
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 17 October 2016
Sec. Microbial Physiology and Metabolism
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.01578
This article is part of the Research Topic Multicellularity, cell-cell interactions and social behavior in bacterial populations View all 33 articles
The metal-reducing bacterium Geobacter sulfurreducens requires the expression of conductive pili to reduce iron oxides and to wire electroactive biofilms, but the role of pilus retraction in these functions has remained elusive. Here we show that of the four PilT proteins encoded in the genome of G. sulfurreducens, PilT3 powered pilus retraction in planktonic cells of a PilT-deficient strain of P. aeruginosa and restored the dense mutant biofilms to wild-type levels. Furthermore, PilT3 and PilT4 rescued the twitching motility defect of the PilT-deficient mutant. However, PilT4 was the only paralog whose inactivation in G. sulfurreducens lead to phenotypes associated with the hyperpiliation of non-retractile mutants such as enhanced adhesion and biofilm-forming abilities. In addition, PilT4 was required to reduce iron oxides. Taken together, the results indicate that PilT4 is the motor ATPase of G. sulfurreducens pili and reveal a previously unrecognized role for pilus retraction in extracellular electron transfer, a strategy that confers on Geobacter spp. an adaptive advantage for metal reduction in the natural environment.
The hallmark of the physiology of bacteria in the family Geobacteraceae is their ability to conserve energy for growth by transferring respiratory electrons to extracellular Fe(III) oxide minerals, a process that requires the expression of conductive type IV pili (T4P) (Reguera et al., 2005). Geobacter T4P are homopolymers of a single pilin subunit (PilA) (Cologgi et al., 2011), a short peptide (61 amino acids in the model representative Geobacter sulfurreducens) that retains the conserved amino-terminal features of type IVa pilins but diverges in amino acid composition and structure at the carboxy-terminus (Feliciano et al., 2012). This also places the Geobacter pilins in an independent line of descent among bacterial type IV pilins and pseudopilins (Reguera et al., 2005). A structural model of the pilus fiber optimized via molecular dynamics revealed the clustering of aromatic amino acids of the pilins once assembled and the formation of axial and transversal pathways with inter-aromatic distances and geometries optimal for multistep charge hopping (Feliciano et al., 2015). Consistent with this mechanism for charge transport, in vitro conductivity measurements of pili purified from G. sulfurreducens demonstrated the thermal dependence of incoherent conductivity (Lampa-Pastirk et al., 2016). Molecular dynamics simulations also revealed that some aromatic side chains can transiently get close to each other (3.5–5 Å), forming aromatic contacts that promote charge transport at rates (about one billion electrons per second along a 1 μm-long pilus at 100 mV) that greatly exceed the respiratory rates of the cell (Lampa-Pastirk et al., 2016). Indeed, an alanine replacement of one of the tyrosines (Y27) involved in the formation of half of the aromatic contacts in the pilus fiber (Feliciano et al., 2015) reduced the carrier mobility, five-fold (Lampa-Pastirk et al., 2016). Similarly, alanine replacements of the pilin's three tyrosines (Tyr3 mutant) reduced the number of aromatic contacts in half (Feliciano et al., 2015) and increased the electrical resistance of the T4P five-fold (Steidl et al., 2016). As a result, a Tyr3 mutant had growth defects in cultures with Fe(III) oxides, with generation times increasing four-fold compared to the wild-type strain (Feliciano et al., 2015).
In addition to their role as electronic conduits between the cell and Fe(III) oxides (Reguera et al., 2005) and uranium (Cologgi et al., 2011), the T4P of G sulfurreducens promote cell-cell aggregation and biofilm formation on various surfaces (Reguera et al., 2006, 2007; Cologgi et al., 2014; Steidl et al., 2016). T4P assembly and conductivity are also required for maximum current generation and electroactivity of anode biofilms grown in microbial electrolysis cells (MECs), which are devices operated with the anode electrode poised at a metabolically oxidizing potential to provide an unlimited electron acceptor for biofilm growth (Steidl et al., 2016). In electroactive biofilms, the T4P work coordinately with matrix-associated c-type cytochromes to transport charges to the underlying electrode until the biofilms grow to a threshold thickness (~10 μm) that limits the rates of electron transfer via the cytochromes (Steidl et al., 2016). The conductive T4P therefore become the primary mechanism to discharge respiratory electrons from cells positioned in the otherwise electron acceptor-limited outer regions of thick biofilms (Steidl et al., 2016).
In contrast to the number of mechanistic studies of pilus conductance, little is known about the components of the Geobacter T4P apparatus and their role in metal respiration and the formation of electroactive biofilms. The expression of T4P is a dynamic process controlled by antagonistic cycles of extension and retraction of the pilus fibers, which are powered by the PilB and PilT ATPases of the pilus apparatus, respectively (Jakovljevic et al., 2008). The genome of G. sulfurreducens contains a pilB and a pilT (pilT4) homolog in the same pil gene cluster where the pilin-encoding gene, pilA, is located (Reguera et al., 2005). The genetic arrangement of this region is similar to the pil region of Myxococcus xanthus (Wall and Kaiser, 1999), another delta-Proteobacterium, and also includes pilC, the gene encoding the PilC protein that couples the coordinated action of PilB and PilT in other bacteria (Takhar et al., 2013). This makes pilB and pilT4 the most obvious candidates to encode for the T4P extension and retraction ATPases in G. sulfurreducens, respectively. Consistent with this prediction, deleting the pilB gene in G. sulfurreducens prevented pili assembly and the growth of biofilms on anode electrodes beyond the threshold thickness (~10 μm) that limits the rates of electron transfer via matrix-associated c-type cytochromes (Steidl et al., 2016). Furthermore, the electrochemical activity of the pilB biofilms was approximately half of that of wild-type anode biofilms grown to the same thickness, consistent with the requirement to produce pili for optimal electron transport to the underlying electrode (Steidl et al., 2016). The role of pilT4 in pilus retraction in G. sulfurreducens remains however unclear, based on the lack of a measurable phenotype of a pilT4 deletion mutant during the reduction of Fe(III) oxides (Reguera et al., 2005). Furthermore, twitching motility in G. sulfurreducens, which requires the retractile properties of the pili, has never been observed using submerged assays with glass coverslips with or without iron coatings (Reguera et al., 2005). Hence, the physiological relevance of pilus retraction in this organism remains uncertain.
Interestingly, the genome of G. sulfurreducens contains three additional pilT genes outside of the pil gene cluster (designated pilT1, pilT2, and pilT3), which could encode for a functional pilus retraction protein. The expression of one or more of these additional PilT proteins could have suppressed any phenotypes associated with the pilT4 mutant in cultures growing with Fe(III) oxides (Reguera et al., 2005), similarly to the compensatory effects that are often observed when components of the respiratory machinery of G. sulfurreducens are inactivated (Kim et al., 2005, 2006, 2008; Leang et al., 2005; Cologgi et al., 2011, 2014; Steidl et al., 2016). It is also possible that Geobacter pili rely on the coordinated action of more than one PilT protein to power pilus retraction under specific conditions. PilU, for example, a well-characterized PilT paralog, is essential for PilT-mediated twitching motility in Pseudomonas aeruginosa (Whitchurch and Mattick, 1994). However, it is dispensable for pilus retraction (Park et al., 2002; Maier et al., 2004) and twitching motility (Maier et al., 2002; Park et al., 2002) in Neisseria gonorrhoeae, which are pili functions that require the activity of a third PilT paralog (PilT2) (Kurre et al., 2012). Further, PilT mediates T4P retraction at low velocity and high force in M. xanthus, but the pili also retract in a PilT-independent mode characterized by its high-velocity and low force, which is presumably powered by one or more of the other four PilT paralogs encoded by genes outside of the pil cluster (Clausen et al., 2009). Hence, the coordinated activity of more than one PilT ATPase may be required for specific PilT-mediated functions in G. sulfurreducens.
In this study, we took a genetic approach to identify functional PilT motors in G. sulfurreducens and, by extension, to investigate if pilus retraction is physiologically relevant in current-harvesting biofilms or during the reduction of Fe(III) oxides. We first expressed each of the pilT genes of G. sulfurreducens in the well-characterized PilT null mutant of P. aeruginosa strain K (Sundin et al., 2002) to assess the ability of the PilT proteins to complement functions in this organism that require PilT-mediated retraction. PilT3 and PilT4 were able to restore all or some of the defects of the pilus retraction deficiency in the heterologous host, respectively. However, PilT4 was the only paralog whose inactivation in G. sulfurreducens resulted in phenotypes expected for a retraction-defective mutant such as increased adhesion and formation of denser biofilms. Furthermore, PilT4 was also required for optimal electron discharge per cell in electroactive biofilms and during the reduction of Fe(III) oxides. The genetic data thus indicates that PilT4 is the main T4P retraction ATPase motor in G. sulfurreducens and supports a previously unrecognized role for PilT4-mediated pilus retraction in the formation and electroactivity of biofilms and during the respiration of Fe(III) oxides. We also discuss the possibility that other PilT paralogs, particularly PilT3, may be involved in fine-tuning pilus dynamics in G. sulfurreducens and the implications that these findings have on the environmental survival of these bacteria.
All the strains of P. aeruginosa used in the heterologous expression experiments (Table 1) and Escherichia coli strains used for cloning and transformation were routinely grown aerobically in Luria-Bertani (LB) medium and incubated at 37°C with gentle agitation. G. sulfurreducens strain PCA was from our culture collection and was used as the wild-type background to construct all of the mutant derivatives (Table 1). All of the Geobacter strains were routinely cultured anaerobically in NB medium (Coppi et al., 2001) supplemented with 0.1% yeast extract and 1 mM cysteine and with 15 mM acetate and 40 mM fumarate (NBAF). When indicated, NBAF was replaced by a modified freshwater FW medium (Cologgi et al., 2011) supplemented with 1 μM Na2SeO4 to stimulate growth (Speers and Reguera, 2012) and with 15 mM acetate as electron donor and 40 mM fumarate as the electron acceptor (FWAF). Fe(III) oxides cultures contained FWA medium and 100 mM poorly crystalline Fe(III) oxides and, when indicated, 4 mM of the metal chelator nitrilotriacetic acid (NTA). Growth in the Fe(III) oxides cultures was measured periodically with the ferrozine method (Stookey, 1970) as the amount of 0.5 N HCl-extractable Fe(II) resulting from the reduction of Fe(III) (Lovley and Phillips, 1986). Unless otherwise indicated, all of the incubations were at 30°C.
The WT strain of G. sulfurreducens was grown in 10 ml of NBAF or FWAF and cultures were incubated either at 30 or 25°C. Exponentially growing cells were then harvested by centrifugation, and the pelleted cells were suspended in 2 ml of TRIzol® reagent (Invitrogen Life Technologies). Extraction of total cellular RNA was performed following manufacturer's recommendations. Genomic DNA was removed with RNase-free DNase I (QIAGEN) and the remaining RNA was purified using a QIAGEN's RNeasy column. Primers were designed to have the same melting temperature and PCR efficiency using the pcrEfficiency open source tool (http://srvgen.upct.es/efficiency.html; Mallona et al., 2011). Complementary strand synthesis was performed in a 20 μl reaction mixture containing 1 μg of total RNA, 10 pmol of gene-specific primer and dNTPs. The mixture was heat-treated at 65°C for 5 min followed by incubation on ice for at least 1 min before adding 1 μl of SuperScript® III reverse transcriptase (Invitrogen) and incubating at 50°C for 60 min. The reaction was inactivated by heating at 70°C for 15 min. PCR-amplification reactions (50 μl, total volume) contained 1 μl of cDNA (serial dilutions from first-strand synthesis reaction), 10 pmol of sense and antisense primers (Table 2), and 25 μl of SYBR green 2X master mix (Applied Biosystems, Life Technologies). qRT-PCR was performed in duplicate reactions under conditions (95°C for 30 s, 50°C for 30 s, and 50 cycles of 1 min at 70°C) that resulted in threshold crossings between 18 and 35 for all of the primer pairs. Fold changes in gene expression of the target genes were calculated in reference to the internal control recA as (Schmittgen and Livak, 2008), where ΔCT is the CT of the gene of interest minus the CT of the recA internal control.
The P. aeruginosa strains and plasmids used for heterologous complementation are listed in Table 1. The wild-type PAK strain, the pilin-deficient PAKΔpilA and pilT-deficient PAKΔpilT derivatives, and plasmids pMMB67EH and pRK2013 were kindly provided by Matthew C. Wolfgang (University of North Carolina School of Medicine). The G. sulfurreducens pilT genes were cloned into the broad host range expression vector pMMB67EH using the In Fusion cloning system (Clontech Laboratories, Inc.). The pMMB67EH plasmids and its derivatives were electroporated in E. coli HB101 carrying the mobilizing helper plasmid pRK2013 and transformants were selected on LB agar containing 30 μg/ml carbenicillin and 50 μg/ml kanamycin. The resulting E. coli donor strains were mated with the PAKΔpilT strain on antibiotic-free LB agar plates for 2 h at 37°C. After the mating, the cells were suspended in 2 ml of LB medium and serially diluted and plated on LB agar containing 500 μg/ml carbenicillin and 25 μg/ml irgasan. The resulting strains were cultured with 300 μg/ml carbenicillin to maintain the plasmid and, when indicated, with 20 μM IPTG to promote the expression of the cloned genes.
T4P from all the P. aeruginosa strains were purified following a previously described protocol (Castric, 1995). Briefly, 100 μl of an LB culture were plated on triplicate LB-agar plates supplemented with 20 μM IPTG and incubated overnight at 37°C. When needed, carbenecillin (30 μg/ml) was included in the LB-agar medium to maintain the pMMB67EH plasmid or any of its derivatives (Table 1). The cells were scraped off the plate and suspended in 1.5 ml of LB medium and 5 μl of the cell suspension was diluted in 2 ml water to measure its OD600 and estimate the cell density. T4P fibers in the cell suspensions were sheared by passing the cells 4 times through a 23-gauge needle and the cells were then removed by centrifugation (2 times at 7500 × g for 15 min at room temperature). The T4P in the supernatant fluids were then precipitated by adding MgCl2 to a final concentration of 100 mM and incubating on ice for 60 min. Once precipitated, the T4P were concentrated by centrifugation (12,000 × g for 15 min at 4°C), suspended in 100 μl of 0.1% SDS, and incubated overnight at 4°C to separate the pili bundles. The T4P content in the samples was quantified using the Bio-Rad protein assay Kit (Bio-Rad Laboratories, Inc.), following the manufacturer's recommendations for the standard procedure in microtiter plates, and its concentration (mg/ml) in each sample was normalized by the sample's cell density (in OD600 units).
T4P purified in the 0.1% SDS solution were diluted appropriately to standardize for the cell density (OD600) and then mixed with an equal volume of Tris-Tricine sample buffer. After incubation at 95°C for 5 min to depolymerize the fibers into the individual pilin subunits, the proteins in 15 μl of sample were separated electrophoretically in a 10–20% Mini-PROTEAN® Tris-Tricine Precast Gel (Bio-Rad). One lane was loaded with 4 μl of the Novex® Sharp Pre-stained Protein Standard (Invitrogen Life Technologies) as standards. The gel was stained overnight with Coomassie Brilliant Blue R-250 Staining Solution (Bio-Rad) and destained overnight with a solution of 50% methanol and 10% acetic acid.
Biofilm assays with strains of P. aeruginosa were performed under static conditions in microtiter plates, essentially as described elsewhere (Merritt et al., 2005) but using M63 minimal medium supplemented with 0.4% arginine as the sole carbon and energy source to stimulate biofilm development over a 24 h period (Caiazza and O'Toole, 2004). The medium was supplemented with carbenecillin (30 μg/ml) to maintain the pMMB67EH plasmid and with IPTG (20 μM) to heterologously express the cloned pilT genes. The biofilm biomass was then stained with 0.1% crystal violet, and allowed to dry before solubilizing the biofilm-associated dye with 95% ethanol and measuring its OD580 (Merritt et al., 2005). To account for technical and biological variability, the biofilm assays included six technical replicates (wells) per strain tested and a minimum of three independent experiments. Outliers were excluded from the compiled biofilm assay results using the Quartile or Fourth-Spread method (Devore, 2000) and the data were plotted as a box-and-whisker graph using the Microsoft Excel software.
For the twitching motility assays, a thin layer (~1 ml) of molten LB medium with 1% agarose and 20 μM IPTG was deposited onto a sterile glass slide and allowed to solidify. The center of the agar was stab-inoculated with overnight LB-20 μM IPTG cultures of the P. aeruginosa strains incubated at 37°C with shaking (180 rpm). When needed, carbenecillin (30 μg/ml) was added to the liquid cultures to maintain the pMMB67EH plasmid. After stab-inoculation, the slide was placed inside a Petri dish surrounded by damp, sterile absorbent paper to maintain the humidity inside the chamber and the dish was then sealed with parafilm to prevent drying. After overnight incubation at 37°C, the areas of growth on the surface of the agar were gently removed with a coverslip and the slides were then stained with 1 ml Coomassie Brilliant Blue R-250 Staining Solution (Bio-Rad) for 2 h. The slides were briefly rinsed with 5 ml of a solution of 50% methanol and 10% acetic acid and then destained in 20 ml of the same solution for 4 h before replacing 15 ml of the solution with an equal volume of a solution of 5% methanol and 7.5% acetic acid and incubating overnight. After destaining, the agar layer on the slides was allowed to dry completely before imaging the stained twitching zones with a scanner.
G. sulfurreducens PCA was used to construct mutants carrying full deletions in each of the four pilT homologs and in the pilB gene, using the primers listed in Table 2. The general procedure was to replace the full length of the wild-type gene with the aaaC1 gentamicin cassette of plasmid pCM351 (Marx and Lidstrom, 2002) in the same gene orientation so as to prevent polar effects on downstream genes. For the construction of the pilT1 and pilT2 deletions, we PCR-amplified regions approximately 500 bp upstream and downstream of the target genes using the Phusion® high fidelity DNA polymerase (New England Biolabs) and cloned the fragments separately into the pCR2.1-TOPO vector (Invitrogen). The downstream fragments were then digested and cloned into the NdeI-XbaI site of pCR2.1-TOPO harboring the upstream fragments. This resulted in plasmids carrying the upstream and downstream regions and an NdeI site in between. The aaaC1 gentamicin-resistance cassette was then PCR-amplified from pCM351, digested with NdeI, and cloned into the NdeI site separating the upstream and downstream fragments. The cloned fragment was excised from the plasmid with EcoRI and XbaI and gel-purified prior to introduction into electrocompetent cells of G. sulfurreducens via electroporation, as described previously (Coppi et al., 2001). The pilT3, pilT4, and pilB deletion mutants were constructed by recombinant PCR (Coppi et al., 2001), using constructs generated by fusing together the upstream region of the target gene, the aaaC1 cassette, and the downstream region in a PCR reaction with the external primers. The final PCR product was gel-purified with a Zymoclean Gel DNA Recovery kit (Zymo Research Co.) and incubated at 70°C for 30 min with one volume of 2X GoTaq Green Master Mix (Promega) to produce a 3′ A-overhang prior to cloning into pCR2.1-TOPO. The cloned fragment was PCR-amplified and gel-purified before electroporation into G. sulfurreducens electrocompetent cells (Coppi et al., 2001). In all cases, transformants were selected on NBAF plates containing gentamicin (5 μg/ml) and confirmed by PCR, DNA sequencing, and Southern blot hybridization, as described previously (Ausubel et al., 1995).
The pCM158 plasmid was electroporated in the pilT4 (ΔpilT4::acc1) and pilB (ΔpilB::acc1) mutant strains to express the Cre-recombinase and excise the acc1 gentamicin-resistance cassette, which is flanked by the loxP sites that the Cre protein recognizes and recombines (Marx and Lidstrom, 2002). After confirming the excision of the gentamicin resistance cassette by PCR, the pCM158 plasmid was cured from the strains by culturing in the absence of kanamycin. The resulting strains, designated ΔpilT4 and ΔpilB, were electroporated with plasmids pRG5-pilT4 and pRG5-pilB to express the wild-type pilT4 and pilB genes, respectively, from the medium-copy number plasmid pRG5 (Kim et al., 2005). All cloning steps used the In-Fusion cloning system (Clontech) and selection of transformants was in agar-solidified NBAF medium with spectinomycin (150–300 μg/ml). The ΔpilT3::acc1 mutation was also introduced in the ΔpilT4 strain to generate the double pilT3 pilT4 mutant.
G. sulfurreducens was grown to mid-exponential phase in FWFA medium and serially transferred (10% v/v) three times before diluting the cells in fresh FWAF medium to an OD600 of 0.04. Two hundred microliters of this cell suspension were dispensed in the wells of a 96-well plate, which was then sealed to prevent evaporation. The plate was incubated at 30°C inside a TECAN Sunrise™ Absorbance Reader (TECAN, Männedorf, Switzerland) housed in an anaerobic glove bag (Coy Labs) and planktonic growth was monitored periodically (OD600) throughout the incubation. After 48 h, the plates were removed from the glove box, the supernatant was decanted, and the biofilms in each well were stained for 20 min with 200 μl of 0.1% crystal violet, gently washed 3 times with 200 μl of ddH2O, and air-dried (Stepanovic et al., 2000). The biofilm-associated dye was released from the biofilms by adding 200 μl of 33% (v/v) acetic acid. After 20 min, the plate was mixed on an orbital shaker and the solubilized dye was transferred to a new microtiter plate to record the OD580.
Loosely bound proteins of the outer membrane of G. sulfurreducens were first mechanically detached from cells grown to mid-exponential phase in FWAF medium. The proteins in the sheared fraction were then separated by SDS-PAGE, as previously described (Cologgi et al., 2011), and those containing heme groups were stained with N, N, N, N-tetramethylbenzidine (Thomas et al., 1976).
The MECs used in these studies were dual-chambered, H-type electrochemical cells equipped with graphite rod (Alfa Aesar,1.27-cm diameter, 99% metals basis, 12 cm2) anode and cathode electrodes, separated with a Nafion membrane (Ion Power, Inc., New Castle, DE), and set up as described previously (Speers and Reguera, 2012). Each chamber was filled with 90 ml of anaerobic mineral DB medium (Speers and Reguera, 2012) and an initial concentration of 1 mM sodium acetate was added to the anode chamber as electron donor to support the growth of the anode biofilms and current production. The WT and mutant strains of G. sulfurreducens used in the MECs experiments were first grown in DB medium with acetate (20 mM) and fumarate (40 mM) at 30°C, harvested by centrifugation, suspended in DB medium, and inoculated into the anode chamber, as previously described (Speers and Reguera, 2012). A VSP potentiostat (BioLogic, Claix, France) was used to set a 0.24-V potential at the anode electrode vs. a 3 M Ag/AgCl reference electrode (Bioanalytical Systems, Inc.) prior to cell inoculation and to monitor current production at the anode electrode throughout the experiment. The anode electrode was removed from the anode chamber when all the acetate was consumed, as indicated by the drop in current production to < 0.1 mA, and the biofilm cells were then differentially stained (green, live cells; red, dead cells) with the BacLight™ viability kit (Invitrogen), following the manufacturer's recommendations. After staining, the anode electrode was placed on a Lab-Tek® coverglass chamber (Nunc) filled with 3 ml of phosphate buffer saline and imaged with a FluoView FV1000 inverted confocal laser scanning microscope (CLSM) (Olympus) equipped with a UPLFLN 40x oil immersion objective (Olympus, NA 1.30). The green fluorescent dye (SYTO 9) was excited at 488 nm and emission was detected with a BA505-525 band pass filter. The red fluorescent dye (propidium iodide) was excited at 543 nm and emission collected with a BA560IF long pass filter. Vertical image stacks of 200 × 200 μm fields were collected every 1 μm and 3-dimensional image projections were generated with the FV10-ASW 3.0 software (Olympus). The biofilm biomass was estimated from the fluorescence emitted by the biofilm cells using the biovolume function of the COMSTAT software (Heydorn et al., 2000).
The genome of G. sulfurreducens contains four genes annotated as pilT (pilT1, GSU0146; pilT2, GSU0230; pilT3, GSU0436; and pilT4, GSU1492) that code for proteins containing the four conserved signature sequences (Walker A and B motifs and Asp and His boxes) of the secretion ATPase protein superfamily that PilT retraction ATPases belong to Savvides (2007). Indeed, all of the PilT paralogs encoded in the genome of G. sulfurreducens contain a Walker A motif with a conserved lysine for ATP-binding, a Walker B motif with a conserved glutamate residue for Mg2+-binding and ATP hydrolysis, and Asp and His boxes with conserved aspartic and histidine residues, respectively, which are essential for PilT activity (Figure 1; Walker et al., 1982; Savvides, 2007; Chiang et al., 2008; Jakovljevic et al., 2008). As shown in Table 3, the putative PilT proteins are 43–52% identical and 61–70% similar to each other, and PilT3 and PilT4 had the highest identities and similarities to the PilT ATPases that power pilus retraction in N. gonorrhoeae (PilTNg), M. xanthus (PilTMx), and P. aeruginosa strain K (PilTPa) (Whitchurch et al., 1991; Wolfgang et al., 1998; Jakovljevic et al., 2008). PilT3, in particular, was 56% identical and 74% similar to PilTPa, whereas PilT4 was 77% identical and 87% similar to PilTMx.
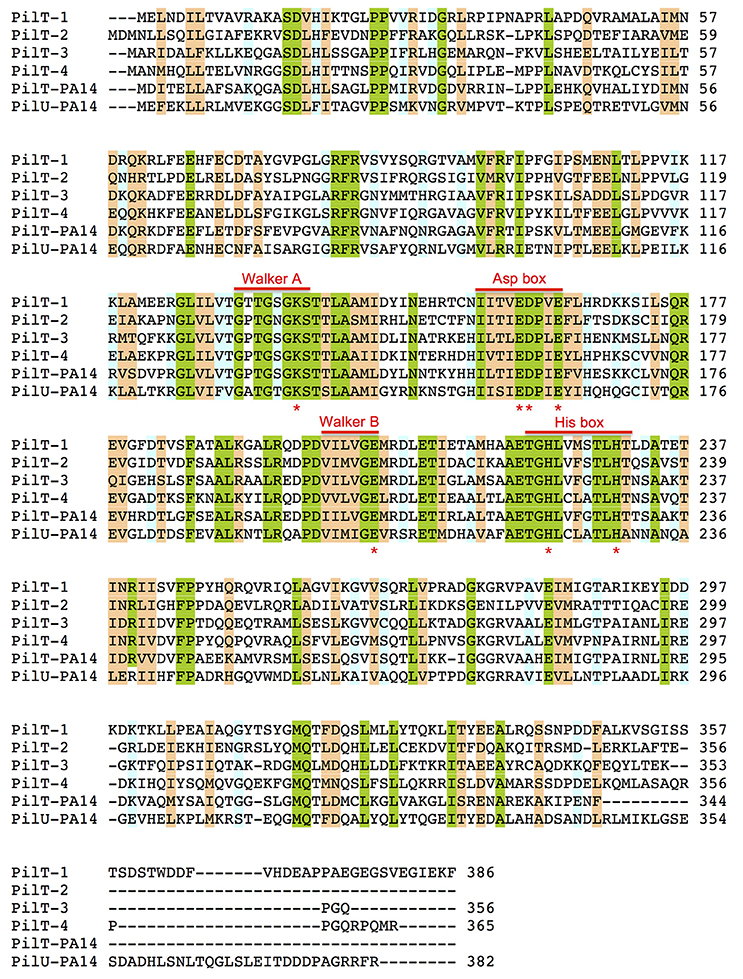
Figure 1. Amino acid sequence alignments of PilT proteins from G. sulfurreducens and P. aeruginosa PA14. Multiple sequence alignment analysis was carried out using Clustal O (1.2.2) program at the UniProt database (http://www.uniprot.org). Shaded areas show positions in the alignment with a single, fully conserved residue (green) or with conservation between groups of strongly (orange) or weakly (blue) similar properties (Gonnet PAM 250 matrix scoring >0.5 or ≤ 0.5, respectively). The conserved Walker A and Walker B motifs and Asp and His boxes of secretion ATPases are indicated. Red stars under the sequence alignment mark highly conserved lysine (Walker A), glutamate (Walker B), aspartate and/or glutamate (Asp box) and histidine (His box) residues that are essential for PilT activity.
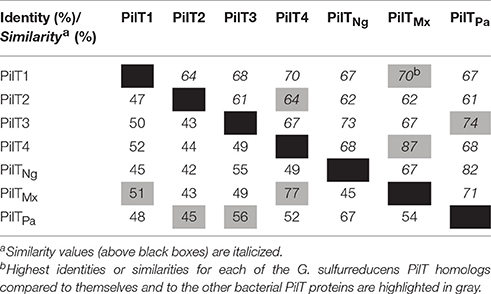
Table 3. Identity and similarity of the putative PilT proteins of G. sulfurreducens (in red) in pairwise comparisons with each other and the PilT's from N. gonorrhoeae (PilTNg), M. xanthus (PilTMx), and P. aeruginosa strain K (PilTPa).
The genetic arrangement of the pilT genes in G. sulfurreducens and its transcriptional expression profile with neighboring genes is shown in Figure 2. pilT1 (GSU0146) is flanked by the recA and recX genes (Figure 2A), a gene arrangement that is conserved in other sequenced genomes in the Geobacteraceae family (Geobacter metallireducens, Geobacter uraniireducens, and Pelobacter carbinolicus). Furthermore, pilT1 was co-transcribed with the recA gene (Figure 2B), consistent with a role for PilT1 in the cellular recombination machinery. The pilT2 gene (GSU0230), on the other hand, is located downstream of a gene (GSU0231) encoding a hypothetical protein (Figure 2A) but transcribed independently from it under the conditions tested (Figure 2B). Downstream of pilT2, and in opposite orientation, is the alkK gene (GSU0229), which encodes a putative medium-chain fatty acid CoA ligase. This arrangement is unique to G. sulfurreducens. Further, pilT2 was not conserved in all of the Geobacter spp. with sequenced genomes. We found, for example, a pilT2 homolog in G. metallireducens (Gmet0260) but none in G. uraniireducens. The lack of conservation of pilT2 in other Geobacter genomes contrasts with the high conservation of the pilin-encoding gene, pilA (Reguera et al., 2005) and argues against its role as the PilT ATPase of the T4P apparatus in these bacteria.
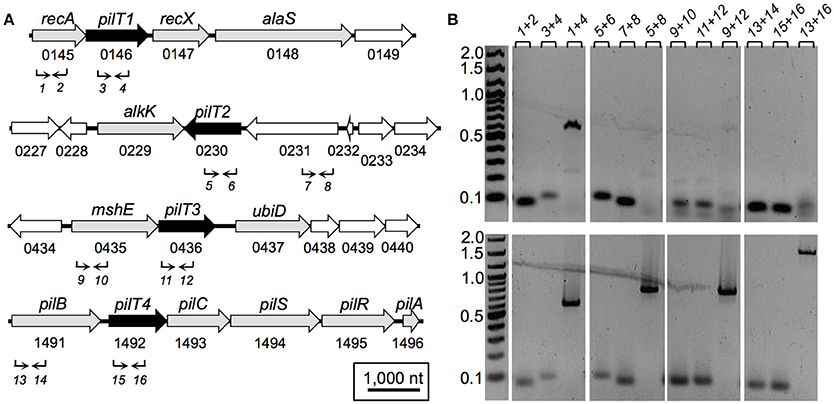
Figure 2. Genetic (A) and transcriptional (B) organization of pilT genes in G. sulfurreducens. (A) Schematic view of the genomic region containing each of the four pilT genes (labeled 1–4, black arrows) and neighboring ORFs (white arrows, hypothetical gene; gray arrows, annotated function). Also shown are gene designations (gene name and ORF number, above and below, respectively) and PCR primers (1–16; Table 2) and direction of amplification (arrows). (B) Agarose gels showing cDNA amplified from each pilT gene region and neighboring genes (top panels) in reference to products amplified from genomic DNA controls (bottom panels). PCR-amplifications used primer sets 1–16, as indicated in (A) and listed on top of the gel panels in (B). Lanes, gene target (primer combination): recA (1+2), pilT1 (3+4), recA-pilT1 (1+4), pilT2 (5+6), GSU0231 (7+8), pilT2-GSU0231 (5+8), mshE (9+10), pilT3 (11+12), mshE-pilT3 (9+12), pilB (13+14), pilT4 (15+16), pilB-pilT4 (13+16).
The pilT3 gene (GSU0436) is downstream of a gene (mshE) encoding a putative mannose-sensitive haemagglutinin (MSHA) biogenesis/general secretory system II protein that is homologous to the pilin polymerization ATPase of MSHA pili in Vibrio cholerae (Jones et al., 2015). The mshE pilT3 arrangement is conserved in the genomes of G. metallireducens and G. uraniireducens. The two genes are unlikely to be co-transcribed in G. sulfurreducens because, although reverse transcription produced a faint band spanning both genes, the band was smaller than in the controls amplified from genomic DNA and thus it was more likely the product of non-specific binding of one or both primers (Figure 2B).
The pilT4 gene (GSU1492) is flanked by genes encoding proteins with predicted or confirmed roles in pili biogenesis and its regulation such as the pilin assembly ATPase PilB (Steidl et al., 2016), the pilin biogenesis protein PilC, the PilS sensor, the PilR transcriptional regulator (Juarez et al., 2009), and the pilin subunit PilA (Reguera et al., 2005; Figure 2A). This genetic arrangement is conserved in other Geobacteraceae (G. metallireducens, G. uraniireducens, and P. carbinolicus) and is similar to the organization of the pil region of M. xanthus (Wall and Kaiser, 1999). The genes flanking pilT4 were also very conserved among Geobacter species. The PilB protein encoded by GSU1491 was, for example, 94% identical and 97% similar to Gmet1393 from G. metallireducens, and 88% identical and 96% similar to Gura2682 from G. uraniireducens. Similarly, the PilC protein encoded by GSU1493 was 89% identical and 95% similar to Gmet1395 from G. metallireducens, and 81% identical and 90% similar to Gura2680 from G. uraniireducens. PilB and PilC proteins are the most highly conserved components of the bacterial T4P apparatus and related systems such as the bacterial type II secretion system and archaeal motility systems and both have been proposed to form the minimal functional motor unit of the pilus (Burrows, 2012). Reverse Transcription (RT)-PCR analysis using a primer set spanning the intergenic pilB pilT4 failed to amplify a full-length product, suggesting that, under the conditions tested, the two genes are not co-transcribed (Figure 2B). Yet, despite their independent transcription, the expression profiles of pilB and pilT4 in both rich (NBAF) and mineral (FWAF) medium were similar and matched well the expression profile of pilA regardless of the incubation temperature (30°C or pili-expressing temperatures of 25°C; (Reguera et al., 2005; Cologgi et al., 2011; Figure 3). Indeed, the pilT4, pilB, and pilA genes were up-regulated under all of the conditions tested, whereas pilT1, pilT2, and pilT3 were all down-regulated. Hence, based on gene and protein homologies, gene clustering, overall conservation in other Geobacter spp., and expression profiles with other pil genes, pilT4 is arguably the most likely candidate to encode for the PilT motor of the T4P apparatus in G. sulfurreducens.
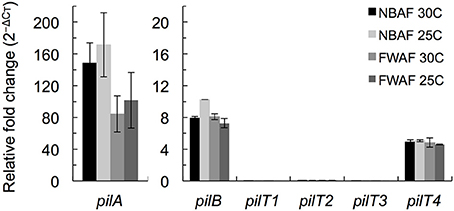
Figure 3. Transcriptional expression analysis of pil genes in G. sulfurreducens by qRT-PCR. The expression levels of pilA, pilB, and the four pilT (pilT1-4) genes of G. sulfurreducens were measured in duplicate samples in reference to recA controls in cultures grown in rich NBAF and mineral FWAF media incubated at pili-inducing (25°C) or non-inducing (30°C) temperatures. There were no statistically significant changes (p < 0.01) in the relative expression of the target genes in reference to the pili-inducing conditions (FWAF 25C) in two-tailed pairwise comparisons using the t-test function of the Microsoft® Excel® software.
We gained insights into the functionality of each of the PilT proteins encoded in the genome of G. sulfurreducens in heterologous complementation experiments that tested the ability of each pilT gene to complement the phenotypes associated with a mutant of P. aeruginosa strain K (PAK) carrying an in-frame deletion in the pilT gene (PAKΔpilT). The inability of the PAKΔpilT mutant to retract the T4P leads to hyperpiliation, a phenotype that can be restored by genetic complementation of the mutation with the wild-type pilT copy in trans (Sundin et al., 2002). PilT3 was the only PilT homolog of G. sulfurreducens that rescued the hyperpiliated phenotype of the PAKΔpilT mutant (p < 0.01), reducing the piliation per cell to WT levels (Figures 4A,B). The hyperpiliation of the PAKΔpilT mutant also increases the adhesion of the cells to surfaces and co-aggregation and promotes the formation of denser biofilms under static conditions, a phenotype that is restored by genetic complementation (Chiang and Burrows, 2003). We observed a similar restoration of the biofilm phenotype when the PAKΔpilT strain was genetically complemented with the pilT3 gene of G. sulfurreducens (Figure 4C). Thus, the PilT3 protein of G. sulfurreducens is able to power pilus retraction and control the levels of piliation and biofilm formation in P. aeruginosa.
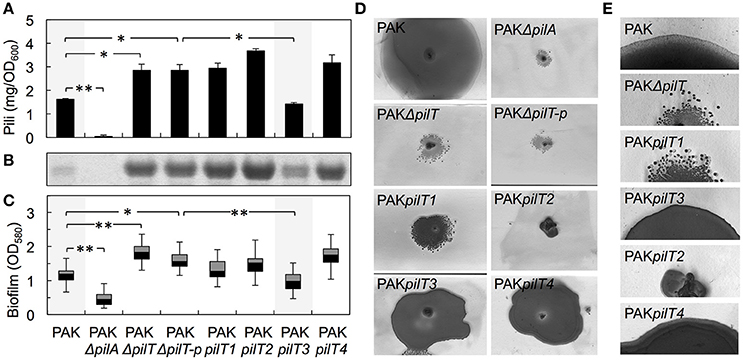
Figure 4. Heterologous expression of G. sulfurreducens pilT genes in the hyperpiliated, non-twitching PAKΔpilT mutant. (A–C) Restoration by pilT3 of WT levels of piliation [shown as purified pili protein (A) and Coomassie-stained pilin protein band after SDS-PAGE (B)] and biofilm density (C) in the PAKΔpilT strain. The pilin-deficient PAKΔpilA and hyperpiliated PAKΔpilT-p strain (carrying the empty pMMB67EH vector) are shown as controls. Data are average and standard deviation of pili protein (mg/OD600) purified from three biological replicates (A) and median and first and third quartiles of biofilm biomass from six biological replicates, each containing 5–8 technical replicates (C). Statistically significant changes in pili content (A) and biofilm biomass (C) were determined in two-tailed pairwise comparisons with the WT or PAKΔpilT-p strains using the t-test function of the Microsoft Excel software (*p < 0.01, **p < 0.001). (D) Twitching motility slide assays showing the Coomassie-stained zones of surface expansion in the wild-type PAK, which are absent in the pilin-deficient PAKΔpilA and non-twitching PAKΔpilT and PAKΔpilT-p strains. Expression of pilT3 and pilT4 (PAKpilT3 and PAKpilT4 strains) restored the twitching motility deficiency of the PAKΔpilT strain. (E) Micrographs showing the zones of expansion on the edge of PAK colonies shown in (D).
Twitching motility in P. aeruginosa requires the activity of PilT and its paralog, PilU (Whitchurch et al., 1991; Whitchurch and Mattick, 1994). This mode of surface translocation involves cycles of pilus extension, adhesion of the pilus tip to the surface, and retraction of the pilus fiber to pull the cell (Skerker and Berg, 2001). The PilT motor hydrolyzes ATP to energize the retraction of the surface-attached pilus and provides the force needed to move the cell forward (Merz et al., 2000). As a result, the inactivation of PilT results in a non-twitching phenotype (Whitchurch and Mattick, 1994). Thus, we also investigated the ability of the PilT proteins of G. sulfurreducens to restore the twitching motility defect of the PAKΔpilT mutant in subsurface twitching motility assays on glass slides (Figure 4D). Interestingly, not only PilT3 but also PilT4 rescued the non-twitching phenotype (Figure 4D). Examination of the colony edges at higher magnification (Figure 4E) revealed the smooth and thin expansion zones in the strains genetically complemented with pilT4, and to a lesser extent with pilT3, that are characteristic of the WT twitching motility phenotype (Semmler et al., 1999). Thus, the heterologous expression studies demonstrated that both PilT3 and PilT4 can function as motor ATPases in P. aeruginosa, but PilT4's activity is restricted to twitching motility.
As in P. aeruginosa, the pili of G. sulfurreducens also promote cell-cell aggregation and biofilm formation (Reguera et al., 2007; Cologgi et al., 2014). Thus, we investigated the biofilm-forming abilities of G. sulfurreducens mutants carrying pilT deletions in reference to the WT strain (Figure 5A). As controls we also included a pili-defective pilB mutant, which makes thin biofilms, and its genetically complemented strain pilB+, which expresses the wild-type pilB gene in trans and restores pili assembly and biofilm formation (Steidl et al., 2016). Deletion of pilT1, pilT2, and pilT3 had no significant effect on biofilm formation in G. sulfurreducens. By contrast, a pilT4 mutant formed denser biofilms on plastic surfaces (p < 0.001), a phenotype associated with hyperpiliation in this bacterium (Reguera et al., 2007; Cologgi et al., 2014). The enhanced biofilm phenotype of the pilT4 mutant was restored in the genetically complemented strain, pilT4+, but not in the control pilT4p strain, which carries the empty pRG5 expression vector in the same pilT4 mutant background (Figure 5A). The enhanced biofilm-forming abilities of the pilT4 mutant cannot be explained by differences in cell growth, because planktonic growth rates for this mutant were comparable to those measured in all of the strains tested (~0.12 OD660/h). Rather, the dense biofilm phenotype observed in the pilT4 mutant is consistent with the increased adhesion and co-aggregation reported for hyperpiliated strains of G. sulfurreducens (Reguera et al., 2007) and other bacteria (Deziel et al., 2001; Chiang and Burrows, 2003).
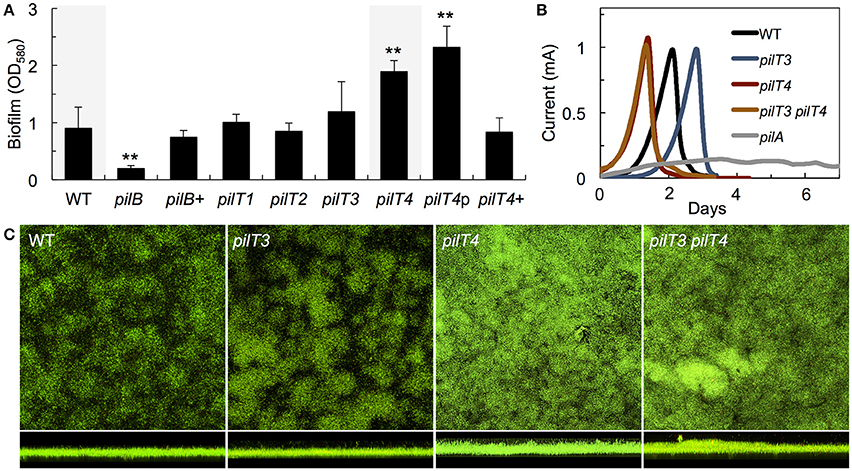
Figure 5. Biofilm formation on plastic surfaces (A) and on poised electrodes (B,C) by WT and pilT mutants of G. sulfurreducens. (A) Biomass of 48-h biofilms of a pilus-deficient pilB mutant, its complemented strain pilB+, and mutants carrying a deletion in one of the pilT paralogs (pilT1-4) relative to the WT strain. The biofilm biomass of a genetically complemented pilT4+ strain and pilT4p control strain carrying the empty plasmid are also shown. Data are the average and standard deviation of four biological replicates, each containing six technical replicates. Statistically significant changes were determined in two-tailed pairwise comparisons with the WT or PAKΔpilT-p strains using the t-test function of the Microsoft® Excel® software (**p < 0.001). (B) Current production by anode biofilms in MECs fed an initial concentration of 1 mM acetate and driven by the WT, pilT3, pilT4, and pilT3 pilT4 strains in reference to the pilin-deficient mutant pilA. (C) Confocal micrographs (200 × 200 μm2) showing top and side view projections of current-producing anode biofilms collected from MECs in (B) and stained with the BacLight viability dies (green, live cells; red, dead cells).
The T4P of G. sulfurreducens are also required for biofilm formation on anode graphite electrodes poised at a metabolically oxidizing potential and for optimal current production in MECs (Reguera et al., 2006; Steidl et al., 2016). Hence, we grew the pilT3 and pilT4 mutants in the anode chamber of a MEC fed with 1 mM acetate and monitored current production in reference to the WT strain (Figure 5B). The pilT4 mutant attached rapidly to the electrode and initiated current production before any other strain. The average attachment or lag phase in triplicate MECs for the pilT4 mutant was, for example, 5 ± 1 h, which is 4–5 times shorter than the WT (24 ±11 h). By contrast, the pilT3 mutant was delayed in attachment (33 ± 3 h average lag phase in triplicate MECs; Figure 5B). Yet once the cells attached and the exponential phase of biofilm growth started (corresponding to the linear phase of current production) the pilT4 and pilT3 mutant biofilms produced current at rates comparable to the WT strain and also reached similar levels of current maxima (Figure 5B). This contrasts with the severe defect in current production of the pilA mutant (Figure 5B), which carries a deletion in the pilin-encoding gene pilA that prevents it from making conductive pili (Reguera et al., 2005; Cologgi et al., 2011) and is also unable to secrete the matrix-associated cytochrome OmcZS required for electron transfer to the underlying electrode (Steidl et al., 2016). Furthermore, coulombic efficiencies in the WT and pilT4 biofilms were similar (80–90%), indicating that on average the WT and pilT4 anode biofilms converted the same amount of acetate into electricity (80–90%). However, the pilT4 mutant accumulated a much greater biofilm biomass on the electrode (23.3 ± 4.2 μm3/μm2) than the WT (11.9 ± 1.5 μm3/μm2) to produce the same amount of current as the WT (Figure 5C). This indicates that, on average, less current was generated per cell in the pilT4 biofilms than in the WT biofilms.
We also ruled out compensatory effects between PilT3 and PilT4 by testing the ability of a pilT3 pilT4 double mutant to colonize and generate current in the MECs. The double mutant was phenotypically indistinguishable from the pilT4 single mutant, having shorter lag phases but otherwise producing current like the WT (Figure 5B) and forming denser biofilms on the anode electrodes (Figure 5C). Hence, the pilT4 mutation was dominant over pilT3, not only rescuing the colonization defect of the pilT3 cells, but also promoting faster attachment and the formation of denser biofilms. This is again consistent with the aggregative nature of hyperpiliated strains of G. sulfurreducens (Reguera et al., 2007; Cologgi et al., 2014), which is expected for cells defective in pilus retraction (Sundin et al., 2002).
As G. sulfurreducens also expresses pili to access and reduce Fe(III) oxides (Reguera et al., 2005), we tested the ability of the pilT3, pilT4, and double pilT3 pilT4 mutants to grow with Fe(III) oxides when provided as sole electron acceptor in reference to the WT strain (Figure 6A). The strains were first grown in Fe(III) oxide media with 4 mM of NTA, a metal chelator that solubilizes the Fe(III) from the Fe(III) oxides and alleviates the need for cells to directly contact the oxide minerals (Reguera et al., 2005). Exponentially growing cells were then transferred to Fe(III) oxides medium without NTA. The chelating activities of the NTA carried over in the inoculum (ca. 0.4 mM) stimulated initial growth in the cultures, as indicated by the low levels of reduced iron (Fe[II]) solubilized by all of the strains during the first 4–5 days (Figure 6A). However, whereas the WT continued to grow exponentially beyond this initial phase with similar doubling times (2.9 ± 0.1 days), the pilT3 mutant plateaued for approximately 4 days before resuming Fe(III) reduction at rates similar to the WT strain (Figure 6A). This contrasts with a Tyr3 mutant, which carries alanine replacements in the pilin's three tyrosines that increase the electrical resistance of the T4P (Steidl et al., 2016) but is able to resume exponential growth without delay after the NTA-dependent phase, albeit at slower rates (doubling time, 10.5 ± 0.8 days). The delay observed in the pilT3 mutant is consistent with a strain that has a transient defect in adhesion, as we previously observed during the colonization of anode electrodes in MECs (Figure 5B). However, the pilT3 defect was chemically rescued in the presence of the metal chelator NTA, which provided a soluble, reducible form of Fe(III) for cellular respiration throughout the experiment (Figure 6B). By contrast, the reductive activities of the pilT4 mutant never fully recovered after the NTA-dependent phase (Figure 6A). Furthermore, as we observed during the colonization of electrodes (Figure 5B), introducing the pilT3 mutation in the pilT4 mutant (pilT3 pilT4 mutant) did not change the pilT4 mutant phenotype. Thus, they are unlikely to have complementary roles or compensate for each other's inactivation. And, as with pilT3, the defects of the single pilT4 and double pilT3 pilT4 mutants were chemically rescued with NTA (Figure 6B).
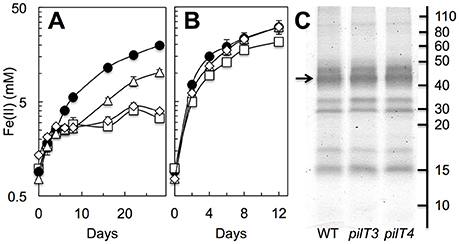
Figure 6. Role of PilT3 and PilT4 in extracellular electron transfer to Fe(III) oxides. (A,B) Fe(III) oxide reduction (measured as acid-extractable Fe[II]) in the absence (A) or presence (B) of the metal chelator NTA (4 mM) by the pilT3 (triangles), pilT4 (squares), and double pilT3 pilT4 (rhomboid) mutants of G. sulfurreducens in reference to the WT (solid circles). Shown are the average and standard deviation of triplicate cultures for each strain. (C) Heme-stained proteins mechanically detached from the outer membrane of the WT, pilT3, and pilT4 strains. The arrow points at the migration of the OmcS cytochrome.
We also ruled out pleiotropic effects of the mutations in c-cytochrome expression by examining the type and abundance of outer membrane c-cytochromes (Figure 6C). There were no measurable differences in the heme-containing proteins mechanically sheared from the outer membrane of the WT, pilT3, and pilT4 strains and all included the OmcS c-cytochrome that is required for optimal reduction of Fe(III) oxides (Mehta et al., 2005). Hence, the Fe(III) oxides experiments support our early conclusion that PilT4 is the retraction ATPase in G. sulfurreducens and highlight a previously unrecognized role for pilus retraction in the respiration of Fe(III) oxides.
The clustering of pilT4 with other pil genes (Figure 2), including the pilin-encoding gene pilA and the gene encoding the pilin assembly ATPase PilB, as well as the similar expression profiles of this pilT paralog and other pil genes (Figure 3) and conservation in Geobacter spp., makes PilT4 the most obvious candidate to power T4P retraction in G. sulfurreducens. We demonstrated its functionality through its ability to restore the twitching motility phenotype of PAKΔpilT, the retraction-deficient mutant of P. aeruginosa strain K (Figures 4D,E). Thus, although PilT4 was less similar (68%) to PilTPa than PilT3 (74%) (Table 3), it was also recruited to the T4P apparatus of P. aeruginosa and it was able to hydrolyze ATP to generate the force needed to power pilus retraction during twitching motility (Merz et al., 2000; Skerker and Berg, 2001; Maier et al., 2002). Yet PilT4 did not restore WT levels of piliation in planktonic cultures (Figures 4A,B) nor did it genetically complement the biofilm phenotype of the PAKΔpilT mutant in static biofilm assays (Figure 4C). Although the exact composition of the T4P apparatus of P. aeruginosa is not known, PilB, PilC, and PilT have been proposed to form the minimal functional motor unit (Burrows, 2012). PilC is predicted to interact with both PilB and PilT in the motor complex to coordinate antagonistic cycles of polymerization and depolymerization, respectively (Takhar et al., 2013). This minimal motor requires an additional component, PilU, for twitching motility (Whitchurch and Mattick, 1994) but not for biofilm formation under static conditions (Chiang and Burrows, 2003). Studies suggest that differences in just a few key residues of the PilT protein can induce conformational changes such that prevent productive interactions with other components of the T4P apparatus (Nakasugi et al., 2007). Hence, the PilT4 conformation may promote interactions with the T4P apparatus of P. aeruginosa formed for twitching motility, which includes PilUPa, but may not be efficiently recruited by the T4P motor unit that powers pilus retraction in planktonic cells and during biofilm formation, which does not include PilUPa.
Consistent with its role as a PilT motor, inactivating the pilT4 gene in G. sulfurreducens led to the formation of the dense biofilms (Figure 5) that are characteristic of the enhanced aggregated behavior of hyperpiliated strains in this bacterium (Reguera et al., 2007; Cologgi et al., 2014). Furthermore, pilT4 cells colonized the surface of anode electrodes faster than the WT strain (Figure 5B). The initial phase of colonization comprises the period whereby planktonic cells attach to the anode to form a saturating monolayer (Marsili et al., 2010). The attached cells then express key biofilm components such as the conductive pili (Reguera et al., 2006; Steidl et al., 2016), the biofilm exopolysaccharide (EPS) (Rollefson et al., 2011), and the matrix-associated c-cytochrome OmcZS (Nevin et al., 2009) and grow and divide exponentially while coupling the oxidation of the electron donor (e.g., acetate) to the reduction of the anode electrode (Marsili et al., 2010). Current production increases linearly during this phase of exponential growth until acetate availability becomes growth-limiting, which marks the beginning of the phase of current deceleration (Speers and Reguera, 2012). Hence, the shorter lag phase observed in the pilT4 mutant is consistent with a mutant with enhanced adhesive properties, as reported for hyperpiliated PilT-deficient strains (Chiang and Burrows, 2003). Hyperpiliated strains of G. sulfurreducens also have enhanced co-aggregation and, as a result, form denser biofilms (Reguera et al., 2007; Cologgi et al., 2014), a phenotype that also matches well the dense anode biofilms observed in the pilT4-driven MECs (Figure 5C).
Interestingly, although more pilT4 cells accumulated on the anode electrodes than in the WT biofilms (Figure 5C), coulombic efficiencies and parameters used to measure the biofilm electroactivity such as rates of current production and maximum current were similar in the pilT4 and WT MECs (Figure 5B). Thus, although the pilT4 biofilms converted the same amount of acetate into current over the same period of time, less current was generated on a per cell basis than in the WT biofilms. The reduced efficiency of individual pilT4 cells in the anode biofilms cannot be explained by defects in c-cytochrome expression, as reported for other strains with pili defects (Juarez et al., 2009; Cologgi et al., 2011), because the profile and expression levels of outer membrane heme-containing proteins was similar in the WT and pilT4 cells (Figure 6C). The conductive pili network works coordinately with c-type cytochromes of the biofilm matrix to maintain optimal rates of electron transfer in anode biofilms (Steidl et al., 2016). The inability of the pilT4 biofilm cells to maintain optimal pilus dynamics through antagonistic cycles of pilus extension and retraction may reduce the number of productive interactions between the pili and electron carriers of the biofilm matrix, thus reducing the rates of electron transfer per cell and requiring the formation of dense biofilms to compensate for the defect. Oral bacteria, which like G. sulfurreducens also use pili to co-aggregate and adhere to surfaces, rely on dynamic cycles of pilus extension and retraction to maintain optimal cell-cell distances needed for effective cell-cell communication and coordinated behavior (Kolenbrander et al., 2010). Cell-cell distance is particularly important for electron transfer in electroactive biofilms, which rely on multistep electron hopping among the electron carriers of the biofilm matrix (Strycharz-Glaven et al., 2011; Yates et al., 2015; Steidl et al., 2016). Hence, PilT4-mediated pilus retraction may be important to maintain optimal cell-cell separation and to promote interactions between the pili and other electron carriers in the biofilm matrix.
Our studies also provide evidence for a role of PilT4-mediated pilus retraction during the respiration of Fe(III) oxides. Inactivation of PilT4 prevented the cells from reducing the oxide minerals unless in the presence of NTA (Figure 6), a metal chelator that alleviates the need for iron-reducers to directly contact the mineral (Lovley et al., 1994; Lovley and Woodward, 1996; Manzella et al., 2013). As a result, NTA chemically rescues defects in pili synthesis (Reguera et al., 2005) and conductivity (Feliciano et al., 2015) in G. sulfurreducens. The inability of the pilT4 mutant to reduce Fe(III) oxides via a direct-contact mechanism contrasts with earlier reports (Reguera et al., 2005) indicating that PilT4 inactivation does not affect Fe(III) oxides reduction. These early studies required extended incubation times (60 days or longer) to unmask mutant phenotypes during the reduction of Fe(III) oxides, which could have selected for strains carrying compensatory mutations (Leang et al., 2005). We minimized the selection of compensatory phenotypes by first growing all of the strains in cultures with Fe(III) oxides in the presence of the metal chelator NTA. Furthermore, some NTA was carried over with the inoculum, which stimulated the initial growth of all the strains until the concentrations of NTA per cell became growth-limiting. Beyond this point, the cells depend on the proper functioning of the pili to bind and reduce the oxides and pili-defective phenotypes are unmasked. The pilT4 mutation caused the most severe defect, as indicated by the inability of the mutant to reduce the Fe(III) oxides beyond the NTA-mediated phase (Figure 6A). However, the mutant phenotype was chemically rescued when an excess NTA was provided as a mediator (Figure 6B) to bypass the cells requirement to use pili to reduce the Fe(III) oxides (Reguera et al., 2005). The conductive pili of G. sulfurreducens bind the mineral particles and function as an electronic conduit between the cell and the minerals during respiration (Reguera et al., 2005). Approximately one third of the reduced Fe(III) in the oxides is solubilized as Fe(II), while the rest remains in mineral form as magnetite particles attached to the pilus fibers (Reguera et al., 2005). The pili also bind the soluble uranyl ion and reduce it to a mononuclear uranium mineral phase, which remains attach to the pili (Cologgi et al., 2011). The attachment of magnetite or reduced uranium mineral particles to the pili prevents the conductive appendages from binding more electron acceptor and could halt respiration. However, PilT-mediated depolymerization of the pilins could allow the cells to retract the T4P and detach the reduced minerals. The pilins could then be recycled in a new round of polymerization to produce “clean” T4P, which are now ready to bind the electron acceptor and discharge metabolic electrons. In this manner, the antagonistic action of PilB and PilT could confer on Geobacter bacteria a competitive advantage for metal respiration. This would be particularly relevant in the natural environments inhabited by Geobacter bacteria, where Fe(III) oxides are often found dispersed in the extracellular medium. The dynamic nature of the T4P could also be critical for the cells to survive in uranium-contaminated environments, as rapid cycles of extension and retraction would promote the binding and reductive precipitation of more soluble uranyl cation, preventing its permeation and mineralization inside the cell envelope and preserving the cell's respiratory functions and viability (Cologgi et al., 2011).
It is important to note that our studies did not rule out the possibility that PilT3, one of the PilT paralogs in G. sulfurreducens, may be required to fine-tune the retraction of T4P under specific conditions. In P. aeruginosa, the PilT paralog PilU is recruited to the T4P apparatus to work coordinately with PilT to power twitching motility (Whitchurch and Mattick, 1994). As a result, mutations in either pilT or pilU lead to hyperpiliation and non-twitching phenotypes (Whitchurch and Mattick, 1994). While the pilT mutant of P. aeruginosa forms denser biofilms under static conditions, the pilU mutant is delayed in biofilm formation only under flow conditions (Chiang and Burrows, 2003). The delayed biofilm-forming abilities of the pilU mutant are similar to the delayed colonization of the anode electrode observed in MECs driven by the pilT3 mutant of G. sulfurreducens (Figure 5B). In MECs, the anode medium is continuously stirred to promote the initial attachment of planktonic cells to the electrode and the diffusion of the electron donor and any other nutrients needed to support the growth of the anode biofilm (Speers and Reguera, 2012). The flow conditions that prevail in the MEC environment are likely to unmask defects from mutants with reduced adherence such as the pilT3 mutant, similarly to how the phenotype of the pilU mutant of P. aeruginosa was unmasked in biofilm assays under flow (Chiang and Burrows, 2003). The reduced adherence of the pilU mutant of P. aeruginosa has been proposed to result from differences in the strength and/or integrity of the T4P (Chiang and Burrows, 2003). However, it is unlikely that T4P integrity was compromised in the pilT3 mutant of G. sulfurreducens because pili-dependent functions such as the structure and electroactivity of anode biofilms was not affected (Figure 5) nor were the rates of Fe(III) oxide reduction affected (Figure 6A). Based on the cross-regulation of EPS production and pili synthesis that may exist in G. sulfurreducens (Steidl et al., 2016), it is possible that the pilT3 mutation negatively influenced the synthesis of the Xap EPS, which Geobacter cells use to attach to positively-charged surfaces such as an anode electrode poised at a positive voltage, as in our MEC studies, or to Fe(III) oxides (Rollefson et al., 2011). The defect may have been transient, because once the cells attached, they grew as biofilms with the same structure and electrochemical activity as the WT (Figures 5B,C). Similarly, the pilT3 mutant had an initial delay during the reduction of Fe(III) oxides (Figure 6A), which also depends on the ability of the cells to produce the EPS to anchor outer membrane c-cytochromes needed for electron transfer to the iron minerals (Rollefson et al., 2011). The coordinated regulation of the assembly of conductive T4P and synthesis of EPS could be analogous to M. xanthus, which uses EPS to trigger the retraction of the pili and relies on the pili dynamics to send feedback information to the Dif chemotaxis pathway to modulate EPS production (Black et al., 2006).
Alternatively, PilT3 may function as an ATPase in a separate apparatus such as a type II secretion system, which also relies on the ATPase activities of PilB- and PilT-like motors to polymerize and depolymerize pseudopilins, respectively, and export proteins across the outer membrane (Vignon et al., 2003). Indeed, pilT3 was located downstream of a pilB-like gene (mshE) and the PilT3 and MshE proteins are highly expressed in Fe(III) oxide cultures of G. sulfurreducens (Ding et al., 2008), suggesting a conserved, coordinated function. PilT-like proteins can also be components of more evolutionary divergent secretion systems such as type IV secretion systems (T4SS), which translocate DNA or proteins across the cell envelope (Sexton et al., 2004). In these systems, PilT-like proteins function as traffic, rather than motor, ATPases, energizing the early steps of biogenesis of the T4SS or the translocation of the substrate (Alvarez-Martinez and Christie, 2009). PilT1, which in G. sulfurreducens is encoded by a gene in the recA operon (Figure 2B), is a likely candidate to function as a traffic ATPase in a T4SS system implicated in DNA uptake, because its expression would be coordinated with the recombination machinery. The function of PilT2, which is encoded by the orphan pilT2 paralog, is more difficult to predict. However, the fact that it is not conserved in other Geobacter spp. points at a role specific to the physiology of G. sulfurreducens.
In summary, we present genetic evidence that identifies PilT4 as the elusive PilT ATPase of the T4P apparatus of G. sulfurreducens and reveals a role for PilT-mediated pilus retraction during cellular respiration, both in electroactive biofilms and during the reduction of Fe(III) oxides. Whether PilT4 also requires the coordinated action of any of the other PilT paralogs, particularly PilT3, remains to be elucidated. Indeed, PilT3 complemented all the PilT functions assayed in P. aeruginosa and showed reduced adherence phenotypes during the colonization of electrodes and reduction of Fe(III) oxides previously linked to retraction defects in other bacteria (Chiang and Burrows, 2003). We cannot rule out that the conductive pili mediate twitching motility in G. sulfurreducens either. Both PilT3 and PilT4 rescued the non-twitching phenotype of a PilT-deficient mutant of P. aeruginosa (Figure 4D). Moreover, the inactivation of pilT4 in G. sulfurreducens produced dense biofilms (Figure 5), a phenotype that could also result from defects in twitching motility (Chiang and Burrows, 2003). Twitching motility was not observed in G. sulfurreducens in submerged assays that used glass coverslips with or without iron coatings (Reguera et al., 2005). Such Fe(III) oxide coatings can be used as sole electron acceptor for respiration (Reguera et al., 2005, 2007) but may not be sufficiently smooth or uniform to promote cell translocation. Hence, assay modifications may be required to better mimic the surfaces and conditions that Geobacter bacteria encounter in the environment and which may select for twitching motility phenotypes. Lastly, direct visualization of pilus retraction could provide valuable insights into the mechanical properties of Geobacter pili that allow them to function as retractile appendages while functioning as electronic conduits during respiration.
AS, BS, JH, and GR designed the experiments and analyzed the data. AS carried out the heterologous expression experiments, with assistance from AG, and phenotypically characterized the Geobacter mutants in microbial electrolysis cells and cultures with Fe(III) oxides. BS constructed the Pseudomonas strains used in the heterologous expression experiments and, with JH, the Geobacter mutant strains; BS also performed the biofilm assays in G. sulfurreducens and JH, the qRT-PCR analyses; together with GR they did comparative genomic and protein analyses. AS, BS, and GR co-wrote the paper. All authors discussed the results and commented on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by grants MCB-1021948 (from the National Science Foundation) and R01 ES017052-03 (from the National Institute of Environmental Health Sciences' Superfund program) to GR. We also acknowledge support from the National Institute of Food and Agriculture of the US Department of Agriculture through Michigan State's AgBioResearch to GR. We are thankful to Dr. Matthew C. Wolfgang for supplying strains and plasmids used in the P. aeruginosa studies and to Dr. Mary E. Lidstrom for providing us with the cre-lox antibiotic marker recycling system.
Alvarez-Martinez, C. E., and Christie, P. J. (2009). Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73, 775–808. doi: 10.1128/MMBR.00023-09
Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., et al. (eds.). (1995). Current Protocols in Molecular Biology. New York, NY: John Wiley & Sons, Inc.
Black, W. P., Xu, Q., and Yang, Z. (2006). Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol. Microbiol. 61, 447–456. doi: 10.1111/j.1365-2958.2006.05230.x
Burrows, L. L. (2012). Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu. Rev. Microbiol. 66, 493–520. doi: 10.1146/annurev-micro-092611-150055
Caccavo, F. Jr., Lonergan, D. J., Lovley, D. R., Davis, M., Stolz, J. F., and McInerney, M. J. (1994). Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl. Environ. Microbiol. 60, 3752–3759.
Caiazza, N. C., and O'Toole, G. A. (2004). SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J. Bacteriol. 186, 4476–4485. doi: 10.1128/JB.186.14.4476-4485.2004
Castric, P. (1995). pilO, a gene required for glycosylation of Pseudomonas aeruginosa 1244 pilin. Microbiology 141(Pt 5), 1247–1254. doi: 10.1099/13500872-141-5-1247
Chiang, P., and Burrows, L. L. (2003). Biofilm formation by hyperpiliated mutants of Pseudomonas aeruginosa. J. Bacteriol. 185, 2374–2378. doi: 10.1128/JB.185.7.2374-2378.2003
Chiang, P., Sampaleanu, L. M., Ayers, M., Pahuta, M., Howell, P. L., and Burrows, L. L. (2008). Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT and PilU. Microbiology 154(Pt 1), 114–126. doi: 10.1099/mic.0.2007/011320-0
Clausen, M., Jakovljevic, V., Sogaard-Andersen, L., and Maier, B. (2009). High-force generation is a conserved property of type IV pilus systems. J. Bacteriol. 191, 4633–4638. doi: 10.1128/JB.00396-09
Cologgi, D. L., Lampa-Pastirk, S., Speers, A. M., Kelly, S. D., and Reguera, G. (2011). Extracellular reduction of uranium via Geobacter conductive pili as a protective cellular mechanism. Proc. Natl. Acad. Sci. U.S.A. 108, 15248–15252. doi: 10.1073/pnas.1108616108
Cologgi, D. L., Speers, A. M., Bullard, B. A., Kelly, S. D., and Reguera, G. (2014). Enhanced uranium immobilization and reduction by Geobacter sulfurreducens biofilms. Appl. Environ. Microbiol. 80, 6638–6646. doi: 10.1128/AEM.02289-14
Coppi, M. V., Leang, C., Sandler, S. J., and Lovley, D. R. (2001). Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 67, 3180–3187. doi: 10.1128/AEM.67.7.3180-3187.2001
Devore, J. L. (2000). Probability and Statistics for Engineering and the Sciences. Pacific Grove, CA: Duxbury Press.
Deziel, E., Comeau, Y., and Villemur, R. (2001). Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183, 1195–1204. doi: 10.1128/JB.183.4.1195-1204.2001
Ding, Y. H., Hixson, K. K., Aklujkar, M. A., Lipton, M. S., Smith, R. D., Lovley, D. R., et al. (2008). Proteome of Geobacter sulfurreducens grown with Fe(III) oxide or Fe(III) citrate as the electron acceptor. Biochim. Biophys. Acta 1784, 1935–1941. doi: 10.1371/journal.pone.0030827
Feliciano, G. T., da Silva, A. J. R., Reguera, G., and Artacho, E. (2012). The molecular and electronic structure of the peptide subunit of Geobacter sulfurreducens conductive pili from first principles. J. Phys. Chem. A 116, 8023–8030. doi: 10.1021/jp302232p
Feliciano, G. T., Steidl, R. J., and Reguera, G. (2015). Structural and functional insights into the conductive pili of Geobacter sulfurreducens revealed in molecular dynamics simulations. Phys. Chem. Chem. Phys. 17, 22217–22226. doi: 10.1039/c5cp03432a
Furste, J. P., Pansegrau, W., Frank, R., Blocker, H., Scholz, P., Bagdasarian, M., et al. (1986). Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48, 119–131. doi: 10.1016/0378-1119(86)90358-6
Heydorn, A., Nielsen, A. T., Hentzer, M., Sternberg, C., Givskov, M., Ersboll, B. K., et al. (2000). Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146(Pt 10), 2395–2407. doi: 10.1099/00221287-146-10-2395
Holmes, D. E., Nevin, K. P., O'Neil, R. A., Ward, J. E., Adams, L. A., Woodard, T. L., et al. (2005). Potential for quantifying expression of the Geobacteraceae citrate synthase gene to assess the activity of Geobacteraceae in the subsurface and on current-harvesting electrodes. Appl. Environ. Microbiol. 71, 6870–6877. doi: 10.1128/AEM.71.11.6870-6877.2005
Jakovljevic, V., Leonardy, S., Hoppert, M., and Søgaard-Andersen, L. (2008). PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J. Bacteriol. 190, 2411–2421. doi: 10.1128/JB.01793-07
Jones, C. J., Utada, A., Davis, K. R., Thongsomboon, W., Zamorano Sanchez, D., Banakar, V., et al. (2015). C-di-GMP regulates motile to sessile transition by modulating MshA pili biogenesis and near-surface motility behavior in Vibrio cholerae. PLoS Pathog. 11:e1005068. doi: 10.1371/journal.ppat.1005068
Juarez, K., Kim, B. C., Nevin, K., Olvera, L., Reguera, G., Lovley, D. R., et al. (2009). PilR, a transcriptional regulator for pilin and other genes required for Fe(III) reduction in Geobacter sulfurreducens. J. Mol. Microbiol. Biotechnol. 16, 146–158. doi: 10.1159/000115849
Kagami, Y., Ratliff, M., Surber, M., Martinez, A., and Nunn, D. N. (1998). Type II protein secretion by Pseudomonas aeruginosa: genetic suppression of a conditional mutation in the pilin-like component XcpT by the cytoplasmic component XcpR. Mol. Microbiol. 27, 221–233. doi: 10.1046/j.1365-2958.1998.00679.x
Kim, B. C., Leang, C., Ding, Y. H., Glaven, R. H., Coppi, M. V., and Lovley, D. R. (2005). OmcF, a putative c-type monoheme outer membrane cytochrome required for the expression of other outer membrane cytochromes in Geobacter sulfurreducens. J. Bacteriol. 187, 4505–4513. doi: 10.1128/JB.187.13.4505-4513.2005
Kim, B. C., Postier, B. L., Didonato, R. J., Chaudhuri, S. K., Nevin, K. P., and Lovley, D. R. (2008). Insights into genes involved in electricity generation in Geobacter sulfurreducens via whole genome microarray analysis of the OmcF-deficient mutant. Bioelectrochemistry 73, 70–75. doi: 10.1016/j.bioelechem.2008.04.023
Kim, B. C., Qian, X., Leang, C., Coppi, M. V., and Lovley, D. R. (2006). Two putative c-type multiheme cytochromes required for the expression of OmcB, an outer membrane protein essential for optimal Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol. 188, 3138–3142. doi: 10.1128/JB.188.8.3138-3142.2006
Kolenbrander, P. E., Palmer Jr, R. J., Periasamy, S., and Jakubovics, N. S. (2010). Oral multispecies biofilm development and the key role of cell–cell distance. Nat. Rev. Microbiol. 8, 471–480. doi: 10.1038/nrmicro2381
Kurre, R., Höne, A., Clausen, M., Meel, C., and Maier, B. (2012). PilT2 enhances the speed of gonococcal type IV pilus retraction and of twitching motility. Mol. Microbiol. 86, 857–865. doi: 10.1111/mmi.12022
Lampa-Pastirk, S., Veazey, J. P., Walsh, K. A., Steidl, R. J., Tessmer, S. H., and Reguera, G. (2016). Thermally activated charge transport in microbial protein nanowires. Sci. Rep. 6:23517. doi: 10.1038/srep23517
Leang, C., Adams, L. A., Chin, K. J., Nevin, K. P., Methe, B. A., Webster, J., et al. (2005). Adaptation to disruption of the electron transfer pathway for Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol. 187, 5918–5926. doi: 10.1128/JB.187.17.5918-5926.2005
Lovley, D. R., and Phillips, E. J. P. (1986). Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Appl. Environ. Microbiol. 51, 683–689.
Lovley, D. R., and Woodward, J. C. (1996). Mechanisms for chelator stimulation of microbial Fe(III)-oxide reduction. Chem. Geol. 132, 19–24. doi: 10.1016/S0009-2541(96)00037-X
Lovley, D. R., Woodward, J. C., and Chapelle, F. H. (1994). Stimulated anoxic biodegradation of aromatic hydrocarbons using Fe(III) ligands. Nature 370, 128–131.
Maier, B., Koomey, M., and Sheetz, M. P. (2004). A force-dependent switch reverses type IV pilus retraction. Proc. Natl. Acad. Sci. U.S.A. 101, 10961–10966. doi: 10.1073/pnas.0402305101
Maier, B., Potter, L., So, M., Long, C. D., Seifert, H. S., and Sheetz, M. P. (2002). Single pilus motor forces exceed 100 pN. Proc. Natl. Acad. Sci. U.S.A. 99, 16012–16017. doi: 10.1073/pnas.242523299
Mallona, I., Weiss, J., and Egea-Cortines, M. (2011). pcrEfficiency: a Web tool for PCR amplification efficiency prediction. BMC Bioinformatics 12:404. doi: 10.1186/1471-2105-12-404
Manzella, M. P., Reguera, G., and Kashefi, K. (2013). Extracellular electron transfer to Fe(III) oxides by the hyperthermophilic archaeon Geoglobus ahangari via a direct contact mechanism. Appl. Environ. Microbiol. 79, 4694–4700. doi: 10.1128/AEM.01566-13
Marsili, E., Sun, J., and Bond, D. R. (2010). Voltammetry and growth physiology of Geobacter sulfurreducens biofilms as a function of growth stage and imposed electrode potential. Electroanalysis 22, 865–874. doi: 10.1002/elan.200800007
Marx, C. J., and Lidstrom, M. E. (2002). Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques 33, 1062–1067. Available online at: http://www.biotechniques.com/multimedia/archive/00010/02335rr01_10092a.pdf
Mehta, T., Coppi, M. V., Childers, S. E., and Lovley, D. R. (2005). Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl. Environ. Microbiol. 71, 8634–8641. doi: 10.1128/AEM.71.12.8634-8641.2005
Merritt, J. H., Kadouri, D. E., and O'Toole, G. A. (2005). Growing and analyzing static biofilms. Curr. Protoc. Microbiol. Chapter 1, Unit 1B 1. doi: 10.1002/9780471729259.mc01b01s00
Merz, A. J., So, M., and Sheetz, M. P. (2000). Pilus retraction powers bacterial twitching motility. Nature 407, 98–102. doi: 10.1038/35024105
Nakasugi, K., Alexova, R., Svenson, C. J., and Neilan, B. A. (2007). Functional analysis of PilT from the toxic cyanobacterium Microcystis aeruginosa PCC 7806. J. Bacteriol. 189, 1689–1697. doi: 10.1128/JB.01640-06
Nevin, K. P., Kim, B. C., Glaven, R. H., Johnson, J. P., Woodard, T. L., Methe, B. A., et al. (2009). Anode biofilm transcriptomics reveals outer surface components essential for high density current production in Geobacter sulfurreducens fuel cells. PLoS ONE 4:e5628. doi: 10.1371/journal.pone.0005628
Park, H. S., Wolfgang, M., and Koomey, M. (2002). Modification of type IV pilus-associated epithelial cell adherence and multicellular behavior by the PilU protein of Neisseria gonorrhoeae. Infect. Immun. 70, 3891–3903. doi: 10.1128/IAI.70.7.3891-3903.2002
Reguera, G., McCarthy, K. D., Mehta, T., Nicoll, J. S., Tuominen, M. T., and Lovley, D. R. (2005). Extracellular electron transfer via microbial nanowires. Nature 435, 1098–1101. doi: 10.1038/nature03661
Reguera, G., Nevin, K. P., Nicoll, J. S., Covalla, S. F., Woodard, T. L., and Lovley, D. R. (2006). Biofilm and nanowire production lead to increased current in microbial fuel cells. Appl. Environ. Microbiol. 72, 7345–7348. doi: 10.1128/AEM.01444-06
Reguera, G., Pollina, R. B., Nicoll, J. S., and Lovley, D. R. (2007). Possible nonconductive role of Geobacter sulfurreducens pilus nanowires in biofilm formation. J. Bacteriol. 189, 2125–2127. doi: 10.1128/JB.01284-06
Rollefson, J. B., Stephen, C. S., Tien, M., and Bond, D. R. (2011). Identification of an extracellular polysaccharide network essential for cytochrome anchoring and biofilm formation in Geobacter sulfurreducens. J. Bacteriol. 193, 1023–1033. doi: 10.1128/JB.01092-10
Savvides, S. N. (2007). Secretion superfamily ATPases swing big. Structure 15, 255–257. doi: 10.1016/j.str.2007.02.003
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Semmler, A. B., Whitchurch, C. B., and Mattick, J. S. (1999). A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 145, 2863–2873. doi: 10.1099/00221287-145-10-2863
Sexton, J. A., Pinkner, J. S., Roth, R., Heuser, J. E., Hultgren, S. J., and Vogel, J. P. (2004). The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J. Bacteriol. 186, 1658–1666. doi: 10.1128/JB.186.6.1658-1666.2004
Skerker, J. M., and Berg, H. C. (2001). Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. U.S.A. 98, 6901–6904. doi: 10.1073/pnas.121171698
Speers, A. M., and Reguera, G. (2012). Electron donors supporting growth and electroactivity of Geobacter sulfurreducens anode biofilms. Appl. Environ. Microbiol. 78, 437–444. doi: 10.1128/AEM.06782-11
Steidl, R., Lampa-Pastirk, S., and Reguera, G. (2016). Mechanistic stratification in electroactive biofilms of Geobacter sulfurreducens mediated by pilus nanowires. Nat. Commun. 7:12217. doi: 10.1038/ncomms12217
Stepanovic, S., Vukovic, D., Dakic, I., Savic, B., and Svabic-Vlahovic, M. (2000). A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 40, 175–179. doi: 10.1016/S0167-7012(00)00122-6
Stookey, L. L. (1970). Ferrozine: a new spectrophotometric reagent for iron. Anal. Chem. 42, 779–781. doi: 10.1021/ac60289a016
Strycharz-Glaven, S. M., Snider, R. M., Guiseppi-Elie, A., and Tender, L. M. (2011). On the electrical conductivity of microbial nanowires and biofilms. Energy Environ. Sci. 4, 4366–4379. doi: 10.1039/c1ee01753e
Sundin, C., Wolfgang, M. C., Lory, S., Forsberg, A., and Frithz-Lindsten, E. (2002). Type IV pili are not specifically required for contact dependent translocation of exoenzymes by Pseudomonas aeruginosa. Microb. Pathog. 33, 265–277. doi: 10.15252/embj.201489096
Takeya, K., and Amako, K. (1966). A rod-shaped Pseudomonas phage. Virology 28, 163–165. doi: 10.1016/0042-6822(66)90317-5
Takhar, H. K., Kemp, K., Kim, M., Howell, P. L., and Burrows, L. L. (2013). The platform protein is essential for type IV pilus biogenesis. J. Biol. Chem. 288, 9721–9728. doi: 10.1074/jbc.M113.453506
Thomas, P. E., Ryan, D., and Levin, W. (1976). An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75, 168–176. doi: 10.1016/0003-2697(76)90067-1
Vignon, G., Kohler, R., Larquet, E., Giroux, S., Prevost, M. C., Roux, P., et al. (2003). Type IV-like pili formed by the type II secreton: specificity, composition, bundling, polar localization, and surface presentation of peptides. J. Bacteriol. 185, 3416–3428. doi: 10.1128/JB.185.11.3416-3428.2003
Walker, J. E., Saraste, M., Runswick, M. J., and Gay, N. J. (1982). Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951.
Wall, D., and Kaiser, D. (1999). Type IV pili and cell motility. Mol. Microbiol. 32, 1–10. doi: 10.1046/j.1365-2958.1999.01339.x
Whitchurch, C. B., Hobbs, M., Livingston, S. P., Krishnapillai, V., and Mattick, J. S. (1991). Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialized protein export system widespread in eubacteria. Gene 101, 33–44. doi: 10.1016/0378-1119(91)90221-V
Whitchurch, C. B., and Mattick, J. S. (1994). Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa. Mol. Microbiol. 13, 1079–1091. doi: 10.1111/j.1365-2958.1994.tb00499.x
Wolfgang, M., Lauer, P., Park, H. S., Brossay, L., Hebert, J., and Koomey, M. (1998). PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29, 321–330. doi: 10.1046/j.1365-2958.1998.00935.x
Keywords: type IV pili, pilus retraction, metal reduction, electroactive biofilms, pilus nanowires
Citation: Speers AM, Schindler BD, Hwang J, Genc A and Reguera G (2016) Genetic Identification of a PilT Motor in Geobacter sulfurreducens Reveals a Role for Pilus Retraction in Extracellular Electron Transfer. Front. Microbiol. 7:1578. doi: 10.3389/fmicb.2016.01578
Received: 01 July 2016; Accepted: 21 September 2016;
Published: 17 October 2016.
Edited by:
José Eduardo González-Pastor, Centro de Astrobiología (CSIC), SpainCopyright © 2016 Speers, Schindler, Hwang, Genc and Reguera. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gemma Reguera, cmVndWVyYUBtc3UuZWR1
†Present Address: Bryan D. Schindler, NSF International, Ann Arbor, MI, USA;
Jihwan Hwang, Department of Microbiology, Pusan National University, Busan, South Korea;
Aycin Genc, Qiagen GmbH, Hilden, Germany
‡These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.