BIT9>3.0.CO;2-U
Guglielmini, J., De La Cruz, F., and Rocha, E. P. (2013). Evolution of conjugation and type IV secretion systems. Mol. Biol. Evol. 30, 315–331. doi: 10.1093/molbev/mss221
Guglielmini, J., Neron, B., Abby, S. S., Garcillan-Barcia, M. P., De La Cruz, F., and Rocha, E. P. (2014). Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 42, 5715–5727. doi: 10.1093/nar/gku194
Gupta, R. S. (1998a). Life’s third domain (Archaea): an established fact or an endangered paradigm? Theor. Popul. Biol. 54, 91–104.
Gupta, R. S. (1998b). Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. 62, 1435–1491.
Gupta, R. S. (2000). The natural evolutionary relationships among prokaryotes. Crit. Rev. Microbiol. 26, 111–131. doi: 10.1080/10408410091154219
Gupta, R. S. (2011). Origin of diderm (Gram-negative) bacteria: antibiotic selection pressure rather than endosymbiosis likely led to the evolution of bacterial cells with two membranes. Antonie Van Leeuwenhoek 100, 171–182. doi: 10.1007/s10482-011-9616-8
Gurung, I., Berry, J. L., Hall, A. M. J., and Pelicic, V. (2017). Cloning-independent markerless gene editing in Streptococcus sanguinis: novel insights in type IV pilus biology. Nucleic Acids Res. 45:e40. doi: 10.1093/nar/gkw1177
Gurung, I., Spielman, I., Davies, M. R., Lala, R., Gaustad, P., Biais, N., et al. (2016). Functional analysis of an unusual type IV pilus in the Gram-positive Streptococcus sanguinis. Mol. Microbiol. 99, 380–392. doi: 10.1111/mmi.13237
Hamberg, Y., Ruimy-Israeli, V., Dassa, B., Barak, Y., Lamed, R., Cameron, K., et al. (2014). Elaborate cellulosome architecture of Acetivibrio cellulolyticus revealed by selective screening of cohesin-dockerin interactions. PeerJ 2:e636. doi: 10.7717/peerj.636
Hayhurst, E. J., Kailas, L., Hobbs, J. K., and Foster, S. J. (2008). Cell wall peptidoglycan architecture in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 105, 14603–14608. doi: 10.1073/pnas.0804138105
Heim, K. P., Crowley, P. J., Long, J. R., Kailasan, S., Mckenna, R., and Brady, L. J. (2014). An intramolecular lock facilitates folding and stabilizes the tertiary structure of Streptococcus mutans adhesin P1. Proc. Natl. Acad. Sci. U.S.A. 111, 15746–15751. doi: 10.1073/pnas.1413018111
Hendrickx, A. P., Budzik, J. M., Oh, S. Y., and Schneewind, O. (2011). Architects at the bacterial surface - sortases and the assembly of pili with isopeptide bonds. Nat. Rev. Microbiol. 9, 166–176. doi: 10.1038/nrmicro2520
Hennequin, C., Janoir, C., Barc, M. C., Collignon, A., and Karjalainen, T. (2003). Identification and characterization of a fibronectin-binding protein from Clostridium difficile. Microbiology 149, 2779–2787. doi: 10.1099/mic.0.26145-0
Henrichsen, J. (1972). Bacterial surface translocation: a survey and a classification. Bacteriol. Rev. 36, 478–503.
Higy, M., Junne, T., and Spiess, M. (2004). Topogenesis of membrane proteins at the endoplasmic reticulum. Biochemistry 43, 12716–12722. doi: 10.1021/bi048368m
Hilleringmann, M., Giusti, F., Baudner, B. C., Masignani, V., Covacci, A., Rappuoli, R., et al. (2008). Pneumococcal pili are composed of protofilaments exposing adhesive clusters of Rrg A. PLOS Pathog. 4:e1000026. doi: 10.1371/journal.ppat.1000026
Holmes, A. R., Mcnab, R., Millsap, K. W., Rohde, M., Hammerschmidt, S., Mawdsley, J. L., et al. (2001). The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence. Mol. Microbiol. 41, 1395–1408. doi: 10.1046/j.1365-2958.2001.02610.x
Holtje, J. V., and Tomasz, A. (1975). Specific recognition of choline residues in the cell wall teichoic acid by the N-acetylmuramyl-L-alanine amidase of Pneumococcus. J. Biol. Chem. 250, 6072–6076.
Huber, C., Ilk, N., Runzler, D., Egelseer, E. M., Weigert, S., Sleytr, U. B., et al. (2005). The three S-layer-like homology motifs of the S-layer protein SbpA of Bacillus sphaericus CCM 2177 are not sufficient for binding to the pyruvylated secondary cell wall polymer. Mol. Microbiol. 55, 197–205. doi: 10.1111/j.1365-2958.2004.04351.x
Hulo, N., Bairoch, A., Bulliard, V., Cerutti, L., De Castro, E., Langendijk-Genevaux, P. S., et al. (2006). The PROSITE database. Nucleic Acids Res. 34, D227–D230. doi: 10.1093/nar/gkj063
Hutchings, M. I., Palmer, T., Harrington, D. J., and Sutcliffe, I. C. (2009). Lipoprotein biogenesis in Gram-positive bacteria: knowing when to hold ’em, knowing when to fold ’em. Trends Microbiol. 17, 13–21. doi: 10.1016/j.tim.2008.10.001
Imam, S., Chen, Z., Roos, D. S., and Pohlschroder, M. (2011). Identification of surprisingly diverse type IV pili, across a broad range of gram-positive bacteria. PLOS ONE 6:e28919. doi: 10.1371/journal.pone.0028919
Janesch, B., Messner, P., and Schaffer, C. (2013). Are the surface layer homology domains essential for cell surface display and glycosylation of the S-layer protein from Paenibacillus alvei CCM 2051T? J. Bacteriol. 195, 565–575. doi: 10.1128/JB.01487-12
Johnson, B. R., Hymes, J., Sanozky-Dawes, R., Henriksen, E. D., Barrangou, R., and Klaenhammer, T. R. (2015). Conserved S-layer-associated proteins revealed by exoproteomic survey of S-layer-forming lactobacilli. Appl. Environ. Microbiol. 82, 134–145. doi: 10.1128/AEM.01968-15
Johnston, C., Martin, B., Fichant, G., Polard, P., and Claverys, J. P. (2014). Bacterial transformation: distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12, 181–196. doi: 10.1038/nrmicro3199
Jolliffe, L. K., Doyle, R. J., and Streips, U. N. (1981). The energized membrane and cellular autolysis in Bacillus subtilis. Cell 25, 753–763. doi: 10.1016/0092-8674(81)90183-5
Jones, C. (2005). Revised structures for the capsular polysaccharides from Staphylococcus aureus Types 5 and 8, components of novel glycoconjugate vaccines. Carbohydr. Res. 340, 1097–1106. doi: 10.1016/j.carres.2005.02.001
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., Mcanulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Jonquieres, R., Bierne, H., Fiedler, F., Gounon, P., and Cossart, P. (1999). Interaction between the protein InlB of Listeria monocytogenes and lipoteichoic acid: a novel mechanism of protein association at the surface of Gram-positive bacteria. Mol. Microbiol. 34, 902–914. doi: 10.1046/j.1365-2958.1999.01652.x
Jonquieres, R., Pizarro-Cerda, J., and Cossart, P. (2001). Synergy between the N- and C-terminal domains of InlB for efficient invasion of non-phagocytic cells by Listeria monocytogenes. Mol. Microbiol. 42, 955–965. doi: 10.1046/j.1365-2958.2001.02704.x
Kainulainen, V., and Korhonen, T. K. (2014). Dancing to another tune-adhesive moonlighting proteins in bacteria. Biology 3, 178–204. doi: 10.3390/biology3010178
Kajikawa, A., Midorikawa, E., Masuda, K., Kondo, K., Irisawa, T., Igimi, S., et al. (2016). Characterization of flagellins isolated from a highly motile strain of Lactobacillus agilis. BMC Microbiol. 16:49. doi: 10.1186/s12866-016-0667-x
Kandaswamy, K., Liew, T. H., Wang, C. Y., Huston-Warren, E., Meyer-Hoffert, U., Hultenby, K., et al. (2013). Focal targeting by human beta-defensin 2 disrupts localized virulence factor assembly sites in Enterococcus faecalis. Proc. Natl. Acad. Sci. U.S.A. 110, 20230–20235. doi: 10.1073/pnas.1319066110
Kaneko, T., Li, L., and Li, S. S. (2008). The SH3 domain–a family of versatile peptide- and protein-recognition module. Front. Biosci. 13, 4938–4952. doi: 10.2741/3053
Kang, H. J., and Baker, E. N. (2012). Structure and assembly of Gram-positive bacterial pili: unique covalent polymers. Curr. Opin. Struct. Biol. 22, 200–207. doi: 10.1016/j.sbi.2012.01.009
Kang, H. J., Coulibaly, F., Clow, F., Proft, T., and Baker, E. N. (2007). Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science 318, 1625–1628. doi: 10.1126/science.1145806
Kang, H. J., Middleditch, M., Proft, T., and Baker, E. N. (2009). Isopeptide bonds in bacterial pili and their characterization by X-ray crystallography and mass spectrometry. Biopolymers 91, 1126–1134. doi: 10.1002/bip.21170
Kankainen, M., Paulin, L., Tynkkynen, S., Von Ossowski, I., Reunanen, J., Partanen, P., et al. (2009). Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc. Natl. Acad. Sci. U.S.A. 106, 17193–17198. doi: 10.1073/pnas.0908876106
Katayama, S., Nozu, N., Okuda, M., Hirota, S., Yamasaki, T., and Hitsumoto, Y. (2009). Characterization of two putative fibronectin-binding proteins of Clostridium perfringens. Anaerobe 15, 155–159. doi: 10.1016/j.anaerobe.2009.03.001
Kawabata, S., Kunitomo, E., Terao, Y., Nakagawa, I., Kikuchi, K., Totsuka, K., et al. (2001). Systemic and mucosal immunizations with fibronectin-binding protein FBP54 induce protective immune responses against Streptococcus pyogenes challenge in mice. Infect. Immun. 69, 924–930. doi: 10.1128/IAI.69.2.924-930.2001
Kern, J., Wilton, R., Zhang, R., Binkowski, T. A., Joachimiak, A., and Schneewind, O. (2011). Structure of surface layer homology (SLH) domains from Bacillus anthracis surface array protein. J. Biol. Chem. 286, 26042–26049. doi: 10.1074/jbc.M111.248070
Kim, S. J., Chang, J., and Singh, M. (2015). Peptidoglycan architecture of Gram-positive bacteria by solid-state NMR. Biochim. Biophys. Acta 1848, 350–362. doi: 10.1016/j.bbamem.2014.05.031
Kinoshita, H., Wakahara, N., Watanabe, M., Kawasaki, T., Matsuo, H., Kawai, Y., et al. (2008). Cell surface glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of Lactobacillus plantarum LA 318 recognizes human A and B blood group antigens. Res. Microbiol. 159, 685–691. doi: 10.1016/j.resmic.2008.07.005
Kleerebezem, M., Boekhorst, J., Van Kranenburg, R., Molenaar, D., Kuipers, O. P., Leer, R., et al. (2003). Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. U.S.A. 100, 1990–1995. doi: 10.1073/pnas.0337704100
Koc, C., Xia, G., Kuhner, P., Spinelli, S., Roussel, A., Cambillau, C., et al. (2016). Structure of the host-recognition device of Staphylococcus aureus phage varphi11. Sci. Rep. 6:27581. doi: 10.1038/srep27581
Koch, A. L., and Doyle, R. J. (1985). Inside-to-outside growth and turnover of the wall of gram-positive rods. J. Theor. Biol. 117, 137–157. doi: 10.1016/S0022-5193(85)80169-7
Kondakova, T., Merlet-Machour, N., Chapelle, M., Preterre, D., Dionnet, F., Feuilloley, M., et al. (2015). A new study of the bacterial lipidome: HPTLC-MALDI-TOF imaging enlightening the presence of phosphatidylcholine in airborne Pseudomonas fluorescens MFAF76a. Res. Microbiol. 166, 1–8. doi: 10.1016/j.resmic.2014.11.003
Konto-Ghiorghi, Y., Mairey, E., Mallet, A., Dumenil, G., Caliot, E., Trieu-Cuot, P., et al. (2009). Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLOS Pathog. 5:e1000422. doi: 10.1371/journal.ppat.1000422
Krishnan, V., Gaspar, A. H., Ye, N., Mandlik, A., Ton-That, H., and Narayana, S. V. (2007). An IgG-like domain in the minor pilin GBS52 of Streptococcus agalactiae mediates lung epithelial cell adhesion. Structure 15, 893–903. doi: 10.1016/j.str.2007.06.015
Kuen, B., Koch, A., Asenbauer, E., Sara, M., and Lubitz, W. (1997). Molecular characterization of the Bacillus stearothermophilus PV72 S-layer gene sbsB induced by oxidative stress. J. Bacteriol. 179, 1664–1670. doi: 10.1128/jb.179.5.1664-1670.1997
Kuroda, A., and Sekiguchi, J. (1991). Molecular cloning and sequencing of a major Bacillus subtilis autolysin gene. J. Bacteriol. 173, 7304–7312. doi: 10.1128/jb.173.22.7304-7312.1991
Kurokawa, K., Kim, M. S., Ichikawa, R., Ryu, K. H., Dohmae, N., Nakayama, H., et al. (2012a). Environment-mediated accumulation of diacyl lipoproteins over their triacyl counterparts in Staphylococcus aureus. J. Bacteriol. 194, 3299–3306. doi: 10.1128/JB.00314-12
Kurokawa, K., Lee, H., Roh, K. B., Asanuma, M., Kim, Y. S., Nakayama, H., et al. (2009). The triacylated ATP binding cluster transporter substrate-binding lipoprotein of Staphylococcus aureus functions as a native ligand for toll-like receptor 2. J. Biol. Chem. 284, 8406–8411. doi: 10.1074/jbc.M809618200
Kurokawa, K., Ryu, K. H., Ichikawa, R., Masuda, A., Kim, M. S., Lee, H., et al. (2012b). Novel bacterial lipoprotein structures conserved in low-GC content gram-positive bacteria are recognized by Toll-like receptor 2. J. Biol. Chem. 287, 13170–13181. doi: 10.1074/jbc.M111.292235
Lai, S. H., Philbrick, W. M., and Wu, H. C. (1980). Acyl moieties in phospholipids are the precursors for the fatty acids in murein lipoprotein of Escherichia coli. J. Biol. Chem. 255, 5384–5387.
Larson, T. R., and Yother, J. (2017). Streptococcus pneumoniae capsular polysaccharide is linked to peptidoglycan via a direct glycosidic bond to beta-D-N-acetylglucosamine. Proc. Natl. Acad. Sci. U.S.A. 114, 5695–5700. doi: 10.1073/pnas.1620431114
Laurenceau, R., Krasteva, P. V., Diallo, A., Ouarti, S., Duchateau, M., Malosse, C., et al. (2015). Conserved Streptococcus pneumoniae spirosomes suggest a single type of transformation pilus in competence. PLOS Pathog. 11:e1004835. doi: 10.1371/journal.ppat.1004835
Laurenceau, R., Pehau-Arnaudet, G., Baconnais, S., Gault, J., Malosse, C., Dujeancourt, A., et al. (2013). A type IV pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae. PLOS Pathog. 9:e1003473. doi: 10.1371/journal.ppat.1003473
Layec, S., Decaris, B., and Leblond-Bourget, N. (2008). Characterization of proteins belonging to the CHAP-related superfamily within the Firmicutes. J. Mol. Microbiol. Biotechnol. 14, 31–40. doi: 10.1159/000106080
Lazzarin, M., Cozzi, R., Malito, E., Martinelli, M., D’onofrio, M., Maione, D., et al. (2015). Noncanonical sortase-mediated assembly of pilus type 2b in group B Streptococcus. FASEB J. 29, 4629–4640. doi: 10.1096/fj.15-272500
Lederberg, J., and McCray, A. T. (2001). ‘Ome sweet’ omics-A genealogical treasury of words. Scientist 15:8.
Lembre, P., Vendrely, C., and Martino, P. D. (2014). Identification of an amyloidogenic peptide from the Bap protein of Staphylococcus epidermidis. Protein Pept. Lett. 21, 75–79. doi: 10.2174/09298665113209990072
LeMieux, J., Woody, S., and Camilli, A. (2008). Roles of the sortases of Streptococcus pneumoniae in assembly of the RlrA pilus. J. Bacteriol. 190, 6002–6013. doi: 10.1128/JB.00379-08
Letunic, I., Doerks, T., and Bork, P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Li, H., and Thanassi, D. G. (2009). Use of a combined cryo-EM and X-ray crystallography approach to reveal molecular details of bacterial pilus assembly by the chaperone/usher pathway. Curr. Opin. Microbiol. 12, 326–332. doi: 10.1016/j.mib.2009.03.002
Linke, C., Young, P. G., Kang, H. J., Bunker, R. D., Middleditch, M. J., Caradoc-Davies, T. T., et al. (2010). Crystal structure of the minor pilin FctB reveals determinants of Group A streptococcal pilus anchoring. J. Biol. Chem. 285, 20381–20389. doi: 10.1074/jbc.M109.089680
Linke-Winnebeck, C., Paterson, N. G., Young, P. G., Middleditch, M. J., Greenwood, D. R., Witte, G., et al. (2014). Structural model for covalent adhesion of the Streptococcus pyogenes pilus through a thioester bond. J. Biol. Chem. 289, 177–189. doi: 10.1074/jbc.M113.523761
Lopez, D., and Kolter, R. (2010). Functional microdomains in bacterial membranes. Genes Dev. 24, 1893–1902. doi: 10.1101/gad.1945010
Lu, Q., Li, S., and Shao, F. (2015). Sweet talk: protein glycosylation in bacterial interaction with the host. Trends Microbiol. 23, 630–641. doi: 10.1016/j.tim.2015.07.003
Lu, Y., Zhang, Y. H., and Lynd, L. R. (2006). Enzyme-microbe synergy during cellulose hydrolysis by Clostridium thermocellum. Proc. Natl. Acad. Sci. U.S.A. 103, 16165–16169. doi: 10.1073/pnas.0605381103
Lupas, A., Engelhardt, H., Peters, J., Santarius, U., Volker, S., and Baumeister, W. (1994). Domain structure of the Acetogenium kivui surface layer revealed by electron crystallography and sequence analysis. J. Bacteriol. 176, 1224–1233. doi: 10.1128/jb.176.5.1224-1233.1994
Macnab, R. M. (2003). How bacteria assemble flagella. Annu. Rev. Microbiol. 57, 77–100. doi: 10.1146/annurev.micro.57.030502.090832
Macnab, R. M. (2004). Type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta 1694, 207–217. doi: 10.1016/j.bbamcr.2004.04.005
Malanovic, N., and Lohner, K. (2016). Antimicrobial peptides targeting gram-positive bacteria. Pharmaceuticals 9:59. doi: 10.3390/ph9030059
Mandlik, A., Swierczynski, A., Das, A., and Ton-That, H. (2008). Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 16, 33–40. doi: 10.1016/j.tim.2007.10.010
Manetti, A. G., Zingaretti, C., Falugi, F., Capo, S., Bombaci, M., Bagnoli, F., et al. (2007). Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol. Microbiol. 64, 968–983. doi: 10.1111/j.1365-2958.2007.05704.x
Marchandin, H., Teyssier, C., Campos, J., Jean-Pierre, H., Roger, F., Gay, B., et al. (2010). Negativicoccus succinicivorans gen. nov., sp. nov., isolated from human clinical samples, emended description of the family Veillonellaceae and description of Negativicutes classis nov., Selenomonadales ord. nov. and Acidaminococcaceae fam. nov. in the bacterial phylum Firmicutes. Int. J. Syst. Evol. Microbiol. 60, 1271–1279. doi: 10.1099/ijs.0.013102-0
Marinelli, P., Pallares, I., Navarro, S., and Ventura, S. (2016). Dissecting the contribution of Staphylococcus aureus alpha-phenol-soluble modulins to biofilm amyloid structure. Sci. Rep. 6:34552. doi: 10.1038/srep34552
Marino, M., Banerjee, M., Jonquieres, R., Cossart, P., and Ghosh, P. (2002). GW domains of the Listeria monocytogenes invasion protein InlB are SH3-like and mediate binding to host ligands. EMBO J. 21, 5623–5634. doi: 10.1093/emboj/cdf558
Marraffini, L. A., and Schneewind, O. (2005). Anchor structure of staphylococcal surface proteins. V. Anchor structure of the sortase B substrate IsdC. J. Biol. Chem. 280, 16263–16271. doi: 10.1074/jbc.M500071200
Matias, V. R., and Beveridge, T. J. (2005). Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol. Microbiol. 56, 240–251. doi: 10.1111/j.1365-2958.2005.04535.x
Matias, V. R., and Beveridge, T. J. (2006). Native cell wall organization shown by cryo-electron microscopy confirms the existence of a periplasmic space in Staphylococcus aureus. J. Bacteriol. 188, 1011–1021. doi: 10.1128/JB.188.3.1011-1021.2006
McCabe, O., Spinelli, S., Farenc, C., Labbe, M., Tremblay, D., Blangy, S., et al. (2015). The targeted recognition of Lactococcus lactis phages to their polysaccharide receptors. Mol. Microbiol. 96, 875–886. doi: 10.1111/mmi.12978
McLean, R. J., Beauchemin, D., and Beveridge, T. J. (1992). Influence of oxidation state on iron binding by Bacillus licheniformis capsule. Appl. Environ. Microbiol. 58, 405–408.
Melville, S., and Craig, L. (2013). Type IV pili in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 77, 323–341. doi: 10.1128/MMBR.00063-12
Merchante, R., Pooley, H. M., and Karamata, D. (1995). A periplasm in Bacillus subtilis. J. Bacteriol. 177, 6176–6183. doi: 10.1128/jb.177.21.6176-6183.1995
Merrigan, M. M., Venugopal, A., Roxas, J. L., Anwar, F., Mallozzi, M. J., Roxas, B. A., et al. (2013). Surface-layer protein A (SlpA) is a major contributor to host-cell adherence of Clostridium difficile. PLOS ONE 8:e78404. doi: 10.1371/journal.pone.0078404
Mesnage, S., Dellarole, M., Baxter, N. J., Rouget, J. B., Dimitrov, J. D., Wang, N., et al. (2014). Molecular basis for bacterial peptidoglycan recognition by LysM domains. Nat. Commun. 5:4269. doi: 10.1038/ncomms5269
Mesnage, S., Fontaine, T., Mignot, T., Delepierre, M., Mock, M., and Fouet, A. (2000). Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19, 4473–4484. doi: 10.1093/emboj/19.17.4473
Mesnage, S., Tosi-Couture, E., and Fouet, A. (1999). Production and cell surface anchoring of functional fusions between the SLH motifs of the Bacillus anthracis S-layer proteins and the Bacillus subtilis levansucrase. Mol. Microbiol. 31, 927–936. doi: 10.1046/j.1365-2958.1999.01232.x
Messner, P., and Sleytr, U. B. (1992). Crystalline bacterial cell-surface layers. Adv. Microb. Physiol. 33, 213–275. doi: 10.1016/S0065-2911(08)60218-0
Mijouin, L., Hillion, M., Ramdani, Y., Jaouen, T., Duclairoir-Poc, C., Follet-Gueye, M. L., et al. (2013). Effects of a skin neuropeptide (substance p) on cutaneous microflora. PLOS ONE 8:e78773. doi: 10.1371/journal.pone.0078773
Mishima, M., Shida, T., Yabuki, K., Kato, K., Sekiguchi, J., and Kojima, C. (2005). Solution structure of the peptidoglycan binding domain of Bacillus subtilis cell wall lytic enzyme CwlC: characterization of the sporulation-related repeats by NMR. Biochemistry 44, 10153–10163. doi: 10.1021/bi050624n
Mishra, N. N., Bayer, A. S., Tran, T. T., Shamoo, Y., Mileykovskaya, E., Dowhan, W., et al. (2012). Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content. PLOS ONE 7:e43958. doi: 10.1371/journal.pone.0043958
Mistou, M. Y., Sutcliffe, I. C., and Van Sorge, N. M. (2016). Bacterial glycobiology: rhamnose-containing cell wall polysaccharides in Gram-positive bacteria. FEMS Microbiol. Rev. 40, 464–479. doi: 10.1093/femsre/fuw006
Mitchell, G. J., Wiesenfeld, K., Nelson, D. C., and Weitz, J. S. (2013). Critical cell wall hole size for lysis in Gram-positive bacteria. J. R. Soc. Interface 10:20120892. doi: 10.1098/rsif.2012.0892
Mitra, S. D., Afonina, I., and Kline, K. A. (2016). Right place, right time: focalization of membrane proteins in gram-positive bacteria. Trends Microbiol. 24, 611–621. doi: 10.1016/j.tim.2016.03.009
Morita, H., Toh, H., Oshima, K., Murakami, M., Taylor, T. D., Igimi, S., et al. (2009). Complete genome sequence of the probiotic Lactobacillus rhamnosus ATCC 53103. J. Bacteriol. 191, 7630–7631. doi: 10.1128/JB.01287-09
Morita, H., Toh, H., Oshima, K., Yoshizaki, M., Kawanishi, M., Nakaya, K., et al. (2011). Complete genome sequence and comparative analysis of the fish pathogen Lactococcus garvieae. PLOS ONE 6:e23184. doi: 10.1371/journal.pone.0023184
Mukherjee, S., and Kearns, D. B. (2014). The structure and regulation of flagella in Bacillus subtilis. Annu. Rev. Genet. 48, 319–340. doi: 10.1146/annurev-genet-120213-092406
Murakami, T., Haga, K., Takeuchi, M., and Sato, T. (2002). Analysis of the Bacillus subtilis spoIIIJ gene and its paralogue gene, yqjG. J. Bacteriol. 184, 1998–2004. doi: 10.1128/JB.184.7.1998-2004.2002
Murphy, E. C., Janulczyk, R., Karlsson, C., Morgelin, M., and Frick, I. M. (2014). Identification of pili on the surface of Finegoldia magna–a gram-positive anaerobic cocci. Anaerobe 27, 40–49. doi: 10.1016/j.anaerobe.2014.03.008
Musyoki, A. M., Shi, Z., Xuan, C., Lu, G., Qi, J., Gao, F., et al. (2016). Structural and functional analysis of an anchorless fibronectin-binding protein FBPS from Gram-positive bacterium Streptococcus suis. Proc. Natl. Acad. Sci. U.S.A. 113, 13869–13874. doi: 10.1073/pnas.1608406113
Nakayama, H., Kurokawa, K., and Lee, B. L. (2012). Lipoproteins in bacteria: structures and biosynthetic pathways. FEBS J. 279, 4247–4268. doi: 10.1111/febs.12041
Nallapareddy, S. R., Singh, K. V., Sillanpaa, J., Garsin, D. A., Hook, M., Erlandsen, S. L., et al. (2006). Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Invest. 116, 2799–2807. doi: 10.1172/JCI29021
Navarre, W. W., and Schneewind, O. (1999). Surface proteins of Gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63, 174–229.
N’Diaye, A., Gannesen, A., Borrel, V., Maillot, O., Enault, J., Racine, P. J., et al. (2017). Substance P and calcitonin gene-related peptide: key regulators of cutaneous microbiota homeostasis. Front. Endocrinol. 8:15. doi: 10.3389/fendo.2017.00015
N’Diaye, A., Mijouin, L., Hillion, M., Diaz, S., Konto-Ghiorghi, Y., Percoco, G., et al. (2016a). Effect of substance P in Staphylococcus aureus and Staphylococcus epidermidis virulence: implication for skin homeostasis. Front. Microbiol. 7:506. doi: 10.3389/fmicb.2016.00506
N’Diaye, A. R., Leclerc, C., Kentache, T., Hardouin, J., Poc, C. D., Konto-Ghiorghi, Y., et al. (2016b). Skin-bacteria communication: involvement of the neurohormone calcitonin gene related peptide (CGRP) in the regulation of Staphylococcus epidermidis virulence. Sci. Rep. 6:35379. doi: 10.1038/srep35379
Necchi, F., Nardi-Dei, V., Biagini, M., Assfalg, M., Nuccitelli, A., Cozzi, R., et al. (2011). Sortase A substrate specificity in GBS pilus 2a cell wall anchoring. PLOS ONE 6:e25300. doi: 10.1371/journal.pone.0025300
Neuhaus, F. C., and Baddiley, J. (2003). A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67, 686–723. doi: 10.1128/MMBR.67.4.686-723.2003
Nielsen, H. (2017). Predicting secretory proteins with SignalP. Methods Mol. Biol. 1611, 59–73. doi: 10.1007/978-1-4939-7015-5_6
Nölling, J., Breton, G., Omelchenko, M. V., Makarova, K. S., Zeng, Q., Gibson, R., et al. (2001). Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183, 4823–4838. doi: 10.1128/JB.183.16.4823-4838.2001
O’Connell Motherway, M., Zomer, A., Leahy, S. C., Reunanen, J., Bottacini, F., Claesson, M. J., et al. (2011). Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. U.S.A. 108, 11217–11222. doi: 10.1073/pnas.1105380108
Oh, S. Y., Lunderberg, J. M., Chateau, A., Schneewind, O., and Missiakas, D. (2017). Genes required for Bacillus anthracis secondary cell wall polysaccharide synthesis. J. Bacteriol. 199, e00613–e00616. doi: 10.1128/JB.00613-16
Oli, M. W., Otoo, H. N., Crowley, P. J., Heim, K. P., Nascimento, M. M., Ramsook, C. B., et al. (2012). Functional amyloid formation by Streptococcus mutans. Microbiology 158, 2903–2916. doi: 10.1099/mic.0.060855-0
Oliveira, H., Melo, L. D., Santos, S. B., Nobrega, F. L., Ferreira, E. C., Cerca, N., et al. (2013). Molecular aspects and comparative genomics of bacteriophage endolysins. J. Virol. 87, 4558–4570. doi: 10.1128/JVI.03277-12
Oliveira, L., Madureira, P., Andrade, E. B., Bouaboud, A., Morello, E., Ferreira, P., et al. (2012). Group B Streptococcus GAPDH is released upon cell lysis, associates with bacterial surface, and induces apoptosis in murine macrophages. PLOS ONE 7:e29963. doi: 10.1371/journal.pone.0029963
Oxaran, V., Ledue-Clier, F., Dieye, Y., Herry, J. M., Pechoux, C., Meylheuc, T., et al. (2012). Pilus biogenesis in Lactococcus lactis: molecular characterization and role in aggregation and biofilm formation. PLOS ONE 7:e50989. doi: 10.1371/journal.pone.0050989
Palmer, S. R., Crowley, P. J., Oli, M. W., Ruelf, M. A., Michalek, S. M., and Brady, L. J. (2012). YidC1 and YidC2 are functionally distinct proteins involved in protein secretion, biofilm formation and cariogenicity of Streptococcus mutans. Microbiology 158, 1702–1712. doi: 10.1099/mic.0.059139-0
Pancholi, V., and Fischetti, V. A. (1992). A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176, 415–426. doi: 10.1084/jem.176.2.415
Pantaleon, V., Soavelomandroso, A. P., Bouttier, S., Briandet, R., Roxas, B., Chu, M., et al. (2015). The Clostridium difficile protease Cwp84 modulates both biofilm formation and cell-surface properties. PLOS ONE 10:e0124971. doi: 10.1371/journal.pone.0124971
Pasztor, L., Ziebandt, A. K., Nega, M., Schlag, M., Haase, S., Franz-Wachtel, M., et al. (2010). Staphylococcal major autolysin (Atl) is involved in excretion of cytoplasmic proteins. J. Biol. Chem. 285, 36794–36803. doi: 10.1074/jbc.M110.167312
Peabody, C. R., Chung, Y. J., Yen, M. R., Vidal-Ingigliardi, D., Pugsley, A. P., and Saier, M. H. Jr. (2003). Type II protein secretion and its relationship to bacterial Type 4 pili and archaeal flagella. Microbiology 149, 3051–3072. doi: 10.1099/mic.0.26364-0
Peabody, M. A., Laird, M. R., Vlasschaert, C., Lo, R., and Brinkman, F. S. (2016). PSORTdb: expanding the bacteria and archaea protein subcellular localization database to better reflect diversity in cell envelope structures. Nucleic Acids Res. 44, D663–D668. doi: 10.1093/nar/gkv1271
Percy, M. G., and Grundling, A. (2014). Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu. Rev. Microbiol. 68, 81–100. doi: 10.1146/annurev-micro-091213-112949
Percy, M. G., Karinou, E., Webb, A. J., and Grundling, A. (2016). Identification of a lipoteichoic acid glycosyltransferase enzyme reveals that GW-domain-containing proteins can be retained in the cell wall of Listeria monocytogenes in the absence of lipoteichoic acid or its modifications. J. Bacteriol. 198, 2029–2042. doi: 10.1128/JB.00116-16
Piepenbrink, K. H., Maldarelli, G. A., Martinez de La Peña, C. F., Dingle, T. C., Mulvey, G. L., Lee, A., et al. (2015). Structural and evolutionary analyses show unique stabilization strategies in the type IV pili of Clostridium difficile. Structure 23, 385–396. doi: 10.1016/j.str.2014.11.018
Pink, D., Moeller, J., Quinn, B., Jericho, M., and Beveridge, T. (2000). On the architecture of the gram-negative bacterial murein sacculus. J. Bacteriol. 182, 5925–5930. doi: 10.1128/JB.182.20.5925-5930.2000
Planchon, S., Chambon, C., Desvaux, M., Chafsey, I., Leroy, S., Talon, R., et al. (2007). Proteomic analysis of cell envelope from Staphylococcus xylosus C2a, a coagulase-negative Staphylococcus. J. Proteome Res. 6, 3566–3580. doi: 10.1021/pr070139+
Planchon, S., Desvaux, M., Chafsey, I., Chambon, C., Leroy, S., Hébraud, M., et al. (2009). Comparative subproteome analyses of planktonic and sessile Staphylococcus xylosus C2a: new insight in cell physiology of a coagulase-negative Staphylococcus in biofilm. J. Proteome Res. 8, 1797–1809. doi: 10.1021/pr8004056
Plaut, R. D., Beaber, J. W., Zemansky, J., Kaur, A. P., George, M., Biswas, B., et al. (2014). Genetic evidence for the involvement of the S-layer protein gene sap and the sporulation genes spo0A, spo0B, and spo0F in Phage AP50c infection of Bacillus anthracis. J. Bacteriol. 196, 1143–1154. doi: 10.1128/JB.00739-13
Pointon, J. A., Smith, W. D., Saalbach, G., Crow, A., Kehoe, M. A., and Banfield, M. J. (2010). A highly unusual thioester bond in a pilus adhesin is required for efficient host cell interaction. J. Biol. Chem. 285, 33858–33866. doi: 10.1074/jbc.M110.149385
Pracht, D., Elm, C., Gerber, J., Bergmann, S., Rohde, M., Seiler, M., et al. (2005). PavA of Streptococcus pneumoniae modulates adherence, invasion, and meningeal inflammation. Infect. Immun. 73, 2680–2689. doi: 10.1128/IAI.73.5.2680-2689.2005
Rakotoarivonina, H., Jubelin, G., Hébraud, M., Gaillard-Martinie, B., Forano, E., and Mosoni, P. (2002). Adhesion to cellulose of the Gram-positive bacterium Ruminococcus albus involves Type 4 pili. Microbiology 148, 1871–1880. doi: 10.1099/00221287-148-6-1871
Rambaran, R. N., and Serpell, L. C. (2008). Amyloid fibrils: abnormal protein assembly. Prion 2, 112–117. doi: 10.4161/pri.2.3.7488
Razin, S., Yogev, D., and Naot, Y. (1998). Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62, 1094–1156.
Regulski, K., Courtin, P., Kulakauskas, S., and Chapot-Chartier, M. P. (2013). A novel type of peptidoglycan-binding domain highly specific for amidated D-Asp cross-bridge, identified in Lactobacillus casei bacteriophage endolysins. J. Biol. Chem. 288, 20416–20426. doi: 10.1074/jbc.M112.446344
Reichmann, N. T., and Grundling, A. (2011). Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol. Lett. 319, 97–105. doi: 10.1111/j.1574-6968.2011.02260.x
Renier, S., Hébraud, M., and Desvaux, M. (2011). Molecular biology of surface colonization by Listeria monocytogenes: an additional facet of an opportunistic Gram-positive foodborne pathogen. Environ. Microbiol. 13, 835–850. doi: 10.1111/j.1462-2920.2010.02378.x
Renier, S., Micheau, P., Talon, R., Hebraud, M., and Desvaux, M. (2012). Subcellular localization of extracytoplasmic proteins in monoderm bacteria: rational secretomics-based strategy for genomic and proteomic analyses. PLOS ONE 7:e42982. doi: 10.1371/journal.pone.0042982
Reunanen, J., Von Ossowski, I., Hendrickx, A. P., Palva, A., and De Vos, W. M. (2012). Characterization of the SpaCBA pilus fibers in the probiotic Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 78, 2337–2344. doi: 10.1128/AEM.07047-11
Richards, F. M. (1977). Areas, volumes, packing and protein structure. Annu. Rev. Biophys. Bioeng. 6, 151–176. doi: 10.1146/annurev.bb.06.060177.001055
Richter, S., Anderson, V. J., Garufi, G., Lu, L., Budzik, J. M., Joachimiak, A., et al. (2009). Capsule anchoring in Bacillus anthracis occurs by a transpeptidation reaction that is inhibited by capsidin. Mol. Microbiol. 71, 404–420. doi: 10.1111/j.1365-2958.2008.06533.x
Rinaudo, C. D., Rosini, R., Galeotti, C. L., Berti, F., Necchi, F., Reguzzi, V., et al. (2010). Specific involvement of pilus type 2a in biofilm formation in group B Streptococcus. PLOS ONE 5:e9216. doi: 10.1371/journal.pone.0009216
Rodriguez Ayala, F., Bauman, C., Bartolini, M., Saball, E., Salvarrey, M., Lenini, C., et al. (2017). Transcriptional regulation of adhesive properties of Bacillus subtilis to extracellular matrix proteins through the fibronectin-binding protein YloA. Mol. Microbiol. 104, 804–821. doi: 10.1111/mmi.13666
Romero, D., Aguilar, C., Losick, R., and Kolter, R. (2010). Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U.S.A. 107, 2230–2234. doi: 10.1073/pnas.0910560107
Romero, D., Vlamakis, H., Losick, R., and Kolter, R. (2011). An accessory protein required for anchoring and assembly of amyloid fibres in B. subtilis biofilms. Mol. Microbiol. 80, 1155–1168. doi: 10.1111/j.1365-2958.2011.07653.x
Romero, D., Vlamakis, H., Losick, R., and Kolter, R. (2014). Functional analysis of the accessory protein TapA in Bacillus subtilis amyloid fiber assembly. J. Bacteriol. 196, 1505–1513. doi: 10.1128/JB.01363-13
Rosch, J. W., Hsu, F. F., and Caparon, M. G. (2007). Anionic lipids enriched at the ExPortal of Streptococcus pyogenes. J. Bacteriol. 189, 801–806. doi: 10.1128/JB.01549-06
Rossez, Y., Wolfson, E. B., Holmes, A., Gally, D. L., and Holden, N. J. (2015). Bacterial flagella: twist and stick, or dodge across the kingdoms. PLOS Pathog. 11:e1004483. doi: 10.1371/journal.ppat.1004483
Roy, H. (2009). Tuning the properties of the bacterial membrane with aminoacylated phosphatidylglycerol. IUBMB Life 61, 940–953. doi: 10.1002/iub.240
Rudolf, M., Tetik, N., Ramos-Leon, F., Flinner, N., Ngo, G., Stevanovic, M., et al. (2015). The peptidoglycan-binding protein SjcF1 influences septal junction function and channel formation in the filamentous Cyanobacterium Anabaena. mBio 6:e00376-15. doi: 10.1128/mBio.00376-15
Salton, M. R. (1967). Structure and function of bacterial cell membranes. Annu. Rev. Microbiol. 21, 417–442. doi: 10.1146/annurev.mi.21.100167.002221
Samuelson, J. C., Chen, M., Jiang, M., Moller, I., Wiedmann, M., Kuhn, A., et al. (2000). YidC mediates membrane protein insertion in bacteria. Nature 406, 637–641. doi: 10.1038/35020586
Sára, M., and Sleytr, U. B. (2000). S-Layer proteins. J. Bacteriol. 182, 859–868. doi: 10.1128/JB.182.4.859-868.2000
Schachtsiek, M., Hammes, W. P., and Hertel, C. (2004). Characterization of Lactobacillus coryniformis DSM 20001T surface protein Cpf mediating coaggregation with and aggregation among pathogens. Appl. Environ. Microbiol. 70, 7078–7085. doi: 10.1128/AEM.70.12.7078-7085.2004
Schäffer, C., Kahlig, H., Christian, R., Schulz, G., Zayni, S., and Messner, P. (1999). The diacetamidodideoxyuronic-acid-containing glycan chain of Bacillus stearothermophilus NRS 2004/3a represents the secondary cell-wall polymer of wild-type B. stearothermophilus strains. Microbiology 145, 1575–1583. doi: 10.1099/13500872-145-7-1575
Schaffer, C., and Messner, P. (2017). Emerging facets of prokaryotic glycosylation. FEMS Microbiol. Rev. 41, 49–91. doi: 10.1093/femsre/fuw036
Schirm, M., Kalmokoff, M., Aubry, A., Thibault, P., Sandoz, M., and Logan, S. M. (2004). Flagellin from Listeria monocytogenes is glycosylated with beta-O-linked N-acetylglucosamine. J. Bacteriol. 186, 6721–6727. doi: 10.1128/JB.186.20.6721-6727.2004
Schirm, M., Schoenhofen, I. C., Logan, S. M., Waldron, K. C., and Thibault, P. (2005). Identification of unusual bacterial glycosylation by tandem mass spectrometry analyses of intact proteins. Anal. Chem. 77, 7774–7782. doi: 10.1021/ac051316y
Schleifer, K. H., and Kandler, O. (1972). Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477.
Schneewind, O., and Missiakas, D. M. (2012). Protein secretion and surface display in Gram-positive bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1123–1139. doi: 10.1098/rstb.2011.0210
Schneewind, O., Model, P., and Fischetti, V. A. (1992). Sorting of protein A to the staphylococcal cell wall. Cell 70, 267–281. doi: 10.1016/0092-8674(92)90101-H
Schneider, J., Klein, T., Mielich-Suss, B., Koch, G., Franke, C., Kuipers, O. P., et al. (2015). Spatio-temporal remodeling of functional membrane microdomains organizes the signaling networks of a bacterium. PLOS Genet. 11:e1005140. doi: 10.1371/journal.pgen.1005140
Schultz, J., Milpetz, F., Bork, P., and Ponting, C. P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864. doi: 10.1073/pnas.95.11.5857
Schwartz, K., Sekedat, M. D., Syed, A. K., O’hara, B., Payne, D. E., Lamb, A., et al. (2014). The AgrD N-terminal leader peptide of Staphylococcus aureus has cytolytic and amyloidogenic properties. Infect. Immun. 82, 3837–3844. doi: 10.1128/IAI.02111-14
Schwartz, K., Syed, A. K., Stephenson, R. E., Rickard, A. H., and Boles, B. R. (2012). Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLOS Pathog. 8:e1002744. doi: 10.1371/journal.ppat.1002744
Schwarz, W. H. (2001). The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56, 634–649. doi: 10.1007/s002530100710
Schwarz-Linek, U., Werner, J. M., Pickford, A. R., Gurusiddappa, S., Kim, J. H., Pilka, E. S., et al. (2003). Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper. Nature 423, 177–181. doi: 10.1038/nature01589
Scotti, P. A., Urbanus, M. L., Brunner, J., De Gier, J. W., Von Heijne, G., Van Der Does, C., et al. (2000). YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19, 542–549. doi: 10.1093/emboj/19.4.542
Shaik, M. M., Maccagni, A., Tourcier, G., Di Guilmi, A. M., and Dessen, A. (2014). Structural basis of pilus anchoring by the ancillary pilin RrgC of Streptococcus pneumoniae. J. Biol. Chem. 289, 16988–16997. doi: 10.1074/jbc.M114.555854
Shankar, V., Baghdayan, A. S., Huycke, M. M., Lindahl, G., and Gilmore, M. S. (1999). Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun. 67, 193–200.
Shockman, G. D., and Barrett, J. F. (1983). Structure, function, and assembly of cell walls of Gram-positive bacteria. Annu. Rev. Microbiol. 37, 501–527. doi: 10.1146/annurev.mi.37.100183.002441
Sibbald, M. J., Ziebandt, A. K., Engelmann, S., Hecker, M., De Jong, A., Harmsen, H. J., et al. (2006). Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol. Mol. Biol. Rev. 70, 755–788. doi: 10.1128/MMBR.00008-06
Siegel, S. D., Reardon, M. E., and Ton-That, H. (2017). Anchoring of LPXTG-like proteins to the gram-positive cell wall envelope. Curr. Top. Microbiol. Immunol. 404, 159–175. doi: 10.1007/82_2016_8
Siezen, R., Boekhorst, J., Muscariello, L., Molenaar, D., Renckens, B., and Kleerebezem, M. (2006). Lactobacillus plantarum gene clusters encoding putative cell-surface protein complexes for carbohydrate utilization are conserved in specific Gram-positive bacteria. BMC Genomics 7:126. doi: 10.1186/1471-2164-7-126
Sigrist, C. J., De Castro, E., Cerutti, L., Cuche, B. A., Hulo, N., Bridge, A., et al. (2013). New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–D347. doi: 10.1093/nar/gks1067
Silhavy, T. J., Kahne, D., and Walker, S. (2010). The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2:a000414. doi: 10.1101/cshperspect.a000414
Sillanpaa, J., Chang, C., Singh, K. V., Montealegre, M. C., Nallapareddy, S. R., Harvey, B. R., et al. (2013). Contribution of individual Ebp Pilus subunits of Enterococcus faecalis OG1RF to pilus biogenesis, biofilm formation and urinary tract infection. PLOS ONE 8:e68813. doi: 10.1371/journal.pone.0068813
Sillanpaa, J., Nallapareddy, S. R., Singh, K. V., Prakash, V. P., Fothergill, T., Ton-That, H., et al. (2010). Characterization of the ebp(fm) pilus-encoding operon of Enterococcus faecium and its role in biofilm formation and virulence in a murine model of urinary tract infection. Virulence 1, 236–246. doi: 10.4161/viru.1.4.11966
Smith, E. J., Corrigan, R. M., Van Der Sluis, T., Grundling, A., Speziale, P., Geoghegan, J. A., et al. (2012). The immune evasion protein Sbi of Staphylococcus aureus occurs both extracellularly and anchored to the cell envelope by binding lipoteichoic acid. Mol. Microbiol. 83, 789–804. doi: 10.1111/j.1365-2958.2011.07966.x
Smith, S. P., and Bayer, E. A. (2013). Insights into cellulosome assembly and dynamics: from dissection to reconstruction of the supramolecular enzyme complex. Curr. Opin. Struct. Biol. 23, 686–694. doi: 10.1016/j.sbi.2013.09.002
Smith, S. P., Bayer, E. A., and Czjzek, M. (2017). Continually emerging mechanistic complexity of the multi-enzyme cellulosome complex. Curr. Opin. Struct. Biol. 44, 151–160. doi: 10.1016/j.sbi.2017.03.009
Solis, N., and Cordwell, S. J. (2011). Current methodologies for proteomics of bacterial surface-exposed and cell envelope proteins. Proteomics 11, 3169–3189. doi: 10.1002/pmic.201000808
Solis, N., and Cordwell, S. J. (2016). Cell shaving and false-positive control strategies coupled to novel statistical tools to profile gram-positive bacterial surface proteomes. Methods Mol. Biol. 1440, 47–55. doi: 10.1007/978-1-4939-3676-2_4
Somarajan, S. R., La Rosa, S. L., Singh, K. V., Roh, J. H., Hook, M., and Murray, B. E. (2015). The fibronectin-binding protein Fnm contributes to adherence to extracellular matrix components and virulence of Enterococcus faecium. Infect. Immun. 83, 4653–4661. doi: 10.1128/IAI.00885-15
Somoskovi, A., Hotaling, J. E., Fitzgerald, M., O’donnell, D., Parsons, L. M., and Salfinger, M. (2001). Lessons from a proficiency testing event for acid-fast microscopy. Chest 120, 250–257. doi: 10.1378/chest.120.1.250
Soohammer, E. L., Eddy, S. R., and Durbin, R. (1997). Pfam: a comprehensive database of protein domain families based on seed alignements. Proteins 28, 405–420. doi: 10.1002/(SICI)1097-0134(199707)28:3<405::AID-PROT10>3.0.CO;2-L
Steen, A., Buist, G., Horsburgh, G. J., Venema, G., Kuipers, O. P., Foster, S. J., et al. (2005). AcmA of Lactococcus lactis is an N-acetylglucosaminidase with an optimal number of LysM domains for proper functioning. FEBS J. 272, 2854–2868. doi: 10.1111/j.1742-4658.2005.04706.x
Steen, A., Buist, G., Leenhouts, K. J., El Khattabi, M., Grijpstra, F., Zomer, A. L., et al. (2003). Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J. Biol. Chem. 278, 23874–23881. doi: 10.1074/jbc.M211055200
Steindl, C., Schaffer, C., Wugeditsch, T., Graninger, M., Matecko, I., Muller, N., et al. (2002). The first biantennary bacterial secondary cell wall polymer and its influence on S-layer glycoprotein assembly. Biochem. J. 368, 483–494. doi: 10.1042/bj20020988
Stern, J., Morais, S., Lamed, R., and Bayer, E. A. (2016). Adaptor scaffoldins: an original strategy for extended designer cellulosomes, inspired from nature. mBio 7:e00083-16. doi: 10.1128/mBio.00083-16
Sullivan, L., Paredes, C. J., Papoutsakis, E. T., and Bennett, G. N. (2007). Analysis of the clostridial hydrophobic with a conserved tryptophan family (ChW) of proteins in Clostridium acetobutylicum with emphasis on ChW14 and ChW16/17. Enzyme Microb. Technol. 42, 29–43. doi: 10.1016/j.enzmictec.2007.07.022
Sutcliffe, I. C. (2010). A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol. 18, 464–470. doi: 10.1016/j.tim.2010.06.005
Sutcliffe, I. C., and Harrington, D. J. (2002). Pattern searches for the identification of putative lipoprotein genes in Gram-positive bacterial genomes. Microbiology 148, 2065–2077. doi: 10.1099/00221287-148-7-2065
Taglialegna, A., Lasa, I., and Valle, J. (2016a). Amyloid structures as biofilm matrix scaffolds. J. Bacteriol. 198, 2579–2588. doi: 10.1128/JB.00122-16
Taglialegna, A., Navarro, S., Ventura, S., Garnett, J. A., Matthews, S., Penades, J. R., et al. (2016b). Staphylococcal Bap proteins build amyloid scaffold biofilm matrices in response to environmental signals. PLOS Pathog. 12:e1005711. doi: 10.1371/journal.ppat.1005711
Tayeb-Fligelman, E., Tabachnikov, O., Moshe, A., Goldshmidt-Tran, O., Sawaya, M. R., Coquelle, N., et al. (2017). The cytotoxic Staphylococcus aureus PSMa3 reveals a cross-alpha amyloid-like fibril. Science 355, 831–833. doi: 10.1126/science.aaf4901
Terrasse, R., Amoroso, A., Vernet, T., and Di Guilmi, A. M. (2015). Streptococcus pneumoniae GAPDH is released by cell lysis and interacts with peptidoglycan. PLOS ONE 10:e0125377. doi: 10.1371/journal.pone.0125377
Tjalsma, H., Antelmann, H., Jongbloed, J. D., Braun, P. G., Darmon, E., Dorenbos, R., et al. (2004). Proteomics of protein secretion by Bacillus subtilis: separating the ”secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68, 207–233. doi: 10.1128/MMBR.68.2.207-233.2004
Tjalsma, H., Bolhuis, A., Jongbloed, J. D., Bron, S., and Van Dijl, J. M. (2000). Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64, 515–547. doi: 10.1128/MMBR.64.3.515-547.2000
Tjalsma, H., Bron, S., and Van Dijl, J. M. (2003). Complementary impact of paralogous Oxa1-like proteins of Bacillus subtilis on post-translocational stages in protein secretion. J. Biol. Chem. 278, 15622–15632. doi: 10.1074/jbc.M301205200
Toh, H., Oshima, K., Nakano, A., Takahata, M., Murakami, M., Takaki, T., et al. (2013). Genomic adaptation of the Lactobacillus casei group. PLOS ONE 8:e75073. doi: 10.1371/journal.pone.0075073
Ton-That, H., and Schneewind, O. (2003). Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 50, 1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x
Torelli, R., Serror, P., Bugli, F., Paroni Sterbini, F., Florio, A. R., Stringaro, A., et al. (2012). The PavA-like fibronectin-binding protein of Enterococcus faecalis, EfbA, is important for virulence in a mouse model of ascending urinary tract infection. J. Infect. Dis. 206, 952–960. doi: 10.1093/infdis/jis440
Tripathi, P., Beaussart, A., Alsteens, D., Dupres, V., Claes, I., Von Ossowski, I., et al. (2013). Adhesion and nanomechanics of pili from the probiotic Lactobacillus rhamnosus GG. ACS Nano 7, 3685–3697. doi: 10.1021/nn400705u
Turner, R. D., Hurd, A. F., Cadby, A., Hobbs, J. K., and Foster, S. J. (2013). Cell wall elongation mode in Gram-negative bacteria is determined by peptidoglycan architecture. Nat. Commun. 4:1496. doi: 10.1038/ncomms2503
Twine, S. M., Paul, C. J., Vinogradov, E., Mcnally, D. J., Brisson, J. R., Mullen, J. A., et al. (2008). Flagellar glycosylation in Clostridium botulinum. FEBS J. 275, 4428–4444. doi: 10.1111/j.1742-4658.2008.06589.x
Twine, S. M., Reid, C. W., Aubry, A., Mcmullin, D. R., Fulton, K. M., Austin, J., et al. (2009). Motility and flagellar glycosylation in Clostridium difficile. J. Bacteriol. 191, 7050–7062. doi: 10.1128/JB.00861-09
Usenik, A., Renko, M., Mihelic, M., Lindic, N., Borisek, J., Perdih, A., et al. (2017). The CWB2 cell wall-anchoring module is revealed by the crystal structures of the s cell wall proteins Cwp8 and Cwp6. Structure 25, 514–521. doi: 10.1016/j.str.2016.12.018
Valguarnera, E., Kinsella, R. L., and Feldman, M. F. (2016). Sugar and spice make bacteria not nice: protein glycosylation and its influence in pathogenesis. J. Mol. Biol. 428, 3206–3220. doi: 10.1016/j.jmb.2016.04.013
Valiente, E., Bouche, L., Hitchen, P., Faulds-Pain, A., Songane, M., Dawson, L. F., et al. (2016). Role of glycosyltransferases modifying type B flagellin of emerging hypervirulent Clostridium difficile lineages and their impact on motility and biofilm formation. J. Biol. Chem. 291, 25450–25461. doi: 10.1074/jbc.M116.749523
van Sorge, N. M., Cole, J. N., Kuipers, K., Henningham, A., Aziz, R. K., Kasirer-Friede, A., et al. (2014). The classical lancefield antigen of group a Streptococcus is a virulence determinant with implications for vaccine design. Cell Host Microbe 15, 729–740. doi: 10.1016/j.chom.2014.05.009
van Wely, K. H. M., Swaving, J., Freudl, R., and Driessen, A. J. M. (2001). Translocation of proteins across the cell envelope of Gram-positive bacteria. FEMS Microbiol. Rev. 25, 437–454. doi: 10.1111/j.1574-6976.2001.tb00586.x
Varga, J. J., Nguyen, V., O’brien, D. K., Rodgers, K., Walker, R. A., and Melville, S. B. (2006). Type IV pili-dependent gliding motility in the Gram-positive pathogen Clostridium perfringens and other Clostridia. Mol. Microbiol. 62, 680–694. doi: 10.1111/j.1365-2958.2006.05414.x
Vinogradov, E., Valence, F., Maes, E., Jebava, I., Chuat, V., Lortal, S., et al. (2013). Structural studies of the cell wall polysaccharides from three strains of Lactobacillus helveticus with different autolytic properties: DPC4571, BROI, and LH1. Carbohydr. Res. 379, 7–12. doi: 10.1016/j.carres.2013.05.020
Vollmer, W., Blanot, D., and De Pedro, M. A. (2008). Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167. doi: 10.1111/j.1574-6976.2007.00094.x
von Ossowski, I., Satokari, R., Reunanen, J., Lebeer, S., De Keersmaecker, S. C., Vanderleyden, J., et al. (2011). Functional characterization of a mucus-specific LPXTG surface adhesin from probiotic Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 77, 4465–4472. doi: 10.1128/AEM.02497-10
Wang, L., Si, W., Xue, H., and Zhao, X. (2017). A fibronectin-binding protein (FbpA) of Weissella cibaria inhibits colonization and infection of Staphylococcus aureus in mammary glands. Cell. Microbiol. 19:e12731. doi: 10.1111/cmi.12731
Wang, P., and Dalbey, R. E. (2011). Inserting membrane proteins: the YidC/Oxa1/Alb3 machinery in bacteria, mitochondria, and chloroplasts. Biochim. Biophys. Acta 1808, 866–875. doi: 10.1016/j.bbamem.2010.08.014
Ward, J. B. (1973). The chain length of the glycans in bacterial cell walls. Biochem. J. 133, 395–398. doi: 10.1042/bj1330395
Ward, J. B. (1981). Teichoic and teichuronic acids: biosynthesis, assembly, and location. Microbiol. Rev. 45, 211–243.
Weidenmaier, C., and Peschel, A. (2008). Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 6, 276–287. doi: 10.1038/nrmicro1861
Wheeler, R., Mesnage, S., Boneca, I. G., Hobbs, J. K., and Foster, S. J. (2011). Super-resolution microscopy reveals cell wall dynamics and peptidoglycan architecture in ovococcal bacteria. Mol. Microbiol. 82, 1096–1109. doi: 10.1111/j.1365-2958.2011.07871.x
White, S. H., and von Heijne, G. (2004). The machinery of membrane protein assembly. Curr. Opin. Struct. Biol. 14, 397–404. doi: 10.1016/j.sbi.2004.07.003
Willing, S. E., Candela, T., Shaw, H. A., Seager, Z., Mesnage, S., Fagan, R. P., et al. (2015). Clostridium difficile surface proteins are anchored to the cell wall using CWB2 motifs that recognise the anionic polymer PSII. Mol. Microbiol. 96, 596–608. doi: 10.1111/mmi.12958
Wisniewski, J. A., and Rood, J. I. (2017). The Tcp conjugation system of Clostridium perfringens. Plasmid 91, 28–36. doi: 10.1016/j.plasmid.2017.03.001
Woese, C. R., Kandler, O., and Wheelis, M. L. (1990). Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U.S.A. 87, 4576–4579. doi: 10.1073/pnas.87.12.4576
Wong, J. E., Alsarraf, H. M., Kaspersen, J. D., Pedersen, J. S., Stougaard, J., Thirup, S., et al. (2014). Cooperative binding of LysM domains determines the carbohydrate affinity of a bacterial endopeptidase protein. FEBS J. 281, 1196–1208. doi: 10.1111/febs.12698
Wooldridge, K. G., and Williams, P. H. (1993). Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol. Rev. 12, 325–348. doi: 10.1111/j.1574-6976.1993.tb00026.x
Xia, G., Maier, L., Sanchez-Carballo, P., Li, M., Otto, M., Holst, O., et al. (2010). Glycosylation of wall teichoic acid in Staphylococcus aureus by TarM. J. Biol. Chem. 285, 13405–13415. doi: 10.1074/jbc.M109.096172
Xu, P., Alves, J. M., Kitten, T., Brown, A., Chen, Z., Ozaki, L. S., et al. (2007). Genome of the opportunistic pathogen Streptococcus sanguinis. J. Bacteriol. 189, 3166–3175. doi: 10.1128/JB.01808-06
Xu, Q., Bayer, E. A., Goldman, M., Kenig, R., Shoham, Y., and Lamed, R. (2004). Architecture of the Bacteroides cellulosolvens cellulosome: description of a cell surface-anchoring scaffoldin and a family 48 cellulase. J. Bacteriol. 186, 968–977. doi: 10.1128/JB.186.4.968-977.2004
Xu, Q., Gao, W., Ding, S. Y., Kenig, R., Shoham, Y., Bayer, E. A., et al. (2003). The cellulosome system of Acetivibrio cellulolyticus includes a novel type of adaptor protein and a cell surface anchoring protein. J. Bacteriol. 185, 4548–4557. doi: 10.1128/JB.185.15.4548-4557.2003
Xu, Q., Mengin-Lecreulx, D., Liu, X. W., Patin, D., Farr, C. L., Grant, J. C., et al. (2015). Insights into substrate specificity of NlpC/P60 cell wall hydrolases containing bacterial SH3 domains. mBio 6:e02327-14. doi: 10.1128/mBio.02327-14
Yahashiri, A., Jorgenson, M. A., and Weiss, D. S. (2015). Bacterial SPOR domains are recruited to septal peptidoglycan by binding to glycan strands that lack stem peptides. Proc. Natl. Acad. Sci. U.S.A. 112, 11347–11352. doi: 10.1073/pnas.1508536112
Yahashiri, A., Jorgenson, M. A., and Weiss, D. S. (2017). The SPOR domain: a widely-conserved peptidoglycan binding domain that targets proteins to the site of cell division. J. Bacteriol. 199:e00118-17. doi: 10.1128/JB.00118-17
Yanagawa, R., Otsuki, K., and Tokui, T. (1968). Electron microscopy of fine structure of Corynebacterium renale with special reference to pili. Jpn. J. Vet. Res. 16, 31–37.
Yasuda, E., Tateno, H., Hirabayashi, J., Iino, T., and Sako, T. (2011). Lectin microarray reveals binding profiles of Lactobacillus casei strains in a comprehensive analysis of bacterial cell wall polysaccharides. Appl. Environ. Microbiol. 77, 4539–4546. doi: 10.1128/AEM.00240-11
Yen, M. R., Peabody, C. R., Partovi, S. M., Zhai, Y., Tseng, Y. H., and Saier, M. H. (2002). Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim. Biophys. Acta 1562, 6–31. doi: 10.1016/S0005-2736(02)00359-0
Yother, J. (2011). Capsules of Streptococcus pneumoniae and other bacteria: paradigms for polysaccharide biosynthesis and regulation. Annu. Rev. Microbiol. 65, 563–581. doi: 10.1146/annurev.micro.62.081307.162944
Yu, X., Jaatinen, A., Rintahaka, J., Hynonen, U., Lyytinen, O., Kant, R., et al. (2015). Human gut-commensalic Lactobacillus ruminis ATCC 25644 displays sortase-assembled surface piliation: phenotypic characterization of its fimbrial operon through in silico predictive analysis and recombinant expression in Lactococcus lactis. PLOS ONE 10:e0145718. doi: 10.1371/journal.pone.0145718
Zdobnov, E. M., and Apweiler, R. (2001). InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848. doi: 10.1093/bioinformatics/17.9.847
Zhao, G., Ali, E., Sakka, M., Kimura, T., and Sakka, K. (2005). Binding of S-layer homology modules from Clostridium thermocellum SdbA to peptidoglycans. Appl. Microbiol. Biotechnol. 23, 1–6.
Zhao, G., Li, H., Wamalwa, B., Sakka, M., Kimura, T., and Sakka, K. (2006). Different binding specificities of S-layer homology modules from Clostridium thermocellum AncA, Slp1, and Slp2. Biosci. Biotechnol. Biochem. 70, 1636–1641. doi: 10.1271/bbb.50699
Ziehe, D., Dunschede, B., and Schunemann, D. (2017). From bacteria to chloroplasts: evolution of the chloroplast SRP system. Biol. Chem. 398, 653–661. doi: 10.1515/hsz-2016-0292
Zoll, S., Patzold, B., Schlag, M., Gotz, F., Kalbacher, H., and Stehle, T. (2010). Structural basis of cell wall cleavage by a staphylococcal autolysin. PLOS Pathog. 6:e1000807. doi: 10.1371/journal.ppat.1000807
Zoll, S., Schlag, M., Shkumatov, A. V., Rautenberg, M., Svergun, D. I., Gotz, F., et al. (2012). Ligand-binding properties and conformational dynamics of autolysin repeat domains in staphylococcal cell wall recognition. J. Bacteriol. 194, 3789–3802. doi: 10.1128/JB.00331-12
Zuber, B., Haenni, M., Ribeiro, T., Minnig, K., Lopes, F., Moreillon, P., et al. (2006). Granular layer in the periplasmic space of gram-positive bacteria and fine structures of Enterococcus gallinarum and Streptococcus gordonii septa revealed by cryo-electron microscopy of vitreous sections. J. Bacteriol. 188, 6652–6660. doi: 10.1128/JB.00391-06
Keywords: Gram-positive bacteria, cell-surface protein, surface proteome, subcellular localization, pili/fimbriae/curli, lipoproteins, LPXTG sortase-dependent proteins, membrane proteins
Citation: Desvaux M, Candela T and Serror P (2018) Surfaceome and Proteosurfaceome in Parietal Monoderm Bacteria: Focus on Protein Cell-Surface Display. Front. Microbiol. 9:100. doi: 10.3389/fmicb.2018.00100
Received: 29 June 2017; Accepted: 16 January 2018;
Published: 14 February 2018.
Edited by:
Awdhesh Kalia, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
Konstantin V. Korotkov, University of Kentucky, United StatesChenggang Wu, McGovern Medical School, United States
Copyright © 2018 Desvaux, Candela and Serror. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mickaël Desvaux, bWlja2FlbC5kZXN2YXV4QGlucmEuZnI=
 Mickaël Desvaux
Mickaël Desvaux Thomas Candela
Thomas Candela Pascale Serror
Pascale Serror