
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 25 January 2018
Sec. Aquatic Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.00007
This article is part of the Research Topic The Responses of Marine Microorganisms, Communities and Ecofunctions to Environmental Gradients View all 22 articles
 Juan Ling1
Juan Ling1 Xiancheng Lin1,2
Xiancheng Lin1,2 Yanying Zhang1,3
Yanying Zhang1,3 Weiguo Zhou1,2
Weiguo Zhou1,2 Qingsong Yang1,2
Qingsong Yang1,2 Liyun Lin1,2
Liyun Lin1,2 Siquan Zeng1,2
Siquan Zeng1,2 Ying Zhang1,2
Ying Zhang1,2 Cong Wang1,2
Cong Wang1,2 Manzoor Ahmad1,2
Manzoor Ahmad1,2 Lijuan Long1
Lijuan Long1 Junde Dong1,3*
Junde Dong1,3*Seagrasses in coral reef ecosystems play important ecological roles by enhancing coral reef resilience under ocean acidification. However, seagrass primary productivity is typically constrained by limited nitrogen availability. Ammonia oxidation is an important process conducted by ammonia-oxidizing archaea (AOA) and bacteria (AOB), yet little information is available concerning the community structure and potential activity of seagrass AOA and AOB. Therefore, this study investigated the variations in the abundance, diversity and transcriptional activity of AOA and AOB at the DNA and transcript level from four sample types: the leaf, root, rhizosphere sediment and bulk sediment of seagrass Thalassia hemprichii in three coral reef ecosystems. DNA and complementary DNA (cDNA) were used to prepare clone libraries and DNA and cDNA quantitative PCR (qPCR) assays, targeting the ammonia monooxygenase-subunit (amoA) genes as biomarkers. Our results indicated that the closest relatives of the obtained archaeal and bacterial amoA gene sequences recovered from DNA and cDNA libraries mainly originated from the marine environment. Moreover, all the obtained AOB sequences belong to the Nitrosomonadales cluster. Nearly all the AOA communities exhibited higher diversity than the AOB communities at the DNA level, but the qPCR data demonstrated that the abundances of AOB communities were higher than that of AOA communities based on both DNA and RNA transcripts. Collectively, most of the samples shared greater community composition similarity with samples from the same location rather than sample type. Furthermore, the abundance of archaeal amoA gene in rhizosphere sediments showed significant relationships with the ammonium concentration of sediments and the nitrogen content of plant tissue (leaf and root) at the DNA level (P < 0.05). Conversely, no such relationships were found for the AOB communities. This work provides new insight into the nitrogen cycle, particularly nitrification of seagrass meadows in coral reef ecosystems.
Many investigations into the effect of ocean acidification (OA) on coral reefs have been conducted (Andersson and Gledhill, 2013), and results indicate that that marine organisms which inhabit the carbonate structures of coral reefs are important and sensitive to OA (Larkum et al., 2006). Albright et al. (2016) and Lough (2016) found that changes to pH in the seawater surrounding natural coral reefs in the southern Great Barrier Reef can significantly affect calcification rates, suggesting that OA may already be altering the growth of coral reefs. By decreasing the ocean pH, OA can affect the calcifying rates of calcifying creatures such as coralline algae with carbonate skeletons (Hendriks et al., 2014; Albright et al., 2016). However, seagrasses can increase the pH of the ambient environment via high photosynthesis rates (Lai et al., 2013; Hendriks et al., 2014). Moreover, Lamb et al. (2017) found that seagrass in coral reef ecosystems can reduce disease levels twofold in comparison with the corals located adjacent to seagrass meadows and corals at paired sites without seagrass.
Seagrass is highly productive and of great ecological importance in the marine environment. For instance, it can provide food, nursery and breeding habitats for other marine organisms inhabiting the ecosystem and nutrients for coral reefs. Nonetheless, available nitrogen is usually the main factor limiting the primary productivity of seagrass because coral reef ecosystems are largely oligotrophic. Nitrification is a key process in the nitrogen cycle in the marine environment. The first and rate-limiting step of nitrification is performed by ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB), both of which are ammonia oxidizers and responsible for converting ammonia to nitrite. However, these microbes have different phylogenetic and physiological features, resulting in significant variations in their abundance, diversity and activity under different environmental conditions. For example, AOA can adapt to a variety of habitats and account for a large proportion of marine and estuary communities (Dang et al., 2008, 2010a,b, 2013; Boyd et al., 2011; Cao et al., 2012; Rusch and Gaidos, 2013). However, Di et al. (2009), Wu et al. (2011), Zhang et al. (2014), Zheng et al. (2014) reported that AOB might play a more significant role in the ammonia oxidation process under certain conditions. Consequently, the relative contribution of AOA and AOB to ammonia oxidation is still in debate.
Zhang J. et al. (2015) suggested that the presence of certain freshwater plants, such as Iris pseudacorus, Thalia dealbata, and Typha orientalis L., affected the ecological characteristics of AOA and AOB significantly by increasing the abundance of ammonia oxidizers in the rhizosphere sediments. For the marine environment, investigations of Moin et al. (2009) and Chen and Gu (2017) revealed that the presence of Spartina alterniflora or S. patens and mangrove roots had a strong influence on the diversity and abundance of AOA and AOB in the coastal area and mangrove ecosystems, respectively. Furthermore, investigations of the ammonia-oxidizing prokaryotes have been conducted extensively in many different marine environments including the Changjiang Estuary, the Jiaozhou Bay, the tropical West Pacific Continental Margin, the Okhotsk Sea, the Sargasso Sea, the Northern South China Sea (Mincer et al., 2007; Nakagawa et al., 2007; Dang et al., 2013; Li and Gu, 2013; Newell et al., 2013; Lipsewers et al., 2014; Cao et al., 2015; Yu et al., 2016).
Regardless, almost all the above-mentioned investigations focused on the seawater and sediment using only DNA as a proxy to assess AOA and/or AOB. Few investigations have focused on ammonia-oxidizing prokaryotes in the aquatic benthic flora, particularly for the seagrass ecosystem (Zhao et al., 2014; Bernhard et al., 2016; Frame et al., 2016; Huang et al., 2016; Mejia et al., 2016; Ettinger et al., 2017). Not to mention the studies of AOA and AOB of seagrass in coral reef ecosystems aimed at revealing the transcriptional activity of relevant functional groups in their natural physiological state. Accordingly, in this study, we prepared archaeal and bacterial amoA gene DNA and cDNA libraries and performed reverse-transcription polymerase chain reaction (PCR) and real-time quantitative PCR (qPCR) to examine the community abundance, diversity and transcriptional activity of ammonia-oxidizing prokaryotes of the seagrass Thalassia hemprichii in the Luhuitou fringing reef, Sanya Bay and Yongxi Island, Xisha Islands. The aims of our investigation were as follows: (i) to evaluate the abundance and diversity of AOA and AOB communities, (ii) to compare community variations within and among the sample types and locations, and (iii) to analyze the transcriptional activity of AOA and AOB.
Seagrass T. hemprichii is one of the most widely distributed seagrass species among tropical southern Indo-Pacific flora, and exists in a monospecific or mixed-species status. The sampling locations were distributed in Sanya Bay (SYT) and Yongxing Island (AT and ST), South China Sea (Supplementary Figure S1). Samples at SYT were collected from the Luhuitou fringing reef (18°12′19″ N, 109°28′27″ E) located in Sanya Bay, Hainan Province. The average atmospheric temperature at this site is 30.74°C, with warm summers (34.75°C) and cold winters (27.20°C). The Luhuitou fringing reef area is under the effect of the northeast monsoon (cold and dry winter and spring) and southwest monsoon (warm and wet summer and autumn) during the East Asian monsoon climate (Wu et al., 2008; Cao et al., 2017). Two other sampling locations, AT (16°50′32″ N, 112°20′41″ E) and ST (16°50′6″ N, 112°22′10″ E), are located on Yongxing Island, which is a reef island formed by the accumulation of white coral skeletal material and shell sand on a reef platform. The annual average temperature on Yongxing Island is 26.5°C. This island is also under the effect of the East Asian monsoon (Shen et al., 2017).
Seagrass meadows in the Luhuitou fringing reef and the Yongxing flat reef are representative of the different styles of seagrass meadows in coral reef ecosystems. T. hemprichii appeared at the Luhuitou fringing reef after the coral reef has degraded, and it is the only seagrass species present. In the Yongxing Islands, the seagrass is found in a mixed-species status, with seagrasses Syringodium isoetifolium, Halodule uninervis, and Halophila ovalis at AT, whereas T. hemprichii was dominant over seagrass H. ovalis at ST. In comparison with the Yongxing flat reef, Luhuitou fringing reef areas show higher nutrient concentration, particularly nitrogen, which was attributed to the increasing anthropogenic activity (Cao et al., 2017).
Samples from the Luhuitou fringing reef and Yongxing flat reef were collected on May 28th and June 1st, 2015, respectively. Sampling was carried out according to the methods of Jensen et al. (2007) at low tide. Plants with surrounding sediment were randomly collected using a spade, and immediately transported in sterilized boxes for subsequent subgrouping in triplicate. Sediment from the plant roots and associated invertebrates from leaves and roots were separated by washing with autoclaved seawater. Bulk sediment was also collected at the same area. All sediment samples were collected in triplicate at each location and thoroughly homogenized using a sterilized spoon. Samples collected from one site were divided into four sections: leaves (L); rhizomes and roots (R); rhizosphere sediment (RS) and bulk sediment (S). All samples for DNA/RNA analysis were stored in sample protectors (TaKaRa, Dalian, China), frozen immediately, and stored at -80°C until further analysis.
Environmental data, samples used for microbial analysis and physiochemical analysis were collected simultaneously. The temperature and salinity of the seawater adjacent to seagrass samples (within 3 cm) was measured using a YSI 6600V2 water quality sonde (YSI, Yellow Springs, OH, United States). Dissolved oxygen (DO) concentrations and pH values were measured using a portable pH/DO Meter (Thermo Fisher Scientific, Inc., Beverly, MA, United States). Inorganic nutrients in seawater, including ammonium, nitrate, nitrite, and phosphate, were measured using standard methods as described previously (Huang et al., 2003). Nitrogen and carbon content of seagrass tissues (L and R) were determined according to Lee et al. (2004), and phosphorus content was analyzed by the colorimetric analysis of phosphate concentration (Fourqurean et al., 1992). Chemical data (Nitrate, ammonium and active phosphorous) of sediments were determined by using standard oceanographic methods (General Administration of Quality Supervision, Inspection and quarantine of the People’s Republic of China, 2002).
DNA and RNA from approximately 1 g of sample (sediment or plant tissue; wet weight) were extracted using the E.Z.N.A® Soil DNA kit and E.Z.N.A® Soil RNA kit (Omega Bio-tek, Norcross, GA, United States) according to the manufacturer’s protocols. Synthesis of cDNA from extracted RNA was performed according to Li and Gu (2013). The nucleic acid concentrations were quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, United States). All qualified DNA and cDNA were stored at -80°C until analysis. For clone library analysis, archaeal and bacterial amoA gene sequences were amplified using the primer sets Arch-amoA F (5′-GGGGTTTCTACTGGTGGT-3′) and Arch-amoA R (5′-CCCCTCKGSAAAGCCTTCTTC-3′) (Francis et al., 2005) and amoA-1F (5′-STAATGGTCTGGCTTAGACG-3′) and amoA-2R (5′-GGGGTTTCTACTGGTGGT-3′) (Rotthauwe et al., 1997), respectively.
PCR reaction mixture for amplifying the amoA gene was prepared in accordance with details described by Hu et al. (2011). The PCR amplification conditions for amoA gene in AOA and AOB were in accordance with previously established protocols (Rotthauwe et al., 1997; Francis et al., 2005; Schmid et al., 2005; Li et al., 2011). The amplification was performed as follows: 5 min at 95°C, followed by 30 cycles of 45 s at 95°C, 60 s at 53°C and 60 s at 72°C, and 10 min at 72°C. For bacterial amoA gene amplification, the PCR conditions were 5 min at 95°C, followed by 30 cycles of 45 s at 95°C, 90 s at 56°C and 60 s at 72°C, and 10 min at 72°C. The PCR products from three reactions were pooled together to minimize PCR amplification bias, purified, and ligated into pMD18-T vector (TaKaRa, Dalian, China) according to the manufacturer’s instructions. Recombinant Escherichia coli cells were inoculated in Luria-Bertani broth containing ampicillin and incubated overnight at 37°C, and the plasmids carrying the target genes were extracted using a MiniBEST Plasmid Purification kit (TaKaRa, Dalian, China). Cloned amoA gene fragments were reamplified using primers M13-F (5′-AGGGTTTTCCCAG-TCACGACG-3′) and RV-R (5′-AGCGGATAACAATTTCACACAGG-3′). The target fragment sizes of archaeal and bacterial amoA genes were 491 and 635 bp, respectively. The PCR products were cloned into the pMD18-T vectors (TaKaRa, Dalian, China). PCR products were screened for the correct size and purity by 1% agarose gel electrophoresis, and clones showing the correct size were sequenced.
Absolute quantification of archaeal and bacterial amoA genes were determined for both DNA and cDNA using qPCR in triplicate reactions with the LightCycler 480 System (Roche Diagnostics, Mannheim, Germany) and the following conditions: 95°C for 30 s, followed by 40 cycles of 30 s at 95°C, 60 s at 56°C, and 60 s at 72°C for archaeal amoA gene, or 95°C for 30 s, followed by 40 cycles of 30 s at 95°C, 60 s at 58°C, and 35 s at 72°C for bacterial amoA gene. To construct standard curves, archaeal and bacterial amoA genes were cloned into the pMD18-T vector (TaKaRa, Dalian, China) and then transformed into E. coli DH5a. The methods were similar to those used for the clone library construction. Recombinant plasmids carrying the target genes were extracted using a TaKaRaMini BEST Plasmid Purification Kit and quantified with a NanoDrop 2000 spectrophotometer. The copy numbers of amoA gene from the extracted plasmids were calculated by the concentrations and average base pairs of the plasmid. Standard curves for the archaeal amoA gene were constructed using standard plasmids obtained from the most dominant genotype clone ARSYTL81(KY794979), and for the bacterial amoA gene from the most dominant genotype clone BRNASYTRS53 (KY795002). Standard curves ranging from 103 to 108 gene copies/μL were obtained using 10-fold serial dilutions of linearized plasmid pMD-18T containing the cloned archaeal and bacterial amoA genes, respectively.
Real-time PCR efficiencies for AOA and AOB for DNA and cDNA were calculated according to E = 10 [-1/slope] (Rasmussen, 2001). The results showed that the amplification efficiencies ranged from 94.5 to 101%, with an R2 of standards higher than 0.99. The specificity of the amplification products was confirmed by melt-curve analysis, and the amplified fragments were checked by electrophoresis in 1.0% gel to confirm the expected sizes of amplicons. The size of archaeal and bacterial amoA genes were 491 and 635 bp, respectively. As the gene copies in the 1 μL of template DNA were determined, the final amoA gene and cDNA abundance of the seagrass and sediment samples were obtained by calculation. The results were expressed as gene copy abundance per gram of sediment or plant tissue (wet weight).
The obtained DNA and cDNA sequences were examined and checked for chimeras using the Check Chimera program of the Ribosomal Database Project (Cole et al., 2007). The operational taxonomic unit (OTU) reads were checked against a local amoA gene database (Ribosomal Database Project FunGene1) and the NCBI database2. Diversity indices were also evaluated using the MOTHUR program (Schloss et al., 2009; Li and Gu, 2013). Diversity statistics, including Shannon–Wiener (H′), Simpson (D) and species richness estimator (SChao1), were calculated. Library coverage (C) was calculated as [1-(n/N)] × 100, where n is the number of OTUs represented by one clone (singletons) and N is the total number of sequences (Good, 1953). Diversity indices and richness estimators are useful statistical methods for comparing the relative complexity of AOA and AOB communities and for assessing the completeness of sample analysis. Reference sequences were selected by comparison with the GenBank database using BLAST, and the closest matches were included in the alignment and phylogenetic analysis with MEGA 6 (JCVI, Rockville, MD, United States) through neighbor-joining trees using Kimura 2-parameter distance with 1000 replicates to produce bootstrap values (Tamura et al., 2013).
One-way statistical analysis of variance (ANOVA) (confidence limit of 95%, P < 0.05) was performed to analyze variables among the three locations. In addition, one-way analysis of similarity (ANOSIM) was performed based on Bray-Curtis distances of AOA and AOB communities among the three sampling locations using the PRIMER v.6 software package (PRIMER-E, Plymouth, WA, United States) (Clark and Gorley, 2006). Moreover, Pearson’s correlation analysis of the abundance and diversity of AOA and AOB with the determined physicochemical parameters (water, tissue and sediment) was analyzed by SPSS v19.0 software (IBM, Inc., Chicago, IL, United States). Multi-Variate Statistical Package (MVSP, version 3.2, Kovach Computing Services, Anglesey) software was used to construct the similarity matrix and dendrograms (Kovach, 1999). In addition, genetic similarities among all clones were calculated by the percent similarity coefficient. Principal coordinate analysis (PCoA) was employed to depict the general ordination patterns of all samples at both DNA and transcript levels. In addition, the weighted pair-group method with arithmetic analysis (WPGMA) was used to generate similarity matrices and dendrograms by percent similarity using MVSP (Kovach, 1999).
Representative sequences of archaeal and bacterial amoA genes for each OTU reported in this study have been deposited in the GenBank database under accession numbers MF796361–MF796384 and MF796347–MF796360, respectively.
The features of four different types of samples from the collection locations were analyzed, and the results are listed in Supplementary Table S1. The pH and salinity of all sampling locations ranged from 8.22–8.39 and 24.70–27.87‰, respectively. The highest DO concentration was recorded at AT, and the lowest at site SYT. The nitrate concentrations for the three locations ranged from 0.014 to 0.025 μM. The pH, salinity and DO concentration at SYT was significantly lower than at AT and ST, though the concentration of nitrite and ammonium were higher at SYT than at AT and ST (P < 0.05). Nonetheless, there was no significant difference in the nitrate concentrations among the three sampling sites (P > 0.05). The highest carbon content was found in the leaves from AT and the lowest value was recorded in the leaves from ST. The nitrogen content, phosphorus content and carbon percentage of roots and leaves from the three different locations were all significantly different (P < 0.05). The values of the bulk sediment environmental parameters (nitrate, ammonium, and active phosphorous concentrations) were all below the limit of detection. As illustrated in Supplementary Table S1, the ammonium and active phosphorous contents of the three RS samples ranged from 1.45 to 5.97 mg/kg and 14 to 15 mg/kg, respectively. Statistically, these three environmental parameters of AT, ST, and SYT rhizosphere sediments were significantly different (P < 0.05).
The abundances of AOA and AOB amoA genes at the DNA and transcript levels as determined by real-time PCR are shown in Figure 1. The abundance of all AOB communities (with the exception of that in sample SYTL) was higher than that of AOA at both the DNA and transcript levels. The abundance of AOA communities at the DNA level ranged from below the detection limit to 5.35 × 106 gene copies per gram of sample (L, R, RS, and S); the abundance of AOB communities at the DNA level was between 7.68 × 105 and 5.74 × 106 gene copies per gram of sample (wet weight). In addition, the abundance of AOA communities at the transcript level was one order of magnitude lower than that at the DNA level. Their abundance at the DNA level ranged from below the detectable levels to 6.45 × 106 copies g-1 per gram of sample, and the abundance of AOB communities had a wide range, from below the limit of detection to 1.15 × 106 copies g-1 per gram of sample. Moreover, the ratio of AOB to AOA at the DNA level was highly variable in all samples. The highest value 42.21 was detected in the SYTRS sample (Supplementary Table S2). The ratio of DNA to cDNA for amoA gene copies for AOA community in sample SYT was greater than 20 and greater than 200 for sample SYTL. However, the values for AOB communities ranged only from 2.04 to 10.74 (Supplementary Table S2). When combing all the samples, the abundance of AOB communities was higher than that of AOA.
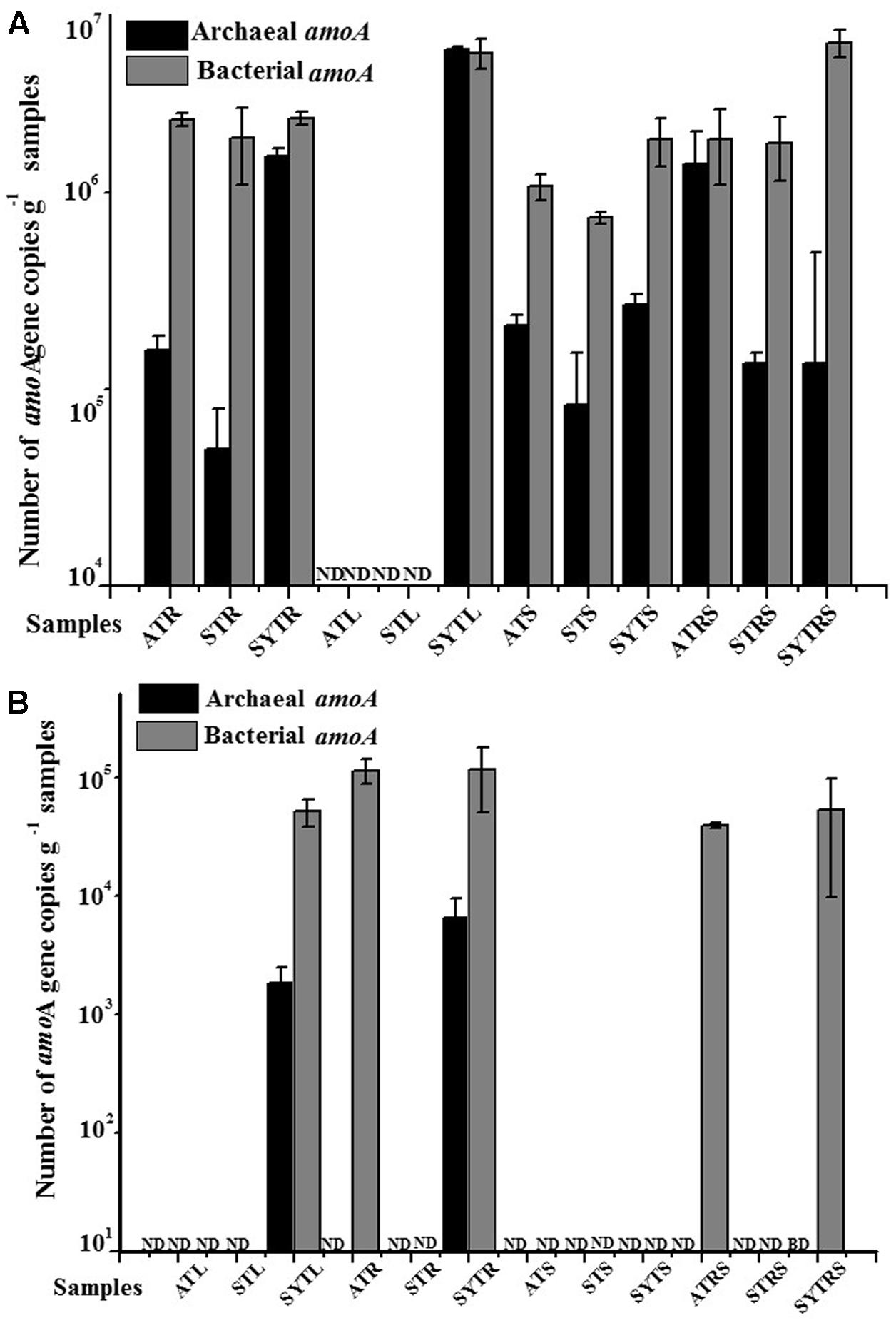
FIGURE 1. Abundance of the archaeal and bacterial amoA genes at the DNA (A) and cDNA (B) levels in the different samples. ND, not determined; BD, below limit of detection.
Bacterial amoA gene libraries from 13 samples (10 DNA-based and 3 cDNA-based) were successfully constructed. Overall, DNA sequences from 342 clones and cDNA sequences from 94 clones for archaea were recovered, while for bacterial amoA gene, there were 255 clones at the DNA level and 228 clones at the transcript level (Supplementary Tables S3, S4). Pearson’s correlation analysis revealed that the abundance of AOA at the DNA level in all RS showed significant positive relationships with the concentrations of seawater ammonium and the nitrogen contents of seagrass roots and leaves (P < 0.05), respectively. However, there was no such relationship between the environmental parameters and the abundance of AOB communities.
The number of sequenced clones differed among samples (ranging from 17 to 47), and they were then used to calculate diversity estimators (Table 1). The coverage, diversity, and richness indices of the nine cDNA-based libraries are summarized in Table 1. The coverage for AOA and AOB at the two different levels ranged from 72.73 to 100% and 94.59 to 100%, respectively (Table 1). Consequently, our results might have reflected the majority of archaeal and bacterial amoA gene clones at the DNA and transcript levels in our samples (Figures 2A,B).
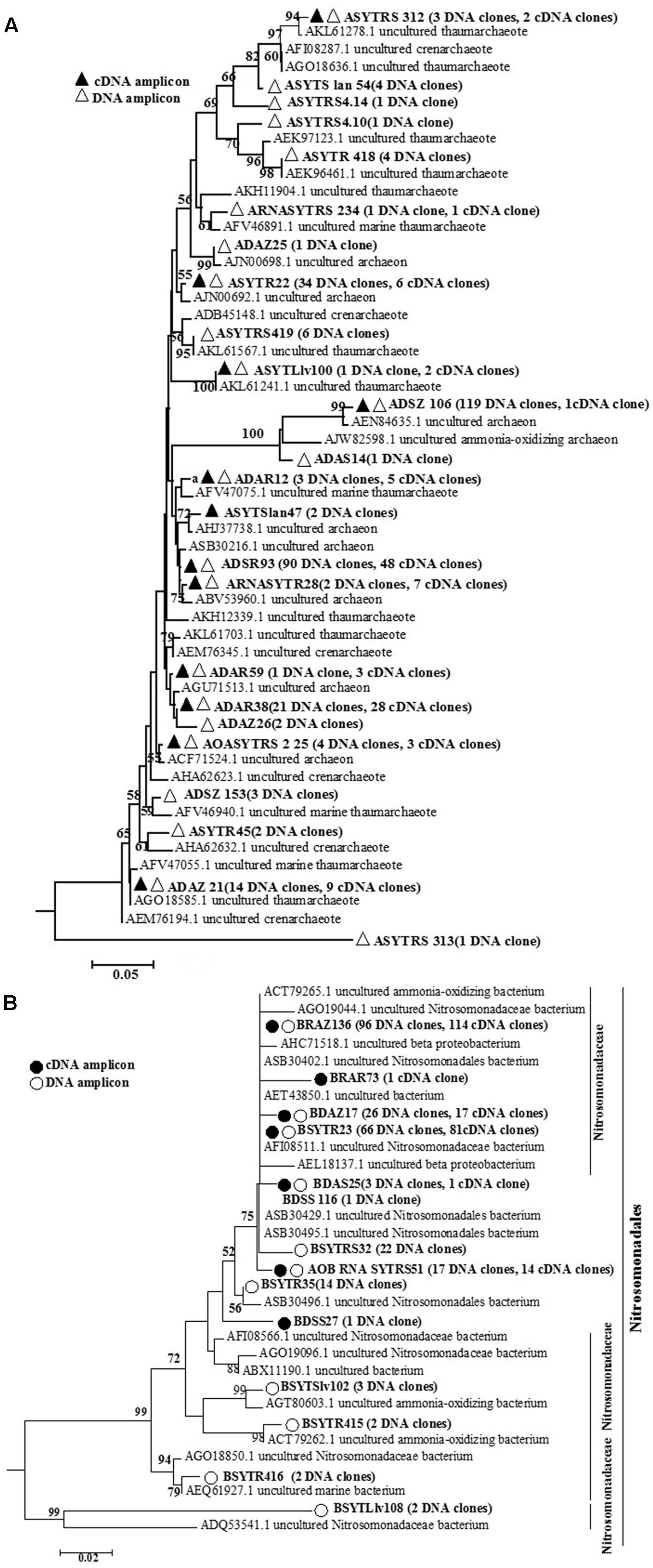
FIGURE 2. Phylogenetic tree constructed using distance and neighbor-joining method for archaeal amoA sequences (A) and bacterial amoA gene sequences (B) translated from cloned archaeal amoA and bacterial amoA gene sequences at the DNA and transcript levels, as recovered from the seagrass Thalassia hemprichii and their closest matches in GenBank from DNA samples and cDNA samples. Bootstrap values greater than 50% of 100 resamplings are shown near the nodes.
The biodiversity and richness indices of AOA and AOB communities at the DNA and cDNA levels are presented in Table 1. Rarefaction analyses were performed for all the bacterial or archaeal amoA gene clone libraries (Supplementary Figure S2). The highest diversity indices for AOB at the two levels were found for SYT. Overall, the indices indicated that AOB were less diverse than those for AOA, and the diversity indices at the DNA level were higher than those at the transcript level.
BLAST results indicated that over 80% of the obtained sequences recovered from this study were related to the sequences of marine sources, such as the marine water column, intertidal and marine sediments, mangrove sediments and marine sponges. The 24 archaeal amoA gene sequences share 87.50–99.48% sequence similarity with the closest GenBank matches. For bacterial amoA gene sequences, similarity of the 14 bacterial amoA genes ranged from 98.36 to 99.79%.
For AOA communities, 24 OTUs and 12 OTUs were found at the DNA and transcript levels, respectively, with 11 OTUs shared at both levels (Figure 2 and Table 1). For AOB communities, the number decreased to 14 OTUs and 7 OTUs, with 6 OTUs shared at the two levels (Figure 2 and Table 1). Based on the phylogenetic analysis, all bacterial amoA gene sequences (256 DNA sequences and 194 cDNA sequences) were mainly grouped into the Nitrosomonadaceae cluster (9 OTUs: 9 DNA sequences and 3 cDNA sequences) (Figure 2B).
The diversity indices showed no significant relationships between the values of H′ and SChao1 with the concentrations of ammonium and phosphate (P > 0.05) for both AOA and AOB communities at the DNA level, respectively. However, our investigation revealed that there was a significant negative relationship between SChao1 and the concentration of nitrite for AOB communities (P < 0.05).
For the archaeal amoA gene libraries, 1 to 11 and 5 to 9 OTUs at the DNA and transcript levels, respectively, were found for different samples. For AOB, only 2 to 5 and 1 to 2 OTUs were obtained for different clones at the DNA and transcript levels, respectively. Some OTUs could be found among almost all the samples, such as ADSR 93 for AOA at the DNA level, sharing approximately 99.37% similarity with an uncultured archaeon clone (KY357274) isolated from mangrove sediment. For AOB at the DNA level, the OTU BSYTR23 exhibited 99.79% similarity with the uncultured bacterial clone HaAOB1 (JN177536) retrieved from marine sponges. However, other OTUs were only detected in ATR, such as the OTU BRAZ73 at the transcript level. The BLAST result for OTU BRAZ73 indicated high similarity to a clone (KC893630) retrieved from the marine sponge Spheciospongia vesparium.
As shown in Supplementary Figure S3, samples obtained from the same location tended to group together regardless of the sample type. For instance, AT and ST samples first grouped with samples obtained from the same location and then grouped together, whereas, SYTS, SYTRS, and SYTR samples shared a high similarity of community composition (Supplementary Figure S3). For the DNA-based analysis, the first two principal coordinates (P1 and P2) could explain 70.40 and 76.77% of the total community variability in PCoA in archaeal and bacterial ammonia oxidizers, respectively (Figures 3A,B). The percentage of variability explained by the first two principal coordinates was 87.10% at the transcript level for AOB (Figure 3C). The WPGMA results were consistent with the PCoA plots (Figure 3). Consequently, the AOA and AOB communities shared higher similarity within the same location than within the same type of samples.
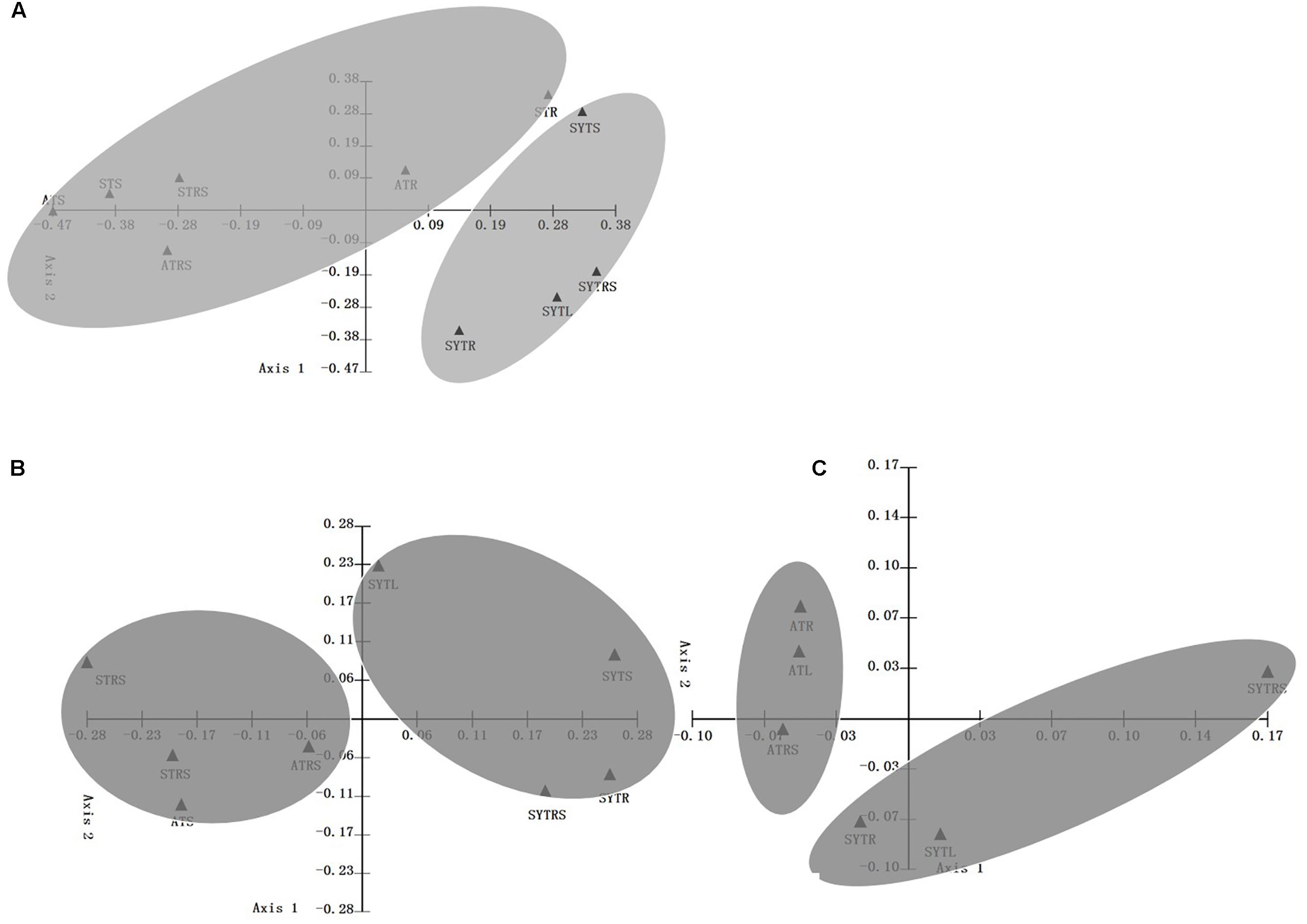
FIGURE 3. Principal coordinate analysis (PCoA) with a weighted UniFrac algorithm using archaeal and bacterial amoA gene sequences recovered from the seagrass T. hemprichii in coral reef ecosystems. Shown are the plots of the first two principal coordinate axes for PCoA and the distribution of AOA (A: DNA level) and AOB (B: DNA level; C: transcript level) (designated with the sampling station names) communities in response to these axes.
All samples collected from the three locations were analyzed by DNA-based and transcript-based approaches. Most of the archaeal and bacterial amoA gene sequences at the DNA level in this study were successfully recovered, whereas a few samples at the transcript level were retrieved. This may be due to low gene copy number or expression of amoA gene in the relatively oligotrophic coral reef ecosystems, resulting in an abundance below the limit of detection. Compared with samples from AT and ST, archaeal and bacterial amoA gene sequences in most samples collected at SYT were successfully recovered. Previous investigations demonstrated that environmental factors, such as ammonia, temperature, salinity, dissolved oxygen and pH, had strong influences on the distribution of AOA and AOB (Li et al., 2011; Cao et al., 2015; Wang et al., 2015). For instance, Li and Gu (2013) showed that the diversity, abundance, and transcriptional activity of AOA and AOB shift in response to N conditions, specifically noting that ammonium amendment increased diversity and a lower nitrite concentration may reduce AOA and AOB diversity. Low temperature also exerted an important effect on the composition of AOA and AOB communities, which exhibited the lowest diversity when exposed to cold water (Urakawa et al., 2008). Different species respond differently to environmental variation, and the results of our investigation revealed the ammonium concentration to be a decisive factor for AOA and AOB community composition.
A stimulation experiment conducted by Prosser and Nicol (2012) suggested that AOA grew faster than AOB at lower ammonia concentrations, as the AOA affinity for ammonium was up to 200-fold that of AOB (Martens-Habbena et al., 2009, 2015). It has also been reported that AOA prefer to inhabit environments with lower ammonia concentrations and have higher amoA gene transcriptional activity than AOB in ammonia-limited water environments. In addition, Takano et al. (2010) discovered that deep-sea archaea adopted the strategy of recycling membrane lipids between growing cells and the surrounding sediment for saving energy to thrive in low ammonium habitats. However, in ammonia-rich areas, AOB communities would be the dominant component and contribute more to ammoxidation (Zhang Y. et al., 2015). Furthermore, plant species and their densities have crucial roles in determining community composition (Wang et al., 2015; Ettinger et al., 2017). Li et al. (2011) found that the presence of mangroves to increase the abundance of AOA and AOB in the mangrove sediment.
The qPCR quantification results presented in this study suggested that bacterial amoA gene abundance in almost all samples was higher than that of the archaeal amoA gene copy number with the exception of sample SYTL (Figure 1). The bacterial amoA gene abundance in the South China Sea was reported to range from 4.24 × 104 to 1.99 × 106 copies per gram of sediment (wet weight), which was consistent with our results (Cao et al., 2012). In addition, Dang et al. (2010a) found that the abundance of β-AOB was much higher than that of the archaeal amoA gene, and Wang et al. (2015) reported that the abundance of AOB at the transcript level was two orders of magnitude higher than that of AOA in a mangrove ecosystem. These findings were in agreement with our results. Furthermore, plants affected the bacterial community composition and activity by competing with rhizosphere microbes for nutrients, such as ammonium, nitrate, urea, and amino acids as nitrogen sources (Skiba et al., 2011). Foreseeably, microbes in rhizosphere sediment may utilize the low molecular weight compounds diffused from plant roots as carbon sources (Philippot et al., 2013). The ratio of β-AOB-amoA/archaeal amoA ranged from 212: 1 to 3090: 1 in deep-sea methane seep sediments of the Okhotsk Sea (Dang et al., 2010b). By comparison, the abundance ratio of AOA to AOB in our study was much lower, ranging from 0.96: 1 to 42.21: 1 (Supplementary Table S2).
The clusters of archaeal amoA gene sequences obtained in this investigation were mainly from uncultured Thaumarchaeota originating from the marine environment. A chemolithoautotrophic marine crenarchaeote has been isolated, and its role in relation to nitrification has been shown to contribute significantly to global nitrogen and carbon cycles (Könneke et al., 2005). In addition, thaumarchaeotes have been found to play a crucial role in nitrification in both marine and terrestrial environments (Leininger et al., 2006; Wuchter et al., 2006; Beman et al., 2008; Erguder et al., 2009; Martens-Habbena et al., 2009). Moreover, Thaumarchaeota accounted for almost 12% of all archaeal sequences retrieved in Checker Reef sediments, and these organisms preferred oxic rather than anoxic sediments (Rusch et al., 2009; Gaidos et al., 2011; Pester et al., 2011). Beman et al. (2007) analyzed AOA communities associated with coral colonies from nine coral species and four different reef locations in the Gulf of California. Their results showed that amoA sequences were broadly distributed phylogenetically and that their closest relatives were related to sequences from coastal/estuarine sediments and oceanic water column sources. Conversely, they obtained no bacterial amoA gene sequences (Beman et al., 2007).
In our study, the most abundant OTU, ADSZ106 (119 clones at the DNA level and 1 clone at the transcript level), shared 93.96% similarity with uncultured archaeon clone S1–24 (KC758384) from saltwater aquaria. The bacterial amoA gene in our investigation was primarily affiliated with the cluster of Nitrosomonadaceae at the DNA and transcript levels (Figures 2A,B), accounting for 71.42% of all OTUs (Figure 2B). Many investigations have showed that most cultured AOB belonged to the family Nitrosomonadaceae, phylum Betaproteobacteria (Koops and Pommerening-Roser, 2001). Moreover, based on the species features, such as affinity for ammonia, and tolerance to salt and nitrite, microbes in this taxon could be further subgrouped into several clusters (Koops et al., 2006).
At the DNA level, the AOA communities were not significantly different between the sampling locations (P > 0.05), while for AOB communities, there were significant difference between SYT and AT (P < 0.05) and between AT and ST (P < 0.05) (Table 2). For all ammonia-oxidizing prokaryotes, some OTUs were universally present in all samples, whereas others occurred in only a few samples. For instance, some unique OTUs, e.g., ASYTRS419, ASYTRS312, and SYTRS313, were detected only in the samples collected at SYT. Zhao et al. (2012) reported many human activities, such as overfishing, reef rock digging and tourism activities in Sanya Bay, and all of these factors in combination with climate change have led to a significant decline in coral cover since the 1960s. The seagrass T. hemprichii gradually colonized under these environmental conditions at that location.

TABLE 2. One-way ANOSIM results for the AOA and AOB communities between the sampling locations Sanya Bay (SYT) and Yongxing Island (AT and ST) at the DNA level.
A positive correlation between amoA gene copy numbers and the potential nitrification rate has been recorded (He et al., 2007). Consequently, quantitative assays targeting amoA gene transcripts were carried out in this study to analyze potential nitrification by AOA and AOB in coral reef ecosystems. The results showed that AOB would contribute more to the first step of nitrification for T. hemprichii (Figure 1). Furthermore, the results of an experiment conducted in the mangrove ecosystem were also consistent with our findings (Cao et al., 2015). However, Feng et al. (2016) obtained conflicting results for a marine sponge, for which the abundance of AOA was much higher than that of AOB at the cDNA level.
In our study, 24 OTUs and 14 OTUs were detected at the DNA level for AOA and AOB, respectively, and the number of OTUs decreased to 12 and 6, respectively, at the transcript level. This may be due to different preferences for ammonia. Hence, under the same condition, some species were dormant or below the PCR sensitivity threshold, whereas others exhibited high activity (Feng et al., 2016). For example, one unique bacterial amoA gene, OTU BDSS27, was detected only at the transcript level, and it was found to be related to uncultured ammonia-oxidizing bacterium clone ML-amoA-0 (FJ652557). A similar circumstance had been reported in the study of a marine sponge (Feng et al., 2016). This could be due to low abundance at the DNA level but high transcriptional activity. The most active amoA gene OTU in AOA communities was ADSR93, which was related to the uncultured archaeon clone GZ16110300849 (KY357274) obtained from mangrove sediments. Moreover, the most active bacterial amoA gene was the OTU BRAZ136 with 96 clones at the DNA level and 114 clones at the transcript level. Its closest BLAST hit was the uncultured bacterial clone HaAOB1 (JN177536) originating from the marine sponge Haliclona sp. collected from China East China at the depth of 20 m. Marine sponges are indispensable components of coral reef ecosystems and can help coral reefs thrive in ocean deserts because they absorb the nutrients from seawater and convert them into food for the reef and other marine organisms. Functional gene expression of ammonia-oxidizing microorganisms would also be altered by the health of their hosts. López-Legentil et al. (2010) reported that amoA gene expression was higher in fatally bleached sponges, whereas different patterns were observed in cyclic bleaching corals. Therefore, a higher abundance of AOB would have a more important function in transforming excess ammonia in the niche to maintain the healthy host.
We herein described the abundance, diversity and transcriptional activity of the AOA and AOB communities of the seagrass T. hemprichii in three coral reef ecosystems at the DNA and cDNA levels. The diversity of AOA communities was higher than that of AOB, though the abundance of AOB communities was greater than that of AOB, and the community compositions of the sampling locations were distinct. As the focuses of this study were community composition and potential AOA and AOB activity, the amount of ammonia oxidized by AOA and AOB for the growth of seagrass, which is crucial for discerning the roles of ammonia-oxidizing microbes in ecosystems, was not determined. Consequently, the pattern for protein expression pattern of the amoA gene product and the 15N-isotope method would be used for elucidating their contributions to the seagrass productivity and their nitrogen transfer pathways for future investigations.
JL, LjL, and JD conceived the research. JL and XL performed the experiments. JL wrote the manuscript. YaZ and MA edited the manuscript. QY, LL, SZ, YiZ, and CW contributed to sampling or data analysis. All authors reviewed and approved the manuscript.
The research was supported by the National Natural Science Foundation of China (41676163, 41406191, 41676107 and 41276114), the Strategic Priority Research Program of the Chinese Academy of Sciences (13020300 and 11020202), National Key Research and Development Program of China (2017YFC0506301), the Guangdong Province Public Welfare Research and Capacity Building Project (2015A020216016), the Science and Technology Planning Project of Guangdong Province, China (2017B030314052), and the China Scholarship Council (201704910156).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Dr. Lauren Hale at the University of Oklahoma for her helpful edits. They thank all of the members of the Tropical Marine Biological Research Station and Xisha/Nansha Ocean Observation and Research Station in Hainan for their work in the field sample collection.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00007/full#supplementary-material
Albright, R., Caldeira, L., Hosfelt, J., Kwiatkowski, L., Maclaren, J. K., Mason, B. M., et al. (2016). Reversal of ocean acidification enhances net coral reef calcification. Nature 531, 362–365. doi: 10.1038/nature17155
Andersson, A. J., and Gledhill, D. K. (2013). Ocean acidification and coral reefs: effects on breakdown, dissolution, and net ecosystem calcification. Annu. Rev. Mar. Sci. 5, 321–348. doi: 10.1146/annurev-marine-121211-172241
Beman, J. M., Popp, B. N., and Francis, C. A. (2008). Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2, 429–441. doi: 10.1038/ismej.2007.118
Beman, J. M., Roberts, K. J., Wegley, L., Rohwer, F., and Francis, C. A. (2007). Distribution and diversity of archaeal ammonia monooxygenase genes associated with corals. Appl. Environ. Microbiol. 73, 5642–5647. doi: 10.1128/AEM.00461-07
Bernhard, A. E., Sheffer, R., Giblin, A. E., Marton, J. M., and Roberts, B. J. (2016). Population dynamics and community composition of ammonia oxidizers in salt marshes after the Deepwater Horizon oil spill. Front. Microbiol. 7:854. doi: 10.3389/fmicb.2016.00854
Boyd, E. S., Lange, R. K., Mitchell, A. C., Havig, J. R., Hamilton, T. L., Lafreniere, M. J., et al. (2011). Diversity, abundance, and potential activity of nitrifying and nitrate-reducing microbial assemblages in a subglacial ecosystem. Appl. Environ. Microbiol. 77, 4778–4787. doi: 10.1128/AEM.00376-11
Cao, D., Cao, W., Yu, K., Wu, G., Yang, J., Su, X., et al. (2017). Evaluation of anthropogenic influences on the Luhuitou fringing reef via spatial and temporal analyses (from isotopic values). J. Geophys. Res. Oceans 122, 4431–4443. doi: 10.1002/2017JC012871
Cao, H., Hong, Y., Li, M., and Gu, J. D. (2012). Community shift of ammonia-oxidizing bacteria along an anthropogenic pollution gradient from the Pearl River Delta to the South China Sea. Appl. Microbiol. Biotechnol. 94, 247–259. doi: 10.1007/s00253-011-3636-1
Cao, H., Zhang, W., Wang, Y., and Qian, P. (2015). Microbial community changes along the active seepage site of one cold seep in the Red Sea. Front. Microbiol. 6:739. doi: 10.3389/fmicb.2015.00739
Chen, J., and Gu, J. D. (2017). Faunal burrows alter the diversity, abundance, and structure of AOA, AOB, anammox and n-damo communities in coastal mangrove sediments. Microb. Ecol. 74, 140–156. doi: 10.1007/s00248-017-0939-5
Cole, J. R., Chai, B., Farris, R. J., Wang, Q., Kulam-Syed-Mohideen, A. S., McGarrell, D. M., et al. (2007). The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35, 169–172. doi: 10.1093/nar/gkl889
Dang, H., Li, J., Chen, R., Wang, L., Guo, L., Zhang, Z., et al. (2010a). Diversity, abundance, and spatial distribution of sediment ammonia-oxidizing Betaproteobacteria in response to environmental gradients and coastal eutrophication in Jiaozhou Bay, China. Appl. Environ. Microb. 76, 4691–4702. doi: 10.1128/AEM.02563-09
Dang, H., Li, J., Zhang, X., Li, T., Tian, F., and Jin, W. (2009). Diversity and spatial distribution of amoA-encoding archaea in the deep-sea sediments of the tropical West Pacific Continental Margin. J. Appl. Microbiol. 106, 1482–1493. doi: 10.1111/j.1365-2672.2008.04109.x
Dang, H., Luan, X., Chen, R., Zhang, X., Guo, L., and Klotz, M. G. (2010b). Diversity, abundance and distribution of amoA-encoding archaea in deep-sea methane seep sediments of the Okhotsk Sea. FEMS Microbiol. Ecol. 72, 370–385. doi: 10.1111/j.1574-6941.2010.00870.x
Dang, H., Zhang, X., Sun, J., Li, T., Zhang, Z., and Yang, G. (2008). Diversity and spatial distribution of sediment ammonia-oxidizing crenarchaeota in response to estuarine and environmental gradients in the Changjiang Estuary and East China Sea. Microbiology 154, 2084–2095. doi: 10.1099/mic.0.2007/013581-0
Dang, H., Zhou, H., Yang, J., Ge, H., Jiao, N., Luan, X., et al. (2013). Thaumarchaeotal signature gene distribution in sediments of the northern South China Sea: an indicator of the metabolic intersection of the marine carbon, nitrogen, and phosphorus cycles? Appl. Environ. Microbiol. 79, 2137–2147. doi: 10.1128/aem.03204-12
Di, H., Cameron, K., Shen, J., Winefield, C., O’Callaghan, M., Bowatte, S., et al. (2009). Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat. Geosci. 2, 621–624. doi: 10.1038/ngeo613
Erguder, T. H., Boon, N., Wittebolle, L., Marzorati, M., and Verstraete, W. (2009). Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol. Rev. 33, 855–869. doi: 10.1111/j.1574-6976.2009.00179.x
Ettinger, C. L., Voerman, S. E., Lang, J. M., Stachowicz, J. J., and Eisen, J. A. (2017). Microbial communities in sediment from Zostera marina patches, but not the Z. marina leaf or root microbiomes, vary in relation to distance from patch edge. Peer J. 27:e3246. doi: 10.7717/peerj.3246
Feng, G., Sun, W., Zhang, F., Karthik, L., and Li, Z. (2016). Inhabitancy of active Nitrosopumilus-like ammonia-oxidizing archaea and Nitrospira nitrite-oxidizing bacteria in the sponge Theonella swinhoei. Sci. Rep. 6:24966. doi: 10.1038/srep24966
Fourqurean, J. W., Zieman, J. C., and Powell, G. V. (1992). Phosphorus limitation of primary production in Florida Bay: evidence from C: N: P ratios of the dominant seagrass Thalassia testudinum. Limnol. Oceanogr. 37, 162–171. doi: 10.4319/lo.1992.37.1.0162
Frame, C. H., Lau, E., Nolan, E. J., Goepfert, T. J., and Lehmann, M. F. (2016). Acidification enhances hybrid N2O production associated with aquatic ammonia-oxidizing microorganisms. Front. Microbiol. 27:2104. doi: 10.3389/fmicb.2016.02104
Francis, C. A., Roberts, K. J., Beman, J. M., Santoro, A. E., and Oakley, B. B. (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U.S.A. 102, 14683–14688. doi: 10.1073/pnas.0506625102
Gaidos, E., Rusch, A., and Ilardo, M. (2011). Ribosomal tag pyrosequencing of DNA and RNA from benthic coral reef microbiota: community spatial structure, rare members and nitrogen-cycling guilds. Environ. Microbiol. 13, 1138–1152. doi: 10.1111/j.1462-2920.2010.02392.x
General Administration of Quality Supervision, Inspection and quarantine of the People’s Republic of China (2002). Marine Sediment Quality (GB 18668-2002). Beijing: Standards Press of China.
Good, I. J. (1953). The population frequencies of species and the estimation of population parameters. Biometrika 40, 237–264. doi: 10.2307/2333344
He, J. Z., Shen, J. P., Zhang, L. M., Zhu, Y. M., Zheng, Y. G., Xu, M. G., et al. (2007). Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ. Microbiol. 9, 2364–2374. doi: 10.1111/j.1462-2920.2007.01358.x
Hendriks, I. E., Olsen, Y. S., Ramajo, L., Basso, L., Steckbauer, A., Moore, T. S., et al. (2014). Photosynthetic activity buffers ocean acidification in seagrass meadows. Biogeosciences 11, 333–346. doi: 10.5194/bg-11-333-2014
Hu, A., Jiao, N., and Zhang, C. L. (2011). Community structure and function of planktonic Crenarchaeota: changes with depth in the South China Sea. Microb. Ecol. 62, 549–563. doi: 10.1007/s00248-011-9866-z
Huang, L., Dong, H., Jiang, H., Wang, S., and Yang, J. (2016). Relative importance of advective flow versus environmental gradient in shaping aquatic ammonium oxidizers near the Three Gorges Dam of the Yangtze River, China. Environ. Microbiol. Rep. 8, 667–674. doi: 10.1111/1758-2229.12420
Huang, L., Tan, Y., Song, X., Huang, X., Wang, H., Zhang, S., et al. (2003). The status of the ecological environment and a proposed protection strategy in Sanya Bay, Hainan Island, China. Mar. Pollut. Bull. 47, 180–186. doi: 10.1016/S0025-326X(03)00070-5
Jensen, S. I., Kühl, M., and Priemé, A. (2007). Different bacterial communities associated with the roots and bulk sediment of the seagrass Zostera marina. FEMS Microbiol. Ecol. 62, 108–117. doi: 10.1111/j.1574-6941.2007.00373.x
Könneke, M., Bernhard, A. E., de la Torre, J. R., Walker, C. B., Waterbury, J. B., and Stahl, D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546. doi: 10.1038/nature03911
Koops, H., Purkhold, U., Pommerening-Röser, A., Timmermann, G., and Wagner, M. (2006). “The lithoautotrophic ammonia-oxidizing bacteria,” in The Prokaryotes Proteobacteria: Alpha and Beta Subclasses, Vol. 5, eds M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (New York, NY: Springer), 778–811.
Koops, H. P., and Pommerening-Roser, A. (2001). Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. 37, 1–9. doi: 10.1111/j.1574-6941.2001.tb00847.x
Kovach, W. L. (1999). MVSP-a Multivariate STATISTICAL PACKAGE FOR WINDOWS, Ver 3.1. Pentraeth: Kovach Computing Services.
Lai, S., Gillis, L. G., Mueller, C., Bouma, T. J., Guest, J. R., Last, K. S., et al. (2013). First experimental evidence of corals feeding on seagrass matter. Coral Reefs 32, 1061–1064. doi: 10.1007/s00338-013-1062-9
Lamb, J. B., van de Water, J. A. J. M., Bourne, D. G., Altier, C., Hein, M. Y., Fiorenza, E. A., et al. (2017). Seagrass ecosystems reduce exposure to bacterial pathogens of humans, fishes, and invertebrates. Science 355, 731–733. doi: 10.1126/science.aal1956
Larkum, A. W. D., Orth, R. J., and Duarte, C. M. (eds) (2006). Seagrasses: Biology, Ecology and Conservation. Dordrecht: Springer.
Lee, K. S., Short, F. T., and Burdick, D. M. (2004). Development of a nutrient pollution indicator using the seagrass, Zostera marina, along nutrient gradients in three New England estuaries. Aquat. Bot. 78, 197–216. doi: 10.1016/j.aquabot.2003.09.010
Leininger, S., Urich, T., Schloter, M., Schwark, L., Qi, J., Nicol, G. W., et al. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442, 806–809. doi: 10.1038/nature04983
Li, M., Cao, H., Hong, Y., and Gu, J. (2011). Spatial distribution and abundances of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in mangrove sediments. Appl. Microbiol. Biotechnol. 89, 1243–1254. doi: 10.1007/s00253-010-2929-0
Li, M., and Gu, J. D. (2013). Community structure and transcript responses of anammox bacteria, AOA, and AOB in mangrove sediment microcosms amended with ammonium and nitrite. Appl. Microbiol. Biotechnol. 97, 9859–9874. doi: 10.1007/s00253-012-4683-y
Lipsewers, Y. A., Bale, N. J., Hopmans, E. C., Schouten, S., Damsté, J. S. S., and Villanueva, L. (2014). Seasonality and depth distribution of the abundance and activity of ammonia oxidizing microorganisms in marine coastal sediments (North Sea). Front. Microbiol. 5:472. doi: 10.3389/fmicb.2014.00472
López-Legentil, S., Erwin, P. M., Pawlik, J. R., and Song, B. (2010). Effects of sponge bleaching on ammonia-oxidizing Archaea: distribution and relative expression of ammonia monooxygenase genes associated with the barrel sponge Xestospongia muta. Microb. Ecol. 60, 561–571. doi: 10.1007/s00248-010-9662-1
Martens-Habbena, W., Berube, P. M., Urakawa, H., de la Torre, J. R., and Stahl, D. A. (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461, 976–979. doi: 10.1038/nature08465
Martens-Habbena, W., Qin, W., Horak, R. E., Urakawa, H., Schauer, A. J., Moffett, J. W., et al. (2015). The production of nitric oxide by marine ammonia-oxidizing archaea and inhibition of archaeal ammonia oxidation by a nitric oxide scavenger. Environ. Microbiol. 17, 2261–2274. doi: 10.1111/1462-2920.12677
Mejia, A. Y., Rotini, A., Lacasella, F., Bookman, R., Thaller, M. C., Shem-Tov, R., et al. (2016). Assessing the ecological status of seagrasses using morphology, biochemical descriptors and microbial community analyses. A study in Halophila stipulacea (Forsk.) Aschers meadows in the northern Red Sea. Ecol. Indic. 60, 1150–1163. doi: 10.1016/j.ecolind.2015.09.014
Mincer, Y. J., Church, M. J., Taylor, L. T., Preston, C., Karl, D. M., and DeLong, E. F. (2007). Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Nakagawa. Environ. Microbiol. 9, 1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x
Moin, N., Nelson, K., Bush, A., and Bernhard, A. (2009). Distribution and diversity of archaeal and bacterial ammonia oxidizers in salt marsh sediments. Appl. Environ. Microbiol. 75, 7461–7468. doi: 10.1128/AEM.01001-09
Nakagawa, T., Mori, K., Kato, C., Takahashi, R., and Tokuyama, T. (2007). Distribution of cold-adapted ammonia-oxidizing microorganisms in the deep-ocean of the northeastern Japan Sea. Microbes Environ. 22, 365–372. doi: 10.1264/jsme2.22.365
Newell, S. E., Fawcett, S. E., and Ward, B. B. (2013). Depth distribution of ammonia oxidation rates and ammonia-oxidizer community composition in the Sargasso Sea. Limnol. Oceanogr. 58, 1491–1500. doi: 10.4319/lo.2013.58.4.1491
Pester, M., Schleper, C., and Wagner, M. (2011). The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr. Opin. Microbiol. 14, 300–306. doi: 10.1016/j.mib.2011.04.007
Philippot, L., Raaijmakers, J., Lemanceau, P., and van der Putten, W. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Prosser, J., and Nicol, G. (2012). Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol. 20, 523–531. doi: 10.1016/j.tim.2012.08.001
Rasmussen, R. (2001). “Quantification on the lightcycler,” in Rapid Cycle Real-Time PCR, Methods and Applications, eds S. Meuer, C. Wittwer, and K. Nakagawara (Heidelberg: Springer), 21–34. doi: 10.1007/978-3-642-59524-0_3
Rotthauwe, J., Witzel, K., and Liesack, W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microb. 63, 4704–4712. doi: 10.1126/science.284.5411.63
Rusch, A., and Gaidos, E. (2013). Nitrogen-cycling bacteria and archaea in the carbonate sediment of a coral reef. Geobiology 11, 472–484. doi: 10.1111/gbi.12048
Rusch, A., Hannides, A. K., and Gaidos, E. (2009). Diverse communities of active Bacteria and Archaea along oxygen gradients in coral reef sediments. Coral Reefs 28, 15–26. doi: 10.1007/s00338-008-0427-y
Schloss, P., Westcott, S., Ryabin, T., Hall, J., Hartmann, M., Hollister, E., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schmid, M. C., Maas, B., Dapena, A., Van De Pas-Schoonen, K., Van De Vossenberg, J., Kartal, B., et al. (2005). Minireview: biomarkers for in situ detection of anaerobic ammonium-oxidizing (Anammox) bacteria. Appl. Environ. Microbiol. 71, 1677–1684. doi: 10.1128/AEM.71.4.1677-1684.2005
Shen, J. W., Johnson, M. E., Fu, F., Wang, Y., and Jin, Y. (2017). Seasonal wind patterns influence the configuration and geomorphology of insular reef systems: Yongxing Island. Xisha Islands, China. Geol. J. 32, 1–13. doi: 10.1002/gj.2925
Skiba, M., George, T., Baggs, E., and Daniell, T. (2011). Plant influence on nitrification. Biochem. Soc. Trans. 39, 275–278. doi: 10.1042/BST0390275
Takano, Y., Chikaraishi, Y., Ogawa, O. N., Nomaki, H., Morono, Y., Inagaki, F., et al. (2010). Sedimentary membrane lipids recycled by deep-sea benthic archaea. Nat. Geosci. 3, 858–861. doi: 10.1038/ngeo983
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Urakawa, H., Tajima, Y., Numata, Y., and Tsuneda, S. (2008). Low temperature decreases the phylogenetic diversity of ammonia-oxidizing archaea and bacteria in aquarium biofiltration systems. Appl. Environ. Microbiol. 74, 894–900. doi: 10.1128/AEM.01529-07
Wang, H., Su, J., Zheng, T., and Yang, X. (2015). Insights into the role of plant on ammonia-oxidizing bacteria and archaea in the mangrove ecosystem. J. Soils Sediments 15, 1212–1223. doi: 10.1007/s11368-015-1074-x
Wu, D., Wang, Y., Lin, X., and Yang, J. (2008). On the mechanism of the cyclonic circulation in the Gulf of Tonkin in the summer. J. Geophys. Res. 113:C09029. doi: 10.1029/2007JC004208
Wu, Y., Lu, L., Wang, B., Lin, X., Zhu, J., Cai, Z., et al. (2011). Long-term field fertilization significantly alters community structure of ammonia-oxidizing bacteria rather than archaea in a paddy soil. Soil Sci. Soc. Am. J. 75, 1431–1439. doi: 10.2136/sssaj2010.0434
Wuchter, C., Abbas, B., Coolen, M. J., Herfort, L., Van Bleijswijk, J., Timmers, P., et al. (2006). Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. U.S.A. 104, 5704–5704. doi: 10.1073/pnas.0701630104
Yu, S., Yao, P., Liu, J., Zhao, B., Zhang, G., Zhao, M., et al. (2016). Diversity, abundance and niche differentiation of ammonia-oxidizing prokaryotes in mud deposits of the eastern China marginal seas. Front. Microbiol. 7:137. doi: 10.3389/fmicb.2016.00137
Zhang, J., Liu, B., Zhou, X., Chu, J., Li, Y., and Wang, M. (2015). Effects of emergent aquatic plants on abundance and community structure of ammonia-oxidising microorganisms. Ecol. Eng. 81, 504–513. doi: 10.1016/j.ecoleng.2015.04.029
Zhang, Y., Chen, L., Dai, T., Sun, R., and Wen, D. (2015). Ammonia manipulates the ammonia oxidizing archaea and bacteria in the coastal sediment water microcosms. Appl. Microbiol. Biotechnol. 99, 6481–6491. doi: 10.1007/s00253-015-6524-2
Zhang, Y., Xie, X., Jiao, N., Hsiao, S. Y., and Kao, S. J. (2014). Diversity and distribution of amoA-type nitrifying and nirS-type denitrifying microbial communities in the Yangtze River estuary. Biogeosciences 11, 2131–2145. doi: 10.5194/bg-11-2131-2014
Zhao, D., Luo, J., Zeng, J., Wang, M., Yan, W., Huang, R., et al. (2014). Effects of submerged macrophytes on the abundance and community composition of ammonia-oxidizing prokaryotes in a eutrophic lake. Environ. Sci. Pollut. Res. 21, 389–398. doi: 10.1007/s11356-013-1909-1
Zhao, M., Yu, K., Zhang, Q., Shi, Q., and Price, G. J. (2012). Long-term decline of a fringing coral reef in the northern South China Sea. J. Coastal Res. 28, 1088–1099. doi: 10.2112/JCOASTRES-D-10-00172.1
Keywords: seagrass, ammonia-oxidizing archaea and bacteria, ammonia monooxygenase subunit A (amoA), cDNA, community structure, coral reef ecosystems
Citation: Ling J, Lin X, Zhang Y, Zhou W, Yang Q, Lin L, Zeng S, Zhang Y, Wang C, Ahmad M, Long L and Dong J (2018) Community Composition and Transcriptional Activity of Ammonia-Oxidizing Prokaryotes of Seagrass Thalassia hemprichii in Coral Reef Ecosystems. Front. Microbiol. 9:7. doi: 10.3389/fmicb.2018.00007
Received: 31 May 2017; Accepted: 04 January 2018;
Published: 25 January 2018.
Edited by:
Hongyue Dang, Xiamen University, ChinaReviewed by:
Wei Xie, Tongji University, ChinaCopyright © 2018 Ling, Lin, Zhang, Zhou, Yang, Lin, Zeng, Zhang, Wang, Ahmad, Long and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junde Dong, ZG9uZ2pkQHNjc2lvLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.