
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 13 November 2017
Sec. Microbial Physiology and Metabolism
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.02205
This article is part of the Research Topic Actinobacteria, a Source of Biocatalytic Tools View all 19 articles
The complex development undergone by Streptomyces encompasses transitions from vegetative mycelial forms to reproductive aerial hyphae that differentiate into chains of single-celled spores. Whereas their mycelial life – connected with spore formation and antibiotic production – is deeply investigated, spore germination as the counterpoint in their life cycle has received much less attention. Still, germination represents a system of transformation from metabolic zero point to a new living lap. There are several aspects of germination that may attract our attention: (1) Dormant spores are strikingly well-prepared for the future metabolic restart; they possess stable transcriptome, hydrolytic enzymes, chaperones, and other required macromolecules stabilized in a trehalose milieu; (2) Germination itself is a specific sequence of events leading to a complete morphological remodeling that include spore swelling, cell wall reconstruction, and eventually germ tube emergences; (3) Still not fully unveiled are the strategies that enable the process, including a single cell’s signal transduction and gene expression control, as well as intercellular communication and the probability of germination across the whole population. This review summarizes our current knowledge about the germination process in Streptomyces, while focusing on the aforementioned points.
Soil microorganisms are exposed to periodic nutrient exhaustions and various abiotic and biotic stresses that inhibit growth. An important survival strategy for many bacteria and fungi in the face of such physiological stresses is the cells’ transition into a dormant state. In the dormant state, cells arrest their growth, discontinue replication and transform themselves into metabolically inactive (or with limited activity), widely resistant forms. Several bacterial clades living in soil, such as Actinomyces, Streptomyces, and Micromonospora, differentiate into dormant fungi-like uninucleoid spores (arthrospores or exospores). Arthrospores significantly differ from the endospores of Bacilli and Clostridia in morphology and function. The endospores exhibit striking resistance to a wide range of environmental stresses, such as heat, desiccation, and ultraviolet radiation (Setlow, 2007; Galperin et al., 2012). Their abilities to survive harsh conditions result from multi-layered surface structures and extremely low water content (Setlow et al., 2006; Henriques and Moran, 2007). The well-studied endospores of Bacillus subtilis contain high levels of dipicolinic acid in chelation with divalent cations (Ca2+) and extremely stable small spore proteins (Setlow et al., 2006). All these features contribute to the overall robust endospore resistance. The streptomycete arthrospores, on the other hand, are more reminiscent of the spores of eukaryotic fungi, possibly due to their convergent evolution in the soil environment. Penicillium species, for example, produce conidiophores (analogous to streptomycete aerial hyphae, see below) that bear individually constricted conidiospores (Foster et al., 1945). Both, streptomycetes and molds produce large numbers of small hydrophobic spores with similar properties; both contain, for example, a relatively thick coat (Briza et al., 1990; Pammer et al., 1992; Neiman, 2005), protective small molecules including sugars (such as trehalose, see below), and heat shock proteins (Wyatt et al., 2013). Spores of these organisms not only guard genetic information during unfavorable conditions, but are also adapted to wind dispersal and may remain airborne for long periods.
Their complex life cycle and the described similarities with eukaryotic fungi make streptomycetes unique organisms within the bacterial kingdom. However, comprehensive information about the diverse features of spore germination in this antibiotic-producing clade is still missing. This review attempts to fill this gap.
The process of germination, when either considering the germination rate of a single spore or the probability of germination within a spore population, varies between different Streptomyces. Some species (mainly S. viridochromogenes and S. granaticolor) exhibit fast and robust germination with nearly all spores germinating. That is also why this organism was subjected to many initial germination experiments (Hirsch and Ensign, 1976a,b; Mikulik et al., 1977; Bobek et al., 2004; Xu and Vetsigian, 2017). In contrast, other species (e.g., S. coelicolor and S. venezuelae) germinate more slowly with a fraction of spores that does not germinate at all. These exhibit more complex germination behavior and possess a fraction of germlings that stop growing soon after germination, probably due to produced inhibitory compounds, such as germicidins and hypnosins, that affect their development (Petersen et al., 1993; Aoki et al., 2007, 2011; Ma et al., 2017; Xu and Vetsigian, 2017). Over time, S. coelicolor became a widely used model streptomycetes whose genome was the first to be resolved and best annotated within the genus (Bentley et al., 2002). Therefore, the later genome-wide expression analyses of germinating spores have also been conducted on this stain (Strakova et al., 2013a,b, 2014; Bobek et al., 2014). Hence, most of the findings mentioned here come from the experimental results obtained with S. coelicolor.
Our knowledge of the complex life of Streptomyces with a certain emphasis on the process of sporulation and spore maturation (together here called as sporogenesis) offers many clues for understanding the readiness of spores for successful germination. The life cycle of Streptomyces, similarly as in the other arthrospore-forming bacteria, encompasses a development of a network of branched hyphae that grow into the substrate thus creating a vegetative mycelium. The subsequent development of aerial hyphae and spores is considered to be the cell’s response to nutrient depletion. The sporulation-specific regulon is activated after repression of sporulation-specific genes by the BldD-c-di-GMP complex is released (Tschowri et al., 2014; Bush et al., 2015). A part of the vegetative mycelium lyses to be used as a surrogate of nutrients. During this developmental phase the activity of secondary metabolism reaches its maximum to synthesize various bioactive compounds, including antibiotics, in its effort to avoid competitive organisms.
The aerial mycelium of Streptomyces is covered with a rodlet layer, a network of pairwise amyloid fibrils (Wildermuth et al., 1971) that consist of chaplin and rodlin proteins (Claessen et al., 2004). This hydrophobic structure lowers the water surface tension thereby enabling the hyphae to grow apically into the air (Elliot and Talbot, 2004; Claessen et al., 2006). The erected sporogenic hyphae are dissected by sporulation septa. The sporulation septation is synchronized with a segregation of chromosomes thus forming unigenomic pre-spore compartments (Ausmees et al., 2007; Wang et al., 2007; Jakimowicz and van Wezel, 2012; Ditkowski et al., 2013). This process is controlled by a family of SsgA-like proteins (SALP) found exclusively in differentiating actinomycetes (Sigle et al., 2015). Their member SsgA is a protein essential for sporulation in S. coelicolor; in concert with SsgB it dynamically controls the assembly of FtsZ rings at septation sites (van Wezel et al., 2000; Keijser et al., 2003; Sevcikova and Kormanec, 2003; Willemse et al., 2011). The role of SsgA is related to the de novo peptidoglycan synthesis; SsgA has been suggested to mark future germination sites and when the protein is overproduced, individual spores generate many germ tubes. SsgC, whose deletion leads to hypersporulation, has been suggested to function as an antagonist of SsgA. The SALP proteins also include SsgD that ensures the development of the thick spore wall and SsgG that controls regular localization of division sites (Noens et al., 2005, 2007; Traag and van Wezel, 2008).
The following process of spore maturation is the transition of the dissected aerial hyphae into the mature ovoid spores. Not much is known about this phase. When spores are only partially mature (harvested at an earlier stage of sporulation), they germinate remarkably faster and more synchronously. On the other hand, the fully maturated spores exhibit less viability and longer lag-phase in conditions suitable for germination (Hirsch and Ensign, 1976a). These differences hint at further but not yet fully understood processes associated with spore maturation, presumably involving the stabilization of nucleic acids and proteins, deactivation of the metabolic apparatus, desiccation, and changes in the cell wall structure. The spore’s wall thickening is dependent on the so called Streptomyces-spore-wall-synthesizing complex (SSSC), a system somewhat similar to the elongasome of rod-shaped bacteria (Kleinschnitz et al., 2011). Members involved in this complex are the cytoskeletal actin-like proteins, MreB and Mbl that cooperate in spore wall synthesis (Mazza et al., 2006; Heichlinger et al., 2011). Maturation is further accompanied by DNA condensation and spore pigmentation, such as the production of gray spore pigment in S. coelicolor (Kelemen et al., 1998). After maturation, the rodlet layer forms a thin, basket-like fibrous sheath around the thickened spore wall (Claessen et al., 2002, 2003; Elliot et al., 2003) and cell wall hydrolases under control of other SALP members, SsgE and SsgF, separate individual spores allowing for their dispersal into the environment (Traag and van Wezel, 2008; Flardh and Buttner, 2009; Haiser et al., 2009).
Dormant spores of streptomycetes are the only haploid state in the development of this multicellular mycelial bacterium. Their main function is to safeguard genetic information throughout unfavorable conditions and consequently spread it into new niches. All the macromolecules that are needed for the future launch of germination have to be pre-synthesized before dormancy. Yet, it remains unclear whether dormant spores possess none or limited metabolism. Despite the commonly held belief that dormant spores are metabolically inactive, Liot and Constant suggested some metabolic activity of dormant spores as their experiments showed that mature spores of S. avermitilis (suspended in Tris-HCl) are able to oxidize atmospheric H2 to supply maintenance energy (Constant et al., 2008; Liot and Constant, 2016). A fascinating example of metabolically active spores comes from another soil-inhabiting actinomycete genus, Actinoplanes, which produces flagellated spores (zoospores) that guided by chemotaxis (sugars, amino acids, aromatic compounds, and mineral ions) are temporarily able to rapidly swim (Palleroni, 1976; Jang et al., 2016).
Generally, other than the thick cell wall, the lack of water in spores ensures their resistance to thermal extremities and to other physical and chemical effects (Kalakoutskii and Agre, 1976). On the other hand, the dehydrated state results in immobility and changed conformation of macromolecules; the condensation of nucleic acids and an inactive form of proteins. The endospores of Bacilli contain a significant amount (20%) of dipicolinic acid (Setlow et al., 2006) that stabilizes their macromolecule structures under dehydrated conditions. In the spores of streptomycetes that lack the Ca-dipicolinate, the stabilization and protection of macromolecules is provided by trehalose, instead. During spore resuscitation, the protective role is transferred to present protein chaperones that help other proteins reobtain their functional conformation (Bobek et al., 2004).
Spores subjected to germination in the presence of the antibiotic rifampicin that inhibits the initiation of transcription were still able to synthesize proteins (termed early proteins) over the experimental period. This result suggested that there is also a pool of stable mRNAs, functional ribosomes, and translational apparatus that have been preserved during spore maturation (Mikulik et al., 2002).
In addition, preserved hydrolytic enzymes are accessible for the initial reconstruction of the cell wall (Haiser et al., 2009). In S. coelicolor, several hydrolases that might take their part in the cell envelope reconstruction (SCO1061, SCO1725, SCO3487, and SCO5466) were shown to be stored throughout spore dormancy and not synthesized de novo during germination (Strakova et al., 2013a). This suggests that cells possess the required enzymatic equipment from the sporulation and/or spore maturation period.
While common opinion has, until recent times, stipulated that spores require external nutrients to launch germination (Flardh and Buttner, 2009), B. subtilis spores were recently shown to germinate even in an environment lacking all germination factors (Sturm and Dworkin, 2015; van Vliet, 2015). The launch of germination was stochastic (discussed below) and depended on the level of a transcription factor involved in spore assembly. In the case of Streptomyces, the probability of germination was suggested to be dependent on an insoluble protein NepA (de Jong et al., 2009). NepA is a structural part of the streptomycete spore wall which was shown to maintain the spore’s dormancy. In the absence of this protein spores germinate faster and more synchronously. Even spores lacking NepA protein were able to produce hyphae (after 3 days) and form mycelial clumps (after 7 days of cultivation) when cultivated in water (de Jong et al., 2009). This would not be possible if the spores did not possess intracellular nutrient sources, such as trehalose (Ranade and Vining, 1993) and polyphosphates (volutin) (Ghorbel et al., 2006; Strakova et al., 2013b) that promote the metabolic restart before the external sugar can be sensed and assimilated. Various strains of Streptomyces have been reported to contain high trehalose levels in their spores. For example in the glucose excess, spores of S. griseus accumulate trehalose that comprises up to 25% of their dry weight (McBride and Ensign, 1987a). Trehalose is also abundant in S. venezuelae spores, whereas glycogen and polyhydroxybutyrate are absent, which stands in contrast to vegetative hyphae (Ranade and Vining, 1993). It has been shown that sporogenic hyphae convert present glycogen to trehalose during the final period of spore maturation in S. brasiliensis (Rueda et al., 2001).
Trehalose is a disaccharide consisting of two glucose molecules that are bound together in α, α-1,1-glycosidic linkage. Trehalose associates in clusters resulting in a large, continuous aggregates formation (Sapir and Harries, 2011). According to the water displacement theory, trehalose present in a system replaces water under desiccation conditions (Sola-Penna and Meyer-Fernandes, 1998). Trehalose is present in various organisms; from Archaea to animals where it may serve as a source of carbon and energy or act as a signaling molecule (Elbein et al., 2003). The unique α, α-1,1-glycosidic linkage makes trehalose non-reducing and therefore unreactive, making it the best intracellular macromolecules stabilizer. Thanks to its ability to make hydrogen bonds with membranes and nucleic acids, as well as its ability to modify the solvation layer of proteins (Sola-Penna and Meyer-Fernandes, 1998), trehalose is capable of protecting cellular membranes, DNA, enzymes (though in their inactive forms), and even whole microorganisms (Yoshinaga et al., 1997; Kandror et al., 2002; Jain and Roy, 2010). The sugar may thus help the cells cope with heat, freezing, desiccation, radiation, and oxidative stresses (Hottiger et al., 1989; Wiemken, 1990; De Virgilio et al., 1994; Yoshinaga et al., 1997; An et al., 2000; Benaroudj et al., 2001; Fillinger et al., 2001). Its multi-protective role is of special need in dormant stages; the sugar is thought to form a gel phase in anhydrous conditions, which prevents aggregation of macromolecules. Subsequent rehydration allows normal restoration of cell metabolism. Consequently, trehalose, as a protective sugar, is quite abundant in bacterial cells, such as those of Mycobacterium, including M. smegmatis and M. tuberculosis (Elbein and Mitchell, 1973), in spores of Myxococcus (McBride and Zusman, 1989) and Streptomyces (Martin et al., 1986), or in spores of eukaryotic yeast and fungi (Trevelyan and Harrison, 1956; Nwaka and Holzer, 1998). Trehalose is also located inside mycobacterial and corynebacterial cell wall structures (Lederer, 1976).
Minimal or no metabolic activity enables the spores to survive harsh conditions such as high temperatures or the presence of antibiotics and thus stay persevered for years. To awake, spores require at least aqueous conditions (Hirsch and Ensign, 1976b). Germination is faster and more convenient, however, in the presence of required nutrients and other germination stimuli (Paredes-Sabja et al., 2011). Despite this, the scattering remains high in the germination rate within a single population. This being the case, Jyothikumar et al. (2008) showed by means of time-lapse microscopy that germination tubes occurred between 3.75 and 7.5 h in their experimental conditions. The known stimuli that consequently lead to a more synchronous development are heat shock (Hirsch and Ensign, 1976a), mechanical disruption of the spore envelopes (Mikulik et al., 1977; Stastna, 1977; Miguelez et al., 1993), or the presence of peptidoglycan residues (Shah et al., 2008). But how may metabolically inert spores detect optimal environmental conditions? As was shown in the case of the B. subtilis spores, germination is launched stochastically, though at much lower frequency in poor environments (Paidhungat and Setlow, 2000; Epstein, 2009; Sturm and Dworkin, 2015). If the environmental conditions remain adverse, for example because of the lack of nutrients, the germinating spore will eventually die (van Vliet, 2015). Thus spores of Saccharomyces cerevisiae, an eukaryotic yeast, that launch germination in a poor glucose solution, without any additional nutrients, are not able to further develop bud emergences in these conditions (Chen et al., 2000). In the optimal environment, however, spores may even stimulate each other’s development. In addition to the spore-forming Streptomyces (Xu and Vetsigian, 2017) and Bacilli (Chen et al., 2006; Sturm and Dworkin, 2015; van Vliet, 2015), the stochastic development from a dormant state was observed in non-sporulating Escherichia coli and M. smegmatis as well (Balaban et al., 2004; Buerger et al., 2012). Such a strategy where successful pioneers wake other fellows up in a favorable environment is probably advantageous and cost-effective from the perspective of the whole population.
Germination is associated with a complete reconstruction of the cell that involves highly accelerating metabolic activity, morphological changes that start with uncoating, and reconstitution of the cellular content. Spore germination is a sequential process and thus can be divided into three distinctive steps (defined by Hardisson et al., 1978): darkening, swelling, and germ tube emergence (Figure 1).
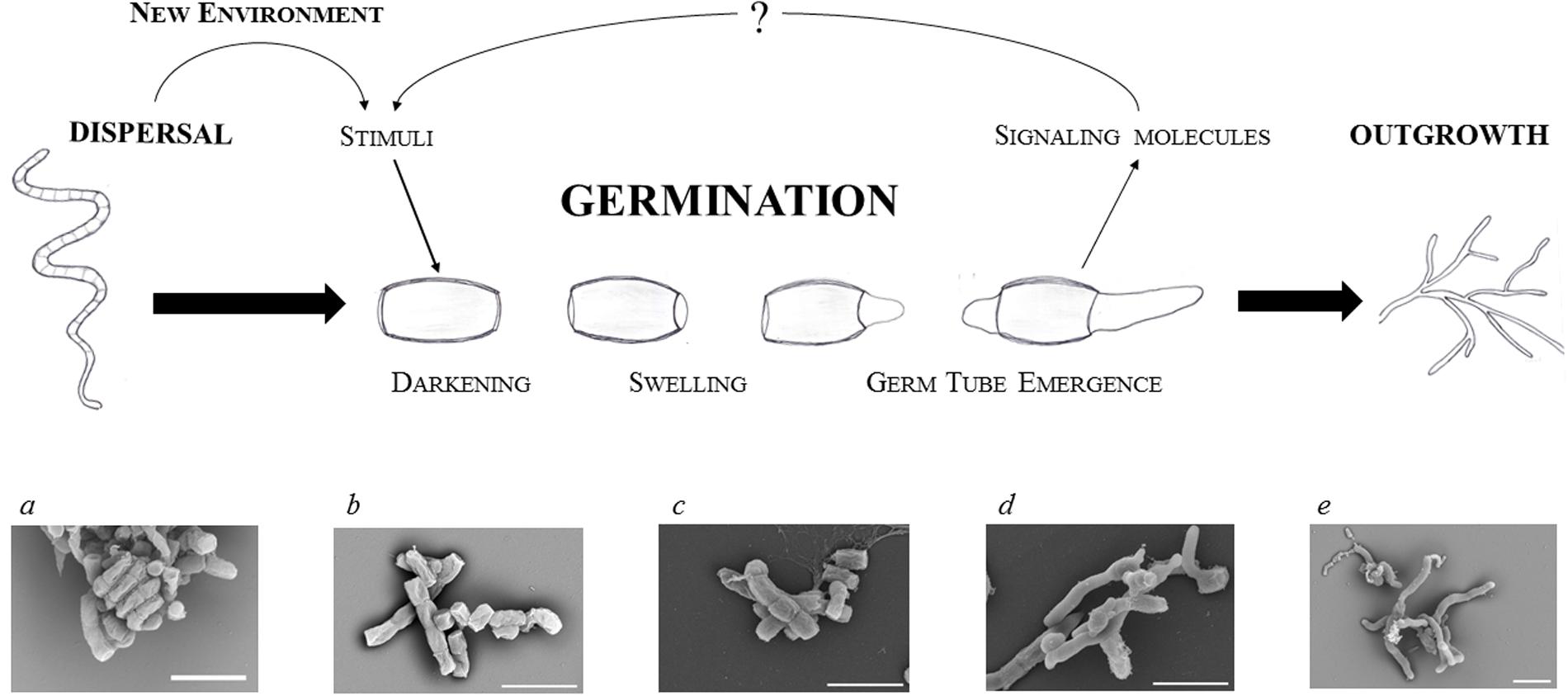
FIGURE 1. Focus on streptomycete spore germination. Schematic illustration of a germinating spore and electron microscopic images of dormant (a,b) and germinating (c–e) spores of S. coelicolor: residue of an aerial hypha with a twisted chain of matured spores (a); dormant spores (b); time-lapse imaging of germinating spores after 2, 4, and 6 h of cultivation (c–e, respectively) in a complex medium (for details see Čihák et al., submitted). The white bars indicate 2.5 μm.
(a) Darkening is associated with cell wall reconstruction
The first step could be described as a transition from pure physical processes, such as osmosis, that progressively activate initial biochemical activities requiring energy sources. During this step spores lose their hydrophobicity resulting in water influx, swelling, and the subsequent loss of heat resistance. These processes change the optical features of spores and allow the cells to re-activate metabolism within the only several minutes after germination initiation. The darkening is caused by the loss of light refraction which requires bivalent cations Ca2+, Mg2+, Mn2+, Zn2+, and Fe2+ (Hardisson et al., 1978; Eaton and Ensign, 1980; Salas et al., 1983). The binding sites for calcium and magnesium are provided by both, the carboxyl units of peptidoglycan and the polyphosphate groups of teichoic acid in the spore wall (Thomas and Rice, 2014). Relatively early the spores undergo an uncoating, during which calcium accumulated in the spore envelope is gradually released (Eaton and Ensign, 1980; Salas et al., 1983). This process is mediated by a calcium-binding protein CabC whose disruption leads to prematurely germinating spores on the spore chain, while an overexpression of the gene delays their development (Wang et al., 2008). During uncoating, spore lysozyme-like hydrolases are reactivated and facilitate changes in spore morphology and optical features. These enzymes provide lysis and facilitate the reconstruction of cell wall peptidoglycan to allow the entrance of external nutrients. Mutants of two cell wall hydrolases, RpfA and SwlA, exhibited slower germination (Haiser et al., 2009). The hydrolases cleave covalent bonds inside the peptidoglycan layer; an act that must be well-coordinated with the new cell wall synthesis. RpfA belongs to a class of so called resuscitation-promoting factors, Rpf, a subgroup of lysozyme-like transglycosylases, which hydrolyze the beta-(1,4)-glycosidic bond between N-acetylglucosamine and N-acetylmuramic acid (Mukamolova et al., 1998b, 2002, 2006; Cohen-Gonsaud et al., 2004; Keep et al., 2006; Telkov et al., 2006; Hett et al., 2008; Haiser et al., 2009; Ruggiero et al., 2009). These muralytic enzymes are known to participate in the restoration of active growth from dormancy not only in Streptomyces but also in other actinobacteria, like Mycobacterium and Micrococcus (Mukamolova et al., 1998a, 2006; Keep et al., 2006). S. coelicolor possesses five Rpf proteins (RpfA to RfpE) (Haiser et al., 2009) whose individual mutants delay germination to varying extents; with RpfA and RfpE having a stronger effect than RpfC and RpfD. This differs to what can be observed in the wild type (Sexton et al., 2015). Although a multiple rpf mutant (with deletion of all five rpf genes) revealed a highly impaired germination, the strain was able to germinate within 12 h. This suggests that the Rpf hydrolase family is highly important but not essential for a smooth germination. The Rpfs family could probably be substituted by other cell wall hydrolases. Three Streptomyces cell wall lytic enzymes (SwlA to SwlC) are examples of such possible candidates that may be responsible for the proper thickness of the dormant spore wall ensuring its heat resistance (Haiser et al., 2009). In this reference one may find an overview of predicted streptomycete cell wall hydrolase genes as well.
(b) Enzymatic activities are recovered during swelling
Continual influx of water causes spores to swell. A dramatic intracellular decline of the trehalose level has been observed in S. griseus during spore swelling (McBride and Ensign, 1987b, 1990). A similar effect was also observed in M. smegmatis (Shleeva et al., 2017), Neurospora (Sussman and Lingappa, 1959; Sussman, 1961), and Dictyostelium discoideum (Roth and Sussman, 1966; Ceccarini, 1967). Consequently, the glucose level increases in swelled spores, suggesting a restored activity of the trehalase enzyme due to an increased concentration of intracellular ATP (McBride and Ensign, 1987b). The hydrolysis of trehalose appears to be an essential step in spore germination (Nwaka and Holzer, 1998). Based on observations of the yeast S. cerevisiae, trehalase possesses an inactive form which is activated by phosphorylation in a cAMP-dependent manner (Thevelein, 1984). The restored glucose subsequently provides an internal energy source for the revived enzymes and initial development. Only after the intracellular concentration of trehalose declines do the protected proteins refold into their active forms with the help of their respective chaperones (Singer and Lindquist, 1998).
The hydration of the cytoplasm supports the re-activation of assorted proteins and ribosomes that are preserved from dormancy (Cowan et al., 2003). Bound chaperones GroEL, Trigger factor and DnaK assist in the reactivation of the proteosynthetic apparatus and nascent proteins (Bobek et al., 2004). These chaperones as well as others (GrpE and peptidyl-prolyl cis–trans isomerases) are constitutively expressed throughout the germination course suggesting that their presence is constantly necessary (Strakova et al., 2013a). Within several minutes of the germination starting, the ribosomes present are fully functional and new proteins are translated from the stable mRNA stock (Strakova et al., 2013b). The proteosynthesis accelerates between 30 and 60 min (Strakova et al., 2013a). At this stage spores are metabolically active and able to utilize primarily internal energy sources, such as trehalose (see below; Elbein, 1974; Crowe et al., 1984), to obtain carbon, nitrogen and energy (Hey-Ferguson et al., 1973; McBride and Ensign, 1987a). The obtained energy is exploited not only for the intensive proteosynthesis (Mikulik et al., 1984, 2008, 2011; Paleckova et al., 2006) but also for the first DNA replication, which occurs just before or simultaneously with the germ tube emergence within about 60 min of germination in S. granaticolor (Mikulik et al., 1977) or 30–60 min later in S. coelicolor (Ruban-Osmialowska et al., 2006; Wolanski et al., 2011). Later the spores are able to detect external nutrient sources and as a response they adjust metabolic pathways. They are capable of this due to the activity of present pleiotropic gene expression regulators acting on both transcriptional (BldD, cyclic AMP-receptor protein Crp, and sigma and anti-sigma factors) and translational (ribonuclease RNase III) levels, as was shown in S. coelicolor (Strakova et al., 2013a).
(c) The vegetative growth starts with germ tube emergence
The eventual germ tube emergence is a microscopically observable event that provides the basis for the apically growing hyphae. The not yet septated tubes rise from the inner wall of spore (Glauert and Hopwood, 1961) and progress through the outer rodlet layer. An important role is carried out by the chaperonin-like protein SsgA that localizes sites of germ tube emergences. This protein is located at the sites of the germ tip appearance (Noens et al., 2007). In S. coelicolor one or two germination tubes appear after 180 min of germination and elongate by apical tip extension. At this developmental point two or three DNA replisomes were demonstrated (Ruban-Osmialowska et al., 2006; Wolanski et al., 2011). The critical determinant of the vegetative growth is the DivIVA protein that localizes at hyphal tips ensuring a new cell wall outgrowth (Flardh, 2003a; Flardh et al., 2012; Hempel et al., 2012; Richards et al., 2012). Interestingly, DivIVA is expressed from early on after spore hydration and its synthesis increases during the whole process of germination (Strakova et al., 2013a). As was also shown, cell division-associated protein FtsZ (SCO2082, Grantcharova et al., 2005), which is the BldD target, and cell growth-associated protein FilP (SCO5396, Flardh, 2003b; Bagchi et al., 2008) are expressed at 300 min of S. coelicolor’s germination. FilP protein, which is associated with DivIVA, accumulates in the tips of young hyphae and is also expressed soon after germination starts (Strakova et al., 2013a). FilP resemble eukaryotic intermediate filaments forming a tangled cytoskeletal network that confers to rigidity and elasticity of hyphae (Kelemen, 2017). The onset of these proteins was previously proposed as the point when the germination phase finalizes (Strakova et al., 2013a).
Several genome-wide studies on gene expression during the streptomycete spore germination have recently been conducted (Bobek et al., 2004, 2014; Piette et al., 2005; Strakova et al., 2013a,b). Furthermore, a biocomputational modeling (Strakova et al., 2013a,b, 2014) was employed to obtain a global view on protein synthesis and to characterize the regulatory networks. The expression profiles of individual proteins from 13 time points throughout the course of germination have been sorted and divided among differential functional groups. A study analyzing the protein synthesis changes between consecutive time intervals revealed that the transcriptional apparatus is indeed highly active from the first 30 min of germination but surprisingly, most of the mRNAs preserved from dormancy are consequently degraded during the rehydration (Strakova et al., 2013a; Bobek et al., 2014). The degradation affects mRNAs from the majority of functional protein groups, i.e., amino acid, carbohydrate, lipid and energy metabolism, proteasomes, and DNA repair pathways. This does not mean, however, that the activity of these functional groups is disabled. For example, besides those mentioned, the degraded mRNAs also encode proteins involved in translation. This occurs despite the intensive protein synthesis, suggesting that the fully active translational apparatus is available from dormancy and is re-activated very rapidly when the germination starts. Subsequently, but still during the first 30 min of germination, the level of most mRNA groups increases again, but now more likely as a relevant response to the available sources and cell requirements under new conditions.
The activity of the transcriptional apparatus is strictly dependent on the set of sigma factors present in distinctive time points. They direct specific developmental tasks, such as the cell wall reconstitution, protein refolding, or the osmotic stress response, and therefore they must be fully accessible at specific points during germination (Bobek et al., 2014). Thus, SigW sigma factor (SCO0632), whose homolog in B. subtilis directs the synthesis of cell wall hydrolases (Helmann, 2006), is already expressed in S. coelicolor during the initial rehydration event, suggesting a similar role in the cell wall reconstruction. At 30 min of germination, a set of other sigma factors is activated, including HrdD and the stress responding ECF family sigma factors SigR, SigE, SigD, and SigH. SigR as well as SigH direct cellular responses to osmotic stress (Kormanec et al., 2000; Kelemen et al., 2001; Viollier et al., 2003; Zdanowski et al., 2006; Shu et al., 2009; Kallifidas et al., 2010; Sevcikova et al., 2010). Moreover, the SigR sigma factor, whose regulon comprise DnaK and ClpB chaperones, ensures control over the process of protein re-folding (Kallifidas et al., 2010). SigE possibly takes part in cell envelope reconstruction control, similarly to SigW in the early phase (Paget et al., 1999; Hong et al., 2002). However, the highest increase in its cellular level during germination is exhibited by S. coelicolor’s SigQ sigma factor (SCO4908; Bobek et al., 2014), whose regulon is now being studied. Most of the sigma factors mentioned are known to respond to stress conditions. Several members of the stress response apparatus that are involved in the antioxidant defense – superoxide dismutase SodF2 (SCO0999), catalase (SCO0379), and thioredoxins TrxA (SCO3889) and TrxA4 (SCO5419) – were also shown to be expressed soon after spore hydration in S. coelicolor (Strakova et al., 2013a). The detoxification system thus reflects the increased metabolic activity. Based on the fact that the stress responding proteins and their regulators (sigma factors) are highly active during germination, it could be proposed that germination itself evokes stress-like conditions, due to the entry of water, oxygen radicals, and salts into the reviving spores.
The ATP level increases approximately 11-fold during germination (Hirsch and Ensign, 1978). Its production is ensured by ATP synthase, whose expression is abolished in the crp- mutant strain (see below). ATP is essential not only as an energy source for the basal metabolism (Eaton and Ensign, 1980) but also for cyclic AMP (cAMP) synthesis (Piette et al., 2005). The cAMP is a signal molecule which accumulates during germination and is bound by the cAMP-receptor protein Crp, an important transcription regulator (Susstrunk et al., 1998; Derouaux et al., 2004). Both Streptomyces mutant strains in which either cya (a gene encoding adenylate cyclase that catalyzes conversion of ATP to cAMP) or the crp gene were deleted exhibited a similar, dramatic defect in germination. The germination defect caused by the missing or inactive Crp system can be explained by the subsequent absence of several members of the affected Crp regulon that play a crucial role in germination. Other than ATP synthase, the Crp regulon encompasses the cell wall hydrolases and chaperones (Piette et al., 2005). Piette et al. (2005) described morphological differences between spores of S. coelicolor wild type and its crp- mutant strains. The spore wall of the crp- strain was twice as thick as that of the wild type strain. This can be related to the suppressed expression of another cell wall hydrolase, SCO5466, in the crp- strain during germination. Besides SCO5466, the expression of the above mentioned RpfA hydrolase is also under regulation of Crp (St-Onge et al., 2015). One may assume that the Cya-cAMP-Crp system might control the muralytic activity of hydrated spores, among other processes, allowing for cell wall reconstruction; an indispensable procedure for successful germination. A differential protein expression analysis between the wild type and crp- strains, provided by the same authors, revealed decreased expression of protein chaperones GroEL1, GroEL2, DnaK that could have a negative effect on protein folding and thus negatively affect the germination in the crp- strain. The crucial role played by the Cya-cAMP-Crp system in germination may however rely on the aforementioned trehalose hydrolysis control.
In natural soil environments, germination of spores and seeds from different organisms is affected by a plethora of external factors. These include population density of organisms of the same species, the presence of rival organisms, and respective intra- or inter-species communication. In laboratory conditions, physiological differences, including inhibition of development or degradation of signaling compounds, were observed when Streptomyces had been cultivated together with B. subtilis or with bacteria containing mycolic acids in their cell walls (Onaka et al., 2011; Traxler et al., 2013).
Germinating spores produce signals that play a role in their own communication framework. The production of an unknown activator that would induce germination under optimal conditions can be expected based on an experiment in which monocultural spores of S. venezualae germinated significantly faster in higher spore densities or in a medium supplemented by its own, earlier germlings (Xu and Vetsigian, 2017). On the other hand, a similar experiment provided by the same authors revealed a reduction of the germinating spores at higher spore densities in S. coelicolor. The existence of germination inhibitors produced by the germlings was firstly shown in S. viridochromogenes (Grund and Ensign, 1985) and later also in S. coelicolor (Song et al., 2006). These germination inhibitors include germicidins, gramicidine S, and hypnosin (Ensign, 1978; Grund and Ensign, 1985; Setlow, 2003; Aoki et al., 2007, 2011). At least four germicidin homologs are produced by Gcs, a type III polyketide synthase in S. coelicolor (Song et al., 2006). The expression of its gene (SCO7221) highly increases at 30 min of germination, as can be found in the whole genome expression data in Strakova et al. (2013a). The hypnosin, whose genetic substantiality is not clear, is bound to the spore surface and released in wet conditions thus probably preventing premature germination of spore chains on aerial mycelia (Xu and Vetsigian, 2017).
Recent analytical technologies allow more detailed investigations into the production of small molecular compounds. Using high-performance liquid chromatography-mass spectrometry, three different compounds – a sesquiterpenoid antibiotic albaflavenone, the polyketide germicidin A, and the chalcone – were detected to be synthesized upon germination in S. coelicolor. The two latter compounds revealed an inhibitory effect on the germination process (Čihák et al., submitted). Germicidin A is known as an autoregulatory germination inhibitor of Streptomyces and other bacteria (Petersen et al., 1993). The compound is expected to be a widespread interspecies regulator affecting germination of various sporogenic organisms. Chalcones, as well as other flavonoids and phenolic compounds, are important signaling molecules in plant–microbe symbioses, being essential for the plant survival (Maxwell et al., 1989; Diaz-Tielas et al., 2012). Although Streptomyces produce flavonoids [e.g., THN and flaviolin by S. coelicolor (Zhao et al., 2005)], the only streptomycete chalcone derivative is known in naringerin biosynthesis, a typical plant secondary metabolite produced by Streptomyces clavuligerus (Alvarez-Alvarez et al., 2015). The presence of different germination inhibitors in S. coelicolor is in accordance with the fact that the germination rate of this strain is much slower and even decreased with a higher spore densities, as mentioned above.
Several streptomycete antibiotics produced during sporulation, such as neomycin in S. fradiae, streptomycin in S. griseus (Barabas and Szabo, 1968), or actinorhodin in S. coelicolor (Čihák et al., submitted) are known to be bound on the spore envelopes and released into the environment during germination. These antibiotics protect the germinating spores against environmental challenges before the internal biosynthetic pathways are re-activated. Some antibiotics, such as cephamycin C and clavulanic acid both synthesized by S. clavuligerus are known to be produced soon after germination (Sanchez and Brana, 1996).
Spore germination in Streptomyces represents an exceptional study model of bacterial cell differentiation that presents a complete transformation of cellular morphology and the restoration of all physiological processes. More detailed knowledge about the structure of dormant spores may provide clues for understanding their readiness to trigger metabolic activity and cell reconstitution.
An interesting area that deserves our attention is the probability of germination and germination control. As discussed in this review, the cell awakening is launched in an aquatic milieu; the probability of awakening differs, possibly depending on the accessibility to nutrients and on other biotic and abiotic factors. Stochastic germination seems to be a widespread strategy of spore-forming organisms that has evolved to ensure the survival of whole populations. From the one-cell perspective, spores represent a safe but non-reproductive state, whereas germination is a risky and irreversible process. From the point of view of the whole population, early germinating pioneers take advantage by occupying vacant niches whereas a hesitant strategy is preferable when conditions become harsh. Therefore, one may assume that the onset of germination is subject to various controlling mechanisms that respond to external signals and activate or suppress the process as is needed. The characterization of these signals and their sensors has not been established to date. The transferred signals are eventually reflected in complete cell reconstruction. This requires the activity of specific genes whose expression is directed by a unique set of transcriptional factors. Characterizing these factors and clarifying their regulons would shed more light on the process by which dormant spores are awakened.
JB wrote the whole manuscript, MČ analyzed the knowledge about metabolic activities of germinating spores, KŠ analyzed the knowledge about transcriptional activities of germinating spores.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are very grateful to Dr. Oldrich Benada and to Dr. Olga Kofronova for electron microscopic pictures of germinating spores. This work was supported by a project of Charles University in Prague: Progress Q26/LF1 to JB, SVV260369 to KŠ, a grant of the Grant Agency of the Charles University (www.cuni.cz/UK-33.html) under contract no. 160214 to KŠ, a project of J. E. Purkinje University: UJEP-SGS-173-07-01 to JB, and by the Czech research infrastructure for systems biology C4SYS (project no. LM2015055) to JB.
Alvarez-Alvarez, R., Botas, A., Albillos, S. M., Rumbero, A., Martin, J. F., and Liras, P. (2015). Molecular genetics of naringenin biosynthesis, a typical plant secondary metabolite produced by Streptomyces clavuligerus. Microb. Cell Fact. 14, 178. doi: 10.1186/s12934-015-0373-7
An, T. Z., Iwakiri, M., Edashige, K., Sakurai, T., and Kasai, M. (2000). Factors affecting the survival of frozen-thawed mouse spermatozoa. Cryobiology 40, 237–249. doi: 10.1006/cryo.2000.2245
Aoki, Y., Matsumoto, D., Kawaide, H., and Natsume, M. (2011). Physiological role of germicidins in spore germination and hyphal elongation in Streptomyces coelicolor A3(2). J. Antibiot. 64, 607–611. doi: 10.1038/ja.2011.59
Aoki, Y., Yoshida, M., Kawaide, H., Abe, H., and Natsume, M. (2007). Isolation and characterization of a spore germination inhibitor from Streptomyces sp. CB-1-1, a phytopathogen causing root tumor of melon. Biosci. Biotechnol. Biochem. 71, 986–992. doi: 10.1271/bbb.60649
Ausmees, N., Wahlstedt, H., Bagchi, S., Elliot, M. A., Buttner, M. J., and Flardh, K. (2007). SmeA, a small membrane protein with multiple functions in Streptomyces sporulation including targeting of a SpoIIIE/FtsK-like protein to cell division septa. Mol. Microbiol. 65, 1458–1473. doi: 10.1111/j.1365-2958.2007.05877.x
Bagchi, S., Tomenius, H., Belova, L. M., and Ausmees, N. (2008). Intermediate filament-like proteins in bacteria and a cytoskeletal function in Streptomyces. Mol. Microbiol. 70, 1037–1050. doi: 10.1111/j.1365-2958.2008.06473.x
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L., and Leibler, S. (2004). Bacterial persistence as a phenotypic switch. Science 305, 1622–1625. doi: 10.1126/science.1099390
Barabas, G., and Szabo, G. (1968). Role of streptomycin in the life of Streptomyces griseus: streptidine-containing fractions in the cell walls of Streptomyces griseus strains. Can. J. Microbiol. 14, 1325–1331. doi: 10.1139/m68-222
Benaroudj, N., Lee, D. H., and Goldberg, A. L. (2001). Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J. Biol. Chem. 276, 24261–24267. doi: 10.1074/jbc.M101487200
Bentley, S. D., Chater, K. F., Cerdeno-Tarraga, A. M., Challis, G. L., Thomson, N. R., James, K. D., et al. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147. doi: 10.1038/417141a
Bobek, J., Halada, P., Angelis, J., Vohradsky, J., and Mikulik, K. (2004). Activation and expression of proteins during synchronous germination of aerial spores of Streptomyces granaticolor. Proteomics 4, 3864–3880. doi: 10.1002/pmic.200400818
Bobek, J., Strakova, E., Zikova, A., and Vohradsky, J. (2014). Changes in activity of metabolic and regulatory pathways during germination of S. coelicolor. BMC Genomics 15:1173. doi: 10.1186/1471-2164-15-1173
Briza, P., Breitenbach, M., Ellinger, A., and Segall, J. (1990). Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae. Genes Dev. 4, 1775–1789. doi: 10.1101/gad.4.10.1775
Buerger, S., Spoering, A., Gavrish, E., Leslin, C., Ling, L., and Epstein, S. S. (2012). Microbial scout hypothesis, stochastic exit from dormancy, and the nature of slow growers. Appl. Environ. Microbiol. 78, 3221–3228. doi: 10.1128/AEM.07307-11
Bush, M. J., Tschowri, N., Schlimpert, S., Flardh, K., and Buttner, M. J. (2015). c-di-GMP signalling and the regulation of developmental transitions in streptomycetes. Nat. Rev. Microbiol. 13, 749–760. doi: 10.1038/nrmicro3546
Ceccarini, C. (1967). The biochemical relationship between trehalase and trehalose during growth and differentiation in the cellular slime mold, Dictyostelium discoideum. Biochim. Biophys. Acta 148, 114–124. doi: 10.1016/0304-4165(67)90285-1
Chen, D., Huang, S. S., and Li, Y. Q. (2006). Real-time detection of kinetic germination and heterogeneity of single Bacillus spores by laser tweezers Raman spectroscopy. Anal. Chem. 78, 6936–6941. doi: 10.1021/ac061090e
Chen, K. C., Csikasz-Nagy, A., Gyorffy, B., Val, J., Novak, B., and Tyson, J. J. (2000). Kinetic analysis of a molecular model of the budding yeast cell cycle. Mol. Biol. Cell 11, 369–391. doi: 10.1091/mbc.11.1.369
Claessen, D., de Jong, W., Dijkhuizen, L., and Wosten, H. A. (2006). Regulation of Streptomyces development: reach for the sky! Trends Microbiol. 14, 313–319. doi: 10.1016/j.tim.2006.05.008
Claessen, D., Rink, R., de Jong, W., Siebring, J., de Vreugd, P., Boersma, F. G., et al. (2003). A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 17, 1714–1726. doi: 10.1101/gad.264303
Claessen, D., Stokroos, I., Deelstra, H. J., Penninga, N. A., Bormann, C., Salas, J. A., et al. (2004). The formation of the rodlet layer of streptomycetes is the result of the interplay between rodlins and chaplins. Mol. Microbiol. 53, 433–443. doi: 10.1111/j.1365-2958.2004.04143.x
Claessen, D., Wosten, H. A., van Keulen, G., Faber, O. G., Alves, A. M., Meijer, W. G., et al. (2002). Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Mol. Microbiol. 44, 1483–1492. doi: 10.1046/j.1365-2958.2002.02980.x
Cohen-Gonsaud, M., Keep, N. H., Davies, A. P., Ward, J., Henderson, B., and Labesse, G. (2004). Resuscitation-promoting factors possess a lysozyme-like domain. Trends Biochem. Sci. 29, 7–10. doi: 10.1016/j.tibs.2003.10.009
Constant, P., Poissant, L., and Villemur, R. (2008). Isolation of Streptomyces sp. PCB7, the first microorganism demonstrating high-affinity uptake of tropospheric H2. ISME J. 2, 1066–1076. doi: 10.1038/ismej.2008.59
Cowan, A. E., Koppel, D. E., Setlow, B., and Setlow, P. (2003). A soluble protein is immobile in dormant spores of Bacillus subtilis but is mobile in germinated spores: implications for spore dormancy. Proc. Natl. Acad. Sci. U.S.A. 100, 4209–4214. doi: 10.1073/pnas.0636762100
Crowe, J. H., Crowe, L. M., and Chapman, D. (1984). Preservation of membranes in anhydrobiotic organisms: the role of trehalose. Science 223, 701–703. doi: 10.1126/science.223.4637.701
de Jong, W., Manteca, A., Sanchez, J., Bucca, G., Smith, C. P., Dijkhuizen, L., et al. (2009). NepA is a structural cell wall protein involved in maintenance of spore dormancy in Streptomyces coelicolor. Mol. Microbiol. 71, 1591–1603. doi: 10.1111/j.1365-2958.2009.06633.x
De Virgilio, C., Hottiger, T., Dominguez, J., Boller, T., and Wiemken, A. (1994). The role of trehalose synthesis for the acquisition of thermotolerance in yeast. I. Genetic evidence that trehalose is a thermoprotectant. Eur. J. Biochem. 219, 179–186. doi: 10.1111/j.1432-1033.1994.tb19928.x
Derouaux, A., Halici, S., Nothaft, H., Neutelings, T., Moutzourelis, G., Dusart, J., et al. (2004). Deletion of a cyclic AMP receptor protein homologue diminishes germination and affects morphological development of Streptomyces coelicolor. J. Bacteriol. 186, 1893–1897. doi: 10.1128/JB.186.6.1893-1897.2004
Diaz-Tielas, C., Grana, E., Reigosa, M. J., and Sanchez-Moreiras, A. M. (2012). The role of peroxidases on the mode of action of chalcone in Arabidopsis roots. Plant Signal. Behav. 7, 1274–1276. doi: 10.4161/psb.21594
Ditkowski, B., Holmes, N., Rydzak, J., Donczew, M., Bezulska, M., Ginda, K., et al. (2013). Dynamic interplay of ParA with the polarity protein, Scy, coordinates the growth with chromosome segregation in Streptomyces coelicolor. Open Biol. 3:130006. doi: 10.1098/rsob.130006
Eaton, D., and Ensign, J. C. (1980). Streptomyces viridochromogenes spore germination initiated by calcium ions. J. Bacteriol. 143, 377–382.
Elbein, A. D. (1974). The metabolism of alpha,alpha-trehalose. Adv. Carbohydr. Chem. Biochem. 30, 227–256. doi: 10.1016/S0065-2318(08)60266-8
Elbein, A. D., and Mitchell, M. (1973). Levels of glycogen and trehalose in Mycobacterium smegmatis and the purification and properties of the glycogen synthetase. J. Bacteriol. 113, 863–873.
Elbein, A. D., Pan, Y. T., Pastuszak, I., and Carroll, D. (2003). New insights on trehalose: a multifunctional molecule. Glycobiology 13, 17R–27R. doi: 10.1093/glycob/cwg047
Elliot, M. A., Karoonuthaisiri, N., Huang, J., Bibb, M. J., Cohen, S. N., Kao, C. M., et al. (2003). The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 17, 1727–1740. doi: 10.1101/gad.264403
Elliot, M. A., and Talbot, N. J. (2004). Building filaments in the air: aerial morphogenesis in bacteria and fungi. Curr. Opin. Microbiol. 7, 594–601. doi: 10.1016/j.mib.2004.10.013
Ensign, J. C. (1978). Formation, properties, and germination of actinomycete spores. Annu. Rev. Microbiol. 32, 185–219. doi: 10.1146/annurev.mi.32.100178.001153
Fillinger, S., Chaveroche, M. K., van Dijck, P., de Vries, R., Ruijter, G., Thevelein, J., et al. (2001). Trehalose is required for the acquisition of tolerance to a variety of stresses in the filamentous fungus Aspergillus nidulans. Microbiology 147(Pt 7), 1851–1862. doi: 10.1099/00221287-147-7-1851
Flardh, K. (2003a). Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol. Microbiol. 49, 1523–1536. doi: 10.1046/j.1365-2958.2003.03660.x
Flardh, K. (2003b). Growth polarity and cell division in Streptomyces. Curr. Opin. Microbiol. 6, 564–571. doi: 10.1016/j.mib.2003.10.011
Flardh, K., and Buttner, M. J. (2009). Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7, 36–49. doi: 10.1038/nrmicro1968
Flardh, K., Richards, D. M., Hempel, A. M., Howard, M., and Buttner, M. J. (2012). Regulation of apical growth and hyphal branching in Streptomyces. Curr. Opin. Microbiol. 15, 737–743. doi: 10.1016/j.mib.2012.10.012
Foster, J. W., Mc, D. L., Woodruff H.B., Stokes, J. L. (1945). Microbiological aspects of penicillin; conidiospore formation in submerged cultures of Penicillium notatum. J. Bacteriol. 50, 365–368.
Galperin, M. Y., Mekhedov, S. L., Puigbo, P., Smirnov, S., Wolf, Y. I., and Rigden, D. J. (2012). Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation-specific genes. Environ. Microbiol. 14, 2870–2890. doi: 10.1111/j.1462-2920.2012.02841.x
Ghorbel, S., Smirnov, A., Chouayekh, H., Sperandio, B., Esnault, C., Kormanec, J., et al. (2006). Regulation of ppk expression and in vivo function of Ppk in Streptomyces lividans TK24. J. Bacteriol. 188, 6269–6276. doi: 10.1128/JB.00202-06
Glauert, A. M., and Hopwood, D. A. (1961). The fine structure of Streptomyces violaceoruber (S. coelicolor). III. The walls of the mycelium and spores. J. Biophys. Biochem. Cytol. 10, 505–516. doi: 10.1083/jcb.10.4.505
Grantcharova, N., Lustig, U., and Flardh, K. (2005). Dynamics of FtsZ assembly during sporulation in Streptomyces coelicolor A3(2). J. Bacteriol. 187, 3227–3237. doi: 10.1128/JB.187.9.3227-3237.2005
Grund, A. D., and Ensign, J. C. (1985). Properties of the germination inhibitor of Streptomyces viridochromogenes spores. J. Gen. Microbiol. 131, 833–847. doi: 10.1099/00221287-131-4-833
Haiser, H. J., Yousef, M. R., and Elliot, M. A. (2009). Cell wall hydrolases affect germination, vegetative growth, and sporulation in Streptomyces coelicolor. J. Bacteriol. 191, 6501–6512. doi: 10.1128/JB.00767-09
Hardisson, C., Manzanal, M. B., Salas, J. A., and Suarez, J. E. (1978). Fine structure, physiology and biochemistry of arthrospore germination in Streptomyces antibioticus. J. Gen. Microbiol. 105, 203–214. doi: 10.1099/00221287-105-2-203
Heichlinger, A., Ammelburg, M., Kleinschnitz, E. M., Latus, A., Maldener, I., Flardh, K., et al. (2011). The MreB-like protein Mbl of Streptomyces coelicolor A3(2) depends on MreB for proper localization and contributes to spore wall synthesis. J. Bacteriol. 193, 1533–1542. doi: 10.1128/JB.01100-10
Helmann, J. D. (2006). Deciphering a complex genetic regulatory network: the Bacillus subtilis sigmaW protein and intrinsic resistance to antimicrobial compounds. Sci. Prog. 89(Pt 3–4), 243–266. doi: 10.3184/003685006783238290
Hempel, A. M., Cantlay, S., Molle, V., Wang, S. B., Naldrett, M. J., Parker, J. L., et al. (2012). The Ser/Thr protein kinase AfsK regulates polar growth and hyphal branching in the filamentous bacteria Streptomyces. Proc. Natl. Acad. Sci. U.S.A. 109, E2371–E2379. doi: 10.1073/pnas.1207409109
Henriques, A. O., and Moran, C. P. Jr. (2007). Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 61, 555–588. doi: 10.1146/annurev.micro.61.080706.093224
Hett, E. C., Chao, M. C., Deng, L. L., and Rubin, E. J. (2008). A mycobacterial enzyme essential for cell division synergizes with resuscitation-promoting factor. PLOS Pathog. 4:e1000001. doi: 10.1371/journal.ppat.1000001
Hey-Ferguson, A., Mitchell, M., and Elbein, A. D. (1973). Trehalose metabolism in germinating spores of Streptomyces hygroscopicus. J. Bacteriol. 116, 1084–1085.
Hirsch, C. F., and Ensign, J. C. (1976a). Heat activation of Streptomyces viridochromogenes spores. J. Bacteriol. 126, 24–30.
Hirsch, C. F., and Ensign, J. C. (1976b). Nutritionally defined conditions for germination of Streptomyces viridochromogenes spores. J. Bacteriol. 126, 13–23.
Hirsch, C. F., and Ensign, J. C. (1978). Some properties of Streptomyces viridochromogenes spores. J. Bacteriol. 134, 1056–1063.
Hong, H. J., Paget, M. S., and Buttner, M. J. (2002). A signal transduction system in Streptomyces coelicolor that activates the expression of a putative cell wall glycan operon in response to vancomycin and other cell wall-specific antibiotics. Mol. Microbiol. 44, 1199–1211. doi: 10.1046/j.1365-2958.2002.02960.x
Hottiger, T., Boller, T., and Wiemken, A. (1989). Correlation of trehalose content and heat resistance in yeast mutants altered in the RAS/adenylate cyclase pathway: is trehalose a thermoprotectant? FEBS Lett. 255, 431–434. doi: 10.1016/0014-5793(89)81139-1
Jain, N. K., and Roy, I. (2010). Trehalose and protein stability. Curr. Protoc. Protein Sci. 59, 4.9.1–4.9.12. doi: 10.1002/0471140864.ps0409s59
Jakimowicz, D., and van Wezel, G. P. (2012). Cell division and DNA segregation in Streptomyces: how to build a septum in the middle of nowhere? Mol. Microbiol. 85, 393–404. doi: 10.1111/j.1365-2958.2012.08107.x
Jang, M. S., Mouri, Y., Uchida, K., Aizawa, S., Hayakawa, M., Fujita, N., et al. (2016). Genetic and transcriptional analyses of the flagellar gene cluster in Actinoplanes missouriensis. J. Bacteriol. 198, 2219–2227. doi: 10.1128/JB.00306-16
Jyothikumar, V., Tilley, E. J., Wali, R., and Herron, P. R. (2008). Time-lapse microscopy of Streptomyces coelicolor growth and sporulation. Appl. Environ. Microbiol. 74, 6774–6781. doi: 10.1128/AEM.01233-08
Kalakoutskii, L. V., and Agre, N. S. (1976). Comparative aspects of development and differentiation in actinomycetes. Bacteriol. Rev. 40, 469–524.
Kallifidas, D., Thomas, D., Doughty, P., and Paget, M. S. (2010). The sigmaR regulon of Streptomyces coelicolor A32 reveals a key role in protein quality control during disulphide stress. Microbiology 156(Pt 6), 1661–1672. doi: 10.1099/mic.0.037804-0
Kandror, O., DeLeon, A., and Goldberg, A. L. (2002). Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc. Natl. Acad. Sci. U.S.A. 99, 9727–9732. doi: 10.1073/pnas.142314099
Keep, N. H., Ward, J. M., Robertson, G., Cohen-Gonsaud, M., and Henderson, B. (2006). Bacterial resuscitation factors: revival of viable but non-culturable bacteria. Cell Mol. Life Sci. 63, 2555–2559. doi: 10.1007/s00018-006-6188-2
Keijser, B. J., Noens, E. E., Kraal, B., Koerten, H. K., and van Wezel, G. P. (2003). The Streptomyces coelicolor ssgB gene is required for early stages of sporulation. FEMS Microbiol. Lett. 225, 59–67. doi: 10.1016/S0378-1097(03)00481-6
Kelemen, G. H. (2017). Intermediate filaments supporting cell shape and growth in bacteria. Subcell. Biochem. 84, 161–211. doi: 10.1007/978-3-319-53047-5_6
Kelemen, G. H., Brian, P., Flardh, K., Chamberlin, L., Chater, K. F., and Buttner, M. J. (1998). Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3 (2). J. Bacteriol. 180, 2515–2521.
Kelemen, G. H., Viollier, P. H., Tenor, J., Marri, L., Buttner, M. J., and Thompson, C. J. (2001). A connection between stress and development in the multicellular prokaryote Streptomyces coelicolor A3(2). Mol. Microbiol. 40, 804–814. doi: 10.1046/j.1365-2958.2001.02417.x
Kleinschnitz, E. M., Heichlinger, A., Schirner, K., Winkler, J., Latus, A., Maldener, I., et al. (2011). Proteins encoded by the mre gene cluster in Streptomyces coelicolor A3(2) cooperate in spore wall synthesis. Mol. Microbiol. 79, 1367–1379. doi: 10.1111/j.1365-2958.2010.07529.x
Kormanec, J., Sevcikova, B., Halgasova, N., Knirschova, R., and Rezuchova, B. (2000). Identification and transcriptional characterization of the gene encoding the stress-response sigma factor sigma(H) in Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 189, 31–38.
Lederer, E. (1976). Cord factor and related trehalose esters. Chem. Phys. Lipids 16, 91–106. doi: 10.1016/0009-3084(76)90001-3
Liot, Q., and Constant, P. (2016). Breathing air to save energy–new insights into the ecophysiological role of high-affinity [NiFe]-hydrogenase in Streptomyces avermitilis. Microbiologyopen 5, 47–59. doi: 10.1002/mbo3.310
Ma, M., Rateb, M. E., Yang, D., Rudolf, J. D., Zhu, X., Huang, Y., et al. (2017). Germicidins H-J from Streptomyces sp. CB00361. J. Antibiot. (Tokyo) 70, 200–203. doi: 10.1038/ja.2016.100
Martin, M. C., Diaz, L. A., Manzanal, M. B., and Hardisson, C. (1986). Role of trehalose in the spores of Streptomyces. FEMS Microbiol. Lett. 35, 49–54. doi: 10.1016/0378-1097(86)90160-6
Maxwell, C. A., Hartwig, U. A., Joseph, C. M., and Phillips, D. A. (1989). A chalcone and two related flavonoids released from alfalfa roots induce nod genes of Rhizobium meliloti. Plant Physiol. 91, 842–847. doi: 10.1104/pp.91.3.842
Mazza, P., Noens, E. E., Schirner, K., Grantcharova, N., Mommaas, A. M., Koerten, H. K., et al. (2006). MreB of Streptomyces coelicolor is not essential for vegetative growth but is required for the integrity of aerial hyphae and spores. Mol. Microbiol. 60, 838–852. doi: 10.1111/j.1365-2958.2006.05134.x
McBride, M. J., and Ensign, J. C. (1987a). Effects of intracellular trehalose content on Streptomyces griseus spores. J. Bacteriol. 169, 4995–5001. doi: 10.1128/jb.169.11.4995-5001.1987
McBride, M. J., and Ensign, J. C. (1987b). Metabolism of endogenous trehalose by Streptomyces griseus spores and by spores or cells of other actinomycetes. J. Bacteriol. 169, 5002–5007. doi: 10.1128/jb.169.11.5002-5007.1987
McBride, M. J., and Ensign, J. C. (1990). Regulation of trehalose metabolism by Streptomyces griseus spores. J. Bacteriol. 172, 3637–3643. doi: 10.1128/jb.172.7.3637-3643.1990
McBride, M. J., and Zusman, D. R. (1989). Trehalose accumulation in vegetative cells and spores of Myxococcus xanthus. J. Bacteriol. 171, 6383–6386. doi: 10.1128/jb.171.11.6383-6386.1989
Miguelez, E. M., Martin, C., Hardisson, C., and Manzanal, M. B. (1993). Synchronous germination of Streptomyces antibioticus spores: tool for the analysis of hyphal growth in liquid cultures. FEMS Microbiol. Lett. 109, 123–129. doi: 10.1111/j.1574-6968.1993.tb06156.x
Mikulik, K., Bobek, J., Bezouskova, S., Benada, O., and Kofronova, O. (2002). Expression of proteins and protein kinase activity during germination of aerial spores of Streptomyces granaticolor. Biochem. Biophys. Res. Commun. 299, 335–342. doi: 10.1016/S0006-291X(02)02606-2
Mikulik, K., Bobek, J., Zikova, A., Smetakova, M., and Bezouskova, S. (2011). Phosphorylation of ribosomal proteins influences subunit association and translation of poly (U) in Streptomyces coelicolor. Mol. Biosyst. 7, 817–823. doi: 10.1039/c0mb00174k
Mikulik, K., Janda, I., Maskova, H., Stastna, J., and Jiranova, A. (1977). Macromolecular synthesis accompanying the transition from spores to vegetative forms of Streptomyces granaticolor. Folia Microbiol. 22, 252–261. doi: 10.1007/BF02877654
Mikulik, K., Janda, I., Weiser, J., Stastna, J., and Jiranova, A. (1984). RNA and ribosomal protein patterns during aerial spore germination in Streptomyces granaticolor. Eur. J. Biochem. 145, 381–388. doi: 10.1111/j.1432-1033.1984.tb08565.x
Mikulik, K., Paleckova, P., Felsberg, J., Bobek, J., Zidkova, J., and Halada, P. (2008). SsrA genes of streptomycetes and association of proteins to the tmRNA during development and cellular differentiation. Proteomics 8, 1429–1441. doi: 10.1002/pmic.200700560
Mukamolova, G. V., Kaprelyants, A. S., Young, D. I., Young, M., and Kell, D. B. (1998a). A bacterial cytokine. Proc. Natl. Acad. Sci. U.S.A. 95, 8916–8921. doi: 10.1073/pnas.95.15.8916
Mukamolova, G. V., Yanopolskaya, N. D., Kell, D. B., and Kaprelyants, A. S. (1998b). On resuscitation from the dormant state of Micrococcus luteus. Antonie Van Leeuwenhoek 73, 237–243. doi: 10.1023/A:1000881918216
Mukamolova, G. V., Murzin, A. G., Salina, E. G., Demina, G. R., Kell, D. B., Kaprelyants, A. S., et al. (2006). Muralytic activity of Micrococcus luteus Rpf and its relationship to physiological activity in promoting bacterial growth and resuscitation. Mol. Microbiol. 59, 84–98. doi: 10.1111/j.1365-2958.2005.04930.x
Mukamolova, G. V., Turapov, O. A., Kazarian, K., Telkov, M., Kaprelyants, A. S., Kell, D. B., et al. (2002). The rpf gene of Micrococcus luteus encodes an essential secreted growth factor. Mol. Microbiol. 46, 611–621. doi: 10.1046/j.1365-2958.2002.03183.x
Neiman, A. M. (2005). Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69, 565–584. doi: 10.1128/MMBR.69.4.565-584.2005
Noens, E. E., Mersinias, V., Traag, B. A., Smith, C. P., Koerten, H. K., and van Wezel, G. P. (2005). SsgA-like proteins determine the fate of peptidoglycan during sporulation of Streptomyces coelicolor. Mol. Microbiol. 58, 929–944. doi: 10.1111/j.1365-2958.2005.04883.x
Noens, E. E., Mersinias, V., Willemse, J., Traag, B. A., Laing, E., Chater, K. F., et al. (2007). Loss of the controlled localization of growth stage-specific cell-wall synthesis pleiotropically affects developmental gene expression in an ssgA mutant of Streptomyces coelicolor. Mol. Microbiol. 64, 1244–1259. doi: 10.1111/j.1365-2958.2007.05732.x
Nwaka, S., and Holzer, H. (1998). Molecular biology of trehalose and the trehalases in the yeast Saccharomyces cerevisiae. Prog. Nucleic Acid Res. Mol. Biol. 58, 197–237. doi: 10.1016/S0079-6603(08)60037-9
Onaka, H., Mori, Y., Igarashi, Y., and Furumai, T. (2011). Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Environ. Microbiol. 77, 400–406. doi: 10.1128/AEM.01337-10
Paget, M. S., Chamberlin, L., Atrih, A., Foster, S. J., and Buttner, M. J. (1999). Evidence that the extracytoplasmic function sigma factor sigmaE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181, 204–211.
Paidhungat, M., and Setlow, P. (2000). Role of ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 182, 2513–2519. doi: 10.1128/JB.182.9.2513-2519.2000
Paleckova, P., Bobek, J., Felsberg, J., and Mikulik, K. (2006). Activity of translation system and abundance of tmRNA during development of Streptomyces aureofaciens producing tetracycline. Folia Microbiol. 51, 517–524. doi: 10.1007/BF02931615
Palleroni, N. J. (1976). Chemotaxis in Actinoplanes. Arch. Microbiol. 110, 13–18. doi: 10.1007/BF00416963
Pammer, M., Briza, P., Ellinger, A., Schuster, T., Stucka, R., Feldmann, H., et al. (1992). DIT101 (CSD2, CAL1), a cell cycle-regulated yeast gene required for synthesis of chitin in cell walls and chitosan in spore walls. Yeast 8, 1089–1099. doi: 10.1002/yea.320081211
Paredes-Sabja, D., Setlow, P., and Sarker, M. R. (2011). Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 19, 85–94. doi: 10.1016/j.tim.2010.10.004
Petersen, F., Zahner, H., Metzger, J. W., Freund, S., and Hummel, R. P. (1993). Germicidin, an autoregulative germination inhibitor of Streptomyces viridochromogenes NRRL B-1551. J. Antibiot. 46, 1126–1138. doi: 10.7164/antibiotics.46.1126
Piette, A., Derouaux, A., Gerkens, P., Noens, E. E., Mazzucchelli, G., Vion, S., et al. (2005). From dormant to germinating spores of Streptomyces coelicolor A3(2): new perspectives from the crp null mutant. J. Proteome Res. 4, 1699–1708. doi: 10.1021/pr050155b
Ranade, N., and Vining, L. C. (1993). Accumulation of intracellular carbon reserves in relation to chloramphenicol biosynthesis by Streptomyces venezuelae. Can. J. Microbiol. 39, 377–383. doi: 10.1139/m93-055
Richards, D. M., Hempel, A. M., Flardh, K., Buttner, M. J., and Howard, M. (2012). Mechanistic basis of branch-site selection in filamentous bacteria. PLOS Comput. Biol. 8:e1002423. doi: 10.1371/journal.pcbi.1002423
Roth, R., and Sussman, M. (1966). Trehalose synthesis in the cellular slime mold Dictyostelium discoideum. Biochim. Biophys. Acta 122, 225–231. doi: 10.1016/0926-6593(66)90064-6
Ruban-Osmialowska, B., Jakimowicz, D., Smulczyk-Krawczyszyn, A., Chater, K. F., and Zakrzewska-Czerwinska, J. (2006). Replisome localization in vegetative and aerial hyphae of Streptomyces coelicolor. J. Bacteriol. 188, 7311–7316. doi: 10.1128/JB.00940-06
Rueda, B., Miguelez, E. M., Hardisson, C., and Manzanal, M. B. (2001). Changes in glycogen and trehalose content of Streptomyces brasiliensis hyphae during growth in liquid cultures under sporulating and non-sporulating conditions. FEMS Microbiol. Lett. 194, 181–185. doi: 10.1111/j.1574-6968.2001.tb09466.x
Ruggiero, A., Tizzano, B., Pedone, E., Pedone, C., Wilmanns, M., and Berisio, R. (2009). Crystal structure of the resuscitation-promoting factor (DeltaDUF)RpfB from M. tuberculosis. J. Mol. Biol. 385, 153–162. doi: 10.1016/j.jmb.2008.10.042
Salas, J. A., Guijarro, J. A., and Hardisson, C. (1983). High calcium content in Streptomyces spores and its release as an early event during spore germination. J. Bacteriol. 155, 1316–1323.
Sanchez, L., and Brana, A. F. (1996). Cell density influences antibiotic biosynthesis in Streptomyces clavuligerus. Microbiology 142(Pt 5), 1209–1220. doi: 10.1099/13500872-142-5-1209
Sapir, L., and Harries, D. (2011). Linking trehalose self-association with binary aqueous solution equation of state. J. Phys. Chem. B 115, 624–634. doi: 10.1021/jp109780n
Setlow, B., Atluri, S., Kitchel, R., Koziol-Dube, K., and Setlow, P. (2006). Role of dipicolinic acid in resistance and stability of spores of Bacillus subtilis with or without DNA-protective alpha/beta-type small acid-soluble proteins. J. Bacteriol. 188, 3740–3747. doi: 10.1128/JB.00212-06
Setlow, P. (2003). Spore germination. Curr. Opin. Microbiol. 6, 550–556. doi: 10.1016/j.mib.2003.10.001
Setlow, P. (2007). I will survive: DNA protection in bacterial spores. Trends Microbiol. 15, 172–180. doi: 10.1016/j.tim.2007.02.004
Sevcikova, B., and Kormanec, J. (2003). The ssgB gene, encoding a member of the regulon of stress-response sigma factor sigmaH, is essential for aerial mycelium septation in Streptomyces coelicolor A3(2). Arch. Microbiol. 180, 380–384. doi: 10.1007/s00203-003-0603-y
Sevcikova, B., Rezuchova, B., Homerova, D., and Kormanec, J. (2010). The anti-anti-sigma factor BldG is involved in activation of the stress response sigma factor sigma(H) in Streptomyces coelicolor A3(2). J. Bacteriol. 192, 5674–5681. doi: 10.1128/JB.00828-10
Sexton, D. L., St-Onge, R. J., Haiser, H. J., Yousef, M. R., Brady, L., Gao, C., et al. (2015). Resuscitation-promoting factors are cell wall-lytic enzymes with important roles in the germination and growth of Streptomyces coelicolor. J. Bacteriol. 197, 848–860. doi: 10.1128/JB.02464-14
Shah, I. M., Laaberki, M. H., Popham, D. L., and Dworkin, J. (2008). A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135, 486–496. doi: 10.1016/j.cell.2008.08.039
Shleeva, M. O., Trutneva, K. A., Demina, G. R., Zinin, A. I., Sorokoumova, G. M., Laptinskaya, P. K., et al. (2017). Free trehalose accumulation in dormant Mycobacterium smegmatis cells and its breakdown in early resuscitation phase. Front. Microbiol. 8:524. doi: 10.3389/fmicb.2017.00524
Shu, D., Chen, L., Wang, W., Yu, Z., Ren, C., Zhang, W., et al. (2009). afsQ1-Q2-sigQ is a pleiotropic but conditionally required signal transduction system for both secondary metabolism and morphological development in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 81, 1149–1160. doi: 10.1007/s00253-008-1738-1
Sigle, S., Ladwig, N., Wohlleben, W., and Muth, G. (2015). Synthesis of the spore envelope in the developmental life cycle of Streptomyces coelicolor. Int. J. Med. Microbiol. 305, 183–189. doi: 10.1016/j.ijmm.2014.12.014
Singer, M. A., and Lindquist, S. (1998). Multiple effects of trehalose on protein folding in vitro and in vivo. Mol. Cell 1, 639–648. doi: 10.1016/S1097-2765(00)80064-7
Sola-Penna, M., and Meyer-Fernandes, J. R. (1998). Stabilization against thermal inactivation promoted by sugars on enzyme structure and function: why is trehalose more effective than other sugars? Arch. Biochem. Biophys. 360, 10–14. doi: 10.1006/abbi.1998.0906
Song, L., Barona-Gomez, F., Corre, C., Xiang, L., Udwary, D. W., Austin, M. B., et al. (2006). Type III polyketide synthase beta-ketoacyl-ACP starter unit and ethylmalonyl-CoA extender unit selectivity discovered by Streptomyces coelicolor genome mining. J. Am. Chem. Soc. 128, 14754–14755. doi: 10.1021/ja065247w
Stastna, J. (1977). A method of rapid wetting and synchronous germination of streptomycete spores. Folia Microbiol. 22, 137–138. doi: 10.1007/BF02881639
St-Onge, R. J., Haiser, H. J., Yousef, M. R., Sherwood, E., Tschowri, N., Al-Bassam, M., et al. (2015). Nucleotide second messenger-mediated regulation of a muralytic enzyme in Streptomyces. Mol. Microbiol. 96, 779–795. doi: 10.1111/mmi.12971
Strakova, E., Bobek, J., Zikova, A., Rehulka, P., Benada, O., Rehulkova, H., et al. (2013a). Systems insight into the spore germination of Streptomyces coelicolor. J. Proteome Res. 12, 525–536. doi: 10.1021/pr300980v
Strakova, E., Bobek, J., Zikova, A., and Vohradsky, J. (2013b). Global features of gene expression on the proteome and transcriptome levels in S. coelicolor during germination. PLOS ONE 8:e72842. doi: 10.1371/journal.pone.0072842
Strakova, E., Zikova, A., and Vohradsky, J. (2014). Inference of sigma factor controlled networks by using numerical modeling applied to microarray time series data of the germinating prokaryote. Nucleic Acids Res. 42, 748–763. doi: 10.1093/nar/gkt917
Sturm, A., and Dworkin, J. (2015). Phenotypic diversity as a mechanism to exit cellular dormancy. Curr. Biol. 25, 2272–2277. doi: 10.1016/j.cub.2015.07.018
Sussman, A. S. (1961). The role of trehalose in the activation of dormant ascospores of neurospora. Q. Rev. Biol. 36, 109–116. doi: 10.1086/403332
Sussman, A. S., and Lingappa, B. T. (1959). Role of trehalose in ascospores of neurospora tetrasperma. Science 130:1343. doi: 10.1126/science.130.3385.1343
Susstrunk, U., Pidoux, J., Taubert, S., Ullmann, A., and Thompson, C. J. (1998). Pleiotropic effects of cAMP on germination, antibiotic biosynthesis and morphological development in Streptomyces coelicolor. Mol. Microbiol. 30, 33–46. doi: 10.1046/j.1365-2958.1998.01033.x
Telkov, M. V., Demina, G. R., Voloshin, S. A., Salina, E. G., Dudik, T. V., Stekhanova, T. N., et al. (2006). Proteins of the Rpf (resuscitation promoting factor) family are peptidoglycan hydrolases. Biochemistry 71, 414–422. doi: 10.1134/S0006297906040092
Thomas, K. J. III, and Rice, C. V. (2014). Revised model of calcium and magnesium binding to the bacterial cell wall. Biometals 27, 1361–1370. doi: 10.1007/s10534-014-9797-5
Traag, B. A., and van Wezel, G. P. (2008). The SsgA-like proteins in actinomycetes: small proteins up to a big task. Antonie Van Leeuwenhoek 94, 85–97. doi: 10.1007/s10482-008-9225-3
Traxler, M. F., Watrous, J. D., Alexandrov, T., Dorrestein, P. C., and Kolter, R. (2013). Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. mBio 4:e00459-13. doi: 10.1128/mBio.00459-13
Trevelyan, W. E., and Harrison, J. S. (1956). Studies on yeast metabolism. 5. The trehalose content of baker’s yeast during anaerobic fermentation. Biochem. J. 62, 177–183. doi: 10.1042/bj0620177b
Tschowri, N., Schumacher, M. A., Schlimpert, S., Chinnam, N. B., Findlay, K. C., Brennan, R. G., et al. (2014). Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158, 1136–1147. doi: 10.1016/j.cell.2014.07.022
van Vliet, S. (2015). Bacterial dormancy: how to decide when to wake up. Curr. Biol. 25, R753–R755. doi: 10.1016/j.cub.2015.07.039
van Wezel, G. P., van der Meulen, J., Kawamoto, S., Luiten, R. G., Koerten, H. K., and Kraal, B. (2000). ssgA is essential for sporulation of Streptomyces coelicolor A3(2) and affects hyphal development by stimulating septum formation. J. Bacteriol. 182, 5653–5662. doi: 10.1128/JB.182.20.5653-5662.2000
Viollier, P. H., Weihofen, A., Folcher, M., and Thompson, C. J. (2003). Post-transcriptional regulation of the Streptomyces coelicolor stress responsive sigma factor, SigH, involves translational control, proteolytic processing, and an anti-sigma factor homolog. J. Mol. Biol. 325, 637–649. doi: 10.1016/S0022-2836(02)01280-9
Wang, L., Yu, Y., He, X., Zhou, X., Deng, Z., Chater, K. F., et al. (2007). Role of an FtsK-like protein in genetic stability in Streptomyces coelicolor A3(2). J. Bacteriol. 189, 2310–2318. doi: 10.1128/JB.01660-06
Wang, S. L., Fan, K. Q., Yang, X., Lin, Z. X., Xu, X. P., and Yang, K. Q. (2008). CabC, an EF-hand calcium-binding protein, is involved in Ca2+-mediated regulation of spore germination and aerial hypha formation in Streptomyces coelicolor. J. Bacteriol. 190, 4061–4068. doi: 10.1128/JB.01954-07
Wiemken, A. (1990). Trehalose in yeast, stress protectant rather than reserve carbohydrate. Antonie Van Leeuwenhoek 58, 209–217. doi: 10.1007/BF00548935
Wildermuth, H., Wehrli, E., and Horne, R. W. (1971). The surface structure of spores and aerial mycelium in Streptomyces coelicolor. J. Ultrastruct. Res. 35, 168–180. doi: 10.1016/S0022-5320(71)80149-1
Willemse, J., Borst, J. W., de Waal, E., Bisseling, T., and van Wezel, G. P. (2011). Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 25, 89–99. doi: 10.1101/gad.600211
Wolanski, M., Wali, R., Tilley, E., Jakimowicz, D., Zakrzewska-Czerwinska, J., and Herron, P. (2011). Replisome trafficking in growing vegetative hyphae of Streptomyces coelicolor A3(2). J. Bacteriol. 193, 1273–1275. doi: 10.1128/JB.01326-10
Wyatt, T. T., Wosten, H. A., and Dijksterhuis, J. (2013). Fungal spores for dispersion in space and time. Adv. Appl. Microbiol. 85, 43–91. doi: 10.1016/B978-0-12-407672-3.00002-2
Xu, Y., and Vetsigian, K. (2017). Phenotypic variability and community interactions of germinating Streptomyces spores. Sci. Rep. 7, 699. doi: 10.1038/s41598-017-00792-7
Yoshinaga, K., Yoshioka, H., Kurosaki, H., Hirasawa, M., Uritani, M., and Hasegawa, K. (1997). Protection by trehalose of DNA from radiation damage. Biosci. Biotechnol. Biochem. 61, 160–161. doi: 10.1271/bbb.61.160
Zdanowski, K., Doughty, P., Jakimowicz, P., O’Hara, L., Buttner, M. J., Paget, M. S., et al. (2006). Assignment of the zinc ligands in RsrA, a redox-sensing ZAS protein from Streptomyces coelicolor. Biochemistry 45, 8294–8300. doi: 10.1021/bi060711v
Zhao, B., Guengerich, F. P., Bellamine, A., Lamb, D. C., Izumikawa, M., Lei, L., et al. (2005). Binding of two flaviolin substrate molecules, oxidative coupling, and crystal structure of Streptomyces coelicolor A3(2) cytochrome P450 158A2. J. Biol. Chem. 280, 11599–11607. doi: 10.1074/jbc.M410933200
Keywords: dormancy, germination, Streptomyces, spore, cell wall, gene expression, metabolism, signaling
Citation: Bobek J, Šmídová K and Čihák M (2017) A Waking Review: Old and Novel Insights into the Spore Germination in Streptomyces. Front. Microbiol. 8:2205. doi: 10.3389/fmicb.2017.02205
Received: 28 August 2017; Accepted: 26 October 2017;
Published: 13 November 2017.
Edited by:
Dirk Tischler, Freiberg University of Mining and Technology, GermanyReviewed by:
Susan Schlimpert, John Innes Centre (BBSRC), United KingdomCopyright © 2017 Bobek, Šmídová and Čihák. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jan Bobek, amFuLmJvYmVrQGxmMS5jdW5pLmN6
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.