- 1Water Research Institute, National Research Council of Italy (IRSA – CNR), Rome, Italy
- 2Dipartimento di Scienze per gli Alimenti, la Nutrizione e l’Ambiente (DeFENS), Università degli Studi di Milano, Milan, Italy
Arsenic (As) is a toxic element released in aquatic environments by geogenic processes or anthropic activities. To counteract its toxicity, several microorganisms have developed mechanisms to tolerate and utilize it for respiratory metabolism. However, still little is known about identity and physiological properties of microorganisms exposed to natural high levels of As and the role they play in As transformation and mobilization processes. This work aims to explore the phylogenetic composition and functional properties of aquatic microbial communities in As-rich freshwater environments of geothermal origin and to elucidate the key microbial functional groups that directly or indirectly may influence As-transformations across a natural range of geogenic arsenic contamination. Distinct bacterial communities in terms of composition and metabolisms were found. Members of Proteobacteria, affiliated to Alpha- and Betaproteobacteria were mainly retrieved in groundwaters and surface waters, whereas Gammaproteobacteria were the main component in thermal waters. Most of the OTUs from thermal waters were only distantly related to 16S rRNA gene sequences of known taxa, indicating the occurrence of bacterial biodiversity so far unexplored. Nitrate and sulfate reduction and heterotrophic As(III)-oxidization were found as main metabolic traits of the microbial cultivable fraction in such environments. No growth of autotrophic As(III)-oxidizers, autotrophic and heterotrophic As(V)-reducers, Fe-reducers and oxidizers, Mn-reducers and sulfide oxidizers was observed. The ars genes, involved in As(V) detoxifying reduction, were found in all samples whereas aioA [As(III) oxidase] and arrA genes [As(V) respiratory reductase] were not found. Overall, we found that As detoxification processes prevailed over As metabolic processes, concomitantly with the intriguing occurrence of novel thermophiles able to tolerate high levels of As.
Introduction
In aquatic environments, arsenic (As) can be found in four oxidation states (+V, +III, 0, -III) showing various levels of toxicity and bioavailability (Rahman and Hassler, 2014). The speciation of As is largely controlled by pH and redox potential (Vu et al., 2003). Arsenate [As(V)] is the predominant form of inorganic arsenic in aqueous aerobic environments, mainly found as H2AsO4- at pH < 6.9 and as HAsO42- at higher pH values. Arsenite [As(III)] prevails under reducing conditions, as uncharged H3AsO30 at pH < 9.2 (Oremland and Stolz, 2003). The occurrence of As in groundwater and surface waters is determined by geothermal inputs and geogenic processes, such as the dissolution of As-bearing minerals in soils and sediments (Smedley and Kinniburgh, 2002; Bissen and Frimmel, 2003; Fendorf et al., 2010).
Microbial activity is linked to the biogeochemistry of arsenic and microorganisms are able to directly catalyze As transformations in natural environments or mediate these processes by coupling with other redox reactions such as nitrate-, sulfate-, manganese-, and iron-reduction (Rittle et al., 1995; Fendorf et al., 2010; Lee, 2013; Zhang et al., 2017). Aquatic microorganisms have evolved different mechanisms to resist to high As concentrations and metabolize it, including sorption, mobilization, precipitation and redox and methylation transformation (Huang, 2014). Microbial resistance to both As(V) and As(III) is based on a detoxification system that reduces As(V) to As(III) through a cytoplasmic arsenate reductase and extrudes the latter from the cellular compartment by means of a membranous As(III) efflux pump (ars system) (Silver and Phung, 2005a,b). This resistance mechanism is widespread in nature and present in bacteria, archaea and eukaryotes (Mukhopadhyay et al., 2002; Stolz et al., 2006). As-resistant bacteria were found to mainly belong to Proteobacteria (Alpha-, Beta- and Gammaproteobacteria), Firmicutes and Actinobacteria (Páez-Espino et al., 2009). As(III) oxidation is also a potential detoxification process found in heterotrophic bacteria such as Herminiimonas arsenicoxydans (Muller et al., 2006).
Besides resistance mechanisms, arsenic metabolisms associated with cell growth were also described. Many aerobic microorganisms are able to oxidize As(III) to As(V) in the periplasm owing to the presence of an arsenite oxidase (aio gene) (Lett et al., 2012). The aio system is phylogenetically and ecologically widespread in bacteria and archaea (van Lis et al., 2013).
Occasionally, As(III) oxidation may be combined with nitrate respiration or integrated into electron transport chain of anoxygenic photosynthesis (Hoeft et al., 2007; Zargar et al., 2012). This anaerobic pathway is catalyzed by the arxAB system, a new arsenite oxidase firstly discovered in Alkalilimnicola ehrlichii strain MLHE-1 (Zargar et al., 2010).
It is well documented that As(III) prevails in geothermal systems, with the rapid oxidation to As(V) occurring in the outflow channels (Wilkie and Hering, 1998; Jackson et al., 2001; Langner et al., 2001; Inskeep et al., 2005). So far, geothermal As-oxidizing bacteria are mainly affiliated to the bacterial phyla Aquificae, Chloroflexi, Deinococcus–Thermus, and to the Archaea domain (Sehlin and Lindström, 1992; Gihring and Banfield, 2001; Donahoe-Christiansen et al., 2004; Tang et al., 2011). Differently from geothermal systems, As(III) oxidizers in mesophilic environments include mainly Alpha-, Beta-, and Gammaproteobacteria (Stolz et al., 2006; van Lis et al., 2013).
Moreover, many microorganisms are able to perform a dissimilatory As(V) reduction using As(V) as electron acceptor and different inorganic (e.g., H2) and organic compounds (e.g., small organic acids, sugars and complex aromatic substrates like benzoate and toluene) as electron donor (Stolz and Oremland, 1999; Kumari and Jagadevan, 2016). This periplasmic anaerobic respiration of As(V) is driven by a membranous arsenate reductase encoded by arr genes (Huang, 2014). As(V) reducers were affiliated to Crenarchaeota, Aquificae, Chrysiogenes, Deferribacteres, Halanaerobacter, and Beta-, Gamma-, Delta-, Epsilonproteobacteria (Stolz et al., 2006). A chemolithotrophic metabolism based on As(V) reduction to As(III) coupled with the oxidation of sulfide to sulfate was shown in strain MLMS-1, anaerobic deltaproteobacterium isolated from an alkaline, hypersaline lake in California (Hoeft et al., 2004). This bacterium was found to be able to grow by disproportionation of monothioarsenates, molecules which contain internally S2- as an electron donor and As(V) as electron acceptor (Planer-Friedrich et al., 2015). Thioarsenates were shown to serve as energy source for Aquificales in the hydrothermal systems of Yellowstone National Park (Haertig and Planer-Friedrich, 2012) and to support, as monothioarsenate, the growth of the aerobic hyperthermophilic bacterium Thermocrinis ruber strain OC14/7/2 (Haertig et al., 2014) and haloalkaliphilic, anoxygenic photosynthetic purple sulfur bacteria (Edwardson et al., 2014).
Recently, the attention is focused on structural and metabolic characteristics (related to arsenic metabolism and resistance) of mixed microbial communities in As-contaminated environments (Escudero et al., 2013; Andres and Bertin, 2016). These studies suggested that As-related genes might help in detecting, monitoring and managing As contamination of either geogenic or anthropogenic origin. However, still little is known about the phylogenetic composition and functional properties of the aquatic microbial communities involved in As transformation processes. Most of the studies performed so far were mainly focused on the microbial cultivable fraction, without exploiting the potential of the overall community to face As contamination.
It is known that the activities of indigenous bacteria in As-rich aquatic environments may control the biogeochemical cycle of this element by mediating other redox reactions. Arsenic mobility in aquatic environments was shown to be controlled by biogeochemical redox transformations of iron and manganese, especially in alluvial plains where organic matter is readily available for biotic reactions (Borch et al., 2010; Jones et al., 2012). Additionally, the involvement of microorganisms in sulfate reduction may directly affect the precipitation of As sulfides species (i.e., realgar) or the As(III) oxidation in geothermal environments (Burton et al., 2013; Rodriguez-Freire et al., 2016). Nitrate-linked microbial transformation were also shown to play an important ecological role in As-contaminated aquatic environments (Oremland and Stolz, 2003).
Our study aimed to evaluate the structural and phylogenetic patterns in the microbiome of As-rich waters of geothermal origin, as those typically found in the Cimino-Vico volcanic area (Central Italy), and to elucidate the key microbial functional groups that directly or indirectly may influence As-transformations across a natural range of geogenic arsenic contamination.
Materials and Methods
Sampling Site
Water samples were collected from the Cimino-Vico volcanic area (Central Italy) (Supplementary Figure S1), a complex hydrogeological system characterized by several perched aquifers and a continuous basal aquifer flowing through volcanites (Baiocchi et al., 2013). In this volcanic region, widespread hydrothermal circulation was reported as underlined by the occurrence of geothermal fields (Dall’aglio et al., 2001; Casentini and Pettine, 2010). The natural occurrence of arsenic is explained by the complexity of the hydrostratigraphy, the structural setting of the area and the related mixing occurring between water circulating in the basal volcanic aquifer and the fluids that rise from depth, all of which characterize the active geothermal system (Angelone et al., 2009; Armiento et al., 2015; Cinti et al., 2015). We selected eight different freshwater sources, differently influenced by the rising of As-rich thermal waters (50–64°C) from a deep aquifer consisting of Mesozoic sedimentary rocks, locally uplifted, fractured, and faulted (Baiocchi et al., 2013). The sampling survey was performed in winter (November, 2015), and samples included (i) thermal waters: one hot spring (SSC) derived from a flowing well 346 m deep and two pools (CAR, PAL) collecting waters from natural hot springs; (ii) groundwaters from four wells (OLI, BEL, ANG, FON); (iii) surface waters sampled 2 m away from the west side shore of the Lake Vico (VICO) directly affected by hydrothermal upwelling.
Water Chemistry
Temperature (T), pH, electrical conductivity (EC), and dissolved oxygen (DO) were measured on-site by field probes (Hach HQ 40d). Sulfides, iron (total and II) were determined by spectrophotometric methods (Casentini et al., 2016). Water samples (50 ml) were directly stored at 4°C for anion analysis by ion chromatography (Dionex DX-120); 50 ml of water were filtered in situ through 0.45 μm cellulose acetate membrane filters (Whatman), acidified with 2% HNO3, and stored at 4°C for cation analysis by ICP-MS equipped with Octapole Reaction System (ORS) (Agilent 7500). Arsenic speciation was assessed by hydride generation-absorption spectrometry (HG-AAS, Perkin Elmer AAnalyst 800). Arsenite determination was carried out using HCl 2% as carrier and reduction to arsine gas was performed with NaBH4 0.4% in acetate buffered samples at pH 4–4.5. Astot was analyzed by HG-AAS prior reduction to As(III) by 5% KI/Acid Ascorbic solution. As(V) concentration was obtained by the difference (details in Casentini et al., 2016).
Prokaryotic Abundance and Community Composition by Flow Cytometry and CARD-FISH
Each water sample was fixed in formaldehyde solution (FA, 1% vol/vol final concentration) and kept at 4°C for a maximum of 24 h. The flow cytometer A50-micro (Apogee Flow System, Hertfordshire, England), equipped with a solid state laser set at 20 mV and tuned to an excitation wave length of 488 nm, was used to characterize microbial communities in fixed samples. The volumetric absolute cell counting was carried out on samples stained with SYBR Green I (1:10,000 dilution; Molecular Probes, Invitrogen). Apogee Histogram Software (v89.0) was used to plot and analyze data; the light scattering signals (forward and side scatters) and the green fluorescence (530/30 nm) were considered for the single cell characterization. Thresholding was set on the green channel and voltages were adjusted to place the background and instrumental noise below the first decade of green fluorescence. Samples were run at low flow rates to keep the number of events below 1000 events s-1. The intensity of green fluorescence emitted by SYBR-positive cells allowed for the discrimination among cell groups exhibiting two different nucleic acid content (cells with Low Nucleic Acid content - LNA; cells with High Nucleic Acid content - HNA) (Amalfitano et al., 2014). The forward scatter signals (FSC expressed in absolute units, AU) can be considered to be relatively proportional to cell size (Tzur et al., 2011), and related to the size variations of LNA and HNA cells.
Further fixed samples (10–200 mL depending on total cell abundance) were filtered on polycarbonate membrane filters (pore size 0.2 μm, 47 mm diameter, Nuclepore) by gentle vacuum (<0.2 bar) then washed with 20 ml of Milli-Q water. The filters were stored in Petri dishes at -20°C until further processing. CARD-FISH analysis was performed following the protocol optimized by Fazi et al. (2007, 2013) using specific rRNA-target HRP-labeled probes (Biomers, Ulm, Germany): EUB338 I-III for Bacteria; ALF968 for Alphaproteobacteria; BET42a for Betaproteobacteria; GAM42a for Gammaproteobacteria; DELTA495 for Deltaproteobacteria; CFX and GNSB for Chloroflexi, LGC354mix for Firmicutes, CF319a for Flavobacteria, PLA46 for Planctomycetes, TM7905 for TM7, HGC69A for Actinobacteria and ARCH915 for Archaea. Details of probes are available at probeBase (Greuter et al., 2016). The stained filter sections were inspected on a Leica DM LB30 epifluorescence microscope (Leica Microsystems GmbH, Wetzlar, Germany) at 1000X magnification. At least 300 cells were counted in > 10 microscopic fields randomly selected across the filter sections. The relative abundance of hybridized cells was estimated as the ratio of hybridized cells to total DAPI-stained cells.
DNA Extraction
For DNA extraction and subsequent PCR applications water samples were filtered (750–1000 ml) through polycarbonate membranes (pore size 0.2 μm, 47 mm diameter, Nuclepore) and immediately stored at -20°C. DNA extraction was performed with PowerSoil® DNA Isolation Kit (MoBio – Carlsbad, CA, United States) by following the manufacturer’s instructions. The quality of extracted DNA (1.6 < A260/280 < 1.8 and A260/230 > 2) was analyzed with a Nanodrop 3300 (Thermo Scientific, Italy). DNA was stored at -20°C in small aliquots.
Bacterial 16S rRNA Gene Pyrosequencing
Bacterial 16S rRNA genes was amplified in triplicate from DNA samples using primers 27F (5′-GAG AGT TTG ATC CTG GTC CAG-3′) and 1495R (5′-CTA CGG CTA CCT TGT TAC GA-3′). Reactions were set up in 25 μL volumes containing 10 ng of DNA, 0.3 μM primers and 1x Taq PCR MAstermix kit (QIAGEN, Hilden, Germany). The thermal protocol included 5 min of denaturation at 95°C followed by 35 cycles of denaturation at 95°C for 1 min, 40 s annealing at 55°C and 1 min and 40 s elongation at 72°C; the final elongation was performed at 72°C for 10 min. Replicated amplicons from the same sample were pooled and purified using the MinElute PCR Purification Kit (QIAGEN) to reach a final concentration of 20 ng μL-1. Pyrosequencing was performed with primer 27F at Molecular Research LP (MRDNA, Shallowater, TX, United States) by bacterial Tag-Encoded FLX Amplicon Pyrosequencing (bTEFAP).
The sequences were processed and analyzed using the QIIME software tools (Caporaso et al., 2010b). Sequences shorter than 200 bp, with barcodes or primer biases, homopolymers and chimeras were filtered out from the dataset. High quality sequences selected for the analysis were trimmed at 400 bp and grouped into Operational Taxonomic Units (OTUs) with 97% similarity with the uclust method (Edgar, 2010) according to the last SILVA SSU Ref database (Quast et al., 2013). Representative sequences for each OTU were selected and aligned using the PyNAST algorithm (Caporaso et al., 2010a), taxonomy was assigned and OTU tables were generated for each sample.
Phylogenetic Analysis
The phylogeny of the majority of reads retrieved from thermal samples (SCC, CAR, PAL) was not successfully assigned with QIIME. Therefore, these sequences were separately compared to the GenBank database with BLASTn. Together with the closest relatives, the sequences were aligned on the MEGA software version 6 (Tamura et al., 2013) using MUSCLE (Edgar, 2004) and the phylogenetic tree was calculated with the Maximum Likelihood method based on the Tamura-Nei model (Tamura and Nei, 1993).
Real-Time Quantification of Arsenic-Related Functional Genes
The screening of functional genes known to be involved in the arsenic cycle was performed by PCR using the Taq98TM Hot Start 2X Master Mix (Lucigen, United States) according to the manufacturer’s instructions. 10 different primer sets were used for the amplification of aioA, arrA, arsB and arsC according to the protocols reported in literature (Supplementary Table S1). In detail, arsenite oxidase gene (aioA) was targeted using aroA#1F - aroA#1R primers according to Karn and Pan (2016); arrAf – arrAr primer set (Malasarn et al., 2004) was used for the amplification of arsenate respiratory reductase gene (arrA). Arsenate cytoplasmic reductase (arsC) was amplified using amlt-42-F/amlt-376-R primers according to Sun et al. (2004). Quantification of arsenite transporter (arsB) was carried out using arsB#1F - arsB#1R primers according to Achour et al. (2007). qPCR reactions were performed using Sso Advanced Universal SYBR Green Supermix (BIO-RAD, United States) according to the manufacturer’s instructions on a CFX96 Touch Real-time PCR detection system. Melting curves were performed for each reaction to confirm the purity of amplified products.
Most Probable Number
Most Probable Number (MPN) counts were determined with triplicate 10-fold dilution series (to 10-9) using liquid selective growth media, inoculated and incubated for 30 days at the same temperature of the original water sample. In particular, MPN was used to quantify aerobic heterotrophs, autotrophic and heterotrophic As(III)-oxidizers, As(V)-reducers, sulfide-oxidizers, sulfate-reducers, nitrate-reducers, iron-oxidizers, iron-reducers and manganese-reducers. For autotrophic and heterotrophic As(III)-oxidizers and As(V)-reducers, 1 ml of water sample was added to 9 ml of basal mineral medium (Corsini et al., 2014) supplemented with sodium arsenite (NaAsO2) or disodium hydrogen arsenate (Na2HAsO4) at a final concentration of 100 mg/L with or without sodium lactate (1 g/L final concentration). Selective growth media for heterotrophs, sulfide-oxidizers and sulfate-reducers were prepared according to Cote and Gherna (1994). Growth media suggested by Lovley (2006) were used to quantify nitrate-, iron- and manganese-reducers with a mix of carbon sources at final concentration of 1g/L (sodium acetate, glucose and lactate in 2:1:1 ratio). The enumeration of iron-oxidizers was performed by using a FeSO4-based mineral medium as described in Barron and Lueking (1990). The growth of As(V)-, Fe-, sulfate-, nitrate- and Mn-reducing bacteria was estimated under anaerobic conditions carefully controlled during either the sampling and laboratory incubation. In particular, samples were dispensed into culture tubes, sealed under N2 atmosphere with butyl rubber stoppers. The reducing growth media were autoclaved and, before inoculation, again stripped with nitrogen under axenic conditions.
MPN (cell/mL) was estimated as described in Sutton (2010). Positive tubes were analyzed at the end of the incubation to confirm the screened metabolisms by evaluating the occurrence of the expected end-products.
Statistical Analysis
The non-parametric Mann–Whitney U-test was applied to verify the statistical difference between thermal and non-thermal waters for all physical, chemical and microbial parameters (Clarke, 1993). The chemical variables with p < 0.05 were incorporated into a Non-metric MultiDimensional Scaling ordination plot (NMDS) in order to graphically synthesize the Euclidean dissimilarity between the two groups of water samples. Chemical and microbial abundance data were then projected onto the NMDS ordination using a vector-fitting procedure, in which the length of the arrow is proportional to the correlation between NMDS-axes and each variable. This method allowed determining the variation pattern of each projected variable discriminating the two groups of waters (Foulquier et al., 2013; Amalfitano et al., 2014). Chemical data were log-transformed, whereas values of cell abundance of the major prokaryotic subgroups, including flow cytometry (LNA and HNA cells) and CARD-FISH data (i.e., major phyla and classes of Proteobacteria) was normalized by log(X+1).
A multi-group SIMilarity PERcentage test (SIMPER), performed on NGS data using the Euclidean similarity measure, was run to identify the microbial phylogenetic groups that were primarily responsible for observed differences between thermal waters and groundwaters.
Non-parametric MANOVA (NPMANOVA) was used to test if microbial taxa obtained from 16S rRNA bar-coded pyrosequencing differed statistically among thermal waters and groundwaters.
Results
Water Chemistry
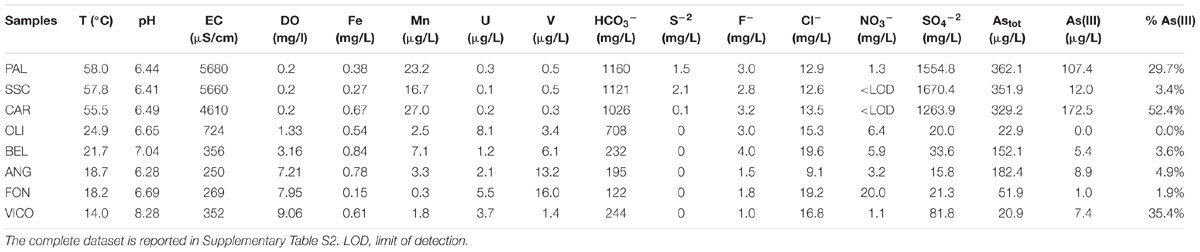
Major physical and chemical parameters (T, pH, EC, DO, HCO3-, SO4-2) differed significantly based on water origin (Table 1). Thermal waters showed high EC (4610-5680 μS/cm), high concentrations of SO4-2 (1264–1670 mg/L), typical of geothermal environments and highest temperature values (55.5–58°C). Groundwaters showed high values of V and U and low values of EC and sulfates. Nitrate concentrations reached the highest value in FON (20 mg/L). Nitrites and phosphates were below detection limits in all samples. Among other ions analyzed (Supplementary Table S2), Sr and Li showed highest values in thermal waters (14270 and 176 μg/L respectively) whereas B varied between 73 (groundwater) and 1206 μg/L (thermal waters). The concentrations of Fe and Mn were generally low and ranged between 0.15 and 0.84 mg Fe/L and 0.3 and 27 μg Mn/L. The highest As concentration was found in thermal waters (329.2–362.1 μg/L). Total arsenic concentrations showed relatively lower concentrations in VICO surface waters (20.9 μg/L) and groundwaters (22.9–182.4 μg/L). As(III) was more concentrated in PAL, CAR and VICO than in groundwaters; the geothermal sample SSC showed less than 4% of As(III). The NMDS ordination allowed visualizing the chemical dissimilarity between thermal and non-thermal waters, also showing the variation pattern of those variables that varied significantly between the two groups of waters (Figure 1).
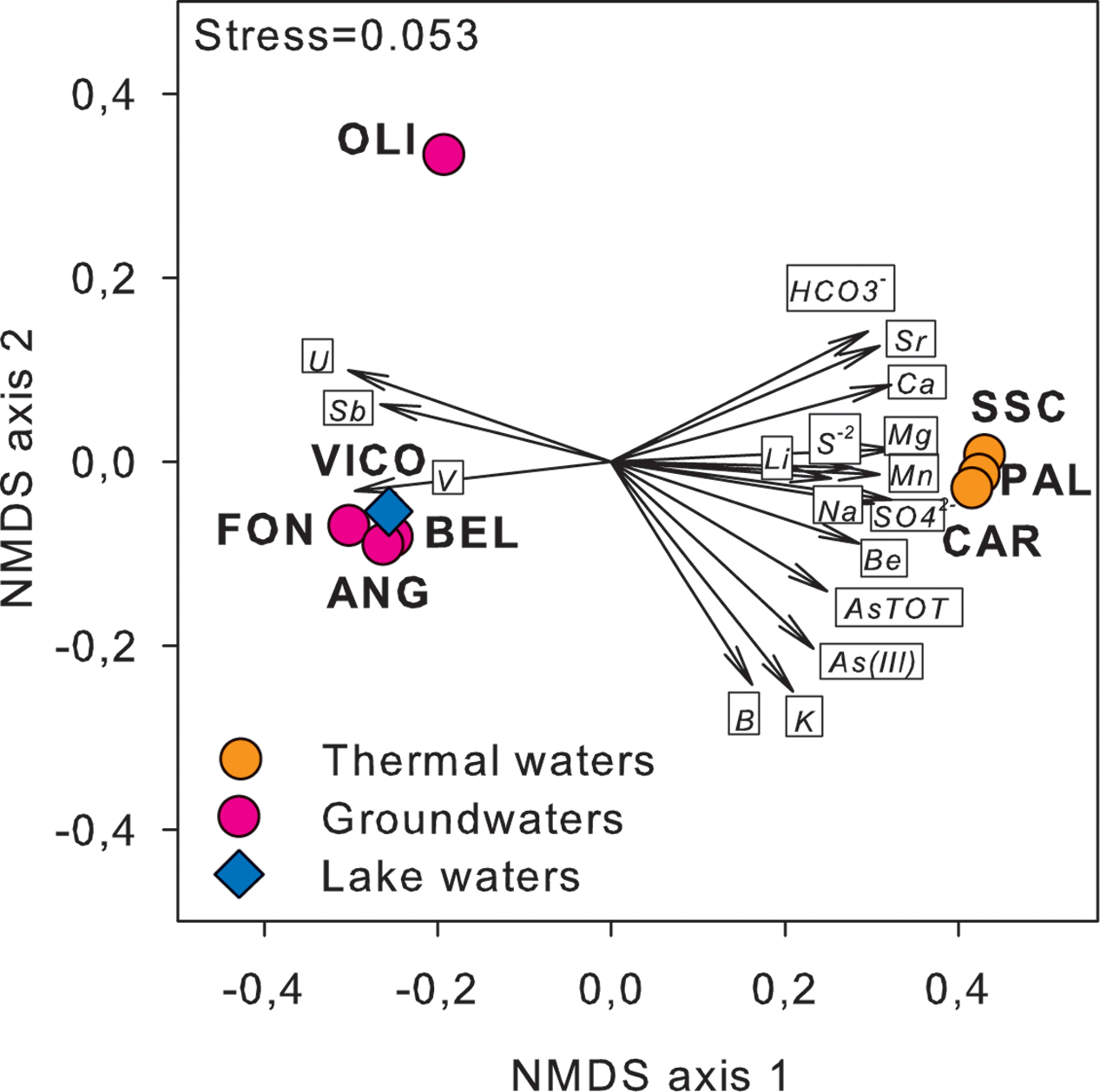
FIGURE 1. NMDS ordination plot, based on Euclidean distance matrix of log-transformed data, showing the variation patterns of chemical variables that varied significantly between thermal and non-thermal waters. The vector length is proportional to the correlation between the NMDS axes and each chemical variable. The stress value (i.e., <0.2) suggests for an accurate representation of the dissimilarity among water samples.
Prokaryotic Cell Abundance and Community Structure
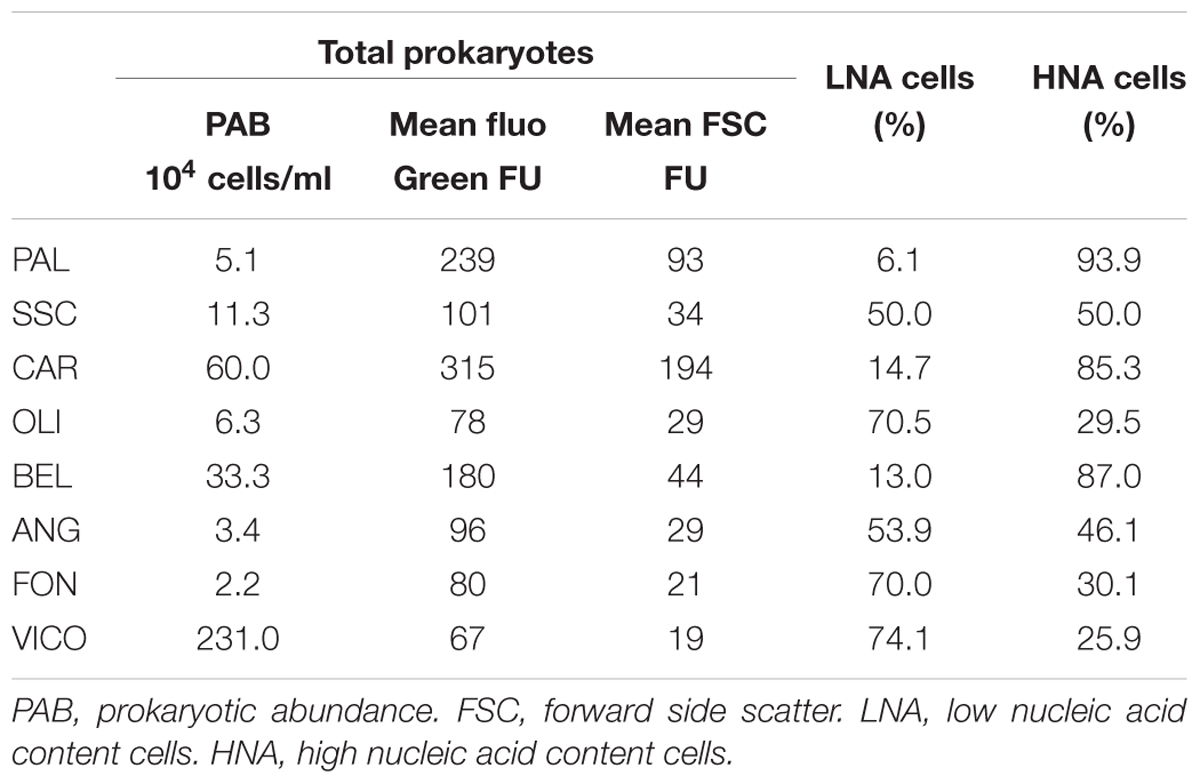
Prokaryotic abundance was higher in lake waters (2.3 × 106 ± 1.9 × 104 cell/ml) and thermal waters (between 5.1 × 104 ± 9.3 × 101 cell/ml and 6 × 105 ± 2.5 × 103 cell/ml) than in groundwaters (between 2.2 × 104 ± 2.6 × 102 cell/ml and 3.3 × 105 ± 4.7 × 103 cell/ml) (Table 2). As assessed by flow cytometry, the ratio between the mean green fluorescence and forward scatter signal intensity of prokaryotes was lower in thermal waters than in non-thermal waters. In groundwaters and surface waters, HNA cells showed lower percentages than those found in thermal waters (26% in lake water and 35.2% in groundwater except BEL with HNA = 87%). On average, the mean fluorescence intensity of the HNA cells was approximately four times higher than that of LNA cells.
As shown in Figure 2, the main microbial components belonged to bacteria domain (from 15% up to 95% of total cells) and were mainly affiliated to Proteobacteria. Alpha- and Betaproteobacteria were the most abundant groups retrieved in groundwaters and surface waters, whereas Gammaproteobacteria dominated in thermal waters (reaching 81.5% of bacterial cells in CAR sample). TM7 division was mainly present in groundwaters reaching the highest abundance in FON sample (1.9 × 103 ± 1.4 × 102 cell/mL). Archaea were found in all screened samples with abundances ranging between 1.9 × 102 ± 0.2 × 101 cell/mL and 6.0 × 104 ± 1.1 × 103 cell/mL (Figure 2).
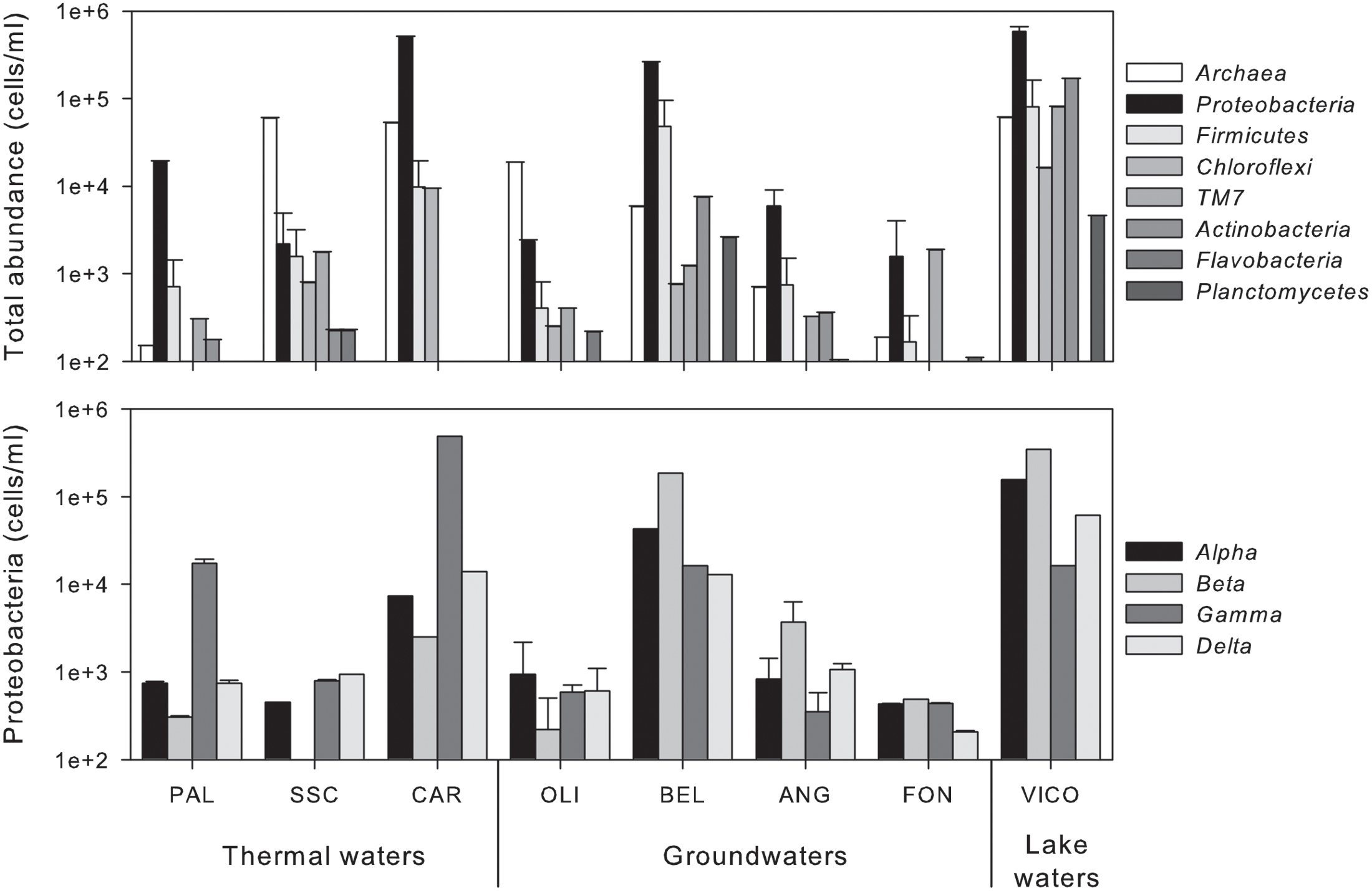
FIGURE 2. Abundance of phylogenetic clusters (Archaea and main phyla within Bacteria) (upper) and single classes within Proteobacteria (lower) in thermal waters, groundwaters and lake waters.
Following a vector-fitting procedure onto the NMDS ordination plot based on chemical dissimilarity, we showed that Archaea, Gammaproteobacteria and HNA cells were relatively more abundant in thermal waters, whereas Betaproteobacteria, Planctomycetes, TM7, Alphaproteobacteria, and LNA cells were more represented in non-thermal waters (Figure 3).
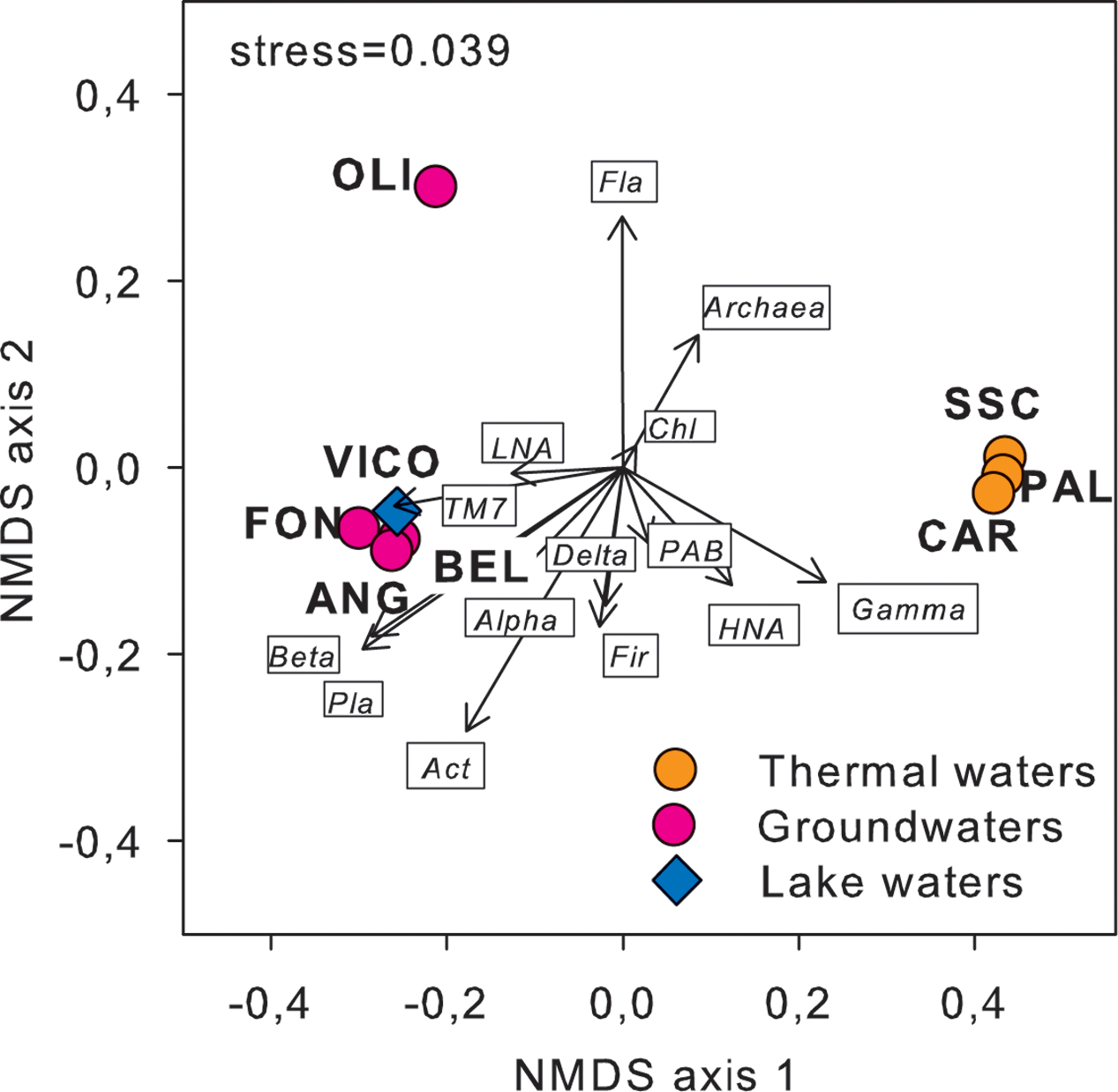
FIGURE 3. Cell abundance variation of the major prokaryotic subgroups, including flow cytometry (LNA and HNA cells) and CARD-FISH data (i.e., major phyla and classes of Proteobacteria), projected onto the NMDS ordination synthesizing the chemical dissimilarity between thermal and non-thermal waters (see Figure 1). The vector length is proportional to the correlation between the NMDS axes and each microbial variable, upon normalization by log(X+1). Act, Actinobacteria; Chl, Chloroflexi; Fir, Firmicutes; Fla, Flavobacteria; Pla, Planctomycetes; Alpha, Alphaproteobacteria; Beta, Betaproteobacteria; Gamma, Gammaproteobacteria; Delta, Deltaproteobacteria; LNA, low nucleic acid content cells; HNA, high nucleic acid content cells; PAB, prokaryotic abundance.
Next Generation Sequencing (NGS)
A total of 71401 ends reads were generated which were assigned into 2988 OTUs. All the resulting 16S rRNA gene fragments were then classified into 24 bacterial phyla, 55 classes, 118 orders, 188 families and 293 genera (Supplementary Table S3). Overall, Proteobacteria was the most abundant phylum (46.5% of total OTUs), followed by Cyanobacteria (12.1%), Bacteroidetes (7.6%), Nitrospirae (7.3%), Firmicutes (1.2%) and Acidobacteria (1.0%). The other 18 phyla represented less than 1.0% of total OTUs. On average, 22.5% of the OTUs showed a very low 16S rDNA identity with known bacterial taxa, with the highest percentage in thermal waters (up to 76.1%).
The relative abundance of OTUs in water samples differed considerably (Figure 4). In surface waters (VICO), Cyanobacteria represented the 94.6% of total OTUs and they were mainly affiliated to Planktothrix genus (90.9%). Nitrospirae, Bacteroidetes and Proteobacteria dominated groundwater samples. Nitrospirae phylum was largely represented by the genus Nitrospira (up to 50.7% of total OTUs). The genus dgA-11 gut group, belonging to Bacteroidetes, was found only in groundwaters (up to 58.6% of OTUs in FON sample). Betaproteobacteria were mainly represented by orders Burkholderiales, Nitrosomonadales, and Rhodocyclales, affiliated to genera Undibacterium and Azoarcus and family Gallionellaceae. Alpha- and Gammaproteobacteria were mainly affiliated to orders Rhizobiales, Sphingomonadales, Rickettsiales, Rhodospirillales and to genus Stenotrophomonas.
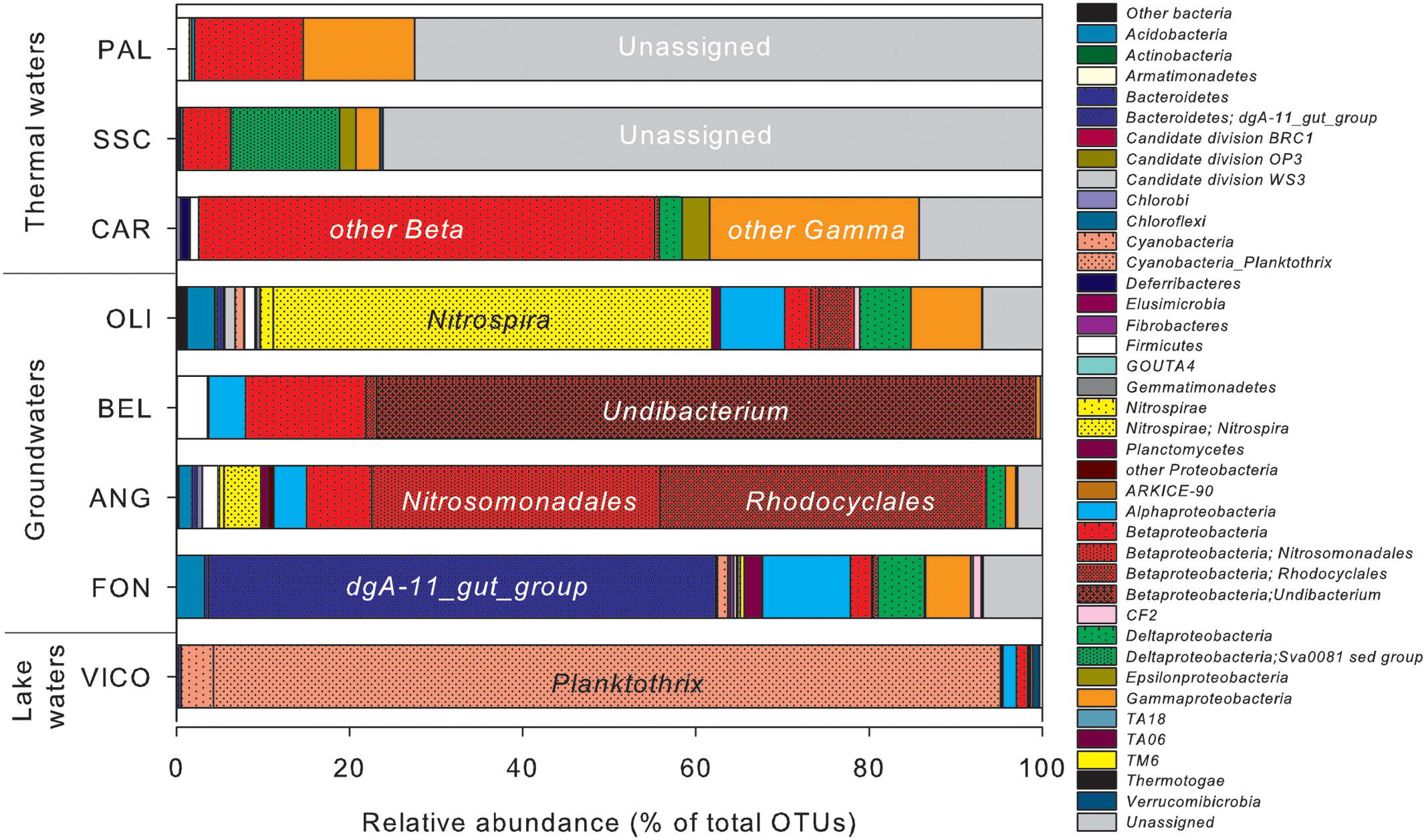
FIGURE 4. Operational taxonomic units (OTUs) relative abundance in water samples estimated by NGS. Clusters making up less than 1% of total composition were classified as ‘other bacteria’ in thermal waters, groundwaters and lake waters.
Proteobacteria was the predominant phylum reported in thermal waters, mainly affiliated to Betaproteobacteria (from 5.3% up to 52.6% of total OTUs) and Gammaproteobacteria (from 2.7% up to 24.2% of total OTUs), but the taxonomic affiliation at lower level classification was not identified. Deltaproteobacteria were mainly retrieved in one thermal sample (SSC) affiliated to the genus Sva0081_sediment_group, belonging to Desulfobacterales.
As assessed by NPMANOVA, thermal waters differed considerably from non-thermal waters. Lake waters were not considered in this analysis, since NGS results highlighted a low diversity in this sample with the predominance of only one genus. According to SIMPER test (Supplementary Table S4), the unassigned portion of OTUs, Betaproteobacteria, Bacteroidia, Nitrospira, Gamma- and Alphaproteobacteria were the first six classes that explained more of the overall dissimilarity between the thermal waters and groundwaters, contributing together > 99% of total variability (Figure 5).
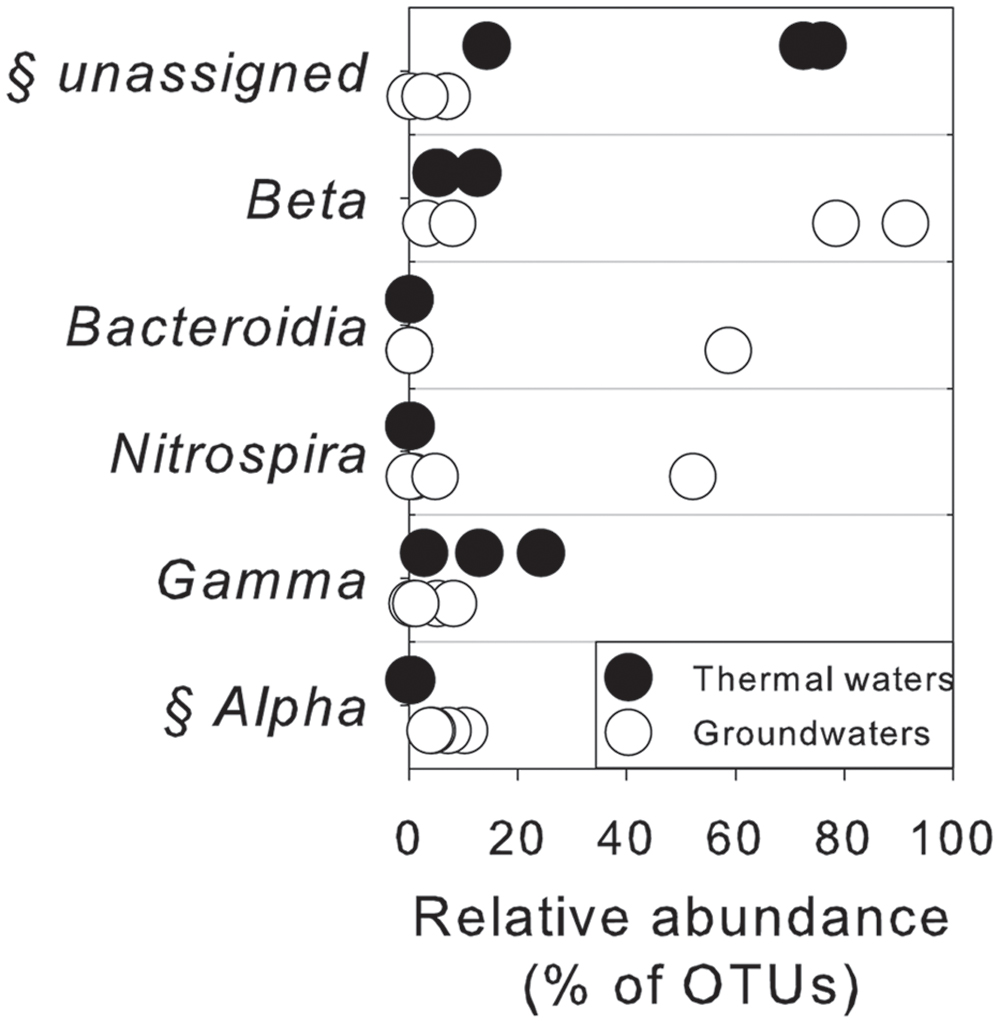
FIGURE 5. Microbiological variables that more significantly explained the overall dissimilarity (according to SIMPER test) between thermal waters and groundwaters at the class level. §, statistically different between two groups of water according to NPMANOVA analysis (p < 0.05).
Phylogenetic Analysis of Unassigned Bacterial 16S rRNA Gene Sequences from Thermal Waters
A number of sequence reads did not exhibit homology to known microbial taxa, which implies the presence in thermal waters of numerous so far unidentified microorganisms. In SSC and PAL, 76.1 and 72.5% of total OTUs, respectively, were not identified using known databases. In CAR, the unassigned portion was around 14.2% of total OTUs. The phylogenetic analysis of thermal samples showed that most of the unassigned 16S rRNA gene sequences were related to Epsilonproteobacteria, Gammaproteobacteria and Firmicutes (Figure 6). Among Epsilonproteobacteria, the sequences were related to the members of Nitratiruptor tergarcus (∼ 90% of similarity). Additionally, the sequences phylogenetically affiliated to the Gammaproteobacteria showed 95% of similarity with Thiofaba tepidiphila. The sequences affiliated with Firmicutes were only distantly related to Caldicellulosiruptor saccharolyticus, Desulfotomaculum thermosapovorans, Thermoanaerobacterium thermosaccharolyticum and Symbiobacterium thermophilum (∼75–80% of similarity). Sequences phylogenetically related to Actinobacteria, Nitrospirae and Betaproteobacteria were also retrieved.
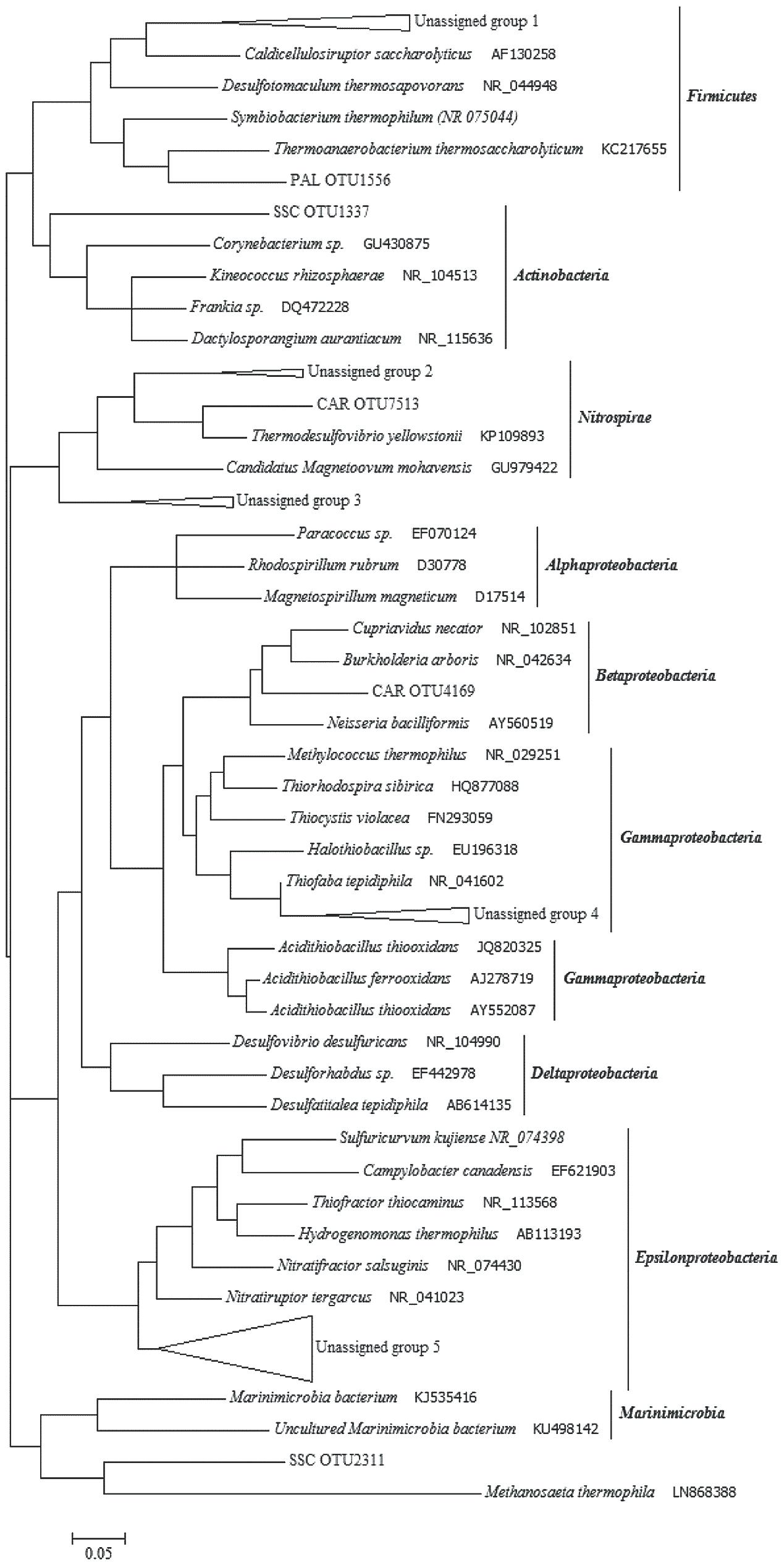
FIGURE 6. Phylogenetic tree constructed with unassigned 16S rRNA gene sequences obtained from the thermal samples. Unassigned group 1 (76 sequences): PAL OTUs 3865/3142/1624/5721/7240; SSC OTU 5116. Unassigned group 2 (17 sequences): CAR OTUs 1743/886/2900. Unassigned group 3 (13 sequences): SSC OTUs 676/3207/8283/6610; Unassigned group 4 (6 sequences): PAL OTUs 5277/848; SSC OTUs 3302/1374/2608; CAR OTU 5062. Unassigned group 5 (58 sequences): SSC OTUs 1776/2215/8205/6801/8038/363/3362/4634/7530/3704/6025/535/4726/3400/5198/6069/43/7956/1858/3460/6393/6056/2124; PAL OTUs 1583/1883.
Most Probable Number
Among the tested microbial functions, only nitrate and sulfate reduction, and heterotrophic As(III)-oxidization were found in all the samples (Figure 7). In particular, nitrate and sulfate reduction were found as main driving metabolisms in groundwaters where these microbial functional groups were retrieved at high abundance (up to ∼ 104 cells/mL). As expected, lower concentrations were found associated to surface waters in which aerobic heterotrophs were mainly observed. In line with the anoxic reaction environment, no aerobic heterotrophs were found in thermal waters. No growth of autotrophic As(III)-oxidizers, As(V)-reducers, Fe-reducers and oxidizers, Mn-reducers and sulfide oxidizers was observed by MPN. The sole As related metabolism found in the samples was related to heterotrophic As(III) oxidation. The abundance of the latter microbial functional group ranged between 0.1 × 101 and 1 × 103 cell/mL (Figure 7).
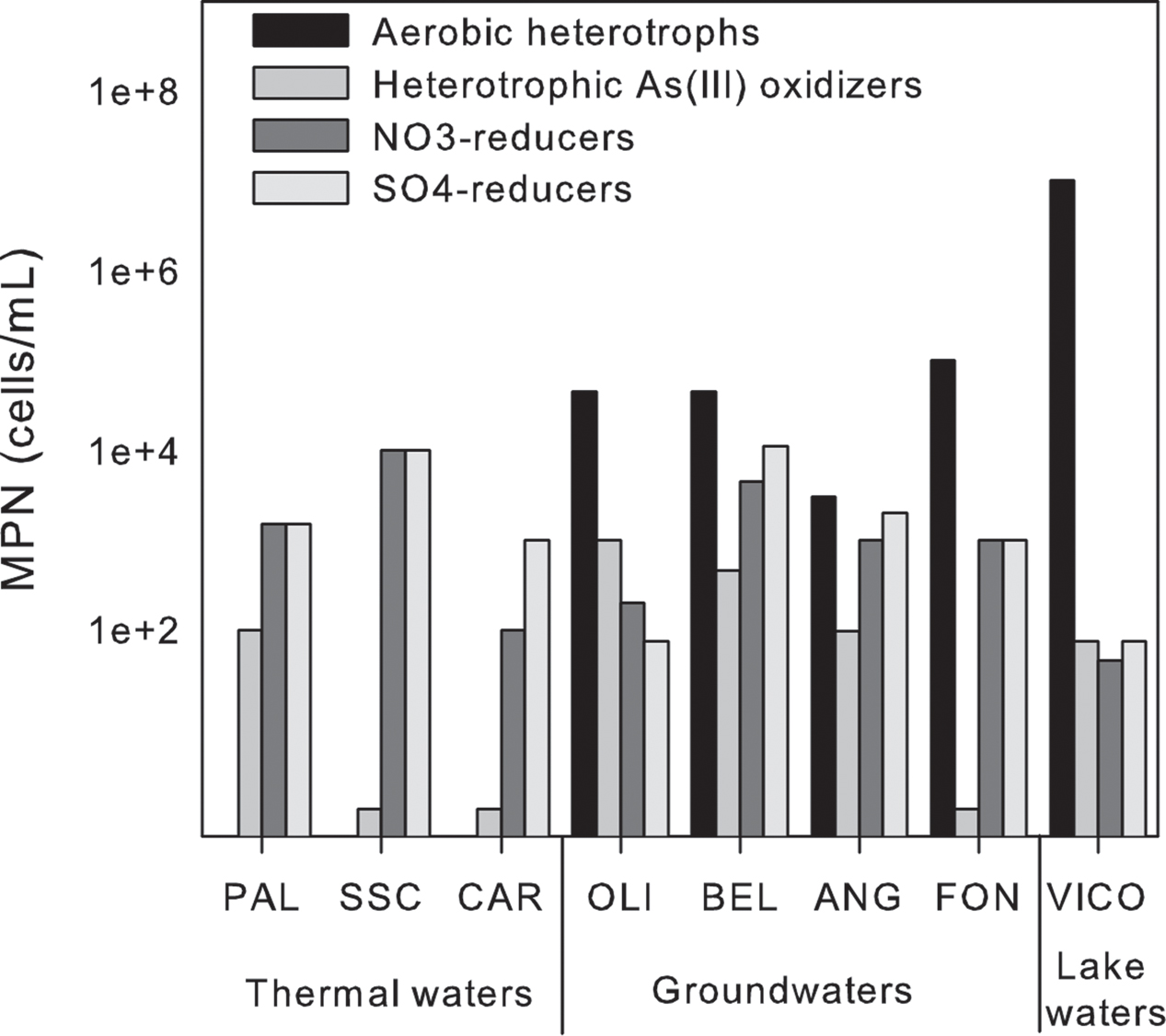
FIGURE 7. Most probable number (MPN) counts of the main microbial functional groups in thermal waters, groundwaters and lake waters.
Real-Time Quantification of Arsenic-Related Functional Genes
A preliminary screening by PCR showed the occurrence of arsB, for arsenite efflux pump, and of cytoplasmic arsenate reductase (arsC) genes in all screened samples, whereas arsenite oxidase (aioA) and arsenate respiratory reductase (arrA) were not amplified with any primer set under any tested condition (Supplementary Table S5). The abundance of arsB and arsC genes was estimated by real-time PCR. arsB and arsC genes were found in all samples at low concentration: arsB genes ranged between 0.3 × 102± 0.01 × 102 and 0.4 × 103 ± 0.1 × 102 gene copies/ml whereas arsC genes were found at concentrations up to 0.4 × 103 ± 0.3 × 102 gene copies/ml except in CAR (<d.l.) (Figure 8).
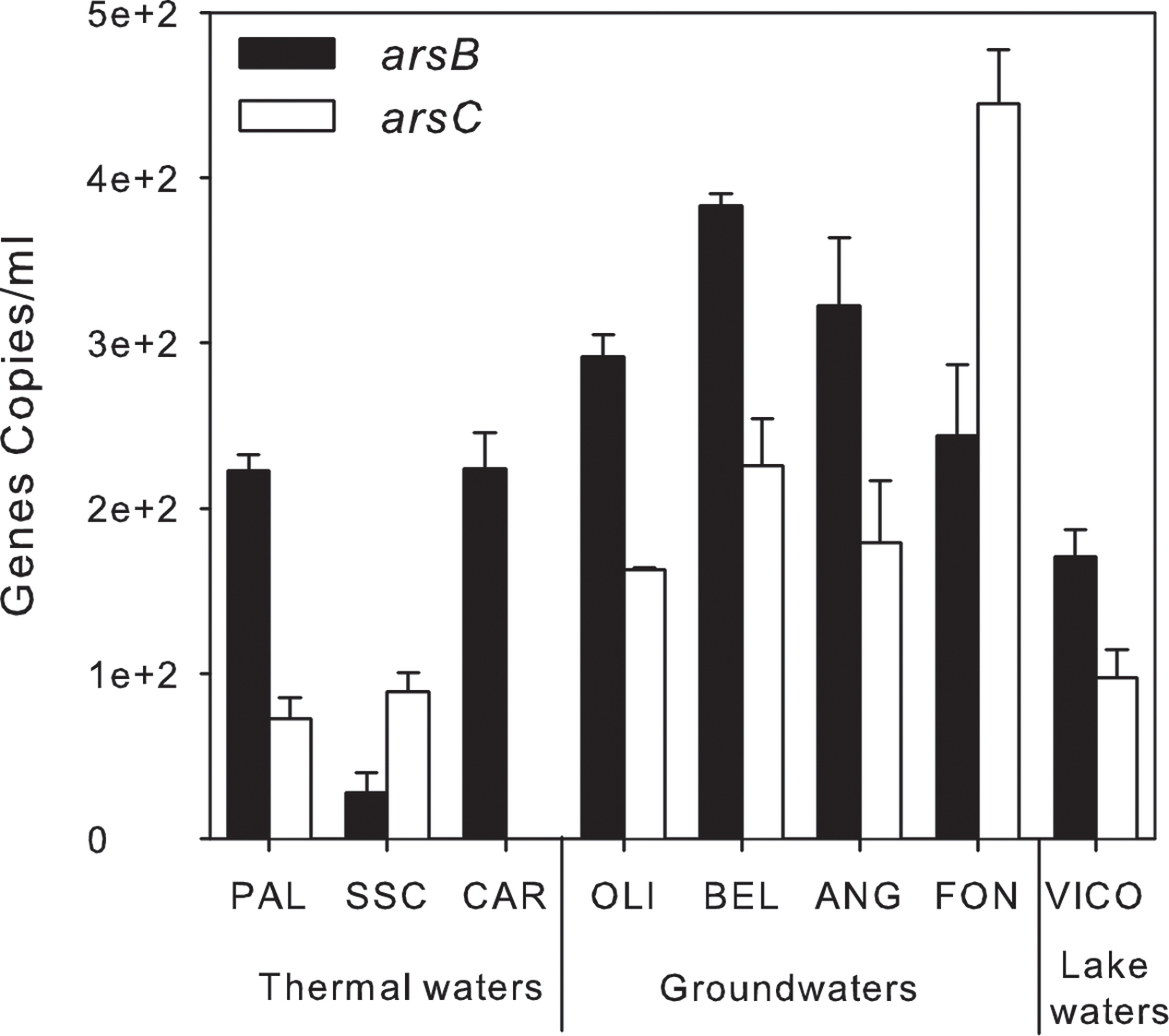
FIGURE 8. Arsenic-related genes abundance (gene copies/ml) estimated by qPCR in thermal waters, groundwaters and lake waters.
Discussion
Arsenic Occurrence and Microbial Community Composition
In this study, As concentrations were in line with previous studies which reported values up to 100 μg/L in groundwaters and 370 μg/L in thermal waters of Cimino-Vico volcanic area (Angelone et al., 2009; Baiocchi et al., 2013; Armiento et al., 2015; Fazi et al., 2016).
In accordance with literature findings reporting a variety of arsenic-resistant and transforming bacterial species in similar environments (Liao et al., 2011; Sarkar et al., 2013; Paul et al., 2015; Bahar et al., 2016), we found the predominance of Proteobacteria in groundwaters. In particular, the presence of Undibacterium and Azoarcus genera in groundwater was often observed in other As-contaminated waters (Griebler and Lueders, 2009; Ghosh and Sar, 2013). Some cultivable representatives of these genera were previously isolated from several environments, including drinking waters, and showed the ability to grow under anoxic condition using nitrate as final electron acceptor (i.e., Azoarcus spp.) (Reinhold-Hurek et al., 2015). The high nitrate concentration observed in FON groundwater indicated that the surficial local aquifer could be impacted by agricultural activities and affected by fecal contamination, as suggested by the high relative abundance of OTUs affiliated to dgA-11 gut group (58.6% of total OTUs), usually isolated from animal gastrointestinal tract and feces (Ozutsumi et al., 2005; Tsukinowa et al., 2008).
Consistently with redox conditions commonly found in geothermal environments (Ballantyne and Moore, 1988), thermal waters were generally characterized by low DO content and high As(III) concentration. The prevalence of oxidized As form, observed only in SSC sample, may indicate oxygen exposure during geothermal water uprising.
The high abundance of HNA cells found in thermal waters was in line with the highly selected microbiomes described by NGS. Indeed, LNA and HNA groups detected by flow cytometry are generally considered as constitutive traits of microbial communities in a variety of environments. Their relative contribution was reported to vary according to ecosystem properties and to reflect different phylotype composition, lifestyles and growth potential of the small genome LNA vs. the large genome HNA cells (Bouvier et al., 2007). HNA are also believed as the most active fraction (Lebaron et al., 2002), even though different phylogenetic affiliations were reported (Vila-Costa et al., 2012; Romina Schiaffino et al., 2013). Detection of HNA and LNA has been reported over a wide range of aquatic ecosystems covering large environmental gradients in bacterial (Troussellier et al., 1999; Gasol and del Giorgio, 2000; Andreatta et al., 2004; Thyssen et al., 2005; Sherr et al., 2006) and archaeal populations (Trigui et al., 2011).
In this study, archaea were found at high abundance in thermal waters, likely due to high temperature and reducing redox conditions (Rogers and Amend, 2005; Antranikian et al., 2017). Moreover, in thermal waters, a dominance of OTUs only distantly related to members affiliated to Epsilonproteobacteria, Gammaproteobacteria and Firmicutes was found, thus indicating the occurrence of bacterial biodiversity so far unexplored. Interestingly, the main cluster of unassigned OTUs (group 5 within Epsilonproteobacteria in Figure 6) was found in SSC sample. Some of the closest relatives are known thermophilic chemolithoautotrophs able to utilize molecular hydrogen as an electron donor and oxygen or nitrate as electron acceptor. Overall, the microbial profiling obtained by NGS analysis revealed in thermal waters the occurrence of microorganisms involved in sulfur and nitrogen biogeochemical cycles (e.g., Sulfuricurvum kujiense, Desulfotomaculum thermosapovorans, Thiofaba tepidiphila, Nitratiruptor tergarcus, Nitratifactor salsuginis, Nitrospira, Azoarcus, Stenotrophomonas). It is worth to noting that, differently from previous studies performed on high-enthalpy geothermal systems (e.g., Yellowstone National Park), the bacterial phyla typically found in such environments (e.g., Aquificae, Chloroflexi, Deinococcus–Thermus) were not retrieved. This is most likely related to the different geochemical conditions occurring at Cimino-Vico volcanic area (e.g., lower T, nearly neutral pH, slightly reducing redox conditions) which may strongly affect the microbial community structure.
Both As species were found in lake waters and they were likely related to redox processes mediated by high organic carbon availability (litter deposition, surface water runoff, 2.5 mg/L DOC) which represents, together with the light, one of the main driving forces of the complex interplay of biological activities occurring in such environment.
Even though the lake is directly affected by hydrothermal upwelling (Agenzia Regionale per la Protezione Ambientale [ARPA], 2012), the microbiological characteristics of Lake Vico were different from other waters analyzed. The abundance of Betaproteobacteria in this aquatic environment showed similar values reported at the same location in previous studies (Davolos and Pietrangeli, 2013; Fazi et al., 2016). Furthermore, the presence of Planktothrix was recurrently observed in this area (Manganelli et al., 2016).
Linking Microbial Metabolic Traits to Arsenic Biogeochemistry
As assessed by the cultivation-dependent approach, sulfate and nitrate reduction and heterotrophic As(III)-oxidation were the main metabolisms occurring in geothermal area. Despite the low concentrations of NO3, the involvement of microbial communities in nitrate-reduction was observed in our samples. This suggested that microbial communities could activate this process as a result of an increase in nitrate concentration due to anthropic contamination (e.g., agricultural activity). Nitrate-linked microbial transformation of As was reported in As-contaminated aquatic environments, and some microorganisms were found to mediate anaerobic As(III) oxidation by coupling to nitrate reduction (Zhang et al., 2017). Differently from alluvial environments (Islam et al., 2004), Fe- and Mn-related metabolisms were not found in our samples, since they were not likely to play an essential role in this geothermal area (Piscopo et al., 2006).
Remarkably, As detoxification processes prevail in groundwater and thermal waters. The detection of arsBC genes is consistent with previous evidences in a variety of environments (Liao et al., 2011; Davolos and Pietrangeli, 2013; Paul et al., 2015). arsB and arsC genes were found simultaneously in most of the samples in line with the assumption generally accepted that their environmental distribution is similar (Davolos and Pietrangeli, 2013; Escudero et al., 2013).
Although heterotrophic As(III) oxidation was revealed by MPN analysis, aioA gene was not detected by PCR using different primer sets. This finding was probably due to the high microbial diversity in our samples and to the primer coverage that may not be sufficient to capture a high gene diversity. It is known that the investigation of As-related functional genes through PCR approaches as well as through metagenome sequencing is currently difficult in ecological studies (Fahy et al., 2015). A wide variety of aioA-like genes exists in natural environments and often these genes show low homology to currently known gene sequences (Hamamura et al., 2009; Jiang et al., 2015). Hence, new primer designs should integrate the most recent sequencing data as well as biochemical data and genetic context (Fahy et al., 2015). The primers in use so far were designed on few microbial strains and were mainly used only on isolates (Inskeep et al., 2007; Quéméneur et al., 2008; Karn and Pan, 2016). Few studies reported the detection of As-related genes in mixed microbial communities in natural environments (Engel et al., 2013; Escudero et al., 2013; Lami et al., 2013), in spite of their presence in different prokaryotic groups such as Proteobacteria, Firmicutes, Chrysiogenetes, Deinococcus-Thermus, Deferribacteres and Chrenarchaeota isolated from a variety of As-rich environments (e.g., mine, arsenical pesticide- or smelter-impacted sites, geothermal sites, geyser, soil and sediments) (Inskeep et al., 2007; Cai et al., 2009; Heinrich-Salmeron et al., 2011; Sultana et al., 2012).
Conclusion
This study provided the structure and composition of the microbiome exposed to a natural range of geogenic As contamination in geothermal waters. Since the occurrence and distribution of arsenic-related genes were not exhaustively explored in waters of geothermal origin, we provided field insights on the metabolic activities and potentialities of microbial communities related to arsenic detoxification, reduction and oxidation. Remarkably, the microbial communities were able to withstand high As levels, without using it for energetic metabolism. Further studies are needed to investigate the occurrence of novel thermophiles able to tolerate high As concentrations in geothermal environments.
Author Contributions
SC conducted CARD-FISH, MPN, qPCR experiments and analyzed the whole set of biological data. SA performed FCM measurements, the statistical data analysis and together with SF contributed to the interpretation of the biomolecular data. SZ, AC, and LC performed the bacterial 16S rRNA gene pyrosequencing and the phylogenetic analysis. BC performed the chemical analysis of the water samples and contributed to the interpretation of chemical data. SR conceived and coordinated the study. All authors contributed to the writing of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The research was support by Fondazione CARIPLO contract No. 2014-1301.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02468/full#supplementary-material
References
Achour, A. R., Bauda, P., and Billard, P. (2007). Diversity of arsenite transporter genes from arsenic-resistant soil bacteria. Res. Microbiol. 158, 128–137. doi: 10.1016/j.resmic.2006.11.006
Agenzia Regionale per la Protezione Ambientale [ARPA] (2012). Piano Della Caratterizzazione Del Lago di Vico. Palermo: Agenzia Regionale per la Protezione Ambientale [ARPA].
Amalfitano, S., Del Bon, A., Zoppini, A., Ghergo, S., Fazi, S., Parrone, D., et al. (2014). Groundwater geochemistry and microbial community structure in the aquifer transition from volcanic to alluvial areas. Water Res. 65, 384–394. doi: 10.1016/j.watres.2014.08.004
Andreatta, S., Wallinger, M. M., Piera, J., Catalan, J., Psenner, R., Hofer, J. S., et al. (2004). Tools for discrimination and analysis of lake bacterioplankton subgroups measured by flow cytometry in a high-resolution depth profile. Aquat. Microb. Ecol. 36, 107–115. doi: 10.3354/ame036107
Andres, J., and Bertin, P. N. (2016). The microbial genomics of arsenic. FEMS Microbiol. Rev. 39, 1–24. doi: 10.1093/femsre/fuv050
Angelone, M., Cremisini, C., Piscopo, V., Proposito, M., and Spaziani, F. (2009). Influence of hydrostratigraphy and structural setting on the arsenic occurrence in groundwater of the Cimino-Vico volcanic area (central Italy). Hydrogeol. J. 17, 901–914. doi: 10.1007/s10040-008-0401-3
Antranikian, G., Suleiman, M., Schäfers, C., Adams, M. W. W., Bartolucci, S., Blamey, J. M., et al. (2017). Diversity of bacteria and archaea from two shallow marine hydrothermal vents from Vulcano Island. Extrempohiles 21, 733–742. doi: 10.1007/s00792-017-0938-y
Armiento, G., Baiocchi, A., Cremisini, C., Crovato, C., Lotti, F., Lucentini, L., et al. (2015). An integrated approach to identify water resources for human consumption in an area affected by high natural arsenic content. Water 7, 5091–5114. doi: 10.3390/w7095091
Bahar, M. M., Megharaj, M., and Naidu, R. (2016). Oxidation of arsenite to arsenate in growth medium and groundwater using a novel arsenite-oxidizing diazotrophic bacterium isolated from soil. Int. Biodeterior. Biodegrad. 106, 178–182. doi: 10.1016/j.ibiod.2015.10.019
Baiocchi, A., Coletta, A., Espositi, L., Lotti, F., and Piscopo, V. (2013). Sustainable groundwater development in a naturally arsenic contaminated aquifer: the case of the Cimino–Vico volcanic area (central Italy). Ital. J. Eng. Geol. Environ. 13, 5–18.
Ballantyne, J. M., and Moore, J. N. (1988). Arsenic geochemistry in geothermal systems. Geochim. Cosmochim. Acta 52, 475–483. doi: 10.1016/0016-7037(88)90102-0
Barron, J. L., and Lueking, D. R. (1990). Growth and maintenance of Thiobacillus ferrooxidans cells. Appl. Environ. Microbiol. 56, 2801–2806.
Bissen, M., and Frimmel, F. H. (2003). Arsenic—A review. Part 1: occurrence, toxicity, speciation, mobility. Acta Hydrochim. Hydrobiol. 31, 9–18. doi: 10.1002/aheh.200390025
Borch, T., Kretzschmar, R., Kappler, A., Van Cappellen, P., Ginder-Vogel, M., Voegelin, A., et al. (2010). Biogeochemical redox processes and their impact on contaminant dynamics. Environ. Sci. Technol. 44, 15–23. doi: 10.1021/es9026248
Bouvier, T., del Giorgio, P. A., and Gasol, J. M. (2007). A comparative study of the cytometric characteristics of High and Low nucleic-acid bacterioplankton cells from different aquatic ecosystems. Environ. Microbiol. 9, 2050–2066. doi: 10.1111/j.1462-2920.2007.01321.x
Burton, E. D., Johnston, S. G., and Planer-Friederich, B. (2013). Coupling of arsenic mobility to sulfur transformations during microbial sulfate reduction in the presence and absence of humic acid. Chem. Geol. 343, 12–24. doi: 10.1016/j.chemgeo.2013.02.005
Cai, L., Liu, G., Rensing, C., and Wang, G. (2009). Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils. BMC Microbiol. 9:4. doi: 10.1186/1471-2180-9-4
Caporaso, J. G., Bittinger, K., Bushman, F. D., DeSantis, T. Z., Andersen, G. L., and Knight, R. (2010a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267. doi: 10.1093/bioinformatics/btp636
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Casentini, B., Falcione, F. T., Amalfitano, S., Fazi, S., and Rossetti, S. (2016). Arsenic removal by discontinuous ZVI two steps system for drinking water production at household scale. Water Res. 106, 135–145. doi: 10.1016/j.watres.2016.09.057
Casentini, B., and Pettine, M. (2010). Effects of desferrioxamine-B on the release of arsenic from volcanic rocks. Appl. Geochem. 25, 1688–1698. doi: 10.1016/j.apgeochem.2010.08.016
Cinti, D., Poncia, P. P., Brusca, L., Tassi, F., Quattrocchi, F., and Vaselli, O. (2015). Spatial distribution of arsenic, uranium and vanadium in the volcanic-sedimentary aquifers of the Vicano-Cimino Volcanic District (Central Italy). J. Geochem. Explor. 152, 123–133. doi: 10.1016/j.gexplo.2015.02.008
Clarke, K. R. (1993). Nonparametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x
Corsini, A., Zaccheo, P., Muyzer, G., Andreoni, V., and Cavalca, L. (2014). Arsenic transforming abilities of groundwater bacteria and the combined use of Aliihoeflea sp. strain 2WW and goethite in metalloid removal. J. Hazard. Mater. 269, 89–97. doi: 10.1016/j.jhazmat.2013.12.037
Cote, R. J., and Gherna, R. L. (1994). “Nutrition and medium,” in Methods for General and Molecular Bacteriology, eds P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Kreig (Washington, DC: American Society for Microbiology), 156–178.
Dall’aglio, M., Giuliano, G., Amicizia, D., et al. (2001). “Assessing drinking water quality in Northern Latium by trace elements analysis,” in Proceedings of the 10th International Symposium on Water-Rock Interaction, ed. R. Cidu (Villasimius: A.A. Balkema), 1063–1066.
Davolos, D., and Pietrangeli, B. (2013). A molecular study on bacterial resistance to arsenic-toxicity in surface and underground waters of Latium (Italy). Ecotoxicol. Environ. Saf. 96, 1–9. doi: 10.1016/j.ecoenv.2013.05.039
Donahoe-Christiansen, J., Imperio, S. D., Jackson, C. R., Inskeep, W. P., and McDermott, T. R. (2004). Arsenite-oxidizing Hydrogenobaculum strain isolated from an acid-sulfate-chloride geothermal spring in Yellowstone National Park. Appl. Environ. Microbiol. 70, 1865–1868. doi: 10.1128/AEM.70.3.1865-1868.2004
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edwardson, C., Planer-Friedrich, B., and Hollibaugh, J. (2014). Transformation of monothioarsenate by haloalkaliphilic, anoxygenic photosynthetic purple sulfur bacteria. FEMS Microbiol. Ecol. 90, 858–868. doi: 10.1111/1574-6941.12440
Engel, A. S., Johnson, L. R., and Porter, M. L. (2013). Arsenite oxidase gene diversity among Chloroflexi and Proteobacteria from EI Tatio Geyser Field, Chile. FEMS Microbiol. Ecol. 83, 745–756. doi: 10.1111/1574-6941.12030
Escudero, L. V., Casamayor, E. O., Chong, G., Pedrós-Alió, C., and Demergasso, C. (2013). Distribution of microbial arsenic reduction, oxidation and extrusion genes along a wide range of environmental arsenic concentrations. PLOS ONE 8:e78890. doi: 10.1371/journal.pone.0078890
Fahy, A., Giloteaux, L., Bertin, P., Le Paslier, D., Méedigue, C., Weissenbach, J., et al. (2015). 16S rRNA and As-related functional diversity: contrasting fingerprints in arsenic-rich sediments from an acid mine drainage. Microb. Ecol. 70, 154–167. doi: 10.1007/s00248-014-0558-3
Fazi, S., Amalfitano, S., Pizzetti, I., and Pernthaler, J. (2007). Efficiency of fluorescence in situ hybridization for bacterial cell identification in temporary river sediments with contrasting water content. Syst. Appl. Microbiol. 30, 463–470. doi: 10.1016/j.syapm.2007.03.003
Fazi, S., Crognale, S., Casentini, B., Amalfitano, S., and Rossetti, S. (2016). The arsenite oxidation potential of native microbial communities from arsenic-rich freshwaters. Microb. Ecol. 72, 25–35. doi: 10.1007/s00248-016-0768-y
Fazi, S., Vázquez, E., Casamayor, E. O., Amalfitano, S., and Butturini, A. (2013). Stream hydrological fragmentation drives bacterioplankton community composition. PLOS ONE 8:e64109. doi: 10.1371/journal.pone.0064109
Fendorf, S., Michael, H. A., and van Geen, A. (2010). Spatial and temporal variations of groundwater arsenic in South and Southeast Asia. Science 328, 1123–1127. doi: 10.1126/science.1172974
Foulquier, A., Volat, B., Neyra, M., Bornette, G., and Montuelle, B. (2013). Long-term impact of hydrological regime on structure and functions of microbial communities in riverine wetland sediments. FEMS Microbiol. Ecol. 85, 211–226. doi: 10.1111/1574-6941.12112
Gasol, J. M., and del Giorgio, P. A. (2000). Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci. Mar. 64, 197–224. doi: 10.3989/scimar.2000.64n2197
Ghosh, S., and Sar, P. (2013). Identification and characterization of metabolic properties of bacterial populations recovered from arsenic contaminated ground water of North East India (Assam). Water Res. 47, 6992–7005. doi: 10.1016/j.watres.2013.08.044
Gihring, T. M., and Banfield, J. F. (2001). Arsenite oxidation and arsenate respiration by a new Thermus isolate. FEMS Microbiol. Lett. 204, 335–340. doi: 10.1111/j.1574-6968.2001.tb10907.x
Greuter, D., Loy, A., Horn, M., and Rattei, T. (2016). probeBase - an online resource for rRNA-targeted oligonucleotide probes and primers: new features 2016. Nucleic Acids Res. 44, D586–D589. doi: 10.1093/nar/gkv1232
Griebler, C., and Lueders, T. (2009). Microbial biodiversity in groundwater ecosystems. Freshw. Biol. 54, 649–677. doi: 10.1111/j.1365-2427.2008.02013.x
Haertig, C., Lohmayer, R., Kolb, S., Horn, M. A., Inskeep, W. P., and Plaanner -Friedrich, B. (2014). Chemolithotropic growth of the aerobic hyperthermopilic bacterium Thermocrinis ruber OC 14/7/2 on monothioarsenate and arsenite. FEMS Microbiol. Ecol. 90, 747–760. doi: 10.1111/1574-6941.12431
Haertig, C., and Planer-Friedrich, B. (2012). Thioarsenate transformation by filamentous microbial mats thriving in an alkaline, sulfidic hot spring. Environ. Sci. Technol. 46, 4348–4356. doi: 10.1021/es204277j
Hamamura, N., Macur, R. E., Korf, S., Ackermann, G., Taylor, W. P., Kozubal, M., et al. (2009). Linking microbial oxidation of arsenic with detection and phylogenetic analysis of arsenite oxidase genes in diverse geothermal environments. Environ. Microbiol. 11, 421–431. doi: 10.1111/j.1462-2920.2008.01781.x
Heinrich-Salmeron, A., Cordi, A., Brochier-Armanet, C., Halter, D., Pagnout, C., Abbaszadeh-fard, E., et al. (2011). Unsuspected diversity of arsenite oxidizing bacteria as revealed by widespread distribution of aoxB gene in prokaryotes. Appl. Environ. Microbiol. 77, 4685–4692. doi: 10.1128/AEM.02884-10
Hoeft, S. E., Blum, J. S., Stolz, J. F., Tabita, F. R., Witte, B., King, G. M., et al. (2007). Alkalilimnicola ehrlichii sp. nov., a novel, arsenite-oxidizing haloalkaliphilic gammaproteobacterium capable of chemoautotrophic or heterotrophic growth with nitrate or oxygen as the electron acceptor. Int. J. Syst. Evol. Microbiol. 57, 504–512. doi: 10.1099/ijs.0.64576-0
Hoeft, S. E., Kulp, T. R., Stolz, J. R., Hollibaugh, J. T., and Oremland, R. S. (2004). Dissimilatory arsenate reduction with sulfide as electron donor: experiments with Mono Lake water and isolation of Strain MLMS-1, a chemoautotrophic arsenate respirer. Appl. Environ. Microbiol. 70, 2741–2747. doi: 10.1128/AEM.70.5.2741-2747.2004
Huang, J. H. (2014). Impact of microorganisms on arsenic biogeochemistry: a review. Water Air Soil Pollut. 225, 1–25. doi: 10.1007/s11270-013-1848-y
Inskeep, W. P., Ackerman, G. G., Taylor, W. P., Kozubal, M., Korf, S., and Macur, E. (2005). On the energetics of chemolithotrophy in nonequilibrium systems: case studies of geothermal springs in Yellowstone National Park. Geobiology 3, 297–317. doi: 10.1111/j.1472-4669.2006.00059.x
Inskeep, W. P., Macur, R. E., Hamamura, N., Warelow, T. P., Ward, S. A., and Santini, J. M. (2007). Detection, diversity and expression of aerobic bacterial arsenite oxidase genes. Environ. Microbiol. 9, 934–943. doi: 10.1111/j.1462-2920.2006.01215.x
Islam, F. S., Gault, A. G., Boothman, C., Polya, D. A., Charnock, J. M., Chatterjee, D., et al. (2004). Role of metal-reducing bacteria in arsenic release from Bengal delta sediments. Nature 430, 68–71. doi: 10.1038/nature02638
Jackson, C. R., Langner, H. W., Donahoe-Christiansen, J., Inskeep, W. P., and McDermott, T. T. (2001). Molecular analysis of microbial community structure in an arsenite-oxidizing acidic thermal spring. Environ. Microbiol. 3, 532–542. doi: 10.1046/j.1462-2920.2001.00221.x
Jiang, D., Li, P., Jiang, Z., Dai, X., Zhang, R., Wang, Y., et al. (2015). Chemolithoautotrophic arsenite oxidation by a thermophilic Anoxybacillus flavithermus strain TCC9-4 from a hot spring in Tengchong of Yunnan, China. Front. Microbiol. 6:360. doi: 10.3389/fmicb.2015.00360
Jones, L. C., Lafferty, B. J., and Sparks, D. L. (2012). Additive and competitive effects of bacteria and Mn oxides on arsenite oxidation kinetics. Environ. Sci. Technol. 46, 6548–6555. doi: 10.1021/es204252f
Karn, S. K., and Pan, X. (2016). Role of Acinetobacter sp. in arsenite As(III) oxidation and reducing its mobility in soil. Chem. Ecol. 32, 460–471. doi: 10.1080/02757540.2016.1157174
Kumari, N., and Jagadevan, S. (2016). Genetic identification of arsenate reductase and arsenite oxidase in redox transformations carried out by arsenic metabolising prokaryotes- A comprehensive review. Chemosphere 163, 400–412. doi: 10.1016/j.chemosphere.2016.08.044
Lami, R., Jones, L. C., Cottrell, M. T., Lafferty, B. J., Ginder-Vogel, M., Sparks, D. L., et al. (2013). Arsenite modifies structure of soil microbial communities and arsenite oxidation potential. FEMS Microbiol. Ecol. 84, 270–279. doi: 10.1111/1574-6941.12061
Langner, H. W., Jackson, C. R., McDermott, T. R., and Inskeep, W. P. (2001). Rapid oxidation of arsenite in a hot spring ecosystem, Yellowstone National Park. Environ. Sci. Technol. 35, 3302–3309. doi: 10.1021/es0105562
Lebaron, P., Servais, P., Baudoux, A. C., Bourrain, M., Courties, C., and Parthuisot, N. (2002). Variations of bacterial-specific activity with cell size and nucleic acid content assessed by flow cytometry. Aquat. Microb. Ecol. 28, 131–140. doi: 10.3354/ame028131
Lee, S.-W. (2013). Enhancement of arsenic mobility by Fe(III)-reducing bacteria from iron oxide minerals. J. Mater. Cycles Waste Manag. 15, 362–369. doi: 10.1007/s10163-013-0132-y
Lett, M. C., Muller, D., Lièvremont, D., Silver, S., and Santini, J. (2012). Unified nomenclature for genes involved in prokaryotic aerobic arsenite oxidation. J. Bacteriol 194, 207–208. doi: 10.1128/JB.06391-11
Liao, V. H. C., Chu, Y. J., Su, Y. C., Hsiao, S. Y., Wei, C. C., Liu, C. W., et al. (2011). Arsenite-oxidizing and arsenate-reducing bacteria associated with arsenic-rich groundwater in Taiwan. J. Contam. Hydrol. 123, 20–29. doi: 10.1016/j.jconhyd.2010.12.003
Lovley, D. (2006). Dissimilatory Fe(II)- and Mn(IV)-reducing prokaryotes. Prokaryotes 2, 635–658. doi: 10.1007/0-387-30742-7_21
Malasarn, D., Saltikov, C. W., Cambpell, K. M., Santini, J. M., Hering, J. G., and Newman, D. K. (2004). arrA is a reliable marker for As(V) respiration. Science 306:455. doi: 10.1126/science.1102374
Manganelli, M., Stefanelli, M., Vichi, S., Andreani, P., Nascetti, G., Scialanca, F., et al. (2016). Cyanobacteria biennal dynamic in a volcanic mesotrophic lake in central Italy: strategies to prevent dangerous human exposures to cyanotoxins. Toxicon 11, 28–40. doi: 10.1016/j.toxicon.2016.03.004
Mukhopadhyay, R., Rosen, B. P., Phung, L. T., and Silver, S. (2002). Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol. Rev. 26, 311–325. doi: 10.1111/j.1574-6976.2002.tb00617.x
Muller, D., Simeonova, D. D., Riegel, P., Mangenot, S., Koechler, S., Lièvremont, D., et al. (2006). Herminiimonas arsenicoxydans sp. nov., a metalloresistant bacterium. Int. J. Syst. Evol. Microbiol. 56, 1765–1769. doi: 10.1099/ijs.0.64308-0
Oremland, R. S., and Stolz, J. F. (2003). The ecology of arsenic. Science 300, 939–944. doi: 10.1126/science.1081903
Ozutsumi, Y., Hayashi, H., Sakamoto, M., Itabashi, H., and Benno, Y. (2005). Culture-independent analysis of fecal microbiota in cattle. Biosci. Biotechnol. Biochem. 69, 1793–1797. doi: 10.1271/bbb.69.1793
Páez-Espino, D., Tamames, J., de Lorenzo, V., and Cánovas, D. (2009). Microbial responses to environmental arsenic. Biometals 22, 117–130. doi: 10.1007/s10534-008-9195-y
Paul, D., Kazy, S. K., Das Banerjee, T., Gupta, A. K., Pal, T., and Sar, P. (2015). Arsenic biotransformation and release by bacteria indigenous to arsenic contaminated groundwater. Bioresour. Technol. 188, 14–23. doi: 10.1016/j.biortech.2015.02.039
Piscopo, V., Barbieri, M., Monetti, V., Pagano, G., Pistoni, S., Ruggi, E., et al. (2006). Hydrogeology of thermal waters in Viterbo area, central Italy. Hydrogeol. J. 14, 1508–1521. doi: 10.1007/s10040-006-0090-8
Planer-Friedrich, B., Härtig, C., Lohmayer, R., Suess, E., McCann, S. H., and Oremland, R. (2015). Anaerobic chemolithotrophic growth of the haloalkaliphilic bacterium strain MLMS-1 by disproportionation of monothioarsenate. Environ. Sci. Technol. 49, 6554–6563. doi: 10.1021/acs.est.5b01165
Quast, C., Pruesse, E., Ylmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
Quéméneur, M., Heinrich-Salmeron, A., Muller, D., Lièvremont, D., Jauzein, M., Bertin, P. N., et al. (2008). Diversity surveys and evolutionary relationships of aoxB Genes in aerobic arsenite-oxidizing bacteria. Appl. Environ. Microbiol. 74, 4567–4573. doi: 10.1128/AEM.02851-07
Rahman, M. A., and Hassler, C. (2014). Is arsenic biotransformation a detoxification mechanism for microorganisms? Aquat. Toxicol. 146, 212–219. doi: 10.1016/j.aquatox.2013.11.009
Reinhold-Hurek, B., Tan, Z., and Hurek, T. (2015). Azoarcus. Bergey’s Manual of Systematics of Archaea and Bacteria. Hoboken, NJ: John Wiley & Sons, 1–19. doi: 10.1002/9781118960608.gbm00994
Rittle, K. A., Drever, J. I., and Colberg, P. J. S. (1995). Precipitation of arsenic during bacterial sulfate reduction. Geomicrobiol. J. 13, 1–11. doi: 10.1080/01490459509378000
Rodriguez-Freire, L., Moore, S. E., Sierra-Alvarez, R., Root, R. A., Chorover, J., and Field, J. A. (2016). Arsenic remediation by formation of arsenic sulfide minerals in a continuous anaerobic bioreactor. Biotechnol. Bioeng. 113, 522–530. doi: 10.1002/bit.25825
Rogers, K. L., and Amend, J. P. (2005). Archaeal diversity and geochemical energy yields in a geothermal well on Vulcano Island, Italy. Geobiology 3, 319–332. doi: 10.1111/j.1472-4669.2006.00064.x
Romina Schiaffino, M., Gasol, J. M., Izaguirre, I., and Unrein, F. (2013). Picoplankton abundance and cytometric group diversity along a trophic and latitudinal lake gradient. Aquat. Microb. Ecol. 68, 231–250. doi: 10.3354/ame01612
Sarkar, S., Kazy, S. K., and Sar, P. (2013). Characterization of arsenic resistant bacteria from arsenic rich groundwater of West Bengal, India. Ecotoxicology 22, 363–376. doi: 10.1007/s10646-012-1031-z
Sehlin, H. M., and Lindström, E. B. (1992). Oxidation and reduction of arsenic by Sulfolobus acidocaldarius strain BC. FEMS Microbiol. Lett. 93, 87–92. doi: 10.1111/j.1574-6968.1992.tb05045.x
Sherr, E. B., Sherr, B. F., and Longnecker, K. (2006). Distribution of bacterial abundance and cell-specific nucleic acid content in the Northeast Pacific Ocean. Deep Sea Res. Part I Oceanogr. Res. Pap. 53, 713–725. doi: 10.1016/j.dsr.2006.02.001
Silver, S., and Phung, L. T. (2005a). Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl. Environ. Microbiol. 71, 599–608. doi: 10.1128/AEM.71.2.599-608.2005
Silver, S., and Phung, L. T. (2005b). A bacterial view of the periodic table: genes and proteins for toxic inorganic ions. J. Ind. Microbiol. Biotechnol. 32, 587–605.
Smedley, P., and Kinniburgh, D. (2002). A review of the source, behavior and distribution of arsenic in natural waters. Appl. Geochem. 17, 517–568. doi: 10.1016/S0883-2927(02)00018-5
Stolz, J. F., Basu, P., Santini, J. M., and Oremland, R. S. (2006). Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 60, 107–130. doi: 10.1146/annurev.micro.60.080805.142053
Stolz, J. F., and Oremland, R. S. (1999). Bacterial respiration of arsenic and selenium. FEMS Microbiol. Rev. 23, 615–627. doi: 10.1111/j.1574-6976.1999.tb00416.x
Sultana, M., Vogler, S., Zargar, K., Schmidt, A. C., Saltikov, C., Seifert, J., et al. (2012). New clusters of arsenite oxidase and unusual bacterial groups in enrichments from arsenic-contaminated soil. Arch. Microbiol. 194, 623–635. doi: 10.1007/s00203-011-0777-7
Sun, Y., Polishchuk, E. A., Radoja, U., and Cullen, W. R. (2004). Identification and quantification of arsC genes in environmental samples by using real-time PCR. J. Microbiol. Methods 58, 335–349. doi: 10.1016/j.mimet.2004.04.015
Sutton, S. (2010). The most probable number method and its uses in enumeration, qualification, and validation. J. Valid. Technol. 16, 35–38.
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tang, K., Barry, K., Chertkov, O., Dalin, E., Han, C. S., Hauser, L. J., et al. (2011). Complete genome sequence of the filamentous anoxygenic phototrophic bacterium Chloroflexus aurantiacus. BMC Genomics 12:334. doi: 10.1186/1471-2164-12-334
Thyssen, M., Lefêvre, D., Caniaux, G., Ras, J., Dugrais, L., Fernandez Ibanez, C., et al. (2005). Spatial distribution of heterotrophic bacteria in the North East Atlantic (POMME study area) during spring 2001. J. Geophys. Res. 110:CO7S16. doi: 10.1029/2004JC002670
Trigui, H., Masmoudi, S., Brochier-Armanet, C., Barani, A., Grégori, G., Denis, M., et al. (2011). Characterization of heterotrophic prokaryote subgroups in the Sfax coastal solar salterns by combining flow cytometry cell sorting and phylogenetic analysis. Extremophiles 15, 347–358. doi: 10.1007/s00792-011-0364-5
Troussellier, M., Courties, C., Lebaron, P., and Servais, P. (1999). Flow cytometric discrimination of bacterial populations in seawater based on SYTO 13 staining of nucleic acids. FEMS Microbiol. Ecol. 29, 319–330. doi: 10.1111/j.1574-6941.1999.tb00623.x
Tsukinowa, E., Karita, S., Asano, S., Wakai, Y., Oka, Y., Furuta, M., et al. (2008). Fecal microbiota of a dugong (Dugong dugong) in captivity at Toba Aquarium. J. Gen. Appl. Microbiol. 54, 25–38. doi: 10.2323/jgam.54.25
Tzur, A., Moore, J. K., Jorgensen, P., Shapiro, H. M., and Kirschner, M. W. (2011). Optimizing optical flow cytometry for cell volume-based sorting and analysis. PLOS ONE 6:e16053. doi: 10.1371/journal.pone.0016053
van Lis, R., Nitschke, W., Duval, S., and Schoepp-Cothenet, B. (2013). Arsenics as bioenergetic substrates. Biochim. Biophys. Acta 1827, 176–188. doi: 10.1016/j.bbabio.2012.08.007
Vila-Costa, M., Gasol, J. M., Sharma, S., and Moran, M. A. (2012). Community analysis of high- and low-nucleic acid-containing bacteria in NW Mediterranean coastal waters using 16S rDNA pyrosequencing. Environ. Microbiol. 14, 1390–1402. doi: 10.1111/j.1462-2920.2012.02720.x
Vu, K. B., Kaminski, M. D., and Nuñez, L. (2003). Review of Arsenic Removal Technologies for Contaminated Groundwaters. Argonne, IL: Argonne National Laboratory.
Wilkie, J. A., and Hering, J. G. (1998). Rapid oxidation of geothermal arsenic(iii) in streamwaters of the Eastern Sierra Nevada. Environ. Sci. Technol. 32, 657–662. doi: 10.1021/es970637r
Zargar, K., Conrad, A., Bernick, D. L., Lowe, T. M., Stolc, V., Hoeft, S., et al. (2012). ArxA, a new clade of arsenite oxidase within the DMSO reductase family of molybdenum oxidoreductases. Environ. Microbiol. 14, 1635–1645. doi: 10.1111/j.1462-2920.2012.02722.x
Zargar, K., Hoeft, S., Oremland, R., and Saltikov, C. W. (2010). Identification of a novel arsenite oxidase gene, arxA, in the haloalkaliphilic, arsenite-oxidizing bacterium Alkalilimnicola ehrlichii strain MLHE-1. J. Bacteriol. 192, 3755–3762. doi: 10.1128/JB.00244-10
Keywords: microbiome, thermal waters, detoxification processes, arsenic-related genes, arsenite, arsenate
Citation: Crognale S, Zecchin S, Amalfitano S, Fazi S, Casentini B, Corsini A, Cavalca L and Rossetti S (2017) Phylogenetic Structure and Metabolic Properties of Microbial Communities in Arsenic-Rich Waters of Geothermal Origin. Front. Microbiol. 8:2468. doi: 10.3389/fmicb.2017.02468
Received: 16 May 2017; Accepted: 28 November 2017;
Published: 12 December 2017.
Edited by:
Télesphore Sime-Ngando, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Antonio Bucci, University of Molise, ItalyChad Saltikov, University of California, Santa Cruz, United States
Copyright © 2017 Crognale, Zecchin, Amalfitano, Fazi, Casentini, Corsini, Cavalca and Rossetti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simona Rossetti, cm9zc2V0dGlAaXJzYS5jbnIuaXQ=
 Simona Crognale
Simona Crognale Sarah Zecchin2
Sarah Zecchin2 Stefano Amalfitano
Stefano Amalfitano Stefano Fazi
Stefano Fazi Anna Corsini
Anna Corsini Lucia Cavalca
Lucia Cavalca Simona Rossetti
Simona Rossetti