- 1403C Althouse Laboratory, Department of Biochemistry and Molecular Biology, The Pennsylvania State University, University Park, PA, USA
- 2Waksman Institute of Microbiology, Rutgers, The State University of New Jersey, Piscataway, NJ, USA
- 3Department of Chemistry and Chemical Biology, Rutgers, The State University of New Jersey, Piscataway, NJ, USA
- 4Department of Chemistry and Biochemistry, Montana State University, Bozeman, MT, USA
For nearly half a century, it was believed that cyanobacteria had an incomplete tricarboxylic acid (TCA) cycle, because 2-oxoglutarate dehydrogenase (2-OGDH) was missing. Recently, a bypass route via succinic semialdehyde (SSA), which utilizes 2-oxoglutarate decarboxylase (OgdA) and succinic semialdehyde dehydrogenase (SsaD) to convert 2-oxoglutarate (2-OG) into succinate, was identified, thus completing the TCA cycle in most cyanobacteria. In addition to the recently characterized glyoxylate shunt that occurs in a few of cyanobacteria, the existence of a third variant of the TCA cycle connecting these metabolites, the γ-aminobutyric acid (GABA) shunt, was considered to be ambiguous because the GABA aminotransferase is missing in many cyanobacteria. In this study we isolated and biochemically characterized the enzymes of the GABA shunt. We show that N-acetylornithine aminotransferase (ArgD) can function as a GABA aminotransferase and that, together with glutamate decarboxylase (GadA), it can complete a functional GABA shunt. To prove the connectivity between the OgdA/SsaD bypass and the GABA shunt, the gadA gene from Synechocystis sp. PCC 6803 was heterologously expressed in Synechococcus sp. PCC 7002, which naturally lacks this enzyme. Metabolite profiling of seven Synechococcus sp. PCC 7002 mutant strains related to these two routes to succinate were investigated and proved the functional connectivity. Metabolite profiling also indicated that, compared to the OgdA/SsaD shunt, the GABA shunt was less efficient in converting 2-OG to SSA in Synechococcus sp. PCC 7002. The metabolic profiling study of these two TCA cycle variants provides new insights into carbon metabolism as well as evolution of the TCA cycle in cyanobacteria.
Introduction
As mankind strives to lower anthropogenic carbon dioxide emissions, our ability to manipulate photosynthetic primary production is critical. Total worldwide primary production has been estimated to be 104.9 Gt C year-1, with roughly equal amounts arising from terrestrial and marine photosynthesis (Field et al., 1998). Cyanobacteria are an essential group of oxygenic photosynthetic organisms that are projected to play a pivotal role in CO2 sequestration and in supplying future renewable energy needs. Owing to their ease of genetic modification, many cyanobacteria have been engineered to improve CO2 capture and light utilization (Ducat et al., 2011; Lan and Liao, 2012; Yu et al., 2013). The capacity to improve these functions depends on how rapidly and completely the downstream steps can utilize the primary metabolic intermediates, thus fundamental studies are needed to understand key metabolic chokepoints and regulatory sites in carbon and energy metabolism (Krishnan et al., 2015b; Qian et al., 2015; Zhang et al., 2015).
The tricarboxylic acid (TCA) cycle is a central metabolic pathway that generates NADH reductant to power cellular growth, repair and homeostasis, as well as providing precursors for the biosynthesis of cellular components (Tang et al., 2011; Zhang and Bryant, 2014). Cyanobacteria were recently shown to have a complete but non-traditional TCA cycle (Zhang and Bryant, 2011). In this variation, 2-oxoglutarate (2-OG) is converted to succinate by 2-oxoglutarate decarboxylase (2-OGDC, OgdA) and succinate semialdehyde dehydrogenase (SSADH, SsaD) (Figure 1). The presence of this bypass in most cyanobacteria not only corrected a long-held misconception that these organisms have an incomplete TCA cycle due to the absence of 2-oxoglutarate dehydrogenase (2-OGDH), but also provided much useful knowledge illustrating the occurrence and physiological functions of TCA variants in these bacteria (Zhang and Bryant, 2011, 2014). Nonetheless, kinetic evidence for flux and regulation of this route remains poorly established. In particular, the involvement of this 2-OGDC/SSADH route under photoautotrophic and photomixotrophic growth conditions in Synechococcus sp. PCC 7002 (hereafter Synechococcus 7002) is so far unavailable. Flux-balance calculations have suggested that minimal flux occurs through the bypass reactions under photoautotrophic growth conditions (Hendry et al., 2016).
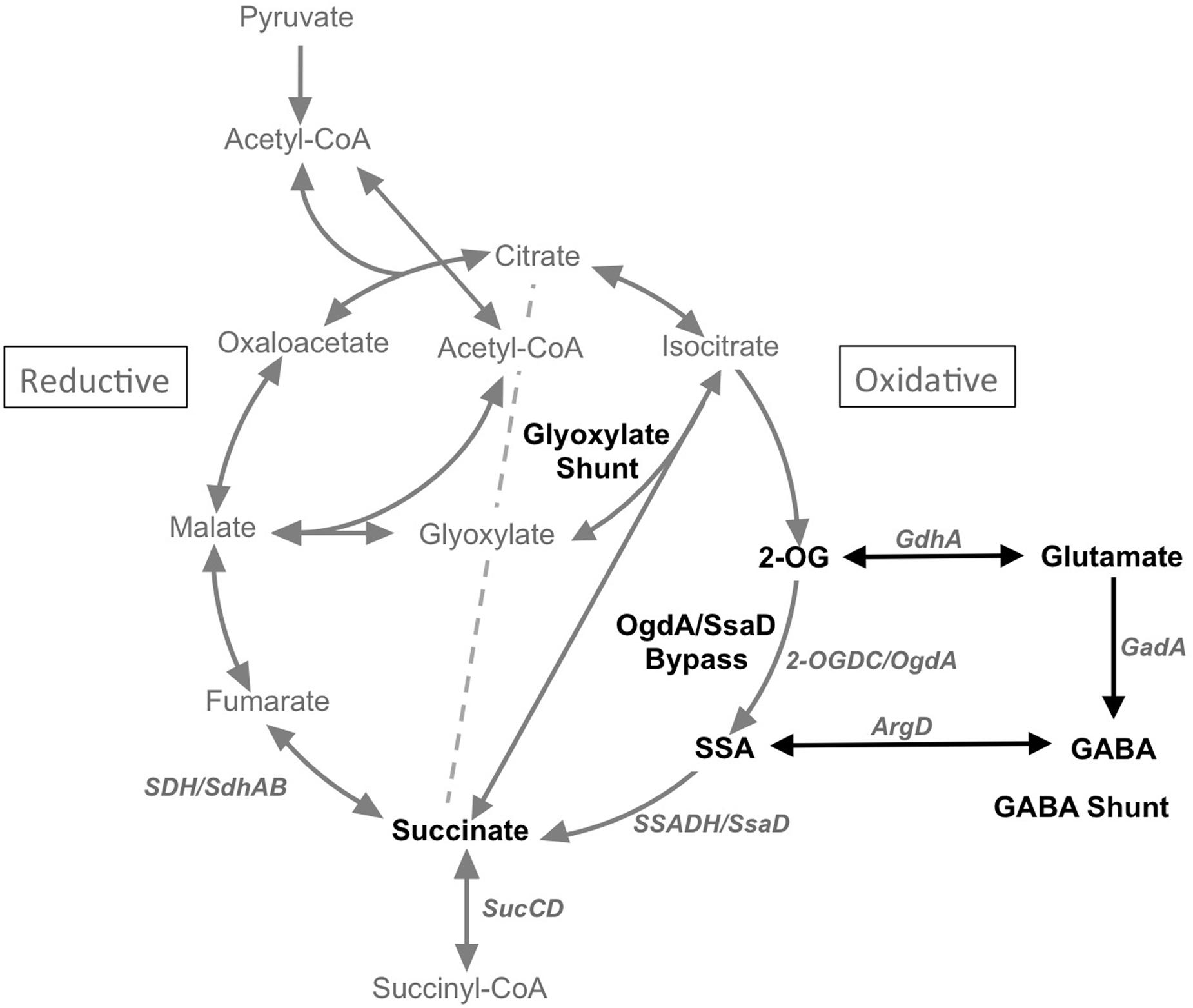
FIGURE 1. Scheme showing the TCA cycle, the glyoxylate cycle and the GABA shunt. 2-OG, 2-oxoglutarate; SSA, succinic semialdehyde; GABA, γ-aminobutyric acid; ArgD, GABA aminotransferase; GadA, glutamate decarboxylase; GdhA, glutamate dehydrogenase; 2-OGDC/OgdA, 2-oxoglutarate decarboxylase; SSADH/SsaD, succinic semialdehyde dehydrogenase.
Other TCA cycle variants are known, including the glyoxylate shunt for acetate assimilation and the γ-amino-butyric acid (GABA) shunt that permits the sensing of nitrogen metabolite levels through glutamate. Based on comparative genomics and biochemical validation, Zhang and Bryant (2015) showed that the glyoxylate shunt only occurs in a few cyanobacteria. They characterized the two enzymes that complete this cycle, isocitrate lyase and malate synthase, in Chlorogloeopsis fritschii PCC 9212 and verified their respective activities. They also showed that this strain can grow in the dark on acetate as carbon and energy source. The introduction of the aceAB genes into Synechococcus 7002 conferred the ability to metabolize acetate in this strain and confirmed the role of the products of these genes in acetate metabolism via the glyoxylate shunt.
The existence of the GABA shunt in cyanobacteria is rather poorly understood. As shown in Figure 1, the GABA shunt includes four enzymes, glutamate dehydrogenase (GdhA), glutamate decarboxylase (GadA), GABA aminotransferase, and succinic semialdehyde dehydrogenase (SSADH/SsaD). The shunt results in the conversion of glutamate to succinate with GABA and succinic semialdehyde (SSA) as intermediates. However, according to comparative genomics, glutamate decarboxylase, which catalyzes the second step in the GABA shunt, is only present in a few cyanobacteria. Additionally, another key enzyme in the GABA shunt, the GABA aminotransferase is also missing in many cyanobacteria. Considering the metabolite links between the GABA shunt and the TCA cycle, both of which can convert 2-OG to succinate via SSA, it is likely that these two pathways are functionally connected. Indeed, it was reported in Bradyrhizobium japonicum that the GABA shunt is functional in a TCA cycle mutant lacking 2-OGDH (Green et al., 2000). A recent study has provided evidence that N-acetylornithine aminotransferase (ArgD) from Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803) also functions as a GABA aminotransferase, which together with glutamate dehydrogenase (GdhA) and glutamate decarboxylase (GadA) form the GABA shunt in cyanobacteria (Xiong et al., 2014). However, biochemical validation of the putative bi-functional N-acetylornithine aminotransferase (ArgD) has not yet been demonstrated.
In this study, a protocol specific for cyanobacteria was developed to extract and analyze intracellular SSA from Synechococcus 7002. Our findings demonstrate the significant involvement of the OgdA/SsaD bypass under dark aerobic growth conditions in Synechococcus 7002, but suggest that there is much lower activity under photoautotrophic conditions. Additionally, we biochemically demonstrated that N-acetylornithine aminotransferase (ArgD) can also function as a GABA aminotransferase. Finally, the gadA gene, encoding glutamate decarboxylase from Synechocystis 6803, was expressed in Synechococcus 7002, which naturally lacks this enzyme. Intracellular metabolite concentrations were measured in this overexpression strain, and potential interactions between the resulting GABA shunt and the TCA cycle were investigated. The findings in this study validate the occurrence of three potential hybrid TCA cycle variants in some cyanobacteria and provide insights into pathway evolution as well as suggestions for future metabolic engineering targets.
Materials and Methods
Strains, Growth Conditions, and Culture Preparation
The strains used in this study, Synechococcus sp. PCC 7002 (WT7002) and Synechocystis sp. PCC 6803 (WT6803), were originally obtained from the Pasteur culture collection (PCC) (Rippka et al., 1979). Routine DNA manipulations were performed with Escherichia coli strain DH5α cells that were grown in Luria-Bertani (LB) medium. All Synechococcus 7002 strains were maintained in liquid medium A+ (Stevens et al., 1973), which contains 10 mM nitrate, and Synechocystis 6803 cells were grown in medium BG-11 (Rippka et al., 1979). The growth media were supplemented with the following antibiotics when appropriate: spectinomycin (50 μg ml-1), kanamycin (100 μg ml-1), gentamycin (20 μg ml-1), and/or erythromycin (20 μg ml-1). Unless noted, cultures were grown photoautotrophically under the following conditions: continuous irradiance of 120 μmol photons m-2 s-1 at 38°C with continuous sparging with 2% (v/v) CO2 in air. Two mutant strains lacking subunits of succinyl-CoA synthase (SucCD): SZ006 and SZ007 (Table 1) could not be grown under the specified high-CO2 sparging conditions. As a result, these two strains were grown by slowly bubbling cultures with 1% (v/v) CO2 in air. Growth of Synechococcus 7002 was measured by following changes in the optical density at 730 nm (OD730 nm) with a spectrophotometer.
To prepare cultures for metabolite analyses, all mutant strains were grown under the same photoautotrophic conditions as a WT7002 control sample until the cultures reached mid-exponential phase and had an OD730 nm of ∼0.6. At this point, an aliquot of cell culture (2 mL) was collected, cells were rapidly harvested by vacuum filtration, and the cells were extracted for metabolomic analysis following the previously described protocol (Bennette et al., 2011). After being sampled for the photoautotrophic metabolomic analysis, cultures were quickly wrapped with aluminum foil and incubated without sparging in a dark growth chamber for 3 h at 38°C. The caps on the culture flasks were left loose so that the cultures inside would remain aerobic. After the 3-h dark aerobic incubation, cultures were sampled for dark aerobic metabolomic analysis following the same protocol (Bennette et al., 2011).
To prepare cultures for the 13C-labeling experiment under photomixotrophic conditions, the WT7002 strain was pre-acclimated to medium A+ supplemented with 5 mM glycerol. 13C3-labeled glycerol was used to conduct this experiment, because Synechococcus 7002 can metabolize exogenous glycerol but not glucose as a reduced carbon source (Shen and Bryant, 1995). Before the experiment, a WT7002 culture (100 mL) was grown photomixotrophically to an OD730 nm of ∼1.0. An aliquot (500 μL) of the 13C3-labeled glycerol stock (1 M) was added immediately after the T0 sample (t = 0 s) was taken. Sampling continued for T1 (t = 0.50 min), T2 (t = 1 min), T3 (t = 10 min), T4 (t = 20 min), and T5 (t = 40 min). Immediately after sample T5 was taken, the culture vessel was wrapped in foil and placed in a dark growth chamber. The purpose of this switch was to identify the carbon source used by Synechococcus 7002 under dark aerobic conditions in the presence of an external reduced carbon source. Sampling was performed under dark aerobic conditions for T6 (t = 45 min), T7 (t = 60 min), and T8 (t = 90 min).
Cloning, Protein Purification, and Identification
Open reading frames sll1641 (gadA), encoding the glutamate decarboxylase, and slr1022 (argD), encoding the N-acetylornithine aminotransferase of Synechocystis 6803, as well as open reading frame SYNPCC7002_A0326 (argD), encoding the N-acetylornithine aminotransferase of Synechococcus 7002, were amplified by polymerase chain reaction (PCR) with Phusion DNA polymerase (New England Biolabs, Ipswich, MA, USA) and cloned into plasmid pAQ1Ex-PcpcBA (Xu et al., 2011). Primer set GADF-GADR was used to amplify gadA; primer set 6803ArgDF-6803ArgDR was used to amplify Synechocystis 6803 argD (argD6803); and primer set 7002ArgDF-7002ArgDR was used to amplify Synechococcus 7002 argD (argD7002) (Supplementary Table S1). An N-terminal His10-tag was introduced into each of the enzymes to facilitate subsequent protein detection and purification.
The resulting plasmids pAQ1Ex-PcpcBA[gadA6803], pAQ1Ex-PcpcBA[argD(6803)], and pAQ1Ex-PcpcBA[argD7002] (Table 1) were transformed into E. coli strain DH5-α and verified by DNA sequencing. Cells were grown overnight in 1 L of Luria-Bertani medium containing 50 μg gentamycin ml-1, harvested by centrifugation at 4°C at 5,000 × g, and washed once with 50 mM Tris-HCl buffer, pH = 8.0. Cells were disrupted by three passages through a chilled French pressure cell operated at 138 MPa. Soluble lysates were obtained by centrifugation at 20,000 × g for 30 min, and were loaded onto a Ni2+-NTA affinity resin (Goldbio, St. Louis, MO, USA). The column was pre-equilibrated with 10 mM imidazole in 50 mM Tris-HCl, pH 8.0 and was washed with 30 mM imidazole in 50 mM Tris-HCl, pH 8.0, 300 mM NaCl. Proteins were eluted stepwise with 50, 100, 150, 200, and 250 mM imidazole in 50 mM Tris-HCl, pH 8.0, 300 mM NaCl. Fractions containing the recombinant proteins were analyzed by polyacrylamide gel electrophoresis in the presence of sodium dodecylsulfate (SDS-PAGE) (Shen and Bryant, 1995), and purified proteins were concentrated by ultrafiltration using Centriprep columns (Millipore, Billerica, MA, USA). Proteins were further analyzed by SDS-PAGE and immunoblotting with commercial polyclonal rabbit antibodies (Rockland, Limerick, PA, USA) to the poly-His10 tag. Each protein was judged to be >90% pure and was also positively identified by tryptic peptide mass fingerprinting as previously described (Zhang and Bryant, 2011). Consistent with the results from mass spectrometry, no contaminating proteins were identified.
Enzymatic Assays
For enzyme assays with Synechocystis 6803 glutamate decarboxylase (GadA6803 = the product of open reading frame sll1641), the reaction mixture (0.2 ml) contained 1 mM glutamate, 50 mM K-phosphate, pH 4.5 and purified enzyme (50 μg); control reaction mixtures were treated identically but without the addition of the purified enzyme. The reaction mixtures were incubated at room temperature (RT) for 1 h, and the GABA produced was detected by using Edman’s reagent as previously described with slight modifications (Tank and Bryant, 2015). An aliquot (30 μl) of the reaction mixture was transferred to a glass test tube and dried by flushing with nitrogen. The dried sample was dissolved in coupling solution (100 μL; mixture containing 50% acetonitrile, 25% pyridine, 10% triethylamine and 15% water, v/v/v/v), and the resulting solution was dried again. The dried sample was redissolved in coupling solution (100 μL), after which 5 μL of phenylisothiocyanate (PITC; Edman’s reagent) was added to the solution, and the coupling reaction was allowed to proceed for 5 min at RT. After the reaction was complete, the liquid sample was dried and the resulting pellet was dissolved in analysis solvent [250 μl, 7:2 (v/v) mixture of HPLC-grade water and acetonitrile]. An aliquot (20 μL) of the solution was injected onto a Shimadzu LC-20AB HPLC system equipped with 254-nm UV detector SPD-20A. Components in the solution were separated on a Kinetex 5-μm C18 100 Å column (15 cm × 4.6 174 mm ID) protected by a SecurityGuard ULTRA cartridge UHPLC C18 for 4.6-mm ID columns 175 (Phenomenex, Torrance, CA, USA). The HPLC analysis method consisted of a 2-solvent gradient (solvents A and B) developed over a 20-min period with a flow rate of 0.5 ml min-1 at 30°C. The initial condition was 100% solvent A (0.14 M sodium acetate, pH 6.2 containing 0.5 mM triethanolamine), which decreased over 10 min to 82.5% and from 10 to 15 min to 0%. Solvent B was a 40:60 (v/v) mixture of HPLC-grade water and acetonitrile. The elution times and concentrations of substrates and products were determined by comparison of results obtained from analyses of individual standard compounds with the same procedure.
For enzyme assays with N-acetylornithine aminotransferase (ArgD), the reaction mixture (0.2 ml) contained 1 mM L-ornithine, 1 mM 2-oxoglutarate, 50 mM K-phosphate, pH 9 and the purified enzyme (50 μg) from Synechocystis 6803 (ArgD6803) or Synechococcus 7002 (ArgD7002). Reaction mixtures were incubated at RT for 1 h, and an aliquot (30 μl) of the reaction mixture was analyzed using the same procedure as described for the characterization of glutamate decarboxylase to detect product formation. Control experiments were performed similarly but without the addition of the purified enzyme. The elution times and concentrations of substrates and products were determined by comparison of results obtained from analyses with purified individual standard compounds.
For enzyme assays with GABA aminotransferase (ArgD), the reaction mixture (0.2 ml) contained 1 mM GABA, 1 mM 2-OG, 50 mM K-phosphate, pH 9 and the purified enzyme (50 μg) from Synechocystis 6803 (ArgD6803) or Synechococcus 7002 (ArgD7002). The mixture was incubated at RT for 1 h, and an aliquot (30 μl) of the reaction mixture was analyzed using the same procedure as described in the enzyme characterization of glutamate decarboxylase to detect product formation. Control experiments were performed similarly but without the addition of the purified enzyme. The elution times and concentrations of substrates and products were determined by comparison of results obtained from analyses of individual standard compounds.
Mutant Construction and Segregation
The coding sequences of ogdA (SynPCC7002_A2770), ssaD(SynPCC7002_A2771), sdhA (SynPCC7002_A2569), sdhB (SynPCC7002_A1094), or the sucCD operon (SynPCC7002_A0890 and SynPCC7002_A0891) were deleted and replaced by a DNA cassette that confers specific antibiotic resistance to produce the corresponding mutant strains of Synechococcus 7002 using the homologous recombination method (Frigaard et al., 2004) (see Table 1; Supplementary Figure S1A). In addition, the ogdA ssaD operon (SynPCC7002_A2770 SynPCC7002_A2771) was deleted in mutant strain SZ004 to generate the triple mutant strain SZ005; and sdhB (SynPCC7002_A1094) as well as the ogdA ssaD operon (SynPCC7002_A2770 SynPCC7002_A2771) were deleted in mutant strain, SZ006, to generate the quadruple mutant, SZ007. Complete segregation of alleles was verified in all cases by PCR with template DNAs derived from the wild-type and mutant strains (see Supplementary Figures S1B–I). The primers used for PCR analyses are listed in Supplementary Table S1. The mutant strains were also verified by DNA sequencing of the resulting PCR amplicons.
In order to construct a Synechococcus 7002 strain expressing the glutamate decarboxylase (GadA6803), the pAQ1Ex-PcpcBA [gadA6803] plasmid was transformed into wild-type Synechococcus 7002 as previously described (Frigaard et al., 2004). The presence of the gadA6803 gene in the resulting strain, denoted SZ008, was confirmed by PCR using primer set GADF-GADR. Immunoblotting was also performed with commercial antibodies (Rockland) to the His10-tag as previously described (Shen and Bryant, 1995) to confirm expression of the recombinant glutamate decarboxylase (Supplementary Figure S2).
To investigate further the metabolic correlation between the GABA shunt and the TCA cycle in cyanobacteria, the gene encoding 2-OGDC (ogdA; SYNPCC7002_A2770) was deleted and replaced by a DNA fragment encoding an antibiotic resistance gene (aphAII, kanamycin resistance), and the resulting strain was denoted as SZ009. Primer sets A2770D1–A2770D2 and A2770D3–A2770D4 (Supplementary Table S1) were used to amplify the upstream and downstream regions of ogdA. Transformation and selection were performed as previously described (Frigaard et al., 2004). Full segregation of the deletion of SYNPCC7002_A2770 was verified by PCR by comparing the products of template DNAs from both the wild-type and the mutant strains using primer set A2770D1–A2770D4 (Supplementary Figure S2). The resulting PCR amplicons were also verified by DNA sequencing.
Intracellular Metabolites Analyses
Metabolite extraction and quantification were performed as previously described (Bennette et al., 2011). Briefly, an aliquot of cell culture (2 mL) was sampled and rapidly vacuum-filtered on a 0.45-μm Nylon membrane filter under dark conditions. The cells adhering to the membrane were immediately quenched in 1.8 mL of ice-cold buffer [80:20 (v/v) MeOH/H2O] in Petri dishes and incubated for 20 min at -20°C. Cell material was then scraped off from the filters, and the solvent and cell material suspension was centrifuged at 14,000 × g at 4°C for 5 min. The supernatant was removed and stored at -80°C. An aliquot of the solvent extract (120 μL) was vacuum-dried (Labconco Centri-Vap Concentrator), and the pellet was resuspended in LC–MS grade water (40 μL) for analysis. The LC–MS analysis system and methods were described previously (Bennette et al., 2011). Extracellular metabolite profiles were analyzed by HPLC according to a previously described protocol (Kumaraswamy et al., 2013).
Determination of Intracellular SSA in Synechococcus 7002
Wild-type Synechococcus 7002 cultures were grown to OD730nm = ∼0.6, and aliquots of the culture (15 mL) were collected before and after incubation under dark aerobic conditions for 3 h. The cells in the culture aliquots were pelleted by centrifugation at 9600 × g at 4°C for 15 min, and the supernatant was discarded. Cell pellets were resuspended in ice-cold, autoclaved water (500 μL), and the suspensions were sonicated 15 times each over ice (with each pulse lasting 10 s) using a Fisher Scientific Model FB-505 sonicator (amplitude, 40%). The sonicated samples were centrifuged at 14,000 × g at 4°C for 5 min. Supernatants (500 μL per sample) were collected and transferred into clean microcentrifuge tubes.
Because SSA is unstable in air (Wermuth, 1979), SSA extracts from cyanobacteria cannot be analyzed by the previously described metabolite isolation method (Bennette et al., 2011), which has an evaporation and concentration step in air. By adapting an SSA derivatization method that was originally used to quantify SSA in liquid samples of human urine (Struys et al., 2005), it is possible to detect and quantify intracellular SSA in the cyanobacterium Synechococcus 7002. SSA was derivatized with 2,4-dinitrophenylhydrazine (DNPH) and detected by LC–MS (Struys et al., 2005). Briefly, DNPH (50 μL of a 3.1 mg mL-1 stock solution prepared in 2.0 M HCl) was added to an aliquot (500 μL) of sample supernatant or SSA calibration standard (concentrations 0, 0.01, 0.05, 0.1, 0.5, and 1 mM), and the reaction mixtures were incubated for 15 min at RT. The SSA derivatives were extracted with ethyl acetate (3.0 mL), and the organic fractions were transferred to clean glass tubes and dried with a stream of nitrogen gas. The dry residue was dissolved in 100% methanol (125 μL), and an aliquot (10 μL) of the liquid sample was injected into an LC–MS/MS system for analysis. Mass transitions were set to be m/z –281.1 →m/z-182.1 and m/z -285.1 →m/z-182.1 for 13C0-SSA (M0) and 13C4-SSA (M4), respectively.
O2 Evolution Rate, Dark Respiration Rate, and Measurement of Chlorophyll a Concentrations
Liquid cultures of WT7002 and mutant strains were grown to mid-exponential phase (OD730 nm = ∼0.6), and cells were harvested to measure their maximal O2 evolution rates under saturating red light (∼750 μmol photons m-2 s-1) and their dark respiration rates. These rates were measured using Clark-type electrodes, employing both commercial (Hansatech) and home-built electrodes (Krishnan et al., 2015a). Sodium bicarbonate (25 mM) was added to the cell suspensions prior to analysis in order to provide excess Ci source to act as the electron acceptor for the photoreactions. Chlorophyll (Chl) a concentrations were measured according to the colorimetric method previously described (Sakamoto and Bryant, 1997).
Results
Identification of SSA-DNPH Derivatives
Based on a comparison of the LC chromatograms of extracts of the WT7002 strain with a 2.5 mM SSA standard (Figures 2A,B), the retention time of SSA-DNPH derivatives was determined to be 14.1 min. The calibration curve was linear over the range 0–1.0 mM for SSA-DNPH derivatives, with a linear regression (R2) of 0.99 (Figure 2B). Deconvolution of the MS/MS spectra of SSA-DNPH showed an intense fragment at m/z -182.1, which corresponds to the loss of the DNPH group from SSA-DNPH.
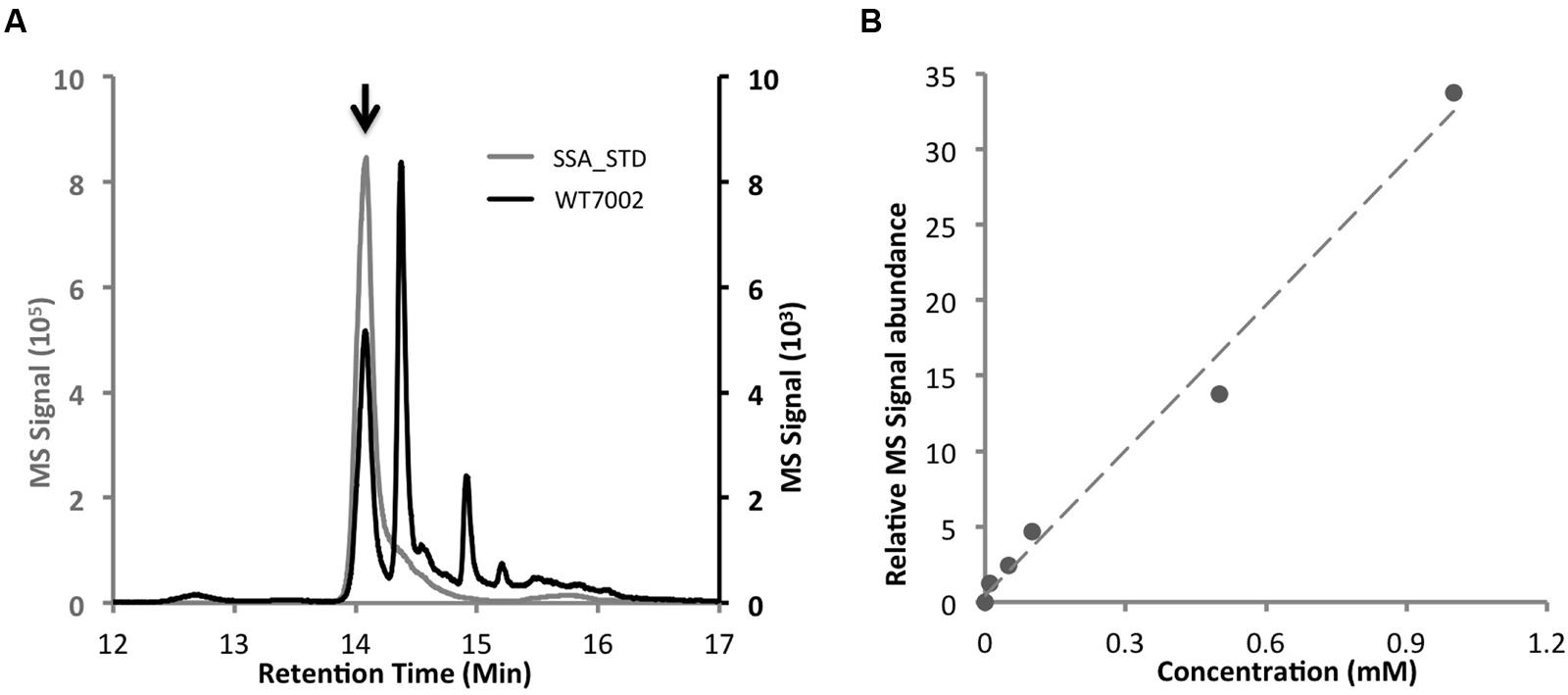
FIGURE 2. LC–MS method to determine intracellular SSA contents. (A) Comparison of chromatograms of WT7002 (black line) and 2.5 mM SSA standard (gray line). Black arrow indicates the peak of SSA. (B) Standard curve of SSA within the concentration range of 0 to 1.0 mM, R2 = 0.99.
13C-Labeling of SSA under Light and Dark Conditions
The level of 13C-labeling of intracellular SSA was determined by measuring the [13C4-SSA]/[13C0-SSA] ratios (M4 /M0) over time (Figure 3A). Other SSA forms with 1, 2, or 3 13C-labeled carbons (M1, M2, and M3) were assayed as well, and it was determined that 13C4-SSA was the clearest and free from interference. Starting with the WT7002 strain under photoautotrophic conditions, the M4/M0 value increased immediately after addition of 13C-labeled glycerol (Figure 3A) and attained a steady-state value of 0.25 at 40 min. The culture was switched to dark aerobic conditions at 40 min and the M4/M0 value slowly decreased over the next 50 min to a value of 0.15 after the light-to-dark transition.
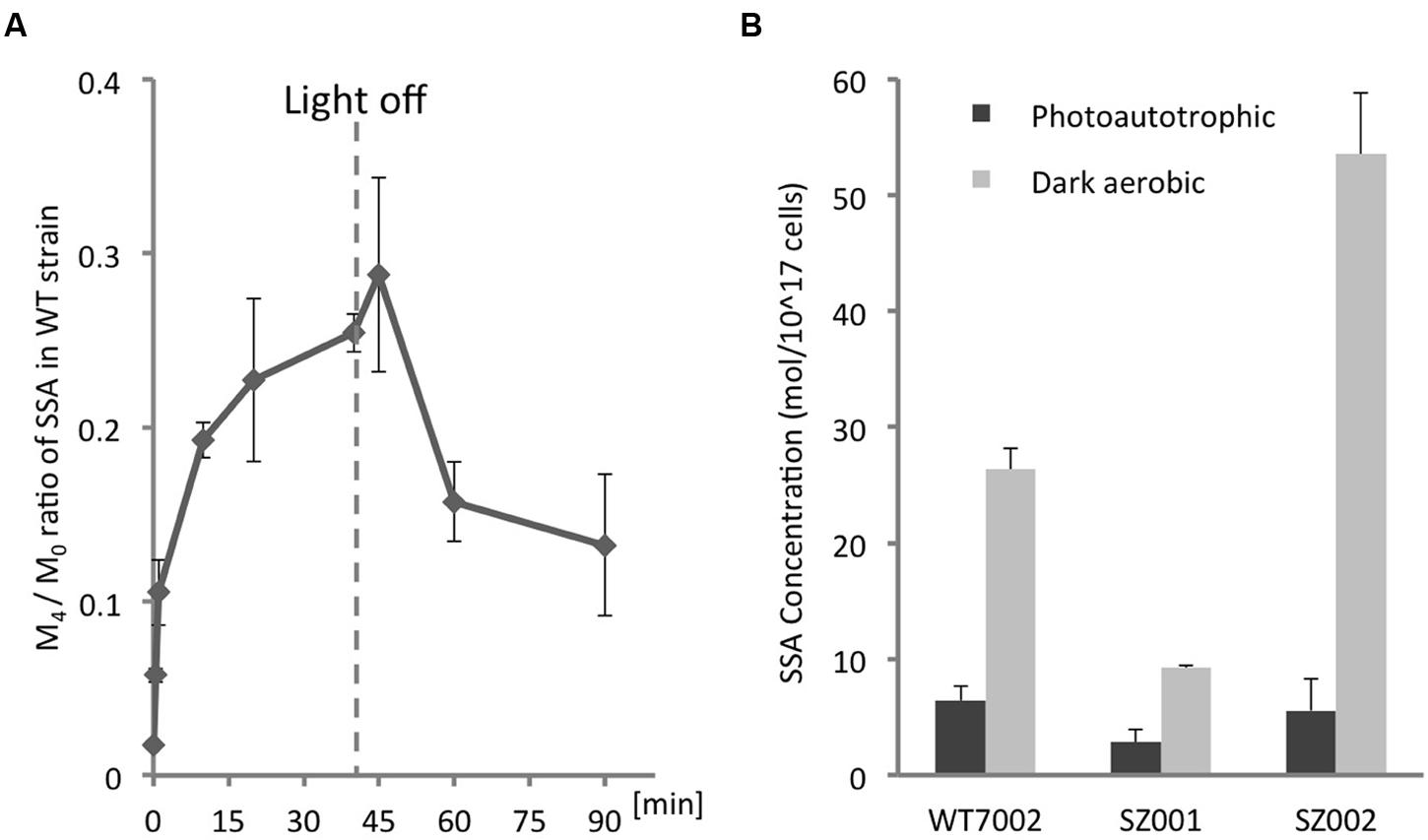
FIGURE 3. Quantification of intracellular SSA. (A)13C-labeling kinetics of SSA under photomixotrophic conditions. The dashed line indicates the time point when the culture was switched from light to dark. (B) Intracellular SSA contents of WT7002 and strains SZ001, and SZ002 under photoautotrophic and dark aerobic conditions. The results are mean values of three biological replicates and error bars represent the standard deviations.
Intracellular Metabolite Analysis of TCA Cycle Mutants
The differences in SSA content among WT7002 and two single mutant strains, SZ001 and SZ002 (see Table 1), were monitored under both photoautotrophic and dark aerobic conditions (Figure 3B). Under photoautotrophic conditions, WT7002 had ∼2-fold more SSA than the SZ001 strain, but had a comparable SSA content to the SZ002 strain. However, after 3 h of dark aerobic incubation and compared to WT7002, the SZ002 strain accumulated ∼2-fold more SSA while the SZ001 mutant had 2.5-fold lower SSA. These results confirm that 2-OGDC (OgdA; product of SynPCC7002_A2770) is functional under both photoautotrophic and respiratory conditions.
Intracellular contents of four TCA cycle metabolites (citrate, 2-OG, succinate, and malate) in the mutant strains were compared to those in the WT7002 strain (Figure 4). Mutants were categorized into three groups according to a terminology as follows: oxidative, reductive and “both branches.” These terms indicate the location of the missing enzyme(s) in the TCA cycle, using citrate-succinate as the dividing line (dashed line in Figure 1, right side: oxidative; left side: reductive). In the two mutant strains in the “oxidative” group, i.e., SZ001, and SZ002 and compared to the levels in the WT7002 strain, the succinate content downstream of these enzymes was significantly reduced under both light and dark conditions. Under photoautotrophic conditions, the 2-OG content in the strain lacking 2-OGDC (OgdA) increased 2.3-fold, as expected, while it remained at a similar level as in the WT7002 strain in the SZ002 mutant strain lacking SSADH. After a 3-h dark aerobic incubation, the 2-OG content in both mutant strains was twofold higher than in strain WT7002. In the succinyl-CoA synthase (sucCD) mutant SZ006, the succinate and malate contents were both reduced to ∼60% of the levels in strain WT7002 under photoautotrophic conditions, and the levels of these compounds decreased further to ∼40% of the levels in WT7002 under dark aerobic conditions.
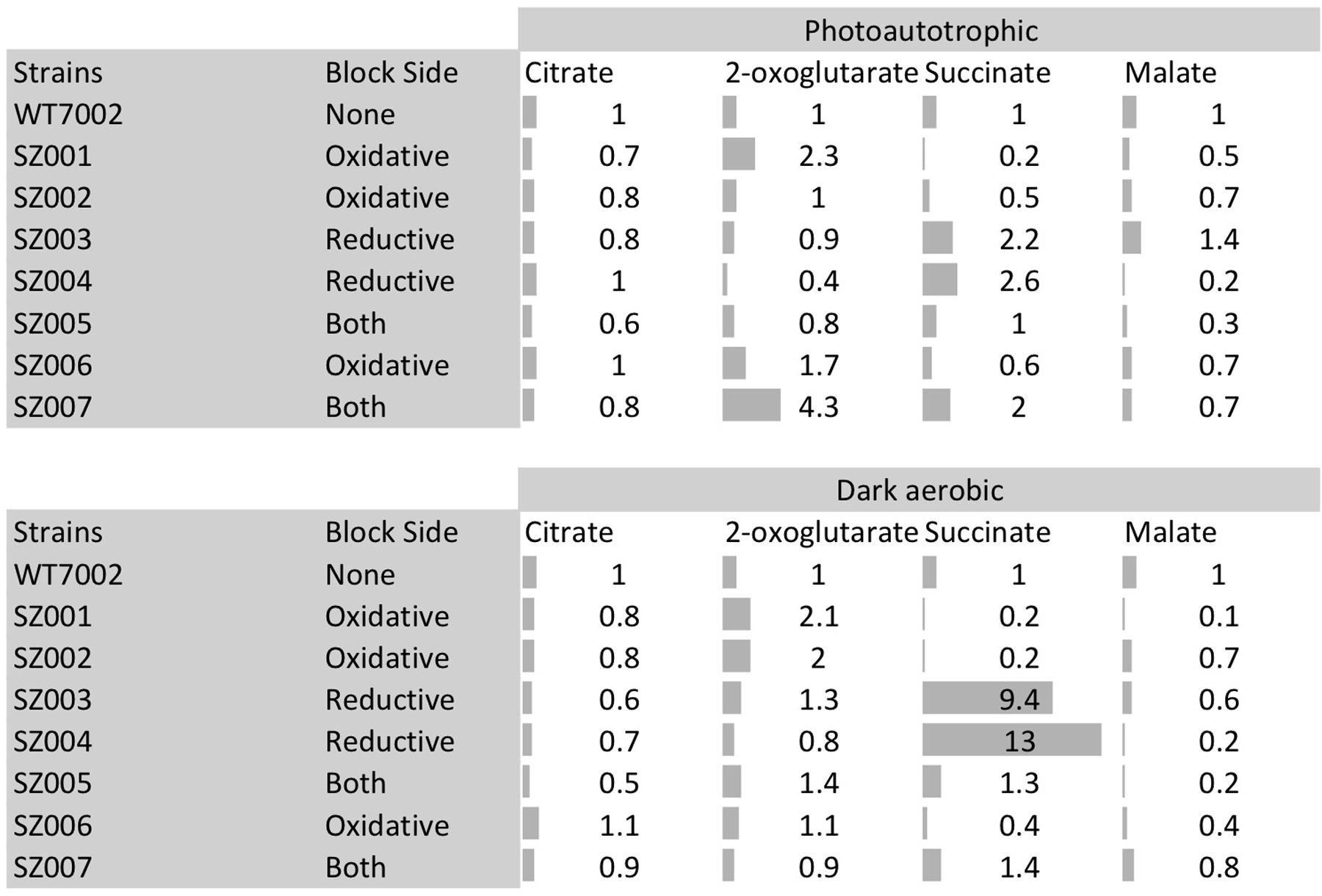
FIGURE 4. Metabolites profiling of TCA cycle metabolites in mutant strains. Category “block side” refers to the side of the TCA cycle (reductive side or oxidative side) that hypothetically prevent the synthesis of succinate due to the corresponding mutation(s). Gray bars indicate the fold changes of these metabolites compared to WT7002 levels.
In the two mutant strains designated as “reductive branch,” SZ003 and SZ004, succinate contents were 2.2- and 2.6-fold higher than in strain WT7002, respectively, under photoautotrophic conditions. Succinate levels in these two strains were substantially higher, 9.4- and 13.0-fold higher, respectively than the in strain WT7002, under dark aerobic conditions. The content of the downstream metabolite, malate, was significantly reduced in the SZ004 strain under both conditions, but it remained at similar levels to WT7002 in the SZ003 strain. In the triple mutant strain, SZ005, designated as “both branches,” 2-OG and succinate remained at similar levels as for WT7002, while the malate content decreased under both conditions. In the quadruple mutant strain, SZ007, designated as “both branches,” 2-OG and succinate were 4.3- and 2.0-fold higher than in WT7002, respectively, under photoautotrophic conditions. The increased pool sizes diminished when the mutant strains were incubated under dark aerobic conditions. In summary, the pattern of the pool size changes is consistent with the operation of a cycle catalyzed by enzymes.
Growth Phenotypes and Succinate Levels in Mutant Strains
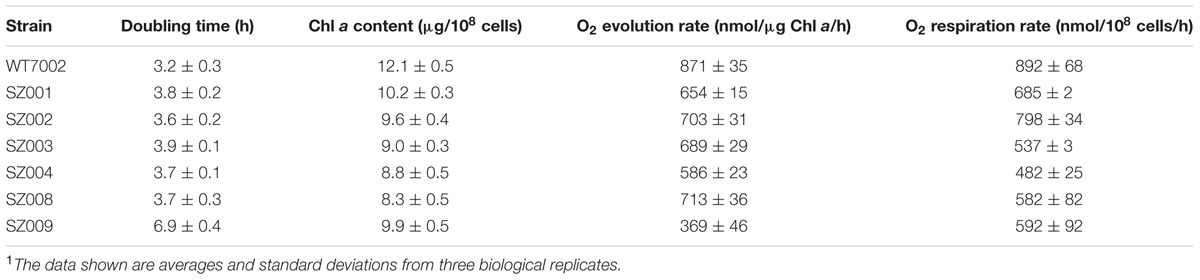
Four single mutant strains (SZ001, SZ002, SZ003, SZ004) showed reduced photoautotrophic growth rates compared to WT7002 (Table 2). The Chl a contents and O2 evolution rates of these mutant strains were all lower than those of WT7002 (Table 2). Dark respiration rates of the SZ003 and SZ004 mutants, which lack a functional succinate dehydrogenase (SDH) enzyme complex, were reduced more than 44% compared to WT7002. Disruptions in the SSA bypass of the oxidative branch caused little or no respiratory deficiency in the corresponding mutant strains (SZ001 and SZ002).
Under both photoautotrophic and dark aerobic respiratory conditions, the succinate pool size increased when either of the succinate dehydrogenase subunits was deleted and decreased when the SSA bypass route was disrupted (Figure 4). These results indicate that, under photoautotrophic and dark aerobic conditions, (1) removal of either subunit of the succinate dehydrogenase complex abolishes the enzyme activity of succinate dehydrogenase; (2) the SSA route is functional in vivo; and (3) the oxidative TCA cycle (operation in the clock-wise direction of Figure 1) together with other potential alternative succinate synthesis pathways are dominant over the reductive TCA branch (counter clock-wise direction of Figure 1) in the synthesis of succinate. In agreement with the present findings, succinate accumulation increased ∼2- to 3-fold in Synechocystis 6803 when 2-OG was added to the growth medium of an sdhB mutant (Cooley et al., 2000; Cooley and Vermaas, 2001).
GABA Shunt Enzymatic Assays
The genes encoding glutamate decarboxylase (gadA6803) and N-acetylornithine aminotransferase (argD6803) of Synechocystis 6803, as well as the N-acetylornithine aminotransferase (argD7002) of Synechococcus 7002 were successfully expressed and the products purified from E. coli as N-terminally poly-[His]10-tagged proteins. The purified proteins were positively immunoreactive with commercial antibodies to the poly-[His]10 tag (Figure 5A), and were also identified by tryptic peptide mass fingerprinting (data not shown).
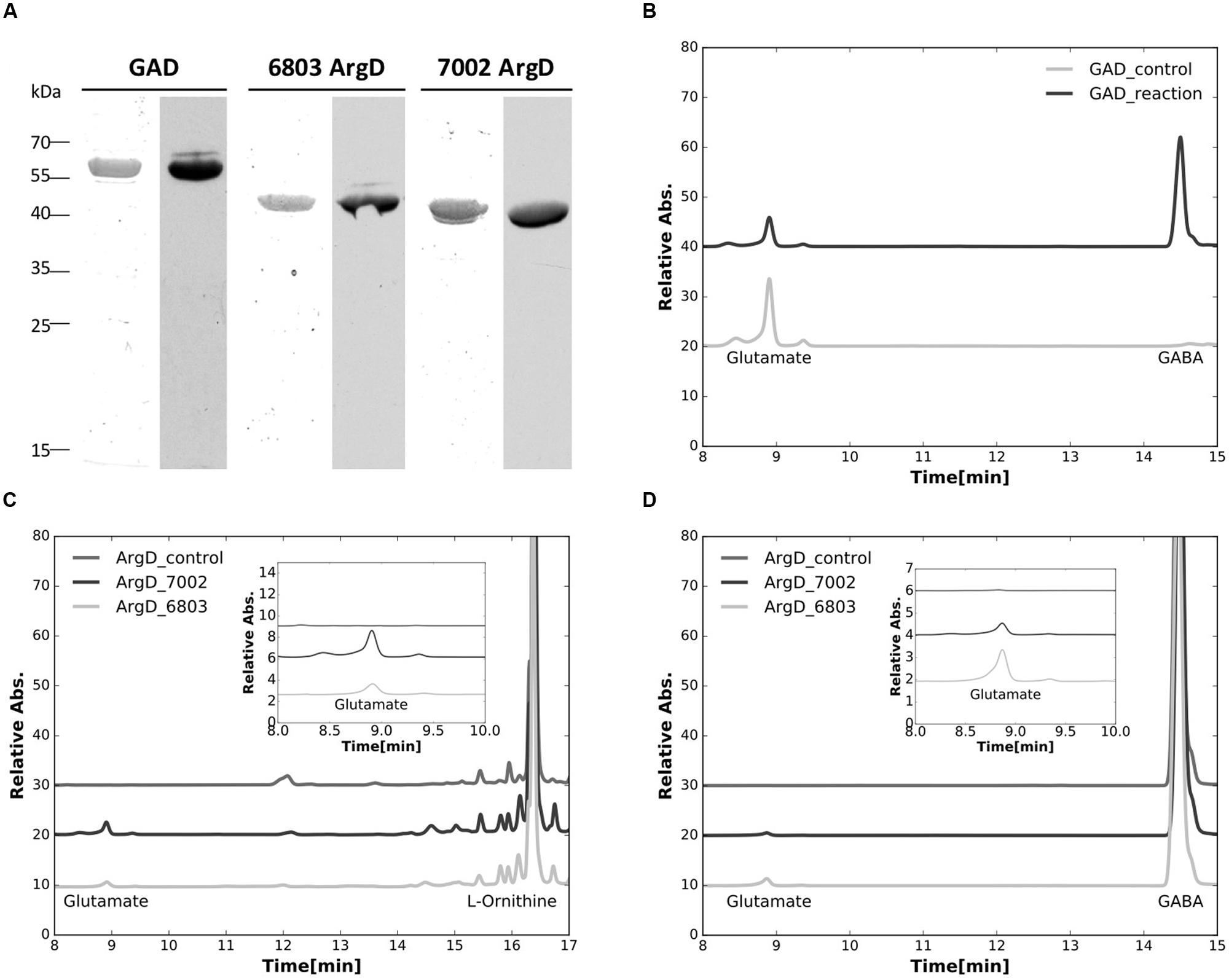
FIGURE 5. Characterizations of purified recombinant proteins. (A) Purified recombinant proteins of the glutamate decarboxylase (GadA), and the N-acetylornithine aminotransferase from both Synechocystis sp. PCC 6803 (ArgD6803) and Synechococcus sp. PCC 7002 (ArgD7002) were analyzed. Left lanes for each enzyme were stained with Coomassie blue and right lanes were detected by immunoblotting with antibodies to the poly-[His]10-tag. (B) HPLC analysis showing the conversion of glutamate to GABA, catalyzed by the purified glutamate decarboxylase (GAD_reaction). (C) HPLC analysis showing the formation of glutamate from L-ornithine catalyzed by the purified N-acetylornithine aminotransferase from both Synechocystis sp. PCC 6803 (ArgD_6803) and Synechococcus sp. PCC 7002 (ArgD_7002). (D) HPLC analysis showing the formation of glutamate from GABA catalyzed by the purified N-acetylornithine aminotransferase from both Synechocystis sp. PCC 6803 (ArgD_6803) and Synechococcus sp. PCC 7002 (ArgD_7002), demonstrating that N-acetylornithine aminotransferase can also function as GABA aminotransferase. Insets in (C,D) represent the enlarged parts of the elution curves from 8 to 10 min to illustrate the changes of the glutamate peaks observed more clearly. It should be noted that the large differences in peak heights occur because the different compounds have very different molar extinction coefficients at the detection wavelength. Control experiments in all of these assays were performed the same way without recombinant proteins added.
The enzymatic activity of glutamate decarboxylase, GadA6803, protein was characterized by confirming the consumption of substrate, glutamate, and parallel formation of GABA (Figure 5B). This result establishes that the gadA gene encodes glutamate decarboxylase and this protein catalyzes the conversion of glutamate to GABA. The enzymatic activity of N-acetylornithine aminotransferase (ArgD6803) was also assayed. When the protein product from argD6803 or argD7002 was incubated with L-ornithine and 2-OG, glutamate was produced (Figure 5C). This shows that these two ArgD proteins have N-acetylornithine aminotransferase activity. Because it has been suggested that N-acetylornithine aminotransferase could also function as GABA aminotransferase (Xiong et al., 2014), we investigated this postulated activity. When the protein product from argD6803 or argD7002 was incubated with GABA and 2-oxoglutarate, glutamate was produced in the reaction mixture (Figure 5D). These biochemical results established that N-acetylornithine aminotransferase is a bifunctional enzyme that has both N-acetylornithine aminotransferase and GABA aminotransferase activities.
Construction of Glutamate Decarboxylase Expression Strains
The foregoing biochemical results validated the in vitro enzymatic activities of the postulated GABA shunt, which is clearly functional in some cyanobacteria, in particular Synechocystis 6803. However, the apparent absence of glutamate decarboxylase in most cyanobacteria (e.g., Synechococcus 7002) suggests that the GABA shunt is not universally present in cyanobacteria (Xiong et al., 2014). To investigate the biochemical role of the GABA shunt in cyanobacterial metabolism, a strain of Synechococcus 7002 expressing glutamate decarboxylase was constructed (SZ008), in which the glutamate decarboxylase (gadA6803) from Synechocystis 6803 was expressed under control of the strong PcpcBA6803 promoter (Xu et al., 2011; Zhou et al., 2014). The presence of the plasmid and the incorporation of the gadA6803 gene into Synechococcus 7002 were verified by PCR amplification of the gadA6803 gene (Supplementary Figure S2A) and DNA sequencing of the amplicon further confirmed the construction of strain SZ2008. Successful expression of glutamate decarboxylase was confirmed by immunoblotting using antibodies against His10-tag, as shown in Supplementary Figure S2B. The gene encoding 2-oxoglutarate decarboxylase (ogdA; SYNPCC7002_A2770) was deleted in strain SZ008 to generate strain SZ009. Complete segregation of the mutant alleles was confirmed by PCR using primer set A2770D1-A2770D4 as shown in Supplementary Figure S2C. Compared to the WT7002 strain, strain SZ008 had a slower growth rate, lower Chl a content, and reduced O2 evolution and consumption (respiration) rates, which are possibly caused by protein over-expression and the resulting metabolic burden in this mutant (Table 2). Interestingly, the SZ009 strain showed further reductions in growth rate and O2 evolution rate, but had similar Chl a contents and O2 consumption (respiration) rate when compared to the SZ008 strain (Table 2).
Metabolic Profiling of Glutamate Decarboxylase Expression Strains
Metabolic profiling of strains SZ008 and SZ009 described above was performed as described in the Materials and Methods. In the SZ008 strain, the 2-OG level decreased to about 10% of the level in WT7002, while the SSA pool was ∼4.4-fold higher than that in WT7002 under photoautotrophic growth conditions. In addition, glutamine and glutamate concentrations were also higher (7.8- and 1.4-fold, respectively) in strain SZ008 than in the WT7002 strain. These results indicated that 2-OG was indeed directed into the GABA shunt, which produced elevated levels of glutamine, glutamate and SSA (Figure 6A).
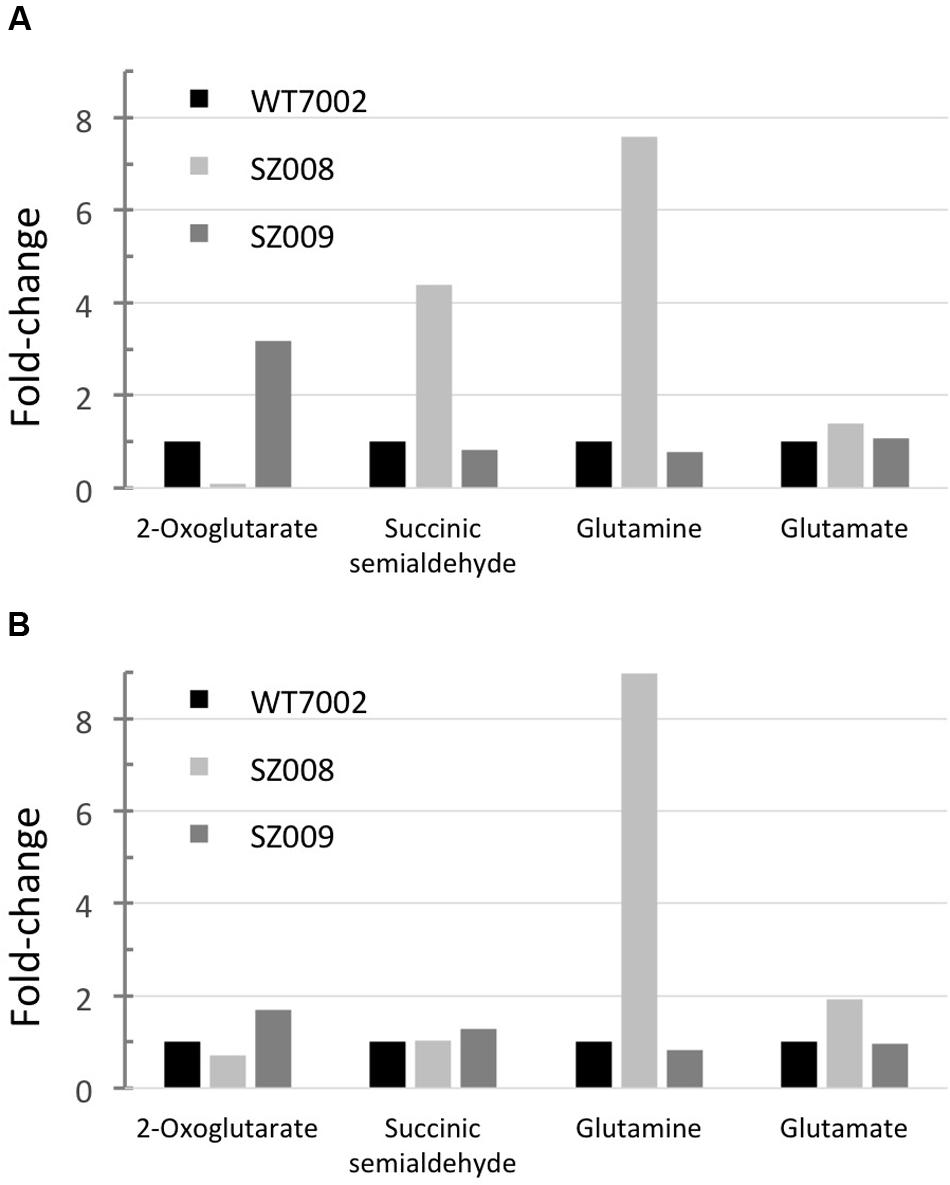
FIGURE 6. Metabolites profiling of GABA shunt mutant strains. Relative metabolites concentrations in strains SZ008 and SZ009, which had been grown under photoautotrophic conditions (A) or dark aerobic conditions (B). Relative concentrations for each metabolite in wild-type Synechococcus 7002 (WT7002) under these growth conditions were set to 1 unit for comparison.
In the SZ009 strain, 2-OG accumulated to a higher level (3.2-fold) than in the WT7002 strain, while SSA, glutamine and glutamate remained at nearly the same levels as in the WT7002 strain under photoautotrophic conditions. Additionally, it was shown that the SSA content was 50% lower than in the SZ001 mutant strain (Figure 2B). Therefore, it is clear that in the SZ009 strain, in which 2-oxoglutarate decarboxylase is absent, the heterologously expressed GadA enables 2-oxoglutarate to be converted to SSA through the reactions of the GABA shunt. However, the reconstructed GABA shunt is less efficient than 2-oxoglutarate decarboxylase in converting 2-OG to SSA, because 2-OG accumulated to a significantly higher level in strain SZ009 (Figure 6A).
After these two strains were incubated under dark aerobic conditions for 3 h, the SSA concentration in strain SZ008 was similar to that in strain WT7002. However, a higher (1.7-fold) amount of 2-OG was present in the SZ009 strain, while the 2-OG concentration in the SZ008 strain decreased to ∼70% of the level in WT7002 (Figure 6B). These metabolite changes imply that the heterologous GABA shunt is not as efficient as the native 2-OGDC/SSADH bypass in converting 2-OG to SSA in Synechococcus 7002, although this could of course result from the heterologous origin and production of GadA6803.
Discussion
These data from this study reveal that during photoautotrophic growth, the TCA cycle functions primarily as a branched pathway for the production of the two essential precursor metabolites, 2-OG and oxaloacetate. Comparison of pool sizes of SSA in WT7002 and the two single-gene knockout mutants (SZ001 and SZ002), which interrupt the 2-OGDC/SSADH bypass route, demonstrate this. When cells respire in the dark, SSA accumulated when SSADH (SsaD) was missing, and SSA levels decreased when OgdA (2-OGDC), the enzyme that produces SSA, was missing. SSA did not accumulate in the SZ002 mutant strain under photoautotrophic conditions, indicating that the SSA route is less active under photoautotrophic conditions than under dark aerobic respiratory conditions. The operation of the TCA cycle as a branched pathway under photoautotrophic conditions is reasonable because cyanobacteria that are fixing CO2 should not be simultaneously be oxidizing acetyl units to CO2 for ATP production, which can be obtained more efficiently from photosynthetic electron transport. The metabolic importance of the SSA route during respiratory metabolism is demonstrated by the metabolite data in Figure 4 and further shown by the 2- and 4-fold increases in the relative abundance of transcripts for ogdA (SynPCC7002_A2770) and ssaD (SynPCC7002_A2771), respectively, under dark aerobic conditions (Ludwig and Bryant, 2011).
Under photomixotrophic conditions, 13C-glycerol was rapidly incorporated into SSA. The M4/M0 ratio (Figure 2A) indicates that at least 20% of the carbon molecules in SSA were labeled within 40 min. This observation demonstrates that the SSA route is a major route for converting 2-OG into downstream metabolites. This result contrasts with a 13C-metabolic flux study in Synechocystis 6803, in which flux through the SSA route was found to be minimal under photomixotrophic conditions (You et al., 2014). One possible explanation for the decreasing percentage of 13C-labeled SSA under dark conditions is that there was an increased influx of 12C-labeled carbon originating from glycogen consumption in the dark. 12C-labeled carbon from glycogen entering the TCA cycle might have diluted the percentage of 13C in the total SSA pool.
Deletion of the OgdA/SsaD bypass route may reduce the availability of some unknown metabolite(s) that are not essential but could be beneficial to the growth of Synechococcus 7002. The knockout mutants of the OgdA/SsaD bypass route (SZ001 and SZ002) exhibit reduced photoautotrophic growth rates as observed previously (Zhang and Bryant, 2011) and as confirmed again in this study (Table 2). Both the reduced Chl a contents and oxygen evolution rates of these knockout mutants appear to be responsible for the reduced photoautotrophic growth. These results demonstrate that prior flux balance analysis (FBA) simulations are incorrect in predicting flux to be negligible through the OgdA/SsaD bypass in cyanobacteria under photoautotrophic conditions (Knoop et al., 2013; Vu et al., 2013; Hendry et al., 2016).
The succinate pool size of the SZ007 mutant was similar to that of the WT7002, which contradicts our expectation that this quadruple knockout mutant should not produce any succinate, because both the SDH and the OgdA/SsaD bypass routes are blocked in this mutant. Three TCA cycle variants produce succinate as an intermediate metabolite: (1) the OgdA/SsaD bypass route; (2) the GABA shunt; and (3) the glyoxylate cycle (Steinhauser et al., 2012). However, proteins required to complete the GABA shunt, namely GadA, and the glyoxylate shunt, isocitrate lyase and malate synthase, are all absent in Synechococcus 7002 (Zhang and Bryant, 2015). Thus, there must be yet another route to produce succinate in Synechococcus 7002. Although, it is well-known that 2-OGDH is absent in all cyanobacteria, succinyl-CoA synthase (encoded by SYNPCC7002_A0890 and SYNPCC7002_A0891) could still convert succinyl-CoA into succinate. Succinyl-CoA can potentially be produced from the degradation pathways for valine, leucine and isoleucine1, although not all of the genes involved in these pathways are annotated yet in Synechococcus 7002. Pool sizes of succinate and malate were significantly reduced in the SZ006 deletion mutant (Figure 4), which supports this hypothesis. Nonetheless, even when the reductive (SDH) and oxidative (OgdA + SsaD) branches of the TCA cycle were blocked, the intracellular succinate concentration in the quadruple mutant strain remained at a comparable level to that in the WT7002 (Figure 4). After searching through the metabolic network of Synechococcus 7002, we found that L-aspartate oxidase (encoded by SYNPCC7002_A0301) catalyzes the conversion of L-aspartate to iminoaspartate, using O2 or fumarate as the electron acceptor. This biochemical reaction yields H2O2 or succinate, depending on the electron acceptor used (Bossi et al., 2002). A more quantitative fluxomic analysis will be needed to identify the flux contributions of SucCD and L-aspartate oxidase routes to the succinate pool in Synechococcus 7002 under both photoautotrophic and dark aerobic respiratory conditions.
The enzymatic and metabolic characterization of the GABA shunt in this study helps to clarify the occurrence of this pathway in cyanobacteria. From the point of view of FBA, the GABA shunt is stoichiometrically identical to the recently discovered TCA cycle variant using OgdA and SsaD. In principle, both variants should result in identical biomass yields if the regulation were identical. However, FBA indicates that the biomass yield using these two pathways is lower than for the conventional cycle using 2-OGDH under respiratory metabolism (Shastri and Morgan, 2005; Knoop et al., 2013). FBA also predicts that autotrophic growth is similarly reduced when metabolites are forced through the 2-OGDH complex or the 2-OGDC/SSADH bypass but not through the GABA shunt. Based on these findings, it has been suggested that the GABA shunt may be an evolutionarily favorable solution to completing the TCA cycle (Nogales et al., 2012). This may also be the reason for the existence of the GABA shunt in some Prochlorococcus and marine Synechococcus species that lack the 2-OGDC/SSADH bypass. However, our data demonstrate that the GABA shunt is actually less efficient in converting 2-oxoglutarate to SSA in the SZ008 strain of Synechococcus 7002. One possible reason might be that heterologous enzyme activity of the expressed glutamate decarboxylase from Synechocystis 6803 is not optimal in Synechococcus 7002.
Compared to the wild-type, 2-OG levels decreased dramatically in the strain expressing gadA, and inactivation of ogdA caused 2-OG levels to increase about threefold under photoautotrophic growth conditions. The 2-OG levels might have decreased in SZ008 because there is a second pathway for the consumption of this intermediate in strain SZ008. Deletion of ogdA in SZ009 eliminates a major route for 2-OG conversion to SSA, which led to an increase in the 2-OG level in those cells. This explanation is supported by an increase in SSA levels when there are two routes for its synthesis, but SSA levels in SZ009 decreased slightly when OgdA was missing under photoautotrophic conditions. In the dark, 2-OG levels showed much smaller differences and no accumulation of SSA was observed in strain SZ008. Glutamate levels were similar in the WT7002 and SZ009 strains but increased slightly in SZ008. The large increase in glutamine in SZ008 in both the light and the dark is more difficult to explain, but it seems unlikely to be directly related to the introduced GadA because the gene encoding this enzyme is present in both SZ008 and SZ009. The glutamine levels likewise are not correlated with the levels of SSA or glutamate. Instead, this increase seems more likely to be due to currently unexplained kinetic or regulatory effects that might be associated with the lower 2-OG levels in the strain overexpressing the gadA gene (SZ008). One possibility is that glutamine synthetase remains active, but the low levels of 2-OG do not allow 2-OG:glutamine amidotransferase (GOGAT) to convert glutamine efficiently into glutamate. Because cells are not nitrogen limited and have a lower 2-OG level, glutamine might accumulate. An explanation for the accumulation of glutamine will require further studies that should probably include transcriptional profiling of these strains.
Genes encoding enzymes for the GABA shunt can be found in the genomes of some cyanobacteria (e.g., Synechocystis sp. PCC 6803, Nostoc sp. PCC 7107, Prochlorococcus marinus str. MIT 9303 and Synechococcus sp. RCC307). However, Prochlorococcus marinus str. MIT 9303 and Synechococcus sp. RCC307 lack the OgdA/SsaD (2-OGDC/SSADH) bypass, and many other Prochlorococcus and marine Synechococcus species seemingly lack the GABA shunt as well as the 2-OGDC/SSADH bypass. These organisms either have yet another alternative bypass or must have a branched (i.e., interrupted) TCA cycle. Similarly, many other cyanobacteria (e.g., Synechococcus 7002) apparently lack the glutamate decarboxylase needed to complete the GABA shunt. Thus, unlike the SSA bypass, the GABA shunt seems to be present in only a few cyanobacteria (Xiong et al., 2014). Considering the mosaic distribution of the complete TCA cycle, the GABA shunt, as well as the glyoxylate cycle in cyanobacteria, the presence of alternative TCA pathways suggests that different options exist for reversible regulation by metabolites. For example, in the case of the GABA pathway, regulation of TCA cycle and hence redox energy production should be possible by nitrogen availability via the shared glutamate/glutamine intermediates.
Conclusion
Cyanobacteria collectively have four different TCA cycle variants: (1) an open, branched-chain pathway; (2) the OGDC/SSADH bypass; (3) the glyoxylate cycle; and (4) the GABA shunt (Zhang and Bryant, 2014). The sporadic distribution of most of these variants among the highly diverse cyanobacteria, as well as the observation that most cyanobacteria have no more than two of these variants, suggests that these variations reflect specialized responses to the specific environmental niches and must confer important metabolic capabilities upon the organisms that possess them. However, as demonstrated here, it is possible to introduce these variants into strains to modify the metabolic capabilities in ways that could be exploited for biotechnological purposes.
Author Contributions
SZ constructed and characterized mutants and wrote the manuscript; XQ developed the analytical procedure for detecting succinic acid semialdehyde, analyzed the metabolites in the mutant strains, and assisted in writing the manuscript. SC assisted in the analysis of the mutants. GD and DB supervised the research and participated in writing the manuscript.
Funding
This work was supported by the National Science Foundation grants MCB-1021725 and MCB-1613022 (to DB) and MCB-1515511 (to GD). The Air Force Office of Scientific Research Grants provided additional funding via grants FA9550-05-1-0365 and FA9550-11-1-0148 (to DB).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01972/full#supplementary-material
Footnotes
References
Bennette, N. B., Eng, J. F., and Dismukes, G. C. (2011). An LC–MS-based chemical and analytical method for targeted metabolite quantification in the model cyanobacterium Synechococcus sp. PCC 7002. Anal. Chem. 83, 3808–3816. doi: 10.1021/ac200108a
Bossi, R. T., Negri, A., Tedeschi, G., and Mattevi, A. (2002). Structure of fad-bound l-aspartate oxidase: insight into substrate specificity and catalysis. Biochemistry 41, 3018–3024. doi: 10.1021/bi015939r
Cooley, J. W., Howitt, C. A., and Vermaas, W. F. (2000). Succinate:quinol oxidoreductases in the cyanobacterium Synechocystis sp. strain PCC 6803: presence and function in metabolism and electron transport. J. Bacteriol. 182, 714–722. doi: 10.1128/JB.182.3.714-722.2000
Cooley, J. W., and Vermaas, W. F. (2001). Succinate dehydrogenase and other respiratory pathways in thylakoid membranes of Synechocystis sp. strain PCC 6803: capacity comparisons and physiological function. J. Bacteriol. 183, 4251–4258. doi: 10.1128/JB.183.14.4251-4258.2001
Ducat, D. C., Way, J. C., and Silver, P. A. (2011). Engineering cyanobacteria to generate high-value products. Trends Biotechnol. 29, 95–103. doi: 10.1016/j.tibtech.2010.12.003
Field, C. B., Behrenfeld, M. J., Randerson, J. T., and Falkowski, P. (1998). Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240. doi: 10.1126/science.281.5374.237
Frigaard, N. U., Sakuragi, Y., and Bryant, D. A. (2004). Gene inactivation in the cyanobacterium Synechococcus sp. PCC 7002 and the green sulfur bacterium Chlorobium tepidum using in vitro-made DNA constructs and natural transformation. Methods Mol. Biol. 274, 325–340. doi: 10.1385/1-59259-799-8:325
Green, L. S., Li, Y., Emerich, D. W., Bergersen, F. J., and Day, D. A. (2000). Catabolism of alpha-ketoglutarate by a sucA mutant of Bradyrhizobium japonicum: evidence for an alternative tricarboxylic acid cycle. J. Bacteriol. 182, 2838–2844. doi: 10.1128/JB.182.10.2838-2844.2000
Hendry, J. I., Prasannan, C. B., Joshi, A., Dasgupta, S., and Wangikar, P. P. (2016). Metabolic model of Synechococcus sp. PCC 7002: prediction of flux distribution and network modification for enhanced biofuel production. Bioresour. Technol. 213, 190–197. doi: 10.1016/j.biortech.2016.02.128
Knoop, H., Gründel, M., Zilliges, Y., Lehmann, R., Hoffmann, S., Lockau, W., et al. (2013). Flux balance analysis of cyanobacterial metabolism: the metabolic network of Synechocystis sp. PCC 6803. PLoS Comput. Biol. 9:e1003081. doi: 10.1371/journal.pcbi.1003081
Krishnan, A., Kumaraswamy, G. K., Vinyard, D. J., Gu, H., Ananyev, G., Posewitz, M. C., et al. (2015a). Metabolic and photosynthetic consequences of blocking starch biosynthesis in the green alga Chlamydomonas reinhardtii sta6 mutant. Plant J. 81, 947–960. doi: 10.1111/tpj.12783
Krishnan, A., Zhang, S., Liu, Y., Tadmori, K. A., Bryant, D. A., and Dismukes, G. C. (2015b). Consequences of ccmR deletion on respiration, fermentation and H2 metabolism in cyanobacterium Synechococcus sp. PCC 7002. Biotechnol. Bioeng. 113, 1448–1459. doi: 10.1002/bit.25913
Kumaraswamy, G. K., Guerra, T., Qian, X., Zhang, S., Bryant, D. A., and Dismukes, G. C. (2013). Reprogramming the glycolytic pathway for increased hydrogen production in cyanobacteria: metabolic engineering of NAD+-dependent GAPDH. Energy Environ. Sci. 6, 3722–3731. doi: 10.1039/C3EE42206B
Lan, E. I., and Liao, J. C. (2012). ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 109, 6018–6023. doi: 10.1073/pnas.1200074109
Ludwig, M., and Bryant, D. A. (2011). Transcription profiling of the model cyanobacterium Synechococcus sp. strain PCC 7002 by next-gen (SOLiD) sequencing of cDNA. Front. Microbiol. 2:41. doi: 10.3389/fmicb.2011.00041
Nogales, J., Gudmundsson, S., Knight, E. M., Palsson, B. O., and Thiele, I. (2012). Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis. Proc. Natl. Acad. Sci. U.S.A. 109, 2678–2683. doi: 10.1073/pnas.1117907109
Qian, X., Kumaraswamy, G. K., Zhang, S., Gates, C., Ananyev, G. M., Bryant, D. A., et al. (2015). Inactivation of nitrate reductase alters metabolic branching of carbohydrate fermentation in the cyanobacterium Synechococcus sp. strain PCC 7002. Biotechnol. Bioeng. 113, 979–988. doi: 10.1002/bit.25862
Rippka, R., Deruelles, J., Waterbury, J. B., Herdman, M., and Stanier, R. Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111, 1–61. doi: 10.1099/00221287-111-1-1
Sakamoto, T., and Bryant, D. A. (1997). Growth at low temperature causes nitrogen limitation in the cyanobacterium Synechococcus sp. PCC 7002. Arch. Microbiol. 169, 10–19. doi: 10.1007/s002030050535
Shastri, A. A., and Morgan, J. A. (2005). Flux balance analysis of photoautotrophic metabolism. Biotechnol. Prog. 21, 1617–1626. doi: 10.1021/bp050246d
Shen, G., and Bryant, D. A. (1995). Characterization of a Synechococcus sp. strain PCC 7002 mutant lacking photosystem I. Protein assembly and energy distribution in the absence of the photosystem I reaction center core complex. Photosynth. Res. 44, 41–53. doi: 10.1007/BF00018295
Steinhauser, D., Fernie, A. R., and Araújo, W. L. (2012). Unusual cyanobacterial TCA cycles: not broken just different. Trends Plant Sci. 17, 503–509. doi: 10.1016/j.tplants.2012.05.005
Stevens, S. E., Patterson, C. O. P., and Myers, J. (1973). The production of hydrogen peroxide by blue-green algae: a survey. J. Phycol. 9, 427–430. doi: 10.1111/j.1529-8817.1973.tb04116.x
Struys, E. A., Jansen, E. E. W., Gibson, K. M., and Jakobs, C. (2005). Determination of the GABA analogue succinic semialdehyde in urine and cerebrospinal fluid by dinitrophenylhydrazine derivatization and liquid chromatography–tandem mass spectrometry: application to SSADH deficiency. J. Inherit. Metab. Dis. 28, 913–920. doi: 10.1007/s10545-005-0111-0
Tang, K.-H., Tang, Y. J., and Blankenship, R. E. (2011). Carbon metabolic pathways in phototrophic bacteria and their broader evolutionary implications. Front. Microbiol. 2:165. doi: 10.3389/fmicb.2011.00165
Tank, M., and Bryant, D. A. (2015). Nutrient requirements and growth physiology of the photoheterotrophic acidobacterium, Chloracidobacterium thermophilum. Front. Microbiol. 6:226. doi: 10.3389/fmicb.2015.00226
Vu, T. T., Hill, E. A., Kucek, L. A., Konopka, A. E., Beliaev, A. S., and Reed, J. L. (2013). Computational evaluation of Synechococcus sp. PCC 7002 metabolism for chemical production. Biotechnol. J. 8, 619–630. doi: 10.1002/biot.201200315
Wermuth, C. G. (1979). A convenient synthesis of (-ethoxy-(-butyrolactone and of succinic semialdehyde. J. Org. Chem. 44, 2406–2408. doi: 10.1021/jo01328a015
Xiong, W., Brune, D., and Vermaas, W. F. (2014). The γ-aminobutyric acid shunt contributes to closing the tricarboxylic acid cycle in Synechocystis sp. PCC 6803. Mol. Microbiol. 93, 786–796. doi: 10.1111/mmi.12699
Xu, Y., Alvey, R. M., Byrne, P. O., Graham, J. E., Shen, G., and Bryant, D. A. (2011). Expression of genes in cyanobacteria: adaptation of endogenous plasmids as platforms for high-level gene expression in Synechococcus sp. PCC 7002. Methods Mol. Biol. 684, 273–293. doi: 10.1007/978-1-60761-925-3_21
You, L., Berla, B., He, L., Pakrasi, H. B., and Tang, Y. J. (2014). 13C-MFA delineates the photomixotrophic metabolism of Synechocystis sp. PCC 6803 under light- and carbon-sufficient conditions. Biotechnol. J. 9, 684–692. doi: 10.1002/biot.201300477
Yu, Y., You, L., Liu, D., Hollinshead, W., Tang, Y. J., and Zhang, F. (2013). Development of Synechocystis sp. PCC 6803 as a phototrophic cell factory. Mar. Drugs 11, 2894–2916. doi: 10.3390/md11082894
Zhang, S., and Bryant, D. A. (2011). The tricarboxylic acid cycle in cyanobacteria. Science 334, 1551–1553. doi: 10.1126/science.1210858
Zhang, S., and Bryant, D. A. (2014). Learning new tricks from an old cycle: the TCA cycle in cyanobacteria, algae and plants. Perspect. Phycol. 1, 73–86. doi: 10.1127/pip/2014/0016
Zhang, S., and Bryant, D. A. (2015). Biochemical validation of the glyoxylate cycle in the cyanobacterium Chlorogloeopsis fritschii strain PCC 9212. J. Biol. Chem. 290, 14019–14030. doi: 10.1074/jbc.M115.648170
Zhang, S., Liu, Y., and Bryant, D. A. (2015). Metabolic engineering of Synechococcus sp. PCC 7002 to produce poly-3-hydroxybutyrate and poly-3-hydroxybutyrate-co-4-hydroxybutyrate. Metab. Eng. 32, 174–183. doi: 10.1016/j.ymben.2015.10.001
Keywords: photosynthesis, TCA cycle, GABA shunt, succinic acid semialdehyde, 2-oxoglutaric acid, Synechococcus sp. PCC 7002, Synechocystis sp. PCC 6803, cyanobacteria
Citation: Zhang S, Qian X, Chang S, Dismukes GC and Bryant DA (2016) Natural and Synthetic Variants of the Tricarboxylic Acid Cycle in Cyanobacteria: Introduction of the GABA Shunt into Synechococcus sp. PCC 7002. Front. Microbiol. 7:1972. doi: 10.3389/fmicb.2016.01972
Received: 03 October 2016; Accepted: 24 November 2016;
Published: 09 December 2016.
Edited by:
Ivan Berg, University of Münster, GermanyReviewed by:
Martin Hagemann, University of Rostock, GermanyJan Zarzycki, Max Planck Institute for Terrestrial Microbiology, Germany
Copyright © 2016 Zhang, Qian, Chang, Dismukes and Bryant. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Donald A. Bryant, ZGFiMTRAcHN1LmVkdQ==
†These authors have contributed equally to this study.
 Shuyi Zhang
Shuyi Zhang Xiao Qian2†
Xiao Qian2† Donald A. Bryant
Donald A. Bryant