- 1School of Biological Sciences, University of Nebraska-Lincoln, Lincoln, NE, United States
- 2Department of Earth and Atmospheric Sciences, University of Nebraska-Lincoln, Lincoln, NE, United States
- 3Lawrence Berkeley National Laboratory, Berkeley, CA, United States
Viruses are the most abundant biological entity on Earth and their interactions with microbial communities are recognized to influence microbial ecology and impact biogeochemical cycling in various ecosystems. While the factors that control the distribution of viruses in surface aquatic environments are well-characterized, the abundance and distribution of continental subsurface viruses with respect to microbial abundance and biogeochemical parameters have not yet been established. In order to begin to understand the factors governing virus distribution in subsurface environments, we assessed microbial cell and virus abundance in groundwater concurrent with groundwater chemistry in a uranium impacted alluvial aquifer adjoining the Colorado River near Rifle, CO. Virus abundance ranged from 8.0 × 104 to 1.0 × 106 mL−1 and exceeded cell abundance in all samples (cell abundance ranged from 5.8 × 104 to 6.1 × 105 mL−1). The virus to microbial cell ratio ranged from 1.1 to 8.1 and averaged 3.0 ± 1.6 with virus abundance most strongly correlated to cell abundance (Spearman's ρ = 0.73, p < 0.001). Both viruses and cells were positively correlated to dissolved organic carbon (DOC) with cells having a slightly stronger correlation (Spearman's ρ = 0.46, p < 0.05 and ρ = 0.54, p < 0.05; respectively). Groundwater uranium was also strongly correlated with DOC and virus and cell abundance (Spearman's ρ = 0.62, p < 0.05; ρ = 0.46, p < 0.05; and ρ = 0.50, p < 0.05; respectively). Together the data indicate that microbial cell and virus abundance are correlated to the geochemical conditions in the aquifer. As such local geochemical conditions likely control microbial host cell abundance which in turn controls viral abundance. Given the potential impacts of viral-mediated cell lysis such as liberation of labile organic matter from lysed cells and changes in microbial community structure, viral interactions with the microbiota should be considered in an effort to understand subsurface biogeochemical cycling and contaminant mobility.
Introduction
Viruses have been identified in every environment where microorganisms are present, often equal to or exceeding microbial cell abundance (Suttle, 2005; Anderson et al., 2013; Knowles et al., 2016). Within the continental subsurface sediments (103–109 cm−3; Engelhardt et al., 2014; Pan et al., 2014; Yanagawa et al., 2014) and groundwater (105–107 mL−1; Kyle et al., 2008; Roudnew et al., 2012) viruses may outnumber cells in situ by as much as 225 to one in the subsurface (Engelhardt et al., 2014). Considering that the continental subsurface harbors an estimated one-third of all microbial life on Earth (Whitman et al., 1998; Kallmeyer et al., 2012), the impact of viruses in terrestrial subsurface biogeochemical cycling is of growing interest (Pan et al., 2014; Wilkins and Fredrickson, 2015). However, we know very little about the role viruses play in subsurface microbial ecology and biogeochemistry. Viruses are obligate intracellular parasites that use the host cell for replication and often lyse the host cell upon release into the environment. Lysogenic viruses may also exist as prophage integrated into host genomes but may be triggered back into a lytic life cycle. As a consequence, virus-mediated cell lysis has the potential to liberate DOC and other nutrients into the surrounding environment contributing to food webs and biogeochemical carbon cycling (Fuhrman, 1999; Wommack and Colwell, 2000; Middelboe and Lyck, 2002; Suttle, 2005, 2007; Weitz and Wilhelm, 2012).
While we recognize that viruses are abundant in aquifers and other subsurface environments, the factors which influence the distribution and abundance of viruses are poorly characterized. In surface aquatic environments, these factors are well characterized such as host cell abundance (Liang et al., 2014; Wigington et al., 2015) and productivity (Maranger and Bird, 1995; Clasen et al., 2008); however the abundance and distribution of continental viruses with respect to subsurface parameters such as microbial abundance and geochemical properties have not yet been established. Since virus replication depends on host cells, factors that alter host microbial growth and productivity will also have an impact on virus production. In surface aquatic microbial ecosystems, the abundance, distribution, and biogeochemical impact of viral infection are not homogenous, but often correspond to the distribution of nutrients accessible in the aqueous environment (Seymour et al., 2006; Dann et al., 2014, 2016; Wang et al., 2016). Chemical factors including DOC have been demonstrated to influence the activity of microorganisms (Peter et al., 2012) and are linked to the distribution of viruses in aquatic environments (Laybourn-Parry et al., 2001; Farnell-Jackson and Ward, 2003). Thus, the distribution of subsurface viruses may also be linked to factors that govern cell distribution, such as carbon, nutrients, and energy in the subsurface. Previous studies have shown that the addition of acetate and an electron acceptor, nitrate, to subsurface sediment stimulated the production of viruses (Pan et al., 2014), suggesting that carbon and electron acceptor availability can influence virus abundance. While stimulation of microbial activity increases virus production, little information exists regarding the distribution of viruses and organic carbon in the shallow subsurface.
Subsurface sediments are geochemically and physically heterogeneous due to deposition and burial of soil horizons and surface derived organic material. Deposition thus forms dispersed organic-rich lenses (Blazejewski et al., 2005, 2009) and is common within alluvial sedimentary environments (Blazejewski et al., 2009; Ricker et al., 2013; Chaopricha and Marín-Spiotta, 2014). As a unique facies type organic-rich deposits represent an important component of subsurface sedimentary systems and contribute to the generation of geochemically reduced zones or hotspots in the subsurface (McClain et al., 2003). These hotspots consist of high concentrations of sediment-associated organic matter in reduced zones that maintain microbial activity and contain elevated concentrations of highly reduced chemical species (Qafoku et al., 2009; Campbell et al., 2012). Together both surface derived and buried organic matter play a significant role influencing microbial activity and biogeochemistry, controlling metal/radionuclide mobility across the upper Colorado River basin (Baker et al., 2000; Janot et al., 2015). One such aquifer is the Rifle alluvial aquifer, a former U.S. Department of Energy uranium ore-processing site near the city of Rifle, CO. Storage of uranium mill tailings at the site resulted in a large resilient groundwater uranium plume (Zachara et al., 2013). Recent research indicates that organic carbon rich regions contribute to geochemically reduced zones that play a role in the persistence of the U plume retaining U as a reduced mineral phase (Campbell et al., 2012; Qafoku et al., 2014; Janot et al., 2015). Uranium reduction to an insoluble mineral form is largely mediated by microbial activity, and as such it is not surprising that the presence of viruses in groundwater within the uranium plume was revealed in metagenomic datasets obtained from this aquifer (Wrighton et al., 2012; Holmes et al., 2015). The activity of viruses has implications for microbially-mediated biogeochemical processes such as metal reduction by directly influencing active populations of metal-reducing microorganisms. While viruses have been identified at this site, studies have not elucidated the abundance and distribution of viruses in the aquifer with respect to host cell abundance and geochemistry. Here, we determined the spatial distribution of microbial cells and viruses in groundwater collected from the Rifle aquifer with respect to groundwater geochemical data. To our knowledge, this is the first report of the spatial distribution of total virus abundance in correlation with aquifer geochemistry. Due to the importance of biogeochemical cycling in subsurface systems and the subsequent impact on the fate and transport of contaminants, understanding factors that control host cell and virus distribution in subsurface systems can help elucidate subsurface biogeochemistry.
Materials and Methods
Study Area
Groundwater was sampled using peristaltic pumps from 20 monitoring wells within a shallow (20–30 ft), unconfined alluvial aquifer adjoining the Colorado River located 0.3 miles east of Rifle, Colorado (USA) (Figure 1). Groundwater flows in a south-southwesterly direction and discharges into the Colorado River. The Holocene-age alluvial sediments consist of sandy gravel and gravelly sand containing silts and clays, characteristic of many alluvial aquifers. Distributed throughout the aquifer are also lenses of naturally reduced sediments containing reduced minerals and high concentrations of organic carbon originating from buried plant material (Campbell et al., 2012; Janot et al., 2015). Groundwater within the aquifer is typically suboxic (<1 mg L−1) and contains spatially varying concentrations of reduced chemical species such as Fe(II) (10–50 μM; Williams et al., 2011). Leaching of U from former stockpiles of ore and mill tailings stored at the site resulted in a persistent plume of groundwater with elevated U concentrations (>100 μg L−1; Zachara et al., 2013).
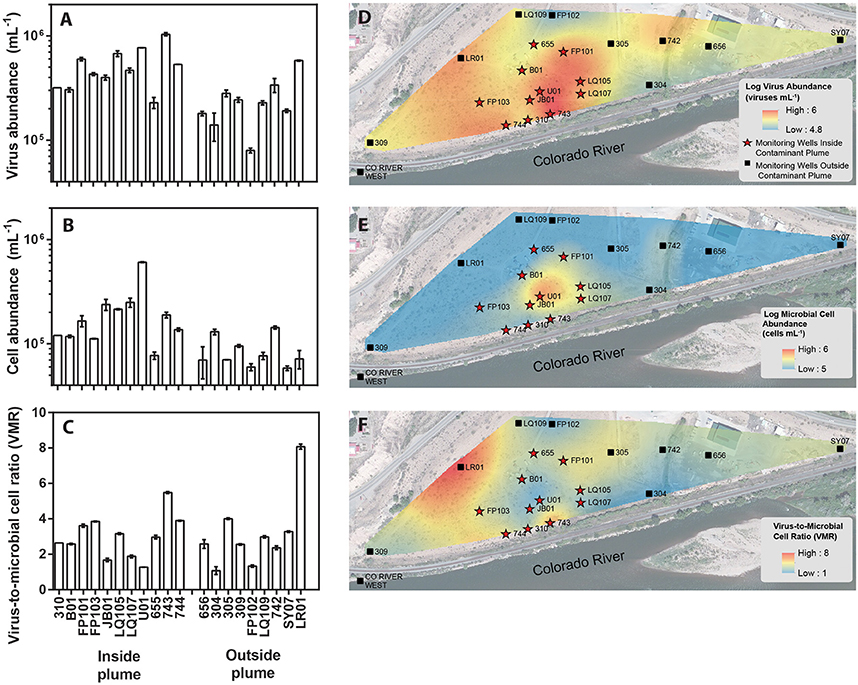
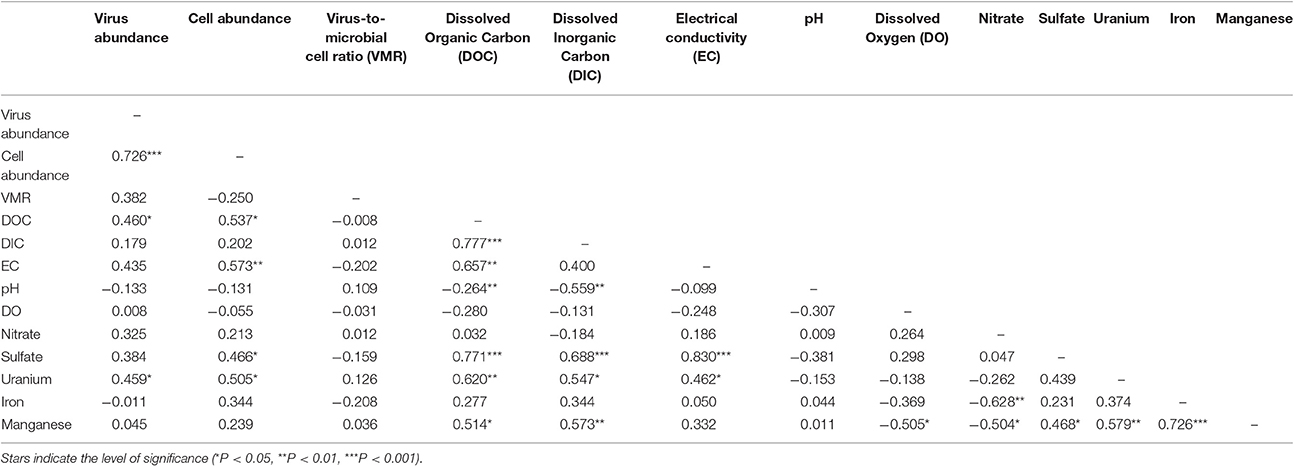
Figure 1. Location of monitoring wells within a uranium contaminated alluvial aquifer located 0.3 miles east of Rifle, Colorado (USA) and adjoining the Colorado River (Well 309 39.52842, −107.774426 and Well SY07 39.529335, −107.770864). Monitoring wells located inside the contaminant plume are denoted with red stars whereas monitoring wells located outside of the contaminant plume are denoted as black boxes. Groundwater virus abundance (A), cell abundance (B), and virus-to-microbial cell ratio (VMR) (C) data collected from monitoring wells across the alluvial aquifer. Error bars denoted standard error of measure for duplicate samples. Spatial interpolation of groundwater viruses (D), cells (E), and virus-to-microbial cell ratio (F) data collected from monitoring wells in the alluvial aquifer depicting spatial distribution. Color gradient from high (red) to low (blue) denotes interpolated values.
The major plume region has been described in prior studies conducted at the Rifle field site (Zachara et al., 2013). For the purpose of this study the boundaries of the plume region were defined where groundwater U concentrations exceeded 100 μg L−1. An elevated groundwater U concentration (171 μg L−1) was measured in well 656 in the eastern portion of the site, but groundwater in this region is geochemically distinct from the major plume with significantly lower groundwater DOC, DIC, and sulfate concentrations. Accordingly, well 656 is excluded from assignment to the major contaminant plume that is located at the center of the field site. Eleven monitoring wells within the uranium contaminated plume and nine monitoring wells outside the plume region were sampled in this study (Figure 1). Each well was approximately 6 m deep. Wells directly associated with regions impacted during prior experimental augmentation (Anderson et al., 2003) were excluded from this study. Further site details have been described elsewhere (Anderson et al., 2003; Vrionis et al., 2005; Zachara et al., 2013).
Data Collection and Processing
Replicate groundwater samples (<50 mL) for geochemical analyses and enumeration of cells and viruses were collected from purged (12 L, ca. 1–1.5 well volumes) wells at a 5 m depth using a peristaltic pump. Groundwater was filtered through 0.45 μm PVDF filters to remove sediment particles for cell enumeration, and through 0.1 μm PVDF filters to remove cells for virus enumeration. In order to reduce background noise and improve filter clarity, samples for virus enumeration were treated with a nuclease, DNase I (10 U mL−1; Danovaro and Middelboe, 2010; Carreira et al., 2015). In this study, viruses are operationally defined as DNase resistant, SYBR Green I fluorescent particles smaller than 0.1 μm and larger than 20 nm. Using this operational definition, defective viruses, gene transfer agents, and other unknown particles may plausibly be included. A maximum cutoff size of 0.1 μm was selected to prevent inclusion of cells smaller than 0.2 μm as was identified by Luef and colleagues at the Rifle site (Luef et al., 2015). To avoid including viruses larger than 0.1 μm in the cellular fraction, a minimum cutoff size of 0.2 μm was used for cell enumeration. A comparison of the sub 0.2 μm fraction and the sub 0.1 μm fraction showed that there was no statistically signficant difference (Figure S1). Thus viruses and cells that are between 0.1 μm to 0.2 μm were not enumerated in this study. Aliquots (1 mL) were preserved for both virus and cell counts by adding glutaraldehyde to a final concentration of 0.5% and incubating 15–30 min at 4°C prior to freezing in liquid N2 (Brussaard, 2009). Samples were packed in dry ice and shipped to the University of Nebraska-Lincoln for storage at −80°C prior to enumeration.
Duplicate samples were thawed for enumeration by epifluorescence microscopy. Viruses were collected on Anodisc filters (0.02 μm), while cells were collected on black polycarbonate filters (0.2 μm). Between 0.5 and 1 mL of sample was passed through each filter. SYBR Green I (400x dilution from original stock) was used to stain the filters (15 min) for enumeration by epifluorescence microscopy. We note that because SYBR Green I binds to dsDNA more efficiently than ssDNA and RNA, the total virus count may be underestimated. After staining and drying, filters were mounted on slides with an anti-fading solution (50% glycerol, 50% phosphate buffered saline, 0.1% p-phenylenediamine). Background fluoresence in groundwater samples was minimal and did not interfere with enumeration (Figure S2). At least 10 fields or 200 particles were enumerated per filter (Patel et al., 2007). For each sample, duplicate field replicates were enumerated for cells and viruses. Blanks of TE buffer were routinely checked to confirm the lack of viral and microbial contaminants on filters.
Groundwater dissolved oxygen (DO) concentrations were measured in situ deploying multi-parameter sondes (YSI Inc., OH) into the well. Groundwater samples were directly filtered (0.45 μm PTFE) into glass vials for analysis of DOC/DIC while samples for anion analysis were filtered directly into HDPE vials. Vials were capped, leaving no headspace, and stored at 4°C prior to analyses (Williams et al., 2011). Aqueous anions (sulfate and nitrate) were measured by ion chromatography (ICS-2100 equipped with AS18 column, Dionex, CA; Kantor et al., 2013). DOC/DIC was measured by combustion catalytic oxidation and NDIR method using a Shimadzu Total Organic Carbon Analyzer (TOC-VCSH; Shimadzu, Corp.). Measurements of DOC will also include cell and viral biomass due to the cutoff used for defining the aqueous fraction (<0.45 μm), however cell and viral biomass do not make a significant portion of any DOC measurement in this study. Using upper limits of 149 fg of C per cell (Vrede et al., 2002) and 106 cells mL−1, no more than 0.15 mg L−1 can come from cellular biomass, which constitutes only a minor fraction of the DOC measured in this study. Viruses, being orders of magnitude smaller than cells, are a negligible component of total measured DOC.
Data Analyses
Spatial interpolation (ArcGIS, Desktop Release 10.1, Environmental Systems Research Institute, Redlands, CA) was used to geographically represent the distribution of viruses and cells as well as geochemical parameters (DOC, DIC, sulfate, dissolved Fe, dissolved Mn, nitrate, and pH) across the alluvial aquifer (Nolan and Weber, 2015). Spline interpolation method (from the ArcGIS Spatial Analyst extension) was selected from among three interpolation methods (kriging, inverse distance weighting, spline) as it resulted in the lowest residual error (Akkala et al., 2010).
Correlation analyses and statistical comparisons were conducted in GraphPad Prism 5.0.3 (GraphPad Software). Significance level was defined at p < 0.05. Analyses involving cell and virus abundances were conducted on log-transformed values. All correlations between all measured parameters were calculated by the Spearman rank correlation method. Statistical comparisons between parameters within the major plume region and outside were conducted by t-test.
Results and Discussion
Groundwater Geochemistry and Virus and Microbial Cell Distribution and Abundance
Groundwater virus abundance ranged from 8.0 × 104 to 1.0 × 106 viruses mL−1 (Figures 1A,D) and exceeded cell abundance (range 6.0 × 104 to 6.1 × 105 cells ml−1; Figures 1B,E) in all 20 monitoring wells. These total abundance values of viruses enumerated in groundwater were similar to results obtained from other groundwater sites (Kyle et al., 2008; Roudnew et al., 2012). Virus abundance in this shallow aquifer was strongly correlated to cell abundance (ρ = 0.73, p < 0.001) (Figure 2C) and is consistent with prior studies comparing virus and cell abundance in aquatic and sedimentary environments including lakes (Maranger and Bird, 1995; Bettarel et al., 2006; de Araújo and Godinho, 2009; Barros et al., 2010), marine waters (Alonso et al., 2001; Pereira et al., 2009), marine surface sediments (Danovaro and Serresi, 2000), marine subsurface sediment (Bird et al., 2001; Engelhardt et al., 2014), and deep granitic groundwater (Kyle et al., 2008). The virus-to-microbial cell ratio (VMR) in this shallow aquifer ranged from 1.1 to 8.1 and averaged 3.0 ± 1.6 (mean ± S.D., n = 20; Figures 1C,F). VMR range observed in groundwater collected from the Rifle aquifer is consistent with another shallow aquifer ranging from 0.4 to 6.1 (Roudnew et al., 2012) but is slightly less than has been observed in deep aquifers (average VMR of 12; Kyle et al., 2008). However, the VMR was notably lower than was measured in water collected from the Colorado River (VMR = 33; Table S1). It should be noted that the VMR in the Colorado River reported in this study is consistent with results from other river systems (Mathias et al., 1995; Jiao et al., 2006; Luef et al., 2007). This result is not surprising as total virus abundance in the river water sample exceeds virus abundance in groundwater. The lower VMR in groundwater relative to the river water sample could be a result of adsorption of viruses to the aquifer alluvium. Free (planktonic) viruses in the groundwater were enumerated in this study so any viruses produced that were adsorbed onto the aquifer alluvium (including clays and reactive minerals) would have been excluded resulting in a lower total virus abundance in the groundwater. The adsorption of viruses to minerals such as clays and iron oxides (Hewson and Fuhrman, 2003; You et al., 2005; Kernegger et al., 2009; Nieto-Juarez and Kohn, 2013) is recognized to reduce planktonic virus abundance. Previous studies have found a correlation between viral abundance and microbial activity in marine waters (Corinaldesi et al., 2003). In subsurface sediments, stimulation of microbial activity was found to result in an increase in VMR (Pan et al., 2014). Thus, the differences in viral abundance/VMR may indicate differences in microbial activity between groundwater and surface water. Together these are plausible reasons that may explain the difference in the abundance of viruses between the river water and groundwater samples.
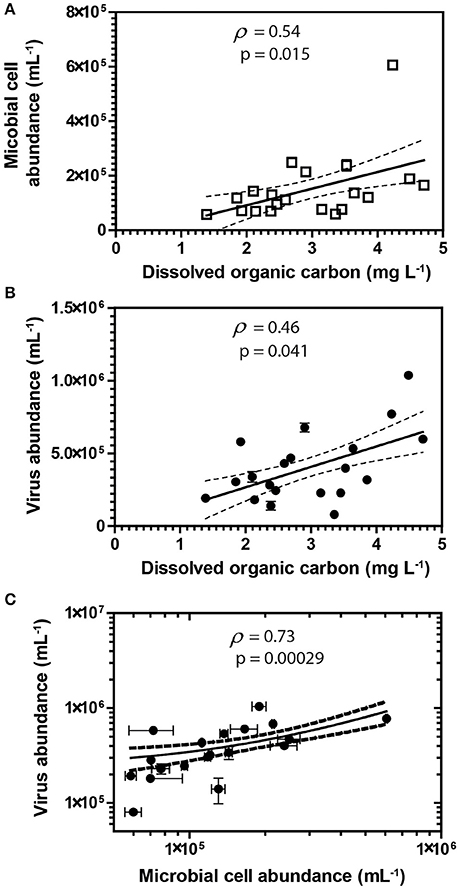
Figure 2. Spearman's rank correlations between DOC and abundances of cells (A) and viruses (B). Spearman's rank correlation between viruses and cells (C) in groundwater samples. Error bars represent the standard deviation of the mean of duplicate measurements. Error bars not visible are smaller than the symbol. Dashed lines represent the 95% confidence interval for the lines of regression presented in the figure.
A correlation between microbial cell abundance, virus abundance, and groundwater geochemistry was observed within the aquifer. Microbial cell abundance in groundwater was significantly higher (Unpaired t-test, p < 0.01) within the uranium plume (2.0 × 105 ± 1.4 × 105 cells mL−1, mean ± S.D., n = 11) compared to cell abundance in groundwater outside of the plume (8.6 × 104 ± 3.1 × 104 cells mL−1, mean ± S.D., n = 9; Table 1, Figures 1B,E). Cell abundance had a strong positive correlation to groundwater DOC (Spearman's ρ = 0.54, p < 0.05), U (Spearman's ρ = 0.51, p < 0.05), and sulfate concentrations (Spearman's ρ = 0.47, p < 0.05). However, it should be noted that groundwater DOC concentrations were also higher within the uranium plume (3.45 ± 0.88 mg L−1; mean ± S.D., n = 11), relative to the concentrations measured outside (2.36 ± 0.60 mg L−1; mean ± S.D., n = 9) of the uranium plume (Table 1, Figure 3A). Elevated concentrations of DOC may arise from buried organic carbon identified at the site which have been demonstrated to be responsible for persistence of the U plume (Campbell et al., 2012; Janot et al., 2015; Boye et al., 2017). Groundwater DOC and U concentrations were also correlated with virus abundance (Spearman's ρ = 0.46, p < 0.05 and ρ = 0.46, p < 0.05, respectively; Figures 2A,B, Table 2). Similar to the distribution of cells in the aquifer, virus abundance in the uranium plume region was statistically higher (Unpaired t-test, p < 0.01), 5.2 × 105 ± 2.4 × 105 viruses mL−1 (mean ± S.D., n = 11), relative to the abundance of viruses outside of the region, 2.5 × 105 ± 1.4 × 105 viruses mL−1 (mean ± S.D., n = 9; Table 1, Figures 1A,D). While virus and cell abundance in groundwater were positively correlated to groundwater DOC concentration, VMR did not exhibit a statistically significant correlation to any geochemical factor (Table 2, Figures 2A,B).
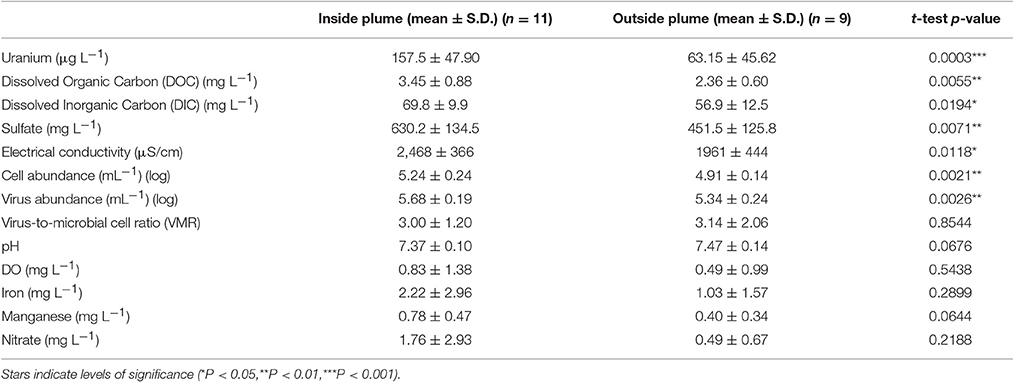
Table 1. Comparison of biotic and geochemical parameters in samples collected from monitoring wells located inside and outside of the contaminant plume.
While there is the potential for hyporheic intrusion of DOC from the Colorado River, elevated DOC concentrations were not observed in samples collected from wells located near the river, nor were other proxies for river water incursion into the aquifer, such as low electrical conductivity (data not shown). While we did not test lability of the DOC, correlations with cell (Spearman's ρ = 0.46, p < 0.05) and virus abundance (Spearman's ρ = 0.54, p < 0.05) strongly suggest that DOC was sufficiently bioavailable to stimulate microbial activity. Microbial activity in groundwater is often stimulated by inputs of DOC (Baker et al., 2000; Sobczak and Findlay, 2002; Findlay et al., 2003; Foulquier et al., 2011; Li et al., 2012). The presence of bioavailable DOC and available electron acceptors may thus provide sufficient energy for stimulation of microbial respiration. Because viruses are reliant on metabolically active hosts for replication, microbial host energy availability favors the production of viruses. Host cell metabolic activity and growth rate has been directly demonstrated to increase virus adsorption rate and decrease the period of time between viral infection and lysis of the host for lytic viruses (Hadas et al., 1997). Lysogenic bacteriophage have also been demonstrated to respond to host cell metabolic activity; control of the lytic and lysogenic pathway is controlled by levels of cAMP, with high energy conditions favoring lysis (Hong et al., 1971; Rolfe et al., 1973). In addition, chronic infections, in which viruses are released without lysis of the host, also produce greater numbers of viruses under higher energy conditions (Brown and Dowell, 1968). This was recently demonstrated in a series of alluvial aquifer sediment microcosms where additions of an energy source, acetate, and electron acceptor, nitrate, not only resulted in the oxidation of organic carbon, but also significant virus production (Pan et al., 2014). This result is consistent with prior studies conducted in surface aquatic environments in which correlation between virus abundance and DOC has been observed (Laybourn-Parry et al., 2001, 2013; Auguet et al., 2005; Holmfeldt et al., 2010; Säwström and Pollard, 2012). Inputs of organic carbon increase microbial activity (Peter et al., 2012), and organic-rich regions are also inferred to have elevated microbial activities (Campbell et al., 2012). As a result elevated microbial activity would result in enhanced virus production and hence higher virus abundance within the plume. Viral production and expression of virus-related genes following acetate biostimulation has been demonstrated previously at the Rifle site (Holmes et al., 2015). Given that stimulated microbial activity will result in the production of viruses (Pan et al., 2014), the elevated abundance of viruses in the plume correlated with organic carbon suggests that there is ongoing microbial activity and virus production in the aquifer.
The consumption of organic carbon in this aquifer has been linked to the reduction of molecular oxygen, nitrate, iron, manganese, uranium, and sulfate as well as fermentation reactions (Wrighton et al., 2012; Anantharaman et al., 2016). While suboxic conditions (DO < 1 mg L−1) predominated throughout most of the plume, oxic conditions were identified in two wells within the center of the plume region: LQ105 (4.3 mg L−1) and U01 (2.6 mg L−1) (Table S2). Dissolved Fe and Mn concentrations suggested the presence of reduced Fe (Fe(II)) and Mn (Mn(II)) and were also substantially lower within these two wells (Table S2, Figure S3). Thus lower Fe and Mn concentrations may be due to oxidative precipitation of Fe or Mn oxide minerals or lack of metal reduction due to oxic conditions in the groundwater. The highest dissolved Fe and Mn concentrations were found in wells located along the central portion of the site closest to the Colorado River (744, 310, 743, JB01, LQ107, 304) (Figure S3). Groundwater U concentrations ranged from 26.5 μg L−1 to 7.4 mg L−1, largely localized to the center of the site (Figure 3B). Nitrate was also low to undetectable in most of the wells throughout the floodplain with the exception of the two oxic monitoring wells LQ105 and U01 (5.89 mg L−1 and 8.25 mg L−1, respectively) (Table S2, Figure 3C). This may reflect operative nitrification processes or lack of denitrification due to oxic conditions in these wells. These geochemical conditions indicate a substantially different redox environment, potentially due to intrusion of dissolved oxygen or nitrate at the capillary fringe (Williams and Oostrom, 2000). The oxic monitoring wells U01 and LQ105 also contained some of the highest cell and virus abundances in the major plume region (Figure 1). This may be expected because O2 mediated respiration is expected to support greater cell abundance and hence greater virus abundance. Sulfate concentrations averaged 630.2 ± 134.5 mg L−1 (mean ± S.D., n = 11) within the plume and 451.5 ± 125.8 mg L−1 outside of the contaminant plume (mean ± S.D., n = 9) (Unpaired t-test P < 0.05; Table 1, Figure 3D). In addition, DIC was also higher within the plume (69.8 ± 9.9 mg L−1, mean ± S.D., n = 11) compared to outside (56.9 ± 12.5 mg L−1, mean ± S.D., n = 9) (Table 1, Figure 3E). Sulfate could also serve as a potential electron acceptor and has been implicated in the generation and precipitation of reduced sulfur phases such as framboidal pyrite, mackinwite, and greigite (Qafoku et al., 2009; Janot et al., 2015) within the aquifer. Elevated virus abundance in the plume region is consistent with prior results demonstrating that bacterial sulfate reduction rates were correlated with viral abundance and distribution in estuarine sediments (Middelboe et al., 2003). As such microbial metabolisms could thus be supported by the dynamic changes in redox conditions that are associated with the influx of oxidants into a carbon-rich reduced system.
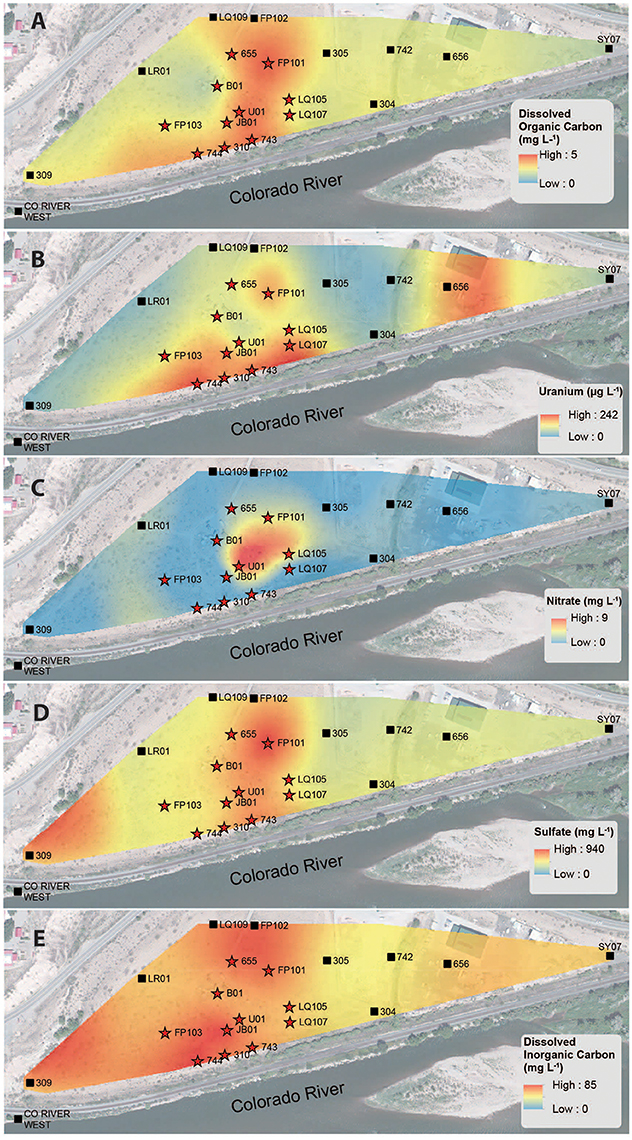
Figure 3. Spatial interpolation of groundwater DOC (A), uranium (B), nitrate (C), sulfate (D), and DIC (E) data collected from monitoring wells in the alluvial aquifer depicting spatial distribution. Color gradient from high (red) to low (blue) denotes interpolated concentrations.
Implications of Viral Activity on Microbially-Mediated Subsurface Biogeochemical Cycling
Here we demonstrate that viruses, cells, and DOC are enriched in the major plume region of the Rifle aquifer. The widespread distribution of viruses at the Rifle aquifer is consistent with prior suggestions that viruses may play a potential role in influencing biogeochemical cycling at the site (Wrighton et al., 2014; Holmes et al., 2015). Interactions between organic carbon, cells, and viruses may be important in riparian aquifers across the upper Colorado River basin where buried organic matter plays an important role in mediating biogeochemical cycles and metal/radionuclide sequestration. Organic carbon availability can promote microbial activity (Baker et al., 2000; Sobczak and Findlay, 2002; Findlay et al., 2003; Foulquier et al., 2011; Li et al., 2012) and, in turn, virus production, which is reflected by elevated virus abundances. Thus, organic rich sediments may potentially represent regions of increased viral activity as a response to higher microbial metabolic activity.
Within the Rifle aquifer, virus mediated cell lysis has been suggested to contribute biologically available organic carbon or to suppress certain taxa responsible for biogeochemically important reactions at the site (Wrighton et al., 2014; Holmes et al., 2015). In addition to lysis, lysogeny is another possible life cycle. Recent proposed models of lysogeny have suggested that at high and low host cell densities lysogenic life cycles may be favored, however with the cell densities encountered in this study (105–106 cells/mL), lytic kill-the-winner dynamics are suggested to be favored (Knowles et al., 2016). Lysis of active members of the microbial community and liberation of organic carbon can both influence biogeochemical cycling. Together these impacts have biogeochemical implications for the long-persisting groundwater U plume located in the Rifle aquifer and other similar aquifers within the upper Colorado River basin. The increase in bioavailable carbon would accelerate cell turnover rates and organic carbon liberation (Middelboe et al., 1996; Noble and Fuhrman, 1999; Middelboe and Lyck, 2002; Eissler et al., 2003), creating labile particulate and dissolved organic carbon thereby providing a source of biologically available carbon (Xu et al., 2013, 2014) subsequently controlling the community from the bottom up. The liberated carbon can increase respiration rates of heterotrophic bacteria (Middelboe and Lyck, 2002), which may include U-reducing bacteria responsible for the precipitation and immobilization of U(IV) (Anderson et al., 2003; Chang et al., 2005; Williams et al., 2011). Alternatively, viruses may also depress rates of biogeochemical transformations by infecting and lysing the organisms responsible for important organic carbon dependent subsurface processes such as denitrification (Burt et al., 1999) or metal reduction (Holmes et al., 2015). Further research is necessary to explore the role that viruses play mediating microbial processes underpinning reactions responsible for the fate and transport of metals/radionuclides impacting groundwater quality.
Author Contributions
DP and KAW contributed to laboratory and statistical analyses of data. KHW and MR contributed to field sample collection and geochemical analyses. JN contributed to the spatial interpolation of data. DP and KAW contributed to the experimental design and data interpretation. All authors contributed to writing of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Funding provided by the U.S. Department of Energy (DOE), Office of Science, Office of Biological and Environmental Research under contracts DE-SC0004113 (University of Nebraska-Lincoln) and USGS (2014NE265G) to KAW and DE-AC02-05CH11231 (Lawrence Berkeley National Laboratory; operated by the University of California) to KHW. This material is partially based upon work supported through the Lawrence Berkeley National Laboratory's Genomes-to-Watershed Scientific Focus Area. DP was supported by National Science Foundation IGERT Fellowship (0903469), NASA Nebraska Space Grant, and the UNL School of Biological Sciences Special Funds.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01199/full#supplementary-material
References
Akkala, A., Devabhaktuni, V., and Kumar, A. (2010). Interpolation techniques and associated software for environmental data. Environ. Prog. Sustain. Energy 29, 134–141. doi: 10.1002/ep.10455
Alonso, M., Jimenez-Gomez, F., Rodriguez, J., and Borrego, J. (2001). Distribution of virus-like particles in an oligotrophic marine environment (Alboran Sea, Western Mediterranean). Microb. Ecol. 42, 407–415. doi: 10.1007/s00248-001-0015-y
Anantharaman, K., Brown, C. T., Hug, L. A., Sharon, I., Castelle, C. J., Probst, A. J., et al. (2016). Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 7:13219. doi: 10.1038/ncomms13219
Anderson, R. E., Brazelton, W. J., and Baross, J. A. (2013). The deep viriosphere: assessing the viral impact on microbial community dynamics in the deep subsurface. Rev. Mineral. Geochem. 75, 649–675. doi: 10.2138/rmg.2013.75.20
Anderson, R. T., Vrionis, H. A., Ortiz-Bernad, I., Resch, C. T., Long, P. E., Dayvault, R., et al. (2003). Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69, 5884–5891. doi: 10.1128/AEM.69.10.5884-5891.2003
Auguet, J., Montanie, H., Delmas, D., Hartmann, H., and Huet, V. (2005). Dynamic of virioplankton abundance and its environmental control in the Charente Estuary (France). Microb. Ecol. 50, 337–349. doi: 10.1007/s00248-005-0183-2
Baker, M. A., Valett, H. M., and Dahm, C. N. (2000). Organic carbon supply and metabolism in a shallow groundwater ecosystem. Ecology 81, 3133–3148. doi: 10.1890/0012-9658(2000)081[3133:OCSAMI]2.0.CO;2
Barros, N., Farjalla, V. F., Soares, M. C., Melo, R. C., and Roland, F. (2010). Virus-bacterium coupling driven by both turbidity and hydrodynamics in an Amazonian floodplain lake. Appl. Environ. Microbiol. 76, 7194–7201. doi: 10.1128/AEM.01161-10
Bettarel, Y., Bouvy, M., Dumont, C., and Sime-Ngando, T. (2006). Virus-bacterium interactions in water and sediment of West African Inland aquatic systems. Appl. Environ. Microbiol. 72, 5274–5282. doi: 10.1128/AEM.00863-06
Bird, D., Juniper, S., Ricciardi-Rigault, M., Martineu, P., Prairie, Y., and Calvert, S. (2001). Subsurface viruses and bacteria in Holocene/Late Pleistocene sediments of Saanich Inlet, BC: ODP holes 1033b and 1034b, Leg 169s. Mar. Geol. 174, 227–239. doi: 10.1016/S0025-3227(00)00152-3
Blazejewski, G. A., Stolt, M. H., Gold, A. J., and Groffman, P. M. (2005). Macro-and micromorphology of subsurface carbon in riparian zone soils. Soil Sci. Soc. Am. J. 69, 1320–1329. doi: 10.2136/sssaj2004.0145
Blazejewski, G. A., Stolt, M. H., Gold, A. J., Gurwick, N., and Groffman, P. M. (2009). Spatial distribution of carbon in the subsurface of riparian zones. Soil Sci. Soc. Am. J. 73, 1733–1740. doi: 10.2136/sssaj2007.0386
Boye, K., Noël, V., Tfaily, M. M., Bone, S. E., Williams, K. H., Bargar, J. R., et al. (2017). Thermodynamically controlled preservation of organic carbon in floodplains. Nat. Geosci. 10, 415–419. doi: 10.1038/ngeo2940
Brown, L. R., and Dowell, C. (1968). Replication of coliphage M-13 I. Effects on host cells after synchronized infection. J. Virol. 2, 1290–1295.
Brussaard, C. D. (2009). “Enumeration of bacteriophages using flow cytometry,” in Bacteriophages, eds M. J. Clokie and A. Kropinski (New York, NY: Humana Press), 97–111.
Burt, T., Matchett, L., Goulding, K., Webster, C., and Haycock, N. (1999). Denitrification in riparian buffer zones: the role of floodplain hydrology. Hydrol. Process. 13, 1451–1463. doi: 10.1002/(SICI)1099-1085(199907)13:10<1451::AID-HYP822>3.0.CO;2-W
Campbell, K. M., Kukkadapu, R. K., Qafoku, N., Peacock, A. D., Lesher, E., Williams, K. H., et al. (2012). Geochemical, mineralogical and microbiological characteristics of sediment from a naturally reduced zone in a uranium-contaminated aquifer. Appl. Geochem. 27, 1499–1511. doi: 10.1016/j.apgeochem.2012.04.013
Carreira, C., Staal, M., Middelboe, M., and Brussaard, C. P. (2015). Counting viruses and bacteria in photosynthetic microbial mats. Appl. Environ. Microbiol. 81, 2149–2155. doi: 10.1128/AEM.02863-14
Chang, Y.-J., Long, P. E., Geyer, R., Peacock, A. D., Resch, C. T., Sublette, K., et al. (2005). Microbial incorporation of 13C-labeled acetate at the field scale: detection of microbes responsible for reduction of U (VI). Environ. Sci. Technol. 39, 9039–9048. doi: 10.1021/es051218u
Chaopricha, N. T., and Marín-Spiotta, E. (2014). Soil burial contributes to deep soil organic carbon storage. Soil Biol. Biochem. 69, 251–264. doi: 10.1016/j.soilbio.2013.11.011
Clasen, J. L., Brigden, S. M., Payet, J. P., and Suttle, C. A. (2008). Evidence that viral abundance across oceans and lakes is driven by different biological factors. Freshw. Biol. 53, 1090–1100. doi: 10.1111/j.1365-2427.2008.01992.x
Corinaldesi, C., Crevatin, E., Del Negro, P., Marini, M., Russo, A., Fonda-Umani, S., et al. (2003). Large-scale spatial distribution of virioplankton in the Adriatic Sea: testing the trophic state control hypothesis. Appl. Environ. Microbiol. 69, 2664–2673. doi: 10.1128/AEM.69.5.2664-2673.2003
Dann, L. M., Mitchell, J. G., Speck, P. G., Newton, K., Jeffries, T., and Paterson, J. (2014). Virio-and bacterioplankton microscale distributions at the sediment-water interface. PLoS ONE 9:e102805. doi: 10.1371/journal.pone.0102805
Dann, L. M., Paterson, J. S., Newton, K., Oliver, R., and Mitchell, J. G. (2016). Distributions of virus-like particles and prokaryotes within microenvironments. PLoS ONE 11:e0146984. doi: 10.1371/journal.pone.0146984
Danovaro, R., and Middelboe, M. (2010). “Separation of free virus particles from sediments in aquatic systems,” in Manual of Aquatic Viral Ecology, eds S. W. Wilhelm, M. G. Weinbauer, and C. A. Suttle (Waco, TX: ASLO), 74–81. doi: 10.4319/mave.2010.978-0-9845591-0-7.74
Danovaro, R., and Serresi, M. (2000). Viral density and virus-to-bacterium ratio in deep-sea sediments of the Eastern Mediterranean. Appl. Environ. Microbiol. 66, 1857–1861. doi: 10.1128/AEM.66.5.1857-1861.2000
de Araújo, M. F., and Godinho, M. J. (2009). Short-term variations of virus-like particles in a tropical lake: relationship with microbial communities (bacteria, ciliates and flagellates). Microbiol. Res. 164, 411–419. doi: 10.1016/j.micres.2007.02.011
Eissler, Y., Sahlsten, E., and Qui-ones, R. A. (2003). Effects of virus infection on respiration rates of marine phytoplankton and microplankton communities. Mar. Ecol. Prog. Ser. 262, 71–80. doi: 10.3354/meps262071
Engelhardt, T., Kallmeyer, J., Cypionka, H., and Engelen, B. (2014). High virus-to-cell ratios indicate ongoing production of viruses in deep subsurface sediments. ISME J. 8, 1503–1509. doi: 10.1038/ismej.2013.245
Farnell-Jackson, E., and Ward, A. (2003). Seasonal patterns of viruses, bacteria and dissolved organic carbon in a riverine wetland. Freshw. Biol. 48, 841–851. doi: 10.1046/j.1365-2427.2003.01052.x
Findlay, S. E., Sinsabaugh, R. L., Sobczak, W. V., and Hoostal, M. (2003). Metabolic and structural response of hyporheic microbial communities to variations in supply of dissolved organic matter. Limnol. Oceanogr. 48, 1608–1617. doi: 10.4319/lo.2003.48.4.1608
Foulquier, A., Malard, F., Mermillod-Blondin, F., Montuelle, B., Dolédec, S., Volat, B., et al. (2011). Surface water linkages regulate trophic interactions in a groundwater food web. Ecosystems 14, 1339–1353. doi: 10.1007/s10021-011-9484-0
Fuhrman, J. A. (1999). Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548. doi: 10.1038/21119
Hadas, H., Einav, M., Fishov, I., and Zaritsky, A. (1997). Bacteriophage T4 development depends on the physiology of its host Escherichia coli. Microbiology 143, 179–185. doi: 10.1099/00221287-143-1-179
Hewson, I., and Fuhrman, J. (2003). Viriobenthos production and virioplankton sorptive scavenging by suspended sediment particles in coastal and pelagic waters. Microb. Ecol. 46, 337–347. doi: 10.1007/s00248-002-1041-0
Holmes, D. E., Giloteaux, L., Chaurasia, A. K., Williams, K. H., Luef, B., Wilkins, M. J., et al. (2015). Evidence of Geobacter-associated phage in a uranium-contaminated aquifer. ISME J. 9, 333–346. doi: 10.1038/ismej.2014.128
Holmfeldt, K., Titelman, J., and Riemann, L. (2010). Virus production and lysate recycling in different sub-basins of the Northern Baltic Sea. Microb. Ecol. 60, 572–580. doi: 10.1007/s00248-010-9668-8
Hong, J.-S., Smith, G. R., and Ames, B. N. (1971). Adenosine 3′: 5′-cyclic monophosphate concentration in the bacterial host regulates the viral decision between lysogeny and lysis. Proc. Natl. Acad. Sci. U.S.A. 68, 2258–2262. doi: 10.1073/pnas.68.9.2258
Janot, N., Lezama Pacheco, J. S., Pham, D. Q., O'Brien, T. M., Hausladen, D., Noël, V., et al. (2015). Physico-chemical heterogeneity of organic-rich sediments in the Rifle aquifer, CO: impact on uranium biogeochemistry. Environ. Sci. Technol. 50, 46–53. doi: 10.1021/acs.est.5b03208
Jiao, N., Zhao, Y., Luo, T., and Wang, X. (2006). Natural and anthropogenic forcing on the dynamics of virioplankton in the Yangtze river estuary. J. Mar. Biol. Assoc. U.K. 86, 543–550. doi: 10.1017/S0025315406013452
Kallmeyer, J., Pockalny, R., Adhikari, R. R., Smith, D. C., and D'Hondt, S. (2012). Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. U.S.A. 109, 16213–16216. doi: 10.1073/pnas.1203849109
Kantor, R. S., Wrighton, K. C., Handley, K. M., Sharon, I., Hug, L. A., Castelle, C. J., et al. (2013). Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. MBio 4, e00708–00713. doi: 10.1128/mBio.00708-13
Kernegger, L., Zweimüller, I., and Peduzzi, P. (2009). Effects of suspended matter quality and virus abundance on microbial parameters: experimental evidence from a large European river. Aquat. Microb. Ecol. 57:161. doi: 10.3354/ame01341
Knowles, B., Silveira, C., Bailey, B., Barott, K., Cantu, V., Cobián-Güemes, A., et al. (2016). Lytic to temperate switching of viral communities. Nature 531, 466–470. doi: 10.1038/nature17193
Kyle, J. E., Eydal, H. S. C., Ferris, F. G., and Pedersen, K. (2008). Viruses in granitic groundwater from 69 to 450m depth of the Aspo hard rock laboratory, Sweden. ISME J. 2, 571–574. doi: 10.1038/ismej.2008.18
Laybourn-Parry, J., Anesio, A. M., Madan, N., and Säwström, C. (2013). Virus dynamics in a large epishelf lake (Beaver Lake, Antarctica). Freshw. Biol. 58, 1484–1493. doi: 10.1111/fwb.12146
Laybourn-Parry, J., Hofer, J. S., and Sommaruga, R. (2001). Viruses in the plankton of freshwater and saline Antarctic lakes. Freshw. Biol. 46, 1279–1287. doi: 10.1046/j.1365-2427.2001.00749.x
Li, D., Sharp, J. O., Saikaly, P. E., Ali, S., Alidina, M., Alarawi, M. S., et al. (2012). Dissolved organic carbon influences microbial community composition and diversity in managed aquifer recharge systems. Appl. Environ. Microbiol. 78, 6819–6828. doi: 10.1128/AEM.01223-12
Liang, Y., Li, L., Luo, T., Zhang, Y., Zhang, R., and Jiao, N. (2014). Horizontal and vertical distribution of marine virioplankton: a basin scale investigation based on a global cruise. PLoS ONE 9:e111634. doi: 10.1371/journal.pone.0111634
Luef, B., Aspetsberger, F., Hein, T., Huber, F., and Peduzzi, P. (2007). Impact of hydrology on free-living and particle-associated microorganisms in a river floodplain system (Danube, Austria). Freshw. Biol. 52, 1043–1057. doi: 10.1111/j.1365-2427.2007.01752.x
Luef, B., Frischkorn, K. R., Wrighton, K. C., Holman, H.-Y. N., Birarda, G., Thomas, B. C., et al. (2015). Diverse uncultivated ultra-small bacterial cells in groundwater. Nat. Commun. 6:6372. doi: 10.1038/ncomms7372
Maranger, R., and Bird, D. F. (1995). Viral abundance in aquatic systems: a comparison between marine and fresh waters. Mar. Ecol. Prog. Ser. 121, 217–226. doi: 10.3354/meps121217
Mathias, C., Kirschner, A., and Velimirov, B. (1995). Seasonal variations of virus abundance and viral control of the bacterial production in a backwater system of the danube river. Appl. Environ. Microbiol. 61, 3734–3740.
McClain, M. E., Boyer, E. W., Dent, C. L., Gergel, S. E., Grimm, N. B., Groffman, P. M., et al. (2003). Biogeochemical hot spots and hot moments at the interface of terrestrial and aquatic ecosystems. Ecosystems 6, 301–312. doi: 10.1007/s10021-003-0161-9
Middelboe, M., Glud, R. N., and Finster, K. (2003). Distribution of viruses and bacteria in relation to diagenetic activity in an estuarine sediment. Limnol. Oceanogr. 48, 1447–1456. doi: 10.4319/lo.2003.48.4.1447
Middelboe, M., Jorgensen, N., and Kroer, N. (1996). Effects of viruses on nutrient turnover and growth efficiency of noninfected marine bacterioplankton. Appl. Environ. Microbiol. 62, 1991–1997.
Middelboe, M., and Lyck, P. G. (2002). Regeneration of dissolved organic matter by viral lysis in marine microbial communities. Aquat. Microb. Ecol. 27, 187–194. doi: 10.3354/ame027187
Nieto-Juarez, J. I., and Kohn, T. (2013). Virus removal and inactivation by iron (hydr)oxide-mediated Fenton-like processes under sunlight and in the dark. Photochem. Photobiol. Sci. 12, 1596–1605. doi: 10.1039/c3pp25314g
Noble, R. T., and Fuhrman, J. A. (1999). Breakdown and microbial uptake of marine viruses and other lysis products. Aquat. Microb. Ecol. 20, 1–11. doi: 10.3354/ame020001
Nolan, J., and Weber, K. A. (2015). Natural uranium contamination in Major U.S. Aquifers linked to nitrate. Environ. Sci. Technol. Lett. 2, 215–220. doi: 10.1021/acs.estlett.5b00174
Pan, D., Watson, R., Wang, D., Tan, Z. H., Snow, D. D., and Weber, K. A. (2014). Correlation between viral production and carbon mineralization under nitrate-reducing conditions in aquifer sediment. ISME J. 8, 1691–1703. doi: 10.1038/ismej.2014.38
Patel, A., Noble, R. T., Steele, J. A., Schwalbach, M. S., Hewson, I., and Fuhrman, J. A. (2007). Virus and prokaryote enumeration from planktonic aquatic environments by epifluorescence microscopy with SYBR green I. Nat. Protoc. 2, 269–276. doi: 10.1038/nprot.2007.6
Pereira, G., Granato, A., Figueiredo, A., and Ebecken, N. (2009). Virioplankton abundance in trophic gradients of an upwelling field. Braz. J. Microbiol. 40, 857–865. doi: 10.1590/S1517-83822009000400017
Peter, S., Koetzsch, S., Traber, J., Bernasconi, S. M., Wehrli, B., and Durisch-Kaiser, E. (2012). Intensified organic carbon dynamics in the ground water of a restored riparian zone. Freshw. Biol. 57, 1603–1616. doi: 10.1111/j.1365-2427.2012.02821.x
Qafoku, N. P., Gartman, B. N., Kukkadapu, R. K., Arey, B. W., Williams, K. H., Mouser, P. J., et al. (2014). Geochemical and mineralogical investigation of uranium in multi-element contaminated, organic-rich subsurface sediment. Appl. Geochem. 42, 77–85. doi: 10.1016/j.apgeochem.2013.12.001
Qafoku, N. P., Kukkadapu, R. K., McKinley, J. P., Arey, B. W., Kelly, S. D., Wang, C., et al. (2009). Uranium in framboidal pyrite from a naturally bioreduced alluvial sediment. Environ. Sci. Technol. 43, 8528–8534. doi: 10.1021/es9017333
Ricker, M. C., Stolt, M. H., Donohue, S. W., Blazejewski, G. A., and Zavada, M. S. (2013). Soil organic carbon pools in riparian landscapes of southern New England. Soil Sci. Soc. Am. J. 77, 1070–1079. doi: 10.2136/sssaj2012.0297
Rolfe, B., Schell, J., Becker, A., Heip, J., Onodera, K., and Schell-Frederick, E. (1973). A colicin-tolerant mutant of Escherichia coli with reduced levels of cyclic AMP and a strong bias towards λ lysogeny. Mol. Gen. Genet. 120, 1–16. doi: 10.1007/BF00332980
Roudnew, B., Seymour, J. R., Jeffries, T. C., Lavery, T. J., Smith, R. J., and Mitchell, J. G. (2012). Bacterial and virus-like particle abundances in purged and unpurged groundwater depth profiles. Groundwater Monit. Remediat. 32, 72–77. doi: 10.1111/j.1745-6592.2011.01393.x
Säwström, C., and Pollard, P. (2012). Environmental influences on virus–host interactions in an Australian subtropical reservoir. Environ. Microbiol. Rep. 4, 72–81. doi: 10.1111/j.1758-2229.2011.00303.x
Seymour, J. R., Seuront, L., Doubell, M., Waters, R. L., and Mitchell, J. G. (2006). Microscale patchiness of virioplankton. J. Mar. Biol. Assoc. U.K. 86, 551–561. doi: 10.1017/S0025315406013464
Sobczak, W. V., and Findlay, S. (2002). Variation in bioavailability of dissolved organic carbon among stream hyporheic flowpaths. Ecology 83, 3194–3209. doi: 10.1890/0012-9658(2002)083[3194:VIBODO]2.0.CO;2
Suttle, C. A. (2007). Marine viruses–major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812. doi: 10.1038/nrmicro1750
Vrede, K., Heldal, M., Norland, S., and Bratbak, G. (2002). Elemental composition (C, N, P) and cell volume of exponentially growing and nutrient-limited bacterioplankton. Appl. Environ. Microbiol. 68, 2965–2971. doi: 10.1128/AEM.68.6.2965-2971.2002
Vrionis, H. A., Anderson, R. T., Ortiz-Bernad, I., O'Neill, K. R., Resch, C. T., Peacock, A. D., et al. (2005). Microbiological and geochemical heterogeneity in an in situ uranium bioremediation field site. Appl. Environ. Microbiol. 71, 6308–6318. doi: 10.1128/AEM.71.10.6308-6318.2005
Wang, C., Wang, Y., Paterson, J. S., Mitchell, J. G., Hu, X., Zhang, H., et al. (2016). Macroscale distribution of virioplankton and heterotrophic bacteria in the Bohai Sea. FEMS Microbiol. Ecol. 92:fiw017. doi: 10.1093/femsec/fiw017
Weitz, J. S., and Wilhelm, S. W. (2012). Ocean viruses and their effects on microbial communities and biogeochemical cycles. F1000 Biol. Rep. 4:17. doi: 10.3410/B4-17
Whitman, W. B., Coleman, D. C., and Wiebe, W. J. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95, 6578–6583. doi: 10.1073/pnas.95.12.6578
Wigington, C. H., Sonderegger, D., Brussaard, C., Buchan, A., Finke, J. F., Fuhrman, J. A., et al. (2015). Re-examination of the relationship between marine virus and microbial cell abundances. Nat. Microbiol. 1, 15024–15024. doi: 10.1038/nmicrobiol.2015.24
Wilkins, M. J., and Fredrickson, J. K. (2015). “Terrestrial subsurface ecosystem,” in Ehrlich's Geomicrobiology, eds H. L. Ehrlich, D. K. Newman and A. Kappler (Boca Raton, FL: CRC Press), 69–95.
Williams, K. H., Long, P. E., Davis, J. A., Wilkins, M. J., N'Guessan, A. L., Steefel, C. I., et al. (2011). Acetate availability and its influence on sustainable bioremediation of uranium-contaminated groundwater. Geomicrobiol. J. 28, 519–539. doi: 10.1080/01490451.2010.520074
Williams, M. D., and Oostrom, M. (2000). Oxygenation of anoxic water in a fluctuating water table system: an experimental and numerical study. J. Hydrol. 230, 70–85. doi: 10.1016/S0022-1694(00)00172-4
Wommack, K. E., and Colwell, R. R. (2000). Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64, 69–114. doi: 10.1128/MMBR.64.1.69-114.2000
Wrighton, K. C., Castelle, C. J., Wilkins, M. J., Hug, L. A., Sharon, I., Thomas, B. C., et al. (2014). Metabolic interdependencies between phylogenetically novel fermenters and respiratory organisms in an unconfined aquifer. ISME J. 8:1452. doi: 10.1038/ismej.2013.249
Wrighton, K. C., Thomas, B. C., Sharon, I., Miller, C. S., Castelle, C. J., VerBerkmoes, N. C., et al. (2012). Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337, 1661–1665. doi: 10.1126/science.1224041
Xu, J., Jing, H., Sun, M., Harrison, P. J., and Liu, H. (2013). Regulation of bacterial metabolic activity by dissolved organic carbon and viruses. J. Geophys. Res. Biogeosciences 118, 1573–1583. doi: 10.1002/2013jg002296
Xu, J., Sun, M., Shi, Z., Harrison, P. J., and Liu, H. (2014). Response of bacterial metabolic activity to riverine dissolved organic carbon and exogenous viruses in estuarine and coastal waters: implications for CO2 emission. PLoS ONE 9:e102490. doi: 10.1371/journal.pone.0102490
Yanagawa, K., Morono, Y., Yoshida-Takashima, Y., Eitoku, M., Sunamura, M., Inagaki, F., et al. (2014). Variability of subseafloor viral abundance at the geographically and geologically distinct continental margins. FEMS Microbiol. Ecol. 88, 60–68. doi: 10.1111/1574-6941.12269
You, Y., Han, J., Chiu, P. C., and Jin, Y. (2005). Removal and inactivation of waterborne viruses using zerovalent iron. Environ. Sci. Technol. 39, 9263–9269. doi: 10.1021/es050829j
Keywords: virus, bacteriophage, dissolved organic carbon, aquifer, subsurface, uranium, groundwater
Citation: Pan D, Nolan J, Williams KH, Robbins MJ and Weber KA (2017) Abundance and Distribution of Microbial Cells and Viruses in an Alluvial Aquifer. Front. Microbiol. 8:1199. doi: 10.3389/fmicb.2017.01199
Received: 22 December 2016; Accepted: 12 June 2017;
Published: 11 July 2017.
Edited by:
David Emerson, Bigelow Laboratory for Ocean Sciences, United StatesReviewed by:
John Senko, University of Akron, United StatesJoanne B. Emerson, Ohio State University Columbus, United States
Copyright © 2017 Pan, Nolan, Williams, Robbins and Weber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karrie A. Weber, a3dlYmVyQHVubC5lZHU=
†Present Address: Donald Pan, Department of Subsurface Geobiological Analysis and Research, Japan Agency for Marine-Earth Science and Technology, Yokosuka, Japan
 Donald Pan
Donald Pan Jason Nolan2
Jason Nolan2 Karrie A. Weber
Karrie A. Weber