- 1Centre for Human Microbiome and Probiotic Research, Lawson Health Research Institute, St. Joseph’s Health Care London, London, ON, Canada
- 2Schulich School of Medicine and Dentistry, Department of Microbiology and Immunology, University of Western Ontario, London, ON, Canada
- 3Schulich School of Medicine and Dentistry, Department of Surgery, University of Western Ontario, London, ON, Canada
Microbiota research often assumes that differences in abundance and identity of microorganisms have unique influences on host physiology. To test this concept mechanistically, germ-free mice are colonized with microbial communities to assess causation. Due to the cost, infrastructure challenges, and time-consuming nature of germ-free mouse models, an alternative approach is needed to investigate host–microbial interactions. Drosophila melanogaster (fruit flies) can be used as a high throughput in vivo screening model of host–microbiome interactions as they are affordable, convenient, and replicable. D. melanogaster were essential in discovering components of the innate immune response to pathogens. However, axenic D. melanogaster can easily be generated for microbiome studies without the need for ethical considerations. The simplified microbiota structure enables researchers to evaluate permutations of how each microbial species within the microbiota contribute to host phenotypes of interest. This enables the possibility of thorough strain-level analysis of host and microbial properties relevant to physiological outcomes. Moreover, a wide range of mutant D. melanogaster strains can be affordably obtained from public stock centers. Given this, D. melanogaster can be used to identify candidate mechanisms of host–microbe symbioses relevant to pathogen exclusion, innate immunity modulation, diet, xenobiotics, and probiotic/prebiotic properties in a high throughput manner. This perspective comments on the most promising areas of microbiota research that could immediately benefit from using the D. melanogaster model.
Drosophila melanogaster Model for Microbiota Studies
Next generation sequencing has increased the popularity and understanding of microbiota (community of microorganisms residing on/in a multicellular organism) contribution to host function in health and disease. Several diseases have been partially attributed to changes in microbiome composition such as Clostridium difficile associated diarrhea, diabetes mellitus, mood disorders, atherosclerosis, and others (Bäckhed et al., 2012). However, microbiota studies often assume differences in identity and abundance of certain host microorganisms promote and/or mitigate disease. This is shown by the growing number of reports on microbiota correlations that lack attempts to demonstrate causality.
A major challenge to experimental design is that the microbiota concept greatly increases the complexity of the reductionist approach in which a single pathogen is responsible for a given disease, as proven by Koch’s postulates (Byrd and Segre, 2016). The gold standard for demonstrating causality uses germ-free mice to demonstrate that a condition of interest is induced following host colonization with microbes associated with the aforementioned condition. However, major barriers to germ-free mouse studies are cost, lengthy study time, technical challenges, and infrastructure demands. These limitations also manifest in germ-free mouse studies often having low experimental sample sizes.
The field of probiotic and prebiotic science faces similar challenges. Probiotics are defined as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (Food and Agriculture Organization of the United Nations, and World Health Organization [FAO/WHO], 2002; Hill et al., 2014). High power clinical trials are required to draw reliable conclusions about probiotic efficacy. However, the cost and time required to perform clinical trials large enough to be informative have left many probiotic claims dependent in vitro studies. This results in scarce evidence as to whether some marketed probiotics are effective in vivo. This issue is further troubled by numerous microbes receiving generalized probiotic claims even though probiotic properties are often conferred in a strain-specific manner.
An alternative approach is needed to investigate microbiota and probiotic interactions in a living host. An ideal model organism would have the following characteristics: high throughput screening capabilities, inexpensive, fast reproduction, and an easily manipulated microbiome. Drosophila melanogaster stands out as an excellent model organism which possesses these qualities, and can allow reliable validation of probiotic effects in a living organism. Furthermore, many of the tools to investigate host–microbe relationships in D. melanogaster are already available due to its rich history in pathogen research (Lemaitre and Hoffmann, 2007).
Compared to the mammalian gastrointestinal tract, the D. melanogaster gut has several major differences, but the overall structure and function are similar (Figure 1). The gastrointestinal physiology, anatomy, and signaling pathways controlling intestinal development, regeneration, and pathology are highly conserved in D. melanogaster (Apidianakis and Rahme, 2011). The details of D. melanogaster gastrointestinal tract are beyond the scope of this article, but can be found in the review cited (Lemaitre and Miguel-Aliaga, 2013).
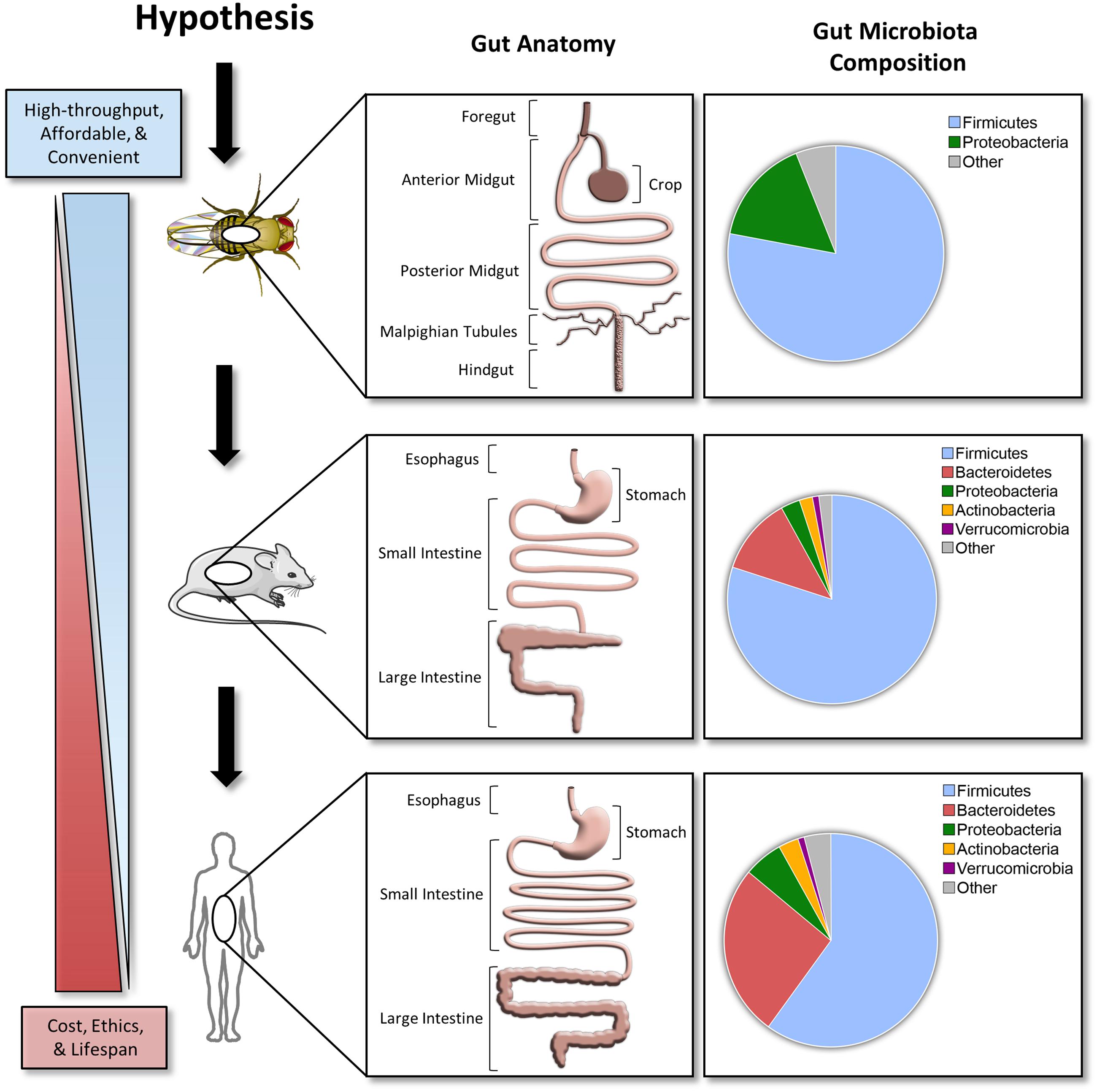
FIGURE 1. A hypothesized pipeline approach investigating host–microbe interactions. Comparisons between the Drosophila melanogaster, C57BL/6 mice, and human gut anatomy and microbiota. Taxonomical distribution data for top phyla in D. melanogaster are based off 454 tag pyrosequencing (Blum et al., 2013). C57BL/6 mice and human data are based off a 2.6 and 4.6 M catalog, respectively (Xiao et al., 2015). Stock clipart images from Servier Medical Art by Servier were used and modified under the Creative Commons Attribution 3.0 Unported License.
The idea of using D. melanogaster for investigating symbiotic modulation of host physiology has been stated by others (Ryu et al., 2010; Storelli et al., 2011; Erkosar et al., 2013; Ma et al., 2015). However, use of the efficient and convenient D. melanogaster model for microbiota research has yet to be implemented widely. A wide range of D. melanogaster strains available in public repositories1 can be derived germ-free and maintained easily without the requirement of expensive animal facilities, equipment, and technicians (Koyle et al., 2016).
Compared to the mouse or human microbiota, the D. melanogaster microbiota is simple with a low microbial diversity (1–30 species) and is typically dominated by Lactobacillus and Acetobacter (Blum et al., 2013; Erkosar et al., 2013; Chaston et al., 2014). This simplified community structure deconvolutes the complexity of deciphering the effect of a given microbial species on the greater community and its host. Similar to the genomics era being initiated with assessment of small and simple genomes, microbiota research could benefit from a simplified model system such as D. melanogaster to deconstruct complex polymicrobial interactions in vivo. Taken together, D. melanogaster experimentation is affordable, convenient, and rarely requires approval by animal ethics review boards. These characteristics make D. melanogaster an ideal high-throughput in vivo model for understanding host–microbiota interactions.
Microbiota-Mediated Pathogen Exclusion
Certain microbes can outcompete others via chemical inhibition, physical and nutritional competitive exclusion, and a variety of other adaptive mechanisms. In particular, the gut microbiota and probiotic organisms are critical for inhibiting intestinal microbial pathogenesis (Reid et al., 2011). However, promising in vitro screens to identify anti-pathogenic gut microbiota isolates are often unsuccessful upon further testing in expensive mammalian models. D. melanogaster has the potential to be an affordable, preliminary in vivo infection model for a variety of bacterial, fungal, and viral pathogens (Apidianakis and Rahme, 2009, 2011). The D. melanogaster infection model has been used to demonstrate pathogen inhibition by the human probiotic L. rhamnosus GG (Blum et al., 2013). This highlights the potential of D. melanogaster as a high throughput screening tool to substantiate in vivo pathogen inhibition claims for human probiotic or microbiota organisms of interest. Specifically, established oral or septic infection models can be used to assess the ability of different microbial communities to prevent pathogen-induced D. melanogaster colonization, persistence, and/or mortality (Neyen et al., 2014). This simplified model system also provides a powerful platform for elucidating ill-defined mechanisms of in vivo microbiota/probiotic-mediated pathogen inhibition (Figure 2).
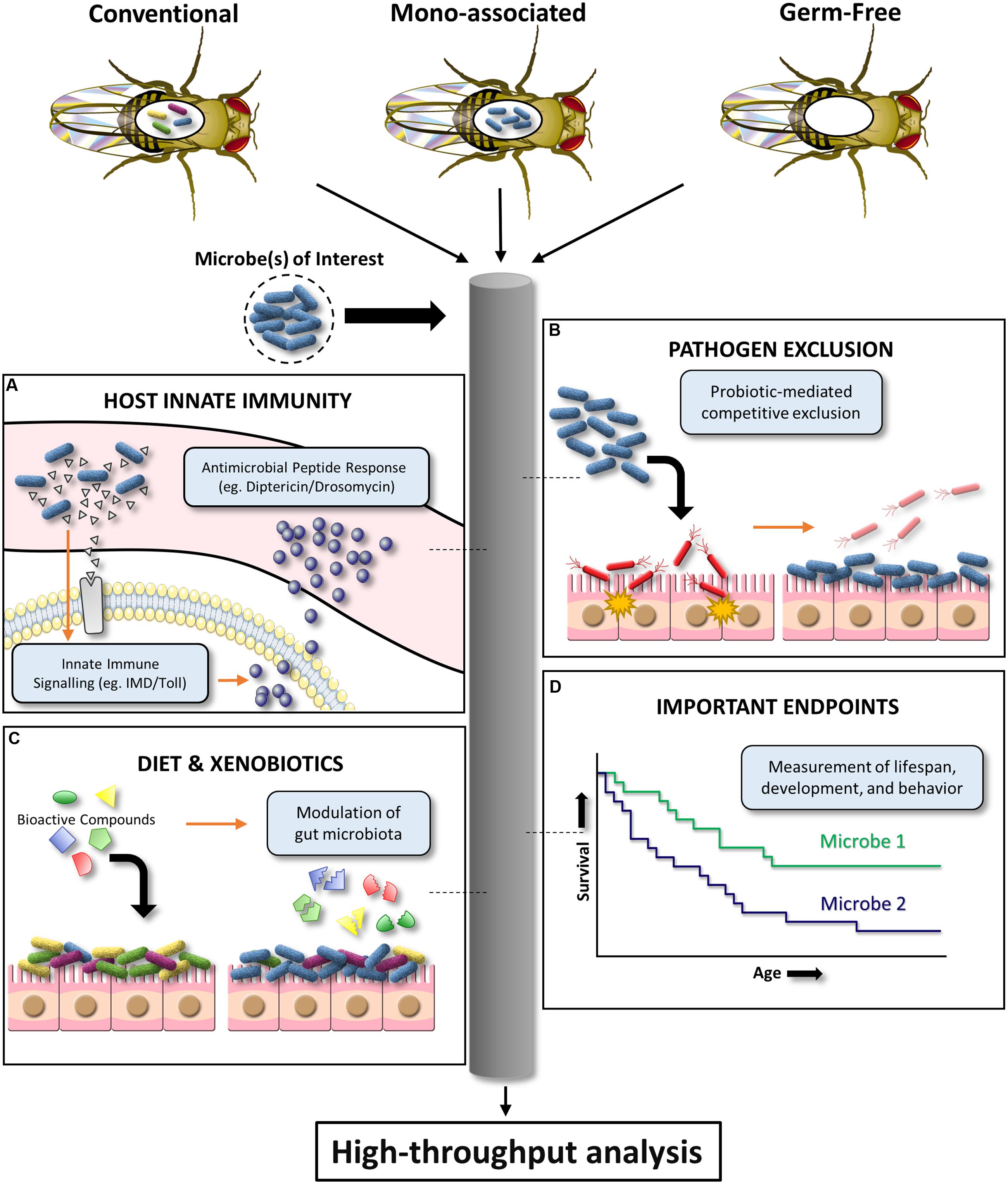
FIGURE 2. Schematic diagram illustrating the high-throughput potential of D. melanogaster to investigate host–microbe interactions. Conventional, mono-associated, and germ-free D. melanogaster can be used to mechanistically dissect the effects of a given microbe on (A) innate immune system signaling, (B) pathogen exclusion, (C) xenobiotic biotransformation and associated effects on the microbiota, and (D) important endpoints such as lifespan, development, and behavior.
Opportunistic infections are another field of study suitable for assessment in D. melanogaster. Gut microbiota commensals are known to become pathogenic under certain circumstances (Fei and Zhao, 2013; Cho et al., 2015). In commonly used experimental models, it is difficult to decipher the causative environmental, microbial, or host factors responsible for inducing this symbiotic-pathogenic shift. For example, the well-known intestinal opportunistic pathogen Clostridium difficile is triggered by an apparent reduction in microbial diversity induced by antibiotic treatment. Replacement of microbial diversity via fecal transplant has been a very effective strategy to overcome this disease, however, failure to identify a specific species or subset of species responsible for resolution of pathogenesis demands further investigation (van Nood et al., 2013). Using the available immunodeficient D. melanogaster stocks as an in vivo model (Neyen et al., 2014), researchers can better decipher the mechanistic triggers that determine how individual species transition from symbiotic-to-pathogenic.
Microbiota Modulation of Host Innate Immunity
Despite lacking an adaptive immune system, D. melanogaster has been a crucial model for innate immunity discoveries. Findings of microbiota-mediated modulation of D. melanogaster local gut immunity (Ryu et al., 2010) provide evidence for using this model to evaluate microbial strain variations in innate immunoregulation. D. melanogaster immune responses to pathogen-associated molecular patterns can largely be separated into two distinct pathways: (i) The Toll pathway which is activated by lysine (Lys)-type peptidoglycan (Gram-positive bacteria) and β-1,3 glucan (fungi); (ii) The immune deficiency (IMD) pathway which is activated by diaminopimelic acid (DAP)-type (Gram-negative and Gram-positive bacilli bacteria) (Lemaitre and Hoffmann, 2007; Buchon et al., 2014). Together, the Toll and IMD pathways are critical for upregulating nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) response genes, such as the antimicrobial peptides Drosomycin and Diptericin (Figure 2). Both D. melanogaster Diptericin and Drosomycin reporter strains are publicly available. Preliminary work has shown that these mutant flies can be used to screen microbes (single or in combination) for in vivo induction or repression of these conserved innate signaling pathways by measuring antimicrobial peptide readouts using a fluorescent microplate reader (for high-throughput interpretation) or microscope (for tissue localization inquiries). However, there are fundamental questions that require further clarification. The upstream transcription factor responsible for regulating Diptericin, Relish, has been shown to have pleiotropic roles in promoting host survival in response to noxious stimuli such as radiation (Karpac et al., 2011), and regulation of cell death (Chinchore et al., 2012). It is currently unknown how Relish modulation by the indigenous microbiota effects host physiology and what aspects of bacteria make this pathway targetable. The availability of IMD and Toll loss- and gain-of-function D. melanogaster mutants will be instrumental for addressing these questions (Neyen et al., 2014). Alternatively, multiplex quantitative RT-PCR can be used to affordably assess the effect of different microbial communities on the global RNA expression of a well-characterized D. melanogaster immunity gene panel (Neyen et al., 2014).
There are several other D. melanogaster assays, although less amendable to high throughput screening, that will be useful for investigating the interplay between microbial symbionts and host innate immunity. Melanin production occurs in the hemolymph (invertebrate equivalent to blood) of D. melanogaster to prevent microbial pathogenesis via encapsulation. Benoit et al. (2017) demonstrated that enteric symbionts of larval D. melanogaster regulated melanin production through activation of a highly conserved hematopoietic pathway. Though this study demonstrated yet another important role of the microbiota in host immunity, the specific microbes and signaling molecules responsible for these interactions have yet to be identified. Simple experiments testing one of the referenced melanization assays in mono-associated versus germ-free D. melanogaster could be used to address this line of inquiry (Neyen et al., 2014; Koyle et al., 2016). In addition, oxidative results in the production of reactive oxygen species (ROS) by Duox in the D. melanogaster gut and is crucial for immunity and microbiota regulation (Ryu et al., 2010). Abnormal commensalism appears to be detrimental to D. melanogaster due to consequential tissue damage associated with Duox overstimulation. The ability to assess ROS production by D. melanogaster in response to diverse microbial communities may provide valuable insights into how the microbiota regulates the balance of immune priming for pathogen eradication versus autoimmunity. Alternatively, evolutionary advancement toward more defensive Wolbachia (obligate endosymbionts) variants in D. melanogaster have been shown to increase resistance to Drosophila C virus (Faria et al., 2016). Thus, D. melanogaster could help to elucidate mechanisms by which symbiotic microbes affect the evolutionary trajectories of their host and improve scientific knowledge of cost-benefit relations associated with these mechanisms (Faria et al., 2015). In summary, D. melanogaster has potential to be used as a screening tool of microbe in vivo innate immunogenicity and as a model organism for evaluating distal site effects of crosstalk between the microbiota and innate immunity.
Interplay of Microbiota with Diet and Xenobiotics
Diet strongly modulates the gut microbiota (David et al., 2014). However, current understanding of how this interaction affects host nutritional status and fitness is still fragmented. D. melanogaster can be used to study diet-microbiota-host interactions due to: (a) their simplistic gut microbiota, (b) consistent feeding behavior regardless of food composition (Partridge et al., 2005), and (c) the simple generation and maintenance of germ-free stocks (Koyle et al., 2016). Wong et al. (2014) took advantage of these favorable characteristics and demonstrated that the D. melanogaster gut microbiota spares dietary B-vitamins on low-yeast diets, promotes protein nutrition, and suppresses lipid storage on high sugar diets. Others have also utilized D. melanogaster to mechanistically assess the life-extending properties of caloric restriction (Lee et al., 2014) and reduction of intestinal bacterial loads during later life stages (Brummel et al., 2004). These studies elude to the crosstalk between diet and the host microbiota as a regulator of important physiological aspects of D. melanogaster physiology such as lifespan (Brummel et al., 2004) and development (Storelli et al., 2011). Future high-throughput studies combining minor modification to D. melanogaster nutritional components and detailed investigation of the host-microbe nutrient interactions hold promise for dietary science. High-throughput dietary assessment in D. melanogaster prior to verification of findings in rodents or humans enables researchers to identify the most promising compounds that would not be feasible in costlier and more time-consuming mouse or human studies. The high proportion Lactobacillus spp.—species of Gram-positive bacteria commonly used as probiotics—in the D. melanogaster gut microbiota makes this model promising for preliminary identification of novel prebiotics.
The gut microbiota can affect the biotransformation of many xenobiotics, but this has been largely overlooked by pharmacology and toxicology. Beyond the direct interaction with xenobiotics, the microbiota has been shown to modulate host xenobiotic metabolic processes (Spanogiannopoulos et al., 2016). Since cytochrome P450 enzymes (major drug metabolism family) vary between species (Sakai et al., 2015), D. melanogaster have rarely been used to assess human-targeted pharmaceuticals. Nevertheless, D. melanogaster offers a platform to screen for microbe-xenobiotic interactions in vivo (Figure 2). In addition, specific in vivo microbe–drug interactions can be tested by mono-associating germ-free D. melanogaster with the microbe of interest followed by exposure to drug(s). This system is particularly useful for assessing the interactions of microbes with environmental toxins or drug-induced toxicity, where lethality, growth impairment, or behavioral abnormalities can be used as high throughput readouts. For instance, sub-chronic pesticide exposure can cause off-target toxicity in humans and wildlife species such as honey bees (Apis mellifera). Microbes can be screened for the ability to mitigate or exacerbate the aforementioned signs of xenobiotic induced-toxicity using the approach exemplified with organophosphate pesticides (Trinder et al., 2016). Preliminary work in our lab using the neonicotinoid insecticide, imidacloprid, suggests D. melanogaster can also be used as a model to identify compounds that induce microbiota changes, and with the help of available mutant stocks also elucidate the mechanism of compound-induced microbiota modification. Therefore, despite obvious limitations to these types of experiments (e.g., transient nature of non-resident microbes in the D. melanogaster intestinal tract), using D. melanogaster to obtain preliminary data from high throughput screens of microbe–drug interactions could save time and resources, as well as provide data with more predictive value (Pandey and Nichols, 2011).
Assessment of Probiotics and Prebiotics on Important Experimental Endpoints
The D. melanogaster microbiota is critical for modulating keystone properties of host health such as development (Storelli et al., 2011), lifespan (Brummel et al., 2004), and behavior (Sharon et al., 2010). These findings suggest that targetable microbiota modulation (e.g., probiotics, prebiotics, diet, antibiotics, etc.) may have predictive validity for understanding how microbiota alteration can influence aspects of human health and disease that are challenging to assess in mammals. In humans, Lactobacillus spp. and Bifidobacterium spp. are the most commonly used probiotic organisms capable of transiting through the harsh gastrointestinal tract to confer their host with physiological benefits. However, many probiotic organisms require further testing to determine strain-specific properties and mechanisms of action. Replication of Lactobacillus plantarum-induced D. melanogaster developmental growth resilience to malnutrition in mice provides evidence for this model’s insect-mammalian translation potential (Schwarzer et al., 2016). D. melanogaster provides a tool to evaluate the effect of extensive permutations of microbiota manipulation on important endpoints such as lifespan, development, and behavior in a cost-effective, timely, and feasible manner (Pandey and Nichols, 2011).
Drosophila melanogaster development progresses through well-defined stages of egg hatching, larval instar growth, pupae formation, and metamorphosis into an imago (adult fruit fly) in approximately 9 days. This rapid generation time enables classical recolonization studies of germ-free D. melanogaster to easily modify the dose and permutations of all microbiota constituents for assessment of their contribution to important host phenotypes singly or in combination. This feat is currently not possible with complex microbiotas such as those found in humans which can have 100–1000 different species of resident bacteria. Using this reductionist approach in D. melanogaster allows for easy identification of keystone species that correlate to host phenotypes of interest.
Prebiotic compounds could be conveniently added to D. melanogaster food media sources (most commonly cornmeal-molasses-yeast agar) and subsequently screened for induction of probiotic effects. In particular, mortality, body size, body weight, flight capability, stress, anesthesia response, activity, aggression, and fecundity are amendable to relatively high throughput screening in adult D. melanogaster (Pandey and Nichols, 2011). Alternatively, larval D. melanogaster can be used to assess mortality, body size, necrosis, and olfaction in a relatively high-throughput fashion (Pandey and Nichols, 2011). It is important to stress that work with D. melanogaster enables researchers to assess the most meaningful experimental endpoint, mortality, without the ethical conundrums of most other animal models. Furthermore, microbial species responsible for inducing host phenotypes of interest can be mechanistically followed-up using the large publicly available repositories (Bloomington Drosophila Stock Center, Kyoto Stock Center, etc.) of mutant D. melanogaster stocks.
Future Directions and Conclusion
The affordable, high throughput, and experimental manipulability of D. melanogaster make it an ideal model for deciphering complex systems biology questions with a more reductionist approach, which would be unfeasible in other models (Supplementary Table 1). For instance, D. melanogaster will be an important tool for understanding how manipulation of multi-variable factors such as drugs, toxins, diet, and microbes effect the host and its associated microbiota. Established D. melanogaster models of human neurologic, cardiovascular, neoplastic, metabolic, and other diseases could potentially be used to study the reported contribution of the microbiota to these conditions. Specifically, the foundational history of D. melanogaster as a neurobiology model provides a wide variety of established methods for investigation of the gut-brain axis. Microbiota-mediated modulation of this axis has been linked to a wide range of neurological diseases including multiple sclerosis, depression, anxiety, and autism.
In addition, chronic human diseases may be partially attributable to early life microbiota acquisition epigenetic priming (Thorburn et al., 2015). The microbiota and its metabolites play a role in epigenetic modifications at critical time points during development that can have long lasting health effects. D. melanogaster appear to be an excellent model to efficiently assess how microbial inheritance could affect development, epigenetic modification, and disease heritability in successive generations. The proposed use of D. melanogaster as an in vivo model for drug discovery with greater predictive validity than in vitro assays suggests that this model could also be used to screen probiotic or prebiotic properties (Pandey and Nichols, 2011). However, experimental design and translation from the D. melanogaster microbiota model will require careful consideration due to major shortcomings of a simplified microbiota (Kostic et al., 2013), lack of adaptive immunity (Apidianakis and Rahme, 2011), and differences in gastrointestinal anatomy and physiology (Lemaitre and Miguel-Aliaga, 2013). In conclusion, D. melanogaster is an underutilized model for deciphering mechanisms of host–microbial symbiotic relationships. The simplified microbiota structure of D. melanogaster makes it amendable for developing tools, techniques, and knowledge required to advance this field.
Author Contributions
MT, BD, JD, and GR conceived ideas for the manuscript. MT and BD drafted the manuscript. MT, BD, JD, and GR revised the manuscript. All authors agree to be accountable for the content of the work.
Funding
This work was funded by the Government of Canada Natural Sciences and Engineering Research Council of Canada (NSERC) (RGPIN-2014-05188).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Dr. Hon S. Leong (University of Western Ontario) and Dr. Sohrab N. Ali (University of Western Ontario) for guidance and resource provision in establishing the D. melanogaster model at our site.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00751/full#supplementary-material
Footnotes
References
Apidianakis, Y., and Rahme, L. G. (2009). Drosophila melanogaster as a model host for studying Pseudomonas aeruginosa infection. Nat. Protoc. 4, 1285–1294. doi: 10.1038/nprot.2009.124
Apidianakis, Y., and Rahme, L. G. (2011). Drosophila melanogaster as a model for human intestinal infection and pathology. Dis. Model. Mech. 4, 21–30. doi: 10.1242/dmm.003970
Bäckhed, F., Fraser, C. M., Ringel, Y., Sanders, M. E., Sartor, R. B., Sherman, P. M., et al. (2012). Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe 12, 611–622. doi: 10.1016/j.chom.2012.10.012
Benoit, J. B., Vigneron, A., Broderick, N. A., Wu, Y., Sun, J. S., Carlson, J. R., et al. (2017). Symbiont-induced odorant binding proteins mediate insect host hematopoiesis. Elife 6:e19535. doi: 10.7554/eLife.19535
Blum, J. E., Fischer, C. N., Miles, J., and Handelsman, J. (2013). Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster. mBio 4:e860-13. doi: 10.1128/mBio.00860-13
Brummel, T., Ching, A., Seroude, L., Simon, A. F., and Benzer, S. (2004). Drosophila lifespan enhancement by exogenous bacteria. Proc. Natl. Acad. Sci. U.S.A. 101, 12974–12979. doi: 10.1073/pnas.0405207101
Buchon, N., Silverman, N., and Cherry, S. (2014). Immunity in Drosophila melanogaster – from microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 14, 796–810. doi: 10.1038/nri3763
Byrd, A. L., and Segre, J. A. (2016). Infectious disease. Adapting Koch’s postulates. Science 351, 224–226. doi: 10.1126/science.aad6753
Chaston, J. M., Newell, P. D., and Douglas, A. E. (2014). Metagenome-wide association of microbial determinants of host phenotype in Drosophila melanogaster. mBio 5:e1631-14. doi: 10.1128/mBio.01631-14
Chinchore, Y., Gerber, G. F., and Dolph, P. J. (2012). Alternative pathway of cell death in Drosophila mediated by NF-κB transcription factor Relish. Proc. Natl. Acad. Sci. U.S.A. 109, E605–E612. doi: 10.1073/pnas.1110666109
Cho, O., Matsukura, M., and Sugita, T. (2015). Molecular evidence that the opportunistic fungal pathogen Trichosporon asahii is part of the normal fungal microbiota of the human gut based on rRNA genotyping. Int. J. Infect. Dis. 39, 87–88. doi: 10.1016/j.ijid.2015.09.009
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., Wolfe, B. E., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. doi: 10.1038/nature12820
Erkosar, B., Storelli, G., Defaye, A., and Leulier, F. (2013). Host-intestinal microbiota mutualism: “learning on the fly”. Cell Host Microbe 13, 8–14. doi: 10.1016/j.chom.2012.12.004
Faria, V. G., Martins, N. E., Magalhães, S., Paulo, T. F., Nolte, V., Schlötterer, C., et al. (2016). Drosophila adaptation to viral infection through defensive symbiont evolution. PLoS Genet. 12:e1006297. doi: 10.1371/journal.pgen.1006297
Faria, V. G., Martins, N. E., Paulo, T., Teixeira, L., Sucena, E., and Magalhaes, S. (2015). Evolution of Drosophila resistance against different pathogens and infection routes entails no detectable maintenance costs. Evolution 69, 2799–2809. doi: 10.1111/evo.12782
Fei, N., and Zhao, L. (2013). An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 7, 880–884. doi: 10.1038/ismej.2012.153
Food and Agriculture Organization of the United Nations, and World Health Organization [FAO/WHO] (2002). Guidelines for the Evaluation of Probiotics in Food. Joint FAO/WHO Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food. Available at: http://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Karpac, J., Younger, A., and Jasper, H. (2011). Dynamic coordination of innate immune signaling and insulin signaling regulates systemic responses to localized DNA damage. Dev. Cell 20, 841–854. doi: 10.1016/j.devcel.2011.05.011
Kostic, A. D., Howitt, M. R., and Garrett, W. S. (2013). Exploring host-microbiota interactions in animal models and humans. Genes Dev. 27, 701–718. doi: 10.1101/gad.212522.112
Koyle, M. L., Veloz, M., Judd, A. M., Wong, A. C., Newell, P. D., Douglas, A. E., et al. (2016). Rearing the fruit fly Drosophila melanogaster under axenic and gnotobiotic conditions. J. Vis. Exp. 113:e54219. doi: 10.3791/54219
Lee, B. C., Kaya, A., Ma, S., Kim, G., Gerashchenko, M. V., Yim, S. H., et al. (2014). Methionine restriction extends lifespan of Drosophila melanogaster under conditions of low amino-acid status. Nat. Commun. 5:3592. doi: 10.1038/ncomms4592
Lemaitre, B., and Hoffmann, J. (2007). The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743. doi: 10.1146/annurev.immunol.25.022106.141615
Lemaitre, B., and Miguel-Aliaga, I. (2013). The digestive tract of Drosophila melanogaster. Annu. Rev. Genet. 47, 377–404. doi: 10.1146/annurev-genet-111212-133343
Ma, D., Storelli, G., Mitchell, M., and Leulier, F. (2015). Studying host-microbiota mutualism in Drosophila: harnessing the power of gnotobiotic flies. Biomed. J. 38, 285–293. doi: 10.4103/2319-4170.158620
Neyen, C., Bretscher, A. J., Binggeli, O., and Lemaitre, B. (2014). Methods to study Drosophila immunity. Methods 68, 116–128. doi: 10.1016/j.ymeth.2014.02.023
Pandey, U. B., and Nichols, C. D. (2011). Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol. Rev. 63, 411–436. doi: 10.1124/pr.110.003293
Partridge, L., Piper, M. D., and Mair, W. (2005). Dietary restriction in Drosophila. Mech. Ageing Dev. 126, 938–950. doi: 10.1016/j.mad.2005.03.023
Reid, G., Younes, J. A., Van der Mei, H. C., Gloor, G. B., Knight, R., and Busscher, H. J. (2011). Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat. Rev. Microbiol. 9, 27–38. doi: 10.1038/nrmicro2473
Ryu, J. H., Ha, E. M., and Lee, W. J. (2010). Innate immunity and gut–microbe mutualism in Drosophila. Dev. Comp. Immunol. 34, 369–376. doi: 10.1016/j.dci.2009.11.010
Sakai, C., Iwano, S., Yamazaki, Y., Ando, A., Nakane, F., Kouno, M., et al. (2015). Species differences in the pharmacokinetic parameters of cytochrome P450 probe substrates between experimental animals, such as mice, rats, dogs, monkeys, and microminipigs, and humans. J. Drug Metab. Toxicol. 5:173. doi: 10.4172/2157-7609.1000173
Schwarzer, M., Makki, K., Storelli, G., Machuca-Gayet, I., Srutkova, D., Hermanova, P., et al. (2016). Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science 351, 853–857. doi: 10.1126/science.aad8588
Sharon, G., Segal, D., Ringo, J. M., Hefetz, A., Zilber-Rosenberg, I., and Rosenberg, E. (2010). Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 107, 20051–20056. doi: 10.1073/pnas.1302753110
Spanogiannopoulos, P., Bess, E. N., Carmody, R. N., and Turnbaugh, P. J. (2016). The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 14, 273–287. doi: 10.1038/nrmicro.2016.17
Storelli, G., Defaye, A., Erkosar, B., Hols, P., Royet, J., and Leulier, F. (2011). Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 14, 403–414. doi: 10.1016/j.cmet.2011.07.012
Thorburn, A. N., McKenzie, C. I., Shen, S., Stanley, D., Macia, L., Mason, L. J., et al. (2015). Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 6:7320. doi: 10.1038/ncomms8320
Trinder, M., McDowell, T. W., Daisley, B. A., Ali, S. N., Leong, H. S., Sumarah, M. W., et al. (2016). Probiotic Lactobacillus rhamnosus reduces organophosphate pesticide absorption and toxicity to Drosophila melanogaster. Appl. Environ. Microbiol. 82, 6204–6213. doi: 10.1128/AEM.01510-16
van Nood, E., Vrieze, A., Nieuwdorp, M., Fuentes, S., Zoetendal, E. G., de Vos, W. M., et al. (2013). Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415. doi: 10.1056/NEJMoa1205037
Wong, A. C., Dobson, A. J., and Douglas, A. E. (2014). Gut microbiota dictates the metabolic response of Drosophila to diet. J. Exp. Biol. 217, 1894–1901. doi: 10.1242/jeb.101725
Keywords: microbiota, microbiome, Drosophila melanogaster, animal model, fruit fly, probiotic, prebiotic, symbiosis
Citation: Trinder M, Daisley BA, Dube JS and Reid G (2017) Drosophila melanogaster as a High-Throughput Model for Host–Microbiota Interactions. Front. Microbiol. 8:751. doi: 10.3389/fmicb.2017.00751
Received: 05 February 2017; Accepted: 12 April 2017;
Published: 28 April 2017.
Edited by:
Omry Koren, Bar-Ilan University, IsraelReviewed by:
François Leulier, École Normale Supérieure de Lyon, FranceMargarida Matos, Universidade de Lisboa, Portugal
Gil Sharon, California Institute of Technology, USA
Copyright © 2017 Trinder, Daisley, Dube and Reid. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gregor Reid, Z3JlZ29yQHV3by5jYQ==
†These authors have contributed equally to this work.
 Mark Trinder
Mark Trinder Brendan A. Daisley
Brendan A. Daisley Josh S. Dube
Josh S. Dube Gregor Reid
Gregor Reid