
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 March 2017
Sec. Food Microbiology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.00486
Adhesion ability of probiotics is the key factor that decides their colonization in the gastrointestinal tract and potential to inhibit pathogens. Therefore, adhesion ability can be considered as a key determinant for probiotic efficacy. Presents study documents the antagonistic activity of viable/untreated, Lithium chloride (LiCl) treated or heat-killed forms of eight probiotic Lactobacillus reuteri strains on the adhesion characteristics of selected pathogens. All strains investigated were able to adhere to Caco-2 cells. L. reuteri strains tested were able to inhibit and displace (P < 0.05) the adhesion of Escherichia coli ATCC25922, Salmonella typhi NCDC113, Listeria monocytogenes ATCC53135, and Enterococcus faecalis NCDC115. The probiotic strain L. reuteri LR6 showed the strongest adhesion and pathogen inhibition ability among the eight L. reuteri strains tested. In addition, the abilities to inhibit and to displace adhered pathogens depended on both the probiotic and the pathogen strains tested suggesting the involvement of various mechanisms. The adhesion and antagonistic potential of the probiotic strains were significantly decreased upon exposure to 5 M LiCl, showing that surface molecules, proteinaceous in nature, are involved. The heat-killed forms of the probiotic L. reuteri strains also inhibited the attachment of selected pathogens to Caco-2 cells. In conclusion, in vitro assays showed that L. reuteri strains, as viable or heat-killed forms, are adherent to Caco-2 cells and are highly antagonistic to pathogens tested in which surface associated proteins play an important role.
Globally, the market of probiotics is growing faster as they have claimed to exert several health promoting effects, including interaction with the immune system, production of antimicrobial substances, enhancement of the mucosal barrier function and competition with enteropathogens for adhesion sites (Boesten and de Vos, 2008; Papadimitriou et al., 2015). There are numerous probiotic genera and species including lactobacilli and bifidobacteria which have been implicated in a number of health promoting functions that affect general health and well-being of the host. Adhesion is considered as a potential biomarker for selection of potential probiotics; as their colonization with extended transit time is extremely crucial for optimal expression of their general as well as specific physiological functions (Duary et al., 2011). Several reports have given special attention to the protective role of probiotics against enteropathogens and the underlying mechanisms (Salminen et al., 1998, 1999; Ouwehand et al., 2002; Ouwehand and Salminen, 2003; Collado et al., 2007). Some of these possible protective mechanisms include competition for nutrients and adhesion sites (Ouwehand and Salminen, 2003) or immune modulation (Schiffrin et al., 1997; Salminen et al., 1998). Thus, the probiotics intervention may provide significant protection against gastrointestinal infection and this would enhance human health.
Lactobacilli have been shown to possess surface adhesins similar to those on bacterial pathogens (Neeser et al., 2000). Several surface-located molecules such as lipoteichoic acid, lectin-like molecules and proteins have been identified as adhesins which interact with their specific receptors displayed on the host cell surface (Martinez et al., 2000; Beganović, 2008; Beganović et al., 2010). Due the importance of probiotics in the prevention of infections, the aim of this study was to assess the antagonistic properties of probiotic strains derived from breast fed human infant feces (Singh et al., 2012). Earlier, we reported that the cell surface proteins play an important role in probiotic activities of the Lactobacillus reuteri strains (Singh et al., 2016). In the present study, the probiotic L. reuteri strains were evaluated for their adhesion and abilities to exclude, displace and compete with selected pathogens using Caco-2 as an experimental model. These experiments were also conducted with LiCl treated and heat-killed forms of L. reuteri strains to check their functional interest and the importance of probiotic cell surface integrity.
Eight L. reuteri strains viz., LR5, LR6, LR9, LR11, LR19, LR20, LR26, and LR34, of fecal origin were selected for this study. The Lactobacillus strains were grown in MRS broth (deMan, Ragosa and Sharp broth; Himedia, Mumbai, India) at 37°C for 18–24 h and maintained as glycerol stocks until further use. From the stock cultures, working cultures were prepared and were propagated twice prior to use by sub-culture in MRS broth. The bacterial pathogens used in this study were Escherichia coli ATCC25922, Salmonella typhi NCDC113, Listeria monocytogenes ATCC53135, Enterococcus faecalis NCDC115 which were maintained in BHI (Brain Heart Infusion) broth.
The Lactobacillus strains were grown overnight (16–18 h) in MRS broth at 37°C and harvested at 5000 g for 10 min. The cells were washed twice in phosphate buffer saline (PBS) and OD600 was adjusted to 1.5 which corresponds to 109 cfu/ml based on calibration curve performed.
The Lactobacillus strains were harvested by centrifugation at 5000 g for 10 min, washed with sterile distilled water and then resuspended in 5 M LiCl for 30 min (Zhang et al., 2013). After LiCl treatment, the cells were washed twice in PBS and OD600 was adjusted to 1.5 which corresponds to 109 cfu/ml.
The Lactobacillus strains were grown overnight (16–18 h) in MRS at 37°C and harvested by centrifugation at 5000 g for 10 min. Then, the cells were washed twice with PBS and OD600 was adjusted to 1.5 which corresponds to 109 cfu/ml. The bacterial suspension was heat killed at 80°C for 10 min in a water bath and stored at -70°C until further use (Ouwehand et al., 2000).
The Caco-2 cell line was procured from the National Center of Cell Science, Pune, India. Cells were routinely grown in Dulbecco‘s modified eagle‘s minimal essential medium (DMEM; Sigma, USA), supplemented with 10% fetal bovine serum (FBS; Sigma, USA), 100 μg streptomycin per ml (Sigma, USA) and 100 U penicillin per ml (Sigma, USA) at 37°C in a 5% CO2 atmosphere. For adhesion and inhibition assays, Caco-2 monolayers were prepared in 6-well tissue culture plates. Cells were inoculated at a concentration of 7 × 104 cells per well to obtain confluence and allowed to differentiate. The culture medium was changed on alternate days, and the last two media changes were without antibiotics.
A 1.0 ml aliquot of the bacterial suspension (viable, heat killed, and LiCl treated lactobacilli; 109 cells) was added to confluent Caco-2 monolayer and incubated for 2 h in a 5% CO2 atmosphere. Following incubation, the Caco-2 monolayers were washed with sterile PBS (pH 7.4), Giemsa-stained and examined microscopically under oil immersion, as described previously by Duary et al. (2011).
The inhibition ability of viable/untreated, LiCl treated or heat-killed forms of L. reuteri strains against pathogens adherence was performed according to procedure described by Zhang et al. (2013) with some modifications. Three different protocols were followed to evaluate the ability of L. reuteri strains (viable, heat inactivated, and LiCl treated) to inhibit pathogen (E. coli ATCC25922, L. monocytogenes ATCC5313, S. typhi NCDC113, and E. faecalis NCDC115) adhesion to Caco-2 cells.
For competition assays, Lactobacillus (live, heat killed, and LiCl treated; approximately 108–109cfu/ml) and pathogens (approximately 107cfu) were co-incubated with Caco-2 monolayer for 2 h. For exclusion assays, Lactobacillus (live, heat killed, and LiCl treated; approximately 108–109cfu/ml) was cultured with Caco-2 monolayer for 1 h. Following 1 h incubation, Caco-2 monolayer was washed three times with PBS (pH 7.4); pathogens (approximately 107cfu) were added and incubated for another 1 h. For displacement assays, pathogens (approximately 107cfu) were cultured with Caco-2 monolayer for 1 h, and then the Lactobacillus (live, heat killed, and LiCl treated) were added and cultured for another 1 h. The monospecies cultures of pathogenic bacteria were used as the controls.
In all the above treatments, non-adhered bacterial cells were removed by washing with PBS (pH 7.4). After washing, the Caco-2 cells were detached by addition of 0.25% (v/v) Trypsin-EDTA solution at 37°C for 5 min and the number of viable adhering E. coli, L. monocytogenes, S. typhi, and E. feacalis were determined by plating on EMB, PALCAM, XLD, and CA agar plates after serial dilutions, respectively.
The results for adhesion and pathogen inhibition are expressed as the mean ± SD of three independent experiments. Statistical analysis was done by StatGraphicPlus software. Data were subjected to a one-way analysis of variance (ANOVA) followed by a Tukey’s post hoc test. Differences were considered statistically significant when P < 0.05.
All the L. reuteri strains adhered to Caco-2 cells albeit at different levels. However, on comparative evaluation, L. reuteri strains LR6, LR20, and LR34 were found to be the most adhesive strains based on their respective adhesion scores, with LR6 being the most adhesive strain among all the strains tested. The adhesion score for other strains tested, i.e., LR5, LR9, LR11, LR19, and LR26 differed significantly. All the L. reuteri test strains were found to be highly adhesive (>100 bacteria/20 microscopic fields) when assessed in Caco-2 cell lines. In comparison, it was also observed that the heat inactivation and LiCl treatment had a marked effect on the adhesion ability of the strains as the adhesion of the strains was significantly (P < 0.05) reduced as shown in Figure 1.
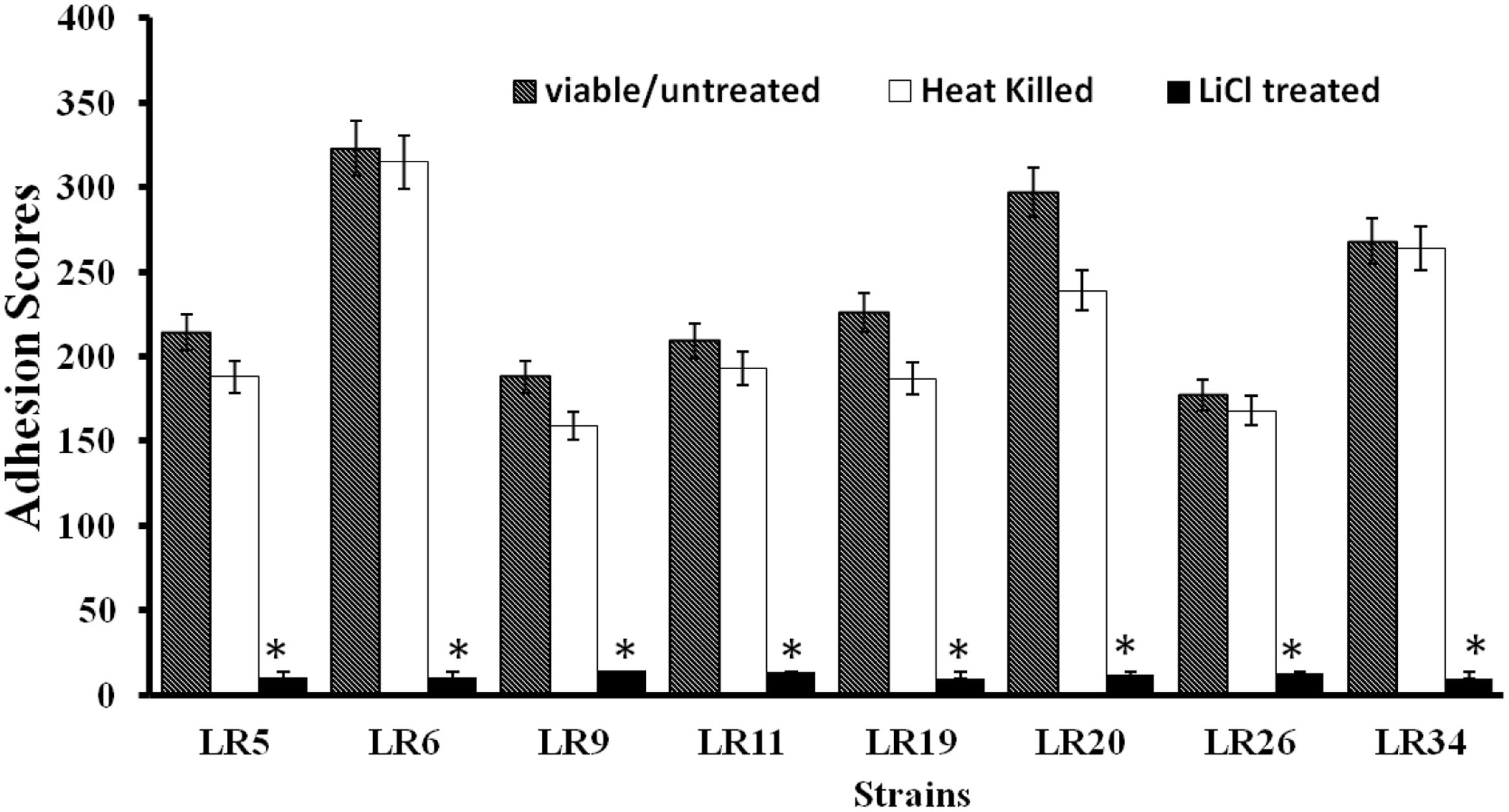
FIGURE 1. Adhesion of differently treated Lactobacillus reuteri strains to Caco-2 cells. ∗Significantly different (P < 0.05) from the untreated control.
Competition assay explained the ability of probiotic strains to compete with pathogens for the adhesion site on epithelial cells. Among the L. reuteri strains tested; LR6, LR9, and LR11 exhibited the maximum inhibition of E. coli ATCC25922. For S. typhi NCDC113, the strains mainly L. reuteri LR6, LR9, LR11, LR19, and LR26 showed the maximum inhibition. Among the tested probiotic L. reuteri strains; LR5, LR6, LR9, LR20, and LR34 inhibited the adhesion of L. monocytogenes ATCC53135 to Caco-2 cells to significant (P < 0.05) levels. For E. feacalis NCDC115, the significant inhibition of adhesion to Caco-2 cells was seen for the strain LR6 and LR9. From the results of competitive assay shown in Table 1, we can conclude that the strain LR6 is the most competitive probiotic strain which can compete strongly with the selected pathogens for the adhesion to epithelial cells.
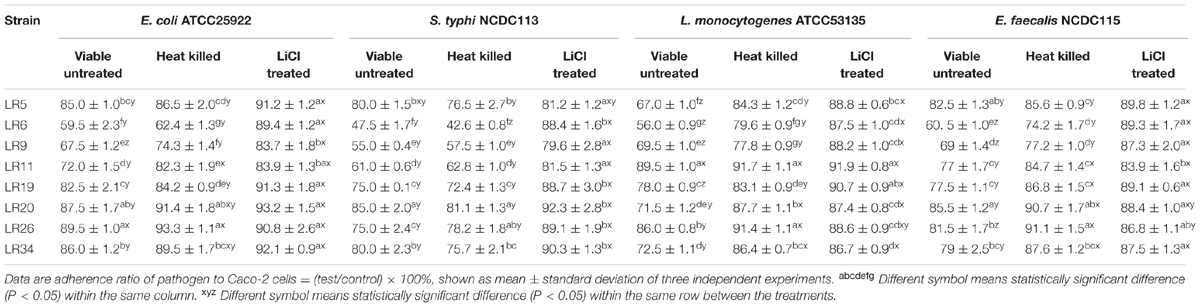
TABLE 1. Competence between pathogens and differently treated probiotic Lactobacillus reuteri strains to adhere to Caco-2 cells.
In competition inhibition assay, it was observed that heat inactivation decreases the ability of L. reuteri strains to compete with selected pathogens for adhesion to Caco-2 cells as compared with their untreated/live forms. The inhibition ability of the heat inactivated forms of L. reuteri strains showed the variability in results ranging from 6.7 ± 1.13% to 37.6 ± 1.07% for E. coli ATCC25922, 18.9 ± 1.32% to 57.4 ± 2.32% for S. typhi NCDC113, 8.3 ± 0.61% to 20.4 ± 1.17% for L. monocytogenes ATCC53135, and 8.9 ± 1.27% to 25.8 ± 1.46% for E. feacalis NCDC115 as shown in Table 1. For heat inactivated forms, the strains LR6, LR9, and LR11 showed the maximum inhibition to adhesion of S. typhi NCDC113. In case of E. coli ATCC25922, the maximum inhibition was exhibited by heat inactivated forms of LR6, LR9, LR11, and LR19, respectively. The strain LR6 also showed the highest inhibition of L. monocytogenes ATCC53135 and E. feacalis NCDC115 adhesion to Caco-2 cells. From the results given in Table 1, it is also evident that the heat inactivated forms of L. reuteri strains were able to compete with pathogens as the results on comparison were found statistically insignificant to their viable forms. However, the ability of the L. reuteri strains to compete with pathogens assayed for adhesion site on Caco-2 decreases significantly (P < 0.05) on LiCl treatment (meant for removal of surface proteins) as depicted in Table 1.
Displacement assay exhibits the potential of the probiotic strains to remove/displace the already adhered pathogen from the epithelial cells. The data depicted that amongst the L. reuteri strains tested, the strain LR6 showed maximum inhibition of E. coli ATCC25922, S. typhi NCDC113, L. monocytogenes ATCC53135, and E. feacalis NCDC115. After 5 M LiCl treatment, the displacement ability of L. reuteri strains against test pathogens were significantly reduced (P < 0.05), shown in Table 2.
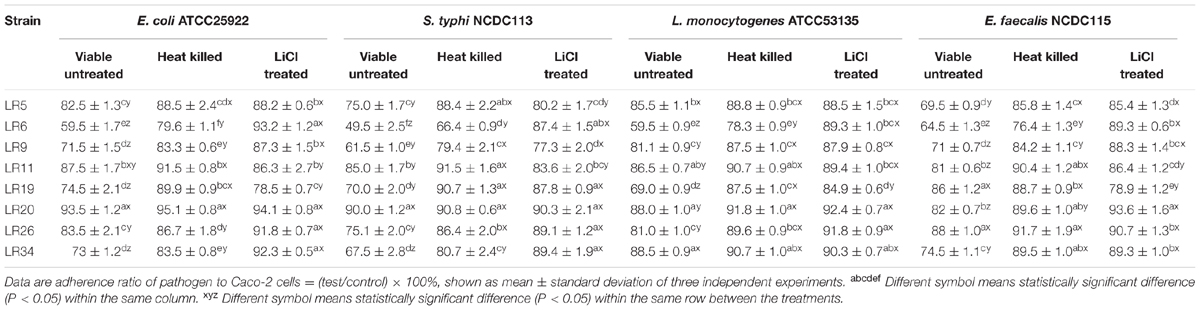
TABLE 2. Displacement of pathogens adhering to Caco-2 cells by differently treated probiotic L. reuteri strains.
The heat inactivated forms of L. reuteri strains showed reduced ability to displace the tested pathogens as compared to their untreated/viable forms. The heat inactivated forms of L. reuteri strains showed the variability in results for percentage displacement ranging from 6.5 ± 1.16% to 40.5 ± 0.99% for E. coli ATCC25922, 10 ± 0.16% to 50.5 ± 2.52% S. typhi NCDC113, 11.5 ± 1.03% to 40.5 ± 1.03% L. monocytogenes ATCC53135, and 12 ± 0.93% to 35.5 ± 0.89% for E. feacalis NCDC115 as shown in Table 2. For heat inactivated forms, the strains LR6 and LR9 showed the maximum displacement for S. typhi NCDC113. In case of E. coli ATCC25922, the maximum displacement was exhibited by heat inactivated forms of LR6. The strain LR6 also showed the highest inhibition of L. monocytogenes ATCC53135 and E. feacalis NCDC115 to Caco-2 cells.
Exclusion assay explains that once the adhesion site is occupied by the probiotic bacteria it becomes unavailable for pathogen. It is evident from the results that the tested strains LR5, LR6, LR9, LR20, and LR26 were able to exclude E. coli ATCC25922 adhesion to significant levels. The significant reduction in E. coli ATCC25922 adhesion to Caco-2 cells was observed for LR5, LR6, LR9, LR20, and LR26. In case of S. typhi NCDC113, the maximum exclusion was showed by strains LR5, LR6, LR9, LR19, and LR26. On the other hand, only LR6 showed the maximum exclusion of L. monocytogenes ATCC53135 from adhesion to caco-2 cells. Similarly, LR6 and LR11 were the only strains which were able to exclude the E. feacalis NCDC115 to significant levels. The data is depicted in Table 3.
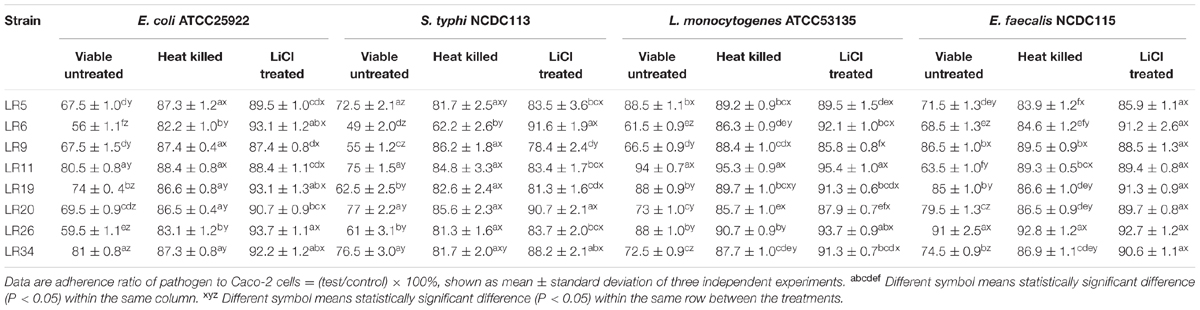
TABLE 3. Exclusion of pathogens from adhesion to Caco-2 cells by differently treated probiotic L. reuteri strains.
The heat inactivated forms of probiotic strains showed significantly reduced exclusion of the pathogens from Caco-2 cells when compared with their untreated viable forms. The exclusion activity of the heat inactivated forms of L. reuteri strains also showed the variability in results ranging from 11.6 ± 1.06% to 17.8 ± 1.36% for E. coli ATCC25922, 13.8 ± 2.25% to 37.8 ± 2.70% S. typhi NCDC113, 4.7 ± 0.75% to 14.3 ± 1.07% L. monocytogenes ATCC53135, 7.2 ± 1.06% to 16.1 ± 1.27% for E. feacalis NCDC115. The strains LR6, LR9, LR20, and LR34 showed the maximum exclusion of L. monocytogenes ATCC53135 in vitro. For E. coli ATCC25922 and S. typhi NCDC113, the maximum exclusion was reported for strain LR6. The strains LR5, LR6, LR19, LR20, and LR34 showed the highest exclusion for E. feacalis NCDC115 in vitro. However, the ability of the L. reuteri strains to exclude pathogens tested decreased significantly (P < 0.05) on LiCl treatment (meant for removal of surface proteins) (Table 3).
Probiotics efficacy is highly dependent on their survival and persistence in gastrointestinal tracts. Therefore, adhesion ability can be considered as a standard biomarker for selecting a potential probiotic (Duary et al., 2011). In the present investigation, we evaluated eight probiotic strains of L. reuteri, previously isolated from breast fed infant feces (Singh et al., 2012), for their potential to adhere Caco-2 cells. The results pertaining to adhesion were recorded in terms of number of bacteria adhering to Caco-2 cell line. On comparative evaluation based on adhesion score, L. reuteri strains LR6, LR20, and LR34 were found to be the most adhesive strains. Adhesion score for all the L. reuteri strains were more than 100 and, therefore, can be regarded as a strongly adhesive to Caco-2 cell lines as per the classification by Jacobsen et al. (1999). Also, the variation observed in adhesion abilities of L. reuteri strains suggests that the trait varies among probiotic strains. This is in complete agreement with the other researchers who also reported that the probiotics ability to adhere is very much strain, species and genus specific (Collado et al., 2007).
Several studies have reported that probiotics compete with pathogens for the adhesion sites, as both probiotics and pathogens possess similar kind of adhesins on their surfaces. Also, the inhibition specifically depends on the probiotic strains and pathogens used as well as the methods of assessment (Chen et al., 2007; Gueimonde et al., 2007). In this study, the probiotic L. reuteri strains were evaluated for their abilities to exclude, compete and displace selected pathogens using Caco-2 as an experimental model. The pathogen adhesion inhibition by probiotic L. reuteri strains showed a high variability and indicated that it was clearly a strain dependent property. The L. reuteri strains tested did not show the same level of inhibition capacity against the pathogens, but they efficiently inhibited the adhesion of pathogenic bacteria to Caco-2 cell in all three assays. The strain LR6 with highest adhesion ability generally showed much higher inhibition of pathogen adhesion to Caco-2 cells, indicating that the pathogen inhibition capacity of probiotic strains may be related to their adhesion ability. Similarly, other workers have also reported the competitive exclusion of enteropathogens by bifidobacteria and lactobacilli (Bernet et al., 1993; Forestier et al., 2001; Lee et al., 2003; Collado et al., 2005, 2007; Weizman et al., 2005; Pham et al., 2009; Wine et al., 2009; Zhang et al., 2013). Meanwhile, the pathogen inhibition ability of the L. reuteri strains did not correlate with the adhesive ability of the strains. In our results, the profile of competition, exclusion and displacement of pathogens by L. reuteri strains were different confirming that the mechanisms of competition, exclusion and displacement might be different. Therefore, it was believed that the differences in competitive exclusion between the strains correlate with the variations in their adhesion ability, possibly due to differences in their surface characteristics. This suggests that the mechanism involved in inhibition is complicated and many factors may be involved.
Generally, adhesion involves the interaction between bacterial associated molecular patterns such as; lipoteichoic acid (Granato et al., 1999), surface layer protein (Chen et al., 2007; Johnson-Henry et al., 2007), peptidoglycan (Van Tassell and Miller, 2011) and their pattern recognition receptors on the host epithelial cells. The surface associated proteinaceous components mediating bacterial adhesion to intestinal epithelial cells have been demonstrated for many Lactobacillus species (Rojas et al., 2002; Roos and Jonsson, 2002; Frece et al., 2005; Chen et al., 2007). In the present study, a significant (P < 0.05) difference was observed on comparing the adhesion ability of untreated and LiCl treated forms of L. reuteri strains, suggesting the importance of the surface associated proteins in adhesion. Also, the ability of the L. reuteri strains to displace, compete and exclude the pathogens from adhesion to caco-2 cells was significantly (P < 0.05) decreased on LiCl treatment. The results are in complete agreement with other workers who reported reduction in binding and adhesion ability of lactobacilli on removal/disruption of surface associated proteins (Sillanpää et al., 2000; Buck et al., 2005; Frece et al., 2005; Chen et al., 2007; Johnson-Henry et al., 2007; Wang et al., 2008; Li et al., 2011; Zhang et al., 2013).
By definition probiotics should be viable in order to exert health benefits. Many researchers have suggested that certain probiotic effects can also be obtained with non-viable probiotics (Ouwehand and Salminen, 1998). Evidences also suggested that non-viable probiotics are less effective which may be attributed to their reduced binding ability than viable probiotics (Conge et al., 1980; De Simone et al., 1987; Kato et al., 1994; Kaila et al., 1995; Perdigon et al., 1995). In this study, a significant reduction in the adhesion and pathogen inhibition abilities of the probiotic L. reuteri strains was observed in heat inactivated forms compared to their viable forms. This suggests that heat treatment inactivate the micro-organisms and also alters their physicochemical properties (El-Nezami et al., 1998). The reduction of adhesion and pathogen inhibition can be explained by the heat sensitive proteinaceous nature of the molecules involved. In contrast, Tareb et al. (2013) reported that heat-killed forms of both Lb. rhamnosus 3698 and Lb. farciminis 3699 exhibited higher adhesion and higher pathogen exclusion potential.
Probiotics intervention is more cost effective and natural approach to preserve intestinal homeostasis and restore the pathogenesis related dysbiosis than antibiotics. The results of this study demonstrate that probiotic strains of L. reuteri tested can exclude, displace and compete with enteropathogens. However, it is important to take into account that these processes studied are highly specific to probiotic and pathogenic strains. This study indicates that strong adhesion ability means greater inhibition activity for probiotic lactobacilli against pathogen, in which surface associated proteins play an important role which further need to be identified and studied. This study also supports the need for further investigations to demonstrate the potential benefits of L. reuteri strains, particularly strain LR6, live or heat-killed, in the food chain.
All the authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Beganović, J. (2008). Application of Proteomics and Other Molecular Methods in the Characterization of Functionality of the Probiotic Bacteria. Ph.D. dissertation, University of Zagreb, Zagreb.
Beganović, J., Guillot, A., van de Guchte, M., Jouan, A., Gitton, C., Loux, V., et al. (2010). Characterization of the insoluble proteome of Lactococcus lactis by SDS-PAGE LC-MS/MS leads to the identification of new markers of adaption of the bacteria to the mouse digestive tract. J. Proteome Res. 9, 677–688. doi: 10.1021/pr9000866
Bernet, M. F., Brassart, D., Nesser, J. R., and Servin, A. L. (1993). Adhesion of human bifidobacterial strains to cultured human intestinal epithelial cells and inhibition of enterophatogen- cell interactions. Appl. Environ. Microbiol. 59, 4121–4128.
Boesten, R. J., and de Vos, W. M. (2008). Interactomics in the human intestine: lactobacilli and bifidobacteria make a difference. J. Clin. Gastroenterol. 42, S163–S167. doi: 10.1097/MCG.0b013e31817dbd62
Buck, B. L., Altermann, E., Svingerud, T., and Klaenhammer, T. R. (2005). Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71, 8344–8351. doi: 10.1128/AEM.71.12.8344-8351.2005
Chen, X. Y., Xu, J. J., Shuai, J. B., Chen, J. S., Zhang, Z. F., and Fang, W. H. (2007). The S-layer proteins of Lactobacillus crispatus strain ZJ001 is responsible for competitive exclusion against Escherichia coli O157:H7 and Salmonella typhimurium. Int. J. Food Microbiol. 115, 307–312. doi: 10.1016/j.ijfoodmicro.2006.11.007
Collado, M. C., Gueimonde, M., Hernández, M., Sanz, Y., and Salminen, S. (2005). Adhesion of selected Bifidobacterium strains to human intestinal mucus and the role of adhesion in enteropathogen exclusion. J. Food Prot. 68, 2672–2678. doi: 10.4315/0362-028X-68.12.2672
Collado, M. C., Surono, I., Meriluoto, J., and Salminen, S. (2007). Indigenous dadih lactic acid bacteria: cell-surface properties and interactions with pathogens. J. Food Sci. 72, M89–M93. doi: 10.1111/j.1750-3841.2007.00294.x
Conge, G. A., Gouache, P., Desormeau-Bedot, J. P., Loisillier, F., and Lemonnier, D. (1980). Comparative effects of a diet enriched in live or heated yogurt on the immune system of the mouse. Reprod. Nutr. Dev. 20, 929–938. doi: 10.1051/rnd:19800603
De Simone, C., Vesely, R., Negri, R., Bianchi, S. B., Zanzoglu, S., Cilli, A., et al. (1987). Enhancement of immune response of murine Peyer’s patches by a diet supplemented with yoghurt. Immunopharmacol. Immunotoxicol. 9, 87–100. doi: 10.3109/08923978709035203
Duary, R. K., Rajput, Y. S., Batish, V. K., and Grover, S. (2011). Assessing the adhesion of putative indigenous probiotic lactobacilli to human colonic epithelial cells. Ind. J. Med. Res. 134, 664–671. doi: 10.4103/0971-5916.90992
El-Nezami, H., KankaanpaÈaÈ, P., Salminen, S., and Ahokas, J. (1998). Physico-chemical alterations enhance the ability of dairy strains of lactic acid bacteria to remove aflatoxin from contaminated media. J. Food Prot. 61, 466–468. doi: 10.4315/0362-028X-61.4.466
Forestier, C., De Champs, C., Vatoux, C., and Joly, B. (2001). Probiotic activities of Lactobacillus casei rhamnosus: in vitro adherence to intestinal cells and antimicrobial properties. Res. Microbiol. 152, 167–173. doi: 10.1016/S0923-2508(01)01188-3
Frece, J., Kos, B., Svetec, I. K., Zgaga, Z., Mrsa, V., and Suskovic, J. (2005). Importance of S-layer proteins in probiotic activity of Lactobacillus acidophilus M92. J. Appl. Microbiol. 98, 285–292. doi: 10.1111/j.1365-2672.2004.02473.x
Granato, D., Perotti, F., Masserey, I., Rouvet, M., Golliard, M., Servin, A., et al. (1999). Cell surface-associated lipoteichoic acid acts as an adhesion factor for attachment of Lactobacillus johnsonii La1 to human enterocyte-like Caco-2 cells. Appl. Environ. Microbiol. 65, 1071–1077.
Gueimonde, M., Margolles, A., de los Reyes-Gavilan, C. G., and Salminen, S. (2007). Competitive exclusion of enteropathogens from human intestinal mucus by Bifidobacterium strains with acquired resistance to bile preliminary study. Int. J. Food Microbiol. 113, 228–232. doi: 10.1016/j.ijfoodmicro.2006.05.017
Jacobsen, C. N., Nielsen, V. R., Hayford, A. E., Møller, P. L., Michaelsen, K. F., Paerregaard, A., et al. (1999). Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl. Environ. Microbiol. 65, 4949–4956.
Johnson-Henry, K. C., Hagen, K. E., Gordonpour, M., Tompkins, T. A., and Sherman, P. M. (2007). Surface-layer protein extracts from Lactobacillus helveticus inhibit enterohaemorrhagic Escherichia coli O157:H7 adhesion to epithelial cells. Cell. Microbiol. 9, 356–367. doi: 10.1111/j.1462-5822.2006.00791.x
Kaila, M., Isolauri, E., Saxelin, M., Arvilommi, H., and Vesikari, T. (1995). Viable versus inactivated Lactobacillus strain GG in acute rotavirus diarrhoea. Arch. Dis. Child. 72, 51–53. doi: 10.1136/adc.72.1.51
Kato, I., Endo, K., and Yokokura, T. (1994). Effects of oral administration of Lactobacillus casei on antitumor responses induced by tumor resection in mice. Int. J. Immunopharmacol. 16, 29–36. doi: 10.1016/0192-0561(94)90116-3
Lee, Y. K., Puong, K. Y., Ouwehand, A. C., and Salminen, S. (2003). Displacement of bacterial pathogens from mucus and Caco-2 cell surface by lactobacilli. J. Med. Microbiol. 52, 925–930. doi: 10.1099/jmm.0.05009-0
Li, P. C., Ye, X. L., and Yang, Y. Q. (2011). Antagonistic activity of Lactobacillus acidophilus ATCC 4356 S-layer protein on Salmonella enterica subsp. enterica serovar Typhimurium in Caco-2 cells. Ann. Microbiol. 62, 905–909. doi: 10.1007/s13213-011-0327-1
Martinez, B., Sillanpaa, J., Smit, E., Korhonen, T. K., and Pouwels, P. H. (2000). expression of cbsA encoding the collagen-binding S-protein of Lactobacillus crispatus JCM5810 in Lactobacillus casei ATCC393. J. Bacteriol. 182, 6857–6861. doi: 10.1128/JB.182.23.6857-6861.2000
Neeser, J. R., Granato, D., Rouvet, M., Servin, A., Teneberg, S., and Karlsson, K. A. (2000). Lactobacillis Johnsonii La1 shares carbohydrate-binding specificities with several enteropathogenic bacteria. Glycobiology 10, 1193–1199. doi: 10.1093/glycob/10.11.1193
Ouwehand, A. C., Isolauri, E., Kirjavainen, P. V., ToÈlkkoÈ, S., and Salminen, S. J. (2000). The mucus binding of Bifidobacterium lactis Bb12 is enhanced in the presence of Lactobacillus GG and Lact. delbrueckii ssp. bulgaricus. Lett. Appl. Microbiol. 30, 10–13. doi: 10.1046/j.1472-765x.2000.00590.x
Ouwehand, A. C., and Salminen, S. (2003). In vitro adhesion assays for probiotics and their in vivo relevance: a review. Microb. Ecol. Health Dis. 15, 175–184. doi: 10.1080/08910600310019886
Ouwehand, A. C., Salminen, S., Tolkko, S., Roberts, P., Ovaska, J., and Salminen, E. (2002). Resected human colonic tissue: new model for characterizing adhesion of lactic acid bacteria. Clin. Diag. Lab. Immunol. 9, 184–186. doi: 10.1128/cdli.9.1.184-186.2002
Ouwehand, A. C., and Salminen, S. J. (1998). The health effects of cultured milk products with viable and non-viable bacteria. Int. Dairy J. 8, 749–758. doi: 10.1016/S0958-6946(98)00114-9
Papadimitriou, K., Zoumpopoulou, G., Foligné, B., Alexandraki, V., Kazou, M., Pot, B., et al. (2015). Discovering probiotic microorganisms: in vitro, in vivo, genetic and omics approaches. Front. Miocrobiol. 6:58. doi: 10.3389/fmicb.2015.00058
Perdigon, G., Alvarez, S., Gobbato, N., de Budeguer, M. V., and de Ruiz Holgado, A. A. P. (1995). Comparative effect of the adjuvant capacity of Lactobacillus casei and lipopolysaccharide on the intestinal secretory antibody response and resistance to Salmonella infection in mice. Food Agric. Immunol. 7, 283–294. doi: 10.1080/09540109509354886
Pham, L. C., van Spanning, R. J., Roling, W. F., Prosperi, A. C., Terefework, Z., Ten Cate, J. M., et al. (2009). Effects of probiotic Lactobacillus salivarius W24 on the compositional stability of oral microbial communities. Arch. Oral Biol. 54, 132–137. doi: 10.1016/j.archoralbio.2008.09.007
Rojas, M., Ascencio, F., and Conway, P. L. (2002). Purification and characterization of a surface protein from Lactobacillus fermentum 104R that binds to porcine small intestinal mucus and gastric mucin. Appl. Environ. Microbiol. 68, 2330–2336. doi: 10.1128/AEM.68.5.2330-2336.2002
Roos, S., and Jonsson, H. A. (2002). A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148, 433–442. doi: 10.1099/00221287-148-2-433
Salminen, S., Bouley, C., Boutron-Ruault, M. C., Cummings, J. H., Franck, A., Gibson, G. R., et al. (1998). Functional food science and gastrointestinal physiology and function. Br. J. Nutr. 80, S147–S171. doi: 10.1079/bjn19980108
Salminen, S., Ouwehand, A. C., Benno, Y., and Lee, Y. K. (1999). Probiotics: how should they be defined? Trends Food Sci. Technol. 10, 107–110. doi: 10.1016/S0924-2244(99)00027-8
Schiffrin, E. J., Brassard, D., Servin, A. L., Rochat, F., and Donnet- Hughes, A. (1997). Immune modulation of blood leukocytes in humans by lactic acid bacteria: criteria for strain selection. Am. J. Clin. Nutr. 66, 515–520.
Sillanpää, J., Martínez, B., Antikainen, J., Toba, T., Kalkkinen, N., Tankka, S., et al. (2000). Characterization of the collagen-binding S-layer protein CbsA of Lactobacillus crispatus. J. Bacteriol. 182, 6440–6450. doi: 10.1128/JB.182.22.6440-6450.2000
Singh, T. P., Kaur, G., Malik, R. K., Schillinger, U., Guigas, C., and Kapila, S. (2012). Characterization of Intestinal Lactobacillus reuteri strains as potential probiotics. Probiotics Antimicrob. Proteins 4, 47–58. doi: 10.1007/s12602-012-9090-2
Singh, T. P., Malik, R. K., and Kaur, G. (2016). Cell surface proteins play an important role in probiotic activities of Lactobacillus reuteri. Nutrire 41, 5. doi: 10.1186/s41110-016-0007-9
Tareb, R., Bernardeau, M., Gueguen, M., and Vernou, J. P. (2013). In vitro characterization of aggregation and adhesion properties of viable and heat-killed forms of two probiotic Lactobacillus strains and interaction with foodborne zoonotic bacteria, especially Campylobacter jejuni. J. Med. Microbiol. 62, 637–649. doi: 10.1099/jmm.0.049965-0
Van Tassell, M. L., and Miller, M. J. (2011). Lactobacillus adhesion to mucus. Nutrients 3, 613–636. doi: 10.3390/nu3050613
Wang, B., Li, Q. R., Li, F. N., Luo, N., Li, Y. S., and Li, N. (2008). Isolation and identification of an adhesive probiotic Lactobacillus strian from human gastrointestinal tract. Chin. J. Biol. 21, 0463–0466.
Weizman, Z., Asli, G., and Alsheikh, A. (2005). Effect of a probiotic infant formula on infections in child care centers: comparison of two probiotic agents. Pediatrics 115, 5–9. doi: 10.1542/peds.2004-1815
Wine, E., Gareau, M. G., Johnson-Henry, K., and Sherman, P. M. (2009). Strain-specific probiotic (Lactobacillus helveticus) inhibition of Campylobacter jejuni invasion of human intestinal epithelial cells. FEMS Microbiol. Lett. 300, 146–152. doi: 10.1111/j.1574-6968.2009.01781.x
Keywords: probiotics, Lactobacillus reuteri, adhesion, antagonistic activity, Caco-2 cells
Citation: Singh TP, Kaur G, Kapila S and Malik RK (2017) Antagonistic Activity of Lactobacillus reuteri Strains on the Adhesion Characteristics of Selected Pathogens. Front. Microbiol. 8:486. doi: 10.3389/fmicb.2017.00486
Received: 12 November 2016; Accepted: 08 March 2017;
Published: 21 March 2017.
Edited by:
Joaquin Bautista-Gallego, Instituto de la Grasa (CSIC), SpainReviewed by:
Cristian Botta, University of Turin, ItalyCopyright © 2017 Singh, Kaur, Kapila and Malik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tejinder P. Singh, ODh0ZWppbmRlcnNpbmdoQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.