- 1Fujian Key Laboratory of Marine Enzyme Engineering, Fuzhou University, Fujian, China
- 2Faculty of Medicine, School of Biomedical Sciences, The Chinese University of Hong Kong, Shatin, Hong Kong
Laccases are a family of copper-containing oxidases with important applications in bioremediation and other various industrial and biotechnological areas. There have been over two dozen reviews on laccases since 2010 covering various aspects of this group of versatile enzymes, from their occurrence, biochemical properties, and expression to immobilization and applications. This review is not intended to be all-encompassing; instead, we highlighted some of the latest developments in basic and applied laccase research with an emphasis on laccase-mediated bioremediation of pharmaceuticals, especially antibiotics. Pharmaceuticals are a broad class of emerging organic contaminants that are recalcitrant and prevalent. The recent surge in the relevant literature justifies a short review on the topic. Since low laccase yields in natural and genetically modified hosts constitute a bottleneck to industrial-scale applications, we also accentuated a genus of laccase-producing white-rot fungi, Cerrena, and included a discussion with regards to regulation of laccase expression.
Introduction
Laccases (EC 1.10.3.2) are a family of copper-containing oxidases found in a variety of bacteria, fungi, insects, and plants (Forootanfar and Faramarzi, 2015). The four copper atoms of a typical laccase molecule are divided into Type 1 (T1), Type 2 (T2), and binuclear Type 3 (T3) Cu sites based on unique spectroscopic features. In the resting enzyme, the four copper ions are in the +2 oxidation state. The T1 and T3 coppers are characterized by absorption at ~600 and 330 nm, respectively, whereas the T2 site lacks strong absorption features. Substrate oxidation occurs at the T1, and electrons are transferred to the T2/T3 trinuclear copper cluster (TNC), where molecular oxygen is reduced to water (Wong, 2009; Jones and Solomon, 2015). Redox potentials of the T1 sites in laccases range from 0.4 to 0.8 V; plant and bacterial laccases (e.g., 0.43 and 0.46 V for Rhus vernicifera and wild-type Bacillus subtilis CotA laccases, respectively) typically have potentials on the low end of this range, whereas fungal laccases have higher redox potentials (0.47–0.79 V) (Forootanfar and Faramarzi, 2015; Jones and Solomon, 2015; Mate and Alcalde, 2015; Pogni et al., 2015). With one-electron oxidation and radical formation, laccases catalyze oxidative coupling or bond cleavage of target compounds (Jeon and Chang, 2013).
Laccases have diverse substrate spectra, which overlap with those of tyrosinase and bilirubin oxidase (Baldrian, 2006; Reiss et al., 2013). They can oxidize a wide range of compounds, such as mono-, di-, poly-, and methoxy-phenols, aromatic and aliphatic amines, hydroxyindoles, benzenethiols, carbohydrates, and inorganic/organic metal compounds (Giardina et al., 2010; Jeon et al., 2012; Karaki et al., 2016). ABTS is the most popular substrate in laccase activity assays, and 2,6-dimethoxyphenol (2,6-DMP), catechol, guaiacol, and syringaldazine are also commonly used. The scope of laccase substrates can be further broadened with the help of redox mediators from natural and synthetic sources, i.e., suitable laccase substrates that can serve as diffusible electron shuttles between enzymes and other compounds (Morozova et al., 2007; Cañas and Camarero, 2010).
Since laccases have wide substrate ranges and use only oxygen as the final electron receptor, they have widespread applications in various industries, such as textile, food, biofuel, organic synthesis, bioremediation, paper and pulp, pharmaceutical, and cosmetic industries (Arora and Sharma, 2010; Majeau et al., 2010; Osma et al., 2010; Kudanga et al., 2011; Jeon et al., 2012; Betancor et al., 2013; Kudanga and Roes-Hill, 2014; Mogharabi and Faramarzi, 2014; Viswanath et al., 2014; Pezzella et al., 2015; Mate and Alcalde, 2016; Senthivelan et al., 2016; Sitarz et al., 2016; Upadhyay et al., 2016). In fact, a few laccase products are already commercially available for food, paper, textile, and other industries (Osma et al., 2010; Rodríguez-Couto, 2012). Laccase-based biocatalysts fit well with the development of industries that are efficient, sustainable, and environment-friendly.
Nonetheless, large-scale applications of laccases are limited by the economy and efficiency of the enzymes (Osma et al., 2010; Strong and Claus, 2011; Singh G. et al., 2015). Efforts have been made to produce large amounts of laccases at lower costs with the use of recombinant organisms or screening for natural hypersecretory strains. Enzyme activity and stability can be improved through immobilization and protein engineering (Pezzella et al., 2015; Upadhyay et al., 2016). The present article is not intended to be a comprehensive review on laccases, instead, it highlights the latest developments in laccase production and applications in bioremediation, especially degradation of emerging micropollutants including antibiotics.
Natural Laccase Producers
Laccase-Producing Fungi
Although, the first discovered laccase came from the exudates of the plant R. vernicifera, laccases of fungal origins have been the most intensively studied. Fungal laccases are implicated in both intra- and extra-cellular physiological processes including delignification, morphogenesis, pigmentation, and pathogenesis (Arora and Sharma, 2010; Kües and Rühl, 2011; Forootanfar and Faramarzi, 2015).
Among fungi, ascomycetes, basidiomycetes, and deuteromycetes can produce laccases, and white-rot basidiomycetes are the most efficient lignin degraders and laccase producers (Rodríguez-Couto and Toca-Herrera, 2007; Arora and Sharma, 2010). Laccases are secreted by white-rot fungi along with other ligninolytic enzymes including manganese peroxidase, lignin peroxidase, and versatile peroxidase, although the specific types of enzymes secreted may differ with the fungus (Wong, 2009; Arora and Sharma, 2010).
Pleurotus ostreatus and Trametes versicolor can be regarded as the model organisms in basic and applied laccase research. Other well-known laccase-producing basidiomycetes include Agaricus bisporus, Cerrena unicolor, Coprinopsis cinerea, Coriolopsis gallica, Cryptococcus neoformans, Cyathus bulleri, Fomes fomentarius, Ganoderma lucidum, Panus rudis, Phlebia radiata, Polyporus brumalis, Pycnoporus cinnabarinus, Pycnoporus sanguineus, Rigidoporus microporus, Schizophyllum commune, as well as various Pleurotus (e.g., P. eryngii, P. florida, P. pulmonarius, and P. sajor-caju) and Trametes (e.g., T. hirsuta, T. pubescens, T. trogii, and T. villosa) species (Baldrian, 2006; Arora and Sharma, 2010; Forootanfar and Faramarzi, 2015).
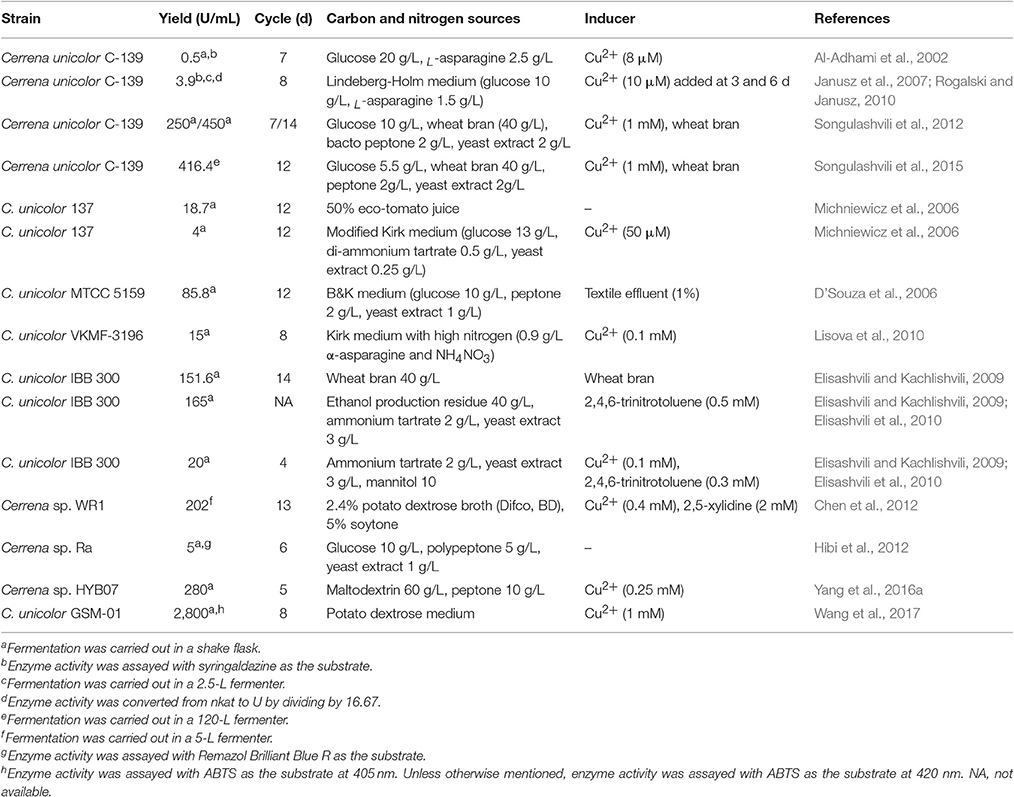
Efforts are still being made to screen naturally-occurring laccase producers with desired laccase yields and properties (Chen et al., 2012; Si et al., 2013; Fang Z. et al., 2015; Iracheta-Cárdenas et al., 2016; Kandasamy et al., 2016; Olajuyigbe and Fatokun, 2017). Laccase yields are variable depending on the species and strain, but most naturally-occurring species appear to be poor laccase producers. However, screening and selection of promising laccase producers from nature, followed by optimization of culture conditions, still constitute a viable and effective approach to obtain organisms with tremendous laccase synthesis ability (Elisashvili and Kachlishvili, 2009). The genus Cerrena with high laccase yields and application potentials deserves attention, and the properties of its laccase can be even more desirable compared to the commercial ones (Chen et al., 2012). However, Cerrena species are relatively less studied, especially compared with Trametes species. C. unicolor, a medicinal mushroom with antitumor and other activities (Mizerska-Dudka et al., 2015; Matuszewska et al., 2016), has been reported as a constitutive laccase producer (Al-Adhami et al., 2002); indeed, for many reported Cerrena strains, an organic inducer is not necessary, but copper ions are beneficial for laccase production. On the contrary, there are also some Cerrena strains that respond to lignocellulosic substrates or aromatic compounds, corroborating that enzyme production varies with the strain and should be characterized for each potentially valuable strain. Laccase production by reported Cerrena species is summarized in Table 1, which is comparable to that by Trametes species (Majeau et al., 2010) or G. lucidum (Postemsky et al., 2017).
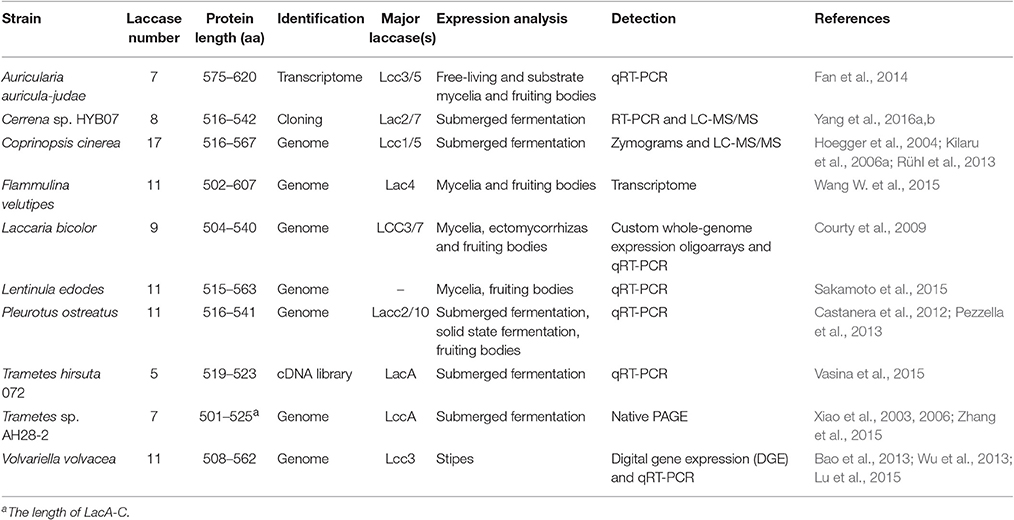
Fungal laccases exist in gene families, and reported basidiomycete laccase gene families contain 5–17 members (Table 2). Laccase isozymes are compared with respect to sequences, phylogenetic relationship, catalytic properties, and expression regulation. Sequence and evolutionary relationship examinations indicate that modern laccase gene families are derived from duplication-divergence events of a small set of ancestral enzymes (Valderrama et al., 2003; Kilaru et al., 2006a; Courty et al., 2009; Kües and Rühl, 2011; Bao et al., 2013; Wang W. et al., 2015). During natural evolution, the laccase paralogs may diversify in their functions, which is supported by numerous biochemical and expression characterization data (Hoegger et al., 2004, 2006; Pezzella et al., 2013; Fan et al., 2014; Yang et al., 2016b). Expression patterns provide valuable information for deducing the physiological roles played by the laccase isoforms. For example, Lcc5 in Auricularia auricula-judae is implicated in the sexual reproduction stage (Fan et al., 2014), LACC10 of P. ostreatus seems to function during vegetative growth (Pezzella et al., 2013), and Lcc3 is possibly involved in stipe elongation of Volvariella volvacea (Lu et al., 2015). Transcriptomic analysis of Flammulina velutipes implies that laccase isozymes are involved in growth and development, such as lignin bioconversion, stipe elongation and pileus formation (Wang W. et al., 2015). Sometimes, functional redundancy among laccase isozymes is suggested (Sakamoto et al., 2015; Wang W. et al., 2015). Furthermore, evidence for in vivo laccase function has been provided by genetic experiments. A siRNA knockdown study demonstrates that Lcc2 in A. bisporus contributes to toxin metabolism and defense against green mold disease (Sjaarda et al., 2015). On the other hand, overexpression of Hypsizygus marmoreus Lcc1 facilitates mycelial growth and fruiting body initiation (Zhang et al., 2015). Indeed, laccase has been developed as a novel screening marker in mushroom breeding (Sun et al., 2014). The levels of secreted laccase activity in edible mushrooms and their growing cycles are closely related, and short growing cycles are accompanied by high laccase activity (Sun et al., 2011).
Other Laccase Producers
Laccases also play diverse physiological roles in plants and bacteria, aside from metabolism of xenobiotics (Dwivedi et al., 2011; Singh et al., 2011; Chandra and Chowdhary, 2015; Forootanfar and Faramarzi, 2015; Wang J. et al., 2015). Plant laccase families are even larger than fungal laccase families. For example, there are at least 22 laccase genes in rice (Oryza sativa) (Huang et al., 2016). The model plant, Arabidopsis thaliana, has 17 members in its laccase gene family, which play several roles in plant growth and development, based on mutant characterization and expression profiling (Cai et al., 2006; Turlapati et al., 2011). Cotton contains 84, 44, and 46 laccase genes in cultivated allotetraploid Gossypium hirsutum and its two progenitor diploids G. arboreum and G. raimondii (Balasubramanian et al., 2016). In opposite to fungal laccases, plant laccases participate in lignin synthesis and therefore can be engineered for energy plant improvement (Wang J. et al., 2015). Plant laccases are also involved in pigmentation (Liang et al., 2006) or pigment breakdown (Fang F. et al., 2015), root elongation (Liang et al., 2006), and responses to external stresses (Cho et al., 2014; Kim et al., 2014).
Bacterial laccases were discovered relatively late compared to plant and fungal laccases (Ausec et al., 2011), but research on bacterial laccases have gained momentum over the past two decades. Bacterial laccases are implicated in various processes ranging from UV protection, pigmentation, metal oxidation, sporulation to xenobiotic degradation (Singh et al., 2011; Chandra and Chowdhary, 2015; Forootanfar and Faramarzi, 2015). Due to the widespread existence and versatility of bacteria, bacterial laccases have higher thermostability, alkaline pH optimum and halotolerance despite their low redox potentials and are valuable functional complements to fungal laccases in dye decolorization, pulp biobleaching, biofuel production as well as various other industrial and biotechnological fields (Santhanam et al., 2011; Singh et al., 2011; Chandra and Chowdhary, 2015; Martins et al., 2015).
Insect laccases are the least characterized of all known laccases. Insect laccases also play important roles in insect physiology such as cuticle sclerotization and melanization (Dittmer and Kanost, 2010; Jeon et al., 2012; Ni and Tokuda, 2013).
Classical and Molecular Breeding for Enhancing Laccase Production
Classical Breeding Approaches
In addition to isolating natural, efficient laccase producers, classical and molecular breeding approaches are also used to increase laccase production. A successful example was provided by N-methyl-N-nitro-N-nitrosoguanidine and ultraviolet light treatments of C. gallica TCK. A mutated strain T906 was obtained, which showed three-fold higher laccase activity than the starting strain and a maximum laccase activity of 303 U/mL after 13 days (Xu et al., 2016). Mating of monokaryotic compatible P. ostreatus strains led to dikaryotic strains with higher laccase activity, and the best one produced 110 U/mL after induction for 8 days (Lettera et al., 2011; del Vecchio et al., 2012). The dikaryotic superiority in laccase activity is derived from non-additive increases in laccase transcription (Castanera et al., 2013). Furthermore, N+ ion implantation has recently been successfully applied to improve laccase production in Paecilomyces sp. WSH-L07 (Liu et al., 2010) and Ceriporiopsis subvermispora (Wang C. et al., 2012).
Heterologous Expression
Heterologous expression is invaluable in obtaining laccase proteins based only on metagenomic sequences (Beloqui et al., 2006; Fang et al., 2011, 2012). Heterologous expression is also important for isolating a laccase from other isozymes, especially when the enzyme is not abundantly expressed or silent. In addition to structural and biochemical characterization, heterologously expressed laccases can be engineered by rational design or directed evolution for enhanced expression, catalytic activity, stability, etc. The readers are referred to the recent reviews on laccase engineering (Rodgers et al., 2010; Alcalde, 2015; Mate and Alcalde, 2015; Pardo and Camarero, 2015). Enzyme resurrection, which is heterologous expression of ancestral enzymes reconstructed based on phylogenetic analysis and inference, is of particular interest (Alcalde, 2015). Ancestral enzymes are likely to have unique and extreme properties, such as greater stability and substrate promiscuity than extant ones, considering characteristics of ancient life (e.g., thermophilic) and generalist-specialist conversion of enzymes during the course of evolution (Risso et al., 2014). White-rot fungi and lignin degradation are dated back to the Permo-Carboniferous period (Floudas et al., 2012), therefore laccase resurrection brings an intriguing and promising toolset to laccase engineering and deserves more research efforts. The resurrected enzymes can then be subjected to further engineering by directed evolution (Alcalde, 2015).
The most common heterologous host is the methylotrophic yeast Pichia pastoris with its inducible alcohol oxidase (AOX1) promoter (Antošovǎ and Sychrová, 2016; Ergün and Çalık, 2016). Other hosts, such as prokaryotes (e.g., E. coli and B. subtilis), yeasts (e.g., Saccharomyces cerevisiae and Yarrowia lipolytica), filamentous fungi (e.g., Trichoderma reesei and Aspergillus niger), and plants (e.g., Nicotiana tabacum and O. sativa), are also used (Piscitelli et al., 2010; Kittl et al., 2012; Liebeton et al., 2014; Mate and Alcalde, 2015; Antošovǎ and Sychrová, 2016). In particular, heterologous laccases were constitutively expressed in basidiomycetes Phanerochaete chrysosporium (Coconi-Linares et al., 2015) and C. cinerea (Muraguchi et al., 2011). Although, filamentous fungi are efficient in protein secretion and have actually given rise to some of the highest recombinant laccase yields reported, genetic techniques are more readily available for yeasts (Piscitelli et al., 2010; Mate and Alcalde, 2015; Antošovǎ and Sychrová, 2016).
Expression systems like P. pastoris are used to produce enzymes at the industrial scale, but ligninolytic enzymes like laccases are notoriously difficult to express heterologously (Gu et al., 2014; Ergün and Çalık, 2016). Summaries of heterologously expressed laccases can be found in recent publications (Kittl et al., 2012; Mate and Alcalde, 2015; Antošovǎ and Sychrová, 2016; Ergün and Çalık, 2016). Occasionally, high recombinant laccase activity is obtained (Hong et al., 2002, 2007; Nishibori et al., 2013), but many recombinant laccases are expressed at levels below 10 U/mL, which can be even lower than that in the native strain (Yang et al., 2016b). An appropriate host is needed for heterologous expression of laccases, but “the best host” remains elusive (Piscitelli et al., 2010; Rivera-Hoyos et al., 2013). Cryptococcus sp. S-2 is a better yeast host than P. pastoris for expression of T. versicolor and Gaeumannomyces graminis laccase genes. The expression advantage is likely due to similar codon usage and GC content of Cryptococcus sp. S-2 with those of T. versicolor and G. graminis (Nishibori et al., 2013). Nonetheless, different codon preferences of the expression host and the gene source fails to explain the variability in production yields between laccase genes derived from the same organism (Piscitelli et al., 2010). Strategies employed to increase laccase production in heterologous systems include promoter and signal peptide selection, protein engineering, codon optimization, and optimization of cultivation medium composition and process. Since different and sometimes controversial results have been recorded, it is still difficult to predict the most promising combination of parameters to maximize heterologous laccase production (Piscitelli et al., 2010; Antošovǎ and Sychrová, 2016).
Homologous Expression
Due to low laccase yields in heterologous hosts, homologous expression in laccase-producing hosts might be of value for promoting laccase production. Homologous laccase expression has been attempted in A. niger (Ramos et al., 2011), C. cinerea (Kilaru et al., 2006b), Gloeophyllum trabeum (Arimoto et al., 2015), P. cinnabarinus (Alves et al., 2004), and T. versicolor (Kajita et al., 2004). For overexpression, the laccase gene is often driven by a strong promoter such as the constitutive glyceraldehyde-3-phosphate dehydrogenase gene (gpd) promoter (Alves et al., 2004; Kajita et al., 2004; Kilaru et al., 2006b; Arimoto et al., 2015) and maltose-induced glucoamylase gene (glaA) promoter (Ramos et al., 2011). In particular, when various basidiomycete promoters were compared, the A. bisporus gpdII promoter is more efficient in driving expression of the homolgous Lcc1 gene in C. cinerea than the C. cinerea tub1 (β-tubulin gene) promoter, Lentinus edodes priA (fruiting body gene) promoter or S. commune Sc3 (hydrophobin gene) promoter (Kilaru et al., 2006b). The native laccase promoter was also used, and a high laccase production level of 1 g/L was achieved in the presence of 40 g/L ethanol after fermentation of transgenic P. cinnabarinus for 24 days (Alves et al., 2004).
Regulation of Laccase Expression
Following successful screening of laccase-producing native hosts, laccase production is improved by fermentation technology development with respect to fermentation type, medium composition, and cultivation parameters (Elisashvili and Kachlishvili, 2009; Forootanfar and Faramarzi, 2015). Enhancing laccase yields is essential to lower production costs and promote industrial applications of the enzyme, which relies on understanding of laccase expression regulation. Numerous publications and reviews have been devoted to expression regulation of laccases (Piscitelli et al., 2011; Janusz et al., 2013).
Expression of laccase isozyme genes is differentially regulated throughout fermentation and in response to medium composition, such as metal ions, xenobiotics as well as nutrient types and levels. Laccase expression analysis has been performed on mRNA and protein levels, and the distinct responses of species, strains as well as genes no doubt paint a complex picture of laccase expression regulation. In accordance, various cis-acting responsive elements have been identified in laccase promoter regions, such as metal response element (MREs), ACE1 copper-responsive transcription factor binding sites (ACE1), xenobiotic response elements (XREs), antioxidant response elements (AREs), heat shock response elements (HSEs), CreA binding sites (CreA), and NIT2 binding sites (NIT2) (Piscitelli et al., 2011; Janusz et al., 2013). Nonetheless, function of most of the putative responsive elements is not experimentally validated, and how they interact with transcription factors remains elusive.
Copper ions are probably the most used inducer in laccase production, and the ACE1 binding site represents the most well-understood regulatory element in the laccase promoter region. Copper ions interact with the transcription factor ACE1 in P. brumalis (Nakade et al., 2013) or CUF1 in C. neoformans (Jiang et al., 2009) to increase laccase expression. In yeast, ACE1 and CUF1 have interchangeable N-terminal copper-fist DNA binding motifs despite opposite roles in maintaining copper homeostasis (Beaudoin et al., 2003). On the other hand, a copper-responsive laccase gene without an orthodox ACE1 binding site within its promoter might be regulated through a nonconventional copper-responsive element or a different mechanism (Yang et al., 2016a). Even when copper ions are not able to induce laccase production, they stabilize the copper-containing catalytic center of the enzyme (Solé et al., 2012), thus contributing to laccase activity. Besides copper, manganese and zinc are also commonly found to stimulate laccase synthesis (Lu and Ding, 2010; Solé et al., 2012; Yang et al., 2016a).
Literature describing laccase induction by xenobiotics, e.g., lignin breakdown products, dyestuffs and organic pollutants, has been accumulating. The effects of organic compounds on laccase production depend on the compound structure, fungal strain, and growth stage, laccase isozyme as well as the culture medium (Elisashvili et al., 2010; Giardina et al., 2010; Lu and Ding, 2010; Piscitelli et al., 2011; Solé et al., 2012; Janusz et al., 2013; Yang Y. et al., 2013). Combinational induction of laccase production by metal ions and organic compounds can be either synergistic (Yang Y. et al., 2013) or antagonistic (Lu and Ding, 2010).
Coculture of laccase-producing strains with other microbes is a natural way to induce laccase production, in the form of either yield increase or induction of new isozymes, and can be more effective than chemical induction. Microbial interactions with laccase inducing effects vary with the strain, but the structure of inducing metabolites and the inducing mechanism remain largely unknown (Zhang et al., 2006; Elisashvili and Kachlishvili, 2009; Flores et al., 2009; Wei et al., 2010; Pan et al., 2014; Li et al., 2016). One proposed mechanism for laccase overproduction in the coculture process is carbon source succession. Li et al. found that glycerol produced from glucose by the yeast Candida sp. is an efficient carbon source for G. lucidum upon glucose deprivation and crucial for laccase overproduction by prolonging laccase secretion time (Li et al., 2011). Phenolics and lysing enzymes produced by opposing microbes have also been suggested to have laccase-inducing ability (Zhang et al., 2006; Wei et al., 2010).
Nutrient types and concentrations have been extensively studied in the context of fungal growth and enzyme secretion. Since basidiomycetes display a wide diversity in their responses, no generalization can be made on the best carbon and nitrogen sources or their optimal concentrations (Elisashvili and Kachlishvili, 2009; Piscitelli et al., 2011; Janusz et al., 2013). Lignocellulosic wastes containing carbohydrates and inducers are often added resulting in benefits such as lower production costs, waste reuse, and laccase production enhancement (Elisashvili and Kachlishvili, 2009; Postemsky et al., 2017).
In many fungi, laccase is produced by secondary metabolism, that is, laccase synthesis is activated by carbon or nitrogen depletion. This no doubt necessitates a long production cycle and encumbers industrial production of laccase. Therefore, a promising commercial laccase producer should produce laccase with high yields and a short fermentation cycle. A recently reported Cerrena sp. HYB07 is an example of such laccase producers (Yang et al., 2016a). Its laccase production is not inhibited by high nutrient levels, which allows biomass accumulation and a quick peak of laccase activity (Table 1). Furthermore, the laccase yield of HYB07 is mostly attributed to the predominantly expressed Lac7. The strength of the Lac7 promoter requires only copper ions and high nutrient concentrations, but not aromatic inducers, making it interesting for recombinant expression of other laccase genes.
Other factors on laccase expression are studied to a lesser extent. Laccase expression could also be regulated by oxidative stress (Yang et al., 2012; Si and Cui, 2013; Fernandez-Alejandre et al., 2016), heat shock (Wang F. et al., 2012), cAMP (Crowe and Olsson, 2001), and calmodulin (Suetomi et al., 2015).
Apparently, crosstalk exists between the internal factors (e.g., fungal strain, growth stage, laccase promoter, etc.) and external factors (metal ions, organic compounds, nutrient sources, and ratios, etc.) influencing laccase synthesis. However, until now, the bulk of the work has only attempted to decipher regulation of laccase expression by an isolated single factor or a few factors, which is undoubtedly a simplification of the complex network controlling laccase expression. The mechanism underlying the regulatory network of laccase expression awaits elucidation.
Laccase Mediators
The efficiency of substrate oxidation by a laccase depends on the difference between the redox potentials of the substrate and the T1 Cu. Due to the lower redox potentials of laccases (≤0.8 V) compared to ligninolytic peroxidases (>1 V) (Wong, 2009; Rivera-Hoyos et al., 2013; Pollegioni et al., 2015; Sitarz et al., 2016), laccases are originally thought to be able to oxidize only the phenolic lignin moiety, with the majority of lignin being non-phenolic and with higher redox potentials. Low-molecular-weight redox mediators are used to expand the laccase substrate range or increase the reaction rate, especially for substrates with higher redox potentials or too large to fit in the enzyme's active site. Commonly used laccase mediators include synthetic mediators such as 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonate) (ABTS) and 1-hydroxybenzotriazole (HBT) as well as natural phenolic mediators such as syringaldehyde (SA) and acetosyringone (AS). Despite the proven efficiency of artificial mediators, natural mediators (believed to be true mediators of fungal laccases in nature) are considered to be alternatives to the artificial ones because they are more economically feasible and environmentally friendly (Cañas and Camarero, 2010). Laccase oxidation of the substrate may proceed differently with a mediator. However, it is not always the case. Malachite green degradation products in the presence and absence of ABTS have been shown to be identical or different, depending on the enzyme (Chhabra et al., 2009; Yang et al., 2015).
Different types of mediators have different catalytic mechanisms. ABTS-mediated substrate oxidation proceeds via an electron transfer route. ABTS is first oxidized to its radical cation (ABTS·+) and then to the di-cation (ABTS2+) with redox potentials of 472 and 885 mV, respectively. Unlike ABTS, an N-OH type mediator (such as HBT and violuric acid) forms the N-oxy radical upon laccase oxidation and subsequent deprotonation; the radical in turn abstracts the benzylic hydrogen atom from the substrate. Similarly, phenolic mediators also follow a radical hydrogen abstraction mechanism, but with the intermediate being a phenoxy radical (Hu et al., 2009; Wong, 2009). The effect of a mediator on laccase oxidation varies with the laccase and substrate and depends on the radicals formed, recyclability of the mediator and stability of the laccase in the presence of the mediator (Morozova et al., 2007; Wong, 2009; Cañas and Camarero, 2010; Pogni et al., 2015). Regardless of the reaction mechanism, mediators incur additional costs, and can cause toxicity (Weng et al., 2013; Becker et al., 2016) and laccase inactivation (Kurniawati and Nicell, 2007; Fillat et al., 2012; Ashe et al., 2016). Although, laccases without the requirement for facilitating mediators, the laccase/mediator system is regarded as a feasible industrial solution, ideal mediators that are cheap, green, effective, stable, recyclable, not toxic, or enzyme-inactivating should be ascertained (Morozova et al., 2007; Kües, 2015).
Laccase Immobilization
Laccases are immobilized for recycling, operational stability, and resistance to application conditions. Immobilization techniques include entrapment, adsorption, covalent binding, self-immobilization as well as combinations of the aforementioned techniques. Activity recovery varies based on the enzyme, the immobilization method of choice, and preparation parameters. Compared with their free counterparts, immobilized laccases are more tolerant to high temperatures and storage and can be reused multiple times (Fernández-Fernández et al., 2013; Asgher et al., 2014), they are also more resistant to inhibitors such as NaCl (Yang et al., 2016c). Immobilization sometimes improves the catalytic activity of laccases (Arsenault et al., 2011; Sinirlioglu et al., 2013; Kumar et al., 2014) despite the common concern of reduced enzyme flexibility, steric hindrance and diffusion limitations (Sheldon, 2011; Talekar et al., 2013). Readers can refer to reviews on preparation and applications of immobilized laccases (Ba et al., 2013; Fernández-Fernández et al., 2013; Asgher et al., 2014).
Laccase Applications in Biodegradation of PPCPs
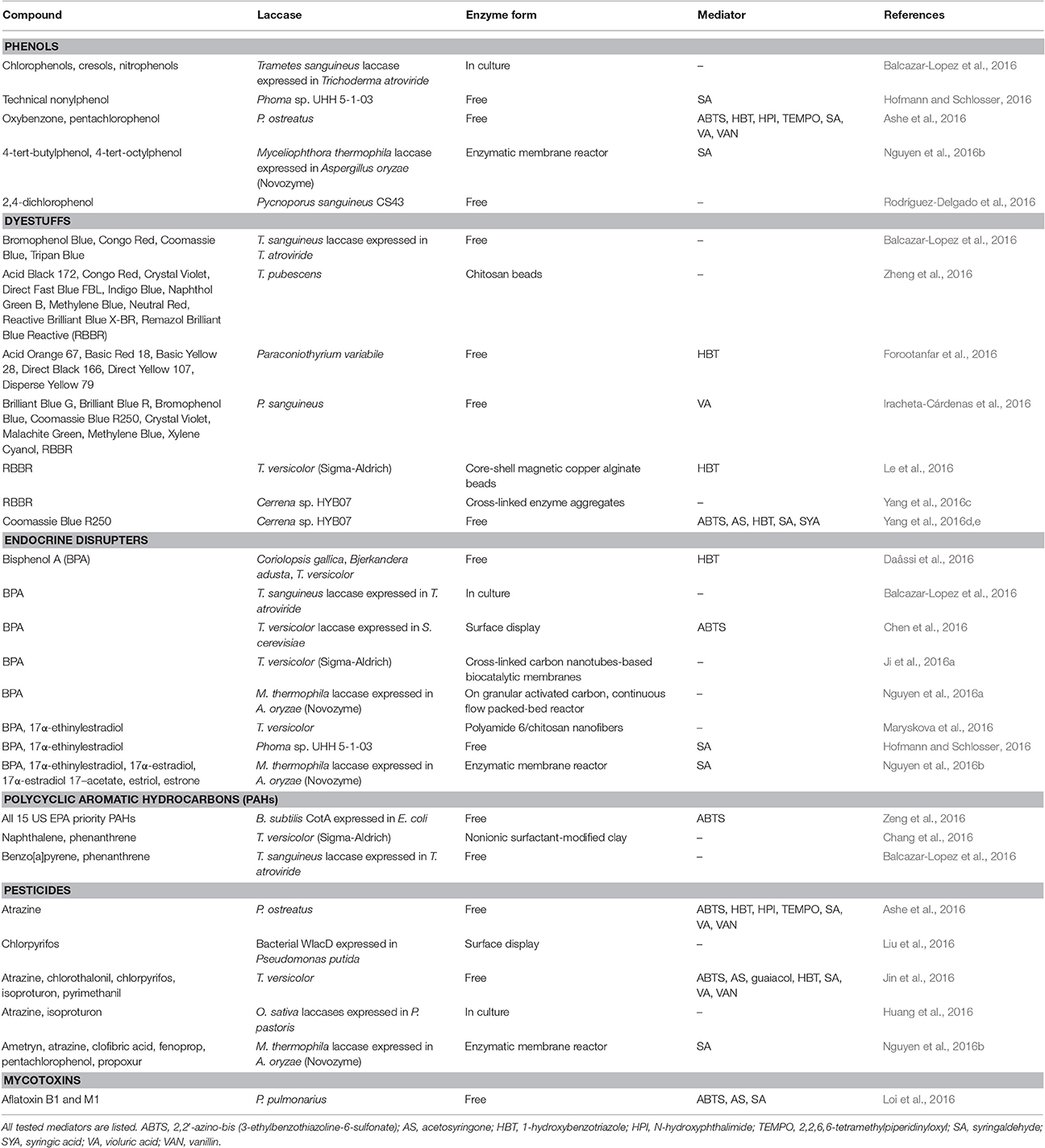
The value of fungi as well as fungal enzymes in pollution control and environment management has been recognized. Examples of environmentally important enzymes comprise hydrolases, laccases, lyases, peroxidases, tyrosinases, and P450 cytochrome monooxidases (Demarche et al., 2012; Yang et al., 2013a; Rao et al., 2014; Kües, 2015; Yadav and Yadav, 2015; Martinkova et al., 2016). The ability of laccases to effectively degrade and detoxify a variety of persistent organic pollutants (POPs) has received considerable attention in the field of bioremediation (Majeau et al., 2010; Strong and Claus, 2011; Gasser et al., 2014; Viswanath et al., 2014; Catherine et al., 2016), and laccases can also be used in enzymatic biosensors for environmental pollution monitoring (Rao et al., 2014). A summary of environmental contaminants as laccase substrates from published research in the year 2016 is provided in Table 3. The contaminants investigated include dyestuffs (Singh R. L. et al., 2015; Sen et al., 2016), polycyclic aromatic hydrocarbons (PAHs) (Librando and Pappalardo, 2013), endocrine disrupters (Cabana et al., 2007; Husain and Qayyum, 2012; Gasser et al., 2014), and pesticides (Maqbool et al., 2016).
Pharmaceuticals and personal care products (PPCPs) are becoming ubiquitous in the environment and are recognized as emerging trace organic contaminants (Onesios et al., 2009; Oulton et al., 2010; Wang and Wang, 2016). Laccases can be employed for their removal (Gasser et al., 2014). Laccases have been used in PPCPs as an ingredient; many products generated by laccases have antimicrobial, anticancer, antioxidant, detoxifying, or other activities (Senthivelan et al., 2016; Upadhyay et al., 2016). Specifically, laccases can be used to synthesize novel antibiotics (Mikolasch et al., 2012, 2016; Pezzella et al., 2015), and laccase-based antimicrobial formulations are considered a safe and green alternative to chemical decontamination (Grover et al., 2013). Nonetheless, the focus of this review lies in the degradation and detoxification of PPCP contaminants with laccases.
Degradation of Antibiotics
Antibiotics constitute one of the most used classes of drugs in the world; they are used in human and veterinary medicine as well as livestock farming. Antibiotics that are not metabolized enter the environment (Larsson, 2014). Conventional water treatment processes cannot effectively remove antibiotics (Oulton et al., 2010), while more efficient advanced treatment methods have disadvantages such as high costs and secondary pollution (Chen et al., 2016). Antibiotics pose health risks by selecting for antibiotic-resistance bacteria (ARB). Antibiotics, ARB, and antibiotic-resistant genes have been detected in soil, sediments, and water bodies including wastewater drinking water and marine water (Thiele-Bruhn, 2003; Kümmerer, 2009; Guo et al., 2014; Larsson, 2014). There has been a fast growth in the literature describing laccase utilization in antibiotic removal, especially within the past 2 years, but this topic has not been properly reviewed.
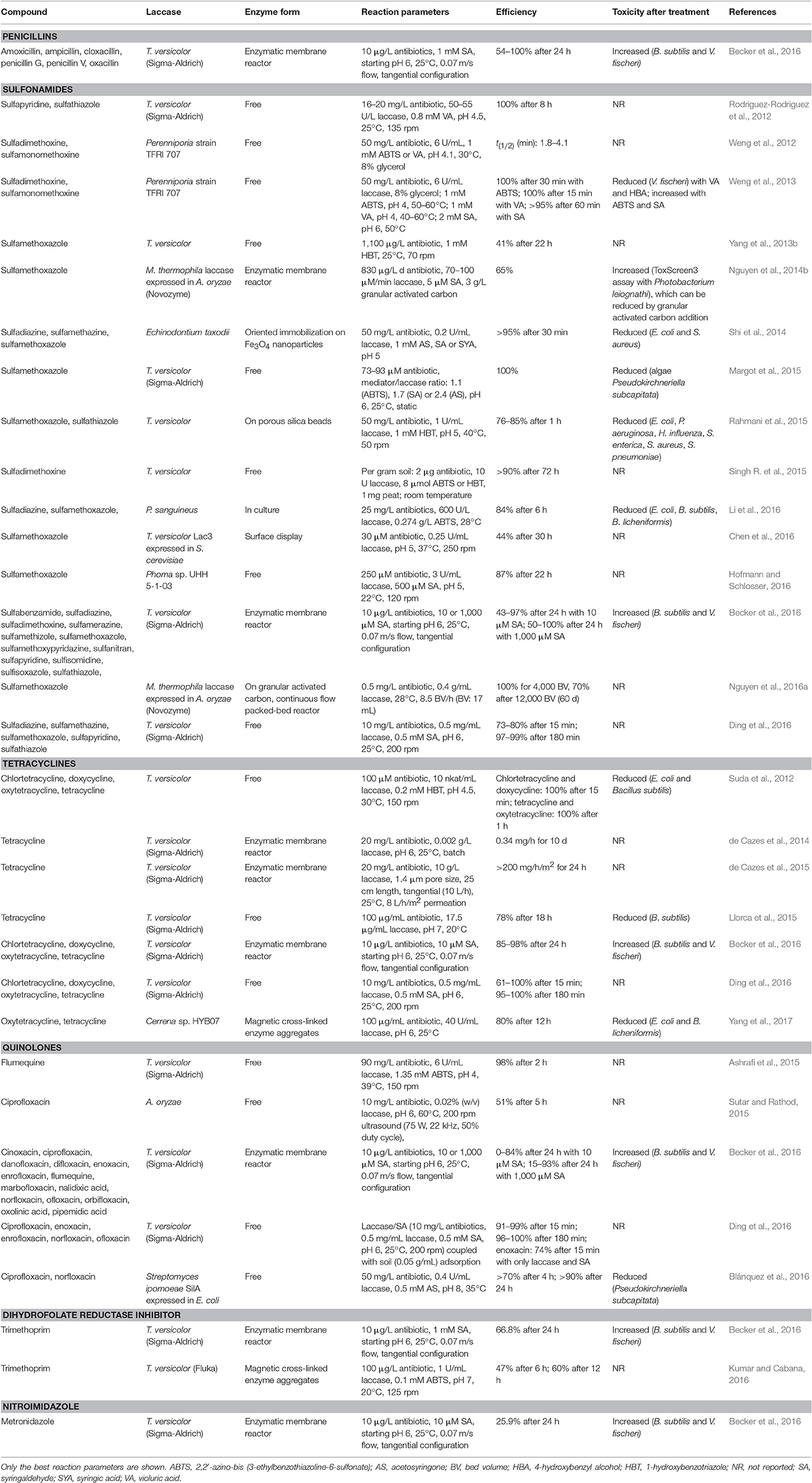
Target antibiotics under investigation include penicillins, tetracyclines, sulfonamides, quinolones and trimethoprim, and sulfamethoxazole and tetracycline are two most studied (Table 4). The removal time ranges from minutes to hours, depending on the laccase, antibiotic and treatment parameters. Mediators such as HBT, ABTS, and SA are often used to enable or accelerate antibiotic conversion by laccases. In fact, significant antibiotic removal within 1 h usually requires involvement of an appropriate mediator (Suda et al., 2012; Weng et al., 2012, 2013; Shi et al., 2014; Ding et al., 2016). Manganese peroxidase was more efficient in tetracycline conversion than laccase, but the addition of HBT can promote laccase catalysis to a rate higher than that of manganese peroxidase (Wen et al., 2010; Suda et al., 2012) although still slower than that of lignin peroxidase (95% degradation efficiency in 5 min; Wen et al., 2009). Interestingly, mediators, i.e., ABTS, SA, and AS, are consumed without observed catalytic activity during degradation of sulfamethoxazole (Margot et al., 2015).
Sulfonamides and tetracyclines are more easily attacked by laccase compared with quinolones (Becker et al., 2016; Ding et al., 2016). This is presumably due to the strong electron donating aromatic amine group in sulfonamides and the phenol group in tetracyclines, which are not found in quinolones (Ding et al., 2016). However, identified tetracycline transformation intermediates suggest that the phenol group is not the primary target for laccase oxidation, and that oxygen addition, demethylation, water elimination reactions occur during laccase treatment (Llorca et al., 2015; Yang et al., 2017). For sulfonamides, increasing electronegativity of the substituents is accompanied by decreased degradation (Yang C. W. et al., 2016). Two sulfonamides, namely sulfapyridine and sulfathiazole, are desulfonated by laccase (Rodriguez-Rodriguez et al., 2012). Covalent cross-coupling of sulfonamides is observed with laccase and mediator SA or AS (Shi et al., 2014; Margot et al., 2015), but not ABTS (Margot et al., 2015). Trimethoprim has 2 amine groups and 3 methoxy groups and is usually administered in combination with sulfamethoxazole. Little (Touahar et al., 2014; Arca-Ramos et al., 2016) to over 60% (Kumar and Cabana, 2016) degradation of this antibiotic without a mediator have been reported. Furthermore, SA at 1,000 μM, but not 10 μM, increases trimethoprim removal from 27 to 67%; nearly complete elimination of sulfamethoxazole is achieved under the same conditions (Becker et al., 2016). Some antibiotics (e.g., penicillins) are unstable in aqueous solutions, and attention should be paid to sample preservation and quantification (Llorca et al., 2014; Becker et al., 2016).
Laccase from T. versicolor, especially the product sold by Sigma-Aldrich, is most frequently used in biodegradation studies of antibiotics as well as other trace organic contaminants. Other laccases include laccases from basidiomycetes Cerrena sp. HYB07, Echinodontium taxodii, Perenniporia strain TFRI 707, and P. sanguineus, from ascomycetes Phoma sp. and Myceliophthora thermophila (recombinantly expressed in Aspergillus oryzae) and from actinobacteria Streptomyces ipomoeae (expressed in E. coli; Table 4). Laccases immobilized by different methods have been used for antibiotic degradation, including enzymatic membrane reactors (Nguyen et al., 2014b; Becker et al., 2016), granular activated carbon (Nguyen et al., 2016a), silica beads (Rahmani et al., 2015), oriented immobilization (Shi et al., 2014), magnetic cross-linked enzyme aggregates (Kumar and Cabana, 2016; Yang et al., 2017), and cell surface display (Chen et al., 2016). In particular, enzymatic membrane reactors (gelatin-ceramic membranes grafted with commercial T. versicolor laccase) in tetracycline degradation have been evaluated in depth with respect to membrane preparation, efficiency, kinetics, and economics (de Cazes et al., 2014, 2015; Abejón et al., 2015a,b). Mathematical cost estimation indicates that the enzymatic process is still economically uncompetitive. Improvements should be made in terms of enzyme kinetics, reactor effective lifetime and regeneration costs (Abejón et al., 2015a). For example, a pore diameter of 1.4 μm, in contrast to 0.2 μm, increases enzyme loading of the membrane reactor, avoids extensive membrane area, and facilitates tetracycline degradation (de Cazes et al., 2015).
Occasionally, laccases do not participate in antibiotic removal by white-rot fungi; for instance, laccase was not responsible for oxytetracycline degradation by P. ostreatus or T. versicolor (Migliore et al., 2012; Mir-Tutusaus et al., 2014) or sulfamethoxazole degradation by aquatic ascomycete Phoma sp. UHH 5-1-03 (Hofmann and Schlosser, 2016). In these cases, other enzymes, such as cytochrome P450, may be resorted to for biodegradation. It should still be pointed out that even when extracellular laccase is not able to directly oxidize sulfamethoxazole, when a mediator is added, significant removal is achieved (Yang et al., 2013b; Hofmann and Schlosser, 2016).
Laccases are also applied in combination with other processes in antibiotic treatment, such as ultrasound (Sutar and Rathod, 2015) and soil adsorption (Ding et al., 2016). The involvement of other processes facilitates degradation of antibiotics, e.g., quinolone antibiotics, which are recalcitrant to laccase oxidation. Laccase can also improve efficiency and stability of antibiotic removal by other organisms. When sulfamethoxazole is the transformed by non-laccase-producing bacteria Alcaligenes faecalis, the efficiency drops when some metabolites such as N4-acetyl-sulfamethoxazole are transformed back to the parent compound. The removal efficiency does not decrease when the coculture of A. faecalis with laccase-producing P. sanguineus is used or when cell-free laccase was added to A. faecalis culture (Li et al., 2016).
Toxicity of antibiotics after laccase treatment is commonly assessed via growth inhibition assay or bioluminescence inhibition test (Table 4). Antibiotic degradation by laccase mostly leads to reduced toxicity. A good example comes from the comparison of the sulfamethoxazole transformation products and their toxicity by A. faecalis with or without exogenous laccase. N-hydroxy sulfamethoxazole (HO-SMX), a toxic and recalcitrant intermediate of sulfamethoxazole, is formed upon A. faecalis treatment. Additional laccase, on the other hand, eliminates HO-SMX along with the toxicity (Li et al., 2016). However, sometimes laccase/mediator-catalyzed antibiotic transformation results in even higher toxicity, and this seems to frequently associate with the mediator SA (Weng et al., 2013; Nguyen et al., 2014b; Becker et al., 2016). It is postulated that the enhanced toxicity can be derived from oxidation of aromatic structures, especially phenols, to quinonoids (Becker et al., 2016).
The majority of studies on antibiotic degradation were carried out in aqueous environments, but there have been a few studies on remediation of soil (Singh R. et al., 2015), river sediment (Chang and Ren, 2015), and sludge (Yang C. W. et al., 2016). Laccase-containing extract from spent mushroom compost of Pleurotus eryngii and extract-containing microcapsules enhanced degradation of three tetracyclines in river sediment (Chang and Ren, 2015) as well as degradation of three sulfonamides in sewage sludge (Yang C. W. et al., 2016). Sulfonamide antibiotics can form stable covalent bonds with humic constituents, and laccase can catalyze unreactive hydroquinone moieties in humic acid to reactive, electrophilic quionone moieties which in turn react with the antibiotic. This will affect the fate, bioactivity, and extractability of sulfonamides in soils (Gulkowska et al., 2012, 2013; Schwarz et al., 2015).
Degradation of Other PPCPs
Besides antibiotics, many other PPCPs are actively evaluated as laccase substrates, including anticonvulsants (e.g., carbamazepine and benzodiazepines) (Ostadhadi-Dehkordi et al., 2012), fungicides (e.g., ketoconazole) (Yousefi-Ahmadipour et al., 2016), anti-inflammatory drugs (e.g., acetaminophen, aspirin, diclofenac, and ketoprofen) (Marco-Urrea et al., 2010a; Ba et al., 2014a; Domaradzka et al., 2015; Singh et al., 2016), antidepressants (e.g., imipramine) (Tahmasbi et al., 2016), lipid regulators (e.g., clofibric acid) (Ji et al., 2016a), biocides (triclosan and chlorophene) (Shi et al., 2016), insect repellents (e.g., N,N-diethyl-m-toluamide) (Tran et al., 2013), and sunscreen agents (e.g., oxybenzone) (Garcia et al., 2011).
Among these PPCPs, diclofenac, carbamazepine, and triclosan are the most investigated (Table 5); triclosan is phenolic and the other two are non-phenolic. Carbamazepine is the most recalcitrant to oxidation by laccase (Yang et al., 2013b; Nguyen et al., 2014a,b; Touahar et al., 2014; Hofmann and Schlosser, 2016; Kumar and Cabana, 2016) or peroxidases (Zhang and Geißen, 2010; Eibes et al., 2011). Carbamazepine contains a strong electron-attracting amide group, which may account for its recalcitrance (Yang et al., 2013b). In contrast, triclosan has a strong electron donating hydroxyl group despite simultaneous presence of electron withdrawing chlorinated groups, which makes it susceptible to laccase oxidation (Garcia-Morales et al., 2015). Chlorine atoms are also found in diclofenac along with aromatic amine (Nguyen et al., 2014c, 2015), but it is more prone to laccase oxidation than carbamazepine.
In addition to removal of pharmaceutical compounds with laccases, laccase-producing fungi, bacteria and actinomycetes are also evaluated. Laccase is at least partially responsible for pollutant degradation (Marco-Urrea et al., 2010b; Tran et al., 2010; Nguyen et al., 2014d; Popa et al., 2014; Boonnorat et al., 2016; Hofmann and Schlosser, 2016; Vasiliadou et al., 2016). On the contrary, other studies failed to established dependence of pharmaceutical removal on extracellular laccase (Marco-Urrea et al., 2009; Jelic et al., 2012; Yang et al., 2013b). Laccase is also used in combination with other enzymes, such as versatile peroxidase and glucose oxidase (Touahar et al., 2014) or tyrosinase (Ba et al., 2014a) in pharmaceutical removal. Other processes, such as adsorption, are also found to improve compound removal when used in combination with laccase biodegradation (Ba et al., 2014b; Nguyen et al., 2014b,d; Xu et al., 2014; Ji et al., 2016a). Horseradish peroxidase is more efficient than laccase in triclosan removal (Melo et al., 2016), and versatile peroxidase has a wider removal spectrum than laccase (Touahar et al., 2014).
Simultaneous laccase degradation of multiple trace organic contaminants demonstrates that phenolic compounds are generally more easily degraded than non-phenolic compounds (Nguyen et al., 2014a,d), which is expected since phenolic compounds are considered natural laccase substrates. Transformation rates may be different if the compounds are present in solutions of single compounds or in mixtures. For example, Margot et al. found that diclofenac removal is enhanced whereas bisphenol A (BPA) and mefenamic acid elimination is decreased in mixtures (Margot et al., 2013b). Nair et al. also found improved degradation of diclofenac in the presence of BPA and 17-α-ethinylestradiol, whereas degradation of the latter two compounds is not affected in the mixture (Nair et al., 2013). Phenolic compounds such as BPA can serve as a mediator for non-phenolic compounds or may facilitate polymerization (Margot et al., 2013b; Nair et al., 2013; Touahar et al., 2014; Ji et al., 2016a). The majority of work on contaminant removal was carried out in buffers; biodegradation efficiency decreases in real wastewaters (Nair et al., 2013; Touahar et al., 2014; Garcia-Morales et al., 2015; Le et al., 2016), which presents a challenge for laccase applications. Real wastewater has elevated pH compared with the optimized buffer system and potential laccase inhibitors (e.g., organic matter, heavy metals, and ions). However, a few studies still achieved efficient pharmaceutical degradation in real wastewaters (Garcia et al., 2011; Ba et al., 2014a,b; Rodríguez-Delgado et al., 2016).
Laccase mediators HBT and SA are most often used in degradation of PPCPs. Degradation improvement upon mediator addition is significant in the enzymatic membrane reactor and limited in batch incubation (Nguyen et al., 2015). Different conversion mechanisms of triclosan, namely oligomerization in the presence of a laccase mediator and bond cleavage followed by dechlorination in the absence of a laccase mediator have been demonstrated (Murugesan et al., 2010). Laccase-mediated triclosan oligomerization has been confirmed, but dechlorination with only laccase has also been reported (Cabana et al., 2011). More studies comparing transformation pathways with or without a mediator should be carried out. Although, laccase most often has detoxifying effects, laccase and mediator pure preparations and mixture are toxic to bioluminescent bacteria (Nguyen et al., 2016b). Furthermore, inclusion of the natural mediator SA, but not synthetic mediator HBT, in laccase treatment of trace organic contaminants elevates effluent toxicity even though the target contaminants showed negligible toxicity at the low concentration (Nguyen et al., 2014a,c, 2016b).
Conclusions
Since the first discovery of laccase over 100 years ago, much has been elucidated about the occurrence, biochemistry, sequences, production, and application potentials of this diverse class of enzymes. We have briefly reviewed some recent developments in laccase research with a focus on production and applications in pharmaceutical degradation. Despite the exciting promise laccases bring, applied research is still mostly performed on the laboratory scale. Emphasis should be placed on augmenting the yields and efficiency and lowering application costs, which constitute bottlenecks in scalable and sustainable applications of laccases. While laccase mediators are widely used to aid laccase-catalyzed oxidation, the transformation metabolites as well as their toxicity should always be analyzed. It is clear that more work needs to be done to realize the full potential of these versatile enzymes.
Author Contributions
JY, TN, JL, and XY wrote the manuscript. JY, WL, and XD compiled the tables.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was funded by Natural Science Foundation of China (31671795) and Fujian Guidance Project (2016Y0059).
References
Abejón, R., Belleville, M. P., and Sanchez-Marcano, J. (2015a). Design, economic evaluation and optimization of enzymatic membrane reactors for antibiotics degradation in wastewaters. Sep. Purif. Technol. 156, 183–199. doi: 10.1016/j.seppur.2015.09.072
Abejón, R., De Cazes, M., Belleville, M. P., and Sanchez-Marcano, J. (2015b). Large-scale enzymatic membrane reactors for tetracycline degradation in WWTP effluents. Water Res. 73, 118–131. doi: 10.1016/j.watres.2015.01.012
Al-Adhami, A. J. H., Bryjak, J., Greb-Markiewicz, B., and Peczyńska-Czoch, W. (2002). Immobilization of wood-rotting fungi laccases on modified cellulose and acrylic carriers. Process Biochem. 37, 1387–1394. doi: 10.1016/S0032-9592(02)00023-7
Alcalde, M. (2015). Engineering the ligninolytic enzyme consortium. Trends Biotechnol. 33, 155–162. doi: 10.1016/j.tibtech.2014.12.007
Alves, A. M., Record, E., Lomascolo, A., Scholtmeijer, K., Asther, M., Wessels, J. G., et al. (2004). Highly efficient production of laccase by the basidiomycete Pycnoporus cinnabarinus. Appl. Environ. Microbiol. 70, 6379–6384. doi: 10.1128/AEM.70.11.6379-6384.2004
Antošovǎ, Z., and Sychrová, H. (2016). Yeast hosts for the production of recombinant laccases: a review. Mol. Biotechnol. 58, 93–116. doi: 10.1007/s12033-015-9910-1
Arca-Ramos, A., Kumar, V. V., Eibes, G., Moreira, M. T., and Cabana, H. (2016). Recyclable cross-linked laccase aggregates coupled to magnetic silica microbeads for elimination of pharmaceuticals from municipal wastewater. Environ. Sci. Pollut. Res. 23, 8929–8939. doi: 10.1007/s11356-016-6139-x
Arimoto, M., Yamagishi, K., Wang, J., Tanaka, K., Miyoshi, T., Kamei, I., et al. (2015). Molecular breeding of lignin-degrading brown-rot fungus Gloeophyllum trabeum by homologous expression of laccase gene. AMB Express 5, 81. doi: 10.1186/s13568-015-0173-9
Arora, D. S., and Sharma, R. K. (2010). Ligninolytic fungal laccases and their biotechnological applications. Appl. Biochem. Biotechnol. 160, 1760–1788. doi: 10.1007/s12010-009-8676-y
Arsenault, A., Cabana, H., and Jones, J. P. (2011). Laccase-based CLEAs: chitosan as a novel cross-linking agent. Enzyme Res. 2011:376015. doi: 10.4061/2011/376015
Asgher, M., Shahid, M., Kamal, S., and Iqbal, H. M. N. (2014). Recent trends and valorization of immobilization strategies and ligninolytic enzymes by industrial biotechnology. J. Mol. Catal. B Enzym. 101, 56–66. doi: 10.1016/j.molcatb.2013.12.016
Ashe, B., Nguyen, L. N., Hai, F. I., Lee, D.-J., van de Merwe, J. P., Leusch, F. D. L., et al. (2016). Impacts of redox-mediator type on trace organic contaminants degradation by laccase: degradation efficiency, laccase stability and effluent toxicity. Int. Biodeterior. Biodegradation 113, 169–176. doi: 10.1016/j.ibiod.2016.04.027
Ashrafi, S. D., Nasseri, S., Alimohammadi, M., Mahvi, A. H., and Faramarzi, M. A. (2015). Optimization of the enzymatic elimination of flumequine by laccase-mediated system using response surface methodology. Desalin. Water Treat. 57, 14478–14487. doi: 10.1080/19443994.2015.1063462
Ausec, L., Zakrzewski, M., Goesmann, A., Schlüter, A., and Mandic-Mulec, I. (2011). Bioinformatic analysis reveals high diversity of bacterial genes for laccase-like enzymes. PLoS ONE 6:e25724. doi: 10.1371/journal.pone.0025724
Ba, S., Arsenault, A., Hassani, T., Jones, J. P., and Cabana, H. (2013). Laccase immobilization and insolubilization: from fundamentals to applications for the elimination of emerging contaminants in wastewater treatment. Crit. Rev. Biotechnol. 33, 404–418. doi: 10.3109/07388551.2012.725390
Ba, S., Haroune, L., Cruz-Morato, C., Jacquet, C., Touahar, I. E., Bellenger, J. P., et al. (2014a). Synthesis and characterization of combined cross-linked laccase and tyrosinase aggregates transforming acetaminophen as a model phenolic compound in wastewaters. Sci. Total Environ. 487, 748–755. doi: 10.1016/j.scitotenv.2013.10.004
Ba, S., Jones, J. P., and Cabana, H. (2014b). Hybrid bioreactor (HBR) of hollow fiber microfilter membrane and cross-linked laccase aggregates eliminate aromatic pharmaceuticals in wastewaters. J. Hazard. Mater. 280, 662–670. doi: 10.1016/j.jhazmat.2014.08.062
Balasubramanian, V. K., Rai, K. M., Thu, S. W., Hii, M. M., and Mendu, V. (2016). Genome-wide identification of multifunctional laccase gene family in cotton (Gossypium spp.); expression and biochemical analysis during fiber development. Sci. Rep. 6:34309. doi: 10.1038/srep34309
Balcazar-Lopez, E., Mendez-Lorenzo, L. H., Batista-Garcia, R. A., Esquivel-Naranjo, U., Ayala, M., Kumar, V. V., et al. (2016). Xenobiotic compounds degradation by heterologous expression of a Trametes sanguineus laccase in Trichoderma atroviride. PLoS ONE 11:e0147997. doi: 10.1371/journal.pone.0147997
Baldrian, P. (2006). Laccases-occurrence and properties. FEMS Microbiol. Rev. 30, 215–242. doi: 10.1111/j.1574-4976.2005.00010.x
Bao, D., Gong, M., Zheng, H., Chen, M., Zhang, L., Wang, H., et al. (2013). Sequencing and comparative analysis of the straw mushroom (Volvariella volvacea) genome. PLoS ONE 8:e58294. doi: 10.1371/journal.pone.0058294
Beaudoin, J., Mercier, A., Langlois, R., and Labbé, S. (2003). The Schizosaccharomyces pombe Cuf1 is composed of functional modules from two distinct classes of copper metalloregulatory transcription factors. J. Biol. Chem. 278, 14565–14577. doi: 10.1074/jbc.M300861200
Becker, D., Varela Della Giustina, S., Rodriguez-Mozaz, S., Schoevaart, R., Barcelo, D., de Cazes, M., et al. (2016). Removal of antibiotics in wastewater by enzymatic treatment with fungal laccase - Degradation of compounds does not always eliminate toxicity. Bioresour. Technol. 219, 500–509. doi: 10.1016/j.biortech.2016.08.004
Beloqui, A., Pita, M., Polaina, J., Martínez-Arias, A., Golyshina, O. V., Zumárraga, M., et al. (2006). Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen: biochemical properties, structural analysis, and phylogenetic relationships. J. Biol. Chem. 281, 22933–22942. doi: 10.1074/jbc.M600577200
Betancor, L., Johnson, G. R., and Luckarift, H. R. (2013). Stabilized laccases as heterogeneous bioelectrocatalysts. ChemCatChem 5, 46–60. doi: 10.1002/cctc.201200611
Blánquez, A., Guillén, F., Rodríguez, J., Arias, M. E., and Hernández, M. (2016). The degradation of two fluoroquinolone based antimicrobials by SilA, an alkaline laccase from Streptomyces ipomoeae. World J. Microbiol. Biotechnol. 32, 52. doi: 10.1007/s11274-016-2032-5
Boonnorat, J., Techkarnjanaruk, S., Honda, R., and Prachanurak, P. (2016). Effects of hydraulic retention time and carbon to nitrogen ratio on micro-pollutant biodegradation in membrane bioreactor for leachate treatment. Bioresour. Technol. 219, 53–63. doi: 10.1016/j.biortech.2016.07.094
Cabana, H., Ahamed, A., and Leduc, R. (2011). Conjugation of laccase from the white rot fungus Trametes versicolor to chitosan and its utilization for the elimination of triclosan. Bioresour. Technol. 102, 1656–1662. doi: 10.1016/j.biortech.2010.09.080
Cabana, H., Jones, J. P., and Agathos, S. N. (2007). Elimination of endocrine disrupting chemicals using white rot fungi and their lignin modifying enzymes: a review. Eng. Life Sci. 7, 429–456. doi: 10.1002/elsc.200700017
Cai, X., Davis, E. J., Ballif, J., Liang, M., Bushman, E., Haroldsen, V., et al. (2006). Mutant identification and characterization of the laccase gene family in Arabidopsis. J. Exp. Bot. 57, 2563–2569. doi: 10.1093/jxb/erl022
Cañas, A. I., and Camarero, S. (2010). Laccases and their natural mediators: biotechnological tools for sustainable eco-friendly processes. Biotechnol. Adv. 28, 694–705. doi: 10.1016/j.biotechadv.2010.05.002
Castanera, R., Omarini, A., Santoyo, F., Pérez, G., Pisabarro, A. G., and Ramírez, L. (2013). Non-additive transcriptional profiles underlie dikaryotic superiority in Pleurotus ostreatus laccase activity. PLoS ONE 8:e73282. doi: 10.1371/journal.pone.0073282
Castanera, R., Péreza, G., Omarini, A., Alfaro, M., Pisabarro, A. G., Faraco, V., et al. (2012). Transcriptional and enzymatic profiling of Pleurotus ostreatus laccase genes in submerged and solid-state fermentation cultures. Appl. Environ. Microbiol. 78, 4037–4045. doi: 10.1128/AEM.07880-11
Catherine, H., Penninckx, M., and Frédéric, D. (2016). Product formation from phenolic compounds removal by laccases: a review. Environ. Technol. Innov. 5, 250–266. doi: 10.1016/j.eti.2016.04.001
Chandra, R., and Chowdhary, P. (2015). Properties of bacterial laccases and their application in bioremediation of industrial wastes. Environ. Sci. Process. Impacts 17, 326–342. doi: 10.1039/C4EM00627E
Chang, B.-V., and Ren, Y.-L. (2015). Biodegradation of three tetracyclines in river sediment. Ecol. Eng. 75, 272–277. doi: 10.1016/j.ecoleng.2014.11.039
Chang, Y. T., Lee, J. F., Liu, K. H., Liao, Y. F., and Yang, V. (2016). Immobilization of fungal laccase onto a nonionic surfactant-modified clay material: application to PAH degradation. Environ. Sci. Pollut. Res. 23, 4024–4035. doi: 10.1007/s11356-015-4248-6
Chen, S.-C., Wu, P.-H., Su, Y.-C., Wen, T.-N., Wei, Y.-S., Wang, N.-C., et al. (2012). Biochemical characterization of a novel laccase from the basidiomycete fungus Cerrena sp. WR1. Protein Eng. Des. Sel. 25, 761–769. doi: 10.1093/protein/gzs082
Chen, Y., Stemple, B., Kumar, M., and Wei, N. (2016). Cell surface display fungal laccase as a renewable biocatalyst for degradation of persistent micropollutants bisphenol A and sulfamethoxazole. Environ. Sci. Technol. 50, 8799–8808. doi: 10.1021/acs.est.6b01641
Chhabra, M., Mishra, S., and Sreekrishnan, T. R. (2009). Laccase/mediator assisted degradation of triarylmethane dyes in a continuous membrane reactor. J. Biotechnol. 143, 69–78. doi: 10.1016/j.jbiotec.2009.06.011
Cho, H. Y., Lee, C., Hwang, S.-G., Park, Y. C., Lim, H. L., and Jang, C. S. (2014). Overexpression of the OsChI1 gene, encoding a putative laccase precursor, increases tolerance to drought and salinity stress in transgenic Arabidopsis. Gene 552, 98–105. doi: 10.1016/j.gene.2014.09.018
Coconi-Linares, N., Ortiz-Vazquez, E., Fernandez, F., Loske, A. M., and Gomez-Lim, M. A. (2015). Recombinant expression of four oxidoreductases in Phanerochaete chrysosporium improves degradation of phenolic and non-phenolic substrates. J. Biotechnol. 209, 76–84. doi: 10.1016/j.jbiotec.2015.06.401
Courty, P. E., Hoegger, P. J., Kilaru, S., Kohler, A., Buée, M., Garbaye, J., et al. (2009). Phylogenetic analysis, genomic organization, and expression analysis of multi-copper oxidases in the ectomycorrhizal basidiomycete Laccaria bicolor. New Phytol. 182, 736–750. doi: 10.1111/j.1469-8137.2009.02774.x
Crowe, J. D., and Olsson, S. (2001). Induction of laccase activity in Rhizoctonia solani by antagonistic Pseudomonas fluorescens strains and a range of chemical treatments. Appl. Environ. Microbiol. 67, 2088–2094. doi: 10.1128/AEM.67.5.2088-2094.2001
Daâssi, D., Prieto, A., Zouari-Mechichi, H., Martínez, M. J., Nasri, M., and Mechichi, T. (2016). Degradation of bisphenol A by different fungal laccases and identification of its degradation products. Int. Biodeterior. Biodegradation 110, 181–188. doi: 10.1016/j.ibiod.2016.03.017
de Cazes, M., Belleville, M. P., Mougel, M., Kellner, H., and Sanchez-Marcano, J. (2015). Characterization of laccase-grafted ceramic membranes for pharmaceuticals degradation. J. Membr. Sci. 476, 384–393. doi: 10.1016/j.memsci.2014.11.044
de Cazes, M., Belleville, M. P., Petit, E., Llorca, M., Rodríguez-Mozaz, S., de Gunzburg, J., et al. (2014). Design and optimization of an enzymatic membrane reactor for tetracycline degradation. Catal. Today 236, 146–152. doi: 10.1016/j.cattod.2014.02.051
Debaste, F., Songulashvili, G., and Penninckx, M. J. (2014). The potential of Cerrena unicolor laccase immobilized on mesoporous silica beads for removal of organic micropollutants in wastewaters. Desalin. Water Treat. 52, 2344–2347. doi: 10.1080/19443994.2013.877851
del Vecchio, C., Lettera, V., Pezzella, C., Piscitelli, A., Leo, G., Birolo, L., et al. (2012). Classical breeding in Pleurotus ostreatus: a natural approach for laccase production improvement. Biocatal. Biotransformation 30, 78–85. doi: 10.3109/10242422.2012.646032
Demarche, P., Junghanns, C., Nair, R. R., and Agathos, S. N. (2012). Harnessing the power of enzymes for environmental stewardship. Biotechnol. Adv. 30, 933–953. doi: 10.1016/j.biotechadv.2011.05.013
Ding, H., Wu, Y., Zou, B., Lou, Q., Zhang, W., Zhong, J., et al. (2016). Simultaneous removal and degradation characteristics of sulfonamide, tetracycline, and quinolone antibiotics by laccase-mediated oxidation coupled with soil adsorption. J. Hazard. Mater. 307, 350–358. doi: 10.1016/j.jhazmat.2015.12.062
Dittmer, N. T., and Kanost, M. R. (2010). Insect multicopper oxidases: diversity, properties, and physiological roles. Insect Biochem. Mol. Biol. 40, 179–188. doi: 10.1016/j.ibmb.2010.02.006
Domaradzka, D., Guzik, U., and Wojcieszyńska, D. (2015). Biodegradation and biotransformation of polycyclic non-steroidal anti-inflammatory drugs. Rev. Environ. Sci. Biotechnol. 14, 229–239. doi: 10.1007/s11157-015-9364-8
D'Souza, D. T., Tiwari, R., Sah, A. K., and Raghukumar, C. (2006). Enhanced production of laccase by a marine fungus during treatment of colored effluents and synthetic dyes. Enzyme Microb. Technol. 38, 504–511. doi: 10.1016/j.enzmictec.2005.07.005
Dwivedi, U. N., Singh, P., Pandey, V. P., and Kumar, A. (2011). Structure–function relationship among bacterial, fungal and plant laccases. J. Mol. Catal. B Enzym. 68, 117–128. doi: 10.1016/j.molcatb.2010.11.002
Eibes, G., Debernardi, G., Feijoo, G., Moreira, M. T., and Lema, J. M. (2011). Oxidation of pharmaceutically active compounds by a ligninolytic fungal peroxidase. Biodegradation 22, 539–550. doi: 10.1007/s10532-010-9426-0
Elisashvili, V., and Kachlishvili, E. (2009). Physiological regulation of laccase and manganese peroxidase production by white-rot Basidiomycetes. J. Biotechnol. 144, 37–42. doi: 10.1016/j.jbiotec.2009.06.020
Elisashvili, V., Kachlishvili, E., Khardziani, T., and Agathos, S. N. (2010). Effect of aromatic compounds on the production of laccase and manganese peroxidase by white-rot basidiomycetes. J. Ind. Microbiol. Biotechnol. 37, 1091–1096. doi: 10.1007/s10295-010-0757-y
Ergün, B. G., and Çalık, P. (2016). Lignocellulose degrading extremozymes produced by Pichia pastoris: current status and future prospects. Bioprocess Biosyst. Eng. 39, 1–36. doi: 10.1007/s00449-015-1476-6
Fan, X., Zhou, Y., Xiao, Y., Xu, Z., and Bian, Y. (2014). Cloning, expression and phylogenetic analysis of a divergent laccase multigene family in Auricularia auricula-judae. Microbiol. Res. 169, 453–462. doi: 10.1016/j.micres.2013.08.004
Fang, F., Zhang, X. L., Luo, H. H., Zhou, J. J., Gong, Y. H., Li, W. J., et al. (2015). An intracellular laccase is responsible for epicatechin-mediated anthocyanin degradation in litchi fruit pericarp. Plant Physiol. 169, 2391–2408. doi: 10.1104/pp.15.00359
Fang, Z., Li, T., Wang, Q., Zhang, X., Peng, H., Fang, W., et al. (2011). A bacterial laccase from marine microbial metagenome exhibiting chloride tolerance and dye decolorization ability. Appl. Microbiol. Biotechnol. 89, 1103–1110. doi: 10.1007/s00253-010-2934-3
Fang, Z., Liu, X., Chen, L., Shen, Y., Zhang, X., Fang, W., et al. (2015). Identification of a laccase Glac15 from Ganoderma lucidum 77002 and its application in bioethanol production. Biotechnol. Biofuels 8, 54. doi: 10.1186/s13068-015-0235-x
Fang, Z. M., Li, T. L., Chang, F., Zhou, P., Fang, W., Hong, Y. Z., et al. (2012). A new marine bacterial laccase with chloride-enhancing, alkaline-dependent activity and dye decolorization ability. Bioresour. Technol. 111, 36–41. doi: 10.1016/j.biortech.2012.01.172
Fernandez-Alejandre, K. I., Flores, N., Tinoco-Valencia, R., Caro, M., Flores, C., Galindo, E., et al. (2016). Diffusional and transcriptional mechanisms involved in laccases production by Pleurotus ostreatus CP50. J. Biotechnol. 223, 42–49. doi: 10.1016/j.jbiotec.2016.02.029
Fernández-Fernández, M., Sanromán, M. Á., and Moldes, D. (2013). Recent developments and applications of immobilized laccase. Biotechnol. Adv. 31, 1808–1825. doi: 10.1016/j.biotechadv.2012.02.013
Fillat, U., Prieto, A., Camarero, S., Martínez, Á. T., and Martínez, M. J. (2012). Biodeinking of flexographic inks by fungal laccases using synthetic and natural mediators. Biochem. Eng. J. 67, 97–103. doi: 10.1016/j.bej.2012.05.010
Flores, C., Vidal, C., Trejo-Hernandez, M. R., Galindo, E., and Serrano-Carreon, L. (2009). Selection of Trichoderma strains capable of increasing laccase production by Pleurotus ostreatus and Agaricus bisporus in dual cultures. J. Appl. Microbiol. 106, 249–257. doi: 10.1111/j.1365-2672.2008.03998.x
Floudas, D., Binder, M., Riley, R., Barry, K., Blanchette, R. A., Henrissat, B., et al. (2012). The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336, 1715–1719. doi: 10.1126/science.1221748
Forootanfar, H., and Faramarzi, M. A. (2015). Insights into laccase producing organisms, fermentation states, purification strategies, and biotechnological applications. Biotechnol. Prog. 31, 1443–1463. doi: 10.1002/btpr.2173
Forootanfar, H., Rezaei, S., Zeinvand-Lorestani, H., Tahmasbi, H., Mogharabi, M., Ameri, A., et al. (2016). Studies on the laccase-mediated decolorization, kinetic, and microtoxicity of some synthetic azo dyes. J. Environ. Health Sci. Eng. 14, 7. doi: 10.1186/s40201-016-0248-9
Garcia, H. A., Hoffman, C. M., Kinney, K. A., and Lawler, D. F. (2011). Laccase-catalyzed oxidation of oxybenzone in municipal wastewater primary effluent. Water Res. 45, 1921–1932. doi: 10.1016/j.watres.2010.12.027
Garcia-Morales, R., Rodriguez-Delgado, M., Gomez-Mariscal, K., Orona-Navar, C., Hernandez-Luna, C., Torres, E., et al. (2015). Biotransformation of endocrine-disrupting compounds in groundwater: bisphenol A, nonylphenol, ethynylestradiol and triclosan by a laccase cocktail from Pycnoporus sanguineus CS43. Water Air Soil Pollut. 226, 251. doi: 10.1007/s11270-015-2514-3
Gasser, C. A., Ammann, E. M., Shahgaldian, P., and Corvini, P. F. (2014). Laccases to take on the challenge of emerging organic contaminants in wastewater. Appl. Microbiol. Biotechnol. 98, 9931–9952. doi: 10.1007/s00253-014-6177-6
Giardina, P., Faraco, V., Pezzella, C., Piscitelli, A., Vanhulle, S., and Sannia, G. (2010). Laccases: a never-ending story. Cell. Mol. Life Sci. 67, 369–385. doi: 10.1007/s00018-009-0169-1
Grover, N., Dinu, C. Z., Kane, R. S., and Dordick, J. S. (2013). Enzyme-based formulations for decontamination: current state and perspectives. Appl. Microbiol. Biotechnol. 97, 3293–3300. doi: 10.1007/s00253-013-4797-x
Gu, C., Zheng, F., Long, L., Wang, J., and Ding, S. (2014). Engineering the expression and characterization of two novel laccase isoenzymes from Coprinus comatus in Pichia pastoris by fusing an additional ten amino acids tag at N-terminus. PLoS ONE 9:e93912. doi: 10.1371/journal.pone.0093912
Gulkowska, A., Krauss, M., Rentsch, D., and Hollender, J. (2012). Reactions of a sulfonamide antimicrobial with model humic constituents: assessing pathways and stability of covalent bonding. Environ. Sci. Technol. 46, 2102–2111. doi: 10.1021/es202272w
Gulkowska, A., Sander, M., Hollender, J., and Krauss, M. (2013). Covalent binding of sulfamethazine to natural and synthetic humic acids: assessing laccase catalysis and covalent bond stability. Environ. Sci. Technol. 47, 6916–6924. doi: 10.1021/es3044592
Guo, X., Li, J., Yang, F., Yang, J., and Yin, D. (2014). Prevalence of sulfonamide and tetracycline resistance genes in drinking water treatment plants in the Yangtze River Delta, China. Sci. Total Environ. 493, 626–631. doi: 10.1016/j.scitotenv.2014.06.035
Hata, T., Shintate, H., Kawai, S., Okamura, H., and Nishida, T. (2010). Elimination of carbamazepine by repeated treatment with laccase in the presence of 1-hydroxybenzotriazole. J. Hazard. Mater. 181, 1175–1178. doi: 10.1016/j.jhazmat.2010.05.103
Hibi, M., Hatahira, S., Nakatani, M., Yokozeki, K., Shimizu, S., and Ogawa, J. (2012). Extracellular oxidases of Cerrena sp. complementarily functioning in artificial dye decolorization including laccase, manganese peroxidase, and novel versatile peroxidases. Biocatal. Agric. Biotechnol. 1, 220–225. doi: 10.1016/j.bcab.2012.03.003
Hoegger, P. J., Kilaru, S., James, T. Y., Thacker, J. R., and Kües, U. (2006). Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 273, 2308–2326. doi: 10.1111/j.1742-4658.2006.05247.x
Hoegger, P. J., Navarro-Gonzalez, M., Kilaru, S., Hoffmann, M., Westbrook, E. D., and Kues, U. (2004). The laccase gene family in Coprinopsis cinerea (Coprinus cinereus). Curr. Genet. 45, 9–18. doi: 10.1007/s00294-003-0452-x
Hofmann, U., and Schlosser, D. (2016). Biochemical and physicochemical processes contributing to the removal of endocrine-disrupting chemicals and pharmaceuticals by the aquatic ascomycete Phoma sp. UHH 5-1-03. Appl. Microbiol. Biotechnol. 100, 2381–2399. doi: 10.1007/s00253-015-7113-0
Hong, F., Meinander, N. Q., and Jönsson, L. J. (2002). Fermentation strategies for improved heterologous expression of laccase in Pichia pastoris. Biotechnol. Bioeng. 79, 438–449. doi: 10.1002/bit.10297
Hong, Y. Z., Zhou, H. M., Tu, X. M., Li, J. F., and Xiao, Y. Z. (2007). Cloning of a laccase gene from a novel basidiomycete Trametes sp. 420 and its heterologous expression in Pichia pastoris. Curr. Microbiol. 54, 260–265. doi: 10.1007/s00284-006-0068-8
Hu, M. R., Chao, Y. P., Zhang, G. Q., Xue, Z. Q., and Qian, S. (2009). Laccase-mediator system in the decolorization of different types of recalcitrant dyes. J. Ind. Microbiol. Biotechnol. 36, 45–51. doi: 10.1007/s10295-008-0471-1
Huang, M. T., Lu, Y. C., Zhang, S., Luo, F., and Yang, H. (2016). Rice (Oryza sativa) laccases involved in modification and detoxification of herbicides atrazine and isoproturon residues in plants. J. Agric. Food. Chem. 64, 6397–6406. doi: 10.1021/acs.jafc.6b02187
Husain, Q., and Qayyum, S. (2012). Biological and enzymatic treatment of bisphenol A and other endocrine disrupting compounds: a review. Crit. Rev. Biotechnol. 33, 260–292. doi: 10.3109/07388551.2012.694409
Iracheta-Cárdenas, M. M., Rocha-Peña, M. A., Galán-Wong, L. J., Arévalo-Niño, K., and Tovar-Herrera, O. E. (2016). A Pycnoporus sanguineus laccase for denim bleaching and its comparison with an enzymatic commercial formulation. J. Environ. Manage. 177, 93–100. doi: 10.1016/j.jenvman.2016.04.008
Janusz, G., Kucharzyk, K. H., Pawlik, A., Staszczak, M., and Paszczynski, A. J. (2013). Fungal laccase, manganese peroxidase and lignin peroxidase: gene expression and regulation. Enzyme Microb. Technol. 52, 1–12. doi: 10.1016/j.enzmictec.2012.10.003
Janusz, G., Rogalski, J., and Szczodrak, J. (2007). Increased production of laccase by Cerrena unicolor in submerged liquid cultures. World J. Microbiol. Biotechnol. 23, 1459–1464. doi: 10.1007/s11274-007-9390-y
Jelic, A., Cruz-Morato, C., Marco-Urrea, E., Sarra, M., Perez, S., Vicent, T., et al. (2012). Degradation of carbamazepine by Trametes versicolor in an air pulsed fluidized bed bioreactor and identification of intermediates. Water Res. 46, 955–964. doi: 10.1016/j.watres.2011.11.063
Jeon, J. R., and Chang, Y. S. (2013). Laccase-mediated oxidation of small organics: bifunctional roles for versatile applications. Trends Biotechnol. 31, 335–341. doi: 10.1016/j.tibtech.2013.04.002
Jeon, J. R., Baldrian, P., Murugesan, K., and Chang, Y. S. (2012). Laccase-catalysed oxidations of naturally occurring phenols: from in vivo biosynthetic pathways to green synthetic applications. Microb. Biotechnol. 5, 318–332. doi: 10.1111/j.1751-7915.2011.00273.x
Ji, C., Hou, J., and Chen, V. (2016a). Cross-linked carbon nanotubes-based biocatalytic membranes for micro-pollutants degradation: performance, stability, and regeneration. J. Membr. Sci. 520, 869–880. doi: 10.1016/j.memsci.2016.08.056
Ji, C., Hou, J., Wang, K., Zhang, Y., and Chen, V. (2016b). Biocatalytic degradation of carbamazepine with immobilized laccase-mediator membrane hybrid reactor. J. Membr. Sci. 502, 11–20. doi: 10.1016/j.memsci.2015.12.043
Jiang, N., Sun, N., Xiao, D., Pan, J., Wang, Y., and Zhu, X. (2009). A copper-responsive factor gene CUF1 is required for copper induction of laccase in Cryptococcus neoformans. FEMS Microbiol. Lett. 296, 84–90. doi: 10.1111/j.1574-6968.2009.01619.x
Jin, X., Yu, X., Zhu, G., Zheng, Z., Feng, F., and Zhang, Z. (2016). Conditions optimizing and application of laccase-mediator system (LMS) for the Laccase-catalyzed pesticide degradation. Sci. Rep. 6:35787. doi: 10.1038/srep35787
Jones, S. M., and Solomon, E. I. (2015). Electron transfer and reaction mechanism of laccases. Cell. Mol. Life Sci. 72, 869–883. doi: 10.1007/s00018-014-1826-6
Kajita, S., Sugawara, S., Miyazaki, Y., Nakamura, M., Katayama, Y., Shishido, K., et al. (2004). Overproduction of recombinant laccase using a homologous expression system in Coriolus versicolor. Appl. Microbiol. Biotechnol. 66, 194–199. doi: 10.1007/s00253-004-1663-x
Kandasamy, S., Muniraj, I. K., Purushothaman, N., Sekar, A., Sharmila, D. J., Kumarasamy, R., et al. (2016). High level secretion of laccase (LccH) from a newly isolated white-rot basidiomycete, Hexagonia hirta MSF2. Front. Microbiol. 7:707. doi: 10.3389/fmicb.2016.00707
Karaki, N., Aljawish, A., Humeau, C., Muniglia, L., and Jasniewski, J. (2016). Enzymatic modification of polysaccharides: mechanisms, properties, and potential applications: a review. Enzyme Microb. Technol. 90, 1–18. doi: 10.1016/j.enzmictec.2016.04.004
Kilaru, S., Hoegger, P., and Kües, U. (2006a). The laccase multi-gene family in Coprinopsis cinerea has seventeen different members that divide into two distinct subfamilies. Curr. Genet. 50, 45–60. doi: 10.1007/s00294-006-0074-1
Kilaru, S., Hoegger, P. J., Majcherczyk, A., Burns, C., Shishido, K., Bailey, A., et al. (2006b). Expression of laccase gene lcc1 in Coprinopsis cinerea under control of various basidiomycetous promoters. Appl. Microbiol. Biotechnol. 71, 200–210. doi: 10.1007/s00253-005-0128-1
Kim, E. Y., Seo, Y. S., Park, K. Y., Kim, S. J., and Kim, W. T. (2014). Overexpression of CaDSR6 increases tolerance to drought and salt stresses in transgenic Arabidopsis plants. Gene 552, 146–154. doi: 10.1016/j.gene.2014.09.028
Kittl, R., Mueangtoom, K., Gonaus, C., Khazaneh, S. T., Sygmund, C., Haltrich, D., et al. (2012). A chloride tolerant laccase from the plant pathogen ascomycete Botrytis aclada expressed at high levels in Pichia pastoris. J. Biotechnol. 157, 304–314. doi: 10.1016/j.jbiotec.2011.11.021
Kudanga, T., and Roes-Hill, M. L. (2014). Laccase applications in biofuels production: current status and future prospects. Appl. Microbiol. Biotechnol. 98, 6525–6542. doi: 10.1007/s00253-014-5810-8
Kudanga, T., Nyanhongo, G. S., Guebitz, G. M., and Burtona, S. (2011). Potential applications of laccase-mediated coupling and grafting reactions: a review. Enzyme Microb. Technol. 48, 195–208. doi: 10.1016/j.enzmictec.2010.11.007
Kües, U. (2015). Fungal enzymes for environmental management. Curr. Opin. Biotechnol. 33, 268–278. doi: 10.1016/j.copbio.2015.03.006
Kües, U., and Rühl, M. (2011). Multiple multi-copper oxidase gene families in basidiomycetes – what for? Curr. Genomics 12, 72–94. doi: 10.2174/138920211795564377
Kumar, V. V., and Cabana, H. (2016). Towards high potential magnetic biocatalysts for on-demand elimination of pharmaceuticals. Bioresour. Technol. 200, 81–89. doi: 10.1016/j.biortech.2015.09.100
Kumar, V. V., Sivanesan, S., and Cabana, H. (2014). Magnetic cross-linked laccase aggregates — bioremediation tool for decolorization of distinct classes of recalcitrant dyes. Sci. Total Environ. 487, 830–839. doi: 10.1016/j.scitotenv.2014.04.009
Kümmerer, K. (2009). Antibiotics in the aquatic environment – a review – Part II. Chemosphere 75, 417–434. doi: 10.1016/j.chemosphere.2008.11.086
Kurniawati, S., and Nicell, J. A. (2007). Efficacy of mediators for enhancing the laccase-catalyzed oxidation of aqueous phenol. Enzyme Microb. Technol. 41, 353–361. doi: 10.1016/j.enzmictec.2007.03.003
Larsson, D. G. (2014). Antibiotics in the environment. Ups. J. Med. Sci. 119, 108–112. doi: 10.3109/03009734.2014.896438
Le, T. T., Murugesan, K., Lee, C. S., Vu, C. H., Chang, Y. S., and Jeon, J. R. (2016). Degradation of synthetic pollutants in real wastewater using laccase encapsulated in core-shell magnetic copper alginate beads. Bioresour. Technol. 216, 203–210. doi: 10.1016/j.biortech.2016.05.077
Lettera, V., Del Vecchio, C., Piscitelli, A., and Sannia, G. (2011). Low impact strategies to improve ligninolytic enzyme production in filamentous fungi: the case of laccase in Pleurotus ostreatus. C. R. Biol. 334, 781–788. doi: 10.1016/j.crvi.2011.06.001
Li, P., Wang, H., Liu, G., Li, X., and Yao, J. (2011). The effect of carbon source succession on laccase activity in the co-culture process of Ganoderma lucidum and a yeast. Enzyme Microb. Technol. 48, 1–6. doi: 10.1016/j.enzmictec.2010.07.005
Li, X., Xu, Q.-M., Cheng, J.-S., and Yuan, Y.-J. (2016). Improving the bioremoval of sulfamethoxazole and alleviating cytotoxicity of its biotransformation by laccase producing system under coculture of Pycnoporus sanguineus and Alcaligenes faecalis. Bioresour. Technol. 220, 333–340. doi: 10.1016/j.biortech.2016.08.088
Liang, M., Davis, E., Gardner, D., Cai, X., and Wu, Y. (2006). Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta 224, 1185–1196. doi: 10.1007/s00425-006-0300-6
Librando, V., and Pappalardo, M. (2013). In silico bioremediation of polycyclic aromatic hydrocarbon: a frontier in environmental chemistry. J. Mol. Graph. Model. 44, 1–8. doi: 10.1016/j.jmgm.2013.04.011
Liebeton, K., Lengefeld, J., and Eck, J. (2014). The nucleotide composition of the spacer sequence influences the expression yield of heterologously expressed genes in Bacillus subtilis. J. Biotechnol. 191, 214–220. doi: 10.1016/j.jbiotec.2014.06.027
Lisova, Z. A., Lisov, A. V., and Leontievsky, A. A. (2010). Two laccase isoforms of the basidiomycete Cerrena unicolor VKMF-3196. Induction, isolation and properties. J. Basic Microbiol. 50, 72–82. doi: 10.1002/jobm.200900382
Liu, J., Tan, L., Wang, J., Wang, Z., Ni, H., and Li, L. (2016). Complete biodegradation of chlorpyrifos by engineered Pseudomonas putida cells expressing surface-immobilized laccases. Chemosphere 157, 200–207. doi: 10.1016/j.chemosphere.2016.05.031
Liu, Z., Zhang, D., Hua, Z., Li, J., Du, G., and Chen, J. (2010). Improvement of laccase production and its properties by low-energy ion implantation. Bioprocess Biosyst. Eng. 33, 639–646. doi: 10.1007/s00449-009-0389-7
Llorca, M., Gros, M., Rodriguez-Mozaz, S., and Barcelo, D. (2014). Sample preservation for the analysis of antibiotics in water. J. Chromatogr. A 1369, 43–51. doi: 10.1016/j.chroma.2014.09.089
Llorca, M., Rodríguez-Mozaz, S., Couillerot, O., Panigoni, K., de Gunzburg, J., Bayer, S., et al. (2015). Identification of new transformation products during enzymatic treatment of tetracycline and erythromycin antibiotics at laboratory scale by an on-line turbulent flow liquid-chromatography coupled to a high resolution mass spectrometer LTQ-Orbitrap. Chemosphere 119, 90–98. doi: 10.1016/j.chemosphere.2014.05.072
Lloret, L., Eibes, G., Lú-Chau, T. A., Moreira, M. T., Feijoo, G., and Lema, J. M. (2010). Laccase-catalyzed degradation of anti-inflammatories and estrogens. Biochem. Eng. J. 51, 124–131. doi: 10.1016/j.bej.2010.06.005
Loi, M., Fanelli, F., Zucca, P., Liuzzi, V. C., Quintieri, L., Cimmarusti, M. T., et al. (2016). Aflatoxin B1 and M1 degradation by Lac2 from Pleurotus pulmonarius and redox mediators. Toxins 8:E245. doi: 10.3390/toxins8090245
Lu, X., and Ding, S. (2010). Effect of Cu2+, Mn2+ and aromatic compounds on the production of laccase isoforms by Coprinus comatus. Mycoscience 51, 68–74. doi: 10.1007/S10267-009-0002-6
Lu, Y., Wu, G., Lian, L., Guo, L., Wang, W., Yang, Z., et al. (2015). Cloning and expression analysis of Vvlcc3, a novel and functional laccase gene possibly involved in stipe elongation. Int. J. Mol. Sci. 16, 28498–28509. doi: 10.3390/ijms161226111
Majeau, J. A., Brar, S. K., and Tyagi, R. D. (2010). Laccases for removal of recalcitrant and emerging pollutants. Bioresour. Technol. 101, 2331–2350. doi: 10.1016/j.biortech.2009.10.087
Maqbool, Z., Hussain, S., Imran, M., Mahmood, F., Shahzad, T., Ahmed, Z., et al. (2016). Perspectives of using fungi as bioresource for bioremediation of pesticides in the environment: a critical review. Environ. Sci. Pollut. Res. 23, 16904–16925. doi: 10.1007/s11356-016-7003-8
Marco-Urrea, E., Pérez-Trujillo, M., Cruz-Morató, C., Caminal, G., and Vicent, T. (2010a). White-rot fungus-mediated degradation of the analgesic ketoprofen and identification of intermediates by HPLC–DAD–MS and NMR. Chemosphere 78, 474–481. doi: 10.1016/j.chemosphere.2009.10.009
Marco-Urrea, E., Perez-Trujillo, M., Cruz-Morato, C., Caminal, G., and Vicent, T. (2010b). Degradation of the drug sodium diclofenac by Trametes versicolor pellets and identification of some intermediates by NMR. J. Hazard. Mater. 176, 836–842. doi: 10.1016/j.jhazmat.2009.11.112
Marco-Urrea, E., Perez-Trujillo, M., Vicent, T., and Caminal, G. (2009). Ability of white-rot fungi to remove selected pharmaceuticals and identification of degradation products of ibuprofen by Trametes versicolor. Chemosphere 74, 765–772. doi: 10.1016/j.chemosphere.2008.10.040
Margot, J., Bennati-Granier, C., Maillard, J., Blánquez, P., Barry, D. A., and Holliger, C. (2013a). Bacterial versus fungal laccase: potential for micropollutant degradation. AMB Express 3:63. doi: 10.1186/2191-0855-3-63
Margot, J., Copin, P.-J., von Gunten, U., Barry, D. A., and Holliger, C. (2015). Sulfamethoxazole and isoproturon degradation and detoxification by a laccase-mediator system: influence of treatment conditions and mechanistic aspects. Biochem. Eng. J. 103, 47–59. doi: 10.1016/j.bej.2015.06.008
Margot, J., Maillard, J., Rossi, L., Barry, D. A., and Holliger, C. (2013b). Influence of treatment conditions on the oxidation of micropollutants by Trametes versicolor laccase. N. Biotechnol. 30, 803–813. doi: 10.1016/j.nbt.2013.06.004
Martinkova, L., Kotik, M., Markova, E., and Homolka, L. (2016). Biodegradation of phenolic compounds by Basidiomycota and its phenol oxidases: a review. Chemosphere 149, 373–382. doi: 10.1016/j.chemosphere.2016.01.022
Martins, L. O., Durao, P., Brissos, V., and Lindley, P. F. (2015). Laccases of prokaryotic origin: enzymes at the interface of protein science and protein technology. Cell. Mol. Life Sci. 72, 911–922. doi: 10.1007/s00018-014-1822-x
Maryskova, M., Ardao, I., Garcia-Gonzalez, C. A., Martinova, L., Rotkova, J., and Sevcu, A. (2016). Polyamide 6/chitosan nanofibers as support for the immobilization of Trametes versicolor laccase for the elimination of endocrine disrupting chemicals. Enzyme Microb. Technol. 89, 31–38. doi: 10.1016/j.enzmictec.2016.03.001
Mate, D. M., and Alcalde, M. (2015). Laccase engineering: from rational design to directed evolution. Biotechnol. Adv. 33, 25–40. doi: 10.1016/j.biotechadv.2014.12.007
Mate, D. M., and Alcalde, M. (2016). Laccase: a multi-purpose biocatalyst at the forefront of biotechnology. Microb. Biotechnol. doi: 10.1111/1751-7915.12422. [Epub ahead print].
Matuszewska, A., Karp, M., Jaszek, M., Janusz, G., Osinska-Jaroszuk, M., Sulej, J., et al. (2016). Laccase purified from Cerrena unicolor exerts antitumor activity against leukemic cells. Oncol. Lett. 11, 2009–2018. doi: 10.3892/ol.2016.4220
Melo, C. F., Dezotti, M., and Marques, M. R. (2016). A comparison between the oxidation with laccase and horseradish peroxidase for triclosan conversion. Environ. Technol. 37, 335–343. doi: 10.1080/09593330.2015.1069897
Michniewicz, A., Ullrich, R., Ledakowicz, S., and Hofrichter, M. (2006). The white-rot fungus Cerrena unicolor strain 137 produces two laccase isoforms with different physico-chemical and catalytic properties. Appl. Microbiol. Biotechnol. 69, 682–688. doi: 10.1007/s00253-005-0015-9
Migliore, L., Fiori, M., Spadoni, A., and Galli, E. (2012). Biodegradation of oxytetracycline by Pleurotus ostreatus mycelium: a mycoremediation technique. J. Hazard. Mater. 215–216, 227–232. doi: 10.1016/j.jhazmat.2012.02.056
Mikolasch, A., Hildebrandt, O., Schluter, R., Hammer, E., Witt, S., and Lindequist, U. (2016). Targeted synthesis of novel α-lactam antibiotics by laccase-catalyzed reaction of aromatic substrates selected by pre-testing for their antimicrobial and cytotoxic activity. Appl. Microbiol. Biotechnol. 100, 4885–4899. doi: 10.1007/s00253-016-7288-z
Mikolasch, A., Manda, K., Schluter, R., Lalk, M., Witt, S., Seefeldt, S., et al. (2012). Comparative analyses of laccase-catalyzed amination reactions for production of novel α-lactam antibiotics. Biotechnol. Appl. Biochem. 59, 295–306. doi: 10.1002/bab.1026
Mir-Tutusaus, J. A., Masis-Mora, M., Corcellas, C., Eljarrat, E., Barcelo, D., Sarra, M., et al. (2014). Degradation of selected agrochemicals by the white rot fungus Trametes versicolor. Sci. Total Environ. 500–501, 235–242. doi: 10.1016/j.scitotenv.2014.08.116
Mizerska-Dudka, M., Jaszek, M., Blachowicz, A., Rejczak, T. P., Matuszewska, A., Osinska-Jaroszuk, M., et al. (2015). Fungus Cerrena unicolor as an effective source of new antiviral, immunomodulatory, and anticancer compounds. Int. J. Biol. Macromol. 79, 459–468. doi: 10.1016/j.ijbiomac.2015.05.015
Mogharabi, M., and Faramarzi, M. A. (2014). Laccase and laccase-mediated systems in the synthesis of organic compounds. Adv. Synth. Catal. 356, 897–927. doi: 10.1002/adsc.201300960
Morozova, O. V., Shumakovich, G. P., Shleev, S. V., and Yaropolov, Y. I. (2007). Laccase-mediator systems and their applications: a review. Appl. Biochem. Microbiol. 43, 523–535. doi: 10.1134/S0003683807050055
Muraguchi, H., Kondoh, M., Ito, Y., and Yanagi, S. O. (2011). Molecular breeding of a novel Coprinopsis cinerea strain possessing a heterologous laccase gene, lccK, driven by a constitutive promoter. Mycoscience 52, 431–435. doi: 10.1007/S10267-011-0122-7
Murugesan, K., Chang, Y. Y., Kim, Y. M., Jeon, J. R., Kim, E. J., and Chang, Y. S. (2010). Enhanced transformation of triclosan by laccase in the presence of redox mediators. Water Res. 44, 298–308. doi: 10.1016/j.watres.2009.09.058
Nair, R. R., Demarche, P., and Agathos, S. N. (2013). Formulation and characterization of an immobilized laccase biocatalyst and its application to eliminate organic micropollutants in wastewater. N. Biotechnol. 30, 814–823. doi: 10.1016/j.nbt.2012.12.004
Nakade, K., Nakagawa, Y., Yano, A., Konno, N., Sato, T., and Sakamoto, Y. (2013). Effective induction of pblac1 laccase by copper ion in Polyporus brumalis ibrc05015. Fungal Biol. 117, 52–61. doi: 10.1016/j.funbio.2012.11.005
Nguyen, L. N., Hai, F. I., Dosseto, A., Richardson, C., Price, W. E., and Nghiem, L. D. (2016a). Continuous adsorption and biotransformation of micropollutants by granular activated carbon-bound laccase in a packed-bed enzyme reactor. Bioresour. Technol. 210, 108–116. doi: 10.1016/j.biortech.2016.01.014
Nguyen, L. N., Hai, F. I., Kang, J., Leusch, F. D. L., Roddick, F., Magram, S. F., et al. (2014a). Enhancement of trace organic contaminant degradation by crude enzyme extract from Trametes versicolor culture: effect of mediator type and concentration. J. Taiwan Inst. Chem. Eng. 45, 1855–1862. doi: 10.1016/j.jtice.2014.03.021
Nguyen, L. N., Hai, F. I., Price, W. E., Kang, J., Leusch, F. D. L., Roddick, F., et al. (2015). Degradation of a broad spectrum of trace organic contaminants by an enzymatic membrane reactor: complementary role of membrane retention and enzymatic degradation. Int. Biodeterior. Biodegradation 99, 115–122. doi: 10.1016/j.ibiod.2014.12.004
Nguyen, L. N., Hai, F. I., Price, W. E., Leusch, F. D., Roddick, F., Ngo, H. H., et al. (2014b). The effects of mediator and granular activated carbon addition on degradation of trace organic contaminants by an enzymatic membrane reactor. Bioresour. Technol. 167, 169–177. doi: 10.1016/j.biortech.2014.05.125
Nguyen, L. N., Hai, F. I., Price, W. E., Leusch, F. D. L., Roddick, F., McAdam, E. J., et al. (2014c). Continuous biotransformation of bisphenol A and diclofenac by laccase in an enzymatic membrane reactor. Int. Biodeterior. Biodegradation 95, 25–32. doi: 10.1016/j.ibiod.2014.05.017
Nguyen, L. N., Hai, F. I., Yang, S., Kang, J., Leusch, F. D. L., Roddick, F., et al. (2014d). Removal of pharmaceuticals, steroid hormones, phytoestrogens, UV-filters, industrial chemicals and pesticides by Trametes versicolor: role of biosorption and biodegradation. Int. Biodeterior. Biodegradation 88, 169–175. doi: 10.1016/j.ibiod.2013.12.017
Nguyen, L. N., van de Merwe, J. P., Hai, F. I., Leusch, F. D., Kang, J., Price, W. E., et al. (2016b). Laccase-syringaldehyde-mediated degradation of trace organic contaminants in an enzymatic membrane reactor: removal efficiency and effluent toxicity. Bioresour. Technol. 200, 477–484. doi: 10.1016/j.biortech.2015.10.054
Ni, J., and Tokuda, G. (2013). Lignocellulose-degrading enzymes from termites and their symbiotic microbiota. Biotechnol. Adv. 31, 838–850. doi: 10.1016/j.biotechadv.2013.04.005
Nishibori, N., Masaki, K., Tsuchioka, H., Fujii, T., and Iefuji, H. (2013). Comparison of laccase production levels in Pichia pastoris and Cryptococcus sp. S-2. J. Biosci. Bioeng. 115, 394–399. doi: 10.1016/j.jbiosc.2012.10.025
Olajuyigbe, F. M., and Fatokun, C. O. (2017). Biochemical characterization of an extremely stable pH-versatile laccase from Sporothrix carnis CPF-05. Int. J. Biol. Macromol. 94, 535–543. doi: 10.1016/j.ijbiomac.2016.10.037
Onesios, K. M., Yu, J. T., and Bouwer, E. J. (2009). Biodegradation and removal of pharmaceuticals and personal care products in treatment systems: a review. Biodegradation 20, 441–466. doi: 10.1007/s10532-008-9237-8
Osma, J. F., Toca-Herrera, J. L., and Rodríguez-Couto, S. (2010). Uses of laccases in the food industry. Enzyme Res. 2010, 1–8. doi: 10.4061/2010/918761
Ostadhadi-Dehkordi, S., Tabatabaei-Sameni, M., Forootanfar, H., Kolahdouz, S., Ghazi-Khansari, M., and Faramarzi, M. A. (2012). Degradation of some benzodiazepines by a laccase-mediated system in aqueous solution. Bioresour. Technol. 125, 344–347. doi: 10.1016/j.biortech.2012.09.039
Otto, B., Beuchel, C., Liers, C., Reisser, W., Harms, H., and Schlosser, D. (2015). Laccase-like enzyme activities from chlorophycean green algae with potential for bioconversion of phenolic pollutants. FEMS Microbiol. Lett. 362:fnv072. doi: 10.1093/femsle/fnv072
Oulton, R. L., Kohn, T., and Cwiertny, D. M. (2010). Pharmaceuticals and personal care products in effluent matrices: a survey of transformation and removal during wastewater treatment and implications for wastewater management. J. Environ. Monit. 12, 1956–1978. doi: 10.1039/c0em00068j
Pan, K., Zhao, N., Yin, Q., Zhang, T., Xu, X., Fang, W., et al. (2014). Induction of a laccase Lcc9 from Coprinopsis cinerea by fungal coculture and its application on indigo dye decolorization. Bioresour. Technol. 162, 45–52. doi: 10.1016/j.biortech.2014.03.116
Pardo, I., and Camarero, S. (2015). Laccase engineering by rational and evolutionary design. Cell. Mol. Life Sci. 72, 897–910. doi: 10.1007/s00018-014-1824-8
Pezzella, C., Guarino, L., and Piscitelli, A. (2015). How to enjoy laccases. Cell. Mol. Life Sci. 72, 923–940. doi: 10.1007/s00018-014-1823-9
Pezzella, C., Lettera, V., Piscitelli, A., Giardina, P., and Sannia, G. (2013). Transcriptional analysis of Pleurotus ostreatus laccase genes. Appl. Microbiol. Biotechnol. 97, 705–717. doi: 10.1007/s00253-012-3980-9
Piscitelli, A., Giardina, P., Lettera, V., Pezzella, C., Sannia, G., and Faraco, V. (2011). Induction and transcriptional regulation of laccases in fungi. Curr. Genomics 12, 104–112. doi: 10.2174/138920211795564331
Piscitelli, A., Pezzella, C., Giardina, P., Faraco, V., and Sannia, G. (2010). Heterologous laccase production and its role in industrial applications. Bioeng. Bugs 1, 252. doi: 10.4161/bbug.1.4.11438
Pogni, R., Baratto, M. C., Sinicropi, A., and Basosi, R. (2015). Spectroscopic and computational characterization of laccases and their substrate radical intermediates. Cell. Mol. Life Sci. 72, 885–896. doi: 10.1007/s00018-014-1825-7
Pollegioni, L., Tonin, F., and Rosini, E. (2015). Lignin-degrading enzymes. FEBS J. 282, 1190–1213. doi: 10.1111/febs.13224
Popa, C., Favier, L., Dinica, R., Semrany, S., Djelal, H., Amrane, A., et al. (2014). Potential of newly isolated wild Streptomyces strains as agents for the biodegradation of a recalcitrant pharmaceutical, carbamazepine. Environ. Technol. 35, 3082–3091. doi: 10.1080/09593330.2014.931468
Postemsky, P. D., Bidegain, M. A., Gonzalez-Matute, R., Figlas, N. D., and Cubitto, M. A. (2017). Pilot-scale bioconversion of rice and sunflower agro-residues into medicinal mushrooms and laccase enzymes through solid-state fermentation with Ganoderma lucidum. Bioresour. Technol. 231, 85–93. doi: 10.1016/j.biortech.2017.01.064
Rahmani, K., Faramarzi, M. A., Mahvi, A. H., Gholami, M., Esrafili, A., Forootanfar, H., et al. (2015). Elimination and detoxification of sulfathiazole and sulfamethoxazole assisted by laccase immobilized on porous silica beads. Int. Biodeterior. Biodegradation 97, 107–114. doi: 10.1016/j.ibiod.2014.10.018
Ramírez-Cavazos, L. I., Junghanns, C., Ornelas-Soto, N., Cárdenas-Chávez, D. L., Hernández-Luna, C., Demarche, P., et al. (2014). Purification and characterization of two thermostable laccases from Pycnoporus sanguineus and potential role in degradation of endocrine disrupting chemicals. J. Mol. Catal. B Enzym. 108, 32–42. doi: 10.1016/j.molcatb.2014.06.006
Ramos, J. A. T., Barends, S., Verhaert, R. M., and Graaff, L. H. D. (2011). The Aspergillus niger multicopper oxidase family: analysis and overexpression of laccase-like encoding genes. Microb. Cell Fact. 10:78. doi: 10.1186/1475-2859-10-78
Rao, M. A., Scelza, R., Acevedo, F., Diez, M. C., and Gianfreda, L. (2014). Enzymes as useful tools for environmental purposes. Chemosphere 107, 145–162. doi: 10.1016/j.chemosphere.2013.12.059
Reiss, R., Ihssen, J., Richter, M., Eichhorn, E., Schilling, B., and Thony-Meyer, L. (2013). Laccase versus laccase-like multi-copper oxidase: a comparative study of similar enzymes with diverse substrate spectra. PLoS ONE 8:e65633. doi: 10.1371/journal.pone.0065633
Risso, V. A., Gavira, J. A., Gaucher, E. A., and Sanchez-Ruiz, J. M. (2014). Phenotypic comparisons of consensus variants versus laboratory resurrections of Precambrian proteins. Proteins 82, 887–896. doi: 10.1002/prot.24575
Rivera-Hoyos, C. M., Morales-Álvarez, E. D., Poutou-Piñales, R. A., Pedroza-Rodríguez, A. M., RodrÍguez-Vázquez, R., and Delgado-Boada, J. M. (2013). Fungal laccases. Fungal Biol. Rev. 27, 67–82. doi: 10.1016/j.fbr.2013.07.001
Rodgers, C. J., Blanford, C. F., Giddens, S. R., Skamnioti, P., Armstrong, F. A., and Gurr, S. J. (2010). Designer laccases: a vogue for high-potential fungal enzymes? Trends Biotechnol. 28, 63–72. doi: 10.1016/j.tibtech.2009.11.001
Rodríguez-Couto, S. (2012). Laccases for denim bleaching: an eco-friendly alternative. Open Text. J. 5, 1–7. doi: 10.2174/1876520301205010001
Rodríguez-Couto, S., and Toca-Herrera, J. L. (2007). Laccase production at reactor scale by filamentous fungi. Biotechnol. Adv. 25, 558–569. doi: 10.1016/j.biotechadv.2007.07.002
Rodríguez-Delgado, M., Orona-Navar, C., García-Morales, R., Hernandez-Luna, C., Parra, R., Mahlknecht, J., et al. (2016). Biotransformation kinetics of pharmaceutical and industrial micropollutants in groundwaters by a laccase cocktail from Pycnoporus sanguineus CS43 fungi. Int. Biodeterior. Biodegradation 108, 34–41. doi: 10.1016/j.ibiod.2015.12.003
Rodriguez-Rodriguez, C. E., Garcia-Galan, M. A., Blanquez, P., Diaz-Cruz, M. S., Barcelo, D., Caminal, G., et al. (2012). Continuous degradation of a mixture of sulfonamides by Trametes versicolor and identification of metabolites from sulfapyridine and sulfathiazole. J. Hazard. Mater. 213–214, 347–354. doi: 10.1016/j.jhazmat.2012.02.008
Rogalski, J., and Janusz, G. (2010). Purification of extracellular laccase from Cerrena unicolor. Prep. Biochem. Biotechnol. 40, 242–255. doi: 10.1080/10826068.2010.488967
Rühl, M., Majcherczyk, A., and Kües, U. (2013). Lcc1 and Lcc5 are the main laccases secreted in liquid cultures of Coprinopsis cinerea strains. Antonie van Leeuwenhoek 103, 1029–1039. doi: 10.1007/s10482-013-9883-7
Sakamoto, Y., Nakade, K., Yoshida, K., Natsume, S., Miyazaki, K., Sato, S., et al. (2015). Grouping of multicopper oxidases in Lentinula edodes by sequence similarities and expression patterns. AMB Express 5, 63. doi: 10.1186/s13568-015-0151-2
Santhanam, N., Vivanco, J. M., Decker, S. R., and Reardon, K. F. (2011). Expression of industrially relevant laccases: prokaryotic style. Trends Biotechnol. 29, 480–489. doi: 10.1016/j.tibtech.2011.04.005
Sathishkumar, P., Mythili, A., Hadibarata, T., Jayakumar, R., Kanthimathi, M. S., Palvannan, T., et al. (2014). Laccase mediated diclofenac transformation and cytotoxicity assessment on mouse fibroblast 3T3-L1 preadipocytes. RSC Adv. 4, 11689–11697. doi: 10.1039/c3ra46014b
Schwarz, J., Knicker, H., Schaumann, G. E., and Thiele-Bruhn, S. (2015). Enzymatic transformation and bonding of sulfonamide antibiotics to model humic substances. J. Chem. 2015, 1–11. doi: 10.1155/2015/829708
Sen, S. K., Raut, S., Bandyopadhyay, P., and Raut, S. (2016). Fungal decolouration and degradation of azo dyes: a review. Fungal Biol. Rev. 30, 112–133. doi: 10.1016/j.fbr.2016.06.003
Senthivelan, T., Kanagaraj, J., and Panda, R. C. (2016). Recent trends in fungal laccase for various industrial applications: an eco-friendly approach - a review. Biotechnol. Bioprocess Eng. 21, 19–38. doi: 10.1007/s12257-015-0278-7
Sheldon, R. A. (2011). Characteristic features and biotechnological applications of cross-linked enzyme aggregates (CLEAs). Appl. Microbiol. Biotechnol. 92, 467–477. doi: 10.1007/s00253-011-3554-2
Shi, H., Peng, J., Li, J., Mao, L., Wang, Z., and Gao, S. (2016). Laccase-catalyzed removal of the antimicrobials chlorophene and dichlorophen from water: reaction kinetics, pathway and toxicity evaluation. J. Hazard. Mater. 317, 81–89. doi: 10.1016/j.jhazmat.2016.05.064
Shi, L., Ma, F., Han, Y., Zhang, X., and Yu, H. (2014). Removal of sulfonamide antibiotics by oriented immobilized laccase on Fe3O4 nanoparticles with natural mediators. J. Hazard. Mater. 279, 203–211. doi: 10.1016/j.jhazmat.2014.06.070
Si, J., and Cui, B.-K. (2013). Study of the physiological characteristics of the medicinal mushroom Trametes pubescens (higher basidiomycetes) during the laccase-producing process. Int. J. Med. Mushrooms 15, 199–210. doi: 10.1615/IntJMedMushr.v15.i2.90
Si, J., Peng, F., and Cui, B. (2013). Purification, biochemical characterization and dye decolorization capacity of an alkali-resistant and metal-tolerant laccase from Trametes pubescens. Bioresour. Technol. 128, 49–57. doi: 10.1016/j.biortech.2012.10.085
Singh, D., Rawat, S., Waseem, M., Gupta, S., Lynn, A., Nitin, M., et al. (2016). Molecular modeling and simulation studies of recombinant laccase from Yersinia enterocolitica suggests significant role in the biotransformation of non-steroidal anti-inflammatory drugs. Biochem. Biophys. Res. Commun. 469, 306–312. doi: 10.1016/j.bbrc.2015.11.096
Singh, G., Bhalla, A., Kaur, P., Capalash, N., and Sharma, P. (2011). Laccase from prokaryotes: a new source for an old enzyme. Rev. Environ. Sci. Biotechnol. 10, 309–326. doi: 10.1007/s11157-011-9257-4
Singh, G., Kaur, K., Puri, S., and Sharma, P. (2015). Critical factors affecting laccase-mediated biobleaching of pulp in paper industry. Appl. Microbiol. Biotechnol. 99, 155–164. doi: 10.1007/s00253-014-6219-0
Singh, R., Sidhu, S. S., Zhang, H., and Huang, Q. (2015). Removal of sulfadimethoxine in soil mediated by extracellular oxidoreductases. Environ. Sci. Pollut. Res. 22, 16868–16874. doi: 10.1007/s11356-015-4893-9
Singh, R. L., Singh, P. K., and Singh, R. P. (2015). Enzymatic decolorization and degradation of azo dyes – a review. Int. Biodeterior. Biodegradation 104, 21–31. doi: 10.1016/j.ibiod.2015.04.027
Sinirlioglu, Z. A., Sinirlioglu, D., and Akbas, F. (2013). Preparation and characterization of stable cross-linked enzyme aggregates of novel laccase enzyme from Shewanella putrefaciens and using malachite green decolorization. Bioresour. Technol. 146, 807–811. doi: 10.1016/j.biortech.2013.08.032
Sitarz, A. K., Mikkelsen, J. D., and Meyer, A. S. (2016). Structure, functionality and tuning up of laccases for lignocellulose and other industrial applications. Crit. Rev. Biotechnol. 36, 70–86. doi: 10.3109/07388551.2014.949617
Sjaarda, C. P., Abubaker, K. S., and Castle, A. J. (2015). Induction of lcc2 expression and activity by Agaricus bisporus provides defence against Trichoderma aggressivum toxic extracts. Microb. Biotechnol. 8, 918–929. doi: 10.1111/1751-7915.12277
Solé, M., Müller, I., Pecyna, M. J., Fetzer, I., Harms, H., and Schlosser, D. (2012). Differential regulation by organic compounds and heavy metals of multiple laccase genes in the aquatic hyphomycete Clavariopsis aquatica. Appl. Environ. Microbiol. 78, 4732–4739. doi: 10.1128/AEM.00635-12
Songulashvili, G., Jimenéz-Tobón, G. A., Jaspers, C., and Penninckx, M. J. (2012). Immobilized laccase of Cerrena unicolor for elimination of endocrine disruptor micropollutants. Fungal Biol. 116, 883–889. doi: 10.1016/j.funbio.2012.05.005
Songulashvili, G., Spindler, D., Jimenez-Tobon, G. A., Jaspers, C., Kerns, G., and Penninckx, M. J. (2015). Production of a high level of laccase by submerged fermentation at 120-L scale of Cerrena unicolor C-139 grown on wheat bran. C. R. Biol. 338, 121–125. doi: 10.1016/j.crvi.2014.12.001
Strong, P. J., and Claus, H. (2011). Laccase: a review of its past and its future in bioremediation. Crit. Rev. Environ. Sci. Technol. 41, 373–434. doi: 10.1080/10643380902945706
Suda, T., Hata, T., Kawai, S., Okamura, H., and Nishida, T. (2012). Treatment of tetracycline antibiotics by laccase in the presence of 1-hydroxybenzotriazole. Bioresour. Technol. 103, 498–501. doi: 10.1016/j.biortech.2011.10.041
Suetomi, T., Sakamoto, T., Tokunaga, Y., Kameyama, T., Honda, Y., Kamitsuji, H., et al. (2015). Effects of calmodulin on expression of lignin-modifying enzymes in Pleurotus ostreatus. Curr. Genet. 61, 127–140. doi: 10.1007/s00294-014-0460-z
Sun, K., Huang, Q., and Gao, Y. (2016). Laccase-catalyzed oxidative coupling reaction of triclosan in aqueous solution. Water Air Soil Pollut. 227, 358. doi: 10.1007/s11270-016-3064-z
Sun, S. J., Liu, J. Z., Hu, K. H., and Zhu, H. X. (2011). The level of secreted laccase activity in the edible fungi and their growing cycles are closely related. Curr. Microbiol. 62, 871–875. doi: 10.1007/s00284-010-9794-z
Sun, S., Li, X., Ruan, L., Zhang, L., and Hu, K. (2014). A novel breeding strategy for new strains of Hypsizygus marmoreus and Grifola frondosa based on ligninolytic enzymes. World J. Microbiol. Biotechnol. 30, 2005–2013. doi: 10.1007/s11274-014-1624-1
Sutar, R. S., and Rathod, V. K. (2015). Ultrasound assisted Laccase catalyzed degradation of Ciprofloxacin hydrochloride. J. Ind. Eng. Chem. 31, 276–282. doi: 10.1016/j.jiec.2015.06.037
Tahmasbi, H., Khoshayand, M. R., Bozorgi-Koushalshahi, M., Heidary, M., Ghazi-Khansari, M., and Faramarzi, M. A. (2016). Biocatalytic conversion and detoxification of imipramine by the laccase-mediated system. Int. Biodeterior. Biodegradation 108, 1–8. doi: 10.1016/j.ibiod.2015.11.029
Talekar, S., Joshi, A., Joshi, G., Kamat, P., Haripurkar, R., and Kambale, S. (2013). Parameters in preparation and characterization of cross linked enzyme aggregates (CLEAs). RSC Adv. 3, 12485–12511. doi: 10.1039/c3ra40818c
Thiele-Bruhn, S. (2003). Pharmaceutical antibiotic compounds in soils – a review. J. Plant Nutr. Soil Sci. 166, 145–167. doi: 10.1002/jpln.200390023
Touahar, I. E., Haroune, L., Ba, S., Bellenger, J.-P., and Cabana, H. (2014). Characterization of combined cross-linked enzyme aggregates from laccase, versatile peroxidase and glucose oxidase, and their utilization for the elimination of pharmaceuticals. Sci. Total Environ. 481, 90–99. doi: 10.1016/j.scitotenv.2014.01.132
Tran, N. H., Hu, J., and Urase, T. (2013). Removal of the insect repellent N,N-diethyl-m-toluamide (DEET) by laccase-mediated systems. Bioresour. Technol. 147, 667–671. doi: 10.1016/j.biortech.2013.08.113
Tran, N. H., Urase, T., and Kusakabe, O. (2010). Biodegradation characteristics of pharmaceutical substances by whole fungal culture Trametes versicolor and its laccase. J. Water Environ. Technol. 8, 125–140. doi: 10.2965/jwet.2010.125
Turlapati, P. V., Kim, K. W., Davin, L. B., and Lewis, N. G. (2011). The laccase multigene family in Arabidopsis thaliana: towards addressing the mystery of their gene function(s). Planta 233, 439–470. doi: 10.1007/s00425-010-1298-3
Upadhyay, P., Shrivastava, R., and Agrawal, P. K. (2016). Bioprospecting and biotechnological applications of fungal laccase. 3 Biotech 6, 15. doi: 10.1007/s13205-015-0316-3
Valderrama, B., Oliver, P., Medrano-Soto, A., and Vazquez-Duhalt, R. (2003). Evolutionary and structural diversity of fungal laccases. Antonie van Leeuwenhoek 84, 289–299. doi: 10.1023/A:1026070122451
Vasiliadou, I. A., Sanchez-Vazquez, R., Molina, R., Martinez, F., Melero, J. A., Bautista, L. F., et al. (2016). Biological removal of pharmaceutical compounds using white-rot fungi with concomitant FAME production of the residual biomass. J. Environ. Manage. 180, 228–237. doi: 10.1016/j.jenvman.2016.05.035
Vasina, D. V., Mustafaev, O. N., Moiseenko, K. V., Sadovskaya, N. S., Glazunova, O. A., Tyurin, A. A., et al. (2015). The Trametes hirsuta 072 laccase multigene family: genes identification and transcriptional analysis under copper ions induction. Biochimie 116, 154–164. doi: 10.1016/j.biochi.2015.07.015
Viswanath, B., Rajesh, B., Janardhan, A., Kumar, A. P., and Narasimha, G. (2014). Fungal laccases and their applications in bioremediation. Enzyme Res. 2014, 1–21. doi: 10.1155/2014/163242
Wang, C., Zhang, L., and Xu, H. (2012). The effects of N+ ion implantation mutagenesis on the laccase production of Ceriporiopsis subvermispora. Biotechnol. Bioprocess Eng. 17, 946–951. doi: 10.1007/s12257-012-0125-z
Wang, F., Guo, C., Wei, T., Zhang, T., and Liu, C.-Z. (2012). Heat shock treatment improves Trametes versicolor laccase production. Appl. Biochem. Biotechnol. 168, 256–265. doi: 10.1007/s12010-012-9769-6
Wang, J., and Wang, S. (2016). Removal of pharmaceuticals and personal care products (PPCPs) from wastewater: a review. J. Environ. Manage. 182, 620–640. doi: 10.1016/j.jenvman.2016.07.049
Wang, J., Feng, J., Jia, W., Chang, S., Li, S., and Li, Y. (2015). Lignin engineering through laccase modification: a promising field for energy plant improvement. Biotechnol. Biofuels 8, 145. doi: 10.1186/s13068-015-0331-y
Wang, S. S., Ning, Y. J., Wang, S. N., Zhang, J., Zhang, G. Q., and Chen, Q. J. (2017). Purification, characterization, and cloning of an extracellular laccase with potent dye decolorizing ability from white rot fungus Cerrena unicolor GSM-01. Int. J. Biol. Macromol. 95, 920–927. doi: 10.1016/j.ijbiomac.2016.10.079
Wang, W., Liu, F., Jiang, Y., Wu, G., Guo, L., Chen, R., et al. (2015). The multigene family of fungal laccases and their expression in the white rot basidiomycete Flammulina velutipes. Gene 563, 142–149. doi: 10.1016/j.gene.2015.03.020.
Wei, F., Hong, Y., Liu, J., Yuan, J., Fang, W., Peng, H., et al. (2010). Gongronella sp. induces overproduction of laccase in Panus rudis. J. Basic Microbiol. 50, 98–103. doi: 10.1002/jobm.200900155
Wen, X., Jia, Y., and Li, J. (2009). Degradation of tetracycline and oxytetracycline by crude lignin peroxidase prepared from Phanerochaete chrysosporium – A white rot fungus. Chemosphere 75, 1003–1007. doi: 10.1016/j.chemosphere.2009.01.052
Wen, X., Jia, Y., and Li, J. (2010). Enzymatic degradation of tetracycline and oxytetracycline by crude manganese peroxidase prepared from Phanerochaete chrysosporium. J. Hazard. Mater. 177, 924–928. doi: 10.1016/j.jhazmat.2010.01.005
Weng, S. S., Ku, K. L., and Lai, H. T. (2012). The implication of mediators for enhancement of laccase oxidation of sulfonamide antibiotics. Bioresour. Technol. 113, 259–264. doi: 10.1016/j.biortech.2011.12.111
Weng, S. S., Liu, S. M., and Lai, H. T. (2013). Application parameters of laccase-mediator systems for treatment of sulfonamide antibiotics. Bioresour. Technol. 141, 152–159. doi: 10.1016/j.biortech.2013.02.093
Wong, D. W. (2009). Structure and action mechanism of ligninolytic enzymes. Appl. Biochem. Biotechnol. 157, 174–209. doi: 10.1007/s12010-008-8279-z
Wu, L., Zhu, G., Chen, M., Wang, H., and Bao, D. (2013). The bioinformatic analyses and the gene expression induced by Cu2+ of 11 laccase homologous genes from Volvariella volvacea. Mycosystema 33, 323–333. doi: 10.13346/j.mycosystema.130231
Xiao, Y. Z., Hong, Y. Z., Li, J. F., Hang, J., Tong, P. G., Fang, W., et al. (2006). Cloning of novel laccase isozyme genes from Trametes sp. AH28-2 and analyses of their differential expression. Appl. Microbiol. Biotechnol. 71, 493–501. doi: 10.1007/s00253-005-0188-2
Xiao, Y. Z., Tu, X. M., Wang, J., Zhang, M., Cheng, Q., Zeng, W. Y., et al. (2003). Purification, molecular characterization and reactivity with aromatic compounds of a laccase from basidiomycete Trametes sp. strain AH28-2. Appl. Microbiol. Biotechnol. 60, 700–707. doi: 10.1007/s00253-002-1169-3
Xu, R., Si, Y., Wu, X., Li, F., and Zhang, B. (2014). Triclosan removal by laccase immobilized on mesoporous nanofibers: strong adsorption and efficient degradation. Chem. Eng. J. 255, 63–70. doi: 10.1016/j.cej.2014.06.060
Xu, R., Tang, R., Zhou, Q., Li, F., and Zhang, B. (2015). Enhancement of catalytic activity of immobilized laccase for diclofenac biodegradation by carbon nanotubes. Chem. Eng. J. 262, 88–95. doi: 10.1016/j.cej.2014.09.072
Xu, X., Feng, L., Han, Z., Luo, S., Wu, A., and Xie, J. (2016). Selection of high laccase-producing Coriolopsis gallica strain T906: mutation breeding, strain characterization, and features of the extracellular laccases. J. Microbiol. Biotechnol. 26, 1570–1578. doi: 10.4014/jmb.1604.04011
Yadav, M., and Yadav, H. S. (2015). Applications of ligninolytic enzymes to pollutants, wastewater, dyes, soil, coal, paper and polymers. Environ. Chem. Lett. 13, 309–318. doi: 10.1007/s10311-015-0516-4
Yang, C. W., Hsiao, W. C., and Chang, B. V. (2016). Biodegradation of sulfonamide antibiotics in sludge. Chemosphere 150, 559–565. doi: 10.1016/j.chemosphere.2016.02.064
Yang, J., Lin, Y., Yang, X., Ng, T. B., Ye, X., and Lin, J. (2017). Degradation of tetracycline by immobilized laccase and the proposed transformation pathway. J. Hazard. Mater. 322, 525–531. doi: 10.1016/j.jhazmat.2016.10.019
Yang, J., Wang, G., Ng, T. B., Lin, J., and Ye, X. (2016a). Laccase production and differential transcription of laccase genes in Cerrena sp. in response to metal ions, aromatic compounds, and nutrients. Front. Microbiol. 6:1558. doi: 10.3389/fmicb.2015.01558
Yang, J., Xu, X., Ng, T. B., Lin, J., and Ye, X. (2016b). Laccase gene family in Cerrena sp. HYB07: sequences, heterologous expression and transcriptional analysis. Molecules 21:1017. doi: 10.3390/molecules21081017
Yang, J., Xu, X., Yang, X., Ye, X., and Lin, J. (2016c). Cross-linked enzyme aggregates of Cerrena laccase: preparation, enhanced NaCl tolerance and decolorization of Remazol Brilliant Blue Reactive. J. Taiwan Inst. Chem. Eng. 65, 1–7. doi: 10.1016/j.jtice.2016.04.025
Yang, J., Yang, X., Lin, Y., Ng, T. B., Lin, J., and Ye, X. (2015). Laccase-catalyzed decolorization of malachite green: performance optimization and degradation mechanism. PLoS ONE 10:e0127714. doi: 10.1371/journal.pone.0127714
Yang, J., Yang, X., Ye, X., and Lin, J. (2016d). Destaining of Coomassie Brilliant Blue R-250-stained polyacrylamide gels with fungal laccase. Anal. Biochem. 493, 27–29. doi: 10.1016/j.ab.2015.10.004
Yang, J., Yang, X., Ye, X., and Lin, J. (2016e). Optimal parameters for laccase-mediated destaining of Coomassie Brilliant Blue R-250-stained polyacrylamide gels. Data Brief 7, 1–7. doi: 10.1016/j.dib.2016.01.029
Yang, S., Hai, F. I., Nghiem, L. D., Price, W. E., Roddick, F., Moreira, M. T., et al. (2013a). Understanding the factors controlling the removal of trace organic contaminants by white-rot fungi and their lignin modifying enzymes: a critical review. Bioresour. Technol. 141, 97–108. doi: 10.1016/j.biortech.2013.01.173
Yang, S., Hai, F. I., Nghiem, L. D., Roddick, F., and Price, W. E. (2013b). Removal of trace organic contaminants by nitrifying activated sludge and wholecell and crude enzyme extract of Trametes versicolor. Water Sci. Technol. 67, 1216–1223. doi: 10.2166/wst.2013.684
Yang, Y., Fan, F., Zhuo, R., Ma, F., Gong, Y., Wan, X., et al. (2012). Expression of the laccase gene from a white rot fungus in Pichia pastoris can enhance the resistance of this yeast to H2O2-mediated oxidative stress by stimulating the glutathione-based antioxidative system. Appl. Environ. Microbiol. 78, 5845–5854. doi: 10.1128/AEM.00218-12
Yang, Y., Wei, F., Zhuo, R., Fan, F., Liu, H., Zhang, C., et al. (2013). Enhancing the laccase production and laccase gene expression in the white-rot fungus Trametes velutina 5930 with great potential for biotechnological applications by different metal Ions and aromatic compounds. PLoS ONE 8:e79307. doi: 10.1371/journal.pone.0079307
Yousefi-Ahmadipour, A., Bozorgi-Koshalshahi, M., Mogharabi, M., Amini, M., Ghazi-Khansari, M., and Faramarzi, M. A. (2016). Laccase-catalyzed treatment of ketoconazole, identification of biotransformed metabolites, determination of kinetic parameters, and evaluation of micro-toxicity. J. Mol. Catal. B Enzym. 133, 77–84. doi: 10.1016/j.molcatb.2016.07.015
Zeng, J., Zhu, Q., Wu, Y., and Lin, X. (2016). Oxidation of polycyclic aromatic hydrocarbons using Bacillus subtilis CotA with high laccase activity and copper independence. Chemosphere 148, 1–7. doi: 10.1016/j.chemosphere.2016.01.019
Zhang, H., Hong, Y. Z., Xiao, Y. Z., Yuan, J., Tu, X. M., and Zhang, X. Q. (2006). Efficient production of laccases by Trametes sp. AH28-2 in cocultivation with a Trichoderma strain. Appl. Microbiol. Biotechnol. 73, 89–94. doi: 10.1007/s00253-006-0430-6
Zhang, J., Chen, H., Chen, M., Ren, A., Huang, J., Wang, H., et al. (2015). Cloning and functional analysis of a laccase gene during fruiting body formation in Hypsizygus marmoreus. Microbiol. Res. 179, 54–63. doi: 10.1016/j.micres.2015.06.005
Zhang, Y., and Geißen, S.-U. (2010). In vitro degradation of carbamazepine and diclofenac by crude lignin peroxidase. J. Hazard. Mater. 176, 1089–1092. doi: 10.1016/j.jhazmat.2009.10.133
Keywords: laccase, production, expression regulation, bioremediation, PPCPs, antibiotics
Citation: Yang J, Li W, Ng TB, Deng X, Lin J and Ye X (2017) Laccases: Production, Expression Regulation, and Applications in Pharmaceutical Biodegradation. Front. Microbiol. 8:832. doi: 10.3389/fmicb.2017.00832
Received: 26 January 2017; Accepted: 24 April 2017;
Published: 16 May 2017.
Edited by:
Octavio Luiz Franco, Universidade Católica de Brasília, BrazilReviewed by:
Susana Rodriguez-Couto, Ikerbasque, SpainGerardo Díaz-Godínez, Autonomous University of Tlaxcala, Mexico
Copyright © 2017 Yang, Li, Ng, Deng, Lin and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Yang, dDA5MTM2QGZ6dS5lZHUuY24=
 Jie Yang
Jie Yang Wenjuan Li
Wenjuan Li Tzi Bun Ng
Tzi Bun Ng Xiangzhen Deng
Xiangzhen Deng Juan Lin1
Juan Lin1