
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 11 July 2017
Sec. Microbial Physiology and Metabolism
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.01273
Staphylococcus aureus is a worldwide pathogen that colonizes the human nasal cavity and is a major cause of respiratory and cutaneous infections. In the nasal cavity, S. aureus thrives with high concentrations of nitric oxide (NO) produced by the innate immune effectors and has available for growth slow-metabolizing free hexoses, such as galactose. Here, we have used deep sequencing transcriptomic analysis (RNA-Seq) and 1H-NMR to uncover how S. aureus grown on galactose, a major carbon source present in the nasopharynx, survives the deleterious action of NO. We observed that, like on glucose, S. aureus withstands high concentrations of NO when using galactose. Data indicate that this resistance is, most likely, achieved through a distinct metabolism that relies on the increased production of amino acids, such as glutamate, threonine, and branched-chain amino acids (BCAAs). Moreover, we found that under NO stress the S. aureus α-acetolactate synthase (ALS) enzyme, which converts pyruvate into α-acetolactate, plays an important role. ALS is proposed to prevent intracellular acidification, to promote the production of BCAAs and the activation of the TCA cycle. Additionally, ALS is shown to contribute to the successful infection of murine macrophages. Furthermore, ALS contributes to the resistance of S. aureus to beta-lactam antibiotics such as methicillin and oxacillin.
Staphylococcus aureus is an opportunistic human pathogen that is persistently found in the epithelial cells of the nasal cavity of patients suffering recurrent sinusitis (Weidenmaier et al., 2012). Even in the absence of symptoms, the staphylococcal nasal carriage is a known risk factor as S. aureus exhibits high capacity to evade and thwart the innate and adaptive immune responses. Moreover, repeated staphylococcal infections can evolve to life-threatening diseases, such as endocarditis, bloodstream infection, and sepsis (Foster, 2005; Thammavongsa et al., 2015). Unfortunately, antibiotic resistant S. aureus strains are commonly found not only in hospital settings but also in the community, and many strains show resistance to the last generation of antibiotics.
Staphylococcus aureus is a facultative anaerobe that besides the respiratory chain has active Embden-Meyerhof-Parnas (EMP), pentose phosphate pathway (PPP), and citric acid cycle (TCA) pathways (Figure 1). As for other pathogens, the synthesis and acquisition of certain nutrients are critical to support the infection capacity of S. aureus, glucose being one of the preferred carbon sources of this bacterium. However, in the nasal cavity the glucose levels have been proposed to be substantially lower than those determined in human plasma, and in this niche, disaccharides and other free hexoses are accessible (Krismer et al., 2014). In particular, galactose is one of the major carbohydrates present in the mucus mucins that coat the nasopharyngeal epithelium of the nasal cavity, and studies done with the nasal colonizer S. pneumoniae showed that: (i) galactose is released by hydrolysis of mucins promoted by bacterial exoglycosidases; (ii) free galactose is abundantly present in the nasal lavage fluid, whether healthy individuals carry or not the pneumococcus; and (iii) glucose was not detected in S. pneumoniae carriers and non-carriers (King, 2010; Blanchette et al., 2016).
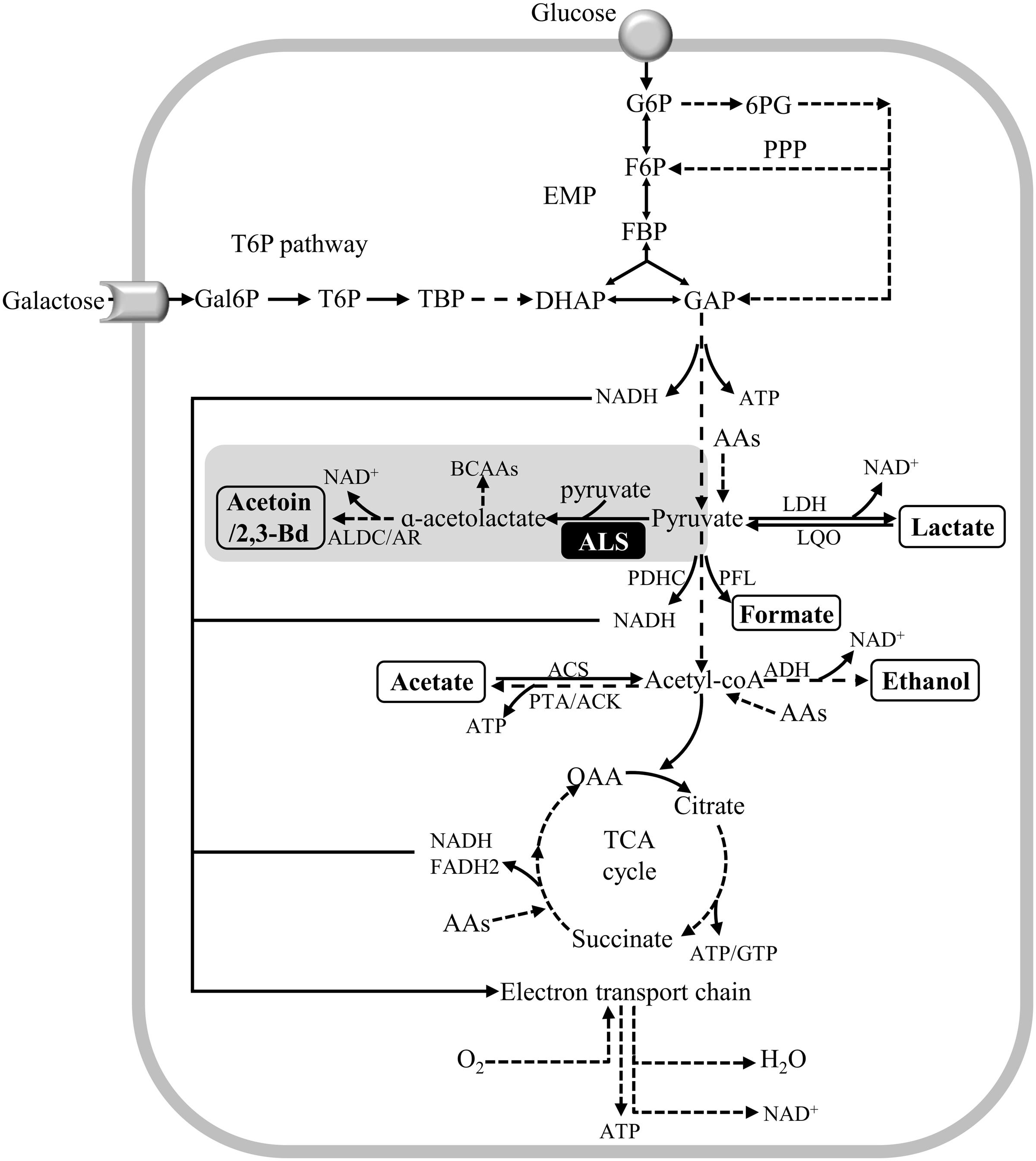
FIGURE 1. Schematic representation of metabolic pathways of Staphylococcus aureus glucose and galactose catabolism. Galactose is catabolized in the tagatose 6-phosphate pathway into the glycolytic intermediates DHAP and GAP. DHAP and GAP originated from galactose and glucose are oxidized to pyruvate in the EMP (glycolysis) pathway, which can be reduced to lactate, α-acetolactate and/or acetyl-CoA. Acetyl-CoA can be converted to acetate, to generate ATP, and ethanol. Alternatively, acetyl-CoA can be used to produce citrate for the citric acid cycle (TCA cycle). Under aerobic conditions, the NADH molecules formed in the EMP and TCA cycle are oxidized in the respiratory chain, thereby restoring the redox balance, and producing ATP. Proposed pathways were reconstructed based on genome information (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi), literature and database surveys (KEGG, MetaCyc). Inside boxes are the end products of pyruvate metabolism. Inside the black box is the ALS enzyme. AAs, amino acids; ACS, acetyl-CoA synthetase; ADH, bifunctional acetaldehyde-CoA/alcohol dehydrogenase; ALDC/AR, α-acetolactate decarboxylase/acetoin reductase; ALS, α-acetolactate synthase; BCAAs, branched-chain amino acids; DHAP, dihydroxyacetone phosphate; FBP, fructose 1,6-bisphosphate; F6P, fructose 6-phosphate; Gal6P, galactose 6-phosphate; GAP, glyceraldehyde 3-phosphate; G6P, glucose 6-phosphate; LDH, lactate dehydrogenase; LQO, lactate-quinone oxidoreductase; PDHC, pyruvate dehydrogenase complex; PFL, pyruvate formate-lyase; 6-PG, 6-phosphogluconate; PPP, pentose phosphate pathway; PTA/ACK, phosphotransacetylase/acetate kinase; OAA, oxaloacetate; TBP, tagatose 1,6-bisphosphate; T6P, tagatose 6-phosphate.
In the nose and paranasal sinuses epithelium, the inducible NO synthase, which is expressed in cells of the innate immune system, generates NO. This molecule has antibacterial and cilia-stimulating properties playing a central role in the host immune defenses (Lanz et al., 2008). The infection of human phagocytes by pathogens like S. aureus stimulates the NO production. The antimicrobial properties of NO and its derivatives, known as the reactive nitrogen species (RNS), rely on several damaging effects (Richardson et al., 2006; Robinson et al., 2014). Yet, in general, pathogens resist NO through the activation of detoxifying enzymes, repairing proteins and complex metabolic adaptations, and S. aureus has an unique ability to thrive in NO rich environments (Gonçalves et al., 2006; Richardson et al., 2008; Tavares et al., 2009; Fuller et al., 2011; Nobre and Saraiva, 2014).
The preferential glycolytic carbon sources of S. aureus in the presence of high NO concentrations are hexoses (e.g., glucose and mannose) (Vitko et al., 2015), and NO-exposed glucose-grown cells of S. aureus use glycolysis (EMP) as the primary central metabolic pathway (Hochgräfe et al., 2008; Richardson et al., 2008; Fuller et al., 2011; Crooke et al., 2013; Vitko et al., 2015; Spahich et al., 2016). In contrast, gluconeogenic or non-glycolytic carbon sources (e.g., glycerol, fatty acids, amino acids) fail to provide resistance to NO, as this compound impairs the respiratory chain and partially inhibits the PDHC and the PFL activities (Richardson et al., 2008; Vitko et al., 2015) (Figure 1). The fluctuating glucose levels occurring in the nasal cavity implies that S. aureus has to resort to other carbon sources, which led us to examine how S. aureus sustains NO stress when utilizing other hexoses available in the nasal niche (King, 2010; Krismer et al., 2014; Blanchette et al., 2016). In particular, galactose is important for the colonization of S. aureus, as studies have shown that: (i) S. aureus binds human nasal galactose rich mucins during colonization (Shuter et al., 1996); (ii) S. aureus binds galactose molecules in host glycoproteins (Sakarya et al., 2010); (iii) the S. aureus tagatose 6-phosphate pathway is essential for the catabolism of lactose/galactose, which are sugars present in milk and derivative products (Miallau et al., 2007); and (iv) the malfunctioning of the S. aureus tagatose 6-phosphate pathway leads to accumulation of the dietary D-galactose causing bacterial cell death (Miallau et al., 2007). While in many bacteria, galactose is predominantly metabolized through the Leloir pathway, in S. aureus the galactose breakdown occurs through the tagatose 6-phosphate pathway. In this bacterium, galactose is transported and phosphorylated via a galactose phosphotransferase system and converted into the glycolytic intermediates DHAP and GAP (Figure 1). In general, the tagatose 6-phosphate pathway is less efficient than the Leloir pathway for the catabolism of galactose, mainly due to the low-affinity for galactose of the phosphotransferase systems and thus, to the slow transport/consumption of the sugar (Neves et al., 2010). In this work, we have analyzed for the first time the effect of NO on cells grown on galactose. We report that although considered a slow-metabolizing sugar, galactose ensures growth of S. aureus in the presence of NO by diverting its metabolism to the production of specific amino acids via activation of several enzymes, namely the ALS. In this work, this enzyme is shown to be important for the protection of S. aureus against NO and antibiotics.
Staphylococcus aureus JE2, a plasmid-cured derivative of the community-associated methicillin-resistant USA300 LAC strain, and the JE2 alsS transposon mutant (NE1397, JE2 alsS::ΦNΣ; EmR) of the Nebraska Transposon Mutant Library (UNMC) were obtained from the Network on Antimicrobial Resistance in S. aureus (NARSA, Chantilly, VA, United States). The alsS mutation was confirmed by PCR according to the Nebraska-UNMC instructions1, and using the following primers: 5′-GCTTTTTCTAAATGTTTTTTAAGTAAATCAAGTAC-3′ and 5′-CATCAAAGTATTGATAATGCTGCG-3′ (alsS-specific).
Routinely, S. aureus was grown aerobically at 37°C and 150 rpm in TSB broth (Difco), and when required, erythromycin was added at 10 μg mL-1. For the transcriptomic and metabolic studies, S. aureus was cultured in chemically defined medium (CDM) containing glucose or galactose as carbon source (20 mM), phosphate buffer, trace minerals, vitamins, nucleobases, nitrogen and sulfur sources and amino acids (Ferreira et al., 2013). The nasal synthetic medium (SNM3), described by Krismer et al. (2014), was also used for the analysis of extracellular metabolites by 1H-NMR. However, to ensure a metabolite concentration detectable by 1H-NMR, glucose was supplemented at higher concentrations (10 mM). All media used for the NMR studies contained no paramagnetic ions to avoid interference with the spectral quality and allow detection of metabolites.
The spermine NONOate (30–40 mM), that was freshly prepared in 0.01 M NaOH and kept protected from light, was used as a NO donor since its half-life (∼39 min at 37°C) is similar to the S. aureus cellular doubling time.
Cultures at an initial OD600 of 0.05–0.1 were prepared by addition to fresh medium of pre-cultured cells harvested at the exponential phase (OD600 of 1). Cells at an OD600 of 0.4, were left untreated or exposed to spermine NONOate (250 μM, Sigma), and growth was monitored hourly.
Staphylococcus aureus JE2 cells grown on galactose-CDM to an OD600 of 0.4 were treated with 250 μM spermine NONOate (or left untreated, the control sample), and harvested after 3 h. Total RNA was isolated from two biologically independent samples following the previously described procedures (Carvalho et al., 2011). RNA samples were analyzed in an Ion ProtonTM Sequencer by the PrimBio company (Primbio Research Institute, Innovation Center, Eagleview, Philadelphia, PA, United States). The transcriptome was analyzed by high-resolution RNA-Seq, and the statistical and functional data analysis was done with the Webserver Pipeline T-Rex (de Jong et al., 2015) and Gene Set Enrichment Analysis for prokaryotes (GSEA-Pro) web tool2. RNA-Seq data was deposited at Gene Expression Omnibus (GEO) under the accession number GSE99563. Genes were considered with a significantly modified expression for adjusted P-values ≤ 0.05 and fold changes of ≥ 2 or ≤ 0.5.
The RNA-Seq data was confirmed by real-time quantitative reverse transcription–polymerase chain reaction (qRT-PCR). For these experiments, S. aureus was grown as described above for the RNA-Seq studies, and the total RNA was extracted from cells with the High Pure RNA Isolation Kit (Roche), and further purified by DNase treatment with the Turbo DNA-freeTM Kit (Ambion®). The absence of chromosomal DNA in RNA samples was confirmed by PCR using oligonucleotides for the S. aureus 16S gene (Supplementary Table S1). RNA concentration and purity was determined by gel electrophoresis and in a NanoDrop 2000c UV-Vis Spectrophotometer (ThermoScientific). Total RNA (∼3 μg) served to synthesize cDNA by means of the Transcriptor High Fidelity cDNA synthesis Kit (Roche). Real-time qRT-PCR reactions containing ∼300 ng cDNA template, 0.5 μM gene-specific oligonucleotides (Supplementary Table S1), H2O PCR-grade and LightCycler® 480 CYBR Green Master Mix in 20 μL were carried out in a Roche LightCycler LC480 Instrument following the LightCycler®480 SYBR Green I Master Kit manufacturer’s instructions (Roche). The expression ratios of the selected genes were normalized relative to S. aureus 16S rRNA reference cDNA, whose expression does not vary under the tested conditions, and calculated by the 2-ΔΔCT method (Livak and Schmittgen, 2001). The data are an average of two independent biological samples.
Cells were grown on galactose-CDM, glucose-CDM and in the nasal synthetic medium to an OD600 = 0.4 and treated with 250 μM spermine NONOate (or left untreated, the control sample), and harvested 1 and 3 h later. After centrifugation (11800 ×g for 2 min) and filtering (0.22 μm Pall filters), the supernatants were stored at -20°C and later used for the 1H-NMR studies. Extracellular metabolites were quantified on a Bruker AVANCE II500 MHz spectrometer (Bruker BioSpin GmbH) operated by TOPSPIN software using a 5 mm BBIXYZ high resolution probe head, at 16°C, and standard Bruker pulse programs. Spectra were referenced to the resonance of externally added trimethylsilyl propionate (TSP), designated at 0 ppm.
Resonances were assigned by addition to the supernatants of pure compounds and comparison with data available in the literature (Carvalho et al., 2011). Concentrations were calculated from the areas of the resonances of the 1H-NMR spectra considering the TSP resonance area, and using an appropriate resonance saturation correction factor (Carvalho et al., 2011). The metabolites concentration was normalized to the OD600, measured for each condition.
Cell cultures (5–10 mL) harvested 3 h after the NO addition were pelleted (11800 ×g, 2 min), immediately flash frozen into liquid nitrogen and stored at -20°C for the next-day analyses. For extraction of NADP+ and NADPH, the pellets were resuspended in 0.4 N HClO4 and 0.4 N KOH, respectively, incubated on ice for 15 min, and heated at 60°C for 7 min. The resulting extracts were cooled on ice for 10 min, and the cells debris were pelleted at 3000 ×g for 10 min for the NADP+ extraction, and at 10,000 ×g for 15 min for NADPH. The extracts were filtered and neutralized by slow addition of NaOH or HCl (2–5 M), and kept on ice (based on Lilius et al., 1979).
NADPH was enzymatically converted to NADP+ following the procedure described by Vaseghi et al. (1999), and the NADP+ concentration was assayed by the enzymatic cycling method (Vaseghi et al., 1999) with minor modifications. Briefly, a reaction mixture containing 0.1 M bicine, 0.1 M nicotinamide, 1 mM PES (phenazine ethosulfate), 4.2 mM MgSO4, 12.7 mM G6P (glucose 6-phosphate) and dinucleotide extracts in a final volume of 250 μL was prepared. The reaction was initiated by addition of glucose 6-phosphate dehydrogenase (G6PDH) and thiazoylyl blue tetrazolium bromide to final concentrations of 2.8 U and 0.25 mM, respectively. Water was used as blank and standard curves were obtained with β-NADP+ (Sigma). Reactions were monitored at 570 nm, for 30 min, using a Multiskan GO Microplate Spectrophotometer (Thermo Fisher Scientific).
Murine macrophages J774A.1 (LGC Promochem) were cultured on 24-well plates, containing Dulbecco’s modified Eagle medium (31966 – DMEM, Gibco) supplemented with 4.5 g L-1 glucose, 110 mg L-1 sodium pyruvate, 862 mg L-1 glutamine, 15 mg L-1 phenol red, 10% fetal bovine serum, and 50 μg mL-1 streptomycin, and incubated in a 5% CO2 containing atmosphere (BINDER CO2 incubator, United States) at 37°C, for 24 h. Prior to infection, macrophages were activated by incubation (5 h) with IFN-γ (1 μg mL-1) and LPS (5 μg mL-1), both from Sigma.
Staphylococcus aureus wild type and alsS mutant were grown aerobically in LB to an OD600 of 0.4–0.5, washed three times with PBS, and resuspended in antibiotic-free DMEM medium (Gibco) to an initial bacterial concentration of about 1 × 107 CFU mL-1. Bacteria were used to infect macrophages at a MOI of ∼5 and, after 30 min the extracellular bacteria were eliminated by a 10 min treatment with DMEM supplemented with 50 μg mL-1 gentamicin. Macrophages were washed with PBS and fresh DMEM (1 mL) was added, followed by a 5 h incubation. When required, 800 μM L-NMMA (Sigma) was added to DMEM to promote inhibition of the murine macrophage inducible NO synthase (iNOS). Macrophages were lysed with 2% saponin, and the number of intracellular bacteria determined by CFU counting.
The NO accumulation in the supernatants of the macrophages cell cultures, which reflects the iNOS activity, was assessed by measuring nitrite by the Griess method, which uses sodium nitrite (Merck) as standard.
Minimal inhibitory concentrations (MICs) were carried out by the broth tube dilution test. Briefly, 96-well plates containing TSB were inoculated to an initial OD600 of 0.01 with exponentially TSB-grown cells of S. aureus wild type and alsS mutant. Beta-lactam antibiotics methicillin and oxacillin were added to bacterial cell suspensions in a concentration range of 2–32 μg mL-1. Plates were incubated at 37°C and 90 rpm, for 18 h. The antibiotic concentration of the first well of the series with no sign of visible growth was considered the MIC value.
Statistical analyses were performed using GraphPad Prism (GraphPad software version 5.01, San Diego, CA, United States). Results were compared using two tailed unpaired Student’s t-tests with a confidence interval of 95%. Growths, metabolite quantification, macrophage assays, and MICs were performed at least in triplicate (at least three biological independent experiments). All data is presented as mean ± standard error of the mean (SEM).
The hexoses, glucose and galactose are two main glycolytic carbon sources that S. aureus has available in the nasal cavity where the NO levels are high. In this work, we have determined the resistance to nitrosative stress of S. aureus grown on galactose and observed that, similar to glucose, spermine-NONOate (250 μM) decreased the growth rate by ∼2-times, and that higher concentrations of spermine-NONOate (500 μM) caused no further decrease (Supplementary Figure S1). These results show that the resistance of S. aureus to high concentrations of NO is similar for cells grown on galactose or on glucose.
To elucidate how S. aureus sustains the nitrosative stress, the metabolic state of S. aureus grown on galactose and exposed to NO was studied by 1H-NMR quantification of the glycolytic carbon source consumption, the citrate uptake from the growth medium to feed the TCA cycle, and the excreted end products, namely those formed downstream of the pyruvate node (Figure 1). For comparison purposes, cells grown on glucose were also analyzed. Thus, supernatants were collected from cells cultured without glycolytic substrate, with either glucose or galactose, treated with 250 μM spermine NONOate, and analyzed 1 and 3 h after the addition of the NO compound (Figures 2A–C). Control samples of untreated cells were collected at the exponential phase as their metabolism remained essentially unchanged due to a quasi steady-state observed at this growth stage (data not shown).
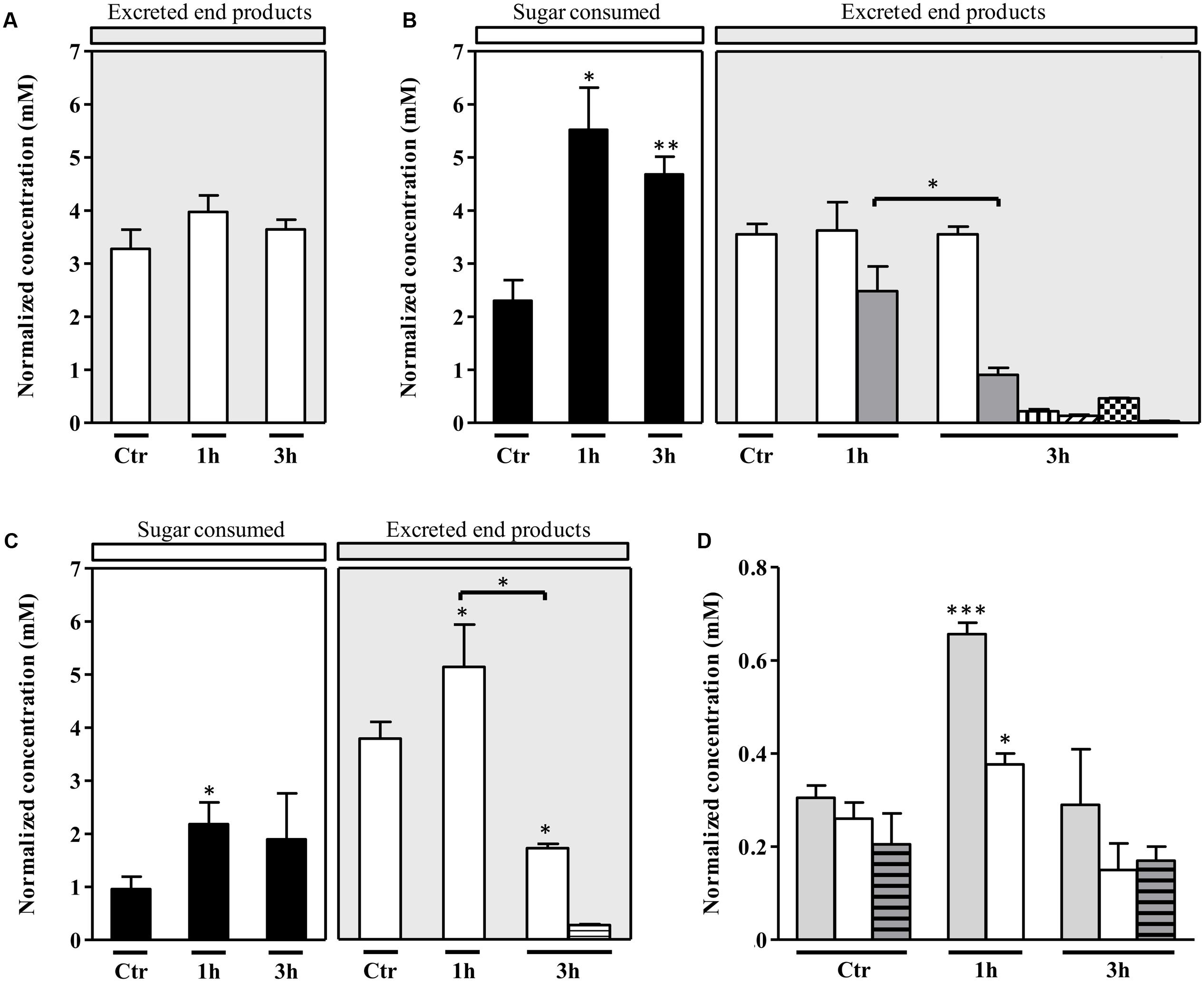
FIGURE 2. Extracellular metabolite profile of S. aureus exposed to NO. Substrate consumed and major end products accumulated by S. aureus grown in CDM and (A) with no glycolytic carbon source, (B) 20 mM glucose, (C) 20 mM galactose, in the absence of NO (control, Ctr) and 1 and 3 h after the addition of NO. Color code of bars of graphs A, B, and C: black, sugar consumed; white, acetate; gray, lactate; horizontally striped, formate; vertically striped, pyruvate; right transversally striped, α-acetolactate; black squares, acetoin and left transversally striped, 2,3-butanediol. (D) Citrate consumption by S. aureus grown with no glycolytic carbon source (light gray bars), on glucose (white bars) and on galactose (gray horizontally striped bars), in the absence of NO (control, Ctr) and treated for 1 and 3 h with NO. The concentrations represented in Y-axis are normalized to OD600. Error bars represent mean ± SEM (n = 3). Asterisks represent statistically significant data relative to the control; ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.0001.
In the absence of a glycolytic substrate, the 1H-NMR profile indicated that S. aureus excreted almost exclusively acetate, in amounts that did not vary significantly upon exposure to NO (Figure 2A).
In S. aureus cells cultured on glucose and stressed with NO, the consumption of glucose was more accentuated at the earlier times of NO treatment (after 1 h) (Figure 2B). Moreover, short exposure of cells to NO induced a considerable accumulation of L-lactate (Figure 2B), together with very small quantities of ethanol (Supplementary Table S2). Between 1 and 3 h after the NO pulse, a small increase of the lactate concentration in the extracellular medium (from 1.6 ± 0.4 mM to 2.4 ± 0.5 mM) (Supplementary Table S2), and a decrease of the normalized lactate concentration was observed (Figure 2B). These alterations may result from the reduction of the lactate dehydrogenase LDH activity and/or higher consumption of lactate by lactate-quinone oxidoreductase LQO (Figure 1) in NO treated cells, as it was also observed by Spahich et al. (2016).
The spectra of supernatants collected after 3 h of NO stress showed peaks for pyruvate, α-acetolactate (generated from the condensation of two molecules of pyruvate), acetoin and 2, 3-butanediol (Figure 2B), the last three being products of ALS (encoded by alsS), α-acetolactate decarboxylase ALDC (encoded by budA) and acetoin reductase AR (encoded by butA) activity, respectively (Figure 1). Interestingly, such a pattern was not observed in cells that were not subjected to NO stress (Figure 2B). In addition, the acetate accumulated in untreated cells, and the end products measured at 1 and 3 h after the NO addition do not account for all the glucose consumed.
The citrate consumption from the glucose-containing medium was also evaluated and the results showed that citrate utilization initially increased after the NO pulse, followed by a decrease after 3 h of stress (Figure 2D). This result may be interpreted considering that at the onset of the NO stress, the carbon flow in the TCA cycle is higher and the pyruvate, resulting from glucose catabolism, is not only converted to end products but is also diverted to the TCA cycle (Figures 1, 2).
On galactose, unstressed S. aureus cells excreted mainly acetate (Figure 2C) and minor amounts of ethanol (Supplementary Table S2). In the presence of NO, cells doubled the galactose consumption and excreted higher amounts of acetate (Figure 2C). For longer times of exposure to NO, the galactose consumption did not vary significantly and a meaningful (P-value, 0.0130) decrease of the acetate produced occurred together with a slight accumulation of formate (Figure 2C). In contrast to what was observed on glucose, cells grown on galactose did not accumulate pyruvate, α-acetolactate, acetoin, and 2,3-butanediol (Figure 2C). Moreover, no production of lactate was detected, which is in agreement with the requirement of glucose for the activation of lctE (encoding LDH) (Crooke et al., 2013).
Also on galactose, cells prolonged exposed to NO showed no major alteration of the total extracellular concentration of acetate [2.7 ± 0.6 mM (1 h) and 2.7 ± 0.8 mM (3 h)] (Supplementary Table S2) and of galactose consumed (Figure 2C), which suggests a metabolic constraint at the level of phosphotransacetylase (PTA)/acetate kinase (ACK) and/or acetate consumption. This impairment may lead either to the conversion of pyruvate into α-acetolactate or to an increase of the TCA cycle flux (Sadykov et al., 2013) (Figure 1). Cells grown on galactose in the absence of stress or NO-treated for 3 h showed similar consumption of extracellular citrate (Figure 2D), which reinforces the hypothesis toward the formation of α-acetolactate through ALS (Figure 1). After 1 h of the NO pulse, cells accumulated citrate (0.11 ± 0.02, concentration normalized to OD600), indicating a reduction of the TCA cycle activity. Additionally, S. aureus cells submitted to NO for 1 h contained an extracellular acetate content higher than that expected on the basis of the galactose consumed (Figure 2C). This result may be due to the utilization of non-glycolytic/gluconeogenic carbon sources such as amino acids (Figure 1). We also observed that after 3 h of the NO pulse, the extracellular content of end products (acetate and formate) did not account for all the galactose consumed.
Overall, our results show that during prolonged exposure to NO, S. aureus grown on galactose exhibits an unprecedented metabolic behavior when compared with that previously reported for cells grown on glucose and long exposed to NO.
To further elucidate the metabolic behavior of S. aureus on galactose during prolonged NO exposure, a RNA-Seq analysis was done using RNA extracted from cells grown on galactose and submitted to 250 μM spermine NONOate for 3 h. Following data acquisition, the RNA-Seq results were analyzed using the T-Rex pipeline web tool http://genome2d.molgenrug.nl/index.php/rnaseq-expression-analysis (de Jong et al., 2015) (Supplementary Table S3), and the data validated by quantitative real-time RT-PCR (Supplementary Table S4).
Exposure of S. aureus to NO altered significantly the expression of approximately 514 genes (fold-differences ≥ 2 and ≤ 0.5 with adjusted P-values ≤ 0.05), which represents approximately 19% of the whole genome. Of these, 213 genes were upregulated (8%) and 301 genes were downregulated (11.4%) (Supplementary Table S3 and Figure S2).
To further interpret the data, a Gene Set Enrichment Analysis (GSEA) of the modified genes was performed, using the pipeline web tool http://pepper.molgenrug.nl/index.php/gsea-pro (GSEA-Pro, MolGen, RUG). This analysis showed that among the upregulated genes involved in central carbon metabolism, the majority are related with metabolic processes and catalytic activities, such as the amino acid and pentose phosphate metabolisms (Supplementary Table S5). The more over-represented classes include genes involved in glycine, serine and threonine metabolism, intracellular trafficking, secretion and vesicular transport and iron transport, storage and homeostasis (Supplementary Table S5).
Amongst the most induced genes belonging to the amino acid transport and metabolism categories were those involved in the synthesis of the gluconeogenic amino acid threonine (Halsey et al., 2017) (thrA, asd, dhoM, thrB, thrC) that were strongly upregulated (∼10-fold). The genes for the synthesis of other gluconeogenic amino acids, such as glycine (gLY1), glutamate (gltB, gltD) and the BCAAs valine and leucine (ilvN) (Supplementary Table S3) were also upregulated, although to a lesser extent. In contrast, NO caused the downregulation of genes involved in respiration, namely those encoding the ATP synthesis, hydrolysis coupled proton transporters, the respiratory electron transport chain and cytochromes (Supplementary Table S3). Other pathways were also altered: (i) the TCA cycle, as judged by the upregulation of the citrate synthase gltA (2.2-fold) gene; and (ii) the PPP, by the induction of the genes glucose 6-phosphate 1-dehydrogenase zwf (2.1-fold), gluconate kinase gntK (2.2-fold) and transaldolase taI (2.9-fold). Overall, these results suggest that cells of S. aureus using galactose and exposed to NO rely on the synthesis of gluconeogenic amino acids and gluconeogenic processes to support growth on the slow-metabolizing galactose.
D-glutamate, generated from L-glutamate by the activity of glutamate racemase (murI), and glycine are cell wall constituents, and BCAAs are precursors of membrane branched-chain fatty acids (BCFA). We observed that genes coding for cell wall, membrane and envelope biogenesis related proteins were significantly induced (Supplementary Table S5). Of note was the induction of genes involved in the synthesis of capsule cap5 (2.1-fold), in adherence and nasopharyngeal colonization, such as sasF (3.9-fold), sdrC (12.3-fold), and sdrD (2.5-fold) (Supplementary Table S3) (Edwards et al., 2012). The modified transcription was also observed for the genes involved in the fatty acid metabolism, namely the fatty acid degradation genes fadXEDBA (23-fold), the fatty acid biosynthesis genes fabHF (4-fold) and fabDG (2.5-fold) (Supplementary Table S3). Altogether, the data suggest that cells of S. aureus using galactose and exposed to NO change their membrane composition to adapt to the new conditions.
The expression of several iron-regulated genes was induced, including that of ftn (14.3-fold) and feoA (4.1-fold) that code for a non-heme ferritin family protein and the ferrous iron transport protein A, respectively, and that of the siderophore biosynthesis gene sbnA (2.2-fold) (Supplementary Table S3). On the contrary, the methionine biosynthesis genes metCFE exhibited down-regulation of approximately 2.5-fold (Supplementary Table S3). Interestingly, the iron-regulated genes and methionine biosynthesis genes that were differentially expressed in S. aureus, when grown on galactose and under prolonged NO stress, exhibited trends similar to those observed in the transcriptome of S. aureus done on samples collected from the anterior nares (Chaves-Moreno et al., 2016).
Several transcriptional regulators had their expression modified, namely TetR, MerR, ArgR, GntR, and DeoR (Supplementary Table S3). The cidR gene was also upregulated by NO, cidR encodes a LysR-type family regulator that controls the cidABC and alsS/budA operons (Supplementary Table S3). In accordance, the upregulation of alsS/budA was observed and confirmed by quantitative real-time RT-PCR (Supplementary Tables S3, S4). Interestingly, our previous work also showed that S. aureus cells grown on glucose and exposed to GSNO had high induction of alsS (∼16-fold) (Nobre and Saraiva, 2013), indicating that ALS confers resistance to NO independently of the sugar source.
Genes involved in the pyruvate metabolism were affected such as the lactate dehydrogenase lctE gene that had its transcription decreased, a pattern that is similar to that observed in cells grown on glucose in the presence of GSNO (Nobre and Saraiva, 2013).
Also downregulated were the genes coding for the PFL (∼3-fold) and eutD encoding phosphotransacetylase (PTA) (∼2-fold), which is the first enzyme of the acetate pathway (Supplementary Table S3 and Figure 1) (Halsey et al., 2017).
Altogether, the modifications induced by prolonged NO exposure on the genes encoding proteins acting downstream of the pyruvate node were consistent with the metabolic profile determined by the 1H-NMR experiments (Figure 2C).
The growth of S. aureus on galactose caused high induction of the cap5A, sdrC, thrA, and fadE genes that are linked to cell wall components, amino acid and fatty acid metabolism. This behavior contrasts with what was observed for cells grown on glucose (Supplementary Table S4), therefore, suggesting a specific galactose-linked response. The alsS/budA operon was upregulated both on galactose- and glucose-grown S. aureus cells subjected to prolonged NO stress (Supplementary Table S4), indicating that alsS is important independently of the glycolytic carbon source utilized. Moreover, this response is related to a prolonged exposure to NO, as galactose-grown cells exposed to 1 h of NO stress caused the downregulation of the alsS and budA genes (0.4 ± 0.0 and 0.6 ± 0.2, respectively), while the expression of these genes did not vary significantly in glucose-grown cells (data not shown).
Taken together, the RNA-Seq and qRT-PCR data indicate that when S. aureus uses galactose and has prolonged exposure to NO stress, alsS is highly upregulated as well as the genes for the biosynthesis of gluconeogenic amino acids, such as threonine. Moreover, the gluconeogenic processes and the biosynthesis of gluconeogenic amino acids are activated.
As mentioned above, the genes involved in the class of amino acid transport and metabolism were highly induced in NO treated S. aureus galactose-grown cells (Supplementary Table S3). Therefore, we performed the quantification by 1H-NMR of the extracellular amino acids, and the results showed that under these conditions cells had a considerable consumption of alanine, and a slightly higher consumption of BCAAs, such as isoleucine and valine (Figure 3), but suffered depletion of glycine, glutamate and threonine (Supplementary Figure S3). It was also seen higher accumulation of isovaleric acid and 2-ketobutyric acid (Supplementary Figure S4), which are products of leucine and valine degradation, respectively, most possibly due to an increased requirement of acetyl-CoA.
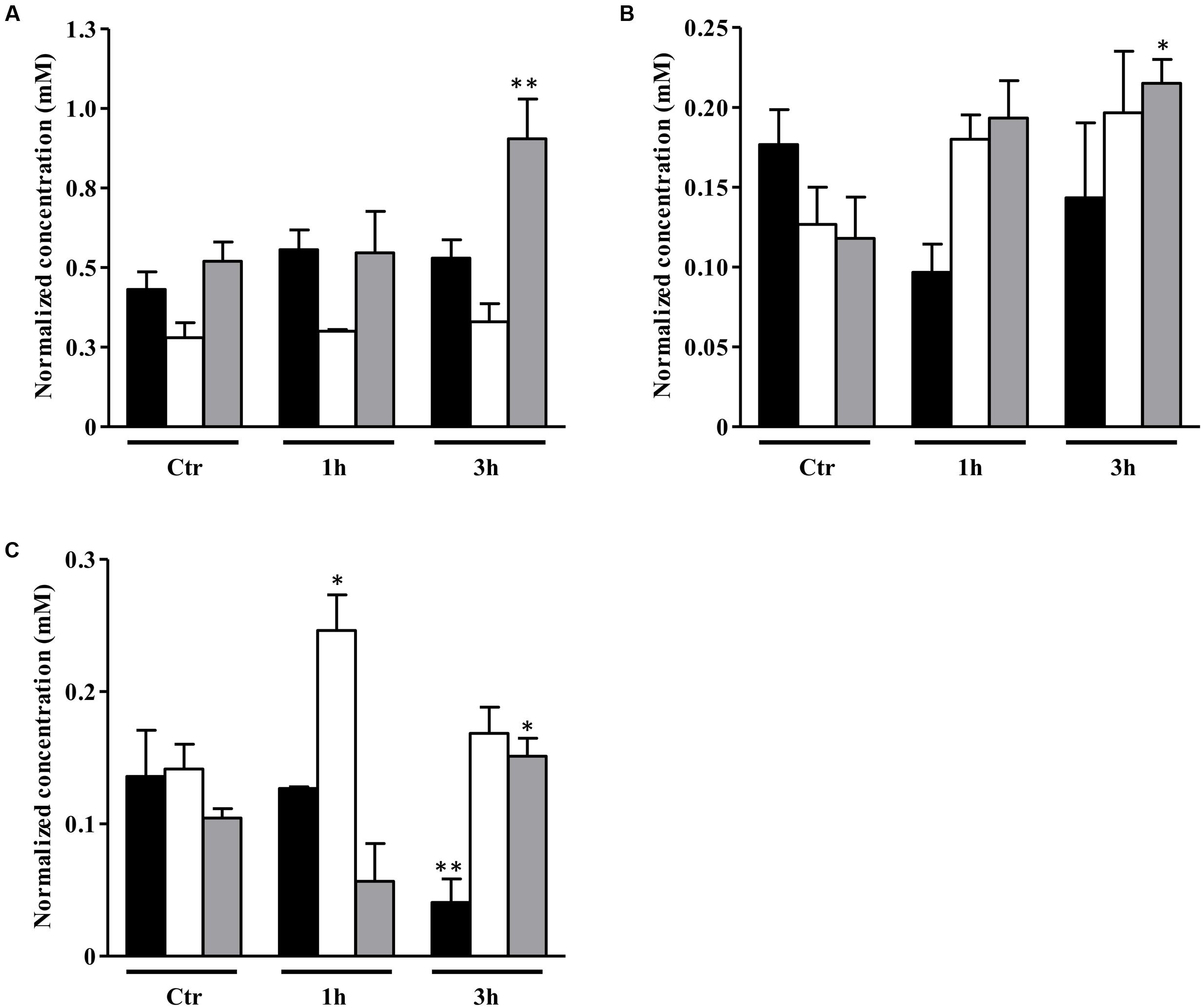
FIGURE 3. Uptake of amino acids by S. aureus cells exposed to NO. Uptake of alanine (A), isoleucine (B), and valine (C) by S. aureus grown in CDM with no glycolytic carbon source (black bar), on glucose (white bar) or on galactose (gray bar), in the absence (Ctr), and treated with NO, for 1 and 3 h. Error bars represent mean ± SEM (n = 3). Asterisks represent statistically significant data relative to the control; ∗P < 0.05 and ∗∗P < 0.01.
Amino acid biosynthesis requires the cofactor NADPH, whose major intracellular source is the PPP that is also used for the production of fatty acids and nucleic acids. A matrix analysis of the RNA-Seq data indicated a high correlation among the PPP genes (zwf SAUSA300_1454, taI SAUSA300_1725, gntK SAUSA300_2443) and amino acid biosynthesis genes (gntR SAUSA300_2444, ilvN SAUSA300_2008, thrA SAUSA300_1225, gLY1 SAUSA300_1214, gltB SAUSA300_0445) (Figure 4). Since the reactions of the PPP are dependent on NADP+, the NADP+/NADPH ratio was determined. Interestingly, following 3 h of NO pulse, cells grown on galactose exhibited a significantly lower NADP+/NADPH ratio when compared with cells using glucose (Figure 5), indicating that the utilization of galactose increases the metabolic activity of the PPP. A similar behavior was reported for S. aureus exposed to oxidative stress, as these cells had higher activity of the PPP and elevated NADPH concentrations (Deng et al., 2014).
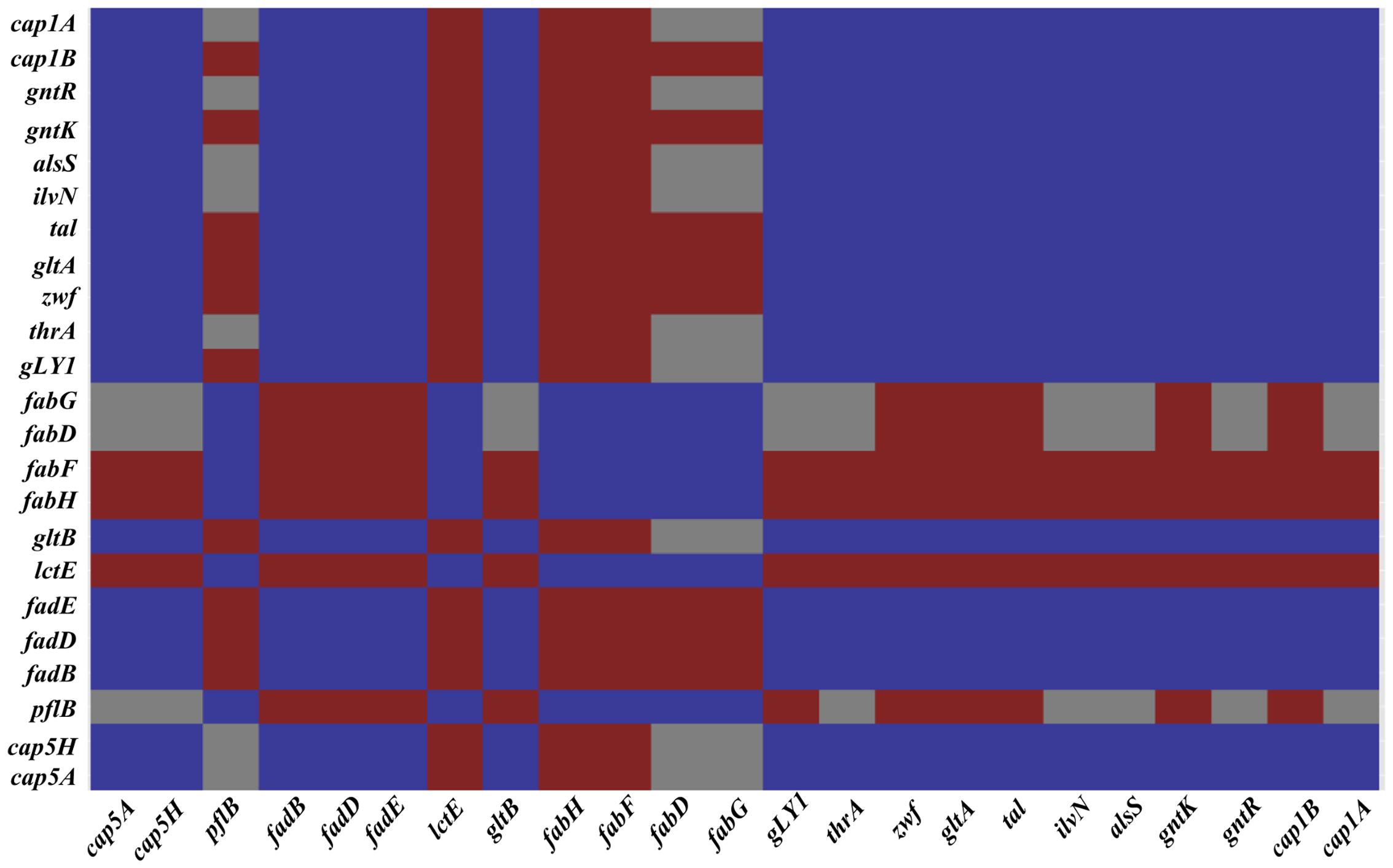
FIGURE 4. A correlation matrix of selected genes. Correlation of selected genes of classes differently expressed on S. aureus cells grown on galactose and under NO stress as determined by the T-Rex pipeline (de Jong et al., 2015). Genes with significant correlation are colored dark blue, genes showing good anti-correlation are colored red and uncorrelated genes are colored gray. SAUSA300_0152 (cap5A), SAUSA300_0159 (cap5H), SAUSA300_2597 (cap1B), SAUSA300_2598 (cap1A), SAUSA300_0220 (pflB), SAUSA300_0226-28 (fadBDE), SAUSA300_0235 (lctE), SAUSA300_0885-6 (fabHF), SAUSA300_1123-4 (fadDG), SAUSA300_1641 (gltA), SAUSA300_1454 (zwf), SAUSA300_1725 (taI), SAUSA300_2443 (gntK), SAUSA300_2166 (alsS), SAUSA300_2444 (gntR), SAUSA300_2008 (ilvN), SAUSA300_1225 (thrA), SAUSA300_1214 (gLY1), and SAUSA300_0445 (gltB).
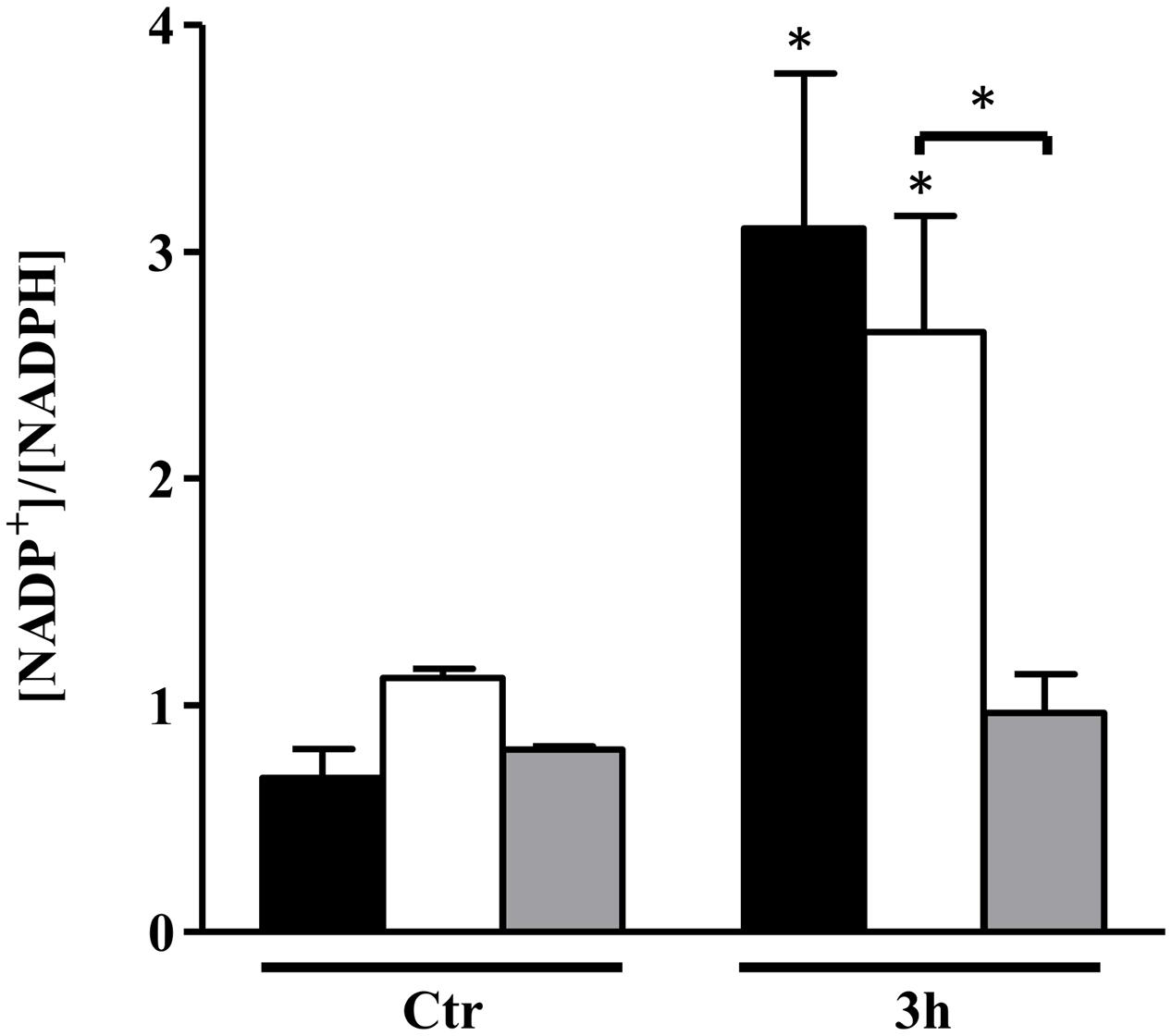
FIGURE 5. NADP+/NADPH ratio of S. aureus NO-stressed cells. S. aureus was grown in CDM with no glycolytic carbon source (black bars), on glucose (white bars), and on galactose (gray bars) in the absence (Ctr) and under NO stress for 3 h. Error bars represent mean ± SEM (n = 3). Asterisks represent statistically significant data relative to the control; ∗P < 0.05.
The metabolic profile of S. aureus suggests that prolonged exposure to NO of cells grown on galactose leads to an increase of the ALS activity. Consistent with these results, we observed higher expression of the alsS gene and of the genes involved in BCAAs biosynthesis whose precursor is alpha-acetolactate, the product of the ALS activity. Thus, we speculated that ALS was most possible one of the enzymes used to replenish the need for amino acids of cells grown on galactose and under extensive NO stress. To test this hypothesis, a S. aureus alsS mutant was studied regarding the growth behavior and metabolic response of stressed galactose-grown cells. For comparison purposes, cells cultured on glucose were also analyzed. The alsS mutant grown on either sugar and treated with NO showed a decrease of the final biomass relative to the parental strain (1.5-fold), which was not observed in the absence of a glycolytic substrate (Figures 6A–C). However, in all cases, the growth rates were not significantly affected (Figures 6A–C), thus suggesting that when using glucose or galactose the strain lacking alsS enters earlier into the post-exponential phase (Figures 6B,C).
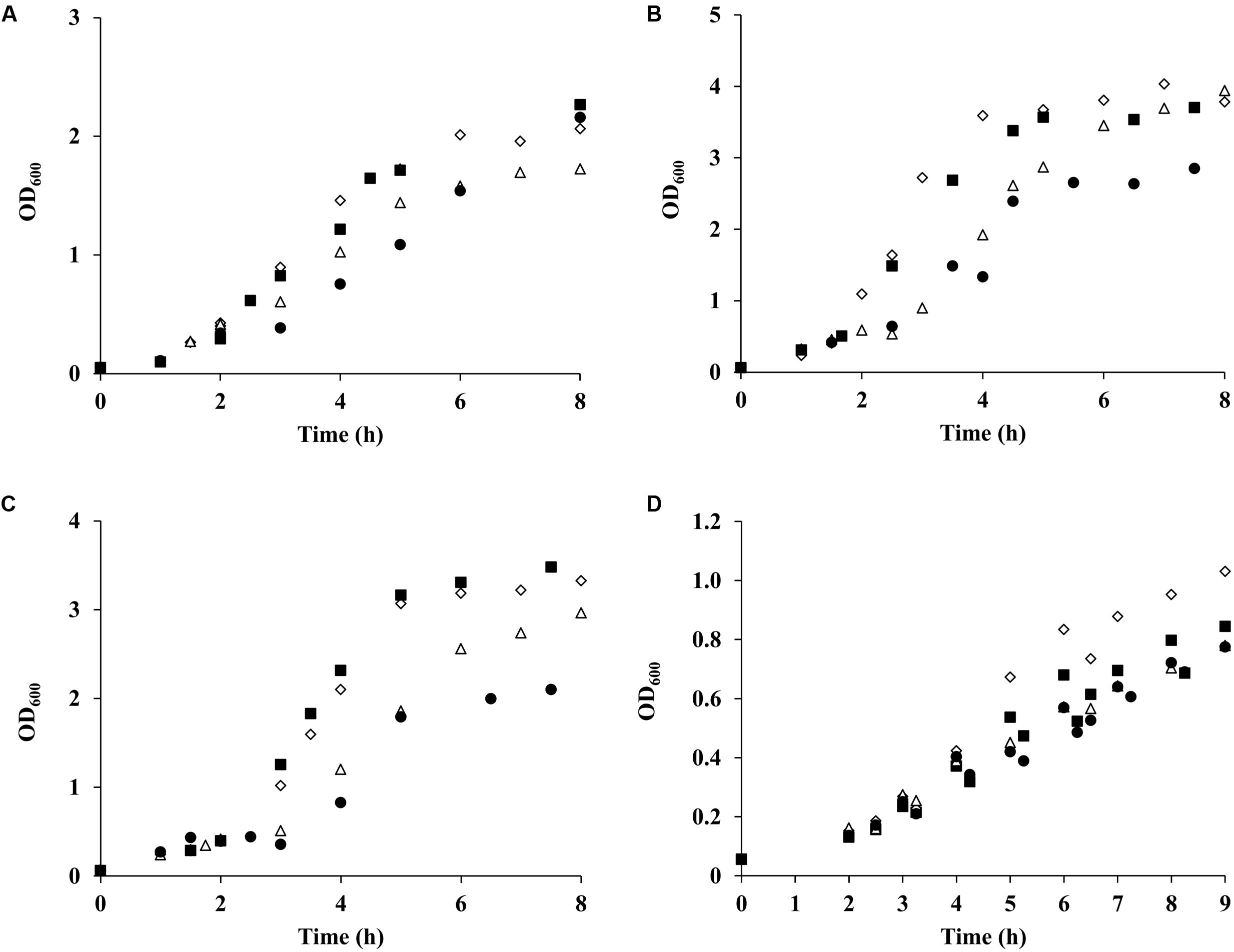
FIGURE 6. Growth of S. aureus wild type and alsS mutant exposed to NO. S. aureus wild type (♢,Δ) and alsS mutant (■,●) were cultured in CDM (A), supplemented with 20 mM glucose (B) or with 20 mM galactose (C), and in nasal-like synthetic medium containing 10 mM glucose (D). Cells were grown under aerobic conditions in the presence (Δ,●) or absence (♢,■) of NO, which was added at OD600 of 0.4. Points represent the average of at least four independent assays.
Concerning the end products profile, the S. aureus alsS mutant grown on a glycolytic carbon source-free medium had only a slight increase of the acetate content relative to that of the wild type independently of the NO treatment (Figure 7A).
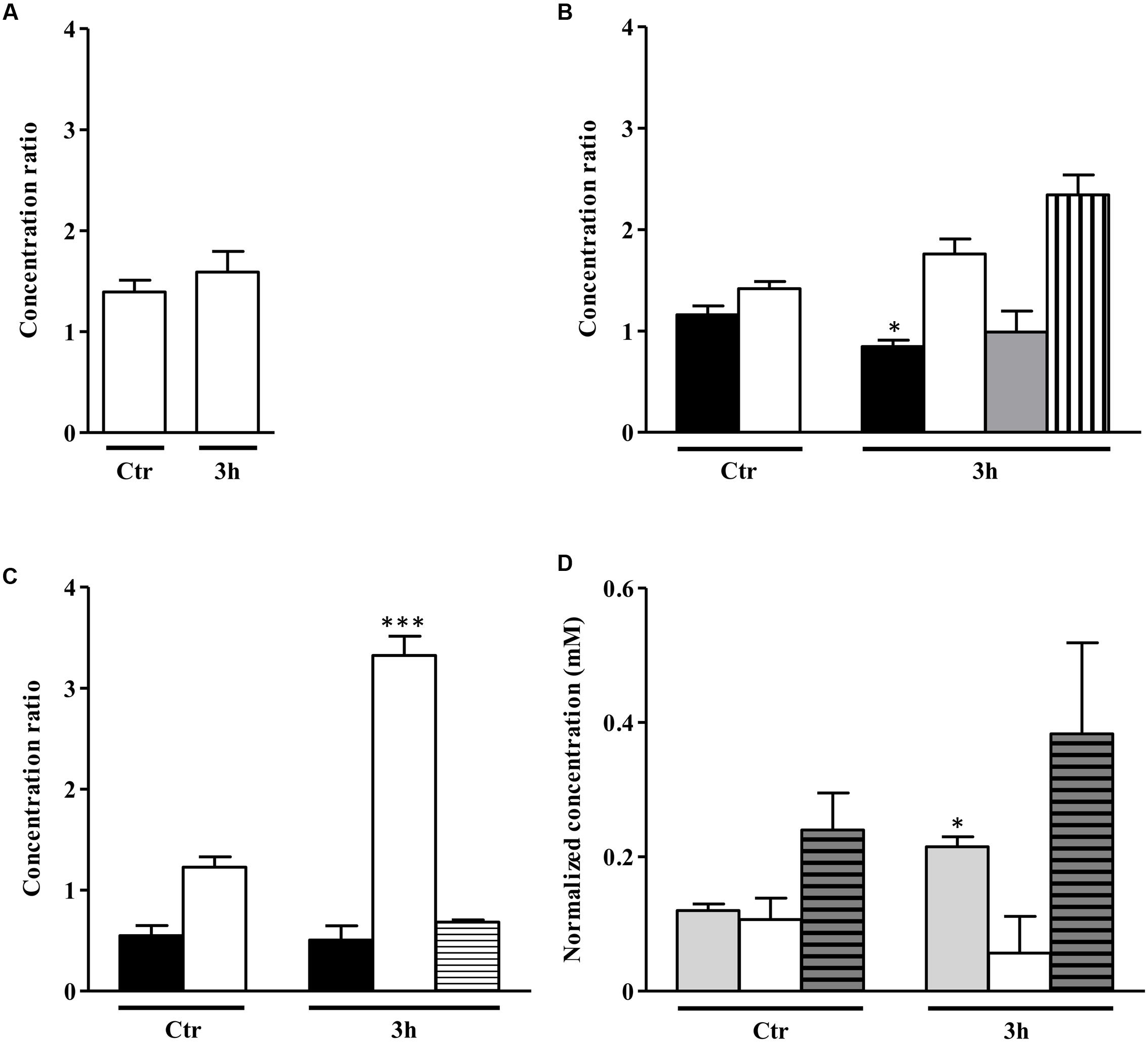
FIGURE 7. Metabolite profile of S. aureus alsS mutant upon exposure to NO. Metabolites evaluated in S. aureus grown in the absence of a glycolytic carbon source (A) and in media containing 20 mM glucose (B) or 20 mM galactose (C), which remained untreated (control, Ctr) or were exposed to NO, for 3 h. Color code of bars of graphs A, B, and C: white, acetate; black, sugar consumed; gray, lactate; horizontally striped, formate; vertically striped, pyruvate. In (D) is depicted the citrate accumulation in cells grown with no glycolytic carbon source (light gray bar), on glucose (white bar) and on galactose (gray horizontally striped bar) untreated (control, Ctr) and exposed to NO for 3 h. For each end product, the ratio between the concentrations in the alsS mutant divided by that of the wild type cells is represented. Error bars represent mean ± SEM (n = 3). Asterisks represent statistically significant data relative to the control; ∗P < 0.05 and ∗∗∗P < 0.0001.
When cultured on glucose, unexposed alsS mutant cells had slightly higher content of acetate than the wild type, and presented minor amounts of lactate (Figure 7B). The end products concentration in the alsS mutant cells fully accounted for all the glucose consumed. In the absence of NO, the alsS mutant produced approximately 5 mM/OD600 of acetate and lactate from the consumption of 2.5 mM/OD600 of glucose. After 3 h of NO stress, the alsS mutant contained approximately two-times more acetate and pyruvate than the wild type, while no accumulation of α-acetolactate, acetoin and 2,3-butanediol was observed, indicating that α-acetolactate is produced by the activity of ALS enzyme (Figure 7B). Moreover, the concentration of the end products accounted all the glucose consumed (∼6.3 mM/OD600 of acetate, 1 mM/OD600 of lactate, and 0.5 mM/OD600 of pyruvate were excreted when 3.9 mM/OD600 of glucose was consumed). This result suggested that the TCA cycle in the alsS mutant is not active, at least to a great extent, which is in agreement with the accumulation of citrate in the extracellular medium rather than consumption (Figure 7D). When compared with the wild type, the 3 h NO-treated glucose-grown alsS mutant cells accumulated extracellularly approximately the double amount of acidic end products (Figure 7B). This data suggest that under these conditions the mutant had a lower intracellular pH level (Thomas et al., 2014), which is consistent with a premature entering into the post-exponential phase (Figure 6B).
On galactose, inactivation of alsS generated cells that in the NO-free environment contained similar levels of acetate, but consumed two-times less galactose than the wild type (Figure 7C). Furthermore, the alsS mutant had an increased amount of acetate and formate end products (4.5 mM/OD600), than those expected from the consumption of 0.5 mM/OD600 of galactose, which contrasts with the total carbon recovery observed on glucose. When exposed to NO, the alsS mutant had a threefold higher accumulation of acetate and a slightly lower amount of formate than the parental strain (Figure 7C). These results indicate that a strain inactivated in alsS and exposed to NO does not produce α-acetolactate and its derivatives, but instead leads to a higher accumulation of acidic end products (Figure 7C).
When treated with NO, S. aureus cells grown on galactose contained no acetoin and 2,3-butanediol. Moreover, the mutation of alsS increased the acetate content. Therefore, the derived compounds of α-acetolactate present in the wild type strain are most probably BCAAs as α-acetolactate is also a known precursor of the BCAAs synthesis (Figure 1) (Goupil-Feuillerat et al., 1997).
Hence, we conclude that under NO stress, the ALS activity plays an important role by preventing extracellular acetate accumulation through the production of α-acetolactate for the biosynthesis of BCAAs, which counteracts the pH stress imposed by the higher content of acetate.
Previously, Krismer et al. (2014) described a nasal synthetic medium that was considered similar to the in vivo environment experienced by S. aureus inside the human nose. Similar to our transcriptome data, the global gene expression analysis of S. aureus grown on the nasal-like medium showed a significant upregulation of amino acid biosynthesis genes of glutamate, valine and leucine, and iron transporters (Krismer et al., 2014). Therefore, metabolic studies on cells grown on the nasal-like synthetic medium were also done. To note that when compared with the CDM, the nasal-like synthetic medium contains lower amount of amino acids (∼2-fold lower concentration), and lacks several amino acids, namely, aspartate, methionine, trans-hydroxyproline, and isoleucine. Furthermore, the medium contains fumaric, maleic, pyruvic and succinic acids and urea, which are absent from the CDM medium. Also, the citric acid and glucose concentrations are lower by 50- and 2-fold, respectively, and no folic acid is present.
We observed that S. aureus wild type grown on the nasal-like synthetic medium accumulated high levels of acetate, which did not account for all the glucose exhausted from the extracellular medium, and consumed small amounts of pyruvate from the extracellular medium (Figure 8A). After 1 h of NO stress, the wild type cells exhibited levels of glucose utilization, similar to those of unstressed cells, and L-lactate accumulation. However, cells accumulated more acetate, had increased consumption of alanine, and no pyruvate was detected (Figures 8B, 9B). Upon prolonged exposure to NO, the lactate levels decreased and pyruvate accumulated (Figure 8C).
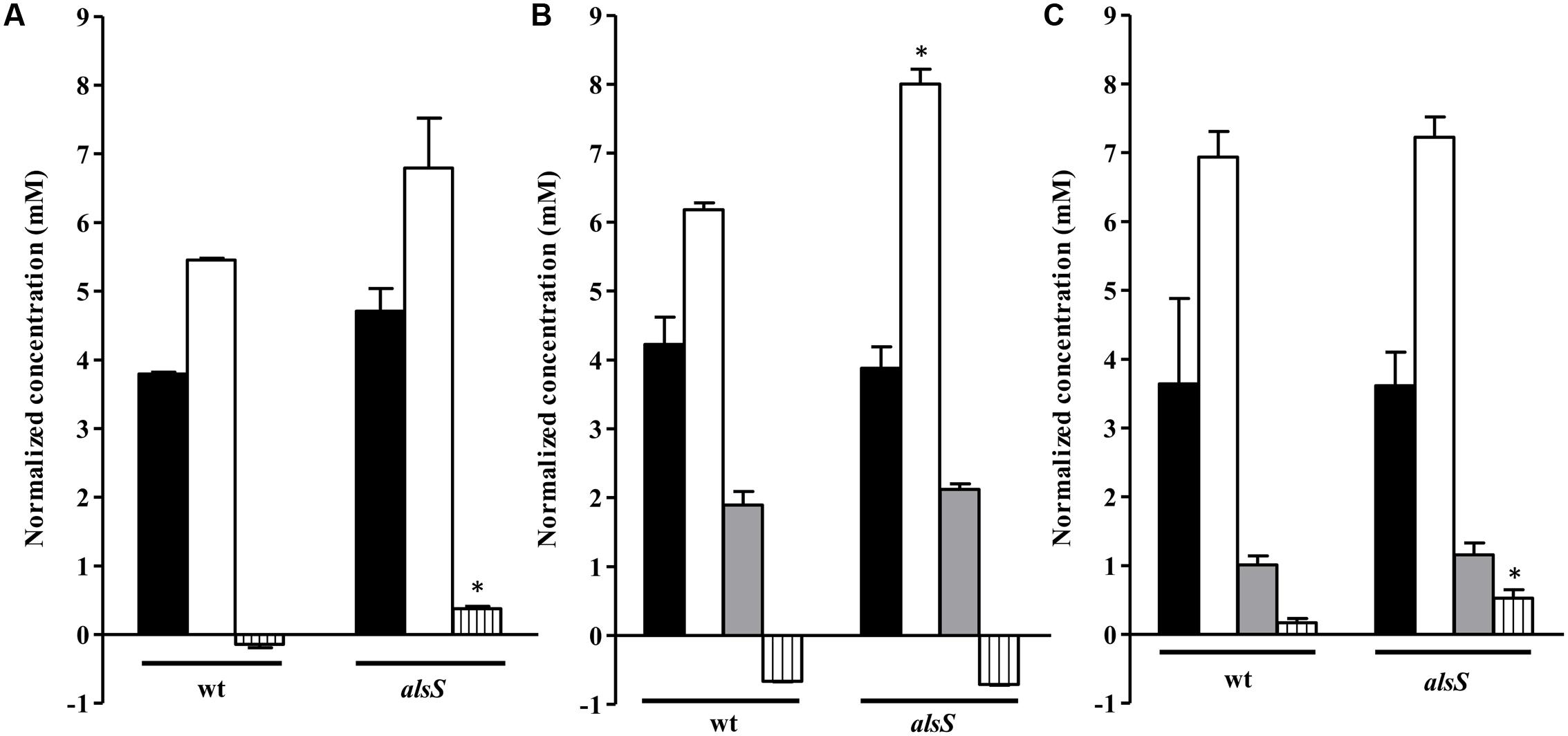
FIGURE 8. Metabolite profile of the alsS mutant grown on nasal synthetic medium exposed to NO. Cells of wild type (wt) and alsS mutant grown in nasal synthetic medium untreated (A) and exposed to NO for 1 h (B) and 3 h (C) were analyzed. Black bar, glucose consumed; white bar, acetate accumulated; gray bar, lactate accumulated; vertically striped bars, pyruvate consumed (below 0) or accumulated (above 0). Error bars represent mean ± SEM (n = 3). Asterisks represent statistically significant data relative to the wild type; ∗P < 0.05.
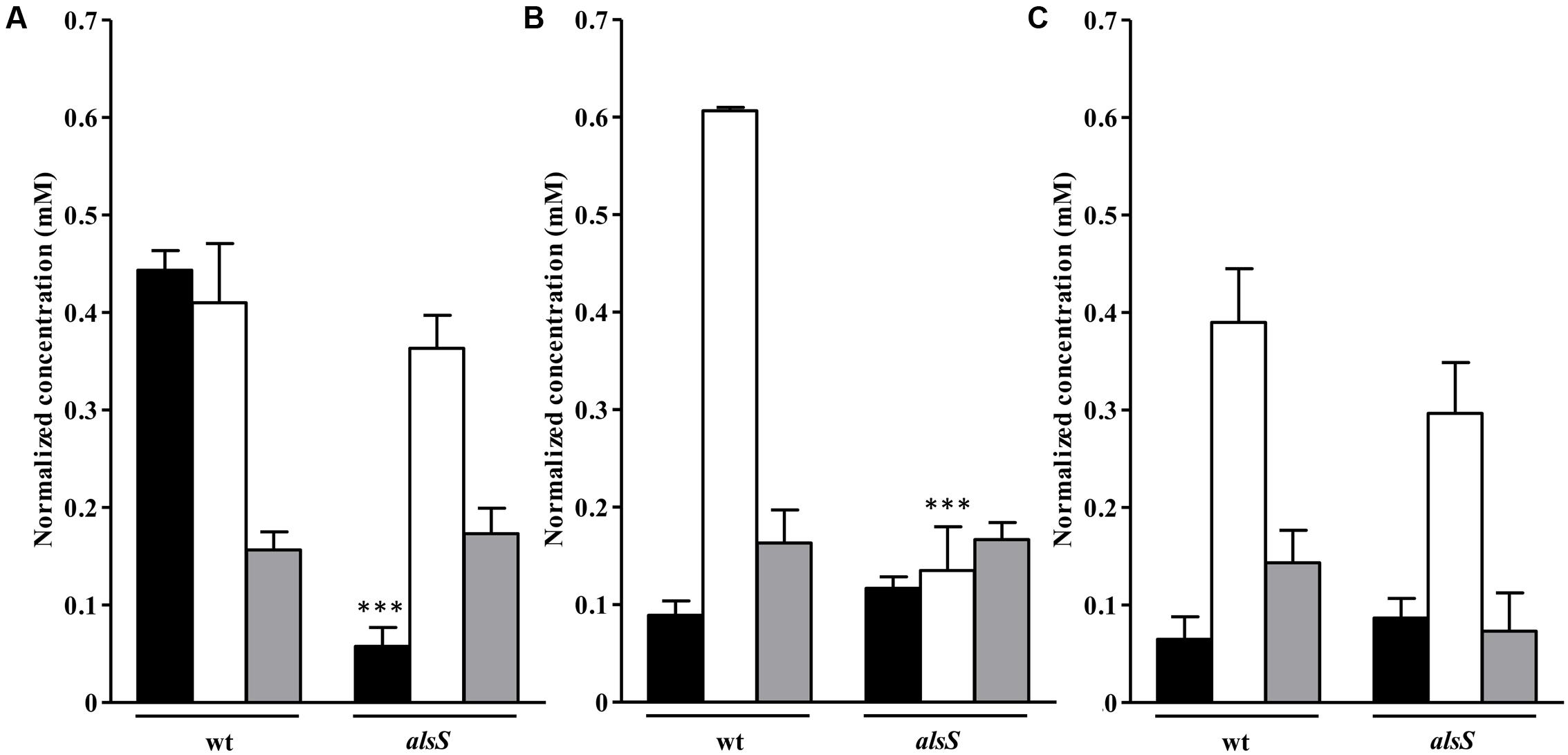
FIGURE 9. Uptake of amino acids and succinate by the S. aureus alsS mutant grown in nasal synthetic medium and subjected to NO. Uptake of valine (black bar), alanine (white bar), and succinate (gray bar) by wild type (wt) and alsS mutant strains, untreated (A) and exposed to NO for 1 h (B) and 3 h (C). Error bars represent mean ± SEM (n = 3). Asterisks represent statistically significant data relative to the wild type; ∗∗∗P < 0.0001.
Cells mutated in alsS showed a 1 mM/OD600 increase in the acetate levels and pyruvate was accumulated instead of being consumed, while in the wild type no accumulation of the ALS, ALDC and AR products was observed (Figure 8A). Moreover, the alsS mutant exhibited reduced viability and a fourfold decrease of the valine consumption (Figures 6D, 9A), suggesting that the ALS activity is required for the consumption of BCAAs, and that α-acetolactate is being utilized for the BCAAs synthesis via ALS (Figure 1).
Cells of the alsS mutant treated with NO for 1 h consumed less alanine and accumulated more acetate extracellularly (Figures 8B, 9B). Furthermore, for longer times of NO exposure, the amount of pyruvate doubled and the consumption of succinate decreased by approximately two-times (Figures 8C, 9C).
Altogether, the NO-stressed S. aureus alsS mutant cells presented a different metabolic pattern on the nasal-like medium, which is most probably due to the quite different medium composition on carbon sources and amino acids relative to CDM.
Taking into consideration that the alsS expression and ALS activity are increased by NO and that inactivation of alsS rendered the strains more sensitive to NO (Supplementary Table S3 and Figures 2, 6B,C), we next determined the viability of the alsS mutant when infecting murine macrophages J774A.1.
Data showed that the alsS mutant is more efficiently killed by macrophages than the wild type (Figure 10). Similar assays were performed with macrophages previously incubated with the mammalian inhibitor of iNOS (L-NMMA), i.e., in macrophages that do not release NO, which was ascertained by the lowered concentration of extracellular nitrite (Supplementary Figure S5). Treatment with the inhibitor reversed the loss of viability of the alsS mutant to levels similar to those of the wild type strain (Figure 10). Hence, we concluded that ALS promotes survival from phagocytosis by protecting S. aureus from the innate immune NO radical.
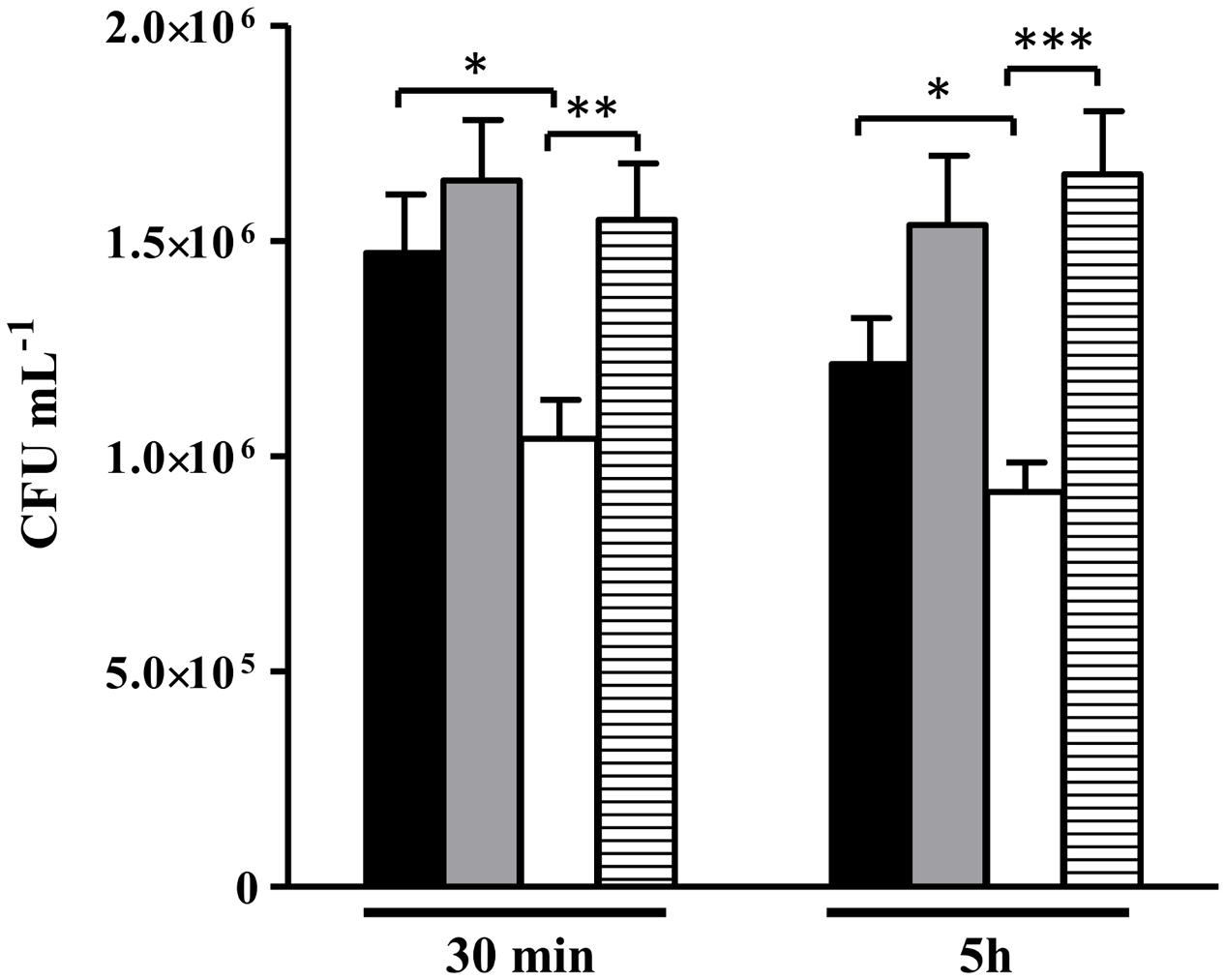
FIGURE 10. Viability of S. aureus alsS mutant contacting with murine macrophages. S. aureus wild type (black and gray bar) and alsS mutant cells (white and striped bar) were incubated with activated macrophages (black and white bar), and with L-NMMA treated macrophages (gray and striped bar). The number of intracellular S. aureus cells were determined at the indicated times. Error bars represent mean ± SEM (n = 3). Asterisks represent statistically significant data; ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.0001.
The matrix analysis of the RNA-Seq data indicated the occurrence of a high correlation between alsS and fatty acid degradation and capsular genes involved in modulation of the cell surface (Figure 4). Thus, we questioned whether ALS could play a role in the resistance of the MRSA to beta-lactam antibiotics, which target the cell wall biosynthesis. For this purpose, we determined for the S. aureus MRSA wild type and alsS mutant the MIC of antibiotics commonly used for the treatment of the pathogen, namely the beta-lactams oxacillin and methicillin. The results showed that inactivation of alsS leads to a twofold decrease in the MIC value, i.e., S. aureus becomes less resistant to these antibiotics (Table 1).
Staphylococcus aureus has been reported to tolerate moderate to high levels of NO when using glucose, one of the carbon sources available in the nasal cavity. In this work, we analyzed the behavior of S. aureus when aerobically grown on galactose, another abundant hexose on the nasal cavity, and exposed to NO, and compared the results with cells cultured on glucose and on a nasal-like medium.
Staphylococcus aureus cultured on galactose exhibited growth rate and biomass yield similar to that observed on glucose, which indicates that the slow-metabolizing galactose also sustains replication of the pathogen under NO stress. However, the metabolism underpinning growth proved to be significantly different. For example, 1 h after the NO stress pulse, cells grown on galactose excreted more acetate than on glucose. Moreover, the increase of acetate excretion and accumulation of extracellular citrate, rather than consumption, indicates a decreased flux of the TCA cycle and respiration as these pathways rely on NO-sensitive enzymes, such as the iron-sulfur containing aconitase and cytochrome oxidase (McCollister et al., 2011; Richardson et al., 2011).
The higher increase of acetate production detected for galactose-grown cells exposed to NO for 1 h provides S. aureus higher ATP levels that are particularly relevant for the growth on slowly metabolized carbon sources as was described for other bacteria (Carvalho et al., 2011). However, prolonged NO stress caused a significant decrease of acetate accumulation and the increased consumption of threonine, glycine, glutamate, and BCAAs from the extracellular medium. Additionally, RNA-Seq data showed upregulation of genes required for the synthesis of threonine, glycine, glutamate, BCAAs, and fatty acids degradation, and acting in the first half of the TCA cycle and the PPP. Consistent with these results, cells had higher content of NADPH, a cofactor that boosts anabolic reactions. In S. typhimurium, the first enzyme of the PPP glucose 6-phosphate dehydrogenase (G6PDH) was shown to provide reducing equivalents in the form of NADPH and to have an important role in the defense against nitrosative stress by maintaining the cellular redox state, regenerating reduced thiols, and repairing oxidative or nitrosative damage (Lundberg et al., 1999). When using galactose, but not glucose under prolonged NO stress, S. aureus showed higher demand of amino acids. Interestingly, similar behavior was also observed for NO-stressed cells grown on the gluconeogenic substrate L-lactate (Spahich et al., 2016).
When S. aureus grows on glucose, the exposure to NO does not alter the amount of excreted acetate but causes the increase of the lactate levels, which decrease under prolonged stress. Although these results apparently differ from those previously reported for S. aureus showing that in cells treated for 4 h with NO there is a strong increase of lactate and acetate is absent, it should be noted that the metabolite production at earlier times was not analyzed and much higher concentrations of NO were used, conditions that completely inhibit the activities of the PDHC and PFL enzymes (Richardson et al., 2008). Moreover, the contrasting effects of NO on S. aureus grown on galactose versus glucose may be a consequence of the slower glycolysis, and such behavior is expected to also occur when S. aureus uses other slow-metabolizing sugars available in its colonization niche.
Transcriptomic analysis revealed that NO induces the expression of the catabolic alsS/budA and butA genes that encode ALS/ALDC and AR proteins. These gene products are generally regarded as involved in the synthesis of non-acidic products acetoin and 2,3-butanediol and regeneration of NAD+ (Goupil-Feuillerat et al., 1997; Thomas et al., 2014). Considering that butanediol fermentation is activated when S. aureus shifts from aerobic to anaerobic growth (Fuchs et al., 2007), the sensitivity of the alsS mutant strain to NO could derive from an indirect consequence of the inhibition of respiration by NO. However, our data do not support this hypothesis, as we did not observe higher expression of the genes that are usually induced when S. aureus grows under anaerobic conditions, such as adhE, adh, narH, narI, narJ, pfl and genes encoding glycolytic enzymes (Fuchs et al., 2007). Moreover, the higher induction of alsS occurred 3 h after the NO pulse, which is when the NO inhibitory effect of respiration is expected to be less prominent considering the half-life of the spermine NONOate (∼40 min).
A previous study reported that the alsS/budA operon is activated by the CidR regulator. The gene encoding CidR was also induced in our RNA-Seq study. CidR has been proposed to be activated by a decrease of the intracellular pH, mainly by accumulation of acetic acid (Thomas et al., 2014). In accordance, increased levels of extracellular acidic levels were observed upon exposure of S. aureus to NO. Moreover, α-acetolactate, acetoin and 2,3-butanediol, which are the products of the ALS and ALDC/AR activities, were observed on glucose-grown cells under prolonged NO stress. The accumulation of butanediol occurred after a short exposure (20 min) to the NO donor MAHMA NONOate (500 μM) for S. aureus COL grown on a glucose-containing medium (Hochgräfe et al., 2008). The absence of α-acetolactate, acetoin and 2,3-butanediol in NO-stressed galactose-grown cells may be due to the higher requirement for BCAAs and the increased levels of NADPH that were observed under these conditions, which shifts the product of ALS activity, α-acetolactate, to the production of BCAAs (Figure 11). Interestingly, it was reported for the Gram-positive Lactococcus lactis that under BCAAs starvation there is a relief of the allosteric activation of ALDC by BCAAs, and of the allosteric inhibition of the BCAAs synthesis pathway (Goupil-Feuillerat et al., 1997). Moreover, BCAAs serve also as precursors for membrane BCFAs and were shown to be important for pH stress tolerance (Kaiser et al., 2016). Alternatively, the requirement for BCAAs may be related with the inhibition by NO of the dihydroxyacid dehydratase enzyme (coded by ilvD and involved in the BCAAs biosynthesis), similarly to what was observed for the dihydroxyacid dehydratase of Escherichia coli (Hyduke et al., 2007). In accordance with the requirement of S. aureus for BCAAs when growing on galactose under prolonged exposure to NO stress, there was a small but significantly higher consumption of the BCAAs valine and isoleucine. The threefold higher accumulation of acetate in the alsS mutant strain suggests that ALS directs the metabolism toward the BCAAs production by fuelling pyruvate resulting from galactose breakdown into α-acetolactate production (Figure 11). BCAAs were shown to be important for the virulence of S. aureus (Kaiser et al., 2016), i.e., under conditions where NO is generated by the host cells. Furthermore, the S. aureus CodY transcriptional regulator, that represses several amino acid biosynthetic pathways when intracellular BCAAs and GTP are available (Majerczyk et al., 2010), has been associated with NO resistance and implicated in virulence (Majerczyk et al., 2010; Grosser et al., 2016). Interestingly, a great number of genes that were transcriptionally modified in our RNA-Seq analysis are regulated by CodY, namely threonine, ilvBN, and glutamate and capsule synthesis genes.
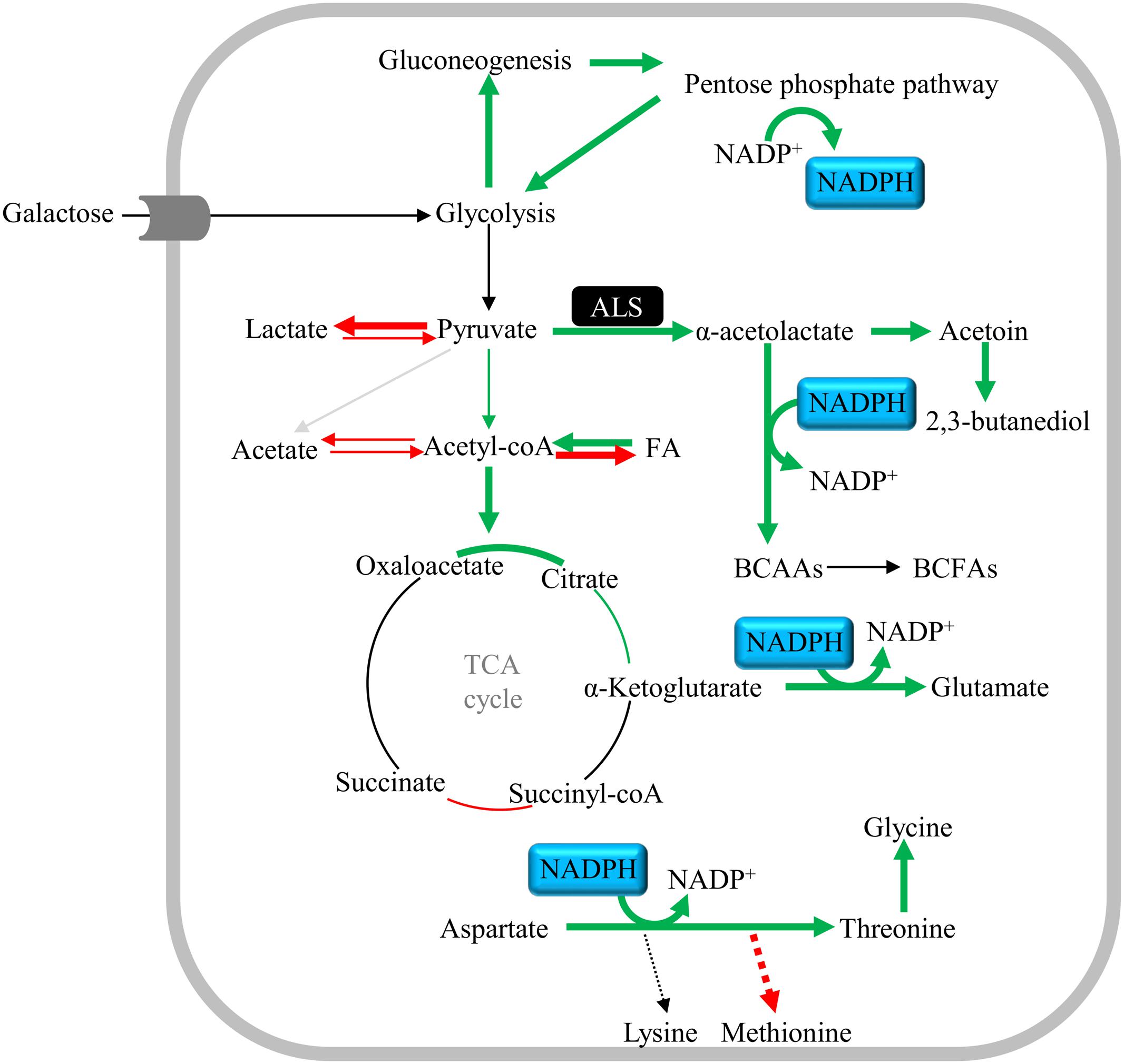
FIGURE 11. Schematic representation of the central metabolic pathways of S. aureus grown on galactose and prolonged exposed to NO. Upregulated and downregulated pathways (cut-off > 2) are denoted with bold green and red arrows, respectively. Narrower green and red arrows indicate significantly altered expression, considering a cut-off between 1.5 and 2. The α-acetolactate synthase (alsS) and the NADP+, NADPH cofactors are highlighted in black and blue, respectively. Proposed pathways were reconstructed based on genome information (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi), literature and database surveys (KEGG, MetaCyc). BCAAs, branched-chain amino acids; BCFAs, branched-chain fatty acids; FA, fatty acids.
Contrary to the wild type, the S. aureus alsS mutant cells did not consume citrate but excreted it to the extracellular medium, which indicates impairment of the TCA cycle. Furthermore, the mutant cells accumulated higher levels of acetate, especially on galactose-containing medium, which may explain the lower survival of the mutant strain (Figure 6C). Consistent with our data, Thomas et al. (2014) reported that the S. aureus alsS/budA mutant had an accrual of extracellular acetate that results in increased cell death linked to ROS accumulation. Moreover, we observed that the valine consumed upon alsS inactivation lowered significantly (data not shown), suggesting that ALS may be required for the consumption of valine, an issue that requires future detailed analysis.
The alsS mutant had lower survival within macrophages, and inhibition of the mammalian NO synthase lifted the growth impairment. This result points to a NO-related protective role for ALS and is in line with the high expression of alsS in samples collected from human nares that contain significant NO levels (Chaves-Moreno et al., 2016).
In the nasal-like medium, the cellular consumption of glucose did not change significantly upon 1 h of NO exposure; however, the pyruvate (present in the medium at high concentrations of 300 μM) was completely utilized and alanine was more consumed. Contrary to what was observed in cells grown in CDM, in the nasal-like medium, pyruvate was consumed instead of glucose to produce lactate for balancing the reducing equivalents (NAD+) (Figures 2B, 8B).
In nasal-like medium, alsS proved also to have an important role as its inactivation causes growth defects. Additionally, the absence of quantifiable amounts of the products of the ALS activity, namely α-acetolactate, acetoin and 2,3-butanediol, indicate a requirement for BCAAs in the nasal medium, which may be due to the absence of isoleucine and the low levels of leucine and valine present in this medium (Krismer et al., 2014).
We show how S. aureus copes with prolonged NO stress when metabolizing a slow-sugar like galactose, that is one of the main hexoses available in the nasal cavity. The following model is therefore proposed (Figure 11): NO promotes the formation of acetyl-CoA, which feeds acetyl molecules into the first half of the TCA cycle, and the synthesis of amino acids (e.g., glutamate, threonine, glycine, BCAAs), which are also boosted by the NADPH produced by the upregulated genes of the PPP. Among the genes products involved in amino acid synthesis, one of the most upregulated is that encoding ALS. Under NO stress, ALS seems to have a dual role by preventing acid stress and promoting the synthesis of BCAAs (Figure 11). Furthermore, this study disclosed the role of ALS in the resistance of S. aureus, by showing that ALS contributes not only to NO resistance but also to the resistance to antibiotics commonly used against S. aureus, such as methicillin and oxacillin.
SC undertook aspects of the experimental work, in the interpretation of the data and writing of the manuscript. AJ, TK, and OK designed and interpreted the RNA-seq experiments. LS designed and interpreted all aspects of the experimental work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was financially supported by Project LISBOA-01-0145-FEDER-007660 (Microbiologia Molecular, Estrutural e Celular) funded by FEDER funds through COMPETE2020 – Programa Operacional Competitividade e Internacionalização (POCI) and national funds through FCT – PTDC/BBB-BQB/0937/2012. SC has a fellowship SFRH/BPD/92687/2013 from Fundação para a Ciência e a Tecnologia (FCT). The NMR data was acquired at CERMAX (Centro de Ressonância Magnética António Xavier) which is a member of the National NMR network. NARSA strain was provided by the network on antimicrobial resistance in S. aureus (http://www.narsa.net/content/default.jsp).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01273/full#supplementary-material
ALS, α-acetolactate synthase; BCAAs, branched-chain amino acids; EMP, Embden-Meyerhof-Parnas; L-NMMA, NG-monomethyl-L-arginine acetate salt; MRSA, methicillin-resistant Staphylococcus aureus; NMR, nuclear magnetic resonance; NO, nitric oxide; PDHC, pyruvate dehydrogenase complex; PFL, pyruvate formate-lyase; PPP, pentose phosphate pathway; RNA-Seq, RNA-Sequencing; TCA, tricarboxylic acid cycle.
Blanchette, K. A., Shenoy, A. T., Milner, J., Gilley, R. P., McClure, E., Hinojosa, C. A., et al. (2016). Neuraminidase A-exposed galactose promotes Streptococcus pneumoniae biofilm formation during colonization. Infect. Immun. 84, 2922–2932. doi: 10.1128/IAI.00277-16
Carvalho, S. M., Kloosterman, T. G., Kuipers, O. P., and Neves, A. R. (2011). CcpA ensures optimal metabolic fitness of Streptococcus pneumoniae. PLoS ONE 6:e26707. doi: 10.1371/journal.pone.0026707
Chaves-Moreno, D., Wos-Oxley, M. L., Jáuregui, R., Medina, E., Oxley, A. P., Pieper, D. H., et al. (2016). Exploring the transcriptome of Staphylococcus aureus in its natural niche. Sci. Rep. 6:33174. doi: 10.1038/srep33174
Crooke, A. K., Fuller, J. R., Obrist, M. W., Tomkovich, S. E., Vitko, N. P., and Richardson, A. R. (2013). CcpA-independent glucose regulation of lactate dehydrogenase 1 in Staphylococcus aureus. PLoS ONE 8:e54293. doi: 10.1371/journal.pone.0054293
de Jong, A., van der Meulen, S., Kuipers, O. P., and Kok, J. (2015). T-REx: transcriptome analysis webserver for RNA-Seq expression data. BMC Genomics 16, 663. doi: 10.1186/s12864-015-1834-4
Deng, X., Liang, H., Ulanovskaya, O. A., Ji, Q., Zhou, T., Sun, F., et al. (2014). Steady-state hydrogen peroxide induces glycolysis in Staphylococcus aureus and Pseudomonas aeruginosa. J. Bacteriol. 196, 2499–2513. doi: 10.1128/JB.01538-14
Edwards, A. M., Massey, R. C., and Clarke, S. R. (2012). Molecular mechanisms of Staphylococcus aureus nasopharyngeal colonization. Mol. Oral Microbiol. 27, 1–10. doi: 10.1111/j.2041-1014.2011.00628.x
Ferreira, M. T., Manso, A. S., Gaspar, P., Pinho, M. G., and Neves, A. R. (2013). Effect of oxygen on glucose metabolism: utilization of lactate in Staphylococcus aureus as revealed by in vivo NMR studies. PLoS ONE 8:e58277. doi: 10.1371/journal.pone.0058277
Foster, T. J. (2005). Immune evasion by staphylococci. Nat. Rev. Microbiol. 3, 948–958. doi: 10.1038/nrmicro1289
Fuchs, S., Pané-Farré, J., Kohler, C., Hecker, M., and Engelmann, S. (2007). Anaerobic gene expression in Staphylococcus aureus. J. Bacteriol. 189, 4275–4289. doi: 10.1128/JB.00081-07
Fuller, J. R., Vitko, N. P., Perkowski, E. F., Scott, E., Khatri, D., Spontak, J. S., et al. (2011). Identification of a lactate-quinone oxidoreductase in Staphylococcus aureus that is essential for virulence. Front. Cell. Infect. Microbiol. 1:19. doi: 10.3389/fcimb.2011.00019
Gonçalves, V. L., Nobre, L. S., Vicente, J. B., Teixeira, M., and Saraiva, L. M. (2006). Flavohemoglobin requires microaerophilic conditions for nitrosative protection of Staphylococcus aureus. FEBS Lett. 580, 1817–1821. doi: 10.1016/j.febslet.2006.02.039
Goupil-Feuillerat, N., Cocaign-Bousquet, M., Godon, J. J., Ehrlich, S. D., and Renault, P. (1997). Dual role of alpha-acetolactate decarboxylase in Lactococcus lactis subsp. lactis. J. Bacteriol. 179, 6285–6293.
Grosser, M. R., Weiss, A., Shaw, L. N., and Richardson, A. R. (2016). Regulatory requirements for Staphylococcus aureus nitric oxide resistance. J. Bacteriol. 198, JB.229–JB.216. doi: 10.1128/JB.00229-16
Halsey, C. R., Lei, S., Wax, J. K., Lehman, M. K., Nuxoll, A. S., Steinke, L., et al. (2017). Amino acid catabolism in Staphylococcus aureus and the function of carbon catabolite repression. MBio 8:e1434-16. doi: 10.1128/MBIO.01434-16
Hochgräfe, F., Wolf, C., Fuchs, S., Liebeke, M., Lalk, M., Engelmann, S., et al. (2008). Nitric oxide stress induces different responses but mediates comparable protein thiol protection in Bacillus subtilis and Staphylococcus aureus. J. Bacteriol. 190, 4997–5008. doi: 10.1128/JB.01846-07
Hyduke, D. R., Jarboe, L. R., Tran, L. M., Chou, K. J. Y., and Liao, J. C. (2007). Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 104, 8484–8489. doi: 10.1073/pnas.0610888104
Kaiser, J. C., Sen, S., Sinha, A., Wilkinson, B. J., and Heinrichs, D. E. (2016). The role of two branched-chain amino acid transporters in Staphylococcus aureus growth, membrane fatty acid composition and virulence. Mol. Microbiol. 102, 850–864. doi: 10.1111/mmi.13495
King, S. J. (2010). Pneumococcal modification of host sugars: a major contributor to colonization of the human airway? Mol. Oral Microbiol. 25, 15–24. doi: 10.1111/j.2041-1014.2009.00564.x
Krismer, B., Liebeke, M., Janek, D., Nega, M., Rautenberg, M., Hornig, G., et al. (2014). Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog. 10:e1003862. doi: 10.1371/journal.ppat.1003862
Lanz, M. J., Prendes, S., Peyrou, N., Toledo, G., and Ferrer, C. M. (2008). Nasal nitric oxide as a noninvasive marker in the antibiotic treatment of acute bacterial sinusitis. J. Allergy Clin. Immunol. 121, 530–531. doi: 10.1016/j.jaci.2007.09.014
Lilius, E. M., Multanen, V. M., and Toivonen, V. (1979). Quantitative extraction and estimation of intracellular nicotinamide nucleotides of Escherichia coli. Anal. Biochem. 99, 22–27. doi: 10.1016/0003-2697(79)90039-3
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2ˆ-ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lundberg, B. E., Wolf, R. E., Dinauer, M. C., Xu, Y., and Fang, F. C. (1999). Glucose 6-phosphate dehydrogenase is required for Salmonella typhimurium virulence and resistance to reactive oxygen and nitrogen intermediates. Infect. Immun. 67, 436–438.
Majerczyk, C. D., Dunman, P. M., Luong, T. T., Lee, C. Y., Sadykov, M. R., Somerville, G. A., et al. (2010). Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192, 2861–2877. doi: 10.1128/JB.00220-10
McCollister, B. D., Hoffman, M., Husain, M., and Vázquez-Torres, A. (2011). Nitric oxide protects bacteria from aminoglycosides by blocking the energy-dependent phases of drug uptake. Antimicrob. Agents Chemother. 55, 2189–2196. doi: 10.1128/AAC.01203-10
Miallau, L., Hunter, W. N., McSweeney, S. M., and Leonard, G. A. (2007). Structures of Staphylococcus aureus D-tagatose-6-phosphate kinase implicate domain motions in specificity and mechanism. J. Biol. Chem. 282, 19948–19957. doi: 10.1074/jbc.M701480200
Neves, A. R., Pool, W. A., Solopova, A., Kok, J., Santos, H., and Kuipers, O. P. (2010). Towards enhanced galactose utilization by Lactococcus lactis. Appl. Environ. Microbiol. 76, 7048–7060. doi: 10.1128/AEM.01195-10
Nobre, L. S., and Saraiva, L. M. (2013). Effect of combined oxidative and nitrosative stresses on Staphylococcus aureus transcriptome. Appl. Microbiol. Biotechnol. 97, 2563–2573. doi: 10.1007/s00253-013-4730-3
Nobre, L. S., and Saraiva, L. M. (2014). Role of the siderophore transporter SirABC in the Staphylococcus aureus resistance to oxidative stress. Curr. Microbiol. 69, 164–168. doi: 10.1007/s00284-014-0567-y
Richardson, A. R., Dunman, P. M., and Fang, F. C. (2006). The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 61, 927–939. doi: 10.1111/j.1365-2958.2006.05290.x
Richardson, A. R., Libby, S. J., and Fang, F. C. (2008). A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science 319, 1672–1676. doi: 10.1126/science.1155207
Richardson, A. R., Payne, E. C., Younger, N., Karlinsey, J. E., Thomas, V. C., Becker, L. A., et al. (2011). Multiple targets of nitric oxide in the tricarboxylic acid cycle of Salmonella enterica serovar typhimurium. Cell Host Microbe 10, 33–43. doi: 10.1016/j.chom.2011.06.004
Robinson, J. L., Adolfsen, K. J., and Brynildsen, M. P. (2014). Deciphering nitric oxide stress in bacteria with quantitative modeling. Curr. Opin. Microbiol. 19, 16–24. doi: 10.1016/j.mib.2014.05.018
Sadykov, M. R., Thomas, V. C., Marshall, D. D., Wenstrom, C. J., Moormeier, D. E., Widhelm, T. J., et al. (2013). Inactivation of the Pta-AckA pathway causes cell death in Staphylococcus aureus. J. Bacteriol. 195, 3035–3044. doi: 10.1128/JB.00042-13
Sakarya, S., Ertugrul, M. B., Öztürk, T., and Gökbulut, C. (2010). Effect of pharynx epithelial cells surface desialylation on receptor-mediated adherence of Staphylococcus aureus. J. Appl. Microbiol. 108, 1313–1322. doi: 10.1111/j.1365-2672.2009.04525.x
Shuter, J., Hatcher, V. B., and Lowy, F. D. (1996). Staphylococcus aureus binding to human nasal mucin. Infect. Immun. 64, 310–318.
Spahich, N. A., Vitko, N. P., Thurlow, L. R., Temple, B., and Richardson, A. R. (2016). Staphylococcus aureus lactate- and malate-quinone oxidoreductases contribute to nitric oxide resistance and virulence. Mol. Microbiol. 100, 759–773. doi: 10.1111/mmi.13347
Tavares, A. F. N., Nobre, L. S., Melo, A. M. P., and Saraiva, L. M. (2009). A novel nitroreductase of Staphylococcus aureus with S-nitrosoglutathione reductase activity. J. Bacteriol. 191, 3403–3406. doi: 10.1128/JB.00022-09
Thammavongsa, V., Kim, H. K., Missiakas, D., and Schneewind, O. (2015). Staphylococcal manipulation of host immune responses. Nat. Rev. Microbiol. 13, 529–543. doi: 10.1038/nrmicro3521
Thomas, V. C., Sadykov, M. R., Chaudhari, S. S., Jones, J., Endres, J. L., Widhelm, T. J., et al. (2014). A central role for carbon-overflow pathways in the modulation of bacterial cell death. PLoS Pathog. 10:e1004205. doi: 10.1371/journal.ppat.1004205
Vaseghi, S., Baumeister, A., Rizzi, M., and Reuss, M. (1999). In vivo dynamics of the pentose phosphate pathway in Saccharomyces cerevisiae. Metab. Eng. 1, 128–140. doi: 10.1006/mben.1998.0110
Vitko, N. P., Spahich, N. A., and Richardson, A. R. (2015). Glycolytic dependency of high-level nitric oxide resistance and virulence in Staphylococcus aureus. MBio 6:e45-15. doi: 10.1128/mBio.00045-15
Keywords: Staphylococcus aureus, nitrosative stress, bacterial metabolism, α-acetolactate synthase (ALS), nuclear magnetic resonance (NMR)
Citation: Carvalho SM, de Jong A, Kloosterman TG, Kuipers OP and Saraiva LM (2017) The Staphylococcus aureus α-Acetolactate Synthase ALS Confers Resistance to Nitrosative Stress. Front. Microbiol. 8:1273. doi: 10.3389/fmicb.2017.01273
Received: 20 April 2017; Accepted: 26 June 2017;
Published: 11 July 2017.
Edited by:
Daniela De Biase, Sapienza Università di Roma, ItalyReviewed by:
James Moir, University of York, United KingdomCopyright © 2017 Carvalho, de Jong, Kloosterman, Kuipers and Saraiva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lígia M. Saraiva, bHN0QGl0cWIudW5sLnB0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.