- 1Key Lab of Urban Environment and Health, Institute of Urban Environment – Chinese Academy of Sciences, Xiamen, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Fujian Provincial Key Laboratory of Agroecological Processing and Safety Monitoring, College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 4State Key Lab of Urban and Regional Ecology, Research Center for Eco-environmental Sciences – Chinese Academy of Sciences, Beijing, China
Arsenite [As(III)] and methylarsenite [MAs(III)] are the most toxic inorganic and methylated arsenicals, respectively. As(III) and MAs(III) can be interconverted in the unicellular cyanobacterium Nostoc sp. PCC 7120 (Nostoc), which has both the arsM gene (NsarsM), which is responsible for arsenic methylation, and the arsI gene (NsarsI), which is responsible for MAs(III) demethylation. It is not clear how the cells prevent a futile cycle of methylation and demethylation. To investigate the relationship between arsenic methylation and demethylation, we constructed strains of Escherichia coli AW3110 (ΔarsRBC) expressing NsarsM or/and NsarsI. Expression of NsarsI conferred MAs(III) resistance through MAs(III) demethylation. Compared to NsArsI, NsArsM conferred higher resistance to As(III) and lower resistance to MAs(III) by methylating both As(III) and MAs(III). The major species found in solution was dimethylarsenate [DMAs(V)]. Co-expression of NsarsM and NsarsI conferred As(III) resistance at levels similar to that with NsarsM alone, although the main species found in solution after As(III) biotransformation was methylarsenate [MAs(V)] rather than DMAs(V). Co-expression of NsarsM and NsarsI conferred a higher level of resistance to MAs(III) than found with expression of NsarsM alone but lower than expression of only NsarsI. Cells co-expressing both genes converted MAs(III) to a mixture of As(III) and DMAs(V). In Nostoc NsarsM is constitutively expressed, while NsarsI is inducible by either As(III) or MAs(III). Thus, our results suggest that at low concentrations of arsenic, NsArsM activity predominates, while NsArsI activity predominates at high concentrations. We propose that coexistence of arsM and arsI genes in Nostoc could be advantageous for several reasons. First, it confers a broader spectrum of resistance to both As(III) and MAs(III). Second, at low concentrations of arsenic, the MAs(III) produced by NsArsM will possibly have antibiotic-like properties and give the organism a competitive advantage. Finally, these results shed light on the role of cyanobacteria in the arsenic biogeochemical cycle.
Introduction
Arsenic is a ubiquitous environmental toxin, and primarily occurs in inorganic forms, including arsenate [As(V)] and arsenite [As(III)] (Zhu et al., 2014). Because of the unavoidable exposure to arsenic, nearly all of the living organisms have arsenic detoxifying systems (Rosen, 2002; Liu et al., 2013). As(III) efflux and oxidation are considered to be efficient detoxification pathways in Bacteria and Archaea (Páez-Espino et al., 2009). As(III) methylation catalyzed by As(III) S-adenosylmethionine methyltransferase (ArsM) has also been shown to confer resistance to As(III) by the eventual production of less toxic pentavalent methylated species, including methylarsonate [MAs(V)], dimethylarsonate [DMAs(V)], trimethylarsine oxide [TMAsO], or volatile methylated arsenicals (Levy et al., 2005; Challenger, 2006; Qin et al., 2006). However, the trivalent methylated arsenic species, methylarsenite [MAs(III)] and dimethylarsenite [DMA(III)], which are the immediate products of As(III) biomethylation (Dheeman et al., 2014; Marapakala et al., 2015), are far more toxic than As(III) (Petrick et al., 2000; Mass et al., 2001). MAs(V) can also be reduced to MAs(III) in some bacteria (Yoshinaga et al., 2011), so other microbes have evolved various processes to detoxify MAs(III) as it is formed. MAs(III), like As(III), is detoxified by oxidation or efflux in some microbes. A NADPH-FMN dependent oxidoreductase (ArsH) and a membrane permease (ArsP) are responsible for MAs(III) oxidation and efflux, respectively (Chen et al., 2015a,b). Furthermore, MAs(III) demethylation catalyzed by a C-As lyase (ArsI) is an effective MAs(III) detoxification pathway (Yoshinaga and Rosen, 2014).
Demethylation reverses methylation by converting methylated arsenic into inorganic arsenic. Since both ArsM and ArsI can use MAs(III) as substrate, they may compete with each other for MAs(III). Intuitively, it would seem that, if both methylation and demethylation occur in the same organism, a futile cycle could be created. Nevertheless, the genes for arsenic methylation and demethylation coexist in some microorganisms. For example, Nostoc can both methylate As(III) and demethylate MAs(III) and has both the NsarsM and NsarsI genes in its chromosome (Yin et al., 2011; Yan et al., 2015). However, it is not known whether coexistence of arsM and arsI creates a futile cycle of arsenic methylation and demethylation or whether it enhances resistances to selected arsenicals.
In cyanobacteria, morphological, physiological and biochemical changes, genomics, and proteomics have been found to be involved in response to arsenic stress (Srivastava et al., 2009; Pandey et al., 2012; Sánchez-Riego et al., 2014). Furthermore, various cyanobacteria were shown to be able to accumulate and resist arsenic by multiple arsenic biotransformations (Yin et al., 2012; Wang et al., 2013). For instance, in Synechocysis sp. PCC 6803, which is the best-understood cyanobacterium on arsenic resistance and metabolism (Huertas et al., 2014), an ars operon (arsBHC) mediates the main arsenic resistance mechanism (López-Maury et al., 2003), and another two arsenate reductases (arsI1 and arsI2) (López-Maury et al., 2009) and an arsenite methyltransferase (arsM) (Yin et al., 2011) were also found to be the resistance determinants. Similarly, Nostoc is also a complex system for analysis of arsenic biotransformations. In addition to methylation and demethylation, Nostoc has other arsenic biotransformation pathways, in particular As(V) reduction (Pandey et al., 2013), As(III) oxidation, and efflux systems. Thus, to investigate the relationship between As(III) methylation and MAs(III) demethylation, we constructed a simplified model using arsenic-hypersensitive Escherichia coli AW3110 strains (ΔarsRBC) expressing NsarsM or/and NsarsI. The effect of co-expression on biotransformations of and resistance to As(III) and MAs(III) was examined. The kinetic parameters of NsArsM for As(III) and MAs(III) and regulations of NsarsM and NsarsI in Nostoc were also evaluated to provide a more comprehensive understanding of the arsenic methylation cycle.
Materials and Methods
Bacterial Strains, Media, and Chemical Reagents
Escherichia coli strains were aerobically cultivated in Lysogeny Broth (LB) medium (Sambrook et al., 1989), supplemented with required antibiotics at 37°C with shaking at 180 rpm. E. coli strain DH5α (Promega, Madison, WI, USA) was used for plasmid construction and replication, and Rosetta (DE3) (Novagen, Madison, WI, USA) was used for protein expression. E. coli strain AW3110 (DE3) [ΔarsRBC; ArsR-repressor, ArsB-As(III) efflux pump, ArsC-As(V) reductase] was used for arsenic resistance and biotransformation assays (Carlin et al., 1995). Nostoc (also known as Anabaena), kindly provided by Professor Wen-Li Chen, Huazhong Agricultural University, was grown in BG11 medium without nitrate and cultured as previously described (Yan et al., 2015). MAs(III) was produced by reduction of MAs(V) using Na2S2O3, Na2S2O5, and H2SO4 (Yoshinaga and Rosen, 2014). All other used reagents were purchased from commercial sources, and were of analytical grade or better.
Construction of E. coli AW3110 Strain Containing NsarsM and NsarsI
NsarsM (alr3095, accession number HQ891148) and NsarsI (alr1104, accession number BAB73061) have been identified in previous studies (Yin et al., 2011; Yan et al., 2015). The NsarsM or NsarsI was cloned into the expression vector pET28a (Novagen, Madison, WI, USA) or pET22b (Novagen, Madison, WI, USA) to generate the plasmid pET28a-NsarsM, pET28a-NsarsI, or pET22b-NsarsI; and the primers used for amplification were listed in Table 1. The two pET28a plasmids were transformed independently into E. coli AW3110 to construct strains expressing NsarsM or NsarsI. The plasmids pET28a-NsarsM and pET22b-NsarsI were co-transformed into E. coli AW3110. Although the two-plasmid system using the same origin of replication are commonly believed not to exist in one E. coli cell, several similar approaches with two incompatible plasmids were successfully used if under the selection pressure of two different antibiotics (Yang et al., 2001; Wang et al., 2007; Zhang et al., 2011). Thus, the strain co-expressing NsarsM and NsarsI was selected by growth on LB agar plate containing 100 μg mL-1 ampicillin, 50 μg mL-1 kanamycin, and 30 μg mL-1 chloramphenicol (pET28a-NsarsM and pET22b-NsarsI plasmids confer kanamycin and ampicillin resistances, respectively).
Western Blot Analysis
Western blots were used to detect expression of NsarsM or/and NsarsI in the E. coli AW3110 strains. E. coli AW3110 cells in exponential phase were induced by 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). The proteins from cell lysate were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; gradient 15%), and transferred to polyvinylidene fluoride (PVDF) membranes (Pall Corporation, East Hills, NY, USA) using the Trans-Blot Turbo Transfer System (Bio-Rad, Hercules, CA, USA). Immunoblot analyses were carried out with anti-His tag (D3I1O, Cell Signaling, Beverly, MA, USA) by incubating at 4°C overnight. Membranes were washed, and incubated with appropriate peroxidase-conjugated secondary antibody. Specific bands were visualized by WesternBright ECL HRP substrate (Advansta, Inc., Menlo Park, CA, USA), and finally scanned on a Kodak image station 4000 mm Pro (Carestream Health, New Haven, CT, USA).
Arsenic Resistance Assays
As(III) and MAs(III) resistance assays of E. coli AW3110 strains bearing pET28a vector, pET28a-NsarsM, pET28a-NsarsI, or pET28a-NsarsM+pET22b-NsarsI plasmids were performed as described previously (Ye et al., 2014; Yan et al., 2015). The cells were cultured in LB medium containing As(III) (0, 10, 30, 50, 70, 90, and 110 μM) at 37°C or in ST medium (10-fold concentrated ST 10-1 medium) (Maki et al., 2006) containing MAs(III) (0, 1, 2, 3, 4, 6, and 8 μM) at 30°C. After incubating for 24 h, optical density at 600 nm (OD600 nm) was measured using an ultraviolet-visible spectrophotometer (UV-6300 double beam spectrophotometer, Mapada, Shanghai, China). In addition, the four E. coli AW3110 strains were treated with 35 μM As(III) and 2 μM MAs(III) at the same time in ST medium, and OD600 nm was monitored at 0, 3, 6, 12, 18, 24, 30 h.
Arsenic Biotransformation in E. coli AW3110 Strains
The IPTG-induced E. coli AW3110 cells were cultured in LB medium containing 25 μM As(III) at 37°C for 24 h or ST 10-1 medium containing 1 μM MAs(III) or 1 μM As(III) and 0.5 μM MAs(III) at 25°C for 1 h. Same amounts of arsenicals were also added to LB medium or ST 10-1 medium without cells as non-inoculated controls. All samples were centrifuged at 13400 g for 2 min, and the supernatants were collected. Arsenic speciation was determined by high-performance liquid chromatography (HPLC, 1200, Agilent Technologies, Palo Alto, CA, USA)-inductively coupled plasma-mass spectrometry (ICP-MS, 7500a, Agilent Technologies, Palo Alto, CA,USA) to analyze arsenic biotransformation.
Purification of NsArsM
The plasmid pET28a-NsarsM was transformed into E. coli strain Rosetta (DE3) for purification of NsArsM. His-tagged NsArsM expression was induced by addition of 1 mM IPTG when OD600 nm of E. coli Rosetta (DE3) cells reached 0.5–0.8. The induced cells were ruptured in a French-press at 10 MPa. NsArsM was purified by Ni-NTA agarose column (Qiagen, Hilden, Germany) according to the manufacturer’s instruction. The purified NsArsM was concentrated by 10-kDa cutoff Amicon Ultrafilter (Millipore, Bedford, MA, USA), and identified using SDS-PAGE.
Enzyme Kinetics of NsArsM
In order to determine kinetic parameters, in vitro As(III) and MAs(III) reaction systems of NsArsM were established according to previous study with a few modifications (Qin et al., 2006). For the methylation reaction, 8 μM purified NsArsM was added in a buffer consisting of 50 mM MOPS buffer (pH 7.5) with 125 mM NaCl, 1 mM S-adenosylmethionine chloride (SAM), 8 mM reduced glutathione (GSH), and the indicated concentrations of As(III) or MAs(III). After the reaction was performed at 37°C for 1 h, the assay was immediately terminated by boiling, and added 3% H2O2 to oxidize arsenic. Enzyme activity was measured by determining arsenic species. The mole equivalents of methyl groups (-CH3) transferred from SAM to arsenic were used to approximate the apparent rates of methylation. Since ArsM catalyzes three separate methylation reactions, the overall rates were lumped together as one mole of SAM methyl groups to methylate one mole of As(III) to MAs, two moles of SAM to methylate As(III) to DMAs, and three moles of SAM to methylate to TMAs (Walton et al., 2003; Wang et al., 2012). Non-linear regression analysis was performed with OriginPro 8.5.
Arsenic Speciation Analysis
Samples were filtered through 0.22 μm filters (Millipore, Bedford, MA, USA), and analyzed by HPLC-ICP-MS using previously established instrument parameters (Zhu et al., 2008). Arsenic species were determined with either a 10-μm PRP-X100 anion exchange column (250 mm × 4.1 mm ID, Hamilton, Reno, NV, USA) eluted isocratically with a mobile phase (pH 6.2) consisting of 10 mM ammonium di-hydrogen phosphate and 10 mM ammonium nitrate (Ye et al., 2014) or a Jupiter 5 μ C18 300A reverse-phase column (250 mm × 4.6 mm, Phenomenex, Torrance, CA, USA) using the mobile phase (pH 5.95) with 3 mM malonic acid, 5 mM tetrabutylammonium hydroxide, and 5% methanol (Yoshinaga and Rosen, 2014). The flow rate for HPLC was 1.0 mL min-1, and ICP-MS was tuned for monitoring of m/z 75 (arsenic). Arsenic species in samples were identified by retention times which were compared with those of the standards. The arsenic was quantified by external calibration curves with peak areas integrated by using WinFASS.
RNA Isolation and Reverse Transcription-Quantitative Real-Time PCR (RT-qPCR)
Nostoc at the mid-exponential growth phase was cultured with or without arsenic for 6 h. Total RNA was extracted from Nostoc cells treated with As(III) (0, 1, 5, 10, 40, and 100 μM) or MAs(III) (0, 0.2, 1, 3, 6, and 12 μM) by using TRIzol reagent (Invitrogen Life Technologies, Gaithersburg, MD, USA) following the manufacturer’s recommendations. Contaminating genomic DNA was removed from total RNA using DNase I (Promega, Madison, WI, USA) at 37°C for 45 min. The total RNA was further purified with RNA clean kit (Tiangen, Beijing, China). About 40 ng purified RNA was used for RT-qPCR with GoTaq® 1-Step RT-qPCR System (Promega, Madison, WI, USA) in a 20-μL volume. The primers for NsarsM and NsarsI were listed in Table 1. qPCR was performed on a LightCycler 480 (Roche Applied Science, Indianapolis, IN, USA). Each reaction was carried out in triplicate with the housekeeping gene rnpB as the internal standard (Vioque, 1992; Latifi et al., 2005). The PCR efficiencies of the targets (NsarsM and NsarsI) and reference (rnpB) were calculated from the slope of their standard curves (E = 10[-1/slope]), respectively, and the relative transcript levels of NsarsM and NsarsI were calculated using the formula (Etarget)ΔCptarget(control-sample) × (Eref)ΔCpref(sample-control) (Pfaffl, 2001).
Results
Gene Expression in E. coli AW3110 Strains
To investigate arsenic resistance and biotransformation when NsArsM and NsArsI co-existed in the same cell, E. coli AW3110 strain co-expressing NsarsM and NsarsI genes was constructed. The E. coli AW3110 strain bearing plasmid pET28a served as negative control. The expression levels of NsarsM and NsarsI in the single or co-expressed E. coli AW3110 cells were estimated by western blot using anti-His antibodies that recognize NsArsM or NsArsI (Figure 1). The results show that both NsarsM and NsarsI were expressed, separately and together. When co-expressed, more NsArsI was produced than NsArsM.
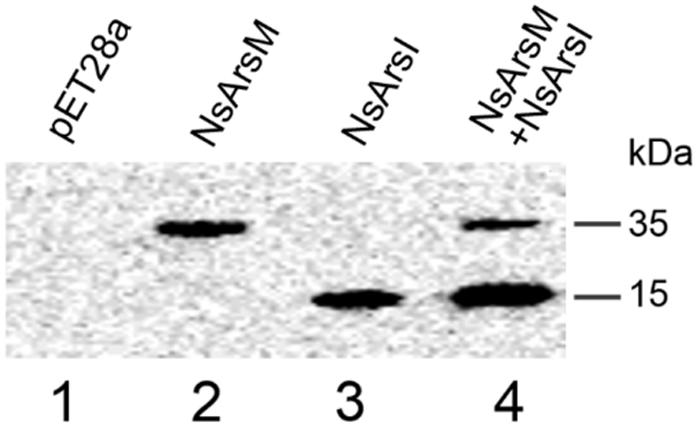
FIGURE 1. Western-blot analysis of NsArsM and NsArsI. lane1 through 4 contained proteins extracted from Escherichia coli. AW3110 cells bearing the empty vector pET28a (lane1) and the following vectors containg the NsarsM and/or NsarsI genes: pET28a-NsarsM (lane2); pET28a-NsarsI (lane3); and pET28a-NsarsM + pET22b-NsarsI (lane4). The blots were probed with anti-His tag, followed by peroxidase-conjugated secondary antibody.
Arsenic Resistance Assays
Resistance assays were conducted using E. coli AW3110 expressing NsarsM and/or NsarsI. When E. coli AW3110 cells were cultured in arsenic-free medium, the growth rates of all four E. coli AW3110 strains were nearly the same (Figure 2). In As(III) resistance assays, the growth of all the E. coli AW3110 cells was stimulated by 10 μM As(III), but inhibited when the As(III) concentration was over 30 μM (Figure 2A). The reason for this apparent growth stimulation by As(III) is not known, but it has been observed in another alga (Zhang et al., 2013). Furthermore, both the E. coli AW3110 cells expressing NsarsM and cells co-expressing NsarsM and NsarsI grew much better than those bearing pET28a or pET28a-NsarsI at 50–110 μM of As(III) (t-test, P < 0.05). For MAs(III) resistance assays, 1 μM MAs(III) inhibited growth of all of E. coli AW3110 cells (Figure 2B). E. coli AW3110 expressing NsarsI grew best at concentrations of MAs(III) between 2 and 8 μM, while E. coli AW3110 expressing NsarsM only grew better than the vector alone at 2 μM MAs(III). E. coli AW3110 co-expressing NsarsM and NsarsI exhibited less resistance to MAs(III) than that expressing NsarsI, and significantly higher resistance than that expressing NsarsM. In addition, the growth rates of E. coli AW3110 exposed to mixed 35 μM As(III) and 2 μM MAs(III) monitored over 30 h (Figure 2C), showed that co-expression of NsarsM and NsarsI conferred higher arsenic resistance than expression of NsarsM, but lower resistance than that of NsarsI.
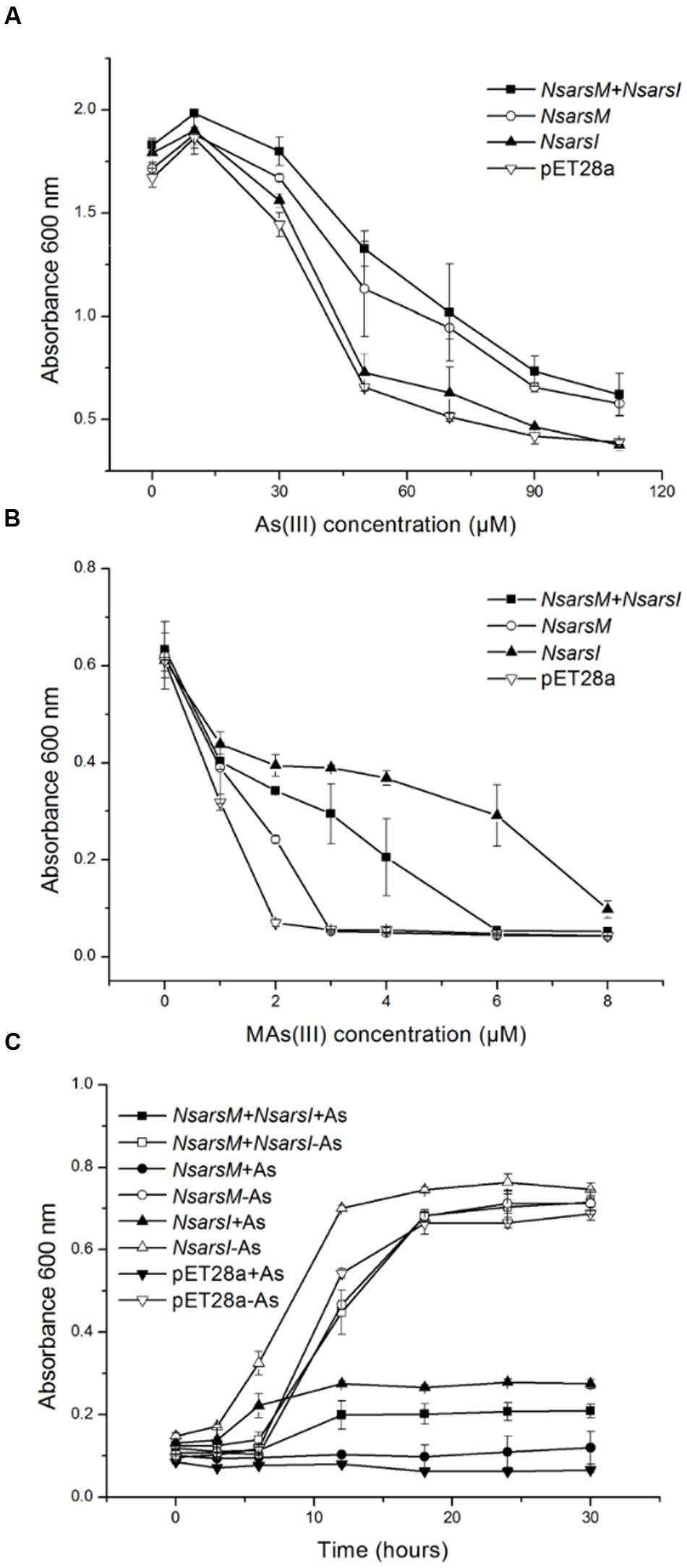
FIGURE 2. Arsenic resistance of E. coli AW3110 bearing pET28a, pET28a-NsarsM, pET28a-NsarsI and pET28a-NsarsM+ pET22b-NsarsI plasmids. The four E. coli AW3110 strains were incubated with the indicated concentrations of As(III) (A) or MAs(III) (B). Cell growth was monitored by spectrophotometry at 600 nm. Filled squares, plasmids pET28a-NsarsM+ pET22b-NsarsI; open circles, plasmid pET28a-NsarsM; filled triangles, plasmid pET28a-NsarsI; open inverted triangles, plasmid pET28a. (C) Sensitivity of the four E. coli AW3110 strains to the mixture of As(III) and MAs(III). The growth curves are shown for LB media containing the mixture of 35 μM As(III) and 2 μM MAs(III) (+As) or without arsenic (-As). Squares, plasmids pET28a-NsarsM+ pET22b-NsarsI; circles, plasmid pET28a-NsarsM; triangles, plasmid pET28a-NsarsI; inverted triangles, plasmid pET28a. Filled symbols, 35 μM As(III) + 2 μM MAs(III); open symbols, 0 μM As(III) + 0 μM MAs(III). The error bars represent the standard error from three independent biological experiments.
Arsenic Biotransformation in E. coli AW3110
To elucidate the biotransformation pathways when NsarsM or/and NsarsI were expressed in E. coli AW3110, arsenic species in As(III) and/or MAs(III)-containing media with or without cells were determined (Figure 3). When treated with 25 μM As(III), E. coli AW3110 bearing pET28a or pET28a-NsarsI did not change arsenic species in the media compared to the non-inoculated control. The detection of As(V) in the control may come from the trace contaminant of reagent and/or oxidation by air. While the cells expressing NsarsM or co-expressing NsarsM and NsarsI transformed As(III) into methylated arsenic (Figure 3A). It is worthwhile to note that the methylated species produced were different for the two strains. DMAs(V) and TMAsO were the predominant arsenic species, and MAs(V) was undetectable in the medium of cells expressing NsarsM. However, MAs(V) was the main arsenic species in the medium culturing E. coli AW3110 co-expressing NsarsM and NsarsI. When treated with 1 μM MAs(III) (Figure 3B), MAs(V) was detected in the controls, and it was probably coming from the spontaneous chemical oxidation of MAs(III) under aerobic conditions. DMAs(V) and As(III) were the primary arsenic species in the culture media of E. coli AW3110 cells expressing NsarsI and NsarsM, respectively. Both As(III) and DMAs(V) were detected in the medium with co-expression AW3110 strain. Similarly, under the exposure to a mixture of As(III) and MAs(III) (Figure 3C), DMAs(V) was only detected in the medium when NsarsM was expressed alone or co-expressed with NsarsI, and the percentage of As(III) increased in the latter.
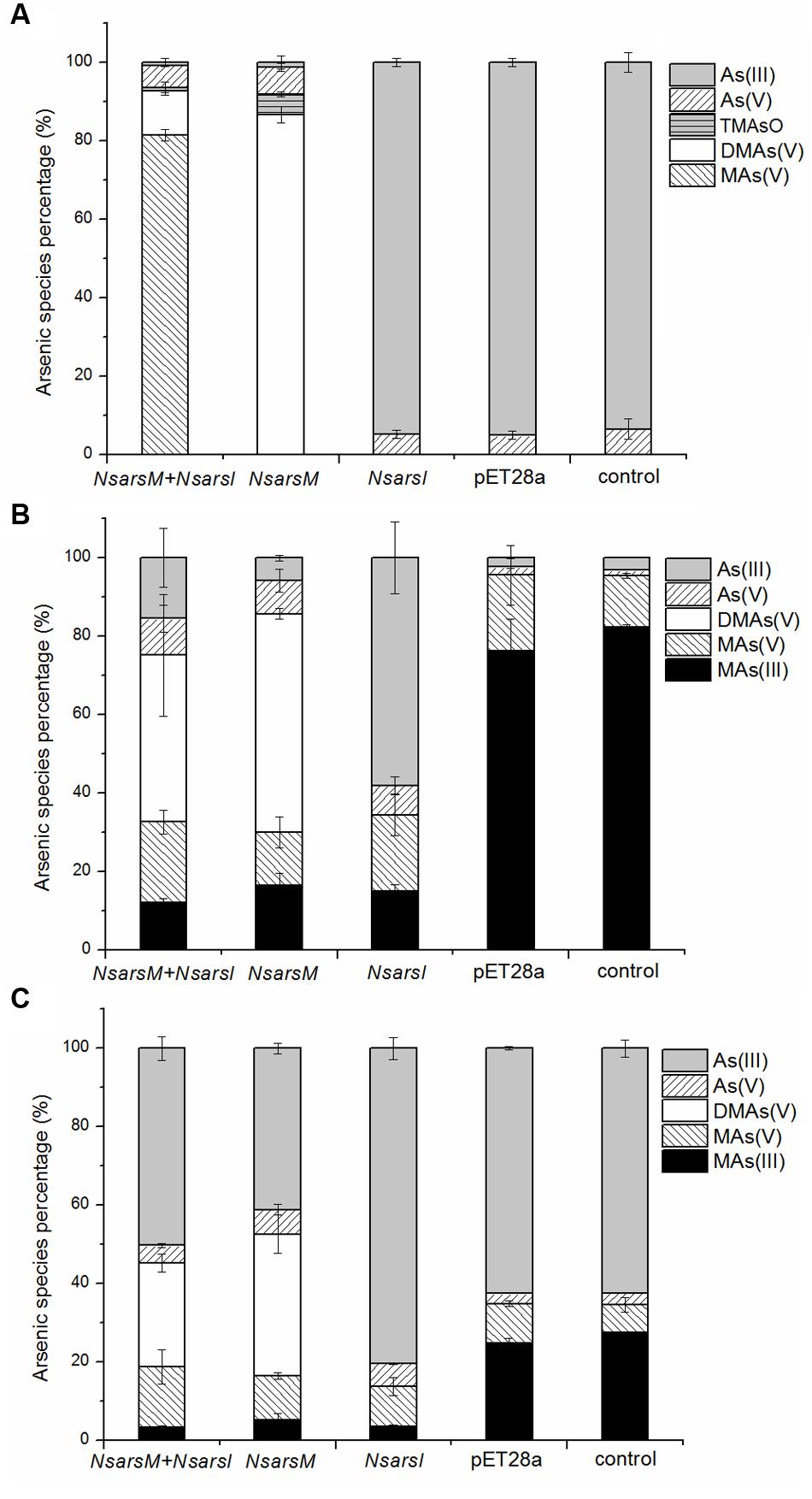
FIGURE 3. The percentage of arsenic species [As(III), gray bars; As(V), left-hatched bars; TMAsO, horizontally hatched bars, DMAs(V), white bars; MAs(V), right-hatched bars; MAs(III), black bars] in media incubated E. coli AW3110 bearing pET28a-NsarsM + pET28a-NsarsI plasmids (NsarsM+NsarsI), pET28a-NsarsM (NsarsM), pET28a-NsarsI (NsarsM) and pET28a vector (pET28a) and non-incubated media (control) exposed to 25 μM As(III) for 24 h (A), 1 μM MAs(III) for 1 h (B), or mixture of 1 μM As(III) and 0.5 μM MAs(III) for 1 h (C). The error bars represent the standard error from three independent biological experiments.
Reaction Kinetics of NsArsM
Apparent kinetic constants were determined with purified NsArsM from the rate of methyl transfer from SAM. The relationship between the substrate As(III) or MAs(III) and the enzyme NsArsM fit conventional Michaelis–Menten kinetics (Figure 4). The affinity for As(III) was sevenfold greater than MAs(III), with a Km of 5 ± 1 μM for As(III) and 37 ± 4 μM for MAs(III). The Vmax for As(III) and MAs(III) were 60 ± 5 pmol CH3 h-1 mg-1 and 167 ± 6 pmol CH3 h-1 mg-1, respectively.
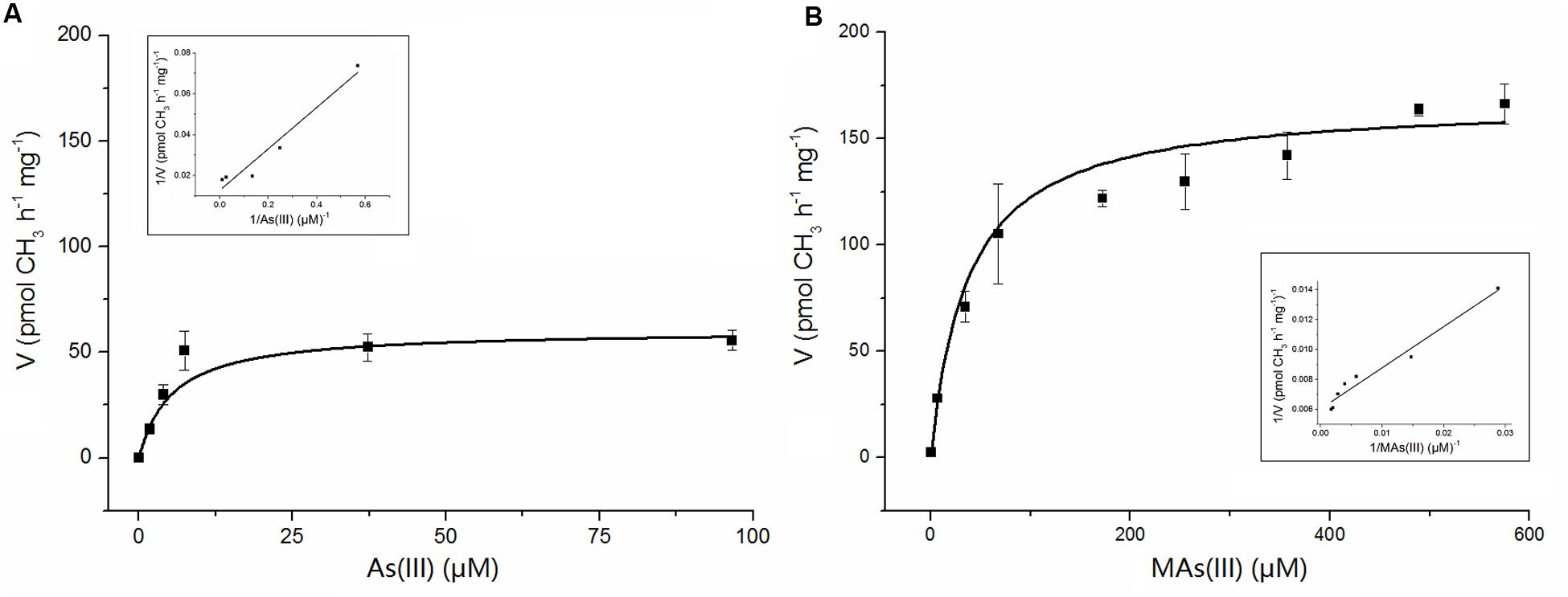
FIGURE 4. The kinetic curves of NsArsM for As(III) (A) and MAs(III) (B) methylation with 1 mM SAM as methyl donor, 8 mM GSH as reductant. Reaction velocity (v) is expressed as pmol CH3 transferred from SAM to arsenic per hour per mg of purified NsArsM. The lines show the least squares fit of Michaelis–Menten equation to the data. The error bars represent the standard error from three independent biological experiments. The insert figure showed the double reciprocal plots of the relation between the concentration of arsenic and the velocity.
Transcription of NsarsM and NsarsI in Nostoc
The transcript levels of NsarsM and NsarsI were analyzed by RT-qPCR after Nostoc exposed to MAs(III) or As(III) at indicated concentrations for 6 h. Nostoc under identical cultivation conditions without arsenic was used as a control. As shown in Figure 5, the transcript levels of NsarsM had no significant difference (P > 0.05) between the Nostoc cultures with and without arsenic. The transcript levels of NsarsI were not significantly increased (P > 0.05) by As(III) less than 40 μM or MAs(III) less than 6 μM, while they were significantly enhanced (P < 0.05) when 40 or 100 μM As(III), or 6 or 12 μM MAs(III) was added in the cultures.
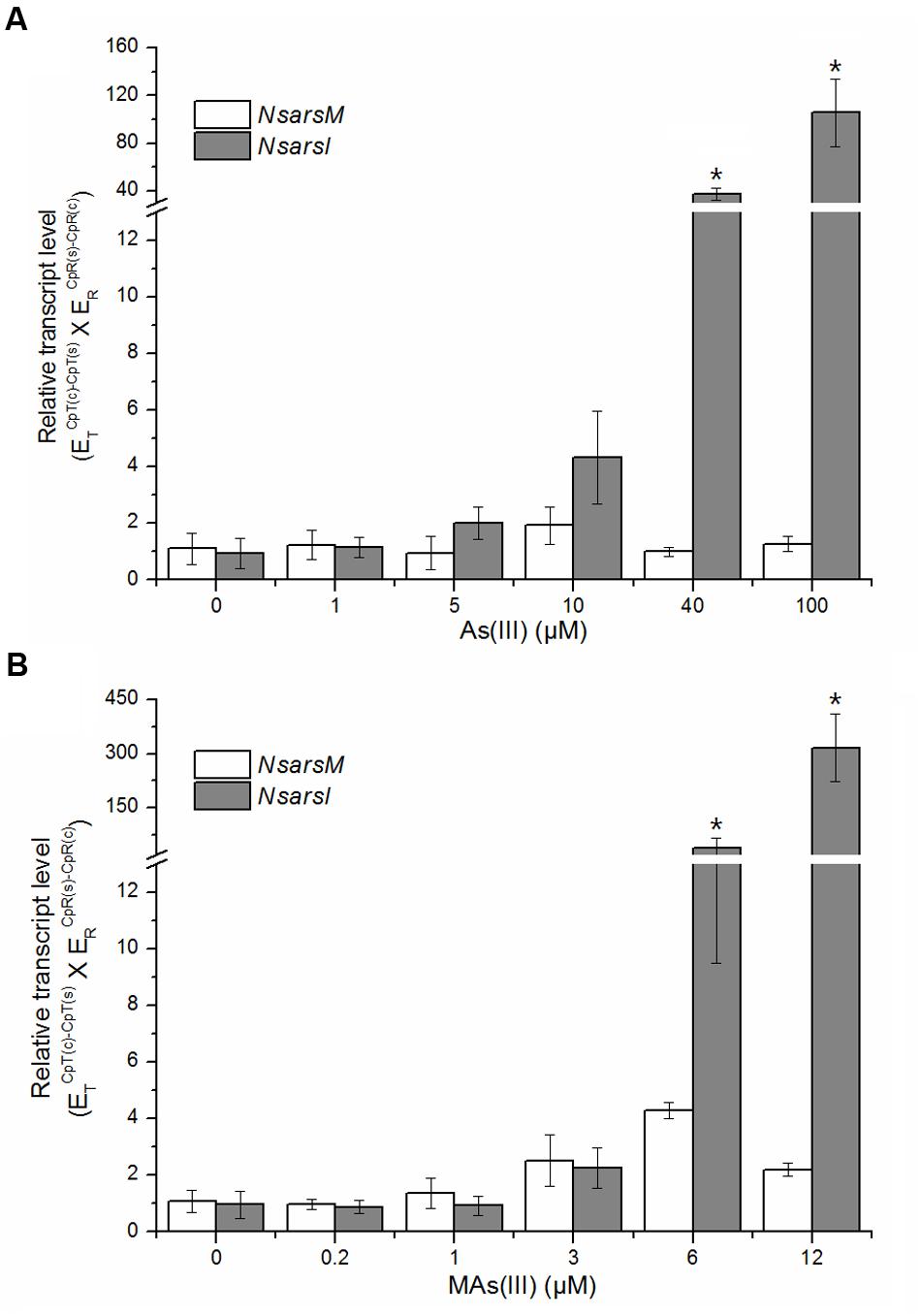
FIGURE 5. Effects of arsenic on the transcript levels of NsarsM (white bars) and NsarsI (gray bars). Mid-exponential Nostoc were exposed to indicated concentrations of As(III) (A) or MAs(III) (B) for 6 h, and NsarsM and NsarsI transcriptions were investigated by RT-qPCR analysis. The error bars indicate the standard error from three independent biological experiments. Asterisks represent significant difference (∗P < 0.05).
Discussion
Arsenic demethylation lowered the efficiency of arsenic methylation in the co-expressing E. coli. The arsenic methylation rate is an important factor to determine whether As(III) methylation is a detoxification mechanism (Rahman and Hassler, 2014). Cells of E. coli over-expressing various arsM genes rapidly methylate As(III) into DMAs(V) or TMAsO, conferring As(III) tolerance (Qin et al., 2006, 2009; Yin et al., 2011). Our results also showed that resistance of E. coli AW3110 expressing NsarsM to As(III) was significantly enhanced by methylating As(III) to DMAs(V) and TMAsO. However, when NsArsM and NsArsI coexisted, As(III) was mainly methylated to MAs(V) rather than DMAs(V). The result suggests that part of MAs dissociates from NsArsM as a methylated trivalent species. In E. coli AW3110 expressing NsarsM, free MAs(III) dissociated from NsArsM could bind to the enzyme again, and was further methylated to DMAs. While NsArsI coexisted with NsArsM, both of them may competitively bind to free MAs(III), MAs(III) released from NsArsM was more likely to be further oxidized to MAs(V). Herein, in addition to the direct transformation of methylated arsenic into inorganic species, competitive binding of ArsM and ArsI to MAs(III) may be another reason that arsenic demethylation limits methylation efficiency.
MAs(III) demethylation was slowed by arsenic methylation when NsarsM and NsarsI were co-expressed in E. coli. The competition between NsArsM and NsArsI with MAs(III) in co-expressed E. coli AW3110 may lead to the decrease of demethylation efficiency. The K0.5 value for MAs(III) of NsArsI was 7.55 μM in our previous study (Yan et al., 2015), and the Km value of NsArsM for MAs(III) was 37 μM in this study. It indicated that MAs(III) has a higher affinity for NsArsI than NsArsM, and MAs(III) preferred to be demethylated rather than be methylated. Therefore, both DMAs(V) and As(III) were detected when E. coli AW3110 co-expressing NsarsM and NsarsI was treated with MAs(III). Even though MAs(III) demethylation was limited, co-expression of NsarsM and NsarsI exhibited higher MAs(III) resistance than single expression of NsarsM due to the high detoxification efficiency of NsArsI for MAs(III).
So far, we demonstrated that arsenic methylation and demethylation may limit each other in E. coli co-expressing NsarsM and NsarsI, and we hypothesized that the regulations of NsarsM and NsarsI may be different to avoid functioning simultaneously, thus entering a futile cycle of methylation and demethylation, in Nostoc. So we analyzed the genome sequences of Nostoc, and found that NsarsM is not adjacent to an arsR, while NsarsI is in an ars operon containing four genes: asr1102 (homolog of arsenite efflux protein; arsB), all1103 (transcriptional regulator; arsR), alr1104 (arsI), and alr1105 (arsenate reductase; arsC) (Pandey et al., 2012). ArsR that regulates the expression of the ars operon is responsive to both As(III) and MAs(III) (Chen et al., 2014). In coincidence with this, the RT-qPCR results showed that NsarsI was induced by higher arsenic concentrations [As(III) at 40 and 100 μM; MAs(III) at 6 and 12 μM], while the expression of NsarsM is constitutive at all arsenic concentrations. Furthermore, the main product was As(V) in Nostoc treated with As(III) up to 100 μM (Yin et al., 2011), suggesting that arsenic methylation may not be the primary pathway of As(III) detoxification. This was also supported by proteomic data which implied arsM seems not to be up-regulated when Nostoc treated with 40 μM As(V) (Pandey et al., 2012). Recently, MAs(III) as the initial product of ArsM was proposed to be a primitive antibiotic produced by organisms at the early Earth (Chen et al., 2015b; Li et al., 2016). Thus, we propose that NsArsM may predominate to produce MAs(III) antibiotic at low concentrations in Nostoc, and since there seems no ArsH in Nostoc (Chen et al., 2015a), NsArsI takes over at high concentrations to protect the cells from excessive MAs(III).
Conclusion
We investigated the relationship between arsenic methylation and demethylation in cells that both arsM and arsI genes coexist. By assessing arsenic resistance and biotransformation by E. coli AW3110 expressing NsarsM or/and NsarsI and both genes’ regulation in Nostoc, we understand better the arsenic detoxification mechanisms in microorganisms.
Author Contributions
JY, Y-GZ, and YY conceived and designed the project. YY, X-MX, and Y-QG did the experiments. YY and X-MX analyzed the data. YY, X-MX, Y-GZ and JY wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work is supported by National Natural Science Foundation of China (21507125 and 31270161) and Natural Science Foundation of Fujian Province (2014J01141).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We thank Dr. Barry P. Rosen for valuable comments on this paper.
References
Carlin, A., Shi, W., Dey, S., and Rosen, B. P. (1995). The ars operon of Escherichia coli confers arsenical and antimonial resistance. J. Bacteriol. 177, 981–986. doi: 10.1128/jb.177.4.981-986.1995
Chen, J., Bhattacharjee, H., and Rosen, B. P. (2015a). ArsH is an organoarsenical oxidase that confers resistance to trivalent forms of the herbicide monosodium methylarsenate and the poultry growth promoter roxarsone. Mol. Microbiol. 96, 1042–1052. doi: 10.1111/mmi.12988
Chen, J., Madegowda, M., Bhattacharjee, H., and Rosen, B. P. (2015b). ArsP: a methylarsenite efflux permease. Mol. Microbiol. 98, 625–635. doi: 10.1111/mmi.13145
Chen, J., Sun, S., Li, C. Z., Zhu, Y. G., and Rosen, B. P. (2014). Biosensor for organoarsenical herbicides and growth promoters. Environ. Sci. Technol. 48, 1141–1147. doi: 10.1021/es4038319
Dheeman, D. S., Packianathan, C., Pillai, J. K., and Rosen, B. P. (2014). Pathway of human AS3MT arsenic methylation. Chem. Res. Toxicol. 27, 1979–1989. doi: 10.1021/tx500313k
Huertas, M. J., López-Maury, L., Giner-Lamia, J., Sánchez-Riego, A. M., and Florencio, F. J. (2014). Metals in cyanobacteria: analysis of the copper, nickel, cobalt and arsenic homeostasis mechanisms. Life 4, 865–886. doi: 10.3390/life4040865
Latifi, A., Jeanjean, R., Lemeille, S., Havaux, M., and Zhang, C. C. (2005). Iron starvation leads to oxidative stress in Anabaena sp. strain PCC 7120. J. Bacteriol. 187, 6596–6598. doi: 10.1128/JB.187.18.6596-6598.2005
Levy, J. L., Stauber, J. L., Adams, M. S., Maher, W. A., Kirby, J. K., and Jolley, D. F. (2005). Toxicity, biotransformation, and mode of action of arsenic in two freshwater microalgae (Chlorella sp. and Monoraphidium arcuatum). Environ. Toxicol. Chem. 24, 2630–2639. doi: 10.1897/04-580R.1
Li, J. J., Pawitwar, S. S., and Rosen, B. P. (2016). The organoarsenical biocycle and the primordial antibiotic methylarsenite. Metallomics 8, 1047–1055. doi: 10.1039/C6MT00168H
Liu, Z., Rensing, C., and Rosen, B. (2013). “Resistance pathways for metalloids and toxic metals,” in Metals in Cells, eds V. Culotta and R. A. Scott (Hoboken, NJ: Wiley & Sons, Inc), 429–442.
López-Maury, L., Florencio, F. J., and Reyes, J. C. (2003). Arsenic sensing and resistance system in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 185, 5363–5371. doi: 10.1128/JB.185.18.5363-5371.2003
López-Maury, L., Sánchez-Riego, A. M., Reyes, J. C., and Florencio, F. J. (2009). The glutathione/glutaredoxin system is essential for arsenate reduction in Synechocystis sp. strain PCC 6803. J. Bacteriol. 191, 3534–3543. doi: 10.1128/JB.01798-08
Maki, T., Watarai, H., Kakimoto, T., Takahashi, M., Hasegawa, H., and Ueda, K. (2006). Seasonal dynamics of dimethylarsenic acid degrading bacteria dominated in Lake Kibagata. Geomicrobiol. J. 23, 311–318. doi: 10.1080/01490450600761953
Marapakala, K., Packianathan, C., Ajees, A. A., Dheeman, D. S., Sankaran, B., Kandavelu, P., et al. (2015). A disulfide-bond cascade mechanism for arsenic(III) S-adenosylmethionine methyltransferase. Acta. Crystallogr. D Struct. Biol. 71, 505–515. doi: 10.1107/S1399004714027552
Mass, M. J., Tennant, A., Roop, B. C., Cullen, W. R., Styblo, M., Thomas, D. J., et al. (2001). Methylated trivalent arsenic species are genotoxic. Chem. Res. Toxicol. 14, 355–361. doi: 10.1021/tx000251l
Páez-Espino, D., Tamames, J., de Lorenzo, V., and Cánovas, D. (2009). Microbial responses to environmental arsenic. Biometals 22, 117–130. doi: 10.1007/s10534-008-9195-y
Pandey, S., Rai, R., and Rai, L. C. (2012). Proteomics combines morphological, physiological and biochemical attributes to unravel the survival strategy of Anabaena sp. PCC7120 under arsenic stress. J. Proteom. 75, 921–937. doi: 10.1016/j.jprot.2011.10.011
Pandey, S., Shrivastava, A. K., Rai, R., and Rai, L. C. (2013). Molecular characterization of Alr1105 a novel arsenate reductase of the diazotrophic cyanobacterium Anabaena sp. PCC7120 and decoding its role in abiotic stress management in Escherichia coli. Plant Mol. Biol. 83, 417–432. doi: 10.1007/s11103-013-0100-x
Petrick, J. S., Ayala-Fierro, F., Cullen, W. R., Carter, D. E., and Aposhian, H. V. (2000). Monomethylarsonous acid (MMA III) is more toxic than arsenite in Chang human hepatocytes. Toxicol. Appl. Pharmacol. 163, 203–207. doi: 10.1006/taap.1999.8872
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic. Acids. Res. 29:e45. doi: 10.1093/nar/29.9.e45
Qin, J., Lehr, C. R., Yuan, C., Le, X. C., McDermott, T. R., and Rosen, B. P. (2009). Biotransformation of arsenic by a Yellowstone thermoacidophilic eukaryotic alga. Proc. Natl. Acad. Sci. U.S.A. 106, 5213–5217. doi: 10.1073/pnas.0900238106
Qin, J., Rosen, B. P., Zhang, Y., Wang, G., Franke, S., and Rensing, C. (2006). Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 103, 2075–2080. doi: 10.1073/pnas.0506836103
Rahman, M. A., and Hassler, C. (2014). Is arsenic biotransformation a detoxification mechanism for microorganisms? Aquat. Toxicol. 146, 212–219. doi: 10.1016/j.aquatox.2013.11.009
Rosen, B. P. (2002). Biochemistry of arsenic detoxification. FEBS. Lett. 529, 86–92. doi: 10.1016/S0014-5793(02)03186-1
Sambrook, J., Fritsh, E., and Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual, 2nd. Edn. New York, NY: Cold Spring Harbor Laboratory Press.
Sánchez-Riego, A. M., López-Maury, L., and Florencio, F. J. (2014). Genomic responses to arsenic in the cyanobacterium Synechocystis sp. PCC 6803. PLoS ONE 9:e96826. doi: 10.1371/journal.pone.0096826
Srivastava, A. K., Bhargava, P., Thapar, R., and Rai, L. C. (2009). Differential response of antioxidative defense system of Anabaena doliolum under arsenite and arsenate stress. J. Basic Microbiol. 49, S63–S72. doi: 10.1002/jobm.200800301
Vioque, A. (1992). Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic. Acids. Res. 20, 6331–6337. doi: 10.1093/nar/20.23.6331
Walton, F. S., Waters, S. B., Jolley, S. L., LeCluyse, E. L., Thomas, D. J., and Styblo, M. (2003). Selenium compounds modulate the activity of recombinant rat AsIII-methyltransferase and the methylation of arsenite by rat and human hepatocytes. Chem. Res. Toxicol. 16, 261–265. doi: 10.1021/tx025649r
Wang, F., Qu, H., Zhang, D., Tian, P., and Tan, T. (2007). Production of 1, 3-propanediol from glycerol by recombinant E. coli using incompatible plasmids system. Mol. Biotech. 37, 112–119. doi: 10.1007/s12033-007-0041-1
Wang, S., Li, X., Song, X., Geng, Z., Hu, X., and Wang, Z. (2012). Rapid equilibrium kinetic analysis of arsenite methylation catalyzed by recombinant human arsenic (+3 oxidation state) methyltransferase (hAS3MT). J. Biol. Chem. 287, 38790–38799. doi: 10.1074/jbc.M112.368050
Wang, Z., Luo, Z., and Yan, C. (2013). Accumulation, transformation, and release of inorganic arsenic by the freshwater cyanobacterium Microcystis aeruginosa. Environ. Sci. Pollut. Res. 20, 7286–7295. doi: 10.1007/s11356-013-1741-7
Yan, Y., Ye, J., Xue, X. M., and Zhu, Y. G. (2015). Arsenic demethylation by a C As lyase in cyanobacterium Nostoc sp. PCC 7120. Environ. Sci. Technol. 49, 14350–14358. doi: 10.1021/acs.est.5b03357
Yang, W., Zhang, L., Lu, Z., Tao, W., and Zhai, Z. (2001). A new method for protein coexpression in Escherichia coli using two incompatible plasmids. Protein Expr. Purif. 22, 472–478. doi: 10.1006/prep.2001.1453
Ye, J., Chang, Y., Yan, Y., Xiong, J., Xue, X. M., Yuan, D., et al. (2014). Identification and characterization of the arsenite methyltransferase from a protozoan, Tetrahymena pyriformis. Aquat. Toxicol. 149, 50–57. doi: 10.1016/j.aquatox.2014.01.028
Yin, X. X., Chen, J., Qin, J., Sun, G. X., Rosen, B. P., and Zhu, Y. G. (2011). Biotransformation and volatilization of arsenic by three photosynthetic cyanobacteria. Plant. Physiol. 156, 1631–1638. doi: 10.1104/pp.111.178947
Yin, X. X., Wang, L., Bai, R., Huang, H., and Sun, G. X. (2012). Accumulation and transformation of arsenic in the blue-green alga Synechocysis sp. PCC6803. Water Air Soil Pollut. 2012, 1183–1190. doi: 10.1007/s11270-011-0936-0
Yoshinaga, M., Cai, Y., and Rosen, B. P. (2011). Demethylation of methylarsonic acid by a microbial community. Environ. Microbiol. 13, 1205–1215. doi: 10.1111/j.1462-2920.2010.02420.x
Yoshinaga, M., and Rosen, B. P. (2014). A CAs lyase for degradation of environmental organoarsenical herbicides and animal husbandry growth promoters. Proc. Natl. Acad. Sci. U.S.A. 111, 7701–7706. doi: 10.1073/pnas.1403057111
Zhang, J. D., Li, A. T., Yu, H. L., Imanaka, T., and Xu, J. H. (2011). Synthesis of optically pure S-sulfoxide by Escherichia coli transformant cells coexpressing the P450 monooxygenase and glucose dehydrogenase genes. J. Ind Microbiol. Biotechnol. 38, 633–641. doi: 10.1007/s10295-010-0809-3
Zhang, S. Y., Sun, G. X., Yin, X. X., Rensing, C., and Zhu, Y. G. (2013). Biomethylation and volatilization of arsenic by the marine microalgae Ostreococcus tauri. Chemosphere 93, 47–53. doi: 10.1016/j.chemosphere.2013.04.063
Zhu, Y. G., Sun, G. X., Lei, M., Teng, M., Liu, Y.-X., Chen, N.-C., et al. (2008). High percentage inorganic arsenic content of mining impacted and nonimpacted Chinese rice. Environ. Sci. Technol. 42, 5008–5013. doi: 10.1021/es8001103
Keywords: arsenic methylation, arsenic demethylation, arsenic resistance, MAs(III) antibiotic, Nostoc sp. PCC 7120
Citation: Yan Y, Xue X-M, Guo Y-Q, Zhu Y -G and Ye J (2017) Co-expression of Cyanobacterial Genes for Arsenic Methylation and Demethylation in Escherichia coli Offers Insights into Arsenic Resistance. Front. Microbiol. 8:60. doi: 10.3389/fmicb.2017.00060
Received: 05 November 2016; Accepted: 10 January 2017;
Published: 24 January 2017.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
Luis López-Maury, IBVF Universidad de Sevilla – CSIC, SpainBP Rosen, Herbert Wertheim College of Medicine, Florida International University, USA
Copyright © 2017 Yan, Xue, Guo, Zhu and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Ye, jye@iue.ac.cn
 Yu Yan
Yu Yan Xi-Mei Xue1
Xi-Mei Xue1 Jun Ye
Jun Ye