
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 February 2017
Sec. Plant Pathogen Interactions
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.00199
This article is part of the Research Topic Plant-Microbe-Insect Interaction: Source for Bio-fertilizers, Bio-medicines and Agent Research View all 43 articles
Medicinal plants are known to harbor potential endophytic microbes, due to their bioactive compounds. In a first study of ongoing research, endophytic bacteria were isolated from two medicinal plants, Hypericum perforatum and Ziziphora capitata with contrasting antimicrobial activities from the Chatkal Biosphere Reserve of Uzbekistan, and their plant-specific traits involved in biocontrol and plant growth promotion were evaluated. Plant extracts of H. perforatum exhibited a remarkable activity against bacterial and fungal pathogens, whereas extracts of Z. capitata did not exhibit any potential antimicrobial activity. Matrix-assisted laser desorption ionization (MALDI) time-of-flight (TOF) mass spectrometry (MS) was used to identify plant associated culturable endophytic bacteria. The isolated culturable endophytes associated with H. perforatum belong to eight genera (Arthrobacter, Achromobacter, Bacillus, Enterobacter, Erwinia, Pseudomonas, Pantoea, Serratia, and Stenotrophomonas). The endophytic isolates from Z. capitata also contain those genera except Arthrobacter, Serratia, and Stenotrophomonas. H. perforatum with antibacterial activity supported more bacteria with antagonistic activity, as compared to Z. capitata. The antagonistic isolates were able to control tomato root rot caused by Fusarium oxysporum and stimulated plant growth under greenhouse conditions and could thus be a cost-effective source for agro-based biological control agents.
Medicinal plants are traditionally used worldwide as remedies for the treatment of various diseases, including asthma, gastrointestinal symptoms, skin disorders, respiratory and urinary problems, and hepatic and cardiovascular disease (Van Wyk and Wink, 2004; Tian et al., 2014). These plants synthesize a diverse array of biologically active compounds (Bajguz, 2007; Cushnie et al., 2014) that are important for them to survive and flourish in the natural environment, including protective functions with respect to abiotic stresses derived from temperature, water status, mineral nutrient supply and to insect pests (Simmonds, 2003; Treutter, 2006; Vardhini and Anjum, 2015). The composition of biologically active compounds of medicinal plants varies widely depending on the plant species, soil type and on their association with microbes (Zhao et al., 2011; Morsy, 2014). These bioactive secondary metabolites synthesized by medicinal plants can also strongly affect plant-associated microbial communities and their physiological functions (Qi et al., 2012; Philippot et al., 2013; Chaparro et al., 2014; reviewed in Köberl et al., 2013). Moreover, plants rely on their microbiome for specific traits and activities, including growth promotion, nutrient acquisition, induced systemic resistance and tolerance to abiotic stress factors (Egamberdieva et al., 2010, 2011; Malfanova et al., 2011; Sessitsch et al., 2013; Berg et al., 2014). Although a vast number of medicinal plants have been well-studied with respect to their phytochemical constitutes and pharmacological properties, their microbiome and the physiological interactions between host and microbes remain poorly understood (Köberl et al., 2014).
The plant-associated microbiome consists of distinct microbial communities living in the roots, shoots and endosphere (Beneduzi et al., 2012; Berg et al., 2014). The rhizosphere of many plants is well-studied and known to be a potential source for selecting beneficial microbes that can positively affect plant health (Weller et al., 2002; Berendsen et al., 2012; Philippot et al., 2013). Hence, understanding the response of microbial communities to alterations in the physiochemical environment of the rhizosphere may provide valuable insights into the microbial ecology of plant-associated bacteria. Köberl et al. (2013) observed a high abundance of antagonistic bacteria in the rhizosphere of the medicinal plants Matricaria chamomilla, Calendula officinalis, and Solanum distichum. The root-associated bacteria of Ajuga bracteosa exhibited a wide range of plant growth promoting activities by producing siderophores and indole acetic acid and exhibiting antioxidant activity (Kumar et al., 2012). Recently, endophytic microorganisms have been under increased investigation due to their intimate interaction with the host (Hardoim et al., 2015); it is believed that the phytochemical constitutes of plants are related either directly or indirectly to endophytic microbes and their interactions with host plants (Chandra, 2012; Qi et al., 2012). Despite first studies of endophytes in medicinal plants (Bharti et al., 2012; López-Fuentes et al., 2012; Miller et al., 2012; El-Deeb et al., 2013; Egamberdieva and Teixeira da Silva, 2015), the potential of medicinal plants is far from exhausted.
Therefore, the current exploratory study was designed to evaluate whether medicinal plants with contrasting antimicrobial activities have an impact on plant-specific traits involved in biocontrol and plant growth promotion of root-associated culturable endophytic bacteria. In first experiments of ongoing research, we studied Ziziphora capitata L. (Field basil) and Hypericum perforatum L. (St John’s wort) from the Chatkal Biosphere Reserve of Uzbekistan, an isolated protected area in Western Tien Shan province, which significantly surpasses other areas with respect to the absolute number of endemic species (Kogure et al., 2004). Z. capitata L. is a medicinal and aromatic plant of the Lamiaceae family, which is traditionally used for the treatment of various ailments, such as heart disease, inflammation, depression, diarrhea, fever, skin disorders, hepatic diseases, and edema (Sonboli et al., 2006). The Ziziphora species are rich in essential oils, flavanoids and sterols (Zhaparkulova et al., 2015). The major component of essential oil found in several species of Ziziphora is pulegone, which has strong antibacterial and antifungal activity (Sonboli et al., 2006), but Z. capitata does not contain pulegone (Ebrahimi et al., 2009). H. perforatum is a species in the family Hypericaceae and is known for analgesic, sedative, antihelmintic, anti-inflammatory, and antibacterial properties (Dall’Agnol et al., 2003). H. perforatum contains a wide range of biological active compounds, such as essential oils, tannins, flavonoids, xanthones, and hyperforin as an antibiotic substance (Jurgenliemk and Nahrstedt, 2002). The crude extracts of H. perforatum exhibited higher antibacterial activity against Gram-positive than Gram-negative bacteria (Sarkisian et al., 2012). The aim of this study was to isolate and characterize endophytic bacteria from two medicinal plants, H. perforatum and Z. capitata, with contrasting antimicrobial activities and evaluate their plant-specific traits involved in biocontrol and plant growth promotion.
Hypericum perforatum (Hypericaceae) and Ziziphora capitata (Lamiaceae) plants were collected during the summer (June 2013, the plant’s flowering stage) from Chatkal Biosphere Reserve of Uzbekistan, western part of Tien Shan mountain (41°08′ N; 69°59′ E). This biosphere reserve is situated in the Tashkent Region within the Chatkal mountain range (1.110–4.000 m above sea level) of the West Tien-Shan Mountains and is unique for its significant role in biodiversity conservation and ethnobotany. The climate is characterized by average annual temperatures ranging from 20 to 25°C with increased annual precipitation from plains to mountains, reaching 700–800 mm.
The aerial parts of H. perforatum and Z. capitata were dried in the laboratory excluding direct sun light at room temperature for 6–7 days and ground into a fine powder by mortar and pestle. Approximately, 10 g of plant powder was extracted with 50 ml of methanol for 24 h in a dark room temperature. Subsequently, the solvent was evaporated in a rotary vacuum evaporator at 40°C and re-suspended in dimethyl sulfoxide (DMSO). The homogenate was filtered through Whatman No. 1 filter paper, centrifuged at 5000 g for 15 min and sterilized by filtration through 0.22-μm sterile filters (Millipore, Bedford, MA, USA). The filtrates were stored at -4°C and used for in vitro screening of antimicrobial activity.
The extracts were individually tested against the following pathogenic microorganisms: Klebsiella oxytoca 6653, K. pneumoniae 40602, K. aerogenes NCTC 8172, Citrobacter freundii 82073, Staphylococcus aureus MRSA 16, Enterococcus faecalis NCTC 775, Providencia rettgeri NCIMB 9570, Pseudomonas aeruginosa NCTC 6749, Escherichia coli NCTC 9001 and Fusarium solani, Fusarium oxysporum, and Alternaria alternata. Reference strains and clinical isolates were obtained from the Department of Microbiology, Manchester Metropolitan University, UK, and the National Culture Type Collection (NCTC), UK. The fungal strains were obtained from the Department of Microbiology and Biotechnology, National University of Uzbekistan. Each plant extract was dissolved in dimethyl sulfoxide (DMSO), sterilized by filtration using a sintered glass filter, and stored at 4°C. The antimicrobial activity of the extracts was tested using the agar well-diffusion method. Microorganisms were grown overnight at 30°C in Mueller-Hinton Broth (Oxoid, Basingstoke, UK) supplemented with 5% horse blood, and 100 μl of suspension containing 106 CFU ml-1 of bacteria was spread on the surface of Mueller-Hinton agar plates. Wells with 6-mm diameters were cut off and filled with 50 μL of each extract (10 mg ml-1). Ampicillin (Sigma-Aldrich, Steinheim, Germany) (0.5 mg ml-1), nystatin (Sigma-Aldrich, Steinheim, Germany) (1 mg ml-1) and DMSO were used as controls. Fungal strains were grown on potato dextrose agar plates (PDA; Difco Laboratories, Detroit, MI, USA) at 28°C for 5 days. Small piece of fungal culture were placed in the middle of Petri plates. Each antimicrobial assay was performed in triplicate. The plates were incubated at an appropriate growth temperature for 2 days for bacterial strains (37°C) and 4 days for fungal strains (30°C). The assessment of antimicrobial activity was based on the measurement of inhibition zones on the agar surface around the well.
Three plants from each species of H. perforatum and Z. capitata including roots (20–30 cm depth) were randomly collected about 1 m apart from each other from an area of 100 m2 in the Chatkal Biosphere Reserve. The whole plants, along with root systems, were wrapped in plastic bags, and brought to the laboratory on same day and immediately stored at 4°C. The isolation of bacterial strains was carried out on the next day to minimize storage effects.
The root systems of the collected plants were separated from the shoots, soil adhering to the roots was removed and roots were carefully washed under running water, taking care to minimize root injury. Three plants of each species were used to determine the number of bacterial colonies cultured from the root tissue. For the bacterial isolation, root tissues were pooled from each of three replicate plants. The roots were surface sterilized by immersion in 70% (v/v) ethanol, following by shaking in 5% (w/v) sodium hypochlorite solution for 5 min. Subsequently, the roots were rinsed in sterile distilled water six times. To test the efficiency of sterilization, the sterile roots were incubated in TSA medium for 2 days at 28°C, and no infestation was observed.
Sterilized roots were weighed aseptically (1 g) and macerated in a mortar employing phosphate buffered saline (PBS) (20 mM sodium phosphate, 150 mM NaCl, pH 7.4) in a laminar air flow cabinet. The extracts were placed in a tube containing 9 ml sterile PBS and shaken with a vortex for 1 min. The supernatant was collected and serially diluted (101–105) in PBS, and 100 μl from appropriate dilutions were spread on Tryptic Soy Agar (TSA, Difco Laboratories, Detroit, MI, USA) plates in triplicate. The plates were incubated at 28°C, and colony forming units (cfu) g-1 root tissue were determined on the third day. A representative number of colonies that exhibited differentiable colony morphologies were picked from plates and were re-streaked for the purification of the isolates. The pure bacterial cultures were preserved on plates at 4°C for the further analyses. In addition all bacterial isolates were stored in Tryptic Soy broth (TSB) (Difco) with 30% glycerol at -80°C.
The identification of bacterial isolates was performed by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) as described previously (Egamberdieva et al., 2016). The sample preparation was performed according to the ethanol/formic acid extraction protocol recommended by Bruker Daltonics (Bremen, Germany) and was described in Egamberdieva et al. (2016). Briefly, the isolates were cultured on TSA medium (Difco Laboratories, Detroit, MI, USA) for 24 h, and approximately 10 mg of cell mass was suspended in 300 μL water (LC–MS CHOMASOLV®; Honeywell) and vortexed to generate a homogenous suspension. The suspension was mixed with 900 μL ethanol (≥99.8% GC; Sigma-Aldrich) and centrifuged. The pellet was resuspended in 50 μL 70% formic acid (v/v) and subsequently carefully mixed with 50 μL acetonitrile. After centrifugation, aliquots of 1 μL supernatant were placed immediately on spots of a MALDI target. Each spot was allowed to dry and subsequently overlaid with 1 μL of matrix (α-ciano-4-hydroxycinnamic acid in 50% aqueous acetonitrile containing 2.5% trifluoroacetic acid). Mass spectra were acquired using a MALDI-TOF MS spectrometer in a linear positive mode (MicroflexTMLT, Bruker Daltonics, Bremen, Germany) in a mass range of 2–20 kDa. A bacterial test standard (BTS, Bruker Daltonics, Bremen, Germany) was used for instrument calibration. The raw spectra were imported into the MALDI BiotyperTM software and then processed and analyzed using standard pattern matching against the reference spectra in the MALDI BiotyperTM reference database (version 3.0, Bruker Daltonics, Bremen, Germany). A calculated matching score (score value) provided a measure of the probability of a correct classification.
The production of IAA (indole 3-acetic acid) was determined as described by Bano and Musarrat (2003). The IAA concentration in culture was calculated using a calibration curve of pure IAA as a standard. The cellulose-degrading ability of bacterial isolates was analyzed by streaking inocula on cellulose (Sigma-Aldrich, St. Louis, MO, USA) Congo-Red agar media as described by Pratima et al. (2012). Furthermore, β-1,3 glucanase activity was tested using the substrate lichenan (Sigma-Aldrich, St. Louis, MO, USA) in top agar plates (Walsh et al., 1995), and protease activity was determined by using 5% skimmed milk agar plates (Brown and Foster, 1970). The production of HCN by bacterial isolates was measured using the protocol described by Castric (1975).
The bacterial isolates were tested in vitro for their antagonistic activities against the following pathogenic fungi: Fusarium oxysporum f.sp. radicis-lycopersici (Forl), F. solani, F. culmorum, Gaeumannomyces graminis pv. tritici (Ggt), Alternaria alternata, and Botrytis cinerea and the oomycete Pythium ultimum. The bacterial isolates were grown in TSB broth for 3 days, and 50-μl bacterial cultures were dropped into a hole of PDA plates (4 mm in diameter). Fungal strains for inoculation were grown in peptone dextrose agar (PDA) plates at 28°C for 5 days. Disks of fresh cultures of the fungus (5 mm diameter) were cut out and placed 2 cm away from the hole filled with bacterial filtrate. The plates were sealed with Parafilm®M and incubated at 28°C in darkness until the fungi had grown over the control plates without bacteria. Antifungal activity was recorded as the width of the zone of growth inhibition between the fungi and the bacteria tested.
Bacterial isolates with antagonistic activity against the majority of tested fungal pathogens, were tested for their ability to control tomato root rot caused by F. oxysporum f.sp. radicis-lycopersici. For the inoculation of soil, F. oxysporum was grown in PDA plates for 5 days. Small pieces of agar from the growing edge of the colony were homogenized and used to inoculate 300 ml of Chapek-Dox medium, which was kept under aeration (110 rpm) at 28°C. After 3 days, the spore suspension was filtrated with sterile glass wool to remove the mycelium. The concentration of spores in the inoculum was adjusted to 107 spores ml-1 by microscopic enumeration with a cell-counting haemocytometer and mixed thoroughly with potting soil to obtain a concentration of approximately 107 spores kg-1 soil. The tomato seeds of the cultivar Fuji Pink (Sakata, Japan) were sterilized by stirring with 70% ethanol for 5 min and in household bleach (adjusted to approximately 5% sodium hypochlorite) for 3 min. Subsequently, the seeds were washed several times with sterile distilled water. After germination in sterile Petri plates, the seeds were placed in a bacterial suspension of 1 × 108 CFU ml-1 prepared as described above and shaken gently for 10 min. The inoculated seeds were sown in plastic pots, and each treatment contained four groups of 24 plants. The plants were grown in a growth chamber under controlled conditions (16 h light, 8 h dark), at temperature light 28°C, dark 20°C and relative humidity 60%. After 3 weeks, the plants were removed from the soil, washed and examined for foot and root rot symptoms as indicated by browning and lesions. Roots without any disease symptoms were classified as healthy.
To test whether bacterial isolates were capable of stimulating plant growth, a pot experiment was conducted in the controlled plant growth chamber. Tomato seeds (Solanum lycopersicum. cv. Fuji Pink, Sakata, Japan) were surface-sterilized as described above. Surface-sterilized seeds were transferred to plastic Petri dishes and germinated for 4 days in a dark room at 25°C. The bacterial isolates were grown overnight in TSB, and 1 ml of each culture was pelleted by centrifugation (10.000 × g for 10 min). Cell pellets were washed with 1 ml PBS, re-suspended in PBS and cell suspensions were adjusted to OD620 nm = 0.1 (0.2 for Bacillus and Arthrobacter) that correspond to a cell density of about 107–108 cells ml-1. Germinated tomato seeds were placed in the bacterial suspension with a sterile forceps and shaken gently. After 10 min, the inoculated seeds were aseptically planted into a plastic pot filled with potting soil (N 250 mg l-1, P 120 mg l-1, K 700 mg l-1, pH 6.0, Floragard GmbH, Germany) to a depth of approximately 1.5 cm. Non-inoculated plants were used as negative controls. Each experiment included six plants per treatment with three replications (total 18 plants) and pots were set-up in a randomized design. Plants were grown in a growth chamber under the conditions described above.
The data were subjected to one-way analysis of variance (ANOVA) in the software package SPSS-22 statistical software (SPSS, Inc., Chicago, IL, USA). Mean comparisons were conducted by the least significant difference (LSD) (P = 0.05) test.
The inhibitory effect of extracts from Z. capitata and H. perforatum, which were tested against diverse enteric pathogens (A. baumanii 60649, K. oxytoca 6653, K. pneumoniae 40602, K. aerogenes NCTC 8172, C. freundii 82073, S. aureus MRSA 16, E. faecalis NCTC 775, Proteus rettgeri NCIMB 9570, P. aeruginosa NCTC 6749, and E. coli NCTC 9001) at a concentration of 10 mg ml-1, resulted in different extents of inhibition (Table 1). The strains A. baumanii 60649, E. coli NCTC 9001, E. faecalis NCTC 775, K. oxytoca 6653, K. pneumoniae 40602, P. aeruginosa NCTC 6749, and S. aureus MRSA 16 were inhibited by the extract of H. perforatum. However, extract of Z. capitata did not exhibit any potential antibacterial activity against the 10 tested pathogens. Extracts of H. perforatum exhibited potential antifungal activity against F. oxysporum and A. alternata, whereas the extract of Z. capitata did not exhibit any inhibitory activity against the tested fungal strains.

TABLE 1. Antimicrobial activity of extracts obtained from Hypericum perforatum and Ziziphora capitataa.
The total number of endophytic bacterial isolates in the root tissue of Z. capitata was significantly higher (4.5 ± 0.8 × 103 CFU g-1 of fresh root tissue) than in H. perforatum roots (2.6 ± 0.71 × 103 CFU g-1 of fresh root tissue). Isolates were chosen randomly from the dilution plates exhibiting different colonial morphology, forms, texture, and color from each plate. A total of 18 bacterial isolates were derived from H. perforatum and 15 isolates from Z. capitata. Taxonomic investigation by MALDI-TOF MS revealed that the majority of strains were identified with secure genus identification and probable species identification (Table 2). The endophytes from the root of H. perforatum were affiliated with nine genera, whereas 14 isolates were identified at the species level. Achromobacter was the predominant genus, which was followed by the genus Pseudomonas. Furthermore, isolates affiliated with the genera Arthrobacter, Bacillus, Erwinia, Pantoea, Serratia, and Stenotrophomonas were found. The most abundant species was Achromobacter piechaudii (S22a, S7a, S7) (Table 2). A total of five bacterial genera were isolated from the root of Z. capitata (Table 2). The most abundant isolates of Z. capitata were also identified as A. piechaudii (M11, M6, M31, M24, M41). Members of the genera Serratia, Stenotrophomonas, and Erwinia were not identified among the endophytes from Z. capitata.
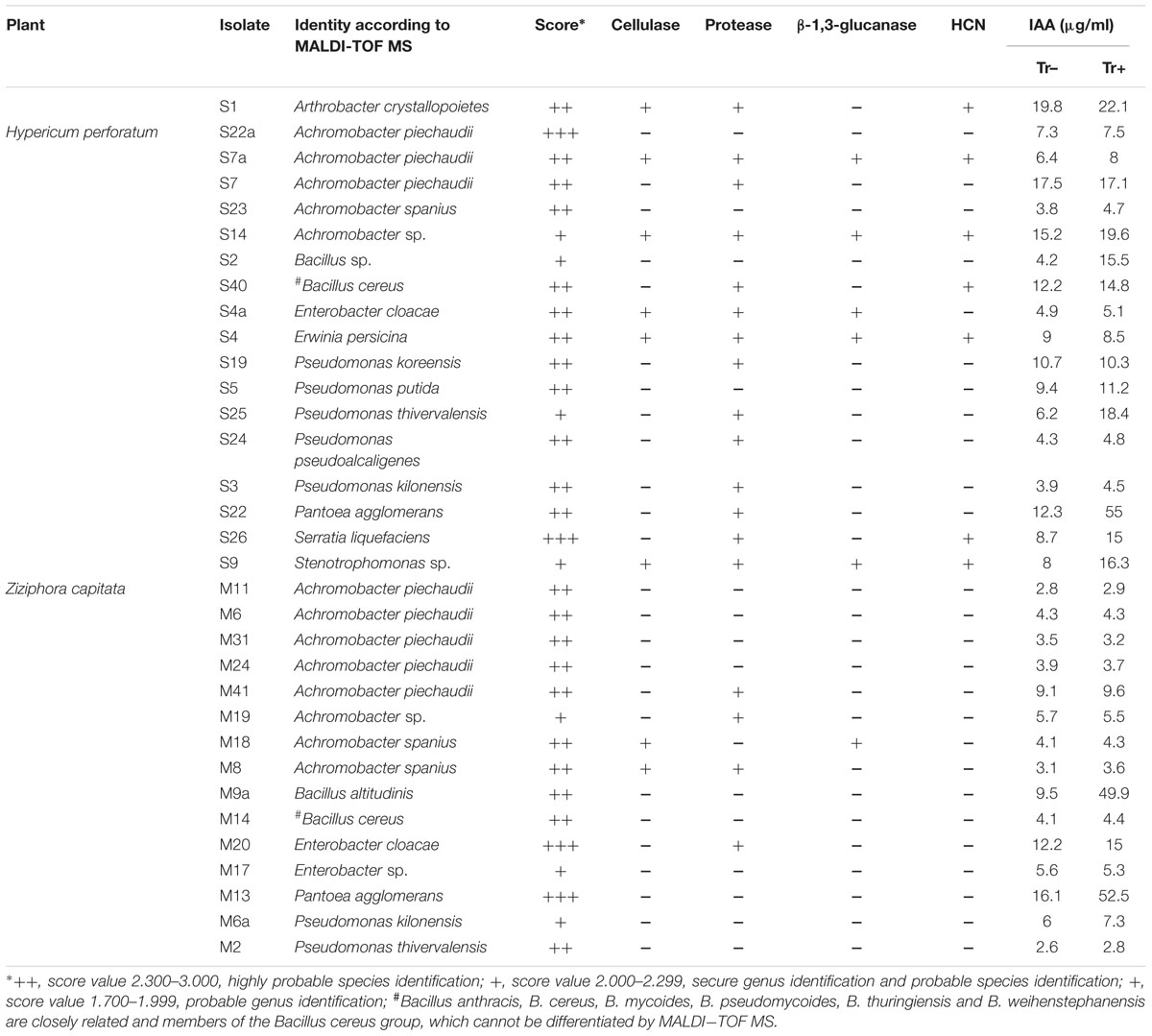
TABLE 2. Matrix-assisted laser desorption ionization (MALDI) biotyper-based identification of culturable endophytic bacteria isolated from the root of Hypericum perforatum and Ziziphora capitata, and traits related to biocontrol and/or plant growth-promoting activity of bacterial strains.
All endophytes isolated from H. perforatum and Z. capitata were screened for multiple plant growth-promoting traits. Most of the bacterial isolates exhibited one or more plant growth-promoting activities. The production of phytohormone IAA by bacterial isolates is presented in Table 2. The highest level of IAA production was observed for Arthrobacter crystallopoietes S1 (19.8 μg ml-1), A. piechaudii S7 (17.5 μg ml-1), Achromobacter sp. S14 (15.2 μg ml-1), Pantoea agglomerans S22 (12.3 μg ml-1), and Bacillus cereus S40 (12.2 μg ml-1), which were isolated from H. perforatum. Two isolates, Enterobacter cloacae M20 and P. agglomerans M13, isolated from Z. capitata also exhibited high IAA production in culture media (12.2 and 16.1 μg ml-1, respectively). The presence of tryptophan did not stimulate auxin production in the majority of the isolates, whereas only four isolates from Z. capitata revealed an increase in IAA synthesis: Bacillus sp. S2 (15.5 μg ml-1), P. agglomerans S22 (μg ml-1), Serratia liquefaciens S26 (15.0 μg ml-1), Stenotrophomonas sp. S9 (16.3 μg ml-1) and two isolates, B. altitudinis M9a (49.9 μg ml-1) and P. agglomerans M13 (52.5 μg ml-1) from H. perforatum (Table 2). All isolates isolated from H. perforatum, except Achromobacter spanius S23, Bacillus sp. S2 and isolate S9, were able to produce one or more cell wall-degrading enzymes. In contrast, only four isolates from Z. capitata (A. piechaudii M41, Achromobacter sp. M19, E. cloacae M20 and isolate M8) were able to produce proteases, and only one isolate, A. spanius M18, produced cellulase and β-1,3-glucanase. HCN was not produced by any isolate from Z. capitata, whereas seven isolates isolated from H. perforatum were able to produce HCN (Table 2).
Antagonistic activity was recorded for endophytes against plant pathogenic fungi F. oxysporum f. sp. radicis-lycopersici, F. solani, F. culmorum, G. graminis pv. tritici, A. alternata, and B. cinerea and the oomycete P. ultimum. As presented in Table 3, all isolates from H. perforatum exhibited antagonistic behavior to one or more of the tested plant pathogenic fungi. The isolates A. crystallopoietes S1, A. piechaudii S7, Pseudomonas koreensis S25, Pseudomonas pseudoalcaligenes S24, and Stenotrophomonas sp. S9 were highly effective against six fungal pathogens and exhibited the highest inhibition of mycelial growth (Figure 1). Among the isolates from Z. capitata, only P. agglomerans M13 exhibited antagonistic activity against five fungal pathogens, but the in vitro inhibition of the mycelium was lower than that of the other isolates. In general, H. perforatum, which exhibited a broad spectrum of antimicrobial activity, supported a higher proportion of antagonistic endophytes compared with Z. capitata.
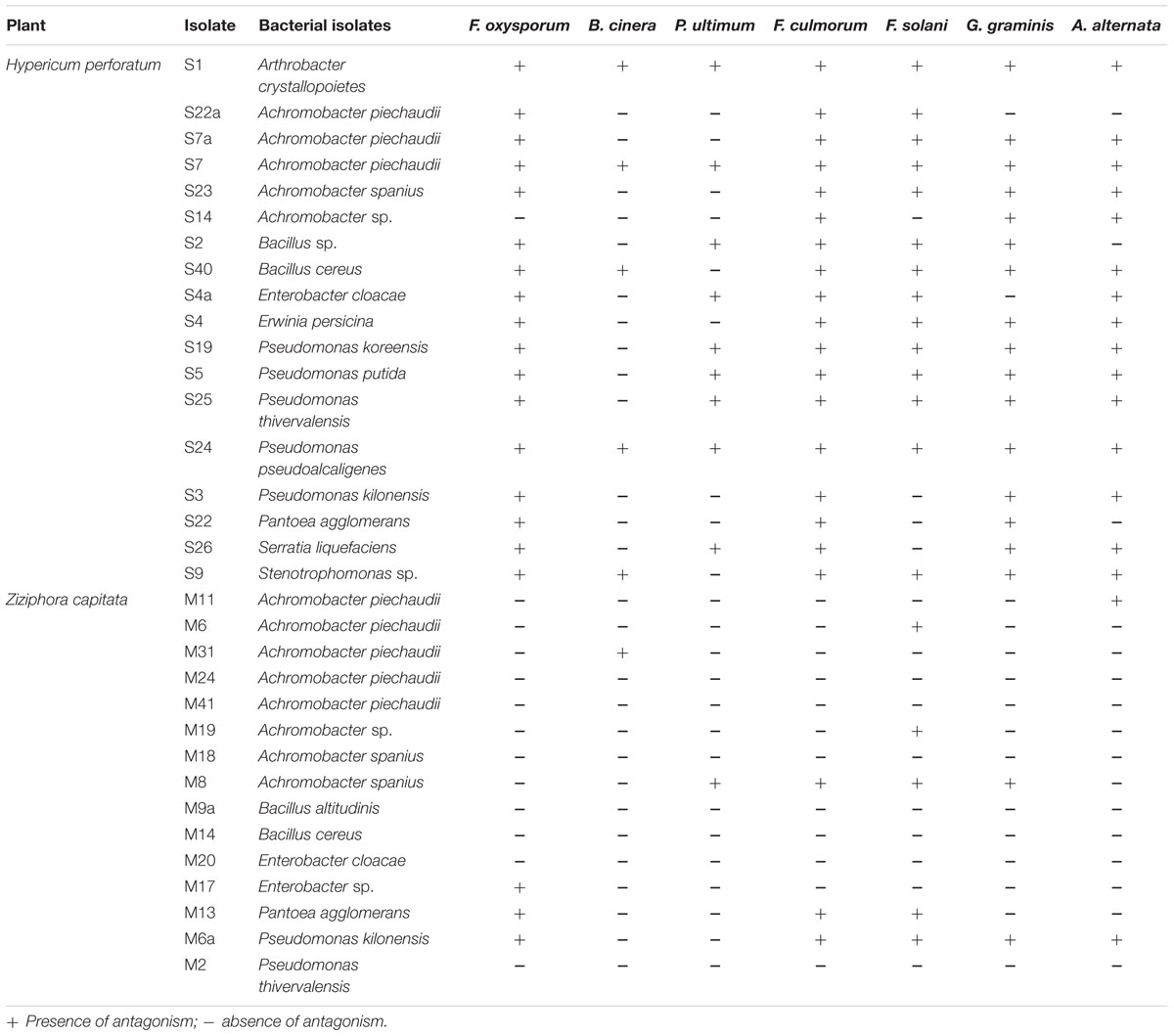
TABLE 3. Antagonistic activity of culturable endophytic bacterial isolates associated with Hypericum perforatum and Ziziphora capitata against soil-borne fungal pathogens.
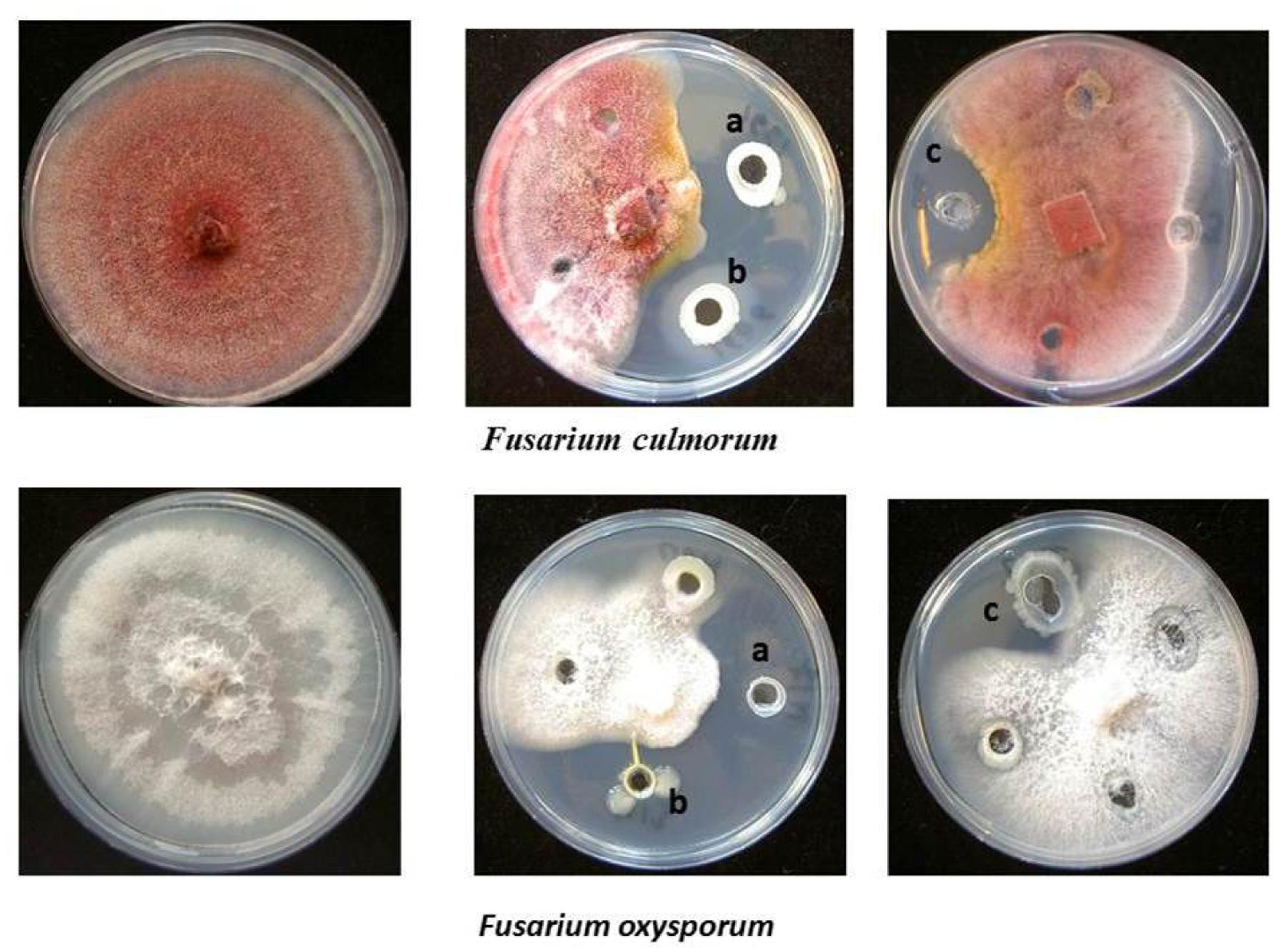
FIGURE 1. Antagonistic activity of endophytic bacterial isolates. (a) Pseudomonas pseudoalcaligenes S24, (b) Stenotrophomonas sp. S9 isolated from Hypericum perforatum, and (c) Enterobacter sp. M17 isolated from Ziziphora capitata against Fusarium culmorum and Fusarium oxysporum.
The bacterial isolates that exhibited antagonistic activity against a wide range of fungal pathogens in vitro were selected to evaluate their ability to suppress tomato foot and root rot caused by F. oxysporum f. sp. radicis-lycopersici in a pot experiment. In non-infested soil, the portion of diseased plants was 2%, whereas in the presence of the pathogen, the portion of plants that exhibited disease symptoms increased to 38% (Figure 2). The selected antagonistic bacterial isolates A. crystallopoietes S1, Bacillus sp. S2, B. cereus S40, P. koreensis S25, S. liquefaciens S26, and Stenotrophomonas sp. S9, exhibited a statistically significant (P < 0.05) disease reduction (up to 9%) compared with Fusarium-infected control plants (Figure 2). Several isolates, namely A. piechaudii S7, Pseudomonas putida S19, Pseudomonas thivervalensis S5, P. pseudoalcaligenes S24, and P. agglomerans M13 reduced disease incident, but the effects were not significant.
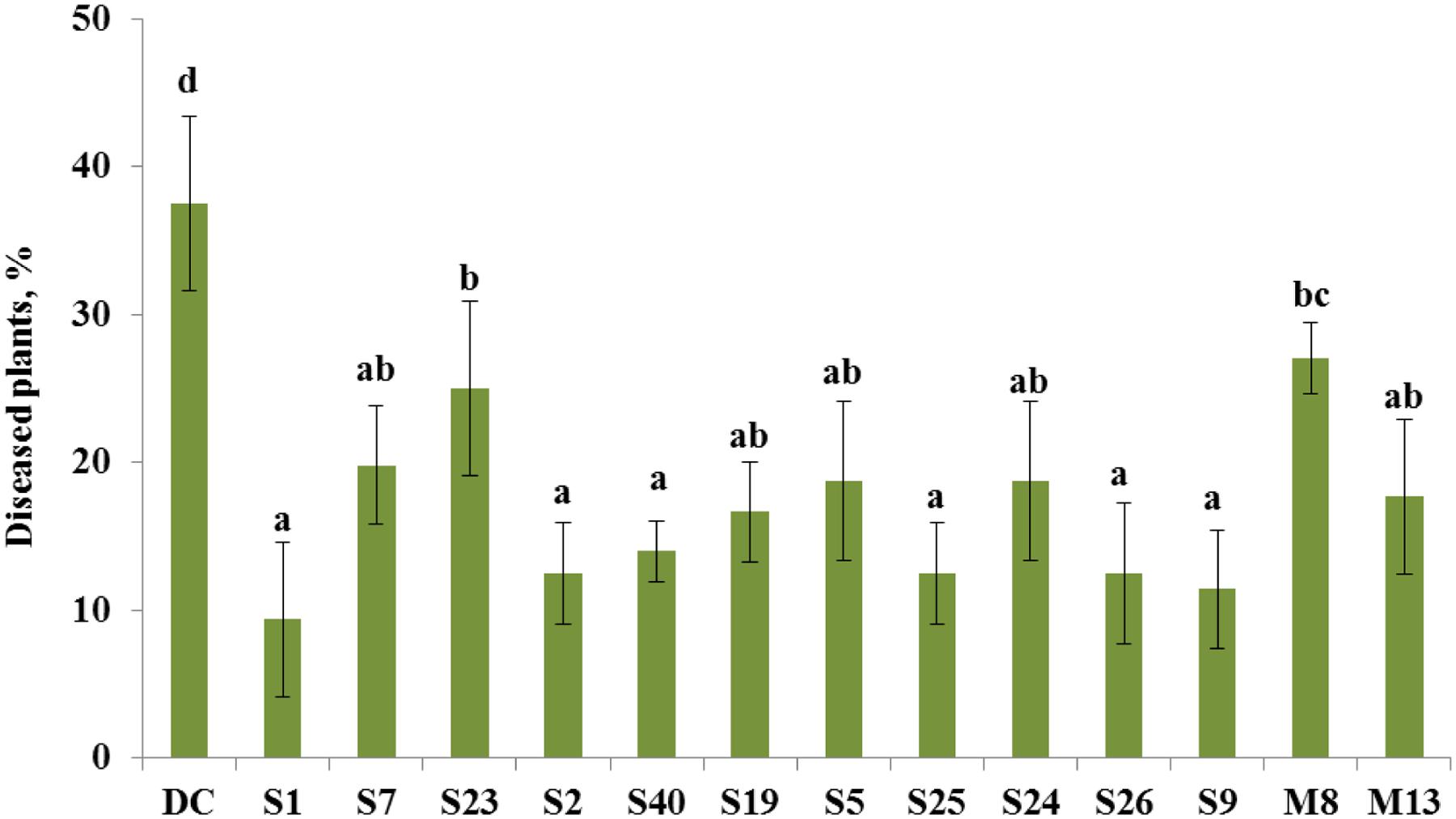
FIGURE 2. Control of tomato foot and root rot caused by F. oxysporum by selected antagonistic endophytic bacteria (Arthrobacter crystallopoietes S1, Achromobacter piechaudii S7, Achromobacter spanius S23, Bacillus sp. S2, Bacillus cereus S40, Pseudomonas putida S19, Pseudomonas thivervalensis S5, Pseudomonas koreensis S25, P. pseudoalcaligenes S24, Serratia liquefaciens S26, Stenotrophomonas sp. S9, A. spanius M8, Pantoea agglomerans M13). DC- disease control (soil infested with F. oxysporum spores), healthy control (no F. oxysporum spores added to the soil) had only 2% diseased plants. Column means marked by different letters indicate significant differences based on Turkey’s HSD test at P < 0.05.
The antagonistic endophytic bacterial isolates were also effective on the growth of tomato plants under controlled conditions (Figure 3). Statistical analysis showed that growth stimulatory effects of the isolates A. crystallopoietes S1, A. spanius S23, Bacillus sp. S2, P. putida S19, and Stenotrophomonas sp. S9 increased plant biomass significantly (P < 0.05) between 30 and 41%. However, four strains namely A. piechaudii S7, B. cereus S40, P. koreensis S25, and P. pseudoalcaligenes S24 reduced plant growth response, leading to a decrease in plant dry biomass between 5.3 and 11.4% (Figure 3).
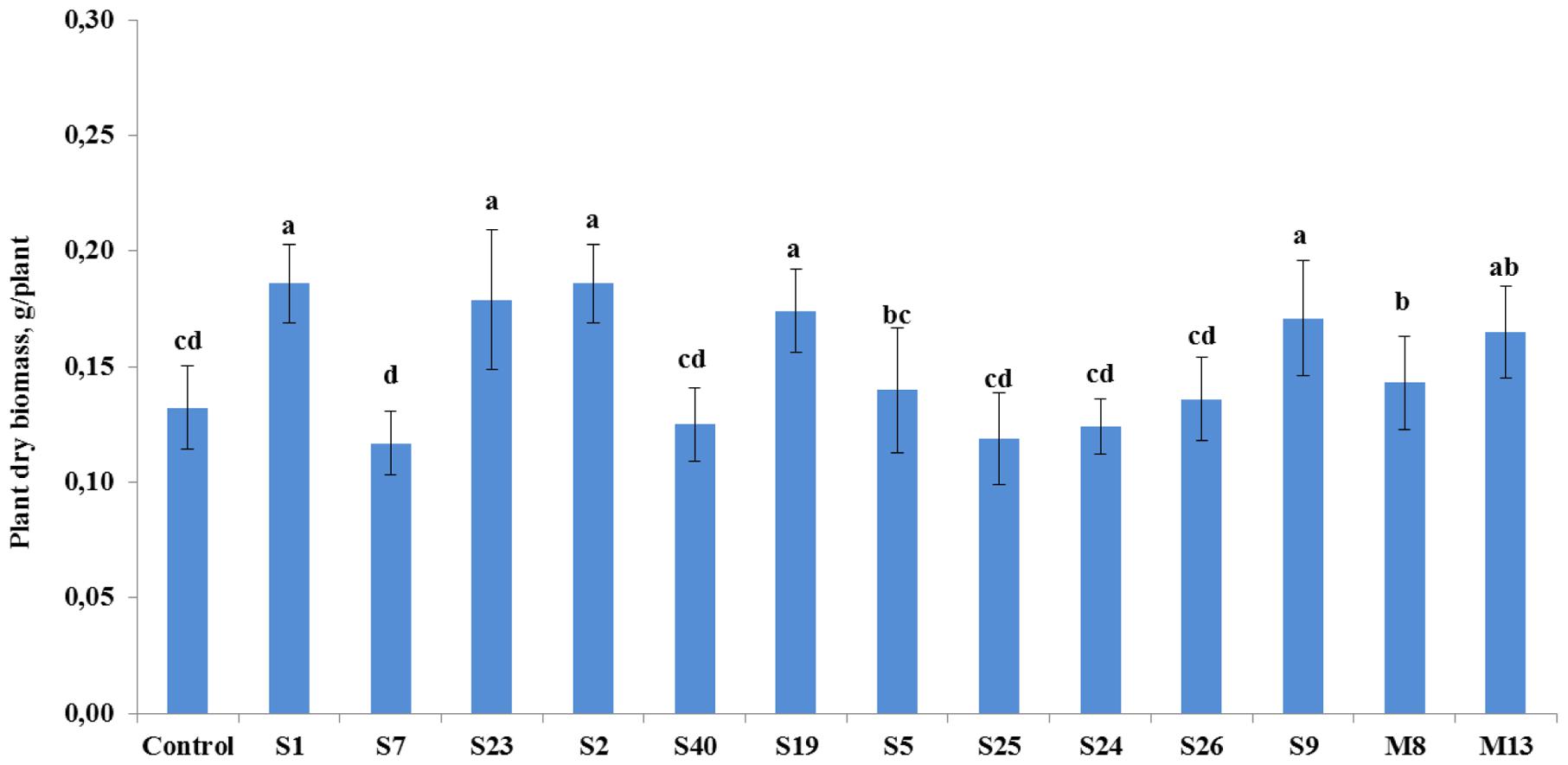
FIGURE 3. The effect of seedling inoculation with selected antagonist endophytic isolates on the dry weight of tomato (A. crystallopoietes S1, A. piechaudii S7, A. spanius S23, Bacillus sp. S2, B. cereus S40, P. putida S19, P. thivervalensis S5, P. koreensis S25, P. pseudoalcaligenes S24, S. liquefaciens S26, Stenotrophomonas sp. S9, A. spanius M8, P. agglomerans M13). Column means marked by different letters indicate significant differences based on Turkey’s HSD test at P < 0.05.
In our study, we analyzed the antimicrobial activity of plant extracts of H. perforatum and Z. capitata, and characterized plant beneficial traits of their associated culturable endophytic bacteria. Both parameters exhibited a relationship – Hypericum plant extracts exhibited greater antimicrobial activity and harbored a higher abundance of endophytes with antagonistic activity than Ziziphora, which lacks antimicrobial activity. In detail, H. perforatum was proved to possess potential antimicrobial activity against a wide range of pathogenic bacteria (A. baumanii, E. coli, E. faecalis, K. oxytoca, K. pneumoniae, P. aeruginosa, S. aureus) as well as fungi (F. oxysporum, A. alternata), whereas the extract of Z. capitata did not exhibit any inhibitory activity against the tested microbes. A similar observation for H. perforatum was reported by Maleš et al. (2006), who found that methanol extracts exhibited strong antibacterial activity against S. aureus, S. epidermidis, E. faecalis, and Bacillus subtilis.
In our study, we observed a lower number of endophytes in H. perforatum compared to Z. capitata that exhibited antibacterial activity. This is consistent with the report of Ahmed et al. (2014), who also reported a smaller microbial population in the rhizosphere of M. chamomilla, which possesses abundant antibacterial activity against pathogenic bacteria (Munir et al., 2014). Our findings suggest that host plants differing in their antibacterial activity exhibited selective effects on physiological properties of endophytes. The understanding of interactions of endophytic bacteria with host plants includes the production of phytohormones, siderophores, and antifungal compounds, which have been well-documented previously by various authors (Berg et al., 2013, 2014; Cho et al., 2015; Egamberdieva et al., 2015a,b). Endophytic bacteria can also improve plant growth by protecting plants against soil-borne diseases or various environmental stresses (Berg et al., 2014; Cao et al., 2014). We have observed that endophytic bacteria associated with both investigated plants exhibited multiple plant beneficial activities, such as the production of IAA, HCN and cell-wall-degrading enzymes. Moreover, the endophytic bacteria associated with H. perforatum demonstrated higher antagonistic activity as compared with endophytes of Z. capitata. This observation is consistent with Gorluk et al. (2009), who also reported a higher proportion of antagonistic endophytes associated with Chelidonium majus L., which is known for its antimicrobial potential (Zuo et al., 2008; Baker and Satish, 2013) against fungal pathogens. Furthermore, it is has been documented that endophytic microbes associated with medicinal plants may produce the same metabolites as their hosts and have been considered a potential source of biologically active metabolites (Mehanni and Safwat, 2010). For example, endophytic species (e.g., Pseudomonas, Bacillus) associated with Aloe vera exhibit antibacterial activity against human pathogenic bacteria, such as S. aureus, Streptococcus pyogenes, P. aeruginosa, and E. coli (Nejatzadeh-Barandoz, 2013), and produce bioactive compounds with antimicrobial activities (Akinsanya et al., 2015).
In our study, endophytic isolates which exhibited antagonistic activity against a wide range of fungal pathogens were evaluated for their capability to suppress tomato foot and root rot caused by F. oxysporum. All selected bacterial isolates of A. crystallopoietes S1, Bacillus sp. S2, B. cereus S40, P. koreensis S25, S. liquefaciens S26, and Stenotrophomonas sp. S9, exhibited statistically significant disease reduction compared with the Fusarium-infected control plants. These observations demonstrate the capability of endophytes to protect plants from soil-borne diseases. In accordance with these results, there is a report of the biological control of Verticillium wilt disease of cotton by endophytic bacteria B. subtilis KDRE 01 and B. megaterium KDRE 25, isolated from the medical plant Sophora alopecuroides (Lin et al., 2013). It has been also reported that Stenotrophomonas maltophilia which is an antagonist against Ralstonia solanacearum significantly suppressed potato brown rot in Egyptian clay soil (Messiha et al., 2007). Moreover, five isolates namely A. crystallopoietes S1, A. spanius S23, Bacillus sp. S2, P. putida S19, and Stenotrophomonas sp. S9 with antifungal activity exhibited enhancement of tomato growth. This finding is consistent with Wei et al. (2014), who also observed an enhanced growth of tomato plants by B. subtilis isolated from the rhizosphere of the traditional Chinese medicinal herb Trichosanthes kirilowii. In another study, endophytic bacteria isolated from a common weed Cassia occidentalis used in several traditional medicines, were able to produce IAA and stimulated growth of mung bean in pot experiments (Arun et al., 2012).
The results from our pilot study of ongoing research provide insights about plant beneficial traits of culturable endophytic bacteria associated with the medicinal plants H. perforatum and Z. capitata with contrasting antimicrobial activities. We observed that H. perforatum with antibacterial activity supported more bacteria with antagonistic activity, as compared to Z. capitata. The antagonistic isolates were able to control tomato root rot caused by F. oxysporum under greenhouse conditions and could be a cost effective source for agro-based biological control agents. However, these findings indicate that further research is necessary to resolve the impact of medicinal plant species with contrasting antimicrobial activity on the endophytic microbial community in more detail, and to identify biological active compounds produced by the hosts and their endophytes.
DE, SW, and GB did experimental design work. DE and UB conducted experiments. PA analyzed the data. DE, SW, UB, and GB wrote the manuscript. All authors read and approved the Manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer SPG and handling Editor declared their shared affiliation, and the reviewer SPG declared a past co-authorship with one of the authors UB to the handling Editor, who ensured that the process met the standards of a fair and objective review.
The authors extend their appreciation to the Deanship of Scientific Research, College of Sciences Research Centre, King Saud University, Riyadh, Saudi Arabia for supporting the project. A Fellowship was provided to DE by the Alexander von Humboldt Foundation.
Ahmed, E. A., Hassan, E. A., El Tobgy, K. M. K., and Ramadan, E. M. (2014). Evaluation of rhizobacteria of some medicinal plants for plant growth promotion and biological control. Ann. Agric. Sci. 59, 273–280.
Akinsanya, M. A., Goh, J. K., Lim, S. P., and Tinga, A. S. Y. (2015). Metagenomics study of endophytic bacteria in Aloe vera using next-generation technology. Genom. Data 6, 159–163. doi: 10.1016/j.gdata.2015.09.004
Arun, B., Gopinath, B., and Sharma, S. (2012). Plant growth promoting potential of bacteria isolated on N free media from rhizosphere of Cassia occidentalis. World J. Microbiol. Biotechnol. 28, 2849–2857. doi: 10.1007/s11274-012-1095-1
Bajguz, A. (2007). Metabolism of brassinosteroids in plants. Plant Physiol. Biochem. 45, 95–107. doi: 10.1016/j.plaphy.2007.01.002
Baker, S., and Satish, S. (2013). Antimicrobial evaluation of fluorescent Pseudomonas sp. inhabiting medicinal plant Annona squamosa L. J. Pure Appl. Microbiol. 7, 1027–1033.
Bano, N., and Musarrat, J. (2003). Characterization of a new Pseudomonas aeruginosa strain NJ-15 as a potential biocontrol agent. Curr. Microbiol. 46, 324–328. doi: 10.1007/s00284-002-3857-8
Beneduzi, A., Ambrosini, A., and Passaglia, L. M. P. (2012). Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35, 1044–1051. doi: 10.1590/S1415-47572012000600020
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berg, G., Grube, M., Schloter, M., and Smalla, K. (2014). Unraveling the plant microbiome: looking back and future perspectives. Front. Microbiol. 5:148. doi: 10.3389/fmicb.2014.00148
Berg, G., Zachow, C., Müller, H., Philipps, J., and Tilcher, R. (2013). Next-generation bio-products sowing the seeds of success for sustainable agriculture. Agronomy 3, 648–656. doi: 10.3390/agronomy3040648
Bharti, P., Bai, S., Seasotiya, L., Malik, A., and Dalal, S. (2012). Antibacterial activity and chemical composition of essential oils of ten aromatic plants against selected bacteria. Int J Drug Dev Res 4, 342–351.
Brown, M. R. W., and Foster, J. H. S. (1970). A simple diagnostic milk medium for Pseudomonas aeruginosa. J. Clin. Path 23, 172–177. doi: 10.1136/jcp.23.2.172
Cao, Y. H., Liu, Y., Yao, S., Li, J. X., Tan, W. Q., and Cheng, C. (2014). Communities diversity of endophytic bacteria from the fruit of Morinda citrifolia (Noni). J. Food Sci. Technol. 32, 39–45.
Castric, P. A. (1975). Hydrogen cyanide, a secondary metabolite of Pseudomonas aeruginosa. Can. J. Microbiol. 21, 613–618. doi: 10.1139/m75-088
Chandra, S. (2012). Endophytic fungi: novel sources of anticancer lead molecules. Appl. Microbiol. Biotechnol. 95, 47–59. doi: 10.1007/s00253-012-4128-7
Chaparro, J. M., Badri, D. V., and Vivanco, J. M. (2014). Rhizosphere microbiome assemblage is affected by plant development. ISME J. 8, 790–803. doi: 10.1038/ismej.2013.196
Cho, S. T., Chang, H. H., Egamberdieva, D., Kamilova, F., Lugtenberg, B., and Kuo, C. H. (2015). Genome analysis of Pseudomonas fluorescens PCL1751: a rhizobacterium that controls root diseases and alleviates salt stress for its plant host. PLOS One 10:e0140231. doi: 10.1371/journal.pone.0140231
Cushnie, T. P. T., Cushnie, B., and Lamb, A. J. (2014). Alkaloids: an overview of their antibacterial, antibiotic-enhancing and antivirulence activities. Int. J. Antim. Agents 44, 377–386. doi: 10.1016/j.ijantimicag.2014.06.001
Dall’Agnol, R., Ferraz, A., Bernardi, A. R., Albring, D., Nör, C., Sarmento, L., et al. (2003). Antimicrobial activity of some Hypericum species. Phytomedicine 10, 511–516. doi: 10.1078/094471103322331476
Ebrahimi, S. N., Hadian, J., and Sonboli, A. (2009). Chemical composition of the essential oil of Ziziphora capitata L. from Iran. J. Essent. Oil Bearing Plants 12, 678–682. doi: 10.1080/0972060X.2009.10643774
Egamberdieva, D., Berg, G., Lindstrom, K., and Rasanen, L. (2010). Root colonising Pseudomonas spp. improve growth and symbiosis performance of fodder galega (Galega orientalis LAM) grown in potting soil. Eur. J. Soil Biol. 46, 269–272. doi: 10.1016/j.ejsobi.2010.01.005
Egamberdieva, D., Jabborova, D., and Berg, G. (2015a). Synergistic interactions between Bradyrhizobium japonicum and the endophyte Stenotrophomonas rhizophila and their effects on growth, nodulation and nutrition of soybean under salt stress. Plant Soil 405, 35–45. doi: 10.1007/s11104-015-2661-8
Egamberdieva, D., Kucharova, Z., Davranov, K., Berg, G., Makarova, N., Azarova, T., et al. (2011). Bacteria able to control foot and root rot and to promote growth of cucumber in salinated soils. Biol. Fertil. Soils 47, 197–205. doi: 10.1007/s00374-010-0523-3
Egamberdieva, D., Li, L., Lindström, K., and Räsänen, L. (2015b). A synergistic interaction between salt tolerant Pseudomonas and Mezorhizobium strains improves growth and symbiotic performance of liquorice (Glycyrrhiza uralensis Fish.) under salt stress. Appl. Microbiol. Biotechnol. 100, 2829–2841. doi: 10.1007/s00253-015-7147-3
Egamberdieva, D., and Teixeira da Silva, J. A. (2015). “Medicinal plant and PGPR: a new frontier for phytochemicals,” in Plant Growth-Promoting Rhizobacteria (PGPR) and Medicinal Plants, eds D. Egamberdieva, S. Shrivastava, and A. Varma (Berlin: Springer Verlag), 287–303.
Egamberdieva, D., Wirth, S., Behrendt, U., Abd-Allah, E. F., and Berg, G. (2016). Biochar treatment resulted in a combined effect on soybean growth promotion and a shift in plant growth promoting rhizobacteria. Front. Microbiol. 7:209. doi: 10.3389/fmicb.2016.00209
El-Deeb, B., Fayez, K., and Gherbawy, Y. (2013). Isolation and characterization of endophytic bacteria from Plectranthus tenuiflorus medicinal plant in Saudi Arabia desert and their antimicrobial activities. J. Plant Interact. 8, 56–64. doi: 10.1080/17429145.2012.680077
Gorluk, A., Rekossz-burlaga, H., and Blaszckyk, M. (2009). Isolation and characterization of bacterial endophytes of Chelidonium majus. Pol. J. Microbiol. 58, 355–361.
Hardoim, P. R., van Overbeek, L. S., Berg, G., Pirttilä, A. M., Compante, S., Campisano, A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/MMBR.00050-14
Jurgenliemk, G., and Nahrstedt, A. (2002). Phenolic compounds from Hypericum perforatum. Planta Med. 68, 88–91. doi: 10.1055/s-2002-20053
Köberl, M., Ramadan, E. M., Adam, M., Cardinale, M., Hallmann, J., Heuer, H., et al. (2013). Bacillus and Streptomyces were selected as broad-spectrum antagonists against soilborne pathogens from arid areas in Egypt. FEMS Microbiol. Lett. 342, 168–178. doi: 10.1111/1574-6968.12089
Köberl, M., Schmidt, R., Ramadan, E. M., Bauer, R., and Berg, G. (2014). The microbiome of medicinal plants: diversity and importance for plant growth, quality and health. Front. Microbiol. 4:400. doi: 10.3389/fmicb.2013.00400
Kogure, K., Yamauchi, I., Tokumura, A., Kondou, K., Tanaka, N., and Takaishi, Y. (2004). Novel antioxidants isolated from plants of the genera Ferula, Inula, Prangos and Rheum collected in Uzbekistan. Phytomedicine 11, 645–651. doi: 10.1016/j.phymed.2003.09.004
Kumar, G., Kanaujia, N., and Bafana, A. (2012). Functional and phylogenetic diversity of root-associated bacteria of Ajuga bracteosa in Kangra valley. Microbiol. Res. 167, 220–225. doi: 10.1016/j.micres.2011.09.001
Lin, T., Zhao, L., Yang, Y., Guan, Q., and Gong, M. (2013). Potential of endophytic bacteria isolated from Sophora alopecuroides nodule in biological control against Verticillium wilt disease. Aust. J. Crop Sci. 7, 139–146.
López-Fuentes, E., Ruiz-Valdiviezo, V. M., Martinez-Romero, E., Gutierrez-Miceli, F. A., Dendooven, L., and Rincon-Rosales, R. (2012). Bacterial community in the roots and rhizosphere of Hypericum silenoides Juss. 1804. Afr. J. Microbiol. Res. 6, 2704–2711. doi: 10.5897/AJMR11.1192
Maleš,Ž, Brantner, A. H., Sović, K., Pilepiæ, K. H., and Plazibat, M. (2006). Comparative phytochemical and antimicrobial investigations of Hypericum perforatum L. subsp. perforatum and H. perforatum subsp. angustifolium (DC.) Gaudin. Acta Pharm. 56, 359–367.
Malfanova, N., Kamilova, F., Validov, S., Shcherbakov, A., Chebotar, V., Tikhonovich, I., et al. (2011). Characterization of Bacillus subtilis HC8, a novel plant-beneficial endophytic strain from giant hogweed. Microbiol. Biotechnol. 4, 523–532. doi: 10.1111/j.1751-7915.2011.00253.x
Mehanni, M. M., and Safwat, M. S. (2010). Endophytes of medicinal plants. Acta Hortic. 854, 31–39. doi: 10.17660/ActaHortic.2010.854.3
Messiha, N. A. S., van Diepeningen, A. D., Farag, N. S., Abdallah, S. A., Janse, J. D., and van Bruggen, A. H. C. (2007). Stenotrophomonas maltophilia: a new potential biocontrol agent of Ralstonia solanacearum, causal agent of potato brown rot. Eur. J. Plant Pathol. 118, 211–225. doi: 10.1007/s10658-007-9136-6
Miller, K. I., Qing, C., Sze, D. M., and Neilan, B. A. (2012). Investigation of the biosynthetic potential of endophytes in traditional Chinese anticancer herbs. PLoS ONE 7:e35953. doi: 10.1371/journal.pone.0035953
Morsy, N. M. (2014). Phytochemical analysis of biologically active constituents of medicinal plants. Main Group Chem. 13, 7–21. doi: 10.3233/MGC-130117
Munir, N., Iqbal, A. S., Altaf, I., Bashir, R., Sharif, N., Saleem, F., et al. (2014). Evaluation of antioxidant and antimicrobial potential of two endangered plant species Atropa Belladonna and Matricaria Chamomilla. Afr. J. Tradit. Complement. Altern. Med. 11, 111–117. doi: 10.4314/ajtcam.v11i5.18
Nejatzadeh-Barandoz, F. (2013). Antibacterial activities and antioxidant capacity of Aloe vera. Org. Med. Chem. Lett. 3, 5. doi: 10.1186/2191-2858-3-5
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Pratima, G., Kalpana, S., and Avinash, S. (2012). Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Int. J. Microbiol. 2012, 5. doi: 10.1155/2012/578925
Qi, X., Wang, E., Xing, M., Zhao, W., and Chen, X. (2012). Rhizosphere and non-rhizosphere bacterial community composition of the wild medicinal plant Rumex patientia. World J. Microbiol. Biotechnol. 28, 2257–2265. doi: 10.1007/s11274-012-1033-2
Sarkisian, S. A., Janssen, M. J., Matta, H., Henry, E., LaPlante, K. L., and Rowley, D. C. (2012). Inhibition of bacterial growth and biofilm production by constituents from Hypericum spp. Phytother. Res. 26, 1012–1016. doi: 10.1002/ptr.3675
Sessitsch, A., Kuffner, M., Kidd, P., Vangronsveld, J., Wenzel, W., Fallmann, K., et al. (2013). The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol. Biochem. 60, 182–194. doi: 10.1016/j.soilbio.2013.01.012
Simmonds, M. S. J. (2003). Flavonoid-insect interactions: recent advances in our knowledge. Phytochemistry 64, 21–30. doi: 10.1016/S0031-9422(03)00293-0
Sonboli, A., Mirjalili, M. H., Hadian, J., Ebrahimi, S. N., and Yousefzadi, M. (2006). Antibacterial activity and composition of the essential oil of Ziziphora clinopodioides subsp. bungeana (Juz.) Rech. F. from Iran. Z. Naturforsch. C 61, 677–680.
Tian, X. R., Feng, G. T., Ma, Z. Q., Xie, N., Zhang, J., Zhang, X., et al. (2014). Three new glycosides from the whole plant of Clematis lasiandra Maxim and their cytotoxicity. Phytochem. Lett. 10, 168–172. doi: 10.1016/j.phytol.2014.09.004
Treutter, D. (2006). Significance of flavonoids in plant resistance: a review. Environ. Chem. Lett. 4, 147–157. doi: 10.1007/s10311-006-0068-8
Vardhini, B. V., and Anjum, N. A. (2015). Brassinosteroids make plant life easier under abiotic stresses mainly by modulating major components of antioxidant defense system. Front. Environ. Sci. 2:67. doi: 10.3389/fenvs.2014.00067
Walsh, G. A., Murphy, R. A., Killeen, G. F., Headon, D. R., and Power, R. F. (1995). Technical note: detection and quantification of supplemental fungal β-glucanase activity in animal feed. J. Anim. Sci. 73, 1074–1076. doi: 10.2527/1995.7341074x
Wei, L. H., Shao, Y., Wan, J. W., Feng, H., Zhu, H., Huang, H. W., et al. (2014). Isolation and characterization of a rhizobacterial antagonist of root-knot nematodes. PLoS ONE 9:e85988. doi: 10.1371/journal.pone.0085988
Weller, D. M., Raaijmakers, J. M., McSpadden Gardner, B. B., and Thomashow, L. S. (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40, 308–348. doi: 10.1146/annurev.phyto.40.030402.110010
Zhao, J., Shan, T., Mou, Y., and Zhou, L. (2011). Plant-derived bioactive compounds produced by endophytic fungi. Mini Rev. Med. Chem. 11, 159–168. doi: 10.2174/138955711794519492
Zhaparkulova, K., Srivedavyasasri, R., Sakipova, Z., and Ross, S. A. (2015). Phytochemical and biological studies on Ziziphora bungeana. Planta Med. 2015, 81–B27.
Keywords: Hypericum perforatum, Ziziphora capitata, endophytic bacteria, plant growth traits, antimicrobial activity, antagonism
Citation: Egamberdieva D, Wirth S, Behrendt U, Ahmad P and Berg G (2017) Antimicrobial Activity of Medicinal Plants Correlates with the Proportion of Antagonistic Endophytes. Front. Microbiol. 8:199. doi: 10.3389/fmicb.2017.00199
Received: 27 August 2016; Accepted: 27 January 2017;
Published: 09 February 2017.
Edited by:
Gero Benckiser, Justus-Liebig-University Giessen, GermanyReviewed by:
Roberta Fulthorpe, University of Toronto Scarborough, CanadaCopyright © 2017 Egamberdieva, Wirth, Behrendt, Ahmad and Berg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dilfuza Egamberdieva, ZWdhbWJlcmRpZXZhQHlhaG9vLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.