
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 20 June 2017
Sec. Extreme Microbiology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.01135
 Tatsuhiko Hoshino1,2*
Tatsuhiko Hoshino1,2* Tomohiro Toki3
Tomohiro Toki3 Akira Ijiri1,2
Akira Ijiri1,2 Yuki Morono1,2
Yuki Morono1,2 Hideaki Machiyama2
Hideaki Machiyama2 Juichiro Ashi4
Juichiro Ashi4 Kei Okamura5
Kei Okamura5 Fumio Inagaki1,2,6
Fumio Inagaki1,2,6Submarine mud volcanoes (SMVs) are formed by muddy sediments and breccias extruded to the seafloor from a source in the deep subseafloor and are characterized by the discharge of methane and other hydrocarbon gasses and deep-sourced fluids into the overlying seawater. Although SMVs act as a natural pipeline connecting the Earth’s surface and subsurface biospheres, the dispersal of deep-biosphere microorganisms and their ecological roles remain largely unknown. In this study, we investigated the microbial communities in sediment and overlying seawater at two SMVs located on the Ryukyu Trench off Tanegashima Island, southern Japan. The microbial communities in mud volcano sediments were generally distinct from those in the overlying seawaters and in the well-stratified Pacific margin sediments collected at the Peru Margin, the Juan de Fuca Ridge flank off Oregon, and offshore of Shimokita Peninsula, northeastern Japan. Nevertheless, in-depth analysis of different taxonomic groups at the sub-species level revealed that the taxon affiliated with Atribacteria, heterotrophic anaerobic bacteria that typically occur in organic-rich anoxic subseafloor sediments, were commonly found not only in SMV sediments but also in the overlying seawater. We designed a new oligonucleotide probe for detecting Atribacteria using the catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH). CARD-FISH, digital PCR and sequencing analysis of 16S rRNA genes consistently showed that Atribacteria are abundant in the methane plumes of the two SMVs (0.58 and 1.5 × 104 cells/mL, respectively) but not in surrounding waters, suggesting that microbial cells in subseafloor sediments are dispersed as “deep-biosphere seeds” into the ocean. These findings may have important implications for the microbial transmigration between the deep subseafloor biosphere and the hydrosphere.
The subseafloor environment is one of the largest biospheres on Earth, with an estimated total abundance of microorganisms of about 2.9 × 1029 cells, which is equivalent to half of the microbial cells worldwide in the oceans (Kallmeyer et al., 2012). Over the past decade, subseafloor life and the deep biosphere have been intensively explored by means of scientific ocean drilling, which has assessed the biosphere extent down to around 2.5 km below the ocean floor (Orcutt et al., 2013; Ciobanu et al., 2014; Inagaki et al., 2015). Furthermore, evidence is accumulating that some subseafloor microbial cells are not dead but rather are physiologically active or revivable to some degree, and their long-term metabolic activity may contribute to global biogeochemical cycles (Morono et al., 2011; Røy et al., 2012; Hoehler and Jørgensen, 2013). The taxonomic composition of microbial communities in subseafloor sedimentary habitats has been extensively studied using a variety of cultivation-dependent and -independent molecular approaches, demonstrating that some predominant microbial taxa, distinct from known water-column communities, widely inhabit sediments at a variety of oceanic locations under similar geochemical and geophysical settings (e.g., Inagaki et al., 2003, 2006; Webster et al., 2004; Biddle et al., 2008). One of the common predominant microbial taxa frequently found in the marine sediment is Atribacteria including members of JS1 and OP9 which are recently revealed to be monophyletic by phylogenomic analyses of single-cell amplified genome (Nobu et al., 2016). Atribacteria are especially predominant in anaerobic, organic carbon replete, and methane-rich marine sediment. Although representatives of this phylum have not been cultivated as yet, several recent studies of single-cell amplified genomes consistently suggested anaerobic heterotrophic metabolism (Carr et al., 2015; Nobu et al., 2016). Although the co-occurrence of Atribacteira with methane (Inagaki et al., 2006), no evidence has been reported that this phylum was directly involved in methane production or consumption. Alternatively, Atribacteira would control methane production in anaerobic marine sediment by primary or secondary fermentation producing fermentation products as acetate or CO2, that can be used as substrates for methanogen (Carr et al., 2015; Nobu et al., 2016).
Among diverse geological and oceanographic settings in the ocean, numerous submarine mud volcanoes (SMVs) have been observed worldwide along the margins of convergent plates, which transport mud, gasses and fluids from several kilometers below the seafloor to the overlying hydrosphere (Milkov, 2000; Kopf, 2002 and references therein). SMVs accordingly serve as conduits for the release of substantial amounts of methane and other hydrocarbons into the water column (Sauter et al., 2006; Wallmann et al., 2006; Sahling et al., 2009), and hence act as natural pipelines in the geosphere. Over the past decades, the microbial communities inhabiting hydrocarbon seeps associated with SMVs have been investigated around the world, including at the Håkon Mosby (Niemann et al., 2006; Jerosch et al., 2007; Losekann et al., 2007), Kazan (Kormas et al., 2008; Pachiadaki et al., 2010), Amsterdam (Pachiadaki et al., 2011), Chefren (Omoregie et al., 2008), and Gulf of Mexico mud volcanoes (Martinez et al., 2006) and at the Nankai Trough (Miyazaki et al., 2009). The studies at SMVs mainly focused on the anaerobic oxidation of methane (AOM) consortia of sulfate-reducing bacteria and methane-oxidizing archaea living in sediments at the sulfate–methane transition zone (SMTZ). Considering that SMVs transport not only solid materials (e.g., muds and breccias) but also gasses and fluids from deep sources (Sauter et al., 2006; Feseker et al., 2008), it is possible that submarine mud volcanism may also transport microbial communities from the deep subseafloor biosphere to the overlying ocean.
Deep-sea hydrothermal vents on the ridge systems have been proposed as “windows to a subsurface biosphere” (Deming and Baross, 1993), and there have been some investigations into the dispersal of subseafloor microbial communities, including mesophilic to hyperthermophilic microbes, through volcanic and hydrothermal vent activity (e.g., Huber et al., 2003, 2006; Takai et al., 2004; Jungbluth et al., 2016). In the hydrothermal systems, however, only surface of vents are habitable zone for microbes due to the very steep temperature gradient up to higher than 300°C. Therefore, deep-sea hydrothermal vents would not be considered as conduits of deep biosphare and hence the geosphere–biosphere interactions at sedimentary geosystems (e.g., oceanic plate subduction zones, forearc sedimentary basins) as well as interactions between the Earth’s surface and deep subseafloor biospheres in the large oceanic province remain largely unknown.
In the present study, we investigated microbial communities inhabiting two SMVs off Tanegashima Island, Japan (Ujiie, 2000), by taking sediment cores from the summit (down to a sediment depth of 3.6 m) and samples of overlying seawater (up to 200–300 m above the summit). Sediment core samples were collected in 2015 using a Navigable Sampling System (see Ashi et al., 2014), which enables pinpoint piston coring with television-monitoring of the seafloor. The concentrations of methane in the water column peaked at 19 and 72 m above the two SMVs, respectively, indicating the presence of a methane plume from the SMV. To identify and quantify members of the microbial communities in mud volcano sediment and seawater samples, we applied molecular analysis of 16S rRNA genes using a second generation sequencer, image-based cell counts, catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH), and microfluidic digital PCR techniques.
Previous acoustic seafloor surveys identified several 10s of SMVs in the northwestern Pacific Ocean at water depths of 1200–3500 m off Tanegashima Island, Japan. For one of these, geochemical information, including stable isotopic compositions of methane and water (i.e., interstitial water in sediment) is available (Nakayama et al., 2010), whereas there are as yet no microbiological studies. In 2015, we investigated the distribution and diversity of microbial communities inhabiting two of the SMVs in this area: MV#1 and MV#14 (Figure 1). MV#1 has a diameter of about 3000 m, a height of 300 m, and is situated at a water depth of 1419 m. MV#14 has a diameter of about 3000 m, a height of 250 m, and is situated at a water depth of 1691 m. Intensive seafloor surveys around MV#1 using the autonomous underwater vehicle Urashima and remotely operated vehicle Hyper-Dolphin (Japan Agency for Marine-Earth Science Technology [JAMSTEC]) showed well-preserved mud-flow channels suggestive of a recent eruption.
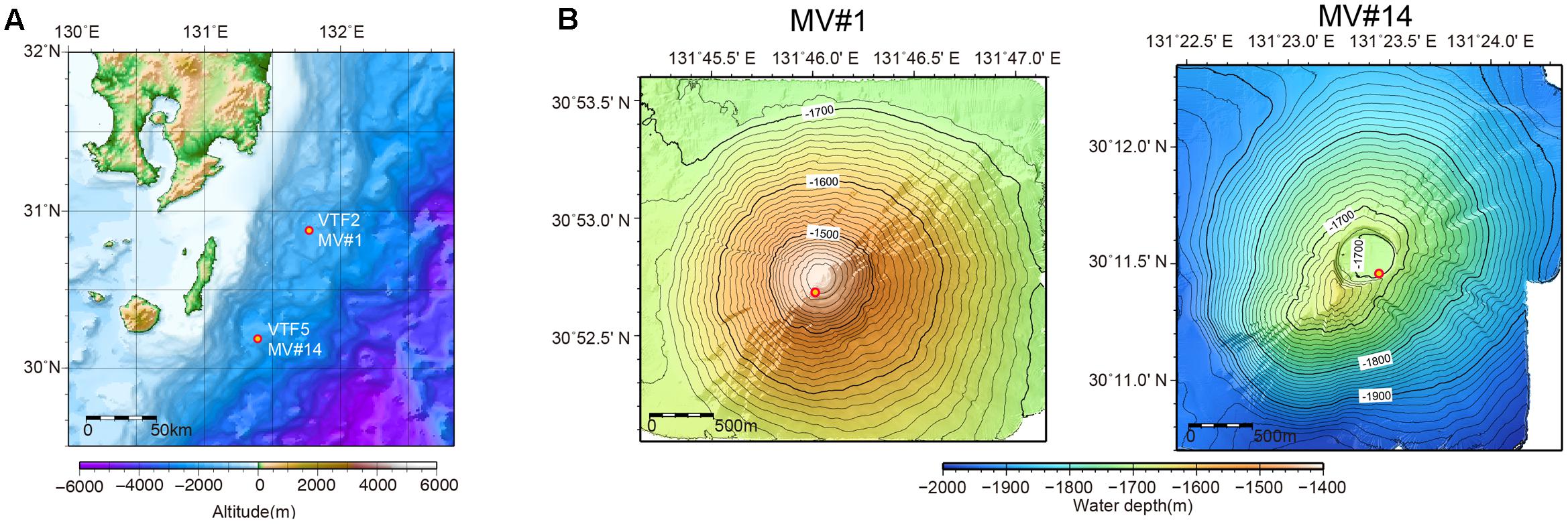
FIGURE 1. (A) Location of submarine mud volcanoes (SMVs) MV#1 and MV#14 along the Ryukyu Trench near Tanegashima Island, Japan. (B) Detailed bathymetric maps of MV#1 and MV#14. Both SMVs are about 3000 m in diameter and 300 m in height. The yellow circles indicate sampling points.
Sediment and seawater samples were obtained at two stations, VTF2 (MV#1) and VTF5 (MV#14), during cruise KH-15-2 of R/V Hakuho Maru in 2015. Sediment samples were collected using the Navigable Sampling System (Atmosphere and Ocean Research Institute, The University of Tokyo). At each station, hydrographic parameters were recorded with a CTD-carousel system (SBE 9, Sea-Bird Scientific, Bellevue, WA, United States), which was deployed with simultaneous water sampling using Niskin bottles (General Oceanics, Inc., Miami, FL, United States). For comparison in the DNA analysis, we used stratified sediment core samples from the Peru Margin, Juan de Fuca ridge flank, and offshore of the Shimokita Peninsula, Japan, which were collected during Ocean Drilling Program (ODP) Leg 201 in 2002, Integrated Ocean Drilling Program (IODP) Expedition 301 in 2004, and shakedown cruise CK06-06 of JAMSTEC’s deep-sea drilling vessel (D/V) Chikyu in 2006, respectively (Table 1). For DNA extraction, all SMV sediments and the stratified sediments were frozen immediately after sampling and stored at -80°C until use. For cell count of the SMVs sediment, 2 g of sediment was collected by using a tip-cut syringe and transferred to a 15-ml falcon tube. Subsequently, 8 ml of 4% paraformaldehyde was added to the sediment for fixation. After overnight fixation at 4°C, the fixed sediment was washed twice with PBS buffer and stored in 50% PBS/ethanol at -20°C.
After core recovery, wet sediment was immediately sampled at 20- to 50-cm intervals and pore water samples were extracted onboard with a stainless-steel squeezer (Manheim and Sayles, 1974). The sulfate content of pore water was analyzed using an ion chromatograph (Dionex ICS-500) with an isocratic carbonate/bicarbonate eluent coupled with suppressed conductivity detection. Analytical precision was estimated to be within 0.4% from repeated measurements of the same samples.
Sediment samples for the determination of methane concentrations were collected using tip-cut plastic syringes immediately after core recovery. The syringe end was tightly capped with a silicone-rubber stopper. The sediment sample was transferred from the syringe into a glass vial in a N2-flushed glove bag, and then the vial was capped with a Teflon-coated rubber septum and aluminum seal for subsequent geochemical analyses. The glass vials were frozen at -20°C and stored until analysis. In a laboratory on shore, the glass vials were heated in an oven at 60°C for 30 min. From the glass vials, 0.1–0.5 mL of the gas phase was extracted with a gas-tight syringe and introduced into a gas chromatograph-isotope ratio monitoring mass spectrometer (Tsunogai et al., 2002). The methane content was calculated by comparing the 44CO2 output of the isotope-ratio-monitoring mass spectrometer with those measured during analyses of a working-standard gas containing 5000 ppm methane in He. The standard deviation of repeated analysis of the laboratory standard gas was <4%.
For quantification of methane in seawater, two replicate samples were collected from each Niskin bottle: two 120-ml serum vials were filled to overflowing after flushing with approximately two vial-volumes to avoid air contamination. The seawater samples were fixed by the addition of 500 μl saturated HgCl2 solution to inhibit microbial activity during storage, sealed with gray butyl-rubber septa and aluminum crimp-caps, and then stored in the dark at 4°C until analysis in the laboratory at the University of the Ryukyus (Okinawa, Japan).
Dissolved methane was analyzed by purge–trap gas chromatography (GC) (GC-2014, Shimadzu) equipped with a flame ionization detector. Hydrocarbons were chromatographically separated on a Porapak Q packed column (80–100 mesh; 2-m length) operated at 40°C using ultra-high-purity He as the carrier gas (Air Liquide, purity 99.9999%). The system was calibrated using a gas standard with a certified methane concentration (9.72 ppm) provided by Japan Fine Products (Kawasaki, Japan). The precision of the method was 10%, as estimated from the standard deviation of replicate analysis of water samples (n = 4).
Cell concentrations in sediment samples were determined using a previously described procedure (Morono et al., 2013). Briefly, after washing fixed sediment with a NaCl solution, a mixture of detergent and methanol was added and the sediment slurry was shaken for 60 min at 500 rpm using a Shake Master (Bio Medical Science, Tokyo, Japan). The slurry was then sonicated and carefully layered onto a high-density cushion consisting of multiple density layers of Nycodenz and sodium polytungstate solutions. Following centrifugation at 10,000 × g for 60 min, the supernatant was carefully transferred to a new vial. The detergent and methanol mixture was added to the remaining pellet, and the mixture was again sonicated and centrifuged with the high-density cushion solution. The supernatants were pooled and the combined recovered suspension was filtered onto a polycarbonate membrane. For seawater samples, 300 mL of formalin-fixed seawater was directly filtered onto a polycarbonate membrane. The number of microbial cells stained by SYBR Green I was determined either using an automated cell counting system with analysis of acquired images using MetaMorph software (Molecular Devices, Sunnyvale, CA, United States) (Morono and Inagaki, 2010), or counted manually under fluorescence microscopy.
DNA was extracted from 5 g wet sediment or 300 mL seawater using the PowerMax Soil DNA Isolation Kit (Qiagen, Tokyo, Japan) according to manufacturer’s instructions. For the seawater samples, soon after recovery of the Niskin bottles 300 mL seawater was filtered using a disposable MicroFunnel 300 Filter Unit with 0.2-μm pore size (Pall Corporation, Tokyo, Japan) and stored at -20°C prior to extraction. For a negative control, 5 mL Milli-Q water (Millipore) was used for extraction in place of sediment or seawater. The extracted DNA solution (approximately 5 mL) from sediment was further concentrated by ethanol precipitation. To increase the DNA yield, 10 μL of linear acrylamide (Thermo Fisher Scientific, Kanagawa, Japan) was added before centrifugation. The DNA precipitate was dissolved in TE buffer (10 mM Tris-HCl, 1.0 mM EDTA; pH 8.0) and stored at -20°C until use.
The abundance 16S rRNA genes was quantified by microfluidic digital PCR (dPCR) using the BioMark Real-time System and 12.765 Digital Array (Fluidigm Corporation, South San Francisco, CA, United States) as described elsewhere (Hoshino and Inagaki, 2012). For each sample, 6 μL of PCR mixture was prepared as follows: 3.0 μL of 2× MightyAmp buffer (TaKaRa Bio, Shiga, Japan), 0.15 μL of Binding Dye sample loading reagent (Fluidigm), 0.12 μL of MightyAmp DNA polymerase (TaKaRa Bio), 0.3 μL of EvaGreen (Biotum, Fremont, CA, United States), 0.015 μL of Rox dye, 0.15 μL each of forward and reverse primer, 1.0 μL of extracted DNA, and 1.115 μL of water. We used the domain-specific primers B27F-B357R and A806F-A958R (Table 2) for the amplification of bacterial and archaeal 16S rRNA genes, respectively.
The V3–V4 hyper-variable region of the 16S rRNA gene, which is often used for analyzing microbial diversity and community structure, was amplified by PCR using universal primers U515F/U806R (Table 2). The 25-μl PCR mixture consists of 0.3 μM of each primer, 1 μl of the template DNA, 1× MightyAmp Buffer Ver.2, and 0.5 μl of MightyAmp DNA polymerase (TaKaRa Bio). The PCR started with 2 min at 98°C followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 55°C for 15 s, and elongation at 68°C for 30 s. After purification of the desired PCR products by agarose gel electrophoresis, index and adapter were added to the purified product during the eight cycles of second-round PCR using KAPA HiFi HotStart Ready mix (Kapa Biosystems) with 250 pg of the purified PCR product as described elsewhere (Caporaso et al., 2012). The indexed PCR products were purified twice by AMPure XP (Beckman Coulter), and then the PCR product was sequenced by the MiSeq platform with MiSeq Reagent Kit v3, 600 cycles (Illumina, San Diego, CA, United States). All the purified PCR products were sequenced in a sequencing run of Miseq and account for two fifth of total clusters. Raw sequences were demultiplexed, and the sequence quality was filtered using the BaseSpace app (Illumina). Further processing, including primer trimming, sequence length screening, taxonomic identification, clustering and diversity analysis, were performed using the ‘mothur’ software package (v. 1.35.0) (Schloss et al., 2009) and USEARCH 64-bit version1. After the screening, the average number of sequence reads per sample was 54,410 (Supplementary Table 2). The operational taxonomic units (OTUs) obtained were used for non-metric multidimensional scaling (nMDS) analysis using the ‘vegan’ package of R software as described elsewhere (Dixon, 2003; Doi and Okamura, 2011) for beta-diversity analysis of microbial communities. To exclude confounding results due to potential contaminants from the extraction process and chemicals used for PCR and sequencing, the top 10 families in the negative controls, and previously reported core-human microbiomes (Li et al., 2013) were arbitrary removed (listed in Supplementary Table 1). The removed top 10 families are generally consistent with laboratory and reagent contaminants in microbiome analyses (Salter et al., 2014). We performed all the analyses in this study without removing those potential contaminant and confirmed that removal of those did not significantly affect the results (Supplementary Figures 1, 2 and Table 2).
To detect and visualize atribacterial cells in microbial communities in the MVs, we designed a new oligonucleotide probe Atri578 (Table 2), which can be used to detect a broad range of atribacterial sequences in the ARB Silva Database by using ARB software and mathFISH (Ludwig et al., 2004; Yilmaz et al., 2011). The coverage of Atri578 to atribacterial sequences determined by TestProbe3.0 with the SILVA Database SSU128 is 70.6% and only two sequences in the outgroup are no-mismtach to Atri578. The horseradish peroxidase (HRP)-labeled Atri578 probe was purchased from Biomers GmbH (Ulm, Germany). The optimal formamide concentration in hybridization buffer was determined by clone-FISH (Schramm et al., 2002) using a synthesized vector (Eurofins Genomics, Tokyo, Japan) containing the full-length 16S rRNA gene sequence of Atribacteria bacterium SCGC AAA255-N14 (ASPC01000002). We tested formamide concentrations of 0% to 60% at 10% intervals, at 35°C and found 10% to be the optimal formamide concentration. Using this concentration, Atribacteria on the membrane filters were detected by CARD-FISH for the water column samples. The sediment samples from SMVs had very low cell number and the number of cells on the filter membrane after cell separation described above was not enough to detect Atribacteria.
Catalyzed reporter deposition-fluorescence in situ hybridization was performed according to a previously described protocol (Hoshino et al., 2008, 2015). Briefly, hybridization was performed in hybridization buffer containing 10% formamide and 0.5 μM of the HRP-labeled Atri578 probe at 35°C for 2 h. After washing away excess probe, the membrane filter was incubated with Alexa555- or Alexa488-labeled tyramide (Thermo Fisher Scientific) at 35°C for 30 min for signal deposition, followed by counterstaining with SYBR Green I or DAPI.
Sequence data obtained in this study have been submitted to the DDBJ database under accession numbers DRA005491 and DRA005492.
Sulfate concentrations in sediment pore water from both SMVs showed maxima around 25–30 mM in the uppermost sediment, with concentrations rapidly decreasing with depth (Figure 2). In sediments deeper than 1 m below the seafloor (mbsf), sulfate was almost depleted at both SMVs. The concentration of methane was generally higher in pore water from MV#1 than MV#14, ranging from 0.5 to 13 mM and from 0.06 to 5.1 mM, respectively. Although measured concentrations varied widely probably because of depressurization of sediment, methane was depleted near the sediment surface at around 1 mbsf. The depth profiles of sulfate and methane indicate that the SMTZ was located at around 1 mbsf at both SMVs. In the overlying water column, the concentration of methane peaked to 0.71 nM at 29 m above MV#1 and to 1.93 nM at 70 m above MV#14. Those peaks indicate the occurrence of methane plume and thus advective flow from the SMVs because methane is immediately oxidized in water column and its concentration almost never reached to that high in normal water column at the similar depth (Toki et al., 2016).
FIGURE 2. Methane (red circles) and sulfate (blue circles) concentrations in sediment pore water at SMVs MV#1 (A) and MV#14 (B).
Cell densities in seawater, based on counts of cells stained with SYBR Green I, were estimated to be in the range of 4.7 × 104 to 1.4 × 105 cells/ml at MV#1 and MV#14, respectively (Figure 3). The gene abundance of 16S rRNA genes in seawater, as sum of archaeal and bacterial 16S rRNA gene (Supplementary Figure 3) quantified by microfluidic dPCR, were overall slightly lower than the cell densities. Cell counts and gene abundance from the seawater above the SMVs were relatively constant throughout the water column at 5 × 103 copies/ml and 6 × 104 cells/ml (Top panel of Figure 3) up to around 400 m above the summit, the upper limit of sample collection.
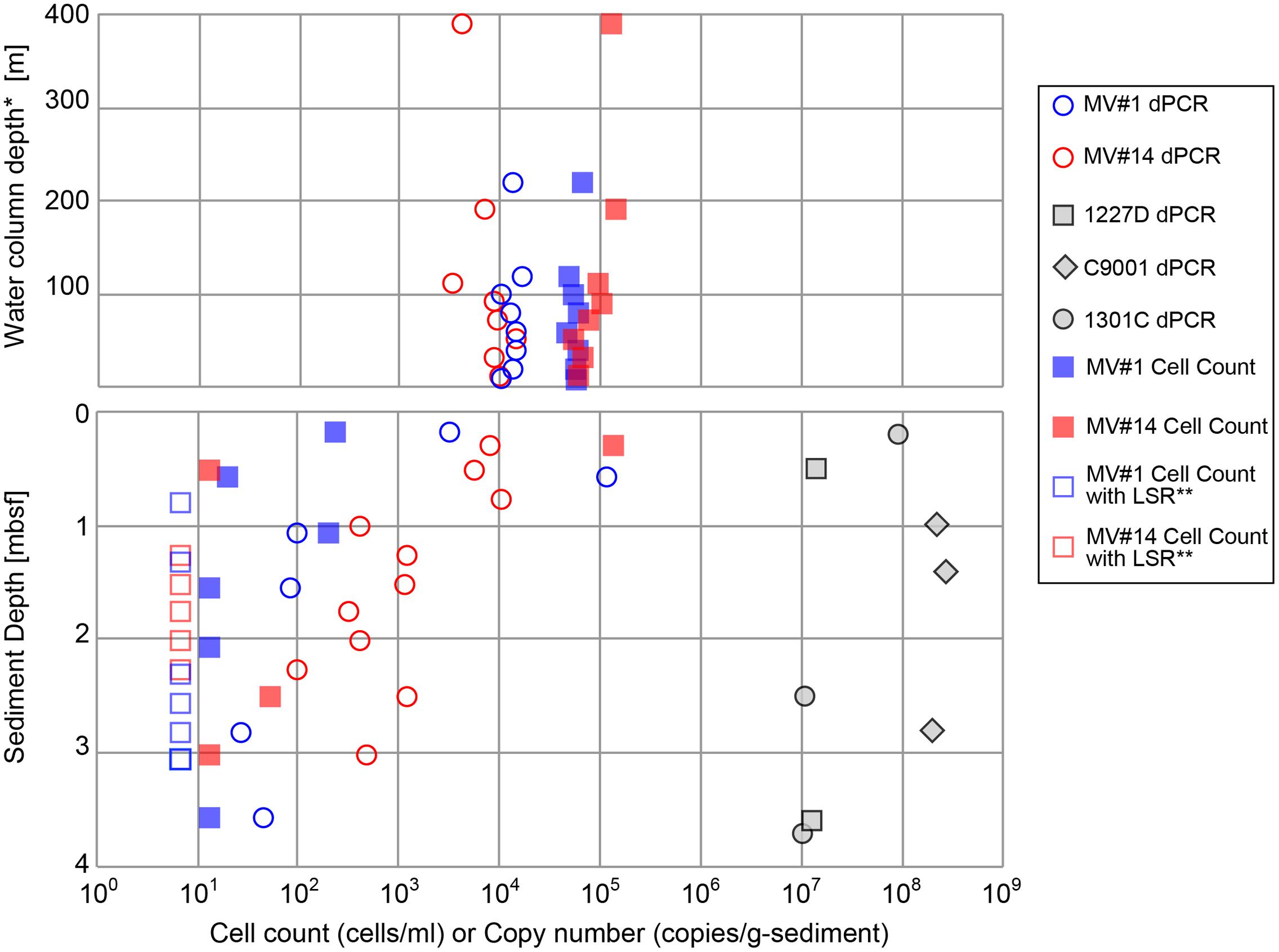
FIGURE 3. Microbial cell counts by SYBR Green I staining, and digital PCR (dPCR) quantification of 16S rRNA gene abundance. dPCR results are the sum of bacterial and archaeal 16S rRNA gene abundance. dPCR results from reference sites from previous drilling expeditions are provided for comparison (see Table 1 for sampling site information). ∗Height above the summit of the SMVs. ∗∗LSR, “low statistical reliability” because of very low, but detectable, cell numbers.
The cell concentrations in the shallowest sediment samples from the seafloor were 2.4 × 102 and 1.3 × 105 cells/g-wet sediment for MV#1 and MV#14, respectively. Cell concentrations at the two SMVs dropped sharply to only 20 and 13 cells/g-wet sediment at 0.58 and 0.51 mbsf, respectively. The 16S rRNA gene abundance for MV#1 in the uppermost sediment sample was 3.3 × 103 copies/g-wet sediment, which is an order of magnitude higher than the cell concentration, whereas the cell counts in the uppermost sediment at MV#14 were higher than the 16S rRNA gene abundance (8.0 × 103 copies/g-wet sediment). The 16S rRNA gene abundance in sediment from depths greater than 1.5 mbsf were 10–100 times the cell concentrations.
Throughout the sediment cores, MV#1 generally had lower 16S rRNA gene abundance than MV#14, and the gene abundance at MV#1 decreased to about 1 × 102 copies at 1.1 mbsf. At MV#14, the 16S rRNA gene abundance gradually decreased with increasing depth to 4.7 × 102 copies/g-wet sediment at 3.0 mbsf, the deepest sediment sampled. The estimated 16S rRNA gene abundance as well as cell concentrations in the deeper mud volcano sediments were markedly lower than those in shallow seafloor sediments and in other stratified subseafloor sediments on Pacific margins collected during previous IODP expeditions. In general, the abundance of 16S rRNA genes in the SMVs varied from 101 to 105 copies/g-wet sediment, which is two to seven orders of magnitude lower than in stratified sedimentary environments (Figure 3).
The cell counts are higher than the gene abundance in the water column in this study, whereas in the sediment the trend was opposite to the water column. The reasons of formar discrepancy can be partially explained by biases as low DNA extraction efficiency or false positive counts of fluorescence-emitting particles (Lloyd et al., 2013). The latter might be caused by low recovery of cells from very sticky texture of the MV sediment. In any case, the results from both methods are consistent with the general trends; (1) Higher microbial abundance in the overlying seawater than mud volcano sediment: (2) Decrease of microbial abundance with sediment depth.
We obtained around 1.6 million sequences in total by Miseq sequencing of SMV sediment and overlying seawater samples (Supplementary Table 2). Phylum-level classification showed the predominance of Proteobacteria in all seawater samples, comprising more than 50% in all sequence libraries (Figure 4). The taxonomic composition of microbial communities in seawater was generally constant at both sites and at different depths. The second most representative phylum was Marinimicrobia (formerly SAR405), comprising 10–14% of the total community, followed by Thaumarchaeota comprising 5–15%.
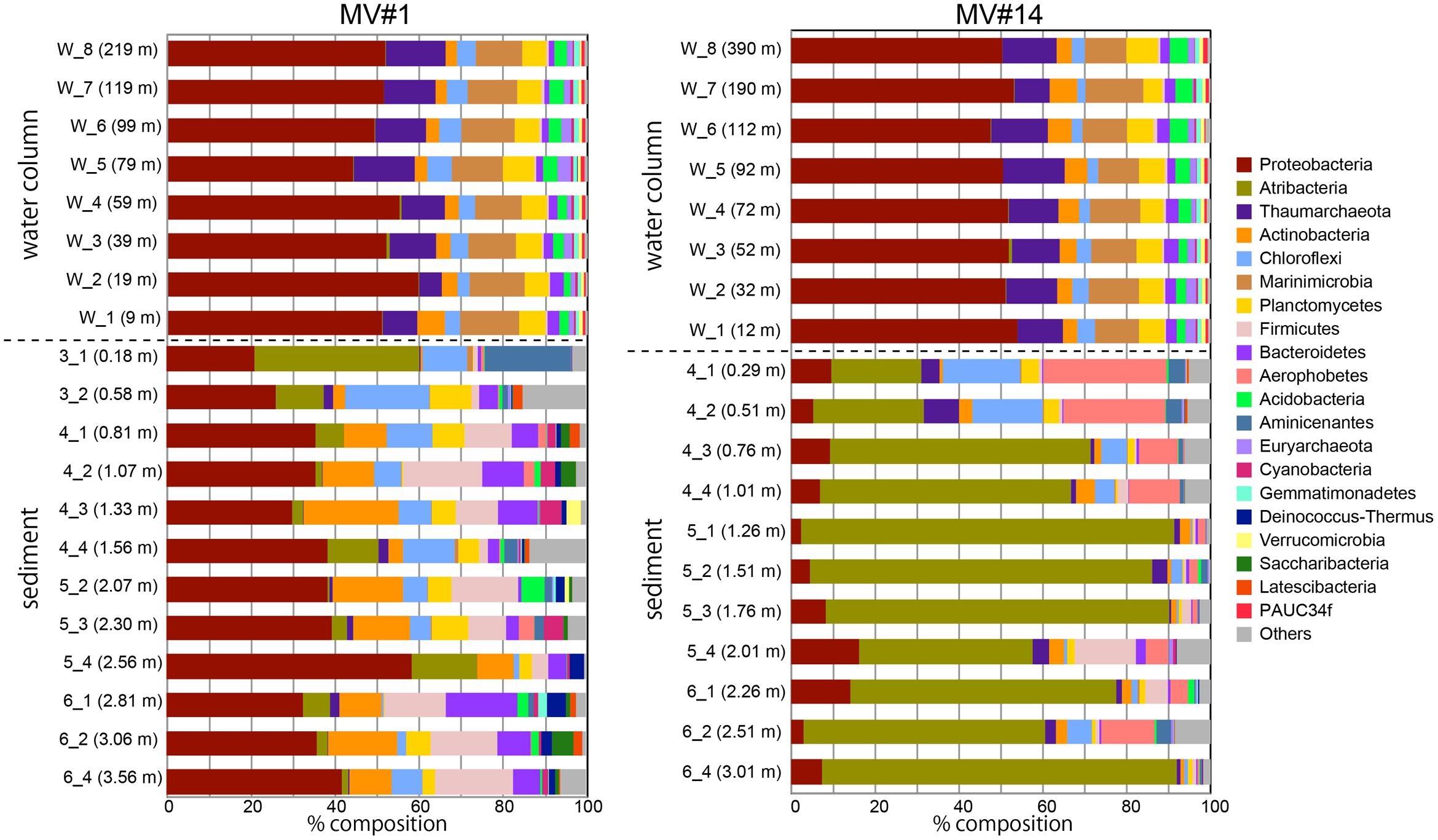
FIGURE 4. Microbial community composition in the sediment and overlying seawater at two SMVs using phylum-level classification. Only the top 20 phyla are shown. Numbers in parentheses show the water column depth (meters above the SMV summit) for water samples (labels that start with “w”) and depth in sediment for sediment samples (labels that start with numbers).
In contrast, the community composition of mud volcano sediments was markedly different from that in the water column, even at the phylum level; for example, Marinimicrobia was nearly absent in sediment samples. The MV community composition was also distinct from those of the reference stratified sediment (Supplementary Figure 4). In addition, the community compositions at the two mud volcano sites were notably different: Proteobacteria was dominant at MV#1, comprising between 21 and 58% of the total, whereas Atribacteria (formerly OP9 and JS1) comprised 21–88 and 0.7–39% of total sequence reads at MV#14 and MV#1, respectively. Some phyla, such as Firmicutes, Aerophobetes (formerly CD12), and Aminicenantes were prevalent only in the sediment samples.
The degree of similarity between microbial communities in sediment and seawater was determined by nMDS analysis applied to OTUs with >97% similarity (Figure 5). Microbial communities in the overlying seawater from 200 to 300 m above the mud volcano summits were generally similar. Compared to the seawater communities, microbial communities in sediments were more diverse, even across very small changes in depth at 2 and 6 mbsf for the SMVs and reference stratified marginal sediments. Microbial communities in stratified sediments were grouped into one cluster despite being collected from geographically distinct sites, whereas the microbial communities in SMV sediments were different from those in both the stratified sediments and in the overlying water column. A gradation in microbial composition is evident from deep to shallow sediment within both SMVs. These trends suggest that physical and chemical conditions may affect microbial composition more than can be explained by the conditions in well-stratified sediment. The nMDS plots (Figure 5) also indicate that shallower SMV sediment communities, especially at MV#14, are similar to those in normal stratified marine sediments. This difference might suggest differences in activity of the SMVs: if MV#1 is more active than MV#14, the seafloor at MV#14 would be covered with marine sediments freshly deposited since the last eruption; consequently, shallower sediments at MV#14 would harbor the microbial communities that typically occur in the surrounding stratified sediments.
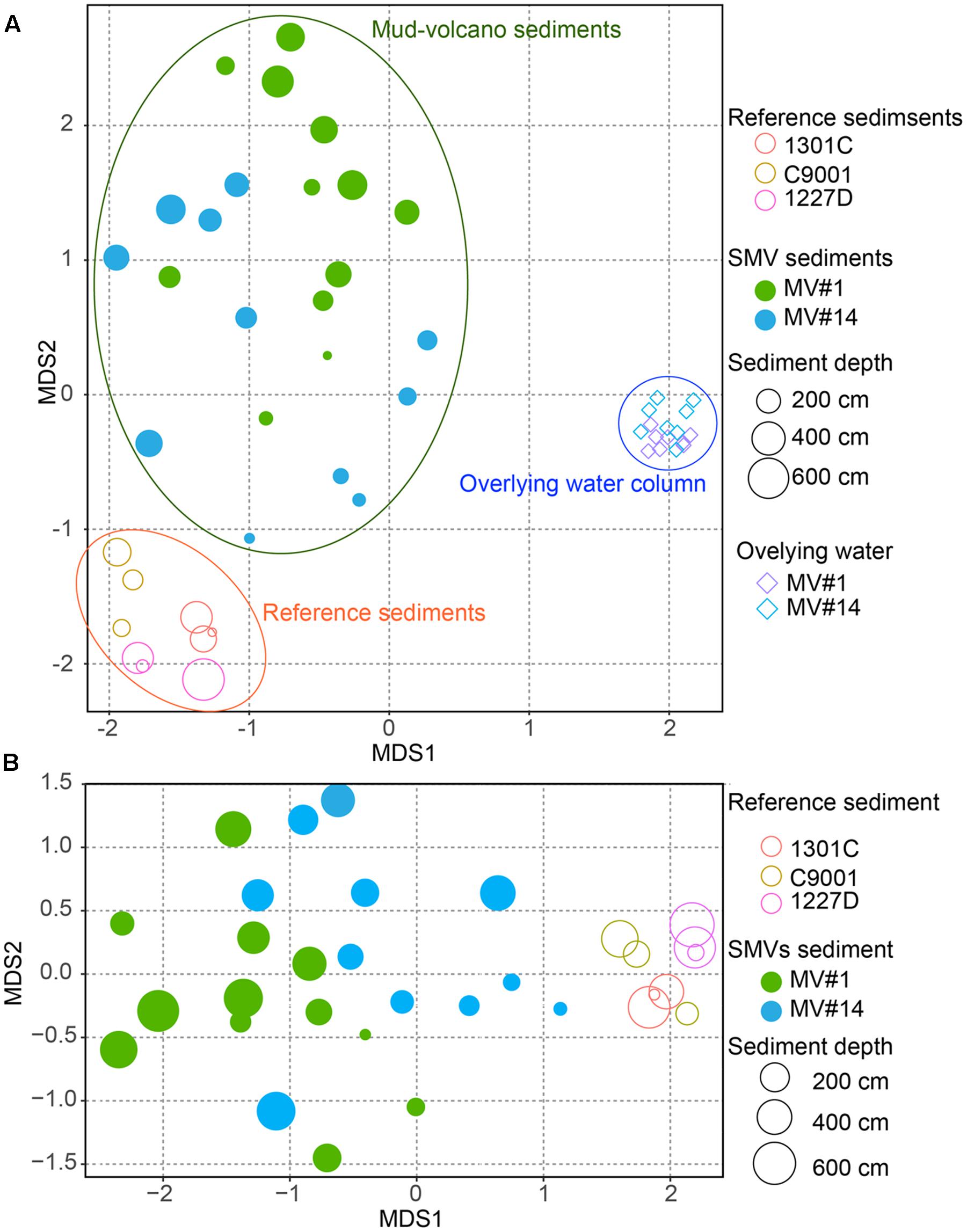
FIGURE 5. Non-metric multidimensional scaling (NMDS) plot of microbial communities from SMV sediment (circles) and the overlying water column (diamonds), with stratified marine sediment communities included for reference, based on operational taxonomic unit (OTU) composition. The size of the circles for sediment samples, including SMV sediment is proportional to sediment depth. (A) NMDS plot of all the samples including the water column. (B) Only SMV and reference sediments.
We identified the common OTUs in all SMV sediment and seawater samples to determine whether mud volcano activity dispersed some of the community members of the subseafloor sedimentary habitats at SMVs to the overlying seawater. We found only one OTU (i.e., OTU-1) common to all of the samples examined for both sites (MV#1 and MV#14). OTU-1 is the most abundant OTU in the SMV sediment samples, comprising 1.4–38 and 22–85% of the sequence reads from MV#1 and MV#14, respectively. OTU-1 sequences are affiliated with Atribacteria, which is well-known as a predominant taxonomic clade of marine sedimentary bacteria (e.g., Inagaki et al., 2006; Webster et al., 2006b; Blazejak and Schippers, 2010); almost all Atribacteria-related sequences detected in this study are included in this OTU (Supplementary Figure 5). The relative abundance of OTU-1-related sequences was generally low in seawater samples, comprising less than 0.1% of total reads. This abundance peaked, however, to more than 0.7% at 39 m (MV#1) and 52 m (MV#14) above the summits of both SMVs and returned to around 0.1% above this depth (Figure 6). Interestingly, the depth profile of methane similarly peaked at 19 m (MV#1) and 72 m (MV#14) above the summit, indicating the presence of Atribacteria in methane plumes emanating from the SMVs.
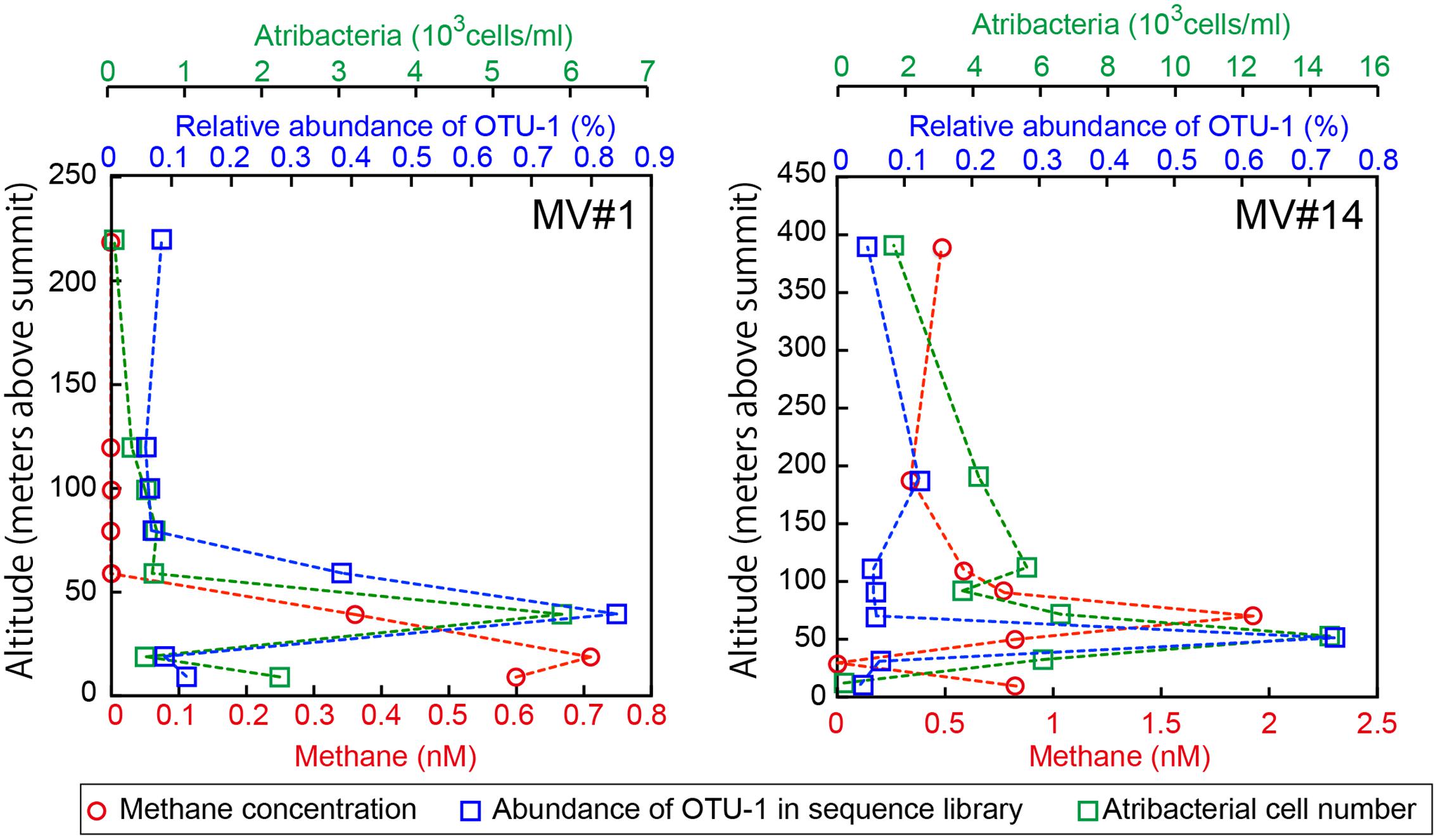
FIGURE 6. Depth profiles of methane concentrations, relative abundance of OTU-1 in the sequence library, and cell numbers of Atribacteria as detected by CARD-FISH using probe Atri578. OTU-1 is the most abundant OTU affiliated with Atribacteria.
Because the relative abundance of OTU-1 in sequence libraries are not necessarily quantitative, we verified the presence and abundance of Atribacteria in seawater samples by CARD-FISH using the newly designed probe Atri578, which specifically targets the Atribacteria-related sequences detected in this study and related sequences in publicly available databases (Figure 7). CARD-FISH analysis showed that Atribacteria in the water column were most abundant at 39 m (MV#1) and 52 m (MV#14) above the respective summits; results overall in good agreement with the relative abundance of OTU-1 sequence. Consequently, we infer that Atribacteria inhabiting the subseafloor sediments at SMVs are dispersed into the overlying seawater along with the methane plume.
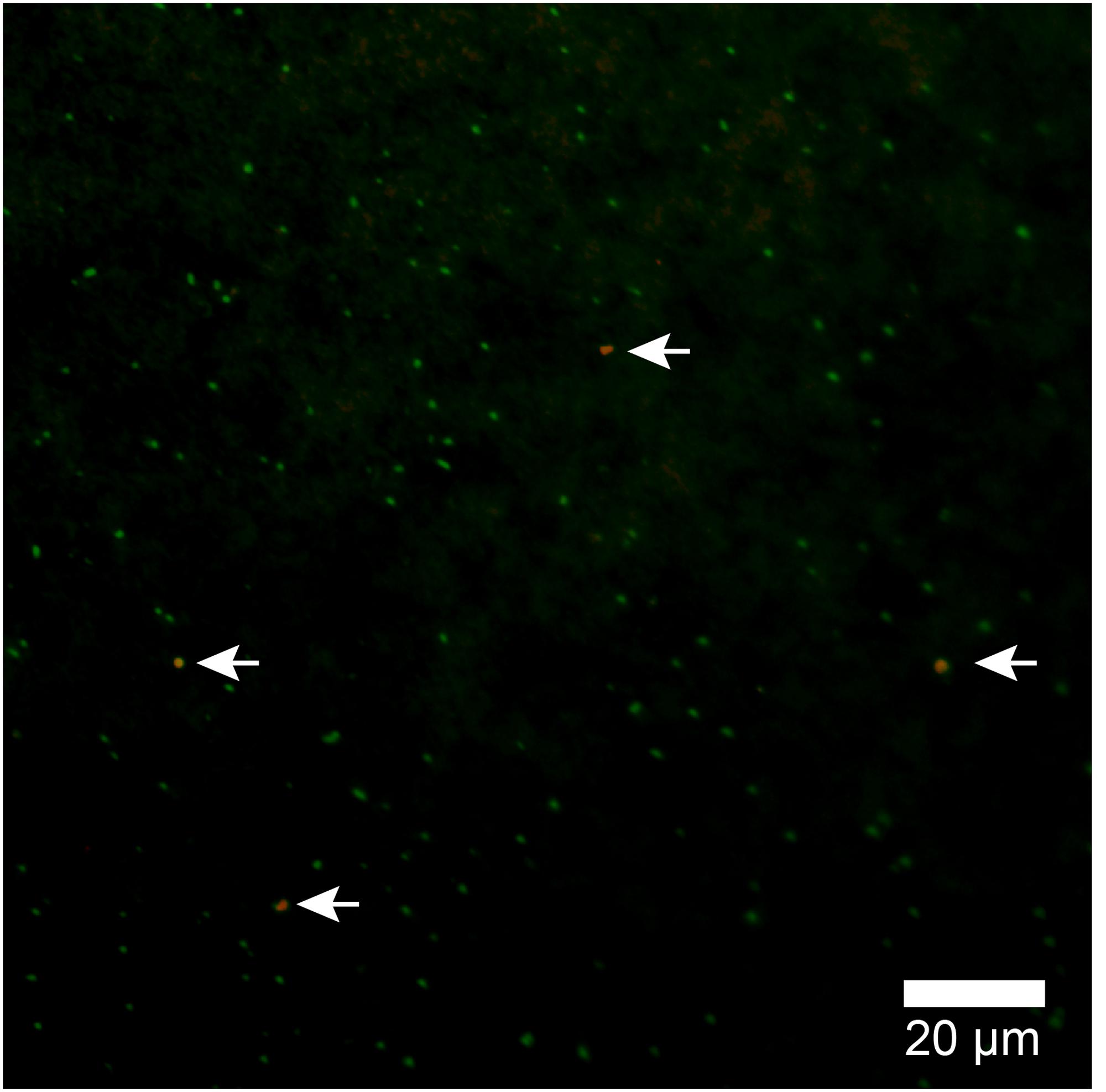
FIGURE 7. Atribacterial cells detected by CARD-FISH using probe Atri578. Alexa555-labeled tyramide was used for horseradish peroxidase (HRP) detection (appears orange) and DNA was counterstained with SYBR Green I. White arrows point to orange cells (Atribacteria); green cells are other microbial cells.
The mechanisms of dispersal of microbial life from the deep biosphere to the Earth’s surface biosphere remains largely unknown. In this study, we observed that deep-biosphere microbial communities are transported from deep subseafloor sedimentary environments to the ocean through the activity of SMVs; specifically, members of the Atribacteria, typically observed in the global subseafloor sedimentary environment, were found to be present in methane plumes above two SMVs. The deep-sea hydrothermal systems have been well-studied and dispersal of thermophilic microbial communities to the ocean has been well-known (Deming and Baross, 1993; Takai et al., 2004; Huber et al., 2006). However, the diffused microbial communities inhabit around the surface of vents due to the high temperature of hydrothemal fluid and those do not come from deep biosphere. In contrast to those systems, little is known about the dispersal and transport of microorganisms from the deep subseafloor sedimentary biosphere to the overlying hydrosphere. A recent molecular ecological study of microbial communities in marine sediments and the overlying seawater showed that some of the OTUs represented by abundant sequences in the sediment are also present but represented by rare sequences in the water column population (Walsh et al., 2016); however, the possible mechanism or the driving force for the dispersal remains unknown. In this study, we examined these scientific questions at two SMVs off Tanegashima Island, Japan.
We found that Atribacteria-related sequences were prevalent throughout the sediment and overlying water column at both SMVs examined. One of the atribacterial phylotypes (i.e., OTU-1) was commonly present in all samples. It is possible that only a very small portion of the sedimentary microbial cells can be diffused from sticky mud volcano sediments into the overlying water column, so we might only be able to detect the most abundant deep biosphere-derived OTUs because of the sensitivity of sequencing analysis. In support of this idea, the second most abundant OTU in the sequence libraries of the sedimentary microbial community (OTU-12, within the Aerophobetes) was found in the water column and showed a profile similar to those of OTU-1 and Atribacteria cell counts (Supplementary Figure 6). These consistent results suggest that there might be no selection of microbial phylotypes in the dispersal process, and hence sedimentary microbes are evenly dispersed to the overlying seawater along with discharging gasses and fluids.
The candidate division JS-1 affiliated with Atribacteria was originally found in the sediment from Japan Sea (Rochelle et al., 1994), and meanwhile the widespread distribution of this group was reported in marine sediments including organic rich deep-sea sediment (Webster et al., 2006a), shallow marine sediment (Oni et al., 2015), tidal flats (Webster et al., 2007) and methane hydrate bearing sediment (Reed et al., 2002; Inagaki et al., 2003, 2006). Their relative abundance often comprised higher than 50% of sequence libraries (Newberry et al., 2004; Inagaki et al., 2006; Carr et al., 2015). Those high abundances of Atribacteria in the methane rich sediments imply that this group was selected in high methane concentrations. In general, sediments of mud volcanoes contain high mathane concentrations and would be suitable enviromnent for Atribacteira as reported in Amsterdam mud volcano (Pachiadaki et al., 2011). The dominance of Atribacteria in MV#1 and MV#14 in this study was consistent with those previous studies.
At the Vodyanitskiy mud volcano in the Black Sea, it has been hypothesized that ANME-1 and ANME-2 archaea are associated with methane bubbles and participate in AOM in the anoxic water column (Schubert et al., 2006). In this study, we detected methane plumes together with atribacterial cells in the oxic water column, which is an environment very different from the deep sedimentary biosphere. We semi-quantitatively compared the frequency of 16S rRNA genes of OTU-1 between seawater and sediment samples, although the number of atribacterial cells in seawater visualized by CARD-FISH could not be directly compared to those in sediment samples, for which estimates were not technically possible because of the low in situ cell densities. Our results showed that, in methane plumes, the highest abundance of OTU-1 in the sequence library approached 1% relative abundance for MV#1 and MV#14 (Figure 6). These abundances are much higher than previously reported, where the highest relative abundance of sedimentary communities in the water column was 0.025% for normal sedimentary environments in the open ocean (Walsh et al., 2016).
The total abundance of 16S rRNA genes determined by microfluidic dPCR in methane plumes was 1.4 × 104 copies/ml in samples from both SMVs. By multiplying the proportion of sequence by the gene abundance, we estimated the density of OTU-1 16S rRNA genes to be 1.2 × 102 copies/ml and 1.1 × 102 copies/ml for MV#1 and MV#14, respectively. The gene abundance in sediment pore water can be similarly estimated under the assumption that bulk density and porosity of the sediment are 1.8 g/ml and 50%, respectively (i.e., using shipboard results available in the Cruise Report of D/V Chikyu training cruise CK09-01, Leg. 12). The estimated abundance of OTU-1 16S rRNA genes in the topmost sediment of MV#1 and MV#14 are 4.4 × 103 and 6.3 × 103 copies/ml pore water, respectively, which are only 37- and 57-fold higher than those in the methane plumes in the water column at 32 and 39 m above the respective SMVs. Given these estimates, we infer that there could be a supply of substantial amounts of the subseafloor microbial community to the overlying seawater, for which we propose the term “deep-biosphere plume.”
Once sedimentary microbial components are released into the water column, it is likely that they are transported via currents as “deep-biosphere seeds” into the ocean, perhaps over long distances, as previously discussed for thermospores found in arctic sediments (Hubert et al., 2009). Nevertheless, at this point, it is difficult to quantify the deep-biosphere seeds, constantly or episodically dispersed from SMVs along with methane and fluids. To expand our understanding of this interesting phenomenon involving geosystem–ecosystem interactions will require continuous monitoring of the geobiological system over a wide spatial range through future deep-sea research and ocean drilling opportunities.
In this study, by using the Atri578 probe targeting Atribacteria for CARD-FISH, we confirmed the presence of sedimentary Atribacteria in methane plumes (Figure 7). The detected atribacterial cells were individualized cocci (diameter approximately 1.0 μm), not forming aggregates or attached to any particulate matter. Our CARD-FISH observations suggest that the Atribacteria are generally free-living in the sediment pore water rather than tightly attached to the minerals. Because CARD-FISH detects rRNA inside individual cells, the detected cells may have been still alive and in their in situ metabolic state. This led us to hypothesize that deep-biosphere seeds of Atribacteria will colonize new habitats on the seafloor and revive to start growing, freshly deposited in a sedimentary habitat after traveling a long distance in the ocean.
Previous studies of the partial genome sequence of Atribacteria indicated that they are heterotrophic anaerobes that ferment organic acids, such as propionate, and produce the fermentation products acetate, ethanol, and CO2 (Carr et al., 2015; Nobu et al., 2016). Their fermentation products may therefore directly or syntrophically support methanogenesis or acetogenesis; that is, the terminal metabolic processes in the anoxic heterotrophic microbial ecosystem. In addition, it is worth noting that there have been no functional genes for methanotrophy so far identified in the known Atribacteria-derived genome sequences. This suggests that there may be little or no biogeochemical contributions from Atribacteria in methane plumes if they are just surviving and drifting in the water column. Their biogeochemical activities would resume when these deep-biosphere seeds are safely redeposited on the seafloor and buried back into the anoxic sedimentary habitat.
The number of cells in MV#1 and MV#14 sediment samples was much lower than in well-stratified sediment core samples from Pacific margin sites. In addition, the shallower mud volcano sediment in this study contained only 102–105 cells/g-wet, which is several orders of magnitude lower than the 108–109 cells/cm3 in very active and water-rich SMVs (e.g., Håkon Mosby mud volcano; Losekann et al., 2007). This implies that cell densities in the deeper zones of MV#1 and MV#14 are possibly much lower than in other, more active SMVs. Because the SMVs near Tanegashima originated from Eocene strata (Ujiie, 2000), it is possible that the low cell concentrations in the mud volcano sediment at this site can be explained by sediment matrices that are too deep and too old to support microbial life, and the geosystem may lack both available electron donors and acceptors. This, however, is still an open question.
In this study, we investigated microbial communities at two SMVs off Tanegashima Island, Japan, using second generation sequencing, microfluidic digital PCR, image-based cell counts, and CARD-FISH techniques. Beta diversity of sediment and water-column microbial communities, as well as cell-count and gene quantification data, consistently indicated that microbial community compositions in mud volcano sediments are very distinct from those in well-stratified marginal sediments and water columns. Sedimentary Atribacteria were present in methane plumes in the overlying water column, suggesting microbial dispersal from the deep sedimentary biosphere to the overlying hydrosphere through submarine mud volcano activity. These findings suggest that a transmigration of “deep-biosphere seeds” may occur between the Earth’s surface and subsurface biospheres.
TH, TT, AI, YM, HM, JA, and FI sampled and pretreated a core sample. HM conducted surveys at the sampling sites. TH carried out molecular works as DNA extraction, sequencing and quantification of 16S rRNA gene, and data analysis. TT, AI, and KO performed measurement of methane and sulfate concentration. TH and YM carried out microscopic works including cell count and CARD-FISH. TH and FI designed of the work and drafted the manuscript, which was critically revised by all the other authors. The final manuscript was approved by all authors.
This work was supported by the Japan Society for the Promotion of Science Grant-in-Aid for Science Research (grant no. 26251041 to FI, grant no. 15K14907 and 17H03956 to TH). This is a contribution to the Deep Carbon Observatory (DCO).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank all crews, drilling team members, and shipboard scientists for core sampling during cruise KH-15-02 of R/V Hakuho Maru, ODP Leg 201, IODP Expedition 301, and shakedown cruise CK06-06 of D/V Chikyu. The authors are grateful for the technical assistance of S. Hashimoto, S. Fukunaga, and T. Terada.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01135/full#supplementary-material
Ashi, J., Sawada, R., Omura, A., and Ikehara, K. (2014). Accumulation of an earthquake-induced extremely turbid layer in a terminal basin of the Nankai accretionary prism. Earth Planets Space 66:51. doi: 10.1186/1880-5981-66-51
Biddle, J. F., Fitz-Gibbon, S., Schuster, S. C., Brenchley, J. E., and House, C. H. (2008). Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc. Natl. Acad. Sci. U.S.A. 105, 10583–10588. doi: 10.1073/pnas.0709942105
Blazejak, A., and Schippers, A. (2010). High abundance of JS-1- and Chloroflexi-related Bacteria in deeply buried marine sediments revealed by quantitative, real-time PCR. FEMS Microbiol. Ecol. 72, 198–207. doi: 10.1111/j.1574-6941.2010.00838.x
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Carr, S. A., Orcutt, B. N., Mandernack, K. W., and Spear, J. R. (2015). Abundant Atribacteria in deep marine sediment from the Adelie Basin, Antarctica. Front. Microbiol. 6:872. doi: 10.3389/fmicb.2015.00872
Ciobanu, M. C., Burgaud, G., Dufresne, A., Breuker, A., Redou, V., Ben Maamar, S., et al. (2014). Microorganisms persist at record depths in the subseafloor of the Canterbury Basin. ISME J. 8, 1370–1380. doi: 10.1038/ismej.2013.250
DeLong, E. F. (1992). Archaea in coastal marine environments. Proc. Natl. Acad. Sci. U.S.A. 89, 5685–5689. doi: 10.1073/pnas.89.12.5685
Deming, J. W., and Baross, J. A. (1993). Deep-sea smokers - windows to a subsurface biosphere. Geochim. Cosmochim. Acta 57, 3219–3230. doi: 10.1016/0016-7037(93)90535-5
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Doi, H., and Okamura, H. (2011). Similarity indices, ordination, and community analysis tests using the software R. Jpn. J. Ecol. 61, 3–20.
Feseker, T., Foucher, J. P., and Harmegnies, F. (2008). Fluid flow or mud eruptions? Sediment temperature distributions on Hakon Mosby mud volcano, SW Barents Sea slope. Mar. Geol. 247, 194–207. doi: 10.1016/j.margeo.2007.09.005
Herlemann, D. P., Labrenz, M., Jurgens, K., Bertilsson, S., Waniek, J. J., and Andersson, A. F. (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571–1579. doi: 10.1038/ismej.2011.41
Hoehler, T. M., and Jørgensen, B. B. (2013). Microbial life under extreme energy limitation. Nat. Rev. Microbiol. 11, 83–94. doi: 10.1038/nrmicro2939
Hoshino, T., and Inagaki, F. (2012). Molecular quantification of environmental DNA using microfluidics and digital PCR. Syst. Appl. Microbiol. 35, 390–395. doi: 10.1016/j.syapm.2012.06.006
Hoshino, T., and Inagaki, F. (2017). Application of stochastic labeling with random-sequence barcodes for simultaneous quantification and sequencing of environmental 16S rRNA genes. PLoS ONE 12:e0169431. doi: 10.1371/journal.pone.0169431
Hoshino, T., Kuratomi, T., Morono, Y., Hori, T., Oiwane, H., Kiyokawa, S., et al. (2015). Ecophysiology of zetaproteobacteria associated with shallow hydrothermal iron-oxyhydroxide deposits in nagahama bay of satsuma Iwo-Jima, Japan. Front. Microbiol. 6:1554. doi: 10.3389/fmicb.2015.01554
Hoshino, T., Yilmaz, L. S., Noguera, D. R., Daims, H., and Wagner, M. (2008). Quantification of target molecules needed to detect microorganisms by fluorescence in situ hybridization (FISH) and catalyzed reporter deposition-FISH. Appl. Environ. Microbiol. 74, 5068–5077. doi: 10.1128/AEM.00208-08
Huber, J. A., Butterfield, D. A., and Baross, J. A. (2003). Bacterial diversity in a subseafloor habitat following a deep-sea volcanic eruption. FEMS Microbiol. Ecol. 43, 393–409. doi: 10.1111/j.1574-6941.2003.tb01080.x
Huber, J. A., Johnson, H. P., Butterfield, D. A., and Baross, J. A. (2006). Microbial life in ridge flank crustal fluids. Environ. Microbiol. 8, 88–99. doi: 10.1111/j.1462-2920.2005.00872.x
Hubert, C., Loy, A., Nickel, M., Arnosti, C., Baranyi, C., Bruchert, V., et al. (2009). A constant flux of diverse thermophilic bacteria into the cold Arctic seabed. Science 325, 1541–1544. doi: 10.1126/science.1174012
Inagaki, F., Hinrichs, K. U., Kubo, Y., Bowles, M. W., Heuer, V. B., Hong, W. L., et al. (2015). DEEP BIOSPHERE. Exploring deep microbial life in coal-bearing sediment down to ∼2.5 km below the ocean floor. Science 349, 420–424. doi: 10.1126/science.aaa6882
Inagaki, F., Nunoura, T., Nakagawa, S., Teske, A., Lever, M., Lauer, A., et al. (2006). Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. U.S.A. 103, 2815–2820. doi: 10.1073/pnas.0511033103
Inagaki, F., Suzuki, M., Takai, K., Oida, H., Sakamoto, T., Aoki, K., et al. (2003). Microbial communities associated with geological horizons in coastal subseafloor sediments from the sea of okhotsk. Appl. Environ. Microbiol. 69, 7224–7235. doi: 10.1128/AEM.69.12.7224-7235.2003
Jerosch, K., Schluter, M., Foucher, J. P., Allais, A. G., Klages, M., and Edy, C. (2007). Spatial distribution of mud flows, chemoautotrophic communities, and biogeochemical habitats at Hakon Mosby Mud Volcano. Mar. Geol. 243, 1–17. doi: 10.1016/j.margeo.2007.03.010
Jungbluth, S. P., Bowers, R. M., Lin, H. T., Cowen, J. P., and Rappe, M. S. (2016). Novel microbial assemblages inhabiting crustal fluids within mid-ocean ridge flank subsurface basalt. ISME J. 10, 2033–2047. doi: 10.1038/ismej.2015.248
Kallmeyer, J., Pockalny, R., Adhikari, R. R., Smith, D. C., and D’hondt, S. (2012). Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. U.S.A. 109, 16213–16216. doi: 10.1073/pnas.1203849109
Kopf, A. J. (2002). Significance of mud volcanism. Rev. Geophys. 40, 1–52. doi: 10.1029/2000RG000093
Kormas, K. A., Meziti, A., Dahlmann, A., Gj, D. E. L., and Lykousis, V. (2008). Characterization of methanogenic and prokaryotic assemblages based on mcrA and 16S rRNA gene diversity in sediments of the Kazan mud volcano (Mediterranean Sea). Geobiology 6, 450–460. doi: 10.1111/j.1472-4669.2008.00172.x
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (Chichester: John Wiley & Sons).
Li, K., Bihan, M., and Methe, B. A. (2013). Analyses of the stability and core taxonomic memberships of the human microbiome. PLoS ONE 8:e63139. doi: 10.1371/journal.pone.0063139
Lloyd, K. G., May, M. K., Kevorkian, R. T., and Steen, A. D. (2013). Meta-analysis of quantification methods shows that archaea and bacteria have similar abundances in the subseafloor. Appl. Environ. Microbiol. 79, 7790–7799. doi: 10.1128/AEM.02090-13
Losekann, T., Knittel, K., Nadalig, T., Fuchs, B., Niemann, H., Boetius, A., et al. (2007). Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby Mud Volcano, Barents Sea. Appl. Environ. Microbiol. 73, 3348–3362. doi: 10.1128/AEM.00016-07
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371. doi: 10.1093/nar/gkh293
Manheim, F. T., and Sayles, F. L. (1974). “Composition and origin of interstitial waters of marine sediments, based on deep sea drill cores,” in The Sea, Marine Chemistry, Vol. 5, ed. E. D. Goldberg (New York, NY: Wiley-Interscience), 527–568.
Martinez, R. J., Mills, H. J., Story, S., and Sobecky, P. A. (2006). Prokaryotic diversity and metabolically active microbial populations in sediments from an active mud volcano in the Gulf of Mexico. Environ. Microbiol. 8, 1783–1796. doi: 10.1111/j.1462-2920.2006.01063.x
Milkov, A. V. (2000). Worldwide distribution of submarine mud volcanoes and associated gas hydrates. Mar. Geol. 167, 29–42. doi: 10.1016/S0025-3227(00)00022-0
Miyazaki, J., Higa, R., Toki, T., Ashi, J., Tsunogai, U., Nunoura, T., et al. (2009). Molecular characterization of potential nitrogen fixation by anaerobic methane-oxidizing archaea in the methane seep sediments at the number 8 Kumano Knoll in the Kumano Basin, offshore of Japan. Appl. Environ. Microbiol. 75, 7153–7162. doi: 10.1128/AEM.01184-09
Morono, Y., and Inagaki, F. (2010). Automatic slide-loader fluorescence microscope for discriminative enumeration of subseafloor life. Sci. Drilling 9, 32–36. doi: 10.5194/sd-9-32-2010
Morono, Y., Terada, T., Kallmeyer, J., and Inagaki, F. (2013). An improved cell separation technique for marine subsurface sediments: applications for high-throughput analysis using flow cytometry and cell sorting. Environ. Microbiol. 15, 2841–2849. doi: 10.1111/1462-2920.12153
Morono, Y., Terada, T., Nishizawa, M., Ito, M., Hillion, F., Takahata, N., et al. (2011). Carbon and nitrogen assimilation in deep subseafloor microbial cells. Proc. Natl. Acad. Sci. U.S.A. 108, 18295–18300. doi: 10.1073/pnas.1107763108
Nakayama, N., Ashi, J., Tsunogai, U., Gamo, T., and Tanahashi, M. (2010). Sources of pore water in a Tanegashima mud volcano inferred from chemical and stable isotopic studies. Geochem. J. 44, 561–569. doi: 10.2343/geochemj.1.0094
Newberry, C. J., Webster, G., Cragg, B. A., Parkes, R. J., Weightman, A. J., and Fry, J. C. (2004). Diversity of prokaryotes and methanogenesis in deep subsurface sediments from the Nankai Trough, Ocean Drilling Program Leg 190. Environ. Microbiol. 6, 274–287. doi: 10.1111/j.1462-2920.2004.00568.x
Niemann, H., Losekann, T., De Beer, D., Elvert, M., Nadalig, T., Knittel, K., et al. (2006). Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 443, 854–858. doi: 10.1038/nature05227
Nobu, M. K., Dodsworth, J. A., Murugapiran, S. K., Rinke, C., Gies, E. A., Webster, G., et al. (2016). Phylogeny and physiology of candidate phylum ‘Atribacteria’ (OP9/JS1) inferred from cultivation-independent genomics. ISME J. 10, 273–286. doi: 10.1038/ismej.2015.97
Omoregie, E. O., Mastalerz, V., De Lange, G., Straub, K. L., Kappler, A., Roy, H., et al. (2008). Biogeochemistry and community composition of iron- and sulfur-precipitating microbial mats at the Chefren mud volcano (Nile Deep Sea Fan, Eastern Mediterranean). Appl. Environ. Microbiol. 74, 3198–3215. doi: 10.1128/AEM.01751-07
Oni, O., Miyatake, T., Kasten, S., Richter-Heitmann, T., Fischer, D., Wagenknecht, L., et al. (2015). Distinct microbial populations are tightly linked to the profile of dissolved iron in the methanic sediments of the Helgoland mud area, North Sea. Front. Microbiol. 6:365. doi: 10.3389/fmicb.2015.00365
Orcutt, B. N., Larowe, D. E., Biddle, J. F., Colwell, F. S., Glazer, B. T., Reese, B. K., et al. (2013). Microbial activity in the marine deep biosphere: progress and prospects. Front. Microbiol. 4:189. doi: 10.3389/fmicb.2013.00189
Pachiadaki, M. G., Kallionaki, A., Dahlmann, A., De Lange, G. J., and Kormas, K. A. (2011). Diversity and spatial distribution of prokaryotic communities along a sediment vertical profile of a deep-sea mud volcano. Microb. Ecol. 62, 655–668. doi: 10.1007/s00248-011-9855-2
Pachiadaki, M. G., Lykousis, V., Stefanou, E. G., and Kormas, K. A. (2010). Prokaryotic community structure and diversity in the sediments of an active submarine mud volcano (Kazan mud volcano, East Mediterranean Sea). FEMS Microbiol. Ecol. 72, 429–444. doi: 10.1111/j.1574-6941.2010.00857.x
Raskin, L., Stromley, J. M., Rittmann, B. E., and Stahl, D. A. (1994). Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60, 1232–1240.
Reed, D. W., Fujita, Y., Delwiche, M. E., Blackwelder, D. B., Sheridan, P. P., Uchida, T., et al. (2002). Microbial communities from methane hydrate-bearing deep marine sediments in a forearc basin. Appl. Environ. Microbiol. 68, 3759–3770. doi: 10.1128/AEM.68.8.3759-3770.2002
Rochelle, P. A., Cragg, B. A., Fry, J. C., Parkes, R. J., and Weightman, A. J. (1994). Effect of sample handling on estimation of bacterial diversity in marine-sediments by 16s ribosomal-RNA gene sequence-analysis. FEMS Microbiol. Ecol. 15, 215–225. doi: 10.1111/j.1574-6941.1994.tb00245.x
Røy, H., Kallmeyer, J., Adhikari, R. R., Pockalny, R., Jørgensen, B. B., and D’hondt, S. (2012). Aerobic microbial respiration in 86-million-year-old deep-sea red clay. Science 336, 922–925. doi: 10.1126/science.1219424
Sahling, H., Bohrmann, G., Artemov, Y. G., Bahr, A., Bruning, M., Klapp, S. A., et al. (2009). Vodyanitskii mud volcano, Sorokin trough, Black Sea: geological characterization and quantification of gas bubble streams. Mar. Petrol Geol. 26, 1799–1811. doi: 10.1016/j.marpetgeo.2009.01.010
Sauter, E. J., Muyakshin, S. I., Charlou, J. L., Schluter, M., Boetius, A., Jerosch, K., et al. (2006). Methane discharge from a deep-sea submarine mud volcano into the upper water column by gas hydrate-coated methane bubbles. Earth Planet. Sci. Lett. 243, 354–365. doi: 10.1016/j.epsl.2006.01.041
Salter, S. J., Cox, M. J., Turek, E. M., Calus, S. T., Cookson, W. O., Moffatt, M. F., et al. (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12:87. doi: 10.1186/s12915-014-0087-z
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schramm, A., Fuchs, B. M., Nielsen, J. L., Tonolla, M., and Stahl, D. A. (2002). Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ. Microbiol. 4, 713–720. doi: 10.1046/j.1462-2920.2002.00364.x
Schubert, C. J., Durisch-Kaiser, E., Holzner, C. P., Klauser, L., Wehrli, B., Schmale, O., et al. (2006). Methanotrophic microbial communities associated with bubble plumes above gas seeps in the Black Sea. Geochem. Geophys. Geosyst. 7:8. doi: 10.1029/2005gc001049
Takai, K., Gamo, T., Tsunogai, U., Nakayama, N., Hirayama, H., Nealson, K. H., et al. (2004). Geochemical and microbiological evidence for a hydrogen-based, hyperthermophilic subsurface lithoautotrophic microbial ecosystem (HyperSLiME) beneath an active deep-sea hydrothermal field. Extremophiles 8, 269–282. doi: 10.1007/s00792-004-0386-3
Toki, T., Chibana, H., Shimabukuro, T., and Yamakawa, Y. (2016). “Investigation of distribution of mud volcanoes in East China Sea using distribution of methane concentrations in seawater,” in Proceedings of the Japan Geoscience Union Meeting 2016, May 22-26, Chiba.
Tsunogai, U., Yoshida, N., and Gamo, T. (2002). Carbon isotopic evidence of methane oxidation through sulfate reduction in sediment beneath seafloor cold seep vents on the seafloor at Nankai Trough. Mar. Geol. 187, 145–160. doi: 10.1016/S0025-3227(02)00263-3
Ujiie, Y. (2000). Mud diapirs observed in two piston cores from the landward slope of the northern Ryukyu Trench, northwestern Pacific Ocean. Mar. Geol. 163, 149–167. doi: 10.1016/S0025-3227(99)00113-9
Wallmann, K., Drews, M., Aloisi, G., and Bohrmann, G. (2006). Methane discharge into the Black Sea and the global ocean via fluid flow through submarine mud volcanoes. Earth Planet Sci. Lett. 248, 545–560. doi: 10.1016/j.epsl.2006.06.026
Walsh, E. A., Kirkpatrick, J. B., Rutherford, S. D., Smith, D. C., Sogin, M., and D’hondt, S. (2016). Bacterial diversity and community composition from seasurface to subseafloor. ISME J. 10, 979–989. doi: 10.1038/ismej.2015.175
Walters, W. A., Caporaso, J. G., Lauber, C. L., Berg-Lyons, D., Fierer, N., and Knight, R. (2011). PrimerProspector: de novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics 27, 1159–1161. doi: 10.1093/bioinformatics/btr087
Webster, G., Parkes, R. J., Cragg, B. A., Newberry, C. J., Weightman, A. J., and Fry, J. C. (2006a). Prokaryotic community composition and biogeochemical processes in deep subseafloor sediments from the Peru Margin. FEMS Microbiol. Ecol. 58, 65–85.
Webster, G., Parkes, R. J., Fry, J. C., and Weightman, A. J. (2004). Widespread occurrence of a novel division of bacteria identified by 16S rRNA gene sequences originally found in deep marine sediments. Appl. Environ. Microbiol. 70, 5708–5713. doi: 10.1128/AEM.70.9.5708-5713.2004
Webster, G., Watt, L. C., Rinna, J., Fry, J. C., Evershed, R. P., Parkes, R. J., et al. (2006b). A comparison of stable-isotope probing of DNA and phospholipid fatty acids to study prokaryotic functional diversity in sulfate-reducing marine sediment enrichment slurries. Environ. Microbiol. 8, 1575–1589.
Webster, G., Yarram, L., Freese, E., Koster, J., Sass, H., Parkes, R. J., et al. (2007). Distribution of candidate division JS1 and other Bacteria in tidal sediments of the German Wadden Sea using targeted 16S rRNA gene PCR-DGGE. FEMS Microbiol. Ecol. 62, 78–89. doi: 10.1111/j.1574-6941.2007.00372.x
Keywords: submarine mud volcano, deep subseafloor biosphere, methane, Atribacteria, digital PCR, CARD-FISH
Citation: Hoshino T, Toki T, Ijiri A, Morono Y, Machiyama H, Ashi J, Okamura K and Inagaki F (2017) Atribacteria from the Subseafloor Sedimentary Biosphere Disperse to the Hydrosphere through Submarine Mud Volcanoes. Front. Microbiol. 8:1135. doi: 10.3389/fmicb.2017.01135
Received: 10 February 2017; Accepted: 06 June 2017;
Published: 20 June 2017.
Edited by:
Jennifer F. Biddle, University of Delaware, United StatesReviewed by:
Casey R. J. Hubert, University of Calgary, CanadaCopyright © 2017 Hoshino, Toki, Ijiri, Morono, Machiyama, Ashi, Okamura and Inagaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tatsuhiko Hoshino, aG9zaGlub3RAamFtc3RlYy5nby5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.