
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 April 2017
Sec. Microbial Physiology and Metabolism
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.00648
 Angela Ruscitto1
Angela Ruscitto1 Kiyonobu Honma1
Kiyonobu Honma1 Vamsee M. Veeramachineni1
Vamsee M. Veeramachineni1 Kiyoshi Nishikawa2
Kiyoshi Nishikawa2 Graham P. Stafford3
Graham P. Stafford3 Ashu Sharma1*
Ashu Sharma1*Tannerella forsythia is a Gram-negative oral anaerobe associated with periodontitis. This bacterium is auxotrophic for the peptidoglycan amino sugar N-acetylmuramic (MurNAc) and likely relies on scavenging peptidoglycan fragments (muropeptides) released by cohabiting bacteria during their cell wall recycling. Many Gram-negative bacteria utilize an inner membrane permease, AmpG, to transport peptidoglycan fragments into their cytoplasm. In the T. forsythia genome, the Tanf_08365 ORF has been identified as a homolog of AmpG permease. In order to confirm the functionality of Tanf_08365, a reporter system in an Escherichia coli host was generated that could detect AmpG-dependent accumulation of cytosolic muropeptides via a muropeptide-inducible β-lactamase reporter gene. In trans complementation of this reporter strain with a Tanf_08365 containing plasmid caused significant induction of β-lactamase activity compared to that with an empty plasmid control. These data indicated that Tanf_08365 acted as a functional muropeptide permease causing accumulation of muropeptides in E. coli and thus suggested that it is a permease involved in muropeptide scavenging in T. forsythia. Furthermore, we showed that the promoter regulating the expression of Tanf_08365 was activated significantly by a hybrid two-component system regulatory protein, GppX. We also showed that compared to the parental T. forsythia strain a mutant lacking GppX in which the expression of AmpG was reduced significantly attenuated in utilizing free muropeptides. In summary, we have uncovered the mechanism by which this nutritionally fastidious microbe accesses released muropeptides in its environment, opening up the possibility of targeting this activity to reduce its numbers in periodontitis patients with potential benefits in the treatment of disease.
Tannerella forsythia is a Gram-negative bacterium strongly associated with severe forms of periodontal disease (Socransky et al., 1998; Tanner and Izard, 2006; Kassebaum et al., 2014), a common inflammatory disease worldwide that affects the soft and hard tissues leading to tooth loss (Kassebaum et al., 2014). The dependence of T. forsythia on exogenous growth factors becamefirst evident in the studies of Tanner et al. (1986) who noted that this bacterium grew on plates only when it was co-streaked with Fusobacterium nucleatum. Subsequently, it was determined that T. forsythia is unable to synthesis its own peptidoglycan precursors and has an absolute requirement for the cell wall constituent N-acetylmuramic acid (MurNAc) as a growth factor (Wyss, 1989). The dependence of the bacterium on exogenous peptidoglycan precursors became clear when it was observed by Wyss (1989) that the growth of bacterium could be rescued in the presence of F. nucleatum co-culture separated by a dialysis membrane. In vivo, T. forsythia at least in part, relies on peptidoglycan fragments released by the cohabiting bacteria during their cell wall recycling in the oral cavity. In Gram-negative bacteria, AmpG-like permeases play an important role in the transport of peptidoglycan (muropeptide) fragments from the periplasm to the cytoplasm, which are then broken down further by AmpD (amidase) and processed intracellularly via a salvage pathway and reenter the peptidoglycan synthesis pathway (Park and Uehara, 2008; Reith and Mayer, 2011; Johnson et al., 2013). AmpG belongs to the major facilitator superfamily (MFS) requiring an active proton motive force to transport GlcNAc-anhMurNAc disaccharide and disaccharide carrying stem peptides, the primary products of the action of lytic glycosylases on peptidoglycan, across the inner membrane of these bacteria (Cheng and Park, 2002). However, little is known of the mechanisms by which T. forsythia is able to utilize peptidoglycan from the environment. During bioinformatics screening, we identified a candidate gene in the bacterial genome coding for a putative muropeptide permease AmpG. Interestingly, this gene (Tanf_08365) is located on an operon that also codes for enzymes that in other bacteria are involved in peptidoglycan recycling as well as protein glycosylation. Interestingly, a recent study reported that this operon, which includes the putative ampG gene Tanf_08365 is highly downregulated in a T. forsythia mutant deficient in the regulatory protein GppX (Niwa et al., 2011). GppX is a unique hybrid two-component system (TCS) regulator comprising of an N-terminal histidine kinase (HK) sensor domain fused to a central receiver and C-terminal response regulator (RR) domain with a putative AraC-like helix-turn-helix DNA binding motif (Niwa et al., 2011). GppX deletion in T. forsythia was shown to pleiotropically affect a range of proteins including the S-layer glycoproteins involved in the virulence of this organism (Niwa et al., 2011). TCSs are signal transduction systems which are comprised of a membrane bound sensor histidine kinase (HK) and a cognate RR (Stock et al., 2000). Upon sensing external stimulus, the sensor kinase autophosphorylates at specific histidine residue and phosphoryl group is then transferred to the cognate RR, and usually enhancing its DNA binding and transcriptional activity (Stock et al., 2000).
Here, we reveal for the first time that Tanf_08365 is a functional AmpG ortholog in T. forsythia that is involved in muropeptide transport and furthermore uncover that its regulation is mediated by the direct interaction of GppX with the promoter region of a large operon involved in muropeptide and MurNAc scavenging.
Escherichia coli strains (Supplementary Table S1) used in this study were grown aerobically at 37°C in Luria–Bertani broth (LB) medium. All cloning experiments were performed using the electrocompetent recA mutant strain E. coli Stellar (Clontech). T. forsythia ATCC 43037 wild-type and ΔgppX (Niwa et al., 2011) mutant strains were grown anaerobically (10% CO2, 10% H2, 80% N2) in Trypticase Soy Broth (TSB).
Escherichia coli ΔampG/ampD double mutant named AR74 was generated by transduction of E. coli ampD mutant TU278 with a P1 lysate from an E. coli ΔampG mutant JW0423-1. E. coli ampD mutant was inoculated in 5 mL LB broth overnight at 37°C. The 1.5 mL of the cell culture was centrifuged at 12, 000 × g for 2 min. The supernatant was discarded and re-suspended in one half the original culture volume in sterile P1 solution (10 mM CaCl2/5 mM MgSO4). One hundred microliters of cell suspension was mixed with varying amounts of lysate (1, 10, 100 μL), incubated for 30 min and then 1 mL of LB broth with 1 M sodium citrate was added. After 1 h incubation at 37°C with gentle shaking, cells were plated on kanamycin plates with 5 mM sodium citrate. The transformed bacteria colonies were selected from the kanamycin plates and were analyzed by PCR to confirm the deletion of ampG and ampD genes. Colonies that gave the expected size PCR products with primers flanking ampG or ampD (AmpG forward, AmpG reverse, AmpD forward, and AmpD reverse) were considered double mutants and used further.
Tanf_08365 open-reading frame (ORF) was amplified from T. forsythia 43037 DNA with primers (ampGNde-F/ampGHind-R) and cloned into plasmid pACY-AR1 at NdeI and HindIII restriction sites to generate the plasmid pAC-Tanf_08635. pACY-AR1 was derived from pACYC184 by inactivating the plasmid backbone HindIII site by the Q5 Site-Directed Mutagenesis Kit (New England Biolabs), and then replacing the tet gene ORF with a short NdeI-HindIII linker using an inverse PCR strategy (In-Fusion, Clontech). DNA sequencing was performed to confirm correct insertion of the linker. For assessing the functionality of Tanf_08365, E. coli ΔampG/ampD mutant AR74 was transformed with pNU305 bearing muropeptide inducible lactamase gene (Lindberg et al., 1987) and pAC-Tanf_08365. Controls consisted of E. coli AR74/pNU305 with pACY-AR1 empty plasmid and lactamase assays were performed as below.
The assays were performed as described previously (Zhang et al., 2010). E. coli strains were grown to an OD600 of 0.1 and 1 μg/mL cefoxitin (Sigma) at 42°C was added to induce cell wall disruption. At 0, 30, 60 min post induction, 10 mL bacterial suspensions were taken, centrifuged for 10 min at 6,000 × g. Cells extracts obtained by brief sonication and centrifugation at 16, 000 × g for 5 min in cold. Lactamase activity of each strain lysate was assayed using the chromogenic nitrocefin substrate (Calbiochem). Briefly, nitrocefin stock solution (500 μg/mL in DMSO) was diluted 10-fold in 0.1 M phosphate, 1 mM EDTA pH 7.0 buffer and 5 μl of diluted solution was then added to 100 μL of cell lysate. After 30 min incubation at 20°C, absorbance was measured at OD486 in a microplate reader.
Total RNA was isolated from bacteria using the RNeasy kit (Qiagen). Single-stranded cDNA was synthesized using reverse transcriptase (Invitrogen Superscript III) and random hexamer primers as per the manufacturer’s protocol. The synthesized cDNA was amplified by PCR with primer sets spanning target genes Tanf_08345-Tanf_08365: region ‘a’ with TF1059F/TF1061R; region ‘b’ with TF1061F/TF1062R; region ‘c’ with TF1062F/TF1063R; region ‘d’ with TF1063F/TF1064R; region ‘e’ with TF1064F/TF1065R, and region ‘f’ with TF1065-TFIF. Primer sequences are listed in Supplementary Table S2.
5′ RLM RACE was performed to identify the transcription start site with the FirstChoice RLM-RACE kit (Ambion). Briefly, 5′ RACE adapter provided in the kit was ligated to RNA isolated by the Qiagen RNeasy Mini Kit with T4 RNA ligase. Reverse transcription reaction was then performed on the ligated RNA by random priming and Reverse Transcriptase. RT reaction then underwent an Outer 5′ RLM-RACE PCR using primers 5′ Outer Primer (provided by kit) and TSS-Outer. This was followed by a second PCR reaction (Inner 5′ RLM RACE) using primers 5′ Race Inner Primer (provided by kit) and TSS-Inner. PCR products were cloned into pGEM-T (Promega) cloning vector and sequenced.
DNA fragment encompassing nucleotides -400 to +1 (transcription start site) of the ampG operon was amplified with primer set ampGProEcoF/ampGProRBam and cloned into plasmid pRS414 into BamH1 and EcoR1 restriction sites to generate recombinant pRS-AmpGpromo. In parallel, a chimeric DNA fragment comprising an IPTG inducible synthetic lac promoter fused in front of a gppX ORF was generated by an overlap PCR strategy. Briefly, a PCR fragment was generated with T. forsythia 43037 DNA as a template and primer set LacGppxF1/GppxXho1. The product of this PCR was used as a template in a second PCR with primer set LacGppx-BamF2/GppxXho1). The PCR product was then cloned into plasmid pACY-AR2 into BamH1 and Xho1 to generate pAC-lacTFgppX. In addition, a construct having deletion of C-terminal helix-turn-helix (HTH) domain encoding fragment was derived from pAC-lacTFgppX by inverse PCR, and named pAC-lacTfgppΔHTH,
β-galactosidase activity was determined as described previously (Miller, 1992). E. coli strains were inoculated in overnight cultures, diluted 1/100 in 10 mL of fresh medium and induced with isopropyl β-D-1-thiogalactopyranoside (IPTG) until grown to mid-log phase. After mid-log phase growth was reached, cultures were incubated on ice for 20 min and pelleted by centrifuging for 10 min at 6,000 rpm. Cell pellets were re-suspended in the same volume of Z buffer (0.06 M Na2HPO4⋅7H2O, 0.04 M NaH2PO4⋅H2O, 0.01 M KCl, 0.001 M MgSO4, and 0.05 M β-mercaptoethanol) and OD600 was taken. Cells were diluted in Z buffer to 1 mL, permeabilized by adding 100 μL chloroform and 50 μL of 0.1% SDS, vortexed, and incubated for 5 min at 28°C. Enzyme activity (Miller Units) was measured by adding 0.2 mL of O-nitrophenyl-β-D-galactoside (ONPG; 4 mg/mL) substrate in phosphate buffer (0.06 M Na2HPO4⋅7H2O, 0.04 M NaH2PO4⋅H2O) to pH 7.0. OD 420 and 550 after sufficient yellow color has been seen. Add 0.5 mL of 1 M Na2CO3 to stop the reaction.
A previously described protocol (Desmarais et al., 2014) was followed for purifying peptidoglycan from F. nucleatum ATCC 25586 cells. F. nucleatum was inoculated in 250 mL of BHI broth and grown to OD600 of 0.6. Cultures was then spun at 5,000 g for 10 min and re-suspended in 3 mL of phosphate buffered saline (PBS). Cell suspension was added to boiling 6 mL 6% sodium dodecyl sulfate (SDS) solution and continued to boil for 3 h, and then left to stir overnight. Next day ultracentrifugation at 400,000 × g for 20 min was continually done until all the SDS was fully removed. The cell wall pellet was then treated with Pronase E (100 μg/mL final concentration) at 60°C for 2 h, ultracentrifuged and treated overnight with muramidase (40 μg/mL final concentration). Muropeptides released after digestion were collected by centrifugation at 15, 000 × g for 10 min at room temperature.
Fluorophore-assisted carbohydrate electrophoresis (FACE) was performed to check the quality of the peptidoglycan fragments as described previously (Young, 1996). Briefly, 5–10 μL of isolated peptidoglycan was dried using centrifugal vacuum evaporator and 5 μL of 0.2 M ANTS in 2.6 M acetic acid and 5 μL of 1 M NaCNBH3 in DMSO was added. The sample was incubated for 37°C for 15–18 h and vacuum centrifuged overnight. The sample was then separated by electrophoresis using a 35% acrylamide gel (Young, 1996). For muropeptide analysis by mass spectrometry, muropeptide oligomers collected after muramidase treatment were concentrated in vacuo to 20 μl and then acidified by addition of 10% formic acid solution. LC-MS analysis was carried out by injecting 7.5 μl of the concentrated muropeptide solution onto a ZORBAX SB-C18 reversed-phase HPLC column (5 μm, 4.6 mm × 150 mm) attached to a Thermo Finnigan LCQ advantage mass spectrometer. The muropeptides were eluted at 200 μl/min using a gradient protocol with 0.1% aqueous formic acid (solvent A) and 0.1% formic acid in acetonitrile (solvent B). The gradient conditions were 0 to 45 min – linear 0 to 20% solvent B; 45 to 90 min – linear 20 to 50% solvent B; 90 to 120 min – linear 50 to 100% solvent B. The resulting LC-MS data was analyzed using Xcalibur QualBrowser program (Thermo-Electron, San Jose, CA, USA).
Tannerella forsythia wild-type and ΔgppX mutant strains were grown in TSB-serum (TSB medium with 5% fetal bovine serum) supplemented with 0.2% muropeptides or 0.2% MurNAc. TSB-serum medium was used as a negative control. Growth was measured at OD600 for 8 days.
Statistical differences were analyzed by ANOVA, and paired comparisons were performed by Tukey’s post hoc test. Statistical analyses were performed with the Prism Software (Graph Pad, San Diego, CA, USA). Data were expressed as mean ± SD and differences were considered to be statistically significant at P < 0.05.
In E. coli and many other bacteria, muropeptides are transported across the inner membrane through an AmpG permease (Park and Uehara, 2008). Given the reliance of T. forsythia on muropeptide scavenging for survival, we set out to identify an AmpG ortholog in T. forsythia. In silico analysis of the T. forsythia ATCC 43037 draft genome (JUET00000000.1)1 indicated Tanf_08365 as a potential MFS protein similar to AmpG permease having 12 transmembrane helices based on a transmembrane helical and topology prediction by the HMMTOP model (Tusnady and Simon, 2001) via the ExPASy server. Tanf_08365 ORF showed 24 and 79% amino acid sequence identity with the AmpG protein of E. coli (accession no. WP_021557614.1) and Bacteroides thetaiotaomicron (accession no. WP_055269099.1), respectively. To assess the function of Tanf_08365 in muropeptide recycling, an E. coli reporter system was generated that relies on the induction of β-lactamase from a reporter plasmid only when a high enough concentration of muropeptides accumulate in the cell via a functional AmpG permease. The reporter E. coli strain is a double mutant lacking the ampG and ampD genes and harbors a plasmid with muropeptide inducible β-lactamase gene. This is enabled by the fact that the native muropeptide uptake gene (ampG) is absent while deletion of the murein amidase (N-acetyl-anhydromuramyl-L-alanine-amidase) gene ampD prevents degradation of muropeptides, enabling accumulation of muropeptides in the cytoplasm. Thus, β-lactamase expression is disabled in this strain unless a functional muropeptide transporter is provided in trans to enable muropeptide accumulation in the cytoplasm. Therefore, this reporter system allowed us to test the function of the heterologous T. forsythia ampG homolog based on the induction of lactamase activity in the reporter E. coli/plasmid strain. To generate this reporter system, an E. coli ΔampD mutant was transduced with a P1 lysate prepared from an E. coli ΔampG mutant to obtain E. coli ΔampG/ΔampD double mutant. This double mutant, named AR74, was then transformed with the pNU305 reporter plasmid expressing β-lactamase from a muropeptide inducible promoter element. A randomly selected positive transformant, named AR74/pNU305, served as the reporter strain. In parallel, the Tet resistance gene of pACYC184 plasmid was replaced with the Tanf_08365 ORF using In-Fusion cloning strategy to generate the recombinant plasmid pAC-Tanf_08635. AR74/pNU305 was transformed with either pAC-Tanf_08635, or the pACY empty vector. E. coli wild-type strain BW25113 and E. coli ΔampD mutant served as positive controls. The β-lactamase induction at various time intervals was determined using the chromogenic substrate nitrocefin by measurement of product at 486 nm. The results showed that when Tanf_08365 (Tf ampG) was provided in trans, the reporter strain expression of β-lactamase was significantly induced in comparison to the empty vector control and the levels were similar to that in the ampD single mutant where the E. coli ampG gene is functional (Figure 1). Together, these data indicated that Tanf_08365 is a functional muropeptide permease AmpG of T. forsythia, and we label Tanf_08365 as Tf AmpG.
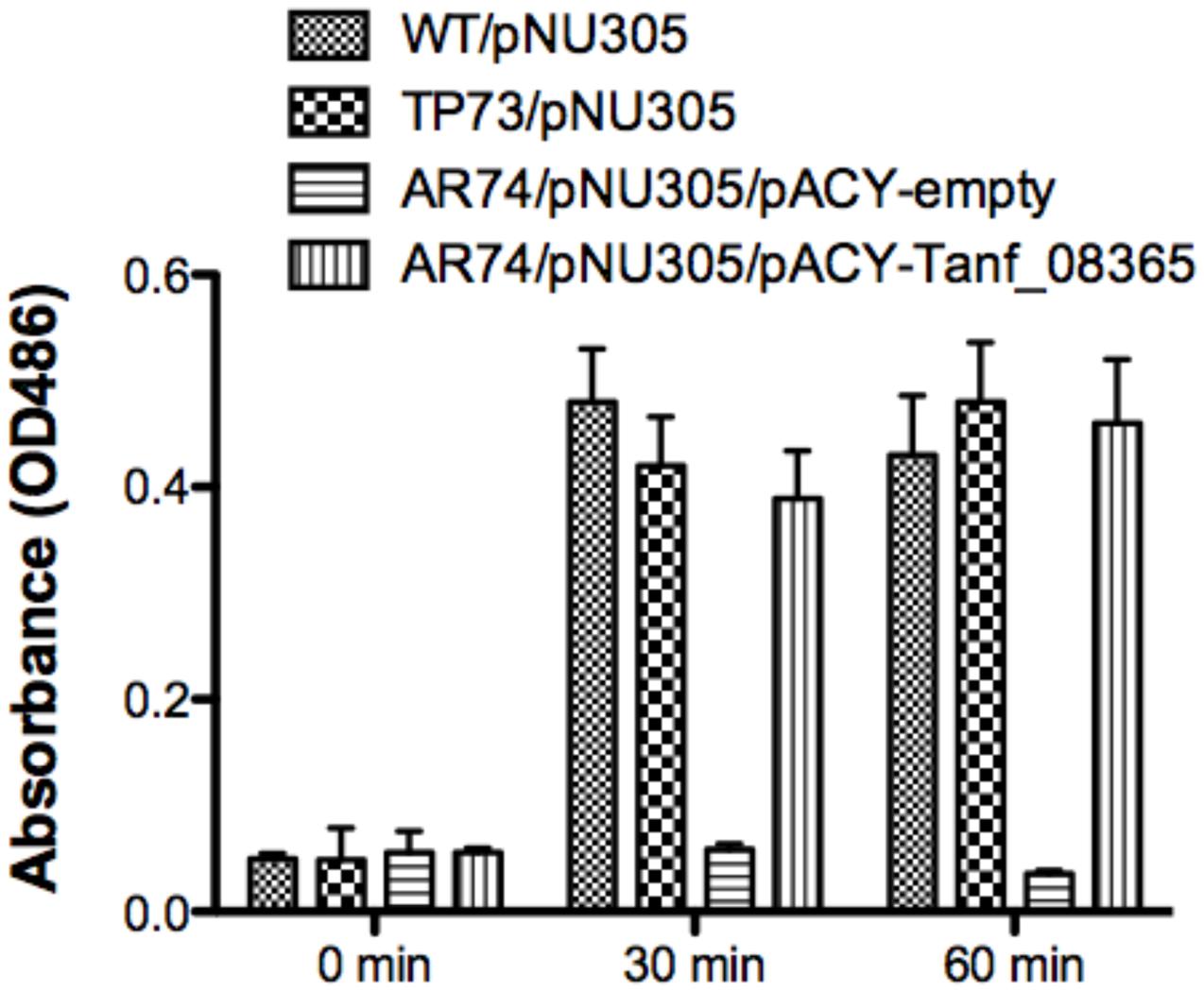
FIGURE 1. Reporter assay for detection of AmpG permease activity. The β-lactamase assay was performed on Escherichia coli strains BW25113 (parental strain), TP73 (ΔampD)/pNU305, AR74 (ΔampG/ΔampD)/pNU305/ pACY-AR1 (empty vector), and AR74/pNU305/pAC-Tanf_08635. Activity was determined by chromogenic nitrocefin substrate induced at 0, 30, 60 min and absorbance was measured at OD486. Data are representative of three independent experiments with similar results. Each value represents the mean (±SD) of three values measured in one representative assay.
We next wanted to ascertain how expression of AmpG might be regulated in T. forsythia. The genetic context of Tf AmpG shows that it is potentially part of a large gene cluster in the chromosome stretching from Tanf_08345-Tanf_08370 in the contig_82 DNA sequence (NZ_JUET01000082) (Figure 2). Interestingly, the Tanf_08345-Tanf_08370 gene cluster is located immediately upstream of a three gene operon Tanf_08375-Tanf_08385 for MurNAc utilization recently identified by our group (Ruscitto et al., 2016). This operon expresses a transporter TfMurT/TfMurK that transports and phosphorylates environmental MurNAc and an enzyme TfMurQ etherase that converts cytoplasm internalized MurNAc-6-P to GlcNAc-6-P for shuttling into glycolytic pathway or peptidoglycan biosynthetic pathway. Previous work by Niwa et al. (2011) showed that transcription of genes associated with the Tanf_08345-Tanf_08370 cluster [which includes the ampG (Tanf_08365) gene studied here] were significantly downregulated in a mutant lacking the GppX regulator protein. To confirm that genes on this cluster indeed form an operon, total RNA from T. forsythia ATCC 43037 cells was extracted and co-transcription of the relevant genes was analyzed using RT-PCR as outlined in Figure 2A. The results showed that Tanf_08345 to Tanf_08370 are transcribed as a single RNA transcript (Figure 2B), since PCR products of the expected size were obtained with primer pairs (Supplementary Table S2) designed to bridge the ends between the ORFs of adjacent genes, and, thus, yielding amplification products only when co-transcription was occurring. In addition, our data showed that the previously identified murTKQ gene cluster formed a contiguous transcription unit with the upstream ampG operon, i.e., from Tanf_08345-Tanf_08385. Additionally, our data defined that murQ is the final gene in this operon since a primer set bridging the region between murQ and Tanf_08390 yielded no PCR product in these experiments (Figure 2B). After establishing that the T. forsythia ampG gene Tanf_08365 (Tf ampG) is part of this operon, the transcription site for the operon was determined; which was located 17 bp upstream (an ‘A’ residue) from the translational start codon of Tanf_08345 (Figure 2C). Given that the genes in the cluster are co-transcribed and that the levels of transcription of all the genes in the cluster are downregulated in a gppX mutant (Niwa et al., 2011), we set out to determine whether the regulation by GppX might be direct. In order to test this we constructed a reporter system where the putative promoter region of this operon encompassing -400 to +1 (TSS) region (5′ of the first gene Tanf_08345) was cloned upstream of a promoterless lacZ ORF in a reporter plasmid pRS414 and placed in E. coli (Simons et al., 1987). In parallel, a synthetic IPTG inducible lac promoter/operator fused to T. forsythia GppX coding fragment with a C-terminal 6xHis tag sequence was obtained by overlap PCR to generate lac-gppX-6xHis chimera Tetracycline resistance gene in pACYC184 was then replaced with this chimeric fragment as described in “Materials and Methods” section. In addition, a chimeric fragment lacking HTH domain of GppX with a 6xHis tag (lac-gppxΔHTH-6xHis) was also cloned into pACYC184. T. forsythia promoter-lacZ and lac-gppX-6His (or lac-gppxΔHTH-6xHis) constructs were placed in the same strain of E. coli and the expression of β-galactosidase with and without IPTG induction was determined with chromogenic ONPG. This allowed us to test the hypothesis that GppX might directly regulate the AmpG containing operon mentioned above. The results showed that the operon promoter is induced sevenfold when expression of GppX is turned on with IPTG. In contrast, in the absence of IPTG the GppX construct fails to elicit this response. A strain that contained vector with IPTG inducible Tet gene as control showed no lacZ expression (negative control). In addition, a reporter strain containing pAC-lacTfgppΔHTH that lacked the DNA binding HTH domain of GppX showed no lacZ expression after induction with IPTG. The western immunoblotting results using anti-His tag antibody showed expected size expressed proteins for each of the chimeric constructs (full length GppX or ΔHTH-GppX) (Supplementary Figure S3); confirming that the reduced promoter induction in the E. coli cells expressing ΔHTH GppX construct is not due to reduced protein expression or degradation of the recombinant protein, but is due to its lack of DNA binding ability. E. coli strain MG1655 was used as a positive control for lacZ responsiveness as it contains the native IPTG inducible lac operon containing β-galactosidase (Figure 3).
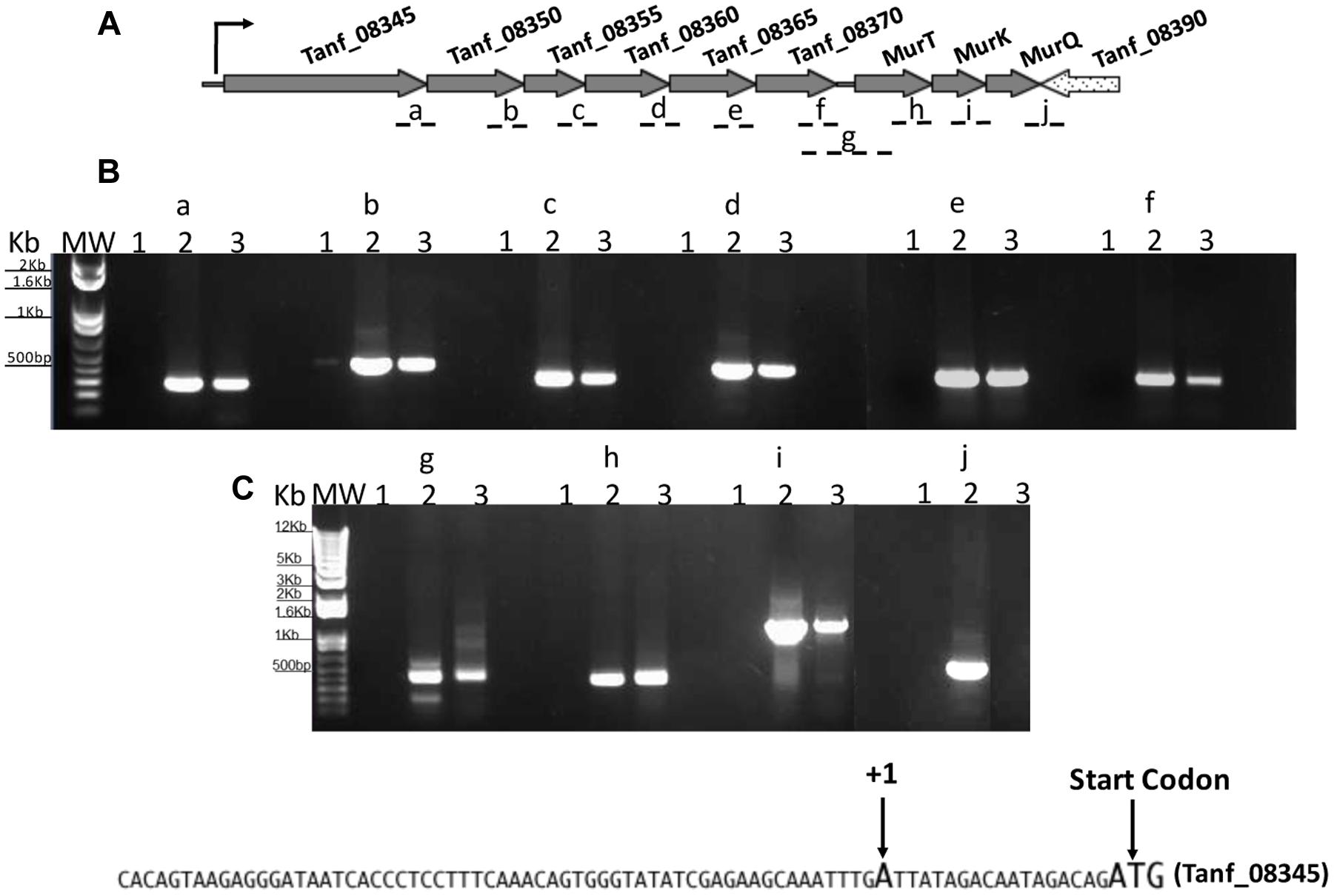
FIGURE 2. Analysis of AmpG and MurNAc utilization operon. RT-PCR analysis with (A) primer sets spanning adjacent genes (fragments a-j). Tanf_08345, xanthane lyase; Tanf_08350 (gtf), glycosyltransferase; Tanf_08355 (gtf), glycosyltransferase; Tanf_08360 (lytB), amidase enhancer precursor; Tanf_08365 (ampG), muropeptide permease; Tanf_08370 (ybbC), conserved hypothetical protein. (B) PCR products were separated on a 1% agarose gel. RNA samples with no reverse transcription reaction as template controls were run in lanes 1, genomic DNA as template in lanes 2, and cDNA as template for each primer set in lanes 3. MW; DNA ladder. (C) DNA sequence showing transcriptional start site determine by 5′RACE.
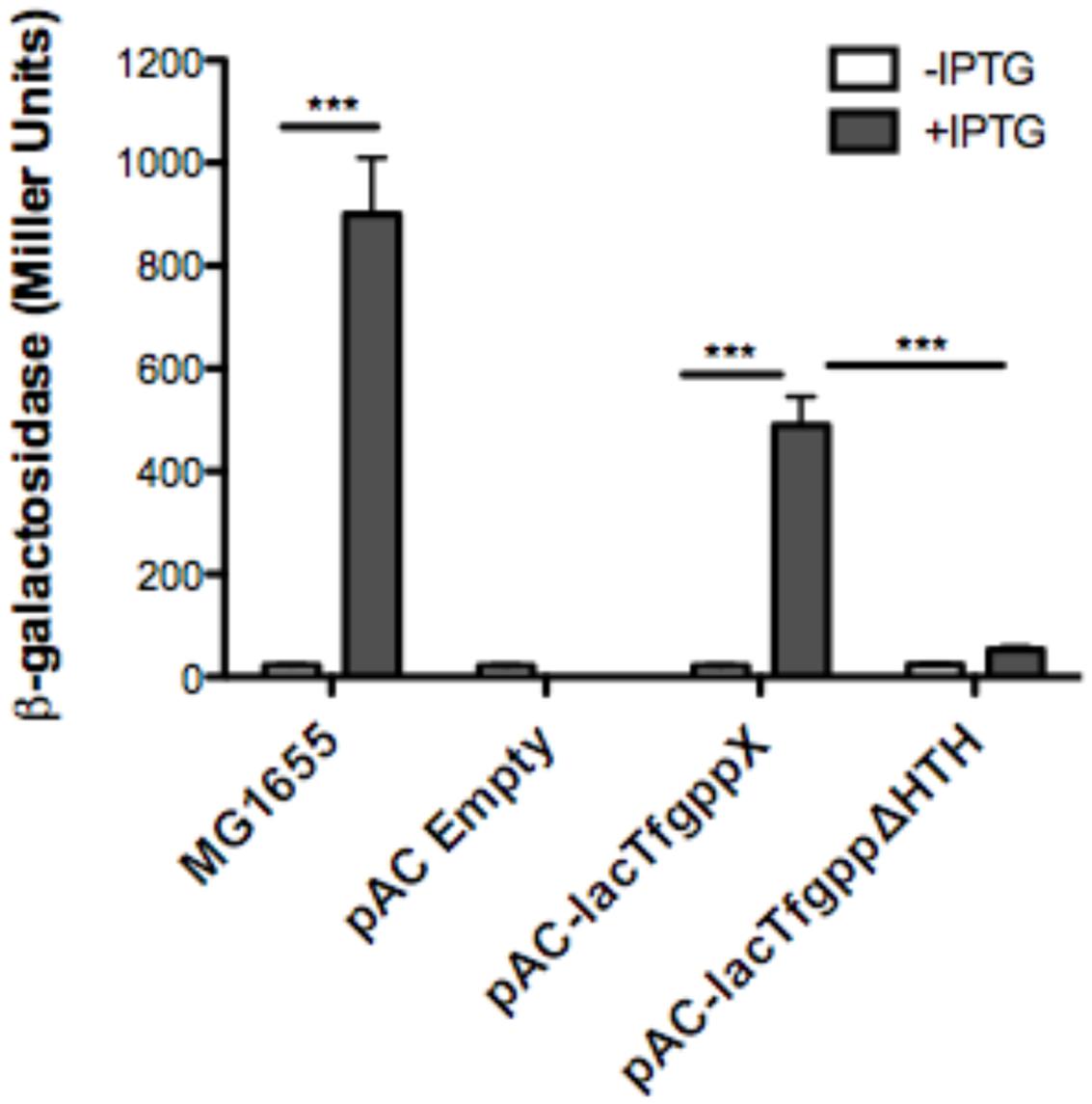
FIGURE 3. Reporter assay to assess GppX as a transcription regulator. The β-galactosidase assay was performed on E. coli strains MG1655 (positive control), pAC-lacTFggpΔHTH (negative control), pACYC184 (empty vector), and pAC-lac-gppX (in trans). Activity was determined by chromogenic O-nitrophenyl-β-D-galactoside (ONPG) substrate with, or without induction of isopropyl β-D-1-thiogalactopyranoside (IPTG) and measured using Miller Units. Each value represents the mean (±SD) of three values measured in one representative assay. Data are representative of three independent experiments with similar results, ∗∗∗P < 0.001.
Tannerella forsythia is auxotrophic for peptidoglycan amino sugar MurNAc, and thus requires exogenous MurNAc for growth in vitro. Our data so far had highlighted that T. forsythia produces a muropeptide transport system encoded by the ampG gene that is part of an operon directly under the control of the GppX transcription factor. We therefore surmised that T. forsythia should not only be able to grow on muropeptides as growth factors but also that this function should be dependent on GppX. In addition, since the genes involved in MurNAc utilization are part of the same operon we predicted that MurNAc utilization in T. forsythia would also be under the control of GppX. As a first step in examining these hypotheses, we set out to examine whether T. forsythia can utilize exogenous muropeptides as these would be readily available in vivo in the oral cavity as byproducts of cell wall recycling or death of cohabiting bacteria. To test this, muropeptides were prepared from F. nucleatum after muramidase digestion as described in the section “Materials and Methods.” The quality and composition of muropeptide fraction was confirmed by subjecting the isolated fraction to FACE and Mass Spectrometry. As shown, a typical ladder-like pattern was observed on a FACE gel (Supplementary Figure S1), and MS analysis (Supplementary Figure S2) confirmed the presence of muropeptide oligomers in the isolated fraction. T. forsythia was grown in broth supplemented with muropeptides in the absence of MurNAc supplementation. The data showed for the first time that T. forsythia grew in broth supplemented with muropeptides prepared from F. nucleatum alone or MurNAc (positive control) (Figure 4).
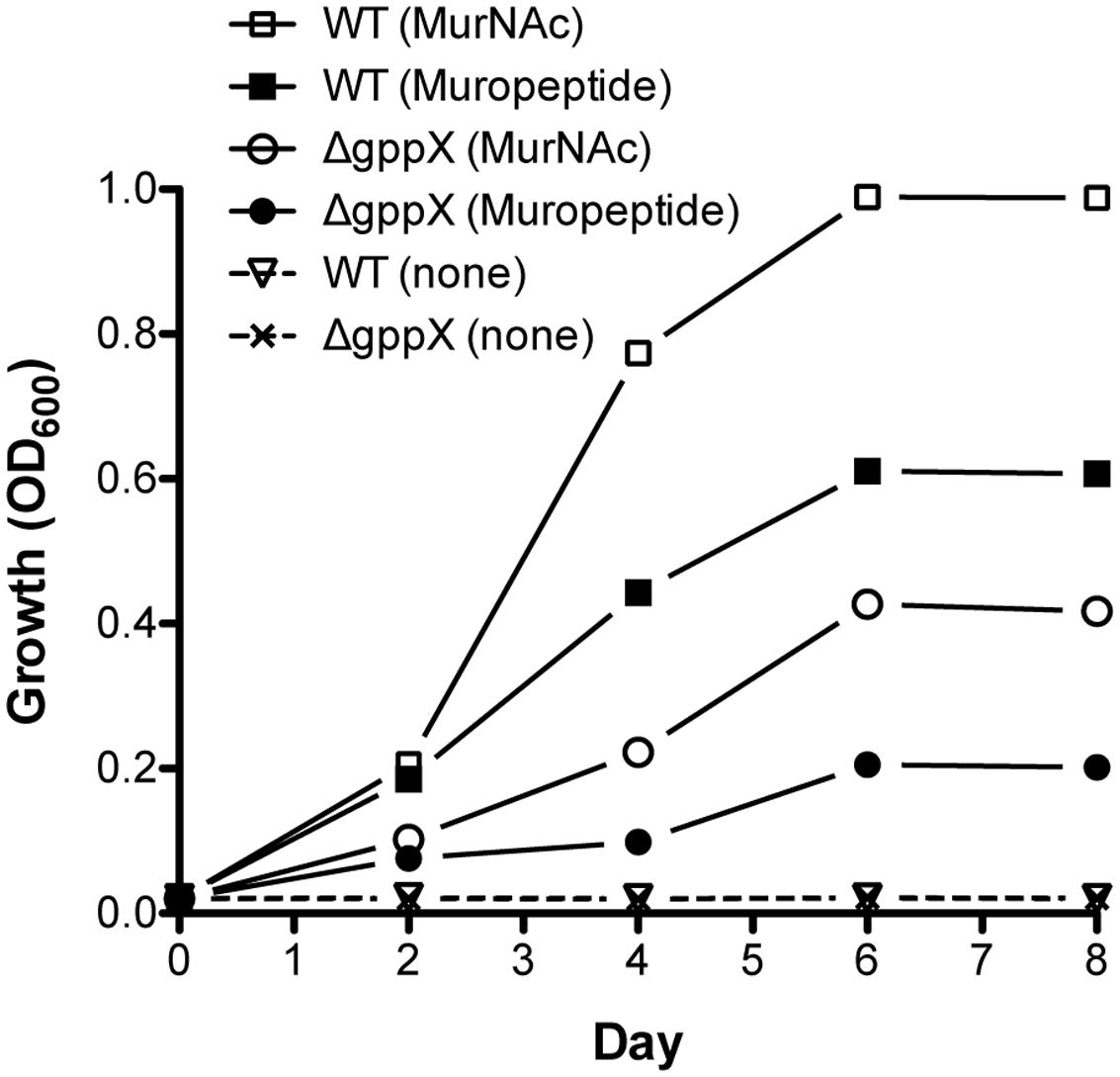
FIGURE 4. Growth of Tannerella forsythia on muropeptides. Growth of T. forsythia wild-type and ΔgppX in M9 liquid medium supplemented with 0.2% muropeptides or 0.2% MurNAc was measured at OD600. Results of one out of three independent cultivations with similar outcome are given.
Since previous data had indicated that GppX controlled the ampG containing operon we set out to now test whether a GppX mutant was deficient in its ability to grow on these muropeptides in this assay. The results showed that in the presence of muropeptides the growth of the wild-type bacteria was significantly higher than that of the mutant strain; reaching fivefold higher OD600 values. Moreover, given that the murTKQ genes for MurNAc utilization are part of a contiguous operon controlled by GppX the growth rate in the presence of MurNAc was also predictably lower for GppX-deficient mutant compared to the wild-type strain. Taken together, the reduction in growth displayed by the ΔgppX mutant strain demonstrated that GppX via takes an important role in the regulation of peptidoglycan transport.
It was first reported by Tanner et al. (1986) that the growth of T. forsythia could be rescued by co-culturing the bacterium with F. nucleatum, which was thought to provide peptidoglycan precursors. Subsequently, Wyss (1989) demonstrated that the growth of T. forsythia could be rescued in the presence of exogenous peptidoglycan amino sugar MurNAc. The reason for T. forsythia’s strict dependence on exogenous MurNAc for growth became evident with the availability of the genome sequence of the organism. The genes for MurA and MurB enzyme homologs involved in the biosynthesis of MurNAc from GlcNAc were found to be absent in the organism (Sharma, 2011). Thus, it is thought that in the hostile oral environment T. forsythia salvages peptidoglycan fragments (muropeptides) released during the cell wall recycling of cohabiting bacteria. The peptidoglycan scavenging by T. forsythia becomes even more relevant given the fact the human host does not make the peptidoglycan amino sugars. Since the AmpG permease in bacteria plays a major role in the recycling of muropeptides (Cheng and Park, 2002), we sought out to identify a functional homolog of AmpG and its regulation in T. forsythia.
This study identified and confirmed that the gene Tanf_08365 in T. forsythia codes for a functional muropeptide permease, which we designate Tf AmpG. Furthermore, we show that AmpG expression in T. forsythia is under the direct control of the GppX (Tanf_13760) regulator (Niwa et al., 2011), a hybrid two-component regulatory protein with an N-terminal HK and a C-terminal RR. A previous study reported that deletion of GppX protein Tanf_13760 resulted in the reduced transcription of glycosylation related operon Tanf_08345-Tanf_08370 in T. forsythia, of which Tanf_08365, the focus of our study here, is part of (Niwa et al., 2011). However, it could not be ascertained from this previous study whether GppX influences the transcription of this operon directly or indirectly. TCSs are known to impact expression of target genes directly or through mediation of collateral regulatory networks (Graham et al., 2002). Our data presented here utilizing a reporter system in E. coli showed that the promoter driving the expression of Tanf_08345-Tanf_08370 operon was directly regulated by GppX in the E. coli heterologous system where a direct regulation and interaction with promoter is the only explanation for the data observed. The direct involvement of GppX interaction with the promoter was further corroborated from the results showing that a construct lacking the DNA binding HTH domain of GppX failed to activate the promoter.
In addition, in this study we provided direct evidence that T. forsythia can utilize exogenous muropeptides and F. nucleatum ATCC 25586 can support T. forsythia growth. As shown, the growth of T. forsythia in muropeptides extracted from F. nucleatum revealed that peptidoglycan fragments can sustain T. forsythia growth. To validate the role of GppX in the regulation of muropeptide transport via AmpG permease, the growth of T. forsythia wild-type and ΔgppX strains were compared. The results showed that the growth of wild-type bacteria was higher than that of the mutant strain. The reduction in growth displayed by the ΔgppX mutant strain demonstrated that peptidoglycan transport in T. forsythia dependent on AmpG permease is regulated by GppX. Furthermore, since the growth of the mutant was not completely abolished on MurNAc or muropeptides suggest that the ampG-containing operon is expressed constitutively at basal levels. These data are line with the previous study showing that gppX mutation leads to down regulation but not complete abolition of expression of the operon (Niwa et al., 2011). Although we were not able to complement the ΔgppX mutant with a functional GppX protein via a strategy involving replacement of gppX::Em locus by a gppX-Cm (chloramphenicol marker attached to wild-type gppX) fragment, our data from the E. coli reporter strain do confirm that GppX protein is involved in the regulation of MurNAc/muropeptide operon. We believe growth attenuation and possibly other changes due to GppX deficiency made bacterial cells less amenable to genetic manipulation. Furthermore, dependence on F. nucleatum peptidoglycan fragments by T. forsythia is likely responsible for the close physical association in the dental plaque and synergy between these two organisms in terms of virulence. Previous studies have shown that T. forsythia and F. nucleatum form synergistic biofilms in vitro (Sharma et al., 2005), in mixed oral infection stetting induce synergistic alveolar bone loss in a mouse oral infection model (Settem et al., 2012), and are present in close proximity to each other in a human subgingival plaque biofilms (Zijnge et al., 2010). In summary, the nutritionally fastidious periodontal pathogen T. forsythia unable to synthesis its own peptidoglycan amino sugar MurNAc scavenges muropeptides from the environment via AmpG-dependent peptidoglycan recycling pathway (Figure 5), whose expression is regulated by a hybrid two-component transcription regulator, GppX. We envision a possibility wherein small molecule inhibitors targeting GppX regulator could be developed as antimicrobial compounds against T. forsythia, and thus as treatment against periodontitis.
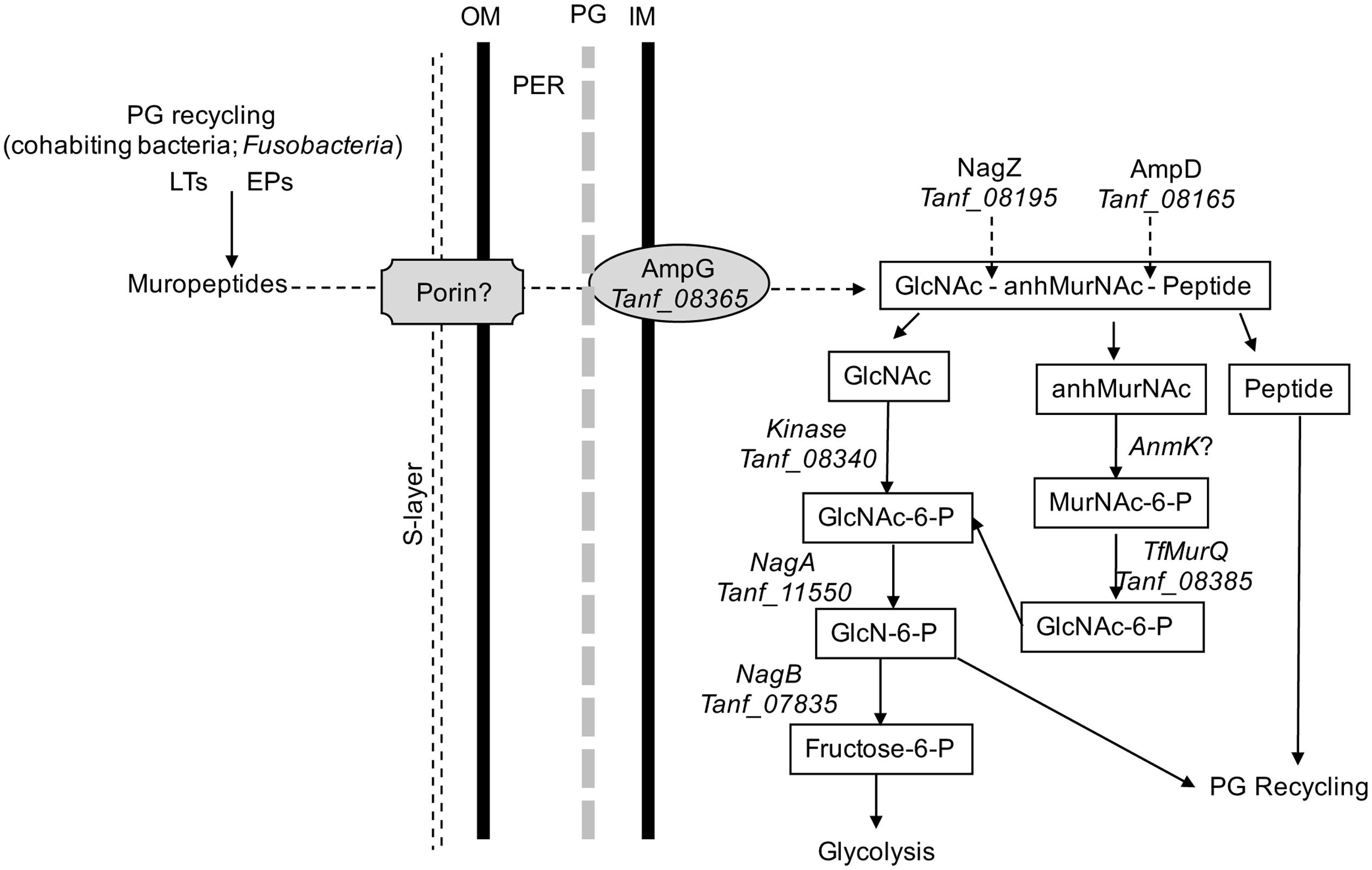
FIGURE 5. Schematic model of T. forsythia peptidoglycan recycling pathway. Abbreviations: anhMurNAc, anhydro-N-acetylmuramic acid; EPs, endopeptidases; GlcNAc, N-acetylglucosamine; GlcNAc-6-P, N-acetylglucosamine-6-phosphate; GlcN-6-P, glucosamine-6-phosphate; IM, inner cytoplasmic membrane; LTs, transglycosylases; MurNAc-6-P, N-acetylmuramic acid 6-phosphate; OM, outer membrane; PER, periplasmic space.
Conceived and designed the experiments: AR and AS. Performed the experiments: AR, KH, and VV. Analyzed the data: AR, KH, VV, KN, GS, and AS. Contributed reagents/analysis tools: KN, VV, and GS. Wrote the paper-original: AR, VV, GS, and AS. Review and editing: AR, GS, and AS.
This work was supported in part by U.S. Public Health grants DE14749 and DE22870 (both to AS). AR was supported by a T32 Training grant (DE023526).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors wish to thank Tsuyoshi Uehara and Christoph Mayer for helpful discussions during the development stages of this study. We thank Hongbaek Cho and Thomas Bernhardt at Harvard Medical School for providing E. coli strains TP73 and TU278.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00648/full#supplementary-material
Cheng, Q., and Park, J. T. (2002). Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J. Bacteriol. 184, 6434–6436. doi: 10.1128/JB.184.23.6434-6436.2002
Desmarais, S. M., Cava, F., de Pedro, M. A., and Huang, K. C. (2014). Isolation and preparation of bacterial cell walls for compositional analysis by ultra performance liquid chromatography. J. Vis. Exp. 15:e51183. doi: 10.3791/51183
Graham, M. R., Smoot, L. M., Migliaccio, C. A., Virtaneva, K., Sturdevant, D. E., Porcella, S. F., et al. (2002). Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. U.S.A. 99, 13855–13860. doi: 10.1073/pnas.202353699
Johnson, J. W., Fisher, J. F., and Mobashery, S. (2013). Bacterial cell-wall recycling. Ann. N. Y. Acad. Sci. 1277, 54–75. doi: 10.1111/j.1749-6632.2012.06813.x
Kassebaum, N. J., Bernabe, E., Dahiya, M., Bhandari, B., Murray, C. J., and Marcenes, W. (2014). Global burden of severe periodontitis in 1990-2010: a systematic review and meta-regression. J. Dent. Res. 93, 1045–1053. doi: 10.1177/0022034514552491
Lindberg, F., Lindquist, S., and Normark, S. (1987). Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundii beta-lactamase. J. Bacteriol. 169, 1923–1928. doi: 10.1128/jb.169.5.1923-1928.1987
Miller, J. H. (1992). A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Plainview, N. Y: Cold Spring Harbor Laboratories.
Niwa, D., Nishikawa, K., and Nakamura, H. (2011). A hybrid two-component system of Tannerella forsythia affects autoaggregation and posttranslational modification of surface proteins. FEMS Microbiol. Lett. 318, 189–196. doi: 10.1111/j.1574-6968.2011.02256.x
Park, J. T., and Uehara, T. (2008). How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 72, 211–227. doi: 10.1128/MMBR.00027-07
Reith, J., and Mayer, C. (2011). Peptidoglycan turnover and recycling in Gram-positive bacteria. Appl. Microbiol. Biotechnol. 92, 1–11. doi: 10.1007/s00253-011-3486-x
Ruscitto, A., Hottmann, I., Stafford, G. P., Schaffer, C., Mayer, C., and Sharma, A. (2016). Identification of a novel N-acetylmuramic acid (MurNAc) transporter in Tannerella forsythia. J. Bacteriol. 198, 3119–3125. doi: 10.1128/JB.00473-16
Settem, R. P., El-Hassan, A. T., Honma, K., Stafford, G. P., and Sharma, A. (2012). Fusobacterium nucleatum and Tannerella forsythia induce synergistic alveolar bone loss in a mouse periodontitis model. Infect. Immun. 80, 2436–2443. doi: 10.1128/iai.06276-11
Sharma, A. (2011). “Genome functions of Tannerella forsythia in bacterial communities,” in Oral Microbial Communities: Genome Inquiry and Interspecies Communication, ed. P. E. Kolenbrander (Washington, DC: American Society for Microbiology), 135.
Sharma, A., Inagaki, S., Sigurdson, W., and Kuramitsu, H. K. (2005). Synergy between Tannerella forsythia and Fusobacterium nucleatum in biofilm formation. Oral Microbiol. Immunol. 20, 39–42. doi: 10.1111/j.1399-302X.2004.00175.x
Simons, R. W., Houman, F., and Kleckner, N. (1987). Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53, 85–96. doi: 10.1016/0378-1119(87)90095-3
Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C., and Kent, R. L. Jr. (1998). Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25, 134–144. doi: 10.1111/j.1600-051X.1998.tb02419.x
Stock, A. M., Robinson, V. L., and Goudreau, P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215. doi: 10.1146/annurev.biochem.69.1.183
Tanner, A. C., and Izard, J. (2006). Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontol. 2000 42, 88–113. doi: 10.1111/j.1600-0757.2006.00184.x
Tanner, A. C. R., Listgarten, M. A., Ebersole, J. L., and Strzempko, M. N. (1986). Bacteroides forsythus sp. nov., a slow growing, fusiform Bacteroides sp. from the human oral cavity. Int. J. Syst. Bacteriol. 36, 213–221. doi: 10.1099/00207713-36-2-213
Tusnady, G. E., and Simon, I. (2001). The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850. doi: 10.1093/bioinformatics/17.9.849
Wyss, C. (1989). Dependence of proliferation of Bacteroides forsythus on exogenous N-acetylmuramic acid. Infect. Immun. 57, 1757–1759.
Young, K. D. (1996). A simple gel electrophoretic method for analyzing the muropeptide composition of bacterial peptidoglycan. J. Bacteriol. 178, 3962–3966. doi: 10.1128/jb.178.13.3962-3966.1996
Zhang, Y., Bao, Q., Gagnon, L. A., Huletsky, A., Oliver, A., Jin, S., et al. (2010). ampG gene of Pseudomonas aeruginosa and its role in beta-lactamase expression. Antimicrob. Agents Chemother. 54, 4772–4779. doi: 10.1128/AAC.00009-10
Keywords: muropeptides, peptidoglycan, Tannerella forsythia, AmpG, GppX
Citation: Ruscitto A, Honma K, Veeramachineni VM, Nishikawa K, Stafford GP and Sharma A (2017) Regulation and Molecular Basis of Environmental Muropeptide Uptake and Utilization in Fastidious Oral Anaerobe Tannerella forsythia. Front. Microbiol. 8:648. doi: 10.3389/fmicb.2017.00648
Received: 24 February 2017; Accepted: 29 March 2017;
Published: 12 April 2017.
Edited by:
Manuel Kleiner, University of Calgary, CanadaReviewed by:
Jayashree Ray, Lawrence Berkeley National Laboratory, USACopyright © 2017 Ruscitto, Honma, Veeramachineni, Nishikawa, Stafford and Sharma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashu Sharma, c2hhcm1hYUBidWZmYWxvLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.