
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 03 November 2017
Sec. Aquatic Microbiology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.02117
 Catherine M. Luria1†
Catherine M. Luria1† Linda A. Amaral-Zettler2,3,4*
Linda A. Amaral-Zettler2,3,4* Hugh W. Ducklow5
Hugh W. Ducklow5 Daniel J. Repeta6
Daniel J. Repeta6 Andrew L. Rhyne7
Andrew L. Rhyne7 Jeremy J. Rich8*
Jeremy J. Rich8*Bacterial consumption of dissolved organic matter (DOM) drives much of the movement of carbon through the oceanic food web and the global carbon cycle. Understanding complex interactions between bacteria and marine DOM remains an important challenge. We tested the hypothesis that bacterial growth and community succession would respond differently to DOM additions due to seasonal changes in phytoplankton abundance in the environment. Four mesocosm experiments were conducted that spanned the spring transitional period (August–December 2013) in surface waters of the Western Antarctic Peninsula (WAP). Each mesocosm consisted of nearshore surface seawater (50 L) incubated in the laboratory for 10 days. The addition of DOM, in the form of cell-free exudates extracted from Thalassiosira weissflogii diatom cultures led to changes in bacterial abundance, production, and community composition. The timing of each mesocosm experiment (i.e., late winter vs. late spring) influenced the magnitude and direction of bacterial changes. For example, the same DOM treatment applied at different times during the season resulted in different levels of bacterial production and different bacterial community composition. There was a mid-season shift from Collwelliaceae to Polaribacter having the greatest relative abundance after incubation. This shift corresponded to a modest but significant increase in the initial relative abundance of Polaribacter in the nearshore seawater used to set up experiments. This finding supports a new hypothesis that starting community composition, through priority effects, influenced the trajectory of community succession in response to DOM addition. As strong inter-annual variability and long-term climate change may shift the timing of WAP phytoplankton blooms, and the corresponding production of DOM exudates, this study suggests a mechanism by which different seasonal successional patterns in bacterial communities could occur.
Marine dissolved organic matter (DOM) represents a large reservoir of carbon that drives a considerable fraction of the oceanic food web (Hedges, 1992). Phytoplankton production is the dominant source of marine organic material, with up to 50% of algal production entering the DOM pool through a variety of mechanisms (Lampert, 1978; Fogg, 1983; Gobler et al., 1997). The resulting DOM pool is a complex mixture of thousands of organic compounds with varying degrees of lability. The labile DOM pool turns over rapidly, within hours to days, supporting the growth of heterotrophic bacteria. The semi-labile pool turns over more slowly, months to years, while the refractory pool turns over very slowly and accounts for the largest fraction of marine DOM (Carlson and Hansell, 2015). Despite ramifications for organic matter and nutrient cycling, the interplay between diverse bacterial assemblages and complex DOM pools is poorly understood.
Shifts in bacterial community composition are associated with phytoplankton blooms (Pinhassi et al., 2004; Teeling et al., 2012; Klindworth et al., 2014; Sperling et al., 2017), perhaps due to changes in DOM availability and phytoplankton composition of the blooms. Previous studies provide conflicting reports of generalist assemblages that can utilize a wide variety of substrates (Mou et al., 2007; Newton et al., 2010; Chronopoulou et al., 2015), as well as communities of specialists that are adapted to take advantage of only certain classes of DOM compounds and respond quickly to disturbance (Cottrell and Kirchman, 2000; Allison and Martiny, 2008; Nelson and Carlson, 2012; Sarmento and Gasol, 2012; Klindworth et al., 2014; Sharma et al., 2014). Nonetheless, some general trends based on broad taxonomic groups have emerged. For example, certain groups of bacteria (e.g., Flavobacteria and Rhodobacteraceae) tend to increase in abundance during phytoplankton blooms, while other groups such as Candidatus Pelagibacter appear better adapted to non-bloom conditions (Williams et al., 2013; Buchan et al., 2014; Voget et al., 2015).
The Western Antarctic Peninsula (WAP) system undergoes an extreme seasonal transition every spring, from almost total darkness to almost continuous sunlight, resulting in a synchronized cascade of environmental changes that culminates in intense phytoplankton blooms, supporting a highly productive food web (Venables et al., 2013). Bacterioplankton activity closely follows the annual phytoplankton cycle supporting the hypothesis that bacterial growth is largely driven by DOM availability (Kirchman et al., 2009; Ducklow et al., 2012; Kim and Ducklow, 2016). Luria et al. (2016) measured seasonal succession in the composition of free-living bacterial communities in nearshore waters of the WAP during a phytoplankton bloom. However, the relationship between phytoplankton-derived DOM and bacterial community succession has not been directly tested in the WAP. Furthermore, the WAP is subject to strong inter-annual variability in sea ice and upper water column dynamics, leading to variability in the timing and magnitude of phytoplankton blooms. Dramatic warming of the WAP region over the last 50 years has reduced sea ice extent and led to earlier sea ice retreat in the spring (Vaughan and Doake, 1996; Meredith and King, 2005; Stammerjohn et al., 2008; Thomas et al., 2009; Saba et al., 2014). It is not known how changes in the timing of sea ice retreat, and subsequently of phytoplankton blooms, may alter bacteria-DOM interactions and carbon cycling.
We conducted a series of DOM-addition mesocosm experiments that spanned the WAP spring transitional period to examine how pre-bloom bacterial communities react to changes in DOM concentration. Because many previous DOM-addition experiments have relied upon relatively simple compounds (e.g., glucose or amino acids), that may serve as poor analogs for the organic carbon utilized by marine bacteria (Straza et al., 2010; Ducklow et al., 2011), we used diatom exudates, a more complex substrate. Our goals were to assess how diatom exudates alter the bacterial community and how these alterations change depending on the timing of the experiment in relation to the progression of the season from winter to late spring. We assumed that DOM additions would stimulate bacterial production and change bacterial community composition and hypothesized that the magnitude of these DOM-driven changes would decrease as the season progressed in relation to phytoplankton biomass based on chlorophyll a (chl a) in the nearshore seawater. This hypothesis is based on previous studies in Antarctica indicating that bacterial communities become less limited by DOM as phytoplankton blooms develop (Ducklow, 2003). Therefore, bacterial communities are likely to be less responsive to DOM additions as phytoplankton blooms develop.
Mesocosms were set up at Palmer Station, Antarctica, using seawater from the station’s intake, located at a depth of 6 m, 16 m from the shore of the station. This source water was monitored during the experimental period (August–December 2013) to determine any changes in environmental parameters that might influence bacterial community responses in mesocosm experiments. Samples for dissolved nutrients (phosphate, silicate, nitrite, and nitrate) and particulate organic carbon and nitrogen (POC and PN) were processed according to Palmer LTER standard protocols1. Briefly, nutrient samples were filtered through combusted 0.7-μm glass fiber filters (Whatman, GE Healthcare Life Sciences, Piscataway, NJ, United States) and frozen at -80°C until analysis on a SEAL AutoAnalyzer 3 (data available at doi: 10.6073/pasta/e893d71c5586769731875d49fde21b1d; Ducklow et al., 2016). Samples for DOC were filtered through combusted glass fiber filters and stored frozen until analyzed by high-temperature catalytic oxidation in a Shimadzu TOC-V, following previously described methods (Carlson et al., 2010). POC and PN samples were collected on combusted 0.7-μm glass fiber filters from 1 to 3 L seawater and were frozen at -80°C until analysis via combustion using a Perkin Elmer 2400 Series II CHNS/O Analyzer. Seawater intake chl a data are routinely collected through the LTER project (data available at doi: 10.6073/pasta/c2df461937789b5e53019dadcd29fc57; Schofield and Vernet, 2016). Bacterial production and abundance were determined as described below.
Dissolved organic matter was collected from large-scale Thalassiosira weissflogii cultures (500 L total) (CCMP1051, National Center for Marine Algae and Microbiota, East Boothbay, ME, United States), a well-characterized marine diatom, grown in f/2 medium (Guillard, 1975). The cultures were grown and harvested in Rhode Island, and the resulting DOM exudates were transported to Antarctica as dried residues. To grow the cultures, seawater for the f/2 medium was drawn from Narragansett Bay and was first passed through 1-μm filters prior to chlorination at 25 ppm for 4 days, followed by passage through successive granulated carbon and 1-μm filters and a 150 W UV sterilizer at a rate of 80 L min-1 with a final pasteurization at 85°C just prior to use. After cooling, seawater was amended with 0.02% v/v each of f/2 “Solution A” and “Solution B” (Fritz Aquatics, Mesquite, TX, United States) and 0.003 g L-1 sodium metasilicate (Na2SiO3). Axenic T. weissflogii cultures (100 L each) were grown in custom, acid-washed polycarbonate containers under 34-watt T8 cool white fluorescent lamps (Philips, Andover, MA, United States). Air enriched with 1800–2000 ppm CO2 and passed through 0.2 μm PTFE filter capsules (Sigma–Aldrich, St. Louis, MO, United States) was bubbled through the containers. The cultures were harvested at 7 days when they had developed dense growth but were still in the exponential growth phase. To create an f/2 “control,” the same volume of f/2 media was treated identically, but was not inoculated with T. weissflogii. This control was to account for any DOM in the f/2-amended seawater itself.
Cells and particulate matter were removed from T. weissflogii cultures via peristaltic pump-driven serial filtration through combusted GF/F filters (Whatman, GE Healthcare Life Sciences, Piscataway, NJ, United States) and acid-washed 0.2 μm polyethersulfone membrane filter capsules (Whatman, GE Healthcare Life Sciences, Piscataway, NJ, United States) using acid-washed platinum-cured silicon tubing (Masteflex, Cole Parmer, Vernon Hills, IL, United States). The filtrate was collected in clean acid-washed polycarbonate containers, acidified by adding sufficient hydrochloric acid to lower the pH to ∼2–3. Solid phase extraction of hydrophilic DOM was performed by pumping the filtrate through C18 columns (Supelco, Discovery C-18; 10 g) at a rate of 50 ml min-1. The columns were prepared by washing with 100 ml methanol followed by 200 ml of ultra high purity water. DOM was recovered by flushing the columns with 100 ml of methanol. The DOM solution was gently warmed to 35°C and was concentrated by vacuum evaporation followed by flushing with nitrogen gas as needed, leaving a dried residue suitable for transport. Prior to mesocosm experiments, the dried DOM was re-dissolved in a 0.2 μm-filtered 10 mM sodium hydroxide solution and added to DOM treated mesocosms to a final concentration of 20 ± 4 μmol L-1 added DOC, based on difference between DOM+ and controls in August–October experiments (Supplementary Table 1).
Mesocosm experiments were conducted in August, September, October, and December 2013 at Palmer Station by filling acid-washed 50 L polycarbonate carboys with seawater from the station’s intake. The filled carboys were divided into two treatments: (1) control (no additions, n = 3) and (2) +DOM (carboys that received diatom exudates; n = 3). An additional f/2 control treatment (uninoculated growth media, n = 3) was included in the October experiment only. The DOC concentration in the f/2 control was not any different than controls without any added DOC. As such, the f/2 control served as an additional no DOC addition control. The mesocosms were incubated for 10 days at 0°C, except in the August experiment when mesocosms were incubated at 3°C. All experiments were conducted under a low-level of continuous light, 1 × 1014 quanta cm-2 s-1, as measured inside the carboys with a hand-held radiometer (Biospherical Instruments, Inc.).
Chlorophyll a (chl a) and phaeopigment, dissolved inorganic nutrients (phosphate, silicate, nitrite, and nitrate), and bacterial production (3H-leucine incorporation rates) were assessed every 48 h for 10 days as previously described (Luria et al., 2016). Bacterial abundance samples were also collected every 48 h and were analyzed by flow cytometry with SYBR® Green I nucleic acid staining (Invitrogen, Carlsbad, CA, United States) on a Guava easyCyte flow cytometer (EMD Millipore, Billerica, MA, United States). Samples for bacterial community composition were collected on days 0, 6, and 10 by filtering ∼2 L water through successive 3.0 μm polycarbonate and 0.22 μm polyethersulfone (EMD Millipore, Billerica, MA, United States) filters. Filters were flash-frozen with liquid N2 and stored at -80°C until further processing. Samples for POC and PN were collected on days 0 and 10 and DOC on days 0, 6, and 10 and analyzed as described above. POC/PN from day 10 in December were lost due to an instrument malfunction. All sampling took place at incubation temperatures.
Sequence libraries were generated as in Eren et al. (2013), with modifications as described in Luria et al. (2016). Briefly, DNA was extracted from cells captured on the 0.22 μm pore size filters from mesocosm experiments [i.e., “free-living” bacterioplankton or any bacteria attached to micro-particles (Busch et al., 2017)] using a DNeasy Plant Mini Kit (Qiagen, Valencia, CA, United States) with an additional bead-beating step. For each DNA sample, the V6 hypervariable region of the 16S rRNA gene was amplified by polymerase chain reaction with custom fusion primers that contained Illumina adaptors and inline barcodes (forward primer) or dedicated indices (reverse primer) (Eren et al., 2013). Size-selected PCR products were quantified and pooled in equimolar amounts prior to sequencing on one lane of an Illumina HiSeq 1000 cycle paired-end run.
Low-quality sequences were filtered from the resulting data by discarding reads without 100% consensus between forward and reverse paired-end sequencing reads (Eren et al., 2013), resulting in more than 16 million sequences across 81 mesocosm samples. OTUs were clustered using Qiime (v 1.9.1; Caporaso et al., 2010) with open reference OTU picking with the default UCLUST method (Edgar, 2010), a minimum cluster size of 2, and a 97% similarity threshold and were assigned Greengenes taxonomy (version 13_8; McDonald et al., 2012). After removing OTUs classified as chloroplasts, rarefied libraries were produced by randomly down-sampling to the smallest library size of 64271 sequences spread among 14751 OTUs. All of our sequence data are MIMARKS-compliant (Supplementary Data Sheet 1) (Yilmaz et al., 2011) and have been deposited in the NCBI Sequence Read Archive under accession number SRP091049 and Bioproject PRJNA344476.
To separate the effects of incubation day in an experiment (i.e., days into incubation period), DOM+ treatment or control, and any differences between experiments on univariate bacterial (i.e., cell abundance or production) parameters, we performed ANOVA on nested linear models using the lm and anova functions in R (Chambers, 1992; Venables and Ripley, 2002; R Development Core Team, 2008). Two linear models are nested if one (the restricted model) is obtained from the other (the full model) by removing a term from the full model. An ANOVA performed on such nested models provides the equivalent of the F test for goodness of fit and indicates whether individual terms improve the model. This provided a statistical test of our hypothesis as the season progressed.
The number of OTUs observed in each library, Shannon’s diversity, and Pielou’s evenness, were calculated using the BiodiversityR package in R (Kindt and Coe, 2005). Differences in overall bacterial community composition between samples were visualized with non-metric multidimensional scaling (NMDS) based on Bray–Curtis similarity using the metaMDS function in the vegan R package (Oksanen et al., 2015). Co-occurrence network analyses based on Pearson’s correlations were used to examine relationships among OTUs and external variables (i.e., experimental treatments and bacterial production). Networks were generated using the Cytoscape CoNet plugin version 1.1.1.beta and were visualized in Cytoscape version 3.4.0 (Shannon et al., 2003; Faust et al., 2012). The R code used for all data analysis and figure production can be accessed at https://github.com/cmluria/DOM.
Our experiments spanned the winter-to-late spring period (August–December 2013), which was characterized by relatively constant environmental conditions (Supplementary Figure 1) compared to the onset of an intense phytoplankton bloom in summer, January 2014 (Luria et al., 2016). During the winter-to-late spring period, there was a slight uptick in leucine incorporation rates (hereafter bacterial production) in October and again in December (Figure 1). These changes in bacterial production corresponded to modest spikes in chl a concentration, suggesting a slight spring bloom (Supplementary Figure 1). To put these changes in context, peak chl a during the January summer bloom was ∼20 μg L-1 and bacterial production was ∼120 pmol L-1 h-1 (Luria et al., 2016). Nevertheless, temporal changes were already underway in the environment during the winter-to-late spring period.
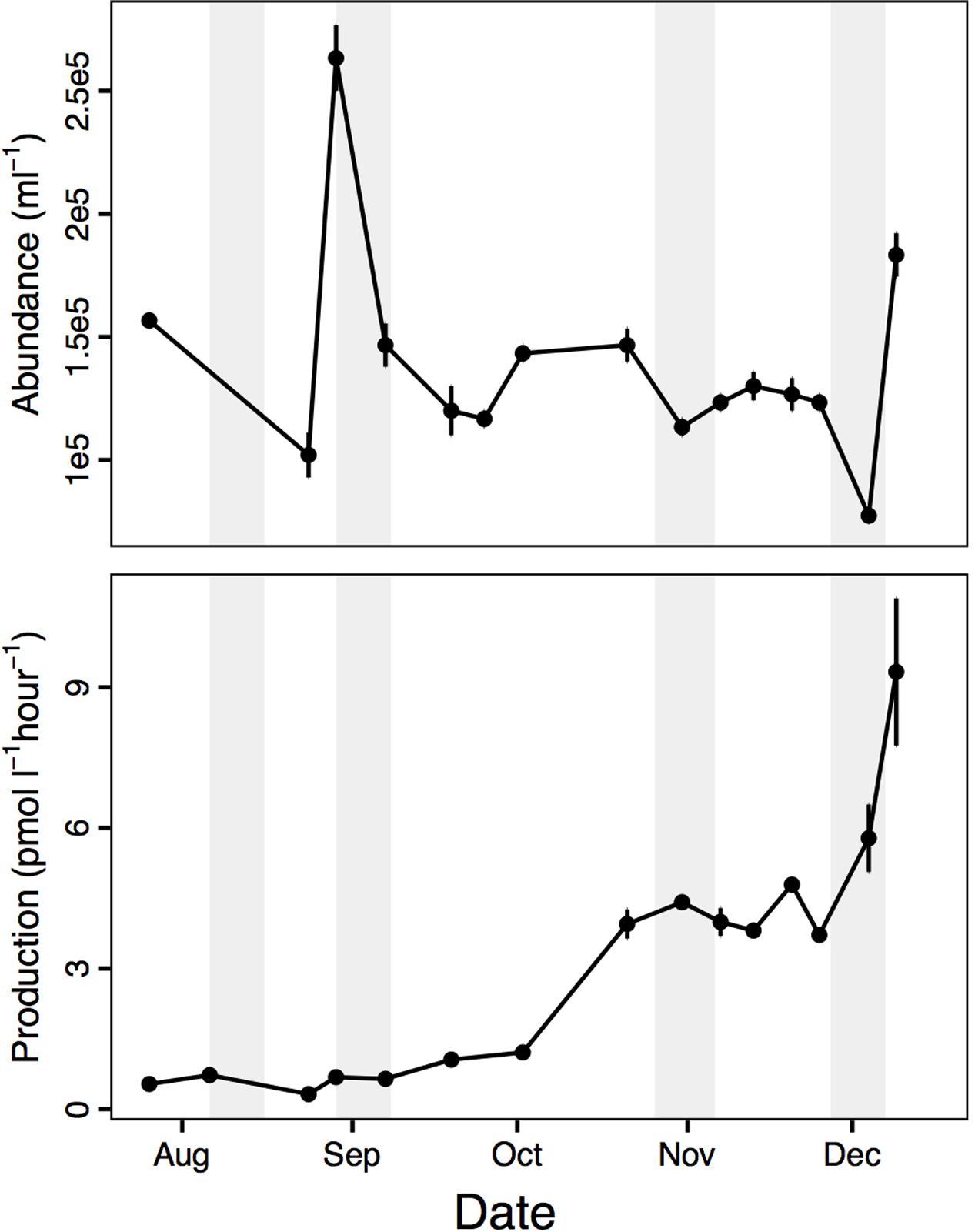
FIGURE 1. Bacterial abundance and leucine incorporation rate (production) in near shore surface water that was used in the dissolved organic matter (DOM) addition mesocosm experiments, August – December 2013 (average ± standard deviation, n = 3). Shaded bars indicate the timing of the four DOM addition experiments.
Bacterial abundance and production increased over time in both control and DOM+ mesocosms, but increases were more rapid in the DOM+ mesocosms than in controls (Figure 2). As samples for bacterial production were not filtered, these measurements included both free-living and particle attached bacteria (and archaea). The DOC concentration in DOM+ or control mesocosms did not change significantly during incubations, indicating that most of the added or ambient DOC was non-labile (Supplementary Table 1). Changes in POC and PN provided further evidence for growth of bacteria during experiments (Supplementary Figure 2). Chl a and inorganic nutrients did not show consistent differences between treatments (Supplementary Figure 3). Based on comparisons of linear models, bacterial abundance was more significantly influenced by DOM treatment (p = 0.0002, F = 15.9) and incubation day (p = 6e-12, F = 76.5) than by what month the experiment was conducted (p = 0.01, F = 3.9) (Supplementary Table 2). Bacterial production was significantly influenced by all three factors (p < 0.005, F > 9). OTU richness of free-living bacteria declined in all experiments regardless of treatment (Figure 2). Evenness and Shannon diversity also declined in most mesocosms (Supplementary Figure 4). Incubation day and month of experiments had a significant effect on richness (p < 0.0001), while any effect of the DOM treatment was too small to pass the p < 0.05 cutoff (p = 0.08). Bacterial community composition in controls and the f/2 treatment were similar (Supplementary Figure 5).
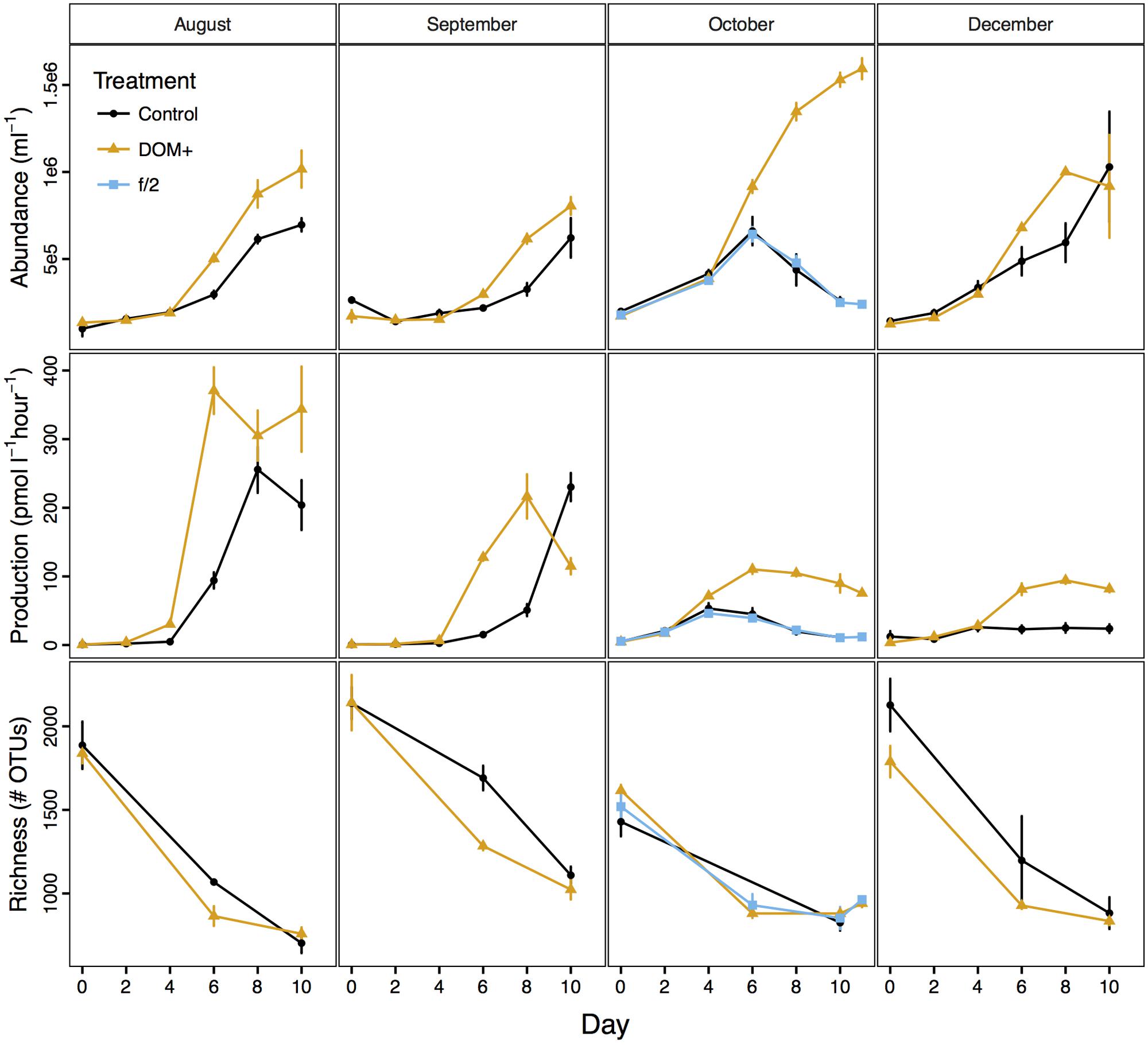
FIGURE 2. Bacterial abundance and production and observed OTU richness over the course of four mesocosm experiments (average ± standard deviation, n = 3).
Non-metric multidimensional scaling based on Bray–Curtis similarity demonstrated changes in overall free-living bacterial community composition across experiments (Figure 3). NMDS axis 1 reflected changes in community composition by incubation day. Changes in controls represent container effects, while changes in DOM+ treatments beyond controls reflect the influence of DOM addition. There were more rapid and greater changes in DOM+ treatments compared to controls. Axis 2 reflected differences in community composition between the start and end of the first two experiments (August/September) compared to the second two experiments (October/December). The grouping by August/September compared to October/December reflected differences in starting community composition between the two periods. The relative change in overall community composition, as reflected in the NMDS, was of similar magnitude across experiments.
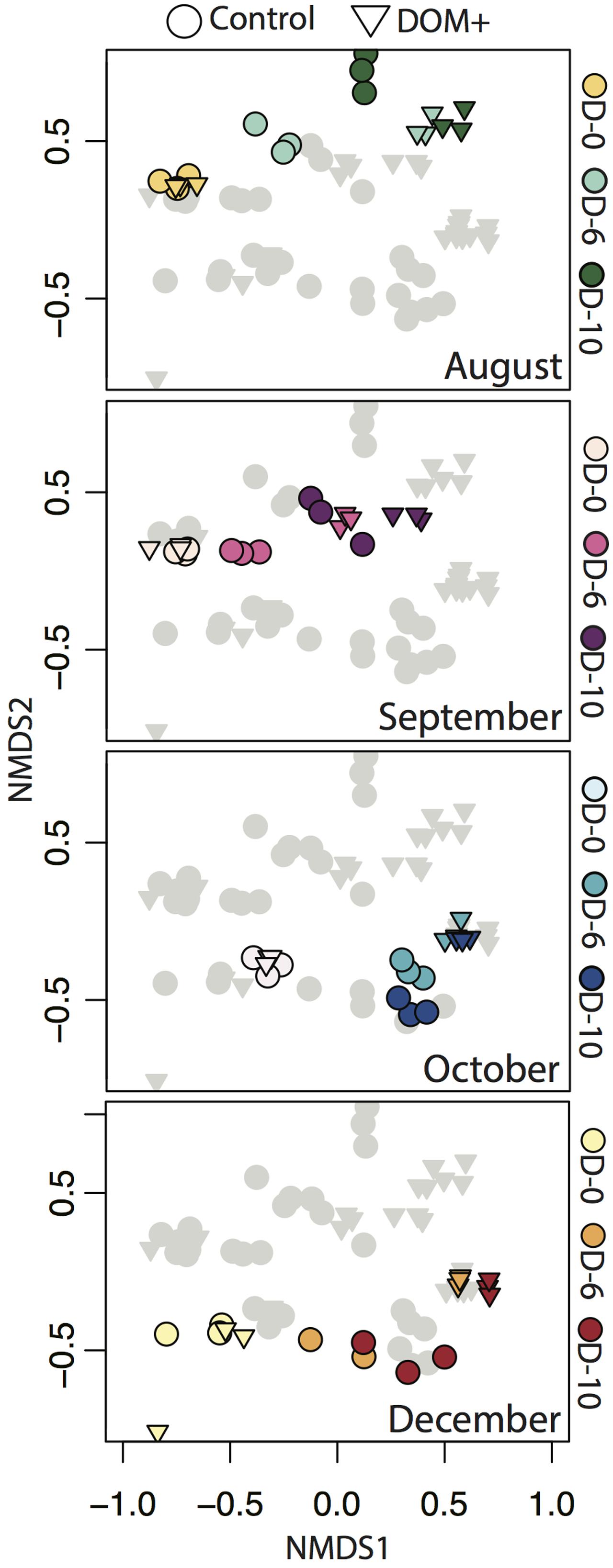
FIGURE 3. Non-metric multidimensional scaling (NMDS) of Bray–Curtis similarity indices based on OTU relative abundance across all four experiments. Each point corresponds to an individual mesocosm replicate (n = 3 per time point per treatment). All samples from all experiments are shown in each panel. Within each panel, a different experiment is highlighted with different colors corresponding to different sample days (0, 6, and 10), with the other experiments shown in gray. Control mesocosm communities are denoted by circles and DOM+ mesocosm communities are denoted by triangles.
The top 12 OTUs of free-living bacteria (by mean relative abundance across all samples) together represented 50–75% of all sequences. In all four experiments, OTUs classified as Pelagibacteraceae, Oceanospirillales, and SAR324 declined over time, while OTUs classified as Collwelliaceae and Polaribacter increased in relative abundance over time (Figure 4). Although the most abundant Collwelliaceae OTU (819278) displayed container effects, it had higher relative abundances in DOM+ mesocosms than controls on day 6 in August/September and on day 6 or 10 in October/December, indicating a response to DOM addition beyond container effects (Figure 4 and Supplementary Figure 6). A different Collwelliaceae OTU (776657) only increased in DOM+ mesocosms and not in controls, indicating a consistent response to DOM addition as the season progressed (Figure 4 and Supplementary Figure 6). The relative abundance of the most abundant Collwelliaceae OTU (819278) and another Collwelliaceae OTU (6644016) shifted between August/September and October/December. This shift corresponded to a smaller but significant in situ increase in the relative abundance of Polaribacter (907828) in October/December compared to August/September, suggesting that the higher starting abundance of Polaribacter (907828) ultimately led to its higher final abundance in October/December. Of the top 12 OTUs, 6 varied in relative abundance by 5% or more at one or more time points between the DOM+ and control communities (Supplementary Figure 6). Rhodobacteraceae (812461), Oceanospirillales (884345), and Pelagibacteraceae (838668) had similar or lower relative abundances in DOM+ mesocosms compared to controls. Polaribacter (907828) displayed both positive and negative effects between DOM+ and controls at different times, but it always showed an increase in relative abundance after the start of experiments, irrespective of treatment (Figure 4).
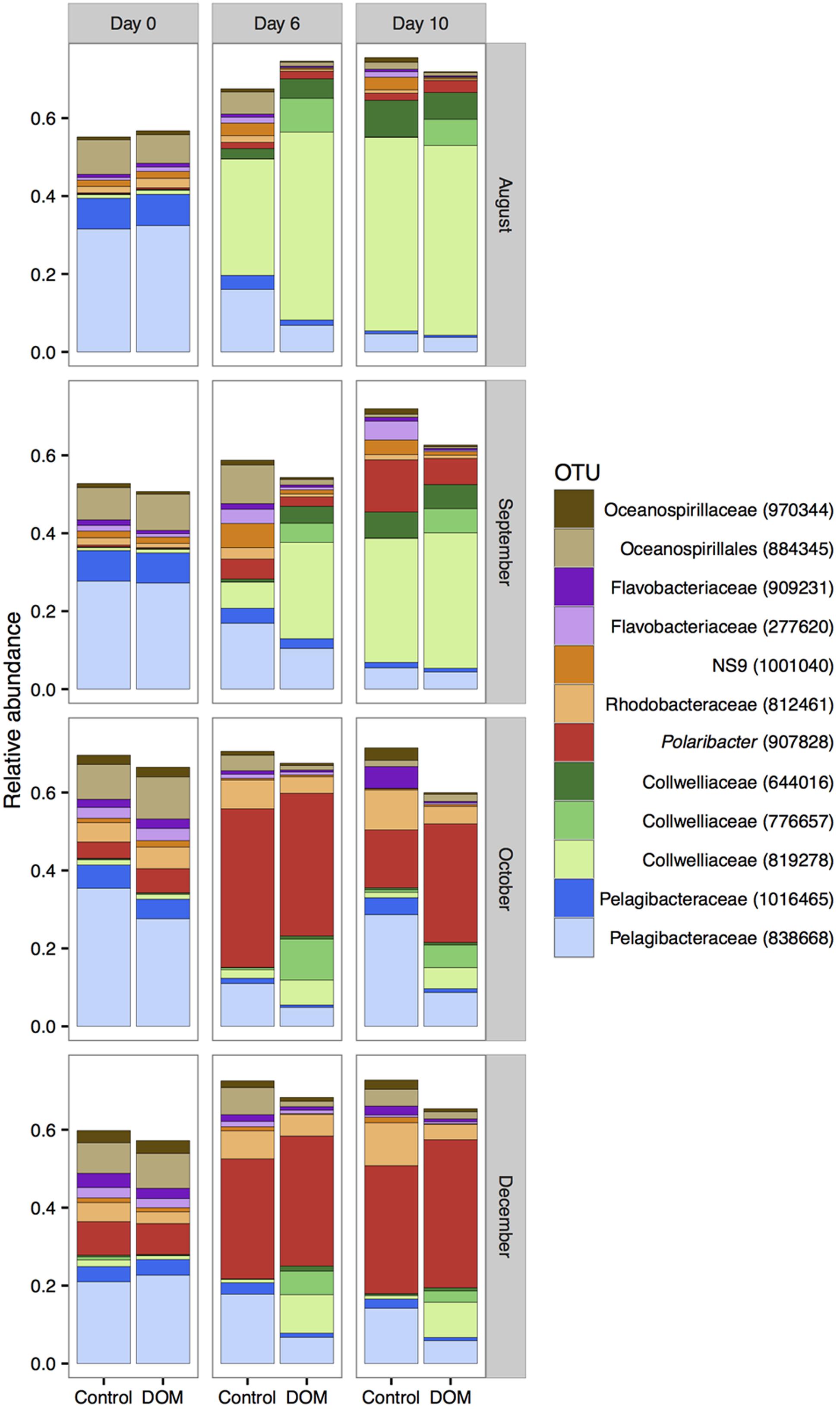
FIGURE 4. Changes in the mean relative abundance of the top 12 OTUs (by mean relative abundance) across all samples (n = 3). Note that the y-axis does not go up to 1 as the top 12 OTUs are shown instead of all OTUs.
We identified a sub-network of OTUs that were significantly associated with either the control group or the DOM treatment (r > 0.5; Figure 5). All of the OTUs that were positively correlated with DOM+ were classified as Gammaproteobacteria, including Collwelliaceae. Different OTUs identified as Collwelliaceae, reflected diversity within this family of bacteria. A few OTUs in this sub-network were negatively correlated with the control. A second sub-network linking OTUs to bacterial production showed that four Collwelliaceae OTUs correlated positively with bacterial production. Alphaproteobacteria OTUs, over half of them classified as Pelagibacteraceae, as well as two Gammaproteobacteria, HTCC2089 and HTCC2188, were negatively correlated with bacterial production (r > 0.7).
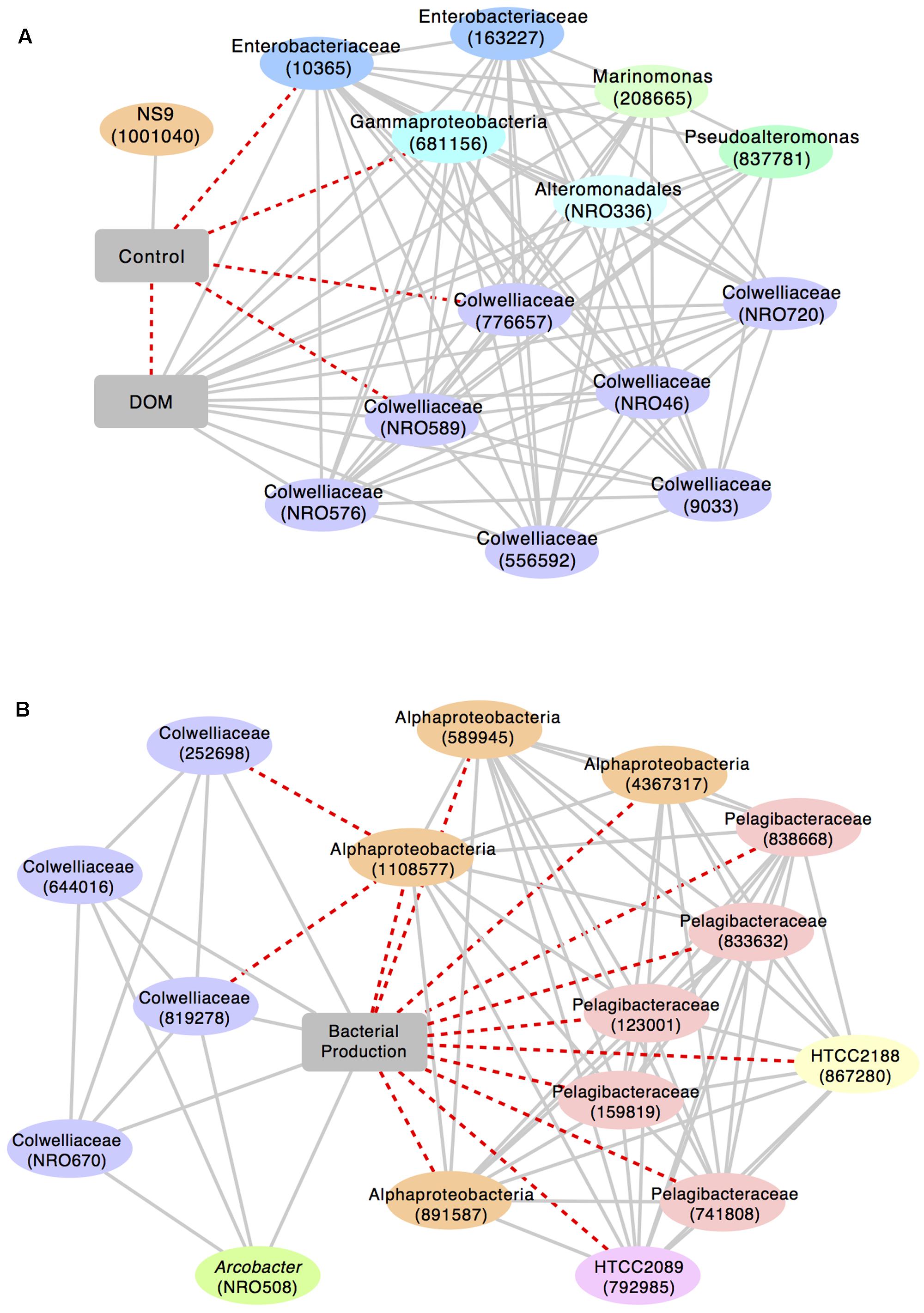
FIGURE 5. Sub-networks of highly correlated OTUs built around (A) treatment level (control vs. DOM+; r > 0.5) and (B) bacterial production (r > 0.7). Solid, gray lines represent positive correlations; dashed, red lines represent negative correlations. Closest taxonomic identification and reference number (in parentheses) are given for each OTU. “NRO” in the reference number indicates a new reference OTU, i.e., an OTU generated through Qiime’s open reference OTU picking pipeline from sequences that did not initially match against the Greengenes database.
Phytoplankton blooms and the resulting release of labile DOM are thought to be a major driver of marine bacterial community composition. Observational studies have shown that bacterial community composition and activity vary during phytoplankton blooms (Pinhassi and Hagström, 2000; Fandino et al., 2001; West et al., 2008; Tada et al., 2011; Teeling et al., 2012; Klindworth et al., 2014; Wemheuer et al., 2015). Similarly, the composition of DOM has been shown to change during phytoplankton blooms (Zhang et al., 2015; Sperling et al., 2017). This body of work suggests that bacterial succession is coupled to different stages of DOM decomposition through changes in relative abundance of bacterial groups with different metabolic strategies and substrate preferences (Cottrell and Kirchman, 2000; Alonso-Sáez and Gasol, 2007; Poretsky et al., 2010; Rinta-Kanto et al., 2012; Sarmento and Gasol, 2012; Teeling et al., 2012). This pattern has been cited as evidence in support of niche partitioning, which may relieve competition between taxa and help explain high microbial diversity (Hutchinson, 1957; Teeling et al., 2012; Alexander et al., 2015). Alternatively, some studies have found that changes in DOM supply only slightly affect bacterial community composition, suggesting that physiological responses of metabolically versatile bacteria may be a factor, in addition to niche partitioning (Kirchman et al., 2004; Rooney-Varga et al., 2005; Rink et al., 2007).
There is growing evidence that labile organic matter availability is a primary factor controlling bacterial growth and community composition in the Southern Ocean (Thingstad and Martinussen, 1991; Kirchman et al., 2009; Ducklow et al., 2012; Kim et al., 2014; Luria et al., 2016). However, directly testing the effects of phytoplankton-derived DOM on bacterial seasonal succession has proven challenging. Mesocosm experiments, like the one we conducted here, have traditionally relied on adding low molecular weight compounds like glucose or amino acids (Havskum et al., 2003; Allers et al., 2007; Gómez-Consarnau et al., 2012). For example, Ducklow et al. (2011) demonstrated that glucose enrichment of WAP seawater reduced bacterial diversity. Using more complex DOM substrates derived from phytoplankton cultures, several recent studies have shown that a wider range of bacterial taxa responds readily to phytoplankton, especially diatom-derived DOM, and that DOM originating from different phytoplankton species stimulates different bacterial phylogenetic groups (Romera-Castillo et al., 2011; Nelson and Carlson, 2012; Sarmento and Gasol, 2012). Conversely, Landa et al. (2014) and Sharma et al. (2014) found that varied natural DOM sources did not have differential effects on bacterial community composition despite variation in carbon quality and quantity. We selected the diatom T. weissflogii as a DOM source. Other species of Thalassiosira are widespread in the WAP region, suggesting that the DOM that we added could be a good analog for DOM exudates for part of the phytoplankton community. However, the quantity and quality of DOM exuded by phytoplankton vary with species and physiological state and DOM from a single source (one species in one growth phase) does not represent the entire spectrum of changes in DOM composition and supply that probably occur during a phytoplankton bloom (Becker et al., 2014; Landa et al., 2014). Similarly, the isolation technique that we used selects for hydrophobic DOM and does not efficiently concentrate very polar compounds. We assume that qualitatively our DOM extracts contained many of the same compounds or types of compounds produced by microbes in seawater. However, we recognize that the amounts and specific details of the compounds will be different. The concentration of DOC that we added is in the range of temporal variation at our study location. Background DOC based on deep-water samples is ∼40 μmol L-1 and nearshore surface water early in spring is around this concentration, consistent with the pattern that we observed in the environment during our study (Supplementary Figure 1). DOC increases and becomes more variable as the season progresses and phytoplankton blooms develop (average ± standard deviation of 51 ± 11 μmol L-1) (Ducklow, 2017), similar to other coastal polar locations (Kirchman et al., 2009).
In our experiments, temporal changes in controls followed those that occurred in DOM treatments, indicating container effects of increasing cell numbers of rapidly growing bacteria of certain taxonomic groups over others (Zobell, 1943; Ferguson et al., 1984; Eilers et al., 2000; Massana et al., 2001). Polaribacter are heterotrophs that target high molecular weight organic matter and increase rapidly during phytoplankton blooms in coastal Antarctic waters (Fernández-Gómez et al., 2013; Williams et al., 2013; Luria et al., 2016). The relative abundance of Polaribacter increased rapidly in our experiments, similar to other incubation experiments of coastal Antarctic waters (Massana et al., 2001; Landa et al., 2016). The specific Polaribacter OTU (907828) that increased in our experiments was also abundant in the environment, reaching peak relative abundance of 20–50% of sequences at our study location, depending on the year (Luria, 2016; Luria et al., 2016). The most abundant Collwelliaceae OTU (819278) in our experiments, showed strong increases in controls, but also consistent increases in DOM treatments over controls. A different Collwelliaceae OTU (810446) increased rapidly in relative abundance in the environment (up to ∼30% of sequences, depending on the year) during the primary phytoplankton bloom in our study area (Luria, 2016; Luria et al., 2016). Based on flow cytometry of bacterioplankton in our study area, Bowman et al. (2017) found an increase in the proportion of high nucleic acid bacteria compared to low nucleic acid bacteria during a phytoplankton bloom, indicating a physiological shift in bacterial communities in addition to taxonomic changes. Colwellia spp. are commonly isolated psychrophiles from polar sea-ice or deep-sea environments and model organisms of psychrophily, reflecting their capacity for rapid growth in cold environments (Deming et al., 1988; Bowman et al., 1997; Methé et al., 2005). Based on culture-independent techniques, Colwellia spp. had a high relative abundance in deep Antarctic coastal waters, associated with sinking Phaeocystis particles (Delmont et al., 2014). Conditions conducive to rapid growth of bacterioplankton occur in Antarctic coastal waters and these types of bacteria were favored in our experiments.
In our experiments, we observed differences in bacterial production and community composition that corresponded to different times during the winter-to-late spring period. Bacterial production in the mesocosms decreased from August/September to October/December, indicating the study period was split into two phases, rather than changing constantly in one direction or another, as we initially hypothesized. Subtle changes in the environment that occurred before our October experiment, as reflected in chl a and bacterial production, might be related to the shift that we observed. Rather than a decrease in magnitude of overall changes in community composition between the two periods, we observed a shift in community composition from Collwelliaceae OTUs (819278 and 6644016) in August/September to Polaribacter (907828) in October/December. This shift appears to be caused by changes in the relative abundance of Polaribacter (907828) in the starting seawater, which seemed to enable Polaribacter (907828) to outcompete the two Collwelliaceae OTUs (819278 and 6644016) in the October/December experiments. Meanwhile, another Collwelliaceae OTU (776657) did not appear to be affected by the increase in relative abundance of Polaribacter (907828). The factors that governed the negative relationship between OTUs are not clear but could be related to competitive interactions, direct antagonism, or differential top-down control (Fuhrman, 2009). Our study suggests that diversity within the Collwelliaceae could have functional implications for interactions among other taxa of bacterioplankton during phytoplankton bloom periods.
Despite longstanding debate about the role of temperature in regulating bacterial production (Pomeroy and Wiebe, 2001; Kirchman et al., 2009), a decade-scale study found that the relationship between bacterial production and temperature varied from year to year and was even negative in some years (Ducklow et al., 2012). In our study, although August mesocosms were incubated at 3°C, changes in community composition between August and September were similar. Ducklow et al. (1999) found no effect of increasing temperature from 2° to 4°C on bacterial production in incubations of Ross Sea water. Differing bacterial mortality rates in the initial source water might have been a factor. Predation in WAP waters has been shown to increase exponentially with temperature and during natural or simulated phytoplankton blooms (Bird and Karl, 1991; Duarte et al., 2005; Garzio et al., 2013). Brum et al. (2016) found that viral abundance and lysogeny increased during the spring-to-summer transition in our study region. Bowman et al. (2017) further suggested the importance of top-down controls in structuring bacterial communities in Antarctic coastal waters in addition to bottom-up controls.
Our initial hypothesis of change in bacterial community responses as the season progressed was grounded in the niche model of community assembly wherein similar environmental conditions (e.g., DOM enrichment) select for the same or similar species from a diverse initial species pool, producing communities with similar structures (Fuhrman et al., 2006). This contrasts with the neutral model of community assembly in which stochastic forces, including growth, dispersal, and mortality are dominant forces in community assembly (Hubbell, 2001). While the differences we observed may be governed by factors that we did not consider (i.e., predation), our findings could be interpreted as priority effects in which variation in the initial relative abundance of species alters final community structure in addition to environmental filtering (Drake, 1990; Chase, 2007; Fukami and Nakajima, 2011; Nemergut et al., 2013). Although priority effects have been demonstrated for sequential colonization of a site by microbes, the application of this concept to slight numerical advantages in a complex initial inoculum is less clear (Warren et al., 2003; Jiang and Patel, 2008; Peay et al., 2010). Strong inter-annual and long-term climate change drives variation in WAP sea ice extent and duration and hence the timing and intensity of phytoplankton blooms (Smith et al., 2008). Subtle changes in bacterial community composition during spring set the stage for the ultimate trajectory of bacterial succession upon DOM addition.
CL, LA-Z, HD, and JR designed and conducted the study. LA-Z and JR provided sequence data for the study. AR and CL cultivated diatoms and DR and CL collected DOM. CL analyzed data and wrote the manuscript with guidance from JR, LA-Z, and HD. All authors approved the final version of the manuscript.
CL was partially funded by the Graduate School and the Department of Ecology and Evolutionary Biology at Brown University and the Brown University-Marine Biological Laboratory Joint Graduate Program. This material is based upon work supported by the National Science Foundation under Grant Nos. ANT-1142114 to LA-Z, OPP-0823101 and PLR-1440435 to HD, and ANT-1141993 to JR. The Gordon and Betty Moore Foundation grant 1711 supported work by DR.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge the help of the following people for providing field and laboratory assistance at Palmer Station as part of this project: Jamie Collins, Sharon Grim, Sean O’Neill, Monica Stegman, Sebastian Vivancos, and Madelyne Willis, as well as the staff at Palmer Station. We thank Annaliese Jones, Leslie Murphy, and Naomi Shelton for conducting laboratory analyses and Doug Rasher for helpful discussion.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02117/full#supplementary-material
Alexander, H., Jenkins, B. D., Rynearson, T. A., and Dyhrman, S. T. (2015). Metatranscriptome analyses indicate resource partitioning between diatoms in the field. Proc. Natl. Acad. Sci. U.S.A. 112, E2182–E2190. doi: 10.1073/pnas.1421993112
Allers, E., Gómez-Consarnau, L., Pinhassi, J., Gasol, J. M., Šimek, K., and Pernthaler, J. (2007). Response of Alteromonadaceae and Rhodobacteriaceae to glucose and phosphorus manipulation in marine mesocosms. Environ. Microbiol. 9, 2417–2429. doi: 10.1111/j.1462-2920.2007.01360.x
Allison, S. D., and Martiny, J. B. (2008). Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. U.S.A. 105(Suppl. 1), 11512–11519. doi: 10.1073/pnas.0801925105
Alonso-Sáez, L., and Gasol, J. M. (2007). Seasonal variations in the contributions of different bacterial groups to the uptake of low-molecular-weight compounds in northwestern Mediterranean coastal waters. Appl. Environ. Microbiol. 73, 3528–3535. doi: 10.1128/AEM.02627-06
Becker, J. W., Berube, P. M., Follett, C. L., Waterbury, J. B., Chisholm, S. W., DeLong, E. F., et al. (2014). Closely related phytoplankton species produce similar suites of dissolved organic matter. Front. Microbiol. 5:111. doi: 10.3389/fmicb.2014.00111
Bird, D. F., and Karl, D. M. (1991). Spatial patterns of glutamate and thymidine assimilation in Bransfield Strait, Antarctica during and following the austral spring bloom. Deep Sea Res. I 38, 1057–1075. doi: 10.1016/0198-0149(91)90096-X
Bowman, J. P., McCammon, S. A., Brown, M. V., Nichols, D. S., and McMeekin, T. A. (1997). Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl. Environ. Microbiol. 63, 3068–3078.
Bowman, J. S., Amaral-Zettler, L., Rich, J., Luria, C., and Ducklow, H. W. (2017). Segmentation of the bacterial community facilitates the prediction of ecosystem function in Arthur Harbor, a highly productive coastal setting along the west Antarctic Peninsula. ISME J. 11, 1460–1471. doi: 10.1038/ismej.2016.204
Brum, J. R., Hurwitz, B. L., Schofield, O., Ducklow, H. W., and Sullivan, M. B. (2016). Seasonal time bombs: dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J. 10, 437–449. doi: 10.1038/ismej.2015.125
Buchan, A., LeCleir, G. R., Gulvik, C. A., and González, J. M. (2014). Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698. doi: 10.1038/nrmicro3326
Busch, K., Endres, S., Iversen, M. H., Michels, J., Nöthig, E.-M., and Engel, A. (2017). Bacterial colonization and vertical distribution of marine gel particles (TEP and CSP) in the arctic Fram Strait. Front. Mar. Sci. 4:166. doi: 10.3389/fmars.2017.00166
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carlson, C. A., and Hansell, D. A. (2015). “DOM sources, sinks, reactivity, and budgets,” in Biogeochemistry of Marine Dissolved Organic Matter, eds D. A. Hansell and C. A. Carlson (Amsterdam: Elsevier), 65–126. doi: 10.1016/B978-0-12-405940-5.00003-0
Carlson, C. A., Hansell, D. A., Nelson, N. B., Siegel, D. A., Smethie, W. M., Khatiwala, S., et al. (2010). Dissolved organic carbon export and subsequent remineralization in the mesopelagic and bathypelagic realms of the North Atlantic basin. Deep Sea Res. II 57, 1433–1445. doi: 10.1016/j.dsr2.2010.02.013
Chambers, J. M. (1992). “Linear models,” in Statistical Models in S, eds J. M. Chambers and T. J. Hastie (Boca Raton, FL: CRC Press), 95–144.
Chase, J. M. (2007). Drought mediates the importance of stochastic community assembly. Proc. Natl. Acad. Sci. U.S.A. 104, 17430–17434. doi: 10.1073/pnas.0704350104
Chronopoulou, P.-M., Sanni, G. O., Silas-Olu, D. I., Meer, J. R., Timmis, K. N., Brussaard, C. P., et al. (2015). Generalist hydrocarbon-degrading bacterial communities in the oil-polluted water column of the North Sea. Microb. Biotechnol. 8, 434–447. doi: 10.1111/1751-7915.12176
Cottrell, M. T., and Kirchman, D. L. (2000). Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low-and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66, 1692–1697. doi: 10.1128/AEM.66.4.1692-1697.2000
Delmont, T. O., Hammar, K. M., Ducklow, H. W., Yager, P. L., Post, A. F., Chafee, M., et al. (2014). Phaeocystis antarctica blooms strongly influence bacterial community structures in the Amundsen Sea polynya. Front. Microbiol. 5:646. doi: 10.3389/fmicb.2014.00646
Deming, J. W., Somers, L. K., Straube, W. L., Swartz, D. G., and Macdonell, M. T. (1988). Isolation of an obligately barophilic bacterium and description of a new genus, Colwellia gen. nov. Syst. Appl. Microbiol. 10, 152–160. doi: 10.1016/S0723-2020(88)80030-4
Drake, J. A. (1990). Communities as assembled structures: do rules govern pattern? Trends Ecol. Evol. 5, 159–164. doi: 10.1016/0169-5347(90)90223-Z
Duarte, C. M., Agustí, S., Vaqué, D., Agawin, N. S. R., Felipe, J., Casamayor, E. O., et al. (2005). Experimental test of bacteria-phytoplankton coupling in the Southern Ocean. Limnol. Oceanogr. 50, 1844–1854. doi: 10.4319/lo.2005.50.6.1844
Ducklow, H. (2003). “Seasonal production and bacterial utilization of DOC in the Ross Sea, Antarctica,” in Biogeochemistry of the Ross Sea, eds G. R. Ditullio and R. B. Dunbar (Hoboken, NJ: Wiley), 143–157.
Ducklow, H. (2017). Data from: Dissolved Organic Carbon (DOC) Taken from Discrete Water Column Samples Collected between October and April at Palmer Station, 2002–2012. Palisades, NY: Lamont Doherty Earth Observatory.
Ducklow, H., Carlson, C., and Smith, W. (1999). Bacterial growth in experimental plankton assemblages and seawater cultures from the Phaeocystis antarctica bloom in the Ross Sea, Antarctica. Aquat. Microb. Ecol. 19, 215–227. doi: 10.3354/ame019215
Ducklow, H., Vernet, M., and Prezelin, B. (2016). Data from: Dissolved Inorganic Nutrients Including 5 Macro Nutrients: Silicate, Phosphate, Nitrate, Nitrite, and Ammonium from Water Column Bottle Samples Collected Between October, and April at Palmer Station, 1991–2015. Palisades, NY: Lamont Doherty Earth Observatory.
Ducklow, H. W., Myers, K. M., Erickson, M., Ghiglione, J. F., and Murray, A. E. (2011). Response of a summertime Antarctic marine bacterial community to glucose and ammonium enrichment. Aquat. Microb. Ecol. 64, 205–220. doi: 10.3354/ame01519
Ducklow, H. W., Schofield, O., Vernet, M., Stammerjohn, S., and Erickson, M. (2012). Multiscale control of bacterial production by phytoplankton dynamics and sea ice along the western Antarctic Peninsula: a regional and decadal investigation. J. Mar. Syst. 98, 26–39. doi: 10.1016/j.jmarsys.2012.03.003
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Eilers, H., Pernthaler, J., Glo, F. O., and Amann, R. (2000). Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66, 3044–3051. doi: 10.1128/AEM.66.7.3044-3051.2000
Eren, A. M., Vineis, J. H., Morrison, H. G., and Sogin, M. L. (2013). A filtering method to generate high quality short reads using Illumina Paired-End technology. PLOS ONE 8:e66643. doi: 10.1371/journal.pone.0066643
Fandino, L., Riemann, L., Steward, G., Long, R., and Azam, F. (2001). Variations in bacterial community structure during a dinoflagellate bloom analyzed by DGGE and 16S rDNA sequencing. Aquat. Microb. Ecol. 23, 119–130. doi: 10.3354/ame023119
Faust, K., Sathirapongsasuti, J. F., Izard, J., Segata, N., Gevers, D., Raes, J., et al. (2012). Microbial co-occurrence relationships in the human microbiome. PLOS Comput. Biol. 8:e1002606. doi: 10.1371/journal.pcbi.1002606
Ferguson, R. L., Buckley, E. N., and Palumbot, A. V. (1984). Response of marine bacterioplankton to differential filtration and confinement. Appl. Environ. Microbiol. 47, 49–55.
Fernández-Gómez, B., Richter, M., Schüler, M., Pinhassi, J., Acinas, S. G., González, J. M., et al. (2013). Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J. 7, 1026–1037. doi: 10.1038/ismej.2012.169
Fogg, G. E. (1983). The ecological significance of extracellular products of phytoplankton photosynthesis. Bot. Mar. 26, 3–14. doi: 10.1515/botm.1983.26.1.3
Fuhrman, J. A. (2009). Microbial community structure and its functional implications. Nature 459, 193–199. doi: 10.1038/nature08058
Fuhrman, J. A., Hewson, I., Schwalbach, M. S., Steele, J. A., Brown, M. V., and Naeem, S. (2006). Annually reoccurring bacterial communities are predictable from ocean conditions. Proc. Natl. Acad. Sci. U.S.A. 103, 13104–13109. doi: 10.1073/pnas.0602399103
Fukami, T., and Nakajima, M. (2011). Community assembly: alternative stable states or alternative transient states? Ecol. Lett. 14, 973–984. doi: 10.1111/j.1461-0248.2011.01663.x
Garzio, L. M., Steinberg, D. K., Erickson, M., and Ducklow, H. W. (2013). Microzooplankton grazing along the Western Antarctic Peninsula. Aquat. Microb. Ecol. 70, 215–232. doi: 10.3354/ame01655
Gobler, C. J., Hutchins, D. A., Fisher, N. S., Cosper, E. M., and Sanudo-Wilhelmy, S. A. (1997). Release and bioavailability of C, N, P, Se, and Fe following viral lysis of a marine chrysophyte. Limnol. Oceanogr. 42, 1492–1504. doi: 10.4319/lo.1997.42.7.1492
Gómez-Consarnau, L., Lindh, M. V., Gasol, J. M., and Pinhassi, J. (2012). Structuring of bacterioplankton communities by specific dissolved organic carbon compounds. Environ. Microbiol. 14, 2361–2378. doi: 10.1111/j.1462-2920.2012.02804.x
Guillard, R. R. L. (1975). Culture of Marine Invertebrate Animals Culture of Phytoplankton for Feeding Marine Invertebrates. New York, NY: Plenum Press, 26–60.
Havskum, H., Thingstad, T. F., Scharek, R., Peters, F., Berdalet, E., Sala, M. M., et al. (2003). Silicate and labile DOC interfere in structuring the microbial food web via algal-bacterial competition for mineral nutrients: results of a mesocosm experiment. Limnol. Oceanogr. 48, 129–140. doi: 10.4319/lo.2003.48.1.0129
Hedges, J. I. (1992). Global biogeochemical cycles: progress and problems. Mar. Chem. 39, 67–93. doi: 10.1016/0304-4203(92)90096-S
Hubbell, S. P. (2001). The Unified Neutral Theory of Biodiversity and Biogeography (MPB-32), Vol. 32. Princeton, NJ: Princeton University Press.
Hutchinson, G. (1957). Concluding remarks. Cold Sprig Harb. Symp. Quant. Biol. 22, 415–427. doi: 10.1101/SQB.1957.022.01.039
Jiang, L., and Patel, S. N. (2008). Community assembly in the presence of disturbance: a microcosm experiment. Ecology 89, 1931–1940. doi: 10.1890/07-1263.1
Kim, H., and Ducklow, H. (2016). A decadal (2002-2014) analysis for dynamics of heterotrophic bacteria in an Antarctic coastal ecosystem: variability and physical and biogeochemical forcings. Front. Mar. Sci. 3:214. doi: 10.3389/fmars.2016.00214
Kim, J.-G., Park, S.-J., Quan, Z.-X., Jung, M.-Y., Cha, I.-T., Kim, S.-J., et al. (2014). Unveiling abundance and distribution of planktonic bacteria and archaea in a polynya in Amundsen Sea, Antarctica. Environ. Microbiol. 16, 1566–1578. doi: 10.1111/1462-2920.12287
Kindt, R., and Coe, R. (2005). Tree Diversity Analysis: A Manual and Software for Common Statistical Methods for Ecological and Biodiversity Studies. Nairobi: World Agroforestry Centre (ICRAF).
Kirchman, D. L., Dittel, A. I., Findlay, S. E., and Fischer, D. (2004). Changes in bacterial activity and community structure in response to dissolved organic matter in the Hudson River, New York. Aquat. Microb. Ecol. 35, 243–257. doi: 10.3354/ame035243
Kirchman, D. L., Morán, X. A. G., and Ducklow, H. (2009). Microbial growth in the polar oceans - role of temperature and potential impact of climate change. Nat. Rev. Microbiol. 7, 451–459. doi: 10.1038/nrmicro2115
Klindworth, A., Mann, A. J., Huang, S., Wichels, A., Quast, C., Waldmann, J., et al. (2014). Diversity and activity of marine bacterioplankton during a diatom bloom in the North Sea assessed by total RNA and pyrotag sequencing. Mar. Genomics 18(Pt B), 185–192. doi: 10.1016/j.margen.2014.08.007
Lampert, W. (1978). Release of dissolved organic carbon by grazing zooplankton. Limnol. Oceanogr. 23, 831–834. doi: 10.3389/fmicb.2012.00302
Landa, M., Blain, S., Christaki, U., Monchy, S., and Obernosterer, I. (2016). Shifts in bacterial community composition associated with increased carbon cycling in a mosaic of phytoplankton blooms. ISME J. 10, 39–50. doi: 10.1038/ismej.2015.105
Landa, M., Cottrell, M. T., Kirchman, D. L., Kaiser, K., Medeiros, P. M., Tremblay, L., et al. (2014). Phylogenetic and structural response of heterotrophic bacteria to dissolved organic matter of different chemical composition in a continuous culture study. Environ. Microbiol. 16, 1668–1681. doi: 10.1111/1462-2920.12242
Luria, C. M. (2016). Drivers of Microbial Community Assembly and Activity in Antarctic Coastal Waters. Ph.D. thesis, Brown University, Providence, RI.
Luria, C. M., Amaral-Zettler, L. A., Ducklow, H. W., and Rich, J. J. (2016). Seasonal succession of bacterial communities in coastal waters of the western Antarctic Peninsula. Front. Microbiol. 7:1731. doi: 10.3389/fmicb.2016.01731
Massana, R., Pedrós-Alió, C., Casamayor, E. O., and Gasol, J. M. (2001). Changes in marine bacterioplankton phylogenetic composition during incubations designed to measure biogeochemically significant parameters. Limnol. Oceanogr. 46, 1181–1188. doi: 10.4319/lo.2001.46.5.1181
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., DeSantis, T. Z., Probst, A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
Meredith, M. P., and King, J. C. (2005). Rapid climate change in the ocean west of the Antarctic Peninsula during the second half of the 20th century. Geophys. Res. Lett. 32, L19604. doi: 10.1029/2005GL024042
Methé, B. A., Nelson, K. E., Deming, J. W., Momen, B., Melamud, E., Zhang, X., et al. (2005). The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc. Natl. Acad. Sci. U.S.A. 102, 10913–10918. doi: 10.1073/pnas.0504766102
Mou, X., Hodson, R. E., and Moran, M. A. (2007). Bacterioplankton assemblages transforming dissolved organic compounds in coastal seawater. Environ. Microbiol. 9, 2025–2037. doi: 10.1111/j.1462-2920.2007.01318.x
Nelson, C. E., and Carlson, C. A. (2012). Tracking differential incorporation of dissolved organic carbon types among diverse lineages of Sargasso Sea bacterioplankton. Environ. Microbiol. 14, 1500–1516. doi: 10.1111/j.1462-2920.2012.02738.x
Nemergut, D. R., Schmidt, S. K., Fukami, T., O’Neill, S. P., Bilinski, T. M., Stanish, L. F., et al. (2013). Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356. doi: 10.1128/MMBR.00051-12
Newton, R. J., Griffin, L. E., Bowles, K. M., Meile, C., Gifford, S., Givens, C. E., et al. (2010). Genome characteristics of a generalist marine bacterial lineage. ISME J. 4, 784–798. doi: 10.1038/ismej.2009.150
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R., et al. (2015). vegan: Community Ecology Package. Available at: https://cran.r-project.org
Peay, K. G., Garbelotto, M., and Bruns, T. D. (2010). Evidence of dispersal limitation in soil microorganisms: isolation reduces species richness on mycorrhizal tree islands. Ecology 91, 3631–3640. doi: 10.1890/09-2237.1
Pinhassi, J., and Hagström, Å. (2000). Seasonal succession in marine bacterioplankton. Aquat. Microb. Ecol. 21, 245–256. doi: 10.3354/ame021245
Pinhassi, J., Sala, M. M., Havskum, H., Peters, F., Guadayol, O., Malits, A., et al. (2004). Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70, 6753–6766. doi: 10.1128/AEM.70.11.6753-6766.2004
Pomeroy, L. R., and Wiebe, W. J. (2001). Temperature and substrates as interactive limiting factors for marine heterotrophic bacteria. Aquat. Microb. Ecol. 23, 187–204. doi: 10.3354/ame023187
Poretsky, R. S., Sun, S., Mou, X., and Moran, M. A. (2010). Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ. Microbiol. 12, 616–627. doi: 10.1111/j.1462-2920.2009.02102.x
R Development Core Team (2008). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rink, B., Seeberger, S., Martens, T., Duerselen, C.-D., Simon, M., and Brinkhoff, T. (2007). Effects of phytoplankton bloom in a coastal ecosystem on the composition of bacterial communities. Aquat. Microb. Ecol. 48, 47–60. doi: 10.3354/ame048047
Rinta-Kanto, J. M., Sun, S., Sharma, S., Kiene, R. P., and Moran, M. A. (2012). Bacterial community transcription patterns during a marine phytoplankton bloom. Environ. Microbiol. 14, 228–239. doi: 10.1111/j.1462-2920.2011.02602.x
Romera-Castillo, C., Sarmento, H., Álvarez-Salgado, X. A., Gasol, J. M., and Marrasé, C. (2011). Net production and consumption of fluorescent colored dissolved organic matter by natural bacterial assemblages growing on marine phytoplankton exudates. Appl. Environ. Microbiol. 77, 7490–7498. doi: 10.1128/AEM.00200-11
Rooney-Varga, J. N., Giewat, M. W., Savin, M. C., Sood, S., LeGresley, M., and Martin, J. (2005). Links between phytoplankton and bacterial community dynamics in a coastal marine environment. Microb. Ecol. 49, 163–175. doi: 10.1007/s00248-003-1057-0
Saba, G. K., Fraser, W. R., Saba, V. S., Iannuzzi, R. A., Coleman, K. E., Doney, S. C., et al. (2014). Winter and spring controls on the summer food web of the coastal West Antarctic Peninsula. Nat. Commun. 5:4318. doi: 10.1038/ncomms5318
Sarmento, H., and Gasol, J. M. (2012). Use of phytoplankton-derived dissolved organic carbon by different types of bacterioplankton. Environ. Microbiol. 14, 2348–2360. doi: 10.1111/j.1462-2920.2012.02787.x
Schofield, O., and Vernet, M. (2016). Data from: Chlorophyll and Phaeopigments from Water Column Samples, Collected at Selected Depths at Palmer Station Antarctica, During Palmer LTER Field Seasons, 1991–2015. New Brunswick, NJ: Rutgers University.
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., and Ramage, D. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sharma, A. K., Becker, J. W., Ottesen, E. A., Bryant, J. A., Duhamel, S., Karl, D. M., et al. (2014). Distinct dissolved organic matter sources induce rapid transcriptional responses in coexisting populations of Prochlorococcus, Pelagibacter and the OM60 clade. Environ. Microbiol. 16, 2815–2830. doi: 10.1111/1462-2920.12254
Smith, R. C., Martinson, D. G., Stammerjohn, S. E., Iannuzzi, R. A., and Ireson, K. (2008). Bellingshausen and western Antarctic Peninsula region: pigment biomass and sea-ice spatial/temporal distributions and interannual variability. Deep Sea Res. II 55, 1949–1963. doi: 10.1016/j.dsr2.2008.04.027
Sperling, M., Piontek, J., Engel, A., Wiltshire, K. H., Niggemann, J., Gerdts, G., et al. (2017). Combined carbohydrates support rich communities of particle-associated marine bacterioplankton. Front. Microbiol. 8:65. doi: 10.3389/fmicb.2017.00065
Stammerjohn, S. E., Martinson, D. G., Smith, R. C., Yuan, X., and Rind, D. (2008). Trends in Antarctic annual sea ice retreat and advance and their relation to el Niño–Southern Oscillation and Southern Annular Mode variability. J. Geophys. Res. 113, C03S90. doi: 10.1029/2007JC004269
Straza, T. R., Ducklow, H. W., Murray, A. E., and Kirchman, D. L. (2010). Abundance and single-cell activity of bacterial groups in Antarctic coastal waters. Limnol. Oceanogr. 55, 2526–2536. doi: 10.1128/AEM.00121-14
Tada, Y., Taniguchi, A., Nagao, I., Miki, T., Uematsu, M., Tsuda, A., et al. (2011). Differing growth responses of major phylogenetic groups of marine bacteria to natural phytoplankton blooms in the western North Pacific Ocean. Appl. Environ. Microbiol. 77, 4055–4065. doi: 10.1128/AEM.02952-10
Teeling, H., Fuchs, B. M., Becher, D., Klockow, C., Gardebrecht, A., Bennke, C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611. doi: 10.1126/science.1218344
Thingstad, T. F., and Martinussen, I. (1991). Are bacteria active in the cold pelagic ecosystem of the Barents Sea? Polar Res. 10, 255–266. doi: 10.3402/polar.v10i1.6744
Thomas, E., Dennis, P., Bracegirdle, T. J., and Franzke, C. (2009). Ice core evidence for significant 100-year regional warming on the Antarctic Peninsula. Geophys. Res. Lett. 36, L20704. doi: 10.1029/2009GL040104
Vaughan, D. G., and Doake, C. S. M. (1996). Recent atmospheric warming and retreat of ice shelves on the Antarctic Peninsula. Nature 379, 328–331. doi: 10.1038/379328a0
Venables, H. J., Andrew, C., and Meredith, M. P. (2013). Wintertime controls on summer stratification and productivity at the western Antarctic Peninsula. Limnol. Oceanogr. 58, 1035–1047. doi: 10.4319/lo.2013.58.3.1035
Venables, W. N., and Ripley, B. D. (2002). Modern Applied Statistics with S, 4th Edn. New York, NY: Springer. doi: 10.1007/978-0-387-21706-2
Voget, S., Wemheuer, B., Brinkhoff, T., Vollmers, J., Dietrich, S., Giebel, H.-A., et al. (2015). Adaptation of an abundant Roseobacter RCA organism to pelagic systems revealed by genomic and transcriptomic analyses. ISME J. 9, 371–384. doi: 10.1038/ismej.2014.134
Warren, P. H., Law, R., and Weatherby, A. J. (2003). Mapping the assembly of protist communities in microcosms. Ecology 84, 1001–1011. doi: 10.1890/0012-9658 (2003)084[1001:MTAOPC]2.0.CO;2
Wemheuer, B., Wemheuer, F., Hollensteiner, J., Meyer, F.-D., Voget, S., and Daniel, R. (2015). The green impact: bacterioplankton response toward a phytoplankton spring bloom in the southern North Sea assessed by comparative metagenomic and metatranscriptomic approaches. Front. Microbiol. 6:805. doi: 10.3389/fmicb.2015.00805
West, N. J., Obernosterer, I., Zemb, O., and Lebaron, P. (2008). Major differences of bacterial diversity and activity inside and outside of a natural iron-fertilized phytoplankton bloom in the Southern Ocean. Environ. Microbiol. 10, 738–756. doi: 10.1111/j.1462-2920.2007.01497.x
Williams, T. J., Wilkins, D., Long, E., Evans, F., DeMaere, M. Z., Raftery, M. J., et al. (2013). The role of planktonic Flavobacteria in processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ. Microbiol. 15, 1302–1317. doi: 10.1111/1462-2920.12017
Yilmaz, P., Kottmann, R., Field, D., Knight, R., Cole, J. R., Amaral-Zettler, L., et al. (2011). Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 29, 415–420. doi: 10.1038/nbt.1823
Zhang, G., Liang, S., Shi, X., and Han, X. (2015). Dissolved organic nitrogen bioavailability indicated by amino acids during a diatom to dinoflagellate bloom succession in the Changjiang River estuary and its adjacent shelf. Mar. Chem. 176, 83–95. doi: 10.1016/j.marchem.2015.08.001
Keywords: 16S rRNA, amplicon sequencing, community assembly, bacterial succession, mesocosms, Collwelliaceae, Polaribacter, phytoplankton exudates
Citation: Luria CM, Amaral-Zettler LA, Ducklow HW, Repeta DJ, Rhyne AL and Rich JJ (2017) Seasonal Shifts in Bacterial Community Responses to Phytoplankton-Derived Dissolved Organic Matter in the Western Antarctic Peninsula. Front. Microbiol. 8:2117. doi: 10.3389/fmicb.2017.02117
Received: 09 June 2017; Accepted: 17 October 2017;
Published: 03 November 2017.
Edited by:
Lasse Riemann, University of Copenhagen, DenmarkReviewed by:
Sara Beier, Leibniz Institute for Baltic Sea Research (LG), GermanyCopyright © 2017 Luria, Amaral-Zettler, Ducklow, Repeta, Rhyne and Rich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linda A. Amaral-Zettler, bGluZGEuYW1hcmFsLXpldHRsZXJAbmlvei5ubA== Jeremy J. Rich, amVyZW15LnJpY2hAbWFpbmUuZWR1
†Present address: Catherine M. Luria, Laboratory of Systems Pharmacology, Harvard Medical School, Boston, MA, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.