
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 October 2017
Sec. Fungi and Their Interactions
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.02058
The regulator of G-protein signaling (RGS) proteins have a conserved RGS domain that facilitates the intrinsic GTPase activity of an activated Gα subunit of heterotrimeric G protein, thereby attenuating signal transduction. Among six predicted RGS proteins in the opportunistic human pathogenic fungus Aspergillus fumigatus, only three (FlbA, GprK, and Rax1) have been studied. The unexplored RgsC composed of the Phox-associated (PXA), RGS, Phox homology (PX), and Nexin_C superfamily domains is highly conserved in many ascomycete fungi, suggesting a crucial role of RgsC in fungal biology. To address this, we have investigated functions of the rgsC gene. The deletion (Δ) of rgsC causes impaired vegetative growth and asexual development coupled with reduced expression of key developmental regulators. Moreover, ΔrgsC results in accelerated and elevated conidial germination regardless of the presence or absence of an external carbon source. Furthermore, ΔrgsC causes reduced conidial tolerance to oxidative stress. In addition, activities and expression of catalases and superoxide dismutases (SODs) are severely decreased in the ΔrgsC mutant. The deletion of rgsC results in a slight reduction in conidial tolerance to cell wall damaging agents, yet significantly lowered mRNA levels of cell wall integrity/biogenesis transcription factors, indicating that RgsC may function in proper activation of cell wall stress response. The ΔrgsC mutant exhibits defective gliotoxin (GT) production and decreased virulence in the wax moth larvae, Galleria mellonella. Transcriptomic studies reveal that a majority of transporters is down-regulated by ΔrgsC and growth of the ΔrgsC mutant is reduced on inorganic and simple nitrogen medium, suggesting that RgsC may function in external nitrogen source sensing and/or transport. In summary, RgsC is necessary for proper growth, development, stress response, GT production, and external nutrients sensing.
Heterotrimeric G-protein (G-protein) signaling plays pivotal roles in sensing and responding to internal/external signals and various stresses. At upstream, a canonical G-protein signaling pathway is typically controlled by three components; G-protein coupled receptors (GPCRs), regulators of G-protein signaling (RGS), and heterotrimeric G proteins composed of α, β, and γ subunits (Lafon et al., 2005; Yu, 2006). RGS proteins harbor a conserved RGS domain that interacts with an activated Gα subunit and modulate the G-protein signaling pathways (Chidiac and Roy, 2003; McCudden et al., 2005). In filamentous fungi, RGS proteins play crucial roles in upstream regulation of vegetative growth, development, secondary metabolism, and virulence (Bayram and Braus, 2012).
In the human pathogenic fungus Aspergillus fumigatus, six genes predicted to encode RGS domain proteins have been identified (flbA, gprK, rgsA, rax1, rgsC, and rgsD). FlbA was shown to attenuate the GpaA (Gα)-dependent signaling pathway (Mah and Yu, 2006). It has been reported that the putative hybrid GPCR-RGS protein GprK also plays an important role in upstream regulation of G-protein signaling and contributes to proper asexual sporulation, gliotoxin (GT) production, and oxidative stress responses (Jung et al., 2016). Recently, Rax1 was shown to positively control vegetative growth and asexual development, and modulate trehalose amount and cell wall melanin levels in conidia, and conidia resistance against hydrogen peroxide (Igbalajobi et al., 2017).
RgsC is similar to Saccharomyces cerevisiae Mdm1 (McConnell et al., 1990), which is required for proper transmission of the nuclei and mitochondria from mother to daughter cells (Fisk and Yaffe, 1997). The Mdm1 protein confers a series of punctate structures distributed throughout the cytoplasm (McConnell and Yaffe, 1992). The mdm1 null mutant fails to transmit mitochondria from the mother cell into the growing bud, simultaneously, suggesting that the Mdm1 protein network has a central function in facilitating organelle inheritance in the budding yeast (McConnell and Yaffe, 1992). However, Mdm1 does not harbor an RGS domain. The domain structure of RgsC of filamentous fungi is quite different from that of the budding yeast. RgsC of filamentous fungi contains the central RGS domain, the C-terminal PhoX homology (PX), and the N-terminal PhoX-associated (PXA) domain identified as a phosphoinositides (PI)-binding motif. The RgsC-type domain architecture has been found in more than 100 eukaryotic proteins with diverse functions (Ponting, 1996; Sato et al., 2001; Xu et al., 2001; Ellson et al., 2002). The PX domain might participate in protein trafficking and signal transduction by binding to PI (Sato et al., 2001). RGS-PX1 is known to play a bifunctional role as a GTPase-activating protein for Gαs and a sorting nexin protein (Zheng et al., 2001). While a potential role of RgsC in coordinating heterotrimeric G-protein signaling, hyphal extension, nuclear positioning, and vesicular trafficking has been speculated in filamentous fungi (Han et al., 2004b), no functional studies have been carried out.
In the present paper, we report the functional characterization of rgsC in A. fumigatus, and present a series of data elucidating the roles of RgsC in governing vegetative growth, asexual sporulation, germination, stress response, GT production, and virulence.
Glucose minimal medium (MMG) and MMG with 0.1% yeast extract (MMY) with appropriate supplements were used for general culture of A. fumigatus strains (Käfer, 1977). For pyrimidine and arginine auxotrophic mutant strain (AF293.6) (Xue et al., 2004), MMY was supplemented with 5 mM uridine, 10 mM uracil (for pyrG1), and 0.1% arginine (for argB1). For liquid submerged culture and phenotypic analyses on air-exposed culture were performed as described previously (Jung et al., 2016). To examine secondary metabolite production, spores of relevant strains were inoculated 50 ml of liquid MMY and incubated at 250 rpm at 37°C for 4 days.
The oligonucleotides used in this study are listed in Supplementary Table 1. The rgsC gene was deleted in A. fumigatus AF293.6 (pyrG1 argB1) strain (Xue et al., 2004). The deletion construct generated employing double-joint PCR (DJ-PCR) (Yu et al., 2004) containing the Aspergillus nidulans selective marker (AnargB+) with the 5′ and 3′ franking regions of the rgsC gene was introduced into the recipient strain AF293.6 (Szewczyk et al., 2006). The selective marker was amplified from A. nidulans FGSC4 genomic DNA with the primer pair oligo 214/oligo 215. The rgsC null mutant was isolated and confirmed by PCR, followed by restriction enzyme digestion (Supplementary Figure 1; Yu et al., 2004). To complement rgsC null mutant, a single joint PCR (SJ-PCR) method was used (Yu et al., 2004). The ORF of rgsC gene with a promoter and terminator was amplified with primer pairs where the 3′ reverse primer carries overlapping sequences with the ptrA gene's 5′ end. Amplification of the ptrA gene was carried out with primer pairs where the 5′ forward primer carries overlapping sequences with rgsC gene's 3′ end. The final amplicon was amplified with the nested primer pair oligo 781/oligo 731 and introduced into a ΔrgsC strain.
To isolate genomic DNA from A. fumigatus, about 106 conidia were inoculated in 2 ml of liquid MMY, and stationary cultured at 37°C for 24 h. The mycelial mat was collected and squeeze-dried, and genomic DNA was isolated as described (Yu et al., 2004). The deletion mutant was confirmed by PCR amplification of the coding region of the gene followed by restriction enzyme digestion of the PCR amplicon. Total RNA isolation was carried out as previously described (Han et al., 2004a; Mah and Yu, 2006). Quantitative RT-PCR (qRT-PCR) assays were performed according to the manufacturer's instruction (Qiagen, USA) using a Rotor-Gene Q (Qiagen, USA). Each run was assayed in triplicate in a total volume of 20 μl containing the RNA template, One Step RT-PCR SYBR Mix (Doctor Protein, Korea), reverse transcriptase, and 10 pmole of each primer (Supplementary Table 1). Reverse transcription was performed at 42°C for 30 min. PCR conditions were 95°C/5 min for one cycle, followed by 95 and 55°C/30 s for 40 cycles. Amplification of one single specific target DNA was checked by melting curve analysis (+0.5°C ramping for 10 s, from 55 to 95°C). The expression ratios were normalized to EF1α expression and calculated according to the ΔΔCt method (Livak and Schmittgen, 2001).
Germination rates were measured as previously described with a slight modification (Ni et al., 2005). To examine germination levels, conidia of WT and mutant were inoculated in 5 ml of liquid MMY, or liquid medium lacking a carbon source, and incubated at 37°C. Levels of germination were examined every 2 h after inoculation under a microscope. Various media were used to assess the roles of RgsC in stress responses. For oxidative stress test, hydrogen peroxide (5 mM), menadione (100 μM), and paraquat (100 μM) were added to the YG media after autoclaving. To assess cell wall stress, Congo red (100 μg/ml), calcofluor white (50 μg/ml), caspofungin (0.1 μg/ml) were added to the YG media after autoclaving. The production of gliotoxin (GT) was determined as described previously (Bok and Keller, 2004). The chloroform extracts were air-dried and resuspended in 100 ml of methanol. Ten micro liter aliquots of each sample were applied to a thin-layer chromatography (TLC) silica plate (Kiesel gel 60, E. Merck). The TLC plate was developed with toluene:ethyl acetae:formic acid (5:4:1, v/v/v) and GT standard was purchased from Sigma (USA). To test alternative nitrogen sources, MMG with nitrogen free salts was used as the base medium. Three grams per liter peptone and yeast extract, 6.0 g/l NaNO3, or 8.13 g/l proline was added, and these media were compared to MMG (NH4Cl).
For catalase and superoxide dismutase (SOD) activity assays, protein was extracted as previous method (Jung et al., 2016). Catalase activity on gels was detected by ferricyanide-negative stain (Wayne and Diaz, 1986) and SOD activity was visualized by inhibition of the reduction of nitro blue tetrazolium (NBT, Sigma) according to the method of Beauchamp and Fridovich (1971).
The insect survival assay was performed as previously described with some modifications (Fuchs et al., 2010; Jung et al., 2016). Briefly, sixth instar Galleria mellonella were infected by injecting the fresh conidia (1 × 105) into the last left pro-leg and incubated at 37°C in the dark for the duration of the experiment. Larvae were checked daily for survival and Kaplan-Meier survival curves were analyzed using the Log-Rank (Mantel-Cox) test for significance (p < 0.01).
The synthesis of target cDNA probes and hybridization were performed using Agilent's Low Input Quick Amp WT Labeling Kit (Agilent Technology, USA) according to the manufacturer's instructions. Briefly, 100 ng total RNA was mixed with WT primer mix and incubated at 65°C for 10 min. cDNA master mix (5 × First strand buffer, 0.1 M DTT, 10 mM dNTP mix, RNase-Out, and MMLV-RT) was prepared and added to the reaction mixture. The samples were incubated at 40°C for 2 h, and then the RT and dsDNA synthesis reactions were terminated by incubating at 70°C for 10 min. The transcription master mix was prepared as directed by the manufacturer's protocol (4 × Transcription buffer, 0.1 M DTT, NTP mix, 50% PEG, RNase-Out, inorganic pyrophosphatase, T7-RNA polymerase, and Cyanine 3/5-CTP). Transcription of dsDNA was performed by adding the transcription master mix to the dsDNA reaction samples and incubating at 40°C for 2 h. Amplified and labeled cRNA was purified and labeled cRNA target was quantified. After checking labeling efficiency, each of cyanine 3-labeled and cyanine 5-labeled cRNA target were mixed, and fragmentation of cRNA was performed by adding 10 × blocking agent and 25 × fragmentation buffer and incubating at 60°C for 30 min. The fragmented cRNA was resuspended with 2 × hybridization buffer and directly pipetted onto assembled MYcroarray.com (A. fumigatus AF293) 30 K Microarray. The arrays hybridized at 57°C for 17 h using an Agilent Hybridization oven (Agilent Technology, USA). The hybridized microarrays were washed as per the manufacturer's washing protocol (Agilent Technology, USA). Hybridization images were analyzed by an Agilent DNA microarray Scanner (Agilent Technology, USA), and the data quantification was performed using Agilent Feature Extraction software 10.7 (Agilent Technology, USA). The average fluorescence intensity for each spot was calculated and local background was subtracted using Gene Pix Pro 6.0 (Axon Instruments, USA). Loess normalization and selection of fold-changed genes were performed using GenoWiz 4.0 (Ocimum biosolutions, India). The data is available in the Gene Expression Omnibus (GEO) at NCBI (the accession number is GSE83200).
The ORF of rgsC of A. fumigatus (AFUA_1G09040) consists of 3,718 bp nucleotides with 1 intron, predicted to encode a 1,216 aa length protein. As shown in Figure 1A, the domain structure of RgsC contains a transmembrane (31–53 aa), PXA (101–290 aa, E-value; 3.34e-27), RGS (419–556 aa, E-value; 8.34e-16), 4 low complexity, and 1 PX domain (863–976 aa, E-value; 2.78e-24). With these protein sequences, we further identified additional RgsC-like proteins in other fungi and carried phylogenetic analyses (Figure 1B). As presented, the A. fumigatus RgsC is closely related to that of Aspergillus fischeri, Aspergillus clavatus, Aspergillus nomius, Aspergillus flavus, Aspergillus niger, A. nidulans, and Penicillium spp., but phylogenetically distinct from RgsC of dimorphic fungi and dermatophytic fungi (Figure 1B). To characterize the rgsC gene, levels of rgsC mRNA at different time points in the life cycle were examined, and found to be low during the early vegetative growth and increased at the later phase of vegetative growth (Figure 1C).
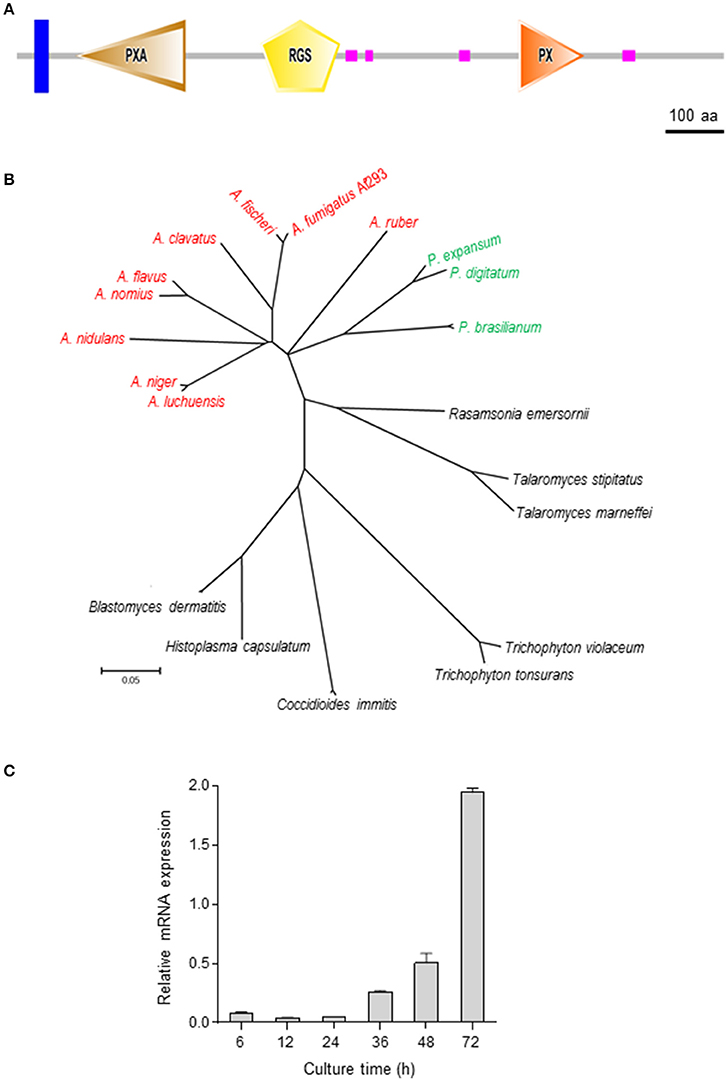
Figure 1. Summary of RgsC in A. fumigatus (A) Schematic presentation of the domain architecture of RgsC in A. fumigatus using SMART (http://smart.embl-heidelberg.de). (B) A phylogenetic tree of the RgsC-like proteins in various fungi was constructed based on the matrix of pair-wise distances between the sequences. (C) Expression of rgsC mRNA during the life cycle of WT.
To characterize functions of rgsC, we generated the ΔrgsC mutant by replacing its ORF with the A. nidulans argB+ marker and we also generated complemented strains (C′) via re-introducing the wild type (WT) allele of rgsC to a deletion strain. Multiple ΔrgsC and C′ strains displaying identical phenotypes were isolated and further examined. When inoculated on solid medium, the rgsC deletion mutant formed a very distinct colony. The color of colony was very faint except a center region and the reverse side of colony was also light compared to WT and C' strains (Figure 2A). Moreover, whereas the colony edge of WT and C′ strains showed abundant conidiophores, the ΔrgsC mutant exhibited a very few number of conidiophores (Figure 2A, right panels). Conidia per plate further demonstrated that asexual spore production in the ΔrgsC mutant (1.8 × 108 conidia/plate) was significantly decreased (p < 0.05) to a level that was only about 70% of WT and C′ strains (Figure 2B). Another noticeable change was that, the deletion of rgsC resulted in a significant reduction (about 85% of WT) in radial colony growth (Figure 2C). Further examination of mRNA levels of key asexual developmental regulators, abaA, brlA, vosA, and wetA in WT and ΔrgsC strains revealed a significantly reduced accumulation of these key developmental activators by the absence of rgsC (Figure 2D). As shown in Figure 2D, accumulation of abaA and wetA mRNAs increased from 12 h, peaked at 36 h, and decreased after 48 h post developmental induction in WT. Accumulation of brlA mRNA increased from 6 h, peaked at 12 h, and decreased after 24 h post developmental induction. However, the deletion of rgsC resulted in significantly low levels (p < 0.05) of these mRNAs at almost all times tested (Figure 2D). These results suggest that RgsC is necessary for proper growth and development in A. fumigatus.
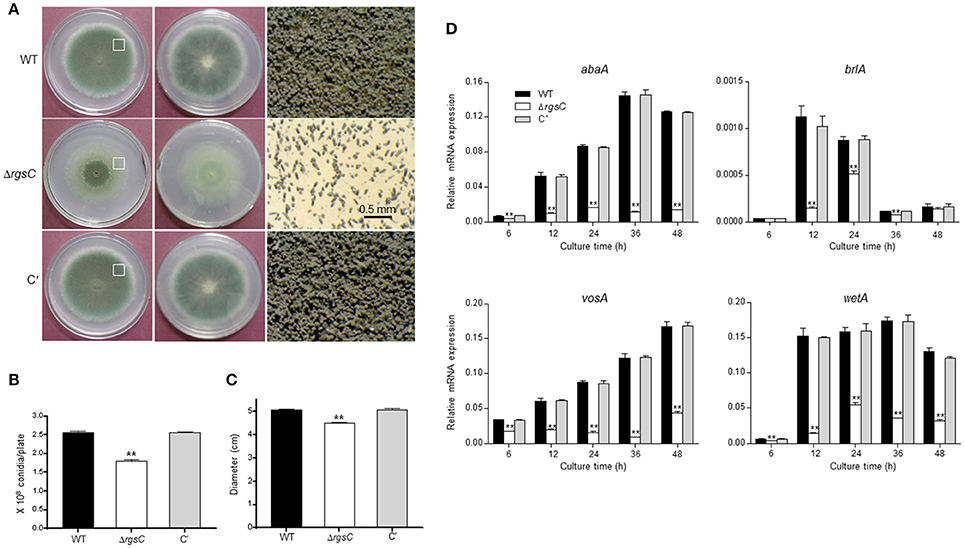
Figure 2. A role of RgsC in growth and development. (A) Colony photographs of WT (AF293), ΔrgsC, and complemented (C′) strains point-inoculated on solid MMY and grown for 3 days (Top: left; Bottom: middle panels). Enlarged photographs from the plate (indicated by the white box) are shown in the right panels with the bar indicating 0.5 mm. (B) Conidia numbers produced by each strain per plate. (C) Colony diameters of WT, ΔrgsC, and C′ strains. (D) mRNA levels of the asexual developmental regulators in WT, ΔrgsC, and C′ strains determined by quantitative real time PCR (qRT-PCR). Fungal cultures were done in liquid MMY and mRNA levels were normalized using the ef1α gene, according to the ΔΔCt method. Data are expressed as the mean ± standard deviation from three independent experiments. Student's t-test: *p < 0.05; **p < 0.01.
G protein signaling plays a positive role in spore germination (Fillinger et al., 2002; Lafon et al., 2005). If RgsC attenuates a G-protein signaling pathway activating germination, the absence of RgsC may result in elevated spore germination. To test this, we first inoculated conidia of WT, ΔrgsC mutant, and C′ strains in liquid MMY and analyzed the kinetics of germ tube emergence. As shown in Figure 3A, WT and C′ strains exhibited about 30% conidial germination at 8 h and near 100% germination at 14 h in liquid submerged culture. On the other hand, the ΔrgsC strain showed about 40% conidial germination at 8 h and near 100% germination at 12 h in liquid medium. To test further, we examined germination rates in the absence of external carbon source by inoculating conidia of WT, ΔrgsC, and C′ strains in liquid MMY (without glucose). After 16 h inoculation, whereas only about 10% of WT and C′ conidia showed germling formation, 40% of the ΔrgsC conidia germinated (Figure 3A), suggesting that RgsC may negatively regulate conidial germination potentially sensing the external carbon source. To investigate whether RgsC mediates sensing of carbon sources, germination of the ΔrgsC mutant conidia in comparison to that of WT and C′ strain conidia were monitored in the presence of various carbon sources. The germination rate was significantly elevated in the rgsC null mutant in all but glucose medium (Figure 3B), suggesting that RgsC is necessary for the proper control of spore germination in response to varying carbon sources.
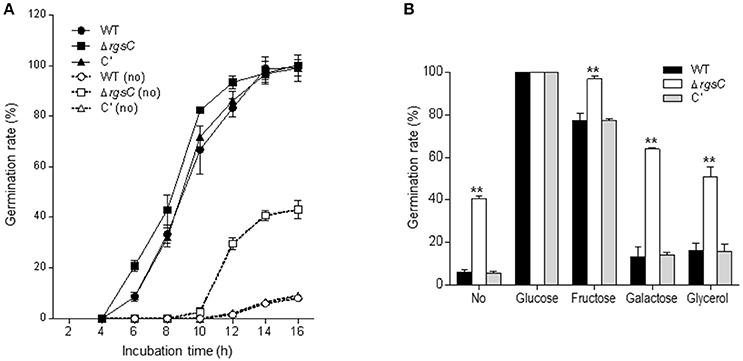
Figure 3. A role of RgsC in spore germination. (A) Kinetics of germ tube outgrowth in A. fumigatus strains when inoculated in liquid MMG at 37°C in the presence or absence (dashed line) of glucose. (B) Conidial germination in response to various carbon sources. Data are expressed as the mean ± standard deviation from three independent experiments. Student's t-test: *p < 0.05; **p < 0.01.
To evaluate functions of RgsC in oxidative stress response, we incubated WT, ΔrgsC, and C′ strains in the presence of H2O2 and the reactive oxygen species (ROS) generating compounds menadione (MD) and paraquat (PQ). As shown in Figure 4A, while the ΔrgsC mutant was hypersensitive to MD, it exhibited a slightly reduced tolerance to H2O2 and PQ. To further investigate the role RgsC, we analyzed activities of the ROS detoxifying enzymes catalase and SOD. Activities of both conidia-specific (CatA) and mycelia-specific (Cat1) catalases were decreased about 5 to 10-fold in the ΔrgsC mutant compared to those of WT and C′ strains (Figure 4B). In A. fumigatus, four genes encoding SODs have been identified and SOD1 and SOD2 were shown to play a major role to detoxify intracellular superoxide anions (Lambou et al., 2010). As catalases activities, activities of SOD1 and SOD2 in the ΔrgsC mutant was only 60 and 20% of WT strain, respectively, suggesting that the reduced tolerance of the ΔrgsC mutant to oxidative stresses could be due to low detoxifying enzymes activities. We then examined whether the absence of rgsC affected mRNA levels of catalases and SODs. We found that mRNA levels of catA, sod1, and sod2 were significantly decreased (p < 0.05) in the ΔrgsC mutant (Figure 4C). Taken together, these results suggest that RgsC is needed for protection against external oxidative stresses.
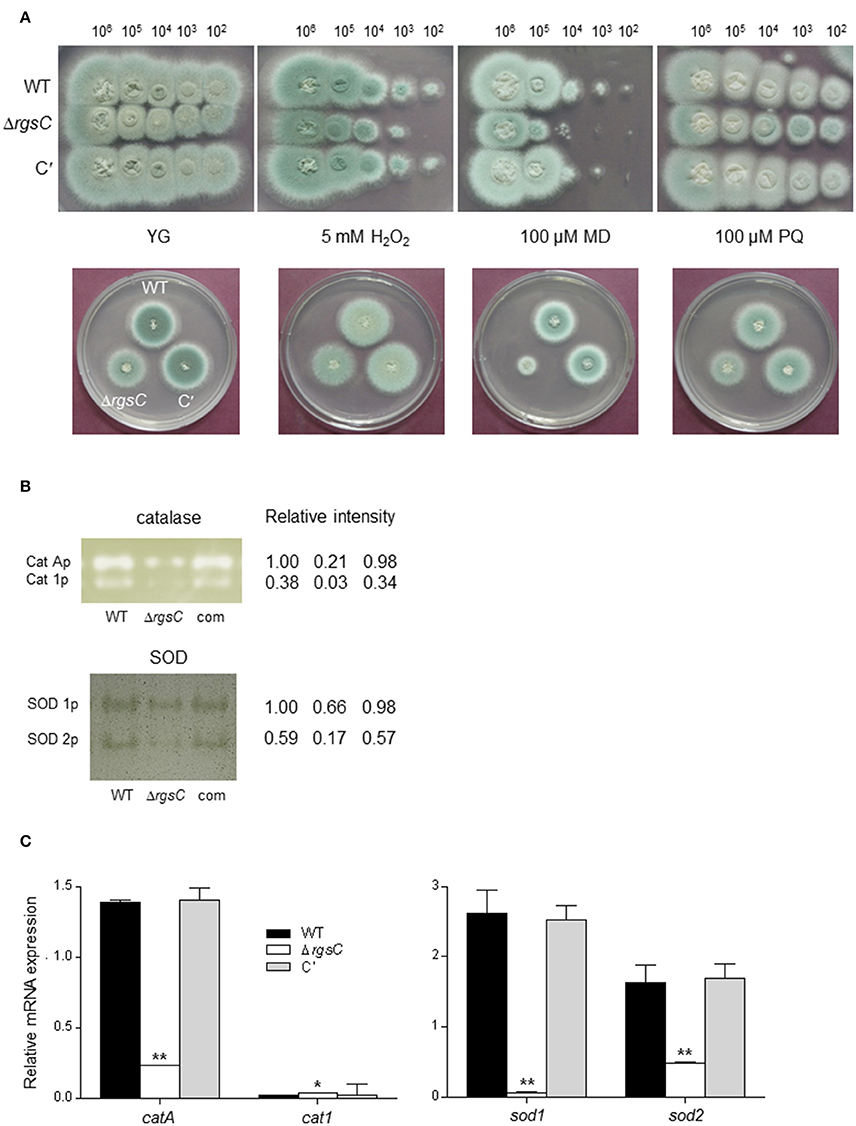
Figure 4. Oxidative stress tests. (A) Radial growth of WT, ΔrgsC, and C′ strains in presence of oxidative stressors H2O2, menadione (MD), or paraquat (PQ) at indicated concentrations following incubation at 37°C for 48 h. (B) Catalases and SODs activities of WT, ΔrgsC, and C′ strains shown in non-denaturing polyacrylamide gels. (C) Levels of catalase and SOD genes' mRNA in WT, ΔrgsC, and C′ strains analyzed by qRT-PCR. Statistical significance was determined by a Student's t-test: *p < 0.05; **p < 0.01.
To examine whether RgsC mediate cell wall stress response, the mutant were exposed to a variety of cell wall damaging compounds including Congo red (100 μg/ml), calcofluor white (50 μg/ml), and caspofungin (0.1 μg/ml). Growth of the ΔrgsC mutant was slightly inhibited by the tested compounds compared to that of WT and C′ strain (Figure 5A), suggesting cell wall biosynthesis and/or integrity may be affected by the deletion of rgsC. We further analyzed mRNA levels of the MADS-box transcription factor rlmA and APSES transcription factor swi4 and swi6 which regulate genes involved in cell wall integrity and biogenesis (Kim et al., 2010; Rocha et al., 2016). In the ΔrgsC mutant it appears that levels of all tested genes' mRNA decreased after 48 h compared to WT (Figure 5B). These results indicate that RgsC is associated with proper cell wall biogenesis and cell wall integrity.
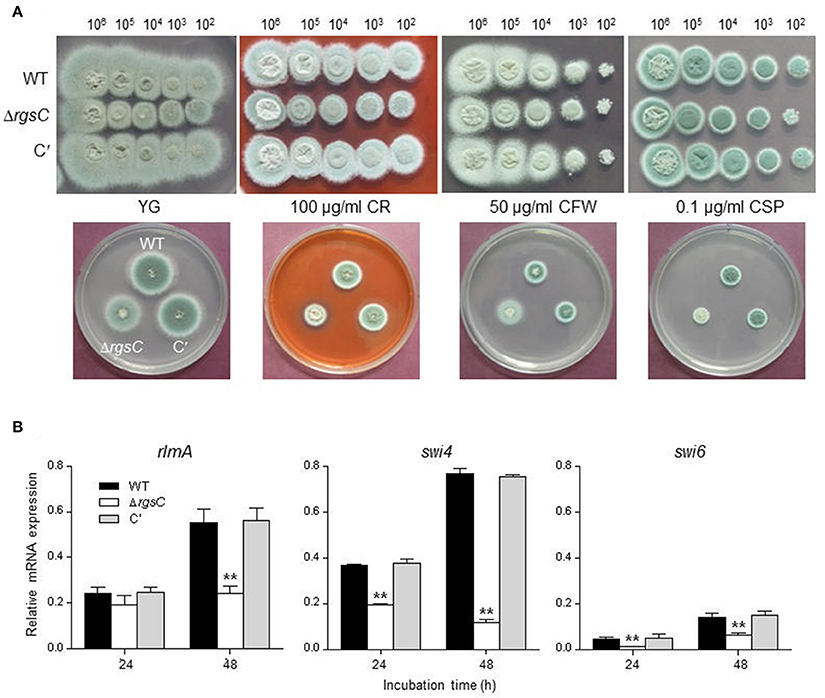
Figure 5. Cell wall stress tests. (A) Radial growth of WT, ΔrgsC, and C′ strains in presence of cell wall damaging agents Congo red (CR), calcofluor white (CFW), or caspofungin (CSP) at indicated concentrations following incubation at 37°C for 48 h. (B) Levels of cell wall integrity and biogenesis genes' mRNA in WT, ΔrgsC, and C′ strains analyzed by qRT-PCR. Statistical significance was determined by a Student's t-test: *p < 0.05; **p < 0.01.
In A. fumigatus, the gliotoxin (GT) production is partially regulated by the asexual developmental activator BrlA (Shin et al., 2015). The deletion of RGSs including FlbA, GprK, and Rax1 resulted in lowered brlA expression and GT production (Mah and Yu, 2006; Jung et al., 2016; Igbalajobi et al., 2017). As the deletion of rgsC resulted in significantly defective conidiation and brlA mRNA levels (Figure 2), we examined levels of GT by TLC. As shown in Figure 6A, the ΔrgsC mutant produced undetectable levels of GT. Then we analyzed mRNA levels of several key GT biosynthetic genes by qRT-PCR using total RNA of WT, mutant, and C' strains. The mRNA levels of the gliM, gliT, and gliZ genes were significantly lower (p < 0.05) in the ΔrgsC mutant than in WT and C′ strains (Figure 6B). We next examined the effect of RgsC on virulence using the G. mellonella larvae survival test. Conidia of WT, ΔrgsC, and C′ strains were inoculated in G. mellonella larvae, and the larvae survival rates were recorded as a function of time. The virulence of ΔrgsC strains in wax moth was significantly reduced compared to WT and C′ strains (Figure 6C). The Log-Rank test revealed that the survival curves of WT and ΔrgsC were significantly different (p < 0.002). These results indicate an important role of RgsC in proper production of GT and the virulence of the fungus.
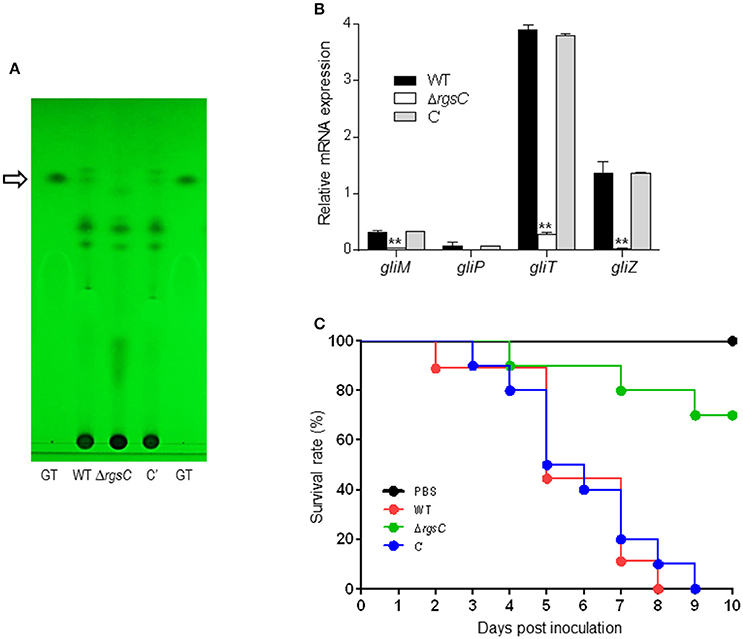
Figure 6. A role of RgsC in GT production and virulence. (A) Determination of GT production in WT, ΔrgsC, and C′ strains. The culture supernatant of each strain was extracted with chloroform and subjected to TLC. The arrow indicates the migration position for the GT standard. (B) qRT-PCR analysis of four gli cluster genes in WT, ΔrgsC, and C′ strains. Statistical differences between WT and mutant strains were evaluated with Student's unpaired t-test. *p < 0.05; **p < 0.01. (C) Survival curves of WT, ΔrgsC, and C′ strains measured using G. mellonella larvae. Note the significant differences (p < 0.002) between WT, ΔrgsC, and C′ strains.
To obtain a more comprehensive insight into the RgsC mediated processes in A. fumigatus, we performed microarray analysis using ΔrgsC and WT cells collected at 12 h post asexual-developmental induction. Two biological replicates showed a high level of correlation (r = 0.885, Figure 7A). As shown in Figure 7B, the hierarchical clustering heat map based on transcriptome analysis showed that a majority of genes are down-regulated in ΔrgsC strain compared to WT. Of the 8,608 probes, 384 genes (4.5%) showed at least 1.5-fold (p < 0.05) differentially expressed, in which 82 genes (1.0%) were up-regulated and 302 genes (3.5%) were down-regulated (Supplementary Table 2). Table 1 lists the genes with increase in expression at least 2.0-fold (p < 0.01) following the deletion of rgsC. The highest up-regulated gene was predicted to encode a conserved hypothetical protein (AFUA_8G06430), with salicylate hydroxylase (AFUA_2G00770) identified as the up-regulated known gene with the maximum fold change in mutant relative to WT. The existence of transcripts corresponding to the conserved hypothetical proteins were first confirmed by qRT-PCR on the same RNA used for the microarray library construction (data not shown). Most of the down-regulated genes were related to nitrogen transport (Table 2), including small oligopeptide transporter (AFUA_2G15240), high affinity nitrate transporter NrtB (AFUA_1G17470), MFS peptide transporter (AFUA_1G12240), ammonium transporter MeaA (AFUA_2G05880), and nitrate transporter CrnA (AFUA_1G12850). These findings led us to test a role for the RgsC in nitrogen source sensing. The ΔrgsC mutant was grown on a variety of nitrogen sources such as, NaNO3, NH4Cl, peptone, proline, and yeast extract. As shown in Figure 8, while there were no differences in growth on organic nitrogen sources, growth of the ΔrgsC mutant was significantly (p < 0.01) restricted with NaNO3, NH4Cl, and proline as a nitrogen source, suggesting that RgsC may play a role in inorganic and simple nitrogen sensing.
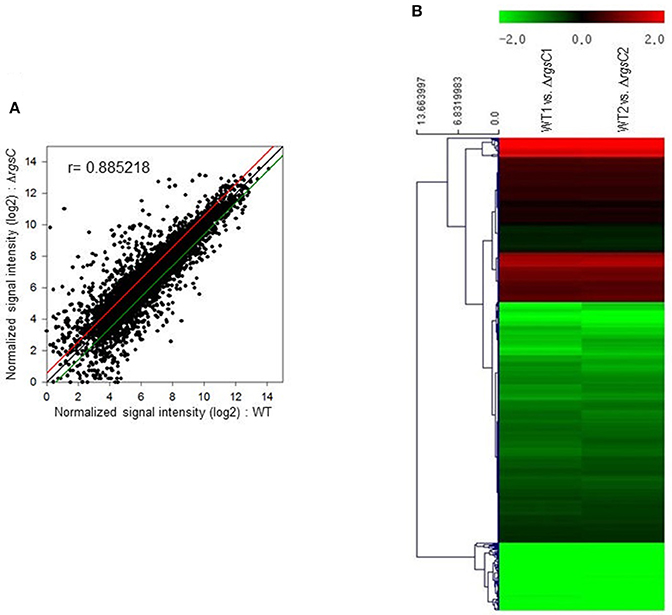
Figure 7. Genome-wide expression correlation between WT and ΔrgsC strains. (A) Linear fitted model showing the correlation between overall gene expression for WT and ΔrgsC strains. The correlation coefficient r is indicated. (B) Heat map illustration of expression level changes between WT and ΔrgsC strains.
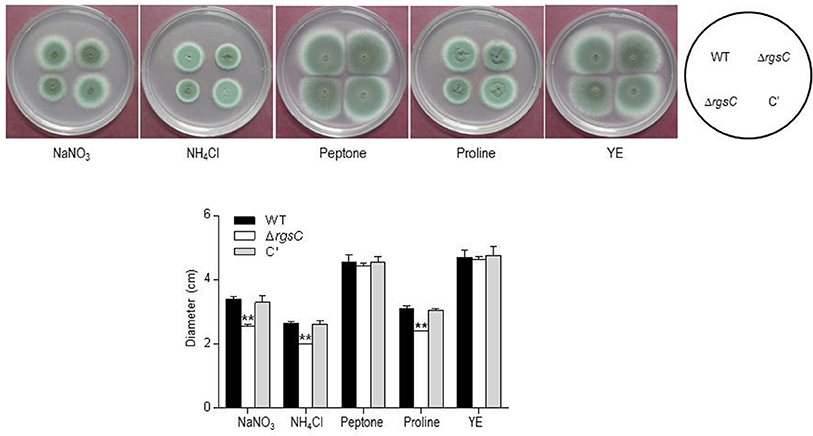
Figure 8. Effect of various nitrogen sources on growth of WT, ΔrgsC, and C′ strains. Note that growth of the mutant was significantly reduced in the presence of NaNO3, NH4Cl, and proline as nitrogen sources compared to WT and C′ strains. Statistical significance was determined by a Student's t-test: *p < 0.05; **p < 0.01.
All life forms are able to sense and respond to various internal and external stimuli, and numerous signaling pathways play an important roles during the cellular processes. G-protein signaling is conserved in all eukaryotes that sense and transmit signals into the cells to amplify appropriate responses (Dohlman et al., 1996). Basic units of the G-protein signaling system typically include a G protein-coupled receptor (GPCR), regulators of G protein signaling (RGS), a heterotrimeric G protein composed of α, β, and γ subunits, and a variety of effectors (Li et al., 2007). RGS proteins are a family of multifunctional signaling regulators having the capacity to bind to activated Gα subunits. Canonical RGS proteins stimulate the intrinsic GTPase activity of the cognate Gα subunits and lead to deactivation of Gα subunits and termination of signaling (De Vries et al., 2000; Siderovski and Willard, 2005). In various fungi, RGSs have been shown to regulate morphogenesis, differentiation, reproduction, toxin production, and virulence (Lengeler et al., 2000; Mah and Yu, 2006; Zhang et al., 2011; Jung et al., 2016; Igbalajobi et al., 2017). Consequently, elucidation of the regulatory mechanisms of RGS proteins is expected to provide a basis for identifying novel targets for controlling human pathogenic fungi.
The five RGS proteins defined in A. nidulans can be grouped into three clades (A, B, and C), where the clade A can further be divided into the sub-clades A-I and A-II. Sub-clade A-I contains 10 RGS proteins and members of A-I all have multiple conserved functional domains, such as, PXA, PX, and Nexin_C (Wang et al., 2013). Based on the domain organization, RgsC of A. fumigatus may belong to the sub-clade A-I. Although it has been speculated that the RgsC-type fungal RGS proteins might function in coordinating G-protein signaling, hyphal extension, nuclear transmission, and organelle transport (Han et al., 2004b), exact function of these proteins are not clear yet.
In the present report, we show several experimental evidence that the RgsC plays a crucial role in governing vegetative growth and asexual development in A. fumigatus. The absence of rgsC results in profound defects in vegetative growth, asexual sporulation, and lowered expression of key asexual developmental regulators (Figure 2). Moreover, overall, the germination rate of the ΔrgsC mutant was significantly higher than that of WT and C′ strains in the absence of carbon source and in the presence of carbon sources other than glucose (Figure 3). It was shown that germination can be induced by various carbon sources by activation of the cAMP/PKA pathway in A. nidulans (Fillinger et al., 2002), implying that RgsC might function in proper control of the cAMP/PKA pathway and spore germination.
A. fumigatus has five catalases (Calera et al., 1997; Paris et al., 2003a) and four SODs (Holdom et al., 2000; Flückiger et al., 2002; Lambou et al., 2010) that can be associated with detoxification of ROS. We investigated the sensitivity of the ΔrgsC mutant against ROS generating compounds and found that the ΔrgsC conidia were significantly more sensitive to compounds tested than the WT and C′ conidia (Figure 4A). Activities of catalases (CatA and Cat1) and SOD (SOD1 and 2) were drastically decreased in the ΔrgsC mutant (Figure 4B). The mRNA levels of catA, sod1, and sod2 in the ΔrgsC conidia were significantly lower than those of the WT and C′ conidia (Figure 4C). Previous studies demonstrated that the deletion of a conidial catalase catA resulted in increased susceptibility of conidia to H2O2, but disruptions of the either mycelial catalases (cat1 or cat 2) did not affect sensitivity to H2O2 (Calera et al., 1997; Paris et al., 2003a,b). These finding suggest that CatA plays a major role in detoxification of H2O2. Sod1 and Sod2 were highly expressed in conidia during growth and both of the Δsod1 and Δsod2 mutants showed hypersensitivity to MD (Lambou et al., 2010). Taken together, RgsC may positively regulate the expression of the key ROS detoxifying enzymes catalases and SODs, conferring proper oxidative stress response.
The ΔrgsC mutant showed increased susceptibility to cell wall disturbing agents such as, CR, CFW, and CSF (Figure 5A). We have investigated mRNA expressions of genes related to cell wall integrity and biogenesis. The cell wall integrity pathway is the primary signaling cascade that controls the synthesis of the fungal cell wall and is highly dependent on the RlmA transcription factor (Rocha et al., 2016). Loss-of-function of rlmA leads to the altered cell wall organization, tolerance to cell wall perturbing agents, and expression of genes encoding cell wall-related proteins (Rocha et al., 2016). The cell cycle transcription factor SBF (Swi4 and Swi6) interacts with the protein kinase C/MAP kinase pathway, which functions in the control of cell wall assembly, thus loss of SBF function leads to a weakened wall (Igual et al., 1996). In the ΔrgsC mutant, levels of rlmA, swi4, and swi6 mRNA decreased after 48 h compared to WT (Figure 5B), suggesting that RgsC may take part in the regulation of cell wall integrity signaling and cell wall assembly pathway. In susceptibility test against azole antifungal agents, there was no significant difference between WT and mutant strains may due to azole antifungal drugs inhibit ergosterol synthetic enzyme (Sheehan et al., 1999) (Supplementary Figure 2).
Biogenesis of gliotoxin (GT) requires activities of the gli gene cluster composed of 13 genes in A. fumigatus (Gardiner and Howlett, 2005). Several studies indicate that GT plays a direct role in aspergillosis virulence in immunocompromised individuals (Gardiner et al., 2005; Lewis et al., 2005; Spikes et al., 2008). In GT biosynthesis, the gliM gene is predicted to encode an o-methyltransferase (Cramer et al., 2006). GliP, a multimodular nonribosomal peptide synthetase, makes the diketopiperazine scaffold of GT (Balibar and Walsh, 2006). The GT oxidoreductase GliT protects the fungus against exogenous GT and is essential for GT biosynthesis (Schrettl et al., 2010; Brakhage, 2013). The gliZ gene controls expression of the remaining genes the gli gene cluster (Bok et al., 2006; Scharf et al., 2012). We found that GT production and expression of GT biosynthetic genes in the ΔrgsC mutant were severely reduced compared to WT and C′ strain (Figure 6), suggesting that RgsC plays a positive role in GT synthesis, likely by conferring proper activation of the asexual developmental regulator brlA. GT suppresses the immune response of G. mellonella larvae by inhibiting the action of haemocytes and thus renders the larvae susceptible (Reeves et al., 2004). The mortality level of the ΔrgsC mutant in wax moth larvae was significantly (p < 0.002) reduced compared to the WT and C′ strains (Figure 6C), which in part may be due to the defective production of GT in the ΔrgsC mutant. Collectively, the data indicate that RgsC-mediated modulation/attenuation of signal transduction pathway(s) is important for proper control of GT biogenesis and virulence of A. fumigatus. While we do not know the target heterotrimeric G protein(s) and/or other signaling elements modulated by RgsC, as RgsC is highly conserved in many pathogenic ascomycete fungi including species of Blastomyces, Histoplasma, and Coccidioides but not found in human, it might be an excellent target for the development of novel antifungal drugs.
Of the 8,608 probes, 384 genes were found to be differentially expressed by the absence of RgsC, and most of them were down-regulated. Intriguingly, most of nitrogen transport-related genes were down-regulated by ΔrgsC, including small oligopeptide transporter, high affinity nitrate transporter NtrB, MFS peptide transporter, ammonium transporter MeaA, nitrate transporter CrnA, amino acid permease Gap1, and MFS drug efflux transporter (Table 2). The results imply that RgsC is needed for proper expression of these genes, and the ΔrgsC mutant might not sense external nitrogen sources effectively. To confirm this, we tested growth of mutant on various nitrogen sources. Growth of the ΔrgsC mutant was significantly (p < 0.01) reduced with NaNO3, NH4Cl, and proline as the nitrogen source (Figure 8). These results suggest that the ΔrgsC mutant fails to sense and/or transport external inorganic and simple nitrogen sources effectively. The key regulator of secondary metabolism LaeA (AFUA_1G14660) was also down-regulated in the ΔrgsC mutant. LaeA represents a global regulator of secondary metabolism and the A. fumigatus ΔlaeA mutant is unable to produce GT (Cramer et al., 2006). Collectively, defective GT production in the ΔrgsC mutant might result from reduced expression of laeA and brlA.
KS and JY conceived and supervised the study; KS and JY designed experiments; YK, IH, and KS performed experiments; KS and JY analyzed data; YK, IH, JY, and KS wrote the manuscript.
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (No. 2017R1A2B4001806) to KS. The work at UW was supported by the Intelligent Synthetic Biology Center of Global Frontier Project funded by the Ministry of Education, Science and Technology (No. 2011-0031955) grants to JY.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer JM and handling Editor declared their shared affiliation.
We thank our lab members for helpful discussions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02058/full#supplementary-material
Supplementary Figure1. Confirmation of WT, ΔrgsC, and complemented (C′) strains. (A) Schematic illustration of the rgsC regions in WT, ΔrgsC, and complemented (C′) strains. (B) PCR amplicons for the three strains. Lane M, molecular weight marker. (C) The HindIII digestion pattern of individual amplicon. While the WT and C′ amplicons are cut into two fragments, the ΔrgsC amplicon remains uncut.
Supplementary Figure2. Clear E-test growth inhibition ellipses for ketoconazole and voriconazole. About 106 conidia were inoculated in YG media containing appropriate supplements and cultured at 37°C for 24 h.
Balibar, C. J., and Walsh, C. T. (2006). GliP, a multimodular nonribosomal peptide synthetase in Aspergillus fumigatus, makes the diketopiperazine scaffold of gliotoxin. Biochemistry 45, 15029–15038. doi: 10.1021/bi061845b
Bayram, O., and Braus, G. H. (2012). Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol. Rev. 36, 1–24. doi: 10.1111/j.1574-6976.2011.00285.x
Beauchamp, C., and Fridovich, I. (1971). Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44, 276–287. doi: 10.1016/0003-2697(71)90370-8
Bok, J. W., Chung, D., Balajee, S. A., Marr, K. A., Andes, D., Nielsen, K. F., et al. (2006). GliZ, a transcriptional regulator of gliotoxin biosynthesis, contributes to Aspergillus fumigatus virulence. Infect. Immun. 74, 6761–6768. doi: 10.1128/IAI.00780-06
Bok, J. W., and Keller, N. P. (2004). LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3, 527–535. doi: 10.1128/EC.3.2.527-535.2004
Brakhage, A. A. (2013). Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 11, 21–32. doi: 10.1038/nrmicro2916
Calera, J. A., Paris, S., Monod, M., Hamilton, A. J., Debeaupuis, J. P., Diaquin, M., et al. (1997). Cloning and disruption of the antigenic catalase gene of Aspergillus fumigatus. Infect. Immun. 65, 4718–4724.
Chidiac, P., and Roy, A. A. (2003). Activity, regulation, and intracellular localization of RGS proteins. Recept. Channels 9, 135–147. doi: 10.3109/10606820308244
Cramer, R. A. Jr., Gamcsik, M. P., Brooking, R. M., Najvar, L. K., Kirkpatrick, W. R., Patterson, T. F., et al. (2006). Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot. Cell 5, 972–980. doi: 10.1128/EC.00049-06
De Vries, L., Fischer, T., Tronchere, H., Brothers, G. M., Strockbine, B., Siderovski, D. P., et al. (2000). Activator of G protein signaling 3 is a guanine dissociation inhibitor for Gai subunits. Proc. Natl. Acad. Sci. U.S.A. 97, 14364–14369. doi: 10.1073/pnas.97.26.14364
Dohlman, H. G., Song, J., Ma, D., Courchesne, W. E., and Thorner, J. (1996). Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein a subunit). Mol. Cell. Biol. 16, 5194–5209. doi: 10.1128/MCB.16.9.5194
Ellson, C. D., Andrews, S., Stephens, L. R., and Hawkins, P. T. (2002). The PX domain: a new phosphoinositide-binding module. J. Cell Sci. 115, 1099–1105.
Fillinger, S., Chaveroche, M. K., Shimizu, K., Keller, N., and d'Enfert, C. (2002). cAMP and ras signalling independently control spore germination in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 44, 1001–1016. doi: 10.1046/j.1365-2958.2002.02933.x
Fisk, H. A., and Yaffe, M. P. (1997). Mutational analysis of Mdm1p function in nuclear and mitochondrial inheritance. J. Cell. Biol. 138, 485–494. doi: 10.1083/jcb.138.3.485
Fückiger, S., Mittl, P. R., Scapozza, L., Fijten, H., Folkers, G., Grutter, M. G., et al. (2002). Comparison of the crystal structures of the human manganese superoxide dismutase and the homologous Aspergillus fumigatus allergen at 2-A resolution. J. Immunol. 168, 1267–1272. doi: 10.4049/jimmunol.168.3.1267
Fuchs, B. B., O'Brien, E., Khoury, J. B., and Mylonakis, E. (2010). Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence 1, 475–482. doi: 10.4161/viru.1.6.12985
Gardiner, D. M., and Howlett, B. J. (2005). Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbiol. Lett. 248, 241–248. doi: 10.1016/j.femsle.2005.05.046
Gardiner, D. M., Waring, P., and Howlett, B. J. (2005). The epipolythiodioxopiperazine (ETP) class of fungal toxins: distribution, mode of action, functions and biosynthesis. Microbiology 151, 1021–1032. doi: 10.1099/mic.0.27847-0
Han, K. H., Seo, J. A., and Yu, J. H. (2004a). A putative G protein-coupled receptor negatively controls sexual development in Aspergillus nidulans. Mol. Microbiol. 51, 1333–1345. doi: 10.1111/j.1365-2958.2003.03940.x
Han, K. H., Seo, J. A., and Yu, J. H. (2004b). Regulators of G-protein signalling in Aspergillus nidulans: RgsA downregulates stress response and stimulates asexual sporulation through attenuation of GanB (Ga) signalling. Mol. Microbiol. 53, 529–540. doi: 10.1111/j.1365-2958.2004.04163.x
Holdom, M. D., Lechenne, B., Hay, R. J., Hamilton, A. J., and Monod, M. (2000). Production and characterization of recombinant Aspergillus fumigatus Cu, Zn superoxide dismutase and its recognition by immune human sera. J. Clin. Microbiol. 38, 558–562.
Igbalajobi, O. A., Yu, J. H., and Shin, K. S. (2017). Characterization of the rax1 gene encoding a putative regulator of G protein signaling in Aspergillus fumigatus. Biochem. Biophys. Res. Commun. 487, 426–432. doi: 10.1016/j.bbrc.2017.04.079
Igual, J. C., Johnson, A. L., and Johnston, L. H. (1996). Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 15, 5001–5013.
Jung, M. G., Kim, S. S., Yu, J. H., and Shin, K. S. (2016). Characterization of gprK encoding a putative hybrid G-protein-coupled receptor in Aspergillus fumigatus. PLoS ONE 11:e0161312. doi: 10.1371/journal.pone.0161312
Kafer, E. (1977). Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19, 33–131. doi: 10.1016/S0065-2660(08)60245-X
Kim, K. Y., Truman, A. W., Caesar, S., Schlenstedt, G., and Levin, D. E. (2010). Yeast Mpk1 cell wall integrity mitogen-activated protein kinase regulates nucleocytoplasmic shuttling of the Swi6 transcriptional regulator. Mol. Biol. Cell. 21, 1609–1619. doi: 10.1091/mbc.E09-11-0923
Lafon, A., Seo, J. A., Han, K. H., Yu, J. H., and D'enfert, C. (2005). The heterotrimeric G-protein GanB(α)-SfaD(β)-GpgA(γ) is a carbon source sensor involved in early cAMP-dependent germination in Aspergillus nidulans. Genetics 171, 71–80. doi: 10.1534/genetics.105.040584
Lambou, K., Lamarre, C., Beau, R., Dufour, N., and Latge, J. P. (2010). Functional analysis of the superoxide dismutase family in Aspergillus fumigatus. Mol. Microbiol. 75, 910–923. doi: 10.1111/j.1365-2958.2009.07024.x
Lengeler, K. B., Davidson, R. C., D'souza, C., Harashima, T., Shen, W. C., Wang, P., et al. (2000). Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64, 746–785. doi: 10.1128/MMBR.64.4.746-785.2000
Lewis, R. E., Wiederhold, N. P., Lionakis, M. S., Prince, R. A., and Kontoyiannis, D. P. (2005). Frequency and species distribution of gliotoxin-producing Aspergillus isolates recovered from patients at a tertiary-care cancer center. J. Clin. Microbiol. 43, 6120–6122. doi: 10.1128/JCM.43.12.6120-6122.2005
Li, L., Wright, S. J., Krystofova, S., Park, G., and Borkovich, K. A. (2007). Heterotrimeric G protein signaling in filamentous fungi. Annu. Rev. Microbiol. 61, 423–452. doi: 10.1146/annurev.micro.61.080706.093432
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mah, J. H., and Yu, J. H. (2006). Upstream and downstream regulation of asexual development in Aspergillus fumigatus. Eukaryot. Cell 5, 1585–1595. doi: 10.1128/EC.00192-06
McConnell, S. J., Stewart, L. C., Talin, A., and Yaffe, M. P. (1990). Temperature-sensitive yeast mutants defective in mitochondrial inheritance. J. Cell. Biol. 111, 967–976. doi: 10.1083/jcb.111.3.967
McConnell, S. J., and Yaffe, M. P. (1992). Nuclear and mitochondrial inheritance in yeast depends on novel cytoplasmic structures defined by the MDM1 protein. J. Cell. Biol. 118, 385–395. doi: 10.1083/jcb.118.2.385
McCudden, C. R., Hains, M. D., Kimple, R. J., Siderovski, D. P., and Willard, F. S. (2005). G-protein signaling: back to the future. Cell. Mol. Life Sci. 62, 551–577. doi: 10.1007/s00018-004-4462-3
Ni, M., Rierson, S., Seo, J. A., and Yu, J. H. (2005). The pkaB gene encoding the secondary protein kinase A catalytic subunit has a synthetic lethal interaction with pkaA and plays overlapping and opposite roles in Aspergillus nidulans. Eukaryot. Cell 4, 1465–1476. doi: 10.1128/EC.4.8.1465-1476.2005
Paris, S., Debeaupuis, J. P., Crameri, R., Carey, M., Charles, F., Prevost, M. C., et al. (2003a). Conidial hydrophobins of Aspergillus fumigatus. Appl. Environ. Microbiol. 69, 1581–1588. doi: 10.1128/AEM.69.3.1581-1588.2003
Paris, S., Wysong, D., Debeaupuis, J. P., Shibuya, K., Philippe, B., Diamond, R. D., et al. (2003b). Catalases of Aspergillus fumigatus. Infect. Immun. 71, 3551–3562. doi: 10.1128/IAI.71.6.3551-3562.2003
Ponting, C. P. (1996). Novel domains in NADPH oxidase subunits, sorting nexins, and PtdIns 3-kinases: binding partners of SH3 domains? Protein Sci. 5, 2353–2357. doi: 10.1002/pro.5560051122
Reeves, E. P., Messina, C. G., Doyle, S., and Kavanagh, K. (2004). Correlation between gliotoxin production and virulence of Aspergillus fumigatus in Galleria melleonella. Mycopathologia 158, 73–79.
Rocha, M. C., Fabri, J. H., Franco de Godoy, K., Alves de Castro, P., Hori, J. I., Ferreira Da Cunha, A., et al. (2016). Aspergillus fumigatus MADS-box transcription factor rlmA is required for regulation of the cell wall integrity and virulence. G3 6, 2983–3002. doi: 10.1534/g3.116.031112
Sato, T. K., Overduin, M., and Emr, S. D. (2001). Location, location, location: membrane targeting directed by PX domains. Science 294, 1881–1885. doi: 10.1126/science.1065763
Scharf, D. H., Heinekamp, T., Remme, N., Hortschansky, P., Brakhage, A. A., and Hertweck, C. (2012). Biosynthesis and function of gliotoxin in Aspergillus fumigatus. Appl. Microbiol. Biotechnol. 93, 467–472. doi: 10.1007/s00253-011-3689-1
Schrettl, M., Carberry, S., Kavanagh, K., Haas, H., Jones, G. W., O'Brien, J., et al. (2010). Self-protection against gliotoxin-a component of the gliotoxin biosynthetic cluster, GliT, completely protects Aspergillus fumigatus against exogenous gliotoxin. PLoS Pathog. 6:e1000952. doi: 10.1371/journal.ppat.1000952
Sheehan, D. J., Hitchcock, C. A., and Sibley, C. M. (1999). Current and emerging azole antifungal agents. Clin. Microbiol. Rev. 12, 40–79.
Shin, K. S., Kim, Y. H., and Yu, J. H. (2015). Proteomic analyses reveal the key roles of BrlA and AbaA in biogenesis of gliotoxin in Aspergillus fumigatus. Biochem. Biophys. Res. Commun. 463, 428–433. doi: 10.1016/j.bbrc.2015.05.090
Siderovski, D. P., and Willard, F. S. (2005). The GAPs, GEFs, and GDIs of heterotrimeric G-protein a subunits. Int. J. Biol. Sci. 1, 51–66. doi: 10.7150/ijbs.1.51
Spikes, S., Xu, R., Nguyen, C. K., Chamilos, G., Kontoyiannis, D. P., Jacobson, R. H., et al. (2008). Gliotoxin production in Aspergillus fumigatus contributes to host-specific differences in virulence. J. Infect. Dis. 197, 479–486. doi: 10.1086/525044
Szewczyk, E., Nayak, T., Oakley, C. E., Edgerton, H., Xiong, Y., Taheri-Talesh, N., et al. (2006). Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1, 3111–3120. doi: 10.1038/nprot.2006.405
Wang, Y., Geng, Z., Jiang, D., Long, F., Zhao, Y., Su, H., et al. (2013). Characterizations and functions of regulator of G protein signaling (RGS) in fungi. Appl. Microbiol. Biotechnol. 97, 7977–7987. doi: 10.1007/s00253-013-5133-1
Wayne, L. G., and Diaz, G. A. (1986). A double staining method for differentiating between two classes of mycobacterial catalase in polyacrylamide electrophoresis gels. Anal. Biochem. 157, 89–92. doi: 10.1016/0003-2697(86)90200-9
Xu, Y., Seet, L. F., Hanson, B., and Hong, W. (2001). The Phox homology (PX) domain, a new player in phosphoinositide signalling. Biochem. J. 360, 513–530. doi: 10.1042/bj3600513
Xue, T., Nguyen, C. K., Romans, A., Kontoyiannis, D. P., and May, G. S. (2004). Isogenic auxotrophic mutant strains in the Aspergillus fumigatus genome reference strain AF293. Arch. Microbiol. 182, 346–353. doi: 10.1007/s00203-004-0707-z
Yu, J. H. (2006). Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans. J. Microbiol. 44, 145–154.
Yu, J. H., Hamari, Z., Han, K. H., Seo, J. A., Reyes-Dominguez, Y., and Scazzocchio, C. (2004). Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–981. doi: 10.1016/j.fgb.2004.08.001
Zhang, P., Su, J., King, M. E., Maldonado, A. E., Park, C., and Mende, U. (2011). Regulator of G protein signaling 2 is a functionally important negative regulator of angiotensin II-induced cardiac fibroblast responses. Am. J. Physiol. Heart. Circ. Physiol. 301, H147–H156. doi: 10.1152/ajpheart.00026.2011
Keywords: RGS, Aspergillus fumigatus, development, stress response, virulence, transcriptome
Citation: Kim Y, Heo I-B, Yu J-H and Shin K-S (2017) Characteristics of a Regulator of G-Protein Signaling (RGS) rgsC in Aspergillus fumigatus. Front. Microbiol. 8:2058. doi: 10.3389/fmicb.2017.02058
Received: 03 August 2017; Accepted: 06 October 2017;
Published: 23 October 2017.
Edited by:
Hector Mora Montes, Universidad de Guanajuato, MexicoReviewed by:
Hiroki Takahashi, Chiba University, JapanCopyright © 2017 Kim, Heo, Yu and Shin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jae-Hyuk Yu, anl1MUB3aXNjLmVkdQ==
Kwang-Soo Shin, c2hpbmtzQGRqdS5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.