- State Key Laboratory of Integrated Management of Pest Insects and Rodents, Institute of Zoology (CAS), Beijing, China
RNA silencing is a conserved mechanism that utilizes small RNAs (sRNAs) to direct the regulation of gene expression at the transcriptional or post-transcriptional level. Plants utilizing RNA silencing machinery to defend pathogen infection was first identified in plant–virus interaction and later was observed in distinct plant–pathogen interactions. RNA silencing is not only responsible for suppressing RNA accumulation and movement of virus and viroid, but also facilitates plant immune responses against bacterial, oomycete, and fungal infection. Interestingly, even the same plant sRNA can perform different roles when encounters with different pathogens. On the other side, pathogens counteract by generating sRNAs that directly regulate pathogen gene expression to increase virulence or target host genes to facilitate pathogen infection. Here, we summarize the current knowledge of the characterization and biogenesis of host- and pathogen-derived sRNAs, as well as the different RNA silencing machineries that plants utilize to defend against different pathogens. The functions of these sRNAs in defense and counter-defense and their mechanisms for regulation during different plant–pathogen interactions are also discussed.
Introduction
Small RNAs (sRNAs) are 20–30 nucleotide (nt)-long non-coding RNA molecules, which are widely present in eukaryotic organisms. It is well established that sRNAs are involved in the regulation of gene expression through a process generally termed RNA silencing. RNA silencing contributes to almost all eukaryotic cellular processes, including preventing the invasion of viruses or transgenes, inhibiting the movement of transposable elements, and regulating developmental and physiological processes (Itaya et al., 2008; Wang et al., 2011a; Castel and Martienssen, 2013; Bond and Baulcombe, 2014; Holoch and Moazed, 2015).
Plant sRNAs are divided into two major classes: microRNAs (miRNAs) and small interfering RNAs (siRNAs). Most miRNAs are 21–24 nt in length and derived from RNAs with imperfectly base-paired hairpin structures (Chen, 2009), while siRNAs are generated from perfectly complementary long dsRNAs (Xie et al., 2004). Plant siRNAs are grouped into four subclasses: trans-acting siRNAs (ta-siRNAs), heterochromatic siRNAs (hc-siRNAs), natural antisense transcript-derived siRNAs (nat-siRNAs), and long siRNAs (lsiRNAs). Proteins, such as Dicer-like proteins (DCLs), HYPONASTIC LEAVES 1 (HYL1), HUA ENHANCER 1 (HEN1), and Serrate (SE) are involved in sRNA biogenesis pathways (Katiyar-Agarwal and Jin, 2010; Rogers and Chen, 2013; Holoch and Moazed, 2015). Some siRNAs require RNA-dependent RNA polymerases (RDRs) and suppressor of gene silencing 3 (SGS3) for amplification (Sijen et al., 2007). After processing and amplification, sRNA duplexes are sorted and loaded into Argonaute (AGO) proteins, and the passenger strand is discarded. In animals, the passenger strand is removed by slicing or unwinding in an ATP-dependent reaction (Liu and Paroo, 2010). In plant, however, the removing mechanism of the passenger strand is still unclear. Matured RNA-induced silencing complexes (RISCs) with the guide strands anneal to its complementary sequence and regulate gene expression at transcriptional and post-transcriptional levels through DNA methylation, chromatin modification, mRNA slicing, mRNA degradation, or translational inhibition (Ghildiyal and Zamore, 2009; Zhang X. et al., 2011).
One of the important functions of RNA silencing is to suppress the infection of pathogens. The RNA silencing machinery of host plants can directly target the genomic RNA and transcripts of viruses, viroids, and virus satellites to suppress their RNA accumulation. However, plants are also susceptible to other pathogens, such as bacteria, fungi, oomycetes, and nemotodes, which unlike viruses, do not replicate or expose their genome in host cells during any part of the infection process. To defeat these pathogens, plants have evolved complicated defense systems, including PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI) (Jones and Dangl, 2006). When successful pathogens evolve new effectors to suppress the host ETI response, plants respond by evolving novel resistance (R) proteins to recognize the effectors and trigger ETI responses in this endless arms race.
Both miRNAs and siRNAs contribute to PTI and ETI by fine-tuning plant hormones and/or silencing the genes involved in pathogen virulence (Navarro et al., 2006; Zhang W. et al., 2011). While host sRNAs play important roles in pathogen resistance, pathogens also encode sRNAs to manipulate host defense responses, as well as mediate pathogen virulence. sRNAs in fungi, oomycetes, and bacteria have been shown to function in promoting pathogen virulence. In fungi and oomycetes, sRNAs are mostly generated from transposable element (TE) regions (Nunes et al., 2011; Vetukuri et al., 2012; Weiberg et al., 2013). Key proteins in the RNA silencing machineries, such as DCLs, AGOs, and RDRs, are also present in these eukaryotic plant pathogens and are involved in the biogenesis and function of some sRNAs (Murata et al., 2007; Vetukuri et al., 2011). However, the biogenesis of sRNAs in fungi is more diverse than in plants. Both DCL-dependent and DCL-independent siRNA biogenesis mechanisms were identified in fungi Neurospora crassa (Lee et al., 2010). Furthermore, at least four different mechanisms that use distinct combinations of proteins, including Dicers, QDE-2, the exonuclease QIP, and an RNAse III domain-containing protein MRPL3, were proposed to be involved in the biogenesis of miRNA-like small RNAs (milRNAs) in N. crassa (Lee et al., 2010). Bacterial non-coding sRNAs are different from sRNAs in eukaryotes (Weiberg et al., 2014). They functionally associate with distinct RNA-binding protein complexes, including the clustered regularly interspaced short palindromic repeat (CRISPR)-associated (Cas) system (CRISPR-Cas) (Fahlgren et al., 2007; Li et al., 2010; Zhang X. et al., 2011; Wiedenheft et al., 2012), the RNA chaperone Hfq (Schu et al., 2015), and CsrA/RsmA (Schu et al., 2015), and regulate the expression of target mRNA through short and impacted base-pair (10–25 nt). Meanwhile, viroids, the smallest known pathogen, which does not code for proteins, have been proposed to encode specific sRNAs that target host genes and result in disease symptoms (Wang et al., 2004). Furthermore, some virus-derived siRNAs (vsiRNAs), which are generated to target viral RNAs, may target host genes, and subsequently mediate the viral disease symptom. Whether viral fitness would be increased by vsiRNAs remains unknown (Qi et al., 2009; Xia et al., 2014). Viruses, oomycetes, and bacteria have RNA silencing suppressors and other effectors that directly inhibit host sRNAs, while some fungi that localize in the intercellular space of plants deliver fungal sRNAs as effectors into plant cells to inhibit the plant PTI response. In this review we will discuss our current understanding of sRNAs in plants and plant pathogens, focusing on their functional differences in plant–pathogen interactions.
Plants Encode sRNAs That Fine-Tune Plant Hormones and Antimicrobial Activity to Defend Against Pathogen Attack
Although a potent immune system is necessary for plants to survive pathogen infections, it also deprives the limited resources available for plant growth and development. Although more studies need to be done, a constitutively active immune system in plants may result in reduced growth and seed yield (Tian et al., 2003; Walters and Heil, 2007). Thus, plant immune responses must be tightly regulated, and one strategy is to generate endogenous sRNAs that silence specific genes involved in plant hormone production or antimicrobial activity. Upon infection, the biogenesis and/or the accumulation of these sRNAs are regulated, which subsequently fine-tune plant hormone levels and the expression of genes involved in plant resistance (Figures 1A and 2A).
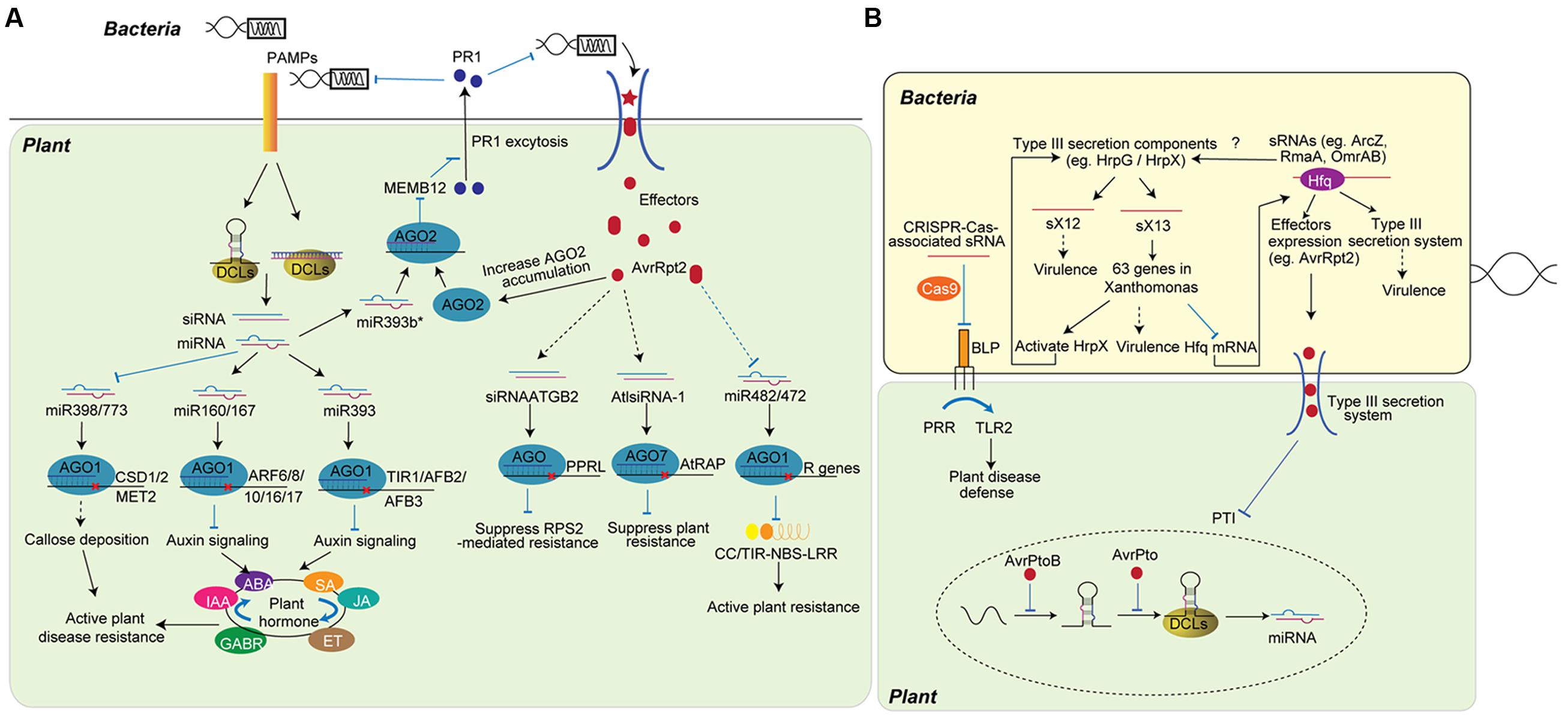
FIGURE 1. Role of sRNAs in plant-bacteria interaction. (A) Plant sRNAs defend bacteria attack by fine-tuning plant hormone and disease resistance activity. Upon the infection, plants detect PAMPs and modulate the accumulation of miRNA and siRNA. miRNAs, such as miR393, miR160, and miR167, regulate disease resistance by fine-tuning plant hormone networks, while other miRNAs regulate the activation of R protein (miR482/miR472) or the slicing of genes inhibiting plant immunity (miR398/miR773). miR393b*, the pairing strand of miR393, increases plant immunity by promoting the exocytosis of antimicrobial protein. siRNAs, including siRNAATGB2 and AtlsiRNA-1, are induced by bacteria effectors and enhance ETI by silencing genes that negatively regulate plant disease resistance. (B) Bacteria non-coding sRNAs (ncRNAs) regulate bacteria gene expression to improve virulence. Through imperfect base-pairing of short regions (10- to 25-nt), bacteria ncRNAs bind to target mRNAs and guide the suppression of genes or proteins that are involved in virulence. ncRNAs can regulate bacteria virulence by inhibiting proteins that trigger host defense (BLP) or affecting the expression of effectors (AvrRpt2). Bacteria effectors translocate into host plant cell and inhibit the regulation of plant sRNA (bottom). The AvrPtoB effector specifically represses the accumulation of miR393 at the transcriptional level, while AvrPto reduces the processing of miR393.
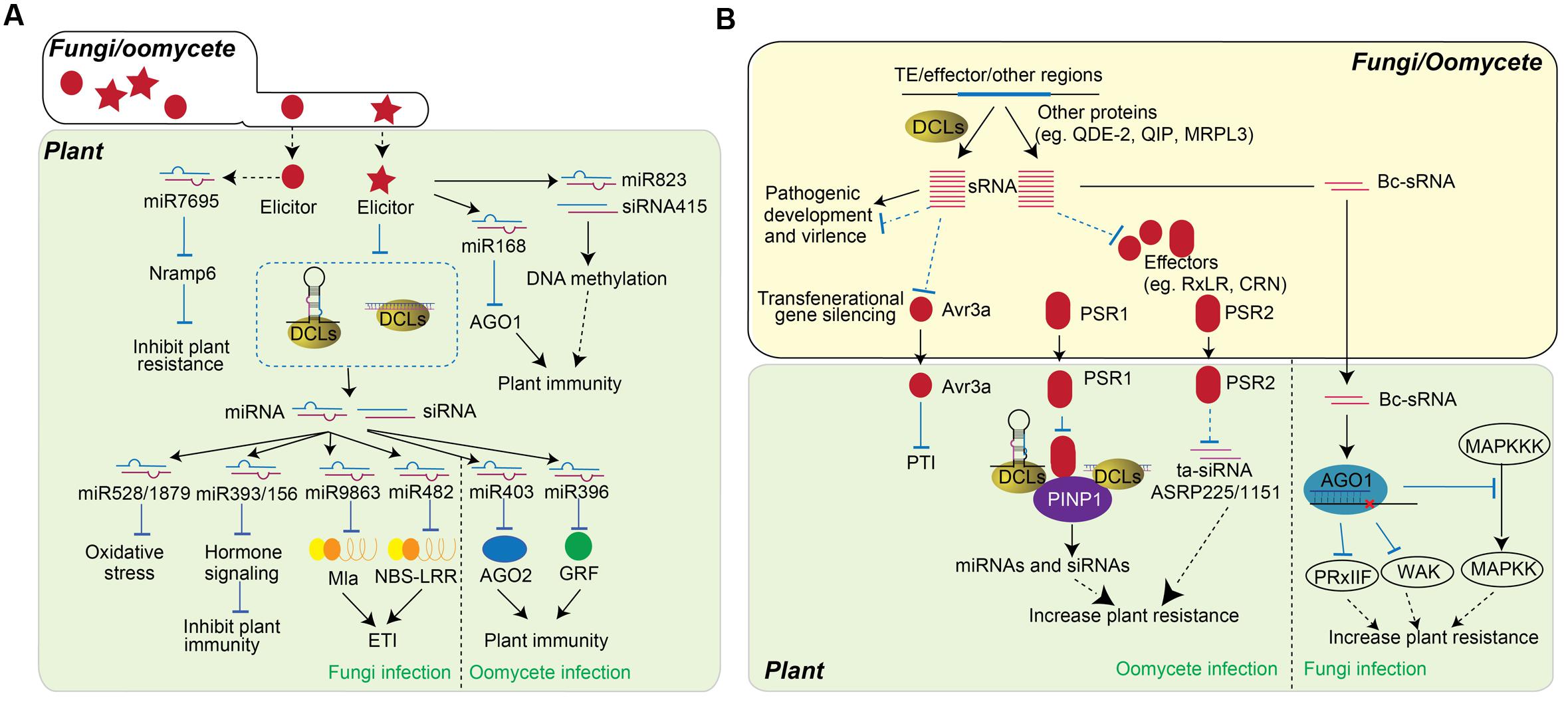
FIGURE 2. Role of sRNAs in plant-fungi/oomycete interaction. (A) Plant sRNAs regulate PTI and ETI in response to fungi or oomycete infection. The infection of fungi (left) and oomycete (right) alters the accumulation of miRNAs, by which changes the expression of genes contribute to plant resistance. Fungi elicitorsor fungi infections triggers the accumulation of some sRNAs, such as miR7695, miR168, miR823 and siRNA415, while miR528, miR1879, miR9863, and miR482 are down-regulated to improve plant resistance. The accumulations of miR403 and miR396 are down-regulated upon oomycete infection. (B) Schematic representation of the function of fungi/oomycete sRNAs in pathogen virulence. sRNAs encoded by fungi and oomycetes are usually generated from TE region, effector coding region, and other regions. These sRNA can be either DCL-dependent or DCL-independent and are involved in the regulation of pathogen development and virulence. In particular, sRNA regulate the expression of effectors, which further influence the accumulation of host miRNA and siRNA. sRNAs generated from Avr3a region of oomycete can transgenerationally change the pathogen virulence. The PSR1 and PSR2 effectors of oomycete are secreted into plant cells and alter host RNA silencing machineries as RNA silencing suppressor to decrease host immunity. On the other hand, fungi sRNAs, Bc-sRNAs, translocate into host cell and utilize plant RNA silencing component to reduce the expression of host immune genes and facilitate fungi infection.
As far as we know, bacteria, fungi, and oomycetes infect plants without direct genome and RNA interaction with the host RNA silencing machineries. To these pathogens, fine-tuning of the plant immune system is critical for host resistance. Various plant miRNAs and siRNAs play critical roles in anti-bacterial resistance (Figure 1A). miR393 is the first miRNA shown to function in anti-bacterial defense. The accumulation of miR393 is up-regulated upon the treatment of the conserved N-terminal part of flagellin, flg22, or the infection of bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000. miR393 enhances host resistance to Pst DC3000 by negatively regulating the expression of F-box auxin receptors, including Transport Inhibitor Response 1 (TIR1), Auxin signaling F-Box proteins 2 (AFB2), and 3 (AFB3) (Navarro et al., 2006). Further studies in rice determined that miR393 is a bona fide stress-related miRNA that is widely involved in plant resistance to other pathogens and abiotic stresses, such as salt and drought (Bian et al., 2012; Xia et al., 2012; Campo et al., 2013). In addition to miR393, miR160 and miR167 also target Auxin response factor (ARF) family transcription factors and are induced by infection with Pst DC3000 hrcC-, a strain with a mutated type III secretion system, to improve plant antibacterial defense (Fahlgren et al., 2007). Further studies uncovered that miR160a and 15 other miRNAs are induced upon flg22 treatment. On the other hand, miR398b, miR773, and 9 other miRNAs are down-regulated upon flg22 treatment (Li et al., 2010). Over-expression of miR398b and miR773 attenuates PTI by repressing flg22 or bacteria-induced callose deposition, indicating miRNAs play important roles in disease resistance. However, the over-expression of miR160, which increases PAMP-induced callose deposition, did not significantly change the basal defense of plant to Pst DC3000 bacteria, suggesting a complicate miRNA regulatory network in plant disease responses (Li et al., 2010). Furthermore, miR393b*, the complementary strand of miR393, is loaded into AGO2 and regulates plant resistance by suppressing the expression of MEMB12. MEMB12 is a Golgi-localized SNARE protein, and its down-regulation leads to increased exocytosis of PR1, which subsequently enhances plant resistance (Zhang X. et al., 2011). Thus, miR393 and miR393b*, two sRNAs generated from a same sRNA duplex, bind AGO1 and AGO2 respectively to regulate distinct hormone pathways and coordinately increase plant immunity (Navarro et al., 2006; Zhang X. et al., 2011). Another interesting finding about miRNA in bacterial defending is that one miRNA can target both negative and positive regulators of immunity depending on the timing and the amplitude of defense responses. miR863-3p improves plant defense by silencing a typical receptor-like pseudokinase1 (ARLPK1) and ARLPK2 during early infection, and negatively regulates defense by silencing SE gene during later infection (Niu et al., 2016).
In response to sRNA-mediated PTI, successful pathogens deliver effectors into host cells to interfere with PTI. For detailed information about the role of pathogen effectors, several reviews are available (Dou and Zhou, 2012; Feng and Zhou, 2012). To counteract pathogen effectors, plants induce ETI. As ETI is more robust and usually triggers a hypersensitive response (HR), the ETI reaction is strictly regulated by siRNAs and miRNAs. siRNA nat-siRNAATGB2, which is specifically induced by Pst DC3000 effector protein AvrRpt2, enhances ETI by suppressing the expression of pentatricopeptide repeats (PPR) protein-like gene (PPRL) and preventing the negative effect of PPRL on the resistance pathway mediated by RPS2, a resistance gene that specifically recognizes effector AvrRpt2 (Katiyar-Agarwal et al., 2006). AtlsiRNA-1, which is also induced by AvrRpt2, improves disease resistance by silencing the expression of AtRAP, a negative regulator of plant disease resistance (Katiyar-Agarwal et al., 2007). In addition to these sRNAs, genome-wide sRNA deep sequencing indicates that the accumulation of more than 20 miRNAs and various nat-siRNAs are significantly altered upon ETI (Zhang M. et al., 2011; Zhang et al., 2012). Some targets of these miRNAs are key genes contributing to the hormone biosynthesis and signaling pathways involved in plant resistance. A TE-siRNA, TE-siR815, generated from the intron of WRKY45-1, represses ST1 and subsequently attenuates WRKY45-mediated resistance to Xanthomonas oryzae pv. Oryzae, which results in the opposite functions of WRKY45-1 and WRKY45-2 (Zhang et al., 2016).
Host sRNAs contribute to ETI not only by regulating the expression of genes involved in plant resistance but also by directly regulating the activation of R proteins. For instance, RPP4 and SNC1, two R genes located in the RPP5 locus, are involved in disease resistance against bacterial and fungal pathogens (Baldrich et al., 2014, 2015). A study demonstrated that these R genes are negatively regulated by RNA silencing. The SNC1 gene was up-regulated in dcl4 and ago1 mutants (Yi and Richards, 2007). When a pathogen interferes with host RNA silencing, it may subsequently disturb the sRNA-mediated inhibition of R genes and activate the function of these R proteins. However, sRNAs complementary to the SNC1 region are not increased in dcl4 and ago1 mutants, suggesting that other sRNAs may contribute to the up-regulation of SNC1 in these mutants (Yi and Richards, 2007). The accumulation of miR482 is decreased in plants infected with Pst DC3000 but not Pst DC3000 hrcC- (Shivaprasad et al., 2012). Further study predicted that miR482 can target mRNAs of 58 coiled-coil, nucleotide-binding site, leucine rich repeat proteins (CC-NBS-LRR). Meanwhile, the production of secondary siRNA, caused by the targeting in a RDR6 dependent manner, may target other mRNAs of a defense-related protein. Thus, upon the infection of virus or bacteria, the accumulation of miR482 is decreased to suppress the miR482-mediated silencing cascade, and subsequently increase the expression of defense-related mRNAs (Shivaprasad et al., 2012). miR482 in cotton was also reported to target more than 10% of NBS-LRR genes and triggers the production of secondary siRNAs. Infection with fungal pathogen Verticillium dahliae down-regulates miR482 accumulation and increases NBS-LRR gene expression in cotton (Zhu et al., 2013). Interestingly, Arabidopsis miR472, which targets RPS5 CC-NBS-LRR genes, modulates both PTI and ETI pathways. Mutation in miR472 results in increased resistance to both Pst DC3000 and Pst DC3000 avrPphB (Boccara et al., 2014).
Host sRNAs also regulate PTI and ETI upon various fungal and oomycete attack (Figure 2A). Magnaportbe oryzae is a rice blast fungus that causes rice blast disease. The accumulation of rice miRNA528/miR1879 is down-regulated by treatment with a M. oryzae elicitor, resulting in up-regulation of their target genes that control oxidative stress (Baldrich et al., 2015). Meanwhile, the accumulation of miR393b/miR156 are also negatively altered upon the elicitor treatment of M. oryzae on rice (Campo et al., 2013). On the other hand, a novel DCL4-processed miRNA, osa-miR7695, was identified in rice to target an alternatively spliced transcript of Nramp6 (Natural resistance associated macrophage protein 6) gene and its overexpression results in enhanced resistance to M. oryzae infection (Campo et al., 2013). Further study identified a group of small RNAs, including miR156, miR165/166, miR170, and miR172 in Arabidopsis that were regulated by elicitors of the fungus Fusarium oxysporum. Particularly, miR168, which is known to regulate plant abiotic responses via control of AGO1, was transcriptionally activated, and its upregulation negatively correlated with AGO1 transcripts (Baldrich et al., 2014). In addition, miR823 and siRNA415, both of which are involved in RNA-directed DNA methylation (RdDM), were also found to be induced by fungal elicitors (Baldrich et al., 2014). The regulation of these sRNAs by fungal elicitors suggests their functions in PTI. Puccinia graminis f.sp. tritici (Bgt) is a powdery mildew fungus that causes devastating disease in wheat, barley, and other plants. Eight different miRNAs, miR159, miR164, miR167, miR171, miR444, miR408, miR1129, and miR1138, that regulate three different defense response processes are significantly induced at the early, but not the late, stage of Bgt infection. Thus, these miRNAs may play a key role in HR at the onset of disease (Gupta et al., 2012). The roles of sRNA in plant immune response were further demonstrated in powdery mildew fungus Blumeria graminis f. sp. hordei (Bgh) (Liu et al., 2014). Mildew resistance locus a (Mla), encoding a group of CC-NBS-LRR proteins that respond to Bgh, are targeted by the miRNA family miR9863. miR9863 was shown to guide the cleavage of Mla1 transcripts in barley, and down-regulate the accumulation of MLA1 protein in the Nicotiana benthamiana expression system. In addition, miR9863 can trigger the biogenesis of 21-nt phased siRNAs (phasiRNAs) and further repress the expression of Mla1. Over-expression of miR9863 specifically attenuates Mla1-mediated cell death and disease resistance (Liu et al., 2014). miR482 in potato can also target NBS-LRR genes. V. dahliae infection down-regulates the accumulation of miR482, which in turn increases NBS-LRR gene expression (Yang et al., 2015). The silencing of NBS-LRR genes by these specific 22-nt miRNAs, and their activation after miRNA down-regulation upon bacteria, fungal, or viral treatments, have been widely studied in different plants (He et al., 2008; Xin et al., 2010; Zhai et al., 2011; Li et al., 2012; Shivaprasad et al., 2012; Zhu et al., 2013; Boccara et al., 2014; Liu et al., 2014; Fei et al., 2015). Phytophthora sojae is a notorious oomycete that infects soybean root and stem. P. sojae infection down-regulates the expression of miR403, a miRNA that targets AGO2, a positive regulator of plant immunity (Guo et al., 2011). Similarly, the accumulation of sRNAs and their targets are also differently regulated in susceptible and resistance soybean cultivars. The expression of miR396 in Solanaceae is down-regulated upon infection with another oomycete, Phytophthora infestans. Over-expression of miR396 resulted in the down-regulation of GRF targets and increased susceptibility to P. infestans (Chen et al., 2015). It is clear that plant sRNAs play a critical role in regulating the expression of genes involved in plant defense and immunity. However, each sRNA has distinct function in plant immune response, and the accumulation and the function of sRNAs are pathogen-dependent. Therefore, in order to obtain the systematic role of RNA silencing in plant resistance, the function of more sRNAs needs to be further investigated.
Plants Utilize sRNAs to Defend Against Pathogen by Directly Targeting on Viral and Viroid Genomes and Transcripts
Viruses and viroids infect plants by replicating their genomes inside the host cells. Post-transcriptional gene silencing (PTGS) was first identified in both transgenes processing and Potato virus X (PVX) infection. sRNAs complementary to the sense transcript of the transgene and the positive strand of PVX were discovered, indicating that sRNAs participate in PTGS transgene silencing and viral defense (Hamilton and Baulcombe, 1999). Further studies revealed that the replication of viruses and viroids, and the folding of their RNA genomes and transcripts, produce dsRNAs that recruit RNA silencing machinery (Ding, 2009).
Viruses contain either single-stranded RNA (ssRNA), double-stranded RNA (dsRNA), ssDNA, or dsDNA genomes (Ding and Voinnet, 2007). During the replication of an ssRNA viral genome, a complementary strand of RNA is synthesized, which forms a long dsRNA with the original viral genome. The dsRNA replicative intermediate forms of ssRNA viruses and the dsRNA genomes of dsRNA viruses can be targeted by host RNA silencing machineries (Figure 3A). Nearly equal amounts of positive and negative strand vsiRNAs without positional bias were derived from Cucumber yellows closterovirus (CuYV), Turnip mosaic potyvirus (TuMV), CMV, Watermelon mosaic virus (WMV), PVX, and Tomato yellow leaf curl virus (TYLCV), all positive ssRNA viruses from different families (Yoo et al., 2004; Ho et al., 2006; Donaire et al., 2009; Wang et al., 2010). It was also shown that vsiRNAs were nearly equally derived from the positive and negative genome of Rice stripe virus (RSV), an ambisense virus with four genomic ssRNAs (Yan et al., 2010). However, more than 80% of vsiRNAs derived from Cymbidium ringspot virus (CymRSV) are generated from the positive strand (Molnar et al., 2005). Similar phenomena are also observed in plants infected with other ssRNA viruses such as TCV, Tobacco mosaic virus (TMV), Tobacco rattle virus (TRV), and Pepper mild mottle virus (PMMoV), in which some positive strand vsiRNAs can account for 97% of total vsiRNAs (Ho et al., 2006; Donaire et al., 2009; Qi et al., 2009). There are no dsRNA intermediate replicative forms for ssDNA and dsDNA viruses. Some vsiRNAs generated from DNA viruses display strong strand bias, indicating that these vsiRNAs may be processed from the structured region of the viral RNA transcripts. 62% of the vsiRNAs match the transcript polarity of Cauliflower mosaic virus (CaMV), a virus from which the commonly used constitutive 35S promoter is derived. Although up to 82% of vsiRNAs are generated from the leader region, these exhibit no strand bias (Blevins et al., 2011). Tomato yellow leaf curl China virus (TYLCCNV) is a Geminiviridae that has an ssDNA genome. Although the vsiRNAs derived from TYLCCNV display site bias, they map nearly equally to the positive and negative genomes (Yang et al., 2011a). Thus, both the dsRNA replicative form and the secondary structure of viral genomes can processed by host RNA silencing machineries. However, the implication of these findings on viral pathogenicity or evolution is still unknown.
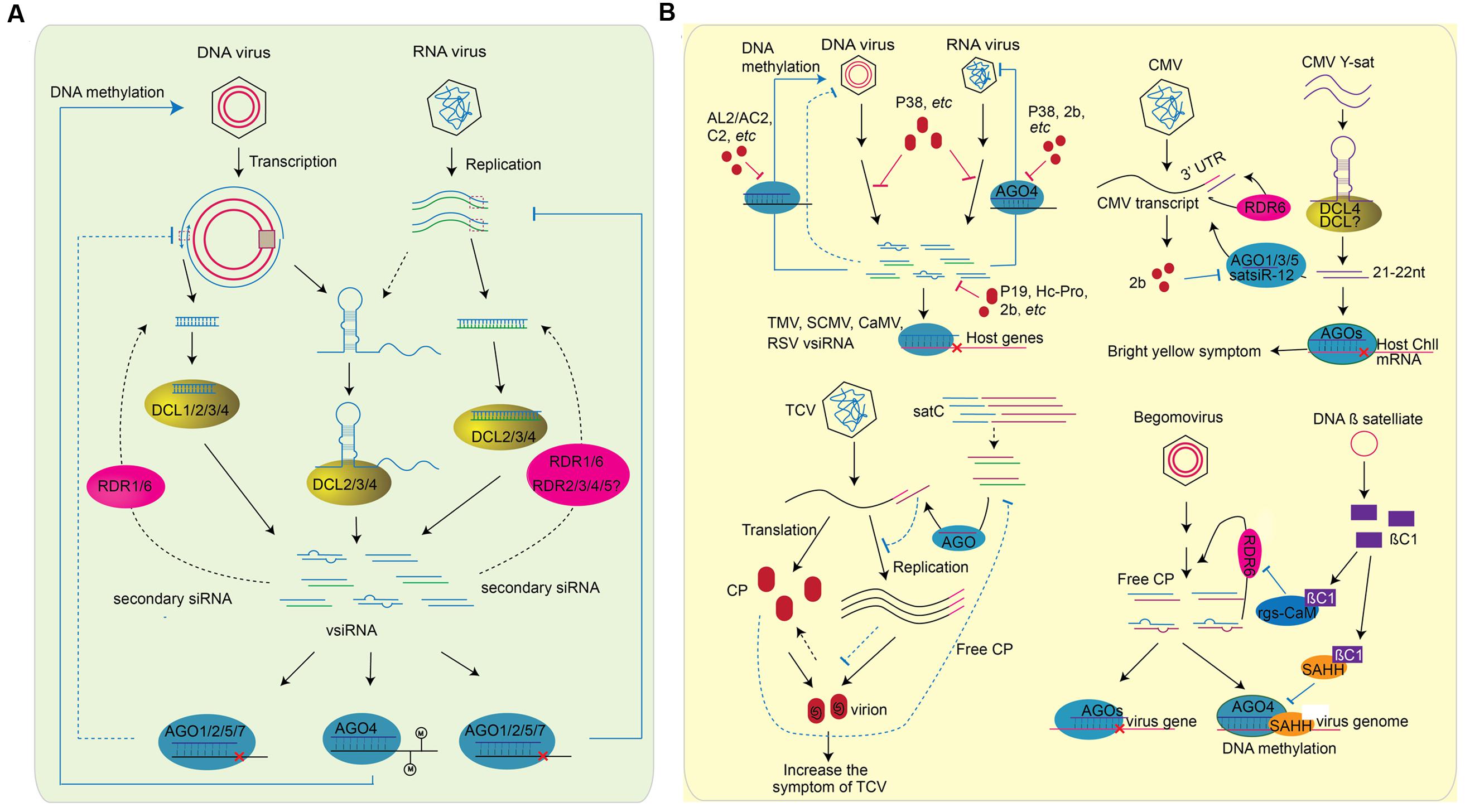
FIGURE 3. Role of sRNAs in plant-virus interaction. RNA silencing inside plant cells can be divided into two parts: (A) Plant generate vsiRNAs, targeting on virus genome directly to defend viral infection. The generation of vsiRNA are slightly different for RNA virus or DNA virus. For RNA virus, the structure region of virus genome, dsRNA replicative intermediate forms of ssRNA viruses, and the dsRNA genomes of dsRNA viruses can be processed by DCL proteins (right). The vsiRNAs of DNA virus, on the other hand, can be processed from the structured region of the transcript and the overlapping region of the bi-direction transcription (left). In both cases, RDR1 and RDR6 are involved in the generation of secondary vsiRNA (shown in blank dash line). After generation, vsiRNAs are loaded into different AGOs and perform the silencing of virus genome. vsiRNAs target on RNA virus to slice the genomic RNA, while perform DNA methylation on the genome of DNA virus. Whether vsiRNA targets on the transcription of DNA virus remains unknown (blue dash line). (B) The counter-defense of virus to plant RNA silencing machinery. As plant generates vsiRNA to silence virus genome, viruses encode suppressors, such as 2b, Hc-Pro, P19, AL2/AC2, P38, and etc., as a counter-defense (left above). The effect of suppressor on RNA silencing include the interfere of DCL slicing, the blocking of methylation, the binding of vsiRNsA, the preventing of RISC assembly, and etc. vsiRNAs encoded by TMV, CMV, CaMV, and RSV can also target the host genes to decrease plant defense. In addition, plant viruses are often accompanied with a variety of subviral RNA/DNAs. These satellite RNA/DNAs affect virus pathogenicity by generating satRNA-derived siRNAs (satsiRNAs). CMV Y satellite (Y-sat) produces a 22-nt satsiRNA that targets Chll, a key gene involved in chlorophyll synthesis, resulting in bright yellow symptom. sat-siR-12, another satsiRNA can loaded into AGO1/3/5 and regulate CMV transcripts accumulation with the function of RDR6. As counter defense, CMV encodes VSR 2b to inhibit the function of AGOs (right above). TCV is often accompanied with a single strand satellite RNA (satC) that is composed of the 3′ end of TCV helper virus (left bottom). Because of the sequence similarity of satsiRNA and the 3′ end of TCV helper virus, the presence of satC-siRNA represses the accumulation of TCV genomic RNA. At the same time, TCV genomic RNA and the CP protein assemble to a virion. CP is a VSR encode by TCV. The down-regulation of TCV transcripts by satC-siRNAs result in the increase of free CP protein, which subsequently suppresses the accumulation of satC-siRNAs (shown in dash line). DNA ß satellites are circular ssDNA that associate with many monopartit begomoviruses. The ßC1 protein encoded by DNAß satellite is a VSR that suppresses TGS by the interaction with SAHH, and PTGS through the interaction with rgs-CaM (right bottom).
Viroids, the smallest pathogen that can replicate in the nucleus or chloroplast, consists of naked, single-stranded, closed circular RNAs with sizes ranging from 250- to 400-nt (Ding, 2009). More than two decades ago, people noticed that Potato spindle tuber viroid (PSTVd) infection results in full methylation of the PSTVd cDNA sequence that is inserted into the tobacco genome (Wassenegger et al., 1994). This methylation occurs by viroid-induced RNA silencing and RdDM. Later studies detected siRNAs in PSTVd-infected tomato and tobacco plants and proved that viroids are the activator and target of RNA silencing (Figure 4A) (Itaya et al., 2001; Papaefthimiou et al., 2001). Viroid-associated siRNAs (vdsiRNAs) of PSTVd are generated from both polarities in the left and right domains. By profiling PSTVd vdsiRNAs through deep sequencing, Itaya et al. (2007) uncovered that PSTVd vdsiRNAs predominately map to the positive strand of the left and right terminal regions, indicating that these sRNAs are generated from the secondary structure of plus-strand RNAs. Some vdsiRNAs are also generated from the negative strand of the central part, indicating they may be processed from the secondary structure of the negative-strand viroid genomic RNA (Itaya et al., 2007). Citrus exocortis viroid (CEVd) replicates in the nucleus and mainly generates 5′-phosphorylated and 3′-methylated vdsiRNAs with positive polarity. Most CEVd vdsiRNAs are located within the right-end domain, suggesting that structured RNA is the main substrate of DCL enzymes (Martin et al., 2007). Avocado sunblotch viroid (ASBVd), Peach latent mosaic viroid (PLMVd), and Chrysanthemum chlorotic mottle viroid (CChMVd) are three viroids that replicate in the chloroplast. CChMVd and PLMVd generate vdsiRNAs from both polarities (de Alba et al., 2002; St-Pierre et al., 2009). ASBVd also generates vdsiRNAs in leaves displaying bleached symptoms (Markarian et al., 2004). Thus, both Pospiviroidae and Avsunviroidae viroid families can produce vdsiRNAs in plants (Ding and Itaya, 2007; Ding, 2009; Hammann and Steger, 2012). The fact that vdsiRNAs can be generated from both the positive and the negative strand of the viroid genome with strand and position bias indicates that vdsiRNAs are predominately processed from the secondary structure of the viroid genomic RNAs. However, it is important to point out that the discoveries of vdsiRNAs may be biased due to the current methods for sRNA cloning.
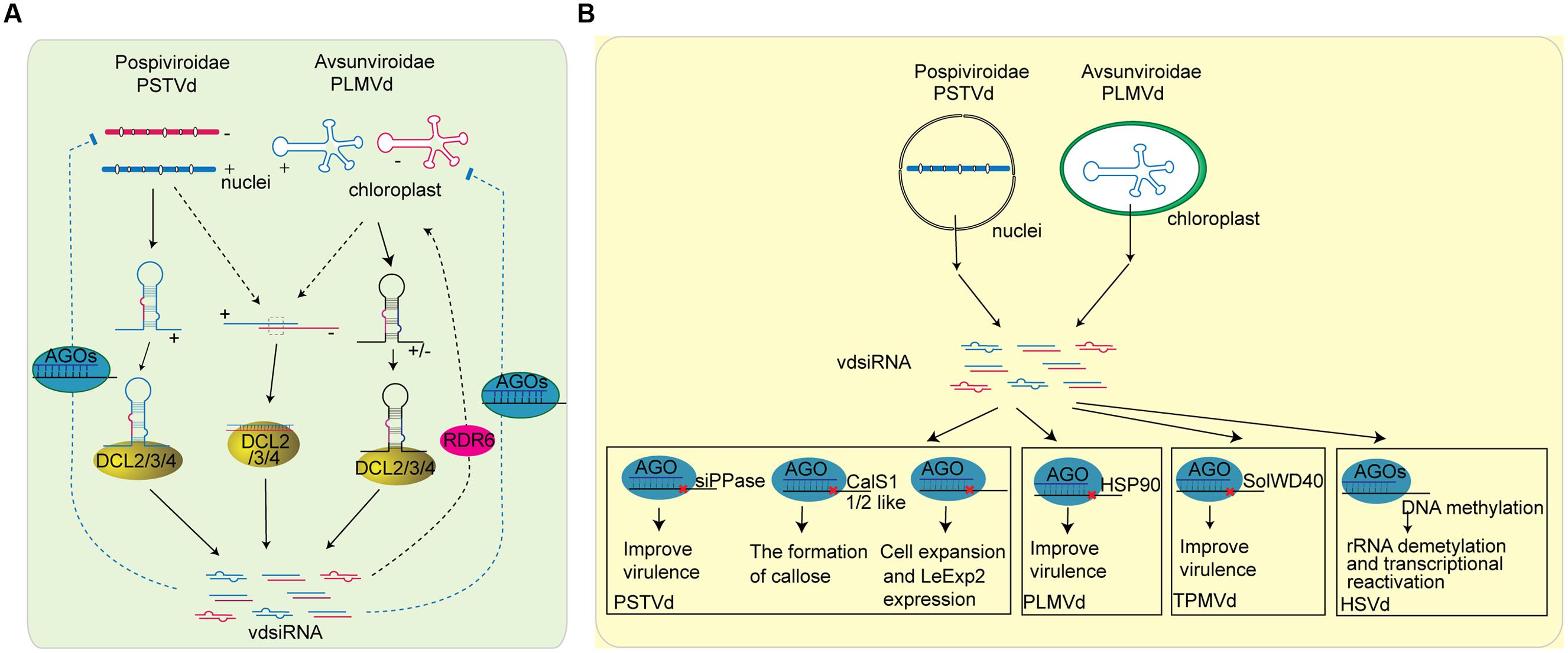
FIGURE 4. Role of sRNAs in plant-viroid interaction. (A) The biogenesis of vdsiRNAs in plant and the possible function of vdsiRNA in plant defense to viroid. PSTVd is mainly found in nucleolus, and its vdsiRNAs predominately map to the positive strand of the left and right terminal regions. It is most likely that PSTVd-vdsiRNAs are generated from the hairpin or stem-loop structure of plus-strand of PSTV transcripts. The secondary structure of PSTVd transcripts are targeted by DCL protein and sliced into vdsiRNA. Another possible source of vdsiRNA are the accidental association of (+) and (-) strand replication, which are further target by DCL protein. On the other hand, PLMVd, viroid that replicate in the chloroplast, generate vdsiRNAs from both polarities. The stem-loop structures of PLMVd are processed by DCL protein to generate vdsiRNAs. Furthermore, some research indicate that vdsiRNAs can be amplified through the activity of RDRs. After generation, vdsiRNA may be loaded into plant AGO proteins and target viroid RNAs. (B) The function of vdsiRNAs in producing viroid symptom. Some of the viroid symptom maybe caused by vdsiRNAs that target host genes. vdsiRNA generated by PSTVd can target on various plant genes including soluble inorganic pyrophosphatase (siPPase) gene, callose synthase genes CalS11-like and CalS12-like, and LeExp2 gene, while PLMVd vdsiRNA has been reported to target HSP90 and trigger signal transduction that eventually leads to viroid disease symptoms. TPMVd vdsiRNA has also been shown to slice the SolWD40 gene. In addition, HSVd vdsiRNAs are involved in TGS by inducing DNA methylation of the promoter region of rRNA genes.
dsRNA inducers are processed by plant DCL proteins to generate sRNAs. Arabidopsis encodes four DCL proteins that generate different sRNAs: DCL1 processes hairpin pri-miRNAs and pre-miRNAs into 21-nt miRNAs; DCL3, DCL4, and DCL2 process long dsRNAs into 24-nt hc-siRNAs, 21-nt siRNAs, and 22-nt siRNAs, respectively. For RNA viruses and viroids, the perfectly paired dsRNA intermediate replication form and the hairpin structure of the single genomic RNA are predominant dsRNA inducers. Indeed, DCL4, DCL2, and DCL3 process ssRNA viruses (e.g., CMV, TuMV, and TCV) into 21-nt, 22-nt and 24-nt vsiRNAs, respectively (Bouche et al., 2006; Deleris et al., 2006; Fusaro et al., 2006; Diaz-Pendon et al., 2007; Garcia-Ruiz et al., 2010). The newly emerging systemic leaves of PSTVd-infected plants only accumulate shorter (21–22-nt) vdsiRNAs, while the older leaves contain both shorter and longer (24-nt) vdsiRNAs (Machida et al., 2007; Schwind et al., 2009). Similar vdsiRNA accumulation patterns are also present in Hop stunt viroid (HSVd)- and Hop latent viroid (HLVd)-infected plants. However, very little is known about the biogenesis of vdsiRNAs. 21-nt, 22-nt, and 24-nt vsiRNAs also accumulate in plants infected with Cabbage Leaf Curl Virus (CalCuV), Beet curly top virus (BCTV), and Pepper golden mosaic virus (PepGMV), which are all ssDNA viruses, and CaMV, a dsDNA virus (Blevins et al., 2006; Rodriguez-Negrete et al., 2009; Raja et al., 2014). 24-nt vsiRNAs are the predominant vsiRNAs produced from DNA viruses. DCL3, DCL4, and DCL2 are required for the accumulation of 24-nt, 21-nt, and 22-nt CalCuV vsiRNAs, respectively. DCL3 and DCL4 are also responsible for accumulation of 24- and 21-nt vsiRNAs derived from BCTV, respectively (Raja et al., 2014). Although the hairpin structure of viral or viroid genomes is one of the main sources of vsiRNA and vdsiRNAs, DCL1-dominant hairpin processing is not involved in vsiRNA and vdsiRNAs accumulation or anti-RNA-viral resistance. However, DCL1, but not DCL4, is required for the accumulation of 21-nt vsiRNAs from CaMV (Blevins et al., 2006).
After the initial processing of dsRNA inducers, the antiviral and antiviroid signals are amplified by host RDRs. Arabidopsis encodes six RDRs, among which the function of RDR1, RDR2, and RDR6 have been well studied. RDR1 is induced by salicyclic acid (SA) treatment and TMV infection in tobacco and Arabidopsis, and a mutation in RDR1 permits efficient multiplication of ssRNA viruses (Xie et al., 2001; Yu et al., 2003; Yang et al., 2004). Furthermore, the Arabidopsis rdr6 mutant is more susceptible to infection with ssRNA and ssDNA viruses (Dalmay et al., 2000; Mourrain et al., 2000; Dalmay et al., 2001; Muangsan et al., 2004). Infection with RSV (a negative ssRNA virus) and RDV (a dsRNA virus) decreases the expression of rice RDR6. Down-regulation of rice RDR6 by antisense transformation results in increased susceptibility to RDV (Jiang L. et al., 2012; Hong et al., 2015). Both RDR1 and RDR6 are required for secondary CMV vsiRNA production in Arabidopsis: RDR1 is required for the production of vsiRNAs from 5′-terminal ends of the genome, while RDR6 is required for the production of vsiRNAs from the 3′-terminal ends (Wang et al., 2010). However, expression of Nicotiana tabacum RDR1 in N. benthamiana plants (which do not encode RDR1) showed that RDR1 suppresses RNA silencing mediated by RDR6 and enhances viral infection in transgenic plants (Ying et al., 2010). In addition, accumulation of HSVd and PSTVd genomic RNAs was higher in RDR6-silenced plants, indicating that RDRs also contribute to anti-viroid resistance (Gomez et al., 2008; Di Serio et al., 2010). Systematic analysis via profiling of vsiRNAs and vdsiRNAs in pathogen infected plants have revealed that sRNAs processed from pathogen genomic RNAs indeed decrease in rdr knock-out mutants or RDR-silenced plants (Gomez et al., 2008; Di Serio et al., 2010; Garcia-Ruiz et al., 2010; Wang et al., 2010; Hong et al., 2015). The decreased accumulation of vsiRNAs and vdsiRNAs and increased susceptibility of rdr mutant plants demonstrate the anti-viral/viroid role of RDRs. Although RDR2 is responsible for 24-nt hc-siRNA accumulation, mutation in RDR2 has little or no effect on the accumulation of vsiRNAs of DNA viruses CalCuV and CaMV (Blevins et al., 2006). Tomato Ty-1 and Ty-3 are alleles of the same gene that encodes RDRs with sequence similarity to Arabidopsis RDR3, RDR4, and RDR5. They are TYLCV resistance genes, and susceptible tomato lines without these loci produce lower levels of TYLCV vsiRNAs and accumulate higher viral titers (Verlaan et al., 2013; Butterbach et al., 2014). However, in Arabidopsis, the antiviral functions of RDR3, RDR4, and RDR5 have not yet been uncovered. Therefore, the function of RDR2 and other RDRs in host-virus/viroid interactions needs to be further explored.
After processing and amplification, vsiRNAs and vdsiRNAs are loaded into AGO proteins to inhibit the replication and movement of viruses and viroids. AGO1 (Morel et al., 2002; Zhang et al., 2006), AGO2 (Takeda et al., 2008; Harvey et al., 2011; Jaubert et al., 2011; Wang X.B. et al., 2011), AGO3 (Schuck et al., 2013), AGO5 (Takeda et al., 2008), AGO7 (Qu et al., 2008), and AGO10 (Garcia-Ruiz et al., 2015) have been shown to bind vsiRNAs or be involved in anti-viral RNA silencing pathways. Recovery from infection with a DNA virus requires the function of host AGO4 (Raja et al., 2008). Thus, the 24-nt vsiRNAs of DNA viruses may associate with AGO4 to methylate the viral genome. A mutant defective in DRB3, a double-stranded RNA binding protein that interacts with DCL3 and AGO4, displays lower methylation of the viral DNA genome and increased hyper susceptibility to germinivirus, further demonstrating the function of the DCL3-AGO4 RdDM pathway in resistance against DNA viruses (Raja et al., 2014). Furthermore, AGO18, a novel AGO that is conserved in monocot plants, is induced by RSV and required for rice antiviral resistance (Wu et al., 2015). In regards to the vdsiRNAs, the 21-nt and 22-nt vdsiRNAs are predominately loaded into AGO1, AGO2, and AGO3 (Minoia et al., 2014); the 24-nt vdsiRNAs are loaded into AGO4, AGO5, and AGO9; while AGO6, AGO7, and AGO10 do not bind vdsiRNAs (Minoia et al., 2014). However, the anti-viroid function of these AGOs needs to be further determined.
Although vdsiRNAs processed by DCLs are loaded into plant AGOs, their regulation of viroid genomes is not well known. PSTVd, CEVd, and CChMVd in plants can be silenced by transgenic dsRNAs or co-inoculated dsRNAs. This silencing is sequence-specific, temperature-dependent and, in some cases, dose-dependent (Vogt et al., 2004; Carbonell et al., 2008; Schwind et al., 2009). However, further studies indicate that viroids may have evolved a mechanism to avoid the silencing of sRNA. Dr. Biao Ding’s group found that PSTVd replicates easily in infected plants even with the present of high accumulation of vdsiRNAs (Itaya et al., 2001). Studies on PSTVd and HSVd show that the circular genome of the viroid is resistant to RNA silencing (Wang et al., 2004; Gomez and Pallas, 2007). A possible explanation is the structured viroid RNA can be processed into active vdsiRNAs, but the viroid RNA is resistant to RISC-mediated degradation due to its secondary structure (Itaya et al., 2007).
Pathogen sRNAs Regulate Pathogen Gene Expression to Increase Virulence
During plant–microbial pathogen interaction, host miRNAs and siRNAs play a role in modulating host immunity while some sRNAs derived from pathogens can decrease host defense or increase pathogen virulence. Fungi, oomycetes, bacteria, viruses, viroids, and satellite RNAs all produce sRNAs that are either similar to or distinct from plant sRNAs (Figures 1B, 2B, 3B, and 4B). During the counter-defense response, these pathogen sRNAs facilitate infection by adjusting pathogen gene expression to increase virulence.
Fungi and oomycetes encode siRNAs that are mainly derived from transposons, inverted, tandem, or other repeat regions, and effector coding regions. These sRNAs display diverse biogenesis pathways, and some require typical RNA silencing components, such as DCLs, AGOs, and RDRs for accumulation (Murata et al., 2007; Lee et al., 2010; Jiang N. et al., 2012; Fahlgren et al., 2013; Qutob et al., 2013; Raman et al., 2013; Weiberg et al., 2013). sRNAs in fungal pathogens have been shown to mediate pathogenic virulence by traveling into host cells and silencing host genes (Figure 2B) (Weiberg et al., 2013). Although there is indirect evidence that links sRNAs to pathogen virulence, the function of sRNAs in pathogen cells has not been well studied. The differential accumulation of M. oryzae sRNAs in vegetative and specialized-infection tissues suggests that sRNAs in M. oryzae may be involved in growth, development, and virulence (Nunes et al., 2011). Moreover, sRNA profiling of M. oryzae identified a set of genes that are transcriptionally regulated by sRNAs. One of these is ACE1, a known avirulence gene that has increased expression in the dcl1 mutant (Raman et al., 2013). The sRNAs in three Phytophthora species, P. infestans, P. sojae and Phytophthora ramorum, were analyzed, and they were predominantly 21-nt and 25-nt long (Fahlgren et al., 2013). The 21-nt sRNAs were found to be derived from gene families including Crinkler (CRN) effectors and type III fibronectins. Some of these 21-nt sRNAs are predicted to target amino acid/auxin permeases, but their exact functions are still unknown (Fahlgren et al., 2013). sRNAs generated from RxLR and CRN effectors loci were also identified. The expression levels of these effectors and the sRNAs, vary in P. infestans strains that differ in virulence, suggesting that these sRNAs may affect the accumulation of effectors, thus alter the virulence (Vetukuri et al., 2012). Some sRNAs map to the tRNA loci of fungi and oomycetes (Nunes et al., 2011; Asman et al., 2014). The biogenesis of these sRNAs requires pathogen DCLs and AGOs. The accumulations of these sRNAs are significantly changed during the infection progress, which suggests that these sRNAs may function in pathogen-host interaction. Moreover, recent study have identified sRNAs associated with P. infestans AGO proteins (Asman et al., 2016). PiAGO1-associated 20–22 nt sRNAs, were generated from genes encoding host cell death-inducing CRN effectors, while 24–26 nt sRNAs, which bound to PiAGO4, were derived mainly from Helitron, Crypton, PiggyBac and Copia transposons. The essential role of PiAGO1 in gene regulation, together with its associated sRNAs, which derived from CRN gene family, implicating 20–22 nt sRNAs may bind to AGO1 to regulate the expression of genes in CRN family and subsequently mediate the pathogen virulence (Asman et al., 2016). In addition, sRNAs that are derived from the effector regions can transgenerationally alter the virulence of the pathogen. Avirulence (Avr) gene Avr3a of P. sojae encodes an effector protein that can be detected by the host R gene. The expression of Avr3a gene in P. sojae attenuated the virulence of plants carrying the R gene Rps3a. Qutob et al. (2013) observed non-Mendelian inheritance of transgenerational gene silencing of Avr3a and gain of virulence in soybean plants. Meanwhile, increased accumulation of 25-nt sRNAs was seen in gene-silenced strains but not in strains with Avr3a mRNA, indicating there is sRNA-associated transgenerational gene silencing (Qutob et al., 2013).
Until now bacteria have not been found to encode typical sRNAs as plants, but they produce 50- to 300-nt non-coding sRNAs (ncRNAs) that regulate the expression of target mRNAs through imperfect base-pairing of short regions (10- to 25-nt) (Figure 1B) (Altuvia, 2007; Weiberg et al., 2014). There is an emerging body of evidence suggesting that ncRNAs are involved in bacterial virulence. Bacterial lipoprotein (BLP) triggers cell activation and host defense through toll-like receptors (TLRs). CRISPR-Cas-associated sRNAs from Francisella novicida guide the Cas9 protein to suppress BLP, which subsequently facilitates evasion of TLR2 (Sampson et al., 2013). The Cas9 system acting with a small, CRISPR/Cas-associated RNA (scaRNA) also controls virulence of Francisella tularensis (Sampson et al., 2013). However, a direct link between the Cas system and plant bacterial pathogenesis has not yet been found. Genome-wide transcriptome analysis has identified 16 intergenic sRNAs and seven cis-encoded antisense sRNAs in the plant pathogen Xanthomonas campestris pv. vesicatoria (Xcv) (Schmidtke et al., 2012). The expression of half of these intergenic sRNAs is controlled by components of the type III secretion system, and some are involved in virulence. The deletion of sX12 delays the development of disease symptoms and HR in pepper plants (Schmidtke et al., 2012). The 115-nt sRNA sX13 regulates 63 genes, which are involved in signal transduction, motility, transcriptional and posttranscriptional regulation, and virulence. Deletion of sX13 strongly delayed development of disease symptoms in susceptible and resistant pepper plants (Schmidtke et al., 2013). However, the function of sX13 is not dependent on Hfq, a hexameric RNA-binding protein that globally interacts with sRNAs to post-transcriptionally regulate gene expression and virulence traits in many animal and plant pathogenic bacteria. Hfq can bind up to 100 sRNAs in Salmonella (Chao and Vogel, 2010). Hfp-dependent sRNAs in Erwinia amylovora were also identified, and 40 of them were found to associate with Hfq. sRNAs ArcZ, RmaA, and OmrAB all contribute to virulence by positively modulating type III secretion system attachment, amylovoran production, and motility (Zeng et al., 2013; Zeng and Sundin, 2014).
Pathogens Encode sRNAs Targeting Host Genes to Improve Virulence
Another strategy of pathogens to counteract host defenses is the production of sRNAs that target host genes to decrease host immune responses. Viroids do not code any protein or peptide and yet are able to replicate, travel cell-to-cell and long distance through phloem, resist plant defense responses, and cause disease in certain hosts (Ding and Itaya, 2007; Ding, 2009). For a long time, the question of how viroids produce disease symptoms without any open reading frames has intrigued scientists. Early studies focus on explaining the molecular mechanism of viroid pathogenesis by determining the interaction of genomic RNA of viroid with host factors, including host proteins or nucleic acids (Navarro et al., 2012a,b). While a few proteins or RNAs were determined to interact with viroid RNA, their roles in viroid pathogenesis is largely inclusive. In recent years, the new hypothesis that viroids cause disease symptoms by producing sRNAs to target host genes was raised and there are many studies supporting this hypothesis (Figure 4B). Over-expression of PSTVd hairpin RNA, which produces sRNAs, results in similar phenotypes as PSTVd infection, suggesting that PSTVd may cause disease symptoms by sRNA-mediated silencing (Wang et al., 2004). Large-scale sequencing uncovered that two genes involved in gibberellin or jasmonic acid biosynthesis contain binding sites for PSTVd vdsiRNAs (Wang et al., 2011b). Moreover, DCL4, which should reduce PSTVd levels by slice or dice its genome RNA to produce vdsiRNA, seems to benefit the accumulation of PSTVd (Dadami et al., 2013). Expression of an artificial miRNA containing the sequence of the PSTVd virulence modulating region down-regulates the expression of a Nicotiana soluble inorganic pyrophosphatase (siPPase) gene and leads to a PSTVd infection phenotype (Eamens et al., 2014). In addition, a recent study showed that single vdsiRNA is able to silence multiple host mRNAs. vdsiRNAs derived from PSTVd can target two callose synthase genes, CalS11-like and CalS12-like, which are essential for the formation of callose. The efficiency of suppression depends on the viroid variants and the target gene (Adkar-Purushothama et al., 2015). PLMVd is a chloroplast-replicating viroid and an insertion of a 12- to 13-nt fragment inhibits chloroplast development (Rodio et al., 2007). Further study uncovered that in Prunus persica, two vdsiRNAs containing the insertion sequence target the chloroplast heat shock protein 90 (HSP90) and triggers signal transduction that eventually leads to viroid disease symptoms (Navarro et al., 2012a). A single U257A change in the PSTVd central conserved region also strongly increases PSTVd virulence by restricting host cell expansion. The lethal phenotype of PSTVd is correlated with the down-regulation of LeExp2 gene expression (Qi and Ding, 2003). It is not clear whether the U257A mutation also produces a novel sRNA that targets some essential host genes that is critical for cell expansion and LeExp2 expression. Furthermore, upon viroid infection, vdsiRNAs generated by Tomato planta macho viroid (TPMVd) targets and slices the SolWD40 gene, the function of which is unknown (Avina-Padilla et al., 2015). Although HSVd genomic RNA is higher in RDR6-silenced plants, the viroid-induced symptoms are absent. Meanwhile, HSVd vdsiRNA accumulation is decreased in RDR6-silenced plants, suggesting that the symptoms of HSVd is dependent on vdsiRNAs (Gomez et al., 2008). The symptom severity of CEVd is also correlated with the level of vdsiRNAs but not the viroid genome level, further supporting that vdsiRNAs are not simply by-pass products of anti-viroid RNA silencing reactions, but they have a purpose in producing disease symptoms karian (Markarian et al., 2004). In addition, there are some evidence that link viroid infection to transcriptional gene silencing (TGS). Wassenegger et al. (1994) discovered that PSTVd cognate DNA sequences were methylated in PTSVD-expressing transgenic tobacco plant, while the T-DNA and the genomic plant DNA remained unaltered. Further studies also demonstrate the correlations between viroid infection and host genes transcriptional alteration. For instance, cucumbers infected with HSVd accumulate high levels of sRNAs derived from ribosomal transcripts, as well as ribosomal RNA (rRNA) precursors. This was caused by altered DNA methylation in the promoter region of rRNA genes, resulting in demethylation and transcriptional reactivation of normally inactive rRNA genes (Martinez et al., 2014). N. benthamiana carrying an HSVd dimeric sequence develops similar phenotype to HSVd-infected plants (Gomez et al., 2008). This plant also accumulates high levels of sRNAs derived from ribosomal transcripts along with a decrease in rDNA methylation, suggesting that this may be a general phenomenon (Castellano et al., 2015). However, the correlation between sRNA accumulation and DNA methylation needs to be further determined.
It is noteworthy that although some studies suggest that symptoms produced by viroids in plants are associated with vdsiRNAs and the RNA silencing machinery, there is no uniform correlation between the levels of vdsiRNA and symptoms (Ding and Itaya, 2007; Ding, 2009; Kovalskaya and Hammond, 2014). Moreover, in contrast to early observation that symptoms similar to those of PSTVd infection were developed in some transgenic tomato lines expressing non-infectious PSTVd hairpin RNA (Wang et al., 2004), no disease symptoms were found in other tomato lines, despite the accumulation of PSTVd hairpin-derived siRNA (Schwind et al., 2009). Whether vdsiRNA indeed results in viroid disease symptoms requires further investigation.
Plant viruses are often accompanied with a variety of subviral RNA/DNAs, which have no or little sequence similarity to plant viruses. Most satellite RNAs do not encode proteins but can significantly alter viral disease symptoms (Figure 3B) (Collmer and Howell, 1992; Simon et al., 2004). More and more studies indicate that the pathogenicity of satellite RNA/DNA may due to host gene silencing induced by satRNA-derived siRNAs (satsiRNAs). CMV Y satellite (Y-sat) causes a bright yellow mosaic phenotype. Replication of Y-Sat is resistant to RNA silencing, but expression of viral suppressors of RNA silencing (VSR) reduces the disease symptoms (Wang et al., 2004). The hairpin structures of CMV satellite RNA are processed by DCL4 and other DCL proteins to form 21-nt and 22-nt satsiRNAs (Du et al., 2007). Y-sat produces a 22-nt satsiRNA that targets Chll, a key gene involved in chlorophyll synthesis, and cleaves Chll mRNA post-transcriptionally, causing the bright yellow mosaic phenotype. Transformation of N. tabacum with a silencing-resistant version of Chll greatly reduces the Y-Sat symptoms (Shimura et al., 2011; Smith et al., 2011). satsiR-12, another satsiRNA generated from SD-CMV satellite RNA, targets the upstream region of the CMV 3′ UTR for slicing. satsiR-12 is loaded into AGO1/2/5 for RDR6-mediated regulation, which can be suppressed by 2b encoded by CMV (Zhu et al., 2011). However, the accumulation of 2b coding subgenomic RNA, RNA4A and 2b proteins is also reduced by SD-CMV satellite RNA, which attenuates the D-CMV yellow symptom in N. benthamiana (Hou et al., 2011). TCV is often accompanied with a single strand satellite RNA, satC, that is composed of the 3′ end of TCV helper virus. The presence of satC represses the accumulation of TCV genomic RNA and virion, which leads to increased levels of free CP proteins. CP is a VSR encoded by TCV that targets the DCL2/4 silencing pathway and suppresses satC accumulation. The satC-mediated enhancement of free CP proteins then increases the symptoms of TCV (Zhang and Simon, 2003; Manfre and Simon, 2008). Thus, sRNAs generated from satellite RNAs produce species-specific disease symptoms by targeting host genes or viral genomes. On the other hand, in the presence of SD-CMV satellite RNA, the infection of CMV-Δ2b lead to high accumulation of satsiRNA, while the accumulation of CMV siRNA was reduced. Thus, the dice and slice of host RNA silencing machinery on SD-CMV satellite RNA may decrease its efficiency on CMV RNAs (Hou et al., 2011). DNA β satellites are circular ssDNA that associate with many monopartit begomoviruses and are essential for viral disease symptoms (Briddon et al., 2001; Jose and Usha, 2003; Cui et al., 2004). The ßC1 protein encoded by DNA β satellite is a VSR that suppresses methylation-mediated TGS and RDR6-mediated PTGS through the interaction of S-adenosyl homocysteine hydrolase (SAHH) and rgs-CaM, which will be discussed later (Cui et al., 2005; Yang et al., 2011b; Li et al., 2014). Thus, viral satellite RNA/DNA can alter the symptoms caused by the helper virus with different sRNA related mechanisms.
Upon infection with a virus, host plants process viral genomic or transcript RNAs into vsiRNAs and load them into RISC complexes to inhibit the amplification and movement of the virus. However, depending on the similarity of vsiRNA-target gene and host genes, some vsiRNAs can target host genes, which subsequently increase viral pathogenicity (Figure 3B). Deep sequencing and bioinformatics studies indicate that 16 TMV vsiRNAs potentially target Arabidopsis genes. Two of these vsiRNAs target and slice transcripts of a polyadenylation specificity factor and an unknown protein similar to translocon-associated protein alpha. The slicing of these two genes only happens upon TMV infection, revealing that they are real vsiRNA targets (Qi et al., 2009). Dozens of Zea mays genes are predicted targets of vsiRNAs encoded by Sugarcane mosaic virus (SCMV). Some vsiRNA targets that contribute to biotic/abiotic stress responses and ribosome biogenesis are down-regulated upon SCMV infection (Xia et al., 2014). In addition, vsiRNAs originating from the leader region of CaMV 35S RNA were found to increase the accumulation of CaMV. Like other vsiRNAs, these leader-derived vsiRNAs are DCL-dependent and subsequently loaded into AGO1 (Blevins et al., 2011). These vsiRNAs may also facilitate CaMV accumulation by suppressing Arabidopsis gene expression. RSV infection causes plant stunting, chlorosis, and other symptoms. A recent study showed that vsiRNAs can be generated from RSV RNA4, and further targeting host gene eIF4A. The infection of RSV down-regulated eIF4A expression. Interestingly, eIF4A suppression by artificial miRNAs leads to rice leaf-twisting and stunting (Shi et al., 2016). Thus, vsiRNAs can directly cause virus pathogenicity, as with vdsiRNAs. Nine chloroplast-related genes (ChRGs) are also down-regulated upon RSV infection and silencing them with artificial miRNAs causes plant chlorosis symptoms, similar to viral infection. However, whether the down-regulation of ChRGs upon RSV infection is also mediated by RSV vsiRNAs need to be further studied (Xia et al., 2014). In contrast to siRNAs, there are relatively few studies done on virus-encoded miRNAs in plants. Studies on Sugarcane streak mosaic virus (SCSMV) and Hibiscus chlorotic ringspot virus (HCRSV) suggest the existence of virus-encoded miRNAs that may target plant genes, but their detailed functions remain unknown (Gao et al., 2012; Viswanathan et al., 2014).
Fungi, omycetes, and bacteria that localize in the intercellular region in the early infection stages can deliver pathogen sRNAs into plant cells to target host genes as counter-defense. Infecting Arabidopsis and Solanum lycopersicum with a destructive fungal plant pathogen, Botrytis cinerea, results in the presence of a set of B. cinerea sRNAs (Bc-sRNAs) in both plants. Among these sRNAs, 73 Bc-sRNAs are able to target host genes in both Arabidopsis and S. lycopersicum. These Bc-sRNAs are processed by fungi DCLs and loaded into a host AGO1 protein to slice host targets. A mutation in Arabidopsis AGO1 reduces the susceptibility of the plant to B. cinerea, and a mutation in B. cinerea DCLs decreases fungi pathogenicity. Multiple Bc-sRNA target genes were identified, including Arabidopsis mitogen-activated protein kinase genes MPK1 and MPK2, a cell wall-associated kinase (WAK), a peroxiredoxin (PRXIIF), and the tomato MPK-kinase kinase 4 (MAPKKK4). Suppression of these genes increases the disease susceptibility of the plant (Weiberg et al., 2013). This is the first study showing that sRNAs from a eukaryotic pathogen mediate pathogen virulence using host RNA silencing machinery; however, it is still unclear how these fungal siRNAs are delivered into plant cells. Pathogen sRNAs have been shown to be delivered into animal cells though RNA transporters. Two membrane-associated RNA transporters, systemic RNAi defective-1 (SID1) and SID2, were identified in C. elegans (Shih and Hunter, 2011; McEwan et al., 2012). However, no membrane-associated RNA transporters have yet been identified in plants.
RNA silencing inhibits the infection, replication, and movement of same viruses at different steps. Thus, pathogens also encode RNA silencing suppressors to decrease the accumulation of sRNAs or inhibit the function of sRNAs (Qi et al., 2004). Many VSRs are viral pathogenicity determinants, indicating that the suppression function is important for pathogenicity. Some VSRs bind viral dsRNA or vsiRNAs and decrease the number of functional sRNAs targeting viral genomes. Other VSRs directly or indirectly target RNA silencing pathway components such as DCLs, RDRs, and AGOs to inhibit the accumulation and function of endogenous miRNAs and siRNAs, thus increasing the severity of infection symptoms (Csorba et al., 2015). It is believed that plant viruses encode multiple VSRs or a multi-functioning VSRs and express them in host cells to counteract host defenses. For examples, the AL2 VSRs encoded by DNA virus CalCuV silences both transcription-dependent PTGS (transcription activation with the interaction with WEL1 and rgs-CaM) and transcription-independent PTGS (ADK inactivation with the interaction with ADK) (Wang et al., 2003; Trinks et al., 2005; Yong Chung et al., 2014). A recent study uncovered that AL2 also reverses TGS by a transcription-activation- and ADK inactivation-independent mechanism (Jackel et al., 2015). Pns10 encoded by RDV can not only bind siRNAs but also down-regulate RDR6 expression to suppress RNA silencing for viral replication and movement (Cao et al., 2005; Ren et al., 2010). While the replication of RNA viruses is suppressed by PTGS, the replication of DNA viruses is inhibited by both PTGS and TGS (Raja et al., 2008). The function of DNA VSR in the accumulation of 24-nt TGS siRNAs and RdDM pathway components has also been determined recently: C2 (also known as AL2 or AC2) inhibits the ADK function and attenuates the degradation of SAMDC1; C4 down-regulates the accumulation of MET1 but not CMT3; Rep represses the expression of MET1 and CMT3; V2 of TYLCV and AC5 of Mungbean yellow mosaic India virus (MYMIV) decreases the methylation of transgenic and endogenous loci by an unknown function; betasatellite βC1 inhibits the activity of SAHH (Wang et al., 2003, 2014; Yang et al., 2011b; Zhang Z. et al., 2011; Rodriguez-Negrete et al., 2013; Li et al., 2015).
Some bacteria and oomycetes also deliver effector proteins into host cells to suppress RNA silencing. Although miR393 is induced upon Pst DC3000 infection, the AvrPtoB effector specifically represses the induction of miR393 at the transcriptional level. AvrPto also reduces miR393 accumulation. However, the accumulation of pri-miR393 is not changed in transgenic plants, which indicates that AvrPto may post-transcriptionally down-regulate the processing of miR393 (Navarro et al., 2008). Oomycete P. sojae encodes two RNA silencing suppressors: PSR1 down-regulates the accumulation of both host miRNAs and siRNAs, while PSR2 specifically decreases the accumulation of host siRNAs (Qiao et al., 2013). Both of them are effector proteins and their over-expression enhances the infection of Phytophthora and viruses. PSR1 interacts with PINP1, a RNA helicase that regulates the accumulation of both miRNAs and siRNAs. The over-expression of PSR1 or the down-regulation of PINP1 impairs the localization of the DCL1 protein complex (Qiao et al., 2015). Another PSR2 protein encoded by P. infestans can also suppress RNA silencing and enhance the plant susceptibility to Phytophthora (Xiong et al., 2014). Thus, the RNA silencing suppressors encoded by oomycetes might be a general counter-defense mechanism. It will be interesting to see whether fungi also deliver effector proteins to inhibit host resistance.
Conclusion
There is an increasing amount of evidence that shows communication occurs between plants and different pathogens via sRNAs. The importance of sRNAs in regulating plant immunity and pathogen virulence allows scientists to utilize and manipulate RNA silencing machinery to improve plant immunity, impair pathogen virulence, and thus increase crop production. RNAi technology has been employed to manipulate plant metabolites, develop plants with improved resistance to environment stresses, and engineer plants to defend against pathogen infections (Koch and Kogel, 2014). In plants, expression of pathogen dsRNAs is widely used for plant resistance to viruses that replicate in plant cells. The different roles of sRNAs have also been demonstrated in anti-fungal, anti-insect, anti-nematode resistance, pointing to the existence of cross-kingdom RNA silencing (Baum et al., 2007; Mao et al., 2007; Nowara et al., 2010; Ibrahim et al., 2011; Koch et al., 2013; Panwar et al., 2013). However, RNA silencing is a complicated system, and there are two sides to the coin. For instance, while plants utilize vsiRNAs to silence viral RNA as a defense strategy, vsiRNAs can also target plant mRNAs to promote viral virulence. The never-ending arms race drives the co-evolution of pathogen and hosts, resulting in the variety of sRNAs and RNAi components. To utilize RNA silencing machinery, further investigation is required to explore this complicated and fascinating sRNA world.
Author Contributions
JH wrote the introduction, the summary of sRNA in plant and virus. MY wrote the summary of sRNA in bacteria and fungi. Both authors contributed to the figures and figure legend. XZ and LL supervised and complemented the writing.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to our colleagues whose studies were unable to be discussed for space restrictions. We would like to thank Dr. Yifan Lii for helpful comments. We would also like to thank Juan Du for figure correction. This work was supported by the Ministry of Agriculture transgenic major projects (2016ZX08010001), the Strategic Priority Research program of the CAS (No. XDB11050700), National Natural Science Foundation of China (No. 31471782, No. 91540116, No. 31622046), and Open research Fund Program of State Key Laboratory of Integrated Pest Management (Chinese IPM1503).
References
Adkar-Purushothama, C. R., Brosseau, C., Giguere, T., Sano, T., Moffett, P., and Perreault, J. P. (2015). Small RNA derived from the virulence modulating region of the potato spindle tuber viroid silences callose synthase genes of tomato plants. Plant Cell 27, 2178–2194. doi: 10.1105/tpc.15.00523
Altuvia, S. (2007). Identification of bacterial small non-coding RNAs: experimental approaches. Curr. Opin. Microbiol. 10, 257–261. doi: 10.1016/j.mib.2007.05.003
Asman, A. K., Fogelqvist, J., Vetukuri, R. R., and Dixelius, C. (2016). Phytophthora infestans Argonaute 1 binds microRNA and small RNAs from effector genes and transposable elements. New Phytol. 211, 993–1007. doi: 10.1111/nph.13946
Asman, A. K., Vetukuri, R. R., Jahan, S. N., Fogelqvist, J., Corcoran, P., Avrova, A. O., et al. (2014). Fragmentation of tRNA in Phytophthora infestans asexual life cycle stages and during host plant infection. BMC Microbiol. 14:308. doi: 10.1186/s12866-014-0308-1
Avina-Padilla, K., Martinez de la Vega, O., Rivera-Bustamante, R., Martinez-Soriano, J. P., Owens, R. A., Hammond, R. W., et al. (2015). In silico prediction and validation of potential gene targets for pospiviroid-derived small RNAs during tomato infection. Gene 564, 197–205. doi: 10.1016/j.gene.2015.03.076
Baldrich, P., Campo, S., Wu, M. T., Liu, T. T., Hsing, Y. I., and San Segundo, B. (2015). MicroRNA-mediated regulation of gene expression in the response of rice plants to fungal elicitors. RNA Biol. 12, 847–863. doi: 10.1080/15476286.2015.1050577
Baldrich, P., Kakar, K., Sire, C., Moreno, A. B., Berger, A., Garcia-Chapa, M., et al. (2014). Small RNA profiling reveals regulation of Arabidopsis miR168 and heterochromatic siRNA415 in response to fungal elicitors. BMC Genomics 15:1083. doi: 10.1186/1471-2164-15-1083
Baum, J. A., Bogaert, T., Clinton, W., Heck, G. R., Feldmann, P., Ilagan, O., et al. (2007). Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. doi: 10.1038/nbt1359
Bian, H., Xie, Y., Guo, F., Han, N., Ma, S., Zeng, Z., et al. (2012). Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol. 196, 149–161. doi: 10.1111/j.1469-8137.2012.04248.x
Blevins, T., Rajeswaran, R., Aregger, M., Borah, B. K., Schepetilnikov, M., Baerlocher, L., et al. (2011). Massive production of small RNAs from a non-coding region of Cauliflower mosaic virus in plant defense and viral counter-defense. Nucleic Acids Res. 39, 5003–5014. doi: 10.1093/nar/gkr119
Blevins, T., Rajeswaran, R., Shivaprasad, P. V., Beknazariants, D., Si-Ammour, A., Park, H. S., et al. (2006). Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 34, 6233–6246. doi: 10.1093/nar/gkl886
Boccara, M., Sarazin, A., Thiebeauld, O., Jay, F., Voinnet, O., Navarro, L., et al. (2014). The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 10:e1003883. doi: 10.1371/journal.ppat.1003883
Bond, D. M., and Baulcombe, D. C. (2014). Small RNAs and heritable epigenetic variation in plants. Trends Cell Biol. 24, 100–107. doi: 10.1016/j.tcb.2013.08.001
Bouche, N., Lauressergues, D., Gasciolli, V., and Vaucheret, H. (2006). An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 25, 3347–3356. doi: 10.1038/sj.emboj.7601217
Briddon, R. W., Mansoor, S., Bedford, I. D., Pinner, M. S., Saunders, K., Stanley, J., et al. (2001). Identification of dna components required for induction of cotton leaf curl disease. Virology 285, 234–243. doi: 10.1006/viro.2001.0949
Butterbach, P., Verlaan, M. G., Dullemans, A., Lohuis, D., Visser, R. G. F., Bai, Y. L., et al. (2014). Tomato yellow leaf curl virus resistance by Ty-1 involves increased cytosine methylation of viral genomes and is compromised by cucumber mosaic virus infection. Proc. Natl. Acad. Sci. U.S.A. 111, 12942–12947. doi: 10.1073/pnas.1400894111
Campo, S., Peris-Peris, C., Sire, C., Moreno, A. B., Donaire, L., Zytnicki, M., et al. (2013). Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 199, 212–227. doi: 10.1111/nph.12292
Cao, X., Zhou, P., Zhang, X., Zhu, S., Zhong, X., Xiao, Q., et al. (2005). Identification of an RNA silencing suppressor from a plant double-stranded RNA virus. J. Virol. 79, 13018–13027. doi: 10.1128/JVI.79.20.13018-13027.2005
Carbonell, A., de Alba, A. E. M., Flores, R., and Gago, S. (2008). Double-stranded RNA interferes in a sequence-specific manner with the infection of representative members of the two viroid families. Virology 371, 44–53. doi: 10.1016/j.virol.2007.09.031
Castel, S. E., and Martienssen, R. A. (2013). RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 14, 100–112. doi: 10.1038/nrg3355
Castellano, M., Martinez, G., Pallas, V., and Gomez, G. (2015). Alterations in host DNA methylation in response to constitutive expression of Hop stunt viroid RNA in Nicotiana benthamiana plants. Plant Pathol. 64, 1247–1257. doi: 10.1111/ppa.12358
Chao, Y., and Vogel, J. (2010). The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 13, 24–33. doi: 10.1016/j.mib.2010.01.001
Chen, L., Luan, Y., and Zhai, J. (2015). Sp-miR396a-5p acts as a stress-responsive genes regulator by conferring tolerance to abiotic stresses and susceptibility to Phytophthora nicotianae infection in transgenic tobacco. Plant Cell Rep. 34, 2013–2025. doi: 10.1007/s00299-015-1847-0
Chen, X. (2009). Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 25, 21–44. doi: 10.1146/annurev.cellbio.042308.113417
Collmer, C. W., and Howell, S. H. (1992). Role of satellite RNA in the expression of symptoms caused by plant viruses. Annu. Rev. Phytopathol. 30, 419–442. doi: 10.1146/annurev.py.30.090192.002223
Csorba, T., Kontra, L., and Burgyan, J. (2015). viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology 479–480:85–103. doi: 10.1016/j.virol.2015.02.028
Cui, X., Li, G., Wang, D., Hu, D., and Zhou, X. (2005). A Begomovirus DNAbeta-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J. Virol. 79, 10764–10775. doi: 10.1128/JVI.79.16.10764-10775.2005
Cui, X., Tao, X. R., Xie, Y., Fauquet, C., and Zhou, X. (2004). A DNA beta associated with tomato yellow leaf curl China virus is required for symptom induction. J. Virol. 78, 13966–13974. doi: 10.1128/Pvi..78.24.13966-13964.2004
Dadami, E., Boutla, A., Vrettos, N., Tzortzakaki, S., Karakasilioti, I., and Kalantidis, K. (2013). DICER-LIKE 4 but not DICER-LIKE 2 may have a positive effect on potato spindle tuber viroid accumulation in Nicotiana benthamiana. Mol. Plant 6, 232–234. doi: 10.1093/mp/sss118
Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D. C. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101, 543–553. doi: 10.1016/s0092-8674(00)80864-8
Dalmay, T., Horsefield, R., Braunstein, T. H., and Baulcombe, D. C. (2001). SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 20, 2069–2078. doi: 10.1093/emboj/20.8.2069
de Alba, A. E. M., Flores, R., and Hernandez, C. (2002). Two chloroplastic viroids induce the accumulation of small RNAs associated with posttranscriptional gene silencing. J. Virol. 76, 13094–13096. doi: 10.1128/Jvi.76.24.13094-13096.2002
Deleris, A., Gallego-Bartolome, J., Bao, J., Kasschau, K. D., Carrington, J. C., and Voinnet, O. (2006). Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313, 68–71. doi: 10.1126/science.1128214
Di Serio, F., Martinez de Alba, A. E., Navarro, B., Gisel, A., and Flores, R. (2010). RNA-dependent RNA polymerase 6 delays accumulation and precludes meristem invasion of a viroid that replicates in the nucleus. J. Virol. 84, 2477–2489. doi: 10.1128/JVI.02336-09
Diaz-Pendon, J. A., Li, F., Li, W. X., and Ding, S. W. (2007). Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19, 2053–2063. doi: 10.1105/tpc.106.047449
Ding, B. (2009). The biology of viroid-host interactions. Annu. Rev. Phytopathol. 47, 105–131. doi: 10.1146/annurev-phyto-080508-081927
Ding, B., and Itaya, A. (2007). Viroid: a useful model for studying the basic principles of infection and RNA biology. Mol. Plant Microbe Interact. 20, 7–20. doi: 10.1094/MPMI-20-0007
Ding, S. W., and Voinnet, O. (2007). Antiviral immunity directed by small RNAs. Cell 130, 413–426. doi: 10.1016/j.cell.2007.07.039
Donaire, L., Wang, Y., Gonzalez-Ibeas, D., Mayer, K. F., Aranda, M. A., and Llave, C. (2009). Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology 392, 203–214. doi: 10.1016/j.virol.2009.07.005
Dou, D., and Zhou, J. M. (2012). Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12, 484–495. doi: 10.1016/j.chom.2012.09.003
Du, Q., Duan, C., Zhang, Z., Fang, Y., Fang, R. X., Xie, Q., et al. (2007). DCL4 targets Cucumber mosaic virus satellite RNA at novel secondary structures. J. Virol. 81, 9142–9151. doi: 10.1128/Jvi.02885-06
Eamens, A. L., Smith, N. A., Dennis, E. S., Wassenegger, M., and Wang, M. B. (2014). In Nicotiana species, an artificial microRNA corresponding to the virulence modulating region of Potato spindle tuber viroid directs RNA silencing of a soluble inorganic pyrophosphatase gene and the development of abnormal phenotypes. Virology 450–451, 266–277. doi: 10.1016/j.virol.2013.12.019
Fahlgren, N., Bollmann, S. R., Kasschau, K. D., Cuperus, J. T., Press, C. M., Sullivan, C. M., et al. (2013). Phytophthora have distinct endogenous small RNA populations that include short interfering and microRNAs. PLoS ONE 8:e77181. doi: 10.1371/journal.pone.0077181
Fahlgren, N., Howell, M. D., Kasschau, K. D., Chapman, E. J., Sullivan, C. M., Cumbie, J. S., et al. (2007). High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2:e219. doi: 10.1371/journal.pone.0000219
Fei, Q., Li, P., Teng, C., and Meyers, B. C. (2015). Secondary siRNAs from medicago NB-LRRs modulated via miRNA-target interactions and their abundances. Plant J. 83, 451–465. doi: 10.1111/tpj.12900
Feng, F., and Zhou, J. M. (2012). Plant-bacterial pathogen interactions mediated by type III effectors. Curr. Opin. Plant Biol. 15, 469–476. doi: 10.1016/j.pbi.2012.03.004
Fusaro, A. F., Matthew, L., Smith, N. A., Curtin, S. J., Dedic-Hagan, J., Ellacott, G. A., et al. (2006). RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 7, 1168–1175. doi: 10.1038/sj.embor.7400837
Gao, R., Liu, P., and Wong, S. M. (2012). Identification of a plant viral RNA genome in the nucleus. PLoS ONE 7:e48736. doi: 10.1371/journal.pone.0048736
Garcia-Ruiz, H., Carbonell, A., Hoyer, J. S., Fahlgren, N., Gilbert, K. B., Takeda, A., et al. (2015). Roles and programming of Arabidopsis ARGONAUTE proteins during Turnip mosaic virus infection. PLoS Pathog. 11:e1004755. doi: 10.1371/journal.ppat.1004755
Garcia-Ruiz, H., Takeda, A., Chapman, E. J., Sullivan, C. M., Fahlgren, N., Brempelis, K. J., et al. (2010). Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip mosaic virus infection. Plant Cell 22, 481–496. doi: 10.1105/tpc.109.073056
Ghildiyal, M., and Zamore, P. D. (2009). Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10, 94–108. doi: 10.1038/nrg2504
Gomez, G., Martinez, G., and Pallas, V. (2008). Viroid-induced symptoms in Nicotiana benthamiana plants are dependent on RDR6 activity. Plant Physiol. 148, 414–423. doi: 10.1104/pp.108.120808
Gomez, G., and Pallas, V. (2007). Mature monomeric forms of Hop stunt viroid resist RNA silencing in transgenic plants. Plant J. 51, 1041–1049. doi: 10.1111/j.1365-313X.2007.03203.x
Guo, N., Ye, W. W., Wu, X. L., Shen, D. Y., Wang, Y. C., Xing, H., et al. (2011). Microarray profiling reveals microRNAs involving soybean resistance to Phytophthora sojae. Genome 54, 954–958. doi: 10.1139/g11-050
Gupta, O. P., Permar, V., Koundal, V., Singh, U. D., and Praveen, S. (2012). MicroRNA regulated defense responses in Triticum aestivum L. during Puccinia graminis f.sp. tritici infection. Mol. Biol. Rep. 39, 817–824. doi: 10.1007/s11033-011-0803-5
Hamilton, A. J., and Baulcombe, D. C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. doi: 10.1126/science.286.5441.950
Hammann, C., and Steger, G. (2012). Viroid-specific small RNA in plant disease. RNA Biol. 9, 809–819. doi: 10.4161/rna.19810
Harvey, J. J., Lewsey, M. G., Patel, K., Westwood, J., Heimstadt, S., Carr, J. P., et al. (2011). An antiviral defense role of AGO2 in plants. PLoS ONE 6:e14639. doi: 10.1371/journal.pone.0014639
He, X., Fang, Y., Feng, L., and Guo, H. (2008). Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett. 582, 2445–2452. doi: 10.1016/j.febslet.2008.06.011
Ho, T., Pallett, D., Rusholme, R., Dalmay, T., and Wang, H. (2006). A simplified method for cloning of short interfering RNAs from Brassica juncea infected with Turnip mosaic potyvirus and Turnip crinkle carmovirus. J. Virol. Methods 136, 217–223. doi: 10.1016/j.jviromet.2006.05.016
Holoch, D., and Moazed, D. (2015). RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16, 71–84. doi: 10.1038/nrg3863
Hong, W., Qian, D., Sun, R., Jiang, L., Wang, Y., Wei, C., et al. (2015). OsRDR6 plays role in host defense against double-stranded RNA virus, Rice Dwarf Phytoreovirus. Sci. Rep. 5:11324. doi: 10.1038/srep11324
Hou, W., Duan, C., Fang, R., Zhou, X., and Guo, H. (2011). Satellite RNA reduces expression of the 2b suppressor protein resulting in the attenuation of symptoms caused by Cucumber mosaic virus infection. Mol. Plant Pathol. 12, 595–605. doi: 10.1111/j.1364-3703.2010.00696.x
Ibrahim, H. M., Alkharouf, N. W., Meyer, S. L., Aly, M. A., Gamal El-Din Ael, K., Hussein, E. H., et al. (2011). Post-transcriptional gene silencing of root-knot nematode in transformed soybean roots. Exp. Parasitol. 127, 90–99. doi: 10.1016/j.exppara.2010.06.037
Itaya, A., Bundschuh, R., Archual, A. J., Joung, J. G., Fei, Z., Dai, X., et al. (2008). Small RNAs in tomato fruit and leaf development. Biochim. Biophys. Acta 1779, 99–107. doi: 10.1016/j.bbagrm.2007.09.003
Itaya, A., Folimonov, A., Matsuda, Y., Nelson, R. S., and Ding, B. (2001). Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Mol. Plant Microbe Interact. 14, 1332–1334. doi: 10.1094/Mpmi.2001.14.11.1332
Itaya, A., Zhong, X., Bundschuh, R., Qi, Y., Wang, Y., Takeda, R., et al. (2007). A structured viroid RNA serves as a substrate for dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. J. Virol. 81, 2980–2994. doi: 10.1128/JVI.02339-06
Jackel, J. N., Buchmann, R. C., Singhal, U., and Bisaro, D. M. (2015). Analysis of geminivirus AL2 and L2 proteins reveals a novel AL2 silencing suppressor activity. J. Virol. 89, 3176–3187. doi: 10.1128/JVI.02625-14
Jaubert, M., Bhattacharjee, S., Mello, A. F., Perry, K. L., and Moffett, P. (2011). ARGONAUTE2 mediates RNA-silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiol. 156, 1556–1564. doi: 10.1104/pp.111.178012
Jiang, L., Qian, D., Zheng, H., Meng, L. Y., Chen, J., Le, W. J., et al. (2012). RNA-dependent RNA polymerase 6 of rice (Oryza sativa) plays role in host defense against negative-strand RNA virus, Rice stripe virus. Virus Res. 163, 512–519. doi: 10.1016/j.virusres.2011.11.016
Jiang, N., Yang, Y. P., Janbon, G., Pan, J., and Zhu, X. D. (2012). Identification and functional demonstration of miRNAs in the fungus Cryptococcus neoformans. PLoS ONE 7:e52734. doi: 10.1371/journal.pone.0052734.
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jose, J., and Usha, R. (2003). Bhendi yellow vein mosaic disease in India is caused by association of a DNA beta satellite with a Begomovirus. Virology 305, 310–317. doi: 10.1006/viro.2002.1768
Katiyar-Agarwal, S., Gao, S., Vivian-Smith, A., and Jin, H. (2007). A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 21, 3123–3134. doi: 10.1101/gad.1595107
Katiyar-Agarwal, S., and Jin, H. (2010). Role of small RNAs in host-microbe interactions. Annu. Rev. Phytopathol. 48, 225–246. doi: 10.1146/annurev-phyto-073009-114457
Katiyar-Agarwal, S., Morgan, R., Dahlbeck, D., Borsani, O., Villegas, A. Jr., Zhu, J. K., et al. (2006). A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. U.S.A. 103, 18002–18007. doi: 10.1073/pnas.0608258103
Koch, A., and Kogel, K. H. (2014). New wind in the sails: improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol. J. 12, 821–831. doi: 10.1111/pbi.12226
Koch, A., Kumar, N., Weber, L., Keller, H., Imani, J., and Kogel, K. H. (2013). Host-induced gene silencing of cytochrome P450 lanosterol C14alpha-demethylase-encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. U.S.A. 110, 19324–19329. doi: 10.1073/pnas.1306373110
Kovalskaya, N., and Hammond, R. W. (2014). Molecular biology of viroid-host interactions and disease control strategies. Plant Sci. 228, 48–60. doi: 10.1016/j.plantsci.2014.05.006
Lee, H. C., Li, L., Gu, W., Xue, Z., Crosthwaite, S. K., Pertsemlidis, A., et al. (2010). Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol. Cell 38, 803–814. doi: 10.1016/j.molcel.2010.04.005
Li, F., Huang, C., Li, Z., and Zhou, X. (2014). Suppression of RNA silencing by a plant DNA virus satellite requires a host calmodulin-like protein to repress RDR6 expression. PLoS Pathog. 10:e1003921. doi: 10.1371/journal.ppat.1003921
Li, F., Pignatta, D., Bendix, C., Brunkard, J. O., Cohn, M. M., Tung, J., et al. (2012). MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. U.S.A. 109, 1790–1795. doi: 10.1073/pnas.1118282109
Li, F., Xu, X., Huang, C., Gu, Z., Cao, L., Hu, T., et al. (2015). The AC5 protein encoded by Mungbean yellow mosaic India virus is a pathogenicity determinant that suppresses RNA silencing-based antiviral defenses. New Phytol. 208, 555–569. doi: 10.1111/nph.13473
Li, Y., Zhang, Q., Zhang, J., Wu, L., Qi, Y., and Zhou, J. M. (2010). Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 152, 2222–2231. doi: 10.1104/pp.109.151803
Liu, J., Cheng, X., Liu, D., Xu, W., Wise, R., and Shen, Q. H. (2014). The miR9863 family regulates distinct Mla alleles in barley to attenuate NLR receptor-triggered disease resistance and cell-death signaling. PLoS Genet. 10:e1004755. doi: 10.1371/journal.pgen.1004755
Liu, Q., and Paroo, Z. (2010). Biochemical principles of small RNA pathways. Annu. Rev. Biochem. 79, 295–319. doi: 10.1146/annurev.biochem.052208.151733
Machida, S., Yamahata, N., Watanuki, H., Owens, R. A., and Sano, T. (2007). Successive accumulation of two size classes of viroid-specific small RNA in potato spindle tuber viroid-infected tomato plants. J. Gen. Virol. 88(Pt. 12), 3452–3457. doi: 10.1099/vir.0.83228-0
Manfre, A. J., and Simon, A. E. (2008). Importance of coat protein and RNA silencing in satellite RNA/virus interactions. Virology 379, 161–167. doi: 10.1016/j.virol.2008.06.011
Mao, Y. B., Cai, W. J., Wang, J. W., Hong, G. J., Tao, X. Y., Wang, L. J., et al. (2007). Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. doi: 10.1038/nbt1352
Markarian, N., Li, H. W., Ding, S. W., and Semancik, J. S. (2004). RNA silencing as related to viroid induced symptom expression. Arch. Virol. 149, 397–406. doi: 10.1007/s00705-003-0215-5
Martin, R., Arenas, C., Daros, J. A., Covarrubias, A., Reyes, J. L., and Chua, N. H. (2007). Characterization of small RNAs derived from Citrus exocortis viroid (CEVd) in infected tomato plants. Virology 367, 135–146. doi: 10.1016/j.virol.2007.05.011
Martinez, G., Castellano, M., Tortosa, M., Pallas, V., and Gomez, G. (2014). A pathogenic non-coding RNA induces changes in dynamic DNA methylation of ribosomal RNA genes in host plants. Nucleic Acids Res. 42, 1553–1562. doi: 10.1093/nar/gkt968
McEwan, D. L., Weisman, A. S., and Hunter, C. P. (2012). Uptake of extracellular double-stranded RNA by SID-2. Mol. Cell 47, 746–754. doi: 10.1016/j.molcel.2012.07.014
Minoia, S., Carbonell, A., Di Serio, F., Gisel, A., Carrington, J. C., Navarro, B., et al. (2014). Specific argonautes selectively bind small RNAs derived from potato spindle tuber viroid and attenuate viroid accumulation in vivo. J. Virol. 88, 11933–11945. doi: 10.1128/JVI.01404-14
Molnar, A., Csorba, T., Lakatos, L., Varallyay, E., Lacomme, C., and Burgyan, J. (2005). Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J. Virol. 79, 7812–7818. doi: 10.1128/JVI.79.12.7812-7818.2005
Morel, J. B., Godon, C., Mourrain, P., Beclin, C., Boutet, S., Feuerbach, F., et al. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14, 629–639. doi: 10.1105/tpc.010358
Mourrain, P., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J. B., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542. doi: 10.1016/S0092-8674(00)80863-6
Muangsan, N., Beclin, C., Vaucheret, H., and Robertson, D. (2004). Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants. Plant J. 38, 1004–1014. doi: 10.1111/j.1365-313X.2004.02103.x
Murata, T., Kadotani, N., Yamaguchi, M., Tosa, Y., Mayama, S., and Nakayashiki, H. (2007). siRNA-dependent and -independent post-transcriptional cosuppression of the LTR-retrotransposon MAGGY in the phytopathogenic fungus Magnaporthe oryzae. Nucleic Acids Res. 35, 5987–5994. doi: 10.1093/nar/gkm646
Navarro, B., Gisel, A., Rodio, M. E., Delgado, S., Flores, R., and Di Serio, F. (2012a). Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 70, 991–1003. doi: 10.1111/j.1365-313X.2012.04940.x.
Navarro, B., Gisel, A., Rodio, M. E., Delgado, S., Flores, R., and Di Serio, F. (2012b). Viroids: How to infect a host and cause disease without encoding proteins. Biochimie 94, 1474–1480. doi: 10.1016/j.biochi.2012.02.020
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., et al. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. doi: 10.1126/science.1126088
Navarro, L., Jay, F., Nomura, K., He, S. Y., and Voinnet, O. (2008). Suppression of the microRNA pathway by bacterial effector proteins. Science 321, 964–967. doi: 10.1126/science.1159505
Niu, D., Lii, Y. E., Chellappan, P., Lei, L., Peralta, K., Jiang, C., et al. (2016). miRNA863-3p sequentially targets negative immune regulator ARLPKs and positive regulator SERRATE upon bacterial infection. Nat. Commun. 7:11324. doi: 10.1038/ncomms11324
Nowara, D., Gay, A., Lacomme, C., Shaw, J., Ridout, C., Douchkov, D., et al. (2010). HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22, 3130–3141. doi: 10.1105/tpc.110.077040
Nunes, C. C., Gowda, M., Sailsbery, J., Xue, M., Chen, F., Brown, D. E., et al. (2011). Diverse and tissue-enriched small RNAs in the plant pathogenic fungus, Magnaporthe oryzae. BMC Genomics 12:288. doi: 10.1186/1471-2164-12-288
Panwar, V., McCallum, B., and Bakkeren, G. (2013). Host-induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the Barley stripe mosaic virus. Plant Mol. Biol. 81, 595–608. doi: 10.1007/s11103-013-0022-7
Papaefthimiou, I., Hamilton, A. J., Denti, M. A., Baulcombe, D. C., Tsagris, M., and Tabler, M. (2001). Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that ave characteristic of post-transcriptional gene silencing. Nucleic Acids Res. 29, 2395–2400. doi: 10.1093/nar/29.11.2395
Qi, X., Bao, F., and Xie, Z. (2009). Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS ONE 4:e4971. doi: 10.1371/journal.pone.0004971
Qi, Y., and Ding, B. (2003). Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA. Plant Cell 15, 1360–1374. doi: 10.1105/tpc.011585
Qi, Y., Zhong, X., Itaya, A., and Ding, B. (2004). Dissecting RNA silencing in protoplasts uncovers novel effects of viral suppressors on the silencing pathway at the cellular level. Nucleic Acids Res. 32:e179. doi: 10.1093/nar/gnh180.
Qiao, Y., Liu, L., Xiong, Q., Flores, C., Wong, J., Shi, J., et al. (2013). Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 45, 330–333. doi: 10.1038/ng.2525
Qiao, Y., Shi, J., Zhai, Y., Hou, Y., and Ma, W. (2015). Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc. Natl. Acad. Sci. U.S.A. 112, 5850–5855. doi: 10.1073/pnas.1421475112
Qu, F., Ye, X., and Morris, T. J. (2008). Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. U.S.A. 105, 14732–14737. doi: 10.1073/pnas.0805760105
Qutob, D., Chapman, B. P., and Gijzen, M. (2013). Transgenerational gene silencing causes gain of virulence in a plant pathogen. Nat. Commun. 4:1349. doi: 10.1038/ncomms2354.
Raja, P., Jackel, J. N., Li, S., Heard, I. M., and Bisaro, D. M. (2014). Arabidopsis double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against geminiviruses. J. Virol. 88, 2611–2622. doi: 10.1128/JVI.02305-13
Raja, P., Sanville, B. C., Buchmann, R. C., and Bisaro, D. M. (2008). Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 82, 8997–9007. doi: 10.1128/JVI.00719-08
Raman, V., Simon, S. A., Romag, A., Demirci, F., Mathioni, S. M., Zhai, J., et al. (2013). Physiological stressors and invasive plant infections alter the small RNA transcriptome of the rice blast fungus, Magnaporthe oryzae. BMC Genomics 14:326. doi: 10.1186/1471-2164-14-326
Ren, B., Guo, Y., Gao, F., Zhou, P., Wu, F., Meng, Z., et al. (2010). Multiple functions of rice dwarf phytoreovirus Pns10 in suppressing systemic RNA silencing. J. Virol. 84, 12914–12923. doi: 10.1128/JVI.00864-10
Rodio, M. E., Delgado, S., De Stradis, A., Gomez, M. D., Flores, R., and Di Serio, F. (2007). A viroid RNA with a specific structural motif inhibits chloroplast development. Plant Cell 19, 3610–3626. doi: 10.1105/tpc.106.049775
Rodriguez-Negrete, E., Lozano-Duran, R., Piedra-Aguilera, A., Cruzado, L., Bejarano, E. R., and Castillo, A. G. (2013). Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol. 199, 464–475. doi: 10.1111/nph.12286
Rodriguez-Negrete, E. A., Carrillo-Tripp, J., and Rivera-Bustamante, R. F. (2009). RNA silencing against geminivirus: complementary action of posttranscriptional gene silencing and transcriptional gene silencing in host recovery. J. Virol. 83, 1332–1340. doi: 10.1128/JVI.01474-08
Rogers, K., and Chen, X. (2013). Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25, 2383–2399. doi: 10.1105/tpc.113.113159
Sampson, T. R., Saroj, S. D., Llewellyn, A. C., Tzeng, Y. L., and Weiss, D. S. (2013). A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 497, 254–257. doi: 10.1038/nature12048
Schmidtke, C., Abendroth, U., Brock, J., Serrania, J., Becker, A., and Bonas, U. (2013). Small RNA sX13: a multifaceted regulator of virulence in the plant pathogen Xanthomonas. PLoS Pathog. 9:e1003626. doi: 10.1371/journal.ppat.1003626
Schmidtke, C., Findeiss, S., Sharma, C. M., Kuhfuss, J., Hoffmann, S., Vogel, J., et al. (2012). Genome-wide transcriptome analysis of the plant pathogen Xanthomonas identifies sRNAs with putative virulence functions. Nucleic Acids Res. 40, 2020–2031. doi: 10.1093/nar/gkr904
Schu, D. J., Zhang, A., Gottesman, S., and Storz, G. (2015). Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J. 34, 2557–2573. doi: 10.15252/embj.201591569
Schuck, J., Gursinsky, T., Pantaleo, V., Burgyan, J., and Behrens, S. E. (2013). AGO/RISC-mediated antiviral RNA silencing in a plant in vitro system. Nucleic Acids Res. 41, 5090–5103. doi: 10.1093/nar/gkt193
Schwind, N., Zwiebel, M., Itaya, A., Ding, B., Wang, M. B., Krczal, G., et al. (2009). RNAi-mediated resistance to Potato spindle tuber viroid in transgenic tomato expressing a viroid hairpin RNA construct. Mol. Plant Pathol. 10, 459–469. doi: 10.1111/j.1364-3703.2009.00546.x
Shi, B., Lin, L., Wang, S., Guo, Q., Zhou, H., Rong, L., et al. (2016). Identification and regulation of host genes related to Rice stripe virus symptom production. New Phytol. 209, 1106–1119. doi: 10.1111/nph.13699
Shih, J. D., and Hunter, C. P. (2011). SID-1 is a dsRNA-selective dsRNA-gated channel. RNA 17, 1057–1065. doi: 10.1261/rna.2596511
Shimura, H., Pantaleo, V., Ishihara, T., Myojo, N., Inaba, J., Sueda, K., et al. (2011). A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog. 7:e1002021. doi: 10.1371/journal.ppat.1002021
Shivaprasad, P. V., Chen, H. M., Patel, K., Bond, D. M., Santos, B. A., and Baulcombe, D. C. (2012). A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24, 859–874. doi: 10.1105/tpc.111.095380
Sijen, T., Steiner, F. A., Thijssen, K. L., and Plasterk, R. H. (2007). Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315, 244–247. doi: 10.1126/science.1136699
Simon, A. E., Roossinck, M. J., and Havelda, Z. (2004). Plant virus satellite and defective interfering RNAs: new paradigms for a new century. Annu. Rev. Phytopathol. 42, 415–437. doi: 10.1146/annurev.phyto.42.040803.140402
Smith, N. A., Eamens, A. L., and Wang, M. B. (2011). Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 7:e1002022. doi: 10.1371/journal.ppat.1002022
St-Pierre, P., Hassen, I. F., Thompson, D., and Perreault, J. P. (2009). Characterization of the siRNAs associated with peach latent mosaic viroid infection. Virology 383, 178–182. doi: 10.1016/j.virol.2008.11.008
Takeda, A., Iwasaki, S., Watanabe, T., Utsumi, M., and Watanabe, Y. (2008). The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 49, 493–500. doi: 10.1093/pcp/pcn043
Tian, D., Traw, M. B., Chen, J. Q., Kreitman, M., and Bergelson, J. (2003). Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423, 74–77. doi: 10.1038/nature01588
Trinks, D., Rajeswaran, R., Shivaprasad, P. V., Akbergenov, R., Oakeley, E. J., Veluthambi, K., et al. (2005). Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J. Virol. 79, 2517–2527. doi: 10.1128/JVI.79.4.2517-2527.2005
Verlaan, M. G., Hutton, S. F., Ibrahem, R. M., Kormelink, R., Visser, R. G. F., Scott, J. W., et al. (2013). The tomato yellow leaf curl virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-class RNA-dependent RNA polymerases. PLoS Genet. 9:e1003399. doi: 10.1371/journal.pgen.1003399
Vetukuri, R. R., Asman, A. K., Tellgren-Roth, C., Jahan, S. N., Reimegard, J., Fogelqvist, J., et al. (2012). Evidence for small RNAs homologous to effector-encoding genes and transposable elements in the oomycete Phytophthora infestans. PLoS ONE 7:e51399. doi: 10.1371/journal.pone.0051399
Vetukuri, R. R., Avrova, A. O., Grenville-Briggs, L. J., Van West, P., Soderbom, F., Savenkov, E. I., et al. (2011). Evidence for involvement of Dicer-like, Argonaute and histone deacetylase proteins in gene silencing in Phytophthora infestans. Mol. Plant Pathol. 12, 772–785. doi: 10.1111/j.1364-3703.2011.00710.x
Viswanathan, C., Anburaj, J., and Prabu, G. (2014). Identification and validation of sugarcane streak mosaic virus-encoded microRNAs and their targets in sugarcane. Plant Cell Rep. 33, 265–276. doi: 10.1007/s00299-013-1527-x
Vogt, U., Pelissier, T., Putz, A., Razvi, F., Fischer, R., and Wassenegger, M. (2004). Viroid-induced RNA silencing of GFP-viroid fusion transgenes does not induce extensive spreading of methylation or transitive silencing. Plant J. 38, 107–118. doi: 10.1111/j.1365-313X.2004.02029.x
Walters, D., and Heil, M. (2007). Costs and trade-offs associated with induced resistance. Physiol. Mol. Plant Pathol. 71, 3–17. doi: 10.1016/j.pmpp.2007.09.008
Wang, B., Li, F., Huang, C., Yang, X., Qian, Y., Xie, Y., et al. (2014). V2 of tomato yellow leaf curl virus can suppress methylation-mediated transcriptional gene silencing in plants. J. Gen. Virol. 95(Pt 1), 225–230. doi: 10.1099/vir.0.055798-0
Wang, H., Hao, L., Shung, C. Y., Sunter, G., and Bisaro, D. M. (2003). Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell 15, 3020–3032. doi: 10.1105/tpc.015180
Wang, M., Bian, X., Wu, L., Liu, L., Smith, N. A., Isenegger, D., et al. (2004). On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. U.S.A. 101, 3275–3280. doi: 10.1073/pnas.0400104101
Wang, X. B., Jovel, J., Udomporn, P., Wang, Y., Wu, Q., Li, W. X., et al. (2011). The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 23, 1625–1638. doi: 10.1105/tpc.110.082305
Wang, X., Wu, Q., Ito, T., Cillo, F., Li, W., Chen, X., et al. (2010). RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 484–489. doi: 10.1073/pnas.0904086107
Wang, Y., Itaya, A., Zhong, X., Wu, Y., Zhang, J., van der Knaap, E., et al. (2011a). Function and evolution of a MicroRNA that regulates a Ca2+-ATPase and triggers the formation of phased small interfering RNAs in tomato reproductive growth. Plant Cell 23, 3185–3203. doi: 10.1105/tpc.111.088013
Wang, Y., Shibuya, M., Taneda, A., Kurauchi, T., Senda, M., Owens, R. A., et al. (2011b). Accumulation of Potato spindle tuber viroid-specific small RNAs is accompanied by specific changes in gene expression in two tomato cultivars. Virology 413, 72–83. doi: 10.1016/j.virol.2011.01.021
Wassenegger, M., Heimes, S., Riedel, L., and Sanger, H. L. (1994). Rna-directed de-novo methylation of genomic sequences in plants. Cell 76, 567–576. doi: 10.1016/0092-8674(94)90119-8
Weiberg, A., Wang, M., Bellinger, M., and Jin, H. (2014). Small RNAs: a new paradigm in plant-microbe interactions. Annu. Rev. Phytopathol. 52, 495–516. doi: 10.1146/annurev-phyto-102313-045933
Weiberg, A., Wang, M., Lin, F. M., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
Wiedenheft, B., Sternberg, S. H., and Doudna, J. A. (2012). RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338. doi: 10.1038/nature10886
Wu, J., Yang, Z., Wang, Y., Zheng, L., Ye, R., Ji, Y., et al. (2015). Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. Elife 4:e05733. doi: 10.7554/eLife.05733
Xia, K., Wang, R., Ou, X., Fang, Z., Tian, C., Duan, J., et al. (2012). OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE 7:e30039. doi: 10.1371/journal.pone.0030039
Xia, Z., Peng, J., Li, Y., Chen, L., Li, S., Zhou, T., et al. (2014). Characterization of small interfering RNAs derived from Sugarcane mosaic virus in infected maize plants by deep sequencing. PLoS ONE 9:e97013. doi: 10.1371/journal.pone.0097013
Xie, Z., Fan, B., Chen, C., and Chen, Z. (2001). An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proc. Natl. Acad. Sci. U.S.A. 98, 6516–6521. doi: 10.1073/pnas.111440998
Xie, Z., Johansen, L. K., Gustafson, A. M., Kasschau, K. D., Lellis, A. D., Zilberman, D., et al. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2:E104. doi: 10.1371/journal.pbio.0020104
Xin, M., Wang, Y., Yao, Y., Xie, C., Peng, H., Ni, Z., et al. (2010). Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 10:123. doi: 10.1186/1471-2229-10-123
Xiong, Q., Ye, W., Choi, D., Wong, J., Qiao, Y., Tao, K., et al. (2014). Phytophthora suppressor of RNA silencing 2 is a conserved RxLR effector that promotes infection in soybean and Arabidopsis thaliana. Mol. Plant Microbe Interact. 27, 1379–1389. doi: 10.1094/Mpmi-06-14-0190-R
Yan, F., Zhang, H. M., Adams, M. J., Yang, J., Peng, J. J., Antoniw, J. F., et al. (2010). Characterization of siRNAs derived from rice stripe virus in infected rice plants by deep sequencing. Arch. Virol. 155, 935–940. doi: 10.1007/s00705-010-0670-8
Yang, L., Mu, X., Liu, C., Cai, J., Shi, K., Zhu, W., et al. (2015). Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J. Integr. Plant Biol. 57, 1078–1088. doi: 10.1111/jipb.12348
Yang, S., Carter, S. A., Cole, A. B., Cheng, N. H., and Nelson, R. S. (2004). A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc. Natl. Acad. Sci. U.S.A. 101, 6297–6302. doi: 10.1073/pnas0304346101
Yang, X., Wang, Y., Guo, W., Xie, Y., Xie, Q., Fan, L., et al. (2011a). Characterization of small interfering RNAs derived from the geminivirus/betasatellite complex using deep sequencing. PLoS ONE 6:e16928. doi: 10.1371/journal.pone.0016928
Yang, X., Xie, Y., Raja, P., Li, S., Wolf, J. N., Shen, Q., et al. (2011b). Suppression of methylation-mediated transcriptional gene silencing by betaC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog. 7:e1002329. doi: 10.1371/journal.ppat.1002329
Yi, H., and Richards, E. J. (2007). A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell 19, 2929–2939. doi: 10.1105/tpc.107.051821
Ying, X., Dong, L., Zhu, H., Duan, C., Du, Q., Lv, D., et al. (2010). RNA-dependent RNA polymerase 1 from Nicotiana tabacum suppresses RNA silencing and enhances viral infection in Nicotiana benthamiana. Plant Cell 22, 1358–1372. doi: 10.1105/tpc.109.072058
Yong Chung, H., Lacatus, G., and Sunter, G. (2014). Geminivirus AL2 protein induces expression of, and interacts with, a calmodulin-like gene, an endogenous regulator of gene silencing. Virology 460–461, 108–118. doi: 10.1016/j.virol.2014.04.034
Yoo, B. C., Kragler, F., Varkonyi-Gasic, E., Haywood, V., Archer-Evans, S., Lee, Y. M., et al. (2004). A systemic small RNA signaling system in plants. Plant Cell 16, 1979–2000. doi: 10.1105/tpc.104.023614
Yu, D., Fan, B., MacFarlane, S. A., and Chen, Z. (2003). Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol. Plant Microbe Interact. 16, 206–216. doi: 10.1094/Mpmi.2003.16.3.206
Zeng, Q., McNally, R. R., and Sundin, G. W. (2013). Global small RNA chaperone Hfq and regulatory small RNAs are important virulence regulators in Erwinia amylovora. J. Bacteriol. 195, 1706–1717. doi: 10.1128/JB.02056-12
Zeng, Q., and Sundin, G. W. (2014). Genome-wide identification of Hfq-regulated small RNAs in the fire blight pathogen Erwinia amylovora discovered small RNAs with virulence regulatory function. BMC Genomics 15:414. doi: 10.1186/1471-2164-15-414
Zhai, J., Jeong, D. H., De Paoli, E., Park, S., Rosen, B. D., Li, Y., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25, 2540–2553. doi: 10.1101/gad.177527.111
Zhang, F., and Simon, A. E. (2003). Enhanced viral pathogenesis associated with a virulent mutant virus or a virulent satellite RNA correlates with reduced virion accumulation and abundance of free coat protein. Virology 312, 8–13. doi: 10.1016/S0042-6822(03)00345-3
Zhang, H., Tao, Z., Hong, H., Chen, Z., Wu, C., Li, X., et al. (2016). Transposon-derived small RNA is responsible for modified function of WRKY45 locus. Nat. Plants 2:16016. doi: 10.1038/nplants.2016.16
Zhang, M., Zhao, H., Xie, S., Chen, J., Xu, Y., Wang, K., et al. (2011). Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc. Natl. Acad. Sci. U.S.A. 108, 20042–20047. doi: 10.1073/pnas.1112186108
Zhang, W., Gao, S., Zhou, X., Chellappan, P., Chen, Z., Zhang, X., et al. (2011). Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol. Biol. 75, 93–105. doi: 10.1007/s11103-010-9710-8
Zhang, X., Xia, J., Lii, Y., Barrera-Figueroa, B., Zhou, X., Gao, S., et al. (2012). Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol. 13, R20. doi: 10.1186/gb-2012-13-3-r20
Zhang, X., Zhao, H., Gao, S., Wang, W., Katiyar-Agarwal, S., Huang, H., et al. (2011). Arabidopsis argonaute 2 regulates innate immunity via miRNA393*-mediated silencing of a golgi-localized SNARE gene, MEMB12. Mol. Cell 42, 356–366. doi: 10.1016/j.molcel.2011.04.010
Zhang, X., Yuan, Y., Pei, Y., Lin, S., Tuschl, T., Patel, D. J., et al. (2006). Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20, 3255–3268. doi: 10.1101/gad.1495506
Zhang, Z., Chen, H., Huang, X., Xia, R., Zhao, Q., Lai, J., et al. (2011). BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 23, 273–288. doi: 10.1105/tpc.110.081695
Zhu, H., Duan, C., Hou, W., Du, Q., Lv, D., Fang, R., et al. (2011). Satellite RNA-derived small interfering RNA satsiR-12 targeting the 3’ untranslated region of Cucumber mosaic virus triggers viral RNAs for degradation. J. Virol. 85, 13384–13397. doi: 10.1128/JVI.05806-11
Keywords: small RNA, RNA silencing, plant immunity, pathogen virulence, plant–pathogen interaction
Citation: Huang J, Yang M, Lu L and Zhang X (2016) Diverse Functions of Small RNAs in Different Plant–Pathogen Communications. Front. Microbiol. 7:1552. doi: 10.3389/fmicb.2016.01552
Received: 25 July 2016; Accepted: 16 September 2016;
Published: 04 October 2016.
Edited by:
Jianzhong Liu, Zhejiang Normal University, ChinaReviewed by:
Kriton Kalantidis, Foundation for Research and Technology Hellas, GreeceYongli Qiao, Institute of Crop Science (CAAS), China
Copyright © 2016 Huang, Yang, Lu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoming Zhang, emhhbmd4bUBpb3ouYWMuY24= Lu Lu, ZG91emlwa3VAZ21haWwuY29t
†These authors have contributed equally to this work.
 Juan Huang
Juan Huang Meiling Yang
Meiling Yang Lu Lu
Lu Lu Xiaoming Zhang
Xiaoming Zhang