
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 19 December 2017
Sec. Microbial Physiology and Metabolism
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.02546
The gram-positive filamentous bacterium Streptomyces is one of the largest resources for bioactive metabolites, particularly antibiotics. Antibiotic production and other metabolic processes are tightly regulated at the transcriptional level. Sigma (σ) factors are components of bacterial RNA polymerases that determine promoter specificity. In Streptomyces, σ factors also play essential roles in signal transduction and in regulatory networks, thereby assisting in their survival in complex environments. However, our current understanding of σ factors in Streptomyces is still limited. In this mini-review, we demonstrate the roles of Streptomyces σ factors, illustrating that these serve as linkers of different metabolic pathways. Further investigations on σ factors may improve our knowledge of Streptomyces physiology and benefit exploitation of Streptomyces resources.
Streptomyces is the largest genus of Actinobacteria and is characterized by complex morphological differentiation and the ability to produce valuable secondary metabolites (reviewed by Demain, 2002; Challis and Hopwood, 2003; Flardh and Buttner, 2009). This genus possesses complex regulatory systems that coordinate various biological processes and allows survival in their habitats. Transcriptional regulation is one of the most extensively studied regulatory mechanisms, with RNA polymerase (RNAP) playing a central role in transcription. Transcription initiation requires a holo-RNAP complex, which is composed of a core-RNAP (α2ββ′ω) and a dissociable sigma (σ) factor that recognizes specific promoter elements and facilitates transcription initiation (reviewed by Browning and Busby, 2004; Paget, 2015). Although the importance of σ factors has been established, further investigations on its regulatory mechanisms are warranted (reviewed by Feklistov et al., 2014; Browning and Busby, 2016). Sigma factors are not only a distinct subunit of RNAPs, but also an essential part of signal transduction systems. Endogenous and exogenous signals regulate the expression and activity of σ factors, thereby modifying the transcriptome (Cho et al., 2001; Takano et al., 2005; Hesketh et al., 2007; Feng et al., 2011). Moreover, various σ factors act as master regulators, regulating more than one biological process (Lee et al., 2005; Kim et al., 2012). In this mini-review, we provide an overview of the functions of σ factors in the life cycle of Streptomyces and illustrate that these also link different metabolic pathways, which benefits further investigation of σ factors and exploitation of Streptomyces resources.
Bacterial σ factors can be divided into two families, namely, σ70 and σ54. The σ54 family, which contains only one member that is distinct from σ70 proteins, has been excluded from this review. The σ70 family is named after the Escherichia coli housekeeping σ70 and consists of four subgroups (1, 2, 3, and 4) based on differential physical functions, phylogenic relationships, and possession of conserved domains σ1 (region 1.1), σ2 (region 1.2, 2.1–2.4), σ3 (region 3.0–3.2), and σ4 (region 4.1–4.2) (Figure 1) (reviewed by Paget, 2015). The number of members in the σ70 family, particularly those in groups 3 and 4, is highly variable among bacterial species. Streptomyces spp. encodes various σ factors to survive in complex environments and coordinate complicated physiological processes. This mini-review focuses on the Streptomyces σ70 family proteins.
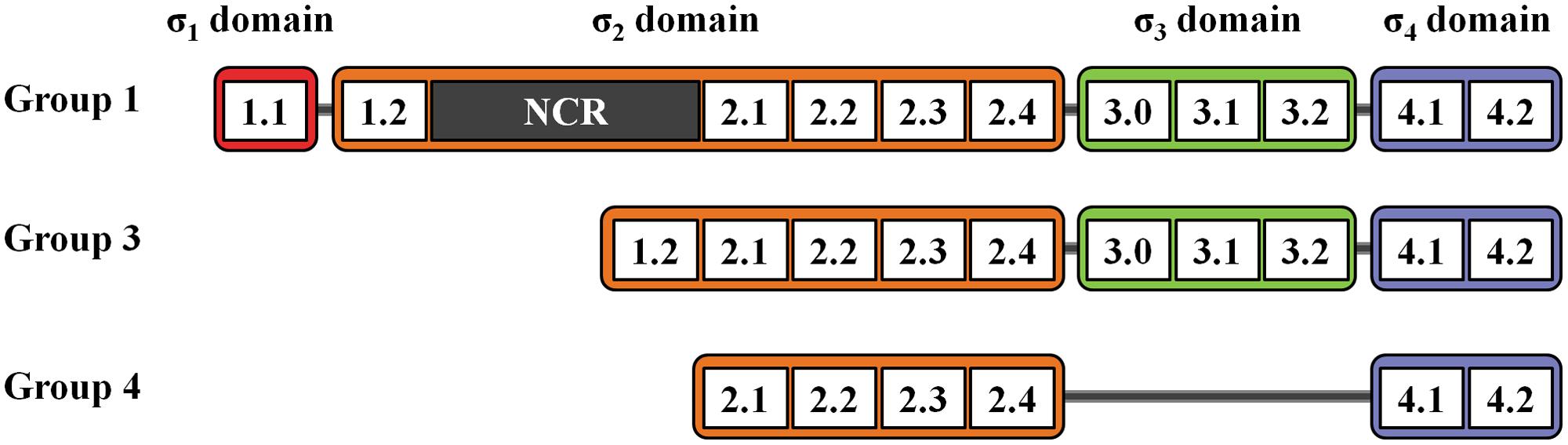
FIGURE 1. σ factor domains in the σ70 family. Domain organization of σ70 family members from groups 1, 3, and 4.
Group 1 σ factors, including E. coli σ70 and its orthologs, possess all four σ domains and a non-conserved region (NCR) of variable length located between regions 1.2 and 2.1 (Figure 1). NCR is involved in core interactions and promoter escape (Leibman and Hochschild, 2007). Group 1 σ factors are responsible for the transcription of essential genes and are thus also called “housekeeping σ factors” (reviewed by Paget, 2015). In Streptomyces, principal housekeeping σ factor (σHrdB) is not only required for essential gene expression, but is also involved in morphological differentiation and secondary metabolism (Fujii et al., 1996). Studies have revealed that σHrdB affects secondary metabolism in at least two ways. First, σHrdB initiates the transcription of pathway-specific regulatory genes that are essential to secondary metabolite biosynthesis. For example, in S. coelicolor, σHrdB regulates the biosynthesis of actinohordin (ACT) and undecylprodigiosin (RED) by initiating the transcription of the pathway-specific regulatory genes actII-ORF4 and redD (Fujii et al., 1996). Similarly, in S. avermitilis, σHrdB initiates the transcription of the pathway-specific regulatory gene aveR to promote the biosynthesis of avermectin (Zhuo et al., 2010). Moreover, the biosynthesis of secondary metabolites requires precursors and energy, which is produced by σHrdB-dependent primary metabolism. Thus, σHrdB mediates the metabolic flux from primary to secondary metabolism.
Because σHrdB connects primary metabolism with secondary metabolism, genetic manipulation of the hrdB gene has been utilized as a strategy in enhancing antibiotic production. The engineering of hrdB can be performed by at least two methods. In the first method, the site-mutated hrdB gene that triggers the upregulation of aveR transcription is introduced into avermectin high-producing strain 3–115, thereby improving avermectin B1a yield by over 50% (Zhuo et al., 2010). In the second method, as the strength of the hrdB promoter (hrdBp) is greater than other commonly used promoters in Streptomyces, the native promoters of the antibiotic biosynthesis genes or pathway-specific regulatory genes are replaced with hrdBp to improve antibiotic yield. For example, nikkomycin yield is significantly enhanced by introducing hrdBp-driven sanG, which encodes the pathway-specific activator of nikkomycin biosynthesis in S. ansochromogenes TH322 (Du et al., 2013).
The structure of group 2 σ factors is highly similar to that of group 1 σ factors, except for the absence of region 1.1. Furthermore, group 2 σ factors are not essential to bacterial growth. The number of group 2 σ factors encoded by bacterial genomes is highly variable (Table 1) (Buttner and Lewis, 1992; Osanai et al., 2008; Battesti et al., 2011). The Streptomyces genome usually contains three group 2 σ factors: σHrdA, σHrdC, and σHrdD (Buttner et al., 1990; Forsman and Granstrom, 1992; Kormanec et al., 1992; Wang et al., 2015). The majority of studies have focused on σHrdD.
Although σHrdD is transcribed during growth, and the σHrdD-RNAP holoenzyme can initiate transcription of genes that are involved in antibiotic biosynthesis (e.g., actII-ORF4, redD) and differentiation (e.g., whiB) in vitro, hrdD mutants do not exhibit changes in secondary metabolism and differentiation (Fujii et al., 1996; Kang et al., 1997), thereby prompting us to examine the biological significance of σHrdD. Earlier studies have determined that hrdD can be induced by stresses such as envelope stress, oxidative damage, and hyperosmolarity (Kang et al., 1997; Paget et al., 2001; Lee et al., 2005), suggesting that σHrdD may be involved in stress response processes. Moreover, in pathogenic actinobacterium, Mycobacterium tuberculosis, group 2 σ factor σMtbB can specifically bind to RbpA, a transcriptional activator without DNA-binding activity, and the activity of σMtbB is stringently dependent on RbpA (Hu et al., 2014). These findings suggest that in Streptomyces, σHrdD may bind to an RbpA-like protein to form a complex, which is required for survival under stress conditions.
Although the expression of group 2 σ factors in Streptomyces may be related to multiple biological processes, knowledge of their biological significance remains limited. E. coli σS, the most extensively studied group 2 σ factor, can affect the expression of about 500 target genes under stress conditions. It is not only involved in general stress responses but also in survival in the stationary phase (Weber et al., 2005; Battesti et al., 2011). Therefore, further investigations are warranted to clarify the underlying regulatory mechanisms of Streptomyces group 2 σ factors.
Group 3 σ factors possess σ2, σ3, and σ4 domains (Figure 1), which are referred to as alternative σ factors that are used to direct RNAP to genes involved in adaptive responses (reviewed by Gruber and Gross, 2003). In Streptomyces, alternative σ factors modulate multiple metabolic pathways in response to intracellular or extracellular signals (Kormanec and Sevcikova, 2002; Viollier et al., 2003; Dalton et al., 2007; Bobek et al., 2014).
σWhiG, an alternative σ factor that regulates differentiation, orchestrates the formation of spores by activating the transcription of three regulatory genes, namely, whiA, whiH, and whiI, which are essential for cell division and spore maturation (Ryding et al., 1998; Ainsa et al., 1999; Kaiser and Stoddard, 2011). In S. chattanoogensis, σWhiG also regulates secondary metabolism. σWhiG promotes natamycin biosynthesis by directly activating the transcription of two natamycin biosynthetic genes, scnC (encodes an aminotransferase) and scnD (encodes a P450 monooxygenase). The deletion of scnC or scnD results in the accumulation of two natamycin analogs, 4,5-deepoxy-natamycinolide and 4,5-deepoxy-natamycin, respectively, but abolishes natamycin biosynthesis (Liu et al., 2015a,b). Besides, whiG expression is regulated by bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP), an important second messenger that imparts global effects in Streptomyces (reviewed by Bush et al., 2015). A transcriptional repressor BldD negatively regulates whiG transcription in a (c-di-GMP)-dependent manner. At the vegetative growth stage, high concentrations of c-di-GMP lead to the formation of BldD-(c-di-GMP) complexes that bind to whiG promoter regions, repressing its transcription. When morphological differentiation starts, c-di-GMP concentrations decline, causing the release of BldD from the target DNA, thereby relieving transcriptional suppression of whiG (Elliot et al., 2001; Tschowri et al., 2014). Therefore, σWhiG responds to the intracellular signal molecule and coordinates morphological differentiation as well as secondary metabolism.
Except for σWhiG, S. coelicolor has nine other alternative σ factors (σB, σF, σG, σH, σI, σK, σL, σM, and σN), all belonging to the σB-like σ factors, which are homologs of Bacillus subtilis σB (Cho et al., 2001; Lee et al., 2004). These σB-like σ factors play important roles in the Streptomyces regulatory system (Kelemen et al., 1996; Kormanec and Sevcikova, 2002; Lee et al., 2005; Mao et al., 2009; Wang et al., 2010). Among the σB-like σ factors, σB has been most extensively investigated. σB can control morphological differentiation and secondary metabolism by directly regulating differentiation-related genes (e.g., dpsA required for spore maturation and whiB that is required for aerial hyphae formation) and antibiotic biosynthetic genes (e.g., redH that encodes a phosphoenolpyruvate-utilizing enzyme and redZ that encodes a pathway-specific regulator for RED) (Lee et al., 2005; Facey et al., 2009, 2011). Furthermore, σB controls morphological differentiation and secondary metabolism by regulating the ppGpp synthesis (Lee et al., 2004). Guanosine tetraphosphate (ppGpp) is an extensively studied signaling nucleotide that is synthesized in nutrient-limited conditions. It interacts directly with RNAP to modulate the transcriptome (reviewed by Hauryliuk et al., 2015). In S. coelicolor, ppGpp affects differentiation and secondary metabolism by modulating the expression of genes required for aerial hyphae formation and pathway-specific regulators (Hesketh et al., 2007). σB influences intracellular ppGpp concentrations by directly regulating the ppGpp synthetase gene relA (Lee et al., 2004).
In addition, σB responds to environmental stress signals and activates the stress-related regulon. First, σB controls osmotic defensive responses by initiating the transcription of genes that are involved in the synthesis and uptake of osmotic compatible solutes (Lee et al., 2005; Fernandez-Martinez et al., 2009), as well as activating two other alternative σ factors that also contribute to osmotic stress responses, namely, σL and σM, in a hierarchical order to strengthen the regulatory network (Lee et al., 2005). Second, σB is involved in oxidative stress responses by promoting cysteine and mycothiol (streptomycetes major thiol buffer) biosynthesis and the expression of catalase (Lee et al., 2005). Finally, the σB regulon includes cold shock proteins, thereby suggesting that it may be involved in cold shock responses (Lee et al., 2005). Interestingly, upon osmotic stress, σB rapidly activates the osmotic defense system and subsequently promotes the transcription of oxidative stress-related genes (Lee et al., 2005), suggesting that σB protects S. coelicolor from subsequent oxidative damage after osmotic shock. These findings indicate that σB governs multiple stress responses in Streptomyces. In summary, σB governs multiple biological processes and acts as a key element in the stress response system, thereby eliciting rapid responses to changes in habitat.
Group 4 σ factors are the most divergent members of the σ70 family and exclusively contain the σ2 (which lacks region 1.2) and σ4 domains (Figure 1). As group 4 σ factors usually correlate with extracytoplasmic stimuli, these are also called extracytoplasmic function (ECF) σ factors (reviewed by Staron et al., 2009; Paget, 2015). The Streptomyces genome encodes various ECF σ factors, which reflects their complex habitats and complicated metabolic processes (Table 1).
Extracytoplasmic function σ factors play essential roles in the stress response system of Streptomyces spp. One of the most extensively studied stress-responsive ECF σ factor is S. coelicolor σR. It is induced by oxidants and translation-inhibiting antibiotics and subsequently activates the oxidative damage reparatory system, ribosome-associated products, and transcriptional regulators (e.g., RbpA) (Paget et al., 2001; Yoo et al., 2016; Lee et al., 2017). Additionally, during thiol-oxidative stress, σR is required to maintain the level and activity of σHrdB, thereby suggesting a crosstalk between ECF σ factor and housekeeping σ factor during stress (Kim et al., 2012). These findings reveal that σR is a master regulator of defense responses against multiple stresses. On the other hand, σR is regulated at multiple levels. First, σR activity is regulated by its cognate anti-σ factor, RsrA. Under normal conditions, RsrA binds to σR and sequesters σR from RNAP. Under stress, RsrA undergoes structural changes that cause the release of σR, which subsequently regulates its regulons (Kang et al., 1999). The expression of σR is also regulated at the transcriptional (promoted by transcriptional activator WblC), translational (repressed by translation initiation factor IF3), and post-translational (degraded by Clp proteases) levels (Kim et al., 2009; Yoo et al., 2016; Feeney et al., 2017). Taken together, σR is a pivotal element in the complex regulatory network of Streptomyces spp.
Extracytoplasmic function σ factors are involved in morphological differentiation. The widely distributed ECF σ factor, σBldN, is required for aerial mycelium formation (Bibb et al., 2000). Studies have revealed that σBldN is responsible for the expression of differentiation-related proteins chaplin, rodlin, and BldM (Bibb et al., 2000, 2012). Deletion of bldN abolishes aerial hyphae formation, thereby causing the “bald” phenotype (Bibb et al., 2000; Yague et al., 2014).
Regulation of secondary metabolism by ECF σ factors is commonly identified among actinomycetes, and some pathway-specific ECF σ factors are located within the antibiotic biosynthesis gene cluster. σMibX in Microbispora corallina is the most extensively studied cluster-situated ECF σ factor that controls the biosynthesis and maturation of microbisporicin (Foulston and Bibb, 2010). Microbisporicin is synthesized by the mib gene cluster, which is composed of six transcriptional units (i.e., mibJYZO, mibQ, mibR, mibXW, mibABCDTUV, and mibEFHSN). The LuxR-family regulator MibR is responsible for the activation of mibABCDTUV, and the ECF σ factor σMibX initiates the transcription of mibJYZO, mibQ, mibR, mibXW, and mibEFHSN. Under normal conditions, the transcriptional activity of σMibX is sequestered by its cognate anti-σ factor MibW, abolishing microbisporicin production. Upon nutrient limitation, cellular ppGpp concentrations increase and subsequently activate mibR transcription in a σMibX-independent manner, followed by the expression of the mibABCDTUV operon, thereby inducing the biosynthesis of immature microbisporicin. Subsequently, the immature antibiotic inactivates MibW, leading to the release of σMibX, which in turn upregulates the expression of mature microbisporicin (Foulston and Bibb, 2011; Fernandez-Martinez et al., 2015). This model illustrates that antibiotics can function as a signal and an ECF-σ factor-(anti-σ factor) complex may play a crucial role in the signaling pathway.
In Streptomyces, the only identified cluster-situated ECF σ factor is σAntA, which has been reported in S. albus. Unlike σMibX, no anti-σ factor regulating the transcriptional activity of σAntA has been reported to date (Seipke et al., 2014). Interestingly, σAntA expression and antimycin production diminish upon differentiation (Seipke et al., 2014), suggesting that the expression/activity of σAntA may be “switched off” by some specific regulatory mechanism that is coupled with differentiation, yet needs further investigation.
In summary, ECF σ factors play essential roles in responding to signals and modulate Streptomyces metabolism. However, compared to various other ECF σ factors in this particular bacterial species, researches investigations on ECF σ factors and the mechanism underlying their response to signals are limited.
Further investigations on ECF σ factors, the corresponding signals, and the relationship between these signals and biological processes are necessary to enrich our understanding of the Streptomyces regulatory network.
Transcriptional regulation is a crucial mechanism in modulating gene expression, and RNAP is the key element in transcription. Sigma factors are the only variable elements of holo-RNAP, indicating that the selection of a particular target gene is mainly dependent on σ factors. In most cases, unlike other transcriptional factors that upregulate or downregulate gene expression, σ factors act as “switches” that turn expression on or off. In this mini-review, we focused on the members of the σ70 family in Streptomyces, illustrating their function and underlying mechanisms.
Streptomyces species undergo complex morphological differentiation and synthesize various secondary metabolites, which are governed by a complex regulatory network. As a crucial component of the transcriptional regulatory system, σ factors fine-tune gene expression and regulate metabolic fluxes. Thus, engineering of specific σ factors is a feasible method of generating high-producing antibiotic strains (Zhuo et al., 2010), which requires detailed and precise knowledge of σ factor function. However, except for very few members, the biological function of most σ factors and whether these are related to secondary metabolism remains unclear. Thus, further investigations on the functions of Streptomyces σ factors should be conducted.
Streptomyces has a subtle signal sensing and transduction network that responds to extracellular and intracellular signals. Recent studies have revealed that the majority of antibiotic biosynthesis gene clusters in Streptomyces are not expressed, and these clusters are described as “silent.” These silent metabolites are not produced under normal conditions, suggesting that special signals are needed to activate them (reviewed by Okada and Seyedsayamdost, 2017). Sigma factors may respond to some special signals and activate these silent antibiotic synthetic gene clusters, thereby extending our arsenal of bioactive metabolites. Hopefully, future investigations on σ factors may improve our understanding of the Streptomyces signal transduction system, thereby promoting antibiotic production and the discovery of novel secondary metabolites.
DS, CL, and JZ conducted the literature study and wrote the draft manuscript. WL edited and revised the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was supported by the National Natural Science Foundation of China (31300054), Fund of the Natural Science Foundation of Jiangsu Province of China (BK20171163), Natural Science Foundation of Xuzhou City (KC15N0014), and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Ainsa, J. A., Parry, H. D., and Chater, K. F. (1999). A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 34, 607–619. doi: 10.1046/j.1365-2958.1999.01630.x
Battesti, A., Majdalani, N., and Gottesman, S. (2011). The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65, 189–213. doi: 10.1146/annurev-micro-090110-102946
Bibb, M. J., Domonkos, A., Chandra, G., and Buttner, M. J. (2012). Expression of the chaplin and rodlin hydrophobic sheath proteins in Streptomyces venezuelae is controlled by σBldN and a cognate anti-sigma factor, RsbN. Mol. Microbiol. 84, 1033–1049. doi: 10.1111/j.1365-2958.2012.08070.x
Bibb, M. J., Molle, V., and Buttner, M. J. (2000). σBldN, an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor A3(2). J. Bacteriol. 182, 4606–4616. doi: 10.1128/JB.182.16.4606-4616.2000
Bobek, J., Strakova, E., Zikova, A., and Vohradsky, J. (2014). Changes in activity of metabolic and regulatory pathways during germination of S. coelicolor. BMC Genomics 15:1173. doi: 10.1186/1471-2164-15-1173
Browning, D. F., and Busby, S. J. (2004). The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2, 57–65. doi: 10.1038/nrmicro787
Browning, D. F., and Busby, S. J. (2016). Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 14, 638–650. doi: 10.1038/nrmicro.2016.103
Bush, M. J., Tschowri, N., Schlimpert, S., Flardh, K., and Buttner, M. J. (2015). c-di-GMP signalling and the regulation of developmental transitions in streptomycetes. Nat. Rev. Microbiol. 13, 749–760. doi: 10.1038/nrmicro3546
Buttner, M. J., Chater, K. F., and Bibb, M. J. (1990). Cloning, disruption, and transcriptional analysis of three RNA polymerase sigma factor genes of Streptomyces coelicolor A3(2). J. Bacteriol. 172, 3367–3378. doi: 10.1128/jb.172.6.3367-3378.1990
Buttner, M. J., and Lewis, C. G. (1992). Construction and characterization of Streptomyces coelicolor A3(2) mutants that are multiply deficient in the nonessential hrd-encoded RNA polymerase sigma factors. J. Bacteriol. 174, 5165–5167. doi: 10.1128/jb.174.15.5165-5167.1992
Challis, G. L., and Hopwood, D. A. (2003). Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. U.S.A. 100(Suppl. 2), 14555–14561. doi: 10.1073/pnas.1934677100
Cho, Y. H., Lee, E. J., Ahn, B. E., and Roe, J. H. (2001). SigB, an RNA polymerase sigma factor required for osmoprotection and proper differentiation of Streptomyces coelicolor. Mol. Microbiol. 42, 205–214. doi: 10.1046/j.1365-2958.2001.02622.x
Dalton, K. A., Thibessard, A., Hunter, J. I. B., and Kelemen, G. H. (2007). A novel compartment, the ‘subapical stem’ of the aerial hyphae, is the location of a sigN-dependent, developmentally distinct transcription in Streptomyces coelicolor. Mor. Microbiol. 64, 719–737. doi: 10.1111/j.1365-2958.2007.05684.x
Demain, A. L. (2002). Prescription for an ailing pharmaceutical industry. Nat. Biotechnol. 20:331. doi: 10.1038/nbt0402-331
Du, D., Zhu, Y., Wei, J., Tian, Y., Niu, G., and Tan, H. (2013). Improvement of gougerotin and nikkomycin production by engineering their biosynthetic gene clusters. Appl. Microbiol. Biotechnol. 97, 6383–6396. doi: 10.1007/s00253-013-4836-7
Elliot, M. A., Bibb, M. J., Buttner, M. J., and Leskiw, B. K. (2001). BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3(2). Mol. Microbiol. 40, 257–269. doi: 10.1046/j.1365-2958.2001.02387.x
Facey, P. D., Hitchings, M. D., Saavedra-Garcia, P., Fernandez-Martinez, L., Dyson, P. J., and Del Sol, R. (2009). Streptomyces coelicolor Dps-like proteins: differential dual roles in response to stress during vegetative growth and in nucleoid condensation during reproductive cell division. Mol. Microbiol. 73, 1186–1202. doi: 10.1111/j.1365-2958.2009.06848.x
Facey, P. D., Sevcikova, B., Novakova, R., Hitchings, M. D., Crack, J. C., Kormanec, J., et al. (2011). The dpsA gene of Streptomyces coelicolor: induction of expression from a single promoter in response to environmental stress or during development. PLOS ONE 6:e25593. doi: 10.1371/journal.pone.0025593
Feeney, M. A., Chandra, G., Findlay, K. C., Paget, M. S. B., and Buttner, M. J. (2017). Translational control of the SigR-directed oxidative stress response in Streptomyces via IF3-mediated repression of a noncanonical GTC start codon. mBio 8:e00815-17. doi: 10.1128/mBio.00815-17
Feklistov, A., Sharon, B. D., Darst, S. A., and Gross, C. A. (2014). Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 68, 357–376. doi: 10.1146/annurev-micro-092412-155737
Feng, W. H., Mao, X. M., Liu, Z. H., and Li, Y. Q. (2011). The ECF sigma factor SigT regulates actinorhodin production in response to nitrogen stress in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 92, 1009–1021. doi: 10.1007/s00253-011-3619-2
Fernandez-Martinez, L., Bishop, A., Parkes, L., Del Sol, R., Salerno, P., Sevcikova, B., et al. (2009). Osmoregulation in Streptomyces coelicolor: modulation of SigB activity by OsaC. Mol. Microbiol. 71, 1250–1262. doi: 10.1111/j.1365-2958.2009.06599.x
Fernandez-Martinez, L. T., Gomez-Escribano, J. P., and Bibb, M. J. (2015). A relA-dependent regulatory cascade for auto-induction of microbisporicin production in Microbispora corallina. Mol. Microbiol. 97, 502–514. doi: 10.1111/mmi.13046
Flardh, K., and Buttner, M. J. (2009). Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7, 36–49. doi: 10.1038/nrmicro1968
Forsman, M., and Granstrom, M. (1992). Mutagenic analysis of the promoter of the Streptomyces fradiae beta-lactamase-encoding gene. Gene 121, 87–94. doi: 10.1016/0378-1119(92)90165-L
Foulston, L., and Bibb, M. (2011). Feed-forward regulation of microbisporicin biosynthesis in Microbispora corallina. J. Bacteriol. 193, 3064–3071. doi: 10.1128/JB.00250-11
Foulston, L. C., and Bibb, M. J. (2010). Microbisporicin gene cluster reveals unusual features of lantibiotic biosynthesis in actinomycetes. Proc. Natl. Acad. Sci. U.S.A. 107, 13461–13466. doi: 10.1073/pnas.1008285107
Fujii, T., Gramajo, H. C., Takano, E., and Bibb, M. J. (1996). redD and actII-ORF4, pathway-specific regulatory genes for antibiotic production in Streptomyces coelicolor A3(2), are transcribed in vitro by an RNA polymerase holoenzyme containing σhrdD. J. Bacteriol. 178, 3402–3405. doi: 10.1128/jb.178.11.3402-3405.1996
Gruber, T. M., and Gross, C. A. (2003). Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57, 441–466. doi: 10.1146/annurev.micro.57.030502.090913
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T., and Gerdes, K. (2015). Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309. doi: 10.1038/nrmicro3448
Hesketh, A., Chen, W. J., Ryding, J., Chang, S., and Bibb, M. (2007). The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol. 8:R161. doi: 10.1186/gb-2007-8-8-r161
Hu, Y., Morichaud, Z., Perumal, A. S., Roquet-Baneres, F., and Brodolin, K. (2014). Mycobacterium RbpA cooperates with the stress-response σB subunit of RNA polymerase in promoter DNA unwinding. Nucleic Acids Res. 42, 10399–10408. doi: 10.1093/nar/gku742
Kaiser, B. K., and Stoddard, B. L. (2011). DNA recognition and transcriptional regulation by the WhiA sporulation factor. Sci. Rep. 1:156. doi: 10.1038/srep00156
Kang, J. G., Hahn, M. Y., Ishihama, A., and Roe, J. H. (1997). Identification of sigma factors for growth phase-related promoter selectivity of RNA polymerases from Streptomyces coelicolor A3(2). Nucleic Acids Res. 25, 2566–2573. doi: 10.1093/nar/25.13.2566
Kang, J. G., Paget, M. S., Seok, Y. J., Hahn, M. Y., Bae, J. B., Hahn, J. S., et al. (1999). RsrA, an anti-sigma factor regulated by redox change. EMBO J. 18, 4292–4298. doi: 10.1093/emboj/18.15.4292
Kelemen, G. H., Brown, G. L., Kormanec, J., Potuckova, L., Chater, K. F., and Buttner, M. J. (1996). The positions of the sigma-factor genes, whiG and sigF, in the hierarchy controlling the development of spore chains in the aerial hyphae of Streptomyces coelicolor A3(2). Mol. Microbiol. 21, 593–603. doi: 10.1111/j.1365-2958.1996.tb02567.x
Kim, M. S., Dufour, Y. S., Yoo, J. S., Cho, Y. B., Park, J. H., Nam, G. B., et al. (2012). Conservation of thiol-oxidative stress responses regulated by SigR orthologues in actinomycetes. Mol. Microbiol. 85, 326–344. doi: 10.1111/j.1365-2958.2012.08115.x
Kim, M. S., Hahn, M. Y., Cho, Y., Cho, S. N., and Roe, J. H. (2009). Positive and negative feedback regulatory loops of thiol-oxidative stress response mediated by an unstable isoform of σR in actinomycetes. Mol. Microbiol. 73, 815–825. doi: 10.1111/j.1365-2958.2009.06824.x
Kormanec, J., Farkasovsky, M., and Potuckova, L. (1992). Four genes in Streptomyces aureofaciens containing a domain characteristic of principal sigma factors. Gene 122, 63–70. doi: 10.1016/0378-1119(92)90032-K
Kormanec, J., and Sevcikova, B. (2002). Stress-response sigma factor σH directs expression of the gltB gene encoding glutamate synthase in Streptomyces coelicolor A3(2). Biochim. Biophys. Acta 1577, 149–154. doi: 10.1016/S0167-4781(02)00409-8
Lee, E. J., Cho, Y. H., Kim, H. S., and Roe, J. H. (2004). Identification of σB-dependent promoters using consensus-directed search of Streptomyces coelicolor genome. J. Microbiol. 42, 147–151.
Lee, E. J., Karoonuthaisiri, N., Kim, H. S., Park, J. H., Cha, C. J., Kao, C. M., et al. (2005). A master regulator σB governs osmotic and oxidative response as well as differentiation via a network of sigma factors in Streptomyces coelicolor. Mol. Microbiol. 57, 1252–1264. doi: 10.1111/j.1365-2958.2005.04761.x
Lee, K. L., Yoo, J. S., Oh, G. S., Singh, A. K., and Roe, J. H. (2017). Simultaneous activation of iron- and thiol-based sensor-regulator systems by redox-active compounds. Front. Microbiol. 8:139. doi: 10.3389/fmicb.2017.00139
Leibman, M., and Hochschild, A. (2007). A sigma-core interaction of the RNA polymerase holoenzyme that enhances promoter escape. EMBO J. 26, 1579–1590. doi: 10.1038/sj.emboj.7601612
Liu, S. P., Yu, P., Yuan, P. H., Zhou, Z. X., Bu, Q. T., Mao, X. M., et al. (2015a). Sigma factor WhiGch positively regulates natamycin production in Streptomyces chattanoogensis L10. Appl. Microbiol. Biotechnol. 99, 2715–2726. doi: 10.1007/s00253-014-6307-1
Liu, S. P., Yuan, P. H., Wang, Y. Y., Liu, X. F., Zhou, Z. X., Bu, Q. T., et al. (2015b). Generation of the natamycin analogs by gene engineering of natamycin biosynthetic genes in Streptomyces chattanoogensis L10. Microbiol. Res. 173, 25–33. doi: 10.1016/j.micres.2015.01.013
Mao, X. M., Zhou, Z., Hou, X. P., Guan, W. J., and Li, Y. Q. (2009). Reciprocal regulation between SigK and differentiation programs in Streptomyces coelicolor. J. Bacteriol. 191, 6473–6481. doi: 10.1128/JB.00875-09
Okada, B. K., and Seyedsayamdost, M. R. (2017). Antibiotic dialogues: induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol. Rev. 41, 19–33. doi: 10.1093/femsre/fuw035
Osanai, T., Ikeuchi, M., and Tanaka, K. (2008). Group 2 sigma factors in cyanobacteria. Physiol. Plant. 133, 490–506. doi: 10.1111/j.1399-3054.2008.01078.x
Paget, M. S. (2015). Bacterial sigma factors and anti-sigma factors: structure, function and distribution. Biomolecules 5, 1245–1265. doi: 10.3390/biom5031245
Paget, M. S., Molle, V., Cohen, G., Aharonowitz, Y., and Buttner, M. J. (2001). Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the σR regulon. Mol. Microbiol. 42, 1007–1020. doi: 10.1046/j.1365-2958.2001.02675.x
Ryding, N. J., Kelemen, G. H., Whatling, C. A., Flardh, K., Buttner, M. J., and Chater, K. F. (1998). A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 29, 343–357. doi: 10.1046/j.1365-2958.1998.00939.x
Seipke, R. F., Patrick, E., and Hutchings, M. I. (2014). Regulation of antimycin biosynthesis by the orphan ECF RNA polymerase sigma factor σAntA. PeerJ 2:e253. doi: 10.7717/peerj.253
Staron, A., Sofia, H. J., Dietrich, S., Ulrich, L. E., Liesegang, H., and Mascher, T. (2009). The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol. Microbiol. 74, 557–581. doi: 10.1111/j.1365-2958.2009.06870.x
Takano, H., Obitsu, S., Beppu, T., and Ueda, K. (2005). Light-induced carotenogenesis in Streptomyces coelicolor A3(2): identification of an extracytoplasmic function sigma factor that directs photodependent transcription of the carotenoid biosynthesis gene cluster. J. Bacteriol. 187, 1825–1832. doi: 10.1128/JB.187.5.1825-1832.2005
Tschowri, N., Schumacher, M. A., Schlimpert, S., Chinnam, N. B., Findlay, K. C., Brennan, R. G., et al. (2014). Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158, 1136–1147. doi: 10.1016/j.cell.2014.07.022
Viollier, P. H., Kelemen, G. H., Dale, G. E., Nguyen, K. T., Buttner, M. J., and Thompson, C. J. (2003). Specialized osmotic stress response systems involve multiple SigB-like sigma factors in Streptomyces coelicolor. Mol. Microbiol. 47, 699–714. doi: 10.1046/j.1365-2958.2003.03302.x
Wang, C., Long, X., Mao, X., Dong, H., Xu, L., and Li, Y. (2010). SigN is responsible for differentiation and stress responses based on comparative proteomic analyses of Streptomyces coelicolor wild-type and sigN deletion strains. Microbiol. Res. 165, 221–231. doi: 10.1016/j.micres.2009.05.003
Wang, L., Gao, C., Tang, N., Hu, S., and Wu, Q. (2015). Identification of genetic variations associated with epsilon-poly-lysine biosynthesis in Streptomyces albulus ZPM by genome sequencing. Sci. Rep. 5:9201. doi: 10.1038/srep09201
Weber, H., Polen, T., Heuveling, J., Wendisch, V. F., and Hengge, R. (2005). Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187, 1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005
Yague, P., Rodriguez-Garcia, A., Lopez-Garcia, M. T., Rioseras, B., Martin, J. F., Sanchez, J., et al. (2014). Transcriptomic analysis of liquid non-sporulating Streptomyces coelicolor cultures demonstrates the existence of a complex differentiation comparable to that occurring in solid sporulating cultures. PLOS ONE 9:e86296. doi: 10.1371/journal.pone.0086296
Yoo, J. S., Oh, G. S., Ryoo, S., and Roe, J. H. (2016). Induction of a stable sigma factor SigR by translation-inhibiting antibiotics confers resistance to antibiotics. Sci. Rep. 6:28628. doi: 10.1038/srep28628
Keywords: Streptomyces, σ factor, primary metabolism, secondary metabolism, morphological differentiation, stress response
Citation: Sun D, Liu C, Zhu J and Liu W (2017) Connecting Metabolic Pathways: Sigma Factors in Streptomyces spp. Front. Microbiol. 8:2546. doi: 10.3389/fmicb.2017.02546
Received: 09 October 2017; Accepted: 07 December 2017;
Published: 19 December 2017.
Edited by:
Michael Sauer, University of Natural Resources and Life Sciences, Vienna, AustriaReviewed by:
Janice Lorraine Strap, University of Ontario Institute of Technology, CanadaCopyright © 2017 Sun, Liu, Zhu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weijie Liu, bGVvbmxpdTIwMTNAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.