
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 23 November 2017
Sec. Infectious Agents and Disease
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.02247
This article is part of the Research Topic Surface Proteins of Gram-Positive Bacteria - Array of Opportunities View all 7 articles
Food and probiotic bacteria, in particular lactic acid bacteria, are ingested in large amounts by humans and are part of the transient microbiota which is increasingly considered to be able to impact the resident microbiota and thus possibly the host health. The lactic acid bacterium Lactococcus lactis is extensively used in starter cultures to produce dairy fermented food. Also because of a generally recognized as safe status, L. lactis has been considered as a possible vehicle to deliver in vivo therapeutic molecules with anti-inflammatory properties in the gastrointestinal tract. One of the key factors that may favor health effects of beneficial bacteria to the host is their capacity to colonize transiently the gut, notably through close interactions with mucus, which covers and protects the intestinal epithelium. Several L. lactis strains have been shown to exhibit mucus-binding properties and bacterial surface proteins have been identified as key determinants of such capacity. In this review, we describe the different types of surface proteins found in L. lactis, with a special focus on mucus-binding proteins and pili. We also review the different approaches used to investigate the adhesion of L. lactis to mucus, and particularly to mucins, one of its major components, and we present how these approaches allowed revealing the role of surface proteins in muco-adhesion.
Lactic acid bacteria (LAB), used as starters in food fermentations and as probiotics, are regularly ingested in large amount by humans. After their ingestion, these bacteria temporarily co-exist with the complex resident gut microbiota. Emerging evidence suggests that this transient microbiota has an impact on composition and metabolism of the gut microbiota and thus possibly on host health (Veiga et al., 2014; Derrien and van Hylckama Vlieg, 2015). Lactococcus lactis is one of the most widely used LAB in starter cultures for manufacturing dairy fermented products. Besides, several natural isolates have been described with beneficial health properties and recombinant L. lactis has been proposed as a delivery vehicle for therapeutic molecules in the gastrointestinal tract (GIT) (Carvalho et al., 2017). Transient colonization of the gut by the ingested bacteria, notably through adhesion to mucus that covers and protects the intestinal epithelium, is expected to favor their beneficial effect on the host. The ability of commensal or pathogenic bacteria to adhere to intestinal mucus glycoconjugates was previously attributed to specific proteins present at the bacterial surface (Kline et al., 2009; Hill, 2012; Juge, 2012; Moonens and Remaut, 2017). Although L. lactis is not a natural inhabitant of the mammalian GIT, proteins able to bind mucins have also been described. This mini-review gives an overview of the beneficial effects reported for L. lactis, the mucus composition and function, the different surface proteins discovered in L. lactis and involved in muco-adhesion and the different complementary approaches and tools used to uncover their role.
The main beneficial effects reported for natural or recombinant L. lactis strains concern their anti-inflammatory potential, making them as possible therapeutic tools in human intestinal bowel diseases. First, several natural L. lactis isolates were shown to possess anti-inflammatory properties in cellular models as well as in vivo in mouse models of intestinal colitis (Nishitani et al., 2009; Luerce et al., 2014; Ballal et al., 2015). Besides, recombinant L. lactis expressing anti-inflammatory molecules (cytokine IL10, anti-oxidant enzymes, or protease inhibitor elafin) efficiently reduce intestinal inflammation symptoms and restore colon homeostasis in mice (Bermudez-Humaran et al., 2013). Alleviation of food allergic manifestations in sensitized mice was also described for L. lactis NCC2287 (Zuercher et al., 2012). Interestingly, a recent study reported that L. lactis present in a fermented milk product was able to modulate the gut microbiota in permissive subjects (rats or humans), and this property was correlated with an increased persistence in the GIT (Zhang et al., 2016).
The mammalian GIT is covered by mucus, a viscoelastic gel that lines and protects the intestinal epithelium, separating it from the lumen content. Mucus functions as a dynamic barrier that is permeable to gasses, water and nutrients, but impermeable to most microorganisms. This substance was long considered to act as a “simple” physical barrier, but is now known to exert other key functions essential for maintaining intestinal homeostasis (Juge, 2012; Ouwerkerk et al., 2013). Mucus covers the intestinal epithelium to a different extent along the GIT. In lower GIT, the small intestine has only a single layer whereas the colon displays a two-layered mucus (Ermund et al., 2013). The inner layer is depicted as essentially sterile in contrast with the outer layer which is highly colonized (Donaldson et al., 2016). The main constituents of mucus are mucins, which are produced, stored and released by goblet cells. Mucins are large glycoproteins in which the glycans constitute more than 80% of the molecular mass. The O-linked glycan chains contain 1–20 sugar residues most commonly attached to the protein backbone through serine or threonine with N-acetylgalactosamine (GalNAc). The chain is elongated with core structures and two potential backbone regions containing N-acetylglucosamine (GlcNAc) and galactose (Gal). Terminal sugars are usually fucose, Gal, GalNAc, or sialic acid residues and both Gal and GlcNAc residues may be sulfated, thus offering a high degree of diversification (Linden et al., 2008). MUC2 is the secreted gel-forming mucin present in the intestine (Johansson et al., 2011). In humans, MUC2 is coated with more than 100 different O-linked glycan chains (Larsson et al., 2009), which notably differ from those in rodents (Thomsson et al., 2012). Mucin oligosaccharides can serve both as binding sites and energy sources for GIT microbes and the difference in the glycan “preferences” of bacteria was suggested to explain host specificity in terms of microbiota (Donaldson et al., 2016).
The cell wall of Gram positive bacteria is made of a thick peptidoglycan sacculus decorated with other glycopolymers (polysaccharides and teichoic acids) as well as proteins (Chapot-Chartier and Kulakauskas, 2014). The cell wall proteins are usually classified based on their mode of attachment to the cell envelope: (i) covalent attachment to peptidoglycan through an LPxTG motif and a sortase A-mediated reaction; (ii) non-covalent binding to cell wall glycopolymers through specific domains (e.g., LysM or SH3 domains); (iii) lipid anchoring in the membrane (lipoprotein); (iv) one or several transmembrane domains.
Among the cell wall proteins, only those protruding at the external surface, that are surface-exposed proteins constituting the surfome (Olaya-Abril et al., 2014), are likely involved in bacterial adhesion to abiotic and biotic surfaces. These proteins can be predicted in silico by specific flow scheme such as Surf G+ (Barinov et al., 2009). At the experimental level, they can be specifically targeted by dedicated proteomic methods such as (i) the “shaving” procedure consisting in proteolytic digestion of surface-exposed proteins on live bacteria and LC-MS/MS identification of released peptides (Olaya-Abril et al., 2014) or (ii) selective labeling with a fluorescent dye followed by 2D gel analysis (Le Marechal et al., 2015).
Although L. lactis is not a typical inhabitant of the mammalian GIT, several surfaces proteins have been previously identified with the ability to bind to mucus/mucins. Most of them belong to the LPxTG-protein family and are encoded by plasmids. Of note, plasmids significantly contribute to the genetic diversity encountered in the L. lactis species (Ainsworth et al., 2014), related to its adaptation to different niches (food, plant, or animal) (Kelly et al., 2010; Garrigues et al., 2013; Kelleher et al., 2017; Laroute et al., 2017). Thus, whereas the laboratory strains, obtained from dairy strains by plasmid and prophage curing, are considered to have a restricted surface proteome (Habimana et al., 2007), the L. lactis surface pan-proteome is probably much richer and more diverse, constituting a potential reservoir of muco-adhesive factors. The proteins described below are likely contributing to muco-adhesion, either through non-specific hydrophobic interactions with mucus components (aggregation factor AggL, protease PrtP) or specific binding to mucin glycans [mucus-binding proteins (MUBs), pilins containing “lectin” domains]. In this latter case, it can be speculated that, in the original ecological niche of L. lactis, i.e., plant, these proteins might play a role in binding sugar motifs at the surface of vegetal cells, and these motifs could be shared by mucin sugars (Meyrand et al., 2013).
Mucus-binding proteins are bacterial surface adhesins with typical signal peptide and C-terminal LPxTG motif. They contain (multiple) Mub domains (around 200 residues in length) and/or MucBP domains (Pfam PF06458, around 50 residues), which have been shown to bind mucins (Juge, 2012). According to structural studies, the Mub domains consist of two modules B1 and B2 with B2 being a MucBP domain (MacKenzie et al., 2009; Etzold et al., 2014). Contrary to the MucBP domain also present in pathogens (Popowska et al., 2017), the Mub domain is predominantly found in LAB and is highly abundant in lactobacilli of the gut microbiota (Boekhorst et al., 2006). The best characterized MUB protein from Lactobacillus reuteri endowed with 14 Mub domains (Roos and Jonsson, 2002) binds mucus via multiple interactions involving terminal sialylated mucin glycans (Etzold and Juge, 2014; Gunning et al., 2016). In L. lactis plasmid free laboratory strains IL1403 and MG1363, only one MUB protein with four Mub domains and devoid of signal sequence, is encoded in the chromosome sequence (Boekhorst et al., 2006). However, these strains do not exhibit muco-adhesive properties (Radziwill-Bienkowska et al., 2017), suggesting that this MUB protein is not expressed or not functional. In contrast, three other MUB proteins found in dairy or vegetal strains were shown to contribute to bacterial adhesion to mucins as tested by different methods and tools (Table 1). These three proteins have different structures (Table 1), with three MucBP domains in MbpL protein from dairy L. lactis BGKP1 (Kojic et al., 2011), two Mub domains in Muc protein from vegetal L. lactis TIL448 (Meyrand et al., 2013) and one MucBP domain in AJ89_07570 protein from dairy L. lactis IBB477 (Radziwill-Bienkowska et al., 2016).
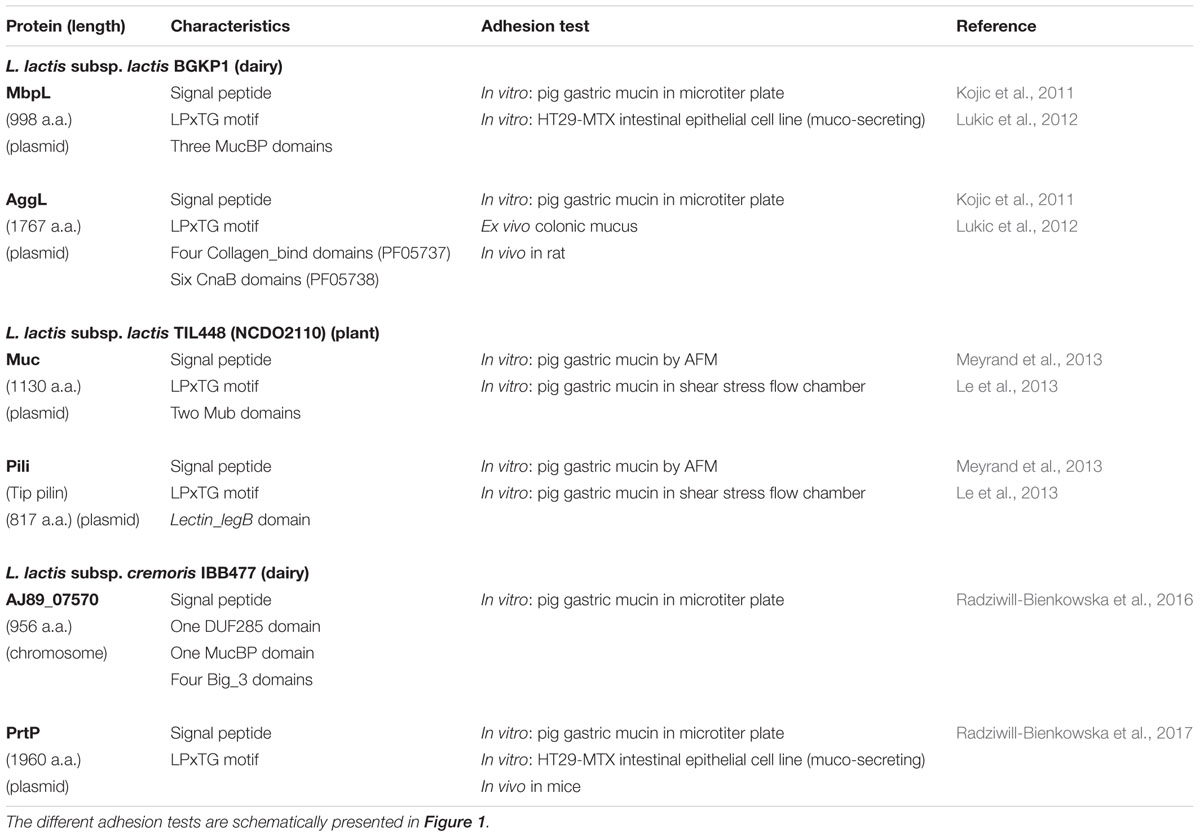
TABLE 1. Surface proteins identified as responsible for muco-adhesive properties in different natural L. lactis strains and corresponding adhesion tests used to probe adhesion to mucus or mucins.
Pili are elongated protein structures of 1–10 nm in diameter and a few μm in length, protruding outside bacterial cells. They were long considered as characteristic of pathogens (Danne and Dramsi, 2012) but have been later discovered in LAB, in probiotic Lactobacillus rhamnosus GG, where they bind human mucins and could explain the strain persistence in human GIT (Kankainen et al., 2009). Pili were also described in L. lactis. Until now, three different gene clusters specifying sortase-dependent heterotrimeric pili have been identified in several L. lactis strains, one chromosomal present in all L. lactis strains (Oxaran et al., 2012) and two plasmid-encoded others in plant (Meyrand et al., 2013) and dairy (Tarazanova et al., 2016) strains respectively. These gene clusters encode three pilins, the major pilin which is polymerized by the sortase C to form the pilus backbone, a minor anchoring pilin and a tip pilin usually endowed with adhesive properties, as well as sortase C (Mandlik et al., 2008; Hendrickx et al., 2011). After synthesis, pili are covalently anchored to peptidoglycan by sortase A (Dieye et al., 2010). Noteworthy, the three identified L. lactis pili gene clusters exhibit limited sequence identity and have different syntheny. The chromosomal pilus gene cluster studied in the laboratory strain IL1403, as well as the plasmid one in L. lactis NCDO712 dairy strain, are not expressed in standard conditions but overexpression allowed pili synthesis (Oxaran et al., 2012; Tarazanova et al., 2016). In contrast, in the plant isolate L. lactis TIL448, the synthesis of pili at the bacterial surface was revealed without overexpression, by a proteomic analysis with the “shaving” approach and the presence of pili was visualized by electron microscopy and atomic force microscopy (AFM). These pili were further shown to contribute, together with the protein Muc, to mucin specific binding (Table 1). Interestingly, the tip pilin is a large protein with an LPxTG motif and containing a lectin domain (PF00139, Lectin_legB domain) that could recognized mucin glycan (Meyrand et al., 2013). Pili were also visualized by electron microcopy in four other L. lactis plant or clinical natural isolates but the genes involved in their synthesis were not identified (Oxaran et al., 2012).
A plasmid-encoded aggregation factor, AggL, was characterized in L. lactis BGKP1. It is a cell-wall anchored protein with a LPxTG motif and seven collagen-binding protein B domains (CnaB domain) and it is responsible for cell surface hydrophobicity and bacterial aggregation phenotype (Kojic et al., 2011). It was shown to contribute to mucus adhesion (Table 1), probably as a result of non-specific hydrophobic interactions with the hydrophobic mucosal surface (Lukic et al., 2012).
The cell-wall anchored proteinase, PrtP, has a crucial role in milk casein degradation required for efficient growth of L. lactis in milk (Siezen, 1999). The presence of PrtP at the surface of L. lactis was shown to modify the cell surface physico-chemical properties, leading to a greater hydrophobicity and increased adhesion to abiotic surfaces (Habimana et al., 2007). In L. lactis IBB477, the cell surface PrtP was shown to contribute to mucin adhesion (Radziwill-Bienkowska et al., 2017) (Table 1).
The different tools and methods used until now to probe adhesion of L. lactis to mucus/mucins are summarized in Figure 1.
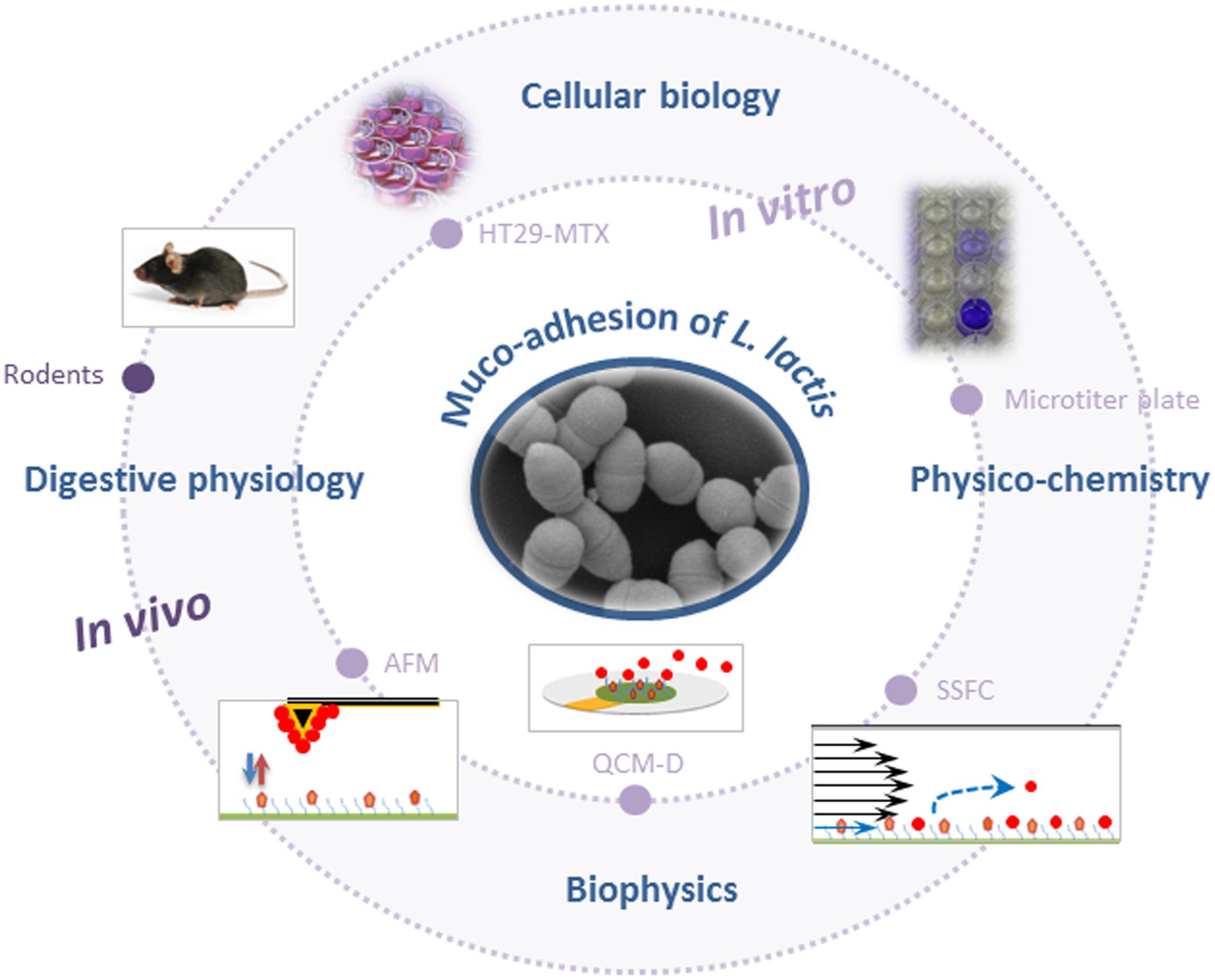
FIGURE 1. Schematic representation of methods and tools used to probe muco-adhesion of Lactococcus lactis (AFM, atomic force microscopy; QCM-D, quartz crystal microbalance with dissipation monitoring; SSFC, shear stress flow chamber). For the sake of clarity, schemes are not to scale.
In vitro models that are most commonly used for evaluation of L. lactis adhesive properties, involve static microtiter plate assays using immobilized commercially available mucin (pig gastric mucin, PGM) and/or gut-related epithelial cell lines (Figure 1). Adherent bacterial cells are quantified using various approaches such as culturing/microscopical enumeration (Kimoto et al., 1999; Meyrand et al., 2013), radiolabeling (Rintahaka et al., 2014), crystal violet staining (Lukic et al., 2012; Radziwill-Bienkowska et al., 2016), or FISH (Radziwill-Bienkowska et al., 2017). The two most commonly used intestinal epithelial cell lines to study LAB adhesion are Caco-2 and HT29, which are originally derived from a human colorectal adenocarcinoma. Their major disadvantage is the lack of substantial mucus production. However, a mucus-secreting subpopulation of goblet cells from HT29 cell population (HT29-MTX) has been obtained after growth adaptation of HT29 cells to anticancer drug methotrexate (MTX) (Lesuffleur et al., 1990) and this cell line is increasingly used to investigate the muco-adhesive properties of L. lactis (Lukic et al., 2012; Radziwill-Bienkowska et al., 2017).
Biophysics-based tools have provided new in vitro insights into the interaction mechanisms between L. lactis and mucins (Figure 1). Interaction forces were quantified at nanoscale by AFM force spectroscopy using a L. lactis-functionalized tip (“lacto-probe”) and PGM-coated surface (Dague et al., 2010). Both non-specific and specific forces (ligand/receptor bonding) were at play in L. lactis adhesion to PGM. For L. lactis IBB477, the percentage of specific adhesive events was high (20%), in comparison with its low-adhesive counterpart L. lactis MG1820 (about 5%) (Le et al., 2011). Furthermore, mucin oligosaccharides were essential in interactions between L. lactis and PGM, as shown with blocking assays. For the first time with living cell probes and mucin, specific interactions were analyzed through kinetic constants: (i) the kinetic dissociation constant Koff was determined with increasing the tip loading rate, which led to a higher adhesion force. The Koff value (0.46 s-1) was consistent with values corresponding to sugar/protein interactions (Le et al., 2011); (ii) the kinetic association constant Kon (3.3 × 102 M-1 s-1) was deduced from enhancing the tip/sample contact time. Furthermore, the high adhesion of IBB477 vs. MG1820 to PGM was confirmed at the multicellular level and under flow field conditions, using real-time quartz crystal microbalance with dissipation monitoring (QCM-D) (Le et al., 2012) and shear stress flow chamber (Radziwill-Bienkowska et al., 2016). Later studies revealed the combined contribution of chromosomal and plasmid-encoded cell-wall proteins (MucBP-Big_3 containing protein and proteinase PrtP respectively) in the muco-adhesive properties of IBB477 (Radziwill-Bienkowska et al., 2016, 2017).
A further work on the plant isolate L. lactis TIL448 enabled to elucidate the L. lactis muco-adhesive phenotype, based on the respective contribution of pili and MUB protein (Le et al., 2013). With AFM force spectroscopy, a high proportion of specific adhesive events to PGM was detected (60%), consistent with the weak shear-flow induced detachment of bacterial cells from the PGM coating. Rupture events were observed in AFM at short (100–200 nm) and long distances (up to 600–800 nm). With AFM force spectroscopy on pili and MUB protein defective mutants, the equivalent role exerted by these two cell surface determinants was established. However, under shear flow, a more critical contribution of MUB protein than pili was found. The importance of pili was further dissected in relation with their nanomechanical properties as probed with optical tweezer (Castelain et al., 2016a,b). AFM blocking assays also revealed that mucin neutral oligosaccharides were involved in adhesion of L. lactis TIL448 to PGM (Le et al., 2013).
A relatively low number of in vivo/ex vivo studies (i.e., biopsy samples) have been conducted to confirm the muco-adhesive phenotype of L. lactis in “real” environmental conditions (Figure 1). Application of such methods seems to be the next step, as shown for lactobacilli (Da Silva et al., 2015; Walsham et al., 2016). Nevertheless, first experimental evidence has been provided for lactococci. In mice, the GFP-labeled L. lactis WH-C1 strain was found to adhere to the gut mucosa (Wang et al., 2011). In another study in rats, in ex vivo and in vivo experiments, AggL protein was found to confer adhesive properties to L. lactis BGKP1 to colonic tissue through non-specific hydrophobic interactions. In contrast, for this particular strain, the MbpL protein did not contribute to bacterial adhesion to colonic tissues but was rather involved in gastric mucin binding (Lukic et al., 2012). More recently, it was shown that the cell-wall proteinase PrtP, albeit contributing to in vitro muco-adhesion of L. lactis IBB477, probably through non-specific interactions, could not confer a selective advantage to this strain in the gut of conventional C57BL/6 mice (Radziwill-Bienkowska et al., 2017).
Certain L. lactis strains synthesize surface proteins with muco-adhesive properties. Although L. lactis is not a natural inhabitant of the mammalian GIT, the main classes of bacterial adhesins, i.e., MUB proteins and pili, allowing mucin glycan recognition, are encoded in the pangenome of the L. lactis species, which includes numerous plasmids. In commensal lactobacilli, MUB proteins and pili promote host mucosae colonization, while in L. lactis in its original ecological niche these proteins might rather play a role in plant tissue colonization. Nevertheless, the lactococcal pangenome appears as a reservoir of novel functions for beneficial gut-targeted activity. Indeed, muco-adhesive L. lactis strains will probably display an augmented fitness in the host GIT, favoring transient colonization, and thus potentially promoting health benefits. Until now, L. lactis muco-adhesion has been mainly studied in vitro with a wide range of tools and approaches on PGM and HT29-MTX cells, that should be completed in the future with in vitro studies on human intestinal mucins and in vivo studies.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ainsworth, S., Stockdale, S., Bottacini, F., Mahony, J., and van Sinderen, D. (2014). The Lactococcus lactis plasmidome: much learnt, yet still lots to discover. FEMS Microbiol. Rev. 38, 1066–1088. doi: 10.1111/1574-6976.12074
Ballal, S. A., Veiga, P., Fenn, K., Michaud, M., Kim, J. H., Gallini, C. A., et al. (2015). Host lysozyme-mediated lysis of Lactococcus lactis facilitates delivery of colitis-attenuating superoxide dismutase to inflamed colons. Proc. Natl. Acad. Sci. U.S.A. 112, 7803–7808. doi: 10.1073/pnas.1501897112
Barinov, A., Loux, V., Hammani, A., Nicolas, P., Langella, P., Ehrlich, D., et al. (2009). Prediction of surface exposed proteins in Streptococcus pyogenes, with a potential application to other Gram-positive bacteria. Proteomics 9, 61–73. doi: 10.1002/pmic.200800195
Bermudez-Humaran, L. G., Aubry, C., Motta, J. P., Deraison, C., Steidler, L., Vergnolle, N., et al. (2013). Engineering lactococci and lactobacilli for human health. Curr. Opin. Microbiol. 16, 278–283. doi: 10.1016/j.mib.2013.06.002
Boekhorst, J., Helmer, Q., Kleerebezem, M., and Siezen, R. J. (2006). Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology 152(Pt 1), 273–280. doi: 10.1099/mic.0.28415-0
Carvalho, R. D. O., do Carmo, F. L. R., de Oliveira Junior, A., Langella, P., Chatel, J. M., Bermudez-Humaran, L. G., et al. (2017). Use of wild type or recombinant lactic acid bacteria as an alternative treatment for gastrointestinal inflammatory diseases: a focus on inflammatory bowel diseases and mucositis. Front Microbiol 8:800. doi: 10.3389/fmicb.2017.00800
Castelain, M., Duviau, M. P., Canette, A., Schmitz, P., Loubiere, P., Cocaign-Bousquet, M., et al. (2016a). The nanomechanical properties of Lactococcus lactis pili are conditioned by the polymerized backbone pilin. PLOS ONE 11:e0152053. doi: 10.1371/journal.pone.0152053
Castelain, M., Duviau, M. P., Oxaran, V., Schmitz, P., Cocaign-Bousquet, M., Loubiere, P., et al. (2016b). Oligomerized backbone pilin helps piliated Lactococcus lactis to withstand shear flow. Biofouling 32, 911–923. doi: 10.1080/08927014.2016.1213817
Chapot-Chartier, M. P., and Kulakauskas, S. (2014). Cell wall structure and function in lactic acid bacteria. Microb. Cell Fact. 13(Suppl. 1), S9. doi: 10.1186/1475-2859-13-S1-S9
Da Silva, S., Robbe-Masselot, C., Raymond, A., Mercade-Loubiere, M., Salvador-Cartier, C., Ringot, B., et al. (2015). Spatial localization and binding of the probiotic Lactobacillus farciminis to the rat intestinal mucosa: influence of chronic stress. PLOS ONE 10:e0136048. doi: 10.1371/journal.pone.0136048
Dague, E., Le, D. T., Zanna, S., Marcus, P., Loubiere, P., and Mercier-Bonin, M. (2010). Probing in vitro interactions between Lactococcus lactis and mucins using AFM. Langmuir 26, 11010–11017. doi: 10.1021/la101862n
Danne, C., and Dramsi, S. (2012). Pili of gram-positive bacteria: roles in host colonization. Res. Microbiol. 163, 645–658. doi: 10.1016/j.resmic.2012.10.012
Derrien, M., and van Hylckama Vlieg, J. E. (2015). Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 23, 354–366. doi: 10.1016/j.tim.2015.03.002
Dieye, Y., Oxaran, V., Ledue-Clier, F., Alkhalaf, W., Buist, G., Juillard, V., et al. (2010). Functionality of sortase A in Lactococcus lactis. Appl. Environ. Microbiol. 76, 7332–7337. doi: 10.1128/AEM.00928-10
Donaldson, G. P., Lee, S. M., and Mazmanian, S. K. (2016). Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32. doi: 10.1038/nrmicro3552
Ermund, A., Schutte, A., Johansson, M. E., Gustafsson, J. K., and Hansson, G. C. (2013). Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am. J. Physiol. Gastrointest. Liver Physiol, 305, G341–G347. doi: 10.1152/ajpgi.00046.2013
Etzold, S., and Juge, N. (2014). Structural insights into bacterial recognition of intestinal mucins. Curr. Opin. Struct. Biol. 28, 23–31. doi: 10.1016/j.sbi.2014.07.002
Etzold, S., Kober, O. I., Mackenzie, D. A., Tailford, L. E., Gunning, A. P., Walshaw, J., et al. (2014). Structural basis for adaptation of lactobacilli to gastrointestinal mucus. Environ. Microbiol. 16, 888–903. doi: 10.1111/1462-2920.12377
Garrigues, C., Johansen, E., and Crittenden, R. (2013). Pangenomics–an avenue to improved industrial starter cultures and probiotics. Curr. Opin. Biotechnol. 24, 187–191. doi: 10.1016/j.copbio.2012.08.009
Gunning, A. P., Kavanaugh, D., Thursby, E., Etzold, S., MacKenzie, D. A., and Juge, N. (2016). Use of atomic force microscopy to study the multi-modular interaction of bacterial adhesins to mucins. Int. J. Mol. Sci. 17, E1854. doi: 10.3390/ijms17111854
Habimana, O., Le Goff, C., Juillard, V., Bellon-Fontaine, M. N., Buist, G., Kulakauskas, S., et al. (2007). Positive role of cell wall anchored proteinase PrtP in adhesion of lactococci. BMC Microbiol. 7:36. doi: 10.1186/1471-2180-7-36
Hendrickx, A. P., Budzik, J. M., Oh, S. Y., and Schneewind, O. (2011). Architects at the bacterial surface - sortases and the assembly of pili with isopeptide bonds. Nat. Rev. Microbiol. 9, 166–176. doi: 10.1038/nrmicro2520
Hill, C. (2012). Virulence or niche factors: what’s in a name? J. Bacteriol. 194, 5725–5727. doi: 10.1128/JB.00980-12
Johansson, M. E., Larsson, J. M., and Hansson, G. C. (2011). The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4659–4665. doi: 10.1073/pnas.1006451107
Juge, N. (2012). Microbial adhesins to gastrointestinal mucus. Trends Microbiol. 20, 30–39. doi: 10.1016/j.tim.2011.10.001
Kankainen, M., Paulin, L., Tynkkynen, S., von Ossowski, I., Reunanen, J., Partanen, P., et al. (2009). Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc. Natl. Acad. Sci. U.S.A. 106, 17193–17198. doi: 10.1073/pnas.0908876106
Kelleher, P., Bottacini, F., Mahony, J., Kilcawley, K. N., and van Sinderen, D. (2017). Comparative and functional genomics of the Lactococcus lactis taxon; insights into evolution and niche adaptation. BMC Genomics 18:267. doi: 10.1186/s12864-017-3650-5
Kelly, W. J., Ward, L. J., and Leahy, S. C. (2010). Chromosomal diversity in Lactococcus lactis and the origin of dairy starter cultures. Genome Biol. Evol. 2, 729–744. doi: 10.1093/gbe/evq056
Kimoto, H., Kurisaki, J., Tsuji, N. M., Ohmomo, S., and Okamoto, T. (1999). Lactococci as probiotic strains: adhesion to human enterocyte-like Caco-2 cells and tolerance to low pH and bile. Lett. Appl. Microbiol. 29, 313–316. doi: 10.1046/j.1365-2672.1999.00627.x
Kline, K. A., Falker, S., Dahlberg, S., Normark, S., and Henriques-Normark, B. (2009). Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5, 580–592. doi: 10.1016/j.chom.2009.05.011
Kojic, M., Jovcic, B., Strahinic, I., Begovic, J., Lozo, J., Veljovic, K., et al. (2011). Cloning and expression of a novel lactococcal aggregation factor from Lactococcus lactis subsp. lactis BGKP1. BMC Microbiol. 11:265. doi: 10.1186/1471-2180-11-265
Laroute, V., Tormo, H., Couderc, C., Mercier-Bonin, M., Le Bourgeois, P., Cocaign-Bousquet, M., et al. (2017). From genome to phenotype: an integrative approach to evaluate the biodiversity of Lactococcus lactis. Microorganisms 5, E27. doi: 10.3390/microorganisms5020027
Larsson, J. M., Karlsson, H., Sjovall, H., and Hansson, G. C. (2009). A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology 19, 756–766. doi: 10.1093/glycob/cwp048
Le, D. T., Guerardel, Y., Loubiere, P., Mercier-Bonin, M., and Dague, E. (2011). Measuring kinetic dissociation/association constants between Lactococcus lactis bacteria and mucins using living cell probes. Biophys. J. 101, 2843–2853. doi: 10.1016/j.bpj.2011.10.034
Le, D. T., Tran, T. L., Duviau, M. P., Meyrand, M., Guerardel, Y., Castelain, M., et al. (2013). Unraveling the role of surface mucus-binding protein and pili in muco-adhesion of Lactococcus lactis. PLOS ONE 8:e79850. doi: 10.1371/journal.pone.0079850
Le, D. T., Zanna, S., Frateur, I., Marcus, P., Loubiere, P., Dague, E., et al. (2012). Real-time investigation of the muco-adhesive properties of Lactococcus lactis using a quartz crystal microbalance with dissipation monitoring. Biofouling 28, 479–490. doi: 10.1080/08927014.2012.688103
Le Marechal, C., Peton, V., Ple, C., Vroland, C., Jardin, J., Briard-Bion, V., et al. (2015). Surface proteins of Propionibacterium freudenreichii are involved in its anti-inflammatory properties. J. Proteomics 113, 447–461. doi: 10.1016/j.jprot.2014.07.018
Lesuffleur, T., Barbat, A., Dussaulx, E., and Zweibaum, A. (1990). Growth adaptation to methotrexate of HT-29 human colon carcinoma cells is associated with their ability to differentiate into columnar absorptive and mucus-secreting cells. Cancer Res. 50, 6334–6343.
Linden, S. K., Sutton, P., Karlsson, N. G., Korolik, V., and McGuckin, M. A. (2008). Mucins in the mucosal barrier to infection. Mucosal Immunol. 1, 183–197. doi: 10.1038/mi.2008.5
Luerce, T. D., Gomes-Santos, A. C., Rocha, C. S., Moreira, T. G., Cruz, D. N., Lemos, L., et al. (2014). Anti-inflammatory effects of Lactococcus lactis NCDO 2118 during the remission period of chemically induced colitis. Gut Pathog. 6, 33. doi: 10.1186/1757-4749-6-33
Lukic, J., Strahinic, I., Jovcic, B., Filipic, B., Topisirovic, L., Kojic, M., et al. (2012). Different roles for lactococcal aggregation factor and mucin binding protein in adhesion to gastrointestinal mucosa. Appl. Environ. Microbiol. 78, 7993–8000. doi: 10.1128/AEM.02141-12
MacKenzie, D. A., Tailford, L. E., Hemmings, A. M., and Juge, N. (2009). Crystal structure of a mucus-binding protein repeat reveals an unexpected functional immunoglobulin binding activity. J. Biol. Chem. 284, 32444–32453. doi: 10.1074/jbc.M109.040907
Mandlik, A., Swierczynski, A., Das, A., and Ton-That, H. (2008). Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 16, 33–40. doi: 10.1016/j.tim.2007.10.010
Meyrand, M., Guillot, A., Goin, M., Furlan, S., Armalyte, J., Kulakauskas, S., et al. (2013). Surface proteome analysis of a natural isolate of Lactococcus lactis reveals the presence of pili able to bind human intestinal epithelial cells. Mol. Cell. Proteomics 12, 3935–3947. doi: 10.1074/mcp.M113.029066
Moonens, K., and Remaut, H. (2017). Evolution and structural dynamics of bacterial glycan binding adhesins. Curr. Opin. Struct. Biol. 44, 48–58. doi: 10.1016/j.sbi.2016.12.003
Nishitani, Y., Tanoue, T., Yamada, K., Ishida, T., Yoshida, M., Azuma, T., et al. (2009). Lactococcus lactis subsp. cremoris FC alleviates symptoms of colitis induced by dextran sulfate sodium in mice. Int. Immunopharmacol. 9, 1444–1451. doi: 10.1016/j.intimp.2009.08.018
Olaya-Abril, A., Jimenez-Munguia, I., Gomez-Gascon, L., and Rodriguez-Ortega, M. J. (2014). Surfomics: shaving live organisms for a fast proteomic identification of surface proteins. J. Proteomics 97, 164–176. doi: 10.1016/j.jprot.2013.03.035
Ouwerkerk, J. P., de Vos, W. M., and Belzer, C. (2013). Glycobiome: bacteria and mucus at the epithelial interface. Best Pract. Res. Clin. Gastroenterol. 27, 25–38. doi: 10.1016/j.bpg.2013.03.001
Oxaran, V., Ledue-Clier, F., Dieye, Y., Herry, J. M., Pechoux, C., Meylheuc, T., et al. (2012). Pilus biogenesis in Lactococcus lactis: molecular characterization and role in aggregation and biofilm formation. PLOS ONE 7:e50989. doi: 10.1371/journal.pone.0050989PONE-D-12-23876
Popowska, M., Krawczyk-Balska, A., Ostrowski, R., and Desvaux, M. (2017). InlL from listeria monocytogenes is involved in biofilm formation and adhesion to mucin. Front. Microbiol. 8:660. doi: 10.3389/fmicb.2017.00660
Radziwill-Bienkowska, J. M., Le, D. T., Szczesny, P., Duviau, M. P., Aleksandrzak-Piekarczyk, T., Loubiere, P., et al. (2016). Adhesion of the genome-sequenced Lactococcus lactis subsp. cremoris IBB477 strain is mediated by specific molecular determinants. Appl. Microbiol. Biotechnol. 100, 9605–9617. doi: 10.1007/s00253-016-7813-0
Radziwill-Bienkowska, J. M., Robert, V., Drabot, K., Chain, F., Cherbuy, C., Langella, P., et al. (2017). Contribution of plasmid-encoded peptidase S8 (PrtP) to adhesion and transit in the gut of Lactococcus lactis IBB477 strain. Appl. Microbiol. Biotechnol. 101, 5709–5721. doi: 10.1007/s00253-017-8334-1
Rintahaka, J., Yu, X., Kant, R., Palva, A., and von Ossowski, I. (2014). Phenotypical analysis of the Lactobacillus rhamnosus GG fimbrial spaFED operon: surface expression and functional characterization of recombinant SpaFED pili in Lactococcus lactis. PLOS ONE 9:e113922. doi: 10.1371/journal.pone.0113922
Roos, S., and Jonsson, H. (2002). A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148(Pt 2), 433–442. doi: 10.1099/00221287-148-2-433
Siezen, R. J. (1999). Multi-domain, cell-envelope proteinases of lactic acid bacteria. Antonie Van Leeuwenhoek 76, 139–155. doi: 10.1023/A:1002036906922
Tarazanova, M., Beerthuyzen, M., Siezen, R., Fernandez-Gutierrez, M. M., de Jong, A., van der Meulen, S., et al. (2016). Plasmid complement of Lactococcus lactis NCDO712 reveals a novel pilus gene cluster. PLOS ONE 11:e0167970. doi: 10.1371/journal.pone.0167970
Thomsson, K. A., Holmen-Larsson, J. M., Angstrom, J., Johansson, M. E., Xia, L., and Hansson, G. C. (2012). Detailed O-glycomics of the Muc2 mucin from colon of wild-type, core 1- and core 3-transferase-deficient mice highlights differences compared with human MUC2. Glycobiology 22, 1128–1139. doi: 10.1093/glycob/cws083
Veiga, P., Pons, N., Agrawal, A., Oozeer, R., Guyonnet, D., Brazeilles, R., et al. (2014). Changes of the human gut microbiome induced by a fermented milk product. Sci. Rep. 4:6328. doi: 10.1038/srep06328
Walsham, A. D., MacKenzie, D. A., Cook, V., Wemyss-Holden, S., Hews, C. L., Juge, N., et al. (2016). Lactobacillus reuteri inhibition of enteropathogenic Escherichia coli adherence to human intestinal epithelium. Front. Microbiol. 7:244. doi: 10.3389/fmicb.2016.00244
Wang, Y., Wang, J., and Dai, W. (2011). Use of GFP to trace the colonization of Lactococcus lactis WH-C1 in the gastrointestinal tract of mice. J. Microbiol. Methods 86, 390–392. doi: 10.1016/j.mimet.2011.06.009
Zhang, C., Derrien, M., Levenez, F., Brazeilles, R., Ballal, S. A., Kim, J., et al. (2016). Ecological robustness of the gut microbiota in response to ingestion of transient food-borne microbes. ISME J. 10, 2235–2245. doi: 10.1038/ismej.2016.13
Keywords: lactic acid bacteria, Lactococcus lactis, mucus, adhesion, surface proteins, pili, mucus-binding protein
Citation: Mercier-Bonin M and Chapot-Chartier M-P (2017) Surface Proteins of Lactococcus lactis: Bacterial Resources for Muco-adhesion in the Gastrointestinal Tract. Front. Microbiol. 8:2247. doi: 10.3389/fmicb.2017.02247
Received: 12 July 2017; Accepted: 31 October 2017;
Published: 23 November 2017.
Edited by:
Thomas Dandekar, University of Würzburg, GermanyReviewed by:
Antonio Colantuoni, University of Naples Federico II, ItalyCopyright © 2017 Mercier-Bonin and Chapot-Chartier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marie-Pierre Chapot-Chartier, bWFyaWUtcGllcnJlLmNoYXBvdC1jaGFydGllckBpbnJhLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.