- 1College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan, China
- 2Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan, China
Pseudomonas lipases are well-studied, but few studies have examined the mechanisms of lipase expression regulation. As a global regulatory protein, PmrA controls the expression of multiple genes such as the Dot/Icm apparatus, eukaryotic-like proteins, and secreted effectors. In this study, the effect of PmrA on expression of the lipase lipA in Pseudomonas aeruginosa PAO1 was investigated by knocking out or overexpressing pmrA, rsmY, and rsmA. PmrA regulated the expression of lipA at both the transcriptional and translational level although translation was the pivotal regulatory mechanism for lipA expression. PmrA also regulated the expression of rsmY. Using gel mobility shift assay and pmrA/rsmY double gene knock-out model, we showed that PmrA directly bound to the promoter sequence of rsmY to regulate lipA expression. Translation of lipA was activated by the PmrA/PmrB system via RsmA. Specifically, the Shine-Dalgarno (SD) sequence located at lipA mRNA was overlapped through combination between RsmA and the AGAUGA sequence, subsequently blocking the 30S ribosomal subunit to the SD sequence, leading to translational inhibition of lipA. Transcriptional repression of RsmY initiated translation of lipA through negative translational regulation of rsmA. In conclusion, this study demonstrated that in P. aeruginosa PAO1, PmrA mainly regulated rsmY expression at a translational level to influence lipA expression. RsmY primarily activated lipA translation via negative translational regulation of rsmA.
Introduction
Lipases (triacylglycerol acylhydrolases, EC 3.1.1.3) are ubiquitous enzymes produced by all animals, plants and microbes. Generally, lipases are vital for catalysis of hydrolysis of esters, which are long-chain fatty acids and glycerol in aqueous solutions. And lipases catalyze other reactions in microaqueous and liposoluble environments such as transesterification, alcoholysis, esterification, aminolysis (Gupta et al., 2004; Angkawidjaja and Kanaya, 2006). The versatility and plasticity of lipases contribute to their extensive application in the production of fine chemicals (Busto et al., 2006; Chang et al., 2007), detergents (Romdhane et al., 2010; Grbavčić et al., 2011), food (Aravindan et al., 2007; Pan et al., 2012), and biodiesel (Yoo et al., 2011; Jin et al., 2013). Lipases from the genera Pseudomonas and Burkholderia have high tolerance to harsh industrial application environments (Tran et al., 2016; Dwivedee et al., 2017). However, Enzyme production is too limited to match the demands of industry because of low lipase expression in original strains.
The lipase genes of Pseudomonas and Burkholderia species are concurrently transcribed with other genes that encode helper proteins (Rosenau and Jaeger, 2000), which are previously known regulators of lipase expression (Frenken et al., 1993). However, according to recent studies, the function of lipase helper proteins is mainly effective lipase folding in the periplasm rather than control of regulation (Rosenau and Jaeger, 2000). Numerous, extensive studies have investigated the transcriptional regulation mechanisms of operons, including lipase genes in the lipAB operon, whose expression is induced by soybean oil in P. alcaligenes (Cox et al., 2001; Krzeslak et al., 2008). Initially, an σ54, RpoN-dependent promoter region and upstream activator sequence (UAS) were recognized in the upstream sequence of the lipAB operon and illustrated (Cox et al., 2001). This result was followed by identification of LipQR, a two-component regulation system of the operon (Krzeslak et al., 2008). There is convincing evidence that after aspartate 52 is phosphorylated, LipR interacts with the UAS of lipAB, is initiated and activates transcription of lipAB, and RpoN is extensively involved in lipase expression (Krzeslak et al., 2012). The transcriptional origin of the lipase operon is downstream of a classical 54-type promoter, with a GG-N10-GC region at transcriptional origins 12–24 in P. alcaligenes M-1 (Nirandarjos et al., 1987; Deretic et al., 1989). These Active components operate together with an UAS of lipase gene that is vital for lipase expression regulation.
The global Gac-Rsm signal transduction system is widely accepted to work effectively in regulating lipase gene expression. GacS-GacA (GacS/A), a two-component regulator system, functions in lipase gene expression in Pseudomonas spp. (Reimmann et al., 1997; Rosenau and Jaeger, 2000; Lalaouna et al., 2012), with high conservation of GacS, a sensor kinase that locates and binds to the inner cytomembrane and is activated via autophosphorylation after recognizing ambiguous signals, and GacA, a cognate response regulator is evoked by phosphorylated GacS via phosphorylation. The transcription of multiple small regulatory RNAs (sRNAs) such as RsmY, RsmX, and RsmZ in P. protegens or RsmY and RsmZ in P. aeruginosa is assumed to be initialized and promoted by activated GacA, based on available models. These sRNAs have multiple single-stranded GGA motifs and high affinities to RNA-binding protein(s) RsmA (and RsmE) from the RsmA-CsrA family. Using titration to identify RNA-binding proteins bound to particular motifs (often GGA or ANGGA) that overlay or are close to the Shine-Dalgarno (SD) sequence of targeted mRNAs, single-stranded GGA motifs can relieve translational repression, so as to enhance translation initiation (Lapouge et al., 2008; Brown, 2010; Sonnleitner and Haas, 2011; Marzi and Romby, 2012). RsmA generally suppresses the two QS systems of P. aeruginosa. Consequently, extracellular products such as lipases are inhibited. RsmA induces expression of lipases via an unidentified mechanism in P. aeruginosa (Heurlier et al., 2004; Gooderham and Hancock, 2008).
As a two-component regulator system, response regulator PmrA, had a great relationship with downstream genes expression, such as the widely investigated multidrug-resistant genes of P. aeruginosa (Ly et al., 2012). In this study, a control group (PS1) and an olive oil-induction group (PS2) were used to study lipase gene regulation. When the growth of the strain was in the stable period, the enzyme activity of the induced group was 3.6 times higher than that of the control group. Then, RNA-seq and proteomics were further employed for the regulatory mechanism analysis. Analyzing associations between RNA-seq [log2 Ratio (PS2/PS1) = 8.8] and proteomic data [Quantition(PS2/PS1) = 2.5] showed that PmrA expression was significantly correlated with lipA expression in P. aeruginosa PAO1. Furthermore, lipase activity was significantly reduced after the PmrA gene was knocked out. Additionally, our preliminary study also inferred that PmrA might interacted with rsmY to control lipase expression in PAO1. Therefore, in this study, the effect of PmrA on lipA expression in P. aeruginosa PAO1 was systematically investigated via the combined strategy of knocking out and overexpressing pmrA, rsmY, and rsmA.
Materials and Methods
Strains and Chemicals
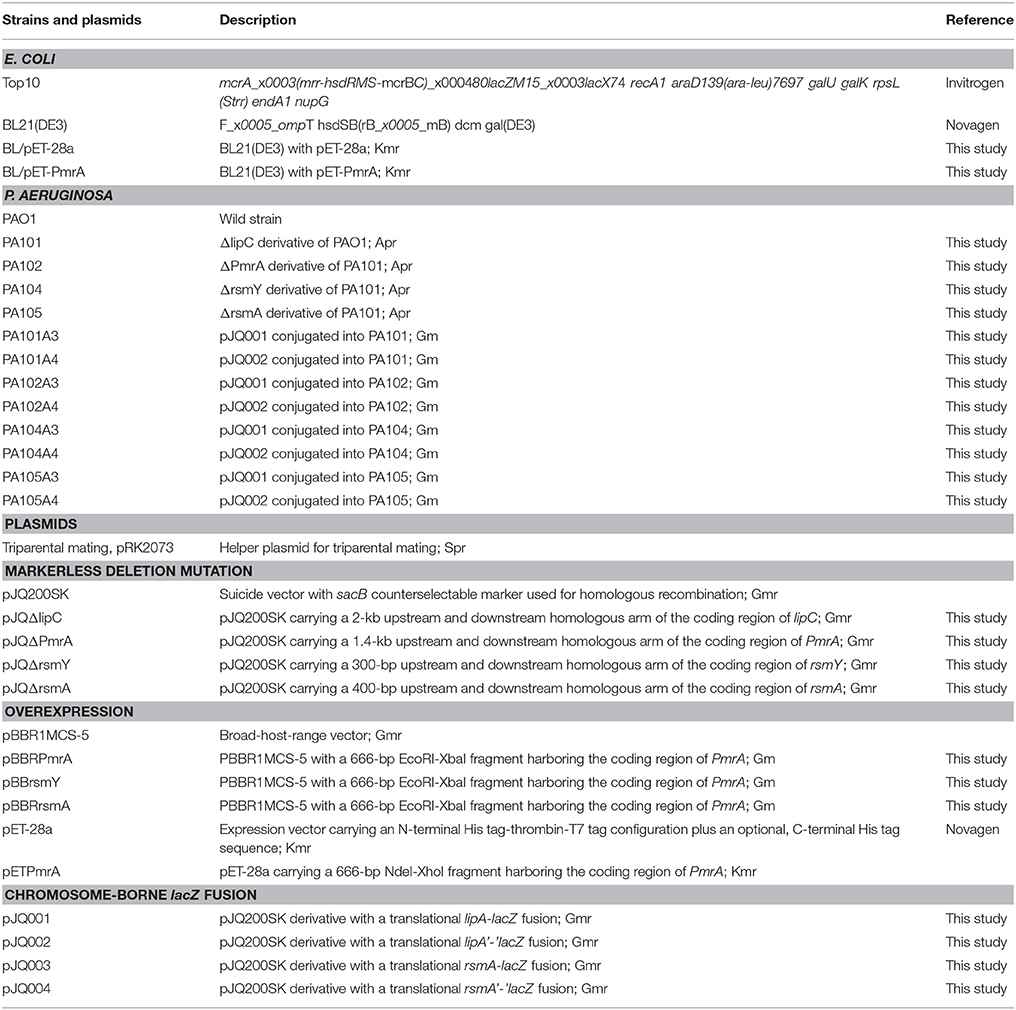
Details of bacterial strains and plasmids used in this study are in Table 1. Propagation of P. aeruginosa strains was in liquid or solid LB or M9 minimal (1.5% agar) medium (0.6% Na2HPO4, 0.3% KH2PO4, 0.1% NH4Cl, 0.05% NaCl, 3 × 10−4% CaCl2, 1 mL 1 mol/L MgSO4·7H2O, 10 mL 20% glucose). Escherichia coli strains were propagated in LB medium (1.5% agar). The antibiotics used were kanamycin (50 μg/mL) and carbenicillin (100 μg/mL) for P. aeruginosa, and ampicillin (100 μg/mL) and tetracycline (25 μg/mL) for E. coli. PrimeSTAR HS DNA polymerase, DNA ligation kits and restriction enzymes were purchased from TaKaRa Biotechnology (Dalian, China). Kits for extracting genomic DNA, DNA gel and preparing plasmids (Omega Bio-Tek, Doraville, GA, USA) were used according to the manufacturer's instructions. DNA fragments were sequenced by Shanghai Sunny Biotechnology (Shanghai, China). Primers were synthesized by Wuhan Anygene Biological Technology (Wuhan, China). Other regular methods were completed according to defined protocols.
Construction of Mutation Plasmid
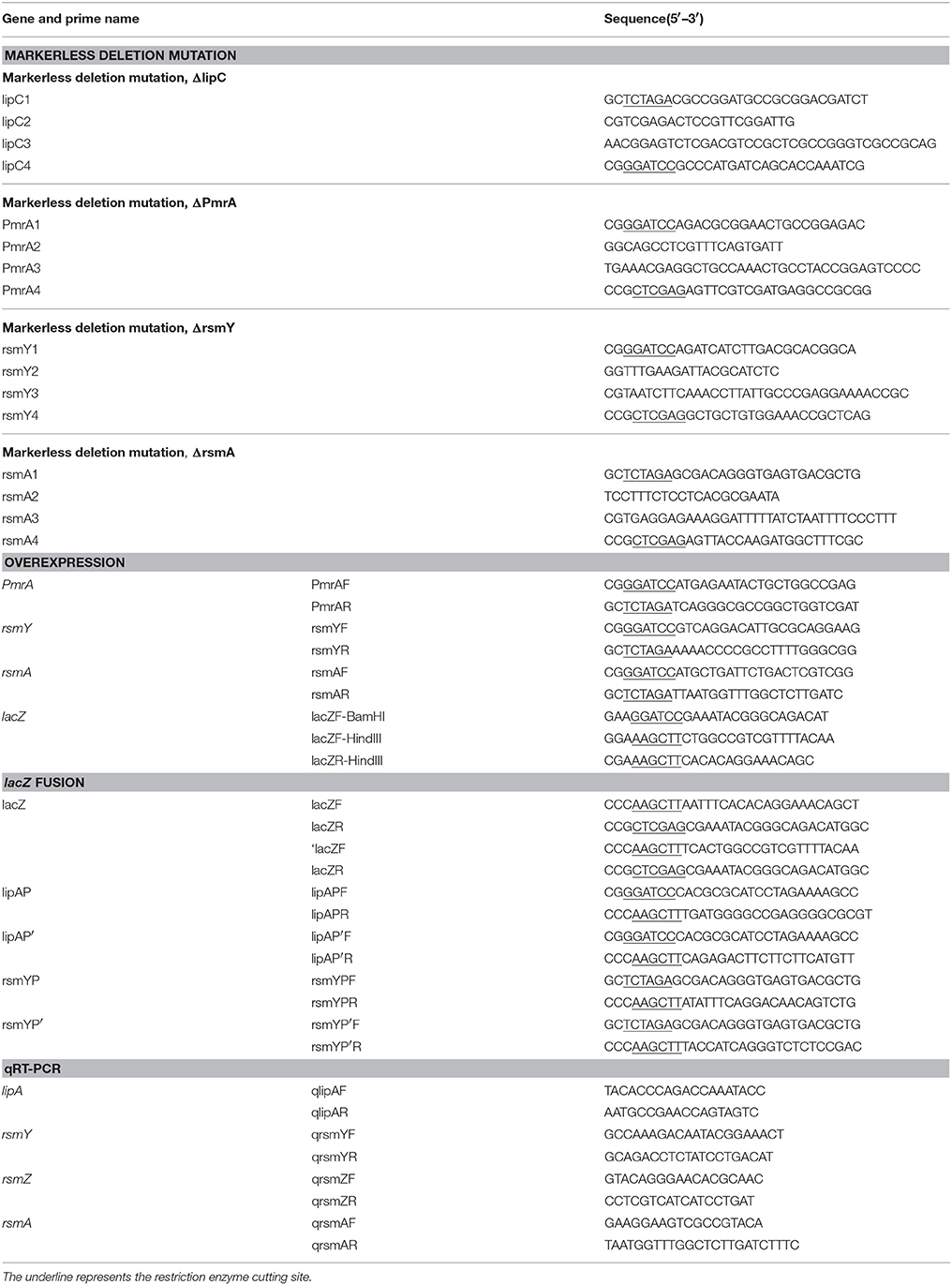
Homologous sequences 1,000 bp upstream and 1,000 bp downstream of lipC were used to produce a markerless deletion-mutant system for the coding area by overlapping PCR. The XbaI-BamHI-digested mutant system and suicide plasmid pJQ200SK (Quandt and Hynes, 1993) were ligated and transformed into E. coli. Transformants were spread on LB medium with 50 μg/ml gentamicin. A PCR-confirmed recombinant plasmid was isolated and sequenced and named as pJQΔlipC (Table 1). Homologous sequences 700 bp upstream and 700 bp downstream of pmrA, 150 bp upstream and 150 bp downstream of RsmY, or 200 bp upstream and 200 bp downstream of rsmA were ligated into the suicide plasmid pJQ200SK to construct pJQΔPmrA, pJQΔRsmY, pJQΔrsmA, respectively.
Construction of Expression Plasmids
BamHI and XbaI were used to excise the coding areas of PmrA (666 bp), rsmY (124 bp), and rsmA (186 bp), which were cloned into plasmid pBBR1MCS-5 to create recombinant plasmids pBBRPmrA, pBBRrsmY, and pBBRrsmA. In a similar method, NdeI and HindIII were used to digest the coding sequences of rsmA (186 bp) to create plasmid pET-rsmA (Table 1).
Construction of LacZ-Fusion Plasmids
To construct translationally and transcriptionally fused lipA-lacZ and lipA'-‘lacZ, amplicons of wild-type lacZ, containing the SD sequence, and ‘lacZ, whose SD sequence as well as the first six codons were absent, were obtained from the genome of E. coli BL21(DE3) by amplification. Amplicons of lipA and lipA' were acquired in a similar method. The lacZ and lipA fragments were digested and cloned into plasmid pJQ200SK for a translationally fused plasmid pJQ001; ‘lacZ and lipA' were digested and cloned into plasmid pJQ200SK for a transcriptionally fused plasmid pJQ002 (Table 1). To construct rsmA-lacZ and rsmA'-‘lacZ translational-transcriptional fusions (Figure 1), amplicons of rsmA and lacZ were digested and cloned into plasmid pJQ200SK to construct translationally fused plasmid pJQ003; amplicons of rsmA' and ‘lacZ were cloned into pJQ200SK for transcriptionally fused plasmid pJQ004 (Table 1). Regular forward primer and specific reverse primers containing targeted replacement sequences were designed to generate single-base substitution mutations in the AGAUGA motif of the lipA mRNA SD sequence (Table 2).
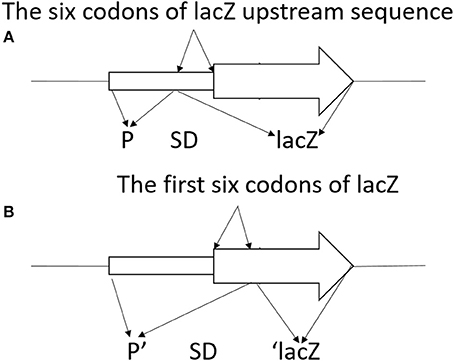
Figure 1. Schematic diagram of lacZ transcriptional fusion (A) and ‘lacZ translational fusion (B). (A) Transcriptional control of lacZ depended on the x promoter (P). Translational product was a hybrid protein containing some upstream amino acids of X coding region and a functional component of β-galactosidase. Promoter strength and effectiveness of inducing translation of the target gene was reflected by the information of the translational fusion. (B) Translational control of ‘lacZ relied on the x promoter (P). Translational product was a hybrid protein containing some upstream amino acids of X coding region and the first codon of X, as well as a large functional component of β-galactosidase at C-terminal. Promoter strength of the corresponding gene was reflected in the information of the translational fusion.
Construction of Strains
Because PAO1 has two main types of lipases—lipA and lipC—we built a mutant to eliminate the disturbance. A double-crossover recombination system with gene substitution was used to construct a mutant with markerless deletions of lipC in plasmid pJQΔlipC (Quandt and Hynes, 1993; Zha et al., 2014), which was named PA101. Similarly, a mutant of PA101 with markerless deletions of the PmrA coding area was named as PA102, a mutant of PA101 with markerless deletions of the rsmY coding area was named as PA104, a mutant of PA101 with markerless deletions of the rsmA coding area was named as PA105, a mutant of PA101 with double markerless deletions of the PmrA/rsmY coding area was constructed, and named PA106. Plasmids were transformed into P. aeruginosa with pRK2073 as a helper plasmid (Leong et al., 1982) by triparental mating.
β-Galactosidase Activity Assays
Miller's method was used to detect β-galactosidase activity in indicated E. coli and P. aeruginosa strains with a chromosomal-fused lacZ on o-nitrophenyl–D-galactopyranoside (Miller, 1972), with normalization of optical density at 600 nm (OD600) to bacterial medium. Data are shown in Miller units. When culturing strains containing pBBR1MCS-5 or pET-28a derivatives, we added isopropyl-D-thiogalactopyranoside (TPTG) (0.1 mM) to the medium for expression induction.
QRT-PCR Analysis
Total RNA was isolated from P. aeruginosa strains without a chromosomal-fused lacZ (OD600 = 2.5) cultured in LB medium at 37°C using an RNA pure Bacteria kit (DNase I) (CWBIO, Beijing, China). In accordance with the protocols, reverse transcription into complementary DNA was performed with RevertAid first-strand cDNA synthesis kits (Thermo Scientific). For qRT-PCR, reactions were 20 mL (10 mL SYBR Green Master Mix, 10 pM each primer, 10 ng final cDNA and specific volume of RNase-free water) for a ABI 7500 Real Time PCR machine (Applied Biosystems, Foster City, CA, USA). Each plate contained three technical replicates. RpoD was the internal control.
Protein Expression and Purification
After amplification using primers PmrAF and PmrAR, PCR products were inserted into pET-28a(+) digested with NdeI and HindIII to add a carboxyl terminal 6-his-tag. Plasmids were transformed into the expression strain E. coli BL21 (DE3) and cultured in LB medium (5 ml) at 37°C overnight. Cells were diluted 1:100 into 1 L LB broth containing ampicillin and cultured. When OD600 was between 0.4 and 0.5, recombinant PmrA was activated with 0.1 mM IPTG at 16°C for 20 h.
Mixtures were centrifuged at 7,000 × g for 20 min at 4°C and resuspended in lysis solution, followed by harvest of 6 L of cells and disruption by a One Shot Cell Disrupter (Constant Systems, Daventry, UK). To isolate insoluble substances, mixtures were passed through 0.45-μm syringe-end filters after centrifuging at 12,000 × g for 20 min at 4°C before loading on a His-tag column. Supernatants were transferred to a Ni-NTA affinity chromatography column (GE Healthcare, Pittsburgh, PA, USA). After balancing the column with lysis buffer, recombinant protein was added and eluted using washing buffer with an imidazole concentration gradient (0, 30, 60, 100, 200, and 500 mM). Washing buffer (30 ml) containing target recombinant proteins was dialyzed in 20 mM Tris-HCl, pH 8.0. Proteins were subjected to SDS-PAGE. Unstained and prestained protein markers (Fermentas, SM0431 and SM0671 Waltham, USA) were applied as references.
Electrophoretic Mobility Shift Assays
Light Shift Chemiluminescent EMSA kits were used for electrophoretic mobility shift assays (EMSAs). DNA fragments of rsmY, rsmZ, and rsmA were synthesized and labeled with biotin; 2 μl biotin-labeled DNA probe was incubated with 3 μl purified PmrA protein (1 mg/ml) in 10 μl binding solution (100 mM HEPES, pH 7.3 with 200 mM KCl, 10 mM MgCl2, and 10 mM DTT) at room temperature for 10 min, to eliminate nonspecific binding of probe and protein. A labeled probe was added and mixed at room temperature for 20 min. For competition experiments, unlabeled probe concentrations were 50, 100, and 150 times the labeled probe concentration. One microliter of binding buffer (10x) was added and mixed immediately. Pre-electrophoresis was performed for 30 min with 0.5 x TBE as the electrophoretic solution, at 80 V. Separated protein-DNA conjugates were electrophoretically transferred to nylon membranes with positive charges (Ambion) using a semidry transfer instrument (Tanon). UV light at 320 nm was used to crosslink transferred PmrA-DNA complexes and free DNA to membranes. A chemiluminescent nucleic acid detection module (Thermo Scientific) was used to detect biotin-labeled bands. To probe DNAs labeled with biotin on nylon membranes, streptavidin-horseradish peroxidase (HRP) conjugates were employed. HRP catalyzed an enhanced luminol-based substrate for optimal block, followed by washing to obtain light-sensitive. Membranes were set in film cassettes in light and exposed to X-ray film for 20–30 s.
Lipase Activity Assay
Supernatant lipase activity was used to detect LipA, an extracellular lipase (Wohlfarth et al., 1992). Mixtures (OD600 2.5) of P. aeruginosa strains without chromosomally fused lacZ were cultured in LB medium at 37°C. Supernatants were collected and lipase activities of p-nitrophenyl caprylate (pNPC) were assessed by spectrophotometry (Yang et al., 2015), with some modifications. The system was 2.9 ml Tris-HCl buffer (50 mM, pH 9.0) and 30 μl pNPC (10 mM pNPC in acetonitrile). After preincubating at 40°C for 5 min, 70 μl supernatant was added. Mixtures were incubated at 40°C for 5 min, followed by centrifugation (12,000 r/m, 2 min, 4°C) and OD410 measurement. The definition of one unit lipase activity was the amount of enzyme needed to release 1 mol p-nitrophenol per minute by 1 ml sample with OD600 1.0. The activity of bacterial lipase was shown in U/ml · OD600.
Statistical Analysis
Two-tailed, unpaired Student's t-tests were used to calculate P-values, with < 0.05 considered statistically significant.
Results
PmrA Activates Transcription and Translation of LipA
To eliminate interference from lipase lipC in studying the regulation of lipase expression, a lipC-mutant strain P101 was constructed. It was found that enzyme activity had little change in the lipC-mutant strain P101 (Figure 2A). Thus, the effect of lipC on the enzyme activity measurement of LipA could be ruled out. To study PmrA in regulation of lipA expression in PAO1, a gene replacement system with plasmid pJQΔPmrA was used to construct the PmrA deletion mutant PA102. A significant reduction in enzyme activity was observed in the PmrA-deletion mutant PA102 (Figure 2A). The results on transcription of lipA by qRT-PCR in PAO1, PA101, and PA102 were consistent with the relative extracellular lipase activities (Figures 2A,B). To further investigate the regulating mechanism of PmrA-activated expression of lipA, expression of chromosome-borne lipA-lacZ and lipA'-‘lacZ fusions was assessed in PA101 and PA102. As a result, PmrA positively regulated the expression of lipA both transcriptionally and translationally, and the latter appeared to be the core regulating mechanism (Figure 2C). To examine the influence of PmrA overexpression on expression of lipA in PA101 and PA102, PmrA was overexpressed in PA101 and PA102 via the PmrA expression plasmid pBBBRPmrA. PA101/pBBR1MCS-5 was the control. Expression of lipA in PA101/pBBRPmrA was higher than in PA101/pBBR1MCS-5 or PA102/pBBR1MCS-5 (Figure 2D). Interestingly, despite the close levels of lipA expression in PA101/pBBRPmrA and PA102/pBBRPmrA, the expression of lipA in PA101/pBBRPmrA was higher than in PA102/pBBRPmrA (Figure 2D). In conclusion, there is a great positive correlation between the expression of lipA and the expression of PmrA, the expression of lipA is regulated by PmrA.
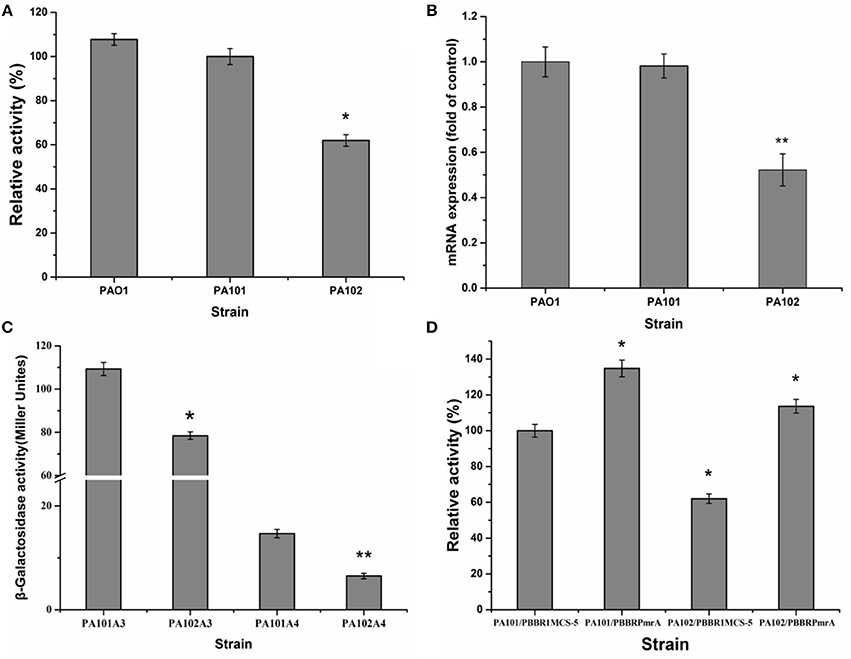
Figure 2. Effect of PmrA on lipA expression. (A) Relative lipase activity in PA01, PA101, and PA102. (B) QRT-PCR of lipA in PA01, PA101, and PA102. (C) Effect of pmrA mutant on expression of translationally fused, chromosome-borne lipA'-‘lacZ (control, PA101A4; pmrA mutant, PA102A4) and transcriptionally fused chromosome-borne lipA-lacZ (control, PA101A3; pmrA mutant, PA102A3). After inoculating β-galactosidase into M9 medium (50 mL), activity of strains was determined in stationary phase. (D) Relative lipase activity in control strain (PA101) and pmrA mutant (PA102). Experiments were in triplicate. *P < 0.05, **P < 0.01 compared with control group.
PmrA Affects RsmY/Z/A Expression
To study how PmrA acted on expression of lipA, the interaction of PmrA with functional genes rsmY, rsmZ, and rsmA was investigated. QRT-PCR was used to determine how pmrA deletion changed transcription of rsmY, rsmZ, and rsmA in PA101 and PA102. Transcription of rsmY and rsmZ was lower in PA102 than in PA101 (Figure 3A), similar to lipA expression in PA102 (Figure 2B). Transcription of rsmA in PA102 was slightly higher than in PA101. To study the interaction between PmrA and rsmY/Z/A, PmrA protein was expressed and the binding site of PmrA protein was explored using EMSAs. PmrA protein was expressed using a purified expression system with pET-28a. EMSAs were used to detect if PmrA protein bound to the rsmY/Z/A promoter sequence (Figures 2B,C). PmrA protein bound directly to the promoter region of rsmY, but not the promoter sequences of rsmZ and rsmA. Overexpression of rsmY, rsmZ, rsmA showed that rsmY promoted expression of lipase (Figures 3A,B), which was partly consistent with the results in Figure 3E. In order to further study the effect of PmrA on the regulation of lipA expression in PAO1, a gene replacement system with plasmid pJQΔRsmY was used to construct mutant PA104, and the double mutant PmrA/rsmY was constructed, and named PA106. The relative lipase activity in PA104/pBBRPmrA was significantly higher than that of PA104/pBBR1MCS-5, the relative lipase activity in PA106/pBBRPmrA was significantly higher than that of PA106/pBBR1MCS-5, and PmrA overexpressed strain of PA106 can greatly compensate for the loss of enzyme activity caused by gene knockout (Figure 3F). It is likely that PmrA is able to regulate lipA expression via rsmY and other unclear pathways.
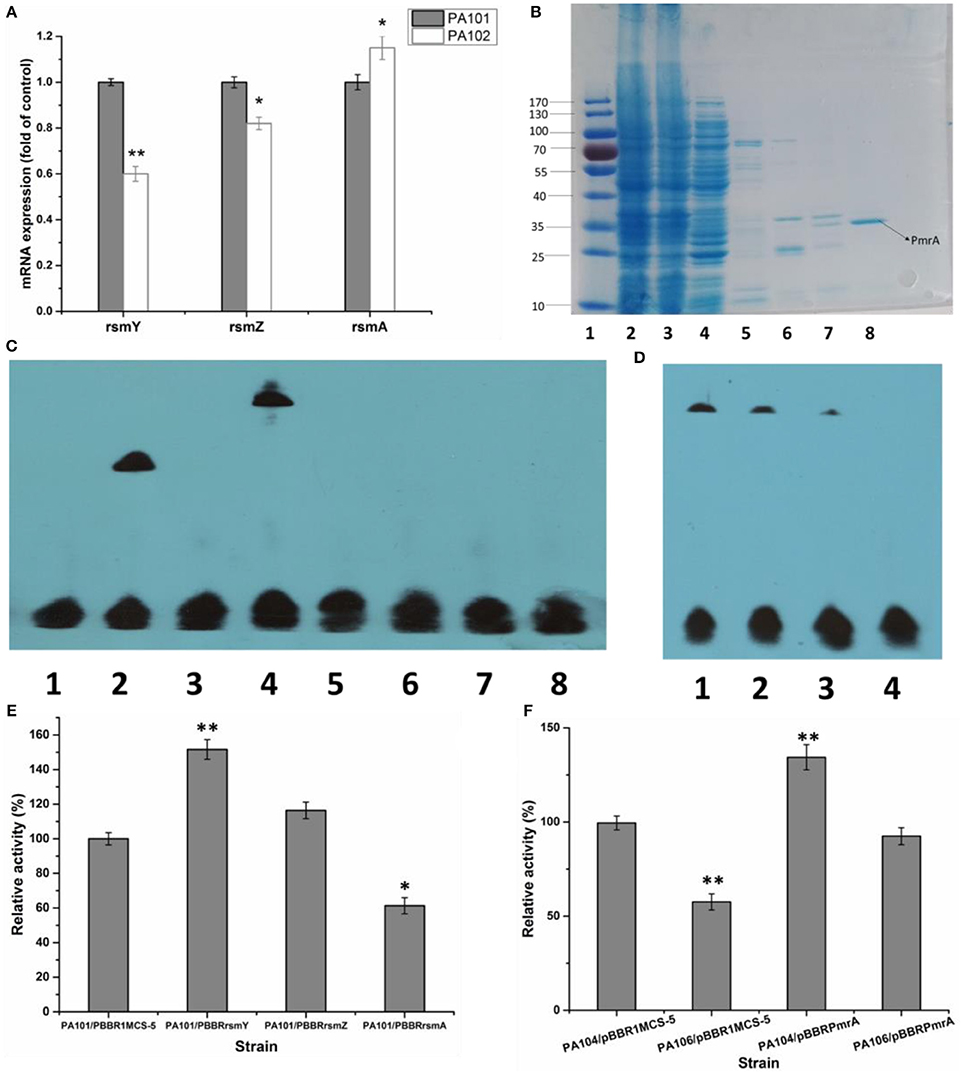
Figure 3. Effect of PmrA overexpression on lipA expression. (A) QRT-PCR of rsmY, rsmZ, and rsmA in control strain (PA101; lane 1) and PmrA mutant strain (PA102; lane 2). Experiments were in triplicate. *P < 0.05, **P < 0.01 compared with control group. (B) SDS PAGE of purified PmrA protein. Lane 1: protein molecular weight marker. Lane 2: whole cell lysate. Lane 3: liquid behind the column. Lane 4: NTA-30. Lane 5: NTA-60. Lane 6: NTA-100. Lane 7: NTA-200. Lane 8: NTA-500. (C) EMSA for binding of PmrA to rsmY/Z/A promoter sequence. Lane 1: negative control (−). Lane 2: positive control (+). Lane 3: rsmY. Lane 4: rsmY+PmrA. Lane 5: rsmZ. Lane 6: rsmZ+PmrA. Lane 7: rsmA. Lane 8: rsmA+PmrA. (D) PmrA protein bound to rsmY sequence, following increases in free rsmY. Ratios of free rsmY to biotin-labeled rsmY were 1:1, 50:1, 100:1, and 150:1 in groups 1 through 4. (E) Relative lipase activity in control strain (PA101) and overexpression strain (bar 1, control strain PA101/pBBR1MCS-5; bar 2, PA101/pBBRrsmY; bar 3, PA101/pBBRrsmZ; bar 4, PA101/pBBRrsmA). (F) Relative lipase activity (bar 1, control strain PA104/pBBR1MCS-5; bar 2, PA106/pBBR1MCS-5; bar 3, PA104/pBBRPmrA; bar 4, PA106/pBBRPmrA). Experiments were completed in triplicate. Experiments were completed in triplicate. *P < 0.05, **P < 0.01, compared with the control.
RsmY and RsmA Differentially Regulate LipA Expression
The translational regulation of the Gac-Rsm signaling pathway largely depends on RNA-combining proteins from the RsmA-CsrA family in various gamma-proteobacteria (Lapouge et al., 2008). For example, the Gac-Rsm regulatory system consists of RsmA and RsmE from the RsmA-CsrA family in P. protegens Pf-5 (Reimmann et al., 2005). To study the effect of RsmY and RsmA on the regulation of lipA expression in PAO1, a gene replacement system with plasmid pJQΔRsmA was used to construct mutant PA105. The expression of chromosomally fused lipA-lacZ (A3) and lipA'-‘lacZ (A4) was evaluated in PA101, PA104, and PA105 to understand the regulatory pathways of RsmY and RsmA on lipA expression. The RsmY-mutant weakly decreased transcription of lipA and reduced expression of lipA at a translational level. Translational control appeared to be the key regulatory pathway (Figure 4A). The RsmA-mutant promoted the translation of lipA and weakly decreased transcription of lipA. Translational control also appeared to be the key regulatory pathway (Figure 4A). The relative lipase activities of PA101/pBBR1MCS-5, PA104/pBBR1MCS-5, PA104/pBBRrsmY, PA105/pBBR1MCS-5, and PA105/pBBRrsmA indicated that the RsmY-mutant decreased expression of lipA. The expression of RsmY in complement counteracted the decrease caused by gene deletion. The RsmA-mutant promoted expression of lipA. The expression of RsmA in complement counteracted the increase caused by gene deletion (Figure 4B). QRT-PCR showed the RsmA-mutant increased lipA transcription, and the RsmY-mutant repressed lipA transcription (Figure 4C). Together, RsmY and RsmA differentially regulated the expression of lipA. RsmY increased lipase expression, and RsmA inhibited lipase expression, exhibiting different regulatory patterns in P. aeruginosa PAO1.
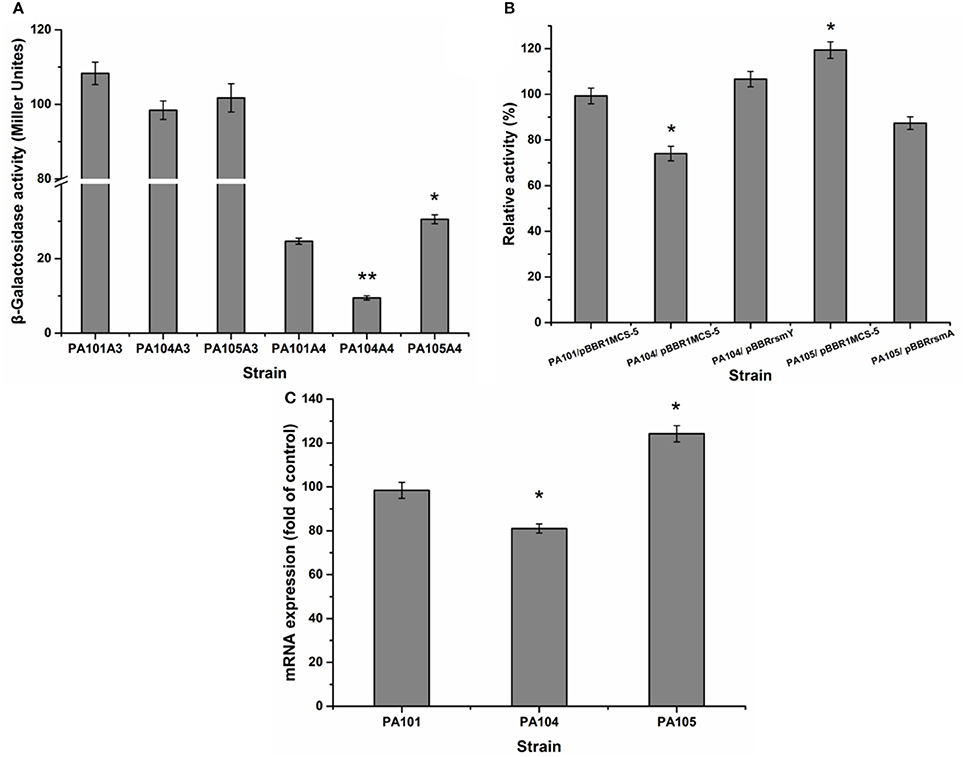
Figure 4. Effect of rsmY and rsmA mutants on lipA expression. (A) Effect of rsmY and rsmA mutants on expression of chromosome-located translationally fused lipA-lacZ (bar 1, control strain PA101A3; bar 2, rsmY-mutant strain PA104A3; bar 3, rsmA-mutant strain PA105A3) and chromosome-located transcriptionally fused lipA'-‘lacZ (bar 4, control strain PA101A4; bar 5, rsmY-mutant strain PA104A4; bar 6, rsmA-mutant strain PA105A4). After inoculating β-galactosidase into M9 medium (50 ml), the activity of strains was determined in stationary phase. (B) Relative lipase activity in PA101/pBBR1MCS-5, PA104/pBBR1MCS-5, PA104/pBBRrsmY, PA105/pBBR1MCS-5, and PA105/pBBRrsmA. (C) Expression of lipA in wild-type strain PA101 (lane 1), PA104 (lane 2) and PA105 (lane 3) by qRT-PCR. Experiments were in triplicate. *P < 0.05, **P < 0.01 compared with control strain.
RsmY Activates LipA Translation by Inhibiting RsmA Translation
To investigate the influence of RsmY on rsmA expression, we analyzed expression of chromosomally fused rsmA'-‘lacZ (C4). Transcription level of rsmA was investigated by qRT-PCR of PA101 and P104. The RsmY-mutant PA104 significantly stimulated expression of rsmA. Expression of RsmY in complement counteracted the increase caused by gene deletion (Figure 5A). By qRT-PCR, the RsmY-mutant repressed rsmA transcription and RsmY transcriptionally regulated rsmA (Figure 5B). The results implied RsmY repressed rsmA translation.
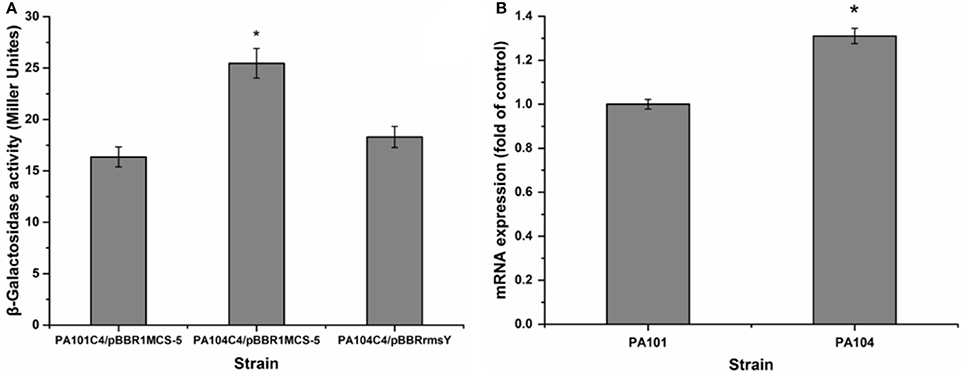
Figure 5. Effect of rsmY-mutant on rsmA expression. (A) Effect of rsmY mutant on expression of chromosome-borne translationally fused rsmA'-‘lacZ (bar 1, control strain PA101C4/pBBR1MCS-5; bar 2, rsmY-mutant strain PA104C4/ pBBR1MCS-5; bar 3, rsmY-overexpression strain PA104C4/pBBRrsmY). (B) rsmA expression in control strain PA101 (lane 1) and rsmY-mutant strain PA104 (lane 2) by qRT-PCR. Experiments were in triplicate. *P < 0.05 compared with control.
RsmA Suppresses LipA Translation by Binding to the SD Sequence of LipA mRNA
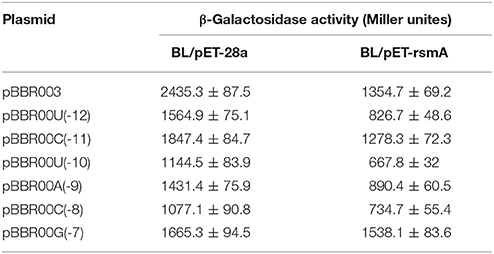
In the literature, RsmE binding to the consensus sequence (A/U)CANGG ANG(U/A) is involved in translation suppression mediated by RsmE by overlapping the SD sequence of targeted mRNA. Binding inhibits recruiting the 30S ribosomal subunit and translation (Schubert et al., 2007; Lapouge et al., 2008; Wang et al., 2013). The protein-RNA interplay between consensus sequence and RsmE has been investigated in P. protegens CHA0, which is the same as RsmE from P. protegens Pf-5 (Reimmann et al., 2005; Schubert et al., 2007). We hypothesized a site of action between RsmA and the SD sequence of lipA. Six single-base mutations at nucleotides U(-12), C(-11), U(-10), A(-9), C(-8), G(-7) [A-U, G-C, A-U, U-A, G-C, A-G] of lipA mRNA were constructed in plasmid-borne lipA'-‘lacZ fusions that were introduced into E. coli BL21(DE3) with plasmid pET-rsmA or pET-28a. The effects of U(-12), C(-11), U(-10), A(-9), C(-8), G(-7) mutations on translational regulation by RsmA are in Table 3. All mutations influenced regulatory control of RsmA. The suppression of lipA'-‘lacZ fusions by RsmA was eradicated after mutating G(-7), even though RsmA expression was affected by all mutations. Thus, RsmA suppressed the translation of lipA via combination with the AGAUGA sequence.
Discussion
Many studies indicate a regulatory role for the PmrA/PmrB two-component system (TCS) in the expression of multiple genes. The PmrA/PmrB TCS conducts bacterial responses to multiple stimuli (Hoch, 2000), and regulates lipopolysaccharide modifications (Chen and Groisman, 2013). Genome-wide microarrays of PmrB and PmrA mutants at exponential and postexponential stages reveal that the PmrA/PmrB TCS affects expression of 279 genes. Affected genes can be categorized into nine groups: type II-secreted proteins, Dot/Icm apparatus and secreted effectors, genes encoding eukaryotic-like proteins, flagellar biosynthesis genes, postexponential phase regulators, metabolic genes, stress response genes, and genes with unknown function (Al-Khodor et al., 2008). However, reports are fewer on the regulation of lipase expression by PmrA/PmrB TCS. To find out if PmrA/PmrB TCS regulates lipase expression in PAO1, the pmrA gene in PA101 was knocked out and the activity of lipase decreased. Overexpression of pmrA in control strain PA101 and mutant PA102 was also established. We found that complementation of pmrA restored the activity of lipase and overexpression of pmrA enhanced the activity of lipase in control strain PA101. Furthermore, after knocking out pmrA, expression of lipA also decreased by qRT-PCR. And the lacZ-fusion experiment proved that PmrA positively regulated the expression of lipA both transcriptionally and translationally, and the latter appeared to be the core regulating mechanism. Thus, our results suggests that there is a great positive correlation between the expression of lipA and the expression of PmrA, the expression of lipA is regulated by PmrA (Figure 2).
In P. aeruginosa PAO1, GacA directly regulates transcription of RsmZ and RsmY to regulate expression of hundreds of genes (Brencic et al., 2009). In P. aeruginosa PAO1, GacA, and RsmA are competitively associated with RsmY to regulate corresponding gene expression (Sorger-Domenigg et al., 2007; Brencic et al., 2009). As a defined two-component system, PmrA may be associated with RsmY to regulate gene expression. We studied if PmrA protein bound to the rsmY/Z/A promoter sequence. EMSA analysis confirmed that the PmrA protein bound directly to the promoter region of rsmY. Together, the results of qRT-PCR, EMSA and the mutant relative lipase activity demonstrated that PmrA is able to regulate lipA expression via rsmY and other unclear pathways (Figure 3).
Analysis of β-galactosidase activity and relative lipase activities in PA101, PA104, and PA105 further indicated that RsmY induced and promoted expression of lipA, while RsmA inhibited expression of lipA. In addition, analyses of β-galactosidase activity and qRT-PCR in PA101 and PA104 indicated that RsmY repressed rsmA transcription. To our knowledge, this is the first clear description of the interaction between PmrA and rsmY, rsmZ, and rsmA (Figure 4).
GGA-motifs are present in P. aeruginosa RsmY RNA and they are suggested to be essential for RsmA binding (Heeb et al., 2004). The rsmY and rsmA mutant strains PA104 and PA105 were established to study the effect of Rsm genes on lipA expression. Differential regulatory mechanisms were observed for expression of lipA regulated by RsmY and RsmA: lipA expression was weakly transcriptionally inhibited, but translationally stimulated by RsmY and translation of lipA was suppressed by RsmA (Figure 4). Generally, Rsm proteins suppress gene expression via translational mechanisms (Lapouge et al., 2008; Romeo et al., 2013). Our findings are consistent with these results.
The specific mechanism of translational activation by Rsm proteins probably lies in their combination with mRNAs to stabilize and facilitate translation initiation, ultimately leading to positive effects on gene expression (Heurlier et al., 2004; Frangipani et al., 2014). In a similar manner, CsrA, an ortholog of the Rsm protein family, activates translation initiation after binding to a highly structured untranslated leader of mRNA by changing RNA structure or interaction with a 5′-end-dependent RNase E cleavage pathway of the transcript (50). RsmZ, an antagonist of RsmA, was identified in P. aeruginosa. RsmZ and RsmY have similar regulatory functions (Heurlier et al., 2004; Brencic et al., 2009). We determined that the mechanism adopted by RsmY to activate lipA translation depended, at least partially, on suppression of rsmA translational expression, in line with the influence of mutant rsmY on rsmA expression (Figure 5A). QRT-PCR results for rsmA also showed an inhibitory effect (Figure 5B).
Rsm proteins suppress translation mainly through combination with corresponding sites in the initially translated area and/or the untranslated leader of targeted mRNAs, either overlapping the cognate SD sequence (Sonnleitner and Haas, 2011; Marzi and Romby, 2012). RsmE from P. protegens CHA0 is reported to be the same as that from P. protegens Pf-5, identifying a consensus sequence of (A/U) CANGGANG (U/A) overlaying the SD sequence of targeted mRNAs (Reimmann et al., 2005; Schubert et al., 2007). We hypothesized a site of action between RsmA and the SD sequence of lipA. The interaction between RsmA and the SD sequence was verified by mutational analysis of the SD sequence in lipA mRNA (Table 3). The results indicated that PmrA activated lipA translation via a combination of RsmA and the SD sequence in lipA mRNA, and the formation of the PmrA-RsmA system, leading to direct translational activation of lipA in P. aeruginosa PAO1.
Conclusion
Transcription of RsmY depended on the presence of PmrA. The function of sRNAs is to relieve translational repression of RNA-binding protein RsmA. RsmY mainly activated lipA translation by inhibiting rsmA translation. However, RsmA inhibited lipA translation by binding to the SD sequence of lipA mRNA. Among these regulatory pathways, the RsmA-mediated pathway had the greatest effect on regulation of lipA expression (Figure 6).
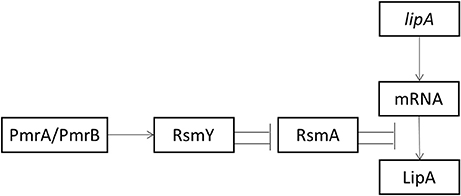
Figure 6. Schematic diagram of activation of lipA expression by the Pmr-Rsm system in P. aeruginosa PAO1. Arrow, positive effect; bar, negative effect.
Author Contributions
WL: Designed the experimental scheme and did most of the preparation and characterizations; ML and LJ: Contributed to SDS-PAGE and protein purification; PW: Helped with the experimental data; WL and YY: Wrote and revised the manuscript; All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was funded by the National Natural Science Foundation of China (grant numbers 31170078 and J1103514), the National High Technology Research and Development Program of China (grant numbers 2011AA02A204 and 2013AA065805). The authors thank Ms. Cheng Hong of the Centre of Analysis and Test, Huazhong University of Science and Technology, for the biodiesel analysis.
References
Al-Khodor, S., Kalachikov, S., Morozova, I., Price, C. T., and Abu Kwaik, Y. (2008). The PmrA/PmrB two-component system of legionella pneumophila is a global regulator required for intracellular replication within macrophages and protozoa. Infect. Immun. 77, 374–386. doi: 10.1128/IAI.01081-08
Angkawidjaja, C., and Kanaya, S. (2006). Family I.3 lipase: bacterial lipases secreted by the type I secretion system. Cell. Mol. Life Sci. 63, 2804–2817. doi: 10.1007/s00018-006-6172-x
Aravindan, R., Anbumathi, P., and Viruthagiri, T. (2007). Lipase applications in food industry. Indian J. Biotechnol. 6, 141–158.
Brencic, A., McFarland, K. A., McManus, H. R., Castang, S., Mogno, I., Dove, S. L., et al. (2009). The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73, 434–445. doi: 10.1111/j.1365-2958.2009.06782.x
Brown, D. (2010). A mathematical model of the Gac/Rsm quorum sensing network in Pseudomonas fluorescens. Biosystems 101, 200–212. doi: 10.1016/j.biosystems.2010.07.004
Busto, E., Gotor-Fernández, V., and Gotor, V. (2006). Kinetic resolution of 4-chloro-2-(1-hydroxyalkyl) pyridines using Pseudomonas cepacia lipase. Nat. Protoc. 1, 2061–2067. doi: 10.1038/nprot.2006.335
Chang, I. A., Kim, I. H., Kang, S. C., Hou, C. T., and Kim, H. R. (2007). Production of 7, 10-dihydroxy-8(E)-octadecenoic acid from triolein via lipase induction by Pseudomonas aeruginosa PR3. Appl. Microbiol. Biotechnol. 74, 301–306. doi: 10.1007/s00253-006-0662-5
Chen, H. D., and Groisman, E. A. (2013). The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu. Rev. Microbiol. 67, 83–112. doi: 10.1146/annurev-micro-092412-155751
Cox, M., Gerritse, G., Dankmeyer, L., and Quax, W. J. (2001). Characterization of the promoter and upstream activating sequence from the Pseudomonas alcaligenes lipase gene. J. Biotechnol. 86, 9–17. doi: 10.1016/S0168-1656(00)00397-7
Deretic, V., Konyecsni, W. M., Mohr, C. D., Martin, D. W., and Hibler, N. S. (1989). Common denominators of promoter control in Pseudomonas and other bacteria. Biotechnology 7, 1250–1254. doi: 10.1038/nbt1289-1249
Dwivedee, B. P., Bhaumik, J., Rai, S. K., Laha, J. K., and Banerjee, U. C. (2017). Development of nanobiocatalysts through the immobilization of Pseudomonas fluorescens lipase for applications in efficient kinetic resolution of racemic compounds. Bioresour. Technol. 239, 464–471. doi: 10.1016/j.biortech.2017.05.050
Frangipani, E., Visaggio, D., Heeb, S., Kaever, V., Cámara, M., Visca, P., et al. (2014). The Gac/Rsm and cyclic-di-GMP signalling networks coordinately regulate iron uptake in Pseudomonas aeruginosa. Environ. Microbiol. 16, 676–688. doi: 10.1111/1462-2920.12164
Frenken, L. G., Bos, J. W., Visser, C., Müller, W., Tommassen, J., and Verrips, C. T. (1993). An accessory gene, lipB, required for the production of active Pseudomonas glumae lipase. Mol. Microbiol. 9, 579–589.
Gooderham, W. J., and Hancock, R. E. (2008). Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 33, 279–294. doi: 10.1111/j.1574-6976.2008.00135.x
Grbavčić, S., Bezbradica, D., Izrael-Živković, L., Avramović, N., Milosavić, N., KaradŽić, I., et al. (2011). Production of lipase and protease from an indigenous Pseudomonas aeruginosa strain and their evaluation as detergent additives: compatibility study with detergent ingredients and washing performance. Bioresour. Technol. 102, 11226–11233. doi: 10.1016/j.biortech.2011.09.076
Gupta, R., Gupta, N., and Rathi, P. (2004). Bacterial lipases: an overview of production, purification and biochemical properties. Appl. Microbiol. Biotechnol. 64, 763–781. doi: 10.1007/s00253-004-1568-8
Heeb, S., Heurlier, K., Valverde, C., Cámara, M., Haas, D., and Williams, P. (2004). Post- transcriptional regulation in pseudomonas spp. via the Gac/Rsm regulatory network. Virulence Gene Regul. 2, 239–255. doi: 10.1007/978-1-4419-9084-68
Heurlier, K., Williams, F., Heeb, S., Dormond, C., Pessi, G., Singer, D., et al. (2004). Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 186, 2936–2945. doi: 10.1128/JB.186.10.2936-2945.2004
Hoch, J. A. (2000). Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3, 165–170. doi: 10.1016/S1369-5274(00)00070-9
Jin, Z., Han, S. Y., Zhang, L., Zheng, S. P., Wang, Y., and Lin, Y. (2013). Combined utilization of lipase-displaying Pichia pastoris whole-cell biocatalysts to improve biodiesel production in co-solvent media. Bioresour. Technol. 130, 102–109. doi: 10.1016/j.biortech.2012.12.020
Krzeslak, J., Gerritse, G., van Merkerk, R., Cool, R. H., and Quax, W. J. (2008). Lipase expression in Pseudomonas alcaligenes is under the control of a two-component regulatory system. Appl. Environ. Microbiol. 74, 1402–1411. doi: 10.1128/AEM.01632-07
Krzeslak, J., Papaioannou, E., Merkerk, R. V., Paal, K. A., Bischoff, R. H., Cool, R. H., et al. (2012). Lipase A gene transcription in Pseudomonas alcaligenes is under control of RNA polymerase r54 and response regulator LipR. FEMS Microbiol. Lett. 329, 146–153. doi: 10.1111/j.1574-6968.2012.02516.x
Lalaouna, D., Fochesato, S., Sanchez, L., Schmitt-Kopplin, P., Haas, D., Heulin, T., et al. (2012). Phenotypic switching in Pseudomonas brassicacearum involves GacS- and GacA-dependent Rsm small RNAs. Appl. Environ. Microbiol. 78, 1658–1665. doi: 10.1128/AEM.06769-11
Lapouge, K., Schubert, M., Allain, F. H., and Haas, D. (2008). Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67, 241–253. doi: 10.1111/j.1365-2958.2007.06042.x
Leong, S. A., Ditta, G. S., and Helinski, D. R. (1982). Heme biosynthesis in Rhizobium. Identification of a cloned gene coding for delta-aminolevulinic acid synthetase from Rhizobium meliloti. J. Biol. Chem. 257, 8724–8730.
Ly, N. S., Yang, J., Bulitta, J. B., and Tsuji, B. T. (2012). Impact of two-component regulatory systems PhoP-PhoQ and PmrA-PmrB on colistin pharmacodynamics in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 56, 3453–3456. doi: 10.1128/AAC.06380-11
Marzi, S., and Romby, P. (2012). RNA mimicry, a decoy for regulatory proteins. Mol. Microbiol. 83, 1–6. doi: 10.1111/j.1365-2958.2011.07911.x
Nirandarjos, J., Saàanchez-Pescador, R., Urdea, M. A., and Covarrubias, A. (1987). The complete nudeotide sequence of the gfnALG operon of Escherichia coli K12. Nucleic Acids Res. 15, 2757–2770. doi: 10.1093/nar/15.6.2757
Pan, X. X., Xu, L., Zhang, Y., Xiao, X., Wang, X. F., Liu, Y., et al. (2012). Efficient display of active Geotrichum sp. lipase on pichia pastoris cell wall and its application as a whole-cell biocatalyst to enrich EPA and DHA in fish oil. J. Agric. Food Chem. 60, 9673–9679. doi: 10.1021/jf301827y
Quandt, J., and Hynes, M. F. (1993). Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127, 15–21. doi: 10.1016/0378-1119(93)90611-6
Reimmann, C., Beyeler, M., Latifi, A., Winteler, H., Foglino, M., Lazdunski, A., et al. (1997). The global activator GacA of Pseudomonas aeruginosa PAO1 positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24, 309–319. doi: 10.1046/j.1365-2958.1997.3291701.x
Reimmann, C., Valverde, C., Kay, E., and Haas, D. (2005). Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J. Bacteriol. 187, 276–285. doi: 10.1128/JB.187.1.276-285.2005
Romdhane, I. B. B., Fendri, A., Gargouri, Y., Gargouri, A., and Belghith, H. (2010). A novel thermoactive and alkaline lipase from Talaromyces thermophilus fungus for use in laundry detergents. Biochem. Eng. J. 53, 112–120. doi: 10.1016/j.bej.2010.10.002
Romeo, T. A., Vakulskas, C., and Babitzke, P. (2013). Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ. Microbiol. 15, 313–324. doi: 10.1111/j.1462-2920.2012.02794.x
Rosenau, F., and Jaeger, K.-E. (2000). Bacterial lipases from Pseudomonas: regulation of gene expression and mechanisms of secretion. Biochimie 82, 1023–1032. doi: 10.1016/S0300-9084(00)01182-2
Schubert, M., Lapouge, K., Duss, O., Oberstrass, F. C., Jelesarov, I., Haas, D., et al. (2007). Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat. Struct. Mol. Biol. 14, 807–813. doi: 10.1038/nsmb1285
Sonnleitner, E., and Haas, D. (2011). Small RNAs as regulators of primary and secondary metabolism in Pseudomonas species. Appl. Microbiol. Biotechnol. 91, 63–79. doi: 10.1007/s00253-011-3332-1
Sorger-Domenigg, T., Sonnleitner, E., Kaberdin, V. R., and Bläsi, U. (2007). Distinct and overlapping binding sites of Pseudomonas aeruginosa Hfq and RsmA proteins on the non-coding RNA RsmY. Biochem. Biophys. Res. Commun. 352, 769–773. doi: 10.1016/j.bbrc.2006.11.084
Tran, D. T., Chen, C. L., and Chang, J. S. (2016). Continuous biodiesel conversion via enzymatic transesterification catalyzed by immobilized Burkholderia lipase in a packed-bed bioreactor. Appl. Energy 168, 340–350. doi: 10.1016/j.apenergy.2016.01.082
Wang, D. P., Lee, S. H., Seeve, C., Yu, J. M., Pierson, L. S., and Pierson, E. A. (2013). Roles of the Gac-Rsm pathway in the regulation of phenazine biosynthesis in Pseudomonas chlororaphis 30-84. Microbiologyopen 2:505. doi: 10.1002/mbo3.90
Wohlfarth, S., Hoesche, C., Strunk, C., and Winkler, U. K. (1992). Molecular genetics of the extracellular lipase of Pseudomonas aeruginosa PAOl. J. Gen. Microbiol. 138, 1325–1335. doi: 10.1099/00221287-138-7-1325
Yang, W., Cao, H., Xu, L., Zhang, H., and Yan, Y. (2015). A novel eurythermic and thermostale lipase LipM from Pseudomonas moraviensis M9 and its application in the partial hydrolysis of algal oil. BMC Biotechnol. 15:94. doi: 10.1186/s12896-015-0214-0
Yoo, H. Y., Simkhada, J. R., Cho, S. S., Park, D. H., Kim, S. W., Seong, C. N., et al. (2011). A novel alkaline lipase from Ralstonia with potential application in biodiesel production. Bioresour. Technol. 102, 6104–6111. doi: 10.1016/j.biortech.2011.02.046
Keywords: lipase, Pseudomonas aeruginosa, mutation, expression, gene regulation
Citation: Liu W, Li M, Jiao L, Wang P and Yan Y (2018) PmrA/PmrB Two-Component System Regulation of lipA Expression in Pseudomonas aeruginosa PAO1. Front. Microbiol. 8:2690. doi: 10.3389/fmicb.2017.02690
Received: 15 October 2017; Accepted: 26 December 2017;
Published: 15 January 2018.
Edited by:
Qiang Wang, Institute of Hydrobiology (CAS), ChinaReviewed by:
Wenli Chen, Huazhong Agricultural University, ChinaXueWei Xu, Second Institute of Oceanography, State Oceanic Administration, China
Copyright © 2018 Liu, Li, Jiao, Wang and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunjun Yan, yanyunjun@hust.edu.cn
 Wu Liu1
Wu Liu1 Yunjun Yan
Yunjun Yan