
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 October 2017
Sec. Aquatic Microbiology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.01985
This article is part of the Research Topic Socio-Ecology of Microbes in a Changing Ocean View all 17 articles
Recent studies applying Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR-MS) showed that the exometabolome of marine bacteria is composed of a surprisingly high molecular diversity. To shed more light on how this diversity is generated we examined the exometabolome of two model strains of the Roseobacter group, Phaeobacter inhibens and Dinoroseobacter shibae, grown on glutamate, glucose, acetate or succinate by FT-ICR-MS. We detected 2,767 and 3,354 molecular formulas in the exometabolome of each strain and 67 and 84 matched genome-predicted metabolites of P. inhibens and D. shibae, respectively. The annotated compounds include late precursors of biosynthetic pathways of vitamins B1, B2, B5, B6, B7, B12, amino acids, quorum sensing-related compounds, indole acetic acid and methyl-(indole-3-yl) acetic acid. Several formulas were also found in phytoplankton blooms. To shed more light on the effects of some of the precursors we supplemented two B1 prototrophic diatoms with the detected precursor of vitamin B1 HET (4-methyl-5-(β-hydroxyethyl)thiazole) and HMP (4-amino-5-hydroxymethyl-2-methylpyrimidine) and found that their growth was stimulated. Our findings indicate that both strains and other bacteria excreting a similar wealth of metabolites may function as important helpers to auxotrophic and prototrophic marine microbes by supplying growth factors and biosynthetic precursors.
The biochemical processes in a living and active prokaryotic cell yield a highly complex blend of metabolites reflecting the catabolic, metabolic and anabolic properties of the organism. On the basis of available genomic information on the metabolic potential, metabolite patterns within the cell, endometabolomics, have been investigated during the recent past in various prokaryotes and as a function of substrate source and growth conditions (Rosselló-Mora et al., 2008; Zech et al., 2009; Frimmersdorf et al., 2010; Paczia et al., 2012; Drüppel et al., 2014). The exometabolome, i.e. the pool of metabolites released into the cell's environment, has also been investigated, showing species- or even ecotype-specific fingerprints as a function of growth stage and conditions (Kell et al., 2005; Villas-Bôas et al., 2006; Paul et al., 2009; Paczia et al., 2012; Romano et al., 2014; Fiore et al., 2015; Johnson et al., 2016).
Most of these studies applied targeted approaches mainly gas chromatography–mass spectrometry (GC–MS), searching for metabolites to be expected from predicted substrate use and metabolic pathways (Villas-Bôas et al., 2006; Zech et al., 2009; Drüppel et al., 2014). Several studies have used Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR-MS) applying non-targeted approaches and some combined both. They found that the diversity of the endo- and in particular the exometabolome is far higher than expected from targeted approaches, yielding several thousand molecular masses (Rosselló-Mora et al., 2008; Brito-Echeverría et al., 2011; Romano et al., 2014; Fiore et al., 2015; Johnson et al., 2016). Besides the expected metabolites, the metabolome obviously includes a surprisingly high proportion of compounds not expected from predicted metabolic pathways. In particular the exometabolome exhibited a wealth of metabolites, many with so far unknown molecular masses and elemental composition (Rosselló-Mora et al., 2008; Kujawinski et al., 2009; Romano et al., 2014; Fiore et al., 2015). There is some evidence that quite a few of these metabolites are released as a result of an overflow metabolism due to growth on an abundant carbon source (Paczia et al., 2012; Romano et al., 2014). However, under less excessive growth conditions, in addition to well-known exometabolites such as signaling compounds (Dickschat, 2010; Hartmann and Schikora, 2012), vitamins (Sañudo-Wilhelmy et al., 2014), siderophores (Mansson et al., 2011), many other unexpected metabolites are released into the environment. The bacterial secretion of several such microbial metabolites, including the plant hormone indole 3-acetic acid (IAA) and vitamin precursors, has been documented (Zhang et al., 2014; Amin et al., 2015; Fiore et al., 2015; Johnson et al., 2016; Paerl et al., 2017). Analyses of dissolved organic matter (DOM) from Sargasso Sea waters detected several compounds with molecular masses known to be released by a strain of the globally abundant SAR11 clade (Kujawinski et al., 2009). The thiamine (vitamin B1) precursor 4-amino-5-hydroxymethyl-2-methylpyrimidine (HMP) was also detected. This precursor is essential for thiamine biosynthesis by members of the SAR11 clade because they lack the gene for its complete biosynthesis (Carini et al., 2014). Thiamine requirements by photoautotrophic eukaryotes can also be met when the phosphorylated form, thiamine diphosphate, is dephosphorylated by a partner. In a bioassay approach Paerl et al. (2015) showed that the thiamine-auxotrophic picoeukaryote Ostreococcus sp. could grow when supplied with thiamine diphosphate in the presence of an Alteromonas strain that exhibit phosphatase activity.
The exchange of metabolites and precursors between microbes appears to be more common because auxotrophy is much wider distributed among microbes than previously assumed (McRose et al., 2014; Garcia et al., 2015; Paerl et al., 2017). Growth of such organisms depends on mutualistic interactions and supply of metabolites by co-occurring microbes. The Black Queen hypothesis applied this phenomenon to explain genome streamlining of prokaryotes by deleting certain metabolic pathways or parts of it as a reaction to leaky metabolic pathways of other microbes, helpers, thus supplying metabolites or precursors as public goods to the auxotrophic prokaryotes (Morris et al., 2012). It has been proposed most recently that gene loss and niche partitioning may be major drivers in the co-evolution of auxotrophs and helpers (Mas et al., 2016). However, considering the metabolites found in the exometabolome of various microbes and the environment in recent studies it appears that exchange of metabolites has larger ramifications than just explaining genome streamlining of selected microbes (Zelezniak et al., 2015; Estrela et al., 2016). We hypothesize that the exometabolome of helpers includes multiple metabolites and precursors, not only vitamins and growth factors, which are beneficial for other microbes. This release may promote growth of auxotrophic organism and even enhance growth of prototrophic microbes because they may not need to allocate energy to synthesize these precursors.
In order to test this hypothesis we assessed the exometabolome of two model bacteria of the marine Roseobacter group, Phaeobacter inhibens DSM 17395 and Dinoroseobacter shibae DSM 16493, growing on either glucose, glutamate and acetate or succinate, by analysis on a 15 Tesla FT-ICR-MS. Phaeobacter inhibens DSM 17395 is a purely heterotrophic bacterium growing in biofilms (Thole et al., 2012; Gram et al., 2015). Strains of this species and the genus Phaeobacter have been found in various marine habitats, such as natural biofilms on solid surfaces and during a bloom of Emiliana huxleyi (Gifford et al., 2014; Gram et al., 2015; Segev et al., 2016; Breider et al., 2017). Dinoroseobacter shibae is photoheterotrophic, grows symbiotically with dinoflagellates (Wagner-Döbler et al., 2010), produces various signaling compounds (Neumann et al., 2013) and has also been found associated to algae in natural phytoplankton blooms (Gifford et al., 2014; Milici et al., 2016; Segev et al., 2016). In order to examine whether identified molecular masses, and more precisely formulas, are also relevant under natural conditions, we screened DOM samples analyzed by FT-ICR-MS from a mesocosm experiment (Osterholz et al., 2015) and the North Sea. It is the first application of such a powerful FT-ICR-MS for exometabolomic studies in combination with the search for molecular formulas in environmental samples thus greatly enhancing the sensitivity and resolution of metabolite identification.
Phaeobacter inhibens DSM 17395 and D. shibae DSM 16493 were first grown on marine broth (MB; Difco MB 2216) medium and afterwards repeatedly (5x) transferred and cultivated in artificial seawater (ASW) medium with the addition of a single organic carbon source. Before every transfer and after centrifugation of the cultures the cell pellets were washed three times with ASW medium. All plastic and glass ware used were rinsed with acidified ultrapure water (MilliQ, pH 2) and all glassware additionally combusted for 3 h at 500°C.
The ASW-medium for the P. inhibens cultures was prepared as described by Zech et al. (2009) but slightly modified by excluding EDTA from the trace element solution. The cultures were supplemented with glucose (5 mM, 30 mM C; ultrapure brand), acetate (30 mM, 60 mM C; ultrapure brand) or glutamate (20 mM, 100 mM C; ultrapure brand). Dinoroseobacter shibae was cultivated in ASW-medium (Soora and Cypionka, 2013) with the same concentrations of glucose as for P. inhibens and of glutamate at 7.5 mM (37.5 mM C). Instead of acetate, succinate was used at an initial concentration of 10 mM (40 mM C; ultrapure brand). Concentrations of the substrates were adjusted due to varying metabolic rate efficiencies to obtain rather equal growth yields as determined by optical density (OD600). Bacteria were cultivated in 500 ml of medium in triplicate 2 liter baffled Erlenmeyer glass flasks at pH 8 at 28°C in the dark on a shaker (100 rpm) and growth was monitored by OD. A sterile flask with media and the respective single carbon source was run as control. Subsamples were withdrawn under laminar flow for the separate analysis of the replicates for dissolved organic carbon, exometabolome-DOM, dissolved free (DFAA) and dissolved combined amino acids (DCAA), dissolved free neutral monosaccharides (DFNCHO), and dissolved combined monosaccharides (DCNCHO) and fatty acids at the start, the lag phase, the mid-exponential and early stationary phase in order to recover the majority of exometabolites released at varying growth conditions. The growth patterns were assessed more precisely than by OD measurements by subsampling every 4–8 h depending on the growth phase patterns for bacterial cell enumeration. Cells were fixed with 2% glutaraldehyde and frozen at −20°C until further analysis.
In order to test the effect of the vitamin B1 precursors HMP (AstaTech inc., Bristol, PA, USA) and HET [4-methyl-5-(β-hydroxyethyl)thiazole; Sigma Aldrich, Munich, Germany] on the growth of the diatoms Thalassiosira pseudonana (CCMP 1335) and Leptocylindrus danicus (CCMP 470) these diatoms were grown axenically in ASW medium in a 12:12 h light dark cycle and illuminated at 70 μE. Instead of B1, the precursors were added at 100 nM final concentration together with vitamins B7 and B12 (100 nM each) and growth was monitored as relative fluorescence against a positive control including all three vitamins at final concentrations of 100 nM each and a negative control including only B7 and B12 at the same concentrations. Axenicity of the diatom cultures was checked microscopically.
Cells of the D. shibae cultures were enumerated by flow cytometry, those of the P. inhibens cultures by epifluorescence microscopy due to the formation of microaggregates. Flow cytometric analyses were done according to Osterholz et al. (2015). Cell aggregates of P. inhibens were dispersed by ultrasonication (5 × 10 s at 15 mV; Bandelin Sonopuls HD 200, Bandelin, Berlin, Germany), filtered onto a 0.2 μm polycarbonate membrane, stained for 30 min with SYBR® Green I and counted by epifluorescence microscopy as described (Lunau et al., 2005). At least 1,000 cells were enumerated per filter.
Aliquots of the cultures were centrifuged at 2,499 g in acid washed and combusted (3 h, 500°C) glass centrifuge tubes. The supernatant was filtered through a 0.22 μm polyethersulfone membrane (Minisart, Sartorius, Göttingen, Germany) and the filtrate stored in combusted 20 ml glass vials at −20°C until further analysis. Concentrations of DFAA and DCAA were analyzed by high performance liquid chromatography (HPLC) after precolumn derivatization with orthophtaldialdehyde (Lunau et al., 2006) and concentrations of DFNCHO and DCNCHO by HPLC and pulsed amperometric detection after desalting (Hahnke et al., 2013). Detection limits for DFAA and DFNCHO were 0.5 and 1.5 nM, respectively. Fatty acid concentrations in the treatments with additions of succinate and acetate were determined by HPLC (Sykam, Fürstenfeldbruck, Germany) equipped with an Aminex HPX-87H column (Biorad, München, Germany) (Graue et al., 2012).
Dissolved organic carbon in the filtrates of the bacterial cultures and of the solid-phase extracted DOM (see below) was quantified as described previously (Osterholz et al., 2014). For FT-ICR-MS analyses, filtrates were acidified to pH 2 (HCl 25% p.a., Carl Roth, Germany), extracted via PPL solid phase cartridges (100 mg; Agilent, Waldbronn, Germany) adapted to a concentration of 15 ppm carbon and analyzed by FT-ICR-MS according to Osterholz et al. (2015). The extraction efficiency increased from 2 to 31% on carbon basis in the course of the experiment, as result of the contrary running substrate availability. Extracted DOM was ionized by soft electrospray ionization (Bruker Apollo, Daltonics, Bremen, Germany) and analyzed in positive and negative mode with a 15 T Solarix FT-ICR-MS (Bruker, Daltonics, Bremen, Germany). For each spectrum 300 scans were accumulated in the mass window of 92 to 2,000 Da. An internal calibration list was generated using Bruker Daltonic Data Analysis software for the calibration of the spectra. FT-ICR-MS instrument performance was verified using a laboratory-internal deep ocean DOM reference sample. Detected mass to charge ratios were processed applying a customized routine Matlab script. Molecular formulas were assigned as described by Koch and Dittmar (2006) to molecular masses with a minimum signal-to-noise ratio of 5 (Koch et al., 2007). From the mass spectrograms of the single time points of each culture that of the respective sterile control was subtracted.
To confirm structures of genome-predicted identified exometabolites, we performed fragmentation experiments using FT-ICR-MS. Therefore, both bacterial strains were cultured again at similar conditions as described above for the exometabolome experiments but the exometabolome was harvested at the time point of the peak concentration of the respective exometabolite. A total of 500 mL was extracted via PPL solid phase cartridges as described above. Extracts were redissolved in a 1:1 MilliQ water/methanol solution at a concentration of 29 ppm carbon and analyzed on the FT-ICR-MS as described above. We selected the exometabolites which were identified as precursors or products of biosynthetic pathways for fragmentation, i.e., 43 of the 107 exometabolites identified in total. However, only seven metabolites had sufficiently high signal intensities in the FT-ICR-MS to perform fragmentation experiments. Limitations to fragmentation experiments include the fact that fragments may not be ionizable and different fragments with rather similar masses may yield overlapping peaks. Exometabolites of interest were isolated in a 1 Da window using the quadrupole unit and collision with argon occurred in the hexapole unit of the FT-ICR-MS. Fragmentation parameters were optimized for each mass with an isolation window ranging from 0.1 to 1 (m/z), collision energy adjusted by applying 10 to 15 mV and 300 to 700 broadband scans were accumulated per run.
Metabolites of P. inhibens and D. shibae, predicted by the genome database BioCyc (Caspi et al., 2012), were listed with their corresponding molecular masses and molecular formulae (MF). All MF calculated from FT-ICR-MS detected masses were scanned against the genome-based metabolite prediction list. Matches were analyzed in more detail, regarding intra and extracellular function identified by previous studies. Putatively identified MF of exometabolites were screened against MF of DOM data sets of naturally derived and North Sea phytoplankton blooms (Osterholz et al., 2015, 2016; Noriega-Ortega et al., in preparation).
Both model strains of the Roseobacter group were grown in batch culture on single carbon sources of three major substrate classes to examine the diversification of the exometabolome as a function of these different substrates. Here we focus on the identified exometabolites whereas the overall exometabolome and its diversity is dealt with in a different publication (Noriega-Ortega et al., in preparation)
Phaeobacter inhibens reached highest growth with maximum ODs of 0.76, 1.04, and 1.96 on acetate, glucose and glutamate respectively, at 68, 56, and 20 h (Figure 1). Respective cell numbers at these time points were 1.4 × 109, 0.6 × 109, and 3.4 × 109 cells mL−1 (see supporting information Figure S1). Growth of D. shibae reached highest ODs of 1.11, 1.03, and 0.90 on succinate, glucose and glutamate respectively, at 23, 65, and 39 h (Figure 1) with corresponding cell numbers of 2.1 × 109, 0.8 × 109, and 3.3 × 109 cells mL−1 respectively (see supporting information Figure S1). Substrate concentrations decreased inversely to growth of both bacteria and went below detection limit in the stationary phase except for the culture of D. shibae growing on glucose (Figure 1).
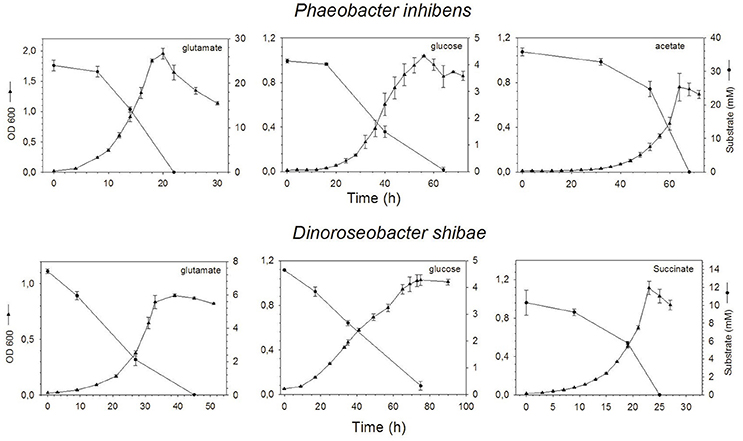
Figure 1. Optical density and concentration of the substrates glutamate, glucose and acetate or succinate over time of P. inhibens (upper) and D. shibae (lower). Note the different scales of the axes.
The analysis of dissolved amino acids within the exometabolome of both strains was biased in the glutamate treatments by the interfering large glutamate peak. Hence, concentrations could only be measured in the stationary phase when glutamate was completely utilized. In the P. inhibens cultures, concentrations of DFAA remained below the detection limit of the HPLC analysis at all growth phases but tryptophan, tyrosine and histidine were detected by the FT-ICR-MS analysis (see below). Concentrations of DCAA in the treatment with acetate remained below 4 μM during the exponential growth phase but reached 235.4 μM in the stationary phase (Figure 2F). In the treatment with glucose, DCAA concentrations increased continuously during the exponential and stationary phase but reached a final concentration of only 15.6 μM (Figure 2E). The glutamate treatment of this culture yielded a concentration of 684.9 μM in the stationary phase (Figure 2D). The substrate source had only a minor influence on the composition of the amino acid pool in the exometabolome. During the exponential and stationary phases aspartate, glutamate, glycine, and alanine constituted the highest mol% of DCAA and together at least 60 mol% (Figure 2, Table S1). The very high DCAA concentrations in the exometabolome may have been a result of cell lysis. To estimate the extent of potential lysis, we made a mass balance of carbon (C) in DCAA and in the bacterial biomass in the stationary phase. DCAA concentrations of 219.8 and 639.0 μM in the treatments with acetate and glutamate in the stationary phase translate into 11.5 and 32.8 mg C L−1, respectively. On the basis of 50 fg C per cell of large bacteria (Simon and Azam, 1989), typical for fast growing cultures, the bacterial numbers of 1.39 × 109 and 3.44 × 109 cells mL−1 in the stationary phase in the acetate and glutamate treatments, respectively, equal 69.5 and 160.9 mg C L−1. Hence, the C bound in DCAA comprises 17.1 and 20.6% of the C bound in bacterial biomass. Therefore, we conclude that at these two conditions, but neither at other conditions nor in the glucose treatments, protein released by lysed cells contributed to the high DCAA concentrations in the stationary phase of the P. inhibens culture. Consequently, these time points were not considered for further exometabolome analyses.
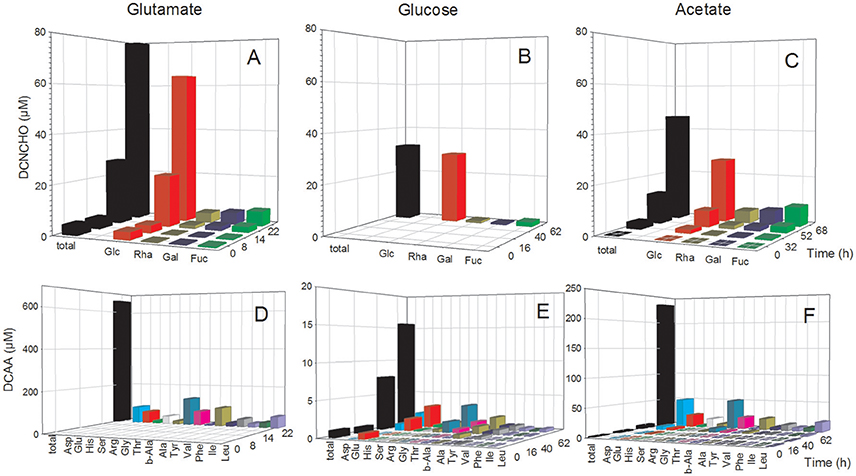
Figure 2. Concentrations of individual neutral monosaccharides and amino acids (AA) bound in total dissolved combined neutral monosaccharides (DCNCHO) and total dissolved combined amino acids (DCAA), respectively, in the P. inhibens cultures. DCNCHO (A–C) and DCAA (D–F) right after inoculation, in the lag, exponential and stationary phase of P. inhibens growing on glutamate (A,D), glucose (B,E) and acetate (C,F). Note the different scales of the parameters in the different panels. Concentrations of DCNCHO in the treatment with glucose are not available until the stationary phase at 62 h due to interference with the HPLC analysis. Concentrations of AA in the glutamate treatment are only available for 22 h in the stationary phase because high glutamate concentrations until the exponential phase interfered with the HPLC analysis.
In the D. shibae cultures, DFAA concentrations were below the detection limit of the HPLC analyses at all growth phases, but tryptophan, tyrosine, phenylalanine and histidine were detected by FT-ICR-MS (see below). Concentrations of DCAA in the exometabolome of the D. shibae cultures continuously increased during the exponential growth phase and reached 12.0 and 14.7 μM in the glucose and succinate treatments in the stationary phase (Figure 3). The glutamate treatment yielded 23.0 μM in the stationary phase (Figure 3D). Glutamate, glycine and alanine dominated the DCAA pool at all substrate conditions and constituted >50 mol% (Table S2). A mass balance of carbon bound in DCAA and the bacterial biomass in the stationary phase of the three treatments showed that DCAA constituted always <1% of the C bound in the biomass of the D. shibae cultures. Hence, cell lysis was minimal in these cultures.
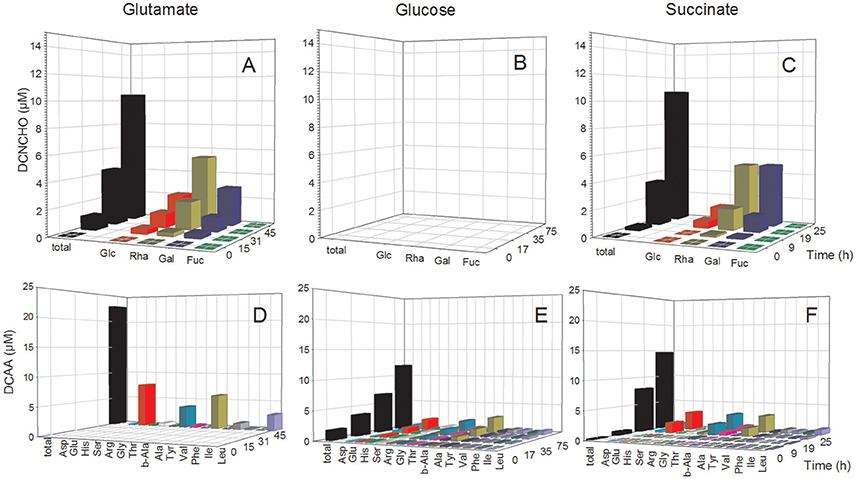
Figure 3. Concentrations of individual neutral monosaccharides and amino acids (AA) bound in total dissolved combined neutral monosaccharides (DCNCHO) and total dissolved combined amino acids (DCAA), respectively, in the D. shibae cultures. DCNCHO (A–C) and DCAA (D–F) right after inoculation, in the lag, exponential and stationary phase of D. shibae growing on glutamate (A,D), glucose (B,E) and succinate (C,F). Concentrations of CHO in the treatment with glucose are not available due to interference with the HPLC analysis. Concentrations of DCAA in the glutamate treatment are only available for 45 h in the stationary phase because high glutamate concentrations until 31 h interfered with the HPLC analysis.
The analysis of dissolved mono- and polysaccharides interfered with the addition of glucose as single substrate source. DFNCHO were not detected in the exometabolome at any growth stage of both strains. Concentrations of DCNCHO increased continuously during growth of both strains on glutamate and acetate or succinate with highest concentrations in the stationary phase (Figures 2, 3). Released DCNCHO in the P. inhibens cultures were greatly dominated by glucose but galactose, rhamnose, and fucose constituted proportions of up to 22 mol% in the treatment with acetate (Figure 2, Table S2). In the D. shibae cultures, DCNCHO concentrations remained lower than in those of P. inhibens (Figure 3). Only glucose, galactose and rhamnose were detected in the exometabolome. In the succinate treatment, rhamnose and galactose dominated whereas in the glutamate treatment galactose became the dominant DCNCHO component in the exponential and stationary phase (Table S2).
Applying the ultrahigh resolution FT-ICR-MS, we detected in total 2767 MF in the exometabolome of P. inhibens and 3,354 MF in that of D. shibae (Figure 4; Tables S3, S4). These numbers include all growth stages and all substrate treatments of both strains, except the stationary phase of P. inhibens grown on glutamate and acetate due to suspected cell lysis. The composition of the exometabolome, but not the exometabolites identified (see below) of both strains varied considerably as a function of the substrate utilized and growth stage (Noriega-Ortega et al., in preparation). Scanning all detected exometabolomic MF against the genome-predicted metabolites of P. inhibens and D. shibae by applying the BioCyc database revealed a match for 67 and 84 exometabolites, respectively (Figure 4). In addition, secondary metabolites known to be produced by P. inhibens and D. shibae were scanned against the exometabolomic MF obtained by FT-ICR-MS, yielding 4 identified compounds in the exometabolome of P. inhibens but none in that of D. shibae (Table 1). Identified exometabolites were further subdivided on the basis of their function and occurrence within the bacterial metabolism (Figure 4). For D. shibae, 35 metabolites were assigned to biosynthetic pathways (Table 1), 16 metabolites to degradation pathways (Table S5) and 33 metabolites originated from spontaneous non-enzymatic chemical-reactions (Table S6). For P. inhibens, 36 metabolites were assigned to biosynthetic pathways (Table 1), 15 to degradation pathways (Table S5), but only 16 derived from spontaneous non-enzymatic chemical reactions (Table S6). In total 43 different exometabolites of biosynthetic pathways were identified of which 28 were present in the exometabolome of both strains. In the D. shibae experiments 19 exametabolites assigned to biosynthetic pathways were detected in >50% of the time points sampled and in the experiments with P. inhibens 16 exometabolites. Alpha-ribazol was detected only at three time points in the exometabolome of D. shibae but in 88% of the time points of the P. inhibens experiments. Pyridoxal-P and histidine were detected only once in the exometabolome of the D. shibae experiments but in 43 and 55% of that of the P. inhibens experiments, respectively. HET was detected only once in the exometabolome of P. inhibens experiments but in 58% of that of the D. shibae experiments. All other exometabolites were detected in 10–50% of the samples analyzed in the experiments of both strains.
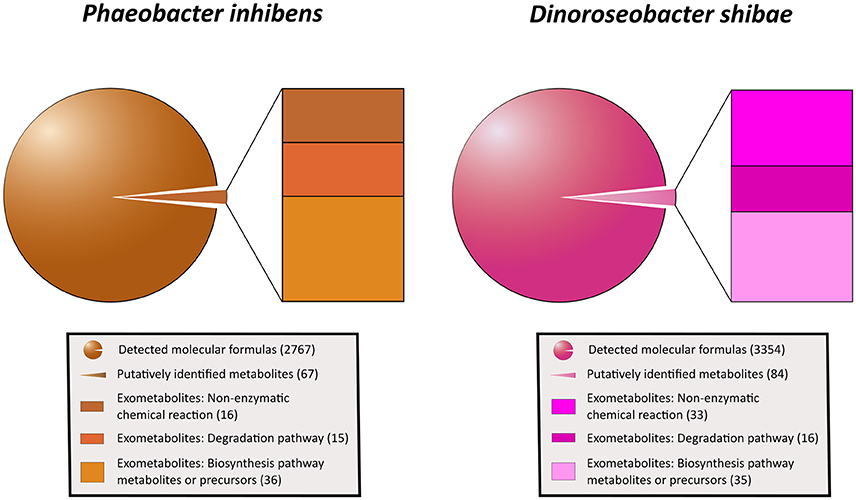
Figure 4. Subdivision of all exometabolites released by P. inhibens and D. shibae during all growth phases. Given (in parenthesis) are total numbers of detected molecular formulas, putatively identified metabolites and those related to non-enzymatic chemical reactions, degradation and biosynthetic pathways and precursors.
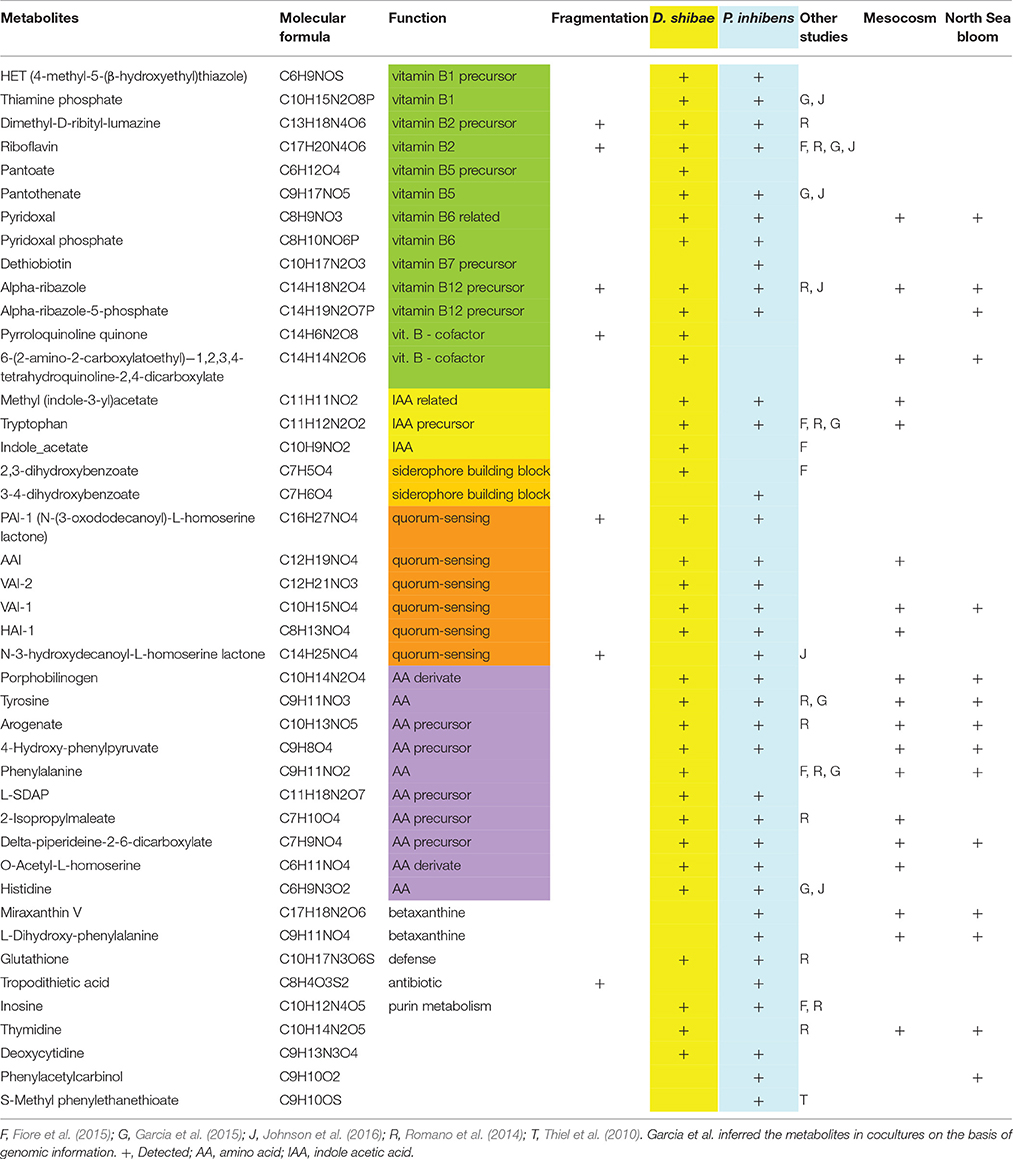
Table 1. Metabolites, their molecular formula, function, fragmentation results detected in the exometabolome of D. shibae and P. inhibens, in other studies, a mesocosm (Osterholz et al., 2015) and in a North Sea phytoplankton bloom (Noriega-Ortega et al., in preparation).
The detailed analysis showed that 12 (34%) and 10 (28%) of the annotated compounds linked to metabolic pathways of D. shibae and P. inhibens, respectively, were B vitamins and/or late precursors of biosynthetic pathways of vitamins B1, B2, B5, B6, B7, and B12 (Figure 5, Table 1). Five (14%) and 6 (18%) of the annotated compounds were putatively quorum sensing-related metabolites and 10 (29%) and 9 (25%) amino acids and precursors of their biosynthetic pathways (Figure 5). Methyl-(indole-3-yl) acetate and tryptophan as precursors in the biosynthetic pathways of IAA were annotated as exometabolites of both organisms whereas IAA was annotated only in that of D. shibae. Altogether, the complete set of annotated compounds was linked to 22 and 20 biosynthetic pathways in D. shibae and in P. inhibens respectively (Table 1).
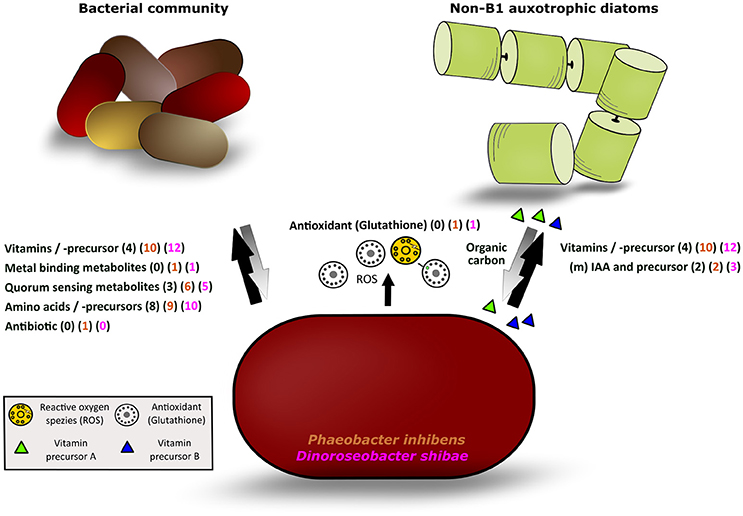
Figure 5. Illustration of the different classes of exometabolites released by P. inhibens and D. shibae and mutualisms with other bacteria and phytoplankton algae. Black arrow: detected metabolites released by both strains; gray arrow: potential response of the bacterial or phytoplankton communities to the bacteria releasing metabolites; in parentheses, the number of exometabolites of each class detected in the natural phytoplankton bloom (black) and released by P. inhibens (brown) and D. shibae (pink) are given.
To further validate the compounds annotated by the genome-based metabolite prediction approach, the respective molecular masses were isolated and fragmented via positive charge collision in the FT-ICR-MS. We confirmed the presence of 7 metabolites in the exometabolomes of D. shibae and P. inhibens respectively, including vitamins, vitamin precursors, acylated homoserine lactones (autoinducers) for quorum sensing and TDA (Table 1). Support for the correct annotation of the compounds was also provided by their detection in other studies applying similar experimental approaches (Table 1). Several identical metabolites were detected by Fiore et al. (2015), Johnson et al. (2016), annotated by Romano et al. (2014), or predicted to be needed exogenously because of lacking biosynthetic pathways in annotated bacterial genomes (Garcia et al., 2015).
In order to test the second part of our hypothesis whether identified precursors indeed enhance growth of prototrophic algae we selected two diatoms, Thalassiosira pseudonana and Leptocylindrus danicus and grew them with the supplementation of HET, HMP, HET, and HMP or B1 and other vitamins they require (B7, B12). Supplementation of HET and HMP resulted in statistically significantly higher growth of both diatoms (p < 0.01; Student's t-test; Table S7). The growth rate and yield of L. danicus was enhanced by 17 and 35%, respectively, relative to a control, whereas the growth rate of T. pseudonana was enhanced by 22% but growth yield remained unaffected (Figure 6, Table S7). In both diatom cultures the growth stimulation differed among the various growth phases. The addition of vitamin B1, surprisingly, resulted in a lower growth stimulation of both diatoms than that of the precursors and only for T. pseudonana the growth rate was significantly higher than that of the control (Figure 6, Table S7).
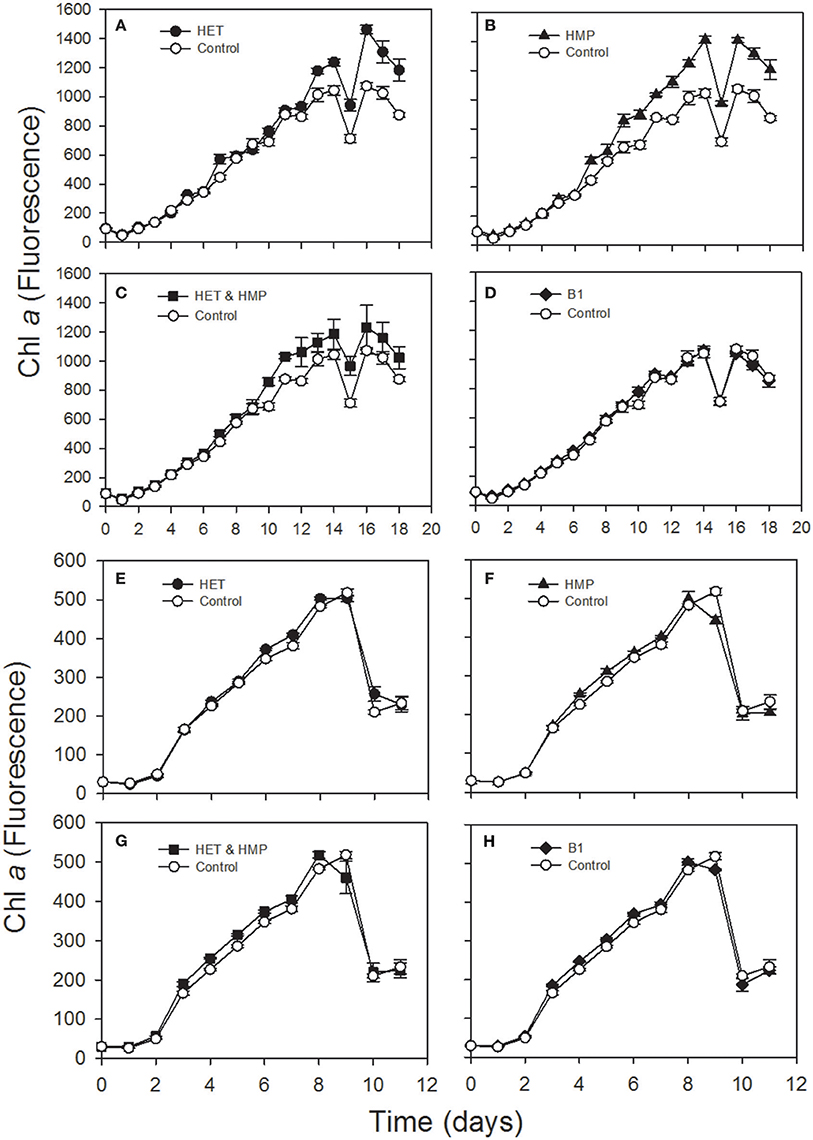
Figure 6. Chlorophyll a fluorescence over time of an axenic culture of the diatoms Leptocylindrus danicus (A–D) and Thalassiosira pseudonana (E–H) supplemented with 4-methyl-5-(β-hydroxyethyl)thiazole (HET; A,E), 4-amino-5-hydroxymethyl-2-methylpyrimidine (HMP; B,F), HET & HMP (C,G) and vitamin B1 (D,H) and a control without any of these supplements.
To examine whether exometabolites produced by the two model strains of the Roseobacter group also occur in natural or naturally-derived DOM, we screened samples of a mesocosm experiment and from phytoplankton blooms in the North Sea. In the DOM of the mesocosm experiment in which a phytoplankton bloom was induced and various bacteria of the Roseobacter group were present (Osterholz et al., 2015), 19 MF were detected which matched those found also in the cultures of both strains. Sixteen and 15 MF were attributed to the cultures of P. inhibens and D. shibae respectively, and 14 occurred in both cultures. Eight MF included amino acids or biosynthetic precursors, three late biosynthetic precursors of vitamins and cofactors, three were autoinducers of quorum sensing and two the plant auxin IAA and its inactive form methyl-IAA (Table 1). In samples from in and outside phytoplankton blooms in the North Sea, 15 MF matched those found in the exometabolomes of both strains. Four of those were biosynthetic precursors of vitamins and cofactors including those detected in the mesocosm experiment, six were amino acids or biosynthetic precursors, matching those detected in the mesocosm experiment, and one was an autoinducer to quorum sensing found also in the mesocosm experiment (Table 1).
Our exometabolome analyses are based predominantly on an untargeted approach using FT-ICR-MS, which enables an ultrahigh resolution of individual molecular masses with a relative but no absolute quantification. The results show that P. inhibens and D. shibae both exhibit distinct exometabolite patterns consisting of 2,767 and 3,354 distinct MF and variations as a function of the carbon source. We were able to identify 2.3 and 2.6% of the exometabolites of P. inhibens and D. shibae, respectively, by comparing detected MF and genome-predicted metabolites. The observation that more than 97% of the MF consist of unknown chemical compounds, also reported in previous studies (Romano et al., 2014; Fiore et al., 2015; Johnson et al., 2016), is surprising and illustrates that the majority of MF comprises compounds is not predicted by genome annotated metabolites. There are indications that some of these compounds are metabolic waste (Fiore et al., 2015) but presumably other biological and physico-chemical reactions contribute to the formation of these compounds. The diversity of the exometabolome and implications for a better understanding of the bacterial processing of organic matter to shape the marine DOM pool are discussed elsewhere (Noriega-Ortega et al., in preparation). Here we focus on the identified exometabolites and the implications these findings have for the understanding of mutualistic interactions among bacteria and algae in marine ecosystems. The detection of seven exometabolites was further supported by fragmentation and we did not find a single case of mismatching fragments. We obtained further support of the correct MF by the genome-based exometabolite prediction. Hence our MF assignment is based on three independent methods and thus appears to be a solid base for discussing their significance in the interplay among marine microbes.
In both strains we identified MF identical to late biosynthetic precursors of and/or the vitamins B1, B2, B5, B6, B7, and B12, IAA and its methylated form, metal-acquisition growth factors, autoinducers for quorum sensing and biosynthetic precursors of several amino acids. Pyridoxal, the dephosphorylated form of vitamin B6, alpha-ribazole, methyl-IAA and 3 autoinducers were also putatively detected in the DOM of a naturally-derived phytoplankton bloom (Osterholz et al., 2015) and the same vitamin precursors, alpha-ribazole-5 phosphate and one of the autoinducers also in the DOM of a North Sea phytoplankton bloom. Most MF identical to biosynthetic precursors of amino acids were also detected in the phytoplankton bloom samples. On the basis of genomic information of both strains and previous data, the vitamins and autoinducers were expected to be present in the exometabolome (Newton et al., 2010; Wagner-Döbler et al., 2010; Thole et al., 2012). However, the putative detection of late precursors of all B vitamins except one, methyl-IAA and five biosynthetic precursors of amino acids in the exometabolome of each strain and in the DOM of the phytoplankton bloom samples was surprising. It showed that these strains and presumably other microorganisms as well release a much greater variety of exometabolites than expected. Even though previous studies found a few of the precursors we detected, such a rich bouquet of biosynthetic precursors has neither been reported before in any bacterial exometabolome nor in the DOM of phytoplankton blooms.
The B2 precursor dimethyl-D-ribityl-lumazine and/or the B12 precursor alpha-ribazole and two amino acid precursors were detected as exometabolites of a Pseudovibrio strain and Ruegeria pomeroyi (Romano et al., 2014; Johnson et al., 2016). The final precursor of the thiazole moiety of vitamin B1, HET, we detected in both strains, is known to be used by several green algae, cryptophytes and dinoflagellates instead of B1 (Lwoff, 1947; Droop, 1958; Turner, 1979). The pyrimidine moiety of vitamin B1 which we did not detect, HMP, has been reported to be released by cyanobacteria, a marine betaproteobacterium and the alga Dunaliella tertiolecta (Carini et al., 2014). Results of previous research provides evidence that HMP and HET can be used in a salvage pathway for biosynthesis of thiamine by various B1-auxotrophic eukaryotic algae and by Pelagibacterales (Turner, 1979; Carini et al., 2014; McRose et al., 2014; Paerl et al., 2015). There is most recent evidence, however, that HMP and unknown HET-related precursors can support growth of B1-auxotrophic microeukaryotic marine algae via various bacteria and that these precursors are present in the open ocean (Paerl et al., 2017). Our results of the growth experiments with T. pseudonana, and L. danicus indicate that HMP and HET also stimulate growth of these vitamin B1 prototrophic coastal diatoms. It was unexpected that the effect was even higher than that of the addition of B1. This result is surprising and implies that L. danicus either lacks a vitamin B1 transporter and/or that in both diatoms these precursors do not only compensate the lacking genetic capabilities of auxotrophic microbes but that HET and HMP have a so far unknown growth-promoting effect on vitamin B1 prototrophic diatoms. If this stimulatory effect on vitamin B1 prototrophic organisms is also true for other microorganisms the release of these precursors has even greater implications for controlling growth of these planktonic communities.
Pantoate as a precursor of vitamin B5 was detected in the exometabolome of D. shibae. It is unknown whether this B5 precursor can be used by marine algae or bacteria. An uptake system for pantoate, however, has been described for Salmonella enterica (Ernst and Downs, 2015) and thus is likely to exist in marine bacteria enabling them to use this precursor. Pyridoxal as the dephosphorylated form of vitamin B6 was detected in the exometabolome of both strains and in the DOM of both phytoplankton blooms. Pyridoxal kinase (EC 2.7.1.35), the enzyme to phosphorylate this inactive form of vitamin B6, is encoded in the genome of many bacteria as documented by a genomic search (https://img.jgi.doe.gov/cgi-bin/mer/main.cgi). If microbes can take up pyridoxal like Sacharomyces cerevisiae (Stolz and Vielreicher, 2003), this inactive form may be a so far neglected source of vitamin B6.
Dethiobiotin, the last stage in the biosynthesis of vitamin B7 (biotin), was detected in the exo-metabolome of P. inhibens. Extracellularly provided dethiobiotin has been demonstrated to cause diverging effects. Exogenous simultaneous supply of biotin and dethiobiotin caused a growth inhibition in several biotin-requiring fungi and bacteria including Sacharomyces, Sordaria and Lactobacilli, and was termed the anti-biotin effect (Dittmer et al., 1944; Lilly and Leonian, 1944). Contrary observations were reported for other fungi and Lactobacilli and a freshwater cyanobacterium, in which biotin auxotrophy was compensated by dethiobiotin addition (Dittmer et al., 1944; Lilly and Leonian, 1944; Bowman and DeMoll, 1993). Hence the significance of released dethiobiotin in marine microbial communities including the mycoplankton is still open and needs further studies.
Alpha-ribazole, a lower ligand building block of vitamin B12, was released by both strains and detected in the phytoplankton bloom samples. The bioactive corrinoid cofactor vitamin B12 has been shown to be a controlling factor of primary production in pelagic ecosystems (Bertrand et al., 2007; Koch et al., 2011). Its cofactor binding, function and catalysis highly depends on the attached lower ligand (Lengyel et al., 1960; Stupperich et al., 1988; Renz, 1999; Yi et al., 2012). It has been shown, however, that the genome of Listeria innocua, lacking the genes for alpha-ribazole biosynthesis, encodes a transporter system of alpha-ribazole, cblT, and is able to synthesize B12 when this moiety is available as an exogenous source (Gray and Escalante-Semerena, 2010). Remodeling of corrinoids, such as vitamin B12, by complementation of the lower ligand via uptake of exogenous compounds appears to be a common phenomenon within microbial communities (Gray and Escalante-Semerena, 2010; Keller et al., 2014; Men et al., 2014). Available 5,6-dimethylbenzimidazole (DMB), an alpha-ribazole precursor, allows major phytoplankton groups to remodel pseudocobalamin, commonly produced by cyanobacteria, into a usable corrinoid cofactor structure (Helliwell et al., 2016). Our results indicate that corrinoid building blocks are exchanged in marine ecosystems and that roseobacters such as D. shibae and P. inhibens may be providers of the most common and bioactive corrinoid cofactor lower ligand.
The auxin IAA has recently been identified to be secreted by various freshwater and marine bacteria and to be an important growth factor of various green algae and a diatom (Bagwell et al., 2014; Zhang et al., 2014; Amin et al., 2015, Fiore et al., 2015). It was also found in a phytoplankton bloom in the Pacific, in a eutrophic lake dominated by cyanobacteria and to be produced by P. inhibens (Zhang et al., 2014; Amin et al., 2015; Segev et al., 2016). We detected IAA in the exometabolome of D. shibae and its precursor tryptophan in the exometabolome of both strains and the naturally derived phytoplankton bloom. Tryptophan has been shown to enhance the production of IAA in P. inhibens (Segev et al., 2016) and also in the haptophyte Emiliania huxleyi (Labeeuw et al., 2016). It thus appears to be important in controlling the production of IAA in these roseobacters and the haptophyte. In both strains and in the naturally derived phytoplankton bloom, we also detected methyl-IAA, a related compound which was neither detected previously in the exometabolome of marine bacteria nor in marine DOM samples. It has been shown in Arabidopsis that methyl-IAA is an inactive form which can be taken up but needs to be demethylated by an esterase to generate the active IAA (Yang et al., 2008). Because of the more hydrophobic form of methyl-IAA as compared to IAA, these authors suggest that it is more easily transported across the cell membrane, possibly even diffuses, thus enhancing the exploitation of the exogenous supply with the subsequent need to demethylate it intracellularly. Another aspect of hydrophobic compounds released into the water is that enhanced hydrophobicity results in a faster supply through this hydrophilic medium thus reducing the time of action and enhancing the accumulation at the target site, e.g. the cell surface (Maier et al., 1994). Hence it is conceivable that the methylated form of IAA in pelagic ecosystems leads to a more efficient use of this important auxin by phytoplankton.
The metal-binding 2,3-dihydroxybenzoate was detected in the exometabolome of D. shibae. This building block of the siderophore enterobactin is known to be secreted by heterotrophic and cyanobacteria under iron-limiting conditions (Young et al., 1967; Byers and Lankford, 1968; Fiore et al., 2015) and to enhance the expression of the 2,3-dihydroxybenzoate-AMP ligase, catalyzing an essential synthesis step toward the formation of enterobactin (Khalil and Pawelek, 2011). Other strains of the Roseobacter group are known to secrete enterobactin but not 2,3-dihydrobenzoate (Hogle et al., 2016). Hence, D. shibae appears to have the potential to provide 2,3-dihydroxybenzoate to marine bacterioplankton communities, thus inducing enterobactin synthesis and favoring iron uptake by itself and other bacteria. A building block of the siderophore petrobactin, 3,4-dihydroxybenzoate, was detected in the exometabolome of P. inhibens and not found before as a bacterial exometabolite. It has been shown that P. inhibens is able to produce enterobactin (Thole et al., 2012). Thus, 3,4-dihydroxybenzoate may have a similar role in metal acquisition as 2,3-dihydroxybenzoate (see above).
Glutathione was detected in the exometabolome of both strains and previously found also in that of a Pseudovibrio strain (Romano et al., 2014). Besides essential intracellular functions, glutathione is a fundamental extracellular protectant for bacteria, in particular as an antioxidant when reactive oxygen species (ROS) are present (Smirnova and Oktyabrsky, 2005; Montoya, 2013; Smirnova et al., 2015). One reason for the presence of glutathione in the exometabolome may be stress caused by the growth conditions and high cell densities and the possible protection against ROS. On the other hand, glutathione has been found in nanomolar concentrations in the oligotrophic north Pacific (Dupont et al., 2006) even though it is rapidly photooxidized (Moingt et al., 2010). Therefore, it is conceivable that bacteria such as our model strains actively excrete glutathione for protection against ROS produced by photochemical DOM oxidation.
Biosynthetic precursors of amino acids were previously reported in bacterial exometabolomes but interpreted as a result of an overflow metabolism (Paczia et al., 2012; Romano et al., 2014). As we found these precursors also in the DOM of phytoplankton blooms, we suggest that they are either actively secreted or released by dividing and growing cells or due to mortality by grazing or viral lysis of bacterial communities and are not a result of an overflow metabolism. They can potentially be used for amino acid biosynthesis by other bacteria but this pathway has yet to be shown. In support of this suggestion, it has recently been shown that bacterial mutants, missing biosynthetic genes in amino acid pathways, have a growth benefit over the wild type when supplied with the respective precursors or amino acids (D'Souza et al., 2014; Waschina et al., 2016). These authors consider amino acid cross-feeding as a specialized evolutionary mechanism of how bacterial subpopulations can receive mutual benefits by saving biosynthetic costs. Auxotrophy of essential amino acids can potentially be conquered by public amino acid goods and even facilitate metabolic interdependency or symbiosis (McCutcheon and Moran, 2007; Garcia et al., 2015). Our findings of several essential and non-essential amino acids and respective biosynthetic precursors in the exometabolome of both strains and in the DOM of the phytoplankton bloom samples suggests that beneficial amino acid cross-feeding also occurs in marine microbial communities.
The release of a wealth of exometabolites by the two model strains indicates that both of them may function as important suppliers of growth factors as well as of biosynthetic precursors, so-called public goods (Morris et al., 2012), to other pro- and eukaryotes in marine ecosystems. Phaeobacter inhibens dwells in biofilms (Gram et al., 2015) and both model strains live in association with microalgae and pelagic environments (Wagner-Döbler et al., 2010; Gifford et al., 2014; Segev et al., 2016) thus suggesting that these exometabolites are released as public goods in biofilm-associated as well as in pelagic marine communities. Biofilms with dense colonization of diverse bacterial communities may include surface-associated habitats but also marine aggregates which often form during phytoplankton blooms (Simon et al., 2002). Bacteria releasing public goods were recently termed Black Queen in the context of the Black Queen hypothesis (BQH; Morris et al., 2012; Morris, 2015), a scenario in which other bacteria benefit from losing genetic traits to synthesize certain growth factors such as vitamins or parts of their biosynthetic pathways when exogenous supply of these compounds is consistently available. Aggregate-associated bacteria acting as Black Queens may also supply free-living pelagic bacteria and phytoplankton algae in the surrounding water with public goods. Genome streamlining features were reported from various pelagic bacteria including Pelagibacterales, the SAR86 clade and Prochlorococcus (Dupont et al., 2012; Carini et al., 2014; Giovannoni et al., 2014) but also from free-living and symbiotic bacteria dwelling in nutrient-rich or constant environments (Van de Guchte et al., 2006; McCutcheon and Moran, 2007; D'Souza et al., 2014). Supply of released compounds as public goods to other microbes is part of a complex network of microbial interactions and there are quite a few public goods in this context, but also private metabolic goods not shared, which act beyond the concept of the BQH (Morris, 2015; Estrela et al., 2016). Our results in fact indicate that the two vitamin B1 prototrophic diatoms benefit from supply by the B1 precursors HET and HMP, a scenario not considered by the BQH. It must also be kept in mind that in a microbial community mutual interactions exist between the primary producers secreting substrates to the (photo)heterotrophic microbes and that different microbes are distinct in their capabilities, e.g., in hydrolyzing polymers. For instance Flavobacteria exhibit a wealth of polysaccharide hydrolyzing enzymes (Teeling et al., 2012) whereas roseobacters are very limited in these polymer-degrading traits (Hahnke et al., 2013). Further, it has been shown that different bacteria, each missing distinct genomic metabolic traits and exhibiting streamlined genomic features, coexist by complementing each other with metabolites for which they are auxotrophic (Garcia et al., 2015). A microbial community with mutual interdependencies between two or among a group of microbes and no unidirectional flows of public and private goods among various bacteria and other microbes appears to be a more suitable model to describe this complex network, a marketplace of microbial metabolites (Figure 5; Zelezniak et al., 2015). Such scenarios presumably characterize the dynamic ecosystems in which both model strains dwell: In tighter or looser association with algae and other bacteria on biofilms or during phytoplankton blooms. Both scenarios exhibit many mutual interactions among a multitude of organisms with non-streamlined as well as streamlined genomes.
All authors designed the experiments. GW and BN carried out the experiments, GW identified the exometabolites and wrote the first draft of the manuscript. MS and GW wrote the final version of the manuscript. All authors critically reviewed and added aspects to the manuscript.
The study was supported by Deutsche Forschungsgemeinschaft within TRR51 Roseobacter.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Kathrin Klapproth, Birgit Kuerzel and Rolf Weinert for help with the sample analyses by FT-ICR-MS and HPLC.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.01985/full#supplementary-material
Amin, S. A., Hmelo, L. R., van Tol, H. M., Durham, B. P., Carlson, L. T., Heal, K. R., et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522, 98–101. doi: 10.1038/nature14488
Bagwell, C. E., Piskorska, M., Soule, T., Petelos, A., and Yeager, C. M. (2014). A diverse assemblage of indole-3-acetic acid producing bacteria associate with unicellular green algae. Appl. Biochem. Biotechnol. 173, 1977–1984. doi: 10.1007/s12010-014-0980-5
Bertrand, E. M., Saito, M. A., Rose, J. M., Riesselman, C. R., Lohan, M. C., et al. (2007). Vitamin B12 and iron colimitation of phytoplankton growth in the Ross Sea. Limnol. Oceanogr. 52, 1079–1093. doi: 10.4319/lo.2007.52.3.1079
Bowman, W. C., and DeMoll, E. (1993). Biosynthesis of biotin from dethiobiotin by the biotin auxotroph Lactobacillus plantarum. J. Bacteriol. 175, 7702–7704. doi: 10.1128/jb.175.23.7702-7704.1993
Breider, S., Freese, H., Spröer, C., Simon, M., Overmann, J., and Brinkhoff, T. (2017). Phaeobacter porticola sp. nov., an antibiotic producing bacterium isolated from a harbor in the southern North Sea. Int. J. System. Evol. Microbiol. 67, 2153–2159. doi: 10.1099/ijsem.0.001879
Brito-Echeverría, J., Lucio, M., López-López, A., Antón, J., Schmitt-Kopplin, P., and Rosselló-Móra, R. (2011). Response to adverse conditions in two strains of the extremely halophilic species Salinibacter ruber. Extremophiles 15, 379–389. doi: 10.1007/s00792-011-0366-3
Byers, B. R., and Lankford, C. E. (1968). Regulation of synthesis of 2,3-dihydroxybenzoic acid in Bacillus subtilis by iron and a biological secondary hydroxamate. Biochim. Biophys. Acta 165, 563–566. doi: 10.1016/0304-4165(68)90244-4
Carini, P., Campbell, E. O., Morré, J., Sa-udo-Wilhelmy, S. A., Thrash, J. C., Bennett, S. E., et al. (2014). Discovery of a SAR11 growth requirement for thiamin's pyrimidine precursor and its distribution in the Sargasso Sea. ISME J. 8, 1727–1738. doi: 10.1038/ismej.2014.61
Caspi, R., Altman, T., Dreher, K., Fulcher, C. A., Subhraveti, P., Keseler, I. M., et al. (2012). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 40, D742–D753. doi: 10.1093/nar/gkr1014
Dickschat, J. S. (2010). Quorum sensing and bacterial biofilms. Nat. Prod. Rep. 27, 343–369. doi: 10.1039/b804469b
Dittmer, K., Melville, D. B., and DU Vigneaud, V. (1944). The possible synthesis of biotin from desthiobiotin by yeast and the anti-biotin effect of desthiobiotin for L. casei. Science 99, 203–205. doi: 10.1126/science.99.2567.203
Droop, M. R. (1958). Requirement for thiamine among some marine and supra-littoral protista. J. Mar. Biol. Assoc. UK 37, 323–329. doi: 10.1017/S0025315400023729
Drüppel, K., Hensler, M., Trautwein, K., Koßmehl, S., Wöhlbrand, L., Schmidt-Hohagen, K., et al. (2014). Pathways and substrate-specific regulation of amino acid degradation in Phaeobacter inhibens DSM 17395 (archetype of the marine Roseobacter clade). Environ. Microbiol. 16, 218–238. doi: 10.1111/1462-2920.12276
D'Souza, G., Waschina, S., Pande, S., Bohl, K., Kaleta, C., and Kost, C. (2014). Less is more: selective advantage can explain the prevalent loss ob biosynthetic genes in bacteria. Evolution 68, 2559–2570. doi: 10.1111/evo.12468
Dupont, C. L., Moffett, J. W., and Ahner, B. A. (2006). Distributions of dissolved and particulate thiols in the sub-artic north Pacific. Deep Sea Res. I 57, 553–566. doi: 10.1016/j.dsr.2006.09.003
Dupont, C. L., Rusch, D. B., Yooseph, S., Lombardo, M. J., Richter, R. A., Valas, R., et al. (2012). Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 6, 1186–1199. doi: 10.1038/ismej.2011.189
Ernst, D. C., and Downs, D. M. (2015). The STM4195 gene product (PanS) transports coenzyme A precursors in Salmonella enterica. J. Bacteriol. 197, 1368–1377. doi: 10.1128/JB.02506-14
Estrela, S., Moris, J. J., and Kerr, B. (2016). Private benefits and metabolic conflicts shape the emergence of microbial interdependencies. Environ. Microbiol. 18, 1415–1427. doi: 10.1111/1462-2920.13028
Fiore, C. L., Longnecker, K., Kido Soule, M. C., and Kujawinski, E. B. (2015). Release of ecologically relevant metabolites by the cyanobacterium Synechococcus elongatus CCMP 1631. Environ. Microbiol. 17, 3949–3963. doi: 10.1111/1462-2920.12899
Frimmersdorf, E., Horatzek, S., Pelnikevich, A., Wiehlmann, L., and Schomburg, D. (2010). How Pseudomonas aeruginosa adapts to various environments: a metabolomic approach. Environ. Microbiol. 12, 1734–1747. doi: 10.1111/j.1462-2920.2010.02253.x
Garcia, S. L., Buck, M., McMahon, K. D., Grossart, H. P., Eiler, A., and Warnecke, F. (2015). Auxotrophy and intrapopulation complementary in the “interactome” of a cultivated freshwater model community. Mol. Ecol. 2, 4449–4459. doi: 10.1111/mec.13319
Gifford, S. M., Sharma, S., and Moran, M. A. (2014). Linking activity and function to ecosystem dynamics in a coastal bacterioplankton community. Front. Microbiol. 5:185. doi: 10.3389/fmicb.2014.00185
Giovannoni, S. J., Cameron Thrash, J., and Temperton, B. (2014). Implications of streamlining theory for microbial ecology. ISME J. 8, 1553–1565. doi: 10.1038/ismej.2014.60
Gram, L., Rasmussen, B. B., Wemheuer, B., Bernbom, N., Hg, Y. Y., Porsby, C., et al. (2015). Phaeobacter inhibens from the Roseobacter clade has an environmental niche as a surface colonizer in harbors. System. Appl. Microbiol. 38, 483–493. doi: 10.1016/j.syapm.2015.07.006
Graue, J., Engelen, B., and Cypionka, H. (2012). Degradation of cyanobacterial biomass in anoxic tidal-flat sediments: a microcosm study of metabolic processes and community changes. ISME J. 6, 660–669. doi: 10.1038/ismej.2011.120
Gray, M. J., and Escalante-Semerena, J. C. (2010). A new pathway for the synthesis of α-ribazole-phosphate in Listeria innocua. Mol. Microbiol. 77, 1429–1438. doi: 10.1111/j.1365-2958.2010.07294.x
Hahnke, S., Brock, N. L., Zell, C., Simon, M., Dickschat, J. S., and Brinkhoff, T. (2013). Physiological diversity of Roseobacter clade bacteria co-occurring during a phytoplankton bloom in the North Sea. Syst. Appl. Microbiol. 36, 39–48. doi: 10.1016/j.syapm.2012.09.004
Hartmann, A., and Schikora, A. (2012). Quorum sensing of bacteria and trans-kingdom interactions of N-acyl homoserine lactones with eukaryotes. J. Chem. Ecol. 38, 704–713. doi: 10.1007/s10886-012-0141-7
Helliwell, K. E., Lawrence, A. D., Holzer, A., Kudahl, U. J., Sasso, S., Kräutler, B., et al. (2016). Cyanobacteria and eukaryotic algae use different chemical variants of vitamin B12. Curr. Biol. 26, 999–1008. doi: 10.1016/j.cub.2016.02.041
Hogle, S. L., Cameron, T. J., Dupont, C. L., Chris, L., and Barbeau, K. A. (2016). Trace metal acquisition by marine heterotrophic bacterioplankton with contrasting trophic strategies. Appl. Environ. Microbiol. 82, 1613–1624. doi: 10.1128/AEM.03128-15
Johnson, W. M., Kido Soule, M. C., and Kujawinski, E. B. (2016). Evidence for quorum sensing and differential metabolite production by a marine bacterium in response to DMSP. ISME J. 10, 2304–2316. doi: 10.1038/ismej.2016.6
Kell, D. B., Brown, M., Davey, H. M., Dunn, W. B., Spasic, I., and Oliver, S. G. (2005). Metabolic footprinting and systems biology: the medium is the message. Nat. Rev. Microbiol. 3, 557–565. doi: 10.1038/nrmicro1177
Keller, S., Ruetz, M., Kunze, C., Kräutler, B., Diekert, G., and Schubert, T. (2014). Exogenous 5,6-dimethylbenzimidazole caused production of a non-functional tetrachloroethene reductive dehalogenase in Sulfurospirillum multivorans. Environ. Microbiol. 16, 3361–3369. doi: 10.1111/1462-2920.12268
Khalil, S., and Pawelek, P. D. (2011). Enzymatic adenylation of 2,3-dihydroxybenzoate is enhanced by a protein-protein interaction between Escherichia coli 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase (EntA) and 2,3-dihydroxybenzoate-AMP ligase (EntE). Biochemistry Mosc. 50, 533–545. doi: 10.1021/bi101558v
Koch, B. P., and Dittmar, T. (2006). From mass to structure: an aromaticity index for highresolution mass data of natural organic matter. Rapid Commun. Mass Spectrom. 20, 926–932. doi: 10.1002/rcm.2386
Koch, B. P., Dittmar, T., Witt, M., and Kattner, G. (2007). Fundamentals of molecular formula assignment to ultrahigh resolution mass data of natural organic matter. Anal. Chem. 79, 1758–1763. doi: 10.1021/ac061949s
Koch, F., Marcoval, M. A., Panzeca, C., Bruland, K. W., Sa-udo-Wilhelmy, S. A., and Gobler, C. J. (2011). The effect of vitamin B12 on phytoplankton growth and community structure in the Gulf of Alaska. Limnol. Oceanogr. 56, 1023–1034. doi: 10.4319/lo.2011.56.3.1023
Kujawinski, E. B., Longkecker, K., Blough, N. V., Del Veccchio, R., Finlay, L., Kitner, J. B., et al. (2009). Identification of possible source markers in marine dissolved organic matter using ultrahigh resolution mass spectrometry. Geochim. Cosmochim. Acta 73, 4384–4399. doi: 10.1016/j.gca.2009.04.033
Labeeuw, L., Khey, J., Bramucci, A. R., Atwal, H., de la Mata, P., Harynuk, J., et al. (2016). Indole-3-acetic acid is produced by Emiliania huxleyi coccolith-bearing cells and triggers a physiological response in bald cells. Front. Microbiol. 7:828. doi: 10.3389/fmicb.2016.00828
Lengyel, P. L., Mazumder, R., and Ochoa, S. (1960). Mammalian methylmalonyl isomerase and vitamin B12 coenzymes. Proc. Natl. Acad. Sci. U.S.A. 46, 1312–1318.
Lilly, V. G., and Leonian, L. H. (1944). The anti-biotin effect of desthiobiotin. Science 99, 205–206. doi: 10.1126/science.99.2567.205
Lunau, M., Lemke, A., Dellwig, O., and Simon, M. (2006). Physical and biogeochemical controls of microaggregate dynamics in a tidally affected coastal ecosystem. Limnol. Oceanogr. 51, 847–859. doi: 10.4319/lo.2006.51.2.0847
Lunau, M., Lemke, A., Walther, K., Martens-Habbena, W., and Simon, M. (2005). An improved method for counting bacteria in samples with high proportions of particle-associated cells by epifluorescence microscopy: Environ. Microbiol. 7, 961–968. doi: 10.1111/j.1462-2920.2005.00767.x
Lwoff, A. (1947). Some aspects of the problem of growth factors for protista. Ann. Rev. Microbiol. 1, 101–114.
Maier, I., Müller, D. G., and Boland, W. (1994). Spermatozoid chemotaxis in Laminaria digitata (Phaeophyceae). III. Pheromone receptor specificity and threshold concentrations. Z. Naturforschung 49c, 601–606.
Mansson, M., Gram, L., and Larsen, T. O. (2011). Production of bioactive secondary metabolites by marine Vibrionaceae. Mar. Drugs 9, 1440–1468. doi: 10.3390/md9091440
Mas, A., Jamshidi, S., van Lagadeuc, Y., Eveillard, D., and Vandenkoornhuyse, P. (2016). Beyond the black queen hypothesis. ISME J. 10, 2085–2091. doi: 10.1038/ismej.2016.22
McCutcheon, J. P., and Moran, N. A. (2007). Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc. Natl. Acad. Sci. U.S.A. 104, 19392–19397. doi: 10.1073/pnas.0708855104
McRose, D., Guo, J., Monier, A., Sudek, S., Wilken, S., Yan, S., et al. (2014). Alternatives to vitamin B1 uptake revealed with discovery of riboswitches in multiple marine eukaryotic lineages. ISME J. 8, 2517–2529. doi: 10.1038/ismej.2014.146
Men, Y., Seth, E. C., Yi, S., Crofts, T. S., Allen, R. H., Taga, M. E., et al. (2014). Identification of specific corrinoids reveals corrinoid modification in dechlorinating microbial communities. Environ. Microbiol. 17, 4873–4884. doi: 10.1111/1462-2920.12500
Milici, M., Deng, Z. L., Tomasch, J., Decelle, J., Wos-Oxley, M., Wang, H., et al. (2016). Co-occurrence analysis of microbial taxa in the Atlantic Ocean reveals high connectivity in the free-living bacterioplankton. Front. Microbiol. 7:649. doi: 10.3389/fmicb.2016.00649
Moingt, M., Bresac, M., Belanger, D., and Amyot, M. (2010). Role of ultra-violet radiation, mercury and copper on the stability of dissolved glutathione in natural and artificial freshwater and saltwater. Chemosphere 80, 1314–1320. doi: 10.1016/j.chemosphere.2010.06.041
Montoya, M. (2013). Bacterial glutathione import. Nat. Struct. Mol. Biol. 20:775. doi: 10.1038/nsmb.2632
Morris, J. J. (2015). Black Queen evolution: the role of leakiness in structuring microbial communities. Trends in Genetics 31, 475–482. doi: 10.1016/j.tig.2015.05.004
Morris, J. J., Lenski, R. E., and Zinser, E. R. (2012). The black queen hypothesis: evolution of dependencies through adaptive gene loss. MBio 3, e00036–e00012. doi: 10.1128/mBio.00036-12
Neumann, A., Patzelt, D., Wagner-Döbler, I., and Schulz, S. (2013). Identification of new N-acylhomoserine lactone signalling compounds of Dinoroseobacter shibae DFL-12T by overexpression of luxI genes. Chembiochem 14, 2355–2361. doi: 10.1002/cbic.201300424
Newton, R. J., Griffin, L. E., Bowles, K. M., Meile, C., Gifford, S., Givens, C. E., et al. (2010). Genome characteristics of a generalist marine bacterial lineage. ISME J. 4, 784–798. doi: 10.1038/ismej.2009.150
Osterholz, H., Dittmar, T., and Niggemann, J. (2014). Molecular evidence for rapid dissolved organic matter turnover in Arctic fjords. Mar. Chem. 160, 1–10. doi: 10.1016/j.marchem.2014.01.002
Osterholz, H., Niggemann, J., Giebel, H. A., Simon, M., and Dittmar, T. (2015). Inefficient microbial production of refractory dissolved organic matter in the ocean. Nat. Commun. 6:7422. doi: 10.1038/ncomms8422
Osterholz, H., Singer, G., Wemheuer, B., Daniel, R., Simon, M., Niggemann, J., et al. (2016). Deciphering associations between dissolved organic molecules and bacterial communities in a pelagic marine system. ISME J. 7, 1717–1730. doi: 10.1038/ismej.2015.231
Paczia, N., Nilgen, A., Lehmann, T., Gätgens, J., Wiechert, W., and Noack, W. S. (2012). Extensive exometabolome analysis reveals extended overflow metabolism in various microorganisms. Microb. Cell Fact. 11:122. doi: 10.1186/1475-2859-11-122
Paerl, R., Bertrand, E., Allen, E., Palenik, B., and Azam, F. (2015). Vitamin B1 ecophysiology of marine picoeukaryotic algae: Strain-specific differences and a new role for bacteria in vitamin cycling. Limnol. Oceanogr. 60, 215–228. doi: 10.1002/lno.10009
Paerl, R. W., Bouget, F. Y., Lozano, J. C., Vergé, V., Schatt, P., Allen, E. E., et al. (2017). Use of plankton-derived vitamin B1 precursors, especially thiazole-related precursor, by key marine picoeukaryotic phytoplankton. ISME J. 11, 753–765. doi: 10.1038/ismej.2016.145
Paul, C., Barofsky, A., Vidoudez, C., and Pohnert, G. (2009). Diatom exudates influence metabolism and cell growth of co-cultured diatom species. Mar. Ecol. Prog. Ser. 389:61–70. doi: 10.3354/meps08162
Renz, P. (1999). “Biosynthesis of the 5,6-dimethylbenzimidazole moiety of cobalamin and of the other bases found in natural corrinoids,” in Chemistry and Biochemistry of B12, ed R. Banerjee (New York, NY: Wiley), 557–575.
Romano, S., Dittmar, T., Bondarev, V., Weber, R. J. M., Viant, M. R., and Schulz-Vogt, H. N. (2014). Exo-metabolome of Pseudovibrio sp. FO-BEG1 analyzed by ultra-high resolution mass spectrometry and the effect of phosphate limitation. PLoS ONE 9:e96038. doi: 10.1371/journal.pone.0096038
Rosselló-Mora, R., Pena, A., Brito-Echeverria, J., Lopez-Lopez, A., Valens-Vadell, M., Frommberger, M., et al. (2008). Metabolic evidence for biogeographic isolation of the extremophilic bacterium Salinibacter ruber. ISME J. 2, 242–253. doi: 10.1038/ismej.2007.93
Sañudo-Wilhelmy, S. A., Gómez-Consarnau, L., Suffridge, C., and Webb, E. A. (2014). The role of B vitamins in marine biogeochemistry. Ann. Rev. Mar. Sci. 6, 339–367. doi: 10.1146/annurev-marine-120710-100912
Segev, E., Wyche, T. E., Kim, K. H., Petersen, J., Ellebrandt, C., Vlamakis, H., Barteneva, N., et al. (2016). Dynamic metabolic exchange governs a marine algal-bacterial interaction. Elife 5:e17473. doi: 10.7554/eLife.17473
Simon, M., and Azam, F. (1989). Protein content and protein synthesis rates of planktonic marine bacteria. Mar. Ecol. Prog. Ser. 51, 201–213. doi: 10.3354/meps051201
Simon, M., Grossart, H. P., Schweitzer, B., and Ploug, H. (2002). Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 28, 175–211. doi: 10.3354/ame028175
Smirnova, G. V., Muzyka, N. G., Ushakov, V. Y., Tyulenev, A. V., and Oktyabrsky, O. N. (2015). Extracellular superoxide provokes glutathione efflux from Escherichia coli cells. Res. Microbiol. 166, 609–617. doi: 10.1016/j.resmic.2015.07.007
Smirnova, G. V., and Oktyabrsky, O. N. (2005). Glutathione in bacteria. Biochemistry 70, 1199–1211. doi: 10.1007/s10541-005-0248-3
Soora, M., and Cypionka, H. (2013). Light enhances survival of Dinoroseobacter shibae during long-term starvation. PLoS ONE 8:e83960. doi: 10.1371/journal.pone.0083960
Stolz, J., and Vielreicher, M. (2003). Tpn1p, the plasma membrane vitamin B6 transporter of Saccharomyces cerevisiae. J. Biol. Chem. 278, 18990–18996. doi: 10.1074/jbc.M300949200
Stupperich, E., Eisiger, H. J., and Krautler, B. (1988). Diversity of corrinoids in acetogenic bacteria. P-cresolylcobamide from Sporomusa ovata, 5-methoxy-6-methylbenzimidazolylcobamide from Clostridium formicoaceticum and vitamin B12 from Acetobacterium woodii. Eur. J. Biochem. 172, 459–464.
Teeling, H., Fuchs, B. M., Becher, D., Klockow, C., Gardebrecht, A., Bennke, C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611. doi: 10.1126/science.1218344
Thiel, V., Brinkhoff, T., Dickschat, J. S., Wickel, S., Grunenberg, J., Wagner-Döbler, I., et al. (2010). Identification and biosynthesis of tropone derivatives and sulfur volatiles produced by bacteria of the marine Roseobacter clade. Org. Biomol. Chem. 8, 234–246. doi: 10.1039/b909133e
Thole, S., Kalhoefer, D., Voget, S., Berger, M., Engelhardt, T., Liesegang, H., et al. (2012). Phaeobacter gallaeciensis genomes from globally opposite locations reveal high similarity of adaptation to surface life. ISME J. 6, 2229–2244. doi: 10.1038/ismej.2012.62
Turner, M. F. (1979). Nutrition of some marine microalgae with special reference to vitamin requirements and utilization of nitrogen and carbon sources. J. Marine Biol. Assoc. UK 59, 535–552. doi: 10.1017/S0025315400045550
Van de Guchte, M., Penaud, S., Grimaldi, C., Barbe, V., Bryson, K., Nicolas, P., et al. (2006). The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. U.S.A. 103, 9274–9279. doi: 10.1073/pnas.0603024103
Villas-Bôas, S. G., Noel, S., Lane, G. A., Attwood, G., and Cookson, A. (2006). Extracellular metabolomics: a metabolic footprinting approach to assess fiber degradation in complex media. Anal. Biochem. 349, 297–305. doi: 10.1016/j.ab.2005.11.019
Wagner-Döbler, I., Baumgart, B., Brinkhoff, T., Buchholz, I., Bunk, B., Cypionka, H., et al. (2010). The complete genome sequence of the algal symbiont Dinoroseobacter shibae – a hitchhiker‘s guide to life in the sea. ISME J. 4, 61–77. doi: 10.1038/ismej.2009.94
Waschina, S., D'Souza, G., Kost, C., and Kaleta, C. (2016). Metabolic network architecture and carbon source determine metabolite production costs. FEBS J. 283, 2149–2163. doi: 10.1111/febs.13727
Yang, Y., Xu, R., Ma, C., Vlot, A. C., Klessig, D. F., and Pichersky, E. (2008). Inactive methyl indole-3-acetic acid ester can be hydrolyzed and activated by several esterases belonging to the AtMES esterase family of Arabidopsis. Plant Physiol. 147, 1034–1045. doi: 10.1104/pp.108.118224
Yi, S., Seth, E. C., Men, Y. J., Allen, R. H., Alvarez-Cohen, L., and Taga, M. E. (2012). Versatility in corrinoid salvaging and remodeling pathways supports the corrinoid-dependent metabolism of Dehalococcoides mccartyi. Appl. Environ. Microbiol. 78, 7745–7752. doi: 10.1128/AEM.02150-12
Young, I. G., Cox, G. B., and Gibson, F. (1967). 2,3-Dihydroxybenzoate as a bacterial growth factor and its route of biosynthesis. Biochim. Biophys. Acta 141, 319–331. doi: 10.1016/0304-4165(67)90106-7
Zech, H., Thole, S., Schreiber, K., Kalhöfer, D., Voget, S., Brinkhoff, T., et al. (2009). Growth phase-dependent global protein and metabolite profiles of Phaeobacter gallaeciensis strain DSM 17395, a member of the marine Roseobacter-clade. Proteomics 9, 3677–3697. doi: 10.1002/pmic.200900120
Zelezniak, A., Andrejev, S., Ponomarova, O. D., Mende, D. R., Bork, P., and Patil, K. R. (2015). Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl. Acad. Sci. U.S.A. 112, 6449–6454. doi: 10.1073/pnas.1421834112
Zhang, F., Harir, M., Moritz, F., Zhang, J., Witting, M., Wu, Y., et al. (2014). Molecular and structural characterization of dissolved organic matter during and post cyanobacterial bloom in Taihu by combination of NMR spectroscopy and FTICR mass spectrometry. Water Res. 57, 280–294. doi: 10.1016/j.watres.2014.02.051
Keywords: roseobacter, DOM, exometabolome, black queen hypothesis
Citation: Wienhausen G, Noriega-Ortega BE, Niggemann J, Dittmar T and Simon M (2017) The Exometabolome of Two Model Strains of the Roseobacter Group: A Marketplace of Microbial Metabolites. Front. Microbiol. 8:1985. doi: 10.3389/fmicb.2017.01985
Received: 02 August 2017; Accepted: 27 September 2017;
Published: 12 October 2017.
Edited by:
Tilmann Harder, University of Bremen, GermanyReviewed by:
Rebecca Case, University of Alberta, CanadaCopyright © 2017 Wienhausen, Noriega-Ortega, Niggemann, Dittmar and Simon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meinhard Simon, bS5zaW1vbkBpY2JtLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.