- 1Department of Pharmaceutical Biology and Biotechnology, Albert-Ludwigs-University of Freiburg, Freiburg im Breisgau, Germany
- 2Department of Pharmaceutical and Medical Chemistry, Albert-Ludwigs-University of Freiburg, Freiburg im Breisgau, Germany
- 3Spemann Graduate School of Biology and Medicine, Albert-Ludwigs-University of Freiburg, Freiburg im Breisgau, Germany
- 4Institute of Biochemistry, Albert-Ludwigs-University of Freiburg, Freiburg im Breisgau, Germany
- 5Department of Pharmaceutical Biology, Philipps-University Marburg, Marburg, Germany
- 6Department of Chemistry and Biology, University of Siegen, Siegen, Germany
Streptomyces diastatochromogenes Tü6028 is known to produce the polyketide antibiotic polyketomycin. The deletion of the pokOIV oxygenase gene led to a non-polyketomycin-producing mutant. Instead, novel compounds were produced by the mutant, which have not been detected before in the wild type strain. Four different compounds were identified and named foxicins A–D. Foxicin A was isolated and its structure was elucidated as an unusual nitrogen-containing quinone derivative using various spectroscopic methods. Through genome mining, the foxicin biosynthetic gene cluster was identified in the draft genome sequence of S. diastatochromogenes. The cluster spans 57 kb and encodes three PKS type I modules, one NRPS module and 41 additional enzymes. A foxBII gene-inactivated mutant of S. diastatochromogenes Tü6028 ΔpokOIV is unable to produce foxicins. Homologous fox biosynthetic gene clusters were found in more than 20 additional Streptomyces strains, overall in about 2.6% of all sequenced Streptomyces genomes. However, the production of foxicin-like compounds in these strains has never been described indicating that the clusters are expressed at a very low level or are silent under fermentation conditions. Foxicin A acts as a siderophore through interacting with ferric ions. Furthermore, it is a weak inhibitor of the Escherichia coli aerobic respiratory chain and shows moderate antibiotic activity. The wide distribution of the cluster and the various properties of the compound indicate a major role of foxicins in Streptomyces strains.
Introduction
Plants, marine organisms, protozoans, fungi, and bacteria produce a wide range of different secondary metabolites. They are not essential for normal growth, development, or reproduction of an organism, but play a secondary role. There have been several discussions about the selective advantage of these natural products for their producers (Firn and Jones, 2000). Secondary metabolites may act as signals for differentiation, as communication molecules, or as weapons to defend against food competitors (Demain and Fang, 2000) and thus they often possess vital functions in their ecological habitat. The genes responsible for the biosynthesis of a compound are often located next to each other in so called biosynthetic gene clusters. The clusters often span more than 100 kb and encode more than 30 genes related to biosynthesis, transport, regulation, self-resistance and modification. Due to their antibiotic, antitumor, cholesterol-lowering, immunosuppressant or antiviral activities, secondary metabolites are invaluable elements of drug discovery research (Vaishnav and Demain, 2011). Approximately 18.000 bioactive secondary metabolites are produced in bacteria, thereof more than 10.000 compounds are synthesized in Streptomyces (Bérdy, 2012). Representatives of the genus Streptomyces have been studied extensively in the last decades (Weber et al., 2015b). A well-known class of bioactive secondary metabolites are polyketides that are assembled by modular megaenzymes called PKSs. The subsequent steps in the assembly process are highly similar to the biosynthesis of fatty acids. A detailed introduction to PKS can be found in Staunton and Weissman (2001). Non-ribosomal peptides (NRP) belong to another important class of bioactive compounds. They are synthesized by non-ribosomal peptide synthetase (NRPS) that share certain characteristics with PKS (see Schwarzer et al., 2003 for details).
The genome size of Streptomyces ranges from 8 to 9 Mb and shows a high GC (>70%) content. After sequencing the first Streptomyces genomes it was noticed that unexpectedly, it contained far more secondary metabolite gene clusters than had been predicted earlier from the numbers of previously identified metabolites (Bentley et al., 2002; Ikeda et al., 2003). Under laboratory conditions Streptomyces and other secondary metabolite producers synthesize only a few compounds, whereas more than twenty different secondary metabolite gene clusters are contained within most genomes.
The presence of similar biosynthetic gene clusters in different strains reflects their evolutionary history through vertical as well as horizontal gene transfer from one organism to another, also across species barriers. Individual genes, sub-clusters or whole clusters can be exchanged (Egan et al., 2001; Metsä-Ketelä et al., 2002; Donadio et al., 2005). Consequently, the secondary metabolites from similar clusters may vary as they are often built up of distinct moieties from functional sub-clusters. Therefore, biosynthetic gene clusters are ideal to study evolutionary routes and to gain knowledge of the metabolite’s importance to a particular strain (Fischbach et al., 2008).
The activation of cryptic biosynthetic gene clusters is one main goal in Streptomyces research to obtain more and novel bioactive compounds to meet the growing requirements of modern medicine. Potential approaches to successfully activate a gene cluster are summarized in the following paragraph.
The cloning and heterologous expression of complete clusters is one strategy to get access to the genetic potential of Streptomyces (Gomez-Escribano and Bibb, 2014). In addition, silent secondary metabolite gene clusters can be activated through genetic manipulation, e.g., by over-expression or deletion of proposed global or specific positive or negative regulatory genes (Makitrynskyy et al., 2013; Gessner et al., 2015). Furthermore, the cultivation of a given strain under different fermentation conditions (Bode et al., 2002) or the co-cultivation with bacterial or fungal strains (Schroeckh et al., 2009) might stimulate the expression of silent clusters.
Streptomyces diastatochromogenes Tü6028 is known to produce the antimicrobial compound polyketomycin, a tetracyclic quinone glycoside (Paululat et al., 1999). Recently, we deleted the oxygenase gene pokOIV in this strain, resulting in a polyketomycin non-producing mutant (Daum et al., 2009). However, new natural products were synthesized in this mutant. These metabolites were named foxicins A–D. Possibly, foxicins are also produced by the wild type strain but only in little amounts.
In this study we report on the purification and structural elucidation of foxicin A. The compound consists of an unusual nitrogen-containing quinone moiety linked to a short fatty acid (Figure 1).
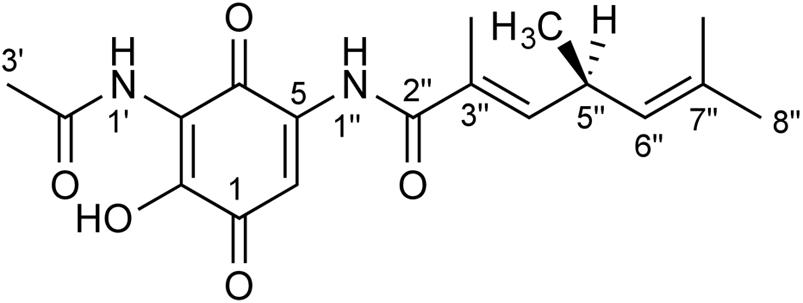
FIGURE 1. Structure of foxicin A. The structure of foxicin A was elucidated as (S)-2-hydroxy-3-(acetylamino)-5-(3″, 5″S, 7″-trimethyl-hepta-3″E, 6″-dienoylamino)-1,4-benzoquinone.
Furthermore, we identified the corresponding gene cluster, encoding a type I polyketide synthase (PKS I) and an NRPS in the genome of S. diastatochromogenes Tü6028. By gene disruption we show that this cluster is responsible for the production of foxicin and we propose the biosynthetic pathway of foxicin A. Similar gene clusters were detected in many other Streptomyces genomes. Nevertheless, as no foxicin-like compound has been described so far, we conclude that these clusters are either not expressed or expressed at a very low level under culture conditions.
Foxicin A shows several interesting biological properties: it acts as siderophore binding ferric ions, it shows antibiotic activity and inhibits respiratory electron transfer. The wide distribution of the cluster and the various properties of the compound indicate a major role of foxicin in Streptomyces strains.
Materials and Methods
Bacterial Culture Condition and Production Analysis
The isolation of polyketomycin of the wild type (wt) strain S. diastatochromogenes Tü6028 has been described previously (Paululat et al., 1999). The mutant S. diastatochromogenes ΔpokOIV (Daum et al., 2009) contains a deletion in pokOIV, a gene encoding an oxygenase involved in the biosynthesis of polyketomycin. Mycelium of the mutant strain S. diastatochromogenes ΔpokOIV was used to inoculate a 300 mL Erlenmeyer flask containing 100 mL of HA medium (yeast extract 0.4%, malt extract 1%, and glucose 0.4% in 1 liter tap water, pH 7.3). The flask was shaken on a rotary shaker (180 rpm) at 28°C. After 48 h, 3 mL of the pre-culture was used to inoculate a second 300 mL Erlenmeyer flask containing 100 mL of HA medium (main culture). After 6 days foxicin A was isolated.
To determine the time dependency of product formation (foxicin A and polyketomycin) the wt strain was grown in 100 mL HA medium for 6 days. Samples were taken after 0, 24, 36, 48, 60, 72, 96, 120, and 144 h of cultivation.
To check the influence of iron on the production of foxicin A, 0.01, 0.1, or 1 mM of FeCl3 or FeSO4 were added to the production media of S. diastatochromogenes ΔpokOIV. The experiment was done in triplicate. The strain was incubated for 6 days and the culture was extracted and analyzed by HPLC. For quantification, the integrals of the corresponding chromatogram peaks were compared.
Isolation of Foxicin A
Mycelium was collected by centrifugation and foxicin A was extracted with acetone (2–3 times the volume of the pellet) by shaking at room temperature for 15 min. After removal of the mycelium by filtration, the extract was evaporated. Finally, this mycelium extract combined with the supernatant was extracted with an equal volume of ethyl acetate. The solvent was removed by evaporation. The crude extract was separated by solid phase chromatography (Oasis HLB 20/35cc) with increasing methanol content (in 10% increments) in the mobile phase. Foxicin A was obtained in the 65 and 70% methanol fraction, foxicin B and C in the 70 and 80% methanol fraction, and foxicin D in the 80% methanol fraction. After thin-layer chromatography in ethyl acetate: formic acid :water (44:3:3) foxicin A (Rf 0.41) was further purified by semi-preparative HPLC (Agilent Technologies), equipped with a Zorbax B-C18 (9.4 × 150 mm) pre-column and Zorbax B-C18 (9.4 mm × 20 mm) main column with acetonitrile + 0.5% acetic acid as buffer A and water + 0.5% acetic acid as buffer B and a flow rate of 2 mL/min. A 3-min washing step with 50% buffer A was followed by a 7-min linear gradient from 50 to 70% of buffer A where the substance was collected. The eluate was dried and resulted in a violet powder. The method was completed by a 4-min delay with 95% buffer A and a 4-min reequilibration step with 50% buffer A.
Analysis of Foxicin by HPLC/MS
For analysis, HPLC-MS equipped with a XBridge C18 (3.5 μm; 20 mm × 4.6 mm) precolumn and a XBridge C18 (3.5 μm; 100 mm × 4.6 mm) main column, an UV/visible light detector and a mass spectrometer (Agilent, 1100 Series) was used. A flow rate of 0.5 mL/min was used. A 1-min washing step with 20% buffer A was followed by a 7-min linear gradient from 20 to 60% buffer A and a 16-min linear gradient ranges from 60 to 95% buffer A. After a 5-min delay the method completed with a 1-min gradient from 95 to 20% buffer A and a 5-min reequilibration step with 20% buffer A.
Structure Elucidation by NMR, VCD, and IR Measurements
The exact mass was analyzed on a MAT 95XL-mass spectrometer (Thermo Electron Corporation). 1H (600 MHz), 13C (150 MHz), and 2D NMR (HSQC, HMBC, 1H-1H COSY) spectra were carried out on a Varian NMR-S600. Chemical shifts are expressed in δ values (ppm), using the correspondent solvent as internal reference (CDCl3: δH = 7.25, δC = 77.0 ppm at T = 25°C or DMSO-d6: δH = 2.50, δC = 39.5 ppm at T = 35°C).
Infrared measurements were carried out on a BRUKER Tensor 27 FT-IR spectrometer equipped with a BRUKER PMA 50 VCD module (Bruker Optik GmbH, Ettlingen). A 100 mM foxicin A solution was prepared in anhydrous CDCl3 and placed in a BaF2 cell with a path length of 110 μm. Experimental spectra (4 cm-1 resolution) represent the average of a 6 h measurement in a rotating cell. IR spectra were corrected by subtraction of the solvent spectrum. VCD spectra were background corrected by solvent subtraction and smoothed by Fourier filtering (8 cm-1 resolution). The aperture of the light source was adjusted to a width of 4 mm. Opus 7.0 software (Bruker Corporation) was used to analyze the spectra.
Conformer Search and Quantum Chemical Calculations
The conformer search was carried out at the MMFF level using Spartan 08 (Wavefunction, Inc., Irvine, CA, USA) and gave a set of 84 possible conformers. The five conformers with the highest population (according to Boltzmann weights calculated in respect to relative energies) account for >99% of the calculated Boltzmann distribution. These conformers were chosen for quantum chemical calculations at the DFT level [B3LYP/6-31+G(d,p)] in Gaussian 09, Revision D.01 (Frisch et al., 2013). All calculations were performed in gas phase, vibrational frequencies were uniformly scaled by an empirical factor of 0.975. Theoretical spectra for each geometry were obtained by adding Lorentzian band shapes (width 6 cm-1) to the calculated IR and VCD intensities. The dissymmetry factor spectrum defined as VCD over IR absorbance was obtained with CDSpecTech (Covington and Polavarapu, 2013, 2014).
Single Crossover of foxBII in S. diastatochromogenes ΔpokOIV
To construct a single crossover of foxBII gene (PKS I/NRPS hybrid gene) an internal 2 kb fragment was amplified (primers GCCGGGAAGCTTGTCCTCTTCGCCTC and GTCGTCGGATCCTGCGC CGCCTCGG). The fragment was cloned into pKC1132 (Bierman et al., 1992) at HindIII/BamHI cloning site and transferred into Escherichia coli ET12567 (dam-, dcm-, hsdS-, cmr) (MacNeil et al., 1992), including conjugation plasmid pUZ8002 (Flett et al., 1997). Positive transformants were selected on LB agar supplemented with kanamycin (30 μg mL-1) and apramycin (50 μg mL-1). The plasmid was transferred into S. diastatochromogenes ΔpokOIV by conjugation. Exconjugants were incubated on MS media supplemented with apramycin.
Bioinformatic Analysis of Foxicin Biosynthetic Gene Cluster and Identification of Similar Biosynthetic Gene Cluster
The draft genome of S. diastatochromogenes Tü6028 was sequenced at the Centrum of Biotechnology, University of Bielefeld (Greule et al., unpublished). Prediction of the gene clusters was performed using antiSMASH 3.01 (Weber et al., 2015a). The sequence of foxicin 57.6 kb hybrid PKS I/NRPS biosynthetic gene cluster was further analyzed and annotated using BLAST2. Similar gene clusters were identified by antiSMASH and BLAST analysis in other Streptomyces strains. The genomes of these strains were analyzed individually by the above mentioned programs.
Phylogenetic Tree of fox Homologous Clusters
A phylogenetic tree was calculated by Clustal Omega (Sievers et al., 2011) using Neighbor-Joining method of foxBII sequence comparison. For better illustration, the tree is shown without distance correction.
Siderophore Chrome Azurol S (CAS) – Assay
Chrome azurol S medium was modified after Schwyn and Neilands (1987). For the CAS medium 60.5 mg CAS, 72.9 mg hexadecyltrimetyl ammonium bromide, and 30.24 g piperazine-1,4-bis-(2-ethanesulfonic acid) were dissolved in 990 mL water and mixed with 10 mL iron (III) solution (1 mM FeCl3 × 6H2O and 10 mM HCl). Purified foxicin A was pipetted to CAS reagent and a color change of the dark blue solution to violet was noted.
Shift of UV/vis Spectra in the Presence of Iron
Foxicin A was dissolved in MeOH. FeCl3 and FeSO4 were added in an amount of 0.5 mM to 100 mM. UV/vis spectra were measured by UviLine 9400 spectrophotometer (SI Analytics).
Isolation of Bacterial Plasma Membranes
Escherichia coli BW25113 cells were grown aerobically (180 rpm) at 37°C in baffled flasks using LB-medium. The cells were harvested by centrifugation (5700 × g, 10 min, 4°C, Rotor JLA 8.1000, Avanti J-26 XP, Beckman Coulter) in the late exponential phase yielding approximately 6.5 g cells/L. All further steps were carried out at 4°C. After centrifugation, 5 g of the cell pellet were resuspended in fourfold volume of buffer 1 (50 mM MES/NaOH, pH 6.0, 50 mM NaCl, 0.1 mM PMSF supplemented with desoxyribonuclease I) and disrupted by passing twice through a French Pressure Cell Press (110 MPa, SLM-Aminco). Cell debris and non-disrupted cells were removed by centrifugation (9500 × g, 20 min, 4°C, Rotor A8.24, RC-5 Superspeed Refrigerated Centrifuge, Sorvall Instruments). The cytoplasmic membranes were obtained from the supernatant by centrifugation at 252000 × g (60 min, 4°C, Rotor 70.1Ti, L8-M Ultrafuge, Beckman). The sediment was suspended in an equal volume (1:1, w/v) of buffer 1 and was used directly or frozen in liquid nitrogen and stored at -80°C.
Determination of NADH Oxidase Activity
The NADH oxidase activity of cytoplasmic membranes was measured with a Clark-type oxygen electrode (RE K1-1, Oxytec) at 30°C. To calibrate the electrode 2 mL 50 mM MES/NaOH, pH 6.0, 50 mM NaCl, 5 mM MgCl2 were deoxygenized by adding sodium dithionite and the signal was set to 237 μM oxygen (Weiss, 1970). Each measurement was performed with 2 mL buffer containing 5 μL of the membrane suspension at 30°C. The reaction was started by adding 1.25 mM NADH. 50–500 μM foxicin A was added to the assay to test its inhibitory action on cell respiration.
Agar Diffusion Assay
Antimicrobial activity of foxicin A was determined by the agar plate diffusion method on a paper disk (6 mm diameter). 100 μg per disk of foxicin A were tested against Streptomyces viridochromogenes Tü57 (Hütter, 1962), Actinokineospora bangkokensis (Intra et al., 2013), Saccharothrix espanaensis (Kong et al., 1998), Bacillus subtilis (Ehrenberg) COHN ATCC6051 (American Type Culture Collection, 18th Edition, 1992), E. coli XL1-Blue, Fusarium verticillioides DSM 62264, Candida parapsilosis DSM 5784, Synechococcus sp. PCC7002 and Synechocystis sp. PCC6803, Mycobacterium smegmatis mc2 155. The compound was dissolved in methanol. After evaporation of the methanol, disks were fixed on the test-plates and incubated over night at 30 or 37°C. Polyketomycin, apramycin, and methanol were used as controls. The following media were used for the assay: LB-medium (tryptone 1%, yeast extract 0.5%, NaCl 0.5%) for E. coli and Bacillus, MS medium (soya flour 2%, mannitol 2%) for actinomycetes strains, BG11-medium (NaNO3 0,15%, K2HPO4 0.004%, MgSO4 × 7H2O 0.0075%, CaCl2 × 2H2O 0.0036%, citric acid 0.0006%, ammonium ferric citrate 0.0006%, EDTA 0.0001%, Na2CO3 0.002%, trace elements 1.0 mL) for cyanobacterial strains, PDA-medium (potato extract 0,4%, dextrose 2%) for Fusarium, YPD-medium (yeast extract 1%, peptone 2%, glucose 2%) for Candida and Middlebrook 7H9 Broth (7H9 broth base 0.52%, 40% glycerol 5.5 mL) for Mycobacterium. For agar plates 2% agar-agar was added to the media.
To assay the herbicidal property, 0.1, 1, and 5 μM of foxicin A was added to 6 mg of Arabidopsis thaliana wt ecotype Wassilewskija seeds that were plated onto plates. The plants were grown for 2 weeks in a phytochamber with long-day conditions (16/8 h), 100 μE m-2 s-1 light intensity and 25°C constant temperature.
To check for H2O2 sensitivity, 2, 5, and 10 μL of 5% H2O2 solution were pipetted on paper disks (6 mm diameter) and placed on MS and TSB culture plates of S. diastatochromogenes WT, ΔpokOIV mutant and ΔpokOIV/foxBII::pKC1132 mutant. The strains were incubated at 28°C for 5 days and inhibition zone was measured.
Cell Viability Assay (MTT-Assay)
The effect on cell viability of foxicin A was tested against cancer cell lines CCRF-CEM, CEM-ADR5000 and Jurkat cells and peripheral blood mononuclear cells (PBMC) using the MTT assay as previously described (Calderón et al., 2014). In brief, cells were seeded in 96-well plates at a density of 4 × 104 cells/well and incubated for 24 h with various concentrations of foxicin A. The chemotherapeutic agent doxorubicin was used as a positive control, and DMSO (0.1%) was the solvent control. The data are expressed as the mean ± SD of three independent experiments.
Results
Production and Isolation of Foxicin A from S. diastatochromogenes Tü6028
Streptomyces diastatochromogenes Tü6028 is known to synthesize polyketomycin, which is produced at high levels after 96 h of cultivation in HA medium. During our studies on polyketomycin biosynthesis we deleted the oxygenase gene pokOIV (Daum et al., 2009). The mutant failed to produce polyketomycin, instead we observed the accumulation of novel compounds, which we named foxicin A, B, C, and D (minor compounds) (Figure 2A). A careful analysis of extracts of the wild type strain showed that foxicins are also produced, but with significantly lower titers. Foxicin production in the wt strain reaches a maximum after 48 h of incubation (Figure 2B). After 96 h less than 5% of the initial foxicin A concentration was detected. In S. diastatochromogenes ΔpokOIV, foxicin production reaches its maximum after 6 days. The mutant also produces fewer spores and visibly lower amounts of melanin, as indicated by the color of the cultivation medium (Figure 2C). Cultivation of S. diastatochromogenes ΔpokOIV in 5 L of HA production medium yielded 9.8 mg of foxicin A (1.96 mg/L) and 3.4 mg of foxicin B (0.68 mg/L), and even lower amounts of foxicins C and D.
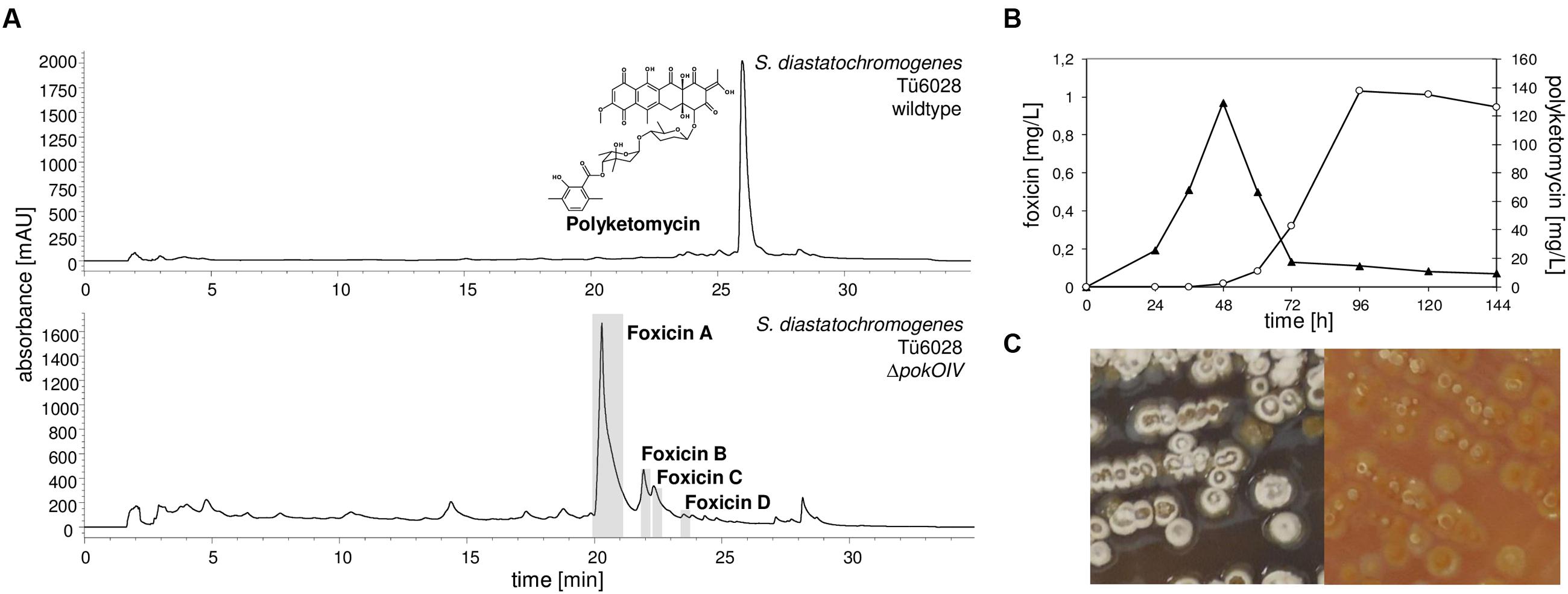
FIGURE 2. Production of polyketomycin and foxicin in Streptomyces diastatochromogenes Tü6028. (A) HPLC chromatogram of S. diastatochromogenes Tü6028 wild type at λ430 nm with polyketomycin structure (top) and of ΔpokOIV mutant at λ320 nm (below); (B) Production of foxicin A (▴) and polyketomycin (O) in S. diastatochromogenes Tü6028 wild type; (C) Morphology of S. diastatochromogenes Tü6028 wild type (left) and ΔpokOIV mutant (right). On the plate, the ΔpokOIV mutant appears yellow and deficient in producing spores and melanins (dark color).
Physiocochemical Properties and Structure Elucidation of Foxicin A
The physicochemical properties of foxicin A are summarized in Supplementary Table S1. Foxicin A is soluble in common organic solvents such as MeOH, CH3CN, CHCl3 and DMSO, but is insoluble in H2O. HR-ESI-MS analysis (m/z found: 346.1532; calculated: 346.1529) revealed the molecular formula as C18H22N2O5. The UV/vis light spectra of the foxicin derivates A–D are similar (Supplementary Figure S1). Foxicin A shows absorption maxima at 246, 280, 316, 390 and a small peak at 462 nm.
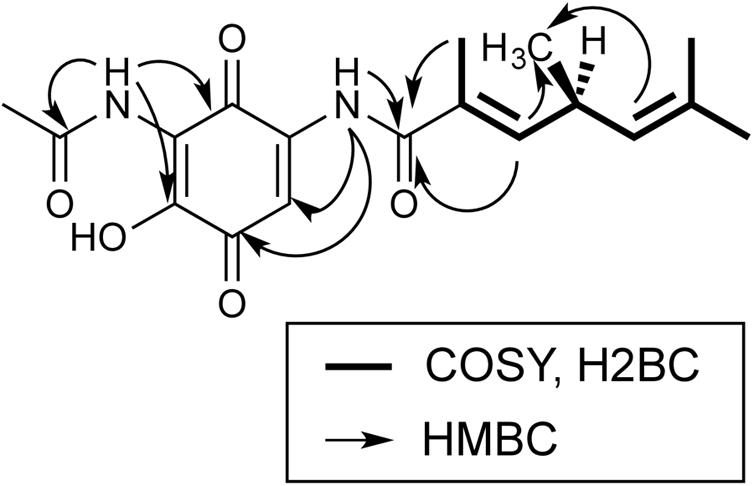
NMR-data of foxicin A were recorded in CDCl3 and DMSO-d6. In Supplementary Tables S2 and S3, NMR assignments of 1D NMR (1H, 13C) and 2D NMR (1H-1H-COSY, HSQC, H2BC, and HMBC) experiments are summarized and Supplementary Figures S2–S14 show the respective spectra. The 13C NMR spectrum shows 18 carbon signals, which could be assigned to five methyl, three methine groups and nine quarternary carbon atoms by the use of HSQC. A 1,4-benzoquinone system was identified from typical carbonyl chemical shifts (δC = 182.3 and δC = 178.6 ppm). The benzoquinone is substituted with a hydroxyl group in position C-2 and an amino-acetate at position C-3, as established by HMBC correlations C-2/6-H, C-2/1′-NH, and C-4/1′-NH. Moreover, an additional side chain is attached at C-5 via an amide functionality, as proven by the HMBC signals C-4/1″-NH and C-6/1″-NH. The side chain contains two double bonds, which are both trisubstituted. One double bond is in conjugation to the amide carbonyl as proven by the HMBC signal C-2″/4″-H and bears a methyl group at C-3″ as shown by the HMBC cross peaks C-2″/3″-CH3, 3″-CH3/4″-H, this double bond is in E configuration as proven by the ROESY signal 3″-CH3/5″-H. The second double bond was determined as a C-6″ = C-7″ double bond with two methyl groups at C-7″ based on HMBC correlations. The two double bonds are connected via the C-5 methine group, which is methyl substituted as established by COSY couplings 4″-H/5″-H, 5″-H/6″-H and H2BC signals C″-5/4″-H and C-5″/6″-H. The important 2D correlations are shown in Figure 3.
Based on our 1- and 2D NMR data, the absolute configuration of foxicin A could not be solved unambiguously. Therefore, we analyzed this compound by comparing vibrational circular dichroism (VCD) and infrared (IR) spectra to spectra from quantum chemical calculations. A molecular model of foxicin A was constructed and subjected to a conformer search algorithm employing molecular mechanics (MMFF). The conformer models were then subjected to a geometry optimization at the DFT level [B3LYP/6-31+G(d,p)] and the relative energies and IR absorbance and VCD intensities were calculated. The comparison of the Boltzmann-weighted average of the spectra calculated for five conformers of foxicin A and the experimental VCD spectrum (Figure 4) allowed for the assignment of the absolute conformation as well as the configuration of foxicin A as (S)-2-hydroxy-3-(acetylamino)-5-(3″, 5″S, 7″-trimethyl-hepta-3″E, 6″-dienoylamino)-1,4-benzoquinone. The structure of foxicin A is shown in Figure 1.
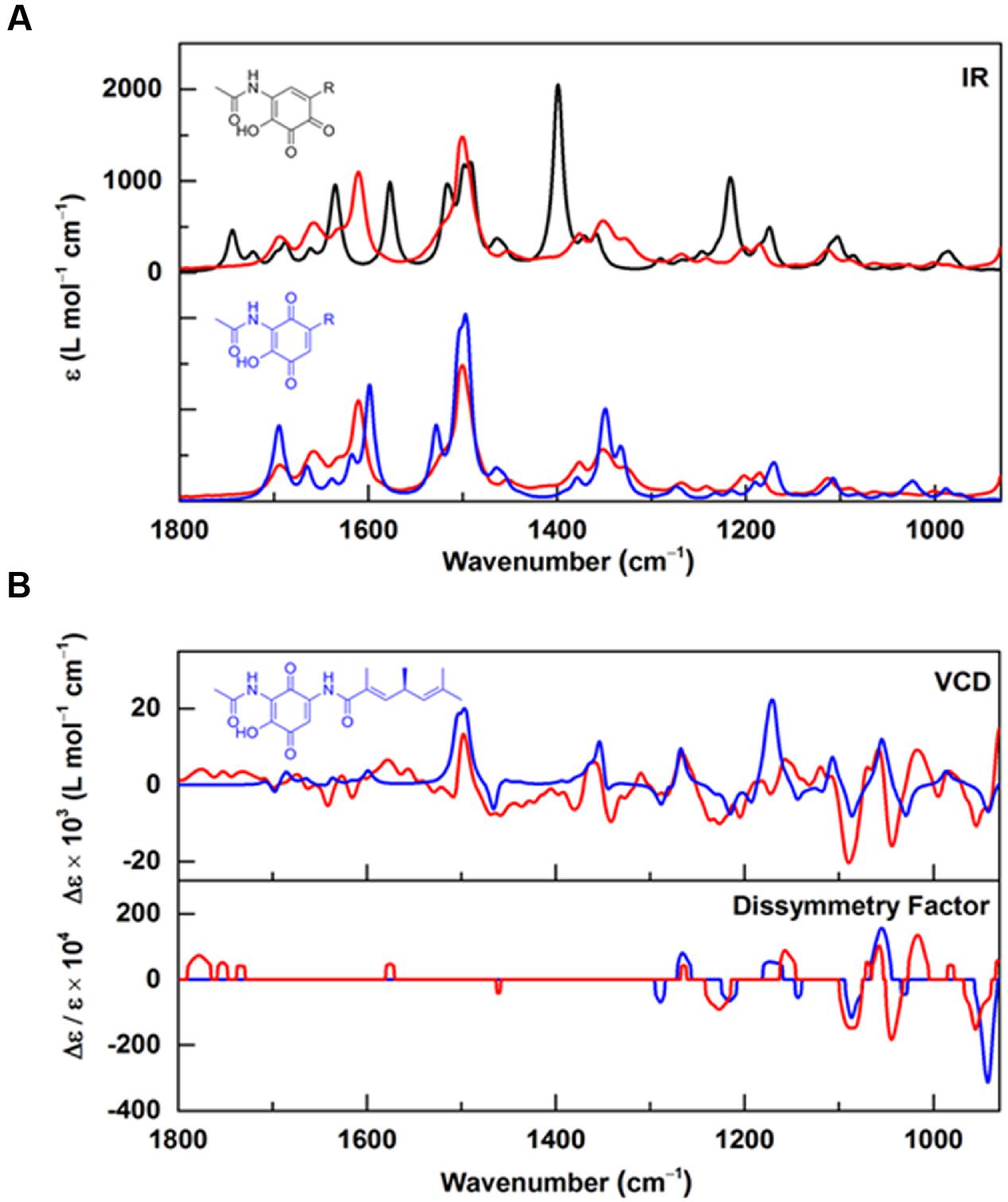
FIGURE 4. Infrared and VCD spectra of foxicin A. (A) Experimental IR spectra recorded for foxicin A (red) overlayed with IR spectra calculated at the B3LYP/6-31+G(d,p) level for the ortho-quinone (black) and for the para-quinone (blue) with considerably better agreement for para (spectra are offset for better comparison); (B) Experimental (red) VCD and dissymmetry factor spectra in comparison to VCD and dissymmetry factor spectra calculated for (S)-foxicin (blue). The good overall agreement allows for assignment of the S-configuration.
Foxicin B has the same mass (346 g/mol) as foxicin A. Foxicin C and foxicin D are only produced in low amounts. Both compounds have a molecular weight of M = 360 g/mol, based on the deprotonated molecular ion peak at m/z 359.2 in the negative ion mode CI-MS spectrum.
Identification and Sequence Analysis of the Biosynthetic Gene Cluster
Due to its chemical structure, foxicin A is likely to be a product of a type I polyketide synthase (PKSI) and a non-ribosomal peptide synthetase (NRPS). Bioinformatics analysis of the 7.9 Mb draft genome sequence of S. diastatochromogenes Tü6028 revealed 23 putative secondary metabolite gene clusters, but only one cluster containing genes with both PKSI- and NRPS function. The cluster was assigned to 57.6 kb with an overall GC content of 72.4%. The annotation analysis revealed 41 open reading frames (ORFs) putatively involved in foxicin A-D biosynthesis (Table 1). The genetic organization of the biosynthetic gene cluster (fox gene cluster) is shown in Figure 5A. The GenBank accession number of the nucleotide sequence is KT440882. In order to verify the correct assignment of the fox gene cluster, we constructed the inactivation plasmid pKC1132_SC_foxBII containing a 2 kb homologous region of foxBII. Conjugation between E. coli ET12567 and S. diastatochromogenes Tü6028 ΔpokOIV and integration of the plasmid into foxBII by single crossover recombination resulted in apramycin-resistant mutants. Integration of the plasmid into foxBII by single crossover recombination was confirmed by PCR. Loss of ability of the mutant strain to produce foxicins confirmed the correct assignment of the fox gene cluster.
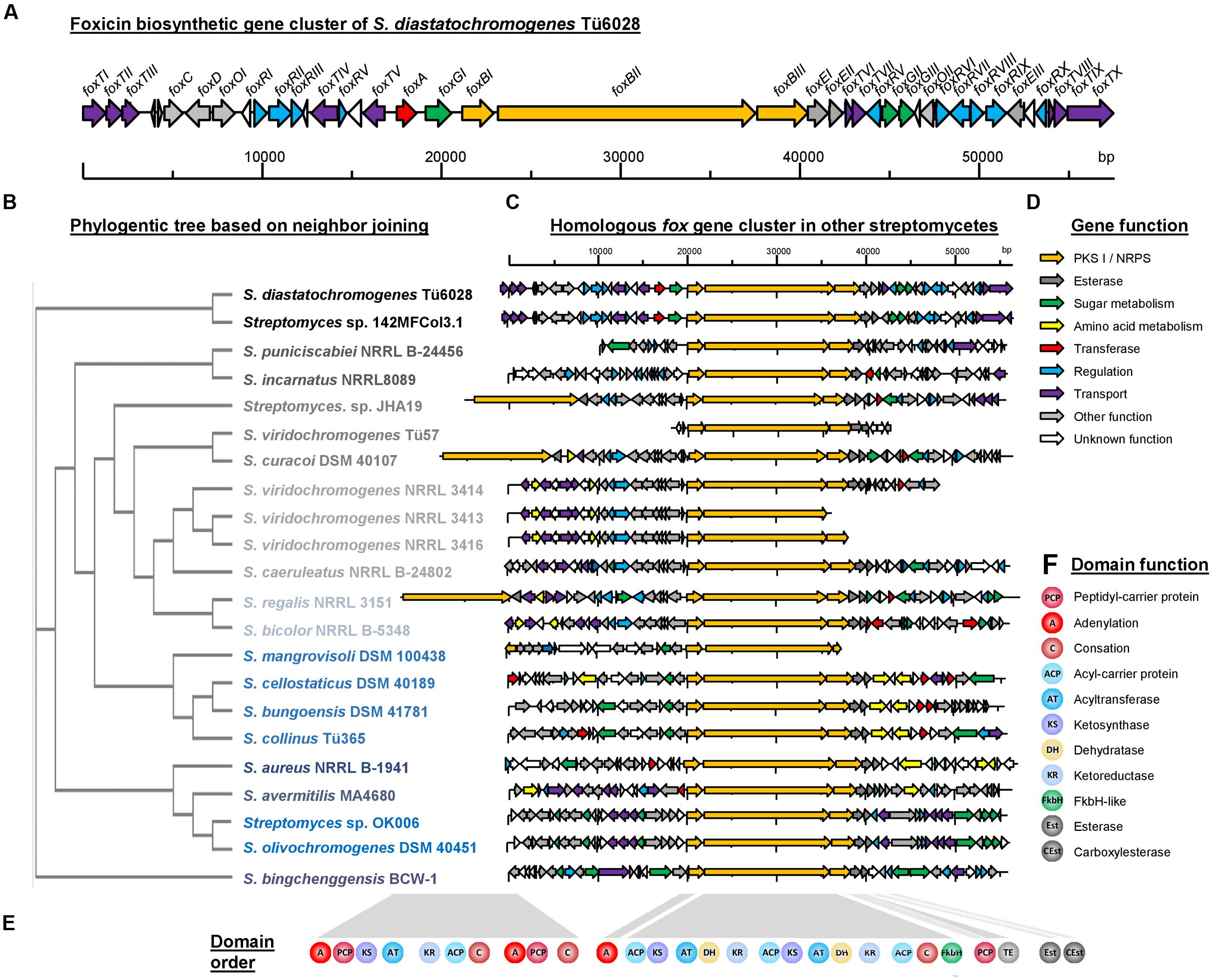
FIGURE 5. Foxicin biosynthetic gene clusters in different Streptomyces strains. (A) Organization of the foxicin biosynthetic gene cluster in S. diastatochromogenes Tü6028. The fox gene cluster spans 57.6 kb and contains 41 ORFs. Genes are indicated by different arrowheads according to their proposed function. (B) Phylogenetic tree based on Neighbor Joining without distance corrections of sequence comparison of foxBII, (C) homologous fox gene clusters in other Streptomyces strains, (D) predicted gene function and (E) domain assembly line of biosynthesis genes and (F) predicted domain function.
Characterization of the Deduced Amino Acid Sequences and Putative Model of the Biosynthesis of Foxicins
Based on bioinformatics analysis of the fox gene cluster of S. diastatochromogenes Tü6028, a putative biosynthetic pathway for foxicins’ biosynthesis is proposed. The 14.4 kb gene foxBII encodes a protein with predicted modular type I polyketide synthase function, with one loading module, two extender modules, a FkbH-like domain, and one condensation domain (C) of a NRPS. The loading module of FoxBII contains only one ACP. Based on the structure of foxicin A we propose isobutyryl-CoA as being the respective starter unit. Module I and II consist of a ketosynthase (KS), acyltransferase (AT), dehydratase (DH), ketoreductase (KR) and an ACP domain, respectively. Both ATs show specificity for methylmalonyl-CoA as predicted using antiSMASH (Weber et al., 2015a), which is in line with the structure of foxicin A.
Foxicin A possesses a double bond between the starter unit and the first extender unit in β,γ-position. Similar structural elements are known from ansamitocin-, rhizoxin-, bacillaen and corallopyronin. It has been shown that special dehydratases or additional shift modules are responsible for the double bond shift in β,γ-position during the biosynthesis of these molecules (Taft et al., 2009; Kusebauch et al., 2010; Moldenhauer et al., 2010; Lohr et al., 2013). In some DH domains, the conserved motif is mutated. The DH domain of module I of the fox cluster shows in contrast to other DH domains a motif of HxxxGxxxxS instead of the conserved HxxxGxxxxP motif, indicating that this domain could introduce the β,γ-double bond during foxicins’ biosynthesis.
The FkbH-like domain is likely to incorporate a glyceryl moiety (Chan et al., 2006; Dorrestein et al., 2006; Sun et al., 2008). The three genes foxGI, foxGII and foxGIII, also located in the fox biosynthetic gene cluster, encode for enzymes known to be involved in sugar metabolism. The proposed functions of the enzymes are glyceraldehyde-3-phosphate dehydrogenase, sugar isomerase and aldolase. Most likely the three of them provide bisphosphoglycerate, which is transferred onto the carrier protein of FoxBIII.
The adenylation domain (A) might be encoded by foxBI, which is separated from foxBIII by the PKS gene foxBII. In silico analysis did not indicate an A domain specificity, but based on the structure of foxicin A, a non-proteinogenic amino acid with two amide groups similar to 2,4-diamino-3-oxobutanoic acid might play a role. This moiety is then most likely linked to the glyceryl-CP.
We propose that the biosynthesis starts with the PKS I of FoxBII and the generated polyketide chain is then transferred onto the amino acid bound to the glyceryl-CP of FoxBIII. Finally, the molecule is cleaved from the enzyme by the thioesterase domain (TE) of FoxBIII, followed by the ring formation. The responsible enzyme for the ring formation is unknown, but with BLAST analysis we identified candidate genes as foxEI or foxEII, with proposed esterase activity. N-acetylation catalyzed by FoxA results ultimately in foxicin A (Figure 6). After cleavage from the enzyme complex foxicin A gets further modified.
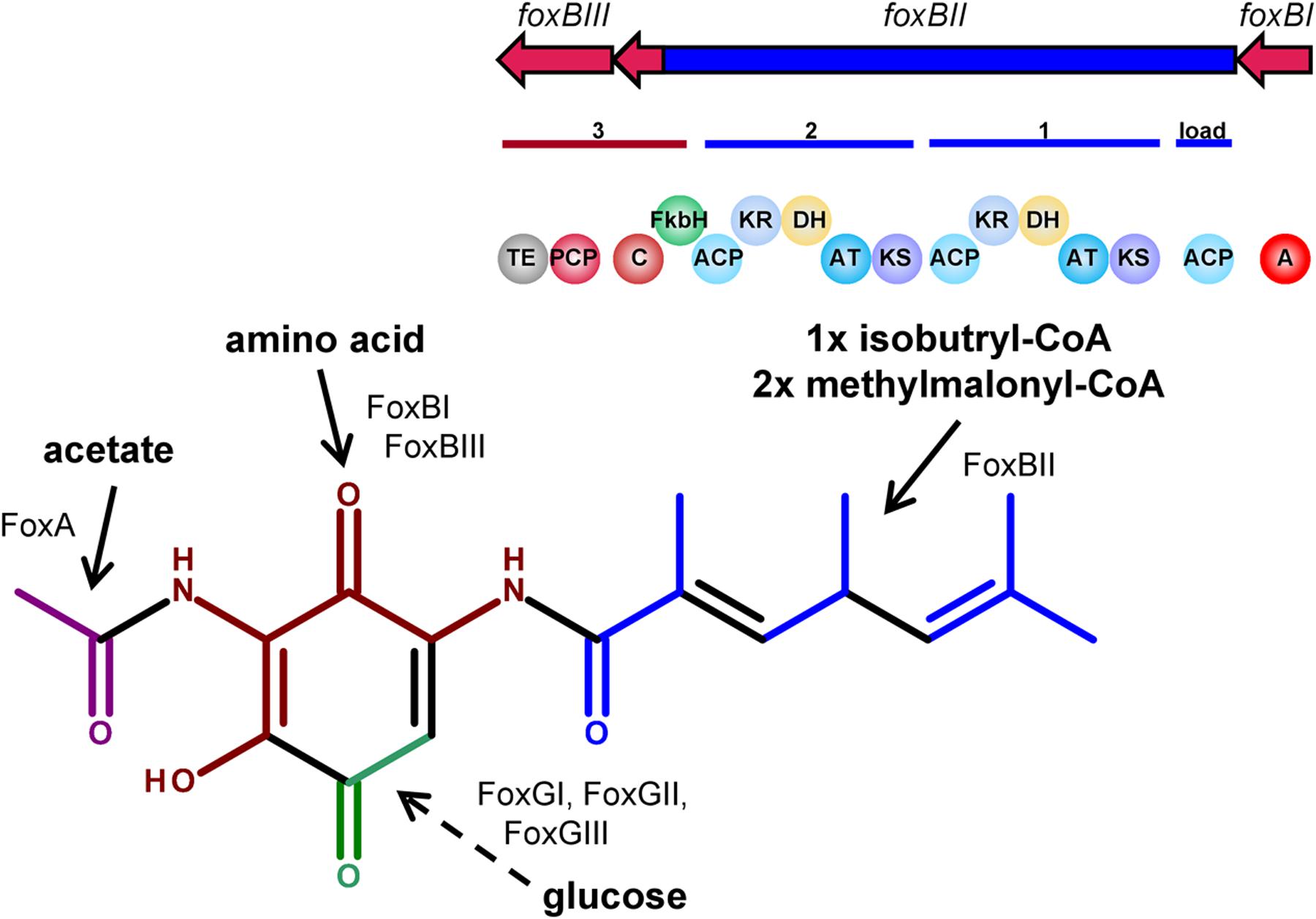
FIGURE 6. Proposed biosynthetic pathway of foxicin. FoxBII is proposed to be involved in the formation of the polyketide side chain (blue), FoxBI and FoxBIII in the incorporation of an amino acid (dark red) and FoxGI-III with FkbH domain of FoxBII in the incorporation of a C2 sugar moiety (green). N-acetylation might be catalyzed by FoxA (magenta).
At each end of the fox gene cluster two ABC transporter genes are located (foxTI-TIII and foxTVIII-X). In addition, foxTIV encodes a transmembrane efflux protein, foxTV an integral membrane transport protein, foxTVI a transporter belonging to the EmrB/QacA family and foxTVII, an export protein. The biosynthetic gene cluster of foxicins also includes 10 regulatory proteins (FoxRI-FoxRX), indicating a complex regulation of the outlined biosynthesis.
Similar fox Biosynthetic Gene Clusters in Various Streptomyces Strains
Similar fox gene clusters were identified in the genomes of 21 additional Streptomyces strains (Figures 5C,D): S. aureus NRRL B-1941 (Doroghazi et al., 2014), S. avermitilis MA4680 (Ikeda et al., 2003), S. bicolor NRRL B-5348 (Doroghazi et al., 2014), S. bingchenggensis BCW-1 (Wang et al., 2010), S. bungoensis DSM 41781, S. caeruleatus NRRL B-24802, S. cellostaticus DSM 40189, S. collinus Tü365 (Rückert et al., 2013), S. curacoi DSM 40107, S. incarnatus NRRL 8089 (Oshima et al., 2015), S. mangrovisoli DSM 100438, S. olivochromogenes DSM 40451, S. puniciscabiei NRRL B-24456, S. regalis NRRL 3151, S. viridochromogenes NRRL 3414, NRRL 3416 and NRRL_3413, S. viridochromogenes Tü57 (Grüning et al., 2013), Streptomyces sp. 142MFCol3.1, Streptomyces sp. JHA19 (Matsunaga et al., 2015) and Streptomyces sp. OK006 (Klingeman et al., 2015). In these strains homologs of foxBI, foxBII, foxBIII, foxEI, and foxEII are located next to each other with high sequence identities of 73% up to 96% (Supplementary Table S4). The closest homologous cluster is located in Streptomyces sp. 142MFCol3.1. A phylogenetic tree based on foxBII sequence analysis is shown in Figure 5B. In all strains the NRPS/PKS I hydride enzyme complex (foxBI – foxBIII) consists of the same series of catalytic domains ([A] – [ACP-KS-AT-DH-KR-ACP-KS-AT-DH-KR-ACP-C-FkbH] – [PCP-TE]) (Figures 5E,F). The strains Streptomyces sp. JHA19, S. curacoi and S. regalis have an additional PKS/NRPS hybrid gene upstream of foxBI-III with [A-PCP-KS-AT-KR-ACP-C-A-PCP-C] catalytic domains. The phylogenetic tree that is based on foxBII illustrates that the biosynthetic gene clusters of these strains do not originate from one clade.
In most of the strains foxEI and foxEII reside adjacent to foxBI-foxBIII indicating that they might play an important role in the biosynthesis of these secondary metabolites. Additionally, in most of the clusters genes from sugar and amino acid metabolism, as well as methyl-/acetyl transferases were identified.
The described genes in the predicted fox clusters do not agree in detail, therefore we assume that the corresponding compounds might exhibit structural differences. Considering the discrete array of catalytic domains, however, we expect similar polyketide tails in all foxicins A–D-like substances. To the best of our knowledge, none of the mentioned strains synthesizes a compound related to foxicins. The responsible gene clusters seem to be silent or have, until now, not been studied. In Supplementary Table S4, an overview of the described Streptomyces strains is shown, as well as their known secondary metabolites and the percentage identity of the identified genes with foxBI, foxBII, foxBIII, foxEI, and foxEII.
Activity of Foxicin A
In order to understand the function of foxicin A we attempted to characterize this molecule in greater detail. By means of LC-MS analysis we could show that Foxicin A production in S. diastatochromogenes is inhibited by ferric ions supplemented to the growth medium. The supplementation of either 1 mM FeCl3 or FeSO4 to the production medium led to a more than 75-fold decrease in foxicin A formation in S. diastatochromogenes ΔpokOIV (Figure 7A). The addition of FeCl3 to foxicin A resulted in a shift of the UV/vis maximum at 320–350 nm (Figure 7B). Using the CAS assay (Schwyn and Neilands, 1987), we observed a color change from blue to violet (Figure 7C), indicating that foxicin A acts as a siderophore.
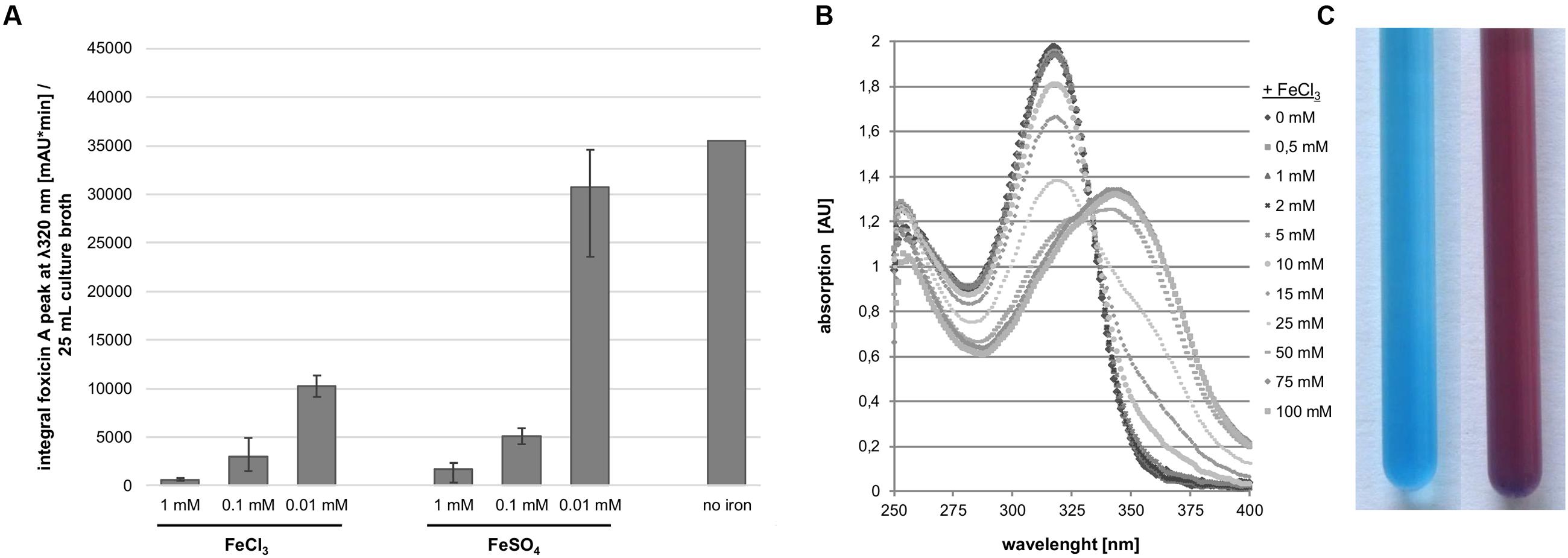
FIGURE 7. Interaction of foxicin with ferric ions. (A) Foxicin production with supplementation of different amounts of ferric ion, calculated integral of the foxicin A peak at 320 nm/25 mL culture; (B) Shift of UV/vis spectrum of foxicin A in presence of FeCl3; (C) CAS-assay of foxicin A, color exchange from blue (left) to violet (right).
As foxicin A contains a quinone moiety similar to molecules involved in various redox processes within the cell (Brunmark and Cadenas, 1989), the compound was tested for its property as an electron acceptor or inhibitor of the respiratory chains. Foxicin A was added to E. coli cytoplasmic membranes and the respiratory chain activity was measured with a Clark-type electrode by starting the reaction with NADH as electron donor. 500 μM foxicin A blocked O2 consumption to 30% indicating its role as a weak inhibitor of the electron transfer chain (Supplementary Table S5). In contrast to other quinones (Kawamukai, 2002) foxicin A does not protect cells from oxidative stress induced by H2O2 (Supplementary Figure S15).
To identify the antibiotic potential of foxicin A, an inhibition zone test was conducted. Therefore, 100 μg foxicin A were applied to paper disks that were transferred to different culture plates. The compound showed moderate activity against Streptomyces viridochromogenes, Saccharothrix espanaensis and the cyanobacterial strains Synechococcus sp. and Synechocystis sp. The tested amount of foxicin A did not visibly influence the growth of the bacterial strains E. coli XL1-Blue, Bacillus subtilis, Mycobacterium smegmatis, the fungal strains Candida parapsilosis and Fusarium verticillioides (Supplementary Table S6) and the plant Arabidopsis thaliana. To determine the effect of foxicin A on cell viability of human cells, leukemia cell lines CCRF-CEM, CEM-ADR5000 and Jurkat cells as well as non-cancer PBMCs were stimulated for 24 h with foxicin A at a concentration range of 0.6–80 μM. However, foxicin A showed no significant effect on cell viability, as measured by using the MTT assay (Supplementary Figure S16).
Discussion
As a result of the deletion of the structural gene pokOIV of polyketomycin biosynthesis, we identified novel secondary metabolites in S. diastatochromogenes Tü6028, named foxicins. In the wild type strain, the gene cluster is expressed only at low levels. In contrast, the ΔpokOIV mutant produces higher amounts of the foxicin derivatives A–D, enabling further investigations of these fascinating compounds. Noteworthy, the mutant strain produces less spores and melanin, indicating a crucial role of polyketomycin for the differentiation and stress response.
The structure of foxicin A could not be completely elucidated by NMR analysis. Eventually, the combination of NMR, IR, and VCD spectra analysis led to the solution of the structure of foxicin A. Foxicin A shows several unusual features. It has a para-quinone moiety, with two amide groups on each site, one is further acetylated and a short fatty acid side chain with non-conjugated double bonds.
The structure of the compound and the encoding genes in the fox cluster suggest a novel biosynthetic pathway. We conclude that foxicins are hybrid compounds with structural elements most probably derived from an amino acid, a C2-moiety derived from sugar metabolism, a polyketide chain of isobutyryl-CoA and methylmalonyl-CoA, and N-acetylation. Although aminoquinones are common structural elements in natural products, none of those that are known is similar to foxicin. Compounds such as N-(3-carboxylpropyl)-5-amino-2-hydroxy-3-tridecyl-1,4-benzoquinone from plant roots of Embelia ribes (Lin et al., 2006), nakijiquinones A-I from marine sponges (Shigemori et al., 1994; Kobayashi et al., 1995; Takahashi et al., 2008, 2009), actinomycin of different Streptomyces strains (Waksman and Gregory, 1954) and the large group of ansamycins (Brufani et al., 1973; Oppolzer and Prelog, 1973; White et al., 1973; Rinehart and Shield, 1976) and mitomycin/porfiromycin (Webb et al., 1962), comprise a 3-amino-1,4-benzoquinone moiety. In few cases, aminoquinones are substituted with an additional amino group such as the Streptomyces products abenquines A–D (Schulz et al., 2011), the antitumor compounds streptonigrin (Rao and Cullen, 1959–1960) and lavendamycin (Doyle et al., 1981), or the fungal pigments lepiotaquinone (Spiteller et al., 2003) or lilacinone (Aulinger et al., 2000) and aminoglycoside antibiotics such as streptomycin. For many of these products, it was shown that the aminoquinone moiety was built by either amination of glucose-6-phosphate (Llewellyn and Spencer, 2006) or the aminoshikimate pathway (Floss et al., 2011).
Quinone moieties are commonly found in natural products. One major group are the ubiquinones, for example, being important electron carriers in respiration and photosynthesis. Furthermore, they are involved in all kinds of redox reactions and play a crucial role as antioxidants (Kawamukai, 2002). Their long isoprenoid chains lead to the ability to penetrate biological membranes. The short polyketide tail of foxicin A indicates that the compound is not located in the membrane. Idebenone, a synthetic quinone with similarities to ubiquinones, but with a much shorter, less lipophilic tail is predominantly active in the cytoplasm and not in cellular membranes. It is a potent antioxidant, prevents lipid peroxidation and protects against ROS-induced damage in multiple systems (Suno and Nagaoka, 1984; Sugiyama et al., 1985; Rauchová et al., 2006). Foxicin A has even a shorter chain than idebenone. The presence of foxicin A in the culture medium supports the assumption that foxicin A activity is not associated with membrane binding.
Like other quinones, foxicin A is able to accept electrons, but does not act as an antioxidant. In vitro, foxicin A inhibits respiratory function in E. coli membranes, without affecting the in vivo viability of the strain. Foxicin A obstructs the growth of other actinomycetes strains, as well as cyanobacterial species. In contrast, the molecule does not visibly influence the growth of Arabidopsis thaliana. Therefore, it may also interact with the photosynthesis machinery, but cannot pass through all types of cell walls. Further studies are needed to support this hypothesis.
The main function of the foxicins is most likely explained by their ability to act as a siderophore. Surprisingly, foxicin A does not inhibit the viability of human cells, even though it is able to bind ions from the medium. Ions are essential for all organisms. The lack of ions, especially ferric ions, often limits the growth of bacteria in their natural habitat. Therefore, siderophores are vital molecules that are released into the medium, and, after scavenging ions, are actively transported back into the cell. The strain S. diastatochromogenes ΔpokOIV produces less foxicins in the presence of ferric ions in the production media. Foxicin A interacts directly with iron as shown by the CAS assay and a shift of the absorbance maximum in the UV/vis spectrum.
Investigations on siderophores are nowadays in the focus of many research groups, in order to obtain new antibacterial compounds, by employing the ‘trojan horse’ strategy. The outer membrane is an important barrier of Gram-negative bacteria, as well as of mycobacteria. Diarra et al. (1996) and Möllmann et al. (2009) have intriguingly shown that the linkage of an antibiotic to a siderophore can lead to facilitated transport via specific transporters into the cell with subsequent death of the pathogens.
The siderophore yersiniabactin has been found in the plague bacterium Yersinia (Pelludat et al., 1998) and in other bacteria such as the nematode symbiont Photorhabdus luminescens (Duchaud et al., 2003), the plant pathogen Pseudomonas syringae, pathogenic strains of E. coli (Bultreys et al., 2006) and even in the Gram-positive marine bacterium Salinispora tropica (Udwary et al., 2007). The ability to synthesize siderophores gives special benefits to a strain and the evolutionary driving force to keep the biosynthetic gene clusters. The foxicin cluster was identified in more than twenty additional Streptomyces strains, which were isolated at different places around the world, indicating an evolutionary early origin. Until now (11/2016) genome sequences of 844 Streptomyces strains are available on NCBI. This means that the cluster is present in about 2.6% of all Streptomyces strains. It is anticipated that further genome sequencing of many more Streptomyces strains will reveal additional homologous fox biosynthetic gene clusters. Surprisingly, the fox cluster was not detected in any other actinomycetes genus. Although horizontal gene transfer is common in the Streptomyces genus, it is unexpected that the cluster is found in that many other strains. In addition, the organization of the structural genes foxBI, foxBII, foxBIII, foxEI, and foxEII remained the same with high sequence similarity. Therefore, we assume that the respective products should only by slightly different to foxicin A-D. In the three strains comprising of additional PKS/NRPS genes, the product might be more complex, e.g., possessing a second polyketide chain. Many of the investigated strains are known producers of secondary metabolites, but not of compounds similar to foxicins. Therefore, the identified clusters seem to be silent in these strains or the compound is produced only in very little amounts. As a consequence, its presence could have been overlooked in routine natural compound screening, as it had been the case for polyketomycin. The high similarity of the clusters indicates an evolutionary driving force to keep the biosynthetic gene clusters in place and consequently a major role of the compounds for Streptomyces strains.
Author Contributions
AG designed and performed experiments, analyzed data, proposed the biosynthesis model and wrote the paper; AG, IP, and SZ purified foxicin A; DD performed CAS assay; MaM and SL conducted and analyzed VCD and IR measurements; SZ, CJ-T, TP, S-ML, MM, and PB interpreted NMR data, SB performed in vitro respiratory assay; CDF conducted MTT assay; AB, IM, and TF administered the experiments; all authors have given approval to the final version of the manuscript.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (RTG 1976) and China Scholarship Council.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Annegret Wilde and Ekaterina Kuchmina (Insitute of Biology III, University of Freiburg) for providing cyanobacteria strain. Thanks to Daniel Álvarez for Arabidopsis-assay. We thank Volker Brecht for the measurements, concerning elucidation of the structure and Dr. Jürgen Wörth for the determination of the exact mass. We are also grateful to technical support of Monika Weber and Elisabeth Welle. Many thanks to Sascha Ferlaino (Pharmaceutical and Medical Chemistry, University of Freiburg) for NMR measurements. We acknowledge the use of the computing resources provided by the Baden-Wuerttemberg HPC Cluster (bwForCluster) for Computational Chemistry.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00221/full#supplementary-material
Abbreviations
A domain, adenylation domain; ACP, acyl carrier protein; C domain, condensation domain; CAS, chrome azurol S; DFT, density functional theory; DH, dehydratase; EtOAc, ethyl acetate; IR, infrared; KR, ketoreductase; KS, ketosynthase; Mbp, mega base pairs; mMFF, molecular mechanics; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; NRPS, non-ribosomal peptide synthetase; PCP, peptidyl carrier protein; PKS, polyketide synthase; VCD, vibrational circular dichroism; wt, wild type.
Footnotes
References
Aulinger, K., Arnold, N., and Steglich, W. (2000). Metabolites of 2-aminophenol from fruit bodies of Lepiota americana (Agaricales). J. Biosci. 55, 481–484. doi: 10.1515/znc-2000-5-628
Bentley, S. D., Chater, K. F., Cerdeño-Tárraga, A.-M., Challis, G. L., Thomson, N. R., James, K. D., et al. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147. doi: 10.1038/417141a
Bérdy, J. (2012). Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. (Tokyo) 65, 441–441. doi: 10.1038/ja.2012.54
Bierman, M., Logan, R., O’Brien, K., Seno, E. T., Rao, R. N., and Schoner, B. E. (1992). Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116, 43–49. doi: 10.1016/0378-1119(92)90627-2
Bode, H. B., Bethe, B., Höfs, R., and Zeeck, A. (2002). Big effects from small changes: possible ways to explore nature’s chemical diversity. Chembiochem 3, 619–627.
Brufani, M., Kluepfel, D., Lancini, G. C., Leitich, J., Mesentsev, A. S., Prelog, V., et al. (1973). The biogenesis of rifamycin S. Helv. Chim. Acta 56, 2315–2323. doi: 10.1002/hlca.19730560718
Brunmark, A., and Cadenas, E. (1989). Redox and addition chemistry of quinoid compounds and its biological implications. Free Radic. Biol. Med. 7, 435–477. doi: 10.1016/0891-5849(89)90126-3
Bultreys, A., Gheysen, I., and de Hoffmann, E. (2006). Yersiniabactin production by Pseudomonas syringae and Escherichia coli, and description of a second yersiniabactin locus evolutionary group. Appl. Environ. Microbiol. 72, 3814–3825. doi: 10.1128/AEM.00119-06
Calderón, C., De Ford, C., Castro, V., Merfort, I., and Murillo, R. (2014). Cytotoxic clerodane diterpenes from Zuelania guidonia. J. Nat. Prod. 77, 455–463. doi: 10.1021/np400672g
Chan, Y. A., Boyne, M. T., Podevels, A. M., Klimowicz, A. K., Handelsman, J., Kelleher, N. L., et al. (2006). Hydroxymalonyl-acyl carrier protein (ACP) and aminomalonyl-ACP are two additional type I polyketide synthase extender units. Proc. Natl. Acad. Sci. U.S.A. 103, 14349–14354. doi: 10.1073/pnas.0603748103
Covington, C., and Polavarapu, P. (2013). Similarity in dissymmetry factor spectra: a quantitative measure of comparison between experimental and predicted vibrational circular dichroism. J. Phys. Chem. A 117, 3377–3386. doi: 10.1021/jp401079s
Covington, C., and Polavarapu, P. (2014). CDSpecTech: Computer Programs for Calculating Similarity Measures of Comparison between Experimental and Calculated Dissymmetry Factors and Differentials. Available at: https://sites.google.com/site/cdspectech1/
Daum, M., Peintner, I., Linnenbrink, A., Frerich, A., Weber, M., Paululat, T., et al. (2009). Organisation of the biosynthetic gene cluster and tailoring enzymes in the biosynthesis of the tetracyclic quinone glycoside antibiotic polyketomycin. Chembiochem 10, 1073–1083. doi: 10.1002/cbic.200800823
Demain, A. L., and Fang, A. (2000). The natural functions of secondary metabolites. Adv. Biochem. Eng. Biotechnol. 69, 1–39.
Diarra, M. S., Lavoie, M. C., Jacques, M., Darwish, I., Dolence, E. K., Dolence, J. A., et al. (1996). Species selectivity of new siderophore-drug conjugates that use specific iron uptake for entry into bacteria. Antimicrob. Agents Chemother. 40, 2610–2617.
Donadio, S., Sosio, M., Stegmann, E., Weber, T., and Wohlleben, W. (2005). Comparative analysis and insights into the evolution of gene clusters for glycopeptide antibiotic biosynthesis. Mol. Genet. Genomics 274, 40–50. doi: 10.1007/s00438-005-1156-3
Doroghazi, J. R., Albright, J. C., Goering, A. W., Ju, K.-S., Haines, R. R., Tchalukov, K. A., et al. (2014). A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 10, 963–968. doi: 10.1038/nchembio.1659
Dorrestein, P. C., Van Lanen, S. G., Li, W., Zhao, C., Deng, Z., Shen, B., et al. (2006). The bifunctional glyceryl transferase/phosphatase OzmB belonging to the HAD superfamily that diverts 1,3-bisphosphoglycerate into polyketide biosynthesis. J. Am. Chem. Soc. 128, 10386–10387. doi: 10.1021/ja0639362
Doyle, T. W., Balitz, D. M., Grulich, R. E., Nettleton, D. E., Gould, S. J., Tann, C., et al. (1981). Structure determination of lavendamycin- a new antitumor antibiotic from Streptomyces lavendulae. Tetrahedron Lett. 22, 4595–4598. doi: 10.1016/S0040-4039(01)82990-7
Duchaud, E., Rusniok, C., Frangeul, L., Buchrieser, C., Givaudan, A., Taourit, S., et al. (2003). The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21, 1307–1313. doi: 10.1038/nbt886
Egan, S., Wiener, P., Kallifidas, D., and Wellington, E. M. (2001). Phylogeny of Streptomyces species and evidence for horizontal transfer of entire and partial antibiotic gene clusters. Antonie Van Leeuwenhoek 79, 127–133. doi: 10.1023/A:1010296220929
Firn, R. D., and Jones, C. G. (2000). The evolution of secondary metabolism - a unifying model. Mol. Microbiol. 37, 989–994. doi: 10.1046/j.1365-2958.2000.02098.x
Fischbach, M. A., Walsh, C. T., and Clardy, J. (2008). The evolution of gene collectives: how natural selection drives chemical innovation. Proc. Natl. Acad. Sci. U.S.A. 105, 4601–4608. doi: 10.1073/pnas.0709132105
Flett, F., Mersinias, V., and Smith, C. P. (1997). High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155, 223–229. doi: 10.1016/S0378-1097(97)00392-3
Floss, H., Yu, T., and Arakawa, K. (2011). The biosynthesis of 3-amino-5-hydroxybenzoic acid (AHBA), the precursor of mC7N units in ansamycin and mitomycin antibiotics: a review. J. Antibiot. (Tokyo) 64, 35–44. doi: 10.1038/ja.2010.139
Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., et al. (2013). Gaussian 09, R-D.01. Wallingford, CT: Gaussian Inc.
Gessner, A., Heitzler, T., Zhang, S., Klaus, C., Murillo, R., Zhao, H., et al. (2015). Changing biosynthetic profiles by expressing bldA in Streptomyces strains. Chembiochem 16, 2244–2252. doi: 10.1002/cbic.201500297
Gomez-Escribano, J. P., and Bibb, M. J. (2014). Heterologous expression of natural product biosynthetic gene clusters in Streptomyces coelicolor: from genome mining to manipulation of biosynthetic pathways. J. Ind. Microbiol. Biotechnol. 41, 425–431. doi: 10.1007/s10295-013-1348-5
Grüning, B. A., Erxleben, A., Hähnlein, A., and Günther, S. (2013). Draft genome sequence of Streptomyces viridochromogenes strain Tü57, producer of avilamycin. Genome Announc. 1:e00384-13. doi: 10.1128/genomeA.00384-13
Hütter, R. (1962). Zur Systematik der Actinomyceten. Arch. Mikrobiol. 43, 23–49. doi: 10.1007/BF00408394
Ikeda, H., Ishikawa, J., Hanamoto, A., Shinose, M., Kikuchi, H., Shiba, T., et al. (2003). Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21, 526–531. doi: 10.1038/nbt820
Intra, B., Matsumoto, A., Inahashi, Y., Omura, S., Takahashi, Y., and Panbangred, W. (2013). Actinokineospora bangkokensis sp. nov., isolated from rhizospheric soil. Int. J. Syst. Evol. Microbiol. 63, 2655–2660. doi: 10.1099/ijs.0.047928-0
Kawamukai, M. (2002). Biosynthesis, bioproduction and novel roles of ubiquinone. J. Biosci. Bioeng. 94, 511–517. doi: 10.1016/S1389-1723(02)80188-8
Klingeman, D. M., Utturkar, S., Lu, T.-Y. S., Schadt, C. W., Pelletier, D. A., and Brown, S. D. (2015). Draft genome sequences of four Streptomyces isolates from the Populus trichocarpa root endosphere and rhizosphere. Genome Announc. 3:e1344-15. doi: 10.1128/genomeA.01344-15
Kobayashi, J., Madono, T., and Shigemori, H. (1995). Nakijiquinones C and D, new sesquiterpenoid quinones with a hydroxy amino acid residue from a marine sponge inhibiting c-erbB-2 kinase. Tetrahedron 51, 10867–10874. doi: 10.1016/0040-4020(95)00661-Q
Kong, F., Zhao, N., Siegel, M. M., Janota, K., Ashcroft, J. S., Koehn, F. E., et al. (1998). Saccharomicins, novel heptadecaglycoside antibiotics effective against multidrug-resistant bacteria. J. Am. Chem. Soc. 120, 13301–13311. doi: 10.1021/ja981641l
Kusebauch, B., Busch, B., Scherlach, K., Roth, M., and Hertweck, C. (2010). Functionally distinct modules operate two consecutive α,β→β,γ double-bond shifts in the rhizoxin polyketide assembly line. Angew. Chemie 122, 1502–1506. doi: 10.1002/ange.200905467
Lin, P., Li, S., Wang, S., Yang, Y., and Shi, J. (2006). A nitrogen-containing 3-alkyl-1,4-benzoquinone and a gomphilactone derivative from Embelia ribes. J. Nat. Prod. 69, 1629–1632. doi: 10.1021/np060284m
Llewellyn, N. M., and Spencer, J. B. (2006). Biosynthesis of 2-deoxystreptamine-containing aminoglycoside antibiotics. Nat. Prod. Rep. 23, 864–874. doi: 10.1039/b604709m
Lohr, F., Jenniches, I., Frizler, M., Meehan, M. J., Sylvester, M., Schmitz, A., et al. (2013). α,β → β,γ double bond migration in corallopyronin A biosynthesis. Chem. Sci. 4:4175. doi: 10.1039/c3sc51854j
MacNeil, D. J., Gewain, K. M., Ruby, C. L., Dezeny, G., Gibbons, P. H., and MacNeil, T. (1992). Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111, 61–68. doi: 10.1016/0378-1119(92)90603-M
Makitrynskyy, R., Ostash, B., Tsypik, O., Rebets, Y., Doud, E., Meredith, T., et al. (2013). Pleiotropic regulatory genes bldA, adpA and absB are implicated in production of phosphoglycolipid antibiotic moenomycin. Open Biol. 3:130121. doi: 10.1098/rsob.130121
Matsunaga, E., Higuchi, Y., Mori, K., Tashiro, K., Kuhara, S., and Takegawa, K. (2015). Draft genome sequence of Streptomyces sp. JHA19, a strain that possesses β-D-galactofuranosidase activity. Genome Announc. 3:e1171-15. doi: 10.1128/genomeA.01171-15
Metsä-Ketelä, M., Halo, L., Munukka, E., Hakala, J., Mäntsälä, P., and Ylihonko, K. (2002). Molecular evolution of aromatic polyketides and comparative sequence analysis of polyketide ketosynthase and 16S ribosomal DNA genes from various streptomyces species. Appl. Environ. Microbiol. 68, 4472–4479. doi: 10.1128/AEM.68.9.4472-4479.2002
Moldenhauer, J., Götz, D. C. G., Albert, C. R., Bischof, S. K., Schneider, K., Süssmuth, R. D., et al. (2010). The final steps of bacillaene biosynthesis in Bacillus amyloliquefaciens FZB42: direct evidence for β,γ dehydration by a trans-acyltransferase polyketide synthase. Angew. Chem. Int. Ed. Engl. 49, 1465–1467. doi: 10.1002/anie.200905468
Möllmann, U., Heinisch, L., Bauernfeind, A., Köhler, T., and Ankel-Fuchs, D. (2009). Siderophores as drug delivery agents: application of the “Trojan Horse” strategy. Biometals 22, 615–624. doi: 10.1007/s10534-009-9219-2
Oppolzer, W., and Prelog, V. (1973). The constitution and configuration of rifamycins B, O, S and SV. Helv. Chim. Acta 56, 2287–2314. doi: 10.1002/hlca.19730560717
Oshima, K., Hattori, M., Shimizu, H., Fukuda, K., Nemoto, M., Inagaki, K., et al. (2015). Draft genome sequence of Streptomyces incarnatus NRRL8089, which produces the nucleoside antibiotic sinefungin. Genome Announc. 3:e00715-15. doi: 10.1128/genomeA.00715-15
Paululat, T., Zeeck, A., Gutterer, J. M., and Fiedler, H. P. (1999). Biosynthesis of polyketomycin produced by Streptomyces diastatochromogenes Tü6028. J. Antibiot. (Tokyo) 52, 96–101. doi: 10.7164/antibiotics.52.96
Pelludat, C., Rakin, A., Jacobi, C. A., Schubert, S., and Heesemann, J. (1998). The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J. Bacteriol. 180, 538–546.
Rao, K. V., and Cullen, W. P. (1959–1960). Streptonigrin, an antitumor substance. I. Isolation and characterization. Antibiot. Annu. 7, 950–953.
Rauchová, H., Vrbackı, M., Bergamini, C., Fato, R., Lenaz, G., Houstek, J., et al. (2006). Inhibition of glycerophosphate-dependent H2O2 generation in brown fat mitochondria by idebenone. Biochem. Biophys. Res. Commun. 339, 362–366. doi: 10.1016/j.bbrc.2005.11.035
Rinehart, K. L., and Shield, L. S. (1976). Chemistry of the ansamycin antibiotics. Fortschr. Chem. Org. Naturst. 33, 231–307.
Rückert, C., Szczepanowski, R., Albersmeier, A., Goesmann, A., Iftime, D., Musiol, E. M., et al. (2013). Complete genome sequence of the kirromycin producer Streptomyces collinus Tü 365 consisting of a linear chromosome and two linear plasmids. J. Biotechnol. 168, 739–740. doi: 10.1016/j.jbiotec.2013.10.004
Schroeckh, V., Scherlach, K., Nützmann, H.-W., Shelest, E., Schmidt-Heck, W., Schuemann, J., et al. (2009). Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. U.S.A. 106, 14558–14563. doi: 10.1073/pnas.0901870106
Schulz, D., Beese, P., Ohlendorf, B., Erhard, A., Zinecker, H., Dorador, C., et al. (2011). Abenquines A–D: aminoquinone derivatives produced by Streptomyces sp. strain DB634. J. Antibiot. (Tokyo) 64, 763–768. doi: 10.1038/ja.2011.87
Schwarzer, D., Finking, R., and Marahiel, M. A. (2003). Nonribosomal peptides: from genes to products. Nat. Prod. Rep. 20, 275–287. doi: 10.1039/b111145k
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Shigemori, H., Madono, T., Sasaki, T., Mikami, Y., and Kobayashi, J. (1994). Nakijiquinones A and B, new antifungal sesquiterpenoid quinones with an amino acid residue from an Okinawan marine sponge. Tetrahedron 50, 8347–8354. doi: 10.1016/S0040-4020(01)85557-5
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Spiteller, P., Arnold, N., Spiteller, M., and Steglich, W. (2003). Lilacinone, a red aminobenzoquinone pigment from Lactarius lilacinus. J. Nat. Prod. 66, 1402–1403. doi: 10.1021/np0303052
Staunton, J., and Weissman, K. J. (2001). Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 18, 380–416. doi: 10.1039/a909079g
Sugiyama, Y., Fujita, T., Matsumoto, M., Okamoto, K., and Imada, I. (1985). Effects of idebenone (CV-2619) and its metabolites on respiratory activity and lipid peroxidation in brain mitochondria from rats and dogs. J. Pharmacobiodyn. 8, 1006–1017. doi: 10.1248/bpb1978.8.1006
Sun, Y., Hong, H., Gillies, F., Spencer, J. B., and Leadlay, P. F. (2008). Glyceryl-S-acyl carrier protein as an intermediate in the biosynthesis of tetronate antibiotics. Chembiochem 9, 150–156. doi: 10.1002/cbic.200700492
Suno, M., and Nagaoka, A. (1984). Inhibition of lipid peroxidation by a novel compound (CV-2619) in brain mitochondria and mode of action of the inhibition. Biochem. Biophys. Res. Commun. 125, 1046–1052. doi: 10.1016/0006-291X(84)91389-5
Taft, F., Brünjes, M., Knobloch, T., Floss, H. G., and Kirschning, A. (2009). Timing of the Δ10,12-Δ11,13 double bond migration during ansamitocin biosynthesis in Actinosynnema pretiosum. J. Am. Chem. Soc. 131, 3812–3813. doi: 10.1021/ja8088923
Takahashi, Y., Kubota, T., Ito, J., Mikami, Y., Fromont, J., and Kobayashi, J. (2008). Nakijiquinones G-I, new sesquiterpenoid quinones from marine sponge. Bioorg. Med. Chem. 16, 7561–7564. doi: 10.1016/j.bmc.2008.07.028
Takahashi, Y., Kubota, T., and Kobayashi, J. (2009). Nakijiquinones E and F, new dimeric sesquiterpenoid quinones from marine sponge. Bioorg. Med. Chem. 17, 2185–2188. doi: 10.1016/j.bmc.2008.10.080
Udwary, D. W., Zeigler, L., Asolkar, R. N., Singan, V., Lapidus, A., Fenical, W., et al. (2007). Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. U.S.A. 104, 10376–10381. doi: 10.1073/pnas.0700962104
Vaishnav, P., and Demain, A. L. (2011). Unexpected applications of secondary metabolites. Biotechnol. Adv. 29, 223–229. doi: 10.1016/j.biotechadv.2010.11.006
Waksman, S. A., and Gregory, F. J. (1954). Actinomycin. II. Classification of organism producing different forms of actinomycin. Antibiot. Chemother. (Northfield) 4, 1050–1056.
Wang, X.-J., Yan, Y.-J., Zhang, B., An, J., Wang, J.-J., Tian, J., et al. (2010). Genome sequence of the milbemycin-producing bacterium Streptomyces bingchenggensis. J. Bacteriol. 192, 4526–4527. doi: 10.1128/JB.00596-10
Webb, J. S., Cosulich, D. B., Mowat, J. H., Patrick, J. B., Broschard, R. W., Meyer, W. E., et al. (1962). The structures of mitomycins A, B and C and porfiromycin–part I. J. Am. Chem. Soc. 84, 3185–3187. doi: 10.1021/ja00875a032
Weber, T., Blin, K., Duddela, S., Krug, D., Kim, H. U., Bruccoleri, R., et al. (2015a). antiSMASH 3.0 - a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243. doi: 10.1093/nar/gkv437
Weber, T., Charusanti, P., Musiol-Kroll, E. M., Jiang, X., Tong, Y., Kim, H. U., et al. (2015b). Metabolic engineering of antibiotic factories: new tools for antibiotic production in actinomycetes. Trends Biotechnol. 33, 15–26. doi: 10.1016/j.tibtech.2014.10.009
Weiss, R. F. (1970). The solubility of nitrogen, oxygen and argon in water and seawater. Deep Sea Res. Oceanogr. Abstr. 17, 721–735. doi: 10.1016/0011-7471(70)90037-9
Keywords: Streptomyces, natural product, foxicin, biosynthetic gene cluster, evolution, siderophore
Citation: Greule A, Marolt M, Deubel D, Peintner I, Zhang S, Jessen-Trefzer C, De Ford C, Burschel S, Li S-M, Friedrich T, Merfort I, Lüdeke S, Bisel P, Müller M, Paululat T and Bechthold A (2017) Wide Distribution of Foxicin Biosynthetic Gene Clusters in Streptomyces Strains – An Unusual Secondary Metabolite with Various Properties. Front. Microbiol. 8:221. doi: 10.3389/fmicb.2017.00221
Received: 28 November 2016; Accepted: 31 January 2017;
Published: 21 February 2017.
Edited by:
Marina G. Kalyuzhanaya, San Diego State University, USAReviewed by:
Joachim Wink, Helmholtz Centre for Infection Research, GermanyCourtney Stairs, Uppsala University, Sweden
Copyright © 2017 Greule, Marolt, Deubel, Peintner, Zhang, Jessen-Trefzer, De Ford, Burschel, Li, Friedrich, Merfort, Lüdeke, Bisel, Müller, Paululat and Bechthold. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andreas Bechthold, YW5kcmVhcy5iZWNodGhvbGRAcGhhcm1hemllLnVuaS1mcmVpYnVyZy5kZQ==
 Anja Greule
Anja Greule Marija Marolt
Marija Marolt Denise Deubel1
Denise Deubel1 Songya Zhang
Songya Zhang Michael Müller
Michael Müller Andreas Bechthold
Andreas Bechthold