- 1Department of Infectious Diseases, University of Georgia, Athens, GA, USA
- 2Center for Vaccines and Immunology, University of Georgia, Athens, GA, USA
- 3Department of Veterinary and Biomedical Sciences, Pennsylvania State University, University Park, PA, USA
Members of the genus Bordetella include human and animal pathogens that cause a variety of respiratory infections, including whooping cough in humans. Despite the long known ability to switch between a within-animal and an extra-host lifestyle under laboratory growth conditions, no extra-host niches of pathogenic Bordetella species have been defined. To better understand the distribution of Bordetella species in the environment, we probed the NCBI nucleotide database with the 16S ribosomal RNA (16S rRNA) gene sequences from pathogenic Bordetella species. Bacteria of the genus Bordetella were frequently found in soil, water, sediment, and plants. Phylogenetic analyses of their 16S rRNA gene sequences showed that Bordetella recovered from environmental samples are evolutionarily ancestral to animal-associated species. Sequences from environmental samples had a significantly higher genetic diversity, were located closer to the root of the phylogenetic tree and were present in all 10 identified sequence clades, while only four sequence clades possessed animal-associated species. The pathogenic bordetellae appear to have evolved from ancestors in soil and/or water. We show that, despite being animal-adapted pathogens, Bordetella bronchiseptica, and Bordetella hinzii have preserved the ability to grow and proliferate in soil. Our data implicate soil as a probable environmental origin of Bordetella species, including the animal-pathogenic lineages. Soil may further constitute an environmental niche, allowing for persistence and dissemination of the bacterial pathogens. Spread of pathogenic bordetellae from an environmental reservoir such as soil may potentially explain their wide distribution as well as frequent disease outbreaks that start without an obvious infectious source.
Introduction
Bacteria of the genus Bordetella are of primary importance in human and veterinary medicine because of their ability to colonize the respiratory tract, causing a wide range of pulmonary and bronchial infections. The common human- and animal-adapted pathogens B. pertussis, B. parapertussis, and B. bronchiseptica are known as the “classical” Bordetella species. B. pertussis is a strictly human pathogen, but B. parapertussis consists of two lineages, one infecting humans and the other infecting sheep (Mattoo and Cherry, 2005). In contrast to these examples of adaptation to a single host, B. bronchiseptica colonizes a variety of animals and even humans (Register et al., 2015), resulting in a broad array of respiratory diseases, from asymptomatic colonization to lethal pneumonia (Goodnow, 1980). Phylogenetic analyses (Musser et al., 1986; Diavatopoulos et al., 2005) and genome comparisons (Parkhill et al., 2003) have revealed that B. pertussis and B. parapertussis represent human-adapted forms of B. bronchiseptica that have evolved independently from a B. bronchiseptica-like ancestor. The genus also contains a number of additional, more recently described species. For example, B. avium (Kersters et al., 1984) causes respiratory disease in birds. B. hinzii (Vandamme et al., 1995) is generally regarded as a non-pathogenic colonizer of the respiratory tract of poultry but some strains were shown to cause disease in turkeys when experimentally inoculated (Register and Kunkle, 2009). Meanwhile, the closely related species B. pseudohinzii colonizes laboratory mice (Ivanov et al., 2015, 2016). B. holmesii (Weyant et al., 1995) causes pertussis-like disease and septicemia in humans (Shepard et al., 2004), and B. bronchialis, B. flabilis, and B. sputigena (Vandamme et al., 2015) were also isolated from human respiratory specimens. In contrast to other bordetellae, B. trematum (Vandamme et al., 1996) and B. ansorpii (Ko et al., 2005) are not associated with respiratory problems but were isolated from human wound infection.
B. petrii, a species originally isolated from a dechlorinating bioreactor enriched by river sediment, represents the first described environmental species within the Bordetella genus (von Wintzingerode et al., 2001). B. petrii strains were also found in marine sponges (Wang et al., 2007), grass root consortia (Wang et al., 2007), and in other environmental samples as members of microbial communities involved in the degradation of aromatic hydrocarbons, such as benzenes (Bianchi et al., 2005; Wang et al., 2007). In apparent conflict with its environmental source, the B. petrii genome contains genes that allow for the synthesis and secretion of factors specifically associated with the virulence of the pathogenic Bordetella sp., including the BvgAS master regulon and filamentous hemagglutinin (Gross et al., 2008). In addition to these environmental sources, B. petrii was also isolated from immunocompromised patients with ear infection, cystic fibrosis and chronic pulmonary disease (Fry et al., 2005; Biederman et al., 2015; Nagata et al., 2015), suggesting broad adaptability of this bacterial species to both environmental conditions and as an opportunistic pathogen of humans and possibly other animals.
Other Bordetella species have been obtained from environments not associated with animal hosts. Ten different bacterial strains were cultured from cotton swabs taken from the plaster wall surface of 1300-year-old mural paintings inside the stone chamber of the Takamatsuzuka Tomb, an ancient circular burial mound in Japan. Taxonomic classification of these isolates revealed three novel species that were then named B. muralis, B. tumulicola, and B. tumbae (Tazato et al., 2015). Isolation of the bacteria from the paintings, but not from the surrounding stone walls, suggested that these species might be involved in the observed biodeterioration of the colorful paintings (Kigawa et al., 2013).
According to their 16S rRNA gene sequences, other environmental bacteria from soil also belong to the genus Bordetella. Interestingly, the majority of those samples originated from contaminated sites such as soil polluted with chlorinated benzenes (Wang et al., 2007), from arctic soils contaminated with polycyclic aromatic hydrocarbons such as oil, diesel fuel or tar (Eriksson et al., 2003), from the sediment of a municipal wastewater plant (Nisola et al., 2010) and from arsenic polluted soils (Cavalca et al., 2010; Bachate et al., 2012). All these observations suggest that members of the Bordetella genus may have the potential for biodegradation of a great variety of organic compounds.
Although these anecdotal findings suggest that members of the Bordetella genus may be found in nature, there is currently no systematic analysis of the occurrence of Bordetella outside human or animal hosts, and the potential impact of environmental isolates on human and animal health is uncertain. Environmental niches of pathogenic Bordetella species have been proposed but not identified. Yet, the ability of Bordetella to survive and persist outside mammalian hosts would allow for its greater dissemination and persistence, and could contribute to a wide distribution of infections and disease, often without an obvious infectious source.
Here, we search the NCBI nucleotide database for 16S ribosomal RNA gene sequences of Bordetella-like microorganisms from various environments and compare them to those of the described species, including the classical bordetellae, to determine their phylogenetic relatedness. We identified 10 clades of related strains, all of which contained samples isolated from environmental sources, though only four clades also contained sequences from animal-associated species. Sequences from environmental samples had a significantly higher genetic diversity and were located closer to the root of the phylogenetic tree than those from animal-associated isolates, suggesting an environmental origin of the genus Bordetella. In addition, we show that the animal-adapted pathogens B. bronchiseptica and B. hinzii grow efficiently in soil extract, indicating that diverse pathogenic bordetellae may have retained the ability to proliferate in the environment.
Materials and Methods
Search for Bordetella 16S rRNA Gene Sequences in the NCBI nt Database
The16S ribosomal gene sequences of Bordetella hinzii strain LMG 13501 (GenBank accession number NR_027537.1); Bordetella holmesii strain ATCC 51541 (NR_121717.1); and Bordetella pertussis strain Tohama I (AF142326.1) were each used as queries for BLAST search (blastn) against the NCBI nr/nt database using the default search parameters with a hitlist size of 5000. From the numerous hits, we excluded sequences of isolates from the known species that are mentioned in the introduction and selected only those that showed higher percentage of similarity to known Bordetella species than to bacteria from any other genus, including Achromobacter. As a control, we ran blastn searches with each of the identified sequences against the NCBI nr/nt database to remove potential false positives. The remaining sequences, all of which were from bacteria obtained from environmental sources, were considered for further analysis. All three searches using 16S rRNA sequences of B. pertussis, B. hinzii, and B. holmesii as queries, respectively, gave consistent results. The accession numbers were then explored for details on sample source, year of isolation, and associated publications. Most sequences were described as Bordetella sp. in the gene description, but some were designated as “uncultured bacterium.”
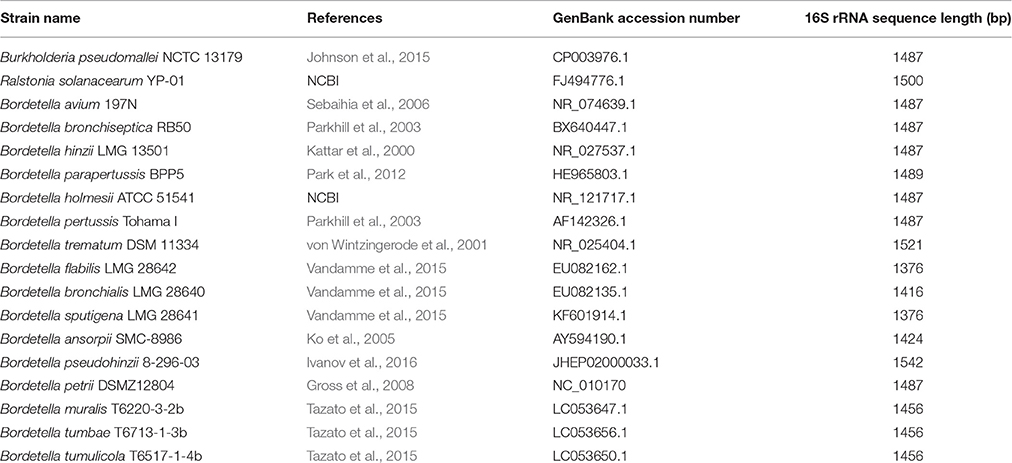
Phylogenetic Analysis and Tree Construction
All 16S rRNA sequences were aligned in Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/), and the alignment was checked manually for consistency. Only sequences containing a 1376 bp gene fragment (near full-length) were used for further analyses. In order to identify the closely related species of environmental Bordetella isolates, the 16S ribosomal RNA gene sequences of members of 16 named Bordetella species were used as references; namely B. pertussis Tohama I, B. parapertussis BPP5, B. bronchiseptica RB50, B. avium 197N, B. hinzii LMG 13501, B. pseudohinzii 8-296-03, B. holmesii ATCC 51541, B. trematum DSM 11334, B. ansorpii SMC-8986, B. bronchialis LMG 28640, B. sputigena LMG 28641, B. flabilis LMG 28642, B. petrii Se-1111R, B. muralis T6220-3-2b, B. tumulicola T6517-1-4b, and B. tumbae T6713-1-3b (Table 1). The 16S rRNA gene sequences of Burkholderia pseudomallei NCTC13179 and Ralstonia solanacearum YP-01 were used as outgroups. The aligned and trimmed sequences (one per unique sequence) were used to generate a Neighbor-joining tree using the Maximum Composite Likelihood algorithm in MEGA (Tamura et al., 2007), and bootstrap support was estimated running 100,000 replications. Nucleotide diversity (Π) within environmental samples and within animal-associated samples were estimated in DnaSP (Librado and Rozas, 2009), and 95% confidence limits (Π95) were estimated using an online confidence limit calculator (https://www.allto.co.uk/tools/statistic-calculators/confidence-interval-for-mean-calculator/).
Soil Sample Collection and Bordetella Growth in Soil Extract
Soils were sampled in April 2016 at two random sites in State College, Pennsylvania, near a suburban park (40°48′40.7″ N 77°53′06.1″ W and 40°48′38.2″ N 77°53′04.2″ W). Each sample was collected to a depth of 20 cm and thoroughly mixed. Fifty grams of each soil sample (100 g total) was placed in a bottle which was filled to 500 ml with sterile PBS. The sample was homogenized by shaking for 10 min, then left to settle for 1 h at room temperature and carefully decanted. The soil-PBS suspension was filter sterilized. Single colonies of B. bronchiseptica strain RB50, B. hinzii strain L60, and B. petrii strain DSMZ12804 were picked from Bordet-Gengou (BG) agar (Difco) plates supplemented with 10% defibrinated Sheep's blood (HemoStat Laboratories, Dixon, CA, USA) and were cultured in liquid Stainer-Scholte medium (Stainer and Scholte, 1970) overnight at 37°C. The Bordetella inocula were prepared as follows. The cultures were centrifuged, resuspended in 1 ml PBS, and the optical density (OD600) was determined. Following five consecutive 10-fold dilutions in 1 ml PBS, 100 μl (= 106-fold dilution) containing ~150 (B. petrii) or 240 bacterial cells (B. hinzii or B. bronchiseptica) were added to 5 ml of the soil extract resulting in starting concentrations of ~30 bacterial cells/ml (B. petrii) and 48 bacterial cells/ml (B. hinzii, B. bronchiseptica). Bacterial numbers were determined by plating an aliquot of each inoculum. The culture tubes were incubated at room temperature (25°C) with shaking. After 24, 48, and 72 h, 100 μl of each culture was plated on BG agar supplemented with 10% defibrinated sheep's blood to determine bacterial numbers. Each experiment was carried out in triplicate. The mean and ± standard error as well as analysis of variance (ANOVA) were conducted using Graphpad Prism version 6.04. The bacterial doubling time was calculated by the formula: doubling time = ln(2)/ln(N(t)/N(0))/t, where N(t) is the number of bacterial cells at time t, N(0) is the number of bacteria at time 0 and t is the time in hours.
Results
Bordetella in the Environment
We mined the NCBI nucleotide databases for Bordetella spp. 16S rRNA gene sequences. The search resulted in a total of 71 Bordetella spp. 16S rRNA gene sequences (Table 2) in addition to those from the named species (Table 1) B. bronchiseptica, B. parapertussis, B. pertussis, B. hinzii, B. pseudohinzii, B. holmesii, B. avium, B. trematum, B. ansorpii, B. flabilis, B. bronchealis, B. sputigena (isolated from samples of human/animal origin), B. petrii, B. tumbae, B. muralis, and B. tumulicola (isolated from environmental samples). The corresponding strains were recovered from different environmental niches (Table 2), including soil (52 strains) and water (11 strains), and from 8 strains associated with plants. The soil samples were of diverse origin, including compost, cave rocks, and metal mines, but the majority were sampled at sites contaminated with oil and several halogenated cyclic hydrocarbons such as chlorinated benzenes or hexachlorocyclohexane. The samples from aquatic environments were also of diverse origin, namely industrial wastewater, a sulfur spring, lake water, surface sea water, and river biofilms. Several samples from plants were isolated from roots and thus at the plant-soil interface (Table 2). Thus, members of the genus Bordetella appear to be widespread across different environmental niches.
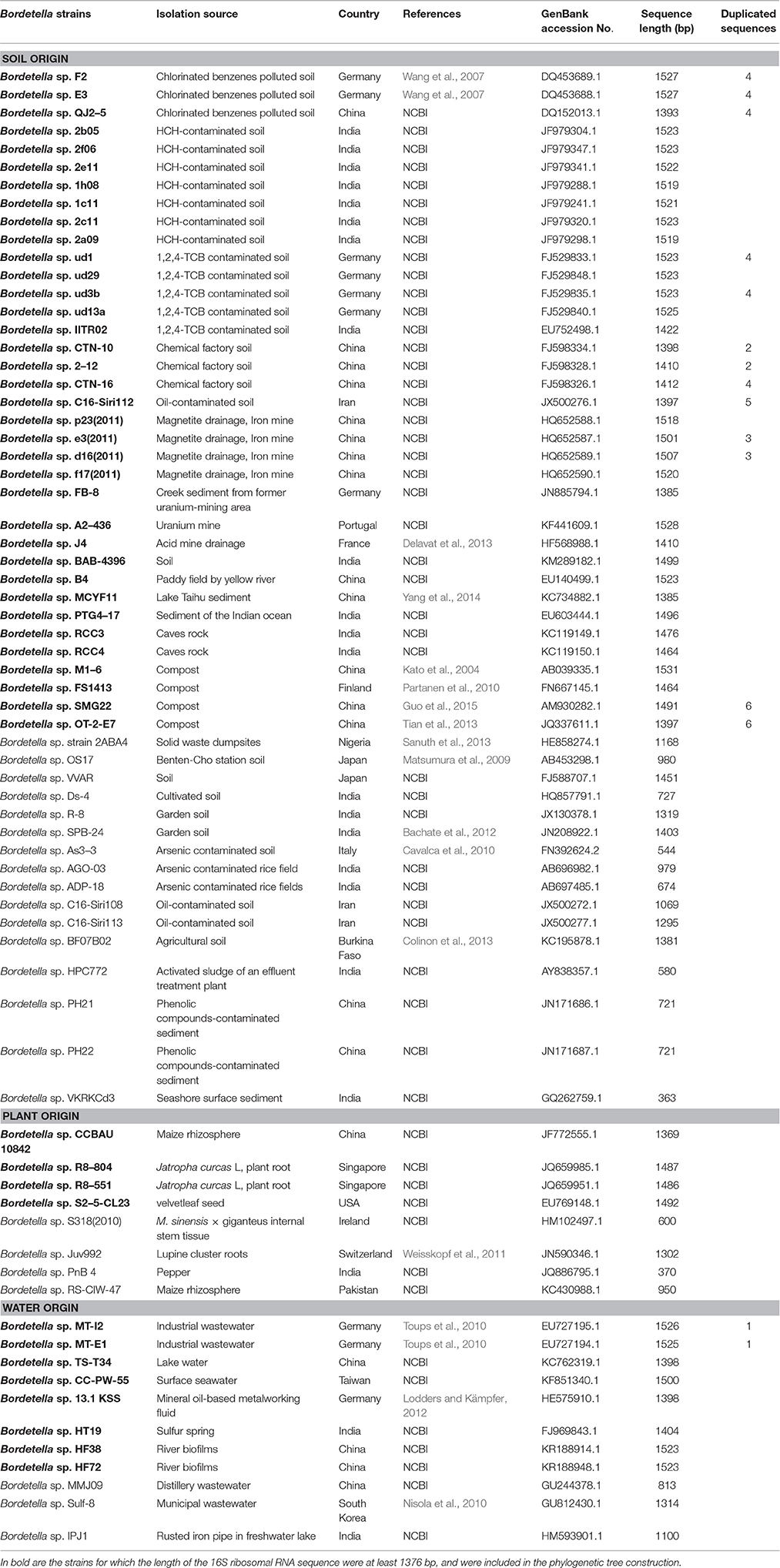
Table 2. Bordetella strains for which the 16S ribosomal RNA sequences were recovered from environmental samples.
16S rRNA Gene Sequence Clades are Associated with Particular Environmental Niches
To relate the environmental isolates to known Bordetella species, we aligned the 16S rRNA gene sequences and constructed a Neighbor-joining tree using the Maximum-likelihood algorithm implemented in MEGA (Tamura et al., 2007). Forty-eight sequences from environmental samples were of sufficient length and used for further analyses (Table 2). Of those, 36 originated from soil (27 haplotypes), eight from aquatic environments (7 haplotypes), and four from plants (4 haplotypes). The tree was rooted with sequences of Burkholderia pseudomallei and Ralstonia solanacearum as outgroups. The Bordetella sequences formed 10 distinct clusters (Figure 1). While most clusters contained at least one described species, such as B. petrii in cluster VI or B. tumbae/B. muralis in cluster V, several Bordetella sequences did not cluster with any described species but rather occupied distinct branches of the tree. These include the two isolates in cluster IV, the isolates from soil samples in clusters VII and X and strains B. sp. CC-PW-55 and B. sp. TS-T34 (cluster IX) isolated from surface sea water and lake water, respectively (Figure 1).

Figure 1. Neighbor-Joining tree based on 16S rRNA gene sequences of animal-associated and environmental strains of Bordetella. The 52 near full-length sequences (1376 bp) formed 10 clades (I–X) of phylogenetically closely related Bordetella isolates/species recovered from soil (brown), water (blue), plants (green) and animals (black). The 16S rRNA gene sequences of the beta-proteobacteria Burkholderia pseudomallei and Ralstonia solanacearum were used as outgroups.
Superimposing the origin of the Bordetella spp. isolates revealed that most of the identified clusters were dominated by sequences of similar environmental/host origin. Thus, cluster I was composed of sequences of B. holmesii and the classical bordetellae (B. bronchiseptica, B. parapertussis, and B. pertussis), all of which were isolated from human and animal infection, but also contained B. sp. HT38 isolated from a river biofilm in China (Figure 1, Table 2). Cluster III contained sequences of species isolated from human respiratory specimen (B. sputigena, B. bronchialis, and B. flabilis) plus an isolate from soil in India. Other clusters either contained, or were dominated by, sequences of environmental origin such as cluster IV (water and soil), cluster V (soil), including the three species recovered from mural paintings B. tumbae, B. tumulicola, and B. muralis; but also B. sp. CCBAU from a maize rhizosphere and B. ansorpii from infection of an immunocompromised patient, and cluster VI (soil, including the environmental species B. petrii). The prominent exception to this pattern, cluster II, contained sequences from animal/human infection (B. avium, B. hinzii, B. pseudohinzii, and B. trematum) as well as from water (B. sp. MT-E1, B. sp. HF27), plant root (B. sp. R8–804, B. sp. R8–551), and soil samples (B. sp. BAB-4396). However, the other clusters were either dominated by animal-associated samples (clusters I and III) or samples of environmental origin (all other clusters).
If the genus Bordetella were of environmental origin, samples isolated from soil and water would be expected to be more diverse and would appear widespread across the tree. Indeed, environmental samples were present in all sequence clusters. In contrast, sequences from animal-associated samples were confined to four clusters, all of which also contained environmental isolates. Three of those four clusters formed a single super clade which originated from one of several clades among sequences from environmental isolates. In contrast, all clusters near the tree root exclusively contained environmental samples, but no animal associated samples (Figure 1). The phylogenetic analyses showed that the genetic diversity was significantly higher in sequences from environmental samples (Π95 = 2.02–2.13%) than in sequences from animal-associated samples (Π95 = 1.30–1.53%). The sequence of branching events within the phylogenetic tree is consistent with an environmental origin of Bordetella and subsequent adaptation of some lineages to animal hosts.
Bordetella bronchiseptica and Bordetella hinzii are Capable of Growing in Soil Extract
Since most environmental Bordetella samples were recovered from soil (and water), we hypothesized that pathogenic, animal-associated species may have retained the ability to thrive in soil as an environmental niche. Therefore, we assessed the ability of B. bronchiseptica strain RB50, B. hinzii strain L60, and B. petrii strain DSMZ12804, to grow in a sterile, homogenized suspension made from soil. Instead of growing pathogenic bordetellae directly on solid soil, we prepared a soil suspension to extract possible nutrients but to avoid solid matter which allowed visual monitoring of bacterial growth and selection of appropriate sampling time points. All three isolates were cultured at room temperature (25°C) with shaking in either liquid soil extract or in Stainer-Scholte (SS) medium as a control. All three species grew fast in SS medium with doubling times of 1.8 ± 0.02 h (B. bronchiseptica), 1.9 ± 0.01 h (B. hinzii), and 1.9 ± 0.02 h (B. petrii), and reached the stationary phase prior to 48 h post-inoculation (Figure 2). As expected from an environmental bacterium, B. petrii strain DSMZ12804 thrived when inoculated into a soil extract, with a doubling time of 7.25 ± 0.24 h (Figure 2). Surprisingly, both B. hinzii strain L60 with a doubling time of 6.4 ± 0.09 h and B. bronchiseptica strain RB50 with a doubling time of 4.0 ± 0.04 h grew in the soil extract faster than B. petrii. Thus, all three species can grow efficiently at 25°C on filter-sterilized soil extract, even though the growth rate was slower than in Stainer-Scholte medium.
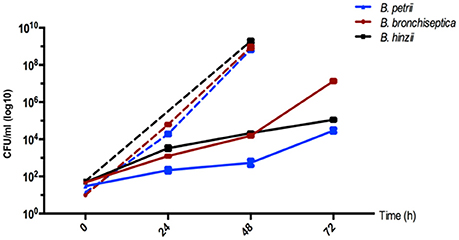
Figure 2. Growth of B. bronchiseptica strain RB50, B. hinzii strain L60, and B. petrii strain DSMZ12804 in soil extract (solid lines) and in Stainer-Scholte medium (dashed lines). All three bacterial species efficiently grow in a sterile-filtered soil suspension suggesting that soil may represent an environmental niche for pathogenic Bordetella species.
Discussion
Bacteria of the genus Bordetella occupy remarkably diverse ecological niches, ranging from soil, water, and plants, to the respiratory tracts of a wide variety of animals including humans. Several environmental Bordetella strains were isolated from soils polluted with oil and oil derivatives (Table 2), including halogenated polycyclic hydrocarbons (Eriksson et al., 2003; Bianchi et al., 2005; Wang et al., 2007). Other strains were found in garden soil, compost, and various sediments suggesting these organisms are quite adaptable to diverse sites. The only sequenced and analyzed genome of an environmental isolate, B. petrii strain DSMZ 12804, revealed a possible genomic basis for substantial metabolic versatility (Gross et al., 2008). The genome encodes multiple auxiliary pathways for the utilization of a variety of nutrients, including pectate, numerous sugar derivatives from degraded plant products and various aromatic compounds. Five of the eight genomic islands that have been identified in this genome contain genes coding for enzymes for the metabolism of aromatic compounds, particularly clusters of genes encoding enzymes of the chlorocatechol pathway, including gene clusters that show high similarity to genes in a 1,2,4-trichlorobenzene-degrading Pseudomonas strain (Gross et al., 2008). The presence of multiple chlorocatechol gene clusters in addition to several different central pathways for aromatic metabolism may provide a competitive advantage for growth in contaminated environments.
Another striking feature of environmental Bordetella isolates is their resistance to heavy metals (Cavalca et al., 2010). Ten out of 52 soil samples (Table 2) were isolated from iron mines (e.g., B. sp. d16, B. sp. f17), from uranium mines (B. sp. FB-8, B. sp. A2–436), or from soil polluted with arsenic (e.g., B. sp. As3–3). Such remarkable metal tolerance is most likely conferred by heavy metal resistance systems. Indeed, the genome of B. petrii strain DSMZ 12804 contains several heavy metal resistance operons on a genomic island absent from the genomes of other sequenced bordetellae, whereas other strains contain different islands of genes. Ultimately, the presence of multiple heavy metal resistance systems may allow environmental Bordetella isolates to thrive in metal rich environments.
Most plant-associated Bordetella strains were recovered from roots (B. sp. R8–804, B. sp. R8–551, B. sp. Juv992) and the rhizosphere at the plant-soil interface (B. sp. CCBAU 10842). Thus, these isolates may in fact represent soil samples or, alternatively, may be involved in interactions with plants at the plant-soil interface. The resemblance between plant responses to bacterial virulence factors and the responses of mammalian immune cells (Berg et al., 2005) serve as evidence that bacteria-plant interactions may have paved the way for bacterial adaptation to animals. In this regard, plant-root isolates B. sp. R8–804 and B. sp. R8–551 from plant roots are closely related to bird pathogens, B. hinzii and B. avium, supporting the view that plants cells could serve as a “training ground” for environmental strains that eventually gain the ability to colonize animal hosts (Berg et al., 2005).
In addition to these plant root isolates, several other environmental isolates were also found to be very closely related to animal-associated pathogens (Figure 1). Interestingly, those strains were isolated from very diverse sources, namely (polluted) soil in India (B. sp. BAB-4396, B. sp. IITR02), industrial waste water (B. sp. MT-E1), and oil-based metal-working emulsion in Germany (B. sp. 13.1 KSS), as well as from river biofilms in China (B. sp. HF38 and B. sp. HF72). The two isolates from a river biofilm in China are of particular interest. The 16S rRNA sequence of one of those (strain HF72) showed 99.56% sequence similarity to that of the human pathogen B. trematum (6 SNPs). According to 16S rRNA gene sequence, the other isolate (B. sp. HF38) is even more closely related (99.78%, three SNPs) to the animal pathogen B. bronchiseptica strain RB50 and the human pathogen B. parapertussis strain 12822, which share an identical sequence in this gene. By this measure, isolate HF38 is as closely related to B. bronchiseptica strain RB50 and B. parapertussis strain 12822 as it is to B. pertussis. This exceptionally close phylogenetic relatedness makes several evolutionary scenarios conceivable. First, isolate B. sp. HF38 may be an environmental, non-pathogenic strain closely related to the animal/human pathogens among the classical bordetellae. Second, this isolate might be a descendant or relative of an ancestor of the classical bordetellae which later became pathogenic after acquisition of several virulence-associated factors, such as pertussis toxin, adenylate cyclase toxin, and dermonecrotic toxin. Third, this isolate may in fact represent a B. bronchiseptica or B. parapertussis strain that naturally survives and/or grows within an environmental reservoir. Although the classical bordetellae have not yet been isolated from outside a mammalian host, our results suggest that animal-pathogenic Bordetella species retain the ability to grow in soil as an environmental niche. This implies that B. bronchiseptica and other species might be found (at least transiently) in soil, for example at farms with suitable animal hosts such as cattle, pig, sheep and horse, or near dog kennels. Interestingly, even fastidious B. pertussis bacteria remained able to be cultured for up to 5 days when spread onto various hospital-setting surfaces such as fabrics, plastics, glass, and paper, and also in several infant foods (Ocklitz and Milleck, 1967). Fourth, B. sp. HF38 as well as other isolates from water and soil may be protected internally by a non-vertebrate host. For example, amoebae are known to host bacteria such as Legionella pneumophila (Molmeret et al., 2005), and amoeba-grown L. pneumophila exhibited radically increased resistance to harsh environmental conditions such as fluctuations in temperature, osmolarity, acidity, as well as to biocides that may facilitate bacterial survival and persistence in the environment (Barker et al., 1995; Abu Kwaik et al., 1997, 1998; Winiecka-Krusnell and Linder, 1999). Amoebae are ubiquitously found in most environments, and shared habitats between amoeba and Bordetella could be an important factor for the persistence of the bacteria. Indeed, our group has shown that the animal-adapted B. bronchiseptica is able to survive and multiply intracellularly in the trophozoites and sori of the amoeba Dictyostelium discoideum before being disseminated with the amoeba spores to novel geographical locations (Bendor et al., in revision). Thus, in addition to our recent data demonstrating that B. bronchiseptica can circulate and efficiently transmit amongst mammals, these data demonstrate that this species can also grow and disseminate efficiently in association with amoebae. These independent but interconnected Bordetella lifecycles allow for disease propagation, transmission, and re-emergence in the absence of an infected animal host.
Strains included in this study were identified as Bordetella spp. based on their 16S rRNA gene sequence. Currently, there are no data available regarding potential pathogenicity of these species. Whole genome sequencing will provide valuable insights into the evolution and ecology of environmental vs. animal-pathogenic bordetellae. Of special interest are environmental isolates closely related to animal pathogens, particularly isolate B. sp. HF38, and analysis of their genomes will reveal whether they are non-pathogenic relatives of known animal pathogens or if they in fact represent environmental reservoirs of B. bronchiseptica or B. parapertussis.
Finally, the majority of environmental B. sp. were recovered from soil samples indicating that soil could be the most frequent natural habitat of bordetellae. Indeed, sequences identified from soil samples were found in 8 of 10 sequence clusters, including samples from compost in cluster X at the root of the tree (Figure 1). The sequence of branching events within the phylogenetic tree, the significantly higher sequence diversity in samples from soil and water than in those from animals, as well as the preserved ability of animal pathogens to grow in soil, suggest an environmental, likely soil-based, origin of the genus Bordetella. Thus, similar to bacteria of the closely related genus Achromobacter, which are of environmental origin but also contain opportunistic pathogens (Li et al., 2013), Bordetella appears to be a bacterium of environmental origin that adapted and became pathogenic via the acquisition of factors mediating specific interactions with animal hosts.
Author Contributions
IHS, BL, and ETH conceived and designed the experiments. IHS and BL performed the experiments and analyzed the data. IHS, BL and ETH wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Holly Vuong, Monica Cartelle Gestal, and Israel Rivera from the Harvill lab for helpful discussions. This work was supported by grants GM113681 and AI116186 by the National Institutes of Health (to ETH).
References
Abu Kwaik, Y., Gao, L. Y., Harb, O. S., and Stone, B. J. (1997). Transcriptional regulation of the macrophage-induced gene (gspA) of Legionella pneumophila and phenotypic characterization of a null mutant. Mol. Microbiol. 24, 629–642. doi: 10.1046/j.1365-2958.1997.3661739.x
Abu Kwaik, Y., Gao, L. Y., Stone, B. J., Venkataraman, C., and Harb, O. S. (1998). Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl. Environ. Microbiol. 64, 3127–3133.
Bachate, S. P., Khapare, R. M., and Kodam, K. M. (2012). Oxidation of arsenite by two ß-proteobacteria isolated from soil. Appl. Microbiol. Biotechnol. 93, 2135–2145. doi: 10.1007/s00253-011-3606-7
Barker, J., Scaife, H., and Brown, M. R. (1995). Intraphagocytic growth induces an antibiotic-resistant phenotype of Legionella pneumophila. Antimicrob. Agents Chemother. 39, 2684–2688. doi: 10.1128/AAC.39.12.2684
Berg, G., Eberl, L., and Hartmann, A. (2005). The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 7, 1673–1685. doi: 10.1111/j.1462-2920.2005.00891.x
Bianchi, F., Careri, M., Mustat, L., Malcevschi, A., and Musci, M. (2005). Bioremediation of toluene and naphthalene: development and validation of a GC-FID method for their monitoring. Ann. Chim. 95, 515–524. doi: 10.1002/adic.200590061
Biederman, L., Rosen, M. R., Bobik, B. S., and Roberts, A. L. (2015). Bordetella petrii recovered from chronic pansinusitis in an adult with cystic fibrosis. IDCases 2, 97–98. doi: 10.1016/j.idcr.2015.09.004
Cavalca, L., Zanchi, R., Corsini, A., Colombo, M., Romagnoli, C., Canzi, E., et al. (2010). Arsenic-resistant bacteria associated with roots of the wild Cirsium arvense (L.) plant from an arsenic polluted soil, and screening of potential plant growth-promoting characteristics. Syst. Appl. Microbiol. 33, 154–164. doi: 10.1016/j.syapm.2010.02.004
Colinon, C., Deredjian, A., Hien, E., Brothier, E., Bouziri, L., Cournoyer, B., et al. (2013). Detection and enumeration of Pseudomonas aeruginosa in soil and manure assessed by an ecfX qPCR assay. J. Appl. Microbiol. 114, 1734–1749. doi: 10.1111/jam.12189
Delavat, F., Lett, M. C., and Lièvremont, D. (2013). Yeast and bacterial diversity along a transect in an acidic, As-Fe rich environment revealed by cultural approaches. Sci. Total Environ. 463–464, 823–828. doi: 10.1016/j.scitotenv.2013.06.023
Diavatopoulos, D. A., Cummings, C. A., Schouls, L. M., Brinig, M. M., Relman, D. A., and Mooi, F. R. (2005). Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog. 1:e45. doi: 10.1371/journal.ppat.0010045
Eriksson, M., Sodersten, E., Yu, Z., Dalhammar, G., and Mohn, W. W. (2003). Degradation of polycyclic aromatic hydrocarbons at low temperature under aerobic and nitrate-reducing conditions in enrichment cultures from northern soils. Appl. Environ. Microbiol. 69, 275–284. doi: 10.1128/AEM.69.1.275-284.2003
Fry, N. K., Duncan, J., Malnick, H., Warner, M., Smith, A. J., Jackson, M. S., et al. (2005). Bordetella petrii clinical isolate. Emerging Infect. Dis. 11, 1131–1133. doi: 10.3201/eid1107.050046
Gross, R., Guzman, C. A., Sebaihia, M., dos Santos, V. A., Pieper, D. H., Koebnik, R., et al. (2008). The missing link: Bordetella petrii is endowed with both the metabolic versatility of environmental bacteria and virulence traits of pathogenic Bordetellae. BMC Genomics 9:449. doi: 10.1186/1471-2164-9-449
Guo, Y., Zhang, J., Yan, Y., Wu, J., Zhu, N., and Deng, C. (2015). Molecular phylogenetic diversity and spatial distribution of bacterial communities in cooling stage during swine manure composting. Asian-Australas. J. Anim Sci. 28, 888–895. doi: 10.5713/ajas.14.0882
Ivanov, Y. V., Linz, B., Register, K. B., Newman, J. D., Taylor, D. L., Boschert, K. R., et al. (2016). Identification and taxonomic characterization of Bordetella pseudohinzii sp. nov. isolated from laboratory-raised mice. Int. J. Syst. Evol. Microbiol. 66, 5452–5459. doi: 10.1099/ijsem.0.001540
Ivanov, Y. V., Shariat, N., Register, K. B., Linz, B., Rivera, I., Hu, K., et al. (2015). A newly discovered Bordetella species carries a transcriptionally active CRISPR-Cas with a small Cas9 endonuclease. BMC Genomics 16:863. doi: 10.1186/s12864-015-2028-9
Johnson, S. L., Bishop-Lilly, K. A., Ladner, J. T., Daligault, H. E., Davenport, K. W., Jaissle, J., et al. (2015). Complete genome sequences for 59 Burkholderia isolates, both pathogenic and near neighbor. Genome Announc. 3:e00159-15. doi: 10.1128/genomeA.00159-15
Kato, S., Haruta, S., Cui, Z. J., Ishii, M., and Igarashi, Y. (2004). Effective cellulose degradation by a mixed-culture system composed of a cellulolytic Clostridium and aerobic non-cellulolytic bacteria. FEMS Microbiol. Ecol. 51, 133–142. doi: 10.1016/j.femsec.2004.07.015
Kattar, M. M., Chavez, J. F., Limaye, A. P., Rassoulian-Barrett, S. L., Yarfitz, S. L., Carlson, L. C., et al. (2000). Application of 16S rRNA gene sequencing to identify Bordetella hinzii as the causative agent of fatal septicemia. J. Clin. Microbiol. 38, 789–794.
Kersters, K., Hinz, K. H., Hertle, A., Segers, P., Lievens, A., Siegmann, O., et al. (1984). Bordetella avium sp. nov., isolated from the respiratory tracts of turkeys and other birds. Int. J. Syst. Bacteriol. 34, 56–70. doi: 10.1099/00207713-34-1-56
Kigawa, R., Sano, C., Nishijima, M., Tazato, N., Kiyuna, T., Hayakawa, N., et al. (2013). Investigation of acetic acid bacteria isolated from the Kitora tumulus in Japan and their involvement in the deterioration of the plaster of the mural paintings. Stud. Conserv. 58, 30–40. doi: 10.1179/2047058412Y.0000000040
Ko, K. S., Peck, K. R., Oh, W. S., Lee, N. Y., Lee, J. H., and Song, J. H. (2005). New species of Bordetella, Bordetella ansorpii sp. nov., isolated from the purulent exudate of an epidermal cyst. J. Clin. Microbiol. 43, 2516–2519. doi: 10.1128/JCM.43.5.2516-2519.2005
Li, X., Hu, Y., Gong, J., Zhang, L., and Wang, G. (2013). Comparative genome characterization of Achromobacter members reveals potential genetic determinants facilitating the adaptation to a pathogenic lifestyle. Appl. Microbiol. Biotechnol. 97, 6413–6425. doi: 10.1007/s00253-013-5018-3
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lodders, N., and Kämpfer, P. (2012). A combined cultivation and cultivation-independent approach shows high bacterial diversity in water-miscible metalworking fluids. Syst. Appl. Microbiol. 35, 246–252. doi: 10.1016/j.syapm.2012.03.006
Matsumura, Y., Hosokawa, C., Sasaki-Mori, M., Akahira, A., Fukunaga, K., Ikeuchi, T., et al. (2009). Isolation and characterization of novel bisphenol-A–degrading bacteria from soils. Biocontrol Sci. 14, 161–169. doi: 10.4265/bio.14.161
Mattoo, S., and Cherry, J. D. (2005). Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin. Microbiol. Rev. 18, 326–382. doi: 10.1128/CMR.18.2.326-382.2005
Molmeret, M., Horn, M., Wagner, M., Santic, M., and Abu Kwaik, Y. (2005). Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71, 20–28. doi: 10.1128/AEM.71.1.20-28.2005
Musser, J. M., Hewlett, E. L., Peppler, M. S., and Selander, R. K. (1986). Genetic diversity and relationships in populations of Bordetella spp. J. Bacteriol. 166, 230–237. doi: 10.1128/jb.166.1.230-237.1986
Nagata, J. M., Charville, G. W., Klotz, J. M., Wickremasinghe, W. R., Kann, D. C., Schwenk, H. T., et al. (2015). Bordetella petrii sinusitis in an immunocompromised adolescent. Pediatr. Infect. Dis. J. 34, 458. doi: 10.1097/INF.0000000000000564
Nisola, G. M., Tuuguu, E., Farnazo, D. M., Han, M., Kim, Y., Cho, E., et al. (2010). Hydrogen sulfide degradation characteristics of Bordetella sp. Sulf-8 in a biotrickling filter. Bioprocess. Biosyst. Eng. 33, 1131–1138. doi: 10.1007/s00449-010-0440-8
Ocklitz, H. W., and Milleck, J. (1967). Die überlebenszeit von pertussisbakterien außerhalb des kranken. experimentelle untersuchungen zur keuchhustenepidemiologie. Zentralblatt Bakteriologie Parasitenkunde Infektionskrankheiten 203, 79–91.
Park, J., Zhang, Y., Buboltz, A. M., Zhang, X., Schuster, S. C., Ahuja, U., et al. (2012). Comparative genomics of the classical Bordetella subspecies: the evolution and exchange of virulence-associated diversity amongst closely related pathogens. BMC Genomics 13:545. doi: 10.1186/1471-2164-13-545
Parkhill, J., Sebaihia, M., Preston, A., Murphy, L. D., Thomson, N., Harris, D. E., et al. (2003). Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35, 32–40. doi: 10.1038/ng1227
Partanen, P., Hultman, J., Paulin, L., Auvinen, P., and Romantschuk, M. (2010). Bacterial diversity at different stages of the composting process. BMC Microbiol. 10:94. doi: 10.1186/1471-2180-10-94
Register, K. B., Ivanov, Y. V., Jacobs, N., Meyer, J. A., Goodfield, L. L., Muse, S. J., et al. (2015). Draft genome sequences of 53 genetically distinct isolates of Bordetella bronchiseptica representing 11 terrestrial and aquatic hosts. Genome Announc. 3:e00152-15. doi: 10.1128/genomeA.00152-15
Register, K. B., and Kunkle, R. A. (2009). Strain-specific virulence of Bordetella hinzii in poultry. Avian Dis. 53, 50–54. doi: 10.1637/8388-070108-Reg.1
Sanuth, H. A., Yadav, A., Fagade, O. E., and Shouche, Y. (2013). epsilon-caprolactam utilization by Proteus sp. and Bordetella sp. Isolated from solid waste dumpsites in Lagos State, Nigeria, first report. Indian J. Microbiol. 53, 221–226. doi: 10.1007/s12088-013-0356-5
Sebaihia, M., Preston, A., Maskell, D. J., Kuzmiak, H., Connell, T. D., King, N. D., et al. (2006). Comparison of the genome sequence of the poultry pathogen Bordetella avium with those of B. bronchiseptica, B. pertussis, and B. parapertussis reveals extensive diversity in surface structures associated with host interaction. J. Bacteriol. 188, 6002–6015. doi: 10.1128/JB.01927-05
Shepard, C. W., Daneshvar, M. I., Kaiser, R. M., Ashford, D. A., Lonsway, D., Patel, J. B., et al. (2004). Bordetella holmesii bacteremia: a newly recognized clinical entity among asplenic patients. Clin. Infect. Dis. 38, 799–804. doi: 10.1086/381888
Stainer, D. W., and Scholte, M. J. (1970). A simple chemically defined medium for the production of phase I Bordetella pertussis. J. Gen. Microbiol. 63, 211–220. doi: 10.1099/00221287-63-2-211
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Tazato, N., Handa, Y., Nishijima, M., Kigawa, R., Sano, C., and Sugiyama, J. (2015). Novel environmental Bordetella species isolated from the plaster wall surface of mural paintings in the Takamatsuzuka tumulus: Bordetella muralis sp. nov., Bordetella tumulicola sp. nov. and Bordetella tumbae sp. nov. Int. J. Syst. Evol. Microbiol. 65, 4830–4838. doi: 10.1099/ijsem.0.000655
Tian, W., Sun, Q., Xu, D. B., Zhang, Z. H., Chen, D., Li, C. Y., et al. (2013). Succession of bacterial communities during composting process as detected by 16S rRNA clone libraries analysis. Int. Biodeterior. Biodegradation 78, 58–66. doi: 10.1016/j.ibiod.2012.12.008
Toups, M., Wübbeler, J. H., and Steinbüchel, A. (2010). Microbial utilization of the industrial wastewater pollutants 2-ethylhexylthioglycolic acid and iso-octylthioglycolic acid by aerobic gram-negative bacteria. Biodegradation 21, 309–319. doi: 10.1007/s10532-009-9302-y
Vandamme, P. A., Peeters, C., Cnockaert, M., Inganäs, E., Falsen, E., Moore, E. R., et al. (2015). Bordetella bronchialis sp. nov., Bordetella flabilis sp. nov. and Bordetella sputigena sp. nov., isolated from human respiratory specimens, and reclassification of Achromobacter sediminum Zhang et al. 2014 as Verticia sediminum gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 65, 3674–3682. doi: 10.1099/ijsem.0.000473
Vandamme, P., Heyndrickx, M., Vancanneyt, M., Hoste, B., De Vos, P., Falsen, E., et al. (1996). Bordetella trematum sp. nov., isolated from wounds and ear infections in humans, and reassessment of Alcaligenes denitrificans Ruger and Tan 1983. Int. J. Syst. Bacteriol. 46, 849–858. doi: 10.1099/00207713-46-4-849
Vandamme, P., Hommez, J., Vancanneyt, M., Monsieurs, M., Hoste, B., Cookson, B., et al. (1995). Bordetella hinzii sp. nov., isolated from poultry and humans. Int. J. Syst. Bacteriol. 45, 37–45. doi: 10.1099/00207713-45-1-37
von Wintzingerode, F., Schattke, A., Siddiqui, R. A., Rösick, U., Göbel, U. B., and Gross, R. (2001). Bordetella petrii sp. nov., isolated from an anaerobic bioreactor, and emended description of the genus Bordetella. Int. J. Syst. Evol. Microbiol. 51, 1257–1265. doi: 10.1099/00207713-51-4-1257
Wang, F., Grundmann, S., Schmid, M., Dörfler, U., Roherer, S., Charles Munch, J., et al. (2007). Isolation and characterization of 1,2,4-trichlorobenzene mineralizing Bordetella sp. and its bioremediation potential in soil. Chemosphere 67, 896–902. doi: 10.1016/j.chemosphere.2006.11.019
Weisskopf, L., Heller, S., and Eberl, L. (2011). Burkholderia species are major inhabitants of white lupin cluster roots. Appl. Environ. Microbiol. 77, 7715–7720. doi: 10.1128/AEM.05845-11
Weyant, R. S., Hollis, D. G., Weaver, R. E., Amin, M. F., Steigerwalt, A. G., O'Connor, S. P., et al. (1995). Bordetella holmesii sp. nov., a new gram-negative species associated with septicemia. J. Clin. Microbiol. 33, 1–7.
Winiecka-Krusnell, J., and Linder, E. (1999). Free-living amoebae protecting Legionella in water: the tip of an iceberg? Scand. J. Infect. Dis. 31, 383–385. doi: 10.1080/00365549950163833
Keywords: Bordetella, environmental strains, ecological niches, extra-host adaptation, environmental origin
Citation: Hamidou Soumana I, Linz B and Harvill ET (2017) Environmental Origin of the Genus Bordetella. Front. Microbiol. 8:28. doi: 10.3389/fmicb.2017.00028
Received: 20 November 2016; Accepted: 05 January 2017;
Published: 24 January 2017.
Edited by:
Tatiana Venkova, The University of Texas Medical Branch at Galveston, USAReviewed by:
Nikolai Ravin, Research Center for Biotechnology (RAS), RussiaLouise Temple, James Madison University, USA
Copyright © 2017 Hamidou Soumana, Linz and Harvill. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bodo Linz, Ym9kby5saW56QHVnYS5lZHU=
Eric T. Harvill, aGFydmlsbEB1Z2EuZWR1
†These authors have contributed equally to this work.
 Illiassou Hamidou Soumana
Illiassou Hamidou Soumana Bodo Linz
Bodo Linz Eric T. Harvill
Eric T. Harvill