- 1Department of Laboratory Medicine, Yeungnam University College of Medicine, Daegu, South Korea
- 2Department of Dental Hygiene, College of Health and Welfare, Silla University, Busan, South Korea
- 3Department of Laboratory Medicine, Kosin University College of Medicine, Busan, South Korea
The worldwide dissemination of carbapenemase-producing Enterobacteriaceae (CPE) has become a major therapeutic concern in clinical settings. Enterobacter cloacae is a major pathogen that causes serious hospital-acquired infections. We investigated the clinical characteristics and molecular mechanisms of the first IMP-4-producing E. cloacae clinical isolates in Korea. Five carbapenemase-producing E. cloacae strains out of 792 E. cloacae clinical isolates, which have been identified at a university hospital in Korea between March 2014 and February 2016, were included in this study. Antimicrobial susceptibilities to imipenem, meropenem, and ertapenem were tested using E-test. Carbapenemase determinant screening, genetic environment, and multilocus sequence typing were conducted using PCR and sequencing analysis. All isolates were not susceptible to at least one of the tested carbapenems and presented highly similar pulsed-field gel electrophoresis (PFGE) patterns, evidencing hospital-wide clonal dissemination. Among all isolates harboring the blaIMP-4 carbapenemase gene, four isolates identified as predominant ST74, also contained blaCMY−2. One strain, designated as rare ST194, carried blaCMY-1. The E. cloacae strain, harboring both blaIMP-4 and blaCMY-1, was resistant to all three tested carbapenems. The blaIMP-4 gene was located on a highly mobile class 1 integron, showing a new form of the blaIMP-4-qacG-aacA4 array. This is the first description of IMP-4-producing E. cloacae strains in Korea. This observation implicates the widespread of blaIMP-4 in Enterobacteriaceae clinical isolates and provides insights into the epidemic potential and clinical therapeutic importance of IMP-4-producing E. cloacae for healthcare-associated infections.
Introduction
The spread of carbapenemase-producing Enterobacteriaceae (CPE) has become a prominent health-care challenge worldwide in the treatment of infectious diseases. Carbapenemases, including Klebsiella pneumoniae carbapenemase (KPC), imipenemase (IMP), New Delhi metallo-β-lactamase (NDM), Verona integron-encoded metallo-β-lactamase (VIM), and oxacillinase (OXA)-48 medicated antibiotic resistance (Nordmann et al., 2011; Shi et al., 2017). IMP-type CPEs have been reported globally (Queenan and Bush, 2007; Tzouvelekis et al., 2012) and have become the predominant form in Australia (Espedido et al., 2008; Leung et al., 2013; Sidjabat et al., 2015) since the first report of IMP-1 from Pseudomonas aeruginosa in Japan (Watanabe et al., 1991). One of the most commonly observed IMP variants was IMP-4 in clinical Enterobacteriaceae isolates (Leung et al., 2013; Hu et al., 2014), which was firstly detected in Hong Kong (Chu et al., 2001). Among more than 11 different species of IMP-4-producing CPE, Enterobacter cloacae has emerged as the predominant species (Sidjabat et al., 2015; Cao et al., 2017). IMP-type E. cloacae isolates have been found in Taiwan (IMP-8), China (IMP-1 and IMP-34), Thailand (IMP-14), Japan (IMP-1 and IMP-11), Spain (IMP-13), United Kingdom (IMP-1), and South Africa (Figure 1; Chen et al., 2009; Shet et al., 2011; Hayakawa et al., 2014; Wang et al., 2015; Osei Sekyere, 2016; Matsumura et al., 2017). IMP-4-producing E. cloacae was particularly reported in Australia and caused clinical outbreaks, which brought greater challenges to infection control (Leung et al., 2013; Chapuis et al., 2016; Pang et al., 2016). The highly mobile class 1 integron facilitates global spread of the blaIMP-4 gene (Espedido et al., 2008; Partridge et al., 2012).
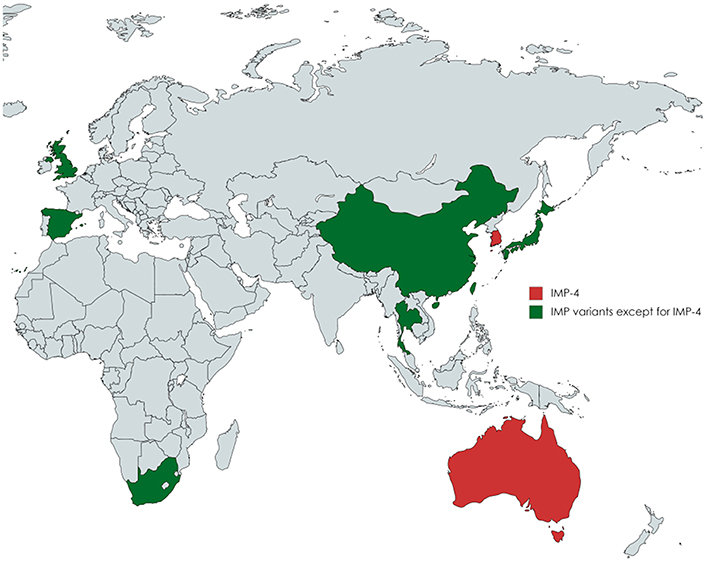
Figure 1. A map showing the global distribution of IMP-producing Enterobacter cloacae isolates. IMP-4-producing E. cloacae strains were especially described in Australia and Korea. IMP-type except for IMP-4 E. cloacae isolates have been reported in Taiwan (IMP-8), China (IMP-1 and IMP-34), Thailand (IMP-14), Japan (IMP-1 and IMP-11), Spain (IMP-13), United Kingdom (IMP-1), and South Africa.
Until now, carbapenem-resistant E. cloacae has rarely been reported in Korea since the initial VIM-2-producing isolate in 2003 (Jeong et al., 2003). Here, we described the clinical characteristics and molecular mechanisms of the first IMP-4-producing E. cloacae clinical isolates in Korea.
Materials and Methods
Bacterial Strains
A total of 792 E. cloacae clinical isolates have been identified at a university hospital in Korea between March 2014 and February 2016. Among the isolates, five carbapenemase-producing E. cloacae strains (0.6%), YUMC1, YUMC2, YUMC3, YUMC4, and YUMC5 were included in this study. The isolates were identified as E. cloacae using the Vitek GNI card (bioMérieux, Marcy l'Étoile, France) and 16S rRNA sequencing (Lane et al., 1985; Mao et al., 2012; Mezzatesta et al., 2012; Jeong et al., 2015). This study was carried out in accordance with the recommendations of Institutional Review Board of Kosin University Gospel Hospital, Busan, Korea; with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. We primarily focused on the analysis of the isolated strains and made our effort to anonymize private information of infected patients.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibilities were determined by the Vitek card AST-N246 (bioMérieux). Carbapenem producers were identified by modified Hodge test on MacConkey agar (Becton, Dickinson and Company, Sparks, MD, USA). The performance of modified Hodge test was reported to be better with MacConkey agar, containing bile compounds, than with Mueller-Hinton agar for screening carbapenemase-producing Gram-negative bacilli (K. Lee et al., 2010). Carbapenemase production was confirmed by KPC+MBL Confirm ID Kit (Rosco Diagnostica, Taastrup, Denmark) using tablets containing meropenem (10 μg) alone or supplemented with dipicolinic acid (1,000 μg), phenylboronic acid (400 μg), and cloxacillin (750 μg), and Mueller-Hinton agar (Oxoid Ltd., Basingstoke, UK). The minimum inhibitory concentrations (MICs) for imipenem, meropenem, and ertapenem were determined using E-test strips (AB Biodisk, Solna, Sweden). The breakpoints were applied according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (Clinical Laboratory Standards Institute, 2016). Double-disk synergy test (DDST) for the detection of extended-spectrum β-lactamases (ESBLs) was also performed according to the CLSI guideline.
Pulsed-Field Gel Electrophoresis
Pulsed-field gel electrophoresis (PFGE) was performed to confirm the clonality of the IMP-4-producing E. cloacae isolates. XbaI (Roche, Mannheim, Germany)-digested genomic DNA was prepared at 37°C for 12–14 h. DNA fragments were separated using a CHEF-DRII System (Bio-Rad, Hercules, CA, USA). Banding patterns were analyzed with InforQuestFP software version 4.5 (Bio-Rad) to generate a dendrogram.
Multilocus Sequence Typing
Multilocus sequence typing (MLST) for seven housekeeping genes, including dnaA, fusA, gyrB, leuS, pyrG, rplB, and rpoB, was conducted. After PCR and sequencing, nucleotide sequences were compared with those in the MLST database (http://pubmlst.org/ecloacae) to identify allelic numbers and sequence types (ST).
Polymerase Chain Reaction and Sequencing
The genomic DNA of five isolates were extracted via the boiling lysis method (L. Chen et al., 2011). The genes for 16S rRNA, carbapenemase, integron components, fluoroquinolones, ESBLs, and plasmid-mediated AmpCs were amplified by polymerase chain reaction (PCR) and sequenced using the primers (Lane et al., 1985; Jeong et al., 2003; Bae et al., 2007, 2011; Mao et al., 2012; Hong et al., 2015) described in Table 1. Briefly, the PCR program was as follows: 94°C denaturation for 5 min, followed by 30 cycles of 94°C denaturation for 30 s, then 55–60°C annealing for 30 s, and subsequently 72°C extension for 30 s, followed by 72°C final extension for 7 min. The amplified products were sequenced and the nucleotide sequences were compared by the Basic Local Alignment Search Tool (BLAST) (https://www.ncbi.nlm.nih.gov/BLAST) (Jeong et al., 2015). Genetic organization of class 1 integron carrying the blaIMP-4 gene cassette of a plasmid was investigated by PCR mapping and sequencing of the regions surrounding the gene using the primers described in Table 1 (Jeong et al., 2003; Bae et al., 2007; Hong et al., 2015). The integron variant was identified using INTEGRALL database (http://integrall.bio.ua.pt/) (Moura et al., 2009).
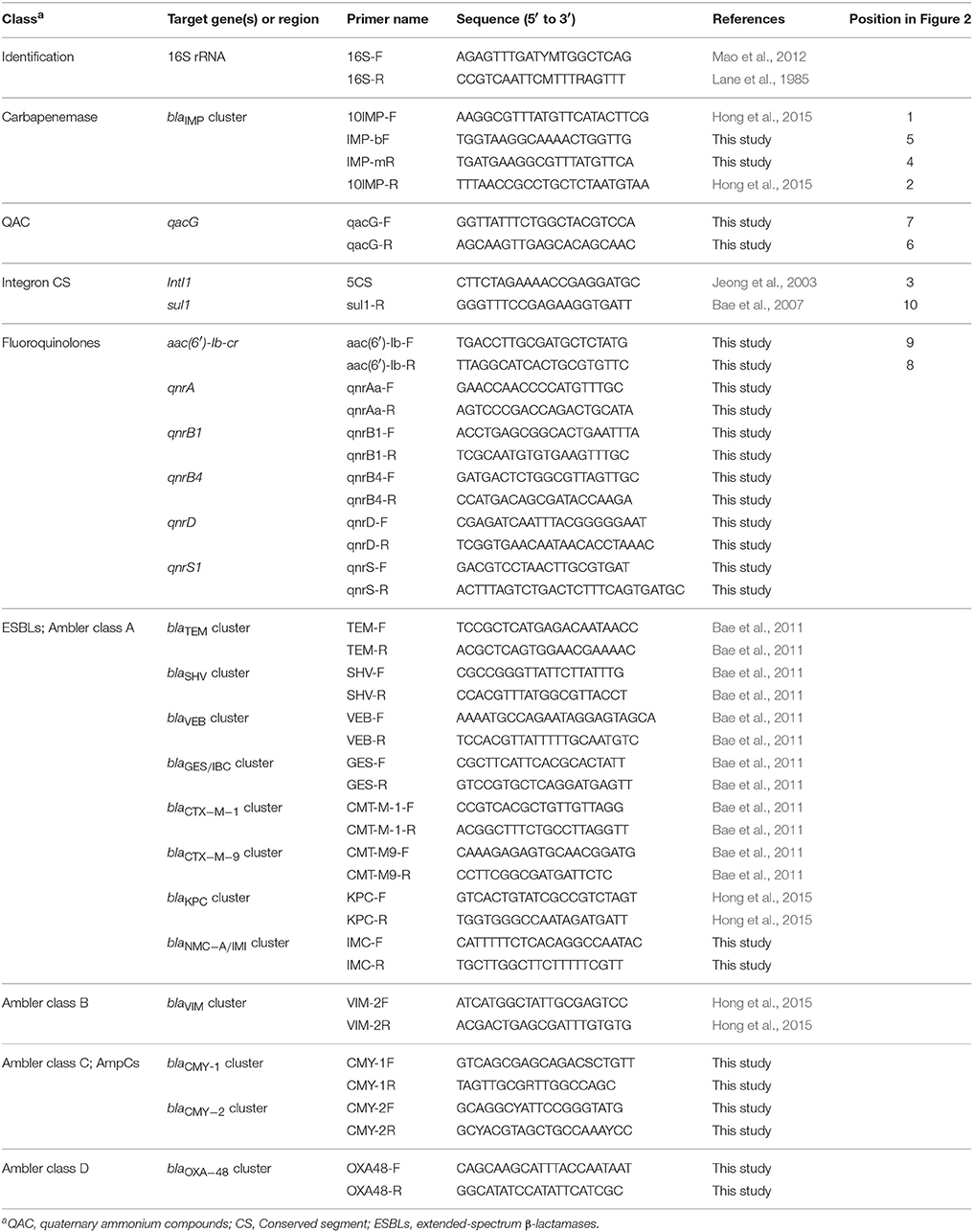
Table 1. Nucleotide sequences of primers used for the identification of species, the detection of resistant genes, and genetic environments in this study.
Nucleotide Sequence Accession Number
Nucleotide sequence data for YUMC2 are available under the GenBank accession number KY884003 and assigned In1456 for class 1 integron based on the INTEGRALL database (http://integrall.bio.ua.pt/) (Moura et al., 2009).
Results
Description of the Patients
The clinical characteristics of the patients infected with five isolates are summarized in Table 2. The carbapenemase-producing E. cloacae strains were isolated from various departments and two of them were recovered from the open wounds in diabetic feet. Most of the patients suffered from underlying diseases such as hypertension, diabetes mellitus and/or cancer causing immunocompromised conditions.
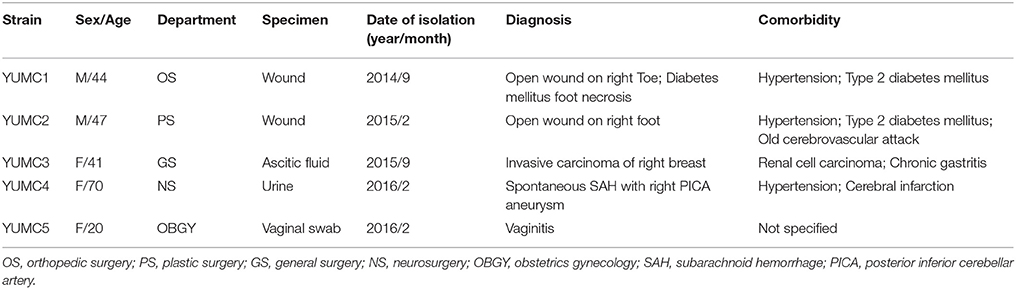
Table 2. Clinical characteristics of the patients infected with IMP-4-producing E. cloacae isolates.
Antimicrobial Susceptibility Profiles
The antimicrobial susceptibility profiles of five E. cloacae isolates with blaIMP-4 are presented in Supplementary Table 1. All five isolates were not susceptible to ampicillin, amoxicillin-clavulanic acid, cephalosporins, and carbapenems, whereas susceptible to amikacin, gentamicin, tigecycline, and ciprofloxacin, except for YUMC2. DDST for ESBL was negative for all five isolates. The antimicrobial susceptibility profiles of the other 787 E. cloacae strains are also summarized in Supplementary Table 2. The overall patterns are similar to those of five IMP-4-producing isolates, except for the carbapenems.
Resistance to Carbapenems
All five isolates were positive as carbapenem producers in the modified Hodge test and KPC+MBL Confirm ID Kit (Rosco Diagnostica). The MICs were determined using E-test strips (AB Biodisk) and the results for imipenem, meropenem, and ertapenem are presented in Table 3. All isolates were not susceptible to at least one of the carbapenems using CLSI breakpoints. Notably, YUMC2 was resistant to all tested carbapenems and had higher MICs than other isolates.
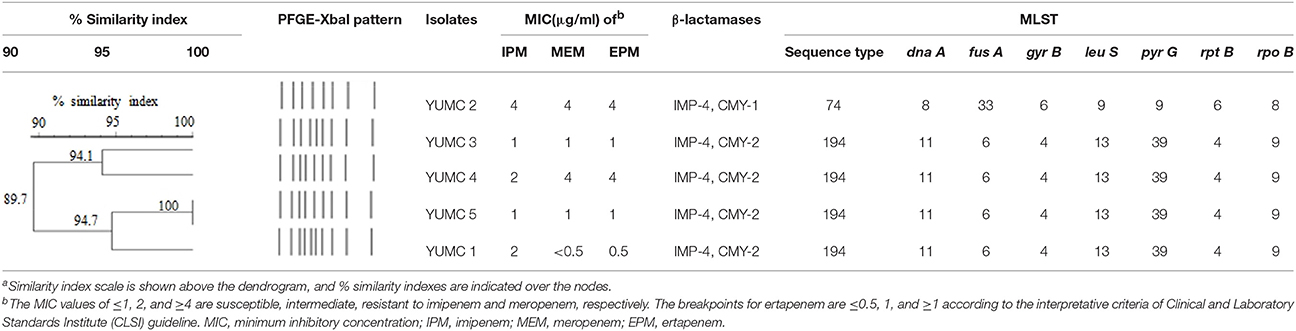
Table 3. Pulsed-field gel electrophoresis (PFGE)-based dendrogram and multilocus sequence typing (MLST) of IMP-4-producing E. cloacae isolatesa.
Clonality of the Isolates
YUMC4 and YUMC5 strains presented identical PFGE patterns and the other isolates also showed highly similar patterns based on the criteria of 85% similarity (Table 3). The strains, isolated same years, presented close relationship.
Sequence Type
The MLST assay assigned the isolates to two STs (Table 3). YUMC2 was assigned to predominant ST74. Four out of the five IMP-4-producing E. cloacae strains were rare ST194, showing significant clonal similarity.
Carbapenemase Genes and Genetic Environment
PCR screening demonstrated the presence of blaIMP-4 in all E. cloacae isolates (Table 3). In addition, YUMC2 was also positive for CMY-1. The other strains contained IMP-4 and CMY-2 simultaneously. The ESBL genes were not detected whereas, aac(6′)-Ib-cr and qnrS1 relevant to fluoroquinolones were found. In this study, blaCMY-1, blaCMY−2, blaIMP−4, aac(6)-Ib-cr, and qnrS1 were identical to previously reported sequences deposited in GenBank database under accession numbers X92508.1, X91840.1, AF244145.1, CP023487.1, and AB187515.1, respectively.
PCR mapping and sequencing generated a 3,585-bp segment that shared 99% identity with E. cloacae pEI1573 (GenBank accession no. JX101693.1) (Partridge et al., 2012). The blaIMP-4-gene was located on class 1 integron In1456, consisted of novel blaIMP-4-qacG2-aacA4 cassette array (Figure 2).
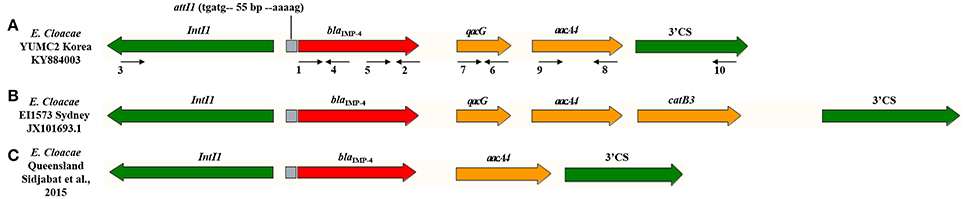
Figure 2. Schematic representation of the class 1 integron gene cassettes bearing the blaIMP-4 genes in E. cloacae isolates. Genes and their directions of transcription are described as broad arrows. The gray box indicates recombination site. The primers, detailed in Table 1, for PCR mapping are depicted as narrow arrows with numbers. The red arrow of blaIMP-4 is related to carbapenemase. The yellow arrow of qacG and aacA4 are associated with resistance to quaternary ammonium compounds and fluoroquinolone, respectively. The 5′ conserved segment (CS) of IntI1 and 3′ CS of sul1 are presented with green arrow. (A) E. cloacae YUMC2 in this study with Genbank accession no. KY884003. (B) E. cloacae EI1573 from Sydney, Australia with Genbank accession no. JX101693.1. (C) E. cloacae from Queensland, Australia reported by Sidjabat et al. (2015).
Discussion
E. cloacae is frequently implicated in serious nosocomial infections with high mortality. Majority of patients were reported to be immunocompromised, similar to our patients (Qureshi et al., 2011). Clinical outbreaks of E. cloacae in the hematology ward, burns unit, and intensive care unit have persisted, despite of concerted infection control to prevent ongoing transmission (Leung et al., 2013; Chapuis et al., 2016; Pang et al., 2016). VIM-2, NDM-1, and IMP-1, frequently found in Asia, have been previously reported mechanisms of carbapenem-resistant E. cloacae in Korea (Jeong et al., 2003; Kim et al., 2015; Lee et al., 2017). Meanwhile, IMP-4-producing E. cloacae isolates have been mainly found in Australia (Peleg et al., 2005; Leung et al., 2013). The first detection of IMP-4 in this study implicates that the plasmid-mediated blaIMP-4 eventually spread in E. cloacae clinical isolates in Korea.
IMP-4 was reported to be strongly active against imipenem and meropenem, with 0.25–16 MIC range (Chu et al., 2001). The MICs of our isolates showed that all five strains were not susceptible to at least one of the carbapenems, including imipenem, meropenem, and ertapenem. Antibiotic resistance profiles of blaIMP-positive Enterobacteriaceae isolates showed 25% resistance, 57% intermediate resistance, and 18% susceptibility to meropenem and 6% resistance, 33% intermediate resistance, and 61% susceptibility to imipenem in a previous study (Dolejska et al., 2016). Natural antibiotic susceptibility of E. cloacae complex to carbapenems were reported to be susceptible (Stock et al., 2001), however, the presence of IMP-4 would influence on the antibiotic profiles.
The antimicrobial susceptibility profiles of E. cloacae isolates in this study were similar to the intrinsic patterns of antibiotics (Mezzatesta et al., 2012). However, 5 strains containing blaIMP-4 were not susceptible to carbapenems and YUMC2 was resistant to ciprofloxacin. The detected genes, aac(6′)-Ib-cr and qnrS1 relevant to fluoroquinolones might be associated with this results. Nevertheless, the cr variant of aac(6′)-Ib confers reduced susceptibility to ciprofloxacin by N-acetylation of its piperazinyl amine (Robicsek et al., 2006), ciprofloxacin resistance was not related to aac(6′)-Ib-cr prevalence (Park et al., 2006). Interestingly, the isolates co-carrying aac(6′)-Ib-cr and qnrS1 were also reported to be sensitive to quinolones (Huang et al., 2012). Therefore, these genes seems to supplement other quinolone resistance mechanisms rather than confer directly to resistance. Although, the aac(6′)-Ib-cr and qnrS1 genes were frequently found to be co-carried with various ESBLs, becoming therapeutic threats (Huang et al., 2012; Mezzatesta et al., 2012), our isolates harbored blaIMP-4 without ESBLs.
The homogeneity of five strains was analyzed using PFGE. Although the strains were isolated from various clinical departments, the high similarity of PFGE patterns of isolates, especially in the same years, might be the evidence of hospital-wide clonal dissemination.
According to MLST results, YUMC2 was designated to ST74, the most predominant clonal lineage with increased epidemic potential based on previous E. cloacae clonality studies (Fernández et al., 2015; Guillard et al., 2015; Izdebski et al., 2015). E. cloacae ST74 had higher carbapenems MICs than other isolates, similar to the results of previous studies, and was assumed to confer with the spread of the resistance to carbapenems (Guillard et al., 2015; Izdebski et al., 2015). The other four IMP-4-producing E. cloacae strains were ST194, presenting significant genetic similarity. To the best of our knowledge, available studies for E. cloacae ST194 were rare, indicating that this is the first report of clinical E. cloacae ST194.
PCR results showed the presence of CMY-1 in YUMC2 and CMY-2 in the other strains as well as IMP-4. Prior studies demonstrated that the most frequently reported AmpC β-lactamase was CMY, consisting of 92.7% of CMY-2 among Enterobacteriaceae isolates in the Asia-Pacific region (Sheng et al., 2013). The combination of blaIMP-4 and blaCMY−2−like was found from one clinical E. cloacae isolate among the CPE in Australia (Sidjabat et al., 2015). In addition, the coexistence of blaIMP-4 and blaCMY-1 in E. cloacae strain was not reported previously and this is the first description of E. cloacae, coproducing IMP-4 and CMY-1 with resistance to all three carbapenems.
When comparing the product of sequencing of our study to E. cloacae pEI1573 (GenBank accession no. JX101693.1) (Partridge et al., 2012), both of the blaIMP-4 genes of our study and pEI1573 were located on class 1 integrons. However, the gene cassettes compositions were slightly different between YUMC2 and pEI1573, containing a reference sequence of typical Australian class 1 integron array (Figure 2). The blaIMP-4-qacG-aacA4-catB3 cassette array of pEI1573 from Sydney, Australia is almost identical to those of pJIBE401 from Sydney index isolate K. pneumoniae (GenBank accession no. AJ609296) (Espedido et al., 2005), pCTX-M3 from Citrobacter freundii in Poland (GenBank accession no. AF550415) (Gołebiewski et al., 2007), and pCTX-M360 from K. pneumoniae in China (GenBank accession no. EU938349) (Zhu et al., 2009). Meanwhile, the class 1 integron of our study consisted of blaIMP-4-qacG-aacA4 and a different array form composed of blaIMP-4-aacA4, which was reported previously from Queensland, Australia (Sidjabat et al., 2015). These cassette arrays, found in diverse isolates with slightly different genetic contexts, suggest movement of the array by homologous recombination and the worldwide dissemination potential of blaIMP-4 gene.
In the respect of epidemiological relationship, the class 1 integrons of Australia and Korea, containing blaIMP-4 genes of E. cloacae isolates, revealed similar gene cassettes, except for catB3 or qacG. Geographically, Australia and Korea are located at the rim of Asian-pacific region. Further, a large-scale transmission of blaIMP-4 of E. cloacae isolates, predominant from of CPE in Australia (Sidjabat et al., 2015), through silver gulls of Australia was previously reported (Dolejska et al., 2016).
In conclusion, we report the first IMP-4-producing E. cloacae strains identified as predominant ST74 and rare ST194 in Korea. Furthermore, it is the first description of blaIMP-4 and blaCMY-1 coexistence and a new class 1 integron cassette array form in Enterobacteriaceae. This finding implicates the emergence of plasmid-mediated blaIMP-4 on the highly mobile class 1 integron in Enterobacteriaceae clinical isolates in Korea with great concern for widespread and therapeutic threats. In addition, it provides insights into the epidemic potential and clinical importance of IMP-4-producing E. cloacae for hospital-acquired infections.
Author Contributions
SJ: analyzed the data, and wrote the manuscript; IKB: designed and performed the experiments, and revised the manuscript; JHL and CHL: helped the experiments and the writing of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (Ministry of Science, ICT & Future Planning) [NRF-2017R1C1B2004597]; and Kosin University College of Medicine [2016-2-1].
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the researchers of Silla University and medical technicians of Yeungnam University College of Medicine for their technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02343/full#supplementary-material
References
Bae, I. K., Jang, S. J., Kim, J., Jeong, S. H., Cho, B., and Lee, K. (2011). Interspecies dissemination of the bla gene encoding PER-1 extended-spectrum beta-lactamase. Antimicrob. Agents Chemother. 55, 1305–1307. doi: 10.1128/AAC.00994-10
Bae, I. K., Lee, Y. N., Lee, W. G., Lee, S. H., and Jeong, S. H. (2007). Novel complex class 1 integron bearing an ISCR1 element in an Escherichia coli isolate carrying the blaCTX-M-14 gene. Antimicrob. Agents Chemother. 51, 3017–3019. doi: 10.1128/AAC.00279-07
Cao, X. L., Cheng, L., Zhang, Z. F., Ning, M. Z., Zhou, W. Q., Zhang, K., et al. (2017). Survey of clinical extended-spectrum beta-lactamase-producing Enterobacter cloacae isolates in a Chinese Tertiary Hospital, 2012-2014. Microb. Drug Resist. 23, 83–89. doi: 10.1089/mdr.2015.0128
Chapuis, A., Amoureux, L., Bador, J., Gavalas, A., Siebor, E., Chrétien, M. L., et al. (2016). Outbreak of extended-spectrum beta-lactamase producing Enterobacter cloacae with high MICs of quaternary ammonium compounds in a hematology ward associated with contaminated sinks. Front. Microbiol. 7:1070. doi: 10.3389/fmicb.2016.01070
Chen, L. R., Zhou, H. W., Cai, J. C., Zhang, R., and Chen, G. X. (2009). Detection of plasmid-mediated IMP-1 metallo-beta-lactamase and quinolone resistance determinants in an ertapenem-resistant Enterobacter cloacae isolate. J. Zhejiang Univ. Sci. B 10, 348–354. doi: 10.1631/jzus.B0820302
Chen, L., Mediavilla, J. R., Endimiani, A., Rosenthal, M. E., Zhao, Y., Bonomo, R. A., et al. (2011). Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (bla KPC) variants. J. Clin. Microbiol. 49, 579–585. doi: 10.1128/JCM.01588-10
Chu, Y. W., Afzal-Shah, M., Houang, E. T., Palepou, M. I., Lyon, D. J., Woodford, N., et al. (2001). IMP-4, a novel metallo-beta-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 45, 710–714. doi: 10.1128/AAC.45.3.710-714.2001
Clinical and Laboratory Standards Institute (2016). M100-S26. Performance Standards for Antimicrobial Susceptibility Testing, 26th Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute.
Dolejska, M., Masarikova, M., Dobiasova, H., Jamborova, I., Karpiskova, R., Havlicek, M., et al. (2016). High prevalence of Salmonella and IMP-4-producing Enterobacteriaceae in the silver gull on Five Islands, Australia. J. Antimicrob. Chemother. 71, 63–70. doi: 10.1093/jac/dkv306
Espedido, B. A., Partridge, S. R., and Iredell, J. R. (2008). bla(IMP-4) in different genetic contexts in Enterobacteriaceae isolates from Australia. Antimicrob. Agents Chemother. 52, 2984–2987. doi: 10.1128/AAC.01634-07
Espedido, B., Iredell, J., Thomas, L., and Zelynski, A. (2005). Wide dissemination of a carbapenemase plasmid among gram-negative bacteria: implications of the variable phenotype. J. Clin. Microbiol. 43, 4918–4919. doi: 10.1128/JCM.43.9.4918-4919.2005
Fernández, J., Montero, I., Martínez, Ó., Fleites, A., Poirel, L., Nordmann, P., et al. (2015). Dissemination of multiresistant Enterobacter cloacae isolates producing OXA-48 and CTX-M-15 in a Spanish hospital. Int. J. Antimicrob. Agents 46, 469–474. doi: 10.1016/j.ijantimicag.2015.07.003
Gołebiewski, M., Kern-Zdanowicz, I., Zienkiewicz, M., Adamczyk, M., Zylinska, J., Baraniak, A., et al. (2007). Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum beta-lactamase gene blaCTX-M-3. Antimicrob. Agents Chemother. 51, 3789–3795. doi: 10.1128/AAC.00457-07
Guillard, T., Cholley, P., Limelette, A., Hocquet, D., Matton, L., Guyeux, C., et al. (2015). Fluoroquinolone resistance mechanisms and population structure of Enterobacter cloacae non-susceptible to ertapenem in North-Eastern France. Front. Microbiol. 6:1186. doi: 10.3389/fmicb.2015.01186
Hayakawa, K., Miyoshi-Akiyama, T., Kirikae, T., Nagamatsu, M., Shimada, K., Mezaki, K., et al. (2014). Molecular and epidemiological characterization of IMP-type metallo-beta-lactamase-producing Enterobacter cloacae in a Large tertiary care hospital in Japan. Antimicrob. Agents Chemother. 58, 3441–3450. doi: 10.1128/AAC.02652-13
Hong, J. S., Kim, J. O., Lee, H., Bae, I. K., Jeong, S. H., and Lee, K. (2015). Characteristics of Metallo-beta-lactamase-producing Pseudomonas aeruginosa in Korea. Infect Chemother. 47, 33–40. doi: 10.3947/ic.2015.47.1.33
Hu, L., Zhong, Q., Shang, Y., Wang, H., Ning, C., Li, Y., et al. (2014). The prevalence of carbapenemase genes and plasmid-mediated quinolone resistance determinants in carbapenem-resistant Enterobacteriaceae from five teaching hospitals in central China. Epidemiol. Infect. 142, 1972–1977. doi: 10.1017/S0950268813002975
Huang, S., Dai, W., Sun, S., Zhang, X., and Zhang, L. (2012). Prevalence of plasmid-mediated quinolone resistance and aminoglycoside resistance determinants among carbapeneme non-susceptible Enterobacter cloacae. PLoS ONE 7:e47636. doi: 10.1371/journal.pone.0047636
Izdebski, R., Baraniak, A., Herda, M., Fiett, J., Bonten, M. J., Carmeli, Y., et al. (2015). MLST reveals potentially high-risk international clones of Enterobacter cloacae. J. Antimicrob. Chemother. 70, 48–56. doi: 10.1093/jac/dku359
Jeong, S. H., Lee, K., Chong, Y., Yum, J. H., Lee, S. H., Choi, H. J., et al. (2003). Characterization of a new integron containing VIM-2, a metallo- beta-lactamase gene cassette, in a clinical isolate of Enterobacter cloacae. J. Antimicrob. Chemother. 51, 397–400. doi: 10.1093/jac/dkg047
Jeong, S., Kim, J. O., Jeong, S. H., Bae, I. K., and Song, W. (2015). Evaluation of peptide nucleic acid-mediated multiplex real-time PCR kits for rapid detection of carbapenemase genes in gram-negative clinical isolates. J. Microbiol. Methods 113, 4–9. doi: 10.1016/j.mimet.2015.03.019
Kim, S. R., Rim, C. B., Kim, Y., Kim, J. W., Song, Y. W., Shin, S. H., et al. (2015). Four cases of carbapenem-resistant enterobacteriaceae infection from january to march in 2014. Korean J. Fam. Med. 36, 191–194. doi: 10.4082/kjfm.2015.36.4.191
Lane, D. J., Pace, B., Olsen, G. J., Stahl, D. A., Sogin, M. L., and Pace, N. R. (1985). Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U.S.A. 82, 6955–6959. doi: 10.1073/pnas.82.20.6955
Lee, J. Y., Hong, Y. K., Lee, H., and Ko, K. S. (2017). High prevalence of non-clonal imipenem-nonsusceptible Enterobacter spp. isolates in Korea and their association with porin down-regulation. Diagn. Microbiol. Infect. Dis. 87, 53–59. doi: 10.1016/j.diagmicrobio.2016.10.004
Lee, K., Kim, C. K., Yong, D., Jeong, S. H., Yum, J. H., Seo, Y. H., et al. (2010). Improved performance of the modified Hodge test with MacConkey agar for screening carbapenemase-producing Gram-negative bacilli. J. Microbiol. Methods 83, 149–152. doi: 10.1016/j.mimet.2010.08.010
Leung, G. H., Gray, T. J., Cheong, E. Y., Haertsch, P., and Gottlieb, T. (2013). Persistence of related bla-IMP-4 metallo-beta-lactamase producing Enterobacteriaceae from clinical and environmental specimens within a burns unit in Australia - a six-year retrospective study. Antimicrob. Resist. Infect. Control 2:35. doi: 10.1186/2047-2994-2-35
Mao, D. P., Zhou, Q., Chen, C. Y., and Quan, Z. X. (2012). Coverage evaluation of universal bacterial primers using the metagenomic datasets. BMC Microbiol. 12:66. doi: 10.1186/1471-2180-12-66
Matsumura, Y., Peirano, G., Motyl, M. R., Adams, M. D., Chen, L., Kreiswirth, B., et al. (2017). Global Molecular Epidemiology of IMP-Producing Enterobacteriaceae. Antimicrob. Agents Chemother. 61:e02729-16. doi: 10.1128/AAC.02729-16
Mezzatesta, M. L., Gona, F., and Stefani, S. (2012). Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol. 7, 887–902. doi: 10.2217/fmb.12.61
Moura, A., Soares, M., Pereira, C., Leitão, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Nordmann, P., Naas, T., and Poirel, L. (2011). Global spread of Carbapenemase-producing Enterobacteriaceae. Emerging Infect. Dis. 17, 1791–1798. doi: 10.3201/eid1710.110655
Osei Sekyere, J. (2016). Current state of resistance to antibiotics of last-resort in South Africa: a review from a public health perspective. Front Public Health 4:209. doi: 10.3389/fpubh.2016.00209
Pang, F., Jia, X. Q., Song, Z. Z., Li, Y. H., Wang, B., Zhao, Q. G., et al. (2016). Characteristics and management of Enterobacteriaceae harboring IMP-4 or IMP-8 carbapenemase in a tertiary hospital. Afr. Health Sci. 16, 153–161. doi: 10.4314/ahs.v16i1.21
Park, C. H., Robicsek, A., Jacoby, G. A., Sahm, D., and Hooper, D. C. (2006). Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 50, 3953–3955. doi: 10.1128/AAC.00915-06
Partridge, S. R., Ginn, A. N., Paulsen, I. T., and Iredell, J. R. (2012). pEl1573 Carrying blaIMP-4, from Sydney, Australia, is closely related to other IncL/M plasmids. Antimicrob. Agents Chemother. 56, 6029–6032. doi: 10.1128/AAC.01189-12
Peleg, A. Y., Franklin, C., Bell, J. M., and Spelman, D. W. (2005). Dissemination of the metallo-beta-lactamase gene blaIMP-4 among gram-negative pathogens in a clinical setting in Australia. Clin. Infect. Dis. 41, 1549–1556. doi: 10.1086/497831
Queenan, A. M., and Bush, K. (2007). Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20, 440–458. doi: 10.1128/CMR.00001-07
Qureshi, Z. A., Paterson, D. L., Pakstis, D. L., Adams-Haduch, J. M., Sandkovsky, G., Sordillo, E., et al. (2011). Risk factors and outcome of extended-spectrum beta-lactamase-producing Enterobacter cloacae bloodstream infections. Int. J. Antimicrob. Agents 37, 26–32. doi: 10.1016/j.ijantimicag.2010.09.009
Robicsek, A., Strahilevitz, J., Jacoby, G. A., Macielag, M., Abbanat, D., Park, C. H., et al. (2006). Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12, 83–88. doi: 10.1038/nm1347
Sheng, W. H., Badal, R. E., and Hsueh, P. R., on behalf of the SMART Program (2013). Distribution of extended-spectrum beta-lactamases, AmpC beta-lactamases, and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal infections in the Asia-Pacific region: results of the study for Monitoring Antimicrobial Resistance Trends (SMART). Antimicrob. Agents Chemother. 57, 2981–2988. doi: 10.1128/AAC.00971-12
Shet, V., Gouliouris, T., Brown, N. M., Turton, J. F., Zhang, J., and Woodford, N. (2011). IMP metallo-beta-lactamase-producing clinical isolates of Enterobacter cloacae in the UK. J. Antimicrob. Chemother. 66, 1408–1409. doi: 10.1093/jac/dkr078
Shi, Z., Zhao, H., Li, G., and Jia, W. (2017). Molecular Characteristics of Carbapenem-Resistant Enterobacter cloacae in Ningxia Province, China. Front. Microbiol. 8:94. doi: 10.3389/fmicb.2017.00094
Sidjabat, H. E., Townell, N., Nimmo, G. R., George, N. M., Robson, J., Vohra, R., et al. (2015). Dominance of IMP-4-producing enterobacter cloacae among carbapenemase-producing Enterobacteriaceae in Australia. Antimicrob. Agents Chemother. 59, 4059–4066. doi: 10.1128/AAC.04378-14
Stock, I., Grüger, T., and Wiedemann, B. (2001). Natural antibiotic susceptibility of strains of the Enterobacter cloacae complex. Int. J. Antimicrob. Agents 18, 537–545. doi: 10.1016/S0924-8579(01)00463-0
Tzouvelekis, L. S., Markogiannakis, A., Psichogiou, M., Tassios, P. T., and Daikos, G. L. (2012). Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25, 682–707. doi: 10.1128/CMR.05035-11
Wang, Y., Lo, W. U., Lai, E. L., Chow, K. H., and Ho, P. L. (2015). Complete sequence of the multidrug-resistant IncL/M plasmid pIMP-HB623 Cocarrying bla IMP-34 and fosC2 in an Enterobacter cloacae strain associated with medical travel to China. Antimicrob. Agents Chemother. 59, 5854–5856. doi: 10.1128/AAC.00375-15
Watanabe, M., Iyobe, S., Inoue, M., and Mitsuhashi, S. (1991). Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35, 147–151. doi: 10.1128/AAC.35.1.147
Zhu, W. H., Luo, L., Wang, J. Y., Zhuang, X. H., Zhong, L., Liao, K., et al. (2009). Complete nucleotide sequence of pCTX-M360, an intermediate plasmid between pEL60 and pCTX-M3, from a multidrug-resistant Klebsiella pneumoniae strain isolated in China. Antimicrob. Agents Chemother. 53, 5291–5293. doi: 10.1128/AAC.00032-09
Keywords: IMP-4, CMY, carbapenem, class 1 integron, Enterobacter cloacae
Citation: Lee JH, Bae IK, Lee CH and Jeong S (2017) Molecular Characteristics of First IMP-4-Producing Enterobacter cloacae Sequence Type 74 and 194 in Korea. Front. Microbiol. 8:2343. doi: 10.3389/fmicb.2017.02343
Received: 11 August 2017; Accepted: 14 November 2017;
Published: 28 November 2017.
Edited by:
Marcelo Tolmasky, California State University, Fullerton, United StatesReviewed by:
Nilton Lincopan, University of São Paulo, BrazilIsabelle Broutin, Centre National de la Recherche Scientifique (CNRS), France
Copyright © 2017 Lee, Bae, Lee and Jeong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seri Jeong, aGVoZWJveEBuYXZlci5jb20=
†These authors have contributed equally to this work.
 Jong Ho Lee
Jong Ho Lee Il Kwon Bae2†
Il Kwon Bae2† Seri Jeong
Seri Jeong