
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 02 June 2017
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.01003
Novel class 1 integrons In1085 and In1086, containing the class D β-lactamase -encoding gene blaOXA, were identified in clinical enterobacterial strains. In this study, we aimed to characterize the genetic contexts of In1085 and In1086, with the goal of identifying putative mechanisms of integron mobilization. Four plasmids, approximately 5.3, 5.3, 5.7, and 6.6 kb, from 71 clinical Enterobacteriaceae strains were found to contain class 1 integrons (In37, In62, In1085, and In1086, respectively). Two of these plasmids, pEco336 and pNsa292, containing In1085 and In1086, respectively, were further characterized by antibiotic susceptibility testing, conjugation experiments, PCR, sequencing, and gene mapping. The OXA-type carbapenemase activities of the parental strains were also assessed. The results revealed that the novel integrons had different genetic environments, and therefore demonstrated diverse biochemical characteristics. Using evolutionary inferences based on the recombination of gene cassettes, we also identified a role for attCaadA16 structural features during attI1 × attC insertion reactions. Our analysis showed that gene cassette insertions in the bottom strand of attCaadA16 in the correct orientation lead to the expression the encoded genes from the Pc promoter. Our study suggests that the genetic features harbored within the integrons are inserted in a discernable pattern, involving the stepwise and parallel evolution of class 1 integron variations under antibiotic selection pressures in a clinical setting.
Gene cassettes (GCs) are small, mobile elements containing one or more genes and recombination sites, and are often contained within larger genetic structures known as integrons. Several cassettes can be inserted into a single integron, where they establish a tandem array (Partridge et al., 2009). Integrons consist of three crucial components: an intI gene, recombination site attI, and a Pc promoter. intI encodes an integrase that mediates the integration and excision of the GCs by site-specific recombination (Barraud and Ploy, 2015). The mechanisms of integration and excision of GCs are well described, with integrations known to occur at attI × attC recombination sites (MacDonald et al., 2006; Loot et al., 2012), and excisions requiring attC × attC recombination sites, which occur in single-stranded sequences and activate the folded bottom strand (bs) (Bouvier et al., 2005, 2009). However, despite their mobility, GC rearrangements resulting in novel GC arrays rarely occur, although it is assumed that integrases could rearrange GCs, generating integron variants, under antibiotic selective pressure (Bouvier et al., 2005, 2009; Barraud and Ploy, 2015).
Generally, integrons are confirmed by the presence of intI and an attI recombination site (Barraud and Ploy, 2015). Integrons can be assigned into three classes (1, 2, and 3) based on their integrase gene sequences, with class 1 integrons most often associated with antimicrobial resistance in clinical bacterial isolates (Huang et al., 2015). By associating with transposons or being plasmid-encoded, integrons may capture genetic structures, express GCs, and mediate their own mobility despite their lack of self-mobility elements (Carattoli, 2003; Mazel, 2006). Transposon- or plasmid-associated integrons containing antibiotic resistance GCs can spread widely among Gram-negative bacteria (Barraud and Ploy, 2015). Integrons can also evolve rapidly through the acquisition, storage, and rearrangement of genes embedded in their GCs, and, by mobilization to conjugative plasmids, they play a crucial role in increasing multidrug resistance amongst clinical bacterial strains (Escudero et al., 2016). In general, integrons from clinical bacterial strains contain less than five GCs (Bennett, 2008), although integrons with up to nine antibiotic resistance genes have been reported (Naas et al., 2001). Presently, more than 130 different GCs (less than 98% identical) containing antibiotic resistance genes have been identified, along with many other GCs containing genes of unknown function (Partridge et al., 2009).
Unlike class D oxacillinases, OXA-type carbapenemase genes were not originally thought to be integrated into integrons as part of GC sequences, and were mainly shown to be encoded by chromosomal genes (Walther-Rasmussen and Høiby, 2006). However, more recent studies have shown that integrons harboring various blaOXA-type genes are present in bacterial plasmids (Wendel et al., 2016). The current study characterizes novel integrons In1085 and In1086 and their surrounding genes within bacterial plasmids, and explores the putative mechanism of integron mobilization.
This study was approved by the Ethics Committee of the Municipal Hospital of Taizhou University, Zhejiang, China, and written informed consent was obtained from each of the participants in accordance with the Declaration of Helsinki. The rights of the research subjects were protected throughout, and we confirm that this study was conducted in our hospital.
The use of human specimens and all related experimental protocols was approved by the Committee on Human Research of the indicated institutions, and was carried out in accordance with approved guidelines.
Four of seventy-one non-redundant multidrug resistant Enterobacteriaceae strains, including Klebsiella pneumoniae Kpn761 (harboring In62), non-typhoidal Salmonella strains Nsa243 and Nsa292 (In37 and In1086, respectively), and Enterobacter cloacae Eco336 (In1085), were recovered from hospitalized patients with clinical infections. The isolates were collected between June 2013 and July 2015, and were primarily assessed for the presence of integrons by antimicrobial susceptibility testing. Bacterial species were identified by 16S rRNA gene sequencing as described previously (Frank et al., 2008). The strains harboring integrons were studied in the study. Escherichia coli TOP10 cells (Invitrogen, Grand Island, NY, United States) were used as a host for cloning experiments, and as a control for susceptibility testing.
The minimum inhibitory concentration (MIC) values of isolates Eco336 and Nsa292 (harboring In1085 and In1086, respectively) for 12 antimicrobial agents, including cephalosporins (cefazolin, ceftazidime, and ceftriaxone), aminoglycosides (netilmicin, tobramycin, and amikacin), carbapenems (ertapenem, meropenem, and imipenem), and quinolones (norfloxacin, ofloxacin, and ciprofloxacin), were determined using the Microscan broth dilution method (Microscan, Renton, WA, United States). The MICs were interpreted in accordance with the Clinical and Laboratory Standards Institute guidelines (Clinical and Laboratory Standards Institute [CLSI], 2015).
Plasmids from four integron-harboring strains were isolated using an AxyPrep Plasmid Miniprep kit (Axygen Biosciences, Beijing, China) according to the manufacturer’s instructions and as described previously (Wang et al., 2014). Two plasmids containing novel integrons In1085 and In1086 were then digested with BamHI (TaKaRa, Dalian, China) and subjected to agarose gel electrophoresis to generate genetic maps.
To characterize the two novel integrons, the relevant BamHI fragments of the two digested plasmids were ligated into the pMD19-T cloning vector (TaKaRa, Dalian, China). The ligation mixtures were electroporated into E. coli TOP10 cells, which were then plated on LB medium with medium supplemented with ampicillin (100 μg/mL) and incubated over-night at 37°C. Plasmids from any resulting aacA4-positive colonies were isolated by PCR amplification and sequencing (Supplementary Table S1), and the inserts were amplified using the primers specified in Supplementary Table S1 and the following thermal cycler conditions: 3 min at 94°C, 30 cycles of 1 min each at 94, 54, and 72°C, followed by 10 min at 72°C. The total reaction volume was 25 μL, and the total eluent volume was 10 μL. Following amplification, the amplicons were separated by gel electrophoresis on a 0.6% agarose gel run at 90 V for 90 min in 0.5 × TBE buffer. Fragments corresponding to plasmid DNA and integrons were recovered, and the initial positions of the relevant genes in the recombinant plasmids were determined according to a previously established method for estimating plasmid DNA sizes (Wang et al., 2003). Thus, the sequences of both integrons, including upstream and downstream genes, were obtained, by PCR amplification and sequencing (Supplementary Table S1), and the genetic structures were mapped and characterized by next generation sequence annotation and genome comparison. Plasmids harboring In1085 and In1086, In1085-TOP10 and In1086-TOP10 were studied, and the values of relevent susceptibility testing were confirmed (Table 1) by conjugation experiments in according with previous report (Wang et al., 2014).
Open reading frames (ORFs) were predicted using RAST1, and further annotated using BLASTP and BLASTN searches2 against the UniProtKB/Swiss-Prot3, and National Center for Biotechnology Information non-redundant4 databases. Annotation of drug resistance genes, mobile elements, and other genes was based on CARD analysis5, the β-lactamase database6, ISfinder7, and INTEGRALL8. Sequence comparisons were performed using BLASTN and CLUSTALW29. Gene organization diagrams were drawn using Inkscape10.
The presence of classes A, B, and D carbapenemase activity in cell extracts from non-typhoidal Salmonella strain Nsa292 and E. cloacae strain Eco336 was determined using a modified CarbaNP test (Chen et al., 2015). Briefly, overnight cultures of each strain grown in Mueller-Hinton (MH) broth were diluted 1:100 into 3 mL of fresh MH broth, and then incubated at 37°C with shaking at 200 rpm to an optical density at 200 nm of 1.0–1.4. When required, ampicillin was used at 200 μg/mL. Bacterial cells were harvested from 2 mL of culture, and cell pellets were washed twice with 20 mM Tris-HCl (pH 7.8). Each cell pellet was resuspended in 500 μL of 20 mM Tris-HCl (pH 7.8), lysed by sonication, and centrifuged at 10,000 × g for 5 min at 4°C. Each 50-μL supernatant (containing the enzymatic bacterial suspension fraction) was mixed with 50 μL of the following substrates (I to V), followed by incubation at 37°C for a maximum of 2 h: substrate I: 0.054% phenol red and 0.1 mM ZnSO4 (pH 7.8); substrate II: 0.054% phenol red, 0.1 mM ZnSO4 (pH 7.8), and 0.6 mg/μL imipenem; substrate III: 0.054% phenol red, 0.1 mM ZnSO4 (pH 7.8), 0.6 mg/μL imipenem, and 0.8 mg/μL tazobactam; substrate IV: 0.054% phenol red, 0.1 mM ZnSO4 (pH 7.8), 0.6 mg/μL imipenem, and 3 mM EDTA (pH 7.8); and substrate V: 0.054% phenol red, 0.1 mM ZnSO4 (pH 7.8), 0.6 mg/μL imipenem, 0.8 mg/μL tazobactam, and 3 mM EDTA (pH 7.8).
The sequences of novel integrons In1085 and In1086 were deposited in the GenBank database under accession numbers KP870111 and KP870112, respectively.
Four strains, including K. pneumoniae strain Kpn761 (harboring In62), non-typhoidal Salmonella strains Nsa292 and Nsa243 (In1086 and In37), and E. cloacae strain Eco336 (In1085), were selected for further study. Non-typhoidal Salmonella Nsa292 and E. cloacae Eco336 showed resistance to aminoglycoside, cephalosporin, and carbapenem antibiotics (Table 1), while the remaining two strains were only used for reference in this study and will be described elsewhere. E. cloacae Eco336 was isolated from the urine of a urological surgery patient, K. pneumoniae Kpn761 (In62) was isolated from a blood culture of an intensive care unit patient, and non-typhoidal Salmonella strains Nsa292 and Nsa243 (In37) were isolated from blood cultures of patients hospitalized with infections.
Following BamHI digestion and ligation into a pMD19-T cloning vector, the four integron-containing recombinant plasmids were transformed into E. coli TOP10 cells. Antibiotic susceptibility test results for the resulting TOP10 cells containing the recombinant plasmids are listed in Table 1. The recombinant plasmids were also electrophoresed to estimate their sizes. The susceptibility test results also indicated that the conjugation experiments were successful (Table 1), and that the observed antibiotic resistance was conferred by plasmid-mediated genes. The electrophoresis results following BamHI digestion indicated that the sizes of the In37, In62, In1085, and In1086 integrons were approximately 5.3, 5.3, 5.7, and 6.6 kb, respectively. Usually, class 1 integrons that integrate within transposons or are encoded on plasmids display regular mobilization and transformation capabilities (Carattoli, 2003; Mazel, 2006). As such, integrons can change from one type to another, with the possibility of generating novel types (Figure 1).
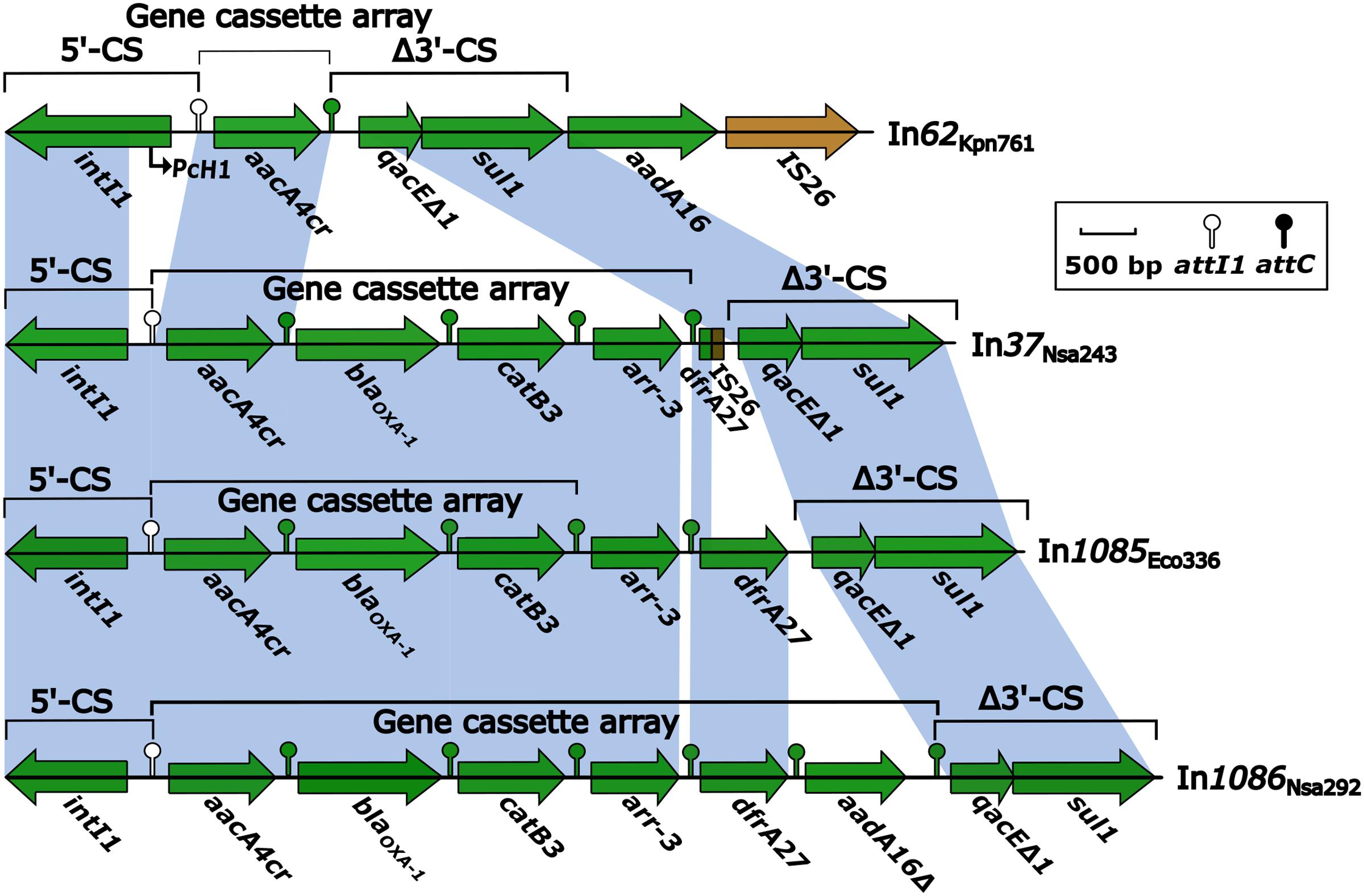
FIGURE 1. Comparison of integron structures based on BamHI digestion of plasmids. Genes are denoted by arrows, and are colored based on gene function classifications. Shaded areas denote regions of homology with >97% nucleotide sequence identity. Only one gene cassette (GC) was identified in In62Kpn761 (In62), as the aadA16 sequence was located beyond the integron. Four GCs were identified in In37, as the remnant of dfrA27 was non-functional. Three functional GCs were identified in In1085, with both GCarr-3 and GCdfrA27 deemed non-functional. Six GCs were identified in In1086, although GCaadA16 was truncated, and the attC sequence was mutated. In37 and In62 sequences have been deposited in GenBank under the accession numbers KR338349 and KR338350, respectively, and will be described in more detail in another study.
Both transformant strains containing the novel integrons, In1085-TOP10 and In1086-TOP10, demonstrated class D carbapenemase activity (data not shown), showing resistance to cephalosporins, carbapenems, and aminoglycosides. However, both strains were susceptible to fluoroquinolones (Table 1). Of the four integrons, In62 apparently represents the most primitive form. It carried two different resistance markers, but only one single-gene GC (GCaacA4cr) (Figure 1). In contrast, In37, In1085, and In1086 contained four or five GCs (Figure 1), containing determinants of at least three different classes of antibiotics. These three larger integrons most likely confer the multidrug resistance phenotype that was demonstrated in the results of antibiotic susceptibility testing of both In1085 and In1086. Interestingly, In37 was almost identical to novel integron In1085, except for the lack of GCdrfA27, which was replaced by non-functional remnants of drfA27 and IS26 in In37 (Figure 1). In addition, a hybrid attC, attCaar-3/catB3, was identified downstream of the GCarr-3 in In1085. This type of hybrid attC is unlikely to be functional for recombination of the GC (GCarr-3). And, instead of the usual 5′-untranslated region (UTR) upstream of the GCdfrA27 in In1085, the upstream attC matched the 5′-UTR of GCarr-3, while the downstream attC was deleted. Therefore, neither GCdfrA27 nor GCarr-3 in In1085 appear to show any integron features, and are likely to be pseudo-integrons. However, the remaining three GCs (GCaacA4cr, GCblaOXA-1, and GCcatB3) do appear to be functional.
In1086 differed from In1085 by the presence of an extra GC, GCaadA16 (Figure 1). Noticeably, GCaadA16 appeared to be truncated, and was thus named GCaadA16Δ. The attC of GCaadA16 also appeared to be mutated to attCqacE/aadA16, rather than the normal attCaadA16 sequence. However, the mutation did not result in any changes in biochemical properties. Thus, In1086 was characterized as containing six GCs. Neither of the novel integrons contained Pc variant sites or 19-bp ORF11 duplicates, but did contain non-functional remnants of intI1 ORFs.
Sequence comparisons revealed a high degree of homology at nucleotide level between Tn6308 (Sun et al., 2016) and pNSA292 (harboring In1085), and between pSA1-like (Soler Bistué et al., 2006) and pECO336 (harboring In1086) (Figure 2). Even greater homology was observed between pNSA292 and pECO336 and pKPS30 (Compain et al., 2014) (Figure 2). pNSA292 shared identity with Tn6308 in both integron regions, except for a lack of GCdfrA27 and GCΔaadA16 in Tn6308, and was confirmed by the high degree of sequence identity between the backbones of Tn6308 and pNSA292 upstream of both integron regions, and at the sap module (Figure 2). pECO336 also showed partial homology to pSA1-like in the integron regions, and to the Tn6309 backbone and sap module in the up- and down-stream integron regions (Figure 2). However, both pNSA292 and pECO336 showed greater homology to pKPS30 in both integron regions (Figure 2).
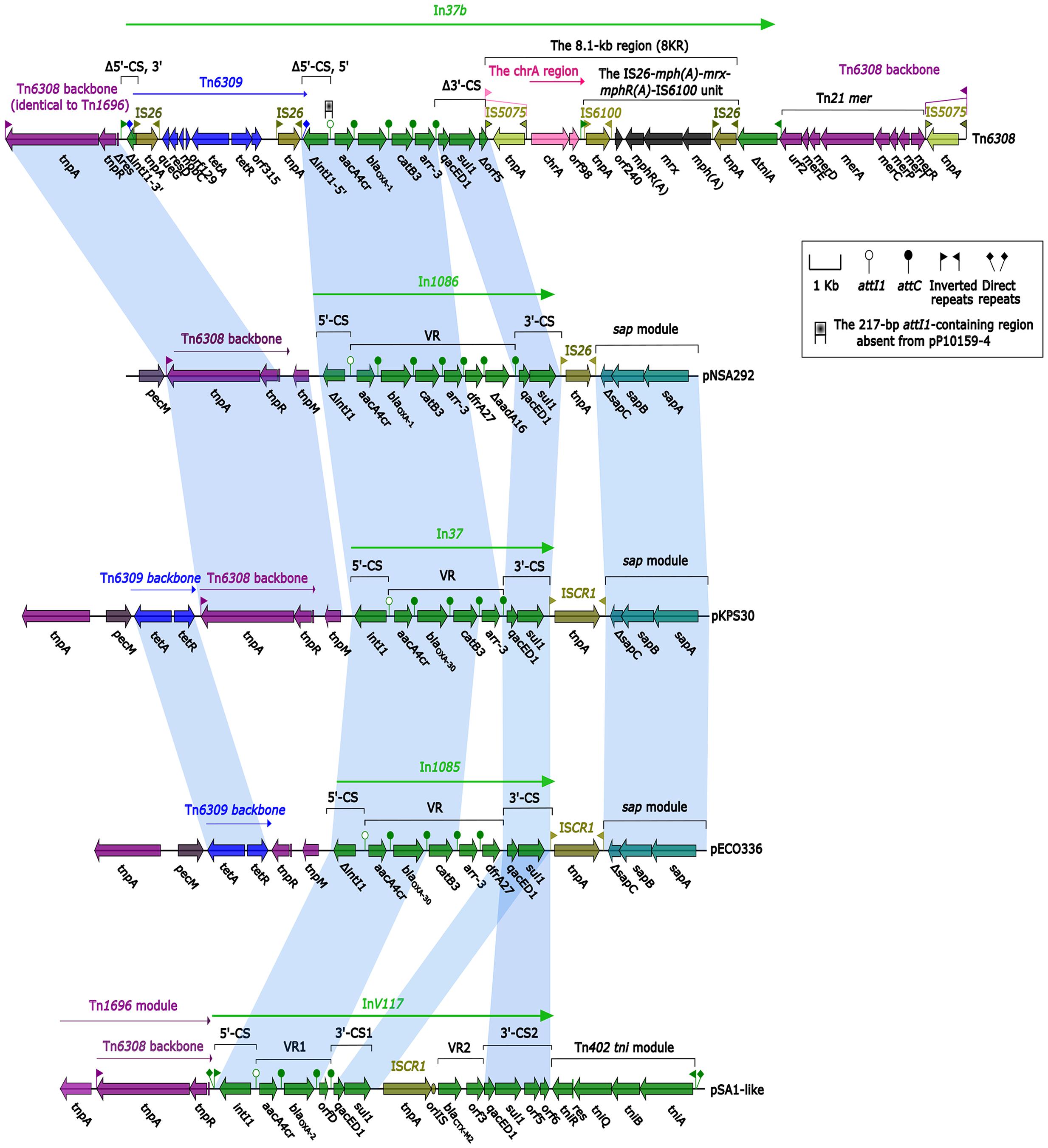
FIGURE 2. Comparison of plasmids and transposons. Genes are denoted by arrows and are colored based on gene function classifications. Shaded areas denote regions of homology with >97% nucleotide sequence identity. Tn6308 was described by Sun et al. (2016), pKPS30 refers to the partial sequence of plasmid KPS30 described by Compain et al. (2014), while pSA1-like shows similarity to pSA1 described by Soler Bistué et al. (2006). pNSA292 and pECO336 are partial plasmids from non-typhoidal Salmonella strain Nsa292 and Enterobacter cloacae strain Eco336, harboring novel integrons In1085 and In1086, respectively.
However, there were a few structural differences between pNSA292 and pECO336, and thus, the plasmids showed different biochemical characteristics. Because of the mobility of transposons and plasmids, we hypothesize that novel integrons In1085 and In1086, located on pEco336 and pNsa292, respectively, have the potential to mobilize. In addition, these integrons may be related to other plasmids and transposons such as Tn6308, Tn6309, Tn1696, pKPS30, and pSA1-like (Figure 2).
Recombination at specific intI1 sites is highly regulated, thereby differing from other reactions mediated by tyrosine recombinases, while recombination at attC sites is much more complicated and variable (Partridge et al., 2009). attCaadA16 comprises two simple sites, each consisting of a pair of conserved “core sites” (7 or 8 bp), designated R″/L″ and L′/R′ (Recchia and Sherratt, 2002; Escudero et al., 2016). R″/L″ sites are separated by a 7-bp spacer, and L′/R′ by a 7- or 8-bp spacer (Figure 3A). R″/L″ and L′/R′ pairs are separated by a central region (Figure 3A). Usually, R″/L″ and L′/R′ are reverse complements of each other, with R″/R′ and L″/L′ generally being complementary, except for the removal of an extra base in the L″ site [G for a bs array and C for a top strand (ts) array] and two bases in the R′ site (A and C for a bs array and T and G for a ts array) (Figure 3B, in bold and italic). The central region is usually a defective inverted repeat, unless there has been a deletion of a single T (bs array) or A (ts array) nucleotide (Figure 3B, in bold and italic). There are two regions in class 1 integrons: stable arrays, including intI1 and the beginning of the attI1 sequence, and variable arrays, involving the end of the attI1 sequence and the GCs. GC integrations take place at attI × attC recombination sites (Figure 3C), and it is essential that the complete coding sequence of the GC is expressed from the promoter, Pc (Figure 3C). If the GC is inserted in the opposite orientation, it will not be expressed (Figure 3C). Only a few integrons have been shown to contain the Pc promoter (Bissonnette et al., 1991; Tolmasky and Crosa, 1993; Biskri and Mazel, 2003; Szekeres et al., 2007; da Fonseca and Vicente, 2012; Guerout et al., 2013), with most GCs, including the stable arrays within novel integrons In1085 and In1086, appearing to be promoterless (Figures 2, 3). Integration of GCs in the opposite orientation would likely hamper the rapid adaptation of integrons (Nivina et al., 2016). Correct orientation of the GCs confirms the selectivity of the integrase toward the bs (Figure 3C) (Nivina et al., 2016). Functional integration does not follow ts recombination, especially if the ts of attC is presented as a substrate (Nivina et al., 2016), because the attC sequence is not conserved, and the interaction between IntI and the folded attC is mostly non-specific (MacDonald et al., 2006). Hence, specific recognition of the bs is the main determinant of successful GC integration. Therefore, determinants required for specific recognition of the bs are likely to be present in the structural features of the variable GC array (Figure 3C). These results can help in determining the formation of integrons from clinical sources.
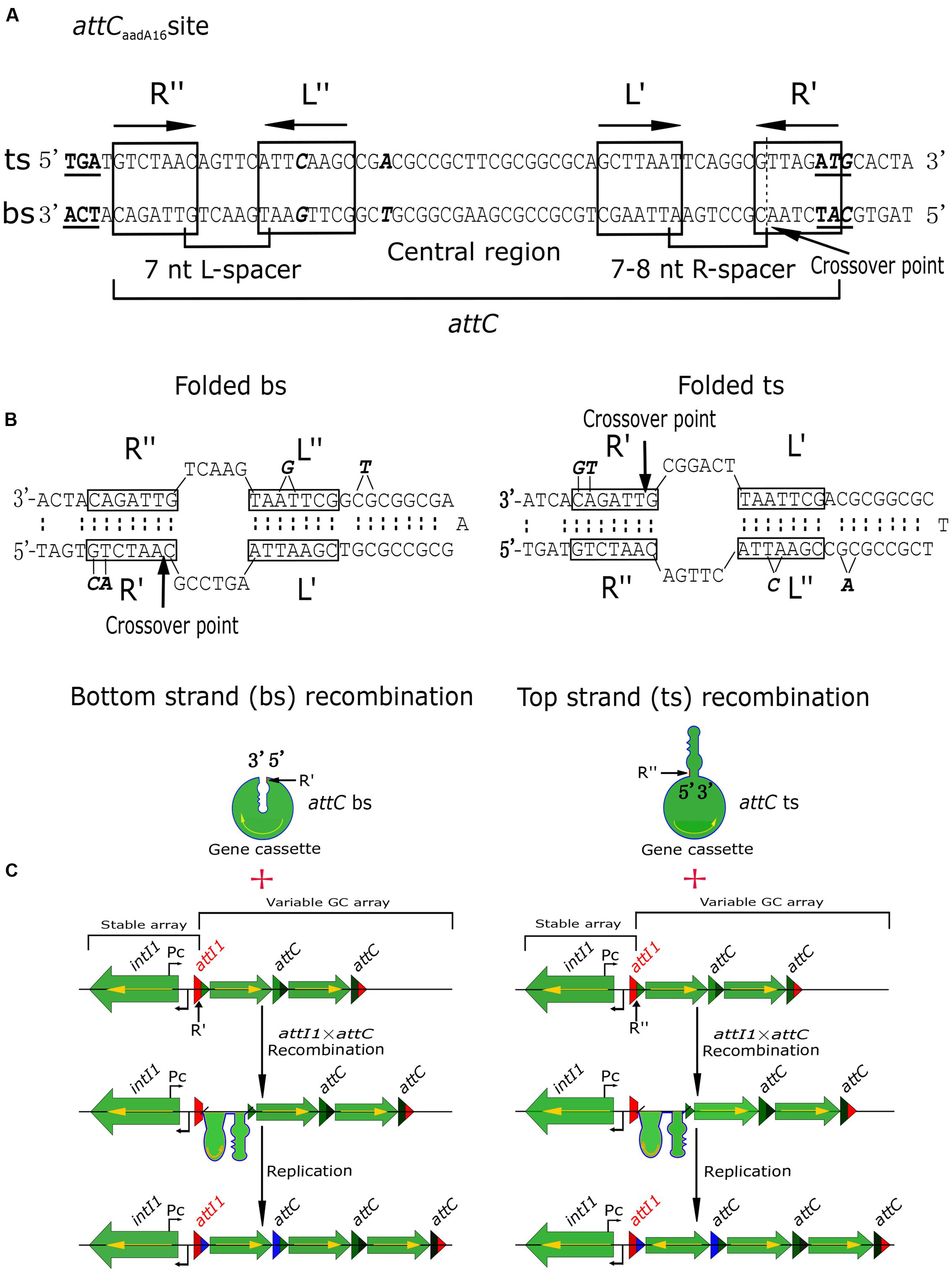
FIGURE 3. Schematic diagrams showing recombination of GCs. (A) Architecture of attCaadA16. The start and stop codons on the top strand (ts) and bottom strand (bs) are in bold and are underlined. Core sites are boxed and labeled, with the end points of the spacers indicated, and the extrahelical bases on the bs and ts shown in bold. The position of the GC “crossover point” is shown as a dotted line, and indicated by an arrow. (B) Folded bs and ts recombinations for the attCaadA16 site showing the deletion of the extrahelical bases. (C) If recombination takes place in the bs of the attC site (R′) and in the bs of attI1, the encoded gene can be expressed from the Pc promoter. However, if recombination occurs in the ts of the attC site (R″) and in the bs of attI1, the GC is inserted in the opposite orientation (yellow arrow) and no gene expression can occur from the Pc promoter.
Novel integrons In1085 and In1086, located on plasmids from clinical bacterial isolates, showed different gene environments. Because of these variations in genetic structure, the two integrons displayed different biochemical characteristics. Using evolutionary inferences for the recombination of the GCs, we also determined that attCaadA16 plays an important role during attI1 × attC insertion reactions, causing preferential recombination of the bs and ensuring that cassettes are inserted in the correct orientation. These findings are evidence of the stepwise and parallel evolution of integrons under antibiotic selection pressures present in clinical settings.
Conception and design of the study: DW. Acquisition of data: DW, WH, JC, DZ, LY, ZY, JF, and ZL. Analysis and interpretation of data: DW, WH, and DZ. Drafting the article: DW. Critical revision: DW, WH, JC, LY, DZ, ZY, JF, and ZL. All authors read and approved the final manuscript.
This work was supported by grants from the Special Key Project of Biosafety Technologies (2016YFC1202600) for the National Major Research and Development Program of China, the Foundation of Department of Science and Technology of Zhejiang Province (2014C33153), the Foundation of Zhejiang Health Department (2014KYB218 and 2017KY717), and the Foundation of Taizhou Science and Technology Bureau(14SF05).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Professor Thomas Jové (INTEGRALL curator), for his designation of the novel In1085 and In1086 integrons, both of which are registered in the INTEGRALL database. English language editing was performed by Edanz (Beijing, China).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01003/full#supplementary-material
Barraud, O., and Ploy, M. C. (2015). Diversity of class 1 integron gene cassette rearrangements selected under antibiotic pressure. J. Bacteriol. 197, 2171–2178. doi: 10.1128/JB.02455-14
Bennett, P. M. (2008). Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br. J. Pharmacol. 153, S347–S357. doi: 10.1038/sj.bjp.0707607
Biskri, L., and Mazel, D. (2003). Erythromycin esterase gene ere(A) is located in a functional gene cassette in an unusual class 2 integron. Antimicrob. Agents Chemother. 47, 3326–3331. doi: 10.1128/aac.47.10.3326-3331.2003
Bissonnette, L., Champetier, S., Buisson, J. P., and Roy, P. H. (1991). Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J. Bacteriol. 173, 4493–4502. doi: 10.1128/jb.173.14.4493-4502.1991
Bouvier, M., Demarre, G., and Mazel, D. (2005). Integron cassette insertion: a recombination process involving a folded single strand substrate. EMBO J. 24, 4356–4367. doi: 10.1038/sj.emboj.7600898
Bouvier, M., Ducos-Galand, M., Loot, C., Bikard, D., and Mazel, D. (2009). Structural features of single-stranded integron cassette attC sites and their role in strand selection. PLoS Genet. 5:e1000632. doi: 10.1371/journal.pgen.1000632
Carattoli, A. (2003). Plasmid-mediated antimicrobial resistance in Salmonella enterica. Curr. Issues Mol. Biol. 5, 113–122.
Chen, Z., Li, H., Feng, J., Li, Y., Chen, X., Guo, X., et al. (2015). NDM-1 encoded by a pNDM-BJ01-like plasmid p3SP-NDM in clinical Enterobacter aerogenes. Front. Microbiol. 6:294. doi: 10.3389/fmicb.2015.00294
Clinical and Laboratory Standards Institute [CLSI] (2015). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement M100– S25. Wayne, PA: CLSI.
Compain, F., Frangeul, L., Drieux, L., Verdet, C., Brisse, S., Arlet, G., et al. (2014). Complete nucleotide sequence of two multidrug-resistant IncR plasmids from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 58, 4207–4210. doi: 10.1128/AAC.02773-13
da Fonseca,É. L., and Vicente, A. C. P. (2012). Functional characterization of a cassette-specific promoter in the class 1 integron-associated qnrVC1 gene. Antimicrob. Agents Chemother. 56, 3392–3394. doi: 10.1128/aac.00113-12
Doublet, B., Praud, K., Bertrand, S., Collard, J. M., Weill, F. X., and Cloeckaert, A. (2008). Novel insertion sequence-and transposon-mediated genetic rearrangements in genomic island SGI1 of Salmonella enterica serovar Kentucky. Antimicrob. Agents Chemother. 52, 3745–3754. doi: 10.1128/aac.00525-08
Escudero, J. A., Loot, C., Parissi, V., Nivina, A., Bouchier, C., and Mazel, D. (2016). Unmasking the ancestral activity of integron integrases reveals a smooth evolutionary transition during functional innovation. Nat. Commun. 7:10937. doi: 10.1038/ncomms10937
Frank, J. A., Reich, C. I., Sharma, S., Weisbaum, J. S., Wilson, B. A., and Olsen, G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74, 2461–2470. doi: 10.1128/aem.02272-07
Guerout, A.-M., Iqbal, N., Mine, N., Ducos-Galand, M., Van Melderen, L., and Mazel, D. (2013). Characterization of the phd-doc and ccd toxin-antitoxin cassettes from Vibrio superintegrons. J. Bacteriol. 195, 2270–2283. doi: 10.1128/jb.01389-12
Harel, J., Lapointe, H., Fallara, A., Lortie, L. A., Bigras-Poulin, M., Larivière, S., et al. (1991). Detection of genes for fimbrial antigens and enterotoxins associated with Escherichia coli serogroups isolated from pigs with diarrhea. J. Clin. Microbiol. 29, 745–752.
Huang, Y. W., Hu, R. M., Lin, Y. T., Huang, H. H., and Yang, T. C. (2015). The contribution of class 1 integron to antimicrobial resistance in Stenotrophomonas maltophilia. Microb. Drug Resist. 21, 90–96. doi: 10.1089/mdr.2014.0072
Loot, C., Ducos-Galand, M., Escudero, J. A., Bouvier, M., and Mazel, D. (2012). Replicative resolution of integron cassette insertion. Nucleic Acids Res. 40, 8361–8370. doi: 10.1093/nar/gks620
Lupp, C., Hancock, R. E. W., and Ruby, E. G. (2002). The Vibrio fischeri sapABCDF locus is required for normal growth, both in culture and in symbiosis. Arch. Microbiol. 179, 57–65. doi: 10.1007/s00203-002-0502-7
MacDonald, D., Demarre, G., Bouvier, M., Mazel, D., and Gopaul, D. N. (2006). Structural basis for broad DNA-specificity in integron recombination. Nature 440, 1157–1162. doi: 10.1038/nature04643
Maynard, C., Fairbrother, J. M., Bekal, S., Sanschagrin, F., Levesque, R. C., Brousseau, R., et al. (2003). Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149: K91 isolates obstained over a 23-year period from pigs. Antimicrob. Agents Chemother. 47, 3214–3221. doi: 10.1128/AAC.47.10.3214-3221.2003
Mazel, D. (2006). Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4, 608–620. doi: 10.1038/nrmicro1462
Mazel, D., Dychinco, B., Webb, V. A., and Davies, J. (2000). Antibiotic resistance in the EcoR collection: integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 44, 1568–1574. doi: 10.1128/AAC.44.6.1568-1574.2000
Naas, T., Mikami, Y., Imai, T., Poirel, L., and Nordmann, P. (2001). Characterization of In53, a class 1 plasmid-and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183, 235–249. doi: 10.1128/JB.183.1.235-249.2001
Nivina, A., Escudero, J. A., Vit, C., Mazel, D., and Loot, C. (2016). Efficiency of integron cassette insertion in correct orientation is ensureed by the interplay of the three unpaired features of attC recombination sites. Nucleic Acids Res. 44, 7792–7803. doi: 10.1093/nar/gkw646
Partridge, S. R., Tsafnat, G., Coiera, E., and Iredell, J. R. (2009). Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33, 757–784. doi: 10.1111/j.1574-6976.2009.00175.x
Recchia, G. D., and Sherratt, D. J. (2002). “Gene acquisition in bacteria by integron-mediated site-specific recombination,”in Mobile DNA II, eds N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (Washington, DC: ASM Press), 162–176.
Soler Bistué, A. J., Martín, F. A., Petroni, A., Faccone, D., Galas, M., Tolmasky, M. E., et al. (2006). Vibrio cholerae InV117, a class 1 integron harbouring aac(6’)-Ib and blaCTX-M-2, is linked to transposition genes. Antimicrob. Agents Chemother. 50, 1903–1907. doi: 10.1128/aac.50.5.1903-1907.2006
Sun, F., Zhou, D., Sun, Q., Luo, W., Tong, Y., Zhang, D., et al. (2016). Genetic characterization of two fully sequenced multi- drug resistant plasmids pP10164-2 and pP10164-3from Leclercia adecarboxylata. Sci. Rep. 23:33982. doi: 10.1038/srep33982
Szekeres, S., Dauti, M., Wilde, C., Mazel, D., and Rowe-Magnus, D. A. (2007). Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol. Microbiol. 63, 1588–1605. doi: 10.1111/j.1365-2958.2007.05613.x
Tolmasky, M. E., and Crosa, J. H. (1993). Genetic organization of antibiotic resistance genes (aac(6’)-Ib, aadA, and oxa9) in the multiresistance transposon Tn1331. Plasmid 29, 31–40. doi: 10.1006/plas.1993.1004
Villa, L., Visca, P., Tosini, F., Pezzella, C., and Carattoli, A. (2002). Composite integron array generated by insertion of an ORF341-type integron within a Tn21-like element. Microb. Drug Resist. 8, 1–8. doi: 10.1089/10766290252913692
Walther-Rasmussen, J., and Høiby, N. (2006). OXA-type carbapenemases. J. Antimicrob. Chemother. 57, 373–383. doi: 10.1093/jac/dki482
Wang, D., Hou, W., Chen, J., Mou, Y., Yang, L., Yang, L., et al. (2014). Characterization of the blaKPC-2 and blaKPC-3 genes and the novel blaKPC-15 gene in Klebsiella pneumoniae. J. Med. Microbiol. 63, 981–987. doi: 10.1099/jmm.0.073841-0
Wang, M., Tran, J. H., Jacoby, G. A., Zhang, Y., Wang, F., and Hooper, D. C. (2003). Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47, 2242–2248. doi: 10.1128/AAC.47.7.2242-2248.2003
Wendel, A. F., Ressina, S., Kolbe-Busch, S., Pfeffer, K. P., and MacKenzie, C. R. (2016). Species diversity of environmental GIM-1-producing bacteria collected during a long-term outbreak. Appl. Environ. Microbol. 82, 3605–3610. doi: 10.1128/AEM.00424-16
Keywords: class 1 integron, In1085, In1086, evolutionary inferences, gene cassettes
Citation: Wang D, Hou W, Chen J, Yang L, Liu Z, Yin Z, Feng J and Zhou D (2017) Characterization of Novel Integrons, In1085 and In1086, and the Surrounding Genes in Plasmids from Enterobacteriaceae, and the Role for attCaadA16 Structural Features during attI1 × attC Integration. Front. Microbiol. 8:1003. doi: 10.3389/fmicb.2017.01003
Received: 03 March 2017; Accepted: 19 May 2017;
Published: 02 June 2017.
Edited by:
Satoru Suzuki, Ehime University, JapanReviewed by:
Juan Wang, University College Dublin, IrelandCopyright © 2017 Wang, Hou, Chen, Yang, Liu, Yin, Feng and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongguo Wang, d2RndHpzQDE2My5jb20= Dongsheng Zhou, ZG9uZ3NoZW5nemhvdTE5NzdAZ21haWwuY29t
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.