- Sécurité et Qualité des Produits d’Origine Végétale, UMR0408, Avignon Université, Institut National de la Recherche Agronomique, Avignon, France
Bacillus cereus is a food-borne pathogen that causes diarrheal disease in humans. After ingestion, B. cereus experiences in the human gastro-intestinal tract abiotic physical variables encountered in food, such as acidic pH in the stomach and changing oxygen conditions in the human intestine. B. cereus responds to environmental changing conditions (stress) by reversibly adjusting its physiology to maximize resource utilization while maintaining structural and genetic integrity by repairing and minimizing damage to cellular infrastructure. As reviewed in this article, B. cereus adapts to acidic pH and changing oxygen conditions through diverse regulatory mechanisms and then exploits its metabolic flexibility to grow and produce enterotoxins. We then focus on the intricate link between metabolism, redox homeostasis, and enterotoxins, which are recognized as important contributors of food-borne disease.
Introduction
Micro-organisms display astonishing abilities to survive and grow in hostile environments. Some have adapted their life cycles to extreme conditions (thermophiles, psychrophiles, halophiles, acidophiles, etc.), but all can marshal temporary adaptation mechanisms to help them survive until conditions have become more favorable. When environmental conditions change, a cell must modify its physiology accordingly to cope with them and survive or reproduce. However, adaptation has limits, which depend on the microorganism and on the environmental variables that have become extreme, upset the cell’s equilibrium, and caused stress. Limits to adaptation depend on the micro-organism’s intrinsic capacity to cope; the physiological responses the cell can call on to address environmental variations include changes in metabolism and/or mechanisms of adaptation and resistance.
Bacillus cereus is a Gram-positive, facultative anaerobe, rod-shaped endospore-forming bacterium that is known to inhabit primarily the soil, where it can complete its saprophytic life cycle (Vilain et al., 2006). As a result of its saprophytic soil life cycle, B. cereus is found in water, vegetables, and many other food ingredients, resulting in the contamination of a wide variety of finished food products. Ingestion of contaminated foods by humans can lead to two types of gastrointestinal infections, both damaging the host epithelium. The emetic type of food poisoning is caused by the ingestion of food containing the toxin cereulide, whereas the diarrheal type depends on the ingestion of B. cereus cells followed by the production of virulence factors in the human small intestine (Stenfors Arnesen et al., 2008; Senesi and Ghelardi, 2010; Ceuppens et al., 2012). B. cereus cells originate from ingested vegetative cells that survive gastric passage (Wijnands et al., 2009) and/or from ingested spores which first adhere to the intestinal mucosa and then germinate. Recent evidence suggests that B. cereus-induced diarrhea is not caused by massive B. cereus proliferation and virulence factor production in the intestinal lumen but by localized growth and virulence factor production at the hosts mucus layer or epithelial surface (Ceuppens et al., 2012). These virulence factors then damage the nearby epithelial cells by pore formation, resulting in microvilli damage and osmotic lysis of the host’s epithelial cells and eventually diarrhea (Beecher et al., 1995; Hardy et al., 2001; Minnaard et al., 2001; Lindback et al., 2004; Ramarao and Lereclus, 2006; Fagerlund et al., 2008). In the vicinity of the epithelial layer, B. cereus is exposed to different oxygen concentrations and different oxidoreduction potentials (ORP; Moriarty-Craige and Jones, 2004; Marteyn et al., 2010a,b) that induce compensatory metabolic pathways in an attempt to maintain the intracellular redox state. The cellular redox status governs the status of redox-sensitive macromolecules and protects against endogenous oxidative stress. Recent studies suggested that virulence factor production by B. cereus is dynamic and shaped by cellular oxidation (Madeira et al., 2015). Thus, there is an intricate link between metabolism, redox homeostasis, and virulence factor production. In the first part of this review, we focus on how microorganisms and B. cereus detect and respond to acid stress, and review the different behavioral, physiological and molecular mechanisms underpinning acid stress adaptation. In the second part, we will begin by describing the basics of B. cereus physiology and will then discuss how metabolism and redox global regulators influence the production of virulence factors under changing oxygen conditions.
Acid Stress Resistance
Acid resistance is especially important for B. cereus that must survive the acidic pH of the stomach – which is 1.5 in the fasting state (Van de Guchte et al., 2002) and rises to 3–5 after ingestion of food (Cotter and Hill, 2003) – before entering and colonizing the small intestines or colon (Stenfors Arnesen et al., 2008). Acid stress is also frequently encountered naturally in many foods, as a result of the use of weak organic acids or short-chain (volatile) fatty acids (FA; e.g., acetic acid, citric acid, and propionic acids) as food preservatives (Alvarez-Ordonez et al., 2010a). Thus, the ability to adapt to an acidified environment is crucial to the virulence of a food-borne pathogen such as B. cereus.
Neutrophilic bacteria have evolved multiple tolerance or resistance mechanisms to prevent cell damage due to acid stress; these are generally referred to as acid tolerance responses (ATRs) and acid resistance mechanisms, respectively. Which system(s) plays the dominant role(s) depends on: (i) the phase of growth of the cells when the ATR is elicited [exponential phase- versus stationary phase- ATR]; and/or (ii) whether certain amino acids are present during exposure to the acidic pH; and/or (iii) whether acidification of the environment results from inorganic or organic acids (Van de Guchte et al., 2002; Alvarez-Ordonez et al., 2010b). B. cereus vegetative cells, like many other bacteria, are able to induce an ATR (Jobin et al., 2002; Thomassin et al., 2006). In addition, it has been shown that B. cereus cells pre-adapted at pH 6.3 coped better with both ethanol stress (12%) and heat stress (49°C) (Browne and Dowds, 2002), suggesting that ATR and/or induction of acid-resistance mechanisms confer cross-protection for other stresses. In this article, we review some model acid-resistant and their related acid resistance mechanisms to understand how B. cereus can adapt to acid stress.
Common Mechanisms of Acid Resistance
Micro-organisms deploy various mechanisms and strategies to address the hostile conditions of low-pH environment, e.g., modification of the architecture and composition of the membrane, change in metabolism and production of alkaline substances, and homeostasis of internal pH (Figure 1).
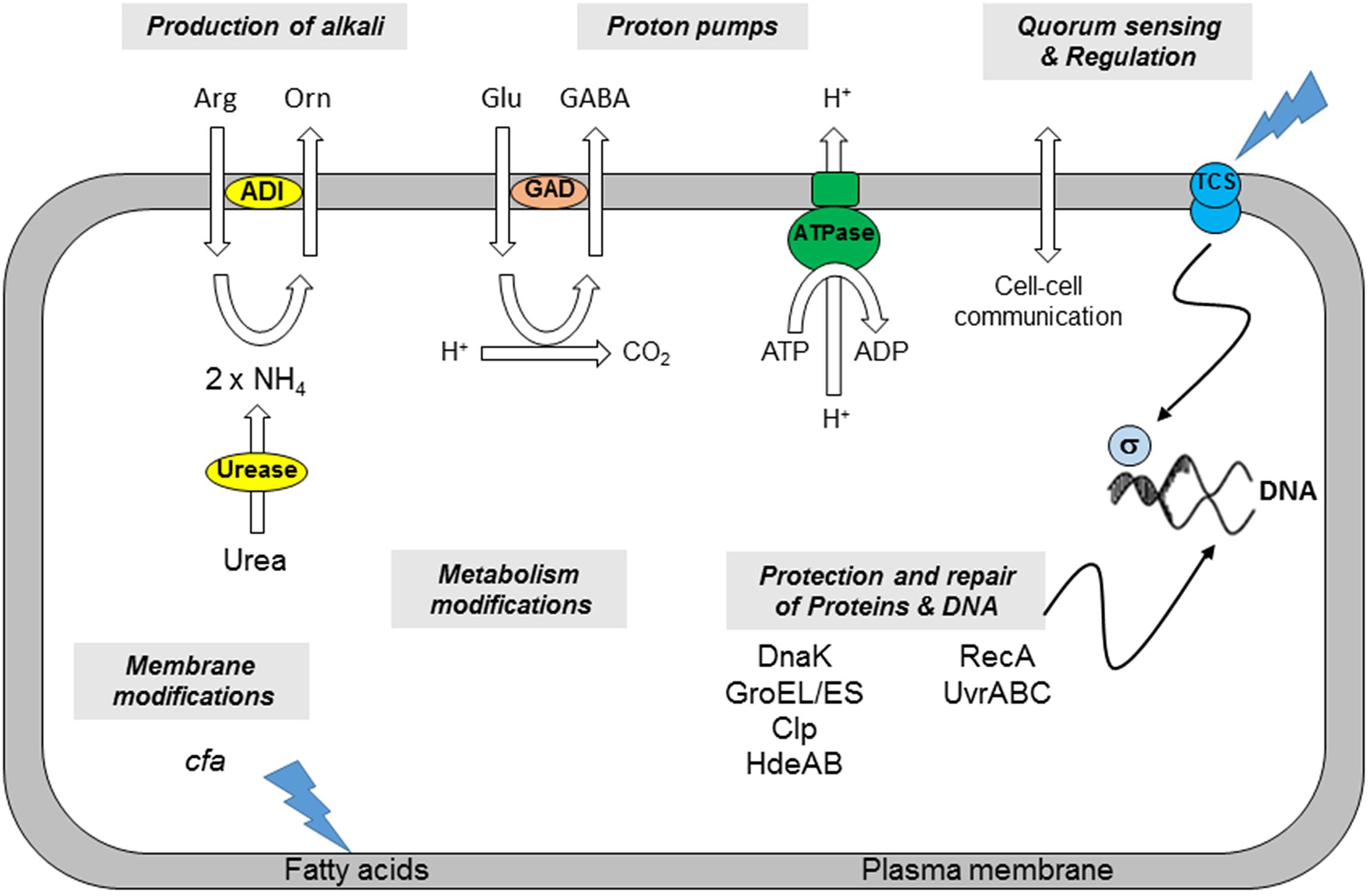
FIGURE 1. Mechanisms involved in B. cereus acid resistance (adapted from Cotter and Hill, 2003). (i) The production of alkali by the ADI or urease system increases the internal pH of the cell. (ii) Proton pumps such as the F1F0-ATPase or that utilized by the GAD system bring about an increase in pHi. (iii) Cell density affects cell-to-cell communication. The involvement of Two Component Systems (TCS) and sigma factors can induce minor or global responses. (iv) Chaperones, proteases, and heat shock proteins (HdeAB) protect cellular proteins or degrade them if damaged. (v) DNA damaged as a consequence of a low pHi can be repaired through the excision of errors or the restarting of stalled replication forks. (vi) Modifications of membrane fatty acids composition affects the properties of the plasma membrane. (vii) Metabolism of the cells may be modified in response to environmental changes.
Modification of the Membrane
The cell membrane of bacteria is in direct contact with external media. It is therefore the first one to be affected by harmful environmental conditions, e.g., an acidic medium. The fluidity of the membrane is important for cells, as it can affect membrane functions such as biochemical reactions, transport systems, and protein secretion. The membrane FA composition is responsible for the maintenance of membrane fluidity, and a number of studies have suggested a relationship between membrane fluidity and stress adaptation. Acid adaptation generally decreased membrane fluidity, and this is likely linked to the overall increase in short-chain saturated FA as observed in Escherichia coli. Salmonella, and Listeria (Kwon and Ricke, 1998a,b; Yuk and Marshall, 2004; Moorman et al., 2008; Alonso-Hernando et al., 2010). However, in some oral bacteria, exposure to acidic condition resulted in increased level of long-chain monounsaturated FA, and fluidity (Fozo and Quivey, 2004; Fozo et al., 2004; Papadimitriou et al., 2007). In E. coli, acid adaptation causes the conversion of a significant proportion of the unsaturated FA to their cyclic derivatives, known as cyclopropane fatty acids (CFA) during the transition from exponential to stationary phase (Merrell and Camilli, 2002; Merrell et al., 2002). The CFAs are formed by CFA synthase, which is encoded by cfa. Defective cfa mutants are unable to produce CFA and are sensitive to low pH (Booth, 2002). Phospholipid composition is also important in E. coli: stationary phase E. coli cells are much more sensitive to acid shock at pH 3 in the absence of the main phospholipid, phosphatidylethanolamine (Canet et al., 2003). Membrane adaptation in response to acid stress has not been yet investigated in B. cereus. However, it has been shown that B. cereus cells increase membrane fluidity by altering membrane FA composition in response to cold and saline stresses (Ultee et al., 2000; De Sarrau et al., 2012).
Production of Alkali
Some bacteria produce alkaline compounds, and specifically ammonia, to neutralize internal pH when exposed to an acidic environment. Ammonia is generated by two systems, the urease and the arginine deiminase systems (ADI; Van de Guchte et al., 2002; Cotter and Hill, 2003). In the urease system, urea is hydrolysed to two molecules of ammonia and one of CO2 by urease. In the arginine deiminase pathway, arginine is catabolized to ornithine with the release of ammonia and CO2. The urease system is more widespread and can protect some oral bacteria against acid-induced changes (Cotter and Hill, 2003; Wilson et al., 2014). Although the catalytic reaction is relatively simple, biogenesis of a functional urease is a highly complex process requiring at least seven genes, which are generally organized in operons [ureABCEFGD, (Marquis and Hager, 2000)]. The expression of bacterial ureases can be constitutive, but more often it is regulated by environmental conditions. Commonly, in enteric bacteria, the presence of urea or limitation for nitrogen can induce urease gene transcription, generally through activation of transcription. Specifically, urease expression is almost completely repressed at neutral pH values, regardless of the limiting nutrient or growth rate. In acidic conditions, the urease genes become rapidly derepressed, and expression then becomes sensitive to carbohydrate availability and rate of growth, with the highest levels of expression under conditions of carbohydrate excess and fast growth rate (Chen and Burne, 1996; Chen et al., 1996; Cotter and Hill, 2003). B. cereus is in contact with the urea present in its diverse habitats, such as soil, human urine, human saliva (2.3–4.1 mM) (Mobley, 2000; Dawes and Dibdin, 2001), stomach (4.8 mM) (Neithercut et al., 1993), blood (1.7–8.3 mM) (Mackay and Mackay, 1927), and some animal foods (e.g., milk), which contain 4.4–6.4 mM urea (Carlsson et al., 1995). Although the activity of urease can play an important role in the life cycle of B. cereus, little information is available on its role in nitrogen metabolism and in acid stress survival in this bacterium. The ADI has also been identified in a broad variety of bacteria, including B. cereus (Van de Guchte et al., 2002; Cotter and Hill, 2003; Senouci-Rezkallah et al., 2011). This system is a three-enzyme pathway that initially converts arginine to citrulline and ammonia via arginine deiminase (encoded by arcA). The citrulline is then transformed into ornithine and carbamyl phosphate by ornithine transcarbamylase (encoded by arcB). The third enzyme in the pathway, carbamate kinase (encoded by arcC), cleaves carbamyl phosphate to ammonia and CO2, concomitantly donating the phosphate to ADP to produce ATP (Griswold et al., 2004). Similar to ureolysis, the net reaction yields two molecules of ammonia and one of CO2, but also provides ATP for growth. Thus, many ADI-positive bacteria can grow with arginine as the sole source of energy. ADI-positive organisms often coordinately regulate the synthesis of an arginine:ornithine antiporter (encoded by arcD) (Cotter and Hill, 2003; Budin-Verneuil et al., 2006). The expression of arcABC operon is induced by low pH or arginine and is suppressed by excess oxygen pressure in Listeria monocytogenes (Ryan et al., 2009). The arcABC operon also contributes to the growth and survival of Lactobacillus plantarum in low-pH environment (De Angelis et al., 2002; Spano et al., 2004). In B. cereus, arginine deiminase gene arcA showed significant up-regulation upon exposure to non-lethal acid shock at pH 5.4–5.5 (Mols et al., 2010a; Senouci-Rezkallah et al., 2011), suggesting that ADI may be of great importance for B. cereus survival in low pH environments.
Homeostasis of Internal pH
Internal pH (pHi) is an important factor in bacterial physiology, and cells regulate its value precisely. The regulation of pHi implies a heightened control of membrane permeability to protons, which can take place via the ion transporters that facilitate proton entry. In general, growth studies have shown that pHi falls with culture pH (Russell, 1991; O’Sullivan and Condon, 1999; Siegumfeldt et al., 2000). In B. cereus, the pHi in continuously cultured cells sampled at equilibrium fell with growth rate, showing that growth rate has an effect on pHi (Thomassin et al., 2006). pHi is disturbed by two main factors: (i) passive movement of protons through the cytoplasmic membrane, and (ii) the production of acids and/or bases in the cytoplasm. Thus, in fermenting bacteria, the accumulation of acidic fermentation products in the cytoplasm can decrease pHi, despite continuous efflux of protons (Russell and Diez-Gonzalez, 1998). The organic acids present in the culture medium can also impede pHi homeostasis, especially when pHi > external pH (pHe). Bacteria that can allow their pHi to decrease, so as to keep a low ΔpH (pHi – pHe), may be more acid-resistant than those that keep their pHi neutral (Siegumfeldt et al., 2000). Non-dissociated organic acids freely cross the permeable membrane lipid bilayer, limiting their accumulation in the cell. In bacteria that maintain a higher pHi, organic acids dissociate in the more alkaline cytoplasm, and accumulate in the cell, halting growth (Mercade et al., 2000). Their dissociation depends on their pKa value, generally less than 5.0. The accumulation of these organic acids in anionic form depends on the pH gradient across the membrane (Russell, 1991). To keep pHi at a value that will conserve the cell’s physiological integrity, bacteria can use many different strategies to control proton flux. These include (i) active transport of protons across the membrane (via the F1F0-ATPase activity), (ii) decarboxylation systems [glutamate decarboxylase (GAD), arginine decarboxylase (AD), and lysine decarboxylase], and (iii) the buffering ability of their cytoplasm.
F1F0-ATPase
A number of studies have demonstrated a role for F1F0-H+ translocating ATPase in pHi homeostasis (Miwa et al., 1997; Kullen and Klaenhammer, 1999; Cotter and Hill, 2003; Fortier et al., 2003). The F0F1-ATPase is a well-established mover of protons across the cell membrane (Figure 2). This complex couples the energy released as protons move into the cell to the generation of ATP from ADP and Pi. The ATPase can also function in the opposite direction, hydrolyzing ATP to pump protons out of the cell. F1F0-ATPase is composed of two protein complexes (Negrin et al., 1980; Takemoto et al., 1981; Cotter and Hill, 2003). The F0 complex, which is integrated in the membrane, allows protons to cross the membrane. The F1 complex bears the catalytic site for the synthesis of ATP. The locus atp codes for the five sub-units α, β, δ, γ, and ε that form the complex F1, and the three subunits a, b and c that compose F0. It has been shown that the expression of atp was induced by an acid pH (Kullen and Klaenhammer, 1999; Quivey et al., 2001; Fortier et al., 2003). In E. coli, F1F0-ATPase acts as a proton pump during acid stress, driving protons out of the cell with a parallel hydrolysis of ATP (Richard and Foster, 2003). The F1F0-ATPase of B. cereus has been isolated from cytoplasmic membranes, purified and characterized. Structurally, B. cereus F1F0-ATPase resembles the enzyme isolated from E. coli and B. subtilis (Voelz, 1964; Banfalvi et al., 1980a,b). However, its enzymatic activity is insensitive to pH, and to N,N’-dicyclohexylcarbodiimide (DCCD), which is known to inhibit the activity of membrane-bound ATPase (Higuti et al., 1992). Recent study showed that B. cereus ATPase activity led to an increase in pHi when cells were exposed to acid stress. Indeed, DCCD had a negative effect on the ability of B. cereus cells to survive and maintain their pHi during acid shock. Furthermore, transcriptional analysis revealed that expression of atpB (encoding b subunit of F1F0-ATPase) was increased in acid-adapted cells compared to non-adapted cells before and after acid shock. These data demonstrate that B. cereus is able to induce an ATR during growth at low pH, depending on the ATPase activity induction and pHi homeostasis (Senouci-Rezkallah et al., 2015).
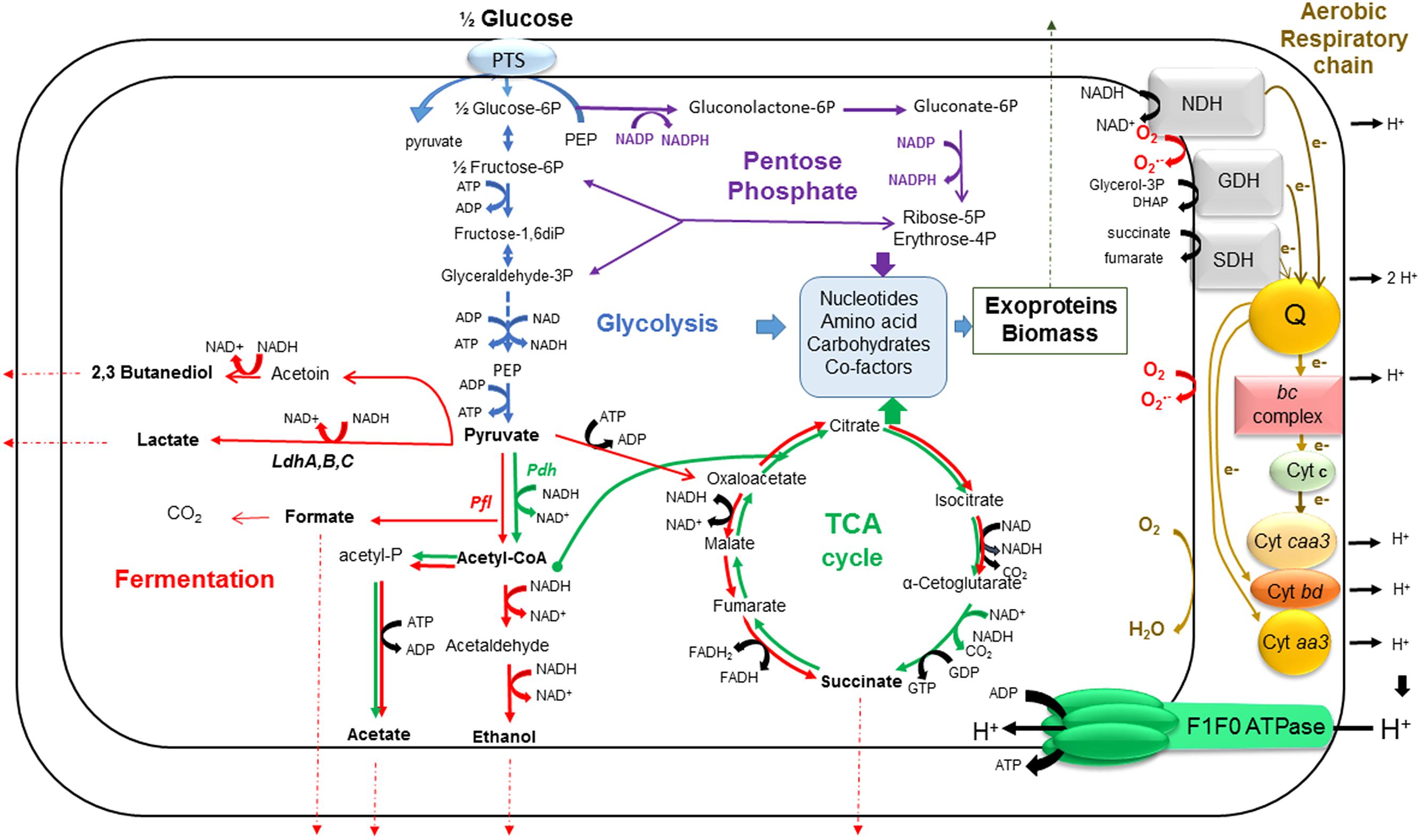
FIGURE 2. A simplified view of B. cereus central metabolism. This schematic shows our current understanding of how glycolysis (blue), the PPP (purple), the fermentation pathways (red), the TCA cycle (green) and aerobic respiratory chain (brown) are interconnected in growing B. cereus cells. The fermentation end products are shown in bold. The electron transport in the respiratory chain goes through dehydrogenation of NADH, as a first step. Electrons are transferred from NADH to menaquinone pool (Q), which serves as an electron carrier, and then, the pathway goes through two main branches. The first is one in which electrons go through the bc complex, cytochrome c and cytochrome oxidase caa3, in turn. The second contains only quinol oxidases (cytochromes bd or aa3). In each branch, electrons are finally accepted by oxygen molecules. The transfer of electrons is coupled to the formation of a proton gradient across the membrane which is tapped by F1F0-ATPase to generate ATP from ADP and Pi. ROS are generated as by-product from this process. Ldh, lactate dehydrogenase; Pdh, pyruvate dehydrogenase; Pfl, pyruvate formate lyase; NDH, NADH:menaquinone reductase; SDH, succinate:menaquinone oxidoreductase; GDH, glycerol-3-P:menaquinone reductase.
Decarboxylation of amino acids
The presence of amino acids (glutamate, lysine, and arginine) in food such as vegetables and dairy products can enhance the ability of bacteria to adapt to and survive acid stress. These amino acids can be decarboxylated by systems composed of one or more decarboxylases, which convert their substrates into amine derivatives and carbon dioxide or bicarbonate, and an antiporter that then exchanges each amino acid for its decarboxylated amine. Decarboxylation of amino acids controls bacterial pH by consuming hydrogen through the decarboxylation reaction. The lysine, arginine and GAD systems predominate in acid tolerance (Booth, 2002). The GAD system has been identified in a variety of bacteria such as E. coli. L. monocytogenes. Shigella flexneri, and Lactococcus lactis (Cotter and Hill, 2003; Cotter et al., 2005; De Biase and Pennacchietti, 2012; Kanjee and Houry, 2013). In E. coli, known components of glutamate-dependent acid resistance include two isoforms of GAD (GadA and GadB) and a putative glutamate: γ-aminobutyric acid (GABA) antiporter called GadC. GadA/GadB is assumed to catalyze the conversion of protonated glutamate to GABA, whereas GadC exports GABA in exchange for a new extracellular glutamate molecule. This process consumes protons in cells, which eventually increases pHi, protecting the cell from the damage caused by acid shock (Richard and Foster, 2003). Three decarboxylases (GadD1, GadD2, and GadD3) and two antiporters (GadD1T1 and GadD2T2) were identified in L. monocytogenes. The GadD2/T2 system was found to be responsible for the survival of the cells in acidic conditions at pH 2.8. GadD1T1 plays a role in growth at moderately acidic pH values (5.1) (Cotter et al., 2005). Gene encoding GAD was identified in B. cereus ATCC 10987, while no gene encoding GABA glutamate exchanger was found. As a result, the gad gene was not up-regulated under low-pH exposure (Mols et al., 2007, 2010a,b). Unlike ATCC 10987, B. cereus ATCC 14579 genome does not contain gad gene. However, glutamate enhanced the resistance of B. cereus ATCC 14579 cells to pH 4.0 acid shock (Senouci-Rezkallah et al., 2011).
The system AD system is composed of a cytoplasmic arginine decarboxylase (AdiA) and an arginine/agmatine antiporter (AdiC). After proton-consuming decarboxylation of arginine by AD to give agmatine in the cell, the agmatine is carried out of the cell by the antiporter in exchange for arginine. The consumption of protons during decarboxylation reduces acidity in the cytoplasm. In E. coli, the presence of arginine during acid shock raised pHi from 3.7 to 4.7, showing that AD could enable pHi homeostasis during acid stress (Richard and Foster, 2004). In S. typhimurium, the AD system is active only under acid growth conditions (Kieboom and Abee, 2006; Alvarez-Ordonez et al., 2010c). In B. cereus, two genes have been annotated as encoding ADs [speA and yaaO, (Ivanova et al., 2003)], but no gene has been annotated as encoding an arginine/agmatine antiporter. However, it has been shown that the presence of arginine improved acid stress resistance of B. cereus cells. It is thus probable that B. cereus utilizes the AD system for surviving in acidic environment (Senouci-Rezkallah et al., 2011).
The lysine decarboxylase system is composed of a decar-boxylase (CadA) and a lysine/cadaverine antiporter (CadB). After decarboxylation of lysine to cadaverine in the cell by proton-consuming lysine decarboxylase, the cadaverine is carried out of the cell by the antiporter in exchange for lysine. The consumption of protons during the decarboxylation reaction again lowers acidity in the cytoplasm, allowing the homeostasis of pHi during acid shock. In E. coli and Vibrio vulnificus, the lysine decarboxylase system is encoded by the cadBA operon. This operon is activated by CadC, and repressed by LysP (Neely and Olson, 1996; Rhee et al., 2005). In Vibrio parahaemolyticus, the expression of cadBA has been shown to increase in the presence of lysine. The mutation of the gene cadA also impaired survival in acid shock conditions relative to the wild-type strain, showing the role of this system in resistance to acid shock by maintenance of pHi (Tanaka et al., 2008). In B. cereus ATCC 14579, the gene encoding the enzyme lysine decarboxylase is yvdD (Ivanova et al., 2003), but the gene encoding the antiporter has not yet been identified. Like glutamate and arginine, the addition of lysine improves B. cereus resistance to acid stress, suggesting a role of the lysine decarboxylase system in this bacterium (Senouci-Rezkallah et al., 2011).
Buffering ability of cytoplasm
Independently of the involvement of the decarboxylation systems, the pHi of a cell can be stabilized by the relatively high buffering ability of the cytoplasm. When acid or alkaline compounds enter the cell, the buffering action of the cytoplasm tends to offset these variations and keep the pHi neutral. Rius and Loren (1998) compared the buffering ability of Bacillus alcalophilus, an alkaliphilic bacterium, with those of B. subtilis and S. aureus, both neutrophilic bacteria. They showed that the buffering ability of B. alcalophilus was influenced by the culture pH and other conditions. In addition, this buffering ability was greater in anaerobic growth conditions than in aerobic growth conditions, clearly showing that the cells have a significantly greater buffering activity in a fermenting medium. In B. cereus, a study showed that cells grown in unregulated batches at pHe 7.0 showed a pHi of 9.0, which fell to 7.9 and 6.2 when the cell growth at pH 7.0 was followed by 1 h incubation at pH 6.3 and 4.6, respectively. It has also been observed that the pHi of cells adapted at pH 6.3 was higher (pHi 6.6) than that of non-adapted cells (pHi 6.1) (Browne and Dowds, 2002). However, in unregulated batch culture, the cell environment is not controlled and several factors, such as growth phase, growth rate, and carbon and oxygen resource availability can therefore also influence pHi.
Cell Density
In addition to responses to several stresses, bacteria are known to regulate diverse physiological processes in a cell density-dependent manner. Cell density-dependent regulation appears to follow a common theme, in which a small, self-generated molecule is exported as the signal for intercellular communication, commonly called quorum sensing. Cell density was found to modulate acid adaptation in Streptococcus mutans log-phase cells, since pre-adapted cells at a higher cell density or from a dense biofilm displayed significantly higher resistance to the killing pH than the cells at a lower cell density (Li et al., 2001). The authors also showed that mutants defective in the comC. comD, or comE genes, which encode a quorum sensing system essential for cell density-dependent induction of genetic competence, had a diminished log-phase ATR. They concluded that optimal development of acid adaptation in S. mutans involves both low pH induction and cell–cell communication. Also in this strain, the gene luxS involved in the signal synthesis in quorum sensing plays a role in the regulation of tolerance to acid stress (Wen and Burne, 2004). The synthesis of LuxS is induced in E. coli on exposure to acetic acid, suggesting that its expression in this organism is induced at low pH (Frees et al., 2003b). Four quorum sensing systems operate in B. cereus and, one of them controls the oxidative stress response (Slamti et al., 2014). Exposure to acid stress induces secondary oxidative stress in B. cereus (Mols and Abee, 2011a). Therefore, the quorum sensing system that controls oxidative stress could be involved in acid adaptation.
Protection and Repair of Proteins and DNA
In an acid stress situation, proteins can undergo modifications, from changes in conformation to complete denaturing, all of which will significantly affect their activity. Denatured proteins are dealt with chaperone proteins, such as DnaK/DnaJ and GroES/EL (Susin et al., 2006). In L. lactis, these chaperones are induced at pH 4.5, and not at pH 5.5, and therefore respond at a certain level of acidity in the medium, or a certain concentration of denatured proteins in the cytoplasm (Frees et al., 2003b). In S. mutans, the expression of dnaK and the quantity of DnaK are higher in acid-adapted cells than in non-adapted cells, and increase in response to an acid shock (Jayaraman et al., 1997), suggesting that the regulation of dnaK by pH is transcriptional in this bacterium. In S. typhimurium DT104, an increase level of DnaK and GroEL was observed in acid-adapted cells (Berk et al., 2005). Exposure of L. lactis to low pH revealed that, in addition to DnaK and GroEL, several heat shock proteins are part of the acid shock response. Among them are ClpE and ClpP (Frees et al., 2003a). The Clp proteins are ATPase-dependent proteases involved in the turn-over of denatured proteins: they are responsible for the rapid degradation of damaged proteins and the regulation of the levels of some proteins in the cell (enzymes and regulators). In B. cereus, DnaK is overproduced in response to acid shock (Browne and Dowds, 2002). Proteins of 66 and 59 kDa, which could be, respectively, DnaK and GroEL, have also been found expressed in stationary phase cells irrespective of pH (Jobin et al., 2002). More recent study showed that chaperone-encoding genes dnaK and groES and protease-encoding gene clpC were up-regulated upon exposure to sublethal acid shocks (Mols et al., 2010b), suggesting that these proteins are involved in acid stress response in this bacterium.
When pHi becomes too acidic, DNA loses purine and pyrimidine units (Cotter and Hill, 2003). First the bases are protonated and then, the glucoside bonds are cleaved. The ultraviolet (UV) excinulease system UvrA-UvrB which is known to repair damage caused to DNA by UV radiation or exposure to many chemical agents (Lage et al., 2003) is associated with low-pH adaptation. The UvrA–UvrB complex identifies the changes in conformation or structure in the damaged DNA. UvrA then dissociates from UvrB, which remains bound to the DNA. UvrC in turn binds to the UvrB-ADN complex and effects the incision of seven nucleotides. UvrD intervenes to remove UvrC and the damaged nucleotides. UvrB remains bound to the DNA until DNA polymerase I synthesizes the excised sequence (Skorvaga et al., 2004). In S. mutans. uvrA is induced by acidity. In addition, a ΔuvrA mutant is more sensitive to growth at pHe 5.0 than the wild-type strain, and when it is pre-incubated at a non-lethal acidic pH, it is unable to survive at pH 3.0. This indicates that UvrA is involved in adaptive response to low pH (Hanna et al., 2001). In Methylobacterium dichloromethanicum, a ΔuvrA mutation significantly limited viability and growth on dichloromethane, when intracellular hydrochloric acid is produced. This dehalogenation produced a genotoxic intermediate compound. However, it is not yet known whether DNA lesions are caused by this reaction product or by the acidity (Kayser et al., 2002). Mutations in the genes recA (coding for a protein involved in homologous recombination and which is a regulator of the SOS response) and uvrB caused a significant decrease in tolerance to low pH in H. pylori (Thompson and Blaser, 1995; Thompson et al., 1998), showing the importance of these DNA repair mechanisms for survival in acid conditions. In B. cereus, the uvrA and uvrB genes are up-regulated upon exposure to pH 5.5 set with lactic acid, suggesting a role of the UvrA–UvrB system in stress acid resistance (Mols et al., 2010b).
Concluding Remarks
Although much has been learned about other bacteria respond to acid stresses, there remains a great deal to discover, making B. cereus acid stress responses a fruitful and important area of future research. In addition, the majority of studies that have analyzed the response of food-borne pathogens to acid stress were performed in presence of oxygen. Anoxia may thus provide attractive condition to study the acid resistance mechanisms in B. cereus and other food-borne pathogens.
Oxygen Sensing and Adaptation to Anoxia
Bacillus cereus is a facultative anaerobic microorganism, i.e., it can survive at various levels of oxygenation. The common terminology to describe the various oxygen conditions, such encountered by B. cereus in food and GI tract is based on a comparison with the atmospheric level, which is 21% (v/v), a level referred to as normoxia. Hypoxia is a condition where oxygen concentration is lower than 21%. No free oxygen is available under anoxia.
B. cereus Central Metabolism and Redox Homeostasis
Whatever the oxygenation conditions, B. cereus catabolizes glucose through glycolysis and, to a lesser extend, through the pentose phosphate pathway (PPP; Zigha et al., 2006). Glycolysis and PPP are fuelled by the phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS), a transport system that cells use to bring glucose into the cytoplasm using energy transferred by PEP (Deutscher et al., 2006; Cao et al., 2011; Warda et al., 2016). Glycolysis produces two molecules of pyruvate per molecule of activated glucose (glucose-6-phosphate, G-6P) and, in the process, reduces two molecules of NAD+ to NADH and produces a net gain of two ATP molecules (Figure 2). In addition to providing ATP by substrate-level phosphorylation, glycolysis is a major source of metabolic intermediates for biosynthetic pathways. The processing of activated glucose (G-6P) through the PPP also produces biosynthetic precursors. Two of these biosynthetic precursors, ribose-5-phosphate, and erythrose-4-phosphate, are essential for the synthesis of nucleotides, histidine, and aromatic amino acids. In addition to providing biosynthetic intermediates, the PPP generates two molecules of NADPH per molecule of G-6P. NADPH provides the reducing power that drives numerous anabolic reactions, including those responsible for the biosynthesis of all major cell components (Spaans et al., 2015). NADPH is also required to maintain and regenerate the cellular detoxifying and anti-oxidative defense systems (Agledal et al., 2010). The antioxidant defense system of B. cereus is constituted by an elaborate, often overlapping network of enzymes, such as superoxide dismutase (SOD), catalase, flavohemoglobin, peroxiredoxins, thioredoxins, among others, and low molecular mass (LMW) thiols such bacillithiol (BSH), coenzyme A (CoASH) and cysteines (Newton et al., 1996; Fang et al., 2013). In addition to their roles in detoxification of ROS, LMW thiols, which are present in millimolar concentrations in the cytoplasm, function as major thiol-redox buffers to maintain the redox state of the cytoplasm (Newton et al., 2009; Loi et al., 2015). The PPP and glycolysis are linked by transketolase and transaldolase that convert ribose-5-phosphate into glyceraldehyde-3-phosphate and fructose-6-phosphate. Fructose-6-phosphate is a precursor for N-acetylglucosamine, which is required for BSH (Helmann, 2011). Increased carbon flow through the PPP is often associated with stressful conditions or infections in Gram-positive pathogens (Eisenreich et al., 2006; Bergman et al., 2007).
In the presence of oxygen, pyruvate is converted to acetyl coenzyme A (Acetyl-CoA) by the pyruvate dehydrogenase complex (Pdh) (Duport et al., 2006). Acetyl-CoA can enter into the tricarboxylic acid cycle (TCA) via a condensation reaction with oxaloacetate that is catalyzed by citrate synthase. Then two carbons are lost as CO2 for every two carbons (i.e., Acetyl-CoA) that enter the TCA. TCA provides three biosynthetic intermediates that are critical for the novo synthesis of many amino acids and porphyrins: oxaloacetate, α-ketoglutarate, and succinate/succinyl-CoA. The reducing equivalents generated by the glycolysis and TCA (NADH and FADH) are reoxidized through the aerobic respiratory chain, resulting in the build-up of a proton motrice force and the subsequent synthesis of ATP by ATP synthase (38 ATP per molecule of consumed glucose). Although it has not been thoroughly studied, the aerobic respiratory chain of B. cereus resembles the respiratory chain of B. subtilis (Contreras-Zentella et al., 2003; Rosenfeld et al., 2005; Garcia et al., 2008; Melo and Teixeira, 2016). It contains two major branches, one quinol oxidase branch (with cytochrome bd or cytochrome aa3 as its terminal oxidase) and one cytochrome oxidase branch (with cytochrome caa3 as its terminal oxidase) (Figure 2). The terminal oxidases catalyze the four-electron reduction of dioxygen to two water molecules. The aerobic respiratory chain is not only a main source of ATP but it also a major source of reactive oxygen species (ROS; Gonzalez-Flecha and Demple, 1995; Messner and Imlay, 2002; Mailloux et al., 2011). ROS generation starts with the formation of a superoxide anion (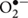 ). Within the respiratory chain, NADH: menaquinone oxidoreductase (complex I) and bc complex (complex III) are generally considered as the main producers of
). Within the respiratory chain, NADH: menaquinone oxidoreductase (complex I) and bc complex (complex III) are generally considered as the main producers of 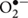 (Figure 2). Dismutation of
(Figure 2). Dismutation of 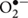 (either spontaneously or through a reaction catalyzed by SODs) produces hydrogen peroxide (H2O2), which in turn may be fully reduced to water or partially reduced to hydroxyl radical (OH∙), one of the strongest oxidants in nature (Imlay, 2013). ROS production serves as a metabolic signal and under normal conditions are quenched by the antioxidant defense system to maintain them to non-toxic levels. However, when released in excess under certain stress conditions such as hypoxia and change in pH that abruptly affect the electron transport chain, ROS can also directly damage cells (Mols et al., 2009, 2010b, 2011).
(either spontaneously or through a reaction catalyzed by SODs) produces hydrogen peroxide (H2O2), which in turn may be fully reduced to water or partially reduced to hydroxyl radical (OH∙), one of the strongest oxidants in nature (Imlay, 2013). ROS production serves as a metabolic signal and under normal conditions are quenched by the antioxidant defense system to maintain them to non-toxic levels. However, when released in excess under certain stress conditions such as hypoxia and change in pH that abruptly affect the electron transport chain, ROS can also directly damage cells (Mols et al., 2009, 2010b, 2011).
When the oxygen concentration drops to a level at which oxygen becomes limiting as a substrate for cytochrome c oxidase, ATP production via oxidative phosphorylation is no longer able to meet cellular demands for ATP. This can be compensated by the activation of glycolytic activity to increase ATP production by substrate-level phosphorylation. However, compared with oxidative phosphorylation, the ATP production through glycolysis alone is much lower. Therefore, glycolytic activity must be strongly up-regulated under hypoxic conditions to generate sufficient ATP. This phenomenon, namely the Pasteur effect requires the efficient recycling of NAD+ from NADH, otherwise glycolysis will become limited by the availability of NAD+. Therefore, the fermentative pathways are induced by activating the expression of key enzymes.
Under anoxia and in absence of external electron acceptor, B. cereus carries out mixed acid-butanediol fermentation (Figure 2). Lactic acid is the major by-product of fermentation (more than 60% of total production) both at neutral and acidic pH (Duport et al., 2004, 2006; Messaoudi et al., 2010; Le Lay et al., 2015). The pyruvate-to-lactate pathway involves three L-lactate dehydrogenases, LdhA, B, and C. It has been shown that LdhA exerted a major control on both B. cereus fermentative growth and enterotoxin production (Laouami et al., 2011). The conversion of pyruvate to acetyl-CoA and formate requires high levels of pyruvate formate lyase (Pfl) compared to Pdh complex, probably to avoid excessive NADH formation under fermentative conditions (Duport et al., 2006). The acetyl-CoA is converted to acetate through the ATP-producing acetate pathway and to ethanol through the NADH-recycling ethanol pathway. At neutral pH, acetate and formate are produced in similar amounts (each accounting for ∼15% of total production), while ethanol, and to higher extend succinate and 2,3 butanediol are minor fermentation products (Duport et al., 2004; Rosenfeld et al., 2005). However, the relative rate of formation of all these glucose by-products is influenced by the ORP of the growth medium (Zigha et al., 2006). At acidic pH, production of 2,3-butanediol highly increased at the expense of acetic acid and succinic acid and to lesser extend lactic acid (Le Lay et al., 2015). During fermentative growth, TCA functions only to supply biosynthetic precursors and is transformed from a cyclic pathway to two oppositely oriented half cycles (Figure 2). In fermenting cells, the direct formation of ROS is abolished by the absence of oxygen. However, several anoxia-specific alterations can promote the oxidative response (Lumppio et al., 2001; Rusnak et al., 2002). The occurrence of an oxidative component in response to oxygen deprivation has been confirmed in B. cereus by microarray studies on the whole genome level and by proteomic studies (Mols and Abee, 2011b; Clair et al., 2012; Madeira et al., 2015).
Under anoxia, B. cereus can growth via nitrate ammonification (Zigha et al., 2006). Nitrate in the human intestine originates both from endogenous synthesis and dietary products rich in nitrate (Tannenbaum et al., 1978; Lidder and Webb, 2013). During nitrate respiration, nitrate is reduced by the respiratory nitrate reductase (NarGHI) to nitrite in B. cereus cells. Nitrite is further reduced to ammonia by a general nitrite reductase (NasDE). Nitrate reduction is coupled to ATP generation through proton motrice force (Rosenfeld et al., 2005). Due to the drastically different ATP yields of respiratory and fermentative processes, B. cereus uses a fine-tuned regulatory system to maintain the most efficient mode of ATP generation under anoxia (Zigha et al., 2006). Nitrate-respiring B. cereus cells produce nitric oxide (NO) as an intermediate product of nitrate reduction to N2O (Kalkowski and Conrad, 1991). The chemical properties of NO make this gas a good candidate for a signaling molecule (Gusarov and Nudler, 2012). Gram-positive bacteria, including B. cereus can also generate NO through a bacterial analog of mammalian NO synthase (bNOS) in presence of oxygen. It has been shown that bNOS from B. subtilis. B. anthracis displayed NO-forming activity dependent on arginine (Adak et al., 2002; Gusarov et al., 2008). The bNOS-mediated NO was implicated in the protection of bacteria against oxidative stress, a variety of antibiotics and other stresses such as acid stress (Tan et al., 2010; Gusarov and Nudler, 2012). NO also regulates growth and pathogenicity of B. anthracis (Popova et al., 2015).
Exoproteins and Virulence Factors Supporting Pathogenesis
Bacillus cereus excretes high level of proteins into the extracellular medium. However, the level of excreted proteins is lower during anaerobic fermentative growth than under aerobic respiratory growth (Madeira et al., 2016a,b). Like transcription and translation, exportation of proteins is an energetically expensive process. Therefore, the decrease of protein excretion in fermentative cells may be attributed to decreased energy availability. Most exoproteins are secreted as precursors with a cleavable N-terminal signal sequence, but a significant fraction is secreted by non-classical pathways, i.e., without signaling peptides and sequence motifs for surface anchoring. Six signal peptide-dependent pathways are currently recognized in Gram-positive bacteria (Schneewind and Missiakas, 2014): the general secretory (Sec) pathway, the twin arginine targeting (Tat) pathway, the fimbrillin-protein exporter, the flagellar export apparatus, the holins, and the ESAT-6/WXG100 secretion system. The Sec pathway is considered the general housekeeping protein translocation system and is essential in B. cereus (Fagerlund et al., 2010; Senesi and Ghelardi, 2010; Senesi et al., 2010). The proteins arising from cellular secretion and other protein export mechanisms are components of the B. cereus exoproteome (Armengaud et al., 2012). B. cereus exoproteome has been the focus of several shotgun proteomic studies (Clair et al., 2010, 2013; Laouami et al., 2014; Madeira et al., 2015, 2016b). These proteomic studies identified up to 377 different exoproteins. Among them 65 putative virulence factors were identified, including 15 toxin-related proteins, 12 motility-related proteins and 36 adhesins and degradative enzymes (Madeira et al., 2015, 2016a,b). These virulence-related proteins represent more than 85% of exoproteins whatever the growth condition (Clair et al., 2010; Madeira et al., 2015). B. cereus exoproteome includes numerous cytoplasmic proteins involved in metabolic pathways (mainly glycolysis) and oxidative stress response. Many of these proteins are conserved in the exoproteome of pathogens. A significant number of these extracellular cytoplasmic proteins have been found to serve two or more functions and are referred as “moonlighting” proteins (Henderson and Martin, 2011; Gotz et al., 2015). Moonlight proteins have been shown to localize at the cell surface and participate in adhesion, colonization and virulence (Henderson and Martin, 2011; Ebner et al., 2016a,b). Surface-associated moonlight proteins have been reported to be reversible and pH dependent (Nelson et al., 2001; Antikainen et al., 2007). Extracellular cytoplasmic proteins are mainly excreted during the stationary growth phase (Yang et al., 2011) and in B. cereus, their time dynamic is negatively correlated to the dynamic of toxin-related proteins, indicating that a specific selection process has to occur (Madeira et al., 2015).
The toxin-related proteins found in B. cereus exoproteome include the lytic components (L1 and L2) and the binding component (B) of Hemolysin BL (Hbl). Hbl requires all three components for full activity (Beecher et al., 1995). Hbl may form a pore similar to other soluble channel-forming proteins in host cell membranes (Madegowda et al., 2008; Stenfors Arnesen et al., 2008). The tripartite Hbl complex is encoded by genes clustered into a polycistronic operon with the transcriptional order hblC. hblD, and hblA (Ryan et al., 1997). An ORF, name hblB, is located immediately downstream of hblCDA in the B. cereus ATCC 14579 genome and is transcribed independently (Clair et al., 2010). hblB encodes HblB’, which is structurally related to the B component of the Hbl complex. Its activity is currently unknown (Clair et al., 2010). NheA, NheB, and NheC are the three components of the non-hemolytic enterotoxin (Nhe) and are encoded by the nheABC operon (Lindback et al., 2004). All three components NheA, NheB, and NheC are required for full toxic activity, although NheC is only expressed in small amounts due to translational repression (Lindback et al., 2004). Nhe is a pore-forming toxin (Didier et al., 2012; Phung et al., 2012). Cytotoxin K (CytK) is a single-component protein toxin and belongs to the family of β-barrel pore-forming toxins (Baida et al., 1999; Lund et al., 2000). CytK possesses dermonecrotic, cytotoxic, and hemolytic activities (Lund et al., 2000; Hardy et al., 2001). HlyI is a thiol-activated cholesterol-binding cytolysin (Kreft et al., 1983; Minnaard et al., 2001; Ramarao and Sanchis, 2013). All the genes encoding Hbl, Nhe, CytK, and HlyI belong to the PlcR virulence regulon (Gohar et al., 2008). Hemolysin II (HlyII) is cytotoxic due to its ability to disrupt cellular and artificial membranes by pore formation (Andreeva et al., 2006). EntFM exhibits three protein–protein interaction SH3 domains and a NlpC/P60 domain that shares similarities with cell wall peptidase. There is controversy over the role of EntFM in B. cereus cytotoxicity, but wide consensus on its role in pathogenicity (Boonchai et al., 2008; Tran et al., 2010). The structurally related proteins EntA, EntB, EntC, and EntD were annotated as “enterotoxin/cell wall-binding proteins” because they possess, in addition to two SH3 domains, an extracellular cell wall-binding 3D domain (Clair et al., 2010). Recent study showed that EntD plays a crucial role in maintaining cell wall structure and that, in the absence of EntD, B. cereus cells are able to reoriente their metabolism to maintain cell wall integrity. Such adaptation program leads to decreased virulence factor production, specifically Nhe and Hbl production (Omer et al., 2015). The functions of EntA, EntB and EntC are currently unknown.
Taken together, the 15 toxin-related proteins found in B. cereus exoproteome represent more than 30% of exoproteins during the exponential growth phase, suggesting that they may have an important cellular function for the producer bacterium (Clair et al., 2010). Toxin-related proteins contain methionine residues that are susceptible to intracellular oxidation in both respiratory and fermenting cells (Madeira et al., 2015). Methionine residues of proteins are known to act as ROS scavengers (Luo and Levine, 2009). High level secretion of toxin-related protein during active growth may thus contribute to the protection of B. cereus cells against cellular oxidation and maintain redox homeostasis by keeping endogenous ROS at bay whatever the oxygen condition.
The regulation of B. cereus toxin gene expression mobilizes a complex machinery (Ceuppens et al., 2011; Jessberger et al., 2015) that includes the virulence regulator PlcR (Salamitou et al., 2000; Gohar et al., 2008) and several transcriptional regulators that coordinately control metabolic and virulence genes such as the CodY repressor (Lindback et al., 2012; Bohm et al., 2016), the ferric uptake regulator Fur (Sineva et al., 2012), the catabolite control protein A [CcpA, (Van der Voort et al., 2008)] and the redox regulators, Fnr, ResD, Rex, and OhrR. All these four redox regulators are able to regulate directly the expression of hbl and nhe after binding to the promoter region of hblCDA and nheABC (Esbelin et al., 2008, 2009; Clair et al., 2013; Laouami et al., 2014). In addition, it was shown that ResD and Fnr form a ternary complex with the virulence regulator PlcR (Esbelin et al., 2012).
Redox Regulators that Coordinate Central Metabolism and Production of Toxin-Related Proteins
A number of classical sensors/regulators are employed by various species of bacteria to sense oxygen changing conditions (Bueno et al., 2012). These sensors/regulators include Fnr, ResDE, Rex, and OhrR (Figure 3). All of them could orchestrate the expression of virulence determinants in B. cereus, both directly, and indirectly, by impacting key metabolic and regulatory circuits.
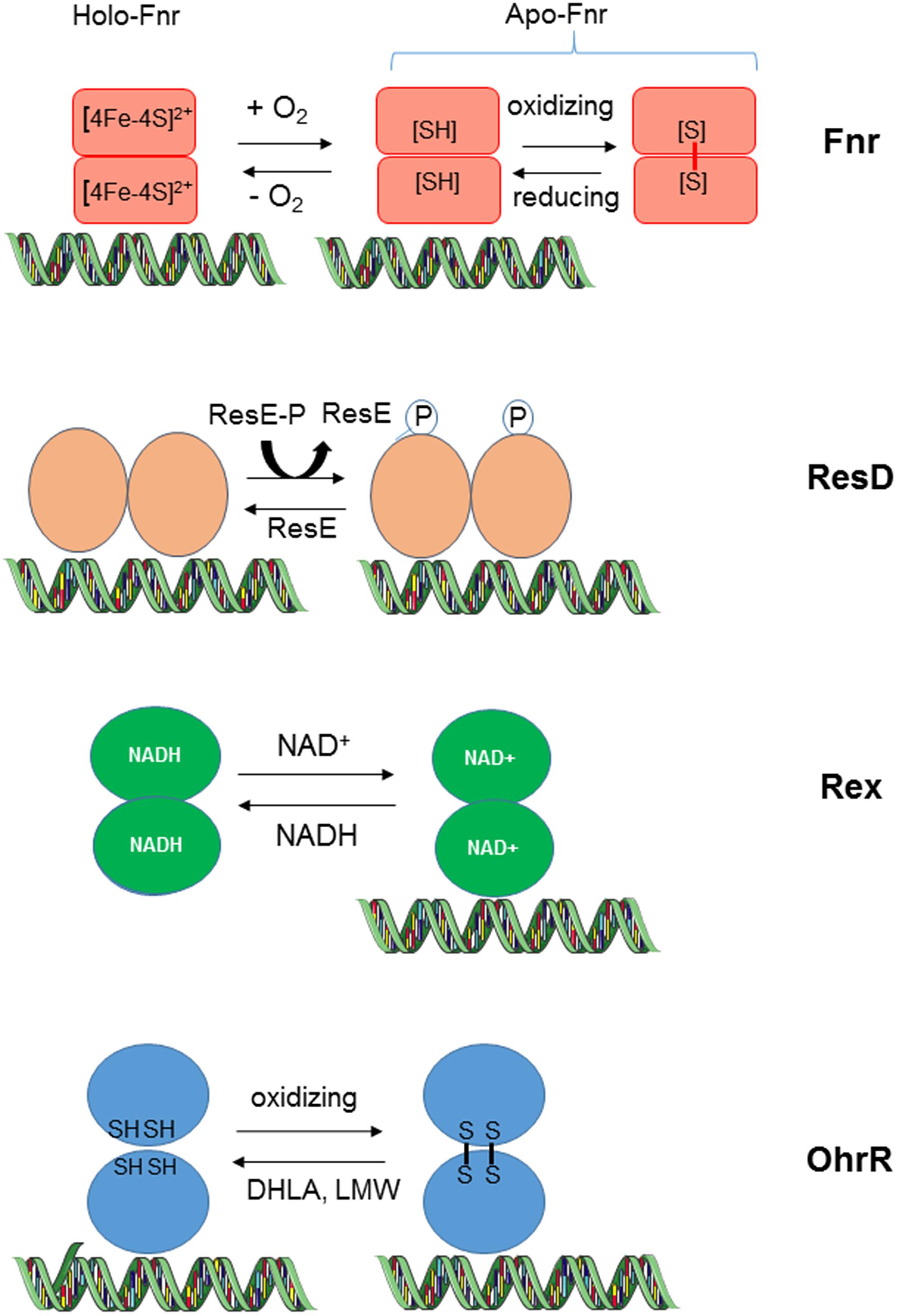
FIGURE 3. Redox-sensing mechanisms of transcriptional regulators in B. cereus. HoloFnr binds one [4Fe-4S]2+ cluster per monomer. This cluster is degraded upon exposition to oxygen. ApoFnr (clusterless) is active as a DNA binding protein under its reduced dimeric form. The oxidized ApoFnr form, which is stabilized by means of one or more SS bonds, is inactive. The thiol-based redox switch mediates a response to oxygen concentration or cellular oxidant (see text). Binding of ResD to DNA is not dependent on ResD phosphorylation status, which is regulated by the kinase and phosphatase activities of ResE. Rex senses the intracellular redox status through changes in the NADH/NAD+ ratio. Unlike the NAD+-bound Rex form, the NADH-bound Rex form is incapable of binding DNA. OhrR senses oxygen concentration or ROS through cysteine residues (see text). OhrR is a non-covalent dimer in its reduced form and a covalent dimer in its oxidized form. Dihydrolipoate (DHLA) and low molecular mass thiols (LMW) participate in the recovery of reduced OhrR from the oxidized form. B. cereus OhrR can bind DNA both under its reduced and oxidized form.
B. cereus Fnr
The B. cereus Fumarate and nitrate reductase regulator Fnr is a member of the Crp/Fnr (cyclic AMP-binding protein/fumarate nitrate reduction regulatory protein) family of helix-turn-helix transcriptional regulators (Korner et al., 2003). Like all the members of the Crp/Fnr family, B. cereus Fnr contains an N-terminal region made up of antiparallel β-strands able to accommodate a nucleotide, and a C-terminal extension with four cysteine residues that coordinate a [4Fe-4S]2+ cluster (Esbelin et al., 2009). Fnr is essentially present in the apo-form (clusterless) in aerobically grown cells, and in the holo-form in anaerobically grown cells (Esbelin et al., 2012). Under aerobiosis, Fnr is able to sense oxygen concentration changing and probably ROS and NO (Jiang et al., 2016) through the oxidation of Cys thiol groups, which link two monomers in an inactive dimer. The inactivation of Fnr by oxygen is reversible. In the current model for Fnr function, the active dimeric forms of Fnr bind to the promoter region of target operons/genes to activate or repress their transcription. Fnr plays a key role within the regulatory cascade governing fermentative pathways in B. cereus because (i) its transcription is strongly induced in fermenting cells and (ii) the inactivation of its gene abolishes fermentative growth. The role of Fnr under anaerobic and aerobic respiratory growth is more moderated (Zigha et al., 2007).
B. cereus ResDE
The ResDE two-component signal transduction system consists of a membrane-bound histidine sensor kinase (ResE) and a cytoplasmic response regulator (ResD). The resD and resE genes compose a transcriptional unit included into a larger operon that comprises resABC; these three genes encode proteins similar to those involved in cytochrome c biogenesis (Duport et al., 2006). The B. cereus resABCDE locus is organized similarly to that in B. subtilis and B. anthracis (Sun et al., 1996; Wilson et al., 2008). The ResDE two-component system regulates the expression of several genes of the fermentative and respiratory pathways in B. cereus. However, it appeared to exert a more important role in anaerobic fermentative pathways (with a more pronounced effect under high reductive conditions) than in aerobic respiratory pathways. Unlike fnr, the resDE mutation did not abolish the fermentative growth of B. cereus, indicating that although it plays an important role, it is not indispensable for B. cereus. The ResDE system is modulated primarily by the autophosphorylation activity of ResE at a conserved histidine residue. The redox signal activating ResE has not been identified in B. cereus but have been postulated to be the redox state of menaquinones in B. subtilis under aerobiosis (Geng et al., 2007). Upon activation, ResE donates a phosphate to its cognate regulator, ResD. The phosphatase activity of ResE controls the level of phosphorylated ResD (ResD∼P). In Bacillus subtilis, phosphatase activity of ResE is regulated by oxygen availability and anaerobic induction of the ResDE regulon is partly due to a reduction of the ResE phosphatase activity during anaerobiosis (Nakano and Zhu, 2001). ResE is the only relevant kinase able to phosphorylate ResD. However, it was proposed that acetyl-phosphate (produced through the acetate pathway) could transfer its phosphate to ResD (Gueriri et al., 2008), linking its phosphorylation state to the metabolic status of the cell. Both phosphorylated and unphosphorylated forms of dimeric B. cereus ResD are able to bind DNA but their DNA binding affinity depends on promoter architecture of the target genes. For example, phosphorylation of ResD, which is higher under anaerobiosis than under aerobiosis enhances its ability to bind to its own promoter and fnr promoter but not to the enterotoxin gene promoters. Both ResD and ResD∼P physically interacts with Fnr and simultaneously bind their target promoter. A model was proposed on which ResD∼P may act as a Fnr co-activator and ResD as a Fnr anti-activator (Esbelin et al., 2012).
B. cereus Rex
Changes in oxygen availability and ORP influence the relative level of dinucleotides NAD+ and NADH in the cells, and such changes are sensed by the transcriptional regulator Rex. The crystal structure of Rex from Thermus aquaticus. Thermus thermophilus in complex with NADH and of B. subtilis Rex without cofactor has been determined (Sickmier et al., 2005; Wang et al., 2008). Rex is composed of two structural domains, an N-terminal domain that adopts a winged helix-turn-helix fold that most likely interacts with DNA, and a C-terminal NADH binding domain. In the complex with NADH, the N-terminal domains pack close to each other in a compact dimer. This conformation of Rex is unable to bind DNA. Rex is thus active as a repressor only when the NAD+/NADH ratio indicates adequate NAD+. By monitoring the NAD+/NADH ratio, Rex helps the cells to regulate pathways that regenerate NAD+. Typically, B. cereus Rex regulates the carbon flow distribution at the pyruvate node by favoring and limiting the carbon flow entry into the NADH-recycling lactate pathway under anoxic and oxic conditions, respectively (Laouami et al., 2014). By controlling this carbon flow, Rex also controls the availability of glycolytic intermediates for macromolecular synthesis as well as supporting NADPH production through different enzymes located in the TCA cycle, glycolysis and PPP (Laouami et al., 2014). In addition to fine-tune the levels of the prooxidant NADH and the antioxidant NADP, Rex regulates directly the synthesis of antioxidant systems like the OhrRA system (Laouami et al., 2014). Rex also regulates directly the expression of fnr. resD, and ldhA. LdhA activity is a critical factor in B. cereus virulence (Laouami et al., 2011).
B. cereus OhrRA
The B. cereus OhrRA (organic hydroperoxide resistance) system comprises a thiol-dependent peroxidase protein (OhrA), which functions as a low ORP sensor under anoxic conditions and a redox-sensing transcriptional regulator (OhrR), which belongs to the MarR family of winged helix-turn-helix DNA binding protein (Panmanee et al., 2006; Dubbs and Mongkolsuk, 2007). The genes encoding OhrR and OhrA form a bicistronic transcriptional unit (Clair et al., 2012). OhrA is usually reported as a protein that detoxifies the organic hydroperoxides (OHP). OHP could result from oxidation of unsaturated FA by molecular oxygen or ROS (Mongkolsuk et al., 1998). This may explain why OhrA is induced under normoxia and low-ORP anoxia where ROS are the by-product of secondary oxidative stress. B. cereus OhrR is an atypical OhrR protein because it contains four cysteine residues at its N-terminal domain while most OhrR regulators contain two cysteines. Like its orthologs in other bacteria, B. cereus OhrR functions as a transcriptional regulator that binds to its own promoter under its dimeric reduced form. However, unlike most of its orthologs, it may function as a repressor and an activator of several metabolism-related genes. OhrR is mainly a non-covalent dimer in its reduced form and a covalent dimer in its oxidized form. Dihydrolipoate and LMW participate in the recovery of reduced OhrR from the oxidized form and thus may control the activity of the redox-sensing OhrR regulator (Clair et al., 2013). Besides to regulate the antioxidant system, which includes OhrA, B. cereus OhrR controls the abundance level of key enzymes of central metabolism. In this way, (i) it modulates the glycolytic flux and restricts B. cereus growth under low ORP fermentative conditions and (ii) it sustains high TCA capacity and limits energy spilling through an overflow metabolism under aerobic respiratory growth.
Interactions among the Redox Regulators
Although each of the global redox sensors senses different signals, they interact with each other and with operon-specific promoters. Transcriptional activation by Fnr is probably the first response to changing oxygen availability. Fnr regulates its own transcription and the transcription of resDE and rex, thereby making ResD and Rex more active/inactive as regulators of both catabolic pathways that supply the metabolic intermediates necessary to synthetize enterotoxins, and enterotoxin gene expression. Because Fnr and ResD interact with each other, the change of ResD level generated by Fnr accentuates or reduces this interaction. In addition, the changes of menaquinone redox state and NAD+/NADH ratio induced by changing oxygen concentrations also affect the activities of ResD and Rex, respectively; this impacts the expression of fnr and their own expression. By regulating the synthesis of the OhrRA system, which is indirectly stimulated by ROS under both normoxia and low-ORP anoxia, Rex also impacts indirectly carbon flow, and enterotoxin synthesis. In conclusion, the regulatory network involving redox sensors is undoubtedly complex but permits the microorganism to coordinate efficiently its central metabolism with enterotoxin production (Figure 4).
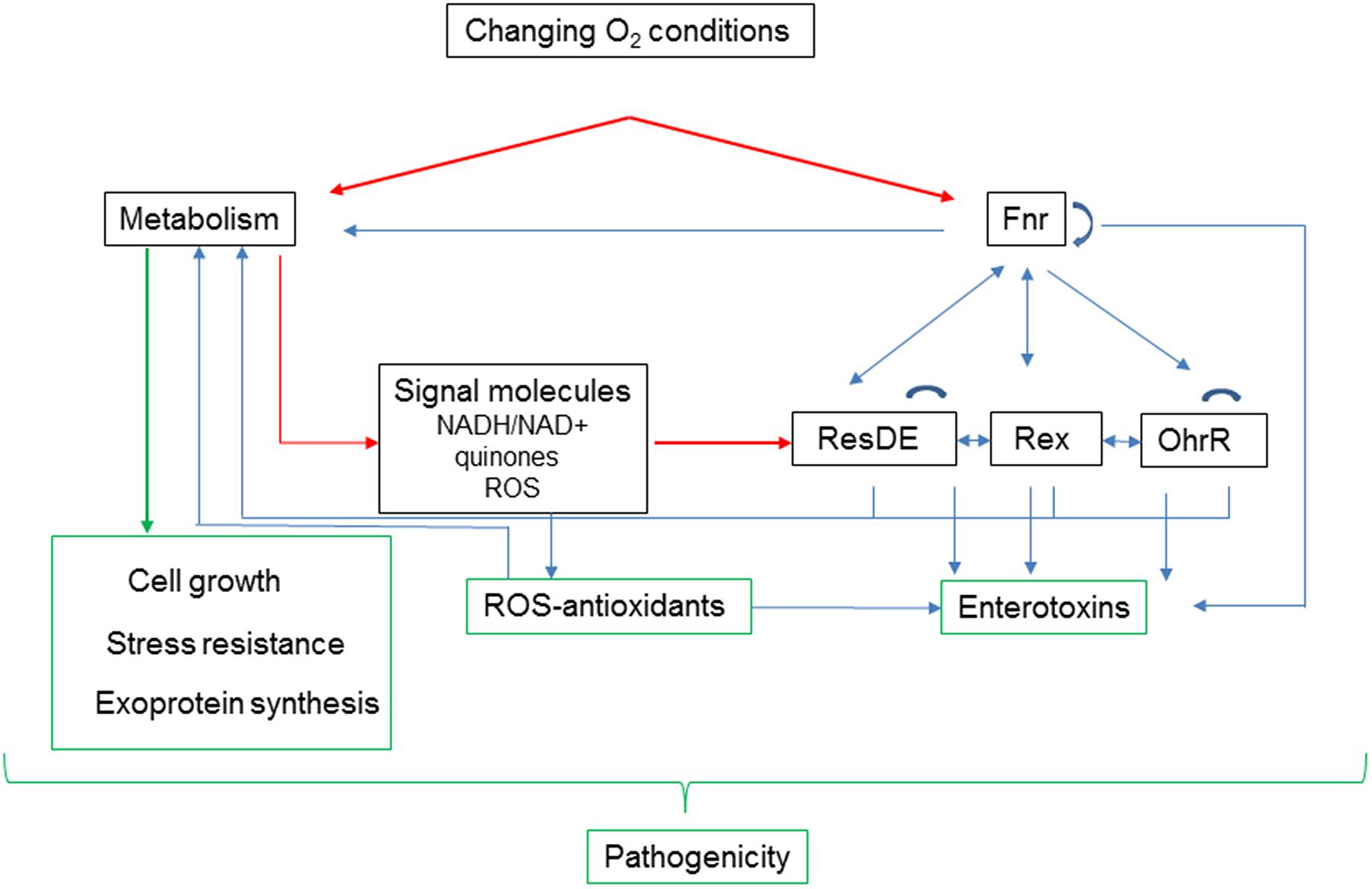
FIGURE 4. Adaptation to changing oxygen availability affects B. cereus pathogenicity at different levels. Oxygen acts directly on activity of Fnr and metabolism. Fnr, ResDE, Rex, and OhrR belong to a redox signaling pathway, which interconnects metabolism with expression of enterotoxins. In addition to its role in removing metabolism-generated ROS, antioxidant status modulates the oxidation status of Met residues in enterotoxins (Madeira et al., 2015). Metabolism also contributes directly to pathogenicity by supporting growth and stress resistance, and by modulating exoprotein synthesis.
Concluding Remarks
Maintaining an appropriate redox balance is essential for B. cereus adaptation to changing oxygen availability, and probably for resistance to acid stress (Liu et al., 2016). Redox homeostasis depends of the antioxidant system, which enables bacterial cells to maintain proteins and other cellular components in active state for metabolism. However, to date, we lack knowledge on the intracellular B. cereus environment, the behavior of redox couples under different environmental conditions, and the mechanisms of sustained redox homeostasis in B. cereus. In particular, a fundamental challenge is to understand how antioxidant-oxidant interactions modulate B. cereus pathogenesis.
Conclusion
Bacillus cereus adaptation to acid and low oxygen environments follows pathways that look quite similar to those that have been examined and described in great detail for other bacteria. However, there are also several clear differences that warrant the further examination of the adaptation mechanisms of B. cereus to its challenging environments from both a fundamental and industrial point of view.
What is clearly lagging in B. cereus research is knowledge of the possible roles that small RNAs (sRNAs) may play in stress responses. Indeed, it is now evident that sRNAs play an essential role in gene regulation under various stress conditions (Babu et al., 2011; Hoe et al., 2013; Miller et al., 2014). Thus, it is crucial that we uncover this important regulatory layer in B. cereus, as this will certainly lead to new insights and refine our understanding of the way in which B. cereus withstands environmental stress. Another issue that requires attention is the importance of cell individuality in responding to stressors. Cells in a bacterial population, even in a very uniform environment, may differ considerably with respect to the genetic program that is operative under these conditions. Such flexibility occurs in B. cereus spores (Van Melis et al., 2014). It would be interesting and important to find out whether culture heterogeneity also plays a role in the responses of B. cereus toward acid stress and low oxygen tension (Pandey et al., 2016). Overall, it is expected that investigations of the stress physiology of B. cereus will continue to be central to understanding its behavior in challenging environments. Omics approaches available today will lead to important discoveries that can be applied in food safety.
Author Contributions
CD wrote the paper. MJ and PS contribute to the writing of the part one of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adak, S., Aulak, K. S., and Stuehr, D. J. (2002). Direct evidence for nitric oxide production by a nitric-oxide synthase-like protein from Bacillus subtilis. J. Biol. Chem. 277, 16167–16171. doi: 10.1074/jbc.M201136200
Agledal, L., Niere, M., and Ziegler, M. (2010). The phosphate makes a difference: cellular functions of NADP. Redox Rep. 15, 2–10. doi: 10.1179/174329210X12650506623122
Alonso-Hernando, A., Alonso-Calleja, C., and Capita, R. (2010). Effects of exposure to poultry chemical decontaminants on the membrane fluidity of Listeria monocytogenes and Salmonella enterica strains. Int. J. Food Microbiol. 137, 130–136. doi: 10.1016/j.ijfoodmicro.2009.11.022
Alvarez-Ordonez, A., Fernandez, A., Bernardo, A., and Lopez, M. (2010a). Acid adaptation sensitizes Salmonella enterica serovar Typhimurium to osmotic and oxidative stresses. Arch. Lebensmittelhyg. 61, 148–152. doi: 10.2376/0003-925X-61-148
Alvarez-Ordonez, A., Fernandez, A., Bernardo, A., and Lopez, M. (2010b). Acid tolerance in Salmonella typhimurium induced by culturing in the presence of organic acids at different growth temperatures. Food Microbiol. 27, 44–49. doi: 10.1016/j.fm.2009.07.015
Alvarez-Ordonez, A., Fernandez, A., Bernardo, A., and Lopez, M. (2010c). Arginine and lysine decarboxylases and the acid tolerance response of Salmonella Typhimurium. Int. J. Food Microbiol. 136, 278–282. doi: 10.1016/j.ijfoodmicro.2009.09.024
Andreeva, Z. I., Nesterenko, V. F., Yurkov, I. S., Budarina, Z. I., Sineva, E. V., and Solonin, A. S. (2006). Purification and cytotoxic properties of Bacillus cereus hemolysin II. Protein Expr. Purif. 47, 186–193. doi: 10.1016/j.pep.2005.10.030
Antikainen, J., Kuparinen, V., Lahteenmaki, K., and Korhonen, T. K. (2007). pH-dependent association of enolase and glyceraldehyde-3-phosphate dehydrogenase of Lactobacillus crispatus with the cell wall and lipoteichoic acids. J. Bacteriol. 189, 4539–4543. doi: 10.1128/JB.00378-07
Armengaud, J., Christie-Oleza, J. A., Clair, G., Malard, V., and Duport, C. (2012). Exoproteomics: exploring the world around biological systems. Expert Rev. Proteomics 9, 561–575. doi: 10.1586/epr.12.52
Babu, M. M., Sridhar, J., and Gunasekaran, P. (2011). Global transcriptome analysis of Bacillus cereus ATCC 14579 in response to silver nitrate stress. J. Nanobiotechnology 9:49. doi: 10.1186/1477-3155-9-49
Baida, G., Budarina, Z. I., Kuzmin, N. P., and Solonin, A. S. (1999). Complete nucleotide sequence and molecular characterization of hemolysin II gene from Bacillus cereus. FEMS Microbiol. Lett. 180, 7–14. doi: 10.1111/j.1574-6968.1999.tb08771.x
Banfalvi, G., Csuzi, S., and Ohlbaum, A. (1980a). DNA-dependent ATPase II from Bacillus cereus. Biochim. Biophys. Acta 615, 262–270. doi: 10.1016/0005-2744(80)90029-7
Banfalvi, G., Ohlbaum, A., Csuzi, S., and Antoni, F. (1980b). Purification and characterization of a DNA-dependent ATPase from Bacillus cereus. Acta Microbiol. Acad. Sci. Hung. 27, 289–297.
Beecher, D. J., Schoeni, J. L., and Wong, A. C. (1995). Enterotoxic activity of hemolysin BL from Bacillus cereus. Infect. Immun. 63, 4423–4428.
Bergman, N. H., Anderson, E. C., Swenson, E. E., Janes, B. K., Fisher, N., Niemeyer, M. M., et al. (2007). Transcriptional profiling of Bacillus anthracis during infection of host macrophages. Infect. Immun. 75, 3434–3444. doi: 10.1128/IAI.01345-06
Berk, P. A., Jonge, R., Zwietering, M. H., Abee, T., and Kieboom, J. (2005). Acid resistance variability among isolates of Salmonella enterica serovar Typhimurium DT104. J. Appl. Microbiol. 99, 859–866. doi: 10.1111/j.1365-2672.2005.02658.x
Bohm, M. E., Krey, V. M., Jessberger, N., Frenzel, E., and Scherer, S. (2016). Comparative bioinformatics and experimental analysis of the intergenic regulatory regions of Bacillus cereus hbl and nhe enterotoxin operons and the impact of CodY on virulence heterogeneity. Front. Microbiol. 7:768. doi: 10.3389/fmicb.2016.00768
Boonchai, N., Asano, S. I., Bando, H., and Wiwat, C. (2008). Study on cytotoxicity and nucleotide sequences of enterotoxin FM of Bacillus cereus isolated from various food sources. J. Med. Assoc. Thai. 91, 1425–1432.
Booth, M. (2002). Discussion on simultaneous intraesophageal impedance and pH measurement of acid and nonacid gastroesophageal reflux: effect of omeprazole. Gastroenterology 122:1545. doi: 10.1053/gast.2002.33376
Browne, N., and Dowds, B. C. (2002). Acid stress in the food pathogen Bacillus cereus. J. Appl. Microbiol. 92, 404–414. doi: 10.1046/j.1365-2672.2002.01541.x
Budin-Verneuil, A., Maguin, E., Auffray, Y., Ehrlich, D. S., and Pichereau, V. (2006). Genetic structure and transcriptional analysis of the arginine deiminase (ADI) cluster in Lactococcus lactis MG1363. Can. J. Microbiol. 52, 617–622. doi: 10.1139/w06-009
Bueno, E., Mesa, S., Bedmar, E. J., Richardson, D. J., and Delgado, M. J. (2012). Bacterial adaptation of respiration from oxic to microoxic and anoxic conditions: redox control. Antioxid. Redox Signal. 16, 819–852. doi: 10.1089/ars.2011.4051
Canet, S., Heyde, M., Portalier, R., and Laloi, P. (2003). Involvement of phospholipids in resistance and adaptation of Escherichia coli to acid conditions and to long-term survival. FEMS Microbiol. Lett. 225, 207–211. doi: 10.1016/S0378-1097(03)00515-9
Cao, Y., Jin, X., Levin, E. J., Huang, H., Zong, Y., Quick, M., et al. (2011). Crystal structure of a phosphorylation-coupled saccharide transporter. Nature 473, 50–54. doi: 10.1038/nature09939
Carlsson, J., Bergstrom, J., and Pehrson, B. (1995). Variations with breed, age, season, yield, stage of lactation and herd in the concentration of urea in bulk milk and individual cow’s milk. Acta Vet. Scand. 36, 245–254.
Ceuppens, S., Rajkovic, A., Hamelink, S., Van De Wiele, T., Boon, N., and Uyttendaele, M. (2012). Enterotoxin production by Bacillus cereus under gastrointestinal conditions and their immunological detection by commercially available kits. Foodborne Pathog. Dis. 9, 1130–1136. doi: 10.1089/fpd.2012.1230
Ceuppens, S., Van De Wiele, T., Boon, N., and Uyttendaele, M. (2011). Gastrointestinal passage of Bacillus cereus. Commun. Agric. Appl. Biol. Sci. 76, 3–6.
Chen, Y. Y., and Burne, R. A. (1996). Analysis of Streptococcus salivarius urease expression using continuous chemostat culture. FEMS Microbiol. Lett. 135, 223–229. doi: 10.1111/j.1574-6968.1996.tb07993.x
Chen, Y. Y., Clancy, K. A., and Burne, R. A. (1996). Streptococcus salivarius urease: genetic and biochemical characterization and expression in a dental plaque streptococcus. Infect. Immun. 64, 585–592.
Clair, G., Armengaud, J., and Duport, C. (2012). Restricting fermentative potential by proteome remodeling: an adaptative strategy evidenced in Bacillus cereus. Mol. Cell. Proteomics 11, M111.013102. doi: 10.1074/mcp.M111.013102
Clair, G., Lorphelin, A., Armengaud, J., and Duport, C. (2013). OhrRA functions as a redox-responsive system controlling toxinogenesis in Bacillus cereus. J. Proteomics 94, 527–539. doi: 10.1016/j.jprot.2013.10.024
Clair, G., Roussi, S., Armengaud, J., and Duport, C. (2010). Expanding the known repertoire of virulence factors produced by Bacillus cereus through early secretome profiling in three redox conditions. Mol. Cell. Proteomics 9, 1486–1498. doi: 10.1074/mcp.M000027-MCP201
Contreras-Zentella, M., Mendoza, G., Membrillo-Hernandez, J., and Escamilla, J. E. (2003). A novel double heme substitution produces a functional bo3 variant of the quinol oxidase aa3 of Bacillus cereus. Purification and paratial characterization. J. Biol. Chem. 278, 31473–31478. doi: 10.1074/jbc.M302583200
Cotter, P. D., and Hill, C. (2003). Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67, 429–453. doi: 10.1128/Mmbr.37.3.429-453.2003
Cotter, P. D., Ryan, S., Gahan, C. G., and Hill, C. (2005). Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl. Environ. Microbiol. 71, 2832–2839. doi: 10.1128/AEM.71.6.2832-2839.2005
Dawes, C., and Dibdin, G. H. (2001). Salivary concentrations of urea released from a chewing gum containing urea and how these affect the urea content of gel-stabilized plaques and their pH after exposure to sucrose. Caries Res. 35, 344–353. doi: 10.1159/000047473
De Angelis, M., Mariotti, L., Rossi, J., Servili, M., Fox, P. F., Rollan, G., et al. (2002). Arginine catabolism by sourdough lactic acid bacteria: purification and characterization of the arginine deiminase pathway enzymes from Lactobacillus sanfranciscensis CB1. Appl. Environ. Microbiol. 68, 6193–6201. doi: 10.1128/AEM.68.12.6193-6201.2002
De Biase, D., and Pennacchietti, E. (2012). Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: function, distribution and biomedical implications of the gadBC operon. Mol. Microbiol. 86, 770–786. doi: 10.1111/mmi.12020
De Sarrau, B., Clavel, T., Clerte, C., Carlin, F., Ginies, C., and Nguyen-The, C. (2012). Influence of anaerobiosis and low temperature on Bacillus cereus growth, metabolism, and membrane properties. Appl. Environ. Microbiol. 78, 1715–1723. doi: 10.1128/AEM.06410-11
Deutscher, J., Francke, C., and Postma, P. W. (2006). How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70, 939–1031. doi: 10.1128/MMBR.00024-06
Didier, A., Dietrich, R., Gruber, S., Bock, S., Moravek, M., Nakamura, T., et al. (2012). Monoclonal antibodies neutralize Bacillus cereus Nhe enterotoxin by inhibiting ordered binding of its three exoprotein components. Infect. Immun. 80, 832–838. doi: 10.1128/IAI.05681-11
Dubbs, J. M., and Mongkolsuk, S. (2007). Peroxiredoxins in bacterial antioxidant defense. Subcell Biochem. 44, 143–193. doi: 10.1007/978-1-4020-6051-9_7
Duport, C., Thomassin, S., Bourel, G., and Schmitt, P. (2004). Anaerobiosis and low specific growth rates enhance hemolysin BL production by Bacillus cereus F4430/73. Arch. Microbiol. 182, 90–95. doi: 10.1007/s00203-004-0688-y
Duport, C., Zigha, A., Rosenfeld, E., and Schmitt, P. (2006). Control of enterotoxin gene expression in Bacillus cereus F4430/73 involves the redox-sensitive ResDE signal transduction system. J. Bacteriol. 188, 6640–6651. doi: 10.1128/JB.00702-06
Ebner, P., Rinker, J., and Gotz, F. (2016a). Excretion of cytoplasmic proteins in Staphylococcus is most likely not due to cell lysis. Curr. Genet. 62, 19–23. doi: 10.1007/s00294-015-0504-z.
Ebner, P., Rinker, J., Nguyen, M. T., Popella, P., Nega, M., Luqman, A., et al. (2016b). Excreted Cytoplasmic proteins contribute to pathogenicity in Staphylococcus aureus. Infect. Immun. 84, 1672–1681. doi: 10.1128/IAI.00138-16
Eisenreich, W., Slaghuis, J., Laupitz, R., Bussemer, J., Stritzker, J., Schwarz, C., et al. (2006). C-13 isotopologue perturbation studies of Listeria monocytogenes carbon metabolism and its modulation by the virulence regulator PrfA. Proc. Natl. Acad. Sci. U.S.A. 103, 2040–2045. doi: 10.1073/pnas.0507580103
Esbelin, J., Armengaud, J., Zigha, A., and Duport, C. (2009). ResDE-dependent regulation of enterotoxin gene expression in Bacillus cereus: evidence for multiple modes of binding for ResD and interaction with Fnr. J. Bacteriol. 191, 4419–4426. doi: 10.1128/JB.00321-09
Esbelin, J., Jouanneau, Y., Armengaud, J., and Duport, C. (2008). ApoFnr binds as a monomer to promoters regulating the expression of enterotoxin genes of Bacillus cereus. J. Bacteriol. 190, 4242–4251. doi: 10.1128/JB.00336-08
Esbelin, J., Jouanneau, Y., and Duport, C. (2012). Bacillus cereus Fnr binds a [4Fe-4S] cluster and forms a ternary complex with ResD and PlcR. BMC Microbiol. 12:125. doi: 10.1186/1471-2180-12-125
Fagerlund, A., Lindback, T., and Granum, P. E. (2010). Bacillus cereus cytotoxins Hbl, Nhe and CytK are secreted via the Sec translocation pathway. BMC Microbiol. 10:304. doi: 10.1186/1471-2180-10-304
Fagerlund, A., Lindback, T., Storset, A. K., Granum, P. E., and Hardy, S. P. (2008). Bacillus cereus Nhe is a pore-forming toxin with structural and functional properties similar to the ClyA (HlyE, SheA) family of haemolysins, able to induce osmotic lysis in epithelia. Microbiology 154, 693–704. doi: 10.1099/mic.0.2007/014134-0
Fang, Z., Roberts, A. A., Weidman, K., Sharma, S. V., Claiborne, A., Hamilton, C. J., et al. (2013). Cross-functionalities of Bacillus deacetylases involved in bacillithiol biosynthesis and bacillithiol-S-conjugate detoxification pathways. Biochem. J. 454, 239–247. doi: 10.1042/BJ20130415
Fortier, L. C., Tourdot-Marechal, R., Divies, C., Lee, B. H., and Guzzo, J. (2003). Induction of Oenococcus oeni H+-ATPase activity and mRNA transcription under acidic conditions. FEMS Microbiol. Lett. 222, 165–169. doi: 10.1016/S0378-1097(03)00299-4
Fozo, E. M., Kajfasz, J. K., and Quivey, R. G. Jr. (2004). Low pH-induced membrane fatty acid alterations in oral bacteria. FEMS Microbiol. Lett. 238, 291–295. doi: 10.1016/j.femsle.2004.07.047
Fozo, E. M., and Quivey, R. G. Jr. (2004). Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Appl. Environ. Microbiol. 70, 929–936. doi: 10.1128/AEM.70.2.929-936.2004
Frees, D., Qazi, S. N., Hill, P. J., and Ingmer, H. (2003a). Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 48, 1565–1578. doi: 10.1046/j.1365-2958.2003.03524.x
Frees, D., Vogensen, F. K., and Ingmer, H. (2003b). Identification of proteins induced at low pH in Lactococcus lactis. Int. J. Food Microbiol. 87, 293–300. doi: 10.1016/S0168-1605(03)00104-1
Garcia, L. M., Contreras-Zentella, M. L., Jaramillo, R., Benito-Mercade, M. C., Mendoza-Hernandez, G., Del Arenal, I. P., et al. (2008). The succinate:menaquinone reductase of Bacillus cereus: characterization of the membrane-bound and purified enzyme. Can. J. Microbiol. 54, 456–466. doi: 10.1139/w08-037
Geng, H., Zhu, Y., Mullen, K., Zuber, C. S., and Nakano, M. M. (2007). Characterization of ResDE-dependent fnr transcription in Bacillus subtilis. J. Bacteriol. 189, 1745–1755. doi: 10.1128/JB.01502-06
Gohar, M., Faegri, K., Perchat, S., Ravnum, S., Okstad, O. A., Gominet, M., et al. (2008). The PlcR virulence regulon of Bacillus cereus. PLoS ONE 3:e2793. doi: 10.1371/journal.pone.0002793
Gonzalez-Flecha, B., and Demple, B. (1995). Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J. Biol. Chem. 270, 13681–13687. doi: 10.1074/jbc.270.23.13681
Gotz, F., Yu, W., Dube, L., Prax, M., and Ebner, P. (2015). Excretion of cytosolic proteins (ECP) in bacteria. Int. J. Med. Microbiol. 305, 230–237. doi: 10.1016/j.ijmm.2014.12.021
Griswold, A., Chen, Y.-Y. M., Snyder, J. A., and Burne, R. A. (2004). Characterization of the arginine deiminase operon of Streptococcus rattus FA-1. Appl. Environ. Microbiol. 70, 1321–1327. doi: 10.1128/AEM.70.3.1321-1327.2004
Gueriri, I., Bay, S., Dubrac, S., Cyncynatus, C., and Msadek, T. (2008). The Pta-AckA pathway controlling acetyl phosphate levels and the phosphorylation state of the DegU orphan response regulator both play a role in regulating Listeria monocytogenes motility and chemotaxis. Mol. Microbiol. 70, 1342–1357. doi: 10.1111/j.1365-2958.2008.06496.x
Gusarov, I., and Nudler, E. (2012). S-nitrosylation signaling in Escherichia coli. Sci. Signal. 5:pe26. doi: 10.1126/scisignal.2003181
Gusarov, I., Starodubtseva, M., Wang, Z. Q., Mcquade, L., Lippard, S. J., Stuehr, D. J., et al. (2008). Bacterial nitric-oxide synthases operate without a dedicated redox partner. J. Biol. Chem. 283, 13140–13147. doi: 10.1074/jbc.M710178200
Hanna, M. N., Ferguson, R. J., Li, Y. H., and Cvitkovitch, D. G. (2001). uvrA is an acid-inducible gene involved in the adaptive response to low pH in Streptococcus mutans. J. Bacteriol. 183, 5964–5973. doi: 10.1128/JB.183.20.5964-5973.2001
Hardy, S. P., Lund, T., and Granum, P. E. (2001). CytK toxin of Bacillus cereus forms pores in planar lipid bilayers and is cytotoxic to intestinal epithelia. FEMS Microbiol. Lett. 197, 47–51. doi: 10.1111/j.1574-6968.2001.tb10581.x
Helmann, J. D. (2011). Bacillithiol, a new player in bacterial redox homeostasis. Antioxid. Redox Signal. 15, 123–133. doi: 10.1089/ars.2010.3562
Henderson, B., and Martin, A. (2011). Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 79, 3476–3491. doi: 10.1128/IAI.00179-11
Higuti, I. H., Stencel, M., Nascimento, K. H., and Nascimento, A. J. (1992). Studies on the ATPase of Bacillus cereus. Cell Biochem. Funct. 10, 237–241. doi: 10.1002/cbf.290100405
Hoe, C. H., Raabe, C. A., Rozhdestvensky, T. S., and Tang, T. H. (2013). Bacterial sRNAs: regulation in stress. Int. J. Med. Microbiol. 303, 217–229. doi: 10.1016/j.ijmm.2013.04.002
Imlay, J. A. (2013). The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 11, 443–454. doi: 10.1038/nrmicro3032
Ivanova, N., Sorokin, A., Anderson, I., Galleron, N., Candelon, B., Kapatral, V., et al. (2003). Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423, 87–91. doi: 10.1038/nature01582
Jayaraman, G. C., Penders, J. E., and Burne, R. A. (1997). Transcriptional analysis of the Streptococcus mutans hrcA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol. Microbiol. 25, 329–341. doi: 10.1046/j.1365-2958.1997.4671835.x
Jessberger, N., Krey, V. M., Rademacher, C., Bohm, M. E., Mohr, A. K., Ehling-Schulz, M., et al. (2015). From genome to toxicity: a combinatory approach highlights the complexity of enterotoxin production in Bacillus cereus. Front. Microbiol. 6:560. doi: 10.3389/fmicb.2015.00560
Jiang, D., Tikhomirova, A., Bent, S. J., and Kidd, S. P. (2016). A discrete role for FNR in the transcriptional response to moderate changes in oxygen by Haemophilus influenzae Rd KW20. Res. Microbiol. 167, 103–113. doi: 10.1016/j.resmic.2015.09.008
Jobin, M. P., Clavel, T., Carlin, F., and Schmitt, P. (2002). Acid tolerance response is low-pH and late-stationary growth phase inducible in Bacillus cereus TZ415. Int. J. Food Microbiol. 79, 65–73. doi: 10.1016/S0168-1605(02)00180-0
Kalkowski, I., and Conrad, R. (1991). Metabolism of nitric oxide in denitrifying Pseudomonas aeruginosa and nitrate-respiring Bacillus cereus. FEMS Microbiol. Lett. 66, 107–111. doi: 10.1111/j.1574-6968.1991.tb04848.x
Kanjee, U., and Houry, W. A. (2013). Mechanisms of acid resistance in Escherichia coli. Annu. Rev. Microbiol. 67, 65–81. doi: 10.1146/annurev-micro-092412-155708
Kayser, M. F., Ucurum, Z., and Vuilleumier, S. (2002). Dichloromethane metabolism and C1 utilization genes in Methylobacterium strains. Microbiology 148, 1915–1922. doi: 10.1099/00221287-148-6-1915
Kieboom, J., and Abee, T. (2006). Arginine-dependent acid resistance in Salmonella enterica serovar Typhimurium. J. Bacteriol. 188, 5650–5653. doi: 10.1128/JB.00323-06
Korner, H., Sofia, H. J., and Zumft, W. G. (2003). Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27, 559–592. doi: 10.1016/S0168-6445(03)00066-4
Kreft, J., Berger, H., Hartlein, M., Muller, B., Weidinger, G., and Goebel, W. (1983). Cloning and expression in Escherichia coli and Bacillus subtilis of the hemolysin (cereolysin) determinant from Bacillus cereus. J. Bacteriol. 155, 681–689.
Kullen, M. J., and Klaenhammer, T. R. (1999). Identification of the pH-inducible, proton-translocating F1F0-ATPase (atpBEFHAGDC) operon of Lactobacillus acidophilus by differential display: gene structure, cloning and characterization. Mol. Microbiol. 33, 1152–1161. doi: 10.1046/j.1365-2958.1999.01557.x
Kwon, Y. M., and Ricke, S. C. (1998a). Induction of acid resistance of Salmonella typhimurium by exposure to short-chain fatty acids. Appl. Environ. Microbiol. 64, 3458–3463.
Kwon, Y. M., and Ricke, S. C. (1998b). Survival of a Salmonella typhimurium poultry isolate in the presence of propionic acid under aerobic and anaerobic conditions. Anaerobe 4, 251–256. doi: 10.1006/anae.1998.0177
Lage, C., de Padula, M., de Alencar, T. A., da Fonseca Goncalves, S. R., da Silva Vidal, L., Cabral-Neto, J., et al. (2003). New insights on how nucleotide excision repair could remove DNA adducts induced by chemotherapeutic agents and psoralens plus UV-A (PUVA) in Escherichia coli cells. Mutat. Res. 544, 143–157. doi: 10.1016/j.mrrev.2003.06.003
Laouami, S., Clair, G., Armengaud, J., and Duport, C. (2014). Proteomic evidences for rex regulation of metabolism in toxin-producing Bacillus cereus ATCC 14579. PLoS ONE 9:e107354. doi: 10.1371/journal.pone.0107354
Laouami, S., Messaoudi, K., Alberto, F., Clavel, T., and Duport, C. (2011). Lactate dehydrogenase A promotes communication between carbohydrate catabolism and virulence in Bacillus cereus. J. Bacteriol. 193, 1757–1766. doi: 10.1128/JB.00024-11
Le Lay, J., Bahloul, H., Serino, S., Jobin, M., and Schmitt, P. (2015). Reducing activity, glucose metabolism and acid tolerance response of Bacillus cereus grown at various pH and oxydo-reduction potential levels. Food Microbiol. 46, 314–321. doi: 10.1016/j.fm.2014.07.007
Li, Y. H., Hanna, M. N., Svensater, G., Ellen, R. P., and Cvitkovitch, D. G. (2001). Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J. Bacteriol. 183, 6875–6884. doi: 10.1128/JB.183.23.6875-6884.2001
Lidder, S., and Webb, A. J. (2013). Vascular effects of dietary nitrate (as found in green leafy vegetables and beetroot) via the nitrate-nitrite-nitric oxide pathway. Br. J. Clin. Pharmacol. 75, 677–696. doi: 10.1111/j.1365-2125.2012.04420.x
Lindback, T., Fagerlund, A., Rodland, M. S., and Granum, P. E. (2004). Characterization of the Bacillus cereus Nhe enterotoxin. Microbiology 150, 3959–3967. doi: 10.1099/mic.0.27359-0
Lindback, T., Mols, M., Basset, C., Granum, P. E., Kuipers, O. P., and Kovacs, A. T. (2012). CodY, a pleiotropic regulator, influences multicellular behaviour and efficient production of virulence factors in Bacillus cereus. Environ. Microbiol. 14, 2233–2246. doi: 10.1111/j.1462-2920.2012.02766.x
Liu, Y., Yang, X., Yin, Y., Lin, J., Chen, C., Pan, J., et al. (2016). Mycothiol protects Corynebacterium glutamicum against acid stress via maintaining intracellular pH homeostasis, scavenging ROS, and S-mycothiolating MetE. J. Gen. Appl. Microbiol. 62, 144–153. doi: 10.2323/jgam.2016.02.001
Loi, V. V., Rossius, M., and Antelmann, H. (2015). Redox regulation by reversible protein S-thiolation in bacteria. Front. Microbiol. 6:187. doi: 10.3389/fmicb.2015.00187
Lumppio, H. L., Shenvi, N. V., Summers, A. O., Voordouw, G., and Kurtz, D. M. J.r. (2001). Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system. J. Bacteriol. 183, 101–108. doi: 10.1128/JB.183.1.101-108.2001
Lund, T., De Buyser, M. L., and Granum, P. E. (2000). A new cytotoxin from Bacillus cereus that may cause necrotic enteritis. Mol. Microbiol. 38, 254–261. doi: 10.1046/j.1365-2958.2000.02147.x
Luo, S., and Levine, R. L. (2009). Methionine in proteins defends against oxidative stress. FASEB J. 23, 464–472. doi: 10.1096/fj.08-118414
Mackay, E. M., and Mackay, L. L. (1927). The concentration of urea in the blood of normal individuals. J. Clin. Invest. 4, 295–306. doi: 10.1172/JCI100124
Madegowda, M., Eswaramoorthy, S., Burley, S. K., and Swaminathan, S. (2008). X-ray crystal structure of the B component of Hemolysin BL from Bacillus cereus. Proteins 71, 534–540. doi: 10.1002/prot.21888
Madeira, J. P., Alpha-Bazin, B., Armengaud, J., and Duport, C. (2015). Time dynamics of the Bacillus cereus exoproteome are shaped by cellular oxidation. Front. Microbiol. 6:342. doi: 10.3389/fmicb.2015.00342
Madeira, J. P., Alpha-Bazin, B., Armengaud, J., Omer, H., and Duport, C. (2016a). Proteome data to explore the impact of pBClin15 on Bacillus cereus ATCC 14579. Data Brief. 8, 1243–1246. doi: 10.1016/j.dib.2016.07.042
Madeira, J. P., Omer, H., Alpha-Bazin, B., Armengaud, J., and Duport, C. (2016b). Deciphering the interactions between the Bacillus cereus linear plasmid, pBClin15, and its host by high-throughput comparative proteomics. J. Proteomics 146, 25–33. doi: 10.1016/j.jprot.2016.06.022
Mailloux, R. J., Lemire, J., and Appanna, V. D. (2011). Metabolic networks to combat oxidative stress in Pseudomonas fluorescens. Antonie Van Leeuwenhoek 99, 433–442. doi: 10.1007/s10482-010-9538-x
Marquis, H., and Hager, E. J. (2000). pH-regulated activation and release of a bacteria-associated phospholipase C during intracellular infection by Listeria monocytogenes. Mol. Microbiol. 35, 289–298. doi: 10.1046/j.1365-2958.2000.01708.x
Marteyn, B., Scorza, F. B., Sansonetti, P. J., and Tang, C. (2010a). Breathing life into pathogens: the influence of oxygen on bacterial virulence and host responses in the gastrointestinal tract. Cell. Microbiol. 13, 171–176. doi: 10.1111/j.1462-5822.2010.01549.x
Marteyn, B., West, N. P., Browning, D. F., Cole, J. A., Shaw, J. G., Palm, F., et al. (2010b). Modulation of Shigella virulence in response to available oxygen in vivo. Nature 465, 355–358. doi: 10.1038/nature08970
Melo, A. M., and Teixeira, M. (2016). Supramolecular organization of bacterial aerobic respiratory chains: from cells and back. Biochim. Biophys. Acta 1857, 190–197. doi: 10.1016/j.bbabio.2015.11.001
Mercade, M., Lindley, N. D., and Loubiere, P. (2000). Metabolism of Lactococcus lactis subsp. cremoris MG 1363 in acid stress conditions. Int. J. Food Microbiol. 55, 161–165. doi: 10.1016/S0168-1605(00)00190-2
Merrell, D. S., and Camilli, A. (2002). Acid tolerance of gastrointestinal pathogens. Curr. Opin. Microbiol. 5, 51–55. doi: 10.1016/S1369-5274(02)00285-0
Merrell, D. S., Hava, D. L., and Camilli, A. (2002). Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol. Microbiol. 43, 1471–1491. doi: 10.1046/j.1365-2958.2002.02857.x
Messaoudi, K., Clavel, T., Schmitt, P., and Duport, C. (2010). Fnr mediates carbohydrate-dependent regulation of catabolic and enterotoxin genes in Bacillus cereus F4430/73. Res. Microbiol. 161, 30–39. doi: 10.1016/j.resmic.2009.11.003
Messner, K. R., and Imlay, J. A. (2002). Mechanism of superoxide and hydrogen peroxide formation by fumarate reductase, succinate dehydrogenase, and aspartate oxidase. J. Biol. Chem. 277, 42563–42571. doi: 10.1074/jbc.M204958200
Miller, E. W., Cao, T. N., Pflughoeft, K. J., and Sumby, P. (2014). RNA-mediated regulation in Gram-positive pathogens: an overview punctuated with examples from the group A Streptococcus. Mol. Microbiol. 94, 9–20. doi: 10.1111/mmi.12742
Minnaard, J., Humen, M., and Perez, P. F. (2001). Effect of Bacillus cereus exocellular factors on human intestinal epithelial cells. J. Food Prot. 64, 1535–1541.
Miwa, T., Esaki, H., Umemori, J., and Hino, T. (1997). Activity of H(+)-ATPase in ruminal bacteria with special reference to acid tolerance. Appl. Environ. Microbiol. 63, 2155–2158.
Mobley, H. L. (2000). UreI-mediated urea transport in Helicobacter pylori: an open and shut case? Trends Microbiol. 8, 346–348. doi: 10.1016/S0966-842X(00)01810-2
Mols, M., and Abee, T. (2011a). Bacillus cereus responses to acid stress. Environ. Microbiol. 13, 2835–2843. doi: 10.1111/j.1462-2920.2011.02490.x
Mols, M., and Abee, T. (2011b). Primary and secondary oxidative stress in Bacillus. Environ. Microbiol. 13, 1387–1394. doi: 10.1111/j.1462-2920.2011.02433.x
Mols, M., Ceragioli, M., and Abee, T. (2011). Heat stress leads to superoxide formation in Bacillus cereus detected using the fluorescent probe MitoSOX. Int. J. Food Microbiol. 151, 119–122. doi: 10.1016/j.ijfoodmicro.2011.08.004
Mols, M., De Been, M., Zwietering, M. H., Moezelaar, R., and Abee, T. (2007). Metabolic capacity of Bacillus cereus strains ATCC 14579 and ATCC 10987 interlinked with comparative genomics. Environ. Microbiol. 9, 2933–2944. doi: 10.1111/j.1462-2920.2007.01404.x
Mols, M., Pier, I., Zwietering, M. H., and Abee, T. (2009). The impact of oxygen availability on stress survival and radical formation of Bacillus cereus. Int. J. Food Microbiol. 135, 303–311. doi: 10.1016/j.ijfoodmicro.2009.09.002
Mols, M., Van Kranenburg, R., Tempelaars, M. H., Van Schaik, W., Moezelaar, R., and Abee, T. (2010a). Comparative analysis of transcriptional and physiological responses of Bacillus cereus to organic and inorganic acid shocks. Int. J. Food Microbiol. 137, 13–21. doi: 10.1016/j.ijfoodmicro.2009.09.027
Mols, M., Van Kranenburg, R., Van Melis, C. C., Moezelaar, R., and Abee, T. (2010b). Analysis of acid-stressed Bacillus cereus reveals a major oxidative response and inactivation-associated radical formation. Environ. Microbiol. 12, 873–885. doi: 10.1111/j.1462-2920.2009.02132.x
Mongkolsuk, S., Praituan, W., Loprasert, S., Fuangthong, M., and Chamnongpol, S. (1998). Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J. Bacteriol. 180, 2636–2643.
Moorman, M. A., Thelemann, C. A., Zhou, S., Pestka, J. J., Linz, J. E., and Ryser, E. T. (2008). Altered hydrophobicity and membrane composition in stress-adapted Listeria innocua. J. Food Prot. 71, 182–185.
Moriarty-Craige, S. E., and Jones, D. P. (2004). Extracellular thiols and thiol/disulfide redox in metabolism. Annu. Rev. Nutr. 2004, 481–509. doi: 10.1146/annurev.nutr.24.012003.132208
Nakano, M. M., and Zhu, Y. (2001). Involvement of ResE phosphatase activity in down-regulation of ResD-controlled genes in Bacillus subtilis during aerobic growth. J. Bacteriol. 183, 1938–1944. doi: 10.1128/JB.183.6.1938-1944.2001
Neely, M. N., and Olson, E. R. (1996). Kinetics of expression of the Escherichia coli cad operon as a function of pH and lysine. J. Bacteriol. 178, 5522–5528.
Negrin, R. S., Foster, D. L., and Fillingame, R. H. (1980). Energy-transducing H+-ATPase of Escherichia coli. Reconstitution of proton translocation activity of the intrinsic membrane sector. J. Biol. Chem. 255, 5643–5648.
Neithercut, W. D., El Nujumi, A. M., and Mccoll, K. E. (1993). Measurement of urea and ammonium concentrations in gastric juice. J. Clin. Pathol. 46, 462–464. doi: 10.1136/jcp.46.5.462
Nelson, D., Goldstein, J. M., Boatright, K., Harty, D. W., Cook, S. L., Hickman, P. J., et al. (2001). pH-regulated secretion of a glyceraldehyde-3-phosphate dehydrogenase from Streptococcus gordonii FSS2: purification, characterization, and cloning of the gene encoding this enzyme. J. Dent. Res. 80, 371–377. doi: 10.1177/00220345010800011301
Newton, G. L., Arnold, K., Price, M. S., Sherrill, C., Delcardayre, S. B., Aharonowitz, Y., et al. (1996). Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 178, 1990–1995.
Newton, G. L., Rawat, M., La Clair, J. J., Jothivasan, V. K., Budiarto, T., Hamilton, C. J., et al. (2009). Bacillithiol is an antioxidant thiol produced in Bacilli. Nat. Chem. Biol. 5, 625–627. doi: 10.1038/nchembio.189
Omer, H., Alpha-Bazin, B., Brunet, J. L., Armengaud, J., and Duport, C. (2015). Proteomics identifies Bacillus cereus EntD as a pivotal protein for the production of numerous virulence factors. Front. Microbiol. 6:1004. doi: 10.3389/fmicb.2015.01004
O’Sullivan, E., and Condon, S. (1999). Relationship between acid tolerance, cytoplasmic pH, and ATP and H+-ATPase levels in chemostat cultures of Lactococcus lactis. Appl. Environ. Microbiol. 65, 2287–2293.
Pandey, R., Vischer, N. O., Smelt, J. P., Van Beilen, J. W., Ter Beek, A., De Vos, W. H., et al. (2016). Intracellular pH response to weak acid stress in individual vegetative Bacillus subtilis cells. Appl. Environ. Microbiol. doi: 10.1128/AEM.02063-16 [Epub ahead of print].
Panmanee, W., Vattanaviboon, P., Poole, L. B., and Mongkolsuk, S. (2006). Novel organic hydroperoxide-sensing and responding mechanisms for OhrR, a major bacterial sensor and regulator of organic hydroperoxide stress. J. Bacteriol. 188, 1389–1395. doi: 10.1128/JB.188.4.1389-1395.2006
Papadimitriou, K., Pratsinis, H., Nebe-Von-Caron, G., Kletsas, D., and Tsakalidou, E. (2007). Acid tolerance of Streptococcus macedonicus as assessed by flow cytometry and single-cell sorting. Appl. Environ. Microbiol. 73, 465–476. doi: 10.1128/AEM.01244-06
Phung, D., Granum, P. E., Dietrich, R., Martlbauer, E., and Hardy, S. P. (2012). Inhibition of cytotoxicity by the Nhe cytotoxin of Bacillus cereus through the interaction of dodecyl maltoside with the NheB component. FEMS Microbiol. Lett. 330, 98–104. doi: 10.1111/j.1574-6968.2012.02538.x
Popova, T. G., Teunis, A., Vaseghi, H., Zhou, W., Espina, V., Liotta, L. A., et al. (2015). Nitric oxide as a regulator of B. anthracis pathogenicity. Front. Microbiol. 6:921. doi: 10.3389/fmicb.2015.00921
Quivey, R. G., Kuhnert, W. L., and Hahn, K. (2001). Genetics of acid adaptation in oral streptococci. Crit. Rev. Oral. Biol. Med. 12, 301–314. doi: 10.1177/10454411010120040201
Ramarao, N., and Lereclus, D. (2006). Adhesion and cytotoxicity of Bacillus cereus and Bacillus thuringiensis to epithelial cells are FlhA and PlcR dependent, respectively. Microbes Infect. 8, 1483–1491. doi: 10.1016/j.micinf.2006.01.005
Ramarao, N., and Sanchis, V. (2013). The pore-forming haemolysins of Bacillus cereus: a review. Toxins (Basel) 5, 1119–1139. doi: 10.3390/toxins5061119
Rhee, J. E., Kim, K. S., and Choi, S. H. (2005). CadC activates pH-dependent expression of the Vibrio vulnificus cadBA operon at a distance through direct binding to an upstream region. J. Bacteriol. 187, 7870–7875. doi: 10.1128/JB.187.22.7870-7875.2005
Richard, H., and Foster, J. W. (2004). Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J. Bacteriol. 186, 6032–6041. doi: 10.1128/JB.186.18.6032-6041.2004
Richard, H. T., and Foster, J. W. (2003). Acid resistance in Escherichia coli. Adv. Appl. Microbiol. 52, 167–186. doi: 10.1016/S0065-2164(03)01007-4
Rius, N., and Loren, J. G. (1998). Buffering capacity and membrane H+ conductance of neutrophilic and alkalophilic gram-positive bacteria. Appl. Environ. Microbiol. 64, 1344–1349.
Rosenfeld, E., Duport, C., Zigha, A., and Schmitt, P. (2005). Characterization of aerobic and anaerobic vegetative growth of the food-borne pathogen Bacillus cereus F4430/73 strain. Can. J. Microbiol. 51, 149–158. doi: 10.1139/w04-132
Rusnak, F., Ascenso, C., Moura, I., and Moura, J. J. (2002). Superoxide reductase activities of neelaredoxin and desulfoferrodoxin metalloproteins. Methods Enzymol. 349, 243–258. doi: 10.1016/S0076-6879(02)49339-1
Russell, J. B. (1991). Resistance of Streptococcus bovis to acetic acid at low pH: relationship between intracellular pH and anion accumulation. Appl. Environ. Microbiol. 57, 255–259.
Russell, J. B., and Diez-Gonzalez, F. (1998). The effects of fermentation acids on bacterial growth. Adv. Microb. Physiol. 39, 205–234. doi: 10.1016/S0065-2911(08)60017-X
Ryan, P. A., Macmillan, J. D., and Zilinskas, B. A. (1997). Molecular cloning and characterization of the genes encoding the L1 and L2 components of hemolysin BL from Bacillus cereus. J. Bacteriol. 179, 2551–2556.
Ryan, S., Begley, M., Gahan, C. G., and Hill, C. (2009). Molecular characterization of the arginine deiminase system in Listeria monocytogenes: regulation and role in acid tolerance. Environ. Microbiol. 11, 432–445. doi: 10.1111/j.1462-2920.2008.01782.x
Salamitou, S., Ramisse, F., Brehelin, M., Bourguet, D., Gilois, N., Gominet, M., et al. (2000). The plcR regulon is involved in the opportunistic properties of Bacillus thuringiensis and Bacillus cereus in mice and insects. Microbiology 146(Pt 11), 2825–2832. doi: 10.1099/00221287-146-11-2825
Schneewind, O., and Missiakas, D. (2014). Sec-secretion and sortase-mediated anchoring of proteins in Gram-positive bacteria. Biochim. Biophys. Acta 1843, 1687–1697. doi: 10.1016/j.bbamcr.2013.11.009
Senesi, S., and Ghelardi, E. (2010). Production, secretion and biological activity of Bacillus cereus enterotoxins. Toxins (Basel) 2, 1690–1703. doi: 10.3390/toxins2071690
Senesi, S., Salvetti, S., Celandroni, F., and Ghelardi, E. (2010). Features of Bacillus cereus swarm cells. Res. Microbiol. 161, 743–749. doi: 10.1016/j.resmic.2010.10.007
Senouci-Rezkallah, K., Jobin, M. P., and Schmitt, P. (2015). Adaptative responses of Bacillus cereus ATCC14579 cells upon exposure to acid conditions involve ATPase activity to maintain their internal pH. Microbiologyopen 4, 1001. doi: 10.1002/mbo3.317
Senouci-Rezkallah, K., Schmitt, P., and Jobin, M. P. (2011). Amino acids improve acid tolerance and internal pH maintenance in Bacillus cereus ATCC14579 strain. Food Microbiol. 28, 364–372. doi: 10.1016/j.fm.2010.09.003
Sickmier, E. A., Brekasis, D., Paranawithana, S., Bonanno, J. B., Paget, M. S., Burley, S. K., et al. (2005). X-ray structure of a Rex-family repressor/NADH complex insights into the mechanism of redox sensing. Structure 13, 43–54. doi: 10.1016/j.str.2004.10.012
Siegumfeldt, H., Bjorn Rechinger, K., and Jakobsen, M. (2000). Dynamic changes of intracellular pH in individual lactic acid bacterium cells in response to a rapid drop in extracellular pH. Appl. Environ. Microbiol. 66, 2330–2335. doi: 10.1128/AEM.66.6.2330-2335.2000
Sineva, E., Shadrin, A., Rodikova, E. A., Andreeva-Kovalevskaya, Z. I., Protsenko, A. S., Mayorov, S. G., et al. (2012). Iron regulates expression of Bacillus cereus hemolysin II via global regulator Fur. J. Bacteriol. 194, 3327–3335. doi: 10.1128/JB.00199-12
Skorvaga, M., Dellavecchia, M. J., Croteau, D. L., Theis, K., Truglio, J. J., Mandavilli, B. S., et al. (2004). Identification of residues within UvrB that are important for efficient DNA binding and damage processing. J. Biol. Chem. 279, 51574–51580. doi: 10.1074/jbc.M409266200
Slamti, L., Perchat, S., Huillet, E., and Lereclus, D. (2014). Quorum sensing in Bacillus thuringiensis is required for completion of a full infectious cycle in the insect. Toxins (Basel) 6, 2239–2255. doi: 10.3390/toxins6082239
Spaans, S. K., Weusthuis, R. A., Van Der Oost, J., and Kengen, S. W. (2015). NADPH-generating systems in bacteria and archaea. Front. Microbiol. 6:742. doi: 10.3389/fmicb.2015.00742
Spano, G., Chieppa, G., Beneduce, L., and Massa, S. (2004). Expression analysis of putative arcA, arcB and arcC genes partially cloned from Lactobacillus plantarum isolated from wine. J. Appl. Microbiol. 96, 185–193. doi: 10.1046/j.1365-2672.2003.02132.x
Stenfors Arnesen, L. P., Fagerlund, A., and Granum, P. E. (2008). From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 32, 579–606. doi: 10.1111/j.1574-6976.2008.00112.x
Sun, G., Birkey, S. M., and Hulett, F. M. (1996). Three two-component signal-transduction systems interact for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19, 941–948. doi: 10.1046/j.1365-2958.1996.422952.x
Susin, M. F., Baldini, R. L., Gueiros-Filho, F., and Gomes, S. L. (2006). GroES/GroEL and DnaK/DnaJ have distinct roles in stress responses and during cell cycle progression in Caulobacter crescentus. J. Bacteriol. 188, 8044–8053. doi: 10.1128/JB.00824-06
Takemoto, L. J., Hansen, J. S., and Hokin, L. E. (1981). Phosphorylation of lens membrane: identification of the catalytic subunit of Na+, K+-ATPase. Biochem. Biophys. Res. Commun. 100, 58–64. doi: 10.1016/S0006-291X(81)80062-9
Tan, M. P., Sequeira, P., Lin, W. W., Phong, W. Y., Cliff, P., Ng, S. H., et al. (2010). Nitrate respiration protects hypoxic Mycobacterium tuberculosis against acid- and reactive nitrogen species stresses. PLoS ONE 5:e13356. doi: 10.1371/journal.pone.0013356
Tanaka, Y., Kimura, B., Takahashi, H., Watanabe, T., Obata, H., Kai, A., et al. (2008). Lysine decarboxylase of Vibrio parahaemolyticus: kinetics of transcription and role in acid resistance. J. Appl. Microbiol. 104, 1283–1293. doi: 10.1111/j.1365-2672.2007.03652.x
Tannenbaum, S. R., Fett, D., Young, V. R., Land, P. D., and Bruce, W. R. (1978). Nitrite and nitrate are formed by endogenous synthesis in the human intestine. Science 200, 1487–1489. doi: 10.1126/science.663630
Thomassin, S., Jobin, M. P., and Schmitt, P. (2006). The acid tolerance response of Bacillus cereus ATCC14579 is dependent on culture pH, growth rate and intracellular pH. Arch. Microbiol. 186, 229–239. doi: 10.1007/s00203-006-0137-1
Thompson, S. A., and Blaser, M. J. (1995). Isolation of the Helicobacter pylori recA gene and involvement of the recA region in resistance to low pH. Infect. Immun. 63, 2185–2193.
Thompson, S. A., Latch, R. L., and Blaser, J. M. (1998). Molecular characterization of the Helicobacter pylori uvr B gene. Gene 209, 113–122. doi: 10.1016/S0378-1119(98)00028-6
Tran, S. L., Guillemet, E., Gohar, M., Lereclus, D., and Ramarao, N. (2010). CwpFM (EntFM) is a Bacillus cereus potential cell wall peptidase implicated in adhesion, biofilm formation, and virulence. J. Bacteriol. 192, 2638–2642. doi: 10.1128/JB.01315-09
Ultee, A., Kets, E. P., Alberda, M., Hoekstra, F. A., and Smid, E. J. (2000). Adaptation of the food-borne pathogen Bacillus cereus to carvacrol. Arch. Microbiol. 174, 233–238. doi: 10.1007/s002030000199
Van de Guchte, M., Serror, P., Chervaux, C., Smokvina, T., Ehrlich, S. D., and Maguin, E. (2002). Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 82, 187–216. doi: 10.1023/A:1020631532202
Van der Voort, M., Kuipers, O. P., Buist, G., De Vos, W. M., and Abee, T. (2008). Assessment of CcpA-mediated catabolite control of gene expression in Bacillus cereus ATCC 14579. BMC Microbiol. 8:62. doi: 10.1186/1471-2180-8-62
Van Melis, C. C., Den Besten, H. M., Nierop Groot, M. N., and Abee, T. (2014). Quantification of the impact of single and multiple mild stresses on outgrowth heterogeneity of Bacillus cereus spores. Int. J. Food Microbiol. 177, 57–62. doi: 10.1016/j.ijfoodmicro.2014.02.015
Vilain, S., Luo, Y., Hildreth, M. B., and Brozel, V. S. (2006). Analysis of the life cycle of the soil saprophyte Bacillus cereus in liquid soil extract and in soil. Appl. Environ. Microbiol. 72, 4970–4977. doi: 10.1128/AEM.03076-05
Voelz, H. (1964). Sites of adenosine triphosphatase activity in bacteria. J. Bacteriol. 88, 1196–1198.
Wang, E., Bauer, M. C., Rogstam, A., Linse, S., Logan, D. T., and Von Wachenfeldt, C. (2008). Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex. Mol. Microbiol. 69, 466–478. doi: 10.1111/j.1365-2958.2008.06295.x
Warda, A. K., Siezen, R. J., Boekhorst, J., Wells-Bennik, M. H., De Jong, A., Kuipers, O. P., et al. (2016). Linking Bacillus cereus genotypes and carbohydrate utilization capacity. PLoS ONE 11:e0156796. doi: 10.1371/journal.pone.0156796
Wen, Z. T., and Burne, R. A. (2004). LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 186, 2682–2691. doi: 10.1128/JB.186.9.2682-2691.2004
Wijnands, L. M., Pielaat, A., Dufrenne, J. B., Zwietering, M. H., and Van Leusden, F. M. (2009). Modelling the number of viable vegetative cells of Bacillus cereus passing through the stomach. J. Appl. Microbiol. 106, 258–267. doi: 10.1111/j.1365-2672.2008.03999.x
Wilson, A. C., Hoch, J. A., and Perego, M. (2008). Virulence gene expression is independent of ResDE-regulated respiration control in Bacillus anthracis. J. Bacteriol. 190, 5522–5525. doi: 10.1128/JB.00312-08
Wilson, C. M., Loach, D., Lawley, B., Bell, T., Sims, I. M., O’toole, P. W., et al. (2014). Lactobacillus reuteri 100-23 modulates urea hydrolysis in the murine stomach. Appl. Environ. Microbiol. 80, 6104–6113. doi: 10.1128/AEM.01876-14
Yang, C. K., Ewis, H. E., Zhang, X., Lu, C. D., Hu, H. J., Pan, Y., et al. (2011). Nonclassical protein secretion by Bacillus subtilis in the stationary phase is not due to cell lysis. J. Bacteriol. 193, 5607–5615. doi: 10.1128/JB.05897-11
Yuk, H. G., and Marshall, D. L. (2004). Adaptation of Escherichia coli O157:H7 to pH alters membrane lipid composition, verotoxin secretion, and resistance to simulated gastric fluid acid. Appl. Environ. Microbiol. 70, 3500–3505. doi: 10.1128/AEM.70.6.3500-3505.2004
Zigha, A., Rosenfeld, E., Schmitt, P., and Duport, C. (2006). Anaerobic cells of Bacillus cereus F4430/73 respond to low oxidoreduction potential by metabolic readjustments and activation of enterotoxin expression. Arch. Microbiol. 185, 222–233. doi: 10.1007/s00203-006-0090-z
Keywords: Bacillus cereus, redox homeostasis, oxygen sensing, acidic pH, metabolism
Citation: Duport C, Jobin M and Schmitt P (2016) Adaptation in Bacillus cereus: From Stress to Disease. Front. Microbiol. 7:1550. doi: 10.3389/fmicb.2016.01550
Received: 28 July 2016; Accepted: 15 September 2016;
Published: 04 October 2016.
Edited by:
Lorena Ruiz, Universidad Complutense de Madrid, SpainReviewed by:
George-John Nychas, Agricultural University of Athens, GreeceFolarin Anthony Oguntoyinbo, University of Lagos, Nigeria
Copyright © 2016 Duport, Jobin and Schmitt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Catherine Duport, Y2F0aGVyaW5lLmR1cG9ydEB1bml2LWF2aWdub24uZnI=
 Catherine Duport
Catherine Duport Michel Jobin
Michel Jobin Philippe Schmitt
Philippe Schmitt