- 1Laboratory for Molecular Biology and Microbial Food Safety, Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, Netherlands
- 2Laboratory Genetic Metabolic Diseases, Academic Medical Center, University of Amsterdam, Amsterdam, Netherlands
Weak organic acids like sorbic and acetic acid are widely used to prevent growth of spoilage organisms such as Bacilli. To identify genes involved in weak acid stress tolerance we screened a transposon mutant library of Bacillus subtilis for sorbic acid sensitivity. Mutants of the rodZ (ymfM) gene were found to be hypersensitive to the lipophilic weak organic acid. RodZ is involved in determining the cell’s rod-shape and believed to interact with the bacterial actin-like MreB cytoskeleton. Since rodZ lies upstream in the genome of the essential gene pgsA (phosphatidylglycerol phosphate synthase) we hypothesized that expression of the latter might also be affected in rodZ mutants and hence contribute to the phenotype observed. We show that both genes are co-transcribed and that both the rodZ::mini-Tn10 mutant and a conditional pgsA mutant, under conditions of minimal pgsA expression, were sensitive to sorbic and acetic acid. Both strains displayed a severely altered membrane composition. Compared to the wild-type strain, phosphatidylglycerol and cardiolipin levels were lowered and the average acyl chain length was elongated. Induction of rodZ expression from a plasmid in our transposon mutant led to no recovery of weak acid susceptibility comparable to wild-type levels. However, pgsA overexpression in the same mutant partly restored sorbic acid susceptibility and fully restored acetic acid sensitivity. A construct containing both rodZ and pgsA as on the genome led to some restored growth as well. We propose that RodZ and PgsA play intertwined roles in membrane homeostasis and tolerance to weak organic acid stress.
Introduction
Weak organic acids (e.g., sorbic-, acetic-, and benzoic- acid) are commonly used preservatives in the food industry since they inhibit the growth of spoilage bacteria, yeasts, and molds (Brul and Coote, 1999; Davidson and Harrison, 2002; Beales, 2004; Brul and Ter Beek, 2010). The acids are most effective at pH conditions close to or below their pKa value. Depending on the lipophilic nature of the compound, the neutral undissociated form of the molecule is able to dissolve in and diffuse over the membrane. The hydrophobic tail of, e.g., lipophilic sorbic acid, can also more permanently insert into the membrane perturbing its structure and interfering with the function of proteins (Sheu and Freese, 1972; Stratford and Anslow, 1998; Chu et al., 2009). Inside the cell the acid dissociates and releases protons to a large extend, since most microorganisms exhibit an intracellular pH (pHi) near neutrality. Consequently, the proton gradient dissipates and, depending on the buffering capacity of the cell, the cytosol may acidify, affecting oxidative phosphorylation, the transport of nutrients, and a number of other metabolic functions (Bauer et al., 2003; Cotter and Hill, 2003; Brul and Ter Beek, 2010; van Beilen et al., 2014). The generation of reactive oxygen species has been described in Saccharomyces cerevisiae and recently has been detected in Bacillus cereus upon weak acid stress (Piper, 1999; Mols et al., 2010), which could damage iron-sulfur clusters, proteins, and DNA. Finally, it has been shown that the accumulation of the anion in the cell can cause a rise in osmolarity and affect cytosolic enzymes (Azukas et al., 1961; York and Vaughn, 1964; Russell, 1992).
Bacillus subtilis is one of the organisms that causes food spoilage and its growth is inhibited by weak organic acids (Eklund, 1983). Previously, we preformed time-resolved transcriptome analysis of B. subtilis sub-lethally stressed with potassium sorbate (KS) to elucidate the sorbic acid adaptive responses of this organism at the molecular level (Ter Beek et al., 2008). The results indicated that sorbic acid induces responses normally seen upon nutrient limitation and alters the expression of many cell envelope-related genes. Upregulation of fatty acid biosynthesis (fab) genes and BkdR-regulated genes indicated the synthesis of longer and more branched lipids. We proposed an adaptation in the fatty acid composition of the B. subtilis plasma membrane as a stress response mechanism, since the sensitivity of cells toward the fab inhibitor cerulenin was reduced in the presence of sorbic acid (Ter Beek et al., 2008). Gene groups regulated by extracytoplasmic function sigma factors SigW and SigX (controlling functions associated with the cell surface and transport) (Mascher et al., 2007) were downregulated, indicating a reduction in cell envelope remodeling and, consequently, a change in cell envelope composition. In addition, similar analyses have been performed by us in later experiments for acetic acid stressed cells in comparison to sorbic acid and the classical uncoupler carbonyl cyanide-m-chlorophenyl hydrazone (CCCP). It was observed that the inhibitory effect of sorbic acid seems to be more focussed on the cell membrane than that of acetic acid and that sorbic acid has an effect on cell physiology that is more akin to a classical uncoupler (Ter Beek et al., 2014; van Beilen et al., 2014). In B. subtilis, PgsA is an essential protein committed to the synthesis of phosphatidylglycerol (PG), which besides being the only essential phospholipid is also the precursor for cardiolipin (CL) and lysyl-phosphatidylglycerol (L-PG) (Matsumoto et al., 2006; Salzberg and Helmann, 2008). Lopez and co-workers have shown that salt-stressed cells increase their CL phospholipid levels and decrease both PG and L-PG levels (López et al., 1998). Sorbic acid was shown to interact with the phospholipid headgroups (Chu et al., 2009) and mutants resistant to uncouplers were found to contain mutations in desaturase, resulting in more unsaturated fatty acids (Krulwich et al., 1990). This further corroborates the notion that the membrane is likely to play a crucial role in weak organic acid sensitivity, especially so since this may help to maintain the proton motive force.
A few years ago, RodZ was discovered as new player in bacterial cell morphogenesis (Gerdes, 2009). RodZ has been described in, among others, Escherichia coli, Caulobacter crescentus and B. subtilis and is reported to interact with the bacterial actin-like MreB cytoskeleton controlling cell shape and cell wall synthesis (Shiomi et al., 2008; Alyahya et al., 2009; Bendezú et al., 2009; White et al., 2010; Dempwolff et al., 2011). Interestingly, in several Bacillus species rodZ (ymfM) lies upstream in the genome of pgsA, encoding phosphatidylglycerol phosphate synthase. Until now, no functional link between RodZ and phospholipid synthesis has been reported. But, since RodZ is also associated with the cell elongation complex (den Blaauwen et al., 2008; Alyahya et al., 2009) a link between the only essential phospholipid and the cell envelop seems obvious as they would need to keep track of each other within a growing cell. Moreover, weak organic acids can act as uncouplers, they may interfere with correct localization of several components of the cytoskeleton (Strahl and Hamoen, 2010; van Beilen et al., 2014).
To elucidate novel weak acid resistance mechanisms in B. subtilis we created a mini-Tn10 transposon library and screened for sorbic acid hypersensitive mutants. We show that inactivation of rodZ by a transposon insertion and reduction in pgsA expression using a conditional mutant lead to a weak acid hypersensitive phenotype. We demonstrate that PgsA depletion contributes primarily to the weak acid sensitivity observed and speculate on a possible link between membrane and cell wall homeostasis through RodZ and PgsA.
Materials and Methods
Strains, Growth Conditions, and Genetic Manipulation
All bacterial strains and plasmids used in this study are listed in Table 1. The B. subtilis strains are derivatives of the laboratory 168 wild-type (WT) lab-strains PB2 (trp2C) or 1A700 (trp2C). E. coli strains XL1-Blue and MC1061 were grown in lysogeny broth (LB) at 37°C. B. subtilis strains were grown in LB, buffered with 80 mM 3-(N-morpholino)propanesulfonic acid (MOPS) at pH 7.4 or 6.4 at 37°C. When required for selection, the following antibiotics were added to the medium at given concentrations: 100 μg/ml ampicillin, 1 μg/ml erythromycin, 100 μg/ml spectinomycin, 10 μg/ml kanamycin.
Standard molecular genetics techniques were used as described by Sambrook et al. (1989). The pDG148 vector was used to overexpress rodZ and pgsA (Stragier et al., 1988). The rodZ gene was PCR amplified from B. subtilis PB2 genomic DNA using the rodZ_FW and rodZ_RV primers (see Supplementary Table S1 of the Supporting Information for the sequences of all used primers). All cloning PCR reactions were performed with Pfu polymerase (Fermentas, Thermo Fisher Scientific). The forward primers for each construct contain a ribosome binding site, which is not present on the plasmid when it is cut with HindIII. The PCR product and vector were digested with HindIII and SalI and ligated with T4 ligase (Fermentas), thus creating pDG-rodZ. For pDG-pgsA, the pgsA gene was amplified using the pgsA_FW and pgsA_RV primers and introduced between the HindIII and SalI sites of pDG148. The combined rodZ-pgsA construct was amplified using the rodZ_FW and pgsA_RV primers and inserted as described above to create pDG-rodZ-pgsA. All constructs were first transformed to chemically competent E. coli MC1061 cells before plasmids were isolated and transformed to competent B. subtilis strains. Plasmids were isolated using a QIAprep Spin Miniprep Kit (Qiagen). Competent B. subtillis cells were obtained and their transformations were performed as described previously (Kunst and Rapoport, 1995). The nucleotide sequence of all newly constructed plasmids was verified by sequencing.
Identification of Sorbic Acid-Susceptible Genes
Two independent transposon mutant libraries in B. subtilis WT strain PB2 were created as previously described using the mini-Tn10 delivery vector pIC333 (Steinmetz and Richter, 1994). The resulting libraries were validated for correct transposition efficiency and randomness of transposition (data not shown). Both constructed mutant libraries have the expected statistical properties according to Petit et al. (1990) and Maguin et al. (1992). The two transposon mutant libraries were subjected to a screen to identify mutants hypersensitive to the presence of sorbic acid. The mutant libraries were plated on 80 mM MOPS-buffered LB agar (pH 7.4). Following overnight incubation at 37°C, cells were transferred by replica plating using sterile velvets to 80 mM MOPS-buffered LB agar of pH 6.4, containing 30 mM potassium sorbate (KS). Next, the same sterile velvets were used to replicate cells on LB plates without KS to verify the successful transfer of cells. The mutants that showed no or minimal growth on KS containing LB agar compared to the plates without sorbate were stored at -80°C until further analysis. Chromosomal DNA from the hypersensitive mutants was isolated using the DNeasy Blood & Tissue Kit (Qiagen). The purified DNA was digested with HindIII. DNA from the restriction reaction was self-ligated using a Ready-To-Go DNA T4 Ligase (Amersham Bioscience). Next, ligated DNA was used directly for transformation of E. coli XL1-BLUE cells (Stratagene). Isolated plasmid DNA was used as a template for sequencing with specific to either end of the transposon and enabling sequencing of the cloned chromosomal DNA. The primers were purchased from Isogen Life Science (see Supplementary Table S1 of the Supporting Information for the sequences) and the sequencing was performed by BaseClear. The sequencing results were aligned to the re-annotated genome sequence of B. subtilis 168 using BLAST at the SubtiList database (Barbe et al., 2009)1 in order to identify the affected gene.
Phase-Contrast Microscopy
Bacillus subtilis strains PB2 (WT) and rodZ transposon mutant ATB012 were grown into the exponential phase in 80 mM MOPS-buffered LB medium of pH 6.4. Cells were immobilized on 1% agarose as described previously by Koppelman et al. (2004), and photographed with a CoolSnap fx (Photometrics) CCD camera mounted on an Olympus BX-60 fluorescence microscope through an UPLANFl 100x/1.3 oil objective (Japan).
Characterization of Stress Sensitivity of Various Mutant Strains
To further characterize the acid sensitivity of the (transposon) mutants the cells were grown on both solid agar plates and in liquid medium containing different concentrations of KS, potassium acetate (KAc) or NaCl. For the conditional pgsA mutant strain (Pspac-pgsA) (Hashimoto et al., 2013), the medium contained additional 0.1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG). Overexpression of rodZ and/or pgsA from the pDG148 vector was induced with 1 mM IPTG in exponentially growing cells 3 h before the start of a stress experiment. For the plating assay on solid medium, the cells were first grown exponentially in 80 mM MOPS-buffered liquid LB medium of pH 6.4, containing the appropriate antibiotics and 0.1 mM IPTG, at 37°C. At an OD600 of 0.2, 10-fold serial dilutions of the cultures were spotted on 80 mM MOPS-buffered LB plates of pH 6.4, containing 1% agar and IPTG if required, together with the indicated stress factors. After 24 h of incubation at 37°C pictures of the plates were taken. For the plating assays, the following end-concentrations of chemicals were used: KS: (15, 30, or 40 mM), KAc (80, 125, or 200 mM), and NaCl (0.7 or 1.4 M). Biologically independent experiments were performed at least three times. To monitor the growth of mutants in liquid media, exponentially growing cultures were grown to an OD600 of 0.4. Then, the cells were twofold diluted in a 96-well micro titer plate containing different concentrations of weak organic acid or NaCl (see below), and IPTG if required. Cells were further cultivated in a FluoStar Optima microtiter plate reader (BMG Labtech) under rigorous shaking at 37°C for 12 h. Cells were exposed to the following stresses: KS (15 mM or 40 mM), KAc (40 mM or 125 mM), and NaCl (0.7 or 1.4 M). All conditions were tested in the microtiter plate reader at least in duplicate, and biologically independent experiments were performed at least three times.
Reverse Transcriptase PCR (RT-PCR)
Cells of B. subtilis wild-type strain PB2 and rodZ transposon mutant strain ATB012 were grown exponentially in 80 mM MOPS-buffered LB medium of pH 6.4. At an OD600 of 0.2, half of the cells were exposed to 3 mM of KS and samples were withdrawn from the stressed and control cultures at 0, 10, 20, 30, 45, and 60 min after the addition of KS. Cultures of PB2 harboring empty pDG148 and pDG148-rodZ were grown exponentially in MOPS-buffered LB medium of pH 6.4 and 1 mM IPTG was added 3 h before sampling. At an OD600 of 0.2 samples were taken every 30 min for 2 h. All samples were snap-frozen in liquid nitrogen and stored at -80°C prior to RNA extraction. Biological independent experiments were performed twice.
Total RNA was isolated using the RNeasy Kit (Qiagen), as described by the manufacturer’s instructions. Total RNA was eluted in sterile RNase/DNase free water (Ambion), concentrations were determined with Nanodrop UV spectroscopy (Ocean Optics), and integrity was analyzed by agarose gel electrophoresis. RNA samples were treated with Turbo DNase (Ambion) to remove genomic DNA as described by the manufacturer.
To see if rodZ and pgsA are part of the same transcriptional unit and possibly the same operon, a reverse transcriptase reaction using the Superscript First Strand kit (Invitrogen) was performed with RNA isolated from the WT strain and a single reverse primer annealing to the sequence of pgsA, named pgsA_Q1_RV. cDNA was amplified for 40 cycles in a Biometra T3000 Thermocycler using the rodZ_Q1_FW and pgsA_Q1_RV primers. The PCR product was analyzed on a 1% agarose gel. The visible band was isolated from the gel using the QIAquick gel extraction kit (Qiagen) and the DNA was sequenced.
To determine the expression levels of rodZ and pgsA in the WT and mutant strains cDNA was synthesized using the Superscript First Strand kit (Invitrogen), using random hexamers. Semi-quantitative real-time PCR analysis was carried out on a 7300 Real-Time PCR system (Applied Biosystems). Primer Express 3.0 software (Applied Biosystems) was used to design specific primers (purchased from Isogen Life Science) for real-time PCR (see Supplementary Table S1 of the Supporting Information). Reactions were carried out in a 20 μl mixture consisting of 1 μl of 3 μM specific primers, 5 μl of 100-fold-diluted cDNA template and SYBR green PCR master mix (Applied Biosystems). The cycling conditions were as follows: 1 cycle at 50°C for 2 min, 1 cycle at 95°C for 10 min, and 40 cycles at 95°C for 15 s and at 60°C for 1 min. Melting curves were used to monitor the specificity of the reaction. RNA of all time points and independent experiments were analyzed with real-time PCR in triplicate. Because the amplification efficiencies of the target and reference genes was tested and found to be approximately equal (not shown), the ΔΔCT method could be used to calculate relative gene expressions(Livak and Schmittgen, 2001). The accA (α subunit of acetyl-CoA carboxylase) and rpsM (small subunit ribosomal protein S13) genes were used as two independent internal controls, since their expression levels were stable during exponential growth, irrespective of sorbic acid stress.
Phospholipid Analysis
Cultures of the different B. subtilis strains were grown exponentially in MOPS-buffered LB medium of pH 6.4. At an OD600 of 0.2, half of each culture was stressed with 5 mM KS and 45 min later 50 ml of each cell culture was harvested by centrifugation (5 min, 4000 rpm) and the pellets were frozen in liquid nitrogen. Three independent biological replicates were assessed.
For HPLC-MS/MS analysis, samples were lyophilized overnight. The total protein concentration of the samples was measured using a BCA Protein assay kit (Thermo Scientific). Cell material corresponding to 1 mg protein was resuspended in 300 μl water and 300 μl 1:1 chloroform-methanol (v/v). The following internal standards (obtained from Avanti Polar Lipids, Inc.) were added: phosphatidylglycerol (PG), lysyl-phosphatidylglycerol (L-PG), phosphatidylethanolamine (PE), and cardiolipin (CL). Each sample including internal standards was shaken vigorously and placed on ice for 15 min, after which it was centrifuged at 1000 ×g for 10 min. The organic layer was transferred into another tube, and the aqueous layer was re-extracted with 3 ml of 2:1 chloroform-methanol (v/v). The combined organic layers were evaporated under a stream of nitrogen at 45°C. The residue was dissolved in 150 μl of 50:45:5 chloroform-methanol-water (v/v/v) (Houtkooper et al., 2006).
The relative abundances of the species in the sample-extracts were determined, using HPLC-MS/MS. The liquid-chromatographic separation was achieved on a modular HPLC system (Surveyor; Thermo Finnigan) consisting of a cooled autosampler set at 12°C, a low-flow quaternary MS pump, and an analytical HPLC column: 2.1 × 250 mm silica column, 5 μm particle diameter (Merck). Phospholipids were separated and eluted with a programmed linear gradient between solution B (chloroform-methanol, 97:3, v/v) and solution A (methanol-water, 85:15, v/v) as described previously (Houtkooper et al., 2006). MS/MS analyses were performed on a TSQ Quantum AM (Thermo Finnigan Corporation, San Jose, CA, USA) operated alternating in the negative- and positive ion electrospray ionization (ESI) mode in consecutive runs. The surface induced collision was set at 10 V; spray voltage was 3600 V and the capillary temperature was 300°C. In the MS/MS experiments argon was used as collision gas at a pressure of 0.5 mtorr; collision energy ranged between 20 and 40 eV for the different optimized transitions. In the negative mode mass spectra of CL molecular species were obtained by continuous scanning between m/z 400–m/z 1000 (2 s/scan). In the positive mode characteristic constant neutral loss (CNL) scans were used to selectively detect specific phospholipids in their corresponding retention time windows: CNL(141) for PE, CNL(172) for PG, and CNL(300.1) for L-PG.
Results
Identification of rodZ::mini-Tn10 as a Sorbic Acid-Hypersensitive Clone
In order to identify stress resistance mechanisms against weak organic acids and potential new antimicrobial targets we screened two independent transposon mutant libraries of B. subtilis 168 lab-strain PB2 for sorbic acid-hypersensitive clones on LB plates of pH 6.4 containing 30 mM potassium sorbate (KS). Around 10.000 clones were screened for sorbic acid sensitivity and resulted in 132 candidates in an initial evaluation. After thorough investigation of these mutants both on solid and in liquid medium, we ended up with four verified clones displaying a hypersensitive phenotype toward KS. All four clones were identified to have a transposon insertion in the rodZ (ymfM) gene (Figure 1A). The mutants containing an interrupted rodZ gene were isolated from both independently constructed transposon libraries and the location of insertion (at basepair 51) was found to be the same for all four mutants.
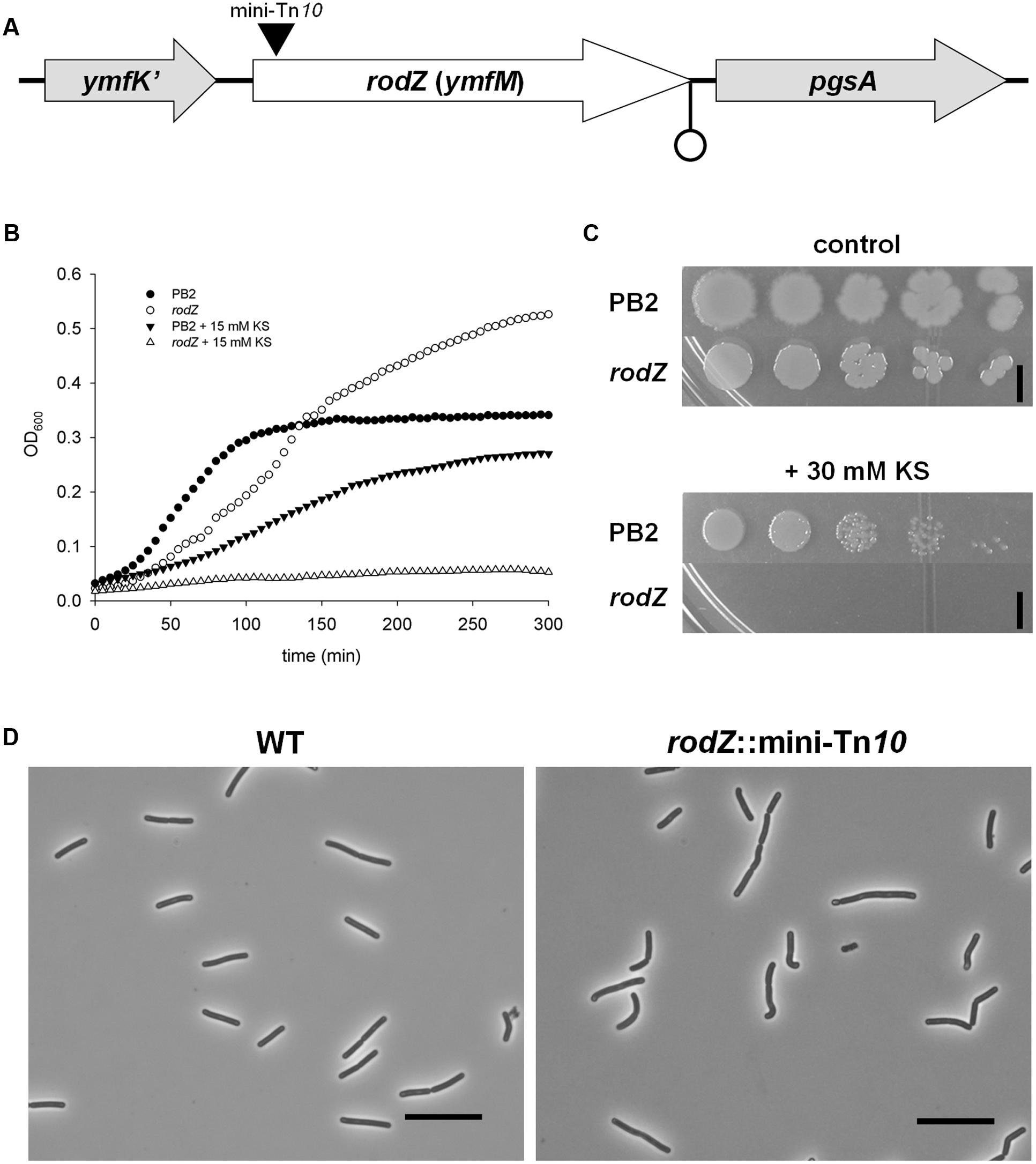
FIGURE 1. Inactivation of rodZ (ymfM) by a transposon leads to a sorbic acid hypersensitive phenotype. (A) Positioning of the rodZ gene between ymfK’ and pgsA in the Bacillus subtilis 168 genome. The black triangle indicates the location of the mini-Tn10 transposon insertion and a possible terminator (due to an inverted repeat) is shown at the end of the rodZ sequence. (B) Growth curves of WT strain PB2 and the rodZ::mini-Tn10 mutant grown in liquid LB medium of pH 6.4 in control conditions (closed and open circles, respectively) or the presence of 15 mM KS (closed downward triangles and open triangles, respectively). (C) Ten-fold dilutions of exponentially growing WT (PB2) and rodZ mutant cells were spotted onto LB plates of pH 6.4 containing no (upper) or 30 mM KS (lower). Plates were photographed after 24 h of growth at 37°C. The black bar indicates 5 mm. (D) Cell morphology of WT (PB2) and rodZ::Tn10 mutant cells as observed with phase contrast microscopy. Phase contrast micrographs of typical examples of cells grown exponentially in liquid cultures are given. Note the curved rod-shape of the rodZ::mini-Tn10 cells. Scale bar: 10 μm.
Sorbic acid hypersensitivity in liquid and on solid media of one of the identified rodZ::mini-Tn10 mutants (strain ATB012) is shown in Figures 1B,C, respectively. In non-stressed liquid cultures of pH 6.4 the transposon mutant lagged a bit behind the WT strain PB2, yet it reached a higher yield than the WT strain after 5 h. However, when stressed with 15 mM KS the rodZ mutant, unlike the WT strain, displayed only a very slow increase in optical density. Although WT strain PB2 was able to grow, the mutant did not form colonies on LB plates of pH 6.4 containing 30 mM KS after overnight incubation at 37°C (Figure 1C, lower). It can be noted that the colonies of mutant strain ATB012 formed on plates are more round when compared to PB2 (Figure 1C, upper). Additionally, the rodZ::Tn10 mutant was found to display an irregular cell morphology, having curved endings and being broader when grown in the exponential phase and examined under the microscope, with wild-type cells being 1.03 ± 0.06 μm and rodZ::Tn10 cells 1.13 ± 0.07 μm (Figure 1D).
rodZ and pgsA Are Co-transcribed
In B. subtilis and in most Gram-positive bacterial genomes that have rodZ, essential gene pgsA lies next to rodZ (Alyahya et al., 2009) (Figure 1A). We hypothesized that rodZ and pgsA are part of the same operon, although the two genes are separated by an inverted repeat which, in spite of the lack of a T-rich tail, may act as a termination sequence (Rasmussen et al., 2009). While we cannot rule out their presence, we did not find indications for additional regulatory elements of pgsA expression. To confirm that rodZ and pgsA are indeed co-transcribed, we performed a reverse transcriptase reaction using one single reverse primer annealing to pgsA and purified RNA of WT strain PB2 as the template. A PCR reaction using specific primers for rodZ and pgsA resulted in one specific product. The sequence of the fragment corresponded exactly with the region between rodZ and pgsA (Supplementary Figure S1 of the Supporting Information).
Additionally we tested whether the mRNA levels of rodZ and/or pgsA were affected by sorbic acid stress. Stressing exponentially growing cells of the WT strain in LB medium of pH 6.4 with 3 mM KS did not significantly change the expression levels of both genes during the first 60 min after KS stress (See Supplementary Excel Data Sheet S1). This observation was in line with our previous studies performing microarrays with 3 mM KS-stressed cells in defined minimal medium of pH 6.4 (Ter Beek et al., 2008).
Stress Profiles of the rodZ Transposon Mutant and a Conditional pgsA Mutant
RT-PCR results indicated a transcriptional link between rodZ and pgsA. Therefore, we decided to further characterize the identified sorbic acid-sensitive rodZ::mini-Tn10 strain, as well as a conditional pgsA mutant (pgsA::Pspac-pgsA) strain by testing their susceptibility to other stresses. Since pgsA is essential, this strain (MHB001), kindly provided by Kouji Matsumoto (Hashimoto et al., 2013), needs a minimal amount of IPTG to be able to grow. The lowest IPTG concentration which sustained exponential growth similar to that of the WT strain in liquid cultures was 0.1 mM in our exponential conditions. Hereby we wanted to identify whether these genes are involved specifically in lipophilic sorbic acid stress, in weak organic acid stress more in general or in generic stress tolerance. Thus we tested their stress sensitivity for the highly water soluble weak organic acid: acetic acid. Although the side chains of sorbic- and acetic acid differ significantly, both have a similar pKa of 4.76. Additionally we tested osmotic stress by the addition of NaCl, thus investigating a form of stress unrelated to weak organic acid stress.
Next to the previously described sorbic acid-hypersensitivity (Figure 1) the results clearly showed also hypersensitivity of the rodZ mutant strain for acetic acid stress on plates when compared to the WT strain (Figure 2A). No colonies were formed on LB plates of pH 6.4 containing 200 mM potassium acetate (KAc). Additionally, the ability of rodZ::mini-Tn10 cells to form colonies was clearly more inhibited by 1.4 M NaCl than of cells from the parent B. subtilis strain. In liquid cultures, the rodZ mutant also displayed clear sensitivity toward KAc and NaCl stress (see Supplementary Figure S2 of the Supporting Information). Interestingly, when grown on solid plates representing the various stress conditions and supplemented with 0.1 mM IPTG, the pgsA mutant strain showed a similar stress profile as that of the rodZ mutant (Figure 2B). Hypersensitivity to sorbic- and acetic acid was observed, and in accordance with previous data, a clear stress sensitivity was detected on plates containing 0.7 M NaCl. The conditional pgsA mutant grown in liquid cultures and supplemented with 0.1 mM IPTG also revealed a clear sensitive phenotype toward KS, KAc, and NaCl (see Supplementary Figure S3 of the Supporting Information). However, the observed phenotypes disappeared when liquid cultures contained 10-fold more (1 mM) IPTG (data not shown).
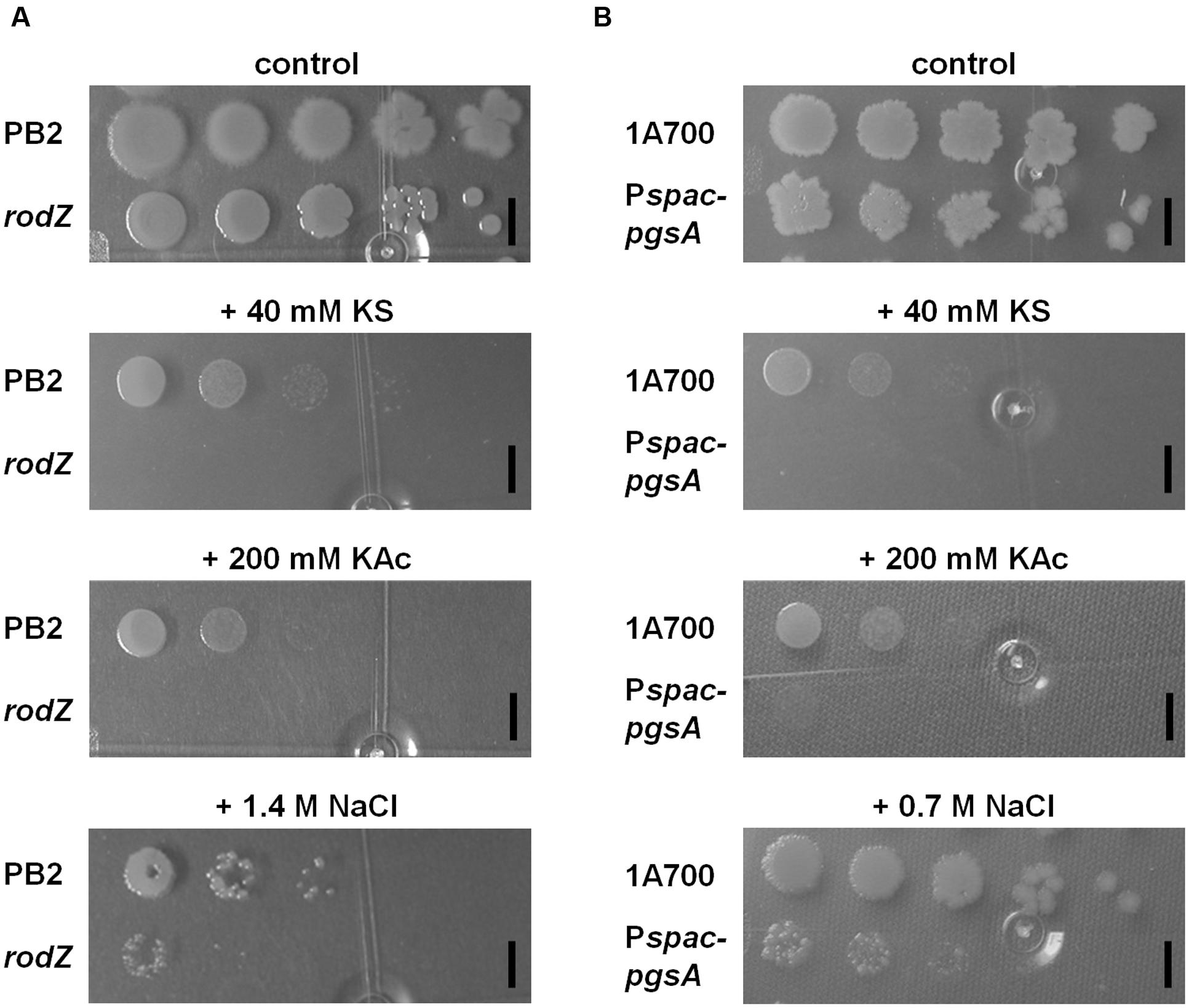
FIGURE 2. Characterization of the rodZ transposon mutant and a conditional pgsA mutant revealed similar (hyper)sensitivity on solid plates for sorbic acid, acetic acid, and NaCl. WT (PB2), rodZ::miniTn10 (ATB012) (A), and WT (1A700), pgsA::Pspac-pgsA (MHB001) (B) were grown exponentially to an OD600 of 0.2 when 10-fold dilutions were spotted onto LB plates of pH 6.4 containing no, 40 mM KS, 200 mM KAc, 0.7, or 1.4 M NaCl. Plates were photographed after 24 h of growth at 37°C. The black bars indicate 5 mm.
The strong overlap in the phenotypes of both the rodZ::mini-Tn10 mutant and the conditional pgsA mutant and the observation that both genes are co-transcribed suggests that the inactivation of rodZ in the transposon mutant gives rise to polar effects. In support of this inference, our RT-PCR data showed ΔΔCT values for pgsA of up to 5 upon comparing rodZ::mini-Tn10 with control cells using accA as reference gene and 3.6 with rpsM as reference (See Supplementary Excel Data Sheet S1). Given the observed phenotypes of both mutants and the involvement of PgsA in crucial membrane phospholipid biosynthesis we next assessed the membrane composition in the rodZ::mini-Tn10 and pgsA::Pspac-pgsA strains.
Phospholipid Composition and Structure in rodZ::mini-Tn10 and pgsA::Pspac-pgsA
Our earlier findings suggested that adaptation to sorbic acid would (in part) be by remodeling of the plasma membrane (Ter Beek et al., 2008). We therefore investigated the effect of sorbic acid on the membrane composition of the WT strain, the rodZ::mini-Tn10 mutant, and the pgsA conditional mutant (pgsA::Pspac-pgsA), as these two mutant strains displayed a similar sensitivity profile. In both the rodZ mutant and in the conditional pgsA mutant strain (under conditions of minimal pgsA expression), the average acyl-chain length of phospholipids was already higher than that of the WT strain when grown without weak organic acid stress. Figure 3 shows the acyl tail length distribution of PG. Similar results were observed for PG-derived phospholipids (CL and L-PG) and PE (see Supplementary Figure S4 of the Supporting Information). Upon KS stress, the most prominently observed phospholipid tail length (2⋅16 = 32 carbon atoms) seemed to increase further in the WT (Figure 3), corroborating our earlier results (Ter Beek et al., 2008). For the conditional pgsA mutant this is less clear, and was not observed in the rodZ mutant.
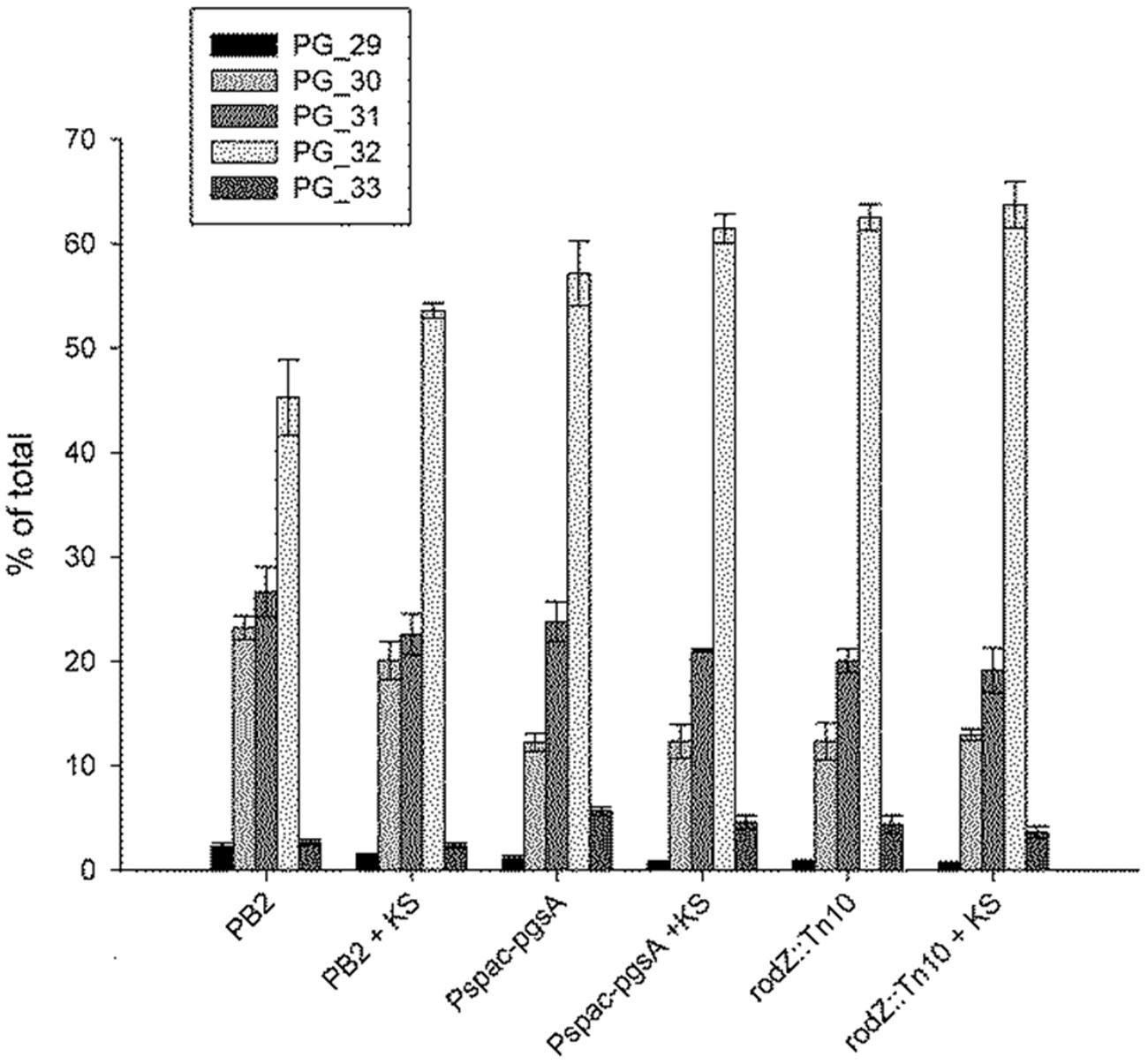
FIGURE 3. Increased average acyl-chain length of phosphatidylglycerol (PG) in the rodZ transposon and conditional pgsA mutant, and increased lengths in the WT strain upon sorbic acid stress. The acyl chain distribution was measured in exponentially growing cells of the WT (PB2), pgsA::Pspac-pgsA and rodZ::mini-Tn10 mutant strains 45 min after treatment with or without 5 mM KS. The PG acyl chain composition is given as a percentage of the total observed PG levels per strain. The number behind PG in the legend indicates the number of carbon atoms per molecule in the acyl chain.
In terms of the presence of different phospholipid classes the main difference between the WT strain and the rodZ::mini-Tn10 strain under non-stressed conditions is a significant decrease in CL and PG phospholipids in the mutant strain (Figure 4). In the conditional pgsA mutant similarly lowered levels of CL and PG phospholipids were observed. Consequently, in both mutant strains the relative PE levels were increased when compared to the levels in the WT strain under control conditions. When the WT strain was exposed to sorbic acid, there was a significant drop observed in the relative L-PG levels (Figure 4), while the relative content of the other phospholipids was hardly affected. This trend was also observed in the conditional pgsA mutant, however, not in the rodZ mutant. Compared to the WT strain, the mutants showed a significant shift in the distribution of the acyl chain lengths (longer) (Figure 3). Also the mutants seems to have a less negative membrane charge due to the significant reduction of PG (-1) and CL (-2) when compared to the WT (Figure 5). Sorbic acid stress does seem to increase the negative net charge on the plasma membrane of WT cells (reduction in L-PG), which may reduce the entry of the sorbate anion. From the data presented it is clear that the phosholipid composition between the two mutants is quite similar, suggesting that the inactivation of rodZ may give rise to polar effects and the hypersensitivity of this mutant strain is perhaps caused by lower expression levels of pgsA.
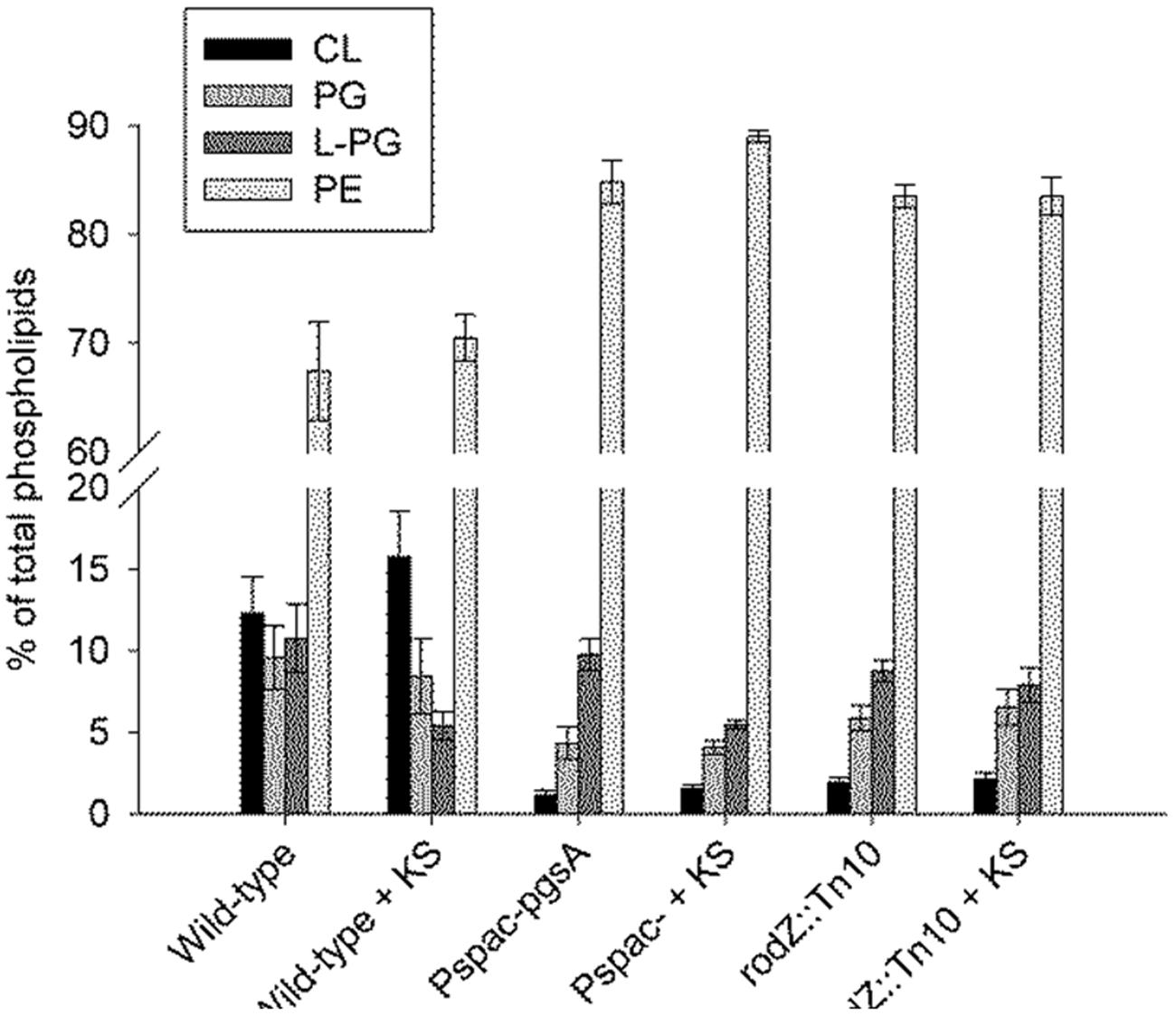
FIGURE 4. Lowered cardiolipin (CL) and phosphatidylglycerol (PG) levels in the rodZ transposon and conditional pgsA mutant, and lowered lysyl-phosphatidylglycerol (L-PG) levels in the WT strain upon sorbic acid stress. The phospholipid composition was measured in exponentially growing cells of the WT (PB2), pgsA::Pspac-pgsA and rodZ::mini-Tn10 mutant strains 45 min after treatment with or without 5 mM KS.
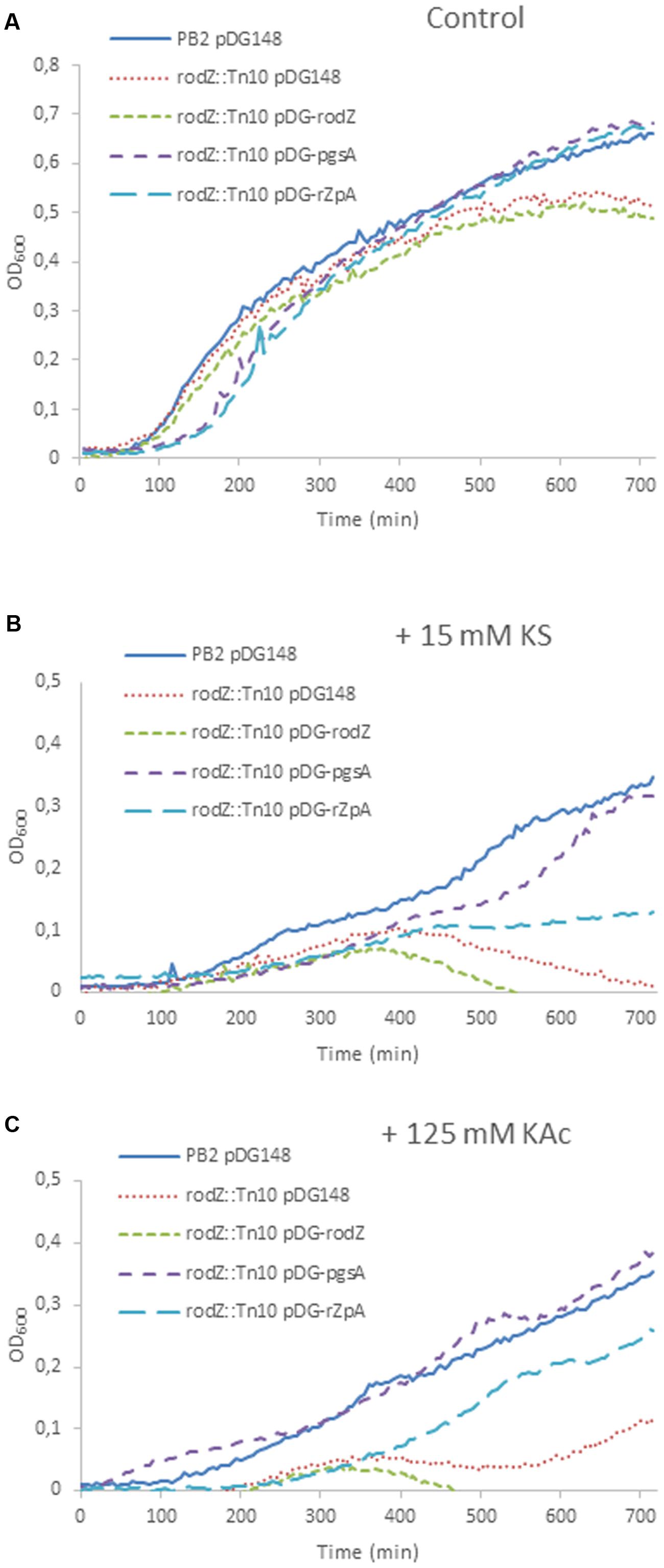
FIGURE 5. Rescue of the weak acid phenotypes by overexpression of pgsA in the rodZ transposon mutant. Shown are growth curves of B. subtilis PB2 pDG148 (solid blue line) and rodZ::Tn10, transformed with pDG148 (red dotted line), pDG-rodZ (green broken line), pDG-pgsA (purple broken line), and pDG-rodZ-pgsA (light-blue broken line) stressed without (A), and with 15 mM KS (B) or 125 mM KAc (C). 1 mM IPTG was added to the pre-cultures of all strains 3 h before the start of the experiments at t = 0 min. to induce overexpression.
Complementation of the rodZ::mini-Tn10 Mutant
We tried to rescue the stress sensitivity of the rodZ transposon mutant by overexpression of rodZ and/or pgsA by introducing the IPTG-inducible plasmid pDG148 (Stragier et al., 1988) containing rodZ and/or pgsA under control of a Pspac promotor. Under control conditions (LB-M, pH = 6.4), the wild-type strain, and mutant strains with pDG148 or pDG-rodZ performed equally during early exponential growth. Mutants containing pDG-pgsA or pDG-rodZ-pgsA displayed a longer lag phase, but ended up with a higher end OD600 (Figure 5A). Weak acid sensitivity of the rodZ::mini-Tn10 mutant with pDG-rodZ (Pspac-rodZ) was not restored (Figure 5B). Complementation with pgsA via pDG-pgsA (Pspac-pgsA) in the rodZ mutant reduced KS stress almost completely. Complementation with the pDG-rodZ-pgsA plasmid (Pspac-rodZ-pgsA) restored some KS sensitivity, but not as much as overexpressing pgsA alone (Figure 5B). When cultured in the presence of 125 mM KAc stress, the overexpression of RodZ and PgsA from pDG-rodZ-pgsA or PgsA alone significantly increased the growth rate of the rodZ::mini-Tn10 mutant strain (Figure 5C). Induction of pDG-rodZ had no effect (Figure 5C).
Discussion
In order to have a direct measure of functional importance for B. subtilis weak organic acid stress resistance we decided to opt for the construction and screening of a transposon mutant library for sorbic acid hypersensitive mutants. Sorbic acid (trans-trans-2,4-hexadienoic acid) is a six-carbon unsaturated fatty acid with a pKa of 4.76 and the acid, or its anionic salt, is commonly utilized by the food industry. We chose to screen for sorbic acid-susceptible genes after pre-growing the transposon library first on LB plates for 24 h. We decided to use rich LB medium in our experiments, so that as many as possible mutants were able to grow with relatively normal rate prior to the screen. Longer incubations revealed many more small colonies emerging on the plates. These slow growers on plates were not used in the screen because they evidently already have severe problems growing in control conditions.
Screening of the library for sorbic acid hypersensitivity led to the identification of a uniquely stress sensitive phenotype in which the transposon was inserted into the rodZ gene (Figures 1B,C). One and the same insertion site was identified in the four discovered clones coming from two independent constructed mutant libraries (Figure 1A). The library was made in B. subtilis WT strain PB2 using the mini-Tn10 delivery vector pIC333 and was designed to increase randomness of the sites of insertion (Steinmetz and Richter, 1994). Perhaps that only a specific insertion into the rodZ gene of B. subtilis resulted in a viable clone with comparable growth rate to the WT strain in control conditions.
Significantly, rodZ lies immediately upstream in the genome of the essential pgsA gene encoding phosphatidylglycerophosphate synthase (Figure 1A). Interestingly, in many other Gram-positive bacteria (e.g., Bacilli, Streptococci, and Stapylococci) rodZ and pgsA are predicted to be part of the same operon (Ermolaeva et al., 2001; Alm et al., 2005; Alyahya et al., 2009). Our data, showing that a transcript containing both genes can be amplified from B. subtilis, corroborates this notion (See Supplementary Figure S1 of the Supporting Information). Moreover, according to several prediction tools, B. subtilis RodZ has one transmembrane domain (amino acids 89 – 113) (Cserzö et al., 1997; Hirokawa et al., 1998) and an Xre-like helix-turn-helix (HTH) motive, commonly seen in DNA binding proteins, in its N-terminal side. Together, these observations initially led us to believe that the RodZ protein might be involved in regulating pgsA expression. However, RT-PCR experiments with overexpression of RodZ in the WT strain failed to show any changes in pgsA mRNA levels (see Supplementary Figure S5 of the Supporting Information). HTH motifs also have been shown to function in DNA replication, RNA metabolism, and protein-protein interactions (Aravind et al., 2005). RodZ was shown to co-localize with components of the cytoskeleton and depend on MreB for its localization (Alyahya et al., 2009) and interaction of RodZ with MreB was shown to be specifically with this HTH motif in Thermotoga maritima (van den Ent et al., 2010). The mutual functional dependence of RodZ and MreB was reinforced by the observation that loss of RodZ, or at least its N-terminal domain, resulted in aberrant localization of MreB and cessation of its movement (van den Ent et al., 2010; Garner et al., 2011).
Since MreB organization also depends on the membrane potential (Strahl and Hamoen, 2010) and weak organic acids lower the proton gradient (by releasing protons in the cell) and may act in certain cases as uncouplers of the membrane potential (van Beilen et al., 2014), the impact of weak acid stress on cell growth may be partially mediated through membrane perturbation effects on the correct localization of MreB and RodZ containing cell shape determining protein complexes. This in turn could lead to a perturbed localization and functioning of PgsA, exacerbating the sorbic acid sensitivity. PgsA normally localizes primarily to the septal membranes in conjunction with cardiolipin and plays an essential role in cell division (Nishibori et al., 2005).
The observation that rodZ and pgsA are part of the same transcript as well as our earlier studies (Ter Beek et al., 2008; van Beilen et al., 2014) suggested to us that the membrane plays a crucial role in weak organic acid stress tolerance. Hence we studied the phospholipid composition of the various strains. The rodZ::mini-Tn10 mutant was shown to contain severely lowered levels of PG and CL (Figure 4). The average acyl chain length of the remaining phospholipids was increased in the rodZ mutant when compared to the WT (Figure 3; Supplementary Figure S4 of the Supporting Information). The same phenomena were observed in the conditional pgsA::Pspac- pgsA mutant that displayed similar sorbic acid-, acetic acid-, and salt-stress sensitivities (Figure 2; Supplementary Figures S2 and S3 of the Supporting Information). Alterations of the cell membrane lipid composition were also found in response to sorbic acid stress where a pronounced increase in acyl chain length and lowering of L-PG was seen most prominently in WT B. subtilis (Figures 3 and 4). An increase in chain length stiffens the membrane and lowers its permeability. On the other hand, the induction of the BkdR-regulated genes (involved in the synthesis of precursor molecules for branched-chain fatty acids (Debarbouille et al., 1999) in sorbic acid-stressed cells (Ter Beek et al., 2008) may indicate increased branching in phospholipids and thereby balancing membrane fluidity levels. Interestingly, Lopez and co-workers have shown that salt-stressed cells increase their CL phospholipid levels and decrease both PG and L-PG levels (López et al., 1998), a similar trend observed in KS-stressed cells (Figure 4). However, they also measured a clear decrease in branched chain fatty lipids in cultures grown in NaCl. Noteworthy is the observation that both mutants hardly seem to change their membrane composition when stressed with KS (Figures 3 and 4). The possible differences in net charge of the membrane between the mutants (significantly reduced PG and CL levels) and the WT may be one explanation for the observed hypersensitivity toward weak organic acid stress. Another explanation might be the inability of the mutants to modify their membrane composition further upon stress, e.g., they cannot adapt further. Besides a potential role for L-PG in weak acid sensitivity, it has strongly been implicated in resistance against cationic antimicrobial peptides (Dunkley et al., 1988; Sohlenkamp et al., 2007; Samant et al., 2009). Alterations in the membrane composition likely pertain to changes in the rate of net proton influx. That is: the rate limiting step for protonophoric uncouplers is the rate with which the anion traverses the liquid-lipid interface (Spycher et al., 2008; Chu et al., 2009; Ter Beek et al., 2014; van Beilen et al., 2014). It is therefore likely that an increase in CL levels in combination with increased acyl-chain length confers increased resistance to uncouplers. On the other hand, this adaptation may conflict with growth at elevated temperatures, but B. subtilis may ultimately strive toward homeo-proton permeability (van de Vossenberg et al., 1999). Alternatively, anion build-up has also been implicated as a stress factor caused by weak organic acid preservatives (Russell, 1992). Both of these mechanisms may interfere with proper cytoskeleton function and put stress on the total cell envelop. If one of these players fails to work in concert, the cell is weakend.
Our data presented here on the rodZ and the conditional pgsA mutant suggest that the phenotypes observed in the rodZ transposon mutant might be primarily the result of polar effects on pgsA expression. The RT-PCR data on pgsA expression levels in the rodZ::mini-Tn10 strain compared to WT PB2 corroborate this conclusion. Finally, overexpression of pgsA in the rodZ::mini-Tn10 background stimulated growth of cells cultured in liquid media in the presence of sorbic and acetic acid. However, overexpression of rodZ in the rodZ transposon mutant did not restore weak acid stress sensitivity to WT levels. These results also indicate that the phenotypes observed in the rodZ transposon mutant can almost solely be ascribed to polar effects on pgsA.
We present here a direct link between phospholipid synthesis and weak acid sensitivity and propose that PgsA plays an important role in membrane homeostasis and tolerance to weak organic acid stress. Future studies are aimed at assessing the membrane permeation efficacy in Bacillus strains with different phospholipid perturbations (Salzberg and Helmann, 2008), by measuring intracellular acidification rates upon addition of various weak organic acids. This can be done with the aid of the pH-sensitive fluorescent protein pHluorin expressed in the lumen of the bacterium. The protocol for this has recently also been established by us in B. subtilis (van Beilen and Brul, 2013; van Beilen et al., 2014; Ter Beek et al., 2014).
Author Contributions
Conceived and designed the experiments: JB, AZ, SB, AB. Performed the experiments: JB, CB, HF, RB, AB. Analyzed the data: JB, CB, AZ, AB. Contributed reagents/materials/analysis tools: WK, FV. Wrote the paper: JB, SB, AB.
Funding
AB was supported by a grant from the Dutch Foundation for Applied Sciences (STW 10431).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Kouji Matsumoto (Saitama University, Japan) is acknowledged for his gift of the MHB001 strain before publication. We are grateful to Tanneke den Blaauwen of the Bacterial Cell Biology group for use of their phase-contrast microscope. We thank Rachna Pandey, Ingar Seeman, Alicia Prats Tur, and Henk van Lenthe for their technical assistance, and Bart J. F. Keijser for initial suggestions on the experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01633
FIGURE S1 | rodZ and pgsA are co-transcribed. cDNA synthesized from RNA of PB2 using the reverse primer for pgsA (pgsA_Q1_RV) was PCR amplified with the ymfM_Q1_FW and pgsA_Q1_RV primers and run on an agarose gel. M: GeneRuler DNA ladder mix (Fermentas); 1: Genomic DNA of B. subtilis WT strain PB2 used as the template (positive control); 2: cDNA made with pgsA_Q1_RV used as the template; 3: No template (primer and RNA control); 4: RNA incubated with RNase A (10 μg/ml) for 20 min at 37°C prior to cDNA reaction (genomic DNA contamination control); 5: No reverse transcriptase control.
FIGURE S2 | Inactivation of rodZ leads to increased sensitivity for acetic acid and salt stress in liquid cultures. Growth curves of WT strain PB2 and the rodZ::mini-Tn10 mutant grown in liquid LB medium of pH 6.4 in control conditions (closed and open circles, respectively) and in the presence of 125 mM KAc (A) or 0.7 M NaCl (B) (closed downward triangles and open triangles, respectively).
FIGURE S3 | Conditional pgsA mutant displays increased sensitivity for sorbic acid, acetic acid, and salt. Growth curves of WT strain 1A700 and the pgsA::Pspac-pgsA conditional mutant grown in liquid LB medium of pH 6.4 in control conditions (closed and open circles, respectively) and in the presence of 15 mM KS (A), 125 mM KAc (B) or 0.7 M NaCl (C) (closed downward triangles and open triangles, respectively).
FIGURE S4 | Acyl tail length distributions of phospholipids measured in WT strain PB2, conditional pgsA mutant, and the rodZ transposon mutant, in the absence and presence of 5 mM potassium sorbate (KS). Acyl chain length distributions of the following phospholipid classes were measured: (A) cardiolipin (CL), (B) lysyl-phosphatidyl glycerol (L-PG), and (C) phosphatidyl glycerol (PE). The number behind the abbreviation indicates the total number of carbon atoms in the acyl chains per molecule.
FIGURE S5 | Overexpression of rodZ did not have an effect on pgsA expression. Relative quantification of gene expression using real-time RT-PCR was determined between B. subtilis WT strain PB2 carrying pDG148 and pDG-rodZ every 30 min until 2 h after induction. Expression of rodZ was induced with 1 mM IPTG at t = 0 min in exponentially growing cells. Data were normalized to accA expression. The error bars indicate the standard deviation.
Footnotes
References
Alm, E. J., Huang, K. H., Price, M. N., Koche, R. P., Keller, K., Dubchak, I. L., et al. (2005). The microbesonline web site for comparative genomics. Genome Res. 15, 1015–1022. doi: 10.1101/gr.3844805
Alyahya, S. A., Alexander, R., Costa, T., Henriques, A. O., Emonet, T., and Jacobs-Wagner, C. (2009). RodZ, a component of the bacterial core morphogenic apparatus. Proc. Natl. Acad. Sci. U.S.A. 106, 1239–1244. doi: 10.1073/pnas.0810794106
Aravind, L., Anantharaman, V., Balaji, S., Babu, M. M., and Iyer, L. M. (2005). The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29, 231–262. doi: 10.1016/j.femsre.2004.12.008
Azukas, J., Costilow, R., and Sadoff, H. (1961). Inhibition of alcoholic fermentation by sorbic acid. J. Bacteriol. 81, 189–194.
Barbe, V., Cruveiller, S., Kunst, F., Lenoble, P., Meurice, G., Sekowska, A., et al. (2009). From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology 155, 1758–1775. doi: 10.1099/mic.0.027839-0
Bauer, B. E., Rossington, D., Mollapour, M., Mamnun, Y., Kuchler, K., and Piper, P. W. (2003). Weak organic acid stress inhibits aromatic amino acid uptake by yeast, causing a strong influence of amino acid auxotrophies on the phenotypes of membrane transporter mutants. Eur. J. Biochem. 270, 3189–3195. doi: 10.1046/j.1432-1033.2003.03701.x
Beales, N. (2004). Adaptation of microorganisms to cold temperatures, weak acid preservatives, low pH, and osmotic stress: a review. Compr. Rev. Food Sci. Food Saf. 3, 1–20. doi: 10.1111/j.1541-4337.2004.tb00057.x
Bendezú, F. O., Hale, C. A., Bernhardt, T. G., and de Boer, P. A. J. (2009). RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 28, 193–204. doi: 10.1038/emboj.2008.264
Boylan, S. A., Rutherford, A., Thomas, S. M., and Price, C. W. (1992). Activation of Bacillus subtilis transcription factor sigma B by a regulatory pathway responsive to stationary-phase signals. J. Bacteriol. 174, 3695–3706.
Brul, S., and Coote, P. (1999). Preservative agents in foods. Mode of action and microbial resistance mechanisms. Int. J. Food Microbiol. 50, 1–17. doi: 10.1016/S0168-1605(99)00072-0
Brul, S., and Ter Beek, A. S. (2010). To kill or not to kill Bacilli: opportunities for food biotechnology. Curr. Opin. Biotechnol. 21, 168–174. doi: 10.1016/j.copbio.2010.03.014
Casadaban, M. J., and Cohen, S. N. (1980). Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138, 179–207. doi: 10.1016/0022-2836(80)90283-1
Chu, S., Hawes, J. W., and Lorigan, G. A. (2009). Solid-state NMR spectroscopic studies on the interaction of sorbic acid with phospholipid membranes at different pH levels. Magn. Reson. Chem. 47, 651–657. doi: 10.1002/mrc.2444
Cotter, P. D., and Hill, C. (2003). Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67, 429–453. doi: 10.1128/MMBR.67.3.429
Cserzö, M., Wallin, E., Simon, I., von Heijne, G., and Elofsson, A. (1997). Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10, 673–676. doi: 10.1093/protein/10.6.673
Davidson, P. M., and Harrison, M. A. (2002). Resistance and adaptation to food antimicrobials, sanitizers, and other process controls. Food Technol. 56, 69–78.
Debarbouille, M., Gardan, R., Arnaud, M., and Rapoport, G. (1999). Role of bkdR, a transcriptional activator of the sigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J. Bacteriol. 181, 2059–2066.
Dempwolff, F., Reimold, C., Reth, M., and Graumann, P. L. (2011). Bacillus subtilis MreB orthologs self-organize into filamentous structures underneath the cell membrane in a heterologous cell system. PLoS ONE 6:e27035. doi: 10.1371/journal.pone.0027035
den Blaauwen, T., de Pedro, M. A., Nguyen-Distèche, M., and Ayala, J. A. (2008). Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 32, 321–344. doi: 10.1111/j.1574-6976.2007.00090.x
Dunkley, E. A., Clejan, S., Guffanti, A. A., and Krulwich, T. A. (1988). Large decreases in membrane phosphatidylethanolamine and diphosphatidylglycerol upon mutation to duramycin resistance do not change the protonophore resistance of Bacillus subtilis. Biochim. Biophys. Acta 943, 13–18. doi: 10.1016/0005-2736(88)90341-0
Eklund, T. (1983). The antimicrobial effect of dissociated and undissociated sorbic acid at different pH levels. J. Appl. Bacteriol. 54, 383–389. doi: 10.1111/j.1365-2672.1983.tb02632.x
Ermolaeva, M. D., White, O., and Salzberg, S. L. (2001). Prediction of operons in microbial genomes. Nucleic Acids Res. 29, 1216–1221. doi: 10.1093/nar/29.5.1216
Garner, E. C., Bernard, R., Wang, W., Zhuang, X., Rudner, D. Z., and Mitchison, T. (2011). Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333, 222–225. doi: 10.1126/science.1203285
Gerdes, K. (2009). RodZ, a new player in bacterial cell morphogenesis. EMBO J. 28, 171–172. doi: 10.1038/emboj.2008.287
Hashimoto, M., Seki, T., Matsuoka, S., Hara, H., Asai, K., Sadaie, Y., et al. (2013). Induction of extracytoplasmic function sigma factors in Bacillus subtilis cells with defects in lipoteichoic acid synthesis. Microbiology 159(Pt. 1), 23–35. doi: 10.1099/mic.0.063420-0
Hirokawa, T., Boon-Chieng, S., and Mitaku, S. (1998). SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378–379. doi: 10.1093/bioinformatics/14.4.378
Houtkooper, R. H., Akbari, H., van Lenthe, H., Kulik, W., Wanders, R. J. A., Frentzen, M., et al. (2006). Identification and characterization of human cardiolipin synthase. FEBS Lett. 580, 3059–3064. doi: 10.1016/j.febslet.2006.04.054
Koppelman, C.-M., Aarsman, M. E. G., Postmus, J., Pas, E., Muijsers, A. O., Scheffers, D.-J., et al. (2004). R174 of Escherichia coli FtsZ is involved in membrane interaction and protofilament bundling, and is essential for cell division. Mol. Microbiol. 51, 645–657. doi: 10.1046/j.1365-2958.2003.03876.x
Krulwich, T. A., Quirk, P. G., and Guffanti, A. A. (1990). Uncoupler-resistant mutants of bacteria. Microbiol. Rev. 54, 52–65.
Kunst, F., and Rapoport, G. (1995). Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177, 2403–2407.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
López, C. S., Heras, H., Ruzal, S. M., Sánchez-Rivas, C., and Rivas, E. A. (1998). Variations of the envelope composition of Bacillus subtilis during growth in hyperosmotic medium. Curr. Microbiol. 36, 55–61. doi: 10.1007/s002849900279
Maguin, E., Duwat, P., Hege, T., Ehrlich, D., and Gruss, A. (1992). New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174, 5633–5638.
Mascher, T., Hachmann, A.-B., and Helmann, J. D. (2007). Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function sigma factors. J. Bacteriol. 189, 6919–6927. doi: 10.1128/JB.00904-07
Matsumoto, K., Kusaka, J., Nishibori, A., and Hara, H. (2006). Lipid domains in bacterial membranes. Mol. Microbiol. 61, 1110–1117. doi: 10.1111/j.1365-2958.2006.05317.x
Mols, M., van Kranenburg, R., Tempelaars, M. H., van Schaik, W., Moezelaar, R., and Abee, T. (2010). Comparative analysis of transcriptional and physiological responses of Bacillus cereus to organic and inorganic acid shocks. Int. J. Food Microbiol. 137, 13–21. doi: 10.1016/j.ijfoodmicro.2009.09.027
Nishibori, A., Kusaka, J., Hara, H., Umeda, M., and Matsumoto, K. (2005). Phosphatidylethanolamine domains and localization of phospholipid synthases in Bacillus subtilis membranes. J. Bacteriol. 187, 2163–2174. doi: 10.1128/JB.187.6.2163-2174.2005
Petit, M. A., Bruand, C., Jannière, L., and Ehrlich, S. D. (1990). Tn10-derived transposons active in Bacillus subtilis. J. Bacteriol. 172, 6736–6740.
Piper, P. W. (1999). Yeast superoxide dismutase mutants reveal a pro-oxidant action of weak organic acid food preservatives. Free Radic. Biol. Med. 27, 1219–1227. doi: 10.1016/S0891-5849(99)00147-1
Rasmussen, S., Nielsen, H. B., and Jarmer, H. (2009). The transcriptionally active regions in the genome of Bacillus subtilis. Mol. Microbiol. 73, 1043–1057. doi: 10.1111/j.1365-2958.2009.06830.x
Russell, J. B. (1992). Another explanation for the toxicity of fermentation acids at low pH: anion accumulation versus uncoupling. J. Appl. Microbiol. 73, 363–370.
Salzberg, L. I., and Helmann, J. D. (2008). Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition. J. Bacteriol. 190, 7797–7807. doi: 10.1128/JB.00720-08
Samant, S., Hsu, F.-F., Neyfakh, A. A., and Lee, H. (2009). The Bacillus anthracis protein MprF is required for synthesis of lysylphosphatidylglycerols and for resistance to cationic antimicrobial peptides. J. Bacteriol. 191, 1311–1319. doi: 10.1128/JB.01345-08
Sambrook, J., Fritsch, E., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Sheu, C. W., and Freese, E. (1972). Effects of fatty acids on growth and envelope proteins of Bacillus subtilis. J. Bacteriol. 111, 516–524.
Shiomi, D., Sakai, M., and Niki, H. (2008). Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J. 27, 3081–3091. doi: 10.1038/emboj.2008.234
Sohlenkamp, C., Galindo-Lagunas, K. A., Guan, Z., Vinuesa, P., Robinson, S., Thomas-Oates, J., et al. (2007). The lipid lysyl-phosphatidylglycerol is present in membranes of Rhizobium tropici CIAT899 and confers increased resistance to polymyxin B under acidic growth conditions. Mol. Plant Microbe Interact. 20, 1421–1430. doi: 10.1094/MPMI-20-11-1421
Spycher, S., Smejtek, P., Netzeva, T. I., and Escher, B. I. (2008). Toward a class-independent quantitative structure–activity relationship model for uncouplers of oxidative phosphorylation. Chem. Res. Toxicol. 21, 911–927. doi: 10.1021/tx700391f
Steinmetz, M., and Richter, R. (1994). Easy cloning of mini-Tn10 insertions from the Bacillus subtilis chromosome. J. Bacteriol. 176, 1761–1763.
Stragier, P., Bonamy, C., and Karmazyn-Campelli, C. (1988). Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell 52, 697–704. doi: 10.1016/0092-8674(88)90407-2
Strahl, H., and Hamoen, L. W. (2010). Membrane potential is important for bacterial cell division. Proc. Natl. Acad. Sci. U.S.A. 107, 12281–12286. doi: 10.1073/pnas.1005485107
Stratford, M., and Anslow, P. A. (1998). Evidence that sorbic acid does not inhibit yeast as a classic “weak acid preservative”. Lett. Appl. Microbiol. 27, 203–206. doi: 10.1046/j.1472-765X.1998.00424.x
Ter Beek, A., Keijser, B. J. F., Boorsma, A., Zakrzewska, A., Orij, R., Smits, G. J., et al. (2008). Transcriptome analysis of sorbic acid-stressed Bacillus subtilis reveals a nutrient limitation response and indicates plasma membrane remodeling. J. Bacteriol. 190, 1751–1761. doi: 10.1128/JB.01516-07
Ter Beek, A., Wijman, J. G. E., Zakrzewska, A., Orij, R., Smits, G. J., and Brul, S. (2014). Comparative physiological and transcriptional analysis of weak organic acid stress in Bacillus subtilis. Food Microbiol. 45(Pt. A), 71–82. doi: 10.1016/j.fm.2014.02.013
van Beilen, J. W. A., and Brul, S. (2013). Compartment-specific pH monitoring in Bacillus subtilis using fluorescent sensor proteins; a tool to analyse the antibacterial effect of weak organic acids. Front. Microbiol. 4:157. doi: 10.3389/fmicb.2013.00157
van Beilen, J. W. A., Teixeira de Mattos, M. J., Hellingwerf, K. J., and Brul, S. (2014). Distinct effects of sorbic acid and acetic acid on the electrophysiology and metabolism of Bacillus subtilis. Appl. Environ. Microbiol. 80, 5918–5926. doi: 10.1128/AEM.01391-14
van de Vossenberg, J. L., Driessen, A. J., da Costa, M. S., and Konings, W. N. (1999). Homeostasis of the membrane proton permeability in Bacillus subtilis grown at different temperatures. Biochim. Biophys. Acta 1419, 97–104. doi: 10.1016/S0005-2736(99)00063-2
van den Ent, F., Johnson, C. M., Persons, L., de Boer, P., and Löwe, J. (2010). Bacterial actin MreB assembles in complex with cell shape protein RodZ. EMBO J. 29, 1081–1090. doi: 10.1038/emboj.2010.9
White, C. L., Kitich, A., and Gober, J. W. (2010). Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol. Microbiol. 76, 616–633. doi: 10.1111/j.1365-2958.2010.07108.x
Keywords: weak organic acids, acetic acid, sorbic acid, Bacillus subtilis, rod Z, pgs A, membrane compositional fluctuations
Citation: van Beilen J, Blohmke CJ, Folkerts H, de Boer R, Zakrzewska A, Kulik W, Vaz FM, Brul S and Ter Beek AT (2016) RodZ and PgsA Play Intertwined Roles in Membrane Homeostasis of Bacillus subtilis and Resistance to Weak Organic Acid Stress. Front. Microbiol. 7:1633. doi: 10.3389/fmicb.2016.01633
Received: 16 May 2016; Accepted: 30 September 2016;
Published: 21 October 2016.
Edited by:
Abram Aertsen, Katholieke Universiteit Leuven, BelgiumReviewed by:
Raivo Vilu, Tallinn University of Technology, EstoniaMarc Heyndrickx, Instituut voor Landbouw en Visserijonderzoek, Belgium
Copyright © 2016 van Beilen, Blohmke, Folkerts, de Boer, Zakrzewska, Kulik, Vaz, Brul and Ter Beek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stanley Brul, cy5icnVsQHV2YS5ubA==
†Present address: Christoph J. Blohmke, Oxford Vaccine Group, Centre for Clinical Vaccinology and Tropical Medicine, The Churchill Hospital, Oxford, UK Hendrik Folkerts, Department of Experimental Hematology, University Medical Center Groningen, University of Groningen, Groningen, Netherlands Anna Zakrzewska, Crucell Vaccine Institute, Janssen Centre of Excellence for Immunoprophylaxis, Leiden, Netherlands Alexander Ter Beek, Dutch DNA Biotech, Utrecht, Netherlands.
‡These authors have contributed equally to this work.
 Johan van Beilen
Johan van Beilen Christoph J. Blohmke1†
Christoph J. Blohmke1† Richard de Boer
Richard de Boer Stanley Brul
Stanley Brul Alexander Ter Beek
Alexander Ter Beek