- 1School of Life Science, Peking University, Beijing, China
- 2Central Laboratory, Peking University School of Stomatology, Beijing, China
- 3Clovis Community College, Fresno, CA, United States
The status of intestinal microbiota is a determinant of host health. However, the alteration of the gut microbiota caused by the innate immune response to virus infection is unclear. Caenorhabditis elegans and its natural virus Orsay provide an excellent model of host–virus interactions. We evaluated the intestinal microbial community complexity of the wild-type N2 and the innate immunity-deficient mutant rde-1 (ne219) strains of C. elegans upon Orsay virus infection. The gut microbiota diversity was decreased in rde-1 (ne219) mutant animals, and a large number of genes were associated with the difference between infected and uninfected rde-1 (ne219) mutant animals. Therefore, this study provides the first evaluation of the alterations caused by Orsay virus on intestinal microbiota in wildtype and innate immunity-deficient animals using C. elegans as the model species. Our findings indicate that virus infection may alters the microbiome in animals with defective immune response.
Introduction
The Orsay virus was discovered in 2011 in Caenorhabditis elegans (C. elegans) nematodes in rotting fruit in Orsay, France. It is the only identified virus capable of naturally infecting C. elegans (Felix et al., 2011). Orsay virus has a bipartite positive-sense RNA genome and a similar genomic organization to nodaviruses, but differs in the presence of an additional open reading frame (ORF) in the RNA2 segment (Felix et al., 2011; Guo et al., 2014). Orsay virus-infected C. elegans display intestinal abnormalities, such as extensive convolutions of the apical intestinal border (Felix et al., 2011). The viral particles are mainly localized to intestinal cells (Franz et al., 2014). Remarkably, Orsay virus infection has little effect on the animal, which continues moving, eating, and producing progeny, albeit at a lower rate. Addition of dead, infected nematodes to healthy nematodes results in transmission of infection, and the presence of viral RNA in the somatic gonad suggests that Orsay virus is transmitted horizontally (Felix et al., 2011). Orsay virus is a useful model for studying host–virus interactions due to its small size, similarity to nodaviruses, tropism to intestinal cells of C. elegans, and its lack of infectivity for humans.
Caenorhabditis elegans is a bacterivore that ingests microbes via its pharynx, in which the process of digestion begins (Avery and Shtonda, 2003). C. elegans has a simple body plan with the intestine as the major body cavity open to the outside environment, so that it can directly sample the environmental microbial community (Balla and Troemel, 2013). Although C. elegans is a well-characterized model organism, little is known about its natural history, particularly its interactions with microbes (Petersen et al., 2015; Berg et al., 2016). Until recently, the intestinal microbiota of C. elegans has been characterized in the laboratory by examining worms from diverse natural soil environments. The results showed that worms harbor diverse gut microbial communities comprising 830 genus-level OTUs, 32 of which were present in all worm populations. Enterobacteriaceae, Burkholderiaceae, Xanthomonadaceae, Pseudomonadaceae, as well as nine other bacterial families were characterized as the worm core gut microbiota (Berg et al., 2016).
The mammalian gut microbiota influences nutrient acquisition, homeostasis stabilization, pathogen colonization, immune responses, and pathogen eradication (Endt et al., 2010; Stecher et al., 2010; Karst, 2016). Accumulating evidence suggests that the normal intestinal flora of C. elegans plays a critical role in maintaining a stable symbiotic relationship and is essential for several host physiological processes (Félix and Duveau, 2012; Portal-Celhay et al., 2012; Montalvokatz et al., 2013; Shapira, 2016). In a previous study, the intestinal microbiota of C. elegans in natural soil was identified to the species level. Two isolates, Bacillus megaterium (B. megaterium) and Pseudomonas mendocina (P. mendocina), were found to confer resistance to the nematode pathogen Pseudomonas aeruginosa (P. aeruginosa). The protection offered by P. mendocina was p38-dependent (Montalvokatz et al., 2013). Another recent study focused on the change in the gut microbial community after Bacillus nematocida (B. nematocida) B16 infection (Niu et al., 2016). Thus, the potential diversity of host–symbiont interactions in the gut is immense.
Furthermore, C. elegans is an important genetic model system for research on aging, development, and host–pathogen interactions. With the recent identification of a natural virus of C. elegans, this host now also represents a model for investigating virus–host interactions, particularly innate antiviral mechanisms. In the absence of a natural virus, previous studies of virus–host responses in C. elegans involved infection of primary cultured cells with vesicular stomatitis virus (VSV) or replication of flock house virus (FHV) in whole animals (Lu et al., 2005; Schott et al., 2005; Wilkins et al., 2005). These studies reported that the RNA interference (RNAi) pathway is involved in antiviral defense in C. elegans and identified genes essential for the RNAi antiviral response.
Orsay virus readily infects laboratory C. elegans mutants defective in RNAi and yields higher levels of viral RNA and obvious disease symptoms, as compared to infection of the corresponding wild-type N2 strain, demonstrating an antiviral role for RNAi in worms (Felix et al., 2011). Immunity to Orsay infection requires the RNAi pathway, consistent with earlier studies (Guo et al., 2012, 2013b; Ashe et al., 2013, 2015). Upon infection of C. elegans by Orsay virus, DRH-1 (Dicer-related RNA helicase-1) recruits Dicer-1 and the dsRNA-binding protein RDE-4 (RNA interference-deficient 4) to recognize the viral dsRNA replication intermediate. Dicer-1, with the aid of its partner RDE-4, cleaves the viral genome into 23 nt viral siRNA duplexes with a 2 nt 3′ overhang (Tabara et al., 2002; Schott et al., 2005; Wilkins et al., 2005; Ding and Voinnet, 2007; Lu et al., 2009; Ashe et al., 2013; Guo et al., 2013a). Duplex viRNAs lose one strand to produce primary viral siRNAs during incorporation into Argonaute protein RDE-1 (RNA interference-deficient 1). Subsequently, primary viRNAs are loaded into RDE-1 containing RISC complexes, which recruit the RNA-dependent RNA polymerase RRF-1 to the viral genome to synthesize secondary viral siRNA (Tabara et al., 2002; Yigit et al., 2006; Sijen et al., 2007; Steiner et al., 2009; Guo et al., 2013b). Secondary viRNAs are 22 nt single-stranded RNAs with G as the 5′-terminal, which are often referred to as 22G RNAs. These secondary siRNAs are then recruited by secondary Argonaute proteins, such as SAGO-2, and act to silence viral transcripts or inhibit virus replication (Aoki et al., 2007; Pak and Fire, 2007; Ding, 2010; Ding and Lu, 2011).
In addition to immune regulation, whether the intestinal microflora is involved in synergistic resistance to virus infection is unclear. The genes involved in the interaction with the intestinal microflora following virus infection virus remains unknown. In this study, we investigated the change in the intestinal microbiota community and the expression of related genes during the antiviral response of C. elegans to Orsay virus using N2 and rde-1 (ne219) mutants.
Materials and Methods
Genetics
The Bristol isolate of C. elegans, N2 strain and the RNAi deficient mutant WM27 rde-1 (ne219) was used in this study. All worm strains were maintained on nematode growth medium (NGM) plates at 20°C and seeded with the Escherichia coli (E. coli) bacteria strain OP50 unless otherwise indicated (Brenner, 1974). Serial passage of both C. elegans N2 and rde-1 (ne219) strain was conducted by transferring one square centimeter of agar with mixed stage animals onto a fresh OP50 seeded NGM plate.
Synchronization of Worm Cultures
Eggs were obtained by incubating freshly starved larvae on new 90 mm diameter seeded NGM plates with basic hypochlorite solution as previously described (Lewis and Fleming, 1995). Synchronous cultures were achieved by allowing the purified eggs to hatch overnight in M9 buffer without bacteria. The larval stage 1 (L1) animals were then washed by M9 buffer and diluted to 1000 worms per ml for Orsay virus infection.
Infectious Filtrate Preparation and Infection Assay
Orsay virus was maintained using the JU1580 isolate of C. elegans at room temperature and the virus filtrate was prepared following a protocol described previously (Felix et al., 2011). To prepare Orsay virus inoculum, infected JU1580 worms were washed off from fresh slightly starved 90 mm plates using M9 buffer, 3 ml per plate. After centrifugation twice at 21000 g for 5 min (4°C), the virus-containing supernatant was then filtered through a 0.22-μm filter unit (Merck Millipore, Germany). For all strains, 10 young adults were inoculated on 55 mm culture plates. At the same time, 40 μl of infectious filtrate was pipetted onto the bacterial lawn. The cultures were incubated at 20°C for 4 days. Infections were performed in four biological replicates.
DNA and RNA Extraction
Four days after infection, all worms were collected in M9 and then divided into two parallels. The worms were treated based on the method described in the literatures (Alegado and Tan, 2008; Niu et al., 2016). The worms were surface sterilized by washes with 100 mM levamisole and incubated in M9 buffer containing 100 mM levamisole and 100 mg/ml gentamicin. Levamisole treatment has been demonstrated to inhibit pharyngeal pumping and defecation. The treatment helps to prevent gentamicin entering into the intestinal lumen and the release of luminal bacteria. After incubation, the adult worms were washed with a levamisole solution to remove the gentamicin and homogenized with M9 containing 1% Triton X-100 to recover bacteria within the worm intestine.
DNA was extracted using the DNeasy® Blood&Tissue Kit (QIAGEN, Germany) according to the manufacturer’s instructions. The genomic DNA was evaluated with a NanoDrop spectrophotometer (Thermo Fisher Scientific, United States), using an A260/A280 ratio between 1.8 and 2.0 as a criterion for quality control and agarose gel electrophoresis, respectively. DNA samples were stored at -20°C until use. Total RNA was prepared using Invitrogen TRIzol (Thermo Fisher Scientific, United States), by following the manufacturer’s instructions.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
cDNA was generated from 1 μg total RNA applying random primers using Superscript III (Thermo Fisher Scientific, United States), and cDNA was diluted to 1:100 for qRT-PCR analysis. qRT-PCR was performed using SYBR Green realtime PCR master Mix (TOYOBO, Japan). The amplification was performed on a CFX96 TouchTM Deep Well Real-Time PCR Detection System (Bio-Rad Laboratories, Inc., United States). Each sample was normalized to ama-1, and then viral RNA1 (primers GW194: 59 ACC TCA CAA CTG CCA TCT ACA and GW195: 59 GAC GCT TCC AAG ATT GGT ATT GGT) levels were compared to those uninfected animals as negative controls.
Single Molecular Fluorescent In Situ Hybridization Assay
Accumulation of Orsay virus and the intestinal cell infection phenotype in N2 and rde-1 (ne219) mutant animals were detected by single molecular fluorescent in situ hybridization (smFISH), which was performed as previously described (Raj et al., 2008; Raj and Tyagi, 2010; Ji and van Oudenaarden, 2012; Franz et al., 2014). We used the set of custom StellarisTM Molecule FISH Probes (Biosearch Technologies, Inc., United States) labeled with Quasars® 670 Dye for the Orsay virus RNA1 molecule in this experiment. The oligonucleotide sequences were reported by Franz et al. (2014). Color images through an Olympus BX51 DIC microscope were acquired by a Canon T1 DSLR connected to an LM Microscope adapter.
DNA and cDNA Library Construction and Sequencing
DNA libraries were constructed according to the standard protocol provided by Illumina. Quality control of libraries were performed before sequencing. Libraries were sequenced using the Illumina Miseq instrument (Illumina, United States) with 2 × 300 base pairs (bp) paired-end (PE) sequencing.
mRNA was concentrated from total RNA and cDNA libraries were constructed using mRNA. After validation and the quality assessment of each library on gel electrophoresis and bioanalyzer (Agilent Technologies, United States), the libraries were sequenced on the Illumina Hiseq platform (Illumina, United States) with 2 × 150bp paired-end (PE) sequencing.
DNA Library Sequencing Data Processing
The raw data generated by sequencing were analyzed using the pipeline tools in QIIME (Caporaso et al., 2010). Sequences were demultiplexed using customized Perl scripts, based on the presence of the unique barcodes assigned to each sample.
Data were de-noised by removing reads with average quality values <30. After removing both the barcodes and primer sequences, the filter-passed reads were clustered into Operational Taxonomic Units (OTUs) at a 97% similarity cutoff using the de novo OTU-picking strategy. Taxonomies were assigned according to the Greengenes database.
Following construction of the OTU table and phylogenetic tree, microbial richness (Chao1, observed species, and phylogenetic diversity (PD)), diversity (Shannon Index and Simpson Index), and evenness (Equitability, Simpson diversity index) estimators were calculated. Besides, microbial structure was evaluated by analyzing weighted UniFrac distance, unweighted UniFrac distance, and Bray–Curtis distance metrics (Lozupone and Knight, 2005). The relative abundances of microbial taxa at the phylum, class, order, family, genus, and species levels were calculated and compared.
Data Processing of RNA Sequencing
Raw sequencing reads were trimmed using the following criteria: (1) reads containing adapters were removed; (2) reads containing greater than 10% N bases were removed; and (3) reads containing greater than 50% low-quality reads (Q ≤ 5) were removed. After which, the resulting filtered reads were mapped and annotated. Gene expression was quantified as Reads Per Kilo bases per Million reads (RPKM) (Mortazavi et al., 2008). DESeq (Anders and Huber, 2010) was used to perform statistical analyses of the gene expression profiles.
Statistical Analysis
The data were further analyzed with the following statistical methods. (1) Differences in alpha- and beta-diversity were evaluated using the non-parametric two-sample t-tests with 1000 permutations. (2) Differences in relative abundances of individual OTUs and microbial taxa (phylum, class, order, family, genus, and species) were analyzed using LEfSe (Segata, 2011) with default parameters [P < 0.05, Linear discriminant analysis (LDA) score > 2.0].
Results
Orsay Virus Replicates in C. elegans Intestinal Cells
To monitor the composition of the intestinal microbiota of C. elegans upon Orsay virus infection, we analyzed 16S ribosomal DNA library data of infected and non-infected worms. The laboratory reference strain N2 and an rde-1 (ne219) mutant deficient in the primary Argonaute protein RDE-1 were used in this study.
Quantitative real-time polymerase chain reaction (qRT-PCR) and single molecular fluorescence in situ hybridization (smFISH) were performed to verify Orsay virus infection of C. elegans. rde-1 (ne219) mutant animals were more sensitive to infection with Orsay virus than were N2 animals, as measured by viral load (Figure 1A). Previous work suggested that C. elegans intestinal cells are structurally altered by Orsay virus infection and the viral particles are mainly localized to intestinal cells (Felix et al., 2011; Franz et al., 2014). Using smFISH, the Orsay virus RNA1 segment was detected in intestinal cells of adult animals. Orsay virus-infected rde-1 (ne219) mutant (rde-1_OrV) animals showed a much greater proportion of infected intestinal cells compared to N2 animals (Figure 1B).
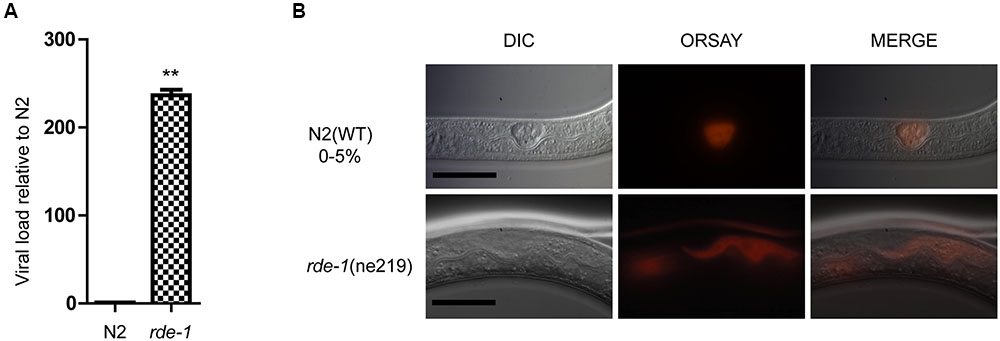
FIGURE 1. Validation of Orsay virus replications in N2 and rde-1 (ne219) mutant. (A) qRT-PCR analysis of viral loads after 4 days Orsay virus infection in N2 and rde-1 (ne219). rde-1 (ne219) mutant animals showed significantly higher levels of Orsay virus RNA compared to N2 animals do (P < 0.01, t-test). (B) Fluorescence in situ hybridization (FISH) staining of Orsay viral RNA1 in Caenorhabditis elegans N2 and rde-1 (ne219) mutant. Viral RNA1 were detected using Quasar® 670 labeled RNA FISH Probe corresponding to the RNA1 of Orsay virus. Red indicates the Orsay viral RNA and all scale bars represent 20 μm.
RDE-1 Deficiency Is Associated with Decreased Microbial Diversity
Orsay-infected N2 (N2_OrV) animals showed no significant difference in intestinal microbial diversity compared with uninfected animals (Figures 2, 3, P > 0.05). According to the weighted UniFrac distance measurements, a clear separation was found between uninfected N2 (N2_mock) animals and uninfected rde-1 (ne219) mutant (rde-1_mock) animals. The weighted UniFrac distance in N2_mock group was greater than in rde-1_mock group, and this demonstrated that the microbial diversity in N2 animals was significantly greater than in rde-1 (ne219) mutant animals. The variation of microbial characteristics was similar for rde-1 (ne219) mutant animals but significantly greater for N2 animals (Figure 2 and Supplementary Table S1). Uninfected N2 animals showed the greatest bacterial community diversity, which was considered to be an indicator of improved health, whereas the rde-1 mutant tended to host similar microbial communities.
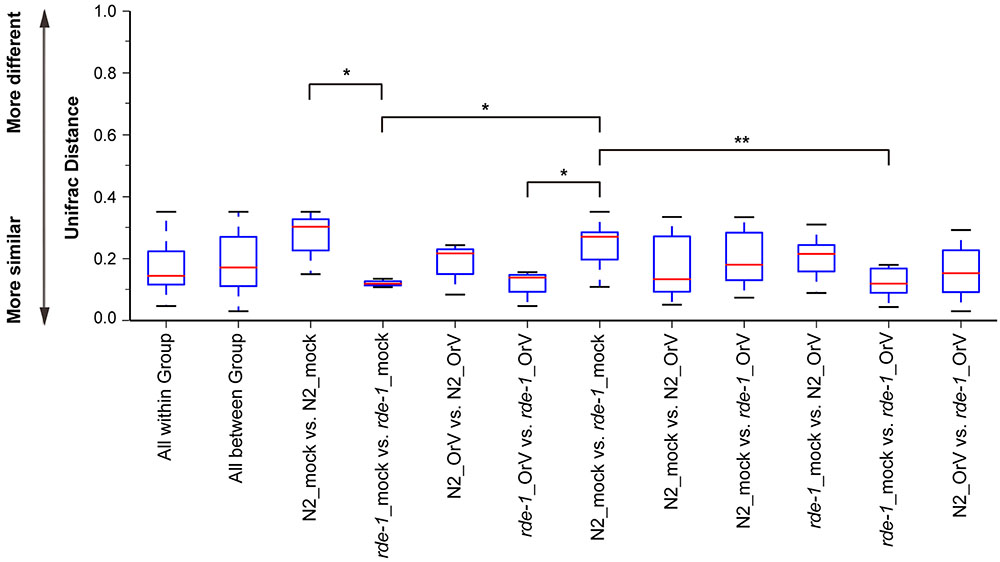
FIGURE 2. Changes of the microbiota diversity due to Orsay infection. The average weighted UniFrac distance values (the beta diversities) of uninfected N2 (N2_mock), Orsay virus-infected N2 (N2_OrV), uninfected rde-1 (ne219) mutants (rde-1_mock) and Orsay virus-infected rde-1 (ne219) (rde-1_OrV). ∗P < 0.05, ∗∗P < 0.01 by one-tailed t-test.
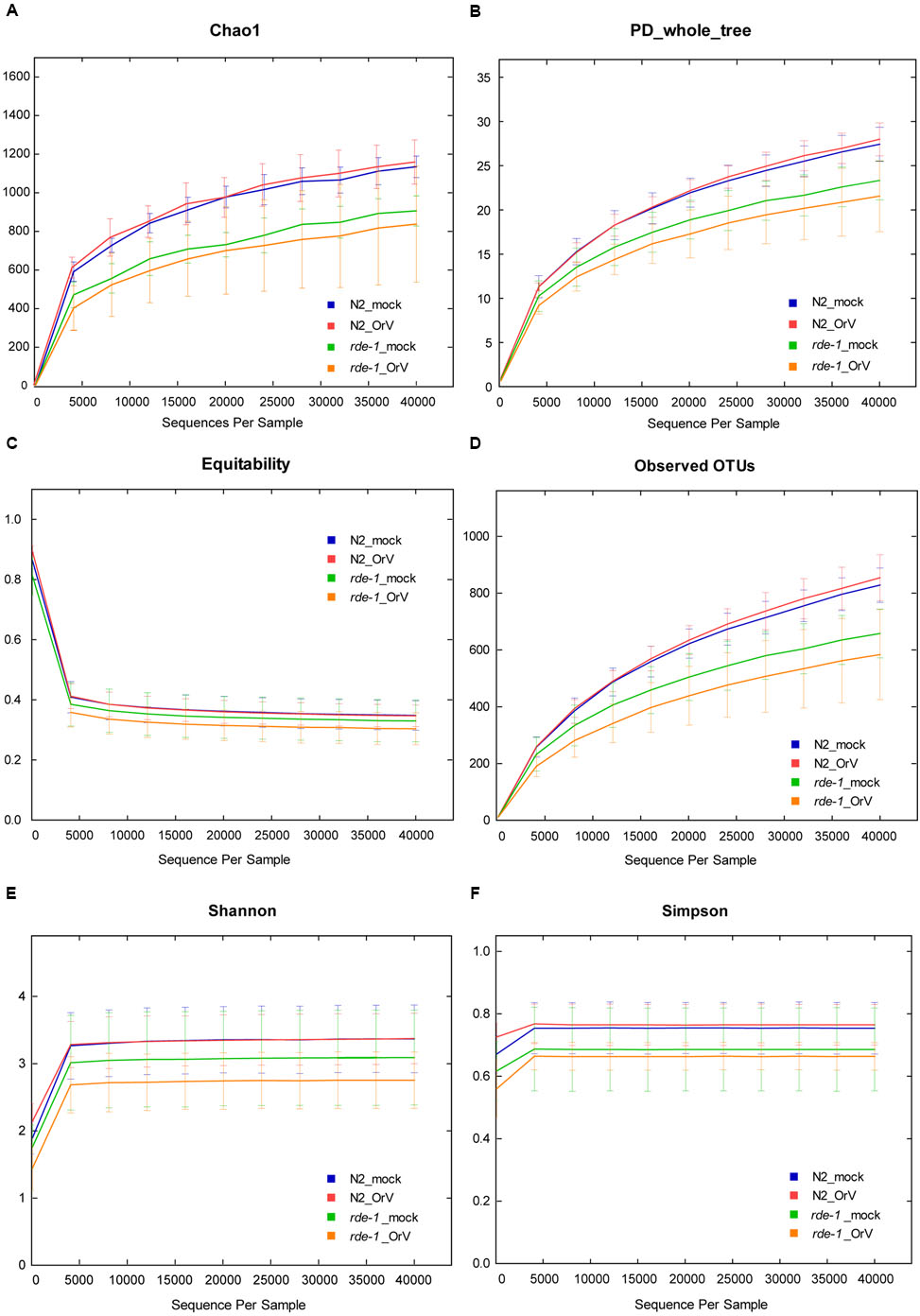
FIGURE 3. Calculation of alpha diversity values for comparison of the total microbial diversity of uninfected and Orsay virus-infected groups of N2 and rde-1 (ne219) mutant. (A) The estimated OTU numbers (Chao1) of N2 were greater than that of rde-1 (ne219) mutants, which especially decreased in Orsay virus-infected rde-1 (ne219) mutants. (B) Phylogenetic diversity (PD) measures of community diversity. (C) Equitability index of the microbiota data for N2 and rde-1 (ne219) mutants. (D) The numbers of observed OTUs decreased in both uninfected and Orsay virus-infected rde-1 (ne219) mutants. (E) Microbial community diversity analysis (Shannon index) showed that the N2 community exhibited greater diversity. (F) Simpson index measures of community diversity.
We analyzed microbial diversity (alpha diversity) to determine whether RDE-1 deficiency and Orsay virus infection were associated with the intestinal microflora of C. elegans. The microbial diversity of N2 animals was not significantly affected by Orsay virus infection, and RDE-1 deficiency was associated with decreased microbial diversity, which decreased further after virus infection. OTU richness did not differ significantly between N2_mock and N2_OrV animals, but was lower in rde-1 (ne219) mutant animals compared with N2 animals. Orsay virus-infected rde-1 (ne219) mutant animals had lower OTU richness than uninfected animals (Figures 3A,D). The Shannon diversity indices showed that rde-1 (ne219) mutant animals tended to harbor fewer diverse bacterial communities than did N2 animals, and that this decrease in diversity was exacerbated by Orsay virus infection (Figure 3E). The equitability index and Simpson index (measures of phylogenetic diversity, PD) indicated that the microbial communities of uninfected N2 animals exhibited the greatest diversity, and those of Orsay virus-infected rde-1 (ne219) mutant animals exhibited the lowest (Figures 3B,C,F). Therefore, N2 animals exhibited higher levels of biodiversity and unevenness estimations compared to RDE-1-deficient animals. This indicated that RNAi plays a role in maintaining microbiomes during virus infection and the defective RNAi response in rde-1 (ne219) animals made the animals unable to maintain their microbiome when challenged with virus infection.
rde-1 (ne219) Mutant and N2 Animals Harbor Distinct Bacterial Communities
To evaluate the response of the gut microbiome to RDE-1 deficiency and virus infection, we compared the microbial clades of infected and uninfected N2 and rde-1 mutant animals using LEfSe. The bacterial compositions differed between uninfected N2 and rde-1 (ne219) mutant animals. In the latter, the gut microbiome was significantly enriched in Actinobacteria, including Leucobacter and Pseudoclavibacter; Proteobacteria, including Acinetobacter and Burkholderia; and Alphaproteobacteria, specifically, Ochrobactrum (Figure 4B and Supplementary Figure S1A). In contrast, the intestinal microbiome of uninfected N2 animals was enriched in Bacteroidetes, including Myroides and Sphingobacterium (Figure 4B and Supplementary Figure S1A).
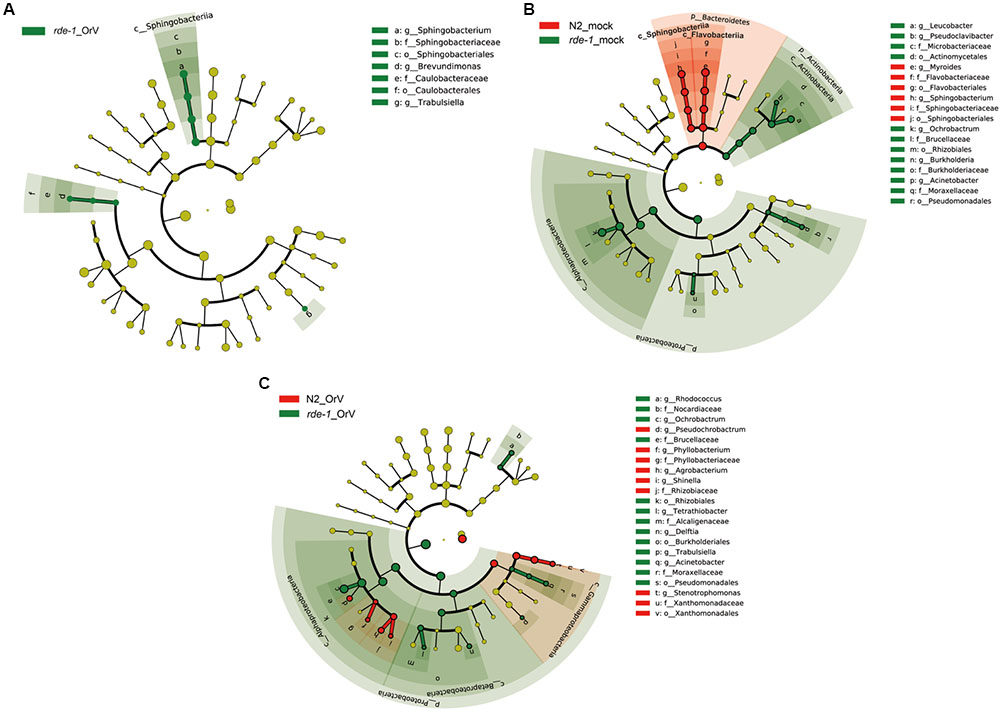
FIGURE 4. rde-1 (ne219) mutant and N2 animals harbor distinct bacterial communities. (A) LEfSe analysis of microbiomes between uninfected and Orsay virus-infected group in rde-1 (ne219) mutants. The taxa that were found at a significantly higher relative abundance in Orsay virus-infected group (green) relative to the uninfected samples are highlighted. (B) LEfSe analysis of microbiomes between uninfected N2 and rde-1 (ne219) mutants. (C) LEfSe analysis of microbiomes between Orsay virus-infected N2 and rde-1 (ne219) mutants. The taxa that were found at a significantly higher relative abundance in N2 (red) and rde-1 (ne219) (green) are highlighted. For the cladogram, yellow circles represent non-significant microbial clades.
To identify the microbial taxa that respond to Orsay virus infection, we examined the microbiomes of infected and uninfected rde-1 (ne219) mutant animals. Orsay virus infection resulted in marked enrichment in the genera Sphingobacterium, Brevundimonas, and Trabulsiella (Figure 4A and Supplementary Figure S1B). However, compared with Orsay virus-infected N2 animals, the relative abundances of Rhodococcus, Ochrobactrum, Tetrathiobacter, Delftia, Trabulsiella, and Acinetobacter were higher in rde-1_OrV animals (Figure 4C and Supplementary Figure S1C), while the relative abundances of Pseudochrobactrum, Phyllobacterium, Agrobacterium, Shinella, and Stenotrophomonas were higher in N2_OrV animals (Figure 4C and Supplementary Figure S1C). These results suggest that Orsay virus infection and RDE-1 deficiency exert marked effects on the composition of the gut microbiome.
Regulation of Viral Response Genes by RDE-1
To assess the relationship between the intestinal microbiome and gene expression in C. elegans after Orsay virus infection, we compared the mRNA profiles of mock and Orsay virus-infected N2 and rde-1 (ne219) animals.
The cluster analysis indicated that a large set of genes was associated with the difference between infected and uninfected rde-1 (ne219) mutant animals (Figure 5A). We searched for genes that were significantly differently expressed (fold-change > 2 and P < 1 × 10-4, Student’s t-test) between uninfected and infected rde-1 (ne219) animals. A total of 144 genes were specifically induced or repressed by Orsay virus infection (Figure 5A and Supplementary Table S2). These 144 genes showed markedly different expression levels; 126 were up-regulated and 18 down-regulated upon infection (Figure 5C and Supplementary Table S2). However, only 17 larger, differently expressed genes were found between uninfected and infected N2 animals; 14 genes were up-regulated and three down-regulated upon infection (Figure 5C and Supplementary Table S3). Gene ontology analysis showed that terms involved in innate immune response or defense response were enriched in the list of upregulated genes in infected compared with uninfected rde-1 (ne219) animals (Figure 5B and Supplementary Figure S2). In contrast, genes downregulated in rde-1 (ne219) animals were mainly involved with lipid metabolism, lipid transport and fatty acid metabolism (Figure 5B and Supplementary Figure S3).
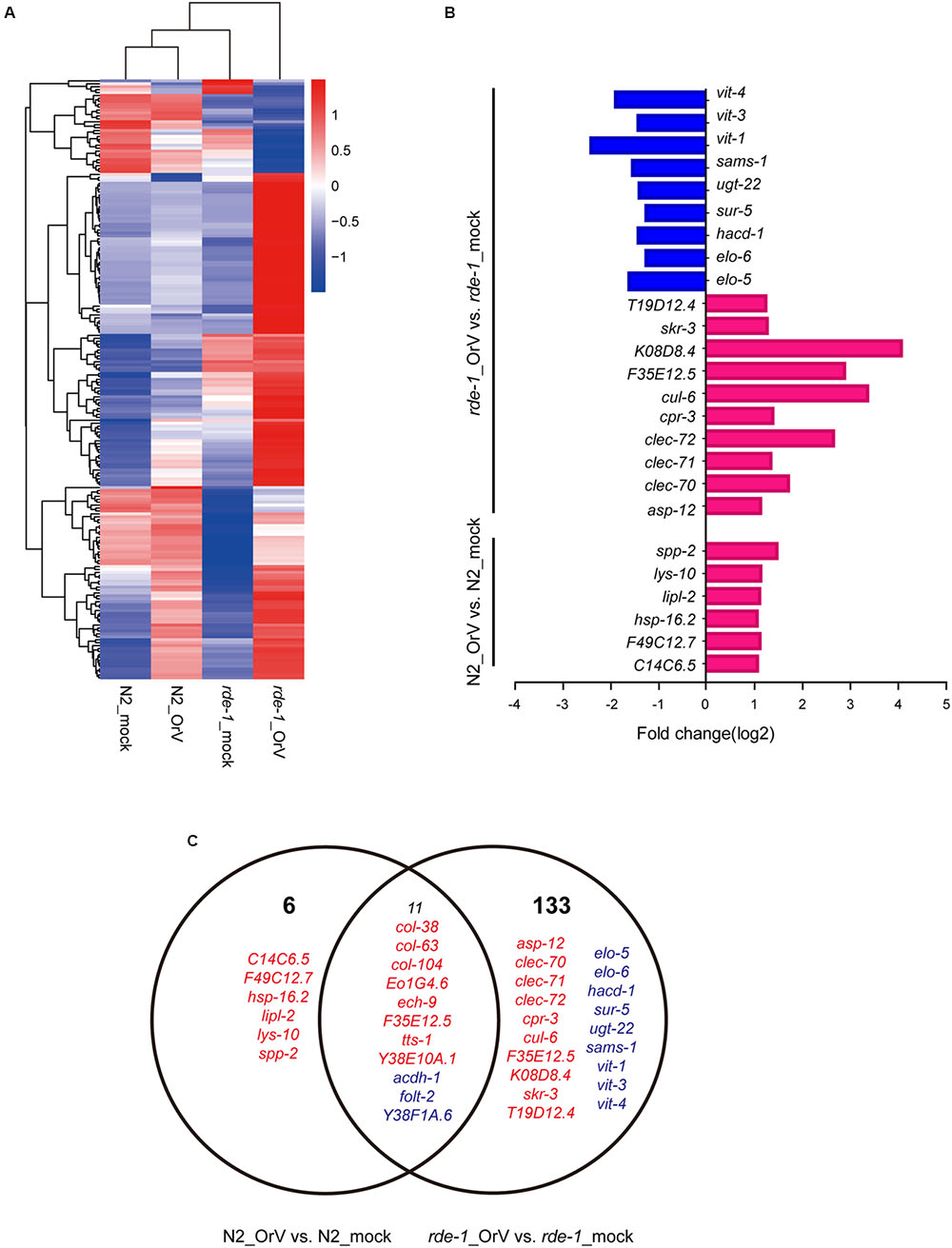
FIGURE 5. Changes in gene expression upon viral infection. (A) The heatmap shows all the genes that changed by more than twofold after infection (t-test < 0.05). Each row shows a different gene. Blue, white, and red indicate expression levels of low, medium, and high gene values, respectively. (B) Gene ontology (GO) and pathway enrichment analysis of predicted targets by differentially expressed mRNAs. Red indicates increased gene expression while blue indicates decreased. (C) The Venn diagram displaying the total number of genes significantly up-and downregulated after Orsay virus-infection in N2 and rde-1 (ne219) mutant samples. Red indicates increased gene expressions, while blue indicates decreased.
Discussion
In this study, we evaluated the complexity of intestinal microbial communities in wildtype and innate immunity-deficient mutant C. elegans upon Orsay virus infection. Gut microbiome diversity was decreased in rde-1 (ne219) mutant animals, and several genes were associated with differences between infected and uninfected rde-1 (ne219) mutant animals. Our findings indicate that the RNAi pathway may play a role in maintaining the microbiome during virus infection.
Orsay virus infection of N2 animals did not significantly influence microbial diversity, while rde-1 deficiency was associated with decreased microbial diversity, which was further reduced by Orsay virus infection. The gut microbiota status is a determining factor of host health. The intestinal microbial diversity of the rde-1 (ne219) mutant was further reduced, suggesting that the mutant animals were unstable and easily disturbed by external factors. Our results are consistent with a previous report of microbial enrichment and complexity in healthy hosts (Niu et al., 2016).
The complex intestinal microbial community is regulated by environmental, diet and host genetic factors (Kau et al., 2011; Sommer and Bäckhed, 2013). The composition, density, and complexity of the intestinal microbiota influence pathogen colonization, immune responses, and pathogen clearance (Endt et al., 2010; Stecher et al., 2010; Portal-Celhay et al., 2012; Cabreiro and Gems, 2013). In this study, we compared the intestinal microbiome in the presence and absence of Orsay virus using LEfSe. Compared to the control group, infected animals had a significantly greater number of OTUs related to the genera Sphingobacterium, Brevundimonas, and Trabulsiella. However, compared with uninfected rde-1 (ne219) mutant animals, Sphingobacterium was enriched in uninfected N2 animals, suggesting that Orsay virus infection and genetic factors influence the microbial community. Sphingobacterium is characterized by high concentrations of sphingophospholipid components (Yabuuchi et al., 1983). Sphingobacterium is abundant in cowpea root nodules and significantly influenced by soil type and plant genotype (Leite et al., 2016). Trabulsiella strains have been found in the termite gut and may play roles in carbohydrate metabolism and aflatoxin degradation. Furthermore, Trabulsiella strains are adapted to their termite host, such as by expressing genes encoding the components of a type VI secretion system (T6SS), which facilitates bacterial competition, colonization, and survival within the host (Chou et al., 2007; Sapountzis et al., 2015; Suman et al., 2016).
To investigate the changes in the intestinal microbiota community and the expression of related genes in C. elegans, we analyzed mRNA profiles of mock and Orsay virus-infected N2 and rde-1 (ne219) mutant animals. Several genes were specifically induced or repressed by Orsay virus infection. acdh-1, a “dietary sensor” (Kawasaki et al., 2013; MacNeil et al., 2013), was significantly down-regulated in Orsay virus-infected N2 animals, and further decreased in rde-1 (ne219) mutant animals. This indicated that stress response genes were stimulated by virus infection and possibly associated with the immune response. In the presence of Orsay virus, genes related to the innate immune response, such as a cluster of C-type lectin (CLEC), were significantly up-regulated in the rde-1 (ne219) mutant animals. CLEC, which contribute to immune specificity in both vertebrates and invertebrates, maintain gut homeostasis by symbiotic microbiome offsets of the gut immunity in mosquitoes (Schulenburg et al., 2008; Pang et al., 2016). The gut microbiome of mosquitoes could induce the expression of CLEC, which coat the bacterial surface to counteract the activity of antimicrobial peptides (AMPs) (Pang et al., 2016). The genome-wide associations of CLEC gene clusters suggest that CLEC genes are associated with human gut microbiome composition and function (Bonder et al., 2016). The up-regulation of clec-70, clec-71, and clec-72 in infected rde-1 (ne219) mutant animals indicated that the CLEC genes may contribute to intestinal microbiome alternation associate with virus infection in C. elegans with defective immune response. Moreover, multiple genes were down-regulated in rde-1 (ne219) mutant animals, which highlights the role of sensor molecules in lipid metabolism, lipid transport and fatty acid metabolism, particularly vitellogenins (VITs). Lipid metabolism is an important pathway associated with aging (Nakamura et al., 1999; Gubert et al., 2016). Besides providing energy for developing embryos, VITs are involved in the stress response in diverse organisms (Fischer et al., 2012). VITs were found to act as acute- phase-proteins and to possess bacterial binding and inhibiting activities in different fish-species (Zhang, 2011; Fischer et al., 2013). VITs increase the stress resistance of C. elegans after Photorhabdus luminescens infection in a manner dependent on the steroid-signaling pathway (Fischer et al., 2013). The down-regulationof vit-1, vit-2, and vit-4 in Orsay virus-infected rde-1 (ne219) mutant animals indicates that VITs may be involved in the interactions among host genetic factors, virus infection and the microbiome.
The intestinal microbiota plays a stimulatory role in viral infection (Karst, 2016). However, the effect on the gut microbiota of virus infection remains unclear, particularly in C. elegans. This study reports for the first time the effect of Orsay virus infection on the intestinal microbiome and expression of related genes of N2 and rde-1 (ne219) mutant C. elegans. Our results demonstrate that intestinal microbiota diversity may be associated with host genes during virus infection. Consistent with a previous report that host genotype affects the intestinal microbiota, our data may help to explain human diseases caused by different genotypes associated with a variety of intestinal microbiome. Furthermore, our results may lead to a better understanding of the mechanism and evolution of the mutualistic relationship between a host and its gut microbial community based on nematode molecular genetics and immunity.
Author Contributions
YG contributed to design all the experiments, bench work and draft the manuscript. ZX contributed to analyze data. SC contributed to smFISH experiment. FC contributed to design all the experiments and draft the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the Caenorhabditis Genetics Center (CGC) for the N2 and WM27 worm strains used in this study and Gina Maduro and Morris F. Maduro for their great help in providing information associated with smFISH. This work would not have been possible without the JU1580 worm strain and the Orsay virus provided by Marie-Anne Felix, Eric Miska, and Dave Wang.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00933/full#supplementary-material
FIGURE S1 | Differentially abundant microbial clades in N2 and rde-1 mutant animals upon Orsay virus infection. (A) Linear discriminant analysis (LDA) score of microbiomes between uninfected N2 and rde-1 mutant. (B) LDA score of microbiomes between Orsay virus-infected N2 and rde-1 mutant. Taxa that are found at a significantly higher relative abundance in N2 (red) and rde-1 (green) are highlighted. (C) LDA score of microbiomes between uninfected and Orsay virus-infected group in rde-1 mutant. Taxa that are found at a significantly higher relative abundance in Orsay viurs-infected group (green) relative to the uninfected samples are highlighted.
FIGURE S2 | Gene ontology (GO) and pathway enrichment analysis of differentially upregulated expression mRNAs in rde-1 mutant upon Orsay virus infection. The vertical axis shows the enriched GO term, and the horizontal axis represents the number of differentially expressed genes in the term. With “*” for significantly enriched GO term.
FIGURE S3 | Gene ontology (GO) and pathway enrichment analysis of differentially downregulated expression mRNAs in rde-1 mutant upon Orsay virus infection. The vertical axis shows the enriched GO term, and the horizontal axis represents the number of differentially expressed genes in the term. With “*” for significantly enriched GO term.
References
Alegado, R. A., and Tan, M. W. (2008). Resistance to antimicrobial peptides contributes to persistence of Salmonella typhimurium in the C. elegans intestine. Cell. Microbiol. 10, 1259–1273. doi: 10.1111/j.1462-5822.2008.01124.x
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Aoki, K., Moriguchi, H., Yoshioka, T., Okawa, K., and Tabara, H. (2007). In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 26, 5007–5019. doi: 10.1038/sj.emboj.7601910
Ashe, A., Bélicard, T., Pen, J. L., Sarkies, P., Frézal, L., Lehrbach, N. J., et al. (2013). A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. Elife 2:e00994. doi: 10.7554/eLife.00994
Ashe, A., Sarkies, P., Pen, J. L., Tanguy, M., and Miska, E. A. (2015). Antiviral RNA interference against orsay virus is neither systemic nor transgenerational in Caenorhabditis elegans. J. Virol. 89, 12035–12046. doi: 10.1128/JVI.03664-14
Avery, L., and Shtonda, B. B. (2003). Food transport in the C. elegans pharynx. J. Exp. Biol. 206, 2441–2457. doi: 10.1242/jeb.00433
Balla, K. M., and Troemel, E. R. (2013). Caenorhabditis elegans as a model for intracellular pathogen infection. Cell. Microbiol. 15, 1313–1322. doi: 10.1111/cmi.12152
Berg, M., Stenuit, B., Ho, J., Wang, A., Parke, C., Knight, M., et al. (2016). Assembly of the Caenorhabditis elegans gut microbiota from diverse soil microbial environments. ISME J. 10, 1998–2009. doi: 10.1038/ismej.2015.253
Bonder, M. J., Kurilshikov, A., Tigchelaar, E. F., Mujagic, Z., Imhann, F., Vila, A. V., et al. (2016). The effect of host genetics on the gut microbiome. Nat. Genet. 48, 1407–1412. doi: 10.1038/ng.3663
Cabreiro, F., and Gems, D. (2013). Worms need microbes too: microbiota, health and aging in Caenorhabditis elegans. EMBO Mol. Med. 5, 1300–1310. doi: 10.1002/emmm.201100972
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chou, J.-H., Chen, W.-M., Arun, A., and Young, C.-C. (2007). Trabulsiella odontotermitis sp. nov., isolated from the gut of the termite Odontotermes formosanus Shiraki. Int. J. Syst. Evol. Microbiol. 57, 696–700. doi: 10.1099/ijs.0.64632-0
Ding, S. W. (2010). RNA-based antiviral immunity. Nat. Rev. Immunol. 10, 632–644. doi: 10.1038/nri2824
Ding, S. W., and Lu, R. (2011). Virus-derived siRNAs and piRNAs in immunity and pathogenesis. Curr. Opin. Virol. 1, 533–544. doi: 10.1016/j.coviro.2011.10.028
Ding, S. W., and Voinnet, O. (2007). Antiviral immunity directed by small RNAs. Cell 130, 413–426. doi: 10.1016/j.cell.2007.07.039
Endt, K., Stecher, B., Chaffron, S., Slack, E., Tchitchek, N., Benecke, A., et al. (2010). The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog. 6:e1001097. doi: 10.1371/journal.ppat.1001097
Felix, M. A., Ashe, A., Piffaretti, J., Wu, G., Nuez, I., Belicard, T., et al. (2011). Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 9:e1000586. doi: 10.1371/journal.pbio.1000586
Félix, M. A., and Duveau, F. (2012). Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol. 10:59. doi: 10.1186/1741-7007-10-59
Fischer, M., Regitz, C., Kahl, M., Werthebach, M., Boll, M., and Wenzel, U. (2012). Phytoestrogens genistein and daidzein affect immunity in the nematode Caenorhabditis elegans via alterations of vitellogenin expression. Mol. Nutr. Food Res. 56, 957–965. doi: 10.1002/mnfr.201200006
Fischer, M., Regitz, C., Kull, R., Boll, M., and Wenzel, U. (2013). Vitellogenins increase stress resistance of Caenorhabditis elegans after Photorhabdus luminescens infection depending on the steroid-signaling pathway. Microbes Infect. 15, 569–578. doi: 10.1016/j.micinf.2013.05.002
Franz, C. J., Renshaw, H., Frezal, L., Jiang, Y., Félix, M.-A., and Wang, D. (2014). Orsay, Santeuil and Le Blanc viruses primarily infect intestinal cells in Caenorhabditis nematodes. Virology 448, 255–264. doi: 10.1016/j.virol.2013.09.024
Gubert, P., Puntel, B., Lehmen, T., Bornhorst, J., Avila, D. S., Aschner, M., et al. (2016). Reversible reprotoxic effects of manganese through DAF-16 transcription factor activation and vitellogenin downregulation in Caenorhabditis elegans. Life Sci. 151, 218–223. doi: 10.1016/j.lfs.2016.03.016
Guo, X., Li, W. X., and Lu, R. (2012). Silencing of host genes directed by virus-derived short interfering RNAs in Caenorhabditis elegans. J. Virol. 86, 11645–11653. doi: 10.1128/JVI.01501-12
Guo, X., Zhang, R., Wang, J., Ding, S. W., and Lu, R. (2013a). Homologous RIG-I-like helicase proteins direct RNAi-mediated antiviral immunity in C. elegans by distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 110, 16085–16090. doi: 10.1073/pnas.1307453110
Guo, X., Zhang, R., Wang, J., and Lu, R. (2013b). Antiviral RNA silencing initiated in the absence of RDE-4, a double-stranded RNA binding protein, in Caenorhabditis elegans. J. Virol. 87, 10721–10729. doi: 10.1128/JVI.01305-13
Guo, Y. R., Hryc, C. F., Jakana, J., Jiang, H., Wang, D., Chiu, W., et al. (2014). Crystal structure of a nematode-infecting virus. Proc. Natl. Acad. Sci. U.S.A. 111, 12781–12786. doi: 10.1073/pnas.1407122111
Ji, N., and van Oudenaarden, A. (2012). “Single molecule fluorescent in situ hybridization (smFISH) of C. elegans worms and embryos,” in Wormbook, ed. The C. elegans Research Community (Pasadena, CA: WormBook). doi: 10.1895/wormbook.1.153.1
Karst, S. M. (2016). The influence of commensal bacteria on infection with enteric viruses. Nat. Rev. Microbiol. 14, 197–204. doi: 10.1038/nrmicro.2015.25
Kau, A. L., Ahern, P. P., Griffin, N. W., Goodman, A. L., and Gordon, J. I. (2011). Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336. doi: 10.1038/nature10213
Kawasaki, I., Jeong, M. H., Yun, Y. J., Shin, Y. K., and Shim, Y. H. (2013). Cholesterol-responsive metabolic proteins are required for larval development in Caenorhabditis elegans. Mol. Cells 36, 410–416. doi: 10.1007/s10059-013-0170-2
Leite, J., Fischer, D., Rouws, L. F., Fernandes-Junior, P. I., Hofmann, A., Kublik, S., et al. (2016). Cowpea nodules harbor non-rhizobial bacterial communities that are shaped by soil type rather than plant genotype. Front. Plant Sci. 7:2064. doi: 10.3389/fpls.2016.02064
Lewis, J. A., and Fleming, J. T. (1995). Basic culture methods. Methods Cell Biol. 48, 3–29. doi: 10.1016/S0091-679X(08)61381-3
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Lu, R., Maduro, M., Li, F., Li, H. W., Broitmanmaduro, G., Li, W. X., et al. (2005). Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 436, 1040–1043. doi: 10.1038/nature03870
Lu, R., Yigit, E., Li, W. X., and Ding, S. W. (2009). An RIG-I-Like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans. PLoS Pathog. 5:e1000286. doi: 10.1371/journal.ppat.1000286
MacNeil, L. T., Watson, E., Arda, H. E., Zhu, L. J., Walhout, and Albertha, J. M. (2013). Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell 153, 240–252. doi: 10.1016/j.cell.2013.02.049
Montalvokatz, S., Huang, H., Appel, M. D., Berg, M., and Shapira, M. (2013). Association with soil bacteria enhances p38-dependent infection resistance in Caenorhabditis elegans. Infect. Immun. 81, 514–520. doi: 10.1128/IAI.00653-12
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. methods 5, 621–628. doi: 10.1038/nmeth.1226
Nakamura, A., Yasuda, K., Adachi, H., Sakurai, Y., Ishii, N., and Goto, S. (1999). Vitellogenin-6 is a major carbonylated protein in aged nematode, Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 264, 580–583. doi: 10.1006/bbrc.1999.1549
Niu, Q., Zhang, L., Zhang, K., Huang, X., Hui, F., Kan, Y., et al. (2016). Changes in intestinal microflora of Caenorhabditis elegans following Bacillus nematocida B16 infection. Sci. Rep. 6:20178. doi: 10.1038/srep20178
Pak, J., and Fire, A. (2007). Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315, 241–244. doi: 10.1126/science.1132839
Pang, X., Xiao, X., Liu, Y., Zhang, R., Liu, J., Liu, Q., et al. (2016). Mosquito C-type lectins maintain gut microbiome homeostasis. Nat. Microbiol. 1, 16023. doi: 10.1038/nmicrobiol.2016.23
Petersen, C., Dirksen, P., and Schulenburg, H. (2015). Why we need more ecology for genetic models such as C. elegans. Trends Genet. 31, 120–127. doi: 10.1016/j.tig.2014.12.001
Portal-Celhay, C., Bradley, E. R., and Blaser, M. J. (2012). Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans. BMC Microbiol. 12:49. doi: 10.1186/1471-2180-12-49
Raj, A., and Tyagi, S. (2010). Detection of individual endogenous RNA transcripts in situ using multiple singly labeled probes. Methods Enzymol. 472, 365–386. doi: 10.1016/S0076-6879(10)72004-8
Raj, A., van den Bogaard, P., Rifkin, S. A., van Oudenaarden, A., and Tyagi, S. (2008). Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879. doi: 10.1038/nmeth.1253
Sapountzis, P., Gruntjes, T., Otani, S., Estevez, J., Costa, R. R. D., Plunkett, G., et al. (2015). The Enterobacterium Trabulsiella odontotermitis presents novel adaptations related to its association with fungus-growing termites. Appl. Environ. Microbiol. 81, 6577–6588. doi: 10.1128/AEM.01844-15
Schott, D. H., Whelan, S. P., Hunter, C. P., and Sharp, P. A. (2005). An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 102, 18420–18424. doi: 10.1073/pnas.0507123102
Schulenburg, H., Hoeppner, M. P., Iii, J. W., and Bornberg-Bauer, E. (2008). Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology 213, 237–250. doi: 10.1016/j.imbio.2007.12.004
Segata, N. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Shapira, M. (2016). Gut microbiotas and host evolution: scaling up symbiosis. Trends Ecol. Evol. 31, 539–549. doi: 10.1016/j.tree.2016.03.006
Sijen, T., Steiner, F. A., Thijssen, K. L., and Plasterk, R. H. (2007). Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315, 244–247. doi: 10.1126/science.1136699
Sommer, F., and Bäckhed, F. (2013). The gut microbiota–masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238. doi: 10.1038/nrmicro2974
Stecher, B., Chaffron, S., Käppeli, R., Hapfelmeier, S., Freedrich, S., Weber, T. C., et al. (2010). Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 6:e1000711. doi: 10.1371/journal.ppat.1000711
Steiner, F. A., Okihara, K. L., Hoogstrate, S. W., Sijen, T., and Ketting, R. F. (2009). RDE-1 slicer activity is required only for passenger-strand cleavage during RNAi in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 16, 207–211. doi: 10.1038/nsmb.1541
Suman, S. K., Dhawaria, M., Tripathi, D., Raturi, V., Adhikari, D. K., and Kanaujia, P. K. (2016). Investigation of lignin biodegradation by Trabulsiella sp. isolated from termite gut. Int. Biodeterior. Biodegradation 112, 12–17. doi: 10.1016/j.ibiod.2016.04.036
Tabara, H., Yigit, E., Siomi, H., and Mello, C. C. (2002). The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-Box helicase to direct RNAi in C. elegans. Cell 109, 861–871. doi: 10.1016/S0092-8674(02)00793-6
Wilkins, C., Dishongh, R., Moore, S. C., Whitt, M. A., Chow, M., and Machaca, K. (2005). RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature 436, 1044–1047. doi: 10.1038/nature03957
Yabuuchi, E., Kanek, T., Yan, I., and Moss, C. W. (1983). Sphingobacterium gen. nov., Sphingobacterium spiritivorum comb. nov., Sphingobacterium multivorum comb. nov., Sphingobacterium mizutae sp. nov., and Flavobacterium indologenes sp. nov. : glucose-nonfermenting gram-negative rods in CDC groups IIK-2 and IIb. Int. J. Syst. Bacteriol. 33, 580–598. doi: 10.1099/00207713-33-3-580
Yigit, E., Batista, P. J., Bei, Y., Pang, K. M., Chen, C. C., Tolia, N. H., et al. (2006). Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127, 747–757. doi: 10.1016/j.cell.2006.09.033
Keywords: intestinal microbiome, innate immunity, rde-1, Orsay virus, Caenorhabditis elegans
Citation: Guo Y, Xun Z, Coffman SR and Chen F (2017) The Shift of the Intestinal Microbiome in the Innate Immunity-Deficient Mutant rde-1 Strain of C. elegans upon Orsay Virus Infection. Front. Microbiol. 8:933. doi: 10.3389/fmicb.2017.00933
Received: 05 March 2017; Accepted: 08 May 2017;
Published: 29 May 2017.
Edited by:
Akio Adachi, Tokushima University, JapanReviewed by:
Chang-Shi Chen, National Cheng Kung University, TaiwanFeng Li, Huazhong Agricultural University, China
Copyright © 2017 Guo, Xun, Coffman and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Chen, Y2hlbmZlbmcyMDExQGhzYy5wa3UuZWR1LmNu
 Yuanyuan Guo
Yuanyuan Guo Zhe Xun
Zhe Xun Stephanie R. Coffman
Stephanie R. Coffman Feng Chen
Feng Chen