
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 05 December 2017
Sec. Infectious Agents and Disease
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.02411
This article is part of the Research Topic Aeromonas: a Jack of All Trades View all 26 articles
Furunculosis, a serious infection caused by the bacterium Aeromonas salmonicida subsp. salmonicida is common in sea-reared rainbow trout production in Denmark. Developing an effective control strategy requires knowledge of the epidemiology, as well as the genomic and virulent variability of the Danish A. salmonicida subsp. salmonicida isolates. To obtain this, the genomes of 101 A. salmonicida subsp. salmonicida, including 99 Danish isolates, one Scottish strain and the type strain NCIMB 1102, were sequenced using the Illumina HiSeq platform. Isolates were de novo assembled, examined for presence of plasmids, virulence and iron acquisition proteins, genomic islands, and antibiotic resistance genes. Single Nucleotide Polymorphisms were aligned and subjected to Bayesian temporal phylogenetic and maximum likelihood tree reconstruction using the published genome of A. salmonicida subsp. salmonicida A449 as reference. Bayesian temporal phylogenetic reconstruction suggests that four major introductions of A. salmonicida subsp. salmonicida into Denmark have occurred. The introductions correlate with the freshwater and subsequent seawater expansion of rainbow trout production. Initial transmission of the bacterium could have been from seawater to freshwater or vice versa, and most minor clades include a mixture of strains from different fresh- and seawater farms. Genomic variation of A. salmonicida subsp. salmonicida mostly appeared to be associated with their plasmids and plasmid encoded virulence factors. Nine A. salmonicida subsp. salmonicida isolates harbored worldwide known antibiotic resistance genes against several antibiotics and there is an indication that 33% of the isolates contained the genomic island AsaGEI1b. These findings not only support the usefulness of whole genome sequencing for genetic studies of homogeneous bacteria in general, but provide novel information about the Danish A. salmonicida subsp. salmonicida population, with implications for vaccine development in efforts to better protect Danish rainbow trout in the future.
Aeromonas salmonicida subsp. salmonicida, the causative agent of a serious infection furunculosis, was first isolated by Emmerich and Weibel (1894) at a German freshwater brown trout hatchery. Although the first rainbow trout (Oncorhynchus mykiss) hatchery in Denmark was already established in 1858 (Christensen, 1980), signs of furunculosis among fish were first described in the 1950s at freshwater rainbow trout farms (Rasmussen, 1964). At this point the Danish freshwater rainbow trout production had begun its massive expansion. In the late 1970s, production was extended to seawater and dry pellet feed was introduced instead of the common wet feed consisting of herring, whiting, sand-eels and other marine fish not used for human consumption (Christensen, 1980). Both actions increased the growth of the Danish rainbow trout production even further.
Currently, it is in the expanded Danish sea-reared rainbow trout production that A. salmonicida subsp. salmonicida is responsible for great financial losses. Despite fish being vaccinated before transfer from freshwater to seawater farms, furunculosis has occurred repeatedly during situations with elevated water temperatures (Larsen and Mellergaard, 1981; Dalsgaard and Madsen, 2000; Pedersen et al., 2008). This situation, along with previous research, has led to the belief that A. salmonicida subsp. salmonicida could be spread from freshwater to the sea by carrier fish that harbor the bacterium without showing any signs of disease (Larsen and Mellergaard, 1981; Dalsgaard and Madsen, 2000). Verifying this would be critical for developing an effective prevention strategy against furunculosis.
There are several genetic aspects of A. salmonicida subsp. salmonicida that could be used for epidemiological purposes and possibly aid in finding an effective prevention strategy against this bacterial species (e.g., Burr et al., 2002; Rasch et al., 2007; Reith et al., 2008; Dallaire-Dufresne et al., 2014; Vincent et al., 2014, 2015; Emond-Rheault et al., 2015a,b). First off, numerous potential virulence factors have been identified in A. salmonicida subsp. salmonicida, including extracellular proteases, lipases, adhesins, and functional secretion systems (Burr et al., 2002; Rasch et al., 2007; Reith et al., 2008; Dallaire-Dufresne et al., 2014). Iron acquisition has also been proven to be an important factor for virulence in almost all bacterial pathogens, including A. salmonicida subsp. salmonicida where it also seems to be linked to survival in aquatic environments (Reith et al., 2008) and to the innate immune response in the host (Ganz, 2009; Lee et al., 2014).
Certain virulence factors such as the A-layer (mainly composed of the VapA protein) in A. salmonicida subsp. salmonicida have a high antigenic conservation due to the A-layer surface structures being in contact with host defenses (Chart et al., 1984) and are encoded on the chromosome. Other virulence factors that are encoded on plasmids have shown a much greater diversity among A. salmonicida subsp. salmonicida (Nash et al., 2006; Vincent et al., 2015, 2016b). Among such belongs the type three secretion system (TTSS) encoded on the plasmid pAsa5 (Stuber et al., 2003; Reith et al., 2008), while the aopP gene that codes for a TTSS effector is present on the plasmid pAsal1 (Fehr et al., 2006). In general, the plasmid content of A. salmonicida subsp. salmonicida has shown to be variable and many new plasmids have been discovered in recent years (Boyd et al., 2003; Fehr et al., 2006; Reith et al., 2008; Vincent et al., 2014, 2016a; Attéré et al., 2015). Along with new plasmids, more antibiotic resistance genes (ARGs), some of which were previously only thought to be present in other bacterial species and ecological niches, have also been found on A. salmonicida subsp. salmonicida plasmids (R plasmids) (e.g., L'Abée-Lund and Sørum, 2001; Sørum et al., 2003; Kadlec et al., 2011; Vincent et al., 2014, 2016a).
Genomic islands (GEI) are another genetic element that has shown to vary among the A. salmonicida subsp. salmonicida population (Emond-Rheault et al., 2015a,b; Long et al., 2016). Emond-Rheault et al. (2015a) identified a new GEI named Aeromonas salmonicida genomic island (AsaGEI) in A. salmonicida subsp. salmonicida. Thus far this GEI has been found in A. salmonicida subsp. salmonicida in five different forms AsaGEI (1a, 1b, 2a, 2b, and 2c) varying in size from 50 to 53 kb, whose presence in A. salmonicida subsp. salmonicida seem to correlate with specific geographical origins of the bacterial isolates (Emond-Rheault et al., 2015a,b; Long et al., 2016).
To have an overview of the local epidemiology and evolution, as well as the variation in genomics and virulence factors of Danish A. salmonicida subsp. salmonicida isolates, a representative collection of 99 Danish A. salmonicida subsp. salmonicida isolates varying in isolation years 1980–2014 and geographical regions, a Scottish strain and the type strain NCIMB 1102 were sequenced using the Illumina HiSeq platform. Sequences of all isolates were de novo assembled and analyzed using the published genome of A. salmonicida subsp. salmonicida A449 (Reith et al., 2008) as reference.
Ninety-Nine Danish A. salmonicida subsp. salmonicida isolates from furunculosis outbreaks between 1980 and 2014 were selected. The collection consisted of 42 A. salmonicida subsp. salmonicida isolated from various freshwater farms, of which 40 were from rainbow trout and two from brown trout (Salmo trutta). Fifty-seven of the A. salmonicida subsp. salmonicida were from rainbow trout at various seawater farms, including 14 isolates (isolated between 1981 and 2014) that belonged to one large seawater farm and nine isolates (isolated between 1989 and 2010) to another large seawater farm named Sj4 and Sj3 respectively in this study. All Aeromonas salmonicida subsp. salmonicida isolates were from diseased fish. The pigment of the colonies isolated from the fish was noted and the adherent “rough” colonies were selected confirmed to belong to the subspecies salmonicida according to methods by Dalsgaard and Madsen (2000). All the isolates were stored in broth culture supplemented with 15–20% glycerol at −80°C for long-term storage (Dalsgaard and Madsen, 2000). The Scottish A. salmonicida subsp. salmonicida strain MT004 from Atlantic salmon (Salmo salar L.) is according to literature isolated around 1980. The A. salmonicida subsp. salmonicida type strain NCIMB 1102 from England was isolated from an Atlantic salmon in year 1962. Extracted genomic DNA from all 101 A. salmonicida subsp. salmonicida was used for sequencing.
When taken out from their long-term storage, all A. salmonicida subsp. salmonicida were grown in Veal Infusion Broth (VIB) (Difco) at 20°C for 48 h and then inoculated on blood agar plates (Colombia agar base (Oxoid) with 5% calf blood at 20°C for 48–72 h. The blood agar plates were checked for haemolysis (clearing zone around the colonies) as well as shape of the colonies. Genomic DNA was extracted from bacterial colonies using a QIAGEN QIAamp DNA mini kit (QIAGEN, Valencia, CA, USA) according to the manufacturer's protocol. DNA quality was determined by NanoDrop ND-1000 (Thermo Scientific, Waltham, MA, USA) and DNA concentration by Qubit 2.0 fluorometer and Quant-iT dsDNA BR kit (Invitrogen, Carlsbad, CA, USA). All DNA extractions were immediately stored at −20°C until further use.
Genomic DNA was prepared for Illumina pair-end sequencing using the Illumina (Illumina, Inc., San Diego, CA) NexteraXT® Guide 150319425031942 following the protocol revision C (http://support.illumina.com/downloads/nextera_xt_sample_preparation_guide_15031942.html). A sample of the pooled NexteraXT Libraries was loaded onto an Illumina HiSeq reagent cartridge using HiSeq Reagent Kit v2 and 500 cycles with a Standard Flow Cell. The libraries were sequenced using an Illumina platform and HiSeq Control Software 2.3.0.3. The genomic DNA of all isolates was pair-end sequenced. Raw sequence data have been submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena) under study accession no.: PRJEB15222. The raw reads were de novo assembled using the assemble pipeline (version 1.0) available from the Center for Genomic Epidemiology (CGE) https://cge.cbs.dtu.dk/services/Assembler/ which is based on the Velvet algorithms for de novo short reads assembly (Zerbino and Birney, 2008). Full genomic data can be retrieved from the Table S1.
Identification of acquired antibiotic resistance genes (ARGs) was performed through assembled genomes using the pipeline ResFinder (version 2.1) (Zankari et al., 2012) available from Center for Genomic Epidemiology (http://cge.cbs.dtu.dk/services/). Threshold for presence of an ARG in an isolate was set to 75% similarity expressed as percent sequence identity (ID) and 60% of alignment length (coverage) of resistance gene.
SNPs were determined by pipeline; CSI phylogeny (Leekitcharoenphon et al., 2012; Kaas et al., 2014) available on the CGE (www.genomicepidemiology.org). The paired-end reads were mapped to the reference chromosome of the French A. salmonicida subsp. salmonicida strain A449 isolated year 1975 from a brown trout (accession number CP000644, chromosome length 4,702,402 bp) using Burrows-Wheeler Aligner (BWA) version 0.7.2 (Li and Durbin, 2009). The “mpileup” module in SAMTools version 0.1.18 (Li et al., 2009) was used to identify SNPs. Qualified SNPs were determined when fulfilling the following criteria: (1) a minimum distance of 10 bps between each SNP, (2) a minimum of 10% of the relative depth at SNP positions, (3) the mapping quality was above 25, (4) the SNP quality was more than 30 and (5) all indels were excluded. The SNPs from each genome were concatenated to a single alignment corresponding to position of the reference genome. The concatenated sequences were subjected to maximum likelihood tree using FasTree (Price et al., 2009).
SNPs were used in Bayesian temporal phylogenetic reconstruction using BEAST (Bayesian Evolutionary Analysis Sampling Trees) version 1.7 (Drummond and Rambaut, 2007; Drummond et al., 2012) to estimate mutation rate and divergence time. Combinations of population size change and molecular clock were evaluated to identify the best-fit model (exponential clock and coalescent Bayesian skyline). The Bayesian temporal tree was constructed using the best-fit model. The BEAST MCMC chains were simulated for 300 million steps and subsampled every 10,000 steps. The final single maximum clade credibility (MCC) was examined using TreeAnnotator (Drummond et al., 2012) with 10% of the MCMC steps discarded as burn-in. Statistical confidence was represented by the 95% highest posterior density (HPD) interval.
The presence of virulence and iron acquisition proteins among all A. salmonicida subsp. salmonicida isolates was analyzed using blastp search (Altschul et al., 1990) with 78 known virulence associated and iron acquisition protein sequences (Table S2) found in the NCBI protein database against the assembled A. salmonicida subsp. salmonicida genomes. Threshold limit for presence of protein in an isolate was set to 65% ID and 60% coverage of protein.
To obtain an indication of the plasmid and GEI content of each A. salmonicida subsp. salmonicida, a blastn search (Altschul et al., 1990) was performed with 17 known A. salmonicida subsp. salmonicida plasmids and five GEI sequences respectively, found in the NCBI database (Table S3) against the assembled A. salmonicida subsp. salmonicida genomes. Threshold limit for indicated presence of plasmid in an isolate was set to 75% ID and 60% coverage of plasmid (except for plasmids pAsa6 and pAsa7) due to the long length of plasmid. For plasmids pAsa6 and pAsa7 the coverage was increased to 100% coverage of plasmid, since pAsa6 is almost identical to the larger plasmid pAsa5 (Najimi et al., 2009) and pAsa7 to pAsa2 (Vincent et al., 2015). Threshold limit for indicated presence of GEIs in an isolate was set to 75% ID and 100% coverage due to the high sequence similarity of the GEIs.
In the chromosome of all the studied A. salmonicida subsp. salmonicida isolates, including the French reference strain A449, a total of 667 SNPs were identified and a SNP matrix was generated, illustrating the SNP difference between each isolate (Table S4). On average, there was a difference of 147 SNPs between the reference strain A449 and the rest of the A. salmonicida subsp. salmonicida isolates. When comparing the average SNP difference of the Scottish isolate and the type strain NCIMB 1102 to the rest of the isolates, the average difference was 115 and 41 SNPs respectively. The two Danish A. salmonicida subsp. salmonicida (Mj2 1990 and Sd8 1992), which were the only isolates from Denmark that were isolated from brown trout instead of rainbow trout, had an average difference of 50 and 42 SNPs respectively when compared to the rest of the isolate collection. The average difference among all the 99 sequenced Danish isolates was 47 SNPs among the Danish isolates from freshwater versus 46 SNPs among the isolates from seawater. The three Danish isolates with the highest average SNP difference were Sj7 1980 with 92 SNPs, Mj12 2014 with 67 SNPs and Mj4 2008 with 61 SNPs (Table S4). Based on the alignment of the 667 SNPs, two trees were constructed: a Bayesian temporal tree (Figure 1) with a Baysian Skyline population size change and an exponential clock rate as the best fit combination model for the A. salmonicida subsp. salmonicida population and a maximum likelihood tree (Figure S1) for topology confirmation. The two trees showed similar topology and the Bayesian tree (Figure 1) was illustrated with obtained genetic information regarding acquired ARGs, virulence and iron acquisition proteins and plasmid and GEI profiles of each A. salmonicida subsp. salmonicida isolate for further analysis.
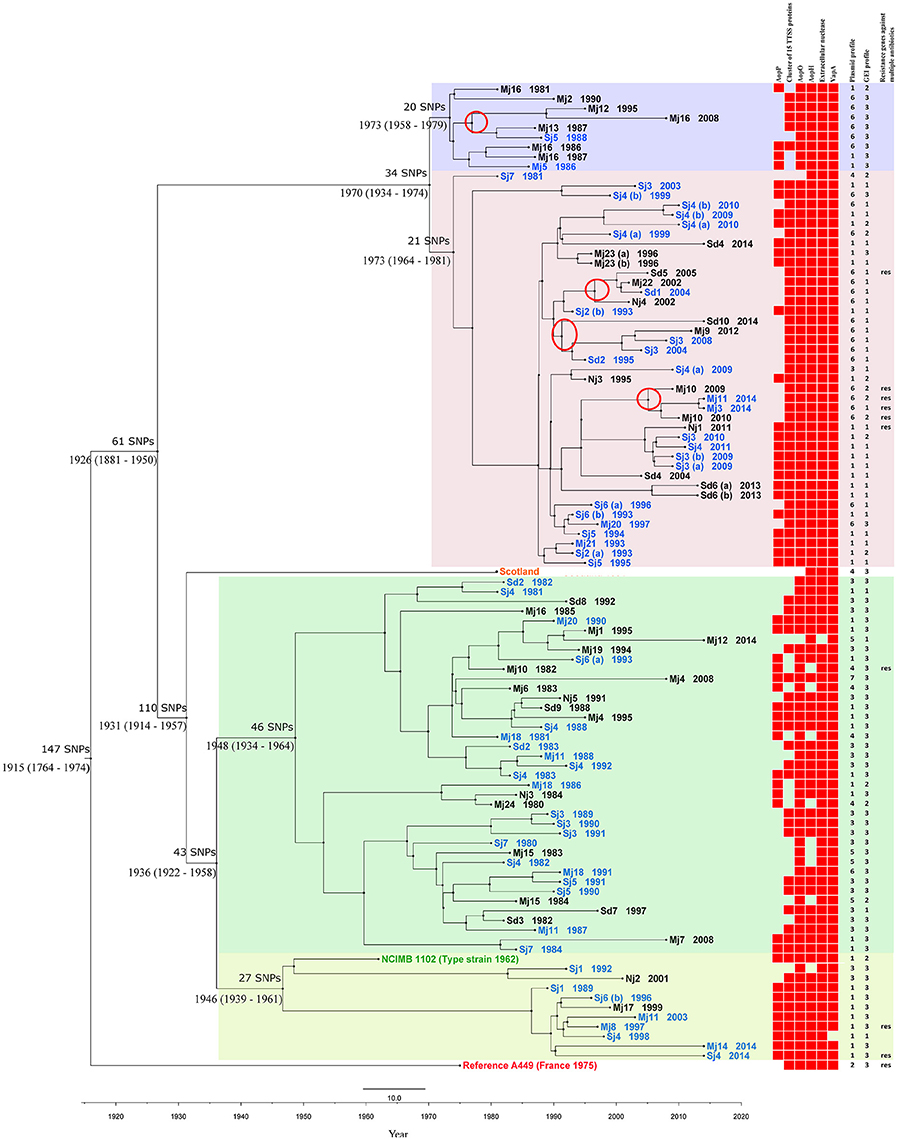
Figure 1. Phylogeny of A. salmonicida subsp. salmonicida. Bayesian temporal phylogenetic tree based on the alignment of 667 SNPs found among the 101 A. salmonicida subsp. salmonicida sequenced isolates and the reference A449. The tree shows the most recent common ancestor of the A. salmonicida subsp. salmonicida isolates dates back to ~1915 (95% HPD interval 1764–1947) and that there have been four main introductions of A. salmonicida subsp. salmonicida in Denmark: ~1973 (95% HPD interval 1958–1979), ~1973 (95% HPD interval 1964–1981), ~1948 (95% HPD interval 1934–1964) and ~1946 (95% HPD interval 1939–1961). The four main clades are each shaded with a color SNP differences between major clades are shown above the estimated year of emergence. The three non-Danish A. salmonicida subsp. salmonicida each have their own color and have the following labels: Scotland, NCIMB 1102 (type strain 1962), Reference A449 (France 1975). The Danish isolates either have a black color (freshwater farms) or a blue color (seawater farms) and they are labeled by region of origin followed by year of isolation. Following abbreviations are used for regions in Denmark: Nj, Northern Jutland; Mj, Central Jutland; Sd, Southern Denmark; Sj, Zealand. A heatmap illustration with information regarding acquired virulence and iron acquisition proteins, ARGs and plasmid and GEI profile numbers of each A. salmonicida subsp. salmonicida isolate is shown to the right of the tree. Presence and absence of protein sequences are illustrated by presence and absence of a red square, respectively. Plasmid profile number is shown and isolates that harbor ARGs against multiple antibiotics are labeled with “res.” Four minor clades marked with a red ring consist solely of isolates without plasmid pAsa3 and pAsal1.
The mutation rate of A. salmonicida subsp. salmonicida isolates was estimated to be 1.93 × 10−7 substitutions/site/year, which corresponds to 0.91 SNPs/genome/year. The most recent common ancestor of the A. salmonicida subsp. salmonicida isolates was estimated to have emerged in ~1915 (95% HPD interval 1764–1947). There are two major clades originating back to ~1926 (95% HPD interval 1881–1950) that each branched out further into two more clades in ~1936 (95% HPD interval 1922–1958) and ~1970 (95% HPD interval 1934–1974) respectively, resulting in roughly four main introductions of A. salmonicida subsp. salmonicida in Denmark: ~1973 (95% HPD interval 1958–1979), ~1973 (95% HPD interval 1964–1981), ~1948 (95% HPD interval 1934–1964), and ~1946 (95% HPD interval 1939–1961). From approximately 1975–1995 the Danish A. salmonicida subsp. salmonicida population experienced a massive clonal expansion. There was a correlation of local geographical transmission among the Danish freshwater isolates grouped together in the upper clade of the tree. There was another transmission link between isolates from a freshwater farm Mj10 and isolates from two seawater farms that had received fish from this farm.
All sequenced A. salmonicida subsp. salmonicida isolates harbored three ARGs against beta-lactam antibiotics encoded on the chromosome (Table 1). Nine Danish A. salmonicida subsp. salmonicida isolates also harbored several other plasmid encoded resistance genes against trimethoprim, sulphonamide and aminoglycoside antibiotics (Tables 1, 2). All three isolates from freshwater farm Mj10 sampled during different years harbored ARGs against several of the above-mentioned antibiotics. The same ARGs against multiple antibiotics were found in isolates sampled from three seawater farms (Mj8 1997, Mj11 2014, and Mj3 2014) located in the same bay that all received fish from the freshwater farm Mj10. The French reference strain A449 also harbored ARGs against beta-lactam, sulphonamide, aminoglycoside, phenicol and tetracycline antibiotics and more resistance genes are described by Reith et al. (2008).
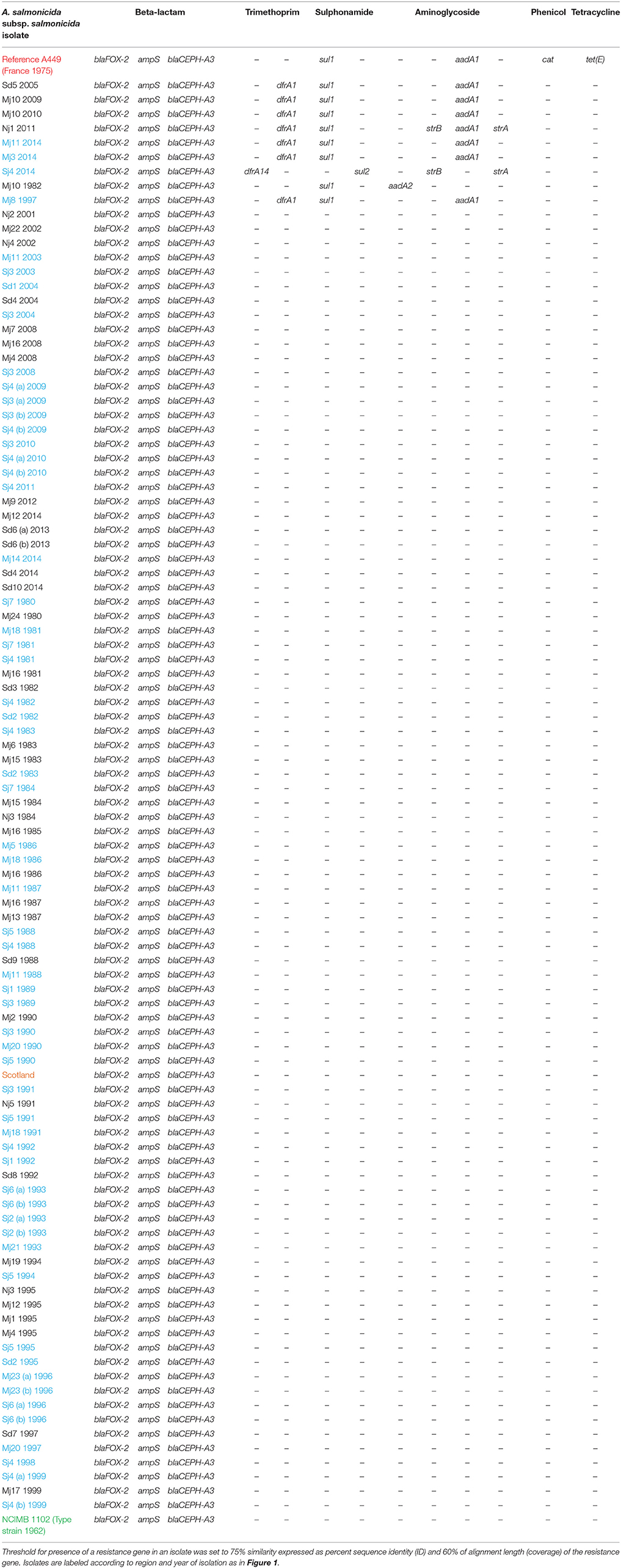
Table 1. Overview of acquired antibiotic resistance genes among the 101 A. salmonicida subsp. salmonicida sequenced isolates and the reference A449.
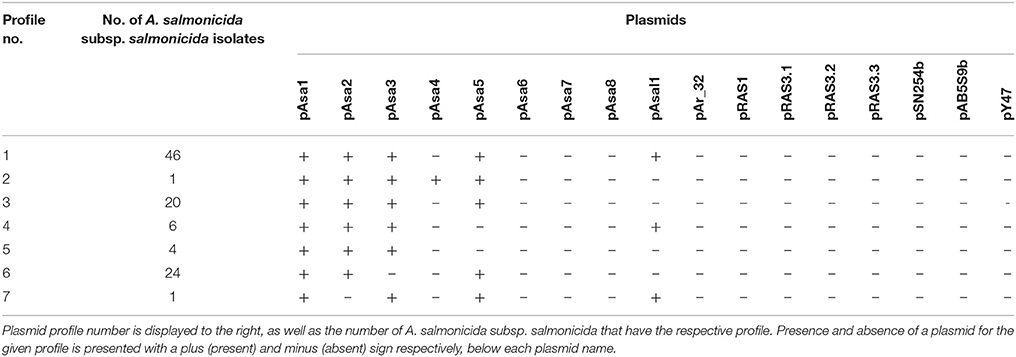
Table 2. Overview of plasmid profiles among the 101 A. salmonicida subsp. salmonicida sequenced isolates and the reference A449.
Out of 78 investigated protein sequences, 22 were considered as absent (<65% ID) in one or more isolates (Figure 2). The T3SS effector protein AopP encoded on plasmid pAsal1 by the aopP gene was absent in 50% of the A. salmonicida subsp. salmonicida isolates, including the reference strain A449. A cluster of 15 T3SS related proteins were absent in 25 isolates. In nine of the 25 isolates, the T3SS putative tyrosine phosphatase AopH and its chaperone that are encoded on pAsa5 and have homologs encoded on pAsa6 were also absent. Three of the isolates were also missing the T3SS putative serine/threonine kinase AopO and its chaperone that are encoded on pAsa5, while isolate Mj12 2014 was missing the extracellular nuclease protein (48% ID) coded by the gene nucH on the chromosome. Isolate Sj4 1998, which is not included in above mentioned group of 25 isolates, did not possess the tetragonal surface virulence array protein VapA (A-layer) encoded on the chromosome. Lastly, the chromosome encoded ABC-type ferric siderophore transporter permease protein only showed 75% ID in all sequenced A. salmonicida subsp. salmonicida as well as the reference strain A449.
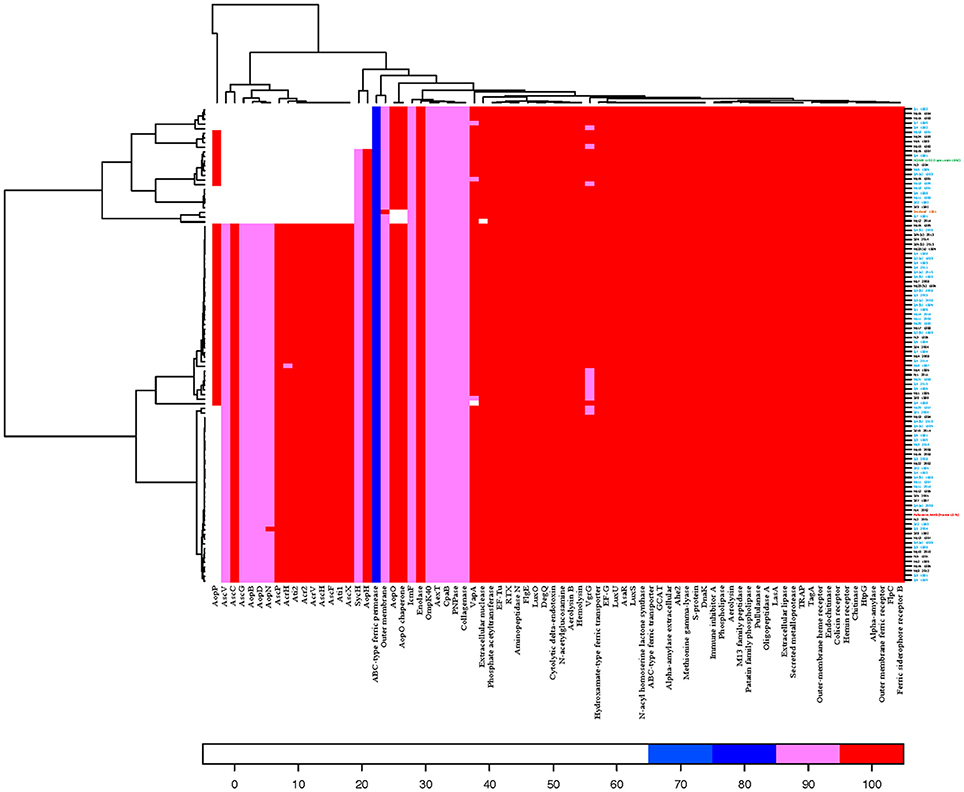
Figure 2. Heatmap illustrating presence and absence of 78 virulence associated and iron acquisition protein sequences found in the NCBI protein database among the 101 A. salmonicida subsp. salmonicida sequenced isolates and the reference A449. Isolates are displayed on the right and sequence protein names on the bottom. Threshold limit for presence of protein in an isolate was set to 65% similarity, expressed as percent sequence identity (ID) and 60% of alignment length (coverage) of the protein sequence. Red color represents > 95% ID, pink color > 85% ID, dark blue > 75% ID, and light blue > 65% ID.
Presence of multiple plasmids was indicated in all examined A. salmonicida subsp. salmonicida isolates (Table S5). Six plasmids seemed present in one or more isolates, while 11 plasmids were not found in any isolates. The only plasmid found in all isolates was pAsa1, although pAsa2 and pAsa5 showed high stability with an indicated presence of 99 and 90% respectively, among the isolates. The two plasmids pAsa3 and pAsal1 seemed present in 76 and 52% of the isolates respectively, while pAsa4 was only found in the reference strain A449. Seven different plasmid profiles were detected among the isolates, with one profile consisting of pAsa1, pAsa2, pAsa3, pAsa5, and pAsal1 representing 46% of the isolates (Table 2).
Ten plasmids are known for harboring ARGs (R plasmids): pAsa4, pAsa7, pAsa8, pAr-32, pRAS1, pRAS3.1, pRAS3.2, pRAS3.3, pSN254b, and pAB5S9b (Table S3) and only plasmid pAsa4 was found in any of the investigated A. salmonicida subsp. salmonicida.
All 51 A. salmonicida subsp. salmonicida isolates, in which the AopP protein encoded on pAsal1 was absent (Figure 2), were missing plasmid pAsal1 (Table S4). Of the 25 A. salmonicida subsp. salmonicida isolates that were missing a cluster of 15 T3SS proteins encoded on pAsa5, 10 also showed no presence of plasmid pAsa5. The remaining 15 isolates all displayed <80% coverage of the pAsa5. All nine A. salmonicida subsp. salmonicida isolates that lacked the protein AopH and its chaperone that are encoded on pAsa5 and have homologs on pAsa6 showed <80% coverage for pAsa5 and showed no indication of pAsa6 presence.
The A. salmonicida subsp. salmonicida isolates indicated a presence of three different GEI profiles (Table S6). Thirty-three percent of the isolates had profile number one, indicating presence of the GEI named AsaGEI1b. Fourteen percent of the isolates had a <80% coverage of AsaGEI1b, indicating a presence of a putative AsaGEI and were given the profile number two. The rest of the isolates had profile number three, as none of the five investigated GEIs were found in these isolates.
A. salmonicida subsp. salmonicida subspecies is known to be a highly homogeneous group that was considered clonal (Wiklund and Dalsgaard, 1998; Garcia et al., 2000; Cunningham and Colquhoun, 2002; Beaz-Hidalgo et al., 2008). The fact that only a total of 667 SNPs were found in the entire 4,702,402 bp long chromosome among the investigated A. salmonicida subsp. salmonicida varying in year of isolation (span of 34 years), geographical region, and host fish species only confirms this further. The highest average SNP difference (average of 147 SNPs) was found between the French reference strain A449 and the remaining A. salmonicida subsp. salmonicida isolates (Table S4), which is not a large difference considering that A449 was isolated in France and from a brown trout, while almost all (97 out of 99) of the Danish isolates were isolated from rainbow trout. When comparing the average SNP difference between the Scottish strain from Atlantic salmon and the rest of the isolates, the results were even lower with an average of 115 SNPs (Table S4). The two Danish A. salmonicida subsp. salmonicida isolated from brown trout (Mj2 1990 and Sd8 1992) and the type strain NCIMB 1102 from Atlantic salmon also grouped together with Danish A. salmonicida subsp. salmonicida from rainbow trout in one of the four major clades (Figure 1) and only have an average difference of 41, 50, and 52 SNPs, which challenges the theory of A. salmonicida subsp. salmonicida genome adapting to the environment of their specific hosts species (Reith et al., 2008). More A. salmonicida subsp. salmonicida isolates from various fish species would, however, need to be sequenced to shed more light on this theory.
Noticeably there appears to be four major A. salmonicida subsp. salmonicida introductions to Denmark, giving rise to four major clades (Figure 1). The two introductions that occurred in ~1973 (95% HPD interval 1958–1979) and ~1973 (95% HPD interval 1964–1981) and gave rise to the two upper clades (Figure 1), seemingly took place right before the massive clonal expansion during 1975–1995, which all four clades underwent. The two introduction points in 1973 and the expansion period of all four clades correspond to the time period where rainbow trout farming in Denmark began expanding out to seawater and intensifying their production (Christensen, 1980). The two bottom clades were introduced further in the past ~1948 (95% HPD interval 1934–1964) and ~1946 (95% HPD interval 1939–1961) respectively and contain A. salmonicida subsp. salmonicida with older isolation years (average year of isolation 1991), than the two upper clades that include A. salmonicida subsp. salmonicida with an average isolation year of 2001 (Figure 1).
The introduction of the two bottom clades ~1948 (95% HPD interval 1934–1964) and ~1946 (95% HPD interval 1939–1961) respectively, correlate with the end of the Second World War and the beginning of an expansion of rainbow trout production in Danish freshwater (Christensen, 1980). When examining the branches of each of the four major clades, there is a possibility that A. salmonicida subsp. salmonicida might have been introduced into freshwater through marine fish wet feed used until the late 1970s (Christensen, 1980). However, the bacterium could also have been transmitted from freshwater to seawater, which is the widespread theory i.e., that A. salmonicida subsp. salmonicida is present in freshwater fish showing no signs of diseases (carriers) and are then transferred out to seawater with the fish, where outbreaks occur during high temperatures (Larsen and Mellergaard, 1981; Dalsgaard and Madsen, 2000; Pedersen et al., 2008).
The local transmission pattern of A. salmonicida subsp. salmonicida among the Danish farms also suggests that transmission of isolates from freshwater to seawater farms have occurred, as exemplified by a minor clade where ARGs were transmitted from a freshwater farm to seawater farms. Isolates from different freshwater farms are moreover mixed with different isolates from seawater in most of the minor clades. Though, in general it is hard to find specific geographical correlations between the fish farms. One of the main causes for this could be the widespread trade of fingerlings for anglers in Denmark throughout the years as well as local trade among fish farmers. There is nonetheless a correlation among the group of freshwater farms isolates in the top clade. Mj13 is located upstream to Mj16 in a stream that runs out into a river named Guden Å. Two other farms (Mj12 and Mj2) are also located at streams that lead out to Guden Å and one of these (Mj12) produces brown trout.
Interestingly, all investigated A. salmonicida subsp. salmonicida isolates possessed three beta-lactam ARGs (Table 1). Since the genes are encoded on the chromosome, it seems that either they have always been a part of the A. salmonicida subsp. salmonicida genome repertoire, or they must have been acquired at least around 67–250 years ago where they all have a most recent common ancestor (Figure 1). Nine Danish A. salmonicida subsp. salmonicida also harbored resistance genes against trimethoprim, sulphonamide and aminoglycoside antibiotics (Table 1), which are all plasmid encoded. Trimethoprim and sulphonamide are also two of the scarce number of antibiotics allowed to be used for treatment of bacterial diseases in Danish fish farms (Dalsgaard and Madsen, 2000). All three A. salmonicida subsp. salmonicida isolates from freshwater farm Mj10, isolated year 1982, 2009, and 2010 harbored resistance genes against at least two of the above-mentioned antibiotics (Table 1). From farm Mj10, fish have always been transferred out to a bay, where several seawater farms are located. In A. salmonicida subsp. salmonicida isolated during 2014 from two of the seawater farms in the bay (Figure 1), the same set of resistance genes were detected as those seen in A. salmonicida subsp. salmonicida from the freshwater farm Mj10 during 2009 and 2010 (Table 1). Fittingly, the two freshwater isolates form a minor clade with the two seawater isolates in the Bayesian temporal tree, according to which the isolates spread from the freshwater to seawater (Figure 1). In seawater farm Mj8, which is also located in the bay, an A. salmonicida subsp. salmonicida isolate from 1997 did harbor ARGs against the mentioned antibiotics as well. However, these ARGs were slightly different than those seen in the Mj10 1982 isolate, where one otherwise would expect the resistance genes had originated (Table 1). This could be associated with the fact that trimethoprim was not licensed for use in Denmark until 1983 (Dalsgaard and Madsen, 2000) and the isolate from 1982 had therefore not acquired ARGs against this antibiotic.
All nine of the above-mentioned isolates showed coverage (< 60%) of at least one of the R plasmids (Table S5), indicating they could have acquired ARGs from the plasmids in the past through horizontal gene transfer and then lost the plasmid. Isolate Sj4 2014 and Nj1 2011 were the only two isolates harboring ARGs strA and strB. Both ARGs are known to be present on the R plasmids pAB5S9b and pSN254b, but none of the investigated isolates in this study showed any coverage of these two plasmids. It is thus most likely that the isolates Sj4 2014 and Nj1 2011 carry the ARGs on new plasmids.
When looking at the phylogeny, it was found that isolates grouped in the two bottom clades were missing a higher amount of virulence associated proteins (average of 1.0 protein per isolate), compared to isolates grouped in the two upper clades where the average absence of virulence associated proteins was 0.6 per isolate (Figure 1). This indicates that the most recent common ancestor of the bottom two clades from ~1936 (95% HPD interval 1922–1958), presumably harbored lower amount of virulence associated proteins and thus gave rise to two older lineages with fewer virulence associated proteins than the more recently introduced two lineages (the two upper clades in the phylogeny tree). Considering these results, it could be suggested that the upper two clades might consist of more virulent A. salmonicida subsp. salmonicida that were introduced from a more recent and more virulent ancestor around 1970.
Due to the important nature of iron acquisition for A. salmonicida subsp. salmonicida (Reith et al., 2008; Ganz, 2009; Lee et al., 2014) and the fact that all the investigated proteins related to iron acquisition were encoded on the chromosome, it was expected that all 101 sequenced A. salmonicida subsp. salmonicida isolates and the A449 reference genome possessed all the investigated iron acquisition proteins (Table S2). Though, it must be noted that the ABC-type ferric siderophore transporter permease protein only showed 75% ID in all 101 A. salmonicida subsp. salmonicida isolates as well as A449.
Most of potential virulence proteins investigated in this study were present in all sequenced A. salmonicida subsp. salmonicida and the reference strain. Though, two proteins encoded on the chromosome and 20 encoded on plasmids were missing in at least one isolate (Table S2). All the 20 plasmid encoded proteins were related to the functional type three secretion system (T3SS). This secretion system is wide spread among Gram-negative bacteria and has several functions, including: disrupting host cells by translocating toxins (effector proteins) into their cytoplasm, preventing phagocytosis by leukocytes, and establishing systemic infection (Burr et al., 2003, 2005; Stuber et al., 2003; Dacanay et al., 2006; Rasch et al., 2007; Dallaire-Dufresne et al., 2014). T3SS is also the only factor proven to be essential for virulence of A. salmonicida subsp. salmonicida, as all in vitro and in vivo studies involving inactivation of T3SS structural proteins in A. salmonicida subsp. salmonicida strains have resulted in non-virulent A. salmonicida subsp. salmonicida mutants (Burr et al., 2002, 2003, 2005; Stuber et al., 2003; Dacanay et al., 2006; Froquet et al., 2007). Nevertheless, among the 20 missing T3SS related proteins in this study were T3SS structural proteins, while all the A. salmonicida subsp. salmonicida in this study are isolated from furunculosis outbreaks, whereby one would assume that all the A. salmonicida subsp. salmonicida isolates are virulent.
There are 36 T3SS encoding genes located on the large plasmid pAsa5 (Reith et al., 2008; Najimi et al., 2009; Tanaka et al., 2012; Vincent et al., 2016b) and 19 of them that were investigated in this study were missing in at least three isolates (Table S2). Though, it is known that pAsa5 becomes unstable under stressful conditions like being subjected to growth in temperature above 22–25°C (Stuber et al., 2003; Tanaka et al., 2012; Dallaire-Dufresne et al., 2014) and is thought to undergo genetic rearrangement resulting in the loss of its T3SS region caused by activation of ISAS11 insertion sequence (IS) elements (Tanaka et al., 2012). This could explain the fact that all the 25 A. salmonicida subsp. salmonicida isolates missing the cluster of T3SS proteins, encoded on pAsa5 in our study still harbored the plasmid, but displayed <80% coverage. The only issue with this justification is that all A. salmonicida subsp. salmonicida cultures in our laboratory are always grown at 20°C, meaning it is unlikely that growth at high temperature triggered the activation of ISAS11. It is unclear what other cause for the rearrangement of pAsa5 could have been.
Unlike most of the T3SS proteins, the AopP protein is encoded on plasmid pAsal1 and the protein was missing in 50% of the A. salmonicida subsp. salmonicida isolates investigated in this study (Figure 2). Interestingly, pAsal1 was present in isolate Sj 1981 and the Scottish isolate, both of which did not harbor the AopP protein. When analyzed using the program BioEdit (Hall, 1999), the isolates did possess the nucleotide sequence for the aopP gene; however, both sequences had an identical frameshift mutation caused by point deletions (data not showed) that presumably lead to an incorrect translation of the AopP protein sequence. Apart from the two mentioned isolates, all A. salmonicida subsp. salmonicida that were missing the AopP protein sequence were also missing the plasmid pAsal1. Previous studies have shown that pAsal1 is lost due to activation of the same insertion sequence as in pAsa5 (Daher et al., 2011; Tanaka et al., 2012; Attéré et al., 2015; Vincent et al., 2016a), though the plasticity of A. salmonicida subsp. salmonicida pAsal1 is complex, as the precise mechanism responsible for loss of pAsal1 remains unknown (Attéré et al., 2015) and at least three larger variants of the plasmid exist: pAsal1B, pAsal1C, and pAsal1D (Trudel et al., 2013; Attéré et al., 2015).
As illustrated on Figure 2, there was otherwise an overall high similarity among all isolates regarding all the chromosome encoded virulence associated protein sequences. The high prevalence and similarity of the A-layer (VapA) in A. salmonicida subsp. salmonicida discovered in this study, along with previous findings of A. salmonicida subsp. salmonicida surface structures being highly conserved (Chart et al., 1984), could also provide valuable knowledge for future vaccine development.
Out of the seven plasmid profiles found in this study, the most abundant profile consisting of pAsa1, pAsa2, pAsa3, pAsa5, and pAsal1 represented 46% of the A. salmonicida isolates (Table 2). In congruence, Nielsen et al. (1993) investigated A. salmonicida subsp. salmonicida from various geographical locations using DNA restriction fragment plasmid profiling and found the same plasmid profile to be the most common one among Danish A. salmonicida subsp. salmonicida isolates, representing 32% of the 57 investigated Danish strains in that study.
Present findings support previous results by Nielsen et al. (1993), Boyd et al. (2003), and Attéré et al. (2015) regarding high stability of plasmid pAsa1 and pAsa2 and instability of plasmid pAsa3 and pAsal1. Attéré et al. (2015) suggested an explanation for the stability of pAsa1 and instability of pAsal1 could be that pAsa1 and pAsa3 encode genes for a type II toxin-antitoxin (TA) system that kills all daughter cells that do not receive the plasmids (Boyd et al., 2003), while the TA system has not been found in plasmids pAsa2 and pAsal1 (Shao et al., 2011). Nevertheless, there is still the issue regarding stability of pAsa2 that does not encode a TA system and the instability of pAsa3 that does have a TA system (Attéré et al., 2015); an issue accentuated by the present findings. It might be possible that some clonal lineages do not acquire pAsa3 and pAsal1 (Attéré et al., 2015), which cannot be ruled out according to our results, where out of 24 A. salmonicida subsp. salmonicida that did not harbor pAsa3 and pAsal1 (plasmid profiles 15–19), 17 clustered together in four minor clades (Figure 1, four minor clades with a red ring).
Finally, it is important to stress that the present results regarding plasmids should only be taken as indications, due to the limitation of the blastn (Altschul et al., 1990) analysis that was used for creating an overview of the plasmid content. To confirm the results, additional experiments would need to be performed, such as PCR with designed primer pairs, each specific to one single plasmid.
Thus far, the majority of A. salmonicida subsp. salmonicida isolates from Europe have been shown to harbor either AsaGEI1b or no GEI (Emond-Rheault et al., 2015a). In this study we not only found an indication of the presence of AsaGEI1b, but also presence of a putative GEI (Table S6). Thus far in other studies, the only isolate from Europe harboring a different GEI was isolate JF3791 from Switzerland, which harbored the variant AsaGEI2b (Emond-Rheault et al., 2015b). Both AsaGEI1a and AsaGEI2a were restricted to North American isolates (Emond-Rheault et al., 2015a), while AsaGEI2c has solely been found in China (Long et al., 2016). Although it is vital to analyze more A. salmonicida subsp. salmonicida before making any conclusions, the overall general notion, is that the GEIs seem to be associated with specific geographical regions over the world and could as such possibly be used as indicators for the geographical origin of an A. salmonicida subsp. salmonicida isolate; a tool that could prove to be valuable in tracking isolates that initiate furunculosis outbreaks around the world (Emond-Rheault et al., 2015a,b; Long et al., 2016). Unfortunately, it seems that the usefulness of GEIs as indicators of the geographical origin of an isolate is limited to continents rather than countries and local regions, as there was no local geographical correlation between those isolates that seemingly harbored AsaGEI1b, the putative GEI and those who did not harbor any GEI in our results (Table S6). Though again it is important to note that the present results regarding GEIs, as with plasmids, should only be taken as indications, due to the limitation of the blastn (Altschul et al., 1990) analysis.
Interestingly 29 out of the total 33 A. salmonicida subsp. salmonicida isolates in our study that had an indication of the presence of AsaGEI1b, were grouped together in the second upper clade (Figure 1). Nine out of the fourteen A. salmonicida subsp. salmonicida that showed an indication of having a putative GEI (Table S6), were also grouped in the second upper clade. It would thus seem that AsaGEI1b or another putative GEI could have been acquired by the ancestor of the clade ~1973 (95% HPD interval 1964–1981) through a horizontal gene transfer and then passed on throughout this clade in the large clonal expansion that took place during 1975–1995 (Figure 1). Even though the GEIs contribution to the host phenotype is unknown (Emond-Rheault et al., 2015a), one could speculate that AsaGEI1b and the putative GEI provide some biological advantage to the isolates since only three out of 41 descendants in the clade (Figure 1) did not seem to harbor a GEI. This is supported by the general idea that GEIs play a crucial role in disseminating genes among bacteria that can aid in their survival and possibly increase virulence (Juhas et al., 2008).
The present findings have provided novel insight into the epidemiology of the disease furunculosis revealing four main introductions in consistency with the historical expansion of the Danish aquaculture production that could have been transmitted either from freshwater to seawater or vice versa. There was also transmission of isolates harboring ARGs from a freshwater farm to seawater farms, supporting the theory of A. salmonicida subsp. salmonicida being spread from freshwater to seawater via carrier fish. The mixture of freshwater and seawater isolates from different farms in almost every minor clade and the lack of geographical connections among farms also indicates that the widespread trade of fingerlings and other fish could have played a role in the local dissemination of A. salmonicida subsp. salmonicida in Denmark. The genome based analysis moreover showed genetic and virulence variability among the highly homogenous A. salmonicida subsp. salmonicida population in Denmark, which consisted of isolates with varying plasmid and GEI profiles and plasmid encoded virulence proteins, especially those related to T3SS. In future studies it would be very interesting to include sequences of other salmonicida subspecies as well as other subspecies to get an idea of the broader phylogenetic distribution of A. salmonicida subsp. salmonicida and we would recommend doing a deeper analysis of plasmids and GEIs, as the purpose in this study was to create an overview of the plasmid and GEI content among the isolates and the present findings can thus only be used as indications. Overall, whole genome sequencing proved to be a highly useful tool for investigating Danish A. salmonicida subsp. salmonicida and presented important new information about this bacterium that could be relevant for possible future vaccine development.
Critically revised the paper, conceived and designed the experiments: ID and FA. Analyzed the data and contributed reagents/materials/analysis tools: SB, PL, ID, and FA. Performed field sampling: ID. Performed the experiments: SB and ID. Wrote the paper: SB and PL.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by The Danish Council for Strategic Research under ProFish project (Grant no. DSF: 11-116252) and National Veterinary Institute (DTU). The authors would like to thank all ProFish project partners. The authors are also grateful to Lisbeth Schade Hansen at the National Veterinary Institute (DTU) and Inge Marianne Hansen at the National Food Institute (DTU) for their technical support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02411/full#supplementary-material
Figure S1. Phylogeny of A. salmonicida subsp. salmonicida. Maximum likelihood tree based on the alignment of 667 SNPs found among the 101 A. salmonicida subsp. salmonicida sequenced isolates and the reference A449. Isolates are labeled as stated in Figure 1.
Table S1. Full genomic data of the 101 sequenced A. salmonicida subsp. salmonicida isolates. Isolates are labeled as stated in Figure 1.
Table S2. Overview of 78 virulence associated and iron acquisition protein sequences found in the NCBI protein database. The following is shown in the table: name of protein sequence, short description, location of the protein coding gene, Genbank accession number and the number of A. salmonicida isolates harboring the given protein sequence. Isolates are labeled as stated in Figure 1.
Table S3. Overview of 17 A. salmonicida subsp. salmonicida plasmids and five genomic islands (GEI) found in the NCBI nucleotide database. The following is shown in the table: name of plasmid or GEI, short description, antibiotic resistance genes encoded on plasmid, length of plasmid or GEI (in base pairs), and Genbank accession number. Isolates are labeled as stated in Figure 1.
Table S4. Single Nucleotide Polymorphism (SNP) matrix of the 101 sequenced A. salmonicida subsp. salmonicida isolates and the reference A449. The matrix shows the SNP difference between each A. salmonicida subsp. salmonicida isolate, with 2 being the lowest and 215 being highest SNP difference among the isolates.
Table S5. Plasmid content of the 101 A. salmonicida subsp. salmonicida sequenced isolates and the reference A449. Numbers under each plasmid name represent percent coverage of that plasmid (in base pairs) for a given isolate. Except for plasmid pAsa6 and pAsa7, if percent coverage is higher than 60%, the plasmid is said to be indicated as present in the isolate and the color of the cell is green. Threshold limit for presence of plasmid in an isolate was set to 75% similarity expressed as percent sequence identity (ID) and 60% of alignment length (coverage) of the plasmid, except for plasmid pAsa6 and pAsa7 due to the high level of identity shared between pAsa5 and pAsa6 and between pAsa2 and pAsa7 respectively. Coverage was set to 100% for plasmids pAsa6 and pAsa7 in order for these plasmids being considered as present in an isolate. Following abbreviations are used for percent coverage: 100, 100% coverage; <100, from 80% up to 99% coverage; <80, from 60% up to 79% coverage; <60, from 10 up to 59% coverage; <10, from 1% up to 9% coverage; 0, 0% coverage. Isolates are labeled as stated in Figure 1.
Table S6. Genomic island (GEI) content of the 101 A. salmonicida subsp. salmonicida sequenced isolates and the reference A449. The GEI content is represented as stated in Table S5 for plasmids pAsa6 and pAsa7. Isolates are labeled as stated in Figure 1.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Attéré, S. A., Vincent, A. T., Trudel, M., Chanut, R., and Charette, S. J. (2015). Diversity and homogeneity among small plasmids of Aeromonas salmonicida subsp. salmonicida linked with geographical origin. Front. Microbiol. 6:1274. doi: 10.3389/fmicb.2015.01274
Beaz-Hidalgo, R., López-Romalde, S., Toranzo, A. E., and Romalde, J. L. (2008). Polymerase chain reaction amplification of repetitive intergenic consensus and repetitive extragenic palindromic sequences for molecular typing of Pseudomonas anguilliseptica and Aeromonas salmonicida. J. Aquat. Anim. Health 20, 75–85. doi: 10.1577/H07-007.1
Boyd, J., Williams, J., Curtis, B., Kozera, C., Singh, R., and Reith, M. (2003). Three small, cryptic plasmids from Aeromonas salmonicida subsp. salmonicida A449. Plasmid 50, 131–144. doi: 10.1016/S0147-619X(03)00058-1
Burr, S. E., Pugovkin, D., Wahli, T., Segner, H., and Frey, J. (2005). Attenuated virulence of an Aeromonas salmonicida subsp. salmonicida type III secretion mutant in a rainbow trout model. Microbiology 151, 2111–2118. doi: 10.1099/mic.0.27926-0
Burr, S. E., Stuber, K., Wahli, T., and Frey, J. (2002). Evidence for a Type III secretion system in Aeromonas salmonicida subsp. salmonicida. J. Bacteriol. 184, 5966–5970. doi: 10.1128/JB.184.21.5966-5970.2002
Burr, S. E., Wahli, T., Segner, H., Pugovkin, D., and Frey, J. (2003). Association of Type III secretion genes with virulence of Aeromonas salmonicida subsp. salmonicida. Dis. Aquat. Organ. 57, 167–171. doi: 10.3354/dao057167
Chart, H., Shaw, D. H., Ishiguro, E. E., and Trust, T. J. (1984). Structural and immunochemical homogeneity of Aeromonas salmonicida lipopolysaccharide. J. Bacteriol. 158, 16–22.
Christensen, N. O. (1980). Fiskesygdomme (Fish diseases) (in Danish). Copenhagen: Den Kgl. Veterinær- og Landbohøjskole.
Cunningham, C. O., and Colquhoun, D. J. (2002). “Molecular diagnosis of Aeromonas salmonicida infections,” in Molecular Diagnosis of Salmonid Diseases, ed C. D. Cunningham (Dordrecht: Kluwer Academic Publishers), 81–99.
Dacanay, A., Knickle, L., Solanky, K. S., Boyd, J. M., Walter, J. A., Brown, L. L., et al. (2006). Contribution of the type III secretion system (TTSS) to virulence of Aeromonas salmonicida subsp. salmonicida. Microbiology 152, 1847–1856. doi: 10.1099/mic.0.28768-0
Daher, R. K., Filion, G., Tan, S. G. E., Dallaire-Dufresne, S., Paquet, V. E., and Charette, S. J. (2011). Alteration of virulence factors and rearrangement of pAsa5 plasmid caused by the growth of Aeromonas salmonicida in stressful conditions. Vet. Microbiol. 152, 353–360. doi: 10.1016/j.vetmic.2011.04.034
Dallaire-Dufresne, S., Tanaka, K. H., Trudel, M. V., Lafaille, A., and Charette, S. J. (2014). Virulence, genomic features, and plasticity of Aeromonas salmonicida subsp. salmonicida, the causative agent of fish furunculosis. Vet. Microbiol. 169, 1–7. doi: 10.1016/j.vetmic.2013.06.025
Dalsgaard, I., and Madsen, L. (2000). Bacterial pathogens in rainbow trout, Oncorhynchus mykiss (Walbaum), reared at Danish freshwater farms. J. Fish Dis. 23, 199–209. doi: 10.1046/j.1365-2761.2000.00242.x
Drummond, A. J., and Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. doi: 10.1186/1471-2148-7-214
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Emmerich, R., and Weibel, E. (1894). Über eine durch bakterien erzeugte seuche unter den forellen (About a disease caused by bacteria among trout) (in German). Arch. für Hyg. 21, 1–21.
Emond-Rheault, J. G., Vincent, A. T., Trudel, M. V., Brochu, F., Boyle, B., Tanaka, K. H., et al. (2015a). Variants of a genomic island in Aeromonas salmonicida subsp. salmonicida link isolates with their geographical origins. Vet. Microbiol. 175, 68–76. doi: 10.1016/j.vetmic.2014.11.014
Emond-Rheault, J. G., Vincent, A. T., Trudel, M. V., Frey, J., Frenette, M., and Charette, S. J. (2015b). AsaGEI2b: a new variant of a genomic island identified in the Aeromonas salmonicida subsp. salmonicida JF3224 strain isolated from a wild fish in Switzerland. FEMS Microbiol. Lett. 362:fnv093. doi: 10.1093/femsle/fnv093
Fehr, D., Casanova, C., Liverman, A., Blazkova, H., Orth, K., Dobbelaere, D., et al. (2006). AopP, a type III effector protein of Aeromonas salmonicida, inhibits the NF-kappaB signalling pathway. Microbiology 152, 2809–2818. doi: 10.1099/mic.0.28889-0
Froquet, R., Cherix, N., Burr, S. E., Frey, J., Vilches, S., Tomas, J. M., et al. (2007). Alternative host model to evaluate Aeromonas virulence. Appl. Environ. Microbiol. 73, 5657–5659. doi: 10.1128/AEM.00908-07
Ganz, T. (2009). Iron in innate immunity: starve the invaders. Curr. Opin. Immunol. 21, 63–67. doi: 10.1016/j.coi.2009.01.011
Garcia, J. A., Larsen, J. L., Dalsgaard, I., and Pedersen, K. (2000). Pulsed-field gel electrophoresis analysis of Aeromonas salmonicida ssp salmonicida. FEMS Microbiol. Lett. 190, 163–166. doi: 10.1016/S0378-1097(00)00322-0
Hall, T. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Juhas, M., van der Meer, J. R., Gaillard, M., Harding, R. M., Hood, D. W., and Crook, D. W. (2008). Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33, 376–393. doi: 10.1111/j.1574-6976.2008.00136.x
Kaas, R. S., Leekitcharoenphon, P., Aarestrup, F. M., and Lund, O. (2014). Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE 9:e104984. doi: 10.1371/journal.pone.0104984
Kadlec, K., von Czapiewski, E., Kaspar, H., Wallmann, J., Michael, G. B., Steinacker, U., et al. (2011). Molecular basis of sulfonamide and trimethoprim resistance in fish-pathogenic aeromonas isolates. Appl. Environ. Microbiol. 77, 7147–7150. doi: 10.1128/AEM.00560-11
L'Abée-Lund, T. M., and Sørum, H. (2001). Class 1 integrons mediate antibiotic resistance in the fish pathogen Aeromonas salmonicida worldwide. Microb. Drug Resist. 7, 263–272. doi: 10.1089/10766290152652819
Larsen, J. L., and Mellergaard, S. (1981). Microbiological and hygienic problems in marine aquaculture: furunculosis and vibriosis in rainbow trout (Salmo gairdneri). Bull. Eur. Ass. Fish Pathol. 1, 29–32.
Lee, J.-H., Pooley, N. J., Mohd-Adnan, A., and Martin, S. A. (2014). Cloning and characterisation of multiple ferritin isoforms in the Atlantic salmon (Salmo salar). PLoS ONE 9:e103729. doi: 10.1371/journal.pone.0103729
Leekitcharoenphon, P., Kaas, R. S., Thomsen, M., Friis, C., Rasmussen, S., and Aarestrup, F. M. (2012). snpTree - a web-server to identify and construct SNP trees from whole genome sequence data. BMC Genomics 13:S6. doi: 10.1186/1471-2164-13-S7-S6
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Long, M., Nielsen, T. K., Leisner, J. J., Hansen, L. H., Shen, Z. X., Zhang, Q. Q., et al. (2016). Aeromonas salmonicida subsp. salmonicida strains isolated from Chinese freshwater fish contain a novel genomic island and possible regional-specific mobile genetic elements profiles. FEMS Microbiol. Lett. 363:fnw190. doi: 10.1093/femsle/fnw190
Najimi, M., Balado, M., Lemos, M. L., and Osorio, C. R. (2009). Genetic characterization of pAsa6, a new plasmid from Aeromonas salmonicida subsp. salmonicida that encodes a type III effector protein AopH homolog. Plasmid 61, 176–181. doi: 10.1016/j.plasmid.2009.01.001
Nash, J. H., Findlay, W. A., Luebbert, C. C., Mykytczuk, O. L., Foote, S. J., Taboada, E. N., et al. (2006). Comparative genomics profiling of clinical isolates of Aeromonas salmonicida using DNA microarrays. BMC Genomics 7:43. doi: 10.1186/1471-2164-7-43
Nielsen, B., Olsen, J. E., and Larsen, J. L. (1993). Plasmid profiling as an epidemiological marker within Aeromonas salmonicida. Dis. Aquat. Organ. 15, 129–135. doi: 10.3354/dao015129
Pedersen, K., Skall, H. F., Lassen-Nielsen, A. M., Nielsen, T. F., Henriksen, N. H., and Olesen, N. J. (2008). Surveillance of health status on eight marine rainbow trout, Oncorhynchus mykiss (Walbaum), farms in Denmark in 2006. J. Fish Dis. 31, 659–667. doi: 10.1111/j.1365-2761.2008.00941.x
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Rasch, M., Kastbjerg, V. G., Bruhn, J. B., Dalsgaard, I., Givskov, M., and Gram, L. (2007). Quorum sensing signals are produced by Aeromonas salmonicida and quorum sensing inhibitors can reduce production of a potential virulence factor. Dis. Aquat. Organ. 78, 105–113. doi: 10.3354/dao01865
Rasmussen, C. J. (1964). Furunkulose hos damørreder (Furunculosis in farmed trout) (in Danish). Scand. J. Vet. Sci. 16, 463–472.
Reith, M. E., Singh, R. K., Curtis, B., Boyd, J. M., Bouevitch, A., Kimball, J., et al. (2008). The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics 9:427. doi: 10.1186/1471-2164-9-427
Shao, Y., Harrison, E. M., Bi, D., Tai, C., He, X., Ou, H. Y., et al. (2011). TADB: A web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 39, D606–D611. doi: 10.1093/nar/gkq908
Sørum, H., L'Abée-Lund, T. M., Solberg, A., and Wold, A. (2003). Integron-containing IncU R plasmids pRAS1 and pAr-32 from the fish pathogen Aeromonas salmonicida. Antimicrob. Agents Chemother. 47, 1285–1290. doi: 10.1128/AAC.47.4.1285-1290.2003
Stuber, K., Burr, S. E., Braun, M., Wahli, T., and Frey, J. (2003). Type III secretion genes in Aeromonas salmonicida subsp. salmonicida are located on a large thermolabile virulence plasmid. J. Clin. Microbiol. 41, 3854–3856. doi: 10.1128/JCM.41.8.3854-3856.2003
Tanaka, K. H., Dallaire-Dufresne, S., Daher, R. K., Frenette, M., and Charette, S. J. (2012). An insertion sequence-dependent plasmid rearrangement in Aeromonas salmonicida causes the loss of the type three secretion system. PLoS ONE 7:e33725. doi: 10.1371/journal.pone.0033725
Trudel, M. V., Tanaka, K. H., and Geneviève, F. (2013). Insertion sequence AS5 (ISAS5) is involved in the genomic plasticity of Aeromonas salmonicida. Mob. Genet. Elem. 3:e25640. doi: 10.4161/mge.25640
Vincent, A. T., Emond-Rheault, J.-G., Barbeau, X., Attéré, S. A., Frenette, M., Lagüe, P., et al. (2016a). Antibiotic resistance due to an unusual ColE1-type replicon plasmid in Aeromonas salmonicida. Microbiol. 162, 942–953. doi: 10.1099/mic.0.000286
Vincent, A. T., Tanaka, K. H., Trudel, M. V., Frenette, M., Derome, N., and Charette, S. J. (2015). Draft genome sequences of two Aeromonas salmonicida subsp. salmonicida isolates harboring plasmids conferring antibiotic resistance. FEMS Microbiol. Lett. 362:fnv002. doi: 10.1093/femsle/fnv002
Vincent, A. T., Trudel, M. V., Freschi, L., Nagar, V., Gagné-Thivierge, C., Levesque, R. C., et al. (2016b). Increasing genomic diversity and evidence of constrained lifestyle evolution due to insertion sequences in Aeromonas salmonicida. BMC Genomics 17, 44. doi: 10.1186/s12864-016-2381-3
Vincent, A. T., Trudel M. V., Paquet V. E., Boyle, B., Tanaka, K. H., Dallaire-Dufresne, S., et al. (2014). Detection of variants of the pRAS3, pAB5S9, and pSN254 plasmids in Aeromonas salmonicida subsp. salmonicida: multidrug-resistance, interspecies exchanges, and plasmid reshaping. Antimicrob. Agents Chemother. 58, 7367–7374. doi: 10.1128/AAC.03730-14
Wiklund, T., and Dalsgaard, I. (1998). Occurrence and significance of atypical Aeromonas salmonicida in non-salmonid and salmonid fish species: a review. Dis. Aquat. Organ. 32, 49–69. doi: 10.3354/dao032049
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: Aeromonas salmonicida subsp. salmonicida, furunculosis, rainbow trout, whole genome sequencing, SNP analysis, BEAST, virulence factors
Citation: Bartkova S, Leekitcharoenphon P, Aarestrup FM and Dalsgaard I (2017) Epidemiology of Danish Aeromonas salmonicida subsp. salmonicida in Fish Farms Using Whole Genome Sequencing. Front. Microbiol. 8:2411. doi: 10.3389/fmicb.2017.02411
Received: 05 September 2017; Accepted: 21 November 2017;
Published: 05 December 2017.
Edited by:
Jonathan Shaw, University of Sheffield, United KingdomReviewed by:
Brian Austin, University of Stirling, United KingdomCopyright © 2017 Bartkova, Leekitcharoenphon, Aarestrup and Dalsgaard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Inger Dalsgaard, aW5kYUB2ZXQuZHR1LmRr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.