
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 09 October 2017
Sec. Evolutionary and Genomic Microbiology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.01966
This article is part of the Research Topic The Good, The Bad and The Ugly: Multiple Roles of Bacteria in Human Life View all 41 articles
Bioaugmentation, or the inoculation of microorganisms (e.g., bacteria harboring the required catabolic genes) into soil to enhance the rate of contaminant degradation, has great potential for the bioremediation of soils contaminated with organic compounds. Regrettably, cell bioaugmentation frequently turns into an unsuccessful initiative, owing to the rapid decrease of bacterial viability and abundance after inoculation, as well as the limited dispersal of the inoculated bacteria in the soil matrix. Genes that encode the degradation of organic compounds are often located on plasmids and, consequently, they can be spread by horizontal gene transfer into well-established, ecologically competitive, indigenous bacterial populations. Plasmid-mediated bioaugmentation aims to stimulate the spread of contaminant degradation genes among indigenous soil bacteria by the introduction of plasmids, located in donor cells, harboring such genes. But the acquisition of plasmids by recipient cells can affect the host’s fitness, a crucial aspect for the success of plasmid-mediated bioaugmentation. Besides, environmental factors (e.g., soil moisture, temperature, organic matter content) can play important roles for the transfer efficiency of catabolic plasmids, the expression of horizontally acquired genes and, finally, the contaminant degradation activity. For plasmid-mediated bioaugmentation to be reproducible, much more research is needed for a better selection of donor bacterial strains and accompanying plasmids, together with an in-depth understanding of indigenous soil bacterial populations and the environmental conditions that affect plasmid acquisition and the expression and functioning of the catabolic genes of interest.
Soils play a vital role in the provision of ecosystem services and harbor one of the most complex and diverse biological communities on Earth (Barrios, 2007). Therefore, the preservation of soil quality/soil health (both terms are often used interchangeably), defined as “the capacity of soil to perform its ecosystem processes and services, while maintaining ecosystem attributes of ecological relevance” (Garbisu et al., 2011), is currently a matter of great priority. Contamination is one of the most important causes of soil degradation. Only in Europe, there are around 2.5 million potentially contaminated sites, with an annual estimated management cost of 6 billion euros (Panagos et al., 2013). Different anthropogenic activities, such as combustion of fossil fuels, incineration, mining, agricultural practices, urbanization, waste disposal, etc. have contributed to the pressing problem of soil contamination (Besser et al., 2009). Among other negative consequences, the presence of contaminants in soil can cause a negative impact on the soil biota, resulting in an altered activity, biomass and/or diversity of soil biological communities (Burges et al., 2015).
Traditionally, a variety of physicochemical methods (e.g., excavation and disposal in landfills, soil washing, chemical oxidation, encapsulation, thermal treatments, incineration, vitrification, solidification, etc.) have been used for soil remediation. However, these physicochemical strategies are often expensive and, many times, reduce the concentration of soil contaminants at the expense of damaging the integrity of the soil ecosystem (Epelde et al., 2009; Gómez-Sagasti et al., 2016).
The main goal of any soil remediation technology must be not only to reduce the concentration of soil contaminants but to restore soil quality (Epelde et al., 2010; Barrutia et al., 2011; Pardo et al., 2014). A variety of soil physicochemical and biological properties (e.g., parameters that provide information on the biomass, activity and diversity of soil microbial communities) (Epelde et al., 2009; Muñoz-Leoz et al., 2013) are often used as indicators of soil quality. It has also been proposed to assess the effectiveness of remediation methods in terms of the recovery of soil ecosystem services and/or attributes of ecological relevance, such as organization, stability, redundancy, etc. (Garbisu et al., 2011; Epelde et al., 2014).
As an alternative to physicochemical treatments, several biological methods of soil remediation, included within the terms bioremediation and phytoremediation, are currently receiving much attention, mainly owing to their lower cost and environmentally friendly character (Juwarkar et al., 2014). Bioremediation, or the use of microorganisms (bacteria, fungi) to break down contaminants, takes advantage of the catabolic capacity of microorganisms to remove contaminants from soil. However, bioremediation is effective only with a limited range of contaminants and contaminant concentrations. In addition, bioremediation techniques might take too long to achieve the desired reduction in the concentration of soil contaminants (Kumavath and Deverapalli, 2013).
In relation to trace elements (a group of non-degradable contaminants of much concern due to their well-known toxicity), microorganisms can only transform them from one oxidation state or organic complex to another (Garbisu et al., 2002). Then, for the biological remediation of metal contaminated soils, metal-accumulating plants (i.e., accumulators and hyperaccumulators) offer many advantages over microbial processes, as these plants can literally extract the toxic metals from the contaminated site through a phytotechnology termed phytoextraction (Barrutia et al., 2009, 2010; Epelde et al., 2010).
Bioremediation has been successfully employed to remediate soils contaminated with organic contaminants, such as aliphatic hydrocarbons, polycyclic aromatic hydrocarbons, polychlorinated biphenyls, organic solvents and so on (Maphosa et al., 2012).
The bioremediation of organic contaminants can be approached by three different strategies: bioattenuation, biostimulation, and bioaugmentation (Figure 1A). Bioattenuation relies on natural processes to maintain the growth and degrading activity of native microbial populations, so that contaminants are biodegraded without human intervention, apart from the monitoring of contaminant dispersal and degradation rates. Instead, the term biostimulation refers to the adjustment of the environmental conditions (e.g., temperature, moisture, aeration, pH, redox potential) and the application of nutrients (e.g., nitrogen, phosphorus) and electron acceptors to contaminated soil, in order to enhance the growth of degrading microbial populations and, then, reduce the concentration of soil contaminants. Finally, bioaugmentation has been defined as the inoculation into contaminated soils of microorganisms with the ability to degrade the target contaminants (Maier, 2000; Heinaru et al., 2005). This inoculation can be performed with only one strain or, alternatively, with a consortium of microbial strains with diverse metabolic capacities. The advantage of using a consortium of different strains is that toxic intermediate products generated by one strain may be degraded by another strain (Heinaru et al., 2005). Apart from inoculating wild strains with the required degradation capacities, laboratory-constructed strains with upgraded catabolic abilities have also been considered for a more efficient bioaugmentation (Mrozik et al., 2011).
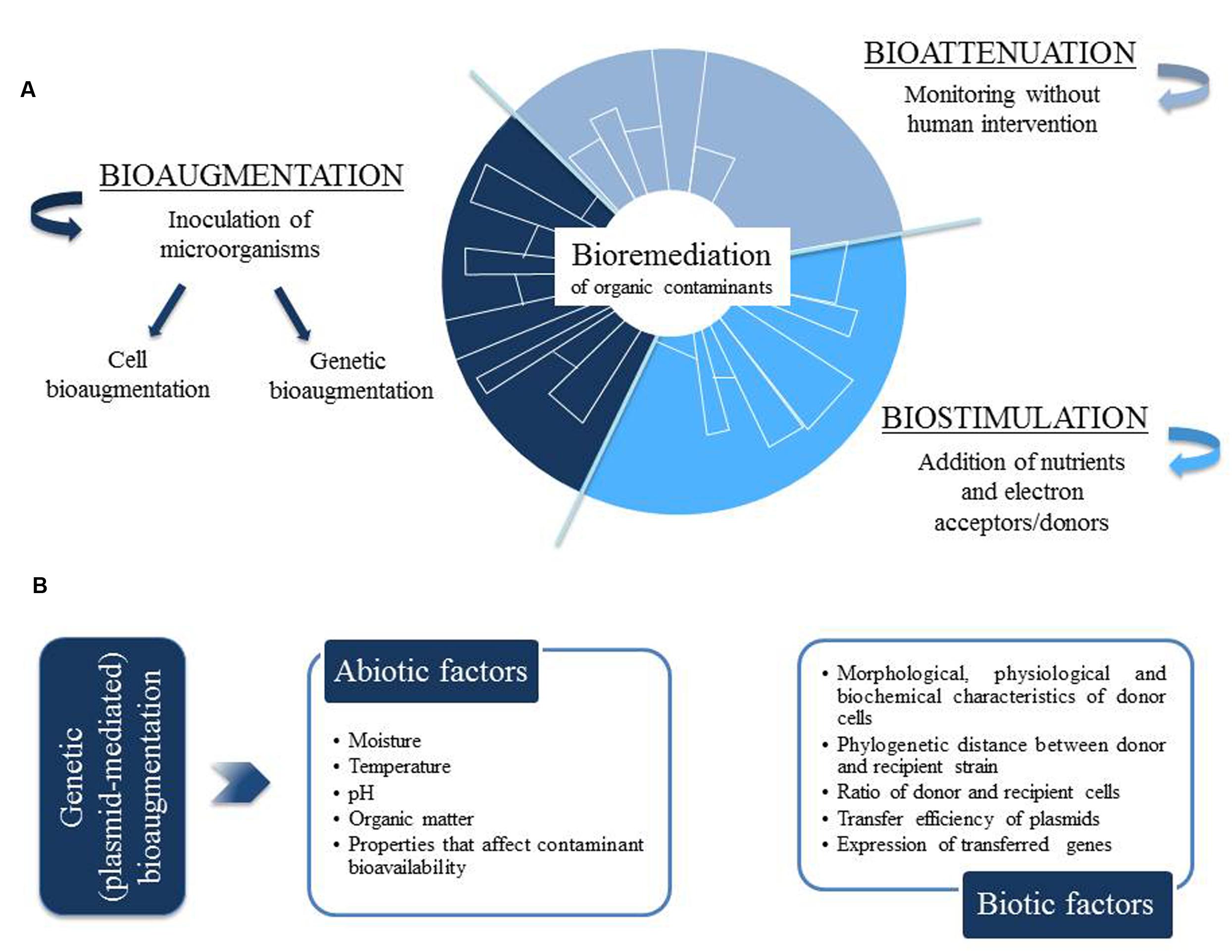
FIGURE 1. (A) Strategies for bioremediation of organic contaminants: bioattenuation, biostimulation and bioaugmentation. (B) Environmental factors affecting the efficiency of plasmid-mediated bioaugmentation.
Iwamoto and Nasu (2001) and El Fantroussi and Agathos (2005) have proposed to apply bioaugmentation in those cases where biostimulation and natural attenuation are proven ineffective. In this regard, in a diesel-contaminated soil, Bento et al. (2005) found bioaugmentation to be more effective for the degradation of the light fraction (C12–C23) of petroleum hydrocarbons than biostimulation. No significant differences were detected between biostimulation and bioaugmentation in relation to the removal of the heavy fraction (C23–C40).
Bioaugmentation can be divided into two different approaches: (i) cell bioaugmentation, which relies on the survival and growth of the inoculated strains to perform the degradation of the target contaminants, and (ii) genetic bioaugmentation, based on the spread of catabolic genes, located in mobile genetic elements (MGEs), into native microbial populations.
However, despite decades of bioremediation research, the real drivers governing the degradation of organic contaminants are still poorly understood (Meckenstock et al., 2015). In order to gain insight into this question, Meckenstock et al. (2015) revisited and challenged current concepts on the controls and limitations of biodegradation, and pointed out some critical research gaps such as, for instance, the role of protozoa and bacteriophages in shaping communities of bacterial degraders and influencing contaminant degradation rates.
Cell bioaugmentation is based on the survival and catabolic activity of inoculated microbial strains (Singh and Ward, 2004). The inoculation of bacteria harboring the necessary metabolic pathways for the degradation of the target contaminants can indeed accelerate the removal of such contaminants and, hence, reduce the time required for the intended bioremediation (Nowak and Mrozik, 2016). Inoculated microbial strains must then compete for energy and resources (e.g., nutrients and electron acceptors) with the autochthonous microbial populations already present in the soil ecosystem. The major drawbacks for the successful application of cell bioaugmentation are the (i) frequently very high mortality of the inoculated microbial strains, due to biotic or abiotic stresses, and (ii) limited dispersal of such strains throughout the soil matrix (Pepper et al., 2002; Quan et al., 2010). Many factors, including cell adhesion to soil organic matter (OM), can strongly limit the distribution of bacteria through the soil matrix. To overcome this limitation, several authors (Wang and Mulligan, 2004; Franzetti et al., 2009) have reported the use of surfactants, foams and adhesion-resistant strains.
Despite these limitations, many studies have supported the potential of cell bioaugmentation for the bioremediation of soils contaminated with organic compounds. Wang et al. (2004) reported an accelerated removal of quinoline after the inoculation of Burkholderia pickettii. Similarly, Mrozik et al. (2011) showed that cell bioaugmentation with Pseudomonas sp. JS150 significantly enhanced phenol degradation in soil, thereby reducing the possibility of formation of phenoxyl radicals (Hanscha et al., 2000). Although the number of Pseudomonas sp. JS150 cells decreased significantly during the first few days, the inoculated bacteria were then able to survive over the experimental period and successfully increased the rate of phenol degradation; actually, phenol biodegradation in soil bioaugmented with Pseudomonas sp. JS150 cells was 68 and 96 days shorter in clay and sandy soil, respectively, in comparison to non-bioaugmented soil (Mrozik et al., 2011).
Genes encoding the degradation of naturally occurring or xenobiotic organic compounds are often located on MGEs, such as plasmids, integrons and transposons. By acquiring these genes through mechanisms of horizontal gene transfer (HGT), recipient bacteria may achieve the capacity to degrade those organic contaminants (Wiedenbeck and Cohan, 2011). HGT allows the exchange of genetic information among bacteria from even distantly related taxonomic groups, thereby allowing bacteria to rapidly adapt to new environmental conditions. Although mutation events can certainly contribute to bacterial adaptation, mutation rates in bacterial populations are generally low. Besides, it is currently assumed that an increased rate of mutations would result in increased death owing to deleterious effects (Martínez et al., 2009).
Out of the three mechanisms of HGT in bacteria (i.e., transformation, transduction and conjugation), conjugation is a most efficient biological process in which genetic information encoded in plasmids is transferred, from donor to recipient bacteria, by direct cell-to-cell contact (Furuya and Lowy, 2006). Bacterial conjugation is known to accelerate the dissemination of resistance to, for instance, antibiotics and heavy metals, as well as to facilitate the distribution of genes involved in the degradation of organic compounds. Nevertheless, the contribution of conjugation to HGT among soil bacteria and the factors involved in the transfer and proliferation of plasmid-containing bacteria in the soil ecosystem are yet not fully understood.
Bacterial adaptation through evolutionary time has been shaped, among other aspects, by the high plasticity of bacterial genomes, which allows bacteria to rearrange and exchange genomic sequences, thus opening the possibility to acquire beneficial traits (Sørensen et al., 2005). As a matter of fact, the loss, rearrangement and acquisition of functional genetic modules can have a vast impact on the extent and speed of the evolutionary adaptation of bacteria (Wozniak and Waldor, 2010; Bertels and Rainey, 2011). MGEs are, to a great extent, responsible for these processes of gene mobility and reorganization, both within genomes (intracellular) and between bacterial cells (intercellular).
Many of the studies on lateral dissemination of genetic material among bacteria have focused on antibiotic and metal resistance. Research on the horizontal transfer of genes associated with the degradation of organic compounds in natural environments, such as the soil ecosystem, is still insufficient to fully understand the mechanisms involved in such process (Christensen et al., 1998; Top et al., 1998; Dejonghe et al., 2000; Aspray et al., 2005; Overhage et al., 2005; Musovic et al., 2010). In any case, some plasmids, such as those implicated in the catabolic pathway of 2,4-dichlorophenoxyacetic acid (2,4-D), have been thoroughly studied (Top et al., 1998; Dejonghe et al., 2000; Newby and Pepper, 2002).
Plasmid transfer between soil bacteria has been contemplated as a promising strategy for the dissemination of catabolic functions within soil bacterial communities (Venkata Mohan et al., 2009; Mrozik and Piotrowska-Seget, 2010). As abovementioned, plasmid-encoded metabolic pathways can be transferred among bacteria, thus playing a critical role in the adaptation of bacteria to different environmental conditions (Reineke, 1998; Sayler and Ripp, 2000). Specifically, HGT has been reported to promote bacterial adaptation to the presence of organic contaminants (Top and Springael, 2003).
The underlying idea behind genetic (plasmid-mediated) bioaugmentation is to stimulate the rate of contaminant degradation by increasing, through HGT, the number and diversity of native bacteria with the capacity to metabolize the target contaminants. In this respect, it must be emphasized that numerous catabolic pathways involved in the degradation of organic contaminants have been identified in MGEs (Top et al., 2002; Jussila et al., 2007).
Genetic (plasmid-mediated) bioaugmentation is defined as a technology in which donor bacteria harboring self-transmissible catabolic plasmids are introduced into the soil matrix in order to enhance, by HGT, the potential and rate of contaminant degradation of existing bacterial populations (Top et al., 2002; Ikuma and Gunsch, 2010, 2012). Compared to cell bioaugmentation, plasmid-mediated bioaugmentation appears a priori a more effective strategy for the bioremediation of organic contaminants, as the bacteria that will eventually degrade the contaminants (i.e., bacteria with the recently acquired plasmids harboring the necessary catabolic genes) are expected to be adapted to live in the soil under remediation. In this manner, one of the main drawbacks for the successful application of cell bioaugmentation, i.e., the low survival of the inoculated microbial strains, appears to be overcome.
For plasmid-mediated bioaugmentation, both an appropriate selection of donor bacteria with the required plasmid and a profound knowledge of native soil bacterial populations are required to increase the probability of an efficient plasmid acquisition and the expression of the catabolic genes of interest.
Many studies on plasmid-mediated bioaugmentation have been published (Table 1). In a microcosm study, Halden et al. (1999) detected an enhanced degradation of 3-phenoxybenzoic acid (3-POB) as a result of the transfer of plasmids pPOB and pD30.9 from Pseudomonas pseudoalcaligenes POB310 (pPOB) and Pseudomonas sp. B13-D5 (pD30.9) and B13ST1 (pPOB) to recipient soil bacteria. Using P. putida as donor strain of two catabolic plasmids (pEMT1 and pJP4), Dejonghe et al. (2000) reported the degradation of 2,4-D in soil under microcosm conditions. These authors investigated the bioaugmentation potential of plasmids pEMT1 and pJP4 in two soil layers (0–30 and 30–60 cm soil depth) differing in physicochemical properties and microbial community structure, finding out a more efficient degradation of 2,4-D in the deeper soil layer where the indigenous microbial communities lacked the ability to catabolize 2,4-D. Under microcosm conditions, Inoue et al. (2012) studied the effect of bioaugmentation with P. putida and Escherichia coli cells, harboring the self-transmissible 2,4-D degradative plasmid pJP4, on the degradation of 2,4-D. These authors found that the number of P. putida and E. coli cells decreased rapidly after their inoculation in a 2,4-D contaminated soil slurry, but the degradation of this contaminant was nevertheless stimulated, most likely due to the occurrence of transconjugants resulting from the transfer of plasmid pJP4. Inoue et al. (2012) concluded that genetic bioaugmentation with P. putida and E. coli cells harboring plasmid pJP4 can stimulate the degradation of 2,4-D in soil without a substantial impact on the soil microbial community, as reflected by the values of parameters which provide information on carbon source utilization (through the use of the well-known BiologTM plates) and nitrogen transformations (nitrate reduction assay, quantification of amoA gene of ammonia-oxidizing bacteria, quantification of nirK and nirS genes of denitrifying bacteria). In sequencing batch reactors, Tsutsui et al. (2013) achieved a complete degradation of 2,4-D by plasmid (pJP4)-mediated bioaugmentation with Cupriavidus necator JMP134 and E. coli HB101 as donor strains. These authors were able to identify the emergence of 2,4-D-degrading transconjugants associated to Achromobacter, Burkholderia, Cupriavidus and Pandoraea.
Pepper et al. (2002) conducted microcosm experiments to enhance the degradation of 3-chlorobenzoate (3-CB) using plasmid pBRC60, which harbors genes for 3-CB mineralization, and Comamonas testosteroni as donor strain. Although they did observe degradation of 3-CB, they could not detect any transfer event of plasmid pBRC60 from C. testosteroni to native soil bacteria.
Miyazaki et al. (2006) isolated a plasmid (pLB1) involved in the dissemination of genes for γ-hexachlorocyclohexane (lindane) degradation. This plasmid, carrying the linB gene, was isolated from Sphingobium japonicum UT26DB and then successfully transferred, under laboratory conditions, from this strain to other α-proteobacterial strains but not to any of the β- or γ-proteobacterial strains tested.
In their study on the transfer of TOL plasmid (also designated pWW0) during bacterial conjugation in vitro and rhizoremediation of oil-contaminated soil in vivo, Jussila et al. (2007) demonstrated the successful transfer of TOL plasmid for toluene degradation from P. putida PaW85 to P. oryzihabitans 29. In rhizosphere microcosms, Mølbak et al. (2007) found that the transfer of plasmid pWW0 from P. putida resulted in transconjugants belonging to Enterobacteria and Pseudomonas. This well-characterized self-transmissible catabolic plasmid, pWW0, was also used by Ikuma and Gunsch (2012) to assess its potential for bioaugmentation in toluene-contaminated soil slurry.
Under laboratory conditions, horizontal transfer of plasmid pGKT2 was successfully carried out by Jung et al. (2011) from Gordonia sp. KTR9 to Gordonia polyisoprenivorans, Rhodococcus jostii RHA1 and Nocardia sp. TW2 strains. These transconjugants showed the ability to use hexahydro-1,3,5-trinitro-1,3,5,-triazine (RDX) as a nitrogen source.
In a contaminated field site located in Cixi, Zhejiang (China), Gao et al. (2015) achieved effective plasmid-mediated bioaugmentation for the degradation of dichlorodiphenyltrichloroethane (DDT) in soil with E. coli TG I (pDOD-gfp) as donor strain. In this study, the catabolic plasmid pDOD from Sphingobacterium sp. D-6 was conjugally transferred to soil bacteria, such as members of Cellulomonas, and accelerated DDT degradation. Different studies have reported the use of the GFP (green fluorescence protein) detection system to monitor plasmid transfer from donor cells to indigenous soil bacteria in soil slurries (Ikuma et al., 2012) and field contaminated soil (Gao et al., 2015).
Filonov et al. (2010) determined transfer frequencies in open soil after inoculation with genetically tagged plasmid-containing naphthalene-degrading P. putida KT2442 and auxotrophic donor BS394 (pNF142::TnMod-OTc) cells, finding out that plasmid pNF142 was transferred to native soil bacteria (mainly to fluorescent pseudomonads) at a frequency of 4 × 10-6/donor cell. After bioaugmentation with E. coli JM109 (pDOC-gfp) strain, Zhang et al. (2012) observed that pDOC plasmid was transferred to native soil bacteria under microcosm conditions, including members of Pseudomonas and Staphylococcus which acquired the capacity to degrade chlorpyrifos (a widely used insecticide). As it is usually the case, the efficiency of this transfer, as measured by the chlorpyrifos degradation efficiency and the number of chlorpyrifos degraders, was influenced by soil type, temperature and moisture content (Zhang et al., 2012).
Finally, attention has also been paid to the use of genetically modified organisms (GMOs) for bioaugmentation. Nonetheless, the deliberate release of GMOs into the environment is subjected to regulatory constraints (Garbisu and Alkorta, 1997; Sayler and Ripp, 2000; Directive 2001/18/EC, 2001). The transfer of catabolic genes between GMOs and wild bacterial strains might facilitate the acquisition and spread of new degradative pathways among indigenous bacterial communities. To this purpose, Massa et al. (2009) engineered the recombinant strain P. putida PaW340/pDH5, constructed by cloning dehalogenase genes from Arthrobacter sp. FG1 in P. putida PaW340, for the degradation of 4-chlorobenzoic acid (CBA) in soil slurry. After inoculation of this recombinant strain into soil slurry, a higher degradation of CBA was observed, compared to the slurry inoculated with pre-adapted cultures of Arthrobacter sp. FG1.
The success of plasmid-mediated bioaugmentation for the bioremediation of contaminated soil relies not only on an efficient transfer of the required plasmid from donor bacteria to soil recipient bacteria, but also on the ability of recipient cells to properly express the plasmid-harbored catabolic genes, so that the desired phenotypic changes (i.e., biodegradation of the target contaminant) can be attained. After plasmid acquisition, the capacity of recipient cells to successfully perform the desired catabolic function depends, among other factors, on their newly acquired competitive abilities and on the alteration of the host’s own competitive abilities (van Rensburg et al., 2012). A thorough understanding of how plasmid acquisition can affect host fitness is fundamental to then achieve the persistence of the introduced plasmid in the recipient cells.
Plasmid acquisition can provide recipient bacteria with a large array of beneficial traits, such as catabolic potential, resistance to antibiotics and/or metals, faster growth, ability to use a wider range of compounds as energy sources, etc. (Top et al., 1998; Riley and Wertz, 2002). In many cases, plasmid-harboring hosts have been found to be competitively fitter than their plasmid-free counterparts (Dionisio et al., 2005; Starikova et al., 2013). Nonetheless, horizontally acquired genes can also function inefficiently in the genomic background of recipient cells (Chou et al., 2011; Park and Zhang, 2012). After all, horizontally acquired genes find themselves immersed in a new metabolic context and their function relies on the host’s machinery. Genetic determinants often encounter the required metabolic “partners” (e.g., substrates, proteins) in the recipient cells, so that the intended changes in the host’s metabolism become possible. Conversely, other times, the required metabolic partners for the proper functioning and regulation of newly acquired genes are missing in the recipient cells. Indeed, the acquisition of plasmids can negatively affect cellular networks in recipient cells and, concomitantly, trigger fitness costs as collateral damage (Bouma and Lenski, 1988; Martínez et al., 2009). Fitness (metabolic) costs derived from plasmid acquisition can be highly variable (De Gelder et al., 2007), as they can originate from a variety of factors, including: (i) energetic costs due to consumption of molecular building blocks and/or energy sources derived from the activity of horizontally acquired regions; (ii) chromosomal disruption by horizontally acquired genes, when such genes are incorporated into the chromosome; (iii) sequestration of cellular processes and associated molecular machinery (e.g., ribosomes) by the horizontally acquired regions; and (iv) plasmid size, since small plasmids can carry only a single accessory determinant but large plasmids can carry more than 10 accessory determinants as well as other genes (Shachrai et al., 2010; Baltrus, 2013; Vogwill and MacLean, 2015).
Fitness costs associated to plasmid acquisition can be offset by benefits derived from the fact that plasmids are ideal biological tools to create genetic variation within bacterial populations. A major benefit from maintaining transferable plasmids derives from the fact that, in this manner, bacterial populations can gain stability against potential environmental changes.
Bacteria with acquired genes can, on the other hand, alleviate fitness costs through compensatory evolution (San Millan et al., 2014). Thus, for instance, bacteria can minimize plasmid-related fitness costs by integrating only the desired plasmid-acquired determinants in the chromosome.
Conjugative plasmids (i) are usually large (they encode genes for the conjugation process itself and for stabilization within the host); (ii) are normally found in low copy number; (iii) appear well maintained over successive generations (Norman et al., 2009; Jung et al., 2011); and (iv) act as fundamental vehicles of HGT (Frost et al., 2005; Thomas and Nielsen, 2005).
In their laboratory study on the capacity of Gordonia sp. KTR9 to transfer plasmid pGKT2 and the associated RDX (hexahydro-1,3,5-trinitro-1,3,5,-triazine) degradation ability to other bacteria, Jung et al. (2011) investigated plasmid stability after HGT from Gordonia sp. KTR9 to G. polyisoprenivorans, R. jostii RHA1 and Nocardia sp. TW2, finding out a marked decrease in plasmid retention after 50 generations with Nocardia sp. TW2, while G. polyisoprenivorans and R. jostii RHA1 transconjugants exhibited retention of pGKT2 plasmid for 100 generations. It was speculated that this decreased stability in Nocardia sp. TW2 might have been caused by a larger metabolic expense incurred by the incorporation of pGKT2 in this strain, compared to the other two bacterial strains (Jung et al., 2011).
Given that positive selection cannot explain the long-term stability of costly plasmids (Hall et al., 2017), the explanation for such long-term stability remains a most challenging task, since segregational loss and the cost of plasmid carriage should drive the loss of plasmids through purifying selection (Hall et al., 2017). In this respect, two evolutionary routes to plasmid stability appear possible (Hall et al., 2017): (i) the evolution of high conjugation rates would allow plasmids to survive as infectious agents through horizontal transmission (Hall et al., 2016; Kottara et al., 2016); and (ii) compensatory evolution to ameliorate the cost of plasmid carriage can weaken purifying selection against the plasmid backbone (Harrison et al., 2015; Porse et al., 2016).
Finally, it must be taken into consideration that plasmids can be classified into incompatibility groups (incompatibility defined as the inability of plasmids sharing similar replication and partition systems to be propagated stably in the same host cell line; in other words, members of each group cannot co-reside within the same bacterial host), such as IncP, IncN, IncW, and IncF. Incompatibility groups have been independently classified in three different genera: there are 27 Inc groups in Enterobacteriaceae, 14 Inc groups in Pseudomonas, and approximately 18 Inc groups in Staphylococcus (Shintani et al., 2015). Plasmids classified in E. coli as IncP and in Pseudomonas as IncP-1 are a well-studied group of plasmids that can carry a variety of phenotypic markers, including antibiotic resistance, metal resistance and the ability to degrade xenobiotics. It has been reported (Popowska and Krawczyk-Balska, 2013) that a detailed analysis of IncP-1 plasmid genomes could provide useful information for the development of effective methods of soil bioremediation. After all, the evolutionary adaptation of microorganisms to the presence and utilization of organic contaminants is often due to plasmids (mainly, from IncP group) that carry genes encoding enzymes involved in the degradation of those compounds. For instance, plasmids IncP-1, IncP-7 and IncP-9 contain genes encoding enzymes required for the degradation of naphthalene, toluene, chlorobenzene, p-toluenesulfonate, 2,4-D, haloacetate and atrazine (Shintani et al., 2010a,b; Popowska and Krawczyk-Balska, 2013). Relevantly, there seems to be a distinction between (i) plasmids that harbor genes for the degradation of naturally occurring compounds and (ii) plasmids that harbor genes for the degradation of xenobiotics (Top et al., 2002): degradation of naturally occurring compounds is often encoded in IncP-2 and IncP-9 plasmids, while the degradation of xenobiotics seems to be encoded by the well-known broad host range IncP-1 plasmids. IncP-1 plasmids are very promiscuous, and this promiscuity appears to play a crucial role in the evolution of new metabolic pathways by recruiting catabolic genes or gene segments from different organisms into a suitable host (Wyndham et al., 1994; Beil et al., 1999).
Therefore, different plasmids potentially useful for plasmid-mediated bioaugmentation (with, for instance, each plasmid harboring a gene encoding a different enzyme involved in the degradation route of a specific contaminant) cannot co-reside within the same host if they belong to the same incompatibility group. Then, if we want to apply different plasmids from the same incompatibility group, each of them harboring a gene for a specific step in the contaminant degradation pathway, they must be applied in different donor cells and, for an effective biodegradation, each plasmid should be transferred to a different recipient cell, decreasing considerably the probability of successful plasmid-mediated bioaugmentation.
The success of both cell and plasmid-mediated bioaugmentation greatly depends on the environmental (abiotic and biotic) conditions present in the soil to be remediated (Cho et al., 2000; Bento et al., 2005; Wolski et al., 2006). In fact, during plasmid-mediated bioaugmentation, environmental factors can play important roles in the (i) transfer efficiency of catabolic plasmids, (ii) expression of horizontally acquired genes and, finally, (iii) contaminant degradation activity (Popa et al., 2011; Ikuma and Gunsch, 2012). In particular, several abiotic factors such as soil moisture, temperature and OM content are known to affect bioaugmentation efficiency (Figure 1B).
Soil moisture can have an effect on plasmid transfer during plasmid-mediated bioaugmentation by affecting the contact between donor and recipient bacteria (Miller et al., 2004; Aminov, 2011). In this respect, Gao et al. (2015) evaluated the effectiveness of plasmid-mediated bioaugmentation for p,p′-DDT degradation at three different soil moisture conditions (40, 60, and 80%), and concluded that 60% moisture content was optimal for maximum plasmid transfer efficiency.
Temperature has been shown to affect plasmid transfer efficiency (Inoue et al., 2005; Zhang et al., 2012). For the enhancement of DDT degradation by plasmid-mediated bioaugmentation with plasmid pDOD, the optimal temperature interval for cell growth and activity of both donor and recipient soil bacteria was established at 25–30°C (Gao et al., 2015). Johnsen and Kroer (2007) found that increasing temperatures resulted in an increase in the transfer of plasmid pRO103 encoding resistance to mercury and tetracycline and partial degradation of 2,4-D.
Regarding soil OM content, in a bioaugmentation laboratory experiment, Greer and Shelton (1992) observed higher rates of mineralization of 2,4-D in soil with a low OM content, compared to soil with a high content of OM. Under laboratory conditions, Kim et al. (2008) found that P. spadix BD-a59 cells were able to degrade BTEX at a slower rate in soil with low OM content than in organic-rich soil. When studying the biodegradation of polychlorinated biphenyls (PCB) in soil under laboratory conditions, Haluška et al. (1995) observed that humic acids affected the survival and activity of the inoculated Alcaligenes xylosoxidans strain, which exhibited maximum survival rates in soil with an intermediate amount of organic carbon and the highest amount of aromatic carbon in humic acids. Highest levels of PCB degradation were found in soil with the highest content of organic carbon and an intermediate amount of aromatic carbon in humic acids (Haluška et al., 1995).
Wang et al. (2014) performed plasmid transfer experiments between soil bacteria, using a TOL-like plasmid carrying the gene encoding for catechol 2,3-dioxygenase, to study some factors (soil depth, soil type, etc.) that could affect the transfer of plasmids, finding out that these factors certainly have a considerable effect on the transfer of the TOL-like plasmid in soil. Concerning soil depth, under microcosm conditions, Wang et al. (2014) found, in general, lower frequencies of plasmid transfer at greater soil depths, a fact most likely due to the often-found gradual decrease in bacterial biomass and activity at increasing soil depths, possibly related to concomitantly decreased oxygen concentrations (Król et al., 2011). Król et al. (2011) reported that oxygen concentration can affect plasmid transfer through an oxygen-related mechanism or indirectly via its impact on cell physiology. When studying the influence of soil type (loamy sand, sandy loam, sandy clay loam, loam) on plasmid transfer, Wang et al. (2014) observed a highest frequency of plasmid transfer in loam soil, probably related to the fact that loam often contains more nutrients and humus than other soil types, and higher values of microbial biomass and metabolic activity (Djokic et al., 2013).
In the same way, the chemical nature, concentration and bioavailability of the contaminants are crucial factors influencing bioaugmentation efficiency (Davis and Madsen, 1996; Stalwood et al., 2005). Sejáková et al. (2009) reported a relationship between pentachlorophenol (PCP) concentration in soil and the number of CFU of the C. testosteroni CCM7530 strain used for bioaugmentation: at a PCP concentration of 100 mg kg-1, the number of C. testosteroni CCM7530 CFUs rapidly increased over 17 days, while, at 10 mg PCP kg-1, the number of CFUs initially decreased until day 7 to then increase until day 17.
In any case, the level of selective pressure required to promote conjugal plasmid transfer depends on the specific contaminant and its concentration, as well as on the specific catabolic plasmid. In soil slurry, Ikuma and Gunsch (2012) observed that environmentally relevant concentrations of toluene might not exert enough pressure for transfer of plasmid TOL from P. putida BBC443 to Serratia marcescens and P. fluorescens cells. In their study on the degradation of 2,4-D, DiGiovanni et al. (1996) observed that this contaminant originated the required selective pressure for conjugal transfer of the intended catabolic plasmids.
Many biotic factors can also affect the success of plasmid-mediated bioaugmentation. Some genetic differences, such as guanine-cytosine (G+C) content and phylogenetic relationship between donor and recipient strain, can negatively affect the expression of the catabolic phenotype following conjugal plasmid transfer, as described by Ikuma and Gunsch (2012). Indeed, for plasmid-mediated bioaugmentation, biological differences between donor and recipient bacterial strains such as, for example, phylogenetic distance (Popa et al., 2011) and plasmid host range (De Gelder et al., 2005; Sorek et al., 2007), can play an important role. In 2,4-D contaminated soils, Newby et al. (2000) studied the bioaugmentation efficiency of two plasmid pJP4-bearing bacteria (the natural host, Ralstonia eutropha JMP134, and a laboratory-generated E. coli strain amenable to donor counterselection, named E. coli D11) and concluded that the correct choice of donor strain is a factor of the utmost importance for bioaugmentation.
Ikuma and Gunsch (2012) indicated that the success of plasmid-mediated bioaugmentation is dependent on: (i) high transfer rates of the catabolic plasmid to as many indigenous bacteria as possible; and (ii) the high expression level of an active contaminant-degrading phenotype in all transconjugants following conjugal plasmid transfer. Then, prior to the bioaugmentation process itself, it is important to characterize potentially recipient soil bacterial communities, paying special attention to dominant taxonomic groups. In the last years, next generation sequencing has provided a more comprehensive analysis of indigenous soil bacterial communities (Walsh, 2000), opening the door to the identification of potential recipient bacterial populations, and therefore a more informed selection of both the donor strain and the plasmid type (Ikuma and Gunsch, 2012).
Other biotic factors, such as competition between inoculated and indigenous bacteria for carbon sources, antagonistic interactions and predation by protozoa and bacteriophages, etc. also play an essential role in bioaugmentation efficiency. The critical factor is the selection of the right bacterial strains (Thompson et al., 2005), since the inoculated strain must be able not only to degrade the target contaminant (or, in the case of plasmid-mediated bioaugmentation, to be able to effectively transfer the catabolic plasmid), but also to successfully compete with indigenous microbial populations and, in general, soil biota. On the other hand, plasmid transfer frequency has been shown to depend on the initial cell density ratio between donor and recipient cells (Pinedo and Smets, 2005; Ikuma et al., 2012).
Morphological, physiological and biochemical characteristics such as, for instance, cell size, growth rate, resource utilization ability, resistance phenotypes, biofilm formation capacity, cell motility, etc. are key traits for bacterial survival and competitiveness. Furthermore, DNA content has a marked influence on bacterial ecophysiological traits (i.e., adaptive traits to environmental changes) affecting, among other aspects, the rate of cell growth (Wickham and Lynn, 1990). Nevertheless, despite the assumption that fitness costs associated to HGT are caused by the need to maintain and replicate the extra-DNA, some studies indicate that they are predominantly due to transcription and translation processes (Bragg and Wagner, 2009; Shachrai et al., 2010).
The capacity of the host to use different carbon substrates before and after plasmid acquisition can provide an estimation of (i) its competitive ability and (ii) changes specifically associated to the plasmid transfer itself. BiologTM plates can be employed to obtain a phenotypic fingerprint of bacterial strains in relation to their capacity to use a variety of carbon sources. Karve et al. (2016) followed phenotypic variations, using Biolog GEN III MicroPlatesTM, to assess functionally relevant consequences of DNA changes.
Antibiotic resistance is probably the most extensively studied bacterial competitive trait. As a consequence of the production of antibiotics by soil microbial populations (D’Costa et al., 2006), soil is thought to be the largest reservoir of antibiotic resistance genes. Owing to fitness costs associated to antibiotic resistance, when bacteria change to an antibiotic-free environment, resistance is expected to disappear (Morosini et al., 2000), according to the assumption that, in the absence of selective pressure, resistant bacteria with a lower fitness will be outcompeted by susceptible counterparts with a higher fitness. However, it seems that bacteria tend to keep the mechanisms of antibiotic resistance, in order to maintain such an advantageous trait in the face of a possible change in environmental conditions (San Millan et al., 2014). Besides, in nature, antibiotics and antibiotic resistance determinants might play a variety of roles (e.g., signaling molecules in quorum sensing and biofilm formation, production of virulence factors, host-parasite interactions) (Sengupta et al., 2013) that justify the preservation of antibiotic resistance determinants in the absence of the selective pressure.
Biofilms are known to protect bacterial cells against antimicrobials (Høiby et al., 2010), predation, oxidative stress (Geier et al., 2008), etc. Biofilms harbor spatially structured bacterial communities where plasmids can be more easily shared through HGT (Jefferson, 2004), facilitating, for instance, the dissemination of catabolic genes. Remarkably, attachment to surfaces by biofilm-associated factors is another cellular function associated to genes present in plasmids (Norman et al., 2009).
Cell motility is a critical aspect for the necessary dispersal of inoculated bacteria toward the target contaminants. Nevertheless, although highly motile bacterial cells, in their search for energy and nutrients, can disperse more easily into the surrounding environment, they also have a higher probability of encountering potential competitors (Reichenbach et al., 2007). In any case, motile bacterial populations, such as swarming bacteria, can more rapidly colonize new niches, with the associated ecological benefits (Verstraeten et al., 2008). Interestingly, there is a complex link between motility and biofilm formation because both processes appear to involve similar components at certain stages and conditions (Verstraeten et al., 2008).
(Gardin and Pauss, 2001; Gentili et al., 2006) have used different strategies of cell encapsulation and immobilization to facilitate inoculation survival, by providing a protective niche and temporary nutrition for the inoculated bacteria. Carrier materials, such as charcoal (Beck, 1991), nylon (Heitkamp and Steward, 1996), chitin, chitosan (Gentili et al., 2006; Chen et al., 2007) and zeolite (Liang et al., 2009) have been used in an attempt to maintain inoculant activity over a sufficiently long period of time after strain inoculation.
It must be taken into consideration that the influence of all these abovementioned abiotic and biotic factors has only been studied in a very limited number of bacterial strains and, in many cases, under controlled simplified environmental conditions, very different from those encountered in the natural environment. Therefore, many more in-depth studies on the impact of abiotic and biotic factors on cell and plasmid-mediated bioaugmentation are needed.
Both cell bioaugmentation and genetic (plasmid-mediated) bioaugmentation have proven effective for the bioremediation of soils contaminated with organic compounds. However, cell bioaugmentation has an important limitation, i.e., the frequently very high mortality of the inoculated microbial strains, due to biotic or abiotic stresses. Then, a priori, plasmid-mediated bioaugmentation appears to have greater potential than cell bioaugmentation, since plasmids can act as gene-messenger biological tools that can transfer the required catabolic genes to indigenous bacterial populations already adapted to the soil under remediation. But for plasmid-mediated bioaugmentation to be successful and reproducible, much more research is needed for a better selection of donor bacterial strains and accompanying plasmids, together with an in-depth understanding of indigenous soil bacterial populations and the environmental conditions that affect plasmid acquisition and the expression and functioning of the catabolic genes of interest. Similarly, further research is required to better understand and then improve the ecological fitness of recipient bacterial strains in the contaminated soil.
CG and IA: Design of the work and the acquisition of the data, writing and revision of the content, approval of the last version and ensuring accuracy and integrity of the work. LE and OG: Acquisition of the data, writing and revision of the content, approval of the last version of the work. EG: Writing and revision of the content, approval of the last version and ensuring accuracy and integrity of the work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work has been supported by the Spanish Ministry of Economy, Industry and Competitiveness (AGL2016-76592-R), and the Interreg SUDOE Programme (PhytoSUDOE-SOE1/P5/EO189). OG was a pre-doctoral student supported by the Basque Government and by the Fundación Biofísica Bizkaia.
Aminov, R. I. (2011). Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2:158. doi: 10.3389/fmicb.2011.0015
Aspray, T. J., Hansen, S. K., and Burns, R. G. (2005). A soil-based microbial biofilm exposed to 2,4-D: bacterial community development and establishment of conjugative plasmid pJP4. FEMS Microbiol. Ecol. 54, 317–327. doi: 10.1016/j.femsec.2005.04.007
Baltrus, D. A. (2013). Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 28, 489–495. doi: 10.1016/j.tree.2013.04.002
Barrios, E. (2007). Soil biota, ecosystem services and land productivity. Ecol. Econ. 64, 269–285. doi: 10.1016/j.ecolecon.2007.03.004
Barrutia, O., Epelde, L., García-Plazaola, J. I., Garbisu, C., and Becerril, J. M. (2009). Phytoextraction potential of two Rumex acetosa L. accessions collected from metalliferous and non-metalliferous sites: effect of fertilization. Chemosphere 74, 259–264. doi: 10.1016/j.chemosphere.2008.09.036
Barrutia, O., Garbisu, C., Epelde, L., Sampedro, M. C., Goicolea, M. A., and Becerril, J. M. (2011). Plant tolerance to diesel minimizes its impact on soil microbial characteristics during rhizoremediation of diesel-contaminated soils. Sci. Total Environ. 409, 4087–4093. doi: 10.1016/j.scitotenv.2011.06.025
Barrutia, O., Garbisu, C., Hernández-Allica, J., García-Plazaola, J. I., and Becerril, J. M. (2010). Differences in EDTA-assisted metal phytoextraction between metallicolous and non-metallicolous accessions of Rumex acetosa L. Environ. Pollut. 158, 1710–1715. doi: 10.1016/j.envpol.2009.11.027
Beck, D. P. (1991). Suitability of charcoal-amended mineral soil as carrier for Rhizobium inoculants. Soil Biol. Biochem. 23, 41–44. doi: 10.1016/0038-0717(91)90160-L
Beil, S., Timmis, K. N., and Pieper, D. H. (1999). Genetic and biochemical analyses of the tec operon suggest a route for evolution of chlorobenzene degradation genes. J. Bacteriol. 181, 341–346.
Bento, F. M., Camargo, F. A. O., Okeke, B. C., and Frankenberger, W. T. (2005). Comparative bioremediation of soils contaminated with diesel oil by natural attenuation, biostimulation and bioaugmentation. Bioresour. Technol. 96, 1049–1055. doi: 10.1016/j.biortech.2004.09.008
Bertels, F., and Rainey, P. B. (2011). Within-genome evolution of REPINs: a new family of miniature mobile DNA in bacteria. PLOS Genet. 7:e1002132. doi: 10.1371/journal.pgen.1002132
Besser, J. M., Brumbaugh, W. G., Allert, A. L., Poulton, B. C., and Schmitt, C. J. (2009). Ecological impacts of lead mining on Ozark streams-toxicity of sediment and pore water. Ecotoxicol. Environ. Saf. 72, 516–526. doi: 10.1016/j.ecoenv.2008.05.013
Bouma, J. E., and Lenski, R. E. (1988). Evolution of a bacteria/plasmid association. Nature 335, 351–352. doi: 10.1038/335351a0
Bragg, J. G., and Wagner, A. (2009). Protein material costs: single atoms can make an evolutionary difference. Trends Genet. 25, 5–8. doi: 10.1016/j.tig.2008.10.007
Burges, A., Epelde, L., and Garbisu, C. (2015). Impact of repeated single-metal and multi-metal pollution events on soil quality. Chemosphere 120, 8–15. doi: 10.1016/j.chemosphere.2014.05.037
Chen, Y. M., Lin, T. F., Huang, C., Lin, J. C., and Hsieh, F. M. (2007). Degradation of phenol and TCE using suspended and chitosan-bead immobilized Pseudomonas putida. J. Hazard Mater. 148, 660–670. doi: 10.1016/j.jhazmat.2007.03.030
Cho, Y. G., Rhee, S. K., and Lee, S. T. (2000). Effect of soil moisture on bioremediation of chlorophenol-contaminated soil. Biotechnol. Lett. 22, 915–919. doi: 10.1023/A:1005612232079
Chou, H. H., Chiu, H. C., Delaney, N. F., Segrè, D., and Marx, C. J. (2011). Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332, 1190–1192. doi: 10.1126/science.1203799
Christensen, B. B., Sternberg, C., Andersen, J. B., Eberl, L., Moller, S., Givskov, M., et al. (1998). Establishment of new genetic traits in a microbial biofilm community. Appl. Environ. Microbiol. 64, 2247–2255.
Davis, J. W., and Madsen, S. (1996). Factors affecting the biodegradation of toluene in soil. Chemosphere 33, 107–130. doi: 10.1016/0045-6535(96)00152-X
D’Costa, V. M., McGrann, K. M., Hughes, D. W., and Wright, G. D. (2006). Sampling the antibiotic resistome. Science 311, 374–377. doi: 10.1126/science.1120800
De Gelder, L., Ponciano, J. M., Joyce, P., and Top, E. M. (2007). Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology 153, 452–463. doi: 10.1099/mic.0.2006/001784-0
De Gelder, L., Vandecasteele, F. P. J., Brown, C. J., Forney, L. J., and Top, E. M. (2005). Plasmid donor affects host range of promiscuous IncP-1β plasmid pB10 in an activated-sludge microbial community. Appl. Environ. Microbiol. 71, 5309–5317. doi: 10.1128/AEM.71.9.5309-5317.2005
Dejonghe, W., Goris, J., El Fantroussi, S., Hofte, M., De Vos, P., Verstraete, W., et al. (2000). Effect of dissemination of 2,4- dichlorophenoxyacetic acid (2,4-D) degradation plasmids on 2,4-D degradation and on bacterial community structure in two different soil horizons. Appl. Environ. Microbiol. 66, 3297–3304. doi: 10.1128/AEM.66.8.3297-3304.2000
DiGiovanni, G. D., Neilson, J. W., Pepper, I. L., and Sinclair, N. A. (1996). Gene transfer of Alcaligenes eutrophus JMP134 plasmid pJP4 to indigenous soil recipients. Appl. Environ. Microbiol. 62, 2521–2526.
Dionisio, F., Conceição, I. C., Marques, A. C., Fernandes, L., and Gordo, I. (2005). The evolution of a conjugative plasmid and its ability to increase bacterial fitness. Biol. Lett. 1, 250–252. doi: 10.1098/rsbl.2004.0275
Directive 2001/18/EC (2001). Directive/18/EC of the European Parliament and of the Council of 12 March 2001 on the Deliberate Release into the Environment of Genetically Modified Organisms and Repealing Council Directive 90/220/EEC. Brussels: European Commission.
Djokic, L., Narancic, T., Biocanin, M., Saljnikov, E., Casey, E., Vasiljevic, B., et al. (2013). Phenol removal from four different natural soil types by Bacillus sp. PS11. Appl. Soil Ecol. 70, 1–8. doi: 10.1016/j.apsoil.2013.04.002
El Fantroussi, S., and Agathos, S. N. (2005). Is bioaugmentation a feasible strategy for pollutant removal and site remediation? Curr. Opin. Microbiol. 8, 268–275. doi: 10.1016/j.mib.2005.04.011
Epelde, L., Becerril, J. M., Alkorta, I., and Garbisu, C. (2009). “Heavy metal phytoremediation: microbial indicators of soil health for the assessment of remediation efficiency,” in The Advances in Applied Bioremediation, eds A. Singh, R. C. Kuhad, and O. P. Ward (Berlin: Springer), 299–313.
Epelde, L., Becerril, J. M., Alkorta, I., and Garbisu, C. (2014). Adaptive long-term monitoring of soil health in metal phytostabilization: ecological attributes and ecosystem services based on soil microbial parameters. Int. J. Phytoremediat. 16, 971–981. doi: 10.1080/15226514.2013.810578
Epelde, L., Becerril, J. M., Kowalchuk, G. A., Deng, Y., Zhou, J., and Garbisu, C. (2010). Impact of metal pollution and Thlaspi caerulescens growth on soil microbial communities. Appl. Environ. Microbiol. 76, 7843–7853. doi: 10.1128/AEM.01045-10
Filonov, A. E., Akhmetov, L. I., Puntus, I. F., Esikova, T. Z., Gafarov, A. B., Kosheleva, I. A., et al. (2010). Horizontal transfer of catabolic plasmids and naphthalene biodegradation in open soil. Microbiology 79, 184–190. doi: 10.1134/S0026261710020098
Franzetti, A., Caredda, P., Ruggeri, C., La Colla, L., Tamburini, E., Papacchini, M., et al. (2009). Potential application of surface active compounds by Gordonia sp. strain BS29 in soil remediation technologies. Chemosphere 75, 801–807. doi: 10.1016/j.chemosphere.2008.12.052
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. 3, 722–732. doi: 10.1038/nrmicro1235
Furuya, E. Y., and Lowy, F. D. (2006). Antimicrobial-resistant bacteria in the community setting. Nat. Rev. Microbiol. 4, 36–45. doi: 10.1038/nrmicro1325
Gao, C., Jin, X., Ren, J., Fang, H., and Yu, Y. (2015). Bioaugmentation of DDT-contaminated soil by dissemination of the catabolic plasmid pDOD. J. Environ. Sci. 27, 42–50. doi: 10.1016/j.jes.2014.05.045
Garbisu, C., and Alkorta, I. (1997). Utilization of genetically engineered microorganisms (GEMs) for bioremediation. J. Chem. Technol. Biotechnol. 74, 599–606. doi: 10.1080/07388550600842794
Garbisu, C., Alkorta, I., and Epelde, L. (2011). Assessment of soil quality using microbial properties and attributes of ecological relevance. Appl. Soil Ecol. 49, 1–4. doi: 10.1016/j.apsoil.2011.04.018
Garbisu, C., Hernández-Allica, J., Barrutia, O., Alkorta, I., and Becerril, J. M. (2002). Phytoremediation: a technology using green plants to remove contaminants from polluted areas. Rev. Environ. Health 17, 173–188. doi: 10.1515/REVEH.2002.17.3.173
Gardin, H., and Pauss, A. (2001). κ-carrageenan/gelatin gel beads for the co-immobilization of aerobic and anaerobic microbial communities degrading 2,4,6-trichlorophenol under air-limited conditions. Appl. Microbiol. Biotechnol. 56, 517–523. doi: 10.1007/s002530000581
Geier, H., Mostowy, S., Cangelosi, G. A., Behr, M. A., and Ford, T. E. (2008). Autoinducer-2 triggers the oxidative stress response in Mycobacterium avium, leading to biofilm formation. Appl. Environ. Microbiol. 74, 1798–1804. doi: 10.1128/AEM.02066-07
Gentili, A. R., Cubitto, M. A., Ferrero, M., and Rodriquéz, M. S. (2006). Bioremediation of crude oil polluted seawater by a hydrocarbon-degrading bacterial strain immobilized on chitin and chitosan flakes. Int. Biodeterior. Biodegrad. 57, 222–228. doi: 10.1016/j.ibiod.2006.02.009
Gómez-Sagasti, M. T., Epelde, L., Alkorta, I., and Garbisu, C. (2016). Reflections on soil contamination research from a biologist’s point of view. Appl. Soil Ecol. 105, 207–210. doi: 10.1016/j.apsoil.2016.04.004
Greer, L. E., and Shelton, D. R. (1992). Effect of inoculant strain and organic matter content on kinetics of 2,4-dichloro-phenoxyacetic acid degradation in soil. Appl. Environ. Microbiol. 58, 1459–1465.
Halden, R. U., Tepp, S., Halden, B. G., and Dwyer, D. F. (1999). Degradation of 3-phenoxybenzoic acid in soil by Pseudomonas pseudoalcaligenes POB310(pPOB) and two modified Pseudomonas strains. Appl. Environ. Microbiol. 65,3354–3359.
Hall, J. P. J., Brockhurst, M. A., Dytham, C., and Harrison, E. (2017). The evolution of plasmid stability: are infectious transmission and compensatory evolution competing evolutionary trajectories. Plasmid 91, 90–95. doi: 10.1016/j.plasmid.2017.04.003
Hall, J. P. J., Wood, A. J., Harrison, E., and Brockhurst, M. A. (2016). Source-sink plasmid transfer dynamics maintain gene mobility in soil bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 113, 8260–8265. doi: 10.1073/pnas.1600974113
Haluška, L., Barančiková, G., Baláž,Š, Dercová, K., Vrana, B., Paz-Weisshaar, M., et al. (1995). Degradation of PCB in different soils by inoculated Alcaligenes xylosoxidans. Sci. Total Environ. 175, 275–285. doi: 10.1016/0048-9697(95)04927-4
Hanscha, C., McKarnsb, S. C., Smith, C. J., and Doolittle, D. J. (2000). Comparative QSAR evidence for a free-radical mechanism of phenol-induced toxicity. Chem. Biol. Interact. 127, 61–72. doi: 10.1016/S0009-2797(00)00171-x
Harrison, E., Guymer, D., Spiers, A. J., Paterson, S., and Brockhurst, M. A. (2015). Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr. Biol. 25, 2034–2039. doi: 10.1016/j.cub.2015.06.024
Heinaru, E., Merimaa, M., Viggor, S., Lehiste, M., Leito, I., Truu, J., et al. (2005). Biodegradation efficiency of functionally important populations selected for bioaugmentation in phenoland oil-polluted area. FEMS Microbiol. Ecol. 51, 363–373. doi: 10.1016/j.femsec.2004.09.009
Heitkamp, M. A., and Steward, W. P. (1996). A novel porous nylon biocarrier for immobilized bacteria. Appl. Environ. Microbiol. 62, 4659–4662.
Høiby, N., Bjarnsholt, T., Givskov, M., Molin, S., and Ciofu, O. (2010). Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35, 322–332. doi: 10.1016/j.ijantimicag.2009.12.011
Ikuma, K., and Gunsch, C. K. (2010). Effect of carbon source addition on toluene biodegradation by an Escherichia coli DH5α transconjugant harboring the TOL plasmid. Biotechnol. Bioeng. 107, 269–277. doi: 10.1002/bit.22808
Ikuma, K., and Gunsch, C. K. (2012). Genetic bioaugmentation as an effective method for in situ bioremediation: functionality of catabolic plasmids following conjugal transfers. Bioengineered 3, 236–241. doi: 10.4161/bioe.20551
Ikuma, K., Holzem, R. M., and Gunsch, C. K. (2012). Impacts of organic carbon availability and recipient bacteria characteristics on the potential for TOL plasmid genetic bioaugmentation in soil slurries. Chemosphere 89, 158–163. doi: 10.1016/j.chemosphere.2012.05.086
Inoue, D., Sei, K., Soda, S., Ike, M., and Fujita, M. (2005). Potential of predominant activated sludge bacteria as recipients in conjugative plasmid transfer. J. Biosci. Bioeng. 100, 600–605. doi: 10.1263/jbb.100.600
Inoue, D., Yamazaki, Y., Tsutsui, T., Sei, K., Soda, S., Fujita, M., et al. (2012). Impacts of gene bioaugmentation with pJP4-harboring bacteria of 2,4-D-contaminated soil slurry on the indigenous microbial community. Biodegradation 23, 263–276. doi: 10.1007/s10532-011-9505-x
Iwamoto, T., and Nasu, M. (2001). Current bioremediation practice and perspective. J. Biosci. Bioeng. 92, 1–8. doi: 10.1016/S1389-1723(01)80190-0
Jefferson, K. K. (2004). What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 236, 163–173. doi: 10.1016/j.femsle.2004.06.005
Johnsen, A. R., and Kroer, N. (2007). Effects of stress and other environmental factors on horizontal plasmid transfer assessed by direct quantification of discrete transfer events. FEMS Microbiol. Ecol. 59, 718–728. doi: 10.1111/j.1574-6941.2006.00230.x
Jung, C. M., Crocker, F. H., Eberly, J. O., and Indest, K. J. (2011). Horizontal gene transfer (HGT) as a mechanism of disseminating RDX-degrading activity among Actinomycete bacteria. J. Appl. Microbiol. 110, 1449–1459. doi: 10.1111/j.1365-2672.2011.04995.x
Jussila, M. M., Zhao, J., Suominen, L., and Lindstróm, K. (2007). TOL plasmid transfer during bacterial conjugation in vitro and rhizoremediation of oil compounds in vivo. Environ. Pollut. 146, 510–524. doi: 10.1016/j.envpol.2006.07.012
Juwarkar, A. A., Misra, R. R., and Sharma, J. K. (2014). “Recent trends in bioremediation,” in Geomicrobiology and Biogeochemistry, eds N. Parmar and A. Singh (Berlin: Springer), 81–100.
Karve, S. M., Tiwary, K., Selveshwari, S., and Dey, S. (2016). Environmental fluctuations do not select for increased variation or population-based resistance in Escherichia coli. J. Biosci. 41, 39–49. doi: 10.1007/s12038-016-9592-2
Kim, J. M., Le, N. T., Chung, B. S., Park, J. H., Bae, J. W., Madsen, E. L., et al. (2008). Influence of soil components on the biodegradation of benzene, toluene, ethylbenzene, and o-, m-, and p-xylenes by the newly isolated bacterium Pseudoxanthomonas spadix BD-a59. Appl. Environ. Microbiol. 74, 7313–7320. doi: 10.1128/AEM.01695-08
Kottara, A., Hall, J. P. J., Harrison, E., and Brockhurst, M. A. (2016). Multi-host environments select for host-generalist conjugative plasmids. BMC Evol. Biol. 16:70. doi: 10.1186/s12862-016-0642-z
Król, J. E., Nguyen, H. D., Rogers, L. M., Beyenal, H., Krone, S. M., and Top, E. M. (2011). Increased transfer of a multidrug resistance plasmid in Escherichia coli biofilms at the air-liquid interface. Appl. Environ. Microbiol. 77, 5079–5088. doi: 10.1128/AEM.00090-11
Kumavath, R. N., and Deverapalli, P. (2013). “Scientific swift in bioremediation: an overview,” in Applied Bioremediation - Active and Passive Approaches, eds Y. B. Patil and P. Rao (Croatia: Intech), 375–388.
Liang, Y., Zhang, X., Dai, D., and Li, G. (2009). Porous biocarrier-enhanced biodegradation of crude oil contaminated soil. Int. Biodeterior. Biodegrad. 63, 80–87. doi: 10.1016/j.ibiod.2008.07.005
Maier, R. M. (2000). “Microorganisms and organic pollutants,” in Environmental Microbiology, eds R. M. Maier, I. L. Pepper, and C. P. Gerba (San Diego, CA: Academic Press), 63–402.
Maphosa, F., Lieten, S. H., Dinkla, I., Stams, A. J., Smidt, H., and Fennell, D. E. (2012). Ecogenomics of microbial communities in bioremediation of chlorinated contaminated sites. Front. Microbiol. 3:351. doi: 10.3389/fmicb.2012.0035
Martínez, J. L., Sánchez, M. B., Martínez-Solano, L., Hernandez, A., Garmendia, L., Fajardo, A., et al. (2009). Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 33, 430–449. doi: 10.1111/j.1574-6976.2008.00157.x
Massa, V., Infantino, A., Radice, F., Orlandi, V., Tavecchio, F., Giudici, R., et al. (2009). Efficiency of natural and engineered bacterial strains in the degradation of 4-chlorobenzoic acid in soil slurry. Int. Biodeterior. Biodegrad. 63, 112–125. doi: 10.1016/j.ibiod.2008.07.006
Meckenstock, R. U., Elsner, M., Griebler, C., Lueders, T., Stumpp, C., Aamand, J., et al. (2015). Biodegradation: updating the concepts of control for microbial cleanup in contaminated aquifers. Environ. Sci. Technol. 49, 7073–7081. doi: 10.1021/acs.est.5b00715
Miller, M. N., Stratton, G. W., and Murray, G. (2004). Effects of soil moisture and aeration on the biodegradation of pentachlorophenol contaminated soil. Bull. Environ. Contam. Toxicol. 72, 101–108. doi: 10.1007/s00128-003-0246-3
Miyazaki, R., Sato, Y., Ito, M., Ohtsubo, Y., Nagata, Y., and Tsuda, M. (2006). Complete nucleotide sequence of an exogenously isolated plasmid, pLB1, involved in gamma-hexachlorocyclohexane degradation. Appl. Environ. Microbiol. 72, 6923–6933. doi: 10.1128/AEM.01531-06
Mølbak, L., Molin, S., and Kroer, N. (2007). Root growth and exudate production define the frequency of horizontal plasmid transfer in the rhizosphere. FEMS Microbiol. Ecol. 59, 167–176. doi: 10.1111/j.1574-6941.2006.00229.x
Morosini, M. I., Ayala, J. A., Baquero, F., Martínez, J. L., and Blazquez, J. (2000). Biological cost of AmpC production for Salmonella enterica serotype Typhimurium. Antimicrob. Agents Chemother. 44, 3137–3143. doi: 10.1128/AAC.44.11.3137-3143.2000
Mrozik, A., Miga, S., and Piotrowska-Seget, Z. (2011). Enhancement of phenol degradation by soil bioaugmentation with Pseudomonas sp. JS150. J. Appl. Microbiol. 111, 1357–1370. doi: 10.1111/j.1365-2672.2011.05140.x
Mrozik, A., and Piotrowska-Seget, Z. (2010). Bioaugmentation as a strategy for cleaning up of soils contaminated with aromatic compounds. Microbiol. Res. 165, 363–375. doi: 10.1016/j.micres.2009.08.001
Muñoz-Leoz, B., Garbisu, C., Charcosset, J. Y., Sánchez-Pérez, J. M., Antigüedad, I., Ruiz, E., et al. (2013). Non-target effects of three formulated pesticides on microbially-mediated processes in a clay-loam soil. Sci. Total Environ. 449, 345–354. doi: 10.1016/j.scitotenv.2013.01.079
Musovic, S., Dechesne, A., Sørensen, J., and Smets, B. F. (2010). A novel assay to assess permissiveness of a soil microbial community toward receipt of mobile genetic elements. Appl. Environ. Microbiol. 76, 4813–4818. doi: 10.1128/AEM.02713-09
Newby, D. T., Gentry, T. J., and Pepper, I. L. (2000). Comparison of 2,4dichlorophenoxyacetic acid degradation and plasmid transfer in soil resulting from bioaugmentation with two different pJP4 donors. Appl. Environ. Microbiol. 66, 3399–3407. doi: 10.1128/AEM.66.8.3399-3407.2000
Newby, D. T., and Pepper, I. L. (2002). Dispersal of plasmid pJP4 in unsaturated and saturated 2,4-dichlorophenoxyacetic acid contaminated soil. FEMS Microbiol. Ecol. 39, 157–164. doi: 10.1111/j.1574-6941.2002.tb00917.x
Norman, A., Hansen, L. H., and Sørensen, S. J. (2009). Conjugative plasmids: vessels of the communal gene pool. Philos. Trans. R Soc. Lond. B Biol. Sci. 364, 2275–2289. doi: 10.1098/rstb.2009.0037
Nowak, A., and Mrozik, A. (2016). Facilitation of co-metabolic transformation and degradation of monochlorophenols by Pseudomonas sp. CF600 and changes in its fatty acid composition. Water Air Soil Pollut. 227, 83. doi: 10.1007/s11270-016-2775-5
Overhage, J., Sielker, S., Homburg, S., Parschat, K., and Fetzner, S. (2005). Identification of large linear plasmids in Arthrobacter spp. encoding the degradation of quinaldine to anthranilate. Microbiology 151, 491–500. doi: 10.1099/mic.0.27521-0
Panagos, P., Van Liedekerke, M., Yigini, Y., and Montanarella, L. (2013). Contaminated sites in Europe: review of the current situation based on data collected through a European network. J. Environ. Public Health 2013, 1–11. doi: 10.1155/2013/158764
Pardo, T., Clemente, R., Epelde, L., Garbisu, C., and Bernal, M. P. (2014). Evaluation of the phytostabilisation efficiency in a trace elements contaminated soil using soil health indicators. J. Hazard Mater. 268, 68–76. doi: 10.1016/j.jhazmat.2014.01.003
Park, C., and Zhang, J. (2012). High expression hampers horizontal gene transfer. Genome Biol. Evol. 4, 523–532. doi: 10.1093/gbe/evs030
Pepper, I. L., Gentry, T. J., Newby, D. T., Roane, T. M., and Josephson, K. L. (2002). The role of cell bioaugmentation and gene bioaugmentation in the remediation of co-contaminated soils. Environ. Health Perspect. 110, 943–946. doi: 10.1289/ehp.02110s6943
Pinedo, C. A., and Smets, B. F. (2005). Conjugal TOL transfer from Pseudomonas putida to Pseudomonas aeruginosa: effects of restriction proficiency, toxicant exposure, cell density ratios, and conjugation detection method on observed transfer efficiencies. Appl. Environ. Microbiol. 71, 51–57. doi: 10.1128/AEM.71.1.51-57.2005
Popa, O., Hazkani-Covo, E., Landan, G., Martin, W., and Dagan, T. (2011). Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res. 21, 599–609. doi: 10.1101/gr.115592.110
Popowska, M., and Krawczyk-Balska, A. (2013). Broad-host-range IncP-1 plasmids and their resistance potential. Front. Microbiol. 4:44. doi: 10.3389/fmicb.2013.00044
Porse, A., Schønning, K., Munck, C., and Sommer, M. O. A. (2016). Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol. Biol. Evol. 33, 2860–2873. doi: 10.1093/molbev/msw163
Quan, X., Tang, H., Xiong, W., and Yang, Z. (2010). Bioaugmentation of aerobic sludge granules with a plasmid donor strain for enhanced degradation of 2,4-dichlorophenoxyacetic acid. J. Hazard Mater. 179, 1136–1142. doi: 10.1016/j.jhazmat.2010.04.00
Reichenbach, T., Mobilia, M., and Frey, E. (2007). Mobility promotes and jeopardizes biodiversity in rock-paper-scissors games. Nature 448, 1046–1049. doi: 10.1038/nature06095
Reineke, W. (1998). Development of hybrid strains for the mineralization of chloroaromatics by patchwork assemble. Annu. Rev. Microbiol. 52, 287–331. doi: 10.1146/annurev.micro.52.1.287
Riley, M. A., and Wertz, J. E. (2002). Bacteriocins: evolution, ecology and application. Annu. Rev. Microbiol. 56, 117–137. doi: 10.1146/annurev.micro.56.012302.161024
San Millan, A., Peña-Miller, R., Toll-Riera, M., Halbert, Z. V., McLean, A. R., Cooper, B. S., et al. (2014). Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat. Commun. 5, 5208. doi: 10.1038/ncomms6208
Sayler, G. S., and Ripp, S. (2000). Field applications of genetically engineered microorganisms for bioremediation processes. Curr. Opin. Biotechnol. 11, 286–289. doi: 10.1016/S0958-1669(00)00097-5
Sejáková, Z., Dercová, K., and Tóthová, L. (2009). Biodegradation and ecotoxicity of soil contaminated by pentachlorophenol applying bioaugmentation and addition of sorbents. World J. Microbiol. Biotechnol. 25, 243–252. doi: 10.1007/s11274-008-9885-1
Sengupta, S., Chattopadhyay, M. K., and Grossart, H. P. (2013). The multifaceted roles of antibiotics and antibiotic resistance in nature. Front. Microbiol. 4:47. doi: 10.3389/fmicb.2013.00047
Shachrai, I., Zaslaver, A., Alon, U., and Dekel, E. (2010). Cost of unneeded proteins in E. coli is reduced after several generations in exponential growth. Mol. Cell 38, 758–767. doi: 10.1016/j.molcel.2010.04.015
Shintani, M., Sanchez, Z. K., and Kimbara, K. (2015). Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front. Microbiol. 31:242. doi: 10.3389/fmicb.2015.00242
Shintani, M., Takahashi, Y., Yamane, H., and Nojiri, H. (2010a). The behavior and significance of degradative plasmids belonging to Inc groups in Pseudomonas within natural environments and microcosms. Microbes Environ. 25, 253–265. doi: 10.1264/jsme2.ME10155
Shintani, M., Yamane, H., and Nojiri, H. (2010b). Behaviour of various hosts of the IncP-7 carbazole-degradative plasmid pCAR1 in artificial microcosms. Biosci. Biotechnol. Biochem. 74, 343–349. doi: 10.1271/bbb.90675
Singh, A., and Ward, O. P. (2004). “Biotechnology and bioremediation An overview,” in Biodegradation and Bioremediation, eds A. Singh and O. P. Ward (Berlin: Springer-Verlag), 1–17.
Sorek, R., Zhu, Y., Creevey, C. J., Francino, M. P., Bork, P., and Rubin, E. M. (2007). Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318, 1449–1452. doi: 10.1126/science.1147112
Sørensen, S. J., Bailey, M., Hansen, L. H., Kroer, N., and Wuertz, S. (2005). Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3, 700–710. doi: 10.1038/nrmicro1232
Stalwood, B., Shears, J., Williams, P. A., and Hughes, K. A. (2005). Low temperature bioremediation of oil-contaminated soil using biostimulation and bioaugmentation with a Pseudomonas sp. from maritime Antarctica. J. Appl. Microbiol. 99, 794–802. doi: 10.1111/j.1365-2672.2005.02678.x
Starikova, I., Al-Haroni, M., Werner, G., Roberts, A., Sørum, V., Nielsen, K. M., et al. (2013). Fitness costs of various mobile genetic elements in Enterococcus faecium and Enterococcus faecalis. J. Antimicrob. Chemother. 68, 2755–2765. doi: 10.1093/jac/dkt270
Thomas, C. M., and Nielsen, K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721. doi: 10.1038/nrmicro1234
Thompson, I. P., Van der Gast, C. J., Ciric, L., and Singer, A. C. (2005). Bioaugmentation for bioremediation: the challenge of strain selection. Environ. Microbiol. 7, 909–915. doi: 10.1111/j.1462-2920.2005.00804.x
Top, E. M., and Springael, D. (2003). The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotechnol. 14, 262–269. doi: 10.1016/S0958-1669(03)00066-1
Top, E. M., Springael, D., and Boon, N. (2002). Catabolic mobile genetic elements and their potential use in bioaugmentation of polluted soils and waters. FEMS Microbiol. Ecol. 42, 199–208. doi: 10.1111/j.1574-6941.2002.tb01009.x
Top, E. M., Van Elsas, E. D., DeSmet, S. N., and Forney, L. J. (1998). Enhancement of 2,4-dichlorophenoxyacetic acid (2,4-D) degradation in soil by dissemination of catabolic plasmids. Antonie Van Leeuwenhoek 73, 87–94. doi: 10.1023/A:1000663619522
Tsutsui, H., Anami, Y., Matsuda, M., Hashimoto, K., Inoue, D., Sei, K., et al. (2013). Plasmid-mediated bioaugmentation of sequencing batch reactors for enhancement of 2,4-dichlorophenoxyacetic acid removal in wastewater using plasmid pJP4. Biodegradation 24, 343–352. doi: 10.1007/s10532-012-9591-4
van Rensburg, E., den Haan, R., Smith, J., van Zyl, W. H., and Görgens, J. F. (2012). The metabolic burden of cellulase expression by recombinant Saccharomyces cerevisiae Y294 in aerobic batch culture. Appl. Microbiol. Biotechnol. 96, 197–209. doi: 10.1007/s00253-012-4037-9
Venkata Mohan, S., Falkentoft, C., Nancharaiah, Y. V., Sturm, B. S. M., Wattiau, P., Wilderer, P. A., et al. (2009). Bioaugmentation of microbial communities in laboratory and pilot scale sequencing batch biofilm reactors using the TOL plasmid. Bioresour. Technol. 100, 1746–1753. doi: 10.1016/j.biortech.2008.09.048
Verstraeten, N., Braeken, K., Debkumari, B., Fauvart, M., Fransaer, J., Vermant, J., et al. (2008). Living on a surface: swarming and biofilm formation. Trends Microbiol. 16, 496–506. doi: 10.1016/j.tim.2008.07.004
Vogwill, T., and MacLean, R. C. (2015). The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol. Appl. 8, 284–295. doi: 10.1111/eva.12202
Walsh, C. (2000). Molecular mechanisms that confer antibacterial drug resistance. Nature 406, 775–781. doi: 10.1038/35021219
Wang, J. L., Mao, Z. Y., Han, L. P., and Qian, Y. (2004). Bioremediation of quinoline-contaminated soil using bioaugmentation in slurry-phase reactor. Biomed. Environ. Sci. 17, 187–195.
Wang, S., and Mulligan, C. N. (2004). An evaluation of surfactant foam technology in remediation of contaminated soil. Chemosphere 57, 1079–1089. doi: 10.1016/j.chemosphere.2004.08.019
Wang, Y., Kou, S., Jiang, Q., Xu, B., Liu, X., Xiao, J., et al. (2014). Factors affecting transfer of degradative plasmids between bacteria in soils. Appl. Soil Ecol. 84, 254–261. doi: 10.1016/j.apsoil.2014.07.009
Wickham, S. A., and Lynn, D. H. (1990). Relation between growth rate, cell size, and DNA content in colpodean ciliates (Ciliophora: Colpodea). Eur. J. Protistol. 25, 345–352. doi: 10.1016/S0932-4739(11)80127-6
Wiedenbeck, J., and Cohan, F. M. (2011). Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 35, 957–976. doi: 10.1111/j.1574-6976.2011.00292.x
Wolski, E. A., Murialdo, S. E., and Gonzales, J. F. (2006). Effect of pH and inoculum size on pentachlorophenol degradation by Pseudomonas sp. Water SA 32, 93–98.
Wozniak, R. A., and Waldor, M. K. (2010). Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8, 552–563. doi: 10.1038/nrmicro2382
Wyndham, R. C., Cashore, A. E., Nakatsu, C. H., and Peel, M. C. (1994). Catabolic transposons. Biodegradation 5, 323–357. doi: 10.1007/BF00696468
Keywords: biodegradation, catabolic plasmid, fitness cost, horizontal gene transfer, soil pollution
Citation: Garbisu C, Garaiyurrebaso O, Epelde L, Grohmann E and Alkorta I (2017) Plasmid-Mediated Bioaugmentation for the Bioremediation of Contaminated Soils. Front. Microbiol. 8:1966. doi: 10.3389/fmicb.2017.01966
Received: 20 May 2017; Accepted: 25 September 2017;
Published: 09 October 2017.
Edited by:
Tatiana Venkova, University of Texas Medical Branch, United StatesReviewed by:
Gloria Del Solar, Consejo Superior de Investigaciones Científicas (CSIC), SpainCopyright © 2017 Garbisu, Garaiyurrebaso, Epelde, Grohmann and Alkorta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Itziar Alkorta, aXR6aS5hbGtvcnRhQGVodS5ldXM=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.