
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 12 December 2017
Sec. Extreme Microbiology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.02474
Extremophilic organisms represent a potentially valuable resource for the development of novel bioprocesses. They can act as a source for stable enzymes and unique biomaterials. Extremophiles are capable of carrying out microbial processes and biotransformations under extremely hostile conditions. Extreme thermoacidophilic members of the well-characterized genus Sulfolobus are outstanding in their ability to thrive at both high temperatures and low pH. This review gives an overview of the biological system Sulfolobus including its central carbon metabolism and the development of tools for its genetic manipulation. We highlight findings of commercial relevance and focus on potential industrial applications. Finally, the current state of bioreactor cultivations is summarized and we discuss the use of Sulfolobus species in biorefinery applications.
Thermophiles gain increasing attention in biotechnological applications due to their potential to expand the thermal range of industrial biotechnology and their unique metabolic capabilities (Littlechild, 2015; Zeldes et al., 2015; Beeler and Singh, 2016; Donati et al., 2016; Basen and Müller, 2017; Straub et al., 2017). In this review, we focus on the well-characterized members of the phylum Crenarchaeota, the extreme thermoacidophilic Archaea belonging to the genus Sulfolobus. Natural habitats of these organisms are solfataric fields all around the world, including the United States, Costa Rica, Mexico, Russia, Japan, China, New Zealand, Germany, Italy, and Iceland. The outstanding characteristic of these organisms, which have been investigated since the 1970s (Supplementary Table S1), is their ability to thrive at extremely low pH and high temperature, unprecedented in Eukaryotes and Bacteria.
Since Sulfolobus spp. can be grown and manipulated under laboratory conditions, they are popular model organisms to study Archaea. Research has been focused on their biology and physiology. Currently, genomics (Bell et al., 2002; Dai et al., 2016), proteomics (Chong and Wright, 2005; Ellen et al., 2010; Pham et al., 2010; Kort et al., 2013), metabolomics (Ulas et al., 2012; Bräsen et al., 2014), composition and function of the archaeal membrane (Albers and Meyer, 2011) and the archaellum (Albers and Jarrell, 2015), as well as interaction with archaeal viruses (Prangishvili et al., 2006) are important fields of research. Nevertheless, there is also growing interest in the utilization of this genus in biotechnological applications and the development of engineered strains to exploit the organisms’ unique characteristics. Sulfolobus spp. are a source of unique enzymes (Littlechild, 2015), biomaterials (Benvegnu et al., 2009; Besse et al., 2015), and metabolic pathways (Bräsen et al., 2014). As most prominent examples, the branched Entner–Doudoroff (ED) pathway (Kouril et al., 2013b) as well as Weimberg and Dahms pathways for the degradation of hexoses and pentoses (Nunn et al., 2010) should be named. These diverse catabolic pathways present a promising field for the exploitation of novel products (Ahmed et al., 2005; Siebers and Schönheit, 2005).
Among the eight Sulfolobus species established in the literature, S. islandicus, S. solfataricus, and S. acidocaldarius are by far the best described members of the genus. While S. islandicus is used as a model organism for comparative genomics and genetics (Reno et al., 2009) and for host–virus interactions (Held and Whitaker, 2009), no type strain has been designated and strains are not commercially available yet. S. solfataricus is the metabolically most diverse species and many catabolic enzymes have been investigated in detail (Bräsen et al., 2014). Unfortunately, this diversity comes along with a significant genetic instability caused by the presence of several hundred mobile elements identified in its genome (Brügger et al., 2002). By contrast, the genome of S. acidocaldarius is much more stable (Chen et al., 2005). This makes S. acidocaldarius interesting for industrial applications, where strain stability is of utmost importance. A phylogenetic tree of the genus Sulfolobus is shown in Figure 1.
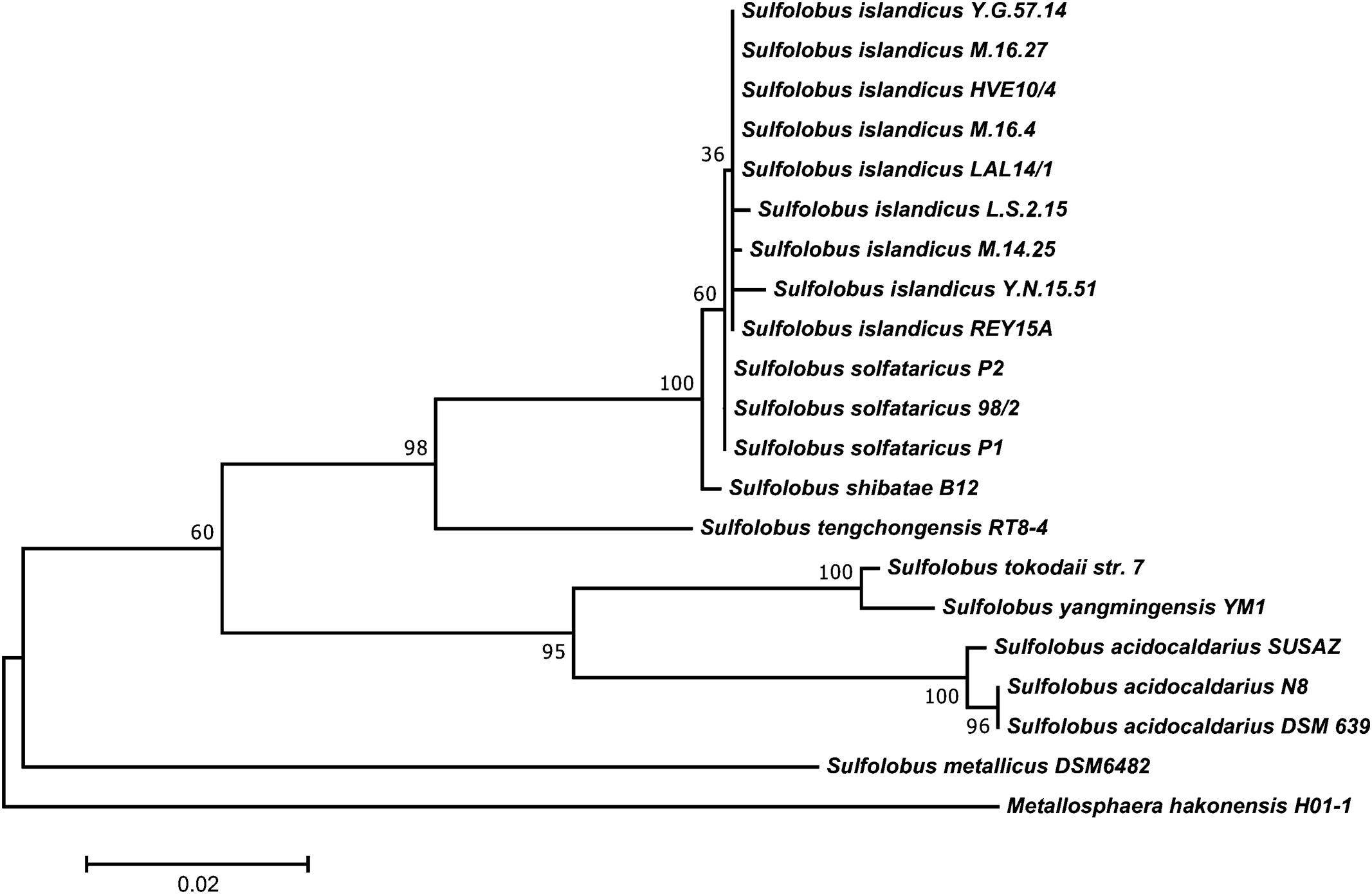
FIGURE 1. Phylogenetic tree of the genus Sulfolobus based on all publicly available 16S rDNA sequences of acknowledged species. The tree was constructed with MEGA 7.0 using the maximum-likelihood method after automated alignment with clustalX2 and manual correction with GeneDoc. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Metallosphaera hakonensis H01-1 was used as out-group.
In this review, we give an overview of the current state of knowledge on carbon metabolism, genetic tools, and fermentation techniques of Sulfolobus spp., describe relevant products, and discuss potential future applications of this genus.
Sulfolobus spp. thrive at pH 2–3 and temperatures around 75–80°C. They are characterized by a chemoorganoheterotrophic lifestyle; however, chemolithoautotrophic growth using sulfur oxidation has been reported for some species (Huber et al., 1992; Schönheit and Schäfer, 1995). All Sulfolobus species exhibit an aerobic lifestyle and for S. solfataricus P2, a preferred growth at lower oxygen concentrations was reported (Grogan, 1989; Simon et al., 2009). The different Sulfolobus strains differ significantly in their metabolic potential. S. solfataricus possesses a broad substrate specificity and uses various sugars such as polysaccharides (e.g., cellulose, starch, dextrin), disaccharides (e.g., maltose and sucrose), hexoses (e.g., D-glucose, D-galactose, D-mannose, and L-fucose), pentoses (e.g., D-arabinose, L-arabinose, D-xylose), aldehydes, alcohols (e.g., ethanol, phenol), sugar acids as well as tryptone, peptides, and amino acids as carbon source (Grogan, 1989; Izzo et al., 2005; Brouns et al., 2006; Joshua et al., 2011; Comte et al., 2013; Wolf et al., 2016; Stark et al., 2017). For S. solfataricus, a genome scale model comprising 718 metabolic and 58 transport/exchange reactions and 705 metabolites was used to simulate growth on 35 different carbon sources (Ulas et al., 2012). While no such modeled data are published for S. acidocaldarius, traditional growth experiments suggest that this species is well adapted to proteolytic growth and can utilize only few other carbon sources such as dextrin, sucrose, D-glucose, D-xylose, and L-arabinose (Grogan, 1989; Joshua et al., 2011). The differences in the metabolic potential are also reflected by the respective genome size of 2.99 Mbp including 200 IS elements for S. solfataricus (She et al., 2001) and of 2.23 Mbp for S. acidocaldarius (Chen et al., 2005). In the following paragraphs, we sum up the knowledge on the central carbohydrate metabolism and give an illustration of these pathways in Figure 2 (hexose and pentose degradation as well as glycogen, trehalose, and pentose formation).
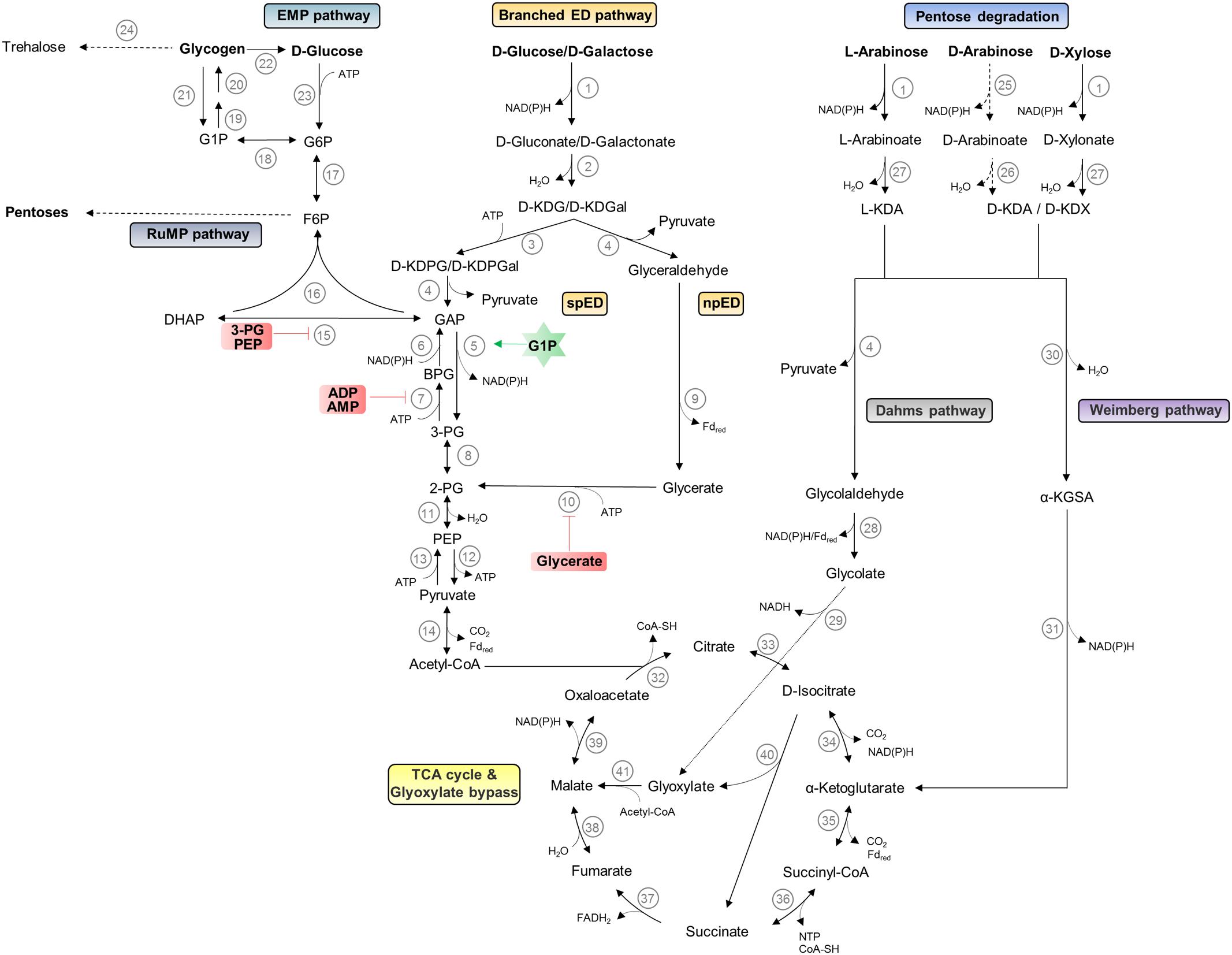
FIGURE 2. Central carbohydrate metabolism in Sulfolobus spp. The pathways for hexose and pentose degradation as well as glycogen, trehalose, and pentose formation are shown. D-arabinose (dashed lines) can only be utilized as carbon source by S. solfataricus and not by S. acidocaldarius. The current understanding of regulation by effectors is indicated by green stars and red boxes for activator and inhibitors, respectively. Enzymes catalyzing different reactions are depicted as numbers: (1) glucose dehydrogenase (broad substrate specificity); (2) gluconate dehydratase; (3) 2-keto-3-deoxygluconate kinase; (4) 2-keto-3-deoxy-(6-phospho) gluconate aldolase (broad substrate specificity); (5) non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase; (6) glyceraldehyde-3-phosphate dehydrogenase; (7) phosphoglycerate kinase; (8) phosphoglycerate mutase; (9) glyceraldehyde:ferredoxin oxidoreductase; (10) glycerate kinase; (11) enolase; (12) pyruvate kinase; (13) phosphoenolpyruvate synthetase; (14) pyruvate:ferredoxin oxidoreductase; (15) triosephosphate isomerase; (16) fructose-1,-6-bisphosphate aldolase/phosphatase; (17) phosphoglucose/phosphomannose isomerase; (18) phosphoglucomutase/phosphomannomutase; (19) NTP-glucose-1-phosphate uridylyltransferase; (20) glycogen synthase; (21) glycogen phosphorylase; (22) glucan-1,4-α-glucosidase; (23) hexokinase; (24) maltooligosyltrehalose synthase/maltooligosyltrehalose trehalohydrolase; (25) D-arabinose dehydrogenase; (26) D-arabinoate dehydratase; (27) L-arabinoate/D-xylonate dehydratase; (28) glycolaldehyde dehydrogenase/glycolaldehyde:ferredoxin oxidoreductase; (29) glycolate dehydrogenase; (30) 2-keto-3-deoxy-arabinoate/xylonate dehydratase; (31) α-ketoglutarate semi-aldehyde dehydrogenase; (32) citrate synthase; (33) aconitase; (34) isocitrate dehydrogenase; (35) α-ketoglutarate:ferredoxin oxidoreductase; (36) succinyl-CoA synthetase; (37) succinate dehydrogenase; (38) fumarase; (39) malate dehydrogenase; (40) isocitrate lyase; (41) malate synthetase. EMP, Embden–Meyerhof–Parnas; ED, Entner–Doudoroff; spED, semi-phosphorylative ED; npED, non-phosphorylative ED; RuMP, reversed ribulose monophosphate; TCA, tricarboxylic acid; G1P, glucose 1-phosphate; G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde 3-phosphate; BPG, 1,3-bisphosphoglycerate; 3-PG, 3-phosphoglycerate; 2-PG, 2-phosphoglycerate; PEP, phosphoenolpyruvate; D-KDG, 2-keto-3-deoxy-D-gluconate; D-KDGal, 2-keto-3-deoxy-D-galactonate; D-KDPG, 2-keto-3-deoxy-6-phosphate-D-gluconate; D-KDPGal, 2-keto-3-deoxy-6-phosphate-D-galactonate; L-KDA, 2-keto-3-deoxy-L-arabinoate; D-KDA, 2-keto-3-deoxy-D-arabinoate; D-KDX, 2-keto-3-deoxy-D-xylonate; α-KGSA, α-ketoglutarate semi-aldehyde.
Like most aerobic bacteria Sulfolobus spp. rely on the ED pathway for carbon degradation; however, in contrast to the classical pathway found, for example, in Pseudomonas species (Entner and Doudoroff, 1952), the archaeal pathway is branched and omits the initial phosphorylation of D-glucose. Instead, the sugar is directly oxidized to D-gluconate and dehydrated to 2-keto-3-deoxygluconate (KDG) as the characteristic intermediate of the pathway. In S. solfataricus KDG is either directly cleaved by the bifunctional aldolase to pyruvate and glyceraldehyde in the non-phosphorylative (np) branch of the ED pathway or first phosphorylated to 2-keto-3-deoxy-6-phosphogluconate (KDPG) and cleaved to pyruvate and glyceraldehyde 3-phosphate (GAP) in the semi-phosphorylated (sp) branch of the ED pathway. In the npED branch, glyceraldehyde is further oxidized and phosphorylated by glyceraldehyde:ferredoxin oxidoreductase and glycerate kinase to 2-phophoglycerate, which enters the lower shunt of the Embden–Meyerhof–Parnas (EMP) pathway (Ahmed et al., 2005). In the spED, GAP is oxidized to 3-phosphoglycerate by a non-phosphorylating GAP dehydrogenase (GAPN), activated by glucose 1-phophate, replacing the classical GAP dehydrogenase (GAPDH) and phosphoglycerate kinase (PGK) couple (GAPDH/PGK). The pathway in S. solfataricus is promiscuous for D-glucose and D-galactose (Lamble et al., 2005). Metabolome analysis of the KDG kinase deletion strain revealed a major function of the spED pathway in providing GAP for gluconeogenesis (Kouril et al., 2013b).
Pyruvate is further oxidatively decarboxylated to acetyl-CoA via the pyruvate:ferredoxin oxidoreductase; the classical pyruvate dehydrogenase complex is absent in Archaea. Acetyl CoA enters the oxidative citric acid cycle and is finally completely oxidized to two molecules of CO2. The substitution of the catabolic GAPDH and PGK couple by GAPN results in no net gain of ATP in the branched ED pathway. Only in the citric acid cycle, the succinyl-CoA synthetase is supposed to provide nucleoside triphosphate (NTP) by substrate level phosphorylation. Therefore, the major energy gain comes from aerobic respiration. The respiratory chain in several members of the Sulfolobales has been studied, and in S. solfataricus as well as in S. acidocaldarius, a branched electron transport chain with three terminal oxidases was reported (Schafer et al., 1999; Auernik and Kelly, 2008). For S. solfataricus the regulation at transcriptome level in response to different oxygen concentrations was demonstrated (Simon et al., 2009).
In Sulfolobus spp., the EMP pathway is only used for gluconeogenesis, although for glycolysis only a functional phosphofructokinase is missing (Kouril et al., 2013a). As key enzymes, especially the classical GAPDH and PGK are only active in the gluconeogenic direction. Further on, a bifunctional, gluconeogenic fructose bisphosphate aldolase/phosphatase (FBPA/ase) catalyzes the one-step formation of fructose 6-phosphate from GAP and dihydroxyacetone phosphate (Say and Fuchs, 2010; Kouril et al., 2013a; Bräsen et al., 2014). Glycogen is formed as carbon storage compound (König et al., 1982) and as source for trehalose formation via the TreY/TreZ pathway [i.e., maltooligosyltrehalose synthase and maltooligosyltrehalose trehalohydrolase (Maruta et al., 1996)]. Trehalose is the only compatible solute reported so far in Sulfolobus spp.
Thus, like in all Archaea, the central carbohydrate metabolism in Sulfolobus spp. is characterized by unusual pathways and enzymes that – moreover – also confer unique regulatory properties. In contrast to the classical bacterial and eukaryotic EMP pathway, the regulation is established at the level of triose phosphates, which seems to be a general feature in (hyper)thermophilic Archaea with optimal growth close to 80°C. Triose phosphates are labile at high temperatures and it was shown that the thermal degradation of these pathway intermediates is a crucial bottleneck for efficient substrate conversion (Kouril et al., 2013a).
In addition, the upper part of the EMP pathway seems to play an important function for pentose generation. In Sulfolobus species, as in most Archaea, the classical pentose phosphate pathway is absent and pentoses are formed from fructose 6-phosphate via the reversed ribulose monophosphate pathway (RuMP) (Soderberg, 2005). The RuMP pathway was previously reported as formaldehyde fixation pathway in methylotrophic bacteria.
Pentose degradation has been studied in S. solfataricus and S. acidocaldarius. For S. solfataricus the D-arabinose degradation was resolved and an oxidative pathway with formation of α-ketoglutarate, which directly enters the citric acid cycle, was demonstrated (Brouns et al., 2006). Later studies revealed that the transporter and degradation pathway is partially promiscuous for L-fucose utilization (Wolf et al., 2016). The D-arabinose and D-xylose pathway merge at the identical intermediates 2-keto-3-deoxy-D-arabionoate (D-KDA) and 2-keto-3-deoxy-D-xylonate (D-KDX). For D-xylose degradation, a branched pathway with an aldolase-dependent branch forming pyruvate and finally glyoxylate (Dahms pathway), which enters the glyoxylate bypass, and an aldolase-independent branch forming the citric acid cycle intermediate α-ketoglutarate (Weimberg pathway) were proposed for S. solfataricus (Nunn et al., 2010). Important for cellulosic biomass conversion the absence of diauxic growth on D-glucose and D-xylose was reported for S. acidocaldarius (Joshua et al., 2011).
In general, the availability of genome scale models, functional genomics, and systems biology approaches for Sulfolobales under different stress and growth conditions in combination with biochemical and genetic studies enabled an in depth insight into metabolism and cellular processes [e.g., growth on L-fucose and casamino acids compared to D-glucose in S. solfataricus (Wolf et al., 2016; Stark et al., 2017)]. The established knowledge forms an important prerequisite for the establishment of Sulfolobus spp. as thermoacidophilic, archaeal platform organisms using metabolic engineering, and synthetic biology approaches for future biotechnological applications.
The lack of genetic tools has been a major drawback for the establishment of archaeal model organisms for basic research and biotech industries. The major problem was that most of the traditionally used antibiotics and resistance cassette genes cannot be used in archaeal phyla and therefore auxotrophies have to be used as selectable markers. However, nowadays very well-developed genetic toolboxes exist for the euryarchaea Thermococcus kodakarensis, Pyrococcus furiosus, Haloferax volcanii, and a number of methanogenic Archaea (Leigh et al., 2011). For Pyrococcus, it has been demonstrated that large gene clusters can be introduced for the production of several compounds (Lipscomb et al., 2014). Also for the genus Sulfolobus, a number of genetic systems have been established (Leigh et al., 2011). Early in the 1990s, the first transformation protocols by electroporation were established for S. solfataricus strains and self-transmissible vectors based on a conjugative plasmid, pNOB8, and the virus SSV1 were developed (Schleper et al., 1992; Elferink et al., 1996). The virus vector-based pMJ0503 was successfully used for the overexpression of tagged proteins in S. solfataricus (Albers et al., 2006). For the expression of proteins in S. islandicus, the plasmid pSeSD1 proved to be very useful (Peng et al., 2012). The first targeted deletion mutants were obtained in a S. solfataricus 98/2 PBL2025, which had a large deletion of 50 kB in the genome including many genes coding for proteins involved in sugar metabolism. As this strain was unable to grow on lactose as single carbon source, the β-galactosidase LacS could be used as marker cassette (Worthington et al., 2003). However, in this case no counterselection could be used to remove the marker cassette and therefore double deletion mutants could not be obtained. In the meantime, three model systems have developed, namely two in S. islandicus strains and one in S. acidocaldarius, which use mainly uracil auxotrophy for the selection and counterselection of mutants (She et al., 2009; Wagner et al., 2012; Zhang and Whitaker, 2012). Whereas the two S. islandicus strains contain a large number of transposable elements, which can lead to large genome rearrangements, the S. acidocaldarius genome is remarkably stable (Chen et al., 2005), which was shown by sequencing several strains isolated from North America, Russia, and Japan (Mao and Grogan, 2012). For S. acidocaldarius currently two uracil auxotrophic mutants are being used, MW001 (Wagner et al., 2012) and MR31 (Reilly and Grogan, 2001). For MW001 a whole set of genetic tools has been established. This includes several plasmids for the construction of markerless deletion mutants or for the insertion of tags into the genome (Wagner et al., 2012). Using these, the glucose ABC transporter of S. solfataricus was ectopically integrated into the MW001 genome and successfully expressed (Wagner et al., 2012). Based on the cryptic plasmid pRN1 from S. islandicus (Zillig et al., 1993), Escherichia coli–Sulfolobus shuttle vectors and expression vectors were established, which enabled the homologous or heterologous expression of tagged proteins of interest (Berkner et al., 2007, 2010). The S. acidocaldarius MW001 genetic system has been successfully used in a number of laboratories and helped to establish S. acidocaldarius as a model crenarchaeon. In a recent achievement, it was possible to harness the endogenous CRISPR/Cas system of S. islandicus for targeted genome editing (Li et al., 2016). This is a great next step in the direction of facilitated and accelerated manipulation of the genus Sulfolobus. Table 1 gives an overview of robust and highly cited expression systems and tools for gene disruption/deletion and genomic integration for the genus Sulfolobus.

TABLE 1. A selection of expression systems and tools for gene disruption/deletion and genomic integration for the genus Sulfolobus.
The availability of potent genetic tools (Wagner et al., 2012; Peng et al., 2017) makes the transfer of heterologous genes to Sulfolobus species possible, allowing to benefit from both the metabolic diversity of S. solfataricus and the stability of S. acidocaldarius. In fact, the simpler, less promiscuous catabolism of S. acidocaldarius is an advantage over S. solfataricus in biotechnological applications, making it much easier to partly knockout metabolic pathways with the aim to redirect substrate fluxes toward a desired product.
To date, extremophiles are exploited as source of thermostable enzymes, so-called extremozymes, for food and feed industry, textile and cleaning industry, pulp and paper industry, but also in scientific research and diagnostics. Starch-hydrolyzing (Elleuche and Antranikian, 2013), (hemi)cellulolytic (Beg et al., 2001; Kuhad et al., 2011), pectinolytic (Sharma et al., 2013), chitinolytic (Chavan and Deshpande, 2013), proteolytic (Li et al., 2013), and lipolytic (Hasan et al., 2006) enzymes are in high demand in industry (Elleuche et al., 2015). Enzymes of Sulfolobus spp. are especially interesting for such applications not only because of their great catalytic diversity, but also mainly due to their superior pH and temperature stability, which comes hand-in-hand with increased resilience toward organic solvents and resistance toward proteolysis (Daniel et al., 1982; Unsworth et al., 2007; Stepankova et al., 2013). However, also tetraether lipids, membrane vesicles with antimicrobial properties, the storage component trehalose, and novel β-galactooligosaccharides are gaining importance nowadays. The most important products are shortly described below and summarized in Table 2.
Stable proteases are of great interest for the industry and a vast number of different proteases from both S. solfataricus (Hanner et al., 1990; Burlini et al., 1992; Colombo et al., 1995; Guagliardi et al., 2002; Gogliettino et al., 2014) and S. acidocaldarius (Fusek et al., 1990; Lin and Tang, 1990) has been described in detail. Condò et al. (1998) described an active, chaperonin-associated aminopeptidase from S. solfataricus MT4. Sommaruga et al. (2014) were able to significantly improve stability and reaction yield of a well-characterized carboxypeptidase also from S. solfataricus MT4 by immobilizing the enzyme on magnetic nanoparticles.
A serine arylesterase from S. solfataricus P1 was expressed. Besides its broad arylesterase activity, it was found to exhibit paraoxonase activity toward organophosphates (Park et al., 2008). With a temperature optimum of 94°C, a half-life of approximately 50 h at 90°C and high stability against detergents, urea and organic solvents, the enzyme has a high potential for industrial applications. An esterase from S. tokodaii strain 7 was expressed in E. coli and in addition to its optimal activity at 70°C remained active in a mixture of water and organic solvents such as acetonitrile and dimethyl sulfoxide (Suzuki et al., 2004).
A small heat shock protein (S.so-HSP20) from S. solfataricus P2 was successfully used to increase the tolerance in response to temperature shocks (50, 4°C) of E. coli cells (Li et al., 2012). The chaperonin Ssocpn, which requires ATP, K+, and Mg2+ but no additional proteins for its function, produced in S. solfataricus GΘ has been shown to yield folded and active protein from denatured materials. For this application, the chaperonin (920 kDa) was retained on an ultrafiltration cell, while the renatured substrates passed through the membrane (Cerchia et al., 2000).
The membrane of extreme thermophilic Archaea is unique in its composition due to its tetraether lipid content. Archaeal lipids are a promising source for liposomes with outstanding temperature and pH stability and tightness against solute leakage. These so-called archaeosomes are potential vehicles for drug, vaccine, and gene delivery (Patel and Sprott, 1999; Krishnan et al., 2000; Benvegnu et al., 2009; Mahmoud et al., 2015). Also the use as components for bioelectronics has been proposed (De Rosa et al., 1994; Hanford and Peeples, 2002). Unfortunately, no such applications using archaeal lipids have been published yet.
Sulfolobus spp. produce an interesting class of antibiotic proteins and peptides which are known under the term archaeocins, or more specifically sulfolobicins (Prangishvili et al., 2000; O’Connor and Shand, 2002; Besse et al., 2015). Sulfolobicins are potent and highly specific growth inhibitors targeting species closely related to the producing organism. Sulfolobicins have been identified as proteins of a size of 20 kDa in S. islandicus (Prangishvili et al., 2000) or heterodimers of 22 kDa per subunit in S. acidocaldarius (Ellen et al., 2011). They are associated with the cell membrane as well as with membrane vesicles of 50–200 nm in diameter. Known producers of sulfolobicins are S. islandicus strain HEN2/2 (Prangishvili et al., 2000), S. acidocaldarius DSM639, S. tokodaii strain 7, and S. solfataricus P2 and P1 (all strains: Ellen et al., 2011). Sulfolobicins are among the most resilient antimicrobial biomolecules withstanding temperatures of 78°C, SDS treatment, a broad pH range from 3 to 10.7, trypsin treatment, and longtime storage (Besse et al., 2015).
Trehalose is crucial for anhydrobiosis in many organisms and is widely used for the preservation of enzymes and antibodies (Ohtake and Wang, 2011). On top of that it serves as a valuable chemical in the food and cosmetics industry (Richards et al., 2002). It is a known metabolite of Sulfolobus spp. and the biosynthetic pathways are identified (Nicolaus et al., 1988; Kobayashi et al., 1996). Since its biosynthesis is regarded to be a stress response, the selective production of trehalose is a promising target for process engineering. The enzymatic capability of S. solfataricus to efficiently produce trehalose was already proven by Lernia et al. (2002): In a cell-free environment, trehalose was produced from dextrins with enzymes from S. solfataricus MT4 in an immobilized bed reactor with a conversion rate of 90%.
A number of applications for enzymes from Sulfolobus spp. in the synthesis of high-value chemicals have been suggested and many innovative processes have been reported: Petzelbauer et al. (2000) developed a high-temperature process for enzymatic hydrolysis of lactose for the generation of novel di- and trisaccharides (Reuter et al., 1999) using β-glycosidases from S. solfataricus MT4 and Pyrococcus furiosus. Sayer et al. (2012) characterized a thermostable transaminase from S. solfataricus P2. This enzyme is part of the non-phosphorylated pathway for serine synthesis which is not described in bacteria, but found in animals and plants (Walsh and Sallach, 1966; Liepman and Olsen, 2001). In S. tokodaii, an L-haloacid dehalogenase was found and characterized by Rye et al. (2009). This enzyme could potentially be used for the chiral production of halo-carboxylic acids which are important precursors in the fine chemical and pharmaceutical industries, as well as for bioremediation. An NAD+/NADH-dependent medium-chain alcohol dehydrogenase with remarkably broad substrate specificity toward primary, secondary, branched as well as cyclic alcohols and their corresponding aldehydes and ketones has been described by Raia et al. (2001). Lactonases have been described both from S. solfataricus MT4 (Merone et al., 2005) and from S. islandicus (Hiblot et al., 2012). These enzymes are attractive for biotechnological and pharmaceutical applications. An aldolase from S. solfataricus P1 catalyzing the reversible C-C bond formation between non-phosphorylated substrates pyruvate and glyceraldehyde to KDG was described by Buchanan et al. (1999). A sterioselective amidase from S. solfataricus MT4 has been described by Scotto d’Abusco et al. (2001).
It is evident that Sulfolobus spp. accommodate a huge variety of high value-added products useful in different fields of research and industry. However, this resource has basically remained untapped until now, due to a lack of proper bioprocessing tools. Of course, many of these products can also be produced recombinantly in mesophilic hosts. Benefits of the heterologous production in mesophilic hosts are much faster growth rates, highly efficient expression, extremely well-developed process technology, and facilitated downstream processing of thermostable proteins, since a considerable amount of host cell proteins can be readily removed via heat precipitation. Nevertheless, the production of proteins difficult to express and products remaining inactive due to differences in the expression and folding machinery, call for protein production in the archaeal host (Eichler and Adams, 2005; Kim and Lee, 2006). Furthermore, certain products are native cell constituents of Sulfolobus spp. (e.g., archaeal membrane containing tetraether lipids), which underlines the need to generate biomass and thus of bioprocess technology.
We are convinced that thermophilic bioprocesses have the potential to compete with conventional bioprocesses, since the drawbacks of typically lower growth rates and protein expression rates can be outweighed by a number of advantages resulting from the elevated process temperature:
(1) Probably the most significant advantage is the reduced risk of contamination. Loss of complete batches or reduced productivity due to chronical basal contamination levels poses serious threats for an economically feasible bioprocess based on mesophiles (Skinner and Leathers, 2004). In case of bioprocesses with Sulfolobus spp. not only the high cultivation temperature, but also the low pH reduce the contamination risk.
(2) While often limited at moderate temperatures, the solubility of substrates is significantly increased at elevated process temperatures (Gray et al., 2007). This is especially crucial in applications where oligomers and polymers are used as substrates, like in waste-to-value processes based on the conversion of lignocellulosic biomass.
(3) Considering energy requirements, a further advantage over mesophilic fermentations is the reduced need for expensive, active cooling of the fermenter in large scales for the removal of excess metabolic heat. Here, high-temperature fermentations benefit from the greater difference between ambient air temperature and fermentation broth (Abdel-Banat et al., 2010).
(4) Expression systems based on so-called cold shock promoters are well known and commercialized for mesophilic hosts (e.g., the pCold expression system from Takara Bio Europe, Saint-Germain-en-Laye, France). Nevertheless, the utilization in large-scale processes is not feasible due to high costs for cooling. In high-temperature processes, cooling is much more cost-efficient due to fast heat transfer. This way, temperature-regulated expression with shifts from growth phase to production phase becomes an option.
(5) The production of volatile compounds like short-chained alcohols benefits from high process temperatures. These compounds can be continuously recovered via the off-gas stream, while no additional separation is required. Furthermore, product inhibition, a common issue when producing toxic substances like alcohols, is prevented (Zeldes et al., 2015).
Although there is a steadily growing interest in the development of extremophilic bioprocesses, no industrial process utilizing Sulfolobus spp. has been developed yet. Doubling times of at least 5–8 h (Brock et al., 1972; Grogan, 1989) and low biomass titers in batch cultures [max. 2 g/L dry cell weight (Schiraldi et al., 1999)] are the main obstacles for establishing efficient bioprocesses. The low biomass titer not only is a severe hindrance for biotechnological applications, but also poses a limitation for basic research because biomass and enzyme production of Sulfolobus spp. in shake flasks is painfully inefficient. As a result, archaeal enzymes are still mainly produced recombinantly in mesophilic hosts like E. coli, despite the aforementioned limitations.
In order to realize a competitive bioprocess, high cell densities in a reasonable time and economically feasible space-time yields must be achieved. This can be done by genetic engineering, optimized nutrient supply, and adjustment of process parameters. On the other hand, for bioconversion reactions, the issue of a low growth rate is not necessarily a neck-breaking drawback, if it is possible to integrate a cell-retention system combined with continuous cultivating. In that case, rather the maximum cell density, which is proportional to the volumetric catalytic activity, is a critical process parameter. However, studies on bioreactor cultivations with Sulfolobus spp. are still scarce.
As shown in Table 3, a high cell density cultivation is only reported for S. shibatae B12. However, it is evident that a sophisticated bioreactor setup including a cell-retention system is needed to realize a competitive bioprocess with Sulfolobus spp. Such a bioreactor setup is exemplarily depicted in Figure 3.

FIGURE 3. Scheme and setup of bioreactor system capable of reaching high cell densities via simultaneously applying a feed and cell-retention strategy. Nutrients can be continuously fed and at the same time spent medium containing metabolites and possibly inhibiting substances is removed via a membrane, while cells are retained.
Remarkably, in none of the fermentations reported to date, defined media were used. Nevertheless, this is of high importance for the generation of platform knowledge and science-based process development. Use of defined media does allow not only the characterization and comparison of the variety of strains, but also the generation of comprehensive process understanding enabling process control and prediction. Furthermore, the use of defined media facilitates the transfer of process knowledge and speeds up process development and optimization. Another aspect worth considering is that bioprocesses that follow good manufacturing practice guidelines call for defined media to avoid batch-to-batch variability. These aspects underline the importance of the substitution of complex carbon sources, like yeast extract or protein hydrolysates, for the application of Sulfolobus spp. in industrial biotechnology for the production of high value-added products. Summarizing, to move Sulfolobus spp. into industrial biotechnology, (1) sophisticated bioreactor solutions and (2) defined media must be available.
Besides being a native source of high value-added products like extremozymes, extreme thermoacidophiles are predestined for the task of sustainably converting lignocellulosic biomass into value-added products due to their resilience toward harsh process conditions and their hemicellulolytic and cellulolytic properties (Turner et al., 2007). S. solfataricus in particular can grow on a very broad range of carbon sources (Grogan, 1989) and harbors a variety of polymer-degrading enzymes such as cellulases (Kufner, 2011), glucoamylases (Kim et al., 2004), alpha-amylases (Haseltine et al., 1996), beta-glucosidases (Moracci et al., 1995), xylanases (Cannio et al., 2004), and xylosidases (Moracci et al., 2000). Optimal growth in a hot, acidic environment means perfect synergy with the state-of-the-art method of substrate pretreatment utilizing high temperature and low pH. Although a variety of concepts for substrate pretreatment exists, the most favored process is the one of dilute sulfuric acid hydrolysis where concentrations of 0.5–1.5% sulfuric acid and temperatures between 120 and 180°C are commonly used (Carvalheiro et al., 2008; Maurya et al., 2015). Thus, pretreated substrate can be utilized in biorefinery applications based on Sulfolobus spp. with little to no need of neutralization and cooling of the medium. During the pretreatment process, a mixture of sugar monomers (mainly D-xylose, D-glucose, D-mannose, and L-arabinose) is released. In contrast to mesophilic hosts like Saccharomyces cerevisiae or E. coli, S. acidocaldarius lacks carbon catabolite repression (Ulas et al., 2012), thus allowing the efficient simultaneous utilization of a variety of sugars.
The combination of broad substrate specificity, lack of carbon catabolite repression, expression of polymer degrading enzymes, and extreme growth conditions make Sulfolobus spp. promising candidates for biorefinery applications. Following this approach, waste streams of the chemical and pulp and paper industry can be converted into value-added products. These processes would greatly benefit from the increased substrate solubility due to high temperatures and low pH. The availability of genetic tools and a broad variety of different strains are the basis for an application of Sulfolobus spp. in the biorefinery – however, the challenge of realizing a competitive bioprocess remains.
There are several reasons to be optimistic with respect to the use of Sulfolobus spp. in biotechnology. Greatly reduced contamination risk, high substrate solubility, adaption to harsh substrate pretreatment conditions, facilitated removal of volatile products, and elimination of cooling costs are benefits of high-temperature processes with Sulfolobus spp. The genus is a source of a broad variety of temperature and acid stable enzymes as well as a producer of unique biomaterials and metabolites. A well-developed genetic toolset makes exploitation of these features possible and emergence of metabolically engineered production strains is reasonable in the near future.
However, there is still a great need for careful bioprocess development. No continuous processes are reported in the literature and sophisticated tools for monitoring and control, like on-line measurement techniques for assessing cell viability, are lacking completely. Furthermore, media development and optimization have largely been neglected. For the establishment of a competitive, long-lasting, or continuous bioprocess, it is mandatory to generate basic process knowledge to be able to understand and control the bioprocess. Thus, we will tackle this challenge to be able to add Sulfolobus spp. as key player in industrial biotechnology in the future.
OS conceived the idea for writing this review. JQ drafted the manuscript, while S-VA and BS contributed the chapters on genetic tools and central carbon metabolism, respectively. LS contributed the figure describing the central carbon metabolism. OS critically reviewed and corrected the manuscript and gave substantial input.
JQ and OS acknowledge funding by Exputec GmbH in the framework of project CrossCat (ERA-IB-15-029). S-VA acknowledges funding from the BMBF (e:Bio initiative, HotSysAPP; 0316188C). LS was supported by the Mercator foundation with a Mercur startup grant (Pr-2013-0010) and by the Federal Ministry of Education and Research (BMBF) (e:Bio initiative, HotSysAPP, 03120078A). BS acknowledge funding by the BMBF (grant nos. 0316188A and 03120078A).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02474/full#supplementary-material
TABLE S1 | Milestones in Sulfolobus research.
Abdel-Banat, B. M. A., Hoshida, H., Ano, A., Nonklang, S., and Akada, R. (2010). High-temperature fermentation: how can processes for ethanol production at high temperatures become superior to the traditional process using mesophilic yeast? Appl. Microbiol. Biotechnol. 85, 861–867. doi: 10.1007/s00253-009-2248-5
Ahmed, H., Ettema, T. J. G., Tjaden, B., Geerling, A. C. M., van der Oost, J., and Siebers, B. (2005). The semi-phosphorylative Entner–Doudoroff pathway in hyperthermophilic archaea: a re-evaluation. Biochem. J. 390, 529–540. doi: 10.1042/BJ20041711
Albers, S.-V., and Driessen, A. J. (2007). Conditions for gene disruption by homologous recombination of exogenous DNA into the Sulfolobus solfataricus genome. Archaea 2, 145–149.
Albers, S.-V., and Jarrell, K. F. (2015). The archaellum: how Archaea swim. Front. Microbiol. 6:23. doi: 10.3389/fmicb.2015.00023
Albers, S.-V., Jonuscheit, M., Dinkelaker, S., Urich, T., Kletzin, A., Tampé, R., et al. (2006). Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl. Environ. Microbiol. 72, 102–111. doi: 10.1128/AEM.72.1.102-111.2006
Albers, S.-V., and Meyer, B. H. (2011). The archaeal cell envelope. Nat. Rev. Microbiol. 9, 414–426. doi: 10.1038/nrmicro2576
Auernik, K. S., and Kelly, R. M. (2008). Identification of components of electron transport chains in the extremely thermoacidophilic crenarchaeon Metallosphaera sedula through iron and sulfur compound oxidation transcriptomes. Appl. Environ. Microbiol. 74, 7723–7732. doi: 10.1128/aem.01545-08
Basen, M., and Müller, V. (2017). “Hot” acetogenesis. Extremophiles 21, 15–26. doi: 10.1007/s00792-016-0873-3
Beeler, E., and Singh, O. V. (2016). Extremophiles as sources of inorganic bio-nanoparticles. World J. Microbiol. Biotechnol. 32:156. doi: 10.1007/s11274-016-2111-7
Beg, Q., Kapoor, M., Mahajan, L., and Hoondal, G. (2001). Microbial xylanases and their industrial applications: a review. Appl. Microbiol. Biotechnol. 56, 326–338. doi: 10.1007/s002530100704
Bell, S. D., Botting, C. H., Wardleworth, B. N., Jackson, S. P., and White, M. F. (2002). The interaction of alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296, 148–151. doi: 10.1126/science.1070506
Benvegnu, T., Lemiègre, L., and Cammas-Marion, S. (2009). New generation of liposomes called archaeosomes based on natural or synthetic archaeal lipids as innovative formulations for drug delivery. Recent Pat. Drug Deliv. Formul. 3, 206–220.
Berkner, S., Grogan, D., Albers, S.-V., and Lipps, G. (2007). Small multicopy, non-integrative shuttle vectors based on the plasmid pRN1 for Sulfolobus acidocaldarius and Sulfolobus solfataricus, model organisms of the (cren-)archaea. Nucleic Acids Res. 35:e88. doi: 10.1093/nar/gkm449
Berkner, S., Wlodkowski, A., Albers, S.-V., and Lipps, G. (2010). Inducible and constitutive promoters for genetic systems in Sulfolobus acidocaldarius. Extremophiles 14, 249–259. doi: 10.1007/s00792-010-0304-9
Besse, A., Peduzzi, J., Rebuffat, S., and Carré-Mlouka, A. (2015). Antimicrobial peptides and proteins in the face of extremes: lessons from archaeocins. Biochimie 118, 344–355. doi: 10.1016/j.biochi.2015.06.004
Bräsen, C., Esser, D., Rauch, B., and Siebers, B. (2014). Carbohydrate metabolism in archaea: current insights into unusual enzymes and pathways and their regulation. Microbiol. Mol. Biol. Rev. 78, 89–175. doi: 10.1128/MMBR.00041-13
Brock, T. D., Brock, K. M., Belly, R. T., and Weiss, R. L. (1972). Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Archiv. Mikrobiol. 84, 54–68. doi: 10.1007/BF00408082
Brouns, S. J. J., Walther, J., Snijders, A. P. L., van de Werken, H. J. G., Willemen, H. L. D. M., Worm, P., et al. (2006). Identification of the missing links in prokaryotic pentose oxidation pathways. J. Biol. Chem. 281, 27378–27388. doi: 10.1074/jbc.M605549200
Brügger, K., Redder, P., She, Q., Confalonieri, F., Zivanovic, Y., and Garrett, R. A. (2002). Mobile elements in archaeal genomes. FEMS Microbiol. Lett. 206, 131–141. doi: 10.1111/j.1574-6968.2002.tb10999.x
Buchanan, C. L., Connaris, H., Danson, M. J., Reeve, C. D., and Hough, D. W. (1999). An extremely thermostable aldolase from Sulfolobus solfataricus with specificity for non-phosphorylated substrates. Biochem. J. 343, 563–570. doi: 10.1042/bj3430563
Burlini, N., Magnani, P., Villa, A., Macchi, F., Tortora, P., and Guerritore, A. (1992). A heat-stable serine proteinase from the extreme thermophilic archaebacterium Sulfolobus solfataricus. Biochim. Biophys. Acta 1122, 283–292. doi: 10.1016/0167-4838(92)90406-4
Cannio, R., Di Prizito, N., Rossi, M., and Morana, A. (2004). A xylan-degrading strain of Sulfolobus solfataricus: isolation and characterization of the xylanase activity. Extremophiles 8, 117–124. doi: 10.1007/s00792-003-0370-3
Carvalheiro, F., Duarte, L. C., and Gírio, F. M. (2008). Hemicellulose biorefineries: a review on biomass pretreatments. J. Sci. Ind. Res. 67, 849–864.
Cerchia, L., Rossi, M., and Guagliardi, A. (2000). An archaeal chaperonin-based reactor for renaturation of denatured proteins. Extremophiles 4, 1–7.
Chavan, S. B., and Deshpande, M. V. (2013). Chitinolytic enzymes: an appraisal as a product of commercial potential. Biotechnol. Prog. 29, 833–846. doi: 10.1002/btpr.1732
Chen, L., Brugger, K., Skovgaard, M., Redder, P., She, Q., Torarinsson, E., et al. (2005). The Genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J. Bacteriol. 187, 4992–4999. doi: 10.1128/JB.187.14.4992-4999.2005
Chong, P. K., and Wright, P. C. (2005). Identification and characterization of the Sulfolobus solfataricus P2 proteome. J. Proteome Res. 4, 1789–1798.
Colombo, S., Toietta, G., Zecca, L., Vanoni, M., and Tortora, P. (1995). Molecular cloning, nucleotide sequence, and expression of a carboxypeptidase-encoding gene from the archaebacterium Sulfolobus solfataricus. J. Bacteriol. 177, 5561–5566. doi: 10.1128/jb.177.19.5561-5566.1995
Comte, A., Christen, P., Davidson, S., Pophillat, M., Lorquin, J., Auria, R., et al. (2013). Biochemical, transcriptional and translational evidences of the phenol-meta-degradation pathway by the hyperthermophilic Sulfolobus solfataricus 98/2. PLOS ONE 8:e82397. doi: 10.1371/journal.pone.0082397
Condò, I., Ruggero, D., Reinhardt, R., and Londei, P. (1998). A novel aminopeptidase associated with the 60 kDa chaperonin in the thermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 29, 775–785. doi: 10.1046/j.1365-2958.1998.00971.x
Dai, X., Wang, H., Zhang, Z., Li, K., Zhang, X., Mora-López, M., et al. (2016). Genome sequencing of Sulfolobus sp. A20 from costa rica and comparative analyses of the putative pathways of Carbon, Nitrogen, and Sulfur metabolism in various sulfolobus strains. Front. Microbiol. 7:1902. doi: 10.3389/fmicb.2016.01902
Daniel, R. M., Cowan, D. A., Morgan, H. W., and Curran, M. P. (1982). A correlation between protein thermostability and resistance to proteolysis. Biochem. J. 207, 641–644. doi: 10.1042/bj2070641
De Rosa, M., Morana, A., Riccio, A., Gambacorta, A., Trincone, A., and Incani, O. (1994). Lipids of the Archaea: a new tool for bioelectronics. Biosens. Bioelectron. 9, 669–675. doi: 10.1016/0956-5663(94)80064-2
Donati, E. R., Castro, C., and Urbieta, M. S. (2016). Thermophilic microorganisms in biomining. World J. Microbiol. Biotechnol. 32:179. doi: 10.1007/s11274-016-2140-2
Eichler, J., and Adams, M. W. W. (2005). Posttranslational protein modification in Archaea. Microbiol. Mol. Biol. Rev. 69, 393–425. doi: 10.1128/MMBR.69.3.393-425.2005
Elferink, M. G. L., Schleper, C., and Zillig, W. (1996). Transformation of the extremely thermoacidophilic archaeon Sulfolobus solfataricus via a self-spreading vector. FEMS Microbiol. Lett. 137, 31–35. doi: 10.1111/j.1574-6968.1996.tb08078.x
Ellen, A. F., Albers, S.-V., and Driessen, A. J. (2010). Comparative study of the extracellular proteome of Sulfolobus species reveals limited secretion. Extremophiles 14, 87–98. doi: 10.1007/s00792-009-0290-y
Ellen, A. F., Rohulya, O. V., Fusetti, F., Wagner, M., Albers, S.-V., and Driessen, A. J. M. (2011). The Sulfolobicin Genes of Sulfolobus acidocaldarius encode novel antimicrobial proteins. J. Bacteriol. 193, 4380–4387. doi: 10.1128/JB.05028-11
Elleuche, S., and Antranikian, G. (2013). “Starch-hydrolyzing enzymes from thermophiles,” in Thermophilic Microbes in Environmental and Industrial Biotechnology, eds T. Satyanarayana, J. Littlechild, and Y. Kawarabayasi (Dordrecht: Springer Netherlands), 509–533. doi: 10.1007/978-94-007-5899-5_20
Elleuche, S., Schäfers, C., Blank, S., Schröder, C., and Antranikian, G. (2015). Exploration of extremophiles for high temperature biotechnological processes. Curr. Opin. Microbiol. 25, 113–119. doi: 10.1016/j.mib.2015.05.011
Entner, N., and Doudoroff, M. (1952). Glucose and gluconic acid oxidation of Pseudomonas saccharophila. J. Biol. Chem. 196, 853–862.
Fusek, M., Lin, X. L., and Tang, J. (1990). Enzymic properties of thermopsin. J. Biol. Chem. 265, 1496–1501.
Gogliettino, M., Riccio, A., Cocca, E., Rossi, M., Palmieri, G., and Balestrieri, M. (2014). A new pepstatin-insensitive thermopsin-like protease overproduced in peptide-rich cultures of Sulfolobus solfataricus. Int. J. Mol. Sci. 15, 3204–3219. doi: 10.3390/ijms15023204
Gray, M. C., Converse, A. O., and Wyman, C. E. (2007). Solubilities of oligomer mixtures produced by the hydrolysis of xylans and corn stover in water at 180°C. Ind. Eng. Chem. Res. 46, 2383–2391. doi: 10.1021/ie060325+
Grogan, D. W. (1989). Phenotypic characterization of the archaebacterial genus Sulfolobus: comparison of five wild-type strains. J. Bacteriol. 171, 6710–6719.
Guagliardi, A., Cerchia, L., and Rossi, M. (2002). An intracellular protease of the crenarchaeon Sulfolobus solfataricus, which has sequence similarity to eukaryotic peptidases of the CD clan. Biochem. J. 368, 357–363. doi: 10.1042/bj20021017
Hanford, M. J., and Peeples, T. L. (2002). Archaeal tetraether lipids. Appl. Biochem. Biotechnol. 97, 45–62.
Hanner, M., Redl, B., and Stöffler, G. (1990). Isolation and characterization of an intracellular aminopeptidase from the extreme thermophilic archaebacterium Sulfolobus solfataricus. Biochim. Biophys. Acta 1033, 148–153. doi: 10.1016/0304-4165(90)90005-H
Hasan, F., Shah, A. A., and Hameed, A. (2006). Industrial applications of microbial lipases. Enzyme Microb. Technol. 39, 235–251. doi: 10.1016/j.enzmictec.2005.10.016
Haseltine, C., Rolfsmeier, M., and Blum, P. (1996). The glucose effect and regulation of alpha-amylase synthesis in the hyperthermophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 178, 945–950. doi: 10.1128/jb.178.4.945-950.1996
Held, N. L., and Whitaker, R. J. (2009). Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ. Microbiol. 11, 457–466. doi: 10.1111/j.1462-2920.2008.01784.x
Hiblot, J., Gotthard, G., Chabriere, E., and Elias, M. (2012). Structural and Enzymatic characterization of the lactonase SisLac from Sulfolobus islandicus. PLOS ONE 7:e47028. doi: 10.1371/journal.pone.0047028
Huber, G., Drobner, E., Huber, H., and Stetter, K. O. (1992). Growth by aerobic oxidation of molecular hydrogen in archaea—a metabolic property so far unknown for this domain. Syst. Appl. Microbiol. 15, 502–504. doi: 10.1016/S0723-2020(11)80108-6
Izzo, V., Notomista, E., Picardi, A., Pennacchio, F., and Di Donato, A. (2005). The thermophilic archaeon Sulfolobus solfataricus is able to grow on phenol. Res. Microbiol. 156, 677–689. doi: 10.1016/j.resmic.2005.04.001
Joshua, C. J., Dahl, R., Benke, P. I., and Keasling, J. D. (2011). Absence of diauxie during simultaneous utilization of glucose and xylose by Sulfolobus acidocaldarius. J. Bacteriol. 193, 1293–1301. doi: 10.1128/JB.01219-10
Kim, M.-S., Park, J.-T., Kim, Y.-W., Lee, H.-S., Nyawira, R., Shin, H.-S., et al. (2004). Properties of a novel thermostable glucoamylase from the hyperthermophilic archaeon Sulfolobus solfataricus in relation to starch processing. Appl. Environ. Microbiol. 70, 3933–3940. doi: 10.1128/AEM.70.7.3933-3940.2004
Kim, S., and Lee, S. B. (2006). Rare codon clusters at 5’-end influence heterologous expression of archaeal gene in Escherichia coli. Protein Expr. Purif. 50, 49–57. doi: 10.1016/j.pep.2006.07.014
Kobayashi, K., Kato, M., Miura, Y., Kettoku, M., Komeda, T., and Iwamatsu, A. (1996). Gene cloning and expression of new trehalose-producing enzymes from the hyperthermophilic archaeum Sulfolobus solfataricus KM1. Biosci. Biotechnol. Biochem. 60, 1882–1885. doi: 10.1271/bbb.60.1882
König, H., Skorko, R., Zillig, W., and Reiter, W.-D. (1982). Glycogen in thermoacidophilic archaebacteria of the genera Sulfolobus, Thermoproteus, Desulfurococcus and Thermococcus. Arch. Microbiol. 132, 297–303. doi: 10.1007/BF00413378
Kort, J. C., Esser, D., Pham, T. K., Noirel, J., Wright, P. C., and Siebers, B. (2013). A cool tool for hot and sour Archaea: proteomics of Sulfolobus solfataricus. Proteomics 13, 2831–2850. doi: 10.1002/pmic.201300088
Kouril, T., Esser, D., Kort, J., Westerhoff, H. V., Siebers, B., and Snoep, J. L. (2013a). Intermediate instability at high temperature leads to low pathway efficiency for an in vitro reconstituted system of gluconeogenesis in Sulfolobus solfataricus. FEBS J. 280, 4666–4680. doi: 10.1111/febs.12438
Kouril, T., Wieloch, P., Reimann, J., Wagner, M., Zaparty, M., Albers, S.-V., et al. (2013b). Unraveling the function of the two Entner–Doudoroff branches in the thermoacidophilic Crenarchaeon Sulfolobus solfataricus P2. FEBS J. 280, 1126–1138. doi: 10.1111/febs.12106
Krahe, M., Antranikian, G., and Märkl, H. (1996). Fermentation of extremophilic microorganisms. FEMS Microbiol. Rev. 18, 271–285. doi: 10.1111/j.1574-6976.1996.tb00243.x
Krishnan, L., Dicaire, C. J., Patel, G. B., and Sprott, G. D. (2000). Archaeosome vaccine adjuvants induce strong humoral, cell-mediated, and memory responses: comparison to conventional liposomes and alum. Infect. Immun. 68, 54–63.
Kufner, K. (2011). Charakterisierung Thermophiler Cellulasen aus Sulfolobus solfataricus und Thermotoga maritima. Available at: https://epub.uni-bayreuth.de/379/ [accessed August 8, 2016].
Kuhad, R. C., Gupta, R., and Singh, A. (2011). Microbial cellulases and their industrial applications. Enzyme Res. 2011:e280696. doi: 10.4061/2011/280696
Lamble, H. J., Theodossis, A., Milburn, C. C., Taylor, G. L., Bull, S. D., Hough, D. W., et al. (2005). Promiscuity in the part-phosphorylative Entner-Doudoroff pathway of the archaeon Sulfolobus solfataricus. FEBS Lett. 579, 6865–6869. doi: 10.1016/j.febslet.2005.11.028
Leigh, J. A., Albers, S.-V., Atomi, H., and Allers, T. (2011). Model organisms for genetics in the domain Archaea: methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol. Rev. 35, 577–608. doi: 10.1111/j.1574-6976.2011.00265.x
Lernia, I. D., Schiraldi, C., Generoso, M., and Rosa, M. D. (2002). Trehalose production at high temperature exploiting an immobilized cell bioreactor. Extremophiles 6, 341–347. doi: 10.1007/s00792-001-0263-2
Li, D.-C., Yang, F., Lu, B., Chen, D.-F., and Yang, W.-J. (2012). Thermotolerance and molecular chaperone function of the small heat shock protein HSP20 from hyperthermophilic archaeon, Sulfolobus solfataricus P2. Cell Stress Chaperones 17, 103–108. doi: 10.1007/s12192-011-0289-z
Li, Q., Yi, L., Marek, P., and Iverson, B. L. (2013). Commercial proteases: present and future. FEBS Lett. 587, 1155–1163. doi: 10.1016/j.febslet.2012.12.019
Li, Y., Pan, S., Zhang, Y., Ren, M., Feng, M., Peng, N., et al. (2016). Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 44:e34. doi: 10.1093/nar/gkv1044
Liepman, A. H., and Olsen, L. J. (2001). Peroxisomal alanine: glyoxylate aminotransferase (AGT1) is a photorespiratory enzyme with multiple substrates in Arabidopsis thaliana. Plant J. 25, 487–498. doi: 10.1046/j.1365-313x.2001.00961.x
Lin, X., and Tang, J. (1990). Purification, characterization, and gene cloning of thermopsin, a thermostable acid protease from Sulfolobus acidocaldarius. J. Biol. Chem. 265, 1490–1495.
Lipscomb, G. L., Schut, G. J., Thorgersen, M. P., Nixon, W. J., Kelly, R. M., and Adams, M. W. W. (2014). Engineering hydrogen gas production from formate in a hyperthermophile by heterologous production of an 18-subunit membrane-bound complex. J. Biol. Chem. 289, 2873–2879. doi: 10.1074/jbc.M113.530725
Littlechild, J. A. (2015). Archaeal enzymes and applications in industrial biocatalysts. Archaea 2015:e147671. doi: 10.1155/2015/147671
Mahmoud, G., Jedelská, J., Strehlow, B., and Bakowsky, U. (2015). Bipolar tetraether lipids derived from thermoacidophilic archaeon Sulfolobus acidocaldarius for membrane stabilization of chlorin e6 based liposomes for photodynamic therapy. Eur. J. Pharm. Biopharm. 95, 88–98. doi: 10.1016/j.ejpb.2015.04.009
Mao, D., and Grogan, D. (2012). Genomic evidence of rapid, global-scale gene flow in a Sulfolobus species. ISME J. 6, 1613–1616. doi: 10.1038/ismej.2012.20
Maruta, K., Mitsuzumi, H., Nakada, T., Kubota, M., Chaen, H., Fukuda, S., et al. (1996). Cloning and sequencing of a cluster of genes encoding novel enzymes of trehalose biosynthesis from thermophilic archaebacterium Sulfolobus acidocaldarius. Biochim. Biophys. Acta 1291, 177–181. doi: 10.1016/S0304-4165(96)00082-7
Maurya, D. P., Singla, A., and Negi, S. (2015). An overview of key pretreatment processes for biological conversion of lignocellulosic biomass to bioethanol. 3 Biotech 5, 597–609. doi: 10.1007/s13205-015-0279-4
Merone, L., Mandrich, L., Rossi, M., and Manco, G. (2005). A thermostable phosphotriesterase from the archaeon Sulfolobus solfataricus: cloning, overexpression and properties. Extremophiles 9, 297–305. doi: 10.1007/s00792-005-0445-4
Moracci, M., Cobucci Ponzano, B., Trincone, A., Fusco, S., De Rosa, M., van Der Oost, J., et al. (2000). Identification and molecular characterization of the first alpha -xylosidase from an archaeon. J. Biol. Chem. 275, 22082–22089. doi: 10.1074/jbc.M910392199
Moracci, M., Nucci, R., Febbraio, F., Vaccaro, C., Vespa, N., La Cara, F., et al. (1995). Expression and extensive characterization of a β-glycosidase from the extreme thermoacidophilic archaeon Sulfolobus solfataricus in Escherichia coli: authenticity of the recombinant enzyme. Enzyme Microb. Technol. 17, 992–997. doi: 10.1016/0141-0229(95)00012-7
Nicolaus, B., Gambacorta, A., Basso, A. L., Riccio, R., De Rosa, M., and Grant, W. D. (1988). Trehalose in Archaebacteria. Syst. Appl. Microbiol. 10, 215–217. doi: 10.1016/S0723-2020(88)80003-1
Nunn, C. E. M., Johnsen, U., Schonheit, P., Fuhrer, T., Sauer, U., Hough, D. W., et al. (2010). Metabolism of pentose sugars in the Hyperthermophilic Archaea Sulfolobus solfataricus and Sulfolobus acidocaldarius. J. Biol. Chem. 285, 33701–33709. doi: 10.1074/jbc.M110.146332
O’Connor, E. M., and Shand, R. F. (2002). Halocins and sulfolobicins: the emerging story of archaeal protein and peptide antibiotics. J. Ind. Microbiol. Biotechnol. 28, 23–31. doi: 10.1038/sj/jim/7000190
Ohtake, S., and Wang, Y. J. (2011). Trehalose: current use and future applications. J. Pharm. Sci. 100, 2020–2053. doi: 10.1002/jps.22458
Park, C. B., and Lee, S. B. (1997). Constant-volume fed-batch operation for high density cultivation of hyperthermophilic aerobes. Biotechnol. Techn. 11, 277–281.
Park, C. B., and Lee, S. B. (1999). Inhibitory effect of mineral ion accumulation on high density growth of the hyperthermophilic archaeon Sulfolobus solfataricus. J. Biosci. Bioeng. 87, 315–319. doi: 10.1016/S1389-1723(99)80038-3
Park, Y.-J., Yoon, S.-J., and Lee, H.-B. (2008). Purification and characterization of a new inducible thermostable extracellular lipolytic enzyme from the thermoacidophilic archaeon Sulfolobus solfataricus P1. J. Mol. Catal. B Enzym. 124, 11–19. doi: 10.1016/j.molcatb.2015.11.023
Patel, G. B., and Sprott, G. D. (1999). Archaeobacterial ether lipid liposomes (Archaeosomes) as novel vaccine and drug delivery systems. Crit. Rev. Biotechnol. 19, 317–357. doi: 10.1080/0738-859991229170
Peng, N., Deng, L., Mei, Y., Jiang, D., Hu, Y., Awayez, M., et al. (2012). A synthetic arabinose-inducible promoter confers high levels of recombinant protein expression in hyperthermophilic archaeon Sulfolobus islandicus. Appl. Environ. Microbiol. 78, 5630–5637. doi: 10.1128/AEM.00855-12
Peng, N., Han, W., Li, Y., Liang, Y., and She, Q. (2017). Genetic technologies for extremely thermophilic microorganisms of Sulfolobus, the only genetically tractable genus of crenarchaea. Sci. China Life Sci. 60, 370–385. doi: 10.1007/s11427-016-0355-8
Petzelbauer, I., Zeleny, R., Reiter, A., Kulbe, K. D., and Nidetzky, B. (2000). Development of an ultra-high-temperature process for the enzymatic hydrolysis of lactose: II. Oligosaccharide formation by two thermostable β-glycosidases. Biotechnol. Bioeng. 69, 140–149.
Pham, T. K., Sierocinski, P., van der Oost, J., and Wright, P. C. (2010). Quantitative proteomic analysis of Sulfolobus solfataricus membrane proteins. J. Proteome Res. 9, 1165–1172. doi: 10.1021/pr9007688
Prangishvili, D., Forterre, P., and Garrett, R. A. (2006). Viruses of the Archaea: a unifying view. Nat. Rev. Microbiol. 4, 837–848. doi: 10.1038/nrmicro1527
Prangishvili, D., Holz, I., Stieger, E., Nickell, S., Kristjansson, J. K., and Zillig, W. (2000). Sulfolobicins, specific proteinaceous toxins produced by strains of the extremely thermophilic Archaeal Genus Sulfolobus. J. Bacteriol. 182, 2985–2988. doi: 10.1128/JB.182.10.2985-2988.2000
Raia, C. A., Giordano, A., and Rossi, M. (2001). “[15] Alcohol dehydrogenase from Sulfolobus solfataricus,” in Hyperthermophilic Enzymes Part B Methods in Enzymology, eds M. W. W. Adams, and R. M. Kelly (Cambridge, MA: Academic Press), 176–195. doi: 10.1016/S0076-6879(01)31056-X
Reilly, M. S., and Grogan, D. W. (2001). Characterization of intragenic recombination in a hyperthermophilic archaeon via conjugational DNA exchange. J. Bacteriol. 183, 2943–2946. doi: 10.1128/JB.183.9.2943-2946.2001
Reno, M. L., Held, N. L., Fields, C. J., Burke, P. V., and Whitaker, R. J. (2009). Biogeography of the Sulfolobus islandicus pan-genome. Proc. Natl. Acad. Sci. U.S.A. 106, 8605–8610. doi: 10.1073/pnas.0808945106
Reuter, S., Rusborg Nygaard, A., and Zimmermann, W. (1999). β-Galactooligosaccharide synthesis with β-galactosidases from Sulfolobus solfataricus, Aspergillus oryzae, and Escherichia coli. Enzyme Microb. Technol. 25, 509–516. doi: 10.1016/S0141-0229(99)00074-5
Richards, A. B., Krakowka, S., Dexter, L. B., Schmid, H., Wolterbeek, A. P. M., Waalkens-Berendsen, D. H., et al. (2002). Trehalose: a review of properties, history of use and human tolerance, and results of multiple safety studies. Food Chem. Toxicol. 40, 871–898. doi: 10.1016/S0278-6915(02)00011-X
Rye, C. A., Isupov, M. N., Lebedev, A. A., and Littlechild, J. A. (2009). Biochemical and structural studies of a l-haloacid dehalogenase from the thermophilic archaeon Sulfolobus tokodaii. Extremophiles 13, 179–190. doi: 10.1007/s00792-008-0208-0
Say, R. F., and Fuchs, G. (2010). Fructose 1,6-bisphosphate aldolase/phosphatase may be an ancestral gluconeogenic enzyme. Nature 464, 1077–1081. doi: 10.1038/nature08884
Sayer, C., Bommer, M., Isupov, M., Ward, J., and Littlechild, J. (2012). Crystal structure and substrate specificity of the thermophilic serine:pyruvate aminotransferase from Sulfolobus solfataricus. Acta Crystallogr. D Biol. Crystallogr. 68, 763–772. doi: 10.1107/S0907444912011274
Schafer, G., Engelhard, M., and Muller, V. (1999). Bioenergetics of the Archaea. Microbiol. Mol. Biol. Rev. 63, 570–620.
Schiraldi, C., Marulli, F., Di Lernia, I., Martino, A., and De Rosa, M. (1999). A microfiltration bioreactor to achieve high cell density in Sulfolobus solfataricus fermentation. Extremophiles 3, 199–204.
Schleper, C., Kubo, K., and Zillig, W. (1992). The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc. Natl. Acad. Sci. U.S.A. 89, 7645–7649. doi: 10.1073/pnas.89.16.7645
Schönheit, P., and Schäfer, T. (1995). Metabolism of hyperthermophiles. World J. Microbiol. Biotechnol. 11, 26–57.
Scotto d’Abusco, A., Ammendola, S., Scandurra, R., and Politi, L. (2001). Molecular and biochemical characterization of the recombinant amidase from hyperthermophilic archaeon Sulfolobus solfataricus. Extremophiles 5, 183–192. doi: 10.1007/s007920100190
Sharma, N., Rathore, M., and Sharma, M. (2013). Microbial pectinase: sources, characterization and applications. Rev. Environ. Sci. Biotechnol. 12, 45–60. doi: 10.1007/s11157-012-9276-9
She, Q., Singh, R. K., Confalonieri, F., Zivanovic, Y., Allard, G., Awayez, M. J., et al. (2001). The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U.S.A. 98, 7835–7840.
She, Q., Zhang, C., Deng, L., Peng, N., Chen, Z., and Liang, Y. X. (2009). Genetic analyses in the hyperthermophilic archaeon Sulfolobus islandicus. Biochem. Soc. Trans. 37, 92–96. doi: 10.1042/BST0370092
Siebers, B., and Schönheit, P. (2005). Unusual pathways and enzymes of central carbohydrate metabolism in Archaea. Curr. Opin. Microbiol. 8, 695–705. doi: 10.1016/j.mib.2005.10.014
Skinner, K. A., and Leathers, T. D. (2004). Bacterial contaminants of fuel ethanol production. J. Ind. Microbiol. Biotechnol. 31, 401–408. doi: 10.1007/s10295-004-0159-0
Simon, G., Walther, J., Zabeti, N., Combet-Blanc, Y., Auria, R., van der Oost, J., et al. (2009). Effect of O2 concentrations on Sulfolobus solfataricus P2. FEMS Microbiol. Lett. 299, 255–260. doi: 10.1111/j.1574-6968.2009.01759.x
Soderberg, T. (2005). Biosynthesis of ribose-5-phosphate and erythrose-4-phosphate in archaea: a phylogenetic analysis of archaeal genomes. Archaea 1, 347–352.
Sommaruga, S., Galbiati, E., Peñaranda-Avila, J., Brambilla, C., Tortora, P., Colombo, M., et al. (2014). Immobilization of carboxypeptidase from Sulfolobus solfataricus on magnetic nanoparticles improves enzyme stability and functionality in organic media. BMC Biotechnol. 14:82. doi: 10.1186/1472-6750-14-82
Stark, H., Wolf, J., Albersmeier, A., Pham, T. K., Hofmann, J. D., Siebers, B., et al. (2017). Oxidative Stickland reactions in an obligate aerobic organism - amino acid catabolism in the Crenarchaeon Sulfolobus solfataricus. FEBS J. 284, 2078–2095. doi: 10.1111/febs.14105
Stepankova, V., Bidmanova, S., Koudelakova, T., Prokop, Z., Chaloupkova, R., and Damborsky, J. (2013). Strategies for stabilization of enzymes in organic solvents. Acs Catal. 3, 2823–2836.
Straub, C. T., Zeldes, B. M., Schut, G. J., Adams, M. W., and Kelly, R. M. (2017). Extremely thermophilic energy metabolisms: biotechnological prospects. Curr. Opin. Biotechnol. 45, 104–112. doi: 10.1016/j.copbio.2017.02.016
Suzuki, Y., Miyamoto, K., and Ohta, H. (2004). A novel thermostable esterase from the thermoacidophilic archaeon Sulfolobus tokodaii strain 7. FEMS Microbiol. Lett. 236, 97–102. doi: 10.1111/j.1574-6968.2004.tb09633.x
Turner, P., Mamo, G., and Karlsson, E. N. (2007). Potential and utilization of thermophiles and thermostable enzymes in biorefining. Microb. Cell Fact. 6:9. doi: 10.1186/1475-2859-6-9
Ulas, T., Riemer, S. A., Zaparty, M., Siebers, B., and Schomburg, D. (2012). Genome-scale reconstruction and analysis of the metabolic network in the hyperthermophilic archaeon Sulfolobus solfataricus. PLOS ONE 7:e43401. doi: 10.1371/journal.pone.0043401
Unsworth, L. D., van der Oost, J., and Koutsopoulos, S. (2007). Hyperthermophilic enzymes - stability, activity and implementation strategies for high temperature applications. FEBS J. 274, 4044–4056. doi: 10.1111/j.1742-4658.2007.05954.x
Wagner, M., van Wolferen, M., Wagner, A., Lassak, K., Meyer, B. H., Reimann, J., et al. (2012). “Versatile genetic tool box for the crenarchaeote Sulfolobus acidocaldarius,” in Recent Advances in Genomic and Genetic Studies in the Archaea, eds F. T. Robb, T. M. Lowe, and Z. Kelman (Lausanne: Frontiers Media), 65.
Walsh, D. A., and Sallach, H. J. (1966). Comparative studies on the pathways for serine biosynthesis in animal tissues. J. Biol. Chem. 241, 4068–4076.
Wolf, J., Stark, H., Fafenrot, K., Albersmeier, A., Pham, T. K., Muller, K. B., et al. (2016). A systems biology approach reveals major metabolic changes in the thermoacidophilic archaeon Sulfolobus solfataricus in response to the carbon source L-fucose versus D-glucose. Mol. Microbiol. 102, 882–908. doi: 10.1111/mmi.13498
Worthington, P., Hoang, V., Perez-Pomares, F., and Blum, P. (2003). Targeted disruption of the α-amylase gene in the hyperthermophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 185, 482–488. doi: 10.1128/JB.185.2.482-488.2003
Zeldes, B. M., Keller, M. W., Loder, A. J., Straub, C. T., Adams, M. W. W., and Kelly, R. M. (2015). Extremely thermophilic microorganisms as metabolic engineering platforms for production of fuels and industrial chemicals. Front. Microbiol. 6:1209. doi: 10.3389/fmicb.2015.01209
Zhang, C., Cooper, T. E., Krause, D. J., and Whitaker, R. J. (2013). Augmenting the genetic toolbox for Sulfolobus islandicus with a stringent positive selectable marker for agmatine prototrophy. Appl. Environ. Microbiol. 79, 5539–5549. doi: 10.1128/AEM.01608-13
Zhang, C., She, Q., Bi, H., and Whitaker, R. J. (2016). The apt/6-Methylpurine counterselection system and its applications in genetic studies of the hyperthermophilic archaeon Sulfolobus islandicus. Appl. Environ. Microbiol. 82, 3070–3081. doi: 10.1128/AEM.00455-16
Zhang, C., and Whitaker, R. J. (2012). A broadly applicable gene knockout system for the thermoacidophilic archaeon Sulfolobus islandicus based on simvastatin selection. Microbiology 158, 1513–1522. doi: 10.1099/mic.0.058289-0
Keywords: Sulfolobus, biotechnology, thermophile, acidophile, bioprocessing, biorefinery
Citation: Quehenberger J, Shen L, Albers S-V, Siebers B and Spadiut O (2017) Sulfolobus – A Potential Key Organism in Future Biotechnology. Front. Microbiol. 8:2474. doi: 10.3389/fmicb.2017.02474
Received: 25 September 2017; Accepted: 28 November 2017;
Published: 12 December 2017.
Edited by:
Don A. Cowan, University of Pretoria, South AfricaReviewed by:
Yutaka Kawarabayasi, Kyushu University, JapanCopyright © 2017 Quehenberger, Shen, Albers, Siebers and Spadiut. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oliver Spadiut, b2xpdmVyLnNwYWRpdXRAdHV3aWVuLmFjLmF0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.