- 1Department of Infectious Diseases, Sir Run Run Shaw Hospital, College of Medicine, Zhejiang University, Hangzhou, China
- 2Key Laboratory of Microbial Technology and Bioinformatics of Zhejiang Province, Hangzhou, China
- 3State Key Laboratory for Diagnosis and Treatment of Infectious Disease, First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
To further characterize the fosB-carrying plasmids of 19 vancomycin-resistant enterococci, the complete sequences of the fosB- and vanA-containing plasmids of Enterococcus faecium (pEMA120) and E. avium (pEA19081) were obtained by single-molecule, real-time sequencing. We found that these two plasmids are essentially identical (99.99% nucleotide sequence identity), which proved the possibility of interspecies transmission. Comparative analysis of the plasmids revealed that the backbone of pEMA120 is 99% similar to a conjugative fosB-negative E. faecium plasmid, pZB18. There is a traE disrupted in the transfer region of pEMA120, in comparison to pZB18 with an intact traE. The difference of their transfer frequencies between pEMA120 and pZB18 suggests this interruption of traE might affect conjugative transfer. Two copies of the fosB gene linked to a tnpA gene, forming an ISL3-like transposon, were found at separate locations within pEMA120, which had not been reported previously. These two fosB-carrying transposons were confirmed to form circular intermediates by inverse PCR. The hybridization of plasmid DNA digested by BsaI, having restriction site within the fosB sequence, demonstrated that the presence of multiple copies of fosB per plasmid is common. The total copy number of the fosB gene as revealed by qRT-PCR did not correlate with fosfomycin MICs or growth rates at sub-MICs of fosfomycin in different transconjugants. From susceptibility tests, the fosB gene, regardless of the copy number, conferred high fosfomycin MICs that ranged from 16384 to 65536 μg/ml. This first complete nucleotide sequence of a plasmid carrying two copies of fosB in VRE suggests that the fosB gene can transfer to multiple loci of plasmids by the ISL3 family transposase TnpA, possibly in the form of circular intermediates, leading to the dissemination of high fosfomycin resistance in VRE.
Introduction
Nowadays, the colonization and infection of vancomycin-resistant enterococci (VRE) is a global public health concern due to the increasing dissemination of these bacteria and limited clinical treatment options (Ranotkar et al., 2014). Fosfomycin, a drug discovered in 1969 that is primarily used to treat uncomplicated urinary tract infections (Keating, 2013), has been shown to be effective against VRE (Superti et al., 2009; Descourouez et al., 2013; Tang et al., 2013). Therefore, fosfomycin has gained attention as a potential therapeutic option for treating VRE infections (Popovic et al., 2010). However, resistance to fosfomycin emerges rapidly, likely because of the high frequency at which fosfomycin-resistant mutants arise (Greenwood, 1990; Hamilton-Miller, 1992). Therefore, additional studies on fosfomycin resistance mechanisms are required to better guide clinical practices. Until now, the fosfomycin resistance enzyme FosB, which catalyzes the Mg(II)-dependent addition of L-cysteine, or bacillithiol, to the epoxide ring of fosfomycin (Thompson et al., 2014), was the only known plasmid-borne fosfomycin-resistance determinant in Enterococcus spp. (Xu et al., 2013; Qu et al., 2014). The fosB gene was reported to be located on a transferable circular intermediate (Xu et al., 2013). Additionally, the fosB gene has also been reported to be present in plasmids of Staphylococcus spp. (99% nucleotide similarity) (Zilhao and Courvalin, 1990; Fu et al., 2016) and in the chromosome of Bacillus subtilis (63% nucleotide similarity) (Cao et al., 2001). In our previous study, we characterized 18 VRE isolates that were resistant to fosfomycin (Qu et al., 2014). The plasmid-encoded fosB gene inserted into the vanA type Tn1546-like transposon, likely accounts substantially for the high fosfomycin resistance rate observed in VRE. This paper reports on the detailed plasmid environment of fosB gene by third-generation sequencing and the formation of circular fosB intermediates. We also discuss the contribution of fosB copy number on the fosfomycin MIC and bacterial growth rate at a sub-MIC level of fosfomycin in the assayed VRE strains.
Materials and Methods
Bacterial Isolates
Enterococcus faecium A120 and 17 additional fosB-carrying, vancomycin resistant E. faecium isolates were characterized in our previous paper (Qu et al., 2014). E. avium 19081 is a clinical isolate that was collected during the same time period from fecal specimens carrying fosB and vanA. Using these isolates as donor strains, fosfomycin resistant transconjugants were obtained by filter mating experiments using E. faecium BM4105RF as the recipient strain (Qu et al., 2014).
Antimicrobial Susceptibility Testing
The MICs of fosfomycin against 19 VRE isolates and their respective E. faecium BM4105RF transconjugants were determined by the agar dilution method. E. faecalis ATCC 29212 was used as the control. The results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) 2015 guidelines (CLSI, 2015).
Conjugation Experiment
Broth mating and filter mating of E. faecium A120 were performed to get the transfer frequency of plasmid pEMA120 using E. faecium BM4105RF as the recipient strain with a donor/recipient ratio of 1:10 as previously described (Zheng et al., 2007).
Bacterial Growth at Sub-MIC Levels
Single colonies were grown at 37°C with shaking at 200 r.p.m. in 2 ml of brain heart infusion (BHI) broth. Independent overnight cultures were diluted (1:1000) and 200 μl was transferred to a 96-well plate that was incubated at 37°C. The turbidity was measured at OD600 using a BioTEK Synergy plate reader (BioTEK, Winooski, VT, United States) every 5 min for 16 h.
A preliminary experiment using isolate A120 was carried out using a fosfomycin concentration gradient from 1/128 the MIC to the MIC. The growth of the 19 transconjugants that were exposed to 1/4 the MIC of fosfomycin was measured and compared with an antibiotic-free control. Each sample was performed in triplicate.
Plasmid Isolation, BsaI Restriction Enzyme Digestion and Southern Blot Hybridization
Plasmids were isolated from the 19 transconjugants using a QIAGEN Plasmid Midi Kit (Qiagen, Germany). The manufacturer’s instructions were followed with the inclusion of the lysozyme treatment (10 mg/mL, 60 min at 37°C) prior to the lysis step with buffer P2 [200 mM NaOH, 1% SDS (w/v)].
The restriction enzyme BsaI (New England Biolabs, United States), for which a target site is present within the fosB gene, was used to digest plasmid DNA. The hybridization probe was designed to bind within the latter half of the fosB gene after BsaI digestion (Supplementary Figure S1A) and was synthesized using the primers fosB-latter-F (5′-GTG GTA TAT GGT TAG CTT TGA ACG AAG-3′) and fosB-latter-R (5′-TGA GGT TTA GCC TCT TTA TAA TAA CTC-3′).
Plasmid DNA was first digested by BsaI at 37°C in a water bath overnight. After the digested DNA was agarose gel electrophoresed for 2 h, the DNA fragments were transferred to a positively charged nylon membrane (Millipore, United States), hybridized with a DIG-labeled fosB-latter specific probe, and then was detected with a NBT/BCIP color detection kit (Roche, Germany).
Quantification of the fosB Gene by Real-Time PCR Amplification
The copy number of fosB was measured via quantitative real-time PCR (qRT-PCR). The chromosomal housekeeping gene purK was chosen as an internal control. The sequences of the fosB and purK primers used are as follows:
Q-fosB-F (5′-CTCAATCTATCTTCTAAACTTCCTG-3′),
Q-fosB-R (5′-CGATTTTGAAGATTGGTATAACTGG-3′),
Q-purK-F (5′-GATATCCAAGATGCGATTGAC-3′), and
Q-purK-R (5′-CTTCTAAAACACAGGTTCCTTCTC-3′).
Quantitative PCR reactions were carried out in a 10 μl reaction that contained 5 μl SYBR® Premix Ex TaqTM PCR kit (Takara Bio, Japan), 2 ng of genomic DNA as template, and 2 pmol of each primer. The Ct values of each sample was measured under appropriate PCR conditions (preheated at 95°C for 5 min; 40 amplification cycles at 95°C for 5 s, 54°C for 30 s and 72°C for 30 s) on an Applied Biosystems ViiATM 7 Dx instrument (Applied Biosystems, United States). Data were calculated based on the 2-ΔΔCt method.
DNA Sequencing and Analysis
Single-molecule, real-time sequencing (Pacific Biosciences, United States) was carried out to complete the whole genome sequencing of E. faecium pEMA120 and E. avium pEA19081 plasmids. The plasmid sequences were validated by PCR, annotated using the RAST server (Aziz et al., 2008) and were supplemented by the BLASTP program. Lastly, the circular map of the pEMA120 and pEA19081 plasmids were generated using the CGview server (Grant and Stothard, 2008). A comparison of pEMA120 and three related plasmids was made with Mauve 2.3.1 and modified by Photoshop 7.0.
Inverse PCR was performed using the primers fosBiF (5′-TGTCAGCCCCTAAAATATCTCT-3′, located within the fosB gene) and fosBiR (5′-GTTTCAAATGTACCTAAAGAACT-3′, located within the tnpA gene) (Xu et al., 2013) with genomic DNA as template. The purified PCR products were cloned into the pMD20-T Vector (Takara Bio, Japan) and were sequenced (Biosune, China).
Results
Plasmid Structure
To further characterize the previously identified fosB-carrying plasmids, the plasmids from two VRE isolates (E. faecium A120 and E. avium 19081) were completely sequenced. Plasmid pEMA120 is 79,797 bp in size with an average G + C content of 33.8%. It contains 87 open reading frames (ORFs), 47 of which encode proteins with homology to proteins with known functions, which can be classified into several functional modules: antibiotic resistance, mobile elements, plasmid stability, replication recombination and repair, and conjugal transfer (Figure 1).
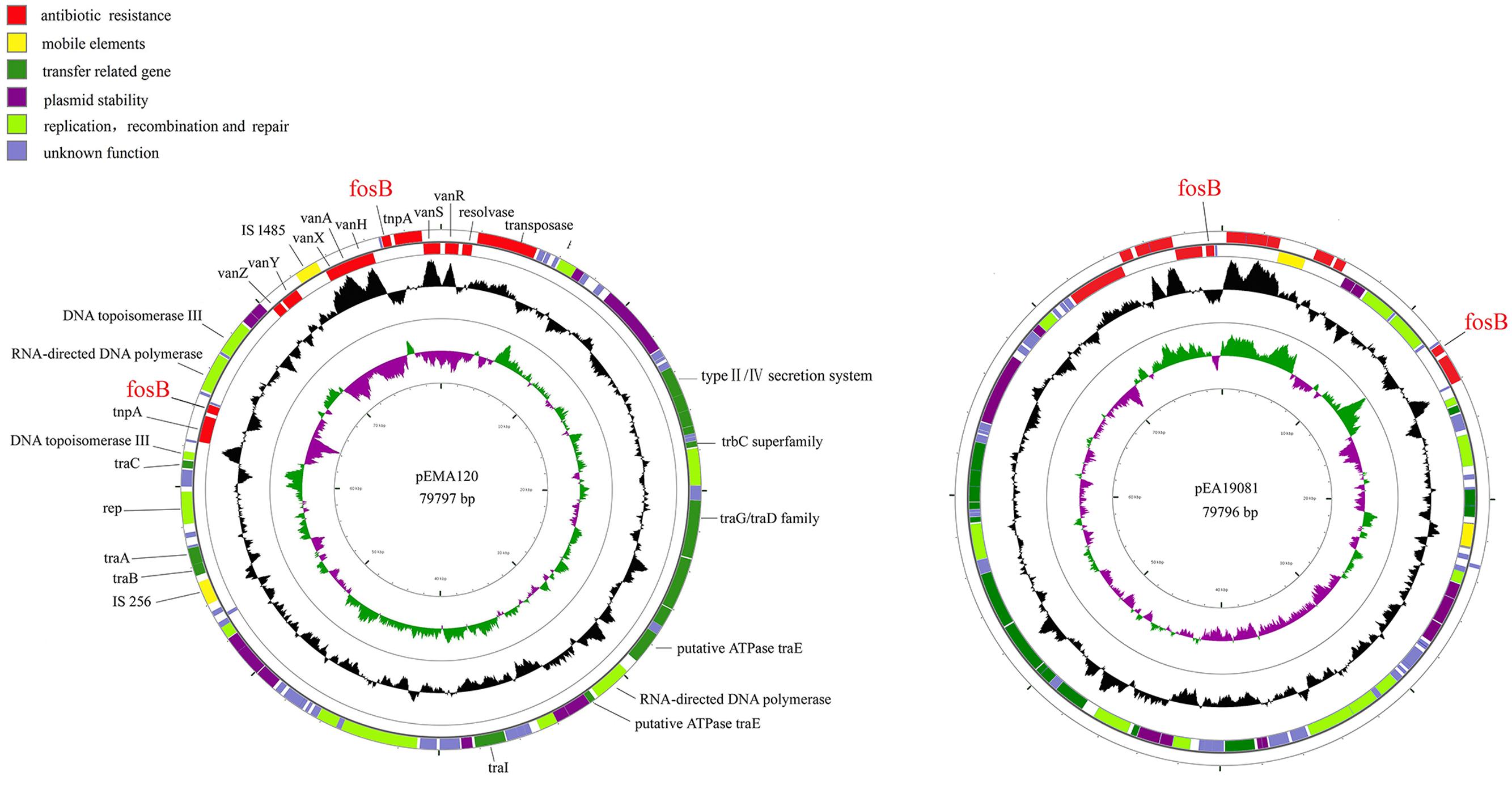
FIGURE 1. Structure of plasmid pEMA120 and pEA19081. The two outer circles represent ORFs in the plus (outside) and minus (inside) orientations, respectively. Functions are color coded as explained in the key. The two inner circles represent the G + C content plotted against the average G + C content of 33.8% (black circle) and GC skew information (green and purple circles).
In the antibiotic resistance region, there are two types of resistance determinants: fosB mediating fosfomycin resistance and vanA-type transposon causing vancomycin resistance. Two copies of fosB gene along with tnpA insert into different position of pEMA120 leading to diverse direct repeats (Figure 2). One fosB gene along with tnpA gene reversely inserts into the vanA-type Tn1546-like transposon between the vanRS and vanHAX intergenic region. Another fosB gene accompanied with tnpA gene is flanked by genes encoding a DNA topoisomerase and a RNA-directed DNA polymerase, which are required for replication, recombination and repair. No same structure was found in existing database, indicating that this is a novel insertion position of the fosB gene.
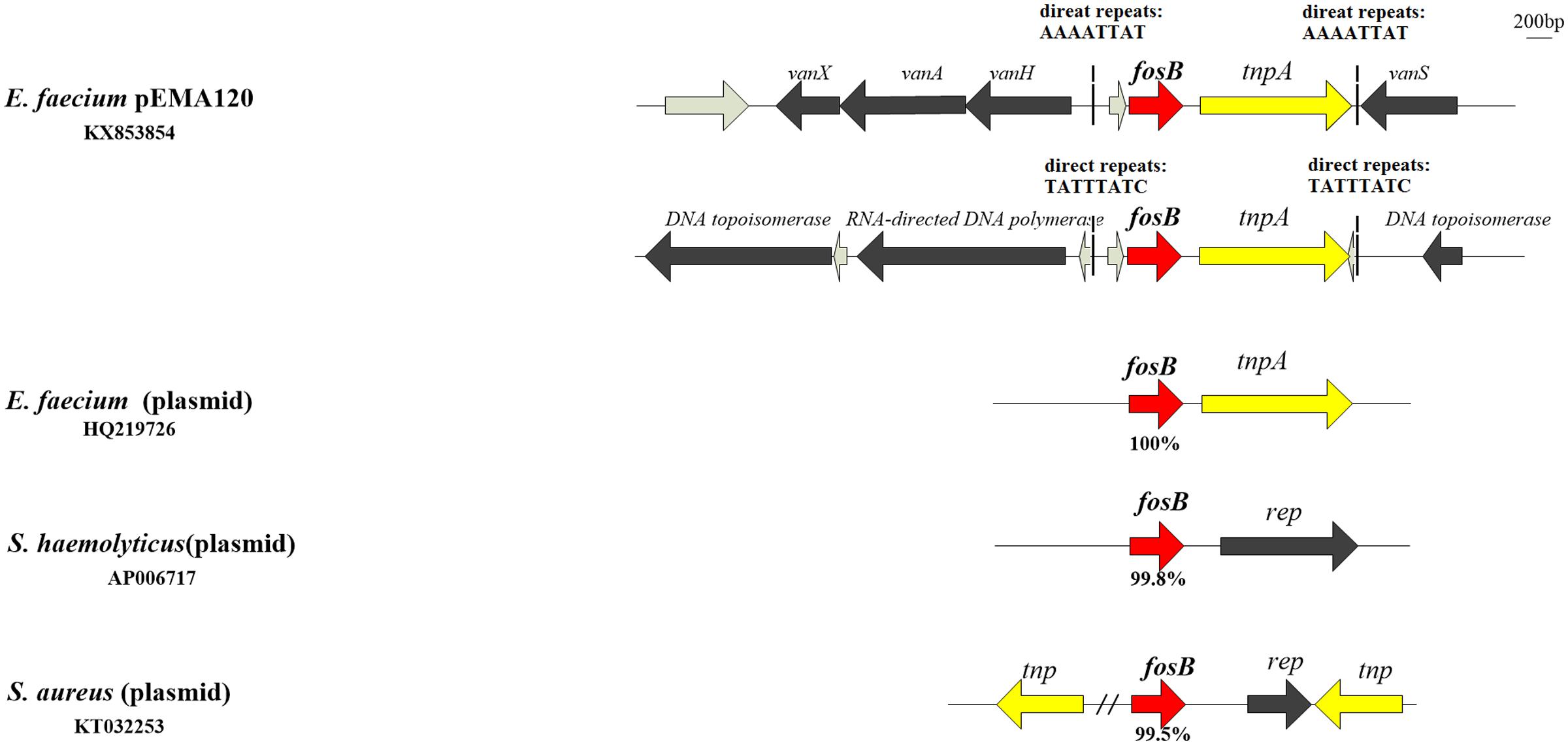
FIGURE 2. The fosB gene and genetic environment of fosB in different species or genera. fosB genes are indicated by red arrows with nucleotide identity below.
The surrounding environment of fosB on pEMA120 was compared with fosB in other species (Figure 2) Plasmid-borne fosB in Staphylococcus haemolyticus and Staphylococcus aureus share 99.8 and 99.5% nucleotide identity with fosB in the plasmid of E. faecium, respectively. As to surrounding environment, there is a rep gene downstream fosB on Staphylococcus spp. plasmid instead of a tnpA gene adjacent to fosB on E. faecium plasmids.
The conjugal transfer region of pEMA120 has some genes that are characteristic of a conjugative plasmid. The genes of ORF60 and ORF61, respectively, show similarity with 43 and 58% identity (at the amino acid level) to traB and traA of the conjugative plasmid pMG1 (Tanimoto and Ike, 2008). There is a predicted interrupted gene comprised of ORF29 and ORF31 that encodes a putative ATPase TraE, which belongs to the motor protein family of the type IV secretion system (T4SS) in enterococci (Chandran et al., 2009; Clewell et al., 2014). The traI gene (ORF38) shows 71% identity at the amino acid level with ORF34 of pHTβ encoding the putative DNA relaxase/nickase, which is needed for the initiation of DNA transfer in conjugative plasmids (Francia et al., 2004).
As regards plasmid replicon typing, ORF64 showed 57% identity at the amino acid level with Rep protein of plasmid pHTβ from E. faecium and showed less than 80% identity, at both nucleotide and amino acid level, with any plasmid replicon type according to the classification system of enterococci (Jensen et al., 2010).
To better study the evolution of pEMA120, we compared it to three similar E. faecium plasmids reported previously (Figure 3). Plasmid pMG1 (GenBank accession no. AB183714) is a gentamicin and kanamycin resistance plasmid capable of highly efficient transfer among enterococci and originally isolated from a clinical strain of E. faecium in Japan, 1998 (Ike et al., 1998). Plasmid pHTβ(GenBank accession no. AB206333) is a vancomycin resistance pMG1-like plasmid which was obtained from a hospital in the United States, 2003 (Tomita et al., 2003). Plasmid pZB18 (GenBank accession no. AB611033.1) acquired from Beijing, 2004 (Zheng et al., 2007) is also a vancomycin resistance conjugative E. faecium plasmid but without conserved traA gene or traB specific for pMG1-like plasmid. The distinctive feature of pEMA120 is the structure of the fosB connected by tnpA, which does not exist in other three plasmids. In general, the backbones of the four plasmids have high homology, which is depicted in the figure as the blue shades. In particular, plasmid pZB18 shares 99% identity and 85% query cover with pEMA120. Plasmid pMG1 and pHTβ contain highly efficient conjugative system, especially the similar oriT and transfer-related regions, which have a high degree of homology with the conjugal transfer region of pEMA120.
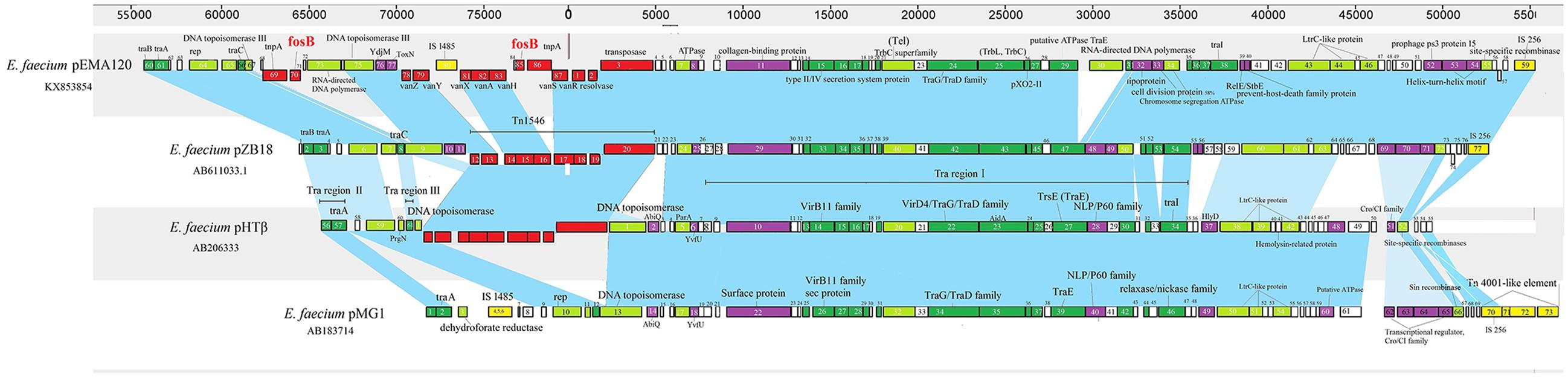
FIGURE 3. Comparison of E. faecium plasmid pEMA120 and three reported E. faecium plasmids pZB18, pHTβ, pMG1. Light blue shades indicate shared regions with a high degree of homology. ORFs are portrayed by rectangles and colored according to their putative functions. Light-green rectangles indicate genes associated with replication, recombination and repair. Genes associated with plasmid conjugal transfers are colored in green. Genes involved in plasmid stability are indicated by purple. Red and yellow rectangles indicate antimicrobial resistance genes and mobile elements genes, respectively. White rectangles indicate genes for hypothetical proteins as well as proteins of unknown function.
As shown in Figure 1, the E. avium plasmid pEA19081 was found to share 99.99% nucleotide sequence identity with the E. faecium plasmid pEMA120 after sequence comparison by BLASTN. Thus, the two plasmids are virtually identical, containing the same two copies of fosB as well as a Tn1546 transposon.
The Effect of Interruption of traE on the Conjugative Transfer
As mentioned in above section, a transfer related gene traE which might play an important role in conjugal transfer process, was disrupted by a gene encoding RNA-directed DNA polymerase. To investigate whether this interruption could affect the conjugative transfer, we compared the transfer frequency of plasmid pEMA120 with a disrupted traE and plasmid pZB18 with an intact traE. From Supplementary Table S1, the transfer frequency of pEMA120 was distinctly lower than that of pZB18, using broth mating or filter mating.
The fosB-Carrying Circular Intermediate
To determine whether fosB gene-containing circular intermediates are present in isolate A120, an inverse PCR was performed using previously reported primers (Xu et al., 2013). As expected, a ∼1 kb band was present after agarose gel electrophoresis, indicating the presence of circular intermediates. However, the sequencing electropherogram had imperfect signals, suggesting that this amplified 0.92-kb PCR product was probably a combination of two or more sequences (Supplementary Figure S2). Thus, TA-based cloning and sequencing was employed to obtain unique sequences. All sequences of PCR products that were ligated into pMD20-T vectors aligned similarly except for two regions. The first region was located 26–31 bp upstream of the fosB gene, showing two variants (ATTTG or TAACAT) (Supplementary Figure S2A), matching the corresponding positions of each fosB-carrying ISL3-like transposon within pEMA120. Another polymorphism region, 5-bp in length, was situated between the two IRs of ISL3-like transposon, i.e., the circle-junction region. Three kinds of sequences were observed, ATTTT, TATTT, or TTATC, as shown in Figure S2B. The complete fosB-transposon circular intermediate is 2425 or 2426 bp in length, consisting of the ISL3-like transposon and a 5-bp intervening DNA sequence (Figure 4). In addition, this fosB-transposon circular intermediate could also be acquired by inverse PCR in the other 18 VRE isolates. However, the circular intermediates could not be detected by southern hybridization (using the fosB-latter specific probe) after gel electrophoresis using the uncut or S1 nuclease digested pEMA120 plasmid (data not shown).
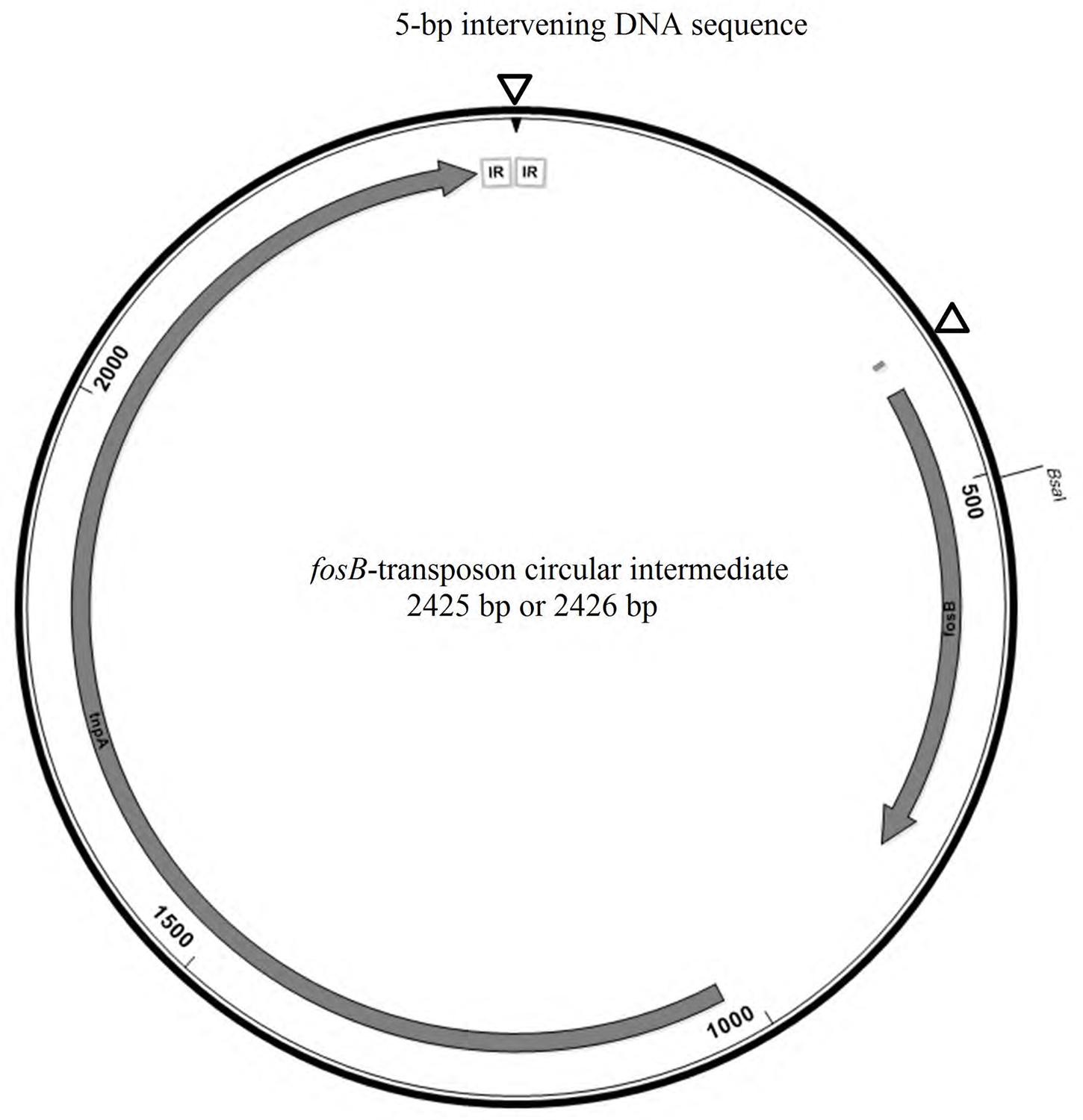
FIGURE 4. The schematic map of fosB-transposon circular intermediate. The polymorphism regions were indicated by triangles. The two IRs sequence was framed in gray.
The fosB Copy Number in Other fosB-Containing Plasmids
Our previous study suggested each of 19 VRE isolates contained one fosB-carrying plasmid by performing S1 nuclease-pulsed-field gel electrophoresis (PFGE) (Qu et al., 2014). The third-generation sequence analysis showed that both pEMA120 and pEA19081 carry two copies of the fosB gene. To determine the fosB copy number in these plasmids from transconjugants of 19 VRE isolates, a special hybridization protocol after a restriction enzyme digestion was used. The principle is depicted in Supplementary Figure S1 using pEMA120 as an example on basis of its clear plasmid sequence.
As Figure 5A shown, the DNA band pattern generated by BsaI digestion showed diverse restriction patterns in the plasmids from the transconjugants of 19 VRE isolates. The hybridization bands of isolate A120 in Figure 5B were well in accordance with the simulating graph of pEMA120 in Supplementary Figure S1, which proved the reliability of BsaI-digestion method to explore the fosB copy number in a plasmid. The isolates IB3, A165, IA110, 19081, A166, 1001, A96, A155, A158, and IA28, which were analyzed but were not shown here, generated very similar two-band signals (16 and 9.5 kb) with isolate A120. This finding suggests that these isolates bear two fosB copies per plasmid. These results were verified by PCR amplification using primers (“between vanSH” and “fosB-2th”) designed to amplify an approximately 3-kb fragment surrounding each fosB (Supplementary Table S2). It was found that all the above isolates contained the two DNA fragments with fosB.
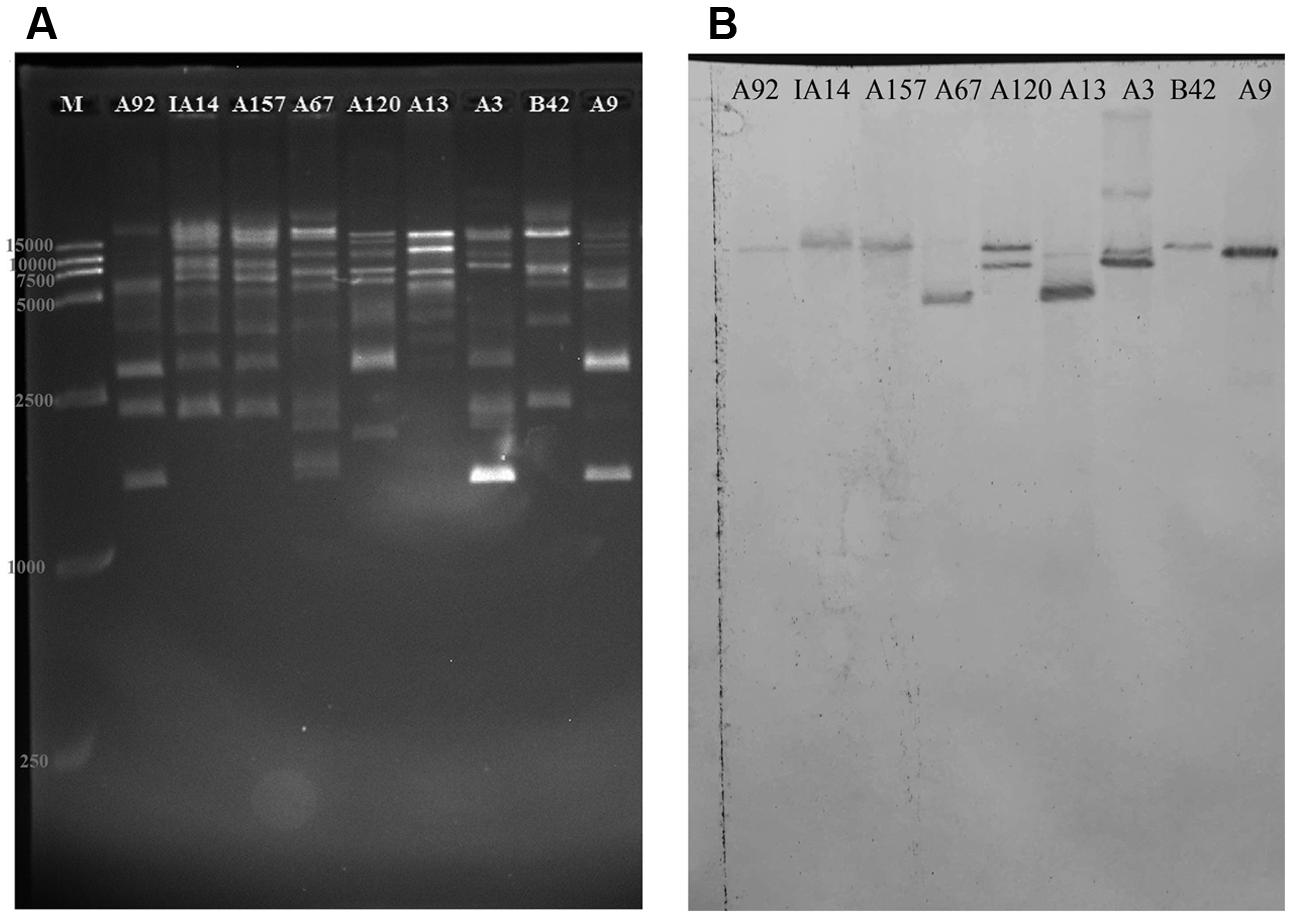
FIGURE 5. Determination of the fosB gene copy number by enzyme BsaI digestion binding hybridization. Shown is (A) agarose (1%) gel electrophoresis in 0.5 × Tris-boric acid-EDTA buffer of BsaI-digested plasmids from the transconjugants of A92, IA14, A157, A67, A120, A13, A3, B42, and A9 stained with Gel Red and visualized under UV light, and (B) Southern blot hybridization with the fosB-latter probe. M is a 15-kb size DNA marker.
The nylon membranes of isolates A67, A13, A9, B42, IA14, A157 and A92 showed a single band, suggesting that there was one fosB copy per plasmid.
With regards to isolate A3, four bands were observed on the hybridization membrane. BsaI-digestion electrophoresis and hybridization blotting of A3 showed the possibility that more than one fosB-carrying plasmid of similar size existed in at least some isolates. The distribution of the four copies of the fosB gene in isolate A3 warrants further investigation using other technologies, such as third-generation sequencing.
The fosB Copy Number Measured by qRT-PCR
To investigate whether fosB copy number has any relevance to the fosfomycin MICs and bacterial growth rate at sub-MIC levels, the fosB copy number in every transconjugant isolate was determined by qRT-PCR. Table 1 shows the total fosB copy number in every isolate, using the housekeeping gene purK as an internal control.
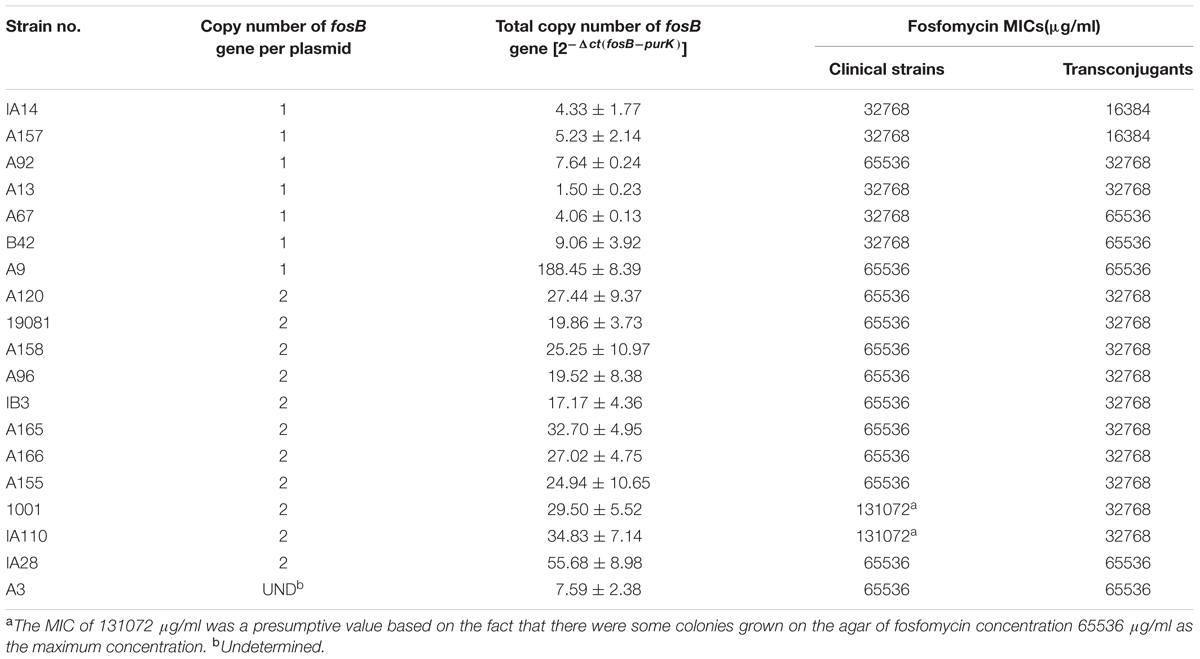
TABLE 1. The fosB copy number and fosfomycin MICs without limitation of 19 vancomycin-resistant enterococci isolates.
The total fosB copy numbers [2-Δct(fosB-purK)] reported in Table 1 range from 17.17 to 55.68 in the isolates with two-copies of fosB per plasmid (transconjugants A165, IA110, 19081, A166, 1001, A96, A155, A158, A120, and IA28). In the A3, A13, A9, A92, B42, IA14, A67, and A157 transconjugants, their total fosB copy number was less than 10, except isolate A9 (188.45).
The Contribution of fosB Copy Number to Fosfomycin MICs
The detailed fosfomycin MICs (without an upper limitation) of 19 VRE isolates and their transconjugants is shown in Table 1. From the susceptibility tests, the MICs of fosfomycin for these isolates and their transconjugants were extremely high, ranging from 16384 to 131,072 μg/ml (Table 1). The MICs of the transconjugants was discussed due to their purer genetic background. As shown in Supplementary Figure S3A, the MIC of fosfomycin did not correlate with the copy number of fosB (Pearson’s R2, 0.1550). For example, although the fosB copy number of isolate A67 and B42 were low, their MICs of fosfomycin were higher than most of the other isolates.
The Effect of fosB Copy Number on Bacterial Growth Rate at Sub-MIC Levels
From a preliminary analysis of isolate A120, fosfomycin concentrations of 1/4 or 1/2 the MIC notably affected the bacterial growth, including a prolonging of the lag phase and a decrease in growth rate and max biomass (data not shown). The growth of the 19 transconjugants following exposure to 1/4 MICs of fosfomycin was measured (Supplementary Table S3), but no correlation was observed between the copy number of fosB and growth rate, maximum optical density (OD600) or lag time (Supplementary Figure S3).
Discussion
Recently, a series of complete genome sequences of Enterococcus have been obtained by high-throughput sequencing (Lam et al., 2012; Boyd et al., 2015; McKenney et al., 2016; Peng et al., 2016). However, the complete sequence of the fosB-carrying Enterococcus plasmid has not been reported. In this study, the complete sequence of a novel E. faecium plasmid containing two copies of fosB was obtained by third-generation DNA sequencing technology, with long read lengths obtained. The coexistence of a vanA transposon and a fosB gene in the same plasmid, which was primarily determined by S1-PFGE in our previous study (Qu et al., 2014), was verified by third-generation sequencing in this report. Furthermore, the fact that E. avium 19081 contained a similar fosB- and vanA-carrying plasmid proved a possibility of interspecies transmission, which could accelerate the dissemination of fosfomycin and vancomycin resistance. The comparison of pEMA120 and three related E. faecium plasmids revealed that their plasmid backbones are relatively conserved. In particular, the fosB-negative E. faecium plasmid pZB18 has a 99% sequence identity with the backbone of pEMA120. It suggested that the two fosB genes along with tnpA and some other sequences, such as IS1485 and a gene encoding a retron-type RNA-directed DNA polymerase, were incorporated into plasmid pZB18 to generate pEMA120. Plasmid pZB18 may have evolved from earlier conjugative plasmids, such as pMG1 (Japan, 1998) and pHTβ (United States, 2003) reflecting a stepwise evolutionary process through several recombination events.
Conjugative transfer is an important means of horizontal spread of antibiotic resistance and an important driver in evolution (Frost et al., 2005; Thomas and Nielsen, 2005; Goessweiner-Mohr et al., 2014). The plasmid pEMA120 was tested to be a conjugative plasmid that can transfer from E. faecium A120 to the recipient strain E. faecium BM4105RF, using filter mating at a rate of 3.4 × 10-4 /donor CFU. The tra region of plasmid pEMA120, of highly homology with that of pZB18, has traB, traA, traC, traI, traG/D, traE and other transfer related genes. It is noteworthy that the gene encoding TraE of plasmid pEMA120 was disrupted, compared with that of plasmid pZB18. The plasmid transfer rate of pZB18 with an intact traE was 7.0 × 10-1 (filter mating) as provided by Zheng et al. (2007), obviously higher than that of plasmid pEMA120. It suggests that the interruption of traE might affect conjugative transfer and would limit the dissemination of this fosB- and vanA-carrying plasmid to some extent compared to plasmid pZB18. However, it cannot be ignored that the transfer rate of 3.4 × 10-4 of pEMA120 still confer high capability of fosB- and vanA co-transmission.
The fosB gene in the E. faecium plasmid has a close phylogenetic relationship with that of Staphylococcus, with more than a 99% shared sequence identity. However, the sequence surrounding the fosB gene is diverse among different genera. Instead of the rep genes that are adjacent to fosB in staphylococcal plasmids (Takeuchi et al., 2005; Fu et al., 2016), there is a tnpA gene (an ISL3-like transposase-encoding gene) downstream of fosB that forms an ISL3-like transposon in enterococcus plasmids, as previously reported (Chen et al., 2014; Qu et al., 2014). This ISL3-like transposon is a 2,516 or 2517-bp element that is flanked by two perfect 25-bp inverse repeats (IRs), which generates two AT-rich 8-bp target duplications upon insertion. ISL3-family elements have been reported to be involved in gene inactivation or transcriptional activation of silent genes in Pseudomonas strains (Kallastu et al., 1998; Christie-Oleza et al., 2008). Recently, Sivertsen et al. (2016) demonstrated that an ISL3-like element insertion upstream of vanHAX mediated a silenced VanA phenotype in enterococci. However, in our study, a similar insertion of the fosB-carrying ISL3-like transposon between vanRS and vanHAX did not cause the transcriptional inactivation of vanA.
In addition to the fosB gene within plasmids, the formation of fosB-carrying circular intermediates that were 2425 or 2426 bp in length was confirmed by inverse PCR in our isolates, as was reported by Xu et al. (2013). Interestingly, two types of circular intermediates with slight sequence differences (26–31 bp) upstream of the fosB gene were observed in the genome of isolate A120, corresponding to the two fosB carrying transposons within plasmid pEMA120. This suggested the two fosB-carrying transposons are not copies but likely result from two separate insertion events. In addition, the junction region of the loop exhibited sequence diversity in a 5-bp DNA sequence between the two IRs of the ISL3-like transposon. The 5-bp intervening DNA sequence was likely derived from the IS flanking regions (Christie-Oleza et al., 2009).
The third generation sequencing has been shown to have advantages in finding multi-copy genes (Guo et al., 2016) because of its genome-wide sequencing scale and high accuracy (McCarthy, 2010). However, the cost of single-molecule, real-time sequencing is high at present. Restriction enzyme digestion and Southern blotting can be a good supplementary tool to explore the copy number of a certain gene in a collection of strains. Here, the hybridization results revealed that the presence of multiple copies of fosB per plasmid was a frequent occurrence in VRE isolates (12 of 19). But this method used in our study had a limitation — if the digested fragments of different fosB copies were in similar sizes, it would be difficult to separate them clearly by agarose gel electrophoresis and distinguish them by Southern blotting. Therefore we also performed quantitative PCR to determine the total copy number of fosB in an isolate. It revealed that the total copy number of fosB in isolates carrying two copies of fosB per plasmid was more than double that of isolates carrying a single copy of fosB per plasmid, possibly due to the higher frequency of fosB-carrying circle formation in the former, as was reported previously (Kallastu et al., 1998). The variable total fosB copy number in different isolates measured by qRT-PCR might be the result of the variety of three factors: fosB copy number per plasmid, plasmid copy number, and probably the amount of fosB-carrying circular intermediates. This complicated existence of fosB could explain why isolate A9 with only one copy of fosB per plasmid has a huge total fosB copy number while A3 is the opposite. Thus, multiple research tools, such as whole-genome sequencing, Southern blotting and quantitative PCR, should be conducted to confirm the distribution and copies of fosB gene in enterococci.
The copy number of some resistance genes has been reported to affect the MICs of the corresponding antibiotics (Bertini et al., 2007). In our study, however, no correlation between the fosB copy number and the fosfomycin MICs was observed. Of note, the fosfomycin MICs observed in this study ranged from 64 to 512 times of 256 μg/ml (the breakpoint of MIC of fosfomycin resistant Enterococcus), far exceeding the normal clinical theoretical dose as well as the maximum serum drug concentration (Cmax) used for intravenous fosfomycin (606 mg/l) (Roussos et al., 2009). In this case, the influence of the fosB copy number on fosfomycin MICs may have minimal practical significance but may be a theoretical reference. However, it is certain that the plasmid-encoded fosB gene, even a single copy, can confer high levels of fosfomycin resistance in enterococci. Subinhibitory concentrations of fosfomycin tended to affect the growth of the bacteria by increasing the lag period and decreasing the growth rate and maximum biomass in our study. However, the copy number of fosB had little correlation with the variance of bacterial growth rate at sub-MIC levels of fosfomycin in our study. That is to say, the increase in fosB copy number did not necessarily help bacterial growth at sub-MIC levels of fosfomycin.
Conclusion
A novel plasmid with two copies of the fosB gene in VRE was first reported in this study. The transfer region and comparative analysis of closely related plasmids suggested that pEMA120 is a pZB18-like conjugative plasmid that has undergone a series of evolutionary changes. The identification of a similar plasmid in E. avium proved the possibility of inter-species dissemination of the fosfomycin resistance. The multiple copies of the fosB gene in plasmids is common in our VRE isolates. However, plasmid-borne fosB, whether single- or multi-copy, can confer a high level fosfomycin resistance. Nevertheless, the fosB ISL3-like transposon, which can translocate to multiple loci in the plasmid, possibly in the form of circular intermediates, may accelerate the dissemination of fosfomycin resistance in VRE.
Author Contributions
Conceived and designed the experiments: YY, YC, TQ, and LS; Performed the experiments: LS, PZ, and KS; Analyzed the data: XH and LS; Wrote the manuscript: YY, LS, TQ, YC, and PZ; All authors read and approved the final manuscript.
Funding
This work was supported by a research grants from the National Natural Science Foundation of China [nos. NSFC81371859 and NSFC81101284].
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01541/full#supplementary-material
References
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bertini, A., Poirel, L., Bernabeu, S., Fortini, D., Villa, L., Nordmann, P., et al. (2007). Multicopy blaOXA-58 gene as a source of high-level resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51, 2324–2328. doi: 10.1128/AAC.01502-06
Boyd, D. A., Levesque, S., Picard, A. C., and Golding, G. R. (2015). Vancomycin-resistant Enterococcus faecium harbouring vanN in Canada: a case and complete sequence of pEfm12493 harbouring the vanN operon. J. Antimicrob. Chemother. 70, 2163–2165. doi: 10.1093/jac/dkv057
Cao, M., Bernat, B. A., Wang, Z., Armstrong, R. N., and Helmann, J. D. (2001). FosB, a cysteine-dependent fosfomycin resistance protein under the control of sigma(W), an extracytoplasmic-function sigma factor in Bacillus subtilis. J. Bacteriol. 183, 2380–2383. doi: 10.1128/JB.183.7.2380-2383.2001
Chandran, V., Fronzes, R., Duquerroy, S., Cronin, N., Navaza, J., and Waksman, G. (2009). Structure of the outer membrane complex of a type IV secretion system. Nature 462, 1011–1015. doi: 10.1038/nature08588
Chen, C., Xu, X., Qu, T., Yu, Y., Ying, C., Liu, Q., et al. (2014). Prevalence of the fosfomycin-resistance determinant, fosB3, in Enterococcus faecium clinical isolates from China. J. Med. Microbiol. 63, 1484–1489. doi: 10.1099/jmm.0.077701-0
Christie-Oleza, J. A., Lanfranconi, M. P., Nogales, B., Lalucat, J., and Bosch, R. (2009). Conjugative interaction induces transposition of ISPst9 in Pseudomonas stutzeri AN10. J. Bacteriol. 191, 1239–1247. doi: 10.1128/JB.01071-08
Christie-Oleza, J. A., Nogales, B., Martin-Cardona, C., Lanfranconi, M. P., Alberti, S., Lalucat, J., et al. (2008). ISPst9, an ISL3-like insertion sequence from Pseudomonas stutzeri AN10 involved in catabolic gene inactivation. Int. Microbiol. 11, 101–110.
Clewell, D. B., Weaver, K. E., Dunny, G. M., Coque, T. M., Francia, M. V., and Hayes, F. (2014). “Extrachromosomal and mobile elements in enterococci: transmission, maintenance, and epidemiology,” in Enterococci: From Commensals to Leading Causes of Drug Resistant Infection, eds M. S. Gilmore, D. B. Clewell, Y. Ike, and N. Shankar (Boston, MA: Massachusetts Eye and Ear Infirmary).
CLSI (2015). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement M100-S25. Wayne, PA: CLSI.
Descourouez, J. L., Jorgenson, M. R., Wergin, J. E., and Rose, W. E. (2013). Fosfomycin synergy in vitro with amoxicillin, daptomycin, and linezolid against vancomycin-resistant Enterococcus faecium from renal transplant patients with infected urinary stents. Antimicrob. Agents Chemother. 57, 1518–1520. doi: 10.1128/AAC.02099-12
Francia, M. V., Varsaki, A., Garcillan-Barcia, M. P., Latorre, A., Drainas, C., and de la Cruz, F. (2004). A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 28, 79–100. doi: 10.1016/j.femsre.2003.09.001
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi: 10.1038/nrmicro1235
Fu, Z., Liu, Y., Chen, C., Guo, Y., Ma, Y., Yang, Y., et al. (2016). Characterization of fosfomycin resistance gene, fosB, in methicillin-resistant Staphylococcus aureus isolates. PLoS ONE 11:e0154829. doi: 10.1371/journal.pone.0154829
Goessweiner-Mohr, N., Arends, K., Keller, W., and Grohmann, E. (2014). Conjugation in gram-positive bacteria. Microbiol. Spectr. 2, LAS–0004–2013. doi: 10.1128/microbiolspec.PLAS-0004-2013
Grant, J. R., and Stothard, P. (2008). The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36, W181–W184. doi: 10.1093/nar/gkn179
Greenwood, D. (1990). Fosfomycin trometamol: activity in vitro against urinary tract pathogens. Infection 18(Suppl. 2), S60–S64. doi: 10.1007/BF01643429
Guo, Q., Ding, B., Jove, T., Stoesser, N., Cooper, V. S., Wang, M., et al. (2016). Characterization of a novel IncHI2 plasmid carrying tandem copies of blaCTX-M-2 in a fosA6-harboring Escherichia coli ST410 strain. Antimicrob. Agents Chemother. 60, 6742–6747. doi: 10.1128/AAC.01173-16
Hamilton-Miller, J. M. (1992). In vitro activity of fosfomycin against ‘problem’ gram-positive cocci. Microbios 71, 95–103.
Ike, Y., Tanimoto, K., Tomita, H., Takeuchi, K., and Fujimoto, S. (1998). Efficient transfer of the pheromone-independent Enterococcus faecium plasmid pMG1 (Gmr) (65.1 kilobases) to Enterococcus strains during broth mating. J. Bacteriol. 180, 4886–4892.
Jensen, L. B., Garcia-Migura, L., Valenzuela, A. J., Lohr, M., Hasman, H., and Aarestrup, F. M. (2010). A classification system for plasmids from enterococci and other Gram-positive bacteria. J. Microbiol. Methods 80, 25–43. doi: 10.1016/j.mimet.2009.10.012
Kallastu, A., Hõrak, R., and Kivisaar, M. (1998). Identification and characterization of IS1411, a new insertion sequence which causes transcriptional activation of the phenol degradation genes in Pseudomonas putida. J. Bacteriol. 180, 5306–5312.
Keating, G. M. (2013). Fosfomycin trometamol: a review of its use as a single-dose oral treatment for patients with acute lower urinary tract infections and pregnant women with asymptomatic bacteriuria. Drugs 73, 1951–1966. doi: 10.1007/s40265-013-0143-y
Lam, M. M., Seemann, T., Bulach, D. M., Gladman, S. L., Chen, H., Haring, V., et al. (2012). Comparative analysis of the first complete Enterococcus faecium genome. J. Bacteriol. 194, 2334–2341. doi: 10.1128/JB.00259-12
McCarthy, A. (2010). Third generation DNA sequencing: pacific biosciences’ single molecule real time technology. Chem. Biol. 17, 675–676. doi: 10.1016/j.chembiol.2010.07.004
McKenney, P. T., Ling, L., Wang, G., Mane, S., and Pamer, E. G. (2016). Complete genome sequence of Enterococcus faecium ATCC 700221. Genome Announc. 4:e00386-16. doi: 10.1128/genomeA.00386-16
Peng, Z., Wang, W., Hu, Y., and Li, F. (2016). Complete genome sequence of Enterococcus hirae R17, a daptomycin-resistant bacterium isolated from retail pork in China. Genome Announc. 4:e00605-16. doi: 10.1128/genomeA.00605-16
Popovic, M., Steinort, D., Pillai, S., and Joukhadar, C. (2010). Fosfomycin: an old, new friend? Eur. J. Clin. Microbiol. Infect. Dis. 29, 127–142. doi: 10.1007/s10096-009-0833-2
Qu, T. T., Shi, K. R., Ji, J. S., Yang, Q., Du, X. X., Wei, Z. Q., et al. (2014). Fosfomycin resistance among vancomycin-resistant enterococci owing to transfer of a plasmid harbouring the fosB gene. Int. J. Antimicrob. Agents 43, 361–365. doi: 10.1016/j.ijantimicag.2013.11.003
Ranotkar, S., Kumar, P., Zutshi, S., Prashanth, K. S., Bezbaruah, B., Anand, J., et al. (2014). Vancomycin-resistant enterococci: troublemaker of the 21st century. J. Glob. Antimicrob. Res. 2, 205–212. doi: 10.1016/j.jgar.2014.04.002
Roussos, N., Karageorgopoulos, D. E., Samonis, G., and Falagas, M. E. (2009). Clinical significance of the pharmacokinetic and pharmacodynamic characteristics of fosfomycin for the treatment of patients with systemic infections. Int. J. Antimicrob. Agents 34, 506–515. doi: 10.1016/j.ijantimicag.2009.08.013
Sivertsen, A., Pedersen, T., Larssen, K. W., Bergh, K., Ronning, T. G., Radtke, A., et al. (2016). A silenced vanA gene cluster on a transferable plasmid caused an outbreak of vancomycin-variable enterococci. Antimicrob. Agents Chemother. 60, 4119–4127. doi: 10.1128/AAC.00286-16
Superti, S., Dias, C. A., and d’Azevedo, P. A. (2009). In vitro fosfomycin activity in vancomycin-resistant Enterococcus faecalis. Braz. J. Infect. Dis. 13, 123–124. doi: 10.1590/S1413-86702009000200010
Takeuchi, F., Watanabe, S., Baba, T., Yuzawa, H., Ito, T., Morimoto, Y., et al. (2005). Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187, 7292–7308. doi: 10.1128/JB.187.21.7292-7308.2005
Tang, H. J., Chen, C. C., Zhang, C. C., Su, B. A., Li, C. M., Weng, T. C., et al. (2013). In vitro efficacy of fosfomycin-based combinations against clinical vancomycin-resistant Enterococcus isolates. Diagn. Microbiol. Infect. Dis. 77, 254–257. doi: 10.1016/j.diagmicrobio.2013.07.012
Tanimoto, K., and Ike, Y. (2008). Complete nucleotide sequencing and analysis of the 65-kb highly conjugative Enterococcus faecium plasmid pMG1: identification of the transfer-related region and the minimum region required for replication. FEMS Microbiol. Lett. 288, 186–195. doi: 10.1111/j.1574-6968.2008.01342.x
Thomas, C. M., and Nielsen, K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721. doi: 10.1038/nrmicro1234
Thompson, M. K., Keithly, M. E., Goodman, M. C., Hammer, N. D., Cook, P. D., Jagessar, K. L., et al. (2014). Structure and function of the genomically encoded fosfomycin resistance enzyme, FosB, from Staphylococcus aureus. Biochemistry 53, 755–765. doi: 10.1021/bi4015852
Tomita, H., Tanimoto, K., Hayakawa, S., Morinaga, K., Ezaki, K., Oshima, H., et al. (2003). Highly conjugative pMG1-Like plasmids carrying Tn1546-Like transposons that encode vancomycin resistance in Enterococcus faecium. J. Bacteriol. 185, 7024–7028. doi: 10.1128/jb.185.23.7024-7028.2003
Xu, X., Chen, C., Lin, D., Guo, Q., Hu, F., Zhu, D., et al. (2013). The fosfomycin resistance gene fosB3 is located on a transferable, extrachromosomal circular intermediate in clinical Enterococcus faecium isolates. PLoS ONE 8:e78106. doi: 10.1371/journal.pone.0078106.t001
Zheng, B., Tomita, H., Xiao, Y. H., Wang, S., Li, Y., and Ike, Y. (2007). Molecular characterization of vancomycin-resistant Enterococcus faecium isolates from mainland China. J. Clin. Microbiol. 45, 2813–2818. doi: 10.1128/JCM.00457-07
Keywords: fosB, fosfomycin resistance, vancomycin resistant enterococci, single-molecule real-time sequencing, copy number
Citation: Sun L, Zhang P, Qu T, Chen Y, Hua X, Shi K and Yu Y (2017) Identification of Novel Conjugative Plasmids with Multiple Copies of fosB that Confer High-Level Fosfomycin Resistance to Vancomycin-Resistant Enterococci. Front. Microbiol. 8:1541. doi: 10.3389/fmicb.2017.01541
Received: 13 March 2017; Accepted: 31 July 2017;
Published: 15 August 2017.
Edited by:
Teresa M. Coque, Instituto Ramón y Cajal de Investigación Sanitaria, SpainReviewed by:
Christopher Morton Thomas, University of Birmingham, United KingdomJose Antonio Escudero, Complutense University of Madrid, Spain
Copyright © 2017 Sun, Zhang, Qu, Chen, Hua, Shi and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunsong Yu, eXZ5czExOUAxNjMuY29t
 Lingyan Sun
Lingyan Sun Ping Zhang
Ping Zhang Tingting Qu
Tingting Qu Yan Chen
Yan Chen Xiaoting Hua
Xiaoting Hua Keren Shi
Keren Shi Yunsong Yu
Yunsong Yu