
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 20 March 2017
Sec. Terrestrial Microbiology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.00409
Manure composting is a significant source of atmospheric methane (CH4) and nitrous oxide (N2O) that are two potent greenhouse gases. The CH4 and N2O fluxes are mediated by methanogens and methanotrophs, nitrifying and denitrifying bacteria in composting manure, respectively, while these specific bacterial functional groups may interplay in CH4 and N2O emissions during manure composting. To test the hypothesis that bacterial functional gene abundances regulate greenhouse gas fluxes in windrow composting systems, CH4 and N2O fluxes were simultaneously measured using the chamber method, and molecular techniques were used to quantify the abundances of CH4-related functional genes (mcrA and pmoA genes) and N2O-related functional genes (amoA, narG, nirK, nirS, norB, and nosZ genes). The results indicate that changes in interacting physicochemical parameters in the pile shaped the dynamics of bacterial functional gene abundances. The CH4 and N2O fluxes were correlated with abundances of specific compositional genes in bacterial community. The stepwise regression statistics selected pile temperature, mcrA and NH4+ together as the best predictors for CH4 fluxes, and the model integrating nirK, nosZ with pmoA gene abundances can almost fully explain the dynamics of N2O fluxes over windrow composting. The simulated models were tested against measurements in paddy rice cropping systems, indicating that the models can also be applicable to predicting the response of CH4 and N2O fluxes to elevated atmospheric CO2 concentration and rising temperature. Microbial abundances could be included as indicators in the current carbon and nitrogen biogeochemical models.
It is of great concern worldwide that gaseous emissions from management of organic solid waste contribute to regional and global-scale environmental processes, such as eutrophication, acidification, and climate change (Naylor et al., 2005; Hou et al., 2015; Owen and Silver, 2015; Pardo et al., 2015). Organic waste management has been identified as an important source of anthropogenic greenhouse gas (GHG) emissions, particularly methane (CH4) and nitrous oxide (N2O) [Intergovernmental Panel on Climate Change (IPCC), 2006]. Global CH4 and N2O emissions contribute considerably to the radiative forcing of the atmosphere, as their global warming potentials are 298 and 25 times that of carbon dioxide (CO2) on mass basis over the 100-year time horizon, respectively [Intergovernmental Panel on Climate Change (IPCC), 2013]. Manures from livestock production account for 30–50% of the global agricultural N2O emissions and 12–41% of the total agricultural CH4 emissions for most countries (Oenema and Tamminga, 2005; Chadwick et al., 2011).
Manure composting is an alternative agricultural strategy for organic waste management that produces organic fertilizer to improve soil structure and fertility in croplands (Larney and Hao, 2007). The CH4 and N2O fluxes from manure composting have been studied extensively, contributing to a comprehensive assessment of CH4 and N2O emissions from manure composting worldwide [Czepiel et al., 1996; Hao et al., 2001; Intergovernmental Panel on Climate Change (IPCC), 2006; Xu et al., 2007; Mulbry and Ahn, 2014; Jiang et al., 2015; Pardo et al., 2015]. Yet, little is known about the interaction between pile physicochemical parameters and bacterial community, which has a key role in CH4 and N2O emissions from manure composting (Sharma et al., 2011; Angnes et al., 2013; Zhang et al., 2015). In particular, few studies have simultaneously focused on quantitative analysis of bacterial community composition and CH4 and N2O fluxes from composting manure (Maeda et al., 2010a,b; Chen et al., 2014; Zhang et al., 2015). Comparative quantitative analysis of specific bacterial functional groups and their interplay in CH4 and N2O emissions during manure composting are still limited.
To date, genes encoding enzymes involved in CH4 and N2O emissions have been targets of choice for studies focusing on functional groups of bacteria. This focus is fundamental for understanding mechanisms of carbon and nitrogen biogeochemistry and strategies for GHGs mitigation (Hu et al., 2015). Morales et al. (2010) illustrated denitrifying gene abundances as proxies for predicting N2O emissions from soils as a response to different long-term land management regimes. Regan et al. (2011) found the evidence that differences in microbial abundances can help explain enhanced N2O emissions in permanent grasslands under elevated atmospheric carbon dioxide. Nevertheless, a trade-off between CH4 and N2O fluxes has frequently been found in rice paddies and manure composting (Hou et al., 2001; Zou et al., 2005; Ahn et al., 2011; Shen et al., 2011; Mulbry and Ahn, 2014), simultaneous comparisons of the abundance of multiple CH4- and N2O-related genes and their interactions would be highly needed, especially when targeting functional bacteria to mitigate GHGs emission from agriculture (Hu et al., 2015).
Manure composting system is suggested as a good study model to examine the role of microbial abundances in shaping dynamics of CH4 and N2O fluxes due to their sensitive responses to changes in pile physicochemical properties, nitrogen transformation, and organic carbon decomposition during composting (Chadwick et al., 2011). Methane is produced by methanogenic organisms during the anaerobic degradation of organic materials; and the final key step, being reduction of CO2 using H2 to generate CH4, is catalyzed by methyl-coenzyme M reductase (MCR, EC 2.8.4.1) (Kim et al., 2008). The highly conserved mcrA gene encoding the α-subunit of MCR has been widely used for analysis and quantification of methanogen communities (Pereyra et al., 2010; Sonoki et al., 2013). The generated CH4 could be oxidized to methanol with the catalysis of particulate membrane bound methane monooxygenase (EC 1.14.13.25) (Xin et al., 2004). The pmoA gene encoding the α-subunit is widely used as the indicator for quantification of the methanotrophs from environmental samples (Wasmund et al., 2009; Sharma et al., 2011). The CH4 flux is the net outcome and combined action of methanogen and methanotrophs that are closely related to changes in physicochemical parameters and environmental factors during manure composting process (Sonoki et al., 2013).
During manure composting, NH4+ generated from amino acids can be oxidized to NO2- by ammonia-oxidizing archaea and bacteria (AOA and AOB, respectively), through ammonia monooxygenase (EC 1.14.99.39, encoding by amoA) and hydroxylamine oxidoreductase. A part of the NO2- could be oxidized to NO3- by nitrite-oxidizing bacteria (NOB) (Maeda et al., 2011). Bacterial denitrification is a biochemical reaction where oxidized forms of nitrogen, including nitrate, nitrite, nitric oxide, and nitrous oxide, are gradually reduced (Wang et al., 2013). The four steps are generally catalyzed by nitrate reductase (encoding by narG), nitrite reductase (nirS/nirK), nitric oxide reductase (norB), and nitrous oxide reductase (nosZ) (Maeda et al., 2011). Eventually, the N2O emission is a result of dynamic balance between N2O production and consumption. In addition, given that a trade-off between CH4 and N2O fluxes has been well documented in windrow composting systems (Hao et al., 2001; Zou et al., 2005; Ahn et al., 2011; Shen et al., 2011; Mulbry and Ahn, 2014), some compositional bacterial genes could be multifunctional as proxies for indicating dynamics of CH4 and N2O fluxes during windrow composting.
We conducted an in situ measurement of CH4 and N2O fluxes from a commercial composting windrow. Molecular techniques were used to quantify the abundances of CH4- and N2O-related functional genes. The main objective of this study is to examine whether bacterial gene abundances can indicate dynamics of CH4 and N2O fluxes during windrow composting. Specifically, we aimed to test three general hypotheses. The first hypothesis stated that changes in pile physicochemical parameters would shape diverse time course patterns of bacterial functional genes abundance during windrow composting, given that bacterial community response variables are sensitive to environmental change. Second, we predicted that some specific physicochemical parameters and compositional bacterial enzymes encoded by relevant genes would be multifunctional as involved both in CH4 and N2O due to a trade-off between CH4 and N2O fluxes during windrow composting. Eventually, as both production and consumption of CH4 and N2O are controlled by the interplay of enzyme encoding bacterial functional genes, we hypothesized that bacterial functional gene abundances could be used as proxies for indicating dynamics of CH4 and N2O fluxes during windrow composting.
The windrow composting experiment was carried out in a commercial organic fertilizer company (Jiangyin Lianye Biological Science and Technology Co., Ltd.), located at Wuxi, Jiangsu province, China. The experiment was initiated on April 26 and completed on June 29, 2014 (65 days). Three replicate commercial-scale compost piles were constructed using a mixture of dairy manure solids and straw bedding with a mixing ratio of 80%:20% on a fresh weight basis. Sawdust used for dairy manure solids was obtained from scraped dairy manure with 75% moisture in a local dairy feedlot. Chopped rice straw collected from local paddy rice fields was used as a bulking agent and a source of carbon. Uniform rectangle windrows were placed on individual platforms to enable determination of windrow mass values in an open-sided but roofed compost. The volume of each compost windrow was set up to be approximately 125 m3 (40 m in length, 2.8 m in width, and 1.1 in height). The composting process can be generally divided into two phases, namely, the bio-oxidative phase with mechanical turning for 25 days (Phase I, April 26 to May 20, 2014) and the cooling and maturing phase without pile turning for 40 days (Phase II, May 21 to June 29, 2014) (Chen et al., 2014). The windrow was mechanically turned using an astraddle compost turner once every 2 days during Phase I, and thereafter the compost piles were moved away for post-maturation without turning. Three compost windrows were treated as experimental replicates. Each windrow along its length was sub-divided into three sections that were treated as three parallel locations for substrate material and gas sampling.
The fluxes of CH4 and N2O during compositing were simultaneously measured using vented chamber technique (Hao et al., 2001; Xin et al., 2004; Zou et al., 2005; Chen et al., 2014). Gas samples were taken once a week except supplementary sampling episodes as needed to capture high flux peaks. To stabilize the disturbance, PVC chamber collar bases (30 cm length × 30 cm width × 25 cm height) were pre-inserted 25 cm into the pile 10–12 h before each gas sampling. The top edge of the collar base exhibits a groove (5 cm in depth) that can be filled with water to seal the rim of the chamber during gas sampling. While taking gas samples, the opaque chamber (30 cm length × 30 cm width × 50 cm height) was placed on the peak of each windrow with rim of the chamber fitted into the groove of collar. Gas samples were extracted from inside the chambers using 60-mL plastic syringes fitted with three-way stopcocks at 0, 5, 10, 20, and 30 min after chamber closure and immediately injected into a 50 mL pre-evacuated Exetainer fitted with butyl rubber septa (Hao et al., 2001; Chen et al., 2014). Gas samples were taken between 0800 and 1000 LST on each sampling day, and they were transported to the laboratory for analysis by gas chromatograph (GC) within a few hours (Zou et al., 2009; Liu et al., 2010).
The mixing ratios of CH4 and N2O were analyzed with a modified GH (Agilent 7890) equipped with a flame ionization detector (FID) and an electron capture detector (ECD) (Zou et al., 2005; Liu et al., 2012). A non-linear fitting approach was adopted to determine the CH4 and N2O fluxes (Hao et al., 2001; Kroon et al., 2008; Chen et al., 2014). On each sampling day, mean of fluxes taken from three parallel sections within each windrow represent flux measurement of the sampling windrows. Average fluxes and standard deviations of CH4 and N2O were calculated from three replicate windrows. Accumulative CH4 and N2O emissions during compositing were sequentially accumulated from the fluxes between every two adjacent intervals of measurements (Zou et al., 2005; Liu et al., 2012).
Real-time quantitative PCR (qPCR) was performed for investigation of the functional microbial community dynamics during the composting process (days 4, 10, 16, 25, 37, 46, 55, 61, and 65). The genes encoding the key enzymes involved in CH4 and N2O emissions included α-subunit of methyl-coenzyme M reductase (mcrA), α-subunit methane monooxygenase (pmoA), ammonia monooxygenase (amoA), nitrate reductase (narG), nitrite reductase (nirK and nirS), nitric oxide reductase (norB), and nitrous oxide reductase (nosZ). The information of these functional genes and the using primers and conditions were referenced in Table 1. According to the manufacturer’s instructions, DNA was extracted from the compost samples that stored at -80°C using the UltraClean soil DNA isolation kit (Mo Bio, USA). Each DNA sample for next-analysis was the mixture of DNA extractions from three parallel sections within each windrow. The concentrations of DNA samples were determined by a Nanodrop (Thermo Scientific, USA). The amplified fragments for each functional gene based on different primers (Table 1) were cloned in pMD 18-T vector and sequenced, the correct clones corresponding to each functional gene were stepwise 10-fold diluted for standard curve preparation. The qPCR amplifications were performed in a total volume of 20 μL using a SYBR@ Premix Ex TaqTM (Takara, China), with reaction mixture consisting of 10 μL SYBR@ Premix Ex Taq, 0.4 μL each primerTM (10 μmol L-1), 0.4 μL ROX reference dye II (50×), 2 μL template DNA, and 6.8 μL sterile water. Amplification was performed triplicate using 7500 System (ABI, USA). The detailed reaction conditions were listed in Table 1. Fluorescence normalization and data analysis were performed with 7500 Fast System SDS software (ABI, USA).
Windrow temperature was measured by inserting the mercury thermometers at 30 cm depth of the pile on each gas sampling day. To examine dynamics of physicochemical parameters of composting material, manure material samples were taken while gas flux sampling (Table 2). Samples were randomly collected from three longitudinal sections and mixture, generating approximately 300 g of subsamples. The collected samples were divided into three parts, two parts were immediately preserved at 4°C until analysis, while the other part was air-dried, passed through a 0.15 mm sieve, and stored in a desiccator as needed for further analysis. The moisture content of different fresh samples was determined by oven-drying to a constant weight at 105°C. The C/N ratio was calculated based on the total organic carbon (TOC) and total nitrogen (TN) contents that were determined by an auto elemental analyzer (Vario EL III, Elementar, Germany). For analysis of the water-soluble fractions of the composting material, the aqueous compost extracts were obtained by shaking of the mixture of 20 g of fresh sample with 200 mL distilled water (1:10 w/v ratio) on a horizontal shaker at 25°C, as described in Castaldi et al. (2008). The pH was performed on aqueous suspensions of the fresh samples (1:10, w/v, compost/water ratio) using a pH electrode (PHS-3C mv/pH detector, Shanghai, China). The NH4+-N, NO3--N, and NO2--N of composting material were extracted with 100 ml 2 M KCl solution at a ratio of 1:20 at 25°C and measured following the three wavelength ultraviolet spectrometry and indophenol blue method, using the ultraviolet spectrophotometer, respectively (HITACHI, U-2900, Japan).
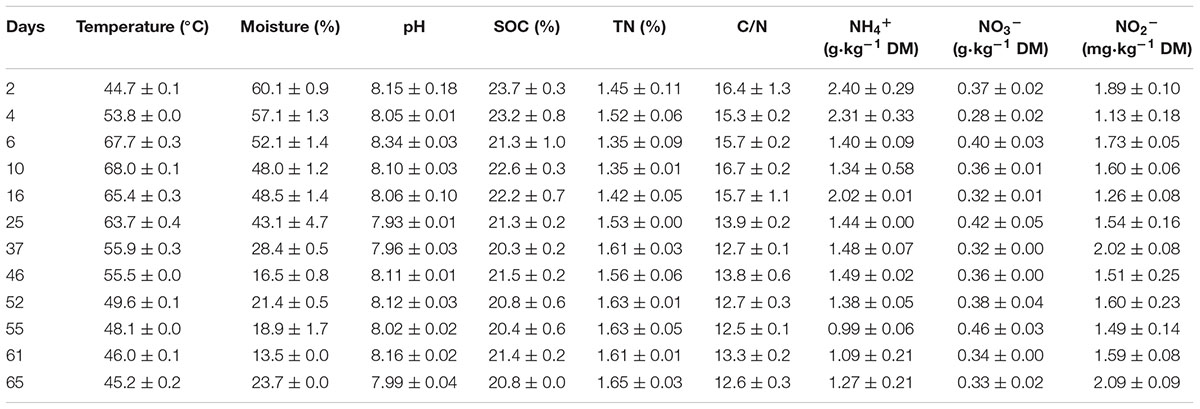
TABLE 2. Changes of physicochemical parameters (mean ± SE, n = 3) during windrow dairy manure composting.
Physicochemical parameters data were expressed as means of replicates based on a dry weight of compost materials. A pairwise correlation was conducted for each pair of variables including CH4 and N2O fluxes, bacterial gene abundance copies, and physicochemical parameters. Bacterial gene abundance copies and CH4 and N2O fluxes were log-transformed for normality and homoscedasticity as needed in statistical analyses. A linear stepwise regression model with the personality of Ordinary Least Squares (OLS) was used to fit CH4 and N2O fluxes by bacterial gene abundance copies and physicochemical parameters. In this method, regression variables are randomly selected based on prior probability, and the randomly selected variables are further screened by stepwise forward regression. Eventually, the forms of model are updated accordingly (Table 3). A t-test was used to examine the statistical significance of parameter estimates in the simulated OLS model.
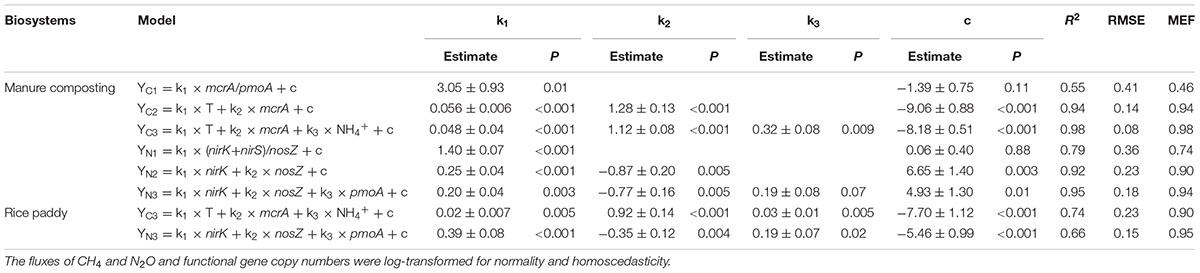
TABLE 3. Modeling CH4 (YC) and N2O (YN) fluxes by coupling functional gene copy numbers with physicochemical parameters during windrow dairy manure composting and the model tested in rice paddies.
To examine whether the simulated OLS models could also be applicable to other environmental systems, the models were tested against field measurements in paddy rice cropping systems under elevated atmospheric CO2 and rising temperature (a T-FACE platform). The field T-FACE platform was established in Kangbo village (31°300N, 120°330E), Guli Township, Changshu Municipality, Jiangsu, China, in 2010. The paddy field soil is a gleyic stagnic anthrosol formed on a clayey lacustrine deposit and cultivated under continuous rice–wheat rotation. The T-FACE platform had 12 octagonal plots, with the inner circle with an area of 25 m2 per plot. The experimental treatments included four experimental treatments with three replicates, namely, one with target atmospheric CO2 up to 500 ppmv (CO2), one with warming of canopy temperature by 1.5–2.0°C above ambient (T), and one with combined CO2 enrichment and warming (CO2+T), and taking an untreated plots with ambient condition as controls (Ambient). Seedlings of a local rice cultivar (Changyou 5) were transplanted into fields on June 20, 2014 and harvested on October 22, 2014. Spacing of hills was 15.3 × 25.4 cm (equivalent to 25.7 hills m2 and resulting in a plant density of 77.1 plants m2) for each experimental plots. All the field plots were under a typical water regime of flooding-midseason drainage-reflooding-moisture irrigation during the rice-growing season. The design of T-FACE platform and field experimental treatments and agricultural practice were detailed in Liu et al. (2014), Cai et al. (2015), and Chen et al. (2016).
In rice paddies, gas flux measurements and soil samples for physicochemical properties and microbial genes abundance analyses were simultaneously taken on July 6, July 21, August 12, August 30, September 10, September 24, and October 22, 2014. The CH4 and N2O fluxes were determined by the static chamber-GC method as shown in our previous studies (Zou et al., 2005; Liu et al., 2012), and the methods for gas flux measurements physicochemical properties and microbial genes abundance analyses were similar to those described in windrow composting experiment.
The three statistics R2 (coefficient of determination), RMSE (root mean square error), and MEF (modeling efficiency) were used for model evaluation (Table 3). All statistical analyses were performed using JMP software version 9.0.2 for Windows (SAS Inst., Cary, NC, USA, 2010).
There was a trade-off between CH4 and N2O fluxes during manure windrow composting (Figure 1). Substantial CH4 emissions occurred primarily during thermophilic Phase I. During Phase I, CH4 fluxes ascended rapidly until the peak fluxes were attained approximately 4 days after the onset of composting. Thereafter, CH4 emission was dramatically decreased by pile turning and then remained lower release rate, which was closely associated with decreases in pile temperature (Table 2). In contrast, N2O fluxes stayed relatively lower during Phase I, and they gradually increased during phase II. Eventually, N2O fluxes were highest by the end of manure compositing (Figure 1).
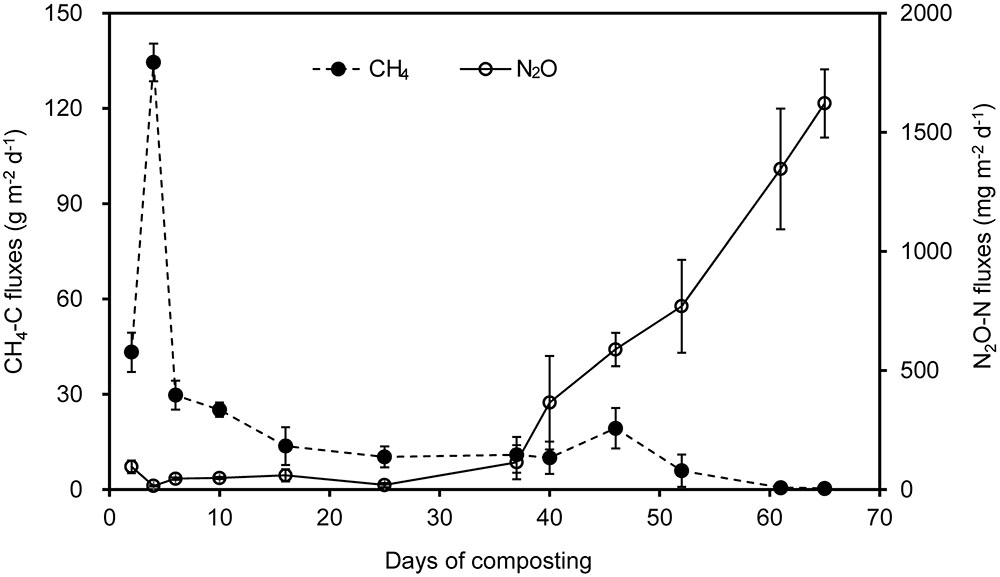
FIGURE 1. Fluxes of CH4 and N2O during a 65-day period of windrow dairy manure composting. Error bars show standard error of the mean of triplicate compost windrows.
Over the 65-day period of manure composting, CH4-C fluxes varied from 0.4 to 134.5 g m-2 d-1, with an average flux of 17.1 g m-2 d-1 (Figure 1). Cumulative CH4 emissions in terms of initial windrow surface area were 1.1 kg m-2, being equivalent to 0.8% of total C in initial manure dry weight (MCF). The fluxes of N2O-N varied within the range of 15.0–95.0 mg m-2 d-1 during Phase I, and rapidly increased from 114.2 to 1621.1 mg m-2 d-1 during Phase II, dedicating to an average of 383.3 mg m-2 d-1 over the whole composting process. Cumulative N2O emissions in terms of initial windrow surface area were 25.1 g m-2, representing 0.18 kg per ton of MDW. The emission factor of N2O (EF, percentage of initial N in manure compost pile emitted as N2O-N) was estimated to be 1.2% for composting windrow.
A contrasting time course pattern of mcrA and pmoA genes abundance was detected during windrow composting. The measured abundances of mcrA were the highest at the onset of composting, being average 6.26 log copy numbers⋅g-1 (Figure 2). Thereafter, the mcrA gene abundance gradually decreased until it remained stable around ∼5.0 log copy numbers⋅g-1. Relative to a smaller variation of mcrA gene abundance, pmoA gene abundance showed larger variation (variation range: 5.11–8.05 log copy numbers⋅g-1) over the composting process. The measured abundance of pmoA decreased in the first week and then kept an ascending trend over the composting process, reaching the greatest abundance by the end of windrow composting (Figure 2).
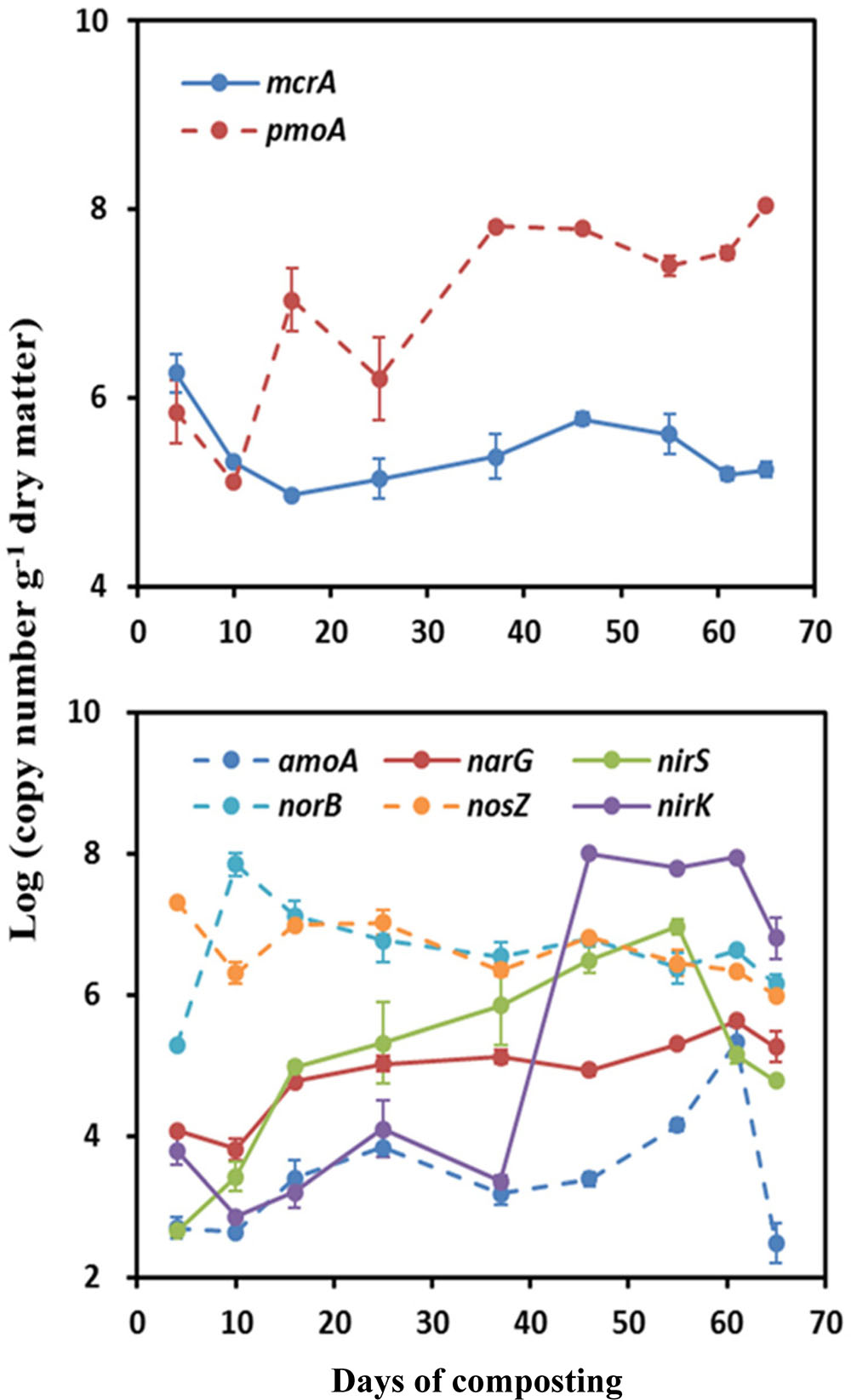
FIGURE 2. Changes in functional gene copy numbers associated with CH4 and N2O emissions during windrow dairy manure composting. Values indicate log-transformed gene copy numbers. Error bars show standard error of the mean of triplicate compost windrows.
For the functional genes involved in N2O emissions, the abundance of amoA, narG, nirK, and nirS genes shared a rising pattern over the composting process, in contrast to a declining trend for norB and nosZ genes abundance (Figure 2). Over the composting process, predominant genes in bacterial community shifted from norB and nosZ genes (6.76–6.91 log copy numbers⋅g-1) during Phase I to nirK gene (6.78 log copy numbers⋅g-1) during Phase II (Figure 2). The amoA gene abundance stayed the lowest (∼3.10 log copy numbers⋅g-1) over the whole windrow composting.
The CH4 fluxes showed strong positive correlations with compost material parameters including moisture, C/N ratio, NH4+-N, and TOC during the composting process (Figure 3). The CH4 fluxes were also positively correlated with mcrA and nosZ gene numbers, but negatively correlated with pmoA and narG gene numbers. For the relative abundance of functional genes group, CH4 fluxes were positively correlated with mcrA/pmoA (r = 0.78, p = 0.01). Besides strong negative correlations between N2O fluxes and pile moisture, temperature and C/N ratio, N2O fluxes showed significant positive correlations with pmoA, narG, and nirK genes abundance, but negatively correlated with nosZ gene abundance (Figure 3). The N2O fluxes were positively correlated with relative abundances of functional gene group (nirK+nirS)/nosZ (r = 0.90, p = 0.001).
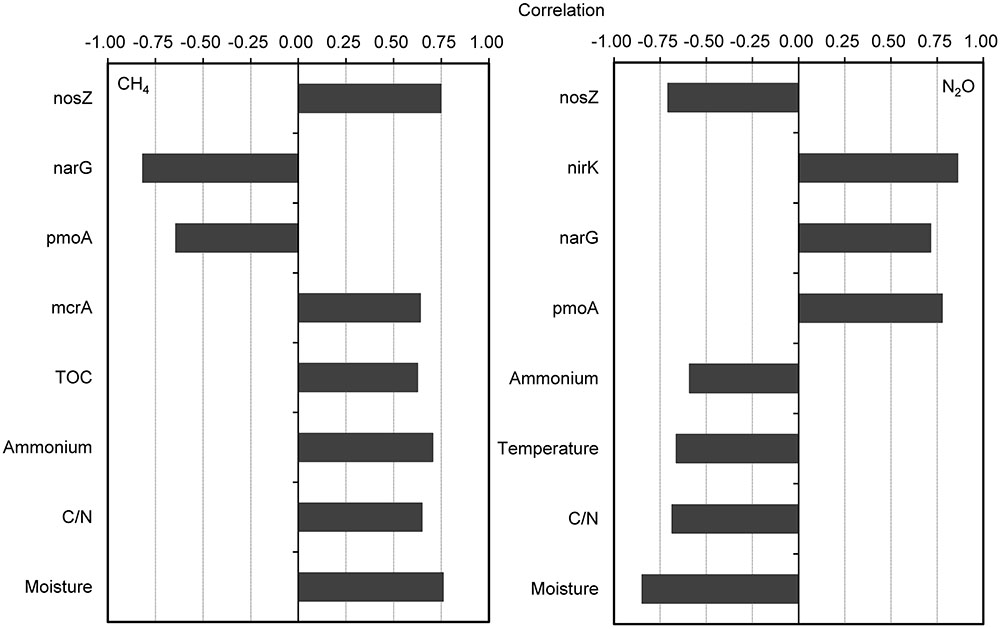
FIGURE 3. Pairwise correlations among all tested variables showing significant probability (p < 0.05) for correlation of CH4 and N2O fluxes with physicochemical parameters and bacterial gene abundances. The gene copy numbers and CH4 and N2O fluxes are log-transformed for normality and homoscedasticity.
During the composting process, TOC, TN, moisture and C/N ratio were significantly correlated with each other, acting as a group to correlate with pmoA, narG, nirK, and nirS genes abundance and CH4 and N2O fluxes (Figures 3, 4). Among functional genes, the abundance of pmoA, narG, and nirS genes were correlated with each other. Besides, the narG gene abundance was correlated with amoA and nirK genes, but amoA gene abundance was not significantly correlated with nirK gene (Figure 4).
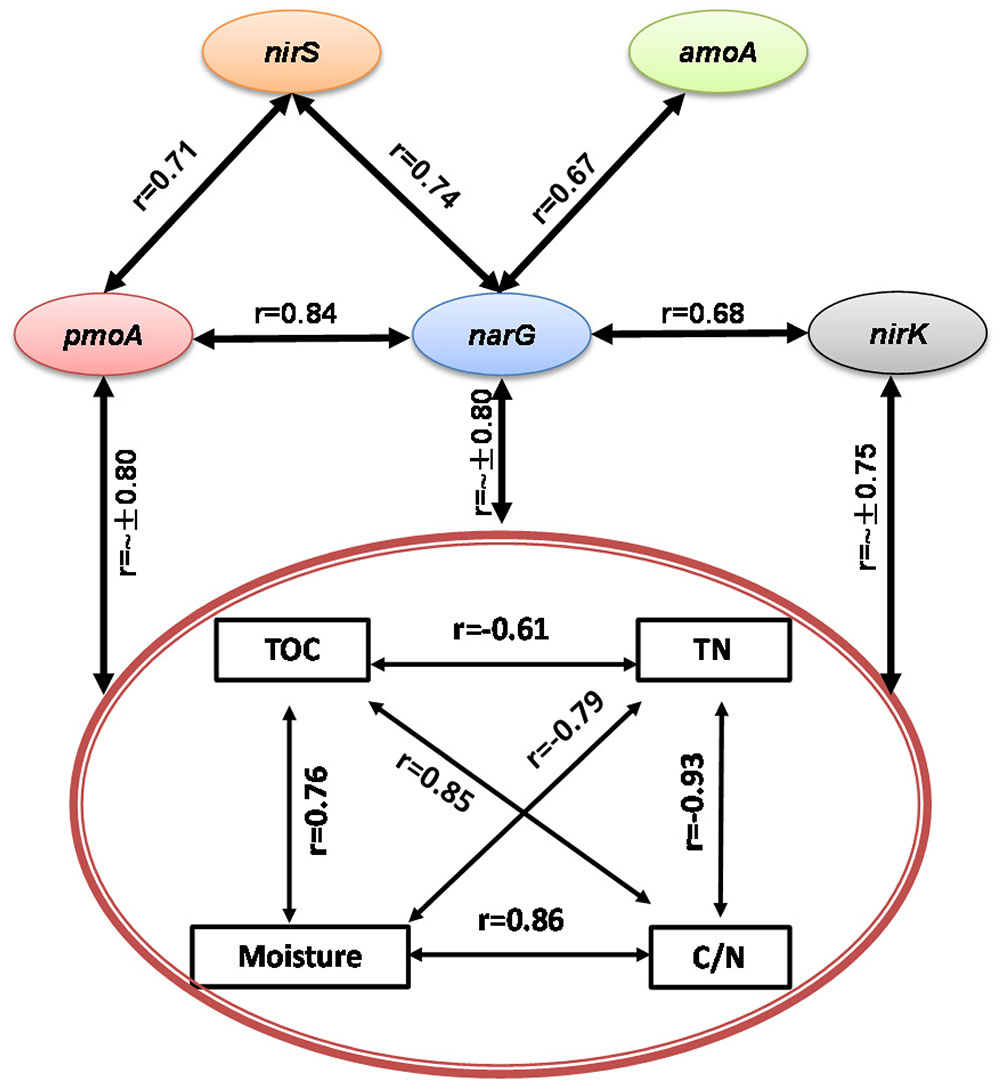
FIGURE 4. Pairwise correlations among all tested variables showing some interacting physicochemical parameters significantly correlated with bacterial gene abundances (p < 0.05).
A stepwise regression analysis was conducted for modeling CH4 and N2O fluxes with bacterial functional gene abundances and physicochemical parameters (Table 3). The model based on bacterial functional genes mcrA/pmoA alone can explain 55% of the variance in CH4 fluxes over the composting process (YC1, Table 3). While integrating mcrA gene abundance together with pile temperature (T), however, the simulated regression model explained as high as 94% of the variance in CH4 fluxes (YC2, Table 3). Furthermore, the regression model including mcrA gene, T and NH4+-N significantly lowered the model error and increased model efficiency, which can almost fully project the time course of CH4 fluxes (YC3, Table 3 and Figure 5). Among the regression models, the model based on pile temperature (T), mcrA and NH4+-N appeared to be the best fit for CH4 flux variance when the statistics R2, P, RMSE, and MEF were comprehensively evaluated (Table 3).
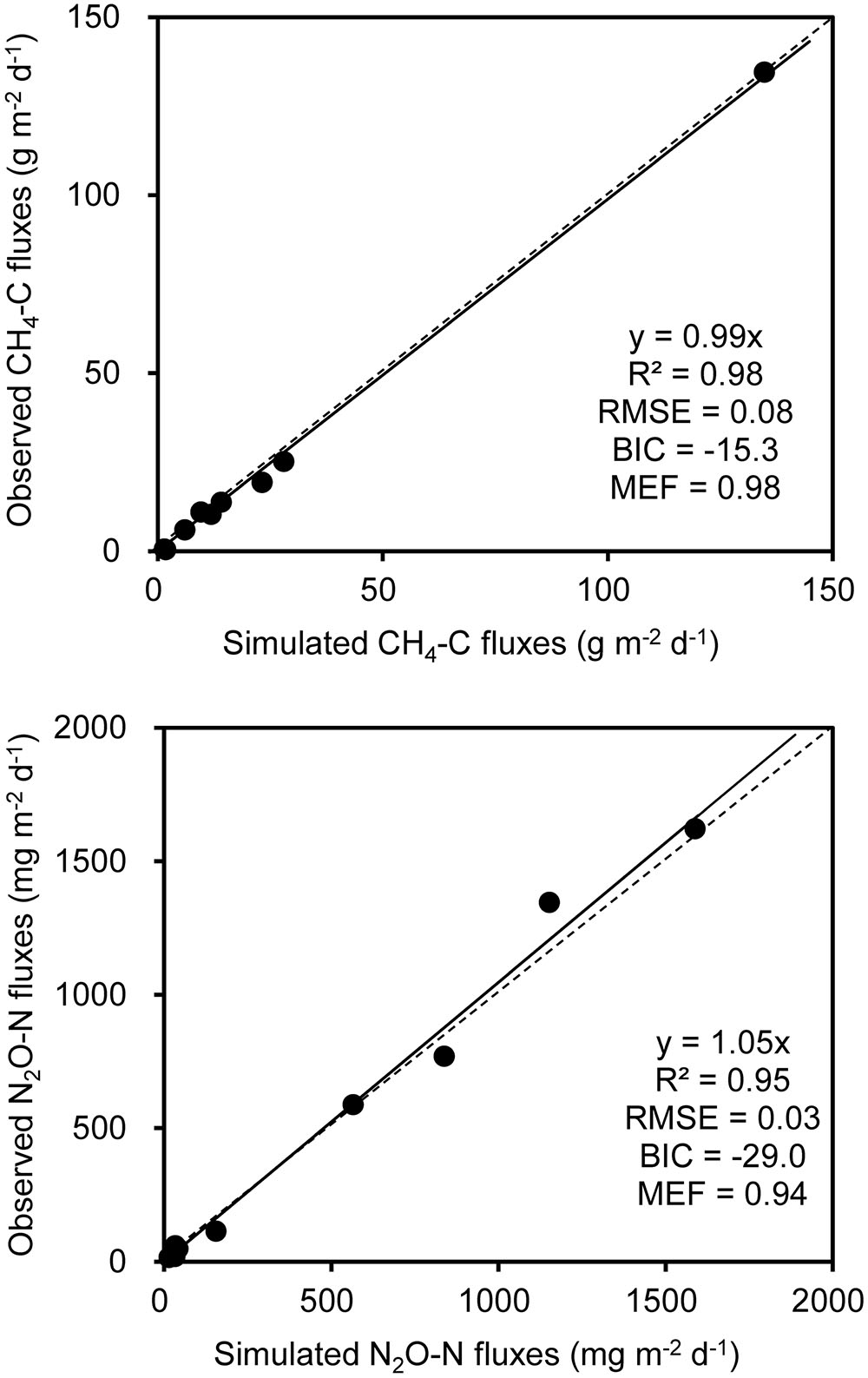
FIGURE 5. Comparison of observed and modeled CH4 and N2O fluxes during dairy manure windrow composting. Estimation of parameters in models for CH4 (YC3) and N2O (YN3) are shown in Table 1. The slope, R2, BIC, the root mean square error (RMSE), and model efficiency (MEF) are indicated at the bottom right of each panel.
As shown in the stepwise regression model, a functional genes group, (nirK+nirS)/nosZ acted as a good proxy for predicting dynamics of N2O fluxes (YN1, Table 1). While taking nirK minus nosZ genes into account but excluding nirS genes, the performance of simulated model was significantly improved, explaining as high as 92% of the variance in N2O fluxes during windrow composting (YN2, Table 1). Besides nirK and nosZ genes, pmoA gene was also responsible for the variance in N2O fluxes as shown in pairwise correlation (Figure 3). Indeed, about 95% of the variance in N2O fluxes can be explained by the model based on linear regression of nirK, nosZ, and pmoA genes abundance (YN3, Table 1). Compared to the YN1 and YN2 models, the YN3 model including pmoA gene abundance as an additional predictor was able to minimize the uncertainty in N2O flux estimates (Table 1 and Figure 5).
The simulated OLS models were also applicable to paddy rice cropping systems (Table 3 and Figures 6, 7). In rice paddies, about 75% of the seasonal variance in CH4 fluxes as a response to elevated atmospheric CO2 concentration and rising temperature can be explained by re-parameterized YC3 model based on linear combination of soil temperature, mcrA and NH4+-N (Figure 7). Similarly, the re-parameterized nirK, nosZ, and pmoA genes abundance in YN3 model can largely reflect seasonal CH4 fluxes response to elevated atmospheric CO2 concentration and rising temperature in paddy rice cropping systems (Figure 7).
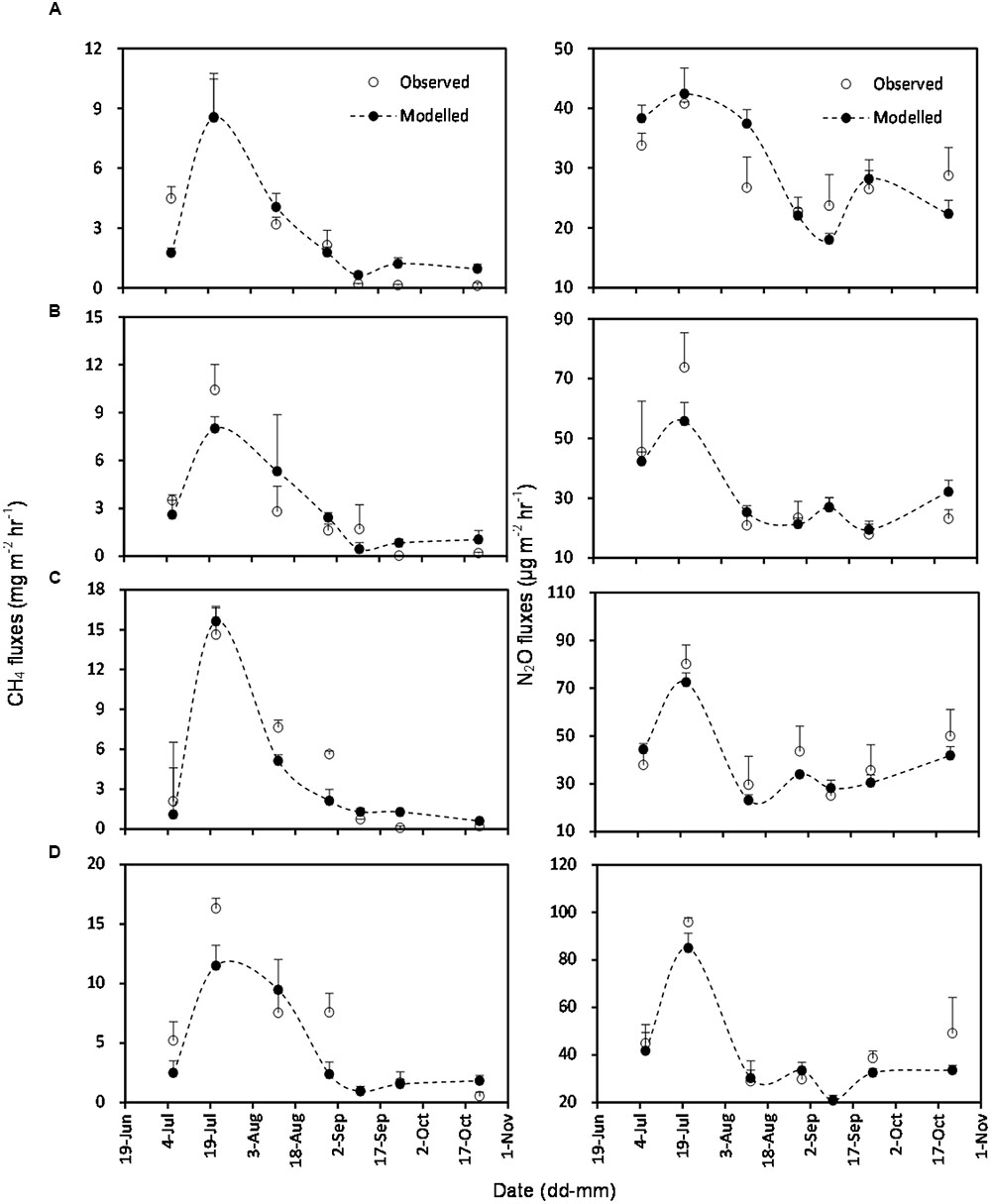
FIGURE 6. Comparison of observed and modeled CH4 and N2O fluxes from paddy rice cropping systems under elevated atmospheric CO2 concentration and rising temperature (T). (A), ambient; (B), elevated CO2; (C), rising temperature (T); (D), elevated CO2 and rising temperature. Error bars show standard error of the mean of data. Overall statistics of model tests are shown in Table 3.
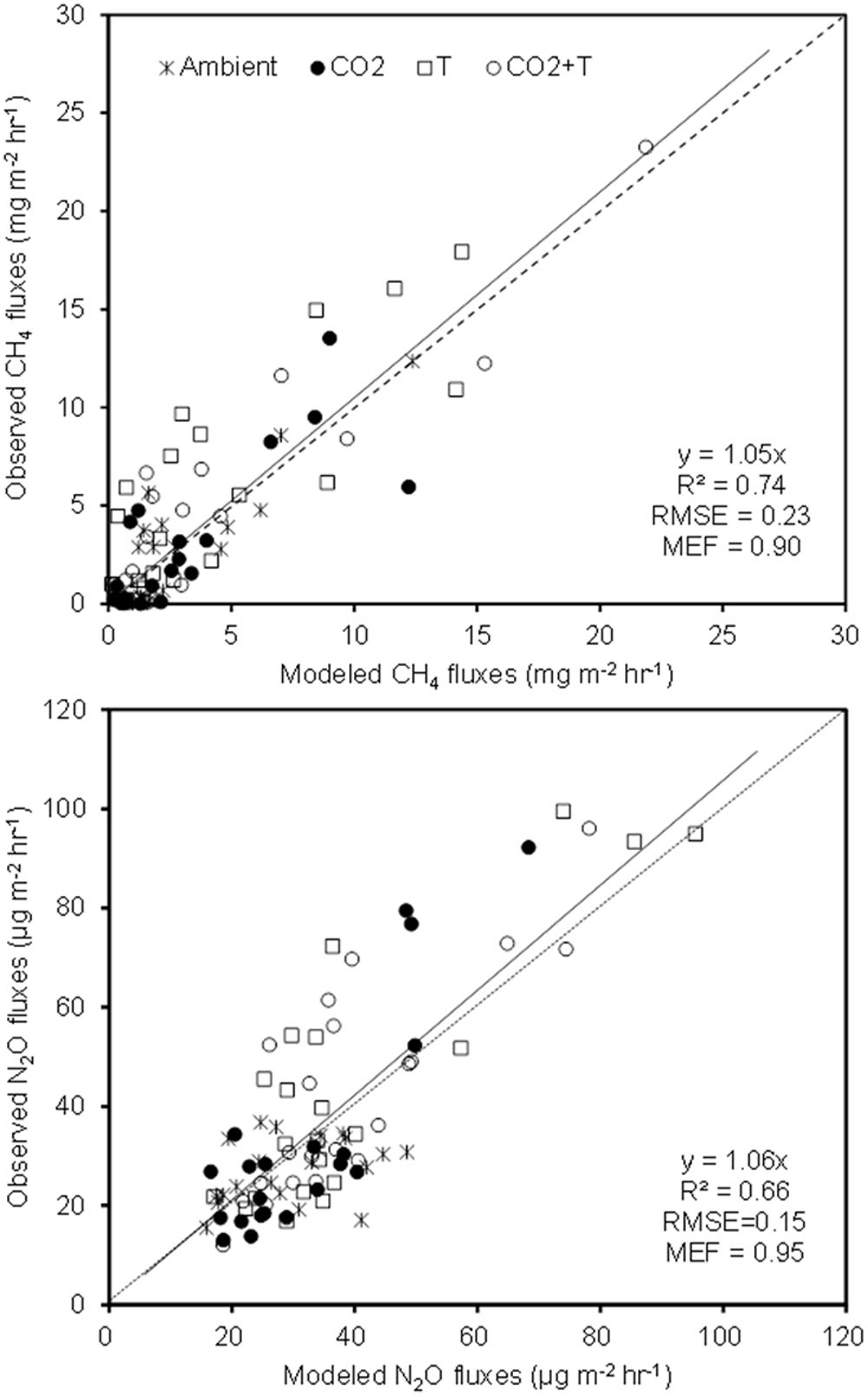
FIGURE 7. The YC3 and YN3 models tested against field measurements in paddy rice cropping systems under elevated atmospheric CO2 and rising temperature (T).
Manure composting has been recognized as an important source of CH4 and N2O to atmosphere [Intergovernmental Panel on Climate Change (IPCC), 2006; Chadwick et al., 2011; Hou et al., 2015; Owen and Silver, 2015; Pardo et al., 2015]. The IPCC guidelines introduce the terms of MCF (methane conversion factor) and EF (emission factor of N for N2O) for accounting CH4 and N2O emissions from manure composting, respectively [Intergovernmental Panel on Climate Change (IPCC), 2006]. In the present study, total CH4 and N2O emissions were estimated to be 1.1 kg m-2 and 25.1 g m-2, being equivalent to a MCF of 0.8% and an EF of 1.2% for composting windrow, respectively. The value of MCF in this study falls well within the IPCC default value range of 0.5–1.5% in composting windrows [Intergovernmental Panel on Climate Change (IPCC), 2006]. The estimated EF (1.2%) in this study is slightly greater than the IPCC default EF of 1.0% [Intergovernmental Panel on Climate Change (IPCC), 2006], but highly close to the recent estimates (mean of EF: 1.2%) based on a summary of available data on composting windrow by Pardo et al. (2015).
Measurements of GHG fluxes showed a trade-off between CH4 and N2O fluxes, as previously found in rice paddy soils and windrow compost (Cai et al., 1997; Hou et al., 2001; Zou et al., 2005; Ahn et al., 2011; Shen et al., 2011). Consistent with previous studies (Hou et al., 2001; Sánchez-Monedero et al., 2010; Sharma et al., 2011; Chen et al., 2014), substantial CH4 emissions occurred mainly in the early stage of manure composting process. In contrast, remarkable N2O emissions were triggered around the middle stage of the composting when pile temperature started to decline and oxygen availability was limited (Fukumoto et al., 2003; Xu et al., 2007; Ahn et al., 2011; Tsutsui et al., 2013; Wang et al., 2013). In addition, norB and nosZ genes were relatively predominant during the early stage of composting (Figure 2), suggesting that much N2O was further transformed into N2 as the final product in denitrification.
Consistent with the first hypothesis prediction, changes in physicochemical parameters shaped different time course patterns of CH4- and N2O-related functional genes (Gödde and Conrad, 1999; Holtan-hartwig et al., 2002; Kandeler et al., 2006; Zhou et al., 2011; Zhang et al., 2015). Correlation analyses showed that the abundance of pmoA, narG, nirK, and nirS genes shared negative correlations with pile temperature (r = ∼-0.70, p < 0.05), moisture, TOC, and C/N ratio, and positive correlations with TN (Figure 4), suggesting that changes in interacting physicochemical parameters in the pile shaped the dynamic pattern of the pmoA, narG, nirK, and nirS genes (Figure 4), in line with the results obtained by Hallin et al. (2009) showing that nitrate reducers and denitrifiers were closely related to soil TOC, TN, and C/N ratio in a 50-year-old fertilization experiment. Similar relationships between methanotrophs abundances and abiotic parameters were found in landfill cover soils (Kumaresan et al., 2009; Lin et al., 2009). Some studies also reported the relationships between denitrifiers and physicochemical parameters, such as correlations of nirK and nirS genes abundance with pile temperature (Zhang et al., 2015), and correlations of narG, nirK and nosZ genes abundance with soil TOC (Kandeler et al., 2006; Zhou et al., 2011).
Significant correlations of nosZ gene abundance with NH4+ and NO2- suggested that nosZ gene abundance dynamics was mainly associated with substrate availabilities. The mcrA and amoA genes abundance did not show significant correlations with any of physicochemical parameters, and amoA gene abundance was, on average, much lower than nitrate reducers and denitrifiers (narG, nirK nirS, norB, and nosZ) abundance, suggesting that denitrification was much stronger than nitrification during windrow composting (Hao et al., 2001). Partially due to pH remaining stable around 8.0 during windrow composting, pile pH did not show significant correlations with any of bacterial functional genes in this study, consistent with Kandeler et al. (2006) but contrary to other previous studies (Deiglmayr et al., 2004; Bárta et al., 2010; Zhang et al., 2015). In addition, NH4+ was slightly correlated with narG, nirK, and nirS genes abundance (p = ∼0.07), while NO3- did not show significant correlations with nitrate reducer and denitrifier abundances, which might suggest that nitrate in pile manure is not important for denitrifiers (Tiedje, 1988; Mergel et al., 2001; Avrahami et al., 2002; Liu et al., 2003; Kandeler et al., 2006; Zhang et al., 2015).
The NH4+ and pmoA gene were involved in CH4 and N2O emissions, respectively, which partially supported the second hypothesis that some specific physicochemical parameters and compositional bacterial enzymes encoded by relevant genes would be multifunctional as involved both in CH4 and N2O. The CH4 fluxes showed a positive correlation with NH4+, and NH4+ was selected as an indicator in the stepwise regression model (Table 3 and Figure 3). A great many studies have revealed that NH4+ has an inhibitory effect on CH4 oxidization through either competition for methane monooxygenase or generation of toxic hydroxylamine and nitrite from ammonium oxidation (Steudler et al., 1989; Bosse et al., 1993; Dunfield and Knowles, 1995; Hanson and Hanson, 1996; Duan et al., 2013; Dam et al., 2014; Karbin et al., 2015), although stimulation effects or no effects of NH4+ on methanotrophs were reported in some other studies (Dunfield et al., 1995; Delgado and Mosier, 1996; Dan et al., 2001; Krüger and Frenzel, 2003; Shrestha et al., 2010; Hu and Lu, 2015). The CH4 fluxes were negatively related with narG but positively related with nosZ genes abundances, which might be due to the significant correlations of narG with pmoA genes (r = 0.84, p = 0.005) and of nosZ genes with NH4+ (r = 0.76, p = 0.02). In addition, N2O fluxes were correlated with pmoA gene abundances, and pmoA gene abundances were included in the regression model (Table 3 and Figure 3), which might suggest denitrification with methane as external carbon source. Some studies reported that aerobic methane-oxidation coupled to denitrification is accomplished by aerobic methanotrophs oxidizing methane and releasing soluble organics that are used by coexisting denitrifiers as electron donors for denitrification (Modin et al., 2007). Indeed, the pmoA, narG, and nirS gene abundances were correlated in this study (Figure 4).
Both mcrA and pmoA genes abundances were correlated with CH4 fluxes, and the balance of mcrA/pmoA genes abundance was selected as a good indicator in the regression model (Table 3 and Figure 3), indicating that both methanogens and methanotrophs played important roles in CH4 fluxes from composting windrow. The N2O fluxes were positively correlated with narG and nirK genes abundance, but negatively correlated with nosZ gene abundance. However, N2O fluxes were not correlated with nirS gene abundances in this study. The nitrite reducers with Cu-containing enzyme encoded by nirK gene are generally believed to be more important than those with cytochrome cd1 nitrite reductase encoded by nirS gene in the nitrite reduction step during manure composting (Yoshida et al., 2009; Bárta et al., 2010; Chen et al., 2010; Zhou et al., 2011; Zhang et al., 2015).
Based on physicochemical and biological variables measurements during the composting and their correlation and regression analyses, we developed a schematic model that explains the dynamics of CH4 and N2O fluxes associated with bacterial functional genes and physicochemical parameters during manure composting (Figure 8). The schematic model shows how the prevalence of bacteria is involved in key steps in the process of CH4 and N2O emissions, and the CH4 and N2O fluxes during windrow composting are controlled by the interplay of enzyme encoding bacterial functional genes (Figure 8). In the schematic model, some physicochemical parameters are correlated with each other and interacting to shape the dynamics of bacterial functional gene abundance. Besides bacterial functional genes are directly involved in CH4 or N2O emissions, CH4 oxidization and denitrification processes interact together, where NH4+ has inhibitory effects on CH4 oxidization and pmoA gene abundance can facilitate denitrification with methane as external carbon source (Figure 8). Some studies stated that the aerobic methanotrophic bacteria are particularly useful for discovering and analyzing diverse mechanisms for nitrification and denitrification processes (Stein and Klotz, 2011; Zhou et al., 2014). By testing against samples in paddy rice cropping systems (Table 3 and Figures 6, 7), the simulated models can also be applicable to predicting seasonal dynamics of CH4 and N2O fluxes as responses to elevated atmospheric CO2 and rising temperature.
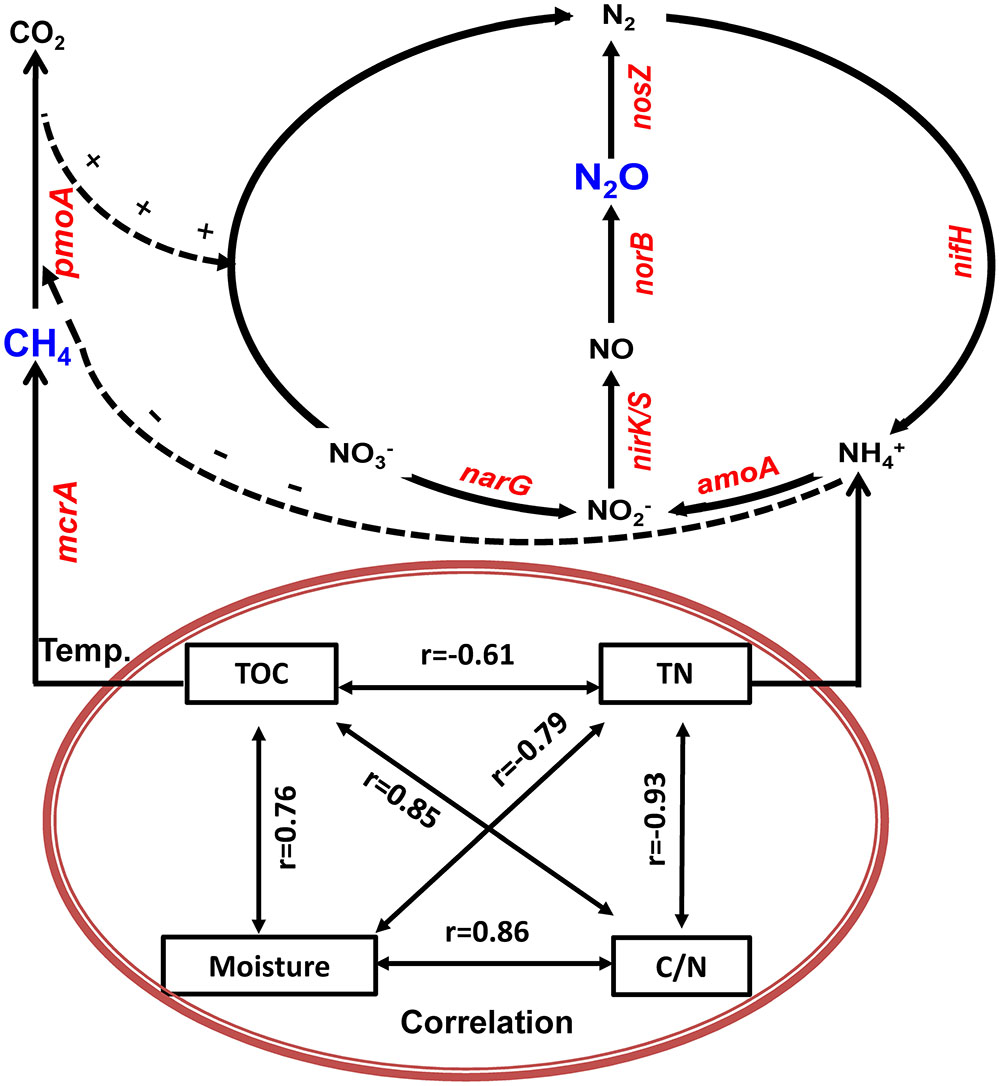
FIGURE 8. Generalized schematic model showing predicted CH4 and N2O fluxes as a function of bacterial functional genes and physicochemical parameters during manure composting. Physicochemical parameters of compost manure material including total organic carbon (TOC), total nitrogen (TN), moisture and carbon/nitrogen ratio (C/N) are correlated with each other, and interacting to shape the dynamics of bacterial functional gene abundance. Solid lines indicate direct connections. Dashed lines refer to indirect connections where ammonium has inhibitory effects on CH4 oxidization, while CH4 oxidization facilitates denitrification with methane as external carbon source.
Bacterial genes key functional to CH4 and N2O fluxes identified in this study may be used as strategies for mitigating GHGs. For example, biochar application can significantly reduce both CH4 and N2O emissions by depressing nirK and mcrA while stimulating nosZ and pmoA genes abundances during manure composting (Sonoki et al., 2013; Wang et al., 2013). We noted that limitations on the use of qPCR for investigation of targeted genes also exist as results of PCR-bias, disturbance by DNA from dead cell, detecting only DNA copy numbers but not RNA transcriptional activity, lacking information regarding detailed community structures of specific microorganisms. The cDNA-based technologies and high throughout strategies, such as reverse transcription quantitative PCR, Illumina sequencing, and Gene Chip, will be very useful for a deeper understanding the characteristics of the functional genes and specific microbial groups, as well as their relationships with GHG emissions.
We presented the quantitative study illustrating interactions between different bacterial activities and their role in controlling CH4 and N2O fluxes as a response to changes in physicochemical parameters during windrow composting. This study also presented the quantitative assessment of CH4 and N2O fluxes based on multiple microbial gene abundances at the functional levels in composting windrow. Additional studies in this area are highly needed to extend such capabilities and allow us to quantitatively address microbial contributions to GHG fluxes from soils and manure management systems. This is particularly important, as it is widely believed that microorganisms play important roles in global carbon and nitrogen biogeochemical cycles, yet they are generally not included in current biogeochemical models for carbon and nitrogen cycles.
SqL and JZ conceived this study. SqL has the main responsibility for microbial sampling and microbial analyses for this study. SwL and QS provided valuable input for the design and data analyses of this study. LS, XG, and YJ performed qPCR analyses and gas sampling. SwL, SqL, and JZ performed the statistical analyses and wrote the paper. All authors edited and approved the final manuscript.
This work was supported by the National Basic Research Program of China (2015CB150502), National Natural Science Foundation of China (NSFC 41225003, 41401321), Fundamental Research Funds for the Central Universities (KYT201404 and KYZ201621, NAU), and Ministry of Education 111 project (B12009) and PADA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Evan Siemann at Rice University for the help in language editing of this manuscript.
Ahn, H. K., Mulbry, W., White, J. W., and Kondrad, S. L. (2011). Pile mixing increases greenhouse gas emissions during composting of dairy manure. Bioresour. Technol. 102, 2904–2909. doi: 10.1016/j.biortech.2010.10.142
Angnes, G., Nicoloso, R. S., da Silva, M. L., de Oliveira, P. A., Higarashi, M. M., Mezzari, M. P., et al. (2013). Correlating denitrifying catabolic genes with N2O and N2 emissions from swine slurry composting. Bioresour. Technol. 140, 368–375. doi: 10.1016/j.biortech.2013.04.112
Avrahami, S., Conrad, R., and Braker, G. (2002). Effect of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers. Appl. Environ. Microbiol. 68, 5685–5692. doi: 10.1128/AEM.68.11.5685-5692.2002
Bárta, J., Applová, M., Vaněk, D., Krištůfková, M., and Šantrůčková, H. (2010). Effect of avalilabe P and phenolics on mineral N release in acidified spruce forest: connection with lignin-degrading enzymes and bacteria and fungal. Biogeochemistry 97, 71–87. doi: 10.1007/s10533-009-9363-3
Bosse, U., Frenzel, P., and Conrad, R. (1993). Inhibition of methane oxidation by ammonium in the surface-layer of a littoral sediment. FEMS Microbiol. Ecol. 13, 123–134. doi: 10.1111/j.1574-6941.1993.tb00058.x
Braker, G., Fesefeldt, A., and Witzel, K. P. (1998). Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64, 3769–3775.
Braker, G., and Tiedje, J. M. (2003). Nitric oxide reductase (norB) genes from pure cultures and environmental samples. Appl. Environ. Microbiol. 69, 3476–3483. doi: 10.1128/AEM.69.6.3476-3483.2003
Cai, C., Yin, X., He, S., Jiang, W., Si, C., Struik, P. C., et al. (2015). Responses of wheat and rice to factorial combinations of ambient and elevated CO2 and temperature in FACE experiments. Glob. Change Biol. 22, 856–874. doi: 10.1111/gcb.13065
Cai, Z., Xing, G., Yan, X., Xu, H., Tsuruta, H., Yagi, K., et al. (1997). Methane and nitrous oxide emissions from rice paddy fields as affected by nitrogen fertilisers and water management. Plant Soil 196, 7–14. doi: 10.1023/A:1004263405020
Castaldi, P., Garau, G., and Melis, P. (2008). Maturity assessment of compost from municipal solid waste through the study of enzyme activities and water-soluble fractions. Waste Manage. 28, 534–540. doi: 10.1016/j.wasman.2007.02.002
Chadwick, D., Sommer, S., Thorman, R., Fangueiro, D., Cardenas, L., Amon, B., et al. (2011). Manure management: implications for greenhouse gas emissions. Anim. Feed. Sci. Technol. 16, 514–531. doi: 10.1016/j.anifeedsci.2011.04.036
Chen, R., Wang, Y., Wei, S., Wang, W., and Lin, X. (2014). Windrow composting mitigated CH4 emissions: characterization of methanogenic and methanotrophic communities in manure management. FEMS Microbiol. Ecol. 90, 575–586. doi: 10.1111/1574-6941.12417
Chen, Z., Luo, X., Hu, R., Wu, M., Wu, J., and Wei, W. (2010). Impact of long-term fertilization on the composition of denitrifier communities based on nitrite reductase analyses in a paddy soil. Microb. Ecol. 60, 850–861. doi: 10.1007/s00248-010-9700-z
Chen, Z., Zhang, J., Xiong, Z., Pan, G., and Müller, C. (2016). Enhanced gross nitrogen transformation rates and nitrogen supply in paddy field under elevated atmospheric carbon dioxide and temperature. Soil Biol. Biochem. 94, 80–87. doi: 10.1016/j.soilbio.2015.11.025
Costello, A. M., and Lidstrom, M. E. (1999). Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 65, 5066–5074.
Czepiel, P., Douglas, E., Harriss, R., and Crill, P. (1996). Measurements of N2O from composted organic wastes. Environ. Sci. Technol. 30, 2519–2525. doi: 10.1021/es950841j
Dam, B., Dam, S., Kim, Y., and Liesack, W. (2014). Ammonium induces differential expression of methane and nitrogen metabolism-related genes in Methylocystis sp. strain SC2. Environ. Microbiol. 16, 3115–3127. doi: 10.1111/1462-2920.12367
Dan, J. G., Krüger, M., Frenzel, P., and Conrad, R. (2001). Effect of a late season urea fertilization on methane emission from a rice field in Italy. Agric. Ecosyst. Environ. 83, 191–199. doi: 10.1016/S0167-8809(00)00265-6
Deiglmayr, K., Philippot, L., Hartwig, U. A., and Kandeler, E. (2004). Structure and activity of the nitrate-reducing community in the rhizosphere of Lolium perenne and Trifolium repens under long-term elevated atmospheric pCO. FEMS Microbiol. Ecol. 49, 445–454. doi: 10.1016/j.femsec.2004.04.017
Delgado, J., and Mosier, A. R. (1996). Mitigation alternatives to decrease nitrous oxides emissions and urea-nitrogen loss and their effect on methane flux. J. Environ. Qual. 25, 1105–1111. doi: 10.2134/jeq1996.00472425002500050025x
Duan, Y. F., Elsgaard, L., and Petersen, S. O. (2013). Inhibition of methane oxidation in a slurry surface crust by inorganic nitrogen: an incubation study. J. Environ. Qual. 42, 507–515. doi: 10.2134/jeq2012.0230
Dunfield, P., and Knowles, R. (1995). Kinetics of inhibition of methane oxidation by nitrate, nitrite and ammonium in a humisol. Appl. Environ. Microbiol. 61, 3129–3135.
Dunfield, P. F., Topp, E., Archambault, C., and Knowles, R. (1995). Effect of nitrogen fertilizers and moisture-content on CH4 and N2O fluxes in a humisol-measurements in the field and intact soil cores. Biogeochemistry 29, 199–222. doi: 10.1007/BF02186048
Fukumoto, Y., Osada, T., Hanajima, D., and Haga, K. (2003). Patterns and quantities of NH3, N2O and CH4 emissions during swine manure composting without forced aeration-effect of compost pile scale. Bioresour. Technol. 89, 109–114. doi: 10.1016/S0960-8524(03)00060-9
Gödde, M., and Conrad, R. (1999). Immediate and adaptational temperature effects on nitric oxide production and nitrous oxide release from nitrification and denitrification in two soils. Biol. Fertil. Soils 30, 33–40. doi: 10.1007/s003740050584
Hallin, S., Jones, C. M., Schloter, M., and Phillippot, L. (2009). Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J. 3, 597–605. doi: 10.1038/ismej.2008.128
Hao, X., Chang, C., Larney, F. J., and Travis, G. R. (2001). Greenhouse gas emissions during cattle feedlot manure composting. J. Environ. Qual. 30, 376–386. doi: 10.2134/jeq2001.302376x
Henry, S., Baudoin, E., López-Gutiérrez, J. C., Martin-Laurent, F., Brauman, A., and Philippot, L. (2004). Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J. Microbiol. Methods 59, 327–335. doi: 10.1016/j.mimet.2004.07.002
Holtan-hartwig, L., Dörsch, P., and Bakken, L. R. (2002). Low tempreture control of soil denitrifying communities: kinetics of N2O production and reduction. Soil Biol. Biochem. 34, 1797–1806. doi: 10.1016/S0038-0717(02)00169-4
Hou, F. S., Milke, M. W., Leung, D. W., and MacPherson, D. J. (2001). Variaions in phytoremediation performance with diesel-contaminated soil. Environ. Technol. 22, 215–222. doi: 10.1080/09593332208618301
Hou, Y., Velthof, G. L., and Oenema, O. (2015). Mitigation of ammonia, nitrous oxide and methane emissions from manure management chains: a meta-analysis and integrated assessment. Glob. Chang. Biol. 21, 1293–1312. doi: 10.1111/gcb.12767
Hu, A., and Lu, Y. H. (2015). The differential effects of ammonium and nitrate on methanotrophs in rice field soil. Soil Biol. Biochem. 85, 31–38. doi: 10.1016/j.soilbio.2015.02.033
Hu, H. W., Chen, D. L., and He, J. Z. (2015). Microbial regulation of terrestrial nitrous oxide formation: understanding the biological pathways for prediction of emission rates. FEMS Microbiol. Rev. 39, 729–749. doi: 10.1093/femsre/fuv021
Intergovernmental Panel on Climate Change (IPCC) (2006). IPCC Guidelines for National Greenhouse Gas Inventories. Kanagawa: IPCC/IGES.
Intergovernmental Panel on Climate Change (IPCC) (2013). Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press.
Jiang, T., Li, G., Tang, Q., Ma, X., Wang, G., and Schuchardt, F. (2015). Effects of aeration method and aeration rate on greenhouse gas emissions during composting of pig feces in pilot scale. J. Environ. Sci. 31, 124–132. doi: 10.1016/j.jes.2014.12.005
Kandeler, E., Deiglmayr, K., Tscherko, D., Bru, D., and Philippot, L. (2006). Abundance of narG, nirS, nirK, and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl. Environ. Microbiol. 72, 5957–5962. doi: 10.1128/AEM.00439-06
Karbin, S., Hagedorn, F., Dawes, M. A., and Niklaus, P. A. (2015). Treeline soil warming does not affect soil methane fluxes and the spatial micro-distribution of methanotrophic bacteria. Soil Biol. Biochem. 86, 164–171. doi: 10.1016/j.soilbio.2015.03.022
Kim, S. Y., Lee, S. H., Freeman, C., Fenner, N., and Kang, H. (2008). Comparative analysis of soil microbial communities and their responses to the short-term drought in bog, fen, and riparian wetlands. Soil Biol. Biochem. 40, 2874–2880. doi: 10.1016/j.soilbio.2008.08.004
Kroon, P. S., Hensen, A., Van den Bulk, W. C. M., Jongejan, P. A. C., and Vermeulen, A. T. (2008). The importance of reducing the systematic error due to non-linearity in N2O flux measurements by static chambers. Nutr. Cycl. Agroecosyst. 82, 175–186. doi: 10.1007/s10705-008-9179-x
Krüger, M., and Frenzel, P. (2003). Effects of N-fertilization on CH4 oxidation and production, and consequences for CH4 emissions from microcosms and rice fields. Glob. Change Biol. 9, 773–784. doi: 10.1046/j.1365-2486.2003.00576.x
Kumaresan, D., Abell, G. C. J., Bodrossy, L., Stralis-Pavese, N., and Murrell, J. C. (2009). Spatial and temporal diversity of methanotrophs in a landfill cover soil are differentially related to soil abiotic factors. Environ. Microbiol. Rep. 5, 398–407. doi: 10.1111/j.1758-2229.2009.00059.x
Larney, F. J., and Hao, X. (2007). A review of composting as a management alternative for beef cattle feedlot manure in southern Alberta. Canada. Bioresour. Technol. 98, 3221–3227. doi: 10.1016/j.biortech.2006.07.005
Lin, B., Monreal, C. M., Tambong, J. T., Miguez, C. B., and Carrasco-Medina, L. (2009). Phylogenetic analysis of methanotrophic communities in cover soils of a landfill in Ontario. Can. J. Microbiol. 55, 1103–1112. doi: 10.1139/w09-069
Liu, S., Qin, Y., Zou, J., and Liu, Q. (2010). Effects of water regime during rice-growing season on annual direct N2O emission in a paddy rice-winter wheat roation system in southeast China. Sci. Total Environ. 408, 906–913. doi: 10.1016/j.scitotenv.2009.11.002
Liu, S., Zhang, L., Liu, Q., and Zou, J. (2012). Fe(III) fertilization mitigating net global warming potential and greenhouse gas intensity in paddy rice-wheat roation systems in China. Environ. Pollut. 164, 73–80. doi: 10.1016/j.envpol.2012.01.029
Liu, X., Tiquia, S. M., Holguin, G., Wu, L., Nold, S. C., Devol, A. H., et al. (2003). Molecular diversity of denitrifying genes in continental margin sediments within the oxygen-deficient zone off the Pacific coast of Mexico. Appl. Environ. Microbiol. 69, 3549–3560. doi: 10.1128/AEM.69.6.3549-3560.2003
Liu, Y., Li, M., Zheng, J., Li, L., Zhang, X., Zheng, J., et al. (2014). Short-term responses of microbial community and functioning to experimental CO2 enrichment and warming in a Chinese paddy field. Soil Biol. Biochem. 77, 58–68. doi: 10.1016/j.soilbio.2014.06.011
López-Gutiérrez, J. C., Henry, S., Hallet, S., Martin-Laurent, F., Catroux, G., and Philippot, L. (2004). Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J. Microbiol. Methods 57, 399–407. doi: 10.1016/j.mimet.2004.02.009
Maeda, K., Hanajima, D., Toyoda, S., Yoshida, N., Morioka, R., and Osada, T. (2011). Microbiology of nitrogen cycle in animal manure compost. Microbiol. Biotechnol. 4, 700–709. doi: 10.1111/j.1751-7915.2010.00236.x
Maeda, K., Morioka, R., Hanajima, D., and Osada, T. (2010a). The impact of using mature compost on nitrous oxide emission and the denitrifier community in the cattle manure composting process. Microb. Ecol. 59, 25–36. doi: 10.1007/s00248-009-9547-3
Maeda, K., Toyoda, S., Shimojima, R., Osada, T., Hanajima, D., Morioka, R., et al. (2010b). Source of nitrous oxide emissions during the cow manure composting process as revealed by isotopomer analysis of and amoA abundance in Betaproteo bacterial ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 76, 1555–1562. doi: 10.1128/AEM.01394-09
Mergel, A., Schmitz, O., Mallmann, T., and Bothe, H. (2001). Relative abundance of denitrifying and dinitrogen-fixing bacteria in layers of a forest soil. FEMS Microbiol. Ecol. 36, 33–42. doi: 10.1016/S0168-6496(01)00113-1
Modin, O., Fukushi, K., and Yamamoto, K. (2007). Denitrification with methane as external carbon source. Water Res. 41, 2726–2738. doi: 10.1016/j.watres.2007.02.053
Morales, S. E., Cosart, T., and Holben, W. E. (2010). Bacterial gene abundances as indicators of greenhouse gas emission in soils. ISME J. 4, 799–808. doi: 10.1038/ismej.2010.8
Mulbry, W., and Ahn, H. (2014). Greenhouse gas emissions during composting of dairy manure: influnce of the timing of pile mixing on total emissions. Biosyst. Eng. 126, 117–122. doi: 10.1016/j.biosystemseng.2014.08.003
Naylor, R., Steinfeld, H., Falcon, W., Galloway, J., Smil, V., Bradford, E., et al. (2005). Agriculture. Losing the links between livestock and land. Science 310, 1621–1622. doi: 10.1126/science.1117856
Oenema, O., and Tamminga, S. (2005). Nitrogen in global animal production and management options for improving nitrogen use efficiency. Sci. China C Life Sci. 48, 871–887.
Owen, J. J., and Silver, W. L. (2015). Greenhouse gas emissions from dairy manure management: a review of field-based studies. Glob. Change Biol. 21, 550–565. doi: 10.1111/gcb.12687
Pardo, G., Moral, R., Aguilera, E., and del Prado, A. (2015). Gaseous emissions from management of solid waste: a systematic review. Glob. Change Biol. 21, 1313–1327. doi: 10.1111/gcb.12806
Pereyra, L. P., Hiibel, S. R., Prieto Riquelme, M. V., Reardon, K. F., and Pruden, A. (2010). Detection and quantification of functional genes of cellulose-degrading, fermentative, and sulfate-reducing bacteria and methanogenic archaea. Appl. Environ. Microbiol. 76, 2192–2202. doi: 10.1128/AEM.01285-09
Regan, K., Kammann, C., Hartung, K., Lenhart, K., Müller, C., Philippot, L., et al. (2011). Can differences in microbial abundances help explain enhanced N2O emissions in a permanent grassland under elevated atmospheric CO2. Glob. Change Biol. 17, 3176–3186. doi: 10.1111/j.1365-2486.2011.02470.x
Rotthauwe, J. H., Witzel, K. P., and Liesack, W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63, 4704–4712.
Sánchez-Monedero, M. A., Serramiá, N., Civantos, C. G., Fernández-Hernández, A., and Roig, A. (2010). Greenhouse gas emissions during composting of two-phase olive mill wastes with different agroindustrial by-products. Chemosphere 81, 18–25. doi: 10.1016/j.chemosphere.2010.07.022
Scala, D. J., and Kerkhof, L. J. (1998). Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol. Lett. 162, 61–68. doi: 10.1016/S0378-1097(98)00103-7
Sharma, R., Ryan, K., Hao, X., Larney, F. J., McAllister, T. A., and Topp, E. (2011). Real-time quantification of mcrA, pmoA for methanogen, methanotroph estimations during composting. J. Environ. Qual. 40, 199–205. doi: 10.2134/jeq2010.0088
Shen, Y., Ren, L., Li, G., Chen, T., and Guo, R. (2011). Influence of aeration on CH4, N2O and NH3 emissions during aerobic composting of a chicken manure and high C/N waste mixture. Waste Manag. 31, 33–38. doi: 10.1016/j.wasman.2010.08.019
Shrestha, M., Shrestha, P. M., Frenzel, P., and Conrad, R. (2010). Effect of nitrogen fertilization on methane oxidation, abundance, community structure, and gene expression of methanotrophs in the rice rhizosphere. ISME J. 4, 1545–1556. doi: 10.1038/ismej.2010.89
Sonoki, T., Furukawa, T., Jindo, K., Suto, K., Aoyama, M., and Sanchez-Monedero, M. A. (2013). Influence of biochar addition on methane metabolism during thermophilic phase of composting. J. Basic Microb. 53, 617–621. doi: 10.1002/jobm.201200096
Stein, L. Y., and Klotz, M. G. (2011). Nitrifying and denitrifying pathways of methanotrophic bacteria. Biochem. Soc. Trans. 39, 1826–1831. doi: 10.1042/BST20110712
Steinberg, L. M., and Regan, J. M. (2009). mcrA-targeted real-time quantitative PCR method to examine methanogen communities. Appl. Environ. Microbiol. 75, 4435–4442. doi: 10.1128/AEM.02858-08
Steudler, P. A., Bowden, R. D., Melillo, J. M., and Aber, J. D. (1989). Influence of nitrogen-fertilization on methane uptake in temperate forest soils. Nature 341, 314–316. doi: 10.1038/341314a0
Tiedje, J. M. (1988). “Ecology of denitrification and of dissimilatory nitrate reduction to ammonium,” in Biology of Anaerobic Microorganisms, ed. A. J. B. Zehnder (New York, NY: John Wiley and Sons, Inc), 179–244.
Tsutsui, H., Fujiwara, T., Matsukawa, K., and Funamizu, N. (2013). Nitrous oxide emission mechanisms during intermittently aerated composting of cattle manure. Bioresour. Technol. 141, 205–211. doi: 10.1016/j.biortech.2013.02.071
Wang, C., Lu, H., Dong, D., Hui, D., Strong, P. J., Hailong, W., et al. (2013). Insight into the effects of biochar on manure composting: evidence supporting the relationship between N2O emission and denitrifying community. Environ. Sci. Technol. 47, 7341–7349. doi: 10.1021/es305293h
Wasmund, K., Kurtboke, D. I., Burns, K. A., and Bourne, D. G. (2009). Microbial diversity in sediments associated with a shallow methane seep in the tropical Timor Sea of Australia reveals a novel aerobic methanotroph diversity. FEMS Microbiol. Ecol. 68, 142–151. doi: 10.1111/j.1574-6941.2009.00667.x
Xin, J. Y., Cui, J. R., Niu, J. Z., Hua, S. F., Xia, C. G., Li, S. B., et al. (2004). Production of methanol from methane by methanotrophic bacteria. Biocatal. Biotransformation 22, 225–229. doi: 10.1080/10242420412331283305
Xu, S., Hao, X., Standford, K., McAllister, T. A., Larney, F. J., and Wang, J. (2007). Greenhouse gas emissions during co-composting of calf mortalities with manure. J. Environ. Qual. 36, 1914–1919. doi: 10.2134/jeq2007.0080
Yoshida, M., Ishii, S., Otsuka, S., and Senoo, K. (2009). Temporal shifts in diversity and quantity of nirS and nirK in a rice paddy field soil. Soil Biol. Biochem. 41, 2044–2051. doi: 10.1016/j.soilbio.2009.07.012
Zhang, L., Zeng, G., Zhang, J., Chen, Y., Yu, M., Lu, L., et al. (2015). Response of denitrifying genes coding for nitrite (nirK or nirS) and nitrous oxide (nosZ) reductases to different physico-chemical parameters during agricultural waste composting. Appl. Microbiol. Biotechnol. 99, 4059–4070. doi: 10.1007/s00253-014-6293-3
Zhou, L., Wang, Y., Long, X. E., Guo, J., and Zhu, G. (2014). High abundance and diversity of nitrite-dependent anaerobic methane-oxidizing bacteria in a paddy field profile. FEMS Microbiol. Lett. 360, 33–41. doi: 10.1111/1574-6968.12567
Zhou, Z. F., Zheng, Y. M., Shen, J. P., Zhang, L. M., and He, J. Z. (2011). Response of denitrification genes nirS, nirK and nosZ to irrigation water quality in a Chinese agricultural soil. Environ. Sci. Pollut. Res. 18, 1644–1652. doi: 10.1007/s11356-011-0482-8
Zou, J., Huang, Y., Jiang, J., Zheng, X., and Sass, R. L. (2005). A 3-year field measurement of methane and nitrous oxide emissions from rice paddies in China: effects of water regime, crop residue, and fertilizer application. Global. Biogeochem. Cycl. 19, GB2012. doi: 10.1029/2004GB002401
Keywords: CH4, carbon and nitrogen biogeochemistry, N2O, bacterial gene abundance, greenhouse gas, statistical model
Citation: Li S, Song L, Gao X, Jin Y, Liu S, Shen Q and Zou J (2017) Microbial Abundances Predict Methane and Nitrous Oxide Fluxes from a Windrow Composting System. Front. Microbiol. 8:409. doi: 10.3389/fmicb.2017.00409
Received: 05 December 2016; Accepted: 27 February 2017;
Published: 20 March 2017.
Edited by:
Marja Tiirola, University of Jyväskylä, FinlandReviewed by:
Kim Yrjälä, University of Helsinki, FinlandCopyright © 2017 Li, Song, Gao, Jin, Liu, Shen and Zou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianwen Zou, and6b3UyMUBuamF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.